Summary
Drug-induced hepatotoxicity is a leading cause of drug withdrawal from the market. High-throughput screening utilizing in vitro liver models is critical for early-stage liver toxicity testing. Traditionally, monolayer human hepatocytes or immortalized liver cell lines (e.g., HepG2, HepaRG) have been used to test compound liver toxicity. However, monolayer-cultured liver cells sometimes lack the metabolic competence to mimic the in vivo condition and are therefore largely appropriate for short-term toxicological testing. They may not, however, be adequate for identifying chronic and recurring liver damage caused by drugs. Recently, several three-dimensional (3D) liver models have been developed. These 3D liver models better recapitulate normal liver function and metabolic capacity. This review describes the current development of 3D liver models that can be used to test drugs/chemicals for their pharmacologic and toxicologic effects, as well as the advantages and limitations of using these 3D liver models for high-throughput screening.
Keywords: high-throughput screening, 3D liver models, in vitro assays, liver organoids, liver spheroids, drug toxicity
In this review article, Yang et al. discuss the current status, challenges, and future promise of three-dimensional liver models for drug development and safety testing. They highlight the implications of cell types from different sources and several 3D culturing approaches for physiologically relevant modeling and high-throughput chemical testing.
Introduction
Approximately one-third of clinically used drugs were removed from the market due to liver damage, indicating that the liver is one of the most frequently adversely affected organs.1 Currently, the US Food and Drug Administration generally requires preclinical testing of any new drug or biological therapeutic “for pharmacologic activity and acute toxicity in animals” prior to initiating human clinical trials, which includes hepatotoxicity testing (https://www.fda.gov/drugs/types-applications/investigational-new-drug-ind-application). Notably, animal testing has been widely used in pharmaceutical and industrial research to predict human toxicity. However, around half of drug candidates induce hepatotoxicity in humans but not in animal models.2,3,4 In fact, many animal models often fail to predict human toxicity, as some toxic effects have been reported in human trials.5,6 For instance, fialuridine, which was investigated as a potential therapy for hepatitis B virus infection, induces a severe toxic reaction characterized by hepatic failure in humans. However, available animal data from mice, rats, dogs, and monkeys showed no indication that the drug would cause liver failure.7 Recently, the US Environmental Protection Agency announced plans to end animal models for chemical and pesticide testing by 2035 (https://www.epa.gov/research/administrator-memo-prioritizing-efforts-reduce-animal-testing-september-10-2019). Therefore, there is an urgent need to develop physiologically relevant human-derived liver models for the hepatotoxic screening of large and continuously expanding chemical and drug libraries.
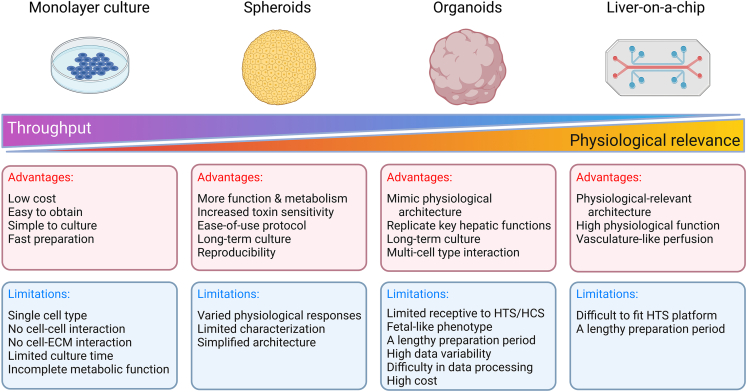
To date, chemical testing has been conducted using primary human hepatocytes (PHHs), liver cancer cell lines, immortalized hepatic cell lines, and stem cell-derived hepatocyte-like cells (HLCs).8 These cells commonly exhibit phenotypic differences from their in vivo counterparts when they are grown in a two-dimensional (2D) monolayer condition, resulting in misinterpretations of pharmacological or toxicological results.9,10,11 Numerous research findings indicate that growing cells in three dimensions (3D) can better recapitulate native physiological conditions than growing the same cell type in 2D. With the goal of recapitulating liver function and metabolic capability, a hepatic model should ideally be performed in 3D culture12,13,14 (Figure 1). Previous studies have demonstrated that 3D liver models, compared to 2D cultures, display higher sensitivity for detecting hepatotoxic chemicals, such as diclofenac and trovafloxacin.15,16,17,18 In addition, in 2D culture, PHHs retain but rapidly degrade hepatocyte function,19 and hepatocellular carcinoma-derived cell lines (e.g., HepG2) do not adequately replicate hepatocyte function.20 The use of 2D cell line systems would mislead one into believing that drug-induced liver injury (DILI) is caused by the compound directly affecting the tested cell while ignoring the effect the compound may have via spatial hepatocyte-hepatocyte, hepatocyte-non-parenchymal cell, or cell-extracellular matrix (ECM) interactions, which occur between either the same cell types or across different cell types. Thus, employing 3D liver models may be a better choice for testing compound toxicity.
Figure 1.
Summary of the advantages and limitations of in vitro liver cellular models for high-throughput screening
Currently, spheroids, organoids, and perfusion-based 3D approaches (e.g., liver-on-a-chip) are the major examples of 3D liver models.8 To capture in vivo conditions, hepatic spheroids and organoids benefit by providing spatial interaction between hepatocytes and either parenchymal cells or non-parenchymal cells or ECMs that aid in hepatocyte maturation.21 Therefore, they may be great platforms for a range of fundamental and translational research applications, including disease modeling, drug screening, gene therapy, revealing microbe-host interactions, and even organ replacement.22 Considering the publication of several comprehensive reviews on 3D liver models,1,2,8,15,22,23,24,25,26,27,28,29,30,31 this review focuses on the current state of research and implementation of human 3D hepatic liver models in drug metabolism and toxicology screening.
Hepatic spheroids
Spheroids are collections of cells that often form the shape of a solid spheroidal structure.8 They may comprise one or more cell types and are capable of mimicking specific functional properties of an organ. Compared with 2D cultures, multicellular spheroids exploit the ability of cells to self-assemble and maintain viability in culture for an extended period of time while maintaining a more hepatocyte-like functional phenotype.32 In addition, some compound metabolites may produce hepatotoxicity in other types of cells in the liver. For example, flucloxacillin can be converted by cytochrome P450 3A4 (CYP3A4) to 5-hydroxymethyl metabolites that are selectively hazardous to biliary epithelial cells but not hepatocytes.33 Spheroids can be assembled from multiple cell types; however, as complexity rises and different cell types contribute to the phenotype or parameter of interest, data processing and interpretation may become increasingly difficult. By contrast, monocellular cultured spheroids enable us to examine the function of single cell types without separating them into their constituent parts, allowing more straightforward metabolic analysis.8
Over the decades, several strategies for spheroid formation have been developed, allowing researchers to utilize spheroids for high-throughput screening. As reviewed in detail by Zhang et al.,34 these strategies can be categorized into scaffold-based and non-scaffold-based systems. Among them, hepatocyte spheroids formed on ultra-low attachment (ULA) plates and hanging drop plates appear to be the most frequently used methods for forming liver spheroids, whereas other 3D liver models (e.g., magnetic beads, scaffolds, and 3D bioprinting) are awaiting additional validation using known hepatotoxic and non-hepatotoxic drugs. The ULA plate is a ready-to-use microplate with a hydrophilic surface that has been neutrally charged. It is used to produce high-quality spheroids and is substantially faster than alternative procedures, such as the agarose method, generally taking only 5–7 days, depending on the cell line.34 While the ULA method utilizes a round and smooth bottom to avoid cell adhesion, the hanging drop method creates spheroids via the culture medium’s air-liquid interface and cellular gravity. Using the hanging drop method, Kelm et al. developed the first liver spheroid utilizing HepG2 cells.35 To further optimize this process, InSphero’s GravityPLUS plates allow a drop of cell suspension (50 μL) to aggregate in the air-media interface at the bottom of the well, resulting in the development of compact spheroids with a diameter variation of less than 5%.36 The spheroids can then be transferred to GravityTRAP plates, which are compatible with high-content imaging equipment and thus enable high-throughput imaging analysis. Following the development of these approaches, high-throughput hepatotoxicological screening has been performed using a variety of liver-related spheroids. The following sections discuss in more detail the characteristics of spheroid models derived from different types of cellular sources.
HepG2 spheroids
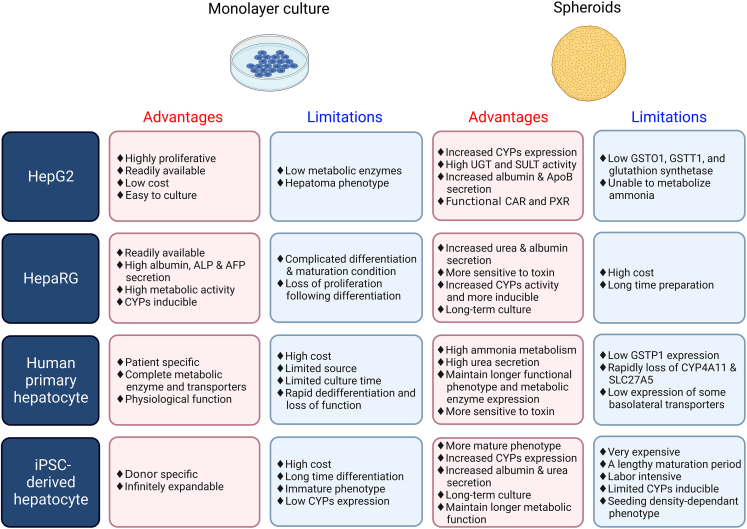
HepG2 is an immortalized human liver cancer cell line obtained from the liver of a 15-year-old Caucasian male with advanced hepatocellular carcinoma.37 Due to its low cost and ease of use, the HepG2 cell line is ideal for high-throughput settings and is commonly employed in preclinical hepatotoxicity screening.38 As a polarized hepatocyte cell line, HepG2 cells exhibit some key characteristics of hepatocytes, including the release of albumin, transferrin, and other plasma proteins. Unfortunately, HepG2 cells are deficient in some liver functions, such as the bulk of CYP450 and phase II metabolic enzyme activity.39 HepG2 spheroids, initially grown in agarose-coated dishes in the early 2000s, have been used in a range of toxicological studies.35 Proteomic analysis revealed that, when compared with monolayer culture, the spheroid model significantly increased the expression of typical hepatic functional markers such as apolipoprotein A-I/II, α-2-HS-glycoprotein, and serum albumin, indicating that HepG2 spheroids outperform their monolayer counterparts in emulating native liver protein expression40 (Figure 2). In addition, within 21 days, HepG2 spheroids produced urea and expressed active CYP450 metabolic enzymes.12 Furthermore, HepG2 cells grown in spheroids preferentially mature and differentiate, whereas cells grown in monolayers undergo consistent extracellular stress (i.e., trypsinization), resulting in an atypical proliferative phenotype.41
Figure 2.
Summary of the advantages and limitations of commonly used cell lines in 2D vs. 3D
Since HepG2 spheroids are easy to generate in the multi-well assay plate, the HepG2 spheroid model was quickly adapted and used in high-throughput toxicity screening.42,43,44 Ramaiahgari et al. devised a hydrogel-based 3D culture platform for hepatotoxic studies, seeding a thousand HepG2 cells per well in a 384-well plate.12 After evaluating a dozen liver toxicants, they observed that spheroids have significantly increased sensitivity to most hepatotoxic substances as compared with 2D culture during repeated compound exposures (21 days). Furthermore, this research group expanded this approach by developing a HepG2 cell line expressing a green fluorescent protein (GFP)-based cellular stress response reporter as a source to generate liver-like spheroids. They then used this spheroid system to evaluate a panel of 33 drugs known to cause DILI.45 By using high-throughput imaging to quantify GFP reporter activity, Hiemstra et al. demonstrated enhanced identification of DILI liability in spheroids following repeated dose treatment when compared with monolayers. In addition, Basharat et al. evaluated the applicability of high-throughput 3D liver spheroid models of HepG2 (C3A) and HepaRG cell lines for the prediction of DILI during early drug development.46 In their study, spheroids were treated with multiple concentrations of each drug in 384-well ULA plates using direct sonic droplet ejection. Using 150 known DILI compounds, they showed that the HepG2 spheroid model could identify 58% DILI-positive compounds and the HepaRG spheroid model could identify 47% DILI drugs in an ATP-based cell viability assay, although both spheroid models had comparable overall accuracy and specificity for DILI prediction.
In general, current screening with HepG2 spheroids is often used to predict long-term hepatotoxicity via repeated dosing. 3D culturing enables these cells to show more physiological function and metabolic capability, as well as the ability to sustain and proliferate for at least 28 days. However, the HepG2 cell line has its limitations when predicting the potential DILI effect of anti-tumor drugs, as this cell line was derived from liver cancer cells, which are sensitive to most anti-proliferative agents. Furthermore, the HepG2 cell line lacks most of the liver’s metabolic capability, resulting in a poor translation of toxicity data from the cell model to the clinical setting. Indeed, HepG2 frequently displays chromosomal abnormalities and oncogenic alterations that improve survival, such as the CYP2C9 point mutation,47,48 and may be a better model for cancer than DILI. Therefore, there is an urgent need to develop more phenotypically suitable in vitro cellular models that can aid in the early detection of compound hepatotoxicity.
Primary human hepatocyte spheroids
PHHs and liver tissue have been the “gold standard” for studying DILI and human-specific toxicities due to their expression of hepatocyte-specific genes following isolation.24 However, PHHs are occasionally scarce, require invasive methods to produce, and often undergo fast dedifferentiation in vitro.15,49 Because of the rapid phenotypic loss of PHHs in these settings, the PHH model is only applicable for acute toxicity assessment when cultured in a monolayer.50 Three-dimensional spheroid models using PHHs and non-parenchymal cells, such as Kupffer (liver resident macrophages) and stellate cells, enable a more physiologically relevant environment. When compared with 2D PHHs, spheroid models retain adequate metabolic activity and have improved sensitivity and specificity for identifying human hepatotoxicants.51 Noticeably, PHH spheroids can form bile canaliculi-like networks and have better drug absorption, distribution, metabolism, and excretion than 2D monolayers.52 They are capable of metabolizing parent compounds to their metabolites, showing the presence of a group of CYP450 enzymes, UDP-glucuronyl transferases, and sulfotransferases. Due to their metabolic capacities, PHH spheroids have been found to be an attractive model for researching xenobiotic metabolism and discovering human-specific metabolites53 (Figure 2).
In comparison to the technique for generating liver organoids (mentioned in the next section), the procedure for generating PHH spheroids is substantially simpler and faster. Bell et al. created PHH aggregates in 7 days using 1,500 cells per well in a 96-well plate to form spheroids with a diameter of 200 μm.52 Using more cells to generate a bigger spheroid may induce intra-spheroid necrosis, leading to a higher background in cell death or viability assays. This time-saving technique permits high-throughput screening using PHH spheroids during the early stages of drug development. In addition, after 5 weeks of culture, PHH spheroids keep their morphology, vitality, and hepatocyte-specific activities while remaining phenotypically stable. These characteristics enable the model to be used to forecast the long-term hepatotoxicity of drugs or chemicals. In Bell et al.’s study, PHH spheroids were treated every other day with each of the five classic hepatotoxicants (amiodarone, bosentan, diclofenac, fialuridine, and tolcapone), and cell viability was assessed on day 2, 8, and 28 separately using ATP content measurements. The half-maximal inhibitory concentration (IC50) of these compounds in the spheroid’s viability assay reached a clinically relevant concentration on day 28, closely reproducing the delayed onset of DILI in vivo.52 The delayed onset of DILI was particularly observed in fialuridine, for which cytotoxicity was identified only with long-term dosage (IC50 > 100 μM at 48 h vs. 0.1 μM at 28 days). Other studies have found that PHH spheroids are an excellent tool to detect cholestatic ability54 and monitor CYP450 metabolism activity.55 In their study, Vorrink et al. employed the 96-well plate-based 3D model to screen 123 drugs with or without direct clinical implications for DILI.56 Using ATP-based viability assay as the sole endpoint, this model accurately distinguished hepatotoxic from nontoxic structural analogues and outperformed all previously published in vitro assays in terms of sensitivity and specificity at significantly lower exposure levels, detecting 69% of all hepatotoxic compounds without producing any false positive results (100% specificity).
Schofield et al. effectively miniaturized and industrialized PHH spheroids into 384-well ULA plates, again utilizing 1,500 cells per well, and compared them with 2D HepG2 cells in a screening of 199 chemicals classified by the US Food and Drug Administration as most-, less-, or no-DILI-concern.38 Notably, 15 new DILI-related chemicals were identified in PHH spheroids through a viability assay. Correspondingly, PHH spheroids testing has largely supplanted the HepG2 monolayer cell viability assay and is being actively used as a part of their early screening approach. To develop a more liver-like 3D model, Proctor et al. co-cultured PHHs and non-parenchymal cells to create spheroid human liver microtissues (hLiMTs) in a 384-well plate using the hanging drop method.57 Using this model, they conducted a 14-day toxicity screening of 110 drugs, including some clinical drugs with DILI-concern. Compared with the results of 2D PHHs, the IC50 values from the hLiMT model, which were attained by an ATP-based cell viability assay, were correlated well with the human plasma concentration (Cmax), allowing for the construction of a “margin of safety” for clinical application. Since the InSphero multiple donor hLiMT model has a more diversified genetic background, it can help reduce bias in models caused by natural variances in human drug-metabolizing enzymes. This study therefore illustrated that the 3D spheroid models enable more precise risk assessment and prediction of drug-induced hepatotoxicity.
New spheroid generation techniques are also currently being developed and applied for toxicology studies. For example, the magnetic beads technique allows for rapid PHH spheroid generation in a 384-well plate (10,000 cells/well) in just 2 days with little functional difference from monolayer PHHs.58 Magnetic spheroids have also been successfully used in an in vitro assay for high-throughput toxicity screening, even though many of their characteristics still need to be determined.59 Similarly, bioprinting has enabled novel strategies to develop liver models and spheroids. Inkjet-based bioprinting, extrusion-based bioprinting, and photocuring-based bioprinting uniquely provide high-fidelity control over both the 3D microstructure and microenvironment of the construct.60 Bioprinting has already been used to successfully develop in vitro 3D liver models consisting of PHHs and non-parenchymal cells (endothelial and stellate cells) embedded in a NovoGel 2.0 hydrogel.61 The 3D liver construct could detect the cytotoxicity of trovafloxacin, a drug whose hepatotoxic potential was not previously identified in 2D models. Nevertheless, bioprinting in high-throughput settings has not been fully achieved as significant barriers, such as the speed of printing, print size, and cell viability, limit the potential usage of bioprinting.
Although the methods for creating spheroids in 1,536-well plates for high-throughput screening have been established,62 the use of PHH spheroids in such large-scale toxicity screening remains to be explored. Apart from the prohibitively high cost and scarcity of PHH, another barrier to employing PHH spheroids on 1,536-well plates is the requirement for constant medium change for long-term drug toxicity studies, which easily causes well-to-well variation. Thus, an adequate approach for changing the medium and repetitive chemical addition is required for the chronic DILI test. Furthermore, because donor-to-donor heterogeneity in PHHs has been demonstrated in in vitro studies,52 hepatocytes from different donors are more appropriate for assessing individual hepatotoxicity risk. Therefore, a homogeneous immortalized liver cell line with equivalent drug metabolic capacities, like differentiated HepaRG cells, will be a better model for toxicological testing.
HepaRG spheroids
To circumvent the disadvantages of PHHs, the HepaRG cell line was recommended as a surrogate cell type for PHHs to establish a 3D liver model for hepatotoxicity testing. Due to their elevated expression of key drug-metabolizing enzymes and transporters (DMETs), HepaRG cells, which are generated from human hepatocellular carcinoma, have emerged as a viable alternative to PHHs.10,63,64 These bipotent progenitor cells are expanded and differentiated into “differentiated” co-cultures of hepatocyte- and cholangiocyte-like cells in 2D culture configurations over a 4-week period.65,66 Recent efforts to cryopreserve “completely differentiated” HepaRG cells have given researchers global access to cells capable of recovering and maintaining drug metabolism and hepatocyte function when cultured in differentiation media.67,68 This is seen clearly through HepaRG’s CYP450 expression and activity, which is comparable to newly isolated PHHs following induction10 (Figure 2). Interestingly, HepaRG monolayer cultures, when compared with HepG2 3D cultures, display superior expression of DMETs.
Several HepaRG 3D models have been reported in recent years and they replicate in vivo-like microenvironments more closely. When compared with HepaRG 2D cultures, these models exhibit improved hepatocyte differentiation, lifespan, and functionality.57,69,70,71 For compound toxicity investigations, HepaRG spheroids were applied in 96- and 384-well ULA plates, which enable spheroid formation in a single step by seeding 1,000 cells per well, resulting in optimal CYP450 enzymatic activity.13 The shape and function of the spheroids can be maintained for up to 28 days. Interestingly, a recent study showed that HepaRG spheroids were slightly less sensitive to hepatotoxic compounds than HepG2 spheroids in a 384-well high-throughput format.46 This could be because HepG2 cells were more susceptible to cell proliferation-inhibiting chemicals than HepaRG cells, which were in a more differentiated condition. Meanwhile, Ott et al. used a unique micromold plate invented by Likarda to rapidly fabricate homogeneous spheroids with a very small diameter.72 Following that, spheroids were transferred to 384-well plates at a density of around 50–70 spheroids per well and incubated for 7 days with a panel of DILI drugs and CYP450 inducers. In comparison to HepaRG 2D culture, the HepaRG 3D model demonstrated greater sensitivity to liver toxins (70% vs. 60%) but retains the same predictability in response to CYP450 inducers.
Although HepaRG spheroids were less responsive to DILI-liability compounds than HepG2 spheroids, HepaRG spheroids appear to be a promising technique for preserving primary hepatic features because of their comparable metabolic enzyme expression levels. Unfortunately, due to the scarcity of published reports on HepaRG spheroids used in screening, it is difficult to estimate the predictability and efficacy of using HepaRG spheroids as a model for assessing other liver toxic parameters in a high-throughput screening format, such as CYP450 drug-metabolizing enzyme activity, mitochondrial state, and cholestatic features. In summary, HepaRG cells may be a more reliable test system than HepG2 cells, notably in terms of expression of DMETs, and may be closer to the physiological relevance of PHHs than other liver cancer cell lines.
iPSC-derived hepatocyte spheroids
Induced pluripotent stem cell (iPSC)-derived hepatocytes (iHeps) have been used to establish 3D liver cell models for toxicity assessment. Established in 2010 by Duncan et al., iHeps provide an alternative to donor livers for primary hepatocyte studies.73 When cultured in 3D conditions, iHeps have improved hepatic properties, although they keep an immature phenotype evident by the expression of fewer mature marker genes17,74,75 (Figure 2).
Many studies have tried to characterize iHeps as suitable 3D models for hepatotoxic screening. Recently, a study compared iHeps with PHHs in 2D and 3D culture conditions. They found that although CYP450 family enzymes have similar basal activities in all culture systems, they can be successfully stimulated only in PHHs.76 In addition, Sirenko et al. reported a comparable outcome between 2D and 3D iHeps in their experiments employing high-content image analysis.77 Interestingly, one study discovered that iHep spheroids were more sensitive to a panel of 24 hepatotoxic substances than HepG2 spheroids,17 while another discovered the opposite for 10 of the 23 chemicals tested.77 When compared with PHH spheroids, iHep spheroids had comparable IC50 values for 12 of 15 hepatotoxic drugs evaluated.78 Another study assessed the sensitivity of seven hepatotoxic substances and observed that PHHs were often more sensitive than the other models.76 These controversial facts, in conjunction with both the dearth of studies on iHep spheroid cultures for toxicity assessment and iHeps’ premature phenotype, make iHeps an unpredictable and inconsistent model for toxicity assessment.
While hepatic differentiation of iPSCs may be a viable technique for in vitro liver modeling, until a comprehensive review of the predictability of iHep spheroids is done, the utility of iHep spheroids for assessing the safety of medications will remain disputed.
Stem cell-derived liver organoid platforms
Liver organoids
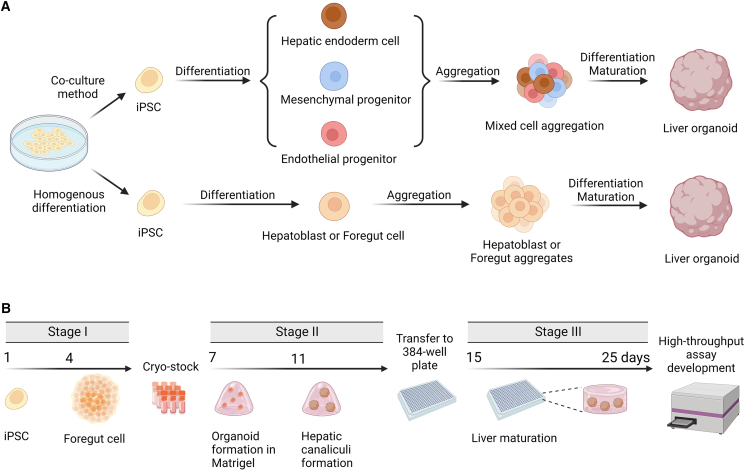
Previously, the term “organoid” referred to a variety of 3D culture systems that exhibit varying degrees of organotypic resemblance to the modeled organ, including distinctly organotypic models derived from stem cells and aggregates of single or multiple terminally differentiated cell types.28,29 In this review, we used Harrison et al.’s definition of “organoid” as an in vitro 3D cellular cluster that undergoes a developmental process autonomously, resulting in organized heterogeneity and complexity, or, more succinctly, self-organized 3D cultures derived from stem cells28 (Figure 3A). Hepatocytes and supportive cells such as Kupffer cells, hepatic stellate cells, cholangiocytes, and endothelial cells, as well as other cell types that mimic hepatocyte functions and structure, can create liver organoids.79,80,81,82 These organoids, which are traditionally derived from iPSCs, are often interchangeably referred to as hepatic organoids, hepatobiliary organoids, or human liver organoids (HLOs). In addition, numerous laboratories have produced and exploited a novel class of organoids to study biliary duct physiology and cholangiopathies.83,84,85 These cholangiocyte organoids exhibit cholangiocyte traits and functions.
Figure 3.
Current strategies for the generation of iPSC-derived liver organoids
(A) The liver organoids were formed by culture of iPSC-differentiated cells of various cell types or homogeneous cell populations that are capable of building cellular constructs with structural and physiological complexity through differentiation.
(B) Schematic representation of the liver organoids generation procedure in the Shinozawa et al.’s study for the 384-well plate assay development.86
The liver organoids consist of a spherical monolayer of epithelium with a hollow-like structure and a wide lumen, mimicking a bile canaliculus.86 This shape is comparable to that of primary hepatic tissue, which has cords of cells with canaliculi resembling chicken wire. Single-cell analysis revealed a 60%–75% parenchymal cell population (hepatocytes) and a 25%–40% non-parenchymal cell population in HLOs,86,87,88 which included Kupffer cells, hepatic stellate cells, cholangiocytes, endothelial cells, and other cell types in the same ratio as in the normal liver.89 Cells in the center of the organoid are exposed to fewer stimuli, imitating the natural liver milieu for chemical processing in vivo.30 HLOs, which are generated from iPSCs, continue to display an immature transcriptome profile compared with adult liver tissue. Nevertheless, the HLCs of HLOs expressed and activated CYP3A4, indicating their potential function in drug metabolism and toxicological screening.87 Intriguingly, a unique characteristic of the fetal liver is that it has a functional drug-metabolizing enzyme system even during the early gestational period, which is remarkably specific to humans but not to other animal species. Accordingly, these iPSC-derived HLOs can be leveraged in drug screening for adverse reaction prediction not only in adults but also in fetuses.90 Thus, this more biologically organotypic in vitro model will be appropriate for drug metabolism and toxicological assessment.
In addition, these organoid-based pathological liver models are effective tools for simulating numerous elements of the in vivo phenotype, particularly at the individual patient level, and can be used to evaluate the bioactivity of compounds for toxicological and preclinical testing. Moreover, organoids produced from stem cells offer advantages in terms of scalability, expansion of cells of a single genotype, and indefinite self-renewal. Utilizing these iPSC-derived organoids in the future will make personalized medicine possible for drug selection, avoiding any potential detrimental effects on the liver of patients.
Whole organoid approaches
Although research on liver organoids has steadily accelerated since 2013, iPSCs utilized to make an organ-like in vitro liver model have not been optimized for a high-throughput screening platform.91 Numerous constraints limit the use of HLOs in high-throughput screening due to their long preparation time, high cost, poor yield, unpredictable inter- and/or intra-batch variability, and a lack of acceptable quantification methods for data processing. Despite the development of protocols for the generation of liver organoids suitable for 96-well or even 384-well formats,92,93 individual organoid variation continues to pose a barrier to the model’s application in the screening field. For example, some organoids contain more stellate-like and macrophage-like cells, while others contain more mesenchymal cells. Cellular variability prevents consistent and reproducible data, an obvious obstacle for high-throughput screening. Even still, Shinozawa et al. successfully extended the use of human HLOs derived from iPSCs to high-throughput compound screening.86 Their groundbreaking study indicates for the first time that HLOs may be miniaturized on a 384-well plate format by improving the differentiation technique to yield HLOs with a more uniform size distribution.86 In their methodology, the HLOs were generated from cryopreserved foregut cells derived from iPSCs, which can decrease differentiation time by approximately a week and allow for the storage of large numbers of foregut cells to boost HLO production86 (Figure 3B). This strategy minimized HLO preparation variance due to batch-to-batch differentiation during comprehensive screening.
Shinozawa et al. treated HLOs with 238 market drugs at multiple concentrations and determined their activities at multiple readouts. They assessed cholestasis, viability, and mitochondrial toxicity, which are three distinct variables used for predicting drug-induced liver toxicity. The sensitivity and specificity for their HLO-based high-throughput toxicity screening (LoT) system, which combined the consideration of the viability assay and the cholyl-lysyl-fluorescein (CLF) assay, were 88.7% and 88.9%, respectively.86,94 The detection methods for both cholestasis and mitochondrial toxicity are imaging-based assays that determine the stained area and intensity of each organoid. The cell viability was determined using a plate reading assay based on luminescence that quantifies the total intracellular ATP content. While employing a uniform organoid can reduce well-to-well variance, their viability data (the whole-well reading signal) needs to be normalized by the total cellular area per well, which is estimated from organoid imaging data. As a result, a high-speed and high-quality image acquisition and processing workflow has been established to obtain high-quality screening data from this complicated in vitro model. This, however, revealed a potential limitation: the variation caused by the non-uniform size of liver organoids affects the magnitude of whole-well luminescent or fluorescent signals. To overcome this hurdle, additional efforts will be required to refine the organoid differentiation procedure and develop an organoid-specific high-throughput testing method.
Dispersed organoid approaches
Zhang et al. established an alternate method for measuring organoid whole-well luminescent or fluorescent signals that decreases well-to-well variation. They dispersed the HLOs using 0.25% trypsin digestion and then plated the cell suspension in a 384-well plate as a monolayer culture to eliminate assay variation caused by using individual liver organoids.88 Their investigation demonstrated that the dispersed HLO technique kept liver cell-specific markers, albumin synthesis, and CYP450 expression 7 days after seeding, suggesting that they retained hepatocyte-like physiological capabilities in the scattered condition. While functional markers of hepatocytes were apparent, the dispersed HLOs lacked spatial cell-cell contact between different cell types, including any type of interaction between liver cells, and consequently failed to depict the liver’s natural milieu since all cells had access to the same stimuli.30 This might result in a failure to detect a drug’s potential toxicity, which requires bioactivation by the collaboration of different liver cell types. In comparison to the co-culture paradigm with 2D liver cells, these dispersed HLO cells still represented liver function more accurately due to their self-differentiation and self-organization. However, the application of this dispersed method is dependent on the detection methods that can be used in the screening field.
Zhang et al. tested 12 liver toxicants at 10 concentrations ranging from 5 nM to 100 μM by employing 3,500 distributed HLO cells in each well of a 384-well plate.88 The viability experiment demonstrated a concentration-response curve specific for hepatocytes with a steady signal throughout the replication wells. Indeed, dispersed cells are more manageable than entire organoids. However, it is unknown if scattered HLO cells can produce a stable readout in other liver functional assays, such as the CYP450 enzyme activity assay, which is used to determine metabolized drug toxicity. In addition, image analysis enabled researchers to identify the morphological profile of the cell populations, meaning that imaging analysis may help to normalize data for this complex in vitro model. Notably, monitoring the population of liver cells and the morphological characteristics of specific cell types may also be used to determine the potential toxicity of drugs and environmental substances processed by liver cells.
Discussion and perspective
In vitro liver models are useful tools for detecting drug/chemical toxicity and can be used to study the mode of action and underlying mechanism. These models have traditionally entailed monolayer cultures of PHHs or immortalized cell lines. However, PHHs rapidly dedifferentiate in vitro, eliminating metabolic competence. Meanwhile, immortalized cells have distinct phenotypic and behavioral traits when compared with in vivo hepatocytes. These limitations complicate their ability to examine metabolic-dependent toxicities and their utility for finding DILI-relevant drugs, particularly during chronic and repeated dose time courses that more closely mimic in vivo DILI.
Toxicity metrics in 3D models
In toxicological testing and drug safety evaluation, the safety margin (IC50/Cmax) is commonly employed as a measure to determine the safety of drugs, with higher values indicating greater safety. Due to the issues connected with the safety margin, however, some toxicologists prefer to use the IC50 value and Cmax directly.95 In the early stages of drug development, the safety of a new chemical is often evaluated by its IC50 value generated from each toxicological test. In the absence of clinical exposure data, the higher the IC50 in toxicological testing, the safer the drug is likely to be in animal testing or clinical trials. Notably, current data indicate that the more complicated in vitro liver models will reflect a greater number of potential toxicology issues, even though most liver toxicology issues will be revealed in assessments of both 2D and 3D models and the IC50 value is comparable. Specifically, the delayed onset of DILI was found in fialuridine on a 3D model with long-term dosing (IC50 > 100 μM at 48 h vs. 0.1 μM at 28 days), but not in 2D testing or animal testing. In addition, 3D cell culture typically outperforms monolayer cell culture in some idiosyncratic DILI estimations that usually require the collaboration of multiple types of liver cells, particularly liver organoids, because the liver organoids system contains several types of liver cells with the same genotype. In addition, the 3D liver model used in the toxicity study was in its early stages, and the methodologies were still in the process of being refined. More information is required to validate the relationship between the IC50 generated from these 3D models in toxicological testing and clinical exposure data. Assessment of drug toxicity in the clinical setting by only using the IC50 remained a difficult task, both experimentally and theoretically.
Key advantages of 3D approaches
There is accumulating evidence that when cells are cultured in 3D conditions, their phenotypic behavior becomes more analogous to that found in in vivo. These beneficial effects could be attributed to the formation of significant cell-to-cell interactions, which have been shown to alter cell signaling and fate.96,97 The ability to sustain 3D cultures for an extended period may also contribute to the formation of a more mature phenotype, as certain cell types (i.e., hepatocytes and cholangiocytes) express metabolic enzymes more abundantly in 3D than in 2D cultures.12,98 This allows researchers to examine and detect chronic drug-induced toxicities through repeated dosing regimens.99 Indeed, numerous toxicological studies utilizing a 3D liver cell model have focused on chronic DILI assessment and the effect of long-term repeated dosing tests,12,45,52 demonstrating the utility of the 3D liver system in the field of long-term drug toxicological assessment. The data generated from liver organoid models is, however, less reproducible compared with the data from 2D cell cultures due to variability in organoid sizes, surface areas and densities, as well as batch-to-batch variation.40 In addition, the current generation of long-term toxicity testing is limited to 384-well plates, even though the technique for creating spheroids in 1,536-well plates is established. This constraint is most likely caused by the difficulty of changing medium in a 1,536-well plate for repeated drug dosing during the culture time as well as the potential for edge effects caused by uneven medium evaporation over prolonged culture durations. Further work will be required to overcome these barriers and promote the 3D liver system in high-throughput drug screening.
Spheroids or organoids?
Indeed, model selection is vital to the effectiveness of the research study. Hepatocyte spheroids, typically formed by solid cell aggregation, are a type of spheroid that is easy to produce and can be precisely manipulated when performing assays. In drug development, a 3D in vitro model may be suitable for general drug screening and toxicity testing. Due to enhanced cell-cell spatial interaction, spheroids often offer more functionality and predictability than monolayer cultures of the same cell type. However, the utility of liver spheroids is often limited by the kind of input cells, such as pure hepatocyte spheroids, which contain only hepatocytes and can be used to assess the toxicity of drugs that directly damage hepatocytes. Incorporation of other liver cell types, such as Kupffer cells and hepatic stellate cells, may contribute to the development of a purpose-specific 3D in vitro liver model, albeit with limited usefulness.
Stem cells frequently develop and self-organize into liver organoids. This results in liver organoids comprised of multiple types of liver cells, including not only traditional liver-supporting cells like Kupffer cells and hepatic stellate cells but also cholangiocytes and other bile duct cells. All these cells can emerge continuously from the proliferation and differentiation of stem cells in organoids, and they can effectively support polarized hepatocytes and form the physiologically relevant liver structure in organoids, as well as promote the proliferation of the zone-specific liver differentiation area from the main organoid body. Due to the cell complexity, liver organoids are not only a good model for assessing compound toxicity directly to hepatocytes, but also a potential model for identifying many other idiosyncratic DILIs, including some immunocyte-related DILIs and some drug-induced cholestasis-related liver damage. Nonetheless, a methodology for evaluating the phenotypes of these DILIs is still in the process of being developed. In addition, liver organoid is an appropriate model for screening and customizing personalized therapies since they can be produced from patient cells for high-throughput testing. In addition, liver organoid can be used as an attractive disease model for drug screening, particularly for rare diseases such as Niemann-Pick Type C (NPC) disease, which contains mutations on the NPC1 or NPC2 gene leading to cholesterol accumulation in the hepatocytes, because they could be generated from a single genetically modified stem cell or patient-derived stem cell. As a result of this progress, liver organoids will be increasingly exploited in the fields of drug development and toxicity evaluation.
Alternative sources for 3D liver models
To generate suitable 3D liver models, the selection of cell types (e.g., HepaRG or PHHs) is critical and should be determined by the purpose of the study. Unfortunately, the utilization of PHHs is limited by insufficient supply, high cost, lot-to-lot variation, and low capacity for in vitro proliferation. However, the use of hepatoma cell lines cannot identify patient-specific toxicities caused by gene polymorphisms, and their tumor background impairs sensitivity to toxic treatments.100 To address these concerns, Wu et al. demonstrated that human hepatocytes may be expanded in vitro to form expandable liver progenitor-like cells (HepLPCs),101 resembling the reversible ductal metaplasia necessary for hepatocyte mass restoration following liver injury.102,103 Furthermore, Wang et al. established an efficient method for producing many human immortalized HepLPCs (iHepLPCs) with the ability to differentiate into mature hepatocytes again under certain culture conditions.104 In addition, the differentiated cells form spheroids in suspension and show hepatocyte functions on par with primary hepatocytes. iHepLPCs have not been used in large-scale screening, but they remain a viable model for toxicological investigations, particularly individualized liver toxicity testing. Indeed, personalized medicine is gaining increasing attention, and this type of model has the potential to improve the prediction of individualized medication effects, hence guiding the personalized use of therapeutic drugs.
iPSC-derived liver organoids in toxicological testing
Recapitulating liver complexity
With the advancement of the technology, attention has been directed toward developing more complex in vitro liver models for toxicological studies, namely iPSC-derived liver organoids. Indeed, the discovery of iPSCs accelerated the development of organoids by resolving challenges associated with the short-term availability of primary cells. These little, irregularly shaped in vitro liver bodies are composed of a variety of cell types, including parenchymal and non-parenchymal supporting cells. The complex structure renders it more mature than monolayer culture or 3D spheroids generated by iHeps, even though it is still a mixture of mature and premature hepatocytes.87 Importantly, many toxins may engage multiple cell types, making them detectable only in these heterogeneous models.8 However, as complexity increases and several cell types contribute to the parameters of interest, data processing and interpretation can become increasingly difficult.
Personalized toxicity testing
To develop personalized treatment, iPSC-based cell differentiation is another viable strategy that could work with different modeling paradigms. Indeed, hepatocytes produced from iPSCs have garnered a lot of interest for toxicity research. Use of iPSCs reprogrammed from patient cells to differentiate into hepatocytes was shown to be a valuable method for evaluating the potential hepatotoxicity of proposed medications to treat patients with genetic variants of known or unknown significance.105 In addition to genetic screening, a small-scale in vitro drug screening employing iPSC-derived patient-specific hepatocytes in monolayer culture or a mini-liver system in 3D mode could be used to help evaluate medication safety and select the optimal drug for personalized use. However, due to the premature phenotype and low level of CYP450 enzyme expression, the use of iHeps in drug toxicology prediction is limited, and there is no significant improvement even in a 3D spheroidal model compared with 2D culture.17,74,75 Thus, improving differentiation of iPSCs into fully mature and functional liver cells in the context of drug-screening platforms remains a work in progress. The cultivation of liver organoids could be a promising method for enabling this individualized toxicity testing or medication screening.
The influence of cell-cell communication
An interesting aspect of iPSC-derived liver organoids is the presence of cell-cell contacts between hepatocytes and non-parenchymal cells, such as fibroblasts and endothelial cells, which was shown to significantly increase hepatocyte function and activity in vitro.106 The roles of cell-cell communication in liver cell activity are complex, and different types of cells support hepatocyte function in distinct ways. Hepatic stellate cells can mimic the matrix environment; bone or adipose mesenchymal stem cells can enhance hepatic function by secreting paracrine cytokines; and vascular endothelial cells can drive angiogenesis and give nutrients.107 Although the mechanisms underpinning liver cell-cell communication are not fully known, extracellular vesicles, such as exosomes, microvesicles or microparticles, and apoptotic bodies play a role.108 The relevance of cell-cell interactions for pharmacological and toxicological testing was demonstrated by a study showing that direct or indirect stimulation of Kupffer cells by toxic substances results in the release of a variety of inflammatory mediators, growth factors, and reactive oxygen species, all of which influence acute hepatocyte injury as well as chronic liver responses and are thus causes of idiosyncratic DILI.109 This kind of DILI will not be detected by general toxicity testing using only hepatocytes as a model, but it will cause harm to humans either during clinical trials or potentially after the drug is approved for use depending on the severity and chronic/cumulative nature of the effects. In addition, 3D culture of hepatocytes can endow the cells with a densely interconnected spatial structure, hence enhancing communication between hepatocytes and facilitating the maintenance of function and activity.110 Therefore, liver organoids produced from stem cells are the most promising future model for drug screening and toxicological testing.
Major challenges to toxicity testing on iPSC-derived organoids
Although there are multiple benefits to the use of HLOs in toxicological testing, there are still obstacles in assay development and implementation that must be solved. The greatest challenge in the creation of toxicological assays was determining how to normalize the data produced by the variance of liver organoids, and this challenge can vary depending on the assay. On the other hand, Shinozawa et al. have demonstrated the feasibility of expanding the use of HLOs produced from iPSCs in the arena of high-throughput drug screening.86 It significantly expands the application of HLOs in toxicology and drug development. Although the uneven form and size of HLOs result in unanticipated fluctuations in many of the routine liver toxicity parameter assessments, imaging-based high-content analysis can assist in normalizing the data in most situations. While high-throughput screening with dispersed HLOs in 384-well plates has the capability of predicting the bioactivity of compounds and can be used for large screening efforts, it still results in decreased CYP450 expression and a lack of critical hepatocyte function,88 indicating that spatial interaction between hepatocytes and other supporting cells is critical for hepatocyte maturation and function.
Zhang et al. paired their HLO cells with a “liver-on-a-chip” system, a 3D in vitro hepatic micro-physiological system aimed at recreating the circumstances of liver tissue on a microscopic scale,111 and showed a greater ability to predict hepatotoxicity than dispersed HLO cells.88 Indeed, numerous studies demonstrated that this perfusion-based in vitro model results in more mature liver function and increased CYP450 enzyme expression when compared with other models, and data indicated that the flow rate and thus the shear stresses imposed on the cell membrane can influence the expression of CYP450 enzymes.112,113,114 In addition, it has been postulated that some types of stress on hepatocytes could result in an increase in the expression level of certain CYP450 enzymes.
Liver-on-a-chip
Beyond liver organoids, in the past few years, the liver-on-a-chip model has attracted great attention in the scientific community because it can simulate in vivo liver conditions and the dynamic physicochemical environment of the liver. Numerous investigations illustrate the efforts toward scaling up such a platform for high-throughput testing. Bircsak et al. designed and validated a microfluidic liver-on-a-chip device for high-throughput hepatotoxicity screening.115 They seeded aggregates of iHeps in the organ channel with endothelial cells and THP-1 monocytes seeded onto the vascular channel of the 96-well Mimetas OrganoPlate 2-lane. Similarly, Chen et al. utilized tumor spheroids, including HepG2 cells, in conjunction with microfluidic chip technology for drug screening.116 Although this technology has been utilized for cancer drug screening, it raises the possibility of further developing and altering these microfluidic platforms for toxicological screening of DILI in the future. Remarkably, Ewart et al. have recently developed a high-throughput Liver-Chip toxicity testing platform that can simultaneously assess and report the hepatotoxic effects of 27 drugs or chemicals on 780 Liver-Chips.117 In their platform, drug toxicity on the liver is monitored based on three criteria: real-time analysis of albumin and alanine aminotransferase production, as well as immunofluorescence imaging of cell morphology using an automated confocal microscope. Taking these three parameters into account, their Liver-Chip platform can achieve 80% sensitivity and 100% specificity in their test for 27 known DILI medications. Recently, at the international conference and exhibition of the Society for Laboratory Automation and Screening, numerous researchers also discussed their efforts to construct a high-throughput perfusion-based liver-on-chip system for drug toxicology screening. With further improvement of this system, further applications are possible.
While there are currently a variety of strategies for increasing the complexity of in vitro models, increased complexity does not always equate to a better model, which is why it is critical to choose an appropriate model to fit one’s experimental design. The future screening for compound liver toxicity will certainly require to use many human physiology-relevant in vitro models for better treatment and prediction of liver toxicity.
Acknowledgments
This study was supported in part by the Intramural Research Program of the National Center for Advancing Translational Sciences, National Institutes of Health. The views expressed in this review are those of the authors and do not necessarily reflect the statements, opinions, views, conclusions, or policies of the National Center for Advancing Translational Sciences, or the US government. Mention of trade names or commercial products does not constitute endorsement or recommendation for use. All figures were created with https://biorender.com.
Declaration of interests
The authors declare no competing interests.
References
- 1.Takebe T., Taniguchi H. Human iPSC-derived miniature organs: a tool for drug studies. Clin. Pharmacol. Ther. 2014;96:310–313. doi: 10.1038/clpt.2014.110. [DOI] [PubMed] [Google Scholar]
- 2.Serras A.S., Rodrigues J.S., Cipriano M., Rodrigues A.V., Oliveira N.G., Miranda J.P. A critical perspective on 3D liver models for drug metabolism and toxicology studies. Front. Cell Dev. Biol. 2021;9:626805. doi: 10.3389/fcell.2021.626805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steigerwald K., Merl S., Kastrati A., Wieczorek A., Vorpahl M., Mannhold R., Vogeser M., Hausleiter J., Joner M., Schömig A., Wessely R. The pre-clinical assessment of rapamycin-eluting, durable polymer-free stent coating concepts. Biomaterials. 2009;30:632–637. doi: 10.1016/j.biomaterials.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Van Norman G.A. Limitations of animal studies for predicting toxicity in clinical trials: is it time to rethink our current approach? JACC. Basic Transl. Sci. 2019;4:845–854. doi: 10.1016/j.jacbts.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olson H., Betton G., Robinson D., Thomas K., Monro A., Kolaja G., Lilly P., Sanders J., Sipes G., Bracken W., et al. Concordance of the toxicity of pharmaceuticals in humans and in animals. Regul. Toxicol. Pharmacol. 2000;32:56–67. doi: 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- 6.Shanks N., Greek R., Greek J. Are animal models predictive for humans? Philos. Ethics Humanit. Med. 2009;4:2. doi: 10.1186/1747-5341-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu D., Nishimura T., Nishimura S., Zhang H., Zheng M., Guo Y.Y., Masek M., Michie S.A., Glenn J., Peltz G. Fialuridine induces acute liver failure in chimeric TK-NOG mice: a model for detecting hepatic drug toxicity prior to human testing. PLoS Med. 2014;11:e1001628. doi: 10.1371/journal.pmed.1001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox C.R., Lynch S., Goldring C., Sharma P. Current perspective: 3D spheroid models utilizing human-based cells for investigating metabolism-dependent drug-induced liver injury. Front. Med. Technol. 2020;2:611913. doi: 10.3389/fmedt.2020.611913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowe C., Gerrard D.T., Jenkins R., Berry A., Durkin K., Sundstrom L., Goldring C.E., Park B.K., Kitteringham N.R., Hanley K.P., Hanley N.A. Proteome-wide analyses of human hepatocytes during differentiation and dedifferentiation. Hepatology. 2013;58:799–809. doi: 10.1002/hep.26414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerets H.H.J., Tilmant K., Gerin B., Chanteux H., Depelchin B.O., Dhalluin S., Atienzar F.A. Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biol. Toxicol. 2012;28:69–87. doi: 10.1007/s10565-011-9208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baxter M., Withey S., Harrison S., Segeritz C.P., Zhang F., Atkinson-Dell R., Rowe C., Gerrard D.T., Sison-Young R., Jenkins R., et al. Phenotypic and functional analyses show stem cell-derived hepatocyte-like cells better mimic fetal rather than adult hepatocytes. J. Hepatol. 2015;62:581–589. doi: 10.1016/j.jhep.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramaiahgari S.C., den Braver M.W., Herpers B., Terpstra V., Commandeur J.N.M., van de Water B., Price L.S. A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies. Arch. Toxicol. 2014;88:1083–1095. doi: 10.1007/s00204-014-1215-9. [DOI] [PubMed] [Google Scholar]
- 13.Ramaiahgari S.C., Waidyanatha S., Dixon D., DeVito M.J., Paules R.S., Ferguson S.S. Three-dimensional (3D) HepaRG spheroid model with physiologically relevant xenobiotic metabolism competence and hepatocyte functionality for liver toxicity screening. Toxicol. Sci. 2017;160:189–190. doi: 10.1093/toxsci/kfx194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliva-Vilarnau N., Vorrink S.U., Ingelman-Sundberg M., Lauschke V.M. A 3D cell culture model identifies wnt/beta-catenin mediated inhibition of p53 as a critical step during human hepatocyte regeneration. Adv. Sci. 2020;7:2000248. doi: 10.1002/advs.202000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y., Shen J.X., Lauschke V.M. Comprehensive evaluation of organotypic and microphysiological liver models for prediction of drug-induced liver injury. Front. Pharmacol. 2019;10:1093. doi: 10.3389/fphar.2019.01093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaskell H., Sharma P., Colley H.E., Murdoch C., Williams D.P., Webb S.D. Characterization of a functional C3A liver spheroid model. Toxicol. Res. 2016;5:1053–1065. doi: 10.1039/c6tx00101g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takayama K., Kawabata K., Nagamoto Y., Kishimoto K., Tashiro K., Sakurai F., Tachibana M., Kanda K., Hayakawa T., Furue M.K., Mizuguchi H. 3D spheroid culture of hESC/hiPSC-derived hepatocyte-like cells for drug toxicity testing. Biomaterials. 2013;34:1781–1789. doi: 10.1016/j.biomaterials.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z., Luo X., Anene-Nzelu C., Yu Y., Hong X., Singh N.H., Xia L., Liu S., Yu H. HepaRG culture in tethered spheroids as an in vitro three-dimensional model for drug safety screening. J. Appl. Toxicol. 2015;35:909–917. doi: 10.1002/jat.3090. [DOI] [PubMed] [Google Scholar]
- 19.Jeffries R.E., Gamcsik M.P., Keshari K.R., Pediaditakis P., Tikunov A.P., Young G.B., Lee H., Watkins P.B., Macdonald J.M. Effect of oxygen concentration on viability and metabolism in a fluidized-bed bioartificial liver using (3)(1)P and (1)(3)C NMR spectroscopy. Tissue Eng. C Methods. 2013;19:93–100. doi: 10.1089/ten.TEC.2011.0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donato M.T., Lahoz A., Castell J.V., Gómez-Lechón M.J. Cell lines: a tool for in vitro drug metabolism studies. Curr. Drug Metabol. 2008;9:1–11. doi: 10.2174/138920008783331086. [DOI] [PubMed] [Google Scholar]
- 21.Gieseck R.L., 3rd, Hannan N.R.F., Bort R., Hanley N.A., Drake R.A.L., Cameron G.W.W., Wynn T.A., Vallier L. Maturation of induced pluripotent stem cell derived hepatocytes by 3D-culture. PLoS One. 2014;9:e86372. doi: 10.1371/journal.pone.0086372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akbari S., Arslan N., Senturk S., Erdal E. Next-generation liver medicine using organoid models. Front. Cell Dev. Biol. 2019;7:345. doi: 10.3389/fcell.2019.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godoy P., Hewitt N.J., Albrecht U., Andersen M.E., Ansari N., Bhattacharya S., Bode J.G., Bolleyn J., Borner C., Böttger J., et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch. Toxicol. 2013;87:1315–1530. doi: 10.1007/s00204-013-1078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soldatow V.Y., Lecluyse E.L., Griffith L.G., Rusyn I. In vitro models for liver toxicity testing. Toxicol. Res. 2013;2:23–39. doi: 10.1039/C2TX20051A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gómez-Lechón M.J., Tolosa L., Conde I., Donato M.T. Competency of different cell models to predict human hepatotoxic drugs. Expet Opin. Drug Metabol. Toxicol. 2014;10:1553–1568. doi: 10.1517/17425255.2014.967680. [DOI] [PubMed] [Google Scholar]
- 26.Kyffin J.A., Sharma P., Leedale J., Colley H.E., Murdoch C., Mistry P., Webb S.D. Impact of cell types and culture methods on the functionality of in vitro liver systems - a review of cell systems for hepatotoxicity assessment. Toxicol. Vitro. 2018;48:262–275. doi: 10.1016/j.tiv.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 27.Underhill G.H., Khetani S.R. Advances in engineered human liver platforms for drug metabolism studies. Drug Metab. Dispos. 2018;46:1626–1637. doi: 10.1124/dmd.118.083295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrison S.P., Baumgarten S.F., Verma R., Lunov O., Dejneka A., Sullivan G.J. Liver organoids: recent developments, limitations and potential. Front. Med. 2021;8:574047. doi: 10.3389/fmed.2021.574047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prior N., Inacio P., Huch M. Liver organoids: from basic research to therapeutic applications. Gut. 2019;68:2228–2237. doi: 10.1136/gutjnl-2019-319256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olgasi C., Cucci A., Follenzi A. iPSC-derived liver organoids: a journey from drug screening, to disease modeling, arriving to regenerative medicine. Int. J. Mol. Sci. 2020;21:6215. doi: 10.3390/ijms21176215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogoke O., Maloy M., Parashurama N. The science and engineering of stem cell-derived organoids-examples from hepatic, biliary, and pancreatic tissues. Biol. Rev. Camb. Phil. Soc. 2021;96:179–204. doi: 10.1111/brv.12650. [DOI] [PubMed] [Google Scholar]
- 32.Edmondson R., Broglie J.J., Adcock A.F., Yang L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014;12:207–218. doi: 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lakehal F., Dansette P.M., Becquemont L., Lasnier E., Delelo R., Balladur P., Poupon R., Beaune P.H., Housset C. Indirect cytotoxicity of flucloxacillin toward human biliary epithelium via metabolite formation in hepatocytes. Chem. Res. Toxicol. 2001;14:694–701. doi: 10.1021/tx0002435. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X., Jiang T., Chen D., Wang Q., Zhang L.W. Three-dimensional liver models: state of the art and their application for hepatotoxicity evaluation. Crit. Rev. Toxicol. 2020;50:279–309. doi: 10.1080/10408444.2020.1756219. [DOI] [PubMed] [Google Scholar]
- 35.Kelm J.M., Timmins N.E., Brown C.J., Fussenegger M., Nielsen L.K. Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol. Bioeng. 2003;83:173–180. doi: 10.1002/bit.10655. [DOI] [PubMed] [Google Scholar]
- 36.Drewitz M., Helbling M., Fried N., Bieri M., Moritz W., Lichtenberg J., Kelm J.M. Towards automated production and drug sensitivity testing using scaffold-free spherical tumor microtissues. Biotechnol. J. 2011;6:1488–1496. doi: 10.1002/biot.201100290. [DOI] [PubMed] [Google Scholar]
- 37.Aden D.P., Fogel A., Plotkin S., Damjanov I., Knowles B.B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature. 1979;282:615–616. doi: 10.1038/282615a0. [DOI] [PubMed] [Google Scholar]
- 38.Schofield C.A., Walker T.M., Taylor M.A., Patel M., Vlachou D.F., Macina J.M., Vidgeon-Hart M.P., Williams A., McGill P.J., Newman C.F., Sakatis M.Z. Evaluation of a three-dimensional primary human hepatocyte spheroid model: adoption and industrialization for the enhanced detection of drug-induced liver injury. Chem. Res. Toxicol. 2021;34:2485–2499. doi: 10.1021/acs.chemrestox.1c00227. [DOI] [PubMed] [Google Scholar]
- 39.Yokoyama Y., Sasaki Y., Terasaki N., Kawataki T., Takekawa K., Iwase Y., Shimizu T., Sanoh S., Ohta S. Comparison of drug metabolism and its related hepatotoxic effects in HepaRG, cryopreserved human hepatocytes, and HepG2 cell cultures. Biol. Pharm. Bull. 2018;41:722–732. doi: 10.1248/bpb.b17-00913. [DOI] [PubMed] [Google Scholar]
- 40.Hurrell T., Lilley K.S., Cromarty A.D. Proteomic responses of HepG2 cell monolayers and 3D spheroids to selected hepatotoxins. Toxicol. Lett. 2019;300:40–50. doi: 10.1016/j.toxlet.2018.10.030. [DOI] [PubMed] [Google Scholar]
- 41.Fey S.J., Wrzesinski K. Determination of drug toxicity using 3D spheroids constructed from an immortal human hepatocyte cell line. Toxicol. Sci. 2012;127:403–411. doi: 10.1093/toxsci/kfs122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivascu A., Kubbies M. Rapid generation of single-tumor spheroids for high-throughput cell function and toxicity analysis. J. Biomol. Screen. 2006;11:922–932. doi: 10.1177/1087057106292763. [DOI] [PubMed] [Google Scholar]
- 43.Das V., Fürst T., Gurská S., Džubák P., Hajdúch M. Reproducibility of uniform spheroid formation in 384-well plates: the effect of medium evaporation. J. Biomol. Screen. 2016;21:923–930. doi: 10.1177/1087057116651867. [DOI] [PubMed] [Google Scholar]
- 44.Hurrell T., Ellero A.A., Masso Z.F., Cromarty A.D. Characterization and reproducibility of HepG2 hanging drop spheroids toxicology in vitro. Toxicol. Vitro. 2018;50:86–94. doi: 10.1016/j.tiv.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 45.Hiemstra S., Ramaiahgari S.C., Wink S., Callegaro G., Coonen M., Meerman J., Jennen D., van den Nieuwendijk K., Dankers A., Snoeys J., et al. High-throughput confocal imaging of differentiated 3D liver-like spheroid cellular stress response reporters for identification of drug-induced liver injury liability. Arch. Toxicol. 2019;93:2895–2911. doi: 10.1007/s00204-019-02552-0. [DOI] [PubMed] [Google Scholar]
- 46.Basharat A., Rollison H.E., Williams D.P., Ivanov D.P. HepG2 (C3A) spheroids show higher sensitivity compared to HepaRG spheroids for drug-induced liver injury (DILI) Toxicol. Appl. Pharmacol. 2020;408:115279. doi: 10.1016/j.taap.2020.115279. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Y., Chen Y., Hu Y., Wang J., Xie X., He G., Chen H., Shao Q., Zeng H., Zhang H. Genomic alterations across six hepatocellular carcinoma cell lines by panel-based sequencing. Transl. Cancer Res. 2018;7:231–239. [Google Scholar]
- 48.Chen J., Raymond K. Identification of CYP2C9∗2 allele in HepG2 cell line. Int. J. Gastrointest. Cancer. 2006;37:79–83. doi: 10.1007/s12029-007-0003-7. [DOI] [PubMed] [Google Scholar]
- 49.Heslop J.A., Rowe C., Walsh J., Sison-Young R., Jenkins R., Kamalian L., Kia R., Hay D., Jones R.P., Malik H.Z., et al. Mechanistic evaluation of primary human hepatocyte culture using global proteomic analysis reveals a selective dedifferentiation profile. Arch. Toxicol. 2017;91:439–452. doi: 10.1007/s00204-016-1694-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lauschke V.M., Vorrink S.U., Moro S.M.L., Rezayee F., Nordling Å., Hendriks D.F.G., Bell C.C., Sison-Young R., Park B.K., Goldring C.E., et al. Massive rearrangements of cellular MicroRNA signatures are key drivers of hepatocyte dedifferentiation. Hepatology. 2016;64:1743–1756. doi: 10.1002/hep.28780. [DOI] [PubMed] [Google Scholar]
- 51.Proctor W.R., Foster A.J., Vogt J., Summers C., Middleton B., Pilling M.A., Shienson D., Kijanska M., Ströbel S., Kelm J.M., et al. Utility of spherical human liver microtissues for prediction of clinical drug-induced liver injury. Arch. Toxicol. 2017;91:2849–2863. doi: 10.1007/s00204-017-2002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bell C.C., Hendriks D.F.G., Moro S.M.L., Ellis E., Walsh J., Renblom A., Fredriksson Puigvert L., Dankers A.C.A., Jacobs F., Snoeys J., et al. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci. Rep. 2016;6:25187. doi: 10.1038/srep25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohkura T., Ohta K., Nagao T., Kusumoto K., Koeda A., Ueda T., Jomura T., Ikeya T., Ozeki E., Wada K., et al. Evaluation of human hepatocytes cultured by three-dimensional spheroid systems for drug metabolism. Drug Metabol. Pharmacokinet. 2014;29:373–378. doi: 10.2133/dmpk.dmpk-13-rg-105. [DOI] [PubMed] [Google Scholar]
- 54.Vilas-Boas V., Gijbels E., Leroy K., Pieters A., Baze A., Parmentier C., Vinken M. Primary human hepatocyte spheroids as tools to study the hepatotoxic potential of non-pharmaceutical chemicals. Int. J. Mol. Sci. 2021;22:11005. doi: 10.3390/ijms222011005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanebratt K.P., Janefeldt A., Vilén L., Vildhede A., Samuelsson K., Milton L., Björkbom A., Persson M., Leandersson C., Andersson T.B., Hilgendorf C. Primary human hepatocyte spheroid model as a 3D in vitro platform for metabolism studies. J. Pharmacol. Sci. 2021;110:422–431. doi: 10.1016/j.xphs.2020.10.043. [DOI] [PubMed] [Google Scholar]
- 56.Vorrink S.U., Zhou Y., Ingelman-Sundberg M., Lauschke V.M. Prediction of drug-induced hepatotoxicity using long-term stable primary hepatic 3D spheroid cultures in chemically defined conditions. Toxicol. Sci. 2018;163:655–665. doi: 10.1093/toxsci/kfy058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gunness P., Mueller D., Shevchenko V., Heinzle E., Ingelman-Sundberg M., Noor F. 3D organotypic cultures of human HepaRG cells: a tool for in vitro toxicity studies. Toxicol. Sci. 2013;133:67–78. doi: 10.1093/toxsci/kft021. [DOI] [PubMed] [Google Scholar]
- 58.Desai P.K., Tseng H., Souza G.R. Assembly of hepatocyte spheroids using magnetic 3D cell culture for CYP450 inhibition/induction. Int. J. Mol. Sci. 2017;18:1085. doi: 10.3390/ijms18051085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Timm D.M., Chen J., Sing D., Gage J.A., Haisler W.L., Neeley S.K., Raphael R.M., Dehghani M., Rosenblatt K.P., Killian T.C., et al. A high-throughput three-dimensional cell migration assay for toxicity screening with mobile device-based macroscopic image analysis. Sci. Rep. 2013;3:3000. doi: 10.1038/srep03000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Magin C.M., Alge D.L., Anseth K.S. Bio-inspired 3D microenvironments: a new dimension in tissue engineering. Biomed. Mater. 2016;11:022001. doi: 10.1088/1748-6041/11/2/022001. [DOI] [PubMed] [Google Scholar]
- 61.Nguyen D.G., Funk J., Robbins J.B., Crogan-Grundy C., Presnell S.C., Singer T., Roth A.B. Bioprinted 3D primary liver tissues allow assessment of organ-level response to clinical drug induced toxicity in vitro. PLoS One. 2016;11:e0158674. doi: 10.1371/journal.pone.0158674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Madoux F., Tanner A., Vessels M., Willetts L., Hou S., Scampavia L., Spicer T.P. A 1536-well 3D viability assay to assess the cytotoxic effect of drugs on spheroids. SLAS Discov. 2017;22:516–524. doi: 10.1177/2472555216686308. [DOI] [PubMed] [Google Scholar]
- 63.Berger B., Donzelli M., Maseneni S., Boess F., Roth A., Krähenbühl S., Haschke M. Comparison of liver cell models using the basel phenotyping cocktail. Front. Pharmacol. 2016;7:443. doi: 10.3389/fphar.2016.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sison-Young R.L.C., Mitsa D., Jenkins R.E., Mottram D., Alexandre E., Richert L., Aerts H., Weaver R.J., Jones R.P., Johann E., et al. Comparative proteomic characterization of 4 human liver-derived single cell culture models reveals significant variation in the capacity for drug disposition, bioactivation, and detoxication. Toxicol. Sci. 2015;147:412–424. doi: 10.1093/toxsci/kfv136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aninat C., Piton A., Glaise D., Le Charpentier T., Langouët S., Morel F., Guguen-Guillouzo C., Guillouzo A. Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab. Dispos. 2006;34:75–83. doi: 10.1124/dmd.105.006759. [DOI] [PubMed] [Google Scholar]
- 66.Guillouzo A., Corlu A., Aninat C., Glaise D., Morel F., Guguen-Guillouzo C. The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem. Biol. Interact. 2007;168:66–73. doi: 10.1016/j.cbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 67.Jackson J.P., Li L., Chamberlain E.D., Wang H., Ferguson S.S. Contextualizing hepatocyte functionality of cryopreserved HepaRG cell cultures. Drug Metab. Dispos. 2016;44:1463–1479. doi: 10.1124/dmd.116.069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mueller D., Krämer L., Hoffmann E., Klein S., Noor F. 3D organotypic HepaRG cultures as in vitro model for acute and repeated dose toxicity studies. Toxicol. Vitro. 2014;28:104–112. doi: 10.1016/j.tiv.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 69.Takahashi Y., Hori Y., Yamamoto T., Urashima T., Ohara Y., Tanaka H. 3D spheroid cultures improve the metabolic gene expression profiles of HepaRG cells. Biosci. Rep. 2015;35:e00208. doi: 10.1042/BSR20150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darnell M., Ulvestad M., Ellis E., Weidolf L., Andersson T.B. In vitro evaluation of major in vivo drug metabolic pathways using primary human hepatocytes and HepaRG cells in suspension and a dynamic three-dimensional bioreactor system. J. Pharmacol. Exp. Therapeut. 2012;343:134–144. doi: 10.1124/jpet.112.195834. [DOI] [PubMed] [Google Scholar]
- 71.Leite S.B., Wilk-Zasadna I., Zaldivar J.M., Airola E., Reis-Fernandes M.A., Mennecozzi M., Guguen-Guillouzo C., Chesne C., Guillou C., Alves P.M., Coecke S. Three-dimensional HepaRG model as an attractive tool for toxicity testing. Toxicol. Sci. 2012;130:106–116. doi: 10.1093/toxsci/kfs232. [DOI] [PubMed] [Google Scholar]
- 72.Ott L.M., Ramachandran K., Stehno-Bittel L. An automated multiplexed hepatotoxicity and CYP induction assay using HepaRG cells in 2D and 3D. SLAS Discov. 2017;22:614–625. doi: 10.1177/2472555217701058. [DOI] [PubMed] [Google Scholar]
- 73.Si-Tayeb K., Noto F.K., Nagaoka M., Li J., Battle M.A., Duris C., North P.E., Dalton S., Duncan S.A. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rashidi H., Luu N.T., Alwahsh S.M., Ginai M., Alhaque S., Dong H., Tomaz R.A., Vernay B., Vigneswara V., Hallett J.M., et al. 3D human liver tissue from pluripotent stem cells displays stable phenotype in vitro and supports compromised liver function in vivo. Arch. Toxicol. 2018;92:3117–3129. doi: 10.1007/s00204-018-2280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meier F., Freyer N., Brzeszczynska J., Knöspel F., Armstrong L., Lako M., Greuel S., Damm G., Ludwig-Schwellinger E., Deschl U., et al. Hepatic differentiation of human iPSCs in different 3D models: a comparative study. Int. J. Mol. Med. 2017;40:1759–1771. doi: 10.3892/ijmm.2017.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qosa H., Ribeiro A.J.S., Hartman N.R., Volpe D.A. Characterization of a commercially available line of iPSC hepatocytes as models of hepatocyte function and toxicity for regulatory purposes. J. Pharmacol. Toxicol. Methods. 2021;110:107083. doi: 10.1016/j.vascn.2021.107083. [DOI] [PubMed] [Google Scholar]
- 77.Sirenko O., Hancock M.K., Hesley J., Hong D., Cohen A., Gentry J., Carlson C.B., Mann D.A. Phenotypic characterization of toxic compound effects on liver spheroids derived from iPSC using confocal imaging and three-dimensional image analysis. Assay Drug Dev. Technol. 2016;14:381–394. doi: 10.1089/adt.2016.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee G., Kim H., Park J.Y., Kim G., Han J., Chung S., Yang J.H., Jeon J.S., Woo D.H., Han C., et al. Generation of uniform liver spheroids from human pluripotent stem cells for imaging-based drug toxicity analysis. Biomaterials. 2021;269:120529. doi: 10.1016/j.biomaterials.2020.120529. [DOI] [PubMed] [Google Scholar]
- 79.Koike H., Iwasawa K., Ouchi R., Maezawa M., Giesbrecht K., Saiki N., Ferguson A., Kimura M., Thompson W.L., Wells J.M., et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature. 2019;574:112–116. doi: 10.1038/s41586-019-1598-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Akbari S., Sevinç G.G., Ersoy N., Basak O., Kaplan K., Sevinç K., Ozel E., Sengun B., Enustun E., Ozcimen B., et al. Robust, long-term culture of endoderm-derived hepatic organoids for disease modeling. Stem Cell Rep. 2019;13:627–641. doi: 10.1016/j.stemcr.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu F., Wu D., Ren Y., Huang Y., Feng B., Zhao N., Zhang T., Chen X., Chen S., Xu A. Generation of hepatobiliary organoids from human induced pluripotent stem cells. J. Hepatol. 2019;70:1145–1158. doi: 10.1016/j.jhep.2018.12.028. [DOI] [PubMed] [Google Scholar]
- 82.Guan Y., Xu D., Garfin P.M., Ehmer U., Hurwitz M., Enns G., Michie S., Wu M., Zheng M., Nishimura T., et al. Human hepatic organoids for the analysis of human genetic diseases. JCI Insight. 2017;2:e94954. doi: 10.1172/jci.insight.94954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sampaziotis F., de Brito M.C., Madrigal P., Bertero A., Saeb-Parsy K., Soares F.A.C., Schrumpf E., Melum E., Karlsen T.H., Bradley J.A., et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat. Biotechnol. 2015;33:845–852. doi: 10.1038/nbt.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ogawa M., Ogawa S., Bear C.E., Ahmadi S., Chin S., Li B., Grompe M., Keller G., Kamath B.M., Ghanekar A. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat. Biotechnol. 2015;33:853–861. doi: 10.1038/nbt.3294. [DOI] [PubMed] [Google Scholar]
- 85.Dianat N., Dubois-Pot-Schneider H., Steichen C., Desterke C., Leclerc P., Raveux A., Combettes L., Weber A., Corlu A., Dubart-Kupperschmitt A. Generation of functional cholangiocyte-like cells from human pluripotent stem cells and HepaRG cells. Hepatology. 2014;60:700–714. doi: 10.1002/hep.27165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shinozawa T., Kimura M., Cai Y., Saiki N., Yoneyama Y., Ouchi R., Koike H., Maezawa M., Zhang R.R., Dunn A., et al. High-fidelity drug-induced liver injury screen using human pluripotent stem cell-derived organoids. Gastroenterology. 2021;160:831–846.e10. doi: 10.1053/j.gastro.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ouchi R., Togo S., Kimura M., Shinozawa T., Koido M., Koike H., Thompson W., Karns R.A., Mayhew C.N., McGrath P.S., et al. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metabol. 2019;30:374–384.e6. doi: 10.1016/j.cmet.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang C.J., O’Meara M.J., Meyer S.R., Huang S., Capeling M.M., Ferrer-Torres D., Childs C.J., Spence J.R., Fontana R.J., Sexton J.Z. A multi-omics human liver organoid screening platform for DILI risk prediction. bioRxiv. 2021 doi: 10.1101/2021.08.26.457824. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Racanelli V., Rehermann B. The liver as an immunological organ. Hepatology. 2006;43:S54–S62. doi: 10.1002/hep.21060. [DOI] [PubMed] [Google Scholar]
- 90.Rane A., Sjöqvist F. Drug metabolism in the human fetus and newborn infant. Pediatr. Clin. 1972;19:37–49. doi: 10.1016/s0031-3955(16)32665-7. [DOI] [PubMed] [Google Scholar]
- 91.Takebe T., Sekine K., Enomura M., Koike H., Kimura M., Ogaeri T., Zhang R.R., Ueno Y., Zheng Y.W., Koike N., et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 92.Thompson W.L., Takebe T. Generation of multi-cellular human liver organoids from pluripotent stem cells. Methods Cell Biol. 2020;159:47–68. doi: 10.1016/bs.mcb.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ramli M.N.B., Lim Y.S., Koe C.T., Demircioglu D., Tng W., Gonzales K.A.U., Tan C.P., Szczerbinska I., Liang H., Soe E.L., et al. Human pluripotent stem cell-derived organoids as models of liver disease. Gastroenterology. 2020;159:1471–1486.e12. doi: 10.1053/j.gastro.2020.06.010. [DOI] [PubMed] [Google Scholar]
- 94.Khetani S.R. Pluripotent stem cell-derived human liver organoids enter the realm of high-throughput drug screening. Gastroenterology. 2021;160:653–655. doi: 10.1053/j.gastro.2020.12.005. [DOI] [PubMed] [Google Scholar]
- 95.Lazic S.E., Williams D.P. Improving drug safety predictions by reducing poor analytical practices. Toxicology Research and Application. 2020;4 doi: 10.1177/2397847320978633. [DOI] [Google Scholar]
- 96.Rubashkin M.G., Ou G., Weaver V.M. Deconstructing signaling in three dimensions. Biochemistry. 2014;53:2078–2090. doi: 10.1021/bi401710d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cozzolino A.M., Noce V., Battistelli C., Marchetti A., Grassi G., Cicchini C., Tripodi M., Amicone L. Modulating the substrate stiffness to manipulate differentiation of resident liver stem cells and to improve the differentiation state of hepatocytes. Stem Cell. Int. 2016;2016:5481493. doi: 10.1155/2016/5481493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Visentin M., Lenggenhager D., Gai Z., Kullak-Ublick G.A. Drug-induced bile duct injury. Biochim. Biophys. Acta, Mol. Basis Dis. 2018;1864:1498–1506. doi: 10.1016/j.bbadis.2017.08.033. [DOI] [PubMed] [Google Scholar]
- 99.Hendriks D.F.G., Hurrell T., Riede J., van der Horst M., Tuovinen S., Ingelman-Sundberg M. Mechanisms of chronic fialuridine hepatotoxicity as revealed in primary human hepatocyte spheroids. Toxicol. Sci. 2019;171:385–395. doi: 10.1093/toxsci/kfz195. [DOI] [PubMed] [Google Scholar]
- 100.Levy G., Bomze D., Heinz S., Ramachandran S.D., Noerenberg A., Cohen M., Shibolet O., Sklan E., Braspenning J., Nahmias Y. Long-term culture and expansion of primary human hepatocytes. Nat. Biotechnol. 2015;33:1264–1271. doi: 10.1038/nbt.3377. [DOI] [PubMed] [Google Scholar]
- 101.Wu H., Zhou X., Fu G.B., He Z.Y., Wu H.P., You P., Ashton C., Wang X., Wang H.Y., Yan H.X. Reversible transition between hepatocytes and liver progenitors for in vitro hepatocyte expansion. Cell Res. 2017;27:709–712. doi: 10.1038/cr.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fu G.B., Huang W.J., Zeng M., Zhou X., Wu H.P., Liu C.C., Wu H., Weng J., Zhang H.D., Cai Y.C., et al. Expansion and differentiation of human hepatocyte-derived liver progenitor-like cells and their use for the study of hepatotropic pathogens. Cell Res. 2019;29:8–22. doi: 10.1038/s41422-018-0103-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tarlow B.D., Pelz C., Naugler W.E., Wakefield L., Wilson E.M., Finegold M.J., Grompe M. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell. 2014;15:605–618. doi: 10.1016/j.stem.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang Z., Li W., Jing H., Ding M., Fu G., Yuan T., Huang W., Dai M., Tang D., Zeng M., et al. Generation of hepatic spheroids using human hepatocyte-derived liver progenitor-like cells for hepatotoxicity screening. Theranostics. 2019;9:6690–6705. doi: 10.7150/thno.34520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang Y.S. Tailored drug therapy for mitigating drug-induced liver injury: is this the era of genetic screening? Per. Med. 2010;7:5–8. doi: 10.2217/pme.09.69. [DOI] [PubMed] [Google Scholar]
- 106.Liu Y., Li H., Yan S., Wei J., Li X. Hepatocyte cocultures with endothelial cells and fibroblasts on micropatterned fibrous mats to promote liver-specific functions and capillary formation capabilities. Biomacromolecules. 2014;15:1044–1054. doi: 10.1021/bm401926k. [DOI] [PubMed] [Google Scholar]
- 107.Wang J., Huang D., Yu H., Cheng Y., Ren H., Zhao Y. Engineered Regeneration; 2022. Developing Tissue Engineering Strategies for Liver Regeneration. [Google Scholar]
- 108.Sato K., Kennedy L., Liangpunsakul S., Kusumanchi P., Yang Z., Meng F., Glaser S., Francis H., Alpini G. Intercellular communication between hepatic cells in liver diseases. Int. J. Mol. Sci. 2019;20:2180. doi: 10.3390/ijms20092180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Roberts R.A., Ganey P.E., Ju C., Kamendulis L.M., Rusyn I., Klaunig J.E. Role of the Kupffer cell in mediating hepatic toxicity and carcinogenesis. Toxicol. Sci. 2007;96:2–15. doi: 10.1093/toxsci/kfl173. [DOI] [PubMed] [Google Scholar]
- 110.Chae S., Hong J., Hwangbo H., Kim G. The utility of biomedical scaffolds laden with spheroids in various tissue engineering applications. Theranostics. 2021;11:6818–6832. doi: 10.7150/thno.58421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Materne E.M., Tonevitsky A.G., Marx U. Chip-based liver equivalents for toxicity testing--organotypicalness versus cost-efficient high throughput. Lab Chip. 2013;13:3481–3495. doi: 10.1039/c3lc50240f. [DOI] [PubMed] [Google Scholar]
- 112.Vinci B., Duret C., Klieber S., Gerbal-Chaloin S., Sa-Cunha A., Laporte S., Suc B., Maurel P., Ahluwalia A., Daujat-Chavanieu M. Modular bioreactor for primary human hepatocyte culture: medium flow stimulates expression and activity of detoxification genes. Biotechnol. J. 2011;6:554–564. doi: 10.1002/biot.201000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rashidi H., Alhaque S., Szkolnicka D., Flint O., Hay D.C. Fluid shear stress modulation of hepatocyte-like cell function. Arch. Toxicol. 2016;90:1757–1761. doi: 10.1007/s00204-016-1689-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang Y., Li J., Pan X., Zhou P., Yu X., Cao H., Wang Y., Li L. Co-culture with mesenchymal stem cells enhances metabolic functions of liver cells in bioartificial liver system. Biotechnol. Bioeng. 2013;110:958–968. doi: 10.1002/bit.24752. [DOI] [PubMed] [Google Scholar]
- 115.Bircsak K.M., DeBiasio R., Miedel M., Alsebahi A., Reddinger R., Saleh A., Shun T., Vernetti L.A., Gough A. A 3D microfluidic liver model for high throughput compound toxicity screening in the OrganoPlate(R) Toxicology. 2021;450:152667. doi: 10.1016/j.tox.2020.152667. [DOI] [PubMed] [Google Scholar]
- 116.Chen Y., Gao D., Liu H., Lin S., Jiang Y. Drug cytotoxicity and signaling pathway analysis with three-dimensional tumor spheroids in a microwell-based microfluidic chip for drug screening. Anal. Chim. Acta. 2015;898:85–92. doi: 10.1016/j.aca.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 117.Ewart L., Apostolou A., Briggs S.A., Carman C.V., Chaff J.T., Heng A.R., Jadalannagari S., Janardhanan J., Jang K.-J., Joshipura S.R., et al. Qualifying a human Liver-Chip for predictive toxicology: performance assessment and economic implications. bioRxiv. 2021 doi: 10.1101/2021.12.14.472674. Preprint at. [DOI] [Google Scholar]