Abstract
Due to heterogenetic-specific nature of the available biomarkers, the incidence of lung adenocarcinoma (LUAD) is on the rise worldwide. Previously reported LUAD-related hub genes were searched from the medical literature via literature mining and were processed to identify few top genes via degree method. Later, a comprehensive in silico methodology was applied on the selected real hub genes to identify their tumor driving, diagnostic, and prognostic roles in LUAD patients with divers clinicopathological variables. Out of total 145 extracted hub genes, six genes including CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1 were identified as real hub genes. The expression analysis showed that all these genes were significantly up-regulated across LUAD samples of different clinicopathological variables. In addition, a variety of unique correlations among the expression and of real hub genes and some other parameters including promoter methylation status, overall survival (OS), genetic changes, tumor purity, and immune cell infiltration have also been explored in the present study. Moreover, via TFS-miRNA-mRNA regulatory network, one important TF (E2F1) and one important miRNAs (hsa-mir-34a-5p) that targeted all the real hub genes were also identified. Finally, a variety of drugs also predicted to be very useful in treating LUAD. The discovery of the real hub genes, TFS-miRNA-mRNA network, and chemotherapeutic drugs associated with LUAD provides new insights into underlying mechanisms and treatment of LUAD overcoming heterogeneity barriers.
Keywords: Lung adenocarcinoma, PubMed, hub gene, miRNA, biomarker
Introduction
Lung cancer is the second most abundantly diagnosed cancer worldwide [1]. It is estimated that more than 2.2 million new lung cancer cases and 1.7 million lung cancer-associated deaths occur per annum around the world [2]. In Pakistan, lung cancer is the third most prevalent cancer accounting for 5.9% new cases of all the cancers [2]. Although LUAD initial growth occurs in the early stage, the progression of this disease is much slower than other cancer subtypes. Moreover, the overall survival of LUAD is also reported to be lower than other cancer subtypes [2]. Recent studies highlighted that an exposure to asbestos, smoking cigarette and exposure to many other types of chemical can enhance the risk of LUAD development [3]. LUAD is a complicated and one of the most deadliest malignancies of the lung periphery where glandular cells secrete mucus and help in breathing [3]. This subtype accounts for more than 40% of lung cancer cases its rate is on the rise currently [5].
A significant improvement has been achieved in LUAD therapy recently, its prognosis is still very low with 18% of 5-year survival rates [6]. We all know that the accurate diagnosis of LUAD is a huge challenge, as it is often detected at the advanced stages [7]. Therefore, an accurate diagnosis of LUAD requires the discovery of new biomarkers that can detect LUAD precisely, increase survival rates, reduce the incidence of tumor invasions, and also enhance the chances of successful therapy [8].
During the last 10 years, researchers around the globe have used next-generation sequencing (NGS) as well as microarray methods to identify novel biomarkers and therapeutic targets in LUAD [9], small sample count however resulted in the significant inter-study inconsistency. In order to address this issue, Gene Expression Omnibus (GEO) database [10] provided the facility to researchers for archiving their expression datasets in GEO database to make it publically available for further integration with other similar datasets via in silico approaches to uncover molecular biomarkers more precisely. Previously, such GEO-based expression datasets of LUAD have been utilized by the earlier studies to discover biomarkers of LUAD [9], but as we know that biomarkers are highly specific biomolecules, and GEO-based LUAD expression datasets consist of cancer patients with different clinical variables, it is clinically impossible to use already identified GEO-based biomarkers in LUAD patients over heterogeneity-barrier.
In the current research, therefore, using a novel integrated approach, we utilized already identified biomarkers (hub genes) from GEO-based expression datasets of LUAD to prioritize a new system of six biomarkers over heterogeneity-barrier.
Materials and methods
Hub genes extraction
PubMed database was effectively searched in our study to find all studies dealing with GEO-expression datasets of LUAD for the identification of hub genes up to December 2021. Following keywords were used in a combination to search the relevant literature: “Hub genes AND Lung adenocarcinoma” or “Hub genes AND Lung neoplasia”. In total, 23 studies out of total 356 appeared studies were selected collectively analyzing 31 GEO expression datasets of LUAD to identify numerous hub genes. Later, all the selected studies were further subjected to extract and combined the reported hub genes for getting a consolidated pool.
Protein-protein interaction (PPI) network, module, and enrichment analysis
For interpreting molecular mechanisms behind LUAD, we used STRING, was used STRING database [11] to construct the information of PPIs network with of not < 0.7. Then, the relationships between hub genes were explored via Cytoscape software (3.8.2) by calculating network properties including the distribution of network node degree, distribution of the shortest path, and proximity to the center [12]. Later, molecular Complex Detection (MCODE) analysis has helped us to find clusters of genes in the constructed PPI network with default cutoff criterions parameters such as “Degree cutoff = 2”, “node score cutoff = 0.2”, “k-core = 2” and “max. depth = 100”. Finally, the real hub genes were chosen via degree method and their enrichment analysis were performed using DAVID tool. A P-value of < 0.05 was selected to show the statistical differences.
Real hub genes expression profiling via UALCAN
The expression analysis of real hub genes were carried out via UALCAN (http://ualcan.path.uab.edu/) [13]. The UALCAN is The Cancer Genome Atlas (TCGA) data set analysis tool. In this study, for expression analysis of real hub genes we used TCGA LUAD dataset consisting of 515 cancerous and 59 normal samples. For statistics, UALCAN used a student t-test and normalized the obtained expression as transcript per million (TPM) reads.
Expression validation of real hub genes
The TIMER [14], GENT2 [15], GEPIA, DriverDBV3 [16], and UALCAN [13] are TCG multi-omics data analysis tools. In this study, we utilized these databases for the expression validation of the real hub genes using new independent LUAD patients’ cohorts.
DriverDBV3 analysis
DriverDBV3 [16] was conducted in this to analyze the promoter methylation levels of real hub genes across LAUD samples paired with normal controls using Pearson correlation analysis.
Prognostic potentials of real hub genes
Kaplan-Meier Plotter and GEPIA tools are widely used for prognostic potentials evaluation of any gene(s) of interest [17]. In the current study, via using these tool, we analyzed the effect of real hub genes expression on the Overall Survival (OS) of LUAD patients. A P-value < 0.05 was chosen as statistically significant.
Genetic alterations in real hub genes
Via cBioPortal, we used TCGA LUAD datasets for analyzing genetic alterations of real hub genes [18]. This online resource is a hub of cancer omics data which includes genetic mutations information, copy number variations, deep amplification, deep deletion, and mRNA expression level information. We used this database for conducting genetic alteration analysis in this study with default settings.
Tumor purity and immune cells analyses
TIMER [19] was conducted in this study to analyze the relationships between tumor purity, immune cells infiltration, and real hub genes expression across LUAD samples. A P-value < 0.05 was chosen as statistically significant.
Constructing TF-miRNA-mRNA network
ENCORI is an openly available public platform developed for the identification of more than 2.5 million TFS-miRNA-mRNA interactions [20,21]. All the target TFS and miRNAs of the real hub genes were screened in ENCORI using default settings. Finally, the TFS-miRNA-mRNA networks of real hub genes were visualized using Cytoscape (version 3.8.2).
Expression profiling of TFS and miRNAs
The expression profiling of real hub genes targeting TFS and miRNAs were also carried out via UALCAN [13] in a LUAD cohort taken from TCGA datasets using default setting.
CancerSEA-based analysis
CancerSEA is an online resource foe exploring relationships among gene(s) of interest and 14 different functional states at a single-cell level across several cancer types [22]. CancerSEA was conducted in this study to investigate relationships between real hub genes and these states at a single-cell level across LUAD. A P-value < 0.05 was chosen as statistically significant.
MuTarget-based analysis
MuTarget is an openly available platform that help the researcher to associate genetic mutations with gene expression across several human cancer types [23]. We conducted MuTarget analysis in this study to identify the mutant genes associated with gene expression alteration of real hub genes in LUAD. A P-value < 0.05 was chosen as statistically significant.
Comparative Toxicogenomics database
Comparative Toxicogenomics database (CTD) [24] was conducted to draw the real hub gene-drug interaction networks via Cytoscape, highlighting different potential drugs capable of decreasing or increasing the expression levels of real hub genes. In clinical application view point, the selected drugs will in the treatment of LUAD.
Results
Extraction of hub genes
In total, 23 studies were shortlisted for hub genes extraction. The selected studies were next subjected to hub genes extraction and finally, after normalizing overlapped genes, we were able to construct a get a brief pool of 145 hub genes from 31 GEO LUAD datasets comprising of 1980 LUAD and 1230 normal samples (Table 1). Without normalization, the original data can be seen in the Table S1.
Table 1.
List of the LUAD microarray expression datasets and the hub genes extracted selected studies
| Dataset | C/N | Source of origin | Extracted hub genes | References |
|---|---|---|---|---|
| GSE118370 | 06/06 | China | ADCY4, S1PR1, FPR2, PPBP, NMU, PF4, GCG, CCNA2, CCNB1, CDC20, CDCA5, CDCA8, FEN1, KIF2C, KPNA2, MCM6, NUSAP1, RACGAP1, RRM2, SPAG5, TOP2A, TPX2, CA4, PECAM1, DNAJB4, AGER, GIMAP6, C10orf54, DOCK4, LRRK2, EPAS1, LDB2, HOXA11-AS, PHACTR2, MSRB3, GHR, PLSCR4, EPB41L2, NPNT, FBXO32, IL6, MMP9, EDN1, FOS, CDK1, CDH1, BIRC5, VWF, UBE2C, CDKN3, CDKN2A, CD34, AURKA, CCNB2, EGR1, UBE2T, PBK, MELK, TNNC1, TPPX2, INS, LPL, HPGDS, DGAT1, UGT1A6, CYP2C9, KIAA0101, BUB1B, PRC1, CEACAM5, NQO1, LCN2, KRT8, EPCAM, ELF3, KRT19, DCN, SERPING1, GNG11, CXCL12, CAV1, DCY8, ADRB2, CALCA, GNGT1, NPSR1, CLDN5, COL1A1, SPP1, UBB, RAC1, ITGB1, SRC, C3, EGFR, TIMP1, GAS6, P4HB, CXCR4, FPR1, LYZ, OCIAD2, ETV4, COL10A, PROM2, MMP11, ABCC3, BAIAP2L1, FABP4, STX11, FHL1, TEK, FMO2, CRYAB, GRK5, TMEM100, DLGAP5, KIF11, RAD51AP1, CDC6, OIP5, NCAPG, CENPF, KIF4A, CDC25A, CDH5, BDNF, SELE, KIF23, PLK1, CBFA2T3, CR2, SEL1L3, TM6SF1, TSPAN32, ITGA6, MAPK11, RASA3, TLR6, AURKB, HMGA2, ASPM, CKS1B, CHRDL1, SPARCL1 | [155-177] |
| GSE68465 | 442/0 | USA | ||
| GSE68571 | 96/0 | USA | ||
| GSE69405 | 52/150 | South Korea | ||
| GSE40791 | 94/100 | USA | ||
| GSE18842 | 72/86 | Spain | ||
| GSE74706 | 18/18 | Germany | ||
| GSE10072 | 135/135 | USA | ||
| GSE29013 | 55/0 | USA | ||
| GSE13213 | 117/0 | Japan | ||
| GSE7670 | 66/59 | Taiwan | ||
| GSE31547 | 30/20 | USA | ||
| GSE136043 | 5/5 | China | ||
| GSE140797 | 7/7 | China | ||
| GSE63459 | 32/33 | USA | ||
| GSE85841 | 8/8 | China | ||
| GSE116959 | 57/11 | France | ||
| GSE75037 | 83/83 | USA | ||
| GSE32863 | 60/60 | USA | ||
| GSE19188 | 78/78 | Netherlands | ||
| GSE33532 | 80/20 | Germany | ||
| GSE85716 | 06/06 | China | ||
| GSE119004 | 23/0 | Canada | ||
| GSE43458 | 84/26 | USA | ||
| GSE6044 | 42/05 | Germany | ||
| GSE62949 | 56/0 | USA | ||
| GSE115002 | 48/56 | China | ||
| GSE40419 | 88/76 | USA | ||
| GSE66727 | 23/0 | USA | ||
| GSE86337 | 10/0 | Australia | ||
| GSE57148 | 7/182 | South Korea | ||
| Total = 31 | Total = 1980/1230 | Total = 145 |
A PPI network, module, and enrichment analysis
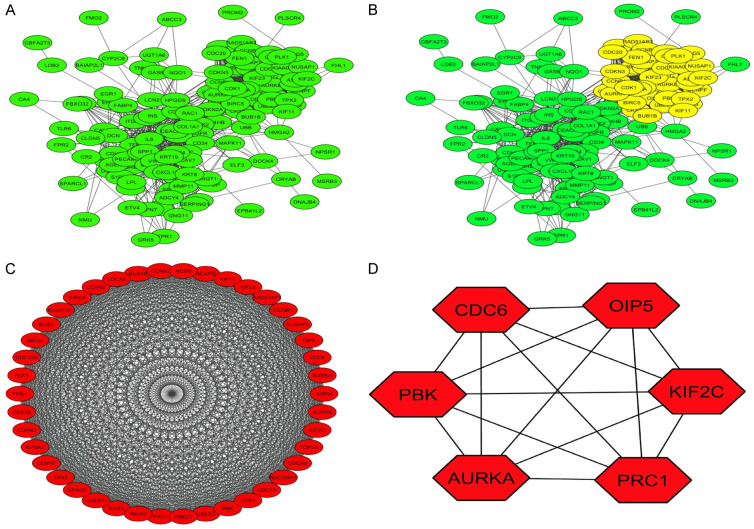
STRING was conducted to get a PPI network of the pooled hub genes. The obtained PPI network was consist of 145 nodes and 2341 edges (Figure 1A). Then, by applying Cytoscape, the MCODE and Cytohubba applications has helped to identified one most significant module and 6 hub genes as the real hub genes (CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1) based on the degree scores (Figure 1B, 1C and Table 2). Later, the enrichment analysis has revealed that real hub are involved in different GO and KEGG terms including Mitotic nuclear division etc. BP terms, Microtubule cytoskeleton etc. CC terms, ATP binding etc. MF terms (Figure 2 and Table 3), and Cell cycle etc. KEGG terms (Figure 2D and Table 4).
Figure 1.
(A) A PPI network showing LUAD-related extracted hub genes from the selected studies. (B) A PPI network showing one significant module, (C) A PPI network of the hub genes identified in the significant module, and (D) Six real hub genes identified via degree method.
Table 2.
List of the real hub genes identified from a PPI network of the extracted 124 LUAD related hub genes
| Sr. No | Name of the gene | MCODE Node Status | MCODE Score |
|---|---|---|---|
| 1 | CDC6 | Clustered | 38.90488 |
| 2 | PBK | Clustered | 38.90488 |
| 3 | AURKA | Clustered | 38.90488 |
| 4 | KIF2C | Clustered | 38.90488 |
| 5 | OIP5 | Clustered | 38.90488 |
| 6 | PRC1 | Clustered | 38.90488 |
Figure 2.
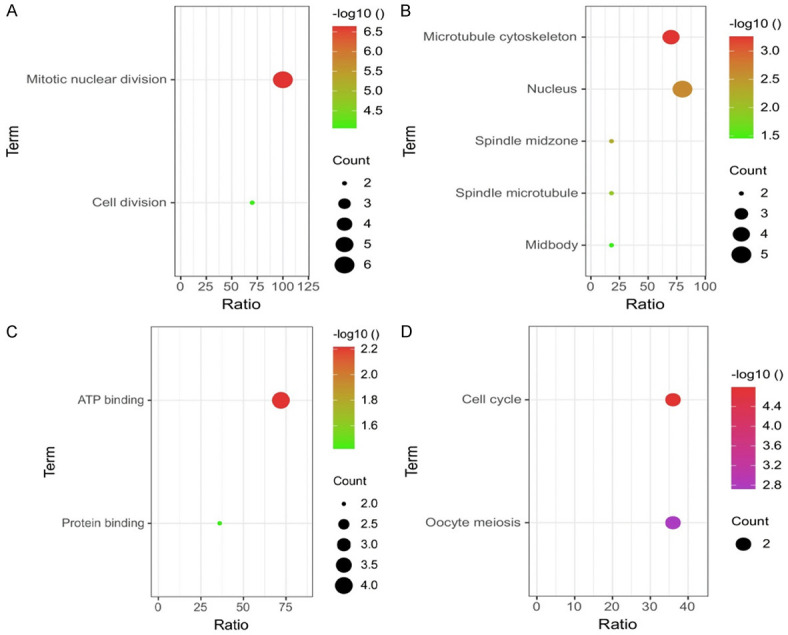
GO and KEGG enrichment analysis of real hub genes. (A) BF, (B) CC, (C) MF, and (D) KEGG analysis. A P-value <0.05 was consider as significant.
Table 3.
Details of the GO analysis of identified hub genes extracted from various GEO LUAD expression microarray datasets
| GO Term | GO ID | Gene count | P-value | Gene name |
|---|---|---|---|---|
| BP | ||||
| GO:0007067 | Mitotic nuclear division | 6 | 2.29554618712754E-7 | PBK, OIP5, KIF2C, CDC6, AURKA |
| GO:0042787 | Cell division | 4 | 8.703225761335279E-5 | OIP5, KIF2C, CDC6, AURKA |
| CC | ||||
| GO:0015630 | Microtubule cytoskeleton | 4 | 5.527753221351057E-4 | PRC1, KIF2C, AURKA |
| GO:0005634 | Nucleus | 5 | 0.0023131822778742635 | PRC1, PBK, OIP5, KIF2C, CDC6, AURKA |
| GO:0051233 | Spindle midzone | 2 | 0.005202617432597425 | CDC6, AURKA |
| GO:0005876 | Spindle microtubule | 2 | 0.012015152655811594 | PRC1, AURKA |
| GO:0030496 | Midbody | 2 | 0.03489913615678372 | CDK1, AURKA |
| MF | ||||
| GO:0005524 | ATP binding | 4 | 0.006046362041106639 | PBK, KIF2C, CDC6, AURKA |
| GO:0005515 | Protein binding | 2 | 0.03814879524822187 | PRC1, PBK, OIP5, KIF2C, CDC6, AURKA |
Table 4.
Details of the KEGG pathway analysis of identified hub genes extracted from various GEO LUAD expression microarray datasets
| Pathway ID | Pathway Name | Gene count | P-value | Gene name |
|---|---|---|---|---|
| hsa04110 | Cell cycle | 2 | 1.616848175824832E-5 | CDC6, AURKA |
| hsa04114 | Oocyte meiosis | 2 | 0.0018887038532489457 | CDC6, AURKA |
Expression analysis and cross validation of real hub genes expression
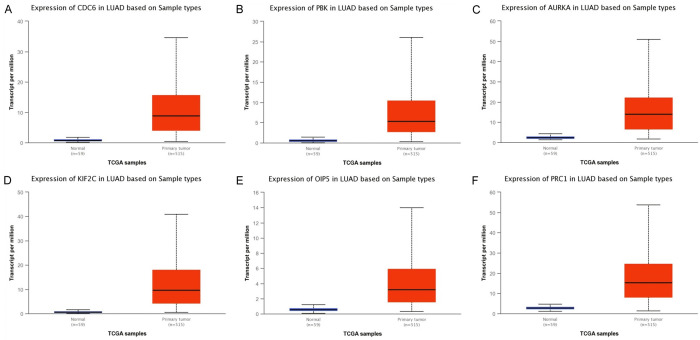
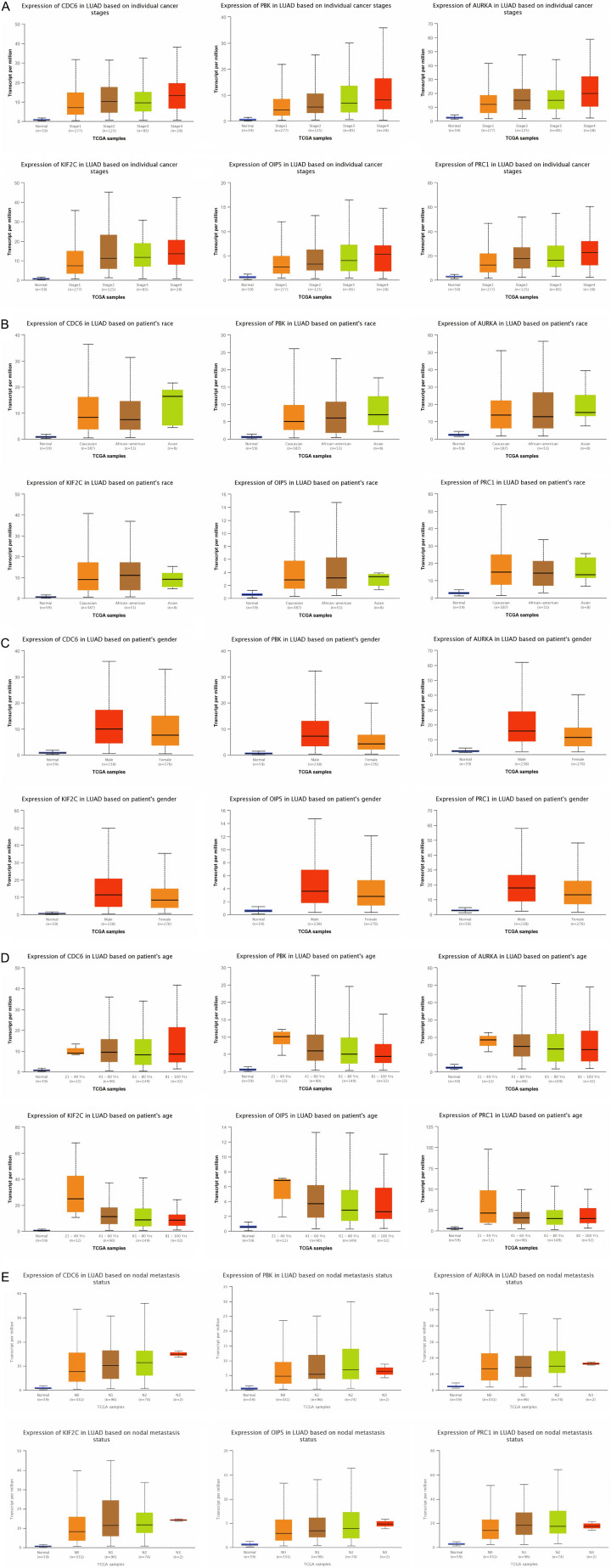
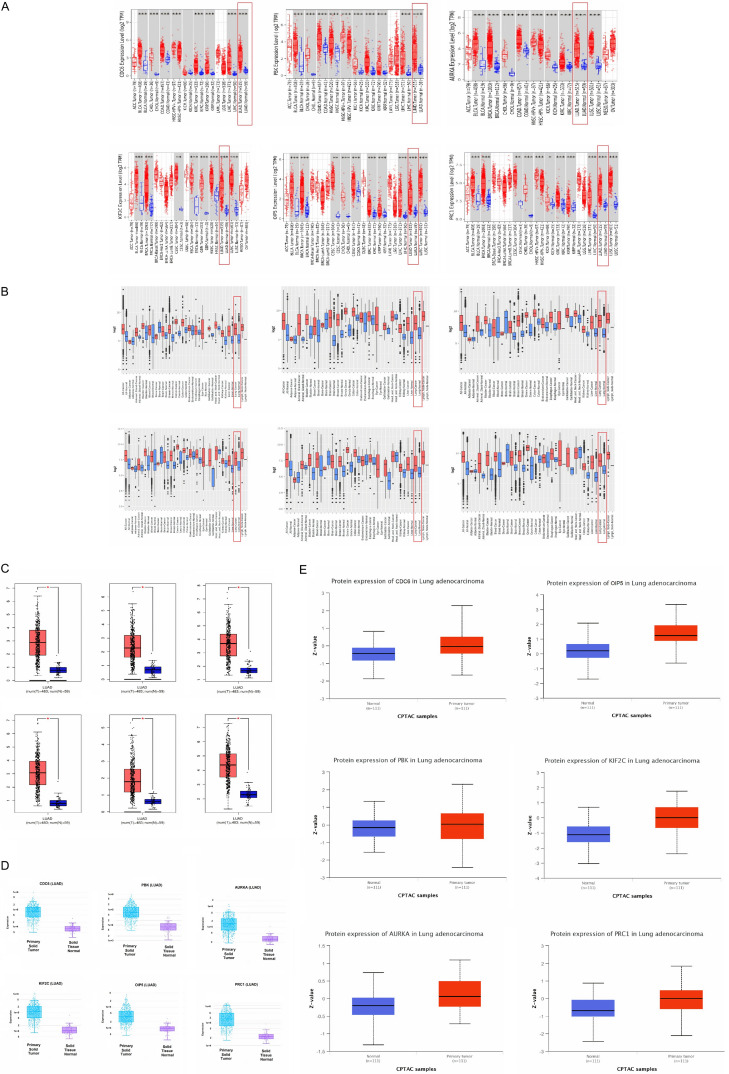
We initially measured real hub genes mRNA expression in 515 LUAD samples paired with 59 normal controls via UALCAN. Results of the analysis have shown the significant (P < 0.05) up-regulation of all the real hub genes (CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1) in LUAD samples of various clinical variables (cancer stage, race, gender, age, and nodal metastasis status) relative to normal controls (Figures 3 and 4). Later, TIMER, GENT2, GEPIA, and DriverDBV2 databases containing 515, 765, 483, and 710 LUAD samples paired with 59, 75, 59, and 111 normal controls, respectively were used to validate the mRNA expression of real hub genes on new independent cohorts. Our validation results showing significant (P < 0.05) up-regulation of all the real hub genes using these addition databases were also in agreement with the expression analysis results of ULACAN (Figure 5A-C). At last, we also validated real hub genes translational expression in 111 LUAD tissues paired with 111 controls via UALCAN. In view of our results, all real hub genes (CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1) were also found significantly (P < 0.05) up-regulated at protein level in LUAD patients relative to controls (Figure 5).
Figure 3.
mRNA expression analysis results of real hub genes in LUAD and normal controls via UALCAN. (A) CDC6, (B) PBK, (C) AURKA, (D), KIF2C, (E) OIP5, and (F) PRC1.
Figure 4.
mRNA and protein expression level validations of the real hub genes in LUAD patients paired with controls via different online expression databases. (A) mRNA expression level validation of real hub genes via TIMER, (B) mRNA expression level validation of real hub genes via GENT2, (C) mRNA expression level validation of real hub genes via GEPIA, (D) mRNA expression level validation of real hub genes via DriverDBV3, and (E) Protein expression level validation of real hub genes via UALCAN.
Figure 5.
Relative mRNA expression analysis results of real hub genes in LUAD patients stratified by different clinicopathological features. (A) Cancer stage (B) Patient’s race, (C) Patient’s gender, (D) Patient’s age, and (E) Nodal metastasis status.
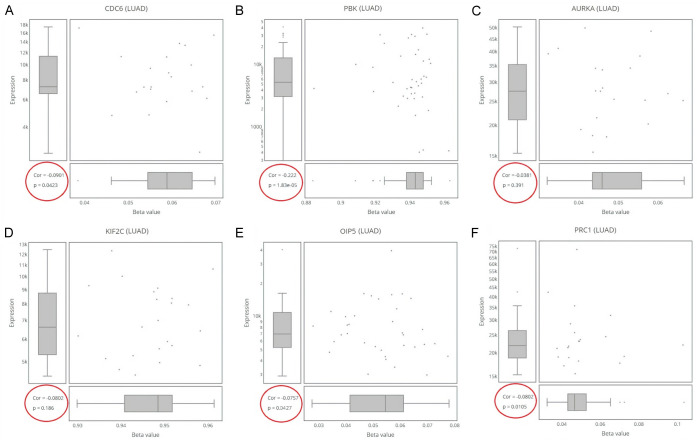
Promoter methylation analysis
In this work, the levels of promoter methylation were assessed via DriverDBV3. In view of our results, all real hub genes (CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1) were found significantly (P < 0.05) hypomethylated relative to controls in LUAD patients (Figure 6).
Figure 6.
Correlation of promoter methylation with mRNA expression of real hub genes in LUAD paired with controls. (A) CDC6, (B) PBK, (C) AURKA, (D), KIF2C, (E) OIP5, and (F) PRC1.
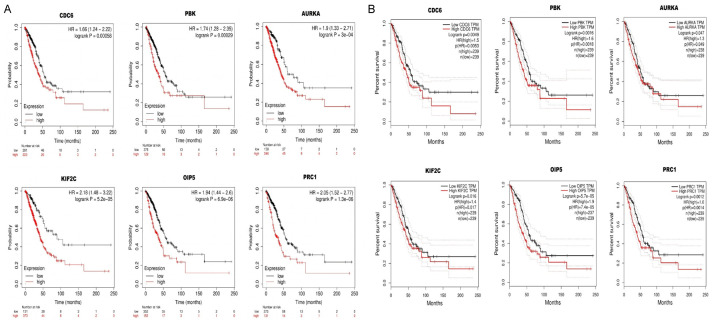
Real hub genes expression and prognosis in LUAD
We next evaluated the prognostic values (OS duration) of real hub genes via Kaplan-Meier Plotter and GEPIA tool. In view of our results, the higher levels of CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1 were found significantly (P < 0.05) linked to the poor OS of the LUAD patients, hence, we speculate these six real hub genes as the good prognostic biomarkers for predicting OS of LUAD patients (Figure 7A, 7B).
Figure 7.
Association between OS and real hub genes expression in LUAD patients. (A) via KM plotter and (B) via GEPIA tool.
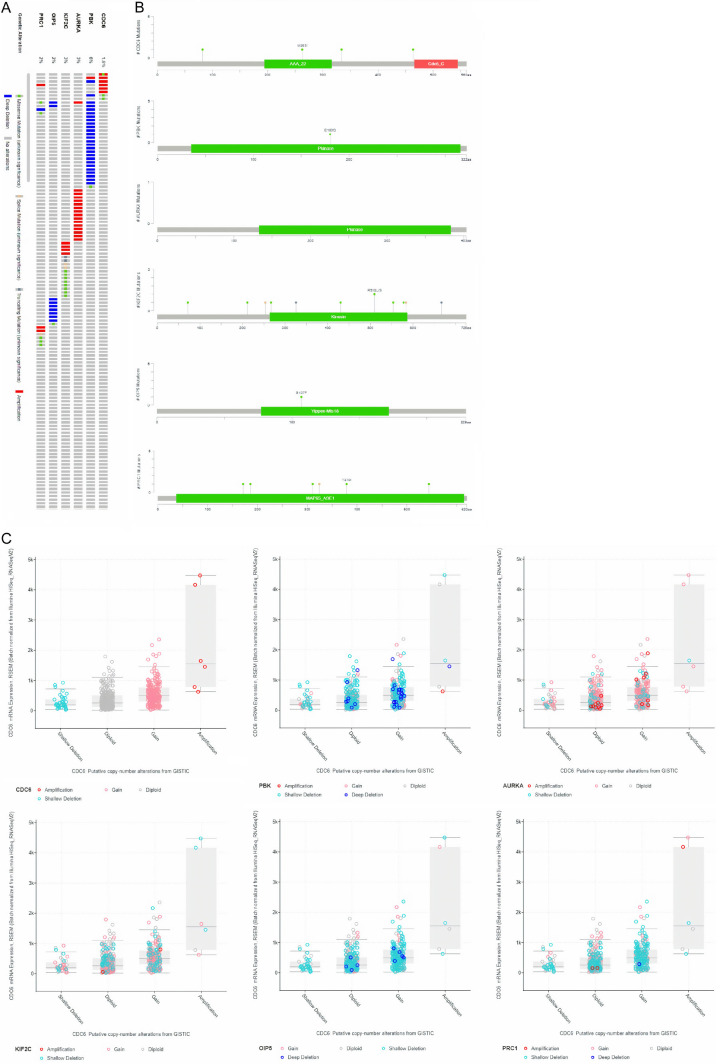
Genetic alterations information of real hub genes
Then, for enquiring genetic alterations in real hub genes, we conducted cBioPortal analysis. As shown in Figure 8A, PBK gene showed highest frequency (6%) of genomic alterations, while AURKA and KIF2C showed the second highest genomic alteration rate of 3%. Meanwhile other genes, including OIP5, PRC1, and CDC6 have shown a genetic alteration frequencies of 2%, 2%, and, 1.6%, respectively, in the analyzed LUAD samples. Furthermore, in PBK, and PRC1, the most frequently observed genetic alterations were deep deletion and missense mutations (Figure 8A), while other remaining real hub genes were enriched in deep amplification alterations only. In addition, we have also observed that most frequent mutation in CDC6 gene (M263I) was lied in AAA_22 domain (Figure 8B), and in PBK and KIF2C, the most frequent mutations including E180Q, and R510L/S were present in their respective Pkinase, and Kinesin domains (Figure 8B), while no mutation was detected in AURKA. However, on the other hand, most frequently seen mutations (S107F and T370I) in OIP5 and PRC1 were found in Yipee-MIs18 and MAP65-A3E1 functional domains, respectively (Figure 8B).
Figure 8.
Frequency and distribution of genomic alterations associated with real hub genes in LUAD. (A) Frequency of genomic alteration, (B) distribution of mutations in protein domains of real hub genes, and (C) types of CNVs in LUAD samples.
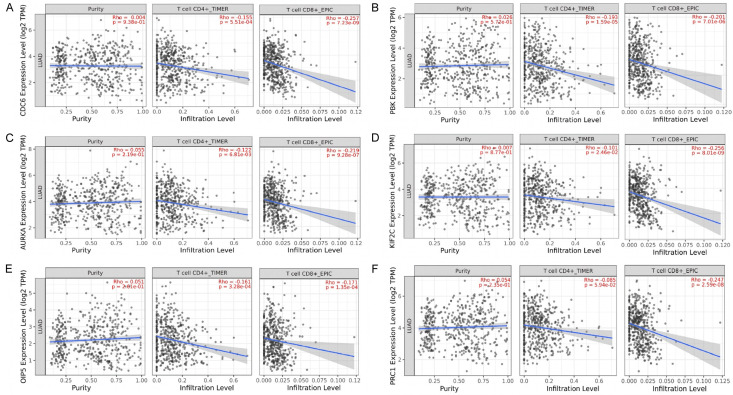
Tumor purity and immune cells infiltration analysis of real hub genes
The correlations among CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1 mRNA expressions and tumor purity, CD8+ T, and CD4+ T immune cells infiltration across LUAD have been analyzed via TIMER database. Our results have revealed the significant positive correlations between the expression of the real hub genes and tumor purity (Figure 9). While notable negative correlations between real hub genes’ expression and CD4+ T and CD8+ T immune cells infiltration levels.
Figure 9.
Correlation analysis of real hub genes expression with tumor purity, CD4+ T, and CD8+ T cells in LUAD. (A) CDC6, (B) PBK, (C) AURKA, (D) KIF2C, (E) OIP5, and (F) PRC1.
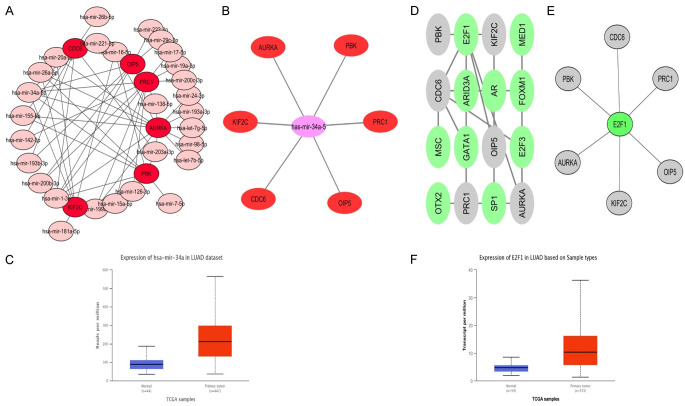
TFS-miRNA-mRNA network
As shown in Figure 10, the TFS-miRNA-mRNA interaction network of real hub genes constructed via ENCORI and Cytoscape is consists of 10 TFS, 28 miRNAs, and 6 mRNAs. Via degree method, one TF (E2F1), and one miRNA (hsa-mir-34a-5p) were identified as top TF and miRNA targeting all 6 real hub genes. Previously, has-mir-34a-5p in axis with LEF1 gene found to be involved in the pathogenesis of CESC [25], while, the role of has-mir-34a-5p miRNA is unclear in LUAD. Moreover, a previous study also revealed that E2F1 promotes EMT by regulating ZEB2 as a transcription factor in lung cancer [26]. The TFS-miRNA-mRNA co-regulatory network here in the current research has highlighted that E2F1 and hsa-mir-34a-5p can also act as the potential inducer of LUAD by dysregulating the identified real hub genes as an E2F1-hsa-mir-124-3p/CDC6/PBK/AURKA/KIF2C/OIP5/PRC1 axis. To further confirm the participation of identified TF and miRNA in LUAD development via dysregulating the real hub genes, we further checked the expression of E2F1 and has-mir-34a-5p in LUAD patients via UALCAN. As a result, a significant up-regulation of E2F1 and has-mir-34a-5p was also observed in LUAD samples than normal samples (Figure 10).
Figure 10.
TF-miRNA-mRNA network analysis of real hub genes in LUAD. (A) miRNAs targeting real hub genes, (B) has-mir-34a-5p miRNA targeting real hub genes, (C) relative expression of has-mir-34a-5p in LUAD and normal controls, (D) TFS targeting real hub genes, (E) E2F1 targeting real hub genes, and (F) relative expression analysis of E2F1 in LUAD samples paired with controls. The pink and grey nodes represent the miRNA, red nodes represent the hub gene, while green node represent the TFS.
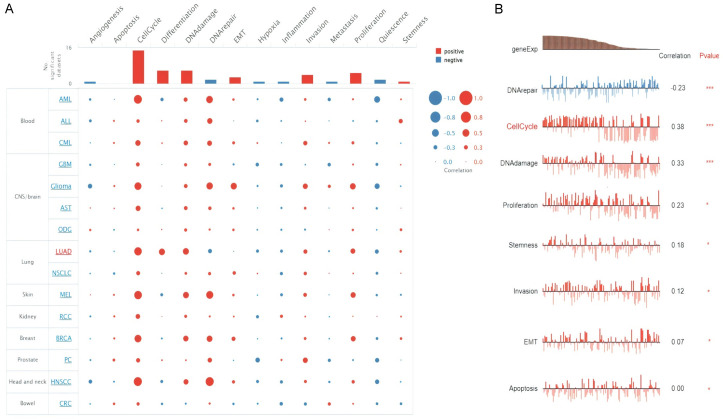
Single-cell functional analysis
CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1 genes further involvement in LUAD at single cell level was explored via CancerSEA database. As a result, the CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1 were revealed to be linked (positively or negatively) with fourteen different states at single cell level in LUAD (Figure 11A). However, real hub genes expression was notably negatively correlated with “DNA repair state, while positively correlated with Cell cycle, DNA Damage, Proliferation, Stemness Invasion, EMT, and Apoptosis states” (Figure 11B).
Figure 11.
Real hub genes and different divers states association in LUAD. (A) Correlation analysis of real hub genes expression with 14 different states in LUAD, and (B) Correlation analysis of real hub genes expression only significant states in LUAD.
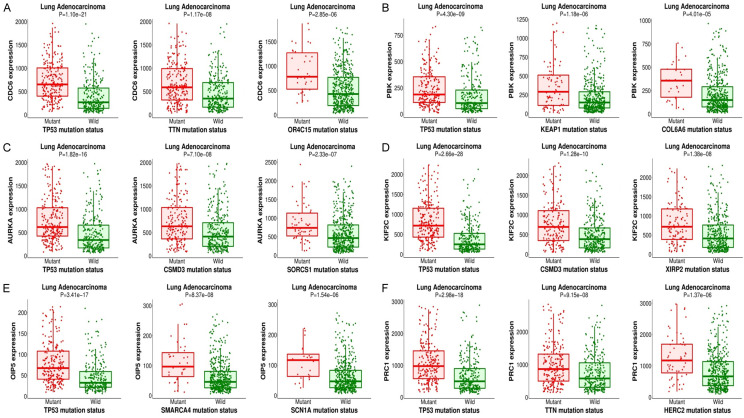
Real hub genes and their correlated different other mutant genes
MuTarget database with default settings “(FC > 1.4 and P < 0.05)” has helped us in this study to select us top 3 mutant genes for each real hub gene. As shown in Figure 12, top 3 mutant genes which positively correlated with the expression of each real hub gene are TP53, TTN, and OR4C15 with CDC6, TP53, KEAP1, and COL6A6 with PBK, TP53, CSMD3, and SORCS1 with AURKA, TP53, CSMD3, and XIR2P with KIF2C, TP53, SMARCA4, and SCN1A with OIP5, and TP53, TTN, and HERC2 with PRC1.
Figure 12.
CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1 genes positively correlated mutant genes in LUAD from MuTarget. (A) with CDC6, (B) with PBK, (C) with AURKA, (D) with KIF2C, (E) with OIP5, and (F) with PRC1.
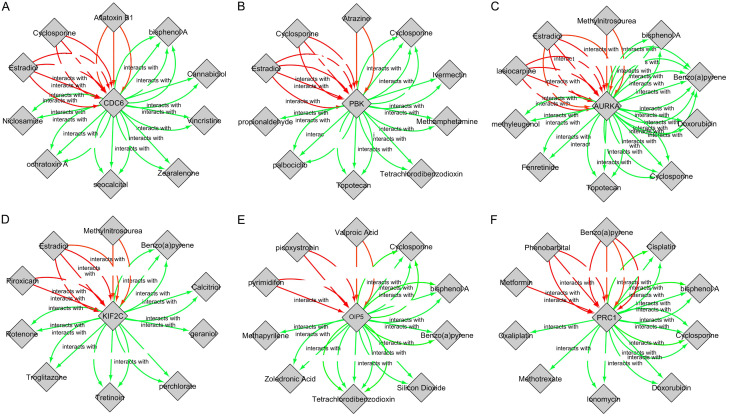
Drug-gene interaction analysis
Through CTD database, several drugs associated with 6 real hub genes were selected, and the relationships between them were visualized via Cytoscape (Figure 13). Finally, the drawn drug-gene interaction network reveled that all real hub genes including CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1 can potentially be regulated by several chemotherapeutic drugs. For instance, aflatoxin B1, and estradiol can elevate CDC6 expression while bisphenol A and cannabidiol can reduce CDC20 expression level (Figure 13).
Figure 13.
Screening of real hub genes-associated chemotherapeutic drugs. (A-F) indicates chemotherapeutic drugs that can decrease or increase the expression of the real hub genes. (A) CDC6-associated chemotherapeutic drugs, (B) PBK-associated chemotherapeutic drugs, (C) AURKA-associated chemotherapeutic drugs, (D) KIF2C-associated chemotherapeutic drugs, (E) OIP5-associated chemotherapeutic drugs, and (F) PRC1-associated chemotherapeutic drugs. Red arrows: drugs that increase the real hub genes expression, Green arrows: drug that decrease the real hub genes expression while the numbers of arrows represent the supported numbers of studies in literature.
Discussion
Previously, many studies have been carried out so far to explore molecular biomarkers and mechanisms behind LUAD for its accurate detection and treatment, the incidence of LUAD and mortality rate of LUAD patients is still increasing worldwide due to the heterogenetic-specific nature of the available biomarkers. In this study, we explored 6 real hub genes including CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1 that can overcome the heterogenetic-specific barrier and can apply as biomarkers in LUAD patients of different clinical variables. Moreover, GO and KEGG enrichment analysis revealed that identified real hub genes were significantly involved in diverse GO and KEGG terms (Figure 2).
CDC6 gene is present on chromosome 17 and encodes the CDC6 protein, which is a member of AAA+ ATPases (ATPases associated with cellular activities) family and play an important role in the initiation of DNA replication [27]. CDC6 usually found in the nucleus of cell, but is translocated to the cytoplasm during S-phase after its phosphorylation by CDKs [28]. When cell division begins, CDC6 mediates the assembly of the pre-replicative complex and maintains it for the loading of minichromosomal maintenance (MCM) proteins on DNA [29]. After the formation of MCM-chromatin complex, CDC6 is no longer needed and undergoes proteasome degradation [30]. Thus, the abnormal expression of CDC6 affects the replication mechanism [31]. Previously, an aberrant expression of CDC6 is significantly associated with breast cancer [32], colorectal cancer progression [33], invasiveness of cervical cancer [34], prostate cancer metastasis [35,36], pulmonary cancer [37], inhibition of apoptotic caspases 3 and 9 in pancreatic cancer [38], hepatocellular carcinoma [39,40], proliferation of esophageal squamous cell carcinoma [41], renal cell carcinoma [42], gastric cancer [43], bladder cancer [44], and osteosarcoma [45].
PBK gene is found on chromosome 8 and encodes a protein PBK of 322 amino acids. The encoded protein is a member of mitogen-activated serine/threonine-protein kinase (MAPKK) family [46,47] and is hard to detect in normal somatic tissues but is found frequently in cancerous tissues promoting cell survival, proliferation, and metastasis [48,49]. Abnormal expression of PBK is found to be associated with different cancers including thyroid carcinoma [50], prostate cancer progression [51,52], ovarian cancer [53,54], cervical cancer by ERK/c-Myc signaling [55], pathogenesis of breast cancer by EMT up-regulation [56] and invasion by TGFβ1-induced NFkB-dependent Snail/Slug [57], glioblastoma [58,59], oxaliplatin resistance in liver cancer [60,61], pancreatic cancer [62], NSCLC proliferation by paclitaxel resistance and inhibition of autophagic cell death [63,64], T-cell leukemia or lymphoma [65], colorectal cancer [66,67], oral squamous cell carcinoma [68], nasopharyngeal carcinoma [69], human endometrial cancer [70], urinary bladder transitional cell cancer [62], multiple myeloma [71], esophageal squamous cell carcinoma [72,73], and gastric cancer [74,75].
AURKA aids in controlling the cell cycle [76]. Various types of cancers have been linked with AURKA overexpression, such as prostate cancer [77], breast cancer [78] colon tumorigenesis [79], gastric cancer [80], head and neck cancer [81], liver metastasis [82], hepatocarcinogenesis [83], bladder cancer [84], NSCLC progression [85], ovarian cells [86], and esophageal cancer [87]. After that, many subsequent studies have been carried out to evaluate the AURKA expression level in LUAD, still, its role is poorly understood.
KIF2C gene encodes for a protein of 792 amino acids which belongs to a kinesin superfamily proteins (KIFs). This protein mainly found in the cell bodies and dendrites of the adult neurons in both the peripheral and central nervous systems [88]. KIF2C gene is consider very crucial for mitotic spindle dynamics, chromosomal segregation during anaphase, and separation of sister chromatids [89]. Dysregulation of KIF2C protein is reported to results in chromosome instability [90] and is also found to be significantly associated with bladder cancer invasion [91], Breast cancer [92,93], mediation of Wnt/β-catenin/mTORC1 signaling [94,95] and MEK/ERK signaling in hepatocellular carcinoma [96], thyroid cancer metastasis via TGF-β1/Smad pathway [97], non-small cell lung cancer (NSCLC) [98,99], colorectal cancer [100,101], gastric cancer [102], endometrial carcinoma [103], regulation of AKT/mTOR pathways in nasopharyngeal carcinoma [104], testicular carcinoma [105], esophageal squamous cell carcinoma [72], platinum-resistant in ovarian cancer [106], and regulation of Wnt/β-catenin pathway in cervical cancer [107].
OIP5 gene is located on chromosome 15 and encodes for a 25 kDa protein belonging to cancer-testis antigens (CTA) family [108]. The members of this family are crucial for the structure and function of kinetochore and centromeric region [109]. In the medical literature, an abnormal expression of OIP5 was linked with proliferation of colorectal and gastric cancer cells [110,111], clear cell and peripheral renal cell carcinoma (CCRCC, PRCC) progression [112,113], acute myeloid leukemia [114], proliferation of lung and esophageal cancers by RAF1 interaction [115], proliferation of thyroid cancer cell by Wnt/β-catenin signaling [116], pancreatic cancer via activation of AGR2/AKT/ERK [117] and miR-186-5p/NGFR signaling pathway [118], breast cancer by regulating GLO1 expression [119] and miR-139-5p/Notch1 pathway [120], NSCLC metastasis by mTOR signaling pathway [121] and miR-140-5p/HDAC7/VEGFA signaling [122], cervical cancer by regulating ROCK1 expression [123], ovarian cancer by mediating miR-128-3p/CCNG1 pathway [124,125], progression of osteosarcoma via miR-137-3p/PTN axis [126], multiple myeloma progression [127], prostate cancer progression via miR-128-3p/SLC7A11 signaling [128], gallbladder cancer [129], head and neck squamous cell carcinoma [130], endometrial cancer by PTEN/AKT pathway [131], Warburg effect in cervical cancer by miR-124-5p/IDH2/HIF-1α signaling [132], proliferation of bladder cancer via miR-217/MTDH pathway [133,134], NPC progression by modulating JAK2/STAT3 [135], and metastasis of glioblastoma [136].
PRC1 gene is mapped on chromosome 15 and encodes for PRC1 protein of 620 amino acids [137]. This gene highly express at early mitosis during S-G2/M phases and reduce when the cell enters into the G1 phase [137]. Expression variations in PRC1 is earlier associated with different human cancers including lung adenocarcinoma [138,139], gastric carcinoma [140], ovarian cancer [141], cholangiocarcinoma [142], liver carcinoma [143,144], prostate cancer [145], breast cancer [93,146], bladder cancer [147], colon cancer [148], osteosarcoma progression [149], human endometrial cancer [150], and nasopharyngeal carcinoma [151]. In this study, we found its significant (P < 0.05) up-regulation in LUAD patients of different clinical characteristics as compared to the normal controls. Taken together the expression profiling of the real hub genes including CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1, we have suggested that up-regulation of these six genes may serve as a potential biomarker in LUAD patients regardless of different clinical characteristics relative to controls.
Moreover, the survival analysis further suggested the application of identified hub genes as the potential prognostic biomarkers in LUAD patients. Next, we observed that real hub genes genetically altered in a small number of LUAD samples and their promoter regions were hypomethylation.
Next, we observed that real hub genes genetically altered in a small number of LUAD samples. Further analysis also revealed that mutations in the real hub genes (CDC6, PBK, KIF2C, OIP5, and PRC1) can change amino acids at distinct positions in the encoded proteins. The correlation analysis among real hub genes expression and promoter methylation levels has shown the expected significant (P < 0.05) negative correlations in LUAD. That is why, we speculated that promoter hypomethylation might be a key factor involved in the up-regulation of real hub genes across LUAD samples.
In the current study, we observed that two immune cells infiltration levels in LUAD could induce differential expression of the CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1 genes. Via TIMER, we found that real hub genes expression are significantly correlated with CD4+ T, CD8+ T immune cells, and tumor purity in LUAD. Some studies have earlier investigated the tumor-associated roles of T cells across LUAD. These studies explored that activated CD8+ T cells in LUAD often elicited type I immune responses, which indicate a favorable prognosis [152,153], however, on the other hand, Th2, and Th17 cells were found to be linked with tumor progression and unfavorable prognosis [154]. The intriguing relationships between tumour purity, CD4+ T, CD8+ T immune cells, and real hub gene expression levels found in this study may inspire fresh approaches to the treatment of LUAD.
We further elucidated that E2F1 TF and hsa-mir-124-3p miRNA target all six real hub genes (CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1). To our knowledge, this study is the first to describe the role of E2F1 and hsa-mir-124-3p in tumorigenesis when combined with CDC6, PBK, AURKA, KIF2C, OIP5, and PRC1 in LUAD. This important knowledge can be applied to the treatment of LUAD to control how real hub genes are expressed.
It was also noticed in the current study that real hub genes’ expressions were positively associated with “Cell cycle, DNA Damage, Proliferation, Stemness Invasion, EMT, and Apoptosis” in LUAD. The function of the identified hub genes in LUAD development is being collectively investigated for the first time in this study.
To find mutant genes that change the expression of real hub genes, we expanded the network of real hub genes in this study using the muTarget platform. TP53, TTN, and OR4C15 mutant genes were correlated with CDC6, TP53, KEAP1, and COL6A6 mutant genes were correlated with PBK, TP53, CSMD3, and SORCS1 mutant genes were correlated with AURKA, TP53, CSMD3, and XIR2P mutant genes were correlated with KIF2C, TP53, SMARCA4, and SCN1A mutant genes were correlated with OIP5, and TP53, TTN, and HERC2 mutant genes were correlated with PRC1. This knowledge could be useful in developing multi-gene and individualized therapeutic strategies for LUAD patients.
Conclusion
Through integrated bioinformatics approach, our study has revealed the 6 real hub genes (hub genes of hub genes) which might play pathogenic roles in LUAD development and can also use as a novel diagnostic and prognostic biomarker for the LUAD patients regardless of heterogeneity barriers. However, more in-depth experimental studies are needed before clinical applications.
Acknowledgements
The authors extend their appreciation to the Researchers Supporting Project number (RSPD2023R725) King Saud University, Riyadh, Saudi Arabia.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Esposito L, Conti D, Ailavajhala R, Khalil N, Giordano A. Lung cancer: are we up to the challenge? Curr Genomics. 2010;11:513–518. doi: 10.2174/138920210793175903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Thandra KC, Barsouk A, Saginala K, Aluru JS, Barsouk A. Epidemiology of lung cancer. Contemp Oncol (Pozn) 2021;25:45–52. doi: 10.5114/wo.2021.103829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 5.Gariani J, Martin SP, Hachulla AL, Karenovics W, Adler D, Soccal PM, Becker CD, Montet X. Noninvasive pulmonary nodule characterization using transcutaneous bioconductance: preliminary results of an observational study. Medicine (Baltimore) 2018;97:e11924. doi: 10.1097/MD.0000000000011924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li R, Han K, Xu D, Chen X, Lan S, Liao Y, Sun S, Rao S. A seven-long non-coding RNA signature improves prognosis prediction of lung adenocarcinoma: an integrated competing endogenous RNA network analysis. Front Genet. 2021;11:625977–625977. doi: 10.3389/fgene.2020.625977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim N, Kim HK, Lee K, Hong Y, Cho JH, Choi JW, Lee J, Suh Y, Ku BM, Eum HH, Choi S, Choi YL, Joung JG, Park WY, Jung HA, Sun JM, Lee SH, Ahn JS, Park K, Ahn MJ, Lee HO. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun. 2020;11:1–15. doi: 10.1038/s41467-020-16164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Zhang Z, Yu Z. Identification of a novel glycolysis-related gene signature for predicting metastasis and survival in patients with lung adenocarcinoma. J Transl Med. 2019;17:423–431. doi: 10.1186/s12967-019-02173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng T, Wei D, Li Q, Yang X, Han Y, Luo Y, Jiang Y. Four novel prognostic genes related to prostate cancer identified using co-expression structure network analysis. Front Genet. 2021;12:584164. doi: 10.3389/fgene.2021.584164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genom Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jézéquel P, Campone M, Gouraud W, Guérin-Charbonnel C, Leux C, Ricolleau G, Campion L. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131:765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 15.Park SJ, Yoon BH, Kim SK, Kim SY. GENT2: an updated gene expression database for normal and tumor tissues. BMC Med Genomics. 2019;12:101. doi: 10.1186/s12920-019-0514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung IF, Chen CY, Su SC, Li CY, Wu KJ, Wang HW, Cheng WC. DriverDBv2: a database for human cancer driver gene research. Nucleic Acids Res. 2016;44:D975–979. doi: 10.1093/nar/gkv1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang DP, Zeng YH, Yuan WQ, Huang XF, Chen SQ, Wang MY, Qiu YJ, Tong GD. Bioinformatics analyses of potential miRNA-mRNA regulatory axis in HBV-related hepatocellular carcinoma. Int J Med Sci. 2021;18:335–346. doi: 10.7150/ijms.50126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, Lee S, Kang B, Jeong D, Kim Y, Jeon HN, Jung H, Nam S, Chung M, Kim JH, Lee I. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46:D380–D386. doi: 10.1093/nar/gkx1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan H, Yan M, Zhang G, Liu W, Deng C, Liao G, Xu L, Luo T, Yan H, Long Z, Shi A, Zhao T, Xiao Y, Li X. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019;47:D900–D908. doi: 10.1093/nar/gky939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagy Á, Győrffy B. muTarget: a platform linking gene expression changes and mutation status in solid tumors. Int J Cancer. 2021;148:502–511. doi: 10.1002/ijc.33283. [DOI] [PubMed] [Google Scholar]
- 24.Mattingly CJ, Colby GT, Forrest JN, Boyer JL. The Comparative Toxicogenomics Database (CTD) Environ Health Perspect. 2003;111:793–795. doi: 10.1289/ehp.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Zhao Y, Lu Q, Fei X, Lu C, Li C, Chen H. MiR-34a-5p inhibits proliferation, migration, invasion and epithelial-mesenchymal transition in esophageal squamous cell carcinoma by targeting LEF1 and inactivation of the Hippo-YAP1/TAZ signaling pathway. J Cancer. 2020;11:3072–3081. doi: 10.7150/jca.39861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T, Chen X, Qiao W, Kong L, Sun D, Li Z. Transcription factor E2F1 promotes EMT by regulating ZEB2 in small cell lung cancer. BMC Cancer. 2017;17:719. doi: 10.1186/s12885-017-3701-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ausiannikava D, Allers T. Diversity of DNA replication in the archaea. Genes (Basel) 2017;8:56. doi: 10.3390/genes8020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piatti S, Böhm T, Cocker JH, Diffley J, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 29.Mizushima T, Takahashi N, Stillman B. Cdc6p modulates the structure and DNA binding activity of the origin recognition complex in vitro. Genes Dev. 2000;14:1631–1641. [PMC free article] [PubMed] [Google Scholar]
- 30.Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci U S A. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liontos M, Koutsami M, Sideridou M, Evangelou K, Kletsas D, Levy B, Kotsinas A, Nahum O, Zoumpourlis V, Kouloukoussa M, Lygerou Z, Taraviras S, Kittas C, Bartkova J, Papavassiliou AG, Bartek J, Halazonetis TD, Gorgoulis VG. Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res. 2007;67:10899–10909. doi: 10.1158/0008-5472.CAN-07-2837. [DOI] [PubMed] [Google Scholar]
- 32.Cabañas Morafraile E, Pérez-Peña J, Fuentes-Antrás J, Manzano A, Pérez-Segura P, Pandiella A, Galán-Moya EM, Ocaña A. Genomic correlates of DNA damage in breast cancer subtypes. Cancers (Basel) 2021;13:2117. doi: 10.3390/cancers13092117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Y, Wang L, Li Z, Wan Z, Shao M, Wu S, Wang G. Potential prognostic and diagnostic values of CDC6, CDC45, ORC6 and SNHG7 in colorectal cancer. OncoTargets Ther. 2019;12:11609. doi: 10.2147/OTT.S231941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy N, Ring M, Heffron CC, King B, Killalea A, Hughes C, Martin CM, McGuinness E, Sheils O, O’leary JJ. p16INK4A, CDC6, and MCM5: predictive biomarkers in cervical preinvasive neoplasia and cervical cancer. J Clin Pathol. 2005;58:525–534. doi: 10.1136/jcp.2004.018895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Z, Cho H, Hampton GM, Theodorescu D. Cdc6 and cyclin E2 are PTEN-regulated genes associated with human prostate cancer metastasis. Neoplasia. 2009;11:66–76. doi: 10.1593/neo.81048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Xu W, Wang T, Yu C, Rao X, Hong X, Wang X. miR-26a inhibits the proliferation and migration of prostate cancer by targeting CDC6. Minerva Med. 2021;112:661–663. doi: 10.23736/S0026-4806.20.06479-4. [DOI] [PubMed] [Google Scholar]
- 37.An C, Liu G, Cheng S, Pang B, Sun S, Zhang Y, Pan Z, Kang X. A pilot study of cdc6 as a biomarker for circulating tumor cells in patients with lung cancer. J Clin Lab Anal. 2020;34:e23245. doi: 10.1002/jcla.23245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Youn Y, Lee JC, Kim J, Kim JH, Hwang JH. Cdc6 disruption leads to centrosome abnormalities and chromosome instability in pancreatic cancer cells. Sci Rep. 2020;10:1–11. doi: 10.1038/s41598-020-73474-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu H, Huang J, Hua S, Liang L, He X, Zhan M, Lu L, Chu J. Interactome analysis of gene expression profiles identifies CDC6 as a potential therapeutic target modified by miR-215-5p in hepatocellular carcinoma. Int J Med Sci. 2020;17:2926. doi: 10.7150/ijms.51145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong DG, Yao FZ. CDC6 is a possible biomarker for hepatocellular carcinoma. Int J Clin Exp Pathol. 2021;14:811. [PMC free article] [PubMed] [Google Scholar]
- 41.Ke Y, Guo W, Huang S, Li Y, Guo Y, Liu X, Jin Y, Ma H. RYBP inhibits esophageal squamous cell carcinoma proliferation through downregulating CDC6 and CDC45 in G1-S phase transition process. Life Sci. 2020;250:117578. doi: 10.1016/j.lfs.2020.117578. [DOI] [PubMed] [Google Scholar]
- 42.Yicong Y, Wang Y, Denglong W, Baoying H. Increased CDC6 expression associates with poor prognosis in patients with clear cell renal cell carcinoma. Front Oncol. 2021;11:666418. doi: 10.3389/fonc.2021.666418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X, Zhang M, Guo Q, Hu X, Zhao Z, Ni L, Liu L, Wang X, Wang Z, Tong D, Chang S, Cao Y, Huang C. MicroRNA-1297 inhibits proliferation and promotes apoptosis in gastric cancer cells by downregulating CDC6 expression. Anti-cancer drugs. 2019;30:803–811. doi: 10.1097/CAD.0000000000000776. [DOI] [PubMed] [Google Scholar]
- 44.Shi X, Chen S, Zhang Y, Xie W, Hu Z, Li H, Li J, Zhou Z, Tan W. Norcantharidin inhibits the DDR of bladder cancer stem-like cells through cdc6 degradation. OncoTargets Ther. 2019;12:4403. doi: 10.2147/OTT.S209907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang W, Yu Y, Liu J, Zhao Q, Wang J, Zhang J, Dang X. Downregulation of Cdc6 inhibits tumorigenesis of osteosarcoma in vivo and in vitro. Biomed Pharmacother. 2019;115:108949. doi: 10.1016/j.biopha.2019.108949. [DOI] [PubMed] [Google Scholar]
- 46.Gaudet S, Branton D, Lue RA. Characterization of PDZ-binding kinase, a mitotic kinase. Proc Natl Acad Sci U S A. 2000;97:5167–5172. doi: 10.1073/pnas.090102397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abe Y, Matsumoto S, Kito K, Ueda N. Cloning and expression of a novel MAPKK-like protein kinase, lymphokine-activated killer T-cell-originated protein kinase, specifically expressed in the testis and activated lymphoid cells. J Biol Chem. 2000;275:21525–21531. doi: 10.1074/jbc.M909629199. [DOI] [PubMed] [Google Scholar]
- 48.Ayllon V, O’connor R. PBK/TOPK promotes tumour cell proliferation through p38 MAPK activity and regulation of the DNA damage response. Oncogene. 2007;26:3451–3461. doi: 10.1038/sj.onc.1210142. [DOI] [PubMed] [Google Scholar]
- 49.Herbert KJ, Ashton TM, Prevo R, Pirovano G, Higgins GS. T-LAK cell-originated protein kinase (TOPK): an emerging target for cancer-specific therapeutics. Cell Death Dis. 2018;9:1089. doi: 10.1038/s41419-018-1131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou J, Dong S, Shi C. Identification of key pathways and biomarkers in anaplastic thyroid cancer using an integrated analysis. J BUON. 2020;25:2690–2699. [PubMed] [Google Scholar]
- 51.Brown-Clay JD, Shenoy DN, Timofeeva O, Kallakury BV, Nandi AK, Banerjee PP. PBK/TOPK enhances aggressive phenotype in prostate cancer via β-catenin-TCF/LEF-mediated matrix metalloproteinases production and invasion. Oncotarget. 2015;6:15594. doi: 10.18632/oncotarget.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma X, Liu J, Wang H, Jiang Y, Wan Y, Xia Y, Cheng W. Identification of crucial aberrantly methylated and differentially expressed genes related to cervical cancer using an integrated bioinformatics analysis. Biosci Rep. 2020;40:BSR20194365. doi: 10.1042/BSR20194365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chuang L, Lyu Y, Liu C. Identification of molecular markers associated with ovarian cancer prognosis using bioinformatics analysis. 2020 doi: 10.1002/jcp.27926. [DOI] [PubMed] [Google Scholar]
- 54.Li C, Lyu Y, Liu C, Yin S. The role of PDZ as a potential prognostic and diagnostic biomarker in ovarian cancer. Cancer Biother Radiopharm. 2022;37:569–579. doi: 10.1089/cbr.2020.4249. [DOI] [PubMed] [Google Scholar]
- 55.Ma H, Han F, Yan X, Qi G, Li Y, Li R, Yan S, Yuan C, Song K, Kong B. PBK promotes aggressive phenotypes of cervical cancer through ERK/c-Myc signaling pathway. J Cell Physiol. 2021;236:2767–2781. doi: 10.1002/jcp.30134. [DOI] [PubMed] [Google Scholar]
- 56.Lee YJ, Park JH, Oh SM. Activation of NF-κB by TOPK upregulates snail/slug expression in TGF-β1 signaling to induce epithelial-mesenchymal transition and invasion of breast cancer cells. Biochem Biophys Res Commun. 2020;530:122–129. doi: 10.1016/j.bbrc.2020.07.015. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Liang F, Zhou Y, Qiu J, Lv Q, Du Z. Sharp Downregulation of hub genes associated with the pathogenesis of breast cancer from ductal carcinoma in situ to invasive ductal carcinoma. Front Oncol. 2021;11:634569. doi: 10.3389/fonc.2021.634569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong C, Fan W, Fang S. PBK as a potential biomarker associated with prognosis of glioblastoma. J Mol Neurosci. 2020;70:56–64. doi: 10.1007/s12031-019-01400-1. [DOI] [PubMed] [Google Scholar]
- 59.Mao P, Bao G, Wang YC, Du CW, Yu X, Guo XY, Li RC, Wang MD. PDZ-binding kinase-dependent transcriptional regulation of CCNB2 promotes tumorigenesis and radio-resistance in glioblastoma. Transl Oncol. 2020;13:287–294. doi: 10.1016/j.tranon.2019.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao H, Yang M, Yang Y, Fang J, Cui Y. PBK/TOPK promotes chemoresistance to oxaliplatin in hepatocellular carcinoma cells by regulating PTEN. Acta Biochim Biophys Sin (Shanghai) 2021;53:584–592. doi: 10.1093/abbs/gmab028. [DOI] [PubMed] [Google Scholar]
- 61.Yang QX, Zhong S, He L, Jia XJ, Tang H, Cheng ST, Ren JH, Yu HB, Zhou L, Zhou HZ, Gong R, Huang AL, Chen J. PBK overexpression promotes metastasis of hepatocellular carcinoma via activating ETV4-uPAR signaling pathway. Cancer Lett. 2019;452:90–102. doi: 10.1016/j.canlet.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 62.Singh PK, Srivastava AK, Dalela D, Rath SK, Goel MM, Bhatt ML. Expression of PDZ-binding kinase/T-LAK cell-originated protein kinase (PBK/TOPK) in human urinary bladder transitional cell carcinoma. Immunobiology. 2014;219:469–474. doi: 10.1016/j.imbio.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 63.Lei B, Qi W, Zhao Y, Li Y, Liu S, Xu X, Zhi C, Wan L, Shen H. PBK/TOPK expression correlates with mutant p53 and affects patients’ prognosis and cell proliferation and viability in lung adenocarcinoma. Hum Pathol. 2015;46:217–224. doi: 10.1016/j.humpath.2014.07.026. [DOI] [PubMed] [Google Scholar]
- 64.Park JH, Park SA, Lee YJ, Park HW, Oh SM. PBK attenuates paclitaxel-induced autophagic cell death by suppressing p53 in H460 non-small-cell lung cancer cells. FEBS Open Bio. 2020;10:937–950. doi: 10.1002/2211-5463.12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ishikawa C, Senba M, Mori N. Mitotic kinase PBK/TOPK as a therapeutic target for adult T-cell leukemia/lymphoma. Int J Oncol. 2018;53:801–814. doi: 10.3892/ijo.2018.4427. [DOI] [PubMed] [Google Scholar]
- 66.Su TC, Chen CY, Tsai WC, Hsu HT, Yen HH, Sung WW, Chen CJ. Cytoplasmic, nuclear, and total PBK/TOPK expression is associated with prognosis in colorectal cancer patients: a retrospective analysis based on immunohistochemistry stain of tissue microarrays. PLoS One. 2018;13:e0204866. doi: 10.1371/journal.pone.0204866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nagano-Matsuo A, Inoue S, Koshino A, Ota A, Nakao K, Komura M, Kato H, Naiki-Ito A, Watanabe K, Nagayasu Y, Hosokawa Y, Takiguchi S, Kasugai K, Kasai K, Inaguma S, Takahashi S. PBK expression predicts favorable survival in colorectal cancer patients. Virchows Archiv. 2021;479:277–284. doi: 10.1007/s00428-021-03062-0. [DOI] [PubMed] [Google Scholar]
- 68.Yu WN, Lin HF, Lee Y, Shia WC, Sung WW, Yeh CM, Lin YM. PBK expression is associated with prognosis of patients with oral squamous cell carcinoma treated with radiotherapy: a retrospective study. Anticancer Res. 2021;41:2177–2182. doi: 10.21873/anticanres.14991. [DOI] [PubMed] [Google Scholar]
- 69.Wang MY, Qi B, Wang F, Lin ZR, Li MY, Yin WJ, Zhu YY, He L, Yu Y, Yang F, Liu JQ, Chen DP. PBK phosphorylates MSL1 to elicit epigenetic modulation of CD276 in nasopharyngeal carcinoma. Oncogenesis. 2021;10:9. doi: 10.1038/s41389-020-00293-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mamoor S. Over-expression of PDZ binding kinase in human endometrial cancer. 2021 [Google Scholar]
- 71.Ota A, Hanamura I, Karnan S, Inaguma S, Takei N, Lam VQ, Mizuno S, Kanasugi J, Wahiduzzaman M, Rahman ML, Hyodo T, Konishi H, Tsuzuki S, Ikeda H, Takami A, Hosokawa Y. Novel interleukin-6 inducible gene PDZ-binding kinase promotes tumor growth of multiple myeloma cells. J Interferon Cytokine Res. 2020;40:389–405. doi: 10.1089/jir.2020.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng L, Li L, Xie J, Jin H, Zhu N. Six novel biomarkers for diagnosis and prognosis of esophageal squamous cell carcinoma: validated by scRNA-seq and qPCR. J Cancer. 2021;12:899–911. doi: 10.7150/jca.50443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ohashi T, Komatsu S, Ichikawa D, Miyamae M, Okajima W, Imamura T, Kiuchi J, Nishibeppu K, Kosuga T, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Tsuda H, Otsuji E. Overexpression of PBK/TOPK contributes to tumor development and poor outcome of esophageal squamous cell carcinoma. Anticancer Res. 2016;36:6457–6466. doi: 10.21873/anticanres.11244. [DOI] [PubMed] [Google Scholar]
- 74.Kwon CH, Park HJ, Choi YR, Kim A, Kim HW, Choi JH, Hwang CS, Lee SJ, Choi CI, Jeon TY, Kim DH, Kim GH, Park do Y. PSMB8 and PBK as potential gastric cancer subtype-specific biomarkers associated with prognosis. Oncotarget. 2016;7:21454. doi: 10.18632/oncotarget.7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ohashi T, Komatsu S, Ichikawa D, Miyamae M, Okajima W, Imamura T, Kiuchi J, Kosuga T, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Tsuda H, Otsuji E. Overexpression of PBK/TOPK relates to tumour malignant potential and poor outcome of gastric carcinoma. Br J Cancer. 2017;116:218–226. doi: 10.1038/bjc.2016.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crane R, Gadea B, Littlepage L, Wu H, Ruderman JV. Aurora A, meiosis and mitosis. Biol Cell. 2004;96:215–229. doi: 10.1016/j.biolcel.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 77.Nouri M, Ratther E, Stylianou N, Nelson CC, Hollier BG, Williams ED. Androgen-targeted therapy-induced epithelial mesenchymal plasticity and neuroendocrine transdifferentiation in prostate cancer: an opportunity for intervention. Front Oncol. 2014;4:370. doi: 10.3389/fonc.2014.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dai ZJ, Kang HF, Wang XJ, Shao YP, Lin S, Zhao Y, Ren HT, Min WL, Wang M, Liu XX. Association between genetic polymorphisms in AURKA (rs2273535 and rs1047972) and breast cancer risk: a meta-analysis involving 37,221 subjects. Cancer Cell Int. 2014;14:91. doi: 10.1186/s12935-014-0091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Takahashi Y, Sheridan P, Niida A, Sawada G, Uchi R, Mizuno H, Kurashige J, Sugimachi K, Sasaki S, Shimada Y, Hase K, Kusunoki M, Kudo S, Watanabe M, Yamada K, Sugihara K, Yamamoto H, Suzuki A, Doki Y, Miyano S, Mori M, Mimori K. The AURKA/TPX2 axis drives colon tumorigenesis cooperatively with MYC. Ann Oncol. 2015;26:935–942. doi: 10.1093/annonc/mdv034. [DOI] [PubMed] [Google Scholar]
- 80.Sehdev V, Katsha A, Arras J, Peng D, Soutto M, Ecsedy J, Zaika A, Belkhiri A, El-Rifai W. HDM2 regulation by AURKA promotes cell survival in gastric cancer. Clin Cancer Res. 2014;20:76–86. doi: 10.1158/1078-0432.CCR-13-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chou CH, Yang NK, Liu TY, Tai SK, Hsu DSS, Chen YW, Chen YJ, Chang CC, Tzeng CH, Yang MH. Chromosome instability modulated by BMI1-AURKA signaling drives progression in head and neck cancer. Cancer Res. 2013;73:953–966. doi: 10.1158/0008-5472.CAN-12-2397. [DOI] [PubMed] [Google Scholar]
- 82.Goos JA, Coupé VM, Diosdado B, Delis-van Diemen PM, Karga C, Beliën JA, Carvalho B, van den Tol MP, Verheul HM, Geldof AA, Meijer GA, Hoekstra OS, Fijneman RJ DeCoDe PET group. Aurora kinase A (AURKA) expression in colorectal cancer liver metastasis is associated with poor prognosis. Br J Cancer. 2013;109:2445–2452. doi: 10.1038/bjc.2013.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Su ZL, Su CW, Huang YL, Yang WY, Sampurna BP, Ouchi T, Lee KL, Wu CS, Wang HD, Yuh CH. A novel AURKA mutant-induced early-onset severe hepatocarcinogenesis greater than wild-type via activating different pathways in zebrafish. Cancers. 2019;11:927. doi: 10.3390/cancers11070927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Long Q, An X, Chen M, Wang N, Sui S, Li Y, Zhang C, Lee K, Wang X, Tian T, Pan Y, Qiu H, Xie F, Deng W, Zheng F, He L. PUF60/AURKA axis contributes to tumor progression and malignant phenotypes in bladder cancer. Front Oncol. 2020;10:568015. doi: 10.3389/fonc.2020.568015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schneider MA, Christopoulos P, Muley T, Warth A, Klingmueller U, Thomas M, Herth FJ, Dienemann H, Mueller NS, Theis F, Meister M. AURKA, DLGAP5, TPX2, KIF11 and CKAP5: five specific mitosis-associated genes correlate with poor prognosis for non-small cell lung cancer patients. Int J Oncol. 2017;50:365–372. doi: 10.3892/ijo.2017.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang C, Yan Q, Hu M, Qin D, Feng Z. Effect of AURKA gene expression knockdown on angiogenesis and tumorigenesis of human ovarian cancer cell lines. Target Oncol. 2016;11:771–781. doi: 10.1007/s11523-016-0436-7. [DOI] [PubMed] [Google Scholar]
- 87.Katsha A, Arras J, Soutto M, Belkhiri A, El-Rifai W. AURKA regulates JAK2-STAT3 activity in human gastric and esophageal cancers. Mol Oncol. 2014;8:1419–1428. doi: 10.1016/j.molonc.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saito N, Okada Y, Noda Y, Kinoshita Y, Kondo S, Hirokawa N. KIFC2 is a novel neuron-specific C-terminal type kinesin superfamily motor for dendritic transport of multivesicular body-like organelles. Neuron. 1997;18:425–438. doi: 10.1016/s0896-6273(00)81243-x. [DOI] [PubMed] [Google Scholar]
- 89.Shimo A, Tanikawa C, Nishidate T, Lin ML, Matsuda K, Park JH, Ueki T, Ohta T, Hirata K, Fukuda M, Nakamura Y, Katagiri T. Involvement of kinesin family member 2C/mitotic centromere-associated kinesin overexpression in mammary carcinogenesis. Cancer Sci. 2008;99:62–70. doi: 10.1111/j.1349-7006.2007.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wagenbach M, Vicente JJ, Ovechkina Y, Domnitz S, Wordeman L. Functional characterization of MCAK/Kif2C cancer mutations using high-throughput microscopic analysis. Mol Biol Cell. 2020;31:580–588. doi: 10.1091/mbc.E19-09-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang C, Li Q, Chen X, Zhang Z, Mou Z, Ye F, Jin S, Jun X, Tang F, Jiang H. Circular RNA circRGNEF promotes bladder cancer progression via miR-548/KIF2C axis regulation. Aging (Albany NY) 2020;12:6865. doi: 10.18632/aging.103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jiang CF, Xie YX, Qian YC, Wang M, Liu LZ, Shu YQ, Bai XM, Jiang BH. TBX15/miR-152/KIF2C pathway regulates breast cancer doxorubicin resistance via promoting PKM2 ubiquitination. Cancer Cell Int. 2021;21:542. doi: 10.1186/s12935-021-02235-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen G, Yu M, Cao J, Zhao H, Dai Y, Cong Y, Qiao G. Identification of candidate biomarkers correlated with poor prognosis of breast cancer based on bioinformatics analysis. Bioengineered. 2021;12:5149–5161. doi: 10.1080/21655979.2021.1960775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wei S, Dai M, Zhang C, Teng K, Wang F, Li H, Sun W, Feng Z, Kang T, Guan X, Xu R, Cai M, Xie D. KIF2C: a novel link between Wnt/β-catenin and mTORC1 signaling in the pathogenesis of hepatocellular carcinoma. Protein Cell. 2021;12:788–809. doi: 10.1007/s13238-020-00766-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao Z, Jia H, Yu F, Guo H, Li B. KIF2C promotes the proliferation of hepatocellular carcinoma cells in vitro and in vivo. Exp Ther Med. 2021;22:1–9. doi: 10.3892/etm.2021.10528. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 96.Ding Q, Jiang C, Zhou Y, Duan J, Lai J, Jiang M, Lin D. Kinesin family member 2C promotes hepatocellular carcinoma growth and metastasis via activating MEK/ERK pathway. Biosci Biotechnol Biochem. 2021;85:2241–2249. doi: 10.1093/bbb/zbab154. [DOI] [PubMed] [Google Scholar]
- 97.Lin Q, Qi Q, Hou S, Chen Z, Jiang N, Zhang L, Lin C. Activation of the TGF-β1/Smad signaling by KIF2C contributes to the malignant phenotype of thyroid carcinoma cells. Tissue Cell. 2021;73:101655. doi: 10.1016/j.tice.2021.101655. [DOI] [PubMed] [Google Scholar]
- 98.Gan H, Lin L, Hu N, Yang Y, Gao Y, Pei Y, Chen K, Sun B. KIF2C exerts an oncogenic role in nonsmall cell lung cancer and is negatively regulated by miR-325-3p. Cell Biochem Funct. 2019;37:424–431. doi: 10.1002/cbf.3420. [DOI] [PubMed] [Google Scholar]
- 99.Xiong L, Lou Y, Wang L. Expressions of Kif2c and Ki-67 in non-small cell lung cancer and their relationship with invasion and metastasis. J Biol Regul Homeost Agents. 2020;34:541–546. doi: 10.23812/19-513-L-8. [DOI] [PubMed] [Google Scholar]
- 100.Ishikawa K, Kamohara Y, Tanaka F, Haraguchi N, Mimori K, Inoue H, Mori M. Mitotic centromere-associated kinesin is a novel marker for prognosis and lymph node metastasis in colorectal cancer. Br J Cancer. 2008;98:1824–1829. doi: 10.1038/sj.bjc.6604379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang X, Hu S, Ji W, Tang Y, Zhang S. Identification of genes associated with clinicopathological features of colorectal cancer. Int J Med Res. 2020;48:0300060520912139. doi: 10.1177/0300060520912139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nakamura Y, Tanaka F, Haraguchi N, Mimori K, Matsumoto T, Inoue H, Yanaga K, Mori M. Clinicopathological and biological significance of mitotic centromere-associated kinesin overexpression in human gastric cancer. Br J Cancer. 2007;97:543–549. doi: 10.1038/sj.bjc.6603905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang W, Gao L, Wang C, Wang S, Sun D, Li X, Liu M, Qi Y, Liu J, Lin B. Combining bioinformatics and experiments to identify and verify key genes with prognostic values in endometrial carcinoma. J Cancer. 2020;11:716. doi: 10.7150/jca.35854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zuo X, Meng P, Bao Y, Tao C, Wang Y, Liu X, Bu Y, Zhu J. Cell cycle dysregulation with overexpression of KIF2C/MCAK is a critical event in nasopharyngeal carcinoma. Genes Dis. 2021 doi: 10.1016/j.gendis.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cao HM, Wan Z, Wang F, Liu ZY, Li XF, Hou JQ. Downregulation of KIF2C and TEKT2 is associated with male infertility and testicular carcinoma. Aging (Albany NY) 2021;13:22898–22911. doi: 10.18632/aging.203583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang W, Zhang W, Hu Y. Identification of keygenes, miRNAs and miRNA-mRNA regulatory pathways for chemotherapy resistance in ovarian cancer. PeerJ. 2021;9:e12353. doi: 10.7717/peerj.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu Y, Wang A, Zhu B, Huang J, Lu E, Xu H, Xia W, Dong G, Jiang F, Xu L. KIF18B promotes tumor progression through activating the Wnt/β-catenin pathway in cervical cancer. OncoTargets and Ther. 2018;11:1707. doi: 10.2147/OTT.S157440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Afsharpad M, Nowroozi MR, Mobasheri MB, Ayati M, Nekoohesh L, Saffari M, Zendehdel K, Modarressi MH. Cancer-testis antigens as new candidate diagnostic biomarkers for transitional cell carcinoma of bladder. Pathol Oncol. 2019;25:191–199. doi: 10.1007/s12253-017-0313-4. [DOI] [PubMed] [Google Scholar]
- 109.Naetar N, Hutter S, Dorner D, Dechat T, Korbei B, Gotzmann J, Beug H, Foisner R. LAP2α-binding protein LINT-25 is a novel chromatin-associated protein involved in cell cycle exit. J Cell Sci. 2007;120:737–747. doi: 10.1242/jcs.03390. [DOI] [PubMed] [Google Scholar]
- 110.Chun HK, Chung KS, Kim HC, Kang JE, Kang MA, Kim JT, Choi EH, Jung KE, Kim MH, Song EY. OIP5 is a highly expressed potential therapeutic target for colorectal and gastric cancers. BMB Rep. 2010;43:349–354. doi: 10.5483/bmbrep.2010.43.5.349. [DOI] [PubMed] [Google Scholar]
- 111.Nakamura Y, Tanaka F, Nagahara H, Ieta K, Haraguchi N, Mimori K, Sasaki A, Inoue H, Yanaga K, Mori M. Opa interacting protein 5 (OIP5) is a novel cancer-testis specific gene in gastric cancer. Ann Surg Oncol. 2007;14:885–892. doi: 10.1245/s10434-006-9121-x. [DOI] [PubMed] [Google Scholar]
- 112.Gong M, Xu Y, Dong W, Guo G, Ni W, Wang Y, Wang Y, An R. Expression of Opa interacting protein 5 (OIP5) is associated with tumor stage and prognosis of clear cell renal cell carcinoma. Acta Histochem. 2013;115:810–815. doi: 10.1016/j.acthis.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 113.Chow MJ, Gu Y, He L, Lin X, Dong Y, Mei W, Kapoor A, Tang D. Prognostic and therapeutic potential of the oip5 network in papillary renal cell carcinoma. Cancers. 2021;13:4483. doi: 10.3390/cancers13174483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yazarloo F, Shirkoohi R, Mobasheri MB, Emami A, Modarressi MH. Expression analysis of four testis-specific genes AURKC, OIP5, PIWIL2 and TAF7L in acute myeloid leukemia: a gender-dependent expression pattern. Med Onco. 2013;30:368. doi: 10.1007/s12032-012-0368-8. [DOI] [PubMed] [Google Scholar]
- 115.Koinuma J, Akiyama H, Fujita M, Hosokawa M, Tsuchiya E, Kondo S, Nakamura Y, Daigo Y. Characterization of an Opa interacting protein 5 involved in lung and esophageal carcinogenesis. Cancer Sci. 2012;103:577–586. doi: 10.1111/j.1349-7006.2011.02167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li Q, Chen W, Luo R, Zhang Z, Song M, Chen W, Yang Z, Yang Y, Guo Z, Yang A. Upregulation of OIP5-AS1 predicts poor prognosis and contributes to thyroid cancer cell proliferation and migration. Mol Ther Nucleic. 2020;20:279–291. doi: 10.1016/j.omtn.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 117.Meng X, Ma J, Wang B, Wu X, Liu Z. Long non-coding RNA OIP5-AS1 promotes pancreatic cancer cell growth through sponging miR-342-3p via AKT/ERK signaling pathway. J Physiol Biochem. 2020;76:301–315. doi: 10.1007/s13105-020-00734-4. [DOI] [PubMed] [Google Scholar]
- 118.Li A, Feng L, Niu X, Zeng Q, Li B, You Z. Downregulation of OIP5-AS1 affects proNGF-induced pancreatic cancer metastasis by inhibiting p75NTR levels. Aging. 2021;13:10688. doi: 10.18632/aging.202847. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 119.Wu Z, Liu Y, Wei L, Han M. LncRNA Oip5-as1 promotes breast cancer progression by regulating mir-216a-5p/glo1. J Surg Res. 2021;257:501–510. doi: 10.1016/j.jss.2020.07.067. [DOI] [PubMed] [Google Scholar]
- 120.Li HC, Chen YF, Feng W, Cai H, Mei Y, Jiang YM, Chen T, Xu K, Feng DX. Loss of the Opa interacting protein 5 inhibits breast cancer proliferation through miR-139-5p/NOTCH1 pathway. Gene. 2017;603:1–8. doi: 10.1016/j.gene.2016.11.046. [DOI] [PubMed] [Google Scholar]
- 121.Yuan L, Feng Y, Li L. Effect of microRNA-653 on biological characteristics of human non-small cell lung cancer cells by targeting OIP5 gene and regulating mTOR signaling pathway. J Mod Oncol. 2018;26:831–837. [Google Scholar]
- 122.Mei J, Liu G, Wang W, Xiao P, Yang D, Bai H, Li R. OIP5-AS1 modulates epigenetic regulator HDAC7 to enhance non-small cell lung cancer metastasis via miR-140-5p. Oncology Lett. 2020;20:7. doi: 10.3892/ol.2020.11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Song L, Wang L, Pan X, Yang C. lncRNA OIP5-AS1 targets ROCK1 to promote cell proliferation and inhibit cell apoptosis through a mechanism involving miR-143-3p in cervical cancer. Braz J Med Biol. 2020;53:344–360. doi: 10.1590/1414-431X20198883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guo L, Chen J, Liu D, Liu L. OIP5-AS1/miR-137/ZNF217 axis promotes malignant behaviors in epithelial ovarian cancer. Cancer Manag Res. 2020;12:6707–6717. doi: 10.2147/CMAR.S237726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu Y, Fu X, Wang X, Liu Y, Song X. Long non-coding RNA OIP5-AS1 facilitates the progression of ovarian cancer via the miR-128-3p/CCNG1 axis. Mol Med Rep. 2021;23:1–12. doi: 10.3892/mmr.2021.12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sun X, Tian C, Zhang H, Han K, Zhou M, Gan Z, Zhu H, Min D. Long noncoding RNA OIP5-AS1 mediates resistance to doxorubicin by regulating miR-137-3p/PTN axis in osteosarcoma. Biomed Pharmacother. 2020;128:110201. doi: 10.1016/j.biopha.2020.110201. [DOI] [PubMed] [Google Scholar]
- 127.Chen J, Liu C, Cen J, Liang T, Xue J, Zeng H, Zhang Z, Xu G, Yu C, Lu Z, Wang Z, Jiang J, Zhan X, Zeng J. KEGG-expressed genes and pathways in triple negative breast cancer: protocol for a systematic review and data mining. Medicine. 2020;99:e19986. doi: 10.1097/MD.0000000000019986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang Y, Guo S, Wang S, Li X, Hou D, Li H, Wang L, Xu Y, Ma B, Wang H. LncRNA OIP5-AS1 inhibits ferroptosis in prostate cancer with long-term cadmium exposure through miR-128-3p/SLC7A11 signaling. Ecotoxicol Environ Saf. 2021;220:112376. doi: 10.1016/j.ecoenv.2021.112376. [DOI] [PubMed] [Google Scholar]
- 129.Li J, Zhang H, Luo H. Long Non-coding RNA OIP5-AS1 contributes to gallbladder cancer cell invasion and migration by miR-143-3p suppression. Cancer Manag Res. 2020;12:12983. doi: 10.2147/CMAR.S278719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kotake Y, Matsunaga N, Wakasaki T, Okada R. OIP5-AS1 Promotes proliferation of non-small-cell lung cancer and head and neck squamous cell carcinoma cells. Int J Genomics Proteomics. 2021;18:543–548. doi: 10.21873/cgp.20279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu Y, Cai X, Cai Y, Chang Y. lncRNA OIP5-AS1 suppresses cell proliferation and invasion of endometrial cancer by regulating PTEN/AKT via sponging miR-200c-3p. J Immunol Res. 2021;2021:4861749. doi: 10.1155/2021/4861749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li L, Ma Y, Maerkeya K, Reyanguly D, Han L. LncRNA OIP5-AS1 regulates the Warburg effect through miR-124-5p/IDH2/HIF-1α pathway in cervical cancer. Front Cell Dev Biol. 2021;9:655018. doi: 10.3389/fcell.2021.655018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang S, Pang S, Wang F, Li X, Zou Q. Long noncoding RNA OIP5-AS1 exhibits oncogenic activity in bladder cancer through miR-217 and MTDH. Eur Rev Med Pharmacol Sci. 2021;25:3211–3220. doi: 10.26355/eurrev_202104_25729. [DOI] [PubMed] [Google Scholar]
- 134.Wang D, Chen Z, Lin F, Wang Z, Gao Q, Xie H, Xiao H, Zhou Y, Zhang F, Ma Y. OIP5 promotes growth, metastasis and chemoresistance to cisplatin in bladder cancer cells. J Cancer. 2018;9:4684. doi: 10.7150/jca.27381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zheng Y, Cui Y, Yang S, Wang Y, Qiu Y, Hu W. Opa interacting protein 5 promotes metastasis of nasopharyngeal carcinoma cells by promoting EMT via modulation of JAK2/STAT3 signal. Eur Rev Med Pharmacol Sci. 2019;23:613–621. doi: 10.26355/eurrev_201901_16875. [DOI] [PubMed] [Google Scholar]
- 136.He J, Zhao Y, Zhao E, Wang X, Dong Z, Chen Y, Yang L, Cui H. Cancer-testis specific gene OIP5: a downstream gene of E2F1 that promotes tumorigenesis and metastasis in glioblastoma by stabilizing E2F1 signaling. Neuro Oncol. 2018;20:1173–1184. doi: 10.1093/neuonc/noy037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jiang W, Jimenez G, Wells NJ, Hope TJ, Wahl GM, Hunter T, Fukunaga R. PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol Cell. 1998;2:877–885. doi: 10.1016/s1097-2765(00)80302-0. [DOI] [PubMed] [Google Scholar]
- 138.Hanselmann S, Wolter P, Malkmus J, Gaubatz S. The microtubule-associated protein PRC1 is a potential therapeutic target for lung cancer. Oncotarget. 2018;9:4985. doi: 10.18632/oncotarget.23577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhan P, Zhang B, Xi GM, Wu Y, Liu HB, Liu YF, Xu WJ, Zhu QQ, Cai F, Zhou ZJ. PRC1 contributes to tumorigenesis of lung adenocarcinoma in association with the Wnt/β-catenin signaling pathway. Mol Cancer. 2017;16:1–15. doi: 10.1186/s12943-017-0682-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang B, Shi X, Xu G, Kang W, Zhang W, Zhang S, Cao Y, Qian L, Zhan P, Yan H. Elevated PRC1 in gastric carcinoma exerts oncogenic function and is targeted by piperlongumine in a p53-dependent manner. J Cell Mol Med. 2017;21:1329–1341. doi: 10.1111/jcmm.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bu H, Li Y, Jin C, Yu H, Wang X, Chen J, Wang Y, Ma Y, Zhang Y, Kong B. Overexpression of PRC1 indicates a poor prognosis in ovarian cancer. Int J Oncol. 2020;56:685–696. doi: 10.3892/ijo.2020.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wang Q, Lu S, Chen Y, He H, Lu W, Lin K. Analysis of transcriptome in the relationship between expression of PRC1 protein and prognosis of patients with cholangiocarcinoma. Int J Med Res. 2021;49:0300060521989200. doi: 10.1177/0300060521989200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen J, Rajasekaran M, Xia H, Zhang X, Kong SN, Sekar K, Seshachalam VP, Deivasigamani A, Goh BKP, Ooi LL. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the Wnt/β-catenin signalling pathway. Gut. 2016;65:1522–1534. doi: 10.1136/gutjnl-2015-310625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen W, Chen M, Zhao Z, Weng Q, Song J, Fang S, Wu X, Wang H, Zhang D, Yang W. ZFP36 Binds With PRC1 to Inhibit Tumor Growth and Increase 5-Fu Chemosensitivity of Hepatocellular Carcinoma. Front Mol Biosci. 2020;7:126. doi: 10.3389/fmolb.2020.00126. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 145.Luo HW, Chen QB, Wan YP, Chen GX, Zhuo YJ, Cai ZD, Luo Z, Han ZD, Liang YX, Zhong WD. Protein regulator of cytokinesis 1 overexpression predicts biochemical recurrence in men with prostate cancer. Biomed Pharmacother. 2016;78:116–120. doi: 10.1016/j.biopha.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 146.Shimo A, Nishidate T, Ohta T, Fukuda M, Nakamura Y, Katagiri T. Elevated expression of protein regulator of cytokinesis 1, involved in the growth of breast cancer cells. Cancer Sci. 2007;98:174–181. doi: 10.1111/j.1349-7006.2006.00381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jia Y, Ding X, Zhou L, Zhang L, Yang X. Mesenchymal stem cells-derived exosomal microRNA-139-5p restrains tumorigenesis in bladder cancer by targeting PRC1. Oncogene. 2021;40:246–261. doi: 10.1038/s41388-020-01486-7. [DOI] [PubMed] [Google Scholar]
- 148.Xu T, Wang X, Jia X, Gao W, Li J, Gao F, Zhan P, Ji W. Overexpression of protein regulator of cytokinesis 1 facilitates tumor growth and indicates unfavorable prognosis of patients with colon cancer. Cancer Cell Int. 2020;20:1–12. doi: 10.1186/s12935-020-01618-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang F, Zhao Q, Liu W, Kong D. CENPE, PRC1, TTK, and PLK4 May Play Crucial Roles in the Osteosarcoma Progression. Technol Cancer Res Treat. 2020;19:1533033820973278. doi: 10.1177/1533033820973278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mamoor S. Over-expression of protein regulator of cytokinesis 1 in human endometrial cancer. Cancer Cell Int. 2020;20:528. doi: 10.1186/s12935-020-01618-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yi L, Ouyang L, Wang S, Li SS, Yang XM. Long noncoding RNA PTPRG-AS1 acts as a microRNA-194-3p sponge to regulate radiosensitivity and metastasis of nasopharyngeal carcinoma cells via PRC1. J Cell Physiol. 2019;234:19088–19102. doi: 10.1002/jcp.28547. [DOI] [PubMed] [Google Scholar]
- 152.Bremnes RM, Busund LT, Kilvær TL, Andersen S, Richardsen E, Paulsen EE, Hald S, Khanehkenari MR, Cooper WA, Kao SC. The role of tumor-infiltrating lymphocytes in development, progression, and prognosis of non-small cell lung cancer. J Thorac Oncol. 2016;11:789–800. doi: 10.1016/j.jtho.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 153.Schalper KA, Brown J, Carvajal-Hausdorf D, McLaughlin J, Velcheti V, Syrigos KN, Herbst RS, Rimm DL. Objective measurement and clinical significance of TILs in non-small cell lung cancer. J Natl Cancer Inst. 2015;107:dju435. doi: 10.1093/jnci/dju435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Marshall EA, Ng KW, Kung SH, Conway EM, Martinez VD, Halvorsen EC, Rowbotham DA, Vucic EA, Plumb AW, Becker-Santos DD. Emerging roles of T helper 17 and regulatory T cells in lung cancer progression and metastasis. Mol cancer. 2016;15:1–15. doi: 10.1186/s12943-016-0551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yu Y, Tian X. Analysis of genes associated with prognosis of lung adenocarcinoma based on GEO and TCGA databases. Medicine. 2020;99:e20183. doi: 10.1097/MD.0000000000020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zeng H, Ji J, Song X, Huang Y, Li H, Huang J, Ma X. Stemness related genes revealed by network analysis associated with tumor immune microenvironment and the clinical outcome in lung adenocarcinoma. Front Genet. 2020;11:549213. doi: 10.3389/fgene.2020.549213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yu DH, Huang JY, Liu XP, Ruan XL, Chen C, Hu WD, Li S. Effects of hub genes on the clinicopathological and prognostic features of lung adenocarcinoma. Oncol Lett. 2020;19:1203–1214. doi: 10.3892/ol.2019.11193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Xu Z, Wu Z, Xu J, Zhang J, Yu B. Identification of hub driving genes and regulators of lung adenocarcinoma based on the gene Co-expression network. Biosci Rep. 2020;40:BSR20200295. doi: 10.1042/BSR20200295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li J, Li Z, Zhao S, Song Y, Si L, Wang X. Identification key genes, key miRNAs and key transcription factors of lung adenocarcinoma. J Thorac Dis. 2020;12:1917–1933. doi: 10.21037/jtd-19-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ni KW, Sun GZ. The identification of key biomarkers in patients with lung adenocarcinoma based on bioinformatics. Math Biosci Eng. 2019;16:7671–7687. doi: 10.3934/mbe.2019384. [DOI] [PubMed] [Google Scholar]
- 161.Bu J, Zhang P, Zhu K, Yan Y, Shi B, Wang J, Xu S. Constructing a global transcriptional regulatory landscape for early non-small cell lung cancer to identify hub genes and key pathways. Aging. 2020;12:17948–17957. doi: 10.18632/aging.103475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li J, Li Q, Su Z, Sun Q, Zhao Y, Feng T, Jiang J, Zhang F, Ma H. Lipid metabolism gene-wide profile and survival signature of lung adenocarcinoma. Lipids Health Dis. 2020;19:222. doi: 10.1186/s12944-020-01390-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dai JJ, Zhou WB, Wang B. Identification of crucial genes associated with lung adenocarcinoma by bioinformatic analysis. Medicine. 2020;99:e23052. doi: 10.1097/MD.0000000000023052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang W, He J, Lu H, Kong Q, Lin S. KRT8 and KRT19, associated with EMT, are hypomethylated and overexpressed in lung adenocarcinoma and link to unfavorable prognosis. Biosci Rep. 2020;40:BSR20193468. doi: 10.1042/BSR20193468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zheng Q, Min S, Zhou Q. Identification of potential diagnostic and prognostic biomarkers for LUAD based on TCGA and GEO databases. Biosci Rep. 2021;41:BSR20204370. doi: 10.1042/BSR20204370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fei H, Chen S, Xu C. Interactive verification analysis of multiple sequencing data for identifying potential biomarker of lung adenocarcinoma. Biomed Res Int. 2020;2020:8931419–8931419. doi: 10.1155/2020/8931419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Deng H, Huang Y, Wang L, Chen M. High expression of UBB, RAC1, and ITGB1 predicts worse prognosis among nonsmoking patients with lung adenocarcinoma through bioinformatics analysis. Biomed Res Int. 2020;2020:2071593. doi: 10.1155/2020/2071593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Guo T, Ma H, Zhou Y. Bioinformatics analysis of microarray data to identify the candidate biomarkers of lung adenocarcinoma. PeerJ. 2019;7:e7313–e7313. doi: 10.7717/peerj.7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Li S, Xuan Y, Gao B, Sun X, Miao S, Lu T, Wang Y, Jiao W. Identification of an eight-gene prognostic signature for lung adenocarcinoma. Cancer Manag Res. 2018;10:3383–3392. doi: 10.2147/CMAR.S173941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Song Y, Tang W, Li H. Identification of KIF4A and its effect on the progression of lung adenocarcinoma based on the bioinformatics analysis. Biosci Rep. 2021;41:BSR20203973. doi: 10.1042/BSR20203973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ma X, Yang S, Jiang H, Wang Y, Xiang Z. Transcriptomic analysis of tumor tissues and organoids reveals the crucial genes regulating the proliferation of lung adenocarcinoma. J Transl Med. 2021;19:368–368. doi: 10.1186/s12967-021-03043-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.He Y, Liu R, Yang M, Bi W, Zhou L, Zhang S, Jin J, Liang X, Zhang P. Identification of VWF as a novel biomarker in lung adenocarcinoma by comprehensive analysis. Front Oncol. 2021;11:639600. doi: 10.3389/fonc.2021.639600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yang X, Feng Q, Jing J, Yan J, Zeng Z, Zheng H, Cheng X. Identification of differentially expressed genes associated with lung adenocarcinoma via bioinformatics analysis. Gen Physiol Biophys. 2021;40:31–48. doi: 10.4149/gpb_2020037. [DOI] [PubMed] [Google Scholar]
- 174.Li S, Li H, Cao Y, Geng H, Ren F, Li K, Dai C, Li N. Integrated bioinformatics analysis reveals CDK1 and PLK1 as potential therapeutic targets of lung adenocarcinoma. Medicine. 2021;100:e26474–e26474. doi: 10.1097/MD.0000000000026474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhong H, Wang J, Zhu Y, Shen Y. Comprehensive analysis of a nine-gene signature related to tumor microenvironment in lung adenocarcinoma. Front Cell Dev Biol. 2021;9:700607. doi: 10.3389/fcell.2021.700607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang K, Zhang M, Wang J, Sun P, Luo J, Jin H, Li R, Pan C, Lu L. A systematic analysis identifies key regulators involved in cell proliferation and potential drugs for the treatment of human lung adenocarcinoma. Front Oncol. 2021;11:737152–737152. doi: 10.3389/fonc.2021.737152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Deng H, Hang Q, Shen D, Zhang Y, Chen M. Low expression of CHRDL1 and SPARCL1 predicts poor prognosis of lung adenocarcinoma based on comprehensive analysis and immunohistochemical validation. Cancer Cell Int. 2021;21:259. doi: 10.1186/s12935-021-01933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.