Abstract
The severe acute respiratory syndrome coronavirus 2, the agent of the ongoing coronavirus disease 2019 (COVID‐19) pandemic, has spread worldwide since it was first identified in November 2019 in Wuhan, China. Since then, progress in pathogenesis linked severity of this systemic disease to the hyperactivation of network of cytokine‐driven pro‐inflammatory cascades. Here, we aimed to identify molecular biomarkers of disease severity by measuring the serum levels of inflammatory mediators in a Brazilian cohort of patients with COVID‐19 and healthy controls (HCs). Critically ill patients in the intensive care unit were defined as such by dependence on oxygen supplementation (93% intubated and 7% face mask), and computed tomography profiles showing ground‐glass opacity pneumonia associated to and high levels of D‐dimer. Our panel of mediators included HMGB1, ATP, tissue factor, PGE2, LTB4, and cys‐LTs. Follow‐up studies showed increased serum levels of every inflammatory mediator in patients with COVID‐19 as compared to HCs. Originally acting as a transcription factor, HMGB1 acquires pro‐inflammatory functions following secretion by activated leukocytes or necrotic tissues. Serum levels of HMGB1 were positively correlated with cys‐LTs, D‐dimer, aspartate aminotransferase, and alanine aminotransferase. Notably, the levels of the classical alarmin HMGB1 were higher in deceased patients, allowing their discrimination from patients that had been discharged at the early pulmonary and hyperinflammatory phase of COVID‐19. In particular, we verified that HMGB1 levels above 125.4 ng/ml is the cutoff that distinguishes patients that are at higher risk of death. In conclusion, we propose the use of serum levels of HMGB1 as a biomarker of severe prognosis of COVID‐19.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Pneumonia and acute respiratory distress syndrome are the major complications of coronavirus disease 2019 (COVID‐19). Severe acute respiratory syndrome coronavirus 2 infection can activate innate and adaptive immune responses and result in massive inflammatory responses later in the disease. These uncontrolled inflammatory responses may lead to local and systemic tissue damage and co‐infections due to intensive care unit environment. This scenario makes it difficult to administer direct treatment of the disease. It is necessary to investigate inflammatory biomarkers that precociously distinguish the worst prognosis.
WHAT QUESTION DID THIS STUDY ADDRESS?
We aimed to identify molecular biomarkers of disease severity in a Brazilian cohort of patients with COVID‐19 by measuring the serum levels of inflammatory mediators as high mobility group box 1 (HMGB1), ATP, tissue factor, PGE2, LTB4, and cys‐LTs at different phases of the disease.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
Our work confirms HMGB1 as an important endogenous danger signal during COVID‐19 and provides evidence that high levels of HMGB1 in the circulation of patients with severe COVID‐19 orchestrates the acute and persistent mediators storm, in association with several other mediators. Indeed, we demonstrated that HMGB1 distinguishes patients that are at higher risk of death during the early hyperinflammatory phase.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
The results suggest that the alarmin HMGB1 could be a severity biomarker for COVID‐19, useful to distinguish patients that are at higher risk of death, and a potential target for innovative therapeutic strategies leading to a direct treatment for severe COVID‐19.
INTRODUCTION
Since the World Health Organization officially declared coronavirus disease 2019 (COVID‐19) as a pandemic, unparalleled efforts were dedicated to investigating the complex pathophysiology responses of the disease. Notwithstanding, the determinants of pathogenic outcome are still objects of intense debate, and further studies are required to identify prognostic biomarkers and potential therapeutic targets. COVID‐19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and present a broad range of symptoms, ranging from asymptomatic and mild cases to severe and critical cases. The latter comprises of 10–20% of symptomatic patients; 15% progress to severe pneumonia, and about 5% are admitted to the intensive care unit (ICU) due to acute respiratory distress syndrome and sepsis/multiple organ failure. 1 The world lethality rate is 2% accounting for more than 5 million deaths worldwide as of early January 2022. 2 , 3 This new disease presents a very particular feature related to inflammatory mediators, cell migration, and activation. 1 , 4 In addition, long‐term sequels have been described post‐COVID‐19. 5 , 6 , 7 , 8 , 9
A series of studies have reported an interindividual variability regarding incidence, severity, and mortality rate of the COVID‐19 outbreak. 10 These variabilities may be partially explained by gender, old age, and comorbidities, such as chronic obstructive pulmonary disease, diabetes, obesity, acute kidney injury, hypertension, cardiovascular diseases, cancer, and high levels of D‐dimer. 11 Chronic comorbidities are clinical risk factors for a fatal outcome associated with coronavirus. Additionally, angiotensin converting enzyme 2 (ACE2) and trans‐membrane protease serine type 2 (TMPRSS2) genetic polymorphisms might account for higher susceptibility and unexpected outcomes of COVID‐19 infections in different populations. 12 As well as individuals with a predisposition to lower production of type I IFN, who are also susceptible to aggravation of the disease. 13
Furthermore, patients with severe COVID‐19 present high levels of interleukin (IL‐1β), IL‐6, interferon γ‐induced protein 10 (IP‐10 or CXCL10), tumoral necrosis factor‐α (TNF‐α), C reactive protein (CRP), lactate dehydrogenase (LDH), and D‐dimer, which are well‐known markers of massive inflammation. 14 An overwhelming immune response aggravates COVID‐19 evolution and worsens clinical outcome, eventually leading to death. Among the broad range of endogenous mediators of inflammation, high mobility group box 1 (HMGB1) is a classic example of an alarmin, that is a nuclear protein released to interstitial spaces following leukocyte activation or cell death‐necrosis. 15 , 16 HMGB1 has a major role in chronic inflammation, being implicated in inflammasome assembly, 17 production of lipid mediators as prostaglandin E2 (PGE2), leukotriene B4 (LTB4), and cys‐LTs, 18 and recruitment of monocytes expressing tissue factor (TF), which contribute for neutrophil extracellular trap formation and consequently to immunothrombosis. 19 In addition, another relevant alarmin adenosine triphosphate (ATP) induces the release of HMGB1 during inflammation, which contributes to amplifying the inflammatory signals. 20
To date, it has been reported that HMGB1 levels in the blood of patients with COVID‐19 have increased, providing circumstantial evidence that this alarmin might contribute to disease outcome. 21 , 22 , 23 Interestingly, HMGB1 increased ACE2 expression on Calu‐3, HepG2, Caco2, and RT4 cells via an end‐product specific receptor (AGER). A different pathway, using TLR4 signaling, is responsible for TNF release by HMGB1. 22 , 24 Added to this, elevated serum levels of S100A8/A9 and HMGB1 correlated with COVID‐GRAM risk scores in critical hospitalized patients. 21
Awareness that HMGB1, lipid mediators, ATP, and TF are implicated in the pathogenesis of vascular and tissue damage during long‐term inflammation has led us to investigate their potential role as biomarkers of disease severity in COVID‐19.
SUBJECTS AND METHODS
Study design and patient selection
This is a prospective and qualitative study in collaboration with Biomedical Research Institute at Marcílio Dias Naval Hospital (MDNH, Rio de Janeiro, Brazil). The Biomedical Research Institute is responsible for the biorepository of COVID‐19's severe cases’ samples. This biorepository is a facility that stores samples of biological materials just for laboratory research. The sera from health donors and patients were collected from March 2020 until December 2020, which are related to the first (SARS‐CoV‐2 Beta variant) and second waves (SARS‐CoV‐2 Gamma variant) of COVID‐19 in Rio de Janeiro city. 25 , 26
The COVID‐19 cohort was established by samples collected from 73 patients randomly chosen (convenience sampling) from the MDNH repository of patients with severe cases of COVID‐19 (322 total cases), with at least two samples collected in different hospitalization days (except for a few patients that we were able to collect three samples), which ranged between 6 and 168 days of hospitalization. The 45 healthy controls (HCs) consisted of volunteer participants recruited by public divulgation of the study, from the community outside the Academy and hospital setting.
All serum samples had the levels of the inflammatory mediators measured. As the timing of COVID‐19 sample collection was very heterogeneous, we decided to analyze the data in two ways. First, for the analysis of the inflammatory mediator levels between HC and COVID‐19 cohorts, we plotted the highest value obtained among the 2–3 samples analyzed from each patient, regardless of the collection time. These data were used for Figures 3 and 4. Second, in order to better understand the timing and amount of cytokine production during the COVID‐19 progression, we evaluated and plotted the cytokine levels according to the course of the disease, determined as following: pulmonary phase with hypoxia from 2 to 11 days after hospitalization; hyper inflammatory early phase from 12 to 25 days after hospitalization; and hyper inflammatory late phase from 26 to 168 days after hospitalization (data in Figure 5).
FIGURE 3.
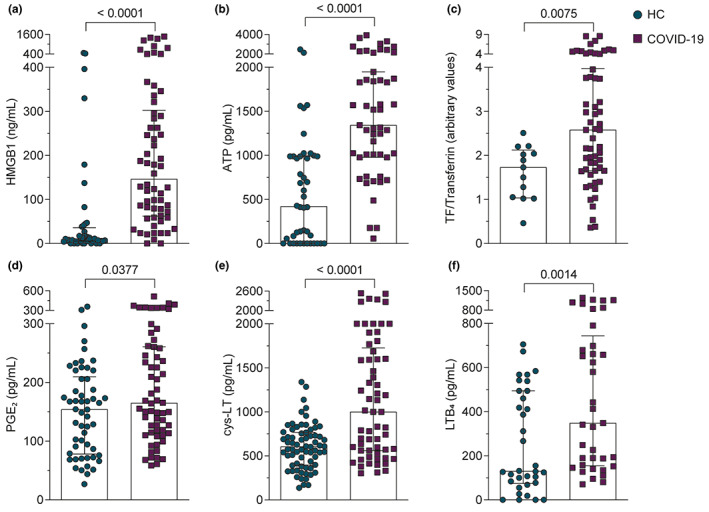
Patients with COVID‐19 present high serum levels of HMGB1, ATP, TF, PGE2, cys‐LT, and LTB4 compared to HCs. (a–f) We plotted the highest serum value obtained after analysis of two or three samples received from each patient with COVID‐19. Mediators were evaluated as described in Methods section. Data were presented as the median with interquartile range and were analyzed by Mann–Whitney test. Any p < 0.05 was considered statistically significant compared to HCs. COVID‐19, coronavirus disease 2019; HCs, healthy controls; TF, tissue factor
FIGURE 4.
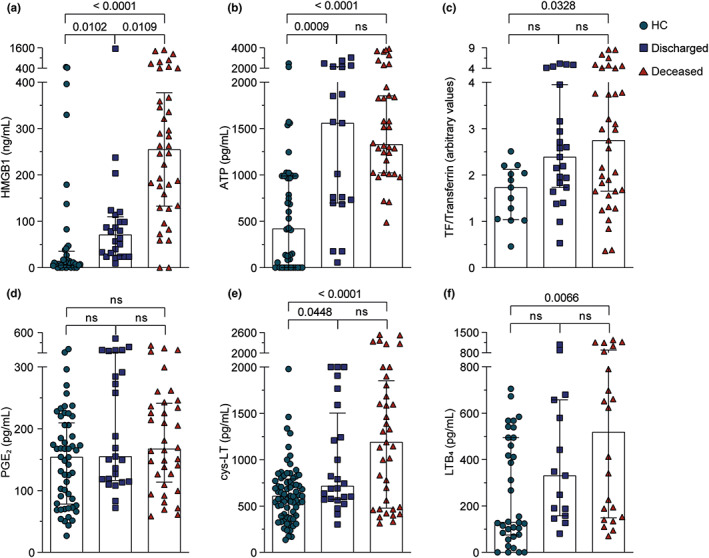
HMGB1 serum levels discriminate deceased from discharged patients with COVID‐19. (a–f) HMGB1, ATP, TF, PGE2, cys‐LT, and LTB4 levels were stratified between discharged and deceased in patients with COVID‐19 and compared to HCs. We plotted the highest serum value obtained after analysis of two or three samples received from each patient with COVID‐19. The data were presented as the median with interquartile range and were analyzed by Kruskal‐Wallis test. Any p < 0.05 was considered statistically significant compared to HCs or discharged groups. COVID‐19, coronavirus disease 2019; HCs, healthy controls; ns, not significant; TF, tissue factor
FIGURE 5.
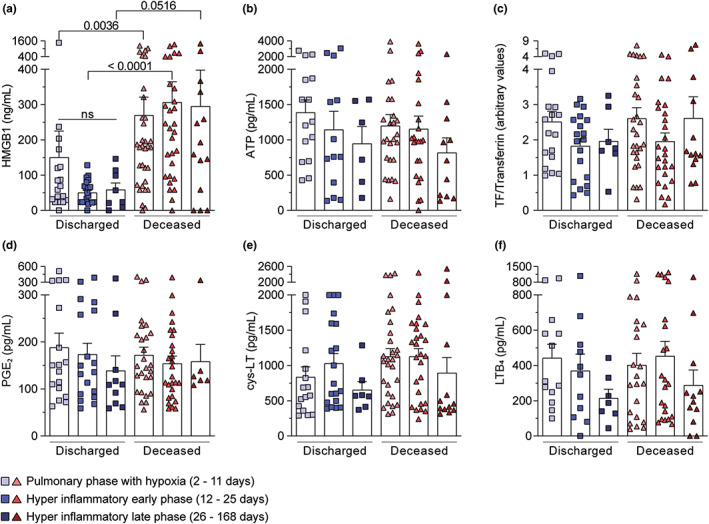
Higher HMGB1 serum levels were observed in the pulmonary phase with hypoxia and hyperinflammatory early phase according to clinical outcome. (a–f) The mediator values were stratified among three temporal phases after hospital admission: pulmonary phase with hypoxia (2–11 days), hyperinflammatory early phase (12–25 days), and hyperinflammatory late phase (26–168 days). Data were presented as mean ± SEM and were analyzed by Kruskal‐Wallis test. ns, not significant. Any p < 0.05 was considered statistically significant compared to the indicated phases
Demographic data, comorbidities, clinical treatments, and biological parameters for patients with COVID‐19 were obtained from medical records. For HCs, these data were collected by anamnesis.
The inclusion criteria for the HC cohort were patients over 18 years old that did not present any flu symptoms in the last 15 days before blood collection. We randomly recruited patients without or with comorbidities. The inclusion criteria for the COVID‐19 cohort were patients over 18 years old that tested positive for SARS‐CoV‐2 by reverse transcriptase‐polymerase chain reaction (RT‐PCR) for at least one respiratory sample. According to the sixth edition of Guidance for Coronavirus Disease 2019: Prevention, Control, Diagnosis, and Management, issued by China's National Health Commission, our cohort is composed only of patients with severe COVID‐19 admitted in the ICU with oxygen supplementation (93% intubated and 7% with face mask), pneumonia with ground‐glass opacity, and high levels of D‐dimer. About the exclusion criteria, the HC participants who revealed detectable levels of anti‐SARS‐CoV‐2 IgM or IgG were excluded from our cohort. Meanwhile, 12 patients with COVID‐19 were excluded because they acquired COVID‐19 after hospitalization for other reasons, such as cancer, risk pregnancy, car, knife, or gun accidents, or for absence of medical records. Thus, the amount of serum samples from patients with COVID‐19 in our cohort was 61 out of 73, as initially randomly chosen from the biorepository of severe cases of COVID‐19.
The clinical treatments of the patients in the ICU consisted of corticoids (dexamethasone and/or methylprednisolone and/or hydrocortisone), anticoagulant (enoxaparin), hydroxychloroquine, antibiotic cocktail (ceftriaxone, azithromycin, linezolid, moxifloxacin, meropenem, vancomycin, teicoplanin, cefepime, levofloxacin, amikacin, piperacillin, and polymyxin B), and sedative drugs for invasive ventilation. The treatment was performed individually according to the disease evolution; about hydroxychloroquine, it was used in the treatment of about 21 patients out of 61 in total.
Ethics statement
The present study was performed in accordance with regulation guidelines and approved by the Institutional Review Board of Ethics Committee of MDNH, where the serum samples analyzed in the present work were collected (32382820.3.0000.5256), and by the Institutional Review Board of Clementino Fraga Filho University Hospital (CFFUH), regulatory body responsible for approving clinical trials conducted in the Federal University of Rio de Janeiro (FURJ) (41002720.7.0000.5257). The written informed consents were obtained from all participants or their legal representatives.
Nucleic acid extraction and cDNA synthesis
Total nucleic acid extractions from nasopharyngeal swab samples were performed using the automated Maxwell System platform (No. AS4500; Promega, Madison, WI). Twenty‐five microliters of total RNA was submitted to cDNA synthesis with high‐capacity cDNA reverse transcription kits (No. 4368814; Thermo Fisher Scientific, Waltham, MA) and stored under −20°C. The extracted RNA was submitted to real‐time RT‐PCR for SARS‐CoV‐2 with of E and RP genes detection using the Fiocruz kit (Biomanguinhos, Rio de Janeiro, Brazil). Reverse transcription and amplification were performed in the QuantStudio 5 real‐time PCR system (Thermo Fisher Scientific).
Clinical samples and isolation of patient sera
Blood samples were collected from hospitalized patients with severe COVID‐19 (SARS‐CoV‐2‐positive by RT–qPCR and serology) admitted to the ICU at MDNH and from HCs (SARS‐CoV‐2‐negative by serology). Whole blood was collected using vacutainer and processed at the day of the collection. Serum samples were collected after centrifugation of whole blood at 1200 g for 10 min at 4°C. The undiluted serum was then transferred to 0.5 ml polypropylene conical tubes, aliquoted, and stored at −80°C for subsequent analysis. Repeated freeze–thaw cycles were avoided.
Enzyme‐linked immunosorbent assay for HMGB1
This assay was performed as described previously. 27 Briefly, the wells of a 96‐well microtiter plate (Greiner Bio‐One, Austria) were coated overnight at 4°C with anti‐HMGB1 mouse monoclonal antibody (No. H9537; Sigma‐Aldrich, St. Louis, MO) in phosphate‐buffered saline (PBS) buffer (8.06 mM sodium phosphate, 1.94 mM potassium phosphate, 2.7 mM KCl, and 137 mM NaCl) at pH 7.4. The plates were blocked for 2 h at 37°C with 1% bovine serum albumin (BSA) in PBS‐Tween (0.05% Tween 20 in PBS), then washed five times with PBS‐T buffer. Similar washing steps were performed at the end of each incubation period. The wells were then incubated with serial dilutions of rHMGB1 (standard curve) ranging from 4000 to 6.25 ng/ml or with patient sera (1:2 in PBS), for 2 h at 37°C. Subsequently, the wells were incubated with rabbit anti‐human rHMGB1 polyclonal antibody diluted in PBS buffer for 1 h at 37°C, and then incubated for 1 h at the same temperature with anti‐rabbit IgG antibody conjugated to horseradish peroxidase (No. W4011; Promega, Madison, WI). The reactions were visualized with OPD (No. P9187; Sigma‐Aldrich) and H2O2 as substrates and 12.5% H2SO4 as quencher. Reactions were monitored by measuring the absorbance at 490 nm in a SpectraMax M5/M5e Multimode Plate Reader (Molecular Devices, San José, CA). The standard curve calculation was performed using the mass value of the serial dilution of rHMGB1 protein against its respective optical density (OD) measurement. The OD measurements were normalized using the mean value of the negative control replicates. Each patient sample was tested just once.
Dosage of systemic levels of ATP
ATP circulating levels were measured in the serum of patients with COVID‐19 and HCs using a luciferase‐based assay kit. The Molecular Probes ATP Determination Kit (No. A22066; Thermo Fisher Scientific) was used according to the manufacturer's instructions. The luminescence of samples plated onto black 96‐well plates was read in a SpectraMax M5/M5e Multimode Reader, and results were expressed as pmol of ATP.
Measurement of lipid mediators
Lipid mediators present in the serum of HCs or SARS‐CoV‐2 infected patients were measured by commercial enzyme‐linked immunosorbent assay (ELISA) kits, according to the manufacturer's instructions (PGE2 No. 514010, LTB4 No. 520111, and cys‐LTs No.501070; Cayman Chemical, Ann Arbor, MI).
Western blot analysis of TF and transferrin
Serum from HCs and patients with COVID‐19 were reduced with β‐mercaptoethanol in sample buffer. Samples were subjected to 10% polyacrylamide gel electrophoresis followed by transfer to nitrocellulose membrane. The immunoblot was performed using anti‐TF (1:1000, No. ab104513; Abcam, Cambridge, UK) and anti‐transferrin as load control (1:10000, No. 82411; Abcam). The bands corresponding to both proteins were quantified using Image J software (National Institutes of Health, Bethesda, MD) and the ratio between TF and transferrin was calculated. The uncut gels images were included as Figure S1.
Measurement of serum anti‐SARS‐CoV‐2 antibodies
For quantitative analysis of anti‐SARS‐CoV‐2 spike protein IgM and IgG antibodies, we performed the S‐UFRJ test, as described previously. 28 Briefly, high binding ELISA plates were coated with 50 μl of SARS‐CoV‐2 spike protein (4 μg/ml in PBS) and incubated overnight. The coating solution was removed and 100 μl of PBS 1% BSA (blocking solution) was added and the plate was incubated at room temperature (RT) for 1–2 h. The blocking solution was removed and 50 μl of patient sera (1:40 in PBS 1% BSA) were added, subsequently, incubated at RT for 2 h. Then, the plate was washed with 150 μl of PBS (5×) and 50 μl of goat anti‐human IgM and IgG (Fc)‐horseradish peroxidase antibody (1:10000; Southern Biotech, Birmingham, AL) were added, and the plate was incubated for 1.5 h at RT. The plate was washed again with 150 μl of PBS (5×) and then treated with tetramethylbenzidine (TMB; 3,3′, 5,5; −tetramethylbenzidine; Scienco Biotech, Brazil) until the reaction was stopped with 50 μl of HCl 1 N. The OD was measured at 450 nm with 655 nm background compensation in a microplate reader (Bio‐Rad Laboratories).
Statistical analysis
A descriptive study of the cohort was conducted, presenting measures of central tendency and dispersion for continuous variables, or relative frequencies for each categorical variable. The Kolmogorov–Smirnov test was used to assess whether the variables were normally distributed. Continuous variables were presented as means ± SDs if normally distributed or as medians with 95% confidence interval (95% CIs) and interquartile ranges in a non‐normally distributed sample. Categorical variables were defined according to better‐ or worse‐expected prognostic values. Continuous variables were compared among multiple groups using analysis of variance followed by the Student–Newman–Keuls test for normally distributed variables or the Kruskal–Wallis test if variables were non‐normally distributed. The nonparametric Mann–Whitney U test was used for univariate comparisons of selected variables according to primary outcomes (discharged vs. deceased). The ability of HMGB1 to predict the final clinical outcome was analyzed using the receiving operating characteristic (ROC) curve and the area under the curve, accompanied by the 95% CI. The optimal cutoff value was determined by the point of maximal sensitivity and specificity, and then used to calculate the associated relative risk and 95% CI of the worst outcome (death). All p values reported are from two‐sided tests. The threshold for significance was set at p = 0.05 and highlighted in the tables and figures. Statistical analyses were performed in R version 3.1.1.
RESULTS
Cohort characteristics
Serum samples from COVID‐19 (n = 61) and HC (n = 45) subjects were obtained between March 23 and December 3, 2020, which were related to the first (SARS‐CoV‐2 Beta variant) and second waves (SARS‐CoV‐2 Gamma variant) of COVID‐19 in Rio de Janeiro city as previously described. 25 Our group did not perform the virus genotyping, we trusted the data released by groups specialized in genotyping circulating viruses. Sixty‐one patients with COVID‐19 admitted to the ICU after 1 to 14 days of symptom onset participated in this study. All subjects in the COVID‐19 cohort looked for medical attention reporting COVID‐19 symptoms for the first time, and they were tested positive for SARS‐CoV‐2 by RT‐PCR (Figure S2). We observed a mean of 6 days of symptoms onset until hospital admission and 38 days of hospitalization, similar data to those observed in the literature (Table 1). 29 , 30 The initial symptoms reported by our patients with COVID‐19 agreed with other studies with a predominance of fever, cough, dyspnea, fatigue, and myalgia (Table 1). HCs did not differ from patients with COVID‐19 related to age (patients with COVID‐19 = 28–86 years old, mean = 62.8 years old; and HCs = 28–79 years old, mean = 65 years old), although, the gender and comorbidity frequencies showed statistical differences between cohorts, despite the comorbidities being similar (Table 1). As expected, our COVID‐19 cohort was mainly composed of male patients (Table 1), corroborating a greater probability of men progressing to a more serious outcome. 12 The most common underlying diseases in our cohort were hypertension (patients = 72.1% vs. HCs = 33.3%), obesity (patients = 44.3% vs. HCs = 17.8%), and diabetes (patients = 29.5% vs. HCs = 13.3%). It is worthy to state that a significant number of patients presented two or more comorbidities (Table 1). The comorbidity frequencies of hypertension, diabetes, chronic lung disease, and vascular disease showed significant differences between the HC and COVID‐19 cohorts, whereas comorbidity frequencies of obesity and chronic heart disease were not statistically different (Table 1). The comorbidities were denied by seven patients in the COVID‐19 cohort and by 14 subjects in the HC group. Patients with COVID‐19 included in our cohort were categorized as severe/critical patients because all were in the ICU, presenting pneumonia and were under mechanical ventilation (57 patients [93%] or oxygen mask support 4 patients [7%]), whereas 60.7% of the patients died (Table 1).
TABLE 1.
Demographics characteristics and outcomes of subjects
| Healthy controls, n = 45 | Severe COVID‐19, n = 61 | p value* | |
|---|---|---|---|
| Age, years, mean ± SD (range) | 64.5 ± 14.4 (28–79) | 64.3 ± 13.2 (29–86) | ns |
| Sex | |||
| Women, n (%) | 27 (60) | 22 (36.1) | 0.014 |
| Men, n (%) | 18 (40) | 39 (63.9) | 0.014 |
| Death, n (%) | – | 37 (60.7) | |
| Onset of symptom to hospital admission, days | – | 6 ± 3.4 (1–14) | – |
| Hospitalization period, days | – | 38 ± 26.17 (6–168) | – |
| Invasive ventilation, n (%) | – | 57 (93.4) | – |
| Presenting symptoms, n (%) | |||
| Fever | – | 41 (67.2) | – |
| Cough | – | 37 (60.7) | – |
| Dyspnea | – | 32 (52.5) | – |
| Myalgia | – | 19 (31.2) | – |
| Fatigue | – | 18 (29.5) | – |
| Rhinorrhea | – | 10 (16.4) | – |
| Loss of smell | – | 10 (16.4) | – |
| Hyperoxia | – | 6 (9.8) | – |
| Headache | – | 6 (9.8) | – |
| Loss of taste | – | 5 (8.2) | – |
| Diarrhea | – | 5 (8.2) | – |
| Nausea and vomiting | – | 3 (4.9) | – |
| Chest pain | – | 3 (4.9) | – |
| Sore throat | – | 2 (3.3) | – |
| Comorbidities, n (%) | 31 (69) | 54 (88.5) | |
| Hypertension | 15 (33.3) | 44 (72.1) | <0.0001 |
| Diabetes | 6 (13.3) | 27 (44.3) | <0.0001 |
| Obesity | 8 (33.3) | 18 (29.5) | ns |
| Vascular disease | 1 (2.2) | 11 (18.0) | <0.0001 |
| Chronic lung disease | 1 (2.2) | 10 (16.4) | 0.018 |
| Chronic kidney disease | – | 9 (14.8) | – |
| Chronic heart disease | 2 (4.4) | 5 (8.2) | ns |
| Malignancy | 3 (6.7) | 4 (6.5) | ns |
| Chronic liver disease | – | 1 (1.6) | – |
Abbreviations: COVID‐19, coronavirus disease 2019; ns, not significant.
p < 0.05 was considered statistically significant compared with healthy controls and patients with severe COVID‐19.
The humoral response of patients with COVID‐19 was evaluated, and as demonstrated, SARS‐CoV‐2 infection caused an enhancement of anti‐S protein IgM and IgG levels above the threshold limit (OD ratio above 2; Figure 1a,b), besides the positive PCR test (Figure S2). The HCs, in addition to the statement of not having contracted COVID‐19, presented low levels (OD ratio under 2) of IgM and IgG anti‐S protein. HCs presenting OD ratio above 2 were excluded from our cohort. Interestingly, we observed some positive PCR‐test patients with COVID‐19 presenting low levels of anti‐S antibodies. This finding was already observed in other studies and probably is due to poor antibody producing individuals, in the same way as elders which have low humoral response. The quantitative antibody response was also evaluated among discharged and deceased patients with severe COVID‐19. The antibody levels were reduced, despite not being statistically different, in the deceased patients compared to discharged patients, but it is still higher than non‐COVID‐19 subjects (Figure 1c,d – bottom graphs). Of note, the few patients with COVID‐19 with low levels of IgM and IgG (values of OD under 2, Figure 1c,d) died, suggesting that they were unable to present a humoral response due to their critical condition.
FIGURE 1.
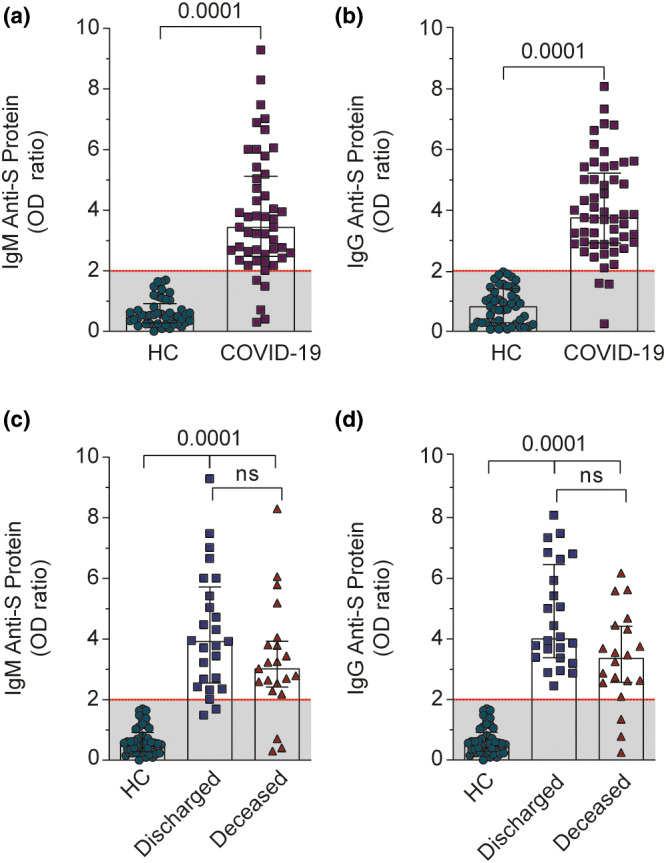
Anti‐S protein immunoglobulin profile of patients with COVID‐19 and HCs. (a, b) Serum IgM and IgG levels of HCs and COVID‐19 groups. We plotted the highest value obtained after analysis of two or three samples received from each patient with COVID‐19. (c, d) Serum IgM and IgG levels from discharged or deceased patients. The data were presented as the median with interquartile range and were analyzed by Mann–Whitney test (b, c) and Kruskal‐Wallis test (d, e). Any p < 0.05 was considered statistically significant compared to HCs. COVID‐19, coronavirus disease 2019; HCs, healthy controls; ns, not significant; OD, optical density
Patients with COVID‐19 present altered biochemical parameters
To further confirm the systemic commitment of patients with COVID‐19 and to correlate with the target inflammatory mediators, we analyzed the data of several organ dysfunction markers from medical records between discharged and deceased patients. The values of the COVID‐19 cohort were evaluated regarding the reference values detached as dashed red lines (Figure 2). One limitation was the fact that all patients with COVID‐19 were critically ill, thus most biochemical markers of severity were similar between discharged and deceased patients. Nevertheless, deceased patients with COVID‐19 presented reduced values of platelets and PO2/FIO2 and increased lactate and CRP levels, compared with discharged patients (Figure 2b,i,j,l). With respect to lactate levels, we did not observe high levels above reference, probably because both groups of patients were being closely monitored to avoid the lactic acidosis.
FIGURE 2.
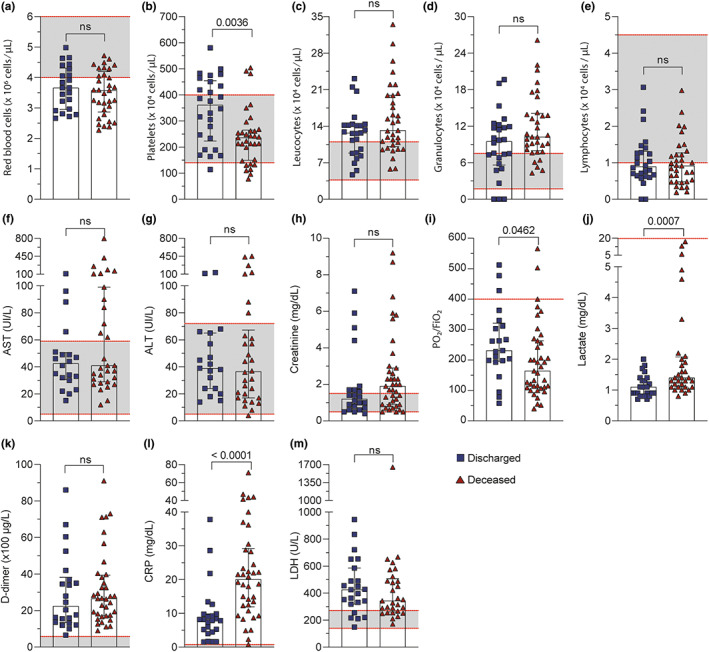
Patients with COVID‐19 display alterations in biochemical parameters according to clinical outcome. Routine laboratory values in the circulation of discharged and deceased patients with COVID‐19. (a–e) Number of red blood cells, platelets, and white blood cells. (f–i) Serum levels of hepatic (AST and ALT), kidneys (creatinine), and lungs (P/F) as injury biomarkers. (j–m) Systemic inflammatory biomarkers (lactate, D‐dimer, CRP, and LDH). The range of references values for all parameters was marked as dashed red line. The data were presented as the median with interquartile range and were analyzed by Mann–Whitney test. Any p < 0.05 was considered statistically significant compared to discharged patients' group. ALT, alanine aminotransferase; AST, aspartate aminotransferase; COVID‐19, coronavirus disease 2019; CRP, C reactive protein; ns, not significant; LDH, lactate dehydrogenase
Regarding other systemic parameters, although they were not significantly different between groups, the number of red blood cells and lymphocytes were below the reference values, whereas total leukocytes and granulocytes were above (Figure 2a,c,d,e). Aspartate aminotransferase (AST), alanine aminotransferase (ALT), and creatinine levels were in the normal range in most patients, having some patients with higher levels (Figure 2f,g,h). D‐dimer and LDH levels of both discharged and deceased patients were way over the reference levels (Figure 2k–m).
High serum levels of HMGB1, ATP, PGE2 , LTB4 , and cys‐LTs, and TF in patients with COVID‐19
Inflammatory storm is well‐described and is one of the main causes of tissue damage and worst prognosis of COVID‐19. 14 Among several mediators involved, we demonstrated that alarmins, as HMGB1 and ATP, were significantly increased in the serum of patients with COVID‐19, compared to HCs (Figures 3a,b). In order to characterize vascular activation, we detected higher levels of TF in patients with COVID‐19 compared to HCs (Figures 3c and S1). Lipid mediators play important roles in acute and late immune response during sepsis. 29 Thus, we also demonstrated increased levels of PGE2, LTB4, and cys‐LTs in the serum of patients with severe COVID‐19 (Figure 3d–f).
Higher levels of HMGB1 in deceased patients with COVID‐19
Further, we correlated those mediators with the primary outcome. HMGB1 was the only mediator showing higher levels (3.6‐fold change) in the deceased group compared to the discharged group (Figure 4a). Even though we could not find statistical differences of cys‐LTs levels between discharged and deceased patients, apparently, patients with fatal outcomes presented higher levels of this lipid mediator; however, the number of patients was not big enough to demonstrate statistical difference (Figure 4e). No differences were noted between discharged and deceased patients regarding serum ATP, TF, PGE2, and LTB4 levels (Figures 4b–d,f).
Higher levels of HMGB1 during distinct phases of COVID‐19: Pulmonary and hyperinflammatory early phases
Looking for a specific marker that could predict the worst prognosis and considering the long period of hospitalization that could direct influence inflammatory response, we stratified the samples of patients with COVID‐19 according to the different phases of the disease in order to evaluate the kinetic of inflammatory mediators, as following: (1) pulmonary phase with hypoxia ‐ from onset of the disease until day 11; (2) hyperinflammatory early phase – period of 12–25 days; and (3) hyperinflammatory late phase period of 26–168 days. These phases were already described in the literature. 30 , 31 HMGB1 was the sole mediator with significantly higher levels in the serum of patients who died compared with discharged patients during the pulmonary phase with hypoxia and in the hyperinflammatory early phase (Figure 5a). None of the other mediators analyzed showed differences.
HMGB1 predicts disease progression
Next, we examined whether the high serum level of HMGB1 correlates with the biochemical data obtained from patient records, or even with the other mediators evaluated in this work. Interestingly, we observed a significant correlation between HMGB1 and cys‐LT (Figure 6a). Furthermore, HMGB1 also correlates with AST and ALT levels (Figures 6b,c), which are markers of hepatic dysfunction. A positive correlation trend between HMGB1 and D‐dimer was observed but did not reach significance (p = 0.09; Figure 6d). We also analyzed correlation with several other markers in our study, but we could not find statistical differences (Figure S3).
FIGURE 6.
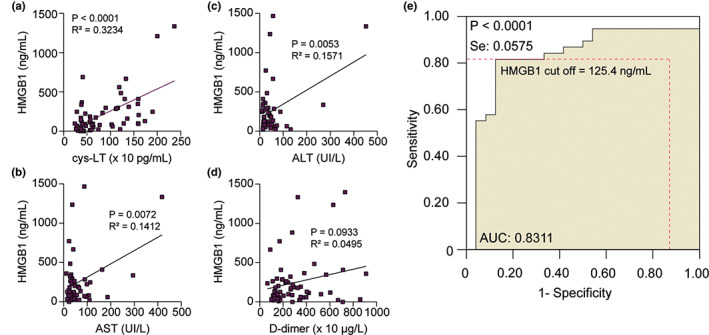
Positive correlations of HMGB1 with cys‐LTs, AST, ALT, and D‐dimer in patients with COVID‐19. (a–d) Spearman's correlation analyses between HMGB1 serum levels peak versus cys‐LTs, AST, ALT, and D‐dimer levels. We plotted the highest value obtained for HMGB1 after analysis of two or three samples received from each patient with COVID‐19. The values for cys‐LT, AST, ALT, and D‐dimer were paired exactly with the same sample chosen for analysis of HMGB1. (e) ROC curve analysis determines the HMGB1 cutoff value (125.4 ng/ml) that predicts the worst clinical outcome (death). The AUC is 0.8311 ± 0.06 and the 95% CI is 0.7185–0.9437. The diagnostic sensitivity and specificity are 81.6% and 87.5%, respectively. ALT, alanine aminotransferase; AST, aspartate aminotransferase; AUC, area under the curve; CI, confidence interval; COVID‐19, coronavirus disease 2019; ROC, receiving operating characteristic
To explore the exact impact of HMGB1 levels on the disease progression, we examined the ROC curves with our data. The ROC cutoff value of serum HMGB1 levels that distinguishes discharged and deceased patients was 125.4 ng/ml. The diagnostic sensitivity and specificity for deceased patients was 81.6% and 87.5%, respectively (Figure 6e). These data demonstrated that values above 125.4 ng/ml predicted disease progression and worst outcome. This finding suggested that serum HMGB1 levels until the twelvth day after hospital admission could be regarded as a biomarker for distinguishing patients with COVID‐19 outcomes.
DISCUSSION
A wide range of host immune responses is triggered by SARS‐CoV‐2, from appropriate and protective to uncontrolled and highly dysfunctional reactions. Regarding the severe cases, the acute phase of COVID‐19 is characterized by immune alterations and a decoupling of the innate and adaptive immunity, which are associated with endothelium dysfunction and uncontrolled inflammatory responses, including lymphopenia and cytokine storm, affecting tissue integrity. 1 , 4 , 14 , 29 These are characteristic of the disease mostly during the first and second waves of the pandemic, due to the fact that it was a new virus and the immune response was not yet trained to respond to this insult. 32 Indeed, the high mortality and disease evolution was resembled among several clinical studies, independently of the circulating SARS‐CoV‐2 variant. 11 Intriguingly, increasing evidence suggest that phenotypic and functional alterations of the immune system persist for a long period after recovery from COVID‐19. 33 , 34 To note, during the same period, when there is no existing antiviral efficient drugs and vaccines, just dexamethasone and tocilizumab have been shown to reduce mortality in patients with COVID‐19. 35 , 36 , 37
Thus, our work is a prospective and qualitative study using a convenience sampling from patients with severe COVID‐19 referring to the first year of the pandemic in Rio de Janeiro city. The main goal of this study was to identify specific molecular biomarkers of disease severity in a Brazilian cohort with COVID‐19. In this report, we provided evidence of high levels of HMGB1, ATP, TF, cys‐LTs, PGE2, and LTB4 in the circulation of patients with severe COVID‐19 compared to HCs. Still, patients who died from COVID‐19 showed higher serum levels of HMGB1, compared to discharged patients, which led us to consider HMGB1 a biomarker of mortality risk in a cohort of hospitalized patients.
The innate immune response, as the first line of defense against the infection, in specific situations may play an outsized role in promoting inflammatory dysfunction, changing metabolic processes, and promoting malfunction of the adaptive immune response, such as T cell exhaustion and inadequate B cells/antibody response. 1 , 38 HMGB1 and ATP stand out among the alarmins increased during COVID‐19 infection. 21 , 22 , 23 , 39 Although the pathogenic role of HMGB1 in COVID‐19 remains to be explored in animal models, studies in culture systems demonstrated that HMGB1 upregulates ACE2 expression, the main SARS‐CoV‐2 entrance on cellular membrane. 22 , 24 Interestingly, ATP may favor increased levels of HMGB1 during COVID‐19, because ATP triggers HMGB1 release by P2X7 receptor activation in a monosodium urate crystals‐induced model of sterile inflammation. 40 In the same line, eicosanoids such as cys‐LTs, PGE2, and LTB4 are also released by ATP and HMGB1 stimuli. 18 , 41 , 42 , 43 In agreement, some data in the literature demonstrated that lipid mediators were increased in patients with severe COVID‐19 and may be associated with poor outcomes. 31 , 44 Meanwhile, the role of eicosanoids in COVID‐19 remains to be characterized. Our data place the lipid mediators, mainly the cys‐LTs, as an interesting target in COVID‐19 in association with ATP and HMGB1. As well‐described, COVID‐19 is a multi‐mediated disease, and we attempt to characterize not just one mediator but the main scenario responsible for promoting the disease.
Molecular analysis revealed that HMGB1 forms complexes with LPS or IL‐1β to enhance immune responses 45 , 46 and ACE2 expression. 24 Although admitting that clinical studies cannot provide conclusive answers to this question, there is an urgent need to identify markers of disease severity in COVID‐19. Following the first report that HMGB1 levels were increased in patients with severe COVID‐19, another study suggested that serum HMGB1 and IL‐6 high levels may serve as prognostic biomarkers, distinguishing patients of unfavorable clinical outcomes. 23 Along similar lines, the serum levels of ATP and activation of CD39/CD73 axis may aggravate COVID‐19 by harnessing cytokine production, inflammasome activation, cell death, and tissue damage. 47
The detection of high TF levels in the serum of patients with severe COVID‐19 raised the possibility that HMGB1 might be implicated in TF‐driven hyperactivation of platelets, perhaps contributing to the thromboembolic complications observed in severe cases. Our cohort studies confirmed that cases of severe COVID‐19 presented high levels of HMGB1, D‐dimers, CRP, low ratio of PO2/FiO2, and lymphopenia. At first sight, these findings suggest that HMGB1, presumably acting as a pleiotropic driver of inflammation, may potentiate TF‐dependent activation of platelets and circulating monocytes. 48
Extending our analysis, we reorganized the data of cytokine levels according to the patient's hospitalization period. This strategy would allow us to evaluate if there was a cytokine release kinetic during the disease, and if the long hospitalization would interfere with this process. We did not observe significant differences of cytokine levels according only to the phases of the disease. But notably, we did observe differences regarding discharged and deceased patients. HMGB1 was the sole cytokine presenting high serum levels during the pulmonary phase with hypoxia and during the hyperinflammatory early phase of patients who died. The implication of this finding must be emphasized because HMGB1 levels measured at the early inflammatory phase of the disease have predictive value for the worst outcome. This may be extremely valuable at the timepoint in which patients are hospitalized showing signs of cytokine storm. Our findings suggest that serum values of HMGB1 above 125.4 ng/ml in critically ill patients with COVID‐19 correlate with the worst outcome and that those patients may be at higher risk of death. Thus, measurements of serum HMGB1 levels may help to instruct pharmacological and medical interventions at early stages of the disease. So, the inhibition of HMGB1 effects during the pulmonary and inflammatory early phases of COVID‐19 could revert ongoing processes skewing the innate and acquired immune response to an efficient and pro‐resolution mode, instead of overwhelming inflammation and immune dysfunction.
Still a positive correlation was observed with high cys‐LTs and ALT/AST levels in the circulation. An exacerbated HMGB1‐cys‐LT axis probably contributes to endothelial cell activation, inflammatory cells recruitment, and long‐term cytokine production by activated leukocytes, leading to systemic complications. These data probably explain the positive correlation between HMGB1 and ALT/AST, which was expected because our group already demonstrated the liver as one of the main sources of systemic HMGB1 during infections. 49 Together, it could partly explain the hepatic dysfunction and alterations in immune metabolism during COVID‐19.
Remarkably, our study suggests a relevant role of HMGB1 in the COVID‐19 evolution. This cytokine stimulates the innate system either by itself or in association with endogenous and exogenous molecules. 46 For example, S100A8/A9 is another host molecule that acts as alarmin, it is also released from dead cells as HMGB1 and ATP. Overproduced S100A8/A9 and HMGB1 in the serum of patients with COVID‐19 were associated with distinct signatures for cytokine storm, and both are poor prognostic indicators. 21 In parallel, we also believe in a partnership between HMGB1 and hypoxia‐inducible factor‐1α (HIF‐1α) favoring the pathogenesis of COVID‐19. As previously demonstrated, HIF‐1α and HMGB1 are released by monocyte and endothelial cells, respectively, under hypoxic conditions observed during COVID‐19. 50 However, whereas HMGB1 promotes inflammatory cytokine production, elevated levels of HIF‐1α repress IRF3 and IRF5 leading to low levels of type I IFN, exactly the undesirable scenario but it is what we observe during SARS‐CoV‐2 infections. Those mediators (S100A8/A9 and HIF‐1α) may act in synergism with HMGB1, and these aspects are under investigation in our clinical trial and in experiments with animal models.
In conclusion, this scenario could explain the pathophysiology initiated by SARS‐CoV‐2 and place HMGB1 as one of the major mediators in triggering an overwhelming inflammatory cascade which impairs the host to wipe out the virus. Therefore, our work suggests that high levels of HMGB1 in the circulation of patients with severe COVID‐19 orchestrates the acute and persistent mediators' storm, in association with several other mediators, which certainly prompts the long‐term consequences already well‐described. ATP, cys‐LTs, PGE2, LTB4, and TF appear to act collaboratively to form an inflammatory platform, a process that is under investigation to better understand the molecular mechanism behind COVID‐19 pathophysiology.
The association of HMGB1 and COVID‐19 has already been documented before by two independent groups. Although, our work adds and ratifies this mediator as a promising biomarker in the Brazilian cohort. This is a pertinent point, because it validates and assures HMGB1 as a promising target to control the immune response in COVID‐19. The well‐done characterization of HMGB1 as a specific biomarker of worst prognostic is necessary to correctly predict relevant clinical outcomes across a variety of diseases and populations.
Limitations
It is important to state that the limited number of participants from a convenience sample provided by MDNH and the inclusion of only severe cases of COVID‐19 in this study did not allow for an expanded robust analysis of different comorbidities, sex, age, or therapeutic treatment. Therefore, larger samples and multicenter studies will be important to corroborate with our initial observations.
AUTHOR CONTRIBUTIONS
A.R.R.V., C.C., and C.F.B. wrote the manuscript. A.R.R.V., C.C., R.V.J., and C.F.B. designed the research. A.R.R.V., C.C., C.F.B., V.S.F.J., M.P., A.C.N.B., N.R.A., L.E.B.S., S.P.C.B., S.N.F., and D.R. performed the research. A.R.R.V., C.C., C.F.B., V.S.F.J., M.P., A.C.N.B., N.R.A., L.E.B.S., S.P.C.B., S.N.F. A.C.O., J.S., and D.A. analyzed the data. D.A., M.R.F., J.S., A.M.V., A.C.O., R.C.S., C.F.B., and L.E.B.S. contributed new reagents/analytical tools.
FUNDING INFORMATION
This work was supported by funding from the Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (Grants SEI‐260003/002696/2020).
CONFLICT OF INTEREST
The authors declared no competing interests for this work.
Supporting information
Figure S1.
ACKNOWLEDGMENTS
We greatly acknowledge Marcia Amorim who participated in this study by collecting blood from the participants of healthy controls, Daniele Cristina Passos da Rocha who performed the initial step of HMGB1 ELISA, and Dr. Marcelo Nunes Afonso Teixeira and Dr. Fernanda Cruz who helped us interpret the medical records of the patients.
Vicentino ARR, Fraga‐Junior VdS, Palazzo M, et al. High mobility group box 1, ATP, lipid mediators, and tissue factor are elevated in COVID‐19 patients: HMGB1 as a biomarker of worst prognosis. Clin Transl Sci. 2023;16:631‐646. doi: 10.1111/cts.13475
REFERENCES
- 1. Schultze JL, Aschenbrenner AC. COVID‐19 and the human innate immune system. Cell. 2021;184(7):1671‐1692. doi: 10.1016/j.cell.2021.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Worldmeters Website . Accessed January 15, 2022. https://www.worldometers.info/coronavirus/
- 3. Worlf Health Organization Website . Accessed January 20, 2022. https://www.who.int/publications/m/item/weekly‐epidemiological‐update‐on‐covid‐19‐‐‐11‐january‐2022
- 4. Sokolowska M, Lukasik ZM, Agache I, et al. Immunology of COVID‐19: mechanisms, clinical outcome, diagnostics, and perspectives—a report of the European academy of allergy and clinical immunology (EAACI). Allergy. 2020;75(10):2445‐2476. doi: 10.1111/all.14462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Willi S, Lüthold R, Hunt A, et al. COVID‐19 sequelae in adults aged less than 50 years: a systematic review. Travel Med Infect Dis. 2021;40:101995. doi: 10.1016/j.tmaid.2021.101995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bellan M, Soddu D, Balbo PE, et al. Respiratory and psychophysical sequelae among patients with COVID‐19 four months after hospital discharge. JAMA Netw Open. 2021;4(1):e2036142. doi: 10.1001/jamanetworkopen.2020.36142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mitrani RD, Dabas N, Goldberger JJ. COVID‐19 cardiac injury: implications for long‐term surveillance and outcomes in survivors. Heart Rhythm. 2020;17(11):1984‐1990. doi: 10.1016/j.hrthm.2020.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sahin AR, Kocyigit BF, Aksu E, et al. Multisystemic Long‐term sequelae of Covid‐19: a review based on the current literature over a year of pandemic experience. Eurasian J Med Oncol. 2021;5(1):6‐19. doi: 10.14744/ejmo.2021.69960 [DOI] [Google Scholar]
- 9. Wang F, Kream RM, Stefano GB. Long‐term respiratory and neurological sequelae of COVID‐19. Med Sci Monit. 2020;26:26. doi: 10.12659/MSM.928996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. doi: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dessie ZG, Zewotir T. Mortality‐related risk factors of COVID‐19: a systematic review and meta‐analysis of 42 studies and 423,117 patients. BMC Infect Dis. 2021;21(1):855. doi: 10.1186/s12879-021-06536-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alshahawey M, Raslan M, Sabri N. Sex‐mediated effects of ACE2 and TMPRSS2 on the incidence and severity of COVID‐19; the need for genetic implementation. Curr Res Transl Med. 2020;68(4):149‐150. doi: 10.1016/j.retram.2020.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Q, Bastard P, Karbuz A, et al. Human genetic and immunological determinants of critical COVID‐19 pneumonia. Nature. 2022;603(7902):587‐598. doi: 10.1038/s41586-022-04447-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang J, Jiang M, Chen X, Montaner LJ. Cytokine storm and leukocyte changes in mild versus severe SARS‐CoV‐2 infection: review of 3939 COVID‐19 patients in China and emerging pathogenesis and therapy concepts. J Leukoc Biol. 2020;108(1):17‐41. doi: 10.1002/JLB.3COVR0520-272R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bonaldi T. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003;22(20):5551‐5560. doi: 10.1093/emboj/cdg516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418(6894):191‐195. doi: 10.1038/nature00858 [DOI] [PubMed] [Google Scholar]
- 17. Deng M, Tang Y, Li W, et al. The endotoxin delivery protein HMGB1 mediates Caspase‐11‐dependent lethality in sepsis. Immunity. 2018;49(4):740‐753.e7. doi: 10.1016/j.immuni.2018.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu T, Barrett NA, Kanaoka Y, et al. Cysteinyl leukotriene receptor 2 drives lung immunopathology through a platelet and high mobility box 1‐dependent mechanism. Mucosal Immunol. 2019;12(3):679‐690. doi: 10.1038/s41385-019-0134-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stark K, Philippi V, Stockhausen S, et al. Disulfide HMGB1 derived from platelets coordinates venous thrombosis in mice. Blood. 2016;128(20):2435‐2449. doi: 10.1182/blood-2016-04-710632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andersson U, Yang H, Harris H. High‐mobility group box 1 protein (HMGB1) operates as an alarmin outside as well as inside cells. Semin Immunol. 2018;38:40‐48. doi: 10.1016/j.smim.2018.02.011 [DOI] [PubMed] [Google Scholar]
- 21. Chen L, Long X, Xu Q, et al. Elevated serum levels of S100A8/A9 and HMGB1 at hospital admission are correlated with inferior clinical outcomes in COVID‐19 patients. Cell Mol Immunol. 2020;17(9):992‐994. doi: 10.1038/s41423-020-0492-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen R, Huang Y, Quan J, et al. HMGB1 as a potential biomarker and therapeutic target for severe COVID‐19. Heliyon. 2020;6(12):e05672. doi: 10.1016/j.heliyon.2020.e05672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sivakorn C, Dechsanga J, Jamjumrus L, et al. High mobility group box 1 and interleukin 6 at intensive care unit admission as biomarkers in critically ill COVID‐19 patients. Am J Trop Med Hyg. 2021;105(1):73‐80. doi: 10.4269/ajtmh.21-0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wei J, Alfajaro MM, DeWeirdt PC, et al. Genome‐wide CRISPR screens reveal host factors critical for SARS‐CoV‐2 infection. Cell. 2021;184(1):76‐91.e13. doi: 10.1016/j.cell.2020.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moreira FRR, D'arc M, Mariani D, et al. Epidemiological dynamics of SARS‐CoV‐2 VOC gamma in Rio de Janeiro, Brazil. Virus Evol. 2021;7(2):1‐12. doi: 10.1093/ve/veab087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Candido DS, Claro IM, de Jesus JG, et al. Evolution and epidemic spread of SARS‐CoV‐2 in Brazil. Science. 2020;369(6508):1255‐1260. doi: 10.1126/SCIENCE.ABD2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Allonso D, Belgrano FS, Calzada N, Guzmán MG, Vázquez S, Mohana‐Borges R. Elevated serum levels of high mobility group box 1 (HMGB1) protein in dengue‐infected patients are associated with disease symptoms and secondary infection. J Clin Virol. 2012;55(3):214‐219. doi: 10.1016/j.jcv.2012.07.010 [DOI] [PubMed] [Google Scholar]
- 28. Alvim RGF, Lima TM, Rodrigues DAS, et al. From a recombinant key antigen to an accurate, affordable serological test: Lessons learnt from COVID‐19 for future pandemics. Biochem Eng J. 2022;186:108537. doi: 10.1016/j.bej.2022.108537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zaid Y, Doré É, Dubuc I, et al. Chemokines and eicosanoids fuel the hyperinflammation within the lungs of patients with severe COVID‐19. J Allergy Clin Immunol. 2021;148(2):368‐380.e3. doi: 10.1016/j.jaci.2021.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Siddiqi HK, Mehra MR. COVID‐19 illness in native and immunosuppressed states: a clinical–therapeutic staging proposal. J Heart Lung Transplant. 2020;39(5):405‐407. doi: 10.1016/j.healun.2020.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin Y, Wu Y, Zhong P, et al. A clinical staging proposal of the disease course over time in non‐severe patients with coronavirus disease 2019. Sci Rep. 2021;11(1):10681. doi: 10.1038/s41598-021-90111-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. González‐Castro A, Cuenca Fito E, Fernandez A, Escudero Acha P, Rodríguez Borregán JC, Peñasco Y. First and second wave of coronavirus‐19 disease: a comparative study in patients hospitalized in an ICU of a third‐level university hospital. Med Intensiva. 2022;46(3):166‐168. doi: 10.1016/j.medin.2021.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davis HE, Assaf GS, McCorkell L, et al. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. EClinicalMedicine. 2021;38:101019. doi: 10.1016/j.eclinm.2021.101019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ryan FJ, Hope CM, Masavuli MG, et al. Long‐term perturbation of the peripheral immune system months after SARS‐CoV‐2 infection. BMC Med. 2022;20(1):26. doi: 10.1186/s12916-021-02228-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tomazini BM, Maia IS, Cavalcanti AB, et al. Effect of dexamethasone on days alive and ventilator‐free in patients with moderate or severe acute respiratory distress syndrome and COVID‐19: the CoDEX randomized clinical trial. JAMA. 2020;324(13):1307‐1316. doi: 10.1001/jama.2020.17021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Selvaraj V, Khan MS, Bavishi C, et al. Tocilizumab in hospitalized patients with COVID‐19: a meta analysis of randomized controlled trials. Lung. 2021;199(3):239‐248. doi: 10.1007/s00408-021-00451-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raslan M, Eslam MS, Sabari NA. Immune system response to COVID‐19. An endless story. Acta Sci Pharm Sci. 2022;6(7):24‐31. [Google Scholar]
- 38. McKechnie JL, Blish CA. The innate immune system: fighting on the front lines or fanning the flames of COVID‐19? Cell Host Microbe. 2020;27(6):863‐869. doi: 10.1016/j.chom.2020.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dorneles GP, Teixeira PC, da Silva IM, et al. Alterations in CD39/CD73 Axis of T cells associated with COVID‐19 severity. J Cell Physiol. 2022;237(8):3394‐3407. doi: 10.1002/jcp.30805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marinho Y, Marques‐da‐Silva C, Santana PT, et al. MSU crystals induce sterile IL‐1β secretion via P2X7 receptor activation and HMGB1 release. Biochim Biophys Acta. 2020;1864(1):129461. doi: 10.1016/j.bbagen.2019.129461 [DOI] [PubMed] [Google Scholar]
- 41. Toki Y, Takenouchi T, Harada H, et al. Extracellular ATP induces P2X7 receptor activation in mouse Kupffer cells, leading to release of IL‐1β, HMGB1, and PGE2, decreased MHC class I expression and necrotic cell death. Biochem Biophys Res Commun. 2015;458(4):771‐776. doi: 10.1016/j.bbrc.2015.02.011 [DOI] [PubMed] [Google Scholar]
- 42. Leclerc P, Wähämaa H, Idborg H, Jakobsson PJ, Harris HE, Korotkova M. IL‐1β/HMGB1 complexes promote the PGE 2 biosynthesis pathway in synovial fibroblasts. Scand J Immunol. 2013;77(5):350‐360. doi: 10.1111/sji.12041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jaulmes A, Thierry S, Janvier B, et al. Activation of sPLA2‐IIA and PGE2 production by high mobility group protein B1 in vascular smooth muscle cells sensitized by IL‐1β. FASEB J. 2006;20(10):1727‐1729. doi: 10.1096/fj.05-5514fje [DOI] [PubMed] [Google Scholar]
- 44. Pérez MM, Pimentel VE, Fuzo CA, et al. Acetylcholine, fatty acid, and lipid mediators are linked to COVID‐19 severity. J Immunol. 2022;209(2):250‐261. doi: 10.4049/jimmunol.2200079 [DOI] [PubMed] [Google Scholar]
- 45. García‐Arnandis I, Guillén MI, Gomar F, Pelletier JP, Martel‐Pelletier J, Alcaraz MJ. High mobility group box 1 potentiates the pro‐inflammatory effects of interleukin‐1β in osteoarthritic synoviocytes. Arthritis Res Ther. 2010;12(4):R165. doi: 10.1186/ar3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hreggvidsdottir HS, Östberg T, Wähämaa H, et al. The alarmin HMGB1 acts in synergy with endogenous and exogenous danger signals to promote inflammation. J Leukoc Biol. 2009;86(3):655‐662. doi: 10.1189/jlb.0908548 [DOI] [PubMed] [Google Scholar]
- 47. Ahmadi P, Hartjen P, Kohsar M, et al. Defining the CD39/CD73 Axis in SARS‐CoV‐2 infection: the CD73‐ phenotype identifies polyfunctional cytotoxic lymphocytes. Cell. 2020;9(8):1750. doi: 10.3390/cells9081750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hottz ED, Azevedo‐Quintanilha IG, Palhinha L, et al. Platelet activation and platelet‐monocyte aggregate formation trigger tissue factor expression in patients with severe COVID‐19. Blood. 2020;136(11):1330‐1341. doi: 10.1182/blood.2020007252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vicentino ARR, Carneiro VC, Allonso D, et al. Emerging role of HMGB1 in the pathogenesis of schistosomiasis liver fibrosis. Front Immunol. 2018;9:1979. doi: 10.3389/fimmu.2018.01979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peng T, Du SY, Son M, Diamond B. HIF‐1α is a negative regulator of interferon regulatory factors: implications for interferon production by hypoxic monocytes. Proc Natl Acad Sci. 2021;118(26):e2106017118. doi: 10.1073/pnas.2106017118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.


