Abstract
An accurate understanding of the evolutionary history of rapidly-evolving viruses like SARS-CoV-2, responsible for the COVID-19 pandemic, is crucial to tracking and preventing the spread of emerging pathogens. However, viruses undergo frequent recombination, which makes it difficult to trace their evolutionary history using traditional phylogenetic methods. In this study, we present a phylogenetic workflow, virDTL, for analyzing viral evolution in the presence of recombination. Our approach leverages reconciliation methods developed for inferring horizontal gene transfer in prokaryotes and, compared to existing tools, is uniquely able to identify ancestral recombinations while accounting for several sources of inference uncertainty, including in the construction of a strain tree, estimation and rooting of gene family trees, and reconciliation itself. We apply this workflow to the Sarbecovirus subgenus and demonstrate how a principled analysis of predicted recombination gives insight into the evolution of SARS-CoV-2. In addition to providing confirming evidence for the horseshoe bat as its zoonotic origin, we identify several ancestral recombination events that merit further study.
Keywords: phylogenetic reconciliation, Sarbecovirus evolution, SARS-CoV-2, viral recombination
1. INTRODUCTION
Phylogenetic analysis of the first available sequence from Wuhan, China placed SARS-CoV-2 in the Sarbecovirus subgenus of Betacoronavirus (Wu et al., 2020), and several subsequent studies have investigated its evolutionary origins (Andersen et al., 2020; Boni et al., 2020; Lytras et al., 2022). SARS-CoV-2 shares 96% sequence similarity to bat Sarbecovirus RaTG13, and the two viruses form a clade distinct from other SARS-related coronaviruses, suggesting that the SARS-CoV-2 lineage may have its zoonotic origins in bats (Zhou et al., 2020). Pangolins have also been suggested as possible hosts (Lam et al., 2020), although later studies have shown that, while pangolins are natural reservoirs of Betacoronaviruses, SARS-CoV-2 likely did not evolve directly from pangolin coronavirus (Boni et al., 2020; Liu et al., 2020; Lytras et al., 2022). Language models have also shown that SARS-CoV-2 is “semantically” closest to bat and next to pangolin (Hie et al., 2021). Such analyses are of biological and societal interest, as identifying the source of the virus may help inform future outbreaks of viruses with zoonotic origins.
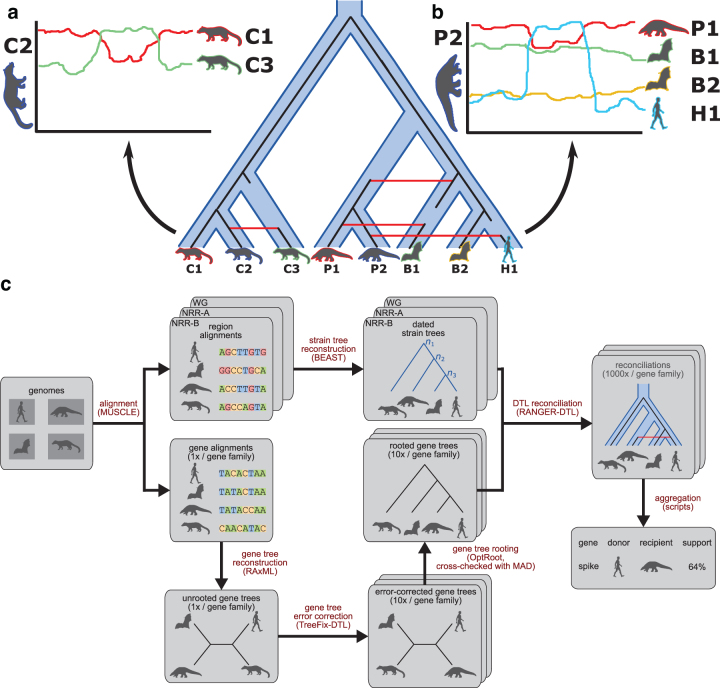
Many viruses, including coronaviruses, undergo frequent recombination (Masters and Perlman, 2013; Forni et al., 2017), which complicates phylogenetic analysis (Patiño-Galindo et al., 2020). Moreover, phylogenetic inference is susceptible to several sources of uncertainty, many of which are exacerbated by recombination between viral genomes. Thus, a common step in the study of viral evolution is to infer recombination, which is commonly done by enumerating triplets of strains and analyzing their sequence similarity (Lole et al., 1999; Martin et al., 2015). This approach works well when recombination occurs infrequently (relative to the rate of evolution) and mostly between extant strains (Fig. 1a). However, as the number of strains grows and recombination occurs multiple times within a lineage, recombination becomes difficult to infer from direct sequence comparison alone (Fig. 1b).
FIG. 1.
virDTL enables inference of ancestral recombination. The figure shows a cartoon example of the virDTL pipeline applied to a toy dataset containing viruses from three civet cats, two pangolins, two bats, and one human. (a) Commonly used tools such as Simplot and RDP are well-suited to inferring recent recombinations between strains of interest, where the recombination signal is clear in the sequence similarity profile. (b) However, in cases where recombination has occurred between ancestral strains, and multiple recombinations have occurred in a single lineage, it becomes significantly more difficult to disentangle the sequence similarity signal to infer all recombinations. (c) Our model-based computational protocol, virDTL, takes into account the entire evolutionary history of a gene family, including several sources of inference uncertainty. A credible strain tree is estimated using nonrecombinant regions of the genome, and multiple gene tree candidates are inferred and error-corrected and reconciled against the strain tree to infer HGTs. In addition to accounting for gene tree topological and rooting uncertainty, we reconcile the same gene tree and species tree multiple times to capture the full landscape of uncertainty in inferring recombination.
Recombination in viruses is similar to gene conversion in that it generally results in the one-sided transfer of genetic material from a donor genome to a recipient genome, rather than an “exchange” of genetic material between the two recombining genomes (Pérez-Losada et al., 2015). Thus, we reasoned that methods for studying horizontal gene transfer (HGT) in prokaryotes could potentially be useful for inferring recombination in viruses. Despite advances in HGT detection (Section 4), these methods have rarely been used to study viral genome evolution or recombination.
In this work, we describe a step-by-step computational protocol, virDTL, for analyzing viral evolution in the presence of recombination. virDTL newly leverages Duplication-Transfer-Loss (DTL) reconciliation, a powerful computational technique used to study HGT in prokaryotes (Gorbunov and Liubetskii, 2009; Tofigh, 2009; Doyon et al., 2010; David and Alm, 2011; Tofigh et al., 2011; Bansal et al., 2012, 2013, 2018; Chen et al., 2012; Stolzer et al., 2012; Szollosi et al., 2012, 2013; Scornavacca et al., 2013, 2015; Libeskind-Hadas et al., 2014; Sjostrand et al., 2014; Jacox et al., 2016; Kordi and Bansal, 2019), to gain insights into viral evolution and recombination (Fig. 1c). In addition, virDTL addresses common sources of HGT inference error and uncertainty under recombination by carefully constructing the strain tree and using resampling and error-correction methods. virDTL addresses some of the key difficulties traditionally associated with viral evolutionary analysis, such as systematic, large-scale identification of ancestral recombination events and precise phylogenetic identification of the recombining strains, and can help virologists and epidemiologists better understand viral evolution and easily infer recombination events.
We demonstrate the utility of virDTL using it to investigate viral recombination in the Sarbecovirus subgenus. Specifically, we ran virDTL on 54 Sarbecovirus genomes from 4 host species, including the novel coronavirus SARS-CoV-2, and assessed its ability to recover recombinations between leaf strains and discover new ancestral recombinations (Section 3). We identify 226 plausible leaf-to-leaf (i.e., between sampled strains) and 362 plausible ancestral HGTs across all gene families and identify 8 well-supported HGTs of potential relevance to SARS-CoV-2 evolution, including 3 in the well-studied spike and nucleocapsid gene families. We use the popular sequence similarity tool SimPlot (Lole et al., 1999) to validate our protocol on a subset of leaf-to-leaf HGTs and explore several case studies where our DTL-reconciliation-based approach enables inference of viral recombination. Among other results, our analysis supports the previously-proposed hypothesis that similarity between the SARS-CoV-2 and pangolin strains arose due to a recombination between the immediate ancestor of SARS-CoV-2 and RaTG13 (i.e., involving the shared parent edge of SARS-CoV-2 and RaTG13 in our strain tree) and an ancestral pangolin viral strain (Boni et al., 2020).
Finally, we identify and discuss the strengths and limitations of the proposed reconciliation-based approach, contrast virDTL with widely-used sequence-similarity based approaches such as SimPlot (Lole et al., 1999) and RDP (Martin et al., 2015), and compare our protocol against two recent approaches used to investigate recombination in coronaviruses using phylogenetic reconciliation, developed in parallel and independently from this work (Fu et al., 2020; Makarenkov et al., 2021) (Section 4).
2. MATERIALS AND METHODS
2.1. Overview of virDTL
The virDTL protocol is designed to infer recombination in viruses while minimizing the impact of key sources of error (Fig. 1c). We describe the key steps below.
-
1.
Strain tree reconstruction and selection: Since viruses are frequently impacted by substantial recombination, virDTL first identifies nonrecombinant or minimally-recombinant genomic regions that could be used to reconstruct credible strain trees. It then further analyzes candidate strain trees to identify a single, minimally recombinant strain tree. virDTL uses BEAST (Suchard et al., 2018) to construct a dated candidate strain tree using DNA sequences from the identified region of the genome.
-
2.
Gene tree reconstruction and error-correction: Gene trees are often impacted by phylogenetic reconstruction error and uncertainty due to lack of sufficient phylogenetic signal. virDTL minimizes the downstream impact of such error and uncertainty by error-correcting the gene tree topologies to match the strain tree unless the sequence data confidently support incongruence. virDTL uses RAxML (Stamatakis, 2014) for initial gene tree construction and TreeFix-DTL (Bansal et al., 2015) for error-correction. virDTL further accounts for topological uncertainty by sampling multiple error-corrected gene trees per gene family for reconciliation analysis.
-
3.
Gene tree rooting: Gene trees reconstructed using standard phylogenetic approaches are unrooted and must be rooted before reconciliation analysis. Since there is often uncertainty in rooting gene trees, virDTL uses multiple gene tree rooting approaches and assesses how the resulting differently rooted gene trees affect support for final evolutionary inferences. virDTL uses OptRoot (Bansal et al., 2018) and Minimum Ancestor Deviation (MAD) rooting (Tria et al., 2017), with OptRoot as the primary rooting method.
-
4.
Phylogenetic reconciliation analysis: To account for ambiguity or uncertainty in phylogenetic reconciliation, virDTL randomly samples many optimal reconciliations per gene tree and aggregates inferences across both reconciliation samples and gene tree samples to identify only well-supported HGTs for each gene family. virDTL uses RANGER-DTL (Bansal et al., 2018) to sample optimal reconciliations.
-
5.
Strain tree dating and evaluation of HGTs: virDTL dates the strain tree so that any HGTs inferred can be evaluated for time-consistency among the participating strains and performs additional analysis to determine if the detected HGTs support the inference of larger recombination events. virDTL uses BEAST (Suchard et al., 2018) to perform strain tree dating.
Next, we first describe our Sarbecovirus dataset and then describe the step-by-step application of virDTL to this dataset.
2.2. Sarbecovirus strain selection
For our analysis we selected 54 strains from the Sarbecovirus subgenus of the Betacoronavirus genus, with 42 strains from bats, 5 from pangolins, 5 from civet cats, and 2 from humans. We limited our strain selection to only the Sarbecovirus subgenus since genomes outside this subgenus, such as MERS-CoV, are generally too divergent from SARS-CoV-2 (Jungreis et al., 2021), and analyses including such distant strains can fail to cleanly identify gene families or can result in phylogenetic artifacts such as long branch attraction. For example, even the closest relative outside the Sarbecovirus subgenus, Hibecovirus Bat Hp-betacoronavirus/Zhejiang2013, shows no detectable homology across ORF6, ORF7a, ORF7b, and ORF8 (Jungreis et al., 2021). Further details on strain selection considerations and a contrast with strain selections in related studies (Boni et al., 2020; Makarenkov et al., 2021) appear in Supplementary Section S1.
2.3. Application of virDTL to the Sarbecovirus dataset
2.3.1. Strain tree reconstruction and dating
Given the importance of strain tree accuracy on the accuracy of HGT inference through phylogenetic reconciliation, we investigated three candidate genomic regions to reconstruct a dated strain tree. As a baseline, we constructed a whole-genome (WG) strain tree based on a WG alignment of the 54 genomes. Since coronaviruses are highly recombinant, we also selected two putative nonrecombinant regions (NRR-B [4000–9000 base pairs] and NRR-A [13,000–18,000 base pairs]) previously identified by Boni et al. (2020).
For each region/WG, we aligned the 54 sequences using Muscle v.3.8.31 (Edgar, 2004) and, following Boni et al. (2020), used BEAST v.1.10.4 (Suchard et al., 2018) to estimate a dated strain tree (see Supplementary Section S1 for details).
The three Sarbecovirus strain trees, corresponding to NRR-A, NRR-B, and WG, each had distinct topologies (Fig. 2). To assess the magnitude of topological divergence between these trees, we computed the normalized unrooted Robinson–Foulds (RF) distance (Robinson and Foulds, 1981) and unrooted subtree prune and regraft (SPR) distance (Whidden and Matsen, 2019) between them (Supplementary Table S2, top rows). Although several important clades appear largely conserved across the three trees (Fig. 2a), the three trees are highly divergent (RF: 0.615–0.788, SPR: 14–19; for reference, the maximum possible RF distance between two trees is 1), suggesting that the Sarbecovirus subgenus is influenced by substantial recombination. This result in turn implies that the WG tree should not be directly used as the strain tree and motivates the need for constructing a reliable strain tree using a nonrecombinant (or minimally recombinant) region.
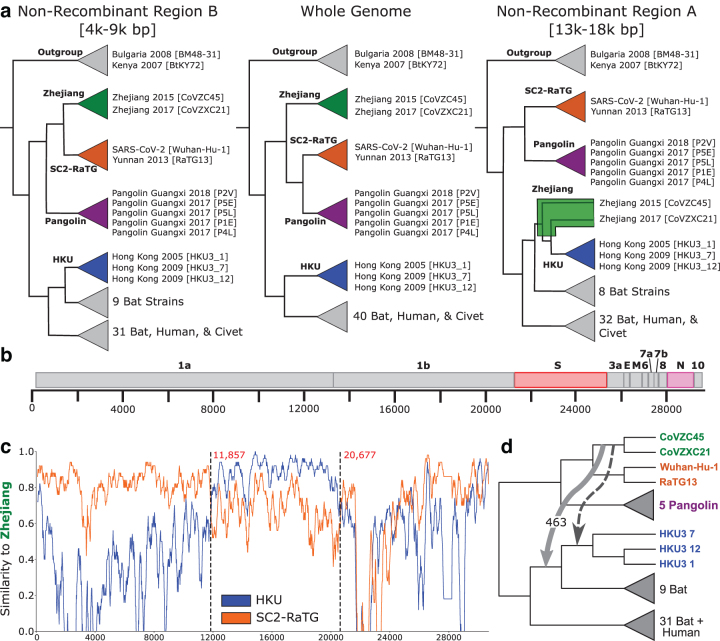
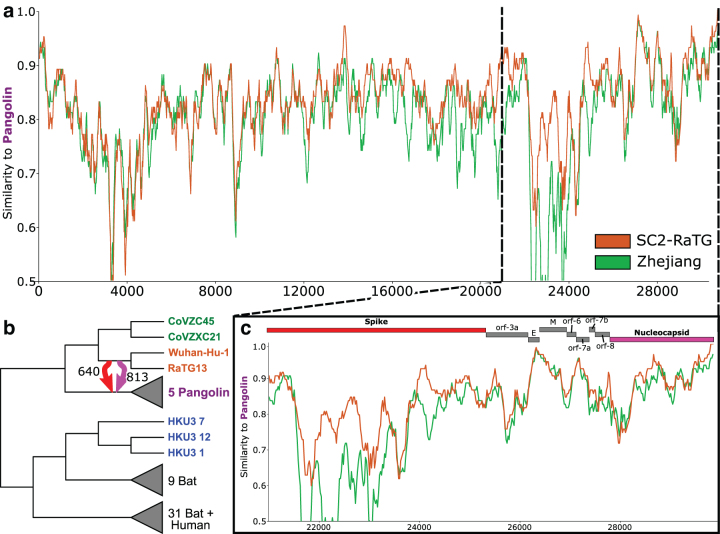
FIG. 2.
Overview of Sarbecovirus genome evolution. (a) We reconstructed three candidate strain trees from the whole genome and two putative nonrecombinant regions A (13,000–18,000 base pairs) and B (4000–9000 base pairs). Their topologies differ substantially, especially in the SARS-CoV-2 lineage, which suggests that the evolution of SARS-CoV-2 was impacted by recombination. We define four clades, Zhejiang (green), SC2-RaTG (orange), Pangolin (purple), and HKU (blue), and show the tree inferred using each region of the genome. (b) The Sarbecovirus genome comprises four well-characterized structural genes which construct the viral spike, envelope, membrane, and nucleocapsid proteins, as well as several open reading frames which encode accessory factors. The spike and nucleocapsid genes are highlighted in red and pink, respectively, as they appear in several ancestral recombinations (Fig. 5). (c) Sequence similarity along the genome using SimPlot. Using Zhejiang clade sequences as query, we compare with the SC2-RaTG and HKU clades. For the majority of the genome, SC2-RaTG is more similar to Zhejiang. Between 11,857 and 20,677 base pairs, HKU is more similar. (d) We find evidence of an HGT from the immediate ancestor of the Zhejiang clade to an ancestor of the HKU clade in ORF1ab. This recombination (light gray) explains the signal shown in the NRR-A tree (a) and SimPlot (c) and is not consistent with the dating of the phylogeny (Supplementary Fig. S2). However, it is not uncommon for inferred HGTs to be off by a single branch due to inference uncertainty. A time-consistent HGT to the ancestor of the three HKU strains (darker gray) similarly explains the signal.
We found a specific instance of recombination that affected the SARS-CoV-2 [Wuhan-Hu-1] lineage to be of particular interest, as it explains a key difference in topology between the trees inferred using NRR-A and NRR-B. Specifically, viral strains CoVZC45 and CoVZXC21 (Zhejiang clade) are placed in a different location in each of the three strain trees (Fig. 2a). In the WG strain tree, the clade containing Wuhan-Hu-1 and RaTG13 (SC2-RaTG clade) and the clade containing the Pangolin viral strains (Pangolin clade) are most closely related, with the Zhejiang clade as the next closest relative. In the NRR-B strain tree, the Zhejiang and SC2-RaTG clades are sisters, with the Pangolin clade as the next closest relative, suggesting a recombination somewhere outside of NRR-B. Finally, in the NRR-A strain tree, the Zhejiang clade does not group with either the SC2-RaTG or Pangolin clades and the two strains instead of group with three viral strains from Hong Kong (HKU clade).
2.3.1.1. Recombination within NRR-A and NRR-B
To assess whether recombination might affect the NRR-A and NRR-B trees, we constructed phylogenies for 1000-base pair windows with a 500-base pair offset along the entire length of the genome. We then computed the average internal pairwise RF and SPR distances between all window trees within 5000-base pair genomic regions and similarly computed the average internal pairwise distance between trees in each nonrecombinant region (Supplementary Table S2, bottom rows). We find that average RF and SPR distances within NRR-B (0.487 and 11.98, respectively) are smaller than within NRR-A (0.595 and 13.82, respectively) and also smaller than all other 5000-base pair regions along the length of the genome (distances ranging between 0.521–0.580 and 12.57–13.60, respectively). Higher average internal RF and SPR distances indicate increased phylogenetic incongruency between windows within each region, suggesting a higher level of recombination within that region.
2.3.1.2. Recombination across NRR-A
We performed further analysis to determine if the discrepancy in NRR-A and NRR-B strain tree topologies is a result of recombination in the putative NRR-A. Specifically, using the NRR-B tree as our viral strain tree, we found evidence for an ancestral HGT between the immediate ancestor of the Zhejiang clade and an ancestor of the HKU clades (Fig. 2d). This HGT is further supported by sequence similarity. Using SimPlot (Lole et al., 1999), we compared a query of Zhejiang 2017 [CoVZC45] against SARS-CoV-2 [Wuhan-Hu-1], Yunnan 2013 [RaTG13], and the three Hong Kong strains HKU3_1, HKU3_7, and HKU3_12. While Zhejiang 2017 is most similar to the SC2-RaTG clade for most of the genome, it is more similar to the HKU clade between 11,857 and 20,677 base pairs, which contains NRR-A (Fig. 2c). We note that this HGT was not inferred using the MAD-rooted gene tree. Nonetheless, the similarity between the Zhejiang and HKU clades in this region indicates that recombination has in fact occurred in NRR-A, making it unsuitable to construct a strain tree using this part of the genome. This finding is consistent with the conclusions of Boni et al. (2020), where they note that the Zhejiang clade needed to be removed to maintain a clean nonrecombinant signal in this region.
Given that (i) the WG tree is generally unreliable as a strain tree for reconciliation analysis due to widespread recombination across the genome, (ii) NRR-A is far less internally consistent than NRR-B and, (iii) a major topological discrepancy in the NRR-A tree is likely the result of an ancestral recombination, we used the NRR-B tree as our viral strain tree for the remainder of our analyses.
2.3.2. Gene tree reconstruction, error-correction, and rooting
The Sarbecovirus genome comprises four well-characterized structural genes, which construct the viral spike, envelope, membrane, and nucleocapsid proteins, and seven open reading frames which act as accessory factors (Fig. 2b). The largest open reading frame, ORF1ab, comprises the replicase–transcriptase complex displayed as two polyproteins (ORF1a and ORF1b), which synthesize 16 nonstructural proteins by 3 viral proteases (Graham et al., 2008; Khailany et al., 2020; Kim et al., 2020). The smaller open reading frames, near the 3′ end of the genomes, encode proteins hypothesized to interact with a diverse array of host biological pathways (Gordon et al., 2020).
We constructed gene trees for each of the 11 gene families (Fig. 2b). While most strains in our dataset were already annotated with genes from all 11 gene families, some were not and some of the unannotated genes had to be extracted using genome alignments. Further details on gene family construction appear in Supplementary Section S1. For each gene family, we aligned nucleotide gene sequences using Muscle v.3.8.31 (Edgar, 2004) and then reconstructed gene trees using RAxML v.8.2.11 (Stamatakis, 2014) using 100 fast bootstrap replicates under a GTR+ Γ substitution model.
We minimized gene tree reconstruction error by error-correcting each RAxML gene tree using TreeFix-DTL (Bansal et al., 2015) with default parameters. TreeFix-DTL is a gene tree error-correction tool that aims to find a “statistically equivalent” gene tree topology that minimizes the DTL reconciliation cost against a given species/strain tree. TreeFix-DTL has been shown to be highly effective in error-correcting gene trees, leading to a substantial reduction in the number of false positive HGTs (Bansal et al., 2015). Since each run of TreeFix-DTL can result in a slightly different estimate of the error-corrected gene tree, we applied TreeFix-DTL 10 times to each RAxML gene tree and used all 10 error-corrected gene trees for each gene family in our analysis. Thus, each gene family is represented not by 1 gene tree but by 10, helping to account for potential uncertainty in inferring gene tree topologies. Note that TreeFix-DTL is only used for gene tree inference, not for reconciliation analysis. Reconciliations are computed in a subsequent step as described in Section 2.3.3.
To account for uncertainty in gene tree rooting, we rooted each error-corrected gene tree using two different methods, OptRoot (Bansal et al., 2018), which seeks a rooting that minimizes the DTL reconciliation cost between the gene tree and species/strain tree, and MAD rooting (Tria et al., 2017), which roots the gene tree at the edge that minimizes the mean relative deviation from the molecular clock. These two rooting methods have been shown to be among the most accurate for prokaryotic gene families (Wade et al., 2020). By default, we report results based on OptRoot-rooted gene trees, but all HGTs are supported by MAD-rooted gene trees unless otherwise stated.
2.3.3. Reconciliation analysis and accounting for HGT inference uncertainty
We reconciled each of the rooted, error-corrected gene trees (10 per gene family) to the NRR-B strain tree using RANGER-DTL 2.0 (Bansal et al., 2018) with default parameters. Since there often exist multiple equally optimal DTL reconciliations of a given gene tree and strain tree (Bansal et al., 2013), we uniformly random sampled (with replacement) 100 optimal reconciliations (per rooting) for each pair of gene and strain trees. Such uniform random sampling makes it possible to assign a support value to each inferred HGT event based on how frequently that event is inferred among all optimal DTL reconciliations. These support values can then be used to distinguish between HGTs that are well supported by DTL reconciliation, despite multiple optima, and those that are not.
3. RESULTS
3.1. Recombination occurs frequently in sarbecoviruses
Recall that we account for topological uncertainty and reconciliation uncertainty by reconstructing 10 error-corrected gene trees per gene family and, for each rooting, sampling 100 optimal DTL reconciliations for each gene tree. Thus, an inferred HGT can have a maximum support of 1000. Using OptRoot-rooted gene trees, we inferred a total of 1530 HGTs, with a support of at least 1, across all gene families. Among these, we consider an HGT to be supported if it is found in at least 100 samples and we identified 588 such HGTs (61.6 percentile). Of these 588 supported HGTs, 226 are leaf-to-leaf, 115 are ancestor-to-ancestor, and 247 involve an ancestral node and a leaf. We also identify the set of 78 highly supported HGTs with support of at least 500 (94.9 percentile), as well as the set of top-25 HGTs, each of which has a support of at least 808 (98.4 percentile). Gene family-specific numbers appear in Supplementary Table S4. As a different rooting of the gene tree may affect the inferred events, we verified that, of the 588 supported HGTs, 441 are also supported using MAD rooting. Of the 78 highly supported HGTs, 71 were also supported using MAD rooting, including all of the top 25 HGTs.
We verified that most highly supported HGTs (support at least 500) are consistent with temporal constraints implied by the divergence times estimated on our strain tree. Note that HGTs that go forward in time can be temporally consistent due to the existence of unsampled strains (Davin et al., 2018), but HGTs cannot go backward in time. Specifically, we found that of the 78 highly supported HGTs, 66 are consistent with the dating implied by the strain tree, including 24 out of the top 25 HGTs (support at least 808). Seven additional HGTs would be time consistent if the donor or recipient was shifted by one branch, leaving only 5 of the 78 events as fully inconsistent. We note that both estimating divergence time and identifying donors and recipients of HGT events can be error prone, and some inconsistency is therefore expected.
While a detailed analysis of all putative HGT events is beyond the scope of this work, we highlight 8 HGTs involving the SARS-CoV-2 lineage, including a recombination in the spike gene between an ancestor of Pangolin viral strains and an ancestor of SARS-CoV-2 and Bat CoV RaTG13. We also validate a subset of inferred leaf-to-leaf HGTs using the recombination detection tool SimPlot (Lole et al., 1999), based on direct sequence comparison, and highlight additional case studies for ancestral HGTs and HGTs with ambiguous direction. In addition, we assess the feasibility of using inferred HGTs to detect larger recombination events spanning multiple genes.
Time-consistent HGTs with ancestral recipients and >500 support are shown in Supplementary Figure S2. We provide a full list of all inferred HGTs (Supplementary Table S3), as well as the full strain tree with all internal nodes labeled (Supplementary Fig. S1).
3.2. Recombination affects the SARS-CoV-2 lineage
Given the interest in understanding SARS-CoV-2 evolution, we used virDTL to search for recombinations involving the SARS-CoV-2 lineage. We inferred six highly-supported HGTs using the default OptRoot-rooted gene trees and two additional highly-supported HGTs using MAD-rooted gene trees (Fig. 3a). Among these events, at least two are transfers into the SARS-CoV-2 lineage and at least one is a transfer from the SARS-CoV-2 lineage; a clear direction of transfer could not be inferred for the remaining five HGTs. All eight HGTs involved ancestors of SARS-CoV-2 (i.e., nonterminal edges leading to SARS-CoV-2) rather than SARS-CoV-2 itself, and all but one are ancestor-to-ancestor HGTs (i.e., HGTs between nonterminal edges).
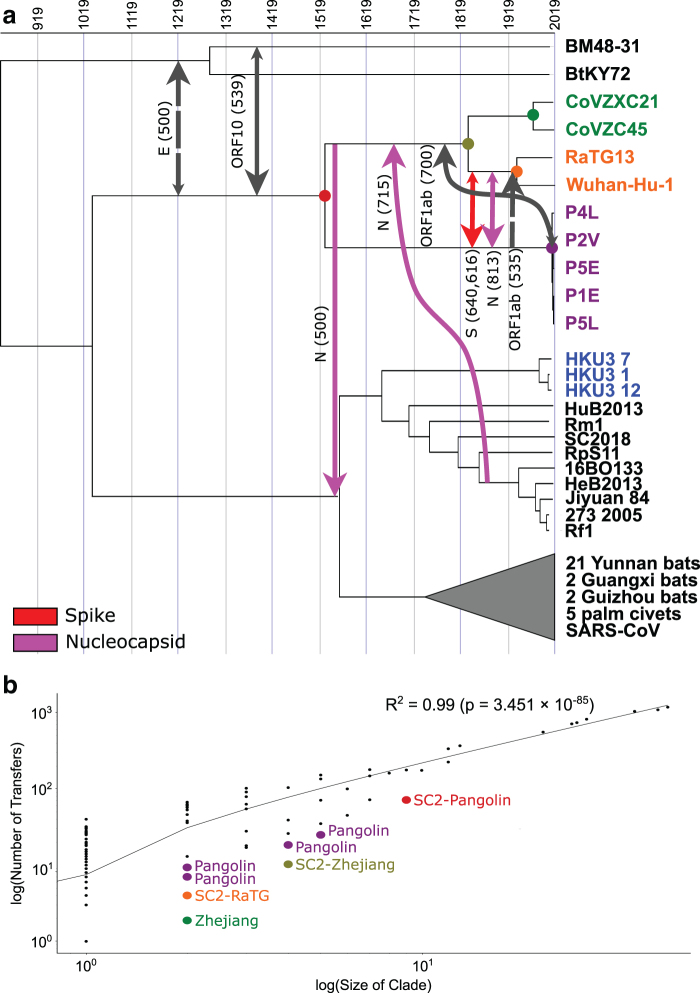
FIG. 3.
HGTs involving the SARS-CoV-2 lineage. (a) We inferred 8 highly supported (with a support of at least 500) HGTs which involve an ancestor of SARS-CoV-2 [Wuhan-Hu-1]. Support values are shown for the OptRoot-rooted gene trees (solid lines) or MAD-rooted gene trees (dashed lines), with one transfer (spike) inferred using both rootings. Smaller arrow heads indicate that there exists an HGT with at least 100 support in the reverse direction using gene trees rooted with either method, suggesting directional uncertainty. (b) We found a strong correlation between the number of leaves in a clade and the number of HGTs identified in that clade (Pearson's R2 = 0.99). However, for every ancestral strain in the SARS-CoV-2 lineage and related clades (highlighted by larger colored points), the number of HGTs in that clade is much lower than would be expected for their size. This paucity of HGTs is likely due to sampling effects, as these strains are more distantly related to the rest of the Sarbecovirus strains in the analysis. MAD, Minimum Ancestor Deviation.
Most prominently, we found evidence for recent recombination in the spike gene family, between the immediate ancestor of Wuhan-Hu-1 (i.e., SARS-CoV-2) and RaTG13 and the immediate ancestor of the pangolin strains. Ignoring directionality, this time-consistent transfer has a support of 1000 using both OptRoot- and MAD-rooted gene trees; that is, it is supported by every gene tree and reconciliation. However, support was roughly evenly split between the two directions, with OptRoot showing support of 640 (360) and MAD showing support of 616 (384) for a transfer from (to) the immediate ancestor of Wuhan-Hu-1 and RaTG13. We discuss a possible cause of this directional uncertainty later in the article (Section 3.4.1.). Despite directional uncertainty, this HGT supports the previously-proposed hypothesis that similarity between SARS-CoV-2 and pangolin strains arose due to recombination rather than pangolins being a possible host (Boni et al., 2020).
We also found evidence for three recombinations in the nucleocapsid gene family. One of these HGTs is similar to the spike gene HGT discussed above. When using OptRoot (MAD), this HGT has a support of 813 (456) from the immediate ancestor of Wuhan-Hu-1 and RaTG13 to the immediate ancestor of the pangolin strains and a support of <100 (444) in the reverse direction. Given the directional uncertainty under MAD, we view the direction of this HGT as uncertain even though our default results using OptRoot suggest that it occurred from the SARS-CoV-2 lineage to the immediate ancestor of the pangolin strains. The second nucleocapsid HGT is a transfer from the immediate ancestor of South Korean, Hebei, Henan, and Hubei bat strains to the immediate ancestor of Wuhan-Hu-1 and Zhejiang strains. This HGT is time consistent within one branch and is highly supported using OptRoot (715) but not MAD (). The third nucleocapsid HGT is a transfer from the immediate ancestor of Wuhan-Hu-1 and Zhejiang strains to the immediate ancestor of SARS-CoV, several Hong Kong and other Asian bat strains, and civet strains. While this HGT is time consistent, it is again only supported by OptRoot (500) and not MAD ().
We also found evidence for recombination in several other gene families. One of these is a time-consistent HGT affecting the ORF1ab gene and is similar to the previously observed HGTs in the spike and nucleocapsid genes, between the immediate ancestor of Wuhan-Hu-1 and RaTG13 and the immediate ancestor of the pangolin strains. While it is not supported by OptRoot-rooted gene trees, it has an undirected support of 1000 using MAD-rooted gene trees. However, this HGT also shows directional uncertainty, with a roughly evenly split support of 535 and 465 in the two directions. In addition, we found two time-consistent transfers between the outgroup of Bulgaria and Kenyan bat strains and the immediate ancestor of Wuhan-Hu-1 and pangolin strains. One of these HGTs occurs in the ORF10 gene and also shows directional uncertainty, with OptRoot showing support of 539 (261) and MAD showing support of 378 (122) for a transfer to (from) the SARS-CoV-2 lineage. A similar HGT occurs in the envelope gene, with OptRoot showing support of 147 (243) and MAD showing support of 500 (<100) for a transfer from (to) the SARS-CoV-2 lineage. Finally, we found a time-inconsistent HGT in ORF1ab from the immediate ancestor of the four pangolin strains to the immediate ancestor of Wuhan-Hu-1 and the Zhejiang strains. We note that we also find several other transfers with lower but still substantial support () that might warrant further investigation (Supplementary Table S3).
Interestingly, by analyzing the donors and recipients of our full list of 588 HGTs supported using OptRoot-rooted gene trees, we found that the ancestors and nearby relatives of the SARS-CoV-2 [Wuhan-Hu-1] genome uniformly undergo recombination less often than the rest of the Sarbecovirus subgenus (Fig. 3b). However, this observation may be an artifact of sampling effects caused by the relatively small number of strains in this clade and because of the low overall diversity among these strains.
3.3. Reconciliation recovers HGTs between leaf strains
After finding evidence for recombination involving the SARS-CoV-2 lineage, we expanded our analysis to the entire Sarbecovirus subgenus. Our analysis identified 226 supported leaf-to-leaf HGTs (≥100 support) and 35 highly supported leaf-to-leaf HGTs (≥500 support). Among the 35 highly supported HGTs, 34 were intrahost HGTs between strains from the same host type (e.g., bat to bat, pangolin to pangolin, and so on), and one was interhost HGTs between strains from different host types (human SARS-CoV to civet C010 in ORF7a). Such leaf-to-leaf HGTs can be orthogonally verified through a SimPlot analysis by choosing the recipient, donor, and sister strains of both recipient and donor, as demonstrated through the following case study.
3.3.1. Case study: spike gene HGT between strains from bats
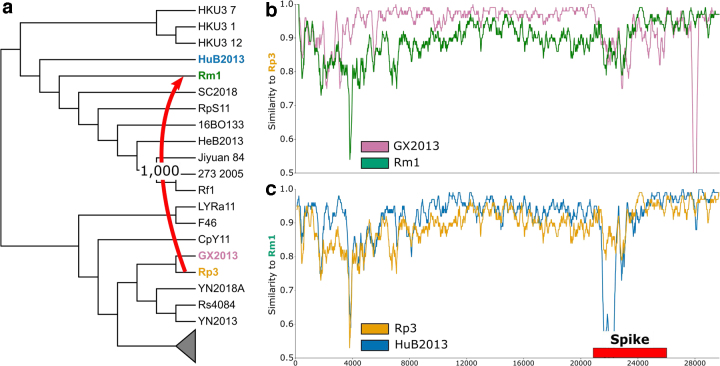
We identified a HGT in the spike gene between the strains Guangxi 2004 [Rp3, donor] and Hubei 2004 [Rm1, recipient] with a support of 1000 (Fig. 4a). For the SimPlot analysis, we selected GX2013 as the sister of Rp3 and HuB2013 as the sister of Rm1. When querying the genome of Rp3, we see high similarity with its sister GX2013 throughout the entire length of its genome with the exception of the spike gene region, where Rp3 is most similar to the recipient Rm1 (Fig. 4b). Reciprocally, when querying the genome of Rm1, we see that sequence similarity with HuB2013 decreases in the region encompassing the spike gene while sequence similarity with Rp3 increases (Fig. 4c). These findings are consistent with a hypothesis in which Rm1 received Rp3's copy of the spike gene. In addition, we observe that Rm1 continues to remain highly similar to Rp3 even beyond the boundary of the spike gene. This observation could indicate a larger multigene recombination event, which was not detected in our reconciliation-based analysis.
FIG. 4.
Highly supported leaf-to-leaf HGTs are consistent with sequence similarity. We present a case study of a leaf-to-leaf HGT between the donor Rp3 (orange) and recipient Rm1 (green). (a) The inferred HGT from Rp3 to Rm1 in the spike gene has a support of 1000, shown on a subtree of the full species tree. (b) Sequence similarity of the donor Rp3 to its sibling GX2013 (purple) and the recipient Rm1. Rp3 and GX2013 are highly similar throughout the length of the genome, and Rm1 is more divergent throughout but equally similar in the spike region. (c) Sequence similarity of the recipient Rm1 to its sibling HuB2013 (blue) and the donor Rp3. Rm1 and HuB2013 are highly similar throughout the length of the genome except in the spike region, where Rm1 has received genetic material from Rp3. Thus, Rp3 and Rm1 are more similar in the spike region.
To further assess the accuracy of recombination events inferred through virDTL, we performed similar SimPlot analyses using the donor, recipient, and recipient-sister strains to orthogonally verify each of the five other highly supported HGTs identified by virDTL in the spike gene. Details of this case study can be found in Supplementary Section S1.
3.4. Reconciliation reveals new ancestral HGTs
While we validate our approach on leaf-to-leaf transfers, the virDTL protocol also enables the inference of ancestral recombination. Our analysis identified 115 supported and 11 highly supported ancestor-to-ancestor HGTs, 113 supported and 14 highly supported ancestor-to-leaf HGTs, and 134 supported and 18 highly supported leaf-to-ancestor HGTs. While ancestor-to-ancestor and leaf-to-ancestor HGTs must correspond to an ancestral HGT, ancestor-to-leaf HGTs may in fact be an HGT from an unsampled leaf to a sampled leaf. Among the 11 highly supported ancestor-to-ancestor HGTs, 5 involve the SARS-CoV-2 lineage, 2 were intrahost HGTs between clades that contain the same host type, and 9 were interhost HGTs between clades that contain different host types. Since SimPlot compares known sequences, it is more difficult to verify ancestral recombination events through the kind of external analysis demonstrated above for leaf-to-leaf HGTs. Despite this limitation, in the following case study, using appropriately chosen descendants of the ancestral donor and recipient, we demonstrate that observed genomic sequence similarity is consistent with the inferred ancestral recombination. However, we note that post facto investigation of inferred ancestral HGTs using sequence similarity is more feasible than discovery of such HGTs from direct sequence comparison alone.
3.4.1. Case study: spike and nucleocapsid HGTs
As previously reported, we identified highly supported HGTs in the spike and nucleocapsid genes between the immediate common ancestor of Wuhan-Hu-1 (i.e., SARS-CoV-2) and RaTG13 (hence SC2-RaTG) and the immediate ancestor of the pangolin strains (Pangolin). The spike gene HGT shows a support value of 640 from SC2-RaTG to Pangolin and 360 in the reverse direction when using OptRoot for gene tree rooting, and 616 and 384, respectively, when using MAD rooting. Likewise, the nucleocapsid HGT has a support of 813 from SC2-RaTG to Pangolin using OptRoot rooting but 444 in the reverse direction when using MAD rooting. Thus, while the analysis clearly shows that recombination occurred between SC2-RaTG and Pangolin in both the spike and nucleocapsid genes, the direction of these HGTs cannot be unambiguously inferred through our analysis. This ambiguity in direction inference is the result of a lack of resolution in the species tree, such that an HGT in either direction between SC2-RaTG and Pangolin may be able to explain the corresponding gene tree topologies. Nonetheless, in this case study we demonstrate how it may sometimes be possible to use sequence similarity to additionally support inferred ancestral HGTs. The SimPlot analysis below also suggests that both the spike and nucleocapsid genes may have been transferred in a single recombination event.
Using Pangolin as the query, we found that the most closely related strain, the immediate ancestor of CoVZC45 and CoVZXC21 (Zhejiang) is more similar for much of the genome (Fig. 5a), but that SC2-RaTG becomes more similar for both the spike and regions of the nucleocapsid gene (Fig. 5c). This finding is consistent with prior literature indicating similarity between the pangolin strains and the SARS-CoV-2 [Wuhan-Hu-1] genome in the spike protein (Lam et al., 2020; Zhang et al., 2020). However, our analysis suggests that this similarity can be accounted for by a recombination between the immediate ancestor of Wuhan-Hu-1 and RaTG13 and the immediate ancestor of the pangolin strains, which is consistent with the findings of Boni et al. (2020). The elevated similarity in parts of the nucleocapsid sequence is likely a result of the same recombination event.
FIG. 5.
Ancestral HGTs are consistent with sequence similarity but difficult to discover from direct sequence comparison alone. We present a case study of an ancestral recombination which is highly supported in both the spike and nucleocapsid gene families, from the immediate ancestor of the SC2-RaTG clade (orange) to the immediate ancestor of the Pangolin clade (purple). (a) For much of the genome, Zhejiang is more similar to the donor SC2-RaTG than the recipient Pangolin. (b) Our analysis infers HGTs from SC2-RaTG to Pangolin in the spike and nucleocapsid genes with supports of 640 and 813, respectively. (c) In the 3′ region of the genome, Pangolin is often more similar to SC2-RaTG, especially in the spike and parts of the nucleocapsid gene families. However, it is difficult to clearly determine from direct sequence comparison alone which gene families have been affected by recombination, especially in ancestral cases such as these where the closest reference relative is the same for both donor and recipient. For this analysis, sequences for ancestral strains were estimated through a majority consensus of their descendants.
3.5. Bidirectional support may suggest a third-party donor
HGT events usually have high support for a single donor-recipient direction, but we found several HGTs that are “bi-directionally supported,” with neither strain appearing as the donor more than 60% of the time. Of the 588 inferred HGTs, 96 are bidirectionally supported, resulting in 48 pairs of strains with roughly equal support for an HGT in either direction in a given gene family. Note that the bidirectionally supported spike HGT between SC2-RaTG and Pangolin, identified above, shows a 640-360 split and would therefore not be counted as bidirectionally supported using the conservative threshold used above.
Bidirectional HGTs may arise due to a lack of resolution in the species tree where an HGT in either direction can explain the gene tree topology equally well, as discussed in the previous case study with the spike and nucleocapsid HGTs between SC2-RaTG and Pangolin, or due to complex HGT scenarios where multiple HGT events occur in quick succession. The case study below demonstrates a case where support for both directions arises when the candidate HGT occurs in quick succession following another HGT from a third party. This case study also highlights a shortcoming of using primarily direct sequence comparison based approaches such as SimPlot and RDP for inferring such complex HGT scenarios. For instance, even the sophisticated RDP tool requires that the user accept or reject proposed recombinations, which inform subsequent proposals. Thus, it lacks the ability to automatically model inference uncertainty and report ambiguous cases. In contrast, our approach explicitly accounts for and reports uncertainty, which can highlight ambiguous cases for further investigation.
3.5.1. Case study: bidirectional HGTs
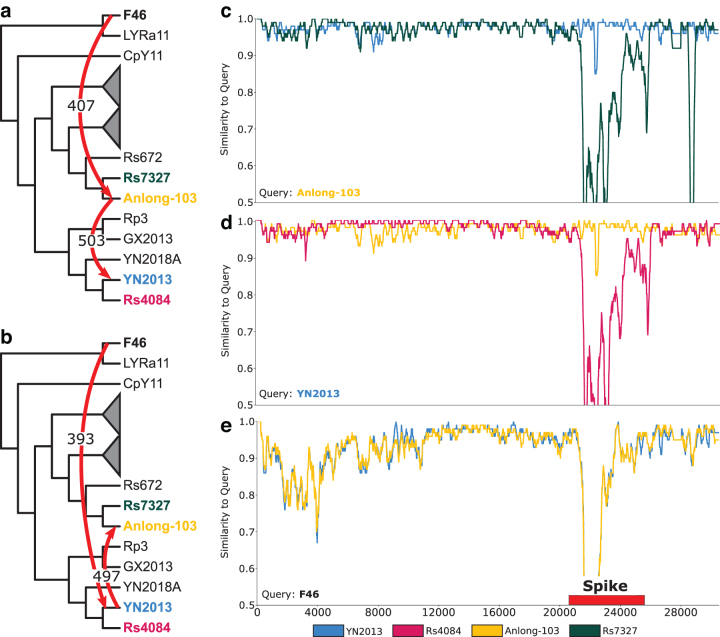
We identified an HGT in the spike gene between the strains Yunnan 2013 [YN2013] and Guizhou 2013 [Anlong-103] (Fig. 6a, b). If we do not consider donor-recipient directionality, this HGT is supported in all 1000 samples. However, support is almost evenly split between each strain as the donor (503 Anlong-103, 497 YN2013). By comparing the sequence similarity of each strain to both each other and its nearest neighbor on the strain tree using SimPlot, we hypothesize that this directional ambiguity can be explained by the presence of a third strain that recombined with one of YN2013 or Anlong-103, which then recombined with the other strain.
FIG. 6.
Pairs of strains with bidirectional HGTs suggest the presence of a third party donor. We present a case study of an inferred HGT between Anlong-103 (yellow) and YN2013 (blue) in the spike gene family, with support of (a) 503 in the forward direction and (b) 497 in the backward direction. Such bidirectional support suggests strong evidence that an HGT occurred and a third party was involved, but ambiguity as to the direction of the HGT. We found support for HGTs (a) from F46 to Anlong-103 (407 support) and (b) from F46 to YN2013 (393 support). (c, d) Show SimPlot analysis demonstrating that both Anlong-103 and YN2013 are significantly different from their respective siblings (RS7327, green, and Rs4084, magenta) in the spike gene. (e) SimPlot analysis shows that both Anlong-103 and YN2013 are equally similar to a putative third-party donor F46 (black).
Using Anlong-103 as the query (Fig. 6c), we found that its sibling Yunnan 2014 [Rs7327] is highly similar along the length of the genome, except in the spike region, where there is a significant drop-off in similarity. In contrast, YN2013 stays highly similar except in the variable-loop region, where there is a slight drop-off in similarity. We observe the same similarity profile using YN2013 as the query with its sibling Yunnan 2012 [Rs4084] (Fig. 6d). In the case of a simple unidirectional HGT event, we would expect only one of these queries to be dissimilar to its sibling.
Indeed, our HGT analysis finds third party HGTs consistent with this interpretation, with Yunnan 2012 [F46] as the third strain. We found unidirectional HGTs between Yunnan 2012 [F46] and both Anlong-103 (407 support, Fig. 6a) and YN2013 (393 support, Fig. 6b). SimPlot analysis supports this interpretation, with both YN2013 and Anlong-103 showing the same similarity drop-off profile to F46 through the spike region (Fig. 6e). The question of which strains recombined first is less clear, but geography and sampling times suggest that F46 and YN2013, both sampled in Yunnan province, could have recombined first followed by the recombination between YN2013 and Anlong-103 (Fig. 6b).
3.6. Recombination occurs across gene boundaries
Our reconciliation-based approach infers HGT events independently for each gene family. However, multiple such “HGTs” may result from a single recombination event across gene boundaries. To investigate this possibility, we assumed a null hypothesis that HGTs are not correlated across genes and assessed the frequency of HGT in adjacent gene families. Specifically, we assessed the p-value of seeing at least t HGTs in a window of w adjacent genes among the inferred donor-recipient pairs (Supplementary Table S5).
In this study, we treat HGTs as undirected edges, with the HGT support values aggregated across both directions to account for areas of directional uncertainty. We establish the null hypothesis that genes are not transferred in groups, assuming a binomial distribution, where the number of trials n is the number of strain pairs in our data set with at least t HGTs, and a success corresponds to a strain pair with at least t HGTs in at least one window of size w. To obtain the null probability of success , we randomly permuted the gene ordering 500,000 times and independently randomly selected a pair of strains. Out of all pairs of strains with at least t HGTs, we calculated as the fraction of those that fit the window condition described above. We then computed the probability of seeing at least k successes from our random permutations of gene order. We investigated several combinations of and rejected the null hypothesis at a significance level of (after Bonferroni correction for seven tests) for , but failed to do so for larger window sizes or more HGTs. This result suggests that HGTs in adjacent pairs of gene families between two strains are likely due to a single recombination event. Thus, it should be possible to combine inferred individual HGTs with gene adjacency information to identify larger recombination regions.
4. DISCUSSION OF virDTL AND RELATED APPROACHES
Many existing analyses of viral recombination often rely on direct sequence comparison alone, using tools such as SimPlot (Lole et al., 1999) and RDP (Martin et al., 2015). While such tools can identify recombinant strains and recombinant regions within those strains, they typically require a combinatorial exploration of query and reference sequences against which to compare the proposed recombinant and are not well suited for detecting ancestral recombinations. Their results are also hard to interpret when the strain lineages being analyzed have been affected by multiple successive recombination events. In addition, they are unable to capture the uncertainty that arises from HGTs occurring in rapid succession in a single lineage. These tools thus work well for investigating recent recombination events in specific strains of interest, but they are difficult to use when one wants to systematically detect ancestral recombination events and precisely identify the recombining ancestral strains.
There have been two recent investigations of HGT and recombination in coronaviruses using phylogenetic reconciliation approaches (Fu et al., 2020; Makarenkov et al., 2021) (performed independently in parallel to the current work). Fu et al. (2020) used DTL reconciliation to infer interhost HGT events using ∼400 coronavirus genomes, including alpha, beta, delta, and gamma coronaviruses from a variety of host species. The authors identified 5 gene clusters that were generally well-conserved among the considered genomes, used their concatenated alignments to reconstruct a coronavirus phylogeny, and reconciled it with gene trees from 20 protein families found in at least 30% of the genomes using the DTL reconciliation software RANGER-DTL (Bansal et al., 2018). The resulting reconciliations were used to identify the host species that were most likely to engage in cross-host-species recombination of coronaviruses. Makarenkov et al. (2021) used phylogenetic techniques to investigate patterns of HGT and recombination in 11 gene families from sarbecoviruses. In particular, the authors use the HGT detection program T-Rex (Boc et al., 2012), based on bipartition dissimilarity between a strain tree and gene trees, to identify partial- and full-gene HGTs. While these investigations illustrate the potential of using phylogenetic reconciliation for studying viral evolution, neither adequately addresses key sources of HGT inference error and uncertainty, likely leading to decreased accuracy and spurious events. For instance, the analysis of Fu et al. (2020) does not account for gene tree error and inference uncertainty, rooting uncertainty, and reconciliation uncertainty.
Our approach also differs significantly from that of Makarenkov et al. (2021), where key differences in methodology lead to several differences in inferred events. For example, Makarenkov et al. (2021) use a single WG phylogeny as their betacoronavirus strain tree, which, as our analysis suggests, has likely been affected by substantial recombination (Section 2.3.1). While we infer transfers of the spike and nucleocapsid genes between the SC2-RaTG clade and the Guangxi pangolin clade, Makarenkov et al. (2021) instead infer transfers between RaTG13 to Guangxi pangolin. While our analyses differ by one branch, Makarenkov et al. (2021) only infer a transfer because they include Guangdong pangolins. That is, their analysis might not have identified a transfer using our set of species, which highlights the potential increased sensitivity of our approach. Makarenkov et al. (2021) also do not infer transfers of the nucleocapsid gene between SC2-Zhejiang and more distant relatives. This discrepancy is likely due to the differences in species tree topology, where Makarenkov et al. (2021) place the Zhejiang clade as the outgroup of the SC2-RaTG-pangolin lineage.
However, based on sequence similarity, it is likely that the Zhejiang clade is a sister clade to SC2-RaTG clade as suggested by the NRR-B species tree. At the same time, Makarenkov et al. (2021) infer several gene transfers that we do not find in our analysis. For example, they postulate partial gene transfers of ORF1ab and membrane genes between the Zhejiang clade and SC2-RaTG clade and complete gene transfers of ORF3a, ORF8, and ORF10 between an ancestor of Guangdong pangolins and Wuhan-Hu-1 and the Zhejiang clade. These events would likely not occur using a nonrecombinant strain tree such as our NRR-B tree, which places the Zhejiang clade as a sister clade to SC2-RaTG. In addition, while Makarenkov et al. (2021) do implicitly consider gene tree inference uncertainty, by considering bootstrap values along gene tree edges to assign support values for inferred HGT events, they do not perform gene tree error-correction, which has been shown to result in significant improvements in downstream HGT inference accuracy (Sjostrand et al., 2014; Bansal et al., 2015; Jacox et al., 2016). Finally, Makarenkov et al. (2021) also use an older HGT detection tool, T-Rex (Boc et al., 2012), which is not based on DTL reconciliation and does not explicitly address HGT inference uncertainty due to multiple optima.
5. CONCLUSION
The emergence of SARS-CoV-2 demonstrates the need to understand how novel pathogens originate by crossing species boundaries and how they adapt through recombination. In this work, we introduce virDTL, a new computational protocol for viral recombination analysis, and use it to provide a more complete picture of the evolutionary history of SARS-CoV-2 in particular and sarbecoviruses in general. A key feature of virDTL is its ability to identify ancestral recombinations and provide support values for each event. virDTL leverages the DTL model and accounts for multiple sources of inference uncertainty, making it a principled, model-based approach and well suited to analyzing rapidly-evolving RNA viruses.
Our analysis of Sarbecovirus evolutionary history lends additional support to the growing body of work that suggests horseshoe bats as the most recent zoonotic origin of the SARS-CoV-2 lineage. Similarity of the ribosome binding domain of the spike protein between SARS-CoV-2 and several pangolin strains has led to the hypothesis of an intermediate pangolin reservoir for the virus (Lam et al., 2020; Zhang et al., 2020). However, our analysis suggests that this similarity is due to a recombination event between the immediate ancestor of SARS-CoV-2 [Wuhan-Hu-1] and RaTG13 and the immediate ancestor of the Guangxi pangolins. Sequence evolution through mutations or recombination with an unsampled strain would account for the divergence of RaTG13 in this region, consistent with the observations of Boni et al. (2020).
Our approach has several limitations that are worth noting. Most importantly, we analyze each gene family separately and thus cannot infer recombination events that affect only parts of genes. Moreover, uncertainty and error in HGT inference and in assigning donors and recipients can make it difficult to infer larger recombination events that affect multiple genes. These limitations can be partially addressed using a window-based analysis, rather than a gene-based analysis, but small windows risk having too little meaningful phylogenetic signal while large windows risk averaging over several different overlapping recombination events. Recently, Lytras et al. (2022) used the recombination detection tool GARD (Kosakovsky Pond et al., 2006) to identify 21 plausible recombination breakpoints in a selection of Sarbecovirus genomes, resulting in 22 putative recombination-free regions. Phylogenies constructed for these 22 regions were then analyzed to identify recombination patterns. A similar approach could be used with virDTL, applying it to identified recombination-free regions rather than to individual gene families.
Another limitation of our approach and analysis is that it ignores low-support HGTs. Low-support HGTs cannot be disregarded altogether, especially when the strains being analyzed contain short genes. Short genes, such as the envelope, ORF7b, and ORF10 gene families in our Sarbecovirus analysis, often have less phylogenetic signal and thus more uncertain gene tree topologies and inferred events. A closer analysis of low-support HGTs, especially those affecting short genes, may thus lead to additional evolutionary insights.
6. DATA ACCESS
All genomic data (Sarbecovirus genomes) underlying this article were downloaded from the NCBI sequence database (NCBI Resource Coordinators, 2018) and are publicly available. The strain trees and gene trees used in our analysis, along with scripts implementing many aspects of the virDTL protocol, are freely available at https://github.com/suz11001/virDTL, with an archival version available on Zenodo at https://doi.org/10.5281/zenodo.5247195
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Irwin Jungreis and Rachel Sealfon for help with dataset assembly and Dong-Hun Lee for helpful discussions on coronavirus evolution.
AUTHORs' CONTRIBUTIONS
All authors agreed that they have read and approved the article.
AUTHOR DISCLOSURE STATEMENT
The authors declare they have no conflicting financial interests.
FUNDING INFORMATION
Sumaira Zaman and Mukul S. Bansal were supported, in part, by NSF grant IIS-1553421. Samuel Sledzieski and Bonnie Berger were supported, in part, by NIH grants R01-GM081871 and R35-GM141861. Yi-Chieh Wu was supported by NSF grant IIS-1751399.
SUPPLEMENTARY MATERIAL
REFERENCES
- Andersen, K.G., Rambaut, A., Lipkin, W.I., et al. . 2020. The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal, M.S., Alm, E.J., and Kellis, M.. 2012. Efficient algorithms for the reconciliation problem with gene duplication, horizontal transfer and loss. Bioinformatics 28, 283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal, M.S., Alm, E.J., and Kellis, M.. 2013. Reconciliation revisited: Handling multiple optima when reconciling with duplication, transfer, and loss. J. Comput. Biol. 20, 738–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal, M.S., Kellis, M., Kordi, M., et al. . 2018. RANGER-DTL 2.0: Rigorous reconstruction of gene-family evolution by duplication, transfer and loss. Bioinformatics 34, 3214–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal, M.S., Wu, Y., Alm, E.J., et al. . 2015. Improved gene tree error correction in the presence of horizontal gene transfer. Bioinformatics 31, 1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boc, A., Diallo, A.B., and Makarenkov, V.. 2012. T-REX: A web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 40, W573–W579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni, M.F., Lemey, P., Jiang, X., et al. . 2020. Evolutionary origins of the SARS-CoV-2 Sarbecovirus lineage responsible for the Covid-19 pandemic. Nat. Microbiol. 5, 1408–1417. [DOI] [PubMed] [Google Scholar]
- Chen, Z., Deng, F., and Wang, L.. 2012. Simultaneous identification of duplications, losses, and lateral gene transfers. IEEE/ACM Trans. Comput. Biol. Bioinform. 9, 1515–1528. [DOI] [PubMed] [Google Scholar]
- David, L.A., and Alm, E.J.. 2011. Rapid evolutionary innovation during an Archaean genetic expansion. Nature 469, 93–96. [DOI] [PubMed] [Google Scholar]
- Davin, A., Tannier, E., Williams, T., et al. . 2018. Gene transfers can date the tree of life. Nat. Ecol. Evol. 2, 904–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon, J., Scornavacca, C., Gorbunov, K.Y., et al. . 2010. An efficient algorithm for gene/species trees parsimonious reconciliation with losses, duplications and transfers, 93–108. In Tannier, E., ed. RECOMB-CG, Volume 6398 of Lecture Notes in Computer Science. Springer, Berlin-Heidelberg. [Google Scholar]
- Edgar, R.C. 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni, D., Cagliani, R., Clerici, M., et al. . 2017. Molecular evolution of human coronavirus genomes. Trends Microbiol. 25, 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., Pistolozzi, M., Yang, X., et al. . 2020. A comprehensive classification of coronaviruses and inferred cross-host transmissions. bioRxiv 2020, 232520. [Google Scholar]
- Gorbunov, K.Y., and Liubetskii, V.A.. 2009. Reconstructing genes evolution along a species tree [in Russian]. Mol. Biol. (Mosk.) 43, 946–958. [PubMed] [Google Scholar]
- Gordon, D.E., Jang, G.M., Bouhaddou, M., et al. . 2020. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, R.L., Sparks, J.S., Eckerle, L.D., et al. . 2008. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 133, 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hie, B., Zhong, E.D., Berger, B., et al. . 2021. Learning the language of viral evolution and escape. Science 371, 284–288. [DOI] [PubMed] [Google Scholar]
- Jacox, E., Chauve, C., Szollosi, G.J., et al. . 2016. eccetera: Comprehensive gene tree-species tree reconciliation using parsimony. Bioinformatics 32, 2056. [DOI] [PubMed] [Google Scholar]
- Jungreis, I., Sealfon, R., and Kellis, M.. 2021. SARS-CoV-2 gene content and covid-19 mutation impact by comparing 44 Sarbecovirus genomes. Nat. Commun. 12, 2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khailany, R.A., Safdar, M., and Ozaslan, M.. 2020. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 19, 100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D., Lee, J., Yang, J., et al. . 2020. The architecture of SARS-CoV-2 transcriptome. Cell 181, 914–921.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordi, M., and Bansal, M.S.. 2019. Exact algorithms for duplication-transfer-loss reconciliation with non-binary gene trees. IEEE/ACM Trans. Comput. Biol. Bioinform. 16, 1077–1090. [DOI] [PubMed] [Google Scholar]
- Lam, T.T., Jia, N., Zhang, Y., et al. . 2020. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583, 282–285. [DOI] [PubMed] [Google Scholar]
- Libeskind-Hadas, R., Wu, Y., Bansal, M.S., et al. . 2014. Pareto-optimal phylogenetic tree reconciliation. Bioinformatics 30, i87–i95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P., Jiang, J., Wan, X., et al. . 2020. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLOS Pathog 16, e1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole, K.S., Bollinger, R.C., Paranjape, R.S., et al. . 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73, 152–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytras, S., Hughes, J., Martin, D., et al. . 2022. Exploring the natural origins of SARS-CoV-2 in the light of recombination. Genome Biol. Evol. 14, evac018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarenkov, V., Mazoure, B., Rabusseau, G., et al. . 2021. Horizontal gene transfer and recombination analysis of SARS-CoV-2 genes helps discover its close relatives and shed light on its origin. BMC Ecol. Evol. 21, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, D.P., Murrell, B., Golden, M., et al. . 2015. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters, P.S., Perlman, S.. 2013. Coronaviridae. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- NCBI Resource Coordinators. 2018. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 46, D8–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patiño-Galindo, J.Á., Filip, I., AlQuraishi, M., et al. . 2020. Recombination and lineage-specific mutations led to the emergence of SARS-CoV-2. bioRxiv 2020, 942748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Losada, M., Arenas, M., Galán, J.C., et al. . 2015. Recombination in viruses: Mechanisms, methods of study, and evolutionary consequences. Infect. Genet. Evol. 30, 296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond, K.S.L., Posada, D., Gravenor, M.B., et al. . 2006. GARD: A genetic algorithm for recombination detection. Bioinformatics 22, 3096–3098. [DOI] [PubMed] [Google Scholar]
- Robinson, D.F., and Foulds, L.R.. 1981. Comparison of phylogenetic trees. Math. Biosci. 53, 131–147. [Google Scholar]
- Scornavacca, C., Jacox, E., and Szollosi, G.J.. 2015. Joint amalgamation of most parsimonious reconciled gene trees. Bioinformatics 31, 841–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scornavacca, C., Paprotny, W., Berry, V., et al. . 2013. Representing a set of reconciliations in a compact way. J. Bioinform. Comput. Biol. 11, 1250025. [DOI] [PubMed] [Google Scholar]
- Sjostrand, J., Tofigh, A., Daubin, V., et al. . 2014. A Bayesian method for analyzing lateral gene transfer. Syst. Biol. 63, 409–420. [DOI] [PubMed] [Google Scholar]
- Stamatakis, A. 2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolzer, M., Lai, H., Xu, M., et al. . 2012. Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics 28, 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard, M.A., Lemey, P., Baele, G., et al. . 2018. Bayesian phylogenetic and phylodynamic data integration using beast 1.10. Virus Evol. 4, vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szollosi, G.J., Boussau, B., Abby, S.S., et al. . 2012. Phylogenetic modeling of lateral gene transfer reconstructs the pattern and relative timing of speciations. Proc. Natl. Acad. Sci. U.S.A. 109, 17513–17518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szollosi, G.J., Tannier, E., Lartillot, N., et al. . 2013. Lateral gene transfer from the dead. Syst. Biol. 62, 386–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofigh, A. 2009. Using Trees to Capture Reticulate Evolution: Lateral Gene Transfers and Cancer Progression. PhD Thesis, KTH Royal Institute of Technology. [Google Scholar]
- Tofigh, A., Hallett, M.T., and Lagergren, J.. 2011. Simultaneous identification of duplications and lateral gene transfers. IEEE/ACM Trans. Comput. Biol. Bioinform. 8, 517–535. [DOI] [PubMed] [Google Scholar]
- Tria, F., Landan, G., and Dagan, T.. 2017. Phylogenetic rooting using minimal ancestor deviation. Nat. Ecol. Evol. 1, 193. [DOI] [PubMed] [Google Scholar]
- Wade, T., Rangel, L.T., Kundu, S., et al. . 2020. Assessing the accuracy of phylogenetic rooting methods on prokaryotic gene families. PLoS One 15, e0232950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whidden, C., and Matsen, F.A.. 2019. Calculating the unrooted subtree prune-and-regraft distance. IEEE/ACM Trans. Comput. Biol. Bioinform. 16, 898–911. [DOI] [PubMed] [Google Scholar]
- Wu, F., Zhao, S., Yu, B., et al. . 2020. A new coronavirus associated with human respiratory disease in china. Nature 579, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T., Wu, Q., and Zhang, Z.. 2020. Probable pangolin origin of SARS-CoV-2 associated with the covid-19 outbreak. Curr. Biol. 30, 1346–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X., Wang, X., et al. . 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.