Abstract
Bacteriophage integrase-directed insertion of transgenic constructs into specific genomic loci has been widely used by Drosophila community. The attP40 landing site located on the second chromosome gained popularity because of its high inducible transgene expression levels. Here, unexpectedly, we found that homozygous attP40 chromosome disrupts normal glomerular organization of Or47b olfactory receptor neuron (ORN) class in Drosophila. This effect is not likely to be caused by the loss of function of Msp300, where the attP40 docking site is inserted. Moreover, the attP40 background seems to genetically interact with the second chromosome Or47b-GAL4 driver, which results in a similar glomerular defect. Whether the ORN phenotype is caused by the neighbouring genes around Msp300 locus in the presence of attP40-based insertions or a second unknown mutation in the attP40 background remains elusive. Our findings tell a cautionary tale about using this popular transgenic landing site, highlighting the importance of rigorous controls to rule out the attP40 landing site-associated background effects.
Keywords: Drosophila, attP40, olfactory receptor neuron, axon terminal organization
Introduction
RNA interference (RNAi)-based genetic screens provide scientists with powerful tools to identify genes involved in various biological processes (Housden et al. 2017). Binary expression systems, such as the GAL4/UAS system, induce the expression of various effectors in the desired cell populations (Brand and Perrimon 1993). In Drosophila carrying transgenes for both cell-type-specific promoter-driven GAL4 (driver) and UAS-RNAi, GAL4 protein binds UAS sites and drives RNAi expression, disrupting the expression and function of the target gene (Brand and Perrimon 1993). As RNAi-based knockdown methods were becoming popular, efforts were initiated to make transgenic libraries of flies carrying UAS-RNAi targeting all the genes in the genome (Dietzl et al. 2007; Ni et al. 2009, 2011; Perkins et al. 2015). These genome-wide libraries were then followed by efforts to generate thousands of GAL4 lines that restrict expression to cellular subpopulations, enabling loss-of-function screens in cells of interest.
Among the RNAi collections, stocks from Transgenic RNAi Project (TRiP) have gained popularity because of their targeted integration of UAS-RNAi transgenes into the genome, efficient expression induced by appropriate GAL4 drivers in different tissues, and high specificity with minimal expected off-target effects (Markstein et al. 2008; Ni et al. 2008; Perkins et al. 2015). To expedite the generation of transgenic libraries, two predetermined chromosomal docking sites were targeted for recombination events that insert UAS-RNAi transgenes: attP40 on the second chromosome and attP2 on the third chromosome (Markstein et al. 2008). With the presence of bacteriophage-originated phiC31 integrase (by co-injection of integrase mRNA or germline-expressing transgenic integrase), the UAS-RNAi construct can be inserted into the corresponding docking sites (Groth et al. 2004; Ni et al. 2008). These two sites, attP40 and attP2, are selected because they exhibit optimal inducible expression levels upon binding with diverse tissue-specific GAL4 drivers (Markstein et al. 2008). Therefore, in addition to the TRiP UAS-RNAi library, many other transgenes, including tissue-specific drivers (GAL4, QF, LexA) and UAS/QUAS/LexAop-effectors/reporters are also routinely integrated into these two landing sites (Zirin et al. 2020).
Given the widespread use of transgenic flies with attP40 and attP2 backbones, and the lesson learned from another popular UAS-RNAi collection with reported non-specific effects due to transgenic docking sites (Green et al. 2014; Vissers et al. 2016), we must be more cognizant of potential phenotypic influences from these genetic backgrounds. Both attP40 and attP2 docking sites are in chromosomal regions populated by many genes. These sites, like any insertion into the genome, can disrupt function of nearby genes. More specifically, the attP40 site is located within one of the large introns of Msp300 gene while attP2 site is inserted in the 5′ untranslated region (UTR) of Mocs1 gene (Larkin et al. 2020). Both Msp300 and Mocs1 have critical biological roles. Specifically, Msp300 is the Drosophila melanogaster orthologue of mammalian Nesprins, which organize postsynaptic cytoskeleton scaffold and are required for stabilization of new synapses (Elhanany-Tamir et al. 2012; Morel et al. 2014; Titlow et al. 2020; Zheng et al. 2020). Mocs1 is involved in Mo-molybdopterin cofactor biosynthetic process and inter-male aggressive behaviours (Gaudet et al. 2011; Ramin et al. 2019). It is unclear how the insertion of various transgenic constructs into attP40 and attP2 docking sites would affect the function of these host genes which may further result in phenotypic defects.
Indeed, recent studies have raised issues related to landing site-associated effects. For example, van der Graaf et al. 2022 showed flies bearing two copies of attP40-derived insertions also show decreased Msp300 transcript levels (van der Graaf et al. 2022). In addition, this study also reported defects in muscle nuclei spacing in larval stages in the attP40 homozygous background, which phenocopies Msp300 mutants (van der Graaf et al. 2022). These results suggest that the attP40 docking site and attP40-based transgenes are insertional mutations of Msp300 gene (van der Graaf et al. 2022). Another study reported that attP40 flies show resistance to cisplatin-induced neuronal damage, compared to the attP2 background (Groen et al. 2022). This study tied the effect to the reduced ND-13A (NADH dehydrogenase 13 kDA subunit, a component of mitochondrial complex I) expression in attP40 homozygous flies (Groen et al. 2022). It is noteworthy that ND-13A flanks the 5′ UTR of Msp300 and is downstream of attP40 docking site. Together, these results imply the integration of attP40 docking site significantly changes the local transcriptional state and interferes with the transcription of surrounding genes.
During a GAL4-driven UAS-RNAi screen for olfactory neuron axon organization, we observed an axon terminal phenotype that is associated with the attP40 background. The phenotype occurs in the flies homozygous for the attP40 docking site alone or with various transgenic insertions, independent of the identity of the transgene. Notably, the phenotype observed in the attP40 background appears to be recessive but is independent of the Msp300 function, possibly implicating other attP40 background mutations nearby or in other locations on the second chromosome. Though the nature of the mutation is unclear, the background effects should be mitigated by designing more rigorous controls to interpret phenotypic data obtained using reagents in concert with the attP40 background.
Materials and methods
Drosophila stocks and genetics
Drosophila were raised in classic molasses media provided by Archon Scientific. For the RNAi screen experiments, flies were raised at 28°C to maximize the knockdown efficiency. Most of the other crosses were also kept at 28°C, except for the experiments shown in Figs. 1, f and g and 3a, which were conducted at room temperature (23°C). After eclosion, the flies are aged for 5–7 days before dissection. In addition to the UAS-RNAi stocks from Bloomington Drosophila Stock Center (listed in Fig. 1b), the following stocks are used: UAS-RFP RNAi attP2 (BDSC# 35785), UAS-beat-Ia RNAi #3 GD1386 (VDRC# 4544), UAS-SMC3 RNAi attP2 (BDSC# 60017), UAS-SMC3 RNAi attP40 (BDSC# 50899), UAS-vtd RNAi attP2 (BDSC# 36786), UAS-vtd RNAi attP40 (BDSC# 65229), attP40 (BDSC# 36304), attP2 (BDSC# 36303), ctrl-gRNA attP40 (BDSC# 67539), UAS-RFP attP40 (BDSC# 32222), UAS-rCD2.RFP attP2 (BDSC# 56179), UAS-rCD2.RFP attP5 (BDSC# 56180), UAS-rCD2.RFP attP40 (BDSC# 56181), Msp300ΔKASH (BDSC# 26781), Msp300MI01145 (BDSC# 53050), Msp300MI00111 (BDSC# 30623), Msp300KG03631 (BDSC# 13024); Or47b-GAL4 (chr2, BDSC#9983), Or47b-GAL4 (chr3, BDSC#9984), Or43a-GAL4 (chr2), Or47a-GAL4 (chr2) (Vosshall et al. 2000; Fishilevich and Vosshall 2005), and Gr21a-GAL4 (chr2) (Scott et al. 2001) are gifts from Dr. Leslie Vosshall; UAS-Syt.GFP (chr2 or chr3), UAS-mCD8.GFP, UAS-RFP are Volkan lab stocks (Barish et al. 2018). The line Or47b-GAL4, Or47a-GAL4, Or43a-GAL4, Gr21a-GAL4, UAS-Syt.GFP/CyO (short for 4xOr-GAL4 > Syt.GFP) was recombined and balanced from the above components.
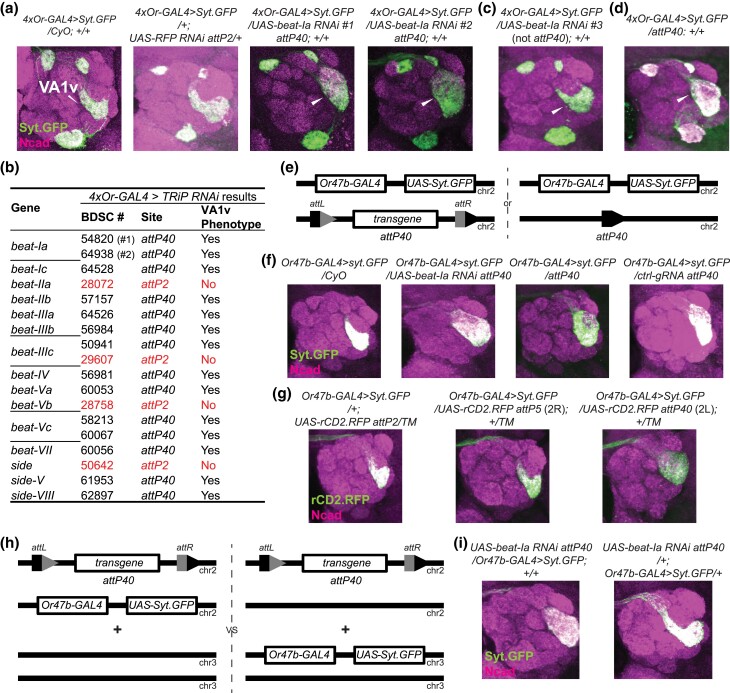
Fig. 1.
Genetic interactions between attP40 and Or47b-GAL4 backgrounds on the second chromosome disrupt the glomerular organization of Or47b ORNs in the antennal lobes. a, c, and d) Confocal images of representative brains from a genetic screen to identify adhesion molecules involved in the glomerular organization of the Drosophila olfactory receptor neuron axon terminals. We crossed a second chromosome containing four different Drosophila olfactory receptor promoter-driven GAL4s (Or47a-GAL4, Or47b-GAL4, Or23a-GAL4, Gr21a-GAL4) together with a UAS-Syt.GFP reporter (4xOr-GAL4 > Syt.GFP) to the indicated UAS-RNAi lines or attP40 background flies. The parental driver chromosome over the CyO balancer was used as a no-RNAi control. The invading Or47b ORN axons are denoted with white arrowheads. b) Summary of the phenotypical results from the genetic screen focusing on beat/side gene families. The Bloomington stock number and the transgenic docking site of each line are also listed. e and f) Schematic in (e) shows the genotype of animals used in (f), where each fly has one copy of the second chromosome carrying an Or47b-GAL4 driver and a UAS-Syt.GFP reporter, and one copy of the indicated second chromosome, either a CyO balancer or attP40 docking site derivatives. attL and attR sites are generated as a result of transgene integration into attP40 docking site. Confocal images of representative brains are shown in (f). g) Confocal brain images of the indicated genotypes. (h and i) Schematic in (h) shows the genotype of animals used in (i), where each fly has one copy of the second chromosome UAS-beat-Ia RNAi transgene inserted into the attP40 docking site, with one copy of Or47b-GAL4 UAS-Syt.GFP, either on the second or third chromosome. Confocal images of representative brains are shown in (i). 10–25 brains were examined in each genotype and the phenotypical penetrance is close to 100% in each attP40-derived group.
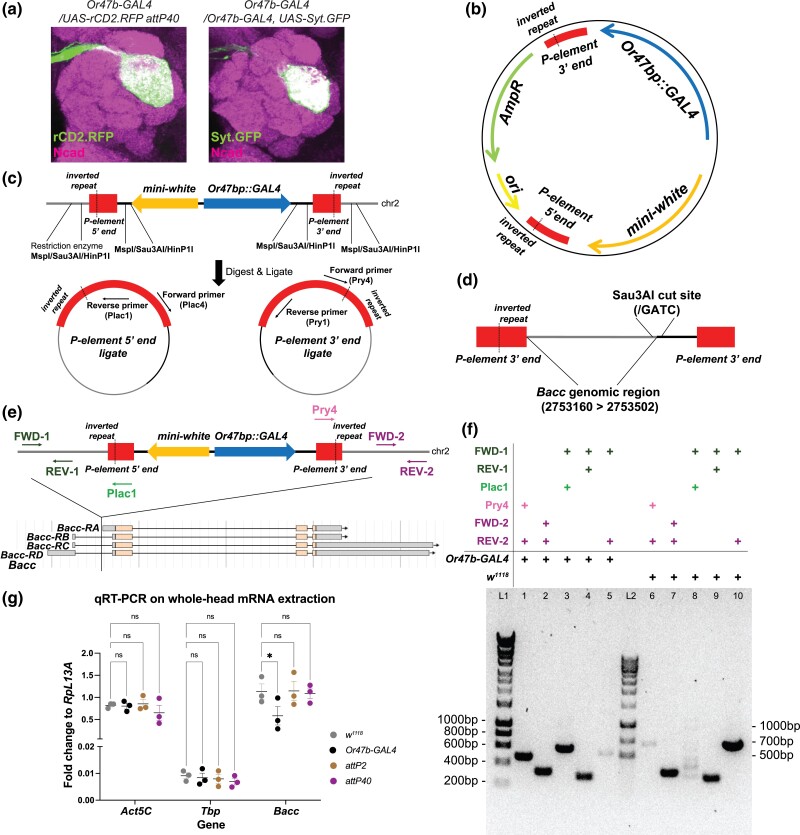
Fig. 3.
Identification of the Or47b-GAL4 transgene insertion site. a) Confocal images of the indicated genotypes suggesting the second chromosome Or47b-GAL4 transgene is accounting for the glomerular expansion phenotype. N = 16 brains in each genotype and the phenotypical penetrance is 100%. b) The structure of the P-element vector containing Or47b-GAL4 transgene (Or47b promoter fused with GAL4 coding sequence, denoted as Or47bp::GAL4 in this and other panels of this figure) and a mini-white selectable marker. c) Schematic illustrating the recovery of genomic sequences flanking P-element insertion by inverse PCR. d) Schematic showing the sequencing results from one ligated genomic DNA template digested by Sau3AI restriction enzyme. A region within Bacc gene was recovered as the immediate sequencing flanking the 3′ end of the inserted P-element containing Or47b-GAL4 transgene. e) Schematic showing the P-element insertion site within the first intron of Bacc gene. The arrows indicate the primers used in a PCR assay to validate this insertion. f) DNA gel showing the amplicons by each indicated primer pairs from Or47b-GAL4 transgenic flies or w1118 control flies. Primer pairs FWD-1/REV-1 and FWD-2/REV-2 can amplify the specific fragments from both Or47b-GAL4 transgenic flies and w1118 control flies as expected because the corresponding sequences exist in both. Primer pairs FWD-1/Plac1 and Pry4/REV-2 only amplify specific products from Or47b-GAL4 transgenic flies while fail to work from w1118 control flies as one primer in each pair targets the sequence of P-element which is not present in w1118 flies. Primer pair FWD-1/REV-2 works in w1118 control flies but fails in Or47b-GAL4 flies as the expected amplicon is too long to be amplified in transgenic animals because of the integration of large P-element sequence. g) qPCR results comparing the indicated gene expression levels normalized by RpL13A transcripts across different genotypes. ns, not significant. *P-value < 0.05 after two-way ANOVA followed by multiple comparison test. Error bars indicate mean ± SEM. N = 3 biological replicates per genotype. Each biological replicate contain 14∼20 fly heads from equal number of males and females.
Immunocytochemistry
Flies were sacrificed in 70% ethanol. Fly brains were then dissected in PBST buffer (0.2% Triton X-100 in 1X PBS), fixed in 4% paraformaldehyde for 30 min, followed by washing with PBST for three 10-min cycles. Brains were incubated in the primary antibody mix at 4°C overnight, followed by three 20-min washes with PBST at room temperature, then incubated in the secondary antibody mix at 4°C overnight. The brains were washed again by three 20-min wash with PBST before being mounted on the slide for imaging. The blocking was done together with each antibody incubation, with 1% natural goat serum mixed with primary and secondary antibodies, respectively. The following primary antibodies were used: 1:1000 rabbit anti-GFP (Invitrogen), 1:20 rat anti-Ncad (DSHB); the following secondary antibodies were used: 1:1000 Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen), 1:200 Alexa Fluor 647 goat anti-rat IgG (Invitrogen); all antibodies are diluted in PBST.
Confocal imaging and phenotypic quantification
Confocal imaging was performed by either Olympus Fluoview FV1000 microscope or Zeiss 880 microscope. Brains were imaged across Z-axis from the posterior side to the most anterior side of the antennal lobes, and all confocal sections were overlayed for phenotypical analysis. The same set of imaging parameters was used between experimental and control groups. The phenotype was qualitatively determined by glomerular morphology, i.e. whether Or47b ORN axons appear in the dorsal antennal lobe region, in contrast to the typical V-shaped glomerulus in wild-type controls. The phenotype shown in Fig. 1 (glomerular expansion) is largely consistent from brain to brain, while the phenotypes shown in Fig. 2, b and e exhibit variability, which were categorized into expansion or dorsal shift. The phenotype was quantified by the percentage of antennal lobes exhibiting each defect among all the brains examined in respective groups. P-value was calculated by two-tailed Fisher's exact test through the built-in functions of GraphPad Prism 9 software.
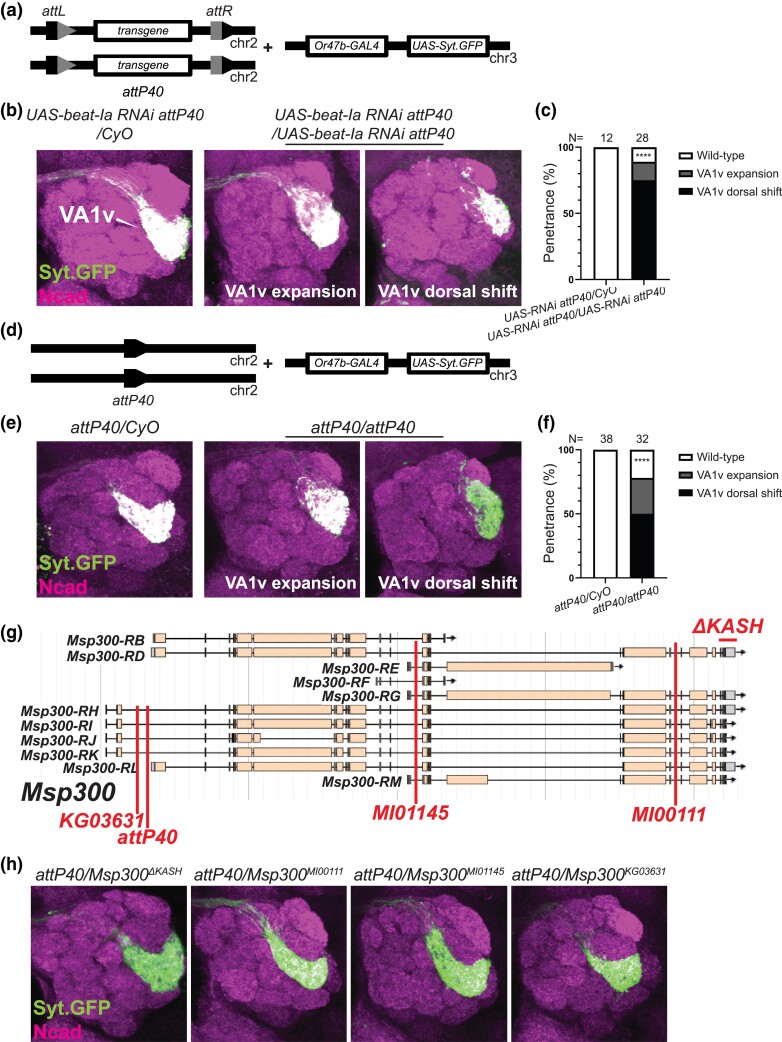
Fig. 2.
Homozygous attP40 chromosome affects glomerular organization of Or47b ORNs independent of the Msp300 function. a–c) Schematic in (a) shows the genotype of animals used in (b), where each fly has one or two copies of the second chromosome UAS-beat-Ia RNAi transgene inserted at the attP40 docking site, with the third chromosome Or47b-GAL4 UAS-Syt.GFP transgenes. Confocal images of representative brains are shown in (b). The percentage of the phenotypes is shown in (c). ****P < 0.0001 after Fisher's exact test. d–f) Schematic in (d) shows the genotype of animals used in (e), where each fly has one or two copies of the second chromosome empty attP40 docking site, with the third chromosome Or47b-GAL4 UAS-Syt.GFP transgenes. Confocal images of representative brains are shown in (e). The percentage of the phenotypes is shown in (f). ****P < 0.0001 after Fisher's exact test. N in (c) and (f) denotes the antennal lobes examined. g) Schematic showing the Msp300 genomic locus, the attP40 docking site, three insertional Msp300 mutations (Msp300MI00111, Msp300MI01145, Msp300KG03631), and one deletion allele (Msp300ΔKASH), each denoted with red lines. h) Confocal images of representative brains of the indicated transheterozygous animals, with the attP40 docking site over the indicated Msp300 alleles. N = 11, 8, 4, 12 brains in each genotype group, from left to right.
Inverse PCR to recover the genomic DNA sequence flanking the Or47b-GAL4 transgenic insertion
Or47b-GAL4 transgene was inserted into an unknown region on the second chromosome by P-element-mediated method (Vosshall et al. 2000). The P-element structure of Or47b-GAL4 transgene was shown in the Fig. 3b. We used previously described inverse PCR method (Huang et al. 2009) to identify the genomic sequence flanking the insertion site. Briefly, genomic DNA was first extracted from 30 Or47b-GAL4 flies (BDSC# 9983), followed by overnight digestion with any of three restriction enzymes, MspI, HinP1I, or Sau3AI (New England BioLabs). Each digest was then ligated by T4 DNA ligase (New England BioLabs) in larger volume (400 μl, see (Huang et al. 2009) for mix details) to promote intramolecular ligation while minimizing intermolecular ligation. Ligation was performed at 16°C overnight. The unknown flanking sequence was then amplified from the ligated genomic DNA by inverse PCR using forward and reverse primers targeting the 3′- or 5′-end sequences of the P-element (Fig. 3c). PCR reaction: 10 μl ligated DNA, 2 μl 5 μM forward and reverse primer mix, 10 μl 5X myTaq reaction buffer, 0.5 μl Taq DNA polymerase. Thermal cycling programme: 3 min initial denaturation at 95°C + 35 cycles (30 s denaturation at 95°C, 1 min annealing at 55°C, 2 min extension at 72°C) + 10 min final extension at 72°C. PCR products were then cleaned with Qiagen QIAquick PCR purification kit followed by commercial DNA sequencing service. Primers are listed in Table 1.
Table 1.
Primers used in this study.
| Experiment | Primer | Sequence (5′ to 3′) | Source |
|---|---|---|---|
| Inverse PCR | For amplify P-element 3′-end flanking region | (Huang et al. 2009) | |
| Pry1 | CCTTAGCATGTCCGTGGGGTTTGAAT | ||
| Pry4 | CAATCATATCGCTGTCTCACTCA | ||
| For amplify P-element 5′-end flanking region | |||
| Plac1 | CACCCAAGGCTCTGCTCCCACAAT | ||
| Plac4 | ACTGTGCGTTAGGTCCTGTTCATTGTT | ||
| Sequencing | For sequencing P-element 3′-end product | (Huang et al. 2009) | |
| Spep1 | GACACTCAGAATACTATTC | ||
| For sequencing P-element 5′-end product | |||
| Sp1 | ACACAACCTTTCCTCTCAACAA | ||
| PCR Validation assay | FWD-1 | GAGCACATATCGGTGGTTAG | This study |
| REV-1 | GGCCCATACAATACACTCAA | ||
| FWD-2 | CGAGAGGCAGTGCTTAAATA | ||
| REV-2 | CTTGAGATCGTCCTTGACAG | ||
| qPCR | Bacc-F | AGGCTCTGGAGGAAATCA | This study |
| Bacc-R | CCGGAACCGTCATCATTATC | ||
| Act5C-F | GGCGCAGAGCAAGCGTGGTA | (Zhao et al. 2020) | |
| Act5C-R | GGGTGCCACACGCAGCTCAT | ||
| RpL13A-F | GCGAGGAGCTGAACCTCTC | ||
| RpL13A-R | GGAAGTGGAATGGACCACGG | ||
| Tbp-F | TAAGCCCCAACTTCTCGATTCC | ||
| Tbp-R | GCCAAAGAGACCTGATCCCC | ||
A genomic region on the second chromosome (starting from 2L: 2753160) within the first intron of Bacc gene was identified (Fig. 3, d and e) as the immediate sequence flanking the inserted P-element 3′ end. To verify this identified genomic insertion site, a PCR assay was designed to amplify the genomic regions from Or47b-GAL4 transgenic flies and w1118 control flies respectively with different primer pairs (Fig. 3, e and f). PCR mix recipe and thermal cycling programmes were the same as abovementioned. Amplicons were also purified and sequenced for validation.
Quantitative reverse transcription-PCR
To measure whether the insertion of Or47b-GAL4 transgene into Bacc gene locus affects its expression, we used the quantitative reverse transcription-PCR (qRT-PCR) to extract and quantify the mRNA levels from whole heads of fruit flies. RNA extraction, cDNA preparation, and qPCR protocols were described previously (Zhao et al. 2020; Deanhardt et al. 2022).
Briefly, for each biological replicate, equal number of male and female heads (7 to 10, 5–7 days old post eclosion) were dissected in RNase-free environment with Trizol, followed by tissue homogenization, cell lysis, and filtration with Qiagen QIAshredder spin column. Three biological replicates were analyzed for each genotype. RNA was then extracted and purified by Qiagen RNeasy Kit per manufacturer's instructions and eluted in 60 μl RNase-free water. Genomic DNA was then removed using Invitrogen TURBO DNA-free Kit per manufacturer's instructions. RNA concentration was measured using NanoDrop after DNase treatment. Reverse transcription was performed by Invitrogen SuperScript IV First-Strand cDNA synthesis Reaction Kit per manufacturer's instructions. Notably, approximately equal amount of template RNA across different samples were added based on RNA concentration.
Lastly, qPCR reactions were run on Roche LightCycler 96 Instrument with FastStart Essential DNA Green Master (2X) in 20 μl volume with technical triplicates per manufacturer's instructions. Thermal cycling programme: 600 s pre-incubation at 95°C + 40 three-step amplification cycles (10 s denaturation at 95°C, 10 s annealing at 55°C, 15 s extension at 72°C with Single acquisition) + Melting Curve (10 s 95°C, 60 s 65°C, 1 s 97°C with 5 Readings/°C). qPCR primers were designed to span two adjacent exons when possible, with target amplicon length of 120 bp. Primer pairs were tested to generate standard curves to evaluate amplification efficiency before being used for expression comparison experiments. Expression levels of the gene of interest were normalized to the house keeping gene RpL13A in each sample by 2−ΔCt method. Two-way ANOVA followed by multiple comparison test was performed in Prism 9 software to determine statistical significance. Primers are listed in Table 1.
Results
We used the Drosophila olfactory receptor neurons (ORNs) as a model to understand the molecular mechanisms underlying neuronal circuit assembly. In Drosophila, each class of ORNs expresses a unique olfactory receptor (Or) gene, and ORN axons target to the brain antennal lobe within class-specific and uniquely positioned synaptic units called glomeruli (Hong and Luo 2014; Barish and Volkan 2015). To identify the molecular players contributing to the glomerular organization of the ORNs, we genetically screened genes encoding cell adhesion molecules whose expression levels increase over pupal development in the antennae (Barish et al. 2018). Among these, beat and side gene families drew our attention because they encode the Ig superfamily proteins, form a heterophilic interacting protein network, and have been previously revealed to be involved in neuronal adhesion (Fambrough and Goodman 1996; Pipes et al. 2001; Sink et al. 2001; de Jong et al. 2005; Siebert et al. 2009; Özkan et al. 2013; Li et al. 2017; Kinold et al. 2021). We obtained a collection of transgenic UAS-RNAi lines from TriP library deposited at the Bloomington Drosophila Stock Center (BDSC) and crossed these lines with an established recombinant chromosome containing four different Or promoter-driven GAL4 transgenes (Or47a-GAL4, Or47b-GAL4, Or23a-GAL4, Gr21a-GAL4, 4xOr-GAL4 for short, Fig. 1 a–d) together with a UAS-Syt.GFP reporter. We examined the knockdown effect of candidate genes on axonal targeting of these four ORN classes. The parent flies with a single copy of the GAL4 drivers showed wild type glomerular organization (Fig. 1a). As an additional control, we also crossed GAL4 driver lines to flies expressing the RNAi against a red fluorescent protein (RFP) mCherry, which also exhibited no apparent defect in glomerular organization (Fig. 1a).
From the screen, we found a strikingly recurrent phenotype, where the axon terminals of Or47b ORNs invade the neighbouring region, leading to an expanded round VA1v glomerulus in contrast to the crescent shape in control brains (Fig. 1a). This phenotype was observed in two independent RNAi lines targeting the same gene, for example, beat-Ia (Fig. 1a). However, screening a list of beat and side family members revealed a pattern for the phenotype, which only correlated with the second chromosome UAS-RNAi transgenes, independent of the gene identity. Figure 1b summarizes the screening results from beat/side gene families. All the RNAi lines inserted at the second chromosome attP40 site yielded the expanded VA1v glomerulus phenotype, whereas none of the RNAi lines inserted at the third chromosome attP2 site showed this defect. Notably, there is one gene, beat-IIIc, with one attP40-derived RNAi line and one attP2-derived RNAi line. Only the attP40 UAS-RNAi insertion gave rise to the phenotype (Fig. 1b). The same phenotype was also observed with randomly selected TRiP UAS-RNAi lines inserted at the attP40 site targeting genes without known roles in ORN development (Supplementary Figure 1, a and b). To test whether this phenotype is caused by specific effects of RNAi-mediated gene knockdown or simply by the presence of attP40-derived insertions, we first crossed the same Or47b-GAL4 driver line to a third UAS-RNAi line from Vienna Drosophila Resource Center (VDRC) targeting beat-Ia, which was generated by random P-element-mediated insertions (Dietzl et al. 2007). This non-attP40 UAS-RNAi line could not reproduce the phenotype obtained by the attP40-derived UAS-RNAi from the TRiP collection (Fig. 1c). In addition, crossing the driver line to an empty attP40 site without any transgenes led to the same glomerular expansion phenotype (Fig. 1d). These results suggest that the Or47b ORN-specific VA1v glomerular defect is independent of the RNAi-based knockdown of the genes examined but caused by an effect from the attP40-derived chromosome.
Since we repeatedly obtained the VA1v glomerular phenotype with the second chromosome Or47b-GAL4-driven UAS-RNAi, we also tested if crossing flies carrying the same Or47b-GAL4 transgene to various attP40 derivatives could result in the same phenotype (Fig. 1e). Compared with the no attP40 control (over a CyO balancer chromosome), the attP40 landing site with and without UAS-RNAi insertion, or a ubiquitous promoter-driven gRNA targeting the QUAS sequence (control gRNA) all produced the same VA1v glomerular defect when crossed to the second chromosome Or47b-GAL4-driven UAS-Syt.GFP (Fig. 1f). We also crossed Or47b-GAL4 UAS-Syt.GFP chromosome to three lines carrying UAS-rCD2.RFP transgenic insertion at three different chromosomal locations, attP2 (on chr3), attP5 (on chr2R), and attP40 (on chr2L). Only Or47b-GAL4 over UAS-rCD2.RFP insertion at attP40 resulted in glomerular expansion phenotype while the insertions at attP2 and attP5 appeared wild type. These results again suggest that the VA1v glomerular defect is uniquely linked to the attP40-associated insertions and is independent of the transgene or other attP landing sites.
The glomerular organization defect could be caused by simply the presence of attP40 insertion or the genetic interaction between the attP40 background and the chromosome carrying the reporter transgene. To distinguish between these possibilities, we examined the animals carrying a single copy of attP40 insertion and a different Or47b-GAL4 UAS-Syt.GFP reporters on the third chromosome. In these animals, VA1v glomerulus appeared normal (Fig. 1, h and i). The observation that a single copy of attP40 is not sufficient to produce a glomerular phenotype indicates that the attP40 effects on VA1v glomerulus are not dominant. Rather, they point to a combinatorial effect of the second chromosome with Or47b-GAL4 UAS-Syt.GFP over the chromosome with the empty or transgene-carrying attP40 docking site on the glomerular phenotype (Fig. 1, h and i). Thus, we conclude that the attP40 chromosome genetically interacts with the second chromosome reporters to disrupt VA1v glomerular organization.
As the attP40 effect appears to be recessive, we next examined if animals homozygous for the attP40 sites display any VA1v glomerular defects. To bypass the glomerular defects arising from the genetic interactions between attP40 and the second chromosome reporters, we used the third chromosome Or47b-GAL4 UAS-Syt.GFP reporter to visualize VA1v glomerulus. Surprisingly, homozygous attP40 derivatives or attP40 empty docking site alone produced strong axon terminal defects (Fig. 2 a–f). In contrast, flies heterozygous for the attP40 site with or without transgenes inserted appeared wild type (Fig. 2 a–f). Most of the brains homozygous for the attP40 site with or without insertions displayed a dorsally positioned VA1v glomerulus (Fig. 2, b and e, middle panels; Fig. 2, c and f), whereas a small proportion also exhibited an expanded glomerulus (Fig. 2, b and e, right panels; Fig. 2, c and f). Given that the attP40 site is located within an intron of Msp300 gene, we posited that it likely disrupts Msp300 function. Msp300 encodes a Nesprin-like protein, which is required for proper positioning of muscle nuclei and neuromuscular junction formation (Elhanany-Tamir et al. 2012; Morel et al. 2014). Single-cell RNA-seq datasets from ORNs also show broad expression of Msp300 across ORN classes (Li et al. 2022). We thus tested if the VA1v glomerular defect is caused by the loss of Msp300 function. We analyzed transheterozygotes of empty attP40 docking site over other mutant alleles of Msp300, such as Msp300ΔKASH (which lacks the KASH domain [Xie and Fischer 2008; Elhanany-Tamir et al. 2012)], Msp300MI00111, Msp300MI01145 [two MIMIC-based alleles predicted to disrupt most splice isoforms of Msp300 transcripts (Venken et al. 2011)], and Msp300KG03631 [a P-element-based insertion which is close to attP40 landing site (Bellen et al. 2004)] (Fig. 2g). However, none of these genetic combinations recapitulated VA1v glomerular phenotype (Fig. 2h). This indicates that the VA1v glomerular defect is independent of the Msp300 function and is likely caused by other genes nearby affected by the attP40 insertion or a second recessive mutation linked to the attP40 docking site.
We next sought to figure out the molecular basis of the genetic interaction between that specific Or47b-GAL4-bearing chromosome and attP40 chromosome. Flies transheterozygous for attP40 (empty or with insertions) over 4xOr-GAL4 UAS-Syt.GFP or Or47b-GAL4 UAS-Syt.GFP robustly exhibit VA1v glomerular phenotype. We infer that the putative genetic lesion is directly caused by or genetically linked to the Or47b-GAL4 transgene for three reasons: (1) a farther second site mutation would likely be lost during meiotic recombination events to generate these stocks; (2) this Or47b-GAL4 recombined with other UAS-reporters, UAS-mCD8.GFP or UAS-RFP, over the attP40 derivatives exhibited the same phenotype (Supplementary Figure 2); and (3) crossing the second chromosome Or47b-GAL4 transgene alone to the UAS-rCD2.RFP reporter inserted at attP40 also reproduced the expanding glomerulus (Fig. 3a), which rules out the confounding effect from UAS-Syt.GFP transgene. Indeed, two copies of this Or47b-GAL4 chromosome also results in similar glomerular phenotypes (Fig. 3a). Or47b-GAL4 transgene was generated by P-element-mediated genomic integration (Fig. 3b) and the exact site of the insertion was not mapped (Vosshall et al. 2000). We used inverse PCR (Huang et al. 2009) to identify the insertion site and determine the gene whose function is potentially disrupted (Fig. 3c). We successfully recovered a piece of genomic sequence immediately flanking the 3′ end of the inserted P-element, which is within the first intron of Bacc gene (chr2L:2753160, Fig. 3, d and e) and also a P-element insertion-enriched region. We performed genomic PCR to further validate the Bacc intronic insertion from both 3′ and 5′ ends of the P-element using different primer pairs targeting the P-element ends and flanking Bacc sequences (Fig. 3e). We amplified the desired DNA fragments from Or47b-GAL4-bearing flies but not from w1118 flies (Fig. 3, e and f). In addition, primer pairs targeting only Bacc sequences flanking the insertion amplified the expected fragment only from w1118 files but not from Or47b-GAL4 flies (Fig. 3, e and f). We next tested if this intronic insertion affects Bacc transcriptional levels using Quantitative Reverse Transcription-PCR (qRT-PCR). We extracted mRNA from whole heads of the adult Or47b-GAL4 homozygotes, as well as homozygous attP2, attP40, and w1118 adults. qRT-PCR results showed that Bacc transcripts normalized to the housekeeping gene RpL13A decrease by ∼two fold in flies homozygous for Or47b-GAL4 but are not significantly altered in attP2 or attP40 animals compared to w1118 controls (Fig. 3g). In contrast, other housekeeping genes Act5C and Tbp remain unchanged across these genotypes (Fig. 3g). These results indicate that: (1) second chromosome Or47b-GAL4 transgene insertion disrupts Bacc gene function and (2) homozygous and transheterozygous combinations of Or47b-GAL4 and attP40 backgrounds likely utilize distinct mechanisms to disrupt VA1v glomerular organization.
Discussion
Here, we found that homozygous attP40 chromosome leads to defective glomerular organization of ORNs. This defect is likely not caused by the loss of Msp300 function, where the attP40 site is inserted. Moreover, the attP40 chromosome genetically interacts with a second chromosome carrying the Or47b-GAL4 transgene, resulting in a similar ORN axon terminal defect. Though the exact genetic reasons and molecular mechanisms are unknown, our finding raises the critical issue with using this popular transgene landing site. Rigorous controls are needed to rule out the attP40-associated background effects, as discussed below.
The genetics underlying the Or47b ORN phenotypes
A recent study reported that flies homozygous for the attP40-derived insertions had 50% reduction in Msp300 transcript levels and phenocopied the defects in larval muscle nuclei clustering in Msp300 mutants (van der Graaf et al. 2022). As homozygotes of the attP40 chromosome are defective in Or47b ORN axon terminal organization, we hypothesized that the attP40-affected Msp300 gene is responsible for the defect. However, this is not the case as attP40 over various Msp300 mutations appeared phenotypically wild type, suggesting the attP40 chromosome may carry an unannotated mutation responsible for Msp300-independent ORN glomerular disorganization.
The attP40 docking site with or without transgene insertions may also disrupt other genes in the vicinity of Msp300. For example, in addition to Msp300, attP40 docking site is flanked on the opposing side by ND-13A, which encodes a component of the mitochondria electron transport chain complex I. Thus, the attP40 docking site alone or with transgene insertions may lead to a variety of phenotypes as a result of disrupted ND-13A. Indeed, Groen et al. reported that attP40 flies exhibit resistance to cisplatin-induced neuronal damage mediated by the reduced expression of ND-13A (Groen et al. 2022). Whether the glomerular defect is dependent on the ND-13A function is beyond the scope of this paper but needs to be tested in the future studies.
Surprisingly, we found transheterozygous animals with an attP40 chromosome over the second chromosome Or47b-GAL4 transgene produced similar but not identical glomerular abnormalities to attP40 homozygotes. Additionally, Or47b-GAL4 homozygotes exhibit comparable phenotypes with Or47b-GAL4/attP40 transheterozygotes. These suggest several possible underlying genetic mechanisms: (1) Or47b-GAL4 and attP40 backgrounds harbour common mutations; (2) Or47b-GAL4 and attP40 backgrounds possess completely separate genetic lesions that genetically interact. The genetic interaction model is favoured due to qualitatively distinguishable phenotypes between Or47b-GAL4/attP40 animals and attP40/attP40 ones. Furthermore, Or47b-GAL4 transgene is inserted into an intron of Bacc gene, which encodes a tyramine-dependent nuclear regulator (Chen et al. 2013), reducing its expression levels by about 50% in homozygotes. No change in transcript levels were observed in attP40 homozygotes. Bacc mRNAs are abundant in brain tissues, comparable to housekeeping genes Act5C and RpL13A (Fig. 3g). Our results imply its potential novel role in ORN axon pathfinding or glomerular patterning. Future functional studies will determine whether the disruption of Bacc expression is causative to the VA1v glomerular phenotype and the mechanisms by which Bacc mutations and their genetic interactors in the attP40 background result in glomerular defects.
Unique genetic sensitivity of VA1v glomerulus architecture
One of the most peculiar observations from our study is that Or47b ORNs seem to be particularly sensitive to changes in genetic background. In fact, VA1v glomerular disruptions are not only restricted to the attP40 background, but can be seen in many other mutants with effects on ORN axon and synapse organization in the antennal lobes (Ang et al. 2003; Yao et al. 2007; Hong et al. 2012; Li et al. 2013; Hueston et al. 2016; Wu et al. 2017; Xie et al. 2019; Hing et al. 2020). In addition to the variability of VA1v glomerular architecture to genetic background effects, pheromone sensing Or47b ORNs and the trichoid at4 sensillum that houses Or47b, Or88a, and Or65a/b/c ORNs are developmentally special. At4 sensillum appears to be a developmentally default state for all trichoid sensilla. For example, loss of transcription factor Rn function, normally expressed in at1 and at3 ORNs, leads to a loss of at1 and at3 sensilla identity, and their conversion to at4 sensillum identity (Li et al. 2013, 2015, 2016). Similarly, Or47b ORNs in at4 sensillum appear to have a default identity, as mutants in Alh, a chromatin factor, result in the conversion of Or88a and Or65a ORNs to Or47b ORN fate (Hueston et al. 2016). In addition to these findings that point to a developmentally special state for pheromone sensing Or47b ORNs or “at” sensilla, the glomeruli targeted by the trichoid ORNs are morphologically plastic. In Drosophila, they are sexually dimorphic, appearing larger in males (Stockinger et al. 2005). In insects such as moths trichoid glomeruli can form separate macro-glomerular complex outside the antennal lobe of male brains (Berg et al. 1998). Given the developmental plasticity of trichoid pheromone sensing ORNs and the developmental ground state of at4 sensilla, Or47b developmental trajectory might be particularly sensitive to genetic background effects to accommodate adaptive developmental, behavioural and evolutionary processes. On the other hand, we only examined Or47b VA1v glomerulus in the attP40 background, and attP40 homozygotes possibly display structural defects in other ORN classes and their glomeruli. Future studies will help identify these phenotypes and the genetic lesions leading to attP40-associated phenotypes.
Addressing genetic background issues when using genetic reagents
To summarize, we found unexpected background effects of the Drosophila attP40 landing site on the ORN glomerular organization. In parallel with other recent studies reporting other phenotypes arising from the attP40 background, ranging from muscle development to neuronal stress responses, such background effects should be seriously considered in using attP40-derived flies. It is recommended to avoid using homozygotes/double-copies of the attP40-based insertions. Researchers should also be aware of the potential genetic interactions between the attP40-bearing chromosome and the other homologous second chromosomes even if it does not contain any attP40 derivatives. Appropriate controls should be applied to override these caveats. For example, when working with GAL4/UAS-effector binary system, it is better to use a GAL4-driven UAS-neutral effector (such as UAS-RNAi against neutral or non-fly genes inserted at the same docking site) as a negative control, rather than the widespread use of GAL4 alone or UAS-effector alone controls. Transgenic rescue of RNAi-based gene knockdowns is not feasible due to targeting of rescue transgenes by the RNAi. Thus, use of full animal mutants or MARCM based clonal mutant analysis should be coupled with RNAi-based phenotypic analyses. Though the underlying genetic reasons remain elusive, studies demonstrated that the attP40 landing site on the second chromosome affects the expression of multiple genes (Groen et al. 2022; van der Graaf et al. 2022). Additional omics-based experiments in the future will be needed to determine all the genetic lesions in attP40 strains that underly many phenotypic defects observed in this background. These studies will also reveal potential genetic alterations associated with glomerular defects, providing new insights into ORN axon pathfinding and glomerular organization.
Supplementary Material
Acknowledgments
We would like to thank Bloomington Drosophila Stock Center and Vienna Drosophila Resource Center for providing all the fly stocks. We thank Duke Light Microscopy Core Facility for help with imaging. We thank Chengcheng Du for help with molecular biology.
Contributor Information
Qichen Duan, Department of Biology, Duke University, Durham, NC 27708, USA.
Rachel Estrella, Department of Biology, Duke University, Durham, NC 27708, USA.
Allison Carson, Department of Biology, Duke University, Durham, NC 27708, USA.
Yang Chen, Department of Biology, Duke University, Durham, NC 27708, USA.
Pelin C Volkan, Department of Biology, Duke University, Durham, NC 27708, USA.
Data availability
The authors affirm that all the data necessary for drawing the conclusions are present in the text, figures, and figure legends. Most of the Drosophila stocks are obtained from Bloomington or Vienna Stock center, with identifiers listed in the Materials and methods section. All the other lines are available upon request.
Supplemental material available at G3 online.
Funding
This study is funded by grant NSF 2006471 and NIH 5R01NS109401 (both to P.C.V.). Q.D. is supported by Duke Biology Department Ph.D. programme.
Author contributions
Q.D. and P.C.V. conceived the study and designed the experiments; Q.D. did most of the experiments with help from R.E., A.C., and Y.C.; Q.D. analyzed the data and prepared the figures; Q.D. and P.C.V. wrote and edited the manuscript.
Literature cited
- Ang LH, Kim J, Stepensky V, Hing H. Dock and Pak regulate olfactory axon pathfinding in Drosophila. Development. 2003;130(7):1307–1316. doi: 10.1242/dev.00356. [DOI] [PubMed] [Google Scholar]
- Barish S, Nuss S, Strunilin I, Bao S, Mukherjee S, et al. Combinations of DIPs and Dprs control organization of olfactory receptor neuron terminals in Drosophila. PLoS Genet. 2018;14(8):e1007560. doi: 10.1371/journal.pgen.1007560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish S, Volkan PC. Mechanisms of olfactory receptor neuron specification in Drosophila. Wiley Interdiscip Rev Dev Biol. 2015;4(6):609–621. doi: 10.1002/wdev.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167(2):761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg B, Almaas T, Bjaalie J, Mustaparta H. The macroglomerular complex of the antennal lobe in the tobacco budworm moth Heliothis virescens: specified subdivision in four compartments according to information about biologically significant compounds. J Comp Physiol A. 1998;183(6):669–682. doi: 10.1007/s003590050290. [DOI] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Chen J, Wang Y, Zhang Y, Shen P. Mutations in bacchus reveal a tyramine-dependent nuclear regulator for acute ethanol sensitivity in Drosophila. Neuropharmacology. 2013;67:25–31. doi: 10.1016/j.neuropharm.2012.10.013. [DOI] [PubMed] [Google Scholar]
- Deanhardt B, Duan Q, Du C, Soeder C, Morlote A, et al. Social experience and pheromone receptor activity reprogram behavioral switch and neuromodulatory gene expression in sensory neurons. bioRxiv 449021. doi: 10.1101/2021.06.18.449021, 16 August2022, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong S, Cavallo JA, Rios CD, Dworak HA, Sink H. Target recognition and synaptogenesis by motor axons: responses to the sidestep protein. Int J Dev Neurosci. 2005;23(4):397–410. doi: 10.1016/j.ijdevneu.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su K-C, Barinova Y, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448(7150):151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Elhanany-Tamir H, Yu YXV, Shnayder M, Jain A, Welte M, et al. Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J Cell Biol. 2012;198(5):833–846. doi: 10.1083/jcb.201204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fambrough D, Goodman CS. The Drosophila beaten path gene encodes a novel secreted protein that regulates defasciculation at motor axon choice points. Cell. 1996;87(6):1049–1058. doi: 10.1016/S0092-8674(00)81799-7. [DOI] [PubMed] [Google Scholar]
- Fishilevich E, Vosshall LB. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005;15(17):1548–1553. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the gene ontology consortium. Brief Bioinform. 2011;12(5):449–462. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EW, Fedele G, Giorgini F, Kyriacou CP. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat Methods. 2014;11(3):222–223. doi: 10.1038/nmeth.2856. [DOI] [PubMed] [Google Scholar]
- Groen CM, Podratz JL, Pathoulas J, Staff N, Windebank AJ. Genetic reduction of mitochondria complex I subunits is protective against cisplatin-induced neurotoxicity in Drosophila. J Neurosci. 2022;42(5):922–937. doi: 10.1523/JNEUROSCI.1479-20.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage φC31. Genetics. 2004;166(4):1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hing H, Reger N, Snyder J, Fradkin LG. Interplay between axonal Wnt5-Vang and dendritic Wnt5-Drl/Ryk signaling controls glomerular patterning in the Drosophila antennal lobe. PLoS Genet. 2020;16(5):e1008767. doi: 10.1371/journal.pgen.1008767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Luo L. Genetic control of wiring specificity in the fly olfactory system. Genetics. 2014;196(1):17–29. doi: 10.1534/genetics.113.154336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Mosca TJ, Luo L. Teneurins instruct synaptic partner matching in an olfactory map. Nature. 2012;484(7393):201–207. doi: 10.1038/nature10926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housden BE, Muhar M, Gemberling M, Gersbach CA, Stainier DY, et al. Loss-of-function genetic tools for animal models: cross-species and cross-platform differences. Nat Rev Genet. 2017;18(1):24–40. doi: 10.1038/nrg.2016.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AM, Rehm EJ, Rubin GM. Recovery of DNA sequences flanking P-element insertions in Drosophila: inverse PCR and plasmid rescue. Cold Spring Harb Protoc. 2009;2009(4):pdb.prot5199. doi: 10.1101/pdb.prot5199. [DOI] [PubMed] [Google Scholar]
- Hueston CE, Olsen D, Li Q, Okuwa S, Peng B, et al. Chromatin modulatory proteins and olfactory receptor signaling in the refinement and maintenance of fruitless expression in olfactory receptor neurons. PLoS Biol. 2016;14(4):e1002443. doi: 10.1371/journal.pbio.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinold JC, Brenner M, Aberle H. Misregulation of Drosophila sidestep leads to uncontrolled wiring of the adult neuromuscular system and severe locomotion defects. Front Neural Circuits. 2021;15:658791. doi: 10.3389/fncir.2021.658791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin A, Marygold SJ, Antonazzo G, Attrill H, dos Santos G, et al. Flybase: updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2020;49(D1):D899–D907. doi: 10.1093/nar/gkaa1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Barish S, Okuwa S, Maciejewski A, Brandt AT, et al. A functionally conserved gene regulatory network module governing olfactory neuron diversity. PLoS Genet. 2016;12(1):e1005780. doi: 10.1371/journal.pgen.1005780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Barish S, Okuwa S, Volkan PC. Examination of endogenous rotund expression and function in developing Drosophila olfactory system using CRISPR-Cas9-mediated protein tagging. G3 (Bethesda). 2015;5(12):2809–2816. doi: 10.1534/g3.115.021857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Ha TS, Okuwa S, Wang Y, Wang Q, et al. Combinatorial rules of precursor specification underlying olfactory neuron diversity. Curr Biol. 2013;23(24):2481–2490. doi: 10.1016/j.cub.2013.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Janssens J, De Waegeneer M, Kolluru SS, Davie K, et al. Fly cell atlas: a single-nucleus transcriptomic atlas of the adult fruit fly. Science. 2022;375(6584):eabk2432. doi: 10.1126/science.abk2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Watson A, Olechwier A, Anaya M, Sorooshyari SK, et al. Deconstruction of the beaten path-sidestep interaction network provides insights into neuromuscular system development. Elife. 2017;6:e28111. doi: 10.7554/eLife.28111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40(4):476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel V, Lepicard S, Rey AN, Parmentier M-L, Schaeffer L. Drosophila nesprin-1 controls glutamate receptor density at neuromuscular junctions. Cell Mol Life Sci. 2014;71(17):3363–3379. doi: 10.1007/s00018-014-1566-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J-Q, Liu L-P, Binari R, Hardy R, Shim H-S, et al. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182(4):1089–1100. doi: 10.1534/genetics.109.103630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J-Q, Markstein M, Binari R, Pfeiffer B, Liu L-P, et al. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods. 2008;5(1):49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J-Q, Zhou R, Czech B, Liu L-P, Holderbaum L, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8(5):405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özkan E, Carrillo RA, Eastman CL, Weiszmann R, Waghray D, et al. An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell. 2013;154(1):228–239. doi: 10.1016/j.cell.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Holderbaum L, Tao R, Hu Y, Sopko R, et al. The transgenic RNAi project at harvard medical school: resources and validation. Genetics. 2015;201(3):843–852. doi: 10.1534/genetics.115.180208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipes GC, Lin Q, Riley SE, Goodman CS. The beat generation: a multigene family encoding IgSF proteins related to the beat axon guidance molecule in Drosophila. Development. 2001;128(22):4545–4552. doi: 10.1242/dev.128.22.4545. [DOI] [PubMed] [Google Scholar]
- Ramin M, Li Y, Chang WT, Shaw H, Rao Y. The peacefulness gene promotes aggression in Drosophila. Mol Brain. 2019;12(1):1. doi: 10.1186/s13041-018-0417-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K, Brady R Jr, Cravchik A, Morozov P, Rzhetsky A, et al. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell. 2001;104(5):661–673. doi: 10.1016/S0092-8674(01)00263-X. [DOI] [PubMed] [Google Scholar]
- Siebert M, Banovic D, Goellner B, Aberle H. Drosophila motor axons recognize and follow a sidestep-labeled substrate pathway to reach their target fields. Genes Dev. 2009;23(9):1052–1062. doi: 10.1101/gad.520509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sink H, Rehm EJ, Richstone L, Bulls YM, Goodman CS. Sidestep encodes a target-derived attractant essential for motor axon guidance in Drosophila. Cell. 2001;105(1):57–67. doi: 10.1016/S0092-8674(01)00296-3. [DOI] [PubMed] [Google Scholar]
- Stockinger P, Kvitsiani D, Rotkopf S, Tirian L, Dickson BJ. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121(5):795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- Titlow J, Robertson F, Järvelin A, Ish-Horowicz D, Smith C, et al. Syncrip/hnRNP Q is required for activity-induced Msp300/nesprin-1 expression and new synapse formation. J Cell Biol. 2020;219(3):e201903135. doi: 10.1083/jcb.201903135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaf K, Srivastav S, Singh P, McNew JA, Stern M. The Drosophila melanogaster attP40 docking site and derivatives are insertion mutations of msp-300. PLoS One. 2022;17(12):e0278598. doi: 10.1371/journal.pone.0278598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011;8(9):737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers JH, Manning SA, Kulkarni A, Harvey KF. A Drosophila RNAi library modulates hippo pathway-dependent tissue growth. Nat Commun. 2016;7(1):10368. doi: 10.1038/ncomms10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000;102(2):147–159. doi: 10.1016/S0092-8674(00)00021-0. [DOI] [PubMed] [Google Scholar]
- Wu B, Li J, Chou YH, Luginbuhl D, Luo L. Fibroblast growth factor signaling instructs ensheathing glia wrapping of Drosophila olfactory glomeruli. Proc Natl Acad Sci U S A. 2017;114(29):7505–7512. doi: 10.1073/pnas.1706533114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Fischer JA. On the roles of the Drosophila KASH domain proteins Msp-300 and klarsicht. Fly (Austin). 2008;2(2):74–81. doi: 10.4161/fly.6108. [DOI] [PubMed] [Google Scholar]
- Xie Q, Wu B, Li J, Xu C, Li H, et al. Transsynaptic fish-lips signaling prevents misconnections between nonsynaptic partner olfactory neurons. Proc Natl Acad Sci U S A. 2019;116(32):16068–16073. doi: 10.1073/pnas.1905832116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Wu Y, Yin C, Ozawa R, Aigaki T, et al. Antagonistic roles of Wnt5 and the Drl receptor in patterning the Drosophila antennal lobe. Nat Neurosci. 2007;10(11):1423–1432. doi: 10.1038/nn1993. [DOI] [PubMed] [Google Scholar]
- Zhao S, Deanhardt B, Barlow GT, Schleske PG, Rossi AM, et al. Chromatin-based reprogramming of a courtship regulator by concurrent pheromone perception and hormone signaling. Sci Adv. 2020;6(21):eaba6913. doi: 10.1126/sciadv.aba6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Buchwalter RA, Zheng C, Wight EM, Chen JV, et al. A perinuclear microtubule-organizing centre controls nuclear positioning and basement membrane secretion. Nat Cell Biol. 2020;22(3):297–309. doi: 10.1038/s41556-020-0470-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirin J, Hu Y, Liu L, Yang-Zhou D, Colbeth R, et al. Large-Scale transgenic Drosophila resource collections for loss- and gain-of-function studies. Genetics. 2020;214(4):755–767. doi: 10.1534/genetics.119.302964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors affirm that all the data necessary for drawing the conclusions are present in the text, figures, and figure legends. Most of the Drosophila stocks are obtained from Bloomington or Vienna Stock center, with identifiers listed in the Materials and methods section. All the other lines are available upon request.
Supplemental material available at G3 online.