Abstract
Genes with sex-biased expression are thought to underlie sexually dimorphic phenotypes and are therefore subject to different selection pressures in males and females. Many authors have proposed that sexual conflict leads to the evolution of sex-biased expression, which allows males and females to reach separate phenotypic and fitness optima. The selection pressures associated with domestication may cause changes in population architectures and mating systems, which in turn can alter their direction and strength. We compared sex-biased expression and genetic signatures in wild and domestic ducks (Anas platyrhynchos), and observed changes of sexual selection and identified the genomic divergence affected by selection forces. The extent of sex-biased expression in both sexes is positively correlated with the level of both dN/dS and nucleotide diversity. This observed changing pattern may mainly be owing to relaxed genetic constraints. We also demonstrate a clear link between domestication and sex-biased evolutionary rate in a comparative framework. Decreased polymorphism and evolutionary rate in domesticated populations generally matched life-history phenotypes known to experience artificial selection. Taken together, our work suggests the important implications of domestication in sex-biased evolution and the roles of artificial selection and sexual selection for shaping the diversity and evolutionary rate of the genome.
Keywords: sex-biased expression, sequence evolution, sexual selection, domestication, duck
1. Introduction
In most sexually reproducing animals, evolutionary conflicts of interest arise whenever males and females interact and their routes to fitness maximization differ, often termed sexual conflict [1–4]. When this conflict revolves around which individuals enjoy priority mating rights, how many offspring are produced, when they are produced, and how much each parent invests into these offspring, sexual selection has also begun to play a role in sexual evolution [5–7]. Sexual selection is selected for traits of one sex, and therefore may be a decisive force in shaping sexual dimorphism [8,9]. We can understand many sexual evolutionary phenomena in a closed-loop way, sexual conflict is the driving force behind sexual selection, and sexual selection shapes phenotypic sexual dimorphism and expression bias, ultimately resolving conflicts of interest between males and females. As mentioned above, sex-biased genes allow each sex to reach separate optima. However, genetic constraints, as a ‘hinderance’, often prevent the resolution of sexual conflict, that is, this evolutionary force seems to be trying to reduce sexual dimorphism, and it also seems to be affecting the sequence characteristics and evolutionary pattern of the genome [10,11]. Concretely, such constraints are generally considered to be related to the diversity and evolution of sex-biased expression, because the biased expression is thought to reduce constraints and thereby enable rapid adaptive evolution and more variation [12,13].
Clearly, a tug-of-war involving many evolutionary factors shapes the unique genetic properties of biased genes. There is no evidence that a directional link exists between sexual selection and sequence evolution, however, proteins encoded by sex-biased genes do show greater amino acid sequence divergence [14,15]. It is foreseeable that as the degree of sex bias (fold change; FC) becomes more extreme, the intensity of sexual selection will increase, while the trend of constraints is the opposite. The existence of genetic signatures such as changes in genetic diversity is the result of a trade-off of multiple evolutionary forces, which means that it is challenging to analyse each evolutionary force independently. When we try to research the populations that already exist in nature, because of the time spans that allow us to observe evolutionary effects, the problem seems to be on the verge of being solved. Domestication has the potential to greatly alter sexual conflict and sexual selection via an altered mating system, and we might expect this to quickly affect sex-biased gene expression and associated population genomic characteristics [16–18]. Both monogamy (having one mate) and polygamy (having several mates) exist in mallard (Anas platyrhynchos), and mate choice is ultimately up to the female. This mating system results in male–male competition for mating opportunities and thus leads to more intense sexual selection in wild populations, although it also exists among females [19,20], males are subjected to strong sexual selection [21]. Duck domestication occurred initially roughly 2200 years ago, and controlled breeding by humans has weakened or eliminated male–male competition, female choice and sperm competition, thereby leading to relaxed sexual selection compared to wild populations [16]. During domestication, genes are expected to experience both high-intensity artificial selection and relaxed sexual selection, it is hard but relative roles and the genetic signatures of these selective forces is yet to be untangled [22].
We chose male and female RNA-seq data from gonadal and liver tissue from one wild breed (mallard duck, MD), one meat-type breed (Pekin duck, PK) and one egg-type breed (Gaoyou duck, GY) to assess the effects of domestication on expression at the early development stage. Both the two domesticated breeds, GY and PK, have experienced a common domestication event, but subsequently they underwent varying sex-specific selection regimens, allowing us to observe the evolutionary effects of artificial selection. These two varying sex-specific selection regimens are caused by artificial selection for different purposes. Many duck breeds (like GY) have been selected explicitly for increased female fecundity, resulting in these breeds that produce numerous and large eggs. Therefore, these breeds result from elevated female-specific selection compared with the mallard ancestor. On the contrary, the meat-type duck (like PK) is artificially selected to be larger in size, more muscular and faster in growth, which corresponds to a stronger male-specific selection compared with the mallard ancestor. Populations that have experienced domestication were selected in order to identify whether the magnitude of sexual selection is associated with sex-biased evolution. Leaning on detailed information on sequence diversity and evolutionary rate, we have determined the evolutionary effect of shaping genome signature intra-breeds and during domestication. Our results provide a clear link between sex-biased gene evolution and domestication through comprehensive population genetic analysis.
2. Results
2.1. Changes of sexual size dimorphism owing to domestication
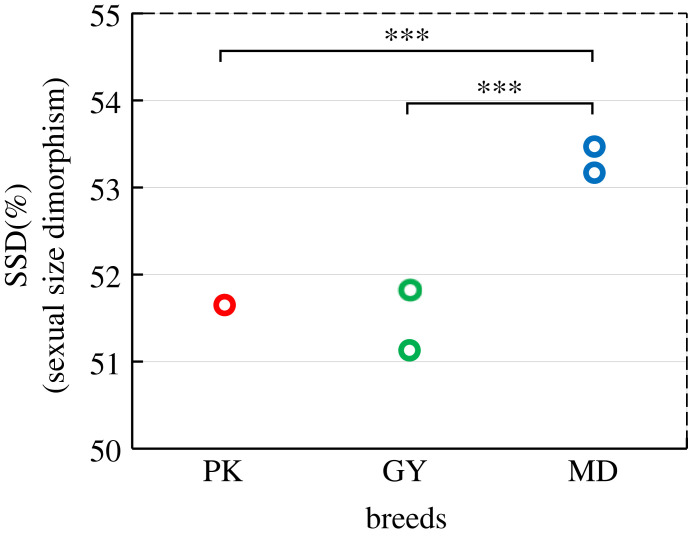
The extent of the sexual size dimorphism (SSD) is thought to be an indicator of sexual selection [23,24], and we, therefore, assessed the potential changes in phenotypic dimorphism associated with domestication. We calculated the SSD parameter of the three breeds (figure 1), and the magnitude of SSD correlates with mean body weight, which is one of the most striking examples of sex differences in animals [25]. Although there are two sets of data from different sources for GY and MD (figure 1; electronic supplementary material, table S3), after a one-to-one test, there are significant SSD differences between the two domestic duck populations and the wild population (PK: 51.65%, GY: 51.13% and 51.82%, MD: 53.17% and 53.47%, p = 0.00067, Z-test). Specifically, MD show more significant male-skewed SSD even though males are larger than females in all observed breeds.
Figure 1.
Sexual size dimorphism (SSD) is associated with domestication in ducks. dimorphism in ducks. SSD is calculated as the ratio between the average body weight of males compared to the total average weight (males and females). Z-tests for two independent samples were used so that individual weight data and sample size were both taken into consideration. Significant difference between SSD of three breeds is indicated (Z-test, *p < 0.05, **p < 0.01, ***p < 0.001).
2.2. Sex-biased expression on the autosomes
We quantified the extent of the sex-biased gene expression in two different tissues (gonad and liver) collected at the embryonic stage (table 1; electronic supplementary material, figure S1), focusing on autosomal loci owing to the complexity of incomplete Z chromosome dosage compensation in birds. We defined sex-bias as greater than 2 FC and a significant difference in expression between the sexes (false discovery rate (FDR) < 0.05). As expected, the gonad showed a higher proportion of sex-biased genes compared to the liver. We observed large differences in the direction of sex-biased expression in the gonad of domesticates, possibly owing to significant differences in sex-specific selection between egg-type and meat-type ducks (Z-test, p = 0.032). Specifically, PK shows a greater proportion of male-biased genes (n = 901, 57% of sex-biased genes), while female-biased genes are more prevalent in GY (n = 1506, 65% of sex-biased genes). For the liver of all three breeds, less than 5% of genes showed a significant sex bias, even in the two domesticated breeds, this proportion is less than 1%. Wild populations showed a significantly greater level of sex-biased expression compared to domestics for both male-biased and female-biased genes (Mann Whitney U-test, p < 0.05) (electronic supplementary material, figure S2).
Table 1.
Description of sex-biased gene expression in the gonad and liver.
| tissue | no. of genes expressed | no. of sex-biased gene of MD |
no. of sex-biased gene of GY |
no. of sex-biased gene of PK |
|||
|---|---|---|---|---|---|---|---|
| total, males, females | proportion (%) | total, males, females | proportion (%) | total, males, females | proportion (%) | ||
| gonad | 11 033 | 2583 1330 1253 | 23.41 | 2316 810 1506 | 20.99 | 1566 901 665 | 14.19 |
| liver | 9189 | 389 160 229 | 4.23 | 67 26 39 | 0.73 | 85 32 53 | 0.93 |
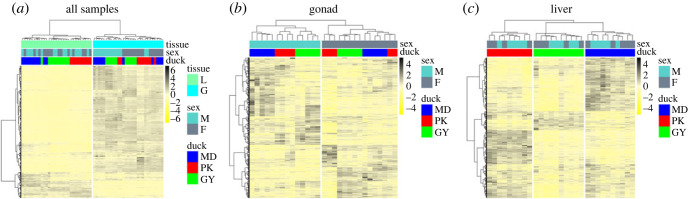
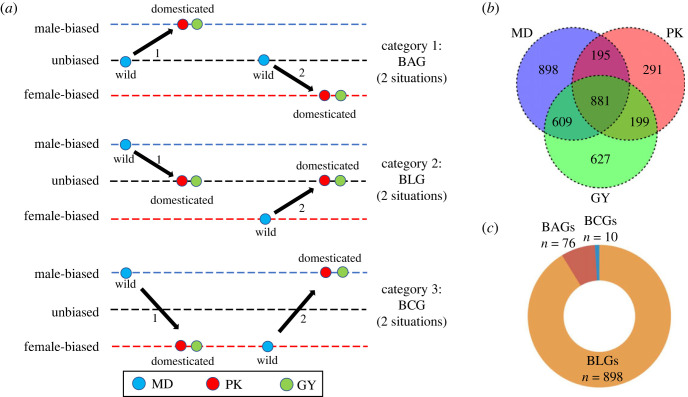
We next used hierarchical clustering of expression levels for all samples of two tissues (figure 2). All samples are clustered by tissue, sex-biased expression was most evident in the gonad, where samples largely cluster first by sex, in contrast to liver samples which clustered first by breed, and there were 881 sex-biased genes in all breeds (figure 3b). Genes that are sex-biased in MD but unbiased in both domestic ducks are termed as bias-lost genes (BLGs) (figure 3a). We observed 898 BLGs in the gonad (figure 3b,c). We observed 76 bias-acquired genes (BAGs), which are unbiased in MD but sex-biased in both domestic ducks, and 10 bias-converted genes (BCGs), which are sex-biased in both three breeds but reverse the direction in domestic ducks (figure 3a,c).
Figure 2.
Heatmaps and hierarchical clustering of gene expression for (a) all samples; (b) gonad; and (c) liver. Shown is the average relative expression for all autosomal expressed genes from two tissues (L, liver; G, gonad) of M (male) and F (female) in three breeds (MD, PK and GY).
Figure 3.
Dynamic changes of sex-biased genes in three breeds. (a) Three categories of sex-biased expression changes among wild duck and domesticated duck. (b) A Venn diagram illustrating statistical results of sex-biased expression in three breeds. (c) The three categories of sex-biased changes and their numbers and proportions.
For the top 10 gene ontology (GO) terms of male-biased pattern, biological processes involved in reproduction, muscle development and embryonic development were identified (electronic supplementary material, table S1). Genes that were universally female-biased were enriched for regulatory processes of reproduction and fear response (electronic supplementary material, table S1). For bias-lost expression, genes were enriched for behaviour and morphogenesis, which could be indicative of relaxed sexual selection (electronic supplementary material, table S2).
2.3. Coding sequence evolution during domestication
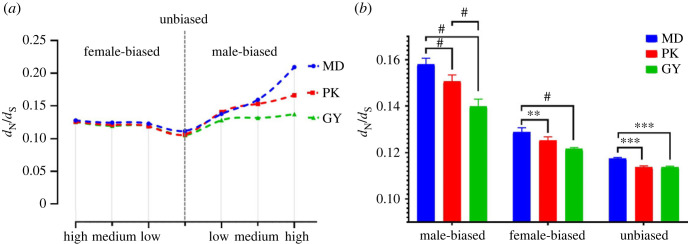
To explore the impact of domestication on sequence evolution rate, and determine whether the overall pattern of selection is distinct in different sex-biased categories, we visualized the relationship between mean dN/dS and the extent of sex-biased expression in gonads of three breeds for male-biased and female-biased genes (figure 4a) [26,27]. We observed that the mean dN/dS of sex-biased genes, especially male-biased genes, was higher within breeds than those genes with unbiased expression. We also observed a strong positive relationship between the evolutionary rate and the level of male bias, that is highly male-biased genes show greater divergence than lowly male-biased genes and female-biased genes. Female-biased genes have a flatter rise in evolutionary speed than male-biased genes.
Figure 4.
Average ratio of nonsynonymous substitutions (dN) to synonymous substitutions (dS) for male-biased, female-biased and unbiased genes in three breeds. (a) Relationship between dN/dS and extent of sex-biased expression in gonads of three breeds, ‘high’, ‘medium’ and ‘low’ below the x-axis are shorthand for ‘high biased expression’, ‘medium biased expression’ and ‘low biased expression’, respectively. (b) Differences in dN/dS among breeds for three biased categories. Displayed significance scores are *p < 0.05, **p < 0.01 and ***p < 0.001.
Among three breeds, all genes in MD universally had higher dN/dS levels than those in two domesticated breeds (figure 4b). Strongly male-biased genes show an elevated dN/dS between breeds, and this divergence of evolution rate has been maximized (MD, 0.2093; PK, 0.1661; GY, 0.1409) when FC is greater than 10 (high male-biased expression). Interestingly, with the increase of male directional FC, the difference in the rate of sequence evolution between the two domesticated groups becomes more obvious. Male-biased genes of all three categories in PK show a higher evolutionary rate than GY, this may be related to relaxed purifying selection or enhanced positive selection.
We also evaluated the coding sequence evolution of BLGs to understand the effects of loss or reduced sexual conflicts following domestication. The dN/dS ratio of BLGs in GY was statistically significantly lower than MD and PK, while the difference between the latter two is not significant (p = 0.19; electronic supplementary material, figure S3).
2.4. Sequence polymorphism of three breeds
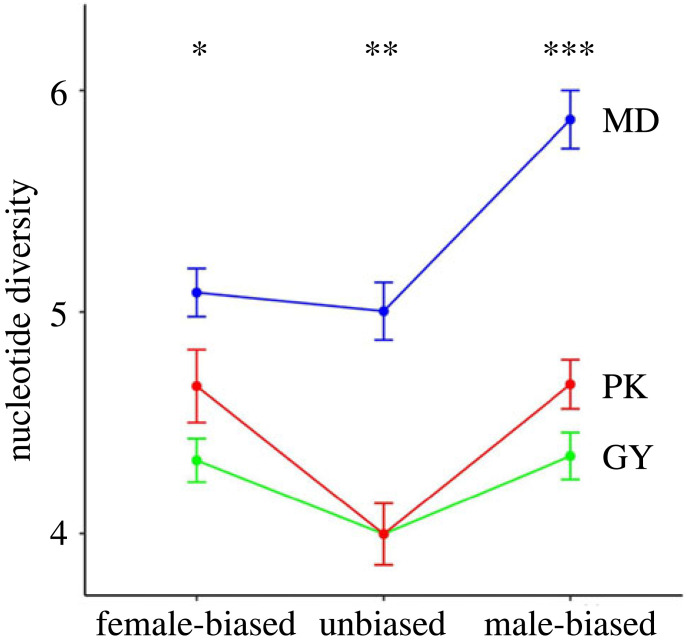
The nucleotide diversity (π), was used as a measure of polymorphism, to reflect the imprint of selection force on sequence diversity before and after domestication (figure 5) [28,29]. Given there are too few sex-biased genes in the liver, and the gonads are subject to strong sexual selection through sperm competition, we assessed the extent of the sequence diversity using the average π value corresponding to different categories of sex-biased genes based on 10 kb sliding windows only in the gonad. Among all sex-biased expression patterns among three breeds, genetic diversity was significantly higher in MD. Specifically, the nucleotide diversity for sex-biased genes was significantly higher than unbiased genes, and this was particularly pronounced for male-biased genes. Interestingly, the two domesticated breeds maintain a high degree of consistency in sequence polymorphism. This low genetic diversity in two domesticated populations highlights the importance of artificial selection in affecting sequence variation.
Figure 5.
Amount of genetic diversity in 10 kb windows explained by the nucleotide diversity (π) for female-biased, unbiased, and male-biased genes expressed in gonads of three breeds. Genetic diversity also significantly varied across three breeds in the same bias categories (Wilcoxon Rank sum test, *p < 0.05, **p < 0.01, ***p < 0.001).
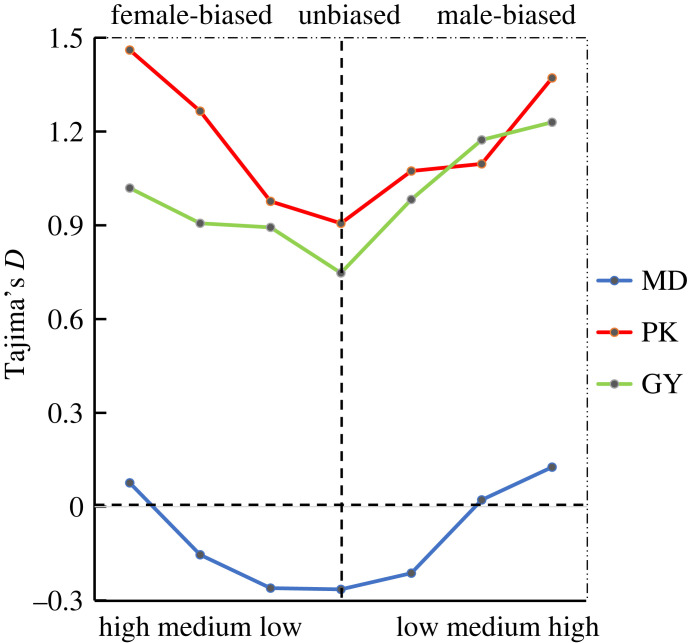
Finally, we assessed Tajima's D for sex-biased genes [30], which summarizes the site-frequency spectrum and reflects several potential evolutionary forces (figure 6). We observe elevated Tajima's D for both male- and female-biased genes across all breeds, although genome-wide estimates vary markedly across breeds.
Figure 6.
Relationship between Tajima's D and extent of sex-biased expression in gonads of MD, PK and GY, ‘high’, ‘medium’ and ‘low’ below the x-axis are shorthand for ‘high biased expression’, ‘medium biased expression’ and ‘low biased expression’, respectively.
3. Discussion
Selection for domestication results in extreme evolutionary pressures, and changes in the mating system, sperm competition and mate choice in domesticates have the potential to vastly shape sex-specific selection. We assessed the potential for domestication to affect sex-specific selection within the genome by assessing the changes in sex-biased gene expression at the embryonic stage, and its sequence properties, in one wild and two domestic duck breeds. Consistent with phenotypic changes in SSD (figure 1), we observe reduced overall proportions of sex-biased genes in the gonad in domestics compared to wild ancestors (table 1). The gonad, as a reproductive tissue, has a higher number of sex-biased genes than the liver as a somatic tissue in all three breeds. This result is consistent with previous research, and can be explained by the fact that the gonads show much greater sexual dimorphism than other tissues [31–33], the result also reflects the decrease of gene expression diversity during domestication events, this effect seems to be common in animals and plants [34]. Our samples were taken from embryonic birds, and the amount of sex-biased gene expression tends to increase during development, with low levels at embryonic stages and high levels in sexually mature adults. Previous studies on birds indicate that 35%∼50% of genes show sex-biased expression in adult gonad which is higher than the ratio in our study (less than 25%) [35,36]. We insisted on used RNA-seq data obtained from the embryonic day 25 gonad for two key reasons. First, we expected to remove interference from acquired factors and environmental effects. Second, female-specific selection in birds is strongest during this developmental time point [35].
The earliest artificial selection may have been unconscious, and subsequent selection drove the emergence of the entire domestication process [37,38]. In addition to purposeful selection, inbreeding of populations and changes in the mating system also occur during and after domestication, and the latter has led to relaxed sexual selection [39,40]. The reduced proportion of sex-biased genes in domestics may result from the removal or reduction in sexual selection. Our results are somewhat different than previous work in Drosophila which has shown that both males and females exhibit transcriptional feminization after the removal of sexual selection [41]. It is not clear whether this discordance reflects fundamental differences in selective pressures, or is a consequence of differences in study design. For example, the Drosophila work was focused on adult samples while our research was based on embryonic tissues [42], and there are major differences through development in the proportion, direction and evolutionary signatures of sex-biased genes in both birds [35] and Drosophila [43].
We estimated the ratio of the nonsynonymous to the synonymous substitution rate (dN/dS) to measure the rate of sequence evolution. Like many previous studies [44–47], we observe obviously elevated rates of sequence evolution in male-biased genes, and, to a lesser extent, the female-biased evolutionary rate also increased compared to unbiased genes (figure 4a,b), and this pattern is similar and clear across all three breeds. Many sex-biased genes, especially the extremely biased ones, often correspond to sex-specific phenotypes or functions and also show narrower expression patterns, so these genes tend to evolve under more sexual selection and less constraint than unbiased genes. If other evolutionary effects are stable, elevated sexual selection increases the rate at which genetic diversity is lost, while relaxed constraint shows faster variant accumulation [48–50]. Thus, it is possible to infer the efficacy of these forces through nucleotide diversity intra-breed. Our intra-breed results suggest that the power of relaxed constraint rather than sexual selection is a major driver that sex-biased genes accumulate variation faster than genes with unbiased expression. Notably, both protein sequence evolutionary rate and genetic diversity of unbiased genes and biased genes differ significantly between the three breeds, this pattern is not breed-specific. Artificial selection will lead to the result that domesticated populations often show low diversity [37], apparently, this force is not just on sex-biased genes. These inter-breed molecular hallmarks suggest that domestication plays a major role in shaping genome diversity and sequence evolution via artificial selection compared with other bias-related selection power (sexual selection and genetic constraint).
Although the inseparable relationship between domestication and relaxed sexual selection is entirely reasonable at the theoretical level, we still found phenotype evidence to prove this crucial link. As an indicator of the intensity of sexual selection, SSD is usually reflected by the male/female weight ratios, and for most birds, including ducks, males are larger on average than females [51]. Our results consistently show the same SSD pattern in which the male is heavier. However, the body size differentiation between the sexes seems to be more extreme in the wild duck population (SSD ratio = 53.14%, p = 0.00067). Assimilation of body size among sexes in domesticated populations shows that the sexual selection maintained by male–male competition or a special mating system is weakened after domestication, which is consistent with our analysis that relaxed sexual selection does exist during domestication. It is worth noting that an effective population ratio between sexes can also reflect sexual selection intensity [52]. Previous studies about the effective sex ratio of mallard pointed to results that males outnumber females [53,54]. The limited supply of females and the resulting competition for mates are the root cause of strong sexual selection in mallards. Predictably, because of domestication and subsequent artificial insemination, this male-skewed mode would decrease and even reverse.
Owing to the consistency of natural conditions and evolutionary processes, the differences in Tajima's D signify the diverse effect of many selection forces including sexual selection. Normally, Tajima's D = 0 under neutrality while D > 0 indicates balancing selection to maintain multiple variants and D < 0 purifying selection or a recent selective sweep [47]. According to the value of Tajima's D, we observed that the category and extent of sex-biased genes determine the net effect of several selection processes. Specifically, female-biased genes with an elevated D show signs of balancing selection or relaxed purifying selection within populations, the homogeneity trend observed in male-biased genes may also indicate similar inferences. An intriguing observation was that the MD population corresponds to a lower level of Tajima's D compared with GY and PK, we had sufficient reasons to infer that this case was not caused by relaxed sexual selection because it also seemed to exist in the whole genome rather than sex-biased genes. This is somewhat surprising that in addition to strongly biased genes, MD showed negative average values for Tajima's D, and this result was contradictory to the high genetic diversity of mallard breeds. This ‘illogicality’ also exists in domesticated ducks whose average Tajima's D is always more than 0.5. This genome-wide extraordinary Tajima's D results may be explained by the difference in the effective population size (Ne) [55], which is much greater in MD than in GY and PK owing to the strong genetic bottleneck associated with domestication. This bottleneck leads to a sudden decrease of Ne in domesticated ducks, and the callback effect of subsequent population expansion events was weak [56]. Another reason may be artificial balancing selection that exists in order to pursue heterosis during the selection of local duck breeds.
The non-adaptive genetic drift, an evolutionary force that cannot be ignored during domestication, makes a fast evolutionary rate at extremely male-biased genes. Previous research conjectured that genetic drift rather than sexual selection promoted sex-biased sequence evolution [14]. In this research, the effect of genetic drift or codon usage bias may be secondary. The faster sequence evolution rate is mainly achieved by the increase of the non-synonymous mutation rate rather than the decrease of the synonymous mutation rate. Given our results and the fact that sexual selection underlies sexual dimorphism, sexual selection and the constraint do affect sequence evolution. Compared with genetic drift and natural selection, sexual selection is directional and only acts on sex-biased genes. Our results indicate that the effect of relaxed sexual selection to shape genomic diversity is counteracted and even reversed by artificial selection during domestication inter-breed. Although our research has determined the role of sexual selection and artificial selection during domestication, several evolutionary forces apparently could push the populations towards a similar direction in this process, which was reflected by the genetic architecture in the genome or transcriptome. For instance, genetic drift and artificial selection can also lead to a decreased level of genetic diversity, while populations subject to balancing selection will show an increase in diversity. Thereby, accurately identifying the effects of a certain evolutionary force using a single detection indicator is hard to achieve. It can be foreseen that these larger datasets, more ideal experimental models, and genetic indicators with multi-parameter and multi-omics, are essential to interpret the complex patterns of evolutionary power and natural variation.
Compared with millions of years of natural selection, short-term domestication has only been thousands of years, since the emergence of human civilization. However, the impact of human intervention on the natural evolution of wild populations is far-reaching. On the one hand, directional selection and small-group captive breeding have reduced biodiversity. On the other hand, the loss of free mating rights has caused the original mating system to be overthrown and rebuilt, which also affects sexual dimorphism and biodiversity. To conclude, we used a combination of a wild breed and broad-time-scale animal models that have undergone a domestication event, to disentangle the evolutionary power that can change genome architecture. Genome-wide evidences proved that although the relaxed sexual selection is not a negligible causal factor for shaping sex-biased diversity, artificial selection mainly explains these signatures of the genome. In addition, evolutionary events such as founder effects, drift, and mixed ancestry undeniably occur during domestication, but equally predictably, these effects are negligible compared to the high-intensity effect of artificial purposeful modification and selection, do not affect our outcome orientation. Our results also implicate that standing genetic variation and sequence evolution within populations represent the outcome of several interacting processes, and in this scenario, domestication and genetic constraints as powerful forces in shaping these special genomic patterns of sex-biased genes under different population dimensions. Although sample size limitations limit the evidence to support only early developmental stages of sex-biased expression, combining similar studies and conjectures, our conclusions should be ‘generally applicable’ throughout the developmental stages of the organism.
4. Material and methods
4.1. Sample collection and sequencing
We obtained fertilized eggs from two domesticated duck breeds. These ducks include one meat-type breed, PK; and one egg-type breed, GY. We also obtained fertilized eggs of MD as wild ducks. The classification of production types follows the description of Animal Genetic Resources in China Poultry. All eggs were kept under standard incubator conditions, qualified eggs were selected by observation of vascular distribution at the early embryonic stage and periodic weighing. For each breed, the left gonad and liver were dissected separately from five male and five female individuals in the laboratory setting where there is an incubator and immediately placed in RNAlater (Invitrogen, Carlsbad, CA, USA) at 25 embryonic days (ed25). At this stage, the duck embryo has developed to the eve of hatching, the liver and many organs have developed to the level of 0 days old, and the gonad is in the rapid development stage. The female-specific selection is strong during this developmental time point [57,58]. RNA was prepared with the Animal Tissue RNA Kit (Qiagen, Hilden, Germany). All tissue samples were sequenced using Illumina NovaSeq 6000 (Illumina Inc., San Diego, CA, USA) as paired-end 150 bp reads at 10× coverage, yielding a total of 685.02 Gb of paired-end reads.
Twenty-four adult individuals of three populations (PK, GY, MD; n = 8) were used to collect whole blood from the brachial veins of ducks by standard venipuncture. The three groups came from three provinces in China separated by more than 1000 km, of which PK came from Beijing, GY came from Jiangsu province, and MD came from Ningxia province. Genomic DNA was extracted using the standard phenol/chloroform method. We sequenced each sample at 5× depth in order to reduce the false-negative rate of variants owing to our strict filter criteria. We randomly selected one individual for 10× coverage, except for the MD, where we sequenced seven individuals at 5 coverage and a random one at 20× coverage. Finally, we generated 159 Gb of paired-end reads. We assessed the quality of both re-sequencing data and RNA-seq data and conducted filtering with fastp v. 0.20. using default parameters.
4.2. RNA-seq and data processing
RNA-seq high-quality reads were mapped to the reference genome of A. platyrhynchos (GenBank Accession GCA_003850225.1) [59] using Hisat2 v. 2.1.0 [60,61]. Transcript abundances for the annotated genes were estimated using StringTie v. 2.1.2 [62,63], where relative expressions are expressed as fragments per kilobase of exon per million mapped reads (FPKM) values. Genes with FPKM of less than 0.5 in all samples were considered not expressed and would be deleted. We deleted all sex chromosome genes because the sex-biased expression is defined based on autosomes. In addition, we also deleted all immune-related genes and the major histocompatibility complex gene family located on duck micro chromosome 17, because these genes have some potential confounding effects independent of sexual conflict [64–66].
4.3. Hierarchical clustering and heatmaps
Hierarchical clustering was performed using the pvclust package [67] of R, with bootstrap resampling (1000 replicates) using Euclidean clustering with complete linkage. Heatmaps were separately generated for all samples, liver and left gonad using log2 average expression of male and female autosomal genes using the R package pheatmap v. 1.0.12.
4.4. Sex-biased gene expression
Normalized FPKM expression counts of autosomal genes for each sex were used to calculate sex bias, with fold-change ratios between males and females, starting at unlogged twofold, and a significance threshold of p < 0.05 after adjusting for multiple testing. The sex-biased genes were further divided into four categories based on fold FC: low bias (2–4 FC), medium bias (4–10 FC) and high bias (greater than 10 FC), resulting in three categories for male-biased genes, three categories for female-biased genes, and an unbiased gene category (less than 2 FC). To further determine the dynamic changes of sex-biased expression, we also identified three classes of genes of autosomal, BLGs, that is, genes that show biased expression in wild breeds are lost in both two domesticated breeds. BAGs refer to genes that only show sex-biased expression in domesticated breeds. BCGs reflect the reversal of sex-biased expression patterns before and after domestication.
4.5. Estimation of sexual size dimorphism
We measured the phenotypes of body weight of 50 (25 males and 25 females) healthy adult PK (12 weeks old) obtained from Beijing Golden Duck Co., Ltd (Beijing, China). Detailed weight data of adult MD and GY are collected from previous studies [68–70], sample information is presented in the electronic supplementary material, table S3.
SSD refers to the differences between males and females of the same species, such as in body size, and weight, and is critical for a better understanding of the dynamic changes of sexual selection. Body size was the mean body mass in grams of adult males and adult females [71], and we calculated (adult male weight/total weight) per cent to visualize SSD, Z-tests for two independent samples were used to test whether the SSD is significantly different between three breeds.
4.6. Population genomics analysis
To assess the polymorphism and sequence evolutionary characteristics that may be caused by dynamic changes in the long-term effects of sexual selection on domestication, we calculated multiple population genetic parameters in the three breeds separately, including both the sex-biased genes and unbiased genes. Compared with the liver and other tissues, the gonads are more sexually dimorphic, and more phenotypic dimorphic tissues within the body exhibit greater levels of transcriptional dimorphism. We therefore only focused on sex-biased genes in the gonad for the next analysis. Nucleotide diversity was used as a measure of sequence diversity. We extract the corresponding gene sets from the reference genome of A. platyrhynchos according to seven sex-biased gene categories (including the unbiased genes). Nucleotide sequence for autosomal genes of three breeds was mapped to the corresponding gene sets using BWA v. 0.7.17 [72]. Picard Tools v. 2.26.0 was used to convert the mapping file from Sam to Bam format. Duplicate reads were removed from individual sample alignments using MarkDuplicates. The Genome Analysis Toolkit v. 4.2.1.0 (GATK) RealignerTargetCreator, and IndelRealigner protocol were used for global realignment of reads around INDELs [73]. We filtered variants both per breed and per individual using GATK according to the stringent filtering criteria. Finally, we obtained polymorphism results under the 10 kb window through VCFtools v. 0.1.13 [74]. The statistical data corresponding to the tail end of genes truncated by 10 kb windows was removed, and then the average of π was extracted. This approach avoids the problem of uncertainty of window statistics' results, especially sex-biased genes that exist discretely in the genome.
Variation in standing genetic variation within populations represents the outcome of several interacting processes, notably balancing selection and purifying selection. To estimate the net effect of these processes, we estimated Tajima's D [30] using VCFtools v. 0.1.13 for sex-biased genes in three breeds.
To further examine the potential explanatory power of relaxed sexual selection resulting from domestication, we extracted the ratio of the non-synonymous to the synonymous substitution rate (dN/dS). Gallus Gallus, as an outgroup, was used to find the orthologous genes. Sex-biased genes would be identified that gene symbols are exactly the same, and these genes also satisfy reciprocal top hits from a BLASTn with an e-value cutoff of 1×10−10 and a minimum percentage identity of 60%. We identified variants of biased and unbiased genes using the previous method, Python scripts were written to calculate dN/dS using VCF files. Specifically, we used the sequencing data of each individual to compare to the reference genome and then get the VCF file. We used the mutation information in the VCF file to deduce the base sequence of each individual target gene by our python script (https://github.com/zhutao1009/dnds), and finally used the aligned fasta sequence file of these genes to calculate the dN/dS of the sequence. The outliers that only appeared in a single breed were removed, and then the average value of dN/dS was calculated.
4.7. Gene ontology
In order to determine the certain biological functions and pathways corresponding to bias-lost genes (BLGs) and sex-biased genes, we conducted a GO enrichment analysis using the clusterProfiler package [75] of R. Genes that have a universal bias pattern across all individuals are regarded as high-confidence male-bias or female-biased genes. To make the enrichment results more meaningful, we use the database human (Homo sapiens) rather than chicken (G. gallus) for annotation. Enrichment was determined using an FDR-corrected significance threshold of 0.05.
Acknowledgements
We thank our colleagues in the Poultry Team at the National Engineering Laboratory for Animal Breeding of China Agricultural University for assistance in sample collection and for helpful comments on the manuscript. We are very grateful to Judith E. Mank for suggesting the manuscript and article structure. The authors declare no conflicts of interest.
Ethics
Our animal experiments were approved by the Animal Care and Use Committee of China Agricultural University (approval ID: XXCB-20090209). All the animals were fed and handled according to the regulations and guidelines established by this committee, and all efforts were made to minimize suffering.
Data accessibility
Whole-genome resequencing data used in this study are available on the NCBI short read archive (Accession PRJNA419832). RNA-seq reads have been deposited in NCBI Sequence Read Archive under accession numbers SRP269397. Data is also available in the electronic supplementary material [76].
Authors' contributions
H.G.: data curation, formal analysis, investigation, validation, writing—original draft, writing—review and editing; L.W.: resources, software; X.L.: resources, software; W.Y.: resources, software; L.Z.: resources, software; Z.Z.: data curation, resources; T.Z.: data curation, resources; Y.J.: resources, software; Y.C.: resources, software; L.Q.: conceptualization, funding acquisition, methodology, project administration, supervision, validation, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
This work was supported by the Beijing Innovation Team of the Modern Agro-industry Technology Research System for Poultry (grant no. BJJQ-G01-2023).
References
- 1.Dapper AL, Wade MJ. 2016. The evolution of sperm competition genes: the effect of mating system on levels of genetic variation within and between species. Evolution 70, 502-511. ( 10.1111/evo.12848) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasimatis KR, Nelson TC, Phillips PC. 2017. Genomic signatures of sexual conflict. J. Hered. 108, 780-790. ( 10.1093/jhered/esx080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsch J, Ellegren H. 2013. The evolutionary causes and consequences of sex-biased gene expression. Nat. Rev. Genet. 14, 83-87. ( 10.1038/nrg3376) [DOI] [PubMed] [Google Scholar]
- 4.Rowe L, Chenoweth SF, Agrawal AF. 2018. The genomics of sexual conflict. Am. Nat. 192, 274-286. ( 10.1086/698198) [DOI] [PubMed] [Google Scholar]
- 5.Andersson M, Iwasa Y. 1996. Sexual selection. Trends Ecol. Evol. 11, 53-58. ( 10.1016/0169-5347(96)81042-1) [DOI] [PubMed] [Google Scholar]
- 6.Hosken DJ, House CM. 2011. Sexual selection. Curr. Biol. 21, R62-R65. ( 10.1016/j.cub.2010.11.053) [DOI] [PubMed] [Google Scholar]
- 7.Kokko H, Brooks R, Jennions MD, Morley J. 2003. The evolution of mate choice and mating biases. Proc. R. Soc. B 270, 653-664. ( 10.1098/rspb.2002.2235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Read AF, Harvey PH. 1989. Validity of sexual selection in birds - reply. Nature 340, 105. ( 10.1038/340105a0) [DOI] [Google Scholar]
- 9.Wong BBM, Candolin U. 2005. How is female mate choice affected by male competition? Biol. Rev. 80, 559-571. ( 10.1017/s1464793105006809) [DOI] [PubMed] [Google Scholar]
- 10.Lande R. 1980. Sexual dimorphism, sexual selection, and adaptation in polygenic characters. Evolution 34, 292-305. ( 10.2307/2407393) [DOI] [PubMed] [Google Scholar]
- 11.Rice WR. 1984. Sex-chromosomes and the evolution of sexual dimorphism. Evolution 38, 735-742. ( 10.2307/2408385) [DOI] [PubMed] [Google Scholar]
- 12.Assis R, Zhou Q, Bachtrog D. 2012. Sex-biased transcriptome evolution in Drosophila. Genome Biol. Evol. 4, 1189-1200. ( 10.1093/gbe/evs093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leichty AR, Pfennig DW, Jones CD, Pfennig KS. 2012. Relaxed genetic constraint is ancestral to the evolution of phenotypic plasticity. Integr. Comp. Biol. 52, 16-30. ( 10.1093/icb/ics049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison PW, Wright AE, Zimmer F, Dean R, Montgomery SH, Pointer MA, Mank JE. 2015. Sexual selection drives evolution and rapid turnover of male gene expression. Proc. Natl Acad. Sci. USA 112, 4393-4398. ( 10.1073/pnas.1501339112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B. 2007. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature 450, 233-237. ( 10.1038/nature06323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gering E, Incorvaia D, Henriksen R, Wright D, Getty T. 2019. Maladaptation in feral and domesticated animals. Evol. Appl. 12, 1274-1286. ( 10.1111/eva.12784) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Driscoll CA, Macdonald DW, O'Brien SJ. 2009. From wild animals to domestic pets, an evolutionary view of domestication. Proc. Natl Acad. Sci. USA 106, 9971-9978. ( 10.1073/pnas.0901586106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Ni P, Li X, Han J, Jakovlic I, Zhang C, Zhao S. 2018. Population size may shape the accumulation of functional mutations following domestication. BMC Evol. Biol. 18, 4. ( 10.1186/s12862-018-1120-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birkhead TR, Atkin L, Moller AP. 1987. Copulation behavior of birds. Behaviour 101, 101-138. (https://www.jstor.org/stable/4534592) [Google Scholar]
- 20.Birkhead TR, Pizzari T. 2002. Postcopulatory sexual selection. Nat. Rev. Genet. 3, 262-273. ( 10.1038/nrg774) [DOI] [PubMed] [Google Scholar]
- 21.Collet JM, Dean RF, Worley K, Richardson DS, Pizzari T. 2014. The measure and significance of Bateman's principles. Proc. R. Soc. B 281, 20132973. ( 10.1098/rspb.2013.2973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright D, Rubin CJ, Barrio AM, Schutz K, Kerje S, Brandstrom H, Kindmark A, Jensen P, Andersson L. 2010. The genetic architecture of domestication in the chicken: effects of pleiotropy and linkage. Mol. Ecol. 19, 5140-5156. ( 10.1111/j.1365-294X.2010.04882.x) [DOI] [PubMed] [Google Scholar]
- 23.Dale J, Dunn PO, Figuerola J, Lislevand T, Szekely T, Whittingham LA. 2007. Sexual selection explains Rensch's rule of allometry for sexual size dimorphism. Proc. R. Soc. B 274, 2971-2979. ( 10.1098/rspb.2007.1043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soulsbury CD, Kervinen M, Lebigre C. 2014. Sexual size dimorphism and the strength of sexual selection in mammals and birds. Evol. Ecol. Res. 16, 63-76. [Google Scholar]
- 25.Szekely T, Freckleton RP, Reynolds JD. 2004. Sexual selection explains Rensch's rule of size dimorphism in shorebirds. Proc. Natl Acad. Sci. USA 101, 12 224-12 227. ( 10.1073/pnas.0404503101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolivar P, Mugal CF, Nater A, Ellegren H. 2016. Recombination rate variation modulates gene sequence evolution mainly via GC-biased gene conversion, not Hill-Robertson interference, in an avian system. Mol. Biol. Evol. 33, 216-227. ( 10.1093/molbev/msv214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellegren H, Parsch J. 2007. The evolution of sex-biased genes and sex-biased gene expression. Nat. Rev. Genet. 8, 689-698. ( 10.1038/nrg2167) [DOI] [PubMed] [Google Scholar]
- 28.Ellegren H, Galtier N. 2016. Determinants of genetic diversity. Nat. Rev. Genet. 17, 422-433. ( 10.1038/nrg.2016.58) [DOI] [PubMed] [Google Scholar]
- 29.Nei M, Li WH. 1979. Mathematical-model for studying genetic-variation in terms of restriction endonucleases. Proc. Natl Acad. Sci. USA 76, 5269-5273. ( 10.1073/pnas.76.10.5269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tajima F. 1989. Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldman TD, Arbeitman MN. 2007. Genomic and functional studies of Drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS Genet. 3, 2278-2295. ( 10.1371/journal.pgen.0030216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mank JE, Hultin-Rosenberg L, Webster MT, Ellegren H. 2008. The unique genomic properties of sex-biased genes: insights from avian microarray data. BMC Genomics 9, 1-4. ( 10.1186/1471-2164-9-148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, Eastman S, Oliver B. 2003. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science 299, 697-700. ( 10.1126/science.1079190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W, et al. 2019. Decrease of gene expression diversity during domestication of animals and plants. BMC Evol. Biol. 19, 19. ( 10.1186/s12862-018-1340-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mank JE, Nam K, Brunstrom B, Ellegren H. 2010. Ontogenetic complexity of sexual dimorphism and sex-specific selection. Mol. Biol. Evol. 27, 1570-1578. ( 10.1093/molbev/msq042) [DOI] [PubMed] [Google Scholar]
- 36.Pointer MA, Harrison PW, Wright AE, Mank JE. 2013. Masculinization of gene expression is associated with exaggeration of male sexual dimorphism. PLoS Genet. 9, e1003697. ( 10.1371/journal.pgen.1003697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diamond J. 2002. Evolution, consequences and future of plant and animal domestication. Nature 418, 700-707. ( 10.1038/nature01019) [DOI] [PubMed] [Google Scholar]
- 38.Conner JK. 2003. Artificial selection: a powerful tool for ecologists. Ecology 84, 1650-1660. ( 10.1890/0012-9658(2003)084[1650:Asaptf]2.0.Co;2) [DOI] [Google Scholar]
- 39.Dapper AL, Wade MJ. 2020. Relaxed selection and the rapid evolution of reproductive genes. Trends Genet. 36, 640-649. ( 10.1016/j.tig.2020.06.014) [DOI] [PubMed] [Google Scholar]
- 40.Swanson WJ, Vacquier VD. 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3, 137-144. ( 10.1038/nrg733) [DOI] [PubMed] [Google Scholar]
- 41.Hollis B, Houle D, Yan Z, Kawecki TJ, Keller L. 2014. Evolution under monogamy feminizes gene expression in Drosophila melanogaster. Nat. Commun. 5, 3482. ( 10.1038/ncomms4482) [DOI] [PubMed] [Google Scholar]
- 42.Mank JE, Ellegren H. 2009. All dosage compensation is local: gene-by-gene regulation of sex-biased expression on the chicken Z chromosome. Heredity 102, 312-320. ( 10.1038/hdy.2008.116) [DOI] [PubMed] [Google Scholar]
- 43.Perry JC, Harrison PW, Mank JE. 2014. The ontogeny and evolution of sex-biased gene expression in Drosophila melanogaster. Mol. Biol. Evol. 31, 1206-1219. ( 10.1093/molbev/msu072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campos JL, Johnston KJA, Charlesworth B. 2018. The effects of sex-biased gene expression and x-linkage on rates of sequence evolution in Drosophila. Mol. Biol. Evol. 35, 655-665. ( 10.1093/molbev/msx317) [DOI] [PubMed] [Google Scholar]
- 45.Dutoit L, Mugal CF, Bolivar P, Wang M, Nadachowska-Brzysk K, Smeds L, Yazdi HP, Gustafsson L, Ellegren H. 2018. Sex-biased gene expression, sexual antagonism and levels of genetic diversity in the collared flycatcher (Ficedula albicollis) genome. Mol. Ecol. 27, 3572-3581. ( 10.1111/mec.14789) [DOI] [PubMed] [Google Scholar]
- 46.Grath S, Parsch J. 2012. Rate of amino acid substitution is influenced by the degree and conservation of male-biased transcription over 50 Myr of Drosophila evolution. Genome Biol. Evol. 4, 346-359. ( 10.1093/gbe/evs012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sayadi A, Barrio AM, Immonen E, Dainat J, Berger D, Tellgren-Roth C, Nystedt B, Arnqvist G. 2019. The genomic footprint of sexual conflict. Nat. Ecol. Evol. 3, 1725-1730. ( 10.1038/s41559-019-1041-9) [DOI] [PubMed] [Google Scholar]
- 48.Huang HT, Rabosky DL. 2015. Sex-linked genomic variation and its relationship to avian plumage dichromatism and sexual selection. BMC Evol. Biol. 15, 10. ( 10.1186/s12862-015-0480-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lao O, Kayser M. 2008. Mechanisms and effects of natural selection. Med. Gen. 20, 308-314. ( 10.1007/s11825-008-0122-y) [DOI] [Google Scholar]
- 50.Weinreich DM, Watson RA, Chao L. 2005. Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evolution 59, 1165-1174. [PubMed] [Google Scholar]
- 51.Ancona S, Liker A, Cristina Carmona-Isunza M, Szekely T. 2020. Sex differences in age-to-maturation relate to sexual selection and adult sex ratios in birds. Evol. Lett. 4, 44-53. ( 10.1002/evl3.156) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ancona S, Denes FV, Krueger O, Szekely T, Beissinger SR. 2017. Estimating adult sex ratios in nature. Phil. Trans. R. Soc. B 372, 20160313. ( 10.1098/rstb.2016.0313) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Humburg DD, Prince HH, Bishop RA. 1978. Social-organization of a mallard population in northern Iowa. J. Wildl. Manage. 42, 72-80. ( 10.2307/3800691) [DOI] [Google Scholar]
- 54.Ohde BR, Bishop RA, Dinsmore JJ. 1983. Mallard reproduction in relation to sex-ratios. J. Wildl. Manage. 47, 118-126. ( 10.2307/3808058) [DOI] [Google Scholar]
- 55.Wiberg RAW, Veltsos P, Snook RR, Ritchie MG. 2021. Experimental evolution supports signatures of sexual selection in genomic divergence. Evol. Lett. 5, 214-229. ( 10.1002/evl3.220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Z, et al. 2018. Whole-genome resequencing reveals signatures of selection and timing of duck domestication. Gigascience 7, giy027. ( 10.1093/gigascience/giy027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S, Bai S, Qin X, Zhang J, Irwin DM, Zhang S, Wang Z. 2019. Comparison of whole embryonic development in the duck (Anas platyrhynchos) and goose (Anser cygnoides) with the chicken (Gallus gallus). Poult. Sci. 98, 3278-3291. ( 10.3382/ps/pez133) [DOI] [PubMed] [Google Scholar]
- 58.Moghadam HK, Pointer MA, Wright AE, Berlin S, Mank JE. 2012. W chromosome expression responds to female-specific selection. Proc. Natl Acad. Sci. USA 109, 8207-8211. ( 10.1073/pnas.1202721109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Z, et al. 2018. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat. Commun. 9, 2648. ( 10.1038/s41467-018-04868-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357-360. ( 10.1038/nmeth.3317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907-915. ( 10.1038/s41587-019-0201-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290-295. ( 10.1038/nbt.3122) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650-1667. ( 10.1038/nprot.2016.095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghosh R, Andersen EC, Shapiro JA, Gerke JP, Kruglyak L. 2012. Natural variation in a chloride channel subunit confers avermectin resistance in C. elegans. Science 335, 574-578. ( 10.1126/science.1214318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hedrick PW. 2011. Population genetics of malaria resistance in humans. Heredity 107, 283-304. ( 10.1038/hdy.2011.16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. 1999. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400, 667-671. [DOI] [PubMed] [Google Scholar]
- 67.Suzuki R, Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540-1542. ( 10.1093/bioinformatics/btl117) [DOI] [PubMed] [Google Scholar]
- 68.Boos M, Zorn T, Le Maho Y, Groscolas R, Robin JP. 2002. Sex differences in body composition of wintering mallards (Anas platyrhynchos): possible implications for survival and reproductive performance. Bird Study 49, 212-218. ( 10.1080/00063650209461268) [DOI] [Google Scholar]
- 69.Janiszewski P, Murawska D, Hanzal V, Gesek M, Michalik D, Zawacka M. 2018. Carcass characteristics, meat quality, and fatty acid composition of wild-living mallards (Anas platyrhynchos L.). Poult. Sci. 97, 709-715. ( 10.3382/ps/pex335) [DOI] [PubMed] [Google Scholar]
- 70.Zhu C, Xu W, Tao Z, Liu H, Song W, Zhang S, Yuan Z, Li H. 2020. Effects of rearing in cages on slaughter performance and intestinal microbiota of Gaoyou ducks (Anas platyrhynchos). J. Agri. Biotechnol. 28, 2200-2208. [Google Scholar]
- 71.Greenwood JG. 2003. Measuring sexual size dimorphism in birds. Ibis 145, E124-E126. ( 10.1046/j.1474-919X.2003.00175.x) [DOI] [Google Scholar]
- 72.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754-1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McKenna A, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297-1303. ( 10.1101/gr.107524.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27, 2156-2158. ( 10.1093/bioinformatics/btr330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu G, Wang L-G, Han Y, He Q-Y. 2012. clusterProfiler: an R Package for comparing biological themes among gene clusters. Omics-J. Integr. Biol. 16, 284-287. ( 10.1089/omi.2011.0118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gu H, et al. 2023. Domestication affects sex-biased gene expression evolution in the duck. Figshare. ( 10.6084/m9.figshare.c.6533391) [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Whole-genome resequencing data used in this study are available on the NCBI short read archive (Accession PRJNA419832). RNA-seq reads have been deposited in NCBI Sequence Read Archive under accession numbers SRP269397. Data is also available in the electronic supplementary material [76].