Abstract
Bi-allelic hypomorphic mutations in DNMT3B disrupt DNA methyltransferase activity and lead to immunodeficiency, centromeric instability, facial anomalies syndrome, type 1 (ICF1). Although several ICF1 phenotypes have been linked to abnormally hypomethylated repetitive regions, the unique genomic regions responsible for the remaining disease phenotypes remain largely uncharacterized. Here we explored two ICF1 patient–derived induced pluripotent stem cells (iPSCs) and their CRISPR-Cas9-corrected clones to determine whether DNMT3B correction can globally overcome DNA methylation defects and related changes in the epigenome. Hypomethylated regions throughout the genome are highly comparable between ICF1 iPSCs carrying different DNMT3B variants, and significantly overlap with those in ICF1 patient peripheral blood and lymphoblastoid cell lines. These regions include large CpG island domains, as well as promoters and enhancers of several lineage-specific genes, in particular immune-related, suggesting that they are premarked during early development. CRISPR-corrected ICF1 iPSCs reveal that the majority of phenotype-related hypomethylated regions reacquire normal DNA methylation levels following editing. However, at the most severely hypomethylated regions in ICF1 iPSCs, which also display the highest increases in H3K4me3 levels and/or abnormal CTCF binding, the epigenetic memory persists, and hypomethylation remains uncorrected. Overall, we demonstrate that restoring the catalytic activity of DNMT3B can reverse the majority of the aberrant ICF1 epigenome. However, a small fraction of the genome is resilient to this rescue, highlighting the challenge of reverting disease states that are due to genome-wide epigenetic perturbations. Uncovering the basis for the persistent epigenetic memory will promote the development of strategies to overcome this obstacle.
In mammals, cytosine methylation occurs approximately at 70%–80% of the CpG dinucleotides in the genome, and to a much lesser extent at non-CG sites (Ramsahoye et al. 2000; Lister et al. 2009). De novo establishment of DNA methylation occurs in embryos around the implantation stage by the combined action of the de novo DNA methyltransferases DNMT3A and DNMT3B, with a greater contribution of DNMT3B (Auclair et al. 2014). In mice the complete inactivation of these two enzymes leads to embryonic or postnatal lethality (Li et al. 1992; Okano et al. 1999), demonstrating their essential role in mammalian development.
In humans, biallelic hypomorphic mutations, mostly in the catalytic domain of DNMT3B, cause the immunodeficiency, centromeric instability, facial anomalies type I (ICF1) syndrome. The main manifestation of this syndrome is a severe form of combined immunodeficiency that leads to lethal infections in early childhood (Maraschio et al. 1988; Weemaes et al. 2013). Additional clinical features at varying degrees of penetrance and severity include intellectual disabilities, neurological defects, facial anomalies, and developmental delay. Although patients carrying DNMT3B variants represent the majority of ICF cases (Hansen et al. 1999; Xu et al. 1999; Weemaes et al. 2013; Simo-Riudalbas et al. 2015), mutations in at least three additional genes can cause ICF syndrome, highlighting the complex genetic heterogeneity of the disease (de Greef et al. 2011; Thijssen et al. 2015). In ICF1 patients, DNA hypomethylation is apparent at specific heterochromatic regions, including the pericentromeric and subtelomeric repeats (Jeanpierre et al. 1993; Yehezkel et al. 2008). The ensuing defect in condensation of the pericentromeric regions of Chromosomes 1, 9, and 16 contributes to the centromeric instability in immune cells, whereas subtelomeric hypomethylation leads to telomere shortening and premature senescence of patient fibroblasts (Gisselsson et al. 2005; Yehezkel et al. 2008).
Despite the identification of ICF syndrome-causative genes, very little is known about the underlying molecular mechanisms and the cascade of pathophysiological events that lead to this severe disease. Particularly, it is yet unknown which of the methylation defects throughout the genome are directly responsible for the aberrant gene expression pattern and the cellular and developmental defects typical of ICF syndrome. DNA methylation and transcriptional studies of ICF1 patient whole blood and lymphoblastoid cell lines (LCLs) identified several key alterations in epigenetic and gene expression profiles (Jin et al. 2008; Gatto et al. 2010, 2017; Heyn et al. 2012). We previously demonstrated that pathogenic DNMT3B variants disrupt the intragenic methylation level and the silencing of alternative and cryptic promoters in several genes involved in the regulation of immune cell function, thereby interfering with the control of transcription initiation (Gatto et al. 2017). This is consistent with recent genome-wide studies describing that intragenic CG methylation of active genes is dependent on DNMT3B activity in both humans and mice (Yang et al. 2014; Baubec et al. 2015; Neri et al. 2017; Dahlet et al. 2020). Despite these important insights, it is difficult to distinguish between the direct genomic targets of DNMT3B during implantation, which constitute the primary methylation defects, and the secondary methylation defects that occur later in development. The latter most probably arise due to genome-wide perturbations of the epigenetic landscape induced by the primary DNA methylation changes.
ICF1 patient-derived induced pluripotent stem cells (ICF1 iPSCs) provide a powerful model for studying the earliest pathogenic events in ICF1 syndrome and for identifying the genomic regions prone to DNA methylation perturbation as a consequence of DNMT3B loss of function (LOF) (Huang et al. 2014; Sagie et al. 2014). Indeed, ICF1 iPSCs retain the characteristic hypomethylation at pericentromeric and subtelomeric repeats (Yehezkel et al. 2008). Consistently, when differentiated into fibroblast-like cells, they recapitulate the abnormal telomere shortening and premature senescence phenotypes, originally described in ICF1 patient fibroblasts (Sagie et al. 2014). DNMT3B residual activity in ICF1 cells is sufficient to permit reprogramming and pluripotency, thereby providing a reliable platform to study the etiology of ICF1 syndrome and the dynamics of epigenetics abnormalities in the context of early development.
By performing homology directed repair (HDR) based gene-editing with CRISPR-Cas9 technology we recently corrected DNMT3B mutations in ICF1 iPSC lines derived from two patients (Toubiana et al. 2019). Focusing on repetitive regions, we revealed that whereas pericentromeric repeats acquired normal methylation levels in corrected iPSCs, DNA methylation at the majority of subtelomeres was only partially restored. Importantly, we found that the chromatin context at subtelomeres is significantly altered in both somatic and patient-derived iPSCs, thus inhibiting full DNA methylation rescue by the corrected DNMT3B. Here, we expand our study of epigenetic abnormalities in ICF1 syndrome iPSCs to the entire genome, and by comparing the patient iPSCs with their corrected isogenic counterparts, we acquire insights into the molecular factors that influence the ability to correct the ICF1 abnormal epigenome.
Results
DNA hypomethylation patterns in different ICF1 iPSCs and their rescue in corrected clones are highly comparable
To extensively evaluate the genome-wide modified epigenetic landscape associated with DNMT3B LOF, we utilized iPSCs derived from pR and pG ICF1 patients and their respective CRISPR-corrected clones (Sagie et al. 2014; Toubiana et al. 2019). The pR patient is homozygous for the p.D817G change in the DNMT3B catalytic domain, whereas the compound heterozygous pG patient carries a p.I41fsX42 variation leading to a premature stop codon in one DNMT3B allele, and a p.S780L change in the catalytic domain of the second allele (Weemaes et al. 2013). Two isogenic clones of pR iPSCs, cR7 and cR35, have biallelic correction of the missense mutation. The cG13 and cG50 iPSC clones have corrected the null mutation in pG iPSCs, and maintain the missense mutation in the catalytic domain of the protein, thus mimicking the genotype of heterozygous carriers that are healthy.
To investigate the effect of the pathogenic DNMT3B variants on DNA methylation levels across the genome, and to identify the regions rescued by the corrected DNMT3B, we performed whole genome bisulfite sequencing (WGBS) in WT, patient and corrected iPSCs. We observed a decrease, albeit mild, of global methylated CG (mCG) levels (from 74.8% in WT-iPSCs to 70.1% in pR iPSCs and 64.6% in pG iPSCs) (Supplemental Fig. S1A). A decrease in non-CG methylation in pR and pG iPSCs was also detected (Supplemental Fig. S1B,C), confirming previous observations in additional ICF1-derived iPSC lines (Huang et al. 2014). Although the global mCG levels in patient iPSCs were not extensively compromised, when we searched for differentially methylated regions (DMRs) between the two ICF1 iPSCs and our internal control WT1 iPSCs we identified 36,876 and 31,854 hypomethylated regions in pR and pG, respectively. To more accurately define the DMRs, we compared the methylation levels of WT1 iPSC to those of an additional WT iPSC from a public data set, which we designated as WT2 iPSC (GSM1385983, derived from human dermal fibroblasts [Ma et al. 2014]). We then filtered out the hypo-DMRs whose methylation level differed between WT1 and WT2 (difference between the ratio values > 0.2). Following this step, we retained 26,031 and 22,727 hypo-DMRs for pR and pG, respectively. A significant fraction of these hypo-DMRs strictly overlapped between the pR and pG samples (8560, P-value < 0.0001; shuffle test), and the majority (62%) were in proximity (within 20 kb), supporting their relevance in the context of disease pathogenesis (Supplemental Fig. S1D). Notably, 30%–40% of hypo-DMRs intersected (or were within a distance of 2 kb) with hypomethylated sites (HMRs) in DNMT3B-depleted human embryonic stem cells 3BKO HUES64 hESCs at early (2–7; 33%) and late (17–22; 40%) passages, and shRNA 3BKD H1 hESCs (31%) (Martins-Taylor et al. 2012; Liao et al. 2015), confirming that these genomic regions are presumably primary targets of DNMT3B (P-adj < 0.0001; shuffle test; Supplemental Fig. S1E).
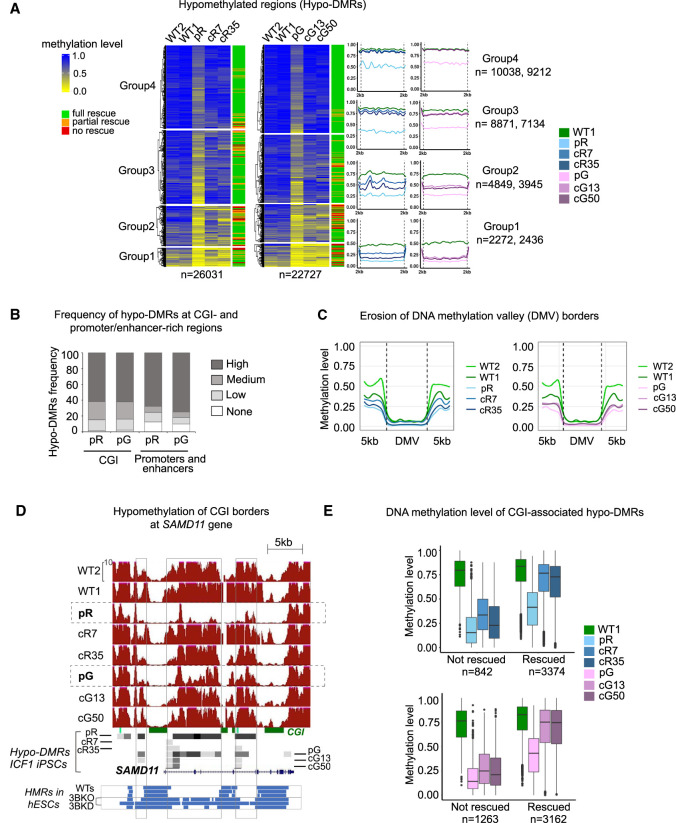
To explore the DNA methylation rescue of these hypo-DMRs, we clustered the hypo-DMRs into four groups based on their DNA methylation levels in patient iPSCs, with Group 1 exhibiting the lowest values compared to the WT controls (Fig. 1A). Overall, the ability to reacquire WT methylation levels following DNMT3B correction was inversely proportional to the degree of hypomethylation in the original ICF1 iPSCs. Hence, whereas the hypo-DMRs in Groups 3 and 4 regained almost full methylation in the corrected clones, the recovery at DMRs with severe hypomethylation (Groups 1 and 2) was highly compromised. To precisely determine the extent of DNA methylation recovery, we measured the number of hypo-DMRs in the corrected compared to WT1 iPSC. We defined the ICF1 iPSC hypo-DMRs as restored when they were no longer differentially hypomethylated or if they were hypermethylated in the corrected clones. Out of the 26,031 and 22,727 hypo-DMRs in pR and pG iPSCs, 74.1% and 75% (19,291 and 17,041) were rescued in both corrected iPSC clones of each patient iPSC (full rescue), respectively (Supplemental Table S3), whereas 18.1% and 13.8% were corrected in only one iPSC clone (partial rescue) and 7.8% and 11.2% remained hypo-DMR in both corrected clones (no rescue). Thus, a minority of hypo DMRs resisted rescue by the restored DNMT3B.
Figure 1.
Hypo-DMRs in ICF1 iPSCs are largely corrected following the restoration of DNMT3B gene activity. (A) Heatmap representation of methylation levels as determined by WGBS (expressed as ratio of the number of Cs over the total number of Cs and Ts) at hypo-DMRs in pR and pG ICF1 iPSCs, their respective corrected clones, and controls WT1 and WT2. The hypo-DMRs were clustered using k-means into four groups based on the methylation level across ICF1 iPSCs. The last column on the right indicates the ICF1 hypo-DMR rescue status: full, partial, and no rescue of ICF1 hypo-DMRs indicate remethylation in both, single, or none of the corrected clones. The profile plots on the right depict the average methylation level of each subgroup of hypo-DMRs and their flanking regions (±2 kb). The numbers of hypo-DMRs in pR and pG iPSCs in each group are indicated on the right. (B) Stacked bar plot representing the proportion of ICF1 hypo-DMRs that are present in genomic regions (1 Mb) categorized based on the number of CGI or promoters/enhancers as high (>10), medium (5–10), low (1–4), or none. (C) Plots of average weighted CG methylation levels across DNA methylation valleys (DMVs) and flanking 5 kb regions in ICF1 and corrected iPSCs clones in comparison to the WT1 and WT2 iPSCs. The 0 to 1 scale at the y-axis denotes the CG methylation level measured in each bin. (D) A genome browser view of CGIs in the SAMD11 gene, representing an example of methylation loss at CGI edges in ICF1 iPSCs compared to WT counterparts. Red tracks denote methylation coverage measured by WGBS in all iPSCs. Gray boxes represent hypo-DMRs detected in patients and corrected iPSCs compared to controls. Four shades of gray from light gray to black indicate differential methylation scores compared to WT1 as follows: −25 to −39, −40 to −59, −60 to −79, and −80 to −100, respectively. The blue tracks at the bottom illustrate the hypomethylated regions (HMRs) in WT human embryonic stem cells (hESCs) from H1, HUES9, and H9 lines, followed by DNMT3B-KO hESCs (early and late passage DNMT3B-KO; 3BKO) and shRNA DNMT3B-KD hESCs (3BKD). (E) Boxplots representing the distribution of methylation levels at CGI-associated hypo-DMRs in ICF1 iPSCs which remain hypomethylated (not rescued) and those that are rescued in the corresponding isogenic clones.
ICF1 DNMT3B variants fail to methylate CGI-rich chromosomal domains
We next asked whether the loci affected by DNMT3B dysfunction, and those that are rescued compared to those that resist rescue, are associated with specific genomic regions. To this end, we explored the distribution of hypo-DMRs across the autosomal chromosomes in the ICF1 iPSCs and their corrected counterparts (Supplemental Fig. S2). Clearly, pR and pG iPSCs display a consistent overlap between the chromosomal distribution of persisting and restored hypo-DMRs. A pattern of distinct, sharp peaks with a higher density at the centromeric/pericentromeric and distal/telomeric regions were detected across almost all chromosomes. Overall, our results were consistent with previous observations in other ICF1 iPSC lines (Huang et al. 2014), and out of 73 large hypomethylated genomic domains identified in the Huang et al. study, 60 and 52 were in common with pR and pG iPSCs, respectively (P-value < 0.001; shuffle test).
Large gene-dense chromosomal regions with elevated hypo-DMR density correlated with high CpG Island (CGI) content and sequences enriched with promoters and enhancers from GeneHancer (GH) Database (Fishilevich et al. 2017) (Fig. 1B; Supplemental Fig. S2; P-value < 1 × 10−5, Poisson regression). We further explored whether DNMT3B LOF affects the status of DNA methylation valleys (DMVs) identified in ES cells (Xie et al. 2013). DMVs are defined as genomic regions devoid of DNA methylation, highly conserved among different cell lines and species. We found that out of 639 DMVs, 223 and 211 DMVs overlap or are in close proximity (±5 kb) with hypo-DMRs in pR and pG iPSCs, and 139 are shared between both patient iPSCs (P-value < 0.001, shuffle test). Similarly, hypomethylation at DMVs is observed in early and late passage of DNMT3B deficient hESCs (426 out of 639 DMVs; P-adj < 0.001; shuffle test). Noticeably, the normally methylated DMV borders were frequently hypomethylated in ICF1 iPSCs (Fig. 1C), leading to expansion of the unmethylated valleys in both directions, as shown at the ICAM5 gene in Supplemental Figure S3A. This indicates that DNMT3B contributes to preserving the methylation state of DMVs borders in normal pluripotent stem cells.
To further investigate whether the erosion of DMV borders may generally affect promoter and intragenic CGIs, we plotted the average methylation profiles of UCSC CGIs intersecting with hypo-DMRs (5706 in pR and 5320 in pG; Supplemental Fig. S3B), and found hypomethylation at CGI-flanking regions (±5 kb). Thus, major hypomethylated elements in ICF1 iPSCs include CGIs and not only the subset of DMVs. Frequently, the nearby hypomethylated CGIs merged into a larger hypomethylated domain, as demonstrated at the SAMD11 gene locus, and visible also in DNMT3B deficient hESCs (Fig. 1D).
We next measured the overall level of methylation recovery at hypo-DMRs localized to CGIs (Fig. 1E). We observed that 60% of this subgroup (3374 and 3162 in pR and pG, respectively) restored normal DNA methylation levels in all corrected clones, whereas 15%–20% were uncorrected (842 and 1263 in pR and pG, respectively). The remaining fraction was partially corrected in only one of the two corrected clones. Out of 2485 CGIs-associated hypo-DMRs commonly hypomethylated in both patient iPSCs, 2038 (82%) fully or partially regained a normal methylation profile in corrected counterparts, and 447 (18%) hypo-DMRs was resistant to remethylation in all four corrected clones. Although the size, density, and position of CGIs (promoter or intragenic) did not influence the ability to regain methylation in corrected iPSCs, we found that the resistant regions were all characterized by the lowest methylation levels identified in patient iPSCs compared to WT1 (Fig. 1E). Collectively, these results indicate that the corrected DNMT3B can fully restore the normal methylation signature only at CGI sites that display milder hypomethylation in patient iPSCs.
We additionally observed that the percentage of hypomethylated GH promoters and enhancers reacquired proper DNA methylation at similar levels as observed at CGIs, amounting to 68% and 67% in both corrected clones of pR iPSC and pG iPSCs, respectively (Supplemental Fig. S3C). The WDR97 gene represents an example of methylation loss at enhancer regions (ENCODE cCRE and GH) in pR and pG iPSCs (Supplemental Fig. S3D). Consistently, we found that the original level of methylation of these regions in patient iPSCs influenced the degree of methylation recovery (Supplemental Fig. S3E). Thus, at gene regulatory regions as well, the loss of DNA methylation beyond a certain threshold, impedes the salvage of normal methylation patterns, despite the recovery of normal DNMT3B enzymatic activity.
Genes with roles in immune response display abnormal promoter and intragenic methylation that is efficiently rescued following gene correction
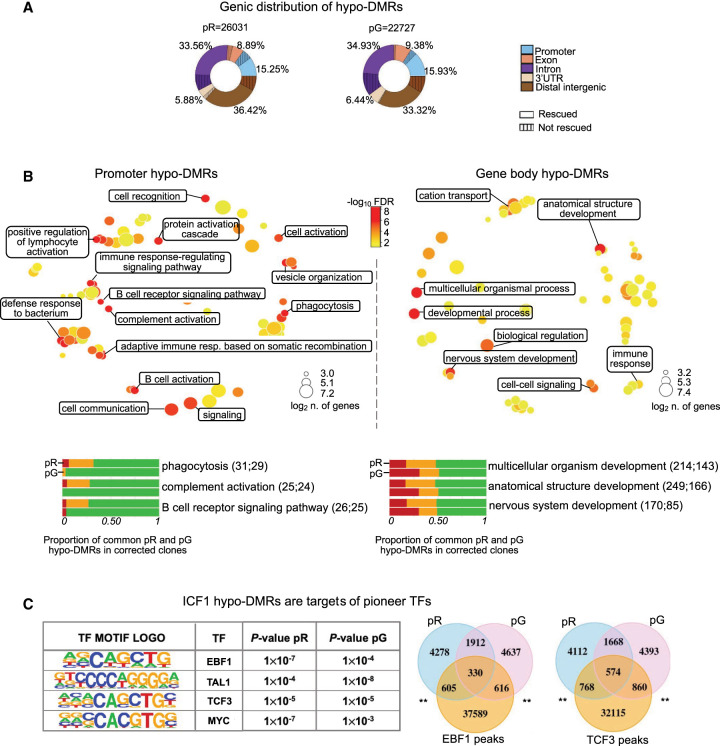
CGI-rich regions, highly correlated with gene coding regions, were the most highly enriched elements in hypo-DMRs across all chromosome arms. Characterization of the specific gene components affected by DNA methylation loss revealed that approximately half of the hypo-DMRs were localized in gene bodies, distributed along all exons, introns, and 3′-UTRs, whereas about 15% mapped to gene promoters and 35% were present in distal intergenic regions (Fig. 2A). The hypo-DMRs common to pR and pG iPSCs (8560) were similarly distributed across these genic features (18% at promoters, 44% at gene body, and 38% at distal intergenic regions). DNA methylation in all these various components was similarly restored following gene correction.
Figure 2.
DNMT3B shapes CG methylation patterns at genes with functions in innate and adaptive immune responses. (A) Donut charts depicting the percentage distribution of hypo-DMRs in pR and pG, annotated to various genic regions. Striped areas: hypo-DMRs resistant to hypomethylation correction. (B, top) Gene Ontology biological processes terms (GO-BP) enriched for genes annotated to promoter (left) and intragenic (right) hypo-DMRs common to both ICF1 iPSCs (pR = 1328 and pG = 2336) are visualized as a multidimensional scaling scatter plots. The color scale represents the −log10 of the adjusted P-value (Benjamini–Hochberg with false discovery ratio correction < 0.01; BH-FDR) of the GO-BP term. The dot size represents the log2 of the number of hypo-DMR associated genes in each GO-BP term. (Bottom) Bar plots depicting the proportion of hypo-DMRs associated with genes enriched in the three most significant categories of GO-BP in promoter and gene-body that are fully (green), partially (orange), or not rescued (red) in corrected iPSCs clones (Supplemental Table S3). For each GO-BP, the upper and lower bars correspond to the hypo-DMRs status in pR and pG, respectively. The number of hypo-DMRs annotated to gene promoter or gene body is indicated in the brackets. (C, left) Motif enrichment analysis of known transcription factor (TF) at hypo-DMRs with reduced DNMT3B binding. The output displays the predicted motif sequence and the P-value significance for ranking the motif enrichment at pR and pG hypo-DMRs. (Right) Venn diagrams representing the overlap between ICF1 hypo-DMRs with enriched target regions of EBF1 and TCF3 TFs (peaks obtained from ENCODE ChIP-seq data sets of WT LCLs), P-adj < 0.001; shuffle test.
To investigate the functions of genes that display hypomethylation at promoters and intragenic regions in ICF1 iPSCs, we performed Gene Ontology (GO) analysis on genes associated with the common hypo-DMRs. Consistent with the immune phenotype present in ICF1 patients, we detected a significant enrichment of biological processes related to the regulation of the immune response. In particular, among genes with promoter hypomethylation we found regulators of both innate and adaptive immunity (Fig. 2B; Supplemental Table S3). Biological processes related to cell fate commitment and embryonic development were predominantly associated with genes hypomethylated at their bodies, consistent with the recognized role of intragenic CGI methylation in developmental regulatory genes (Jeziorska et al. 2017; Lee et al. 2017). Following DNMT3B correction, the majority of the immune response-related hypo-DMRs fully restored normal DNA methylation patterns, whereas the hypo-DMRs positioned in or at the vicinity of genes associated with developmental processes showed relatively lower rescue in the corrected clones (Fig. 2B).
The functional categories of the hypomethylated genes enriched in ICF1 iPSCs, resemble those detected in LCLs derived from two unrelated ICF1 patients (Jin et al. 2008; Gatto et al. 2017). When comparing the ICF1 iPSCs and LCLs, we detected a significant overlap between hypo-DMRs in both cell types (pR and pG compared to WGBS-ICF1p1 and RRBS-ICF1p1 and ICF1p2; P-adj < 0.001; shuffle test) (Supplemental Fig. S4A). Similarly, hypomethylated regions in ICF1 iPSC overlapped, or were positioned within a distance of maximum 2 kb, with those identified in ICF1 blood samples (Velasco et al. 2018) (Supplemental Fig. S4B; P-adj < 0.001; shuffle test). These results support the notion that a significant fraction of the abnormal methylation detected in ICF1 somatic cells arises during de novo establishment of DNA methylation around the implantation stage.
Highly relevant to the agammaglobulinemia characterizing ICF syndrome patients, we detected hypomethylation of the immunoglobulin heavy chain (IGH) genes in the distal region of Chromosome 14q in ICF1 iPSCs, similar to what is seen in DNMT3B-depleted hESCs (Supplemental Fig. S4C). This suggests that DNMT3B plays a specific role in the intricate regulation of the IGH locus. Following gene editing, this region recovered the majority of DNA methylation, thereby predicting that gene correction might reverse the epigenetic defects in this chromosomal domain and restore normal immunoglobulin expression in ICF1 patient blood cells.
Beyond the IGH locus, hypomethylation was detected in a large number of genes required in immune response, hematopoietic differentiation, lymphocyte activation, B cell receptor signaling, T cell costimulation, T cell receptor signaling pathways, the complement cascade, and cell adhesion and migration (Supplemental Table S3). Collectively, we identified genes that are early direct targets of DNMT3B-dependent de novo methyltransferase activity, whose hypomethylation might be functionally relevant in the context of the ICF immunodeficiency phenotype. The hypomethylation patterns at the immune-related genes and their regulatory regions are frequently mirrored in ICF1 LCLs, suggesting that these methylation profiles in somatic ICF1 cells are premarked early during embryonic development.
Additionally relevant to ICF1 abnormal phenotypes, motif enrichment analysis for transcription factor (TF) binding sites at ICF1 iPSCs hypo-DMRs (which also showed decreased DNMT3B binding; see below) highlighted binding motifs for pioneer TFs important for hematopoietic differentiation and B cell development, such as TCF3 and EBF1 (Lin et al. 2010), TAL1 (also known as SCL; Real et al. 2012) and MYC (Laurenti et al. 2008) (Fig. 2C; Supplemental Table S3). When we compared the hypo-DMRs with published TF ChIP-seq data sets in WT hematopoietic stem and progenitor cells (HSPCs; Huang et al. 2016) and LCLs (ENCODE), we found a significant overlap between the hypo-DMRs in pR and pG ICF1 iPSCs and the corresponding TF peaks (Fig. 2C; P-adj < 0.001; shuffle test). Collectively, these findings imply that abnormal hypomethylation at these TF binding sites might affect their recruitment at the relevant time point during cell differentiation, thereby interfering with the normal regulation of tissue-specific gene expression programs during development of ICF1 embryos.
Transcriptome profiling reveals minor changes in ICF1 iPSCs
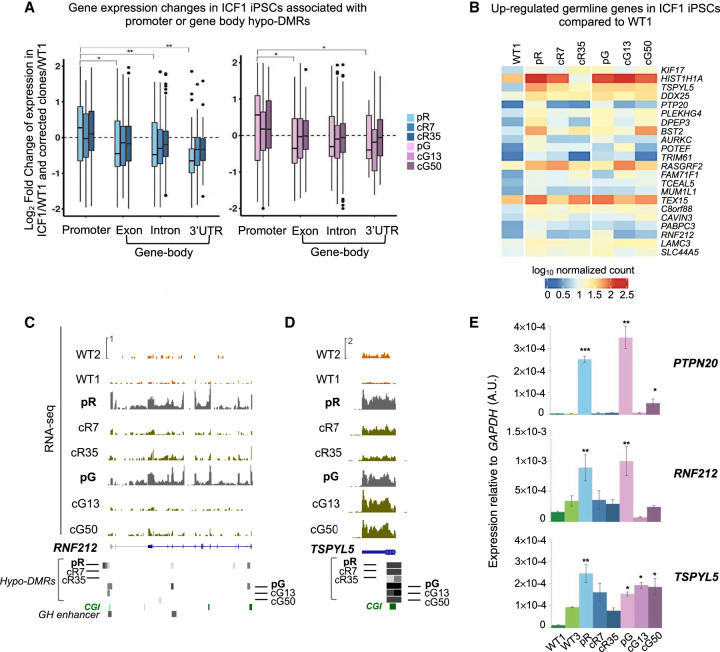
To investigate hypomethylation related gene expression deregulation, we performed RNA-seq for WT1, ICF1, and corrected iPSCs and compared the data sets of gene expression and DNA methylation. Changes in gene expression levels were limited to 344 genes that were differentially expressed in both ICF1 iPSCs compared to WT1 iPSCs (DE; posterior probability [pp] > 0.9; Supplemental Fig. S5A; Supplemental Table S4). Among the deregulated genes in each ICF1 iPSC, 25% (pR) and 27% (pG) were either fully, partially, or slightly rescued (green bars) in the respective corrected clones (Supplemental Fig. S5B). The majority of hypo-DMRs (85% in pR and 70% in pG) associated with these rescued genes regained normal methylation in the corrected clones (Supplemental Fig. S5C).
Notably, out of the large group of genes hypomethylated at regulatory regions (CGIs, GH promoters and enhancers; 3178 and 3319 in pR and pG, respectively), only a limited fraction demonstrated altered expression in ICF1 iPSCs (173 in pR and 144 in pG, pp > 0.9), and we found no enrichment of these genes within the group of genes demonstrating the most severe hypomethylation (Supplemental Fig. S5D). Promoter hypomethylation is most commonly correlated with gene up-regulation (Fig. 3A; Supplemental Table S4). Indeed, genes associated with germline functions were ectopically activated in both ICF1 iPSCs compared to WT1 iPSCs, and regained partial or full methylation in the corrected clones, as exemplified by the RNF212 and PTPN20 genes (Fig. 3B,C,E). The CGI-associated hypo-DMR of RNF212 was rescued in the corrected clones of both ICF1 iPSCs, and consistently, its abnormal expression was reversed in all four clones (Fig. 3C,E). The recovery of proper gene expression levels was highly correlated with the degree of DNA methylation recovery, which in turn depended on the severity of hypomethylation in ICF1 iPSCs. One such example is the TSPYL5 gene which exhibits high-score hypo-DMRs in ICF1 iPSCs. Persisting hypomethylation in the corrected clones of pG iPSCs is clearly correlated with sustained high expression levels, whereas partial correction of hypomethylation in the cR35 clone restored transcription to levels close to normal (Fig. 3D,E). These results indicate that preserving promoter CGI methylation is crucial for the repression of germline genes and, more importantly, that the recovery of normal DNA methylation patterns enables the reestablishment of normal expression levels.
Figure 3.
Transcriptional deregulation associated with promoter and intragenic hypo-DMRs. (A) Boxplot representation of the log2 fold change of the differentially expressed genes (pp > 0.8) with hypo-DMRs associated with regulatory elements (CGIs and GH promoters/enhancers) in pR (n = 471) and pG (n = 332) iPSCs and their corrected clones compared to WT1 iPSCs. The x-axis shows the gene feature category, and the y-axis denotes the log2FC of the genes in each ICF1 iPSCs versus WT1 iPSCs. The statistical significance of the differences in the log2FC of genes with hypo-DMRs annotated to promoters compared with genes containing hypo-DMRs annotated to other gene features was calculated using a nonparametric two samples two-sided Wilcoxon test with BH-FDR correction: (*) FDR < 0.01; (**) FDR < 0.001. (B) Heatmap representing the expression levels of germline-specific genes hypomethylated and up-regulated in ICF1 iPSCs. The scale denotes the log10 of normalized counts of genes compared between ICF1 iPSCs and corrected iPSCs to WT1 iPSCs. (C,D) Genome browser views of the expression profiles of the hypomethylated germline-specific genes RNF212 and TSPYL5. The coverage tracks display the RNA-seq expression level of the genes in WT1 and WT2 internal and external controls, ICF1, and corrected iPSCs. Gray boxes represent hypo-DMRs in ICF1 and corrected iPSC clones compared to WT1 and WT2. (E) RT-qPCR of the up-regulated RNF212, PTPN20, TSPYL5 germline genes in WT, ICF1, and corrected iPSCs. Each bar represents the mean of the relative expression compared to the expression of GAPDH in the same sample. The RT-qPCR data are presented as mean ± SD from independent triplicates, each amplified twice. Statistical analyses were performed using a one-tail two-sample Student's t-test compared to WT: (*) P-value < 0.05, (**) P-value < 0.01, (***) P-value < 0.001.
However, taken together these findings demonstrate that DNA hypomethylation is insufficient by itself to disrupt global gene expression, and the major effect of the epigenetic abnormalities present in stem cells is likely postponed to later developmental stages at which the affected genes are normally expressed. Consistently, in CD43+ hematopoietic progenitors (HPCs) differentiated from ICF1 iPSCs, we observed that out of 658 genes up- and down-regulated in both pR and pG HPCs (pp > 0.95), 364 were associated to hypo-DMRs identified in undifferentiated ICF1 iPSCs (pR and/or pG) (Supplemental Fig. S5E; Supplemental Table S4). Gene Ontology analysis of the 658 deregulated genes showed enrichment of functions related to regulation of hematopoiesis and immune system processes, which are relevant to ICF syndrome abnormal phenotypes (Supplemental Fig. S5F).
The corrected DNMT3B fails to bind regions with the highest abnormal increases in H3K4me3
We previously showed that subtelomeric regions with persisting hypomethylation in the corrected ICF1 iPSCs were unable to recruit and bind the restored DNMT3B protein (Toubiana et al. 2019). Therefore, we proceeded to determine the genome-wide DNMT3B binding profile in ICF1 iPSCs and corrected clones compared to WT1 iPSCs by ChIP-seq. Among the 19,706 DNMT3B enriched peaks detected in WT1 iPSCs, we observed significantly reduced binding (P-adj < 0.0001; paired Wilcoxon test) in both pR and pG iPSCs (Supplemental Fig. S6A; see also ChIP-seq data analysis in Supplemental Material; Supplemental Fig. S6B,C). This reduction cannot be attributed to lower levels of cellular DNMT3B in ICF1 iPSCs, as DNMT3B levels are comparable between WT, ICF1, and corrected iPSCs, including pG-related iPSCs that carry one DNMT3B null allele (Toubiana et al. 2019). Similarly, DNMT3A protein levels are comparable between the various iPSCs (Supplemental Fig. S6D). In corrected clones, DNMT3B binding was recovered at 74% in pR and 85% in pG of the affected regions. Thus, although the majority of the genomic targets acquired proper DNMT3B binding following mutation correction, up to 25% of the affected regions failed to do so. This suggests that similarly to subtelomeres, the recruitment of the corrected enzyme to these specific regions is impeded.
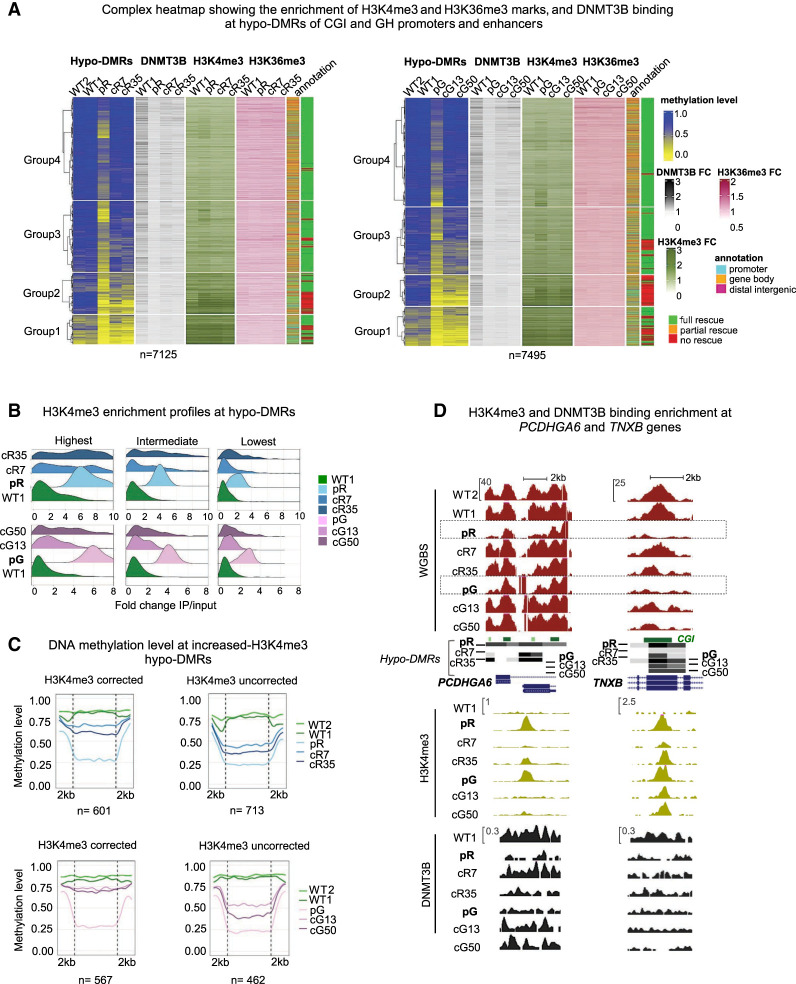
Epigenetic memory, exemplified by elevated H3K4me3 levels, was demonstrated previously at the subtelomeric regions with persisting hypomethylation. To assess whether epigenetic memory can similarly interfere with DNA methylation rescue at nonrepetitive regions throughout the genome, we compared the epigenetic landscape of rescued and unrescued hypo-DMRs, associated with the CGIs or GH promoters/enhancers. These hypo-DMRs were merged after removing duplicates, and analyzed for enrichment profiles of H3K4me3, H3K36me3 histone marks and DNMT3B binding obtained by ChIP-seq in WT1, ICF1 iPSCs and their corrected counterparts (Fig. 4A). The hypo-DMRs were divided in the complex heatmap into four groups based on their methylation level in ICF1 iPSCs. The hypo-DMRs of Groups 1 and 2 exhibited the lowest levels of DNA methylation in ICF1 iPSCs and a limited recovery of both DNA methylation and DNMT3B binding in the corrected clones. In contrast, the hypo-DMRs in Groups 3 and 4, demonstrated almost full rescue of DNA methylation levels and DNMT3B binding in the corrected clones (Fig. 4A). Groups 1 and 2 hypo-DMRs were enriched for promoters and showed the highest enrichment of H3K4me3 in all iPSCs compared to Group 3 and 4 hypo-DMRs that were mostly annotated to gene bodies. Overall, we observed that regions with the lowest methylation levels in ICF1 iPSCs correlated with the highest increase in the H3K4me3 mark (P-value < 0.0001; correlation t-test rPearson = −0.92 in pR and −0.91 in pG).
Figure 4.
Hypo-DMRs persist at regions with the highest aberrant increase of H3K4me3. (A) A complex heatmap showing the integration of ChIP-seq data of DNMT3B binding, and H3K4me3 and H3K36me3 marks with methylation levels of hypo-DMRs at CGI and GH elements. The gene annotation of the hypo-DMRs and their status of correction are depicted on the right. The hypo-DMRs are divided into four clusters based on Groups 1–4 derived from Figure 1A. FC = Fold change (IP/input) of the ChIP-seq enrichment calculated at the hypo-DMRs. (B) Ridgeline plots depicting the distribution of the fold change (FC) of H3K4me3 enrichment at increased H3K4me3 differentially enriched regions (DERs) proximal to ICF1 hypo-DMRs. The increased H3K4me3 DERs in pR and pG were divided into three groups based on the FC ranking levels in ICF1 iPSCs, and the FC was calculated for the respective corrected clones at the DERs. (C) Plots of average CG methylation levels across hypo-DMRs associated with increased H3K4me3 DERs in ICF1 iPSCs. The left panels include DERs in which the H3K4me3 levels are restored to normal levels in both corrected clones of pR (top) and pG (bottom), whereas the right panels include increased H3K4me3 DERs which persist in corrected clones of pR and pG. (D) Genome browser views of representative regions characterized by abnormal increase of H3K4me3 levels at hypo-DMRs in ICF1 iPSCs. The H3K4me3 levels are fully or partially reverted in corrected clones together with hypomethylation at the PCDHGA6 gene, whereas high H3K4me3 levels persist at the TNXB gene. Green and black tracks represent H3K4me3 and DNMT3B binding enrichment levels.
In contrast to H3K4me3, the levels of H3K36me3 at hypo-DMRs were less affected in ICF1 iPSCs compared to WT iPSCs (Fig. 4A). This suggests that DNMT3B LOF does not strongly affect the activity of SETD2 histone H3K36 methyltransferase in ICF1 iPSCs and is consistent with the observations that deposition of H3K36me3 by SETD2 is an event upstream of DNMT3B mediated methylation (Baubec et al. 2015; Tan et al. 2019; Masalmeh et al. 2021).
Noticeably, many of the H3K4me3 abnormally enriched regions in ICF1 iPSCs, particularly in Groups 3 and 4 restored normal levels of this histone mark following correction of DNMT3B mutations (Fig. 4A). These regions generally overlap with those demonstrating rescued DNA methylation levels (green in right column of the heatmap) and recovery of DNMT3B binding in the corrected clones. To quantify the association between hypomethylation and increased H3K4me3 enrichment in ICF1 iPSCs, we intersected the 26,031 and 22,727 hypo-DMRs in pR and pG iPSCs, respectively (Fig. 1A) with genomic regions exhibiting a significant increase in H3K4me3 levels (increased DERs) in pR (n = 2823) and pG (n = 1178) ICF1 iPSCs. We detected 1442 and 1081 hypo-DMRs overlapping, or in close proximity (±2 kb), to increased H3K4me3 DERs in pR and pG, respectively (P-adj < 0.0001; shuffle test). On average, the significantly increased H3K4me3 DERs (2823 and 1178) were characterized by very low DNA methylation levels (≤0.25) in patient iPSCs (Supplemental Fig. S6E), indicating that H3K4 trimethylation significantly increases at regions where DNA methylation is reduced below a certain threshold.
To explore the effect of DNMT3B correction on this mark, we divided the hypo-DMRs overlapping with increased H3K4me3 DERs into three groups based on the H3K4me3 enrichment level in ICF1 iPSCs. We found that hypo-DMRs with the highest H3K4me3 increase in ICF1 iPSC are the most resistant to changes in this histone modification following DNMT3B correction (Fig. 4B). Out of 1442 hypo-DMRs in pR and 1081 in pG, 713 and 462 hypo-DMRs remained highly enriched with this mark, whereas 601 and 567 hypo-DMRs fully restored normal H3K4me3 levels in the corrected clones (partial rescue occurred at 128 hypo-DMRs in pR and 52 in pG iPSCs) (Fig. 4C). Notably, when we computed the methylation level for each hypo-DMRs in the subgroup that retained abnormally high H3K4me3 levels, we observed that they show a significantly less regain of DNA methylation in comparison to the subgroup that corrected its H3K4me3 levels (P-adj < 0.0001; Welch's t-test). Thus, a subset of genomic regions can overcome the aberrant H3K4me3 increase and recruit the corrected DNMT3B protein to reestablish the normal DNA methylation pattern, whereas others are impervious to such a correction.
The correlation between H3K4me3 levels and DNA methylation restoration can be exemplified at specific genic regions. The PCDHGA6 gene from the protocadherin gene cluster contains hypo-DMRs with abnormally high H3K4me3 levels in ICF1 iPSCs. H3K4me3 levels were reverted back to normal in cR7, cG13, and cG50 corrected clones, and simultaneously, the corrected DNMT3B was recruited and normal DNA methylation levels were restored. However, in the cR35 clone, where the H3K4me3 barrier was not entirely removed, the DNA hypomethylation was not corrected. Another example is the region of the TNXB gene in which H3K4me3 levels are abnormally high and persisting in all iPSCs, and accordingly, DNMT3B binding and DNA methylation levels were not restored (Fig. 4D).
Abnormal CTCF binding interferes with restoration of DNA methylation
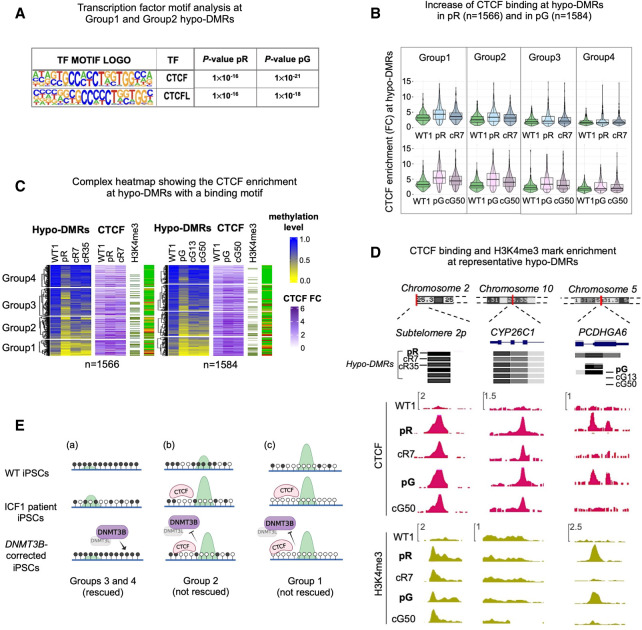
We hypothesized that DNMT3B recruitment may be impeded by factors that have affinity to regions with high H3K4me3 levels. To explore this possibility, we used the HOMER suite to perform motif enrichment analysis for TF binding sites at all hypo-DMRs showing persistent loss of DNMT3B binding in combination with the highest H3K4me3 and lowest DNA methylation levels (belonging to Groups 1 and 2 shown in Fig. 1A). We additionally analyzed hypo-DMRs from Groups 3 and 4 showing recovery of DNMT3B binding and normal DNA methylation levels. Noticeably, we observed within hypo-DMRs of Groups 1 and 2 a significant enrichment of binding motifs for CTCF and its paralogous factor CTCFL at a relatively higher frequency (14%; P-value < 1 × 10−17; HOMER analysis) than within hypo-DMRs of Groups 3 and 4 (7%; P-value < 1 × 10−3) (Fig. 5A; Supplemental Table S5). This analysis suggested that the failure to restore DNA methylation at hypo-DMRs from Groups 1 and 2 might be promoted by the abnormal binding of the CTCF chromatin insulator in ICF1 iPSCs, that in turn might impede the recruitment of the corrected DNMT3B protein.
Figure 5.
Abnormal CTCF binding at a small fraction of hypo-DMRs contributes to persisting hypomethylation. (A) Motif enrichment analysis of known TFs at hypo-DMRs with reduced DNMT3B binding and increased H3K4me3 levels. The panel corresponds to motifs enriched at the subset of hypo-DMRs corresponding to Groups 1 and 2 in pR (n = 1177) and pG (n = 1230) iPSCs, as shown in Figure 1B. (B) Violin and boxplot representation of CTCF enrichment (IP/input) calculated across hypo-DMRs with a CTCF HOMER motif in WT1, ICF1 iPSCs, and one of their corrected clones (cR7 for pR and cG50 for pG). To quantify the statistical significance of the increased CTCF enrichment in Groups 1 and 2 compared to Groups 3 and 4, we calculated the difference between ICF1(FC) – WT(FC) and corrected clone (FC) – WT1(FC) for each group and applied nonparametric paired Wilcoxon test with one-sided alternative and BH-FDR correction. We then computed the effect size between the differences (for pR, G1 = 0.38; G2 = 0.26; G3 = 0.14; G4 = 0.03; for pG, G1 = 0.63; G2 = 0.48; G3 = 0.35; G4 = 0.11). (C) A complex heatmap showing the integration of methylation level and CTCF ChIP-seq enrichment (FC) across pR and pG ICF1 hypo-DMRs containing CTCF binding motifs. The presence of H3K4me3 enriched peaks proximal to hypo-DMRs (within 1 kb) and the status of hypo-DMR correction is depicted on the right. The proportion of H3K4me3 enriched peaks out of the total number of hypo-DMRs in each group is higher in Group 1 (pR = 137 [40%]; pG = 145 [51%]) and Group 2 (pR = 140 [31%]; pG = 150 [50%]) compared to Group 3 (pR = 98 [20%]; pG = 85 [17%]) and Group 4 (pR = 46 [13%]; pG = 50 [12%]); one-sided Fisher's exact test: Group 1 versus Group 3 and versus Group 4 (P-adj < 0.0001), and Group 2 versus Group 3 (P-adj < 0.001) and Group 4 (P-adj < 0.0001). (D) Genome browser views of representative regions characterized by abnormal increase of CTCF binding, as observed by ChIP-seq in WT, ICF1, and corrected iPSCs (cR7 for pR and cG50 for pG). Green tracks represent H3K4me3 enrichment levels in iPSCs. (E) Graphical representation of hypo-DMRs identified in ICF1 iPSCs and clustered in Groups 1–4 based on their DNA hypomethylation level, as reported in Figure 4A: (a) Rescued hypo-DMRs of Groups 3 and 4, showing high methylation and low H3K4me3 levels (green peak) in WT iPSCs, mild hypomethylation and H3K4me3 increase in ICF1 iPSCs, and regain of normal DNA methylation and H3K4me3 levels upon restoration of DNMT3B activity by gene editing; (b) unrescued hypo-DMRs of Group 2, showing initial intermediate/high DNA methylation levels and H3K4me3-enrichment in WT iPSCs, severe hypomethylation, high H3K4me3 increase in ICF1 iPSCs and/or abnormal CTCF binding, which persist in the corrected clones; (c) unrescued hypo-DMRs of Group 1, showing initial low DNA methylation level and high H3K4me3-enrichment in WT iPSCs, severe hypomethylation, high H3K4me levels and/or abnormal CTCF binding in ICF1 iPSCs, which are resistant to correction as in Group 2.
We next evaluated the binding of CTCF in WT, ICF1, and corrected iPSCs clones (cR7 for pR and cR50 for pG) by ChIP-seq. A higher increase of CTCF binding was observed at hypo-DMRs (1566 and 1584 sites in pR and pG) encompassing the HOMER recognition site of CTCF belonging to Groups 1 and 2 compared to Groups 3 and 4 (Fig. 5B; paired Wilcoxon test with one side alternative and effect size on the difference between ICF1 and WT iPSCs, and corrected and WT iPSCs), indicating that the severely hypomethylated regions were more likely to abnormally recruit CTCF protein. Following correction of DNMT3B, CTCF binding persisted to a high degree at the subset of hypo-DMRs of Groups 1 and 2. This supports the notion that persistent binding of CTCF interferes with the recruitment of the restored DNMT3B and consequently with DNA methylation recovery.
To further explore the association between CTCF binding and DNA methylation we examined the methylation levels in subgroups of hypo-DMRs ordered based on the degree of CTCF enrichment in ICF1 iPSCs (Fig. 5C). We found that the degree of CTCF binding was negatively correlated with the DNA methylation level at the 1566 and 1584 hypo-DMRs (P-value < 0.0001, correlation t-test; rPearson = −0.59, pR and −0.69, pG). When we analyzed H3K4me3 levels at all hypo-DMRs containing a CTCF motif, we found that two thirds of them (1132 in pR and 1162 in pG) lacked H3K4me3 peaks in ICF1 iPSCs. However, the small fraction of hypo-DMRs enriched with both CTCF and H3K4me3 included a higher proportion of regions belonging to Groups 1 and 2 and resistant to remethylation (Fig. 5C). Among the persistent hypomethylated regions in corrected clones, characterized by a concomitant increase in CTCF binding and H3K4me3 levels, we detected subtelomere 2p, which is severely hypomethylated in patient iPSCs, as we previously described (Toubiana et al. 2019). Persistent CTCF binding in corrected clones was observed also at resistant hypo-DMRs regions unassociated with H3K4me3 peaks in ICF1 iPSCs, as shown for the CYP26C1 gene (Fig. 5D; Supplemental Fig. S6F).
We then explored rescued hypo-DMRs with increased CTCF binding. We examined the hypo-DMRs associated with PCDHGA6 gene, which showed overlapping CTCF recognition sites. At this locus, CTCF bound the hypomethylated PCDHGA6 gene only in patient iPSCs and was absent in the corrected clones coinciding with DNA methylation recovery (Fig. 5D; Supplemental Fig. S6F). Altogether, these data demonstrate that an abnormal binding of CTCF, alone or in combination with H3K4me3, can obstruct the binding of the restored DNMT3B to a fraction of hypomethylated regions in corrected iPSCs (Fig. 5E).
Discussion
ICF1 patient-derived iPSCs mimic the initial developmental stage at which the mutated DNMT3B fails to properly methylate its genomic targets. Therefore, they can be used to identify genomic regions affected early in development, and follow their effects in ICF1 somatic cells.
Correction of the pathogenic DNMT3B variants in ICF1 iPSCs enables the evaluation of the capacity to restore the normal DNA methylation patterns. Previously, we demonstrated that correction of two ICF1 iPSCs fully restored the normal methylation levels at pericentromeric satellite DNA (Toubiana et al. 2019), whose hypomethylation is a hallmark of ICF syndrome (Jeanpierre et al. 1993). In contrast, abnormal DNA hypomethylation persisted at several subtelomeric regions, indicating that certain genomic regions resist de novo methylation by the corrected DNMT3B. Here we expanded our study to the nonrepetitive fraction of the genome to characterize the abnormal epigenetic landscape of the ICF1 iPSCs and their isogenic corrected clones, aiming to understand the molecular consequences of DNMT3B LOF throughout the entire genome and the feasibility of rescuing an abnormal epigenome arising in a human genetic disease. This is, to our knowledge, the first attempt to evaluate whether the sole correction of an inherited enzymatic deficiency leading to an epigenetic disorder, suffices to rescue genome-wide epigenetic perturbations.
While taking into account that several of the ICF phenotypes may be caused by disruption of chromatin/nuclear organization due to pericentromeric and subtelomeric hypomethylation, we searched for disrupted methylation patterns elsewhere in the genome that could be linked to the phenotypes of ICF1 syndrome. Our comprehensive analysis detected highly comparable patterns of DNA hypomethylation in both ICF1 iPSCs, highlighting preferential targets of DNMT3B across the genome. At the chromosomal level, in addition to the pericentromeric regions, and distal regions of several chromosomes, we detected large domains of CGI-rich regions and gene clusters characterized by loss of DNA methylation. Comparing the methylation defects in ICF1 iPSCs with early passage DNMT3B knockout (3BKO) hESCs, we identified regions representing primary and direct DNMT3B targets methylated during the embryonic wave of de novo methylation. Possibly, the ICF1 hypo-DMRs uniquely overlapping with those detected in late passage 3BKO hESCs might represent genomic regions/genes at which DNMT3B plays a predominant role in DNA methylation maintenance. However, it should be noted that the DNMT3B variants in ICF1 cells sustain some residual activity, and therefore are predicted to have a milder effect compared to complete inactivation of the protein.
Recent observations suggest that in stem and progenitor cells, regions separated by dozens of mega bases may be anchored in “grand canyons” (Zhang et al. 2020). These anchorage points, known also as DMVs, are devoid of DNA methylation, and highly conserved among different cell lines and species (Xie et al. 2013). Deletion of Dnmt3a in mouse hematopoietic stem cells led to the erosion of DNA methylation at valleys (Jeong et al. 2014). Notably, DNMT3B LOF in ICF1 iPSCs similarly affects methylation levels at DMVs edges, thus leading to DMV expansion. This was similarly observed at a significant number of CGIs. Hence, DNMT3B contributes to preserving the methylation status and borders of DMVs and CGIs in WT pluripotent stem cells.
In addition to heterochromatic regions, ICF1 is also characterized by DNA methylation changes at genic regions that lead to expression deregulation (Jin et al. 2008, 2010; Ehrlich et al. 2008). When we studied gene regions in ICF1 iPSCs we frequently observed abnormally low DNA methylation levels in proximity to gene regulatory elements, such as CGI-rich promoters, as well as at enhancer-like regions. Consistently, we observed a significant overlap between the hypo-DMRs identified in ICF1 iPSCs and those described in ICF1 patient peripheral blood and derived LCLs (Gatto et al. 2017; Velasco et al. 2018). However, gene expression was only mildly affected in ICF1 iPSCs, and the pluripotent state was undisrupted (Sagie et al. 2014). We did detect up-regulation of several lineage-commitment genes, suggesting that DNMT3B-mediated DNA methylation may prevent their ectopic expression in pluripotent stem cells. We also found down-regulated genes associated with intragenic hypomethylation. However, we cannot determine unequivocally whether the gene body hypomethylation observed at genes with reduced expression in ICF1 iPSCs is a consequence or a cause of the reduced expression.
When scrutinizing the genes specifically hypomethylated in ICF1 iPSCs, we observed enrichment of genes involved in transcriptionally altered pathways in ICF1 LCLs, such as B cell receptor signaling, T cells costimulation, cytokine signaling in the immune system and receptor tyrosine kinase signaling (Gatto et al. 2017). Thus, we hypothesize that the major transcriptional consequences of aberrant hypomethylation in pluripotent embryonic cells, are postponed to later developmental stages at which the affected genes are normally activated, and additional required factors are present, or at an aberrant temporal window in the case of precocious gene activation. Supporting this hypothesis, we find that in hematopoietic progenitor cells derived from ICF1 patient iPSCs, genes that display aberrant expression are abnormally hypomethylated at the undifferentiated state.
A highly relevant example is the immunoglobulin IGH gene cluster, which is hypomethylated in ICF1 iPSCs and in ICF1 LCLs, but transcriptionally affected only in ICF1 LCLs and peripheral blood. Within the IGH cluster, the immunoglobulin IGHA1, IGHG1, IGHG3 genes are aberrantly silenced in ICF1 LCLs, whereas the expression of IGHM and IGHD genes is variably affected (Ehrlich 2001; Jin et al. 2008; Gatto et al. 2017). These transcriptional defects may contribute to the hypogammaglobulinemia caused by the defective terminal B cell differentiation, which is the major cause of immunodeficiency in ICF syndrome patients (Blanco-Betancourt et al. 2004; Weemaes et al. 2013). Loss of ZBTB24 and Lsh/HELLS functions, responsible for ICF2 and ICF4 subtypes, respectively, has also been associated with specific defects in immunoglobulin class-switch recombination and consequently immunoglobulin production and isotype balance (He et al. 2020; Helfricht et al. 2020). However, the exact molecular link between hypomethylation at the IGH cluster and IGHG and IGHA transcriptional repression in ICF1-4 patients is still unknown. Supporting this notion are recent studies in mice that demonstrate that methylation patterns at the majority of cis-elements at the immunoglobulin heavy chain locus, including promoters, enhancers, and insulators, are established early in development and faithfully maintained independently of B cell activation or transcription initiation at the constant genes (Oudinet et al. 2019).
Acquisition of hematopoietic and lymphopoietic cell identity requires strict epigenetic control to ensure stepwise activation of lineage-specific gene expression programs. An increasing body of evidence suggests that gene expression changes that direct hematopoietic and B cell differentiation require cross talk between TFs and the epigenetic machinery, including DNA methylation changes. Several TFs that play key roles in hematopoietic and B cell fate decisions, such as TCF3, EBF1 (Lin et al. 2010), TAL1 (Real et al. 2012), and MYC (Laurenti et al. 2008), act as pioneer factors by binding DNA methylated regions and inducing local chromatin accessibility and demethylation of naive chromatin (Boller et al. 2016; Lio et al. 2016; Li et al. 2018). Here, we found that a significant fraction of hypo-DMRs in ICF1 iPSC coincides with the target regions of these TFs in hematopoietic stem and progenitor cells and LCLs. We thus suggest that disruption of the normal methylation at target regions in ICF1 iPSCs affects TF binding during cell differentiation, and interferes with the normal regulation of lineage-specific gene expression programs. Alternatively, abnormal TF binding to these sites potentially caused by tissue-specific TF up-regulation might prevent them from being methylated in patient cells during development. Further studies of ICF1 iPSCs differentiated towards hematopoietic and lymphoid progenitors are required to fully investigate this aspect.
The recent development of genome editing tools promotes new strategies for treating genetic diseases. Although in many genetic diseases the repair or restoration of an affected enzyme is likely to rescue the disease phenotype, it is unclear whether this is achievable in the case of a genetic disease in which the enzymatic disorder affects the epigenetic landscape throughout the genome, and thus the expression of numerous genes. Whether the multiple epigenetic changes triggered by hypomethylation, as in ICF syndrome, are reversible, remains unclear. Because iPSCs model the earliest developmental stage at which DNA methylation is disrupted, we took advantage of this platform to discover whether the rescue of the molecular abnormalities in ICF1 syndrome is feasible. Here, we demonstrated that upon CRISPR-Cas9 correction of the DNMT3B disease-causing variants, the majority (75%) of the genomic targets regained DNMT3B binding, and respectively, the majority of the hypomethylated sites in ICF1 iPSCs fully or partially restored WT iPSC methylation patterns. These restored regions included the IGH locus, and several immune-related genes with functions in the innate and adaptive immune responses, thereby indicating that these genes are premarked by DNMT3B activity at a very early stage of development. Nevertheless, up to 10% of the affected regions failed to regain DNA methylation in both corrected clones, and 15% were corrected in only one iPSC clone, suggesting that the recruitment of the corrected enzyme to these specific regions is impeded by the epigenetic memory of the disease state in the parental cells.
Our multi-omic approach enabled us to identify the intertwined action of several factors that inhibit the recruitment of the corrected DNMT3B to the unrescued target regions (Fig. 5E). First and foremost, these regions are characterized by the most severe hypomethylation in patient iPSCs, which in most cases, also display the highest abnormal increase of H3K4me3. Notably, at hypo-DMRs in which H3K4me3 was elevated to a milder degree, the corrected DNMT3B protein could overcome the abnormal H3K4me3 levels and restore normal H3K4me3 and DNA methylation levels. Thus, there appears to be a certain threshold of H3K4me3 enrichment beyond which DNMT3B fails to bind its target regions and reestablish normal chromatin characteristics. This notion is further confirmed by our previous results demonstrating that pharmacological inhibition of MLL methyltransferase/WDR5 interaction in corrected iPSCs reduces H3K4me3 and partially increases DNA methylation at correction-resistant subtelomeres (Toubiana et al. 2019). The hypomethylated regions associated with the highest abnormal H3K4me3 levels mostly correspond to areas with active and permissive chromatin features, such as promoters and gene regulatory regions, which are characterized by relatively high H3K4me3 levels also in WT iPSCs. Accordingly, centromeric and pericentromeric regions that retain normal H3K4me3 levels in ICF1 iPSCs despite their hypomethylation, efficiently restore DNA methylation profiles in the corrected clones.
Additional factors besides H3K4me3 enrichment are predicted to inhibit DNMT3B binding. In this context, we found that a fraction of regions with persisting DNA hypomethylation showed a significant increase in CTCF binding. CTCF is categorized as a DNA methylation–sensitive TF, its occupancy across the genome is strongly linked with CG methylation levels, and its binding blocks de novo DNA methylation (Bell and Felsenfeld 2000; Wang et al. 2012; Hashimoto et al. 2017). However, beyond abnormal CTCF binding and H3K4me3 enrichment, additional factors are predicted to inhibit DNA methylation recovery, such as TET enzymes and/or methylation-sensitive TFs. Further studies will be necessary to characterize the contribution of these additional factors.
In conclusion, analysis of remethylation-resistant genomic loci in gene corrected ICF1 iPSCs, sheds light on the potentialities and limitations in repairing the abnormal epigenome of ICF1 syndrome. Restoration of normal methylation level at immunoglobulin genes and other immune-related genes is predicted to correct their expression, which is severely affected in patient lymphocytes, thereby potentially ameliorating the immunodeficiency phenotype. Additional relevant chromosomal regions undergoing full methylation correction are the centromeric and pericentromeric regions. Based on these observations, we might expect that the chromosomal aberrations of Chr 1, 16, and 9, which represent the cytological phenotype of ICF patients, may also be corrected. However, to provide evidence of these improvements, these phenotypes are required to be evaluated in terminally differentiated cells.
The limitations on the ability of the corrected enzyme to fully carry out its normal activity are caused by the abnormal epigenetic landscape of ICF1 chromatin. Full characterization of these abnormalities will assist in designing future strategies for alleviating the obstacles preventing the full recovery of normal DNA methylation patterns and the associated epigenetic features in ICF1 cells.
Methods
Cells and cell culture conditions
In this study we utilized iPSCs from two unrelated ICF1 patients, pR (female) and pG (male), their isogenic corrected clones (cR7 and cR35, cG13 and CG50) obtained following CRISPR-Cas9 correction of DNMT3B mutations, and control UN1-22 iPSCs, defined as WT1 iPSCs (Shinnawi et al. 2015; Toubiana et al. 2019). An additional control iPSC line (UWWC1-DS2U from WiCell repository) defined as WT3 iPSCs, was utilized for gene expression qPCR experiments. The iPSC lines were cultured on Geltrex LDEV-Free Reduced Growth Factor Basement Membrane Matrix (Gibco A1413302) coated plates and maintained in StemMACS iPS-Brew XF, human medium (Miltenyi Biotec 130-104-368). Cells were passaged every 4–6 d. Cells were dissociated from plates with StemPro Accutase Cell Dissociation Reagent (Gibco A1110501), and following each passage, 1× of RevitaCell Supplement (100×) (Gibco A2644501), was added to the media. The identity of iPSCs was verified by Sanger sequencing of the patient-specific mutations and of the silent mutations incorporated during the editing process into the DNMT3B gene. All cells were negative in a screen for mycoplasma.
Hematopoietic progenitor cells (HPCs) were obtained from WT and ICF iPSCs through a feeder-free differentiation protocol (StemDiff Hematopoietic Kit, Stem Cell Technologies 05310). The cell population expressing the pan hematopoietic marker CD43 (current and HGNC-approved symbol is SPN) (eBioscience eBio84-3C1, PE) was analyzed for PTPRC (previously known as CD45) (eBioscience HI30, FITC) and CD34 (eBioscience 4H11, APC) expression, and sorted by FACS (BD FACS ARIAIII cell sorter) for bulk RNA-seq experiments.
Whole genome bisulfite sequencing (WGBS) and data processing
We extracted genomic DNA from WT1 iPSC, ICF1 iPSCs, and their respective corrected clones, using the Wizard Genomic DNA Purification Kit (Promega A1125). Approximately 100 ng of genomic DNA was used for the WGBS experiment. The bisulfite-treated library preparation and the 100 bp length paired-end (PE) sequencing were performed at Genomix4Life S.r.l (Salerno, Italy) using the Illumina NextSeq 500 system. After adaptor trimming and quality filtering with cutadapt (Martin 2011) we used Bismark (Krueger and Andrews 2011) to align the PE reads (Supplemental Table S1) to the bisulfite-converted hg38 reference assembly and extracted the cytosine methylation calls. The methylation level was expressed as the ratio of the number of Cs over the total number of Cs and Ts at base resolution and across the regions of interest. We employed the methylKit R package to identify the DMRs (Akalin et al. 2012). Hypomethylated DMRs (hypo-DMRs; 1 kb-size) were detected by setting a threshold for the difference in methylation percentage (meth.diff score) > 25%; adjusted P-value (q) < 0.01 and level = “hypo”, when individually comparing ICF1 iPSCs and corrected clones to WT1 iPSCs. Then, we filtered the lists of hypo-DMRs by removing from downstream analyses the hypo-DMRs, where the internal control WT1 iPSCs showed a difference over 0.2 in the methylation ratio compared to WT2, an additional control iPSC corresponding to the human dermal fibroblasts-derived iPSC downloaded from the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) accession number GSM1385983 (Ma et al. 2014). The ICF1 hypo-DMRs were defined as rescued in corrected clones if, compared with the internal control WT1, they had (i) meth.diff score < 25%, level = ”hypo” and/or q > 0.01; or (ii) meth.diff > 25% and level = ”hyper”. For detailed descriptions and further downstream analyses, see the Supplemental Material.
Chromatin immunoprecipitation sequencing (ChIP-seq) and data processing
We performed ChIP to identify DNMT3B, H3K4me3, H3K36me3, and CTCF enriched regions in WT1, ICF1 iPSCs and their corrected clones (for CTCF only, one corrected clone for each patient iPSCs, cR7 and cG50). 3 × 106 cells were fixed twice for 40 min in 20 mM ethylene glycol bis (succinimidyl succinate) (EGS), and for 10 min in 1% formaldehyde solution to promote crosslinking. Excessive formaldehyde was quenched with 125 mM glycine solution and cells were washed twice with PBS. Cells were then lysed in a solution containing 10 mM Tris-HCl pH 8, 100 mM NaCl, 10 mM EDTA, 0.25% Triton X-100, and cOmplete Protease Inhibitor Cocktail (Roche PICS 11697498001). Nuclei were lysed in a solution of 1% SDS containing 50 mM HEPES-KOH (pH 7.5), 150 mM NaCl, 1% SDS, 2 mM EDTA, 1% Triton X-100, 0.1% NaDOC and PICS. After centrifugation, the chromatin pellet was resuspended in the same solution containing 0.1% SDS. For ChIP with the DNMT3B and CTCF antibody, sonication was conducted using the Covaris S220 apparatus (12 cycles, 40” on, 20” off, intensity 4). Sonicated chromatin was incubated overnight at 4°C with Magna ChIP Protein A Magnetic Beads (Merck Millipore 16661) and anti-DNMT3B antibody (Diagenode C15410218) and anti-CTCF antibody (Abcam ab70303). Decrosslinking from beads was performed in a 1% SDS solution containing Tris-HCl pH 8 and 10 mM EDTA for 2 h at 69°C with agitation. Decrosslinking of the DNA-protein complex was performed by adding Proteinase K (20 ng/mL) for 2 h at 62°C with agitation. DNA was then purified using Expin PCR SV (GeneAll 103–102). ChIP with antibodies for histone marks (H3K4me3, ab8580; H3K36me3, ab9050, Abcam) was performed as previously described (Matarazzo et al. 2004). Immunoprecipitated samples and corresponding mock samples were used for ChIP-qPCR (Supplemental Table S2). ChIP libraries (200–500 bp fragment sizes) were generated from two biological IP replicates, one input for DNMT3B, one input per IP for CTCF and one pooled input for histone marks (H3K4me3 and H3K36me3) per iPSC sample. Single-end (SE) sequencing performed on the Illumina HiSeq 2500 system at the Technion Genome Center (Haifa, Israel) produced reads of desired length from each IP and input (Supplemental Table S1). The preprocessing steps included quality analysis and trimming of sequenced SE reads, alignment to the hg38 reference assembly using Bowtie 2 aligner (Langmead and Salzberg 2012) and conversion of deduplicated BAM reads to BED files for peak calling. We used the SICER2 tool (Zang et al. 2009) to call enriched IP peaks versus their input and to identify DERs in ICF1 iPSCs and their corrected clones compared to WT1 iPSCs. Increased H3K4me3 DERs identified in ICF1 iPSCs were defined as rescued in corrected iPSCs if not detected in the list of increased H3K4me3 DERs compared to WT1 or if called as decreased H3K4me3 DERs in this comparison. The rescue of decreased DNMT3B DERs in corrected iPSCs was measured following a similar criterion. Average ChIP-seq enrichment profiles and heatmaps were generated using ngs.plot (Shen et al. 2014). For a detailed description and further downstream analyses, see Supplemental Material.
RNA sequencing and data processing
Total RNA extraction from iPSCs was performed using TRI Reagent (Sigma-Aldrich T9424), according to the manufacturer's protocol. RNA was quantified using NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific) and the absorbance ratio 260/280 was measured to assess the RNA purity. Extracted RNA was further purified from genomic DNA using the TURBO DNA-free Kit (Invitrogen AM1907). We tested the absence of residual DNA, using RNA as a template to amplify a genomic region (MYOD1 gene). The quality and integrity of RNA was assessed by measuring the RNA Integrity Number (RIN) with the Bioanalyzer instrument (Agilent). Strand-specific libraries for RNA-seq analysis of iPSCs were prepared according to Illumina's instructions and the libraries were sequenced at the IGA (Udine, Italy) using the Illumina HiSeq 2500 platform. We obtained PE reads of 125 bp length from two technical replicates per each iPSC sample (Supplemental Table S1). First, we performed quality control to remove low-quality PE reads and adapter sequences and then we aligned the sequences to the hg38 reference assembly using HISAT2 (Kim et al. 2019) and reverse-strand specific parameters. Next, we quantified gene expression as read counts using the featureCounts function from Rsubread package (Liao et al. 2019), and identified the differentially expressed genes (DE) using the NOIseq R package (Tarazona et al. 2015), after normalization and batch effect removal. We applied a threshold of pp > 0.9 to call DE genes and divided them into up- and down-regulated DE genes based on the log2 fold change in expression versus WT1 (±log2FC). For the analyses including the integration of gene expression with DNA methylation (Supplemental Fig. S5D; Fig. 3A) we relaxed the threshold, which defines genes as DE, to pp > 0.8. DE genes identified in ICF1 iPSCs were defined as fully rescued if their posterior probability in the comparison versus WT1 was pp < 0.5 in both clones, as partially rescued if pp < 0.5 in only in one clone, and as slightly rescued if in both corrected clones 0.5 < pp < 0.8. For a detailed description of RNA-seq experiments performed in hematopoietic progenitor cells differentiated from patient and WT iPSCs, see Supplemental Material.
Statistical analysis
Specific statistical tests performed in genome-wide studies are described in figure legends and the “Results” section. P-values were adjusted with Benjamini–Hochberg method with FDR < 0.01 (BH-FDR) and the significant P-adjusted (P-adj) values were represented as follows: (*) P-adj < 0.01; (**) P-adj < 0.001; (***) P-adj < 0.0001. The data analyzed using RT-qPCR and ChIP-qPCR is presented as mean ± SD from independent triplicates, each amplified twice. Statistical analyses were performed using a one-tail two-sample Student's t-test; (*) P-value < 0.05, (**) P-value < 0.01, (***) P-value < 0.001. In the case of RT-qPCR, statistical significance was calculated comparing each ICF1 or corrected iPSCs to each WT (WT1 and WT3) iPSCs separately and the least significant values were reported in the corresponding figures.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE197925.
Supplementary Material
Acknowledgments
We are grateful to Miriam Gagliardi for her valuable help in the initial phases of the study and for the critical reading of the manuscript. We thank Dario Righelli and Ankit Verma for their advice about the application of bioinformatic tools for next-generation sequencing (NGS) data analysis of iPSC samples, and Vincenzo Lullo for his help in carrying out the ChIP experiments. We acknowledge Sequentia Biotech for their contribution to the analysis of RNA-seq data from the HPC samples. We thank the FACS facility of the Institute of Genetics and Biophysics “Adriano Buzzati Traverso,” CNR. We are grateful to Daniel Kornitzer and Andrea Riccio for their comments on the manuscript and for helpful discussions. This research was funded by (i) the Telethon (GGP15209) and PON/MISE (2014-2020 FESR F/050011/ 01-02/X32) to M.R.M.; (ii) the Israel Science Foundation (grant number 1362/17) to S.S.; (iii) the PRIN MUR (20179J2P9J) to C.A. V.P.K. and M.K. acknowledge INCIPIT Innovative Life Science PhD Program in South Italy (cofunded by Marie Sklodowska Curie Action) n. 665403, for funding her PhD scholarship. B.M. is grateful to POR Campania FSE 2014/2020 for funding her PhD scholarship. S.T. is grateful to The Edmond de Rothschild Foundation (IL) for funding her PhD scholarship.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276986.122.
Competing interest statement
The authors declare no competing interests.
References
- Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE. 2012. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 13: R87. 10.1186/gb-2012-13-10-r87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auclair G, Guibert S, Bender A, Weber M. 2014. Ontogeny of CpG island methylation and specificity of DNMT3 methyltransferases during embryonic development in the mouse. Genome Biol 15: 545. 10.1186/s13059-014-0545-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, Akalin A, Schübeler D. 2015. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520: 243–247. 10.1038/nature14176 [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485. 10.1038/35013100 [DOI] [PubMed] [Google Scholar]
- Blanco-Betancourt CE, Moncla A, Milili M, Jiang YL, Viegas-Péquignot EM, Roquelaure B, Thuret I, Schiff C. 2004. Defective B-cell-negative selection and terminal differentiation in the ICF syndrome. Blood 103: 2683–2690. 10.1182/blood-2003-08-2632 [DOI] [PubMed] [Google Scholar]
- Boller S, Ramamoorthy S, Akbas D, Nechanitzky R, Burger L, Murr R, Schübeler D, Grosschedl R. 2016. Pioneering activity of the C-terminal domain of EBF1 shapes the chromatin landscape for B cell programming. Immunity 44: 527–541. 10.1016/j.immuni.2016.02.021 [DOI] [PubMed] [Google Scholar]
- Dahlet T, Argüeso Lleida A, Al Adhami H, Dumas M, Bender A, Ngondo RP, Tanguy M, Vallet J, Auclair G, Bardet AF, et al. 2020. Genome-wide analysis in the mouse embryo reveals the importance of DNA methylation for transcription integrity. Nat Commun 11: 3153. 10.1038/s41467-020-16919-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Greef JC, Wang J, Balog J, den Dunnen JT, Frants RR, Straasheijm KR, Aytekin C, van der Burg M, Duprez L, Ferster A, et al. 2011. Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Am J Hum Genet 88: 796–804. 10.1016/j.ajhg.2011.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. 2001. DNA methyltransferase 3B mutations linked to the ICF syndrome cause dysregulation of lymphogenesis genes. Hum Mol Genet 10: 2917–2931. 10.1093/hmg/10.25.2917 [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Sanchez C, Shao C, Nishiyama R, Kehrl J, Kuick R, Kubota T, Hanash SM. 2008. ICF, an immunodeficiency syndrome: DNA methyltransferase 3B involvement, chromosome anomalies, and gene dysregulation. Autoimmunity 41: 253–271. 10.1080/08916930802024202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Iny Stein T, Rosen N, Kohn A, Twik M, Safran M, et al. 2017. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database 2017: bax028. 10.1093/database/bax028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto S, Della Ragione F, Cimmino A, Strazzullo M, Fabbri M, Mutarelli M, Ferraro L, Weisz A, D'Esposito M, Matarazzo MR. 2010. Epigenetic alteration of microRNAs in DNMT3B-mutated patients of ICF syndrome. Epigenetics 5: 427–443. 10.4161/epi.5.5.11999 [DOI] [PubMed] [Google Scholar]
- Gatto S, Gagliardi M, Franzese M, Leppert S, Papa M, Cammisa M, Grillo G, Velasco G, Francastel C, Toubiana S, et al. 2017. ICF-specific DNMT3B dysfunction interferes with intragenic regulation of mRNA transcription and alternative splicing. Nucleic Acids Res 45: 5739–5756. 10.1093/nar/gkx163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D, Shao C, Tuck-Muller CM, Sogorovic S, Pålsson E, Smeets D, Ehrlich M. 2005. Interphase chromosomal abnormalities and mitotic missegregation of hypomethylated sequences in ICF syndrome cells. Chromosoma 114: 118–126. 10.1007/s00412-005-0343-7 [DOI] [PubMed] [Google Scholar]
- Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM. 1999. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci 96: 14412–14417. 10.1073/pnas.96.25.14412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Wang D, Horton JR, Zhang X, Corces VG, Cheng X. 2017. Structural basis for the versatile and methylation-dependent binding of CTCF to DNA. Mol Cell 66: 711–720.e3. 10.1016/j.molcel.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Ren J, Xu X, Ni K, Schwader A, Finney R, Wang C, Sun L, Klarmann K, Keller J, et al. 2020. Lsh/HELLS is required for B lymphocyte development and immunoglobulin class switch recombination. Proc Natl Acad Sci 117: 20100–20108. 10.1073/pnas.2004112117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfricht A, Thijssen PE, Rother MB, Shah RG, Du L, Takada S, Rogier M, Moritz J, IJspeert H, Stoepker C, et al. 2020. Loss of ZBTB24 impairs nonhomologous end-joining and class-switch recombination in patients with ICF syndrome. J Exp Med 217: e20191688. 10.1084/jem.20191688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Vidal E, Sayols S, Sanchez-Mut JV, Moran S, Medina I, Sandoval J, Simó-Riudalbas L, Szczesna K, Huertas D, et al. 2012. Whole-genome bisulfite DNA sequencing of a DNMT3B mutant patient. Epigenetics 7: 542–550. 10.4161/epi.20523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Wu Z, Liu Z, Hu G, Yu J, Chang KH, Kim K-P, Le T, Faull KF, Rao N, et al. 2014. Selective demethylation and altered gene expression are associated with ICF syndrome in human-induced pluripotent stem cells and mesenchymal stem cells. Hum Mol Genet 23: 6448–6457. 10.1093/hmg/ddu365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liu X, Li D, Shao Z, Cao H, Zhang Y, Trompouki E, Bowman TV, Zon LI, Yuan G-C, et al. 2016. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev Cell 36: 9–23. 10.1016/j.devcel.2015.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanpierre M, Turleau C, Aurias A, Prieur M, Ledeist F, Fischer A, Viegas-Pequignot E. 1993. An embryonic-like methylation pattern of classical satellite DNA is observed in ICF syndrome. Hum Mol Genet 2: 731–735. 10.1093/hmg/2.6.731 [DOI] [PubMed] [Google Scholar]
- Jeong M, Sun D, Luo M, Huang Y, Challen GA, Rodriguez B, Zhang X, Chavez L, Wang H, Hannah R, et al. 2014. Large conserved domains of low DNA methylation maintained by Dnmt3a. Nat Genet 46: 17–23. 10.1038/ng.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeziorska DM, Murray RJS, De Gobbi M, Gaentzsch R, Garrick D, Ayyub H, Chen T, Li E, Telenius J, Lynch M, et al. 2017. DNA methylation of intragenic CpG islands depends on their transcriptional activity during differentiation and disease. Proc Natl Acad Sci 114: E7526–E7535. 10.1073/pnas.1703087114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B, Tao Q, Peng J, Soo HM, Wu W, Ying J, Fields CR, Delmas AL, Liu X, Qiu J, et al. 2008. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet 17: 690–709. 10.1093/hmg/ddm341 [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572. 10.1093/bioinformatics/btr167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, Knoepfler PS, Cheng P-F, MacDonald HR, Eisenman RN, et al. 2008. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell 3: 611–624. 10.1016/j.stem.2008.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-M, Lee J, Noh K-M, Choi W-Y, Jeon S, Oh GT, Kim-Ha J, Jin Y, Cho S-W, Kim Y-J. 2017. Intragenic CpG islands play important roles in bivalent chromatin assembly of developmental genes. Proc Natl Acad Sci 114: E1885–E1894. 10.1073/pnas.1613300114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926. 10.1016/0092-8674(92)90611-F [DOI] [PubMed] [Google Scholar]
- Li R, Cauchy P, Ramamoorthy S, Boller S, Chavez L, Grosschedl R. 2018. Dynamic EBF1 occupancy directs sequential epigenetic and transcriptional events in B-cell programming. Genes Dev 32: 96–111. 10.1101/gad.309583.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, Akopian V, Gifford CA, Donaghey J, Galonska C, et al. 2015. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat Genet 47: 469–478. 10.1038/ng.3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2019. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 47: e47. 10.1093/nar/gkz114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. 2010. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol 11: 635–643. 10.1038/ni.1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lio C-W, Zhang J, González-Avalos E, Hogan PG, Chang X, Rao A. 2016. Tet2 and Tet3 cooperate with B-lineage transcription factors to regulate DNA modification and chromatin accessibility. eLife 5: e18290. 10.7554/eLife.18290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo Q-M, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322. 10.1038/nature08514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Morey R, O'Neil RC, He Y, Daughtry B, Schultz MD, Hariharan M, Nery JR, Castanon R, Sabatini K, et al. 2014. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature 511: 177–183. 10.1038/nature13551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraschio P, Zuffardi O, Dalla Fior T, Tiepolo L. 1988. Immunodeficiency, centromeric heterochromatin instability of chromosomes 1, 9, and 16, and facial anomalies: the ICF syndrome. J Med Genet 25: 173–180. 10.1136/jmg.25.3.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Martins-Taylor K, Schroeder DI, LaSalle JM, Lalande M, Xu R-H. 2012. Role of DNMT3B in the regulation of early neural and neural crest specifiers. Epigenetics 7: 71–82. 10.4161/epi.7.1.18750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masalmeh RHA, Taglini F, Rubio-Ramon C, Musialik KI, Higham J, Davidson-Smith H, Kafetzopoulos I, Pawlicka KP, Finan HM, Clark R, et al. 2021. De novo DNA methyltransferase activity in colorectal cancer is directed towards H3K36me3 marked CpG islands. Nat Commun 12: 694. 10.1038/s41467-020-20716-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarazzo MR, Lembo F, Angrisano T, Ballestar E, Ferraro M, Pero R, De Bonis ML, Bruni CB, Esteller M, D'Esposito M, et al. 2004. In vivo analysis of DNA methylation patterns recognized by specific proteins: coupling CHIP and bisulfite analysis. BioTechniques 37: 666–8, 670, 672–3. 10.2144/04374DD02 [DOI] [PubMed] [Google Scholar]
- Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato C, Basile G, Maldotti M, Anselmi F, Oliviero S. 2017. Intragenic DNA methylation prevents spurious transcription initiation. Nature 543: 72–77. 10.1038/nature21373 [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257. 10.1016/S0092-8674(00)81656-6 [DOI] [PubMed] [Google Scholar]
- Oudinet C, Braikia F-Z, Dauba A, Santos JM, Khamlichi AA. 2019. Developmental regulation of DNA cytosine methylation at the immunoglobulin heavy chain constant locus. PLoS Genet 15: e1007930. 10.1371/journal.pgen.1007930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. 2000. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci 97: 5237–5242. 10.1073/pnas.97.10.5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real PJ, Ligero G, Ayllon V, Ramos-Mejia V, Bueno C, Gutierrez-Aranda I, Navarro-Montero O, Lako M, Menendez P. 2012. SCL/TAL1 regulates hematopoietic specification from human embryonic stem cells. Mol Ther 20: 1443–1453. 10.1038/mt.2012.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagie S, Ellran E, Katzir H, Shaked R, Yehezkel S, Laevsky I, Ghanayim A, Geiger D, Tzukerman M, Selig S. 2014. Induced pluripotent stem cells as a model for telomeric abnormalities in ICF type I syndrome. Hum Mol Genet 23: 3629–3640. 10.1093/hmg/ddu071 [DOI] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E. 2014. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15: 284. 10.1186/1471-2164-15-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnawi R, Huber I, Maizels L, Shaheen N, Gepstein A, Arbel G, Tijsen AJ, Gepstein L. 2015. Monitoring human-induced pluripotent stem cell-derived cardiomyocytes with genetically encoded calcium and voltage fluorescent reporters. Stem Cell Reports 5: 582–596. 10.1016/j.stemcr.2015.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simo-Riudalbas L, Diaz-Lagares A, Gatto S, Gagliardi M, Crujeiras AB, Matarazzo MR, Esteller M, Sandoval J. 2015. Genome-wide DNA methylation analysis identifies novel hypomethylated non-pericentromeric genes with potential clinical implications in ICF syndrome. PLoS One 10: e0132517. 10.1371/journal.pone.0132517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan HK, Wu C-S, Li J, Tan ZH, Hoffman JR, Fry CJ, Yang H, Di Ruscio A, Tenen DG. 2019. DNMT3B shapes the mCA landscape and regulates mCG for promoter bivalency in human embryonic stem cells. Nucleic Acids Res 47: 7460–7475. 10.1093/nar/gkz520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S, Furió-Tarí P, Turrà D, Pietro AD, Nueda MJ, Ferrer A, Conesa A. 2015. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res 43: e140. 10.1093/nar/gkv711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijssen PE, Ito Y, Grillo G, Wang J, Velasco G, Nitta H, Unoki M, Yoshihara M, Suyama M, Sun Y, et al. 2015. Mutations in CDCA7 and HELLS cause immunodeficiency–centromeric instability–facial anomalies syndrome. Nat Commun 6: 7870. 10.1038/ncomms8870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubiana S, Gagliardi M, Papa M, Manco R, Tzukerman M, Matarazzo MR, Selig S. 2019. Persistent epigenetic memory impedes rescue of the telomeric phenotype in human ICF iPSCs following DNMT3B correction. eLife 8: e47859. 10.7554/eLife.47859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco G, Grillo G, Touleimat N, Ferry L, Ivkovic I, Ribierre F, Deleuze J-F, Chantalat S, Picard C, Francastel C. 2018. Comparative methylome analysis of ICF patients identifies heterochromatin loci that require ZBTB24, CDCA7 and HELLS for their methylated state. Hum Mol Genet 27: 2409–2424. 10.1093/hmg/ddy130 [DOI] [PubMed] [Google Scholar]
- Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, et al. 2012. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res 22: 1680–1688. 10.1101/gr.136101.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weemaes CMR, van Tol MJD, Wang J, van Ostaijen-ten Dam MM, van Eggermond MCJA, Thijssen PE, Aytekin C, Brunetti-Pierri N, van der Burg M, Graham Davies E, et al. 2013. Heterogeneous clinical presentation in ICF syndrome: correlation with underlying gene defects. Eur J Hum Genet 21: 1219–1225. 10.1038/ejhg.2013.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. 2013. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153: 1134–1148. 10.1016/j.cell.2013.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G-L, Bestor TH, Bourc'his D, Hsieh C-L, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Péquignot E. 1999. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402: 187–191. 10.1038/46052 [DOI] [PubMed] [Google Scholar]
- Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. 2014. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 26: 577–590. 10.1016/j.ccr.2014.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehezkel S, Segev Y, Viegas-Péquignot E, Skorecki K, Selig S. 2008. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum Mol Genet 17: 2776–2789. 10.1093/hmg/ddn177 [DOI] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. 2009. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25: 1952–1958. 10.1093/bioinformatics/btp340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jeong M, Huang X, Wang XQ, Wang X, Zhou W, Shamim MS, Gore H, Himadewi P, Liu Y, et al. 2020. Large DNA methylation nadirs anchor chromatin loops maintaining hematopoietic stem cell identity. Mol Cell 78: 506–521.e6. 10.1016/j.molcel.2020.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.