Abstract
The purpose of this study was to examine possible pathways by which genetic risk associated with externalizing is transmitted in families. We used molecular data to disentangle the genetic and environmental pathways contributing to adolescent externalizing behavior in a sample of 1,111 adolescents (50% female; 719 European and 392 African ancestry) and their parents from the Collaborative Study on the Genetics of Alcoholism. We found evidence for genetic nurture such that parental externalizing polygenic scores were associated with adolescent externalizing behavior, over and above the effect of adolescents’ own externalizing polygenic scores. Mediation analysis indicated that parental externalizing psychopathology partly explained the effect of parental genotype on children’s externalizing behavior. We also found evidence for evocative gene-environment correlation, whereby adolescent externalizing polygenic scores were associated with lower parent-child communication, less parent-child closeness, and lower parental knowledge, controlling for parental genotype. These effects were observed among participants of European ancestry but not African ancestry, likely due to the limited predictive power of polygenic scores across ancestral background. These results demonstrate that in addition to genetic transmission, genes influence offspring behavior through the influence of parental genotypes on their children’s environmental experiences, and the role of children’s genotypes in shaping parent-child relationships.
Keywords: adolescent externalizing, polygenic score, genetic nurture, gene-environment correlation, parenting
Externalizing behaviors or characteristics refer to a constellation of behaviors characterized by disinhibition and antagonism, which can manifest in a variety of psychiatric disorders, including substance use disorders (SUDs), attention-deficit/hyperactivity disorder (ADHD), and conduct disorder (CD; Achenbach, 1966; Lahey et al., 2008). Externalizing behavior and disorders are common (Hamdi & Iacono, 2014) and associated with substantial cost for affected individuals, their families, and society at large (Case & Deaton, 2017; Richmond-Rakerd et al., 2020). Evidence from twin and family studies suggests that externalizing behaviors are strongly influenced by genetic factors, which account for approximately 50–80% of the variation in externalizing spectrum phenotypes (Barr & Dick, 2020) and moderately influenced by environmental factors shared within families (Krueger et al., 2002). Thus, both genetic and environmental factors are critical in the intergenerational transmission of externalizing behaviors.
Interrogation of the specific mechanisms by which environmental factors influence externalizing is complicated by the entangled nature of genetic and environmental influences, as genetic and environmental influences are not independent. Such relationships are termed gene-environment correlations (Jaffee & Price, 2007; Rutter & Silberg, 2002), and manifest in three types: passive, evocative (reactive), and active. In passive gene-environment correlation, genetically related parents pass on genes and also provide an environment influenced by their shared genes, both of which may influence the child’s behaviors. For example, parents may pass on genes that are involved in the development of externalizing behaviors and parenting in a way that is shaped by their own externalizing dispositions. In evocative gene-environment correlation, individuals’ genetic predispositions evoke a response from others, such as when child’s genetically influenced characteristics influence a parent to change their parenting practices in response to that behavior. In active gene-environment correlation, individuals select and shape their environments based on their genotype. For example, a child who has inherited a genetic propensity for externalizing behavior may actively choose to be in peer groups that encourage such behaviors. Of particular interest to the current study, passive gene-environment correlations arise when parents directly pass down genes involved in a given trait/behavior while also creating familial environments that are influenced by the same genetic factors and may confer additional risk (Scarr & McCartney, 1983). Classic twin and adoption studies provide ample evidence of gene-environment correlations (Hambrick & Tucker-Drob, 2015; Jaffee & Price, 2007; Jang et al., 2001; Kendler & Baker, 2007; Klahr & Burt, 2014; Reuben et al., 2016). For example, genes that are directly transmitted to the child that influence temperamental characteristics may also influence the home environment. Lemery-Chalfant and colleagues (2013) found evidence for a genetically mediated association between temperament and home environment such that children higher in effortful control also lived in home environments that were less chaotic.
In recent years, molecular genetic approaches, in combination with advancement in gene identification efforts, have made it possible to disentangle direct and indirect pathways of genetic effects within families. Using measured genotype data, such as genome-wide polygenic scores (PGS), we can disentangle the direct effect of genes transmitted to the offspring from the indirect effects of genes on shaping the environments that may further influence the child’s behavior. Genome-wide polygenic scores (PGS) represent an individual’s aggregate genetic loading for a given trait/behavior using molecular genetic data (Wray et al., 2014). This approach uses results from large discovery genome-wide associations studies (GWAS) to calculate a personalized measure of genetic risk for individuals in the target sample. PGS are then calculated by summing the number of risk-enhancing alleles present across the genome, weighted by the effect size, based upon a discovery GWAS (Bogdan et al., 2018). We can use these polygenic scores to examine the pathways by which genetic risk is transmitted in families.
In their seminal study, Kong and colleagues (2018) used a virtual parent design to demonstrate that alleles that influence educational attainment, but that were not transmitted to the offspring, nevertheless influenced offspring’s educational attainment. They termed the effect of non-transmitted alleles on offspring outcome “genetic nurturance”. Genetic nurture effects refer to the notion that parents’ genotypes, even those not transmitted to their children, play a role in influencing children’s behaviors and outcomes via genetically-influenced socio-environmental pathways (Bates et al., 2018; Kong et al., 2018). This effect has been replicated across different measures of educational attainment in multiple samples (Armstrong-Carter et al., 2020; Bates et al., 2018; de Zeeuw et al., 2020; Wang et al., 2021; Willoughby et al., 2021). Subsequent studies have investigated specific environmental factors that may explain the genetic nurture effects. For example, parental IQ (Willoughby et al., 2021), family socioeconomic status (Armstrong-Carter et al., 2020; Willoughby et al., 2021), and maternal health (Armstrong-Carter et al., 2020) have been shown to mediate the association between parental educational attainment PGS and offspring educational outcomes.
Although a growing list of studies has examined evidence of genetic nurture influencing academic and educational outcomes, fewer studies have sought to disentangle the genetic and environmental influences on externalizing characteristics using PGS. Saunders and colleagues (2021) found that a parental PGS for smoking initiation explained unique variance in offspring frequency of tobacco and alcohol use over and above the offspring’s own PGS, providing evidence of genetic nurturance. Further, the association between parental PGS and offspring substance use was mediated in part by parental substance use and family socioeconomic status. In contrast, de Zeeuw et al. (2020) found no evidence of genetic nurture influencing ADHD, which they attributed to the consistent finding that ADHD is not strongly influenced by the shared environment. Additionally, extant studies of genetic nurturing effects on externalizing related phenotypes have focused on individual externalizing behaviors/disorders in isolation, limiting our ability to determine the extent to which these effects similarly operate on a broad range of externalizing behaviors. This is particularly important given evidence of cross-phenotype prediction (Saunders et al., 2021) which may suggest that polygenic scores for individual externalizing traits capture genetic risk for multiple externalizing phenotypes. Many behaviors and disorders across the externalizing spectrum share an underlying genetic liability (Kendler et al., 2003; Krueger et al., 2002; Young et al., 2000).
In the present study, we used family-based data from the ancestrally diverse Collaborative Study on the Genetics of Alcoholism (COGA) sample to test the pathways through which genetic risk is transmitted in families. We used PGS derived from a recent multivariate GWAS of externalizing (Karlsson Linnér et al., 2021) to examine evidence for the role of genetic nurture on adolescent externalizing behavior. This multivariate GWAS identified 579 conditionally independent loci associated with a latent externalizing factor. Polygenic scores derived from the latent externalizing factor were associated with externalizing factor scores in two independent samples and explained ~9–10% of the variance in externalizing behaviors/problems (Karlsson Linnér et al., 2021). Figure 1 provides a conceptual model of our study, adapted from Saunders et al. (2021), by linking parental genotype and offspring externalizing through both direct genetic transmission and indirect effect of the parental genotype via the environment. We tested whether there were direct and indirect effects of parental polygenic scores on offspring externalizing in adolescence, and examined whether aspects of the home environment mediate genetic risk for externalizing across generations.
Figure 1.
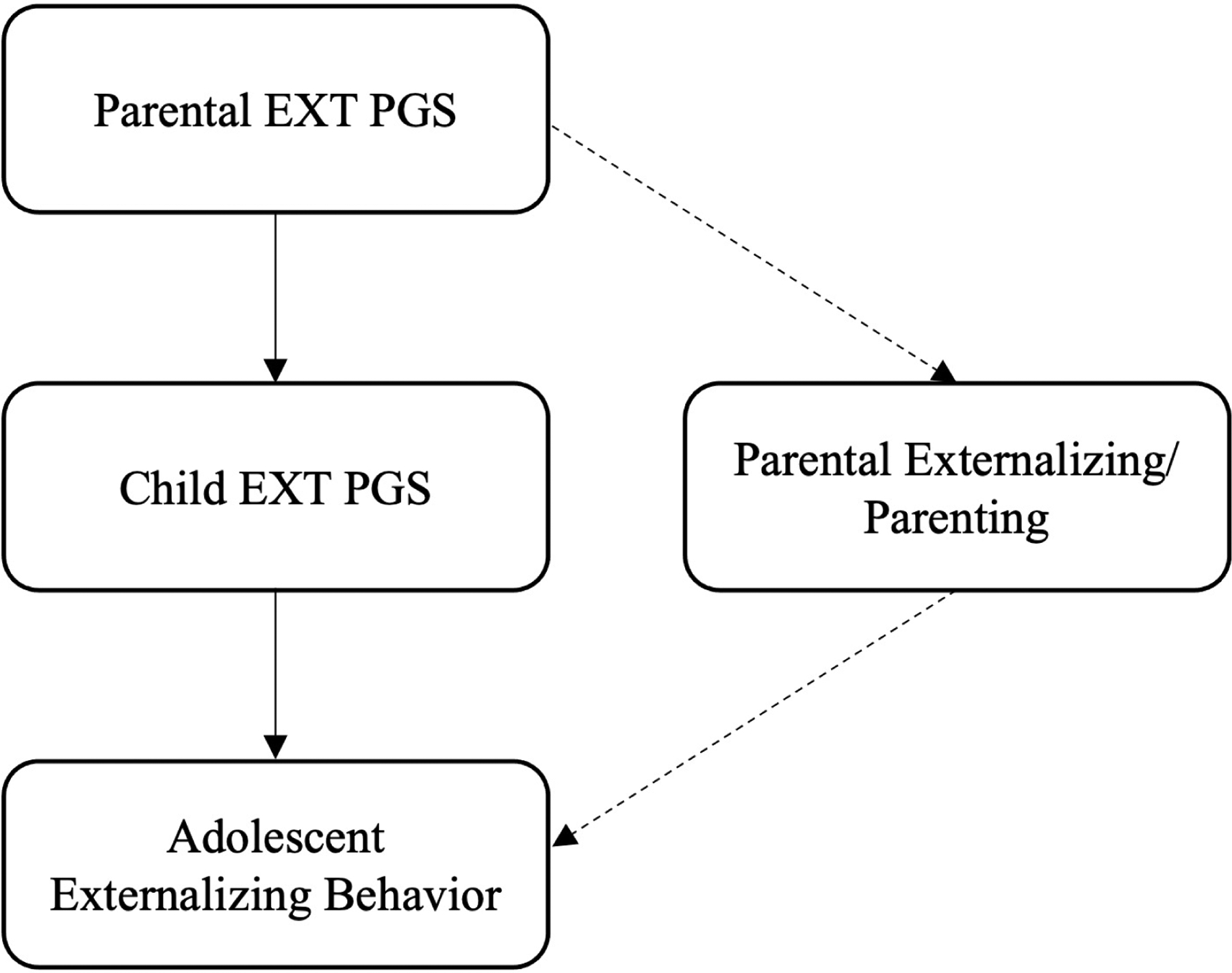
Conceptual model, adapted from Saunders et al. (2021), linking parental genotype and adolescent externalizing behavior. Solid lines represent direct genetic transmission. Dashed lines represent indirect effect of parental genotype on adolescent externalizing behavior through parental externalizing and parenting.
It has been consistently demonstrated that parenting is genetically influenced, with both parental and offspring genotype playing an important role (Avinun & Knafo, 2014; Klahr & Burt, 2014; Wertz et al., 2019). We focused on parental externalizing psychopathology and parenting behaviors as plausible mechanisms linking parental polygenic scores to offspring externalizing behaviors. Investigation of the potential mediating role that parenting and parent-child relationship plays is of particular importance given that parenting behaviors have been consistently linked to childhood and adolescent externalizing problems (Pinquart, 2017; Rothbaum & Weisz, 1994) and are targets of efficacious interventions designed to reduce childhood and adolescent externalizing behaviors (Tully & Hunt, 2016).
In this study, we tested three pre-registered hypotheses (https:/osf.io/ehjy3):
Parental externalizing PGS will be associated with offspring externalizing behavior, after accounting for direct genetic effect (i.e., the effect of adolescent externalizing PGS). The effect of parental externalizing PGS on offspring externalizing behavior independent of offspring’s own PGS would provide evidence of genetic nurture.
Parental externalizing PGS will be associated with parents’ own history of externalizing psychopathology and parenting (i.e., parental involvement, parent-child communication, parent-child closeness, and parental knowledge).
Parental externalizing psychopathology and parenting will partially explain the unique association between parental externalizing PGS and adolescent externalizing behavior.
Methods
Participants came from the Collaborative Study on the Genetics of Alcoholism (COGA) Prospective Study. COGA is a diverse, multi-site, multi-generational family-based study of genetic and environmental factors for alcohol use disorders (Begleiter et al., 1995). Probands (individuals with alcohol dependence) were ascertained through alcohol treatment programs at seven sites in the United States. In addition, a group of non-ascertained comparison families were recruited from the same communities. Beginning in 2004, a sample of adolescent and young adult offspring (ages 12–22) of prior adult COGA participants were recruited into the COGA Prospective Study (Bucholz et al., 2017) to study the development of alcohol use disorders and related problems (Dick et al., 2014). Prospective Study participants were interviewed at enrollment and followed up at approximately biennial intervals. The Institutional Review Board at all sites approved this study, and written consent (and assent for adolescents) was obtained from all participants.
The present study made use of data from the COGA Prospective Study. We included adolescents (aged 12–17) for whom the following apply: (1) had GWAS data available, (2) completed the adolescent version of the Semi-Structured Assessment for the Genetics of Alcoholism for Children (C-SSAGA; Bucholz et al., 1994; Kuperman et al., 2013) during their baseline and first follow-up (approximately two years after the baseline) assessments, (3) were under age 18 at their first follow-up assessment, (4) were of European or African ancestry, as determined by genetic ancestry principal component analysis, and (5) had genotypic data from at least one biological parent.
This strategy resulted in an analytic sample of 1,111 adolescents (50% female) from a total of 739 nuclear families (from 443 COGA extended families). Specifically, this sample included 719 European (EA; Mage = 12.97, SD = 1.13 at baseline assessment; 50% female) and 392 African (AA; Mage = 13.01, SD = 1.12 at baseline assessment; 50% female) ancestry participants. Among the EA sample, parental genomic data were available for 664 (92%) of the mothers and 466 (65%) of the fathers. Among the AA analytic sample, parental genomic data were available for 355 (91%) of the mothers and 176 (45%) of the fathers.
This is a relatively early adolescent sample, as 47.5% of the sample was aged 12 at their baseline assessment. The interval between adolescents’ baseline assessment and their first follow-up assessment was, on average, 2.08 years apart (SD = .55).
Measures
Adolescents completed the C-SSAGA, a comprehensive interview that assesses demographic and home environmental factors, as well as alcohol use and externalizing related behaviors (Bucholz et al., 1994) at the baseline and first follow-up assessment.
Adolescent Externalizing Behaviors.
We created an externalizing behavior/disorder composite measure based on both non-clinical and clinical indicators measured in the first follow-up (when offspring were, on average, aged 15) C-SSAGA interview, which has demonstrated reliability and validity (Bucholz et al., 1994; Hesselbrock et al., 1999): (1) alcohol use, (2) marijuana use, (3) cigarette use, (4) DSM-IV clinical criterion counts (American Psychiatric Association, 1994) of conduct disorder (CD) criteria, and (5) DSM-IV oppositional defiant disorder (ODD criterion counts). Rather than using the more severe clinical-level substance use problems as sole indices of the externalizing behaviors, we used developmentally-appropriate substance use variables to capture more variability in externalizing behavior in our sample of young adolescents. Alcohol use was measured in the C-SSAGA by asking individuals to rate the frequency of past-year drinking on a 12-point scale from 1 (about 1 to 2 days a year) to 12 (every day). Non-drinkers were coded as zero. Marijuana use was assessed by asking participants to report the number of times they used marijuana in the last 12 months. Cigarette use was coded as 1 = past-year use, and 0 = no cigarette use in the past year. We include DSM-IV CD and ODD symptoms because they index externalizing behaviors and problems in youth. Descriptive statistics for externalizing indicators are summarized in Table 1. The composite measure was created by standardizing each indicator and averaging across the indices.
Table 1.
Descriptive statistics for adolescent externalizing indicators
| Indicators | Mean | SD |
|---|---|---|
| Alcohol use | .88 | 1.81 |
| Marijuana use | 12.38 | 57.49 |
| Cigarette usea | 8.6% | - |
| Conduct disorder criterion count | .65 | 1.20 |
| Oppositional defiant disorder criterion count | .92 | 1.53 |
Note. Abbreviations: SD = standard deviation.
For binary variable, proportion of response category = 1 were reported.
Alcohol use was measured by asking individuals to report the frequency of past-year use on a 12-point scale. Non-drinkers were coded as zero. Marijuana use was assessed by asking participants to report the number of times they used marijuana in the last 12 months. Cigarette use was coded as 1 = past-year use, and 0 = no cigarette use in the past year.
Parenting Measures.
Data on parenting behaviors came from items included in the Home Environment section of the adolescents’ baseline C-SSAGA interview that assesses characteristics and features of the home environment and family relationships (e.g., parent-child), adapted into the C-SSAGA from the Home Environment Interview for Children (Reich et al., 1988; Reich & Earls, 1982). These scales have been used in previous COGA studies (Su et al., 2018).
Adolescents were asked to report on their relationship with the two people who play major parent roles in their life at home. They were asked to indicate the nature of the relationships for whom they were reporting using a freeform question, “For this part of the interview, I’d like you to tell me the two people who play the major parent roles in your life at home. It could be your biological mother and father, a stepmother or stepfather, or another relative such as grandparents.” Some adolescents indicated non-biological mother (n = 63) or non-biological father (n= 324) as their parental figures. Because our analyses incorporate parental genotypic data from biological parents, adolescent-reported parenting behaviors from non-biological parents as their parental figures were set to missing.
Parental Involvement.
Parental involvement was assessed using 5 questions. These questions asked adolescents whether their mother/father figure helped them with schoolwork, chores, fun activities, shopping, and making plans. Response options were 0 (no) and 1 (yes). Scores were summed to create maternal and paternal involvement. Cronbach’s alphas were .60 for maternal and .61 for paternal involvement.
Parent-Child Communication.
Parent-child communication was assessed using 3 questions. These items asked adolescents whether they and their mother/father figure talked about news, problems/worries, and other topics such as movies, friends, or anything else. Response options were 0 (no) and 1 (yes). Scores were summed to create mother-child and father-child communication. Cronbach’s alphas were .56 for mother-child and .58 for father-child communication.
Parent-Child Closeness.
Parent-child closeness was assessed using 2 questions. Adolescents were asked how well they got along with their mother/father figure most of the time, and the response options ranged from 1 (poor) to 4 (excellent). The second item asked how close participants felt to their father/mother figure, and response options ranged from 1 (not at all close) to 3 (very close). Scores were standardized and averaged across the two items to create variables indexing mother-child and father-child closeness. Cronbach’s alphas were .63 for mother-child and .69 father-child closeness.
Parental Knowledge.
Parental knowledge was assessed via participants’ responses to three questions (how much their parent figures know about their plans, their interests, and where and with whom they spend time when not at home) adapted from Chassin et al. (1993), and included as part of the C-SSAGA. Responses were rated on a 4-point scale from 1 (always) to 4 (rarely). Items were reversed coded and averaged, and higher scores indicated higher parental knowledge. Cronbach’s alpha was .70.
Parental Externalizing Disorder Composite.
Data on parental history of externalizing related disorders came from parents’ SSAGA interviews (Bucholz et al., 1994; Hesselbrock et al., 1999) from earlier phases of the COGA study. Among the EA analytic sample, parental SSAGA data were available for 666 (93%) of the mothers (age at last assessment, M = 33.72 years, SD = 7.05) and 511 (71%) of the fathers (age at last assessment, M = 36.33 years, SD = 7.39). Among the AA analytic sample, parental SSAGA data were available for 355 (91%) of the mothers (age at last assessment, M = 32.00 years, SD = 6.76) and 184 (47%) of the fathers (age at last assessment, M = 33.72 years, SD = 7.90).
The parental externalizing-disorder composite was created based on lifetime DSM-IV (American Psychiatric Association, 1994) alcohol dependence and abuse criteria, antisocial personality disorder criteria, and drug (cocaine, marijuana, sedatives, stimulants, opiates, and other substances) dependence and abuse criteria. Criterion counts were obtained from the SSAGA interviews (Bucholz et al., 1994; Hesselbrock et al., 1999). For participants for whom multiple assessments were available, data from the interview in which they endorsed the greatest number of alcohol dependence criterion counts were used. Bivariate correlations between externalizing indicators ranged between .54 and .84. Component scores from a principal component analysis were extracted to index externalizing disorders. The first principal component accounted for 68% of the common variance among the externalizing disorder variables with the following loadings: alcohol dependence, 0.86; alcohol abuse, 0.86; antisocial behavior, 0.80; drug dependence, 0.82; drug abuse, 0.79 (detailed in Salvatore et al., 2015).
Genotyping and Externalizing Polygenic Scores.
Participants’ DNA samples were genotyped using the Illumina Human1M array (Illumina, San Diego, CA), the Illumina Human OmniExpress 12V1 array (Illumina), the Illumina 2.5M array (Illumina) or the Smokescreen genotyping array (Biorealm LLC, Walnut, CA; Baurley et al., 2016). Samples were imputed to 1000 Genomes using the cosmopolitan reference panel (Phase 3) using SHAPEIT2 (Delaneau et al., 2013) and Minimac3 (Das et al., 2016) within each array. Variants with non-A/T or C/G alleles, missing rates <5%, minor allele frequencies >3%, and Hardy-Weinberg equilibrium p-values >0.0001 were used for imputation. Imputed variants with information scores <0.30, missing rate >25%, minor allele frequency <1%, or Hardy-Weinberg equilibrium (p < 10−6) were excluded from analysis. A full description of data processing, quality control, and imputation is available elsewhere (Lai et al., 2019; Lai et al., 2022).
To avoid population stratification (Cardon & Palmer, 2003), we conducted our analyses separately by ancestry group. Genetic ancestry principal components were computed from genetic variants using Eigenstrat (Price et al., 2006). These principal components reflect continuous variation in allele frequencies, which represent ancestral differences. Individuals were assigned an ancestry classification (European, African, or Other) based on the first two principal components (Lai et al., 2019). Our analyses included participants of European (EA) and African (AA) ancestry, the two largest groups in COGA.
Genetic risk for externalizing problems was assessed by constructing polygenic scores (PGS), which are aggregate measures of the number of risk alleles individuals carry, weighted by effect sizes from GWAS summary statistics. We used summary statistics from discovery GWAS to construct genome-wide polygenic scores using PRS-CS (Ge et al., 2019). This approach employs a Bayesian regression and continuous shrinkage method to correct for the non-independence among nearby SNPs in the genome (i.e., linkage disequilibrium or LD).
For EA participants, we calculated externalizing polygenic scores using estimates from the recent multivariate genomic analysis results of externalizing behaviors/ problems (including alcohol problems, ADHD, lifetime cannabis use, age of first sexual intercourse, number of sexual partners, general risk tolerance, and lifetime smoking initiation) with an effective sample size of ~1.5 million individuals of European ancestry, from the international Externalizing Consortium (Karlsson Linnér et al., 2021).
For AA participants, an ancestry-matched GWAS similar to the one utilized in those of European ancestries is currently unavailable. Thus, we derived the PGS based on the weights from the discovery GWAS of European ancestry individuals (Karlsson Linnér et al., 2021). We recognize the limitation of this approach, particularly in view of prior evidence that PGS are most predictive when individuals in the discovery GWAS and the target sample are matched on ancestral background (Martin et al., 2017; Peterson et al., 2019); the Discussion section further elaborates on these issues.
To account for population stratification, we regressed each polygenic score on the first 10 genetic ancestry principal components (PC1-PC10) and saved the residualized PGS. Analyses were conducted separately by ancestry. We used standardized, residualized polygenic scores in subsequent analyses.
Analytic Strategy
We first calculated parental externalizing polygenic scores, parental externalizing disorder composite, and parenting related measures as the mean of mother and father on a given variable. In instances where information was only available for one parent, the available score was used. We employed this “midparent” approach for two reasons: (1) to make maximal use of the available data in order to maximize the sample size, and (2) because we made no specific hypotheses regarding maternal versus paternal genetic nurture effects. This strategy is also consistent with other prior studies of genetic nurturance (e.g., Saunders et al., 2021; Willoughby et al., 2021).
We used linear regression analyses to examine the associations between parental externalizing polygenic score and adolescent externalizing polygenic score in predicting adolescent externalizing behavior. We also used linear regression models to test the associations between parental externalizing polygenic score and adolescent externalizing polygenic score in predicting parental externalizing and parenting measures (i.e., parental involvement, parent-child communication, parent-child closeness, and parental knowledge). Analyses were conducted separately for each parenting measure. Covariates included adolescent age and sex. These association analyses were conducted in Mplus version 8.3 (Muthén & Muthén, 1998–2017) with the CLUSTER command, which accounts for the nesting of individuals within families.
We next examined the possible pathways through which parental externalizing polygenic scores may be associated with adolescent externalizing behavior, controlling for adolescent externalizing polygenic scores. Covariates included adolescent age and sex. We conducted mediation analyses using the mediation package (Tingley et al., 2020) for R (R Development Core Team, 2019) with clustered standard errors at the family level. These mediation analyses tested the extent to which the association between parental externalizing polygenic score and adolescent externalizing behavior is explained by parental externalizing psychopathology and parenting measures (i.e., parental involvement, parent-child communication, parent-child closeness, and parental knowledge). Each potential mediator was tested in a separate model. Confidence intervals for indirect effects were calculated via the bootstrap method based on 5000 replications.
We conducted all analyses separately by ancestry to minimize the issue of population stratification. In view of our pre-registered hypotheses, we used a p-value threshold of p < .05 for inference criteria.
Results
Table 2 presents the descriptive statistics and zero-order correlations between the study variables for each ancestry group, separately.
Table 2.
Descriptive statistics and zero-order correlations among study variables for European ancestry and African ancestry samples
| Measure | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EA | AA | EA | AA | EA | AA | EA | AA | EA | AA | EA | AA | EA | AA | EA | AA | |
| 1. Adolescent EXT PGS | – | – | ||||||||||||||
| 2. Parental EXT PGS | .55 | .51 | – | – | ||||||||||||
| 3. Adolescent Externalizing | .23 | .01 | .19 | .03 | – | – | ||||||||||
| 4. Parental Externalizing | .30 | .06 | .35 | .11 | .30 | .10 | – | – | ||||||||
| 5. Parental Involvement | −.06 | −.04 | −.03 | −.07 | −.13 | −.13 | −.05 | −.05 | – | – | ||||||
| 6. Parent-child Communication | −.09 | −.05 | −.05 | −.02 | −.10 | −.09 | −.07 | .03 | .45 | .38 | – | – | ||||
| 7. Parent-child Closeness | −.12 | −.05 | −.09 | −.12 | −.25 | −.15 | −.15 | −.04 | .39 | .33 | .35 | .40 | – | – | ||
| 8. Parental Knowledge | −.12 | .00 | −.07 | −.05 | −.38 | −.26 | −.18 | −.07 | .28 | .30 | .30 | .33 | .44 | 0.39 | – | – |
| Mean | 0 | 0 | 0 | 0 | −0.04 | 0.09 | 0.06 | 0.10 | 3.95 | 3.96 | 2.22 | 2.23 | 0.11 | 0.06 | 3.49 | 3.27 |
| SD | 1 | 1 | 0.84 | 0.89 | 0.69 | 0.56 | 0.84 | 0.89 | 1.03 | 1.09 | 0.88 | 0.87 | 0.69 | 0.8 | 0.57 | 0.73 |
Note. Abbreviations: SD = standard deviation. EXT PGS = Externalizing polygenic score. EA = participants of European ancestry. AA = participants of African ancestry. Correlations and descriptive statistics for the AA sample are presented in the grey shaded columns. PGS residualized on first 10 genetic ancestry principal components. In the AA sample, EXT PGS was constructed based on the weights from the discovery GWAS of European ancestry individuals.
Bolded correlations indicate p < .05
In the EA sample, parental EXT PGS was corelated with their own externalizing behavior, adolescent externalizing behavior, and parent-child closeness, but not with other aspects of parenting. Adolescent EXT PGS was correlated with their own externalizing behavior, and negatively correlated with all aspects of parenting except parental involvement (i.e., higher externalizing genetic risk was associated with less parent-child communication, closeness, and knowledge). Parental externalizing behavior was correlated with lower parent-child closeness and lower parental knowledge. Adolescent externalizing behavior was correlated negatively with all parenting measures.
In the AA sample, parental EXT PGS was correlated with their own externalizing behavior, but was not correlated with adolescents’ externalizing behavior. Adolescent EXT PGS was not correlated with any parenting measure. Parental EXT PGS was correlated with lower parent-child closeness, but not other aspects of parenting. Adolescent externalizing behavior was correlated with lower parental involvement, lower parent-child closeness, and lower parental knowledge.
Analyses of genetic nurture effect
To examine genetic nurture effects, we present the results from the model testing the effect of adolescent EXT PGS on externalizing behavior without the inclusion of parental genotype, which would lead genetic nurture effects to be assumed in the estimate of adolescent EXT PGS if it existed. We then fit models that included both adolescent EXT PGS and parental EXT PGS, which tests the effect of parental EXT PGS that uniquely predicts their children’s externalizing behavior in a way that is not explained by adolescent’s own EXT PGS. A unique effect from parental EXT PGS in the combined model would provide evidence of genetic nurture. Table 3 summarizes the results from these analyses for each ancestry group, separately.
Table 3.
Associations between adolescent and parental externalizing polygenic scores in predicting adolescent externalizing behavior in the European and African ancestry samples
| Adolescent PGS alone | Adolescent and Parental PGS combined model | |||||
|---|---|---|---|---|---|---|
| Adolescent PGS | Parental PGS | |||||
| β | [95% CI] | β | [95% CI] | β | [95% CI] | |
| European Ancestry | .23 | [.15, .31] | .18 | [.09, .26] | .09 | [.01, .17] |
| African Ancestry | .00 | [−.09, .09] | −.01 | [−.12, .09] | .02 | [−.12, .16] |
Note. PGS = Externalizing polygenic score. All PGS residualized on first 10 genetic ancestry principal components. Standardized coefficients along with 95% confidence interval are shown. Adolescent and parental PGS combined model indicates that both adolescent and parental PGS were included in the same model. All models controlled for adolescent sex and adolescent age.
Bold indicates p < .05
Among EA participants, adolescent EXT PGS predicted the adolescent’s externalizing behavior (β = .23, [95% CI .15, .31]) and accounted for 5% of the variance after we controlled adolescent sex and age. In the combined model, parental EXT PGS added incremental R2 to adolescent EXT PGS in predicting adolescent externalizing behavior, raising total variance accounted for in outcome to 6%. Parental EXT PGS was associated with adolescent externalizing behavior (β = .09, [95% CI .01, .17], over and above adolescent’s own EXT PGS (β = .18, [95% CI .09, .26]. These results indicate that parental EXT PGS uniquely predicted their children’s externalizing behavior beyond the variance associated with adolescents’ own EXT PGS, providing evidence for genetic nurture.
Among AA participants, adolescent EXT PGS was not associated with adolescent externalizing behavior. In the combined model, neither parental EXT PGS nor adolescent EXT PGS was associated with adolescent externalizing behavior.
Associations between parental and adolescent polygenic scores with parental externalizing behavior and parenting
Table 4 summarizes the associations of parental EXT PGS and adolescent EXT PGS with parental externalizing behavior and parenting, separately by ancestry.
Table 4.
Associations between parental and adolescent externalizing polygenic scores for parental externalizing and parenting measures in the European and African ancestry samples
| Parental PGS alone | Parental and Adolescent PGS combined model | |||||
|---|---|---|---|---|---|---|
| Parental PGS | Adolescent PGS | |||||
| EA sample | β | [95% CI] | β | [95% CI] | β | [95% CI] |
| Parental Externalizing | .35 | [.25,.44] | .26 | [.15, .37] | .15 | [.07, .24] |
| Parental Involvement | −.03 | [−.10, .04] | .00 | [−.09, .10] | −.06 | [−.16, .05] |
| Parent-child Communication | −.06 | [−.14, .03] | −.01 | [−.11, .09] | −.09 | [−.17, −.01] |
| Parent-child Closeness | −.09 | [−.17, .00] | −.03 | [−.13, .08] | −.10 | [−.21, −.00] |
| Parental Knowledge | −.07 | [−.14, .00] | −.00 | [−.09, .09] | −.12 | [−.22, −.02] |
| Parental PGS alone | Parental and Adolescent PGS combined model | |||||
| Parental PGS | Adolescent PGS | |||||
| AA sample | β | [95% CI] | β | [95% CI] | β | [95% CI] |
| Parental Externalizing | .10 | [−.13, 0.32] | .09 | [−.13, .31] | .02 | [−.10, .13] |
| Parental Involvement | −.07 | [−.16, .03] | −.06 | [−.18, .05] | −.01 | [−.11, .10] |
| Parent-child Communication | −.01 | [−.13, .10] | .01 | [−.11, .14] | −.05 | [−.17, .06] |
| Parent-child Closeness | −.12 | [−.22, −.01] | −.13 | [−.25, −.01] | .03 | [−.07, .13] |
| Parental Knowledge | −.03 | [−.14, .07] | −.05 | [−.17, .07] | .04 | [−.07, .16] |
Note. PGS = Externalizing polygenic score. PGS residualized on first 10 genetic ancestry principal components. EA = participants of European ancestry. AA = participants of African ancestry. Standardized coefficients along with 95% confidence interval (CI) are shown. Parental and adolescent PGS combined models indicate that both adolescent and parental PGS were included in the same models. All models controlled for adolescent sex and adolescent age. Parental externalizing and each parenting measure were tested in separate models.
Bold indicates p ≤ .05
We first tested associations between parental EXT PGS and parents’ own externalizing behavior and aspects of parenting. Among EA participants, parental EXT PGS was associated with their own externalizing behavior. Higher parental EXT PGS also predicted lower parent-child closeness, providing evidence of gene-environment correlation. We next tested the associations between adolescent EXT PGS with parental externalizing behavior and aspects of parenting, adjusting for parental EXT PGS. Adolescent EXT PGS uniquely predicted parental externalizing behavior over and above the effect of parental EXT PGS, suggesting that the child’s genotype further influences externalizing behavior in the parent. Higher adolescent EXT PGS was also associated with poorer parent-child communication, lower parent-child closeness, and lower parental knowledge. There was no evidence of associations between parental EXT PGS and parenting measures after controlling for adolescent EXT PGS. These results provide evidence for evocative gene-environment correlation, whereby the child’s genotype influences parent-child relationship characteristics.
Among AA participants, parental EXT PGS was not associated with their own externalizing behavior. Parental EXT PGS was associated with lower parent-child closeness, but it was not associated with other aspects of parenting. The effect of parental EXT PGS on parent-child closeness remained significant after controlling for adolescent EXT PGS. Adolescent EXT PGS was not associated with any aspects of parenting examined.
Analyses of genetic nurture effect through parental externalizing and parenting
We next tested whether the association between parental EXT PGS and adolescent externalizing behavior was mediated by parental externalizing psychopathology and aspects of parenting. Mediators were tested in separate models. Table 5 summarizes the models testing whether indirect effects of parental EXT PGS (i.e., genetic nurture effects) are mediated via parental externalizing or parenting measures. Among EA participants, there was evidence that parental EXT PGS had an indirect genetic effect on adolescent externalizing behavior through parental externalizing behavior. This indirect effect remained significant after correcting the multiple tests using a 10% false discovery rate (Benjamini & Hochberg, 1995). Parental externalizing accounted for 68% of the variance in the unique association between parental EXT PGS and adolescent externalizing behavior, controlling for the effect of adolescent EXT PGS. There was no evidence that parental EXT PGS had indirect effects on adolescent externalizing behavior through any aspects of parenting examined.
Table 5.
Indirect effects of parental EXT PGS mediated through parental externalizing and parenting measures, controlling for adolescent EXT PGS in the European ancestry sample
| Mediator | β | [95% CI] | proportion mediated |
|---|---|---|---|
| Parental Externalizing | .06 | [.03, .11] | 68% |
| Parental Involvement | −.00 | [−.01, .01] | - |
| Parent-child Communication | .00 | [−.01, .01] | - |
| Parent-child Closeness | .01 | [−.02, .03] | - |
| Parental Knowledge | −.00 | [−.03, .03] | - |
Note. EXT PGS = externalizing polygenic score. Standardized coefficients along with 95% confidence interval for indirect effects are shown. Mediators were tested in separate models as potential mediators linking parental EXT PGS and adolescent externalizing, controlling for adolescent EXT PGS.
Bold coefficient indicates p < .05
Among AA participants, since there was no evidence of genetic nurture effect, we did not proceed in testing whether parental externalizing behavior and parenting mediated the association between parental EXT PGS and adolescent externalizing behavior.
Discussion
The conventional interpretation of genetic risk is that it is passed from parents to children through direct genetic transmission. However, intergenerational transmission of externalizing behavior could reflect both direct and indirect genetic influences. Parents can pass risk-increasing genetic variants to their children, but parental genotypes may also influence children’s outcomes by impacting the rearing environments they provide (Jaffee & Price, 2007; Kendler & Baker, 2007; Rutter & Silberg, 2002). Leveraging genome-wide association data and using molecular genetic data from parents and offspring, we tested the extent to which parental genotypes influence adolescent externalizing behavior through the environment.
In European ancestry families, we found evidence consistent with our hypothesis that parents’ genotypes, both transmitted and non-transmitted to offspring, are associated with adolescent externalizing behavior. By incorporating parental genotypes and child genotypes into models predicting adolescent externalizing behavior, our results show that parental externalizing polygenic scores were uniquely associated with adolescent externalizing behavior, above and beyond the effect of adolescent externalizing polygenic scores. This indicates that genetic associations with externalizing behavior reflect both direct and indirect genetic influences. The association between parental externalizing polygenic score and adolescent externalizing behavior, independent of the genetic risk that is directly transmitted, provides evidence for genetic nurture. Our results add to the growing evidence highlighting the importance of both direct and indirect genetic effects (Bates et al., 2018; Kong et al., 2018; Saunders et al., 2021; Wang et al., 2021). The fact that parental externalizing polygenic scores predicted adolescent externalizing behavior over and above the effect of the adolescent’s externalizing polygenic scores suggests that some environmental factors associated with parental externalizing polygenic loading influence adolescent externalizing behavior.
Second, we found evidence for gene-environment correlation. Consistent with our hypothesis, parental externalizing polygenic score was associated with specific parenting/parent-child relationship characteristics that have been linked to adolescent externalizing behavior. Specifically, parents’ polygenic loading for externalizing was associated with less parent-child closeness in both European and African ancestry families. However, parental externalizing polygenic score was not associated with other aspects of parenting examined. Because we had genetic data from both parents and adolescents, we conducted additional analyses examining whether different forms of gene-environment correlation operate in the family at the same time. By including both parental and adolescent externalizing polygenic scores in a combined model in predicting parenting, we found a pattern of results consistent with evocative gene-environment correlation. In European ancestry families, controlling for parental externalizing polygenic scores, higher adolescent externalizing polygenic scores were associated with poorer parent-child communication, less parent-child closeness, and lower parental knowledge. Twin studies have repeatedly shown the presence of evocative effects on parenting in adolescence (Klahr & Burt, 2014; Marceau et al., 2013; McGue et al., 2005; Neiderhiser et al., 2007; Neiderhiser et al., 2004), and longitudinal studies of children compellingly demonstrate the influence of child behavior on parenting across time (Lansford et al., 2018). Our results support evocative gene-environment correlations between adolescents’ genetic predisposition and parenting, highlighting the role of children’s genotypes in shaping parenting and features of parent-child relationship in adolescence.
Third, in European ancestry families, we found evidence consistent with our hypothesis that parental externalizing psychopathology mediated the association between parental externalizing polygenic score and adolescent externalizing behavior, after accounting for adolescent externalizing polygenic score. Family and adoption studies have shown that parental externalizing behaviors represent both genetic and environmental risk for offspring (Kendler et al., 2015; Kendler et al., 2012; Keyes et al., 2008). Using parental and adolescent polygenic scores, we built on prior evidence from latent genetic studies to show that genetic nurture effects on adolescent externalizing behavior, while relatively small in magnitude, are explained, in part, by parents’ own externalizing behavior. This demonstrates that parental externalizing represents a form of environmental risk for children. Our result suggests that environmental interventions that focus on treating externalizing related disorders in parents, and promotion of recovery, may help reduce intergenerational transmission of genetic risk for externalizing behavior.
We did not find evidence for potential environmental mediation of genetic nurture effects for adolescent externalizing behavior through the other parenting characteristics studied here (parental involvement, parent-child communication, parent-child closeness, parental knowledge). This was partly driven by the fact that there was generally no association between parental externalizing polygenic scores and the parenting variables examined (with the exception of parent-child closeness). The lack of association between parental genotype and parenting could partly reflect that our parenting measures were adolescent-report data. Thus, our measures of the parenting/parent-child relationship tapped adolescent perceptions of parenting, which may explain why they were more strongly correlated with the child’s own genotype. Moreover, prior evidence that parenting mediates the indirect genetic effects on child behavior comes measures of parenting earlier in development (Armstrong-Carter et al., 2020; Wertz et al., 2020). It is possible that genetic nurture processes via parenting might be more salient at earlier stages of development. Identifying other psychosocial factors and processes that may serve as the mediating pathways of indirect genetic risk is clearly needed.
For the most part, we did not find evidence for genetic nurture or associations between polygenic loading for externalizing or parenting in our participants of African ancestry. It is important to note that these null effects are most likely attributable to the limited predictive power of polygenic scores derived from European ancestry individuals in samples of non-European ancestry individuals that arises due to different LD patterns and/or possible different causal effects (Martin et al., 2019). A challenge with incorporating genetic risk scores into developmental and biomedical studies is that creating polygenic scores in diverse populations is not straightforward (Wang, Tsuo, Kanai, Neale, & Martin, 2022). Populations of non-European ancestry, particularly those of African ancestry, have been historically excluded from GWAS across fields (Popejoy & Fullerton, 2016), and this lack of diversity in genomic research limits our ability to create valid polygenic scores in follow-up studies with diverse samples. Rather than limiting our analyses to focus on participants of European ancestry, we used GWAS results from European ancestry discovery samples to create polygenic scores for the African ancestry sample in our study. Although this approach is more inclusive, we recognize that it is not ideal because the mismatch in ancestry between the discovery and target samples make the polygenic scores less accurate and less predictive, creating difficulties in interpreting null findings associated with polygenic scores. However, we believe it is a scientific and ethical imperative to include individuals of African ancestry in genetic research as the exclusion of historically underrepresented individuals from health research has the potential to further perpetuate health disparities (Davis, 2021). Predictive power of polygenic scores in non-European ancestry groups will improve as large-scale GWAS in diverse populations becomes available. Additionally, because we conducted analyses separately by ancestry group to accommodate the inclusion of polygenic scores, our study is not informative about how socioenvironmental factors may operate differently across the ancestry groups, and it does not provide information on any ethnic-racial differences in externalizing behaviors or parenting. Clearly, concerted efforts to have adequate representation of diversity in genetic research is important, and efforts are underway to increase the diversity of participants in genetic research (Hindorff et al., 2018), such as the All of Us Research Program. There are also various new methods under development (e.g., Liang et al., 2022; Ruan et al., 2022) to account for population allele frequency and LD pattern differences across populations to improve cross-ancestry portability of genetic risk information. While these new methodological approaches are likely to help improve accuracy and generalizability of polygenic scores in diverse populations, prioritizing diversity in genetic research is key in fundamentally addressing the problems and ensuring that all racial/ethnic groups benefit from advances related to genetic findings in an equitable manner (Martin et al., 2019).
Limitations
Our results should be interpreted within the context of the following limitations. First, COGA is a high-risk sample, with individuals from extended families enriched for alcohol use disorders. Findings may not be generalizable to other samples recruited and ascertained with different risk profiles. Second, measures of parenting/parent-child relationship were reported from the adolescent’s perspective. It is possible that adolescents’ externalizing behavior or their genotype impacted their perception of parent-child relationship characteristics (Kendler & Baker, 2007), and these parenting measures may not objectively reflect parental behavior. Capturing parenting as a multidimensional construct with multi-informant data is an important next step. Third, assessments of parental externalizing disorders occurred years prior to the adolescent interview. Thus, they represent parents’ lifetime assessments of externalizing psychopathology, rather than externalizing behavior measured contemporaneously with adolescent reports. Fourth, at present, the amount of variance accounted for by polygenic score for the externalizing phenotype remains modest. In addition, in our genetic nurturance analyses, the mediating effect via parental externalizing psychopathology was of small magnitude, though comparable to the genetic nurturance analyses for substance use phenotypes reported in Saunders et al. (2021). Future studies should explore and identify other possible environmental mediators of genetic nurture effects. Finally, our results are observational in nature and should not be interpreted as causal pathways.
Conclusion
The present study adds to the literature by demonstrating that parental genetic predispositions toward externalizing influence adolescent externalizing behavior not only through direct genetic transmission but also via genetic nurturance. Our findings illustrate that parental externalizing polygenic scores uniquely predicted their adolescent’s externalizing behavior beyond the variance explained by children’s own externalizing polygenic scores. In addition, parental externalizing psychopathology partly explained the relationship between parental externalizing polygenic score and adolescent externalizing behavior. This supports the idea that parental externalizing psychopathology represents both genetic and environmental risk. Our results also indicate associations between adolescent genotype and self-reported parenting, providing some evidence for evocative gene-environment correlation. Results of the present study underscore the complex interplay between genes and environments in contributing to intergenerational risk for externalizing behavior. Genetic and environmental influences should be considered together to understand parent and child behavior in the development of psychopathology.
Acknowledgements
The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, T. Foroud; Scientific Director, A. Agrawal; Translational Director, D. Dick, includes eleven different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, T. Foroud, Y. Liu, M. Plawecki); University of Iowa Carver College of Medicine (S. Kuperman, J. Kramer); SUNY Downstate Health Sciences University (B. Porjesz, J. Meyers, C. Kamarajan, A. Pandey); Washington University in St. Louis (L. Bierut, J. Rice, K. Bucholz, A. Agrawal); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield, R. Hart, J. Salvatore); The Children’s Hospital of Philadelphia, University of Pennsylvania (L. Almasy); Virginia Commonwealth University (D. Dick); Icahn School of Medicine at Mount Sinai (A. Goate, P. Slesinger); and Howard University (D. Scott). Other COGA collaborators include: L. Bauer (University of Connecticut); J. Nurnberger Jr., L. Wetherill, X. Xuei, D. Lai, S. O’Connor, (Indiana University); G. Chan (University of Iowa; University of Connecticut); D.B. Chorlian, J. Zhang, P. Barr, S. Kinreich, G. Pandey (SUNY Downstate); N. Mullins (Icahn School of Medicine at Mount Sinai); A. Anokhin, S. Hartz, E. Johnson, V. McCutcheon, S. Saccone (Washington University); J. Moore, Z. Pang, S. Kuo (Rutgers University); A. Merikangas (The Children’s Hospital of Philadelphia and University of Pennsylvania); F. Aliev (Virginia Commonwealth University); H. Chin and A. Parsian are the NIAAA Staff Collaborators. We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting- Kai Li, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
The Externalizing Consortium: Principal Investigators: Danielle M. Dick, Philipp Koellinger, K. Paige Harden, Abraham A. Palmer. Lead Analysts: Richard Karlsson Linnér, Travis T. Mallard, Peter B. Barr, Sandra Sanchez-Roige. Significant Contributors: Irwin Waldman.
The Externalizing Consortium has been supported by the National Institute of Alcohol Abuse and Alcoholism (R01AA015146 - administrative supplement), and the National Institute on Drug Abuse (R01DA050721). Additional funding for investigator effort has been provided by K02AA018755, U10AA008401, P50AA022537, as well as a European Research Council Consolidator Grant (647648 EdGe). The content is solely the responsibility of the authors and does not necessarily represent the official views of the above funding bodies. The Externalizing Consortium would like to thank the following for making its research possible: 23andMe, Add Health, Vanderbilt University Medical Center’s BioVU, Collaborative Study on the Genetics of Alcoholism (COGA), the Psychiatric Genomics Consortium’s Substance Use Disorders working group, UK10K Consortium, UK Biobank, and Philadelphia Neurodevelopmental Cohort.
Financial Support
This study was funded by the National Institutes of Health through the National Institute on Alcohol Abuse and Alcoholism (U10AA008401) and the National Institute on Drug Abuse (R01DA050721).
Footnotes
Conflicts of Interest: None.
References
- Achenbach TM (1966). The classification of children’s psychiatric symptoms: A factor-analytic study. Psychological Monographs: General and Applied, 80(7), 1–37. doi: 10.1037/h0093906 [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. (1994). Diagnostic and Statistical Manual of Mental Disorders (4th ed.). Washington, DC: American Psychiatric Association. [Google Scholar]
- Armstrong-Carter E, Trejo S, Hill LJB, Crossley KL, Mason D, & Domingue BW (2020). The Earliest Origins of Genetic Nurture: The Prenatal Environment Mediates the Association Between Maternal Genetics and Child Development. Psychological Science, 31(7), 781–791. doi: 10.1177/0956797620917209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avinun R, & Knafo A (2014). Parenting as a Reaction Evoked by Children’s Genotype: A Meta-Analysis of Children-as-Twins Studies. Personality and Social Psychology Review, 18(1), 87–102. doi: 10.1177/1088868313498308 [DOI] [PubMed] [Google Scholar]
- Barr PB, & Dick DM (2020). The Genetics of Externalizing Problems. In de Wit H & Jentsch JD (Eds.), Recent Advances in Research on Impulsivity and Impulsive Behaviors (pp. 93–112). Cham: Springer International Publishing. [Google Scholar]
- Bates TC, Maher BS, Medland SE, McAloney K, Wright MJ, Hansell NK, … Gillespie NA (2018). The Nature of Nurture: Using a Virtual-Parent Design to Test Parenting Effects on Children’s Educational Attainment in Genotyped Families. Twin Research and Human Genetics, 21(2), 73–83. doi: 10.1017/thg.2018.11 [DOI] [PubMed] [Google Scholar]
- Baurley JW, Edlund CK, Pardamean CI, Conti DV, & Bergen AW (2016). Smokescreen: A targeted genotyping array for addiction research. BMC Genomics, 17, 145. doi: 10.1186/s12864-016-2495-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begleiter H, Reich T, Hesselbrock V, Porjesz B, Li T-K, Schuckit MA, … Rice JP (1995). The collaborative study on the genetics of alcoholism. Alcohol Health and Research World, 19, 228–228. [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, & Hochberg Y (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological), 57(1), 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bogdan R, Baranger DAA, & Agrawal A (2018). Polygenic Risk Scores in Clinical Psychology: Bridging Genomic Risk to Individual Differences. Annual Review of Clinical Psychology, 14(1), 119–157. doi: 10.1146/annurev-clinpsy-050817-084847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, … Schuckit MA (1994). A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. Journal of Studies on Alcohol, 55(2), 149–158. doi: 10.15288/jsa.1994.55.149 [DOI] [PubMed] [Google Scholar]
- Bucholz KK, McCutcheon VV, Agrawal A, Dick DM, Hesselbrock VM, Kramer JR, … Porjesz B (2017). Comparison of Parent, Peer, Psychiatric, and Cannabis Use Influences Across Stages of Offspring Alcohol Involvement: Evidence from the COGA Prospective Study. Alcoholism, Clinical and Experimental Research, 41(2), 359–368. doi: 10.1111/acer.13293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, & Palmer LJ (2003). Population stratification and spurious allelic association. The Lancet, 361(9357), 598–604. doi: 10.1016/S0140-6736(03)12520-2 [DOI] [PubMed] [Google Scholar]
- Case A, & Deaton A (2017). Mortality and morbidity in the 21(st) century. Brookings Papers on Eonomic Activity, 2017, 397–476. doi: 10.1353/eca.2017.0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassin L, Pillow DR, Curran PJ, Molina BSG, & Barrera M Jr. (1993). Relation of parental alcoholism to early adolescent substance use: A test of three mediating mechanisms. Journal of Abnormal Psychology, 102(1), 3–19. doi: 10.1037/0021-843X.102.1.3 [DOI] [PubMed] [Google Scholar]
- Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, Stambolian D, Loh P-R, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, … Fuchsberger C (2016). Next-generation genotype imputation service and methods. Nature Genetics, 48(10), 1284–1287. 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LK (2021). Human genetics needs an antiracism plan. Scientific American. Retrieved from https://www.scientificamerican.com/article/human-genetics-needs-an-antiracism-plan/ [Google Scholar]
- Delaneau O, Howie B, Cox AJ, Zagury J-F, & Marchini J (2013). Haplotype estimation using sequencing reads. American Journal of Human Genetics, 93(4), 687–696. 10.1016/j.ajhg.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zeeuw EL, Hottenga J-J, Ouwens KG, Dolan CV, Ehli EA, Davies GE, … van Bergen E (2020). Intergenerational Transmission of Education and ADHD: Effects of Parental Genotypes. Behavior Genetics, 50(4), 221–232. doi: 10.1007/s10519-020-09992-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Cho SB, Latendresse SJ, Aliev F, Nurnberger JI, Edenberg HJ, … Kuperman S (2014). Genetic influences on alcohol use across stages of development: GABRA2 and longitudinal trajectories of drunkenness from adolescence to young adulthood. Addiction Biology, 19(6), 1055–1064. doi: 10.1111/adb.12066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Chen C-Y, Ni Y, Feng Y-CA, & Smoller JW (2019). Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nature Communications, 10(1), 1776. doi: 10.1038/s41467-019-09718-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambrick DZ, & Tucker-Drob EM (2015). The genetics of music accomplishment: Evidence for gene–environment correlation and interaction. Psychonomic Bulletin & Review, 22(1), 112–120. doi: 10.3758/s13423-014-0671-9 [DOI] [PubMed] [Google Scholar]
- Hamdi NR, & Iacono WG (2014). Lifetime prevalence and co-morbidity of externalizing disorders and depression in prospective assessment. Psychological Medicine, 44(2), 315–324. doi: 10.1017/S0033291713000627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, & Hesselbrock V (1999). A validity study of the SSAGA-a comparison with the SCAN. Addiction, 94(9), 1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Bonham VL, Brody LC, Ginoza M, Hutter CM, Manolio TA, & Green ED (2018). Prioritizing diversity in human genomics research. Nature Reviews Genetics, 19(3), 175–185. 10.1038/nrg.2017.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffee SR, & Price TS (2007). Gene–environment correlations: a review of the evidence and implications for prevention of mental illness. Molecular Psychiatry, 12(5), 432–442. doi: 10.1038/sj.mp.4001950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang KL, Vernon PA, Livesley WJ, Stein MB, & Wolf H (2001). Intra- and extra-familial influences on alcohol and drug misuse: a twin study of gene–environment correlation. Addiction, 96(9), 1307–1318. doi: 10.1046/j.1360-0443.2001.969130710.x [DOI] [PubMed] [Google Scholar]
- Karlsson Linnér R, Mallard TT, Barr PB, Sanchez-Roige S, Madole JW, Driver MN, … Dick DM (2021). Multivariate analysis of 1.5 million people identifies genetic associations with traits related to self-regulation and addiction. Nature Neuroscience, 24(10), 1367–1376. doi: 10.1038/s41593-021-00908-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, & Baker JH (2007). Genetic influences on measures of the environment: a systematic review. Psychological Medicine, 37(5), 615–626. doi: 10.1017/S0033291706009524 [DOI] [PubMed] [Google Scholar]
- Kendler KS, Ji J, Edwards AC, Ohlsson H, Sundquist J, & Sundquist K (2015). An Extended Swedish National Adoption Study of Alcohol Use Disorder. JAMA Psychiatry, 72(3), 211–218. doi: 10.1001/jamapsychiatry.2014.2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA, Myers J, & Neale MC (2003). The Structure of Genetic and Environmental Risk Factors for Common Psychiatric and Substance Use Disorders in Men and Women. Archives of General Psychiatry, 60(9), 929–937. doi: 10.1001/archpsyc.60.9.929 [DOI] [PubMed] [Google Scholar]
- Kendler KS, Sundquist K, Ohlsson H, Palmér K, Maes H, Winkleby MA, & Sundquist J (2012). Genetic and Familial Environmental Influences on the Risk for Drug Abuse: A National Swedish Adoption Study. Archives of General Psychiatry, 69(7), 690–697. doi: 10.1001/archgenpsychiatry.2011.2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes M, Legrand LN, Iacono WG, & McGue M (2008). Parental Smoking and Adolescent Problem Behavior: An Adoption Study of General and Specific Effects. American Journal of Psychiatry, 165(10), 1338–1344. doi: 10.1176/appi.ajp.2008.08010125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahr AM, & Burt SA (2014). Elucidating the etiology of individual differences in parenting: A meta-analysis of behavioral genetic research. Psychological Bulletin, 140(2), 544–586. doi: 10.1037/a0034205 [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Frigge ML, Vilhjalmsson BJ, Young AI, Thorgeirsson TE, … Stefansson K (2018). The nature of nurture: Effects of parental genotypes. Science, 359(6374), 424–428. doi: 10.1126/science.aan6877 [DOI] [PubMed] [Google Scholar]
- Krueger RF, Hicks BM, Patrick CJ, Carlson SR, Iacono WG, & McGue M (2002). Etiologic connections among substance dependence, antisocial behavior and personality: Modeling the externalizing spectrum. Journal of Abnormal Psychology, 111(3), 411–424. doi: 10.1037/0021-843X.111.3.411 [DOI] [PubMed] [Google Scholar]
- Kuperman S, Chan G, Kramer JR, Wetherill L, Bucholz KK, Dick D, … Schuckit M (2013). A Model to Determine the Likely Age of an Adolescent’s First Drink of Alcohol. Pediatrics, 131(2), 242. doi: 10.1542/peds.2012-0880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahey BB, Rathouz PJ, Van Hulle C, Urbano RC, Krueger RF, Applegate B, … Waldman ID (2008). Testing Structural Models of DSM-IV Symptoms of Common Forms of Child and Adolescent Psychopathology. Journal of Abnormal Child Psychology, 36(2), 187–206. doi: 10.1007/s10802-007-9169-5 [DOI] [PubMed] [Google Scholar]
- Lai D, Johnson EC, Colbert S, Pandey G, Chan G, Bauer L, … & Foroud T (2022). Evaluating risk for alcohol use disorder: Polygenic risk scores and family history. Alcoholism: Clinical and Experimental Research, 46(3), 374–383. 10.1111/acer.14772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai D, Wetherill L, Bertelsen S, Carey CE, Kamarajan C, Kapoor M, … Foroud T (2019). Genome‐wide association studies of alcohol dependence, DSM-IV criterion count and individual criteria. Genes, Brain & Behavior, 18(6). doi: 10.1111/gbb.12579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansford JE, Rothenberg WA, Jensen TM, Lippold MA, Bacchini D, Bornstein MH, … Al-Hassan SM (2018). Bidirectional Relations Between Parenting and Behavior Problems From Age 8 to 13 in Nine Countries. Journal of Research on Adolescence, 28(3), 571–590. doi: 10.1111/jora.12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemery-Chalfant K, Kao K, Swann G, & Goldsmith HH (2013). Childhood temperament: passive gene-environment correlation, gene-environment interaction, and the hidden importance of the family environment. Development and Psychopathology, 25(1), 51–63. doi: 10.1017/S0954579412000892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Pividori M, Manichaikul A, Palmer AA, Cox NJ, Wheeler HE, & Im HK (2022). Polygenic transcriptome risk scores (PTRS) can improve portability of polygenic risk scores across ancestries. Genome Biology, 23(1), 1–18. 10.1186/s13059-021-02591-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau K, Horwitz BN, Narusyte J, Ganiban JM, Spotts EL, Reiss D, & Neiderhiser JM (2013). Gene-Environment Correlation Underlying the Association Between Parental Negativity and Adolescent Externalizing Problems. Child Development, 84(6), 2031–2046. doi: 10.1111/cdev.12094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, … Kenny EE (2017). Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. The American Journal of Human Genetics, 100(4), 635–649. doi: 10.1016/j.ajhg.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, & Daly MJ (2019). Clinical use of current polygenic risk scores may exacerbate health disparities. Nature Genetics, 51(4), 584–591. doi: 10.1038/s41588-019-0379-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Elkins I, Walden B, & Iacono WG (2005). Perceptions of the Parent-Adolescent Relationship: A Longitudinal Investigation. Developmental Psychology, 41(6), 971–984. doi: 10.1037/0012-1649.41.6.971 [DOI] [PubMed] [Google Scholar]
- Muthén K, & Muthén BO (1998–2017). Mplus User’s Guide (8th ed.). Los Angeles, CA: Authors. [Google Scholar]
- Neiderhiser JM, Reiss D, Lichtenstein P, Spotts EL, & Ganiban J (2007). Father-adolescent relationships and the role of genotype-environment correlation. Journal of Family Psychology, 21(4), 560–571. doi: 10.1037/0893-3200.21.4.560 [DOI] [PubMed] [Google Scholar]
- Neiderhiser JM, Reiss D, Pedersen NL, Lichtenstein P, Spotts EL, Hansson K, … Ellhammer O (2004). Genetic and environmental influences on mothering of adolescents: a comparison of two samples. Developmental Psychology, 40(3), 335–351. doi: 10.1037/0012-1649.40.3.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RE, Kuchenbaecker K, Walters RK, Chen C-Y, Popejoy AB, Periyasamy S, … Duncan LE (2019). Genome-wide Association Studies in Ancestrally Diverse Populations: Opportunities, Methods, Pitfalls, and Recommendations. Cell, 179(3), 589–603. doi: 10.1016/j.cell.2019.08.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinquart M (2017). Associations of parenting dimensions and styles with externalizing problems of children and adolescents: An updated meta-analysis. Developmental Psychology, 53(5), 873–932. doi: 10.1037/dev0000295 [DOI] [PubMed] [Google Scholar]
- Popejoy AB, & Fullerton SM (2016). Genomics is failing on diversity. Nature, 538(7624), 161–164. doi: 10.1038/538161a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, & Reich D (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics, 38(8), 904–909. doi: 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- R Development Core Team. (2019). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Retrieved from https://www.R-project.org/ [Google Scholar]
- Reich W, Earls F, & Powell J (1988). A Comparison of the Home and Social Environments of Children of Alcoholic and Non-alcoholic Parents. British Journal of Addiction, 83(7), 831–839. doi: 10.1111/j.1360-0443.1988.tb00518.x [DOI] [PubMed] [Google Scholar]
- Reich W, & Earls FJ (1982). The Home Environment Interview for Children, child and parent versions. Washington University. St. Louis. [Google Scholar]
- Reuben JD, Shaw DS, Neiderhiser JM, Natsuaki MN, Reiss D, & Leve LD (2016). Warm Parenting and Effortful Control in Toddlerhood: Independent and Interactive Predictors of School-Age Externalizing Behavior. Journal of Abnormal Child Psychology, 44(6), 1083–1096. doi: 10.1007/s10802-015-0096-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond-Rakerd LS, D’Souza S, Andersen SH, Hogan S, Houts RM, Poulton R, … Moffitt TE (2020). Clustering of health, crime and social-welfare inequality in 4 million citizens from two nations. Nature Human Behaviour, 4(3), 255–264. doi: 10.1038/s41562-019-0810-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbaum F, & Weisz JR (1994). Parental caregiving and child externalizing behavior in nonclinical samples: A meta-analysis. Psychological Bulletin, 116(1), 55–74. doi: 10.1037/0033-2909.116.1.55 [DOI] [PubMed] [Google Scholar]
- Ruan Y, Lin Y-F, Feng Y-CA, Chen C-Y, Lam M, Guo Z, Stanley Global Asia Initiatives, He L, Sawa A, Martin AR, Qin S, Huang H, & Ge T (2022). Improving polygenic prediction in ancestrally diverse populations. Nature Genetics. 10.1038/s41588-022-01054-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M, & Silberg J (2002). Gene-environment interplay in relation to emotional and behavioral disturbance. Annual Review of Psychology, 53(1), 463. doi: 10.1146/annurev.psych.53.100901.135223 [DOI] [PubMed] [Google Scholar]
- Salvatore JE, Aliev F, Bucholz K, Agrawal A, Hesselbrock V, Hesselbrock M, … Dick DM (2015). Polygenic Risk for Externalizing Disorders: Gene-by-Development and Gene-by-Environment Effects in Adolescents and Young Adults. Clinical Psychological Science, 3(2), 189–201. doi: 10.1177/2167702614534211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders GRB, Liu M, Vrieze S, McGue M, & Iacono WG (2021). Mechanisms of parent–child transmission of tobacco and alcohol use with polygenic risk scores: Evidence for a genetic nurture effect. Developmental Psychology, 57(5), 796–804. doi: 10.1037/dev0001028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarr S, & McCartney K (1983). How People Make Their Own Environments: A Theory of Genotype → Environment Effects. Child Development, 54(2), 424–435. doi: 10.2307/1129703 [DOI] [PubMed] [Google Scholar]
- Su J, Kuo SI, Aliev F, Guy MC, Derlan CL, Edenberg HJ, … Dick DM (2018). Influence of Parental Alcohol Dependence Symptoms and Parenting on Adolescent Risky Drinking and Conduct Problems: A Family Systems Perspective. Alcoholism, Clinical and Experimental Research, 42(9), 1783–1794. doi: 10.1111/acer.13827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingley D, Yamamoto T, Hirose K, Keele L, & Imai K (2020). mediation: Causal Mediation Analysis (Version 4.5.0) Retrieved from https://cran.r-project.org/web/packages/mediation/
- Tully LA, & Hunt C (2016). Brief Parenting Interventions for Children at Risk of Externalizing Behavior Problems: A Systematic Review. Journal of Child and Family Studies, 25(3), 705–719. doi: 10.1007/s10826-015-0284-6 [DOI] [Google Scholar]
- Wang B, Baldwin JR, Schoeler T, Cheesman R, Barkhuizen W, Dudbridge F, … Pingault J-B (2021). Robust genetic nurture effects on education: A systematic review and meta-analysis based on 38,654 families across 8 cohorts. The American Journal of Human Genetics, 108(9), 1780–1791. doi: 10.1016/j.ajhg.2021.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tsuo K, Kanai M, Neale BM, & Martin AR (2022). Challenges and opportunities for developing more generalizable polygenic risk scores. Annual Review of Biomedical Data Science, 5. 10.1146/annurev-biodatasci-111721-074830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz J, Belsky J, Moffitt TE, Belsky DW, Harrington H, Avinun R, … Caspi A (2019). Genetics of nurture: A test of the hypothesis that parents’ genetics predict their observed caregiving. Developmental Psychology, 55(7), 1461–1472. doi: 10.1037/dev0000709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz J, Moffitt TE, Agnew‐Blais J, Arseneault L, Belsky DW, Corcoran DL, … Richmond-Rakerd LS (2020). Using DNA From mothers and children to study parental investment in children’s educational attainment. Child Development, 91(5), 1745–1761. doi: 10.1111/cdev.13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby EA, McGue M, Iacono WG, Rustichini A, & Lee JJ (2021). The role of parental genotype in predicting offspring years of education: evidence for genetic nurture. Molecular Psychiatry, 26(8), 3896–3904. doi: 10.1038/s41380-019-0494-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Lee SH, Mehta D, Vinkhuyzen AAE, Dudbridge F, & Middeldorp CM (2014). Research Review: Polygenic methods and their application to psychiatric traits. Journal of Child Psychology and Psychiatry, 55(10), 1068–1087. 10.1111/jcpp.12295 [DOI] [PubMed] [Google Scholar]
- Young SE, Stallings MC, Corley RP, Krauter KS, & Hewitt JK (2000). Genetic and environmental influences on behavioral disinhibition. American Journal of Medical Genetics, 96(5), 684–695. [DOI] [PubMed] [Google Scholar]


