Abstract
We report the genome-editing of an existing iPSC line carrying the London mutation in APP (V717I) into an iPSC line in which the pathogenic mutation was corrected. The resulting isogenic iPSC line maintained pluripotent stem cell morphology, a normal karyotype, expression of pluripotency markers and the ability to differentiate into the three germ-layers in vitro.
1. Resource utility
The amyloid beta precursor protein (APP) V717I London mutation is a dominant inherited mutation that causes early onset Alzheimer’s disease. APP mutation carriers typically present with an onset at the age of 45 to 60 years (Bateman et al., 2011). Using isogenic cell pairs with and without the disease-causing mutation allows a controlled assessment of APP (V717I) in iPSCs and progeny.
2. Resource details
The iPSC line F16574 was previously generated from one individual with a mutation in V717I APP (London mutation) (Karch et al., 2018). These iPSCs were generated from fibroblasts reprogrammed by nucleofection of episomal vectors containing OCT4, SOX2, KLF4, L-MYC, LIN28, and shRNA against p53 with selection of multiple clones and subsequent characterisation of pluripotency and genomic integrity (Karch et al., 2018). Here, we used the parental iPSC line F16574 clone 3 for genome editing of the APP locus and correction of the disease-causing mutation V717I allowing the generation of isogenic iPSC lines for APP mutation (UOMELBi002-A) (Table 1, Fig. 1A). One clone was subsequently isolated, expanded and recharacterized as above. The CRISPR/Cas9-edited iPSC line showed the typical human pluripotent stem cell-morphology and expressed markers of pluripotency TRA-1-60 and OCT4 (Fig. 1A). Quantification by flow cytometry analysis demonstrated 81.3% and 78.3% of live cells were positive for TRA-1-60 and OCT4 respectively (Fig. 1B). Gene editing correction of the APP V717I mutation was confirmed by Sanger DNA sequencing in UOMELBi002-A by comparing to the parental FA1657 clone 3 line (Fig. 1C). A silent DNA mutation at the PAM sequence (codon TTA instead TTG both encoding for Leucine) was included to enhance homology directed repair efficiency, allow identification of targeted allele by Sanger sequencing and to prevent recutting of the edited allele. Genomic integrity was assessed by copy number variation analysis of the parental iPSCs and the CRISPR/Cas edited iPSCs, represented with log R ratio and B allele frequency (Fig. 1D). This analysis confirmed the absence of deletions, insertions and aneuploidies. Of note, balanced rearrangements cannot be detected by this method. Cell identity was confirmed by PCR-based fingerprinting system using short tandem repeat (STR) profiling of samples (data not shown). The iPSCs were also able to differentiate into the three germ layers, as demonstrated by positive immunostaining for endodermal (alpha-fetoprotein, AFP), mesodermal (smooth muscle actin, SMA) and ectodermal (NESTIN) markers following embryoid body formation (Fig. 1E).
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
|
| |||
| Morphology | Phase contrast brightfield morphology pictures | Normal | Fig. 1 panel A |
| Phenotype | Qualitative analysis of pluripotency Immunochemistry | Expression of TRA-1-60 and OCT4 | Fig. 1 panel A |
| Quantitative analysis (Flow cytometry] | TRA-1-60: 78.3%; OCT4: 81.3% | Fig. 1 panel B | |
| Genotype | CNV array Illumina HumanCore Beadchip array which contains over 300,000 informative SNPs with a median spacing of 5.8 kb. |
46, Resolution
450–500. Sex chromosomes were cropped to mantain blinding of gender identity (available upon request) |
Fig. 1 panel D |
| Identity | STR analysis | 10 sites tested, all sites matched between parental and isogenic cell lines | Submitted, in archive with journal |
| Mutation analysis (IF APPLICABLE) | Sanger Sequencing | Assessment of rs63750264 status confirmed (A/G) in parental and UOMELBi002-A cell line (G/G) | Fig. 1 panel C |
| Southern Blot OR WGS | NA | NA | |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by luminescence were Negative |
Reading B/A ratio; UOMELBi002-A = 0.59 F16574 clone 3 = 0.50 Positive control = 3.78 Negative control = 0.34 |
| Differentiation potential | Direct differentiation, STEMdiff™ Trilineage Differentiation Kit (Stemcell technologies) | Expression of AFP, SMA and NESTIN by immunostaining | Fig. 1 panel E |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | NA | NA |
| Genotype additional info (OPTIONAL) | Blood group genotyping HLA tissue typing |
NA
NA |
NA
NA |
Fig. 1.
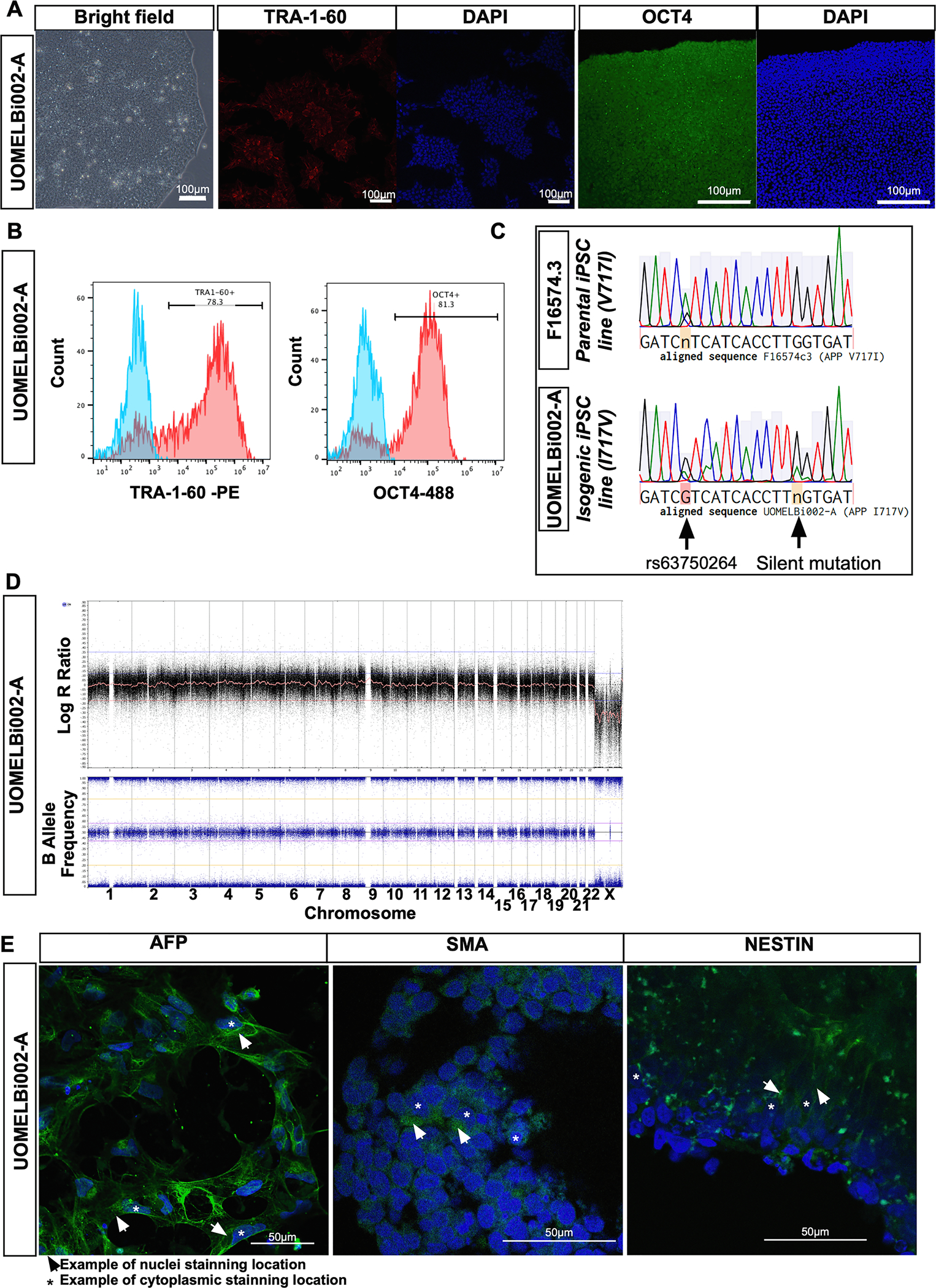
Characterization of UOMELBi002-A.
3. Materials and methods
3.1. Ethics
All experimental work performed in this study was approved by the Human Research Ethics committees of the University of Melbourne (1545394) with the requirements of the National Health & Medical Research Council of Australia (NHMRC) and conformed with the Declaration of Helsinki (McCaughey et al., 2016).
3.2. iPSC culture
The iPSCs were maintained in StemFlex medium (Gibco) using 6-well plates pre-coated with vitronectin (Stemcell Technologies). Media was changed every second day and cells were passaged with ReleSR (Stemcell Technologies) on a weekly basis when colonies reached 80% confluency.
3.3. Generation of isogenic lines
Genome editing was performed with the CRISPR/Cas9 system in combination with a single-stranded DNA (ssDNA, Table 2) to guide the single nucleotide correction of the APP V717I mutation by homologous recombination. Single guide (sg) RNA sequence was designed as described by Zhang’s laboratory (Ran et al., 2013) and selected based on the highest on-target and off-target score (Doench et al., 2016; Hsu et al., 2013). The APP SNP rs63750264 allele (A) of the parental iPSC F16574 clone 3 line was genetically modified to generate isogenic lines with a homozygous G/G nucleotide with the designed sgRNA and the ssDNA (Table 2). The ribonucleoprotein (RNP) complex consisting of Cas9 protein and sgRNA (containing a tracrRNA labelled with a red fluorophore ATTO 550) was assembled in Duplex buffer (all from Integrated DNA Technologies). RNP complex was subsequently transfected into dissociated iPSCs by electroporation (1200 V, 30 ms, 1 pulse) with the Neon transfection system (Invitrogen). After electroporation the cells were immediately plated onto vitronectin-plated 6 well plates containing Stemflex medium supplemented with 10 μM ROCK inhibitor (RevitaCell, Gibco). After 48 h, ATTO 550 positive / DAPI negative cells were sorted by flow cytometry (BD) (Fig. 1S) and plated at low density for clonal selection (1505 cells into 1 well of a six-well plate). Cells were screened for SNP editing by PCR and Sanger sequencing (Australian Genome Research Facility), from 96 clones that were analysed only one clone was re-plated at low density for sub-clonal selection. One colony out of ninety-six was subsequently dissociated for sub-clonal selection, as to obtain a pure edited population from one out of eight sub-clones analysed. The resultant isogenic lines were named UOMELBi002-A.
Table 2.
Reagents details.
| Antibodies used for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
|
| |||
| Antibody | Dilution | Company Cat # and RRID | |
|
| |||
| Pluripotency Markers | mouse anti-TRA-1–60 | 1:200 | Invitrogen Cat#MA1-023-PE, RRID:AB_2536704 |
| Pluripotency Markers | mouse anti-OCT3/4 | 1:80 | Santa Cruz Biotechnology Cat# sc-5279, RRID:AB_628051 |
| Differentiation Markers | mouse anti-SMA | 1:500 | R and D Systems Cat# MAB1420, RRID:AB_262054 |
| Differentiation Markers | mouse anti-NESTIN | 1:1000 | Abcam Cat# ab22035, RRID:AB_446723 |
| Differentiation Markers | mouse anti-AFP | 1:1000 | Millipore Cat# ST1673–100UG, RRID:AB_10697987 |
| Secondary antibodies | Alexa Fluor 568 Goat Anti-Mouse IgG | 1:1000 | Thermo Fisher Scientific Cat# A-11031, RRID:AB_144696 |
| Secondary antibodies | Alexa Fluor 488 Goat Anti-Mouse IgG | 1:1000 | Thermo Fisher Scientific Cat# A-11029, RRID:AB_2534088 |
| Primers | |||
| Target | Forward/Reverse primer (5′–3′) | ||
| Targeted mutation | sgRNA, rs63750264 | TGGTGATGCTGAAGAAGAAA | |
| Targeted mutation | ssDNA, rs63750264 | GTGCAATCATTGGACTCATGGTGGGCGGTGTTGTCATAGCGACAGTGATCgTCATCACCTTaGTGATGCTGAAGAAGAAACAGTACACATCCATTCATCATGGTGTGGTGGA | |
| Genotyping | PCR, rs63750264 | Forward (GTCACACATCAGGGCTCAGAGT) | |
| Genotyping | PCR, rs63750264 | Reverse (AACCCAAGCATCATGGAAGCAC) | |
| Genotyping | Sequencing, rs63750264 | Forward (CCTCATCCAAATGTCCCCTGCA) | |
3.4. Virtual karyotype
Copy number variation analysis of isogenic iPSCs was performed using Illumina Infinium CoreExome-24 v1.1, performed by the Victorian Clinical Genetics Services (VCGS, Melbourne, Australia).
3.5. Cell identity
Short tandem repeat (STR) profiling of samples was performed by PCR-based fingerprinting using Promega GenePrint 10 system, and performed by the Australian Genome Research Facility (AGRF, Melbourne, Australia).
3.6. Differentiation to the three-germ layer
Embryoid bodies were generated using a tri-lineage differentiation kit (Stem Cell Technologies). Germ layer differentiation was assessed by immunochemistry.
3.7. Immunocytochemistry
Cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton-X (Sigma). Immunocytochemistry was performed using the following primary antibodies: mouse anti-TRA-1-60-PE (Invitrogen), mouse anti-OCT3/4 (Santa Cruz Biotechnology), smooth muscle actin (R&D Systems), mouse anti-NESTIN (Abcam) or rabbit anti-alpha-fetoprotein (Sigma-Aldrich) (Table 2). Cells were then immunostained with isotype-specific secondary antibodies (Alexa Fluor 568 or 488, Life Technologies, Table 2). Nuclei were counterstained using DAPI (Sigma-Aldrich) and mounted in Vectashield (Vector Labs). Specificity of the staining was verified by the absence of staining in negative controls consisting of the appropriate negative control immunoglobulin fraction (Dako). Images were acquired on a Zeiss AxioImager M2 fluorescent microscope or LMS 880 confocal microscope using ZEN software (Zeiss).
3.8. Flow cytometry analysis
Cells were dissociated into single cells with ReleSR and incubated with a fixable viability dye (Miltenyi). Then cells were fixed and permeabilized with the Inside stain Kit (Miltenyi). Cells were incubated with primary antibodies mouse anti-TRA-1-60-PE (Invitrogen) and mouse anti-OCT3/4 (Table 2), following incubation with isotype-specific secondary antibody (Alexa Fluor 488, Life Technologies, Table 2). Unstained cells were used as negative controls. Cells were analysed by flow cytometry analysis (Cytoflex S, Beckman Coulter).
3.9. Mycoplasma testing
Mycoplasma test was performed using the MycoAlert kit (Lonza) following the manufacturer’s instruction.
Supplementary Material
Resource Table.
| Unique stem cell line identifier | UOMELBi002-A |
|---|---|
|
| |
| Alternative name(s) of stem cell line | F16574c3_A_A1_G8 |
| Institution | The University of Melbourne |
| Contact information of distributor | Dr. Damian Hernández, damian.hernandez@unimelb.edu.au |
| Type of cell line | iPSC |
| Origin | human |
| Additional origin info | Age: Blinded for publication due to risk of unblinding; available upon request Sex: Blinded for publication due to risk of unblinding; available upon request Ethnicity if known: Blinded for publication due to risk of unblinding; available upon request |
| Cell Source | iPSC |
| Clonality | Clonal |
| Method of reprogramming | Episomal for parental iPSC |
| Genetic Modification | YES |
| Type of Modification | Gene Correction and silent mutation in one single allele (Heterozygous mutation). |
| Associated disease | Alzheimer’s disease |
| Gene/locus | APP London mutation (V717I, rs63750264) in exon 17 of APP. (GRCh38; Chr21:25891784 G > A) |
| Method of modification | CRISPR Cas9 |
| Name of transgene or resistance | NA |
| Inducible/constitutive system | NA |
| Date archived/stock date | NA |
| Cell line repository/bank | NA |
| Ethical approval | All experimental work performed in this study was approved by the University of Melbourne (1545394) with the requirements of the National Health & Medical Research Council of Australia (NHMRC) and conformed with the Declaration of Helsinki. |
Acknowledgments
We gratefully acknowledge the altruism of the participants and their families and the contributions of the DIAN research and support staff at each of the participating sites for their contributions to this study. The DIAN Expanded Registry welcomes contact from any families or treating clinicians interested in research about autosomal dominant familial Alzheimer’s disease. Data collection and sharing for this project were supported by The Dominantly Inherited Alzheimer’s Network (DIAN; grant UF1AG032438) funded by the National Institute on Aging (NIA), the German Center for Neurodegenerative Diseases (DZNE), and Raul Carrea Institute for Neurological Research (FLENI). Partial support was provided by the Research and Development Grants for Dementia from Japan Agency for Medical Research and Development (AMED) and by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI). We also acknowledge the Biological Optical Microscopy Platform and the Melbourne Cytometry Platform (Melbourne Brain Centre Node) at the University of Melbourne for technical assistance.
This research was supported by grants from the Yulgilbar Alzheimer’s Research Program, the DHB Foundation, Dementia Australia, the Brain Foundation, a National Health and Medical Research Council Senior Research Fellowship (AP, 1154389), and the University of Melbourne.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
DIAN Consortium Full Name and Credentials
Sarah Adams, MS; Ricardo Allegri, PhD; Aki Araki; Nicolas Barthelemy, PhD; Randall Bateman, MD; Jacob Bechara, BS; Tammie Benzinger, MD, PhD; Sarah Berman, MD, PhD; Courtney Bodge, PhD; Susan Brandon, BS; William (Bill) Brooks, MBBS, MPH; Jared Brosch, MD, PhD; Jill Buck, BSN; Virginia Buckles, PhD; Kathleen Carter, PhD; Lisa Cash, BFA; Charlie Chen, BA; Jasmeer Chhatwal, MD, PhD; Patricio Chrem Mendez, MD; Jasmin Chua, BS; Helena Chui, MD; Laura Courtney, BS; Carlos Cruchaga, PhD; Gregory S Day, MD; Chrismary DeLa-Cruz, BA; Darcy Denner, PhD; Anna Diffenbacher, MS; Aylin Dincer, BS; Tamara Donahue, MS; Jane Douglas, MPh; Duc Duong, BS; Noelia Egido, BS; Bianca Esposito, BS; Anne Fagan, PhD; Marty Farlow, MD; Becca Feldman, BS, BA; Colleen Fitzpatrick, MS; Shaney Flores, BS; Nick Fox, MD; Erin Franklin, MS; Nelly Joseph-Mathurin, PhD; Hisako Fujii, PhD; Samantha Gardener, PhD; Bernardino Ghetti, MD; Alison Goate, PhD; Sarah Goldberg, MS, LPC, NCC; Jill Goldman, MS, MPhil, CGC; Alyssa Gonzalez, BS; Brian Gordon, PhD; Susanne Gräber-Sultan, PhD; Neill Graff-Radford, MD; Morgan Graham, BA; Julia Gray, MS; Emily Gremminger, BA; Miguel Grilo, MD; Alex Groves; Christian Haass, PhD; Lisa Häsler, MSc; Jason Hassenstab, PhD; Cortaiga Hellm, BA; Elizabeth Herries, BA; Laura Hoechst-Swisher, MS; Anna Hofmann, MD; Anna Hofmann; David Holtzman, MD; Russ Hornbeck, MSCS, MPM; Yakushev Igor, MD; Ryoko Ihara, MD; Takeshi Ikeuchi, MD; Snezana Ikonomovic, MD; Kenji Ishii, MD; Clifford Jack, MD; Gina Jerome, MS; Erik Johnson, MD, PHD; Mathias Jucker, PhD; Celeste Karch, PhD; Stephan Käser, PHD; Kensaku Kasuga, MD; Sarah Keefe, BS; William (Klunk, MD, PHD; Robert Koeppe, PHD; Deb Koudelis, MHS, RN; Elke Kuder-Buletta, RN; Christoph Laske, PhD; Allan Levey, MD, PHD; Johannes Levin, MD; Yan Li, PHD; Oscar Lopez MD, MD; Jacob Marsh, BA; Ralph Martins, PhD; Neal Scott Mason, PhD; Colin Masters, MD; Kwasi Mawuenyega, PhD; Austin McCullough, PhD Candidate; Eric McDade, DO; Arlene Mejia, MD; Estrella Morenas-Rodriguez, MD, PhD; John Morris, MD; James Mountz, MD; Cath Mummery, PhD; N eelesh Nadkarni, MD, PhD; Akemi Nagamatsu, RN; Katie Neimeyer, MS; Yoshiki Niimi, MD; James Noble, MD; Joanne Norton, MSN, RN, PMHCNS-BC; Brigitte Nuscher; Ulricke Obermüller; Antoinette O’Connor, MRCPI; Riddhi Patira MD; Richard Perrin, MD, PhD; Lingyan Ping, PhD; Oliver Preische, MD; Alan Renton, PhD; John Ringman, MD; Stephen Salloway, MD; Peter Schofield, PhD; Michio Senda, MD, PhD; Nicholas T Seyfried, D. Phil; Kristine Shady, BA, BS; Hiroyuki Shimada, MD, PhD; Wendy Sigurdson, RN; Jennifer Smith, PhD; Lori Smith, PA-C; Beth Snitz, PhD; Hamid Sohrabi, PhD; Sochenda Stephens, BS, CCRP; Kevin Taddei, BS; Sarah Thompson, PA-C; Jonathan Vöglein, MD; Peter Wang, PhD; Qing Wang, PhD; Elise Weamer, MPH; Chengjie Xiong, PhD; Jinbin Xu, PhD; Xiong Xu, BS, MS.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2021.102373.
Data availability
Data will be made available on request.
References
- Bateman RJ, Aisen PS, De Strooper B, Fox NC, Lemere CA, Ringman JM, Salloway S, Sperling RA, Windisch M, Xiong C, 2011. Autosomal-dominant Alzheimer’s disease: a review and proposal for the prevention of Alzheimer’s disease. Alzheimer’s Res. Ther. 3 (1), 1. 10.1186/alzrt59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE, 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34 (2), 184–191. 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F, 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31 (9), 827–832. 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch CM, Hernández D, Wang J-C, Marsh J, Hewitt AW, Hsu S, Norton J, Levitch D, Donahue T, Sigurdson W, Ghetti B, Farlow M, Chhatwal J, Berman S, Cruchaga C, Morris JC, Bateman RJ, Pébay A, Goate AM, 2018. Human fibroblast and stem cell resource from the dominantly inherited Alzheimer network. Alzheimer’s Res. Therapy 10 (1). 10.1186/s13195-018-0400-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughey T, Liang H, Chen C, Fenwick E, Rees G, Wong RB, Vickers J, Summers M, MacGregor C, Craig J, Munsie M, Pébay A, Hewitt A, 2016. An interactive multimedia approach to improving informed consent for induced pluripotent stem cell research. Cell Stem Cell 18 (3), 307–308. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8 (11), 2281–2308. 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.


