Significance
Predicting and optimizing protein–ligand affinity is a central problem in small-molecule drug design. The significant increase in the throughput of structural biology provides an exciting source of data that characterizes protein–ligand interactions. However, the missing piece is connecting these data to predictive models for ligand design. Here, we develop a machine learning framework that extracts physically meaningful descriptors of protein–ligand complexes and relates them to bioactivity. Our approach outperforms ligand-based or structure-based methods on data from a large-scale open science drug discovery campaign against the main protease of SARS-CoV-2. Moreover, we prospectively deployed our method to discover potent main protease inhibitors. Our method provides the key bridge that unlocks the power of high-throughput structural biology in drug design.
Keywords: machine learning, drug design, crystallography
Abstract
A common challenge in drug design pertains to finding chemical modifications to a ligand that increases its affinity to the target protein. An underutilized advance is the increase in structural biology throughput, which has progressed from an artisanal endeavor to a monthly throughput of hundreds of different ligands against a protein in modern synchrotrons. However, the missing piece is a framework that turns high-throughput crystallography data into predictive models for ligand design. Here, we designed a simple machine learning approach that predicts protein–ligand affinity from experimental structures of diverse ligands against a single protein paired with biochemical measurements. Our key insight is using physics-based energy descriptors to represent protein–ligand complexes and a learning-to-rank approach that infers the relevant differences between binding modes. We ran a high-throughput crystallography campaign against the SARS-CoV-2 main protease (MPro), obtaining parallel measurements of over 200 protein–ligand complexes and their binding activities. This allows us to design one-step library syntheses which improved the potency of two distinct micromolar hits by over 10-fold, arriving at a noncovalent and nonpeptidomimetic inhibitor with 120 nM antiviral efficacy. Crucially, our approach successfully extends ligands to unexplored regions of the binding pocket, executing large and fruitful moves in chemical space with simple chemistry.
Predicting protein–ligand affinity is a longstanding challenge that underpins computer-aided drug design. The challenge often lies in designing chemical modifications which would significantly improve the potency of a weakly potent starting point (hit-to-lead) or finding chemotypes that maintain potency while designing away other liabilities (lead optimization). Established medicinal chemistry heuristics focus on making interpretable and modest chemical changes, iteratively “morphing” the ligand to optimize interactions and explore unknown binding pockets (1, 2). Significant acceleration can be realized if this iterative process is replaced by methods which suggest large and synthetically facile changes to the ligand to lead to a significant increase in potency, motivating a computational approach to ligand design.
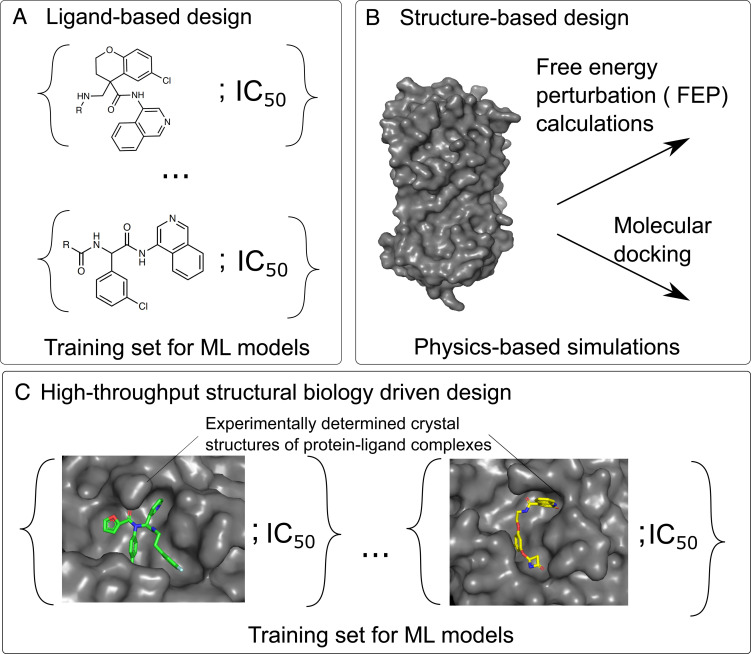
The plethora of computational methods in the literature can be organized in terms of available information they make use of. Ligand-based approaches (Fig. 1A) derive information only from the chemical identity of ligands which are binding to the protein and focus on learning the relationship between the chemical structure of the ligand and its activity. Such methods, however, are circumscribed by the problem of extrapolation: The model cannot extrapolate to regions of the binding site which are not already explored by molecules in the dataset, nor to unexplored interactions between novel chemotypes and the binding site.
Fig. 1.
Overview of approaches used in computer-aided drug design. Conventional approaches predict the activity of ligands (A) by learning the relationships between the molecular structure of a molecule and its activity or (B) through physics-based modeling using only the structure of the target and relying on methods such as free-energy perturbation (FEP) calculations or molecular docking. (C) Here, we demonstrated a strategy that exploits high-throughput crystallographic characterization of protein–ligand complexes to predict ligand affinity.
Structure-based approaches ameliorate this limitation by taking the protein structure into account and explicitly model the protein–ligand interactions (Fig. 1B). However, rigorous methods, such as free-energy perturbations (FEP) or alchemical free-energy calculations, generally require substantial computational resources and are constrained by the quality of the approximate forcefield (3–5). In practice, these approaches are typically used to compute relative free-energy changes of small modifications to predefined scaffolds (6, 7), as convergence time and error both increase with the size of change relative to the starting ligand. To reduce computational cost, empirical scoring functions such as docking (8–11) have been developed. While docking can identify hits from virtually screening large libraries (12, 13), it is typically not used in ligand optimization as the correlation between predicted and experimentally measured protein–ligand interaction energy is often weak.
The rapid acceleration in the throughput of structural biology unlocks a new source of data (Fig. 1C). Historically, protein structure determination was laborious; thus, on a particular target, there were only a handful of cocrystallized ligands reported in the literature. Although databases such as the Protein Data Bank (14) could be mined to parametrize docking algorithms (15, 16), this necessitates training on diverse classes of proteins with varied protein–ligand affinity measurement techniques, introducing noise and dataset bias (17). The synergy between modern robotic techniques for crystallization and crystal soaking (18), automated data analysis pipelines (19), and modern synchrotron infrastructure has increased the monthly throughput to up to hundreds of ligands against a target (20). However, the missing piece of the puzzle is a framework that can turn high-throughput crystallographic data into predictive models for ligand design.
In this paper, we present a machine learning approach that relates high-throughput crystallography data, represented as empirical energy terms, to measured bioactivity. We used this to accelerate the COVID Moonshot initiative (21), an open science consortium that reported over 200 protein–ligand complexes against SARS-CoV-2 main protease (MPro) with associated potency (IC50) measurements. Retrospective validation shows that our method outperforms ligand-based and structure-based approaches. We prospectively designed one-step library syntheses, improving the potency of two distinct micromolar hits by over 10-fold and arrived at a lead compound with 120 nM antiviral efficacy. Crucially, our designed inhibitors gain potency by extending to unsampled regions of the binding site, illustrating the ability of our model to generalize via the incorporating physical interactions.
Results and Discussion
Energy-Based Model Is Generalizable Across Chemical Space.
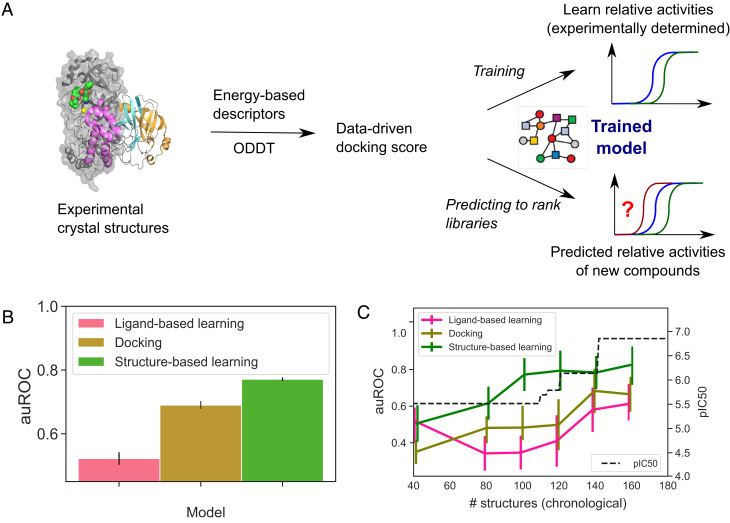
To describe protein–ligand structures as a fixed-length vector for downstream machine learning (Fig. 2A), we turn to the literature on empirical scoring functions. We use the terms of empirical energy function—hydrophobic, electrostatic, hydrogen bonding, etc.—as descriptors of the structures. Our hypothesis is that while empirical energy terms capture different aspects of protein–ligand interactions, how these interactions stack up to yield the free energy of binding depends on binding-site–specific variables such as binding-site flexibility. We further hypothesize that those protein-specific corrections are learnable from our dataset, comprising high-throughput structural biology data and associated bioactivity. To fix ideas, in our approach, we featurize all the protein–ligand complexes using the Open Drug Discovery Toolkit (ODDT) (22) and extract the Autodock Vina descriptors to represent the structures (Fig. 2A). The terms of the descriptor corresponded to affinity score, two Gaussian steric interaction terms, a repulsion term, hydrophobic interactions, and hydrogen interactions, as has been described in detail elsewhere (23, 24). Instead of predicting IC50 values directly, we focused our attention on a learn-to-rank approach that predicts the pairwise comparison of the ligands, choosing a cutoff of 0.5 log10 units for classifying one compound as more active than another (Fig. 2A). The threshold was chosen to match typical assay error. This approach allows us to combine qualitative (potency below measurable) and quantitative measurements and forces the model to ignore irrelevant experimental noise by ensuring that it is ranking only structures with demonstrably different bioactivity (25).
Fig. 2.
Structure-based learning outperforms docking and ligand-based machine learning in relative affinity predictions. (A) To build a model that captures the relationship between the crystal structure of a protein–ligand complex and the activity of the ligand, we extracted Autodock Vina descriptors from each crystal structure and deployed it to predict the relative potency of ligands. (B) Comparison of this approach (green) with ligand-based learning (pink) and docking (yellow) using data from the COVID Moonshot campaign and a classification task that evaluates pairwise comparisons between the activities of ligands using a cutoff of 0.5 log10 units for classifying one compound as more active than another. The average auROC scores were computed using a scaffold split, with the error bars corresponding to SE of the mean. Performance on individual scaffolds is shown in Dataset 4. (C) Time-split evaluation of our model. Our approach maintains its predictive power when trained on older, less potent molecules and asked to rank the affinities of newer more potent molecules (green line). Error bars correspond to SE of the mean. The black line tracks the potency of the most potent molecule discovered at that point.
Specifically, we applied this approach to the high-throughput structural biology campaign against the SARS-CoV-2 MPro, an essential protein in viral replication and a validated target for anticoronavirus therapeutics (26–29). All MPro clinical candidates to date are peptidomimetics inhibiting via a covalent mechanism, which are generally suboptimal for drug development. We launched the COVID Moonshot, an open science initiative aiming to develop noncovalent small-molecule oral antiviral (21). The campaign obtained 236 structures of noncovalent inhibitors binding to the MPro. Of these ligands, 94 had IC50 below 50 μM. To the best of our knowledge, COVID Moonshot is the only openly accessible dataset with over 100 structures of different ligands against a single target with associated bioactivity measurement; as such, our model evaluation will focus on this dataset.
To implement the models and evaluate their performance in ranking novel ligands, we use a scaffold-split approach where an entire scaffold is held out from training and placed in the test set (SI Appendix, Fig. S1). There are four salient chemical scaffolds in the dataset: aminopyridine-like, isoquinoline, benzotriazole, and quinolone (Materials and Methods and SI Appendix, Fig. S2) with 123, 44, 19, and 15 structures, respectively. For the compounds in the left-out test set, the features were extracted from docked structures (the ligands had been docked to the active site using OpenEye’s FRED hybrid docking mode as implemented in the “Classic OEDocking” floe on the Orion online platform; Materials and Methods) instead of the experimental crystal structures. This is because when deploying the model as a prioritization tool for synthesis and screening as we will do later in this work, experimental structural information is inaccessible, and the docked structure serves as its approximation. To find a well-performing model architecture, we explored a variety of traditional machine learning models (logistic regression, k-nearest neighbors, extra tree, and random forest) and performed a hyperparameter search for all (Materials and Methods). We found an optimized random forest-based architecture to show good performance across all of the chemical series both when the area under the receiver operating characteristic curve (auROC; Dataset 1) or the area under the precision-recall curve (auPRC; Dataset 2) was used as the performance metric. The high performance values both globally as well as for each of the chemical series separately illustrate that our approaches have the potential to accurately rank unseen ligands without the requirement to have any structures from that specific scaffold as part of the training set.
Structural Data Are Salient to Model Performance.
To understand the impact of experimental structural biology data, we consider two alternative models: i) a ligand-based model that relied only on the use of ligand-based descriptors providing no information about the protein-crystal structure and ii) a model that used a docked structure instead of the measured crystallographic structure. Specifically, for the former case, we featurized the ligands using Morgan fingerprints (30), implemented through the use of the RDKit package. For the latter case, we docked ligands to the active site using OpenEye’s FRED hybrid docking modes described above. For consistency, the same model architecture (random forest) was used for all cases with the hyperparameters tuned separately for each model.
Fig. 2B shows that our approach which incorporates experimental structural data outperforms both docking-based and ligand-based models, highlighting the importance of high-throughput structural biology. The auROC values correspond to the average values across the four chemical series. The elevated performance of the docking-based model over the ligand-based one likely stems from the fact that the model does not rely solely on ligand-based input but also incorporates information about the protein–ligand interactions.
Finally, with access to both experimentally determined and docked structures of the protein–ligand complexes, we set out to examine in more detail how the accuracy of the docking step affects model performance. To this effect, we first noted that the docked and experimental structures differed on average by around 1.5 Å as quantified by heavy atom root mean squared displacement (RMSD; SI Appendix, Fig. S3), with there being no significant difference between the four chemical series (P < 0.01 using the Mann–Whitney test). We then trained models identically to the docking-based learning approach but constrained training data to these docked structures that closely matched the real experimental structures as quantified by the RMSD values. We found the performance to decline when a larger mismatch between the two types of data was allowed (SI Appendix, Fig. S4), in particular for the aminopyridine series, which was the worst performing chemical series when all data were included (Dataset 4). In parallel, we noticed that performance was generally best when the validation data were restricted to cases where there was a good match between docked and crystal structures (SI Appendix, Fig. S5). Taken together, these results suggest that good agreement between the experimental and the docked structures helps to ensure good predictive capabilities, likely because fully energy-driven approaches, such as docking, aim to be generic and have thus limited capability to generalize effectively to every target. The integration of real protein-specific data, in this case, through crystallographic protein–ligand structures, enables the model to learn protein-specific information in parallel with global interactions. The results further suggest that pose prediction and energy prediction should be considered as dual tasks, and an approach that combines high-throughput crystallography with a high-throughput biological assay can supply both sources of data to train docking scoring function.
Pretraining on Larger Protein–Ligand Complex Datasets Does Not Increase Performance But Improves the Robustness of the Predictions.
With the size of the experimental data here being modest (around 200 crystal structures with parallel activity measurements), we next set out to investigate whether the predictive power of the structure-driven modeling strategy increases even further by first pretraining a model on a larger dataset of experimentally determined protein–ligand structures and then fine-tuning the model to our data. Such transfer-learning approaches are actively pursued in the context of a variety of areas from image processing to speech recognition, natural language processing, and protein function prediction (31). Specifically, in the context of protein–ligand binding, fine-tuning has been shown to increase performance on specific protein families after having first trained a universal protein–ligand binding prediction model (32). To evaluate possible gains for our task, we turned to the latest version (v2020) of the PDBBind (33) and its refined dataset that had filtered all the protein–ligand complexes in the PDB Database with parallel activity measurements down to a set of 5318 complexes with well-resolved structures and good-quality measurements on the binding data as described in detail by Liu et al. (34).
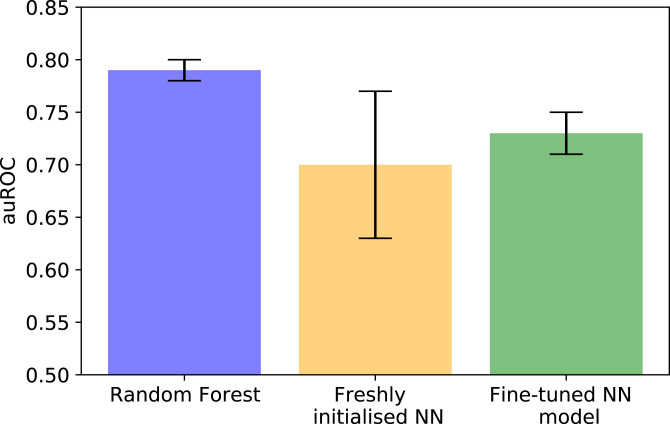
We used the ODDT package (22) to extract the descriptors for all of the structures and, similarly to before, trained a model that would learn to rank the structures based on the relative activity of the ligands. Specifically, we compared three cases: i) our previously found top-performing model (random forest), ii) a freshly initialized neural network, and iii) a neural network pretrained on PDBBind (Fig. 3). To compare the approaches reliably, we performed 15 different runs and calculated the average performance across these. Our results suggested that the freshly initialized neural network, on average, did not outperform the random forest-based architecture, which is likely a direct effect of it being challenging to train highly generalizable deep learning networks in a low-data regime. Pretraining on PDBBind followed by fine-tuning to our data also did not result in a model that would outperform the random forest-based approach, but it did allow us to substantially reduce the variability in performance compared to a case where a similar neural network-based architecture was initialized freshly.
Fig. 3.
Effect of pretraining on PDBBind on model performance. Area under the receiver–operator characteristic curve (auROC) when learning the relative binding affinities of ligands to a SARS-CoV-2 main protease using a random forest-based architecture (blue), a neural network-based architecture (yellow), and by using PDBBind to train a model before fine-tuning it (green). The error bars correspond to the SD in the auROC value across 15 different runs. Fine-tuning improves robustness compared to a freshly initialized neural network, but global performance remains below what was achieved with a random forest-based architecture.
Model Maintains Performance Throughout the Campaign.
Having demonstrated that our proposed strategy can reliably rank ligands by potency even when outside the chemical space that it encountered during training (i.e., for a new scaffold), we next explore how its utility varies through the campaign and what is the amount of structural data required for efficient performance. As a drug discovery campaign progresses, more knowledge about the chemical attributes that determine the binding of a ligand to its target is gathered, and the designs are honed accordingly. Therefore, trying to predict the potency of molecules tested earlier in the campaign with molecules tested later in the campaign as the training set is much easier (and less useful) than the converse, i.e., hindsight is usually much more accurate than foresight.
To examine the predictive capability of the structure-driven ligand prioritization approach as the campaign progresses, we use a time-split strategy. We ordered the compounds by the time when they had been tested and used only the structures available until that specific time point for training. We first note that in the course of the campaign, the potency of the molecules increased by many orders of magnitude (Fig. 2C, black line). To avoid susceptibility of the model to memorize the specifics of a particular scaffold, we kept all aminopyridine-like and isoquinoline-like molecules in the training data while using benzotriazole-like and quinolone-like ligands for validation. Fig. 2C (green line) shows that our structure-based model remains predictive when trained on molecules tested early in the campaign and deployed to rank molecules tested later. This is in contrast to the model that relied on ligand-based or docking-based input (Fig. 2C, pink and dark yellow lines).
Finally, from these data, we can interrogate that our proposed approach performs effectively (auROC value above 0.7) when only around 100 crystal structures are available to train the model. Crucially, this is a throughput that could be achieved in a modern synchrotron (20) on a monthly timescale, illustrating that our proposed strategy has the potential to be exploited routinely in the context of drug discovery campaigns.
Model-Guided Library Synthesis Discovers Potent Leads.
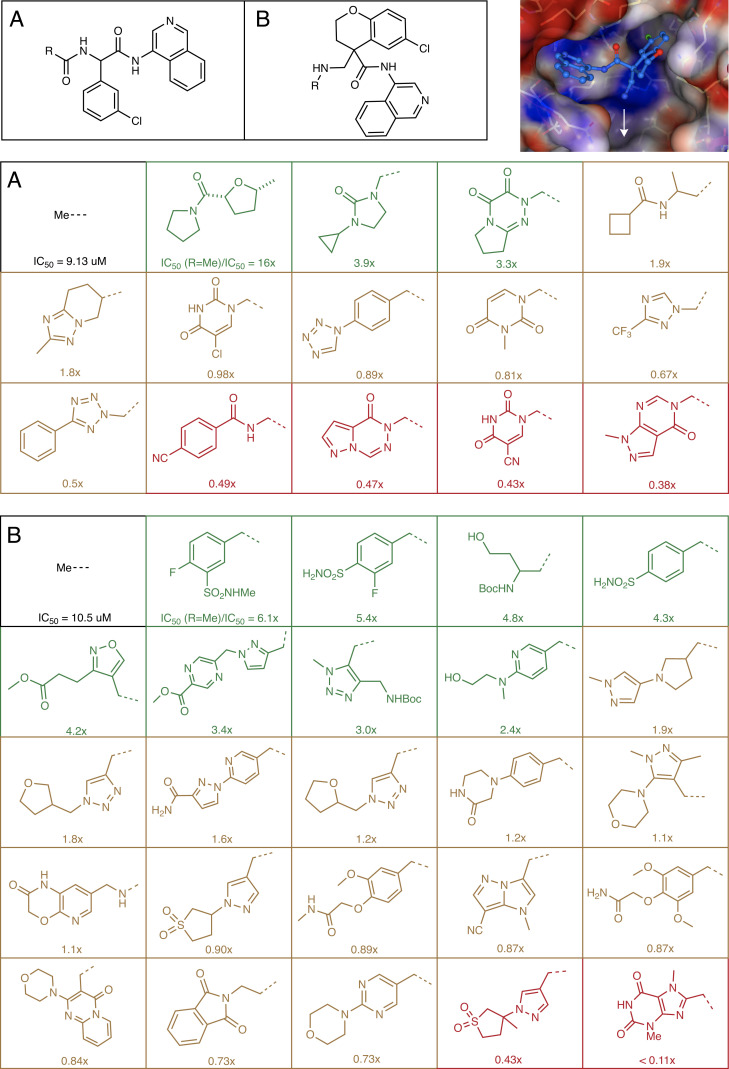
To apply our model to lead discovery, we need to generate protein–ligand structures for unseen ligands. Starting from two hits with an amine handle reported by the COVID Moonshot Consortium (35), chosen because they have detectable potency and ease of synthetic access, we generate a virtual library that is synthesizable in a single reaction step using amide formation (Fig. 4A) and reductive amination (Fig. 4B). The library design is motivated by structural data, aiming to extend the hit into the unoccupied P1’ binding pocket (Fig. 4, Top Right). Using the Manifold platform (postera.ai/manifold), we select carboxylic acid and aldehydes that are in-stock building blocks in Enamine (a synthetic chemistry CRO with one of the largest building block collections onsite) with the building blocks further filter based on predicted reactivity and the final compound having clog P < 3. In total, there are 15,720 compounds in the amide virtual library and 2,664 compounds in the reductive amination library.
Fig. 4.
Structure-enabled machine learning identified multiple, more potent ligands in a small number of synthesized compounds. We applied the model to design a reductive amination library (A) and amidation library (B), which is screened against the main protease inhibition using a fluorescence assay. The library is designed to extend into the untapped P1’ binding pocket of the active site. The structure shown is Mpro-x11498 from Diamond/XChem (35).
We then generated a predicted binding pose by constrained docking into the binding site using existing structural data in the isoquinoline series as the constraints (Materials and Methods) and used our trained structure-based learn-to-rank model to rank the ligands in this virtual library. We used a random forest-based model trained on the full dataset with hyperparameters fixed to the set that was found to work best across all the four chemical series (Fig. 2B; { nestimators = 100, max_depth = 3, max_features = 100, min_samples_leaf = 2}. Specifically, each of the docked poses is ranked against the top 5 most potent noncovalent binders in the dataset (SI Appendix, Fig. S6) and the mean of the five predictions estimated to generate the final ranking. Top 18 compounds from the final ranking were selected from the amide formation library with 15 successfully synthesized and top 32 from the reductive amination library with 25 successfully synthesized. We have included all the chemical structures in Dataset 5. This 80% success rate in synthesis could be improved on by considering reaction yield as part of the library design, which could be done using a mechanistic descriptor (36) or building block-based reactivity score (37).
The successfully synthesized 40 compounds were then assessed for inhibition of Mpro activity using a biochemical assay with a fluorescence-based readout (25, 35) (Dataset 6). Fig. 4 A and B shows that around 30% of the library has potency that is that greater than 2x compared to the reference. These compounds are all substantial changes to the hit, in some cases doubling the atom count, and reaching the ligand into unknown regions of the binding site while remaining a low logP. The high hit rate suggests that the model can accurately prioritize these chemotypes. The inclusion of more and less potent compounds from this round of testing could be integrated with the training data in the next iteration to further improve the predictive capabilities of the model.
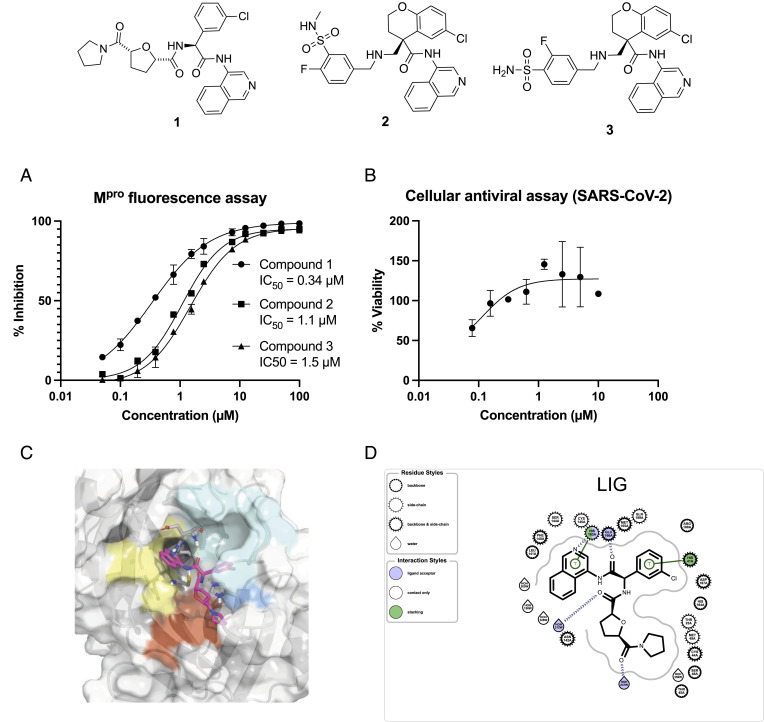
Finally, we characterized the most potent leads in vitro and in cells after having resolved the enantiomers/diastereoisomers. Fig. 5A shows that our top enantiopure compound, compound 1, achieved nM potency in the florescence Mpro assay. Compound 1 was further profiled in the SARS-CoV-2 antiviral assay (CPE assay in Vero E6 cells; Fig. 5B), attaining EC50 = 120 nM. Compound 1 displayed no cytotoxicity effect against Vero E6 cells at 10 μM. We experimentally determined the structure of its protein–ligand complex, confirming that the hit compound reaches into P1’ pocket (Fig. 5 C and D). Table 1 summarizes the in vitro ADME properties of compound 1, showing that it is largely unbound in plasma; it exhibits excellent in vitro metabolic stability in human and rat but suffers from poor permeability. Compound 1 is a potential starting point for the development of antiviral therapeutics.
Fig. 5.
Lead compounds are starting points for antiviral development. (A) Top hits reported in Fig. 4 are purified and assayed in an enzymatic assay for their activity against the SARS-CoV-2 main protease. The 90% CI for enzymatic assays of compound 1 [0.26, 0.42] μM, compound 2 [0.98, 1.2] μM, and compound 3 [1.4, 1.6] μM; for viral assay: [0.019, 0.83] μM. (B) Compound 1, with nM potency, is further profiled in the SARS-CoV-2 CPE assay in Vero E6 cells and found to show no cytotoxic effect at 10 μM. (C) Crystallographic structure of compound 1 bound to Mpro (structure P1477_0A) (35) and (D) 2D map of salient protein–ligand interactions obtained using OpenEye.
Table 1.
Compound 1 exhibits promising in vitro ADME properties
| Summary of biochemical and ADME properties of 1 | |
|---|---|
| IC50 (μ M) | 0.34 |
| EC50 (VeroE6, SARS-CoV-2) (μ M) | 0.12 |
| Cytotoxicity CC50 (VeroE6) (μ M) | > 10 |
| log D | 0.90 |
| PPB (human, rat) | 68.5%, 58.7% |
| MDCK-MDR1 Papp A-to-B (10−6 cm/s) | 0.20 |
| MDCK-MDR1 Papp B-to-A (10−6 cm/s) | 0.60 |
| MDCK-LE Papp A-to-B (10−6 cm/s) | 0.30 |
| Liver hepatocytes CLint (human, rat) (μg/min/106 cells) | 4.40, 4.00 |
| Liver hepatocytes t1/2 (human, rat) (min) | 158, 172 |
| Liver microsome CLint (human, rat) (μl/min/mg) | < 10.0, < 10.0 |
| Liver microsome t1/2 (human, rat) (min) | > 139, > 139 |
The table shows the measured lipophilicity, plasma protein binding, permeability, and metabolic stability of the compound.
Conclusion
With the crystal structures of protein–ligand complexes being acquired at an increasing throughput, here, we show how these data can be used to power an approach to computational ligand design by using physics-inspired empirical energy terms as a descriptor of the protein–ligand complex. We focus on the COVID Moonshot Initiative, which reported an unprecedentedly rich dataset of 200 ligands for which both their activity and structure of binding to the main protease of SARS-CoV-2 had been determined. We developed a machine learning model that learned the relationship between the multidimensional docking score extracted from the crystal structure and the relative bioactivity of ligands. The approach maintained a high and robust performance (auROC of 0.79), even when making predictions outside the training scaffold. It also yielded powerful results in a prospective campaign, increasing the potency of hit compounds by more than 10x with simple chemistry that extends the hits to an unsampled region of the binding site. Our approach arrived at a lead compound with 120 nM antiviral efficacy.
Materials and Methods
Dividing the Molecules by Chemical Series.
In order to reliably estimate the performance of our developed model, we performed the train:test splits in a scaffold-stratified manner. To this effect, four distinct scaffold categories were defined—aminopyridine-like, isoquinoline, benzotriazole, and quinolone—and each compound was classified as belonging to one of them by using SMARTS to define chemical substructures. SI Appendix, Fig. S5 shows representative examples of each chemical series, highlighting with the chemical substructure that gives rise to the name.
Model Development.
All models explored in this paper utilized a learn-to-rank approach where the data were divided into a training set and a test set with compounds from the chemical series always kept in the same group. After this split, all possible pairs between compounds in each of the two sets were generated. For each pair, the difference in their pIC50 value as well as between the descriptors evaluated as has been illustrated in SI Appendix, Fig. S1. Compounds that were determined to be inactive were included when forming pairs, and they were all allocated activities equal to the highest measured activity values.
To find optimally performing models, extra tree classifier, random forest, k-nearest neighbors, and neural network-based architectures were considered. All the models were trained using the scikit-learn package (38), and the list of hyperparameters considered for each architecture are shown in Dataset 3. The optimal set of hyperparameters for each model was arrived to be monitoring the average auROC value across the four chemical series.
Pretraining on PDBBind.
The PDBBind (33) refined dataset, v2020; 5318 structures; construction described by Liu et al. (34) was used to train a learn-to-rank predictor linking the activity of a ligand to the descriptors extracted from the corresponding protein–ligand complex structure. We trained networks of multiple linear layers with the Relu activation function using Adam optimizer and dropout regularization at each layer. The model hyperparameters (number of layers, dimensions of the layers, the learning rate, and the dropout probability) were chosen via the hyperband algorithm (a variation of random parameter searching) on a random 3% of the data. After training on the data for 20 epochs, the final linear layer of the neural network was reinitialized for fine-tuning on the SARS-CoV-2 data, while the weight parameters for other layers were kept fixed. The impact of transfer learning was evaluated by making a copy of the model and completely reinitialized the model weights before fitting directly to the SARS-CoV-2 data.
Docking Experiments.
We redocked all compounds synthesized by The COVID Moonshot Consoritium against x2908 structure reported by Diamond XChem. We use the “Classic OEDocking” floe v0.7.2 as implemented in the Orion 2020.3.1 Academic Stack (OpenEye Scientific). Omega was used to enumerate conformations (and expand stereochemistry) with up to 500 conformations. FRED was used for docking in HYBRID mode using the x2908 bound ligand. The docked poses are available on GitHub.
Fluorescence MPro Inhibition Assay.
The method is described previously (35). Compounds were seeded into assay-ready plates (Greiner 384 low volume, cat 784900) using an Echo 555 acoustic dispenser, and DMSO was back-filled for a uniform concentration in assay plates (DMSO concentration maximum 1%). Screening assays were performed in duplicate at 20 μM and 50 μM. Hits of greater than 50% inhibition at 50 μM were confirmed by dose–response assays. Dose–response assays were performed in 12-point dilutions of 2-fold, typically beginning at 100 μM. Highly active compounds were repeated in a similar fashion at lower concentrations beginning at 10 μM or 1 μM. Reagents for the Mpro assay were dispensed into the assay plate in 10-μl volumes for a final volume of 20 μM.
Final reaction concentrations were 20 mM HEPES pH7.3, 1.0 mM TCEP, 50 mM NaCl, 0.01% Tween-20, 10% glycerol, 5 nM Mpro, and 375 nM fluorogenic peptide substrate [5-FAM]-AVLQSGFR-[Lys(Dabcyl)]-K-amide. Mpro was preincubated for 15 min at room temperature with the compound before the addition of the substrate and a further 30-min incubation. Protease reaction was measured in a BMG Pherastar FS with a 480/520 ex/em filter set. Raw data were mapped and normalized to high (Protease with DMSO) and low (No Protease) controls using Genedata Screener software. Normalized data were then uploaded to CDD Vault (Collaborative Drug Discovery). Dose–response curves were generated for IC50 using nonlinear regression with the Levenberg–Marquardt algorithm with minimum inhibition = 0% and maximum inhibition=100%.
SARS-CoV-2 Antiviral Assay.
The method is described previously (35). SARS-CoV-2 (GISAID accession EPI_ISL_406862) was kindly provided by Bundeswehr Institute of Microbiology, Munich, Germany. Virus stocks were propagated (4 passages) and tittered on Vero E6 cells. Handling and working with SARS-CoV-2 virus were conducted in a BSL3 facility in accordance with the biosafety guidelines of the Israel Institute for Biological Research (IIBR). Vero E6 cells were plated in 96-well plates and treated with compounds in medium containing 2% fetal bovine serum. The assay plates containing compound dilutions and cells were incubated for 1 h at a temperature of 37 °C temperature prior to adding multiplicity of infection (MOI) 0.01 of viruses. Viruses were added to the entire place. This included control wells that included the virus but not the test compound or the Remdesivir drug used as positive control. After 72-h incubation, the viral cytopathic effect (CPE) inhibition assay was measured with XTT reagent. Three replicate plates were used. We note that compounds were assayed as enantiomers or diastereoisomers in this initial triage.
Chemical Synthesis.
All compounds were purchased from Enamine and are available in their catalog without any restriction/exclusivity. Enamine catalog ID of the compounds assayed and a description of the synthesis protocol as provided by Enamine are provided in SI Appendix.
In Vitro ADME Assays.
All assays were provided as in-kind contributions by Novartis International AG to the COVID Moonshot Consortium and summarized in (35).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (DOCX)
Acknowledgments
The research leading to these results has received funding from the Schmidt Science Fellows program in partnership with the Rhodes Trust (KLS). Research reported in this publication was supported in part by NIAID of the National Institutes of Health under award number U19AI171399. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The Chodera laboratory receives or has received funding from multiple sources, including the NIH, the NSF, the Parker Institute for Cancer Immunotherapy, Relay Therapeutics, Entasis Therapeutics, Silicon Therapeutics, EMD Serono (Merck KGaA), AstraZeneca, Vir Biotechnology, Bayer, XtalPi, Interline Therapeutics, the Molecular Sciences Software Institute, the Starr Cancer Consortium, the Open Force Field Consortium, Cycle for Survival, a Louis V. Gerstner Young Investigator Award, and the Sloan Kettering Institute. A complete funding history for the Chodera lab can be found at http://choderalab.org/funding. A.A.L. holds a Royal Society University Research Fellowship.
Author contributions
K.L.S. and A.A.L. designed research; K.L.S., W.M., D.F., M.B., H.B., A.B.-S., and T.C. performed research; K.L.S., W.M., D.F., M.B., N.L., F.V.D. and J.D.C. contributed new reagents/analytic tools; K.L.S., W.M., and M.B. analyzed data; and K.S. and A.A.L. wrote the paper.
Competing interests
K.L.S. is a consultant to and owns equity in Transition Bio. N.L. is on the SAB of Monte Rosa Therapeutics and Larkspur Biosciences and is a consultant for FoRx therapeutics and Outrun therapeutics. J.D.C. is on the Scientific Advisory Board of OpenEye, Interline Therapeutics, Redesign Science, and Ventus Therapeutics and owns equity in Interline Therapeutics and Redesign Science. A.A.L. is a cofounder and shareholder of PostEra Inc. and Byterat Ltd, and A.A.L. has substantial stock ownership in PostEra Inc. F.V.D.’s Small Research Facility at U. Oxford provides paid-for research collaboration services to A.A.L.’s company PostEra Inc.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Alpha A. Lee, Email: aal44@cam.ac.uk.
Collaborators: Matthew C. Robinson, Nir London, Efrat Resnick, Daniel Zaidmann, Paul Gehrtz, Rambabu N. Reddi, Ronen Gabizon, Haim Barr, Shirly Duberstein, Hadeer Zidane, Khriesto Shurrush, Galit Cohen, Leonardo J. Solmesky, Alpha Lee, Andrew Jajack, Milan Cvitkovic, Jin Pan, Ruby Pai, Emily Grace Ripka, Luong Nguyen, Mikhail Shafeev, Tatiana Matviiuk, Oleg Michurin, Eugene Chernyshenko, Vitaliy A. Bilenko, Serhii O. Kinakh, Ivan G. Logvinenko, Kostiantyn P. Melnykov, Victor D. Huliak, Igor S. Tsurupa, Marian Gorichko, Aarif Shaikh, Jakir Pinjari, Vishwanath Swamy, Maneesh Pingle, Sarma BVNBS, Anthony Aimon, Frank von Delft, Daren Fearon, Louise Dunnett, Alice Douangamath, Alex Dias, Ailsa Powell, Jose Brandao Neto, Rachael Skyner, Warren Thompson, Tyler Gorrie-Stone, Martin Walsh, David Owen, Petra Lukacik, Claire Strain-Damerell, Halina Mikolajek, Sam Horrell, Lizbé Koekemoer, Tobias Krojer, Mike Fairhead, Elizabeth M. MacLean, Andrew Thompson, Conor Francis Wild, Mihaela D. Smilova, Nathan Wright, Annette von Delft, Carina Gileadi, Victor L. Rangel, Chris Schofield, Eidarus Salah, Tika R. Malla, Anthony Tumber, Tobias John, Ioannis Vakonakis, Anastassia L. Kantsadi, Nicole Zitzmann, Juliane Brun, J. L. Kiappes, Michelle Hill, Karolina D Witt, Dominic S Alonzi, Laetitia L Makower, Finny S. Varghese, Gijs J. Overheul, Pascal Miesen, Ronald P. van Rij, Jitske Jansen, Bart Smeets, Susana Tomésio, Charlie Weatherall, Mariana Vaschetto, Hannah Bruce Macdonald, John D. Chodera, Dominic Rufa, Matthew Wittmann, Melissa L. Boby, Michael Henry, William G. Glass, Peter K. Eastman, Joseph E. Coffland, David L. Dotson, Ed J. Griffen, Willam McCorkindale, Aaron Morris, Robert Glen, Jason Cole, Richard Foster, Holly Foster, Mark Calmiano, Rachael E. Tennant, Jag Heer, Jiye Shi, Eric Jnoff, Matthew F.D. Hurley, Bruce A. Lefker, Ralph P. Robinson, Charline Giroud, James Bennett, Oleg Fedorov, St Patrick Reid, Melody Jane Morwitzer, Lisa Cox, Garrett M. Morris, Matteo Ferla, Demetri Moustakas, Tim Dudgeon, Vladimír Pšenák, Boris Kovar, Vincent Voelz, Anna Carbery, Alessandro Contini, Austin Clyde, Amir Ben-Shmuel, Assa Sittner, Boaz Politi Einat B. Vitner, Elad Bar-David, Hadas Tamir, Hagit Achdout, Haim Levy, Itai Glinert, Nir Paran, Noam Erez, Reut Puni, Sharon Melamed, Shay Weiss, Tomer Israely, Yfat Yahalom-Ronen, Adam Smalley, Vladas Oleinikovas, John Spencer, Peter W. Kenny, Walter Ward, Emma Cattermole, Lori Ferrins, Charles J. Eyermann, Bruce F. Milne, Andre S. Godoy, Gabriela D. Noske, Glaucius Oliva, Rafaela S. Fernandes, Aline M. Nakamura, Victor O. Gawriljuk, Kris M. White, Briana L. McGovern, Romel Rosales, Adolfo Garcia-Sastre, Daniel Carney, Edcon Chang, Kumar Singh Saikatendu, Laura Vangeel Johan Neyts, Kim Donckers, Dirk Jochmans, Steven De Jonghe, Gregory R. Bowman, Bruce Borden, Sukrit Singh, Andrea Volkamer, Jaime Rodriguez-Guerra, Gwen Fate, Storm Hassell Hart, Vitaliy A. Bilenko, Serhii O. Kinakh, Ivan G. Logvinenko, Kostiantyn P. Melnykov, Victor D. Huliak, Igor S. Tsurupa, Kadi L Saar, Benjamin Perry, Laurent Fraisse, Peter Sjö, Pascale Boulet, Sophie Hahn, Charles Mowbray, Lauren Reid, Paul Rees, Qiu Yu Judy Huang, Sarah N Zvornicanin, Ala M. Shaqra, Nese Kurt Yilmaz, Celia A. Schiffer, Ivy Zhang, Iván Pulido, Charlie Tomlinson, Jenny C. Taylor, Tristan Ian Croll, and Lennart Brwewitz
Data, Materials, and Software Availability
Source code and data required to reproduce this study are available at https://github.com/kadiliissaar/ligand_design_structural_biology.
Supporting Information
References
- 1.Topliss J. G., Utilization of operational schemes for analog synthesis in drug design. J. Med. Chem. 15, 1006–1011 (1972). [DOI] [PubMed] [Google Scholar]
- 2.Awale M., Hert J., Guasch L., Riniker S., Kramer C., The playbooks of medicinal chemistry design moves. J. Chem. Inf. Mod. 61, 729–742 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Sherborne B., et al. , Collaborating to improve the use of free-energy and other quantitative methods in drug discovery. J. Comput.-Aided Mol. Des. 30, 1139–1141 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Abel R., Wang L., Harder E. D., Berne B., Friesner R. A., Advancing drug discovery through enhanced free energy calculations. Acc. Chem. Res. 50, 1625–1632 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Schindler C. E., et al. , Large-scale assessment of binding free energy calculations in active drug discovery projects. J. Chem. Inf. Mod. 60, 5457–5474 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Schindler C. E., et al. , Large-scale assessment of binding free energy calculations in active drug discovery projects. J. Chem. Inf. Mod. 60, 5457–5474 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Gapsys V., et al. , Large scale relative protein ligand binding affinities using non-equilibrium alchemy. Chem. Sci. 11, 1140–1152 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.G. Wang, W. Zhu, Molecular docking for drug discovery and development: A widely used approach but far from perfect (2016). [DOI] [PubMed]
- 9.S. S. Schweiker, S. M. Levonis, Navigating the intricacies of molecular docking (2019). [DOI] [PubMed]
- 10.Pinzi L., Rastelli G., Molecular docking: Shifting paradigms in drug discovery. Int. J. Mol. Sci. 20, 4331 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.R. N. Dos Santos, L. G. Ferreira, A. D. Andricopulo, “Practices in molecular docking and structure-based virtual screening” in Computational Drug Discovery and Design (Springer, 2018), pp. 31–50. [DOI] [PubMed]
- 12.Lyu J., et al. , Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorgulla C., et al. , An open-source drug discovery platform enables ultra-large virtual screens. Nature 580, 663–668 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berman H. M., et al. , The protein data bank. Nucleic Acids Res. 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballester P. J., Mitchell J. B., A machine learning approach to predicting protein-ligand binding affinity with applications to molecular docking. Bioinformatics 26, 1169–1175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. T. McNutt et al., GNINA 1.0: Molecular docking with deep learning. J. Chem. 13, 1–20 (2021). [DOI] [PMC free article] [PubMed]
- 17.Sieg J., Flachsenberg F., Rarey M., In need of bias control: Evaluating chemical data for machine learning in structure-based virtual screening. J. Chem. Inf. Mod. 59, 947–961 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Douangamath A., et al. , Achieving efficient fragment screening at XChem facility at diamond light source. J. Vis. Exp.: Jove, e62414 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Pearce N. M., et al. , A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density. Nat. Commun. 8, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Douangamath A., et al. , Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 11, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chodera J., Lee A. A., London N., von Delft F., Crowdsourcing drug discovery for pandemics. Nat. Chem. 12, 581–581 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Wójcikowski M., Zielenkiewicz P., Siedlecki P., Open drug discovery toolkit (ODDT): A new open-source player in the drug discovery field. J. Chem. 7, 1–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quiroga R., Villarreal M. A., Vinardo: A scoring function based on autodock vina improves scoring, docking, and virtual screening. PloS One 11, e0155183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trott O., Olson A. J., Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris A., et al. , Discovery of SARS-CoV-2 main protease inhibitors using a synthesis-directed de novo design model. Chem. Commun. 57, 5909–5912 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S. H., An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: Peptidomimetics and small molecule chemotherapy. J. Med. Chem. 59, 6595–6628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ullrich S., Nitsche C., The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 30, 127377 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cannalire R., Cerchia C., Beccari A. R., Di Leva F. S., Summa V., Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: State of the art and future opportunities. J. Med. Chem. 65, 2716–2746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh A. K., Brindisi M., Shahabi D., Chapman M. E., Mesecar A. D., Drug development and medicinal chemistry efforts toward SARS-coronavirus and COVID-19 therapeutics. Chem. Med. Chem. 15, 907–932 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers D., Hahn M., Extended-connectivity fingerprints. J. Chem. Inf. Mod. 50, 742–754 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Niu S., Liu Y., Wang J., Song H., A decade survey of transfer learning (2010–2020). IEEE Trans. Artif. Intell. 1, 151–166 (2020). [Google Scholar]
- 32.Imrie F., Bradley A. R., van der Schaar M., Deane C. M., Protein family-specific models using deep neural networks and transfer learning improve virtual screening and highlight the need for more data. J. Chem. Inf. Mod. 58, 2319–2330 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Liu Z., et al. , PDB-wide collection of binding data: Current status of the PDBbind database. Bioinformatics 31, 405–412 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Liu Z., et al. , Forging the basis for developing protein-ligand interaction scoring functions. Acc. Chem. Res. 50, 302–309 (2017). [DOI] [PubMed] [Google Scholar]
- 35.H. Achdout et al., Open science discovery of oral non-covalent SARS-CoV-2 main protease inhibitor therapeutics. bioRxiv pp. 2020–10 (2021).
- 36.Haas B. C., Goetz A. E., Bahamonde A., McWilliams J. C., Sigman M. S., Predicting relative efficiency of amide bond formation using multivariate linear regression. Proc. Natl. Acad. Sci. U.S.A. 119, e2118451119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grygorenko O. O., et al. , Generating multibillion chemical space of readily accessible screening compounds. Iscience 23, 101681 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.L. Buitinck et al., “API design for machine learning software: Experiences from the scikit-learn project” in ECML PKDD Workshop: Languages for Data Mining and Machine Learning (2013), pp. 108–122.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (DOCX)
Data Availability Statement
Source code and data required to reproduce this study are available at https://github.com/kadiliissaar/ligand_design_structural_biology.