Highlights
-
•
Expression of eEF1A1 and eEF1A2 variants is tissue-specific and mutually exclusive.
-
•
eEF1A1 has an extended shape in solution while eEF1A2 is a compact protein.
-
•
eEF1A1 and eEF1A2 may have distinct non-translational functions.
-
•
eEF1B complex is a trimer in solution encompassing six guanine exchange sites.
-
•
eEF1B and/or its subunits may function in non-translational processes.
Keywords: Translation elongation, Multiprotein complex, Protein spatial organization, eEF1A1, eEF1A2, eEF1B
Abstract
The eEF1 family of mammalian translation elongation factors is comprised of the two variants of eEF1A (eEF1A1 and eEF1A2), and the eEF1B complex. The latter consists of eEF1Bα, eEF1Bβ, and eEF1Bγ subunits. The two eEF1A variants have similar translation activity but may differ with respect to their secondary, “moonlighting” functions. This variability is underlined by the difference in the spatial organization of eEF1A1 and eEF1A2, and also possibly by the differences in their post-translational modifications. Here, we review the data on the spatial organization and post-translation modifications of eEF1A1 and eEF1A2, and provide examples of their involvement in various processes in addition to translation. We also describe the structural models of eEF1B subunits, their organization in the subcomplexes, and the trimeric model of the entire eEF1B complex. We discuss the functional consequences of such an assembly into a complex as well as the involvement of individual subunits in non-translational processes.
1. Introduction
The eEF1A1 and eEF1A2 variants of eEF1A, and the eEF1Bα, eEF1Bβ and eEF1Bγ subunits of the eEF1B complex comprise the eEF1 family of mammalian translation elongation factors. A key role of the eEF1 family in protein biosynthesis is the delivery of aminoacyl–tRNA to а corresponding codon of the mRNA-programmed ribosome. Though the basic principles of eukaryotic eEF1A function resemble those of prokaryotic EF1A (also known as EF-Tu), there are some important differences in their actions [1]. One reason for this may be the compartmentalization of the translation apparatus that serves as a structural basis for tRNA/aminoacyl–tRNA channeling during protein synthesis in mammalian cells. eEF1 is a central partner in this process, and transfers aminoacyl–tRNA from aminoacyl–tRNA synthetase to the ribosome, and then a deacylated tRNA back to aminoacyl–tRNA synthetase [2]. Also, the members of the eEF1 family were found to interact with several non-translational proteins, and are probably involved in carrying out other independent moonlighting functions in the cell. Both eEF1A1 and eEF1A2 have been implicated in the development of numerous human diseases through their enhancement of oncogenesis, the blockade of apoptosis, and increased viral pathogenesis. All these features have prompted an increasing interest in the eEF1 family. In this review, we focus on the structure of the eEF1 family members as the basis for understanding their involvement in mammalian translation and other cellular functions.
2. eEF1A1 and eEF1A2
eEF1A1 was considered to be the only mammalian analogue of bacterial EF-Tu factor until the tissue-specific eEF1A2 variant in rat and human tissues was discovered by Eugenia Wang's [3] and Brian Clark's [4] labs, respectively. The variants were 97% homologous and 92% similar, suggesting their full functional resemblance. The expression of eEF1A1 and eEF1A2 was mutually exclusive. eEF1A2 was found in muscle and brain tissue, whereas eEF1A1 was expressed in all other tissue types. The EEF1A1 and EEF1A2 genes were assigned to different human chromosomes [5]. The developmental role of eEF1A1/eEF1A2 substitution in neuronal tissue and muscle was illuminated by the detection of a wasted mutation in mice, disabling the expression of the EEF1A2 gene, which is normally observed in postnatal mouse development [6]. eEF1A2 was described as an oncogene in ovarian and lung cancer [7, 8] whereas it is absent in corresponding healthy tissue. This raised the question of the selective regulation of eEF1A1 and eEF1A2 expression, which may be disrupted by cancer. We concluded that the uncontrolled increase of the eEF1A2 expression in cancerous tissue may result from the loss of miRNA-mediated control. Indeed, we found that miR-663 and miR-744 negatively influenced the amount of eEF1A2 in breast cancer cells, and inhibited their proliferation. Apart from indicating the role of these microRNA as anticancer agents, this suggests that one of the possible mechanisms of tissue-specific eEF1A2 suppression could be related to miRNA [9]. This is especially important, considering the recently observed co-expression of eEF1A1 and eEF1A2 mRNAs in mature hippocampal neurons and the predominant expression of eEF1A1 mRNA in dendrites [10].
What is so specific about the eEF1A variants with 92% similar amino acids sequences, which makes it impossible to replace one with another? One way to answer this question is to physically examine differences between the eEF1A1 and eEF1A2 proteins. A comparison of the physical properties of the eEF1A1 and eEF1A2 proteins became possible after development of the procedures of their isolation in preparative quantities from mammalian tissues [11, 12], as the mammalian proteins expressed in E. coli were not functionally active. The differential scanning calorimetry values of transition temperature, half-width, and enthalpy change for the eEF1A1 and eEF1A2 proteins suggested that the latter had a more compact state. Subsequent CD measurements of eEF1A1 and eEF1A2 displayed the difference in their near-UV region spectra, indicating the difference in the tertiary-structure organization of the proteins [13]. A comparison of the surface hydrophobicity of the protein variants revealed that eEF1A1 displayed more hydrophobic properties than eEF1A2 [14]. In line with this, small-angle X-ray scattering, and analytical ultracentrifugation showed eEF1A1’s increased ability to self-associate. Also, small-angle X-ray scattering measurements at various urea concentrations revealed a lower resistance of the A1 protein structure to denaturation by urea [14]. All these data permitted us to conclude that the eEF1A2 protein is more compact and stable than eEF1A1. Therefore, eEF1A2, rather than eEF1A1, was chosen for structural studies by X-ray crystallography. eEF1A2 crystals were successfully obtained [15], and the X-ray structure of the dimeric form of eEF1A2 was solved [16]. The first X-ray structure of mammalian eEF1A revealed major differences in the organization of its nucleotide-binding domain, when compared to bacterial analogue EF-Tu, namely, a lack of involvement of a Mg2+ ion in stabilizing GDP. The availability of the structure of yeast eEF1A in the complex with nucleotide exchange factor eEF1Bα [17] permitted the modeling of structural rearrangements of eEF1A during the nucleotide exchange process. Understanding the intramolecular movements of eEF1A allowed us to suggest that the first step of eEF1A*GDP dissociation from the 80S ribosome is the rotation of its nucleotide-binding domain, induced by GTP hydrolysis [16]. The structure obtained proved to be useful for modeling eEF1A2*GDP*aminoacyl–tRNA/tRNA complexes that may contribute to an understanding the overall translation elongation control in eukaryotic cells [18] and mechanisms of the aminoacyl–tRNA/tRNA channeling process [19], [20], [21].
Along with substitutions of certain amino acid residues, the structures of eEF1A1 and eEF1A2 may differ in the number of post-translational modifications. Our earlier observation that more compact eEF1A2 may be less modified post-translationally than eEF1A1 [22] is consistent with the recent data. The PhosphositePlus database [23] mentions twenty seven phosphorylated, nineteen acetylated, and twenty three ubiquitinated amino acid residues in eEF1A1, and twelve phosphorylated, ten acetylated, and seven ubiquitinylated residues in eEF1A2 (accessed July 26, 2022). Here, the sites with more than three independently confirmed modifications were considered. These data may provide independent evidence that the spatial structure of eEF1A1 is more “open,” as it is more accessible to the enzymes that catalyze various post-translational modifications.
Interestingly, the number and location of methylated amino acid residues in eEF1A1 and eEF1A2 were quite similar, which may be explained by the localization of methylated residues on the surfaces of eEF1A1 and eEF1A2 proteins. In 1989, William Merrick's lab identified five methylated residues of rabbit eEF1A1 [24], but the mammalian eEF1A methylation studies practically came to a standstill for twenty five years, until corresponding methyltransferases were identified (for a review see [25, 26]). Consequently, it became clear that eEF1A is the only known target of these enzymes. Dimethylation of Lys55 was found to promote tumorigenesis by increasing translation efficiency [27], probably by stabilizing a certain conformation of the nucleotide binding site, caused by the strengthening of cation-π interaction with neighboring aromatic residues [28]. A possible, important role of other methylated lysine residues in both variants of eEF1A is not yet known. Recently we verified whether methylation may have an effect on the interaction of eEF1A2 with the subunits of nucleotide exchange complex eEF1B [29]. We produced a set of eEF1A2 mutants in which potentially methylated Lys-residues were replaced with Arg. BRET analysis and pull-down experiments have shown no difference in the interaction of eEF1Bα, eEF1Bβ, and eEF1Bγ subunits of eEF1B with eEF1A2 and its unmethylable mutants in HEK293 cells [29]. This suggests that the methylation of Lys35, Lys55, Lys79, Lys165, and Lys318 does not influence the interaction of eEF1A2 with the eEF1B complex.
We have demonstrated that in contrast to eEF1A1, eEF1A2 interacts with the SH2 domains of Grb2, RasGAP, Shc and the C-terminal part of Shp2, and with the SH3in domains of Crk, Fgr, Fyn, and phospholipase C-gamma1 [30]. Tyrosine phosphorylation normally promotes the interactions of signaling proteins via SH2 domains, while for SH3 domains, the post-translational modification prevents interaction. The specific phosphorylated tyrosine residues responsible for this interaction remain unidentified. A recent theoretical analysis led to the interesting conclusion that all Tyr-residues, reported to be phosphorylated in eEF1A2, cannot be phosphorylated in a crystal dimer structure, owing to chemical constraints [28]. The straightforward explanation for this apparent contradiction is the difference in the spatial structures of an individual monomer, and a monomer that is part of a dimer. Besides, it seems possible that the crystal and in-solution structures of the dimer differ. Also, we cannot exclude the existence of a set of various solution conformations of both monomeric and dimeric forms of eEF1A2. Finally, the 56th and 177th Tyr-residues found to be phosphorylated, according to the PhosphoSitePlus database, were not included in the analysis [28], for unknown reasons. A possible conformational variety of eEF1A variants should be particularly investigated in the future studies. If we consider Ser-and Thr, there were seventeen additional phosphorylated residues in eEF1A1, and three additional phosphorylated residues in eEF1A2, compared to the opposite isoform (no search limits in PhosphositePlus were imposed). It is worth mentioning an eEF1A1-specific phosphorylation pattern in domain 1, including S157, T158, S163, S175, and T176, which may be linked to the increased structural lability of eEF1A1. Prospective roles of eEF1A phosphorylation and other post-translational modifications of the protein were described by Mills and Gago [28].
Both eEF1A1 and eEF1A2 promoted actin bundling. The dimerization of eEF1A2 in the crystal permitted us to specify a requirement for a dimeric structure of eEF1A to interact with F-actin. Peculiarly, removing the N-terminal part of eEF1A completely eradicated eEF1A's actin-bundling property, despite this element not being necessary to the interaction of eEF1A with actin [31]. On the other hand, according to the crystal structure, the N-terminal part was important for the formation of eEF1A dimers [16], which is why it has been concluded that the dimers are responsible for actin bundling. The model of actin-bundle formation in the presence of eEF1A dimers was advanced, and it was suggested that domain 3 of each monomer in the eEF1A dimer interacts with different actin chains [31]. Subsequent modeling and molecular dynamic simulation studies showed that it is specifically the C-terminal 445 to 463 α-helical region of each eEF1A2 monomer that is most probably involved in actin bundling [32]. In contrast to eEF1A1, eEF1A2 induced formation of short and thick actin bundles [33], which was eventually explained by the different contacts of the actin-interacting C-terminal region, triggered by the presence of a penultimate glycine residue in eEF1A2, rather than in eEF1A1 [32]. Interestingly, in mice, the overexpression of eEF1A2—but not eEF1A1—leads to cytoskeleton rearrangements that may contribute to corticospinal axon repair following injury [34]
There is an indication that eEF1A2 mutant with four eEF1A2-specific phosphomimetic substitutions in domain 3 (S342E, S358E, S393E, and S445E) loses both F-actin binding and bundling activities, and shows increased mobility in spines and dendrites [35]. Ser 445 is an element of the C-terminal region that was predicted to provide eEF1A2–actin interaction, which suggests that its phosphorylation may be most crucial for preventing the eEF1A2–actin interaction. On the other hand, Ser 358 is the only residue of the four mentioned, whose phosphorylation has been confirmed experimentally (PhosphositePlus, accessed July 26, 2022). Also, the exclusive phosphorylation of Ser 454 in eEF1A1 may have an effect on the eEF1A1-induced shape of actin bundles. However, eEF1A1 may also have a different mechanism for regulating its binding to actin. We have found that Ca2+/calmodulin is capable of binding eEF1A1 [36], and interferes with its tRNA-binding and actin-bundling activities in vitro, whereas eEF1A2 showed no signs of interacting with calmodulin [33]. As Ca2+/calmodulin dislocates tRNA from its complex with eEF1A1, and suppresses the actin-bundling activity of the latter, it is possible to hypothesize that the replacing eEF1A1 by eEF1A2 in muscle, myocardial, and neuronal tissues is necessary for protecting ribosomal polypeptide elongation from the effect of quick changes in [Ca2+] observed in these tissues [37,38] .
Our further investigation of the possible functional effects of structural differences between eEF1A1 and eEF1A2 revealed that eEF1A1 rather than eEF1A2 interacted with the Sgt1 (suppressor of G2 allele of Skp1) protein. The interaction sites were identified, and we modeled a protein–protein complex that demonstrated both the shape and charge complementarities of the eEF1A1–Sgt1 interface, stabilized by a few salt bridges [39]. Sgt1 is involved in antiviral defense systems [40], whereas eEF1A interacts with viral RNA to enhance virus performance [41]. Therefore, we suggested that Sgt1 can compete with viral RNA, disrupting the eEF1A1–RNA complex, and provided in vitro evidence that such a mechanism may be functional [39].
Apart from eEF1A's involvement in viral propagation, including SARS-CoV-2 [42] and HIV1 [43], and cytoskeleton rearrangement, which may also be virus-related [44], it may be linked to a few other non-translational processes. The eEF1A variants participate in protein renaturation [45], contribute to spermatogenesis [46], are involved in cell-cycle progression [47], apoptosis [48], and autophagy [49], participate in the heat shock response [50] and protect against toxin-induced neuronal death [51]. They are associated with carcinogenesis [52], myocardial hypertrophy [53], Parkinson's disease [54], Alzheimer's disease [55], fatal familial insomnia [56], and neurodevelopmental disorders [57], may exacerbate insulin resistance [58], and act as an antigen for the autoantibodies production in type 1 diabetes [59]. We realize that a full understanding of the exact role of the eEF1A variants in non-translational cellular processes and diseases still has a way to go, but recent progress in structural and proteomics studies gives us hope that there will be rapid progress toward this goal.
3. eEF1B
Mammalian complex eEF1B comprises three subunits – eEF1Bα (EEF1B2), eEF1Bβ (EEF1D), and eEF1Bγ (EEF1G) – according to the established nomenclature [60]. In the UniProtKB database, eEF1Bα is described as Elongation factor 1-beta (EF1B_human, accession number P24534), eEF1Bβ as Elongation factor 1-delta (EF1D_human, accession number P29692), and eEF1Bγ as Elongation factor 1-gamma (EF1G_human, accession number P26641). eEF1Bα and eEF1Bγ have been well-preserved during evolution, whereas eEF1Bβ is a metazoan-specific protein [61]. The N-terminal domains of all eEF1B subunits are responsible for their involvement in the complex, whereas the C-terminal domains of eEF1Bα and eEF1Bβ carry out nucleotide exchange activity [62]. The complete spatial structures of the subunits are not known, however, there are some structural data on their domains.
The part of the human eEF1Bα C-terminal domain was resolved by NMR [63]. The unpublished structure of the human eEF1Bα N-terminal domain, crystallized in the complex with the eEF1Bγ N-terminal domain, is present in PDB (5DQS). The structure of the human eEF1Bβ C-terminal fragment was determined by NMR [64], whereas the unpublished structure of the human eEF1Bβ N-terminal domain fragment complexed with the eEF1Bγ N-terminal domain is available in PDB (5JPO). Also known are the NMR-derived structure of the 19 kDa C-terminal fragment of human eEF1Bγ [65], the crystal structure of the individual N-terminal domain of yeast eEF1Bγ [66], and the aforementioned structures of the human eEF1Bγ N-terminal domain complexed with its eEF1Bα or eEF1Bβ counterparts.
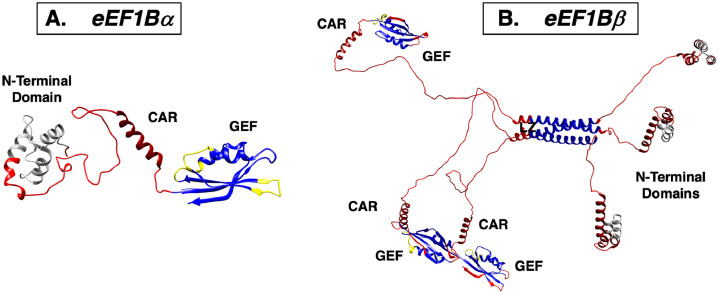
We have found that human eEF1Bα possesses an elongated shape in solution [67]. The structures of the folded N- and C-terminal domains of eEF1Bα are known, and represent rigidly structured elements of this protein. We used hydrogen–deuterium exchange coupled with mass spectrometry (HDX–MS) [68] to show that, in contrast, the linker and central acidic region are highly dynamic [69]. Hence, eEF1Bα has a structure wherein two structured domains are connected by a long dynamic linker (Fig. 1A).
Fig. 1.
Structural dynamic organization of (A) eEF1Bα and (B) eEF1Bβ subunits of the eEF1B complex. The structural models are depicted using previously published PDB file [69] and are colored in accordance with the dynamic data from the H/D exchange experiments. Abbreviation of the structural domains: GEF – guanine nucleotide exchange factor, LZ – leucine zipper motif, CAR – central acidic region. Rigidly structured regions with high protection from H/D exchange are indicated in blue, dynamic regions with weak and no protection from H/D exchange are indicated in yellow and red, respectively. CAR region of both proteins and the N-terminal domain of eEF1Bβ that display no protection from H/D exchange but were modeled to have α-helical organization are shown in dark red. Gray color indicates the region of eEF1Bα and eEF1Bβ which undergo additional protection from H/D exchange upon interaction with eEF1Bγ subunit. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
This may contribute to the general dynamic properties of eEF1Bα. Interestingly, N-terminally truncated eEF1Bα showed increased nucleotide exchange activity, which was unexpected, considering that catalytic activity of eEF1Bα resides in its C-terminal part. A conventional role of the N-terminal domain of eEF1Bα is to bind eEF1Bγ. Addition of eEF1Bγ has a similarly positive effect on the activity of full-sized eEF1Bα. All these data led to the conclusion that the N-terminal domain negatively influences eEF1A binding to the C-terminal catalytic domain of the individual eEF1Bα subunit, owing to the overall flexibility of the molecule, whereas the formation of a stable complex of the eEF1Bγ and eEF1Bα N-terminal domains establishes a way to overcome this inhibitory effect [67]. It may have some regulatory consequences in cancer cells, where eEF1Bα is present in both unbound and bound to eEF1B statuses. Interestingly, the self-inhibitory action of a distant domain of a protein was previously observed in a different member of eukaryotic translation machinery: methionyl-tRNA synthetase [70,71].
We have revealed that the N-terminal protein-binding domain of eEF1Bβ (1–77 residues) is a monomer in solution [72]. This eEF1Bβ fragment has been found to bind eEF1Bγ in an equimolar ratio. The secondary structure of eEF1Bβ(1–77) is mostly α-helical, with a portion of disordered region. The flexibility of this entity was confirmed by rapid hydrogen/deuterium exchange for all eEF1Bβ(1–77) peptides measured with HDX–MS. Taking into account a computational modeling of eEF1Bβ(1–77), we suggested the existence of several conformation states of eEF1Bβ(1–77), each composed of three labile α-helices connected by flexible linkers.
Together, the data suggest that the protein-binding domain of eEF1Bβ shows a highly dynamic spatial organization, which may be needed for interaction with eEF1Bγ or other protein partners [72]. Further research was directed at elucidating the exact oligomeric status of the eEF1Bβ molecule, as the purified protein was seen to form oligomers in solution [73]. The analytical ultracentrifugation in sedimentation velocity mode, and the sedimentation equilibrium data showed that recombinant eEF1Bβ self-associates in a stable trimer with a highly elongated shape [69]. In the subsequent experiments we unequivocally attributed a trimer-forming role to a heptad repeat that contains six leucine residues that occupy every seventh position, and create a hydrophobic path along the helix. The trimeric coiled-coil conformation of the leucine-zipper motif region was successfully modeled [69]. The internal dynamics of the eEF1Bβ trimer were estimated by HDX–MS. The leucine-zipper motif region and most of the guanine exchange (GEF) domain were shown to be rigidly structured. In contrast, the N-terminal domain, linker, and central acidic region were demonstrated to be unprotected, indicating highly dynamic structures. An atomistic model of full-sized eEF1Bβ was developed by combining a homology modeling approach with HDX–MS and analytical ultracentrifugation data (Fig. 1B) [69].
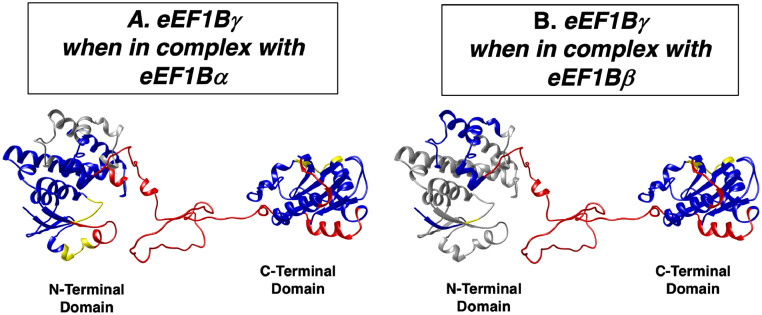
As mentioned above, the crystal and NMR-derived structures of the individual eEF1Bγ domains are known. We combined the homology-modeling approach and HDX analysis to develop an atomistic model that shows that eEF1Bγ is a non-globular protein with a moderately elongated shape, which consists of two rigidly structured domains connected by a long, highly dynamic linker (Fig. 2) [69]. The eEF1Bγ protein is known for its elevated self-aggregation capacity [74,75], so it is rather difficult to estimate its actual oligomeric state in vitro and in vivo. The sedimentation velocity experiments revealed that the minor and major eEF1Bγ sedimenting species had masses close to the monomeric and dimeric forms of the factor, whereas analytical ultracentrifugation in the sedimentation equilibrium mode showed a mixture of monomeric and variously-sized oligomeric forms of eEF1Bγ in solution [69].
Fig. 2.
Structural dynamic organization of eEF1Bγ subunit. The structural model of each subunit is depicted using previously published PDB file [69] and are colored in accordance with the dynamic data from the H/D exchange experiments. Rigidly structured regions with high protection from H/D exchange are indicated in blue, dynamic regions with weak and no protection from H/D exchange are indicated in yellow and red, respectively. Gray color indicates the region of eEF1Bγ that undergoes additional protection from H/D exchange upon interaction with eEF1Bα (A) and eEF1Bβ (B). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
Until now, there was no information about the structure of the whole eEF1B complex, except for low-resolution EM images of ValRS/EF-1 complexes published in 2005 [76]. At approximately that time, a few biochemical studies by various laboratories time proposed various structural models of the eEF1B complex. One of the first models was developed by the Wim Möller lab, and suggested that the N-terminal domains of eEF1Bα and eEF1Bβ bind to the eEF1Bγ subunit to form the eEF1Bαβγ complex, whereas each of their C-terminal domains binds one molecule of eEF1A [77]. The structural role of the leucine-zipper motif of eEF1Bβ in a possible dimerization of eEF1Bαβγ was first proposed by the Jean-Pierre Waller lab [78], whereas Minella et al. hypothesized about the trimerization of an eEF1Bαβγ monomer [79]. Charlotte Knudsen's lab demonstrated the independent binding of eEF1Bα and eEF1Bβ to eEF1Bγ, using a three-hybrid yeast system [80]. Jolinda Traugh suggested the dimerization of eEF1Bγ, with eEF1Bα and eEF1Bβ binding to various molecules of eEF1Bγ in the eEF1Bαβγ complexes, which, in turn, could be dimerized via the eEF1Bα subunit [81].
We have determined the regions of the subunits involved in the interaction within the eEF1B complex by using HDX–MS. A whole N-terminal domain of eEF1Bα (Fig. 1A) and the only 19-amino-acid-long region of eEF1Bβ (Fig. 1B) increased their protection against deuterium incorporation when interacting with eEF1Bγ [69]. eEF1Bγ is known to bind eEF1Bα and eEF1Bβ concurrently [80], which makes it interesting to understand how eEF1Bγ binds both proteins at the same time. As expected, according to HDX–MS, the N-terminal part of eEF1Bγ participated in the interaction with both proteins. Amino acid residues 144–161 and 170–190 participated in the interaction with eEF1Bα (Fig. 2A), whereas most of the remaining peptides of the N-terminal region were more strongly protected when they interacted with eEF1Bβ (Fig. 2B). The C-terminal domain and the linker region of eEF1Bγ were not involved in this interaction. The same interaction patterns for the eEF1Bα, eEF1Bβ, and eEF1Bγ proteins were observed for the whole eEF1B complex [69].
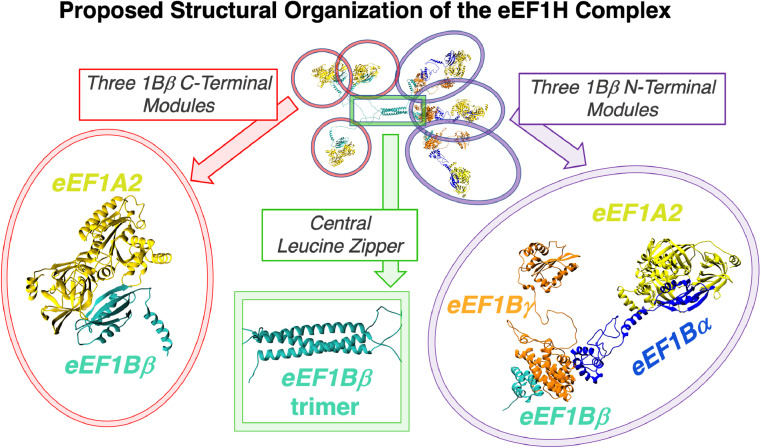
The stoichiometry of the binary eEF1Bαγ and eEF1Bβγ and ternary eEF1Bαβγ complexes was studied by analytical ultracentrifugation in various modes. eEF1Bαγ revealed the presence of eEF1Bαγ and eEF1B(αγ)2 forms. eEF1Bβγ displayed peaks that corresponded to the molecular masses of heterotrimer eEF1B(βγ)3 and heterohexamer eEF1B(βγ)6. Sedimentation velocity analysis of the ternary eEF1Bαβγ complex indicated the presence of the major species corresponding to the heterotrimeric eEF1B(αβγ)3, whereas sedimentation equilibrium analysis revealed the presence of the heterotrimer–heterohexamer mixture in solution. We concluded that the α, β, and γ subunits of eEF1B preferentially associate in a stable heterotrimeric complex with the possibility of further oligomerization in vitro, mediated by the eEF1Bγ subunit [69]. The existence of a heterotrimeric eEF1B (αβγ)3 complex indicates the presence of six guanine nucleotide exchanging domains within one complex, which raises the question of whether they function concurrently. We found that eEF1B(αβγ)3 was saturated by eEF1A2 in a 1:6 molar ratio, demonstrating that one molecule of the eEF1B complex can successfully bind up to six molecules of eEF1A2 [69]. The supercomplex of eEF1B and eEF1A has historically been called the heavy complex, or eEF1H. The model of spatial organization of eEF1H is shown in Fig. 3.
Fig. 3.
Structural organization of the eEF1H complex. eEF1Bα, eEF1Bβ and eEF1Bγ subunits are painted in blue, green and orange, respectively. eEF1А2 is painted in yellow. The structural model of the eEF1B complex is depicted using previously published PDB file [69]. The structural model of eEF1А2 was developed using the PDB structures 4C0S and 6RA9. The proposed structure is assembled upon a template of a trimer of eEF1Bβ subunits, held together by a Central Leucine Zipper region (green rectangle). The C-terminal domain of each eEF1Bβ subunit interacts with one eEF1A2 molecule (pink ovals). The N-terminal domain of each eEF1Bβ subunit interacts directly with one eEF1Bγ subunit, with the latter engaging a complex of eEF1Bα subunit and another eEF1A2 molecule (purple ovals). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
An obvious role of the stable trimeric eEF1B complex may be to increase the local concentration of the GEF domains, which may be important for maintaining efficient GDP/GTP exchange in the eEF1A molecules in the vicinity of the translating ribosomes. The question whether eEF1Bα and eEF1Bβ show identical functions in vivo remains unanswered. An exclusive role of eEF1Bβ may be to serve as a sole nucleotide exchange factor for eEF1A interacting with valyl-tRNA synthetase and forming valyl–tRNA–eEF1A–GTP complexes [82] which does not exclude eEF1Bβ interaction with eEF1A molecules that are not routed to valyl-tRNA synthetase. In turn, eEF1Bα may be a preferential nucleotide exchange factor for eEF1A2 in neurons. Neuronal and muscle-tissue-specific eEF1A2 depends more significantly on nucleotide exchange factors than does eEF1A1 [67]. Local translation of specific mRNAs in different regions of neurons is well known, thus the presence of trimeric eEF1B in the neuronal translational compartments may be especially important to supporting efficient translation of specific mRNAs. There is also an indication of a possibility of localized translation in a sarcomere [83].
Recently-published translatomes of synapse/axon- and neuron-body-enriched sections of hippocampi [84] revealed that the distribution of newly synthesized eEF1A2 among these fractions was different than the distribution of eEF1Bβ and eEF1Bγ. However, the distribution of eEF1Bα still followed eEF1A2-like trends. If the glial and neuronal cells were compared, eEF1A1, eEF1Bβ, and eEF1Bγ showed very similar glial preference, whereas eEF1A2 demonstrated neuronal localization, and eEF1Bα occupied an intermediate position. Additional observation that favored possible eE1A2–eEF1Bα coupling came from the study of eEF1B subunit expression in the brain and liver throughout late embryonic and postnatal mouse development [85]. The amount of eEF1Bα in late embryonic tissue was very low. However, it increased, especially in brain tissue, and stabilized during postnatal development, to some extent resembling the changes in the expression profile of eEF1A2 in the mouse brain. In contrast, eEF1Bβ was abundantly expressed in late embryonic tissue, whereas the amount rapidly decreased and stabilized after 10 days of post-embryonic brain and liver development. Interestingly, the phosphomymetic eEF1A2 mutants of 342, 358, 393, and 445 serine residues showed a significant reduction in the interaction with eEF1Bα in the spines of hippocampal neurons and in the soma of mouse neuroblasts Neuro-2A cell line, with no significant effects on translation [35]. The idea that eEF1Bα may be a local nucleotide exchange factor of eEF1A2*GDP is not new (see [85] for discussion). A plausible explanation is that some portion of just-synthesized eEF1Bα does not enter the eEF1B complex, and in some cases may serve as a local nucleotide exchange factor for eEF1A2*GDP. In any case, these findings raise a question concerning the extent of integrity of the whole eEF1B complex in vivo.
We addressed the existence of individual subunits of the eEF1B complex in the organism by studying their expression level in various human cancer tissues. The changes in the expression of various eEF1 subunits in human cancers, compared to those in the tumor-surrounding tissue, were found to be unbalanced. The independent overexpression of at least one eEF1 component was observed in 72% of 25 clinical samples of cardioesophageal cancer [86] and 52% of 25 samples of lung cancer [87]. Peculiarly, a concomitant cancer-related increase of both eEF1Bβ and eEF1Bγ was found in four cases of cardioesophageal cancer and five cases of lung carcinomas whereas, no cases of a coordinated increase in eEF1Bα and eEF1Bγ were revealed. The coordinated up-regulation of eEF1Bβ and eEF1Bγ was also found in oral squamous cell carcinoma [88]. An associated increase in eEF1Bβ and eEF1Bγ expression was noticed in the prenatal (compared to postnatal) mouse brain and liver [85]. These data hint at the possibility of the separate functioning of pools of individual eEF1Bα, and the eEF1Bβ–eEF1Bγ subcomplex in brain tissue. However, if eEF1Bγ was not present in the complex with eEF1Bα, it would not overcome the self-inhibitory effect of the latter, described above. In this case, the guanine nucleotide exchange activity of eEF1Bα decreased, and the GDP-bound form of the neuron-specific eEF1A2 variant may accumulate in the cell, perhaps fulfilling some non-translational role specific to eEF1A2*GDP.
Immunohistochemical analysis of eEF1B subunits in normal and cancerous tissue indicated the possibility of their non-overlapping localization. In normal cardioesophageal tissue, localization of eEF1Bα, eEF1Bβ and eEF1Bγ subunits was nuclear-cytoplasmic, whereas in the cancerous tissue the only eEF1Bγ subunit was revealed in the nucleus [86]. eEF1Bα was present in the nuclei of normal lung cells, eEF1Bβ was found in the nuclei of lung carcinoma cells, and eEF1Bγ showed nuclear localization in both normal and carcinomatous tissues [87]. However, all three subunits of eEF1B were present in both the cytoplasm and the nucleus of lung carcinoma cell line A549, suggesting cell/tissue-specific effects [87].
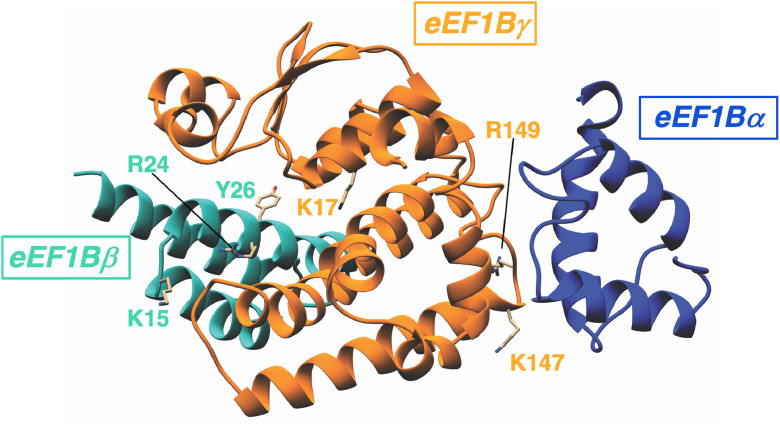
Hence, a pool of individual subunits can exist separately from the eEF1B complex in human tissues. We believe that this is mainly the result of the impossibility of newly synthesized subunits entering the complex, which could be modulated by their post-translational modification. Given the known regions of interaction between the subunits [69], we have analyzed data on modifications of eEF1B subunits presented by the PhosphositePlus database (accessed 18 July 2022). Five phosphorylated residues were found in the N-terminal domain of eEF1Bα, whereas in eEF1Bγ the post-translational modifications were found only in the interaction region comprising residues 144–161. Of those, no modifications in eEF1Bα, and the modifications of K147 and R149 in eEF1Bγ are promising candidates, as they may affect eEF1Bγ–eEF1Bα interaction, according to the eEF1B structure [69]. Unexpectedly heavy phosphorylation of the linker region of eEF1Bα was detected, where 8 residues were found to be phosphorylated in the short region comprising 79 to 95 residues. The phosphorylation of the linker may directly or indirectly influence the overall structural dynamics of eEF1Bα, and affect the self-inhibitory interaction between N- and C-terminal parts of the protein in its non-complexed state [67].
In eEF1Bβ, a short region comprising 11 to 29 residues interacts with eEF1Bγ [69]. Remarkably, it contains a few modified residues, including phosphorylated tyrosines, monomethylated arginine, acetylated or ubiquitylated lysines. From these, modifications of K15, R24, and Y26 may have an effect on eEF1Bβ binding to eEF1Bγ. So far, in eEF1Bγ, sixteen post-translationally modified residues were found in its extended region, interacting with eEF1Bβ, however the only K17 modification may influence the eEF1Bγ–eEF1Bβ interaction, according to the eEF1B structure [69].
All the residues whose modification may influence the interaction of eEF1Bα and eEF1Bβ with eEF1Bγ are depicted in Fig. 4. It should be noted that the PhosphositePlus database reports on post-translational modifications of the subunits most likely involved in the eEF1B complex rather than the free forms. One may suggest that more sites are open to modification in the individual subunits than in complexed ones. To address this question, one needs to conduct special studies of the eEF1B subunit modifications under conditions where a portion of individual subunits is increased, as in some human cancer tissues.
Fig. 4.
Structural model of the interface of the N-terminal domains of eEF1Bα, eEF1Bβ and eEF1Bγ subunits in the eEF1B complex. The eEF1Bα, eEF1Bβ and eEF1Bγ domains are painted in blue, green and orange, respectively. The indicated residues can undergo posttranslational modifications, potentially influencing protein-protein interactions in the complex. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
Individual eEF1B subunits or their subcomplexes can participate in translation, albeit with less efficiency in the case of individual eEF1Bα. However, several processes that show little or no relation to translation are known to be linked to eEF1B subunits [62]. For instance, eEF1Bα may be involved in repair of DNA double-strand breaks [89,90] and cytoskeleton rearrangements [91]. eEF1Bβ is related to adipose tissue inflammation and diabetes [92,93]. eEF1Bγ negatively regulates the motor-mediated transport of membrane organelles along microtubules [94]. It is involved in the retrograde transport from Golgi apparatus to endoplasmic reticulum [95] and microRNAs transfer to extracellular fluid [96]. It is not clear yet whether eEF1Bα, eEF1Bβ, and eEF1Bγ fulfill these and other functions in an individual or complex state.
To understand a variety of non-translational processes coupled to eEF1Bβ and eEF1Bγ subunits in human cells, we identified their possible protein partners by co-immunoprecipitation with subsequent mass spectrometric analysis. Several bioinformatic approaches were used to classify the biological functions of the partners [97], [98], [99], [100]. Based on the similarity of the processes in which the subunits are involved, we could predict that the eEF1Bβ–eEF1Bγ complex is involved in cell-cycle regulation, chromatin and nucleosome remodeling, mRNA splicing and turnover. Individual eEF1Bβ may be linked to DNA replication and repair, adipose tissue biology, epithelial-to-mesenchymal transition, microRNA turnover, protein chaperoning and degradation. Individual eEF1Bγ may be coupled to viral RNA transcription, oxidative stress response, cytoskeleton–membrane linking, and cellular trafficking. We do not consider this list to be in any way complete, however, it gives some idea of what to expect from the further development of eEF1B studies.
4. Concluding remarks
In this short review, we present a summary of our knowledge of the eEF1 family of mammalian translation elongation factors, attained through years of studies by several generations of researchers. We analyzed the data on spatial organization and post-translation modifications of eEF1A1 and eEF1A2, and provided examples of their various involvements in a variety of non-translational processes. The structural models of eEF1B subunits, their organization in the subcomplexes, and the trimeric structural model of the whole eEF1B complex are described. The possibility of the individual subunits’ involvement in some non-translational processes is also discussed.
We believe that this description of the structure and possible functions of the members of the eEF1 family during and beyond translation will encourage further studies. These could include investigating the role of post-translational modifications of eEF1 proteins, deciphering the structural organization of eEF1A1 and eEF1A2 protein variants, research on non-translational functions of the members of the eEF1 group, and identification of auxiliary members of this group, together with further study of their possible synergistic actions in health and disease.
Funding
The studies of the Ukrainian laboratory was supported by the National Research Foundation of Ukraine (project 2020.02/0028); the “Genomic, molecular and cell foundations for development of the innovation biotechnologies” program of the National Academy of Sciences of Ukraine; and the Mobility Program 2022–2024 of the NAS of Ukraine and Polish Academy of Sciences.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
BSN is an AUFF–Ukraine Research Fellow at Aarhus University, Denmark funded by the Aarhus University Research Foundation. He thanks S.R. Keiding, L. Holm, and V.M. Sorensen for their ongoing support. We would like to thank C.R. Knudsen for useful comments on the manuscript. We thank Prof. Alexey Ladokhin for valuable advice on presentation of the illustrations.
Data Availability
No data was used for the research described in the article.
References
- 1.El'skaya A.V., Negrutskii B.S., Shalak V.F., Vislovukh A.A., Vlasenko D.O., Novosylna A.V., Lukash T.O., Veremieva M.V. Specific features of protein biosynthesis in higher eukaryotes. Biopolym. Cell. 2013;29:177–187. doi: 10.7124/bc.000818. [DOI] [Google Scholar]
- 2.Negrutskii B.S., El'skaya A.V. Eukaryotic translation elongation factor 1 alpha: structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog. Nucleic Acid Res. Mol. Biol. 1998;60:47–78. doi: 10.1016/s0079-6603(08)60889-2. [DOI] [PubMed] [Google Scholar]
- 3.Lee S., Francoeur A.M., Liu S., Wang E. Tissue-specific expression in mammalian brain, heart, and muscle of S1, a member of the elongation factor-1 alpha gene family. J. Biol. Chem. 1992;267:24064–24068. doi: 10.1016/S0021-9258(18)35946-5. [DOI] [PubMed] [Google Scholar]
- 4.Knudsen S.M., Frydenberg J., Clark B.F., Leffers H. Tissue-dependent variation in the expression of elongation factor-1 alpha isoforms: isolation and characterisation of a cDNA encoding a novel variant of human elongation-factor 1 alpha. Eur. J. Biochem. 1993;215:549–554. doi: 10.1111/j.1432-1033.1993.tb18064.x. [DOI] [PubMed] [Google Scholar]
- 5.Lund A., Knudsen S.M., Vissing H., Clark B., Tommerup N. Assignment of human elongation factor 1alpha genes: EEF1A maps to chromosome 6q14 and EEF1A2 to 20q13.3. Genomics. 1996;36:359–361. doi: 10.1006/geno.1996.0475. [DOI] [PubMed] [Google Scholar]
- 6.Chambers D.M., Peters J., Abbott C.M. The lethal mutation of the mouse wasted (wst) is a deletion that abolishes expression of a tissue-specific isoform of translation elongation factor 1alpha, encoded by the Eef1a2 gene. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4463–4468. doi: 10.1073/pnas.95.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anand N., Murthy S., Amann G., Wernick M., Porter L.A., Cukier I.H., Collins C., Gray J.W., Diebold J., Demetrick D.J., Lee J.M. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat. Genet. 2002;31:301–305. doi: 10.1038/ng904. [DOI] [PubMed] [Google Scholar]
- 8.Jia L., Ge X., Du C., Chen L., Zhou Y., Xiong W., Xiang J., Li G., Xiao G., Fang L., Li Z. EEF1A2 interacts with HSP90AB1 to promote lung adenocarcinoma metastasis via enhancing TGF-β/SMAD signalling. Br. J. Cancer. 2021;124:1301–1311. doi: 10.1038/s41416-020-01250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vislovukh A., Kratassiouk G., Porto E., Gralievska N., Beldiman C., Pinna G., El'skaya A., Harel-Bellan A., Negrutskii B., Groisman I. Proto-oncogenic isoform A2 of eukaryotic translation elongation factor eEF1 is a target of miR-663 and miR-744. Br. J. Cancer. 2013;108:2304–2311. doi: 10.1038/bjc.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wefers Z., Alecki C., Huang R., Jacob-Tomas S., Vera M. Analysis of the Expression and Subcellular Distribution of eEF1A1 and eEF1A2 mRNAs during Neurodevelopment. Cells. 2022:11. doi: 10.3390/cells11121877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shalak V.F., Budkevich T.V., Negrutskil B.S., El'skaia A.V. A fast and effective method for purification of elongation factor 1 alpha from rabbit liver. Ukr. Biochem. J. 1997;69:104–109. [PubMed] [Google Scholar]
- 12.Kahns S., Lund A., Kristensen P., Knudsen C.R., Clark B.F., Cavallius J., Merrick W.C. The elongation factor 1 A-2 isoform from rabbit: cloning of the cDNA and characterization of the protein. Nucleic Acids Res. 1998;26:1884–1890. doi: 10.1093/nar/26.8.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novosylna O.V., Timchenko A.A., Tiktopulo E.I., Serdyuk I.N., Negrutskii B.S., El`skaya A.V. Characterization of physical properties of two isoforms of translation elongation factor 1A. Biopolym. Cell. 2007;23:386–390. doi: 10.7124/bc.000777. [DOI] [Google Scholar]
- 14.Timchenko A.A., Novosylna O.V., Prituzhalov E.A., Kihara H., El'skaya A.V., Negrutskii B.S., Serdyuk I.N. Different oligomeric properties and stability of highly homologous A1 and proto-oncogenic A2 variants of mammalian translation elongation factor eEF1. Biochemistry. 2013;52:5345–5353. doi: 10.1021/bi400400r. [DOI] [PubMed] [Google Scholar]
- 15.Yaremchuk A., Shalak V.F., Novosylna O.V., Negrutskii B.S., Crepin T., El'skaya A.V., Tukalo M. Purification, crystallization and preliminary X-ray crystallographic analysis of mammalian translation elongation factor eEF1A2. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012;68:295–297. doi: 10.1107/S1744309112000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crepin T., Shalak V.F., Yaremchuk A.D., Vlasenko D.O., McCarthy A., Negrutskii B.S., Tukalo M.A., El'skaya A.V. Mammalian translation elongation factor eEF1A2: x-ray structure and new features of GDP/GTP exchange mechanism in higher eukaryotes. Nucleic Acids Res. 2014;42:12939–12948. doi: 10.1093/nar/gku974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen G.R., Pedersen L., Valente L., Chatterjee I., Kinzy T.G., Kjeldgaard M., Nyborg J. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Balpha. Mol. Cell. 2000;6:1261–1266. doi: 10.1016/s1097-2765(00)00122-2. [DOI] [PubMed] [Google Scholar]
- 18.Negrutskii B., Vlasenko D., Mirande M., Futernyk P., El'skaya A. mRNA-Independent way to regulate translation elongation rate in eukaryotic cells. IUBMB Life. 2018;70:192–196. doi: 10.1002/iub.1724. [DOI] [PubMed] [Google Scholar]
- 19.Negrutskii B.S., Deutscher M.P. Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc. Natl. Acad. Sci. U. S. A. 1991;88:4991–4995. doi: 10.1073/pnas.88.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrushenko Z.M., Budkevich T.V., Shalak V.F., Negrutskii B.S., El'skaya A.V. Novel complexes of mammalian translation elongation factor eEF1A.GDP with uncharged tRNA and aminoacyl-tRNA synthetase. Implications for tRNA channeling. Eur. J. Biochem. 2002;269:4811–4818. doi: 10.1046/j.1432-1033.2002.03178.x. [DOI] [PubMed] [Google Scholar]
- 21.Petrushenko Z.M., Negrutskii B.S., Ladokhin A.S., Budkevich T.V., Shalak V.F., El'skaya A.V. Evidence for the formation of an unusual ternary complex of rabbit liver EF-1alpha with GDP and deacylated tRNA. FEBS Lett. 1997;407:13–17. doi: 10.1016/s0014-5793(97)00242-1. [DOI] [PubMed] [Google Scholar]
- 22.Negrutskii B., Vlasenko D., El'skaya A. From global phosphoproteomics to individual proteins: the case of translation elongation factor eEF1A. Expert Rev. Proteomics. 2012;9:71–83. doi: 10.1586/epr.11.71. [DOI] [PubMed] [Google Scholar]
- 23.Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dever T.E., Costello C.E., Owens C.L., Rosenberry T.L., Merrick W.C. Location of seven post-translational modifications in rabbit elongation factor 1 alpha including dimethyllysine, trimethyllysine, and glycerylphosphorylethanolamine. J. Biol. Chem. 1989;264:20518–20525. doi: 10.1016/S0021-9258(19)47093-2. [DOI] [PubMed] [Google Scholar]
- 25.Hamey J.J., Wilkins M.R. Methylation of Elongation Factor 1A: where, Who, and Why? Trends Biochem. Sci. 2018;43:211–223. doi: 10.1016/j.tibs.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Jakobsson M.E., Małecki J., Falnes P.Ø. Regulation of eukaryotic elongation factor 1 alpha (eEF1A) by dynamic lysine methylation. RNA Biol. 2018;15:314–319. doi: 10.1080/15476286.2018.1440875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu S., Hausmann S., Carlson S.M., Fuentes M.E., Francis J.W., Pillai R., Lofgren S.M., Hulea L., Tandoc K., Lu J., Li A., Nguyen N.D., Caporicci M., Kim M.P., Maitra A., Wang H., Wistuba I.I., Porco J.A.J., Bassik M.C., Elias J.E., Song J., Topisirovic I., Van Rechem C., Mazur P.K., Gozani O. METTL13 Methylation of eEF1A Increases Translational Output to Promote Tumorigenesis. Cell. 2019;176:491–504.e21. doi: 10.1016/j.cell.2018.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mills A., Gago F. On the Need to Tell Apart Fraternal Twins eEF1A1 and eEF1A2, and Their Respective Outfits. Int. J. Mol. Sci. 2021:22. doi: 10.3390/ijms22136973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porubleva L.V., Kolesnik D.L., El'skaya A.V., Negrutskii B.S. Methylation of human elongation factor eef1a2 is not essential for eef1a2-eef1b interaction. Biopolym. Cell. 2020;36:254–263. doi: 10.7124/bc.000A31. [DOI] [Google Scholar]
- 30.Panasyuk G., Nemazanyy I., Filonenko V., Negrutskii B., El'skaya A.V. A2 isoform of mammalian translation factor eEF1A displays increased tyrosine phosphorylation and ability to interact with different signalling molecules. Int. J. Biochem. Cell Biol. 2008;40:63–71. doi: 10.1016/j.biocel.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vlasenko D.O., Novosylna O.V., Negrutskii B.S., El'skaya A.V. Truncation of the A, A∗, A′ helices segment impairs the actin bundling activity of mammalian eEF1A1. FEBS Lett. 2015;589:1187–1193. doi: 10.1016/j.febslet.2015.03.030. [DOI] [PubMed] [Google Scholar]
- 32.Carriles A.A., Mills A., Muñoz-Alonso M.-.J., Gutiérrez D., Domínguez J.M., Hermoso J.A., Gago F. Structural Cues for Understanding eEF1A2 Moonlighting. Chembiochem. 2021;22:374–391. doi: 10.1002/cbic.202000516. [DOI] [PubMed] [Google Scholar]
- 33.Novosylna O., Doyle A., Vlasenko D., Murphy M., Negrutskii B., El'Skaya A. Comparison of the ability of mammalian eEF1A1 and its oncogenic variant eEF1A2 to interact with actin and calmodulin. Biol. Chem. 2017;398:113–124. doi: 10.1515/hsz-2016-0172. [DOI] [PubMed] [Google Scholar]
- 34.Romaus-Sanjurjo D., Saikia J.M., Kim H.J., Tsai K.M., Le G.Q., Zheng B. Overexpressing eukaryotic elongation factor 1 alpha (eEF1A) proteins to promote corticospinal axon repair after injury. Cell Death Discov. 2022;8:390. doi: 10.1038/s41420-022-01186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendoza M.B., Gutierrez S., Ortiz R., Moreno D.F., Dermit M., Dodel M., Rebollo E., Bosch M., Mardakheh F.K., Gallego C. The elongation factor eEF1A2 controls translation and actin dynamics in dendritic spines. Sci. Signal. 2021:14. doi: 10.1126/scisignal.abf5594. [DOI] [PubMed] [Google Scholar]
- 36.Kanibolotsky D.S., Novosyl'na O.V., Abbott C.M., Negrutskii B.S., El'skaya A.V. Multiple molecular dynamics simulation of the isoforms of human translation elongation factor 1A reveals reversible fluctuations between “open” and “closed” conformations and suggests specific for eEF1A1 affinity for Ca2+-calmodulin. BMC Struct. Biol. 2008;8:4. doi: 10.1186/1472-6807-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Acharya A., Nemade H., Rajendra Prasad K., Khan K., Hescheler J., Blackburn N., Hemmersbach R., Papadopoulos S., Sachinidis A. Live-Cell Imaging of the Contractile Velocity and Transient Intracellular Ca(2+) Fluctuations in Human Stem Cell-Derived Cardiomyocytes. Cells. 2022:11. doi: 10.3390/cells11081280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin Y., Li L.-.L., Nie W., Liu X., Adler A., Xiao C., Lu F., Wang L., Han H., Wang X., Gan W.-.B., Cheng H. Brain activity regulates loose coupling between mitochondrial and cytosolic Ca(2+) transients. Nat. Commun. 2019;10:5277. doi: 10.1038/s41467-019-13142-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novosylna O., Jurewicz E., Pydiura N., Goral A., Filipek A., Negrutskii B., El'skaya A. Translation elongation factor eEF1A1 is a novel partner of a multifunctional protein Sgt1. Biochimie. 2015;119:137–145. doi: 10.1016/j.biochi.2015.10.026. [DOI] [PubMed] [Google Scholar]
- 40.Peart J.R., Lu R., Sadanandom A., Malcuit I., Moffett P., Brice D.C., Schauser L., Jaggard D.A.W., Xiao S., Coleman M.J., Dow M., Jones J.D.G., Shirasu K., Baulcombe D.C. Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10865–10869. doi: 10.1073/pnas.152330599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li D., Wei T., Abbott C.M., Harrich D. The unexpected roles of eukaryotic translation elongation factors in RNA virus replication and pathogenesis. Microbiol. Mol. Biol. Rev. 2013;77:253–266. doi: 10.1128/MMBR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White K.M., Rosales R., Yildiz S., Kehrer T., Miorin L., Moreno E., Jangra S., Uccellini M.B., Rathnasinghe R., Coughlan L., Martinez-Romero C., Batra J., Rojc A., Bouhaddou M., Fabius J.M., Obernier K., Dejosez M., Guillén M.J., Losada A., Avilés P., Schotsaert M., Zwaka T., Vignuzzi M., Shokat K.M., Krogan N.J., García-Sastre A. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science. 2021;371:926–931. doi: 10.1126/science.abf4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li D., Rawle D.J., Wu Z., Jin H., Lin M.-.H., Lor M., Abbott C.M., Harrich D. eEF1A demonstrates paralog specific effects on HIV-1 reverse transcription efficiency. Virology. 2019;530:65–74. doi: 10.1016/j.virol.2019.01.023. [DOI] [PubMed] [Google Scholar]
- 44.Snape N., Li D., Wei T., Jin H., Lor M., Rawle D.J., Spann K.M., Harrich D. The eukaryotic translation elongation factor 1A regulation of actin stress fibers is important for infectious RSV production. Virol. J. 2018;15:182. doi: 10.1186/s12985-018-1091-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lukash T.O., Turkivska H.V., Negrutskii B.S., El'skaya A.V. Chaperone-like activity of mammalian elongation factor eEF1A: renaturation of aminoacyl-tRNA synthetases. Int. J. Biochem. Cell Biol. 2004;36:1341–1347. doi: 10.1016/j.biocel.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 46.Tash J.S., Chakrasali R., Jakkaraj S.R., Hughes J., Smith S.K., Hornbaker K., Heckert L.L., Ozturk S.B., Hadden M.K., Kinzy T.G., Blagg B.S.J., Georg G.I. Gamendazole, an orally active indazole carboxylic acid male contraceptive agent, targets HSP90AB1 (HSP90BETA) and EEF1A1 (eEF1A), and stimulates Il1a transcription in rat Sertoli cells. Biol. Reprod. 2008;78:1139–1152. doi: 10.1095/biolreprod.107.062679. [DOI] [PubMed] [Google Scholar]
- 47.Sivan G., Aviner R., Elroy-Stein O. Mitotic modulation of translation elongation factor 1 leads to hindered tRNA delivery to ribosomes. J. Biol. Chem. 2011;286:27927–27935. doi: 10.1074/jbc.M111.255810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y., Jiang S., Yang P.-.Y., Zhang Y.-.F., Li T.-.J., Rui Y.-.C. EF1A1/HSC70 Cooperatively Suppress Brain Endothelial Cell Apoptosis via Regulating JNK Activity. CNS Neurosci. Ther. 2016;22:836–844. doi: 10.1111/cns.12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prommahom A., Dharmasaroja P. Effects of eEF1A2 knockdown on autophagy in an MPP(+)-induced cellular model of Parkinson's disease. Neurosci. Res. 2021;164:55–69. doi: 10.1016/j.neures.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 50.Vera M., Pani B., Griffiths L.A., Muchardt C., Abbott C.M., Singer R.H., Nudler E. The translation elongation factor eEF1A1 couples transcription to translation during heat shock response. Elife. 2014;3:e03164. doi: 10.7554/eLife.03164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chalorak P., Dharmasaroja P., Meemon K. Downregulation of eEF1A/EFT3-4 Enhances Dopaminergic Neurodegeneration After 6-OHDA Exposure in C. elegans Model. Front. Neurosci. 2020;14:303. doi: 10.3389/fnins.2020.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hassan M.K., Kumar D., Naik M., Dixit M. The expression profile and prognostic significance of eukaryotic translation elongation factors in different cancers. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0191377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grund A., Szaroszyk M., Korf-Klingebiel M., Malek Mohammadi M., Trogisch F.A., Schrameck U., Gigina A., Tiedje C., Gaestel M., Kraft T., Hegermann J., Batkai S., Thum T., Perrot A., Dos Remedios C., Riechert E., Völkers M., Doroudgar S., Jungmann A., Bauer R., Yin X., Mayr M., Wollert K.C., Pich A., Xiao H., Katus H.A., Bauersachs J., Müller O.J., Heineke J. TIP30 counteracts cardiac hypertrophy and failure by inhibiting translational elongation. EMBO Mol. Med. 2019;11:e10018. doi: 10.15252/emmm.201810018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khwanraj K., Madlah S., Grataitong K., Dharmasaroja P. Comparative mRNA Expression of eEF1A Isoforms and a PI3K/Akt/mTOR Pathway in a Cellular Model of Parkinson's Disease. Parkinsons. Dis. 2016;2016 doi: 10.1155/2016/8716016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beckelman B.C., Day S., Zhou X., Donohue M., Gouras G.K., Klann E., Keene C.D., Ma T. Dysregulation of Elongation Factor 1A Expression is Correlated with Synaptic Plasticity Impairments in Alzheimer's Disease. J. Alzheimers. Dis. 2016;54:669–678. doi: 10.3233/JAD-160036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frau-Méndez M.A., Fernández-Vega I., Ansoleaga B., Blanco Tech R., Carmona Tech M., Antonio Del Rio J., Zerr I., Llorens F., José Zarranz J., Ferrer I. Fatal familial insomnia: mitochondrial and protein synthesis machinery decline in the mediodorsal thalamus. Brain Pathol. 2017;27:95–106. doi: 10.1111/bpa.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McLachlan F., Sires A.M., Abbott C.M. The role of translation elongation factor eEF1 subunits in neurodevelopmental disorders. Hum. Mutat. 2019;40:131–141. doi: 10.1002/humu.23677. [DOI] [PubMed] [Google Scholar]
- 58.Guo M., Li Y., Wang Y., Li Z., Li X., Zhao P., Li C., Lv J., Liu X., Du X., Chen Z. eEF1A2 exacerbated insulin resistance in male skeletal muscle via PKCβ and ER stress. J. Endocrinol. 2020;244:25–40. doi: 10.1530/JOE-19-0051. [DOI] [PubMed] [Google Scholar]
- 59.Koo B.K., Chae S., Kim K.M., Kang M.J., Kim E.G., Kwak S.H., Jung H.S., Cho Y.M., Choi S.H., Park Y.J., Shin C.S.C.H., Jang H.C., Shin C.S.C.H., Hwang D., Yi E.C., Park K.S. Identification of novel autoantibodies in type 1 diabetic patients using a high-density protein microarray. Diabetes. 2014;63:3022–3032. doi: 10.2337/db13-1566. [DOI] [PubMed] [Google Scholar]
- 60.Clark B.F.C., Grunberg-Manago M., Gupta N.K., Hershey J.W.B., Hinnebusch A.G., Jackson R.J., Maitra U., Mathews M.B., Merrick W.C., Rhoads R.E., Sonenberg N., Spremulli L.L., Trachsel H., Voorma H.O. Prokaryotic and eukaryotic translation actors: international Union of Biochemistry and Molecular Biology (IUBMB) Biochimie. 1996;78:1119–1122. doi: 10.1016/S0300-9084(97)86738-7. [DOI] [Google Scholar]
- 61.Sourd F.Le, Boulben S., Bouffant R.Le, Cormier P., Morales J., Belle R., Mulner-Lorillon O. eEF1B: at the dawn of the 21st century. Biochim. Biophys. Acta. 2006;1759:13–31. doi: 10.1016/j.bbaexp.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 62.Negrutskii B. Non-translational Connections of eEF1B in the Cytoplasm and Nucleus of Cancer Cells. Front. Mol. Biosci. 2020;7:56. doi: 10.3389/fmolb.2020.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pérez J.M., Siegal G., Kriek J., Hård K., Dijk J., Canters G.W., Möller W. The solution structure of the guanine nucleotide exchange domain of human elongation factor 1beta reveals a striking resemblance to that of EF-Ts from Escherichia coli. Structure. 1999;7:217–226. doi: 10.1016/s0969-2126(99)80027-6. [DOI] [PubMed] [Google Scholar]
- 64.Wu H., Wang C., Gong W., Wang J., Xuan J., Perrett S., Feng Y. The C-terminal region of human eukaryotic elongation factor 1Bδ. J. Biomol. NMR. 2016;64:181–187. doi: 10.1007/s10858-016-0012-6. [DOI] [PubMed] [Google Scholar]
- 65.Vanwetswinkel S., Kriek J., Andersen G.R., Güntert P., Dijk J., Canters G.W., Siegal G. Solution structure of the 162 residue C-terminal domain of human elongation factor 1Bgamma. J. Biol. Chem. 2003;278:43443–43451. doi: 10.1074/jbc.M306031200. [DOI] [PubMed] [Google Scholar]
- 66.Jeppesen M.G., Ortiz P., Shepard W., Kinzy T.G., Nyborg J., Andersen G.R. The crystal structure of the glutathione S-transferase-like domain of elongation factor 1Bgamma from Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:47190–47198. doi: 10.1074/jbc.M306630200. [DOI] [PubMed] [Google Scholar]
- 67.Trosiuk T.V., Shalak V.F., Szczepanowski R.H., Negrutskii B.S., El'skaya A.V. A non-catalytic N-terminal domain negatively influences the nucleotide exchange activity of translation elongation factor 1Balpha. FEBS J. 2016;283:484–497. doi: 10.1111/febs.13599. [DOI] [PubMed] [Google Scholar]
- 68.Engen J.R. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal. Chem. 2009;81:7870–7875. doi: 10.1021/ac901154s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bondarchuk T.V., Shalak V.F., Lozhko D.M., Fatalska A., Szczepanowski R.H., Liudkovska V., Tsuvariev O.Y., Dadlez M., El'skaya A.V., Negrutskii B.S. Quaternary organization of the human eEF1B complex reveals unique multi-GEF domain assembly. Nucleic Acids Res. 2022 doi: 10.1093/nar/gkac685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Havrylenko S., Legouis R., Negrutskii B., Mirande M. Methionyl-tRNA synthetase from Caenorhabditis elegans: a specific multidomain organization for convergent functional evolution. Protein Sci. 2010;19:2475–2484. doi: 10.1002/pro.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaminska M., Shalak V., Mirande M. The appended C-domain of human methionyl-tRNA synthetase has a tRNA-sequestering function. Biochemistry. 2001;40:14309–14316. doi: 10.1021/bi015670b. [DOI] [PubMed] [Google Scholar]
- 72.Bondarchuk T.V., Lozhko D.M., Shalak V.F., Fatalska A., Szczepanowski R.H., Dadlez M., Negrutskii B.S., El'skaya A.V. The protein-binding N-terminal domain of human translation elongation factor 1Bβ possesses a dynamic α-helical structural organization. Int. J. Biol. Macromol. 2019;126:899–907. doi: 10.1016/j.ijbiomac.2018.12.220. [DOI] [PubMed] [Google Scholar]
- 73.Bondarchuk T.V., Shalak V.F., Negrutskii B.S., El'skaya A.V. Leucine-zipper motif is responsible for self-association of translation elongation factor 1Bβ. Biopolym. Cell. 2016;32:9–20. doi: 10.7124/bc.000907. [DOI] [Google Scholar]
- 74.Achilonu I., Siganunu T.P., Dirr H.W. Purification and characterisation of recombinant human eukaryotic elongation factor 1 gamma. Protein Expr. Purif. 2014;99:70–77. doi: 10.1016/j.pep.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 75.Trosiuk T.V., Liudkovska V.V., Shalak V.F., Negrutskii B.S., El'Skaya A.V. Structural dissection of human translation elongation factor 1Bγ (eEf1Bγ): expression of full-length protein and its truncated forms. Biopolym. Cell. 2014;30:96–106. doi: 10.7124/BC.000887. [DOI] [Google Scholar]
- 76.Jiang S., Wolfe C.L., Warrington J.A., Norcum M.T. Three-dimensional reconstruction of the valyl-tRNA synthetase/elongation factor-1H complex and localization of the delta subunit. FEBS Lett. 2005;579:6049–6054. doi: 10.1016/j.febslet.2005.09.062. [DOI] [PubMed] [Google Scholar]
- 77.Janssen G.M.C., Van Damme H.T.F., Kriek J., Amons R., Moller W. The subunit structure of elongation factor 1 from Artemia. Why two α- chains in this complex? J. Biol. Chem. 1994;269:31410–31417. doi: 10.1016/s0021-9258(18)31709-5. [DOI] [PubMed] [Google Scholar]
- 78.Bec G., Kerjan P., Waller J.P. Reconstitution in vitro of the valyl-tRNA synthetase-elongation factor (EF) 1 beta gamma delta complex. Essential roles of the NH2-terminal extension of valyl-tRNA synthetase and of the EF-1 delta subunit in complex formation. J. Biol. Chem. 1994;269:2086–2092. doi: 10.1016/S0021-9258(17)42139-9. [DOI] [PubMed] [Google Scholar]
- 79.Minella O., Mulner-Lorillon O., Bec G., Cormier P., Bellé R. Multiple phosphorylation sites and quaternary organization of guanine-nucleotide exchange complex of elongation factor-1 (EF-1betagammadelta/ValRS) control the various functions of EF-1alpha. Biosci. Rep. 1998;18:119–127. doi: 10.1023/a:1020140527930. [DOI] [PubMed] [Google Scholar]
- 80.Mansilla F., Friis I., Jadidi M., Nielsen K.M., Clark B.F.C., Knudsen C.R. Mapping the human translation elongation factor eEF1H complex using the yeast two-hybrid system. Biochem. J. 2002;365:669–676. doi: 10.1042/BJ20011681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sheu G.T., Traugh J.A. A structural model for elongation factor 1 (EF-1) and phosphorylation by protein kinase CKII. Mol. Cell. Biochem. 1999;191:181–186. doi: 10.1023/A:1006802125856. [DOI] [PubMed] [Google Scholar]
- 82.Negrutskii B.S., Shalak V.F., Kerjan P., El'skaya A.V., Mirande M. Functional interaction of mammalian valyl-tRNA synthetase with elongation factor EF-1alpha in the complex with EF-1H. J. Biol. Chem. 1999;274:4545–4550. doi: 10.1074/jbc.274.8.4545. [DOI] [PubMed] [Google Scholar]
- 83.Mansilla F., Dominguez C.A.G., Yeadon J.E., Corydon T.J., Burden S.J., Knudsen C.R. Translation elongation factor eEF1A binds to a novel myosin binding protein-C-like protein. J. Cell. Biochem. 2008;105:847–858. doi: 10.1002/jcb.21880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Glock C., Biever A., Tushev G., Nassim-Assir B., Kao A., Bartnik I., Dieck S.Tom, Schuman E.M. The translatome of neuronal cell bodies, dendrites, and axons. Proc. Natl. Acad. Sci. U. S. A. 2021:118. doi: 10.1073/pnas.2113929118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao Y., Portela M., Janikiewicz J., Doig J., Abbott C.M. Characterisation of translation elongation factor eEF1B subunit expression in mammalian cells and tissues and co-localisation with eEF1A2. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0114117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Veremieva M., Khoruzhenko A., Zaicev S., Negrutskii B., El'skaya A. Unbalanced expression of the translation complex eEF1 subunits in human cardioesophageal carcinoma. Eur. J. Clin. Invest. 2011;41:269–276. doi: 10.1111/j.1365-2362.2010.02404.x. [DOI] [PubMed] [Google Scholar]
- 87.Veremieva M., Kapustian L., Khoruzhenko A., Zakharychev V., Negrutskii B., El'skaya A. Independent overexpression of the subunits of translation elongation factor complex eEF1H in human lung cancer. BMC Cancer. 2014;14:913. doi: 10.1186/1471-2407-14-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Flores I.L., Kawahara R., Miguel M.C.C., Granato D.C., Domingues R.R., Macedo C.C.S., Carnielli C.M., Yokoo S., Rodrigues P.C., Monteiro B.V.B., Oliveira C.E., Salmon C.R., Nociti F.H.J., Lopes M.A., Santos-Silva A., Winck F.V., Coletta R.D., Paes Leme A.F. EEF1D modulates proliferation and epithelial-mesenchymal transition in oral squamous cell carcinoma. Clin. Sci. (Lond). 2016;130:785–799. doi: 10.1042/CS20150646. [DOI] [PubMed] [Google Scholar]
- 89.Dilworth D., Gong F., Miller K., Nelson C.J. FKBP25 participates in DNA double-strand break repair. Biochem. Cell Biol. 2020;98:42–49. doi: 10.1139/bcb-2018-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xue R., Peng Y., Han B., Li X., Chen Y., Pei H. Metastasis suppressor NME1 promotes non-homologous end joining of DNA double-strand breaks. DNA Repair (Amst) 2019;77:27–35. doi: 10.1016/j.dnarep.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 91.Dilworth D., Gudavicius G., Xu X., Boyce A.K.J., O'Sullivan C., Serpa J.J., Bilenky M., Petrochenko E.V., Borchers C.H., Hirst M., Swayne L.A., Howard P., Nelson C.J. The prolyl isomerase FKBP25 regulates microtubule polymerization impacting cell cycle progression and genomic stability. Nucleic Acids Res. 2018;46:2459–2478. doi: 10.1093/nar/gky008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li J., Feng B., Nie Y., Jiao P., Lin X., Huang M., An R., He Q., Zhou H.E., Salomon A., Sigrist K.S., Wu Z., Liu S., Xu H. Sucrose Nonfermenting-Related Kinase Regulates Both Adipose Inflammation and Energy Homeostasis in Mice and Humans. Diabetes. 2018;67:400–411. doi: 10.2337/db17-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiang Y.-.L., Ning Y., Ma X.-.L., Liu Y.-.Y., Wang Y., Zhang Z., Shan C.-.X., Xu Y.-.D., Yin L.-.M., Yang Y.-.Q. Alteration of the proteome profile of the pancreas in diabetic rats induced by streptozotocin. Int. J. Mol. Med. 2011;28:153–160. doi: 10.3892/ijmm.2011.696. [DOI] [PubMed] [Google Scholar]
- 94.Serpinskaya A.S., Tuphile K., Rabinow L., Gelfand V.I. Protein kinase Darkener of apricot and its substrate EF1gamma regulate organelle transport along microtubules. J. Cell Sci. 2014;127:33–39. doi: 10.1242/jcs.123885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Esposito A.M., Kinzy T.G. The eukaryotic translation elongation Factor 1Bgamma has a non-guanine nucleotide exchange factor role in protein metabolism. J. Biol. Chem. 2010;285:37995–38004. doi: 10.1074/jbc.M110.160887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hagiwara K., Katsuda T., Gailhouste L., Kosaka N., Ochiya T. Commitment of Annexin A2 in recruitment of microRNAs into extracellular vesicles. FEBS Lett. 2015;589:4071–4078. doi: 10.1016/j.febslet.2015.11.036. [DOI] [PubMed] [Google Scholar]
- 97.Kapustian L.M., Dadlez M., Negrutskii B.S. Non-canonical interactions of the β subunit of the translation elongation complex eEF1B and analysis of their possible functional role. Biopolym. Cell. 2016;32:347–358. doi: 10.7124/bc.00092F. [DOI] [Google Scholar]
- 98.Kapustian L.M., Dadlez M., Negrutskii B.S. Protein partners of the eEF1Bβ subunit of the translation elongation complex eEF1B in the nuclear fraction of human lung carcinoma cells. Biopolym. Cell. 2017;33:243–255. doi: 10.7124/bc.00095D. [DOI] [Google Scholar]
- 99.Kapustian L.M., Lysetsky I.L., Bondarchuk T.V., Novosylna O.V., Negrutskii B.S. Mass-spectrometric and bioinformatic analysis of eEF1Bγ interactome in the cytoplasmic fraction of A549 cells. Biopolym. Cell. 2018;34:292–302. doi: 10.7124/bc.000982. [DOI] [Google Scholar]
- 100.Kapustian L.M., Lysetsky I.L., Bondarchuk T.V., Novosylna O.V., Negrutskii B.S. Analysis of eEF1Bγ interactome in the nuclear fraction of A549 human lung adenocarcinoma cells. Biopolym. Cell. 2019;35:268–287. doi: 10.7124/bc.000A0A. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.