Abstract
Metabolic health and immune function are intimately connected via diet and the microbiota. Nearly 90% of all immune cells in the body are associated with the gastrointestinal tract and these immune cells are continuously exposed to a wide range of microbes and microbial‐derived compounds, with important systemic ramifications. Microbial dysbiosis has consistently been observed in patients with atopic dermatitis, food allergy and asthma and the molecular mechanisms linking changes in microbial populations with disease risk and disease endotypes are being intensively investigated. The discovery of novel bacterial metabolites that impact immune function is at the forefront of host‐microbe research. Co‐evolution of microbial communities within their hosts has resulted in intertwined metabolic pathways that affect physiological and pathological processes. However, recent dietary and lifestyle changes are thought to negatively influence interactions between microbes and their host. This review provides an overview of some of the critical metabolite‐receptor interactions that have been recently described, which may underpin the immunomodulatory effects of the microbiota, and are of relevance for allergy, asthma and infectious diseases.
Keywords: AhR, GPCRs, metabolites, microbiome, nuclear receptors
Abbreviations
- AhR
Aryl Hydrocarbon Receptor
- ARNT
AhR Nuclear Translocator
- CaS
Calcium‐Sensing Receptor
- COVID‐19
Coronavirus Disease 2019
- FXR
Farnesoid X Receptor
- GPCR
G Protein‐Coupled Receptor
- IFN
Interferon
- ILC
Innate Lymphoid Cell
- LXR
Liver X Receptor
- MAIT
Mucosal‐Associated Invariant T cells
- NR
Nuclear Receptor
- PPAR
Peroxisome Proliferator‐Activated Receptors
- PXR
Pregnane X Receptor
- SCFAs
Short‐Chain Fatty Acids
- TLR
Toll‐Like Receptor
- TSLP
thymic stromal lymphopoietin SARS‐CoV‐2 – Severe Acute Respiratory Syndrome Coronavirus 2
- VDR
Vitamin D Receptor
1. INTRODUCTION
Human mucosal surfaces and body cavities harbour diverse communities of commensal microbes that play essential roles in regulation of host metabolic responses, epithelial barrier function, immune education and immune regulation. 1 , 2 , 3 , 4 Microbial‐derived factors are integral components of the molecular circuitry that regulate immune and metabolic functions required for host physiology and survival. These host effects are partially induced by activation of host pattern recognition receptors to microbial‐derived danger signals, but increasingly the role of secreted bacterial metabolites in shaping host immune function is being recognized. 5 , 6 , 7 , 8 Immunoregulatory bacterial metabolites can trigger host G protein‐coupled receptors (GPCRs), aryl hydrocarbon receptor (AhR), nuclear hormone receptors such as the farnesoid X receptor (FXR) or can directly modulate gene expression through epigenetic mechanisms.
Importantly, many immunoregulatory bacterial metabolites are derived from their metabolism of dietary ingredients (e.g. fibre, tryptophan), linking diet and lifestyle to protection from immune‐mediated disorders via microbial mechanisms. Microbial fermentation of dietary components in vivo potentially generates thousands of molecules, some of which regulate immune and metabolic functions. These in turn are thought to protect against aberrant inflammatory processes or hypersensitive responses, but also promote effector immune responses that efficiently eliminate pathogens, such as SARS‐CoV‐2. 9 , 10 , 11 While individual microbes, individual dietary components and individual metabolites are certainly important, the overall community functional capacity and community metabolic outputs that underpin interactions with the host immune system are perhaps more relevant with regard to understanding disease risk. A low‐risk microbe‐diet configuration may generate sufficient levels of several regulatory metabolites that are associated with protection from aberrant inflammatory responses. In contrast, a high‐risk microbe‐diet configuration may consistently generate multiple pro‐inflammatory metabolites that may contribute to a higher risk of inappropriate immune reactivity (this model is illustrated in Figure 1). Indeed, recent changes in dietary habits and microbiota composition, especially evident in obese individuals, have resulted in reduced levels of immune regulatory metabolites that are expected and evolutionarily hardwired into immune cell decision‐making processes. 12 The lack of immune regulatory molecules may lead to a hypersensitive immune system that does not respond appropriately thus leading to a chronic state of inflammation that culminates in organ damage and disease for an increasing number of people. 13 , 14 , 15 , 16 , 17 In this review, we will describe the immune cell receptor systems that recognize secreted microbial‐derived metabolites and highlight their relevance to immune‐mediated disorders. We have deliberately excluded pattern recognition receptors from this review as these have been extensively reviewed elsewhere. 18
FIGURE 1.
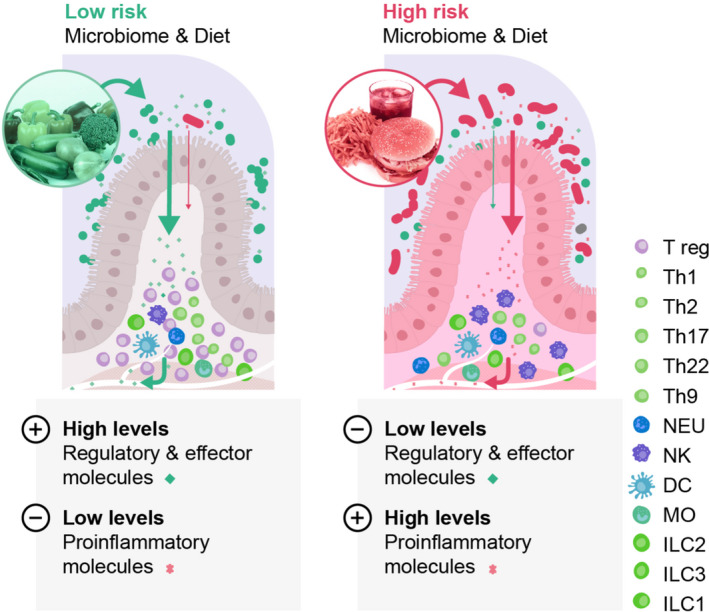
Schematic model illustrating the concept of a high‐risk versus a low‐risk configuration of microbiota‐dietary interactions that influence risk of immune‐mediated disorders
2. DIETARY EFFECTS ON MICROBE‐HOST IMMUNE INTERACTIONS
Malnutrition is well recognized as an important risk factor for poor immunological responses, especially related to infectious diseases. However, obesity has more recently also been identified as a risk factor for severity of infectious diseases, including influenza and coronavirus. In addition, obesity is associated with a range of chronic inflammatory non‐communicable diseases, such that obesity is now associated with higher mortality worldwide compared with being underweight. 19 A simple excess in calories does not explain these negative effects on the immune system. Rather, the transition to reduced diversity, low‐quality, highly processed, high‐energy diets have altered the metacommunity, its processes that underpin assembly and activity of the human microbiome and, consequently, increased risk of inappropriate and uncontrolled immune responses that damage host tissues and function. 20 Indeed, weight loss is associated with improvements in microbiota diversity and intestinal permeability, suggesting the negative effects associated with obesity are reversible, at least in part. 21 , 22 , 23
Consumption of a higher diversity of fruits, vegetables and fermented foods are associated with a reduced risk of atopic disorders and asthma in children, potentially mediated in part by microbial‐derived butyrate and propionate. 24 , 25 Children of mothers who consumed lower levels of fruits, vegetables and yoghurt during pregnancy, had a similar risk of allergic outcomes as children with the filaggrin gene mutation, demonstrating that diet was just as important as a genetic risk allele for prevention of this immune‐mediated disorder. 26 In addition, adults who more regularly consumed plant‐based or pescatarian diets seemed to have lower odds of developing severe COVID‐19. 14 , 27 , 28 However, the specific plant‐based substrates (e.g. fibre type, fatty acids, polyphenols) that are responsible for these positive associations are unknown. A number of studies have focussed on supplementing intake of fibre to compensate for shortfalls in regular consumption. However, one recent study in human volunteers showed that high fibre consumption did not result in the expected microbiota and immune benefits, potentially due to a lack of pre‐existing microbes that can process non‐digestible carbohydrates into immune modulatory metabolites. 29 In addition, there are many different types of fibres and the optimal fibre‐bacterial strain combinations that generate immune‐modulatory compounds are not yet known. 30 , 31
It is thought that a sufficiently large microbial community of diverse genomic inputs allows buffering and redundancy in case certain community members are lost, thereby maintaining important metabolic and host protective functions. Microbial communities that could quickly, and appropriately, shift their functional repertoire in response to diet change would have subsequently enhanced human dietary flexibility and survival. However, recent surveys of microbial composition from industrialized human cohorts are significantly different compared with traditional cohorts, with many species disappearing from industrialized populations. 32 , 33 For example, Lactobacillus reuteri (L. reuteri) was regularly detected in multiple human studies performed around the 1960s and is still found today in many individuals living in rural Papua New Guinea but is absent in all control samples from individuals in the USA. 34 Overall, species diversity and richness has been shown to be reduced by about one‐third in Americans compared with Malawians or Amerindians. 35 Importantly, antibiotic‐driven depletion of the gut microbiome in humans, with associated impacts on secondary bile acid and tryptophan metabolism, disrupts the induction of antibody responses to influenza vaccination possibly by driving inflammatory signalling in innate immune cells in a manner consistent with age‐associated changes in immune responses. 36 Interestingly, altered tryptophan metabolism and bile acid metabolism were also recently shown to be associated with elevated levels of multiple pathobionts and pro‐inflammatory cytokines in patients with severe and fatal outcomes to SARS‐CoV‐2 infection. 9 In contrast, less severe clinical outcomes to infection were associated with SCFAs, IL‐17A and clusters of microbes with previously recognized immune regulatory effects (e.g. Ruminococcus, Roseburia, Bifidobacterium and Faecalibacterium). Disturbed tryptophan metabolism and increased levels of alarmins such as thymic stromal lymphopoietin (TSLP) remained evident in Long COVID patients linking microbial dysbiosis with significant metabolic reprogramming, impaired epithelial barrier function and ineffective immune responses. 37 , 38 , 39 , 40 , 41 Alterations in microbial tryptophan and bile acid metabolism have also been described in subgroups of patients with asthma or food allergy. 42
3. IMMUNOREGULATORY BACTERIAL METABOLITES
Due to the complexity of the human microbiome and its vast coding potential, there are likely many bacterially produced molecules that can influence immune cell activity and immune regulation. Indeed, one systematic analysis identified over 3000 small molecule biosynthetic gene clusters within the human microbiome. 43 Remarkably, the majority of these gene clusters have never been studied or even previously described.
One example that clearly illustrates the metabolic promiscuity underpinning co‐evolution of host and microbial metabolism is tryptophan. 44 , 45 , 46 Microbial‐derived tryptophan metabolites such as indole‐3‐acetic acid, indole‐3‐propionic acid and indole‐3‐aldehyde trigger host AhR protective responses such as IL‐10, IL‐22 and type I IFNs secretion that improve the epithelial barrier and limit local inflammatory responses in murine models. 47 , 48 , 49 Microbial‐derived indoles can also inhibit TH17 CD4 T cell differentiation through interactions with RORγt. In addition, the microbiota‐derived metabolites taurine, histamine and spermine were shown to modulate NLRP6 inflammasome signalling, epithelial IL‐18 secretion, and anti‐microbial peptide (AMP) profiles in mice. 50 Treatment of human dendritic cells with the bacterial‐derived 12,13‐dihydroxy‐9Z‐octadecenoic acid (12,13‐diHOME) reduced anti‐inflammatory cytokine secretion and the number of Treg cells, suggesting that this metabolite impedes immune tolerance. 51 In contrast, short‐chain fatty acids (SCFAs) are potent immunomodulators that promote IL‐10 secretion by dendritic cells, influence Treg numbers and effectiveness, influence bone marrow haematopoiesis, reduce effector T cell activity, improve epithelial barrier, and inhibit mast cell and ILC2 activation. 52 , 53 , 54 , 55 Fibre consumption or SCFA administration in murine models protects against colitis, inflammatory arthritis, respiratory syncytial virus infection and allergic diseases. SCFAs exert effects on the host immune system via binding to G protein‐coupled receptors (GPCRs) such as GPR41, GPR43 and GPR109A, and via epigenetic modifications. Lastly, nicotinamide production by Akkermansia muciniphila was shown to be protective in a murine model of amyotrophic lateral sclerosis. 56
In addition to bacterial‐derived metabolites binding to immunoregulatory receptors, unconventional lymphocyte subpopulations can directly recognize microbial‐derived metabolites as antigens. For example, human immature intrathymic mucosal‐associated invariant T (MAIT) cells recognize bacterial‐derived 5‐(2‐oxopropylideneamino)‐6D‐ribitylaminouracil (5‐OP‐RU) that acts as an antigen and stimulator for the MAIT cells, via a major histocompatibility complex Class Ib molecule MR1. 57 This finding not only demonstrated that microbial‐derived metabolites control development of mucosal targeted T cells but also challenges our traditional distinction between exogenous and self‐antigens. Similarly, another unconventional T cell lineage, invariant natural killer T cells (iNKT cells) respond to bacterial glycolipid antigens. 58 The functional objectives of T cell responses to the microbiota are not completely understood and are likely context‐dependent. However, the malleability of T cells in response to microbiota metabolism presents an opportunity to edit T cellularity, identity and functionality by utilizing microbiota‐driven molecular pathways to promote human health.
For the remainder of this review, we will focus in more detail on the three host receptor systems that have been best studied for mediating bacterial‐derived metabolite effects on immune cell function and activity. These receptors are located on the cell membrane (GPCRs), within the cell cytoplasm (aryl hydrocarbon receptor) or within the cell nucleus (nuclear receptors).
4. G PROTEIN‐COUPLED RECEPTORS
GPCRs are involved in mediating a large range of cellular events in response to external stimuli and serve as means of communication between the external and internal environments of the cell. 59 GPCRs are an extensive family of cell‐surface proteins that represent the largest family of membrane proteins in the human genome. 60 Over 800 GPCRs have been identified to date but many remain orphan receptors as their ligands and function are still unknown. 61 GPCRs share a common structure that is characterized by 7 transmembrane domains, an extracellular amino terminus and an intracellular carboxyl terminus. 62 The transmembrane domains of GPCRs are composed of 25–35 amino acid residues and are alpha‐helical in conformation, connected to adjacent transmembrane domains via alternating intracellular and extracellular loops. 63 Upon ligand binding, the GPCR becomes activated and stabilized in a conformation that allows it to interact with its associated heterotrimeric G protein. This binding allows the GDP bound to the Gα subunit of the G protein to be exchanged for GTP. 64 The binding of GTP to the Gα subunit causes the G protein to dissociate into its Gα and Gβγ subunit components that subsequently initiate distinct downstream signalling activities (Figure 2). The intracellular signalling cascade induced by G protein activation can include cAMP or phosphatidylinositol pathways, depending on the α subunit type (Gαs, Gαi/o, Gαq or Gα12/13). Studies have identified multiple microbes and microbial‐derived metabolites that can interact with human GPCRs as agonists or antagonists. 65 , 66 , 67 Their α subunit type binding is summarized in Figure 3.
FIGURE 2.
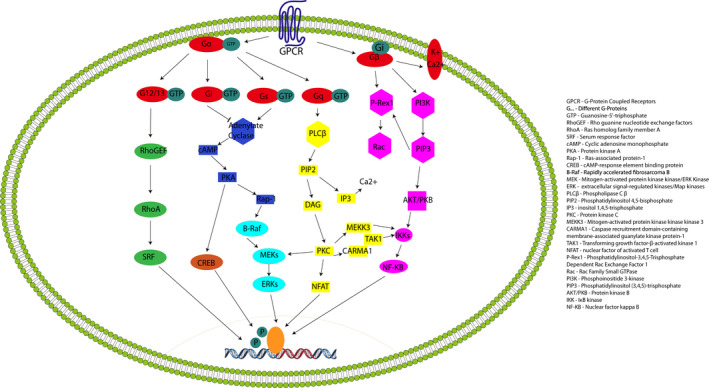
GPCR signalling. Following stimulation by their specific ligands, a conformational change causes the G‐Proteins to disengage from the GPCR and create a signal cascade dependent on the Gα subunit that is activated
FIGURE 3.
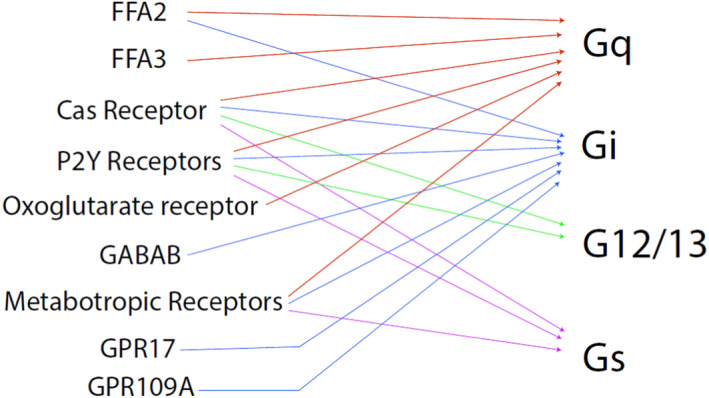
Gα subunit usage by different GPCRs
GPR41 and GPR43 (also known as free fatty acid receptor 3 [FFA3] and FFA2, respectively) are activated by bacterial‐produced SCFAs. 68 SCFAs are some of the most well‐studied bacterial metabolites and consist of fatty acids containing aliphatic tails of <6 carbon atoms. 69 These carboxylic acids, which include acetate, butyrate and propionate, are produced as end‐products from the bacterial fermentation of complex carbohydrates and the digestion of protein and peptides that avoid being digested and absorbed in the small intestine. 70 The highest concentrations of short‐chain fatty acids are found in the proximal colon. 71 Here, these metabolites can cross the intestinal epithelium and enter the cell by diffusion or are absorbed using specific transporters and can be transported into the bloodstream to travel to organs and tissues distant to the gut. 69 , 72 SCFA activation of murine and human neutrophils via GPR43 triggers chemotaxis, as well as promoting superoxide and IL‐1β production. 73 , 74 Murine ILC3 upregulate IL‐1R via GPR43 leading to IL‐22 production following activation by IL‐1β. 75 Dendritic cell GPR43 activation results in the production of BAFF and A2ALD1a2, which support B cell IgA secretion in mice. 76 , 77 Type 2 macrophages produce increased TNFα levels and demonstrated increased bactericidal activity following GPR43 activation. 78 GPR109a has been shown to be activated by bacterial‐derived butyrate and niacin. 79 GPR109a activation promotes anti‐inflammatory properties in macrophages and dendritic cells in murine models and enables them to induce differentiation of Treg cells and IL‐10‐producing Tr1 cells. 80
The four known histamine receptors (H1R‐H4R) are GPCRs and have been shown to be triggered by bacterial‐derived histamine. 7 , 81 Activation of H2R can influence a range of immune cell subsets, including mast cell degranulation, the response of dendritic cells to microbial ligands, iNKT cell responses to lipid antigens, proliferation and cytokine production from T cells and antibody secretion by B cells. 82 , 83 , 84 , 85 Human TH1 and TH2 cells preferentially express H1R and H2R, respectively, which influences cytokine secretion and proliferation. 86 The H4R receptor modulates chemotaxis in dendritic cells and regulates the activation of T cells. 87 , 88 An increased abundance of histamine secreting microbes, especially Morganella morganii, was observed within the gut of adult asthma patients, while histamine secretion from gut microbes could influence immune responses in the murine lung. 89 , 90 In addition to histamine, a wide range of microbes can produce biogenic amines and polyamines within the human gut. 91 The calcium‐sensing (CaS) receptor has been shown to be stimulated by spermine, spermidine and putrescine and these bacterial‐derived metabolites can influence airway inflammation in murine models. 92 , 93 The CaS receptor and GPR139 can also be activated by L‐tryptophan, which can be produced by microbes. 94 , 95 , 96
The inhibitory neurotransmitter gamma‐aminobutyric acid (GABA) is an agonist of the metabotropic GPCR GABAB (GPR51) and can be produced by bacterial species. 97 , 98 The activation of GABAB on human leukocytes has been shown to interfere with chemotaxis induced by chemokine receptors and reduce production of TNF‐α by macrophages. 99 All metabotropic glutamate receptors have been shown to interact with L‐Glutamic Acid, which can be produced by bacteria. 100 , 101 Activation of metabotropic receptors on murine mast cells influences gene expression of pro‐inflammatory molecules such as IL‐6 and CCL2. 102 Human T cell adhesion, chemotactic migration, cytokine secretion and gene expression are also affected by the activation of metabotropic receptors. 103
GPR17 can be stimulated by the nucleotide sugars uridine diphosphate, UDP‐galactose and UDP glucose, which can be produced by bacteria. 104 , 105 Knockout of GPR17 has been shown to influence TH2, TH17 and TH1 cytokine expression in murine models. 106 The P2Y14 receptor (GPR105) has also been shown to be activated by the same nucleotide sugars as well as bacterial‐derived UDP‐glucuronic acid and UDP N‐acetyl‐glucosamine. 107 , 108 , 109 , 110 P2Y14 activation has a chemoattractant effect on monocytes/macrophages, as well as on human neutrophils and dendritic cells. 111 , 112 P2Y14 can also influence degranulation of human mast cells and suppresses T cell proliferation. 113 , 114 Bacterial‐derived uridine diphosphate and uridine triphosphate can interact with multiple different P2Y receptors. 115 The activation of P2Y receptors promotes chemotaxis and efferocytosis (the identification, isolation and engulfment of apoptotic cells) in human macrophages, neutrophils and dendritic cells by acting as “find me” signals and upregulating expression of phagocytotic receptors. 116 , 117 P2Y11R also blocks neutrophil apoptosis. 118
Succinate is an important metabolic intermediate in human cells and microbes. 119 The succinate receptor (GPR91) has been shown to influence the chemotaxis of human immature DCs while also playing a role in enhancing the production of the pro‐inflammatory cytokines TNF‐α and IL‐1β. 120 , 121 Succinate receptor knockout murine mast cells showed a hyperreactive phenotype, with effects in a contact dermatitis model. 122
The G2A receptor, also known as GPR132, is activated by microbiota‐encoded N‐acyl amides. 123 This receptor is highly expressed in murine macrophages and lymphoid tissue and is a member of the stress‐inducible ovarian cancer G protein‐coupled receptor 1 family of GPCR. 124 The endogenous ligand for the G2A receptor is unknown; however, the microbiota‐encoded N‐acyl amide N‐3‐hydroxypalmitoyl glycine, commonly referred to as Commendamide, has been shown to activate it. 123 The G2A receptor has been implicated in murine models of both autoimmune disease and atherosclerosis. 65
The oxoglutarate receptor (GPR99) is activated by α‐ketoglutaric acid, which can be bacterial‐derived. 125 , 126 α‐ketoglutaric acid has been shown to have a positive effect on histone demethylation, resulting in the promotion of T cell differentiation but inhibits murine M1 and enhances M2 polarization. 127 , 128
5. ARYL HYDROCARBON RECEPTOR
The aryl hydrocarbon receptor (AhR) is a ligand‐activated cytosolic receptor, which belongs to Pern‐Arnt‐Sim (PAS) superfamily. 129 It was originally identified as a sensor of xenobiotic chemicals, in particular, aromatic (aryl) hydrocarbons from which the receptor derives its name. 130 When AhR is in a latent or non‐DNA‐binding state, it is present in the cytoplasm and associated with the 90 kDa molecular heat shock protein 90 (hsp90), p23 and hepatitis B virus X‐associated protein (XAP2/AIP/Ara9). 131 Hsp90 masks AhR's nuclear localization sequence (NLS) which retains AhR in the cytoplasm and it chaperones a high‐affinity ligand‐binding conformation of the AhR. 132 When AhR binds with its ligand, the AhR/Hsp90 complex translocate to the nucleus where Hsp90 is exchanged for the partner protein AhR nuclear translocator (ARNT). 133 The nuclear‐localized, ligand‐bound AhR/ARNT heterodimer recognizes and promotes transcription from dioxin‐ or xenobiotic‐responsive elements (DREs/XREs), typified by the xenobiotic metabolizing enzymes cytochrome P4501A1 and the glutathione S‐transferase Ya subunit. 134 The aryl hydrocarbon receptor repressor (AhRR), a basic helix–loop–helix (bHLH/PAS) protein is also recognized by AHR/ARNT. 135 The AhRR shows high sequence similarity with the AhR in the N‐terminal region but diverges significantly at the C‐terminus and does not have ligand binding or transactivation domains. It is thought that in vivo the AhRR is expressed in response to AhR ligands and subsequently functions to down‐regulate AhR target genes by sequestering ARNT and competing for DRE sequences in the promoters of target genes. 135 Along with recognition of DREs by the AhR/ARNT complex it also interacts with other transcriptional regulators to control the expression of target genes (non‐canonical pathway). For example, NF‐κB induces AhR expression and AhR subsequently regulates NF‐κB signalling. 136 AhR activation and signalling are summarized in Figure 4.
FIGURE 4.
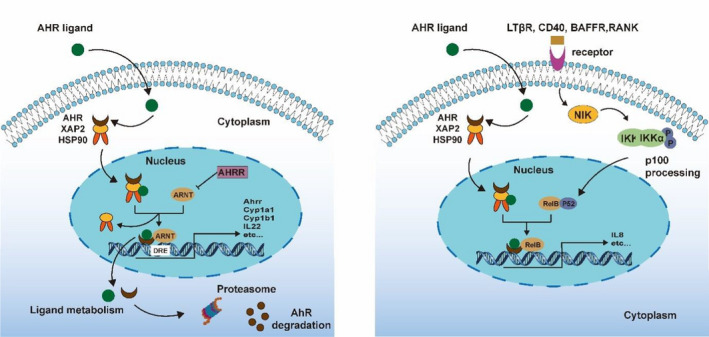
Mechanisms of canonical (left panel) and non‐canonical (right panel) AhR signalling
The best described microbial‐derived metabolites that activate AhR are the indole products of microbial tryptophan metabolism. These include indole, tryptamine, skatole, indole‐3‐pyruvate, indole‐3‐lactate, indole‐3‐acrylate, indole‐3‐propionate, indole‐3‐acetamide, indole‐3‐acetate, indole‐3‐ethanol, indole‐3‐aldehyde and indole‐3‐acetaldehyde. 137 In addition, bacterial virulence factors and quorum sensors (e.g. phenazines, naphthoquinone and malassezin) can activate the AhR. 138 , 139 Most recently, the SCFA butyrate has also been shown to activate the AhR. 140 A list of microbial species known to secrete AhR ligands is shown in Table 1.
TABLE 1.
Microbial species associated with production of AhR ligands
| Bacteria | AhR ligands |
| Lactobacillus reuteri | Indole derivative 47 , 141 |
| Lactobacillus murinus | Indole derivative 141 |
| Lactobacillus taiwanensis | Indole derivative 141 |
| Bacillus alvei | Indole derivative 142 |
| Clostridium novyi | Indole derivative 143 |
| Clostridium limosum | Indole derivative 144 |
| Clostridium tetani | Indole derivative 143 |
| Corynebacterium acnes | Indole derivative 145 |
| Enterococcus faecalis | Indole derivative 146 |
| Bacteroides thetaiotaomicron | Indole derivative 147 |
| Bacteroides sp. | Indole derivative 148 |
| Citrobacter sp. | Indole derivative 149 |
| Escherichia coli | Indole derivative 150 |
| Flavobacterium sp. | Indole derivative 151 |
| Fusobacterium sp. | Indole derivative 152 |
| Haemophilus influenza | Indole derivative 153 |
| Kleibsella planticola | Indole derivative 154 |
| Shigella flexneri | Indole derivative 155 |
| Vibrio cholera | Indole derivative 156 |
| Lactobacillus bulgaricus OLL1181 | Not yet identified AhR ligands 157 |
| Kleibsella pneumonia | Indirubin, Indigo 158 |
| Malassezia | Trypthantrin 159 |
| Propionibacterium freudenreichii ET‐3 | 1,4‐dihydroxy‐2‐naphtoic acid 160 |
| Malassezia | Malassezin 159 |
| Malassezia | Indirubin 159 |
| Providencia stuartii | Indirubin 158 |
AhR is mainly expressed within barrier organs such as the skin, intestine, lung and associated immune cells, where AhR plays an important role in development and regulation of both innate and adaptive immunity. Multiple murine models suggest an important role for the AhR in maintaining the functional properties of Foxp3 Tregs, in particular the T cell immunoglobin and ITIM domain (TIGIT) + Foxp3+ Treg cells that express the highest levels of AhR relative to other Treg populations. 161 , 162 , 163 IL‐27 drives AhR expression in IL‐10 producing Tr1 cells where AhR cooperates with STAT3 to induce expression of the immunoregulatory ectonucleotidase CD39. 164 , 165 , 166 In addition, AhR plays a role in transdifferentiation of murine TH17 cells into Tregs. 167 However, AhR can also enhance TH17 cell differentiation and IL‐22 secretion suggesting that specific ligands may have divergent effects within different microenvironments and cell differentiation stages. 168 Indeed, AhR‐deficient murine dendritic cells fail to promote Treg cell differentiation but instead drive TH17 cell generation in vitro. 169 Importantly, AhR activation has been shown to be involved in the induction of murine CD4 + CD8αα + double‐positive intraepithelial lymphocytes (DP IELs), which display regulatory functions associated with the induction of tolerance to dietary antigens. 170 Similarly, AhR signalling contributes to the persistence of murine tissue‐resident CD8+ memory cells (TRMs) within the skin, supporting their protective role against microbial challenge. 171
6. NUCLEAR RECEPTORS
Nuclear receptors (NRs) are a family of ligand‐activated transcription factors that regulate numerous physiological processes such as metabolism, reproduction, inflammation, as well as the circadian rhythm. NRs sense changes in lipid metabolite levels to drive differential gene expression, producing distinct physiologic effects. 172 NRs share a common architecture of an N‐terminal ligand‐independent activation domain (AF2) followed by a DNA‐binding domain (DBD), a flexible hinge region (Hinge) and a ligand‐binding domain (LBD) composed of 12 α‐helices (α1‐α12). 173 NRs that respond to bacterial‐derived ligands are summarized in Figure 5.
FIGURE 5.
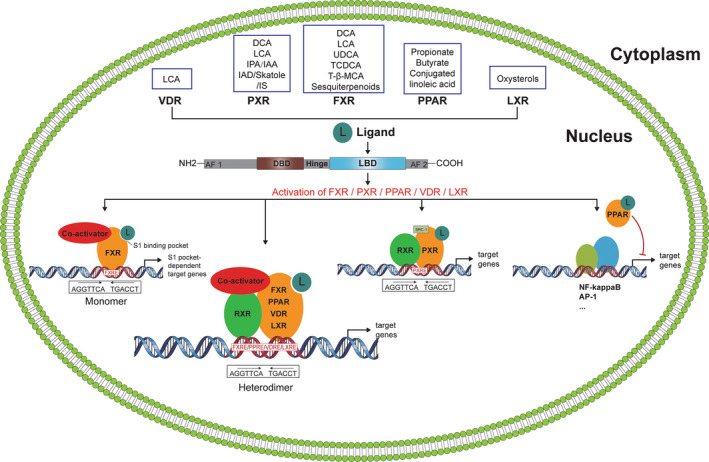
The different nuclear receptors are illustrated
Farnesoid X receptor (FXR) is a bile acid‐activated nuclear receptor. 174 As a ligand‐activated transcription factor, FXR binds to DNA either as a monomer or as a heterodimer with retinoid X receptor (RXR, NR2B1) to regulate the transcription of target genes. 175 FXR is expressed within the human intestinal tract but also at other sites including the lung. 176 , 177 Bacterial modified secondary bile acids, such as deoxycholic acid and lithocholic acid, are FXR agonists, while supernatants from in vitro bacterial cultures also possess FXR ligands. 178 , 179 FXR‐RARα signalling in murine mucosal dendritic cells supported food allergen‐specific IgE and IgG1 production, while allergic diarrhoea can be attenuated by FXR agonists due to their antisecretory effects in colonic epithelial cells. 180 , 181 Ursodeoxycholic acid treatment of OVA‐sensitized mice prior to OVA aerosol challenge significantly reduced eosinophilic airway inflammation via FXR signalling in dendritic cells. 182 Similarly, the FXR ligand chenodeoxycholic acid attenuates murine allergic airway inflammation by inhibiting TH2 cytokine secretion. 183
Pregnane X receptor (PXR), a critical xenobiotic‐sensing nuclear receptor, is expressed predominantly in the gastrointestinal tract and liver, but has also been shown to play an important role in murine and human skin where activation by environmental pollutants comprises epidermal barrier function favouring a TH2/Th17 response that resembles atopic dermatitis. 184 , 185 Bacterial‐modified bile acids are PXR ligands, while bacterial‐derived tryptophan metabolites such as indole‐3‐acetamide and indole‐3‐acetic acid also activate PXR with significant effects on epithelial barrier function in the murine gut. 186 , 187 , 188 PXR‐deficient mice exhibit an exaggerated T lymphocyte proliferation with increased CD25 expression. Furthermore, PXR‐deficient lymphocytes produce more IFN‐γ and less IL‐10. 189
Peroxisome proliferator‐activated receptors (PPARs) form a small family of ligand‐activated transcription factors belonging to the nuclear receptor superfamily, which consists of 3 isotypes that are known to be involved in diverse biologic processes, including immune and inflammatory responses. 190 Microbial linoleic acid‐derived fatty acids and conjugated linoleic acids are potent PPARα and PPARγ agonists. 191 , 192 More recently, butyrate and propionate have been identified as PPARγ agonists in human intestinal epithelial cells. 193 PPARγ signalling is important in regulating murine T cell‐mediated responses by inhibiting effector T cell responses, while supporting Treg cells. 194 , 195 In addition, PPARγ signalling suppresses mast cell activation and TLR‐induced NF‐κB phosphorylation in human dendritic cells. 196 , 197 , 198
Vitamin D receptor (VDR) is a nuclear hormone receptor and transcription factor expressed in a variety of tissues, including the intestines, adipose tissue and liver, as well as many immune cell subsets. 199 In addition to vitamin D, the bacterial‐derived secondary bile acid lithocholic acid is a physiological VDR ligand, which has been shown to influence human and murine TH1 lymphocyte activation. 200 , 201 Interestingly, gut microbiota composition was significantly altered in intestinal epithelial VDR conditional knockout mice, while a genome‐wide association study in humans identified associations between overall microbial variation and individual taxa with the VDR gene. 202 , 203
Liver X receptors (LXR)α and LXR β are members of the nuclear receptor superfamily that regulate the expression of genes involved in lipid, glucose and cholesterol metabolism and homeostasis. 204 While LXR has been well described as a negative regulator of murine macrophage cytokine secretion, no microbial ligands have been discovered to date that are selective LXR ligands. 205
7. CONCLUSIONS AND FUTURE DIRECTIONS
The studies described in this review demonstrate that the different types of metabolites generated by microbes in the gut can have a profound impact on immune regulatory and immune effector functions. The structural similarities and overlaps that exist between microbiota‐derived metabolites and endogenous signalling molecules are surprising, but this allows for relatively uncomplicated signalling molecules produced by the microbiome to impact more complex cellular interactions within the host. The immune response to bacteria should not be considered simply as a form of host defence but also represents a variety of intimate interactions with important symbiotic physiological effects on the host. Virtually all known cells with immunological functions (e.g. dendritic cells, epithelial cells, ILCs, T regulatory cells, effector lymphocytes, NKT cells and B cells) can be influenced directly or indirectly by microbial factors. However, interactions between the host and microbiota are almost certainly bidirectional, with species‐ and strain‐specific behaviours shaped by the genetic background and microenvironmental niche in which they occur. Microbial factors are clearly evolutionarily hardwired into the molecular circuitry governing immune cell decision‐making processes, but we have only discovered a relatively small number of metabolites thus far that contribute to this intimate and sophisticated inter‐kingdom dialogue. In addition to identifying the metabolites themselves, there are also major gaps in our understanding of the mechanisms that integrate microbiota‐derived signals into host immune pathways.
While BMI and (un)healthy dietary patterns have consistently been shown to be related to specific microbiota configurations, many of the strongest associations have been with poorly characterized microbes and often there is little or no information available on their specific molecular interactions within the host. In addition, due to the low resolution of dietary questionnaire data, the complexity of dietary patterns, nutrient–nutrient interactions and incomplete knowledge on the clustering of healthy/less healthy food items, it is often challenging to disentangle the independent associations of single foods with microbial species. Thus, future studies should endeavour to investigate beyond correlations to provide a functional basis for understanding causation. This is critically needed as in many cases it remains unclear whether and, if so, to what extent patterns of microbial taxonomical or functional dysbiosis actually drives rather than merely reflects associated patterns of immune reactivity.
Identifying the missing metabolites critical for immune development and regulation will substantially impact our understanding of the communications platform used by the immune system to interact with our microenvironment and will provide us with new tools to modify this communication when necessary to improve immune health. Indeed, given the malleability of the human microbiome, its integration into the immune system and its responsiveness to diet, makes it a highly attractive target for therapeutic and nutritional intervention. New studies focussed on understanding how specific dietary interventions impact the microbiota to generate immunoregulatory metabolites will be critical to develop effective diets that improve human immune health and prevent aberrant inflammatory responses.
AUTHOR CONTRIBUTIONS
BF, LY, RS, SM, NL and LOM contributed to drafting the manuscript. All authors read, reviewed and agreed the final version of this manuscript.
CONFLICT OF INTEREST
LOM is a consultant to PrecisionBiotics and has received research funding from GSK and Chiesi. LOM has participated in speaker's bureau for Nestle, Nutricia, Reckitt and Abbott. None of the other authors report any conflict of interest.
ACKNOWLEDGEMENTS
The authors are supported by a Science Foundation Ireland research center grant 12/RC/2273_P2. The authors thank Gil Costa (https://www.gilcosta.com) for support in the generation of graphical illustrations accompanying this Review. Open access funding provided by IReL.
Forde B, Yao L, Shaha R, Murphy S, Lunjani N, O’Mahony L. Immunomodulation by foods and microbes: Unravelling the molecular tango. Allergy. 2022;77:3513‐3526. doi: 10.1111/all.15455
REFERENCES
- 1. Ivanov II, Tuganbaev T, Skelly AN, Honda K. T cell responses to the microbiota. Annu Rev Immunol. 2022;40:559‐587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lunjani N, Ahearn‐Ford S, Dube FS, Hlela C, O'Mahony L. Mechanisms of microbe‐immune system dialogue within the skin. Genes Immun. 2021;22(5–6):276‐288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walter J, O'Mahony L. The importance of social networks‐An ecological and evolutionary framework to explain the role of microbes in the aetiology of allergy and asthma. Allergy. 2019;74(11):2248‐2251. [DOI] [PubMed] [Google Scholar]
- 4. Akdis CA. Does the epithelial barrier hypothesis explain the increase in allergy, autoimmunity and other chronic conditions? Nat Rev Immunol. 2021;21:739‐751. [DOI] [PubMed] [Google Scholar]
- 5. Liwinski T, Zheng D, Elinav E. The microbiome and cytosolic innate immune receptors. Immunol Rev. 2020;297:207‐224. [DOI] [PubMed] [Google Scholar]
- 6. Hosseinkhani F, Heinken A, Thiele I, Lindenburg PW, Harms AC, Hankemeier T. The contribution of gut bacterial metabolites in the human immune signaling pathway of non‐communicable diseases. Gut Microbes. 2021;13:1‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barcik W, Wawrzyniak M, Akdis CA, O'Mahony L. Immune regulation by histamine and histamine‐secreting bacteria. Curr Opin Immunol. 2017;48:108‐113. [DOI] [PubMed] [Google Scholar]
- 8. Trujillo J, Lunjani N, Ryan D, O'Mahony L. Microbiome‐immune interactions and relationship to asthma severity. J Allergy Clin Immunol. 2022;149(2):533‐534. [DOI] [PubMed] [Google Scholar]
- 9. Albrich WC, Ghosh TS, Ahearn‐Ford S, et al. A high‐risk gut microbiota configuration associates with fatal hyperinflammatory immune and metabolic responses to SARS‐CoV‐2. Gut Microbes. 2022;14(1):2073131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wallimann A, Magrath W, Pugliese B, et al. Butyrate inhibits osteoclast activity in vitro and regulates systemic inflammation and bone healing in a murine osteotomy model compared to antibiotic‐treated mice. Mediat Inflamm. 2021;2021:8817421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lunjani N, Satitsuksanoa P, Lukasik Z, Sokolowska M, Eiwegger T, O'Mahony L. Recent developments and highlights in mechanisms of allergic diseases: microbiome. Allergy. 2018;73(12):2314‐2327. [DOI] [PubMed] [Google Scholar]
- 12. Michalovich D, Rodriguez‐Perez N, Smolinska S, et al. Obesity and disease severity magnify disturbed microbiome‐immune interactions in asthma patients. Nat Commun. 2019;10(1):5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Plovier H, Cani PD. Microbial impact on host metabolism: Opportunities for novel treatments of nutritional disorders? Microbiol Spectr. 2017;5(3). [DOI] [PubMed] [Google Scholar]
- 14. Merino J, Joshi AD, Nguyen LH, et al. Diet quality and risk and severity of COVID‐19: a prospective cohort study. Gut. 2021;70(11):2096‐2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. GBD 2015 Obesity Collaborators . Health effects of overweight and obesity in 195 countries over 25 years. N Engl J Med. 2017;377:13‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Depommier C, Everard A, Druart C, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof‐of‐concept exploratory study. Nat Med. 2019;25(7):1096‐1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sokolowska M, Frei R, Lunjani N, Akdis CA, O'Mahony L. Microbiome and asthma. Asthma Res Pract. 2018;4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kucuksezer UC, Ozdemir C, Akdis M, Akdis CA. Influence of innate immunity on immune tolerance. Acta Med Acad. 2020;49(2):164‐180. [DOI] [PubMed] [Google Scholar]
- 19. Yumuk V, Tsigos C, Fried M, et al. European guidelines for obesity management in adults. Obes Facts. 2015;8:402‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Venter C, Greenhawt M, Meyer RW, et al. EAACI position paper on diet diversity in pregnancy, infancy and childhood: novel concepts and implications for studies in allergy and asthma. Allergy. 2020;75(3):497‐523. [DOI] [PubMed] [Google Scholar]
- 21. Cotillard A, Kennedy SP, Kong LC, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585‐588. [DOI] [PubMed] [Google Scholar]
- 22. Liou AP, Paziuk M, Luevano JM, Machineni S, Turnbaugh PJ, Kaplan LM. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med. 2013;5:178ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koutoukidis DA, Jebb SA, Zimmerman M, et al. The association of weight loss with changes in the gut microbiota diversity, composition, and intestinal permeability: a systematic review and meta‐analysis. Gut Microbes. 2022;14(1):2020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roduit C, Frei R, Ferstl R, et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy. 2019;74(4):799‐809. [DOI] [PubMed] [Google Scholar]
- 25. Venter C, Palumbo MP, Glueck DH, et al. The maternal diet index in pregnancy is associated with offspring allergic diseases: the healthy start study. Allergy. 2022;77(1):162‐172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Venter C, Palumbo MP, Sauder KA, et al. Associations between child filaggrin mutations and maternal diet with the development of allergic diseases in children. Pediatr Allergy Immunol. 2022;33:e13753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jagielski P, Łuszczki E, Wnęk D, et al. Associations of nutritional behavior and gut microbiota with the risk of COVID‐19 in healthy Young adults in Poland. Nutrients. 2022;14(2):350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim H, Rebholz CM, Hegde S, et al. Plant‐based diets, pescatarian diets and COVID‐19 severity: a population‐based case‐control study in six countries. BMJ Nutr Prev Health. 2021;4:257‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wastyk HC, Fragiadakis GK, Perelman D, et al. Gut‐microbiota‐targeted diets modulate human immune status. Cell. 2021;184(16):4137‐4153.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gill SK, Rossi M, Bajka B, Whelan K. Dietary fibre in gastrointestinal health and disease. Nat Rev Gastroenterol Hepatol. 2021;18(2):101‐116. [DOI] [PubMed] [Google Scholar]
- 31. Venter C, Meyer RW, Greenhawt M, et al. Role of dietary fiber in promoting immune health‐An EAACI position paper. Allergy. 2022;77:3185‐3198. doi: 10.1111/all.15430 [DOI] [PubMed] [Google Scholar]
- 32. Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet‐induced extinctions in the gut microbiota compound over generations. Nature. 2016;529(7585):212‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fragiadakis GK, Smits SA, Sonnenburg ED, et al. Links between environment, diet, and the hunter‐gatherer microbiome. Gut Microbes. 2019;10(2):216‐227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol. 2011;65:411‐429. [DOI] [PubMed] [Google Scholar]
- 35. Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222‐227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hagan T, Cortese M, Rouphael N, et al. Antibiotics‐driven gut microbiome perturbation alters immunity to vaccines in humans. Cell. 2019;178(6):1313‐1328.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lunjani N, Albrich WC, Suh N, et al. Higher levels of bacterial DNA in serum associate with severe and fatal COVID‐19. Allergy. 2022;77:1312‐1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yeoh YK, Zuo T, Lui GC, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID‐19. Gut. 2021;70:698‐706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zuo T, Zhang F, Lui GCY, et al. Alterations in gut microbiota of patients with COVID‐19 during time of hospitalization. Gastroenterology. 2020;159:944‐955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ahearn‐Ford S, Lunjani N, McSharry B, et al. Long‐term disruption of cytokine signalling networks is evident in patients who required hospitalization for SARS‐CoV‐2 infection. Allergy. 2021;76(9):2910‐2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sadlier C, Albrich WC, Neogi U, et al. Metabolic rewiring and serotonin depletion in patients with postacute sequelae of COVID‐19. Allergy. 2022;77:1623‐1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Crestani E, Harb H, Charbonnier LM, et al. Untargeted metabolomic profiling identifies disease‐specific signatures in food allergy and asthma. J Allergy Clin Immunol. 2020;145(3):897‐906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Donia MS, Cimermancic P, Schulze CJ. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402‐1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. 2018;23(6):716‐724. [DOI] [PubMed] [Google Scholar]
- 45. Gheorghe CE, Martin JA, Manriquez FV, Dinan TG, Cryan JF, Clarke G. Focus on the essentials: tryptophan metabolism and the microbiome‐gut‐brain axis. Curr Opin Pharmacol. 2019;48:137‐145. [DOI] [PubMed] [Google Scholar]
- 46. Zelante T, Puccetti M, Giovagnoli S, Romani L. Regulation of host physiology and immunity by microbial indole‐3‐aldehyde. Curr Opin Immunol. 2021;70:27‐32. [DOI] [PubMed] [Google Scholar]
- 47. Zelante T, Lannitti RG, Cunha C, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin‐22. Immunity. 2013;39:372‐385. [DOI] [PubMed] [Google Scholar]
- 48. Powell DN, Swimm A, Sonowal R, et al. Indoles from the commensal microbiota act via the AHR and IL‐10 to tune the cellular composition of the colonic epithelium during aging. Proc Natl Acad Sci U S A. 2020;117:21519‐21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Swimm A, Giver CR, DeFilipp Z, et al. Indoles derived from intestinal microbiota act via type I interferon signaling to limit graft‐versus‐host disease. Blood. 2018;132:2506‐2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Levy M, Thaiss CA, Zeevi D, et al. Microbiota‐modulated metabolites shape the intestinal microenvironment by regulating NLRP6 Inflammasome signaling. Cell. 2015;163(6):1428‐1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Levan SR, Stamnes KA, Lin DL, et al. Elevated faecal 12,13‐diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat Microbiol. 2019;4(11):1851‐1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Trompette A, Gollwitzer ES, Yadava K, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20(2):159‐166. [DOI] [PubMed] [Google Scholar]
- 53. Antunes KH, Fachi JL, de Paula R, et al. Microbiota‐derived acetate protects against respiratory syncytial virus infection through a GPR43‐type 1 interferon response. Nat Commun. 2019;10(1):3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zheng L, Kelly CJ, Battista KD, et al. Microbial‐derived butyrate promotes epithelial barrier function through IL‐10 receptor‐dependent repression of claudin‐2. J Immunol. 2017;199:2976‐2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thio CL, Chi PY, Lai AC, Chang YJ. Regulation of type 2 innate lymphoid cell‐dependent airway hyperreactivity by butyrate. J Allergy Clin Immunol. 2018;142:1867‐1883.e12. [DOI] [PubMed] [Google Scholar]
- 56. Blacher E, Bashiardes S, Shapiro H, et al. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature. 2019;572(7770):474‐480. [DOI] [PubMed] [Google Scholar]
- 57. Legoux F, Bellet D, Daviaud C, et al. Microbial metabolites control the thymic development of mucosal‐associated invariant T cells. Science. 2019;366(6464):494‐499. [DOI] [PubMed] [Google Scholar]
- 58. An D, Oh SF, Olszak T, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123‐133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Katritch V, Cherezov V, Stevens RC. Structure‐function of the G protein–coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531‐556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G‐protein‐coupled receptors. Nature. 2013;494(7436):185‐194. [DOI] [PubMed] [Google Scholar]
- 61. Tang XL, Wang Y, Li DL, Luo J, Liu MY. Orphan G protein‐coupled receptors (GPCRs): biological functions and potential drug targets. Acta Pharmacol Sin. 2012;33(3):363‐371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kobilka BK. G protein coupled receptor structure and activation. Biochim Biophys Acta Biomembr. 2007;1768(4):794‐807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rosenbaum D, Rasmussen S, Kobilka B. The structure and function of G‐protein‐coupled receptors. Nature. 2009;459(7245):356‐363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Weis W, Kobilka B. The molecular basis of G protein–coupled receptor activation. Annu Rev Biochem. 2018;87:897‐919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cohen LJ, Esterhazy D, Kim SH, et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature. 2017;549:48‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chen H, Nwe PK, Yang Y, et al. A forward chemical genetic screen reveals gut microbiota metabolites that modulate host physiology. Cell. 2019;177(5):1217‐1231.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Colosimo DA, Kohn JA, Luo PM, et al. Mapping interactions of microbial metabolites with human G‐protein‐coupled receptors. Cell Host Microbe. 2019;26:273‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Brown A, Goldsworthy S, Barnes A, et al. The orphan G protein‐coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312‐11319. [DOI] [PubMed] [Google Scholar]
- 69. Liu P, Wang Y, Yang G, et al. The role of short‐chain fatty acids in intestinal barrier function, inflammation, oxidative stress, and colonic carcinogenesis. Pharmacol Res. 2021;9:105420. [DOI] [PubMed] [Google Scholar]
- 70. Macfarlane S, Macfarlane GT. Regulation of short‐chain fatty acid production. Proc Nutr Soc. 2003;62(1):67‐72. [DOI] [PubMed] [Google Scholar]
- 71. Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short‐chain fatty acids in health and disease. Adv Immunol. 2014;121:91‐119. [DOI] [PubMed] [Google Scholar]
- 72. Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30(6):492‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Maslowski K, Vieira A, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282‐1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lind S, Holdfeldt A, Mårtensson J, et al. Functional selective ATP receptor signaling controlled by the free fatty acid receptor 2 through a novel allosteric modulation mechanism. FASEB J. 2019;33:6887‐6903. [DOI] [PubMed] [Google Scholar]
- 75. Sutton C, Lalor S, Sweeney C, Brereton C, Lavelle E, Mills K. Interleukin‐1 and IL‐23 induce innate IL‐17 production from γδ T cells, amplifying Th17 responses and autoimmunity. Immunity. 2009;31:331‐341. [DOI] [PubMed] [Google Scholar]
- 76. Yang W, Xiao Y, Huang X, et al. Microbiota metabolite short‐chain fatty acids facilitate mucosal adjuvant activity of cholera toxin through GPR43. J Immunol. 2019;203:282‐292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wu W, Sun M, Chen F, Yao S, Liu Z, Cong Y. Microbiota metabolite short chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Gastroenterology. 2017;152:S199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bolognini D, Dedeo D, Milligan G. Metabolic and inflammatory functions of short‐chain fatty acid receptors. Curr Opinion Endocrine Metabolic Res. 2021;16:1‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ganapathy V, Thangaraju M, Prasad PD, Martin PM, Singh N. Transporters and receptors for short‐chain fatty acids as the molecular link between colonic bacteria and the host. Curr Opin Pharmacol. 2013;13:869‐874. [DOI] [PubMed] [Google Scholar]
- 80. Singh N, Gurav A, Sivaprakasam S, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40(1):128‐139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Coton E, Coton M. Multiplex PCR for colony direct detection of gram‐positive histamine‐ and tyramine‐producing bacteria. J Microbiol Methods. 2005;63:296‐304. [DOI] [PubMed] [Google Scholar]
- 82. Meiler F, Zumkehr J, Klunker S, Rückert B, Akdis C, Akdis M. In vivo switch to IL‐10–secreting T regulatory cells in high dose allergen exposure. J Exp Med. 2008;205:2887‐2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Frei R, Ferstl R, Konieczna P, et al. Histamine receptor 2 modifies dendritic cell responses to microbial ligands. J Allergy Clin Immunol. 2013;132:194‐204. [DOI] [PubMed] [Google Scholar]
- 84. Ferstl R, Frei R, Barcik W, et al. Histamine receptor 2 modifies iNKT cell activity within the inflamed lung. Allergy. 2017;72:1925‐1935. [DOI] [PubMed] [Google Scholar]
- 85. Ferstl R, Frei R, Schiavi E, et al. Histamine receptor 2 is a key influence in immune responses to intestinal histamine‐secreting microbes. J Allergy Clin Immunol. 2014;134:744‐746. [DOI] [PubMed] [Google Scholar]
- 86. Jutel M, Watanabe T, Klunker S, et al. Histamine regulates T‐cell and antibody responses by differential expression of H1 and H2 receptors. Nature. 2001;413(6854):420‐425. [DOI] [PubMed] [Google Scholar]
- 87. Gutzmer R, Diestel C, Mommert S, et al. Histamine H4 receptor stimulation suppresses IL‐12p70 production and mediates chemotaxis in human monocyte‐derived dendritic cells. J Immunol. 2005;174:5224‐5232. [DOI] [PubMed] [Google Scholar]
- 88. Dunford PJ, O'Donnell N, Riley JP, Williams KN, Karlsson L, Thurmond RL. The histamine H4 receptor mediates allergic airway inflammation by regulating the activation of CD4+ T cells. J Immunol. 2006;176:7062‐7070. [DOI] [PubMed] [Google Scholar]
- 89. Barcik W, Pugin B, Brescó MS, et al. Bacterial secretion of histamine within the gut influences immune responses within the lung. Allergy. 2019;74(5):899‐909. [DOI] [PubMed] [Google Scholar]
- 90. Barcik W, Pugin B, Westermann P, et al. Histamine‐secreting microbes are increased in the gut of adult asthma patients. J Allergy Clin Immunol. 2016;138:1491‐1494. [DOI] [PubMed] [Google Scholar]
- 91. Pugin B, Barcik W, Westermann P, et al. A wide diversity of bacteria from the human gut produces and degrades biogenic amines. Microb Ecol Health Dis. 2017;28:1353881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Rogers AC, McDermott FD, Mohan HM, O'Connell PR, Winter DC, et al. The effects of polyamines on human colonic mucosal function. Eur J Pharmacol. 2015;764:157‐163. [DOI] [PubMed] [Google Scholar]
- 93. Wawrzyniak M, Groeger D, Frei R, et al. Spermidine and spermine exert protective effects within the lung. Pharmacol Res Perspect. 2021;9(4):e00837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nøhr AC, Shehata MA, Palmer D, et al. Identification of a novel scaffold for a small molecule GPR139 receptor agonist. Sci Rep. 2019;9(1):3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhao X, Xian Y, Wang C, et al. Calcium‐sensing receptor‐mediated L‐tryptophan‐induced secretion of cholecystokinin and glucose‐dependent insulinotropic peptide in swine duodenum. J Vet Sci. 2018;19(2):179‐187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nyeste L, Pécs M, Sevella B, Holló J. Production of L‐tryptophan by microbial processes. Adv Biochem Eng Biotechnol. 1983;26:176‐202. [PubMed] [Google Scholar]
- 97. Bowery NG. GABAB receptor pharmacology. Annu Rev Pharmacol Toxicol. 1993;33:109‐147. [DOI] [PubMed] [Google Scholar]
- 98. Phuengjayaem S, Kuncharoen N, Booncharoen A, Ongpipattanakul B, Tanasupawat S. Genome analysis and optimization of γ‐aminobutyric acid (GABA) production by lactic acid bacteria from plant materials. J Gen Appl Microbiol. 2021;67(4):150‐161. [DOI] [PubMed] [Google Scholar]
- 99. Duthey B, Hübner A, Diehl S, Boehncke S, Pfeffer J, Boehncke WH. Anti‐inflammatory effects of the GABA(B) receptor agonist baclofen in allergic contact dermatitis. Exp Dermatol. 2010;19(7):661‐666. [DOI] [PubMed] [Google Scholar]
- 100. Pilc A, Ossowska K. Metabotropic glutamate receptors. Amino Acids. 2007;32(2):165‐167. [DOI] [PubMed] [Google Scholar]
- 101. Zareian M, Ebrahimpour A, Bakar FA, et al. A glutamic acid‐producing lactic acid bacteria isolated from Malaysian fermented foods. Int J Mol Sci. 2012;13(5):5482‐5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Alim MA, Grujic M, Ackerman PW, et al. Glutamate triggers the expression of functional ionotropic and metabotropic glutamate receptors in mast cells. Cell Mol Immunol. 2021;18:2383‐2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ganor Y, Besser M, Ben‐Zakay N, Unger T, Levite M. Human T cells express a functional ionotropic glutamate receptor GluR3, and glutamate by itself triggers integrin‐mediated adhesion to laminin and fibronectin and chemotactic migration. J Immunol. 2003;170:4362‐4372. [DOI] [PubMed] [Google Scholar]
- 104. Marucci G, Dal Ben D, Lambertucci C, et al. The G protein‐coupled receptor GPR17: overview and update. ChemMedChem. 2016;11:2567‐2574. [DOI] [PubMed] [Google Scholar]
- 105. Xiong ZQ, Kong LH, Lai PF, et al. Genomic and phenotypic analyses of exopolysaccharide biosynthesis in Streptococcus thermophilus S‐3. J Dairy Sci. 2019;102:4925‐4934. [DOI] [PubMed] [Google Scholar]
- 106. Maekawa A, Xing W, Austen KF, Kanaoka Y. GPR17 regulates immune pulmonary inflammation induced by house dust mites. J Immunol. 2010;185(3):1846‐1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Rafehi M, Müller CE. Tools and drugs for uracil nucleotide‐activated P2Y receptors. Pharmacol Ther. 2018;190:24‐80. [DOI] [PubMed] [Google Scholar]
- 108. Fricks IP, Carter RL, Lazarowski ER, Harden TK. Gi‐dependent cell signaling responses of the human P2Y14 receptor in model cell systems. J Pharmacol Exp Ther. 2009;330(1):162‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Coyne MJ, Fletcher CM, Reinap B, Comstock LE. UDP‐glucuronic acid decarboxylases of Bacteroides fragilis and their prevalence in bacteria. J Bacteriol. 2011;193(19):5252‐5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rodríguez‐Díaz J, Rubio‐Del‐Campo A, Yebra MJ. Regulatory insights into the production of UDP‐N‐acetylglucosamine by lactobacillus casei. Bioengineered. 2012;3(6):339‐342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Xu J, Morinaga H, Oh D, et al. GPR105 ablation prevents inflammation and improves insulin sensitivity in mice with diet‐induced obesity. J Immunol. 2012;189(4):1992‐1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sesma JI, Kreda SM, Steinckwich‐Besancon N, et al. The UDP‐sugar‐sensing P2Y(14) receptor promotes rho‐mediated signaling and chemotaxis in human neutrophils. Am J Physiol Cell Physiol. 2012;303(5):C490‐C498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Gao ZG, Wei Q, Jayasekara MP, Jacobson KA. The role of P2Y(14) and other P2Y receptors in degranulation of human LAD2 mast cells. Purinergic Signal. 2013;9(1):31‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Scrivens M, Dickenson JM. Functional expression of the P2Y14 receptor in murine T‐lymphocytes. Br J Pharmacol. 2005;146(3):435‐444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Nishimura A, Sunggip C, Oda S, Numaga‐Tomita T, Tsuda M, Nishida M. Purinergic P2Y receptors: molecular diversity and implications for treatment of cardiovascular diseases. Pharmacol Ther. 2017;180:113‐128. [DOI] [PubMed] [Google Scholar]
- 116. Kim GT, Hahn KW, Sohn KY, Yoon SY, Kim JW. PLAG enhances macrophage mobility for efferocytosis of apoptotic neutrophils via membrane redistribution of P2Y2. FEBS J. 2019;286:5016‐5029. [DOI] [PubMed] [Google Scholar]
- 117. Chen Y, Corriden R, Inoue Y, et al. ATP release guides neutrophil chemotaxis via P2Y2 and A3 receptors. Science. 2006;314(5806):1792‐1795. [DOI] [PubMed] [Google Scholar]
- 118. Vaughan KR, Stokes L, Prince LR, et al. Inhibition of neutrophil apoptosis by ATP is mediated by the P2Y11 receptor. J Immunol. 2007;179(12):8544‐8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wang K, Liao M, Zhou N, et al. Parabacteroides distasonis alleviates obesity and metabolic dysfunctions via production of succinate and secondary bile acids. Cell Rep. 2019;26(1):222‐235. [DOI] [PubMed] [Google Scholar]
- 120. Rubic T, Lametschwandtner G, Jost S, et al. Triggering the succinate receptor GPR91 on dendritic cells enhances immunity. Nat Immunol. 2008;9(11):1261‐1269. [DOI] [PubMed] [Google Scholar]
- 121. Trauelsen M, Rexen Ulven E, Hjorth SA, et al. Receptor structure‐based discovery of non‐metabolite agonists for the succinate receptor GPR91. Mol Metab. 2017;6(12):1585‐1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Rubić‐Schneider T, Carballido‐Perrig N, Regairaz C, et al. GPR91 deficiency exacerbates allergic contact dermatitis while reducing arthritic disease in mice. Allergy. 2017;72(3):444‐452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Cohen LJ, Kang HS, Chu J, et al. Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist. Proc Natl Acad Sci U S A. 2015;112(35):E4825‐E4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Johnson LE, Elias MS, Bolick DT, Skaflen MD, Green RM, Hedrick CC. The G protein–coupled receptor G2A: involvement in hepatic lipid metabolism and gallstone formation in mice. Hepatology. 2008;48(4):1138‐1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Omede A, Zi M, Prehar S, et al. The oxoglutarate receptor 1 (OXGR1) modulates pressure overload‐induced cardiac hypertrophy in mice. Biochem Biophys Res Commun. 2016;479(4):708‐714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ogata K, Osugi M. Production of α‐Ketoglutaric acid from salicylic acid by bacteria. Agric Biol Chem. 1966;30(4):416‐417. [Google Scholar]
- 127. Liu PS, Wang H, Li X, et al. α‐Ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat Immunol. 2017;18(9):985‐994. [DOI] [PubMed] [Google Scholar]
- 128. Chisolm DA, Savic D, Moore AJ, et al. CCCTC‐binding factor translates interleukin 2‐ and α‐Ketoglutarate‐sensitive metabolic changes in T cells into context‐dependent gene programs. Immunity. 2017;47(2):251‐267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Gu YZ, Hogenesch JB, Bradfield CA. The PAS superfamily: sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol. 2000;40:519‐561. [DOI] [PubMed] [Google Scholar]
- 130. Stevens EA, Mezrich JD, Bradfield CA. The aryl hydrocarbon receptor: a perspective on potential roles in the immune system. Immunology. 2009;127(3):299‐311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Pollenz RS, Sattler CA, Poland A. The aryl hydrocarbon receptor and aryl hydrocarbon receptor nuclear translocator protein show distinct subcellular localizations in Hepa 1c1c7 cells by immunofluorescence microscopy. Mol Pharmacol. 1994;45(3):428‐438. [PubMed] [Google Scholar]
- 132. Ikuta T, Tachibana T, Watanabe J, Yoshida M, Yoneda Y, Kawajiri K. Nucleocytoplasmic shuttling of the aryl hydrocarbon receptor. J Biochem. 2000;127(3):503‐509. [DOI] [PubMed] [Google Scholar]
- 133. Lees MJ, Whitelaw ML. Multiple roles of ligand in transforming the dioxin receptor to an active basic helix‐loop‐helix/PAS transcription factor complex with the nuclear protein Arnt. Mol Cell Biol. 1999;19(8):5811‐5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Kleman MI, Overvik E, Poellinger L, Gustafsson JA. Induction of cytochrome P4501A isozymes by heterocyclic amines and other food‐derived compounds. Princess Takamatsu Symp. 1995;23:163‐171. [PubMed] [Google Scholar]
- 135. Baba T, Mimura J, Gradin K, et al. Structure and expression of the ah receptor repressor gene. J Biol Chem. 2001;276(35):33101‐33110. [DOI] [PubMed] [Google Scholar]
- 136. Vogel CF, Matsumura F. A new cross‐talk between the aryl hydrocarbon receptor and RelB, a member of the NF‐kappaB family. Biochem Pharmacol. 2009;77(4):734‐745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Vyhlídalová B, Krasulová K, Pečinková P, et al. Gut microbial catabolites of tryptophan are ligands and agonists of the aryl hydrocarbon receptor: a detailed characterization. Int J Mol Sci. 2020;21(7):2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Moura‐Alves P, Fae K, Houthouys E, et al. AhR sensing of bacterial pigments regulate antibacterial defence. Nature. 2014;512:387‐392. [DOI] [PubMed] [Google Scholar]
- 139. Moura‐Alves P, Puyskens A, Stinn A, et al. Host monitoring of quorum sensing during Pseudomonas aeroginosa infection. Science. 2019;366:1472. [DOI] [PubMed] [Google Scholar]
- 140. Marinelli L, Martin‐Gallausiaux C, Bourhis JM, Béguet‐Crespel F, Blottière HM, Lapaque N. Identification of the novel role of butyrate as AhR ligand in human intestinal epithelial cells. Sci Rep. 2019;9(1):643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Lamas B, Richard ML, Leducq V, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. 2016;22:598‐605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Roth CW, Hoch JA, DeMoss RD. Physiological studies of biosynthetic indole excretion in Bacillus alvei. J Bacteriol. 1971;106:97‐106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Brazier JS, Duerden BI, Hall V, et al. Isolation and identification of clostridium spp. from infections associated with the injection of drugs: experiences of a microbiological investigation team. J Med Microbiol. 2002;51:985‐989. [DOI] [PubMed] [Google Scholar]
- 144. Elsden SR, Hilton MG, Waller JM. The end products of the metabolism of aromatic amino acids by clostridia. Arch Microbiol. 1976;107:283‐288. [DOI] [PubMed] [Google Scholar]
- 145. Voss JG. Differentiation of two groups of Corynebacterium acnes. J Bacteriol. 1970;101:392‐397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Schleifer KH, Kilpper‐Bälz R, Kraus J, Gehring F. Relatedness and classification of Streptococcus mutans and ‘mutans‐like’ streptococci. J Dent Res. 1984;63:1047‐1050. [DOI] [PubMed] [Google Scholar]
- 147. Tannock GW. Characteristics of Bacteroides isolates from the cecum of conventional mice. Appl Environ Microbiol. 1977;33:745‐750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. DeMoss RD, Moser K. Tryptophanase in diverse bacterial species. J Bacteriol. 1969;98:167‐171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Booth EV, McDonald S. A new group of enterobacteria, possibly a new Citrobacter sp. J Med Microbiol. 1971;4:329‐336. [DOI] [PubMed] [Google Scholar]
- 150. Li G, Young KD. Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology. 2013;159:402‐410. [DOI] [PubMed] [Google Scholar]
- 151. Pickett MJ. Methods for identification of flavobacteria. J Clin Microbiol. 1989;27:2309‐2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Langworth BF. Fusobacterium necrophorum: its characteristics and role as an animal pathogen. Bacteriol Rev. 1977;41:373‐390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Kilian M. A taxonomic study of the genus Haemophilus, with the proposal of a new species. J Gen Microbiol. 1976;93:9‐62. [DOI] [PubMed] [Google Scholar]
- 154. Liu Y, Mee BJ, Mulgrave L. Identification of clinical isolates of indole‐positive Klebsiella spp., including Klebsiella planticola, and a genetic and molecular analysis of their beta‐lactamases. J Clin Microbiol. 1997;35:2365‐2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Rezwan F, Lan R, Reeves PR. Molecular basis of the indole‐negative reaction in Shigella strains: extensive damages to the tna operon by insertion sequences. J Bacteriol. 2004;186:7460‐7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Nuidate T, Tansila N, Saengkerdsub S, Kongreung J, Bakkiyaraj D, Vuddhakul V. Role of indole production on virulence of vibrio cholerae using galleria mellonella larvae model. Indian J Microbiol. 2016;56:368‐374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Takamura T, Harama D, Matsuoka S, et al. Activation of the aryl hydrocarbon receptor pathway may ameliorate dextran sodium sulfate‐induced colitis in mice. Immunol Cell Biol. 2010;88:685‐689. [DOI] [PubMed] [Google Scholar]
- 158. Dealler SF, Hawkey PM, Millar MR. Enzymatic degradation of urinary indoxyl sulfate by Providencia stuartii and Klebsiella pneumoniae causes the purple urine bag syndrome. J Clin Microbiol. 1988;26:2152‐2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Magiatis P, Pappas P, Gaitanis G, et al. Malassezia yeasts produce a collection of exceptionally potent activators of the ah (dioxin) receptor detected in diseased human skin. J Invest Dermatol. 2013;133:2023‐2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Fukumoto S, Toshimitsu T, Matsuoka S, et al. Identification of a probiotic bacteria‐derived activator of the aryl hydrocarbon receptor that inhibits colitis. Immunol Cell Biol. 2014;92:460‐465. [DOI] [PubMed] [Google Scholar]
- 161. Goettel JA, Gandhi R, Kenison JE, et al. AHR activation is protective against colitis driven by T cells in humanized mice. Cell Rep. 2016;17(5):1318‐1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Kaye J, Piryatinsky V, Birnberg T, et al. Laquinimod arrests experimental autoimmune encephalomyelitis by activating the aryl hydrocarbon receptor. Proc Natl Acad Sci U S A. 2016;113(41):E6145‐E6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Joller N, Lozano E, Burkett PR, et al. Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity. 2014;40(4):569‐581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Mascanfroni ID, Takenaka MC, Yeste A, et al. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1‐α. Nat Med. 2015;21(6):638‐646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Apetoh L, Quintana FJ, Pot C, et al. The aryl hydrocarbon receptor interacts with c‐Maf to promote the differentiation of type 1 regulatory T cells induced by IL‐27. Nat Immunol. 2010;11(9):854‐861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Wu HY, Quintana FJ, da Cunha AP, et al. In vivo induction of Tr1 cells via mucosal dendritic cells and AHR signaling. PLoS One. 2011;6(8):e23618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Gagliani N, Amezcua Vesely MC, Iseppon A, Brockmann L, Xu H, et al. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature. 2015;523(7559):221‐225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Quintana FJ, Basso AS, Iglesias AH, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453(7191):65‐71. [DOI] [PubMed] [Google Scholar]
- 169. Nguyen NT, Kimura A, Nakahama T, et al. Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine‐dependent mechanism. Proc Natl Acad Sci U S A. 2010;107:19961‐19966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Cervantes‐Barragan L, Chai JN, Tianero MD, et al. Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα+ T cells. Science. 2017;357(6353):806‐810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Zaid A, Mackay LK, Rahimpour A, et al. Persistence of skin‐resident memory T cells within an epidermal niche. Proc Natl Acad Sci U S A. 2014;111(14):5307‐5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Weikum ER, Xu L, Ortlund EA. The nuclear receptor superfamily: a structural perspective. Protein Sci. 2018;27:1876‐1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269‐1304. [DOI] [PubMed] [Google Scholar]
- 174. Forman BM, Goode E, Chen J, et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687‐693. [DOI] [PubMed] [Google Scholar]
- 175. Wang YD, Chen WD, Moore D, et al. FXR: a metabolic regulator and cell protector. Cell Res. 2008;18:1087‐1095. [DOI] [PubMed] [Google Scholar]
- 176. Friedman ES, Li Y, Shen TCD, et al. FXR‐dependent modulation of the human small intestinal microbiome by the bile acid derivative Obeticholic acid. Gastroenterology. 2018;155:1741‐1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Wu JN, Chen JR, Chen JL. Role of farnesoid X receptor in the pathogenesis of respiratory diseases. Can Respir J. 2020;2020:9137251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Parkssteven DJ, Blanchardrandy G. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365‐1368. [DOI] [PubMed] [Google Scholar]
- 179. Zhang X, Osaka T, Tsuneda S. Bacterial metabolites directly modulate farnesoid X receptor activity. Nutr Metab. 2015;12:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Wu R, Yuan X, Li X, et al. The bile acid‐activated retinoic acid response in dendritic cells is involved in food allergen sensitization. Allergy. 2022;77:483‐498. [DOI] [PubMed] [Google Scholar]
- 181. Mroz MS, Keating N, Ward JB, et al. Farnesoid X receptor agonists attenuate colonic epithelial secretory function and prevent experimental diarrhoea in vivo. Gut. 2014;63:808‐817. [DOI] [PubMed] [Google Scholar]
- 182. Willart MAM, van Nimwegen M, Grefhorst A, et al. Ursodeoxycholic acid suppresses eosinophilic airway inflammation by inhibiting the function of dendritic cells through the nuclear farnesoid X receptor. Allergy. 2012;67:1501‐1510. [DOI] [PubMed] [Google Scholar]
- 183. Shaik FB, Panati K, Narasimha VR, Narala VR. Chenodeoxycholic acid attenuates ovalbumin‐induced airway inflammation in murine model of asthma by inhibiting the T(H)2 cytokines. Biochem Biophys Res Commun. 2015;463(4):600‐605. [DOI] [PubMed] [Google Scholar]
- 184. Kliewer SA, Moore JT, Wade L, et al. An orphan nuclear receptor activated by Pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73‐82. [DOI] [PubMed] [Google Scholar]
- 185. Elentner A, Schmuth M, Yannoutsos N, et al. Epidermal overexpression of xenobiotic receptor PXR impairs the epidermal barrier and triggers Th2 immune response. J Investig Dermatol. 2018;138:109‐120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Carazo A, Hyrsova L, Dusek J, et al. Acetylated deoxycholic (DCA) and cholic (CA) acids are potent ligands of pregnane X (PXR) receptor. Toxicol Lett. 2017;265:86‐96. [DOI] [PubMed] [Google Scholar]
- 187. Illés P, Krasulová K, Vyhlídalová B, et al. Indole microbial intestinal metabolites expand the repertoire of ligands and agonists of the human pregnane X receptor. Toxicol Lett. 2020;334:87‐93. [DOI] [PubMed] [Google Scholar]
- 188. Venkatesh M, Mukherjee S, Wang H, et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and toll‐like receptor 4. Immunity. 2014;41:296‐310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Dubrac S, Elentner A, Ebner S, Horejs‐Hoeck J, Schmuth M. Modulation of T lymphocyte function by the Pregnane X receptor. J Immunol. 2010;184:2949‐2957. [DOI] [PubMed] [Google Scholar]
- 190. Desvergne B, Wahli W. Peroxisome proliferator‐activated receptors: Nuclear control of metabolism. Endocr Rev. 1999;20(5):649‐688. [DOI] [PubMed] [Google Scholar]
- 191. Goto T, Kim JI, Furuzono T, et al. 10‐oxo‐12(Z)‐octadecenoic acid, a linoleic acid metabolite produced by gut lactic acid bacteria, potently activates PPARγ and stimulates adipogenesis. Biochem Biophys Res Commun. 2015;459:597‐603. [DOI] [PubMed] [Google Scholar]
- 192. Bassaganya‐Riera J, Viladomiu M, Pedragosa M, et al. Probiotic bacteria produce conjugated linoleic acid locally in the gut that targets macrophage PPAR γ to suppress colitis. PLoS One. 2012;7(2):e31238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Nepelska M, de Wouters T, Jacouton E, et al. Commensal gut bacteria modulate phosphorylation‐dependent PPARγ transcriptional activity in human intestinal epithelial cells. Sci Rep. 2017;7:43199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Carregaro V, Napimoga MH, Peres RS, et al. Therapeutic treatment of arthritic mice with 15‐deoxy Δ12,14‐prostaglandin J2 (15d‐PGJ2) ameliorates disease through the suppression of Th17 cells and the induction of CD4+CD25−FOXP3+ Cells. Med Inf. 2016;2016:9626427. doi: 10.1155/2016/9626427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Wang W, Zhu Z, Zhu B, Ma Z. Peroxisome proliferator‐activated receptor–γ agonist induces regulatory T cells in a murine model of allergic rhinitis. Otolaryngol Head Neck Surg. 2011;144(4):506‐513. [DOI] [PubMed] [Google Scholar]
- 196. Zhang Y, Li X, Fang S, et al. Peroxisome proliferator‐activated receptor γ agonist suppresses mast cell maturation and induces apoptosis. Mol Med Rep. 2017;16:1793‐1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Yao L, Gu Y, Jiang T, Che H. Inhibition effect of PPAR‐γ signaling on mast cell‐mediated allergic inflammation through Down‐regulation of PAK1/ NF‐κB activation. Int Immunopharmacol. 2022;108:108692. doi: 10.2139/ssrn.3977785 [DOI] [PubMed] [Google Scholar]
- 198. Appel S, Mirakaj V, Bringmann A, Weck MM, Grünebach F, Brossart P. PPAR‐γ agonists inhibit toll‐like receptor‐mediated activation of dendritic cells via the MAP kinase and NF‐κB pathways. Blood. 2005;106(12):3888‐3894. [DOI] [PubMed] [Google Scholar]
- 199. Sun J. Dietary vitamin D, vitamin D receptor, and microbiome. Curr Opin Clin Nutr Metab Care. 2018;21(6):471‐474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Ishizawa M, Matsunawa M, Adachi R, et al. Lithocholic acid derivatives act as selective vitamin D receptor modulators without inducing hypercalcemia. J Lipid Res. 2008;49:763‐772. [DOI] [PubMed] [Google Scholar]
- 201. Pols TWH, Puchner T, Korkmaz HI, Vos M, Soeters MR, de Vries CJM. Lithocholic acid controls adaptive immune responses by inhibition of Th1 activation through the vitamin D receptor. PLoS One. 2017;12(5):e0176715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Wu S, Zhang YG, Lu R, et al. Intestinal epithelial vitamin D receptor deletion leads to defective autophagy in colitis. Gut. 2015;64(7):1082‐1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Wang J, Thingholm LB, Skiecevičienė J, et al. Genome‐wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2016;48(11):1396‐1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Venkateswaran A, Laffitte BA, Joseph SB, et al. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXRα. PNAS. 2000;97:12097‐12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Joseph S, Castrillo A, Laffitte B, et al. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med. 2003;9:213‐219. [DOI] [PubMed] [Google Scholar]


