Abstract
Bladder cancer (BC) is a clinical challenge worldwide with late clinical presentation, poor prognosis, and low survival rates. Traditional cystoscopy and tissue biopsy are routine methods for the diagnosis, prognosis, and monitoring of BC. However, due to the heterogeneity and limitations of tumors, such as aggressiveness, high cost, and limited applicability of longitudinal surveillance, the identification of tumor markers has attracted significant attention in BC. Over the past decade, liquid biopsies (e.g., blood) have proven to be highly efficient methods for the discovery of BC biomarkers. This noninvasive sampling method is used to analyze unique tumor components released into the peripheral circulation and allows serial sampling and longitudinal monitoring of tumor progression. Several liquid biopsy biomarkers are being extensively studied and have shown promising results in clinical applications of BC, including early detection, detection of microscopic residual disease, prediction of recurrence, and response to therapy. Therefore, in this review, we aim to provide an update on various novel blood-based liquid biopsy markers and review the advantages and current limitations of liquid biopsy in BC therapy. The role of blood-based circulating tumor cells, circulating tumor DNA, cell-free RNA, exosomes, metabolomics, and proteomics in diagnosis, prognosis, and treatment monitoring, and their applicability to the personalized management of BC, are highlighted.
Keywords: Bladder cancer, Liquid biopsy, Circulating tumor cells, Circulating tumor DNA, Cell-free RNA, Exosomes, Metabolomics, Proteomics, Clinical application
Introduction
Bladder cancer (BC) poses a huge social burden, with a global yearly occurrence of more than 430,000 cases, resulting in nearly 170,000 deaths annually [1]. It is a highly heterogeneous malignancy, especially in advanced stages [2]. BC can present as non-muscle-invasive bladder cancer (NMIBC), muscle-invasive bladder cancer (MIBC), or metastatic disease incidents, each with different molecular drivers. NMIBC accounts for approximately 75% of all BC types, and at least half of patients with NMIBC have a high rate of recurrence and disease progression to MIBC within 5 years [3]. Additionally, MIBC has a high propensity to spread to lymph nodes and other organs, and approximately half of patients with MIBC develop metastases and die within 3 years [4, 5]. The high morbidity and mortality of BC is in part associated with the lack of effective early diagnosis and prognostic approaches. Therefore, early diagnosis and lifelong surveillance are clinically important to improve the long-term survival of patients with BC.
Currently, invasive cystoscopy and tissue biopsy remain the gold standard for BC identification and surveillance. However, the drawbacks of this method, such as sampling bias, invasiveness, and difficulty in sampling deep tumors, limit its use in large-scale screening. Additionally, urine cytology represents another diagnostic possibility but is limited by poor sensitivity, especially for low-grade tumors [6, 7]. Other imaging tests are limited to BC staging in clinical practice owing to increased ionizing radiation and/or cost, and there is often a delay in identifying recurrence and metastasis with imaging [8]. Therefore, there is an urgent need for biomarkers with high specificity and sensitivity that can be applied to BC.
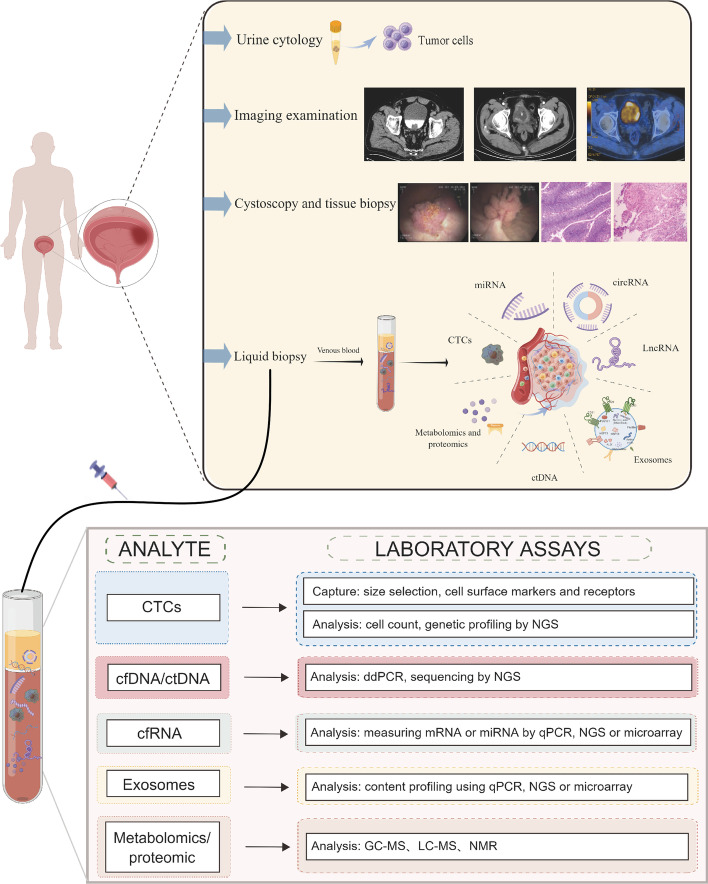
Recently, genomic profiles of blood and body fluids based on liquid biopsies have been shown to be highly correlated with the genomic profiles of tumors [9–11]. DNA fragments containing tumor-specific alterations, including information about point mutations, DNA methylation, and copy number variation, are released into circulation by tumors. The dynamic assessment of specific molecular markers by liquid biopsy allows for real-time monitoring of tumors, thus facilitating the selection of targeted therapies. In contrast to cystoscopy and tissue biopsy, liquid biopsy is a holistic examination of blood and body fluids, reflecting the overall expression of tumor cells [12]. Figure 1 summarizes the current testing methods applied to BC and the laboratory analytical techniques of liquid biopsy [13, 14].
Fig. 1.
Overview of clinical examination methods for bladder cancer and laboratory analytical techniques for liquid biopsies. Routine imaging tests used for bladder cancer detection and diagnosis include X-ray, CT, and PET–CT. Cystoscopy and tissue biopsy are the gold standard for the diagnosis of bladder cancer. Urine cytology is available as a complementary test. Liquid biopsy is emerging as a promising method. Liquid biopsy involves the collection and analysis of five different tumor components from peripheral blood samples: CTCs, cell-free nucleic acids (cfDNA/ctDNA, cfRNA), exosomes, and metabolomics and proteomics. Tumor components are then captured and analyzed in peripheral blood samples using appropriate laboratory assays. CT computed tomography, PET positron emission computed tomography, CTCs circulating tumor cells, ctDNA circulating tumor deoxyribonucleic acid, cfDNA cell-free DNA, cfRNA cell-free RNA
Liquid biopsy is based on the detection of circulating tumor cells (CTCs) and “cell-free” nucleic acids in blood and body fluids, including urine, saliva, tears, sweat, amniotic fluid, cerebrospinal and pleural fluids, and cervicovaginal secretions, to detect and quantify targets of interest [13, 14]. Significant progress has been made in the detection of urinary biomarkers for BC, but it is currently limited by its low sensitivity and specificity [15, 16]. Since blood is in contact with most tumors, liquid biopsies mainly involve blood sampling. Liquid biopsies contribute to better characterization of BC by identifying tumor cells or tumor DNA that is released into the bloodstream with tumor progression or regression [17]. Meanwhile, unlike urine, the blood-based biomarkers or liquid biopsies can be applied to all BC, but may be particularly useful after cystectomy for curative purposes, providing a strategy for detecting minimal residual disease (MRD) prior to conventional imaging tests. The highly sensitive technique of blood testing provides detail of tumor characteristics, and blood-based cancer genomics may not be limited by sampling frequency, tumor availability, or the presence of a clinically significant disease compared with tissue sampling.
Historically, it is difficult to determine potentially recurrent or successfully cured patients with BC after surgery. This limitation has resulted in the unnecessary exposure of successfully cured patients to the toxicity of adjuvant therapy, whereas others with residual disease may not receive potentially beneficial treatment until disease progression can be detected by imaging. A comprehensive understanding of the subgroup of patients predicted to become resistant to cisplatin is essential for patient treatment modalities [18]. The advent of immunotherapy, especially immune checkpoint inhibitors (ICIs), has revolutionized the landscape of BC systemic therapy over the past 5 years. However, ICIs provide durable immune memory in only a minority of patients [19]; additionally, most patients exhibit poor or no response to ICIs, and those who initially respond eventually develop resistance and disease progression. Moreover, ICIs are expensive and impose a significant financial burden on patients. The occurrence of immune-related adverse effects can be fatal and may lead to permanent disease [20]. Therefore, there is a need to use predictive biomarkers of response to ICIs to assess early efficacy and toxicity of drugs and to better understand resistance mechanisms to guide therapeutic decisions. Studies have shown that liquid biopsies can be applied to comprehensively examine tumor genome and monitor genetic variation dynamically during treatment [21–23]. Overall, liquid biopsy can help in the selection of patients for immunotherapy.
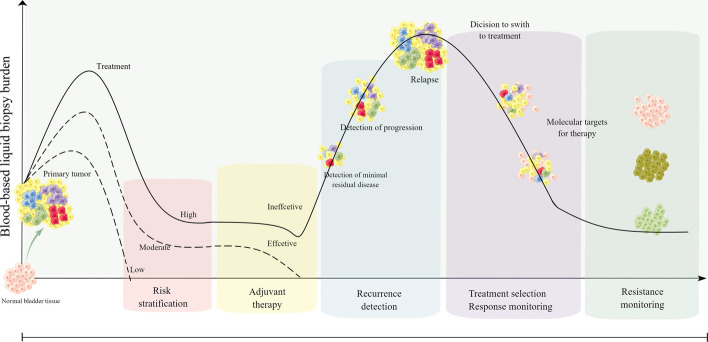
Currently, liquid biopsies usually detect CTCs, exosomes, circulating tumor deoxyribonucleic acid (ctDNA), microRNAs (miRNAs), long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), extracellular vesicles (EVs), tumor-educated platelets, proteins, and metabolites [23–25]. New evidence for the clinical utility of liquid biopsies from noninvasive blood tests in BC continues to emerge and may provide a genomic profile that can be utilized in patients with BC and guide their treatment in some ways (Fig. 2). The purpose of this review is to discuss the latest information on liquid biopsy components and their clinical utility for early diagnosis of BC, prognosis, monitoring of cancer treatment outcomes, detection of early recurrence, and individualized treatment selection.
Fig. 2.
Overview of the clinical use of liquid biopsy in bladder cancer. Sample analysis of bladder cancer biomarkers early in the disease process allows for early diagnosis and risk stratification. After receiving surgery, liquid biopsy can detect minimal residual disease as a prognostic indicator and allow early detection of recurrent disease. During treatment, liquid biopsy can enhance longitudinal surveillance through its noninvasive approach, thus enabling continuous sampling. In addition, liquid biopsies have the advantage of capturing the entire tumor genome, which can help identify novel genetic markers for targeted therapy and detect treatment resistance
CTC
The concept of CTCs was first described by the Australian scholar Ashworth in 1869, and it was defined as tumor cells shed from primary tumors or metastatic lesions and cleared by the lymphatic or circulatory system [26]. To date, most studies on CTCs have focused on the exploration of CTCs in the blood circulation, which are few in number (up to a few hundred per milliliter), depending on the available detection/isolation techniques and the definition of CTCs used.
After release from the primary tumor, the process by which CTCs survive in the circulatory system and spread to remote organs requires overcoming several obstacles, with less than 0.01% of CTCs released into the circulation eventually surviving and producing metastases [27]. First, solid tumors shed tumor cells that normally cross the endothelium into the circulation via epithelial–mesenchymal transition (EMT), a highly dynamic process of epithelial to quasi-mesenchymal cell transformation with enhanced metastatic potential and invasive capacity and resistance to some therapeutic strategies [28, 29]. Thereafter, most CTCs experience necrosis or apoptosis owing to environmental challenges in the blood, such as hemodynamic shear or immune system attack, with only a small percentage of CTCs interacting closely with cancer-associated fibroblasts, neutrophils or platelets, eventually evading the immune system and surviving [30–32]. Lastly, CTCs must evade the defenses of the natural immune system and recognition by natural killer cells [33, 34].
Since most cancers are epithelial in nature, the most commonly used marker for CTCs is epithelial cell adhesion molecule (EpCAM), a “universal” marker of cancer [35]. Several studies have shown the predictive value of EpCAM-based CTC assays in breast cancer and non-small cell lung cancer, which are cancers that strongly express EpCAM [36, 37]. Moreover, high-grade advanced stages of BC were significantly associated with EpCAM expression, and EpCAM expression was associated with poor overall survival. The strong association of EpCAM expression with high-grade tumors suggests a possible role in tumor progression, making EpCAM a potential target for antibody-mediated therapy [38]. Additionally, EpCAM was found to be a viable imaging target for the diagnosis of lymph node metastases in migratory cell carcinoma of the bladder. Perioperative examination of these metastases could improve disease staging and increase the rate of complete resection of MIBC lymph node metastases. Overall, these findings indicate that EpCAM-positive CTCs could be a robust and sensitive biomarker [39].
CTCs detected via liquid biopsy contain the complete genomic and transcriptomic information of cancer cells, making CTCs suitable clinical biomarkers [40]. The application of CTC in tumors has many advantages, including the analysis of genetic information or tumor morphology, which can improve patient classification and stratification, effective monitoring of patient response, and improved clinical prognosis [33, 41]. Additionally, CTCs can be selectively recovered from patients’ blood, amplified in vitro, and used to generate relevant animal models, enabling drug sensitivity assessment, biomarker discovery, and personalization of targeted therapies. These features suggest that blood detection of CTCs may be important for the clinical management of BC, facilitating the identification of tumor heterogeneity, potential micrometastases, and tumor evolution over time [42–44].
Over the last two decades, emerging isolation techniques have enabled the biological study of CTCs and promote their use in clinical applications for screening, treatment response monitoring, and prognostic assessment of cancer. Techniques for isolating CTCs include both physical property-based and biologic property-based approaches. The physical separation of CTCs for enrichment is based on the differences between CTCs and blood cells in size (filter-based devices), density, deformability, and charge (electrophoresis) [45]. Biological property-based techniques include immunoaffinity methods that rely on antibody–antigen interactions, in which CTCs are enriched positively by EpCAM and interstitial material (vimentin) or negatively by the leukocyte-specific antigen CD45 to remove unwanted leukocytes [46]. The CellSearch system is the most frequently used technique for the detection of CTCs and it is an immunoaffinity-based isolation strategy for identifying CTCs on the basis of EpCAM-positive expression [47]. However, a previous study found that only about 32% of patients with BC were EpCAM positive [48]. Moreover, CTCs could not be detected in the blood of patients with nonmetastatic BC [49]. Therefore, the application of CellSearch in BC may be limited in the case of patients with low expression of CTCs. However, some novel techniques, such as nanotechnology-based techniques and microfluidic chips, have been developed and are expected to be examined in clinical studies [50, 51]. Despite the advantages and disadvantages between the different methods, the effective integration of such technologies may facilitate CTC research in several aspects, particularly in terms of in-depth analyses and potential clinical utilization. Although the application of CTC assays for BC still has technical challenges, they are being evaluated in clinical trials as tools for predictive and response biomarkers and MRD surveillance.
CTCs in BC diagnosis/prognosis
CTCs are released into the circulation during the early cancer stages; therefore, the detection of CTCs could have considerable clinical application for the early diagnosis of several types of BC. One study detected CTCs by tumor-specific ligand PCR and evaluated their utility in the diagnosis of BC. The sensitivity and specificity of CTC quantitative assay for BC based on high expression of folate receptor-α were 82.1% and 61.9, with an area under the receiver operating characteristic curve (AUC) of 0.819 [52]. In another study, after next-generation sequencing (NGS) of matched BCs and CTCs, KMT2C mutations were detected among those commonly observed in CTCs and corresponding primary tumors [53]. In contrast, the KMT2C gene is frequently observed to be mutated in patients with MIBC [54], indicating the potential clinical value of CTCs in the early identification of high-risk BC.
Several studies have evidenced that the presence of CTCs is associated with increased disease recurrence, post-radical cystectomy (RC) cancer-specific survival (CSS) and overall survival (OS). The detection of CTCs is a strong predictor of disease progression and poor prognosis in early BC [55–61]. A recent metaanalysis of 30 studies showed that the presence of CTCs in peripheral blood is an independent predictor of poor prognosis in patients with BC [62]. Similarly, the presence of CTCs has been shown to be an independent predictor of disease progression to muscle-invasive disease in a large and homogeneous group of patients with high-risk NMIBC, with a 75% predictive value [56]. Lin et al. used the CellSearch system to analyze CTCs in 188 patients with BC, and found that approximately one-quarter had detectable CTC prior to RC, and CTC-positive patients treated with RC (with or without adjuvant chemotherapy) had poorer progression free survival (PFS), CSS, and OS compared with CTC-negative patients [56].
It was reported that preoperative detection of any CTCs predicts a higher risk of recurrence and poorer CSS in RC-treated patients with clinically non-metastatic BC compared with CTC-negative patients [58]. Similarly, in a recent study, Beije et al. sought to explore the predictive value of CTCs in the prognostic value of patients with non-metastatic MIBC after RC. They found a significant decrease in OS, CSS, and recurrence-free survival (RFS) in patients with positive CTCs [63]. In addition, Soave et al. investigated the potential role of CTC status on adjuvant chemotherapy (AC) management decisions in patients with BC after RC, and found that the presence of CTC was associated with poorer PFS, CSS, and OS in patients not on AC, indicating that CTCs may be useful in decisions for or against AC [61]. Gazzaniga et al. found that CTCs were detectable in approximately 18% of NMIBC, and that CTC levels could be used to distinguish patients at high risk of recurrence from those at high risk for progression, facilitating the early identification of candidate patients for adjuvant therapy after RC [59]. Additionally, Osman et al. found that patients with urea/EGFR-positive CTCs after RC had a higher risk of recurrence [64].
Several studies have detected CTCs in high-risk or progressive BC, and have reported a correlation between CTCs and tumor stage, lymph node metastasis, and survival [42, 65–68]. CTCs were identified in 19% of patients with advanced BC undergoing RC (with or without perioperative chemotherapy), and the presence of CTCs was associated with increased risk of disease progression; moreover, imaging metastatic disease was found to be significantly correlated with the presence of CTCs [65]. Additionally, higher levels of CTCs were detected in MIBC compared with NMIBC, suggesting that these cells may predominate during tumor muscle infiltration and disease progression [66]. Gradilone et al. found that CTCs could be isolated in the blood of 24 of 54 (44%) patients with T1G3 BC, and that CTC positivity was an independent prognostic factor (p < 0.001) of disease-free survival (DFS) [67]. Similarly, Nicolazzo et al. concluded that the presence of CTCs significantly affected disease-free survival and tumor-specific survival (p < 0.0001) on the basis of the findings of a long-term follow-up study of patients with T1G3 BC [68].
The identification of CTCs is considered a promising complementary tool for risk stratification of BC recurrence and progression, which is essential for the development of optimal surveillance strategies for patients after diagnosis [69–71]. Busetto et al. evaluated 155 patients with BC with pathologically confirmed T1G3 with preoperative CTCs, and observed a strong correlation between the presence of CTCs and time to first recurrence and time to progression, which could be used as a prognostic marker for risk stratification in patients with NMIBC [69]. Nicolazzo et al. demonstrated that the presence of even a single CTC in the peripheral blood of patients with BC indicates a high risk of local recurrence and/or disease progression in patients with ultra-high risk NMIBC [70]. Additionally, CTC counts were found to be significantly higher in the MIBC group than in the NMIBC group, and mortality was significantly higher in patients with high-grade BC than in patients with low-grade BC [71]. These findings indicate that CTC can accurately predict high-risk subgroups of patients, thus allowing longitudinal long-term monitoring of BC.
CTCs as predictor of treatment response in BC
CTCs may be sensitive biomarkers in assessing chemotherapy response, which could improve the timing of RC in clinical care or changing chemotherapy regimens [58, 63, 72]. Winters et al. conducted a prospective trial using the CellSearch system to examine CTCs in patients with BC treated with cisplatin chemotherapy, and observed a decrease in CTC counts after chemotherapy [72]. Moreover, the absence of CTC in patients with MIBC has been associated with reduced risk of recurrence after cystectomy, as well as tumor-specific death; however, CTC-positive patients with MIBC may benefit more from neoadjuvant chemotherapy [63].
Furthermore, the HER2 status of CTCs was not consistent with the HER2 status of the corresponding BC, suggesting that a subgroup of patients with BC with HER2-positive CTCs may benefit from HER2-targeted therapy. The presence of HER2-negative CTCs in patients with HER2-positive primary tumors may account for the failure of HER2-targeted therapy in a significant number of patients [58].
A recent study showed that baseline and serial assessment of programmed death-ligand 1 (PD-L1) using CTC monitoring may be an effective tool to guide treatment selection in candidate patients for PD-L1 inhibitors after Bacillus Calmette–Guérin (BCG) failure [73]. Serial CTC assays of whole blood could be used to assess the efficacy of checkpoint inhibitor immunotherapies, which are active in a certain percentage of CTC-positive patients and usually have durable responses [74]. Studies regarding blood-based CTCs as potential biomarkers for BC were summarized in Table 1.
Table 1.
Studies regarding blood-based CTCs as potential biomarkers for bladder cancer
| Authors (year) | Sample type | No. of patients | Laboratory technique | Clinical application | Detection rate | Refs. |
|---|---|---|---|---|---|---|
| Qi et al. (2014) | Serum | 120 | ELISA, RT-qPCR | Diagnostic biomarker | AUC = 81.9%, sensitivity = 82.1%, specificity = 61.9% | [52] |
| Kim et al. (2020) | Peripheral blood | 20 | Whole exome sequencing | Discrimination of MIBC from NMIBC and healthy individuals | KMT2C mutations were detected in 20% of CTCs | [53] |
| Gazzaniga et al. (2014) | Peripheral blood | 102 | CellSearch System | Predict prognosis | DFS (p = 0.005), PFS (p = 0.004) | [56] |
| Soave et al. (2017) | Peripheral blood | 188 | CellSearch System | Predict prognosis | RFS (p < 0.001), CSS (p < 0.001) | [57] |
| Rink et al. (2012) | Peripheral blood | 100 | CellSearch System | Predict prognosis | OS (p = 0.003), CSS (p = 0.002), RFS (p < 0.001) | [58] |
| Gazzaniga et al. (2012) | Peripheral blood | 44 | CellSearch System | Predict prognosis | TFR (p < 0.001) | [59] |
| Rink et al. (2011) | Peripheral blood | 50 | CellSearch System | Predict prognosis | OS (p = 0.001), CSS (p < 0.001), PFS (p < 0.001) | [60] |
| Soave et al. (2017) | Peripheral blood | 226 | CellSearch System | Predict prognosis | OS (p < 0.001), CSS (p < 0.001), RFS (p < 0.001) | [61] |
| Beije et al. (2022) | Peripheral blood | 273 | CellSearch System | Predict treatment response | CTC-positive patients treated with NAC have longer survival times | [63] |
| Osman et al. (2004) | Peripheral blood | 62 | Nested RT-PCR | Monitoring recurrence | Positive predict value = 79%, negative predict value = 50% | [64] |
| Abrahamsson et al. (2017) | Peripheral blood | 88 | CellSearch System | Predict disease progression | Disease progression (p = 0.049) | [65] |
| Haga et al. (2020) | Peripheral blood | 26 | FISHMAN-R system | Predict disease progression | More CTC detected in progressive BC (p = 0.01) | [66] |
| Gradilone et al. (2010) | Peripheral blood | 54 | RT-PCR, CELLection Dynabeads | Monitoring recurrence | DFS (p < 0.001) | [67] |
| Nicolazzo et al. (2017) | Peripheral blood | 54 | RT-PCR, CELLection Dynabeads | Predict prognosis | DFS (p < 0.0001), CSS (p < 0.0001) | [68] |
| Busetto et al. (2017) | Peripheral blood | 155 | CellSearch System | Predict prognosis | TFR (p < 0.0001), TTP (p < 0.0001) | [69] |
| Nicolazzo et al. (2019) | Peripheral blood | 102 | CellSearch System | Predict prognosis | TFR (p < 0.001), TSR (p < 0.001), TTP (p < 0.001) | [70] |
| Fu et al. (2021) | Peripheral blood | 48 | Microfluidic-Assay System | Early risk stratification | More CTC detected in progressive BC (p = 0.024) | [71] |
| Winters et al. (2015) | Peripheral blood | 31 | CellSearch System | Predict treatment response | Evaluate chemotherapy response: CTC declined after chemotherapy | [72] |
| Nicolazzo et al. (2021) | Peripheral blood | 20 | CellSearch System, ScreenCell | Treatment monitoring | Serial evaluation of CTCs can guide treatment selection for suitable PD-L1 inhibitors after BCG failure | [73] |
| Anantharaman et al. (2016) | Peripheral blood | 25 | Algorithmic analysis | Predict treatment response | High PD-L1+ /CD45− CTC burden and low burden of apoptotic CTCs had worse overall survival | [74] |
CTCs Circulating tumor cells, AUC area under the receiver operating characteristics curve, OS Overall survival, CSS Cancer-specific survival, RFS Recurrence-free survival, DFS Disease free survival, PFS Progression free survival, TFR time to first recurrence, TTP time to progression, TSR time to second recurrence, NAC neoadjuvant chemotherapy, BCG Bacillus Calmette–Guérin, PD-L1 Programmed death-ligand 1
Circulating non-coding RNAs (ncRNAs) in BC
Blood ncRNAs can act as key functional components or regulatory molecules of gene expression in a variety of cancers, playing a role in tumorigenesis, cell differentiation, proliferation, inhibition of angiogenesis, metastasis, and apoptosis. Since miRNAs, lncRNAs, and circRNAs represent the most studied ncRNA types to date, we will briefly describe their roles in BC here. Table 2 summarizes the role of blood-based ncRNAs in BC. miRNAs are short ncRNAs, with an average length of 22 nucleotides. Normal and tumor cells secrete miRNAs into various body fluids, including plasma, urine, and vaginal secretions [75]. In blood, miRNAs can be bound to specific ribonucleoprotein complexes, platelets, or packaged in EVs, such as exosomes, to avoid degradation and to obtain higher stability [75]. There are two different strategies for cell-free miRNA studies in blood: (1) screening of differentially expressed miRNAs by microarray analysis or RNA sequencing, or (2) analysis of candidate miRNAs selected from published tumor tissue data. miRNAs are more rapidly biogenic and activated and highly stable, which may give miRNAs diagnostic and prognostic predictive aspects for BC potential.
Table 2.
Studies regarding circulating ncRNAs in blood as potential biomarkers for bladder cancer
| ncRNA | Authors (year) | Sample type | Significant miRNA expression | Study design | Detection method and reference method | Clinically relevant findings | Refs. |
|---|---|---|---|---|---|---|---|
| miRNA | |||||||
| Feng et al. (2014) | Plasma | miR-99a↓ |
• 50 patients • 50 controls |
• RT-qPCR • RNU6 |
Diagnostic marker: no information concerning diagnostic accuracy | [76] | |
| Feng et al. (2014) | Plasma | • miR-19a↑ |
• 50 patients • 50 controls |
• RT-qPCR • RNU6 |
Diagnostic marker: no information concerning diagnostic accuracy | [77] | |
| Adam et al. (2013) | Plasma |
• miR-200b↑ • miR-33b↓ • miR-92b↓ |
• 20 patients • 18 controls |
• RT-qPCR • RNU6 |
• 89% accuracy for detecting BC • 92% accuracy for distinguishing invasive BC from other cases • 100% accuracy for distinguishing MIBC from controls • 79% accuracy for three-way classification between MIBC, NIMBC, and controls |
[78] | |
| Wang et al. (2019) | Serum | • miRNA‑373↑ |
• 55 patients • 45 controls |
• RT-qPCR • RNU6 |
Diagnosis of BC (AUC = 0.847) | [79] | |
| Motawi et al. (2016) | Plasma |
• miR-92a↓ • miR-100↓ • miR-143↓ |
• 70 patients • 62 controls |
• RT-qPCR • RNU6 |
• Diagnosis of BC with miR-92a (AUC = 0.940, DS = 97.1%, DSp = 76.7%) • Diagnosis of BC with miR-100 (AUC = 0.820, DS = 90%, DSp = 66.7%) • Diagnosis of BC with miR-143 (AUC = 0.915, DS = 78.6%, DSp = 93.3%) |
[80] | |
| Yang et al. (2015) | Serum | • miR-210↑ |
• 168 patients • 104 controls |
• RT-qPCR • RNU6 |
• Diagnosis of BC (AUC = 0.898, DS = 97.6%, DSp = 69.2%) • Prognostic marker: monitor tumor dynamics |
[81] | |
| Fang et al. (2016) | Plasma | • miR-205↑ |
• 89 patients • 56 controls |
• RT-qPCR • RNU6 |
• Diagnosis of BC (AUC = 0.950, DS = 76.4%, DSp = 96.4%) • Differentiation of NIMBC from MIBC (AUC = 0.668, DS = 57.1%, DSp = 77.0%) |
[82] | |
| Du et al. (2015) | Plasma |
• miR-497↓ • miR-663b↑ |
• Discovery: pools of 10 patients and 10 controls • Validation: training set with 56 patients and 60 controls, and test set with 109 patients and 115 controls |
• Discovery: TaqMan array • Validation: RT-qPCR |
Diagnosis of cancer with the two combined miRNAs (AUC = 0.711, DS = 69.7%, DSp = 69.6%) | [83] | |
| Lian et al. (2018) | Serum | • miR-409-3p↓ |
• Discovery: 140 patients, 139 healthy controls • Validation: 140 patients, 139 controls |
• Discovery: TaqMan array • Validation: RT-qPCR |
Diagnostic marker: no information concerning diagnostic accuracy | [84] | |
| Jiang et al. (2015) | Serum |
• miR-152↑ • miR-1486-3p↑ • miR-3187-3p↓ • miR-15b-5p↓ • miR-27a-3p↓ • miR-30a-5p ↓ |
• Discovery: pooled samples from 10 patients with NMIBC, 10 patients with MIBC, 10 healthy controls • Validation: training set 120 patients and 120 controls, and validation set 110 patients and 110 controls |
• Discovery: sequencing • Validation: RT-qPCR |
• Diagnosis of cancer with all miRNAs (AUC = 0.899, DS = 80%, DSp = 89%) • Differentiation of NIMBC from MIBC (AUC = 0.841, DS = 90%, DSp: 66.4%) • Prognostic marker set miR-152 and miR-3187-3p: recurrence-free survival for NIMBC, not for MIBC |
[85] | |
| Jiang et al. (2016) | Serum |
• miR-422a-3p (↑ in NMIBC, ↓in MIBC) • miR-486-3p↓ • miR-103a-3p (↑ in NMIBC, ↓in MIBC) • miR-27a-3p↓ |
• Discovery: pooled samples from 6 patients with NMIBC, 6 patients with MIBC, 6 healthy controls • Validation: training set 40 MIBC, 40 NMIBC, and 40 controls, and validation set 90 MIBC and 168 NMIBC |
• Discovery: sequencing • Validation: RT-qPCR |
• Diagnosis of cancer with the four combined miRNAs (training set: AUC = 0.894, validation set: AUC = 0.880) • Prognostic marker: miR-486-3p and miR-103a-3p were associated with OS of MIBC |
[86] | |
| Usuba et al. (2019) | Serum |
• miR-6087↓ • miR-6724-5p↑ • miR-3960↓ • miR-1343-5p↓ • miR-1185–1-3p↑ • miR-6831-5p↑ • miR-4695-5p↓ |
• Discovery: 196 patients with BC, 50 controls, 240 other types of cancers • Validation set: 196 patients with BC, 50 controls, 240 other types of cancers |
• Discovery: TaqMan array • Validation: RT-qPCR |
Diagnosis of cancer with the seven combined miRNAs (AUC = 0.97, DS = 95%, DSp = 87%) | [87] | |
| LncRNA | |||||||
| Duan et al. (2016) | Serum |
• lncRNA-MEG↓ • lncRNA- SNHG16↑ • lncRNA-MALAT1↑ |
• Discovery: pooled samples from 80 patients with BC and 80 healthy controls • Validation: training set 120 patients with BC, 68 patients with benign disease, 52 healthy controls, validation set 100 patients with BC, 52 patients with benign disease, 48 healthy controls |
• RT-qPCR • GAPDH |
• Diagnosis of cancer with three miRNAs (AUC = 0.899, DS = 80%, DSp = 89%) • Prognostic marker: low expression of MEG3 was associated with poor recurrence-free survival |
[90] | |
| Luo et al. (2019) | Plasma | • lncRNA CASC11↑ |
• 89 patients • 62 controls |
• RT-qPCR • RNU6 |
Diagnosis of BC (AUC = 0.899) | [91] | |
| Zhou et al. (2020) | Serum | • lncRNA LUCAT1↑ |
• 30 patients • 30 controls |
• RT-qPCR • GAPDH |
Diagnostic marker: no information concerning diagnostic accuracy | [92] | |
| Li et al. (2019) | Plasma | • lncRNA TUC338↑ |
• 56 patients • 56 controls |
• RT-qPCR • 18S |
Diagnosis of BC (AUC = 0.924) | [93] | |
| Zhang et al. (2020) | Serum | • lncRNA PCAT6↑ |
• 106 patients • 106 controls |
• RT-qPCR • GAPDH |
• Diagnosis of BC (AUC > 8.0) • Prognostic marker: high expression of lncRNA PCAT6 was associated with poor OS and PFS |
[94] | |
| circRNA | |||||||
| Pan et al. (2019) | Serum |
• circ-FARSA↑ • circ-SHKBP1↑ • circ-BANP↑ |
• 70 patients • 70 controls |
• RT-qPCR • β-actin |
• Diagnosis of cancer with combined lncRNA UCA1 and circSHKBP1 (AUC = 0.804) • Combined circFARSA and circBANP distinguishing recurrent from non-recurrent patients (AUC = 0.737) |
[95] | |
| Chi et al. (2019) | Serum | • hsa_circ_0000285↓ |
• 197 patients • 97 controls |
• RT-qPCR • β-actin |
• Diagnostic marker: no information concerning diagnostic accuracy • Treatment monitoring: low expression of hsa_circ_0000285 was associated with poor chemosensitivity • Prognostic marker: low expression of hsa_circ_0000285 was associated with poor survival rate |
[96] | |
| Xu et al. (2018) | Blood | • circPTK2↑ |
• 40 patients • 40 controls |
• RT-qPCR • GAPDH |
Diagnostic marker: no information concerning diagnostic accuracy | [97] |
ncRNA non-coding RNA, miRNA microRNA, circRNA circular RNA, lncRNA long non-coding RNA, BC bladder cancer, MIBC muscle-invasive bladder cancer, NMIBC non-muscle-invasive bladder cancer, AUC area under the receiver operating characteristics curve, DS diagnostic sensitivity, DSp diagnostic specificity, RT-qPCR reverse-transcription quantitative polymerase chain reaction. ↑ upregulated, ↓ downregulated
Many studies have explored the diagnostic, prognostic, and therapeutic potential of circulating miRNAs as potential biomarkers and therapeutic targets in BC. Feng et al. investigated plasma miR-19a and miR-99a in 100 patients with BC and healthy blood donors, respectively [76, 77]. As in tumor tissue, miR-19a expression was significantly increased in BC plasma, whereas miR-99a expression was significantly decreased, suggesting the potential of miR-19a and miR-99a as potential diagnostic markers for BC. Similarly, another study reported that plasma miR-200b expression was upregulated in patients with MIBC. In contrast, plasma miR-33b and miR-92b were downregulated in plasma from patients with BC and negatively correlated with pathological stage [78]. Recently, a study found that serum miRNA-373 levels were significantly higher in patients with BC compared with healthy controls and may serve as a potential diagnostic marker for BC (AUC was 0.847). In addition, miRNA-373 overexpression may promote BC cell proliferation, migration, and invasion [79].
Moreover, a study by Motawi et al. demonstrated that plasma miR-92a, miR-100, and miR-143 may be promising novel circulating biomarkers for clinical testing in BC [80]. The sensitivity and specificity of miR-92a calculated in this study were 97.1% and 76.7%, respectively, with an AUC value of 0.940. MiR-100 had a sensitivity and specificity of 90% and 66.7%, respectively, with an AUC value of 0.82. MiR-143 had a sensitivity and specificity of 78.6% and 93.3%, respectively, with an AUC value of 0.915. Similarly, another study observed that serum miR-210 expression was significantly elevated in patients with BC with a sensitivity of 97.6%, specificity of 69.2%, and AUC value of 0.898. Also, the authors found that serum miR-210 levels increased with increasing disease stage and grade. In addition, serum miR-210 expression was significantly lower in paired postoperative samples, whereas miR-210 expression was elevated in most patients with recurrent BC [81]. Similarly, a study found that plasma miR-205 levels were significantly higher in patients with BC than in normal controls, with a sensitivity of 76.4%, specificity of 96.4% and AUC: 0.950. Also, miR-205 could have the potential to be used as a biomarker to differentiate MIBC from NMIBC [82].
Using high-throughput TaqMan analysis, Du et al. identified miR-497 and miR-663b with significant differential expression by quantitative PCR for plasma miRNA analysis and two-stage validation of candidate miRNA expression. When integrating miR-497 and miR-663b, the AUC, sensitivity, and specificity were 0.711, 69.7%, and 69.6%, respectively [83]. Similarly, another study similarly used high-throughput TaqMan analysis to profile serum miRNAs to predict the risk of NMIBC. After validation in two independent cohorts (NMIBC: n = 280, control: n = 278), miR-409-3p was confirmed to be significantly associated with NMIBC [84].
Several groups developed a blood-based miRNA panel to improve the diagnostic efficacy of BC. Jiang et al. performed genome-wide serum miRNA analysis by MiSeq sequencing and established a panel of six miRNAs for the diagnosis of BC (miR-152, miR-3187-3p, miR-30a-5p, miR-27a-3p, miR-15b-5p, and miR-148b-3p). This miRNA model obtained high accuracy in this training set (sensitivity: 90.0%, specificity: 90.0%, AUC: 0.956) and was confirmed in the validation cohort (AUC: 0.899, sensitivity: 80.0%, specificity: 89.09%). Also, they found that serum miR-152 and miR-3187-3p expression was strongly associated with recurrence of NMIBC [85]. In the next step, they also defined a panel of four miRNAs (miR-422a-3p, miR-486-3p, miR-103a-3p, and miR-27A-3p) that predicted MIBC with high diagnostic accuracy (AUC: 0.89, sensitivity: 90.99%, specificity: 72.97%) and that was validated in an independent cohort (AUC: 0.88; sensitivity: 90.0%; specificity: 70.06%). Also, they found that miR-486-3p and miR-103a-3p were independently associated with OS [86]. A subsequent study found that a seven-miRNA panel (miR-6087, miR-6724-5p, miR-3960, miR-1343-5p, miR-1185–1-3p, miR-6831-5p, and miR-4695-5p) could be used as biomarkers for the specific and early detection of BC, with the highest accuracy for distinguishing BC from noncancerous tumors and other types of tumors (AUC: 0.97, sensitivity: 95%, specificity: 87%) [87].
In addition to miRNAs, several other non-coding RNAs, including lncRNAs and circRNAs, have been extensively studied and are associated with a variety of biological functions, including acting as microRNA sponges, RNA-binding proteins, regulating transcription, and encoding peptides [88, 89]. In view of BC, the utility of lncRNAs and circRNAs as prognostic, diagnostic, and therapeutic biomarkers was investigated [90–97]. Due to lack of space, their roles in BC were summarized in Table 2.
Circulating tumor DNA (ctDNA) in BC
All tumor cells release DNA into the circulatory system, with cell-free DNA (cfDNA) constituting a major portion of released DNA circulating in the bloodstream [98]. Cancer cell-derived mutant cfDNA contains ctDNA, which consists of small nucleic acids released from necrotic or apoptotic tumor cells and circulating tumor cells, usually in the form of double-stranded fragments [99, 100]. The half-life of plasma ctDNA is short, estimated to be < 2 h, indicating that plasma ctDNA levels are in a state of dynamic and real-time change [101]. Although the half-life is relatively short, the rate of release of cancer DNA fragments is relatively constant owing to rapid cell turnover within tumors [102], providing real-time information on tumors in a longitudinal study, with multiple time points. ctDNA possesses several cancer-associated molecular features, such as single nucleotide mutations [103], altered methylation [104], and tumor-derived viral sequences [105], which may distinguish ctDNA from normal circulating cfDNA. There is a correlation between ctDNA levels and tumor size and staging, and analysis of ctDNA can help identify tumor-specific abnormalities that can be used in a highly specific detection strategy [106]. ctDNA levels also vary considerably with tumor staging, surgery, and chemotherapy [107]. Moreover, the collection of ctDNA from blood is noninvasive and can be useful for early cancer detection and identification of microscopic residual disease after treatment, in addition to cancer screening and prognosis prediction [108, 109]. ctDNA retains relatively intact genetic information when tumor cells enter the circulatory system after apoptosis or necrosis, clearly presenting all mutations throughout the patient’s tumor progression [108]. Therefore, the use of ctDNA could help address the issue of tumor heterogeneity, allowing the genotyping of multiple somatic aberrations across a wide genomic space in a single comprehensive assay [110]. Despite the pathological differences found between tissue and ctDNA-based mutation testing, ctDNA is frequently used to inform clinical decisions in the diagnosis of advanced disease, particularly in cases where biopsies are difficult to obtain or the primary location of the cancer is unknown [111]. Considering the heterogeneity of BC and the evolutionary pressure to intervene in treatment, ctDNA-based profiles are more reflective of advanced tumor burden than histology. However, ctDNA is challenging to detect, owing to its small percentage (sometimes < 0.01%), compared with non-tumorigenic cfDNA [112].
Currently, the main techniques for ctDNA detection in blood include quantitative polymerase chain reaction (qPCR) and digital polymerase chain reaction (dPCR), both of which allow the quantitative assessment of candidate genes and are rapid, cost-effective, and highly sensitive and specific. Additionally, both methods are suitable for the detection of certain simple mutations; however, the application of both methods is limited by the fact that they can only be used to monitor known genomic changes [113, 114]. To overcome this problem, next-generation sequencing (NGS)-based technologies are increasingly being applied to detect ctDNA, including a wide range of methods from small targeted panels to whole genome sequencing (WGS). None of these NGS techniques require any a priori molecular mutation knowledge, and they allow for the detection of known and unknown mutations, as well as hard-to-detect alterations, for example, gene fusions and copy number changes [102]. Additionally, NGS techniques can characterize personalized cancer genetic profiles, which could help in the development of personalized drugs. However, these techniques are expensive, time-consuming, and require more robust bioinformatics support [115]. In the era of precision oncology, the use of ctDNA analysis represents a paradigm shift in the use of genomic biomarkers, with considerable impact on clinical practice [116].
Compared with tissue-based tumor DNA analysis, plasma ctDNA is more convenient, easier and faster to obtain, and less invasive. Despite potential heterogeneity, tumor-specific genomic alterations detected in plasma ctDNA and tumor tissue have a fairly high concordance rate [117, 118]. These advantages facilitate the adoption of ctDNA as the companion diagnostic test to detect certain mutations required for the implementation of targeted therapies and for the serial surveillance of tumor progression [119]. An increase in ctDNA levels has been shown to occur prior to radiological progression [120, 121]. Additionally, the high somatic mutation rate in BC enhances the potential application of ctDNA, as fewer genes or targeted regions can provide the necessary information. Currently, ctDNA has a wide range of clinical applications in BC, including cancer screening, treatment response and drug resistance monitoring, and disease recurrence monitoring.
ctDNA for BC screening and diagnosis
Several studies have shown that promoter methylation, leading to epigenetic inactivation of tumor suppressor genes, has diagnostic potential for BC. Analysis of preoperative plasma samples of patients with BC showed a significant association between ctDNA methylation status and aberrant methylation of tumor suppressor genes [122–126]. A study reported highly specific detection of methylated p16 DNA in the serum of patients with BC with sensitivity, specificity, and positive predictive values of 22.6, 95, and 98%, respectively [122]. Similarly, another study reported the presence of methylated p16 DNA sequences in the plasma of patients with BC and the absence of methylated ctDNA in healthy individuals [123]. These results suggest that methylated p16 can be used as a tumor marker for the diagnosis of BC. Additionally, a small cohort study involving 45 patients found that hypermethylation of serum ctDNA was detected in 59% of cases, and the hypermethylation of APC, GSTP1, or TIG1 differentiated patients with BC from healthy individuals, with 80% sensitivity and 93% specificity [124]. Similarly, methylation of the promoter of tumor suppressor CDH13 gene was observed in the serum of 30.7% of patients with BC, with a higher frequency in patients with advanced high-grade BC [125]. Moreover, a prospective multicenter study involving 227 patients with BC showed that serum ctDNA methylation is a useful noninvasive biomarker for differentiating patients with BC from healthy individuals (62% sensitivity, 89% specificity) [126]. Although the identification of aberrant gene promoter methylation could facilitate the detection of malignancies, its utility is still limited by the low concentrations of extracted ctDNA available for analysis. However, noninvasive testing for cancer-related alterations in DNA methylation is feasible, and this can be a source of information for liquid biopsies.
Other approaches involve identifying gene mutations rather than methylation changes in ctDNA. For examples, alterations in oncogenes are common in BC, with the most studied being FGFR3, PIK3CA, ERBB2, and EGFR [127]. Additionally, potential mutations were identified in the ctDNA of 51 patients with metastatic BC (including RB1, CDKN2A, and ERBB2), MAPK/ERK or PI3K/AKT/mTOR pathway-associated mutations, and chromatin remodeling-associated mutations [2]. Although most studies were limited by small sample sizes, all authors concluded that ctDNA analysis by liquid biopsy has potential applications as a prognostic biomarker in clinical settings.
ctDNA as predictor of detecting recurrence and determining prognosis in BC
To date, the strongest evidence regarding the prognostic utility of liquid biopsy has been for ctDNA. CtDNA testing can be a valuable tool for early identification of disease recurrence after radical cystectomy (RC) or triple therapy for MIBC, which could facilitate early systemic treatment. In a previous study, ctDNA was collected at different timepoints before, during, and after treatment in 17 patients with MIBC who received neoadjuvant chemotherapy (NAC) and underwent surgery, and it was found that detection of ctDNA reflected persistence of disease, with a 83% sensitivity and 100% specificity for predicting recurrence [128]. Additionally, ctDNA levels were found to be significantly higher in patients who experienced metastatic recurrence or disease progression after cystectomy than in those who remained disease-free, with liquid biopsy results predating imaging evidence of recurrence by 101 days [129]. Moreover, additional ctDNA analysis after cystectomy has been shown to accurately predict metastatic recurrence, with 100% sensitivity and 98% specificity [130].
Others have used NGS to detect chromosomal breakpoints in 12 patients with NMIBC, and they subsequently designed droplet digital PCR (ddPCR) assays for these variants and found that somatic variants were detected in longitudinally collected plasma samples and that ctDNA levels decreased in patients after treatment, with high levels of ctDNA predicting disease progression. ctDNA can be detected early (before disease progression or recurrence, or before clinical manifestations). Moreover, 67% of patients with progressive disease have detectable levels of ctDNA in their plasma several months before evidence of clinical progression [131].
In addition, ctDNA can be widely detected in metastatic BC and has outstanding clinical application value [2, 131, 132]. In a recent study in a large mUC cohort comparing blood-based “liquid” biopsies with patient-matched tumor tissue, ctDNA provided a more representative genomic snapshot of disease. ctDNA analysis can identify genetic alterations such as FGFR3, ERCC2, ERBB2, and TMB, which were previously proposed as biomarkers of BC treatment response. Cost-effective and minimally invasive approaches to identify them will enable biomarker-driven stratification of patients in clinical trials, as well as real-world application of precision oncology [118].
The analysis of tumor-associated genomic alterations in ctDNA assays reflects an important approach for biomarker discovery and prognosis in patients with advanced malignancies [133–135]. A previous study reported disease progression using ddPCR to determine ctDNA for FGFR3 and PI3KCA hotspot mutations. It was found that tumor FGFR3 and PI3KCA mutations were significantly associated with subsequent disease recurrence, and that the expression of genes with multiple hotspot mutations could improve the prognostic monitoring performance of ctDNA [133]. Sequential ctDNA analysis can monitor dynamic changes in the frequency of genomically altered variant alleles and can predict disease progression and guide precise medication administration in patients with advanced BC [134]. In a recent retrospective study, ctDNA was found to identify clinically relevant molecular abnormalities, particularly, alterations in the BRCA-1 DNA repair-associated (BRCA1) and Raf-1 proto-oncogene (RAF1) genes appeared to be negatively associated with clinical outcomes [134].
The association between specific ctDNA genomic mutations and BC prognosis provides a new reference and basis for personalized BC treatment [136]. An association between the number of genomic alterations detected in ctDNA and the rate of disease control after receiving immunotherapy has also been suggested. In a recent randomized phase III trial involving 581 patients with uroepithelial carcinoma who had undergone surgery, ctDNA-positive patients in the atezolizumab treatment group were found to have improved disease-free survival and overall survival compared with the observation group [137]. Another study examined mutations in plasma ctDNA in 82 patients with NMIBC who underwent transurethral resection of the urinary bladder (TUR) and received immunotherapy, and found that higher somatic variants were independent predictors of recurrence after immunotherapy after bladder TUR. These findings suggest that ctDNA analysis based on targeted sequencing is a promising method for predicting disease recurrence in patients with NMIBC undergoing bladder TUR and immunotherapy [138].
ctDNA as predictor of monitoring response to treatment
Quantitative changes in ctDNA levels have shown potential as an early indicator of treatment efficacy. Monitoring ctDNA scores during treatment has been reported to be useful in predicting treatment response in metastatic uroepithelial carcinoma. Treatment-responsive patients exhibited rapidly decreasing ctDNA scores, whereas non-response patients exhibited stable or increasing scores, indicating that ctDNA could be used to identify responsive patients prior to imaging [139]. Moreover, dynamic changes in ctDNA during chemotherapy in patients with BC showed a superior association with patient prognosis [140]. In contrast, patients with ctDNA clearance indicated a response to chemotherapy, and could provide additional cycles of chemotherapy before cystectomy [130]. A recent study showed that ctDNA can be used as a marker for MRD to predict response to adjuvant immunotherapy in patients with BC [137]. Overall, ctDNA analysis could be used to detect MRD and monitor treatment effects at early timepoints when patients can benefit from change in treatment regimen.
Additionally, an association between the number of genomic alterations detected in ctDNA and response to immunotherapy has been observed [134, 141–143]. Changes in the frequency of ctDNA variant alleles early in treatment were found to identify checkpoint inhibitor monotherapy non-responders, thus helping to adjust treatment decisions. ctDNA levels are a predictor of long-term benefits of BC immunotherapy [142]. A previous study showed that sequential ctDNA analysis can be used to monitor dynamic changes in the frequency of genomically altered variant alleles and to predict treatment response. Serial ctDNA testing is a promising option for dynamically monitoring ICI treatment effects, predicting outcomes, and guiding adaptive treatment modalities [134]. One or more genomic alterations in ctDNA, mainly TP53, TERT, and BRCA1/BRCA2, were detected in 95% and 100% of patients before and after treatment with ICI, respectively, and genomic alterations in these oncogenic drivers were strongly associated with tumor resistance in advanced uroepithelial carcinoma [143]. Studies regarding blood-based ctDNAs as potential biomarkers for BC were summarized in Table 3.
Table 3.
Studies regarding circulating DNAs in blood as potential biomarkers for bladder cancer
| Authors (year) | Sample type | No. of patients | Laboratory technique | Clinical application | Detection rate | Refs. |
|---|---|---|---|---|---|---|
| Valenzuela et al. (2002) | Serum | 135 | Methylation-specific PCR | Diagnostic biomarker | AUC = 95%, sensitivity = 22.6%, specificity = 98% | [122] |
| Domínguez et al. (2002) | Plasma | 27 | 5–4520 kit, QIAamp Blood kit, PCR | Diagnostic biomarker | Detected in 40% patients | [123] |
| Ellinger et al. (2008) | Serum | 45 | Restriction endonuclease-based assay, qRT-PCR | Increase the accuracy of the diagnosis of BC | Sensitivity = 80%, specificity = 93% | [124] |
| Lin et al. (2011) | Serum | 168 | Methylation-specific PCR | Diagnostic biomarker | Detected in 30.7% patients, higher in advanced BC | [125] |
| Hauser et al. (2013) | Serum | 227 | Methylation-specific PCR | Discrimination of patients with BC from healthy individuals | Sensitivity = 62%, specificity = 89% | [126] |
| Vandekerkhove et al. (2017) | Plasma | 51 | Targeted and exome sequencing | Revealing aggressive mutations in metastatic BC | 95% of patients harboring deleterious alterations | [2] |
| Patel et al. (2017) | Plasma | 17 | TAm-Seq, WGS | Monitoring recurrence | Positive predict value = 100%, negative predict value = 85.7% | [128] |
| Birkenkamp-Demtröder et al. (2018) | Plasma | 60 | WES, ddPCR | Monitoring recurrence | Earlier recurrence detection compared with imaging | [129] |
| Christensen et al. (2019) | Plasma | 68 | WES, ultra-deep sequencing |
• Predict metastatic recurrence • Monitoring of therapeutic efficacy |
• Sensitivity = 100%, specificity = 98%; • Changes in ctDNA during chemotherapy in high-risk patients correlated with disease recurrence (p = 0.023) |
[130] |
| Birkenkamp-Demtröder et al. (2016) | Plasma | 12 | NGS, ddPCR | Predicts disease progression and residual disease | Disease progression (p = 0.032) | [131] |
| Vandekerkhove et al. (2021) | Plasma | 104 | WES, QIAGEN DNeasy Blood and Tissue Kit | Predict prognosis | OS (p = 0.01), PFS (p = 0.02) | [118] |
| Shohdy et al. (2022) | Plasma | 182 | NGS, WES | Predict disease progression | OS (p = 0.03) | [134] |
| Grivas et al. (2019) | Blood | 124 | Exon sequencing | Predict prognosis | OS (p = 0.07), FFS (p = 0.016) | [135] |
| Powles et al. (2021) | Plasma | 581 | WES, multiplex PCR |
• Predict recurrence • Predict treatment response |
• DFS (p < 0.0001) • ctDNA can be used as a marker for MRD to predict response to adjuvant immunotherapy |
[137] |
| Zhang et al. (2021) | Plasma | 82 | Targeted sequencing | Predict prognosis | DFS (p = 0.0146) | [138] |
| Sundahl et al. (2019) | Blood | 9 | RT-PCR | Response monitoring | Predicting treatment response in metastatic uroepithelial carcinoma prior to imaging | [139] |
| Khagi et al. (2017) | Blood | 69 | NGS | Predict treatment response | ctDNA-determined hypermutated states predict improved response, PFS, and OS after checkpoint inhibitor therapy | [141] |
| Raja et al. (2018) | Plasma | 29 | Targeted sequencing | Predict treatment response | Changes in the frequency of ctDNA variant alleles early in treatment were found to identify checkpoint inhibitor monotherapy non-responders | [142] |
| Ravi et al. (2022) | Blood | 45 | NGS | Treatment monitoring | Detection of one or more genomic alterations in ctDNA before and after ICI treatment is associated with tumor resistance in advanced uroepithelial carcinoma | [143] |
ctDNA circulating tumor deoxyribonucleic acid, AUC area under the receiver operating characteristics curve, WGS whole genome sequencing, WES whole-exome sequencing, NGS next-generation sequencing, PCR polymerase chain reaction, ddPCR droplet digital PCR, OS overall survival, PFS progression free survival, DFS disease free survival, FFS failure-free survival, MRD minimal residual disease, ICIs immune checkpoint inhibitors
Exosomes
Exosomes are a subtype of a broad class of EVs, ranging from 40 to 100 nm in diameter, which are thought to be lipid bilayer membrane vesicles actively released by most cells and capable of stable circulation in body fluids [144, 145]. Exosomes are released by both normal and tumor cells, and are present in various body fluids, such as urine, plasma, saliva, and tears [146]. The biogenesis and release of exosomes is a complex multistep process involving plasma membrane invagination, formation of multivesicular bodies, and exosome secretion. Several studies have shown that donor cell-derived bioactive molecules are enriched in exosomes, indicating the key role of exosomes in the exchange of genetic information [147]. Exosomes carry specific proteins (CD9, CD81, CD63, ALIX, TSG101, and HSP70), nucleic acids (DNA, miRNA, lncRNA, and circRNA), lipids, and metabolites with cancer information, making them potential cancer diagnostic biomarkers. Moreover, their specific cup-like features can be identified by electron microscopy [147–149]. Moreover, their specific cup-like features can also be identified by electron microscopy [150].
Exosomes play key roles in many pathophysiological processes, including immune responses, cardiovascular diseases, pregnancy disorders, and cancer [151, 152]. Exosomes are involved in EMT, angiogenesis, invasion and metastasis, evasion of immune surveillance, and drug resistance during tumorigenesis and progression, with promising clinical applications. Additionally, exosomes can be designed to deliver different therapeutic loads, including short interfering RNAs, antisense oligonucleotides, chemotherapeutic agents, and immunomodulators, indicating their potential application in tumor therapy [147]. In addition to their therapeutic potential, exosome-based liquid biopsies highlight their potential application in early cancer diagnosis, progression, prognosis, and response to therapy [153, 154]. Current reviews on BC exosomes focus on miRNAs, lncRNAs, and circRNAs.
Exosomes possess potential applications in precision tumor therapy, owing to their unique biological properties. However, the isolation and enrichment of exosomes from complex biological components is crucial for basic research and clinical translation. To date, some methods have been developed for exosome isolation and enrichment, with varying output and purity [155, 156]. Traditional separation methods include ultracentrifugation, size-exclusion chromatography, sedimentation techniques, and density gradient/buffered centrifugation. Recently, novel techniques, such as immunoaffinity assays, lipid-based separation techniques, microbeads/microfluidic chips, and thermophoresis, have made the enrichment of exosomes fast and easy. However, there is need to standardize the classification and extraction methods of exosomes from different body fluids to facilitate clinical use of exosomes.
Exosomes in BC diagnosis
Serum exosomes have been examined as noninvasive stable and sensitive biomarkers for the early diagnosis of BC. Exosomes reveal information about living tumor cells, and possess promising opportunities for the early detection of lesions [157–161]. Elsharkawi et al. observed an increase in the serum exosome levels of patients with BC with increasing stages of the disease. Moreover, patients with BC were successfully differentiated from healthy individuals on the basis of serum BC exosome levels, with 100% specificity and 82.4% sensitivity [160]. Similarly, Wang et al. observed an upregulation in exosomal lncRNA H19 levels in patients with BC compared with healthy subjects, indicating that exosomal lncRNA H19 could be a novel minimally invasive diagnostic biomarker [157]. Moreover, a previous study showed that hypoxic BC cells secreted lncRNA-rich uroepithelial carcinoma-associated 1 (UCA1) exosomes to remodel the tumor microenvironment and promote tumor growth and progression, indicating that serum exosomal lncRNA UCA1 could be a potential diagnostic biomarker of BC [159]. Furthermore, Zheng et al. found that exosomal lncRNA PTENP1 levels could be used to successfully differentiate patients with BC from healthy individuals, with an AUC value of 0.743 [161].
Some studies have also explored the combined application of exosomal non-coding RNAs in the diagnosis of BC. For example, a diagnostic model based on the combination of three serum exosomal lncRNAs (PCAT-1, UBC1, and SNHG16) was highly accurate for the early diagnosis of BC, with an AUC of 0.826, sensitivity of 80%, and specificity of 75%, which were significantly higher than the results of urine cytology [158].
Exosomes in BC prognosis
Recently, exosomal miRNAs have been found to play important roles in tumorigenesis and progression [162–164]. Exosomal miRNAs regulate communication and multiple signaling pathways between BC cells and other cells, and cancer cell-derived exosomes have been associated with BC progression and metastasis through enriched specific miRNAs [163]. Exosomal miR-21 from BC cells has been reported to promote BC cell invasion and migration by polarizing the PI3K/AKT signaling pathway in tumor-associated macrophages [165]. Additionally, the exosome-based miR-139-5p delivery system was found to help inhibit the proliferation, migration, and invasive potential of BC in vitro and in vivo, presenting potential innovative options to curb carcinogenesis [166].
Several studies have shown that blood-based exosomes can provide novel noninvasive prognostic biomarkers for BC. Yin et al. found that miR-663b levels were elevated in plasma exosomes of patients with BC compared with healthy controls. Exosomal miR-663b from BC cells promotes BC cell proliferation and tumor progression by mediating the function of ERF [167]. Yan et al. observed higher levels of miR-4644 in plasma exosomes from patients with BC compared with patients without BC; additionally, exosomal miR-4644 directly targets and downregulates the expression of UBIAD1-containing protein 1 (UBIA prenyltransferase domain-containing protein 1) and promotes BC cell proliferation, migration and invasion [168]. Conversely, two recent studies demonstrated that both miR-375-3p and miR-133b were significantly lower in BC serum, and inhibited cell proliferation and metastasis, but promoted apoptosis in both in vivo and in vitro models, serving as suppressors and potential therapeutic targets of BC [169, 170]. Furthermore, Sabo et al. confirmed that low expression of miR-185-5p and miR-106a-5p, or high expression of miR-10b-5p, in exosomes from BC serum was significantly associated with shorter survival [171].
Additionally, studies have shown that exosomal lncRNAs play a crucial role in cell proliferation and invasion, lymphangiogenesis, drug resistance, and prognostic assessment in BC [158, 161, 172–177]. A research group found that cancer-associated fibroblast-derived exosome-mediated LINC00355 not only promotes cisplatin resistance in BC cells, but is also involved in regulating BC cell proliferation and invasion [172, 173]. Exosome-mediated lncRNA LINC01133 can inhibit BC suppression of cell viability, proliferation, migration, and invasion ability by regulating the Wnt signaling pathway, which may provide a novel target for BC therapy [174]. High expression of lncRNA UBC1 in serum exosomes is associated with a high recurrence rate of NMIBC, and is an independent risk factor for recurrence-free survival in NMIBC [158]. Zheng et al. found that exosomes from normal cells transferred PTENP1 into BC cells and reduced the invasive and migratory capacity of the cells in vitro and reduced BC progression both in vivo and in vitro [161].
The prognosis of patients with BC with lymph node metastases is extremely poor in clinical practice. It has been reported that exosomes can act as a vehicle of communication between the lymphatic system and BC, potentially remodeling the lymphatic system laterally by transferring epigenetic and genetic information [178]. Chen et al. identified an exosomal lncRNA, called lymph node metastasis-associated transcript 2 (LNMAT2), which was significantly elevated in BC serum samples [177]. Additionally, higher LNMAT2 expression levels were detected in serum exosomes of patients with BC with lymph node metastasis compared with patients without lymph node metastasis. Moreover, exosomes secreted by BC cells are internalized by human lymphatic endothelial cells, which triggers the upregulation of Prox1-expressing exosomes through hnRNPA2B1 recruitment and increases H3K4 trimethylation of the Prox1 promoter, leading to lymphangiogenesis and lymph node metastasis in BC cells [176]. Furthermore, serum exosome LNMAT2 overexpression is also associated with shorter OS in patients with BC [177].
Exosomal circRNAs are abundant and stably present in tumors, and are important for intercellular communication [179]. Recent studies using high-throughput sequencing have revealed that several exosomal circRNAs can serve as biomarkers for cancer diagnosis, invasion, and drug resistance [180, 181]. It was found that blood exosome-derived circTRPS1 from patients with BC can regulate intracellular activity homeostasis and CD8+ T cell depletion through the circTRPS1/miR141-3p/GLS1 axis, enhance BC cell proliferation and invasiveness, and impair antitumor immune function in the BC microenvironment [182]. Recent studies found that circPRMT5 is upregulated in serum exosomes of patients with BC, and plays a key role in the EMT and/or invasiveness of BC cells; moreover, high expression of circPRMT5 is positively correlated with metastasis and/or poorer survival in patients with BC, making it a potential therapeutic target for patients with BC [183].
Furthermore, it was found that the CT10 regulator kinase (CRK) promoted the expression of ErbB2 in BC cells, and these tumor proteins were transferred via exosomes to receptor cells in target organs, contributing to the distant metastasis of BC. These findings indicate that CRK intervention with exosomal ErbB2 blockade may be an effective therapeutic strategy for patients with advanced and metastatic BC with ErbB2 overexpression [184]. Studies regarding blood-based exosomes as potential biomarkers for BC were summarized in Table 4.
Table 4.
Studies regarding blood-based exosomes as potential biomarkers for bladder cancer
| Authors (year) | Sample type | Functional component | No. of patients | Laboratory technique | Clinical application | Detection rate | Refs. |
|---|---|---|---|---|---|---|---|
| Wang et al. (2018) | Serum | H19 | 104 | ExoQuick exosome purification kit | Diagnostic biomarker | AUC = 85.1%, sensitivity = 74.07%, specificity = 78.8% | [157] |
| Zhang et al. (2019) | Serum | Three-lncRNA panel (PCAT-1, UBC1, and SNHG16) | 520 | ExoQuick exosome purification kit | Diagnostic biomarker | AUC = 0.857 in training set, and 0.826 in validation set | [158] |
| Xue et al. (2017) | Serum | lncRNA-UCA1 | 60 | ExoQuick exosome purification kit | Diagnostic biomarker | AUC = 87.83%, sensitivity = 80%, specificity = 83.33% | [159] |
| Elsharkawi et al. (2019) | Serum | Tumor-derived exosomes | 82 | ExoQuantTM overall exosome capture and quantification assay kit, ELISA | Diagnostic biomarker | Sensitivity = 82.4%, specificity = 100% | [160] |
| Zheng et al. (2018) | Plasma | lncRNA-PTENP1 | 110 | ExoQuick exosome purification kit | Diagnostic biomarker, predict disease progression | AUC = 74.3%, sensitivity = 65.4%, specificity = 84.2%, predict disease progression (p < 0.05) | [161] |
| Yin et al. (2020) | Plasma | miR-663b | 122 | ExoQuick‐TC Exosome Precipitation Solution | Predict disease progression | Disease progression (p < 0.05) | [167] |
| Yan et al. (2020) | Plasma | LINC00355 | 57 | miRNA microarray | Predict disease progression | Disease progression (p < 0.05) | [168] |
| Cai et al. (2020) | Serum | miR-133b | 11 | Total exosome isolation reagent | Predict disease progression | Disease progression (p < 0.05) | [170] |
| Sabo et al. (2020) | Plasma | miR-126-3p | 93 | Deep sequencing | Predict prognosis | OS (p < 0.05) | [171] |
| Zhang et al. (2019) | Serum | LINC00355 | 520 | ExoQuick exosome purification kit | Predict prognosis | RFS (p = 0.01) | |
| Chen et al. (2020) | Serum | circRNA hsa_circ_0051443 | 592 | FRET, CD spectroscopy | Predict prognosis | OS (p < 0.05) | [181] |
| Yang et al. (2022) | Serum | circTRPS1 | 90 | ExoQuick Exosome Precipitation Solution | Predict prognosis | OS (p = 0.01) | [182] |
| Chen et al. (2018) | Serum | circRNA-PRMT5 | 119 | Electron microscopy | Predict prognosis | OS (p = 0.028) | [183] |
miRNA microRNA, circRNA circular RNA, lncRNA long non-coding RNA, AUC area under the receiver operating characteristics curve, ELISA enzyme linked immunosorbent assay, OS overall survival, RFS recurrence-free survival
Blood-based metabolomics and proteomics
Among the molecular and analytical techniques used to identify biomarkers, modern metabolomics technologies have made significant advances in characterizing and differentiating patients with BC from control subjects, identifying signature metabolites, and revealing disease biology and potential therapeutic targets. Metabolomics attempts to use the metabolic profile of cancer to assess disease risk, enabling real-time documentation and monitoring of disease stages, prediction of prognosis, recurrence or progression, and assessment of response or resistance to therapy [185]. Metabolomics is a comprehensive analysis of small molecule metabolites and can provide critical information on cancer stage [186, 187].
Metabolomics is a functional readout of tissue biochemistry that better reflects phenotypic status than other methods, such as transcriptomics and proteomics. There is increasing evidence that alterations in tumor metabolism may lead to local immunosuppression in the tumor microenvironment. Recently, metabolomic techniques are increasingly being utilized to identify potential biomarkers for cancer detection and monitoring. Serum/plasma is usually unaffected by exogenous factors and therefore varies little between individuals [188]. The analysis of sera from patients with BC allows the quantification of surface adhesion molecules, proteins, lipids, and amino acids. The chromatographic properties and mass spectra obtained provide features that can be compared with known metabolomics libraries, resulting in a “fingerprint” that becomes a biomarker. Metabolomics can provide a better understanding of the biological mechanisms of each tumor phenotype, thus promoting precision medicine and personalizing treatment for patients with different cancer types.
The main techniques used in metabolomics include gas chromatography–mass spectrometry (GC–MS), liquid chromatography–mass spectrometry (LC–MS), and nuclear magnetic resonance (NMR) methods for the detection of compounds in any biological fluid or cellular/tissue liquid-phase extracts [189, 190]. GC–MS and LC–MS are the two most widely used analytical techniques in metabolomics and can be used to identify compounds with a unique quality of several thousand features [191]. Among them, LC–MS-based metabolomics has become increasingly popular, owing to the high coverage of metabolites, further improving detection sensitivity and data reliability in cancer metabolomic studies [192]. NMR is a versatile method that can be used for biological samples in liquid, solid, or gaseous form without prior processing [193]. Other techniques for metabolite analysis are more limited, such as matrix-assisted laser desorption/ionization mass spectrometry imaging and magnetic resonance spectroscopy imaging. The increased sensitivity, specificity, and development of metabolomics-related technologies may help to identify good metabolic biomarkers, which can be applied in clinical settings.
Despite considerable improvements in genomics- and metabolomics-based strategies, the importance of proteomic-based analysis in liquid biopsies for cancer cannot be overemphasized. Recent advances in clinical proteomic techniques offer promising opportunities for the study of proteins in plasma/serum [194]. Since all tissues of the body are perfused by blood, each cell leaves a “trace” of its constituent proteins and other compounds in the circulatory system. Blood-based proteomics studies and links protein expression profiles to specific disease phenotypes, and identifies potential biomarkers reflecting the physiological or pathological state of the body, which may aid early disease detection.
Recently, separation techniques, such as electrophoresis, reversed-phase liquid chromatography, ion exchange and size-exclusion chromatography, and immune-enrichment or depletion methods, have been used to isolate or concentrate proteins or peptides, followed by mass spectrometry in combination with advanced computer software, making it possible to accurately identify, characterize, and quantify proteomic data [195]. Mass spectrometry-based proteomics can be divided into data-intensive discovery-based proteomics and targeted proteomics. The former allows unbiased analysis of the proteome on a global scale, whereas the latter provides highly specific quantitative detection of selected candidates in biological samples. To date, blood-based proteomics studies have provided considerable information on BC-associated proteomic changes, providing a better understanding of the intrinsic changes in the proteome of the blood during BC.
Role of blood-based metabolomics and proteomics in BC diagnosis
Metabolomics and proteomics may help identify biomarkers for the diagnosis of BC. Serum is recognized for biomarker studies because it is constantly perfused in tissues and may pick up disturbed metabolites and proteins secreted and shed from cells and tissues. A previous study showed that patients with BC had lower serum levels of citric acid, glycine, isoleucine/leucine, lactate, and tyrosine, and higher levels of lipids and glucose compared with healthy individuals [196]. Additionally, NMR spectroscopy-based serum metabolomics showed that a combination of dimethylamine (DMA), glutamine, and malonic acid accurately isolated high-grade BC from low-grade BC, with good external validation (96% sensitivity and 94% specificity) [197]. Moreover, metabolites involved in malignant proliferation, immune escape, differentiation, apoptosis, and invasion of cancer cells were identified by ultra-high performance liquid chromatography (UHPLC) combined with Q-TOF mass spectrometry, and three combinations of serum metabolites, namely inosine, N1acetyl-N2-formyl-5-methoxykynurenine, and PS (O-18:0/0:0), showed good properties in predicting high-level BC [198].
Compared to urine analysis, proteomic studies using blood are rare. However, it was found that the proteomic pattern of serum samples from patients with BC versus controls was an effective screening tool for identifying diagnostic BC, with 96.4% sensitivity and 86.5% specificity, even in low stage low grade tumors [199]. Bansal et al. analyzed 80 serum samples from healthy individuals and patients with low-grade and high-grade BC using bidirectional gel electrophoresis, and identified five differentially expressed proteins, including S100A8 and S100A9, which accurately distinguished between BC (low-grade and high-grade) and healthy controls, with an AUC value of 0.946 [200]. Similarly, abnormally expressed serum levels of S100A4, S100A8, S100A9, CA-I, and annexin V proteins were found to serve to be effective protein biomarkers for BC diagnosis based on the analysis of 160 serum samples from 52 healthy individuals, 55 presurgical, and 53 postsurgical patients with BC [201].
Role of blood-based metabolomics and proteomics in BC disease progression
Metabolomics and proteomics can help identify potential biomarkers of high aggressiveness in patients with BC [202–204]. Certain metabolic pathways may be associated with the regulation of chemosensitivity and immunotherapy in BC and may serve as potential biomarkers for identifying high-risk patients [205, 206]. A previous metabolomic showed that a set of genetic features, specifically COMT, IYD, and TTL, play potentially important roles in the progression of BC in smokers [207].
Minami et al. demonstrated that tumor-associated proteins S100A8 and S100A9 are associated with BC staging, grading and CSS, and S100A8 expression is an important predictor of BC recurrence [208]. Similarly, a total of nine differentially expressed glycoproteins in another study, including haptoglobin, transferrin, and IgM glyco-variants, or various cleavage products of fibrinogen and complement C3b, may be promising in the monitoring of BC disease progression [209]. Studies regarding blood-based metabolomics and proteomics as potential biomarkers for BC were summarized in Table 5.
Table 5.
Studies regarding circulating metabolomics and proteomics in blood as potential biomarkers for bladder cancer
| Authors (year) | Sample type | No. of patients | Laboratory technique | Clinical application | Detection rate | Refs. |
|---|---|---|---|---|---|---|
| Cao et al. (2012) | Serum | 112 | 1H NMR measurements | Discrimination of patients with BC from healthy individuals | Detected abnormal serum metabolic profiles in 100% of patients | [196] |
| Bansal et al. (2013) | Serum | 99 | 1H NMR measurements | Diagnostic biomarker | Sensitivity = 94%, specificity = 97% | [197] |
| Tan et al. (2017) | Serum | 172 | UHPLC coupled with Q-TOF MS | Diagnostic biomarker | AUC = 0.961 | [198] |
| Schwamborn et al. (2009) | Serum | 248 | MALDI-TOF–MS | Diagnostic biomarker | Sensitivity = 96.4%, specificity = 86.5% | [199] |
| Bansal et al. (2014) | Serum | 90 | MALDI-TOF–MS | Diagnostic biomarker | AUC = 0.946 (S100A8 and S100A9) | [200] |
| Bansal et al. (2016) | Serum | 160 | ELISA, MARS | Diagnostic biomarker | AUC = 0.957 (S100A9) | [201] |
| Amara et al. (2019) | Serum | 87 | LC–MS | Predict disease progression | OS (p = 0.0065) | [207] |
| Minami et al. (2010) | Serum | 2 | Reversed-phase high-performance liquid chromatography | Predict prognosis | RFS (p = 0.026), CSS (p = 0.041) | [208] |
| Lemańska-Perek et al. (2019) | Plasma | 6 | MALDI-TOF–MS | Predict disease progression | Increasing abundance in progressing BC | [209] |
AUC area under the receiver operating characteristics curve, ELISA enzyme linked immunosorbent assay, OS overall survival, RFS recurrence-free survival, CSS cancer-specific survival, NMR nuclear magnetic resonance, LC–MS liquid chromatography–mass spectrometry, Q-TOF MS quadrupole time of flight mass spectrometry, UHPLC ultra-high performance liquid chromatography, MALDI-TOF–MS matrix-assisted laser desorption/ionization time of flight mass spectrometry
Challenges facing liquid biopsy in BC
Liquid biopsies represent a promising strategy in biomarker research. Despite the considerable potential implications, a range of technical, biological, and clinical challenges need to be addressed before liquid biomarkers can be adopted in clinical practice. Limitations such as the lack of standardized assays and the high cost of liquid biopsies hinder the potential translation of liquid biomarkers into clinical practice. Additionally, most studies evaluating liquid biopsy techniques have been limited by small sample sizes, indicating the need for large-scale higher-quality studies to validate previous findings before routine clinical application.
CTC analysis can provide unique insights into tumor heterogeneity; however, CTC count during early-stage disease is low and may be undetected. Moreover, different tumor regions in a single patient may not show the same CTC propensity. Therefore, it is unclear whether CTC analysis could cause potential biological bias in intratumor heterogeneity. Although several techniques for CTC detection have emerged, their sensitivity and specificity still require further improvement. High-throughput sequencing can facilitate CTC-based multiomics studies, which can further benefit clinical applications, but can also contribute to the difficulty and complexity of data analysis [210].
The advantage of ctDNA analysis over CTC detection is that plasma/serum samples can be stored for a longer period of time and are easily collected before final analysis, making ctDNA an attractive option for multicenter studies. Currently, ctDNA analysis is a major method for assessing BC heterogeneity, without sampling bias of tissue biopsy. However, most studies on BC liquid biopsies have focused on patients with advanced disease. The use of ctDNA for early detection may be highly limited due to the low detection rate of early cancers, and therefore there is a need to develop techniques with higher sensitivity and to obtain sufficient sample volumes to detect trace amounts of ctDNA. Additionally, the sensitivity, specificity, and reproducibility of ctDNA assays, and the optimization and standardization of preanalytical conditions, require further studies [211].
Exosomes are a novel means of intercellular information exchange and play an important role in the tumor microenvironment. However, the heterogeneity and number of exosomes in body fluids may be a drawback to their utility as biomarkers, as these may yield false negatives or positives in cancer diagnosis. Moreover, further studies are necessary to develop and optimize methods for loading bioactive substances into exosomes. Additionally, extensive studies on the safety, targeting ability, and efficacy of exosomes are necessary to facilitate the successful application of exosomes as targets or drug delivery tools in BC.
Before fully evaluating cfRNA for clinical applications, the sensitivity of cfRNA for cancer detection in early-stage cancer needs to be determined. This is because cfRNA is unstable and highly fragmented. Targeted assays such as hybridization capture or amplicon sequencing should be used, which will allow for more sensitive quantification of cfRNA.
Furthermore, BC metabolic profiles are mainly characterized by alterations in metabolites associated with energy metabolic pathways, amino acid metabolism, and fatty acid metabolism, which are important for cell proliferation and maintenance of cellular redox homeostasis. However, the lack of standardized sample collection methods, variations in metabolite analysis methods, environmental stress, and food intake of patients severely influence metabolome composition, all of which contribute to differences in metabolomic profiles obtained from different laboratories. The analysis of changes in serum protein profiles in patients with BC is an emerging area of liquid biopsy that can reveal complex interactions between tumor tissue and the surrounding vascular microenvironment. However, additional clinical trials are still needed to validate previous proteomic data. Despite these challenges, metabolomics and proteomics show great clinical promise. Figure 3 provides a summary of the main advantages, disadvantages, and clinical applications of liquid biopsy components represented by CTCs, ctDNA, cfRNA, exosomes, and metabolomics and proteomics.
Fig. 3.
Comparison of the major liquid biopsy methods in bladder cancer and their main advantages, limitations, clinical applications, and future directions
Conclusion and future perspectives
Management strategy for patients with BC
Despite the high accuracy of cystoscopy and pathology biopsy in detecting BC, there are multiple limitations to their application for screening BC, including complications associated with invasive biopsy, patient anxiety, and financial burden. Although not currently considered a standard tool for the confirmation and diagnosis of BC, liquid biopsy could be a promising and effective alternative to traditional invasive sampling methods, especially in cases where tissue samples are not available. Additionally, it may be reasonable to defer invasive cystoscopy in patients with low risk of recurrence on the basis of preliminary liquid biopsy, which could reduce patients’ pain. The use of circulating biomarkers and liquid biopsies in real-time assessment of BC during treatment could provide useful information for treatment strategy and prevent the human and economic costs associated with delaying treatment. Combining molecular alterations at different levels (genomic, transcriptomic, and proteomic) can improve the accuracy of diagnosis, prognosis, and prediction of BC, and is a key prerequisite for successful individualized treatment strategies.
However, several key questions still need to be carefully addressed in the future to accelerate the adoption of biomarkers into clinical practice. (1) What biomarkers can help in the early detection of BC in high-risk populations? (2) What are the minimum requirements for biomarkers suitable to complement current routine screening for overall satisfactory accuracy? (3) Does liquid biopsy have the ability to differentiate between benign and malignant tumor? (4) What potential solutions can bridge the gap between basic research and clinical translation of these biomarkers?
Future perspectives for precision treatment of BC
Over the past decade, precision medicine has evolved considerably. Liquid biopsy has revolutionized the field of clinical oncology considerably by facilitating tumor sampling, continuous monitoring through repeat sampling, the development of personalized treatment plans, and the screening for treatment resistance. Although liquid biopsy technology is still evolving, its noninvasive nature promises to revolutionize the precision treatment of BC. Additionally, we hypothesize that a shift from single to multiple markers could improve BC diagnosis, monitoring, and treatment. Active collaboration between clinicians, industrial scientists, and biologists is necessary.
Noninvasive liquid biopsy technology holds promise for clinical applications in the early diagnosis of BC, accurate drug administration, real-time documentation and monitoring of disease evolution, and prognostic assessment. At the same time, it is critical to conduct more multicenter, large-scale clinical trials to verify its clinical effectiveness.
Acknowledgements
We thank Figdraw as the figures in this article were drawn by Figdraw. We also thank Editage (www.editage.cn) for assistance with English discourse revision and editing.
Abbreviations
- BC
Bladder cancer
- MIBC
Muscle-invasive bladder cancer
- NMIBC
Non-muscle-invasive bladder cancer
- CTCs
Circulating tumor cells
- ICIs
Immune checkpoint inhibitors
- cfRNA
Cell-free RNA
- ctDNA
Circulating tumor deoxyribonucleic acid
- miRNAs
MicroRNAs
- lncRNAs
Long non-coding RNAs
- circRNAs
Circular RNAs
- EVs
Extracellular vesicles
- EMT
Epithelial-mesenchymal transition
- EpCAM
Epithelial cell adhesion molecule
- cfDNA
Cell-free DNA
- NGS
Next-generation sequencing
- WGS
Whole genome sequencing
- LC–MS
Liquid chromatography–mass spectrometry
- GC–MS
Gas chromatography–mass spectrometry
- AUC
Area under the receiver operating characteristics curve
- OS
Overall survival
- CSS
Cancer-specific survival
- RFS
Recurrence-free survival
- DFS
Disease-free survival
- PFS
Progression free survival
- TFR
Time to first recurrence
- TSR
Time to second recurrence
- TTP
Time to progression
- NMR
Nuclear magnetic resonance
- UCA1
Uroepithelial carcinoma-associated 1
- UHPLC
Ultra-high performance liquid chromatography
- NAC
Neoadjuvant chemotherapy
- TUR
Transurethral resection of the urinary bladder
- RC
Radical cystectomy
- LNMAT2
Lymph node metastasis-associated transcript 2
- CRK
CT10 regulator kinase
- MRD
Minimal residual disease
- PD-L1
Programmed death-ligand 1
Author contributions
X.N.C. designed the study, helped to create tables and figures, and revised the manuscript. Z.Q.X. and B.T.L. designed the study and revised the manuscript. S.J.L. and K.R.X. designed the study, wrote the initial manuscript draft, and created tables and figures. S.P. and Y.W. created tables, figures, and helped draft the manuscript. J.Y.Z., Z.Y.L., and X.F.L. helped revise the manuscript and tables. All authors read and approved the final manuscript.
Funding
This work was supported by Key Research and Development Joint Program of Liaoning Province (grant nos. 2020JH 2/10300148, 2020JH 2/10300137), and 345 Talent Project of Shengjing Hospital of China Medical University (grant no. M0176).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shijie Li, Kerong Xin, Shen Pan, and Yang Wang contributed equally to this work.
Contributor Information
Bitian Liu, Email: liu_bitian@163.com.
Zhenqun Xu, Email: zqxu@cmu.edu.cn.
Xiaonan Chen, Email: chenxn@cmu.edu.cn.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Vandekerkhove G, Todenhöfer T, Annala M, Struss WJ, Wong A, Beja K, Ritch E, Brahmbhatt S, Volik SV, Hennenlotter J, et al. Circulating tumor DNA reveals clinically actionable somatic genome of metastatic bladder cancer. Clin Cancer Res. 2017;23:6487–6497. doi: 10.1158/1078-0432.CCR-17-1140. [DOI] [PubMed] [Google Scholar]
- 3.Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer. 2015;15:25–41. doi: 10.1038/nrc3817. [DOI] [PubMed] [Google Scholar]
- 4.Babjuk M, Böhle A, Burger M, Capoun O, Cohen D, Compérat EM, Hernández V, Kaasinen E, Palou J, Rouprêt M, et al. EAU Guidelines on non-muscle-invasive urothelial carcinoma of the bladder: Update 2016. Eur Urol. 2017;71:447–461. doi: 10.1016/j.eururo.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 5.Kartolo A, Kassouf W, Vera-Badillo FE. Adjuvant immune checkpoint inhibition in muscle-invasive bladder cancer: Is it ready for prime time? Eur Urol. 2021;80:679–681. doi: 10.1016/j.eururo.2021.07.019. [DOI] [PubMed] [Google Scholar]
- 6.Lotan Y, Roehrborn CG. Sensitivity and specificity of commonly available bladder tumor markers versus cytology: results of a comprehensive literature review and meta-analyses. Urology. 2003;61(109–118):118. doi: 10.1016/s0090-4295(02)02136-2. [DOI] [PubMed] [Google Scholar]
- 7.Yafi FA, Brimo F, Steinberg J, Aprikian AG, Tanguay S, Kassouf W. Prospective analysis of sensitivity and specificity of urinary cytology and other urinary biomarkers for bladder cancer. Urol Oncol. 2015;33:25–66. doi: 10.1016/j.urolonc.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Galgano SJ, Porter KK, Burgan C, Rais-Bahrami S. The role of imaging in bladder cancer diagnosis and staging. Diagnostics (Basel) 2020;10:89. doi: 10.3390/diagnostics10090703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerner C, Costigliola V, Golubnitschaja O. Multiomic patterns in body fluids: technological challenge with a great potential to implement the advanced paradigm of 3P medicine. Mass Spectrom Rev. 2020;39:442–451. doi: 10.1002/mas.21612. [DOI] [PubMed] [Google Scholar]
- 10.Crocetto F, Barone B, Ferro M, Busetto GM, La Civita E, Buonerba C, Di Lorenzo G, Terracciano D, Schalken JA. Liquid biopsy in bladder cancer: State of the art and future perspectives. Crit Rev Oncol Hematol. 2022;170:103577. doi: 10.1016/j.critrevonc.2022.103577. [DOI] [PubMed] [Google Scholar]
- 11.Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics. 2019;13:34. doi: 10.1186/s40246-019-0220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geeurickx E, Hendrix A. Targets, pitfalls and reference materials for liquid biopsy tests in cancer diagnostics. Mol Aspects Med. 2020;72:100828. doi: 10.1016/j.mam.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Ferro M, La Civita E, Liotti A, Cennamo M, Tortora F, Buonerba C, Crocetto F, Lucarelli G, Busetto GM, Del GF, et al. Liquid biopsy biomarkers in urine: a route towards molecular diagnosis and personalized medicine of bladder cancer. J Pers Med. 2021;11:78. doi: 10.3390/jpm11030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lone SN, Nisar S, Masoodi T, Singh M, Rizwan A, Hashem S, El-Rifai W, Bedognetti D, Batra SK, Haris M, et al. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol Cancer. 2022;21:79. doi: 10.1186/s12943-022-01543-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koguchi D, Matsumoto K, Shiba I, Harano T, Okuda S, Mori K, Hirano S, Kitajima K, Ikeda M, Iwamura M. Diagnostic potential of circulating tumor cells, urinary microRNA, and urinary cell-free dna for bladder cancer: A review. Int J Mol Sci. 2022;23:89. doi: 10.3390/ijms23169148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matuszczak M, Kiljańczyk A, Salagierski M. A liquid biopsy in bladder cancer-the current landscape in urinary biomarkers. Int J Mol Sci. 2022;23:8. doi: 10.3390/ijms23158597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lodewijk I, Dueñas M, Rubio C, Munera-Maravilla E, Segovia C, Bernardini A, Teijeira A, Paramio JM, Suárez-Cabrera C. Liquid biopsy biomarkers in bladder cancer: A current need for patient diagnosis and monitoring. Int J Mol Sci. 2018;19:6. doi: 10.3390/ijms19092514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanmalar M, Abdul SS, Kamri N, Said N, Jamil A, Kuppusamy S, Mun KS, Bradley DA. Raman spectroscopy biochemical characterisation of bladder cancer cisplatin resistance regulated by FDFT1: a review. Cell Mol Biol Lett. 2022;27:9. doi: 10.1186/s11658-022-00307-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins RW, Barbie DA, Flaherty KT. Mechanisms of resistance to immune checkpoint inhibitors. Br J Cancer. 2018;118:9–16. doi: 10.1038/bjc.2017.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramos-Casals M, Brahmer JR, Callahan MK, Flores-Chávez A, Keegan N, Khamashta MA, Lambotte O, Mariette X, Prat A, Suárez-Almazor ME. Immune-related adverse events of checkpoint inhibitors. Nat Rev Dis Primers. 2020;6:38. doi: 10.1038/s41572-020-0160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nabet BY, Esfahani MS, Moding EJ, Hamilton EG, Chabon JJ, Rizvi H, Steen CB, Chaudhuri AA, Liu CL, Hui AB, et al. Noninvasive early identification of therapeutic benefit from immune checkpoint inhibition. Cell. 2020;183:363–376. doi: 10.1016/j.cell.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, Fu Y, Zhu B, Wang J, Zhang B. Predictive biomarkers of immune checkpoint inhibitors-related toxicities. Front Immunol. 2023;2020:11. doi: 10.3389/fimmu.2020.02023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang HM, Li HX. Tumor heterogeneity and the potential role of liquid biopsy in bladder cancer. Cancer Commun (Lond) 2021;41:91–108. doi: 10.1002/cac2.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heitzer E, Haque IS, Roberts C, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20:71–88. doi: 10.1038/s41576-018-0071-5. [DOI] [PubMed] [Google Scholar]
- 25.Ramalingam N, Jeffrey SS. Future of liquid biopsies with growing technological and bioinformatics studies: opportunities and challenges in discovering tumor heterogeneity with single-cell level analysis. Cancer J. 2018;24:104–108. doi: 10.1097/PPO.0000000000000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasseur A, Kiavue N, Bidard FC, Pierga JY, Cabel L. Clinical utility of circulating tumor cells: an update. Mol Oncol. 2021;15:1647–1666. doi: 10.1002/1878-0261.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okajima W, Komatsu S, Ichikawa D, Miyamae M, Ohashi T, Imamura T, Kiuchi J, Nishibeppu K, Arita T, Konishi H, et al. Liquid biopsy in patients with hepatocellular carcinoma: Circulating tumor cells and cell-free nucleic acids. World J Gastroenterol. 2017;23:5650–5668. doi: 10.3748/wjg.v23.i31.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 29.Kang Y, Massagué J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Rejniak KA. Circulating tumor cells: when a solid tumor meets a fluid microenvironment. Adv Exp Med Biol. 2016;936:93–106. doi: 10.1007/978-3-319-42023-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garrido-Navas C, de Miguel-Perez D, Exposito-Hernandez J, Bayarri C, Amezcua V, Ortigosa A, Valdivia J, Guerrero R, Garcia PJ, Lorente JA, Serrano MJ. Cooperative and Escaping Mechanisms between Circulating Tumor Cells and Blood Constituents. Cells (Basel) 2019;8:897. doi: 10.3390/cells8111382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen DX, Bos PD, Massagué J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 33.Zhong X, Zhang H, Zhu Y, Liang Y, Yuan Z, Li J, Li J, Li X, Jia Y, He T, et al. Circulating tumor cells in cancer patients: developments and clinical applications for immunotherapy. Mol Cancer. 2020;19:15. doi: 10.1186/s12943-020-1141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gires O, Pan M, Schinke H, Canis M, Baeuerle PA. Expression and function of epithelial cell adhesion molecule EpCAM: where are we after 40 years? Cancer Metastasis Rev. 2020;39:969–987. doi: 10.1007/s10555-020-09898-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mederer T, Elsner F, Robold T, Großer C, Neu R, Ried M, Bleicher S, Schamberger T, Blochberger I, Hofmann HS, Klein CA. EpCAM-positive disseminated cancer cells in bone marrow impact on survival of early-stage NSCLC patients. Lung Cancer. 2022;167:73–77. doi: 10.1016/j.lungcan.2022.02.008. [DOI] [PubMed] [Google Scholar]
- 37.Spizzo G, Went P, Dirnhofer S, Obrist P, Simon R, Spichtin H, Maurer R, Metzger U, von Castelberg B, Bart R, et al. High Ep-CAM expression is associated with poor prognosis in node-positive breast cancer. Breast Cancer Res Treat. 2004;86:207–213. doi: 10.1023/B:BREA.0000036787.59816.01. [DOI] [PubMed] [Google Scholar]
- 38.Brunner A, Prelog M, Verdorfer I, Tzankov A, Mikuz G, Ensinger C. EpCAM is predominantly expressed in high grade and advanced stage urothelial carcinoma of the bladder. J Clin Pathol. 2008;61:307–310. doi: 10.1136/jcp.2007.049460. [DOI] [PubMed] [Google Scholar]
- 39.van der Fels C, Rosati S, de Jong IJ. EpCAM Expression in Lymph Node Metastases of Urothelial Cell Carcinoma of the Bladder: A Pilot Study. Int J Mol Sci. 2017;18:78. doi: 10.3390/ijms18081802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faugeroux V, Lefebvre C, Pailler E, Pierron V, Marcaillou C, Tourlet S, Billiot F, Dogan S, Oulhen M, Vielh P, et al. An accessible and unique insight into metastasis mutational content through whole-exome sequencing of circulating tumor cells in metastatic prostate cancer. Eur Urol Oncol. 2020;3:498–508. doi: 10.1016/j.euo.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 41.Zhao L, Wu X, Zheng J, Dong D. DNA methylome profiling of circulating tumor cells in lung cancer at single base-pair resolution. Oncogene. 2021;40:1884–1895. doi: 10.1038/s41388-021-01657-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang R, Xia J, Wang Y, Cao M, Jin D, Xue W, Huang Y, Chen H. Co-expression of stem cell and epithelial mesenchymal transition markers in circulating tumor cells of bladder cancer patients. Onco Targets Ther. 2020;13:10739–10748. doi: 10.2147/OTT.S259240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar A, Golani A, Kumar LD. EMT in breast cancer metastasis: an interplay of microRNAs, signaling pathways and circulating tumor cells. Front Biosci (Landmark Ed) 2020;25:979–1010. doi: 10.2741/4844. [DOI] [PubMed] [Google Scholar]
- 45.Vona G, Estepa L, Béroud C, Damotte D, Capron F, Nalpas B, Mineur A, Franco D, Lacour B, Pol S, et al. Impact of cytomorphological detection of circulating tumor cells in patients with liver cancer. Hepatology. 2004;39:792–797. doi: 10.1002/hep.20091. [DOI] [PubMed] [Google Scholar]
- 46.Sajay BN, Chang CP, Ahmad H, Khuntontong P, Wong CC, Wang Z, Puiu PD, Soo R, Rahman AR. Microfluidic platform for negative enrichment of circulating tumor cells. Biomed Microdevices. 2014;16:537–548. doi: 10.1007/s10544-014-9856-2. [DOI] [PubMed] [Google Scholar]
- 47.Agerbæk MØ, Bang-Christensen SR, Yang MH, Clausen TM, Pereira MA, Sharma S, Ditlev SB, Nielsen MA, Choudhary S, Gustavsson T, et al. The VAR2CSA malaria protein efficiently retrieves circulating tumor cells in an EpCAM-independent manner. Nat Commun. 2018;9:3279. doi: 10.1038/s41467-018-05793-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Went PT, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, Dirnhofer S. Frequent EpCam protein expression in human carcinomas. Hum Pathol. 2004;35:122–128. doi: 10.1016/j.humpath.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 49.Naoe M, Ogawa Y, Morita J, Omori K, Takeshita K, Shichijyo T, Okumura T, Igarashi A, Yanaihara A, Iwamoto S, et al. Detection of circulating urothelial cancer cells in the blood using the Cell Search System. Cancer. 2007;109:1439–1445. doi: 10.1002/cncr.22543. [DOI] [PubMed] [Google Scholar]
- 50.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Li M, Gao X, Chen Y, Liu T. Nanotechnology in cancer diagnosis: progress, challenges and opportunities. J Hematol Oncol. 2019;12:137. doi: 10.1186/s13045-019-0833-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qi F, Liu Y, Zhao R, Zou X, Zhang L, Li J, Wang Y, Li F, Zou X, Xia Y, et al. Quantitation of rare circulating tumor cells by folate receptor α ligand-targeted PCR in bladder transitional cell carcinoma and its potential diagnostic significance. Tumour Biol. 2014;35:7217–7223. doi: 10.1007/s13277-014-1894-0. [DOI] [PubMed] [Google Scholar]
- 53.Kim TM, Yoo JS, Moon HW, Hur KJ, Choi JB, Hong SH, Lee JY, Ha US. Distinct mutation profiles between primary bladder cancer and circulating tumor cells warrant the use of circulating tumors cells as cellular resource for mutation follow-up. BMC Cancer. 2020;20:1203. doi: 10.1186/s12885-020-07684-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, Hinoue T, Laird PW, Hoadley KA, Akbani R, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2018;174:1033. doi: 10.1016/j.cell.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raimondi C, Gradilone A, Gazzaniga P. Circulating tumor cells in early bladder cancer: insight into micrometastatic disease. Expert Rev Mol Diagn. 2014;14:407–409. doi: 10.1586/14737159.2014.908119. [DOI] [PubMed] [Google Scholar]
- 56.Gazzaniga P, de Berardinis E, Raimondi C, Gradilone A, Busetto GM, De Falco E, Nicolazzo C, Giovannone R, Gentile V, Cortesi E, Pantel K. Circulating tumor cells detection has independent prognostic impact in high-risk non-muscle invasive bladder cancer. Int J Cancer. 2014;135:1978–1982. doi: 10.1002/ijc.28830. [DOI] [PubMed] [Google Scholar]
- 57.Soave A, Riethdorf S, Dahlem R, Minner S, Weisbach L, Engel O, Fisch M, Pantel K, Rink M. Detection and oncological effect of circulating tumour cells in patients with variant urothelial carcinoma histology treated with radical cystectomy. Bju Int. 2017;119:854–861. doi: 10.1111/bju.13782. [DOI] [PubMed] [Google Scholar]
- 58.Rink M, Chun FK, Dahlem R, Soave A, Minner S, Hansen J, Stoupiec M, Coith C, Kluth LA, Ahyai SA, et al. Prognostic role and HER2 expression of circulating tumor cells in peripheral blood of patients prior to radical cystectomy: a prospective study. Eur Urol. 2012;61:810–817. doi: 10.1016/j.eururo.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 59.Gazzaniga P, Gradilone A, de Berardinis E, Busetto GM, Raimondi C, Gandini O, Nicolazzo C, Petracca A, Vincenzi B, Farcomeni A, et al. Prognostic value of circulating tumor cells in nonmuscle invasive bladder cancer: a Cell Search analysis. Ann Oncol. 2012;23:2352–2356. doi: 10.1093/annonc/mdr619. [DOI] [PubMed] [Google Scholar]
- 60.Rink M, Chun FK, Minner S, Friedrich M, Mauermann O, Heinzer H, Huland H, Fisch M, Pantel K, Riethdorf S. Detection of circulating tumour cells in peripheral blood of patients with advanced non-metastatic bladder cancer. Bju Int. 2011;107:1668–1675. doi: 10.1111/j.1464-410X.2010.09562.x. [DOI] [PubMed] [Google Scholar]
- 61.Soave A, Riethdorf S, Dahlem R, von Amsberg G, Minner S, Weisbach L, Engel O, Fisch M, Pantel K, Rink M. A nonrandomized, prospective, clinical study on the impact of circulating tumor cells on outcomes of urothelial carcinoma of the bladder patients treated with radical cystectomy with or without adjuvant chemotherapy. Int J Cancer. 2017;140:381–389. doi: 10.1002/ijc.30445. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Z, Fan W, Deng Q, Tang S, Wang P, Xu P, Wang J, Yu M. The prognostic and diagnostic value of circulating tumor cells in bladder cancer and upper tract urothelial carcinoma: a meta-analysis of 30 published studies. Oncotarget. 2017;8:59527–59538. doi: 10.18632/oncotarget.18521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beije N, de Kruijff IE, de Jong JJ, Klaver SO, de Vries P, Jacobs R, Somford DM, Te SE, van der Heijden AG, Alfred WJ, et al. Circulating tumour cells to drive the use of neoadjuvant chemotherapy in patients with muscle-invasive bladder cancer. ESMO Open. 2022;7:100416. doi: 10.1016/j.esmoop.2022.100416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Osman I, Kang M, Lee A, Deng FM, Polsky D, Mikhail M, Chang C, David DA, Mitra N, Wu XR, et al. Detection of circulating cancer cells expressing uroplakins and epidermal growth factor receptor in bladder cancer patients. Int J Cancer. 2004;111:934–939. doi: 10.1002/ijc.20366. [DOI] [PubMed] [Google Scholar]
- 65.Abrahamsson J, Aaltonen K, Engilbertsson H, Liedberg F, Patschan O, Rydén L, Sjödahl G, Gudjonsson S. Circulating tumor cells in patients with advanced urothelial carcinoma of the bladder: Association with tumor stage, lymph node metastases, FDG-PET findings, and survival. Urol Oncol. 2017;35:606–609. doi: 10.1016/j.urolonc.2017.05.021. [DOI] [PubMed] [Google Scholar]
- 66.Haga N, Tsubouchi K, Maruta H, Koguchi T, Hoshi S, Ogawa S, Akaihata H, Hata J, Kojima Y. Increase in circulating tumor cells in invasive bladder cancer after transurethral resection of bladder tumor. Anticancer Res. 2020;40:4299–4307. doi: 10.21873/anticanres.14432. [DOI] [PubMed] [Google Scholar]
- 67.Gradilone A, Petracca A, Nicolazzo C, Gianni W, Cortesi E, Naso G, Vincenzi B, Cristini C, De Berardinis E, Di Silverio F, et al. Prognostic significance of survivin-expressing circulating tumour cells in T1G3 bladder cancer. Bju Int. 2010;106:710–715. doi: 10.1111/j.1464-410X.2009.09130.x. [DOI] [PubMed] [Google Scholar]
- 68.Nicolazzo C, Busetto GM, Del GF, Sperduti I, Giannarelli D, Gradilone A, Gazzaniga P, de Berardinis E, Raimondi C. The long-term prognostic value of survivin expressing circulating tumor cells in patients with high-risk non-muscle invasive bladder cancer (NMIBC) J Cancer Res Clin Oncol. 2017;143:1971–1976. doi: 10.1007/s00432-017-2449-8. [DOI] [PubMed] [Google Scholar]
- 69.Busetto GM, Ferro M, Del GF, Antonini G, Chung BI, Sperduti I, Giannarelli D, Lucarelli G, Borghesi M, Musi G, et al. The Prognostic Role of Circulating Tumor Cells (CTC) in High-risk Non-muscle-invasive Bladder Cancer. Clin Genitourin Cancer. 2017;15:e661–e666. doi: 10.1016/j.clgc.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 70.Nicolazzo C, Busetto GM, Gradilone A, Sperduti I, Del GF, Loreni F, Cortesi E, de Berardinis E, Gazzaniga P, Raimondi C. Circulating tumor cells identify patients with super-high-risk non-muscle-invasive bladder cancer: updated outcome analysis of a prospective single-center trial. Oncologist. 2019;24:612–616. doi: 10.1634/theoncologist.2018-0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fu G, Cheng KS, Chen A, Xu Z, Chen X, Tian J, Xu C, Sun Y, Neoh KH, Dai Y, et al. Microfluidic assaying of circulating tumor cells and its application in risk stratification of urothelial bladder cancer. Front Oncol. 2021;11:701298. doi: 10.3389/fonc.2021.701298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winters B, James A, Lee J, Kho J, Morrissey C, Wright J. Chemotherapeutic effects on circulating tumor cells in bladder cancer. Int J Urol. 2015;22:612–613. doi: 10.1111/iju.12758. [DOI] [PubMed] [Google Scholar]
- 73.Nicolazzo C, de Berardinis E, Gazzaniga P. Liquid biopsy for predicting Bacillus Calmette-Guérin unresponsiveness in non-muscle-invasive bladder cancer. Eur Urol Oncol. 2021;4:124–125. doi: 10.1016/j.euo.2020.09.003. [DOI] [PubMed] [Google Scholar]
- 74.Anantharaman A, Friedlander T, Lu D, Krupa R, Premasekharan G, Hough J, Edwards M, Paz R, Lindquist K, Graf R, et al. Programmed death-ligand 1 (PD-L1) characterization of circulating tumor cells (CTCs) in muscle invasive and metastatic bladder cancer patients. BMC Cancer. 2016;16:744. doi: 10.1186/s12885-016-2758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mai S, Zhang Z, Mi W. Upregulation of circ_PVT1 and circ_001569 indicate unfavorable prognosis in colorectal cancer. Ann Clin Lab Sci. 2021;51:55–60. [PubMed] [Google Scholar]
- 76.Feng Y, Kang Y, He Y, Liu J, Liang B, Yang P, Yu Z. microRNA-99a acts as a tumor suppressor and is down-regulated in bladder cancer. BMC Urol. 2014;14:50. doi: 10.1186/1471-2490-14-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feng Y, Liu J, Kang Y, He Y, Liang B, Yang P, Yu Z. miR-19a acts as an oncogenic microRNA and is up-regulated in bladder cancer. J Exp Clin Cancer Res. 2014;33:67. doi: 10.1186/s13046-014-0067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adam L, Wszolek MF, Liu CG, Jing W, Diao L, Zien A, Zhang JD, Jackson D, Dinney CP. Plasma microRNA profiles for bladder cancer detection. Urol Oncol. 2013;31:1701–1708. doi: 10.1016/j.urolonc.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Y, Xu Z, Wang X. miRNA-373 promotes urinary bladder cancer cell proliferation, migration and invasion through upregulating epidermal growth factor receptor. Exp Ther Med. 2019;17:1190–1195. doi: 10.3892/etm.2018.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Motawi TK, Rizk SM, Ibrahim TM, Ibrahim IA. Circulating microRNAs, miR-92a, miR-100 and miR-143, as non-invasive biomarkers for bladder cancer diagnosis. Cell Biochem Funct. 2016;34:142–148. doi: 10.1002/cbf.3171. [DOI] [PubMed] [Google Scholar]
- 81.Yang Y, Qu A, Liu J, Wang R, Liu Y, Li G, Duan W, Fang Q, Jiang X, Wang L, et al. Serum miR-210 contributes to tumor detection, stage prediction and dynamic surveillance in patients with bladder cancer. PLoS ONE. 2015;10:e135168. doi: 10.1371/journal.pone.0135168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fang Z, Dai W, Wang X, Chen W, Shen C, Ye G, Li L. Circulating miR-205: a promising biomarker for the detection and prognosis evaluation of bladder cancer. Tumour Biol. 2016;37:8075–8082. doi: 10.1007/s13277-015-4698-y. [DOI] [PubMed] [Google Scholar]
- 83.Du M, Shi D, Yuan L, Li P, Chu H, Qin C, Yin C, Zhang Z, Wang M. Circulating miR-497 and miR-663b in plasma are potential novel biomarkers for bladder cancer. Sci Rep. 2015;5:10437. doi: 10.1038/srep10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lian J, Lin SH, Ye Y, Chang DW, Huang M, Dinney CP, Wu X. Serum microRNAs as predictors of risk for non-muscle invasive bladder cancer. Oncotarget. 2018;9:14895–14908. doi: 10.18632/oncotarget.24473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang X, Du L, Wang L, Li J, Liu Y, Zheng G, Qu A, Zhang X, Pan H, Yang Y, Wang C. Serum microRNA expression signatures identified from genome-wide microRNA profiling serve as novel noninvasive biomarkers for diagnosis and recurrence of bladder cancer. Int J Cancer. 2015;136:854–862. doi: 10.1002/ijc.29041. [DOI] [PubMed] [Google Scholar]
- 86.Jiang X, Du L, Duan W, Wang R, Yan K, Wang L, Li J, Zheng G, Zhang X, Yang Y, Wang C. Serum microRNA expression signatures as novel noninvasive biomarkers for prediction and prognosis of muscle-invasive bladder cancer. Oncotarget. 2016;7:36733–36742. doi: 10.18632/oncotarget.9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Usuba W, Urabe F, Yamamoto Y, Matsuzaki J, Sasaki H, Ichikawa M, Takizawa S, Aoki Y, Niida S, Kato K, et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci. 2019;110:408–419. doi: 10.1111/cas.13856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Foruzandeh Z, Zeinali-Sehrig F, Nejati K, Rahmanpour D, Pashazadeh F, Seif F, Alivand MR. CircRNAs as potent biomarkers in ovarian cancer: a systematic scoping review. Cell Mol Biol Lett. 2021;26:41. doi: 10.1186/s11658-021-00284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu Z, Liu X, Liu L, Deng H, Zhang J, Xu Q, Cen B, Ji A. Regulation of lncRNA expression. Cell Mol Biol Lett. 2014;19:561–575. doi: 10.2478/s11658-014-0212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Duan W, Du L, Jiang X, Wang R, Yan S, Xie Y, Yan K, Wang Q, Wang L, Zhang X, et al. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget. 2016;7:78850–78858. doi: 10.18632/oncotarget.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luo H, Xu C, Le W, Ge B, Wang T. lncRNA CASC11 promotes cancer cell proliferation in bladder cancer through miRNA-150. J Cell Biochem. 2019;120:13487–13493. doi: 10.1002/jcb.28622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou Y, Song X, Li X, Li H, Peng Y. Serum LUCAT1 implicates the pathogenesis of muscle-invasive bladder cancer via targeting miR-199a-5p and miR-199b-5p. J Mol Histol. 2020;51:583–591. doi: 10.1007/s10735-020-09907-3. [DOI] [PubMed] [Google Scholar]
- 93.Li G, Zhang Y, Mao J, Hu P, Chen Q, Ding W, Pu R. lncRNA TUC338 is a potential diagnostic biomarker for bladder cancer. J Cell Biochem. 2019;120:18014–18019. doi: 10.1002/jcb.29104. [DOI] [PubMed] [Google Scholar]
- 94.Zhang D, Du D, Yi S, Li X. LncRNA PCAT6: A potential biomarker for diagnosis and prognosis of bladder cancer. Ann Diagn Pathol. 2020;49:151642. doi: 10.1016/j.anndiagpath.2020.151642. [DOI] [PubMed] [Google Scholar]
- 95.Pan J, Xie X, Li H, Li Z, Ren C, Ming L. Detection of serum long non-coding RNA UCA1 and circular RNAs for the diagnosis of bladder cancer and prediction of recurrence. Int J Clin Exp Pathol. 2019;12:2951–2958. [PMC free article] [PubMed] [Google Scholar]
- 96.Chi BJ, Zhao DM, Liu L, Yin XZ, Wang FF, Bi S, Gui SL, Zhou SB, Qin WB, Wu DM, Wang SQ. Downregulation of hsa_circ_0000285 serves as a prognostic biomarker for bladder cancer and is involved in cisplatin resistance. Neoplasma. 2019;66:197–202. doi: 10.4149/neo_2018_180318N185. [DOI] [PubMed] [Google Scholar]
- 97.Xu ZQ, Yang MG, Liu HJ, Su CQ. Circular RNA hsa_circ_0003221 (circPTK2) promotes the proliferation and migration of bladder cancer cells. J Cell Biochem. 2018;119:3317–3325. doi: 10.1002/jcb.26492. [DOI] [PubMed] [Google Scholar]
- 98.Song P, Wu LR, Yan YH, Zhang JX, Chu T, Kwong LN, Patel AA, Zhang DY. Limitations and opportunities of technologies for the analysis of cell-free DNA in cancer diagnostics. Nat Biomed Eng. 2022;6:232–245. doi: 10.1038/s41551-021-00837-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang CC, Du M, Wang L. Bioinformatics Analysis for Circulating Cell-Free DNA in Cancer. Cancers (Basel) 2019;11:805. doi: 10.3390/cancers11060805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kumaki Y, Olsen S, Suenaga M, Nakagawa T, Uetake H, Ikeda S. Comprehensive genomic profiling of circulating Cell-Free DNA distinguishes Focal MET amplification from aneuploidy in diverse advanced cancers. Curr Oncol. 2021;28:3717–3728. doi: 10.3390/curroncol28050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheng ML, Pectasides E, Hanna GJ, Parsons HA, Choudhury AD, Oxnard GR. Circulating tumor DNA in advanced solid tumors: Clinical relevance and future directions. CA Cancer J Clin. 2021;71:176–190. doi: 10.3322/caac.21650. [DOI] [PubMed] [Google Scholar]
- 102.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
- 103.Freidin MB, Freydina DV, Leung M, Montero FA, Nicholson AG, Lim E. Circulating tumor DNA outperforms circulating tumor cells for KRAS mutation detection in thoracic malignancies. Clin Chem. 2015;61:1299–1304. doi: 10.1373/clinchem.2015.242453. [DOI] [PubMed] [Google Scholar]
- 104.Balgkouranidou I, Chimonidou M, Milaki G, Tsarouxa EG, Kakolyris S, Welch DR, Georgoulias V, Lianidou ES. Breast cancer metastasis suppressor-1 promoter methylation in cell-free DNA provides prognostic information in non-small cell lung cancer. Br J Cancer. 2014;110:2054–2062. doi: 10.1038/bjc.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Campitelli M, Jeannot E, Peter M, Lappartient E, Saada S, de la Rochefordière A, Fourchotte V, Alran S, Petrow P, Cottu P, et al. Human papillomavirus mutational insertion: specific marker of circulating tumor DNA in cervical cancer patients. PLoS ONE. 2012;7:e43393. doi: 10.1371/journal.pone.0043393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Husain H, Velculescu VE. Cancer DNA in the circulation: the liquid biopsy. JAMA. 2017;318:1272–1274. doi: 10.1001/jama.2017.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sato KA, Hachiya T, Iwaya T, Kume K, Matsuo T, Kawasaki K, Abiko Y, Akasaka R, Matsumoto T, Otsuka K, Nishizuka SS. Individualized mutation detection in circulating Tumor DNA for monitoring colorectal tumor burden using a cancer-associated gene sequencing panel. PLoS ONE. 2016;11:e146275. doi: 10.1371/journal.pone.0146275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang YH, Song Z, Hu XY, Wang HS. Circulating tumor DNA analysis for tumor diagnosis. Talanta. 2021;228:122220. doi: 10.1016/j.talanta.2021.122220. [DOI] [PubMed] [Google Scholar]
- 109.Kurtz DM, Soo J, Co TKL, Alig S, Chabon JJ, Sworder BJ, Schultz A, Jin MC, Scherer F, Garofalo A, et al. Enhanced detection of minimal residual disease by targeted sequencing of phased variants in circulating tumor DNA. Nat Biotechnol. 2021;39:1537–1547. doi: 10.1038/s41587-021-00981-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.De Mattos-Arruda L, Weigelt B, Cortes J, Won HH, Ng C, Nuciforo P, Bidard FC, Aura C, Saura C, Peg V, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25:1729–1735. doi: 10.1093/annonc/mdu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pessoa LS, Heringer M, Ferrer VP. ctDNA as a cancer biomarker: A broad overview. Crit Rev Oncol Hematol. 2020;155:103109. doi: 10.1016/j.critrevonc.2020.103109. [DOI] [PubMed] [Google Scholar]
- 112.Ignatiadis M, Dawson SJ. Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Ann Oncol. 2014;25:2304–2313. doi: 10.1093/annonc/mdu480. [DOI] [PubMed] [Google Scholar]
- 113.Ehlert T, Tug S, Brahmer A, Neef V, Heid F, Werner C, Jansen-Winkeln B, Kneist W, Lang H, Gockel I, Simon P. Establishing PNB-qPCR for quantifying minimal ctDNA concentrations during tumour resection. Sci Rep. 2017;7:8876. doi: 10.1038/s41598-017-09137-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Oxnard GR, Paweletz CP, Sholl LM. Genomic analysis of plasma cell-free DNA in patients with cancer. Jama Oncol. 2017;3:740–741. doi: 10.1001/jamaoncol.2016.2835. [DOI] [PubMed] [Google Scholar]
- 115.Lin C, Liu X, Zheng B, Ke R, Tzeng CM. Liquid Biopsy, ctDNA Diagnosis through NGS. Life (Basel) 2021;11:890. doi: 10.3390/life11090890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Deveson IW, Gong B, Lai K, LoCoco JS, Richmond TA, Schageman J, Zhang Z, Novoradovskaya N, Willey JC, Jones W, et al. Evaluating the analytical validity of circulating tumor DNA sequencing assays for precision oncology. Nat Biotechnol. 2021;39:1115–1128. doi: 10.1038/s41587-021-00857-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, Collisson EA, Divers SG, Hoon DS, Kopetz ES, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS ONE. 2015;10:e140712. doi: 10.1371/journal.pone.0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vandekerkhove G, Lavoie JM, Annala M, Murtha AJ, Sundahl N, Walz S, Sano T, Taavitsainen S, Ritch E, Fazli L, et al. Plasma ctDNA is a tumor tissue surrogate and enables clinical-genomic stratification of metastatic bladder cancer. Nat Commun. 2021;12:184. doi: 10.1038/s41467-020-20493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Malapelle U, Sirera R, Jantus-Lewintre E, Reclusa P, Calabuig-Fariñas S, Blasco A, Pisapia P, Rolfo C, Camps C. Profile of the Roche cobas® EGFR mutation test v2 for non-small cell lung cancer. Expert Rev Mol Diagn. 2017;17:209–215. doi: 10.1080/14737159.2017.1288568. [DOI] [PubMed] [Google Scholar]
- 120.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, Gale D, Forshew T, Mahler-Araujo B, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 121.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Valenzuela MT, Galisteo R, Zuluaga A, Villalobos M, Núñez MI, Oliver FJ, Ruiz DAJ. Assessing the use of p16(INK4a) promoter gene methylation in serum for detection of bladder cancer. Eur Urol. 2002;42(622–628):628–630. doi: 10.1016/s0302-2838(02)00468-2. [DOI] [PubMed] [Google Scholar]
- 123.Domínguez G, Carballido J, Silva J, Silva JM, García JM, Menéndez J, Provencio M, España P, Bonilla F. p14ARF promoter hypermethylation in plasma DNA as an indicator of disease recurrence in bladder cancer patients. Clin Cancer Res. 2002;8:980–985. [PubMed] [Google Scholar]
- 124.Ellinger J, El KN, Heukamp LC, Matthews S, Cubukluoz F, Kahl P, Perabo FG, Müller SC, von Ruecker A, Bastian PJ. Hypermethylation of cell-free serum DNA indicates worse outcome in patients with bladder cancer. J Urol. 2008;179:346–352. doi: 10.1016/j.juro.2007.08.091. [DOI] [PubMed] [Google Scholar]
- 125.Lin YL, Sun G, Liu XQ, Li WP, Ma JG. Clinical significance of CDH13 promoter methylation in serum samples from patients with bladder transitional cell carcinoma. J Int Med Res. 2011;39:179–186. doi: 10.1177/147323001103900119. [DOI] [PubMed] [Google Scholar]
- 126.Hauser S, Kogej M, Fechner G, Pezold VON, J, Vorreuther R, Lümmen G, Müller SC, Ellinger J, Serum DNA hypermethylation in patients with bladder cancer: results of a prospective multicenter study. Anticancer Res. 2013;33:779–784. [PubMed] [Google Scholar]
- 127.Millis SZ, Bryant D, Basu G, Bender R, Vranic S, Gatalica Z, Vogelzang NJ. Molecular profiling of infiltrating urothelial carcinoma of bladder and nonbladder origin. Clin Genitourin Cancer. 2015;13:e37–e49. doi: 10.1016/j.clgc.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 128.Patel KM, van der Vos KE, Smith CG, Mouliere F, Tsui D, Morris J, Chandrananda D, Marass F, van den Broek D, Neal DE, et al. Association of plasma and urinary mutant DNA with clinical outcomes in muscle invasive bladder cancer. Sci Rep. 2017;7:5554. doi: 10.1038/s41598-017-05623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Birkenkamp-Demtröder K, Christensen E, Nordentoft I, Knudsen M, Taber A, Høyer S, Lamy P, Agerbæk M, Jensen JB, Dyrskjøt L. Monitoring treatment response and metastatic relapse in advanced bladder cancer by liquid biopsy analysis. Eur Urol. 2018;73:535–540. doi: 10.1016/j.eururo.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 130.Christensen E, Birkenkamp-Demtröder K, Sethi H, Shchegrova S, Salari R, Nordentoft I, Wu HT, Knudsen M, Lamy P, Lindskrog SV, et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra-deep sequencing of plasma cell-free DNA in patients with urothelial bladder carcinoma. J Clin Oncol. 2019;37:1547–1557. doi: 10.1200/JCO.18.02052. [DOI] [PubMed] [Google Scholar]
- 131.Birkenkamp-Demtröder K, Nordentoft I, Christensen E, Høyer S, Reinert T, Vang S, Borre M, Agerbæk M, Jensen JB, Ørntoft TF, Dyrskjøt L. Genomic alterations in liquid biopsies from patients with bladder cancer. Eur Urol. 2016;70:75–82. doi: 10.1016/j.eururo.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 132.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224r. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Corcoran RB, Chabner BA. Application of Cell-free DNA Analysis to Cancer Treatment. N Engl J Med. 2018;379:1754–1765. doi: 10.1056/NEJMra1706174. [DOI] [PubMed] [Google Scholar]
- 134.Shohdy KS, Villamar DM, Cao Y, Trieu J, Price KS, Nagy R, Tagawa ST, Molina AM, Sternberg CN, Nanus DM, et al. Serial ctDNA analysis predicts clinical progression in patients with advanced urothelial carcinoma. Br J Cancer. 2022;126:430–439. doi: 10.1038/s41416-021-01648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Grivas P, Lalani AA, Pond GR, Nagy RJ, Faltas B, Agarwal N, Gupta SV, Drakaki A, Vaishampayan UN, Wang J, et al. Circulating tumor DNA alterations in advanced urothelial carcinoma and association with clinical outcomes: a pilot study. Eur Urol Oncol. 2020;3:695–699. doi: 10.1016/j.euo.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 136.Green EA, Li R, Albiges L, Choueiri TK, Freedman M, Pal S, Dyrskjøt L, Kamat AM. Clinical utility of cell-free and circulating tumor DNA in kidney and bladder cancer: a critical review of current literature. Eur Urol Oncol. 2021;4:893–903. doi: 10.1016/j.euo.2021.04.005. [DOI] [PubMed] [Google Scholar]
- 137.Powles T, Assaf ZJ, Davarpanah N, Banchereau R, Szabados BE, Yuen KC, Grivas P, Hussain M, Oudard S, Gschwend JE, et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature. 2021;595:432–437. doi: 10.1038/s41586-021-03642-9. [DOI] [PubMed] [Google Scholar]
- 138.Zhang J, Dai D, Tian J, Li L, Bai J, Xu Y, Wang Z, Tang A. Circulating tumor DNA analyses predict disease recurrence in non-muscle-invasive bladder cancer. Front Oncol. 2021;11:657483. doi: 10.3389/fonc.2021.657483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sundahl N, Vandekerkhove G, Decaestecker K, Meireson A, De Visschere P, Fonteyne V, De Maeseneer D, Reynders D, Goetghebeur E, Van Dorpe J, et al. Randomized phase 1 trial of pembrolizumab with sequential versus concomitant stereotactic body radiotherapy in metastatic urothelial carcinoma. Eur Urol. 2019;75:707–711. doi: 10.1016/j.eururo.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 140.de Kruijff IE, Beije N, Martens J, de Wit R, Boormans JL, Sleijfer S. Liquid biopsies to select patients for perioperative chemotherapy in muscle-invasive bladder cancer: a systematic review. Eur Urol Oncol. 2021;4:204–214. doi: 10.1016/j.euo.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 141.Khagi Y, Goodman AM, Daniels GA, Patel SP, Sacco AG, Randall JM, Bazhenova LA, Kurzrock R. Hypermutated circulating tumor DNA: correlation with response to checkpoint inhibitor-based immunotherapy. Clin Cancer Res. 2017;23:5729–5736. doi: 10.1158/1078-0432.CCR-17-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Raja R, Kuziora M, Brohawn PZ, Higgs BW, Gupta A, Dennis PA, Ranade K. Early Reduction in ctDNA Predicts Survival in patients with lung and bladder cancer treated with durvalumab. Clin Cancer Res. 2018;24:6212–6222. doi: 10.1158/1078-0432.CCR-18-0386. [DOI] [PubMed] [Google Scholar]
- 143.Ravi P, Ravi A, Riaz IB, Freeman D, Curran C, Mantia C, McGregor BA, Kilbridge KL, Pan CX, Pek M, et al. Longitudinal evaluation of circulating tumor DNA using sensitive amplicon-based next-generation sequencing to identify resistance mechanisms to immune checkpoint inhibitors for advanced urothelial carcinoma. Oncologist. 2022;27:e406–e409. doi: 10.1093/oncolo/oyac037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yáñez-Mó M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 146.Shen J, Zhu X, Fei J, Shi P, Yu S, Zhou J. Advances of exosome in the development of ovarian cancer and its diagnostic and therapeutic prospect. Onco Targets Ther. 2018;11:2831–2841. doi: 10.2147/OTT.S159829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:897. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Oliveira GJ, Zigon E, Rogers G, Davodian D, Lu S, Jovanovic-Talisman T, Jones J, Tigges J, Tyagi S, Ghiran IC. Detection of Extracellular Vesicle RNA Using Molecular Beacons. Science. 2020;23:100782. doi: 10.1016/j.isci.2019.100782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yu W, Hurley J, Roberts D, Chakrabortty SK, Enderle D, Noerholm M, Breakefield XO, Skog JK. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. 2021;32:466–477. doi: 10.1016/j.annonc.2021.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 151.Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 152.Shao J, Jin Y, Shao C, Fan H, Wang X, Yang G. Serum exosomal pregnancy zone protein as a promising biomarker in inflammatory bowel disease. Cell Mol Biol Lett. 2021;26:36. doi: 10.1186/s11658-021-00280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tong Y, Liu X, Xia D, Peng E, Yang X, Liu H, Ye T, Wang X, He Y, Xu H, et al. Biological roles and clinical significance of exosome-derived noncoding RNAs in bladder cancer. Front Oncol. 2021;11:704703. doi: 10.3389/fonc.2021.704703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lu L, Huang J, Mo J, Da X, Li Q, Fan M, Lu H. Exosomal lncRNA TUG1 from cancer-associated fibroblasts promotes liver cancer cell migration, invasion, and glycolysis by regulating the miR-524-5p/SIX1 axis. Cell Mol Biol Lett. 2022;27:17. doi: 10.1186/s11658-022-00309-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mateescu B, Kowal EJ, van Balkom BW, Bartel S, Bhattacharyya SN, Buzás EI, Buck AH, de Candia P, Chow FW, Das S, et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J Extracell Vesicles. 2017;6:1286095. doi: 10.1080/20013078.2017.1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ramirez MI, Amorim MG, Gadelha C, Milic I, Welsh JA, Freitas VM, Nawaz M, Akbar N, Couch Y, Makin L, et al. Technical challenges of working with extracellular vesicles. Nanoscale. 2018;10:881–906. doi: 10.1039/C7NR08360B. [DOI] [PubMed] [Google Scholar]
- 157.Wang J, Yang K, Yuan W, Gao Z. Determination of serum exosomal H19 as a noninvasive biomarker for bladder cancer diagnosis and prognosis. Med Sci Monit. 2018;24:9307–9316. doi: 10.12659/MSM.912018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang S, Du L, Wang L, Jiang X, Zhan Y, Li J, Yan K, Duan W, Zhao Y, Wang L, et al. Evaluation of serum exosomal LncRNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med. 2019;23:1396–1405. doi: 10.1111/jcmm.14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xue M, Chen W, Xiang A, Wang R, Chen H, Pan J, Pang H, An H, Wang X, Hou H, Li X. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer. 2017;16:143. doi: 10.1186/s12943-017-0714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Elsharkawi F, Elsabah M, Shabayek M, Khaled H. Urine and serum exosomes as novel biomarkers in detection of bladder cancer. Asian Pac J Cancer Prev. 2019;20:2219–2224. doi: 10.31557/APJCP.2019.20.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zheng R, Du M, Wang X, Xu W, Liang J, Wang W, Lv Q, Qin C, Chu H, Wang M, et al. Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer. 2018;17:143. doi: 10.1186/s12943-018-0880-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Di W, Zhang W, Zhu B, Li X, Tang Q, Zhou Y. Colorectal cancer prompted adipose tissue browning and cancer cachexia through transferring exosomal miR-146b-5p. J Cell Physiol. 2021;236:5399–5410. doi: 10.1002/jcp.30245. [DOI] [PubMed] [Google Scholar]
- 163.Takahashi RU, Prieto-Vila M, Hironaka A, Ochiya T. The role of extracellular vesicle microRNAs in cancer biology. Clin Chem Lab Med. 2017;55:648–656. doi: 10.1515/cclm-2016-0708. [DOI] [PubMed] [Google Scholar]
- 164.Liang Z, Liu L, Gao R, Che C, Yang G. Downregulation of exosomal miR-7-5p promotes breast cancer migration and invasion by targeting RYK and participating in the atypical WNT signalling pathway. Cell Mol Biol Lett. 2022;27:88. doi: 10.1186/s11658-022-00393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lin F, Yin HB, Li XY, Zhu GM, He WY, Gou X. Bladder cancer cell-secreted exosomal miR-21 activates the PI3K/AKT pathway in macrophages to promote cancer progression. Int J Oncol. 2020;56:151–164. doi: 10.3892/ijo.2019.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jia Y, Ding X, Zhou L, Zhang L, Yang X. Mesenchymal stem cells-derived exosomal microRNA-139-5p restrains tumorigenesis in bladder cancer by targeting PRC1. Oncogene. 2021;40:246–261. doi: 10.1038/s41388-020-01486-7. [DOI] [PubMed] [Google Scholar]
- 167.Yin X, Zheng X, Liu M, Wang D, Sun H, Qiu Y, Chen J, Shi B. Exosomal miR-663b targets Ets2-repressor factor to promote proliferation and the epithelial-mesenchymal transition of bladder cancer cells. Cell Biol Int. 2020;44:958–965. doi: 10.1002/cbin.11292. [DOI] [PubMed] [Google Scholar]
- 168.Yan L, Li Q, Sun K, Jiang F. MiR-4644 is upregulated in plasma exosomes of bladder cancer patients and promotes bladder cancer progression by targeting UBIAD1. Am J Transl Res. 2020;12:6277–6289. [PMC free article] [PubMed] [Google Scholar]
- 169.Li Q, Huyan T, Cai S, Huang Q, Zhang M, Peng H, Zhang Y, Liu N, Zhang W. The role of exosomal miR-375-3p: A potential suppressor in bladder cancer via the Wnt/尾-catenin pathway. Faseb J. 2020;34:12177–12196. doi: 10.1096/fj.202000347R. [DOI] [PubMed] [Google Scholar]
- 170.Cai X, Qu L, Yang J, Xu J, Sun L, Wei X, Qu X, Bai T, Guo Z, Zhu Y. Exosome-transmitted microRNA-133b inhibited bladder cancer proliferation by upregulating dual-specificity protein phosphatase 1. Cancer Med. 2020;9:6009–6019. doi: 10.1002/cam4.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sabo AA, Birolo G, Naccarati A, Dragomir MP, Aneli S, Allione A, Oderda M, Allasia M, Gontero P, Sacerdote C, et al. Small Non-Coding RNA profiling in plasma extracellular vesicles of bladder cancer patients by next-generation sequencing: expression levels of miR-126–3p and piR-5936 Increase with Higher Histologic Grades. Cancers (Basel) 2020;12:78. doi: 10.3390/cancers12061507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Luo G, Zhang Y, Wu Z, Zhang L, Liang C, Chen X. Exosomal LINC00355 derived from cancer-associated fibroblasts promotes bladder cancer cell resistance to cisplatin by regulating miR-34b-5p/ABCB1 axis. Acta Biochim Biophys Sin (Shanghai) 2021;53:558–566. doi: 10.1093/abbs/gmab023. [DOI] [PubMed] [Google Scholar]
- 173.Zhang Y, Luo G, You S, Zhang L, Liang C, Chen X. Exosomal LINC00355 derived from cancer-associated fibroblasts promotes bladder cancer cell proliferation and invasion by regulating miR-15a-5p/HMGA2 axis. Acta Biochim Biophys Sin (Shanghai) 2021;53:673–682. doi: 10.1093/abbs/gmab041. [DOI] [PubMed] [Google Scholar]
- 174.Yang H, Qu H, Huang H, Mu Z, Mao M, Xie Q, Wang K, Hu B. Exosomes-mediated transfer of long noncoding RNA LINC01133 represses bladder cancer progression via regulating the Wnt signaling pathway. Cell Biol Int. 2021;45:1510–1522. doi: 10.1002/cbin.11590. [DOI] [PubMed] [Google Scholar]
- 175.Liu Q. The emerging roles of exosomal long non-coding RNAs in bladder cancer. J Cell Mol Med. 2022;26:966–976. doi: 10.1111/jcmm.17152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yan L, Wang P, Fang W, Liang C. Cancer-associated fibroblasts-derived exosomes-mediated transfer of LINC00355 regulates bladder cancer cell proliferation and invasion. Cell Biochem Funct. 2020;38:257–265. doi: 10.1002/cbf.3462. [DOI] [PubMed] [Google Scholar]
- 177.Chen C, Luo Y, He W, Zhao Y, Kong Y, Liu H, Zhong G, Li Y, Li J, Huang J, et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest. 2020;130:404–421. doi: 10.1172/JCI130892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Park RJ, Hong YJ, Wu Y, Kim PM, Hong YK. Exosomes as a communication tool between the lymphatic system and bladder cancer. Int Neurourol J. 2018;22:220–224. doi: 10.5213/inj.1836186.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Seimiya T, Otsuka M, Iwata T, Shibata C, Tanaka E, Suzuki T, Koike K. Emerging roles of exosomal circular RNAs in cancer. Front Cell Dev Biol. 2020;8:568366. doi: 10.3389/fcell.2020.568366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, Ma P, Jiang H, Wu X, Shu Y, Xu T. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 2020;19:112. doi: 10.1186/s12943-020-01208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, Chen L, Liu Q, He P, Ye Y. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–128. doi: 10.1016/j.canlet.2020.01.022. [DOI] [PubMed] [Google Scholar]
- 182.Yang C, Wu S, Mou Z, Zhou Q, Dai X, Ou Y, Chen X, Chen Y, Xu C, Hu Y, et al. Exosome-derived circTRPS1 promotes malignant phenotype and CD8+ T cell exhaustion in bladder cancer microenvironments. Mol Ther. 2022;30:1054–1070. doi: 10.1016/j.ymthe.2022.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ, et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin Cancer Res. 2018;24:6319–6330. doi: 10.1158/1078-0432.CCR-18-1270. [DOI] [PubMed] [Google Scholar]
- 184.Yoshida K, Tsuda M, Matsumoto R, Semba S, Wang L, Sugino H, Tanino M, Kondo T, Tanabe K, Tanaka S. Exosomes containing ErbB2/CRK induce vascular growth in premetastatic niches and promote metastasis of bladder cancer. Cancer Sci. 2019;110:2119–2132. doi: 10.1111/cas.14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kumar A, Misra BB. Challenges and opportunities in cancer metabolomics. Proteomics. 2019;19:e1900042. doi: 10.1002/pmic.201900042. [DOI] [PubMed] [Google Scholar]
- 186.Graça G, Lau CE, Gonçalves LG. Exploring cancer metabolism: applications of metabolomics and metabolic phenotyping in cancer research and diagnostics. Adv Exp Med Biol. 2020;1219:367–385. doi: 10.1007/978-3-030-34025-4_19. [DOI] [PubMed] [Google Scholar]
- 187.Schmidt DR, Patel R, Kirsch DG, Lewis CA, Vander HM, Locasale JW. Metabolomics in cancer research and emerging applications in clinical oncology. Cancer J Clin. 2021;71:333–358. doi: 10.3322/caac.21670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Walsh MC, Brennan L, Malthouse JP, Roche HM, Gibney MJ. Effect of acute dietary standardization on the urinary, plasma, and salivary metabolomic profiles of healthy humans. Am J Clin Nutr. 2006;84:531–539. doi: 10.1093/ajcn/84.3.531. [DOI] [PubMed] [Google Scholar]
- 189.Li Y, Ruan Q, Li Y, Ye G, Lu X, Lin X, Xu G. A novel approach to transforming a non-targeted metabolic profiling method to a pseudo-targeted method using the retention time locking gas chromatography/mass spectrometry-selected ions monitoring. J Chromatogr A. 2012;1255:228–236. doi: 10.1016/j.chroma.2012.01.076. [DOI] [PubMed] [Google Scholar]
- 190.Du H, Rao Y, Liu R, Deng K, Guan Y, Luo D, Mao Q, Yu J, Bo T, Fan Z, et al. Proteomics and metabolomics analyses reveal the full spectrum of inflammatory and lipid metabolic abnormalities in dyslipidemia. Biomed Chromatogr. 2021;35:e5183. doi: 10.1002/bmc.5183. [DOI] [PubMed] [Google Scholar]
- 191.Liu Y, Lu S, Zhao L, Dong X, Zhu Z, Jin Y, Chen H, Lu F, Hong Z, Chai Y. Effects of glaucocalyxin A on human liver cancer cells as revealed by GC/MS- and LC/MS-based metabolic profiling. Anal Bioanal Chem. 2018;410:3325–3335. doi: 10.1007/s00216-018-0996-8. [DOI] [PubMed] [Google Scholar]
- 192.Lee SH, Jeong S, Lee J, Yeo IS, Oh MJ, Kim U, Kim S, Kim SH, Park SY, Kim JH, et al. Glycomic profiling of targeted serum haptoglobin for gastric cancer using nano LC/MS and LC/MS/MS. Mol Biosyst. 2016;12:3611–3621. doi: 10.1039/C6MB00559D. [DOI] [PubMed] [Google Scholar]
- 193.Crook AA, Powers R. Quantitative NMR-Based Biomedical Metabolomics: Current Status and Applications. Molecules. 2020;25:7. doi: 10.3390/molecules25215128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Thadikkaran L, Siegenthaler MA, Crettaz D, Queloz PA, Schneider P, Tissot JD. Recent advances in blood-related proteomics. Proteomics. 2005;5:3019–3034. doi: 10.1002/pmic.200402053. [DOI] [PubMed] [Google Scholar]
- 195.Pan S, Brentnall TA, Chen R. Proteomics analysis of bodily fluids in pancreatic cancer. Proteomics. 2015;15:2705–2715. doi: 10.1002/pmic.201400476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Cao M, Zhao L, Chen H, Xue W, Lin D. NMR-based metabolomic analysis of human bladder cancer. Anal Sci. 2012;28:451–456. doi: 10.2116/analsci.28.451. [DOI] [PubMed] [Google Scholar]
- 197.Bansal N, Gupta A, Mitash N, Shakya PS, Mandhani A, Mahdi AA, Sankhwar SN, Mandal SK. Low- and high-grade bladder cancer determination via human serum-based metabolomics approach. J Proteome Res. 2013;12:5839–5850. doi: 10.1021/pr400859w. [DOI] [PubMed] [Google Scholar]
- 198.Tan G, Wang H, Yuan J, Qin W, Dong X, Wu H, Meng P. Three serum metabolite signatures for diagnosing low-grade and high-grade bladder cancer. Sci Rep. 2017;7:46176. doi: 10.1038/srep46176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Schwamborn K, Krieg RC, Grosse J, Reulen N, Weiskirchen R, Knuechel R, Jakse G, Henkel C. Serum proteomic profiling in patients with bladder cancer. Eur Urol. 2009;56:989–996. doi: 10.1016/j.eururo.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 200.Bansal N, Gupta A, Sankhwar SN, Mahdi AA. Low- and high-grade bladder cancer appraisal via serum-based proteomics approach. Clin Chim Acta. 2014;436:97–103. doi: 10.1016/j.cca.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 201.Bansal N, Gupta AK, Gupta A, Sankhwar SN, Mahdi AA. Serum-based protein biomarkers of bladder cancer: A pre- and post-operative evaluation. J Pharm Biomed Anal. 2016;124:22–25. doi: 10.1016/j.jpba.2016.02.026. [DOI] [PubMed] [Google Scholar]
- 202.van der Knaap JA, Verrijzer CP. Undercover: gene control by metabolites and metabolic enzymes. Genes Dev. 2016;30:2345–2369. doi: 10.1101/gad.289140.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Crispo F, Condelli V, Lepore S, Notarangelo T, Sgambato A, Esposito F, Maddalena F, Landriscina M. Metabolic dysregulations and epigenetics: a bidirectional interplay that drives tumor progression. Cells (Basel) 2019;8:56. doi: 10.3390/cells8080798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hao Y, Li D, Xu Y, Ouyang J, Wang Y, Zhang Y, Li B, Xie L, Qin G. Investigation of lipid metabolism dysregulation and the effects on immune microenvironments in pan-cancer using multiple omics data. BMC Bioinformatics. 2019;20:195. doi: 10.1186/s12859-019-2734-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Aggen DH, Drake CG. Biomarkers for immunotherapy in bladder cancer: a moving target. J Immunother Cancer. 2017;5:94. doi: 10.1186/s40425-017-0299-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lee MY, Yeon A, Shahid M, Cho E, Sairam V, Figlin R, Kim KH, Kim J. Reprogrammed lipid metabolism in bladder cancer with cisplatin resistance. Oncotarget. 2018;9:13231–13243. doi: 10.18632/oncotarget.24229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Amara CS, Ambati CR, Vantaku V, Badrajee PD, Donepudi SR, Ravi SS, Arnold JM, Putluri V, Chatta G, Guru KA, et al. Serum metabolic profiling identified a distinct metabolic signature in bladder cancer smokers: a key metabolic enzyme associated with patient survival. Cancer Epidemiol Biomarkers Prev. 2019;28:770–781. doi: 10.1158/1055-9965.EPI-18-0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Minami S, Sato Y, Matsumoto T, Kageyama T, Kawashima Y, Yoshio K, Ishii J, Matsumoto K, Nagashio R, Okayasu I. Proteomic study of sera from patients with bladder cancer: usefulness of S100A8 and S100A9 proteins. Cancer Genomics Proteomics. 2010;7:181–189. [PubMed] [Google Scholar]
- 209.Lemańska-Perek A, Lis-Kuberka J, Lepczyński A, Dratwa-Chałupnik A, Tupikowski K, Kątnik-Prastowska I, Ożgo M. Potential plasma biomarkers of bladder cancer identified by proteomic analysis: a pilot study. Adv Clin Exp Med. 2019;28:339–346. doi: 10.17219/acem/79296. [DOI] [PubMed] [Google Scholar]
- 210.Lin D, Shen L, Luo M, Zhang K, Li J, Yang Q, Zhu F, Zhou D, Zheng S, Chen Y, Zhou J. Circulating tumor cells: biology and clinical significance. Signal Transduct Target Ther. 2021;6:404. doi: 10.1038/s41392-021-00817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.van Dessel LF, Beije N, Helmijr JC, Vitale SR, Kraan J, Look MP, de Wit R, Sleijfer S, Jansen MP, Martens JW, Lolkema MP. Application of circulating tumor DNA in prospective clinical oncology trials - standardization of preanalytical conditions. Mol Oncol. 2017;11:295–304. doi: 10.1002/1878-0261.12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.