Significance
Complex behavior involves the collaborative processing of multiple brain regions across 3D-space in the whole brain. Histaminergic neurons, which send its fibers to all major brain regions, is believed to regulate many physiological and pathological processes. However, the understanding of the precise 3D structural pattern of histaminergic projections is limited. Here, we reconstructed the whole-brain 3D precise structure of histaminergic projections in the mouse brain from the whole population and individual neuron perspectives, in which we quantitatively analyzed the distribution characteristics of histaminergic somata and fibers. More importantly, the direct evidence of differences between individual histaminergic neuron was provided in this work. Our findings build a solid foundation for future research studies of histaminergic neural circuits and pharmacological targets.
Keywords: histaminergic network, histaminergic neurons, whole-brain map, fMOST
Abstract
Histamine is a conserved neuromodulator in mammalian brains and critically involved in many physiological functions. Understanding the precise structure of the histaminergic network is the cornerstone in elucidating its function. Herein, using histidine decarboxylase (HDC)-CreERT2 mice and genetic labeling strategies, we reconstructed a whole-brain three dimensional (3D) structure of histaminergic neurons and their outputs at 0.32 × 0.32 × 2 μm3 pixel resolution with a cutting-edge fluorescence microoptical sectioning tomography system. We quantified the fluorescence density of all brain areas and found that histaminergic fiber density varied significantly among brain regions. The density of histaminergic fiber was positively correlated with the amount of histamine release induced by optogenetic stimulation or physiological aversive stimulation. Lastly, we reconstructed a fine morphological structure of 60 histaminergic neurons via sparse labeling and uncovered the largely heterogeneous projection pattern of individual histaminergic neurons. Collectively, this study reveals an unprecedented whole-brain quantitative analysis of histaminergic projections at the mesoscopic level, providing a foundation for future functional histaminergic study.
The central histaminergic system is believed to regulate a multitude of physiological functions: sleep–wake cycle (1, 2), feeding behavior (3, 4), cognitive (5, 6) and motor functions (7, 8), and pathological process, including sleep disorders (9, 10), cognitive dysfunctions (11) and Alzheimer’s disease (12), Parkinson’s disease (13), multiple sclerosis (14), and schizophrenia (15, 16). Histaminergic neurons are the exclusive resource of neuronal histamine, which are located only in the tuberomammillary nucleus (TMN) of the posterior hypothalamus and send neuronal fibers to nearly every major brain region. Unlike classic neurotransmitters (gamma-aminobutyric acid and glutamate), histamine is rarely transmitted through synapses and instead is predominantly released via varicosities beaded on histaminergic axons, quickly binding to various surrounding histamine receptors (17); a process called volume transmission or paracrine (18). Therefore, establishing a comprehensive map of the histaminergic network, focusing not simply on axon terminals that usually form a synapse but instead on all axonal fibers, would be a milestone for functional research of histamine.
Previous studies have explored the distribution of histaminergic fibers in rat brains using immunohistochemical staining (19–22). However, due to limitations in the labeling tools and imaging technology, these studies have not fully demonstrated the distribution of histaminergic neurons and their axon fibers at the whole-brain 3D level. Furthermore, although thorough, careful, and objective work has been made in previous studies, it is inevitable to miss some information by presenting the research results by fiber density grade. On the other hand, a question remains in the histaminergic research, whether the histaminergic network is a whole center or distinct subpopulations. Early study used retrograde virus to explore the histaminergic distribution and found that histaminergic neurons in different subregions of TMN can project to all major downstream regions (23, 24). However, later studies gave different views (25); some researchers found that different external stimulations can activate different subsets and numbers of histaminergic neurons in TMN (26, 27), which indicated functional heterogeneity of histaminergic neuron subpopulations. What’s more, it has been proved by the microdialysis experiment that the increased levels of histamine in the downstream brain regions are different when histaminergic neurons are activated by different pharmacological reagents (28–30), suggesting the receptor heterogeneity of histaminergic neurons. But the individual fine morphological structure characteristics of histaminergic neurons still need an objective display and analysis. Recently, the development of the fluorescence microoptical sectioning tomography (fMOST) system has enabled researchers to explore the precise characteristics of specific neurons and their projections on a mesoscopic scale (31, 32).
In the present study, we established a comprehensive whole-brain 3D map of histaminergic neurons and projection in mice through using HDC-CreERT2 mice, genetic labeling strategies, and fMOST reconstruction. This whole-brain 3D map may produce a profound influence on the future structural and functional studies for the histaminergic circuit.
Results
Whole-Brain Visualization and Quantification of Histaminergic Neurons.
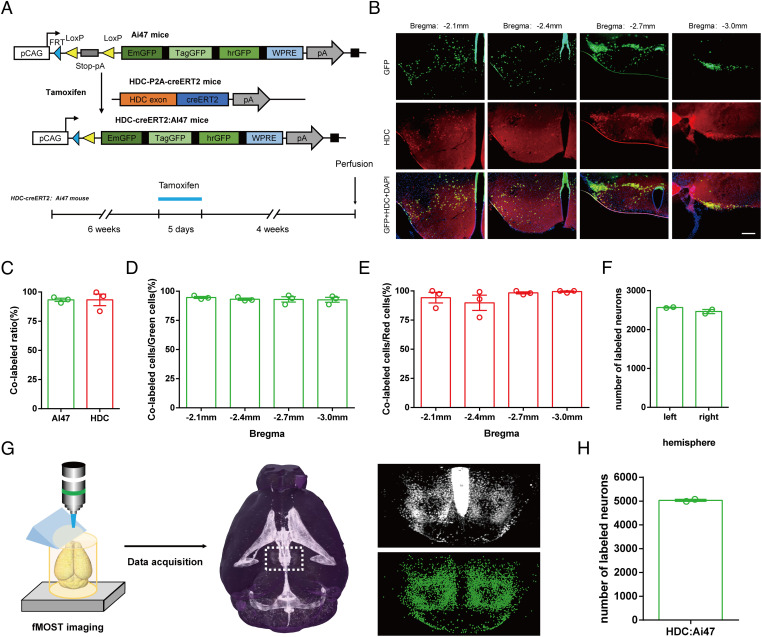
To genetically label histaminergic neurons across the whole mouse brain, we crossed tamoxifen-inducible HDC-CreERT2 mice, generated via 2A-CreERT2 expression box insertion upstream of the termination codon tag HDC through CRISPR/Cas9, with Ai47 reporter mice containing green fluorescent protein (EmGFP), TagGFP2, and hrGFP genes with stop sequences in the same expression frame (32), generating HDC-CreERT2:Ai47 mice. To avoid the possible influence of histaminergic neurons during embryonic development, we injected tamoxifen (100 mg/kg) into 6-wk-old HDC-CreERT2:Ai47 mice for 5 consecutive days, allowing the three GFP genes to be expressed in a Cre-dependent manner over the next 4 wk (Fig. 1A). We found that GFP+ neurons were predominantly expressed in the posterior hypothalamus, as in previous studies (33); surprisingly, a few neurons were also found in other brain regions, such as the suprachiasmatic nucleus and the substantia nigra, compact part (SI Appendix, Fig. S1A). Additionally, we observed a bright fluorescent signal at the edge of the ventricles (SI Appendix, Fig. S1B), which may be related to message exchanges between the central histaminergic system and cerebrospinal fluid (34) or influenced by the peripheral histaminergic system. To confirm the accuracy of histaminergic neuron genetic labeling, we performed immunostaining against HDC on brain slices from HDC-CreERT2:Ai47 mice at several locations in the posterior hypothalamus and other GFP+ regions. We found that most GFP+ neurons in the posterior hypothalamus (93.29 ± 1.41%, n = 3) expressed HDC and that most HDC+ neurons (93.27 ± 4.94%, n = 3) also expressed GFP (Fig. 1 B–E) from the anterior to posterior sections of this brain region, convincingly suggesting that the specificity and penetrance of GFP expression in the genetic-labeled mice. No immunohistochemical HDC signals were found in other brain regions containing only a few GFP+ neurons, indicating that these neurons are likely not real histaminergic neurons (SI Appendix, Fig. S1A). One plausible explanation is the migration of histaminergic neurons during fetal development (17). Although we utilized the ERT2-tamoxifen system to mitigate this potential problem, slight effects were induced by a minimal quantity of estrogen in male mice. Alternatively, one previous study found that the suprachiasmatic nucleus, a nonposterior hypothalamus area expressing GFP in our study, contained histamine-immunoreactivity neurons which did not express HDC in both mice and rats, potentially through an uncharacterized uptake mechanism (35), highlighting another possible mechanism for nonhypothalamus GFP expression. Collectively, these findings exhibit that histaminergic neurons in adult mice brains do concentrate in the posterior hypothalamus.
Fig. 1.
Whole-brain histaminergic neurons in the TMN of HDC-CreERT2:Ai47 mice. (A) Generation of HDC-CreERT2:Ai47 transgenic mice. Breeding Ai47 mice with HDC-CreERT2 mice yielded HDC-CreERT2:Ai47 mice, in which GFP is expressed specifically in HDC-positive neurons after injection of tamoxifen (100 mg/kg, i.p., 5 d). (B) Representative immunohistochemistry of expression specificity and penetrance in HDC-CreERT2:Ai47 mice on coronal sections with different distances from bregma. (Scale bars: 200 μm.) (C) Colocalization analysis of GFP and HDC in all sections of the posterior hypothalamus from 3 HDC-CreERT2:Ai47 mice. (D and E) Expression specificity (D) and penetrance (E) of coronal sections with different distances from bregma in HDC-CreERT2:Ai47 mice. (F) Count numbers of histaminergic neurons in the posterior hypothalamus of 2 HDC-CreERT2:Ai47 mouse bilateral hemispheres. (G) Main steps of data generation, acquisition, and processing in fMOST imaging. Raw data were analyzed by NeuroGPS system and manual correction. (H) Total numbers of labeled histaminergic neurons in the posterior hypothalamus of 2 HDC-CreERT2:Ai47 mice.
In order to quantify the exact number of histaminergic neurons in mice brains and study their spatial distribution characteristics, we acquired over 5,000 brain slices to reconstruct whole-brain GFP+ neurons of each HDC-CreERT2:Ai47 mouse at a resolution of 0.32 × 0.32 × 2 μm3 via an fMOST system. A NeuroGPS system was used to determine the number of GFP+ neurons at an integrated 1 × 1 × 2 μm3 pixel resolution in posterior hypothalamic slices (SI Appendix, Fig. S1C). We then accurately counted the number of fluorescent somata by manual correction (Movie S1) and displayed them as 3D neuronal information points (Fig. 1G), revealing that there are ~5,000 histaminergic neurons (5,030.0 ± 42.5, n = 2, Fig. 1H) in the brain of an adult male mouse. The number of labeled neurons in each hemisphere is ~2,500 (2,566.0 ± 11.0 for the left side, 2,464 ± 53.5 for the right side, n = 2, Fig. 1F), which reflects the symmetry of histaminergic distribution.
Whole-Brain Visualization and Quantification of Histaminergic Fibers.
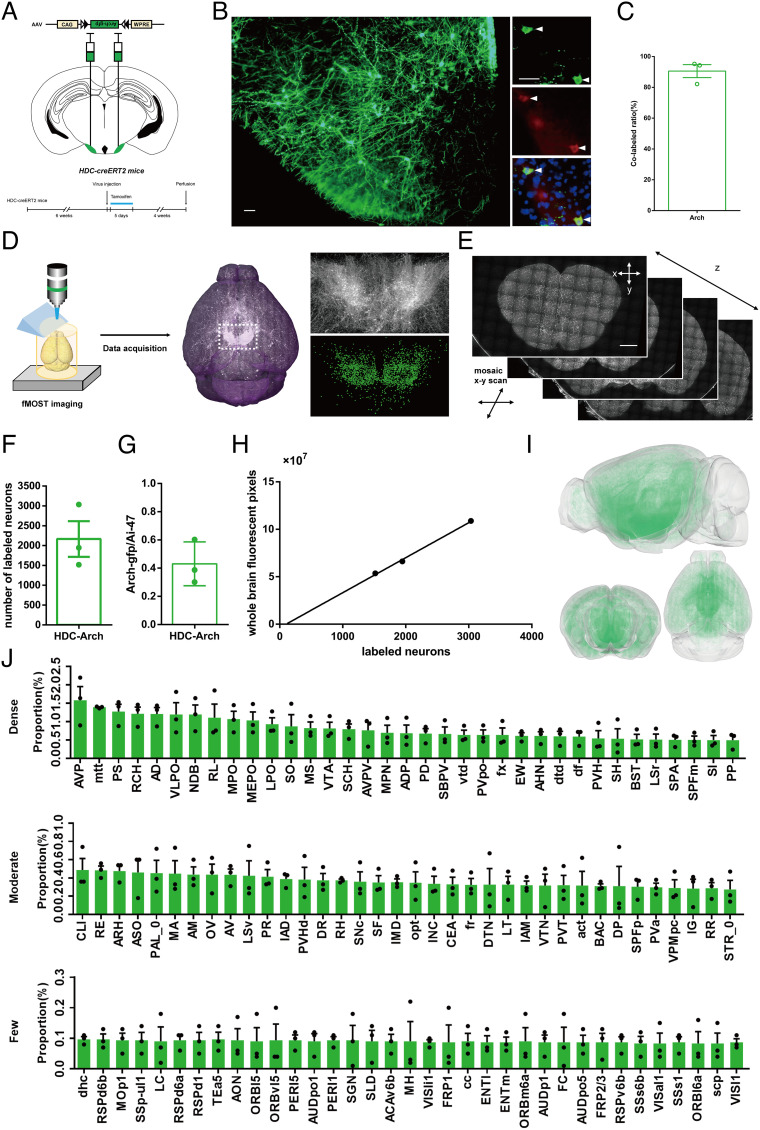
Although the spatial information of histaminergic somata acquired by genetic labeling is accurate and reliable, even after 6 mo of tamoxifen injection in HDC-CreERT2:Ai47 mice we observed no obvious fluorescently labeled axons in brain areas outside the posterior hypothalamus; GFP expression was minimal even on the histaminergic axons near the somata (SI Appendix, Fig. S1D). One plausible explanation is that the three GFP genes in the Ai47 reporter mice are predominantly expressed in the cellular bodies of neurons, rather than their axons. To map the whole-brain histaminergic projections, we need to find a way to clearly label the somata and axons of histaminergic neurons. To that end, we tested several Cre-dependent viruses with GFP, including AAV-Ef1α-DIO-ChR2-eYFP, AAV-CAG-DIO-ArchT-GFP, and AAV-Ef1α-DIO-GCaMP6s. Notably, AAV-CAG-DIO-ArchT-GFP was capable of expressing bright fluorescent proteins in both the somata and axons of histaminergic neurons, with different densities observed across nearly every brain region (Fig. 2 A and B and SI Appendix, Fig. S2A), while AAV-Ef1α-DIO-ChR2-eYFP and AAV-Ef1α-DIO-GCaMP6s only expressed at the injection site (SI Appendix, Fig. S2 B and C). To confirm the labeling accuracy of histaminergic neurons by virus injection, we performed immunostaining against HDC on brain slices collected from HDC-Arch-GFPTMN mice at the injection sites. We found that most GFP+ neurons (90.56 ± 4.22%, n = 3) expressed HDC (Fig. 2 B and C), indicating that the virus selected was working in a Cre-dependent manner.
Fig. 2.
Virus-labeled histaminergic neurons and downstream circuits in HDC-Arch-GFPTMN mice. (A) Experimental scheme and timeline for generation of HDC-Arch-GFPTMN mice. AAV-CAG-FLEX-ArchT-GFP was injected into each side of the TMN of HDC-CreERT2 adult mice. (B) Representative immunohistochemical pictures of expression specificity in HDC-Arch-GFPTMN mice; GFP and HDC double-positive neurons are indicated by arrowheads. (Scale bars: 50 μm.) (C) Expression specificity analysis of GFP in HDC-Arch-GFPTMN mice. (D and E) Main steps of data generation, acquisition (D), and processing (E) in fMOST imaging. Raw data were analyzed by NeuroGPS system and manual correction. (Scale bars: 1 mm.) (F) Count numbers of fluorescently labeled neurons in the posterior hypothalamus of HDC-Arch-GFPTMN mice. (G) The ratio of fluorescent neurons in HDC-Arch-GFPTMN mice to the number of histaminergic neurons in the whole-brain. (H) Relationship between virus-labeled neurons and whole-brain fluorescent pixels (10 × 10 × 10 μm resolution) from 3 HDC-Arch-GFPTMN mice. (I) Fluorescent-labeled information visualization of registered HDC-Arch-GFPTMN mouse brain (Top: sagittal view; Bottom: coronal and horizontal views). (J) Fluorescence fiber density value of various brain regions in 3 HDC-Arch-GFPTMN mice. Top 35 brain regions in the groups “Dense,” “Moderate,” and “Few” are shown, respectively. See the details and list of abbreviations in SI Appendix, Table S1. All data are presented as mean ± SEM.
To acquire complete information on GFP-labeled histaminergic axons, we then reconstructed the whole-brain GFP dataset of each HDC-Arch-GFPTMN mouse at a pixel resolution of 0.32 × 0.32 × 2 μm3 through the fMOST system (Fig. 2 D and E and Movie S2). We obtained the whole-brain 3D structure of histaminergic neurons’ axonal distribution in HDC-Arch-GFPTMN mice. However, there are inherent morphological differences in different mouse brains, and in the process of sampling, preprocessing, and slicing, brains will be further, differently deformed. As such, it is difficult to use the raw 3D data for unified quantitative analysis. To solve this problem, we employed a standardization process on the sample mouse brain. We manually drew the 3D structures of tissue borders and landmarks to make alignment with the Allen Reference Atlas Common Coordinate Framework (CCF v3.0) (36) to standardize the spatial information of sample brains, from which we acquired the spatial segmentation of all brain regions in each standardized mouse brain. We then utilized these processes to generate standardized 3D fluorescence information for each HDC-Arch-GFPTMN mouse (Fig. 2I and SI Appendix, Fig. S3 A and B).
Because histaminergic neurons send axonal fibers to nearly every brain region, it is difficult to describe their distribution characteristics. To solve this challenge, we calculated the density of fluorescent-labeled histaminergic axonal fibers in all brain regions using standard segmentation information. For example, there were 3,823,000 pixels with 10 × 10 × 10 μm3 resolution in the standard ventrolateral preoptic nucleus (VLPO), and 69,148 of them were fluorescently labeled in HDC-Arch-GFPTMN mouse NO. 192979’s brain; so, in this sample, the fluorescence density value of histaminergic neuron axonal fibers in the VLPO is 1.81%. We determined the histaminergic fiber density values of all brain regions of three HDC-Arch-GFPTMN mice (SI Appendix, Table S1). Considering the severe interference of histaminergic neuron somata fluorescence on the calculation of fluorescence density value, we excluded brain regions containing histaminergic neuron somata in the follow-up analysis (SI Appendix, Table S2). We then divided the histaminergic downstream nuclei into three categories according to their fluorescence density value: Dense (value ≥ 0.5%), Moderate (0.1% ≤ value < 0.5%), and Few (value < 0.1%). Using this classification, 35 brain regions were dense, 204 brain regions were moderate, and 326 brain regions were few. All brain regions in the Dense group and some of the top in the Moderate and Few groups are shown in a histogram (Fig. 2J).
As the number of labeled histaminergic neurons in each HDC-Arch-GFPTMN mouse is different (Fig. 2 F and G), the absolute fluorescence density differs between individuals. Accordingly, we quantified the whole-brain fluorescent pixels of each mouse to confirm the virus expression stability in the whole-brain axons. We determined that the number of whole-brain fluorescent pixels correlated linearly with the number of virus-labeled histaminergic neurons in mice (Fig. 2H). Further, to verify whether there were significant differences in the projection trends of histaminergic neurons in different HDC-Arch-GFPTMN mice, we calculated the ratio of pixel numbers in the 35 brain regions with the highest total fluorescent pixels, to the number of fluorescence pixels in the whole brain. The results showed no significant differences in labeled histaminergic neuron projection trends of across different HDC-Arch-GFPTMN mice (SI Appendix, Fig. S3C).
According to the quantitative analysis results, we find that after excluding the virus injection area, the densest area of histaminergic fibers in the whole brain was still the hypothalamus, with the cerebellum exhibiting the lowest fiber density. Most hypothalamus brain regions belong to the Dense group: the anteroventral preoptic nucleus, VLPO, medial preoptic area, median preoptic nucleus, lateral preoptic area, anteroventral periventricular nucleus, medial preoptic nucleus, anterodorsal preoptic nucleus (SI Appendix, Fig. S4B), posterodorsal preoptic nucleus, periventricular hypothalamic nucleus, preoptic part (SI Appendix, Fig. S4C), retrochiasmatic area, supraoptic nucleus, suprachiasmatic nucleus, subparaventricular zone, anterior hypothalamic nucleus, and paraventricular hypothalamic nucleus (SI Appendix, Fig. S4E). All areas of the cerebellum have fiber densities close to 0%, whether in the cerebellar cortex or nuclei. The pallidus, part of the cerebral nuclei, exhibit the second most abundant fiber distribution in the whole mouse brain, including nuclei with Dense fiber networks such as the diagonal band nucleus, medial septal nucleus (MS) (SI Appendix, Fig. S4A), bed nuclei of the stria terminalis (BST), and substantia innominate (SI Appendix, Fig. S4D). The other section of the cerebral nuclei, the striatum, has Moderate fiber networks in most of its nuclei with two exceptions: the caudoputamen (SI Appendix, Fig. S4A), which has the largest volume but the fewest fiber density within, and the lateral septal nucleus rostral (rostroventral) part (SI Appendix, Fig. S4A), the only nucleus with Dense fiber networks in the striatum. All parts of the cerebral cortex show Few to Moderate fiber networks, including the isocortex, olfactory areas, hippocampal formation, and cortical subplate (SI Appendix, Fig. S4 E–H); there are no significant differences between sublayers of cerebral cortex nuclei. The thalamus exhibits nuclei with Few to Moderate fiber distribution with some notable exceptions: the anterodorsal nucleus (SI Appendix, Fig. S4E), subparafascicular area, subparafascicular nucleus, magnocellular section (SI Appendix, Fig. S4F), and peripeduncular nucleus (SI Appendix, Fig. S4G). A Dense network of fibers traversed the rostral linear nucleus raphe, ventral tegmental area, and Edinger–Westphal nucleus (SI Appendix, Fig. S4G) while most other midbrain brain regions have Moderate fiber density. The hindbrain exhibits low fiber density, with only a few regions in the pons showing Moderate fiber networks. Fiber densities in the fiber tracts are disparate, most parts exhibit Few fiber density, some Moderate, and several Dense, including the mammillothalamic tract, columns of the fornix, dorsal fornix networks (SI Appendix, Fig. S4 D and F), ventral tegmental decussation, and dorsal tegmental decussation (SI Appendix, Fig. S4H). The densities of the histaminergic fibers in different brain regions are summarized in SI Appendix, Table S3.
Correlation Between Histamine Release and Histaminergic Fiber Density.
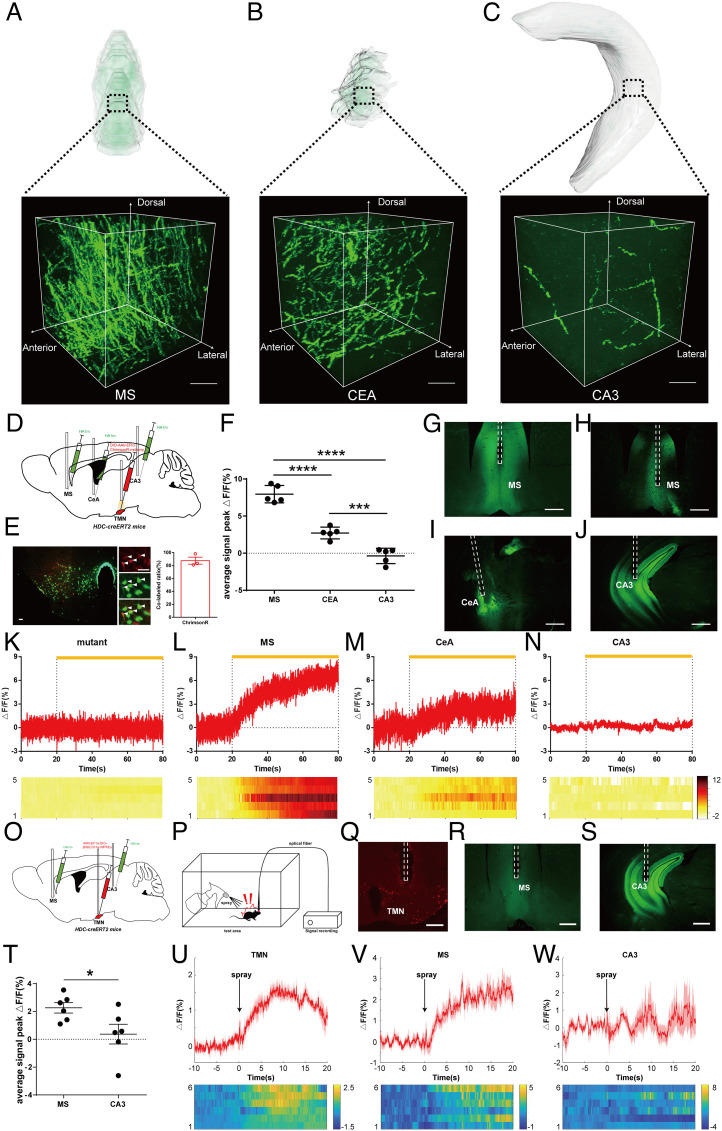
Then, we questioned whether there is a functional relationship between fiber density and histamine secretion in various histaminergic downstream areas. First, we selected three histaminergic downstream nuclei, including MS in the Dense group, central amygdalar nucleus (CeA) in the Moderate group, and field CA3 (CA3) in the Few group, whose detailed 3D reconstruction images are shown at 200 × 200 × 200 μm3 with 0.32 × 0.32 × 2 μm3 pixel resolution (Fig. 3 A–C). We then performed functional correlation experiments using optogenetics combined with GRABHA histamine sensors. HA1m is a type of GRABHA sensor which only displays green fluorescence when it is combined with histamine under 473-nm blue light excitation. As such, it is a suitable method to detect histamine fluctuations in the histaminergic downstream through fluorescence HA1m signal changes. Considering potential histaminergic axonal activation induced by 473-nm blue light during downstream fluorescence signal recording, we chose to use ChrimsonR (37), an optogenetic opsin responding to 589-nm yellow light, to activate histaminergic neurons. We injected the AAV-hSyn-DIO-ChrimsonR-mCherry optogenetic virus into the right TMN nucleus and the AAV-hSyn-HA1m virus into the MS, ipsilateral CeA, and CA3 nucleus of adult HDC-CreERT2 mice (Fig. 3D). Following injection, the light-sensitive protein ChrimsonR was efficiently expressed in HDC+ neurons (87.48 ± 5.38%, n = 3; Fig. 3E), and the GRABHA sensor HA1m was expressed in all types of neurons located in the MS, CeA, and CA3 regions (Fig. 3 H–J). We then activated histaminergic neurons with the 589-nm yellow light (monophasic square-wave pulses, 10 Hz, 10 ms/pulse, 600 pulses) and concurrently recorded signal changes of the MS, CeA, and CA3, respectively, through optic fibers embedded in these brain regions of freely moving mice.
Fig. 3.
Positive correlation between histamine release and histaminergic fiber density in downstream brain regions. (A–C) Representative detailed 3D reconstruction images of MS (A), CeA (B), and CA3 (C): 200 × 200 × 200 μm3 with 0.32 × 0.32 × 2 μm3 pixel resolution. (Scale bars: 100 μm.) (D) Experimental scheme for generating HDC-ChrimsonR-mCherryTMN-HA1mMS, CeA, CA3 mice. AAV-hSyn-DIO-ChrimsonR-mCherry was injected into the right TMN of HDC-CreERT2 adult mice, and GRABHA was injected into the MS, right CeA, and right CA3 simultaneously. (E) Characterization of the injection site and coexpression analysis of expression specificity of HDC:Ai47-ChrimsonR-mCherryTMN mice, GFP, and mCherry double-positive neurons are indicated by arrowheads. (Scale bars: 50 μm.) (F) Average signal peak ΔF/F of 3 tested brain regions. (G) Representative image of the MS from an HDC-ChrimsonR-mCherryTMN-HAmutantMS mouse. (H–J) Representative images of the MS (H), CeA (I), and CA3 (J) from an HDC-ChrimsonR-mCherryTMN-HA1mMS, CeA, CA3 mouse. (Scale bars: 200 μm.) (K) Fiber photometry of MS histamine concentration by HAmutant sensor during stimulation by 589-nm yellow light. (L–N) Fiber photometry of MS (L), CeA (M), and CA3 (N) histamine concentration by HA1m sensor during stimulation by 589-nm yellow light in tested mouse. The signals of 5 mice are illustrated in the heatmap; color scale indicates ΔF/F and warmer colors indicate higher fluorescence signal. (O) Experimental scheme for generating HDC-jRGECO1αTMN-HA1mMS, CA3 mice. rAAV-EF1α-DIO-jRGECO1α was injected into the right TMN of HDC-CreERT2 adult mice, and GRABHA was injected into the MS, right CeA, and right CA3 simultaneously. (P) Experimental scheme for functional test of HA1m signal in aversive response of mice. (Q–S) Representative images of the TMN (Q), MS (R), and CA3 (S) from an HDC-jRGECO1αTMN-HA1mMS, CeA, CA3 mouse. (Scale bars: 200 μm.) (T) Average signal peak ΔF/F of MS and CA3. (U) Fiber photometry of TMN fluorescent signal during aversive response. (V and W) Fiber photometry of MS (V) and CA3 (W) histamine concentration by HA1m sensor during aversive response in tested mice. The signals of 6 mice are illustrated in the heatmap; color scale indicates ΔF/F and warmer colors indicate higher fluorescence signal. Aversive response: suddenly spray a stream of water on the back of the freely moving mouse. TMN, tuberomammillary nucleus; MS, medial septal nucleus; CeA, central amygdalar nucleus; CA3, field CA3.
While activating histaminergic neurons, we detected a significant increase in signal response in the MS (average signal peak ΔF/F: 7.95 ± 0.53%, Fig. 3L) but only a moderate increase in the ipsilateral CeA (average signal peak ΔF/F: 2.72 ± 0.35%, Fig. 3M), and almost no change—even a slight decrease—in the ipsilateral CA3 (average signal peak ΔF/F: −0.37 ± 0.46%, Fig. 3N). When we injected the GRABHA control virus HAmutant sensor, there was no signal response under 589-nm stimulation (Fig. 3 G and K). These results demonstrated that the higher the density of histaminergic axonal fibers, the more histamine is released in the downstream brain region when the histaminergic neurons are activated, highlighting a correlation between histaminergic fiber density and histamine secretion in the histaminergic downstream nuclei out of the TMN.
We further explored the relationship between the histaminergic fiber density and histamine concentration in different brain regions during physiological behavior. It is well-known that histaminergic signaling is closely related to the aversive response of rodents (38–41). We chose the MS and CA3, which represent the Dense and Few groups, respectively, for the experiment. We injected the rAAV-EF1α-DIO-jRGECO1α (42) virus into the right TMN nucleus and the AAV-hSyn-HA1m virus into the MS and right CA3 nucleus of adult HDC-CreERT2 mice (Fig. 3 O and Q–S) to record histaminergic fluorescent signal of TMN and HA1m signal of the MS and CA3. We then performed a spray of water (as an aversive stimulation) on the back of tested mouse and concurrently recorded fluorescent signal changes of the TMN, MS, and CA3 (Fig. 3P). We detected a more significant signal increase in the MS (average signal peak ΔF/F: 2.26 ± 0.38%, Fig. 3 T and V) than that in the CA3 (average signal peak ΔF/F: 0.37 ± 0.71%, Fig. 3 T and W) while histaminergic neurons are activated by aversive stimulation (Fig. 3U). These results suggested that the release of histamine in different downstream areas has a potential relationship with histaminergic fiber density in the physiological functions related to the histaminergic system.
Whole-Brain 3D Reconstruction of Single Histaminergic Neuron by Sparse Labeling.
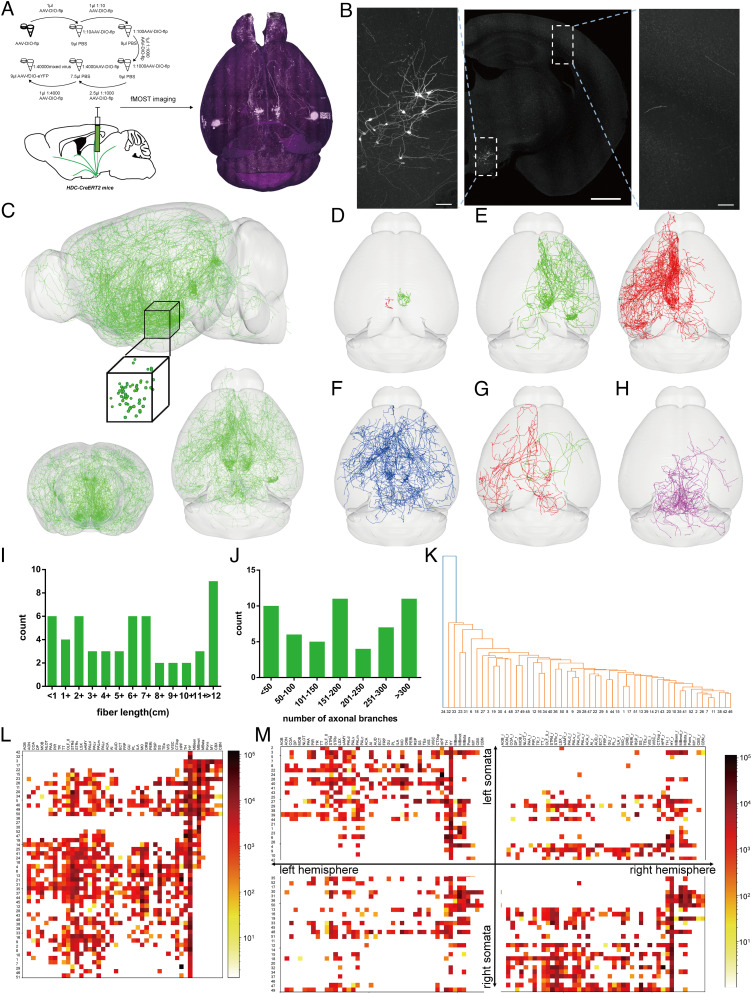
Previous studies have made different conjectures about the projection pattern of individual histaminergic neurons (26, 28, 43). To further uncover the projection pattern of individual histaminergic neurons, we performed fMOST reconstruction of sparse-labeled histaminergic neurons. rAAV-EF1α-DIO-flp-WPRE-hGH (diluted in PBS at 1:40,000) and rAAV-nEF1α-fDIO-eYFP-eYFP-WPRE-hGH were infused into bilateral TMN of HDC-CreERT2 mice (Fig. 4A). Following infusion, eYFP was efficiently expressed in HDC+ neurons (91.9 ± 1.7%, n = 2; SI Appendix, Fig. S6 B–D), and only a small number of HDC+ neurons were labeled by eYFP (7.08 ± 0.42%; SI Appendix, Fig. S6 B–D). To acquire fine morphological information on eYFP-labeled histaminergic axons, we then reconstructed the whole-brain GFP dataset of every single HDC-AAV-DIO-flp+AAV-EF1α-fDIO-eYFPTMN mouse at a pixel resolution of 0.32 × 0.32 × 1 μm3 through the fMOST system, and clearly identifiable histaminergic nerve fibers can be found in both TMN and other brain regions (Fig. 4 A and B). A total of 60 well-separated histaminergic neurons were randomly selected from the bilateral TMN of three tested mice, and the reconstructed 3D data of somata and axons were shown after arborization tracing and landmark registration (Fig. 4C). Out of the 60 neurons, 52 have long projection structures. Their axon length and branch number analysis were shown, which reflects the structure complexity of histaminergic neurons and the common differences between them (Fig. 4 I and J). Besides, a small number of neurons only has very limited fibers located in the local TMN (8 out of 60, Fig. 4D).
Fig. 4.
Whole-brain reconstruction of sparse-labeled single histaminergic neuron. (A) Main steps of data generation and acquisition in fMOST imaging of sparse-labeled single histaminergic neuron. (B) Representative images of the fluorescence-labeled neurons and fibers from an HDC-DIO-flp+fDIO-eYFPTMN mouse. (C) 3D views of 60 fully reconstructed histaminergic neurons from HDC-AAV-DIO-flp+AAV-EF1α-fDIO-eYFPTMN mice (Top: sagittal view with the cube representing the somatic location; Bottom: coronal and horizontal views). (Scale bar: 1 mm for Top, 100 μm for Bottom.) (D) Horizontal view of histaminergic neurons only projected to the local part of TMN. (E) Horizontal view of histaminergic neurons mainly projected to the anterior right and left hemisphere. (F) Horizontal view of histaminergic neurons mainly projected to both hemispheres. (G) Horizontal view of histaminergic neurons mainly projected to the contralateral hemisphere. (H) Horizontal view of histaminergic neurons mainly projected to the posterior part of the brain. (I and J) Interval distribution of all reconstructed histaminergic neuron fiber lengths (I) and axonal branch number (I). (K) Cluster analysis diagram of long-projecting neurons based on the detailed fiber distribution information. Every single branch indicates a different subgroup. (L and M) Heatmap shows the fiber length of long-projecting neurons in all major brain regions (L) and each hemisphere (M). Warmer colors indicate higher fiber length value.
We tried to classify them into different categories according to differences of axonal distribution in all major brain regions. However, the classification cannot be realized due to the large dispersion of distribution characteristics with different neurons (Fig. 4K). Here, we showed the axonal fiber density of 52 histaminergic neurons in major bilateral brain regions (Fig. 4 L and M). As we can see, most selected neurons mainly project to the ipsilateral hemisphere (38 out of 52, Fig. 4E), some of them also project to the contralateral hemisphere (13 out of 52, Fig. 4F), and only a few mainly project to the contralateral hemisphere (6 out of 52, Fig. 4G), which reflects the ipsilateral projection characteristics of histaminergic neurons. Moreover, there are several selected neurons that mainly project to the posterior part of the brain but hardly pass through the anterior brain areas (7 out of 52, Fig. 4H), which indicates the potential difference between the forward- and backward-projecting histaminergic neurons. Therefore, there are obvious differences in the fiber structure of histaminergic neurons, and we can roughly group them from the main projection direction. However, due to the complexity of their structural characteristics, we cannot complete the typical cluster analysis, and other ways to understand them are needed.
Whole-Brain Collateral Projection Pattern of Histaminergic Neurons.
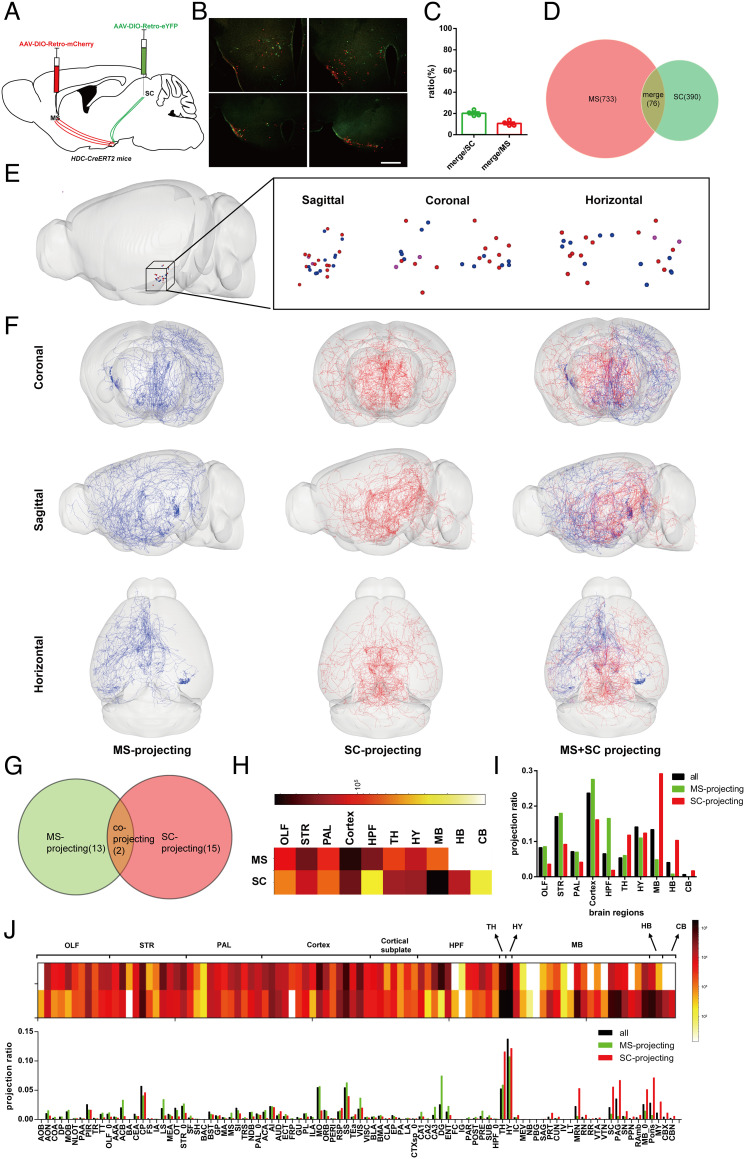
Firstly, we used retrograde tracing viruses to explore the distribution of histaminergic neurons that projected to different downstream brain regions. Previous studies had speculated on the characteristics of the projection pathway of histaminergic nerve fibers, especially the different pathways of rostral and caudal sides (19, 20, 24), but there was no direct evidence to show the difference of their projections. To verify this speculation, we intend to select two brain regions that meet the following two requirements: 1) They are both located on the midline of the coronal plane. 2) They are located in the anterior and posterior sides of TMN, respectively. After careful investigation and consideration, we selected two brain regions the MS and superior colliculus (SC) where histamine is considered to mediate different functions. Histamine signaling in the MS is considered to be related to feeding (44), emotion (39), learning, and memory (45), while in the SC, it is considered to be related to fear reaction of escape behavior (46) and visual function (47). We then injected AAV-DIO-Retro-mCherry into the MS and AAV-DIO-Retro-eYFP into the SC of HDC-CreERT2 mice at the same time (Fig. 5A). After retrograde tracing, there were mCherry and eYFP-labeled neurons in the TMN (Fig. 5B). We found that both of them were distributed in anterior and posterior parts of the TMN, but only a small part of them was colabeled (20.16 ± 1.31% of eYFP-labeled neurons, 10.66 ± 1.22% of mCherry-labeled neurons, n = 4; Fig. 5 C and D), which reveals the projection heterogeneity of different histaminergic neurons. To verify whether such differences exist only between the anterior and posterior downstream brain regions, we then explored the distribution of histaminergic neurons project to nuclei both located in the anterior part of the brain: the MS and agranular insular area, posterior part (AIp). AAV-DIO-Retro-mCherry was injected into the MS, and AAV-DIO-Retro-eYFP was injected into the bilateral AIp of HDC-CreERT2 mice (SI Appendix, Fig. S5A). After 4 wk, there were mCherry- and eYFP-labeled neurons in anterior and posterior parts of the TMN (SI Appendix, Fig. S5B), and some parts of them were colabeled (36.04 ± 3.38% of eYFP-labeled neurons, 21.14 ± 4.04% of mCherry-labeled neurons, n = 4; SI Appendix, Fig. S5 C and D), which reveals the general projection heterogeneity of different histaminergic neurons.
Fig. 5.
Whole-brain 3D structure for collateral projection analysis of MS/SC-projecting neurons. (A) Experimental scheme for generating HDC-Retro-mCherryMS-Retro-GFPSC mice. AAV-DIO-Retro-mCherry was injected into the MS of HDC-CreERT2 mice, and AAV-DIO-Retro-GFP was injected into the SC simultaneously. (B) Representative coronal sections showing Retro virus-labeled histaminergic somata in an HDC-Retro-mCherryMS-Retro-GFPSC mouse with several distance from bregma. (Scale bar: 400 μm.) (C) Co-label ratio of GFP and mCherry-labeled somata of 4 HDC-Retro-mCherryMS-Retro-GFPSC mice. (D) Venn diagram shows the number of all GFP and mCherry-labeled somata of 4 HDC-Retro-mCherryMS-Retro-GFPSC mice. (E) A 3D view of 26 MS- or SC-projecting histaminergic neuron somata (Left: sagittal view of the somatic location in the whole brain; Right: sagittal, coronal, and horizontal views of the somatic location). (F) A 3D visualization of whole-brain fiber distribution of MS- or SC-projecting neurons from different views. (G) Venn diagram shows the overlap of MS- and SC-projecting neurons. (H) Fiber length of MS- or SC-projecting neurons in the major brain areas are illustrated in the heatmap, and warmer colors indicate higher fiber length value. (I) Fiber length ratio (relevant brain areas/whole brain) of different groups in the major brain areas (black column: fiber length ratio of all 52 long projection neurons; green column: fiber length ratio of MS-projecting neurons; red column: fiber length ratio of SC-projecting neurons). (J, Top) Fiber length of MS- or SC-projecting neurons in the whole brain are illustrated in the heatmap, and warmer colors indicate higher fiber length value (top row: MS-projecting; bottom row: SC-projecting). (J, Bottom) Fiber length ratio (relevant brain areas/whole brain) of different groups in the whole brain (black column: fiber length ratio of all 52 long-projection neurons; green column: fiber length ratio of MS-projecting neurons; red column: fiber length ratio of SC-projecting neurons).
Next, we tested whether there was a significant difference between the somata position and whole-brain fiber distribution of MS and SC-projecting neurons. First, the 3D position of MS and SC-projecting neuron somata were shown in the Allen Brain Atlas (Fig. 5E), and there was no specific location preference of any group. We then reconstructed whole-brain 3D data of all MS-projecting neurons (13 out of 52 neurons) and SC-projecting neurons (15 out of 52) and found that MS-projecting neurons more likely project to the anterior part of the brain, while SC-projecting neurons mainly focus on the posterior areas (Fig. 5 F and G). Through major brain area fiber density analysis, we found that compared with all reconstructed neurons, MS-projecting neurons tend to project to the cortex and hippocampal formation, while SC-projecting neuron fibers are concentrated in the thalamus, midbrain, hindbrain, and cerebellum (Fig. 5 H and I). Based on a more detailed analysis, more information could be discovered. For example, in the cortex, MS-projecting neurons show more axonal innervation of the anterior cingulate area, the somatomotor areas (MO), and the visual areas (VIS) than SC-projecting neurons. In the hippocampal formation, almost all brain nuclei receive more axonal innervation from MS-projecting neurons, while the dentate gyrus (DG) receives extremely more than others do, which indicates a close relationship between DG and MS as histaminergic downstream areas. In addition, in the midbrain, a brain area dominated by SC projection neurons, the midbrain reticular nucleus and the periaqueductal gray showed significantly more fiber density ratio than other midbrain areas (Fig. 5J). Therefore, histaminergic neurons do have clusters that are not determined by the spatial position of their somata, but their projection characteristics are very complex and have similarities and differences with each other, which cannot be easily analyzed by the traditional clustering method.
Discussion
Whole-Brain 3D Mapping of Histaminergic Neurons and Outputs.
3D reconstruction data and counting approaches are essential in developing a comprehensive understanding of the histaminergic network (48). In the present study, we provide a comprehensive atlas of the histaminergic projections through genetic labeling, virus infusion, and fMOST reconstruction. For the whole-brain study of histaminergic neurons, we imaged all GFP+ neurons and counted them through a NeuroGPS system and manual correction. In the posterior hypothalamus, the only source of histaminergic neurons, ~93% GFP-labeled neurons coexpressed HDC and ~93% of the HDC+ neurons were labeled by GFP, demonstrating reliable specificity and penetrance in the crossed mice. The total number of GFP-labeled neurons was ~5,000, which is higher than previous studies using immunohistochemical staining and Axiophot microscope counting on wild-type mice brain sections (3,508 ± 703) (49). This discrepancy may be due to the more comprehensive histaminergic neuron counting achieved by the whole-brain 3D reconstruction data in concert with NeuroGPS analysis and manual correction. Due to rare GFP expression on crossed mice axons, we utilized virus infusion to elucidate the whole-brain histaminergic nerve fibers. The number of fluorescence-labeled histaminergic neurons differs between mice; however, we demonstrated herein that whole-brain histaminergic nerve fiber distribution trends across all mice were nearly identical (Fig. 2H and SI Appendix, Fig. S3D), meaning that the number of virus-labeled neurons is suitable for whole-brain histaminergic projection analysis. The overall histaminergic nerve fiber distribution characteristics of the whole mouse brain, including the densest area (hypothalamus) and the thinnest areas (hindbrain and cerebellum), roughly agree with previous rat brain histaminergic network studies (19, 20). However, through more precise analysis and brain region segmentation, we have elucidated more details and reached some conclusions which differ from previous studies.
First, due to a lack of quantitative information of fiber density, the distribution characteristics of the histaminergic network were previously shown only in density grades through comparisons with data from photographed immunofluorescence samples (20). Although thorough, careful, and objective work has been carried out, it is still not accurate and comprehensive enough (in 3D level) to become a guideline for current histaminergic research; in our study, the whole-brain distribution of histaminergic nerve fibers was instead quantified by objective, accurate fluorescence density values, which can better meet the requirements of future histamine research. Second, although brain structures were manually delineated with careful considerations in previous studies, problems such as discontinuous spatial results and lack of information in some subbrain regions still exist inevitably. To solve these challenges, we make alignment with the Allen Reference Atlas to standardize the spatial information of sample brains. This enabled us to obtain comprehensive quantifiable atlas of whole-brain histaminergic projections as well as some details not explored in previous studies. For example, the cerebral cortex is a large and complex part of the brain; however, in previous studies, the distribution characteristics of the histaminergic nerve fibers in the cortex have been presented as unified and homogeneous, with the cerebral cortex only divided into the neocortex, piriform cortex, and entorhinal cortex and no further distinction between different cortexes, let alone differences between various sublayers of the same cortex, explored. In our study, all regions and sublayers of the cerebral cortex were precisely segmented. We found that there were substantial differences in fiber density across different cortical regions. The differences in fiber density between the different sublayers of the same cortical nucleus were generally very small, which may indicate that the histaminergic neural network has no clear layer specificity in cortex regulation.
3D Mapping of Histaminergic Outputs: An Update from Traditional Concept.
Previous findings differ in fiber distribution in hippocampal formation. Some conclude that the fiber densities of the DG and Ammon’s horn are very low and that there is no difference between them (19), while others believe that the fiber density of the DG is slightly higher (20). We found that the DG molecular layer has a very low fiber density (0.01%) similar to Ammon’s horn (0.02%), while the fiber density of other DG parts is higher (0.05%). The fimbria of the hippocampus has been previously thought to have medium fiber density (19, 20), but our study found that it has only a low-grade fiber network (0.08%). Amygdalar nuclei are complex structures including several subregions where results differ in previous studies. Some researchers believe that both the basolateral amygdalar (BLA) and central amygdalar nuclei have medium-density fiber networks (19), while others think that their fiber distribution is very low (20). Our study found the fiber distribution of basolateral amygdalar to be low level (0.08%), while the CeA was medium level (0.33%). The caudoputamen is the largest brain region in the cerebral nuclei, believed to have a moderate fiber density in one study (19) and a low density in another (20); our results revealed a low-grade fiber network (0.07%). A previous study concluded that most areas of the thalamus had no HA-IR immunoreactivity (20), leading to a lack of fiber density information in the literature for many thalamic nuclei. One study did report the histaminergic nerve fiber density characteristics of various thalamic nuclei; although, many of their conclusions significantly differ from ours. In their findings, the three subareas of the anterior nucleus of the thalamus (ANT), the anterodorsal nucleus (AD), anteromedial nucleus (AM), and anteroventral nucleus of thalamus (AV), were thought to have moderate to dense histaminergic fibers, with no significant differences between them (19). However, we found that AD had substantially more dense fiber distribution than the AM or AV (AD: 1.21%, AM: 0.44%, and AV: 0.43%), which may indicate a specific role for the histaminergic network in regulating the different subregions of the thalamic nuclei. Additionally, the lateral dorsal nucleus of thalamus (LD) and mediodorsal nucleus of thalamus (MD) have been previously considered to have medium to dense fiber distributions (19), an assertion which might be overestimated according to our findings (LD: 0.07%, MD: 0.15%). Conversely, the zona incerta has been generally considered to have a very low density of histaminergic nerve fibers (19), which the present study highlights as a clear underestimation (0.21%). In previous studies, the substantia nigra (SN) in the midbrain has typically not been finely subdivided; further, its fiber density grade has been inconsistently reported, varying between very low (19) or medium grade (20). We determined that the compact part of the substantia nigra (SNc) showed significantly higher histaminergic fiber density than the reticular part of substantia nigra (SNr), which may be indicative of the characteristics of histaminergic neural circuits in motor regulation (SNc: 0.36%, SNr: 0.16%). The dorsal nucleus raphe (DR) is a nucleus-rich region in serotonergic neurons in the midbrain and was believed previously to have almost no histaminergic nerve fiber distribution (19, 20). Our study confirmed that the DR has moderate fiber density (0.38%), which could provide a structural basis for future comprehensive studies of monoamine nervous system interactions. The histaminergic nerve fiber distribution differences in mice brains between the present and previous studies are summarized in SI Appendix, Table S4.
Functional Relevance Correlated with Histaminergic Fiber Density.
It is well-known that histaminergic nerve fibers rarely contact downstream cells through synapses and that fibers instead produce histamine through volume transmission, regulating downstream cells via histamine receptor binding (17). Although the whole-brain histaminergic network has been studied repeatedly, no direct evidence has previously been put forward for a relationship between the density of downstream histaminergic nerve fibers and the amount of histamine secreted. In this study, we performed functional testing through optogenetics, fiber photometry recording, and the specific histamine probe GRABHA in selected histaminergic downstream areas. In the brain nuclei we tested, histamine secretion was positively correlated with fiber density under both optogenetic activation and physiological stimulation, indicating that the whole-brain histaminergic map has significant functional relevance. Interestingly, after GRABHA injection, the CA3—the brain region with the lowest density of histaminergic nerve fibers—exhibited significantly higher basal fluorescence values than other brain regions, which could indicate that the neurons in this region have particularly good binding efficiency with the probe virus or a particularly high basal histamine concentration. However, the histaminergic concentration of CA3 remained essentially the same when histaminergic neurons were optogenetically activated, suggesting that the density of histaminergic nerve fibers in other brain regions is indeed related to histaminergic neuron regulation. On the other hand, the HA1m fluorescent signal of CA3 shows an unstable change in physiological aversive response. Considering the stable trend of fiber photometry change in histaminergic neurons, this may indicate that the change of histamine concentration in the CA3 under physiological aversive responses is affected by other unknown pathways.
Brain histamine is believed to have an important role in regulating numerous functions including sleep–wake rhythm, sensory and motor function, cognition, learning and memory, and feeding; meanwhile, its role in neuropsychiatric disorders including sleep disorders, cognitive disorders, and other diseases has also been carefully summarized by many review articles (16, 17, 34, 50). Neuronal histamine is an important source of histamine in the central nervous system (CNS). In our work, we carefully analyzed the quantitative distribution of histaminergic nerve fibers in the whole mouse brain. And combined with previous works of brain histamine function, it may provide reasonable directions for the functional study of histamine in different regions of the brain. According to previous studies, histamine mediates its effects on wakefulness or sleep disorders at least by regulating basal forebrain cholinergic system (51, 52), VLPO (53, 54), glutamatergic neurons of the thalamus (55, 56), mesopontine tegmentum (57), and serotonergic neurons in the dorsal raphe nucleus (58). Almost all of them have at least moderate histaminergic fiber density, suggesting that they are suitable for more functional research on neural circuits. In particular, VLPO, one of the most important brain nuclei that control the activity of histaminergic neurons (59, 60), is very worthy of further research on potential closed-loop projection mechanisms. Brain histamine seems to promote motor functions overall (34), but there are still some studies that have come to different conclusions (61–63), which may be due to the different roles histamine plays in motor function in different brain regions, including globus pallidus (64), medial septal nucleus (39), nucleus accumbens (65), and substantia nigra (66). All of them have at least moderate histaminergic fiber density. It is well-known that brain histamine has an important role in cognition, learning, and memory, as well as Alzheimer's disease (16, 50). Histamine and histamine receptors in different brain regions have been extensively studied for their involvement in cognitive and memory-related processes, mainly including hippocampal CA1 (67–69) and CA3 (70, 71), BLA (72, 73), vermis (74), and nucleus basalis magnocellularis (75, 76). Interestingly, most of these brain regions only have very low histaminergic fiber density, which may indicate that these brain regions may be regulated by present unknown indirect pathways or nonneuronal histamine. The close relationship between brain histamine and feeding behavior was well-summarized recently (77). Brain histamine may regulate the feeding behavior through acting on the lateral hypothalamus (LH) (78), BST (79), PVT (80), and MS (44), which all have dense histaminergic fiber distribution. There are few studies that directly analyze the functional histaminergic circuits (44, 81, 82), but we believe that the future research on the histaminergic circuit will greatly benefit from this work.
Heterogeneous Morphology in Individual Histaminergic Neurons.
The fine structure of individual histaminergic neurons has always been a mystery in histaminergic research (26, 28, 43). Here, we performed whole-brain tracing and reconstruction experiments of 60 sparse-labeled histaminergic neurons after precise imaging. The general structural differences between different histaminergic neurons are reflected by the total fiber length, the number of axonal branches, and the fiber length distribution in different brain regions (Fig. 4 I, J, L, and M). The results of cluster analysis for long projection histaminergic neurons according to projection patterns (Fig. 4K) indicated a special clustering mode of them, neither a whole center nor clearly separated subgroups. To confirm the manifestation of this pattern, we manually selected seven neurons with very close fiber paths at the beginning of departure from the TMN and found that they projected to different downstream brain regions after a certain distance (SI Appendix, Fig. S7 A and B). We also conducted a retrospective study on the upstream histaminergic neurons of the MS and SC and found that there were significant differences in their upstream according to the reconstructed data. Moreover, there were many differences in the main collateral projection dominant areas between them, while their somata distribution had no obvious spatial location preference. The clustering of the histaminergic neural network may be understood in a way that is not so strict and focuses on a part of important regulation targets. Such clustering exists objectively, but it cannot be listed one-by-one in parallel. When taking different downstream brain regions as the comparison object, comparative analysis can be carried out, so as to provide a theoretical basis for analyzing the histaminergic circuits responsible for different functions.
Taken together, the whole-brain histaminergic projection atlas established in this study has the potential to be a framework for future functional brain histaminergic studies; this atlas contains information not only regarding the somata but also the distribution characteristics of the histaminergic neurons’ projection fibers. Our study will facilitate understanding and parsing the functional role of histaminergic nerve fibers in different mouse brain areas and may eventually shed light on histamine’s role in CNS disorders.
Materials and Methods
Animals.
For the histaminergic neuron reconstruction experiments HDC-CreERT2:Ai47 double-positive adult mice were used. HDC-CreERT2:Ai47 mice were generated by crossing HDC-CreERT2 mice (2A-CreERT2 expression box insertion upstream of the termination codon tag HDC via CRISPR/Cas9 technology; generated by Shanghai Model Organisms, Shanghai, China), with Ai47 reporter mice (presented by Zilong Qiu, Chinese Academy of Sciences, Shanghai, China). In the histaminergic fiber reconstruction experiments and functional testing, HDC-CreERT2 adult mice were used. Mice were housed in the vivarium with a 12-h light/dark cycle: 22 ± 1 °C and 55 ± 5% humidity with food and water given ad libitum. Animal experiments were performed in accordance with the guidelines of the Animal Advisory Committee of Zhejiang University and the US NIH Guidelines for the Care and Use of Laboratory Animals. All procedures were approved by the Animal Advisory Committee of Zhejiang University.
Viruses.
Details are given in SI Appendix.
Stereotactic Injections.
Details are given in SI Appendix.
Stereotactic Surgery.
Details are given in SI Appendix.
Functional Test.
The fiber photometry system (Nanjing Thinkertech) used a 488-nm diode laser (OBIS 488LS; Coherent), reflected by a dichroic mirror (MD498, Thorlabs) and coupled into a 0.23-mm 0.37 NA optical fiber using a 10× objective lens (Olympus) and fiber launch (Thorlabs). The laser intensity at the interface between the fiber tip and the animal ranged from 0.01 to 0.03 mW, to minimize bleaching. The HA1m fluorescence was bandpass filtered (MF525-39, Thorlabs) and collected by a photomultiplier tube (R3896, Hamamatsu). An amplifier (C7319, Hamamatsu) was used to convert the photomultiplier tube current output to voltage signals, which were then further filtered through a low-pass filter (40 Hz cut-off). The analog voltage signals were digitized at 500 Hz and recorded. To facilitate environmental adaptation before testing, post virus injection and surgery mice were carefully placed inside a clean square plastic frame and allowed to move freely for 30 min. For recording of histamine signal after histaminergic neuron activation, optic fibers link the mouse to the fiber photometry system and a 589-nm laser pulse generator. During the test, the HA1m fluorescence of the MS, right CeA, or right CA3 was recorded for 60 s without laser stimulation; subsequently, a 594-nm laser (10 Hz, 10 ms pulse, 60 s) was applied to the right TMN of the mice and the HA1m fluorescence was recorded. For recording of histamine signal after water spray stimulation, optic fibers link the mouse to the fiber photometry system. During the test, the HA1m fluorescence of the MS or right CA3 was recorded for 60 s with freely moving mice; subsequently, a spray of water was sprayed on the back of the mice, and the HA1m fluorescence was recorded. Photometry data were exported as MATLAB Mat files for further analysis. We segmented the data based on individual laser stimulation trials and derived the values of fluorescence change (ΔF/F) by calculating (F−F0)/F0. ΔF/F values are presented via heatmaps or average plots.
fMOST Tissue Preparation.
All histological procedures were performed as has been described in a previous study (83). Details are given in SI Appendix.
Whole-Brain Imaging.
Details are given in SI Appendix.
Image Preprocessing.
Details are given in SI Appendix.
Visualization and Reconstruction.
Details are given in SI Appendix.
Cell Counting and Registration.
The methods of cell counting and registration have been described elsewhere (84, 85). Briefly, for GFP-labeled neuron counting, the GFP-labeled dataset of the posterior hypothalamus was sampled at 1 × 1 × 2 μm3 using the Cell Counter ImageJ plug-in and then automatically identified by NeuroGPS which saved the data in an SWC format. These data were then manually corrected in Amira. We used the BrainsMapi for brain structure registration; the dataset of reconstructed brain was rotated and resampled at a resolution of 10 × 10 × 10 μm3 for registering in the template dataset of Allen CCF v3.0 (86). Then, we manually segmented several brain regions as landmarks by Amira software. Finally, we registered these results by ANTs tools (87) and loaded the outline of the mouse brain and the results into Amira simultaneously to generate figures.
Single Neural Structure Tracing.
We used GTree software (88) with TDat plugin (Thermo Fisher Amira) (89) through human–computer interaction to trace and reconstruct the single neural structure. The reconstruction of each neuron usually starts from the cell body; we identified a soma as the seed point and then navigated through the image data blocks to connect the signal node to the seed point until all the branches are traversed. To improve the tracing efficiency, we chose semiautomatic tracing at sparsely distributed long-distance fibers. Briefly, automatic reconstruction calculation is performed first, and then, a skilled annotator manually checks the results of the automatic calculation. If potential errors (when fluorescent signal is too dense or too weak to be automatically traced) are found, manual correction is carried out immediately. To minimize subjective errors, when an annotator finds it difficult to judge, it is recorded as a special event. Then three skilled annotators work independently on the reconstruction of each neuron. Finally, a neuroanatomy expert is responsible for checking all special events and acceptance of the results. All reconstructed results were saved as SWC files. MATLAB (2017a, MathWorks) was used to calculate the morphological parameters of the reconstructed neurons.
Immunohistochemistry.
Details are given in SI Appendix.
Statistics.
Details are given in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
The location of histaminergic neuron somata from anterior to posterior TMN in one of the tested mice.
The 3D fluorescent raw data of whole-brain histaminergic neural circuits from one of the tested mice.
Acknowledgments
This project was supported by grants from The National Key R&D program of China (2021ZD0202803) and the National Natural Science Foundation of China (U21A20418, 82022071 and 81872844).
Author contributions
Y.W. and Z.C. designed research; W.L., L.X., and Y.Z. performed research; M.L., H.D., Y.L., and Q.L. contributed new reagents/analytic tools; W.L., S.A., M.Z., A.L., H.G., and G.P. analyzed data; W.H., H.G., and Q.L. contributed to discussion; and W.L., W.H., Y.W., and Z.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Yoshikawa T., Nakamura T., Yanai K., Histaminergic neurons in the tuberomammillary nucleus as a control centre for wakefulness. Br. J. Pharmacol. 178, 750–769 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K., Lin J. S., Sakai K., Neuronal activity of histaminergic tuberomammillary neurons during wake-sleep states in the mouse. J. Neurosci. 26, 10292–10298 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dietrich M. O., Horvath T. L., Feeding signals and brain circuitry. Eur. J. Neurosci. 30, 1688–1696 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Zendehdel M., Baghbanzadeh A., Aghelkohan P., Hassanpour S., Central histaminergic system interplay with suppressive effects of immune challenge on food intake in chicken. Br. Poult. Sci. 57, 271–279 (2016). [DOI] [PubMed] [Google Scholar]
- 5.de Almeida M. A., Izquierdo I., Memory facilitation by histamine. Arch. Int. Pharmacodyn. Ther. 283, 193–198 (1986). [PubMed] [Google Scholar]
- 6.Onodera K., Yamatodani A., Watanabe T., Wada H., Neuropharmacology of the histaminergic neuron system in the brain and its relationship with behavioral disorders. Prog. Neurobiol. 42, 685–702 (1994). [DOI] [PubMed] [Google Scholar]
- 7.Inoue I., et al. , Impaired locomotor activity and exploratory behavior in mice lacking histamine H1 receptors. Proc. Natl. Acad. Sci. U.S.A. 93, 13316–13320 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li B., et al. , Histamine increases neuronal excitability and sensitivity of the lateral vestibular nucleus and promotes motor behaviors via HCN channel coupled to H2 receptor. Front. Cell. Neurosci. 10, 300 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishino S., et al. , Decreased CSF histamine in narcolepsy with and without low CSF hypocretin-1 in comparison to healthy controls. Sleep 32, 175–180 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franco P., et al. , Impaired histaminergic neurotransmission in children with narcolepsy type 1. CNS Neurosci. Ther. 25, 386–395 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bardgett M. E., Davis N. N., Schultheis P. J., Griffith M. S., Ciproxifan, an H3 receptor antagonist, alleviates hyperactivity and cognitive deficits in the APP Tg2576 mouse model of Alzheimer’s disease. Neurobiol. Learn. Mem. 95, 64–72 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zlomuzica A., et al. , Neuronal histamine and cognitive symptoms in Alzheimer’s disease. Neuropharmacology 106, 135–145 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Rinne J. O., et al. , Increased brain histamine levels in Parkinson’s disease but not in multiple system atrophy. J. Neurochem. 81, 954–960 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Tuomisto L., Kilpelainen H., Riekkinen P., Histamine and histamine-N-methyltransferase in the CSF of patients with multiple sclerosis. Agents Actions 13, 255–257 (1983). [DOI] [PubMed] [Google Scholar]
- 15.Prell G. D., et al. , Histamine metabolites in cerebrospinal fluid of patients with chronic schizophrenia: Their relationships to levels of other aminergic transmitters and ratings of symptoms. Schizophr. Res. 14, 93–104 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Shan L., Bao A. M., Swaab D. F., The human histaminergic system in neuropsychiatric disorders. Trends Neurosci. 38, 167–177 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Panula P., Nuutinen S., The histaminergic network in the brain: Basic organization and role in disease. Nat. Rev. Neurosci. 14, 472–487 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Takagi H., et al. , Histaminergic axons in the neostriatum and cerebral cortex of the rat: A correlated light and electron microscopic immunocytochemical study using histidine decarboxylase as a marker. Brain Res. 364, 114–123 (1986). [DOI] [PubMed] [Google Scholar]
- 19.Inagaki N., et al. , Organization of histaminergic fibers in the rat brain. J. Comp. Neurol. 273, 283–300 (1988). [DOI] [PubMed] [Google Scholar]
- 20.Panula P., Pirvola U., Auvinen S., Airaksinen M. S., Histamine-immunoreactive nerve fibers in the rat brain. Neuroscience 28, 585–610 (1989). [DOI] [PubMed] [Google Scholar]
- 21.Vanhala A., Yamatodani A., Panula P., Distribution of histamine-, 5-hydroxytryptamine-, and tyrosine hydroxylase-immunoreactive neurons and nerve fibers in developing rat brain. J. Comp. Neurol. 347, 101–114 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Watanabe T., et al. , Distribution of the histaminergic neuron system in the central nervous system of rats; a fluorescent immunohistochemical analysis with histidine decarboxylase as a marker. Brain Res. 295, 13–25 (1984). [DOI] [PubMed] [Google Scholar]
- 23.Ericson H., Blomqvist A., Kohler C., Origin of neuronal inputs to the region of the tuberomammillary nucleus of the rat brain. J. Comp. Neurol. 311, 45–64 (1991). [DOI] [PubMed] [Google Scholar]
- 24.Köhler C., Swanson L. W., Haglund L., Wu J. Y., The cytoarchitecture, histochemistry and projections of the tuberomammillary nucleus in the rat. Neuroscience 16, 85–110 (1985). [DOI] [PubMed] [Google Scholar]
- 25.Provensi G., Passani M. B., Blandina P., Different peas in the same pod: The histaminergic neuronal heterogeneity. Curr. Top. Behav. Neurosci. 59, 303–327 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Miklos I. H., Kovacs K. J., Functional heterogeneity of the responses of histaminergic neuron subpopulations to various stress challenges. Eur. J. Neurosci. 18, 3069–3079 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Umehara H., Mizuguchi H., Fukui H., Identification of a histaminergic circuit in the caudal hypothalamus: An evidence for functional heterogeneity of histaminergic neurons. Neurochem. Int. 61, 942–947 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Blandina P., Munari L., Provensi G., Passani M. B., Histamine neurons in the tuberomamillary nucleus: A whole center or distinct subpopulations? Front. Syst. Neurosci. 6, 33 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munari L., Provensi G., Passani M. B., Blandina P., Selective brain region activation by histamine H receptor antagonist/inverse agonist ABT-239 enhances acetylcholine and histamine release and increases c-Fos expression. Neuropharmacology 70, 131–140 (2013). [DOI] [PubMed] [Google Scholar]
- 30.De Luca R., et al. , Mechanisms of N-oleoyldopamine activation of central histaminergic neurons. Neuropharmacology 143, 327–338 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Li A., et al. , Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain. Science 330, 1404–1408 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Li X., et al. , Generation of a whole-brain atlas for the cholinergic system and mesoscopic projectome analysis of basal forebrain cholinergic neurons. Proc. Natl. Acad. Sci. U.S.A. 115, 415–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panula P., Yang H. Y., Costa E., Histamine-containing neurons in the rat hypothalamus. Proc. Natl. Acad. Sci. U.S.A. 81, 2572–2576 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haas H., Panula P., The role of histamine and the tuberomamillary nucleus in the nervous system. Nat. Rev. Neurosci. 4, 121–130 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Michelsen K. A., et al. , Histamine-immunoreactive neurons in the mouse and rat suprachiasmatic nucleus. Eur. J. Neurosci. 22, 1997–2004 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Wang Q., et al. , The Allen Mouse Brain common coordinate framework: A 3D reference atlas. Cell 181, 936–953.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klapoetke N. C., et al. , Independent optical excitation of distinct neural populations. Nat. Methods 11, 338–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santos N. R., Huston J. P., Brandão M. L., Blockade of histamine H2 receptors of the periaqueductal gray and inferior colliculus induces fear-like behaviors. Pharmacol. Biochem. Behav. 75, 25–33 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Zarrindast M.-R., Moghimi M., Rostami P., Rezayof A., Histaminergic receptors of medial septum and conditioned place preference: D1 dopamine receptor mechanism. Brain Res. 1109, 108–116 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Alvarez E. O., Banzán A. M., Histamine in dorsal and ventral hippocampus. II. Effects of H1 and H2 histamine antagonists on exploratory behavior in male rats. Physiol. Behav. 37, 39–45 (1986). [DOI] [PubMed] [Google Scholar]
- 41.Ruarte M. B., Orofino A. G., Alvarez E. O., Hippocampal histamine receptors and conflictive exploration in the rat: Studies using the elevated asymmetric plus-maze. Braz. J. Med. Biol. Res. 30, 1451–1461 (1997). [DOI] [PubMed] [Google Scholar]
- 42.Dana H., et al. , Sensitive red protein calcium indicators for imaging neural activity. Elife 5, e12727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ericson H., Watanabe T., Köhler C., Morphological analysis of the tuberomammillary nucleus in the rat brain: Delineation of subgroups with antibody against L-histidine decarboxylase as a marker. J. Comp. Neurol. 263, 1–24 (1987). [DOI] [PubMed] [Google Scholar]
- 44.Xu L., et al. , An H2R-dependent medial septum histaminergic circuit mediates feeding behavior. Curr. Biol. 32, 1937–1948.e5 (2022). [DOI] [PubMed] [Google Scholar]
- 45.Flood J. F., Uezu K., Morley J. E., Effect of histamine H2 and H3 receptor modulation in the septum on post-training memory processing. Psychopharmacology (Berl) 140, 279–284 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Santos N. R., Huston J. P., Brandão M. L., Escape behavior under tonic inhibitory control of histamine H(2)-receptor mediated mechanisms in the midbrain tectum. Behav. Brain Res. 124, 167–175 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Nakagawa Y., et al. , Marked increase in [3H](R) alpha-methylhistamine binding in the superior colliculus of visually deprived rats after unilateral enucleation. Brain Res. 643, 74–80 (1994). [DOI] [PubMed] [Google Scholar]
- 48.Benes F. M., Lange N., Two-dimensional versus three-dimensional cell counting: A practical perspective. Trends Neurosci. 24, 11–17 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Valko P. O., et al. , Increase of histaminergic tuberomammillary neurons in narcolepsy. Ann. Neurol. 74, 794–804 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Hu W., Chen Z., The roles of histamine and its receptor ligands in central nervous system disorders: An update. Pharmacol. Ther. 175, 116–132 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Vu M. T., Du G., Bayliss D. A., Horner R. L., TASK channels on basal forebrain cholinergic neurons modulate electrocortical signatures of arousal by histamine. J. Neurosci. 35, 13555–13567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cecchi M., Passani M. B., Bacciottini L., Mannaioni P. F., Blandina P., Cortical acetylcholine release elicited by stimulation of histamine H1 receptors in the nucleus basalis magnocellularis: A dual-probe microdialysis study in the freely moving rat. Eur. J. Neurosci. 13, 68–78 (2001). [PubMed] [Google Scholar]
- 53.Williams R. H., et al. , Optogenetic-mediated release of histamine reveals distal and autoregulatory mechanisms for controlling arousal. J. Neurosci. 34, 6023–6029 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y.-W., Li J., Ye J.-H., Histamine regulates activities of neurons in the ventrolateral preoptic nucleus. J. Physiol. 588, 4103–4116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Groenewegen H. J., Berendse H. W., The specificity of the “nonspecific midline and intralaminar thalamic nuclei”. Trends Neurosci. 17, 52–57 (1994). [DOI] [PubMed] [Google Scholar]
- 56.Matsumoto N., Minamimoto T., Graybiel A. M., Kimura M., Neurons in the thalamic CM-Pf complex supply striatal neurons with information about behaviorally significant sensory events. J. Neurophysiol. 85, 960–976 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Lin J. S., Hou Y., Sakai K., Jouvet M., Histaminergic descending inputs to the mesopontine tegmentum and their role in the control of cortical activation and wakefulness in the cat. J. Neurosci. 16, 1523–1537 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brown R. E., Sergeeva O. A., Eriksson K. S., Haas H. L., Convergent excitation of dorsal raphe serotonin neurons by multiple arousal systems (orexin/hypocretin, histamine and noradrenaline). J. Neurosci. 22, 8850–8859 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sherin J. E., Elmquist J. K., Torrealba F., Saper C. B., Innervation of histaminergic tuberomammillary neurons by GABAergic and galaninergic neurons in the ventrolateral preoptic nucleus of the rat. J. Neurosci. 18, 4705–4721 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng J., et al. , The interaction between the ventrolateral preoptic nucleus and the tuberomammillary nucleus in regulating the sleep-wakefulness cycle. Front. Neurosci. 14, 615854 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clapham J., Kilpatrick G. J., Thioperamide, the selective histamine H3 receptor antagonist, attenuates stimulant-induced locomotor activity in the mouse. Eur. J. Pharmacol. 259, 107–114 (1994). [DOI] [PubMed] [Google Scholar]
- 62.Akhtar M., Uma Devi P., Ali A., Pillai K. K., Vohora D., Antipsychotic-like profile of thioperamide, a selective H3-receptor antagonist in mice. Fundam. Clin. Pharmacol. 20, 373–378 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Hudzik T. J., et al. , Assessment of the abuse liability of ABT-288, a novel histamine H receptor antagonist. Psychopharmacology (Berl) 228, 187–197 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Ionov I. D., et al. , Activation of pallidal H receptors induces catalepsy in Wistar rats: A regulatory role of CRF receptors. Behav. Brain Res. 428, 113881 (2022). [DOI] [PubMed] [Google Scholar]
- 65.Bristow L. J., Bennett G. W., Biphasic effects of intra-accumbens histamine administration on spontaneous motor activity in the rat; a role for central histamine receptors. British J. Pharmacol. 95, 1292–1302 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anichtchik O. V., Rinne J. O., Kalimo H., Panula P., An altered histaminergic innervation of the substantia nigra in Parkinson’s disease. Exp. Neurol. 163, 20–30 (2000). [DOI] [PubMed] [Google Scholar]
- 67.da Silva W. C., Bonini J. S., Bevilaqua L. R. M., Izquierdo I., Cammarota M., Histamine enhances inhibitory avoidance memory consolidation through a H2 receptor-dependent mechanism. Neurobiol. Learn. Mem. 86, 100–106 (2006). [DOI] [PubMed] [Google Scholar]
- 68.da Silveira C. K. B., Furini C. R. G., Benetti F., Monteiro S. d. C., Izquierdo I., The role of histamine receptors in the consolidation of object recognition memory. Neurobiol. Learn. Mem. 103, 64–71 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Fabbri R., et al. , Memory retrieval of inhibitory avoidance requires histamine H1 receptor activation in the hippocampus. Proc. Natl. Acad. Sci. U.S.A. 113, E2714–E2720 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yanovsky Y., Haas H. L., Histamine increases the bursting activity of pyramidal cells in the CA3 region of mouse hippocampus. Neurosci. Lett. 240, 110–112 (1998). [DOI] [PubMed] [Google Scholar]
- 71.Alvarez E. O., Ruarte M. B., Banzan A. M., Histaminergic systems of the limbic complex on learning and motivation. Behav. Brain Res. 124, 195–202 (2001). [DOI] [PubMed] [Google Scholar]
- 72.Garrido Zinn C., et al. , Major neurotransmitter systems in dorsal hippocampus and basolateral amygdala control social recognition memory. Proc. Natl. Acad. Sci. U.S.A. 113, E4914–E4919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benetti F., da Silveira C. K. B., Rosa J., Izquierdo I., Histamine acting on the basolateral amygdala reverts the impairment of aversive memory of rats submitted to neonatal maternal deprivation. Behav. Brain Res. 278, 83–89 (2015). [DOI] [PubMed] [Google Scholar]
- 74.Gianlorenço A. C. L., Canto-de-Souza A., Mattioli R., Intra-cerebellar microinjection of histamine enhances memory consolidation of inhibitory avoidance learning in mice via H2 receptors. Neurosci. Lett. 557, 159–164 (2013). [DOI] [PubMed] [Google Scholar]
- 75.Privou C., Li J. S., Hasenöhrl R. U., Huston J. P., Enhanced learning by posttrial injection of H1-but not H2-histaminergic antagonists into the nucleus basalis magnocellularis region. Neurobiol. Learn. Mem. 71, 308–324 (1999). [DOI] [PubMed] [Google Scholar]
- 76.Orsetti M., Ferretti C., Gamalero R., Ghi P., Histamine H3-receptor blockade in the rat nucleus basalis magnocellularis improves place recognition memory. Psychopharmacology (Berl) 159, 133–137 (2002). [DOI] [PubMed] [Google Scholar]
- 77.Xu L., Lin W., Zheng Y., Wang Y., Chen Z., The diverse network of brain histamine in feeding: Dissect its functions in a circuit-specific way. Curr. Neuropharmacol., 10.2174/1570159X21666221117153755 (2022). [DOI] [PMC free article] [PubMed]
- 78.Yao L., et al. , The dual orexin receptor antagonist, DORA-22, lowers histamine levels in the lateral hypothalamus and prefrontal cortex without lowering hippocampal acetylcholine. J. Neurochem. 142, 204–214 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Cumming P., Damsma G., Fibiger H. C., Vincent S. R., Characterization of extracellular histamine in the striatum and bed nucleus of the stria terminalis of the rat: An in vivo microdialysis study. J. Neurochem. 56, 1797–1803 (1991). [DOI] [PubMed] [Google Scholar]
- 80.Jørgensen E. A., Knigge U., Watanabe T., Warberg J., Kjaer A., Histaminergic neurons are involved in the orexigenic effect of orexin-A. Neuroendocrinol. 82, 70–77 (2005). [DOI] [PubMed] [Google Scholar]
- 81.Zhang X. Y., et al. , Targeting presynaptic H3 heteroreceptor in nucleus accumbens to improve anxiety and obsessive-compulsive-like behaviors. Proc. Natl. Acad. Sci. U.S.A. 117, 32155–32164 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qi Z.-X., et al. , Histamine bidirectionally regulates intrinsic excitability of parvalbumin-positive neurons in lateral globus pallidus and promotes motor behavior. Br. J. Pharmacol., 10.1111/bph.16010 (2022). [DOI] [PubMed] [Google Scholar]
- 83.Gong H., et al. , High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nat. Commun. 7, 12142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peng J., et al. , A quantitative analysis of the distribution of CRH neurons in whole mouse brain. Front. Neuroanat. 11, 63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ni H., et al. , A robust image registration interface for large volume brain atlas. Sci. Rep. 10, 2139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kuan L., et al. , Neuroinformatics of the Allen Mouse Brain connectivity atlas. Methods 73, 4–17 (2015). [DOI] [PubMed] [Google Scholar]
- 87.Avants B. B., Epstein C. L., Grossman M., Gee J. C., Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12, 26–41 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou H., et al. , GTree: An open-source tool for dense reconstruction of brain-wide neuronal population. Neuroinformatics 19, 305–317 (2021). [DOI] [PubMed] [Google Scholar]
- 89.Li Y., et al. , TDat: An efficient platform for processing petabyte-scale whole-brain volumetric images. Front. Neural Circuits 11, 51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
The location of histaminergic neuron somata from anterior to posterior TMN in one of the tested mice.
The 3D fluorescent raw data of whole-brain histaminergic neural circuits from one of the tested mice.
Data Availability Statement
All study data are included in the article and/or SI Appendix.