Abstract
The intestinal lumen harbors a diverse consortium of microorganisms that participate in reciprocal crosstalk with intestinal immune cells and with epithelial and endothelial cells, forming a multi-layered barrier that enables the efficient absorption of nutrients without an excessive influx of pathogens. Despite being a lung-centered disease, severe coronavirus disease 2019 (COVID-19) affects multiple systems, including the gastrointestinal tract and the pertinent gut barrier function. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can inflict either direct cytopathic injury to intestinal epithelial and endothelial cells or indirect immune-mediated damage. Alternatively, SARS-CoV-2 undermines the structural integrity of the barrier by modifying the expression of tight junction proteins. In addition, SARS-CoV-2 induces profound alterations to the intestinal microflora at phylogenetic and metabolomic levels (dysbiosis) that are accompanied by disruption of local immune responses. The ensuing dysregulation of the gut-lung axis impairs the ability of the respiratory immune system to elicit robust and timely responses to restrict viral infection. The intestinal vasculature is vulnerable to SARS-CoV-2-induced endothelial injury, which simultaneously triggers the activation of the innate immune and coagulation systems, a condition referred to as “immunothrombosis” that drives severe thrombotic complications. Finally, increased intestinal permeability allows an aberrant dissemination of bacteria, fungi, and endotoxin into the systemic circulation and contributes, to a certain degree, to the over-exuberant immune responses and hyper-inflammation that dictate the severe form of COVID-19. In this review, we aim to elucidate SARS-CoV-2-mediated effects on gut barrier homeostasis and their implications on the progression of the disease.
Keywords: COVID-19, SARS-CoV-2, Intestinal barrier, Dysbiosis, Immunothrombosis, Gut-lung axis
Core Tip: Severe coronavirus disease 2019 (COVID-19) is associated with a multi-layered disruption of gut barrier integrity. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) inflicts direct cytopathic or indirect immune-mediated injury to intestinal epithelial and endothelial cells and enhances paracellular permeability by downregulating tight junction proteins. SARS-CoV-2 induces profound gut microbiome alterations accompanied by dysregulation of mucosal immune responses. Gut dysbiosis attenuates, through the gut-lung axis, the ability of the respiratory immune system to elicit vigorous responses to contain SARS-CoV-2. Additionally, intestinal barrier dysfunction promotes endothelial activation and predisposes to detrimental COVID-19-related thrombotic complications. Finally, bacterial translocation and endotoxemia contribute to over-exuberant immune responses and hyper-inflammation in severe COVID-19.
INTRODUCTION
The emergence of the novel, pathogenic, and highly transmissible severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causing coronavirus disease 2019 (COVID-19), triggered an unprecedented public health crisis with profound socioeconomic sequelae. In most cases, COVID-19 is associated with mild-to-moderate symptoms that mainly involve the respiratory tract. However, in a subset of patients, COVID-19 may progress into a more severe disease plagued with complications such as pneumonia, acute respiratory distress syndrome (ARDS), coagulopathy, myocarditis, hepatic injury, renal dysfunction, sepsis, multiple organ failure, or even death[1]. These detrimental effects are considered to be driven by aberrant activation of the host’s immune system in response to viral invasion and proliferation into the pulmonary parenchyma[2]. In particular, the virus-laden pneumonocytes secrete excessive amounts of pro-inflammatory mediators and chemoattractant molecules, such as interleukins (IL-1β, IL-6, IL-7, IL-8, IL-12), tumor necrosis factor alpha (TNF-α), interferons (IFN-γ, IFN-λ), macrophage inflammatory protein-1 alpha (MIP-1α), monocyte chemoattractant protein-1 (MCP-1), and granulocyte colony stimulating factor. Subsequently, the recruitment and activation of innate and adaptive immune cells into the lungs further aggravate tissue injury and inflammation[2]. In parallel, the systemic dissemination of this “cytokine storm” precipitates overactivation of the immune system beyond the sites of infection and elicits hyperinflammatory responses that impair the function of several organs including the heart, kidneys, liver, nervous system, and gastrointestinal (GI) tract[3].
The GI system harbors an enormous interface that directly communicates with the external environment of the body and fulfills multifaceted functions. The GI mucosa serves as a semi-permeable membrane, allowing the efficient absorption of water, electrolytes, and nutrients while, in parallel, preventing the influx of xenobiotics, intraluminal microbiota, microbial components, or other inflammatory stimuli into the organism[4]. This subtle balance is maintained thanks to intestinal barrier function, which, apart from providing a physical barrier, regulates complex immune system responses and mediates the intricate crosstalk with the gut microbiome[5]. The integrity of the intestinal barrier can be compromised in many acute or chronic pathological conditions, leading to increased bacterial translocation and excessive penetration of pro-inflammatory signals. This dysfunction is associated with infectious complications and the establishment of a systemic pro-inflammatory status that can exacerbate or accelerate the pathophysiological processes of the underlying disease[6].
In this review, we summarize the deleterious effects of SARS-CoV-2 infection on gut barrier homeostasis. Subsequently, we discuss the mechanisms that explain how intestinal barrier dysfunction might drive severe COVID-19 or induce detrimental complications.
GI INVOLVEMENT AND THE MECHANISMS OF INTESTINAL INFECTION IN COVID-19
SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA virus with a genome of approximately 30000 nucleotides that encodes 29 proteins, including 4 structural proteins: spike (S), nucleocapsid (N), membrane (M), and envelope (E) proteins. The S protein consists of the S1 subunit, which interacts with angiotensin-converting enzyme 2 (ACE2), and the S2 subunit which promotes membrane fusion[7,8]. The principal route of transmission of SARS-CoV-2 is via respiratory droplets or aerosols from person-to-person, and its main target is type II alveolar epithelial cells (AEC2s). The entry of SARS-CoV-2 is primarily mediated by the attachment of the S glycoprotein with ACE2 on the cellular surface, a process facilitated by proteolytic cleavage at the S1/S2 boundary by host proteins. Transmembrane protease serine 2 (TMPRSS2) is a significant host protease that mediates the cleavage of S2 protein, leading to conformational changes that precipitate viral entry through membrane fusion[7,8]. Intriguingly, ACE2 and TMPRSS2 are widely co-expressed on the membrane of intestinal epithelial cells (IECs) at a level comparable or even higher to that of the AEC2s, rendering the GI tract a potential target for SARS-CoV-2 infection[9].
Multiple studies have demonstrated that the development of GI-related symptoms is a common extrapulmonary manifestation, affecting up to one-fifth of patients with COVID-19[10-12]. According to a meta-analysis of 108 studies comprising 17776 COVID-19 patients, the pooled prevalence estimates of GI symptoms were: 21% for loss of appetite, 13% for diarrhea, 8% for nausea or vomiting, and 4% for abdominal pain, while derangement of liver function tests was observed in 24% of the participants[10]. Whether the occurrence of GI symptoms is associated with COVID-19 severity or outcomes has been a field of debate. Most meta-analyses support that GI-symptoms and predominantly abdominal pain tend to appear at higher rates among patients with severe disease[10,13-15]; however, these symptoms are not always predictive of mortality or intensive care unit (ICU) admission[16-18]. Importantly, viral RNA can be detected in the stool of about half of COVID-19 patients and, especially, in those with GI-related symptoms[19,20]. Viral rectal shedding appears to persist long after RNA clearance in respiratory samples (mean duration: 21.8 d vs 14.7 d)[20], while RNA concentration in the stool may be as high as 107 copies/g, exceeding even that in pharyngeal swabs in some cases[21]. The combination of this particularly high RNA load in the stool and the long-lasting viral presence in fecal samples strongly suggests that SARS-CoV-2 invades and proliferates in the intestinal tract. Indeed, COVID-19 elicits acute inflammatory responses in the gut, as documented by increased fecal calprotectin levels[22]. Accordingly, Livanos et al[18] provided direct evidence of direct infection of IECs by SARS-CoV-2 in vivo, using immunofluorescence staining and electron microscopy. Viral subgenomic mRNA (sgmRNA) is transcribed in infected cells, but is not encapsulated into virions, which means that sgmRNA is vulnerable to enzymatic degradation by intestinal ribonucleases. Therefore, the detection of sgmRNA in the stool of COVID-19 patients also indicates that the virus can actively and specifically replicate in the gut[21].
Although the data above corroborate the concept of SARS-CoV-2 tropism for the GI system, the routes and mechanisms of infection remain less clarified. At least theoretically, the feasibility of the fecal-oral route of transmission is supported by prolonged viral excretion in the feces. Nevertheless, although SARS-CoV-2 retains viability in stool for 1-2 d at room temperature, this is accompanied by a profound 5-log loss of its infectivity[23]. In addition, ingested SARS-CoV-2 needs to overcome the harsh gastric acidity, resist the detergent effects of bile acids in the duodenum, and avoid degradation by digestive enzymes to preserve its infectivity and spread into the intestinal epithelia[24]. The uninterrupted function of these host defense mechanisms hinders the fecal-oral transmission of enveloped viruses, such as SARS-CoV-2[25]. Besides, several lines of evidence support that intestinal infection by SARS-CoV-2 could occur via the bloodstream[19,26]. The virus replicating in pulmonary alveolar epithelial cells or in ciliary cells of the nasal cavity is capable of breaching the basement membrane and causing viremia[27]. The GI tract comprises an extended network of capillaries harboring vascular epithelial cells, which are potential targets of circulating SARS-CoV-2. Thereafter, the proliferating virus is released from the infected cells into the gut, where it can infect neighboring enterocytes or reenter the circulation to generate new cycles of infection[27,28].
THE INTESTINAL BARRIER FUNCTION IN HEALTH
The gut barrier function is equipped with three major lines of defense that conjointly serve its complex purpose[5]. First, the mechanical barrier consists of tightly linked columnal IECs, the overlying mucus layer, and the capillary endothelial cells of the submucosa[29]. Tight junctions (TJs) and adherens junctions (AJs) are transmembrane multiprotein complexes that confer structural stability to the mechanical barrier and engender the establishment of cell polarity. TJs connect the most apical of the lateral surfaces of the adjacent epithelial or endothelial cells (kissing points) and form channels that regulate the selective diffusion of ions and salutes through the paracellular space. AJs are more basal than TJs and contribute to the establishment and maintenance of cell-cell adhesion[30,31]. The mucus layer comprises a hydrated network of polymers, predominantly highly glycosylated mucin proteins, as well as several immune regulators, such as antimicrobial proteins (AMPs) and secretory immunoglobulin A molecules. The mucus lubricates the luminal contents and serves as a physical barrier against digestive enzymes, proliferating microorganisms, microbial components and byproducts, food-associated toxins, or other inflammatory stimuli[32]. In addition, it nurtures a thriving biofilm of microorganisms, the gut microbiome, which typifies the second mechanism of protection, the biological barrier[29,32]. Luminal microbiota are indispensable for the fermentation of indigestible carbohydrates, a process that provides the gut with short-chain fatty acids (SCFAs), mainly consisting of acetate, propionate, and butyrate. Apart from being an important source of energy for host cells, SCFAs exhibit significant anti-inflammatory and anti-tumor effects and participate in various host signaling pathways, contributing to intestinal barrier integrity and metabolic homeostasis The commensal bacteria regulate choline bioavailability, promote the enterohepatic circulation of bile acids, and synthesize vitamin K and group B vitamins. In parallel, normal intestinal flora restrains the overgrowth of harmful microorganisms, a phenomenon referred to as colonization resistance, through nutrient antagonism[33]. Moreover, the microbiome-derived pathogen-associated molecular patterns (PAMPs) are recognized by pattern recognition receptors (PRRs) expressed by intestinal immune cells, indicating a relentless reciprocal dialogue between the microbiota and the intestinal immune system[6]. This leads to the third line of defense, that is, the immune barrier, which encompasses the gut-associated lymphoid tissue (GALT), effector and regulatory T cells (Tregs), immunoglobulin A (IgA)-secreting B (plasma) cells, innate lymphoid cells (ILCs), as well as macrophages and dendritic cells (DCs) of the lamina propria[5]. The cells of innate immunity carry an armamentarium of PRRs such as toll-like receptors (TLRs), nucleotide oligomerization domain-like receptors (NLRs), and retinoic acid-inducible gene-like receptors (RLRs) that recognize molecular pathogen-associated molecular patterns (PAMPs) or damaged-associated MPs (DAMPs) and orchestrate the well-tuned responses of the adaptive arm of the immune system.
The continuous crosstalk of the immune cells with the gut microbiome sustains the delicate balance between tolerance to beneficial bacteria and immunosurveillance against pathogenic species[6]. Commensal-derived signals and metabolites are recognized by myeloid cells in the lamina propria and orchestrate innate and adaptive immune responses[34]. Under homeostatic conditions, innate immune cells, such as macrophages and DCs, obtain a regulatory phenotype that promotes the secretion of anti-inflammatory molecules, i.e., IL-10 and TGF-β. The production of IL-22 by type 3 ILCs supports tissue homeostasis and epithelial barrier integrity[35]. Properly-regulated antigen presenting cells (APCs) remain in the GALT or migrate to the mesenteric lymph nodes, where priming of naïve CD4+ T cells occurs, and stimulate Treg cell activity that plays a central role in the suppression of intestinal inflammation. In this immunomodulatory milieu, B cell activation and effective class-switching generate large numbers of IgA-secreting plasma cells that serve multiple functions, including protection against infection and maintenance of gut microbiome homeostasis[35].
THE INTESTINAL BARRIER FUNCTION IN COVID-19
SARS-CoV-2 disrupts the intestinal mechanical barrier
SARS-CoV-2 can invade and propagate in IECs by using the vastly expressed ACE2 and TMPRSS2 receptors on those cells[18,36]. In a mouse model, ACE2 was found to be a key inducer of intestinal stem cell proliferation and differentiation under pathologic conditions, while ACE2 deficiency was associated with a significant reduction of mucin-2 expression[37]. Accordingly, ACE2 knockout mice exhibited gut barrier dysfunction with subsequent leakage of bacterial components into the circulation[38]. Therefore, it could be hypothesized that interference of SARS-CoV-2 with ACE2 signaling could destabilize the mechanical barrier by interrupting the renewal of epithelial cells or by compromising mucus composition. Mucins create a protective matrix covering the epithelium and inhibit viral invasion, presumably via steric hindrance[39]. Even though evident macroscopic alterations were usually negligible, infection of the human small bowel by SARS-CoV-2 was associated with villi blunting and an aberrant accumulation of activated intraepithelial CD8+ T cells in the epithelium[36]. Cell trafficking could be driven by direct infection of IECs or could be the result of systemic immune activation due to COVID-19. In any case, the recruitment of intraepithelial CD8+ T cells enhanced the apoptotic process of IECs, as demonstrated by an upsurge in cleaved caspase-3+ apoptotic epithelial cells. This event was accompanied by a regenerative response of the epithelium marked by an increase in Ki67+ proliferating epithelial cells that extended beyond their typical localization in the crypts and occupied the villus compartment[36]. Evidently, SARS-CoV-2 can inflict a deleterious impact on the mechanical barrier through dysregulation of the balance between cell apoptosis and proliferation.
The function and integrity of epithelia and endothelia greatly depend on TJs, reflecting a putative mechanism of intestinal barrier injury in COVID-19 patients. The fundamental transmembrane proteins that frame TJs include the family of claudins (central regulators of paracellular permeability encompassing 26 members in humans), the junctional adhesion molecules, and the three junctional MARVEL domain proteins, i.e., occludin, tricellulin, and MARVEL domain-containing protein 3. The most prominent intracellular junctional plaque components are zonula occludens (ZO) proteins (ZO1, ZO2, and ZO3), cingulin, and protein associated with LIN7 1 (PALS1)[40]. These proteins function as adaptor proteins or cytoskeletal linkers and participate in multiple extracellular and intracellular signaling pathways. Indeed, impairment of TJs occurs early in the course of COVID-19 and might represent the first hit in a multistage model of the disease[41]. SARS-CoV-2 infection decreases the expression of TJ proteins, such as occludin, claudin 5, and ZO-1 in vivo, and undermines the coherence of TJs between neighboring endothelial cells of the brain. As a result, TJs are haphazardly distributed, irregular, or gapped throughout the vascular endothelial layer, leading to derangement of the function of the blood-brain barrier[42]. Cryo-electron microscopy and in silico modeling analyses have shown that SARS-CoV-2 uses its envelope (E) protein to interact with the TJ-associated PALS1 protein[43,44]. According to in vitro models of lung injury, the interplay between SARS E protein and PALS1 interrupts intracellular trafficking of E-cadherin, delays the formation of TJs and AJs, and affects epithelial polarity[45]. In a proof-of-concept study, Guo et al[46] developed a biomimetic human gut-on-chip model that reconstructs basic elements of the gut barrier, as it consists of IECs, endothelial, and mucin-producing cells under normal fluid flow and closely reproduces the pathophysiological processes of intestinal SARS-CoV-2 infection. Interestingly, IECs exhibit particularly high susceptibility to SARS-CoV-2 infection. Viral inoculation induces the dispersal of the physiological distribution of mucus-secreting cells and a profound reduction of both E-cadherin expression in the epithelium and VE-cadherin in the endothelium, which delineate serious impairment of AJs in the corresponding structures. As a consequence, widespread destruction of the villus-like complexes along with severe morphological remodeling of the vascular endothelium was observed[46]. IL-6 is a prominent mediator of inflammation and a reliable biomarker of disease severity in SARS-CoV-2 infection that was found to increase, in a sustained manner, endothelial permeability in a mouse model[47,48]. Administration of IL-6 induced vascular leakage and disruption of junctional localization of VE-cadherin and ZO-1 via Janus kinase-mediated signal transducer and activator of transcription 3 phosphorylation and de novo protein synthesis[48].
Serum levels of endotoxin and ZO-1 were significantly increased in patients with COVID-19-related pneumonia on admission compared to healthy controls. Importantly, endotoxemia is positively correlated with certain markers of inflammation, such as C-reactive protein (CRP) and ferritin[49]. Endotoxins are complex lipopolysaccharides (LPS), integral parts of the membrane of gram-negative bacteria, and potent drivers of inflammation. Indeed, endotoxemia can occur as a result of gram-negative bacteremia; however, endotoxemia is most commonly caused by a compromised gut barrier[50]. ZO-1 is a peripheral membrane scaffolding protein and a basic constituent of TJs that fulfills versatile functions including establishment of cell-cell adhesion, modulation of the paracellular barrier, regulation of cell migration and angiogenesis, and induction of mucosal repair processes[51,52]. Previous studies have confirmed that serum ZO-1 represents a reliable biomarker of disrupted paracellular permeability, as it inversely correlates with intestinal ZO-1 expression in diverse pathologic conditions[53,54]. These results are consistent with another study, in which severe SARS-CoV-2 infection presented key features of gut barrier dysfunction in tandem with increased intestinal permeability. Specifically, patients with severe COVID-19 presented with endotoxemia and higher serum levels of zonulin, occludin, and regenerating family member 3 alpha, indicating severe impairment of the intestinal epithelial barrier[55].
SARS-CoV-2-induced gut microbiome dysbiosis
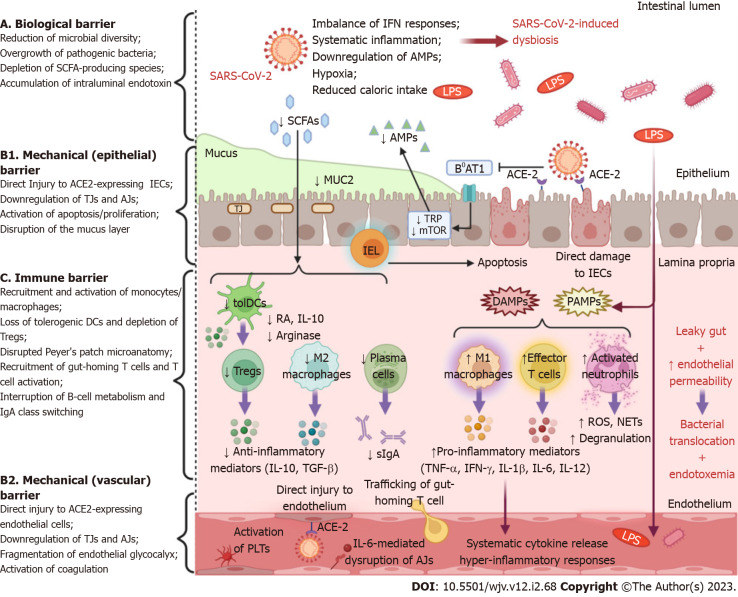
In the course of evolution, hosts and their microbial ecosystems have mutually developed, forging an intimate relationship of interdependence. Dysbiosis refers to alterations in the composition, quantity, or distribution of the gut microbiome. This condition is characterized by a predominance of pathogenic species and has been linked to the pathophysiology of numerous intestinal and extraintestinal disorders[56]. There is a growing body of data suggesting that COVID-19 is associated with drastic alterations of the normal intestinal flora, even when removing the confounding effect of antibiotics (Figure 1)[57-59]. In addition, SARS-CoV-2-induced dysbiosis appears to persist long after the resolution of symptoms and might be a predisposing factor for long-term complications in COVID-19 patients[57,58,60].
Figure 1.
Schematic representation of intestinal barrier dysfunction in severe acute respiratory syndrome coronavirus 2 infection. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is associated with a multifaceted dysfunction of the gut barrier as it exerts detrimental effects on all of its major levels of defense. A: Systemic inflammation, an imbalance of interferon (IFN) responses, hypoxia or low blood flow, and reduced caloric intake, due to coronavirus disease 2019-associated cachexia, contribute to intestinal microbiome alterations. Gut dysbiosis is characterized by an overgrowth of opportunistic pathogens, which are a source of harmful substances, e.g., endotoxin, peptidoglycan, β-glucan, as well as a depletion of commensal microorganisms, which synthesize beneficial metabolites such as short-chain fatty acids; B: SARS-CoV-2 induces either direct cytopathic injury to angiotensin-converting enzyme 2 (ACE2)-expressing intestinal epithelial cells (IECs) and endothelial cells, or indirect immune-mediated damage, or both. Moreover, the secretion of mucin glycoproteins, the fundamental element of the mucus layer, is severely impaired. In addition, the virus undermines the expression of tight junction and adherens junction proteins, leading to increased paracellular permeability and structural destabilization of the epithelium and endothelium. Viral interference with ACE2 signaling attenuates the activity of the amino acid transporter B0AT1, which is necessary for intestinal absorption of tryptophan (TRP). As a result, TRP depletion downregulates the mammalian target of rapamycin pathway, which promotes the expression of antimicrobial peptides. The overwhelming intraepithelial CD8+ lymphocytes trigger the apoptosis of IECs, which is followed by reactive regeneration. In parallel, disease-activated and apoptotic endothelial cells are potent drivers of neutrophil/monocyte recruitment, platelet aggregation, and coagulation activation; C: Commensal-derived metabolites modulate innate immune responses by inducing tolerogenic dendritic cells and M2-polarized macrophages. SARS-CoV-2-induced dysbiosis eliminates these immunoregulatory effects, inhibits T regulatory (Treg) cell activity, and suppresses the secretion of anti-inflammatory cytokines such as interleukin 10 (IL-10) and transforming growth factor beta. The release of damage-associated molecular patterns from injured cells and the influx of pathogen-associated molecular patterns, as a result of leaky gut, orchestrate the recruitment and activation of innate immune cells that secrete pro-inflammatory mediators (IFN-γ, tumor necrosis factor alpha, IL-1β, IL-6, IL-12). Subsequently, this pro-inflammatory microenvironment precipitates the derangement of adaptive immune responses, as demonstrated by increased trafficking of gut-homing T cells and effector T cell predominance. Furthermore, interruption of B-cell metabolism, plasma cell differentiation, and immunoglobulin A (IgA) class switching impede the effective secretion of protective dimeric IgA. This multi-layered disruption of intestinal barrier integrity allows the dissemination of intraluminal bacteria and endotoxin into the mesenteric lymphatic vessel or/and into the systemic circulation (created with biorender.com). IEL: Intraepithelial lymphocyte; LPS: Lipopolysaccharides; MUC2: Mucin 2; NETs: Neutrophil extracellular traps; PLTs: Platelets; RA: Retinoic acid; ROS: Reactive oxygen species; sIgA: Secretory immunoglobulin A; SCFAs: Short-chain fatty acids.
The mechanisms underlying COVID-19-related dysbiosis have not yet been fully elucidated. The interactions between SARS-CoV-2 and the ACE2 receptor can influence the composition of gut microbiota by interfering with the secretion of AMPs. The activity of the amino acid transporter B0AT1, which mediates the intestinal uptake of tryptophan, depends on ACE2 signaling[61]. Tryptophan regulates AMP production through the mammalian target of rapamycin pathway[62]. Therefore, tryptophan depletion due to ACE2 blockade can interrupt AMP production, and thus, perturb the intraluminal microbial community. Commensal bacteria, in turn, contribute to mucosal homeostasis by regulating ACE2 expression in the gut[63]. The release of pro-inflammatory cytokines, particularly TNF-α, during respiratory infections has a powerful anorexigenic effect via hypothalamic action. The ensuing reduction in caloric intake and dietary fibers disturbs the composition of the gut microbiota and the production of their metabolites. In animal models of respiratory syncytial virus (RSV) and influenza infection, the neutralization of anorexigenic cytokines prevented weight loss and mitigated gut microbiome alterations[64]. Alternatively, SARS-CoV-2 infection can cause dysbiosis through an imbalance of systemic or intestinal IFNs[65]. In this regard, in an influenza mouse model, the microbiome was amenable to significant changes via an IFN-type I-dependent mechanism[66]. Hypoxia is a serious feature of severe COVID-19 and hypoxic stress could be an important instigator of dysbiosis by dysregulation of hypoxia-inducible factor signaling[67]. Finally, local epithelial injury, which results in leaky gut and DAMP secretion, might disrupt the immune control of microbial homeostasis and could further aggravate the dysbiotic state[68].
In their recently published meta-analysis, Farsi et al[69] offered a thorough synthesis of the gut microbiota changes in COVID-19 patients. At the phylum level, dysbiosis is typified by a decrease in the Firmicutes to Bacteroidetes ratio. More specifically, COVID-19 is associated with a decrease in important butyrate-producing bacteria, including Faecalibacterium and Roseburia[69-71]. The genus Roseburia contributes to mucosal integrity and colonic motility and exerts significant anti-inflammatory effects by modulating IL-10 production[72]. Similarly, Faecalibacterium prausnitzii is a valuable gut symbiont with recognized anti-inflammatory effects in IBD patients via inhibition of the nuclear factor kappa B pathway[73]. The gut microbiome of COVID-19 patients is also depleted of several other beneficial genera such as Alistipes, Eubacterium, and Bifidobacterium[69]. The genus Eubacterium consists of a phylogenetically diverse group of SCFA-producing bacteria that contribute to bile acid metabolism and exhibit compelling associations with intestinal health[74]. Bifidobacterium strains prompt Treg responses, induce tolerogenic DC phenotypes, and demonstrate vigorous antimicrobial and antiviral activity[75]. Of note, their ability to suppress gut dysbiosis and promote mucosal homeostasis has paved the way for researchers to investigate the therapeutic efficacy of Bifidobacterium-based probiotic preparations in several diseases, including irritable bowel syndrome, inflammatory bowel disease (IBD), or COVID-19[76-78]. On the other hand, the COVID-19-derived gut microbiome is relatively enriched in opportunistic pathogens such as Streptococcus, Bacteroides, Rothia, Veillonella, Actinomyces, and Eggerthella[69,70]. Although the genus Ruminococcus is generally underrepresented, certain species such as Ruminococcus gnavus and Ruminococcus torques are significantly more abundant in COVID-19 patients. This is not surprising considering that Ruminococcus gnavus and Ruminococcus torques are harmful bacteria that degrade mucin glycans to harvest energy, secrete pro-inflammatory mediators, and are involved in IBD pathogenesis[79,80].
The degree of microbiome alterations correlates well with COVID-19 severity, and identification of early patterns of dysbiosis could lead to a microbiome-based stratification of patients according to their risk of progressing to severe COVID-19[81]. Indeed, the commensal genera Faecalibacterium and Roseburia are depleted in the gut microbiome of patients with critical disease[57,82]. The baseline abundance of Clostridium ramosum and Clostridium hathewayi is also associated with COVID-19 severity and could represent risk factors for portal vein thrombosis[69,82]. The genus Enterococcus is overrepresented in COVID-19 patients who necessitate ICU admission or developed bloodstream infections, whereas, surprisingly, the gut microbiome of other critically ill non-COVID-19 patients is devoid of this bacterium[83]. The reduction of the Firmicutes/Bacteroidetes ratio is indicative of severe disease, which is mainly attributed to the depletion of fiber-utilizing bacteria, namely Faecalibacterium prausnitzii, Phocaeicola plebeius, and Prevotella[84]. These findings are consistent with an interesting study exploring the role of gut microbiota as predictors of disease severity[81]. A lower Firmicutes/Bacteroidetes ratio, a higher prevalence of Proteobacteria, and an exhaustion of commensal butyrate-producing microorganisms are more evident in severely ill patients, while a lower bacterial diversity, defined by the Shannon diversity index, is identified as a prognostic biomarker of disease severity[81]. In agreement with this, the fecal microbiome of patients requiring mechanical ventilation has demonstrated low bacterial richness as assessed by Shannon or Chao1 indices[81,85,86]. Implementation of a multiomics approach to decipher the dysregulation of metabolic and microbial signatures during COVID-19 could provide a basis for the development of novel microbiome-targeted therapeutics[87].
SARS-CoV-2 deranging mucosal immune system responses
The mucosal immune system is the largest immunologically aware organ in the body, committed to maintaining the equilibrium between active protection against pathogens and immune-tolerance to commensal microorganisms, dietary substances, and self-antigens. The gut-derived metabolites orchestrate immune cell responses and differentiation and impart a critical role in mucosal homeostasis[56,88]. In particular, the transcriptional “education” of innate immune cells is strongly influenced by intestinal microbiota metabolites[56]. Butyrate, for example, induces M2-like polarization of macrophages, which shapes an immunomodulatory milieu by increasing the expression of arginase 1 and suppressing TNF-α, IL-6, IL-12b, and nitric oxide synthase 2[89]. As regards adaptive immunity, butyrate enhances transcription of the forkhead box P3 gene in naïve T cells through inhibition of histone deacetylases, and thereby, expands Treg cell populations[90]. The capsular polysaccharide A, which originates from the prominent human symbiont fragilis, promotes the proliferation of Treg cells in the lamina propria and shapes a pro-inflammatory microenvironment rich in IL-10[91]. Vitamin A or RA, produced by the gut commensal Bifidobacterium infantis, enhances the expression of aldehyde dehydrogenase 1 family member A2 encoding retinal dehydrogenase 2 in resident DCs of the mucosa. Subsequently, gut-modulated DCs secrete high levels of RA that drive naïve T cell differentiation into Treg cells[92]. Treg cell generation in the intestinal mucosa is also triggered by β-glucan polysaccharides deriving from the cell surface of Bifidobacterium bifidum[93]. In addition, B cell metabolism and differentiation in mucosal and systemic tissues are regulated by gut-derived SCFAs, emphasizing the significance of symbionts in effective antibody production[94]. Apparently, the depletion of SCFA-producing microorganisms or other beneficial species as well as gut metabolome modifications might have a detrimental impact on mucosal immunity in the course of COVID-19.
Defensins are prominent members of the AMP family with multifaceted immunomodulatory functions and broad antimicrobial and antiviral activity. Defensins provide protection against SARS-CoV-2 infection not only by maintaining gut microbiome homeostasis but also by inhibiting viral fusion via interference with ACE2 receptors[95,96]. Dysregulation in the expression of various defensin genes was evident following infection with COVID-19[97]. IFNs are multipotent cytokines of innate immunity with crucial role in the containment of viral infections. The proliferation of SARS-CoV-2 in human gut cells is effectively inhibited by type I (IFN-α, IFN-β) and type III IFNs (IFN-λs); however, type III IFNs elicit a more profound and long-lasting antiviral effect[98,99]. In order to escape immune surveillance, SARS-CoV-2 has developed strategies to proliferate stealthily into cells without eliciting strong IFN responses[65]. Alternatively, SARS-CoV-2 can interfere with IFN-γ (type II IFN) signaling to boost its infectively in the gut. In human colonic organoids, IFN-γ drives cellular differentiation towards ACE2-expessing epithelial cells, which are highly susceptible to SARS-CoV-2 infection[100]. Moreover, IFN-γ acts synergistically with TNF-α to instigate inflammatory cell death and tissue damage[101].
Elevated levels of plasma markers of inflammation and immune activation are hallmarks of severe COVID-19. Similarly, the expression of genes encoding pro-inflammatory mediators, including TNF-α, IL-6, chemokine (C-C motif) ligand 5 (CCL5), chemokine (C-X-C motif) ligand 1 (CXCL1), CXCL10, CXCL11, and CSF-3 were upregulated in digestive tissues in both in vitro and in vivo models of SARS-CoV-2 infection[46,102]. Compared to uninfected individuals, patients with COVID-19 have increased fecal levels of pro-inflammatory cytokines such as IL-8 and IL-18, whereas IL-23 is higher in patients with severe disease[103,104]. Strikingly, analysis of human ileal biopsies has demonstrated that several pro-inflammatory genes including IFNG, CXCL8, CXCL2, and IL1B are downregulated. This pro-inflammatory response is associated with milder symptoms and improved outcomes, revealing an immunomodulatory function of the GI in controlling SARS-CoV-2 infection[18]. Nevertheless, molecular events in severe COVID-19, due to increased bacterial translocation and systemic immune activation, might eliminate the immunoregulatory functions of the GI. Indeed, post-mortem evaluation of COVID-19 patients’ intestinal tissues revealed the overexpression of TNF-α and IL-10, which exerts antithetical effects[105]. The presence of IL-10 in the gut and lung may have immunosuppressive effects by inhibiting the expression of the human leukocyte antigen DR isotype (HLA-DR) on APCs[105]. Depletion of HLA-DR expression on monocytes is a major characteristic of immune dysregulation in COVID-19 patients who develop severe respiratory failure[106].
Essentially, changes in the expression of cytokines and downstream dysregulation of their signaling networks mirror alterations in the composition of immune cell populations of the lamina propria. Imaging mass cytometric analysis showed that the intestinal tract of deceased patients with COVID-19 accommodated higher numbers of CD11b+ macrophages, CD11c+ DCs, natural killer T cells, and B cells compared to healthy controls[105]. Another study, which evaluated post-mortem tissues of COVID-19 patients, demonstrated severe disorganization of ileal Peyer's patches with loss of B cell/T cell zonation and depletion of the germinal center. In addition, impaired interactions between B and T cells, an enhanced number of follicular macrophages, the reduction of CD27+ memory B cells, and downregulation of CD74 expression on B cells were reported[107]. Livanos et al[18] in their study, which supports an anti-inflammatory function of the GI in SARS-CoV-2 infection, showed that the lamina propria of COVID-19 patients was depleted of conventional DCs and plasmacytoid DCs, whereas effector CD4+ and CD8+ T cells as well as tissue resident memory T cells were increased. Lehmann et al[36]. revealed that the activation and proliferation of antigen-experienced intraepithelial CD8+ T cells into the intestinal mucosa was associated with epithelial barrier dysfunction in COVID-19 patients, while monocytes and macrophages of the duodenal mucosa expressed markers indicative of their recent recruitment from the circulation. In agreement, levels of CCL25, a gut homing marker, were increased in the sera of COVID-19 patients, suggesting that intestinal inflammation might result from CCL5/CCR9-mediated trafficking of gut-specific T cells into the mucosa[108,109]. Humoral immune responses are vital for counterattacking viral invasion through the production of neutralizing antibodies. Secretory dimeric IgA is the predominant mucosal antibody and an integral component of the immunological barrier[6]. IgA antibody overproduction dominates the early pre-specific humoral response to SARS-CoV-2 infection, while virus-specific IgA antibodies display more robust neutralizing capacity compared to their monomeric IgG counterparts[110]. Interestingly, the levels of mucosal SARS-CoV-2-specific IgA are inversely correlated with age[111]. Therefore, it can be inferred that the inability of B cells to mount an effective IgA response contributes to excessive viral propagation in the course of severe COVID-19[112].
THE GUT-LUNG AXIS IN COVID-19
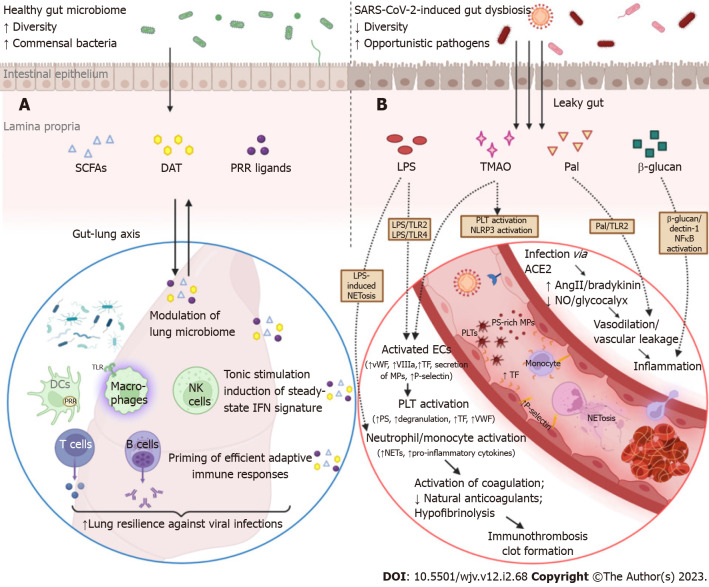
As mentioned above, homeostasis of the gut microbiome can be immensely affected in the course of respiratory tract infections such as COVID-19. Conversely, intestinal microbiota plays an important role in fine-tuning the systemic immune system and eliciting efficient antiviral responses to address lung infections[113-115]. Gut bacterial components and metabolites can enter portal circulation or mesenteric lymphatics, which drain to the cisterna chyli first, then to the thoracic duct, and finally to the left subclavian vein. Intriguingly, the pulmonary vascular bed is the first to interact with the mesenteric lymph, implicating the importance of gut-derived signals in shaping lung immune responses in health as well as in driving ARDS in critically ill patients[116]. Under normal conditions, commensal-associated stimuli provide an indispensable for optimal antiviral activity, tonic activation of the host’s innate immunity through their impact on alveolar macrophages, resident DCs, and lung epithelial cells (Figure 2)[117]. In contrast, germ-free mice are unable to evoke strong innate and adaptive immune responses and, thus, experience feeble control of viral infections and unfavorable outcomes[117,118]. Although the intestinal and respiratory tracts are anatomically distinct compartments, their mucosal immune cells and microbial communities configure a bidirectional “gut-lung” axis cross-talk that is highly pertinent to COVID-19 pathogenesis.
Figure 2.
Overview of the sequelae of gut barrier dysfunction in severe coronavirus disease 2019: dysregulation of lung immune responses and establishment of a prothrombotic state. A: Although the intestinal and respiratory tracts are anatomically distinct compartments, their mucosal immune cells and microbial ecosystems participate in a bidirectional immunological crosstalk (gut-lung axis). An intact intestinal barrier is pivotal in maintaining lung microbiome homeostasis and fine-tuning the respiratory immune system to elicit potent antiviral responses in the case of infection. Commensal bacteria provide tonic stimulation (through the production of pattern-recognition receptor-ligands, desaminotyrosine, short-chain fatty acids, etc.) of the epithelial, stromal, and innate immune cells of the lungs and modulate the steady-state interferon-signature, which is essential for suppressing the early phase of viral proliferation. In addition, gut-derived signals and metabolites orchestrate the effective priming of adaptive immune responses by inducing the differentiation of virus-specific CD4+ and CD8+ T cells and antibody-secreting plasma cells, which are responsible for viral control and clearance in the later stages of infection. In coronavirus disease 2019 (COVID-19), gut barrier dysfunction and depletion of symbiotic microorganisms eliminate the aforementioned immunomodulatory effects and compromise the ability of the respiratory immune system to effectively contain severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection; B: The gut vascular bed has a massive endothelial surface that is susceptible to SARS-CoV-2 infection. SARS-CoV-2 can inflict direct injury to angiotensin-converting enzyme 2 (ACE2)-expressing endothelial cells; interruption of ACE2 signaling can dysregulate both the renin-angiotensin and kinin-kallikrein systems, leading to vascular leakage. The activated or apoptotic endothelial cells release phosphatidylserine-rich endothelial microparticles and secrete large amounts of tissue factor, VIIIa, von Willebrand factor, and other procoagulant cofactors. Circulating platelets accumulate at sites of vascular injury, adhere to each other, and become activated, leading to further secretion of prothrombotic substances. Overexpression of adhesion molecules, such as P-selectin, facilitates the recruitment and activation of monocytes and other leukocytes, including neutrophil extracellular trap (NET)-producing neutrophils. COVID-19-associated immunothrombosis refers to this concurrent aberrant activation of the innate immune and coagulation systems, which predisposes to serious thrombotic complications. This vicious cycle can be further exacerbated by gut barrier dysfunction. Low-grade endotoxemia, due to increased intestinal permeability, enhances the activation of endothelial cells and platelets by inducing lipopolysaccharide (LPS)/toll-like receptor 2 (TLR2) and LPS/TRL4 downstream signaling pathways. In parallel, LPS is a potent driver of NET formation. Several other bacterial lipoproteins, such as peptidoglycan-associated lipoprotein (Pal) or Pam3Cys, aggravate vascular leakage and precipitate thrombus formation through TLR2 activation. Moreover, translocation of fungal components, such as β-glucan, could directly stimulate leukocytes and promote inflammation by binding to the Dectin-1 receptor and activating the nuclear factor-κB pathway. Finally, gut dysbiosis is associated with trimethylamine N-oxide overproduction, which is a recognized risk factor for clotting events as it enhances platelet hyperresponsiveness, endothelial dysfunction, and NLR family pyrin domain containing 3 inflammasome activation (created with biorender.com). AngII: Angiotensin II; DCs: Dendritic cells; ECs: Endothelial cells; NK cells: Natural killer cells; NO: Nitric oxide; PLT: Platelet; DAT: Desaminotyrosine; SCFAs: Short-chain fatty acids; PRRs: Pattern recognition receptors.
The mechanisms through which intestinal flora reinforces lung resilience against viral invasion have only recently begun to be unraveled with the assistance of murine models. The gut microbiota orchestrates the steady-state IFN signature in lung stromal cells, which protects against early influenza virus proliferation by driving the expression of IFN-α/β receptor[113]. In line with this, when mice were exposed to antibiotics, their macrophage-related type I and type II IFN responses were severely impaired, resulting in the uncontrolled spread of systemic lymphocytic choriomeningitis virus or mucosal (influenza virus) infection. In a murine model of influenza infection, desaminotyrosine (DAT), a degradation product of dietary flavonoids, induced an efficient amplification of the IFN type I loop, which suppressed viral gene expression and airway epithelial damage. Notably, a distinct human-derived commensal, Clostridium orbiscindens, produced DAT and rescued antibiotically-treated mice from viral infection[119]. An outer membrane glycolipid originating from the gut symbiont Bacteroides fragilis protected mice against viral infection through the induction and systemic release of IFN-β by colonic DCs[120]. Acetate, another metabolite derived from the gut microbiome, is able to enter the circulation, enhance IFN-β signal transduction via stimulation of the G-protein-coupled receptor 43 in pulmonary epithelial cells, and thus limit severe RSV infection[114]. Moreover, depletion of commensal bacteria precipitated significant epigenetic modifications at the level of mononuclear phagocytes residing in non-mucosal lymphoid organs. As a result, these cells were unable to induce type I IFN secretion, which led to suboptimal NK cell priming and poor antiviral responses[121]. Admittedly, an imbalance of IFN signaling is an inherent characteristic of the pathogenesis of respiratory tract infections, including SARS-CoV-2-associated pneumonia[122]. Early severe COVID-19 is governed by overwhelming IFN-α responses as well as NK cell functional exhaustion, which is manifested by abnormal expression of interferon-stimulated genes[123]. Therefore, it is plausible to assume that metabolites and signals stemming from luminal bacteria could influence the early response to SARS-CoV-2 infection by altering the IFN signature and compromising innate immunocompetence in the lungs.
Regulation of the mucosal immune system of the respiratory tract, which supervises airway colonization, depends on reciprocal signaling with the gut. The respiratory tract is not sterile; instead, it harbors a unique microbial ecosystem and its role in homeostasis and disease is being increasingly recognized with the advent of culture-independent molecular techniques[124]. Alterations of the lung microbiome signature are associated with clinical outcomes in critically ill patients infected with SARS-CoV-2[125]. The immunomodulatory potential of airway bacteria was exemplified by a pathogen-free murine model of influenza virus. Colonization of the upper respiratory tract by Staphylococcus aureus induces the recruitment of peripheral macrophages into the alveoli and their polarization toward an M2-like phenotype via TLR2 signaling. As a result, Staphylococcus aureus creates an anti-inflammatory pulmonary milieu that attenuated immune-mediated injury and prevented lethal influenza infection[126].
The robust and timely priming of adaptive immunity is necessary to contain SAR-CoV-2 infection. Delayed activation of adaptive immune responses and depletion of virus-specific T cells are hallmarks of severe or fatal COVID-19[127]. Interestingly, multiple studies have revealed that the gut microbiome is actively involved in shaping adaptive immunity in the respiratory tract. In a murine model of West Nile virus infection, exposure to antibiotics impaired the ability to elicit potent T cell responses, decreased the number of virus-specific CD8+ T cells, and led to worse disease outcomes[118]. Ichinohe et al[115] demonstrated that intestinal microflora regulates an even broader spectrum of adaptive immunity responses in the respiratory mucosa. More specifically, neomycin-sensitive commensal bacteria were essential for the induction of effective humoral responses and the generation of virus-specific CD4+ and CD8+ T cells in the lungs. Normal intestinal flora provided signals that maintained a steady-state IL-1β, pro-IL-18, and NLR family pyrin domain containing 3 (NLRP3) expression. Antibiotic-induced immunodeficiency is attributed to impaired inflammasome activation, abnormal activation and distribution of respiratory DCs, and inadequate DC migration to draining lymph nodes of the lung[115]. In this setting, Gauguet et al[128] demonstrated that the presence of segmented filamentous bacteria in the murine microbiome was vital for priming strong Th17 immunity responses and IL-22 secretion in the respiratory system. In response to fiber supplementation, the gut microbiome generates large amounts of SCFAs, which hinders influenza-induced lung injury through a dual mechanism concerning both arms of immunity[129]. First, SCFAs enhances the cellular metabolism and the effector functions of CD8+ T cells in the respiratory tract. Second, SCFAs induce an alternative activation of macrophages, which exhibit the limited ability to express the chemokine CXCL1. As a result, SCFA-modulated macrophages reduce early neutrophil infiltration and subsequent injury in the airways[129]. In a randomized controlled trial, non-hospitalized patients with symptomatic COVID-19 were allocated 1:1 to groups receiving an oral probiotic formulation or placebo for 30 d. Remarkably, probiotic supplementation was associated with higher rates of complete remission, decreased nasopharyngeal viral levels, and shorter duration of symptoms by inducing vigorous virus-specific IgM and IgG antibody responses.
In summary, a well-preserved intestinal barrier function, harboring a diverse consortium of commensal bacteria, provides the essential signals for appropriate and effective modulation of immune system responses in the lungs. Regardless of being the cause or the result of severe SARS-CoV-2 infection, gut dysbiosis is not a silent bystander but an active orchestrator of dysregulated immune responses in the respiratory tract. Modulation of gut microbiota represents an emerging therapeutic intervention to mitigate immune-mediated lung injury and improve COVID-19 outcomes[130].
GUT BARRIER DYSFUNCTION INFLAMING COVID-19-ASSOCIATED COAGULOPATHY
COVID-19-associated coagulopathy (CAC) is a life-threatening condition that can lead to arterial thromboembolism (ATE), such as acute coronary syndrome and cerebrovascular accident, or venous thromboembolism (VTE), which manifests as deep vein thrombosis or pulmonary embolism[131]. Mild thrombocytopenia, prolonged prothrombin time, and increased serum levels of fibrinogen, CRP, P-selectin, and d-dimers have all been linked to clotting events in COVID-19 patients; these deviations become more pronounced as the disease progresses[131,132]. A recent meta-analysis, comprised of more than 90000 patients, concluded that hospitalized COVID-19 patients have a significantly increased 90-d risk of VTE in comparison to hospitalized individuals with influenza virus (9.5% vs 5.3%), and that this peril persists even after the breakthrough of effective vaccines[133]. COVID-19-related thromboembolic events are major burdens of morbidity and mortality, and their incidence increases with disease severity, affecting up to one third of patients in ICUs[134-136]. The pathophysiology underlying CAC is multifactorial, encompassing endothelial injury, over-exuberant immune responses, and overt dysregulation of coagulation and fibrinolytic pathways, which collectively result in a procoagulant state[131]. The activation of these mechanisms can be further aggravated by defects in intestinal barrier integrity due to SARS-CoV-2 infection[27,137].
The dysfunction of the endothelium is a hallmark of COVID-19, representing a common feature in multiple clinical manifestations of the disease such as thromboembolic events, neurological complications, and renal dysfunction[138,139]. The intestinal tract accommodates an enormous vascular endothelial surface consisting of a monolayer of squamous endothelial cells. Under physiological conditions, endothelial cells regulate the vascular tone and secrete anticoagulant and antiplatelet agents that preclude clotting events[140]. Human endothelial cells express the key cofactors, namely ACE2 and TMPRSS2, which are exploited by SARS-CoV-2 to invade its target cells[141]. There is evidence of viral inclusions in endothelial cells and mononuclear cell infiltrates in the walls of small vessels, as well as markers of endothelial cell apoptosis[142]. SARS-CoV-2-induced dysfunction of gut microvasculature and fragmentation of the endothelial glycocalyx eliminate these protective effects, promote vasoconstriction due to depletion of endothelium-derived nitric oxide, and drive platelet activation and fibrin formation[131]. Indeed, mesenteric thrombosis is not uncommon in critically ill patients with COVID-19, while focal ischemic lesions, inflammation of the endothelium, vessel wall edema, microhemorrhage, and microthrombi are frequent findings in resected bowel segments[27,143-145]. SARS-CoV-2-mediated dysregulation of ACE2 signaling in intestinal vascular endothelium might create a prothrombotic microenvironment through a dual mechanism. First, ACE2 catalyzes the conversion of angiotensin (Ang II) to angiotensin 1-7 (Ang 1-7), which confers important antithrombotic and immunoregulatory effects by binding to G-protein coupled Mas receptors. Therefore, ACE2 depletion attenuates the Ang 1-7 downstream pathway and shifts the balance in favor of Ang II, which binds to its cognate receptor and exerts harmful prothrombotic and inflammatory effects[146]. Second, the kinin-kallikrein system is also directly modulated by ACE and ACE2. Thus, the ACE/ACE2 imbalance can induce a “kinin storm” and amplify vascular permeability, cell migration, platelet activation, and oxidative stress[131,147].
There are data supporting the hypothesis that SARS-CoV-2 inflicts damage on endothelial cells (endotheliitis) via both direct cytopathic and indirect immune-mediated mechanisms[148]. Activated or apoptotic intestinal endothelial cells retract their margins, release endothelial microparticles (MPs), and abolish their ability to confine phosphatidylserine (PS) into the inner layer of the cellular membrane[27,149]. In this setting, microRNA expression in endothelial-derived extracellular vesicles has been associated with cerebrovascular events in COVID-19 patients by compromising the function of the blood-brain barrier[150,151]. Endothelial MPs and PS-positive filopods support the formation of the prothrombinase complex along with activated coagulation factors Va and Xa and thereby catalyze a pivotal step of the coagulation cascade, that is, the proteolytic activation of thrombin[27,152]. In parallel, exposure of subendothelial tissues and tissue factor (TF) to plasma procoagulants triggers the extrinsic coagulation pathway[132]. Damaged endothelial cells sustain this hypercoagulable milieu and promote platelet aggregation by secreting large amounts of factor VIII and von Willebrand factor (VWF), respectively[153-156]. Thrombotic microangiopathy is further exacerbated due to relative deficiency in metalloproteinase ADAM metallopeptidase with thrombospondin type 1 motif 13 activity in COVID-19 patients, which leads to insufficient VWF cleavage and enhanced platelet-vessel wall interactions[157]. Numerous other endothelial dysfunction markers, such as circulating endothelial cells, soluble (s)E-selectin, soluble thrombomodulin, and soluble intercellular adhesion molecule 1 are significantly increased in COVID-19 patients receiving ICU care[153,155,158]. Furthermore, critically ill patients demonstrate functional exhaustion of natural anticoagulants (protein C, protein S, and antithrombin) and develop anti-phospholipid antibodies at high rates[154]. This procoagulant state is accompanied by suboptimal fibrinolytic potential and remodeling of the clot structure, which displays a denser fibrin network as well as thinner and shorter fibrin fibers. In severe COVID-19, hypofibrinolysis is dictated by an upregulation of plasminogen activator inhibitor-1 and its stabilizing cofactor vitronectin, which reduce plasminogen generation, despite concurrent elevations of tissue plasminogen activator[159].
Immunothrombosis illustrates the intricate cross-talk between the innate immune system and the coagulation pathway, which aims to locally contain an infection by facilitating recognition and eradication of invading pathogens. An aberrant activation of immunothrombosis is associated with severe thrombotic complications in SARS-CoV-2 infection[160]. Endothelium expresses a variety of adhesion and chemoattractant molecules, promoting the recruitment of monocytes and neutrophils at sites of injury. The release of DAMPs and PAMPs stimulates innate immune cells, which subsequently express TF and pro-inflammatory mediators in large amounts and precipitates the formation of neutrophil extracellular traps (NETs)[156]. NETs are web-like structures consisting of DNA complexed with histones, bactericidal enzymes, complement factors, and coagulants. NETosis is a potent driver of immunothrombosis in COVID-19 and contributes to the procoagulant state in IBD through various mechanisms: cleavage of natural anticoagulants, direct activation of the contact-dependent and extrinsic pathways of coagulation, and aggregation of platelets[156,161]. In turn, activated platelets overexpress P-selectin and other adhesion molecules, which enhance their interactions with monocytes and NETs[162]. Through the secretion of their intracellular prothrombotic granule substances and externalization of their PS-rich membrane, platelets trigger and sustain thromboinflammation[131].
There is evidence that this vicious cycle could be further aggravated by gut barrier dysfunction. In particular, Oliva et al[163] reported that serum LPS and zonulin were increased in hospitalized COVID-19 patients and showed that endotoxemia was an independent predictor of in-hospital thrombotic complications. Zonulin is involved in the modulation of gut permeability by orchestrating the disassembly of intercellular TJ[53]. In this regard, serum LPS positively correlated with zonulin, supporting that low-grade endotoxemia precipitating thrombosis in COVID-19 originates from the gut[163]. In addition, thrombogenesis in patients with endotoxemia was associated with the TLR4-dependent activation of platelets[163]. In agreement, LPS/TLR4 and LPS/TLR2 downstream signaling pathways have been previously implicated in procoagulant conditions by promoting endothelial cell and platelet activation[164]. Several other bacterial lipoproteins, such as Pam3Cys and peptidoglycan-associated lipoprotein, promote vascular leakage and thrombus formation through TLR2 activation[165]. In a murine model, microbial translocation and release of related patterns as a result of gut barrier leakage enhanced platelet pro-aggregating capacity and thrombus growth by inducing VWF synthesis; this effect was mediated via TLR2 activation in the hepatic endothelium[166]. Restoration of endothelial function through L-arginine supplementation was followed by improved outcomes in hospitalized patients with COVID-19[167]. L-arginine, a semi-essential amino acid, stimulates endothelium relaxation by serving as a substrate for the synthesis of nitric oxide by NOS. It also plays a role in immunomodulation by controlling T cell proliferation[168]. The importance of gut microbiome homeostasis in preventing immunothrombosis and inflammatory damage was evidenced in a model of acute mesenteric ischemia/reperfusion injury. More specifically, commensal bacteria mitigated LPS-induced NETosis by providing tonic stimulation of the neutrophil-intrinsic TLR4 downstream signaling[169]. Furthermore, perturbation of intestinal microflora due to SARS-CoV-2 infection and the ensuing depletion of SCFAs might enhance thromboinflammation by eliminating the inhibitory effects of butyrate on the LPS/TLR4/NF-κB pro-inflammatory pathway[170]. Alternatively, dysregulation of microorganisms involved in the metabolism of trimethylamine and its metabolite TMA N-oxide (TMAO) could contribute to the hypercoagulative state in CAC[171]. TMA is generated by gut bacterial metabolism of choline, carnitine, and betaine and is rapidly converted into TMAO in the liver. TMAO is a recognized risk factor for clotting events by enhancing platelet hyperresponsiveness, endothelial dysfunction, and NLRP3 inflammasome activation[171,172]. Indeed, a recent study revealed significant alterations in metabolites pertaining to the choline/TMAO and carnitine/TMAO pathways in COVID-19 patients; these aberrations were associated with disease symptoms and severity[173].
GUT BARRIER DYSFUNCTION: AN UNDERAPPRECIATED DRIVER OF SYSTEMIC INFLAMMATION
The critical form of COVID-19 is governed by multi-layered immune system dysregulation and hyper-inflammatory responses. Overexpression of pro-inflammatory cytokines (IL-1, IL-6, TNF-α) and chemokines (MCP-1, MIP-1β, CCL5), as well as excessive oxidative stress and hyperactivation of the complement (C3a) and coagulation (d-dimer) systems are hallmarks of severe COVID-19[55,174]. In parallel, SARS-CoV-2 infection is accompanied by a tremendous influx of microbial components and metabolites into the systemic circulation due to intestinal barrier dysfunction. Several studies have shown that critically ill patients with COVID-19 present with endotoxemia and elevated plasma levels of zonulin and LPS-binding protein (LBP), which are surrogate markers of leaky gut[55,59,108,174-177]. Additional indices of disrupted intestinal permeability, such as fatty acid binding protein 2, an intracellular protein specifically expressed in IECs, and peptidoglycan, a core element of the cell wall in Gram-positive bacteria, were also increased in COVID-19 cases[178]. Furthermore, patients with severe COVID-19 as well as those with SARS-CoV-2 post-acute sequelae (PASC) had higher circulating levels of β-glucan, a fungal cell wall polysaccharide, indicating persistent fungal translocation[55,175,179]. Interestingly, the presence of circulating bacteriome in critically ill COVID-19 patients without evidence of secondary infections further corroborates the assumption of gut-derived bacterial translocation in severe SARS-CoV-2-infection[175]. Essentially, exuberant translocation precipitates microbial-mediated myeloid activation as demonstrated by increased serum levels of sCD14 and myeloperoxidase, which correspond to monocyte and neutrophil inflammation, respectively[55,174]. Multiple studies have revealed strong and unequivocal correlations between intestinal barrier dysfunction and biomarkers of inflammation and/or immunological activation, implicating a detrimental role of gut barrier defects in triggering or enhancing hyper-inflammatory responses in severe SARS-CoV-2 infection[55,59,108,163,174,177]. More specifically, Giron et al[55] reported that LBP, β-glucan, and zonulin levels were all significantly associated with higher systemic levels of IL-6. During PASC, β-glucan could directly stimulate leukocytes and promote inflammation by binding to the Dectin-1 receptor and activating NF-κB pathway[179]. In hospitalized COVID-19 patients with cardiac involvement, NLRP3 inflammasome activation and subsequent IL-18 and IL-1Ra secretion were linked with circulating LBP levels[108]. In line with this, Sun et al[59] demonstrated that plasma levels of LBP were associated with inflammation biomarkers (CRP, IL-6, IL-8) as well as with changes in relative frequencies of lymphocytes and neutrophils. Endotoxemia was significantly associated with TNF-α, CCL5, and MIP-1β in another cohort of COVID-19 inpatients, whereas sCD14 was negatively associated with TGF-β[174].
Furthermore, gut microbiome alterations, which become more pronounced as disease progresses, showed solid correlations with markers of inflammation and tissue injury. In particular, gut dysbiosis coincided with derangements in the serum levels of IL-10, TNF-α, CRP, erythrocyte sedimentation rate, aspartate aminotransferase, and lactate dehydrogenase in patients with COVID-19[60]. A negative correlation was found between CRP and the symbiotic microorganisms Faecalibacterium prausnitzii and Clostridium butyricum, which perish in critically ill patients[60]. The abundance of commensal bacteria (Lachnospiraceae, Eubacterium ventriosum, Faecalibacterium prausnitzii) was followed by an increased number of CD4+ T cells, CD8+ T cells, and NK cells[69]. Conversely, opportunistic pathogens, which are commonly overrepresented in the COVID-19-related gut microbiome, such as Bacteroides dorei and Akkermansia muciniphila, were positively correlated with pro-inflammatory cytokines, including IL-1β, IL-6, and IL-8[69]. Accordingly, the predominance of the emerging pathogen, Burkholderia contaminans, was accompanied by T cell anergy and complement activation[59]. In parallel, the prevalence of pathogenic species in SARS-CoV-2-induced gut dysbiosis predisposes to bacteremia, leading to secondary microbial bloodstream infections, and thus, to an increased risk of serious complications[178,180]. It should be noted that dysbiosis induces multifaceted derangements in microbial-mediated metabolic functions, including regulation of amino acid, lipid, and carbohydrate metabolism, that could further contribute to worsening outcomes. In fact, severe COVID-19 disease is characterized by a drastic shift in the composition of gut-modulated biologically active molecules that engage in inflammation signaling and immune system activation[55,59]. The long-lasting alteration of the microbial signature due to SARS-CoV-2, characterized by the predominance of pathogenic species and activation of pro-inflammatory pathways, perpetuates intestinal inflammation and could lead to long-term complications, such as tumorigenesis and colorectal cancer[60,181]. Systemic immune activation in COVID-19, which is at least partially fueled by leaky gut, has been linked to all of the severe manifestations of the disease, including cytokine storm syndrome, ARDS, renal failure, cardiovascular events, thromboembolic disease, and neurological manifestations[182]. The co-existence of intestinal barrier dysfunction in individuals with underlying diseases, such as obesity, diabetes, colorectal cancer, or immunosuppression, could exacerbate endotoxemia and the consequent immune system overactivation, predisposing to a more severe disease course[182,183].
CONCLUSION
Although the respiratory tract is the main target organ of SARS-CoV-2 infection, severe COVID-19 is considered a complex disorder affecting multiple systems. The development of GI-related symptoms, long-lasting fecal shedding of viral RNA, and identification of the virus in human intestinal tissues have brought to the spotlight the potential effects of the GI system in COVID-19 pathophysiology. The integrity of the intestinal barrier is a sine qua non for the accomplishment of the diverse digestive and immunomodulatory functions of the GI tract. In this setting, SARS-CoV-2 is capable of inducing deleterious effects on the gatekeepers of paracellular transport, i.e., TJs, as well as on intestinal epithelial and endothelial cells through direct ACE2-dependent or indirect immune-mediated mechanisms, or both. Apart from dismantling the mechanical structures of the mucosa, COVID-19 is accompanied by profound alterations of the intestinal microflora at taxonomic and functional levels that are associated with disease severity and the host’s immune system activation. Furthermore, SARS-CoV-2 hijacks innate immune responses, principally through interference with IFN signaling, and, thus, leads to inappropriate trafficking and activation of virus-specific T and B cells. In turn, this multi-layered disruption of the gut barrier can exacerbate the underlying immunopathology of COVID-19 or precipitate serious complications.
The gut and the lungs, albeit anatomically distinct, participate in a bi-directional immunological crosstalk through their respective microbes and immune cells. A well-tuned intestinal barrier harboring a diverse community of commensal microorganisms is pivotal in modulating lung immune responses and the lung microbiome. Therefore, gut dysbiosis impairs the ability to prime vigorous immune responses in the respiratory tract to effectively contain viral infections such as SARS-CoV-2. Moreover, the gut vascular bed provides an enormous endothelial surface susceptible to SARS-CoV-2-mediated injury. Disease-activated and apoptotic endothelial cells are potent drivers of neutrophil/monocyte recruitment, platelet aggregation, and coagulation activation. In parallel, dysbiosis, endotoxemia, and systemic hyperimmune reactions shape a procoagulant state within the gut microvasculature that possibly contributes to extraintestinal thrombotic complications or ARDS pathogenesis, which are common manifestations of severe COVID-19. A compromised gut barrier allows an excessive influx of intraluminal microbiota into otherwise sterile extraintestinal compartments. This systemic dissemination of microbial constituents and metabolites contributes, to a certain extent, to immune system activation and hyper-inflammatory responses that govern the severe form of COVID-19. Preexisting comorbidities plagued with impaired intestinal permeability, such as obesity, diabetes, cirrhosis, and autoimmune disorders, might act synergistically with SARS-CoV-2 to further aggravate endotoxemia and endotoxin-mediated immune activation, predisposing to a more complicated disease course. Modulation of the gut barrier function emerges as a promising intervention to prevent or alleviate severe COVID-19 and related complications.
Footnotes
Conflict-of-interest statement: All the authors report no relevant conflicts of interest for this article.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Peer-review started: October 12, 2022
First decision: November 3, 2022
Article in press: January 16, 2023
Specialty type: Virology
Country/Territory of origin: Greece
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Rotondo JC, Italy; Santulli G, United States S-Editor: Gao CC L-Editor: Filipodia P-Editor: Gao CC
Contributor Information
Efthymios P Tsounis, Division of Gastroenterology, Department of Internal Medicine, Medical School, University Hospital of Patras, Patras 26504, Greece.
Christos Triantos, Division of Gastroenterology, Department of Internal Medicine, Medical School, University Hospital of Patras, Patras 26504, Greece.
Christos Konstantakis, Division of Gastroenterology, Department of Internal Medicine, Medical School, University Hospital of Patras, Patras 26504, Greece.
Markos Marangos, Division of Infectious Diseases, Department of Internal Medicine, Medical School, University of Patras, University Hospital of Patras, Patras 26504, Greece.
Stelios F Assimakopoulos, Division of Infectious Diseases, Department of Internal Medicine, Medical School, University of Patras, University Hospital of Patras, Patras 26504, Greece. sassim@upatras.gr.
References
- 1.Osuchowski MF, Winkler MS, Skirecki T, Cajander S, Shankar-Hari M, Lachmann G, Monneret G, Venet F, Bauer M, Brunkhorst FM, Weis S, Garcia-Salido A, Kox M, Cavaillon JM, Uhle F, Weigand MA, Flohé SB, Wiersinga WJ, Almansa R, de la Fuente A, Martin-Loeches I, Meisel C, Spinetti T, Schefold JC, Cilloniz C, Torres A, Giamarellos-Bourboulis EJ, Ferrer R, Girardis M, Cossarizza A, Netea MG, van der Poll T, Bermejo-Martín JF, Rubio I. The COVID-19 puzzle: deciphering pathophysiology and phenotypes of a new disease entity. Lancet Respir Med. 2021;9:622–642. doi: 10.1016/S2213-2600(21)00218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang L, Liu S, Liu J, Zhang Z, Wan X, Huang B, Chen Y, Zhang Y. COVID-19: immunopathogenesis and Immunotherapeutics. Signal Transduct Target Ther. 2020;5:128. doi: 10.1038/s41392-020-00243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu B, Huang S, Yin L. The cytokine storm and COVID-19. J Med Virol. 2021;93:250–256. doi: 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.König J, Wells J, Cani PD, García-Ródenas CL, MacDonald T, Mercenier A, Whyte J, Troost F, Brummer RJ. Human Intestinal Barrier Function in Health and Disease. Clin Transl Gastroenterol. 2016;7:e196. doi: 10.1038/ctg.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Assimakopoulos SF, Scopa CD, Vagianos CE. Pathophysiology of increased intestinal permeability in obstructive jaundice. World J Gastroenterol. 2007;13:6458–6464. doi: 10.3748/wjg.v13.i48.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Assimakopoulos SF, Triantos C, Maroulis I, Gogos C. The Role of the Gut Barrier Function in Health and Disease. Gastroenterology Res. 2018;11:261–263. doi: 10.14740/gr1053w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rotondo JC, Martini F, Maritati M, Caselli E, Gallenga CE, Guarino M, De Giorgio R, Mazziotta C, Tramarin ML, Badiale G, Tognon M, Contini C. Advanced Molecular and Immunological Diagnostic Methods to Detect SARS-CoV-2 Infection. Microorganisms. 2022;10 doi: 10.3390/microorganisms10061193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hikmet F, Méar L, Edvinsson Å, Micke P, Uhlén M, Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol. 2020;16:e9610. doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorrell RD, Dougherty MK, Barash EL, Lichtig AE, Clayton SB, Jensen ET. Gastrointestinal and hepatic manifestations of COVID-19: A systematic review and meta-analysis. JGH Open. 2021;5:107–115. doi: 10.1002/jgh3.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao R, Qiu Y, He JS, Tan JY, Li XH, Liang J, Shen J, Zhu LR, Chen Y, Iacucci M, Ng SC, Ghosh S, Chen MH. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2020;5:667–678. doi: 10.1016/S2468-1253(20)30126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sultan S, Altayar O, Siddique SM, Davitkov P, Feuerstein JD, Lim JK, Falck-Ytter Y, El-Serag HB AGA Institute. AGA Institute Rapid Review of the Gastrointestinal and Liver Manifestations of COVID-19, Meta-Analysis of International Data, and Recommendations for the Consultative Management of Patients with COVID-19. Gastroenterology. 2020;159:320–334.e27. doi: 10.1053/j.gastro.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry BM, de Oliveira MHS, Benoit J, Lippi G. Gastrointestinal symptoms associated with severity of coronavirus disease 2019 (COVID-19): a pooled analysis. Intern Emerg Med. 2020;15:857–859. doi: 10.1007/s11739-020-02329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng W, Qi K, Ye M, Zheng L, Liu X, Hu S, Zhang W, Tang W, Xu J, Yu D, Wei Y. Gastrointestinal symptoms are associated with severity of coronavirus disease 2019: a systematic review and meta-analysis. Eur J Gastroenterol Hepatol. 2022;34:168–176. doi: 10.1097/MEG.0000000000002072. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi Y, Wagatsuma K, Nojima M, Yamakawa T, Ichimiya T, Yokoyama Y, Kazama T, Hirayama D, Nakase H. The characteristics of gastrointestinal symptoms in patients with severe COVID-19: a systematic review and meta-analysis. J Gastroenterol. 2021;56:409–420. doi: 10.1007/s00535-021-01778-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shehab M, Alrashed F, Shuaibi S, Alajmi D, Barkun A. Gastroenterological and hepatic manifestations of patients with COVID-19, prevalence, mortality by country, and intensive care admission rate: systematic review and meta-analysis. BMJ Open Gastroenterol. 2021;8 doi: 10.1136/bmjgast-2020-000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Li Y, Zhang Y, Liu Y. Are gastrointestinal symptoms associated with higher risk of Mortality in COVID-19 patients? BMC Gastroenterol. 2022;22:106. doi: 10.1186/s12876-022-02132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livanos AE, Jha D, Cossarini F, Gonzalez-Reiche AS, Tokuyama M, Aydillo T, Parigi TL, Ladinsky MS, Ramos I, Dunleavy K, Lee B, Dixon RE, Chen ST, Martinez-Delgado G, Nagula S, Bruce EA, Ko HM, Glicksberg BS, Nadkarni G, Pujadas E, Reidy J, Naymagon S, Grinspan A, Ahmad J, Tankelevich M, Bram Y, Gordon R, Sharma K, Houldsworth J, Britton GJ, Chen-Liaw A, Spindler MP, Plitt T, Wang P, Cerutti A, Faith JJ, Colombel JF, Kenigsberg E, Argmann C, Merad M, Gnjatic S, Harpaz N, Danese S, Cordon-Cardo C, Rahman A, Schwartz RE, Kumta NA, Aghemo A, Bjorkman PJ, Petralia F, van Bakel H, Garcia-Sastre A, Mehandru S. Intestinal Host Response to SARS-CoV-2 Infection and COVID-19 Outcomes in Patients With Gastrointestinal Symptoms. Gastroenterology. 2021;160:2435–2450.e34. doi: 10.1053/j.gastro.2021.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Cen M, Hu M, Du L, Hu W, Kim JJ, Dai N. Prevalence and Persistent Shedding of Fecal SARS-CoV-2 RNA in Patients With COVID-19 Infection: A Systematic Review and Meta-analysis. Clin Transl Gastroenterol. 2021;12:e00343. doi: 10.14309/ctg.0000000000000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 22.Effenberger M, Grabherr F, Mayr L, Schwaerzler J, Nairz M, Seifert M, Hilbe R, Seiwald S, Scholl-Buergi S, Fritsche G, Bellmann-Weiler R, Weiss G, Müller T, Adolph TE, Tilg H. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut. 2020;69:1543–1544. doi: 10.1136/gutjnl-2020-321388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan KH, Sridhar S, Zhang RR, Chu H, Fung AY, Chan G, Chan JF, To KK, Hung IF, Cheng VC, Yuen KY. Factors affecting stability and infectivity of SARS-CoV-2. J Hosp Infect. 2020;106:226–231. doi: 10.1016/j.jhin.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esseili MA. Great escape: how infectious SARS-CoV-2 avoids inactivation by gastric acidity and intestinal bile. Gut. 2022 doi: 10.1136/gutjnl-2021-326624. [DOI] [PubMed] [Google Scholar]
- 25.Bushman FD, McCormick K, Sherrill-Mix S. Virus structures constrain transmission modes. Nat Microbiol. 2019;4:1778–1780. doi: 10.1038/s41564-019-0523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng W, Bao L, Gao H, Xiang Z, Qu Y, Song Z, Gong S, Liu J, Yu P, Qi F, Xu Y, Li F, Xiao C, Lv Q, Xue J, Wei Q, Liu M, Wang G, Wang S, Yu H, Chen T, Liu X, Zhao W, Han Y, Qin C. Ocular conjunctival inoculation of SARS-CoV-2 can cause mild COVID-19 in rhesus macaques. Nat Commun. 2020;11:4400. doi: 10.1038/s41467-020-18149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, Jing H, Wang C, Wang Y, Zuo N, Jiang T, Novakovic VA, Shi J. Intestinal Damage in COVID-19: SARS-CoV-2 Infection and Intestinal Thrombosis. Front Microbiol. 2022;13:860931. doi: 10.3389/fmicb.2022.860931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu F, Chen F, Ou Z, Fan Q, Tan X, Wang Y, Pan Y, Ke B, Li L, Guan Y, Mo X, Wang J, Luo C, Wen X, Li M, Ren P, Ke C, Li J, Lei C, Tang X, Li F. A compromised specific humoral immune response against the SARS-CoV-2 receptor-binding domain is related to viral persistence and periodic shedding in the gastrointestinal tract. Cell Mol Immunol. 2020;17:1119–1125. doi: 10.1038/s41423-020-00550-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vancamelbeke M, Vermeire S. The intestinal barrier: a fundamental role in health and disease. Expert Rev Gastroenterol Hepatol. 2017;11:821–834. doi: 10.1080/17474124.2017.1343143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Assimakopoulos SF, Papageorgiou I, Charonis A. Enterocytes' tight junctions: From molecules to diseases. World J Gastrointest Pathophysiol. 2011;2:123–137. doi: 10.4291/wjgp.v2.i6.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng W, Takeichi M. Adherens junction: molecular architecture and regulation. Cold Spring Harb Perspect Biol. 2009;1:a002899. doi: 10.1101/cshperspect.a002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroeder BO. Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep (Oxf) 2019;7:3–12. doi: 10.1093/gastro/goy052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. Role of the normal gut microbiota. World J Gastroenterol. 2015;21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aggeletopoulou I, Konstantakis C, Assimakopoulos SF, Triantos C. The role of the gut microbiota in the treatment of inflammatory bowel diseases. Microb Pathog. 2019;137:103774. doi: 10.1016/j.micpath.2019.103774. [DOI] [PubMed] [Google Scholar]
- 35.Okumura R, Takeda K. Maintenance of gut homeostasis by the mucosal immune system. Proc Jpn Acad Ser B Phys Biol Sci. 2016;92:423–435. doi: 10.2183/pjab.92.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehmann M, Allers K, Heldt C, Meinhardt J, Schmidt F, Rodriguez-Sillke Y, Kunkel D, Schumann M, Böttcher C, Stahl-Hennig C, Elezkurtaj S, Bojarski C, Radbruch H, Corman VM, Schneider T, Loddenkemper C, Moos V, Weidinger C, Kühl AA, Siegmund B. Human small intestinal infection by SARS-CoV-2 is characterized by a mucosal infiltration with activated CD8(+) T cells. Mucosal Immunol. 2021;14:1381–1392. doi: 10.1038/s41385-021-00437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu W, Ou X, Liu X, Zhang S, Gao X, Cheng H, Zhu B, Yan J. ACE2 contributes to the maintenance of mouse epithelial barrier function. Biochem Biophys Res Commun. 2020;533:1276–1282. doi: 10.1016/j.bbrc.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duan Y, Prasad R, Feng D, Beli E, Li Calzi S, Longhini ALF, Lamendella R, Floyd JL, Dupont M, Noothi SK, Sreejit G, Athmanathan B, Wright J, Jensen AR, Oudit GY, Markel TA, Nagareddy PR, Obukhov AG, Grant MB. Bone Marrow-Derived Cells Restore Functional Integrity of the Gut Epithelial and Vascular Barriers in a Model of Diabetes and ACE2 Deficiency. Circ Res. 2019;125:969–988. doi: 10.1161/CIRCRESAHA.119.315743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biering SB, Sarnik SA, Wang E, Zengel JR, Leist SR, Schäfer A, Sathyan V, Hawkins P, Okuda K, Tau C, Jangid AR, Duffy CV, Wei J, Gilmore RC, Alfajaro MM, Strine MS, Nguyenla X, Van Dis E, Catamura C, Yamashiro LH, Belk JA, Begeman A, Stark JC, Shon DJ, Fox DM, Ezzatpour S, Huang E, Olegario N, Rustagi A, Volmer AS, Livraghi-Butrico A, Wehri E, Behringer RR, Cheon DJ, Schaletzky J, Aguilar HC, Puschnik AS, Button B, Pinsky BA, Blish CA, Baric RS, O'Neal WK, Bertozzi CR, Wilen CB, Boucher RC, Carette JE, Stanley SA, Harris E, Konermann S, Hsu PD. Genome-wide bidirectional CRISPR screens identify mucins as host factors modulating SARS-CoV-2 infection. Nat Genet. 2022;54:1078–1089. doi: 10.1038/s41588-022-01131-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zihni C, Mills C, Matter K, Balda MS. Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol. 2016;17:564–580. doi: 10.1038/nrm.2016.80. [DOI] [PubMed] [Google Scholar]
- 41.Tian W, Zhang N, Jin R, Feng Y, Wang S, Gao S, Gao R, Wu G, Tian D, Tan W, Chen Y, Gao GF, Wong CCL. Immune suppression in the early stage of COVID-19 disease. Nat Commun. 2020;11:5859. doi: 10.1038/s41467-020-19706-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang RC, Huang K, Zhang HP, Li L, Zhang YF, Tan C, Chen HC, Jin ML, Wang XR. SARS-CoV-2 productively infects human brain microvascular endothelial cells. J Neuroinflammation. 2022;19:149. doi: 10.1186/s12974-022-02514-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Maio F, Lo Cascio E, Babini G, Sali M, Della Longa S, Tilocca B, Roncada P, Arcovito A, Sanguinetti M, Scambia G, Urbani A. Improved binding of SARS-CoV-2 Envelope protein to tight junction-associated PALS1 could play a key role in COVID-19 pathogenesis. Microbes Infect. 2020;22:592–597. doi: 10.1016/j.micinf.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chai J, Cai Y, Pang C, Wang L, McSweeney S, Shanklin J, Liu Q. Structural basis for SARS-CoV-2 envelope protein recognition of human cell junction protein PALS1. Nat Commun. 2021;12:3433. doi: 10.1038/s41467-021-23533-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teoh KT, Siu YL, Chan WL, Schlüter MA, Liu CJ, Peiris JS, Bruzzone R, Margolis B, Nal B. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol Biol Cell. 2010;21:3838–3852. doi: 10.1091/mbc.E10-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo Y, Luo R, Wang Y, Deng P, Song T, Zhang M, Wang P, Zhang X, Cui K, Tao T, Li Z, Chen W, Zheng Y, Qin J. SARS-CoV-2 induced intestinal responses with a biomimetic human gut-on-chip. Sci Bull (Beijing) 2021;66:783–793. doi: 10.1016/j.scib.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santa Cruz A, Mendes-Frias A, Oliveira AI, Dias L, Matos AR, Carvalho A, Capela C, Pedrosa J, Castro AG, Silvestre R. Interleukin-6 Is a Biomarker for the Development of Fatal Severe Acute Respiratory Syndrome Coronavirus 2 Pneumonia. Front Immunol. 2021;12:613422. doi: 10.3389/fimmu.2021.613422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alsaffar H, Martino N, Garrett JP, Adam AP. Interleukin-6 promotes a sustained loss of endothelial barrier function via Janus kinase-mediated STAT3 phosphorylation and de novo protein synthesis. Am J Physiol Cell Physiol. 2018;314:C589–C602. doi: 10.1152/ajpcell.00235.2017. [DOI] [PubMed] [Google Scholar]
- 49.Assimakopoulos SF, Mastronikolis S, DE Lastic AL, Aretha D, Papageorgiou D, Chalkidi T, Oikonomou I, Triantos C, Mouzaki A, Marangos M. Intestinal Barrier Biomarker ZO1 and Endotoxin Are Increased in Blood of Patients With COVID-19-associated Pneumonia. In Vivo. 2021;35:2483–2488. doi: 10.21873/invivo.12528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hurley JC, Nowak P, Öhrmalm L, Gogos C, Armaganidis A, Giamarellos-Bourboulis EJ. Endotoxemia as a diagnostic tool for patients with suspected bacteremia caused by gram-negative organisms: a meta-analysis of 4 decades of studies. J Clin Microbiol. 2015;53:1183–1191. doi: 10.1128/JCM.03531-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuo WT, Zuo L, Odenwald MA, Madha S, Singh G, Gurniak CB, Abraham C, Turner JR. The Tight Junction Protein ZO-1 Is Dispensable for Barrier Function but Critical for Effective Mucosal Repair. Gastroenterology. 2021;161:1924–1939. doi: 10.1053/j.gastro.2021.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tornavaca O, Chia M, Dufton N, Almagro LO, Conway DE, Randi AM, Schwartz MA, Matter K, Balda MS. ZO-1 controls endothelial adherens junctions, cell-cell tension, angiogenesis, and barrier formation. J Cell Biol. 2015;208:821–838. doi: 10.1083/jcb.201404140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fasano A. Intestinal permeability and its regulation by zonulin: diagnostic and therapeutic implications. Clin Gastroenterol Hepatol. 2012;10:1096–1100. doi: 10.1016/j.cgh.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sapone A, de Magistris L, Pietzak M, Clemente MG, Tripathi A, Cucca F, Lampis R, Kryszak D, Cartenì M, Generoso M, Iafusco D, Prisco F, Laghi F, Riegler G, Carratu R, Counts D, Fasano A. Zonulin upregulation is associated with increased gut permeability in subjects with type 1 diabetes and their relatives. Diabetes. 2006;55:1443–1449. doi: 10.2337/db05-1593. [DOI] [PubMed] [Google Scholar]
- 55.Giron LB, Dweep H, Yin X, Wang H, Damra M, Goldman AR, Gorman N, Palmer CS, Tang HY, Shaikh MW, Forsyth CB, Balk RA, Zilberstein NF, Liu Q, Kossenkov A, Keshavarzian A, Landay A, Abdel-Mohsen M. Plasma Markers of Disrupted Gut Permeability in Severe COVID-19 Patients. Front Immunol. 2021;12:686240. doi: 10.3389/fimmu.2021.686240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17:219–232. doi: 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- 57.Reinold J, Farahpour F, Fehring C, Dolff S, Konik M, Korth J, van Baal L, Hoffmann D, Buer J, Witzke O, Westendorf AM, Kehrmann J. A Pro-Inflammatory Gut Microbiome Characterizes SARS-CoV-2 Infected Patients and a Reduction in the Connectivity of an Anti-Inflammatory Bacterial Network Associates With Severe COVID-19. Front Cell Infect Microbiol. 2021;11:747816. doi: 10.3389/fcimb.2021.747816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Q, Mak JWY, Su Q, Yeoh YK, Lui GC, Ng SSS, Zhang F, Li AYL, Lu W, Hui DS, Chan PK, Chan FKL, Ng SC. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut. 2022;71:544–552. doi: 10.1136/gutjnl-2021-325989. [DOI] [PubMed] [Google Scholar]
- 59.Sun Z, Song ZG, Liu C, Tan S, Lin S, Zhu J, Dai FH, Gao J, She JL, Mei Z, Lou T, Zheng JJ, Liu Y, He J, Zheng Y, Ding C, Qian F, Chen YM. Gut microbiome alterations and gut barrier dysfunction are associated with host immune homeostasis in COVID-19 patients. BMC Med. 2022;20:24. doi: 10.1186/s12916-021-02212-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, Chung AC, Cheung CP, Tso EY, Fung KS, Chan V, Ling L, Joynt G, Hui DS, Chow KM, Ng SSS, Li TC, Ng RW, Yip TC, Wong GL, Chan FK, Wong CK, Chan PK, Ng SC. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, Wild B, Camargo SM, Singer D, Richter A, Kuba K, Fukamizu A, Schreiber S, Clevers H, Verrey F, Rosenstiel P, Penninger JM. ACE2 Links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao Y, Chen F, Wu W, Sun M, Bilotta AJ, Yao S, Xiao Y, Huang X, Eaves-Pyles TD, Golovko G, Fofanov Y, D'Souza W, Zhao Q, Liu Z, Cong Y. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 2018;11:752–762. doi: 10.1038/mi.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edwinson A, Yang L, Chen J, Grover M. Colonic expression of Ace2, the SARS-CoV-2 entry receptor, is suppressed by commensal human microbiota. Gut Microbes. 2021;13:1984105. doi: 10.1080/19490976.2021.1984105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Groves HT, Higham SL, Moffatt MF, Cox MJ, Tregoning JS. Respiratory Viral Infection Alters the Gut Microbiota by Inducing Inappetence. mBio. 2020;11 doi: 10.1128/mBio.03236-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guimarães Sousa S, Kleiton de Sousa A, Maria Carvalho Pereira C, Sofia Miranda Loiola Araújo A, de Aguiar Magalhães D, Vieira de Brito T, Barbosa ALDR. SARS-CoV-2 infection causes intestinal cell damage: Role of interferon's imbalance. Cytokine. 2022;152:155826. doi: 10.1016/j.cyto.2022.155826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deriu E, Boxx GM, He X, Pan C, Benavidez SD, Cen L, Rozengurt N, Shi W, Cheng G. Influenza Virus Affects Intestinal Microbiota and Secondary Salmonella Infection in the Gut through Type I Interferons. PLoS Pathog. 2016;12:e1005572. doi: 10.1371/journal.ppat.1005572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gopal AB, Chakraborty S, Padhan PK, Barik A, Dixit P, Chakraborty D, Poirah I, Samal S, Sarkar A, Bhattacharyya A. Silent hypoxia in COVID-19: a gut microbiota connection. Curr Opin Physiol. 2021;23:100456. doi: 10.1016/j.cophys.2021.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sencio V, Machado MG, Trottein F. The lung-gut axis during viral respiratory infections: the impact of gut dysbiosis on secondary disease outcomes. Mucosal Immunol. 2021;14:296–304. doi: 10.1038/s41385-020-00361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Farsi Y, Tahvildari A, Arbabi M, Vazife F, Sechi LA, Shahidi Bonjar AH, Jamshidi P, Nasiri MJ, Mirsaeidi M. Diagnostic, Prognostic, and Therapeutic Roles of Gut Microbiota in COVID-19: A Comprehensive Systematic Review. Front Cell Infect Microbiol. 2022;12:804644. doi: 10.3389/fcimb.2022.804644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gu S, Chen Y, Wu Z, Gao H, Lv L, Guo F, Zhang X, Luo R, Huang C, Lu H, Zheng B, Zhang J, Yan R, Zhang H, Jiang H, Xu Q, Guo J, Gong Y, Tang L, Li L. Alterations of the Gut Microbiota in Patients With Coronavirus Disease 2019 or H1N1 Influenza. Clin Infect Dis. 2020;71:2669–2678. doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao J, Wang C, Zhang Y, Lei G, Xu K, Zhao N, Lu J, Meng F, Yu L, Yan J, Bai C, Zhang S, Zhang N, Gong Y, Bi Y, Shi Y, Chen Z, Dai L, Wang J, Yang P. Integrated gut virome and bacteriome dynamics in COVID-19 patients. Gut Microbes. 2021;13:1–21. doi: 10.1080/19490976.2021.1887722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamanai-Shacoori Z, Smida I, Bousarghin L, Loreal O, Meuric V, Fong SB, Bonnaure-Mallet M, Jolivet-Gougeon A. Roseburia spp.: a marker of health? Future Microbiol. 2017;12:157–170. doi: 10.2217/fmb-2016-0130. [DOI] [PubMed] [Google Scholar]
- 73.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mukherjee A, Lordan C, Ross RP, Cotter PD. Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microbes. 2020;12:1802866. doi: 10.1080/19490976.2020.1802866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Konieczna P, Akdis CA, Quigley EM, Shanahan F, O'Mahony L. Portrait of an immunoregulatory Bifidobacterium. Gut Microbes. 2012;3:261–266. doi: 10.4161/gmic.20358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yao S, Zhao Z, Wang W, Liu X. Bifidobacterium Longum: Protection against Inflammatory Bowel Disease. J Immunol Res. 2021;2021:8030297. doi: 10.1155/2021/8030297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yuan F, Ni H, Asche CV, Kim M, Walayat S, Ren J. Efficacy of Bifidobacterium infantis 35624 in patients with irritable bowel syndrome: a meta-analysis. Curr Med Res Opin. 2017;33:1191–1197. doi: 10.1080/03007995.2017.1292230. [DOI] [PubMed] [Google Scholar]
- 78.Gautier T, David-Le Gall S, Sweidan A, Tamanai-Shacoori Z, Jolivet-Gougeon A, Loréal O, Bousarghin L. Next-Generation Probiotics and Their Metabolites in COVID-19. Microorganisms. 2021;9 doi: 10.3390/microorganisms9050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 80.Henke MT, Kenny DJ, Cassilly CD, Vlamakis H, Xavier RJ, Clardy J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn's disease, produces an inflammatory polysaccharide. Proc Natl Acad Sci U S A. 2019;116:12672–12677. doi: 10.1073/pnas.1904099116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moreira-Rosário A, Marques C, Pinheiro H, Araújo JR, Ribeiro P, Rocha R, Mota I, Pestana D, Ribeiro R, Pereira A, de Sousa MJ, Pereira-Leal J, de Sousa J, Morais J, Teixeira D, Rocha JC, Silvestre M, Príncipe N, Gatta N, Amado J, Santos L, Maltez F, Boquinhas A, de Sousa G, Germano N, Sarmento G, Granja C, Póvoa P, Faria A, Calhau C. Gut Microbiota Diversity and C-Reactive Protein Are Predictors of Disease Severity in COVID-19 Patients. Front Microbiol. 2021;12:705020. doi: 10.3389/fmicb.2021.705020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zuo T, Zhang F, Lui GCY, Yeoh YK, Li AYL, Zhan H, Wan Y, Chung ACK, Cheung CP, Chen N, Lai CKC, Chen Z, Tso EYK, Fung KSC, Chan V, Ling L, Joynt G, Hui DSC, Chan FKL, Chan PKS, Ng SC. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology. 2020;159:944–955.e8. doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gaibani P, D'Amico F, Bartoletti M, Lombardo D, Rampelli S, Fornaro G, Coladonato S, Siniscalchi A, Re MC, Viale P, Brigidi P, Turroni S, Giannella M. The Gut Microbiota of Critically Ill Patients With COVID-19. Front Cell Infect Microbiol. 2021;11:670424. doi: 10.3389/fcimb.2021.670424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Khan M, Mathew BJ, Gupta P, Garg G, Khadanga S, Vyas AK, Singh AK. Gut Dysbiosis and IL-21 Response in Patients with Severe COVID-19. Microorganisms. 2021;9 doi: 10.3390/microorganisms9061292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mazzarelli A, Giancola ML, Farina A, Marchioni L, Rueca M, Gruber CEM, Bartolini B, Ascoli Bartoli T, Maffongelli G, Capobianchi MR, Ippolito G, Di Caro A, Nicastri E, Pazienza V INMI COVID-19 study group. 16S rRNA gene sequencing of rectal swab in patients affected by COVID-19. PLoS One. 2021;16:e0247041. doi: 10.1371/journal.pone.0247041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rueca M, Fontana A, Bartolini B, Piselli P, Mazzarelli A, Copetti M, Binda E, Perri F, Gruber CEM, Nicastri E, Marchioni L, Ippolito G, Capobianchi MR, Di Caro A, Pazienza V. Investigation of Nasal/Oropharyngeal Microbial Community of COVID-19 Patients by 16S rDNA Sequencing. Int J Environ Res Public Health. 2021;18 doi: 10.3390/ijerph18042174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.He F, Zhang T, Xue K, Fang Z, Jiang G, Huang S, Li K, Gu Z, Shi H, Zhang Z, Zhu H, Lin L, Li J, Xiao F, Shan H, Yan R, Li X, Yan Z. Fecal multi-omics analysis reveals diverse molecular alterations of gut ecosystem in COVID-19 patients. Anal Chim Acta. 2021;1180:338881. doi: 10.1016/j.aca.2021.338881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang B, Zhang L, Wang Y, Dai T, Qin Z, Zhou F. Alterations in microbiota of patients with COVID-19: potential mechanisms and therapeutic interventions. Signal Transduct Target Ther. 2022;7:143. doi: 10.1038/s41392-022-00986-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scott NA, Andrusaite A, Andersen P, Lawson M, Alcon-Giner C, Leclaire C, Caim S, Le Gall G, Shaw T, Connolly JPR, Roe AJ, Wessel H, Bravo-Blas A, Thomson CA, Kästele V, Wang P, Peterson DA, Bancroft A, Li X, Grencis R, Mowat AM, Hall LJ, Travis MA, Milling SWF, Mann ER. Antibiotics induce sustained dysregulation of intestinal T cell immunity by perturbing macrophage homeostasis. Sci Transl Med. 2018;10 doi: 10.1126/scitranslmed.aao4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 91.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 92.Konieczna P, Ferstl R, Ziegler M, Frei R, Nehrbass D, Lauener RP, Akdis CA, O'Mahony L. Immunomodulation by Bifidobacterium infantis 35624 in the murine lamina propria requires retinoic acid-dependent and independent mechanisms. PLoS One. 2013;8:e62617. doi: 10.1371/journal.pone.0062617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Verma R, Lee C, Jeun EJ, Yi J, Kim KS, Ghosh A, Byun S, Lee CG, Kang HJ, Kim GC, Jun CD, Jan G, Suh CH, Jung JY, Sprent J, Rudra D, De Castro C, Molinaro A, Surh CD, Im SH. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3(+) regulatory T cells. Sci Immunol. 2018;3 doi: 10.1126/sciimmunol.aat6975. [DOI] [PubMed] [Google Scholar]
- 94.Kim M, Qie Y, Park J, Kim CH. Gut Microbial Metabolites Fuel Host Antibody Responses. Cell Host Microbe. 2016;20:202–214. doi: 10.1016/j.chom.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu C, Wang A, Marin M, Honnen W, Ramasamy S, Porter E, Subbian S, Pinter A, Melikyan GB, Lu W, Chang TL. Human Defensins Inhibit SARS-CoV-2 Infection by Blocking Viral Entry. Viruses. 2021;13 doi: 10.3390/v13071246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang C, Wang S, Li D, Wei DQ, Zhao J, Wang J. Human Intestinal Defensin 5 Inhibits SARS-CoV-2 Invasion by Cloaking ACE2. Gastroenterology. 2020;159:1145–1147.e4. doi: 10.1053/j.gastro.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Al-Bayatee NT, Ad'hiah AH. Human beta-defensins 2 and 4 are dysregulated in patients with coronavirus disease 19. Microb Pathog. 2021;160:105205. doi: 10.1016/j.micpath.2021.105205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stanifer ML, Kee C, Cortese M, Zumaran CM, Triana S, Mukenhirn M, Kraeusslich HG, Alexandrov T, Bartenschlager R, Boulant S. Critical Role of Type III Interferon in Controlling SARS-CoV-2 Infection in Human Intestinal Epithelial Cells. Cell Rep. 2020;32:107863. doi: 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Metz-Zumaran C, Kee C, Doldan P, Guo C, Stanifer ML, Boulant S. Increased Sensitivity of SARS-CoV-2 to Type III Interferon in Human Intestinal Epithelial Cells. J Virol. 2022;96:e0170521. doi: 10.1128/jvi.01705-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heuberger J, Trimpert J, Vladimirova D, Goosmann C, Lin M, Schmuck R, Mollenkopf HJ, Brinkmann V, Tacke F, Osterrieder N, Sigal M. Epithelial response to IFN-γ promotes SARS-CoV-2 infection. EMBO Mol Med. 2021;13:e13191. doi: 10.15252/emmm.202013191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Karki R, Sharma BR, Tuladhar S, Williams EP, Zalduondo L, Samir P, Zheng M, Sundaram B, Banoth B, Malireddi RKS, Schreiner P, Neale G, Vogel P, Webby R, Jonsson CB, Kanneganti TD. Synergism of TNF-α and IFN-γ Triggers Inflammatory Cell Death, Tissue Damage, and Mortality in SARS-CoV-2 Infection and Cytokine Shock Syndromes. Cell. 2021;184:149–168.e17. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiao L, Li H, Xu J, Yang M, Ma C, Li J, Zhao S, Wang H, Yang Y, Yu W, Wang J, Yang J, Long H, Gao J, Ding K, Wu D, Kuang D, Zhao Y, Liu J, Lu S, Liu H, Peng X. The Gastrointestinal Tract Is an Alternative Route for SARS-CoV-2 Infection in a Nonhuman Primate Model. Gastroenterology. 2021;160:1647–1661. doi: 10.1053/j.gastro.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tao W, Zhang G, Wang X, Guo M, Zeng W, Xu Z, Cao D, Pan A, Wang Y, Zhang K, Ma X, Chen Z, Jin T, Liu L, Weng J, Zhu S. Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18. Med Microecol. 2020;5:100023. doi: 10.1016/j.medmic.2020.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Britton GJ, Chen-Liaw A, Cossarini F, Livanos AE, Spindler MP, Plitt T, Eggers J, Mogno I, Gonzalez-Reiche AS, Siu S, Tankelevich M, Grinspan LT, Dixon RE, Jha D, van de Guchte A, Khan Z, Martinez-Delgado G, Amanat F, Hoagland DA, tenOever BR, Dubinsky MC, Merad M, van Bakel H, Krammer F, Bongers G, Mehandru S, Faith JJ. Limited intestinal inflammation despite diarrhea, fecal viral RNA and SARS-CoV-2-specific IgA in patients with acute COVID-19. Sci Rep. 2021;11:13308. doi: 10.1038/s41598-021-92740-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang C, Xu J, Wang S, Pan S, Zhang J, Han Y, Huang M, Wu D, Yang Q, Yang X, Yang Y, Shu T, Zou X, Li R, Luo Y, Yao R, Wang Y, Qiu Y, Zhang DY, Yao Q, Yan Y, Zhou X, Shang Y. Imaging Mass Cytometric Analysis of Postmortem Tissues Reveals Dysregulated Immune Cell and Cytokine Responses in Multiple Organs of COVID-19 Patients. Front Microbiol. 2020;11:600989. doi: 10.3389/fmicb.2020.600989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Giamarellos-Bourboulis EJ, Netea MG, Rovina N, Akinosoglou K, Antoniadou A, Antonakos N, Damoraki G, Gkavogianni T, Adami ME, Katsaounou P, Ntaganou M, Kyriakopoulou M, Dimopoulos G, Koutsodimitropoulos I, Velissaris D, Koufargyris P, Karageorgos A, Katrini K, Lekakis V, Lupse M, Kotsaki A, Renieris G, Theodoulou D, Panou V, Koukaki E, Koulouris N, Gogos C, Koutsoukou A. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Trevelin SC, Pickering S, Todd K, Bishop C, Pitcher M, Garrido Mesa J, Montorsi L, Spada F, Petrov N, Green A, Shankar-Hari M, Neil SJD, Spencer J. Disrupted Peyer's Patch Microanatomy in COVID-19 Including Germinal Centre Atrophy Independent of Local Virus. Front Immunol. 2022;13:838328. doi: 10.3389/fimmu.2022.838328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hoel H, Heggelund L, Reikvam DH, Stiksrud B, Ueland T, Michelsen AE, Otterdal K, Muller KE, Lind A, Muller F, Dudman S, Aukrust P, Dyrhol-Riise AM, Holter JC, Trøseid M. Elevated markers of gut leakage and inflammasome activation in COVID-19 patients with cardiac involvement. J Intern Med. 2021;289:523–531. doi: 10.1111/joim.13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wurbel MA, McIntire MG, Dwyer P, Fiebiger E. CCL25/CCR9 interactions regulate large intestinal inflammation in a murine model of acute colitis. PLoS One. 2011;6:e16442. doi: 10.1371/journal.pone.0016442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sterlin D, Mathian A, Miyara M, Mohr A, Anna F, Claër L, Quentric P, Fadlallah J, Devilliers H, Ghillani P, Gunn C, Hockett R, Mudumba S, Guihot A, Luyt CE, Mayaux J, Beurton A, Fourati S, Bruel T, Schwartz O, Lacorte JM, Yssel H, Parizot C, Dorgham K, Charneau P, Amoura Z, Gorochov G. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci Transl Med. 2021;13 doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cervia C, Nilsson J, Zurbuchen Y, Valaperti A, Schreiner J, Wolfensberger A, Raeber ME, Adamo S, Weigang S, Emmenegger M, Hasler S, Bosshard PP, De Cecco E, Bächli E, Rudiger A, Stüssi-Helbling M, Huber LC, Zinkernagel AS, Schaer DJ, Aguzzi A, Kochs G, Held U, Probst-Müller E, Rampini SK, Boyman O. Systemic and mucosal antibody responses specific to SARS-CoV-2 during mild vs severe COVID-19. J Allergy Clin Immunol. 2021;147:545–557.e9. doi: 10.1016/j.jaci.2020.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Roy K, Agarwal S, Banerjee R, Paul MK, Purbey PK. COVID-19 and gut immunomodulation. World J Gastroenterol. 2021;27:7925–7942. doi: 10.3748/wjg.v27.i46.7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bradley KC, Finsterbusch K, Schnepf D, Crotta S, Llorian M, Davidson S, Fuchs SY, Staeheli P, Wack A. Microbiota-Driven Tonic Interferon Signals in Lung Stromal Cells Protect from Influenza Virus Infection. Cell Rep. 2019;28:245–256.e4. doi: 10.1016/j.celrep.2019.05.105. [DOI] [PubMed] [Google Scholar]
- 114.Antunes KH, Fachi JL, de Paula R, da Silva EF, Pral LP, Dos Santos AÁ, Dias GBM, Vargas JE, Puga R, Mayer FQ, Maito F, Zárate-Bladés CR, Ajami NJ, Sant'Ana MR, Candreva T, Rodrigues HG, Schmiele M, Silva Clerici MTP, Proença-Modena JL, Vieira AT, Mackay CR, Mansur D, Caballero MT, Marzec J, Li J, Wang X, Bell D, Polack FP, Kleeberger SR, Stein RT, Vinolo MAR, de Souza APD. Microbiota-derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nat Commun. 2019;10:3273. doi: 10.1038/s41467-019-11152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Assimakopoulos SF, Triantos C, Thomopoulos K, Fligou F, Maroulis I, Marangos M, Gogos CA. Gut-origin sepsis in the critically ill patient: pathophysiology and treatment. Infection. 2018;46:751–760. doi: 10.1007/s15010-018-1178-5. [DOI] [PubMed] [Google Scholar]
- 117.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, Wherry EJ, Artis D. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Thackray LB, Handley SA, Gorman MJ, Poddar S, Bagadia P, Briseño CG, Theisen DJ, Tan Q, Hykes BL Jr, Lin H, Lucas TM, Desai C, Gordon JI, Murphy KM, Virgin HW, Diamond MS. Oral Antibiotic Treatment of Mice Exacerbates the Disease Severity of Multiple Flavivirus Infections. Cell Rep. 2018;22:3440–3453.e6. doi: 10.1016/j.celrep.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Steed AL, Christophi GP, Kaiko GE, Sun L, Goodwin VM, Jain U, Esaulova E, Artyomov MN, Morales DJ, Holtzman MJ, Boon ACM, Lenschow DJ, Stappenbeck TS. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science. 2017;357:498–502. doi: 10.1126/science.aam5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stefan KL, Kim MV, Iwasaki A, Kasper DL. Commensal Microbiota Modulation of Natural Resistance to Virus Infection. Cell. 2020;183:1312–1324.e10. doi: 10.1016/j.cell.2020.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, Lienenklaus S, Weiss S, Staeheli P, Aichele P, Diefenbach A. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 122.Akamatsu MA, de Castro JT, Takano CY, Ho PL. Off balance: Interferons in COVID-19 Lung infections. EBioMedicine. 2021;73:103642. doi: 10.1016/j.ebiom.2021.103642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Krämer B, Knoll R, Bonaguro L, ToVinh M, Raabe J, Astaburuaga-García R, Schulte-Schrepping J, Kaiser KM, Rieke GJ, Bischoff J, Monin MB, Hoffmeister C, Schlabe S, De Domenico E, Reusch N, Händler K, Reynolds G, Blüthgen N, Hack G, Finnemann C, Nischalke HD, Strassburg CP, Stephenson E, Su Y, Gardner L, Yuan D, Chen D, Goldman J, Rosenstiel P, Schmidt SV, Latz E, Hrusovsky K, Ball AJ, Johnson JM, Koenig PA, Schmidt FI, Haniffa M, Heath JR, Kümmerer BM, Keitel V, Jensen B, Stubbemann P, Kurth F, Sander LE, Sawitzki B Deutsche COVID-19 OMICS Initiative (DeCOI), Aschenbrenner AC, Schultze JL, Nattermann J. Early IFN-α signatures and persistent dysfunction are distinguishing features of NK cells in severe COVID-19. Immunity. 2021;54:2650–2669.e14. doi: 10.1016/j.immuni.2021.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yagi K, Huffnagle GB, Lukacs NW, Asai N. The Lung Microbiome during Health and Disease. Int J Mol Sci. 2021;22 doi: 10.3390/ijms221910872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sulaiman I, Chung M, Angel L, Tsay JJ, Wu BG, Yeung ST, Krolikowski K, Li Y, Duerr R, Schluger R, Thannickal SA, Koide A, Rafeq S, Barnett C, Postelnicu R, Wang C, Banakis S, Pérez-Pérez L, Shen G, Jour G, Meyn P, Carpenito J, Liu X, Ji K, Collazo D, Labarbiera A, Amoroso N, Brosnahan S, Mukherjee V, Kaufman D, Bakker J, Lubinsky A, Pradhan D, Sterman DH, Weiden M, Heguy A, Evans L, Uyeki TM, Clemente JC, de Wit E, Schmidt AM, Shopsin B, Desvignes L, Li H, Zhang B, Forst CV, Koide S, Stapleford KA, Khanna KM, Ghedin E, Segal LN. Microbial signatures in the lower airways of mechanically ventilated COVID-19 patients associated with poor clinical outcome. Nat Microbiol. 2021;6:1245–1258. doi: 10.1038/s41564-021-00961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang J, Li F, Sun R, Gao X, Wei H, Li LJ, Tian Z. Bacterial colonization dampens influenza-mediated acute lung injury via induction of M2 alveolar macrophages. Nat Commun. 2013;4:2106. doi: 10.1038/ncomms3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gauguet S, D'Ortona S, Ahnger-Pier K, Duan B, Surana NK, Lu R, Cywes-Bentley C, Gadjeva M, Shan Q, Priebe GP, Pier GB. Intestinal Microbiota of Mice Influences Resistance to Staphylococcus aureus Pneumonia. Infect Immun. 2015;83:4003–4014. doi: 10.1128/IAI.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Trompette A, Gollwitzer ES, Pattaroni C, Lopez-Mejia IC, Riva E, Pernot J, Ubags N, Fajas L, Nicod LP, Marsland BJ. Dietary Fiber Confers Protection against Flu by Shaping Ly6c(-) Patrolling Monocyte Hematopoiesis and CD8(+) T Cell Metabolism. Immunity. 2018;48:992–1005.e8. doi: 10.1016/j.immuni.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 130.Chen J, Vitetta L. Modulation of Gut Microbiota for the Prevention and Treatment of COVID-19. J Clin Med. 2021;10 doi: 10.3390/jcm10132903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Conway EM, Mackman N, Warren RQ, Wolberg AS, Mosnier LO, Campbell RA, Gralinski LE, Rondina MT, van de Veerdonk FL, Hoffmeister KM, Griffin JH, Nugent D, Moon K, Morrissey JH. Understanding COVID-19-associated coagulopathy. Nat Rev Immunol. 2022;22:639–649. doi: 10.1038/s41577-022-00762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Loo J, Spittle DA, Newnham M. COVID-19, immunothrombosis and venous thromboembolism: biological mechanisms. Thorax. 2021;76:412–420. doi: 10.1136/thoraxjnl-2020-216243. [DOI] [PubMed] [Google Scholar]
- 133.Lo Re V 3rd, Dutcher SK, Connolly JG, Perez-Vilar S, Carbonari DM, DeFor TA, Djibo DA, Harrington LB, Hou L, Hennessy S, Hubbard RA, Kempner ME, Kuntz JL, McMahill-Walraven CN, Mosley J, Pawloski PA, Petrone AB, Pishko AM, Driscoll MR, Steiner CA, Zhou Y, Cocoros NM. Association of COVID-19 vs Influenza With Risk of Arterial and Venous Thrombotic Events Among Hospitalized Patients. JAMA. 2022;328:637–651. doi: 10.1001/jama.2022.13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kollias A, Kyriakoulis KG, Lagou S, Kontopantelis E, Stergiou GS, Syrigos K. Venous thromboembolism in COVID-19: A systematic review and meta-analysis. Vasc Med. 2021;26:415–425. doi: 10.1177/1358863X21995566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jiménez D, García-Sanchez A, Rali P, Muriel A, Bikdeli B, Ruiz-Artacho P, Le Mao R, Rodríguez C, Hunt BJ, Monreal M. Incidence of VTE and Bleeding Among Hospitalized Patients With Coronavirus Disease 2019: A Systematic Review and Meta-analysis. Chest. 2021;159:1182–1196. doi: 10.1016/j.chest.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Agarwal G, Hajra A, Chakraborty S, Patel N, Biswas S, Adler MK, Lavie CJ. Predictors and mortality risk of venous thromboembolism in patients with COVID-19: systematic review and meta-analysis of observational studies. Ther Adv Cardiovasc Dis. 2022;16:17539447221105013. doi: 10.1177/17539447221105013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pastorelli L, Dozio E, Pisani LF, Boscolo-Anzoletti M, Vianello E, Munizio N, Spina L, Tontini GE, Peyvandi F, Corsi Romanelli MM, Vecchi M. Procoagulatory state in inflammatory bowel diseases is promoted by impaired intestinal barrier function. Gastroenterol Res Pract. 2015;2015:189341. doi: 10.1155/2015/189341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sardu C, Gambardella J, Morelli MB, Wang X, Marfella R, Santulli G. Hypertension, Thrombosis, Kidney Failure, and Diabetes: Is COVID-19 an Endothelial Disease? J Clin Med. 2020;9 doi: 10.3390/jcm9051417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gambardella J, Santulli G. What is linking COVID-19 and endothelial dysfunction? Eur Heart J Cardiovasc Pharmacother. 2021;7:e2–e3. doi: 10.1093/ehjcvp/pvaa145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.van Hinsbergh VW. Endothelium--role in regulation of coagulation and inflammation. Semin Immunopathol. 2012;34:93–106. doi: 10.1007/s00281-011-0285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gambardella J, Kansakar U, Sardu C, Messina V, Jankauskas SS, Marfella R, Maggi P, Wang X, Mone P, Paolisso G, Sorriento D, Santulli G. Exosomal miR-145 and miR-885 regulate thrombosis in COVID-19. J Pharmacol Exp Ther. 2022 doi: 10.1124/jpet.122.001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Quinaglia T, Shabani M, Breder I, Silber HA, Lima JAC, Sposito AC. Coronavirus disease-19: The multi-level, multi-faceted vasculopathy. Atherosclerosis. 2021;322:39–50. doi: 10.1016/j.atherosclerosis.2021.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Peshevska-Sekulovska M, Boeva I, Sekulovski M, Zashev M. Gastrointestinal Ischemia—Stumbling Stone in COVID-19 Patients. Gastroenterol Insigh . 2022;13:206–217. [Google Scholar]
- 144.Kaafarani HMA, El Moheb M, Hwabejire JO, Naar L, Christensen MA, Breen K, Gaitanidis A, Alser O, Mashbari H, Bankhead-Kendall B, Mokhtari A, Maurer L, Kapoen C, Langeveld K, El Hechi MW, Lee J, Mendoza AE, Saillant NN, Parks J, Fawley J, King DR, Fagenholz PJ, Velmahos GC. Gastrointestinal Complications in Critically Ill Patients With COVID-19. Ann Surg. 2020;272:e61–e62. doi: 10.1097/SLA.0000000000004004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schurink B, Roos E, Radonic T, Barbe E, Bouman CSC, de Boer HH, de Bree GJ, Bulle EB, Aronica EM, Florquin S, Fronczek J, Heunks LMA, de Jong MD, Guo L, du Long R, Lutter R, Molenaar PCG, Neefjes-Borst EA, Niessen HWM, van Noesel CJM, Roelofs JJTH, Snijder EJ, Soer EC, Verheij J, Vlaar APJ, Vos W, van der Wel NN, van der Wal AC, van der Valk P, Bugiani M. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe. 2020;1:e290–e299. doi: 10.1016/S2666-5247(20)30144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Carvalho PR, Sirois P, Fernandes PD. The role of kallikrein-kinin and renin-angiotensin systems in COVID-19 infection. Peptides. 2021;135:170428. doi: 10.1016/j.peptides.2020.170428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Qin Z, Liu F, Blair R, Wang C, Yang H, Mudd J, Currey JM, Iwanaga N, He J, Mi R, Han K, Midkiff CC, Alam MA, Aktas BH, Heide RSV, Veazey R, Piedimonte G, Maness NJ, Ergün S, Mauvais-Jarvis F, Rappaport J, Kolls JK, Qin X. Endothelial cell infection and dysfunction, immune activation in severe COVID-19. Theranostics. 2021;11:8076–8091. doi: 10.7150/thno.61810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.He Z, Si Y, Jiang T, Ma R, Zhang Y, Cao M, Li T, Yao Z, Zhao L, Fang S, Yu B, Dong Z, Thatte HS, Bi Y, Kou J, Yang S, Piao D, Hao L, Zhou J, Shi J. Phosphotidylserine exposure and neutrophil extracellular traps enhance procoagulant activity in patients with inflammatory bowel disease. Thromb Haemost. 2016;115:738–751. doi: 10.1160/TH15-09-0710. [DOI] [PubMed] [Google Scholar]
- 150.Gambardella J, Coppola A, Izzo R, Fiorentino G, Trimarco B, Santulli G. Role of endothelial miR-24 in COVID-19 cerebrovascular events. Crit Care. 2021;25:306. doi: 10.1186/s13054-021-03731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mone P, Gambardella J, Wang X, Jankauskas SS, Matarese A, Santulli G. miR-24 Targets the Transmembrane Glycoprotein Neuropilin-1 in Human Brain Microvascular Endothelial Cells. Noncoding RNA. 2021;7 doi: 10.3390/ncrna7010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gao C, Xie R, Yu C, Ma R, Dong W, Meng H, Zhang Y, Si Y, Zhang Z, Novakovic V, Kou J, Bi Y, Li B, Gilbert GE, Zhou J, Shi J. Thrombotic Role of Blood and Endothelial Cells in Uremia through Phosphatidylserine Exposure and Microparticle Release. PLoS One. 2015;10:e0142835. doi: 10.1371/journal.pone.0142835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Goshua G, Pine AB, Meizlish ML, Chang CH, Zhang H, Bahel P, Baluha A, Bar N, Bona RD, Burns AJ, Dela Cruz CS, Dumont A, Halene S, Hwa J, Koff J, Menninger H, Neparidze N, Price C, Siner JM, Tormey C, Rinder HM, Chun HJ, Lee AI. Endotheliopathy in COVID-19-associated coagulopathy: evidence from a single-centre, cross-sectional study. Lancet Haematol. 2020;7:e575–e582. doi: 10.1016/S2352-3026(20)30216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang Y, Cao W, Jiang W, Xiao M, Li Y, Tang N, Liu Z, Yan X, Zhao Y, Li T, Zhu T. Profile of natural anticoagulant, coagulant factor and anti-phospholipid antibody in critically ill COVID-19 patients. J Thromb Thrombolysis. 2020;50:580–586. doi: 10.1007/s11239-020-02182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Vassiliou AG, Keskinidou C, Jahaj E, Gallos P, Dimopoulou I, Kotanidou A, Orfanos SE. ICU Admission Levels of Endothelial Biomarkers as Predictors of Mortality in Critically Ill COVID-19 Patients. Cells. 2021;10 doi: 10.3390/cells10010186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bonaventura A, Vecchié A, Dagna L, Martinod K, Dixon DL, Van Tassell BW, Dentali F, Montecucco F, Massberg S, Levi M, Abbate A. Endothelial dysfunction and immunothrombosis as key pathogenic mechanisms in COVID-19. Nat Rev Immunol. 2021;21:319–329. doi: 10.1038/s41577-021-00536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Martinelli N, Montagnana M, Pizzolo F, Friso S, Salvagno GL, Forni GL, Gianesin B, Morandi M, Lunardi C, Lippi G, Polati E, Olivieri O, De Franceschi L. A relative ADAMTS13 deficiency supports the presence of a secondary microangiopathy in COVID 19. Thromb Res. 2020;193:170–172. doi: 10.1016/j.thromres.2020.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Guervilly C, Burtey S, Sabatier F, Cauchois R, Lano G, Abdili E, Daviet F, Arnaud L, Brunet P, Hraiech S, Jourde-Chiche N, Koubi M, Lacroix R, Pietri L, Berda Y, Robert T, Degioanni C, Velier M, Papazian L, Kaplanski G, Dignat-George F. Circulating Endothelial Cells as a Marker of Endothelial Injury in Severe COVID -19. J Infect Dis. 2020;222:1789–1793. doi: 10.1093/infdis/jiaa528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Whyte CS, Simpson M, Morrow GB, Wallace CA, Mentzer AJ, Knight JC, Shapiro S, Curry N, Bagot CN, Watson H, Cooper JG, Mutch NJ. The suboptimal fibrinolytic response in COVID-19 is dictated by high PAI-1. J Thromb Haemost. 2022;20:2394–2406. doi: 10.1111/jth.15806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Shaw RJ, Bradbury C, Abrams ST, Wang G, Toh CH. COVID-19 and immunothrombosis: emerging understanding and clinical management. Br J Haematol. 2021;194:518–529. doi: 10.1111/bjh.17664. [DOI] [PubMed] [Google Scholar]
- 161.Gillot C, Favresse J, Mullier F, Lecompte T, Dogné JM, Douxfils J. NETosis and the Immune System in COVID-19: Mechanisms and Potential Treatments. Front Pharmacol. 2021;12:708302. doi: 10.3389/fphar.2021.708302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hottz ED, Azevedo-Quintanilha IG, Palhinha L, Teixeira L, Barreto EA, Pão CRR, Righy C, Franco S, Souza TML, Kurtz P, Bozza FA, Bozza PT. Platelet activation and platelet-monocyte aggregate formation trigger tissue factor expression in patients with severe COVID-19. Blood. 2020;136:1330–1341. doi: 10.1182/blood.2020007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Oliva A, Cammisotto V, Cangemi R, Ferro D, Miele MC, De Angelis M, Cancelli F, Pignatelli P, Venditti M, Pugliese F, Mastroianni CM, Violi F. Low-Grade Endotoxemia and Thrombosis in COVID-19. Clin Transl Gastroenterol. 2021;12:e00348. doi: 10.14309/ctg.0000000000000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hasan RA, Koh AY, Zia A. The gut microbiome and thromboembolism. Thromb Res. 2020;189:77–87. doi: 10.1016/j.thromres.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shin HS, Xu F, Bagchi A, Herrup E, Prakash A, Valentine C, Kulkarni H, Wilhelmsen K, Warren S, Hellman J. Bacterial lipoprotein TLR2 agonists broadly modulate endothelial function and coagulation pathways in vitro and in vivo. J Immunol. 2011;186:1119–1130. doi: 10.4049/jimmunol.1001647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jäckel S, Kiouptsi K, Lillich M, Hendrikx T, Khandagale A, Kollar B, Hörmann N, Reiss C, Subramaniam S, Wilms E, Ebner K, Brühl MV, Rausch P, Baines JF, Haberichter S, Lämmle B, Binder CJ, Jurk K, Ruggeri ZM, Massberg S, Walter U, Ruf W, Reinhardt C. Gut microbiota regulate hepatic von Willebrand factor synthesis and arterial thrombus formation via Toll-like receptor-2. Blood. 2017;130:542–553. doi: 10.1182/blood-2016-11-754416. [DOI] [PubMed] [Google Scholar]
- 167.Fiorentino G, Coppola A, Izzo R, Annunziata A, Bernardo M, Lombardi A, Trimarco V, Santulli G, Trimarco B. Effects of adding L-arginine orally to standard therapy in patients with COVID-19: A randomized, double-blind, placebo-controlled, parallel-group trial. Results of the first interim analysis. EClinicalMedicine. 2021;40:101125. doi: 10.1016/j.eclinm.2021.101125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Adebayo A, Varzideh F, Wilson S, Gambardella J, Eacobacci M, Jankauskas SS, Donkor K, Kansakar U, Trimarco V, Mone P, Lombardi A, Santulli G. l-Arginine and COVID-19: An Update. Nutrients. 2021;13 doi: 10.3390/nu13113951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ascher S, Wilms E, Pontarollo G, Formes H, Bayer F, Müller M, Malinarich F, Grill A, Bosmann M, Saffarzadeh M, Brandão I, Groß K, Kiouptsi K, Kittner JM, Lackner KJ, Jurk K, Reinhardt C. Gut Microbiota Restricts NETosis in Acute Mesenteric Ischemia-Reperfusion Injury. Arterioscler Thromb Vasc Biol. 2020;40:2279–2292. doi: 10.1161/ATVBAHA.120.314491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Luo QJ, Sun MX, Guo YW, Tan SW, Wu XY, Abassa KK, Lin L, Liu HL, Jiang J, Wei XQ. Sodium butyrate protects against lipopolysaccharide-induced liver injury partially via the GPR43/ β-arrestin-2/NF-κB network. Gastroenterol Rep (Oxf) 2021;9:154–165. doi: 10.1093/gastro/goaa085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M, Sartor RB, McIntyre TM, Silverstein RL, Tang WHW, DiDonato JA, Brown JM, Lusis AJ, Hazen SL. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell. 2016;165:111–124. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Boini KM, Hussain T, Li PL, Koka S. Trimethylamine-N-Oxide Instigates NLRP3 Inflammasome Activation and Endothelial Dysfunction. Cell Physiol Biochem. 2017;44:152–162. doi: 10.1159/000484623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Israr MZ, Ibrahim W, Salzano A, Sarmad S, Wilde MJ, Cordell RL, Greening NJ, Brightling CE, Siddiqui S, Suzuki T EMBER consortium. Association of gut-related metabolites with respiratory symptoms in COVID-19: A proof-of-concept study. Nutrition. 2022;96:111585. doi: 10.1016/j.nut.2021.111585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Teixeira PC, Dorneles GP, Santana Filho PC, da Silva IM, Schipper LL, Postiga IAL, Neves CAM, Rodrigues Junior LC, Peres A, Souto JT, Fonseca SG, Eller S, Oliveira TF, Rotta LN, Thompson CE, Romão PRT. Increased LPS levels coexist with systemic inflammation and result in monocyte activation in severe COVID-19 patients. Int Immunopharmacol. 2021;100:108125. doi: 10.1016/j.intimp.2021.108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sirivongrangson P, Kulvichit W, Payungporn S, Pisitkun T, Chindamporn A, Peerapornratana S, Pisitkun P, Chitcharoen S, Sawaswong V, Worasilchai N, Kampunya S, Putcharoen O, Thawitsri T, Leelayuwatanakul N, Kongpolprom N, Phoophiboon V, Sriprasart T, Samransamruajkit R, Tungsanga S, Tiankanon K, Lumlertgul N, Leelahavanichkul A, Sriphojanart T, Tantawichien T, Thisyakorn U, Chirathaworn C, Praditpornsilpa K, Tungsanga K, Eiam-Ong S, Sitprija V, Kellum JA, Srisawat N. Endotoxemia and circulating bacteriome in severe COVID-19 patients. Intensive Care Med Exp. 2020;8:72. doi: 10.1186/s40635-020-00362-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Oliva A, Miele MC, Di Timoteo F, De Angelis M, Mauro V, Aronica R, Al Ismail D, Ceccarelli G, Pinacchio C, d'Ettorre G, Mascellino MT, Mastroianni CM. Persistent Systemic Microbial Translocation and Intestinal Damage During Coronavirus Disease-19. Front Immunol. 2021;12:708149. doi: 10.3389/fimmu.2021.708149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kumar NP, Venkataraman A, Hanna LE, Putlibai S, Karthick M, Rajamanikam A, Sadasivam K, Sundaram B, Babu S. Systemic Inflammation and Microbial Translocation Are Characteristic Features of SARS-CoV-2-Related Multisystem Inflammatory Syndrome in Children. Open Forum Infect Dis. 2021;8:ofab279. doi: 10.1093/ofid/ofab279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Prasad R, Patton MJ, Floyd JL, Fortmann S, DuPont M, Harbour A, Wright J, Lamendella R, Stevens BR, Oudit GY, Grant MB. Plasma Microbiome in COVID-19 Subjects: An Indicator of Gut Barrier Defects and Dysbiosis. Int J Mol Sci. 2022;23 doi: 10.3390/ijms23169141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Giron LB, Peluso MJ, Ding J, Kenny G, Zilberstein NF, Koshy J, Hong KY, Rasmussen H, Miller GE, Bishehsari F, Balk RA, Moy JN, Hoh R, Lu S, Goldman AR, Tang HY, Yee BC, Chenna A, Winslow JW, Petropoulos CJ, Kelly JD, Wasse H, Martin JN, Liu Q, Keshavarzian A, Landay A, Deeks SG, Henrich TJ, Abdel-Mohsen M. Markers of fungal translocation are elevated during post-acute sequelae of SARS-CoV-2 and induce NF-κB signaling. JCI Insight. 2022;7 doi: 10.1172/jci.insight.160989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Venzon M, Bernard-Raichon L, Klein J, Axelrad JE, Hussey GA, Sullivan AP, Casanovas-Massana A, Noval MG, Valero-Jimenez AM, Gago J, Wilder E Yale IMPACT Research Team, Thorpe LE, Littman DR, Dittmann M, Stapleford KA, Shopsin B, Torres VJ, Ko AI, Iwasaki A, Cadwell K, Schluter J. Gut microbiome dysbiosis during COVID-19 is associated with increased risk for bacteremia and microbial translocation. Res Sq. 2021 [Google Scholar]
- 181.Mozaffari SA, Salehi A, Mousavi E, Zaman BA, Nassaj AE, Ebrahimzadeh F, Nasiri H, Valedkarimi Z, Adili A, Asemani G, Akbari M. SARS-CoV-2-associated gut microbiome alteration; A new contributor to colorectal cancer pathogenesis. Pathol Res Pract. 2022;239:154131. doi: 10.1016/j.prp.2022.154131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Assimakopoulos SF, Eleftheriotis G, Lagadinou M, Karamouzos V, Dousdampanis P, Siakallis G, Marangos M. SARS CoV-2-Induced Viral Sepsis: The Role of Gut Barrier Dysfunction. Microorganisms. 2022;10 doi: 10.3390/microorganisms10051050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Alhumaid S, Al Mutair A, Busubaih JS, Al Dossary N, Alsuliman M, Baltyour SA, Alissa I, Al Hassar HI, Al Aithan NA, Albassri HA, AlOmran SA, ALGhazal RM, Busbaih A, Alsalem NA, Alagnam W, Alyousef MY, Alseffay AU, Al Aish HA, Aldiaram A, Al Eissa HA, Alhumaid MA, Bukhamseen AN, Al Mutared KM, Aljwisim AH, Twibah AM, AlSaeed MM, Alkhalaf HA, ALShakhs FM, Koritala T, Al-Tawfiq JA, Dhama K, Rabaan AA, Al-Omari A. Colorectal cancer in patients with SARS-CoV-2: a systematic review and meta-analysis. Infect Agent Cancer. 2022;17:49. doi: 10.1186/s13027-022-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]