Abstract
Background
Type 2 Diabetes (T2D) diagnosis is based solely on glycaemia, even though it is an endpoint of numerous dysmetabolic pathways. Type 2 Diabetes complexity is challenging in a real‐world scenario; thus, dissecting T2D heterogeneity is a priority. Cluster analysis, which identifies natural clusters within multidimensional data based on similarity measures, poses a promising tool to unravel Diabetes complexity.
Methods
In this review, we scrutinize and integrate the results obtained in most of the works up to date on cluster analysis and T2D.
Results
To correctly stratify subjects and to differentiate and individualize a preventive or therapeutic approach to Diabetes management, cluster analysis should be informed with more parameters than the traditional ones, such as etiological factors, pathophysiological mechanisms, other dysmetabolic co‐morbidities, and biochemical factors, that is the millieu. Ultimately, the above‐mentioned factors may impact on Diabetes and its complications. Lastly, we propose another theoretical model, which we named the Integrative Model. We differentiate three types of components: etiological factors, mechanisms and millieu. Each component encompasses several factors to be projected in separate 2D planes allowing an holistic interpretation of the individual pathology.
Conclusion
Fully profiling the individuals, considering genomic and environmental factors, and exposure time, will allow the drive to precision medicine and prevention of complications.
Keywords: big data, cluster analysis, diabetes, machine learning
Abbreviations
- BMI
body mass index
- CAD
coronary artery disease
- CKD
chronic kidney disease
- CV
cardiovascular
- DKD
diabetic kidney disease
- eGFR
estimated glomerular filtration rate
- GRS
genetic risk score
- HOMA‐B
homeostatic model assessment for beta‐cell function
- HOMA‐IR
homeostatic model assessment for insulin resistance
- MARD
mild age‐related diabetes
- ML
machine learning
- MOD
mild obesity‐related diabetes
- MR
Mendelian randomization
- MRI
magnetic resonance imaging
- NAFLD
nonalcoholic fatty liver disease
- OAD
oral antidiabetic drugs
- OGTT
oral glucose tolerance test
- PAM
partition around medoids
- PD
prediabetes
- SAID
severe autoimmune diabetes
- SIDD
severe insulin‐deficient diabetes
- SIRD
severe insulin‐resistant diabetes
- SNPs
single nucleotide polymorphisms
- SOM
self‐organizing maps
- T1D
type 1 diabetes mellitus
- T2D
type 2 diabetes mellitus
- UACR
urine albumin–creatinine ratio
1. INTRODUCTION
In diabetes, glucose metabolism is affected due to individual or simultaneous changes in insulin secretion, action or metabolism. Diabetes is diagnosed based on glycaemia and cut‐off values were defined based on the presence of microvascular complications, namely retinopathy. 1 However, dysglycaemia, or the glucose altered metabolism, is not an all‐or‐nothing phenomenon, and on the contrary, it occurs continuously. Prediabetes (PD) is a less severe hyperglycemic state that depicts a higher risk of progression to diabetes. Importantly, individuals with PD can develop diabetes complications, whereas others with diabetes may never develop them, showing the limitations of the current clinical classification. 2 Therefore, glycemic levels are not sufficient to inform about the onset and severity of the condition.
Notwithstanding all investment in diabetes, specifically in Type 2 diabetes mellitus (T2D), it is still one of the main noncommunicable diseases, and its mortality increased by 70% since 2000. 3 T2D is extremely heterogenous, 4 , 5 both in its initial presentation and complications' development, which is crucial to explain the sustained morbidity and increased mortality attributable to this condition. 3 , 6 The empirical individualisation of therapy in diabetes dates back to 19th century, 7 , 8 and is still practised. The latest therapeutic guidelines for T2D include several recent drugs that are giving better results regarding cardiometabolic complications 9 and start to have an increased focus on the patient's comorbidities. 10 The concept of precision medicine has been proposed, aiming at defining the most effective approach for a similar group of patients regarding genetic, environmental, lifestyle and clinical factors, among others. 6 However, further advances in the ability to define precise therapies for diabetes also depend on the acquired knowledge regarding the heterogeneity of the condition.
As early as 1965, two major groups were acknowledged in diabetes pathophysiology: insulin‐resistant and insulin‐deficient individuals. 11 The two pathophysiological mechanisms associated with these groups were assumed to be related to two main organs: insulin secretion impairment in the pancreas; and insulin resistance in the skeletal muscle. Since then, much more complexity was added to diabetes pathophysiology, especially to T2D. 12 More recently, it has been shown that other organs and factors, such as the lung and microbiome, can impact on T2D onset and progression. 13 , 14 , 15 Additionally, it is currently accepted that T2D aetiology encompasses thousands of low impactful genes, as well as environmental and lifestyle factors, that interact with each other. 16
Glucose metabolism is part of an intricate metabolic network where carbohydrates, lipids and other metabolic pathways should be considered as a whole and, when affected, result in dysmetabolism and/or haemodynamic alterations. Thus, depending on the affected mechanisms, diabetes can appear in distinct dysmetabolic contexts. Interestingly, there are lipodystrophic phenotypes in which the inability of white adipose tissue to expand, despite diverse BMI values, causes ectopic fat deposition. 17 These subjects are exposed to atherogenic dyslipidemia, 18 and in the liver, the development of fatty liver may progress to steatohepatitis 19 that can be further impacted by different adipose tissue amounts and functions. Despite showing similar patterns regarding hyperglycaemia and hyperlipidemia, subjects with lipodystrophy might require distinct treatment. 20 Another example relates to diabetes and hypertension bidirectional association. Both conditions have several common pathophysiological mechanisms, namely hyperinsulinemia, increased sympathetic nervous activity, activation of the renin–angiotensin–aldosterone system, endothelial dysfunction, etc. 21 The onset of hypertension in subjects with Type 1 diabetes (T1D) has been related to the onset of kidney dysfunction; however, in subjects with T2D, it can appear before 22 and they can show a prehypertensive profile some years earlier. 23 The causal association of T2D in hypertension was depicted in a Mendelian Randomization (MR) study but does not explain the onset of T2D in hypertensive subjects. 24 However, a higher incidence of T2D in hypertensive subjects as compared with normotensive subjects is evident. 24 The above‐described complexity, although easy to understand in concept, is very hard to demonstrate and tackle in clinical practice. Dissecting and understanding T2D heterogeneity is a priority to reverse the current scenario. 25
To tackle the overly complex clinical challenges, involving multiple aetiological factors, organs and mechanisms, classical statistical analyses are frankly insufficient. Recent progress in memory and computation power allowed for the development and implementation of more complex algorithms, including a collection of tools that can learn from data, named machine learning (ML). Specifically, cluster analyses, using unsupervised learning algorithms (algorithms that deal with observations that do not have a label to learn from 26 ), are promising tools to unravel diabetes complexity.
We will critically review distinct cluster analysis methodologies currently used to study diabetes and integrate results from different studies. Since all analyses aimed at understanding diabetes/T2D pathophysiology, we anticipate their conclusions to fit as pieces in a puzzle. Finally, we suggest a model that can be applied to diabetes precision medicine and from a wider perspective to dysmetabolism overall.
2. ADVANCEMENT OF DIABETES MANAGEMENT—TRAVELLING ON THE ROAD TO PRECISION MEDICINE
The word diabetes (‘to go through’ or siphon) is attributed to Apollonius of Memphis in Greece around 250 BC. However, its clinical description and some complications date back to 3500 years ago in Egypt. 27 Interestingly, two types of diabetes—congenital and late onset—and their relationship to heredity, obesity, sedentariness and diet, were already recognized in medical treatments in ancient India. 8 , 28 At the time diabetes resulted in death and preventing it was the main goal. Additionally, complications of diabetes, such as peripheral neuropathy, gangrene and erectile dysfunction, were described by an Arab doctor, Avicenna (AD 960–1037). 27 Centuries later Matthew Dobson (1732–1784) and Michel Chevreul (1786–1889), through the application of chemistry to diagnosis, identified glucose as the sugar that was increased both in urine and serum of these patients. 8 Arguing that glucose appeared in the urine because the body was unable to assimilate it, Dobson considered diabetes a systemic disease rather than kidney disease, as it was considered until then. 28 These findings led to the research on the metabolism of carbohydrates. However, insulin was not yet available and treatments were based on individualisation of diets, rest or other lifestyle changes, 7 unable to prevent death from acute complications. Neurological complications were also quite frequent, the association of neuropathy, vascular disease, plantar ulcers and gangrene with diabetes was also described, rising the hypothesis that microvascular disease was the cause of some complications. 28
In 1921–22 Banting and Best isolated insulin, one of the great discoveries in medicine, which has allowed most people with insulin‐dependent diabetes to be treated to this day. On the contrary, it led to the distinction of T1D, in which people needed insulin, from T2D, in which insulin was present but ineffective. 27 Since the problem in question was hyperglycaemia, other therapeutic strategies would be developed based on glycemic control. 27 In the 1950s, the first sulfonylurea appeared—the first oral antidiabetic drug (OAD) for people with T2D. 29 Metformin, the most used OAD, appeared a few years later with its mechanism of action only recently fully understood. 30 Since then, other groups have been made available as the involvement of other organs and mechanisms is known. 10 , 12 , 29 In a paradigm of therapy, which in the meantime has become evidence‐based clinical guidelines began to be published, with the main therapeutic focus on glycemic control. 31 It was also recognized that the reduction in complications implied simultaneous treatment of other diseases that represent risk factors for the same complications, such as dyslipidemia and hypertension. 31
The aetiologic classification of diabetes recognizes several types besides Type 1, Type 2 and gestational diabetes. 1 The recognition that there is still a high degree of heterogeneity leads to an effort to adapt the numerous drugs with distinct mechanisms to the patients who benefit most from them. 32 Weight control, hypertension and dyslipidemia, among others, have gained increasing relevance along with glycemic control. 10 Nowadays, these diseases are recognized as comorbidities but treated as independent conditions.
3. CLUSTER ANALYSIS
Cluster analysis is an ML methodology that uses a group of algorithms that can deal with nonlabelled data, named unsupervised learning (Figure 1). Cluster analysis aims to stratify population observations' in natural groups/clusters without needing a priori categorization. Within each cluster observations, similarity is maximized whilst minimized between clusters. 33
FIGURE 1.
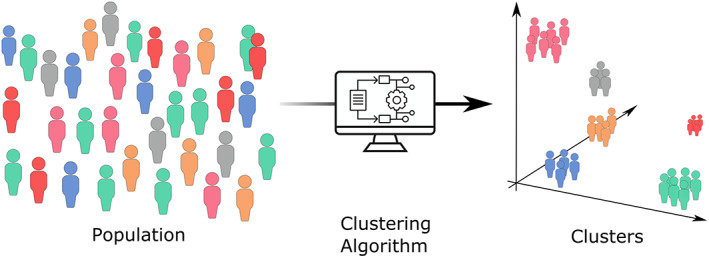
Cluster analysis scheme. A heterogenous population regarding characteristics of interest is stratified by a chosen algorithm that places them in a hyperplane, differentiating natural homogenous groups.
Distinct clustering algorithms have advantages and drawbacks related to computation time, the need for an a priori knowledge regarding the number of groups, and cluster shape in a multidimensionality space that they can find (Table 1). 26 In (dys)glycaemia, specifically in the resolution of T2D heterogeneity, one should consider several parameters with distinct and specific characteristics (e.g. genes, environmental factors, biochemical analysis, omics, etc.). Therefore, it is natural that the best result is obtained using an ensemble of algorithms.
TABLE 1.
Clustering algorithms used in diabetes studies
| Hierarchical | Partitioning |
|---|---|
|
Cluster analysis workflow implies taking several decisions (e.g. choosing the algorithm, variables to inform the cluster, similarity and distance measures, etc.). When algorithms are not able to find the best number of clusters (Table 2), there is the need to determine a priori a number of clusters. 34 Still, different measures can give a distinct optimal number of clusters and therefore should be carefully selected and interpreted. Of note, the found groups should be clinically relevant. Furthermore, aside from finding natural groups in data, cluster analysis is a powerful tool in data exploration and visualization. In the context of (dys)glycaemia heterogeneity, by profiling the found groups, we can explore what characterizes them, posing a promising tool to explore and tackle (dys)glycaemia complexity.
TABLE 2.
Advantages and drawbacks of clustering algorithms (Adapted from 26 )
| Clustering Algorithm | Advantages | Disadvantages |
|---|---|---|
| Hierarchical |
|
|
| k‐means |
|
|
| PAM |
|
|
| SOM |
|
|
| b‐NMF |
|
|
Abbreviations: PAM, partition around medoids; SOM, self‐organizing maps.
4. CLUSTER ANALYSIS ALGORITHM IMPACT ON FOUNDED CLUSTERS
To perform a cluster analysis, impactful decisions must be made: inclusion and exclusion criteria, choice of variables and the algorithm to perform the analysis, among others. Additionally, indexes that define the best number of clusters and distance metrics have to be selected. 26 Cluster analysis used to date to tackle T2D and dysmetabolism have a dissimilar methodology that must be considered when interpreting and integrating the results (Table 2). 35 , 36 , 37 , 38
Hierarchical clustering and k‐means are two of the most well‐known clustering algorithms. Agglomerative hierarchical clustering 26 is a simple algorithm that hierarchically joins nested clusters in a bottom‐up way, with its agglomerative process visualized in a dendrogram. This process does not need the prespecification of the optimal number of clusters, though it requires an a posteriori cut‐off to define them. Furthermore, data can be analysed at different cut‐off values, allowing us to understand how observations aggregate. However, it can only find clusters with specific shapes, it gives distinct solutions depending on the chosen aggregation methodology to join the observations and has a high computation cost. 26 k‐means is a simple and efficient algorithm. Besides not dealing well with categorical variables, the final solution is highly impacted by its random initialization, requires an a priori specification of the number of clusters, and importantly, it is prone to find spherical clusters, even if this is not their natural shape. 26 The latter can limit its use. Partition around medoids (PAM) is a k‐medoids algorithm, that is less sensitive to noise than k‐means, but with a higher computational cost. 26
k‐means, PAM and hierarchical clustering have been used mainly when few parameters are used to tackle T2D. 39 To perform more complex analyses, self‐organizing maps (SOMs) and topological‐based analyses have proven to be more efficient and able to find clusters that have nonspherical shapes. 26 , 40
Hierarchical SOMs, followed by hierarchical clustering, 41 have been used to solve multiple intricate problems, including clustering analysis of T1D complications. 40 SOM is a neural network‐based algorithm, which maps observations to neurons in a grid that at the end will represent the cluster (cluster centroid). 42 In summary, the first algorithm allows data dimensionality reduction, whereas the second enables the stratification and understanding of how the units agglomerate together. Aside from dealing with large and complex data, SOMs can find different cluster formats. Nonetheless, it has drawbacks as requiring too many parameters to be set and optimized, its computational cost and the number of clusters must be set a priori. 42 Network analysis is a graph‐based method that assesses subjects (nodes) in relation to each other (edges). 36
The abovementioned algorithms are classified as hard clustering algorithms, i.e. they group the population to assign one subject only to one cluster. Contrarily, soft clustering uses algorithms that define the probability of one observation belonging to distinct clusters 43 , 44 ; thus, one subject can belong to multiple clusters at a given time. Despite computational cost and convergence drawbacks, soft clustering algorithms are extremely useful when an item can belong to more than one cluster, as is the case of clustering T2D‐related genes/SNPs and mechanisms. 38
5. POPULATION AND PARAMETER SET TO RESOLVE TYPE 2 DIABETES
Clusters analyses to resolve T2D heterogeneity are also diverse regarding the analysed population and set of parameters used to inform the cluster, 40 , 43 , 44 , 45 thus impacting on the groups found. Methodological heterogeneity reveals the authors' distinct perspectives on diabetes definition, where it stands within the wider dysmetabolism concept, and the number and type of parameters that allows a precision medicine approach to T2D.
Although T2D is classically considered an affection of glucose metabolism, glucose metabolism occurs integrated with other substrates'. 45 Glucose metabolism‐related parameters though informing about groups with different conditions, do not give a broader perspective on metabolism nor account for the overall metabolic heterogeneity. T2D impact relies mainly on its complications' development that, in turn, relate to other factors. 46 Herein, we distinguish aetiological factors (e.g. time, genes, environmental factors, lifestyle factors), pathophysiological mechanisms (e.g. overall or organ‐specific insulin resistance, insulin secretion, overall or organ‐specific insulin clearance), other dysmetabolic comorbidities (e.g. hypertension, dyslipidemia), biochemical and other internal environment factors present in the organism or that the organism is exposed to, that is its milieu (e.g. glycaemia, insulinemia, free fatty acids, blood pressure, body weight).
Ahlqvist et al performed a cluster analysis on a population of individuals recently diagnosed with diabetes. 35 The analysis considered six clinical parameters: the presence of GAD antibodies (GADA), age at diagnosis, HbA1c, BMI, HOMA‐IR and HOMA‐B. The analysis does not rely only on glycaemia nor on insulin levels. Nevertheless, the population solely includes individuals that were diagnosed based on current criteria. The authors found five optimal clusters using the silhouette index and hierarchical clustering. One of these clusters, named severe autoimmune diabetes (SAID), included GADA+ subjects. Afterwards, GADA+ subjects were excluded and the k‐means algorithm was used to define the other 4 clusters: severe insulin‐deficient diabetes (SIDD); severe insulin‐resistant diabetes (SIRD); mild obesity‐related diabetes (MOD); and mild age‐related diabetes (MARD). These clusters were replicated in other northern European cohorts. 35 In brief, SAID subjects showed an early‐onset condition, low BMI and poor metabolic control. Subjects in the SIDD cluster were similar to SAID but GADA; these subjects showed a higher risk of having diabetic retinopathy. A variant in human leukocyte antigen (HLA) locus (rs2854275) was found to be associated with SAID but not with SIDD.
Interestingly, Zaharia et al showed that, in a German population, individuals that were GADA‐ at baseline could be GADA+ after 5 years, determining that for better classification of autoimmune diabetes other antibodies should be used. 47 SIRD cluster included individuals with marked insulin resistance, high BMI and a high prevalence of nonalcoholic fatty liver disease (NAFLD). Of note, this cluster also revealed to have the highest β‐cell function. Additionally, individuals in the SIRD cluster were at the highest risk of developing chronic kidney disease (CKD) and diabetic kidney disease (DKD, defined by persistent macroalbuminuria), despite proper glycemic control. Finally, subjects in MOD showed higher values of BMI but not insulin resistance, whereas MARD subjects were older, with only modest metabolic affection, and were not associated with the evaluated diabetes complications. These last two clusters included most of the population and still have a considerable proportion of subjects with diabetes complications. Furthermore, not all diabetes complications were evaluated. In fact, it has been suggested that borderline diabetes is associated with an increased risk of dementia and Alzheimer's disease, which is potentiated when hypertension is present. Regarding gene loci, rs7903146 (a TCF7L2 SNP associated with T2D) was associated with SIDD, MOD and MARD; whereas rs10401969 (a TM6sf2 gene variant associated with NAFLD) was associated with SIRD but not with MOD. 35 The abovementioned four subgroups of T2D have been overall replicated, using the same methodology as Ahlqvist et al., in distinct geographical locations and ethnicities. This further confirms the already known association of diabetes with younger subjects, with lower BMI and more insulin deficiency in Asian and Indian populations. 48 , 49 Moreover, 23% of subjects changed cluster in the 5‐year follow‐up. 47 Particularly, people in the insulin‐deficient cluster (SIDD) were changed to clusters with better insulin secretion (MOD and MARD).
Li et al. performed a topology‐based cluster analysis of 2552 T2D subjects from several ethnicities, informed by 73 mixed features from electronic medical records derived from clinical data. 36 These features included biochemical and clinical parameters besides glycaemia, thus approaching T2D in a wider (dys)metabolic context. This was a landmark study and one of the first studies to show the ability to deal with a high number of variables to stratify subjects with T2D. However, the stratification results depend on the parameters selected to inform the cluster rather than the chosen population. It is not clear whether the authors have found three diabetes subtypes or three subtypes of patients that have diabetes, considering the highly mixed chosen parameters to inform the analysis that also included several disease codes and medications. The chosen methodology renders it difficult to validate it in different populations.
To extend clusters' evaluation to subjects with normoglycaemia and PD, we accounted for age as a surrogate of time exposition, anthropometry and biochemical parameters (glycaemia, insulin, c‐peptide and free fatty acids) in three‐time points of an OGTT. 37 In this study, we used a hierarchical SOM, followed by a hierarchical clustering algorithm. Subjects were then profiled concerning the abovementioned parameters and several mechanism's surrogate indexes, including overall and tissue‐specific insulin resistance, insulin secretion, insulin clearance, NAFLD and glomerular filtration rate (GFR). The sample had a limited number of subjects with nontreated T2D. Nonetheless, none of the subjects had diabetes 5 years earlier. In this work, we found two main clusters: one that includes subjects with a median metabolic phenotype compared with the overall population; and the other with elevated insulin resistance and insulin secretion. However, these 2 clusters were highly heterogeneous when they were evaluated for a higher number of clusters. For example, despite the presence of a main insulin‐resistant group that comprised subjects with normoglycaemia and dysglycaemia, it included subgroups that could be differentiated by their adipose tissue insulin resistance. Moreover, even though groups with lower estimated GFR (eGFR) were insulin‐resistant, not all insulin‐resistant groups showed this association. Additionally, we found that clusters including individuals with normo/dysglycaemia and low eGFR could be further profiled and showed insulin resistance and NAFLD. Consistently, other groups have also shown that both high insulin resistance and NAFLD are related to kidney dysfunction in subjects with or without T2D. 50 In Ahlqvist et al. the group of individuals that had the highest risk of developing CKD/DKD, even considering proper glycemic control, was the most insulin‐resistant one. 35
Furthermore, these subjects had the lowest GFR at baseline (when they had less than 12 months from diagnosis) in the German Diabetes Study cohort. 47 The impact of insulin resistance and NAFLD on GFR seems to be, at least partially, independent from glycaemia. Importantly, both conditions can be associated with hyperinsulinemia and insulin is a known trigger and a target of kidney (dys)function that may have a role in the pathophysiology of T2D. 51 Interestingly, the heterogeneity of affected mechanisms was not exclusive to people with T2D, including also subjects with PD and normoglycaemia. Our work would benefit from being validated in other cohorts. Nevertheless, we highlight that T2D diagnosis should consider other parameters besides glycaemia. In fact, glucose level impact is differently perceived by each individual. Therefore, it should include subjects with different ranges of glycemic values together with other parameters.
An interesting complementary approach to dissect T2D heterogeneity is the use of genetic markers. Reasoning that genetic variants remain constant despite disease progression and treatment, unlike clinical variables, thus being more likely to reveal T2D causal mechanisms, a cluster analysis including T2D gene‐traits associations, including 94 genetic variants and 47 traits was performed. 38 Aside from genetic data the analysis was informed with clinical parameters, including surrogate indexes of insulin secretion and insulin resistance, as well as lipid parameters, that allowed for the identification of other insulin resistance‐related groups. Importantly, in this work b‐NMF, a soft clustering algorithm was used, allowing an SNP to be associated with more than one mechanism and one cluster. The authors identified five clusters of genetic loci‐traits associations: two with variant‐trait associations related to reduced β‐cell function, distinct in pro‐insulin levels; and three insulin resistance‐related, namely obesity‐mediated, lipodystrophy‐like fat distribution and disrupted liver lipid metabolism. Of note, this is also a potentially complex approach. As more than 100 loci were already found to be associated with T2D, each one with a very slight impact on the increased risk of the disease and in dysmetabolism aetiology, we should consider, along with genetic factors, their interactions with environmental and lifestyle factors. Interestingly, Udler et al. evaluated the Genetic Risk Score (GRS) association with relevant outcomes in each cluster. Coronary artery disease (CAD) was mostly associated with lipodystrophy and Beta‐cell clusters. The beta‐cell cluster was also associated with ischaemic stroke. Increased blood pressure was only associated with lipodystrophy cluster, which also showed an association with higher urine albumin–creatinine ratio (UACR). Liver/Lipid cluster was associated with decreased renal function and diminished UACR. GRS outcomes were validated in T2D cohorts by profiling subjects' characteristics in top quantile GRS's subjects. 38
More recently, Wagner et al. focussed on a german population considered at risk of developing diabetes based on BMI, previous history and family history (TUEF/TULIP cohort). 52 Besides OGTT‐based measures reflecting blood glucose, insulin resistance and insulin secretion, liver, subcutaneous and visceral fat values measured by MRI and HDL levels, polygenic risk scores for diabetes were also included. The defined six clusters were then evaluated in a larger cohort (Whitehall II). However, to assign the latter individuals to the clusters, the authors used less profiling variables, still based on OGTT measurements. The authors reported a relocation rate of only 60% in the original cohort, which suggests that MRI fat measurements do not appear to be superior to measurements such as BMI and waist circumference. 37 Importantly, progression to diabetes, CKD, CV events and all‐cause mortality were assessed. 52 Consistent with our findings, 37 Wagner et al. demonstrated that pathophysiological affection is already present before diabetes diagnosis. 52 Within the six defined clusters, three that were older and/or more obese showed higher glycaemia (clusters 3, 5 and 6); one related to insulin deficiency and raised genetic risk (cluster 3); and two with insulin resistance (clusters 5 and 6). Cluster 6 showed a dissociation of both risks of progression to diabetes and CKD in Whitehall II cohort. However, considering that GFR is not depicted in TULIP/TULIF and CKD progression models in Whitehall II were not adjusted to GFR at the baseline these results should be carefully interpreted. Cluster 4 is consistent with a metabolically healthy obese profile that includes younger subjects than the most dysmetabolic groups and did not show protected profile overtime, namely regarding CV events. In fact, although clusters in TULIP/TULIF cohort differ in intima‐media thickness, in the Whitehall II cohort, the clusters did not differ in CV outcomes risk, after adjustment for BMI and age, except for Cluster 2 that had a protected profile. Considering the relevance of CV events in diabetes, this highlights the importance of an enriched milieu to better stratification. 37
6. WHAT CAN WE LEARN FROM CLUSTER ANALYSIS?
Insulin secretion and resistance have been included in parameters informing cluster analyses. However, there can be different mechanisms that lead to insulin deficiency and resistance. 37 It has been suggested that insulin resistance can be considered a defensive mechanism against elevated insulin secretion due to a highly nutritional load in a sensitive β‐cell. 53 In distinct cluster analysis, most of the groups found to be insulin‐resistant were the ones with the highest insulin secretion. 35 , 37 Nevertheless, the amount of circulating insulin depends not only on the cells' secretion capacity but on overall insulin metabolism and on insulin clearance. 54 Changes in insulin clearance have also been linked to hyperinsulinemia. 37 , 54 Insulin resistance has been associated with age and BMI. Interestingly, in work by Alqhvist et al., MARD and MOD groups differ from the SIRD in that they are less fat or younger, respectively, showing better metabolic control. 35
Several questions remain to be clarified concerning the mechanisms leading to insulin resistance. One concerns the mechanisms through which age and BMI impact on insulin resistance and whether this implies a different therapeutic approach. Secondly, in the setting of insulin resistance, it is known the association between liver and adipose tissue but whether insulin resistance develops through distinct pathways, implying distinct therapeutic approaches, remains elusive. Thirdly, when it comes to diabetes complications, the majority of the results were obtained using patients undergoing treatments, which may, in its turn, promote the complication's onset. 55 Finally, cluster analysis showed the association between GFR and albuminuria with insulin‐resistant states 40 , 45 , 52 , 56 ; however, the presence of an association does not necessarily imply homogeneity between clusters, when it comes to kidney function, making this an aetiological factor of the uttermost importance in diabetes stratification.
Udler using SNP and traits, in addition to HOMA‐IR and HOMA‐B, namely lipid profile, found three groups of insulin‐resistant subjects that showed involvement of different mechanisms and organs. 38 We and others have shown that distinct insulin resistance patterns can be present in subjects with normoglycaemia and PD. 37 , 52
Altogether these support the view that, in order to stratify subjects to differentiate a preventive or therapeutic approach to diabetes, one should inform the cluster analysis with more parameters reflecting other mechanisms metabolites and factors (e.g. lipids, blood pressure, insulin). Additionally, diabetes pathophysiology occurs continuously and people without diabetes can already have diabetes complications, hinting at different susceptibilities to glycemic levels. This may be due to concomitant exposure to other factors such as hypertension or dyslipidemia or due to the common underlying pathophysiologic mechanisms.
7. NEW MODELS FOR AN APPROACH TO DIABETES IN PRECISION MEDICINE
Cluster analysis is contributing to uncover the heterogeneity of diabetes. 35 , 37 , 38 , 47 However, its superiority over simple predictive models (e.g. predicting complications such as renal dysfunction) is being questioned. 56
McCarthy proposed the palette model to resolve T2D heterogeneity. 57 The model defined component planes, such as mechanisms, aetiological factors and others, that can be affected, comparing them to a palette hue. The characterization of subjects by their component planes places them in a bidimensional plane where the path from normoglycaemia to diabetes can be assessed for each individual. Importantly this model includes subjects with normoglycaemia and dysglycaemia, which have different affected mechanisms. Ahlqvist et al. suggested a model based on the assumption that there is a dominant pathway that gives at least to the majority of patients with diabetes a well‐defined ‘palette colour’. 58 Additionally, few clinical parameters render larger groups.
In our view, a precision medicine model to approach diabetes must consider glycaemia and glucose metabolism, as well as other substrates and factors, that impact on dysglycaemia and/or diabetes complications onset and progression. Diabetes complications occur for different values of glycaemia, impacted by the metabolic context of the individual. In fact, dysmetabolic factors interaction might potentiate the risk for specific conditions, as is the case of glycaemia and blood pressure interaction in the development of Alzheimer's disease. 55 Finally, the model must be holistic and applicable to different ethnicities. There are ethnicities that show a higher risk for the onset of T2D at younger ages and for lower BMI. 48 Interestingly, subjects with an Asian genetic background seem to have diminished insulin secretory capacity, but one cannot exclude the environmental and culture‐related factors.
We propose to paint another picture, the integrative model (Figure 2). We consider that the approach can only be attained by being detailed in the metabolic characterization of the individuals and by placing it in a wider context of dysmetabolism. Thus, we consider the path from normometabolism to dysmetabolism, in which dysglycaemia is one axis among other factors that can impact on complications onset/progression and organ dysfunction. Therefore, the metabolic condition of each subject is approached in an integrated way. Also, we differentiate three types of components: aetiological factors, mechanisms and milieu. Each encompasses several factors or axis that are projected in separate 2D planes. We postulate that, by deeply profiling a subject for one type of component, we can place him in the corresponding plan. Furthermore, we postulate that we can predict where the individual is in one plan by knowing the others. Ultimately it will allow placing each individual in a last plan where his metabolic state is known. It is natural that there are groups in the data. However, given the possible combinations of affected mechanisms and organs, it is clear that their number is too high for human understanding.
FIGURE 2.
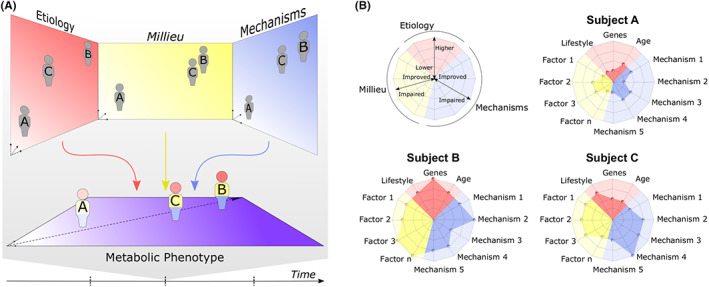
Integrative model of diabetes. (A) Subjects are deeply characterized regarding aetiological factors (including genes, lifestyle and environmental factors), underlying physiopathological mechanisms and metabolic and haemodynamic factors that they are exposed to. They are placed correspondingly onto the aetiology, mechanisms and milieu plan. The location of a subject in each plan can be predicted by knowing their position in the others. Ultimately, aetiology, mechanisms and milieu project the subject onto the metabolic phenotype plan where its health condition is assessed also considering diabetes complications as nephropathy, retinopathy and cardiovascular complications. Each subject path through time in the metabolic phenotype plan can be analysed but also predicted, leveraging therapeutic and preventive strategies. (B) Aetiology, mechanisms and milieu for each subject can be summarized and more easily visible on a radarplot.
This model differs from McCarthy's palette model in two main points: (1) it considers the path to dysmetabolism and not to hyperglycaemia; (2) it separates the different aetiological factors from the affected mechanisms and from the internal environment to which the person is exposed on different levels (Figure 3). The different planes are thus projected among themselves, giving us the possibility to know one when we fully evaluate the others. This differentiation can be relevant to prioritize the clinical approach to the individual and to delineate distinct integrated therapeutic and preventive strategies to be adopted in the different planes that nonetheless should be validated in clinical studies.
FIGURE 3.
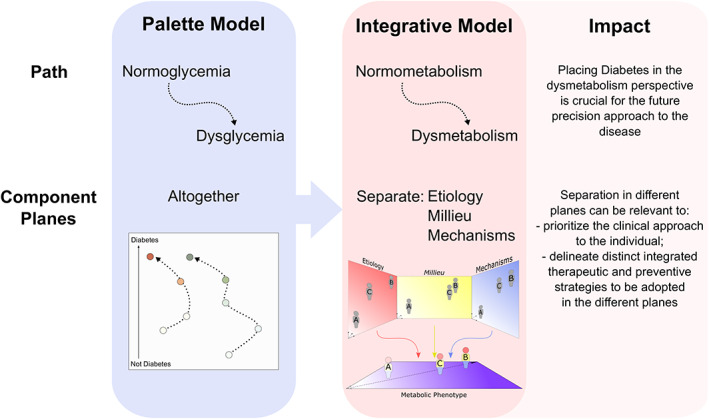
From the palette model to the proposed integrative model. The integrative model that we propose was based on the McCarthys' palette model 57 but differs essentially in the path and in the component planes of the model.
Currently, in therapeutic individualization, therapy is first prescribed to hyperglycaemia and then adapted according to the individual characteristics of each patient. By contrast, in precision medicine, the therapeutic approach is chosen after assigning the patient to a group that already considers the individual specificities. For example, in the individualized treatment of T2D, a subject without atherosclerotic disease or CKD but with hypertension and poorly controlled glycaemia, when on metformin, can be medicated with one of five drugs (DPP4, GLP‐1, SGLT‐2, thiazolidinediones, sulfonylureas). This will be chosen by each doctor considering some characteristics of the patient, such as weight. In addition, an antihypertensive is associated. In real life, situations are not so clear as in the guidelines. For instance, what to do with a patient with T2D on metformin, with good glycemic control (average HbA1c 6.8%) but with evidence of early DKD and without other metabolic risk factors? How intensive and with which agents should he be treated to have the best health outcome? Is it better to use an SGLT‐2 inhibitor or/and start an ACE2 inhibitor? Is this the best treatment for all patients in this condition? Or what to do with another patient with 15 years of T2D, mostly with poor control (HbA1c >8.5%) under different antihyperglycemic medication, without other risk factors or evidence of diabetes complications? Should we keep trying to put him in a good track of glycemic control? For what purpose? In a precision medicine approach, he would first be assigned to a group of people sharing common features of the overall metabolic condition, already accounting with all his specificities (including milieu, mechanisms and aetiological factors) for which the optimal treatment of that group would be already tested, defined and can then be prescribed for that individual.
In order to train and validate this theoretical model, datasets that consider the overall metabolism and deep phenotyping subjects in the distinct proposed planes are needed. Ultimately this model may be implemented in a decision support system that predicts where people are in their overall metabolism. This would assign the individual to a homogeneous group, eventually unravelling his metabolic footprint.
8. CONCLUSION
Precision medicine allows tailoring an approach or treatment to different individuals. In other words, a population is stratified into similar groups, considering relevant characteristics to the condition (e.g. T2D). Doing so for each group an appropriate therapeutic approach is defined. Although precision medicine approaches can make use of genetic data, they can also be based on many other types of clinical data. Observed complexity is solved with the help of mathematical algorithms that stratify individuals into groups by similarity.
In the era of omics and digital health, in which we can extract and deal with thousands of features and use them to tailor care to diabetes, it is not prudent to limit cluster analysis to a few already preestablished common mechanisms. Furthermore, these new strategies allow us to deal with blood glucose levels as a continuum, together with the overall milieu, surpassing the artificial glycaemia‐based cut‐off approach. By fully profiling subjects regarding genomics, environmental factors and time exposition, we will be able to know which mechanism(s) is(are) affected and is(are) responsible for a dysmetabolic condition. This enables the use of drugs in a precise manner and the discovery of new ones. Additionally, the prevention of complications, such as cardiovascular events, may be earlier and more effective. The great big challenge will be identifying which features are relevant to consider precise care and gather the data to perform these analyses. In a global village such as our world, we should gather robust clinical data working in a worldwide consortium.
CONFLICT OF INTEREST
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ACKNOWLEDGEMENTS
This work was supported by ‘Fundação para a Ciência e a Tecnologia’—FCT to AP (PD/BD/136887/2018), MPM (PTDC/MEC‐MET/29314/2017 and PTDC/BIM‐MET/2115/2014), iNOVA4Health (UIDB/Multi/04462/2020); by the European Commission Marie Skłodowska‐Curie Action H2020 (grant agreement n. 734719); by FEDER and Lisboa2020 and Alentejo2020; and by the Sociedade Portuguesa de Diabetologia.
Pina AF, Meneses MJ, Sousa‐Lima I, Henriques R, Raposo JF, Macedo MP. Big data and machine learning to tackle diabetes management. Eur J Clin Invest. 2023;53:e13890. doi: 10.1111/eci.13890
REFERENCES
- 1. Soh SB, Topliss D. Classification and laboratory diagnosis of diabetes mellitus. Diabetes Care. 2014;27:S5‐S10. [DOI] [PubMed] [Google Scholar]
- 2. Kumar R, Nandhini LP, Kamalanathan S, Sahoo J, Vivekanadan M. Evidence for current diagnostic criteria of diabetes mellitus. World J Diabetes. 2016;7:396‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hunter WB. Diabetes as a public health problem. N C Med J. 1950;11:289‐292. [PubMed] [Google Scholar]
- 4. Fajans SS, Cloutier MC, Crowther RL. Clinical and etiologic heterogeneity of idiopathic diabetes mellitus. Diabetes. 1978;27:1112‐1125. [DOI] [PubMed] [Google Scholar]
- 5. Tuomi T, Santoro N, Caprio S, Cai M, Weng J, Groop L. The many faces of diabetes: a disease with increasing heterogeneity. Lancet. 2014;383:1084‐1094. [DOI] [PubMed] [Google Scholar]
- 6. Fradkin JE, Hanlon MC, Rodgers GP. NIH precision medicine initiative: implications for diabetes research. Diabetes Care. 2016;39:1080‐1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Joslin EP. Apollinaire Bouchardat 1806‐1886. Diabetes. 1952;1:490‐491. [Google Scholar]
- 8. Schneider T. Diabetes through the ages: a salute to insulin. South African Med J. 1972;46:1394‐1400. [PubMed] [Google Scholar]
- 9. Sattar N. Advances in the clinical management of type 2 diabetes: a brief history of the past 15 years and challenges for the future. BMC Med. 2019;17:2‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Care D, Suppl SS. Pharmacologic approaches to glycemic treatment: standards of medical care in diabetesd 2021. Diabetes Care. 2021;44:S111‐S124. [DOI] [PubMed] [Google Scholar]
- 11. World Health Organization . Diabetes mellitus. Report of a WHO expert committee; 1965. [PubMed]
- 12. DeFronzo RA. From the triumvirate to the ominous octet: a new paradigm for the treatment of type 2 diabetes mellitus. Diabetes. 2009;58:773‐795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang BY, Qian ZM, Li S, et al. Ambient air pollution in relation to diabetes and glucose‐homoeostasis markers in China: a cross‐sectional study with findings from the 33 communities Chinese health study. Lancet Planet Health. 2018;2:e64‐e73. [DOI] [PubMed] [Google Scholar]
- 14. Teeter JG, Riese RJ. Cross‐sectional and prospective study of lung function in adults with type 2 diabetes: the atherosclerosis risk in communities (ARIC) study. Diabetes Care. 2008;31:e82. [DOI] [PubMed] [Google Scholar]
- 15. Gurung M, Li Z, You H, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020;51:102590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cuschieri S. The genetic side of type 2 diabetes – a review. Diabetes Metab Syndr Clin Res Rev. 2019;13:2503‐2506. [DOI] [PubMed] [Google Scholar]
- 17. Rother KI, Brown RJ. Novel forms of lipodystrophy. Diabetes Care. 2013;36:2142‐2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kinzer AB, Shamburek RD, Lightbourne M, Muniyappa R, Brown RJ. Advanced lipoprotein analysis shows atherogenic lipid profile that improves after Metreleptin in patients with lipodystrophy. J Endocr Soc. 2019;3:1503‐1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Polyzos SA, Perakakis N, Mantzoros CS. Fatty liver in lipodystrophy: a review with a focus on therapeutic perspectives of adiponectin and/or leptin replacement. Metabolism. 2019;96:66‐82. [DOI] [PubMed] [Google Scholar]
- 20. Stefan N. Causes, consequences, and treatment of metabolically unhealthy fat distribution. Lancet Diabetes Endocrinol. 2020;8:616‐627. [DOI] [PubMed] [Google Scholar]
- 21. Colussi GL, Da Porto A, Cavarape A. Hypertension and type 2 diabetes: lights and shadows about causality. J Hum Hypertens. 2020;34:91‐93. [DOI] [PubMed] [Google Scholar]
- 22. Van Buren PN, Toto R. Hypertension in diabetic nephropathy: epidemiology, mechanisms, and management. Adv Chronic Kidney Dis. 2011;18:28‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsimihodimos V, Gonzalez‐villalpando C, Meigs JB, et al. Coprediction and Time Trajectories. 71:422–428. 10.1161/HYPERTENSIONAHA.117.10546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun D, Zhou T, Heianza Y, et al. Type 2 diabetes and hypertension: a study on bidirectional causality. Circ Res. 2019;124:930‐937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chung WK, Erion K, Florez JC, et al. Precision medicine in diabetes: a consensus report from the American Diabetes Association (ADA) and the European Association for the Study of diabetes (EASD). Diabetes Care. 2020;43:1617‐1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pina A, Macedo MP, Henriques R. Clustering clinical data in R. In: Matthiesen R, ed. Mass Spectrometry Data Analysis in Proteomics. Springer New York; 2020:309‐343. [DOI] [PubMed] [Google Scholar]
- 27. Lasker SP, Mclachlan CS, Wang L, et al. Discovery, treatment and management of diabetes. J Diabetol. 2010;1:1‐8. [Google Scholar]
- 28. Ahmed AM. History of diabetes mellitus. Saudi Med J. 2002;23:373‐378. [PubMed] [Google Scholar]
- 29. White JR. A brief history of the development of diabetes medications. Diabetes Spectr. 2014;27:82‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rena G, Pearson ER, Sakamoto K. Molecular mechanism of action of metformin: old or new insights? Diabetologia. 2013;56:1898‐1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Of S, Carediabetes M. Updates to the standards of medical Care in Diabetes‐2018. Diabetes Care. 2018;41:2045‐2047. [DOI] [PubMed] [Google Scholar]
- 32. Meneses MJ, Silva BM, Sousa M, Sá R, Oliveira P, Alves M. Antidiabetic drugs: mechanisms of action and potential outcomes on cellular metabolism. Curr Pharm des. 2015;21:3606‐3620. [DOI] [PubMed] [Google Scholar]
- 33. Jain AK, Murty MN, Flynn PJ. Data clustering: a review. ACM Comput Surv. 1999;31:264‐323. [Google Scholar]
- 34. Xu S, Qiao X, Zhu L, Zhang Y, Xue C, Li L. Reviews on determining the number of clusters. Appl Math Inf Sci. 2016;10:1493‐1512. [Google Scholar]
- 35. Ahlqvist E, Storm P, Käräjämäki A, et al. Novel subgroups of adult‐onset diabetes and their association with outcomes: a data‐driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018;6:361‐369. [DOI] [PubMed] [Google Scholar]
- 36. Li L, Cheng WY, Glicksberg BS, et al. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci Transl Med. 2015;7:311ra174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pina AF, Patarrão RS, Ribeiro RT, et al. Metabolic footprint, towards understanding type 2 diabetes beyond glycemia. J Clin Med. 2020;9:2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Udler MS, Kim J, von Grotthuss M, et al. Type 2 diabetes genetic loci informed by multi‐trait associations point to disease mechanisms and subtypes: a soft clustering analysis. PLoS Med. 2018;15:e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xing L, Peng F, Liang Q, et al. Clinical characteristics and risk of diabetic complications in data‐driven clusters among type 2 diabetes. Front Endocrinol (Lausanne). 2021;12:617628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Toppila I. Identifying Novel Phenotype Profiles of Diabetic Complications and their Genetic Components Using Machine Learning Approaches; 2016. Aalto University. [Google Scholar]
- 41. Vesanto J, Alhoniemi E. Clustering of the self‐organizing map. IEEE Trans Neural Netw. 2000;11:586‐600. [DOI] [PubMed] [Google Scholar]
- 42. Kohonen T. The self‐organizing map. Proc IEEE. 1990;78:1464‐1480. [Google Scholar]
- 43. Dunn JC. A fuzzy relative of the ISODATA process and its use in detecting compact well‐separated clusters. J Cybern. 1973;3:32‐57. [Google Scholar]
- 44. Devarajan K. Nonnegative matrix factorization: an analytical and interpretive tool in computational biology. PLoS Comput Biol. 2008;4:e1000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cohain AT, Barrington WT, Jordan DM, et al. An integrative multiomic network model links lipid metabolism to glucose regulation in coronary artery disease. Nat Commun. 2021;12:547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saeedi P, Salpea P, Karuranga S, et al. Mortality attributable to diabetes in 20–79 years old adults, 2019 estimates: results from the international diabetes federation diabetes atlas, 9th edition. Diabetes Res Clin Pract. 2020;162:108086. [DOI] [PubMed] [Google Scholar]
- 47. Zaharia OP, Strassburger K, Strom A, et al. Risk of diabetes‐associated diseases in subgroups of patients with recent‐onset diabetes: a 5‐year follow‐up study. Lancet Diabetes Endocrinol. 2019;7:684‐694. [DOI] [PubMed] [Google Scholar]
- 48. Zou X, Zhou X, Zhu Z, Ji L. Novel subgroups of patients with adult‐onset diabetes in Chinese and US populations. Lancet Diabetes Endocrinol. 2019;7:9‐11. [DOI] [PubMed] [Google Scholar]
- 49. Anjana RM, Baskar V, Nair ATN, et al. Novel subgroups of type 2 diabetes and their association with microvascular outcomes in an Asian Indian population: a data‐driven cluster analysis: the INSPIRED study. BMJ Open Diabetes Res Care. 2020;8:e001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Byrne CD, Targher G. NAFLD as a driver of chronic kidney disease. J Hepatol. 2020;72:785‐801. [DOI] [PubMed] [Google Scholar]
- 51. Pina AF, Borges DO, Meneses MJ, et al. Insulin: trigger and target of renal functions. Front Cell Dev Biol. 2020;8:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wagner R, Heni M, Tabak AG, et al. Pathophysiology‐based subphenotyping of individuals at elevated risk for type 2 diabetes. Nat Med. 2021;27:49‐57. [DOI] [PubMed] [Google Scholar]
- 53. Nolan CJ, Prentki M. Insulin resistance and insulin hypersecretion in the metabolic syndrome and type 2 diabetes: time for a conceptual framework shift. Diabetes Vasc Dis Res. 2019;16:118‐127. [DOI] [PubMed] [Google Scholar]
- 54. Borges DO, Patarrão RS, Ribeiro RT, et al. Loss of postprandial insulin clearance control by insulin‐degrading enzyme drives dysmetabolism traits. Metabolism. 2021;118:154735. doi: 10.1016/j.metabol.2021.154735 [DOI] [PubMed] [Google Scholar]
- 55. Xu W, Qiu C, Winblad B, Fratiglioni L. The effect of borderline diabetes on the risk of dementia and Alzheimer's disease. Diabetes. 2007;56:211‐216. [DOI] [PubMed] [Google Scholar]
- 56. Dennis JM, Shields BM, Henley WE, Jones AG, Hattersley AT. Disease progression and treatment response in data‐driven subgroups of type 2 diabetes compared with models based on simple clinical features: an analysis using clinical trial data. Lancet Diabetes Endocrinol. 2019;8587:1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McCarthy MI. Painting a new picture of personalised medicine for diabetes. Diabetologia. 2017;60:793‐799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ahlqvist E, Prasad RB, Groop L. Subtypes of type 2 diabetes determined from clinical parameters. Diabetes. 2020;69:2086‐2093. [DOI] [PubMed] [Google Scholar]
- 59. Udler MS, Kim J, von Grotthuss M, et al. Clustering of Type 2 diabetes genetic loci by multi‐trait associations identifies disease mechanisms and subtypes. bioRxiv. 2016. doi: 10.1101/319509 [DOI] [PMC free article] [PubMed] [Google Scholar]


