Summary
Quantifying animal behavior is important for biological research. Identifying behaviors is the prerequisite of quantifying them. Current computational tools for behavioral quantification typically use high-level properties such as body poses to identify the behaviors, which constrains the information available for a holistic assessment. Here we report LabGym, an open-source computational tool for quantifying animal behaviors without this constraint. In LabGym, we introduce “pattern image” to represent the animal’s motion pattern, in addition to “animation” that shows all spatiotemporal details of a behavior. These two pieces of information are assessed holistically by customizable deep neural networks for accurate behavior identifications. The quantitative measurements of each behavior are then calculated. LabGym is applicable for experiments involving multiple animals, requires little programming knowledge to use, and provides visualizations of behavioral datasets. We demonstrate its efficacy in capturing subtle behavioral changes in diverse animal species.
Keywords: behavior, automatic categorization, automatic quantification, deep learning, machine learning, deep neural network, rat, mouse, Drosophila
Graphical abstract
Highlights
-
•
LabGym accurately mimics experimenters’ categorization of behavior across species
-
•
“Pattern images” effectively provide spatiotemporal information for the analysis
-
•
Removing background to focus on the animals leads to efficient behavioral analysis
-
•
LabGym outputs diverse quantitative measures after behavioral categorization
Motivation
Identifying and quantifying animal behaviors are important for biological research. Current automatic tools for this purpose are either specialized in one aspect, such as tracking, or using simplified high-level properties (e.g., body poses) to identify a behavior, which constrains the information available for a holistic assessment. In addition, many current computational tools require users to have computer programming skills to use, limiting their accessibility. A user-friendly tool is needed for both identifying behaviors by holistic assessments and quantifying diverse aspects of the behaviors. Therefore, we developed LabGym to fill this gap.
Hu et al. report an open-source, user-friendly software for analyzing user-defined animal behaviors. This tool not only identifies behavioral types but also quantifies them. It integrates multi-animal tracking, pattern recognition, and customizable deep learning to assess behavior holistically, making the analysis efficient and accurate.
Introduction
Quantitative measurements of animal behavior are important for many branches of biology and biomedical research.1,2,3,4 Identifying a behavior is a prerequisite to quantifying it. In most current computational tools for behavioral quantification, identifying a given behavior relies largely on tracking a few “high-level” properties such as the position of body parts in space (poses) and their speed of motion. Thus, data-rich videos and complex movements are simplified to skeletons and relative positions in space of body parts.5,6,7,8,9,10,11,12,13,14,15,16,17,18,19 For example, JAABA uses instantaneous speed, positions, and areas that are derived from the outlines of the animals’ bodies to identify a set of behaviors in mice and flies.6 Similarly, B-SOiD uses body part positions computed by DeepLabCut20 to generate speed, angles, and body lengths; behavioral information is then extracted from these data.17 MotionMapper uses postural properties to identify behaviors in adult Drosophila,7 and SALAM/LARA evaluates several specific contour properties for behavior detection in Drosophila larvae.5
While tracking these high-level properties is useful for summarizing the quantitative changes of a behavior, relying on them to identify a given behavior has limitations. First, these properties need to be defined a priori for identifying certain behaviors, which may fail to generalize to other behavior types. For example, movement speed can be used to identify locomotion but is useless for distinguishing facial grooming from body grooming during which the animal is immobile. Second, the limited number of high-level properties typically underrepresent the information needed to capture the complexity of animal behavior; and defining a large set of properties that encompass every aspect of a behavior a priori is not only computationally inefficient but also might be impossible. Finally, significant information loss likely occurs when using high-level properties, which makes the behavior identification less accurate. For example, a tool that only relies on postures would miss the information on the body color and textures, which can also be important for behavioral identifications.
Recent advancements in deep learning offer new approaches for behavior analysis. For example, DeepLabCut uses deep convolutional neural networks21,22 to enable accurate tracking of the positional changes of the animal body over time,20 but on its own does not identify or quantify any behavior. As a result, additional post hoc tools, such as B-SOiD,17 are required to identify and quantify a behavior based on the outputs of the tracking tools. Other deep-learning-based tools, including DeepEthogram,23 focus on behavior classification but do not provide quantitative measurements of the body kinematics during a behavior (e.g., motion speed). SIPEC classifies the behaviors and also tracks body key points.24 However, it requires manual labeling for the tracking, and the tracking is still a step away from the calculated measurements on the behavior. Moreover, these tools are computationally expensive and require powerful graphics processing units (GPUs) to run.
Here, we report a computational tool, LabGym, for quantifying user-defined animal behaviors without these limitations. When people manually analyze behaviors in a video, they typically ignore irrelevant information (such as static background) and make holistic observations of the behaving animal by considering both spatiotemporal changes and the overall movement pattern to categorize the behavior. Inspired by this human cognitive process, we designed LabGym to ignore static backgrounds, track multiple animals, accurately categorize behavior by holistic assessment, and then calculate quantitative measurements of each behavior. The technical innovations in LabGym include an approach for efficiently evaluating the animal’s movement patterns and a method of effective background subtraction. Moreover, LabGym provides users a way to generate stand-alone, visualizable behavior examples that can be shared across the research community as ground truth (i.e., the examples with assigned true labels)25,26,27 or for benchmarking different algorithms. Finally, LabGym has several features that improve user-friendliness and accessibility, which include a graphical user interface (GUI) requiring no programming knowledge to use, the deep neural networks that can be customized through the GUI, and no requirement of GPUs to run.
We show that LabGym precisely captures subtle changes in user-defined behaviors in animals ranging from soft-bodied invertebrates to rodents. We further demonstrate its strength by comparing it with the existing standards.
Results
The LabGym pipeline
LabGym was designed to efficiently and accurately identify user-defined behavioral types for each animal in an experimental session, and then provide quantitative measurements of each behavioral type. The open-source code and a full manual of LabGym are in its GitHub page (https://github.com/umyelab/LabGym).
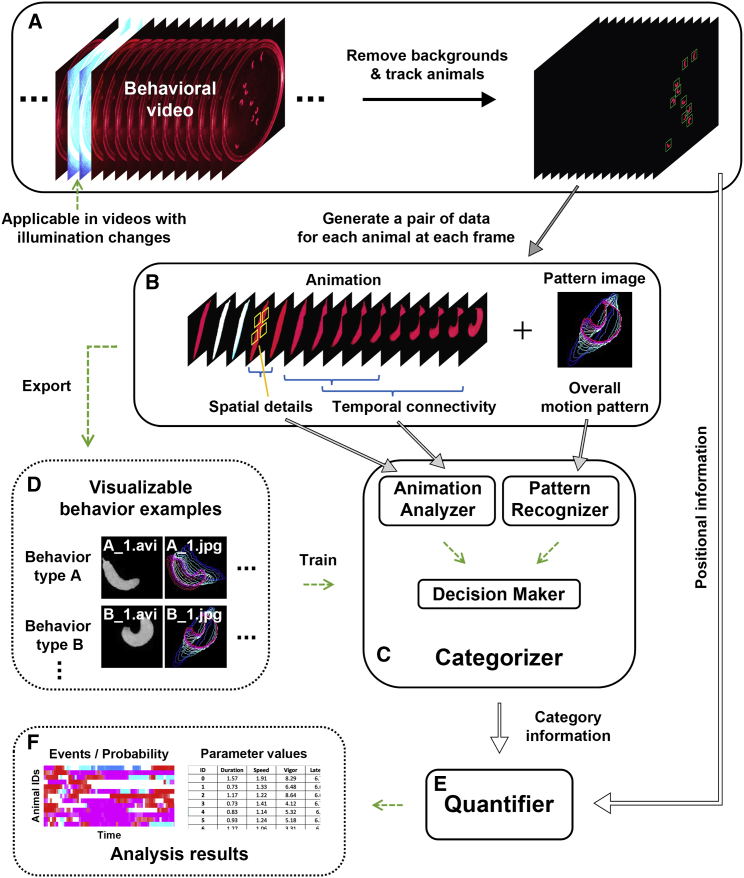
LabGym first uses an effective method to remove background in each video frame and tracks each animal within the video (Figure 1A). At each frame of the video, a module termed the “Categorizer” determines which behavior an animal performs during a time window that spans from the current frame back to a user-defined number of frames prior (“time window”). To achieve this, two types of data are extracted at each video frame for each animal (Figure 1B and Data S1, video 1): (1) an animation (“animation”) that spans the specified time window, and (2) the imprint of the positional changes of the animal’s body (including body parts) over time in the animation, which we term “pattern image.” These two pieces of data are then analyzed by the Categorizer that comprises three submodules: the “Animation Analyzer,” the “Pattern Recognizer,” and the “Decision Maker” (Figure 1C). The Animation Analyzer analyzes all spatiotemporal details in each animation, whereas the Pattern Recognizer evaluates the animal’s overall motion pattern in the paired pattern image (see details in “The categorizer assesses holistic behavioral information”). The Decision Maker integrates the outputs from the Animation Analyzer and Pattern Recognizer to determine the behavior type.
Figure 1.
The pipeline of LabGym
(A) Static backgrounds are removed in each video frame and individual animals are tracked.
(B) At each frame of the video, LabGym generates a pair of data for each animal, which comprises a short animation spanning a user-defined time window and its paired pattern image representing the movement pattern of the animal within the animation.
(C) The Categorizer uses both the animations and pattern images to categorize behaviors during the user-defined time window (i.e., the duration of the animation). The Categorizer comprises three submodules, the “Animation Analyzer,” the “Pattern Recognizer,” and the “Decision Maker.” The Animation Analyzer analyzes all spatiotemporal details in the animation for each animal whereas the Pattern Recognizer evaluates the overall movement patterns in the paired pattern image. The Decision Maker integrates the outputs from both the Animation Analyzer and the Pattern Recognizer to determine which user-defined behavioral category is present in the animation for each animal.
(D) The animations and pattern images can be exported to build visualizable, sharable behavior examples for training the Categorizer.
(E and F) After the behavioral categorizations, the quantification module (“Quantifier”) uses information from the behavioral categories and the animal foregrounds to compute specific quantitative measurements for different aspects of each behavior.
See also Data S1.
The Categorizer needs to be trained on behavior examples of ground truth (i.e., user-labeled behavior examples). A behavior example consists of an animation and its paired pattern image, both of which are generated by LabGym (Figure 1B). Users then label the behavior examples by sorting them into folders according to their behavior types and naming the folder to indicate the behavior types. This way, users establish a ground truth behavior dataset that can be used to train a Categorizer (Figure 1D).
After behavior categorizations, the quantification module called the “Quantifier” (Figure 1E) uses information from the animal foregrounds to compute various quantitative measurements for each behavior (e.g., duration, speed), which are stored in individual spreadsheets (Figure 1F). The output also includes temporal raster plots of behavioral events (and their probabilities) and annotated videos for visualization of behavioral categorization.
Effective background subtraction ensures efficient analyses
Behavioral experiments often involve behavior-irrelevant backgrounds—such as the enclosures for holding the animals or changes in illumination during optogenetic manipulation—that render analyses inaccurate and inefficient. To address these issues, we designed a module in LabGym that removes backgrounds but retains all the pixels representing the animals in each frame (i.e., foreground; see also below in “The categorizer assesses holistic behavioral information” for options that incorporate background information).
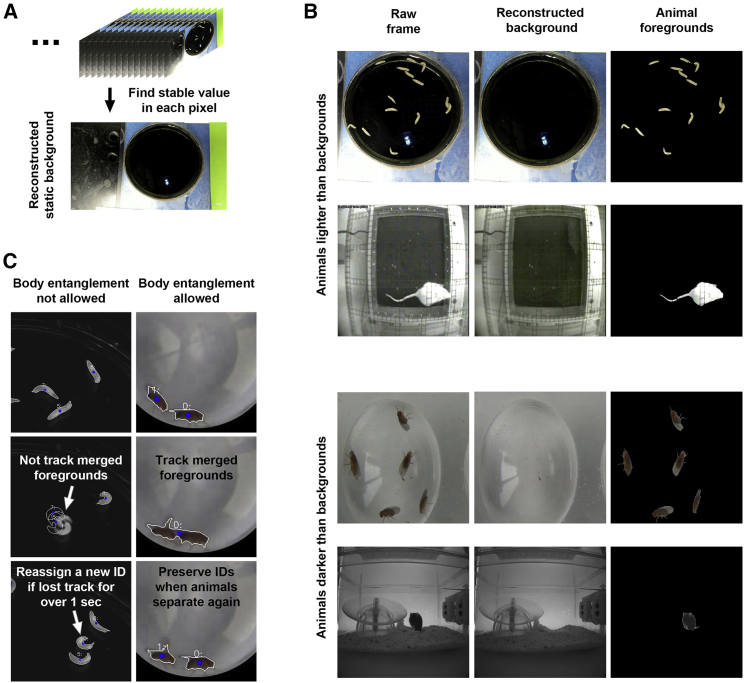
We reasoned that backgrounds in experimental conditions are often static and that the pixel value in each background location would be stable over time (Figure S1A). Therefore, we developed a method to find the stable value of each pixel and use these values to reconstruct the static background (Figure 2A and STAR Methods). The static background is then subtracted from each frame to obtain the foreground, which represents the animal (Figure 2B). Compared with two state-of-the-art background subtraction methods, Local SVD Binary Pattern (LSBP)28 and GSoC29 (implemented in Google Summer of Code 2017), our stable-value detection method reconstructed much cleaner backgrounds (with fewer traces of the animals) on behavioral videos of multiple animals and in diverse experimental settings (Figures 2B and S1B). Notably, animals are reliably tracked by LabGym even in videos with unstable or shifting illuminations (Data S1, video 2) because the background during illumination changes (e.g., during optogenetic stimulation) can be reconstructed separately.
Figure 2.
LabGym removes backgrounds and tracks multiple animals
(A) The stable value of each pixel is used to reconstruct the static background for a video.
(B) Examples of video frames showing that animal foregrounds are clearly presented after removing the static backgrounds that are constructed by our method of stable-value detection.
(C) Examples of video frames showing that LabGym tracks multiple animals performing either non-interactive or social behavior. Left: non-interactive behavior, in which animal (larva) contact is not allowed; right: social interaction in which animal (adult fly) contact is allowed.
The tracking of each animal over time is performed based on the assumption that the Euclidean distance between the centers of the same animal in consecutive frames are always the smallest among all Euclidean distances between each possible pair of animal centers. Each tracked animal is assigned by a unique identity (ID) that links to a matrix for storing all information about that animal over time.
Multiple animals in the same enclosure sometimes collide with each other, which can cause tracking errors. To make the analysis fully automatic, LabGym excludes animals from tracking if users choose not to allow animal body entanglements (Figure 2C, Data S1, video 3, and STAR Methods). This ensures that the analysis is only performed on reliably tracked individual animals. If body entanglements are allowed, LabGym is also useful to identify social behaviors in which animals have body contact (Figure 2C, Data S1, video 4, and STAR Methods). In this scenario, merged animals are analyzed as one.
The Categorizer assesses holistic behavioral information
We built the Categorizer with three submodules: Animation Analyzer, Pattern Recognizer, and Decision Maker (Figure S2 and STAR Methods), each of which consists of neural networks that suit the design goals. Importantly, the two subnetworks—the Animation Analyzer and the Pattern Recognizer—were designed to analyze two types of behavioral representations that complement each other: one is an animation and the other is an image that contains the imprints of the contours of the animal’s whole body and body parts (e.g., limbs) at different time points. This ensures accurate categorizations of behavior through holistic assessment.
Animal behavior contains changes in both spatial and temporal dimensions. The Animation Analyzer first uses time-distributed convolutional layers to analyze raw pixel values that represent the animal in each frame of an animation to learn the frame-wise spatial details. It then uses recurrent layers to compute temporal connectivity among these frame-wise spatial details, in order to learn about how they are organized along the temporal axis (Figure S2A).
During a behavior, the positional changes of the animal body over time can form a pattern that is unique to the behavior. However, Animation Analyzer might not be sensitive enough to capture this motion pattern. Therefore, we introduced “pattern image” to present the overall motion pattern of a behavior and designed Pattern Recognizer that comprises convolutional layers to analyze the pattern image (Figure S2B). In each pattern image, the contours of the animal body (including body parts) in each frame of the animation were superimposed with gradually changing colors to indicate their temporal sequences. This shows the animal’s overall movement pattern during the animation in a single 2-dimensional image to allow for efficient analyses.
The Decision Maker uses a concatenating layer to integrate the outputs from both the Animation Analyzer and the Pattern Recognizer and passes the integrated information to fully connected (dense) layers for discriminating behavioral categories (Figure S2C).
The background of the behavior videos sometimes can be useful for identifying the behaviors. For example, bedding in a mouse’s mouth can indicate nest building and, thus, including the bedding in the animations can facilitate the categorization of nest building behavior (Figure S3A). Therefore, in LabGym we provide users with an option of whether to include the background in the animations. Note that we did not include background in any animation used in this study since the background is irrelevant to the behaviors in this study.
The motion patterns of whole-body contours are useful for distinguishing behaviors like curling versus uncoiling in Drosophila larvae, wing extension versus abdomen bending in adult flies, or walking versus rearing in rats (Figure S3B). However, they are not useful for distinguishing behaviors like facial grooming versus sniffing, or ear grooming versus sitting still in mice (Figure S3C). In these behaviors, the motion patterns of specific body parts (e.g., limbs or noses) are the key to behavior categorizations. Therefore, we provide users an option in LabGym of whether to show the movement of body parts in the pattern images. It is noteworthy that LabGym automatically identifies the contours of all the body parts without the need of manual labeling or model training, which differs conceptually from the approaches that tracks body parts with deep learning.
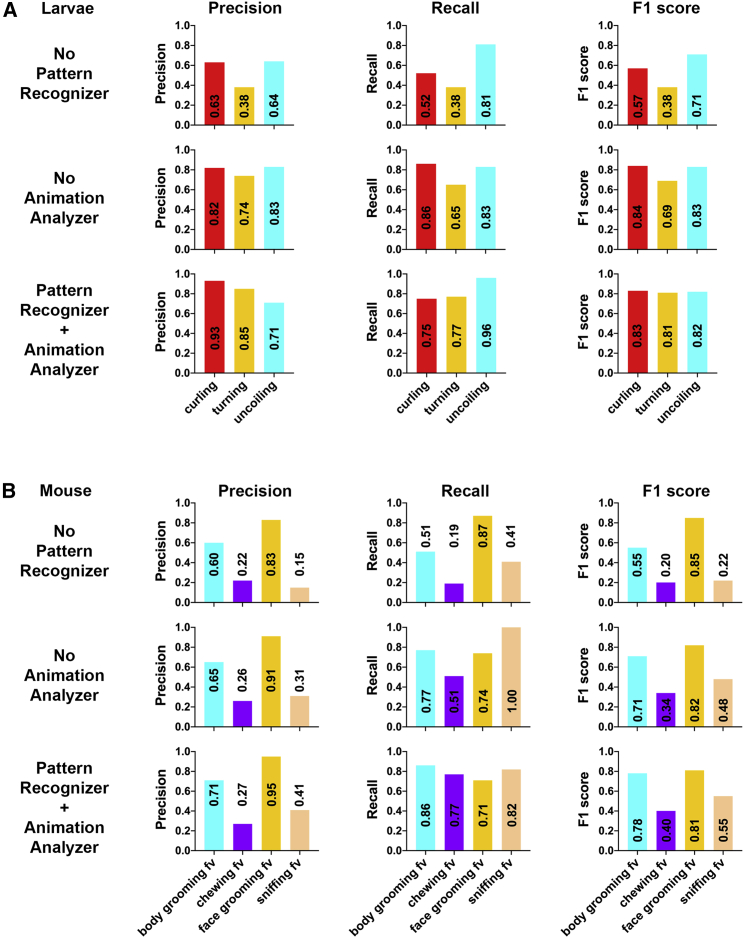
To test whether combining the Animation Analyzer with the Pattern Recognizer achieves better performance than using either alone, we performed the “ablation” experiment. In this experiment, we compared the behavior categorization accuracy of three Categorizers, one with Pattern Recognizer removed, one with Animation Analyzer removed, and one containing both. The three Categorizers were trained and tested on the same training (1,596 pairs of examples) and testing datasets (3,120 pairs of examples). As all the other experiments in this study, the training examples and testing examples for the ablation experiments were generated by LabGym from different videos and animals and are publicly available (STAR Methods). Specifically, the training examples were from over 100 different video recordings of ∼300 different larvae, 10 different rats, and three different mice; the testing examples were from videos of ∼200 different larvae, eight different rats, and two different mice. We chose three larval behaviors and four mouse behaviors to test the three Categorizers because these behaviors are visually similar and thus challenging for the Categorizers to distinguish (Data S1, video 5). We assessed the precisions, recalls (sensitivity), and F1 score (weighted precision and sensitivity, STAR Methods) of the three Categorizers for behavioral categorizations. The Categorizer containing both the Animation Analyzer and Pattern Recognizer significantly outperformed the Animation Analyzer alone across most behaviors (Figure 3), demonstrating the necessity of including the Pattern Recognizer in the Categorizer to achieve higher accuracy in behavioral categorizations. Notably, the Categorizer with Pattern Recognizer alone performed better than that with the Animation Analyzer, indicating the efficacy of using Pattern Recognizer to analyze the pattern images.
Figure 3.
The inclusion of Pattern Recognizer for analyzing the Pattern images enhances the accuracy of the Categorizer
(A and B) Bars showing the test metrics (precisions, recalls, and F1 scores) for three Categorizers: one without Pattern Recognizer, one without Animation Analyzer, and one with both. Test results of datasets from Drosophila larvae (A) and mice (B) are shown. In all three Categorizers, the input shapes and the complexity levels are the same for Pattern Recognizer (8 × 8 × 3, level 1 for larva dataset; 32 × 32 × 3, level 3 for mouse dataset), and Animation Analyzer (8 × 8 × 1 × 15, level 1 for larva; 32 × 32 × 1 × 14, level 3 for mouse). Seven videos for larvae (10 larvae per video) and two videos for mice (one mouse per video) that were not used for generating the training datasets were used to generate the testing data (animations and their paired pattern images). The data with visibly missing body parts (e.g., missing limbs/heads) were excluded (56 pairs, which is 1.76% of the total data generated), because they were caused by occasional, false tracking (often due to changes in the background) and might negatively affect the performance of the Categorizer. Such performance decline is not because of any issues of the Categorizer and might bias the testing of Categorizers. A total of 795 pairs (for larvae) and 2,325 pairs (mice) of data were randomly selected and then were sorted by experimenters into different folders under user-defined behavioral names (building ground truth testing datasets). The ground truth testing datasets were then used to test the categorization metrics of the three Categorizers.
The Categorizer accurately categorizes user-defined behaviors across species
Since the behavior types for the Categorizer to recognize are defined by users, rather than pre-defined in the program, the Categorizer can identify various behaviors across diverse animal species. To demonstrate this, we used LabGym to study relatively simple behaviors in Drosophila larvae and behaviors of increasing complexity in rodents. We first used LabGym to generate and sort separate behavioral datasets for Drosophila larvae, rats, and mice (Data S1, videos 6–8). The larva dataset (5,251 pairs of examples) contains the fewest categories with sufficient examples to cover almost all the variations of each behavior; the rat dataset (3,864 pairs of examples) contains a larger number of categories and more complex behaviors that encompass subtle distinctions between categories; the mouse dataset (4,869 pairs of examples) contains additional categories of even more complex behaviors.
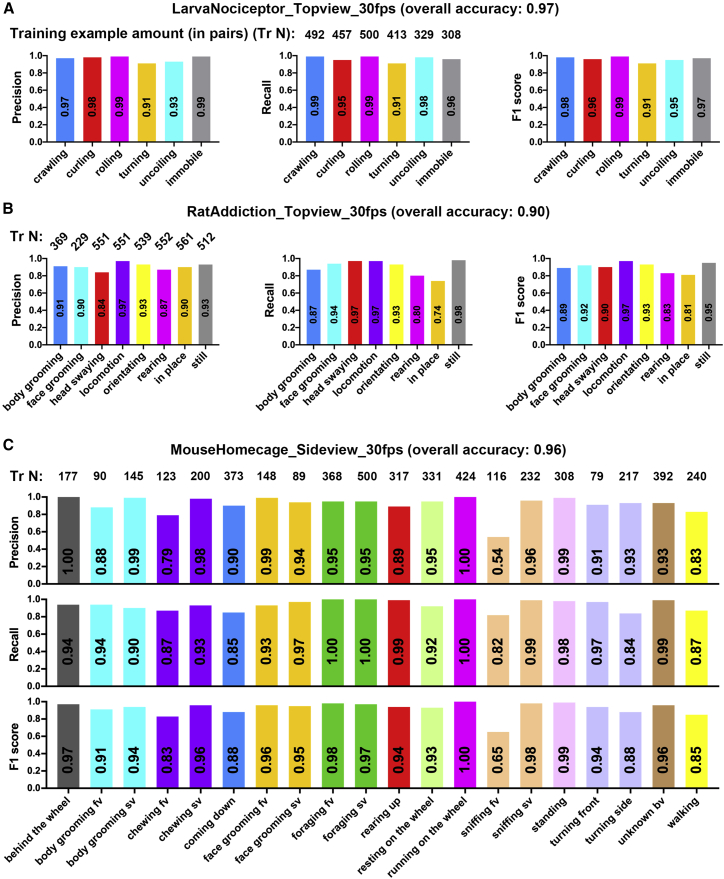
The Categorizer is customizable with different input shapes (input frame/image width/height, colored/gray channel, and timestep) and different levels of complexity (the network structure and the number of layers) to address behaviors of various complexity. To provide a reference for customizing the Categorizer, which is a trial-and-error process, we trained several Categorizers with different complexity for each species and determined the most suitable ones based on their overall accuracy of behavioral categorizations (Table S1 and STAR Methods). The selected Categorizer for each dataset was named as LarvaNociceptor_Topview_30fps (LarvaN), RatAddiction_Topview_30fps (RatA), and MouseHomecage_Sideview_30fps (MouseH). These three Categorizers were then tested on 19,643 pairs of testing examples in total.
Larval behaviors are relatively simple. The LarvaN with simple complexity achieved 97% accuracy in frame-wise categorizations of six different behaviors elicited by noxious stimuli (Figure 4A and Data S1, video 9).
Figure 4.
Accurate frame-wise categorizations of various user-defined behaviors by different Categorizers
(A–C) Bars showing the test metrics (overall accuracy, precisions, recalls, and F1 scores) for LarvaN (A), RatA (B), and MouseH (C) in video analysis. Seven videos for larvae (10 larvae per video), 10 videos for rats (one rat per video), and two videos for mice (one mouse per video) that were not used for generating the training datasets were used to generate the testing data (animations and their paired pattern images). A total of 2,162 pairs (for larvae), 4,828 pairs (rats), and 12,653 pairs (mice) of data were randomly selected and then were sorted by experimenters into different folders under user-defined behavioral names (building ground truth testing datasets).
Rats show complex behaviors and the differences among them can be extremely subtle due to the recording or experimental conditions. Despite these challenges, the RatA achieves 90% accuracy in frame-wise categorizations of eight types of visually similar behaviors such as body grooming versus face grooming, or head swaying versus in place (Figure 4B and Data S1, video 10).
We next examined 20 types of user-defined behaviors of mice in their home cage. The MouseH achieved 96% accuracy in frame-wise categorizations on these behaviors with as few as 79 training examples for some categories (Figure 4C and Data S1, video 11). Although both were used for categorizing rodent behaviors, MouseH trained on pattern images including the body parts performed better than RatA trained on those excluding body parts (Figures 4B and 4C). This indicates that including the body parts in pattern images can boost the learning efficiency of the Categorizer.
Due to the data-driven nature of deep learning, the accuracy of the Categorizer largely depends on the amount, diversity, and labeling accuracy of the training examples. Therefore, the performance of the Categorizers can be continuously improved over time with the accumulation of well-selected training examples.
The Quantifier provides quantitative measurements that are sufficient for capturing subtle changes in diverse aspects of each behavior
To quantitatively measure the behavior intensity and the body kinematics during a behavior, we designed a quantification module in LabGym called “Quantifier.” The Quantifier calculates 14 parameters to quantify a given behavior: count, latency, duration, angle, speed (total traveling distance divided by time), velocity (the shortest distance from the beginning to the end divided by time), acceleration/velocity reduction, distance, intensity (area), intensity (length), magnitude (area), magnitude (length), vigor (area), and vigor (length) (Figure S4 and STAR Methods). Additional parameters can be added in future versions of LabGym. The definitions of these parameters are as follows:
-
•
The count is the summary of the behavioral frequencies, which is the occurrence number of a behavior within the entire duration of analysis. Consecutive single occurrences (at a single frame) of the same behavior are considered as one count.
-
•
The latency is the summary of how soon a behavior starts, which is the time starting from the beginning of the analysis to the time point that the behavior occurs for the first time.
-
•
The duration is the summary of how persistent a behavior is, which is the total time of a behavior within the entire duration of analysis.
-
•
The angle is the movement direction (against to the animal body axis) of the animal during a behavior episode, which is the mean of all the included angle () between animal body axis and the movement direction during the time window (tw) for categorizing the behavior.
-
•
The speed is the summary of how fast the animal moves when performing a behavior, which is the total distance traveled (can be back and forth) (d) (between the two centers of mass of the animal) during the time window (tw) for categorizing the behavior divided by tw.
-
•
The velocity is the summary of how efficient the animal’s movement is when performing a behavior, which is the shortest distance between the start and the end positions (dt) (between the two centers of mass of the animal) divided by the time (t) that such displacement takes place.
-
•
The acceleration/velocity reduction is the summary of how fast the animal’s velocity changes while performing a behavior, which is the difference between maximum velocity (vmax) and minimum velocity (vmin) divided by the time (tv) that such velocity change takes place.
-
•
The distance is the total distance traveled of the animal by performing a behavior within the entire duration of analysis.
-
•
The intensity (area)/intensity (length) is the summary of how intense a behavior is, which is the accumulated proportional changes of the animal body area (a)/length (L) between frames divided by the time window for categorizing the behaviors (tw) when performing a behavior.
-
•
The magnitude (area)/magnitude (length) is the summary of the motion magnitude, which is the maximum proportional change in animal body area (a) or length (L) when performing a behavior.
-
•
The vigor (area)/vigor (length) is the summary of how vigorous a behavior is, which is the magnitude (area)/magnitude (length) divided by the time (ta or tl) that such a change takes place.
To test how well the Quantifier performs in quantifying behaviors in diverse animal species, we applied LabGym to experiments in larvae, rats, and mice.
Drosophila larva behavior
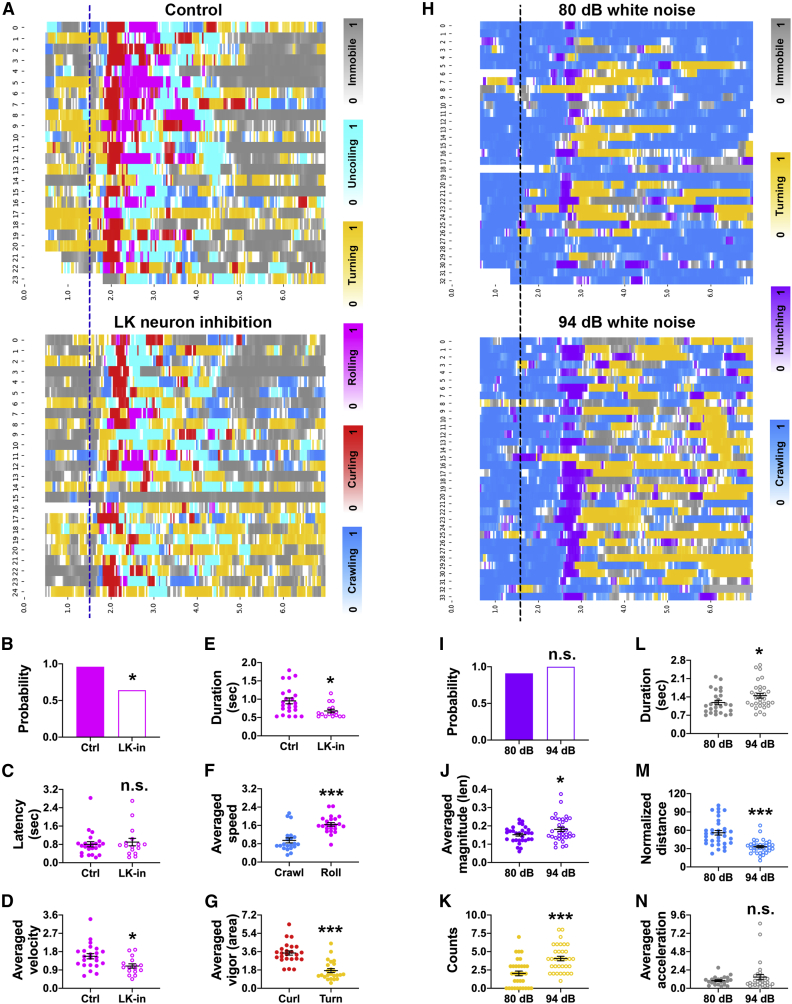
Previous studies show that optogenetic inhibition of larval leucokinin (LK) neurons slightly impaired larval behaviors elicited by nociceptor activation.30 In these experiments, both the subtle changes in behaviors and the shifting illumination caused by optogenetic manipulations posed challenges for automatic behavioral analysis. We thus tested whether LabGym was able to capture the behavioral changes in these experiments (Data S1, video 12). Compared with the control group, larvae in the LK inhibition group showed a modest reduction in the probability of body rolling behavior, which is a characteristic nociceptive response31,32 (Figures 5A and 5B). No change was observed in the latency of the response onset after optogenetic stimulation of nociceptors (Figure 5C). Moreover, LK inhibition caused a slight yet significant reduction in rolling velocity (Figure 5D) and duration (Figure 5E). We also compared several quantitative measurements of different behaviors and found that the results indeed reflect the ethological meaning of these behaviors. For example, rolling achieves higher speed than crawling (Figure 5F), supporting the notion that rolling was a more efficient way than crawling to escape from harm.31,32 Similarly, despite their spatial similarity, curling was more vigorous than turning (Figure 5G), echoing the fact that larvae typically perform curling under noxious stimuli but tend to turn in in response to milder situation like gentle touch or exploration.31,32,33,34 Thus, LabGym is sufficiently sensitive to detect modest effects of in vivo neuronal inhibition on behavior, and it does so on videos with shifting illumination.
Figure 5.
The Quantifier captures subtle changes in the behavior of Drosophila larvae
(A and H) Raster plots showing the frame-wise categorizations by: (A) LarvaN on control (w; LK-GAL4/+; TrpA1-QF, QUAS-ChR2T159C, UAS-jRCaMP1b/+) and LK neuron inhibition (w; LK-GAL4/+; TrpA1-QF, QUAS-ChR2T159C, UAS-GtACR1/+) groups after nociceptors were optogenetically activated (blue dashed line indicates the stimulation onset); (H) LarvaMechanosensor_Topview_30fps on larvae (Canton S) under different intensities of sound stimuli (black dashed line indicates the stimulation onset). Color bars: behavioral categories (color intensities indicate the behavioral probabilities). The x axis represents the time and y axis represents the larval IDs.
(B–G) Parameters calculated by the Quantifier for different behaviors elicited by optogenetic activation of nociceptors. The colors match those for behaviors shown in (A). Data in (F) and (G) are from the control group in (A).
(I–N) Parameters calculated by the Quantifier for different behaviors elicited by sound stimulation. The colors match those in (H). Probability in (B) and (I) is the fraction of responders (behavioral count > 0) in total animals. n.s., p > 0.05; ∗p < 0.05; ∗∗∗p < 0.001 (Fisher exact test for B and I; unpaired t test for C–G and J–N). Error bars represent standard error of the mean.
When Drosophila larvae are exposed to sound, they display a startle response that is characterized by the cessation of crawling (“freezing”), retraction of their heads (“hunching”), and excessive turning movements.35 To test whether LabGym could identify the subtle differences in behaviors elicited by different intensities of sound, we trained another Categorizer (LarvaMechanosensor_TopView_30fps) with examples of sound-elicited behaviors in Drosophila larvae and used it for analysis (Data S1, video 13). This Categorizer has the same complexity as LarvaN. Compared with larvae exposed to 80 dB white noise, those exposed to 94 dB showed a similar hunching probability (Figures 5H and 5I), but the magnitude of head retraction during hunching was enhanced (Figure 5J). Furthermore, larvae exposed to 94 dB showed more frequent turning (Figure 5K) and prolonged freezing (i.e., immobility; Figure 5L), which resulted in significant reductions in crawling distance (Figure 5M). However, when larvae froze, the reduction in the velocity of their movement was similar between the two groups (Figure 5N). Thus, LabGym was able to capture quantitative differences in the behaviors induced by auditory stimuli of different intensities.
Rodent behavior
It is well-established that drugs like amphetamine, produce dose-dependent increases in psychomotor activity, and that the intensity of these behaviors increases (i.e., sensitizes) with repeated drug exposure.36,37 Psychomotor activity in rats encompasses a wide range of behaviors that include rearing, hyper-locomotion, and stereotyped, repetitive head movements (i.e., stereotypy).36,37
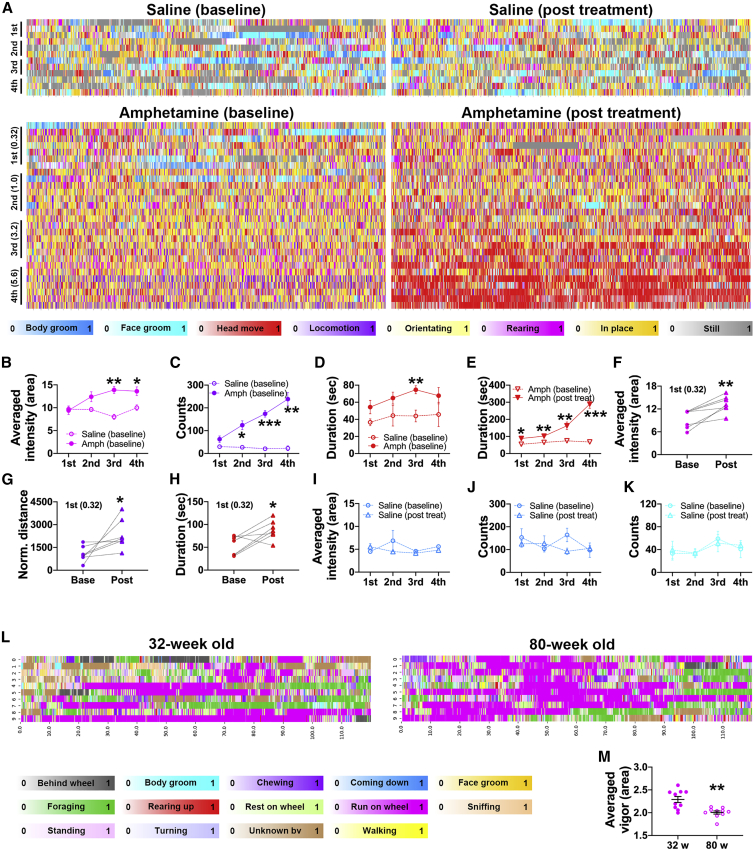
LabGym automatically captured dose-dependent changes in the intensity and kinematics of the psychomotor activity (Figure 6A and Data S1, video 14). These include the intensity of rearing (Figure 6B), the counts and the traveling distance of locomotion (Figures 6C and 6G), and the duration of head movements (persistent head movements indicating stereotypy; Figure 6D), following systemic amphetamine administration. Impressively, LabGym even captured stereotypy interspersed with small bursts of locomotion—an indication of a strong psychomotor response at high dose of amphetamine (e.g., Figures 6A and 6C, 5.6 mg/kg dose group in amphetamine baseline group). LabGym also captured quantitative changes in these behaviors across the development of sensitization, in which repeated injection of amphetamine resulted in more intense behavioral responses to the same dose of drug (Figures 6E–6H). In contrast, the intensity of spontaneously occurring behaviors such as body/face grooming were stable across time in rats treated with repeated saline (Figures 6I–6K).
Figure 6.
The Quantifier captures quantitative changes in rodent behaviors
(A) Raster plots showing the frame-wise categorizations by RatA of behavior after saline or amphetamine injection. Plots at the left (baseline) show responses to the first exposure to amphetamine across increasing doses (baseline; 0.32, 1.0, 3.2, and 5.6 mg/kg, i.p.); Plots at the right show responses to increasing doses of amphetamine after repeated amphetamine treatment (post treatment; 21 days). Animals in the saline group received repeated saline throughout. Color bars: behavioral categories (intensities indicate the behavioral probabilities). x axis: time (each tick is 10-s); y axis: each row represents an individual rat. These same colors are used to indicate behavioral categories in (B)–(K).
(B–D) The dose-response curves of rearing intensity (B), locomotion counts (C), and head movement duration (D) following the first exposure to amphetamine or saline (baseline). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 (unpaired t test between saline and amphetamine groups).
(E) Dose-response curves for head movement duration at baseline and after repeated amphetamine exposure (post treatment, i.e., sensitization). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 (unpaired t test between baseline and post treatment groups).
(F–H) Within subject comparison of rearing intensity (F), locomotion distance (G) and the head movement duration (H) in response to the first 0.32 mg/kg amphetamine at baseline and after repeated amphetamine treatment. ∗p < 0.05; ∗∗p < 0.01 (paired t test).
(I–K) Dose-response curves of the intensity and counts of body grooming (I and J) and the duration of face grooming (K) in in rats repeatedly treated with saline.
(L) Raster plots showing the frame-wise categorizations by MouseH for a younger (32-week) or an older (80-week) mouse. Color bars: behavioral categories (intensities indicate the behavioral probabilities). x axis: time (in seconds); y axis: different random 2-min time window. Multiple (>100) 2-min time windows were randomly selected from a 4-h recording for each mouse, and those containing wheel-running behavior (targeted time window) were sorted out. Ten targeted time windows for each mouse were randomly selected for analysis.
(M) The running vigor of the two mice in (L). ∗∗p < 0.01 (unpaired t test).
All error bars represent standard error of the mean.
When an animal ages, its ethogram might not change, yet the quantitative measurements of its behaviors (e.g., running speed) may decline. LabGym not only categorized all the behaviors of interest for both younger (32-week) and older mice (80-week) in a cage with a running wheel (Figure 6L and Data S1, video 15), but also captured the decline in running vigor of the older versus the young mice by a parameter (vigor_area) that summarizes the areal changes per unit time of the animal’s body (Figure 6M and STAR Methods).
Taken together, these results demonstrate that the Quantifier reliably and precisely captures quantitative changes in behaviors across species.
GUI with optional settings for user-friendly usage
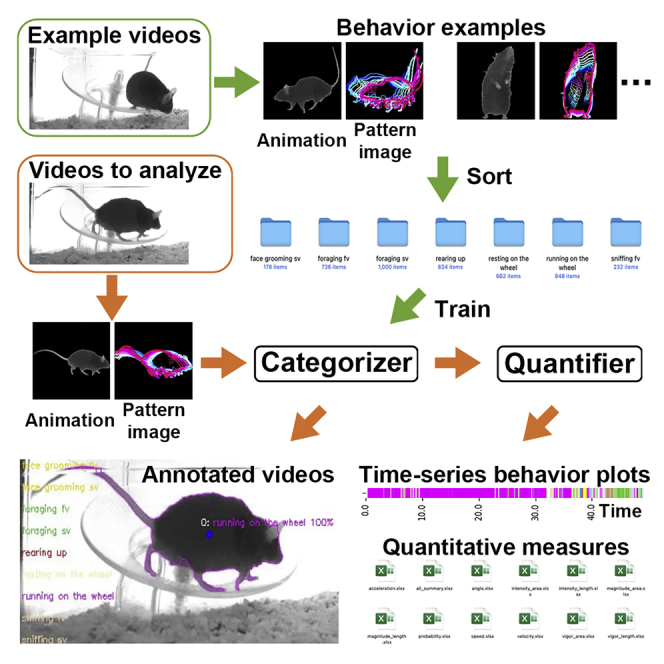
To make LabGym adaptable to various experimental settings and accessible to users without requiring a computer programming background, we implemented a GUI (Figure S5A). This includes four functional units: Generate Behavior Examples, Train Categorizers, Test Categorizers, and Analyze Behaviors.
The “Generate Behavior Examples” unit is for batch processing of videos to generate visualizable behavior examples (animations and paired pattern images). Users can select and sort these examples into different folders according to their behavior types. Users can then name these folders with the behavior names and use them to train the Categorizers in “Train Categorizers” unit, or to test a trained Categorizer in “Test Categorizers” unit. The “Analyze Behaviors” unit is for batch processing of videos for behavioral analysis. It produces an annotated video (Data S1, videos 9–15), a raster plot of behavioral events (displayed in user-defined colors), and their occurring probabilities (by color intensities) for each animal in one batch of analysis (Figures 5A, 5H, 6A, and 6L), and spreadsheets in Excel or CSV format containing individual behavioral parameters for each behavior of each animal.
Notably, the flexible options in the GUI of LabGym, especially the customizable input shapes and the complexity of Categorizers, allow it to achieve fast processing speed in network training and in analyzing behaviors of various complexity on commonly used laptops that do not have GPUs (STAR Methods).
Benchmark comparison with the existing standards
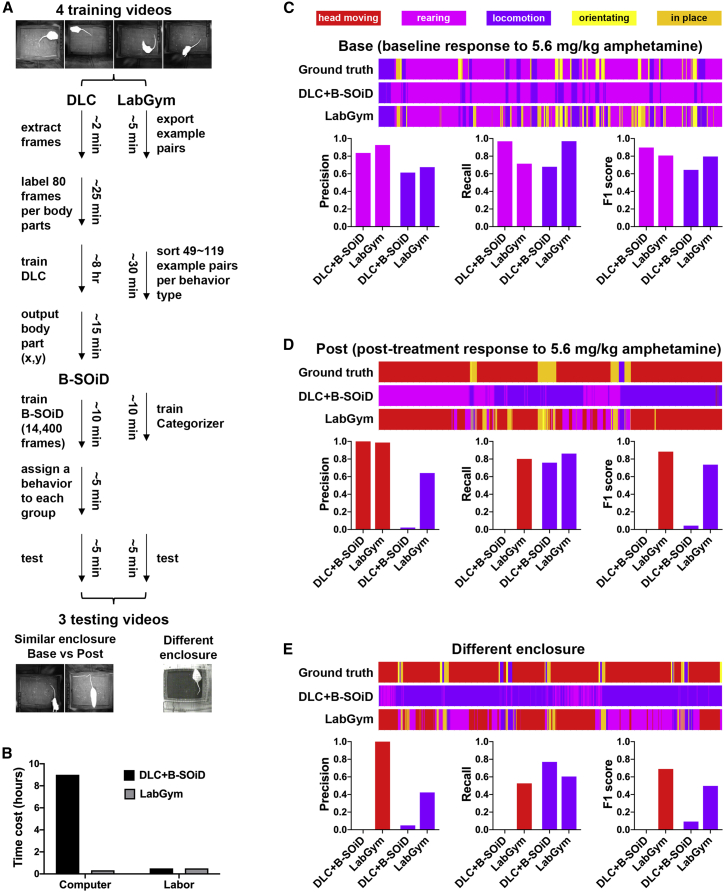
We compared the performance of LabGym against that of the combination of DeepLabCut20 and B-SOiD17 (“DLC + B-SOiD”), an exciting approach within the field of behavioral neuroscience. We compare LabGym to “DLC + B-SOiD” because both tools track animals, classify user-defined behaviors, provide quantitative measurements of kinematics in a behavior, and have a GUI for accessibility to users without requiring computer programming knowledge.
Both “DLC + B-SOiD” and LabGym can be iteratively optimized by adding more training data and refining the data labeling. Therefore, the goal of comparisons here is not to explore how to modify the training datasets to achieve their best performance, but to compare their performance given the same training resources. To achieve this, for each tool we used the same computer, used the same videos to generate training datasets, and set the time for manual labeling and sorting to a fixed 30-min. We then compared the performance of each approach on the same testing dataset, which was distinct from the training data (Figure 7A).
Figure 7.
Benchmark comparison between the existing standards and LabGym
(A) The workflow of the benchmark comparison between the combination of DeepLabCut and B-SOiD (DLC + B-SOiD) and LabGym, which was performed on the same computer that has a 3.6 GHz Intel Core i9-9900K CPU, a 64 GB memory, and an NVIDIA GeForce RTX 2080 GPU. The setting of Categorizer in LabGym: Animation Analyzer: level 1 with input shape 8 × 8 × 1 × 20; Pattern Recognizer: level 2 with input shape 16 × 16 × 3.
(B) The total time spent for DLC + B-SOiD and LabGym to finish the workflow in (A).
(C and D) Comparisons of frame-wise behavior classifications between the computer software and the ground truth (expert human annotation) in two testing videos: baseline (C) and post treatment (D). The raster plots show frame-wise behavior events with the x axis indicating the time (1 min). Different colors indicate different behavior types.
(E) Comparison of frame-wise behavior classifications between the computer software and the ground truth (expert human annotation) in generalization to a different testing enclosure. The raster plots show frame-wise behavior events with the x axis indicating the time (1 min). The colors match those for behaviors shown in (C).
Specifically, the training dataset was composed of four videos (2-min duration at 30 fps) containing a range of rat behavior from the psychomotor activity experiments described above. The labor for “DLC + B-SOiD” was spent on labeling animal body parts in extracted frames for tracking (∼25 min) and assigning behavior names for behavior classification (∼5 min), while that in LabGym was spent on sorting behavior examples for behavior classification (∼30 min). The labelers are experienced users for each approach. The testing dataset contained three videos (1-min duration at 30 fps per video).
-
(1)
A video that was randomly selected and trimmed from “Amphetamine (baseline)” in Figure 6A, in which the rats were exposed to 5.6 mg/kg amphetamine for the first time. The rat in this video showed psychomotor activities such as frequent rearing and locomotion.
-
(2)
A video that was randomly selected and trimmed from “Amphetamine (post treatment)” in Figure 6A, in which the rats were exposed to 5.6 mg/kg amphetamine after experiencing repeated amphetamine exposure (i.e., a sensitizing regimen). The rat in this video showed intensified psychomotor activity such as frequent stereotypy (repeated head moving).
-
(3)
A video from Ferrario et al.36 in which testing occurred in a different enclosure than that used in the training videos. This video was used to test how well each approach can be applied to experiment settings (such as enclosure, camera, and lighting conditions) that are completely different from those used for training.
Comparisons were performed on two key aspects: the total time cost and the end accuracy of identifying behaviors of interest. We first input the four training videos into DLC to extract frames and labeled six body parts in each frame. We then used these labeled frames to train a DLC model. This resulted in only 2.83 pixels of error in evaluation, indicating a successful training (Figure S5B). Next, we ran the trained DLC model on the four training videos to get the frame-wise positions of the six body parts. These outputs were then fed into B-SOiD to train a B-SOiD model for behavior classification. B-SOiD uses an unsupervised approach to form behavior groups and outputs video examples for each group, which is a convenient way for users to determine behavior type for each group and supervise the behavior classification. We tested different numbers of behavior groups and inspected the video examples for each group. We found that a 5-group model showed well-separated behavioral clusters (Figure S5C) and provided meaningful and consistent behavior type for each group. Using more groups resulted in too many different groups that turned out to be the same behavior that was expressed in different ways, or a single group containing video examples of different behaviors. We then labeled each group as a single behavior type based on the most frequently observed behavior among the videos in that group (Figure S5D). Finally, we ran the trained DLC model on the three testing videos to produce the frame-wise body part position output and fed that into the trained B-SOiD model for behavior classification.
While “DLC + B-SOiD” requires manual labeling and network training for tracking the animal body parts, LabGym saves this step; users only need to specify a time window in a video for background extraction to enable the tracking. We inputted the four training videos into LabGym to generate behavior examples and then sorted these examples into different folders according to their behavior types. Next, we input these folders into LabGym to train a Categorizer and ran LabGym on the three testing videos for behavior classification.
The total time to set up and run “DLC + B-SOiD” on all training and testing videos was about 9 h, while that for LabGym was less than 1 h (Figure 7B). To compare the end accuracy of behavioral categorizations with each tool, we first manually annotated the behavior types in each frame of each testing video (1,800 frames per video). We then compared the frame-wise behavior classification outputs of B-SOiD and LabGym with the hand-annotated data. We found that both “DLC + B-SOiD” and LabGym accurately identified rearing and locomotion in the amphetamine baseline video (Figure 7C and Data S2, video 16); however, “DLC + B-SOiD” failed to identify head movements post amphetamine treatment video, whereas LabGym achieved good accuracy in identifying this behavior (Figure 7D and Data S2, video 17).
When the testing was applied to a video made in a different enclosure with different camera and lighting conditions, LabGym was still able to correctly identify the behaviors of psychomotor activity with only a modest drop in accuracy (Figure 7E and Data S2, video 18). In contrast, “DLC + B-SOiD” was not able to identify these behaviors at all (Figure 7E).
It is possible that “DLC + B-SOiD” is able to accurately identify the “head movement” behavior in the testing videos if additional training data were provided. Alternatively, it is possible that the high-level properties (body length, angle, and speed) used by B-SOiD to form behavioral groups might not be useful for distinguishing head movements from other behaviors. Nevertheless, the purpose here was to directly compare approaches using the same training resource. Both approaches were able to accurately identify rearing, but only LabGym was able to generalize to another experimental setting, and capture behaviors specific to psychostimulant drug effects.
Discussion
LabGym opens opportunities for automatic, high-throughput, targeted, and customizable analyses of behaviors without the cost of commercial products, which will greatly facilitate the process of mechanistic discoveries in biological research. The flexibility and trainability of this tool allows it to be applied to a very broad variety of species, behaviors, and questions, and to be iteratively refined toward a particular set of conditions over time. The visualizable behavioral datasets generated by LabGym will also benefit research in neuroscience, behavioral science, and computer science by helping to standardize procedures within and across research groups, and by providing long-term references for future studies to build on.
Innovations in LabGym
First, to the input of the deep neural network we added the “pattern image” to represent the animal’s motion pattern of both the whole body and body parts efficiently and precisely. We then designed the Pattern Recognizer in Categorizer to analyze the pattern image. The results from the ablation experiments demonstrate that the inclusion of “pattern image” greatly enhances the categorization accuracy of the Categorizer.
Second, we developed a method for effective subtraction of image background that outperforms state-of-the-art background subtraction approaches. This method enables accurate tracking of animal bodies without the need for manual labeling and model training, which significantly saves the human labor and accelerates the analysis.
Third, unlike other tools using fixed and complex neural network structures, such as ResNet50, the users of LabGym can customize the input shapes and the complexity of the neural network (the Categorizer) through the GUI for behavioral categorization. This allows LabGym to suit behavior data with different complexity and enables fast running speed in even common laptops without GPUs.
Last, LabGym provides a GUI and does not require users to have programming knowledge to achieve the goal of quantifying the behaviors of their interest. All that users need to do is to recognize and sort the animations and their paired pattern images that are generated by LabGym as examples of a given behavior to “teach” LabGym to identify the behavior. The behavior is then automatically quantified.
LabGym provides a way for users to generate visualizable behavioral datasets
In the computer vision field, high-quality datasets, such as ImageNet,38 have been established to facilitate the training of deep neural networks and to benchmark different algorithms. In neuroscience, however, such benchmark datasets for behaviors in model organisms are largely absent. A source of this problem is the lack of tools that can help researchers to generate standardized, visualizable behavioral datasets that are easy to label and to share. Although transfer learning that applies acquired network weights to new categorization problems can potentially be used to compensate for the lack of species-specific behavioral datasets, its efficiency relies on the similarity between existing datasets used for pretraining the networks and the new data.39 Current tools using transfer learning for behavioral categorizations23 used network weights that were acquired from human behavioral datasets such as Kinetics-700.40 However, human behaviors are intrinsically different from the behaviors of model organisms, especially the soft-bodied ones. Therefore, species-specific behavioral datasets are still in critical demand for accurate behavioral categorizations in model organisms.
LabGym addresses this issue by providing a way for users to generate visualizable behavioral datasets. Importantly, the well-selected behavioral datasets, if shared in the research community, not only ensure the accurate and reproducible executions of categorization criteria across different experimenters or trials, but also provide benchmarks for comparing different algorithms, and references for future studies to build on.
Limitations of the study
First, although we tried our best to make the training resource for each tool as close as possible in benchmarking LabGym with the existing standard, the training data for each tool was different. This is because the data formats in these tools are different. DLC uses images and B-SOiD uses the time-series (x, y) positions of body parts derived from DLC, while LabGym uses pairs of animations and pattern images. It is empirically difficult to use the same training data for each approach. Moreover, the labor spent in annotating the training data in each approach was also intrinsically different. Considering this, we used the same labeling time for each approach in the comparison.
Second, although LabGym does not have restriction on the kind of background, enclosure, or camera angle needed, these aspects need to be static during the period for behavior analysis. Moreover, the level of illumination needs to be stable during analysis. Furthermore, animals need to move around in relation to the background for LabGym to differentiate them from the background. Thus, a time window during which animals are moving is needed for background extraction in LabGym. These limitations can potentially be addressed by implementing mask region-based convolutional neural networks (mask R-CNN), which is useful for segmenting animals from backgrounds that continuously change over time.41
Third, the current version of LabGym does not track the positions of the limbs, although the motion sequences of limb movement can be shown in the pattern image for accurate behavior identification.
Last, the current version of LabGym is not optimized for analyzing social behaviors. In social behaviors, animals have body contacts. The animal identity reassignment after animal re-separation might not be accurate. The issue might also be addressed by implementing mask R-CNN in future versions of LabGym.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| D-Amphetamine hemisulfate salt | Sigma Aldrich | Cat# 51-63-8 |
| All-trans-Retinal | AG Scientific | Cat#116-31-4 |
| Experimental models: Organisms/strains | ||
| Rat: Sprague Dawley | Envigo; Indianapolis IN | N/A |
| Mouse: C57BL/6J | The Jackson Laboratory | RRID:IMSR_JAX:000664 |
| Drosophila: TrpA1-QF | Bloomington Drosophila Stock Center | 36345 |
| Drosophila: QUAS-ChR2T159C-HA | Bloomington Drosophila Stock Center | 52260 |
| Drosophila: LK-GAL4 | Bloomington Drosophila Stock Center | 51993 |
| Drosophila: UAS-GtACR1-YFP | Bloomington Drosophila Stock Center | 92983 |
| Drosophila: UAS-jRCaMP1b | Bloomington Drosophila Stock Center | 63793 |
| Drosophila: Canton S | Bloomington Drosophila Stock Center | 64349 |
| Software and algorithms | ||
| LabGym | This study | https://github.com/umyelab/LabGym; https://doi.org/10.5281/zenodo.7579381 |
| Python 3.9 | The Python Software Foundation | https://www.python.org/; RRID:SCR_008394 |
| Prism 9 | GraphPad | RRID:SCR_002798 |
| NumPy | The NumPy community | https://numpy.org/; RRID:SCR_008633 |
| OpenCV | The OpenCV community | https://opencv.org/; RRID:SCR_015526 |
| SciPy | The SciPy community | https://scipy.org/; RRID:SCR_008058 |
| Matplotlib | The Matplotlib community | https://matplotlib.org/; RRID:SCR_008624 |
| scikit-learn | The scikit-learn community | https://scikit-learn.org/; RRID:SCR_002577 |
| scikit-image | The scikit-image community | https://scikit-image.org/; RRID:SCR_021142 |
| MoviePy | The MoviePy Developers | https://zulko.github.io/moviepy/ |
| Pandas | The Pandas community | https://pandas.pydata.org; RRID:SCR_018214 |
| Seaborn | The Seaborn community | https://seaborn.pydata.org/; RRID:SCR_018132 |
| Tensorflow | The Tensorflow community | https://www.tensorflow.org/; RRID:SCR_016345 |
| wxPython | The wxPython Developers | https://docs.wxpython.org/wx.grid.Grid.html |
Resource availability
Lead contact
Further information and requests should be directed to and will be fulfilled by the lead contact, Bing Ye (bingye@umich.edu).
Materials availability
This study did not generate new reagents.
Experimental model and subject details
Drosophila
Both male and female foraging third-instar larvae were used. All experiments were done on age- and size-matched larvae. The fly strains are in Key resources table.
Rodents
All rodent procedures were approved by University of Michigan Institutional Animal Care and Use Committee and are consistent with NIH guidelines.
Adult male Sprague-Dawley rats (∼55 days old at the start of the experiment) were pair-housed on reverse 12/12 h light/dark cycle throughout, food and water were available ad libitum and rats were tested in red light conditions during the dark phase of the cycle (Envigo; Indianapolis, IN).
Adult male C57BL/6J mice (one animal aged ∼80 weeks old at the start of the video recordings, one animal aged ∼32 weeks old at the start of the video recordings) were single housed in cages within a humidity and temperature-controlled vivarium and kept on a 12/12 h light/dark cycle (lights on 6 a.m.) with ad libitum access to food and water. At the time of video recording, both mice were single-housed in a cage with bedding, 1/4 Nestlet (Ancare, Bellmore, NY) for nest building, and a Mouse Igloo and Fast Trac running wheel (Bio-Serv, Flemington, NJ), located within a custom-made white PVC foam sheet (American Plastics Solutions, Saline, MI) enclosure, located within a humidity and temperature-controlled room. Animals had ad libitum access to food and water during the video recordings and were kept on a 12/12 h light/dark cycle (lights on 6 a.m.).
Methods details
Overall implementation
LabGym was written in Python programming language (version 3.9).42 Python libraries used in LabGym are: Numpy,43 Scipy,44 scikit-learn,45 scikit-image,46 MoviePy, Matplotlib,47 OpenCV,29 Pandas,48 Seaborn,49 Tensorflow,50 wxPython.51
Computation hardware
The computational procedures in this study were performed on the following 4 computers. (1) a MacBook Pro 13 inch (early 2015 model) with Mac OS Big Sur (or later), 3.1 GHz Dual-Core Intel Core i7 processor, 16 GB 1867 MHz DDR3 memory, and Intel Iris Graphics 6100 1536 MB graphics. (2) HP Z4 Workstation with Windows 10 Enterprise for Workstations, 3.2 GHz Intel Xeon W-2104 processor, 32GB 2666 MHz DDR4, and NVIDIA GeForce GTX 1080 Ti. (3) HP Z4 G4 Workstation with Windows 10 Pro for Workstations, 3.6 GHz Intel Xeon W-2133 processor, 32GB 2666 MHz DDR4, and NVIDIA Quadro P2000. (4) DESKTOP-G2HRLHT with Windows 10 Pro for Workstations, 3.6 GHz Intel Core i9-9900K CPU, 64 GB 2666 MHz DDR4, and NVIDIA GeForce RTX 2080.
Remove backgrounds and track multiple animals
Reconstruct the static backgrounds for a video
To reconstruct the static background of a video, the stable intensity value over time for each pixel, denoted as Pstable, was identified by looking for a sliding time window tstable of 100 frames (about 3 s if the fps is 30) during which the value of this pixel was stable. Pstable is the mean intensity during tstable, which is obtained by searching through the entire duration of the video. Specifically, for a video in which the animals were generally lighter than the background, it was identified when the sum of the mean and SD of pixel values in the time window was the smallest (Equation 1). For a video in which the animals were generally darker than the background, tstable was the time window in which the sum of the mean and SD of the inverted pixel values was the smallest (Equation 1).
| (Equation 1) |
where t represents 100-frame time windows sliding over the video, std(t) is the SD of the pixel values during the time window t.
In the less likely case that the animals were partially lighter or darker than the background, Pstable was the mean of this pixel over the entire duration of the video. In addition, if the illumination was generally stable in the entire video, then we simply used the extremum value (the minimum for animals lighter than the background, and the maximum for animals darker than the background) to achieve similar results as using Pstable while keeping the computational cost low. Therefore, we provide users with the option to specify whether the illumination in the video is stable.
The Pstable for the time when the illumination changes (for example, the optogenetic manipulation period) was identified separately when the mean value of all the pixels in the current frame was 1.2-fold greater than that in the first frame.
Track the animals
The foreground (all the pixels of an animal) was obtained by subtracting background in each frame. The approximate area of single animal was estimated by averaging the total area of all the animals over the animal number. Each animal is a matrix of [identity, [contours], [centers], [behavioral information], …]. We assumed that the Euclidean distance between the centers of the same animal in consecutive frames is always the smallest among the distances between all the center pairs. In this way, all the animals in current frames are linked to the same animals in previous frames and the new information of the same animals is added into the animal matrices. Since the animals might be undetectable in some frames, the algorithm examined the entire frames from the beginning to the end of the analysis, rather than only examined the consecutive frames, to minimize the probability of not being tracked.
Multiple animals in the same enclosure sometimes collide with each other, which might cause false tracking. To make the analysis fully automatic, LabGym excludes animals from tracking if they do not meet the user-defined criteria for the number of entangled animals to be allowed for analysis. For example, if entanglement is not allowed, the merged foreground of two or more entangled animal would be temporarily excluded from tracking until they separate again. The animal that is lost track for over 1 s will be reassigned by a new ID/matrix and the previous ID/matrix for this animal will be deactivated. Any ID/matrix that is deactivated for longer than 20% of the entire analysis duration is considered as disappeared and is automatically excluded from the analysis, which ensures that the analysis is only performed on the reliably tracked animals. When the entanglement is allowed, merged foreground will always be tracked and animal IDs/matrices can be preserved when they separate again.
Generation of animations and pattern images
Generate animations
The animation of an animal is a set of blobs over a user-defined time window. Each blob is a cropped video frame containing a single animal. The animal foreground in a blob is masked by the animal contour and the area inside the contour in the original frame and the background is set to pixel value of 0. Users also have the option to include backgrounds in the animations since the environmental factors sometimes are key to behavioral categorization.
Generate pattern images
The pattern image for each animation was generated by drawing all animal contours (the whole body and the body parts) within the animation onto a black background with gradual changing colors to indicate the temporal sequences of the contours. The contours of inner body parts were identified using the ‘Canny edge detection’ and ‘find contours’ functions in OpenCV library after preprocessing of each frame. The SD of each pixel value in the detected edges over the user-defined time window (the duration of its paired animation) was calculated and thresholded to determine whether this pixel belongs to the edges of a body part that has movement during the time window. The threshold of the SD is user definable. The sizes of pattern images are the same as the frame in their paired animations. Users can also choose whether to show animals’ body parts in the pattern images, depending on their needs.
The design and implementation of Categorizer
Animation Analyzer
Animation Analyzer consists of convolutional blocks warped with time distributed layers and followed by recurrent layers (long short-term memory, LSTM).21,50,52 It takes 4-dimentional tensors (timestep, width, height, color channel) as inputs, in which the timestep, width/height and color channel are all user definable. The timestep of the input tensors is the number of frames in an animation (the length of an animation). Ideally the length should precisely match the variable durations of different behaviors so that each animation only contains a single behavior. However, since deep neural networks require their input shape to be consistent during the analysis, the durations for all the animations need to be the same. We did not use zero paddings to arbitrarily make the input shape consistent because we wanted the Animation Analyzer to learn to ignore those frames mixed with behaviors of nontargeted categories, which is a practical scenario in behavioral analyses. To achieve the best efficiency in training the Categorizer, the duration of the animations should be the shortest time for experimenters to distinguish all the behavioral categories. Animation Analyzer implements two different architectures: VGG-like53 and ResNet-like.54 There are 7 different complexity levels of Animation Analyzer for user to choose to fit different datasets. Levels 1, 2, 3 or 4 is VGG-like architecture with 2, 5, 9, or 13 convolutional layers (Conv2D), respectively. Each convolutional layer is followed by a batch normalization layer (BN). Max pooling layers (MaxPooling2D) are added after the second BN in level 1, after the second and fifth BN in level 2, after the second, fifth and ninth BN in level 3, or after the second, fifth, 9th and 13th BN in level 4. Levels 5, 6, or 7 is ResNet18, ResNet34 or ResNet50 architecture. After the convolutional operations, the outputs are flattened into 1-dimentional (1D) vectors at each time step and then are passed to LSTM. The outputs of LSTM are passed to the Decision Maker submodule.
To make LabGym applicable to behavioral datasets of various complexities, the frame sizes and color channels of the input tensors are user definable. Therefore, the network architectures in Animation Analyzer are provided with various options, from simple 2-layer VGG-like to complex 50-layer ResNet50 architectures. The number of filters in the first convolution layer is determined by the height or width dimension of input tensors (the height and width of the input tensor is the same), and then doubled after each max pooling layer. The number of filters in the LSTM is the same as that of the last convolution layer.
Pattern Recognizer
Pattern Recognizer consists of convolutional blocks and takes 3-dimentional tensors (width, height, color channel) as inputs, in which both the width and height are user definable. The color channel is fixed to 3 (Red, Green, and Blue; RGB) since the colors in the pattern images indicate the temporal sequences of animal motions. There are also 7 different complex levels of Pattern Recognizer for the user to choose from to suit different datasets. The architecture for each level in Pattern Recognizer is the same as that for each level in Animation Analyzer, except that there is no time-distributed wrapper in Pattern Recognizer.
Decision Maker
Decision Maker consists of a concatenation layer and two fully connected (dense) layers. The concatenation layer merges the outputs from both Animation Analyzer and Pattern Recognizer into a single 1D vector and outputs to the first dense layer. The first dense layer then outputs to the second dense layer in which a SoftMax function is used for computing the probabilities of all the behavioral categories. The number of nodes in the first dense layer of Decision Maker is determined by the complexity levels of Animation Analyzer or Pattern Recognizer, whichever is higher. This association allows the Decision Maker to adapt based on the structure of the other two modules. A Batch Normalization layer and a Dropout layer (dropout rate is set to 0.5) are added before the second dense layer. The number of nodes in the second dense layer is set to the number of behavioral categories. At each frame during the analysis, the output of Decision Maker for an animal is a matrix of probabilities for all the behavioral categories and the behavioral category with the highest probability is determined to be the behavior that the animal performs. The probabilities for all the behaviors at each frame are exported to Microsoft Excel files for the usage as users see fit.
Train Categorizers
The loss function used to train Categorizers is either binary crossentropy if the number of behavioral categories is two, or categorical crossentropy if the number of behavioral categories is three or more. Stochastic gradient descent (SGD)55 with an initial learning rate of 1 × 10−4 is used for optimizer in training. The learning rate decreases by a factor of 0.2 if the validation loss stops decreasing (decreasing by < 0.001 is considered as ‘stop decreasing’) for 2 training epochs, and the training stops if the validation loss stops decreasing for 4 training epochs. The model is automatically updated after a training epoch reaches a minimal validation loss. The batch size in training is 8, 16, or 32 if the number of validation data is less than 5,000, between 5,000 and 50,000, or over 50,000, respectively. The ratio of training and validation data split is 0.8:0.2.
To enhance the training efficacy and the generalizability of the trained models, we developed 9 different data augmentation methods that apply the same random alterations to both an animation and its paired pattern image. Notably, during data augmentation, all the alterations of frames/images are performed only on animal foregrounds and their contours. The absolute dimensions of frames/images and the black backgrounds are unchanged. These methods are performed in combination and users have the options to choose which method to perform. If all the data augmentation methods are applied, the amount of data can be expanded to 47 times of its original amount.
-
1.
Random rotation: both the animations and their paired pattern images are rotated with a random angle in ranges between 10°–50°, 50°–90°, 90°–130°, 130°–170°, 30°–80° and 100°–150° (the latter two ranges are used to be combined with random brightness changes).
-
2.
Horizontal flipping: both the animations and their paired pattern images are flipped horizontally.
-
3.
Vertical flipping: both the animations and their paired pattern images are flipped vertically.
-
4.
Random brightening: the animations have brightness increase in ranges between 30 and 80.
-
5.
Random dimming: the animations have brightness decrease in ranges between 30 and 80.
-
6.
Random shearing: both the animations and their paired pattern images are sheared with a random factor in ranges between −0.21 and -0.15 and 0.15–0.21.
-
7.
Radom rescaling: both the animations and their paired pattern images are rescaled in either their widths or heights with a random ratio in a range between 0.6–0.9.
-
8.
Random deletion: one or two frames in the animations will be randomly deleted and replaced with black images of the same dimensions.
Select suitable categorizers for different behavioral datasets
The Categorizer can be customized into different complexity to address behavioral datasets with different complexity. The complexity of Categorizer is determined by two factors: the level of Animation Analyzer/Pattern Recognizer and the size of input frame/image. The former determines how deep whereas the latter determines how wide the Categorizer is (see the design and implementation of categorizer). There are 7 levels of Animation Analyzer/Pattern Recognizer for user to choose and the input frame size is completely defined by users. The general principle to guide the selection of suitable Categorizer for each behavioral dataset is Occam’s razor, which means that if the performances of two Categorizers are comparable, the simpler one is better. Therefore, we started from Categorizers with the simple complexity and gradually increased the complexity until their performance are satisfying, for all 3 behavioral datasets.
The behaviors in the larva dataset are distinguishable from the sequential changes of their body shapes during the behaviors. Besides, the resolution in the animations is insufficient to show details of the larva body. Therefore, we chose to downsize the frame in animations into 8 × 8 (gray scale) for Animation Analyzer so that it can focus on only the body shapes without learning too many irrelevant details. The pattern images for larva behaviors contain more details such as the lines of different colors indicating the temporal sequences of the behaviors. Therefore, we chose 32 × 32 (RGB scale) as the input image size of Pattern Recognizer. We started from the level 1 of Animation Analyzer and level 2 of Pattern Recognizer, which already achieved excellent overall accuracy on the larva nociceptive behavior subset. When we increased the complexity of the Categorizer to level 1 Animation Analyzer and level 3 Pattern Recognizer, the accuracy decreased, indicating the potential overfitting of the Categorizer. Therefore, we stopped at level 1 Animation Analyzer and level 2 Pattern Recognizer as the suitable Categorizer for the larva dataset (LarvaN).
To select the suitable Categorizer for the rat dataset, we chose 16 × 16 or 32 × 32 (gray scale) for the input frame size of Animation Analyzer and 32 × 32 or 64 × 64 (RGB scale) for the input image size of Pattern Recognizer. This was because the differences among different behaviors in the rat dataset are more subtle than those in the larva dataset and we wanted the Categorizer to learn more details. The overall validation accuracy stopped increasing at level 4/4 for Animation Analyzer/Pattern Recognizer but was still not ideal (0.8), indicating that the unsatisfying accuracy was not because of low complexity of the Categorizer. Note that due to the data-driven nature of deep-learning based tools, the accuracy of Categorizer also largely relies on the quality of the training dataset such as the amount and diversity of the data, and the labeling accuracy. Therefore, we refined the rat dataset by discarding some examples that were ambiguous for categorization by the labeler. We then trained the Categorizer of level 4/4 for Animation Analyzer (32 × 32 × 1)/Pattern Recognizer (64 × 64 × 3) on the refined rat dataset, which achieved significantly improved overall accuracy and was determined as the suitable Categorizer for the rat dataset (RatA).
To select the suitable Categorizer for the mouse dataset, we included all the categories in which the number of examples (the number of animations and pattern images in pair) is greater than 50. Since the animations in the mouse dataset contain details that are key to categorizations of the behaviors, such as the movement of paws/noses, we chose 32 × 32 or 64 × 64 (gray scale) for Animation Analyzer and 32 × 32 or 64 × 64 (RGB scale) for Pattern Recognizer. We found that level 4/4 for Analyzer (64 × 64 × 1)/Pattern Recognizer (64 × 64 × 3) achieved satisfying overall accuracy and determined it as the suitable Categorizer for the mouse dataset (MouseH).
Criteria for labeling the behavioral categories
Drosophila larva
Crawling: A larva performs one of the following actions: 1) peristalsis 2) moving with body mostly straight 3) moving along the direction mostly aligned with the body axis. The crawling results in positional changes of larval body that can be clearly seen in the pattern image.
Curling: A larva performs ‘C’-shape bending and both the head and tail bend approximately at the same time from a straight, resting body shape.
Hunching: A larva retracts only the head.
Rolling: A larva moves with a curling body shape along the anterior-posterior body axis, which causes lateral positional changes in the larval body that can be seen in the pattern image.
Turning: A larva performs a body bending that is not in ‘C’-shape (body bending that is not curling).
Uncoiling: A larva performs the actions in temporal sequences that are opposite to curling or turning (from a coiling body shape to a straight one).
Immobile: A larva does not move or only has subtle movements.
Mouse
Behind the wheel: A mouse is positioned behind the running wheel.
Body grooming: A mouse licks its body, below the neck.
Chewing: A mouse chews on food pellets.
Coming down: A mouse lowers from a standing pose on its hindpaws to a sitting pose on both forepaws and hindpaws.
Crawling: A mouse crawls under the running wheel.
Face grooming: A mouse uses its paws to groom the face, head, mouth and/or ear.
Foraging: A mouse walks with its snout on the cage bedding, digs through the cage bedding with its forepaws, or investigates the cage bedding with its forepaws while stationary.
Hind paw grooming: A mouse uses its hindpaws to groom its face or body.
Jumping onto the wheel: A mouse jumps onto the running wheel.
Nest building: A mouse uses its mouth to build a nest or reposition nestlet material in the homecage.
Rearing up: A mouse rises from a sitting position with both forepaws and hindpaws on the cage bedding, to a standing position with only the hindpaws on the cage bedding.
Resting on the wheel: A mouse remains stationary on the running wheel.
Running on the wheel: A mouse runs on the running wheel.
Sleeping: A mouse sleeps in the homecage.
Sniffing: A mouse repeatedly sniffs the air, with the snout repeating superior and inferior movements while pointed at the cage walls or the lid of the homecage.
Standing: A mouse remains in a standing posture on both hindpaws.
Turning: A mouse turns to show a frontal view, a side profile view, or a back view.
Unknown bv: A mouse is performing some behavior while only its back is visible to the camera. The behavior is unidentifiable, unless the mouse is rearing up, coming down, or standing.
Walking: A mouse walks on the bedding (not on the running wheel).
Rat
Body grooming: A rat grooms its body.
Face grooming: A rat grooms its face.
Head swaying (moving): A rat sways its head side to side for more than one repeats.
Locomotion: A rat proceeds forward.
Orientating: A rat turns or repositions.
Rearing: A rat rears up.
In place: A rat rests in place with some small movements.
Still: A rat is completely motionless.
Behavioral experiments
Drosophila larval behaviors responding to nociceptor stimulation
Nociceptors of late third instar larvae were activated optogenetically TrpA1-QF > QUAS-ChR2T159C transgenes. To stimulate TrpA1-QF > QUAS-ChR2T159C, 50 μW/mm2 blue light was applied for 2.5 s. GtACR1 was expressed in LK neurons to inhibit these neurons.30 GtACR1-mediated optogenetic inhibition was done with 2.5 s of amber light (590 nm). All LEDs used were from LUXEON StarLED. Embryos were collected on standard cornmeal containing 500 μM all-trans-retinal (ATR) (A.G. Scientific).
For each trial in larval behaviors, we placed 10–15 larvae on a ∅ 35-mm or 90-mm plate covered with a thin layer of water. The larvae behaviors were recorded by using a webcam (Logitech) at a resolution of 1280 × 720 or 864 × 480 resolution (30 fps) in the MP4 or AVI format.
Drosophila larval behaviors responding to sound stimuli
Late third instar larvae were exposed to white noise at 80 or 94 dB SPL (0.2–20 kHz) for 5 s. The larvae were tested on a black agar plate to increase the contrast between the background and the foreground for a better signal-to-noise ratio. All larvae were raised in a custom-made sound-resistant box for 5–7 days at room temperature to minimize sound exposure, and were recorded with a Logitech 4K Brio webcam (30 fps) at 1080 × 1080 resolution in the MP4 format.
Drosophila adult courtship behaviors
To demonstrate the potential capacity of LabGym in analyzing social behaviors, we used some videos of Drosophila courtship behaviors from a previous report.56 In these experiments, a wild-type male (Oregon R) was paired with either one or 4 females (Oregon R or w1118) in a mating chamber.
Psychomotor sensitization in rats
All rodent procedures were approved by University of Michigan Institutional Animal Care and Use Committee and are consistent with NIH guidelines.
Adult male Sprague-Dawley rats (∼55 days old at the start of the experiment) were pair-housed on reverse 12/12 h light/dark cycle throughout, food and water were available ad libitum and rats were tested in red light conditions during the dark phase of the cycle (Envigo; Indianapolis, IN). All testing occurred in custom made chambers (13″W x 26.5″L x 24″T) equipped with overhead video cameras (fps:30; frame size: 928 × 480; video format:.MP4). Behavior was recorded throughout each session. Rats were first habituated to the testing chambers and injection procedure; they were placed in the chamber for 40 min, given an i.p. injection of saline (0.9%, 1 mL/kg) and returned to their home cage after an additional 40 min (4 sessions, 1/day). Rats were then split into two groups given i.p. injections of either saline (N = 3) or d-amphetamine sulfate dissolved in saline (Sigma Aldrich, N = 7). On each injection day rats were placed in the chambers for 40 min prior to injection. On the first day, responses to increasing doses of d-amphetamine were determined within session (0.32, 1.0, 3.2, and 5.6 mg/kg, i.p), as previously described.57 The following day rats began a sensitization regimen in which they were given increasing concentrations of d-amphetamine (0.5, 2, 4, 5, 6, 0.5, 6, 6, 0.5 mg/kg, i.p.) with one injection given per day, adapted from.58 The within session dose response was then re-assessed after 20–21 days of withdrawal. Rats in the saline group received saline throughout. The rat videos used for demonstration of tracking in LabGym were sample video in previous reports.36,37
Mouse
All rodent procedures were approved by University of Michigan Institutional Animal Care and Use Committee and are consistent with NIH guidelines.
Adult male C57BL/6J mice (one animal aged ∼80 weeks old at the start of the video recordings, one animal aged ∼32 weeks old at the start of the video recordings) were single housed in cages within a humidity and temperature-controlled vivarium and kept on a 12/12 h light/dark cycle (lights on 6 a.m.) with ad libitum access to food and water. At the time of video recording, both mice were single-housed in a cage with bedding, 1/4 Nestlet (Ancare, Bellmore, NY) for nest building, and a Mouse Igloo and Fast Trac running wheel (Bio-Serv, Flemington, NJ), located within a custom-made white PVC foam sheet (American Plastics Solutions, Saline, MI) enclosure, located within a humidity and temperature-controlled room. Animals had ad libitum access to food and water during the video recordings and were kept on a 12/12 h light/dark cycle (lights on 6 a.m.). Behavior was recorded with a Basler acA1300-200μm camera, equipped with a Basler C123-0418-5M lens and a NIR Bandpass Infrared Filter (Machine Vision Store, St. Paul, MN). InfraRed lighting (850 nm, LEDLightsWorld) diffused by a 1/4” x 13.5″ x 24″ sheet of white acrylic plexiglass (estreetplastics, Royse City, Tx) was used to illuminate the homecage as well as to provide the background for all video recordings. Free home-cage behavior was recorded in 4-h segments as.mkv files, at a resolution of 1280 × 1024, 30 fps, with an exposure time of 6500 μs.
Quantification and statistical analysis
Test Categorizer in practice
LabGym provides two convenient ways of validation on how Categorizers perform. First, the copies of behavioral videos with full annotations of the behaviors assist the visualization of both the tracking and the behavioral categorizations. Second, for a more solid test, users can use Generate Behavior Examples functional unit in the GUI of LabGym to generate unsorted behavior examples (an animation and its paired pattern image) and sort them into different categories (into folders under the behavior names) to build a ground-truth testing dataset. Users can then use this ground-truth testing dataset in Test Categorizers functional unit to test a trained Categorizer. In this way, the users are blinded to the predictions of the Categorizer to be tested and thus the validation is unbiased. In this study, all tests for the in-practice performance of the LarvaN, RatA and MouseH were performed through this approach.
We used the following performance metrics in testing Categorizers: precision (Equation 2), recall (Equation 3), f1 score (Equation 4), overall accuracy (Equation 5).
| (Equation 2) |
| (Equation 3) |
| (Equation 4) |
| (Equation 5) |
Calculate behavioral parameters
In the current version of LabGym, 14 behavioral parameters are computed based on the information of behavioral categories and animal foregrounds: acceleration/velocity reduction, angle, count, distance, duration, intensity (area), intensity (length), latency, magnitude (area), magnitude (length), speed, velocity, vigor (area) and vigor (length).
-
1.
The angle is the movement direction (against to the animal body axis) of the animal during a behavior, which is the mean of all the included angle () between animal body axis and the movement direction during the time window (t) for categorizing the behavior (Equation 6).
| (Equation 6) |
-
2.
The count is the summary of the behavioral frequencies, which is the occurrence number of a behavior within the entire duration of analysis. Consecutive single occurrences (at a single frame) of the same behavior are considered as one count.
-
3.
The distance is the total distance traveled of the animal by performing a behavior within the entire duration of analysis.
-
4.
The duration is the summary of how persistent a behavior is, which is the total time of a behavior within the entire duration of analysis.
-
5.
The intensity (area)/intensity (length) is the summary of how intense a behavior is, which is the accumulated proportional changes of the animal body area (a)/length (L) between frames divided by the time window for categorizing the behaviors (t) when performing a behavior (Equations 7 and 8).
| (Equation 7) |
| (Equation 8) |
-
6.
The latency is the summary of how soon a behavior starts, which is the time starting from the beginning of the analysis to the time point that the behavior occurs for the first time.
-
7.
The magnitude (area)/magnitude (length) is the summary of the motion magnitude, which is the maximum proportional change in animal body area (a) or length (L) when performing a behavior (Equations 9 and 10).
| (Equation 9) |
| (Equation 10) |
-
8.
The speed is the summary of how fast the animal moves when performing a behavior, which is the total distance traveled (can be back and forth) (d) (between the two centers of mass of the animal) during the time window for categorizing the behavior divided by the time window (Equation 11).
| (Equation 11) |
-
9.
The velocity is the summary of how efficient the animal’s movement is when performing a behavior, which is the shortest distance between the beginning and end location (dt) (between the two centers of mass of the animal) divided by the time (toccuring) that such displacement takes place (Equation 12).
| (Equation 12) |
-
10.
The acceleration/velocity reduction is the summary of how fast the animal’s velocity changes while performing a behavior, which is the difference between maximum velocity (vmax) and minimum velocity (vmin) divided by the time (toccuring) that such velocity change takes place (Equation 13).
| (Equation 13) |
-
11.
The vigor (area)/vigor (length) is the summary of how vigorous a behavior is, which is the magnitude (area)/magnitude (length) divided by the time (toccuring) that such a change takes place (Equations 14 and 15).
| (Equation 14) |
| (Equation 15) |
Statistics
Each statistical test was indicated in the according figure legend.
Acknowledgments
We thank Drs. Daniel Leventhal, Hanchuan Peng, Guoqiang Yu, and Ruonan Li for helpful comments on earlier versions of the manuscript. This research was supported by National Institutes of Health (NIH) to B.Y. (R01NS104299 and R01EB028159), and C.R.F. (R01DK106188, R01DK115526, R01DA044204, R01DK130246). Drosophila Stocks from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH.
Author contributions
Y.H. and B.Y. conceived the project. Y.H. designed and programmed LabGym with the guidance of J.Z. and B.Y. Y.H., K.I., H.W., C.R.F., A.G., and B.W. performed the behavioral experiments in Drosophila larvae (Y.H, K.I., and H.W.), rats (C.R.F.), and mice (A.G. and B.W.). Y.H., K.I., H.W., and Y.X. generated and labeled the dataset for larval behaviors (Y.H., K.I., H.W., and Y.X.), rat behaviors (Y.H.), and mouse behaviors (Y.H. and A.G.). Y.H. performed the computational analyses and the validation of behavioral categorizations with the assistance from C.R.F., A.G., B.W., K.I., and H.W. Y.H, A.B.M., and R.B.I. performed the benchmark comparison between LabGym and DeepLabCut + B-SOiD. Y.H., C.R.F., B.W., J.Z., and B.Y. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: February 24, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2023.100415.
Contributor Information
Yujia Hu, Email: henryhu@umich.edu.
Jie Zhou, Email: jzhou@niu.edu.
Bing Ye, Email: bingye@umich.edu.
Supplemental information
Data and code availability
-
•
All the data reported in this paper will be shared by the lead contact upon request. The behavioral datasets generated and analyzed in the current study is deposited for free access (https://drive.google.com/drive/folders/1OISUkm8An4isLFzBtj7BilI2Jp-J6jEU?usp=sharing).
-
•
All original code has been deposited at Zenodo and GitHub and is publicly available. The DOI and GitHub website are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Egnor S.E.R., Branson K. Computational analysis of behavior. Annu. Rev. Neurosci. 2016;39:217–236. doi: 10.1146/annurev-neuro-070815-013845. [DOI] [PubMed] [Google Scholar]
- 2.Datta S.R., Anderson D.J., Branson K., Perona P., Leifer A. Computational neuroethology: a call to action. Neuron. 2019;104:11–24. doi: 10.1016/j.neuron.2019.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altimus C.M., Marlin B.J., Charalambakis N.E., Colón-Rodriquez A., Glover E.J., Izbicki P., Johnson A., Lourenco M.V., Makinson R.A., McQuail J., et al. The next 50 Years of neuroscience. J. Neurosci. 2020;40:101–106. doi: 10.1523/Jneurosci.0744-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira T.D., Shaevitz J.W., Murthy M. Quantifying behavior to understand the brain. Nat. Neurosci. 2020;23:1537–1549. doi: 10.1038/s41593-020-00734-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denisov G., Ohyama T., Jovanic T., Zlatic M. 2013. Model-based Detection and Analysis of Animal Behaviors Using Signals Extracted by Automated Tracking; pp. 175–181. [Google Scholar]
- 6.Kabra M., Robie A.A., Rivera-Alba M., Branson S., Branson K. JAABA: interactive machine learning for automatic annotation of animal behavior. Nat. Methods. 2013;10:64–67. doi: 10.1038/Nmeth.2281. [DOI] [PubMed] [Google Scholar]
- 7.Berman G.J., Choi D.M., Bialek W., Shaevitz J.W. Mapping the stereotyped behaviour of freely moving fruit flies. J. R. Soc. Interface. 2014;11:20140672. doi: 10.1098/rsif.2014.0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiltschko A.B., Johnson M.J., Iurilli G., Peterson R.E., Katon J.M., Pashkovski S.L., Abraira V.E., Adams R.P., Datta S.R. Mapping sub-second structure in mouse behavior. Neuron. 2015;88:1121–1135. doi: 10.1016/j.neuron.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klibaite U., Berman G.J., Cande J., Stern D.L., Shaevitz J.W. An unsupervised method for quantifying the behavior of paired animals. Phys. Biol. 2017;14:015006. doi: 10.1088/1478-3975/aa5c50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graving J.M., Chae D., Naik H., Li L., Koger B., Costelloe B.R., Couzin I.D. DeepPoseKit, a software toolkit for fast and robust animal pose estimation using deep learning. Elife. 2019;8:e47994. doi: 10.7554/eLife.47994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereira T.D., Aldarondo D.E., Willmore L., Kislin M., Wang S.S.H., Murthy M., Shaevitz J.W. Fast animal pose estimation using deep neural networks. Nat. Methods. 2019;16:117–125. doi: 10.1038/s41592-018-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bala P.C., Eisenreich B.R., Yoo S.B.M., Hayden B.Y., Park H.S., Zimmermann J. Automated markerless pose estimation in freely moving macaques with OpenMonkeyStudio. Nat. Commun. 2020;11:4560. doi: 10.1038/s41467-020-18441-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klibaite U., Shaevitz J.W. Paired fruit flies synchronize behavior: uncovering social interactions in Drosophila melanogaster. PLoS Comput. Biol. 2020;16:e1008230. doi: 10.1371/journal.pcbi.1008230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilsson S.R., Goodwin N.L., Choong J.J., Hwang S., Wright H.R., Norville Z.C., Tong X., Lin D., Bentzley B.S., Eshel N. Simple Behavioral Analysis (SimBA)–an open source toolkit for computer classification of complex social behaviors in experimental animals. bioRxiv. 2020 doi: 10.1101/2020.04.19.049452. Preprint at. [DOI] [Google Scholar]
- 15.Wiltschko A.B., Tsukahara T., Zeine A., Anyoha R., Gillis W.F., Markowitz J.E., Peterson R.E., Katon J., Johnson M.J., Datta S.R. Revealing the structure of pharmacobehavioral space through motion sequencing. Nat. Neurosci. 2020;23:1433. doi: 10.1038/s41593-020-00706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn T.W., Marshall J.D., Severson K.S., Aldarondo D.E., Hildebrand D.G.C., Chettih S.N., Wang W.L., Gellis A.J., Carlson D.E., Aronov D., et al. Geometric deep learning enables 3D kinematic profiling across species and environments. Nat. Methods. 2021;18:564–573. doi: 10.1038/s41592-021-01106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu A.I., Yttri E.A. B-SOiD, an open-source unsupervised algorithm for identification and fast prediction of behaviors. Nat. Commun. 2021;12:5188. doi: 10.1038/s41467-021-25420-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall J.D., Aldarondo D.E., Dunn T.W., Wang W.L., Berman G.J., Ölveczky B.P. Continuous whole-body 3D kinematic recordings across the rodent behavioral repertoire. Neuron. 2021;109:420–437.e8. doi: 10.1016/j.neuron.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segalin C., Williams J., Karigo T., Hui M., Zelikowsky M., Sun J.J., Perona P., Anderson D.J., Kennedy A. The Mouse Action Recognition System (MARS) software pipeline for automated analysis of social behaviors in mice. Elife. 2021;10:e63720. doi: 10.7554/eLife.63720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathis A., Mamidanna P., Cury K.M., Abe T., Murthy V.N., Mathis M.W., Bethge M. DeepLabCut: markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 2018;21:1281–1289. doi: 10.1038/s41593-018-0209-y. [DOI] [PubMed] [Google Scholar]
- 21.Fukushima K. Neocognitron - a self-organizing neural network model for a mechanism of pattern-recognition unaffected by shift in position. Biol. Cybern. 1980;36:193–202. doi: 10.1007/Bf00344251. [DOI] [PubMed] [Google Scholar]
- 22.LeCun Y., Bengio Y. The Handbook of Brain Theory and Neural Networks. Vol. 3361. 1995. Convolutional networks for images, speech, and time series; p. 1995. [Google Scholar]
- 23.Bohnslav J.P., Wimalasena N.K., Clausing K.J., Dai Y.Y., Yarmolinsky D.A., Cruz T., Kashlan A.D., Chiappe M.E., Orefice L.L., Woolf C.J., Harvey C.D. DeepEthogram, a machine learning pipeline for supervised behavior classification from raw pixels. Elife. 2021;10:e63377. doi: 10.7554/eLife.63377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marks M., Qiuhan J., Sturman O., von Ziegler L., Kollmorgen S., von der Behrens W., Mante V., Bohacek J., Yanik M.F. Deep-learning-based identification, tracking, pose estimation and behaviour classification of interacting primates and mice in complex environments. Nat. Mach. Intell. 2022;4:331–340. doi: 10.1038/s42256-022-00477-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 26.Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Roh Y., Heo G., Whang S.E. A survey on data collection for machine learning: a Big data-AI integration perspective. IEEE Trans. Knowl. Data Eng. 2021;33:1328–1347. doi: 10.1109/Tkde.2019.2946162. [DOI] [Google Scholar]
- 28.Guo L., Xu D., Qiang Z. Background subtraction using local SVD binary pattern. IEEE Comput Soc Conf. 2016:1159–1167. doi: 10.1109/Cvprw.2016.148. [DOI] [Google Scholar]
- 29.Bradski G. The OpenCV library. Dr. Dobb's J. 2000;120 122–25. [Google Scholar]
- 30.Hu Y., Wang C., Yang L., Pan G., Liu H., Yu G., Ye B. A neural basis for categorizing sensory stimuli to enhance decision accuracy. Curr. Biol. 2020;30:4896–4909.e6. doi: 10.1016/j.cub.2020.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang R.Y., Zhong L., Xu Y., Johnson T., Zhang F., Deisseroth K., Tracey W.D. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr. Biol. 2007;17:2105–2116. doi: 10.1016/j.cub.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tracey W.D., Wilson R.I., Laurent G., Benzer S. painless, a Drosophila gene essential for nociception. Cell. 2003;113:261–273. doi: 10.1016/S0092-8674(03)00272-1. [DOI] [PubMed] [Google Scholar]
- 33.Lahiri S., Shen K., Klein M., Tang A., Kane E., Gershow M., Garrity P., Samuel A.D.T. Two alternating motor programs drive navigation in Drosophila larva. PLoS One. 2011;6:e23180. doi: 10.1371/journal.pone.0023180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jovanic T., Schneider-Mizell C.M., Shao M., Masson J.B., Denisov G., Fetter R.D., Mensh B.D., Truman J.W., Cardona A., Zlatic M. Competitive disinhibition mediates behavioral choice and sequences in Drosophila. Cell. 2016;167:858–870.e19. doi: 10.1016/j.cell.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W., Yan Z., Jan L.Y., Jan Y.N. Sound response mediated by the TRP channels NOMPC, NANCHUNG, and INACTIVE in chordotonal organs of Drosophila larvae. Proc. Natl. Acad. Sci. USA. 2013;110:13612–13617. doi: 10.1073/pnas.1312477110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrario C.R., Gorny G., Crombag H.S., Li Y., Kolb B., Robinson T.E. Neural and behavioral plasticity associated with the transition from controlled to escalated cocaine use. Biol. Psychiatry. 2005;58:751–759. doi: 10.1016/j.biopsych.2005.04.046. [DOI] [PubMed] [Google Scholar]
- 37.Ellinwood E.H., Jr., Balster R.L. Rating the behavioral effects of amphetamine. Eur. J. Pharmacol. 1974;28:35–41. doi: 10.1016/0014-2999(74)90109-5. [DOI] [PubMed] [Google Scholar]
- 38.Deng J., Dong W., Socher R., Li L.J., Li K., Li F.F. Cvpr: 2009 IEEE Conference on Computer Vision and Pattern Recognition. 1–4. 2009. ImageNet: a large-scale hierarchical image database; pp. 248–255. [DOI] [Google Scholar]
- 39.Yosinski J., Clune J., Bengio Y., Lipson H. How transferable are features in deep neural networks. Adv. Neural. Inf. Process. Syst. 2014;27 [Google Scholar]
- 40.Carreira J., Noland E., Hillier C., Zisserman A. A short note on the kinetics-700 human action dataset. arXiv. 2019 doi: 10.48550/arXiv.1907.06987. Preprint at. [DOI] [Google Scholar]
- 41.He K., Gkioxari G., Dollár P., Girshick R. 2017. Mask R-Cnn; pp. 2961–2969. [DOI] [PubMed] [Google Scholar]
- 42.Van Rossum G., Drake F.L. 2000. Python Reference Manual (iUniverse Indiana) [Google Scholar]
- 43.Harris C.R., Millman K.J., van der Walt S.J., Gommers R., Virtanen P., Cournapeau D., Wieser E., Taylor J., Berg S., Smith N.J., et al. Array programming with NumPy. Nature. 2020;585:357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 46.van der Walt S., Schönberger J.L., Nunez-Iglesias J., Boulogne F., Warner J.D., Yager N., Gouillart E., Yu T., scikit-image contributors scikit-image: image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. doi: 10.1109/Mcse.2007.55. [DOI] [Google Scholar]
- 48.McKinney W. 2010. Data Structures for Statistical Computing in python; pp. 51–56. [Google Scholar]
- 49.Waskom M., Botvinnik O., O'Kane D., Hobson P., Lukauskas S., Gemperline D.C., Augspurger T., Halchenko Y., Cole J.B., Warmenhoven J. 2017. Mwaskom/Seaborn: V0. 8.1. [Google Scholar]
- 50.Abadi M., Agarwal A., Barham P., Brevdo E., Chen Z., Citro C., Corrado G.S., Davis A., Dean J., Devin M. Tensorflow: large-scale machine learning on heterogeneous distributed systems. arXiv. 2016 doi: 10.48550/arXiv.1603.04467. Preprint at. [DOI] [Google Scholar]
- 51.Rappin N., Dunn R. Manning Publications); 2006. wxPython in Action. [Google Scholar]
- 52.Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 53.Simonyan K., Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv. 2014 doi: 10.48550/arXiv.1409.1556. Preprint at. [DOI] [Google Scholar]
- 54.He K., Zhang X., Ren S., Sun J. IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [DOI] [Google Scholar]
- 55.Ruder S. An overview of gradient descent optimization algorithms. arXiv. 2016 doi: 10.48550/arXiv.1609.04747. Preprint at. [DOI] [Google Scholar]
- 56.Hu Y., Han Y., Wang X., Xue L. Aging-related neurodegeneration eliminates male courtship choice in Drosophila. Neurobiol. Aging. 2014;35:2174–2178. doi: 10.1016/j.neurobiolaging.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 57.Robinson M.J.F., Burghardt P.R., Patterson C.M., Nobile C.W., Akil H., Watson S.J., Berridge K.C., Ferrario C.R. Individual differences in cue-induced motivation and striatal systems in rats susceptible to diet-induced obesity. Neuropsychopharmacology. 2015;40:2113–2123. doi: 10.1038/npp.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferrario C.R., Robinson T.E. Amphetamine pretreatment accelerates the subsequent escalation of cocaine self-administration behavior. Eur. Neuropsychopharmacol. 2007;17:352–357. doi: 10.1016/j.euroneuro.2006.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All the data reported in this paper will be shared by the lead contact upon request. The behavioral datasets generated and analyzed in the current study is deposited for free access (https://drive.google.com/drive/folders/1OISUkm8An4isLFzBtj7BilI2Jp-J6jEU?usp=sharing).
-
•
All original code has been deposited at Zenodo and GitHub and is publicly available. The DOI and GitHub website are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.