Abstract
Host immunity is classically divided into “innate” and “adaptive.” While the former has always been regarded as the first, rapid, and antigen-nonspecific reaction to invading pathogens, the latter represents the more sophisticated and antigen-specific response that has the potential to persist and generate memory. Recent work however has challenged this dogma, where murine studies have successfully demonstrated the ability of innate immune cells (monocytes and macrophages) to acquire antigen-specific memory to allogeneic major histocompatibility complex (MHC) molecules. The immunoreceptors so far identified that mediate innate immune memory are the paired immunoglobulin-like receptors (PIRs) in mice, which are orthologous to human leukocyte immunoglobulin-like receptors (LILRs). These receptor families are mainly expressed by the myelomonocytic cell lineage, suggesting an important role in the innate immune response. In this review, we will discuss the role of immunoglobulin-like receptors in the development of innate immune memory across species.
Keywords: Leukocyte immunoglobulin-like receptors, Paired immunoglobulin-like receptors, Evolution, Monocyte, Innate immune memory
Introduction
Traditionally, host immunity has been classified into “innate” and “adaptive” (Buchmann 2014; Medzhitov 2001). Adaptive immunity is mediated by lymphocytes that possess highly diverse antigen receptors generated via somatic gene rearrangement, while innate immunity is mediated by lymphoid and myeloid cells that utilize germline-encoded receptors. Although adaptive immunity plays a pivotal role in host protection against a wide range of pathogens and tumors, it is also a critical driver of autoimmunity and allograft rejection. Hence, for decades, most of the focus in the literature has been on understanding the mechanisms regulating the development of adaptive immune responses, which are common to all jawed vertebrates and generate long-lasting protection (memory) against previously encountered antigens. Recently, innate immunity, mediated primarily by cells of myeloid origin such as monocytes, macrophages, and dendritic cells, has gained more attention as it represents a conserved ancient defense system that is shared among almost all animal species, and importantly, is an essential trigger of subsequent adaptive immune responses. Myeloid cells, particularly dendritic cells, play a central role in bridging innate and adaptive immune responses via antigen presentation to T lymphocytes.
Monocytes along with other phagocytic cells represent a first line of defense in the innate immune system (Medzhitov 2001). The presence of an ancestral equivalent for them in invertebrates highlights their importance across species. For instance, invertebrates possess hemocytes that resemble the monocyte-macrophage lineage. Hemocytes, as the main immunosurveillance cells of invertebrates, phagocytose and engulf invading pathogens through a sophisticated recognition system, parallel to the pattern recognition receptors (PRR) which include Toll-like receptor (TLR) and scavenger receptor systems in vertebrate animals. Those ancestral monocytic relatives are involved in apoptosis and wound healing as well (Vlisidou and Wood 2015). Classically, monocytes are considered progenitors of mature phagocytic cells, dendritic cells, and macrophages. However, they can also be professional antigen-presenting cells, linking the innate and adaptive immune responses.
Monocytes are armed with an extensive arsenal of immune receptors and mediators that enable them to exert their actions (Medzhitov 2001; Nie et al. 2018; Ifrim et al. 2014). Monocytes possess pattern recognition receptors (PRRs) such as TLRs that recognize molecules common to microbes called pathogen-associated molecular patterns (PAMPs) as well as molecules released upon tissue damage and known as danger-associated molecular patterns (DAMPs) (Medzhitov 2001; Nie et al. 2018). PRRs are germline-encoded, are selected over evolutionary time, and tend to be conserved across Vertebrate phyla (Bagheri and Zahmatkesh 2018). In addition, monocytes are equipped with a family of immunomodulatory receptors called paired immunoglobulin-like receptors (PIRs) in mice and leukocyte immunoglobulin-like receptors (LILRs) in humans, most of which bind to MHC class I molecules, suggesting a role for monocytes in self-non-self-recognition. In this review, we will discuss the role of these receptors in regulating monocyte function and the acquisition of allo-specific innate immune memory.
Origins and heterogeneity of monocytes
Monocytes are derived from common myeloid progenitors (CMP) in the bone marrow during hematopoiesis (Guilliams et al. 2018; Geissmann et al. 2010; Martinez et al. 2009). CMP are also the precursors of other myeloid cells such as the granulocytes and dendritic cells. Monocytes were traditionally considered the sole progenitors of tissue macrophages (Geissmann et al. 2010), but recent evidence has shown that most of the resident macrophages originate from a separate yolk sac precursor during embryogenesis (Geissmann et al. 2008). This refuted the classical dogma and limited monocytes’ role more narrowly to precursors of macrophages and dendritic cells during inflammation (Geissmann et al. 2010).
Advanced flow cytometry has allowed subtyping of human monocytes into three phenotypically distinct groups based on the differential expression of the cell surface antigens CD14 and CD16 on MHC class II (HLA-DR)+ cells (Boyette et al. 2017). Classical monocytes (CD14+CD16−) account for the majority of monocytes, whereas non-classical (CD14lowCD16+) and intermediate (CD14+CD16 +) monocyte subtypes account for no more than 20% of the monocyte pool (Passlick et al. 1989). Classical monocytes are proinflammatory cells that are equipped with a wide range of chemokine receptors (CCR1, CCR2, and CCR5) and secrete inflammatory cytokines and chemokines (IL6, IL8, CCL2, CCL3, and CCL5). In addition, these cells have the potential to generate monocyte-derived dendritic cells and macrophages (Günther et al. 2019; Gren et al. 2015). On the other hand, intermediate monocytes are the most efficient of all monocytes at antigen processing and presentation (Wong et al. 2011; Lee et al. 2017). Interestingly, cells of this subgroup can simultaneously secrete proinflammatory mediators such as TNFa and IL-6, and the anti-inflammatory cytokine IL-10 upon TLR activation (Wong et al. 2011; Skrzeczyńska-Moncznik et al. 2008; Belge et al. 2002). This dual function places intermediate monocytes at the crossroad of immune modulation. Although non-classical monocytes possess some antigen-presenting capacity, they differentiate into macrophages and have been shown to play a role in wound healing (Schmidl et al. 2014).
A similar division of monocytes is present in the mouse, where classical monocytes—also known as inflammatory monocytes— are defined as Ly6ChiCX3CR1intCCR2+CD62L+CD43low, while non-classical monocytes—also known as patrolling monocytes—are Ly6ClowCX3CR1hiCCR2lowCD62L−CD43+ (Shortman and Liu 2002). Although they do not patrol the peripheral vasculature, murine intermediate monocytes (Ly6CintCX3CR1hiCCR2lowC62L−CD43+) possess many of the proinflammatory characteristics of their classical counterparts. (Wolf et al. 2019).
Recently, single-cell RNA sequencing analysis of human monocytes demonstrated that intermediate monocytes are not homogeneous but comprise four distinct subpopulations, two of which express genes shared with classical and non-classical monocytes and the other two distinctly express genes involved in cell cycle regulation, cell trafficking, and cytotoxicity (Villani et al. 2017).
Trained immunity
Trained immunity and its involvement in the ability of innate cells to remember previous insult gained momentum recently as it represents a new field in immunology that requires further investigation to understand its implication on health and disease states. “Priming” of immune cells I response to an insult represents a primitive form of enhanced immune memory-like response, first identified in plants. In “priming,” an innate process mainly seen in invertebrate and slightly different from “training,” the initial exposure/stimulus induces changes in the immune functional status of the responding cells such as macrophages by activating the transcriptional machinery. However, as opposite to training, these inducible changes become intrinsic to the exposed cells which never return to their initial functional status prior to priming and upon secondary exposure exhibit a synergistic immune response (Foster et al. 2007; Divangahi et al. 2021). Whereas in training, following clearance of the exposure or insult, immune cells return to their basal functional level, keeping their trained epigenetic material. However, upon secondary exposure to the same or related antigen, the trained cells will activate their epigenetically enhanced immune functions and generate a stronger immune response (Divangahi et al. 2021).
Trained immunity is “bestowed” onto hematopoietic stem cells and innate immune cells through cytokine-dependent, long-term epigenetic changes (histone modifications and DNA methylation), leading to a stronger and faster innate immune response (Netea et al. 2016; Domínguez-Andrés et al. 2020). However, this form of enhanced immunity is not specific to the elicitor. For instance, human in vivo BCG vaccination induced changes at the transcriptomic level of the hematopoietic stem cells of healthy individuals, upregulating genes that also enhanced monocyte responsiveness to unrelated antigens such as Candida albicans β-glucan, which persisted for up to 90 days post-vaccination (Cirovic et al. 2020). Along the same lines, trained immunity occurs in monocytes, represented by an enhanced activation state of monocytes that can persist for weeks following the resolution of the insult due to epigenetic changes and metabolic reprogramming (Netea et al. 2016; Bowdish et al. 2007; Kurtz 2005). These epigenetic changes alter gene expression by inducing chromatin conformational changes through histone modifications. This epigenetic alteration confers to trained monocytes a metabolic activation state comparable to the more differentiated activated macrophages, suggesting an enhancement of their responsiveness to process an immune response (Netea et al. 2016; Bowdish et al. 2007; Kurtz 2005). Indeed, monocytes trained with β-glucan were shown to switch to the aerobic glycolytic pathway by reducing their basal respiration, increasing their glucose uptake and by-production of lactate, and raising their oxidation state (Cheng et al. 2014). β-Glucan training upregulated the expression of glycolytic enzyme genes such as hexokinase and pyruvate kinase. This training was mediated by the dectin-1–Akt–mTOR–HIF-1α pathway. For instance, administration of metformin, inhibitor of mTOR pathway, to Candia-infected mice abrogated its protective effect against disseminated candidemia (Cheng et al. 2014). Furthermore, BCG-vaccinated individuals exhibit upregulation glycolytic rate-limiting enzymes in their circulating monocytes (Arts et al. 2016). Studies have also shown that epigenetic modifications and reprogramming of monocytes can result in tolerance. For instance, a continuous exposure to LPS prevented monocytes from being involved in inflammation, suggesting a role in controlling the inflammatory process and preventing bystander cellular and tissue damage (Seeley and Ghosh 2017). These newly identified functions of monocytes place them at the heart of fine-tuning of the innate response (Bordon 2014). However, it is important to note that although trained immunity shares the enhanced activation property with adaptive immune memory in lymphoid cells, it lacks the antigen specificity that defines adaptive memory (Bordon 2014; Ferro et al. 2019).
Innate immune receptors
The initiation of an immune response requires the recognition of non-self along with inflammatory stimuli and is carried out primarily by dendritic cells, monocytes, and various phagocytic cells through germline-encoded PRRs that recognize PAMPs and DAMPs (Kawai and Akira 2010). Interactions of myelomonocytic cells with other members of the innate or even adaptive immune compartments, with each other, and with non-immune cells however are mediated by a wide array of germline-encoded receptors and surface molecules that modulate immune responses (Domínguez-Andrés and Netea 2020). Murine studies have shown that monocyte functions related to inducing tolerance, initiating rejection cascades, immunogenicity, and vaccine memory are influenced by paired receptor families that belong to the immunoglobulin superfamily (IgSF) (Domínguez-Andrés and Netea 2020). A given paired receptor family contains both activating and inhibitory members that, together, modulate the monocyte response. As we will elaborate below, some of these receptors confer specificity to allogeneic non-self and endow memory to monocytes above and beyond what has been described in the context of trained immunity.
The human leukocyte immunoglobulin‑like receptors (LILRs): genetic and structural insights
The human LILR family is a germline-encoded family of paired receptors that modulate immune responses by fine-tuning inhibitory and stimulatory signals. They belong to the ancestral C2 IgSF, which can be traced back to invertebrates (Nie et al. 2018) where receptors such as the Down syndrome cell adhesion molecules (DSCAMs) have been identified (Gourbal et al. 2018; Litman et al. 2010). In 1997, Samaridis and Colonna identified two novel cDNA that encode for transmembrane proteins expressed by myeloid and lymphoid cells, related to the bovine Fcy2R, human KIR and human FcaR, and murine gp49. The first identified receptors were called immunoglobulin-like transcript 1 which is equivalent to LILR A2 and immunoglobulin-like transcript 2 which is the same as LILR B1 (Samaridis and Colonna 1997). In the same year, Colonna et al. added ILT 3 to the family of LILRs. This receptor was expressed by myeloid cells and exhibit an inhibitory function by activating the inhibitory SHP-1 pathway (Cella et al. 1997). LILR genes are located in the leukocyte receptor complex (LRC) region on human chromosome 19q13.4, with a length of 1 Mb (Martin et al. 2002; Hirayasu and Arase 2015; Barrow and Trowsdale 2008) (Fig. 1). It harbors up to 45 gene sequences that encode for 30 different Ig-like receptors including the LILRs and killer cell Ig-like receptors (KIRs) (Martin et al. 2002). The LILR cluster in the LRC region consists of two inverted and duplicated clusters, centromeric and telomeric, separated by the LRC-encoded novel gene (LENG) sequence (Martin et al. 2002). Each of these clusters extends over 150 kb and are in proximity to the closely related KIR genes. LILR clusters collectively encode for 13 genes (Martin et al. 2002). The syntenic mouse chromosome 7 houses the mouse LRC region that extends from the murine activator receptor 1 (MAR-1) to the ribosomal protein s9 gene (Martin et al. 2002). The murine genes encoding for the paired immunoglobulin-like receptors (PIRs), orthologs of LILRs, are located at the centromeric region of the mouse LRC (Martin et al. 2002). The homology between human LILRs and murine PIRs suggests their divergence from a common ancestral origin (Hogan et al. 2012).
Fig. 1.
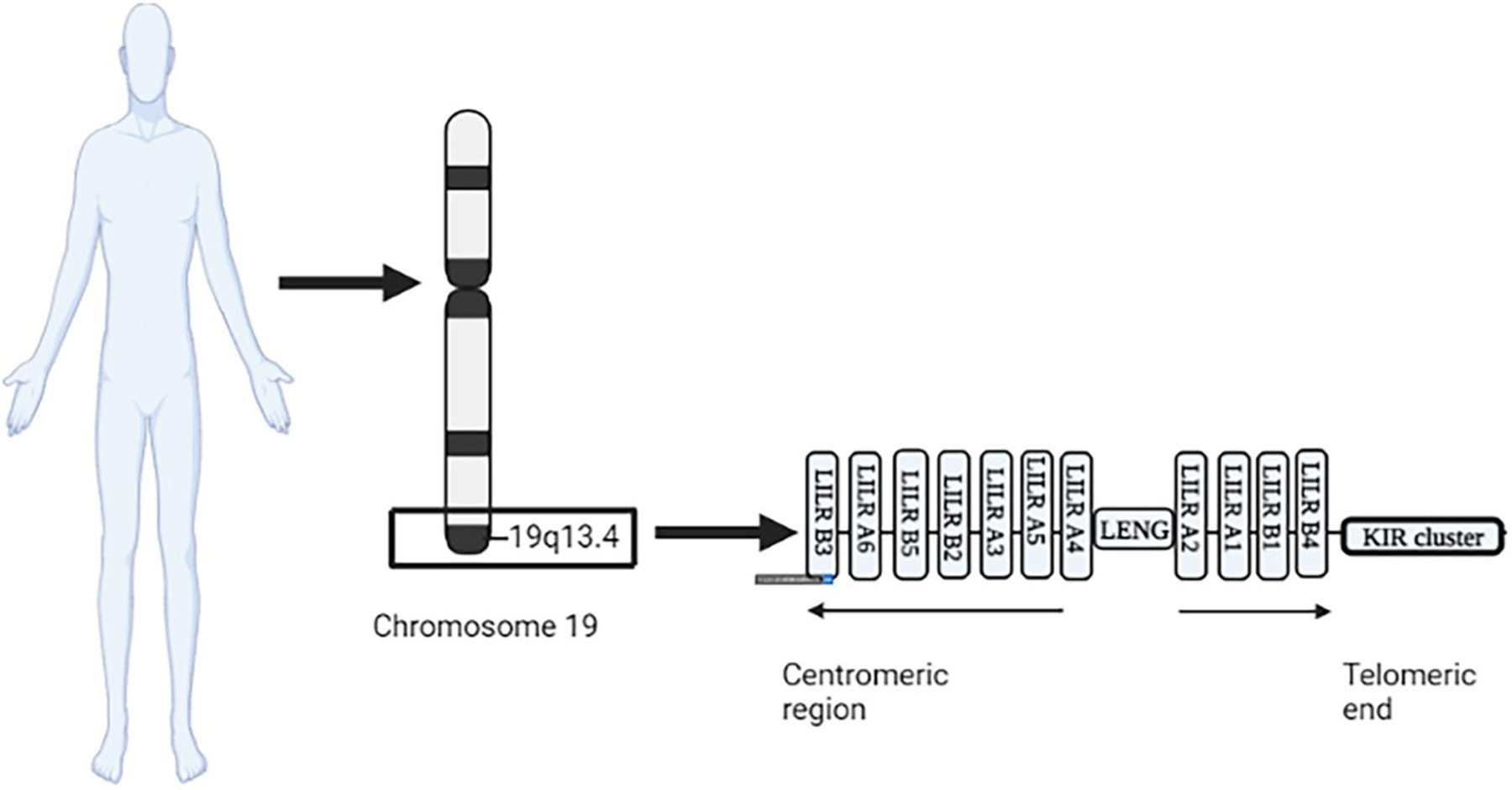
Leukocyte immunoglobulin-like receptor loci within the leukocyte receptor complex on human chromosome 19
LILR family can be classified into two distinct groups based on their immunomodulatory functions: the inhibitory LILR subfamily B and the activating LILRA family. LILRs are expressed mainly on myelomonocytic cells. Although they are closely related to the KIR system, the latter is almost exclusively expressed by natural killer (NK) cells. The MHC class I molecules are the main known ligands for both LILRs and KIRs. The human LILR locus encodes for 5 inhibitory receptors (LILRB1–5), five activating receptors (LILR A1, 2, 4, 5, and 6), one secreted molecule (LILRA3), and two pseudogenes. This locus is considerably polymorphic contributing to the high diversity and variability within these receptor populations with LILRB3, LILRB4, and LILRA6 having at least 15 significant allelic variations. The receptors consist of four Ig domains, a stem region, a transmembrane domain, and a cytoplasmic tail except for LILRA1 and LILRB4 which only express two Ig domains. LILRAs and LILRBs share homologous C-2-type Ig-like extracellular domains but have distinct transmembrane and cytoplasmic regions. For instance, LILRAs are characterized by a short truncated cytoplasmic tail with positively charged arginine residues spread in their transmembrane domains allowing for potential interaction with tyrosine kinase activation motif bearing Fc receptor common gamma chain (FcRg) to initiate a stimulatory downstream cascade. In contrast, LILRBs possess longer cytoplasmic tails containing multiple immunoreceptor tyrosine-based inhibition motifs (ITIMs) which can be coupled with phosphatases to provide an inhibitory signal. LILRs bind to various ligands that range from classical and non-classical MHC class I (the best characterized ligands), pathogen-associated proteins, and host immunomodulatory molecules such as angiopontin-like 4 protein. ITIM/ITAM system involvement in transduction of various immune receptor signaling is well established in multiple innate as well as adaptive processes including the KIR, BCR, TCR, and FcR signaling pathways. This wide range of ligands suggests a fundamental immunomodulatory role assumed by the myelomonocytic family via interactions with LILR primate that may have evolved over time (Halaby and Mornon 1998).
The localization of human and murine immunoglobulin-like receptor genes on the highly plastic and dynamic leukocyte receptor complex allowed their diversification and rapid evolution by various processes including duplication, deletion, and rearrangement involving exon shuffling events from a common ancient ancestral organization of paired inhibitory and activating immunoglobulin-like receptor gene. This common ancestral origin was supported by the identification of various immunoglobulin-like receptors in different species as distant relative of the LILR and PIR systems in the phylogenic tree of vertebrates. Comparative analysis of various mammals and birds including chicken revealed a surprising structural and functional diversity of their paired immunoglobulin-like receptors (Martin et al. 2002; Viertlboeck et al. 2005; Fayngerts et al. 2007). Beyond mammalian and birds, relatives to the human and murine paired immunoglobulin-like receptors were identified in amphibians, particularly in tropical clawed frogs (Silurana tropicalis immunoglobulin-like receptor (silrs)) on chromosome 7 in a LRC-like region (Guselnikov et al. 2008). This further suggests an ancient ancestral synteny of the leukocyte immunoglobulin-like receptors (Wang et al. 2021). In addition, the identification of novel and related immune type receptors that possess immunoglobulin-like ectodomains in channel fish underlines the importance of conserving such innate immune receptor system during species evolution (Stafford et al. 2006). Although it is well established now that LILR and PIR and their relatives in mammalians engage MHC class I molecules among other ligands, Stafford et al. revealed that channel fish Ictalurus punctatus leukocyte immune-type receptor D1 and D2 ectodomains harbor a putative MHC class 1 binding site that is very similar to MHC a3 biding site of LILR B3 (Stafford et al. 2007). These findings suggest that although immunoglobulin-like receptors are extremely genetically diverse and dynamic, they might share a functional connection that is conserved across species. This conserve function could highlight the involvement of the innate receptors in the innate allorecognition mechanism that still require further investigations.
KIRs: close relatives to LILRs
Discovered few years prior to LILRs, killer cell immunoglobulin-like receptors (KIRs) are close relatives to the immunoglobulin-like receptors. Both house in the LRC complex on chromosome 19. Similarly to LILRs, KIRs are transmembrane glycoproteins expressed on NK cell primarily and some T cell subsets, which belong to the Ig superfamily and display a unique interaction with various MHC class I molecules (Campbell and Purdy 2011; Pende et al. 2019). The KIR family encodes for 14 genes (2DL1–5, 3DL1–L3, 2DS1–S5, and 3DS1) that are characteristically highly polymorphic when compared to their LILR relative (Campbell and Purdy 2011). These receptors can propagate either an activating or inhibitory cascade depending on the intracellular signaling pathway they induce upon ligand engagement. These receptors vary by the number of their extracellular Ig domains (2D [2 Ig domains] vs. 3D [3 Ig domains) and the length of their cytoplasmic tails (L [long] vs S [short]) (Marsh et al. 2003). All inhibitory KIRs possess a long cytoplasmic tail containing ITIM motifs that recruits tyrosine phosphatases similar to their LILRB relatives (MacFarlane and Campbell 2006). However, short cytoplasmic domains propagate their intracellular signal via the transmembrane signaling adaptor protein DAP12 (MacFarlane and Campbell 2006). KIR2DL4 is an exception to the long/short signaling dichotomy, where although it has a log cytoplasmic tail, it mediates its intracellular signaling via ITAM containing FcεRI-γ adaptor protein instead of DAP 12 (Goodridge et al. 2003). Individual KIRs bind to various distinct MHC class I molecules. However, the inhibitory KIRs exhibit a higher affinity to MHC class I molecules than their activating counterparts (Stewart et al. 2005).
LILR polymorphism and MHC class I engagement
Although LILR represents a somehow conserved genomic organization, recent literature reveals some evidence of LILR polymorphism. LILRA3, LILRB3, and LILR A6 consist the most polymorphic and variable of the family with 11, 14, and 11 different allelic variations detected respectively for each (Young et al. 2001). To add to this allelic diversification, LILRs are extremely susceptible to mRNA alternative splicing that allow to extend the pool of these receptors. Alternative splicing in the LILR mRNA results in the production of soluble isoforms for their membrane-bound receptors, allowing for a more complex immunomodulatory role. For instance, soluble LILRB4, referred to as sLILR B4, was detected in the sera of patients with melanoma and pancreatic and colorectal adenocarcinomas (Suciu-Foca et al. 2007). The production of soluble variants represents a mechanism by which tumors evade the host immune response. For example, sLILR B4 was shown to induce CD4 cell anergy and CD8 differentiation into regulatory CD25 + cells, preventing allogenic tumor graft rejection in humanized SCID mice (Suciu-Foca et al. 2007). This polymorphism is further illustrated by the copy number variation, which is characteristic of LIlR A3, LILR A6, and LILR B3. This mechanism is driven by various cellular processes such as sequence deletion, non-allelic homologous recombination, or crossing over between non-allelic sequences.
Despite that LILRs exhibit a broad binding specificity to MHC class I molecules when compared to TCR and KIR, variation in the binding affinity of LILRs to various MHC class I molecules has been recently correlated with HIV infection progression and outcomes.
Similar to their neighboring relative KIRs, LILRs recognize and bind to various MHC class I molecules. However, KIRs exhibit some degree of antigen specificity but to a lesser extent than TCR (Peruzzi et al. 1996). Colonna demonstrated that LILRB1 recognize HLA -A, B, and G1 with a relatively broad specificity (Colonna et al. 1997). The engagement of LILRB1 with HLA class I members allowed the identification of other LILRs that bind MHC class 1 molecules. This resulted in the classification of members of LILR family into two subgroups depending on their binding ability to MHC class I. LILRA1, LILRA2, LILRA3, LILRB1, and LILR B2 were found to engage MHC class I, and they are referred to as group 1 receptors. However, this binding to HLA class 1 molecules exhibited allelic variations. For instance, LILRB1 exhibits a lower affinity for a subset of HLA-A alleles and LILR B2 displays a lower affinity for some HLA-B variants, whereas HLA A1 and A3 bind preferentially to HLA-C. LILRB 2 has been shown to bind through D1-D2 immunoglobulin domains to the alpha 3 and B2m subunits of the MHC-peptide complex. However, D1-D2 of LILR B1 strongly binds to B2m subunit. LILR binding variability does not always correlate with HLA polymorphism, raising the possibility of the presence of other factors that can contribute to this binding variability. This interaction can be mediated through binding involving the D3-D4 immunoglobulin domains. MHC class 1-bound peptides, indeed, were shown to influence LILR-MHC interaction. For example, the binding strength of LILR B2 to HLA B2705 -HIV Gag KK10 depends on the amino acid sequence of the bound KK10 peptide (Lichterfeld et al. 2007).
LILR interaction with MHC class I modulating the immune response is still not well established when compared with the KIR system and MHC-TCR axis. Although MHC-TCRs represent the most sophisticated of all three systems as they are highly antigen specific. KIRs were found to exhibit some antigen specificity in their MHC recognition. Three-dimensional crystal analysis, however, highlights that both KIR and TCR interact with the MHC class 1 molecule involving the top helices and the exposed regions of the MHC bound peptides. To add, various studies demonstrate that KIR binding to HLA class 1 molecules depends on the amino acid sequence of the presented peptide, For example, a single amino acid mutation in the peptide binding site of HLA B 2705 led to variable degrees of protection form HLA B 2705 specific NK clones toxicity (Malnati et al. 1995).
The interaction of LILRs with various HLA class I molecules suggests a role of these receptors in shaping the immune response against some viral and bacterial infections and their progression and outcomes, and the pathogenesis of autoimmunity and tumorigenesis.
LILR functions
Upon binding to their respective ligands and interaction with signaling receptors, LILRs initiate an intracellular signaling cascade that serves as a switch, activating or inhibiting different aspects of the immune response such as cytokine production, cellular proliferation, antigen presentation, and apoptosis. LILRBs possess characteristically 2 to 4 ITIMs in their cytoplasmic tails. Ligand binding induces changes in the receptor 3D conformation along with phosphorylation of ITIM’s tyrosine by Src kinases. Phosphorylated tyrosine residues initiate a cascade of events by recruiting the inositol phosphatase SHIP and tyrosine phosphatases SHP-1/2 preferentially based on the amino acid sequence of the ITIM domain. It is well known that ITIM negatively regulates immune cells. For instance, SHP-1 can dephosphorylate a number of intracellular immune mediators: the activated immunoreceptor tyrosine-based activation motif (ITAM) which plays a central role in the signaling transduction cascade of LILRAs, Src proto-oncogene non-receptor tyrosine kinase (Src), spleen tyrosine kinases, zeta-chain-associated protein kinase 70 KD (ZAP70), Lck/Yes-related novel protein tyrosine kinase (Lyn), phosphatidylinositol-4-phosphate 3 kinase (PI3K), phospholipase C gamma (PLC-gamma), or Vav 1 guanine nucleotide exchange factor (vav1). Although LILRAs have a truncated cytoplasmic tail, their positively charged arginine containing transmembrane domain allows their interaction with FcRg which mediates their intracellular effect via ITAM. Ligand engagement with their corresponding LILRA induces the activation for Src kinases which subsequently phosphorylate the tyrosine residues within the ITAM. Consequently, the recruitment and activation of Syk or ZAP-70 tyrosine kinases would propagate a cascade of events resulting in the activating immune functions.
The immune inhibitory functions of LILRBs have gained increased interest lately since they provide basis for an innate checkpoint that modulates adaptive immune functions. For instance, LILRB1, which is widely expressed on cellular descendants of the myeloid lineage, bind with quite high affinity to various HLA class I molecules (HLA-A/B/C) and the non-classical HLA class I molecules E and G. These ligands allow LILRB1 to interact with various types of immune and non-immune cells. It has been shown that cross-linked anti-LILRB1 monoclonal antibodies inhibit antigen-specific T cell proliferation along with increased production of immune downregulatory cytokines such as IL-10 and TGF-B and decrease in proinflammatory cytokines such as IFNg, whereas monoclonal antibody blockade of LILRB1 allowed T cells to proliferate at higher rate and produce higher levels of proinflammatory cytokines in response to a specific antigen (Saverino et al. 2002). Interestingly, the binding of LILRB1 to decidual maternal macrophages to HLA-G, which is ubiquitously expressed by fetal chorionic membrane cytotrophoblasts, prevents macrophage activation and inhibits the alloreactive anti-fetal T cell response (Petroff et al. 2002). Such role provides an insight into the possible involvement of LILRB1 on myelo-phagocytes in peripheral tolerance. In tandem to this inhibitory function of LILRB1, LILRB2 binds preferentially to HLA class I molecules, co-ligating with FcgRI (CD64) to inhibit the phosphorylation of Syk molecules and FcRg and prevent intracellular calcium influx, suggesting a role in downmodulating the signals induced by IgG, IgE, and IgA receptors (Fanger et al. 1998). In addition, treatment of cardiac allografts with agents that upregulate LILRB2 and LILRB4 on the donor antigen-presenting cells leads to peripheral tolerance (Manavalan et al. 2003). Although little is known about the role of LILRB3 in monocyte function, antibodies directed against this receptors led to the killing of antigen-presenting cells via complement and antibody-mediated cellular cytotoxicity in the setting of graft-vs-host disease (Pfistershammer et al. 2009). On the other hand, LILRB4 can bind to HLA-DR, an MHC class II molecule, inhibiting the influx of calcium into monocyte and therefore increasing the threshold of their activation (Cella et al. 1997). LILRB5 has been shown to bind to HLA-B7 and HLA-B27. Function of LILRB 5 is still not completely understood in monocyte activation (Zhang et al. 2015).
The functions of LILRAs are not extensively studied and understood. LILRA1 was found to bind HLA-C but with a lower affinity than the inhibitory counterparts LILRB1 and LILRB2 (Jones et al. 2011). LILRA2 was suggested to be involved in anti-microbial responses as it was found to bind to microbially leaved immunoglobulins with no affinity to any HLA molecule (Hirayasu et al. 2016).
LILRs and their involvement in the anti‑viral response
Since infections almost always activate the adaptive immune system by antigen presentation on MHC class I, LILRs, by virtue of their binding to MHC class I, were thought to play a role in shaping the anti-microbial immune response. For instance, it has been shown that neonatal sepsis induces the upregulation of inhibitory LILR B2, B3, and B4 along with the activating soluble isoform LILR A3. This suggests a role of LILR in not only influencing the immune response but also influencing the disease symptomatology as well (Smith et al. 2014). Furthermore, trivalent influenza vaccination was demonstrated to upregulate the expression of LILR A1, A2, A3, A6, and B1 by myelomonocytic cells at day 3 post vaccination, suggesting a role of these LILRs in amplifying and optimizing an adequate vaccine-specific antibody response (Nakaya et al. 2011).
A particular important insight on the role of LILRs in modulating anti-viral response has been emerging in the setting of HIV-1 infection and various disease outcomes. LILR expression signature varies significantly dependent on the chronicity of infection, onset time, treatment, and elite controller status (Coindre et al. 2018; Huang et al. 2010). For example, HIV-positive patients in the chronic phase of the infection exhibit an upregulation of LILR B1 and B2 (O’Connor et al. 2007; Anfossi et al. 2004). This upregulation is primarily mediated by the IL-10 (Vlad et al. 2003). Various studies have highlighted the association between different HLA class 1 molecules and either HIV infection control or progression to AIDS. For instance, HLA -B27 and B57 were associated with a slower progression of HIV infection, whereas HLA -B35 with rapid progression (Kaslow et al. 1996; Carrington and O’Brien 2003). Interestingly, these HLA variants show differential binding affinity to LILR B2, where HLA-B27 exhibits a lower affinity to this inhibitory receptor and HLA B35 demonstrates a higher affinity to LILR B2 (Huang et al. 2009). These findings suggest that a weaker affinity to the inhibitory LILR B2 prevents dendritic cell inhibition and therefore results in a more efficient anti-HIV response. In a cohort of HIV-positive patients, G. Yu et al. sought to determine the effect of HLA-1 haplotypes and their binding affinity on LILRs on the disease progression. Their data elucidated the fact that LILR B2, rather than LILR B1, binding strength to various HLA class 1 haplotypes correlates with disease control and viral load (Bashirova et al. 2014).
LILR-MHC class I interactions can influence the outcome and progression of other infectious processes. For example, patients with HLA-B 27 have been shown to clear hepatitis C infection spontaneously. Based on findings in HIV model, HLA-B27 weak interaction with LILR B2 might provide a mechanistic insight into this protective feature (Bengsch et al. 2009).
LILR and autoimmunity
The role of immunoglobulin-like receptors in controlling and maintaining self-tolerance has been under extensive research since the binding of LILRs to classical and non-classical HLA class 1 molecules renders them a putative therapeutic agents and diagnostic markers of diseases (Zhang et al. 2017). For instance, Ujikie et al. 2002 demonstrated that PIRB is essential for B cell suppression, dendritic cell maturation and differentiation, and balancing humoral th2 and cellular Th1 responses (Ujike et al. 2002). He demonstrated that Pirb−/− mice display hypersensitivity in their splenic B2 cells upon BCR ligation, mount a significantly more ample humoral response upon challenge with TNP-LPS, and produce significantly higher level of interleukin 4 and markedly reduced level of IFN gamma upon immunization with T-dependent antigens (Ujike et al. 2002). In addition, treatment of collagen induced arthritis mice with HLA-G, a non-classical MHC class 1 molecule known with its binding affinity to the human LILRB1 and B2, and reduced arthritic symptoms such as swelling via engagement of PIR B (Kuroki et al. 2013). These early murine studies and the well-established affinity of LILR to various MHC class I made them a possible culprit in the pathogenesis of multiple autoimmune diseases (Zhang et al. 2017). In fact, altered expression of distinct LILRBs has been associated with multiple human autoimmune pathologies such as multiple sclerosis, SLE, and rheumatoid arthritis. Patients with SLE have been shown to have a significantly compromised LILR B1 inhibitory activity on T cell activation and reduced expression of LILR B1 on B cells (Monsiváis-Urenda et al. 2007). Phenotypic analysis of monocyte-derived DC of SLE patients revealed that in vitro treatment of DC with IL-10 confers a tolerogenic phenotype to these DCs by upregulating the expression of CD69, LILRB1, and CD32 (Figueroa-Vega et al. 2006). Furthermore, the soluble LILR A3 expression correlates strongly and positively with RA severity (An et al. 2010). Because of its variable affinity to HLA-B27, LILRB2 has been suggested to contribute to the pathogenesis of ankylosing spondylitis (SA) (Kollnberger et al. 2002).
LILR relatives in invertebrates
Although there is no record of any ortholog for LILRs in invertebrates, distant relatives of this family of receptors were identified in multiple invertebrates and protochordates (Hernández Prada et al. 2006), for instance, DSCAMs (Brümmendorf and Lemmon 2001). DSCAMs were first described in Drosophila melanogaster as cell adhesion molecules important in neural wiring, but homologs were identified in other members of the invertebrate family as well (Brümmendorf and Lemmon 2001). DSCAMS and other orthologs in invertebrates have a characteristic structure consisting of a signal peptide, ten Ig domains, up to 6 fibronectin type III domains, a transmembrane domain, and a cytoplasmic tail (Brümmendorf and Lemmon 2001). Since these receptors were found later to be expressed by hemocytes and phagocytes, they were shown to contribute to protection against a wide range of pathogens (Brümmendorf and Lemmon 2001). Alternative splicing, driven by pathogen exposure, plays a crucial role in the distinctive hypervariability of these receptors, generating a broad repertoire of adhesion molecules with varying specificities towards multiple microbial antigens. In other words, challenge with a particular antigen elicits antigen-specific splicing leading to the production of receptor molecules with specific affinity for the elicitor. For instance, challenging Anopheles gambiae with E. coli or Pseudomonas veronii produces distinct splice forms of DSCAM that are only specific for E. coli and P. veronii, respectively, and different from the isoforms induced by S. aureus exposure or those produced in exposure-naïve insects (Dong et al. 2006). In addition, engineering DSCAM molecules in vitro by varying exon combinations created forms that had different specificities of binding (Brites and Pasquier 2015; Watson et al. 2005). DSCAM receptors are important for induction of phagocytosis in invertebrates (Brümmendorf and Lemmon 2001; Watson et al. 2005). For instance, treating hemocytes of Drosophila with specific RNAi or anti-DSCAM antibodies reduced their phagocytic capacity (Brites and Pasquier 2015).
Mammalian and bird LILR orthologs
In an attempt to find a relative or an ortholog for the human FcαR, the paired immunoglobulin-like receptors (PIRs) were identified in mice (Ben Baruch-Morgenstern et al. 2014). The PIR system is encoded on the murine chromosome 7 in a region syntenic with the human chromosome 19q13.4 (Fig. 2) that houses the LRC (Ben Baruch-Morgenstern et al. 2014; Takai 2005). These receptors are expressed on the surface of multiple immune cells such as macrophages, dendritic cells, and B cells but not expressed by NK or T cells in the mouse (Ben Baruch-Morgenstern et al. 2014; Takai 2005). Similar to the human LILRs, PIRs can be further classified into 2 subgroups: the first is activating (PIR-A) and the second propagates an inhibitory signal intracellularly (PIR-B) (Takai 2005). However, murine receptors have 6 Ig domains as opposed to the 2–4 Ig domains in their human relatives (Takai 2005). Additionally, mice possess only one inhibitory receptor and at least eight activating receptors (Ben Baruch-Morgenstern et al. 2014; Takai 2005; Pereira et al. 2004). PIR-B is characterized by a cytoplasmic tail containing four immunoreceptor tyrosine kinase motifs (Ben Baruch-Morgenstern et al. 2014). As opposed to PIR-B, PIR-A possesses a short cytoplasmic tail that is devoid of any ITIMs (Takai 2005) (Fig. 3). However, the highly hydrophobic transmembrane domain of PIR-A harbors a positively charged arginine residue allowing the association of these receptors with the FcRg chain, which is essential not only for the expression of these receptors on the cellular membrane but to generate an intracellular stimulatory cascade (Ben Baruch-Morgenstern et al. 2014). In mouse, both receptors bind to MHC class I with variable affinity, similar to their human orthologs, hinting to a pivotal role of these receptors in self/non-self-recognition mechanisms by the innate compartment of the immune system (Takai 2005).
Fig. 2.
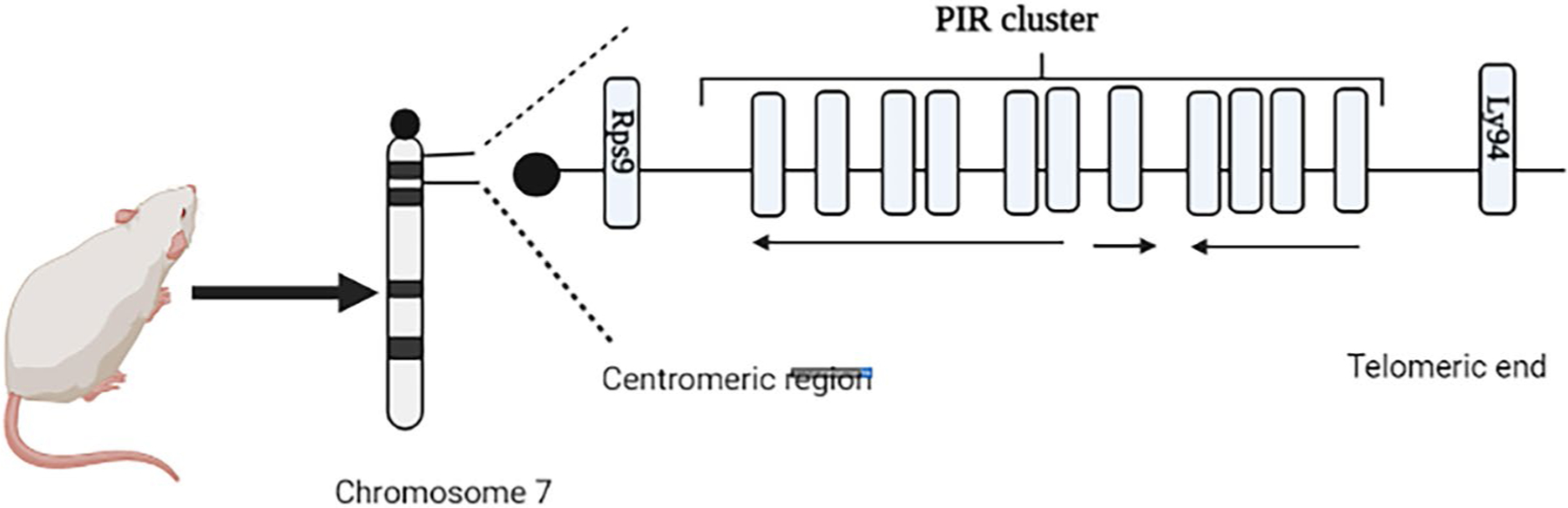
Murine paired immunoglobulin-like receptor cluster within the leukocyte receptor complex on mouse chromosome 7
Fig. 3.
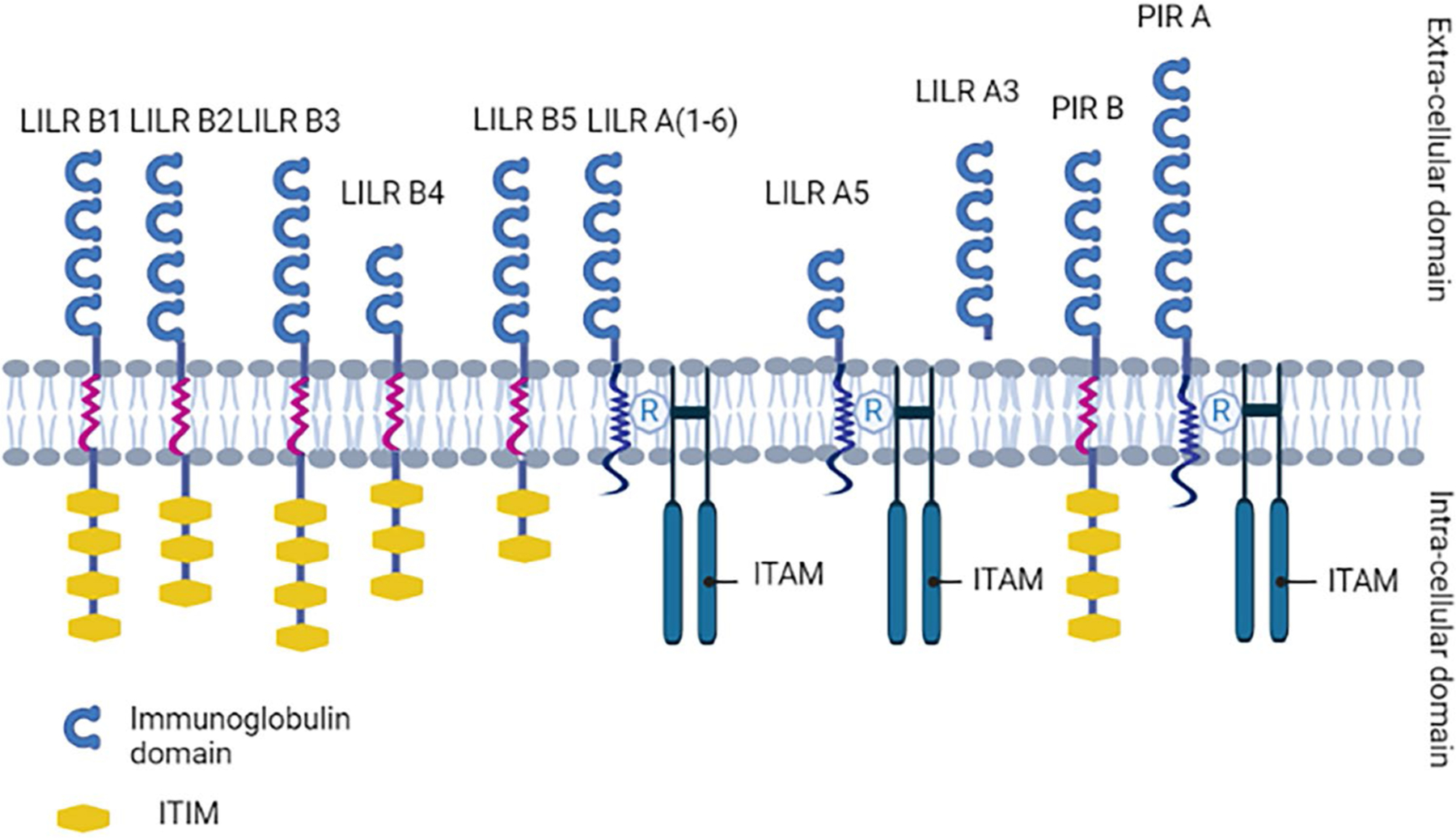
LILR and PIR structure
Orthologs to the PIR and LILRs were described in other rodents (Dennis et al. 1999), non-human primates (Magalhães and Church 2007), gray seals (Hammond et al. 2009), goats (Schwartz et al. 2019), pigs (Schwartz et al. 2019), and chicken (Nikolaidis et al. 2005). These all share a dichotomous functional distinction of inhibitory and activating receptors with some structural variability. For instance, chicken immunoglobulin-like receptors, also referred to as CHIR, have only 2 ectoplasmic Ig domains (Nikolaidis et al. 2005). It is worth mentioning that chicken, in addition to their activating and inhibitory immunoglobulin-like receptors, express a particular Ig-like receptors that seem to be bi-functional (Arnon et al. 2008). CHIR ABs combine a positively charged transmembrane region similar to their activating counterpart and ITIM, ITSM, and YXXM containing motifs which allow the potential interactions with inhibitory intracellular mediators (Viertlboeck et al. 2005). Similar to their immunoglobulin receptor relatives in various species, CHIRs bind to molecules of MHC class I primarily (Viertlboeck et al. 2005; Arnon et al. 2008; Meziane et al. 2019). However, CHIR AB1, expressed on B cells, monocytes, macrophages, and NK cells, exhibits a specific interaction with the Fc portion of IgY, the avian counterpart of the mammalian immunoglobulin IgG (Arnon et al. 2008).
Insights into immune memory in plants
Studies in plant and invertebrate models showed the acquisition of immunological memory against previously encountered microbial pathogens, suggesting that innate immune systems can develop memory programming following cleared or persistent insults in order to preserve the host’s integrity (Netea et al. 2016; Bowdish et al. 2007). For instance, “systemic acquired response” (SAR) is a plant-specific phenomenon that has been extensively studied. This concept provided basis for the memory potential of the innate immune system (Kachroo and Robin 2013). The molecular mechanisms of such a response have been identified and well described in the literature (Kachroo and Robin 2013). However, this mechanism is driven by epigenetic modifications of effector genes, leading to silencing or activation of specific loci. This epigenetic “reprograming” or “priming” leads to faster and stronger secondary immune responses against previously encountered environmental offenders or insults. In addition, such immunological adaptations were shown to be passed down to the progeny, a concept called “transgenerational SAR.” For instance, progeny of Pseudomonas-infected Arabidopsis parents were primed for salicylic acid-dependent defenses as compared to the progeny of healthy controls (Luna et al. 2012). Transgenerational SAR suggests that these epigenetic modifications are rendered intrinsic and heritable, providing long-term immunity essential for optimize symbiotic relationships in the ecosystem (Luna and Ton 2012; Nyholm and Graf 2012). This transfer of immunity across generation serves a key evolutionary role, allowing the offspring to possess advantageous defense mechanisms to better adapt to their surroundings with the immunological experiences of their primed parents (Melillo et al. 2018). Despite the effort made to understand the molecular interaction in their immune systems, no relatives of the paired immunoglobulin-like systems found in animal kingdom have been identified in plants (Hirayasu and Arase 2015).
Innate immune memory in the invertebrate realm
Investigations of immunological memory in invertebrates date back to the early 1980s in the work of Cooper and Roch. In their experiments, they successfully demonstrated that the innate immune system of earthworms, in the absence of adaptive immune cells (T and B lymphocytes), can recognize allogenic tissues and generate a strong and fast secondary immune response upon allograft rechallenge. They did not only show that earthworms have the capacity to recognize and reject grafts from a different earthworm but also show that rechallenging recipient earthworms with tissues from previous donors lead to faster rejection of the grafts (Cooper and Roch 1986). This work provided one of the earliest evidences demonstrating the capacity of the innate immune system to establish specific memory in an invertebrate. Additionally, results of this work suggested that the concept of allorecognition predates the emergence of the adaptive immune system, confronting the old immunology dogma that allorecognition and memory are characteristic features of the adaptive immune system. Further characterization of the invertebrate allorecognition system was shown in the colonial cnidarian Hydractinia symbiolongicarpus. Adjacent colonies that are compatible (with at most one allelic variation in their allorecognition genotype) successfully fuse together and share vasculature, whereas colonies that have distinct alleles at both allorecognition loci were rejected and dissolved (Melillo et al. 2018; Lakkis et al. 2008; Powell et al. 2007). Similar findings were reported in Porifera species (sponge C. diffusa) (Hildemann et al. 1979), Echinodermata species (sea urchin) (Coffaro and Hinegardner 1977), and Tunicata species (Scofield et al. 1982). Natural transplantation of compatible colonies of tunicates suggested their possession of a complex immune system that has the ability to discriminate between self and non-self. Although they represent one of the most primitive invertebrates, tunicates have been shown to possess a complex mammalian-like hematopoietic system. For instance, Botryllus schlosseri, a colonial tunicate, has hematopoietic stem cells that have the potential to give rise to progenitor and myeloid-like and lymphoid-like populations (Rosental et al. 2018). Worth mentioning, B. schlosseri’s myeloid cells were responsible for allogenic phagocytosis of rejected incompatible colonies (Rosental et al. 2018). Morula cells expressing high level of peroxidase were demonstrated to be the mediator of cytotoxic rejection (Rosental et al. 2018). In addition to natural transplantation immunity, evidence was accumulating to elucidate the innate immune system capability to establish a memory response against pathogens. For instance, Anopheles gambiae mosquitoes exposed a second time to their natural parasite Plasmodium falciparum mount a stronger immune response manifested by increased circulating granulocytes as compared to the primary response (Rodrigues et al. 2010). Drosophila melanogaster has also been shown to mount a stronger and faster immune response upon re-exposure to lethal dose of Streptococcus pneumonia and Beauveria bassina (Pham et al. 2007). This primed immune response is dependent on TLR pathways which was demonstrated by loss of immunity against Streptococcus re-infection in Toll knockout flies (Pham et al. 2007).
Vertebrate innate immune memory
Accumulation of evidence on the ability of the innate immune system to establish a memory response in plants and invertebrates suggests that memory is a conserved or convergent process in the evolution of the immune system that is not restricted to the adaptive immune response alone (Netea et al. 2019). This opened the door to exploring this aspect of innate immunity in vertebrates.
Plethora of data in the literature highlights the ability of NK cells, member of the innate immune system, to acquire adaptive feature such as antigen-specific response and generation of memory. Here, we will emphasize work done in the realm of myeloid cells. The first evidence of the ability of NK cells to generate a specific memory-like response was elucidated by Fairchild and Gorbachev 2001 using a heptan-induced contact hypersensitivity mice (Gorbachev and Fairchild 2001). In 2006, Van Adrian et al. demonstrated that NK cells can generate a long-lived, antigen-specific recall immune response independently of T and B cells (O’Leary et al. 2006). Rag 2−/− mice which were initially sensitized to DNFB on days 0 and 1 demonstrated a significant hypersensitivity response upon rechallenge with the water-soluble analog of DNFB, 2,4 dinitrobenzene sulfonic acid. NK cells were accumulating in the heptan-challenged tissue upon re-exposure (O’Leary et al. 2006). To further elucidate the role of NK cells in mediating such a memory-like response, SCID mice (lacking T and B cells with dysfunctional NK cells) and Rag 2−/− Il 2rg−/− mice (lacking T, B, and NK cells) failed to mount a hypersensitivity response upon re-exposure to heptan (O’Leary et al. 2006). The adoptive transfer of pooled NK cells from heptan-sensitized Rag 2−/−mice to Rag2−/− Il2rg−/− results in vigorous hypersensitivity reaction upon rechallenge with the heptan122. Interestingly, adoptive transfer of NK cells from sensitized donor liver, rather than other organs, is enough to initiate a hypersensitivity reaction upon re-exposure to heptan (Paust et al. 2010). To add, memory-like NK subpopulation express the chemokine receptor CXCR6 that is essential for NK homing to the liver and acquisition of memory features (Paust et al. 2010). The same group demonstrated that NK cells have the capacity to mount a specific recall response in the form of a more effective and faster delayed hypersensitivity reaction against various viruses such as influenza, VSV, and HIV (Paust et al. 2010). The first evidence of Nk cell antivital memory phenotype was demonstrated in murine cytomegalovirus infection (Sun et al. 2009). The viral glycoprotein m 157, restricted to the MCMV-infected cells, is recognized by Ly49H receptor on NK cells and induces Ly49 H + NK cell activation and expansion in the face of MCMV infection. These NK cells follow a T cell behavior by contracting into a small long-lived and self-regenerating memory population that expand upon reactivation. MCMV rechallenge of previously sensitized mice led to a more efficient and a faster response of NK-specific cells (Sun et al. 2009). Similarly, human cytomegalovirus induces expansion of activating NKG2C receptor expressing NK cells (Gumá et al. 2004). Humans with deletion of one allele of KLRC2 which encodes for NKG2C receptors fail to expand NK cells during cytomegalovirus infection (Goodier et al. 2014). Although expansion of CD56 dim/CD16bright/NKG2C + K cells was detected in patients infected with HIV1, HCV, HBV, and EBV, those patients were all previously infected with HCMV, suggesting that this phenomenon is CMV specific rather than a random immune activation post viral infections (Bengsch et al. 2009; Béziat et al. 2012; Maria and Moretta 2008; Hendricks et al. 2014). Optimal natural killer cell memory necessitates not only antigen recognition but a proinflammatory cytokine signal (Sun et al. 2012). Though, cytokines are not required for NK cell maintenance at steady state (Sun et al. 2012). For instance, MCMV infection of mixed bone marrow chimeric mice, in which half of their hematopoietic cells express the IL12 R2 and the other half lack this receptor subunit, results in almost five-fold expansion of wild-type IL12 RB2 expressing natural killer cells compared to the deficient NK cells 7 days post infection in spleen and other nonlymphoid organs (Sun et al. 2012). Adoptive transfer of purified Ly49H + NK cells derived from a wild-type mouse and IL12rB−/− mice in equal numbers to a Dap 12 knockout mice (which do not express Ly49H on their NK cells) revealed 10–20- fold increase in the WT Lys49H + K cells when compared with IL12rB-deficient NK cells post MCMV infection. Further, adoptive transfer experiments showed than wild-type NK cells that express IL12 R B subunit were isolated 1 month after CMV infection, suggesting the essential role of the cytokine signal in NK memory generation (Sun et al. 2012). The memory-like behavior of NK cells can be mediated by proinflammatory cytokines such as IL12, IL-15, and IL-18 in an antigen-independent manner as opposed to the CMV-specific memory and in keeping with innate training feature. Inoculation of NK cells which are preactivated in vitro with IL12, IL-15, and IL-18 into Rag 2−/− mice and restimulation with cytokines lead to a higher expression of IFN gamma by NK cells which persist up to 12 weeks (Cooper et al. 2009).
However, the memory behavior of myeloid cells was limited to trained immunity, and not until recently, the ability of these cells to generate a specific memory phenotype was ventured. One of the first attempts at exploring memory-like phenomena in the vertebrate innate immune system was perhaps taken by Tribouly back in 1978 (Tribouley et al. 1978). Tribouly showed that vaccination of nude athymic mice, which lack T lymphocytes, with BCG provided protection against secondary infection with Candida albicans or Schistosoma mansoni (Tribouley et al. 1978). He suggested that the mechanism underlying this protective process is at least partially independent of T lymphocytes (Tribouley et al. 1978). In 1992, Vont’s Wout further showed that activated macrophages mediated this protection against re-infection with Candida albicans in BCG-vaccinated mice (van ‘t Wout et al. 1992). Further, Bistoni showed that athymic mice pretreated with the attenuated PCA-2 strain of C. albicans were protected against infections with virulent CA2 strain of C. albicans (Bistoni et al. 1988). These early findings shed light on the capacity of innate cells (non-T cells) to acquire one memory feature, heightened response upon re-exposure, but not another, specificity to the first encounter.
The innate immune system plays an important role early in the immune cascade that leads to allograft rejection (Kreisel et al. 2011). A self and non-self awareness by the innate cells is crucial to generate a response against the allograft. However, the mechanism of allorecognition by the innate immune system remained unclear for some time. Recognition of self by innate immune system seems to be independent of the activation of PRRs such as the TLRs, which are widely expressed by myeloid cells and are robust mediators of the anti-microbial response (Zhao et al. 2020; Abou-Daya and Oberbarnscheidt 2021). TLR signaling has been shown to be crucial for minor histocompatibility antigen-mismatched rejection (Goldstein et al. 2003), but is dispensable for the rejection of MHC-mismatched allografts (Tesar et al. 2004). The first insight on the ability of the innate immune system to sense and mount a response against non-self in a TLR-independent manner was provided by differential response to allogenic and syngeneic splenocyte immunization of RAG−/− mice that are devoid of T and B lymphocytes (Zecher et al. 2009). Only recipients of allogenic splenocytes demonstrated a delayed hypersensitivity like response. Further, adoptive transfer experiments and depletion experiments highlighted that monocytes were the cellular mediators of the innate allo-response in an MHC-independent fashion (Zecher et al. 2009; Oberbarnscheidt et al. 2014). Mouse genetic mapping studies comparing the monocyte response to congenic donors led to the identification of signal regulatory protein alpha (SIRPa) as a significant non-MHC allodeterminant (Barclay and Berg 2014). SIRPa belongs to the Ig superfamily, is polymorphic, and is expressed on monocytes and binds to the ubiquitous non-polymorphic CD47. SIRPa propagates an inhibitory response, whereas CD47 is stimulatory. SIRPa variation between donor and recipient leads to differential affinity to CD47 and therefore altering the activation status of the monocyte (Dai et al. 2017). Under homeostatic condition, a balance between stimulatory and inhibitory signaling is established. This balance is skewed towards activation when an allogenic tissue introduces a non-self SIRPalpha (Dai et al. 2017)—leading to monocyte differentiation to potent antigen-presenting dendritic cells.
In addition to their ability to recognize non-self, monocytes have the capacity to generate a memory response to previously encountered allo-antigens. Recently, our group successfully demonstrated an allo-specific, innate memory response in the setting of transplantation (Dai et al. 2020). Dai et al. took a murine experimental approach to investigate the role of PIRs in innate myeloid cell memory responses during allo-immunity. To achieve such aim, the authors establish an allogeneic mouse model using B6.RAG−/−.gcKO (H2b) mice as the host to study monocyte and macrophage memory responses independently of T, B, and NK cells. The mice were injected with irradiated splenocytes from syngeneic (B6) or allogeneic (Balb/c or C3H) mice followed by a bone marrow graft from a Balb/c allogeneic donor. Interestingly, the authors observed significantly high number of monocyte-derived dendritic cells (Mo-DCs) in the host mice immunized with Balb/c splenocytes compared to those immunized with syngeneic B6 or third-party allogeneic C3H splenocytes. Inflammatory Ly6Chi monocytes mediated the memory response. Monocyte allorecognition required an MHC-dependent and MHC-independent dual signal during the priming stage to generate a memory response. Blocking PIRA or genetic deletion of Pira prevented monocytes from acquiring memory. Further experiments showed that kidney allografts were able to survive for a long period of time in Pira−/− mice but were rapidly rejected in the inhibitory Pirb−/− mice in keeping with the inhibitory immunomodulatory function of PIRB. This elegant experimental approach showed that monocytes can recognize non-self MHC class I molecules and generate memory against them via interaction with the PIR system (Dai et al. 2020).
Concluding remarks
Based on mouse studies, understanding the mechanistic mysteries of innate immune memory in the myeloid lineage is starting to evolve. Multiple LILRs, the human orthologs of the mouse PIRs, have been shown to bind preferentially to MHC class 1 molecules, suggesting a possible involvement in monocytic allorecognition and development of innate immune memory in humans. The immunomodulatory function of LILRs is underlined by the functional dichotomy of this system of receptors between inhibitory and activating signals. This will allow LILRs on monocytes to orchestrate the immune response by integrating different signals and resulting in desirable but opposite responses such as innate immune memory to non-self and innate immune tolerance to self.
References
- Abou-Daya KI, Oberbarnscheidt MH (2021) Innate allorecognition in transplantation. J Heart Lung Transplant 40(7):557–561. 10.1016/j.healun.2021.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anfossi N, Doisne JM, Peyrat MA et al. (2004) Coordinated expression of Ig-like inhibitory MHC class I receptors and acquisition of cytotoxic function in human CD8+ T cells. J Immunol 173(12):7223–7229. 10.4049/jimmunol.173.12.7223 [DOI] [PubMed] [Google Scholar]
- An H, Chandra V, Piraino B et al. (2010) Soluble LILRA3, a potential natural antiinflammatory protein, is increased in patients with rheumatoid arthritis and is tightly regulated by interleukin 10, tumor necrosis factor-alpha, and interferon-gamma. J Rheumatol 37(8):1596–1606. 10.3899/jrheum.091119 [DOI] [PubMed] [Google Scholar]
- Arnon TI, Kaiser JT, West AP et al. (2008) The crystal structure of CHIR-AB1, a primordial avian classical Fc receptor. J Mol Biol 381(4):1012–1024. 10.1016/j.jmb.2008.06.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts RJW, Carvalho A, La Rocca C et al. (2016) Immunometabolic Pathways in BCG-Induced Trained Immunity. Cell Rep 17(10):2562–2571. 10.1016/j.celrep.2016.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri M, Zahmatkesh A (2018) Evolution and species-specific conservation of toll-like receptors in terrestrial vertebrates. Int Rev Immunol 37(5):217–228. 10.1080/08830185.2018.1506780 [DOI] [PubMed] [Google Scholar]
- Barclay AN, Van den Berg TK (2014) The interaction between signal regulatory protein alpha (SIRPα) and CD47: structure, function, and therapeutic target. Annu Rev Immunol 32:25–50. 10.1146/annurev-immunol-032713-120142 [DOI] [PubMed] [Google Scholar]
- Barrow AD, Trowsdale J (2008) The extended human leukocyte receptor complex: diverse ways of modulating immune responses. Immunol Rev 224(1):98–123. 10.1111/j.1600-065X.2008.00653.x [DOI] [PubMed] [Google Scholar]
- Bashirova AA, Martin-Gayo E, Jones DC et al. (2014) LILRB2 Interaction with HLA Class I Correlates with Control of HIV-1 Infection. Barsh GS, ed. PLoS Genet 10(3):e1004196. 10.1371/journal.pgen.1004196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Baruch-Morgenstern N, Shik D, Moshkovits I et al. (2014) Paired immunoglobulin-like receptor A is an intrinsic, self-limiting suppressor of IL-5-induced eosinophil development. Nat Immunol 15(1):36–44. 10.1038/ni.2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belge KU, Dayyani F, Horelt A et al. (2002) The Proinflammatory CD14+CD16+DR++ Monocytes Are a Major Source of TNF. J Immunol 168(7):3536–3542. 10.4049/jimmunol.168.7.3536 [DOI] [PubMed] [Google Scholar]
- Bengsch B, Thimme R, Blum HE (2009) Role of host genetic factors in the outcome of hepatitis C virus infection. Viruses 1(2):104–125. 10.3390/v1020104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béziat V, Dalgard O, Asselah T et al. (2012) CMV drives clonal expansion of NKG2C+ NK cells expressing self-specific KIRs in chronic hepatitis patients. Eur J Immunol 42(2):447–457. 10.1002/eji.201141826 [DOI] [PubMed] [Google Scholar]
- Bistoni F, Verducci G, Perito S et al. (1988) Immunomodulation by a low-virulence, agerminative variant of Candida albicans. Further evidence for macrophage activation as one of the effector mechanisms of nonspecific anti-infectious protection. J Med Vet Mycol 26(5):285–299. 10.1080/02681218880000401 [DOI] [PubMed] [Google Scholar]
- Bordon Y (2014) Innate memory training. Nat Rev Immunol 14(11):713–713. 10.1038/nri3759 [DOI] [PubMed] [Google Scholar]
- Bowdish DME, Loffredo MS, Mukhopadhyay S, Mantovani A, Gordon S (2007) Macrophage receptors implicated in the “adaptive” form of innate immunity. Microbes Infect 9(14–15):1680–1687. 10.1016/j.micinf.2007.09.002 [DOI] [PubMed] [Google Scholar]
- Boyette LB, Macedo C, Hadi K et al. (2017) Phenotype, function, and differentiation potential of human monocyte subsets. Zissel G, ed. PLoS One 12(4):e0176460. 10.1371/journal.pone.0176460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brites D, Du Pasquier L (2015) Somatic and Germline Diversification of a Putative Immunoreceptor within One Phylum: Dscam in Arthropods. In: Hsu E, Du Pasquier L, eds. Pathogen-Host Interactions: Antigenic Variation v. Somatic Adaptations. Results and Problems in Cell Differentiation Springer Int Publish; 131–158. 10.1007/978-3-319-20819-0_6 [DOI] [PubMed] [Google Scholar]
- Brümmendorf T, Lemmon V (2001) Immunoglobulin superfamily receptors: cis-interactions, intracellular adapters and alternative splicing regulate adhesion. Curr Opin Cell Biol 13(5):611–8. University of Miami. Accessed 17 Aug 2021. 10.1016/s0955-0674(00)00259-3. https://scholarship.miami.edu/esploro/outputs/bookReview/Immunoglobulin-superfamily-receptors-cis-interactions-intracellular-adapters-and-alternative-splicing-regulate-adhesion/991031562746502976 [DOI] [PubMed] [Google Scholar]
- Buchmann K (2014) Evolution of Innate Immunity: Clues from Invertebrates via Fish to Mammals. Front Immunol 5:459. 10.3389/fimmu.2014.00459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KS, Purdy AK (2011) Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology 132(3):315–325. 10.1111/j.1365-2567.2010.03398.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington M, O’Brien SJ (2003) The influence of HLA genotype on AIDS. Annu Rev Med 54:535–551. 10.1146/annurev.med.54.101601.152346 [DOI] [PubMed] [Google Scholar]
- Cella M, Döhring C, Samaridis J et al. (1997) A novel inhibitory receptor (ILT3) expressed on monocytes, macrophages, and dendritic cells involved in antigen processing. J Exp Med 185(10):1743–1751. 10.1084/jem.185.10.1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SC, Quintin J, Cramer RA et al. (2014) mTOR- and HIF-1α–mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345(6204):1250684. 10.1126/science.1250684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirovic B, de Bree LC, Groh L et al. (2020) BCG Vaccination in Humans Elicits Trained Immunity via the Hematopoietic Progenitor Compartment. Cell Host & Microbe 28(2):322–334.e5. 10.1016/j.chom.2020.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffaro KA, Hinegardner RT (1977) Immune response in the sea urchin Lytechinus pictus. Science 197(4311):1389–1390. 10.1126/science.331476 [DOI] [PubMed] [Google Scholar]
- Coindre S, Tchitchek N, Alaoui L et al. (2018) Mass Cytometry Analysis Reveals the Landscape and Dynamics of CD32a+ CD4+ T Cells From Early HIV Infection to Effective cART. Front Immunol 9:1217. 10.3389/fimmu.2018.01217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M, Navarro F, Bellón T et al. (1997) A Common Inhibitory Receptor for Major Histocompatibility Complex Class I Molecules on Human Lymphoid and Myelomonocytic Cells. J Exp Med 186(11):1809–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper EL, Roch P (1986) Second-set allograft responses in the earthworm Lumbricus terrestris. Kinetics and Characteristics Transplantation 41(4):514–520 [PubMed] [Google Scholar]
- Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM (2009) Cytokine-induced memory-like natural killer cells | PNAS . Proc Natl Acad Sci USA 106(6):1915–1919. Accessed Nov 2021. 10.1073/pnas.0813192106. https://www.pnas.org/content/106/6/1915?ijkey=1d4c6d00b2da9afc55def73f10ec3eccbff2964d&keytype2=tf_ipsecsha [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Friday AJ, Abou-Daya KI et al. (2017) Donor SIRPα polymorphism modulates the innate immune response to allogeneic grafts. Sci Immunol 2(12):eaam6202. 10.1126/sciimmunol.aam6202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Lan P, Zhao D et al. (2020) PIRs mediate innate myeloid cell memory to nonself MHC molecules. Science 368(6495):1122–1127. 10.1126/science.aax4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Magalhães JP, Church GM (2007) Analyses of human-chimpanzee orthologous gene pairs to explore evolutionary hypotheses of aging. Mech Ageing Dev 128(5–6):355–364. 10.1016/j.mad.2007.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maria A, Moretta L (2008) NK cell function in HIV-1 infection. Curr HIV Res 6(5):433–440. 10.2174/157016208785861221 [DOI] [PubMed] [Google Scholar]
- Dennis G, Stephan RP, Kubagawa H, Cooper MD (1999) Characterization of Paired Ig-Like Receptors in Rats. J Immunol 163(12):6371–6377 [PubMed] [Google Scholar]
- Divangahi M, Aaby P, Khader SA et al. (2021) Trained immunity, tolerance, priming and differentiation: distinct immunological processes. Nat Immunol 22(1):2–6. 10.1038/s41590-020-00845-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Andrés J, Fanucchi S, Joosten LAB, Mhlanga MM, Netea MG (2020) Advances in understanding molecular regulation of innate immune memory. Curr Op in Cell Bio 63:68–75. 10.1016/j.ceb.2019.12.006 [DOI] [PubMed] [Google Scholar]
- Domínguez-Andrés J, Netea MG (2020) The specifics of innate immune memory. Science 368(6495):1052–1053. 10.1126/science.abc2660 [DOI] [PubMed] [Google Scholar]
- Dong Y, Taylor HE, Dimopoulos G (2006) AgDscam, a hypervariable immunoglobulin domain-containing receptor of the anopheles gambiae innate immune system. PLoS Biol 4(7):e229. 10.1371/journal.pbio.0040229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanger NA, Cosman D, Peterson L, Braddy SC, Maliszewski CR, Borges L (1998) The MHC class I binding proteins LIR-1 and LIR-2 inhibit Fc receptor-mediated signaling in monocytes. Eur J Immunol 28:3423–3434. Accessed 1 Sept 2021. [DOI] [PubMed] [Google Scholar]
- Fayngerts SA, Najakshin AM, Taranin AV (2007) Species-specific evolution of the FcR family in endothermic vertebrates. Immunogenetics 59(6):493–506. 10.1007/s00251-007-0208-8. | SpringerLink. Accessed 2 Nov 2021. https://link.springer.com/article/10.1007%2Fs00251-007-0208-8 [DOI] [PubMed] [Google Scholar]
- Ferro K, Peuß R, Yang W, Rosenstiel P, Schulenburg H, Kurtz J (2019) Experimental evolution of immunological specificity. Proc Natl Acad Sci U S A 116(41):20598–20604. 10.1073/pnas.1904828116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Vega N, Galindo-Rodríguez G, Bajaña S et al. (2006) Phenotypic analysis of IL-10-treated, monocyte-derived dendritic cells in patients with systemic lupus erythematosus. Scand J Immunol 64(6):668–676. 10.1111/j.1365-3083.2006.01849.x [DOI] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R (2007) Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447(7147):972–978. 10.1038/nature05836 [DOI] [PubMed] [Google Scholar]
- Geissmann F, Auffray C, Palframan R et al. (2008) Blood monocytes: distinct subsets, how they relate to dendritic cells, and their possible roles in the regulation of T-cell responses. Immunol Cell Biol 86(5):398–408. 10.1038/icb.2008.19 [DOI] [PubMed] [Google Scholar]
- Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K (2010) Development of Monocytes, Macrophages, and Dendritic Cells. Science 327(5966):656–661. 10.1126/science.1178331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DR, Tesar BM, Akira S, Lakkis FG (2003) Critical role of the Toll-like receptor signal adaptor protein MyD88 in acute allograft rejection. J Clin Invest 111(10):1571–1578. 10.1172/JCI17573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier MR, White MJ, Darboe A et al. (2014) Rapid NK cell differentiation in a population with near-universal human cytomegalovirus infection is attenuated by NKG2C deletions. Blood 124(14):2213–2222. 10.1182/blood-2014-05-576124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge JP, Witt CS, Christiansen FT, Warren HS (2003) KIR2DL4 (CD158d) genotype influences expression and function in NK cells. J Immunol 171(4):1768–1774. 10.4049/jimmunol.171.4.1768 [DOI] [PubMed] [Google Scholar]
- Gorbachev AV, Fairchild RL (2001) Induction and regulation of T-cell priming for contact hypersensitivity. Crit Rev Immunol 21(5):451–472 [PubMed] [Google Scholar]
- Gourbal B, Pinaud S, Beckers GJM, Van Der Meer JWM, Conrath U, Netea MG (2018) Innate immune memory: An evolutionary perspective. Immunol Rev 283(1):21–40. 10.1111/imr.12647 [DOI] [PubMed] [Google Scholar]
- Gren ST, Rasmussen TB, Janciauskiene S, Håkansson K, Gerwien JG, Grip O (2015) A Single-Cell Gene-Expression Profile Reveals Inter-Cellular Heterogeneity within Human Monocyte Subsets. PLoS One 10(12):e0144351. 10.1371/journal.pone.0144351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M, Mildner A, Yona S (2018) Developmental and Functional Heterogeneity of Monocytes. Immunity 49(4):595–613. 10.1016/j.immuni.2018.10.005 [DOI] [PubMed] [Google Scholar]
- Gumá M, Angulo A, Vilches C, Gómez-Lozano N, Malats N, López-Botet M (2004) Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 104(12):3664–3671. 10.1182/blood-2004-05-2058 [DOI] [PubMed] [Google Scholar]
- Günther P, Cirovic B, Baßler K et al. (2019) A Rule-Based Data-Informed Cellular Consensus Map of the Human Mononuclear Phagocyte Cell Space 658179. 10.1101/658179 [DOI]
- Guselnikov SV, Ramanayake T, Erilova AY et al. (2008) The Xenopus FcR family demonstrates continually high diversification of paired receptors in vertebrate evolution. BMC Evol Biol 8:148. 10.1186/1471-2148-8-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaby DM, Mornon JPE (1998) The immunoglobulin superfamily: An insight on its tissular, species, and functional diversity. J Mol Evol 46(4):389–400. 10.1007/PL00006318 [DOI] [PubMed] [Google Scholar]
- Hammond JA, Guethlein LA, Abi-Rached L, Moesta AK, Parham P (2009) Evolution and Survival of Marine Carnivores Did Not Require a Diversity of Killer Cell Ig-Like Receptors or Ly49 NK Cell Receptors. J Immunol 182(6):3618–3627. 10.4049/jimmunol.0803026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks DW, Balfour HH, Dunmire SK, Schmeling DO, Hogquist KA, Lanier LL (2014) Cutting edge: NKG2C(hi)CD57+ NK cells respond specifically to acute infection with cytomegalovirus and not Epstein-Barr virus. J Immunol 192(10):4492–4496. 10.4049/jimmunol.1303211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández Prada JA, Haire RN, Allaire M et al. (2006) Ancient evolutionary origin of diversified variable regions demonstrated by crystal structures of an immune-type receptor in amphioxus. Nat Immunol 7(8):875–882. 10.1038/ni1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildemann WH, Johnson IS, Jokiel PL (1979) Immunocompetence in the lowest metazoan phylum: transplantation immunity in sponges. Science 204(4391):420–422. 10.1126/science.441730 [DOI] [PubMed] [Google Scholar]
- Hirayasu K, Arase H (2015) Functional and genetic diversity of leukocyte immunoglobulin-like receptor and implication for disease associations. J Hum Genet 60(11):703–708. 10.1038/jhg.2015.64 [DOI] [PubMed] [Google Scholar]
- Hirayasu K, Saito F, Suenaga T et al. (2016) Microbially cleaved immunoglobulins are sensed by the innate immune receptor LILRA2. Nat Microbiol 1(6):16054. 10.1038/nmicrobiol.2016.54 [DOI] [PubMed] [Google Scholar]
- Hogan L, Bhuju S, Jones DC et al. (2012) Characterisation of Bovine Leukocyte Ig-like Receptors. PLoS One 7(4):e34291. 10.1371/journal.pone.0034291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Goedert JJ, Sundberg EJ et al. (2009) HLA-B*35-Px-mediated acceleration of HIV-1 infection by increased inhibitory immunoregulatory impulses. J Exp Med 206(13):2959–2966. 10.1084/jem.20091386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Burke PS, Cung TDH et al. (2010) Leukocyte immunoglobulin-like receptors maintain unique antigen-presenting properties of circulating myeloid dendritic cells in HIV-1-infected elite controllers. J Virol 84(18):9463–9471. 10.1128/JVI.01009-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifrim DC, Quintin J, Joosten LAB et al. (2014) Trained Immunity or Tolerance: Opposing Functional Programs Induced in Human Monocytes after Engagement of Various Pattern Recognition Receptors. Papasian CJ, ed. Clin Vaccine Immunol 21(4):534–545. 10.1128/CVI.00688-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DC, Kosmoliaptsis V, Apps R et al. (2011) HLA class I allelic sequence and conformation regulate leukocyte Ig-like receptor binding. J Immunol 186(5):2990–2997. 10.4049/jimmunol.1003078 [DOI] [PubMed] [Google Scholar]
- Kachroo A, Robin GP (2013) Systemic signaling during plant defense. Curr Opin Plant Biol 16(4):527–533. 10.1016/j.pbi.2013.06.019 [DOI] [PubMed] [Google Scholar]
- Kaslow RA, Carrington M, Apple R et al. (1996) Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med 2(4):405–411. 10.1038/nm0496-405 [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S (2010) The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11(5):373–384. 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- Kollnberger S, Bird L, Sun MY et al. (2002) Cell-surface expression and immune receptor recognition of HLA-B27 homodimers. Arthritis Rheum 46(11):2972–2982. 10.1002/art.10605 [DOI] [PubMed] [Google Scholar]
- Kreisel D, Sugimoto S, Zhu J et al. (2011) Emergency granulopoiesis promotes neutrophil-dendritic cell encounters that prevent mouse lung allograft acceptance. Blood 118(23):6172–6182. 10.1182/blood-2011-04-347823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki K, Hirose K, Okabe Y et al. (2013) The long-term immunosuppressive effects of disulfide-linked HLA-G dimer in mice with collagen-induced arthritis. Hum Immunol 74(4):433–438. 10.1016/j.humimm.2012.11.060 [DOI] [PubMed] [Google Scholar]
- Kurtz J (2005) Specific memory within innate immune systems. Trends Immunol 26(4):186–192. 10.1016/j.it.2005.02.001 [DOI] [PubMed] [Google Scholar]
- Lakkis FG, Dellaporta SL, Buss LW (2008) Allorecognition and chimerism in an invertebrate model organism. Organogenesis 4(4):236–240. 10.4161/org.4.4.7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Tam H, Adler L, Ilstad-Minnihan A, Macaubas C, Mellins ED (2017) The MHC class II antigen presentation pathway in human monocytes differs by subset and is regulated by cytokines. PLoS One 12(8):e0183594. 10.1371/journal.pone.0183594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichterfeld M, Kavanagh DG, Williams KL et al. (2007) A viral CTL escape mutation leading to immunoglobulin-like transcript 4–mediated functional inhibition of myelomonocytic cells. J Exp Med 204(12):2813–2824. 10.1084/jem.20061865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litman GW, Rast JP, Fugmann SD (2010) The origins of vertebrate adaptive immunity. Nat Rev Immunol 10(8):543–553. 10.1038/nri2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E, Bruce TJA, Roberts MR, Flors V, Ton J (2012) Next-generation systemic acquired resistance. Plant Physiol 158(2):844–853. 10.1104/pp.111.187468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E, Ton J (2012) The epigenetic machinery controlling transgenerational systemic acquired resistance. Plant Signal Behav 7(6):615–618. 10.4161/psb.20155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane AW, Campbell KS (2006) Signal transduction in natural killer cells. Curr Top Microbiol Immunol 298:23–57. 10.1007/3-540-27743-9_2 [DOI] [PubMed] [Google Scholar]
- Malnati MS, Peruzzi M, Parker KC et al. (1995) Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science 267(5200):1016–1018. 10.1126/science.7863326 [DOI] [PubMed] [Google Scholar]
- Manavalan JS, Rossi PC, Vlad G et al. (2003) High expression of ILT3 and ILT4 is a general feature of tolerogenic dendritic cells. Transpl Immunol 11(3):245–258. 10.1016/S0966-3274(03)00058-3 [DOI] [PubMed] [Google Scholar]
- Marsh SGE, Parham P, Dupont B et al. (2003) Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Immunogenetics 55(4):220–226. 10.1007/s00251-003-0571-z [DOI] [PubMed] [Google Scholar]
- Martin AM, Kulski JK, Witt C, Pontarotti P, Christiansen FT (2002) Leukocyte Ig-like receptor complex (LRC) in mice and men. Trends Immunol 23(2):81–88. 10.1016/s1471-4906(01)02155-x [DOI] [PubMed] [Google Scholar]
- Martinez FO, Helming L, Gordon S (2009) Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol 27:451–483. 10.1146/annurev.immunol.021908.132532 [DOI] [PubMed] [Google Scholar]
- Medzhitov R (2001) Toll-like receptors and innate immunity. Nat Rev Immunol 1(2):135–145. 10.1038/35100529 [DOI] [PubMed] [Google Scholar]
- Melillo D, Marino R, Italiani P, Boraschi D (2018) Innate Immune Memory in Invertebrate Metazoans: A Critical Appraisal. Front Immunol 9:1915. 10.3389/fimmu.2018.01915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meziane EK, Potts ND, Viertlboeck BC et al. (2019) Bi-Functional Chicken Immunoglobulin-Like Receptors With a Single Extracellular Domain (ChIR-AB1): Potential Framework Genes Among a Relatively Stable Number of Genes Per Haplotype. Front Immunol 10:2222. 10.3389/fimmu.2019.02222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsiváis-Urenda A, Niño-Moreno P, Abud-Mendoza C et al. (2007) Analysis of expression and function of the inhibitory receptor ILT2 (CD85j/LILRB1/LIR-1) in peripheral blood mononuclear cells from patients with systemic lupus erythematosus (SLE). J Autoimmun 29(2–3):97–105. 10.1016/j.jaut.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Nakaya HI, Wrammert J, Lee EK et al. (2011) Systems Biology of Seasonal Influenza Vaccination in Humans. Nat Immunol 12(8):786–795. 10.1038/ni.2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Joosten LAB, Latz E et al. (2016) Trained immunity: A program of innate immune memory in health and disease. Science 352(6284):aaf1098–aaf1098. 10.1126/science.aaf1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Schlitzer A, Placek K, Joosten LAB, Schultze JL (2019) Innate and Adaptive Immune Memory: an Evolutionary Continuum in the Host’s Response to Pathogens. Cell Host Microbe 25(1):13–26. 10.1016/j.chom.2018.12.006 [DOI] [PubMed] [Google Scholar]
- Nie L, Cai SY, Shao JZ, Chen J (2018) Toll-Like Receptors, Associated Biological Roles, and Signaling Networks in Non-Mammals. Front Immunol 9:1523. 10.3389/fimmu.2018.01523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaidis N, Makalowska I, Chalkia D, Makalowski W, Klein J, Nei M (2005) Origin and evolution of the chicken leukocyte receptor complex. PNAS 102(11):4057–4062. 10.1073/pnas.0501040102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Graf J (2012) Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat Rev Microbiol 10(12):815–827. 10.1038/nrmicro2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberbarnscheidt MH, Zeng Q, Li Q et al. (2014) Non-self recognition by monocytes initiates allograft rejection. J Clin Invest 124(8):3579–3589. 10.1172/JCI74370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor GM, Holmes A, Mulcahy F, Gardiner CM (2007) Natural Killer cells from long-term non-progressor HIV patients are characterized by altered phenotype and function. Clin Immunol 124(3):277–283. 10.1016/j.clim.2007.05.016 [DOI] [PubMed] [Google Scholar]
- O’Leary JG, Goodarzi M, Drayton DL, von Andrian UH (2006) T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol 7(5):507–516. 10.1038/ni1332 [DOI] [PubMed] [Google Scholar]
- Passlick B, Flieger D, Ziegler-Heitbrock HW (1989) Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood 74(7):2527–2534 [PubMed] [Google Scholar]
- Paust S, Gill HS, Wang BZ et al. (2010) Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol 11(12):1127–1135. 10.1038/ni.1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende D, Falco M, Vitale M et al. (2019) Killer Ig-Like Receptors (KIRs): Their Role in NK Cell Modulation and Developments Leading to Their Clinical Exploitation. Front Immunol 10:1179. 10.3389/fimmu.2019.01179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peruzzi M, Wagtmann N, Long EO (1996) A p70 killer cell inhibitory receptor specific for several HLA-B allotypes discriminates among peptides bound to HLA-B*2705. J Exp Med 184(4):1585–1590. 10.1084/jem.184.4.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham LN, Dionne MS, Shirasu-Hiza M, Schneider DS (2007) A Specific Primed Immune Response in Drosophila Is Dependent on Phagocytes. PLoS Pathog 3(3):e26. 10.1371/journal.ppat.0030026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira S, Zhang H, Takai T, Lowell CA (2004) The Inhibitory Receptor PIR-B Negatively Regulates Neutrophil and Macrophage Integrin Signaling. J Immunol 173(9):5757–5765. 10.4049/jimmunol.173.9.5757 [DOI] [PubMed] [Google Scholar]
- Petroff MG, Sedlmayr P, Azzola D, Hunt JS (2002) Decidual macrophages are potentially susceptible to inhibition by class Ia and class Ib HLA molecules. J Reprod Immunol 56(1):3–17. 10.1016/S0165-0378(02)00024-4 [DOI] [PubMed] [Google Scholar]
- Pfistershammer K, Lawitschka A, Klauser C et al. (2009) Allogeneic disparities in immunoglobulin-like transcript 5 induce potent antibody responses in hematopoietic stem cell transplant recipients. Blood 114(11):2323–2332. 10.1182/blood-2008-10-183814 [DOI] [PubMed] [Google Scholar]
- Powell AE, Nicotra ML, Moreno MA, Lakkis FG, Dellaporta SL, Buss LW (2007) Differential Effect of Allorecognition Loci on Phenotype in Hydractinia symbiolongicarpus (Cnidaria: Hydrozoa). Genetics 177(4):2101–2107. 10.1534/genetics.107.075689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues J, Brayner FA, Alves LC, Dixit R, Barillas-Mury C (2010) Hemocyte Differentiation Mediates Innate Immune Memory in Anopheles gambiae Mosquitoes. Science 329(5997):1353–1355. 10.1126/science.1190689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosental B, Kowarsky M, Seita J et al. (2018) Complex mammalian-like haematopoietic system found in a colonial chordate. Nature 564(7736):425–429. 10.1038/s41586-018-0783-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaridis J, Colonna M (1997) Cloning of novel immunoglobulin superfamily receptors expressed on human myeloid and lymphoid cells: Structural evidence for new stimulatory and inhibitory pathways. Eur J Immunol 27(3):660–665. 10.1002/eji.1830270313 [DOI] [PubMed] [Google Scholar]
- Saverino D, Merlo A, Bruno S, Pistoia V, Grossi CE, Ciccone E (2002) Dual Effect of CD85/Leukocyte Ig-Like Receptor-1/Ig-Like Transcript 2 and CD152 (CTLA-4) on Cytokine Production by Antigen-Stimulated Human T Cells. J Immunol 168(1):207–215. 10.4049/jimmunol.168.1.207 [DOI] [PubMed] [Google Scholar]
- Schmidl C, Renner K, Peter K et al. (2014) Transcription and enhancer profiling in human monocyte subsets. Blood 123(17):e90–e99. 10.1182/blood-2013-02-484188 [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Sanderson ND, Bickhart DM, Smith TPL, Hammond JA (2019) The Structure, Evolution, and Gene Expression Within the Caprine Leukocyte Receptor Complex. Front Immunol 10:2302. 10.3389/fimmu.2019.02302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield VL, Schlumpberger JM, West LA, Weissman IL (1982) Protochordate allorecognition is controlled by a MHC-like gene system. Nature 295(5849):499–502. 10.1038/295499a0 [DOI] [PubMed] [Google Scholar]
- Seeley JJ, Ghosh S (2017) Molecular mechanisms of innate memory and tolerance to LPS. J Leukoc Biol 101(1):107–119. 10.1189/jlb.3MR0316-118RR [DOI] [PubMed] [Google Scholar]
- Shortman K, Liu YJ (2002) Mouse and human dendritic cell subtypes. Nat Rev Immunol 2(3):151–161. 10.1038/nri746 [DOI] [PubMed] [Google Scholar]
- Skrzeczyńska-Moncznik J, Bzowska M, Loseke S, Grage-Griebenow E, Zembala M, Pryjma J (2008) Peripheral blood CD14high CD16+ monocytes are main producers of IL-10. Scand J Immunol 67(2):152–159. 10.1111/j.1365-3083.2007.02051.x [DOI] [PubMed] [Google Scholar]
- Smith CL, Dickinson P, Forster T et al. (2014) Identification of a human neonatal immune-metabolic network associated with bacterial infection. Nat Commun 5:4649. 10.1038/ncomms5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford JL, Bengtén E, Du Pasquier L et al. (2006) A novel family of diversified immunoregulatory receptors in teleosts is homologous to both mammalian Fc receptors and molecules encoded within the leukocyte receptor complex. Immunogenetics 58(9):758–773. 10.1007/s00251-006-0134-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford JL, Bengtén E, Du Pasquier L, Miller NW, Wilson M (2007) Channel catfish leukocyte immune-type receptors contain a putative MHC class I binding site. Immunogenetics 59(1):77–91. 10.1007/s00251-006-0169-3 [DOI] [PubMed] [Google Scholar]
- Stewart CA, Laugier-Anfossi F, Vély F et al. (2005) Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A 102(37):13224–13229. 10.1073/pnas.0503594102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suciu-Foca N, Feirt N, Zhang QY et al. (2007) Soluble Ig-Like Transcript 3 Inhibits Tumor Allograft Rejection in Humanized SCID Mice and T Cell Responses in Cancer Patients. J Immunol 178(11):7432–7441. 10.4049/jimmunol.178.11.7432 [DOI] [PubMed] [Google Scholar]
- Sun JC, Madera S, Bezman NA, Beilke JN, Kaplan MH, Lanier LL (2012) Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J Exp Med 209(5):947–954. 10.1084/jem.20111760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Beilke JN, Lanier LL (2009) Adaptive immune features of natural killer cells. Nature 457(7229):557–561. 10.1038/nature07665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai T (2005) Paired immunoglobulin-like receptors and their MHC class I recognition. Immunology 115(4):433–440. 10.1111/j.1365-2567.2005.02177.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar BM, Zhang J, Li Q, Goldstein DR (2004) TH1 immune responses to fully MHC mismatched allografts are diminished in the absence of MyD88, a toll-like receptor signal adaptor protein. Am J Transplant 4(9):1429–1439. 10.1111/j.1600-6143.2004.00544.x [DOI] [PubMed] [Google Scholar]
- Tribouley J, Tribouley-Duret J, Appriou M (1978) Effect of Bacillus Callmette Guerin (BCG) on the receptivity of nude mice to Schistosoma mansoni. C R Seances Soc Biol Fil 172(5):902–904 [PubMed] [Google Scholar]
- Ujike A, Takeda K, Nakamura A, Ebihara S, Akiyama K, Takai T (2002) Impaired dendritic cell maturation and increased TH2 responses in PIR-B−/− mice. Nat Immunol 3(6):542–548. 10.1038/ni801 [DOI] [PubMed] [Google Scholar]
- van ‘t Wout JW, Poell R, van Furth R (1992) The role of BCG/PPD-activated macrophages in resistance against systemic candidiasis in mice. Scand J Immunol 36(5):713–719. 10.1111/j.1365-3083.1992.tb03132.x [DOI] [PubMed] [Google Scholar]
- Viertlboeck BC, Habermann FA, Schmitt R, Groenen MAM, Pasquier LD, Göbel TW (2005) The Chicken Leukocyte Receptor Complex: A Highly Diverse Multigene Family Encoding at Least Six Structurally Distinct Receptor Types. J Immunol 175(1):385–393. 10.4049/jimmunol.175.1.385 [DOI] [PubMed] [Google Scholar]
- Villani AC, Satija R, Reynolds G et al. (2017) Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356(6335):eaah4573. 10.1126/science.aah4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad G, Piazza F, Colovai A et al. (2003) Interleukin-10 induces the upregulation of the inhibitory receptor ILT4 in monocytes from HIV positive individuals. Hum Immunol 64(5):483–489. 10.1016/s0198-8859(03)00040-5 [DOI] [PubMed] [Google Scholar]
- Vlisidou I, Wood W (2015) Drosophila blood cells and their role in immune responses. FEBS J 282(8):1368–1382. 10.1111/febs.13235 [DOI] [PubMed] [Google Scholar]
- Wang J, Belosevic M, Stafford JL (2021) Identification of distinct LRC- and Fc receptor complex-like chromosomal regions in fish supports that teleost leukocyte immune-type receptors are distant relatives of mammalian Fc receptor-like molecules. Immunogenetics 73(1):93–109. 10.1007/s00251-020-01193-3 [DOI] [PubMed] [Google Scholar]
- Watson FL, Püttmann-Holgado R, Thomas F et al. (2005) Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science 309(5742):1874–1878. 10.1126/science.1116887 [DOI] [PubMed] [Google Scholar]
- Wolf AA, Yáñez A, Barman PK, Goodridge HS (2019) The Ontogeny of Monocyte Subsets. Front Immunol 10:1642. 10.3389/fimmu.2019.01642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KL, Tai JJY, Wong WC et al. (2011) Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood 118(5):e16–e31. 10.1182/blood-2010-12-326355 [DOI] [PubMed] [Google Scholar]
- Young NT, Canavez F, Uhrberg M, Shum BP, Parham P (2001) Conserved organization of the ILT/LIR gene family within the polymorphic human leukocyte receptor complex. Immunogenetics 53(4):270–278. 10.1007/s002510100332 [DOI] [PubMed] [Google Scholar]
- Zecher D, van Rooijen N, Rothstein DM, Shlomchik WD, Lakkis FG (2009) An innate response to allogeneic nonself mediated by monocytes. J Immunol 183(12):7810–7816. 10.4049/jimmunol.0902194 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hatano H, Shaw J et al. (2015) The Leukocyte Immunoglobulin-Like Receptor Family Member LILRB5 Binds to HLA-Class I Heavy Chains. PLoS One 10(6):e0129063. 10.1371/journal.pone.0129063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Mai S, Chen HM et al. (2017) Leukocyte immunoglobulin-like receptors in human diseases: an overview of their distribution, function, and potential application for immunotherapies. J Leukoc Biol 102(2):351–360. 10.1189/jlb.5MR1216-534R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Abou-Daya KI, Dai H, Oberbarnscheidt MH, Li XC, Lakkis FG (2020) Innate Allorecognition and Memory in Transplantation. Front Immunol 11:918. 10.3389/fimmu.2020.00918 [DOI] [PMC free article] [PubMed] [Google Scholar]


