Abstract
Cancer progression is influenced by junctional adhesion molecule (JAM) family members. The relationship between JAM family members and different types of cancer was examined using The Cancer Genome Atlas dataset. mRNA levels of the F11R (F11 receptor) in tumours were inversely correlated to the expression of JAM‐2 and JAM‐3. This relationship was unique to breast cancer (BCa) and was associated with poor prognosis (p = 0.024, hazard ratio = 1.44 [1.05–1.99]). A 50‐gene molecular signature (prediction analysis of microarray 50) was used to subtype BCa. F11R mRNA expression significantly increased in human epidermal growth factor receptor 2 (HER2)‐enriched (p = 0.0035) and basal‐like BCa tumours (p = 0.0005). We evaluated F11R protein levels in two different compositions of BCa subtype patient tissue array cohorts to determine the relationship between BCa subtype and prognosis. Immunohistochemistry staining revealed that a high F11R protein level was associated with poor overall survival (p < 0.001; Taipei Medical University [TMU] cohort, p < 0.001; Kaohsiung Veterans General Hospital [KVGH] cohort) or disease‐free survival (p < 0.001 [TMU cohort], p = 0.034 [KVGH cohort]) in patients with BCa. Comparison of F11R levels in different subtypes revealed the association of poor prognosis with high levels of F11R among luminal (p < 0.001 [TMU cohort], p = 0.027 [KVGH cohort]), HER2 positive (p = 0.018 [TMU cohort], p = 0.037 [KVGH cohort]), and triple‐negative (p = 0.013 [TMU cohort], p = 0.037 [KVGH cohort]) BCa. F11R‐based RNA microarray analysis and Ingenuity Pathway Analysis were successful in profiling the detailed gene ontology of triple‐negative BCa cells regulated by F11R. The EP300 transcription factor was highly correlated with F11R in BCa (R = 0.51, p < 0.001). By analysing these F11R‐affected molecules with the L1000CDs datasets, we were able to predict some repurposing drugs for potential application in F11R‐positive BCa treatment.
Keywords: F11R, breast cancer, EP300, survival curve, transcriptomics
Introduction
Breast cancer (BCa) is globally responsible for 11.7% of cases (approximately 2.3 million cases) and 6.9% of deaths (0.68 million cases) from a total of 19.3 million cancer cases [1]. There are four widely recognised and accepted subtypes of BCa according to the expression of oestrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). They are luminal (types A and B), HER2‐enriched, and basal‐like (the so‐called triple negative) [2]. Both luminal A and luminal B tumours are ER and PR positive, with low Ki67 expression in luminal A tumours and high Ki67 expression in luminal B tumours. The HER2‐enriched subtype is HER2‐positive and ER/PR‐negative, whereas triple‐negative BCa (TNBC) is negative for ER/PR/HER2 [2, 3, 4, 5]. A high Ki67 index makes HER2‐enriched and TNBC more aggressive than luminal cancers [4, 6]. Luminal BCa represents 70–80% of all BCa cases, followed by HER2‐overexpressing cancers (10–20%) and TNBC (10–15%) [7, 8]. The advent of targeted therapies for BCa with specific receptor expression (ER, PR, or HER2) has led to significantly improved disease‐free survival (DFS) rates [9]. Conversely, the lack of ER, PR, and HER2 in TNBC hinders HER2‐targeted interventions [10]. Therefore, it is important to develop new biomarkers and therapeutic strategies for TNBC.
The F11 receptor (F11R), also known as junctional adhesion molecule‐A (JAM‐A), is a member of the immunoglobulin superfamily of cell adhesion receptors [11, 12, 13, 14, 15, 16, 17]. F11R exerts a variety of functions in various pathological tissues, and a correlation has been found between F11R levels and prognosis outcomes in multiple cancers [18, 19, 20, 21, 22, 23, 24]. Initially, F11R was found to be expressed in normal mammary epithelium, but downregulated in metastatic BCa [25]. In later studies, higher F11R expression was associated with poorer outcomes in BCa [26, 27]. Furthermore, high F11R expression has been linked to HER2 expression in BCa tissues, although there were fewer cases of HER2 overexpression in that study [28]. The discrepancy between previous studies on F11R expression and its correlation with prognosis may be attributed to the heterogeneity of BCa. Therefore, a better understanding of the relationship between F11R levels and BCa subtype will lead to a more accurate prognosis.
In total, 542 cases from two cohorts of patients were examined in this study to confirm the association between F11R expression and the clinical outcome of BCa. In addition to the observation that F11R has a unique function in BCa compared to other family members (JAM‐2 and JAM‐3), we also investigated the prognostic value of F11R in BCa, its limitations, and the application of therapeutic strategies in various BCa subtypes.
Materials and methods
Evaluation of the JAM family and clinical correlation
To analyse the correlation between different members of the JAM family or genes and various cancer types, several databases were used to analyse the related clinical values as described previously [29]. The databases and analysis tools included TIMER 2.0 (http://timer.cistrome.org/) [30], Kaplan–Meier plotter (https://kmplot.com/analysis/index.php?p=background) [31], GEPIA 2.0 (http://gepia2.cancer-pku.cn/#index) [32], PrognoScan (http://dna00.bio.kyutech.ac.jp/PrognoScan/) [33], and UCSC XENA (https://xenabrowser.net/) [34]. The correlation between the JAM family and BCa was analysed based on the included references and detailed operation tutorials for each database website. TIMER 2.0 was used to determine the distribution of intermolecular genes in different cancer types. JAM family expression in BCa was based on XENA and GEPIA 2.0. The consequences of each probe/gene for patient prognosis were analysed using the Kaplan–Meier plotter and PrognoScan. Correlation analysis between genes was performed using GEPIA 2.0. All statistical values were calculated and provided by the statistical algorithms formulated on each website.
Clinical data and samples from BCa patients
This study collected 542 clinical specimens from two different cohorts of women with primary BCa. The initial cohort consisted of 302 women diagnosed with breast ductal carcinoma between 1991 and 1999 through the archives of the Department of Pathology at Kaohsiung Veterans General Hospital (KVGH). From 1998 to 2008, 240 resections of BCa were performed at Wan Fang Hospital of Taipei Medical University (TMU). All data were analysed anonymously, and the identities of the participants were not disclosed. Tissue microarrays (TMAs) were constructed using BCa specimens as described previously [35]. Clinical records and tissue samples were collected in accordance with the Institutional Review Board protocols at TMU (reference number WFH‐IRB‐99049, TMU cohort) and the Institutional Review Board at the KVGH (reference number VGHKS12‐CT2‐07). Using TMA blocks, 4‐μm‐thick sections were generated. Clinical records, including hormone receptor status, tumour recurrence, and survival, were used to obtain information on the clinical pathology and follow‐up. Traditionally, this approach has been used as a measure of patient survival. Overall survival (OS) time was calculated from the time the treatment started to the time of the patient's last follow‐up or death. DFS was defined as the duration of time after treatment during which no sign of recurrence or relapse of cancer was found. Each study participant was monitored for at least 60 months (5 years), and sometimes for as long as 250 months, or until death.
Immunohistochemistry staining to determine F11R protein levels in human tissues
Immunohistochemistry (IHC) staining of samples was performed as previously described [35, 36]. The sections were deparaffinised, rehydrated, blocked, and antigen retrieval was performed in Tris‐EDTA buffer (pH 9.0). Sections were incubated with anti‐F11R antibody (Cat. H00050848‐M01, 1:1000 dilution; Abnova, Taipei, Taiwan) overnight at 4 °C. After washing, the sections were incubated with anti‐mouse probe (MACH 1 Universal HRP Detection system; Biocare Medical, Pacheco, CA, USA) at room temperature for 30 min, followed by incubation with horseradish peroxidase‐conjugated secondary antibody for another 30 min. Immunoreactivity was revealed by the reaction with 3,3′‐diaminobenzidine. Sections were counterstained with haematoxylin and mounted.
Evaluation of IHC staining scores
Pathologists (CLC and JSC) who were blinded to the clinical parameters independently reviewed and scored the TMA sections stained with F11R. The staining pattern of F11R showed distinct membrane staining across tumour regions. The intensity of tumour regions with positive staining was used to semi‐quantitatively score F11R expression levels. According to the staining intensity, 0 indicated no staining, 1 weakly perceptible membrane staining, 2 moderate complete membrane staining, and 3 strong complete membrane staining. An expression score of 0–1 and 2–3 indicated low and high expression of F11R, respectively. There was no major discrepancy in the interpretation of IHC results. Variations in scoring in some cases were reviewed and consensus was reached by the two pathologists for final interpretation.
DNA microarray‐based molecular interaction network
HDQ‐P1 and CAL‐120 cells were overexpressing F11R cDNA (OHu24113; GenScript, Piscataway, NJ, USA) and an empty vector via TOOLSFect transfection reagent TTM‐TF01 (BIOTOOLS, Taipei, Taiwan). A TOOLSQuant II Fast RT Kit KRT‐BA06‐2 (BIOTOOLS, Taipei, Taiwan) was used to generate all cDNAs. Related gene expression levels were analysed using the Human Genome U133 Plus 2.0 Array. Raw data were exported from GreenSpring software, and the molecular network was analysed using Ingenuity Pathway Analysis (IPA) software (https://analysis.ingenuity.com/pa/installer/select).
Simulation of potential drugs
The L1000CDs website (https://maayanlab.cloud/L1000CDS2/#/index/6194cd57d99ec600506d5c0c) was employed to analyse potential drugs against the F11R‐genetic profile. The microarray analysis results were submitted to the L1000CDs website for reverse mimic analysis.
Statistical analyses
Chi‐square test for categorical data and Student's t‐test for continuous data were used to analyse the relationships between clinicopathological characteristics and F11R expression. DFS and OS curves were calculated using the Kaplan–Meier method, and the differences between the expression groups were evaluated using the log‐rank test. Univariate and multivariate analyses were performed using a Cox proportional hazards model to identify the significant prognostic factors of OS and DFS. Statistical significance was set at p < 0.05.
Results
BCa survival is correlated with the expression of F11R
To comprehensively examine the expression of F11R in different cancers, we conducted pan‐cancer analysis to determine whether gene expression correlates with tumour characteristics in The Cancer Genome Atlas (TCGA) database. The relative expression of F11R in cancers of different types is shown in Figure 1A. The expression of F11R in cancer tissues was compared with that in normal tissues. Several cancers, including BCa, displayed a marked increase in F11R expression. Currently, the JAM family comprises of F11R, JAM‐2, and JAM‐3. We also compared the expression levels of JAM‐2 and JAM‐3 in other cancers (supplementary material, Figure S1A). Notably, an increase in F11R was associated with bladder urothelial carcinoma (BLCA), BCa, cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), lung squamous cell carcinoma (LUSC), and uterine corpus endometrial carcinoma (UCEC), and a decrease in JAM‐2/JAM‐3. To determine whether these trends in tumour and normal cases are relevant to OS, a correlation analysis of OS and F11R was performed. The results in supplementary material, Figure S1B revealed that poor survival in BCa was positively correlated only with high F11R expression (hazard ratio [HR] = 1.44 [1.05–1.99], p = 0.024). A heatmap analysis of F11R expression in individual cases revealed that F11R expression was generally lower in normal tissues than in tumour tissues, and JAM‐2/JAM‐3 expression was higher in normal tissue (Figure 1B and supplementary material, Table S1). A review of different database sources, including GEPIA 2.0, also indicated that F11R was highly expressed in BRCA, in contrast to JAM‐2 and JAM‐3 (Figure 1C). These findings indicate that the F11R was unique among the other members of JAM and that BCa patients with higher F11R gene expression had a poorer prognosis in terms of OS.
Figure 1.
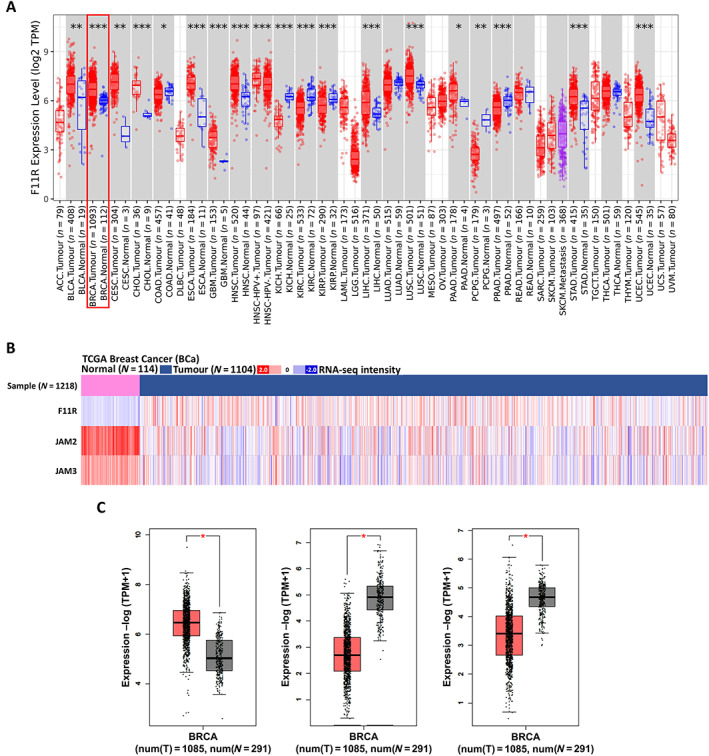
Distribution of JAM members within BCa. (A) Expression of F11R in cancer types. Analysed data from TIMER 2.0 website. (B) Heatmap illustrating the expression of JAM members in BCa. The data were obtained from XENA website. (C) Boxplots illustrating the expression of the JAM gene family in the BCa normal and tumour groups. Data were analysed using the GEPIA 2.0 website (*p < 0.05; **p < 0.01; ***p < 0.001).
F11R expression is specific to BCa subtypes
Relapse is one of the reasons that cancer treatment is challenging. To understand the relationship between JAM and BCa, the correlation of JAM family expression and recurrence‐free survival was analysed (Figure 2A). Comparison of the prognostic endpoints of the JAM family using clinical data from multiple sources of BCa consistently revealed the association of F11R with a higher survival risk in BCa patients, including relapse‐free survival (RFS), disease‐specific survival (DSS), and DFS. Patients with JAM‐2 or JAM‐3 had better prognosis (Figure 2B).
Figure 2.
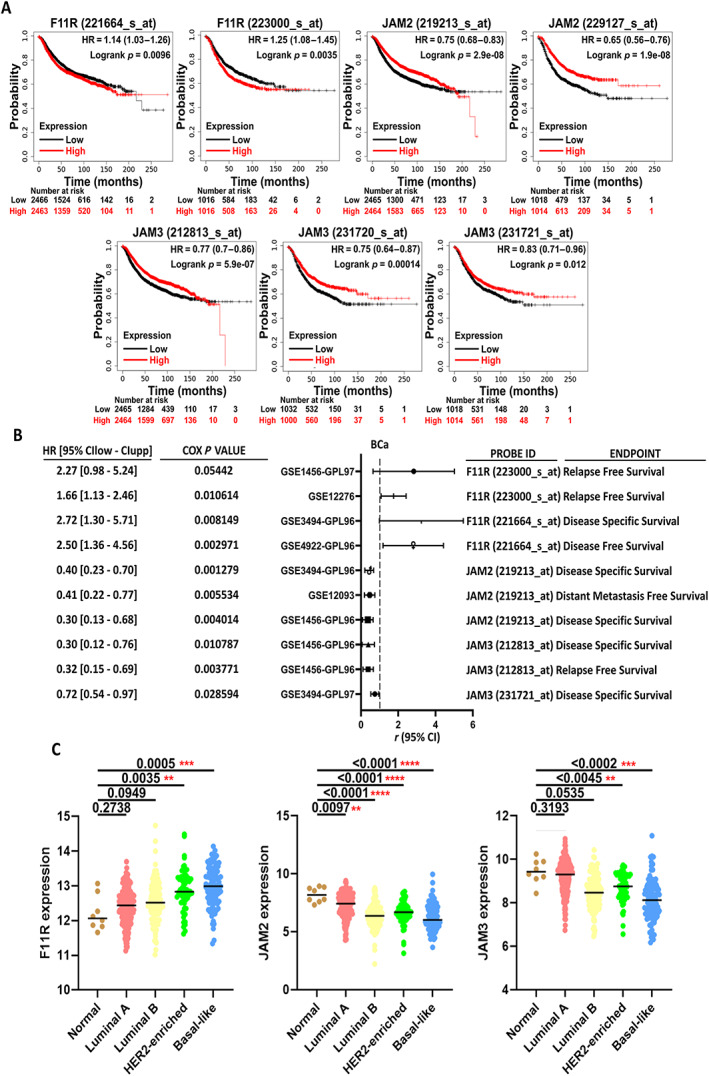
F11R and JAM‐2/JAM‐3 are inversely related to BCa prognosis. (A) Kaplan–Meier survival curves for JAM members in BCa with recurrence‐free survival. Data were analysed using the Kaplan–Meier plotter website. (B) HRs for JAM members with different BCa prognosis features under different clinical scenarios. The data were obtained from the PrognoScan website. (C) Distribution of JAM members in the different types of BCa. The data were analysed using the XENA website (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0001).
We also examined the correlation between the JAM family and BCa subtyping. The expression levels of F11R in TCGA‐BCa datasets were analysed in accordance with the subtype defined by the 50‐gene molecular signature (prediction analysis of microarray 50 [PAM50]) [37] (Figure 2C and supplementary material, Table S2). Expression of F11R was significantly higher in patients with HER2‐enriched and basal‐like subtypes of BCa. The opposite trends were observed for JAM‐2 and JAM‐3. Further analysis revealed no correlation between the expression levels of F11R and JAM‐2/JAM‐3, whereas the expression levels of JAM2 and JAM3 were highly correlated in BCa (supplementary material, Figure S2). These results indicate that F11R is unique and has clinical prognostic and subtype classification value in the diagnosis of BCa.
BCa tissues with high F11R expression are significantly associated with TNBC subtype, and tumour recurrence is associated with high expression levels of F11R
Because BCa has a heterogeneous composition, the statistical correlation between protein expression levels and diagnosis results or pathological features may vary depending on the composition of the cohort. Consequently, we used BCa cohorts from two different sources to compare the relationship between F11R protein levels and prognosis in the BCa subtype classification.
Of the 240 patients in the TMU cohort, 222 had complete records of ER/PR/HER2 status. In total, 130 luminal (58%), 55 HER2‐enriched (25%), and 37 TNBC (17%) cases were identified. The majority (72%, 164 of 227) of patients had early‐stage (I–II) tumours, while 27% (63 of 227) had late‐stage (III‐IV) tumours. Half (51%, 113 of 222) of the patients had lymph node involvement, and 18% (44 of 245) developed tumour recurrences or distant metastases following treatment. The KVGH cohort included 302 patients; ER/PR/HER2 information was available for 282. The 282 patients comprised 181 luminal‐type cases (65%), 52 HER2‐enriched cases (18%), and 49 TNBC cases (17%). Of these, 67% of the patients (201 of 302) had early‐stage tumours (I–II) and 33% (101 of 302) had late‐stage tumours (III–IV). Over half (57%, 174 of 302) the patients had lymph node involvement, and 41% (125 of 302) developed tumour recurrences or distant metastases after treatment.
The expression of F11R in TMA samples containing primary BCa samples from the two cohorts was determined using IHC staining. A low level of F11R expression was observed in normal mammary epithelium (Figure 3A). Cancer tissues varied in their expression of F11R from no staining to strong membranous staining (Figure 3A). The intensity and percentage of positively stained cells were used to divide the F11R expression into low and high groups as detailed in the ‘Materials and methods’ section. Table 1 summarises the associations between F11R expression and clinicopathological features. In the TMU and KVGH cohort, 73 (30%) and 114 (38%) cases with high F11R expression were identified, respectively. The F11R levels did not differ significantly with age, tumour stage, or lymph node status. In the TMU cohort, elevated levels of F11R were associated with increased tumour grade (p = 0.001). This association was not evident in the KVGH cohort (p = 0.078). High levels of F11R expression were associated with increased tumour size (T1 + T2 versus T3 + T4; p = 0.035) in the KVGH cohort, but not in the TMU cohort.
Figure 3.
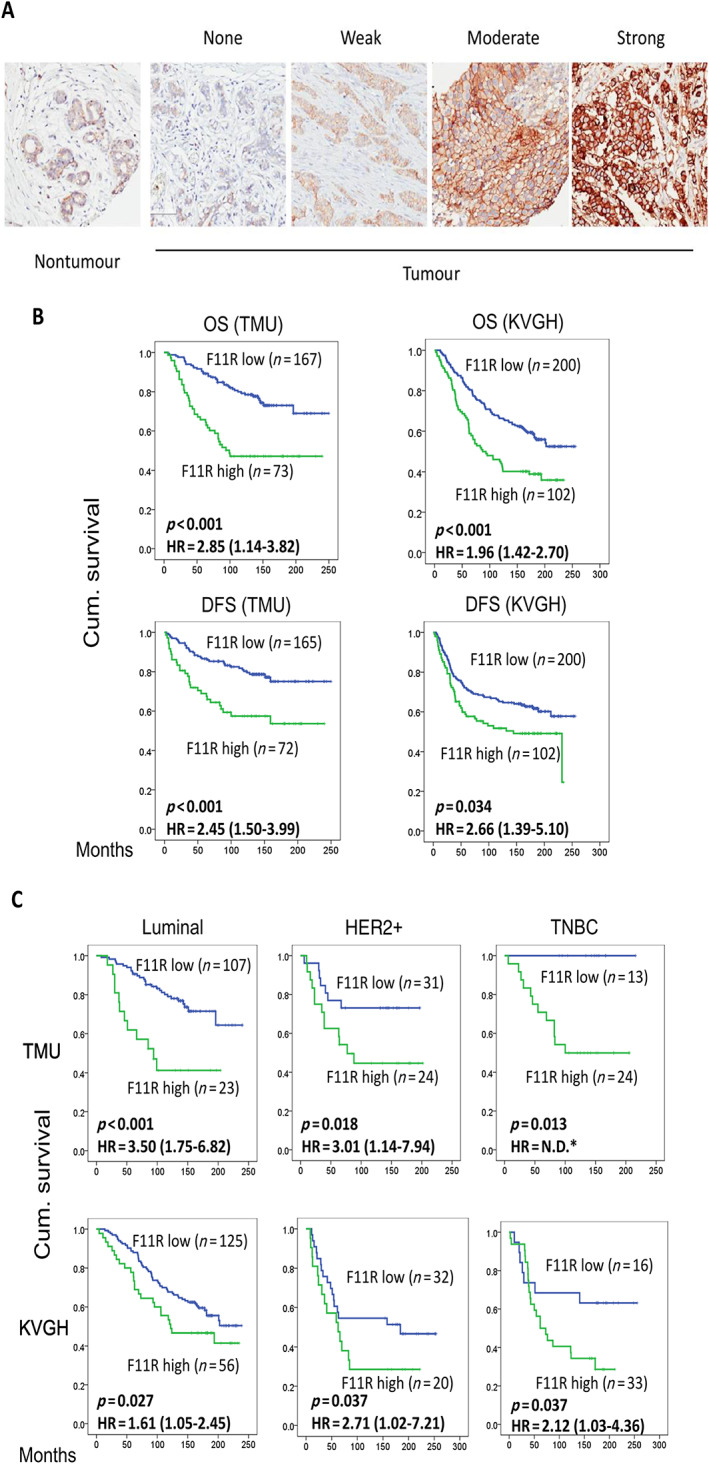
Relationship between F11R protein levels and prognosis in BCa subtype classification. (A) Grading of immunohistochemical staining for F11R between tumour and non‐tumour. (B) Correlation between expression of F11R and OS or DFS in different BCa tissues. (C) OS according to F11R and different BCa subtype classifications. *HR was not determined since all the cases in the low F11R group are censored.
Table 1.
Clinical and pathological characteristics and F11R expression in BCa patients
| Cohort | TMU | KVGH | ||||||
|---|---|---|---|---|---|---|---|---|
| F11R | F11R | |||||||
| Variables | N | Low (n = 167) | High (n = 73) | P value | N | Low (n = 188) | High (n = 114) | P value |
| Age | 240 | 0.565 | 302 | 0.683 | ||||
| <50 years | 73 (71.6) | 29 (28.4) | 87 (63.5) | 50 (36.5) | ||||
| >50 years | 94 (68.1) | 44 (31.9) | 101 (61.2) | 64 (38.8) | ||||
| Stage | 217 | 0.069 | 302 | 0.593 | ||||
| I + II | 112 (71.8) | 44 (28.2) | 123 (61.2) | 78 (38.8) | ||||
| III + IV | 36 (59.0) | 25 (41.0) | 65 (64.4) | 36 (35.6) | ||||
| Grade | 214 | 0.001 | 302 | 0.078 | ||||
| I | 24 (88.9) | 3 (11.1) | 8 (80.0) | 2 (20.0) | ||||
| II | 86 (74.8) | 29 (25.2) | 146 (64.6) | 80 (35.4) | ||||
| III | 39 (54.2) | 33 (54.8) | 34 (51.5) | 32 (48.5) | ||||
| Tumour size | 219 | 0.787 | 302 | 0.035 | ||||
| T1 + T2 | 124 (68.9) | 56 (31.1) | 173 (64.3) | 96 (35.7) | ||||
| T3 + T4 | 26 (66.7) | 13 (33.3) | 15 (45.5) | 18 (54.5) | ||||
| Lymph node status | 213 | 0.184 | 302 | 0.752 | ||||
| N0 | 76 (72.4) | 29 (27.6) | 81 (63.3) | 47 (36.7) | ||||
| N1–N3 | 69 (63.9) | 39 (36.1) | 107 (61.5) | 67 (38.5) | ||||
| ER status | 233 | <0.001 | 278 | <0.001 | ||||
| Negative (0–10%) | 47 (51.1) | 45 (48.9) | 51 (47.7) | 56 (52.3) | ||||
| Positive (>10%) | 115 (81.6) | 26 (18.4) | 120 (70.2) | 51 (29.8) | ||||
| PR status | 233 | <0.001 | 286 | 0.002 | ||||
| Negative (0–10%) | 80 (58.4) | 57 (41.6) | 67 (51.5) | 63 (48.5) | ||||
| Positive (>10%) | 82 (85.4) | 14 (14.6) | 108 (69.2) | 48 (30.8) | ||||
| HER2 status (I) | 222 | 0.060 | 293 | 0.497 | ||||
| 0–2+ | 125 (74%) | 51 (26%) | 137 (62.6) | 82 (37.4) | ||||
| 3+ | 26 (58%) | 20 (42%) | 43 (58.1) | 31 (41.9) | ||||
| Subtype | 222 | <0.001 | 282 | <0.001 | ||||
| Luminal A + B | 107 (82%) | 23 (18%) | 125 (69.1) | 56 (30.9) | ||||
| HER2 | 31 (56%) | 24 (44%) | 32 (61.5) | 20 (38.5) | ||||
| Triple‐negative | 13 (36%) | 24 (64%) | 16 (32.7) | 33 (67.3) | ||||
| Recurrence | 237 | 0.001 | 302 | 0.101 | ||||
| No | 130 (75.6) | 42 (24.4) | 117 (66.1) | 60 (33.9) | ||||
| Yes | 35 (53.8) | 30 (46.2) | 71 (56.8) | 54 (43.2) | ||||
Interestingly, the expression of F11R was inversely correlated with ER and PR expression (p < 0.002) in both cohorts. The percentage of high F11R expression was 26% in TMU patients with 0 to 2+ HER2 expression, but increased to 42% in patients with 3+ HER2 expression (Table 1), although no statistical significance was noted (p = 0.060). F11R status was determined in 209 cases of the TMU cohort. In both cohorts, the percentage of high F11R expression was low in luminal cancer (18% in TMU and 31% in KVGH) and increased to approximately 40% in HER2‐enriched tumours (44% in TMU and 39% in KVGH). High expression of F11R predominated in TNBC (64% in TMU and 67% in KVGH, p < 0.001). Tumour recurrence was significantly correlated with high expression of F11R (p = 0.001, Table 1; HR = 2.45 [1.50–3.99], Table 2) in the TMU cohort, but did not reach significance in the KVGH cohort (p = 0.01, Table 1; HR = 2.66 [1.39–5.10], Table 3).
Table 2.
OS and DFS of BCa patients in the TMU cohort by univariate and multivariate analyses
| Variables | Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| OS | DFS | OS | DFS | |||||
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | |
| Age | ||||||||
| <50 years | 1 | 1 | 1 | 1 | ||||
| ≥50 years | 2.23 (1.36–3.66) | 0.001 | 1.23 (0.75–2.02) | 0.417 | 1.76 (1.01–3.07) | 0.047 | 1.10 (0.62–1.95) | 0.757 |
| Tumour T status | ||||||||
| T1 + T2 | 1 | 1 | 1 | 1 | ||||
| T3 + T4 | 2.02 (1.22–3.37) | 0.007 | 2.51 (1.47–4.30) | 0.001 | 1.17 (0.55–2.48) | 0.691 | 1.01 (0.47–2.17) | 0.980 |
| Lymph node metastasis | ||||||||
| No | 1 | 1 | 1 | 1 | ||||
| Yes | 2.22 (1.39–3.55) | 0.001 | 3.77 (2.10–6.76) | <0.001 | 1.87 (0.94–3.73) | 0.077 | 2.89 (1.27–6.54) | 0.011 |
| AJCC stage | ||||||||
| I + II | 1 | 1 | 1 | 1 | ||||
| III + IV | 1.67 (1.33–2.10) | <0.001 | 2.00 (1.56–2.56) | <0.0.001 | 1.41 (0.98–2.03) | 0.068 | 1.43 (0.96–2.12) | 0.078 |
| ER status | ||||||||
| Negative (0–10%) | 1 | 1 | 1 | 1 | ||||
| Positive (>10%) | 0.81 (0.51–1.27) | 0.350 | 1.00 (0.60–1.64) | 0.938 | 1.09 (0.54–2.20) | 0.803 | 1.20 (0.55–2.65) | 0.648 |
| PR status | ||||||||
| Negative (0–10%) | 1 | 1 | 1 | 1 | ||||
| Positive (>10%) | 0.70 (0.44–1.12) | 0.137 | 0.76 (0.46–1.26) | 0.293 | 0.80 (0.40–1.62) | 0.537 | 0.83 (0.38–1.79) | 0.627 |
| HER2 status | ||||||||
| 0–2+ | 1 | 1 | 1 | 1 | ||||
| 3+ | 1.85 (1.09–3.12) | 0.022 | 2.07 (1.19–3.60) | 0.010 | 1.06 (0.55–2.03) | 0.860 | 1.25 (0.61–2.55) | 0.538 |
| F11R | ||||||||
| Low | 1 | 1 | 1 | 1 | ||||
| High | 2.85 (1.14–3.82) | <0.001 | 2.45 (1.50–3.99) | <0.001 | 3.32 (1.90–5.80) | <0.001 | 2.69 (1.43–5.05) | 0.002 |
AJCC, American Joint Committee on Cancer, eight edition.
Table 3.
Univariate and multivariate analyses of BCa patients in the KVGH cohort
| Variables | Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| OS | DFS | OS | DFS | |||||
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P alue | |
| Age | ||||||||
| <50 years | 1 | 1 | 1 | 1 | ||||
| ≥50 years | 1.60 (1.15–2.22) | 0.005 | 1.31 (0.75–2.78) | 0.349 | 1.70 (1.20–2.41) | 0.003 | 1.26 (0.87–1.83) | 0.226 |
| Tumour T status | ||||||||
| T1 + T2 | 1 | 1 | 1 | 1 | ||||
| T3 + T4 | 2.58 (1.67–4.00) | <0.001 | 2.03 (1.14–3.60) | 0.015 | 1.40 (0.86–2.29) | 0.172 | 0.99 (0.56–1.74) | 0.974 |
| Lymph node metastasis | ||||||||
| No | 1 | 1 | 1 | 1 | ||||
| Yes | 2.42 (1.70–3.45) | <0.001 | 1.93 (1.10–3.38) | 0.023 | 1.40 (0.88–2.24) | 0.195 | 2.02 (1.18–3.45) | 0.010 |
| AJCC stage | ||||||||
| I + II | 1 | 1 | 1 | 1 | ||||
| III + IV | 2.65 (1.92–3.66) | <0.001 | 1.55 (0.89–2.71) | 0.121 | 2.09 (1.33–3.29) | 0.001 | 2.30 (1.43–3.70) | 0.001 |
| ER status | ||||||||
| Negative (0–10%) | 1 | 1 | 1 | 1 | ||||
| Positive (>10%) | 0.64 (0.46–0.89) | 0.007 | 1.68 (0.95–2.99) | 0.077 | 0.64 (0.37–1.10) | 0.103 | 0.56 (0.29–1.08) | 0.083 |
| PR status | ||||||||
| Negative (0–10%) | 1 | 1 | 1 | 1 | ||||
| Positive (>10%) | 0.75 (0.54–1.04) | 0.081 | 1.31 (0.73–2.34) | 0.362 | 1.32 (0.77–2.27) | 0.308 | 1.50 (0.79–2.87) | 0.219 |
| HER2 status | ||||||||
| 0–2+ | 1 | 1 | 1 | 1 | ||||
| 3+ | 1.53 (1.07–2.18) | 0.019 | 2.66 (1.39–5.10) | 0.003 | 1.12 (0.75–1.66) | 0.579 | 1.05 (0.68–1.63) | 0.824 |
| F11R | ||||||||
| Low | 1 | 1 | 1 | 1 | ||||
| High | 1.96 (1.42–2.70) | <0.001 | 2.66 (1.39–5.10) | 0.003 | 1.92 (1.37–2.68) | <0.001 | 1.39 (0.95–2.02) | 0.089 |
AJCC, American Joint Committee on Cancer, eighth edition.
F11R in BCa reflects poor prognosis, tumour recurrence, and is independently prognostic
The prognostic value of F11R expression was evaluated using Kaplan–Meier analysis and log‐rank test for BCa. In both the TMU and KVGH cohorts, patients with high expression of F11R had a shorter OS (p < 0.001, HR = 2.85 [1.14–3.82] and 1.96 [1.42–2.70], respectively) and shorter DFS (p < 0.001, HR = 2.45 [1.50–3.99] in TMU and p = 0.034, HR = 2.66 [1.39–5.10] in KVGH) than those displaying lower expression (Figure 3B). Since the expression of F11R is associated with BCa subtypes (Table 1), further analysis by stratifying the patients by BCa subtypes (luminal, HER2‐enriched, and TNBC) revealed high expression of F11R as an adverse prognostic indicator for all three subtypes (Figure 3C; p < 0.05). Even though there were many fewer cases with high F11R expression in luminal‐type BCa than with low F11R expression, elevated expression of F11R was associated with a shorter OS (Figure 3C). In the TNBC group of the TMU cohort, patients with low F11R expression did not survive the follow‐up period. Overall, F11R expression was a useful prognostic factor for individual subtypes of BCa in multiple cohorts.
Cox proportional hazards regression analysis was performed to determine the prognostic factors associated with BCa. When univariate analyses of both cohorts were performed for OS, age, tumour T status, lymph node status, tumour stage, HER2, and the expression of F11R were significantly associated with poor outcomes (p < 0.05; Tables 2 and 3). The expression of F11R was a significant prognostic marker for multivariate analysis, in addition to age (for both cohorts) and tumour stage (for the KVGH cohort; Tables 2 and 3). In the multivariate analysis of the TMU cohort, the risk of mortality was 3.32‐fold higher for those with high F11R expression (95% confidence interval [CI] 1.90–5.80; p < 0.001). For univariate DFS analysis, T status, lymph node status, tumour stage, HER2 status, and F11R expression were associated with a shorter DFS (p < 0.05). A multivariate analysis of the TMU cohort showed that lymph node metastasis and F11R expression were associated with short DFS (p < 0.05). Univariate DFS analysis revealed 2.45‐fold (95% CI 1.50–3.99; p < 0.001) and 2.66‐fold (95% CI 1.39–5.10; p = 0.003) increases in cancer recurrence when F11R was overexpressed (95% CI 1.50–3.99; p < 0.001), as measured by multivariate analysis in the TMU and KVGH cohort, respectively. Also, in the multivariate analysis, the number of recurrences increased by 2.69‐fold (95% CI 1.43–5.05; p = 0.002) in the TMU cohort. The findings suggest that F11R expression is one of the most crucial biomarkers for poor outcomes (endpoint OS) in patients with BCa as well as for poor RFS.
Involvement of F11R in molecular mechanisms of TNBC
Considering that the pathological features suggest a major role for F11R in metastasis (Tables 1 and 2), it is still unclear how F11R functions in TNBC. Thus, using microarrays, we attempted to elucidate the possible mechanism of F11R‐mediated BCa progression. Using the CCLE database (supplementary material Table S3), we first compared F11R expression levels in BCa‐related cell lines. Using cell lines that express low levels of F11R, stable clones overexpressing F11R, such as CAL‐120_F11R (134.35‐fold) and HDQ‐P1_F11R (127.61‐fold), were established in breast carcinomas and breast ductal carcinoma, respectively (supplementary material, Table S4). Notably, CAL‐120 and HDQ‐P1 are classified as TNBC subtypes, with HDQ‐P1 being basal‐like and CAL‐120 being mesenchymal‐like [38]. The microarray and Venn diagram results showed that approximately 24,673 probes were consistently regulated by F11R in CAL‐120 and HDQ‐P1 cells (Figure 4A and supplementary material, Table S4). Furthermore, by incorporating these co‐regulated genes into IPA, we were able to profile the molecular interaction network (Figure 4B) and signalling pathways (Figure 4C and supplementary material, Table S5) involved in F11R regulation. Of note, these gene ontologies include cell growth and movement‐related molecular networks. In particular, F11R may mediate cell migration by regulating epithelial adhesion junction signalling and epithelial–mesenchymal transition (EMT) through growth factors (Figure 4C). F11R was also associated with HER2‐signalling in BCa, in accordance with previous observations that the expression of F11R regulates HER2 transcription activation via FOXA1 and HER2 protein stability to increase BCa malignancy [30, 39, 40]. However, a clear understanding of the crosstalk and mechanisms involved in F11R and HER2 signalling between different subtypes of cancer is still lacking. In accordance with clinical findings, the results of the above microarray analysis implicate F11R in cellular EMT changes that contribute to migration, and indicate that metastasis may be influenced by these signalling pathways.
Figure 4.
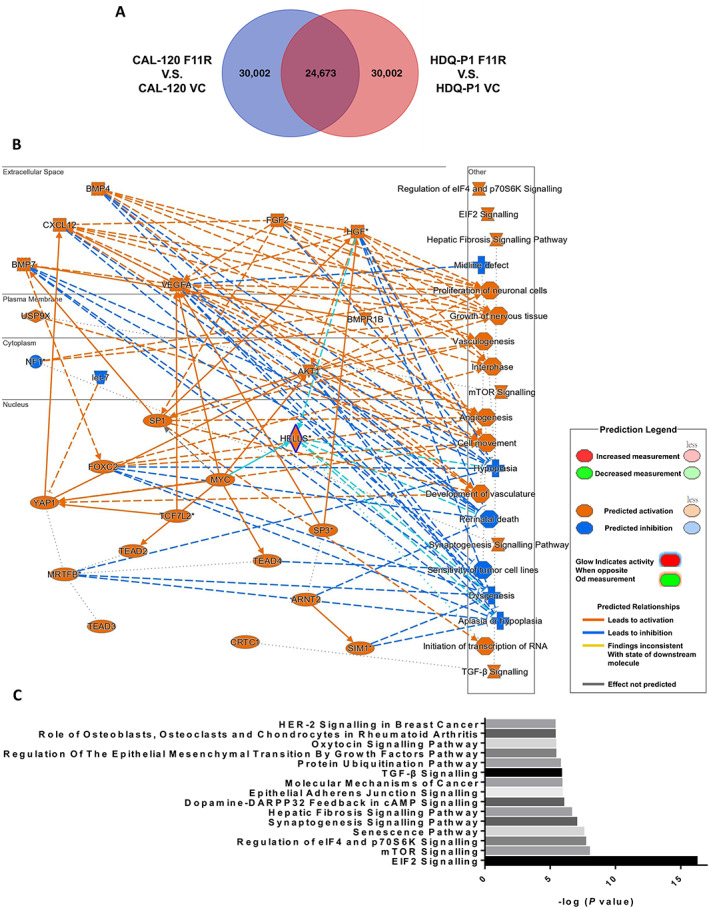
Prediction of molecular biological events using F11R‐based transcriptomics in TNBC cells. (A) Venn diagram of significant changes in targets following microarray analysis of the F11R overexpression model in CAL120 and HDQ‐P1 cells. (B) Networks identified by IPA of F11R‐overexpressing cell models of BCa cells. (C) Potential signalling pathways predicted using IPA that may be affected by F11R overexpression. Data were analysed using IPA software.
Relationship between F11R and the EP300 axis in development of TNBC metastasis
To further identify the most crucial genes that F11R primarily regulates to influence cell movement during EMT, supplementary material, Figure S3 highlights key genes involved in the regulation of EMT by growth factor pathways that are impacted by F11R. A number of these genes showed positive correlations with F11R in BCa by >±0.3 Spearman's correlation. They included GSK3B, MAPK1, SMAD4, PTPN11, OCLN, CDC42, MTOR, TGFBR1, RHOA, and IL6R (Figure 5A). Expression levels of these genes in CAL‐120 and HDQ‐P1 cells were elevated when F11R was overexpressed. The 10 genes chosen as signature molecules also exhibited a positive correlation with F11R (Figure 5B). Analysis of the downstream regulators showed that EP300 was the most regulated gene among all the transcription factors, with a Spearman's correlation >±0.3 (supplementary material, Table S6). In spite of SMAD4, KLF11, and YAP1 having high Spearman's correlation, EP300 had a higher P value than the others. Among those regulated by EP300, EMT‐related genes included GSK3B, RHOA, and TGFBR1 (Figure 5C). EP300 was highly correlated with F11R, GSK3B, RHOA, and TGFBR1 in BRACA (Figure 5D). Additionally, CCLE dataset analysis indicated that F11R and EP300 were also highly correlated in breast carcinoma or breast ductal carcinoma cells carcinoma or breast ductal carcinoma cells (supplementary material, Figure S4 and Table S7). In BCa, a clinical correlation indicated that patients with F11R and downstream effectors of EP300 had a worse prognosis, particularly with downstream effectors, such as GSK3B, RHOA, and TGFB1 (supplementary material, Figure S5).
Figure 5.
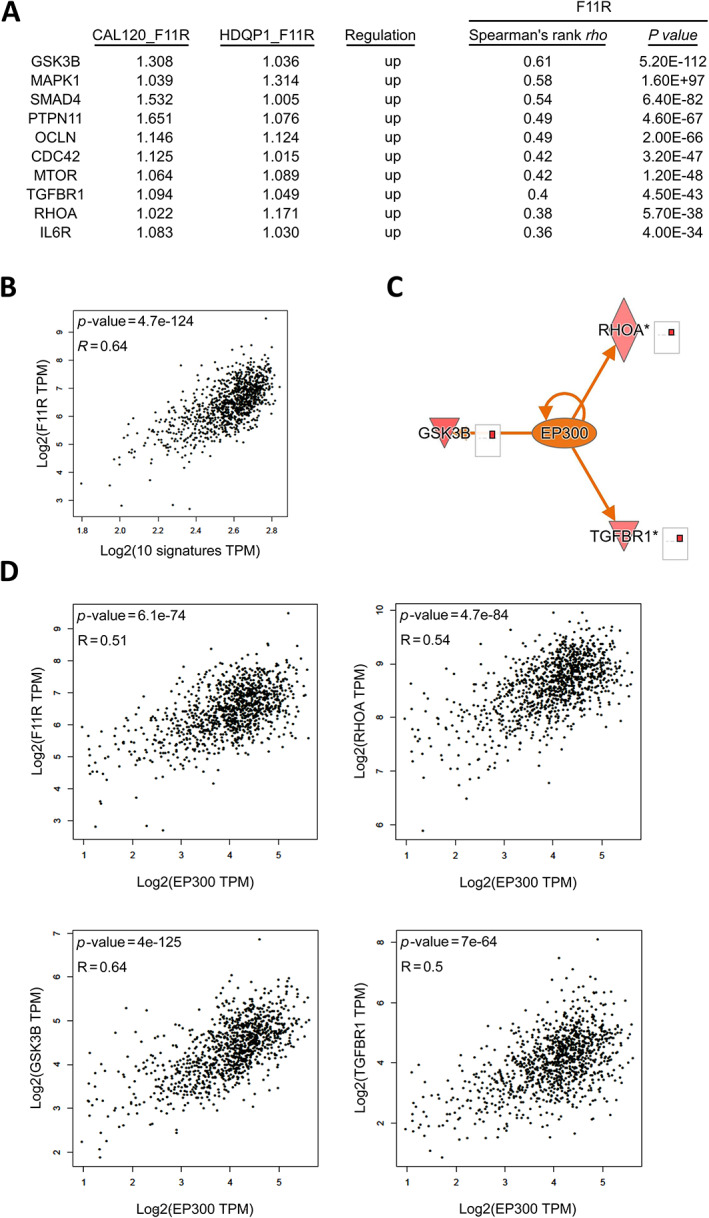
The F11R/EP300 axis is involved in the regulation of cell motility in TNBC. (A) Correlation of selected EMT‐related molecules with F11R in BCa. (B) EMT‐selected molecules and F11R gene as a signature of BCa. (C) Upstream effectors of EP300 after the overexpression of F11R. (D) Relationship between EP300 and downstream effectors in BCa. Data were analysed using IPA software.
EP300 plays an oncogenic role in BCa malignancy and may also contribute to drug resistance [38], which may be related to F11R regulation. To identify potential treatments based on the F11R genetic profile, drug repurposing analysis was performed (Figure 6 and supplementary material, Table S8). Among these candidates, Tyrphostin AG 1478, a tyrosine kinase inhibitor of epidermal growth factor receptor, may act as a downstream effector of TGFB1 when inhibiting F11R. Therefore, targeting F11R‐related genes using Tyrphostin AG 1478 may result in improved TNBC treatment. Accordingly, F11R may play a role in the regulation of the EMT by the growth factor pathway via EP300, including GSK3B, RHOA, and TGFBR1, consequently contributing to the metastasis of BCa.
Figure 6.
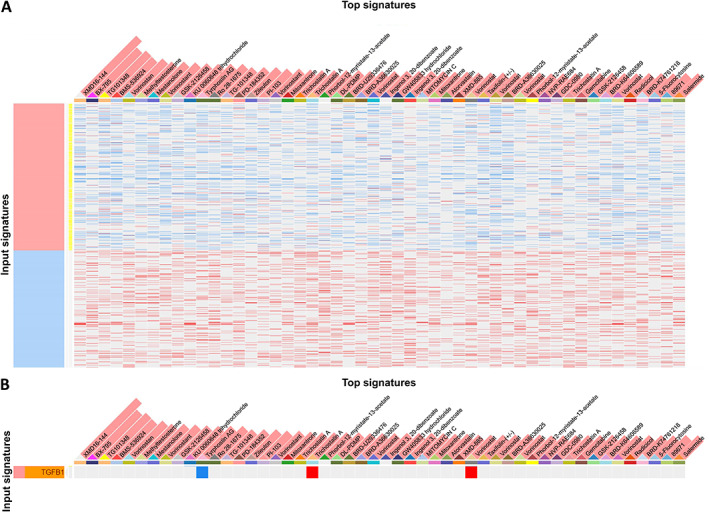
Simulation of drug repurposing based on F11R genetic profiles. (A) Illustration of the correlation between gene expression and drug candidates in a heatmap. (B) Tyrphostin AG 1478 is a potential candidate to inhibit TGFB1. Data were analysed using the L1000CDs website.
Discussion
Many biomarkers used to categorise specific cancers have proven beneficial for understanding cancer and for developing clinical diagnostic methods that allow for more effective treatment. Based on the use of markers such as ER, PR, and HER2, BCa is currently classified as luminal, HER2‐enriched, and TNBC. However, the molecular regulation of these subtypes is not yet fully understood. Therefore, identifying specific markers to aid diagnosis and treatment is extremely important.
In this study, a pan‐cancer approach was used to analyse the expression levels of the JAM family. Several types of cancer express the F11R gene in combination with the other members of the JAM family (Figure 2A), which has also been previously reported [26, 27]. Notably, we confirmed that F11R expression was elevated in BCa and was unique among other members of the JAM family in terms of its associated prognostic characteristics. F11R has been related to many clinical outcomes associated with BCa [26, 27, 41]. Intriguingly, the results indicated that JAM‐2 and JAM‐3 are inversely related to F11R in terms of OS. Comparing pan‐cancer analysis, BLCA, BCa, CESC, LUSC, and UCEC showed similar trends (supplementary material, Figure S1A). In particular, such a relationship was positively correlated with OS, and a poor prognosis was associated with the F11R gene in patients (supplementary material, Figure S1B). TCGA‐BCa data also indicated that, in normal tissues, F11R exhibited a lower distribution than JAM‐2 or JAM‐3 (Figure 1B). Comparing the differences between normal and tumour populations also revealed that, in contrast to JAM‐2 or JAM‐3, F11R was expressed to a significantly higher level in the tumour group than in the normal group (Figure 1C). Accordingly, the difference between normal and tumour can explain why only F11R is associated with poor recurrence‐free survival of BCa, whereas JAM‐2 or JAM‐3 is not (Figure 2A). A review of different clinical data also indicated a similar situation, suggesting that high expression of F11R results in a low RFS, DSS, or DFS of patients (Figure 2A,B), while JAM‐2/JAM‐3 is associated with low risk. However, it should be noted that the relationship between JAM family members and BCa subtypes has not been systematically evaluated. In this study, a series of comparisons was made between different BCa subtypes based on the expression levels of JAM genes. Both in silico and IHC results showed that F11R was significantly increased in luminal, HER2‐enriched and basal‐like subtypes (Figure 2C), demonstrating that F11R is associated with OS with luminal, HER2, and TNBC (Figure 3C). In contrast to F11R, patients with any BCa subtype showed lower levels of JAM‐2/JAM‐3 expression. Similar trends were observed in patients with HER2‐enriched or basal‐like tumours. However, no correlation was evident between the distributions of F11R and JAM‐2/JAM‐3 among the inverse indicators (supplementary material, Figure S2). It is noteworthy that there was a strong correlation between JAM‐2 and JAM‐3 (R = 0.8). However, no studies have demonstrated a possible relationship or biological function between JAM‐2 and JAM‐3.
Clinically, we evaluated the expression of F11R protein and its correlation with the clinicopathological characteristics and prognosis of patients with BCa in two independent cohorts (TMU and KVGH). Both cohorts showed similar OS and DFS trends when categorised by F11R expression (Figure 3B). The survival trends in the three subtypes of BCa were also similar (Figure 3C). The correlation of F11R with pathological characteristics revealed a similar tendency in ER/PR status and subtypes, but showed variations in grade/tumour size and recurrence between these two cohorts (Table 1). In the TMU cohort, F11R was associated with OS and DFS in both univariate and multivariate analyses (Table 2). In the KVGH cohort, the same results were observed, except for DFS, in multivariate analysis (p = 0.089, Table 3). The TMU cohort was collected in later years (from 1998 to 2008) from northern Taiwan, while the KVGH cohort was collected earlier (1991–1999) from southern Taiwan. Perhaps due to advances in BCa treatment (surgery and/or adjuvant therapies) with time, these two cohorts presented the aforementioned variations when analysed in this study. A difference in the survival profile of these two cohorts can be seen in Figure 3B,C. The TMU cohort had better OS than the KVGH cohort. Based on these observations, we decided not to combine them in a single analysis. Instead, we presented them individually to show that while most of the characteristics are similar in the F11R analysis, they still have certain uniqueness.
TNBC is one of the most difficult subtypes to treat, with metastasis as one of the main reasons. Considering that there is no detailed analysis to profile how F11R functions in TNBC cells, we simulated the possible molecular mechanism between F11R and TNBC cells through microarray profiling analysis. Gene ontology analysis revealed that cells overexpressing F11R activated genes related to cell growth and motility (Figure 4B). Analysis of canonical pathways also revealed the major signalling mechanisms involved in F11R (Figure 4C). Based on the findings of this study, it is evident that F11R plays a role in several processes related to cell motility, including tight junction signalling, regulation of EMT by growth factors, HER2 signalling in BCa, and ER signalling, which are associated with BCa tumorigenesis. This is consistent with recent studies suggesting that HER2‐induced resistance to treatment is mediated by HER3 and F11R in luminal and basal‐like BCa cell lines [41]. Overexpression of F11R can also be observed in similar cells, which regulates integrin signalling and influences cell motility [42]. Our observations (Figure 4C) are consistent with these prior results. Here, we demonstrate that F11R can exhibit consistent signalling regulation in both basal‐like and mesenchymal‐like TNBC cells. Thus, F11R appears to be a therapeutic target that may be cross‐subtype applicable, in accordance with IHC results.
F11R is involved in EMT‐related signalling in cells, which is consistent with the features observed in pathological studies. At the molecular level, we observed that EMT was regulated by growth factors via the F11R signalling pathway (supplementary material, Figure S3). Molecules highly correlated with F11R included GSK3B, MAPK1, SMAD4, PTPN11, OCLN, CDC42, MTQR, TGFBR1, RHOA, and IL6R (Figure 5A). Several of these molecules were highly correlated with F11R in BCa (Figure 5B) and have previously been implicated in BCa progression [43, 44, 45, 46]. These molecules may serve as cluster signatures and are highly correlated with F11R in BCa. With a more detailed analysis of upstream regulators, we identified EP300 as a novel factor involved in F11R regulation (Figure 5C). EP300 expression has been consistently linked to a wide range of biological processes, including cell growth, metastasis, and stemness properties of TNBC [47]. IPA indicated that EP300 regulates RHOA, GSK3B, and TGFBR1. Analysis of the correlations revealed a strong correlation between EP300 and F11R, RHOA, GSK3B, and TGFBR1 (Figure 5D). Similar correlations were observed among the different subtypes of BCa cell lines (supplementary material, Figure S4). A number of studies have demonstrated the connection between RHOA, GSK3B, and TGFBR1 and the development of metastatic disease in BCa [43, 44, 45, 46]. The findings suggest that crosstalk between F11R and TGFRB1 signalling may also be mediated by EP300. The precise mechanism still needs to be determined.
Recently, inhibitors of F11R have been identified. A soluble antagonistic peptide 4D specific for the F11R/JAM‐A system was developed to inhibit the activity of thymosin beta 4 and transforming growth factor‐beta‐mediated F11R tight junctions in BCa microenvironments [48]. An F11R expression‐based screening of natural products identified the antibiotic tetrocarcin‐A as effective at inhibiting F11R expression to modulate tumour growth in BCa [49]. However, to date, there has been no comprehensive evaluation of the efficacy of different subtypes of BCa. It remains to be determined whether antagonistic peptides or natural products targeting F11R can inhibit the F11R‐E300 axis in TNBC models.
The collective findings of the present study indicate the feasibility of using expression levels of F11R in patient specimens in conjunction with specific biomarkers of PAM50 for the prognosis and assessment of BCa subtypes. Moreover, as the interpretation between the molecular networks demonstrated, F11R participates in BCa progression through molecular mechanisms and can be used as a potential target for future drug design in BCa therapy.
Author contributions statement
CLC, MH, CHL, CYF and YCC conceived and designed the study. CHL and CYF developed the methodology. CLC, PJL, LPG, JSC and MH acquired data (acquired and managed patients, provided facilities, etc.). CHL, CYF and YCC analysed and interpreted data (e.g. statistical analysis, biostatistics, computational analysis). CHL and CYF drafted the manuscript. All authors critically revised the manuscript for important intellectual content. CLC, YCC and MH supervised the study. All authors read and approved the final manuscript.
Ethical approval and consent to participate
In the present study, tissue samples were collected and reviewed by the Institutional Review Boards at Kaohsiung Veterans General Hospital (VGHKS12‐CT9‐07; Kaohsiung, Taiwan) and Wan Fang Hospital of Taipei Medical University (WFH‐IRB‐99049; Taipei, Taiwan). Before any tissue samples were collected during the planned treatment, all patients gave their informed consent. In this study, all procedures were performed in accordance with the ethical standards of the institutional research committee and/or the national research committee and with the 1964 Helsinki Declaration, and later amendments, or comparable ethical standards.
Supporting information
Figure S1. Countertrends in F11R and JAM members with respect to cancer prognosis
Figure S2. Correlation between members of JAM and BCa
Figure S3. F11R overexpression activates EMT signalling in BCa
Figure S4. Correlation between F11R and EP300 in BCa cells
Figure S5. Overall survival prognosis correlation between F11R and EP300 downstream effectors in BCa
Table S1. Expression of the JAM genes in TCGA‐BCa patients
Table S2. Expression of the JAM family in patients
Table S3. Relative expression levels of F11R in BCa cells
Table S4. Microarray values of overexpressed F11R in BCa cells
Table S5. Impact of F11R on signalling hallmarks
Table S6. Regulators of EMT signalling involved in F11R
Table S7. Relative expression levels of F11R and EP300 in BCa cells
Table S8. Potential candidates derived from drug repurposing simulations based on F11R expression profiles
Acknowledgements
For the Affymetrix microarray and Aperio digital pathology analyses, we would like to express our sincere appreciation to GRC Instrument Core Facilities.
This study was supported by Ministry of Science and Technology (MOST‐110‐2320‐B‐010‐008‐MY2), Yen Tjing Ling Medical Foundation (CI‐111‐9) and Veterans General Hospitals and University System of Taiwan Joint Research Program (VGHUST111‐G3‐3‐2) to CYC and Ministry of Science and Technology (MOST‐110‐2320‐B‐038‐074) to CLC.
No conflicts of interest were declared.
Contributor Information
Yu‐Chan Chang, Email: jameskobe0@gmail.com.
Chi‐Long Chen, Email: chencl@tmu.edu.tw.
Michael Hsiao, Email: mhsiao@gate.sincia.edu.tw.
Data availability statement
Detailed data are included in the manuscript and its supplementary information files.
References
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021; 71: 209–249. [DOI] [PubMed] [Google Scholar]
- 2. Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature 2000; 406: 747–752. [DOI] [PubMed] [Google Scholar]
- 3. Goldhirsch A, Wood WC, Coates AS, et al. Strategies for subtypes – dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol 2011; 22: 1736–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harbeck N, Penault‐Llorca F, Cortes J, et al. Breast cancer. Nat Rev Dis Primers 2019; 5: 66. [DOI] [PubMed] [Google Scholar]
- 5. Kavarthapu R, Anbazhagan R, Dufau ML, et al. Crosstalk between PRLR and EGFR/HER2 signaling pathways in breast cancer. Cancer 2021; 13: 4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001; 98: 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eroles P, Bosch A, Pérez‐Fidalgo JA, et al. Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat Rev 2012; 38: 698–707. [DOI] [PubMed] [Google Scholar]
- 8. Sweeney C, Bernard PS, Factor RE, et al. Intrinsic subtypes from PAM50 gene expression assay in a population‐based breast cancer cohort: differences by age, race, and tumor characteristics. Cancer Epidemiol Biomarkers Prev 2014; 23: 714–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Verma S, Miles D, Gianni L, et al. Trastuzumab emtansine for HER2‐positive advanced breast cancer. N Engl J Med 2012; 367: 1783–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bianchini G, Balko JM, Mayer IA, et al. Triple‐negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 2016; 13: 674–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang TW, DeMarco RA, Mrsny RJ, et al. Characterization of huJAM: evidence for involvement in cell–cell contact and tight junction regulation. Am J Physiol Cell Physiol 2000; 279: C1733–C1743. [DOI] [PubMed] [Google Scholar]
- 12. Mandell KJ, Babbin BA, Nusrat A, et al. Junctional adhesion molecule 1 regulates epithelial cell morphology through effects on beta 1 Integrins and Rap1 activity. J Biol Chem 2005; 280: 11665–11674. [DOI] [PubMed] [Google Scholar]
- 13. Sobocka MB, Sobocki T, Banerjee P, et al. Cloning of the human platelet F11 receptor: a cell adhesion molecule member of the immunoglobulin superfamily involved in platelet aggregation. Blood 2000; 95: 2600–2609. [PubMed] [Google Scholar]
- 14. Ostermann G, Weber KSC, Zernecke A, et al. JAM‐1 is a ligand of the beta(2) integrin LFA‐1 involved in transendothelial migration of leukocytes. Nat Immunol 2002; 3: 151–158. [DOI] [PubMed] [Google Scholar]
- 15. Naik UP, Ehrlich YH, Kornecki E. Mechanisms of platelet activation by a stimulatory antibody – cross‐linking of a novel platelet receptor for monoclonal‐antibody F11 with the Fc‐gamma‐RII receptor. Biochem J 1995; 310: 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kornecki E, Walkowiak B, Naik UP, et al. Activation of human platelets by a stimulatory monoclonal antibody. J Biol Chem 1990; 265: 10042–10048. [PubMed] [Google Scholar]
- 17. Ebnet K. Junctional adhesion molecules (JAMs): cell adhesion receptors with pleiotropic functions in cell physiology and development. Physiol Rev 2017; 97: 1529–1554. [DOI] [PubMed] [Google Scholar]
- 18. Laukoetter MG, Nava P, Lee WY, et al. JAM‐A regulates permeability and inflammation in the intestine in vivo. J Exp Med 2007; 204: 3067–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vetrano S, Rescigno M, Cera MR, et al. Unique role of junctional adhesion molecule‐A in maintaining mucosal homeostasis in inflammatory bowel disease. Gastroenterology 2008; 135: 173–184. [DOI] [PubMed] [Google Scholar]
- 20. Magara K, Takasawa A, Osanai M, et al. Elevated expression of JAM‐A promotes neoplastic properties of lung adenocarcinoma. Cancer Sci 2017; 108: 2306–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kakuki T, Kurose M, Takano K, et al. Dysregulation of junctional adhesion molecule‐A via p63/GATA‐3 in head and neck squamous cell carcinoma. Oncotarget 2016; 7: 33887–33900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gutwein P, Schramme A, Voss B, et al. Downregulation of junctional adhesion molecule‐A is involved in the progression of clear cell renal cell carcinoma. Biochem Biophys Res Commun 2009; 380: 387–391. [DOI] [PubMed] [Google Scholar]
- 23. Huang JY, Xu YY, Sun Z, et al. Low junctional adhesion molecule A expression correlates with poor prognosis in gastric cancer. J Surg Res 2014; 192: 494–502. [DOI] [PubMed] [Google Scholar]
- 24. Fong D, Spizzo G, Mitterer M, et al. Low expression of junctional adhesion molecule A is associated with metastasis and poor survival in pancreatic cancer. Ann Surg Oncol 2012; 19: 4330–4336. [DOI] [PubMed] [Google Scholar]
- 25. Naik MU, Naik TU, Suckow AT, et al. Attenuation of junctional adhesion molecule‐A is a contributing factor for breast cancer cell invasion. Cancer Res 2008; 68: 2194–2203. [DOI] [PubMed] [Google Scholar]
- 26. McSherry EA, McGee SF, Jirstrom K, et al. JAM‐A expression positively correlates with poor prognosis in breast cancer patients. Int J Cancer 2009; 125: 1343–1351. [DOI] [PubMed] [Google Scholar]
- 27. Murakami M, Giampietro C, Giannotta M, et al. Abrogation of junctional adhesion molecule‐A expression induces cell apoptosis and reduces breast cancer progression. PLoS One 2011; 6: e21242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brennan K, McSherry EA, Hudson L, et al. Junctional adhesion molecule‐A is co‐expressed with HER2 in breast tumors and acts as a novel regulator of HER2 protein degradation and signaling. Oncogene 2013; 32: 2799–2804. [DOI] [PubMed] [Google Scholar]
- 29. Li CH, Chan MH, Chang YC. The role of fructose 1,6‐bisphosphate‐mediated glycolysis/gluconeogenesis genes in cancer prognosis. Aging (Albany NY) 2022; 14: 3233–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor‐infiltrating immune cells. Cancer Res 2017; 77: e108–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lanczky A, Gyorffy B. Web‐based survival analysis tool tailored for medical research (KMplot): development and implementation. J Med Internet Res 2021; 23: e27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large‐scale expression profiling and interactive analysis. Nucleic Acids Res 2019; 47: W556–W560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mizuno H, Kitada K, Nakai K, et al. PrognoScan: a new database for meta‐analysis of the prognostic value of genes. BMC Med Genomics 2009; 2: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu J, Sanborn JZ, Benz S, et al. The UCSC cancer genomics browser. Nat Methods 2009; 6: 239–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fang CY, Lin YH, Chen CL. Overexpression of AKR1B10 predicts tumor recurrence and short survival in oral squamous cell carcinoma patients. J Oral Pathol Med 2019; 48: 712–719. [DOI] [PubMed] [Google Scholar]
- 36. Fang CY, Liew PL, Chen CL, et al. High HMGA2 expression correlates with reduced recurrence‐free survival and poor overall survival in oral squamous cell carcinoma. Anticancer Res 2017; 37: 1891–1899. [DOI] [PubMed] [Google Scholar]
- 37. Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009; 27: 1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Asaduzzaman M, Constantinou S, Min H, et al. Tumour suppressor EP300, a modulator of paclitaxel resistance and stemness, is downregulated in metaplastic breast cancer. Breast Cancer Res Treat 2017; 163: 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cruz RGB, Madden SF, Brennan K, et al. A transcriptional link between HER2, JAM‐A and FOXA1 in breast cancer. Cell 2022; 11: 735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leech AO, Vellanki SH, Rutherford EJ, et al. Cleavage of the extracellular domain of junctional adhesion molecule‐A is associated with resistance to anti‐HER2 therapies in breast cancer settings. Breast Cancer Res 2018; 20: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cruz RGB, Madden SF, Richards CE, et al. Human epidermal growth factor receptor‐3 expression is regulated at transcriptional level in breast cancer settings by junctional adhesion molecule‐a via a pathway involving beta‐catenin and FOXA1. Cancer 2021; 13: 871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McSherry EA, Brennan K, Hudson L, et al. Breast cancer cell migration is regulated through junctional adhesion molecule‐A‐mediated activation of Rap1 GTPase. Breast Cancer Res 2011; 13: R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu ZY, Chu SZ, Yao S, et al. CD74 interacts with CD44 and enhances tumorigenesis and metastasis via RHOA‐mediated cofilin phosphorylation in human breast cancer cells. Oncotarget 2016; 7: 68303–68313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yan D, Avtanski DB, Saxena NK, et al. Leptin‐induced epithelial–mesenchymal transition in breast cancer cells requires beta‐catenin activation via Akt/GSK3‐dependent and MTA1/Wnt1‐dependent pathways. J Biol Chem 2012; 287: 8598–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang XP, Jiang GY, Sun MF, et al. Cytosolic THUMPD1 promotes breast cancer cells invasion and metastasis via the AKT‐GSK3‐Snail pathway. Oncotarget 2017; 8: 13357–13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dragoi D, Krattenmacher A, Mishra VK, et al. Twist1 induces distinct cell states depending on TGFBR1‐activation. Oncotarget 2016; 7: 30396–30407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ring A, Kaur P, Lang JE. EP300 knockdown reduces cancer stem cell phenotype, tumor growth and metastasis in triple negative breast cancer. BMC Cancer 2020; 20: 1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bednarek R, Selmi A, Wojkowska D, et al. Functional inhibition of F11 receptor (F11R/junctional adhesion molecule‐A/JAM‐A) activity by a F11R‐derived peptide in breast cancer and its microenvironment. Breast Cancer Res Treat 2020; 179: 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vellanki SH, Cruz RGB, Jahns H, et al. Natural compound tetrocarcin‐A downregulates junctional adhesion molecule‐A in conjunction with HER2 and inhibitor of apoptosis proteins and inhibits tumor cell growth. Cancer Lett 2019; 440–441: 23–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Countertrends in F11R and JAM members with respect to cancer prognosis
Figure S2. Correlation between members of JAM and BCa
Figure S3. F11R overexpression activates EMT signalling in BCa
Figure S4. Correlation between F11R and EP300 in BCa cells
Figure S5. Overall survival prognosis correlation between F11R and EP300 downstream effectors in BCa
Table S1. Expression of the JAM genes in TCGA‐BCa patients
Table S2. Expression of the JAM family in patients
Table S3. Relative expression levels of F11R in BCa cells
Table S4. Microarray values of overexpressed F11R in BCa cells
Table S5. Impact of F11R on signalling hallmarks
Table S6. Regulators of EMT signalling involved in F11R
Table S7. Relative expression levels of F11R and EP300 in BCa cells
Table S8. Potential candidates derived from drug repurposing simulations based on F11R expression profiles
Data Availability Statement
Detailed data are included in the manuscript and its supplementary information files.


