Key Words: circ_0002538, circRNA sequencing, competing endogenous RNAs, demyelination, diabetic peripheral neuropathy, miR-138-5, myelination, plasmolipin, protein profiling, Schwann cells
Abstract
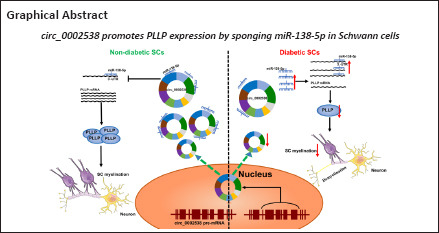
Circular RNAs (circRNAs) play a vital role in diabetic peripheral neuropathy. However, their expression and function in Schwann cells in individuals with diabetic peripheral neuropathy remain poorly understood. Here, we performed protein profiling and circRNA sequencing of sural nerves in patients with diabetic peripheral neuropathy and controls. Protein profiling revealed 265 differentially expressed proteins in the diabetic peripheral neuropathy group. Gene Ontology indicated that differentially expressed proteins were mainly enriched in myelination and mitochondrial oxidative phosphorylation. A real-time polymerase chain reaction assay performed to validate the circRNA sequencing results yielded 11 differentially expressed circRNAs. circ_0002538 was markedly downregulated in patients with diabetic peripheral neuropathy. Further in vitro experiments showed that overexpression of circ_0002538 promoted the migration of Schwann cells by upregulating plasmolipin (PLLP) expression. Moreover, overexpression of circ_0002538 in the sciatic nerve in a streptozotocin-induced mouse model of diabetic peripheral neuropathy alleviated demyelination and improved sciatic nerve function. The results of a mechanistic experiment showed that circ_0002538 promotes PLLP expression by sponging miR-138-5p, while a lack of circ_0002538 led to a PLLP deficiency that further suppressed Schwann cell migration. These findings suggest that the circ_0002538/miR-138-5p/PLLP axis can promote the migration of Schwann cells in diabetic peripheral neuropathy patients, improving myelin sheath structure and nerve function. Thus, this axis is a potential target for therapeutic treatment of diabetic peripheral neuropathy.
Introduction
Diabetes mellitus is a major global health concern affecting more than 9% of the global population, and this is expected to increase over time (Feldman et al., 2019a). The most common complication of diabetes mellitus is diabetic peripheral neuropathy (DPN), which affects approximately 50% of people with diabetes during their lifetime (Pop-Busui et al., 2017). DPN is the key initiating factor of diabetic foot conditions that can lead to nontraumatic lower limb amputation, which can seriously reduce the quality of life and patient life expectancy (Feldman et al., 2019a; Selvarajah et al., 2019). DPN is characterized by pain, paresthesia, and loss of sensation, and is associated with axon atrophy, demyelination, weakened regenerative potential, and the loss of peripheral nerve fibers (Farmer et al., 2012). Although several therapeutic approaches have been introduced in clinical practice, the current DPN treatment has only been found to relieve some symptoms with limited effects (Singh et al., 2014). Current studies have found that the occurrence and development of DPN are largely caused by hyperglycemia, insulin deficiency, and dyslipidemia. However, the molecular mechanisms that lead to demyelination and neurological dysfunction remain unclear. Therefore, clarification of the molecular mechanism that promotes DPN initiation and development has important clinical significance and may lead to more effective treatments for DPN.
Circular RNAs (circRNAs) are a recently characterized type of noncoding RNA. They play a key role in the occurrence and development of many diseases and are highly evolutionarily conserved, stable, and tissue-specific (Zhang et al., 2019; Shi et al., 2020). circRNAs are involved in the modification of transcription or posttranscriptional gene expression, and their mode of action includes protein binding, translation, and microRNA (miRNA) sponges (Wang et al., 2020a). circRNA sequencing in spinal cord tissue and dorsal root ganglia of DPN mice revealed 135 and 15 differentially expressed circRNAs (Zhang et al., 2020; He et al., 2021), respectively, which were associated with the occurrence and development of neuronal abnormalities. However, the characteristics and functions of circRNAs in Schwann cells (SCs) in DPN remain unclear.
In the present study, we used circRNA sequencing and protein profiling analyses of nerve tissues from humans with or without DPN to explore the onset and developmental mechanisms of DPN. circ_0002538 is a circRNA derived from Kelch-like family member 8 (KLHL8) with downregulated expression in circRNA sequencing of nerves from patients with DPN, whose function has not previously been characterized. Moreover, we investigated the role of circ_0002538 in the development of DPN in vitro and in vivo.
Methods
Ethics statement
This study was approved by the Ethics Committee of Tongji Medical College, Huazhong University of Science and Technology (approval No. IEC 2021-S085, approved on March 31, 2021), and informed consent was obtained from each patient. All animal study protocols were approved by the Animal Care Committee of Huazhong University of Science and Technology (No. 2020-S2665, approved on December 1, 2020). The timeline of the experiment was shown in Figure 1.
Figure 1.
Schematic diagram illustrating the timeline of the experiment.
DPN: Diabetic peripheral neuropathy; STZ: streptozotocin.
Patient tissue specimens
Sural nerve tissues and skin tissues were collected from 29 patients who underwent lower limb amputation at the Union Hospital and Liyuan Hospital of Huazhong University of Science and Technology from 2014 to 2020. The DPN diagnoses were based on a history of diabetes, typical symptoms, abnormal nerve conduction, and the exclusion of neuropathy with causes other than diabetes (Pop-Busui et al., 2017; Feldman et al., 2019b). For diabetic patients without nerve conduction data, we confirmed the diagnosis of DPN by performing a skin biopsy to assess intraepidermal nerve fiber density and utilizing transmission electron microscopy (HT7700, Hitachi, Hitachi, Japan) to confirm neuropathy in the peripheral nerves (Holland et al., 1997). Individuals diagnosed with the following diseases were excluded from the study: neuropathic deficits caused by other diseases, severe peripheral vascular disease, a history of major amputation, other serious chronic medical diseases, or alcohol and drug abuse.
Under a microscope, the epineurium of the sural nerve tissues in the distal calf was stripped, and the nerve bundles were drawn out and immediately snap-frozen in liquid nitrogen for further research. Skin tissues 10 cm above the lateral malleolus were collected for immunofluorescence staining of protein gene product 9.5. The intraepidermal nerve fiber density was calculated according to a previously described method (Vlcková-Moravcová et al., 2008).
Protein profiling analysis
Total proteins were extracted from three pairs of sural nerves from the patients with DPN and individuals without DPN using a protein lysis solution (4% sodium dodecyl sulfate, 100 mM Tris HCl, pH 7.6). We then performed proteomic profiling using the tandem mass tag labeling system (Thermo Fisher Scientific, Waltham, MA, USA). We used a Q Exactive Plus high-resolution mass spectrometer (Thermo Fisher Scientific) to perform tandem mass tag quantitative proteomic analysis, and the software programs Mascot 2.6 (Matrix Science, Boston, MA, USA) and Proteome Discoverer 2.1 (Thermo Fisher Scientific) for library identification and quantitative analysis, respectively (false discovery rate < 0.01).
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analysis
The differentially expressed proteins or mRNAs were further analyzed via Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis for functional prediction. We used GO analysis to annotate the cell components and biological processes based on the GO resource (http://www.geneontology.org), and pathway analysis to explore the enrichment of different pathways based on the KEGG database (http://www.genome.jp/kegg). The protein–protein interaction network analysis was based on the STRING database (https://string-db.org) and visualized using Cytoscape 3.7.2 (Shannon et al., 2003).
circRNA sequencing analysis
The sequencing libraries were constructed as described in a previous report (Lu et al., 2020). Briefly, the total RNA of the aforementioned three pairs of peripheral nerves was prepared using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The RNA integrity number was evaluated using the Agilent 2200 TapeStation (Agilent Technologies, Eugene, OR, USA), and all RNA samples with an RNA integrity number above 7.0 were subjected to further circRNA sequencing analysis. Before constructing the circRNA sequencing libraries, we used the Epicentre Ribo-Zero rRNA Removal Kit (Illumina, San Diego, CA, USA) to remove ribosomal RNA from the RNA samples, and incubated 40 U RNase R (Epicenter, Madison, WI, USA) with the total RNA at 37°C for 3 hours to remove linear RNA. The libraries were sequenced using the HiSeq-3000 sequencing platform, and we examined the differentially expressed circRNA between the sural nerves from patients with DPN and tissues from individuals without DPN using DESeq2 software (v 2.11.40.2; Bioconductor, Inc.).
Cell culture and treatments
We isolated the primary SCs from human sural nerves (three donors were randomly selected from each group), as previously described, to examine the impaired function of SCs from DPN patients (Wang et al., 2020b). Briefly, the sural nerves were cut into 5-mm-long sections after the epineurium had been stripped and predegenerated in SC culture medium for 10 days. Next, the nerve segments were cut into 2-mm3 pieces and transferred to a mixture containing Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific), 10% fetal calf serum, 0.125% type IV collagenase (Sigma-Aldrich, St. Louis, MO, USA), 1.25 U/mL dispase II (Solaribo, Beijing, China), and 1% penicillin-streptomycin to digest for 18–20 hours. The cells were cultured in SC medium (ScienCell, Carlsbad, CA, USA). The SCs used in the other experiments were purchased from ScienCell Research Laboratories and cultured in SC medium containing 5% fetal calf serum. We added oxidized low-density lipoprotein (ox-LDL, BioVision, Exton, PA, USA) to the culture medium to mimic diabetic conditions. After growing to confluent or subconfluent cell layers, the SCs were cultured for another 6 days to examine plasmolipin (PLLP) expression as previously described (Gillen et al., 1996). SCs were identified via immunofluorescence staining with S100 calcium binding protein B and glial fibrillary acidic protein. HEK293 cells (ACC305, DMSZ, Braunschweig, Lower Saxony, Germany, RRID: CVCL_0045) were cultured in high glucose Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum and 1% penicillin/streptomycin. The cells were cultured at 37°C in a humidified atmosphere containing 5% CO2.
Real-time polymerase chain reaction
We extracted the total RNA from the sural nerves and cells using TRIzol reagent (TaKaRa, Kyoto, Japan). The genomic DNA was isolated using a TIANamp Genomic DNA Kit (TianGen Biotech, Beijing, China) according to the manufacturer’s instructions. The RNA samples were then reverse transcribed into complementary DNA (cDNA) using the PrimeScript™ RT Reagent (TaKaRa, RR036A). We performed real-time polymerase chain reactions (RT-PCRs) using a 7500 Real-time PCR System (Applied Biosystems, Carlsbad, CA, USA) with the Universal SYBR Green Master Mix (4913914001; Roche, Shanghai, China). β-Actin was used as an internal control. The RT-PCR protocol was as follows: one cycle of 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Gene expression was quantified using the 2–ΔΔCt method (Livak and Schmittgen, 2001). For circRNA, the total RNA was reverse transcribed to cDNA using the PrimeScript™ RT reagent kit (TaKaRa, RR037A). We used convergent and divergent primers to detect the expression of linear RNA and circRNA transcripts. The primers are shown in Additional Table 1.
Additional Table 1.
Primers used in this study
| Name | Sequence (5’-3’) |
|---|---|
| circRNA | |
| hsa_circ_0004001 | Forward: TGACGAACATCACAGTACATTGG |
| Reverse: AAGGTGCGTTCATCACGTTTT | |
| hsa_circ_0001847 | Forward: ACAATCAGATGGCACCAGGGA |
| Reverse: TCCAAGCCCCTTTGAGTCCAT | |
| hsa_circ_0000137 | Forward: TTGAGGCTGTTGTTCAGAGTGT |
| Reverse: ACAGAGTCATCCCCAGAAGCA | |
| hsa_circ_0005019 | Forward: CTGGAGCCTGGTGAGAACTT |
| Reverse: CAGATGTGTCAGAACCCTCACT | |
| hsa_circ_0000711 | Forward: AGGTAGCCGAGGGGCAGTAA |
| Reverse: GTGGTAAGCAAAGTGGTGTGGT | |
| hsa_circ_0001897 | Forward: GCTGGCCTTGGGAGGTTATTTA |
| Reverse: GCCCACTGTCATCCAAGAAGAA | |
| hsa_circ_0004896 | Forward: CTAACCACCGCCGAGAACGA |
| Reverse: TGTCACCTGGGCGGAAACTC | |
| hsa_circ_0006156 | Forward: AAGGGCCATAGTGGTGGAAGT |
| Reverse: GCTTGGGGGATAACACTCAGGA | |
| hsa_circ_0000471 | Forward: CACACAAAGACCTCCTCCTCC |
| Reverse: GCTTGTTTTGCTGTACCCATCT | |
| hsa_circ_0001647 | Forward: GTCTGAGTTTACCTGAAAGGGATA |
| Reverse: ATGCCTGTACTTCATCACCTG | |
| hsa_circ_0087960 | Forward: GTAGTTCTGGGGCGTGTTCA |
| Reverse: TAGGTGGATGGGGAGCTTCA | |
| hsa_circ_0004374 | Forward: ACACCAGCATACTTTGCCTCA |
| Reverse: CACATTTAGGACAGCGCAGC | |
| hsa_circ_0020433 | Forward: ACAAAGTCATCGCTGCCAAAG |
| Reverse: CGGCTGAAAGGGAATGAAATGC | |
| hsa_circ_0024604 | Forward: AAAAGGCAACAACAGCACCAGC |
| Reverse: CAAAACCCACTCAACTGCCATTGT | |
| hsa_circ_0005615 | Forward: ACCCTTTACCTGGAGCAAACCA |
| Reverse: TTTGGAGCTGAAACGATGGTGAC | |
| hsa_circ_0040823 | Forward: ATCGGAGAAGACGGACAGGT |
| Reverse: AGTCGGATTCTGTGATGCCA | |
| hsa_circ_0007715 | Forward: ACGGAGGCTCCAGAGACTACTA |
| Reverse: TTGTCTGACGACTTGCCTGC | |
| hsa_circ_0003781 | Forward: TGAGTACGACCCCGAGGACA |
| Reverse: CTCATCGGTGGCAGCGTAGT | |
| hsa_circ_0001824 | Forward: AAGTGACGGGATCCGGAAAG |
| Reverse: TCAGCAGCCAGTTTTTCAATGT | |
| hsa_circ_0002882 | Forward: GGGAGAGTTTGGAGCTGTGAT |
| Reverse: TCCTTCAGCTCTTCACTGATGC | |
| hsa_circ_0001819 | Forward: CCTGGTAGGACAAGCGACTCTC |
| Reverse: GCAGCATGATTTGGTCCCAC | |
| hsa_circ_0008394 | Forward: TGAACACTAGTCTGAATGTATACCG |
| Reverse: ACGAATGAAGCCTCGTGTGG | |
| hsa_circ_0006535 | Forward: CATGCTGAGCTTTGCCAGAGAC |
| Reverse: GCAATCTCCTGTTGGCTGGC | |
| hsa_circ_0002538 | Forward: AAAAGGCAACAACAGCACCAGC |
| Reverse: CAAAACCCACTCAACTGCCATTGT | |
| hsa_circ_0002538 convergent | Forward: ACCTTCTGCCTTCTCTCTACCCT |
| Reverse: GCTGTTGTTGCCTTTTCCCCTT | |
| mRNA (5’-3’) | |
| SERINC5 | Forward: GGAGGCTTGGTTTTGATGGCA |
| Reverse: CCGAGTGTGGCTGTCGATTTT | |
| GJC3 | Forward: TTGTGCTTCTGGGTTTGGGGA |
| Reverse: TGGGAGGCTATCGGTTGCTTT | |
| PLP1 | Forward: CATCACCTATGCCCTGACCGT |
| Reverse: AGGCAATAGACTGGCAGGTGG | |
| PRX | Forward: GGTGGCCAAGCTGAACATCCA |
| Reverse: AGGAGAACTCGACGTCAACAGG | |
| MPZ | Forward: AGAGGAGGCTCAGTGCTATGG |
| Reverse: CAGCTTTGGTGCTTCTGCTGT | |
| KLHL8 | Forward: CGTGGAGGAGTTGGCTCTGTT |
| Reverse: CCTGCTCTTCGCTGACCCATT | |
| GAPDH | Forward: ATCCACAGTCTTCTGGGTGGC |
| Reverse: TCCTGGAACAGCAAAACAAGGC | |
| PLLP | Forward: CTTTAACATCAGCGCCACCGTT |
| Reverse: ACCAAACACGCAAAGAACGAGG | |
| P-Actin | Forward: CAGCCTTCCTTCCTGGGCAT |
| Reverse: GGGCAGTGATCTCCTTCTGCAT | |
| P-Actin-divergent | Forward: AAATCGTGCGTGACATTAAGGAGA |
| Reverse: CATACCCCTCGTAGATGGGCA | |
| U3 | Forward: TGTAGAGCACCGAAAACCACG |
| Reverse: CAGCCAAGCAACGCCAGA | |
| miRNA (5’-3’) | |
| hsa-miR-138-5p | AGCTGGTGTTGTGAATCAGG |
| miR-3714 | GAAGGCAGCAGTGCTCCCCTGT |
| U6 | Forward: CTCGCTTCGGCAGCACA |
| Reverse: AACGCTTCACGAATTTGCGT | |
| miR-138-5p stem-loop | RT: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCGGCCT |
| Forward: GCGAGCTGGTGTTGTGAATC | |
| Reverse: AGTGCAGGGTCCGAGGTATT | |
| U6 stem-loop RT | RT: GTCGTATCGACTGCAGGGTCCGAGGTATTCGCAGTCGATACGACAAAATATG |
| Forward: AGCACATATACTAAAATTGGAACGAT | |
| Reverse: ACTGCAGGGTCCGAGGTATT |
circRNAs: circular RNAs; SERINC5: serine incorporator 5; PLLP: plasmolipin; GJC3: gap junction protein gamma 3; PLP1: proteolipidprotein 1; PRX: periaxin; MPZ: myelin protein zero; KLHL8: Kelch-like family member 8; GAPDH: glyceraldehyde-3-phosphatedehydrogenase; U3: small nucleolar U3 RNA.
Sanger sequencing
We conducted Sanger sequencing to verify the back-splicing position of circ_0002538. The total RNA was extracted from the SCs and reverse transcribed into cDNA. circ_0002538 was amplified with divergent primers and 2× Taq Master Mix (Vazyme, Nanjing, Jiangsu, China) using qPCR. The qPCR protocol was one cycle of 95°C for 5 minutes followed by 34 cycles of 95°C for 30 seconds, 52°C for 30 seconds, and 72°C for 30 seconds. Then, the base sequences of the products were determined using Sanger sequencing and compared with the data in circBase (http://circrna.org/).
Nuclear and cytoplasmic separation assay
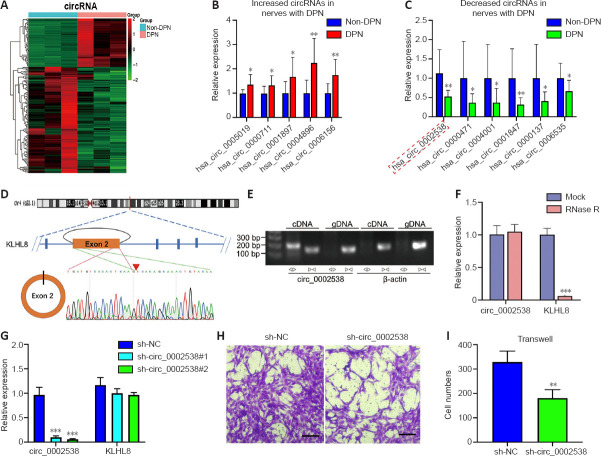
To detect the cellular localization of circRNAs, we extracted RNA from nuclear and cytoplasmic fractions using a cytoplasmic and nuclear RNA isolation kit (Norgen Biotek, Ontario, Canada) according to the manufacturer’s protocol. The relative expression levels of circ_0002538 in the nucleus and cytoplasm were detected via RT-PCR. We used GAPDH and U6 small nuclear RNA as internal controls.
Digestion with RNase R
For RNase R digestion, 10 μg of total RNA was incubated with 2 U/μg RNase R (BioVision, Milpitas, CA, USA) at 37°C for 30 minutes. RNAs treated with the same process without RNase R were the mock group. The expression levels of KLHL8 and circ_0002538 were determined via RT-PCR.
Plasmid construction and stable transfection
circ_0002538 cDNA was synthesized by Tsingke Biological Technology (Wuhan, China) and cloned into the GV689 vector (Shanghai GeneChem Co., Ltd., Shanghai, China) to construct overexpression plasmids. Short hairpin RNA (shRNA) for circ_0002538 was designed using the CircInteractome tool and cloned into the GV493 vector (Shanghai GeneChem Co., Ltd.) to construct silencing plasmids. The plasmids for the overexpression and knockdown of PLLP were designed and synthesized by Shanghai Gene Chemical Co., Ltd. Then, the constructed plasmids were packaged into lentivirals (LVs) by Shanghai Gene Chemical Co., Ltd. and cell transfection was performed according to the manufacturer’s instructions. The transfected cells were incubated with 2 μg/mL of puromycin (BIOFOX, Nantong, China) for 5 days, and the surviving cells were used as stable transfectants.
Oligonucleotide transfection
miRNA mimics, miRNA inhibitors, and corresponding negative control oligonucleotides were synthesized by RiboBio (Guangzhou, China). The sequences used are listed in Additional Table 2. Transfection was carried out using a PECTTM CP Transfection kit (RiboBio) with a final concentration of 50 nM for miRNA mimics and 100 nM for miRNA inhibitors, according to the manufacturer’s protocol.
Additional Table 2.
Nucleic acid sequences used in this study
| Nucleic acid | Sequences (5’-3’) |
|---|---|
| sh NC | Sense: TTCTCCGAACGTGTCACGT |
| Antisense: ACGTGACACGTTCGGAGAA | |
| sh1 circ_0002538 | Sense: GTCACACTCAAGTCACAGCAA |
| Antisense: TTGCTGTGACTTGAGTGTGAC | |
| sh2 circ_0002538 | Sense: ACTCAAGTCACAGCAAACTGT |
| Antisense: ACAGTTTGCTGTGACTTGAGT | |
| biotin-miR NC | TTTGTACTACACAAAAGTACTG |
| biotin-miR-138-5p mimic | AGCTGGTGTTGTGAATCAGGCCG |
| biotin-circ_0002538 NC | GAACTCTGTGATGTCACACTCAAGTCACAGCAAACTGTACAATGGCAG |
| biotin-circ_0002538 | CTGCCATTGTACAGTTTGCTGTGACTTGAGTGTGACATCACAGAGTTC |
| mimics NC | Sense: UUUGUACUACACAAAAGUACUG |
| Antisense: AAACAUGAUGUGUUUUCAUGAC | |
| miR-138-5p mimics | Sense: AGCUGGUGUUGUGAAUCAGGCCG |
| Antisense: UCGACCACAACACUUAGUCCGGC | |
| Inhibitor miR-NC | CAGUACUUUUGUGUAGUACAAA |
| Inhibitor miR-138-5p | AGCTGGTGTTGTGAATCAGG |
NC: Normal control; sh circ_0002538: short hairpin RNA for circ_0002538; sh NC: normal control for short hairpin RNA.
Transwell assay
SC migration was determined using a Transwell chamber (8-μm pore size, Corning, Corning city, NY, USA) according to the manufacturer’s protocol. Approximately 2 × 104 cells suspended in 200 μL of serum-free medium were added to the upper chamber, and a total of 650 μL of Schwann medium containing 5% fetal calf serum was added to the lower chamber as a chemical attractant. After a 24-hour incubation period, we evaluated cell migration by counting the number of migrated cells on the lower surface of the chamber in at least five random fields.
Western blot analysis
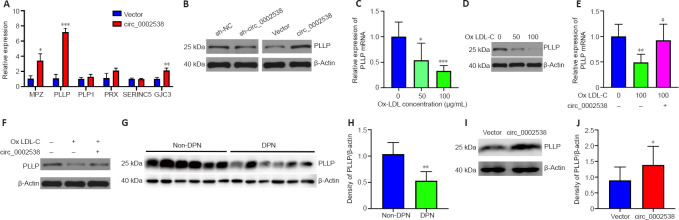
We tested the expression levels of PLLP protein in SCs and neural tissues via a western blot analysis. The protein was extracted using a radioimmunoprecipitation assay lysis buffer, supplemented with 1% protease inhibitor. Equal amounts of protein (30 μg) were separated in a 10% sodium dodecyl sulfate-polyacrylamide gel and then transferred to polyvinylidene fluoride membranes (Millipore, Darmstadt, Germany). The membranes were blocked in 5% (w/v) bovine serum albumin (Aladdin, Shanghai, China) before incubation with the primary antibodies at 4°C overnight. Then, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:5000, Aspen Biotechnology Co., Ltd., Wuhan, China, Cat# AS1107) for 1 hour at room temperature and visualized using a BioSpectrum Imaging System (UVP, Upland, CA, USA) with the Immobilon ECL substrate kit (Millipore, Darmstadt, Germany). We used primary antibodies specific to PLLP (rabbit, 1:700, Cusabio, Houston, TX, USA, Cat# CSB-PA896501LA01HU). All tests were repeated three times, and the typical images were provided.
RNA pulldown assay
To detect the combination of circRNAs and miRNAs, we performed RNA pulldown assays with biotinylated probes according to the manufacturer’s protocol (MCE, Shanghai, China, Cat# HY-K0208). In brief, the biotinylated probe or nonsense control probe (RiboBio) was incubated with M-280 streptavidin magnetic beads (MCE) at room temperature for 2 hours to generate probe-coated beads. Approximately 1 × 107 SCs were crosslinked with 1% paraformaldehyde and then neutralized with 1.25 M glycine. Next, these cells were harvested, lysed, and incubated with probe-coated magnetic beads at 4°C overnight. After being washed, the RNA complexes bound to the beads were eluted and extracted using an Rneasy Mini Kit (Qiagen, Hilden, Germany). Then, the abundance of circRNA or miRNA was evaluated via RT-PCR.
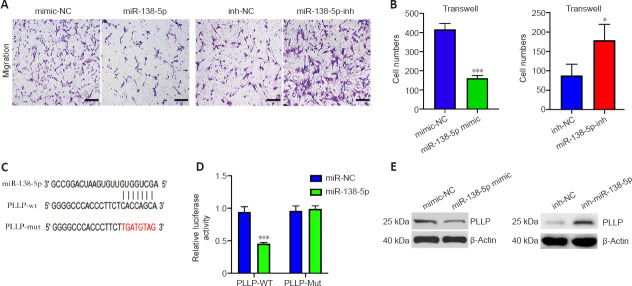
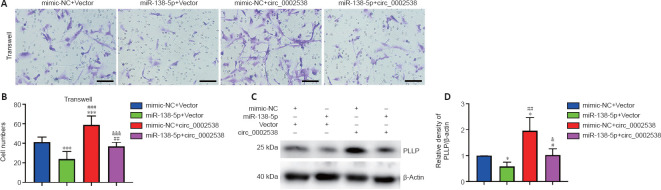
Dual-luciferase reporter assay
We predicted the binding sites of miR-138-5p targeting circ_0002538 and PLLP using RNAhybrid (Rehmsmeier et al., 2004) and TargetScan (McGeary et al., 2019), respectively. The wild-type or mut-circ_0002538 fragment was cloned into the downstream of the luciferase reporter gene of the pMIR-report vector (Promega, Madison, WI, USA), while wild-type or mut-PLLP fragment was inserted into the downstream of the hRluc (Renilla) reporter gene of the psi-check2 vector (Promega). The corresponding plasmid and miRNA mimic were cotransfected into HEK293T cells (5 × 104) seeded in a 12-well plate using Lipofectamine 2000 (Thermo Fisher Scientific). The firefly and Renilla luciferase activity of the cells was quantified using a Dual Luciferase Reporter System Kit (E1910, Promega) according to the manufacturer’s instructions.
Prediction of miRNAs targeting circ_0002538 or PLLP
We made predictions regarding the miRNAs that target circ_0002538 or PLLP to ascertain the connection between circ_0002538 and PLLP. The prediction process was conducted by RiboBio (Guangzhou, China). For PLLP, miRNAs predicted by at least three databases (miRDB, miRTarBase, miRWalk, and TargetScan) were considered candidates (Dweep et al., 2011; McGeary et al., 2019; Chen and Wang, 2020; Huang et al., 2020). For circ_0002538, miRNAs predicted by at least two databases (RNAhybrid, miRanda, and TargetScan) were considered candidates (Rehmsmeier et al., 2004; McGeary et al., 2019). We used a Venn diagram to find the common miRNAs (Hulsen et al., 2008).
Induction of diabetes
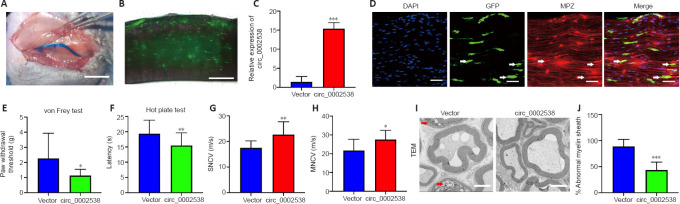
Due to the high similarity to human and the stability in genes, mice were used to explore circ_0002538 function in vivo (Perlman, 2016). Sex is one factor influencing variations in diabetes induction. As estrogen interferes with streptozotocin (STZ) action, female animals are less sensitive to the diabetogenic action of STZ than male animals. Further, male mice are more commonly used in neuroscience research (Beery and Zucker, 2011). As a result, we chose to use male animals for our study. Compared with other age groups, rodents aged 8–9 weeks show maximal induction of diabetes (Goyal et al., 2016). Thus, we used rodents in this age group. The induction of diabetes was conducted as previously described (Wang et al., 2020b). Briefly, a total of 60 male (8-week-old) C57BL/6J mice (specific-pathogen-free level, SiPeiFu, Beijing, China, SCXK2019-0010) were intraperitoneally injected with STZ (Sigma-Aldrich) at a dose of 50 mg/kg for 5 consecutive days. Subsequently, 45 mice had fasting blood glucose levels of 16.7 mM or higher and were thus diagnosed with diabetes (Wang et al., 2020b). Forty mice with significantly increased mechanical and thermal thresholds were diagnosed with DPN (Fan et al., 2020). We randomly selected one side of the sciatic nerve to be injected with circ_0002538 (circ_0002538 group) and injected the other side with LV-vector (vehicle group, n = 40).
Surgery and lentiviral vector injection
We injected a LV-vector into the sciatic nerve of the mice with DPN, as previously described (Tannemaat et al., 2008). Briefly, after exposure and isolation of the sciatic nerve, 2.5 µL of lentiviral solution (6 × 106 TU LV-circ_0002538 or LV-GFP vector) was injected into the distal peroneal and tibial branches of the sciatic nerve through the epineurium using a 10-µL Hamilton syringe (Hamilton Co., Reno, NV, USA). Fast Green (Sigma-Aldrich) at a final concentration of 0.1% was added to the lentiviral solution to monitor the injection process and ensure that there was no obvious leakage. A 2.5-µL lentiviral solution containing 6 × 106 TU LV-sh-PLLP or LV-vector was injected into the sciatic nerve of normal mice to determine the role of PLLP. The epineurium at the injection site was repaired with 10-0 nylon sutures under an operating microscope (Xintian Medical Instrument Co., LTD, Zhenjiang, China).
Behavioral testing and electrophysiology
Eight weeks after diabetic induction, we assessed thermal and mechanical nociceptive thresholds via double-blind trials. Before the nociceptive behavior test, the mice were acclimated to the environment for at least half an hour. Mechanical allodynia was assessed using von Frey filaments (Danmic Aesthesio, Campbell, CA, USA), as described previously (Xu et al., 2015; Pan et al., 2019). A brisk withdrawal or flinching of the paw was considered a positive response. The inter-test interval between the two sides of the plantar hind paw was more than 15 minutes, and the 50% force withdrawal threshold was determined for the plantar hind paws using the “up-and-down” method (Chaplan et al., 1994). The thermal nociceptive threshold was assessed using the hot plate test (Masocha et al., 2016). A mouse was placed in a Plexiglas cylinder on a hot plate (Model 7280, Ugo Basile, Gemonio, Italy), and the time required for the stimulus to elicit behavioral changes (such as paw licking, stomping, and withdrawal of the hindpaw) was recorded.
At 8 weeks post-surgery, we evaluated the nerve conduction velocity of the sciatic nerve as a sign of DPN. The sciatic nerve conduction velocity was measured via orthodromic recording techniques, as described previously (Ii et al., 2005; Baum et al., 2016; Wang et al., 2020b). The sensory nerve conduction velocity and motor nerve conduction velocity were calculated using an electromyograph (Nicolet, Madison, WI, USA) according to a previous method (Ii et al., 2005).
Hematoxylin and eosin staining, immunofluorescence analysis
We conducted hematoxylin and eosin (HE) staining to evaluate the intraepidermal nerve fiber density of skin samples from diabetic and non-diabetic individuals. The samples were collected and fixed in paraformaldehyde (4%) within 2 hours of amputation, then dehydrated and embedded in paraffin. Four-micron-thick slices of skin were prepared and subjected to HE (Bioyear, Wuhan, China) to examine subcutaneous nerves in the skin.
We used protein gene product 9.5 to evaluate the number of subcutaneous nerves in the skin samples. Glial fibrillary acidic protein and S100 calcium binding protein B were used to characterize primary SCs extracted from the sural nerves. We used myelin protein zero (MPZ) to locate SCs in the sciatic nerves of the DPN mice. The mice were sacrificed 8 weeks after the operation, and the bioluminescence of green fluorescent protein (GFP)-expressing cells was detected via fluorescence microscopy (Olympus, Tokyo, Japan). Then, the sciatic nerve tissues were collected for morphological analysis. For immunofluorescence analyses, we incubated primary antibodies against protein gene product 9.5 (rabbit,1:300, Proteintech, Wuhan, China, Cat# 14730-1-AP, RRID: AB_2210497), glial fibrillary acidic protein (rabbit, 1:400, Abcam, Carlsbad, CA, USA, Cat# ab68428, RRID: AB_1209224), S100 calcium binding protein B (rabbit, 1:200, Abcam, Cat# ab52642, RRID: AB_882426), and MPZ (rabbit, 1:200, Abcam, Cat# ab183868, RRID: AB_2895675) overnight at 4°C. On the second day, we incubated goat anti-rabbit secondary antibody (Fluor® 488, 1:400, Abcam, Cat# ab150077) at 37°C for 1 hour. We used 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (Biosharp, Wuhan, China, Cat# BL105A) to stain the cell nuclei. Fifteen-micrometer-thick frozen sections of nerve tissues were stained with MPZ. Images were obtained using a fluorescence microscope (Olympus, Tokyo, Japan), with at least three visual fields for each sample.
Transmission electron microscopy
The collected nerves were cut into 5-mm long sections, prefixed in 2.5% glutaraldehyde for 30 minutes, and then postfixed in 1% osmium tettroxide for 1 hour. After dehydration and embedding in epoxy resin, ultrathin sections (60 µm) were prepared and stained with uranyl acetate and lead citrate. Images were captured under a transmission electron microscope (HT7700, Hitachi), and 15 random images were captured for each sample.
Statistical analysis
According to previous methods (Charan and Kantharia, 2013), we determined a minimum sample size of 35 mice. Considering the potential for unexpected death in the experiment and the failure of the STZ-induced diabetes model, we used a sample size of 60.
The data are expressed as the mean ± standard deviation (SD), median (interquartile range (IQR)), or number (%). P values were obtained using the paired t-test, independent-samples t-test, or Fisher’s exact test (normal distribution) combined with the Mann-Whitney U test (nonnormal distribution) or one-way analysis of variance with Tukey’s post hoc test (more than two groups). P < 0.05 was considered significant, and all statistical analyses were performed using Graphpad Prism 8.0 (GraphPad Software, San Diego, CA, USA, www.graphpad.com).
Results
Characteristics of patients and confirmation of DPN
Twenty-nine patients from two tertiary teaching hospitals were recruited for the study. The median age of the DPN group was 60.0 years (IQR: 56.0–67.0 years) and that of the non-DPN group was 63.5 years (IQR: 55.75–65.0 years). The calf skin and sural nerve were intact in all patients when undergoing amputation. Detailed patient information is provided in Additional Table 3. Because some of the patients had not undergone nerve conduction studies, which is the gold standard for diagnosing DPN, we attempted to verify the diagnosis using other indicators. HE staining revealed a decreased number of subcutaneous nerves in the skin of the lateral malleolus in the DPN group (Additional Figure 1A (1.7MB, tif) and B (1.7MB, tif) ), which was confirmed by protein gene product 9.5 staining of axons (Additional Figure 1C (1.7MB, tif) and D (1.7MB, tif) ). Furthermore, the numbers of axons and intact myelin sheaths were decreased in the nerves of the DPN group, as shown by transmission electron microscopy (Additional Figure 1E (1.7MB, tif) and F (1.7MB, tif) ). We thus confirmed DPN in the collected diabetic peripheral nerves.
Additional Table 3.
Basic characteristics of patients included in the study
| Variables | Non-diabetic donators Diabetic donators | P-value | |
|---|---|---|---|
| Number | 14 | 15 | NA |
| Age (yr) | 63.5 (55.75-65.0) | 60.0 (56.0-67.0) | 0.78 |
| Female [n (%)] | 4 (29) | 4 (27) | NA |
| BMI (kg/m2) | 24.22 (23.35-26.23) | 24.36 (23.1-25.265) | 0.55 |
| SBP (mmHg) | 133.5 (123.75-140) | 138 (126-150.5) | 0.27 |
| DBP (mmHg) | 78.5 (73.5-84.25) | 82 (70.5-88) | 0.82 |
| FBG (mM) | 5.8 (5.49-6.345) | 11.3 (8.1-14.375) | <0.0001 |
| HbA1c (%) | NA | 7.2 (6.8-7.35) | NA |
| Total cholesterol (mM) | 4.165 (3.52-4.72) | 3.66 (3.14-5.32) | 0.61 |
| Triglyceride (mM) | 1.29 (1.09-1.565) | 1.39 (1.11-1.56) | 0.91 |
| Creatinine (^M) | 67.4 (47.4-76.5) | 71.8 (67.3-96.2) | 0.07 |
| BUN (mM) | 5.27 (3.56-6.27) | 5.49 (4.15-7.29) | 0.31 |
| HDL-C (mM) | 1.09 (0.765-1.16) | 0.79 (0.72-0.87) | 0.40 |
| LDL-C (mM) | 2.69 (1.96-3) | 2.56 (1.58-3.93) | 0.42 |
Data are median (IQR) or number (%), unless otherwise specified. P-values comparing patients with or without DPN were obtained by the independent-samples t-test or Fisher’s exact test. BMI: Body mass index; BUN: blood urea nitrogen; DBP: diastolic blood pressure; FBG: fasting blood glucose; HbA1c: glycated hemoglobin; HDL-C: high-density lipoprotein cholesterol; IQR: interquartile range; LDL-C: low-density lipoprotein cholesterol; NA: not applicable; SBP: systolic blood pressure.
Impaired myelination and SC migration in the peripheral nerves of the DPN group
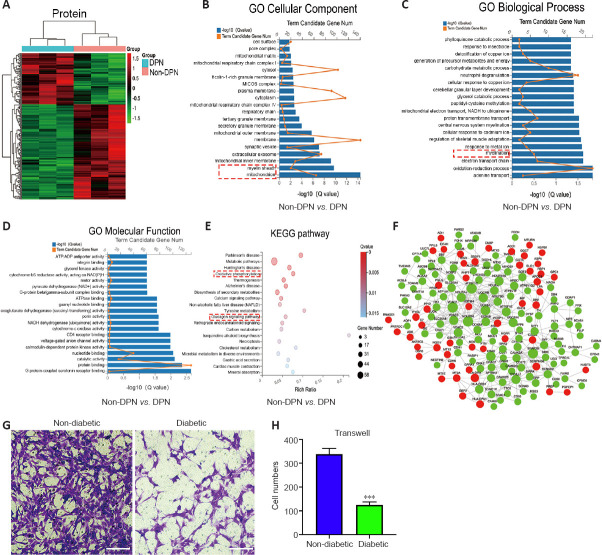
Protein profiling analyses were performed on three pairs of peripheral nerves in the DPN and non-DPN groups. A total of 5353 proteins were identified, and 265 proteins were significantly [P < 0.05, |fold change (FC)| ≥ 1.3] differentially expressed in the DPN group (Additional Table 4 (452KB, pdf) ), as shown by the hierarchical cluster analysis (Figure 2A). GO cellular component analysis indicated that the differentially expressed proteins were mainly found in the mitochondrion and myelin sheath (Figure 2B and Additional Table 5). GO biological process analysis showed that 390 terms were significantly enriched, among which myelination was potentially related to DPN (Figure 2C and Additional Table 6 (486.4KB, pdf) ). The proteins related to myelination were serine incorporator 5, PLLP, gap junction protein gamma 3, proteolipid protein 1, periaxin, and MPZ. GO molecular function analysis showed significant enrichment in G protein-coupled serotonin receptor binding and protein binding (Figure 2D and Additional Table 7 (555.4KB, pdf) ). KEGG pathway analysis revealed that 77 pathways were significantly enriched, among which oxidative phosphorylation and the glucagon signaling pathways were potentially related to DPN (Figure 2E and Additional Table 8). Figure 2F shows a protein–protein interaction network constructed according to the differentially expressed proteins and showing the interactions among these proteins. These results indicate that abnormal myelination might play an important role in the pathogenesis of DPN.
Figure 2.
Protein profiling analysis and the detection of SC function in DPN.
(A) Hierarchical clustering analyses of differentially expressed proteins in the non-DPN vs. DPN group (n = 3). (B) GO cellular component analysis of differentially expressed proteins. The red dotted box highlights the cellular components of interest. (C) GO biological process analysis of differentially expressed proteins. The red dotted box highlights the biological processes of interest. (D) GO molecular function analysis of differentially expressed proteins. (E) KEGG pathway analysis of differentially expressed proteins. The red dotted box highlights the pathways of interest. (F) The PPI network based on the STRING database showed the interactions between differentially expressed proteins. The green and red nodes represent proteins with decreased and increased expression, respectively. (G, H) Transwell assays indicated that the migrating number of SCs in the diabetic group was lower compared with that in the non-diabetic group. Scale bars: 100 μm. All bar graphs represent the average of three independent replicates, and the error bars are the SD. ***P < 0.001, vs. non-diabetic (independent-sample t-test). DPN: Diabetic peripheral neuropathy; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; PPI: protein-protein interaction.
Additional Table 5.
GO cellular component analysis of differentially expressed proteins
| Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Term | Candidat | Term | Total | |||||||
| Candidate e Gene | Gene | Gene | ||||||||
| GO C Term ID | GO_C Term Desc | GO C Term Level1 | GO C Term Level2 Gene | Num | Num | Num | Num | Rich Ratio | P value | Q value |
| GO:0005739 | mitochondrion | cellular_component | organelle | 64 | 255 | 1926 | 25187 | 0.033229 | 9.47E-18 | 3.36E-15 |
| GO:0043209 | myelin sheath | cellular_component | cell part | 20 | 255 | 246 | 25187 | 0.081301 | 9.73E-13 | 1.73E-10 |
| GO:0005743 | mitochondrial inner membrane | cellular_component | organelle | 28 | 255 | 565 | 25187 | 0.049558 | 5.14E-12 | 6.08E-10 |
| GO:0070062 | extracellular exosome | cellular_component | organelle | 73 | 255 | 3513 | 25187 | 0.02078 | 7.01E-10 | 6.22E-08 |
| GO:0008021 | synaptic vesicle | cellular_component | organelle | 14 | 255 | 162 | 25187 | 0.08642 | 1.21E-09 | 8.61E-08 |
| GO:0016020 | membrane | cellular_component | membrane | 141 | 255 | 9539 | 25187 | 0.014781 | 1.08E-08 | 6.37E-07 |
| GO:0005741 | mitochondrial outer membrane | cellular_component | organelle | 15 | 255 | 244 | 25187 | 0.061475 | 3.24E-08 | 1.64E-06 |
| GO:0030667 | secretory granule membrane | cellular_component | organelle | 10 | 255 | 144 | 25187 | 0.069444 | 2.22E-06 | 9.85E-05 |
| GO:0070821 | tertiary granule membrane | cellular_component | organelle | 8 | 255 | 97 | 25187 | 0.082474 | 6.57E-06 | 2.59E-04 |
| GO:0070469 | respiratory chain | cellular_component | membrane | 6 | 255 | 61 | 25187 | 0.098361 | 3.54E-05 | 0.001256 |
| GO:0005751 | mitochondrial respiratory chain | cellular_component | organelle | 4 | 255 | 22 | 25187 | 0.181818 | 6.50E-05 | 0.002099 |
| GO:0005737 | cytoplasm | cellular_component | cell part | 115 | 255 | 8450 | 25187 | 0.013609 | 7.81E-05 | 0.002309 |
| GO:0005886 | plasma membrane | cellular_component | membrane | 91 | 255 | 6315 | 25187 | 0.01441 | 9.80E-05 | 0.002676 |
| GO:0061617 | MICOS complex | cellular_component | organelle | 255 | 10 | 25187 | 0.3 | 1.17E-04 | 0.002961 | |
| GO:0101003 | ficolin-1-rich granule membrane | cellular_component | organelle | 6 | 255 | 78 | 25187 | 0.076923 | 1.42E-04 | 0.003356 |
| GO:0005829 | cytosol | cellular_component | cell part | 102 | 255 | 7429 | 25187 | 0.01373 | 2.03E-04 | 0.004498 |
| GO:0005747 | mitochondrial respiratory chain | cellular_component | organelle | 5 | 255 | 60 | 25187 | 0.083333 | 3.55E-04 | 0.007407 |
| GO:0005759 | mitochondrial matrix enclosed lumen | cellular_component | membrane- | 14 | 255 | 481 | 25187 | 0.029106 | 4.27E-04 | 0.008414 |
| GO:0046930 | pore complex | cellular_component | membrane | 3 | 255 | 18 | 25187 | 0.166667 | 7.48E-04 | 0.013972 |
| GO:0009986 | cell surface | cellular_component | cell | 20 255 | 900 | 25187 | 0.022222 | 9.01E-04 | 0.015988 | |
| GO:0000275 | mitochondrial proton- transporting ATP synthase complex, catalytic core F(1) | cellular_component | organelle | 2 | 255 | 5 | 25187 | 0.4 | 0.001001 | 0.016916 |
| GO:0048471 | perinuclear region of cytoplasm | cellular_component | cell part | 20 | 255 | 914 | 25187 | 0.021882 | 0.001085 | 0.01751 |
| GO:0005768 | endosome | cellular_component | organelle | 17 | 255 | 724 | 25187 | 0.023481 | 0.001211 | 0.017915 |
| GO:0045202 | synapse | cellular_component | synapse | 17 | 255 | 723 | 25187 | 0.023513 | 0.001193 | 0.017915 |
| GO:0001401 | mitochondrial sorting and assembly machinery complex | cellular_component | organelle | 2 | 255 | 6 | 25187 | 0.333333 | 0.001491 | 0.019776 |
| GO:0031225 | anchored component of membrane | cellular_component | membrane | 7 | 255 | 165 | 25187 | 0.042424 | 0.001504 | 0.019776 |
| GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1) | cellular_component | membrane | 2 | 255 | 6 | 25187 | 0.333333 | 0.001491 | 0.019776 |
| GO:0030054 | cell junction | cellular_component | cell junction | 21 | 255 | 1016 | 25187 | 0.020669 | 0.001652 | 0.020218 |
| GO:0030666 | endocytic vesicle membrane | cellular_component | organelle | 6 | 255 | 123 | 25187 | 0.04878 | 0.001609 | 0.020218 |
| GO:0042584 | chromaffin granule membrane | cellular_component | organelle | 2 | 255 | 7 | 25187 | 0.285714 | 0.002073 | 0.023744 |
| GO:0045254 | pyruvate dehydrogenase complex | cellular_component | cell part | 2 | 255 | 7 | 25187 | 0.285714 | 0.002073 | 0.023744 |
| GO:0044306 | neuron projection terminus | cellular_component | cell part | 3 | 255 | 26 | 25187 | 0.115385 | 0.002245 | 0.024901 |
| GO:0030315 | T-tubule | cellular_component | cell part | 4 | 255 | 55 | 25187 | 0.072727 | 0.002334 | 0.025113 |
| GO:0043190 | ATP-binding cassette (ABC) transporter complex | cellular_component | membrane | 2 | 255 | 8 | 25187 | 0.25 | 0.002746 | 0.027854 |
| GO:1990246 | uniplex complex | cellular_component | organelle | 2 | 255 | 8 | 25187 | 0.25 | 0.002746 | 0.027854 |
| GO:0030424 | axon | cellular_component | cell part | 12 | 255 | 466 | 25187 | 0.025751 | 0.002996 | 0.028964 |
| GO:0042645 | mitochondrial nucleoid enclosed lumen | cellular_component | membrane- | 4 | 255 | 59 | 25187 | 0.067797 | 0.003019 | 0.028964 |
| GO:0030658 | transport vesicle membrane | cellular_component | organelle | 5 | 255 | 98 | 25187 | 0.05102 | 0.003233 | 0.030206 |
| GO:0042734 | presynaptic membrane | cellular_component | membrane | 5 | 255 | 100 | 25187 | 0.05 | 0.003527 | 0.032102 |
| GO:0008091 | spectrin | cellular_component | organelle | 2 | 255 | 10 | 25187 | 0.2 | 0.004355 | 0.037706 |
| GO:0032592 | integral component of mitochondrial membrane | cellular_component | organelle | 2 | 255 | 10 | 25187 | 0.2 | 0.004355 | 0.037706 |
| GO:0043005 | neuron projection | cellular_component | cell part | 2 | 12 255 | 496 | 25187 | 0.024194 | 0.004908 | 0.041487 |
| GO:0005921 | gap junction | cellular_component | cell junction | 3 | 255 | 35 | 25187 | 0.085714 | 0.005285 | 0.042658 |
| GO:0014731 | spectrin-associated cytoskeleton | cellular_component | organelle | 2 | 255 | 11 | 25187 | 0.181818 | 0.005287 | 0.042658 |
| GO:0005834 | heterotrimeric G-protein complex | cellular_component | membrane | 3 | 255 | 36 | 25187 | 0.083333 | 0.005723 | 0.045144 |
| GO:0043204 | perikaryon | cellular_component | cell part | 6 | 255 | 160 | 25187 | 0.0375 | 0.005897 | 0.045509 |
| GO:0009898 | cytoplasmic side of plasma membrane | cellular_component | membrane | 4 | 255 | 76 | 25187 | 0.052632 | 0.007447 | 0.05508 |
| GO:0031982 | vesicle | cellular_component | organelle | 7 | 255 | 220 | 25187 | 0.031818 | 0.007313 | 0.05508 |
| GO:0005586 | collagen type III trimer | cellular_component | extracellular region | 1 | 255 | 1 | 25187 | 1 | 0.010124 | 0.061968 |
| GO:0009295 | nucleoid | cellular_component | part nucleoid | 1 | 255 | 1 | 25187 | 1 | 0.010124 | 0.061968 |
| GO:0020003 | symbiont-containing vacuole | cellular_component | organelle | 1 | 255 | 1 | 25187 | 1 | 0.010124 | 0.061968 |
| GO:0020005 | symbiont-containing vacuole membrane | cellular_component | organelle | 1 | 255 | 1 | 25187 | 1 | 0.010124 | 0.061968 |
| GO:0031305 | integral component of mitochondrial inner membrane | cellular_component | organelle | 3 | 255 | 44 | 25187 | 0.068182 | 0.010005 | 0.061968 |
| GO:0033017 | sarcoplasmic reticulum membrane | cellular_component | organelle | 3 | 255 | 43 | 25187 | 0.069767 | 0.009392 | 0.061968 |
| GO:0034358 | plasma lipoprotein particle | cellular_component | extracellular region part | 1 | 255 | 1 | 25187 | 1 | 0.010124 | 0.061968 |
| GO:0034466 | chromaffin granule lumen | cellular_component | organelle | 1 | 255 | 1 | 25187 | 1 | 0.010124 | 0.061968 |
| GO:0099189 | postsynaptic spectrin- associated cytoskeleton | cellular_component | organelle | 1 | 255 | 1 | 25187 | 1 | 0.010124 | 0.061968 |
| GO:1990031 | pinceau fiber | cellular_component | cell part | 1 | 255 | 1 | 25187 | 1 | 0.010124 | 0.061968 |
| GO:0005614 | interstitial matrix | cellular_component | extracellular region | 2 | 255 | 16 | 25187 | 0.125 | 0.011158 | 0.064935 |
| GO:0016471 | vacuolar proton- transporting V-type ATPase complex | cellular_component | part organelle | 2 255 | 16 | 25187 | 0.125 | 0.011158 | 0.064935 | |
| GO:0099738 | cell cortex region | cellular_component | cell part | 2 | 255 | 16 | 25187 | 0.125 | 0.011158 | 0.064935 |
| GO:0031226 | intrinsic component of plasma membrane | cellular_component | membrane | 3 | 255 | 47 | 25187 | 0.06383 | 0.011981 | 0.068601 |
| GO:0005740 | mitochondrial envelope | cellular_component | organelle | 2 | 255 | 17 | 25187 | 0.117647 | 0.012562 | 0.070785 |
| GO:0030672 | synaptic vesicle membrane | cellular_component | organelle | 4 | 255 | 90 | 25187 | 0.044444 | 0.013298 | 0.073764 |
| GO:0005750 | mitochondrial respiratory chain | cellular_component | organelle | 2 | 255 | 20 | 25187 | 0.1 | 0.017204 | 0.092537 |
| GO:0015629 | actin cytoskeleton | cellular_component | organelle | 8 | 255 | 321 | 25187 | 0.024922 | 0.017011 | 0.092537 |
| GO:0005922 | connexin complex | cellular_component | membrane | 2 | 255 | 22 | 25187 | 0.090909 | 0.020641 | 0.096417 |
| GO:0005954 | calcium- and calmodulin-dependent protein kinase complex | cellular_component | cell part | 1 | 255 | 2 | 25187 | 0.5 | 0.020146 | 0.096417 |
| GO:0008076 | voltage-gated potassium channel complex | cellular_component | membrane | 4 | 255 | 99 | 25187 | 0.040404 | 0.018269 | 0.096417 |
| GO:0009353 | mitochondrial oxoglutarate dehydrogenase complex | cellular_component | membrane-enclosed lumen | 1 | 255 | 2 | 25187 | 0.5 | 0.020146 | 0.096417 |
| GO:0019008 | molybdopterin synthase complex | cellular_component | cell part | 1 | 255 | 2 | 25187 | 0.5 | 0.020146 | 0.096417 |
| GO:0032473 | cytoplasmic side of mitochondrial outer membrane | cellular_component | organelle | 1 | 255 | 2 | 25187 | 0.5 | 0.020146 | 0.096417 |
| GO:0060987 | lipid tube | cellular_component | protein-containing complex | 1 | 255 | 2 | 25187 | 0.5 | 0.020146 | 0.096417 |
| GO:0097180 | serine protease inhibitor complex | cellular_component | protein-containing complex | 1 | 255 | 2 | 25187 | 0.5 | 0.020146 | 0.096417 |
| GO:0098688 | parallel fiber to Purkinje cell synapse | cellular_component | synapse | 2 | 255 | 22 | 25187 | 0.090909 | 0.020641 | 0.096417 |
| GO:0099160 | postsynaptic intermediate filament cytoskeleton | cellular_component | organelle | 1 | 255 | 2 | 25187 | 0.5 | 0.020146 | 0.096417 |
| GO:0005868 | cytoplasmic dynein complex | cellular_component | organelle | 2 | 255 | 23 | 25187 | 0.086957 | 0.022458 | 0.103542 |
| GO:0010008 | endosome membrane | cellular_component | membrane | 8 | 255 | 340 | 25187 | 0.023529 | 0.023063 | 0.104967 |
| GO:0031201 | SNARE complex | cellular_component | membrane | 3 | 255 | 62 | 25187 | 0.048387 | 0.02503 | 0.112479 |
| GO:0034361 | very-low-density lipoprotein particle | cellular_component | extracellular region part | 2 | 255 | 25 | 25187 | 0.08 | 0.026281 | 0.116623 |
| GO:0031410 | cytoplasmic vesicle | cellular_component | organelle | 14 | 255 | 776 | 25187 | 0.018041 | 0.027302 | 0.119657 |
| GO:0005945 | 6-phosphofructokinase complex | cellular_component | cell part | 1 | 255 | 3 | 25187 | 0.333333 | 0.030068 | 0.120195 |
| GO:0016021 | integral component of membrane | cellular_component | membrane | 86 | 255 | 7102 | 25187 | 0.012109 | 0.030133 | 0.120195 |
| GO:0020018 | ciliary pocket membrane | cellular_component | organelle | 1 | 255 | 3 | 25187 | 0.333333 | 0.030068 | 0.120195 |
| GO:0043296 | apical junction complex | cellular_component | cell junction | 2 | 255 | 26 | 25187 | 0.076923 | 0.028284 | 0.120195 |
| GO:0045177 | apical part of cell | cellular_component | cell | 4 | 255 | 113 | 25187 | 0.035398 | 0.028042 | 0.120195 |
| GO:0060201 | clathrin-sculpted acetylcholine transport vesicle membrane | cellular_component | organelle | 1 | 255 | 3 | 25187 | 0.333333 | 0.030068 | 0.120195 |
| GO:0098560 | cytoplasmic side of late endosome membrane | cellular_component | membrane | 1 | 255 | 3 | 25187 | 0.333333 | 0.030068 | 0.120195 |
| GO:0099503 | secretory vesicle | cellular_component | organelle | 1 | 255 | 3 | 25187 | 0.333333 | 0.030068 | 0.120195 |
| GO:0034774 | secretory granule lumen | cellular_component | organelle | 5 | 255 | 172 | 25187 | 0.02907 | 0.031036 | 0.122421 |
| GO:0005753 | mitochondrial proton-transporting ATP synthase complex | cellular_component | organelle | 2 | 255 | 28 | 25187 | 0.071429 | 0.032467 | 0.122613 |
| GO:0005783 | endoplasmic reticulum | cellular_component | organelle | 27 | 255 | 1831 | 25187 | 0.014746 | 0.031832 | 0.122613 |
| GO:0016459 | myosin complex | cellular_component | organelle | 3 | 255 | 68 | 25187 | 0.044118 | 0.031747 | 0.122613 |
| GO:0034704 | calcium channel | cellular_component | membrane | 2 | 255 | 28 | 25187 | 0.071429 | 0.032467 | 0.122613 |
| GO:0030425 | dendrite | cellular_component | cell part | 11 | 255 | 582 | 25187 | 0.0189 | 0.035725 | 0.132106 |
| GO:0098685 | Schaffer collateral - CA1 synapse | cellular_component | synapse | 4 | 255 | 122 | 25187 | 0.032787 | 0.035692 | 0.132106 |
| GO:0005846 | nuclear cap binding complex | cellular_component | cell part | 1 | 255 | 4 | 25187 | 0.25 | 0.039889 | 0.134861 |
| GO:0016529 | sarcoplasmic reticulum | cellular_component | organelle | 3 | 255 | 74 | 25187 | 0.040541 | 0.03931 | 0.134861 |
| GO:0030027 | lamellipodium | cellular_component | cell part | 6 | 255 | 245 | 25187 | 0.02449 | 0.039092 | 0.134861 |
| GO:0030478 | actin cap | cellular_component | organelle | 1 | 255 | 4 | 25187 | 0.25 | 0.039889 | 0.134861 |
| GO:0034518 | RNA cap binding complex | cellular_component | protein-containing complex | 1 | 255 | 4 | 25187 | 0.25 | 0.039889 | 0.134861 |
| GO:0034686 | integrin alphav-beta8 complex | cellular_component | membrane | 1 | 255 | 4 | 25187 | 0.25 | 0.039889 | 0.134861 |
| GO:0042588 | zymogen granule | cellular_component | organelle | 1 | 255 | 4 | 25187 | 0.25 | 0.039889 | 0.134861 |
| GO:0060203 | clathrin-sculpted glutamate transport vesicle membrane | cellular_component | organelle | 1 | 255 | 4 | 25187 | 0.25 | 0.039889 | 0.134861 |
| GO:0098559 | cytoplasmic side of early endosome membrane | cellular_component | membrane | 1 | 255 | 4 | 25187 | 0.25 | 0.039889 | 0.134861 |
| GO:0042613 | MHC class II protein complex | cellular_component | membrane | 3 | 255 | 76 | 25187 | 0.039474 | 0.042016 | 0.140714 |
| GO:0045121 | membrane raft | cellular_component | membrane | 7 | 255 | 317 | 25187 | 0.022082 | 0.043252 | 0.1435 |
| GO:0031307 | integral component of mitochondrial outer membrane | cellular_component | organelle | 2 | 255 | 33 | 25187 | 0.060606 | 0.043886 | 0.144256 |
| GO:0005947 | mitochondrial alpha- ketoglutarate dehydrogenase complex | cellular_component | membrane- enclosed lumen | 1 | 255 | 5 | 25187 | 0.2 | 0.049611 | 0.151825 |
| GO:0014704 | intercalated disc | cellular_component | cell junction | 3 | 255 | 81 | 25187 | 0.037037 | 0.049178 | 0.151825 |
| GO:0016342 | catenin complex | cellular_component | membrane | 2 | 255 | 35 | 25187 | 0.057143 | 0.048812 | 0.151825 |
| GO:0033557 | Slx1-Slx4 complex enclosed lumen | cellular_component | membrane- | 1 | 255 | 5 | 25187 | 0.2 | 0.049611 | 0.151825 |
| GO:0070032 | synaptobrevin 2-SNAP- 25-syntaxin-1a- complexin I complex | cellular_component | membrane | 1 | 255 | 5 | 25187 | 0.2 | 0.049611 | 0.151825 |
| GO:0070083 | clathrin-sculpted monoamine transport vesicle membrane | cellular_component | organelle | 1 | 255 | 5 | 25187 | 0.2 | 0.049611 | 0.151825 |
| GO:0098857 | membrane microdomain | cellular_component | membrane | 1 | 255 | 5 | 25187 | 0.2 | 0.049611 | 0.151825 |
| GO:1990726 | Lsm1-7-Pat1 complex | cellular_component | protein-containing complex | 1 | 255 | 5 | 25187 | 0.2 | 0.049611 | 0.151825 |
Additional Table 8.
KEGG pathway analysis of differentially expressed proteins
| Term | Total | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| KEGG | Candidat | Candidat Term | Total | |||||||
| Pathway | KEGG Pathway Term | KEGG Pathway | e Gene | e Gene | Gene | Gene | Rich | |||
| Term ID | KEGG Pathway Term Desc | Level1 | Term Level2 | Num | Num | Num | Num | Ratio | P value | Q value |
| 5012 | Parkinson’s disease | Human Diseases | Neurodegenerative diseases | 20 | 187 | 209 | 15870 | 0.09569 | 5.64E-13 | 1.60E-10 |
| 1100 | Metabolic pathways | Metabolism | Global and overview rn r* k—\ r» | 58 | 187 | 1923 | 15870 | 0.03016 | 4.81E-12 | 6.81E-10 |
| 5016 | Huntington’s disease | Human Diseases | maps Neurodegenerative diseases | 19 | 187 | 283 | 15870 | 0.06714 | 1.03E-09 | 9.72E-08 |
| 190 | Oxidative phosphorylation | Metabolism | Energy metabolism | 15 | 187 | 195 | 15870 | 0.07692 | 1.04E-08 | 7.33E-07 |
| 4714 | Thermogenesis | Organismal Systems | Environmental adaptation | 17 | 187 | 318 | 15870 | 0.05346 | 2.27E-07 | 1.28E-05 |
| 5010 | Alzheimer’s disease | Human Diseases | Neurodegenerative diseases | 15 | 187 | 256 | 15870 | 0.05859 | 3.76E-07 | 1.77E-05 |
| 1110 | Biosynthesis of secondary metabolites | Metabolism | Global and overview | 21 | 187 | 558 | 15870 | 0.03763 | 2.73E-06 | 1.10E-04 |
| 4020 | Calcium signaling pathway | Environmental Information Processing | maps Signal transduction | 12 | 187 | 256 | 15870 | 0.04688 | 5.27E-05 | 0.00186 |
| 4932 | Non-alcoholic fatty liver disease (NAFLD) | Human Diseases | Endocrine and metabolic diseases | 11 | 187 | 250 | 15870 | 0.044 | 1.89E-04 | 0.00594 |
| 350 | Tyrosine metabolism | Metabolism | Amino acid metabolism | 5 | 187 | 48 | 15870 | 0.10417 | 2.44E-04 | 0.00629 |
| 4922 | Glucagon signaling pathway | Organismal Systems | Endocrine system | 8 | 187 | 138 | 15870 | 0.05797 | 2.30E-04 | 0.00629 |
| 4723 | Retrograde endocannabinoid signaling | Organismal Systems | Nervous system | 9 | 187 | 180 | 15870 | 0.05 | 2.86E-04 | 0.00675 |
| 1200 | Carbon metabolism | Metabolism | Global and overview | 9 | 187 | 189 | 15870 | 0.04762 | 4.10E-04 | 0.00892 |
| 950 | Isoquinoline alkaloid biosynthesis | Metabolism | maps Biosynthesis of other secondary metabolites | 3 | 187 | 14 | 15870 | 0.21429 | 5.33E-04 | 0.01005 |
| 4217 | Necroptosis | Cellular Processes | Cell growth and /-J /~\ r* -I- L-\ death | 10 | 187 | 236 | 15870 | 0.04237 | 4.97E-04 | 0.01005 |
| 4979 | Cholesterol metabolism | Organismal Systems | Digestive system | 5 | 187 | 60 | 15870 | 0.08333 | 6.96E-04 | 0.01231 |
| 1120 | Microbial metabolism in diverse environments | Metabolism | Global and overview maps | 10 | 187 | 250 | 15870 | 0.04 | 7.77E-04 | 0.01267 |
| 4971 | Gastric acid secretion | Organismal Systems | Digestive system | 6 | 187 | 93 | 15870 | 0.06452 | 8.06E-04 | 0.01267 |
| 4260 | Cardiac muscle contraction | Organismal Systems | Circulatory system | 6 | 187 | 97 | 15870 | 0.06186 | 0.001 | 0.01497 |
| 4978 | Mineral absorption | Organismal Systems | Digestive system | 5 | 187 | 66 | 15870 | 0.07576 | 0.00108 | 0.01522 |
| 4728 | Dopaminergic synapse | Organismal Systems | Nervous system | 8 | 187 | 177 | 15870 | 0.0452 | 0.0012 | 0.01616 |
| 4725 | Cholinergic synapse | Organismal Systems | Nervous system | 7 | 187 | 145 | 15870 | 0.04828 | 0.00166 | 0.02136 |
| 1130 | Biosynthesis of antibiotics | Metabolism | Global and overview rn r* k—\ r» | 11 | 187 | 327 | 15870 | 0.03364 | 0.00175 | 0.02154 |
| 4730 | Long-term depression | Organismal Systems | maps Nervous system | 5 | 187 | 75 | 15870 | 0.06667 | 0.00191 | 0.0225 |
| 360 | Phenylalanine metabolism | Metabolism | Amino acid metabolism | 3 | 187 | 22 | 15870 | 0.13636 | 0.0021 | 0.02284 |
| 4218 | Cellular senescence | Cellular Processes | Cell growth and | 10 | 187 | 287 | 15870 | 0.03484 | 0.00218 | 0.02284 |
| 4921 | Oxytocin signaling pathway | Organismal Systems | death Endocrine system | 8 | 187 | 194 | 15870 | 0.04124 | 0.00214 | 0.02284 |
| 5031 | Amphetamine addiction | Human Diseases | Substance dependence | 5 | 187 | 86 | 15870 | 0.05814 | 0.00347 | 0.03401 |
| 5216 | Thyroid cancer | Human Diseases | Cancers: Specific types | 4 | 187 | 53 | 15870 | 0.07547 | 0.00348 | 0.03401 |
| 4720 | Long-term potentiation | Organismal Systems | Nervous system | 5 | 187 | 88 | 15870 | 0.05682 | 0.00383 | 0.03593 |
| 4916 | Melanogenesis | Organismal Systems | Endocrine system | 6 | 187 | 127 | 15870 | 0.04724 | 0.00394 | 0.03593 |
| 4022 | cGMP-PKG signaling pathway | Environmental Information Processing | Signal transduction | 8 | 187 | 228 | 15870 | 0.03509 | 0.00569 | 0.05033 |
| 4514 | Cell adhesion molecules (CAMs) | Environmental Information Processing | Signaling molecules and interaction | 9 | 187 | 279 | 15870 | 0.03226 | 0.00591 | 0.05068 |
| 790 | Folate biosynthesis | Metabolism | Metabolism of cofactors and vitamins | 3 | 187 | 33 | 15870 | 0.09091 | 0.00677 | 0.05544 |
| 3320 | PPAR signaling pathway | Organismal Systems | Endocrine system | 5 | 187 | 101 | 15870 | 0.0495 | 0.00686 | 0.05544 |
| 4216 | Ferroptosis | Cellular Processes | Cell growth and /-J /~\ r* -I- L-\ death | 4 | 187 | 65 | 15870 | 0.06154 | 0.00723 | 0.0568 |
| 4911 | Insulin secretion | Organismal Systems | Endocrine system | 5 | 187 | 104 | 15870 | 0.04808 | 0.00774 | 0.0592 |
| 4621 | NOD-like receptor signaling pathway | Organismal Systems | Immune system | 8 | 187 | 247 | 15870 | 0.03239 | 0.00904 | 0.06734 |
| 4972 | Pancreatic secretion | Organismal Systems | Digestive system | 5 | 187 | 111 | 15870 | 0.04505 | 0.0101 | 0.07331 |
| 4745 | Phototransduction - fly | Organismal Systems | Sensory system | 3 | 187 | 39 | 15870 | 0.07692 | 0.01078 | 0.07625 |
| 30 | Pentose phosphate pathway | Metabolism | Carbohydrate metabolism | 3 | 187 | 41 | 15870 | 0.07317 | 0.01236 | 0.08032 |
| 52 | Galactose metabolism | Metabolism | Carbohydrate metabolism | 3 | 187 | 41 | 15870 | 0.07317 | 0.01236 | 0.08032 |
| 4212 | Longevity regulating pathway - worm | Organismal Systems | Aging | 5 | 187 | 117 | 15870 | 0.04274 | 0.01249 | 0.08032 |
| 5034 | Alcoholism | Human Diseases | Substance dependence | 7 | 187 | 208 | 15870 | 0.03365 | 0.01174 | 0.08032 |
| 4912 | GnRH signaling pathway | Organismal Systems | Endocrine system | 5 | 187 | 118 | 15870 | 0.04237 | 0.01292 | 0.08125 |
| 4713 | Circadian entrainment | Organismal Systems | Environmental adaptation | 5 | 187 | 120 | 15870 | 0.04167 | 0.01381 | 0.08497 |
| 480 | Glutathione metabolism | Metabolism | Metabolism of other amino acids | 4 | 187 | 79 | 15870 | 0.05063 | 0.01413 | 0.08507 |
| 5230 | Central carbon metabolism in cancer | Human Diseases | Cancers: Overview | 4 | 187 | 81 | 15870 | 0.04938 | 0.01536 | 0.09059 |
| 4360 | Axon guidance | Organismal Systems | Development | 7 187 | 223 | 15870 | 0.03139 | 0.01665 | 0.09474 | |
| 4925 | Aldosterone synthesis and secretion | Organismal Systems | Endocrine system | 5 | 187 | 126 | 15870 | 0.03968 | 0.01674 | 0.09474 |
| 51 | Fructose and mannose metabolism | Metabolism | Carbohydrate metabolism | 3 | 187 | 47 | 15870 | 0.06383 | 0.01786 | 0.09536 |
| 4371 | Apelin signaling pathway | Environmental Information Processing | Signal transduction | 6 | 187 | 175 | 15870 | 0.03429 | 0.0176 | 0.09536 |
| 4721 | Synaptic vesicle cycle | Organismal Systems | Nervous system | 4 | 187 | 84 | 15870 | 0.04762 | 0.01734 | 0.09536 |
| 4261 | Adrenergic signaling in cardiomyocytes | Organismal Systems | Circulatory system | 6 | 187 | 185 | 15870 | 0.03243 | 0.02245 | 0.11763 |
| 250 | Alanine, aspartate and glutamate metabolism | Metabolism | Amino acid metabolism | 3 | 187 | 52 | 15870 | 0.05769 | 0.02333 | 0.12003 |
| 10 | Glycolysis / Gluconeogenesis | Metabolism | Carbohydrate metabolism | 4 | 187 | 94 | 15870 | 0.04255 | 0.02506 | 0.12664 |
| 4640 | Hematopoietic cell lineage | Organismal Systems | Immune system | 6 | 187 | 194 | 15870 | 0.03093 | 0.02751 | 0.1366 |
| 20 | Citrate cycle (TCA cycle) | Metabolism | Carbohydrate metabolism | 3 | 187 | 58 | 15870 | 0.05172 | 0.03095 | 0.14129 |
| 71 | Fatty acid degradation | Metabolism | Lipid metabolism | 3 | 187 | 57 | 15870 | 0.05263 | 0.0296 | 0.14129 |
| 4066 | HIF-1 signaling pathway | Environmental Information Processing | Signal transduction | 5 | 187 | 148 | 15870 | 0.03378 | 0.03089 | 0.14129 |
| 4726 | Serotonergic synapse | Organismal Systems | Nervous system | 5 | 187 | 147 | 15870 | 0.03401 | 0.03012 | 0.14129 |
| 5214 | Glioma | Human Diseases | Cancers: Specific types | 4 | 187 | 100 | 15870 | 0.04 | 0.03054 | 0.14129 |
| 1230 | Biosynthesis of amino acids | Metabolism | Global and overview rn r* k—\ r» | 4 | 187 | 104 | 15870 | 0.03846 | 0.03455 | 0.15501 |
| 4974 | Protein digestion and absorption | Organismal Systems | maps Digestive system | 4 | 187 | 105 | 15870 | 0.0381 | 0.0356 | 0.15501 |
| 5030 | Cocaine addiction | Human Diseases | Substance dependence | 3 | 187 | 61 | 15870 | 0.04918 | 0.0352 | 0.15501 |
| 330 | Arginine and proline metabolism | Metabolism | Amino acid metabolism | 3 | 187 | 63 | 15870 | 0.04762 | 0.03819 | 0.15716 |
| 340 | Histidine metabolism | Metabolism | Amino acid metabolism | 2 | 187 | 27 | 15870 | 0.07407 | 0.03997 | 0.15716 |
| 520 | Amino sugar and nucleotide sugar metabolism | Metabolism | Carbohydrate metabolism | 3 | 187 | 64 | 15870 | 0.04688 | 0.03973 | 0.15716 |
| 4012 | ErbB signaling pathway | Environmental Information Processing | Signal transduction | 4 | 187 | 109 | 15870 | 0.0367 | 0.03999 | 0.15716 |
| 4727 | GABAergic synapse | Organismal Systems | Nervous system | 4 | 187 | 108 | 15870 | 0.03704 | 0.03886 | 0.15716 |
| 5110 | Vibrio cholerae infection | Human Diseases | Infectious diseases: Bacterial | 3 | 187 | 62 | 15870 | 0.04839 | 0.03668 | 0.15716 |
| 5166 | HTLV-I infection | Human Diseases | Infectious diseases: Viral | 10 | 187 | 447 | 15870 | 0.02237 | 0.03928 | 0.15716 |
| 4722 | Neurotrophin signaling pathway | Organismal Systems | Nervous system | 5 | 187 | 160 | 15870 | 0.03125 | 0.04105 | 0.15728 |
| 4970 | Salivary secretion | Organismal Systems | Digestive system | 4 | 187 | 110 | 15870 | 0.03636 | 0.04113 | 0.15728 |
| 4015 | Rap1 signaling pathway | Environmental Information Processing | Signal transduction | 7 | 187 | 273 | 15870 | 0.02564 | 0.04316 | 0.16287 |
| 1212 | Fatty acid metabolism | Metabolism | Global and overview rn r* \ r. | 3 | 187 | 69 | 15870 | 0.04348 | 0.0479 | 0.17797 |
| 4614 | Renin-angiotensin system | Organismal Systems | maps Endocrine system | 2 | 187 | 30 | 15870 | 0.06667 | 0.04842 | 0.17797 |
KEGG: Kyoto Encyclopedia of Genes and Genomes.
Myelin is composed of SCs, which are indispensable for the physiological functions of peripheral nerves (Salzer, 2015). Previously, impaired SC migration was reported to contribute to the abnormal myelination and demyelination of peripheral nerves (Anliker et al., 2013; Yi et al., 2019). Thus, we compared the function of SCs from nerves in the DPN and control groups. The primary SCs isolated from the peripheral nerves exhibited a long spindle shape under an optical microscope (Additional Figure 2A (1.1MB, tif) ). These were confirmed via positive immunofluorescence staining of S100 calcium binding protein B and glial fibrillary acidic protein (Additional Figure 2B (1.1MB, tif) ). Cell migration assays showed significantly impaired migration of SCs derived from patients with DPN (Figure 2G and H).
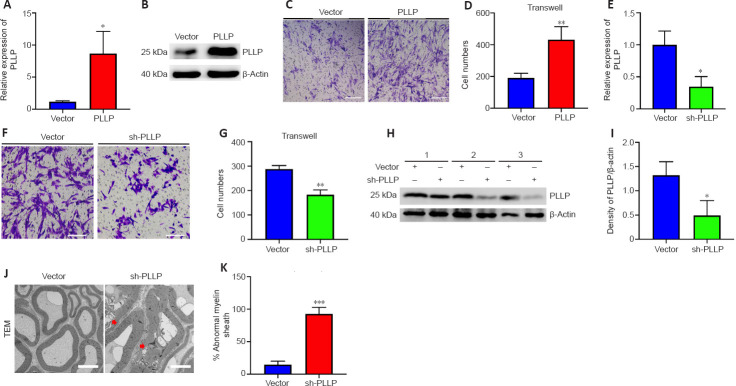
Characterization of circ_0002538 and its function in SCs
We performed circRNA sequencing for the three pairs of peripheral nerves to uncover their characteristics in the development of DPN. In diabetic peripheral nerves, we identified a total of 15637 circRNAs. A total of 169 circRNAs showed significantly (P < 0.01, q < 0.05, readings ≥ 50, FC ≥ 2) dysregulated expression in the DPN group: 116 circRNAs had significantly downregulated expression and 53 circRNAs had significantly upregulated expression (Additional Table 9). The differentially expressed circRNAs (DEcircRNAs) were directly displayed by hierarchical cluster analysis (Figure 3A). The DEcircRNAs were verified using RT-PCR, and the results showed that six circRNAs with downregulated expression and five with upregulated expression were confirmed in the DPN group (Figure 3B and C). These DEcircRNAs may play an important role in the pathogenesis of DPN.
Additional Table 9.
The DEcircRNAs analyzed in this study were selected from the results of circRNA sequencing analysis
| circName | gene symbol | CZL-FC-B | FFL-FQ-B | YYC-FC-B | CCK-FC | WCF-FC-BX | CZG-FC-BS | log2(Fold_change) | p-value | q-value | circBase | Web |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa_circ:chr11:120276827-120278532 | ARHGEF12 | 319 | 435 | 636 | 9234 | 2564 | 4666 | 2.710491179 | 1.26E-10 | 2.22E-08 | hsa_circ_0024604 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0024604 |
| hsa_circ:chr9:134381501-134381840 | POMT1 | 330 | 683 | 739 | 8179 | 4331 | 6055 | 2.598539216 | 2.77E-20 | 6.19E-17 | hsa_circ_0001897 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001897 |
| hsa_circ:chr10:128806996-128810638 | DOCK1 | 80 | 136 | 217 | 1933 | 1163 | 1073 | 2.468022558 | 9.41E-15 | 6.13E-12 | hsa_circ_0020433 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0020433 |
| hsa_circ:chr13:24823615-24826000 | SPATA13 | 63 | 90 | 66 | 1552 | 398 | 310 | 2.347737447 | 2.78E-06 | 0.000109 | hsa_circ_0003040 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0003040 |
| hsa_circ:chr16:68155890-68160513 | ENSG00000261864.1,NFATC3 | 140 | 127 | 170 | 1522 | 1492 | 944 | 2.331800087 | 3.09E-14 | 1.79E-11 | hsa_circ_0000711 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000711 |
| hsa_circ:chr15:101775287-101775782 | CHSY1 | 113 | 161 | 107 | 2302 | 673 | 706 | 2.278883636 | 1.92E-05 | 0.000503 | hsa_circ_0005019 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0005019 |
| hsa_circ:chr19:1271328-1272050 | CIRBP | 115 | 273 | 289 | 2890 | 1394 | 1281 | 2.184967412 | 8.65E-11 | 1.73E-08 | hsa_circ_0007715 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0007715 |
| hsa_circ:chr3:171965323-171969331 | FNDC3B | 358 | 1139 | 926 | 8645 | 4811 | 4483 | 2.039068321 | 3.77E-06 | 0.000138 | hsa_circ_0006156 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0006156 |
| hsa_circ:chr5:171482592-171484477 | STK10 | 86 | 172 | 117 | 1260 | 705 | 793 | 2.001937464 | 8.13E-08 | 5.36E-06 | hsa_circ_0001555 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001555 |
| hsa_circ:chr1:14042036-14075982 | PRDM2 | 82 | 83 | 105 | 809 | 562 | 480 | 1.919088074 | 9.56E-08 | 6.13E-06 | NA | NA |
| hsa_circ:chr16:88061089-88071617 | BANP | 169 | 147 | 192 | 1461 | 1166 | 846 | 1.906629396 | 4.70E-10 | 6.56E-08 | hsa_circ_0040823 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0040823 |
| hsa_circ:chr10:99196948-99197507 | EXOSC1 | 172 | 254 | 482 | 2531 | 1820 | 1364 | 1.872337049 | 2.45E-10 | 3.76E-08 | hsa_circ_0004896 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0004896 |
| hsa_circ:chr15:41036245-41037457 | RMDN3 | 70 | 133 | 158 | 1171 | 517 | 624 | 1.838657173 | 1.16E-06 | 5.33E-05 | hsa_circ_0004942 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0004942 |
| hsa_circ:chr15:93540187-93541851 | CHD2 | 60 | 55 | 125 | 426 | 490 | 464 | 1.794259793 | 2.85E-05 | 0.000704 | hsa_circ_0000655 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000655 |
| hsa_circ:chr16:68155890-68157024 | ENSG00000261864.1,NFATC3 | 133 | 99 | 174 | 1039 | 779 | 652 | 1.764900392 | 1.28E-07 | 7.93E-06 | hsa_circ_0005615 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0005615 |
| hsa_circ:chr19:41754419-41754725 | AXL | 211 | 297 | 597 | 3127 | 1340 | 1883 | 1.741241582 | 6.40E-08 | 4.27E-06 | hsa_circ_0002882 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0002882 |
| hsa_circ:chr8:131164982-131181313 | ASAP1 | 542 | 363 | 544 | 4679 | 1996 | 2298 | 1.71720344 | 2.15E-08 | 1.69E-06 | hsa_circ_0001824 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001824 |
| hsa_circ:chr16:11114050-11154879 | CLEC16A | 57 | 63 | 55 | 435 | 309 | 308 | 1.712257159 | 0.000168 | 0.002957 | hsa_circ_0000672 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000672 |
| hsa_circ:chr21:48063447-48064400 | PRMT2 | 143 | 220 | 351 | 1972 | 1321 | 843 | 1.711577476 | 5.98E-08 | 4.07E-06 | hsa_circ_0003781 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0003781 |
| hsa_circ:chr22:34157358-34252790 | LARGE1 | 110 | 143 | 250 | 1555 | 527 | 832 | 1.708406728 | 5.51E-06 | 0.000187 | hsa_circ_0063019 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0063019 |
| hsa_circ:chr9:131271155-131277918 | GLE1 | 75 | 101 | 141 | 1006 | 413 | 462 | 1.706348705 | 1.24E-05 | 0.000357 | hsa_circ_0002675 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0002675 |
| hsa_circ:chr12:57059988-57064148 | PTGES3 | 69 | 86 | 84 | 589 | 276 | 526 | 1.702531372 | 0.00013 | 0.00244 | hsa_circ_0027089 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0027089 |
| hsa_circ:chr21:46275125-46281186 | PTTG1IP | 357 | 285 | 333 | 2777 | 1239 | 1859 | 1.698755826 | 1.32E-07 | 8.12E-06 | hsa_circ_0001200 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001200 |
| hsa_circ:chr6:43023283-43024183 | MRPL2 | 57 | 66 | 131 | 629 | 367 | 418 | 1.69182392 | 3.44E-05 | 0.000814 | hsa_circ_0001608 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001608 |
| hsa_circ:chr12:123983091-123984083 | RILPL1 | 236 | 520 | 520 | 3230 | 1977 | 1984 | 1.666769005 | 1.52E-08 | 1.25E-06 | hsa_circ_0007552 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0007552 |
| hsa_circ:chr1:154207067-154207767 | UBAP2L | 104 | 216 | 266 | 1431 | 644 | 1087 | 1.640288416 | 8.60E-06 | 0.00027 | NA | NA |
| hsa_circ:chr14:103918255-103923549 | MARK3 | 157 | 139 | 150 | 1321 | 530 | 727 | 1.617693132 | 1.54E-05 | 0.000426 | hsa_circ_0033475 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0033475 |
| hsa_circ:chr1:27269151-27269556 | NUDC | 256 | 302 | 550 | 2609 | 1519 | 1684 | 1.595422136 | 6.40E-09 | 5.99E-07 | hsa_circ_0005087 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0005087 |
| hsa_circ:chr3:153912433-153935747 | ARHGEF26 | 73 | 92 | 85 | 526 | 246 | 556 | 1.584230606 | 0.000715 | 0.008975 | NA | NA |
| hsa_circ:chr12:124904503-124915333 | NCOR2 | 88 | 118 | 162 | 1099 | 509 | 376 | 1.544679115 | 7.38E-05 | 0.001525 | hsa_circ_0029308 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0029308 |
| hsa_circ:chr1:215759838-215768813 | KCTD3 | 146 | 310 | 261 | 1709 | 765 | 1238 | 1.531205242 | 2.13E-05 | 0.000548 | hsa_circ_0005521 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0005521 |
| hsa_circ:chr15:41361768-41362745 | INO80 | 87 | 91 | 116 | 621 | 421 | 468 | 1.527105285 | 5.26E-05 | 0.00115 | hsa_circ_0007489 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0007489 |
| hsa_circ:chr3:119222379-119222868 | TIMMDC1 | 628 | 987 | 1512 | 7770 | 4093 | 3672 | 1.486728502 | 8.43E-09 | 7.53E-07 | hsa_circ_0008394 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0008394 |
| hsa_circ:chr11:124517261-124518071 | SIAE | 145 | 58 | 148 | 631 | 502 | 583 | 1.46585348 | 0.000785 | 0.009691 | hsa_circ_0000367 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000367 |
| hsa_circ:chr8:103372299-103373854 | UBR5 | 700 | 859 | 1621 | 6211 | 4483 | 4183 | 1.449358147 | 7.72E-10 | 1.01E-07 | hsa_circ_0001819 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001819 |
| hsa_circ:chr19:47767860-47768203 | CCDC9 | 671 | 1253 | 1708 | 7592 | 4454 | 4462 | 1.379543586 | 7.66E-08 | 5.07E-06 | hsa_circ_0000944 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000944 |
| hsa_circ:chr12:120592774-120593523 | GCN1 | 254 | 214 | 353 | 1843 | 877 | 1084 | 1.356950971 | 1.22E-05 | 0.000355 | hsa_circ_0000448 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000448 |
| hsa_circ:chr3:47139445-47147610 | SETD2 | 355 | 449 | 487 | 2582 | 2034 | 1387 | 1.354151656 | 2.39E-07 | 1.36E-05 | hsa_circ_0001290 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001290 |
| hsa_circ:chr14:23378692-23380612 | RBM23 | 667 | 977 | 1913 | 7053 | 3121 | 5100 | 1.342548268 | 7.29E-06 | 0.000234 | hsa_circ_0000524 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000524 |
| hsa_circ:chr8:131164982-131193126 | ASAP1 | 416 | 319 | 408 | 2441 | 1681 | 1141 | 1.297037309 | 1.40E-05 | 0.000395 | hsa_circ_0008934 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0008934 |
| hsa_circ:chr20:47570093-47580435 | ARFGEF2 | 154 | 158 | 286 | 1158 | 673 | 740 | 1.293570365 | 5.27E-05 | 0.001151 | hsa_circ_0003998 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0003998 |
| hsa_circ:chr15:80412670-80415142 | ZFAND6 | 527 | 568 | 743 | 3469 | 1985 | 2536 | 1.284851683 | 4.57E-07 | 2.43E-05 | hsa_circ_0000643 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000643 |
| hsa_circ:chr2:160025761-160027316 | TANC1 | 180 | 223 | 267 | 1150 | 1009 | 737 | 1.277151485 | 1.78E-05 | 0.000475 | hsa_circ_0056810 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0056810 |
| hsa_circ:chr3:196118684-196120490 | UBXN7 | 142 | 174 | 237 | 1046 | 654 | 651 | 1.255873553 | 6.77E-05 | 0.001419 | hsa_circ_0005051 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0005051 |
| hsa_circ:chr18:9524592-9525849 | RALBP1 | 1003 | 1152 | 2868 | 10330 | 4744 | 5270 | 1.235638445 | 1.91E-05 | 0.000502 | hsa_circ_0005158 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0005158 |
| hsa_circ:chr11:108137898-108138069 | ATM | 179 | 204 | 289 | 1390 | 602 | 838 | 1.228247142 | 0.000216 | 0.003586 | hsa_circ_0007694 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0007694 |
| hsa_circ:chr9:33971649-33973235 | UBAP2 | 657 | 1419 | 1442 | 5858 | 4652 | 3689 | 1.195953494 | 6.18E-06 | 0.000205 | hsa_circ_0001851 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001851 |
| hsa_circ:chr2:72958136-72960247 | EXOC6B | 287 | 311 | 549 | 2065 | 941 | 1492 | 1.170419692 | 0.000174 | 0.003028 | hsa_circ_0001030 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001030 |
| hsa_circ:chr3:56600622-56601081 | CCDC66 | 322 | 286 | 510 | 1987 | 1398 | 1019 | 1.138112107 | 4.21E-05 | 0.000963 | hsa_circ_0001312 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001312 |
| hsa_circ:chr3:197592294-197593090 | LRCH3 | 194 | 209 | 359 | 1218 | 729 | 916 | 1.113625109 | 0.000336 | 0.005019 | hsa_circ_0008439 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0008439 |
| hsa_circ:chr2:168920010-168986268 | STK39 | 275 | 623 | 721 | 3239 | 1381 | 1516 | 1.077885846 | 0.000968 | 0.011373 | hsa_circ_0005882 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0005882 |
| hsa_circ:chr2:209209835-209212747 | PIKFYVE | 221 | 327 | 371 | 1433 | 1151 | 829 | 1.054921911 | 0.000252 | 0.004053 | hsa_circ_0001097 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001097 |
| hsa_circ:chr2:153431650-153437563 | FMNL2 | 556 | 825 | 1057 | 3816 | 2604 | 2419 | 1.038670032 | 1.03E-05 | 0.000312 | NA | NA |
| hsa_circ:chr5:142434004-142437312 | ARHGAP26 | 527 | 543 | 586 | 456 | 476 | 468 | -1.063382894 | 0.000916 | 0.010935 | hsa_circ_0074371 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0074371 |
| hsa_circ:chr11:46098305-46113774 | PHF21A | 928 | 1052 | 1333 | 995 | 1093 | 678 | -1.084886764 | 4.59E-05 | 0.001024 | hsa_circ_0000296 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000296 |
| hsa_circ:chr5:65284463-65290692 | ERBIN | 794 | 640 | 1888 | 996 | 1003 | 629 | -1.087183343 | 0.000669 | 0.00854 | hsa_circ_0001492 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001492 |
| hsa_circ:chr8:62593527-62596747 | ASPH | 1413 | 1936 | 2315 | 2148 | 1127 | 1320 | -1.146691876 | 3.42E-06 | 0.000127 | hsa_circ_0084615 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0084615 |
| hsa_circ:chr3:157839892-157841780 | RSRC1 | 333 | 362 | 550 | 331 | 321 | 292 | -1.186546656 | 0.000256 | 0.004091 | hsa_circ_0001355 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001355 |
| hsa_circ:chr4:128995615-128999117 | LARP1B | 288 | 242 | 369 | 301 | 240 | 148 | -1.247937024 | 0.000787 | 0.009699 | hsa_circ_0001438 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001438 |
| hsa_circ:chr11:77394755-77404656 | RSF1 | 541 | 609 | 1027 | 614 | 535 | 430 | -1.254991191 | 5.89E-06 | 0.000197 | hsa_circ_0000344 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000344 |
| hsa_circ:chr10:32197100-32199491 | ARHGAP12 | 850 | 564 | 874 | 589 | 560 | 521 | -1.289193851 | 3.98E-05 | 0.000919 | hsa_circ_0000231 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000231 |
| hsa_circ:chr8:48308936-48320523 | SPIDR | 236 | 385 | 371 | 384 | 177 | 187 | -1.289993629 | 0.000696 | 0.008758 | hsa_circ_0001798 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001798 |
| hsa_circ:chr9:16727795-16738483 | BNC2 | 502 | 322 | 648 | 342 | 400 | 297 | -1.303581784 | 0.000208 | 0.003485 | hsa_circ_0008732 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0008732 |
| hsa_circ:chr2:9083316-9098771 | MBOAT2 | 783 | 991 | 1306 | 839 | 729 | 587 | -1.329282379 | 1.20E-07 | 7.47E-06 | hsa_circ_0007334 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0007334 |
| hsa_circ:chr12:120995085-120995485 | RNF10 | 402 | 505 | 580 | 284 | 487 | 231 | -1.373430304 | 0.000227 | 0.003722 | hsa_circ_0028899 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0028899 |
| hsa_circ:chr10:71243447-71244971 | TSPAN15 | 201 | 543 | 588 | 248 | 341 | 258 | -1.395283832 | 0.000598 | 0.007849 | hsa_circ_0002758 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0002758 |
| hsa_circ:chr12:70193989-70195501 | RAB3IP | 633 | 939 | 1455 | 742 | 555 | 598 | -1.445561613 | 1.99E-07 | 1.16E-05 | hsa_circ_0000419 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000419 |
| hsa_circ:chr2:24357989-24369956 | FAM228B | 389 | 521 | 717 | 494 | 268 | 297 | -1.44910951 | 3.08E-06 | 0.000118 | hsa_circ_0000982 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000982 |
| hsa_circ:chr17:57430576-57430887 | YPEL2 | 415 | 523 | 518 | 393 | 331 | 243 | -1.45137057 | 4.95E-06 | 0.000171 | hsa_circ_0005600 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0005600 |
| hsa_circ:chr17:30310018-30315516 | SUZ12 | 246 | 265 | 363 | 238 | 191 | 140 | -1.459103606 | 5.79E-05 | 0.00125 | hsa_circ_0002629 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0002629 |
| hsa_circ:chr2:191523884-191537878 | NAB1 | 218 | 174 | 400 | 157 | 215 | 115 | -1.468917338 | 0.000695 | 0.008753 | hsa_circ_0002024 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0002024 |
| hsa_circ:chr3:196118684-196129890 | UBXN7 | 5879 | 6433 | 9687 | 4835 | 5200 | 3721 | -1.473332214 | 1.01E-11 | 2.63E-09 | hsa_circ_0001380 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001380 |
| hsa_circ:chr6:82920531-82922510 | IBTK | 217 | 163 | 457 | 157 | 235 | 97 | -1.528497514 | 0.000981 | 0.011495 | hsa_circ_0002041 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0002041 |
| hsa_circ:chr15:62299507-62306191 | VPS13C | 557 | 1016 | 942 | 555 | 472 | 491 | -1.536923666 | 4.72E-07 | 2.48E-05 | hsa_circ_0000607 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000607 |
| hsa_circ:chr2:240929491-240946787 | NDUFA10 | 240 | 263 | 316 | 182 | 160 | 151 | -1.556620967 | 3.99E-05 | 0.00092 | hsa_circ_0001118 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001118 |
| hsa_circ:chr17:63739186-63746842 | CEP112 | 231 | 220 | 285 | 166 | 144 | 133 | -1.563739774 | 7.63E-05 | 0.001566 | hsa_circ_0002910 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0002910 |
| hsa_circ:chr4:87685746-87689129 | PTPN13 | 448 | 382 | 838 | 275 | 361 | 301 | -1.569552881 | 8.09E-06 | 0.000257 | hsa_circ_0007948 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0007948 |
| hsa_circ:chr15:76152219-76165909 | UBE2Q2 | 235 | 303 | 372 | 306 | 132 | 126 | -1.578701729 | 6.23E-05 | 0.001327 | NA | NA |
| hsa_circ:chr1:107866904-107867544 | NTNG1 | 135 | 227 | 433 | 108 | 134 | 177 | -1.586753878 | 0.000925 | 0.011016 | hsa_circ_0002286 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0002286 |
| hsa_circ:chr8:25265499-25266456 | DOCK5 | 197 | 155 | 386 | 178 | 114 | 128 | -1.591595366 | 0.000197 | 0.003357 | hsa_circ_0007618 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0007618 |
| hsa_circ:chr8:42812237-42819617 | HOOK3 | 504 | 280 | 548 | 203 | 331 | 231 | -1.595558057 | 8.55E-05 | 0.00172 | hsa_circ_0006376 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0006376 |
| hsa_circ:chr5:145634506-145638156 | RBM27 | 142 | 174 | 323 | 118 | 147 | 90 | -1.603683599 | 0.000366 | 0.005392 | hsa_circ_0006087 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0006087 |
| hsa_circ:chr20:35532560-35533906 | SAMHD1 | 147 | 285 | 320 | 138 | 151 | 131 | -1.611456748 | 0.000156 | 0.002811 | hsa_circ_0060221 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0060221 |
| hsa_circ:chr14:80163973-80271533 | NRXN3 | 466 | 706 | 1022 | 368 | 437 | 392 | -1.61911495 | 3.73E-07 | 2.03E-05 | hsa_circ_0032812 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0032812 |
| hsa_circ:chr1:9991949-9994918 | LZIC | 929 | 1146 | 1706 | 803 | 821 | 508 | -1.629900769 | 5.59E-10 | 7.54E-08 | hsa_circ_0000014 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000014 |
| hsa_circ:chr17:49340635-49346265 | UTP18 | 546 | 322 | 588 | 343 | 252 | 248 | -1.638251504 | 3.23E-06 | 0.000122 | hsa_circ_0002789 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0002789 |
| hsa_circ:chr13:21305980-21306260 | EEF1AKMT1 | 208 | 172 | 572 | 194 | 172 | 138 | -1.642222482 | 0.000137 | 0.002537 | hsa_circ_0003285 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0003285 |
| hsa_circ:chr12:109509417-109511337 | USP30 | 207 | 316 | 495 | 265 | 189 | 114 | -1.661513009 | 1.53E-05 | 0.000425 | hsa_circ_0028094 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0028094 |
| hsa_circ:chr21:34804484-34805178 | IFNGR2 | 359 | 487 | 801 | 379 | 249 | 268 | -1.663057865 | 1.47E-07 | 8.93E-06 | hsa_circ_0001185 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001185 |
| hsa_circ:chr18:51797730-51800460 | POLI | 333 | 147 | 305 | 141 | 138 | 156 | -1.67484072 | 0.000324 | 0.004882 | hsa_circ_0007180 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0007180 |
| hsa_circ:chr18:39607407-39629569 | PIK3C3 | 149 | 149 | 203 | 115 | 93 | 73 | -1.675462109 | 0.000252 | 0.004052 | hsa_circ_0007765 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0007765 |
| hsa_circ:chr1:155646339-155649303 | YY1AP1 | 2356 | 3540 | 5429 | 1894 | 2148 | 1856 | -1.685354126 | 3.27E-11 | 7.30E-09 | hsa_circ_0014606 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0014606 |
| hsa_circ:chr10:17746430-17747740 | STAM | 169 | 183 | 371 | 100 | 101 | 161 | -1.691618912 | 0.000374 | 0.005482 | hsa_circ_0008311 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0008311 |
| hsa_circ:chr4:17963526-17974508 | LCORL | 167 | 212 | 161 | 137 | 123 | 55 | -1.695139251 | 0.00062 | 0.008076 | hsa_circ_0069285 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0069285 |
| hsa_circ:chr7:156758964-156759786 | NOM1 | 198 | 174 | 224 | 111 | 148 | 67 | -1.71930518 | 0.000266 | 0.004207 | hsa_circ_0004210 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0004210 |
| hsa_circ:chr21:16386665-16415895 | NRIP1 | 714 | 326 | 806 | 421 | 311 | 273 | -1.725063678 | 2.17E-06 | 9.04E-05 | hsa_circ_0004771 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0004771 |
| hsa_circ:chr8:52773405-52773806 | PCMTD1 | 208 | 300 | 498 | 198 | 84 | 223 | -1.729606809 | 0.000145 | 0.002661 | hsa_circ_0001801 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001801 |
| hsa_circ:chr17:28011581-28030080 | SSH2 | 323 | 364 | 556 | 123 | 269 | 224 | -1.731580628 | 4.70E-05 | 0.001042 | hsa_circ_0000754 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000754 |
| hsa_circ:chr6:69943182-69949118 | ADGRB3 | 323 | 457 | 1022 | 389 | 312 | 213 | -1.734214663 | 8.51E-07 | 4.11E-05 | hsa_circ_0076952 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0076952 |
| hsa_circ:chr1:67356837-67371058 | WDR78 | 1412 | 2109 | 2693 | 1131 | 1452 | 672 | -1.736691757 | 1.60E-09 | 1.91E-07 | hsa_circ_0006677 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0006677 |
| hsa_circ:chr13:61013822-61041513 | TDRD3 | 355 | 326 | 514 | 135 | 245 | 212 | -1.758880801 | 1.71E-05 | 0.000465 | hsa_circ_0003441 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0003441 |
| hsa_circ:chr15:59204762-59209198 | SLTM | 1416 | 1922 | 2225 | 1377 | 835 | 741 | -1.771539504 | 1.54E-13 | 7.28E-11 | hsa_circ_0000605 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000605 |
| hsa_circ:chr16:69729039-69729282 | NFAT5 | 143 | 149 | 213 | 146 | 66 | 60 | -1.778319789 | 0.000239 | 0.00388 | hsa_circ_0006845 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0006845 |
| hsa_circ:chr9:123593609-123595734 | PSMD5 | 174 | 282 | 304 | 227 | 84 | 98 | -1.780625243 | 5.56E-05 | 0.001203 | hsa_circ_0088300 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0088300 |
| hsa_circ:chr12:50488220-50490755 | SMARCD1 | 115 | 118 | 198 | 105 | 58 | 60 | -1.781723407 | 0.00039 | 0.005678 | hsa_circ_0006535 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0006535 |
| hsa_circ:chr11:61133517-61135470 | TMEM138 | 189 | 146 | 182 | 135 | 81 | 64 | -1.7955221 | 0.000178 | 0.003096 | hsa_circ_0002058 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0002058 |
| hsa_circ:chr11:74500671-74528759 | RNF169 | 110 | 121 | 172 | 64 | 54 | 78 | -1.808809005 | 0.000729 | 0.0091 | hsa_circ_0006705 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0006705 |
| hsa_circ:chr18:12999420-13019205 | CEP192 | 221 | 102 | 184 | 70 | 115 | 72 | -1.815986154 | 0.000885 | 0.010634 | hsa_circ_0000831 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000831 |
| hsa_circ:chr1:65830318-65831879 | DNAJC6 | 295 | 481 | 347 | 160 | 205 | 189 | -1.834491757 | 6.26E-06 | 0.000207 | hsa_circ_0002454 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0002454 |
| hsa_circ:chr6:42571326-42574389 | UBR2 | 112 | 163 | 185 | 66 | 99 | 58 | -1.836536466 | 0.00037 | 0.005439 | hsa_circ_0003177 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0003177 |
| hsa_circ:chr6:163876311-163899928 | QKI | 1657 | 1249 | 1307 | 782 | 652 | 692 | -1.859275235 | 1.03E-09 | 1.30E-07 | hsa_circ_0005328 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0005328 |
| hsa_circ:chr1:46105882-46108171 | GPBP1L1 | 328 | 293 | 612 | 254 | 169 | 167 | -1.860555838 | 1.06E-07 | 6.68E-06 | hsa_circ_0008774 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0008774 |
| hsa_circ:chr13:41400642-41411021 | TPTE2P5 | 245 | 394 | 782 | 293 | 190 | 169 | -1.887843272 | 3.14E-07 | 1.74E-05 | hsa_circ_0030049 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0030049 |
| hsa_circ:chr14:73614503-73614814 | PSEN1 | 320 | 237 | 641 | 234 | 190 | 134 | -1.887868333 | 7.37E-07 | 3.65E-05 | hsa_circ_0003848 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0003848 |
| hsa_circ:chr7:77214860-77230123 | PTPN12 | 495 | 455 | 833 | 414 | 225 | 217 | -1.897059838 | 1.89E-09 | 2.20E-07 | hsa_circ_0003764 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0003764 |
| hsa_circ:chr1:155408118-155429689 | ASH1L | 328 | 269 | 304 | 285 | 97 | 92 | -1.897198961 | 2.31E-05 | 0.000588 | hsa_circ_0003247 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0003247 |
| hsa_circ:chr11:130130751-130131824 | ZBTB44 | 608 | 571 | 848 | 383 | 334 | 251 | -1.89787129 | 5.36E-11 | 1.13E-08 | hsa_circ_0002484 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0002484 |
| hsa_circ:chr1:8601273-8617582 | RERE | 290 | 681 | 464 | 171 | 272 | 208 | -1.931936853 | 3.90E-06 | 0.000142 | hsa_circ_0002158 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0002158 |
| hsa_circ:chr4:3088666-3109150 | HTT | 404 | 453 | 842 | 234 | 347 | 162 | -1.957630898 | 8.89E-08 | 5.81E-06 | hsa_circ_0001392 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001392 |
| hsa_circ:chr4:88116476-88116842 | KLHL8 | 1033 | 1215 | 1827 | 722 | 706 | 411 | -1.963351471 | 9.68E-14 | 5.22E-11 | hsa_circ_0002538 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0002538 |
| hsa_circ:chr2:61749746-61761038 | XPO1 | 959 | 665 | 1209 | 544 | 502 | 253 | -1.984531006 | 4.67E-10 | 6.56E-08 | hsa_circ_0001017 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001017 |
| hsa_circ:chr17:57808782-57816308 | VMP1 | 361 | 318 | 726 | 154 | 211 | 214 | -1.988030719 | 4.03E-07 | 2.19E-05 | hsa_circ_0006508 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0006508 |
| hsa_circ:chr7:35707044-35712888 | HERPUD2 | 340 | 387 | 699 | 234 | 218 | 163 | -1.993507775 | 1.99E-09 | 2.28E-07 | hsa_circ_0001696 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001696 |
| hsa_circ:chr2:72945232-72960247 | EXOC6B | 1431 | 2095 | 3069 | 1431 | 1054 | 480 | -2.001467344 | 1.03E-11 | 2.63E-09 | hsa_circ_0009043 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0009043 |
| hsa_circ:chr6:13579683-13584457 | SIRT5 | 146 | 288 | 318 | 129 | 100 | 97 | -2.004710089 | 2.56E-06 | 0.000103 | hsa_circ_0007218 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0007218 |
| hsa_circ:chr4:25334805-25335610 | ZCCHC4 | 156 | 207 | 280 | 133 | 77 | 73 | -2.022388568 | 2.96E-06 | 0.000115 | hsa_circ_0001398 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001398 |
| hsa_circ:chr5:176370336-176385155 | UIMC1 | 461 | 772 | 1120 | 353 | 216 | 351 | -2.106445561 | 5.79E-10 | 7.74E-08 | hsa_circ_0001558 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001558 |
| hsa_circ:chrX:84322133-84329397 | APOOL | 145 | 198 | 269 | 118 | 83 | 53 | -2.112907281 | 2.78E-06 | 0.000109 | NA | NA |
| hsa_circ:chr17:40652725-40653322 | ATP6V0A1 | 254 | 223 | 372 | 146 | 113 | 90 | -2.113586009 | 5.38E-08 | 3.71E-06 | hsa_circ_0008179 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0008179 |
| hsa_circ:chr9:86293356-86301070 | UBQLN1 | 189 | 163 | 310 | 107 | 79 | 80 | -2.115649688 | 1.13E-06 | 5.25E-05 | hsa_circ_0087357 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0087357 |
| hsa_circ:chr1:24840804-24841057 | RCAN3 | 206 | 210 | 329 | 183 | 56 | 77 | -2.123426738 | 4.46E-06 | 0.000158 | hsa_circ_0003553 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0003553 |
| hsa_circ:chr2:203329532-203332412 | BMPR2 | 484 | 444 | 797 | 269 | 218 | 201 | -2.125658637 | 6.93E-12 | 1.84E-09 | hsa_circ_0003218 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0003218 |
| hsa_circ:chr19:5604594-5604947 | SAFB2 | 442 | 373 | 995 | 333 | 227 | 158 | -2.128204396 | 1.52E-09 | 1.83E-07 | hsa_circ_0000880 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000880 |
| hsa_circ:chr1:231672959-231678357 | TSNAX,TSNAX-DISC1 | 201 | 158 | 286 | 100 | 97 | 62 | -2.140515735 | 1.59E-06 | 6.94E-05 | hsa_circ_0004834 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0004834 |
| hsa_circ:chr15:65471272-65472542 | CLPX | 299 | 355 | 497 | 176 | 139 | 139 | -2.146513558 | 5.34E-10 | 7.32E-08 | hsa_circ_0004374 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0004374 |
| hsa_circ:chr9:96233423-96261168 | FAM120A | 197 | 224 | 255 | 107 | 106 | 60 | -2.162766439 | 6.63E-07 | 3.31E-05 | hsa_circ_0001875 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001875 |
| hsa_circ:chr1:155408118-155408859 | ASH1L | 364 | 271 | 580 | 97 | 204 | 141 | -2.190087424 | 4.80E-07 | 2.52E-05 | hsa_circ_0000137 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000137 |
| hsa_circ:chr15:52073241-52075025 | TMOD2 | 437 | 1044 | 1579 | 504 | 311 | 321 | -2.194525839 | 1.25E-10 | 2.21E-08 | hsa_circ_0005566 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0005566 |
| hsa_circ:chr9:86294690-86301070 | UBQLN1 | 151 | 147 | 200 | 56 | 78 | 50 | -2.242152547 | 8.23E-06 | 0.00026 | hsa_circ_0008207 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0008207 |
| hsa_circ:chr2:148730308-148733544 | ORC4 | 365 | 388 | 689 | 135 | 252 | 126 | -2.242823654 | 1.62E-08 | 1.33E-06 | hsa_circ_0001074 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001074 |
| hsa_circ:chr10:112356156-112358048 | SMC3 | 491 | 172 | 529 | 212 | 188 | 62 | -2.264025134 | 3.61E-06 | 0.000134 | hsa_circ_0000260 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000260 |
| hsa_circ:chr13:28748409-28752072 | PAN3 | 183 | 153 | 260 | 88 | 65 | 62 | -2.294869535 | 4.05E-07 | 2.19E-05 | hsa_circ_0004372 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0004372 |
| hsa_circ:chr4:170523159-170523829 | NEK1 | 176 | 347 | 97 | 96 | 63 | 79 | -2.306289829 | 5.38E-05 | 0.001169 | NA | NA |
| hsa_circ:chr18:19345733-19359646 | MIB1 | 323 | 351 | 426 | 217 | 120 | 77 | -2.315777256 | 1.85E-09 | 2.18E-07 | hsa_circ_0000835 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000835 |
| hsa_circ:chr17:1264386-1265302 | YWHAE | 262 | 157 | 384 | 85 | 136 | 57 | -2.321810669 | 1.49E-06 | 6.58E-05 | hsa_circ_0007643 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0007643 |
| hsa_circ:chr3:119222379-119236162 | TIMMDC1 | 210 | 263 | 481 | 60 | 167 | 80 | -2.342166994 | 1.90E-06 | 8.04E-05 | hsa_circ_0001330 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001330 |
| hsa_circ:chr9:113734353-113735838 | LPAR1 | 2936 | 4423 | 8134 | 1906 | 1654 | 1478 | -2.365204654 | 2.86E-21 | 1.12E-17 | hsa_circ_0087960 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0087960 |
| hsa_circ:chr13:33091994-33101669 | N4BP2L2 | 2837 | 2329 | 4577 | 940 | 1585 | 707 | -2.369990899 | 7.91E-09 | 7.15E-07 | hsa_circ_0000471 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000471 |
| hsa_circ:chr16:69404386-69406258 | TERF2 | 498 | 542 | 898 | 247 | 245 | 159 | -2.374102438 | 1.49E-14 | 8.94E-12 | NA | NA |
| hsa_circ:chr18:76953183-76974038 | ATP9B | 252 | 454 | 450 | 113 | 102 | 158 | -2.391382487 | 1.05E-08 | 9.19E-07 | hsa_circ_0003275 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0003275 |
| hsa_circ:chr13:61013822-61034674 | TDRD3 | 244 | 203 | 403 | 115 | 104 | 65 | -2.395858265 | 5.94E-09 | 5.69E-07 | hsa_circ_0004245 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0004245 |
| hsa_circ:chr12:12397196-12397589 | LRP6 | 188 | 294 | 547 | 165 | 98 | 62 | -2.473349321 | 5.41E-09 | 5.28E-07 | hsa_circ_0000378 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000378 |
| hsa_circ:chr2:201721405-201721708 | CLK1 | 309 | 194 | 574 | 194 | 85 | 64 | -2.506600637 | 2.21E-08 | 1.72E-06 | hsa_circ_0004001 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0004001 |
| hsa_circ:chr6:144858718-144860579 | UTRN | 582 | 657 | 729 | 197 | 171 | 234 | -2.508556777 | 1.32E-13 | 6.45E-11 | hsa_circ_0001647 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001647 |
| hsa_circ:chr9:33953283-33963789 | UBAP2 | 347 | 516 | 608 | 128 | 222 | 97 | -2.509558617 | 2.45E-10 | 3.76E-08 | hsa_circ_0001847 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001847 |
| hsa_circ:chr2:9083316-9114564 | MBOAT2 | 372 | 604 | 682 | 185 | 198 | 106 | -2.583299781 | 1.02E-13 | 5.31E-11 | NA | NA |
| hsa_circ:chr3:119222379-119232566 | TIMMDC1 | 424 | 751 | 949 | 204 | 277 | 109 | -2.644168253 | 9.51E-13 | 3.54E-10 | hsa_circ_0066875 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0066875 |
| hsa_circ:chr6:79770195-79770535 | PHIP | 194 | 377 | 456 | 69 | 136 | 68 | -2.660814144 | 6.00E-09 | 5.69E-07 | hsa_circ_0003810 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0003810 |
| hsa_circ:chr7:158552177-158557544 | ESYT2 | 666 | 659 | 1374 | 205 | 250 | 196 | -2.794191361 | 1.93E-18 | 2.51E-15 | hsa_circ_0001776 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001776 |
| hsa_circ:chr2:167304122-167328955 | SCN7A | 456 | 1950 | 1151 | 123 | 303 | 386 | -2.832543078 | 0.000396 | 0.005733 | NA | NA |
| hsa_circ:chr8:71071740-71075089 | NCOA2 | 342 | 417 | 675 | 121 | 154 | 57 | -2.914152365 | 1.76E-13 | 8.09E-11 | hsa_circ_0001810 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001810 |
| hsa_circ:chr4:144464662-144465125 | SMARCA5 | 5007 | 5266 | 8372 | 1224 | 1054 | 1807 | -2.924894033 | 1.23E-13 | 6.19E-11 | hsa_circ_0001445 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0001445 |
| hsa_circ:chr17:67151160-67152066 | ABCA10 | 327 | 365 | 1210 | 184 | 153 | 73 | -2.95426875 | 2.63E-12 | 7.90E-10 | NA | NA |
| hsa_circ:chr8:71126138-71128999 | NCOA2 | 243 | 465 | 708 | 116 | 107 | 69 | -3.050636317 | 1.82E-15 | 1.43E-12 | NA | NA |
| hsa_circ:chr1:48821342-48825442 | SPATA6 | 1753 | 2831 | 3554 | 736 | 508 | 480 | -3.050955648 | 2.72E-37 | 4.25E-33 | hsa_circ_0008202 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0008202 |
| hsa_circ:chr10:61844360-61845011 | ANK3 | 397 | 537 | 1227 | 154 | 115 | 95 | -3.31743446 | 4.15E-21 | 1.30E-17 | NA | NA |
| hsa_circ:chr6:139264650-139265759 | REPS1 | 420 | 387 | 825 | 82 | 91 | 98 | -3.322250921 | 4.80E-19 | 9.39E-16 | hsa_circ_0004368 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0004368 |
| hsa_circ:chr11:18312989-18314523 | HPS5 | 462 | 281 | 521 | 56 | 61 | 87 | -3.405443326 | 5.65E-15 | 3.84E-12 | hsa_circ_0000280 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0000280 |
| hsa_circ:chr12:121220458-121222396 | SPPL3 | 427 | 685 | 958 | 59 | 80 | 87 | -3.918139861 | 1.22E-25 | 9.58E-22 | hsa_circ_0003472 | http://www.circbase.org/cgi-bin/singlerecord.cgi?id=hsa_circ_0003472 |
Figure 3.
Characterization of circ_0002538 and its function in SCs.
(A) Hierarchical clustering analyses of DEcircRNAs (n = 3). (B, C) RT-PCR verified five circRNAs with upregulated expression and six circRNAs with downregulated expression, and the results were consistent with the RNA-seq data (n = 12). The red dotted box highlights the circRNA of interest. Y-axis: Fold changes in circRNA expression compared with the non-diabetic group. *P < 0.05, **P < 0.01, vs. non-diabetic group (independent-sample t-test). (D) Schematic diagram showing that circ_0002538 was formed by the circularization of KLHL8 exon 2. The red arrow represents the “head-to-tail” splicing site of circ_0002538, confirmed by Sanger sequencing. (E) We used divergent primers and convergent primers to amplify circ_0002538 in cDNA and gDNA. We used β-actin as a negative control. (F) circ_0002538 and KLHL8 mRNA in SCs were detected via RT-PCR after incubation with or without RNase R. Y-axis: fold changes in RNA expression compared with the mock group. ***P < 0.001, vs. mock group (independent-sample t-test). (G) circ_0002538 and KLHL8 mRNA levels were evaluated in the sh-circ_0002538-transfected SCs via RT-PCR. Y-axis: fold changes in RNA expression compared with the sh-NC group. ***P < 0.001, vs. sh-NC group. (H, I) The migrating number of SCs in the sh-circ_0002538 group was lower than that in the sh-NC group in the Transwell assays. **P < 0.01, vs. sh-NC group (independent-sample t-test). Scale bars: 100 μm. All bar graphs represent the average of at least three independent replicates, and the error bars are the SD. cDNA: Complementary DNA; DPN: diabetic peripheral neuropathy; gDNA: genomic DNA; KLHL8: Kelch-like family member 8; sh-circ_0002538: short hairpin RNA for circ_0002538; sh-NC: normal control for short hairpin RNA.
Figure 5.
circ_0002538 regulates the expression of PLLP in vitro and in vivo.
(A) We examined the mRNA expression of the myelination-related genes SERINC5, PLLP, GJC3, PLP1, PRX, and MPZ in the circ_0002538-overexpressing SCs. Y-axis: fold changes in mRNA expression compared with the vector group. *P < 0.05, **P < 0.01, ***P < 0.001, vs. vector group (independent-sample t-test). (B) We used western blot analysis to evaluate the effect of circ_0002538 on PLLP in SCs. (C, D) We examined PLLP expression in the SCs cultured in ox-LDL via RT-PCR and western blotting. Y-axis: fold changes in PLLP mRNA expression compared with the 0 μg/mL group. *P < 0.05, ***P < 0.001, vs. 0 μg/mL group (one-way analysis of variance and Tukey’s post hoc test). (E, F) We evaluated the effect of circ_0002538 on PLLP in SCs cultured with ox-LDL via RT-PCR and western blotting. Y-axis: fold changes in PLLP mRNA expression compared with the control group. **P < 0.01, vs. control group; #P < 0.05, vs. SCs cultured in ox-LDL (one-way analysis of variance and Tukey’s post hoc test). (G, H) We tested PLLP protein expression in peripheral nerve tissues from patients with or without DPN via western blotting (n = 6). Y-axis: Fold changes in PLLP protein expression normalized to β-actin compared with the vector group. **P < 0.01 (independent-sample t-test). (I) The overexpression of circ_0002538 increased the protein expression level of PLLP in the sciatic nerve of mice with DPN (n = 3). (J) Quantification of PLLP by the densitometry of protein bands. *P < 0.05, vs. vector group (paired t-test). All bar graphs represent the average of three independent replicates, and the data are given as the mean ± SD. DPN: Diabetic peripheral neuropathy; GJC3: gap junction protein gamma 3; MPZ: myelin protein zero; ox-LDL-C: oxidized low-density lipoprotein cholesterol; PLLP: plasmolipin; PLP1: proteolipid protein 1; PRX: periaxin; SERINC5: serine incorporator 5; sh-circ_0002538: short hairpin RNA for circ_0002538; sh-NC: normal control for short hairpin RNA.fffc
To further investigate the function of DEcircRNAs in DPN, we focused on circRNA circ_0002538, which showed a 2.14-FC decrease in expression in the DPN group compared with the non-DPN group. circ_0002538 is formed by head-to-tail splicing of exon 2 of the KLHL8 gene, which is located on chromosome 4 (q22.1) (Figure 3D). Sanger sequencing verified the head-to-tail splicing, which was consistent with the data in circBase (Figure 3D). circ_0002538 could be amplified by RT-PCR using divergent primers in cDNA but not in genomic DNA (Figure 3E). circ_0002538 was barely altered after incubation with RNase R comparing to the mock group (Figure 3F), which further confirmed that circ_0002538 has a loop structure.
We confirmed that circ_0002538 expression was decreased in DPN tissues (Figure 3C). Then, we transfected LV-circ_0002538-shRNA into SCs to simulate the pathological process of SCs during DPN. shRNA significantly reduced circ_0002538 expression without affecting the KLHL8 mRNA expression (Figure 3G). We chose sh-circ_0002538 #2 in the following experiments because it had a high inhibitory efficiency compared with the other shRNAs. Migration assays revealed that the knockdown of circ_0002538 impeded the migration of SCs (Figure 3H and I). We further validated the effects of circ_0002538 in the circ_0002538-overexpressing SCs. The expression level of circ_0002538 in these stable overexpression cells was substantially increased, while there was no change in the KLHL8 mRNA level (Additional Figure 3A (777KB, tif) ). Migration assays revealed that the overexpression of circ_0002538 increased the number of SCs that migrated to the lower chamber (Additional Figure 3B (777KB, tif) and C). These findings indicate that circ_0002538 was involved in regulating SC migration in vitro.
Overexpression of circ_0002538 improves the neuropathic phenotype and symptoms of DPN
To further assess the role of circ_0002538 in DPN in vivo, we injected circ_0002538 LV into mice with DPN (Figure 4A). We used a fluorescence microscope to examine GFP-positive cells in the sciatic nerve at the 8th week after surgery, and found that injection of the LV-vector led to long-term transgene expression in the sciatic nerve (Figure 4B). RT-PCR revealed that circ_0002538 expression in the circ_0002538 group was higher than that in the vector group (Figure 4C). Immunofluorescence showed that GFP-positive cells also expressed MPZ protein in the circ_0002538 overexpression group, indicating that circ_0002538 was stably expressed in SCs (Figure 4D).
Figure 4.
Overexpression of circ_0002538 improves demyelination and symptoms of DPN.
(A) Intraoperative images showing the sciatic nerve after the injection of lentiviral solution. Scale bar: 1500 μm. (B) Eight weeks after the injection of lentiviral solution. We observed green fluorescence in the sciatic nerve under a fluorescence microscope. Scale bar: 200 μm. (C) Eight weeks after the injection of LV-circ_0002538, we examined the mRNA expression level of circ_0002538 in the sciatic nerve via RT-PCR (n = 4). Y-axis: fold changes in circ_0002538 expression compared with the vector side. (D) Immunofluorescence staining of MPZ showed that GFP+ cells also expressed MPZ. The arrows indicate the co-localized regions. (E, F) Eight weeks after the injection of lentiviral solution, the mechanical (E) and thermal (F) nociceptive thresholds were evaluated in the circ_0002538 group and the vector group (n = 20). (G, H) SNCV and MNCV were measured in the circ_0002538 group and the control group (n = 20). (I) The number of abnormal myelin sheaths in the circ_0002538 group, detected by transmission electron microscopy, was lower than that in the vector group (n = 4). Arrows indicate abnormal myelin sheaths. Scale bars: 5 μm. (J) Quantification of the ratio of myelin abnormalities in I. The data are given as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, vs. vector side (paired t-test). DAPI: 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride; GFP: green fluorescent protein; MNCV: motor nerve conduction velocity; MPZ: myelin protein zero; SNCV: sensory nerve conduction velocity; TEM: transmission electron microscope.
To further examine the effect of circ_0002538 on the signs and symptoms of DPN in vivo, we conducted behavioral tests and neurophysiological measurements. Compared with the control vector group, the circ_0002538 group showed improved thermal and mechanical thresholds (Figure 4E and F). Electrophysiological records showed that compared with those of the control group, the sensory and motor nerve conduction velocities of the circ_0002538 group were significantly increased (Figure 4G and H). These results demonstrated that the upregulation of circ_0002538 expression improved the function of the sciatic nerve in diabetic mice with DPN. Transmission electron microscopy revealed that the percentage of abnormal myelin sheaths, which manifested as myelin infoldings, vacuolization, and uneven thickness, increased in the DPN group but significantly decreased in the circ_0002538 group (Figure 4I and J). These results suggest that the overexpression of circ_0002538 ameliorated the symptoms of DPN by improving myelination.
Overexpression of circ_0002538 increases PLLP expression
To examine the effect of circ_0002538 on myelination-related proteins, we detected the expression of serine incorporator 5, PLLP, gap junction protein gamma 3, proteolipid protein 1, periaxin, and MPZ in the circ_0002538-overexpressing SCs because protein profiling indicated that these molecules are dysregulated in DPN. RT-PCR showed that circ_0002538 regulated the expression of PLLP, gap junction protein gamma 3, and proteolipid protein 1, and PLLP showed the greatest FC (Figure 5A). Western blotting further revealed that knocking down circ_0002538 led to the downregulation of PLLP expression (Figure 5B left). Accordingly, the overexpression of circ_0002538 increased PLLP protein expression in SCs (Figure 5B right). These results confirmed that circ_0002538 could regulate the expression of PLLP.
To simulate diabetic conditions, we added ox-LDL to the culture medium. RT-PCR revealed decreased PLLP expression in the ox-LDL-cultured SCs. We used 100-μg/mL ox-LDL in the following experiments because it produced a more significant effect (Figure 5C and D). RT-PCR showed that the overexpression of circ_0002538 increased PLLP expression in the SCs cultured with ox-LDL. This was further confirmed by western blotting (Figure 5E and F). We also investigated PLLP expression in the nerve tissues from the patients with DPN via western blots. PLLP expression was significantly downregulated in the nerve tissues of the patients with DPN compared with those without DPN (Figure 5G and H). In addition, the administration of circ_0002538 LV significantly increased the expression of PLLP in the sciatic nerve of the mice with DPN compared with the administration of control LV (Figure 5I and J). These results indicated that circ_0002538 could regulate the expression of PLLP in vitro and in vivo.
PLLP regulates SC migration and myelination
To further verify the role of PLLP in SCs, we transfected a lentiviral vector containing the PLLP gene into SCs. RT-PCR showed that the PLLP overexpression cells had significantly increased PLLP expression, which was further confirmed by the western blotting results (Figure 6A and B). We performed a mRNA-sequencing analysis of the SCs transduced with the LV carrying either PLLP or the control vector. A total of 23448 mRNAs were identified, and 1671 mRNAs met the filtering criteria (P < 0.05, FC ≥ 2) (Additional Table 10 (2MB, pdf) ). The filtered mRNAs were further analyzed using GO analysis for functional prediction (Additional Tables 11 (441.8KB, pdf) –13). GO biological process analysis showed that the filtered mRNAs were significantly enriched in neutrophil migration, regulation of neutrophil migration, positive regulation of neutrophil migration, and positive regulation of leukocyte migration, indicating that PLLP might be related to cell migration (Additional Figure 4A (2.9MB, tif) ). Transwell assays confirmed that the overexpression of PLLP significantly increased SC migration (Figure 6C and D). We further validated the role of PLLP by knocking it down. RT-PCR revealed that PLLP expression was decreased in PLLP knockdown SCs (Figure 6E). Transwell assays showed that the knockdown of PLLP effectively inhibited SC migration (Figure 6F and G). These results indicate that PLLP affects SC migration.
Figure 6.
PLLP regulates SC migration and myelination.
(A, B) RT-PCR and western blotting showed that PLLP expression was increased in the SCs transfected with LV-PLLP. Y-axis: fold changes in mRNA expression compared with the vector group. *P < 0.05, vs. vector group (independent-sample t-test). (C, D) Transwell assays revealed that the number of migrating SCs in the PLLP group was greater than that in the vector group. **P < 0.01, vs. vector group (independent-sample t-test). Scale bars: 100 μm. (E) RT-PCR analysis showed that the mRNA expression of PLLP was decreased in PLLP knockdown SCs. Y-axis: fold changes in mRNA expression compared with the vector group. *P < 0.05, vs. vector group (independent-sample t-test). (F, G) The number of migrating SCs in the sh-PLLP group was low compared with that in the vector group in the Transwell assays. **P < 0.01, vs. vector group (independent-sample t-test). Scale bars: 100 μm. (H) Eight weeks after the injection of LV-vector or LV-sh-PLLP, PLLP expression was examined via western blotting (n = 3). (I) Quantification of PLLP by the densitometry of protein bands. *P < 0.05, vs. vector group (paired t-test). (J) The number of abnormal myelin sheaths in the sh-PLLP group, measured via transmission electron microscopy, was higher than that in the vector group (n = 4). Arrows point to abnormal myelin sheaths. Scale bars: 5 μm. (K) Quantification of the ratio of myelin abnormalities in J. The data are given as the mean ± SD. **P < 0.01, vs. vector group (paired t-test). LV: Lentivirus; PLLP: plasmolipin; sh-PLLP: short hairpin RNA for PLLP; TEM: transmission electron microscope.
Additional Table 13.
GO molecular function analysis of filtered mRNAs
| ONTOLOGY | GOID | Description | GeneRatio | BgRatio | pvalue | p.adjust | geneID | Count |
|---|---|---|---|---|---|---|---|---|
| MF | GO:0005125 | cytokine activity | 23/646 | 218/17258 | 7.74E-06 | 0.00276242 | CXCL11/TNFSF10/TNFSF13/IL1A/IL1B/INHA/EBI3/EDN1/NRG1/AREG/IL6/IL7/LIF/CXCL6/CXCL8/CXCL1/CXCL3/CXCL5/CCL7/TNFRSF11B/CSF3/CCL20/IL12A | 23 |
| MF | GO:0048018 | receptor ligand activity | 37/646 | 454/17258 | 8.15E-06 | 0.00276242 | CXCL11/TNFSF10/TNFSF13/IL1A/IL1B/INHA/EBI3/EDN1/ADA2/ERFE/EREG/PTHLH/SFRP2/NRG1/NRG2/NRG4/AREG/FNDC5/AMH/IL6/IL7/LIF/CXCL6/CXCL8/CXCL1/CXCL3/CXCL5/SPX/UCN/GHRL/CCL7/TNFRSF11B/PSPN/CSF3/CCL20/EPHA7/IL12A | 37 |
| MF | GO:0030545 | receptor regulator activity | 38/646 | 483/17258 | 1.38E-05 | 0.00276242 | CXCL11/TNFSF10/TNFSF13/IL1A/IL1B/INHA/EBI3/EDN1/ADA2/ERFE/EREG/PTHLH/SFRP2/NRG1/NRG2/NRG4/AREG/FNDC5/AMH/IL6/IL7/LIF/CXCL6/CXCL8/CXCL1/CXCL3/CXCL5/WFIKKN1/SPX/UCN/GHRL/CCL7/TNFRSF11B/PSPN/CSF3/CCL20/EPHA7/IL12A | 38 |
| MF | GO:0045236 | CXCR chemokine receptor binding | 6/646 | 16/17258 | 1.56E-05 | 0.00276242 | CXCL11/CXCL6/CXCL8/CXCL1/CXCL3/CXCL5 | 6 |
| MF | GO:0008009 | chemokine activity | 8/646 | 46/17258 | 0.00027339 | 0.02748107 | CXCL11/CXCL6/CXCL8/CXCL1/CXCL3/CXCL5/CCL7/CCL20 | 8 |
| MF | GO:0022836 | gated channel activity | 26/646 | 332/17258 | 0.00033158 | 0.02748107 | CHRNE/CHRNA10/TRPV4/KCND1/KCNG2/KCNE3/KCNQ5/ANO1/ASIC3/JPH2/GRID2/GRIK5/GABRA2/GABRA5/SCN1A/SCN3B/SCN3A/P2RX6/GRIN2C/GRIN2D/GRIN3A/PIEZO2/CLIC6/CLIC2/CATSPER1/GPR89B | 26 |
| MF | GO:0005231 | excitatory extracellular ligand-gatedion channel activity | 8/646 | 48/17258 | 0.0003702 | 0.02748107 | CHRNE/CHRNA10/GRID2/GRIK5/P2RX6/GRIN2C/GRIN2D/GRIN3A | 8 |
| MF | GO:0005230 | extracellular ligand-gated ionchannel activity | 10/646 | 75/17258 | 0.00046517 | 0.02748107 | CHRNE/CHRNA10/GRID2/GRIK5/GABRA2/GABRA5/P2RX6/GRIN2C/GRIN2D/GRIN3A | 10 |
| MF | GO:0043178 | alcohol binding | 10/646 | 75/17258 | 0.00046517 | 0.02748107 | TRPC4/RBP4/RBP1/APOE/NR1H3/SYP/CETP/LRAT/PROM2/OSBP2 | 10 |
| MF | GO:0022824 | transmitter-gated ion channelactivity | 8/646 | 50/17258 | 0.00049322 | 0.02748107 | CHRNE/CHRNA10/GRID2/GRIK5/GABRA2/GRIN2C/GRIN2D/GRIN3A | 8 |
| MF | GO:0022835 | transmitter-gated channel activity | 8/646 | 50/17258 | 0.00049322 | 0.02748107 | CHRNE/CHRNA10/GRID2/GRIK5/GABRA2/GRIN2C/GRIN2D/GRIN3A | 8 |
| MF | GO:0005520 | insulin-like growth factor binding | 6/646 | 28/17258 | 0.00050026 | 0.02748107 | HTRA4/WISP1/IGFBP5/KAZALD1/ITGB4/ITGA6 | 6 |
| MF | GO:0004970 | ionotropic glutamate receptoractivity | 5/646 | 19/17258 | 0.00054341 | 0.02748107 | GRID2/GRIK5/GRIN2C/GRIN2D/GRIN3A | 5 |
| MF | GO:0005234 | extracellularly glutamate-gated ionchannel activity | 5/646 | 19/17258 | 0.00054341 | 0.02748107 | GRID2/GRIK5/GRIN2C/GRIN2D/GRIN3A | 5 |
| MF | GO:0005201 | extracellular matrix structuralconstituent | 10/646 | 78/17258 | 0.00063874 | 0.03014845 | COL14A1/HAPLN3/COL19A1/TECTA/ENAM/ELN/CHI3L1/COL5A3/COL4A3/MATN3 | 10 |
| MF | GO:0005216 | ion channel activity | 30/646 | 425/17258 | 0.00069167 | 0.03025376 | CHRNE/CHRNA10/TRPC4/TRPV4/KCND1/KCNG2/KCNE3/KCNQ5/LRRC8E/SLC40A1/ANO1/ASIC3/JPH2/GRID2/GRIK5/ABCC9/GABRA2/GABRA5/SCN1A/SCN3B/SCN3A/P2RX6/GRIN2C/GRIN2D/GRIN3A/PIEZO2/CLIC6/CLIC2/CATSPER1/GPR89B | 30 |
| MF | GO:0015267 | channel activity | 32/646 | 466/17258 | 0.00074207 | 0.03025376 | CHRNE/CHRNA10/TRPC4/TRPV4/KCND1/KCNG2/KCNE3/KCNQ5/LRRC8E/SLC40A1/ANO1/ASIC3/JPH2/GRID2/GRIK5/ABCC9/GABRA2/GABRA5/SCN1A/SCN3B/SCN3A/BCL2A1/P2RX6/GJB7/GRIN2C/GRIN2D/GRIN3A/PIEZO2/CLIC6/CLIC2/CATSPER1/GPR89B | 32 |
| MF | GO:0022803 | passive transmembrane transporteractivity | 32/646 | 467/17258 | 0.00076916 | 0.03025376 | CHRNE/CHRNA10/TRPC4/TRPV4/KCND1/KCNG2/KCNE3/KCNQ5/LRRC8E/SLC40A1/ANO1/ASIC3/JPH2/GRID2/GRIK5/ABCC9/GABRA2/GABRA5/SCN1A/SCN3B/SCN3A/BCL2A1/P2RX6/GJB7/GRIN2C/GRIN2D/GRIN3A/PIEZO2/CLIC6/CLIC2/CATSPER1/GPR89B | 32 |
| MF | GO:0022838 | substrate-specific channel activity | 30/646 | 435/17258 | 0.00100299 | 0.03737468 | CHRNE/CHRNA10/TRPC4/TRPV4/KCND1/KCNG2/KCNE3/KCNQ5/LRRC8E/SLC40A1/ANO1/ASIC3/JPH2/GRID2/GRIK5/ABCC9/GABRA2/GABRA5/SCN1A/SCN3B/SCN3A/P2RX6/GRIN2C/GRIN2D/GRIN3A/PIEZO2/CLIC6/CLIC2/CATSPER1/GPR89B | 30 |
| MF | GO:0015245 | fatty acid transporter activity | 4/646 | 13/17258 | 0.00106137 | 0.03757264 | MFSD2A/SLCO2A1/FABP3/SLC27A5 | 4 |
| MF | GO:0008083 | growth factor activity | 15/646 | 162/17258 | 0.00113973 | 0.03842505 | INHA/ADA2/EREG/NRG1/NRG2/NRG4/AREG/AMH/IL6/IL7/LIF/CXCL1/PSPN/CSF3/IL12A | 15 |
| MF | GO:0042379 | chemokine receptor binding | 8/646 | 58/17258 | 0.00135494 | 0.04360441 | CXCL11/CXCL6/CXCL8/CXCL1/CXCL3/CXCL5/CCL7/CCL20 | 8 |
| MF | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity | 4/646 | 14/17258 | 0.0014422 | 0.04439455 | B3GALT2/B3GALT1/B3GALT5/B3GNT4 | 4 |
| MF | GO:0005261 | cation channel activity | 23/646 | 314/17258 | 0.00174445 | 0.05146139 | CHRNE/CHRNA10/TRPC4/TRPV4/KCND1/KCNG2/KCNE3/KCNQ5/SLC40A1/ANO1/ASIC3/JPH2/GRIK5/ABCC9/SCN1A/SCN3B/SCN3A/P2RX6/GRIN2C/GRIN2D/GRIN3A/PIEZO2/CATSPER1 | 23 |
| MF | GO:0005319 | lipid transporter activity | 13/646 | 141/17258 | 0.0024669 | 0.06573686 | SLC10A4/RBP4/TMEM30B/APOE/MFSD2A/SLCO2A1/ABCB4/FABP3/SPNS2/CETP/PITPNM3/SLC27A5/ATP10B | 13 |
| MF | GO:0048531 | beta-1,3-galactosyltransferaseactivity | 4/646 | 16/17258 | 0.00247041 | 0.06573686 | B3GALT2/B3GALT1/B3GALT5/B3GNT4 | 4 |
| MF | GO:0005126 | cytokine receptor binding | 20/646 | 266/17258 | 0.00250691 | 0.06573686 | CXCL11/TNFSF10/TNFSF13/IL1A/IL1B/INHA/EBI3/AMH/IL6/IL7/LIF/CXCL6/CXCL8/CXCL1/CXCL3/CXCL5/CCL7/CSF3/CCL20/IL12A | 20 |
| MF | GO:0008066 | glutamate receptor activity | 5/646 | 27/17258 | 0.00294555 | 0.07448045 | GRID2/GRIK5/GRIN2C/GRIN2D/GRIN3A | 5 |
| MF | GO:0019838 | growth factor binding | 12/646 | 130/17258 | 0.00352661 | 0.08609797 | HTRA4/WISP1/FLT4/A2M/ACVR1C/WFIKKN1/S100A13/IGFBP5/KAZALD1/HAP1/ITGB4/ITGA6 | 12 |
| MF | GO:0004222 | metalloendopeptidase activity | 11/646 | 115/17258 | 0.00389625 | 0.09195151 | PAPLN/MMP3/MMP8/TMPRSS6/KEL/ADAMTS14/ADAMTS19/MMP10/TLL2/ADAM28/ADAM33 | 11 |
| MF | GO:0001664 | G-protein coupled receptor binding | 19/646 | 260/17258 | 0.00432576 | 0.09879484 | C3/CXCL11/RAMP1/EDN1/ADA2/WNT16/CXCL6/CXCL8/CXCL1/CXCL3/CXCL5/UCN/NECAB2/GHRL/GNAZ/CCL7/CCL20/GPRC5B/ITGB4 | 19 |
| MF | GO:0015485 | cholesterol binding | 6/646 | 43/17258 | 0.00503575 | 0.11141589 | APOE/NR1H3/SYP/CETP/PROM2/OSBP2 | 6 |
| MF | GO:0015276 | ligand-gated ion channel activity | 12/646 | 139/17258 | 0.00602901 | 0.12554528 | CHRNE/CHRNA10/ASIC3/JPH2/GRID2/GRIK5/GABRA2/GABRA5/P2RX6/GRIN2C/GRIN2D/GRIN3A | 12 |
| MF | GO:0022834 | ligand-gated channel activity | 12/646 | 139/17258 | 0.00602901 | 0.12554528 | CHRNE/CHRNA10/ASIC3/JPH2/GRID2/GRIK5/GABRA2/GABRA5/P2RX6/GRIN2C/GRIN2D/GRIN3A | 12 |
| MF | GO:0061135 | endopeptidase regulator activity | 14/646 | 177/17258 | 0.00682657 | 0.13526148 | C3/SERPINE3/SERPINF1/PAPLN/SERPINB12/BIRC3/SFRP2/NLRC4/A2M/WFIKKN1/COL4A3/SPINT2/SPINT1/HMSD | 14 |
| MF | GO:0086080 | protein binding involved inheterotypic cell-cell adhesion | 3/646 | 11/17258 | 0.0068777 | 0.13526148 | DSC2/CXADR/NFASC | 3 |
| MF | GO:0005496 | steroid binding | 9/646 | 92/17258 | 0.00748345 | 0.13953101 | PAQR5/HSD11B2/ESR1/APOE/NR1H3/SYP/CETP/PROM2/OSBP2 | 9 |
| MF | GO:0005244 | voltage-gated ion channel activity | 15/646 | 198/17258 | 0.00768603 | 0.13953101 | KCND1/KCNG2/KCNE3/KCNQ5/ANO1/SCN1A/SCN3B/SCN3A/GRIN2C/GRIN2D/GRIN3A/CLIC6/CLIC2/CATSPER1/GPR89B | 15 |
| MF | GO:0022832 | voltage-gated channel activity | 15/646 | 198/17258 | 0.00768603 | 0.13953101 | KCND1/KCNG2/KCNE3/KCNQ5/ANO1/SCN1A/SCN3B/SCN3A/GRIN2C/GRIN2D/GRIN3A/CLIC6/CLIC2/CATSPER1/GPR89B | 15 |
| MF | GO:0004867 | serine-type endopeptidase inhibitoractivity | 9/646 | 93/17258 | 0.00801867 | 0.14030831 | SERPINE3/SERPINF1/PAPLN/SERPINB12/A2M/WFIKKN1/SPINT2/SPINT1/HMSD | 9 |
| MF | GO:0004497 | monooxygenase activity | 9/646 | 94/17258 | 0.00858253 | 0.14030831 | CYP4F3/CYP2W1/CYP4X1/AGMO/FMO2/FMO3/CYP11A1/CYP1A1/CYP27A1 | 9 |
| MF | GO:0032934 | sterol binding | 6/646 | 48/17258 | 0.00866409 | 0.14030831 | APOE/NR1H3/SYP/CETP/PROM2/OSBP2 | 6 |
| MF | GO:0005178 | integrin binding | 10/646 | 111/17258 | 0.00874837 | 0.14030831 | ICAM5/WISP1/SFRP2/NRG1/MADCAM1/CXADR/COL4A3/VWF/LCP1/ITGA6 | 10 |
| MF | GO:0005542 | folic acid binding | 3/646 | 12/17258 | 0.0089179 | 0.14030831 | FTCD/GNMT/FTCDNL1 | 3 |
| MF | GO:0031994 | insulin-like growth factor I binding | 3/646 | 12/17258 | 0.0089179 | 0.14030831 | IGFBP5/ITGB4/ITGA6 | 3 |
| MF | GO:0099094 | ligand-gated cation channel activity | 8/646 | 80/17258 | 0.01002526 | 0.15430186 | CHRNE/CHRNA10/ASIC3/JPH2/GRIK5/GRIN2C/GRIN2D/GRIN3A | 8 |
| MF | GO:0008237 | metallopeptidase activity | 14/646 | 186/17258 | 0.01035614 | 0.15600316 | PAPLN/MMP3/MMP8/TMPRSS6/KEL/ADAMTS14/ADAMTS19/MMP10/TLL2/ADAM28/ADAM33/CPB2/CPA5/CPA4 | 14 |
| MF | GO:0043225 | ATPase-coupled aniontransmembrane transporter activity | 3/646 | 13/17258 | 0.01127486 | 0.16630417 | ABCC2/ABCC6/ABCC9 | 3 |
| MF | GO:0004866 | endopeptidase inhibitor activity | 13/646 | 171/17258 | 0.0122209 | 0.17657951 | C3/SERPINE3/SERPINF1/PAPLN/SERPINB12/BIRC3/NLRC4/A2M/WFIKKN1/COL4A3/SPINT2/SPINT1/HMSD | 13 |
| MF | GO:0098631 | cell adhesion mediator activity | 5/646 | 38/17258 | 0.01307493 | 0.18514099 | DSC2/MADCAM1/CXADR/EPCAM/NFASC | 5 |
| MF | GO:0019841 | retinol binding | 3/646 | 14/17258 | 0.01395649 | 0.19374899 | RBP4/RBP1/LRAT | 3 |
| MF | GO:0005507 | copper ion binding | 6/646 | 55/17258 | 0.01644908 | 0.22396053 | SNAI3/IL1A/AOC2/SCO2/SOD3/S100A13 | 6 |
| MF | GO:0005548 | phospholipid transporter activity | 6/646 | 56/17258 | 0.01785861 | 0.23656451 | TMEM30B/MFSD2A/ABCB4/CETP/PITPNM3/ATP10B | 6 |
| MF | GO:0030414 | peptidase inhibitor activity | 13/646 | 180/17258 | 0.01804306 | 0.23656451 | C3/SERPINE3/SERPINF1/PAPLN/SERPINB12/BIRC3/NLRC4/A2M/WFIKKN1/COL4A3/SPINT2/SPINT1/HMSD | 13 |
| MF | GO:0046873 | metal ion transmembranetransporter activity | 26/646 | 449/17258 | 0.01887147 | 0.24292727 | SLC10A4/SLC11A1/CHRNA10/TRPC4/TRPV4/KCND1/KCNG2/KCNE3/KCNQ5/SLC40A1/ASIC3/JPH2/GRIK5/SLC5A2/SLC1A2/ABCC9/SLC34A3/SCN1A/SCN3B/SCN3A/GRIN2C/GRIN2D/GRIN3A/ATP2C2/CLDN16/CATSPER1 | 26 |
| MF | GO:0048038 | quinone binding | 3/646 | 16/17258 | 0.0203141 | 0.25682829 | VKORC1/AOC2/NDUFS7 | 3 |
| MF | GO:0043492 | ATPase activity, coupled tomovement of substances | 10/646 | 129/17258 | 0.02311831 | 0.28715375 | ATP6V1C2/ABCB6/ABCB4/ABCC2/ABCC6/ABCC9/ATP6V0C/ATP6AP1L/ATP10B/ATP2C2 | 10 |
| MF | GO:0035250 | UDP-galactosyltransferase activity | 4/646 | 30/17258 | 0.02462161 | 0.30016121 | B3GALT2/B3GALT1/B3GALT5/B3GNT4 | 4 |
| MF | GO:0042625 | ATPase coupled ion transmembranetransporter activity | 7/646 | 77/17258 | 0.02501343 | 0.30016121 | ATP6V1C2/ABCC2/ABCC6/ABCC9/ATP6V0C/ATP6AP1L/ATP2C2 | 7 |
| MF | GO:0005272 | sodium channel activity | 5/646 | 45/17258 | 0.02573771 | 0.30370495 | ASIC3/GRIK5/SCN1A/SCN3B/SCN3A | 5 |
| MF | GO:0022853 | active ion transmembranetransporter activity | 10/646 | 132/17258 | 0.02659957 | 0.30856562 | SLC10A4/ATP6V1C2/SLC1A2/ABCC2/ABCC6/ABCC9/SLC34A3/ATP6V0C/ATP6AP1L/ATP2C2 | 10 |
| MF | GO:0042626 | ATPase activity, coupled totransmembrane movement ofsubstances | 9/646 | 114/17258 | 0.02730206 | 0.30856562 | ATP6V1C2/ABCB6/ABCB4/ABCC2/ABCC6/ABCC9/ATP6V0C/ATP6AP1L/ATP2C2 | 9 |
| MF | GO:0098632 | cell-cell adhesion mediator activity | 4/646 | 31/17258 | 0.02745711 | 0.30856562 | DSC2/CXADR/EPCAM/NFASC | 4 |
| MF | GO:0016820 | hydrolase activity, acting on acidanhydrides, catalyzingtransmembrane movement ofsubstances | 9/646 | 116/17258 | 0.03010289 | 0.33136558 | ATP6V1C2/ABCB6/ABCB4/ABCC2/ABCC6/ABCC9/ATP6V0C/ATP6AP1L/ATP2C2 | 9 |
| MF | GO:0005544 | calcium-dependent phospholipidbinding | 5/646 | 47/17258 | 0.03042198 | 0.33136558 | RPH3AL/C2CD4D/SYT17/SYTL3/ANXA8L1 | 5 |
| MF | GO:0030594 | neurotransmitter receptor activity | 8/646 | 99/17258 | 0.03218147 | 0.34196627 | CHRNE/CHRNA10/GRID2/GRIK5/GABRA2/GRIN2C/GRIN2D/GRIN3A | 8 |
| MF | GO:0016289 | CoA hydrolase activity | 3/646 | 19/17258 | 0.03236122 | 0.34196627 | NUDT7/THEM5/ACOT1 | 3 |
| MF | GO:0061134 | peptidase regulator activity | 14/646 | 216/17258 | 0.03297354 | 0.34331273 | C3/SERPINE3/SERPINF1/PAPLN/SERPINB12/BIRC3/SFRP2/NLRC4/A2M/WFIKKN1/COL4A3/SPINT2/SPINT1/HMSD | 14 |
| MF | GO:0008378 | galactosyltransferase activity | 4/646 | 34/17258 | 0.03709021 | 0.36985726 | B3GALT2/B3GALT1/B3GALT5/B3GNT4 | 4 |
| MF | GO:0008395 | steroid hydroxylase activity | 4/646 | 34/17258 | 0.03709021 | 0.36985726 | CYP2W1/CYP11A1/CYP1A1/CYP27A1 | 4 |
| MF | GO:0016709 | oxidoreductase activity, acting onpaired donors, with incorporation orreduction of molecular oxygen,NAD(P)H as one donor, andincorporation of one atom of oxygen | 4/646 | 34/17258 | 0.03709021 | 0.36985726 | CYP4F3/FMO2/FMO3/CYP1A1 | 4 |
| MF | GO:0005179 | hormone activity | 9/646 | 121/17258 | 0.03796825 | 0.37335449 | INHA/EDN1/ERFE/PTHLH/FNDC5/AMH/SPX/UCN/GHRL | 9 |
| MF | GO:0015399 | primary active transmembranetransporter activity | 9/646 | 123/17258 | 0.04147331 | 0.38661999 | ATP6V1C2/ABCB6/ABCB4/ABCC2/ABCC6/ABCC9/ATP6V0C/ATP6AP1L/ATP2C2 | 9 |
| MF | GO:0015405 | P-P-bond-hydrolysis-driventransmembrane transporter activity | 9/646 | 123/17258 | 0.04147331 | 0.38661999 | ATP6V1C2/ABCB6/ABCB4/ABCC2/ABCC6/ABCC9/ATP6V0C/ATP6AP1L/ATP2C2 | 9 |
| MF | GO:0001614 | purinergic nucleotide receptoractivity | 3/646 | 21/17258 | 0.04204765 | 0.38661999 | P2RY6/P2RY2/P2RX6 | 3 |
| MF | GO:0016502 | nucleotide receptor activity | 3/646 | 21/17258 | 0.04204765 | 0.38661999 | P2RY6/P2RY2/P2RX6 | 3 |
| MF | GO:0046961 | proton-transporting ATPase activity,rotational mechanism | 3/646 | 21/17258 | 0.04204765 | 0.38661999 | ATP6V1C2/ATP6V0C/ATP6AP1L | 3 |
To verify the effect of PLLP on peripheral nerve myelination in vivo, we injected sh-PLLP LV into the mouse sciatic nerve. Western blotting revealed that PLLP was decreased in the PLLP knockdown group compared with the control vector group (Figure 6H and I). The ratio of myelin abnormalities was strongly increased in the PLLP knockdown group, as shown by transmission electron microscopy (Figure 6J and K). These results indicate that PLLP might regulate myelination in peripheral nerves.
circ_0002538 serves as a sponge for miR-138-5p in SCs
The most common function of circRNAs is to act as sponges for miRNAs, thus regulating downstream target genes. We located circ_0002538 in cellular components via nuclear and cytoplasmic separation experiments. RT-PCR analysis showed that circ_0002538 was predominantly localized in the cytoplasm (Figure 7A), indicating that it might target specific miRNAs to regulate PLLP expression. Forty-eight candidate miRNAs were predicted to bind to PLLP and 130 candidate miRNAs were predicted to bind to circ_0002538 (Additional Tables 14 and 15). After overlapping the candidate miRNAs of PLLP and the candidate miRNAs of circ_0002538, only two miRNAs (miR-138-5p and miR-3714) were found (Figure 7B). We conducted pulldown assays using the biotinylated circ_0002538 probe to verify the interaction between circ_0002538 and the two candidate miRNAs. The circ_0002538 probe effectively pulled down circ_0002538 (Figure 7C), and miR-138-5p was significantly enriched in the circ_0002538 probe sponge complex, while miR-3714 was not significantly enriched (Figure 7D). RT-PCR and agarose gel electrophoresis confirmed that the miR-138-5p probe could prominently pull down circ_0002538 (Figure 7E and F). We further verified this interaction using a dual-luciferase reporter assay. A schematic model showed the putative binding site of circ_0002538 and miR-138-5p (Figure 7G). Luciferase reporter assays demonstrated that miR-138-5p decreased the luciferase activity of HEK293T cells in the wild-type circ_0002538 group but had no effect in the mutant group (Figure 7H), demonstrating the direct binding between circ_0002538 and miR-138-5p in SCs. Taken together, these data demonstrate that circ_0002538 acts as a miRNA sponge for miR-138-5p in SCs.
Figure 7.
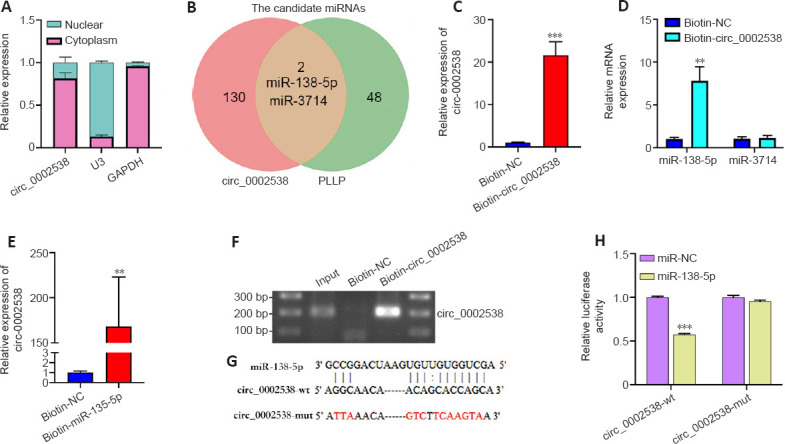
circ_0002538 acts as a sponge for miR-138-5p in SCs.
(A) Nuclear and cytoplasmic separation assays detecting the localization of circ_0002538 in SCs. Y-axis: proportion of nuclear and cytoplasmic RNA to total RNA. (B) Venn diagram showing the overlap of circ_0002538 candidate miRNAs and PLLP candidate miRNAs. (C) circ_0002538 was pulled down in SC lysates by the biotin-circ_0002538 probe and detected via RT-PCR. The relative level of circ_0002538 was normalized to the input. Y-axis: fold changes in circ_0002538 expression compared with the biotin-NC group. ***P < 0.001, vs. Biotin-NC group (independent-sample t-test). (D) miR-138-5p was pulled down by the biotin-circ_0002538 probe, while miR-3714 was not, as shown by the RT-PCR. Y-axis: fold changes in miRNAs expression compared with the biotin-NC group. **P < 0.01, vs. Biotin-NC group (independent-sample t-test). (E, F) circ_0002538 was pulled down in SC lysates by the biotin-miR-138-5p probe, as shown by RT-PCR. The relative level of circ_0002538 was normalized to the input. Y-axis: fold changes in circ_0002538 expression compared with the biotin-NC group. **P < 0.01, vs. Biotin-NC group (independent-sample t-test). (G) The miR-138-5p binding site of circ_0002538 was predicted via RNAhybrid. The mutant sequences are marked in red. (H) Dual-luciferase reporter assays of HEK293T cells cotransfected with miR-138-5p mimics, circ_0002538 wild-type (circ_0002538-wt), or circ_0002538 mutant type (circ_0002538-mut) plasmids. Y-axis: relative luciferase activity compared with the miR-NC + circ_0002538-wt group. ***P < 0.001, vs. miR-NC group (one-way analysis of variance and Tukey’s post hoc test). All bar graphs represent the average of three independent replicates, and the error bars are the SD. GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; mut: mutant type; NC: normal control; PLLP: plasmolipin; U3: small nucleolar U3 RNA; wt: wild-type.
Additional Table 14.
Candidate miRNAs binding to circ_0002538 predicted by RNAhybrid, miRanda and TargetScan
| circRNA | miRNA | miRanda | targetscan | RNAhybrid |
|---|---|---|---|---|
| hsa_circ_0002538 | hsa-miR-519e-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3659 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4659a-3p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-548q | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-let-7g-3p | 1 | 1 | 0 |
| hsa_circ_0002538 | hsa-miR-449c-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3691-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-513c-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-376b-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-298 | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6858-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6501-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-1910-3p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-548as-3p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6826-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-211-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-181c-3p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-145-3p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4677-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4650-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6831-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-548ay-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6770-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6879-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-3689a-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-3154 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-1185-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4632-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-3689b-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-6873-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-6758-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3127-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4446-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-489-3p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4436b-3p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-7978 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3927-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6878-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-4458 | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-4451 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4453 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4456 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6827-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-450b-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-204-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-130a-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4301 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-197-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3670 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-574-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-877-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4504 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-376a-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6777-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-589-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3913-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3118 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4299 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-1304-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3162-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-127-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-let-7a-2-3p | 1 | 1 | 0 |
| hsa_circ_0002538 | hsa-miR-5197-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-146b-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-185-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-4769-3p | 1 | 1 | 0 |
| hsa_circ_0002538 | hsa-miR-22-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-10526-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3925-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3059-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6817-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-2116-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-892c-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4689 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6885-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-605-3p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6735-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-4659b-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4306 | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-4658 | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3610 | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-616-5p | 1 | 1 | 0 |
| hsa_circ_0002538 | hsa-miR-6782-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-548ag | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-1267 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3689e | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-4661-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6808-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-384 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4659b-3p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-3907 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6511a-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6884-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-616-3p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-138-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6780a-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-2117 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3164 | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3714 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-142-3p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-6808-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3680-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-4786-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-744-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4695-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-4758-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6880-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-3132 | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-146a-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6856-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-8070 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4423-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-136-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6755-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6745 | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-4289 | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-134-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6832-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-2682-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6893-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-576-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6760-5p | 1 | 0 | 1 |
| hsa_circ_0002538 | hsa-miR-507 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-3935 | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-499a-3p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-4732-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-942-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-6790-5p | 1 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-1238-5p | 0 | 1 | 1 |
| hsa_circ_0002538 | hsa-miR-7843-3p | 0 | 1 | 1 |
circ_0002538: hsa_circ_0002538; circRNA: circular RNAs; miRNA: microRNA.
Additional Table 15.
The candidate miRNAs binding to PLLP predicted by miRDB, miRTarBase, miRWalk and TargetScan
| miRNA | ID | GENE | miRDB | miRTarBase | miRWalk | TargetScan |
|---|---|---|---|---|---|---|
| hsa-miR-6785-5p | MIMAT0027470 | PLLP | 1 | 1 | 1 | 1 |
| hsa-miR-1258 | MIMAT0005909 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-1273e | MIMAT0018079 | PLLP | 0 | 1 | 1 | 1 |
| hsa-miR-1289 | MIMAT0005879 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-1295b-5p | MIMAT0022293 | PLLP | 0 | 1 | 1 | 1 |
| hsa-miR-138-5p | MIMAT0000430 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-1470 | MIMAT0007348 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-149-3p | MIMAT0004609 | PLLP | 1 | 1 | 1 | 0 |
| hsa-miR-181a-2-3p | MIMAT0004558 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-1827 | MIMAT0006767 | PLLP | 0 | 1 | 1 | 1 |
| hsa-miR-185-3p | MIMAT0004611 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-186-3p | MIMAT0004612 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-18a-5p | MIMAT0000072 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-25-5p | MIMAT0004498 | PLLP | 0 | 1 | 1 | 1 |
| hsa-miR-302f | MIMAT0005932 | PLLP | 0 | 1 | 1 | 1 |
| hsa-miR-30c-1-3p | MIMAT0004674 | PLLP | 0 | 1 | 1 | 1 |
| hsa-miR-3122 | MIMAT0014984 | PLLP | 0 | 1 | 1 | 1 |
| hsa-miR-3714 | MIMAT0018165 | PLLP | 0 | 1 | 1 | 1 |
| hsa-miR-3909 | MIMAT0018183 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-3910 | MIMAT0018184 | PLLP | 0 | 1 | 1 | 1 |
| hsa-miR-3937 | MIMAT0018352 | PLLP | 0 | 1 | 1 | 1 |
| hsa-miR-3975 | MIMAT0019360 | PLLP | 0 | 1 | 1 | 1 |
| hsa-miR-4251 | MIMAT0016883 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-4283 | MIMAT0016914 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-4291 | MIMAT0016922 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-4473 | MIMAT0019000 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-450a-1-3p | MIMAT0022700 | PLLP | 0 | 1 | 1 | 1 |
| hsa-miR-454-3p | MIMAT0003885 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-4667-3p | MIMAT0019744 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-4671-3p | MIMAT0019753 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-4673 | MIMAT0019755 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-4728-5p | MIMAT0019849 | PLLP | 1 | 1 | 1 | 0 |
| hsa-miR-4768-5p | MIMAT0019920 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-512-3p | MIMAT0002823 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-548az-3p | MIMAT0025457 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-582-3p | MIMAT0004797 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-6504-3p | MIMAT0025465 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-6509-3p | MIMAT0025475 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-6513-5p | MIMAT0025482 | PLLP | 0 | 1 | 1 | 1 |
| hsa-miR-654-3p | MIMAT0004814 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-6739-5p | MIMAT0027379 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-6799-5p | MIMAT0027498 | PLLP | 0 | 1 | 1 | 1 |
| hsa-miR-6829-5p | MIMAT0027558 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-6883-5p | MIMAT0027666 | PLLP | 1 | 1 | 1 | 0 |
| hsa-miR-7113-5p | MIMAT0028123 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-7162-5p | MIMAT0028234 | PLLP | 1 | 0 | 1 | 1 |
| hsa-miR-887-5p | MIMAT0026720 | PLLP | 0 | 1 | 1 | 1 |
| hsa-miR-940 | MIMAT0004983 | PLLP | 0 | 1 | 1 | 1 |
PLLP: Plasmolipin.
miR-138-5p inhibits the migration of SCs by targeting PLLP
To investigate the function of miR-138-5p, we transfected miR-138-5p mimic or inhibitor into SCs. In the migration assays, the number of SCs that migrated to the lower chamber was significantly reduced after transfection with the miR-138-5p mimics. In contrast, the miR-138-5p inhibitor enhanced SC migration (Figure 8A and B). Then, we used a dual-luciferase reporter assay to determine whether miR-138-5p could bind to PLLP to regulate its expression. Figure 8C shows the predicted binding sites and mutated sites of miR-138-5p on the 3′UTR of PLLP. The overexpression of miR-138-5p significantly weakened the relative Rluc activity of the wild-type plasmids but not the mutant plasmids (Figure 8D), suggesting that miR-138-5p could directly bind to the PLLP 3′UTR and block its activity. Western blot analysis further demonstrated that the miR-138-5p mimics significantly reduced PLLP protein expression, while the miR-138-5p inhibitors increased PLLP protein expression (Figure 8E). These results revealed that miR-138-5p could strongly suppress SC migration by targeting PLLP.
Figure 8.
miR-138-5p inhibits SC migration by targeting PLLP.
(A) The number of migrating SCs in the miR-138-5p group was less than that in the mimic-NC group via the Transwell assays. Further, the number of migrating SCs in the miR-138-5p-inh group was more than that in the inh-NC group. Scale bars: 100 μm. (B) Quantification of the number of migrating cells in A. *P < 0.05, ***P < 0.001, vs. mimic-NC or inh-NC group (independent-sample t-test). (C) The potential binding site of miR-138-5p on the 3’UTR of PLLP mRNA. The mutant sequences are marked in red. (D) Dual-luciferase reporter assays of HEK293T cells cotransfected with miR-138-5p mimics, PLLP wild-type (PLLP-wt), or PLLP-mutant type (PLLP-mut) plasmids. Y-axis: Relative luciferase activity compared with the miR-NC + PLLP-Wt group. ***P < 0.001, vs. miR-NC group (one-way analysis of variance and Tukey’s post hoc test). (E) We tested PLLP expression via western blotting in the SCs transfected with miR-138-5p mimics or miR-138-5p inhibitor. All bar graphs represent the average of three independent replicates, and the error bars are the SD. inh: Inhibitor; mut: mutant type; NC: normal control; PLLP: plasmolipin; wt: wild-type.
miR-138-5p reverses the effect of circ_0002538 on SCs
We demonstrated that circ_0002538 could sponge miR-138-5p and that miR-138-5p could inhibit SC migration by targeting PLLP. Subsequently, we explored whether circ_0002538 could regulate PLLP through miR-138-5p. The SCs cotransfected with the miR-138-5p mimics and circ_0002538 exhibited decreased migration compared with the SCs transfected with circ_0002538 only (Figure 9A and B), which indicated that ectopic expression of miR-138-5p could partially eliminate the promoting effect of circ_0002538. Western blot analysis showed that the SCs cotransfected with the miR-138-5p mimic and circ_0002538 exhibited reduced PLLP expression compared with the SCs transfected with circ_0002538 only (Figure 9C and D). The above results demonstrated that circ_0002538 regulated SC migration in part by sponging miR-138-5p and subsequently influencing PLLP expression.
Figure 9.
miR-138-5p reverses the circ_0002538-mediated promotion of SCs.
(A) Transwell analysis revealed that circ_0002538 promoted SC migration, but its effect was partially neutralized by the overexpression of miR-138-5p. Scale bars: 100 μm. (B) Quantification of the number of migrating cells in (A). All bar graphs represent the average of three independent replicates, and error bars are the SD. ***P < 0.001, vs. mimic-NC + vector group; ##P < 0.01, ###P < 0.001, vs. miR-138-5p + vector group; &&&P < 0.001, vs. mimic-NC + circ_0002538 group (one-way analysis of variance and Tukey’s post hoc test). (C) Western blot analyses showed that the overexpression of circ_0002538 increased PLLP protein expression, while the ectopic expression of miR-138-5p could partially eliminate this effect. (D) Quantification of PLLP by the densitometry of protein bands. *P < 0.05, vs. mimic-NC + vector group; #P < 0.05, ##P < 0.01, vs. miR-138-5p + vector group; &P < 0.05, vs. mimic-NC + circ_0002538 group (one-way analysis of variance and Tukey’s post hoc test). PLLP: Plasmolipin; NC: normal control.
Discussion
DPN is the most common complication of diabetes, and thus represents a major burden to healthcare systems and society worldwide (Selvarajah et al., 2019). Few studies have been used circRNA sequencing to study the etiology of human DPN. Although nontraumatic amputations are mainly caused by DPN, the actual number of calf amputations each year is not high, limiting the availability of sural nerve samples from individuals with DPN. We collected peripheral nerve tissues from individuals with or without DPN and performed circRNA sequencing and protein profiling. We verified the results of circRNA sequencing and further showed that circ_0002538 could ameliorate symptoms in diabetic mice with DPN by promoting the migration and myelination of SCs. Therefore, our data indicate that the overexpression of circ_0002538 may be a promising treatment for patients with DPN.
Transcriptomic alterations often occur during the pathogenesis and progression of diseases. Previous studies have identified hundreds of differentially expressed genes in patients with static or progressive diabetic neuropathy that are functionally enriched in pathways, including the regulation of axonogenesis and lipid metabolism (Hur et al., 2011). A microarray analysis of the dorsal root ganglia of diabetic rats found that DE mRNAs with downregulated expression were significantly enriched in various biological processes, including myelination, peripheral nervous system myelination, axon guidance, and the regulation of axon production (Guo et al., 2018). Further, aberrantly expressed mRNAs in SCs isolated from the sciatic nerves of diabetic rats were enriched in downregulated biological processes related to myelination, axonogenesis, and axon development (Wang et al., 2020b). In this study, we identified 265 proteins with dysregulated expression in peripheral nerves from DPN patients that were enriched in myelination. SCs provide protection and nutritional support to enable myelinated axons to maintain normal physiological functions, and impaired SC function eventually leads to axonal loss (Dey et al., 2013). Therefore, we focused on the influence of SCs on DPN. We evaluated the function of SCs from patients with diabetes and found that these SCs had reduced migration, consistent with the results of previous studies (Gumy et al., 2008; Jia et al., 2018).
Although circRNAs were originally thought to be byproducts of abnormal splicing events (Cocquerelle et al., 1993), recent studies have shown that certain circRNAs are involved in some important physiological processes. However, the role of circRNAs in the SCs of DPN has rarely been examined, especially in human DPN. Zhang et al. (2020) reported 15 DEcircRNAs in the dorsal root ganglia between wild-type mice and mice with diabetes mellitus. Liu et al. (2019) reported that mmu_circRNA_006636 could relieve high glucose-induced apoptosis and autophagy in RSC96 cells. In our study, 116 circRNAs had downregulated expression and 53 circRNAs had upregulated expression in DPN. Among them, 11 circRNAs were verified, of which circ_0000711 and circ_0006156 were previously reported to play important roles in tumors (Li et al., 2018; Hong et al., 2019; Chen et al., 2020; He et al., 2020), but none were found to be involved in neuropathy. The functions of most verified DEcircRNAs are still unclear. Therefore, more research is needed to explore the potential roles of noncoding RNAs in human DPN.
One of the most common and important functions of circRNAs is to act as competing endogenous RNAs that sequester miRNAs through their binding sites and then modulate the activity of miRNAs on their target genes (Salmena et al., 2011). Although the function of circ_0002538 has not previously been characterized, we found decreased circ_0002538 expression in the nerves of patients with DPN. Further, we found that the overexpression of circ_0002538 improved the symptoms of DPN in diabetic mice. Transmission electron microscopy demonstrated that the administration of circ_0002538 decreased the number of damaged myelin sheaths in DPN, indicating that circ_0002538 might help repair damaged myelin sheaths by improving myelination. According to the GO biological process analysis, the proteins with dysregulated expression identified using protein profiling were significantly enriched in the myelination process, indicating that circ_0002538 improved DPN by regulating myelination-related proteins. The expression of myelination-related proteins was detected in the circ_0002538-overexpressing SCs, which demonstrated that circ_0002538 could regulate the expression of PLLP. Based on the computational predictions and experimental validation of candidate miRNAs binding circ_0002538 and PLLP, we selected miR-138-5p for the construction of competing endogenous RNAs. The circ_0002538-miR-138-5p-PLLP axis was demonstrated using RNA pulldown assays, dual luciferase assays, and a mouse model of DPN. We further verified that circ_0002538 could competitively adsorb miR-138-5p to antagonize its suppression of PLLP.
DPN is involved in deleterious changes in peripheral nerves, such as myelin damage (Cermenati et al., 2012). The myelin sheath is a multilayer membrane produced by SCs that allows efficient transmission of nerve impulses. PLLP has been found to assemble myelin membrane precursor domains via its ability to attract liquid-ordered lipids between the Golgi complex and plasma membrane (Yaffe et al., 2015), and PLLP expression was found to be elevated in nerve stumps following axotomy (Bosse et al., 2003). However, the characteristics and functions of PLLP have not been examined in DPN. In our research, we found that PLLP regulated the migration of SCs, which is an important step preceding myelination and remyelination of the peripheral nervous system (Anliker et al., 2013). Impaired or delayed SC migration contributes to abnormal myelination and demyelination of peripheral nerves (Anliker et al., 2013; Yi et al., 2019). These data are consistent with our finding that silencing PLLP can lead to impaired SC migration and peripheral nerve demyelination in mice. PLLP expression was decreased in diabetic mice with DPN. The increased expression of PLLP, mediated by the overexpression of circ_0002538, improved demyelination. Therefore, we concluded that circ_0002538 and PLLP might play important roles in DPN, and thus might be useful in the development of treatments for demyelinating diseases.
This study had several limitations. First, the number of nerve samples used for sequencing and verification was relatively small. Second, to minimize the influence of other cells, we only used nerve bundles for sequencing and subsequent verification. However, we still cannot completely exclude the influence of other components in peripheral nerves, such as axons, fibroblasts, endothelial cells, and inflammatory cells. Their effects on DPN are the subjects of further research. Third, as circRNAs can interact with different proteins or be translated in a way that mediates their biological roles, further research is needed to identify more circRNAs related to the pathogenesis of DPN. Finally, although we validated the protective effects of circ_0002538 in mice and found an improvement in the neuropathic phenotype and symptoms of DPN, the therapeutic effects on humans need to be verified.
In conclusion, this study reported the results of circRNA sequencing and protein profiling of peripheral nerves from individuals with DPN. As a result, we verified 11 DEcircRNAs in the DPN and control groups. Furthermore, our study demonstrated that circ_0002538 expression was downregulated in patients with DPN and that increased expression of circ_0002538 improved the symptoms of diabetic mice with DPN. Mechanistically, circ_0002538 regulated SC migration and myelination, at least in part, through the miR-138-5p/PLLP axis. Collectively, our study illuminated the key role of the circ_0002538/miR-138-5p/PLLP axis in DPN. Our results provide new insight into the mechanisms and potential treatments for DPN.
Additional files:
Additional Table 1: Primers used in this study.
Additional Table 2: Nucleic acid sequences used in this study.
Additional Table 3: Basic characteristics of patients included in the study.
Additional Table 4 (452KB, pdf) : The differentially expressed proteins analyzed in this study were selected from the results of protein profiling analysis with fold change (FC) > 1.3 and P < 0.05.
The differentially expressed proteins analyzed in this study were selected from the results of protein profiling analysis
Additional Table 5: GO cellular component analysis of differentially expressed proteins.
Additional Table 6 (486.4KB, pdf) : GO biological process analysis of differentially expressed proteins.
GO biological process analysis of differentially expressed proteins
Additional Table 7 (555.4KB, pdf) : GO molecular function analysis of differentially expressed proteins.
GO molecular function analysis of differentially expressed proteins
Additional Table 8: KEGG pathway analysis of differentially expressed proteins.
Additional Table 9: The DEcircRNAs analyzed in this study were selected from the results of circRNA sequencing analysis with FC > 2.0, P<0.01, q < 0.05 and readings ≥ 50.
Additional Table 10 (2MB, pdf) : The differentially expressed mRNAs analyzed in this study were selected from the results of mRNA sequencing analysis.
The differentially expressed mRNAs analyzed in this study were selected from the results of mRNA sequencing analysis
Additional Table 11 (441.8KB, pdf) : GO biological process analysis of filtered mRNAs.
GO biological process analysis of filtered mRNAs
Additional Table 12: GO cellular component analysis of filtered mRNAs.
Additional Table 12.
GO cellular component analysis of filtered mRNAs
| ONTOLOGY | GOID | Description | GeneRatio | BgRatio | pvalue | p.adjust | geneID | Count |
|---|---|---|---|---|---|---|---|---|
| CC | GO:0005578 | proteinaceous extracellular matrix | 28/683 | 365/18399 | 0.00024621 | 0.07164718 | COL14A1/COL17A1/HAPLN3/COL19A1/SERPINF1/TECTA/MMP3/MMP8/FLRT1/WISP1/ENAM/FRAS1/WNT16/FREM3/ELN/CHI3L1/COL5A3/COL4A3/ADAMTS14/ADAMTS19/MMP10/VWF/LRRN3/TNFRSF11B/KAZALD1/MFAP4/MATN3/ITGA6 | 28 |
| CC | GO:0034702 | ion channel complex | 23/683 | 288/18399 | 0.00049174 | 0.07164718 | CHRNE/CHRNA10/TRPC4/KCND1/KCNG2/KCNE3/KCNQ5/LRRC8E/ANO1/GRID2/GRIK5/ABCC9/GABRA2/GABRA5/SCN1A/SCN3B/SCN3A/GRIN2C/GRIN2D/GRIN3A/CLIC6/CLIC2/CATSPER1 | 23 |
| CC | GO:1902495 | transmembrane transportercomplex | 25/683 | 325/18399 | 0.0004987 | 0.07164718 | CHRNE/CHRNA10/TRPC4/KCND1/KCNG2/KCNE3/KCNQ5/LRRC8E/ANO1/GRID2/GRIK5/ABCB6/ABCC9/GABRA2/GABRA5/SCN1A/SCN3B/SCN3A/GRIN2C/GRIN2D/GRIN3A/UQCC3/CLIC6/CLIC2/CATSPER1 | 25 |
| CC | GO:1990351 | transporter complex | 25/683 | 332/18399 | 0.00068024 | 0.07329627 | CHRNE/CHRNA10/TRPC4/KCND1/KCNG2/KCNE3/KCNQ5/LRRC8E/ANO1/GRID2/GRIK5/ABCB6/ABCC9/GABRA2/GABRA5/SCN1A/SCN3B/SCN3A/GRIN2C/GRIN2D/GRIN3A/UQCC3/CLIC6/CLIC2/CATSPER1 | 25 |
| CC | GO:0101003 | ficolin-1-rich granule membrane | 8/683 | 60/18399 | 0.00160995 | 0.13877781 | SLC11A1/SERPINB12/NCKAP1L/ENPP4/ATP6V0C/CD93/PKP1/NFASC | 8 |
| CC | GO:0098802 | plasma membrane receptorcomplex | 15/683 | 177/18399 | 0.00253432 | 0.18204886 | CHRNE/CHRNA10/RAMP1/CARD11/GRID2/GRIK5/ACVR1C/IL6/TLR1/GRIN2C/GRIN2D/GRIN3A/ITGB4/ITGB8/ITGA6 | 15 |
| CC | GO:0005788 | endoplasmic reticulum lumen | 21/683 | 298/18399 | 0.00388676 | 0.23931312 | COL14A1/C3/COL17A1/COL19A1/EBI3/EDN1/CYP2W1/ENAM/COL25A1/APOE/MGAT4A/BCHE/IL6/COL5A3/COL4A3/GHRL/CES1/MELTF/IGFBP5/MATN3/IL12A | 21 |
| CC | GO:0017146 | NMDA selective glutamatereceptor complex | 3/683 | 11/18399 | 0.00672195 | 0.32590943 | GRIN2C/GRIN2D/GRIN3A | 3 |
| CC | GO:0034706 | sodium channel complex | 4/683 | 21/18399 | 0.00680553 | 0.32590943 | GRIK5/SCN1A/SCN3B/SCN3A | 4 |
| CC | GO:0043235 | receptor complex | 21/683 | 325/18399 | 0.0101341 | 0.43677987 | CHRNE/ADRB2/CHRNA10/RAMP1/CARD11/GRID2/GRIK5/FLT4/ACVR1C/IL6/GABRA2/GABRA5/NR1H3/PLXNA4/TLR1/GRIN2C/GRIN2D/GRIN3A/ITGB4/ITGB8/ITGA6 | 21 |
| CC | GO:0001518 | voltage-gated sodium channelcomplex | 3/683 | 14/18399 | 0.0136496 | 0.47861007 | SCN1A/SCN3B/SCN3A | 3 |
| CC | GO:0044420 | extracellular matrix component | 10/683 | 120/18399 | 0.01391592 | 0.47861007 | COL14A1/COL17A1/SERPINF1/FRAS1/FREM3/ELN/COL5A3/COL4A3/MFAP4/ITGA6 | 10 |
| CC | GO:0097060 | synaptic membrane | 19/683 | 295/18399 | 0.01443603 | 0.47861007 | CHRNE/CHRNA10/SNCAIP/TMEM108/BAALC/ANK1/GRID2/GRIK5/GABRA2/GABRA5/SYP/CNKSR2/NDUFS7/GRIN2C/GRIN2D/GRIN3A/DLGAP2/EPHA7/RIMS1 | 19 |
| CC | GO:0030285 | integral component of synapticvesicle membrane | 3/683 | 15/18399 | 0.01659897 | 0.50912388 | TMEM163/GABRA2/SYP | 3 |
| CC | GO:0033267 | axon part | 13/683 | 181/18399 | 0.01771893 | 0.50912388 | SERPINF1/L1CAM/TMEM108/MGARP/NRG1/ANK1/GRIK5/SLC1A2/SCN1A/SYP/UCN/HAP1/NFASC | 13 |
| CC | GO:0030673 | axolemma | 3/683 | 16/18399 | 0.01987621 | 0.53431783 | NRG1/ANK1/SLC1A2 | 3 |
| CC | GO:0045211 | postsynaptic membrane | 15/683 | 225/18399 | 0.02107518 | 0.53431783 | CHRNE/CHRNA10/TMEM108/BAALC/ANK1/GRID2/GRIK5/GABRA2/GABRA5/CNKSR2/GRIN2C/GRIN2D/GRIN3A/DLGAP2/EPHA7 | 15 |
| CC | GO:0030175 | filopodium | 8/683 | 96/18399 | 0.02629834 | 0.58544602 | TRPV4/ACPP/PPP1R9A/CXADR/LCP1/ACTA2/CD302/ITGA6 | 8 |
| CC | GO:0034703 | cation channel complex | 14/683 | 212/18399 | 0.02711827 | 0.58544602 | TRPC4/KCND1/KCNG2/KCNE3/KCNQ5/GRIK5/ABCC9/SCN1A/SCN3B/SCN3A/GRIN2C/GRIN2D/GRIN3A/CATSPER1 | 14 |
| CC | GO:0008328 | ionotropic glutamate receptorcomplex | 5/683 | 46/18399 | 0.02716687 | 0.58544602 | GRID2/GRIK5/GRIN2C/GRIN2D/GRIN3A | 5 |
| CC | GO:0044304 | main axon | 6/683 | 63/18399 | 0.0290586 | 0.59639312 | NRG1/ANK1/SLC1A2/SCN1A/UCN/NFASC | 6 |
| CC | GO:0098878 | neurotransmitter receptorcomplex | 5/683 | 48/18399 | 0.03196607 | 0.62624432 | GRID2/GRIK5/GRIN2C/GRIN2D/GRIN3A | 5 |
| CC | GO:0034707 | chloride channel complex | 5/683 | 49/18399 | 0.03455175 | 0.64746977 | ANO1/GABRA2/GABRA5/CLIC6/CLIC2 | 5 |
| CC | GO:0070820 | tertiary granule | 11/683 | 163/18399 | 0.0408761 | 0.67918917 | SLC11A1/MMP8/SERPINB12/NCKAP1L/ENPP4/CXCL1/ATP6V0C/METTL7A/CD93/PKP1/NFASC | 11 |
| CC | GO:0032809 | neuronal cell body membrane | 3/683 | 21/18399 | 0.04118619 | 0.67918917 | FLRT1/KCNE3/GABRA5 | 3 |
| CC | GO:0044298 | cell body membrane | 3/683 | 21/18399 | 0.04118619 | 0.67918917 | FLRT1/KCNE3/GABRA5 | 3 |
| CC | GO:0005581 | collagen trimer | 7/683 | 88/18399 | 0.04507474 | 0.67918917 | COL14A1/COL17A1/COL19A1/COL25A1/COL5A3/COL4A3/C1QL3 | 7 |
| CC | GO:0016528 | sarcoplasm | 6/683 | 70/18399 | 0.0452783 | 0.67918917 | ANK1/JPH2/FABP3/CLEC18B/GSTM2/MRVI1 | 6 |
| CC | GO:0098563 | intrinsic component of synapticvesicle membrane | 3/683 | 22/18399 | 0.04641385 | 0.67918917 | TMEM163/GABRA2/SYP | 3 |
| CC | GO:0070382 | exocytic vesicle | 11/683 | 167/18399 | 0.04727535 | 0.67918917 | TRIM9/SNCAIP/TMEM163/SLC18A2/SLC40A1/SYN2/STX11/GABRA2/SYP/SYTL3/HAP1 | 11 |
Additional Table 13: GO molecular function analysis of filtered mRNAs.
Additional Table 14: Candidate miRNAs binding to circ_0002538 predicted by RNAhybrid, miRanda and TargetScan.
Additional Table 15: The candidate miRNAs binding to PLLP predicted by miRDB, miRTarBase, miRWalk and TargetScan.
Additional Figure 1 (1.7MB, tif) : Confirmation of DPN in the collected peripheral nerve tissues.
Confirmation of DPN in the collected peripheral nerve tissues.
(A, B) HE staining showed that the number of subcutaneous nerves (arrows) in the skin 10 cm above the lateral malleolus of patients with diabetes was decreased compared with that of patients without diabetes. The arrows point to subcutaneous nerves. Scale bars: 200 µm; 100 µm (high-magnification images). (C, D) The IF micrographs showed IENFD (arrows) in the skin of patients with diabetes was decreased than that of patients without diabetes. The images on the right are the high-magnification images in the square of the images on the left. The arrows pointed to PGP9.5 positive nerve fibers. Scale bars: 50 µm; 25 µm (high-magnification images). (E, F) TEM showed that the number of axons and intact myelin sheaths were decreased in the sural nerves of patients with diabetes. Arrows indicate abnormal myelin sheaths. Scale bars: 10 µm (left); 1 µm (right). Data are expressed as mean ± SD (n = 10). **P < 0.01, **P < 0.001, vs. no-diabetes (independent-sample t-test). HE: Hematoxylin and eosin; IENFD: intraepidermal nerve fiber density; IF: immunofluorescence staining; PGP9.5: protein gene product 9.5; TEM: transmission electron microscopy.
Additional Figure 2 (1.1MB, tif) : Identification of SCs isolated from sural nerves of patients.
Identification of SCs isolated from sural nerves of patients.
(A) The isolated SCs exhibited a long spindle shape under an optical microscope. Scale bar: 200 µm. (B) The positive SC markers, S100B (Fluor® 488) and GFAP (Fluor® 488), indicated that the isolated cells were SCs. Scale bars: 50 µm. DAPI: 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride; GFAP: glial fibrillary acidic protein; IF: immunofluorescence; S100B: S100 calcium binding protein B.
Additional Figure 3 (777KB, tif) : Overexpression of circ_0002538 promoted SC migration.
Overexpression of circ_0002538 promotes SC migration.
(A) As assessed by RT-PCR, circ_0002538 was increased in circ_0002538-overexpressing SCs, while KLHL8 mRNA did not change significantly. Y-axis: fold changes of RNA expressions compared with the vector group. (B, C) Migration assays showed that overexpression of circ_0002538 increased the number of SCs that migrated to the lower chamber. Scale bars: 100 µm. All bar graphs represent the average of three independent replicates, and the error bars are the SD. **P < 0.01, ***P < 0.001, vs. vector group (independent-sample t-test). KLHL8: Kelch-like family member 8; RT-PCR: Real-time polymerase chain reaction; SCs: Schwann cells.
Additional Figure 4 (2.9MB, tif) : The filtered mRNAs in the mRNA sequencing results of the PLLP-overexpressing SCs and the control SCs were further analyzed with GO enrichment analysis.
The filtered mRNAs in the mRNA-sequencing results of the PLLP-overexpressing SCs and the control SCs were further analyzed with GO enrichment analysis.
(A) GO biological process analysis. The red dotted box highlighted the interesting biological process. (B) GO cellular component analysis. (C) GO molecular function analysis. GO: Gene Ontology; PLLP: plasmolipin; SCs: Schwann cells.
Footnotes
Funding: This work was supported by the National Natural Science Foundation of China, Nos. 81772094 (to ZBC), 81974289 (to ZBC); the Key Research and Development Program of Hubei Province, No. 2020BCB031 (to ZBC); and Natural Science Foundation of Hubei Province, No. 2020CFB433 (to YTL).
Conflicts of interest: The authors declare no competing interests.
Availability of data and materials: All data generated or analyzed during this study are included in this published article and its supplementary information files.
C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Yu J, Song LP; T-Editor: Jia Y
References
- 1.Anliker B, Choi JW, Lin ME, Gardell SE, Rivera RR, Kennedy G, Chun J. Lysophosphatidic acid (LPA) and its receptor, LPA1 , influence embryonic schwann cell migration, myelination and cell-to-axon segregation. Glia. 2013;61:2009–2022. doi: 10.1002/glia.22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baum P, Kosacka J, Estrela-Lopis I, Woidt K, Serke H, Paeschke S, Stockinger M, Klöting N, Blüher M, Dorn M, Classen J, Thiery J, Bechmann I, Toyka KV, Nowicki M. The role of nerve inflammation and exogenous iron load in experimental peripheral diabetic neuropathy (PDN) Metabolism. 2016;65:391–405. doi: 10.1016/j.metabol.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Beery AK, Zucker I. Sex bias in neuroscience and biomedical research. Neurosci Biobehav Rev. 2011;35:565–572. doi: 10.1016/j.neubiorev.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosse F, Hasse B, Pippirs U, Greiner-Petter R, Müller HW. Proteolipid plasmolipin:localization in polarized cells, regulated expression and lipid raft association in CNS and PNS myelin. J Neurochem. 2003;86:508–518. doi: 10.1046/j.1471-4159.2003.01870.x. [DOI] [PubMed] [Google Scholar]
- 5.Cermenati G, Abbiati F, Cermenati S, Brioschi E, Volonterio A, Cavaletti G, Saez E, De Fabiani E, Crestani M, Garcia-Segura LM, Melcangi RC, Caruso D, Mitro N. Diabetes-induced myelin abnormalities are associated with an altered lipid pattern:protective effects of LXR activation. J Lipid Res. 2012;53:300–310. doi: 10.1194/jlr.M021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53:55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 7.Charan J, Kantharia ND. How to calculate sample size in animal studies? J Pharmacol Pharmacother. 2013;4:303–306. doi: 10.4103/0976-500X.119726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen KH, Pan JF, Chen ZX, Pan D, Gao T, Huang M, Huang JN. Effects of hsa_circ_0000711 expression level on proliferation and apoptosis of hepatoma cells. Eur Rev Med Pharmacol Sci. 2020;24:4161–4171. doi: 10.26355/eurrev_202004_20996. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Wang X. miRDB:an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 11.Dey I, Midha N, Singh G, Forsyth A, Walsh SK, Singh B, Kumar R, Toth C, Midha R. Diabetic Schwann cells suffer from nerve growth factor and neurotrophin-3 underproduction and poor associability with axons. Glia. 2013;61:1990–1999. doi: 10.1002/glia.22570. [DOI] [PubMed] [Google Scholar]
- 12.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database:prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Fan B, Li C, Szalad A, Wang L, Pan W, Zhang R, Chopp M, Zhang ZG, Liu XS. Mesenchymal stromal cell-derived exosomes ameliorate peripheral neuropathy in a mouse model of diabetes. Diabetologia. 2020;63:431–443. doi: 10.1007/s00125-019-05043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farmer KL, Li C, Dobrowsky RT. Diabetic peripheral neuropathy:should a chaperone accompany our therapeutic approach? Pharmacol Rev. 2012;64:880–900. doi: 10.1124/pr.111.005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldman EL, Callaghan BC, Pop-Busui R, Zochodne DW, Wright DE, Bennett DL, Bril V, Russell JW, Viswanathan V. Diabetic neuropathy. Nat Rev Dis Primers. 2019a;5:41. doi: 10.1038/s41572-019-0092-1. [DOI] [PubMed] [Google Scholar]
- 16.Feldman EL, Callaghan BC, Pop-Busui R, Zochodne DW, Wright DE, Bennett DL, Bril V, Russell JW, Viswanathan V. Diabetic neuropathy. Nat Rev Dis Primers. 2019b;5:42. doi: 10.1038/s41572-019-0097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillen C, Gleichmann M, Greiner-Petter R, Zoidl G, Kupfer S, Bosse F, Auer J, Müller HW. Full-lenth cloning, expression and cellular localization of rat plasmolipin mRNA, a proteolipid of PNS and CNS. Eur J Neurosci. 1996;8:405–414. doi: 10.1111/j.1460-9568.1996.tb01223.x. [DOI] [PubMed] [Google Scholar]
- 18.Goyal SN, Reddy NM, Patil KR, Nakhate KT, Ojha S, Patil CR, Agrawal YO. Challenges and issues with streptozotocin-induced diabetes - A clinically relevant animal model to understand the diabetes pathogenesis and evaluate therapeutics. Chem Biol Interact. 2016;244:49–63. doi: 10.1016/j.cbi.2015.11.032. [DOI] [PubMed] [Google Scholar]
- 19.Gumy LF, Bampton ET, Tolkovsky AM. Hyperglycaemia inhibits Schwann cell proliferation and migration and restricts regeneration of axons and Schwann cells from adult murine DRG. Mol Cell Neurosci. 2008;37:298–311. doi: 10.1016/j.mcn.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Guo G, Liu Y, Ren S, Kang Y, Duscher D, Machens HG, Chen Z. Comprehensive analysis of differentially expressed microRNAs and mRNAs in dorsal root ganglia from streptozotocin-induced diabetic rats. PLoS One. 2018;13:e0202696. doi: 10.1371/journal.pone.0202696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He J, Wang HB, Huang JJ, Zhang L, Li DL, He WY, Xiong QM, Qin ZS. Diabetic neuropathic pain induced by streptozotocin alters the expression profile of non-coding RNAs in the spinal cord of mice as determined by sequencing analysis. Exp Ther Med. 2021;22:775. doi: 10.3892/etm.2021.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He YX, Ju H, Li N, Jiang YF, Zhao WJ, Song TT, Ren WH. Association between hsa_circ_0006156 expression and incidence of gastric cancer. Eur Rev Med Pharmacol Sci. 2020;24:3030–3036. doi: 10.26355/eurrev_202003_20667. [DOI] [PubMed] [Google Scholar]
- 23.Holland NR, Stocks A, Hauer P, Cornblath DR, Griffin JW, McArthur JC. Intraepidermal nerve fiber density in patients with painful sensory neuropathy. Neurology. 1997;48:708–711. doi: 10.1212/wnl.48.3.708. [DOI] [PubMed] [Google Scholar]
- 24.Hong Y, Qin H, Li Y, Zhang Y, Zhuang X, Liu L, Lu K, Li L, Deng X, Liu F, Shi S, Liu G. FNDC3B circular RNA promotes the migration and invasion of gastric cancer cells via the regulation of E-cadherin and CD44 expression. J Cell Physiol. 2019;234:19895–19910. doi: 10.1002/jcp.28588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang HY, Lin YC, Li J, Huang KY, Shrestha S, Hong HC, Tang Y, Chen YG, Jin CN, Yu Y, Xu JT, Li YM, Cai XX, Zhou ZY, Chen XH, Pei YY, Hu L, Su JJ, Cui SD, Wang F, et al. miRTarBase 2020:updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020;48:D148–154. doi: 10.1093/nar/gkz896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hur J, Sullivan KA, Pande M, Hong Y, Sima AA, Jagadish HV, Kretzler M, Feldman EL. The identification of gene expression profiles associated with progression of human diabetic neuropathy. Brain. 2011;134:3222–3235. doi: 10.1093/brain/awr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ii M, Nishimura H, Kusano KF, Qin G, Yoon YS, Wecker A, Asahara T, Losordo DW. Neuronal nitric oxide synthase mediates statin-induced restoration of vasa nervorum and reversal of diabetic neuropathy. Circulation. 2005;112:93–102. doi: 10.1161/CIRCULATIONAHA.104.511964. [DOI] [PubMed] [Google Scholar]
- 29.Jia L, Chopp M, Wang L, Lu X, Szalad A, Zhang ZG. Exosomes derived from high-glucose-stimulated Schwann cells promote development of diabetic peripheral neuropathy. FASEB J. 2018;32:fj201800597R. doi: 10.1096/fj.201800597R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Ni S, Zhou C, Ye M. The expression profile and clinical application potential of hsa_circ_0000711 in colorectal cancer. Cancer Manag Res. 2018;10:2777–2784. doi: 10.2147/CMAR.S172388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Chen X, Yao J, Kang J. Circular RNA ACR relieves high glucose-aroused RSC96 cell apoptosis and autophagy via declining microRNA-145-3p. J Cell Biochem. 2019 doi: 10.1002/jcb.29568. doi:10.1002/jcb.29568. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Lu S, Zhu N, Guo W, Wang X, Li K, Yan J, Jiang C, Han S, Xiang H, Wu X, Liu Y, Xiong H, Chen L, Gong Z, Luo F, Hou W. RNA-Seq revealed a circular RNA-microRNA-mRNA regulatory network in hantaan virus infection. Front Cell Infect Microbiol. 2020;10:97. doi: 10.3389/fcimb.2020.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masocha W, Kombian SB, Edafiogho IO. Evaluation of the antinociceptive activities of enaminone compounds on the formalin and hot plate tests in mice. Sci Rep. 2016;6:21582. doi: 10.1038/srep21582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGeary SE, Lin KS, Shi CY, Pham TM, Bisaria N, Kelley GM, Bartel DP. The biochemical basis of microRNA targeting efficacy. Science. 2019;366:eaav1741. doi: 10.1126/science.aav1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan Z, Li GF, Sun ML, Xie L, Liu D, Zhang Q, Yang XX, Xia S, Liu X, Zhou H, Xue ZY, Zhang M, Hao LY, Zhu LJ, Cao JL. MicroRNA-1224 splicing circularRNA-Filip1l in an Ago2-dependent manner regulates chronic inflammatory pain via targeting Ubr5. J Neurosci. 2019;39:2125–2143. doi: 10.1523/JNEUROSCI.1631-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perlman RL. Mouse models of human disease:An evolutionary perspective. Evol Med Public Health. 2016;2016:170–176. doi: 10.1093/emph/eow014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pop-Busui R, Boulton AJ, Feldman EL, Bril V, Freeman R, Malik RA, Sosenko JM, Ziegler D. Diabetic neuropathy:a position statement by the American Diabetes Association. Diabetes Care. 2017;40:136–154. doi: 10.2337/dc16-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis:the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salzer JL. Schwann cell myelination. Cold Spring Harb Perspect Biol. 2015;7:a020529. doi: 10.1101/cshperspect.a020529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selvarajah D, Kar D, Khunti K, Davies MJ, Scott AR, Walker J, Tesfaye S. Diabetic peripheral neuropathy:advances in diagnosis and strategies for screening and early intervention. Lancet Diabetes Endocrinol. 2019;7:938–948. doi: 10.1016/S2213-8587(19)30081-6. [DOI] [PubMed] [Google Scholar]
- 43.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape:a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi L, Sun BL, Zhang SY, Zhang YQ. Biological function of circular RNA and its role in tissue repair. Zhongguo Zuzhi Gongcheng Yanjiu. 2020;24:2770–2774. [Google Scholar]
- 45.Singh R, Kishore L, Kaur N. Diabetic peripheral neuropathy:current perspective and future directions. Pharmacol Res. 2014;80:21–35. doi: 10.1016/j.phrs.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Tannemaat MR, Eggers R, Hendriks WT, de Ruiter GC, van Heerikhuize JJ, Pool CW, Malessy MJ, Boer GJ, Verhaagen J. Differential effects of lentiviral vector-mediated overexpression of nerve growth factor and glial cell line-derived neurotrophic factor on regenerating sensory and motor axons in the transected peripheral nerve. Eur J Neurosci. 2008;28:1467–1479. doi: 10.1111/j.1460-9568.2008.06452.x. [DOI] [PubMed] [Google Scholar]
- 47.Vlcková-Moravcová E, Bednarík J, Dusek L, Toyka KV, Sommer C. Diagnostic validity of epidermal nerve fiber densities in painful sensory neuropathies. Muscle Nerve. 2008;37:50–60. doi: 10.1002/mus.20889. [DOI] [PubMed] [Google Scholar]
- 48.Wang A, Toma MA, Ma J, Li D, Vij M, Chu T, Wang J, Li X, Xu Landén N. Circular RNA hsa_circ_0084443 is upregulated in diabetic foot ulcer and modulates keratinocyte migration and proliferation. Adv Wound Care (New Rochelle) 2020a;9:145–160. doi: 10.1089/wound.2019.0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C, Xu X, Chen J, Kang Y, Guo J, Duscher D, Yang X, Guo G, Ren S, Xiong H, Yuan M, Jiang T, Machens HG, Chen Z, Chen Y. The construction and analysis of lncRNA-miRNA-mRNA competing endogenous RNA network of Schwann cells in diabetic peripheral neuropathy. Front Bioeng Biotechnol. 2020b;8:490. doi: 10.3389/fbioe.2020.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu ZZ, Kim YH, Bang S, Zhang Y, Berta T, Wang F, Oh SB, Ji RR. Inhibition of mechanical allodynia in neuropathic pain by TLR5-mediated A-fiber blockade. Nat Med. 2015;21:1326–1331. doi: 10.1038/nm.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yaffe Y, Hugger I, Yassaf IN, Shepshelovitch J, Sklan EH, Elkabetz Y, Yeheskel A, Pasmanik-Chor M, Benzing C, Macmillan A, Gaus K, Eshed-Eisenbach Y, Peles E, Hirschberg K. The myelin proteolipid plasmolipin forms oligomers and induces liquid-ordered membranes in the Golgi complex. J Cell Sci. 2015;128:2293–2302. doi: 10.1242/jcs.166249. [DOI] [PubMed] [Google Scholar]
- 52.Yi S, Liu Q, Wang X, Qian T, Wang H, Zha G, Yu J, Wang P, Gu X, Chu D, Li S. Tau modulates Schwann cell proliferation, migration and differentiation following peripheral nerve injury. J Cell Sci. 2019;132:jcs222059. doi: 10.1242/jcs.222059. [DOI] [PubMed] [Google Scholar]
- 53.Zhang HH, Zhang Y, Wang X, Yang P, Zhang BY, Hu S, Xu GY, Hu J. Circular RNA profile in diabetic peripheral neuropathy:analysis of coexpression networks of circular RNAs and mRNAs. Epigenomics. 2020;12:843–857. doi: 10.2217/epi-2020-0011. [DOI] [PubMed] [Google Scholar]
- 54.Zhang SB, Lin SY, Liu M, Liu CC, Ding HH, Sun Y, Ma C, Guo RX, Lv YY, Wu SL, Xu T, Xin WJ. CircAnks1a in the spinal cord regulates hypersensitivity in a rodent model of neuropathic pain. Nat Commun. 2019;10:4119. doi: 10.1038/s41467-019-12049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The differentially expressed proteins analyzed in this study were selected from the results of protein profiling analysis
GO biological process analysis of differentially expressed proteins
GO molecular function analysis of differentially expressed proteins
The differentially expressed mRNAs analyzed in this study were selected from the results of mRNA sequencing analysis
GO biological process analysis of filtered mRNAs
Confirmation of DPN in the collected peripheral nerve tissues.
(A, B) HE staining showed that the number of subcutaneous nerves (arrows) in the skin 10 cm above the lateral malleolus of patients with diabetes was decreased compared with that of patients without diabetes. The arrows point to subcutaneous nerves. Scale bars: 200 µm; 100 µm (high-magnification images). (C, D) The IF micrographs showed IENFD (arrows) in the skin of patients with diabetes was decreased than that of patients without diabetes. The images on the right are the high-magnification images in the square of the images on the left. The arrows pointed to PGP9.5 positive nerve fibers. Scale bars: 50 µm; 25 µm (high-magnification images). (E, F) TEM showed that the number of axons and intact myelin sheaths were decreased in the sural nerves of patients with diabetes. Arrows indicate abnormal myelin sheaths. Scale bars: 10 µm (left); 1 µm (right). Data are expressed as mean ± SD (n = 10). **P < 0.01, **P < 0.001, vs. no-diabetes (independent-sample t-test). HE: Hematoxylin and eosin; IENFD: intraepidermal nerve fiber density; IF: immunofluorescence staining; PGP9.5: protein gene product 9.5; TEM: transmission electron microscopy.
Identification of SCs isolated from sural nerves of patients.
(A) The isolated SCs exhibited a long spindle shape under an optical microscope. Scale bar: 200 µm. (B) The positive SC markers, S100B (Fluor® 488) and GFAP (Fluor® 488), indicated that the isolated cells were SCs. Scale bars: 50 µm. DAPI: 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride; GFAP: glial fibrillary acidic protein; IF: immunofluorescence; S100B: S100 calcium binding protein B.
Overexpression of circ_0002538 promotes SC migration.
(A) As assessed by RT-PCR, circ_0002538 was increased in circ_0002538-overexpressing SCs, while KLHL8 mRNA did not change significantly. Y-axis: fold changes of RNA expressions compared with the vector group. (B, C) Migration assays showed that overexpression of circ_0002538 increased the number of SCs that migrated to the lower chamber. Scale bars: 100 µm. All bar graphs represent the average of three independent replicates, and the error bars are the SD. **P < 0.01, ***P < 0.001, vs. vector group (independent-sample t-test). KLHL8: Kelch-like family member 8; RT-PCR: Real-time polymerase chain reaction; SCs: Schwann cells.
The filtered mRNAs in the mRNA-sequencing results of the PLLP-overexpressing SCs and the control SCs were further analyzed with GO enrichment analysis.
(A) GO biological process analysis. The red dotted box highlighted the interesting biological process. (B) GO cellular component analysis. (C) GO molecular function analysis. GO: Gene Ontology; PLLP: plasmolipin; SCs: Schwann cells.