Abstract
Proteins and RNAs are fundamental parts of biological systems, and their interactions affect many essential cellular processes. Therefore, it is crucial to understand at a molecular and at a systems level how proteins and RNAs form complexes and mutually affect their functions. In the present mini-review, we will first provide an overview of different mass spectrometry (MS)-based methods to study the RNA-binding proteome (RBPome), most of which are based on photochemical cross-linking. As we will show, some of these methods are also able to provide higher-resolution information about binding sites, which are important for the structural characterisation of protein–RNA interactions. In addition, classical structural biology techniques such as nuclear magnetic resonance (NMR) spectroscopy and biophysical methods such as electron paramagnetic resonance (EPR) spectroscopy and fluorescence-based methods contribute to a detailed understanding of the interactions between these two classes of biomolecules. We will discuss the relevance of such interactions in the context of the formation of membrane-less organelles (MLOs) by liquid–liquid phase separation (LLPS) processes and their emerging importance as targets for drug discovery.
Keywords: cross-linking, mass spectrometry, protein-RNA complexes, protein-RNA interactions
Introduction
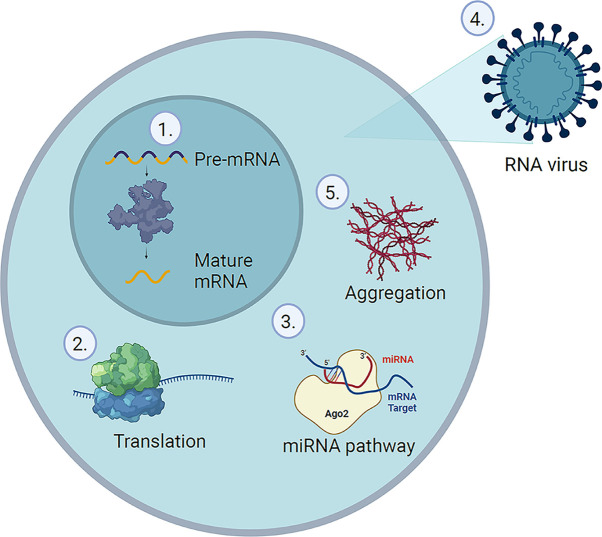
Protein–RNA interactions are involved in many cellular processes, including, but not limited to RNA maturation, stability, translation, and host defence [1] (Figure 1). RNA-binding proteins (RBPs) play a crucial role in these fundamental cellular functions. Consequently, mutations that lead to aberrant RBPs or RBP-binding sites can have severe pathological implications. A typical eukaryotic cell contains tens to hundreds of thousands of protein and RNA species, whose dynamic interactions generate a huge number of transient and stable protein–RNA complexes. In the past, the number of existing RBPs has been significantly underestimated [1–3]. However, the recent improvements in RNA sequencing and mass spectrometry (MS) have facilitated the identification of RBPs from individual samples and up to the proteome-wide scale, defining the RNA-binding proteome (RBPome) [2,4].
Figure 1. Examples for protein–RNA interactions in the cell.
(1) Spliceosome, (2) ribosome, (3) miRNA-binding proteins, (4) RBPs related to viral replication, and (5) oligomers and aggregates of RBPs.
Approaches to study the RBPome
Protein–RNA interactions can be studied from two angles, either by identifying proteins that are bound to a specific RNA or by identifying all RNAs that are bound to a specific protein. Nowadays, most methods are based on UV cross-linking (XL) of such ribonucleoproteins (RNPs) coupled to RNA sequencing or MS. Despite an increasing number of methodologies to study RBPs by MS, most follow a very similar, three-step strategy. In brief, the sample of interest is cross-linked by applying irradiation with UV light at 254 nm to covalently link RNA and protein at their contact site at virtually zero length distance. In a second step, the cross-linked molecules (RNA–protein conjugates or adducts) are enriched. This enrichment can be achieved by many different means and will be discussed in more detail below. Finally, the enriched sample is further purified and cleaned for MS analysis.
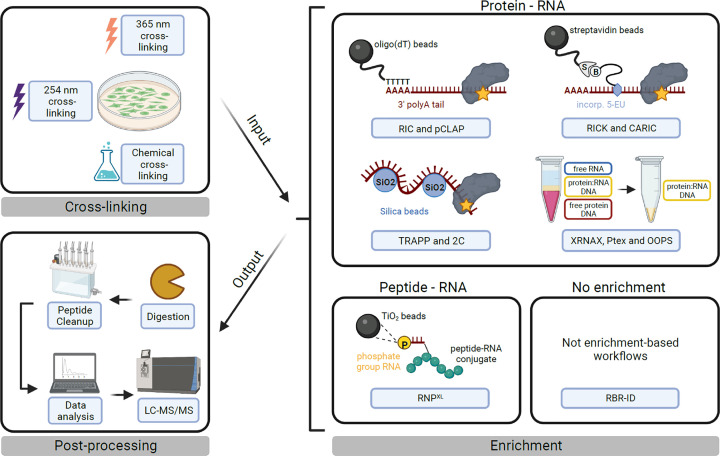
In the last decade, many research groups have successfully developed protocols based on UV cross-linking MS to not only study single RNPs but also the RBPome of different organisms and cell types. Those methods can be divided into six main groups based on their enrichment strategy of RNA–protein or RNA–peptide conjugates (Figure 2):
The family of RNA interactome capture (RIC) protocols comprises a variety of more specialised workflows (vRIC [5], qRIC [6], eRIC [7], cRIC [8], RBDmap [9], serIC [10]) as well as peptide cross-linking and affinity purification (pCLAP [11]), which are all based on the enrichment of poly-adenylated RNA with oligo(dT) beads. This step is highly biased towards mRNAs, but allows stringent enrichment conditions due to the very specific and strong interaction between polyA tails and the beads.
RICK (RNA interactome using click chemistry) and CARIC (click chemistry-assisted RIC) utilise a non-native base, 5-ethynyluridine (5-EU) [12,13]. 5-EU is incorporated in newly transcribed RNA, modified by azide-biotin in a click chemistry reaction, and can be pulled down with streptavidin beads. Due to the high-affinity interaction between biotin and streptavidin, this approach also allows for stringent enrichment conditions.
RNAXL further enriches for RNA–peptide conjugates with titanium dioxide solid-phase extraction after UV cross-linking and protein digestion with trypsin. Since the enrichment of RNA conjugates occurs at the peptide rather than the protein level, such a sample can identify the binding position of RNA and protein at single amino acid resolution as will be discussed in the next chapter [14].
The RBR-ID method does not include a dedicated enrichment step, but compares peptide intensities between cross-linked samples prepared with 4-thiouridine(4-SU)-labelled RNA and non-cross-linked samples. The mass shift introduced by covalently bound RNA will lead to a decrease in signal for successfully cross-linked peptides between the two samples [15].
2C (silica-based solid-phase extraction), TRAPP (total RNA-associated protein purification), and RBS-ID utilise silica beads/membranes for nucleic acid enrichment [16,17].
The family of liquid–liquid phase extraction methods – orthogonal organic phase separation (OOPS), phenol–toluene extraction (PTex), and XRNAX (and recently developed derivatives, e.g., PPE, photoCAX, and targeted RNA–protein identification using OOPS (TROOPS)) – isolate protein–RNA conjugates based on their physiochemical properties [18–23]. Protein–RNA conjugates share properties of RNA and protein. During liquid–liquid phase extraction in an aqueous-organic (e.g., phenol/chloroform) system, cross-linked material will form a layer in between the organic and aqueous phase. This interface layer can be extracted and further purified. A recently developed method that might be interesting, not from a systems-wide, but from a biochemical perspective is TROOPS. TROOPS is based on a previously published liquid–liquid phase extraction method to generate an intermediate, RBP-RNA-XL enriched sample that is used as input for a pulldown of a cross-linked RNA of interest [23].
Figure 2. The protein–RNA UV cross-linking workflow.
Schematic overview of the three steps – cross-linking, enrichment, and postprocessing – of an RBPome-wide UV cross-linking MS experiment. Cross-linking: RNPs can be cross-linked directly in cells (as depicted) using UV light, most commonly at 254 nm for native RNA, at 365 nm for 4-SU modified RNA or – less commonly – using chemicals. Note that cross-linking may also be performed in solution on lysates or purified complexes. Enrichment: this step is highly variable between protocols and can be split into six different modes of enrichment as described in the text. An alternative classification of protocols may be based on the input and/or output material for enrichment, which can be either protein–RNA (cross-links are represented as yellow stars) or peptide–RNA conjugates (as highlighted above the boxes). Although helpful, such a classification is practically more difficult to make, as RNA and protein digestion steps can be sequential or uncoupled. Postprocessing: postprocessing of enriched conjugates includes digestion with proteases and RNases (this step is dependent on the protocol used), clean-up of the digestion products, data acquisition by liquid chromatography tandem mass spectrometry (LC-MS/MS) and data analysis. Data analysis may be performed with conventional MS software or specialised XL-MS software tools (e.g., RNPXL) [14].
Differences between existing protocols summarised above and methodological details of MS analysis are described elsewhere in detail [24–27], including tables with aggregated information about the above-mentioned methods [26,27].
Although UV-XL-MS is currently the gold standard to study RBPomes, it has some major disadvantages to consider, including low cross-linking efficiency (estimated at <5%) and differences in reactivity depending on protein and nucleotide sequences, the relative orientation of amino acid side chains and nucleobases at the interaction site [28,29]. To overcome the low cross-linking efficiency of native RNA, a range of chemically modified bases, most prominently 4-SU, have been introduced; they are more photoreactive and significantly boost cross-linking efficiency. Another strategy that tries to overcome nucleobase preferences and the potentially low sample penetration of UV light uses chemical cross-linking reagents. These are often bifunctional chemicals that react with peptides on the one end and RNA on the other end. A comprehensive list of chemical cross-linkers can be found in the review by Fabris and co-workers [30].
From RBPome to structural read-out
In the past decade, the characterisation of RBPomes deriving from different samples and species has led to many insights into the world of RNA biology. RBPome-wide studies have catalogued an inventory of hundreds to thousands of RBPs in different species (4300 RBPs in human) (https://rbpbase.shiny.embl.de/) [2,4]. These data enable the collection of broad information about RBPs, frequently revealing insights into their structural organisation, like for instance the lack of canonical RNA-binding domains (RBDs) and the binding of RNA to unstructured regions in proteins [9]. Apart from that, many proteins that have been identified in RBPome-wide studies do not have a function known to be involved in RNA biology. Beckmann et al. named this group of RBPs ‘enigmRBPs’ [4], which includes many metabolic enzymes, particularly in the glycolysis pathway [31,32].
Although protein abundance is an important parameter in the cellular context, not only protein quantity but also protein localisation, post-translational modifications (PTMs) and conformational changes are vital for correct protein function. In the case of RBPs, binding to their target RNA and associated changes in, e.g., protein conformation add an additional layer of information that needs to be disentangled in order to understand a protein’s function or an observed phenotype.
Multiple high-throughput approaches that utilise UV cross-linking to characterise the interaction between RNA and protein molecules down to single amino acid resolution have been described in the literature. RNA-sequencing-based approaches focus on extracting bound RNA from RNPs and perform sequencing on the recovered RNA. Regions that bind to the protein(s) will show reduced coverage by sequencing due to covalently linked amino acids at the binding site. Such differences in RNA coverage can be used for RNA footprinting, but require access to sequencing infrastructure. Trendel et al. proposed another approach that can be directly applied to MS-generated datasets. Their method takes advantage of the open modification search (OMS) strategy, which can identify virtually any peptide modification that results in a mass shift. An example for a widely used OMS software is MSFragger [33]. By treating RNA residues as a peptide PTM, OMS does not only identify whether a peptide is cross-linked to RNA but it also proposes the RNA composition (although not necessarily the correct nucleotide order). Additional postprocessing tools such as PTM-Shepherd can in some cases narrow down the binding site to a single amino acid [34]. Similarly, RBS-ID can precisely identify the RNA-binding sites at a single amino acid resolution by performing a complete digestion of the cross-linked RNA using hydrofluoric acid. This strategy focuses on uridine as the most commonly cross-linked base and has identified almost 2000 binding sites at single amino acid resolution with conventional software (‘closed search’) [35].
Among the specific software tools for RNA cross-linking data, Urlaub and co-workers developed RNPXL [14]. For the identification of cross-linking sites, it is necessary to enrich peptide–RNA conjugates rather than protein–RNA conjugates. Metal oxides such as titanium dioxide bind compounds containing phosphate groups (e.g., on the RNA backbone or on phosphorylated amino acid) in a pH-dependent manner [36]. A similar strategy is employed by iTRAPP, which relies on a two-step enrichment protocol, TRAPP and TiO2, and the cross-linking search engine called xiSEARCH [16]. RBDmap is a modified version of RIC that applies sequential digestion and polyA-tail enrichment of the cross-linked protein–RNA complexes to determine the region of the protein that is involved in RNA binding [9]. Binding positions at a peptide level are also defined by pCLAP and RBR-ID methods [11,37]. Although these studies only identified binding regions in the small fraction of RBPs that are known today, they highlight the impact of UV cross-linking MS on structural biology, as in some cases 50% of the binding sites were found in intrinsically disordered regions (IDRs), which are difficult to model by conventional structural methods (discussed further below) [9].
A dedicated workflow that allows the characterisation of RNA–protein interactions of individual complexes or complex subunits is CLIR-MS (cross-linking of isotope-labelled RNA coupled to MS) [38]. Similar to RNPXL, CLIR-MS is based on the enrichment of peptide–RNA conjugates with metal oxides and allows the identification of RNA–protein-binding interfaces down to the single amino acid level. In addition, incorporation of stable isotopes at specific positions or regions of the RNA sequence allows the determination of the binding sites on the RNA. Thus, CLIR-MS is very well suited for follow-up studies of proposed RBPs without additional structural information, if recombinant protein is available.
CLIR-MS has initially been used to pinpoint interaction sites between PTBP1 and one of its natural RNA targets, a part of the internal ribosomal entry site of encephalomyocarditis virus [38]. Recent applications include interactions between the SARS-CoV-2 nucleocapsid protein and s2m element, part of the viral RNA genome (discussed below) [39], and the RNA-binding properties of the ubiquitin-like domain of SF3A1 [40].
Studying the RBPome in a disease context
Based on their involvement in cellular housekeeping processes, mutated RBPs might be expected to cause system-wide effects. However, changes in structure and function of RBPs can also induce tissue-specific effects, which make them interesting (tissue-specific) therapeutic targets for drug discovery [2].
So far, few RBPome-wide studies have addressed the impact of tumorigenesis on the RBPome or vice versa. Mestre-Farràs et al. performed RIC experiments on both non-cancerous and metastatic cell lines and found that the RBPome significantly changes its RNA-binding activity in cancerous cell lines [41]. Specifically, the authors showed that RNA binding is required in the case of PDIA6, an ER-lumen chaperone, for its tumorigenic properties. Interestingly, PDIA6 was found to bind to RNA via a C-terminally located IDR. Other studies have exploited available RBPome datasets partially generated by UV cross-linking MS to find cancer-related RBPs or biomarkers for multiple cancer types by large-scale bioinformatic analyses [42–44]. Although these studies only make use of the experimentally determined RBP annotations, they highlight the potential of RBPome data for hypothesis generation and target selection for further functional studies.
The effect of viral infections on the host RBPome has also been investigated in a series of studies [5,8,45]. Modified protocols of RIC (cRIC and vRIC) were applied to show that approximately 25% and 33% of the host cell RBPome undergo remodelling upon infection with SINV or SARS-CoV-2, respectively. Comparison of the RNA interactomes between viruses and host cells revealed a broad overlap of 60%, suggesting that different viruses may share common host RBP targets [5,8]. Additionally, Kim et al. developed VIR-CLASP, another method to study RBPome interactions between viral RNPs and the host cell [45].
Future large-scale RNA interactome screens may thus offer candidate RBPs as new therapeutic targets in the oncology and infectious disease areas, among others. However, applications towards drug discovery require detailed structural characterisation of target proteins and protein–RNA complexes. This cannot be achieved by MS alone, but requires input from structural biology and biophysics techniques, which will be discussed in the following.
Structural biology and biophysics of RBPs
The majority of RBPs possess a modular architecture, with the key components being globular RBDs, such as RNA recognition motif, K homology domain, zinc finger, and others [46,47]. Each RBD is characterised by its distinct specificity and affinity in the RNA-binding process [48]. As some RBD types are characterised by a low RNA sequence specificity (vide cold-shock domain [49]), they often co-occur (along with high specificity domains) in order to enhance the specificity and affinity of RNA binding [46]. These domains are flanked and connected by linkers that are often characterised by a high degree of conformational flexibility, and thus referred to as IDRs. Longer linkers allow the independent recognition of more than one RNA motif by the connected domains, while shorter linkers can mediate joint RNA binding across domains [46,48]. However, in recent years, it has become clear that some linkers may be directly involved in RNA interactions on their own [50–52]. Upon RNA binding, the disordered linker may adapt its structure via induced folding, called disorder-to-order transition [52]. This can result in a rearrangement and proper positioning of two RBDs flanked by transitioning linkers and thus the formation of a larger RNA interaction surface of a protein [53].
IDRs show poor RNA sequence specificity, and their interaction is thought to be electrostatically driven [54], as they often consist of hydrophilic residues and carry a large net charge [52]. Frequently, they also feature repeats of arginine/serine (RS repeats), arginine/glycine (RG repeats), short linear motifs, low complexity sequences, and electronegative clusters [46,52,55]. These distinctive patterns reduce the informational content of the linker sequence; however, they do play a role in the variety of biochemical and physiological processes, such as RNA metabolism, binding, folding, and transportation [52,56]. IDRs are also rich in PTM sites and are therefore essential for the regulation and control of many cellular processes. In RBPs, arginine (methylation), tyrosine, and serine (phosphorylation) are the most frequently modified amino acids. For example, it was shown that methylation of DDX4 and FUS/TLS RBPs reduced the formation of membrane-less organelles (MLOs) by these proteins [57,58]. Moreover, long homorepeats of single amino acids may be present, such as poly-A or poly-G, poly-K/R or poly-N/Q [59]. While being involved in many physiological events, both low complexity domains (LCDs) and long homorepeats have also been found to facilitate liquid–liquid phase separation (LLPS) events [48,52,59,60]. As a result, MLOs are formed as higher-order assemblies of RBPs and RNA due to the coacervation of protein–RNA assemblies from the surrounding solvent [61].
These large biomolecular condensates (Figure 3) can be characterised as thermodynamically reversible, highly dynamic, and liquid-like, as they are prone to dissociate upon increase in temperature or salt concentration, pH changes, and can undergo fusion and shear force deformation [54,62]. The membrane-less architecture of liquid compartments that are separated from the cyto- or nucleoplasm allows a rapid and reversible exchange of their content with the plasma surrounding and provides a distinct microenvironment and physical constraints [48,54]. Typical MLOs consisting of RBPs and RNA include cytoplasmic processing bodies (P-bodies, PBs), stress granules (SGs), and P-granules, but also nucleoplasmic Cajal bodies, nucleoli, nuclear speckles, and paraspeckles [48,52]. MLOs are involved in a myriad of cellular processes, ranging from protein synthesis, RNA metabolism, transportation, and regulation to stress response and cell signalling [52,63]. They form rapidly under stress conditions (temperature, pH, starvation) as a response to environmental stimuli. Furthermore, MLOs were shown in vitro to undergo a spontaneous transition to more solid-like structures, such as hydrogels and fibrils [54,64].
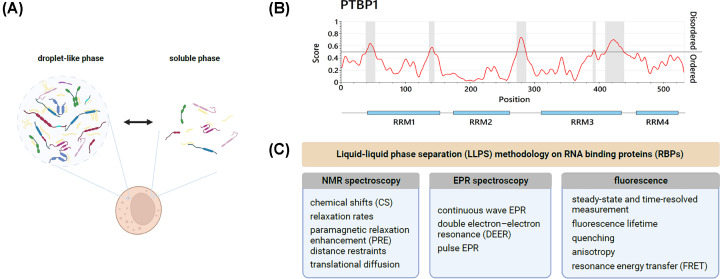
Figure 3. Liquid-liquid phase separation of RNA-binding proteins.
(A) Schematic representation of coexisting, reversible droplet-like (condensed), and soluble (dispersed) phases in the cytoplasm. (B) Prediction of disordered protein regions of the RBP PTBP1. Identified regions (highlighted in grey) are mainly located in flexible linkers, flanking RBDs. Generated using IUPred3 [65]. (C) A summary of commonly exploited methods to study LLPS phenomena involving RBPs in vitro.
The mechanisms of formation of liquid droplets remain largely unknown. So far, the majority of studies predominantly put an emphasis on the role of proteins in LLPS, neglecting the RNA contribution. As it was previously mentioned, amino acid sequence motifs were shown to govern the coacervation of assemblies, for example, tyrosines and arginines in FUS [57] or aromatic residues in hnRNPA1 [66]; unspecific hydrophobic, electrostatic, and hydrogen-bonding interactions were also found to contribute [66,67]. Interestingly, glycine-rich motifs enhance fluidity of liquid droplets while glutamine and serine residues promote their hardening [57]. On the other hand, RNA itself was found to undergo LLPS on its own and serves as a nucleation core in the formation of YB-1-rich SGs.
Biophysical properties of coacervated structures largely depend on RNA characteristics such as length, sequence, and secondary structure [68]. So far, LLPS has been studied in vitro on several RBPs, such as DDX4, hnRNPA2, CAPRIN1, FUS, TDP-43, TIA-1 [52,66], DHH1 [69], PTBP1 [70], SARS-CoV-2 nucleocapsid protein [67], and many others. As the concept of LLPS gained widespread attention over the last few years, six databases collecting the reports of proteins and RNAs undergoing phase separation have been established, namely LLPSDB, PhaSePro, PhaSepDB, DrLLPS, RNAGranuleDB, and RNAPhaSep [68].
Because of the dynamic and conformationally flexible behaviour of LLPS assemblies, the proper choice of research methods for characterising them remains non-trivial. In in vitro studies, nuclear magnetic resonance (NMR) spectroscopy has taken the lead as it provides detailed information on structure, transient interactions, and molecular motions across different timescales, even of highly dynamic macromolecular regions [61]. In comparison, other structural biology techniques like cryoelectron microscopy and X-ray crystallography have the disadvantage of providing only a static picture of analysed molecules and a poor insight into disordered regions of proteins [69]. Notably, NMR-based experiments elucidated the importance of unspecific and transient interactions between proteins and RNAs (for FUS, DDX4, hnRNPA1), the role of specific interactions and dimerisation (for TDP-43) and PTMs (for TDP-43, FUS family proteins, hnRNPA2) in liquid droplet formation [57,60,61,66]. NMR methods are, however, limited by protein size, show increased redundancy due to the presence of tandem repeats and provide little insight into the overall shape and mobility of LLPS droplets [71]. Electron paramagnetic resonance (EPR) spectroscopy is, in contrast, not limited by these factors. So far, it has been successfully used to determine the intramolecular distance distribution in liquid droplets of FUS [72] and the role of oligomerisation in phase separation of TDP-43 [73]. EPR-derived detailed structural information requires cryogenic temperatures; however, this raises concerns about possible structural changes of formed droplets and their macromolecular components [66].
Various fluorescence techniques, such as anisotropy, quenching, Förster resonance energy transfer (FRET) or steady-state and time-resolved measurements, have been widely utilised to provide insight into many LLPS features, like formation, dynamics, and compactness of FUS and TDP-43 droplets [57,60,74]. Altogether, these spectroscopic techniques provide complementary information about protein–RNA interactions, especially for dynamic and often transient LLPS systems, contributing to further exploitation of these interactions and their manipulation in biological systems.
In addition, MS-based methods can deliver information on interactions within droplet-like condensates, taking into account those driven by IDRs as well. Cross-linking MS of protein–protein interactions has been successfully exploited so far in a few cases of, e.g., FUS and SARS-CoV-2 nucleocapsid protein [75–77], while applications of protein–RNA cross-linking in connection with LLPS have not yet been reported in the literature.
RBPs and drug discovery
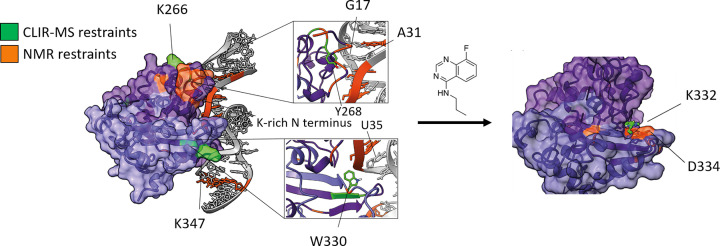
Drug development relies on detecting compounds that can specifically bind to target molecules and may be potential candidates for development as a medical treatment. Benchmark techniques to study molecules that may also inhibit or interfere with protein–RNA interactions specifically include NMR-based screening, X-ray crystallography, and structural MS. In addition, computational techniques have been proven to be an excellent tool, reducing the time and cost of drug discovery by predicting the interactions between a protein and a ligand. Finding the suitable ligand is based on a virtual screening of molecular libraries, which contain data obtained by the above-mentioned traditional techniques. High-throughput screening is performed based on algorithms that use protein properties and binding site information to select the suitable molecules with desired protein specificity and affinity [78,79]. In addition to this, molecular-docking simulations allow protein–ligand interaction predictions at the structural and thermodynamic level [80]. Li et al. moved one step further by introducing MONN, a deep-learning tool, which explores the mechanisms behind protein–ligand interactions and predicts the binding sites between the two molecules [81]. However, despite the recent advances in bioinformatics, certain features of interactions still need to be further explored in order to obtain reliable computational data, such as including flexible protein regions in high-throughput docking or including protein–ligand-binding kinetic rate calculations [82]. Combining experimental data, including cross-linking data in the form of distance restraints, might be a particularly promising direction to overcome the deficiencies of purely computational methods. This was recently shown for the SARS-CoV-2 nucleocapsid protein in complex with the viral RNA element, s2m [39] (Figure 4).
Figure 4. Fragment screening to find inhibitors for the interaction between SARS-CoV-2 nucleocapsid protein and the s2m element of the viral RNA.
A computational model of the protein–RNA complex was generated based on information obtained from CLIR-MS and NMR spectroscopy (left), and small-molecule fragments were identified that bind to either the protein (shown here on the right) or the RNA part of the complex. Reproduced from [39].
As mentioned above, RBPs regulate various biological functions of RNA and vice versa; not only in physiological but also in disease-affected pathological conditions, and can be related to many human diseases, such as cancer, autoimmune diseases, neurodegenerative and cardiovascular diseases, as well as viral infection and virus pathogenesis [83–88]. Consequently, RBPs could be potential therapeutic targets for drug development [89]. However, RNPs are conformationally highly dynamic systems, which pose many challenges for drug discovery. Strategies to approach this assignment could be developed based on three objectives:
Drugs that directly target protein–RNA interactions. These could include compounds that react either with the protein or with the RNA molecule by accessing their binding sites and blocking protein–RNA interactions [89–91], as exemplified by recent work on the N protein of SARS-CoV-2 (Figure 4).
Drugs that render the protein or the RNA dysfunctional. N protein, for instance, plays a key role in the life cycle of the virus and therefore inducing N aggregation could inhibit viral replication [92,93].
Drugs that can decrease the levels of pathological protein aggregates as in the case of neurodegenerative diseases [84].
Furthermore, the exact mechanism on which the drug’s action is based on should be fully understood and it should ideally remain an irreversible process. Cell permeability, efficient drug delivery, and tissue distribution are some drug characteristics that should be taken into consideration when a drug is designed [84,94,95]. Additionally, given that delivering the drug to the desired organ or tissue can be challenging, the use of artificial intelligence and computer modelling seems to be a way to address these issues [96]. All of the factors are particularly challenging when targeting the RNA part of protein–RNA interactions.
Conclusions
As we have shown, a diverse set of experimental and computational tools exists to characterise the interactions between proteins and RNA. MS-based methods have seen an enormous growth in recent years, taking advantage of the high sensitivity of this method and its ability to deal with samples of high complexity. However, it is apparent that other methods will continue to play a crucial role to obtain a comprehensive picture of the protein–RNA interactome. We believe that the integration of different methods will soon provide us with a better understanding even of complex phenomena such as LLPS processes in vitro and in vivo. This should form the basis for further biological and biomedical discoveries, including new directions towards the treatment of diseases.
Summary
Cross-linking methods in combination with MS detection have emerged as a key player to characterise individual protein–RNA complexes and the RBPome.
MS-based methods are complementing structural biology and biophysical techniques to study protein–RNA interactions.
An increased understanding of the role of protein–RNA interactions in health and disease suggests that an integration of MS- and non-MS-based techniques will provide crucial insights into fundamental biological processes such as LLPS and a new strategy for drug discovery target selection and characterisation.
Acknowledgements
Figures 1–3 were created with BioRender.com.
Abbreviations
- 4-SU
4-thiouridine
- 5-EU
5-ethynyluridine
- CARIC
click chemistry-assisted RNA interactome capture
- CLIP
cross-linking and immunoprecipitation
- CLIR-MS
cross-linking of isotope-labelled RNA coupled to mass spectrometry
- EPR
electron paramagnetic resonance
- ER
endoplasmic reticulum
- FRET
Förster resonance energy transfer
- FUS/TLS
fused in sarcoma/translocated in liposarcoma
- IDR
intrinsically disordered region
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- LLPS
liquid–liquid phase separation
- MLO
membrane-less organelle
- MS
mass spectrometry
- NMR
nuclear magnetic resonance
- N protein
nucleocapsid protein
- OMS
open modification search
- OOPS
orthogonal organic-phase separation
- pCLAP
peptide cross-linking and affinity purification
- PTM
post-translational modification
- RBD
RNA-binding domain
- RBP
RNA-binding protein
- RBPome
RNA-binding proteome
- RIC
RNA interactome capture
- RICK
RNA interactome using click chemistry
- RNP
ribonucleoprotein
- SARS-CoV-2
severe acute respiratory syndrome coronavirus type 2
- SINV
sindbis virus
- TRAPP
total RNA-associated protein purification
- TROOPS
targeted RNA–protein identification using OOPS
Contributor Information
Maria Bikaki, Email: bikaki@imsb.biol.ethz.ch.
Alexander Leitner, Email: leitner@imsb.biol.ethz.ch.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
Work on protein–RNA interactions in the authors’ laboratory was funded by the Swiss National Science Foundation through the general project funding scheme [grant numbers 310030_200679, 205321_204920]; the National Research Programme “Covid-19” [grant number 4078P0_198253]; and the National Centre for Competence in Research RNA & Disease [grant number 51NF40-205601].
Open Access
Open access for this article was enabled by the participation of ETH Zurich in an all-inclusive Read & Publish agreement with Portland Press and the Biochemical Society.
Author Contribution
Conceptualisation: M.B. and A.L.; Visualisation: B.S., I.S., and M.B., Writing – original draft: all the authors; Writing – review and editing: all the authors.
References
- 1.Gerstberger S., Hafner M. and Tuschl T. (2014) A census of human RNA-binding proteins. Nat. Rev. Genet. 15, 829–845 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gebauer F., Schwarzl T., Valcárcel J. and Hentze M.W. (2021) RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 22, 185–198 10.1038/s41576-020-00302-y [DOI] [PubMed] [Google Scholar]
- 3.Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C.et al. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 10.1016/j.cell.2012.04.031 [DOI] [PubMed] [Google Scholar]
- 4.Hentze M.W., Castello A., Schwarzl T. and Preiss T. (2018) A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327–341 10.1038/nrm.2017.130 [DOI] [PubMed] [Google Scholar]
- 5.Kamel W., Noerenberg M., Cerikan B., Chen H., Järvelin A.I., Kammoun M.et al. (2021) Global analysis of protein-RNA interactions in SARS-CoV-2-infected cells reveals key regulators of infection. Mol. Cell 81, 2851–2867 10.1016/j.molcel.2021.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vieira-Vieira C.H., Dauksaite V., Sporbert A., Gotthardt M. and Selbach M. (2022) Proteome-wide quantitative RNA-interactome capture identifies phosphorylation sites with regulatory potential in RBM20. Mol. Cell 82, 2069–2083 10.1016/j.molcel.2022.03.024 [DOI] [PubMed] [Google Scholar]
- 7.Perez-Perri J.I., Rogell B., Schwarzl T., Stein F., Zhou Y., Rettel M.et al. (2018) Discovery of RNA-binding proteins and characterization of their dynamic responses by enhanced RNA interactome capture. Nat. Commun. 9, 4408 10.1038/s41467-018-06557-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Moreno M., Noerenberg M., Ni S., Järvelin A.I., González-Almela E. and Lenz C.E. (2019) System-wide profiling of RNA-binding proteins uncovers key regulators of virus infection. Mol. Cell 74, 196–211 10.1016/j.molcel.2019.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castello A., Fischer B., Frese C.K., Horos R., Alleaume A.-M., Foehr S.et al. (2016) Comprehensive identification of RNA-binding domains in human cells. Mol. Cell 63, 696–710 10.1016/j.molcel.2016.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conrad T., Albrecht A.-S., de Melo Costa V.R., Sauer S., Meierhofer D. and Ørom U.A. (2016) Serial interactome capture of the human cell nucleus. Nat. Commun. 7, 11212 10.1038/ncomms11212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullari M., Lyon D., Jensen L.J. and Nielsen M.L. (2017) Specifying RNA-binding regions in proteins by peptide cross-linking and affinity purification. J. Proteome Res. 16, 2762–2772 10.1021/acs.jproteome.7b00042 [DOI] [PubMed] [Google Scholar]
- 12.Bao X., Guo X., Yin M., Tariq M., Lai Y., Kanwal S.et al. (2018) Capturing the interactome of newly transcribed RNA. Nat. Methods 15, 213–220 10.1038/nmeth.4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang R., Han M., Meng L. and Chen X. (2018) Transcriptome-wide discovery of coding and noncoding RNA-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 115, E3879–E3887 10.1073/pnas.1718406115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramer K., Sachsenberg T., Beckmann B.M., Qamar S., Boon K.-L., Hentze M.W.et al. (2014) Photo-cross-linking and high-resolution mass spectrometry for assignment of RNA-binding sites in RNA-binding proteins. Nat. Methods 11, 1064–1070 10.1038/nmeth.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He J.-X., Fei Z.-C., Fu L., Tian C.-P., He F.-C., Chi H.et al. (2022) A modification-centric assessment tool for the performance of chemoproteomic probes. Nat. Chem. Biol. 18, 904–912 10.1038/s41589-022-01074-8 [DOI] [PubMed] [Google Scholar]
- 16.Shchepachev V., Bresson S., Spanos C., Petfalski E., Fischer L., Rappsilber J.et al. (2019) Defining the RNA interactome by total RNA‐associated protein purification. Mol. Syst. Biol. 15, e8689 10.15252/msb.20188689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asencio C., Chatterjee A. and Hentze M.W. (2018) Silica-based solid-phase extraction of cross-linked nucleic acid–bound proteins. Life Sci. Alliance 1, e201800088 10.26508/lsa.201800088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Queiroz R.M., Smith T., Villanueva E., Marti-Solano M., Monti M., Pizzinga M.et al. (2019) Comprehensive identification of RNA–protein interactions in any organism using orthogonal organic phase separation (OOPS). Nat. Biotechnol. 37, 169–178 10.1038/s41587-018-0001-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urdaneta E.C., Vieira-Vieira C.H., Hick T., Wessels H.-H., Figini D., Moschall R.et al. (2019) Purification of cross-linked RNA-protein complexes by phenol-toluol extraction. Nat. Commun. 10, 990 10.1038/s41467-019-08942-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trendel J., Schwarzl T., Horos R., Prakash A., Bateman A., Hentze M.W.et al. (2019) The human RNA-binding proteome and its dynamics during translational arrest. Cell 176, 391–403 10.1016/j.cell.2018.11.004 [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y., Xu Y., Skaggs T.H., Ferreira J.F.S., Chen X. and Sandhu D. (2022) Plant phase extraction (PPE): a novel method for enhanced discovery of RNA-binding proteome in plants. bioRxiv 10.1101/2022.06.02.494555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo H., Tang W., Liu H., Zeng X., Ngai W.S.C., Gao R.et al. (2022) Photocatalytic chemical crosslinking for profiling RNA–protein interactions in living cells. Angew. Chem., Int. Ed. Engl. 61, e202202008 10.1002/anie.202202008 [DOI] [PubMed] [Google Scholar]
- 23.Kerr A.G., Wang Z., Wang N., Kwok K.H.M., Jalkanen J., Ludzki A.et al. (2022) The long noncoding RNA ADIPINT regulates human adipocyte metabolism via pyruvate carboxylase. Nat. Commun. 13, 2958 10.1038/s41467-022-30620-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nechay M. and Kleiner R.E. (2020) High-throughput approaches to profile RNA-protein interactions. Curr. Opin. Chem. Biol. 54, 37–44 10.1016/j.cbpa.2019.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarnowski C.P., Bikaki M. and Leitner A. (2022) Cross-linking and mass spectrometry as a tool for studying the structural biology of ribonucleoproteins. Structure 30, 441–461 10.1016/j.str.2022.03.003 [DOI] [PubMed] [Google Scholar]
- 26.Van Ende R., Balzarini S. and Geuten K. (2020) Single and combined methods to specifically or bulk-purify RNA–protein complexes. Biomolecules 10, 1160 10.3390/biom10081160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith J.M., Sandow J.J. and Webb A.I. (2021) The search for RNA-binding proteins: a technical and interdisciplinary challenge. Biochem. Sci. Trans. 49, 393–403 10.1042/BST20200688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vieira-Vieira C.H. and Selbach M. (2021) Opportunities and challenges in global quantification of RNA-protein interaction via UV cross-linking. Front. Mol. Biosci. 8, 669939 10.3389/fmolb.2021.669939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knörlein A., Sarnowski C.P., de Vries T., Stoltz M., Götze M., Aebersold R.et al. (2022) Nucleotide-amino acid π-stacking interactions initiate photo cross-linking in RNA-protein complexes. Nat. Commun. 13, 2719 10.1038/s41467-022-30284-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scalabrin M., Dixit S.M., Makshood M.M., Krzemien C.E. and Fabris D. (2018) Bifunctional cross-linking approaches for mass spectrometry-based investigation of nucleic acids and protein-nucleic acid assemblies. Methods 144, 64–78 10.1016/j.ymeth.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beckmann B.M., Horos R., Fischer B., Castello A., Eichelbaum K., Alleaume A.-M.et al. (2015) The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 6, 10127 10.1038/ncomms10127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trendel J., Schwarzl T., Horos R., Prakash A., Bateman A., Hentze M.W.et al. (2019) The human RNA-Binding proteome and its dynamics during translational arrest. Cell 176, 391–403 10.1016/j.cell.2018.11.004 [DOI] [PubMed] [Google Scholar]
- 33.Kong A.T., Leprevost F.V., Avtonomov D.M., Mellacheruvu D. and Nesvizhskii A.I. (2017) MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry–based proteomics. Nat. Methods 14, 513–520 10.1038/nmeth.4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geiszler D.J., Kong A.T., Avtonomov D.M., Yu F., Leprevost F.d.V. and Nesvizhskii A.I. (2021) PTM-Shepherd: analysis and summarization of post-translational and chemical modifications from open search results. Mol. Cell. Proteomics 20, 100018 10.1074/mcp.TIR120.002216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bae J.W., Kwon S.C., Na Y., Kim V.N. and Kim J.-S. (2020) Chemical RNA digestion enables robust RNA-binding site mapping at single amino acid resolution. Nat. Struct. Mol. Biol. 27, 678–682 10.1038/s41594-020-0436-2 [DOI] [PubMed] [Google Scholar]
- 36.Leitner A. (2010) Phosphopeptide enrichment using metal oxide affinity chromatography. Trends Anal. Chem. 29, 177–185 10.1016/j.trac.2009.08.007 [DOI] [Google Scholar]
- 37.He C., Sidoli S., Warneford-Thomson R., Tatomer D.C., Wilusz J.E., Garcia B.A.et al. (2016) High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol. Cell 64, 416–430 10.1016/j.molcel.2016.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dorn G., Leitner A., Boudet J., Campagne S., von Schroetter C., Moursy A.et al. (2017) Structural modeling of protein–RNA complexes using crosslinking of segmentally isotope-labeled RNA and MS/MS. Nat. Methods 14, 487–490 10.1038/nmeth.4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Padroni G., Bikaki M., Novakovic M., Wolter A.C., Rüdisser S.H., Gossert A.D.et al. (2022) A hybrid structure determination approach to investigate the druggability of the nucleocapsid protein of SARS-CoV-2. bioRxiv 10.1101/2022.09.15.507991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Vries T., Martelly W., Campagne S., Sabath K., Sarnowski C.P., Wong J.et al. (2022) Sequence-specific RNA recognition by an RGG motif connects U1 and U2 snRNP for spliceosome assembly. Proc. Natl. Acad. Sci. U.S.A. 119, e2114092119 10.1073/pnas.2114092119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mestre-Farràs N., Guerrero S., Bley N., Rivero E., Coll O., Borràs E.et al. (2022) Melanoma RBPome identification reveals PDIA6 as an unconventional RNA-binding protein involved in metastasis. Nucleic. Acids. Res. 50, 8207–8225 10.1093/nar/gkac605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z., Tang W., Yuan J., Qiang B., Han W. and Peng X. (2020) Integrated analysis of RNA-binding proteins in glioma. Cancers 12, 892 10.3390/cancers12040892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z.-L., Li B., Luo Y.-X., Lin Q., Liu S.-R., Zhang X.-Q.et al. (2018) Comprehensive genomic characterization of RNA-binding proteins across human cancers. Cell Rep. 22, 286–298 10.1016/j.celrep.2017.12.035 [DOI] [PubMed] [Google Scholar]
- 44.Ming R., Li X., Wang E., Wei J., Liu B., Zhou P.et al. (2022) The prognostic signature of head and neck squamous cell carcinoma constructed by immune-related RNA-binding proteins. Front. Oncol. 12, 795781 10.3389/fonc.2022.795781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim B., Arcos S., Rothamel K., Jian J., Rose K.L., McDonald W.H.et al. (2020) Discovery of widespread host protein interactions with the pre-replicated genome of CHIKV Using VIR-CLASP. Mol. Cell 78, 624–640 10.1016/j.molcel.2020.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corley M., Burns M.C. and Yeo G.W. (2020) How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol. Cell 78, 9–29 10.1016/j.molcel.2020.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuschel K., Helwig M., Hüttelmaier S., Heckl D., Klusmann J.-H. and Hoell J.I. (2020) RNA-binding proteins in acute leukemias. Int. J. Mol. Sci. 21, 3409 10.3390/ijms21103409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Helder S., Blythe A.J., Bond C.S. and Mackay J.P. (2016) Determinants of affinity and specificity in RNA-binding proteins. Curr. Opin. Struct. Biol. 38, 83–91 10.1016/j.sbi.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 49.Heinemann U. and Roske Y. (2021) Cold-shock domains-abundance, structure, properties, and nucleic-acid binding. Cancers 13, 190 10.3390/cancers13020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loughlin F.E., Lukavsky P.J., Kazeeva T., Reber S., Hock E.-M., Colombo M.et al. (2019) The solution structure of FUS bound to RNA reveals a bipartite mode of RNA recognition with both sequence and shape specificity. Mol. Cell 73, 490–504 10.1016/j.molcel.2018.11.012 [DOI] [PubMed] [Google Scholar]
- 51.Ozdilek B.A., Thompson V.F., Ahmed N.S., White C.I., Batey R.T. and Schwartz J.C. (2017) Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic. Acids. Res. 45, 7984–7996 10.1093/nar/gkx460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balcerak A., Trebinska-Stryjewska A., Konopinski R., Wakula M. and Grzybowska E.A. (2019) RNA–protein interactions: disorder, moonlighting and junk contribute to eukaryotic complexity. Open Biol. 9, 190096 10.1098/rsob.190096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ottoz D.S. and Berchowitz L.E. (2020) The role of disorder in RNA binding affinity and specificity. Open Biol. 10, 200328 10.1098/rsob.200328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uversky V.N. (2019) Intrinsically disordered proteins and their “mysterious”(meta) physics. Front. Phys. 7, 10 10.3389/fphy.2019.00010 [DOI] [Google Scholar]
- 55.Santiago-Frangos A., Kavita K., Schu D.J., Gottesman S. and Woodson S.A. (2016) C-terminal domain of the RNA chaperone Hfq drives sRNA competition and release of target RNA. Proc. Natl. Acad. Sci. U.S.A. 113, E6089–E6096 10.1073/pnas.1613053113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Busa V.F., Rector M.J. and Russell R. (2017) The DEAD-box protein CYT-19 uses arginine residues in its C-tail to tether RNA substrates. Biochemistry 56, 3571–3578 10.1021/acs.biochem.7b00362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hofweber M. and Dormann D. (2019) Friend or foe-post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 294, 7137–7150 10.1074/jbc.TM118.001189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Csizmok V. and Forman-Kay J.D. (2018) Complex regulatory mechanisms mediated by the interplay of multiple post-translational modifications. Curr. Opin. Struct. Biol. 48, 58–67 10.1016/j.sbi.2017.10.013 [DOI] [PubMed] [Google Scholar]
- 59.Darling A.L. and Uversky V.N. (2017) Intrinsic disorder in proteins with pathogenic repeat expansions. Molecules 22, 2027 10.3390/molecules22122027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conicella A.E., Zerze G.H., Mittal J. and Fawzi N.L. (2016) ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure 24, 1537–1549 10.1016/j.str.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murthy A.C. and Fawzi N.L. (2020) The (un) structural biology of biomolecular liquid-liquid phase separation using NMR spectroscopy. J. Biol. Chem. 295, 2375–2384 10.1074/jbc.REV119.009847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou H.-X., Nguemaha V., Mazarakos K. and Qin S. (2018) Why do disordered and structured proteins behave differently in phase separation? Trends Biochem. Sci 43, 499–516 10.1016/j.tibs.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bratek‐Skicki A., Pancsa R., Meszaros B., Van Lindt J. and Tompa P. (2020) A guide to regulation of the formation of biomolecular condensates. FEBS J. 287, 1924–1935 10.1111/febs.15254 [DOI] [PubMed] [Google Scholar]
- 64.Liu J., Liu X., Zhang T., Huang J., Tang B.Z. and Chau Y. (2022) Metastability of biological matter in liquid phase separation. Authorea Preprints 10.22541/au.165272514.42816852/v1 [DOI] [Google Scholar]
- 65.Erdős G., Pajkos M. and Dosztányi Z. (2021) IUPred3: prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic. Acids. Res. 49, W297–W303 10.1093/nar/gkab408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emmanouilidis L., Esteban-Hofer L., Jeschke G. and Allain F.H.-T. (2021) Structural biology of RNA-binding proteins in the context of phase separation: What NMR and EPR can bring? Curr. Opin. Struct. Biol. 70, 132–138 10.1016/j.sbi.2021.07.001 [DOI] [PubMed] [Google Scholar]
- 67.Perdikari T.M., Murthy A.C., Ryan V.H., Watters S., Naik M.T. and Fawzi N.L. (2020) SARS‐CoV‐2 nucleocapsid protein phase‐separates with RNA and with human hnRNPs. EMBO J. 39, e106478 10.15252/embj.2020106478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu H., Fu H., Cui T., Ning L., Shao H., Guo Y.et al. (2022) RNAPhaSep: a resource of RNAs undergoing phase separation. Nucleic. Acids. Res. 50, D340–D346 10.1093/nar/gkab985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heinrich S. and Hondele M. (2022) Probing liquid–liquid phase separation of RNA-binding proteins in vitro and in vivo. Meth. Mol. Biol. 2537, 307–333 10.1007/978-1-0716-2521-7_18 [DOI] [PubMed] [Google Scholar]
- 70.Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J.et al. (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Uversky V.N. and Dunker A.K. (2012) Multiparametric analysis of intrinsically disordered proteins: looking at intrinsic disorder through compound eyes. Anal. Chem. 84, 2096–2104 10.1021/ac203096k [DOI] [PubMed] [Google Scholar]
- 72.Emmanouilidis L., Esteban-Hofer L., Damberger F.F., de Vries T., Nguyen C.K., Ibanez L.F.et al. (2021) NMR and EPR reveal a compaction of the RNA-binding protein FUS upon droplet formation. Nat. Chem. Biol. 17, 608–614 10.1038/s41589-021-00752-3 [DOI] [PubMed] [Google Scholar]
- 73.Babinchak W.M., Haider R., Dumm B.K., Sarkar P., Surewicz K., Choi J.-K.et al. (2019) The role of liquid–liquid phase separation in aggregation of the TDP-43 low-complexity domain. J. Biol. Chem. 294, 6306–6317 10.1074/jbc.RA118.007222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamad N., Yoneda R., So M., Kurokawa R., Nagata T. and Katahira M. (2021) Non-coding RNA suppresses FUS aggregation caused by mechanistic shear stress on pipetting in a sequence-dependent manner. Sci. Rep. 11, 9523 10.1038/s41598-021-89075-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boczek E.E., Fürsch J., Niedermeier M.L., Jawerth L., Jahnel M., Ruer-Gruß M.et al. (2021) HspB8 prevents aberrant phase transitions of FUS by chaperoning its folded RNA-binding domain. eLife 10, e69377 10.7554/eLife.69377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jack A., Ferro L.S., Trnka M.J., Wehri E., Nadgir A., Nguyenla X.et al. (2021) SARS-CoV-2 nucleocapsid protein forms condensates with viral genomic RNA. PLoS Biol. 19, e3001425 10.1371/journal.pbio.3001425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu S., Ye Q., Singh D., Cao Y., Diedrich J.K., Yates J.R. 3rdet al. (2021) The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat. Commun. 12, 502 10.1038/s41467-020-20768-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Akinlalu A.O., Chamundi A., Yakumbur D.T., Afolayan F.I.D., Duru I.A., Arowosegbe M.A.et al. (2021) Repurposing FDA-approved drugs against multiple proteins of SARS-CoV-2: an in silico study. Sci. African 13, e00845 10.1016/j.sciaf.2021.e00845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Julio A.R. and Backus K.M. (2021) New approaches to target RNA binding proteins. Curr. Opin. Chem. Biol. 62, 13–23 10.1016/j.cbpa.2020.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumar S. and Kumar S. (2019) Molecular docking: a structure-based approach for drug repurposing. In Silico Drug Design, pp. 161–189, Academic Press/Elsevier, London: 10.1016/B978-0-12-816125-8.00006-7 [DOI] [Google Scholar]
- 81.Li S., Wan F., Shu H., Jiang T., Zhao D. and Zeng J. (2020) MONN: a multi-objective neural network for predicting compound-protein interactions and affinities. Cell Syst. 10, 308–322 10.1016/j.cels.2020.03.002 [DOI] [Google Scholar]
- 82.Cavasotto C.N., Aucar M.G. and Adler N.S. (2019) Computational chemistry in drug lead discovery and design. Int. J. Quant. Chem. 119, e25678 10.1002/qua.25678 [DOI] [Google Scholar]
- 83.Conlon E.G. and Manley J.L. (2017) RNA-binding proteins in neurodegeneration: mechanisms in aggregate. Genes Dev. 31, 1509–1528 10.1101/gad.304055.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hyun S. and Shin D. (2021) Chemical-mediated targeted protein degradation in neurodegenerative diseases. Life 11, 607 10.3390/life11070607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Bruin R.G., Rabelink T.J., van Zonneveld A.J. and van der Veer E.P. (2017) Emerging roles for RNA-binding proteins as effectors and regulators of cardiovascular disease. Eur. Heart J. 38, 1380–1388 [DOI] [PubMed] [Google Scholar]
- 86.Wang Y., Wang Y., Luo W., Song X., Huang L., Xiao J.et al. (2020) Roles of long non-coding RNAs and emerging RNA-binding proteins in innate antiviral responses. Theranostics 10, 9407 10.7150/thno.48520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vos P.D., Leedman P.J., Filipovska A. and Rackham O. (2019) Modulation of miRNA function by natural and synthetic RNA-binding proteins in cancer. Cell. Mol. Life Sci. 76, 3745–3752 10.1007/s00018-019-03163-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mohibi S., Chen X. and Zhang J. (2019) Cancer the ‘RBP’eutics–RNA-binding proteins as therapeutic targets for cancer. Pharmacol. Therapeutics 203, 107390 10.1016/j.pharmthera.2019.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu P. (2020) Inhibition of RNA-binding proteins with small molecules. Nat. Rev. Chem. 4, 441–458 10.1038/s41570-020-0201-4 [DOI] [PubMed] [Google Scholar]
- 90.Warner K.D., Hajdin C.E. and Weeks K.M. (2018) Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug Discov. 17, 547–558 10.1038/nrd.2018.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Costales M.G., Childs-Disney J.L., Haniff H.S. and Disney M.D. (2020) How we think about targeting RNA with small molecules. J. Med. Chem. 63, 8880–8900 10.1021/acs.jmedchem.9b01927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X.et al. (2020) Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B 10, 1228–1238 10.1016/j.apsb.2020.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Peng Y., Du N., Lei Y., Dorje S., Qi J., Luo T.et al. (2020) Structures of the SARS‐CoV‐2 nucleocapsid and their perspectives for drug design. EMBO J. 39, e105938 10.15252/embj.2020105938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang S., Huang W., Ren L., Ju X., Gong M., Rao J.et al. (2022) Comparison of viral RNA–host protein interactomes across pathogenic RNA viruses informs rapid antiviral drug discovery for SARS-CoV-2. Cell Res. 32, 9–23 10.1038/s41422-021-00581-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Corey D. (2017) Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat. Neurosci. 20, 497–499 10.1038/nn.4508 [DOI] [PubMed] [Google Scholar]
- 96.Roberts T.C., Langer R. and Wood M.J. (2020) Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 19, 673–694 10.1038/s41573-020-0075-7 [DOI] [PMC free article] [PubMed] [Google Scholar]