Abstract
Amyotrophic Lateral Sclerosis (ALS) is a heterogenous neurodegenerative disorder affecting motor neurons and voluntary muscle control1. ALS heterogeneity includes the age of manifestation, the rate of progression, and the anatomical sites of symptom onset. Disease-causing mutations in specific genes have been identified and define different subtypes of ALS1. Despite several ALS-associated genes have been shown to affect immune functions2, whether specific immune features account for ALS heterogeneity is poorly understood. ALS4 is characterized by juvenile-onset and slow progression3. ALS4 patients show motor difficulties by their 30s, and most of them require walking assistance devices by their 50s. ALS4 is caused by mutations in the gene SETX. Using Setx knock-in (KI) mice carrying the ALS4 causative L389S mutation, we discovered an immunological signature consisting of clonally expanded, terminally differentiated effector memory (TEMRA) CD8 T cells in the central nervous system and blood of KI animals. Increased frequencies of antigen-specific CD8 T cells in KI mice mirror motor neuron disease progression, and correlate with anti-glioma immunity. Furthermore, bone marrow transplantation experiments indicate a key role of the immune system in ALS4 neurodegeneration. In ALS4 patients, clonally expanded TEMRA CD8 T cells circulate in the peripheral blood. Our results provide evidence of an antigen-specific CD8 T cell response in ALS4, which can serve to unravel disease mechanisms and as a potential biomarker of disease state.
Amyotrophic Lateral Sclerosis (ALS) is the most common form of motor neuron disease (MND). While the principal pathological manifestation of ALS is the death of motor neurons in the motor cortex and spinal cord1, several reports have highlighted a link between some disease-causing genes and the immune system. Studies in mice expressing a mutant pathogenic SOD1 (Superoxide Dismutase 1, mSOD1) have shown that deletion of the transgene in the myeloid compartment or in astrocytes slows disease progression4,5, and suggested the contribution of peripheral immune cells in neurodegeneration6. The C9ORF72 (Chromosome 9 open reading frame 72) locus harbors a G4C2 intronic repeat polymorphism that, through disease-linked repeat expansion, is the most common cause of familial ALS. C9ORF72 is mainly expressed by splenic CD11b+ cells and microglia, in which it controls inflammatory responses7,8. Knock-out (KO) of C9orf72 in mice results in a generalized autoimmune phenotype8–10. Mutations in TBK1 (TANK-binding kinase 1) and other genes with a known role in autophagy and innate immune signaling, have been recently associated with ALS11. Interestingly, TBK1 deficiency in dendritic cells (DCs) also leads to autoimmunity in mice12.
ALS4 is caused by autosomal dominant mutations in the SETX gene, which encodes for the Senataxin protein, a ubiquitously expressed nuclear ATP-dependent DNA/RNA helicase 13,14. Loss of function studies have shown that Senataxin is implicated in RNA metabolism and type I interferon (IFN-I) responses upon infection13,14. Whether SETX missense mutations that cause ALS4 have a role in immunity is unknown. In this study we set out to investigate this question.
Immunophenotyping of KI mice
We first tested whether mutated forms of SETX linked to ALS4 might lead to inflammatory dysfunctions. For this, we utilized Setx-L389S+/− knock-in (hereinafter called KI) mice, which carry the most common ALS4-associated mutation, L389S, into the highly conserved endogenous mouse gene, and develop motor abnormalities starting at the age of 8 months (Extended Data Fig.1a and15). We differentiated bone marrow derived dendritic cells (BMDCs) from Setx KI, wild-type (WT), knock-out (KO) and heterozygous (HET) littermates and infected them in vitro with the Gram negative bacterium Citrobacter rodentium, which is a natural rodent pathogen that induces a strong IFN-I response16. By measuring the expression of inflammatory genes by real-time quantitative polymerase chain reaction (RT-qPCR) (Extended Data Fig.1b), we found that, consistently with previous report14 Setx KO cells showed a 4 to 5-fold increase in Il6 and Ifnb transcripts as compared to their HET counterparts, while the expression level of inflammatory genes detected in KI and WT BMDCs was almost identical.
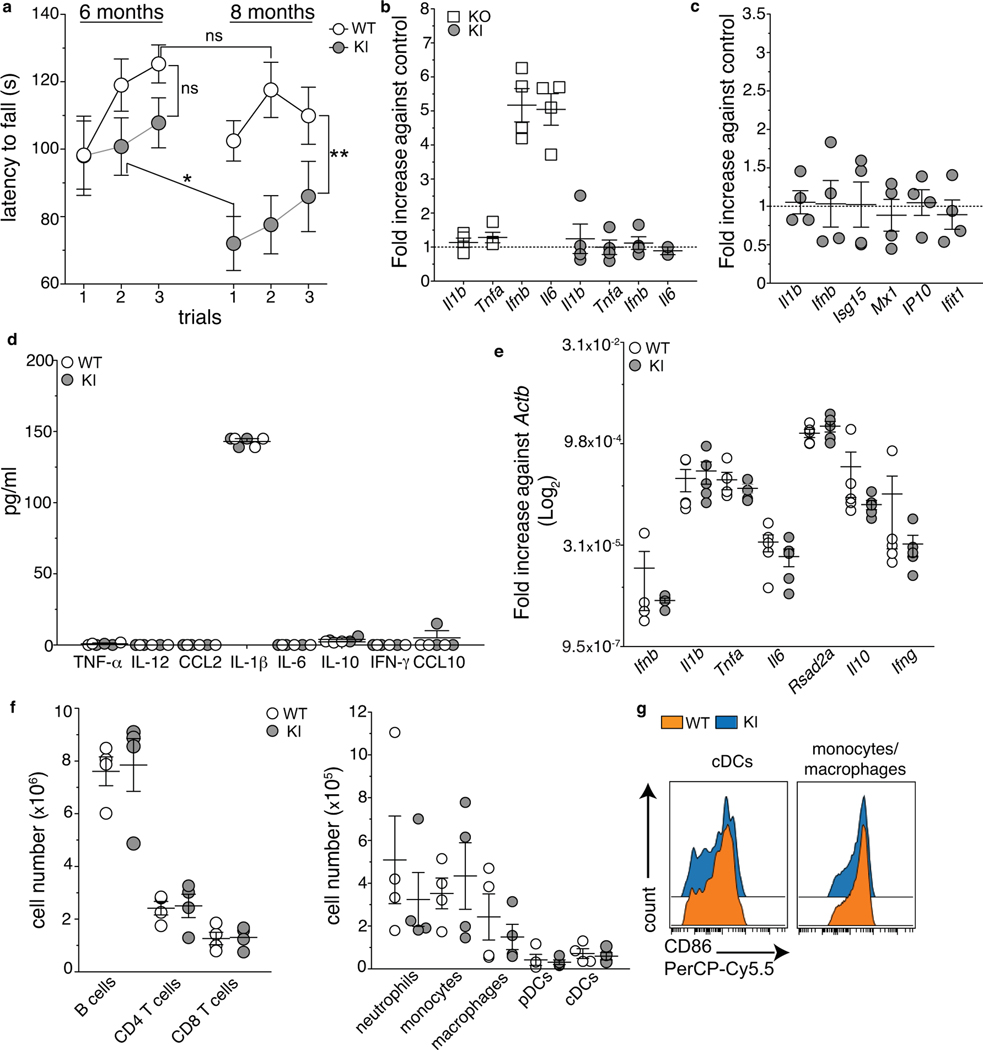
Recent work has reported that lack of expression of the mouse C9orf72 ortholog leads to a systemic hyperactive IFN-I response caused by impaired regulation of the stimulator of interferon genes (STING)7. In addition, ALS-causative mutations in the (TAR)-DNA binding Protein (TDP-43), the mislocalization of which in the cytoplasm of motor neurons is a hallmark of most forms of ALS -including ALS415, cause aberrant IFN-I responses via the STING pathway17. Based on these reports, we stimulated BMDCs from WT and KI mice with the STING activator cyclic guanosine monophosphate–adenosine monophosphate (cGAMP), and measured the expression of inflammatory genes by RT-qPCR to determine whether this pathway was dysregulated in ALS4 mice (Extended Data Fig.1c). We detected similar levels of expression of Il1b, Ifnb and interferon stimulated genes (ISG) in WT and KI cells upon cGAMP treatment (Extended Data Fig.1c). Therefore, the L389S mutation does not impair Setx activity in regulating IFN-I responses triggered by infection or STING activation. Furthermore, we did not detect any abnormal production of inflammatory cytokines in either serum or spinal cord of Setx KI mice when comparing disease-affected and control animals (Extended Data Fig.1d–e). Also, no changes in myeloid and lymphoid subsets were observed after flow cytometry analysis of splenocytes from KI and WT littermates (Extended Data Fig.1f–g). Finally, high-throughput autoantibody screening did not reveal increased production of autoantibodies in symptomatic KI animals compared to WT controls (Extended Data Fig.2, Supplementary Table 1). Taken together, our results exclude a general hyper-responsive inflammatory phenotype in KI mice.
Activation of CD8 T cells in murine ALS4
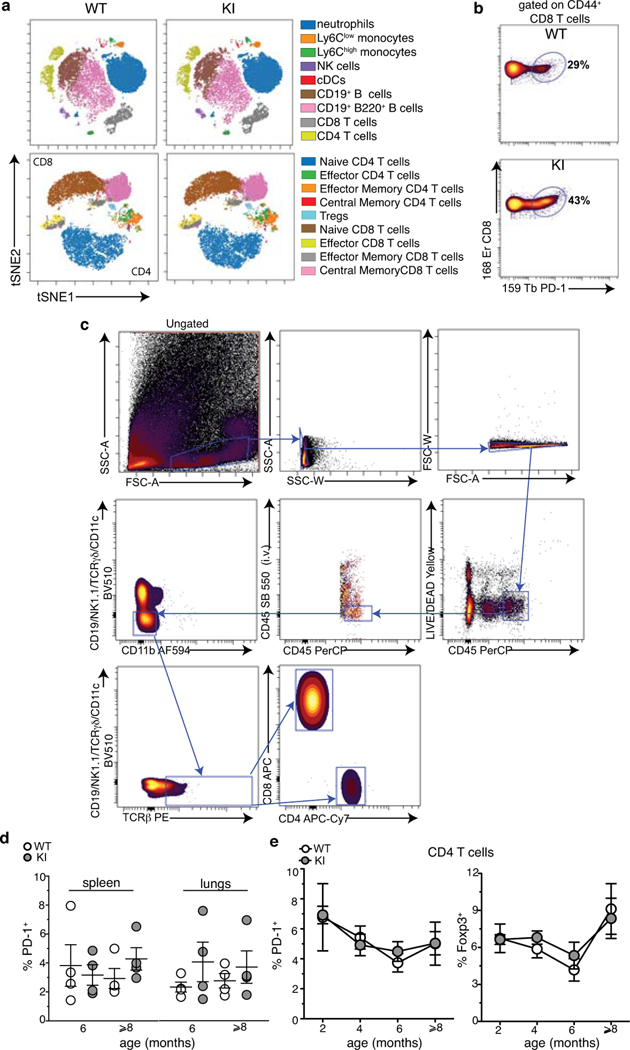
In neurodegenerative diseases, activated CD4 and CD8 T cells are found in the peripheral blood and in the central nervous system (CNS) not only in patients affected by Multiple Sclerosis (MS), which has a clear immunological component, but also, as shown recently, in Alzheimer’s disease (AD), Parkinson’s disease (PD), and Lewy body dementia18–21. To unveil whether this was also the case in mice expressing Setx-L389S, we performed a deep immune-phenotyping by single cell mass cytometry (or CyTOF, cytometry by time of flight) in the blood of KI and WT mice using an antibody panel detecting all major subsets of immune cells. No major differences were detected among immune cell subsets and mouse groups (Extended Data Fig.3a). However, we noticed an increased frequency of activated (i.e. CD44+) CD8 T cells expressing the marker Programmed cell death protein 1 (PD-1) in KI animals (Extended Data Fig.3b). Further characterization by spectral flow cytometry (Extended Data Fig.3c) in pre-symptomatic (2, 4 and 6 month old) and symptomatic (older than 8 months) KI mice revealed a significant increase in the frequency of PD-1+ CD8 T cells in the spinal cord, brain and blood (2 to 5 folds; Fig.1a,b), but not in the spleen or lungs (Extended Data Fig.3d), as compared to age-matched WT animals. Interestingly, the frequency of PD-1+ CD8 T increased in the CNS earlier than in the blood, and shortly before disease onset, suggesting a correlation between the accumulation of PD-1+ CD8 T cells and the manifestation of neurodegenerative symptoms (Fig.1b). No differences in the expression of PD-1 or other immune activation markers were detected in CD4 T cells, and the frequency of Foxp3+ regulatory T cells (Tregs) was not altered between genotypes (Extended Data Fig.3e).
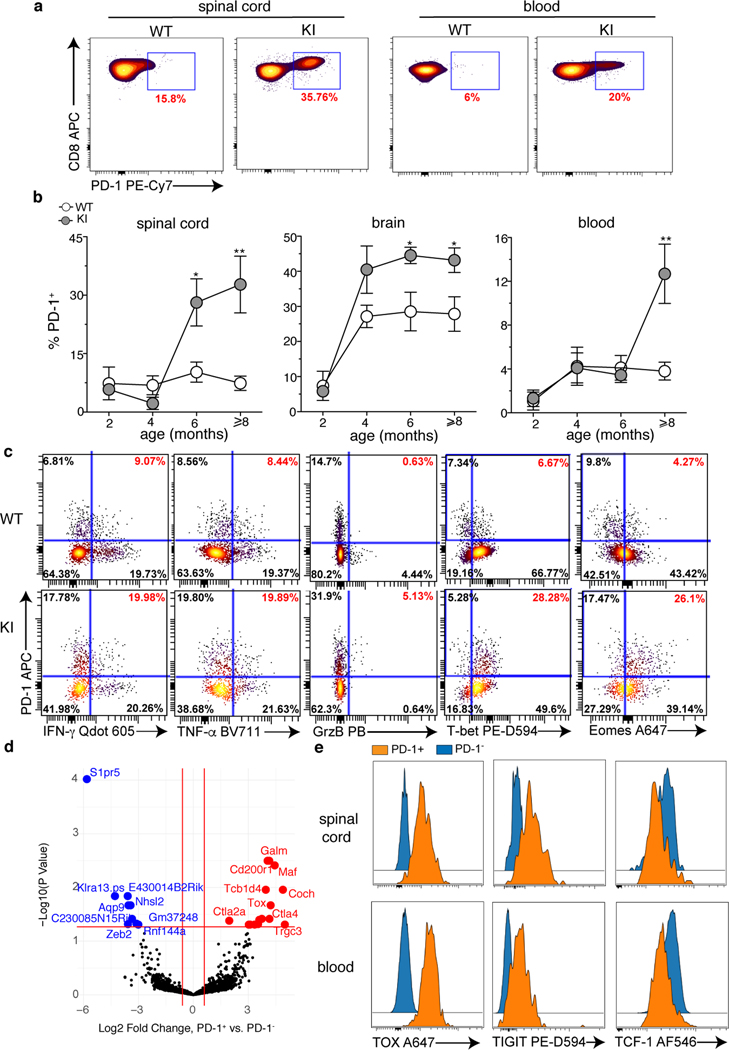
Fig. 1. A CD8 T cell signature in SetxL389S+/− KI mice.
a, Representative dot plots of the frequency of PD-1+ CD8 T cells in the spinal cord (left) and the peripheral blood (right) of 10 month old WT and KI mice. b, Frequencies of PD-1+ CD8 T cells in the spinal cord (left), brain (middle) and peripheral blood (right) of WT and KI mice at the indicated ages. Data show 3 independent experiments, n=8 (brain and blood, 8 months) and n=6 individual mice/group. For spinal cord and brain of 2 months old animals, 2 mice/experiment were pooled, n=6; mean +/− SEM. p-value calculated using the two sided Mann Whitney test (spinal cord: 6 months *p=0.034, >8 months **p=0.0087; brain: 6 months *p=0.043, >8 months *p=0.0258; blood: **p=0.0015). c, Expression of indicated effector molecules and transcription factors by PD-1+ CD8 T cells from the spinal cord of 10 month old mice after ex vivo re-stimulation with phorbol myristate acetate (PMA) and ionomycin. Dot plots show 2 concatenated experiments out of 3 independent experiments (n=6, 2 mice were pooled/experiment). Cells are gated on total CD8 T cells. d, Differential expression analysis of PD-1+ and PD-1− CD8 T cells sorted from the blood of 10–12 month old KI mice, n=3 individual mice. e, Expression of indicated markers by PD-1+ and PD-1− CD8 T cells isolated from the spinal cord and blood of 10–12 month old KI mice. Histograms are representative of 2 independent experiments (n=6, blood TIGIT and TOX, n=5, spinal cord TIGIT and TOX, n=4 TCF-1 individual mice/group). In (d, e), both PD-1+ and PD-1− are gated on CD44+CD62L−CD8 T cells. BV=brilliant violet; PB=pacific blue; PE-D594=PE-Dazzle 594; AF= alexa fluor.
Phenotype of PD-1+ CD8 T cells
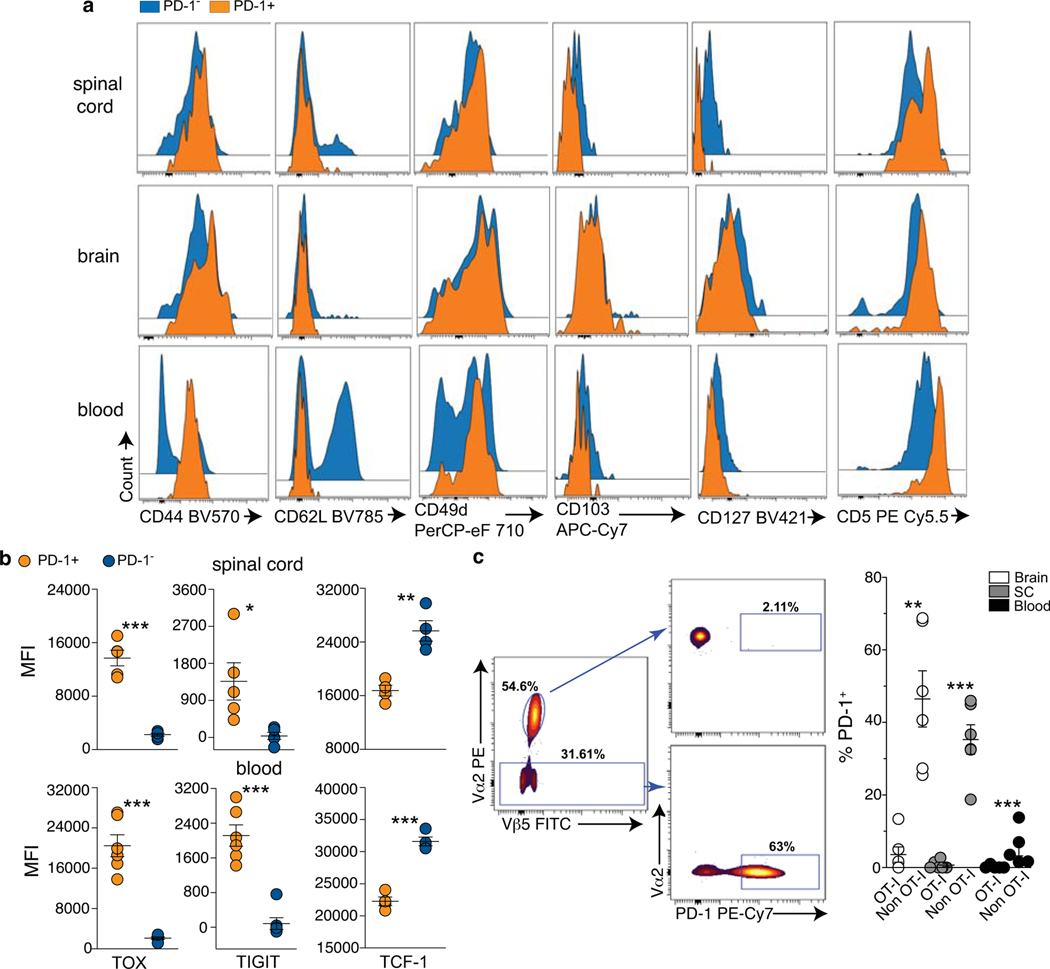
PD-1 is a marker of activation and exhaustion22, and the presence of non-functional PD-1+ T cells has been suggested in a mouse model of AD23. However, PD-1+ CD8 T cells from the spinal cord of KI mice were still able to produce the effector molecules IFN-γ, tumor necrosis (TNF)-α, and a minority of these cells, approximately (5%), also Granzyme B (GrzB) following short ex-vivo restimulation (Fig.1c). These results, together with a profile characterized by high expression of the transcription factors T-bet (T-box expressed in T-cells), Eomes (Eomesodermin), the activation marker CD44, and no expression of CD62L, suggest a functional and effector memory phenotype (Fig.1c, Extended Data Fig.4a). Additionally, while their expression of CD103, a marker of tissue-resident T cells24, was low, PD-1+ CD8 T cells were positive for CD49d (alpha4 chain of the very late antigen 4, VLA-4), consistent with their homing to the CNS25,26 (Extended Data Fig.4a). Interestingly, PD-1+ CD8 T cells showed high levels of expression of CD5 (a marker of T cell receptor, TCR, activation in mice)27, and downregulated the expression of the IL-7 receptor (CD127), which is involved in the maintenance of memory T cells in the absence of their cognate antigen28 (Extended Data Fig.4a).
To further understand the nature of PD-1+ CD8 T cells, we performed a SMART (Switching Mechanism At the end of the 5’-end of the RNA Transcript)-sequencing analysis of the transcriptome of effector memory CD8 T cells expressing or not PD-1 from the blood of symptomatic KI animals (Fig.1d, Supplementary Table 2). Amongst the transcripts differentially expressed between these two populations, we detected upregulation of Tox (Thymocyte Selection Associated High Mobility Group Box) in PD-1+ as compared to PD-1− CD8 T cells (Fig.1d, Supplementary Table 2). Highest levels of TOX expression are triggered by chronic TCR stimulation, and define TEMRA CD8 T cells, a subset of terminally differentiated, effector memory cells with potent effector functions that circulate in the human blood29. TEMRA cells are associated with senescence, and chronic viral infections (such as cytomegalovirus, CMV, and Epstein Barr Virus, EBV), but also with autoimmune and neurodegenerative diseases such as MS, rheumatoid arthritis (RA), and AD18,30,31. In TEMRA CD8 T cells, TOX expression is accompanied by upregulation of inhibitor receptors such as PD-1 and TIGIT (T cell immunoglobulin and ITIM domain), while inversely correlates with the expression of the transcription factor T cell factor 1 (TCF-1)29. Interestingly, this signature is reported in CNS- and pancreatic-derived autoreactive CD8 T cells in mice32,33. Flow cytometry analysis in the spinal cord and the blood demonstrated that KI PD-1+ CD8 T cells when compared to their PD-1− counterpart, can also be distinguished as TIGIThighTOXhighTCF-1low cells (Fig.1e, Extended Data Fig.4b), accordingly with a TEMRA-like signature.
Clonal expansion of PD-1+ CD8 T cells
Depending on the physiologic context, the activation of CNS-associated T cells can be either dependent18,34 or independent35–37 on TCR engagement. To determine whether the generation of PD-1+ CD8 T cells was dependent on TCR or cytokine stimulation, we employed two complementary approaches. First, we cell sorted effector memory (CD44+ CD62L−) PD-1+ and PD-1− CD8 T cells from the CNS of KI animals to sequence their TCR-Vβ repertoire by ImmunoSEQ assay (Fig.2a, Supplementary Table 3). Strikingly, PD-1+, but not PD-1−, CD8 T cells displayed pronounced clonal expansion with one clone comprising more than 60% of the productive T-cell repertoire in one of the KI mice analyzed and two clones comprising ~30% and ~20% of the productive T-cell repertoire in the other two animals (Fig.2a, left). Consistently, both the maximum frequency of productive rearrangements (i.e. rearrangements that can produce a functional TCR) and the Simpson clonality38 were higher in PD-1+ as compared to PD-1− CD8 T cells (Fig.2a, right). Since TCR sequences are unique to individual T cells, large numbers of T cells expressing the same TCR is evidence of clonal expansion in response to an antigen.
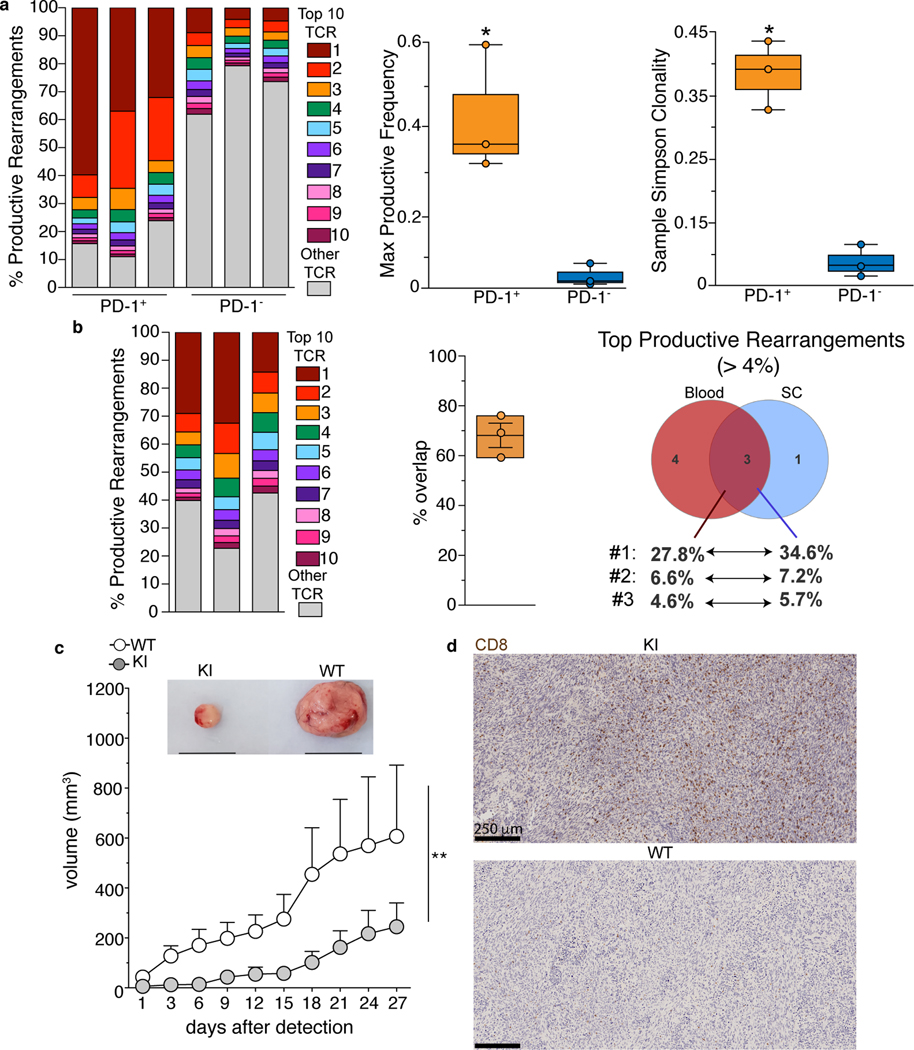
Fig. 2. Activation of CD8 T cells is antigen-dependent and correlates with increased anti-glioma immune response.
(a,b) PD-1+ and PD-1− CD8 T cells from the CNS (a), and PD-1+ CD8 T cells from blood and spinal cord (b) were isolated by cell sorting from 9 to 12 month old KI mice. Frequencies of productive rearrangements for the top 10 TCRβ sequences in each individual mouse are shown in color, while the sum of the other sequences is shown in grey. Each bar represents an individual animal, n=3. a, right, Comparison of maximum TCR productive frequencies and Simpson clonality between PD-1+ and PD-1− CD8 T cells. n=3; whiskers=max and min values, centre of the box=median, bounds of the box=first and third percentile. p-value calculated using unpaired two-sided t test (*p=0.025). b, middle, Percentage of sequences overlapping between blood and spinal cord in the 3 animals. whiskers=max and min values, centre of the box=mean, bounds of the box=SD. b, right Venn diagram showing the overlap between the top (> 4%) productive TCR sequences from PD-1+ CD8 T cells isolated form blood and spinal cord in mouse 1. (c, d) Ten-twelve month old KI and WT mice were subcutaneously injected in the flank with tumor-derived neural stem cells. c, Kinetics of tumor growth and representative images (top, black line=1 cm) of tumors from the indicated genotype at day 27 post-detection. Two independent experiments, n=8 WT and 12 KI mice; mean+/− SEM. p-value calculated using a two-tailed Wilcoxon matched-pairs signed rank test (**p=0.002). d, Representative scans of CD8 staining inside the glioma (3 mice/group from 2 independent experiments).
We further sequenced the TCR repertoire of PD-1+ CD8 T cells from the peripheral blood of symptomatic KI mice and compared them to their spinal cord-derived counterpart from the same animal (Fig.2b, Supplementary Table 3) . Although less extensive than in the CNS, clonal expansion was also present in PD-1+ CD8 T cells from the blood, with one TCR sequence comprising from 15 to 30% of the entire Vβ repertoire (Fig.2b, left). Notably, we found an overlap between the most expressed TCR sequences in blood and spinal cord (Fig. 2b, middle and right, Supplementary Table 3), suggesting that PD-1+ CD8 T cells in the CNS (re)circulated through the blood.
In parallel, we crossed KI to OT-I mice, which carry a transgenic Vα2Vβ5 TCR recognizing an epitope derived from the chicken protein ovalbumin (OVA), which is not expressed in KI animals (Extended Data Fig.4c). Analyses in the CNS and blood of 10 month old KIxOT-I mice showed that very few OT-I (Vα2highVβ5+) cells were able to express PD-1 (Extended Data Fig.4c). In contrast, we found high frequencies of PD-1+ cells amongst non-TCR transgenic CD8 T cells, the activation of which is not restricted to the exogenous antigen OVA.
Therefore, our data strongly supports the hypothesis of an antigen-driven mechanism of CD8 T cell activation in contrast to a secondary non-specific response. This phenomenon could be either a concause or an effect of the neurodegenerative process.
Anti-tumor immunity in KI mice
Increased frequencies of PD-1+ CD8 T cells are detected in KI mice specifically in the CNS, suggesting that the antigen(s) driving the expansion of these cells are tissue-specific. To confirm our hypothesis, we tested 2 models of anti-tumor immunity in symptomatic KI mice and WT controls. Our rationale was that the presence of already activated autoreactive CD8 T cells would protect KI animals against cancer cells expressing their cognate self-antigen. We thus injected subcutaneously in the flank WT and KI animals with tumor-derived neural stem cells (NSCs) overexpressing PDGFB (Platelet Derived Growth Factor Subunit B) and deficient for p53 (tumor protein P53) and Nf1 (Neurofibromin 1), which induce high grade glioma in mice39 (Fig.2c,d). A second group of WT and KI littermates was similarly injected with the B16-F10 melanoma cell line (Extended Data Fig.5a). Interestingly, the size of glioma was significantly smaller in KI than in WT mice, while melanoma grew comparably in both groups (Fig.2c, Extended Data Fig.5a). Of note, higher infiltration of CD8 T cells was detected in glioma harvested from KI than WT controls (Fig.2d, Extended Data Fig.5b). Comparison of TCR-Vβ sequences from total tumor-infiltrating CD8 T cells and spinal cord-derived PD-1+ CD8 T cells from KI mice showed that at least one of top 10 productive rearrangements were shared between both tissues (Supplementary Table 3). We concluded that KI mice express a population of activated CD8 T cells, the repertoire of which is skewed toward antigens of CNS origin.
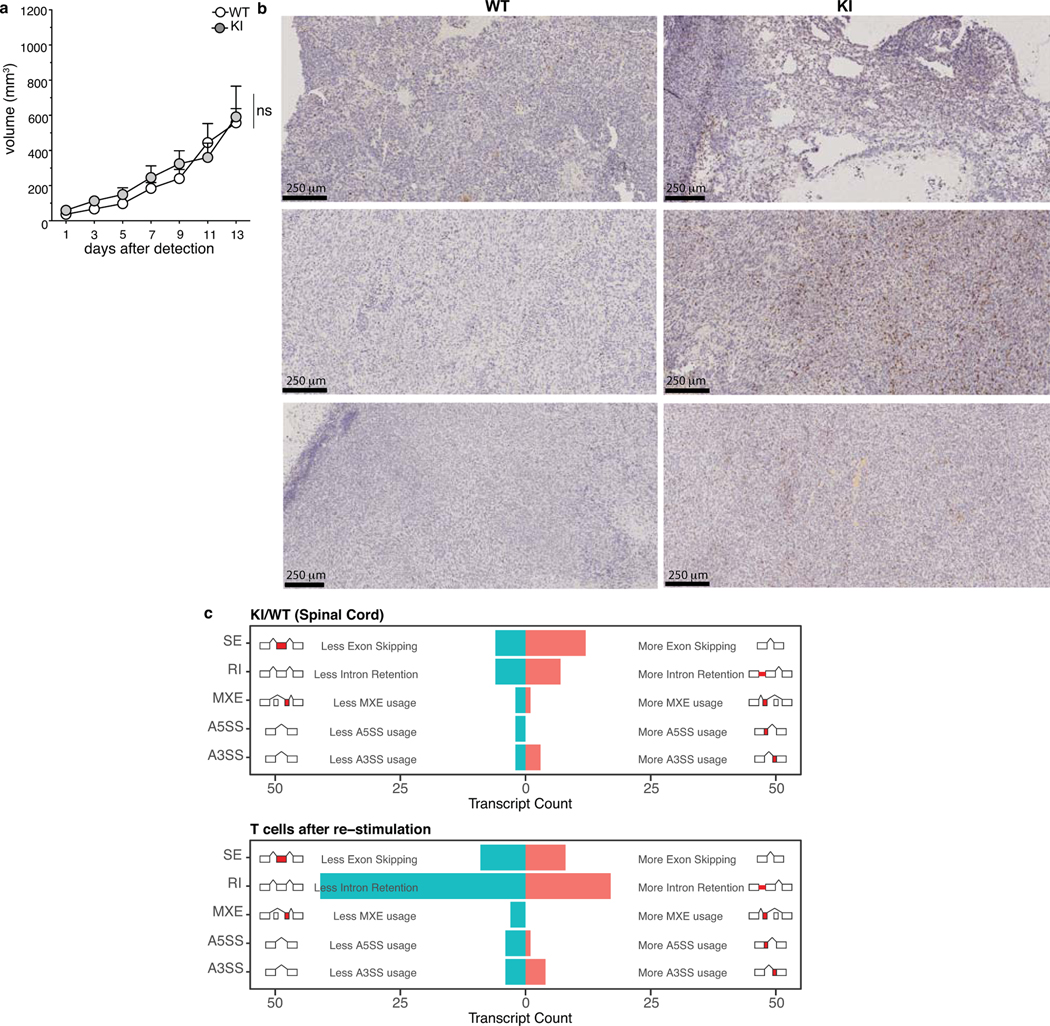
Since alternative splicing can be a source of neo-antigens in cancer40, and is increased in autoimmune diseases41, we analyzed whether symptomatic KI mice were enriched in their spinal cord for alternatively spliced genes that would generate untolerized neoantigens (Extended Data Fig 5c, Supplementary Table 4). Our data exclude the presence of a generalized splicing defect in KI animals, when compared to WT littermates, and thus the generation of neoantigens from aberrant transcripts as mechanism of tolerance break.
The immune system contributes to MND
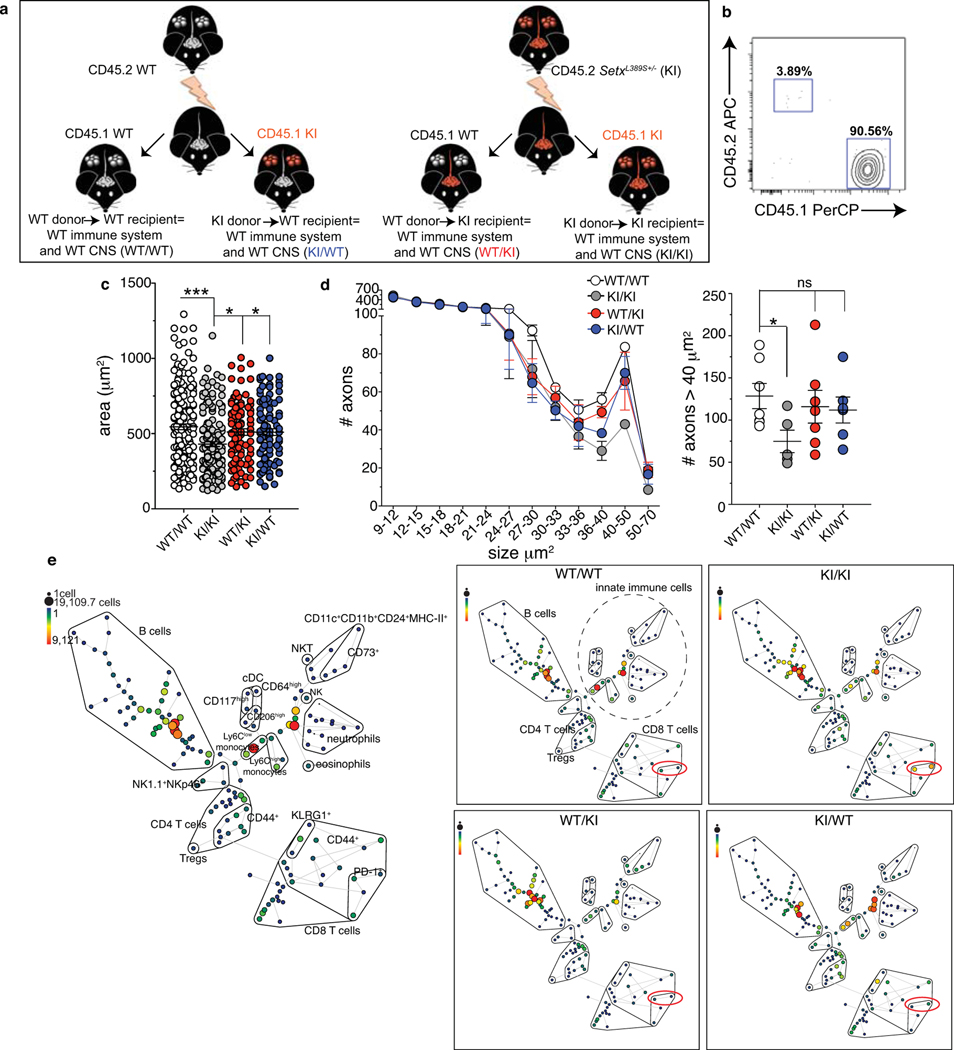
Our results point toward an immune component in ALS4 pathology. To assess the contribution of the immune system to motor impairment, and in particular the role of CD8 T cells, we performed bone marrow transplantation experiments27. We opted for this strategy instead of depleting CD8 T cells or using mice deficient for CD8 T cells or the major histocompatibility complex (MHC)-I because prolonged or complete absence of T cells causes several immunological and neurological defects that would generate confounding results36,42. We thus irradiated WT and KI mice and reconstituted them with bone marrow cells from congenic CD45 1.1 or 1.2 WT or KI animals. We generated 4 groups of chimeras, in which the mutation L389S was restricted to the hematopoietic system (KI bone marrow cells transplanted into WT mice, KI/WT group) or to the non-hematopoietic system, including the CNS (WT bone marrow cells transplanted into KI mice, WT/KI group) or expressed in both (KI/KI group) or in none (WT/WT group) (Fig.3, Extended Data Fig.6a,b). We started testing chimeric mice 10 months after reconstitution for MND by behavioral tests and histology. First, we measured their latency to fall from an accelerating rotarod: KI/KI chimera displayed significant motor incoordination as compared to the other groups, and in a manner similar to non-chimeric KI mice (Fig.3b). In KI/KI compared to WT/WT chimera, histological analyses showed a decrease in the size of motor neurons in the ventral horn of the lumbar spinal cord and a reduction in the number of large caliber axons (> 40 μm2) in the sciatic nerve (Extended Data Fig.6c,d), demonstrating that irradiation and bone marrow transplantation in KI/KI chimera did not prevent the neurodegenerative phenotype caused by Setx-L389S expression. Notably, we noticed that chimeric mice, in which the L389S mutation was restricted to non-hematopoietic (WT/KI) or hematopoietic (KI/WT) compartments performed similarly on the accelerating rotarod to WT controls, showing no signs of motor incoordination (Fig. 3b). Consistently, no significant differences were found in the size of motor neurons nor in the number of large caliber axons among WT/WT, WT/KI and KI/WT chimera (Extended Data Fig.6c,d). We concluded that the expression of the pathological form of Senataxin in the hematopoietic compartment is necessary for the development of the motor phenotype in mice, supporting the idea that dysfunction in both the CNS and the hematopoietic/immune system contribute to disease ontogeny or progression.
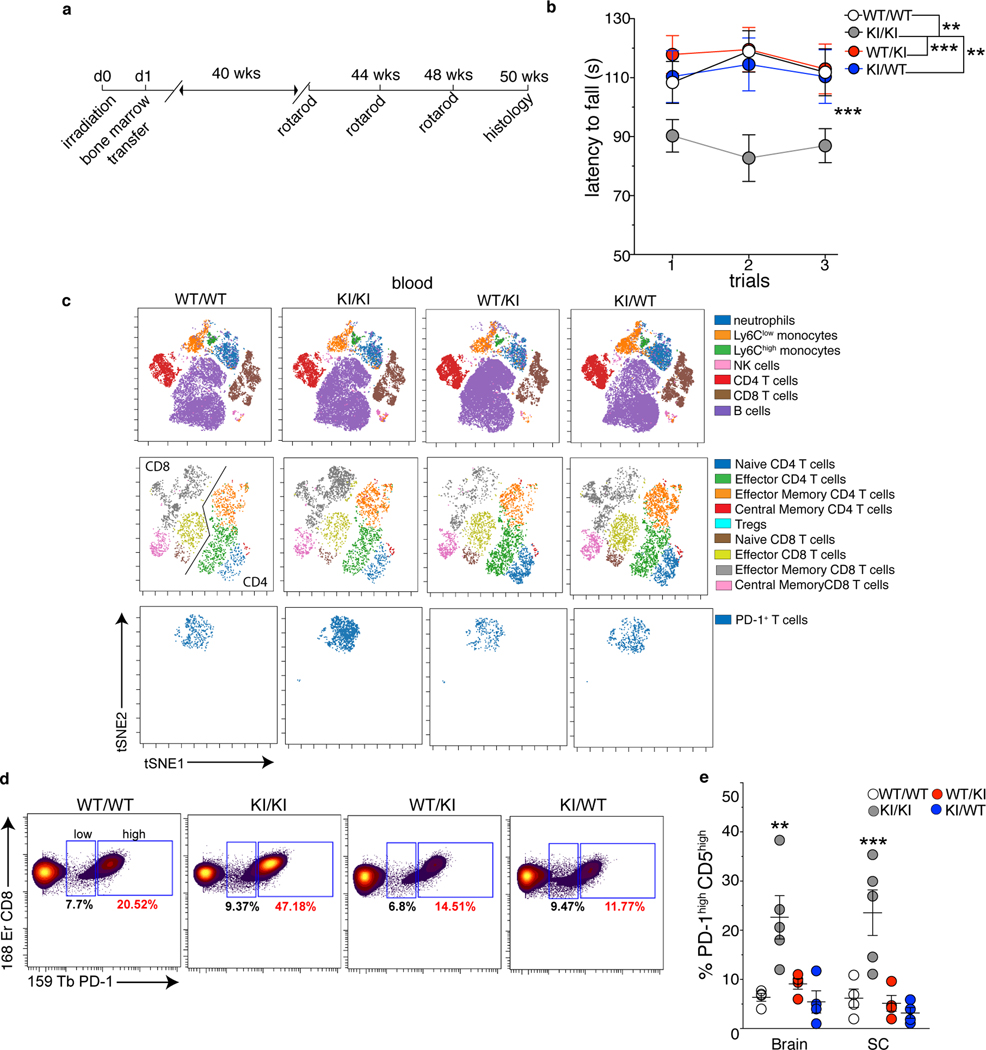
Fig. 3. The immune and the central nervous systems are both involved in MND.
8–10 week old CD45.2 WT and KI mice were subjected to hematopoietic cell-ablative doses of irradiation and reconstituted with bone marrow cells from congenic CD45.1 or 1.2 WT or KI animals. After 10 months of reconstitution, chimera were analyzed. a, Experimental schematic. b, Average time to latency before falling from an accelerating rotarod (pool of 3 independent experiments, n=21 (WT/WT), =15 (KI/KI), =20 (KI/WT), =25 (WT/KI) individual mice/group; mean +/− SEM). p-value calculated using a one-way repeated measures ANOVA test (***p=0.0003) and Bonferroni’s post-test. (c,d) Twelve months after reconstitution, peripheral blood cells from two mice of the indicated group were pooled and analyzed by mass cytometry. c, t-SNE plots of FlowSOM metaclusters after gating on total CD45high (top), TCRβ+ (middle) and CD8+ PD-1+ (bottom) cells. d, Frequency of CD8 T cells expressing PD-1 in the blood. e, Frequency of PD-1+ CD5high CD8 T cells from the SC and brain of the indicated chimeric mice 12 months after reconstitution (2 independent experiments, n=5 mice/group; mean +/− SEM). p-value calculated using one-way ANOVA and Dunnett’s post-test (**p=0.0025; ***p=0.0009).
To link our observation in chimeric mice to an immune dysfunction, we immunophenotyped all groups of chimeric mice. Strikingly, mass cytometry analysis confirmed that the major difference among immune cell types and mouse groups was an increase in the frequency of PD-1+ CD8 T cells in the blood of KI/KI chimera as compared to the other groups (Fig. 3c,d, Extended Data Fig.6e). Furthermore, the frequency of PD-1+ CD8 T cells expressing high levels of CD5 was 3 to 5-folds higher in the brain and the spinal cord of disease-affected compared to unaffected chimera (Fig.3e). As in non-chimeric KI mice, we did not detect significant dysfunctions in the innate immune system or a general increase in the secretion of autoantibodies between KI/KI and asymptomatic chimeric mice (Extended Data Fig.6e and 7). Thus, our data suggest that increased frequency of activated CD8 T cells correlates with disease development.
T cells in ALS1 and ALS6 mice
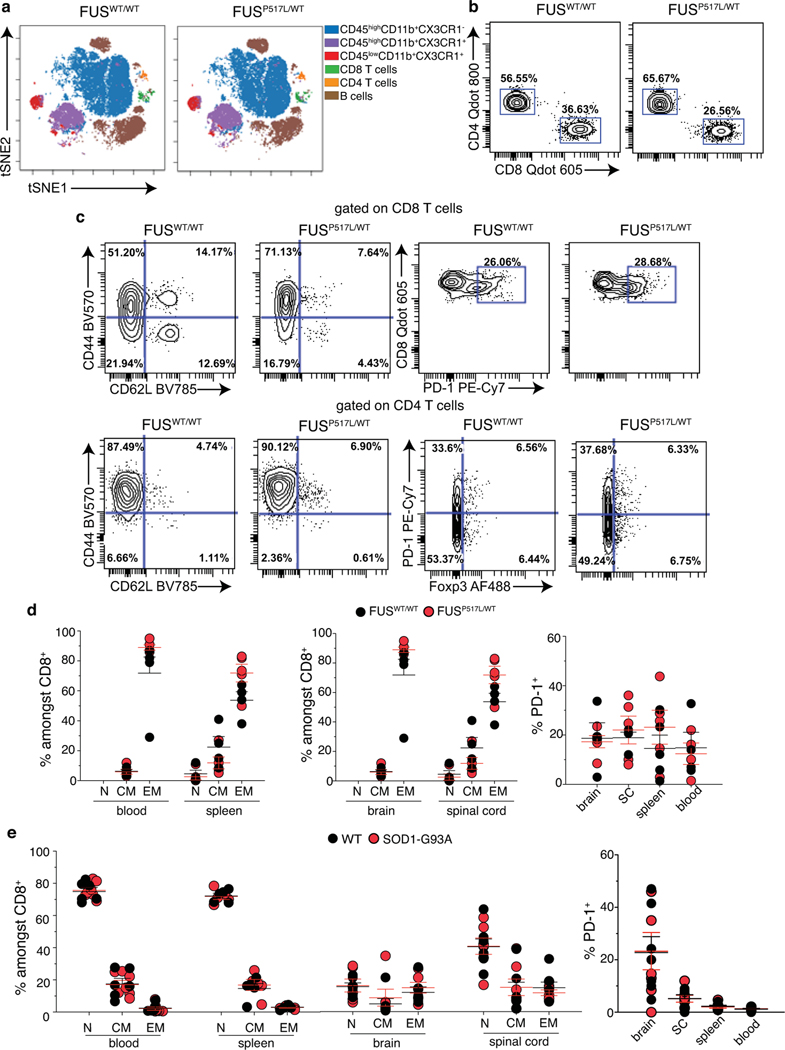
To investigate whether aberrant activation of CD8 T cells occurs in other forms of ALS, we profiled FusP517L/WT mice, which express a mutation in the Fus (Fused in sarcoma) gene that mimics a variant found in ALS6 patients43, and SOD1-G93A transgenic mice carrying a G93A mutant form of human SOD144,45. In contrast to Setx KI mice, we did not detect any significant dysfunction in either CD4 or CD8 T cells by spectral flow cytometry in blood, spleen, brain and spinal cord of symptomatic 2 year old FusP517L/WT and FusWT/WT animals (Extended Data Fig.8a–d). Similarly, increased frequencies of activated T cells was not found in the spleen, blood and CNS of symptomatic 3–4 month old SOD1-G393A when compared to non-carrier controls (Extended Data Fig.8e). Our data indicate that accumulation of activated CD8 T cells is not an immune-signature common to all mouse models of ALS.
CD8 T cell subsets in ALS4 patients
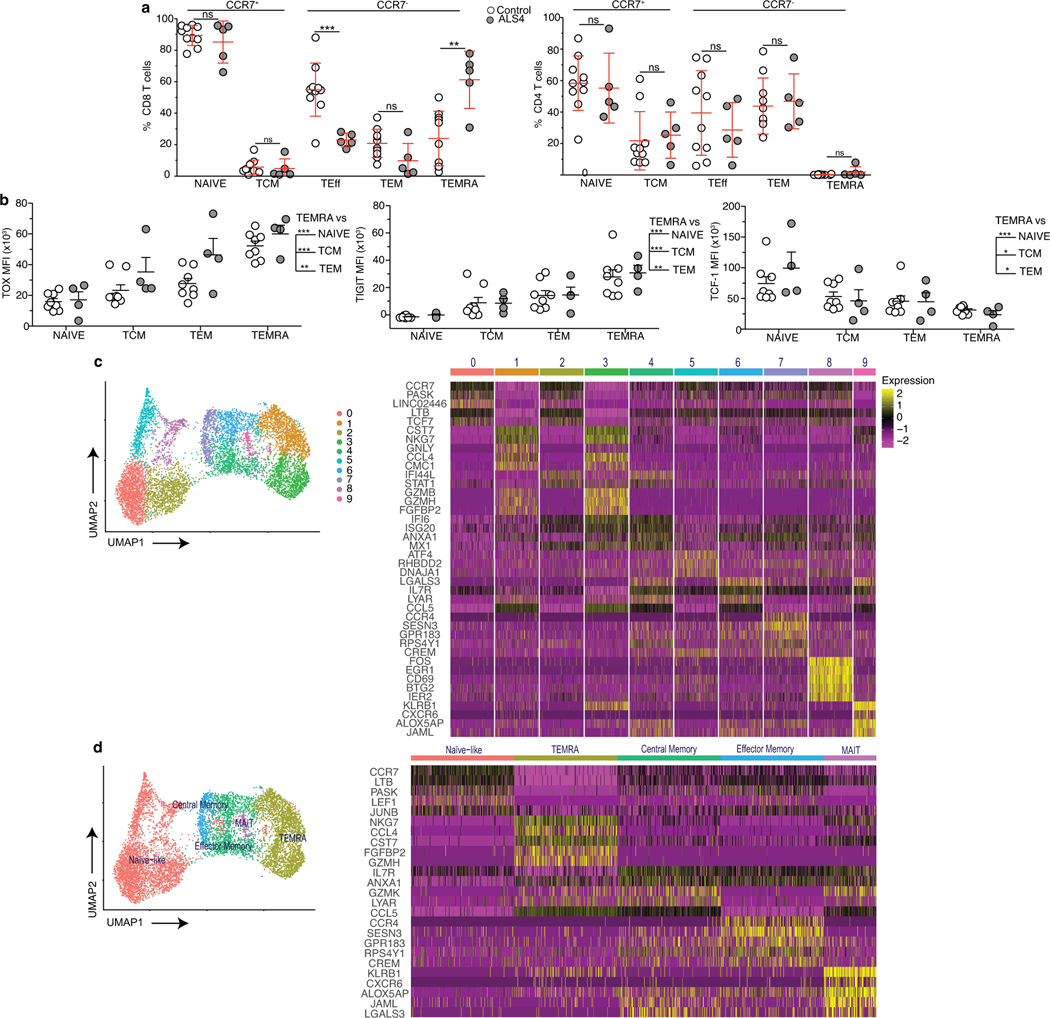
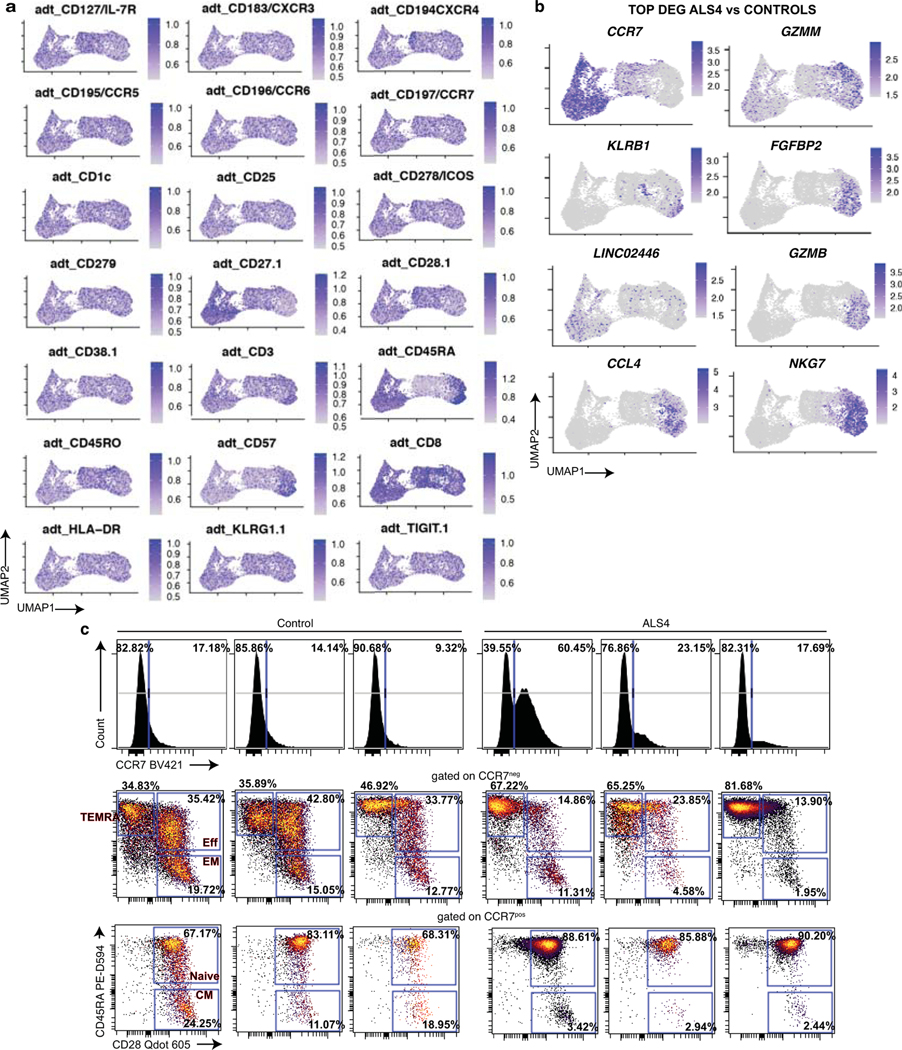
To assess whether increased frequencies of activated T cells similarly exist in patients carrying the SETX L389S mutation, we analyzed peripheral blood mononuclear cells (PBMCs) from unrelated ALS4 patients and age-matched healthy individuals (Supplementary Table 5) by spectral flow cytometry. Notably, based on the expression of the C-C chemokine receptor type 7 (CCR7) and the two human isoforms CD45RA and CD45RO46, we found that subsets of CD8 T cells were altered in patients as compared to healthy controls (Fig.4a, Extended Data Fig.9a, left). While the frequencies of effector and memory CD8 T cells were all decreased (2 to 3-folds), there was an enrichment in CD45RA+ CD28− CD27− (TEMRA) cells (3-fold increase) in patients. TEMRA CD8 T cells in the peripheral blood are also characterized by TOX, TIGIT and TCF-1 expression levels (Extended Data Fig.9b and 29). Thus, aberrant frequencies of a subset of terminally differentiated CD8 T cells are detected in both mouse and human ALS4.
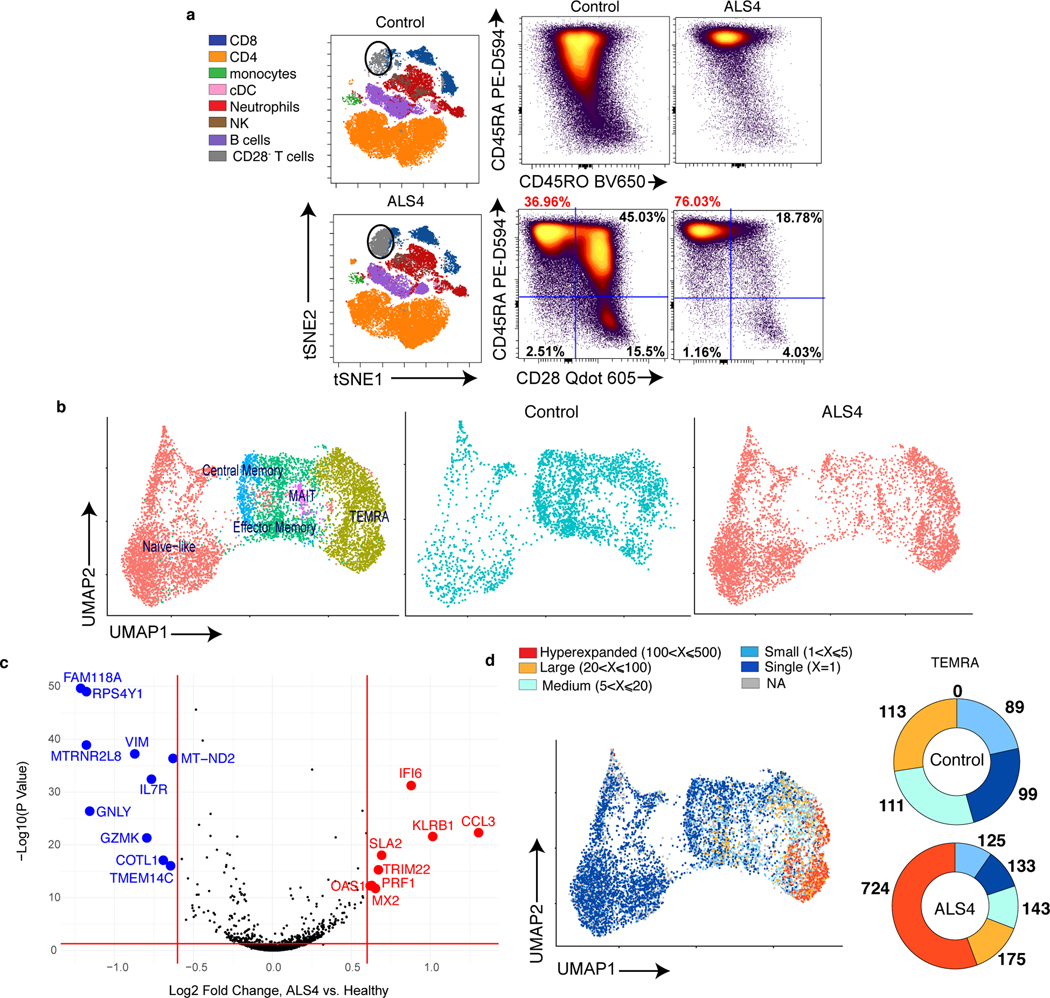
Fig. 4. Clonally expanded CD8 T cells with a TEMRA phenotype are increased in the peripheral blood of ALS4 patients.
a, Total PBMCs from ALS4 patients and age-matched controls were analyzed by spectral flow cytometry. Left, tSNE plots of FlowSOM metaclusters. The circular gates highlight a metacluster corresponding to a subset of CD8 T cells. Right, Dot plots displaying the frequency of CD45RA and CD45RO expression among CD8 T cells and of CD45RA and CD28 expression after gating on CCR7− CD8 T cells. Data shows concatenated data from 1 out of 2 independent experiments, n=5 (ALS4 patients) and n=10 (healthy controls). b-d, CD8 T cells purified from PBMCs of 3 ALS4 patients and 3 age-matched controls were analyzed by CITE-seq. b, Gene-expression analysis was conducted on all 6 samples and clustered by UMAP. Left, Identification of CD8 T cell subsets using a mixture of all groups is shown. Middle and right, Overlay of CD8 T cells from control or ALS4 patients on cellular subsets clustered by UMAP. c, Differential expression analysis of TEMRA CD8 T cells from ALS patients and healthy controls. d, Single-cell TCR analysis overlaid on UMAP projections of identified clusters showing TCRαβ clonality for each CD8 T cell subset (left) and donut plots depicting the number of TEMRA CD8 T cells expressing the same TCRαβ sequence (right).
We did not detect significant differences in the CD4 T cell compartment (Fig.4a, left, Extended Data Fig.9a, right), again paralleling what observed in our mouse model.
In order to characterize CD8 T cell subsets, and in particular TEMRA, from ALS4 patients and controls, we performed Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-Seq) coupling single cell RNA and TCR sequencing of purified CD8 T cells from PBMCs of 3 patients and 3 age-matched healthy individuals. By combining RNA and surface protein expression followed by multidimensional reduction using the Uniform Manifold Approximation and Projection (UMAP), we could identify 9 clusters corresponding to different CD8 T cell subsets: naïve/naïve like (clusters 0, 2, 5, 8), central memory (cluster 7), effector memory (clusters 4 and 6), TEMRA (clusters 1 and 3), and mucosal associated invariant T (MAIT) cells (cluster 9) (Fig.4b, Extended Data Fig.9c,d and 10a, Supplementary Table 6). In agreement with our flow cytometry data, ALS4 patients exhibited a higher proportion of TEMRA, and a decreased frequency of effector/effector memory cells, as compared to controls (Fig.4b). Furthermore, downregulation of IL7R (as seen in mice) and CCR7, and upregulation of GZMM and GZMB (encoding for Granzyme M and B, respectively), KLRB1 (Killer Cell Lectin Like Receptor B1), NKG7 (Natural Killer Cell Granule Protein 7), FGFBP2 (Fibroblast Growth Factor Binding Protein 2) and CCL4 (Chemokine (C-C motif) ligands 4), all of which correlate with a TEMRA phenotype47, are among the top deregulated genes in ALS4 CD8 T cells (Extended Data Fig.10b, Supplementary Table 7). The proportion of naïve cells seemed to be increased as well; however as shown by flow cytometry, analyses of CD8 T cells from the same samples used for CITE-seq demonstrated that only 1 patient had an abnormally high frequency of CCR7+CD45RA+CD28+ cells, while the TEMRA signature was consistent among all ALS4 donors (Extended Data Fig.10c). Comparison of peripheral TEMRA in controls and patients also revealed increased expression of genes related to TEMRA activation, such as KLRB1 and perforin (PRF1) (Fig.4c, Supplementary Table 7). Therefore, despite PD-1 is not consistently upregulated in patients, increased frequencies of CD8 T cells with a TEMRA phenotype is a common signature in murine and human ALS4.
Clonal analysis in ALS4 patients
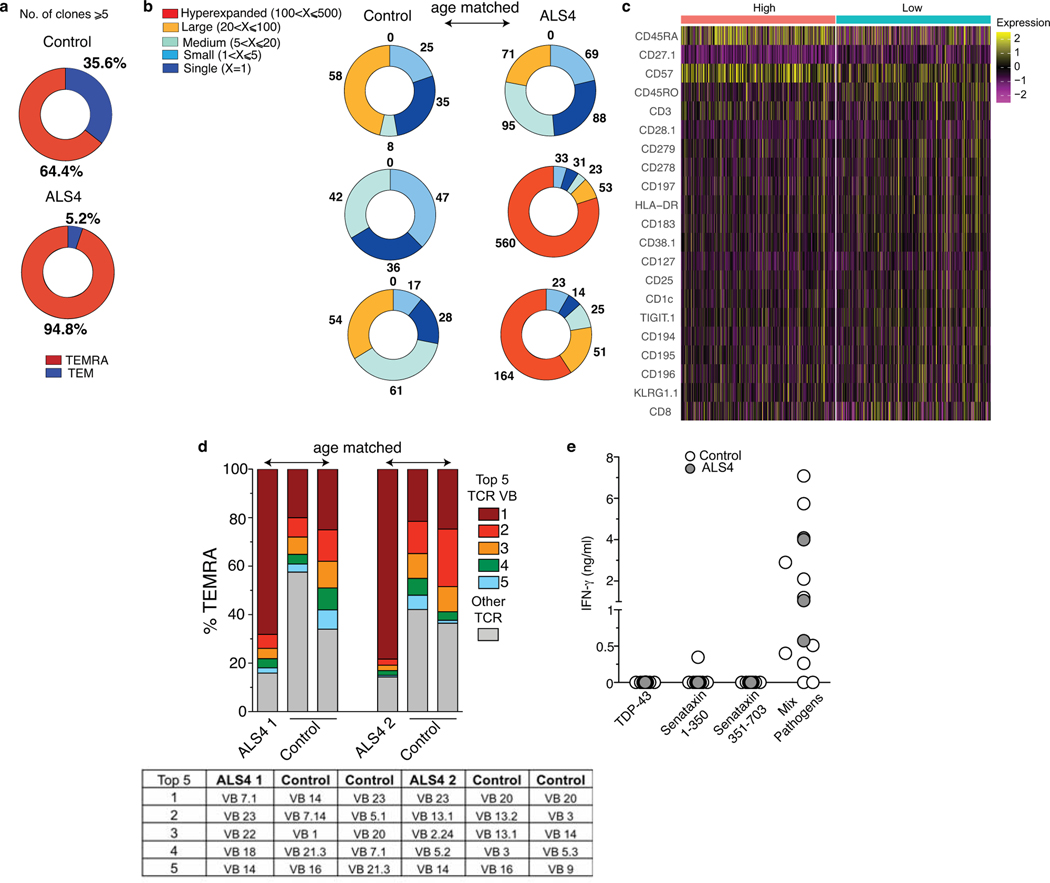
We further examined the clonality of the distinct CD8 T cell subsets by combining surface protein and RNA expression with TCR sequencing. Interestingly, more than 90% of expanded clones (≥5) are TEMRA cells in ALS4 patients, in comparison to ~ 60% in healthy controls (Fig. 4d, Extended Data Fig.11a, Supplementary Table 8). Strikingly, by matching UMAP coordinates from RNA and TCR analyses, we observed a colocalization between the most expanded clones, i.e. CD8 T cells in which the same TCR sequence was shared by more than 20 cells, and the TEMRA subset that we found enriched in patients (Fig.4d). We quantified this clonal expansion and found that over half of all TEMRA cells comprised of highly expanded clones in patients (Fig.4d, Extended Data Fig.11b). Hyperexpanded (>100 clones) TEMRA cells were detected in 2 of the 3 patients, with one clone reaching as high as 407 cells (Extended Data Fig.11b, Supplementary Table 8). No hyperexpanded clones were found in other CD8 T cell subsets or in control patients (Fig.4d, Extended Data Fig.11b). Comparison of surface protein expression from our CITE-seq analysis in “Low” and “High” (>20) expanded clones from ALS4 patients revealed a stronger TEMRA phenotype in the latter, as indicated by CD27 downregulation and CD45RA and CD57 upregulation (Extended Data Fig.11c).
We further confirmed our clonality data by analyzing the TCR-Vβ expression in TEMRA CD8 T cells from PBMCs by flow cytometry, using a commercial kit that detects 24 different specificities (i.e. ~70% coverage of the normal human TCR-Vβ repertoire) (Extended Data Fig.11d). Consistent with our TCR sequencing analysis, we observed that the same TCR-Vβ sequence comprised 68–78% of the detected repertoire in ALS4 patients while in controls it never exceeded 25%. Notably, these experiments were conducted in a separate cohort of patients and age-matched healthy controls recruited a year after the first cohort.
Comparison of our TCR sequencing data with published human TCR sequences of known antigenic specificity48–50 excluded that the most expanded clones in ALS4 patients were generated in response to viral antigens from CMV and EBV (Supplementary Table 9). Accordingly, ex vivo re-stimulation using a pool of 80 MHC-I-restricted peptide epitopes from common agents of human infections did not show increased levels of IFN-γ production in PBMCs from ALS4 patients as compared to age-matched controls (Extended Data Fig.11e). Additionally, using this same experimental setting, no T cell-mediated IFN-γ response was detected against the self-proteins Senataxin or TDP-43 (Extended Data Fig.11e). This latter result indicates either that highly clonally expanded CD8 T cells in ALS4 patients are not generated in response to self-antigens derived from Senataxin or TDP-43, or that the low affinity of the autoreactive TCR for its cognate antigen prevents the detection of CD8 T cell responses in our experimental conditions, consistent with other reports51.
In conclusion, our data support a model in which expansion of TEMRA CD8 T cells of autoreactive origin is associated with ALS4 in both mice and patients.
Histological analyses of ALS4 patients
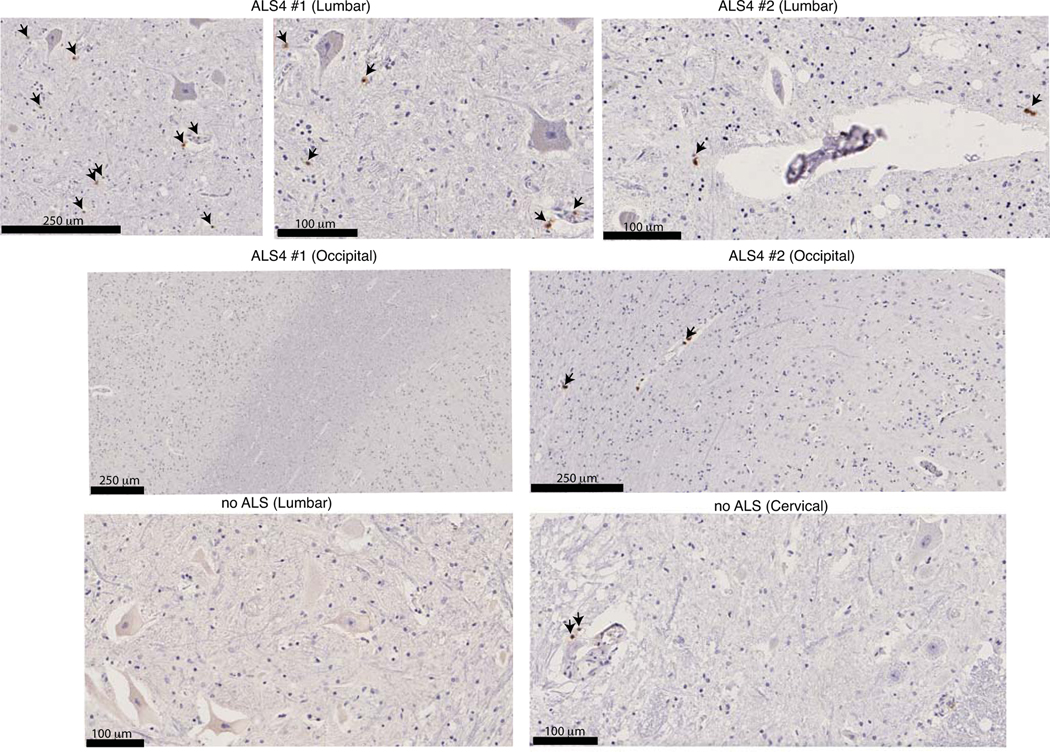
To link the CD8 T cell phenotype to the pathology of ALS4 in patients, we examined post-mortem spinal cord sections from 2 ALS4 patients. Even if the small number of patients and cells did not allow a quantitative analysis, we clearly localized CD8 T cells in the ventral horn of lumbar spinal cord sections from ALS4 patients (Extended Data Fig.12). Of note, CD8 T cells were not detected in the parenchyma of spinal cords from non-ALS controls, or of brain cortical regions from the same ALS4 patients, while as expected, a CD8 staining was present in the blood vessels of all tissues and human samples (Extended Data Fig.12).
Discussion
We discovered an immune-signature characterized by activated antigen-specific CD8 T cells in ALS4. In ALS4 mice, clonally expanded CD8 T cells are detected in the spinal cord, brain and peripheral blood. In ALS4 patients, a similar immune-signature is observed in the peripheral blood. We did not have access to the cerebrospinal fluids from ALS4 patients; however in AD, clonally expanded TEMRA CD8 T cells in peripheral blood of patients were shown to be also present in the intrathecal space18. Additionally, we localized CD8 T cells in the ventral horn of lumbar spinal cord sections from two ALS4 patients.
While the source of the antigen, self or non-self, ubiquitously expressed or tissue-specific, and its mechanism of generation can raise several hypotheses, our data indicate the presence of an activated immune response directed against self-antigens of CNS origin. In fact, i- our mice are housed in pathogen-free condition, ii- we did not detect increased T cell responses against the most common agents of human infection, and iii- symptomatic KI mice showed increased resistance and CD8 T cell response against glioma, but not melanoma, than WT animals.
Based on the knowledge that cell death can promote autoimmunity27,52, and having ruled out , albeit not formally excluded, mechanisms involving antigens generated by alternative splicing and infection, our data support a model in which neurodegenerative cell death favors the presentation of self-antigens to CD8 T cells. The neurodegenerative process likely starts in a cell-autonomous way, since neurons isolated from Setx KI mice are more prone to die ex vivo than their counterpart from WT animals15. Furthermore, our experiments using bone marrow chimeric mice show that SetxL389S needs to be expressed in the CNS to generate disease. On the other hand, the frequency of autoreactive CD8 T cells positively correlates with motor impairment, raising the question whether and how activated CD8 T cells contribute in return to the neurodegenerative process. Interestingly, WT CD8 T cells from KI recipients do not expand or differentiate into PD-1+ cells at the same level as KI CD8 T cells, suggesting that the mutation could intrinsically promote the loss of T cell tolerance, similarly to what reported in the immunopathogenesis of RA53. The identification of the antigen(s) triggering CD8 T cell activation and the generation of TCR transgenic CD8 T cells specific to such antigen(s) will be necessary to establish the role of these cells in disease onset and progression.
Our findings add ALS4 to growing evidence for an unexpected role of T cells in regulating physiological and pathological events in the CNS18,19,34,35,37,54. Specifically, the CNS-derived CD8 T cell signature described here for ALS4 is yet-to-be identified in any other MND. Lastly, our data suggest that adaptive immune cells could be a potential biomarker to monitor ALS4 progression in patients.
Methods
Mice and mouse-related methods
Five week to two year old WT (C57Bl/6J), CD45.1 (C57BL/6.Ly5.1), OT-I (C57BL/6-Tg(TcraTcrb)1100Mjb/J), and hemizygous and no carrier SOD1-G93A (B6SJL-Tg(SOD1*G93A)1Gur/J) were purchased from The Jackson Laboratories. Setx deficient mice were generated as described13, and kindly provided by Martin F. Lavin (University of Queensland, Brisbane, Australia). Setx-L389S+/− mice, a kind gift from A. R. La Spada, were previously described15. All strains were bred in the mouse facility of the Icahn School of Medicine at Mount Sinai (ISMMS). FusWT/WT and FusP517L/WT mice were generated by the laboratory of N. A. Shneider43, and bred in the mouse facility of the Columbia University, NY, USA. Mice were all housed under standard conditions at a temperature of 22°C and a 12 h light/dark cycle with ad libitum access to standard food and water.
Chimeric mice were generated using 8–10 week old mice after two rounds of lethal irradiation with 600 rads 6 h apart. Twenty-four hours later, irradiated mice were reconstituted by intravenous injection of bone marrow (1×106 to 4×106 cells) isolated from indicated strains of mice. Chimeric mice were studied starting at 10 months after bone marrow transplantation.
All mice were kept under specific pathogen-free conditions in the animal care facility at the ISMMS. Littermate mice, WT and KI, males and females, were studied at the indicated weeks of age. Numbers of males and females were matched between WT and KI groups. We favored the use of males versus females, since KI males show an earlier and more reproducible phenotype, a phenomenon also observed in human patients3,55. Mice were processed identically throughout the whole experiment (housed on the same shelf in the same room, and all procedures performed at the same time). Investigators were not blinded to sample identity for this study, except for rotarod and histological analyses.
Animal numbers were empirically determined as the minimum needed to obtain statistical significance and validate reproducibility, accordingly with our IACUC approved protocol. Animal care, handling, genotyping and experiments were approved by Institutional Animal Care and Use Committee (IACUC) of ISMMS and Columbia University.
Human blood collection and PBMC isolation
Human research was approved by and conducted in compliance with the Program for protection of Human Subjects, Institutional Review Board of the ISMMS, US (studies # 19–00613 and 21–01861) and the Comitato Etico Milano Area 3, Italy (approval number: 308–19052021).
Patients were recruited by Mount Sinai Health System and at the Centro Clinico Nemo (between November 1st, 2019 through May 31st, 2021) and signed an informed consent. Clinical and demographic data of recruited patients are detailed in Supplementary Table 5. Unrelated healthy controls were as close as possible sex and age-matched to ALS4 patients. Buffy coats from de-identified healthy controls were provided by the New York Blood Bank.
From each patient, blood was drawn into a Cell Preparation Tube with sodium heparin (BD Vacutainer). Peripheral blood mononuclear cells (PBMCs) were isolated by density centrifugation using Ficoll (Ficoll Hystopaque; Sigma) or Vacutainer CPT Mononuclear Cell Preparation Tubes (BD Bioscience), according to the manufacturer’s protocol. PBMCs were aliquoted and frozen at −80°C in FBS (Gibco) containing 10% DMSO (sodium dodecyl sulfate, Sigma). Frozen vials of PBMCs were thawed and washed with culture medium (i.e. RPMI 1640, from Gibco, supplemented with 10% FBS, 100 μg/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 10 mM HEPES, and 1 nM sodium pyruvate, all from Sigma) containing 2 mg/ml of DNAse I (Roche)56. Cells were resuspended at the density of 2×106/ml in medium containing 0.1 mg/ml of DNAse I and left in culture for 18 hours in an incubator at 37ºC and 5% CO2 before analysis.
Ex vivo stimulation of PBMCs
PBMCs from ALS4 patients and controls were stimulated with pools of peptides as described19, with some modifications. Briefly, 5×105 cells were resuspended in RPMI 1640 supplemented with 5% human AB serum (Sigma), 100 μg/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 10 mM HEPES, and 1 nM sodium pyruvate, and 20 U/mL interleukin-2 (R&D), and then stimulated with 2 μg/ml of peptides or left untreated for 6 days in an incubator at 37ºC and 5% CO2. Medium was added to the cell culture every two days to maintain cell viability.
To detect pathogen-specific responses, we used a CEFX Ultra SuperStim Pool MHC-I Subset (JPT Peptides), a pool of 80 known peptide epitopes against the following infectious agents: Clostridium tetani, Coxsackievirus B4, Haemophilus influenza, Helicobacter pylori, Human adenovirus 5, Human herpesvirus 1, Human herpesvirus 2, Human herpesvirus 3, Human herpesvirus 4, Human herpesvirus 5, Human herpesvirus 6, Human papillomavirus, Influenza A, JC polyomavirus, Measles virus, Rubella virus, Toxoplasma gondii, Vaccinia virus.
To detect T cell responses against human proteins, overlapping 15mers were synthesized by JPT Peptides by ultra-high throughput SPOT technology. For TDP-43, PBMCs were stimulated with an unique pool of 106 peptides. For Senataxin, the synthesis of 703 peptides was necessary to cover the entire protein sequence. Peptides were then aliquoted in 4 sub-pools of ~175 peptides each. PBMCs were separately stimulated with a mix of sub-pools 1 and 2 (peptides 1–350) and 3 and 4 (peptides 351–703).
IFN-γ production was measured in culture medium after 6 days of stimulation using a Human IFN-gamma DuoSet ELISA (R&D), according to manufacturer’s instructions. For each individual sample, concentrations were measured using Gen5 3.11 and normalized to the corresponding unstimulated condition to account for unspecific IFN-γ secretion.
In vitro infection and stimulation
Bone marrow (BM) cells were isolated by flushing the long bones with PBS. BM-derived GM-CSF DC cultures were grown in 24 well plates as previously described57 in RPMI 1640 supplemented with 40 pg/ml recombinant GM-CSF (BioLegend) and 5% FBS, plus 100 μg/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, 10 mM HEPES, 1 nM sodium pyruvate, 1× MEM nonessential amino acids, and 2.5 μM β-mercaptoethanol (all Sigma). After 5 days of culture, BMDCs were harvested and replated with MOI (multiplicity of infection) =10 of Citrobacter rodentium (DBS100, ATCC) or incubated with 5 μg/ml of the 2’3’-cGAMP (cyclic [G(2’,5’)pA(3’,5’)p]) (Invivogen). For infection, bacteria were grown to exponential phase in lysogeny broth (LB) medium, then washed and resuspended in PBS.
Two hours post-treatment, culture medium was removed and cells resuspended in 1 ml of TRIzol Reagent (Life Technologies).
Quantitative reverse transcription PCR (RT-qPCR)
Spinal cords were homogenized in 1 ml of TRIzol Reagent (Life Technologies) using a mechanical homogenizer. RNA separation and isolation were performed using chloroform and isopropanol (both from Sigma), respectively, according to manufacturer’s instructions.
RNA from BMDCs was isolated using a Direct-zol RNA kit (Zymo Research), according to manufacturer’s instructions.
cDNA was in vitro transcribed using a High-Capacity cDNA RT Kit (Thermo Fisher Scientific). qPCR was performed using the iTaq Universal SYBR Green One-Step Kit (Bio-Rad) according to manufacturer’s instructions. Data were collected and analyzed using a LightCycler 480 Software release 1.5.1.62 SP3.
The statistical significance of all pairwise comparisons in qPCR assays’ change in cycling threshold (ΔCT) values was determined with a two-tailed Student’s t test.
Sequences of primers used for qPCR were as follows:
Actb forward, 5′-TTACGGATGTCAACGTCACAGTTC;
Actb reverse, 5′-ACTATTGG- CAACGAGCGGTTC;
Il6 forward, 5′-TGAGATCTACTCGGCAAACCTAGTG;
Il6 reverse, 5′-CTTCGTAGAGAACAACATAAGTCAGATACC;
Il1b forward, 5′-GCAACTGTTCCTGAACTCAACT;
Il1b reverse, 5′-ATCTTTTGGGGTCCGTCAACT;
Tnfa forward, 5’-AGTGAAGTGCTGGCAACCAC;
Tnfa reverse, 5’-GAGGAAGGCCTAAGGTCCAC;
Ifnb forward, 5’-GGAAAGATTGACGTGGGAGAT;
Ifnb reverse, 5’-CAGGCGTAGCTGTTGTACTT;
Rsad2a forward, 5’-TGGTTCAAGGACTATGGGGAGT;
Rsad2a reverse, 5’-AGCCACCTTGTAATCCCTGC;
Il10 forward, 5’-AGCCGGGAAGACAATAACTG;
Il10 reverse, 5’-GGAGTCGGTTAGCAGTATGTTG;
Ifng forward, 5’-TGCTGATGGGAGGAGATGTCTAC;
Ifng reverse, 5’-TTTCTTTCAGGGACAGCCTGTTAC;
Isg15 forward, 5’-CACAGTGATGCTAGTGGTACAG;
Isg15 reverse, 5’-CTGCGTCAGAAAGACCTCATAG;
Ifit1 forward, 5’-CGTAGCCTATCGCCAAGATTTA;
Ifit1 reverse, 5’-AGCTTTAGGGCAAGGAGAAC;
Mx1 forward, 5’-GACCATAGGGGTCTTGACCAA;
Mx1 reverse, 5’-AGACTTGCTCTTTCTGAAAAGCC;
Ip10 forward, 5’-TGCTGGGTCTGAGTGGGACT;
Ip10 reverse, 5’-CCTATGGCCCTCATTCTCAC.
Isolation of murine cells
Mice were euthanized by intraperitoneal injection of sodium pentobarbital and then intracardiacally perfused with 30 ml of Phosphate Buffer Saline (Corning). Blood were collected in PBS with 0.5 mM of Ethylenediaminetetraacetic acid (EDTA, Boston BioProducts, Inc.). Cells from brain, spinal cord, meninges, spleen and lungs were dissociated in RPMI 1640 medium, 2% FBS, 1 mg/ml of Collagenase D (Roche) and 0.5 mg/ml of DNAse I at 37°C for 30 min with mild shaking. Tissue suspensions were then filtered through a 70-μm cell strainer (BD Falcon). Myelin and debris from brain and spinal cord were eliminated using a 30% Percoll (GE Healthcare) gradient. After 20 min of centrifugation at 560g cell pellets were washed twice with PBS.
For spleen and blood, red blood cells were lysed in 1 and 5 ml of RBC Lysis Buffer (Affymetrix eBioscience), respectively.
Flow cytometry analyses and related reagents
For surface staining, cells were suspended in PBS containing 2% FBS, anti-mouse or anti-human CD16/32 (Biolegend) and 0.1% NaN3. Super Bright Complete Staining Buffer (eBioscience) was added when appropriate, according to the manufacturer’s protocol.
For intracellular staining, cells were fixed in Fixation/Permeabilization buffer (eBioscience) and stained in Perm/ Wash buffer (eBioscience).
For cytokine detection, cells were stimulated for 4 h with a Cell Stimulation Cocktail, PMA/Ionomycin (Biolegend) containing Brefeldin A in an incubator at 37ºC and 5% CO2.
T cells were gated as live CD45+CD19−NK1.1−TCRγδ−CD11b−TCRβ+. To exclude blood contaminations in the brain and spinal cords, 3 min before perfusion mice were injected intravenously with an anti-CD45 antibody conjugated with a fluorophore different from the one used in the antibody cocktail for flow cytometry analysis, as described58. Cells positive for the CD45 antibody injected intravenously were then excluded from the analysis.
Dead cells were discriminated in all experiments using LIVE/DEAD Fixable Dead Cell stain kit (Molecular Probes by Life technologies).
The following antibodies were used:
anti-mouse TruStain FcX PLUS (anti-mouse CD16/32) Antibody (clone S17011E, Biolegend, Catalog # 156603, 1:200), anti-mouse CD11b (clone M1/70, BioLegend, Catalog # 101245, 101254, 101222, 1:700), CD11c (clone N418, BioLegend, Catalog # 117365, 117303, 1:200), CD19 (clone 6D5, Catalog # BioLegend, 115545, 1:400), TCRgammadelta (clone GL3, BioLegend, Catalog # 118131, 1:400), NK1.1 (clone PK136, BioLegend, Catalog # 108737, 1:400), CX3CR1 (clone SA011F11, BioLegend, Catalog # 149039, 149005, 1:200), CD45 (clone 30-F11, BioLegend, Catalog # 103129, 103166, 103111,103115, 1:400 ), TCRbeta(clone H57–597, BioLegend, Catalog # 109229, 109207, 1:200 ), CD4 (clone GK1.5, BioLegend, Catalog # 100411, 100413, 100449, 1:400; clone RM4–5, Invitrogen, Catalog # 58–0042-82, Q22165, 1:400 ), CD8beta (clone 53–5.8, BioLegend, Catalog # 140410, 140415, 1:400), CD8alpha (clone 53–6.7, BioLegend, Catalog # 100709, 1:400), PD-1 (clone 29F.1A12, BioLegend, Catalog # 135215, 135209, 1:200), TNF-alpha (clone MP6-XT22, BioLegend, Catalog # 506349, 506321, 1:200), Tbet (clone 4B10, BioLegend, Catalog # 644807, 644827, 1:200), GrzB (clone GB11, BioLegend, Catalog # 515407, 1:100), IFN-gamma (clone XMG1.2, BioLegend, Catalog # 505803, 505845, 505821, 1:200), Eomes (clone W17001A, BioLegend, Catalog # 157703, 1:200), CD44 (clone IM7, BioLegend, Catalog # 103054, 103025, 103011, 103037, 1:400), CD62L (clone MEL-14, BioLegend, Catalog #104440, 104405, 1:400), CD49d (clone R1–2, Miltenyi Biotec, Catalog # 130–103-852, 1:100; clone R1–2, Biolegend, Catalog # 103625, 103613, 1:200), CD103 (clone 2E7, BioLegend, Catalog # 121431, 121415, 1:200 ), CD127 (clone A7R34, BioLegend, Catalog # 135021, 135009, 135031, 135023, 1:200 ), CD5 (clone 53–7.3, BioLegend, Catalog # 100609, 1:1000), Valpha2 (clone B201, eBioscience, Catalog # 12–5812-82, 1:200), Vbeta 5.1, 5.2 (clone MR9–4, BD Biosciences, Catalog # 51–01354L, 1: 100), Ly6C (clone HK1.4, BioLegend, Catalog # 128005, 1:200), Foxp3 (clone 150D, BioLegend, Catalog # 320011, 1:100), PDCA-1 (clone 129c1, BioLegend, Catalog # 127107, 1:200), F4/80 (clone BM8, BioLegend, Catalog # 123109, 1:200), Ly6G (clone 1A8, BD Biosciences, Catalog # 127653, 1:200), CD86 (clone GL-1, BioLegend, Catalog # 105027, 1:100), IA/IE (clone M5/114.15.2, BioLegend, Catalog # 107617, 1:1000), CD45.1 (clone A20, BioLegend, Catalog # 110725, 1:200), CD45.2 (clone 104, BioLegend, Catalog # 109813, 1:200), H2Kb/H2Db (clone 28–8-6, BioLegend, Catalog # 114605, 114613, 1:100), CD69 (clone H1.2F3, BioLegend, Catalog # 104539, 1:200), KLRG1 (clone 2F1/KLRG1, BioLegend, Catalog # 138409, 1:200), CD122 (clone 5H4, BD Biosciences, Catalog # 564924, 1:200), CD25 (clone 3C7, BioLegend, Catalog # 102021, 1:200), Fas (clone SA367H8, BioLegend, Catalog # 152609, 1:100), CCR6 (clone 29–2L17, BioLegend, Catalog # 129823, 1:200), PD-L1 (clone 10F.9G22, BioLegend, 124323, 1:200), TIM3 (clone 3MT3–23, BioLegend, Catalog # 119705, 1:200), VCAM1 (clone 429, BioLegend, Catalog # 105717, 1:200), CD28 (clone 37.51, BioLegend, Catalog # 102105, 1:200), CTLA4 (clone UC10–4B9, BioLegend, Catalog # 106317, 1:200), Perforin (clone S16009A, BioLegend, Catalog # 154305, 1:200), CCL2 (clone 2H5, BioLegend, Catalog # 505909, 1:100), CCR2 (clone SA203G11, BioLegend, Catalog # 150627, 1:200), TIGIT (clone Vstm3, Biolegend, Catalog # 142109, 1:100).
anti-mouse/human TOX (clone TXRX10, eBioscience, Catalog # 12–6502-80, 50–6502-80, 1:100), TCF-1/TCF-7 (clone C.225.7, Invitrogen, Catalog # MA5–14965, 1:100).
anti-human TruStain FcX (Fc Receptor Blocking Solution, BioLegend, catalog # 422301, 1:200), anti-human CCR7 (clone 2-L1-A, BD Biosciences, Catalog # 566744, 1:200), CD56 (clone 5.1H11, BioLegend, Catalog # 362519, 1:200), PD-1 (clone EH12.1, BD Biosciences, Catalog # 566175, 1:200), HLADR (clone L243, BioLegend, Catalog # 307637, 1:200), CD28 (clone CD28.2, BioLegend, Catalog # 302904, 1:200), CD8 (clone 3B5, Invitrogen, Catalog # Q22157, 1:200; clone SK1, Biolegend, Catalog # 344731, 1:200), CD69 (clone FN50, BioLegend, Catalog # 310913, 1:200), CD38 (clone HIT2, BioLegend, Catalog # 303507, 1:200), CD20 (clone 2H7, BioLegend, Catalog # 302333, 1:200), CD45RO (clone UCHL1, BioLegend, Catalog # 304231, 1:400), CD49d (clone (NF10, BioLegend, Catalog # 304331, 1:200), CD19 (clone H1B19, BioLegend, Catalog # 302261, 1:200), Fas (clone DX2, BioLegend, Catalog # 305647, 1:200), CD14 (clone MφP-9, BD Biosciences, Catalog # 566466),CD16 (clone CB16, eBioscience, Catalog # 46–0168-41, 1:200), CD3 (clone OKT3, BioLegend, Catalog # 317308, 1:200; clone S4.1, Invitrogen, Catalog # Q10484, 1:200), CD45RA (clone HI100, BioLegend, Catalog # 304145, 1:400), CD4 (clone SK3, eBioscience), CCR6 (clone G034E3, BioLegend), CD127 (clone A019D5, BioLegend, Catalog # 351317, 1:100), CD27 (clone M-T271, BioLegend, Catalog # 356415, 1:200), CXCR3 (clone G025H7, Biolegend, Catalog # 353721, 1:200), CD5 (clone UCHT2, BD Biosciences, Catalog # 564648, 1:400), CD44 (clone C44Mab-5, BioLegend, Catalog # 397509, 1:200), CD25 (clone M-A251, BioLegend, Catalog # 356131, 1:200), KLRG1 (clone 2F1/KLRG1, BioLegend, Catalog # 138421, 1:200), CD57 (clone QA1704, BioLegend, Catalog # 393327, 1:200), TIM3 (clone F38–2E2, BioLegend, Catalog # 345011, 1:100), CD64 (clone 10.1, BioLegend, Catalog # 305013 , 1:200), CD24 (clone ML5, Biolegend, Catalog # 311105, 1:200), TIGIT (clone MBSA43, eBioscience, Catalog # 25–9500-41, 1:100).
For unconjugated antibodies, we used PE Streptavidin Conjugate (BioLegend, Catalog # 405203, 1:500) or Qdot 605 Streptavidin Conjugate (Invitrogen, Catalog # Q10123MP, 1:500), and Alexa Fluor 546 donkey anti-rabbit IgG (H+L) (Life Technologies, Catalog # A10040, 1:2000).
Analysis of TCR Vbeta expression in human PBMCs by flow cytometry was performed using the Beta Mark TCR Vbeta Repertoire Kit (Beckman Coulter, Inc, Catalog # IM3497), according to the manufacturer’s protocol.
Data were acquired using the spectral cytometer Aurora (Cytek Bioscience) with 4 or 5 lasers and SpectroFlo 2.0. Analyses were performed with Cytobank Software 9.0 (Cytobank, Inc) and FCS Express 7 (De Novo Software).
Histology
Lumbar sections of spinal cords and sciatic nerves were fixed overnight in 4% PAF (paraformaldehyde). Spinal cords were additionally dehydrated in 30% sucrose (Sigma) for 1 hour before processing. Embedding in paraffin blocks and staining were conducted by the Biorepository and Pathology Dean’s CoRE at the ISMMS. Serially paraffin sections of 6–8 μm (n 4 sections per animal) of the spinal cord were stained with a rabbit anti-mouse polyclonal ChAT (Choline Acetyltransferase, a marker of motor neurons) serum (Millipore, Catalog # AB143, 1:100).
Glioma were harvested from mice and fixed overnight in 10% formalin solution (Sigma). Embedding in paraffin blocks and staining were conducted by the Biorepository and Pathology Dean’s CoRE at the ISMMS. Paraffin sections of 8 μm were stained using a rat anti-mouse CD8 (clone 4SM16, eBioscience, Catalog # 14–0195-82, 1:150) Tissue sections of paraffin embedded tissues from ALS4 and control patients were provided by The Johns Hopkins ALS Postmortem Core, at the Johns Hopkins School of Medicine, Baltimore, MD, and stained using a rabbit anti-human CD8 (clone SP57, Roche, Catalog # 05493846001, purchased pre-diluted from the vendor) by the Biorepository and Pathology Dean’s CoRE at the ISMMS.
Slides were digitally scanned and analyzed using NDP.view2.
For sciatic nerves, ultrathin sections of 1 μm thick were generated and stained with 1% toluidine blue for 30 sec by The Duke Electron Microscopy Service of the Duke University Pathology Core. Images were generated using a Keyence BZ-X710 microscope and Keyence BZ-X Analyzer Software 1.3.1.1. Axon number, perimeter and area were counted as described below.
Axon counting by Gaussian filters
We applied Gaussian filters to automatically count the number of cells per image. A Gaussian filter is an approximation of the Gaussian Function (eq1) to reduce noises and blur regions of an image object. Each pixel of the targeted region is multiplied by an odd sized symmetric kernel.
| (eq 1) |
Where Eq1 is the 2-D version of Gaussian function, (x,y) are the coordinates of each pixel, and is the standard deviation of the Gaussian kernel.
To obtain the total number of living cells, we first converted the colored image into grayscale, we then applied Gaussian filters with a standard deviation of 3 on the Gaussian kernel. Given that living cells exhibited higher brightness on the image, a threshold of pixel value of 100 was adopted to detect living cells. We then calculated each cell’s area (μm2) and perimeter (μm).
Mouse SMART-seq v4 and TCR Sequencing
For TCR sequencing in the CNS, brains and spinal cords from 10 month-old KI mice were pooled. After a 30% Percoll gradient, myelin was further eliminated using a Myelin Removal Beads II (Miltenyi Biotec), according to the manufacturer’s protocol. For TCR and SMART-seq, blood was collected by PBS perfusion in PBS with 0.5 mM EDTA and red blood cells were lysed as described above.
Cell suspensions from CNS, spinal cord and blood were enriched in T cells by Pan T Cell Isolation Kit II (Miltenyi Biotec), according to the manufacturer’s protocol. CD8 T cells were then sorted into CD44+ CD62L− PD-1+ and PD-1− using a 5 lasers BD FACSAria (BD Bioscience) with FACSDiva 8.0 (BD Bioscience).
For TCR sequencing in glioma, tumor-infiltrating immune cells were isolated as described59. Briefly, after perfusion with PBS, tumors from KI mice at day 27 post-tumor detection were minced into small pieces and meshed using a rubber plunger of a syringe through a cell strainer. Mononuclear cells were then isolated by density centrifugation using Ficoll-Plaque PREMIUM 1.084 (Cytiva) and CD8 T cells sorted using a 3 lasers BD FACSAria with FACSDiva 8.0 (BD Bioscience).
For TCR sequencing , genomic DNA from sorted cells was extracted using a QIAamp DNA Micro Kit (Qiagen) and sent for analysis. CDR3b regions were amplified and sequenced by Adaptive Biotechnologies Corp (Seattle, WA) using the ImmunoSEQ assay. Data were analyzed using the ImmunoSEQ ANALYZER 3.0.
For SMART-seq, 500 CD44+ CD62L− PD-1+ and PD-1− CD8 T cells from the blood were sorted directly into nuclease free water (final volume = 9.5 μl) and processed by the NGS Platform of the Center for Advanced Genomics Technology at ISMMS. Cell samples were passed to whole-transcriptome library preparation using the Takara Bio SMART-Seq v4 PLUS Kit following the manufacturer’s instructions. Briefly, cDNA synthesis from low amounts of RNA was enabled through Takara Bio’s SMART technology. 3’ ends of cDNA were then adenylated prior to ligation with adapters utilizing unique dual indices (96 UDIs) to barcode samples to allow for efficient pooling and high throughput sequencing. Libraries were enriched with PCR, with all samples undergoing 16 cycles of amplification prior to purification and pooling for sequencing. Completed libraries were quantified using Quant-iT reagents and equimolar pools were generated and sequenced on the NextSeq 550 using a high output 150 cycle flow cell generating paired end 75 bp reads and approximately 30 million reads per sample.
SMART-seq analysis
Reads were aligned to the Mus Musculus GRCm39 reference genome using STAR (v2.6.0a)60. Counts were summarized using the featureCount command from Subread (v2.0.3)61. Counts were used to generate a DGEList object using the DGEList function in edgeR (v3.32.1)62 and converted to counts per million using the cpm function. Genes with fewer than 1 cpm in each library were filtered and libraries were normalized with the trimmed mean of M values (TMM) method using the calcNormFactors function. Differential expression between PD-1+ and PD-1− groups was calculated using limma (v3.46.0)63. To do so, the filtered and normalized DGEList object was transformed to log2 counts per million and the mean-variance relationship was computed using the voom function. Following this, a linear model was fitted using the lmFit function and empirical Bayes statistics were calculated using the eBayes function. The differentially expressed genes between PD-1+ and PD-1− groups were visualized using ggplot2 (v3.3.3) and ggrepel (v0.9.1).
Cell isolation for splicing analysis
After intracardiac perfusion, spinal cords from 10–12 month old KI and WT mice were dissociated in Hank’s Balanced Salt Solution (HBSS) medium (Gibco), 1 mg/ml of DNAse I and 2 mg/ml of papain (Worhington) at 37°C for 30 min with mild shaking. Tissue suspensions were then filtered through a 70-μm cell strainer. Cell debris were eliminated after resuspension in HBSS 3.5% Bovine Serum Albumin (BSA, Sigma) and centrifuged at 120 g for 10 min. Cell pellets were resuspended in HBSS, layered over a 1.06 g/ml solution of Optiprep (Sigma). After centrifugation at 900g for 20 min, cells were collected in the upper layer, washed and resuspended in 1 ml of TRIzol.
Splenocytes from 5–6 week old WT animals were dissociated in RPMI 2% FBS, 0.5 mg/ml of Collagenase D and 0.2 mg/ml of DNAse I at 37°C for 30 min with shaking. After red blood cell lysis, CD8 T cell were negatively enriched using a Dynabeads Untouched Mouse CD8 T Cells Kit (Invitrogen), according to the manufacturer’s protocol. After staining, naïve CD62L+ CD44− CD8 T cells were sorted using a 5 lasers BD FACSAria with FACSDiva 8.0 (BD Bioscience). Naïve CD8 T cells (105) were resuspended in complete RPMI containing 4 μg/ml of purified anti-mouse CD28 (clone 37.51, Biolegend, Catalog # 102101) and stimulated in 48 well plates coated with 20 μg/ml of purified anti-mouse CD3 (clone 2C11, Biolegend, Catalog # 100201) in an incubator at 37ºC and 5% CO2. Recombinant human IL-2 (10 U/ml, R&D) was added to the culture at the end of day 1. After 3 days of culture, cells were left untreated or washed and additionally restimulated for 1 hour using a Cell Stimulation Cocktail, PMA/Ionomycin (Biolegend). In vitro activated CD8 T cells were then washed and resuspended in 1 ml TRIzol.
RNA separation and isolation were performed using chloroform and isopropanol, as described above.
RNAseq and splicing analysis
To prepare RNA-sequencing libraries, DNAse-treated (TURBO DNA-free Kit, Thermofisher scientific) RNA from the spinal cord samples (300 ng) and T cell samples (30 ng) were used. Libraries were prepared using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB) following manufacturer’s recommendations. Final libraries were quantified and sizing was determined using the High Sensitivity DNA Assay reagents and chip in the Agilent 2100 Bioanalyzer System and the Qubit 1X dsDNA HS Assay Kit respectively. Individual libraries were then pooled and sequenced using 75bp paired end on the NextSeq 550 using the NextSeq 500/550 High Output Kit.
For analysis, adaptor removal was performed with cutadapt64 and base-quality trimming to remove 3′read sequences if more than 20 bases with Q < 20 were present. Paired-end reads were mapped to the mus musculus (mm10) reference genomes with STAR. Summaries of gene-counts were performed using featureCounts65. The numeric matrix of raw read counts was then used for differential gene expression analysis with the Bioconductor Limma package63 after removing genes with less than 50 total reads across all samples or of less than 200 nucleotides in length. Normalization factors were computed on the filtered data matrix using the weighted trimmed mean of M-values (TMM) method, followed by voom66 mean-variance transformation in preparation for Limma linear modeling. Gene set enrichment analyses were performed with clusterProfiler67.
Alternative splicing analysis was perfomed using rMATS 4.1.068. Downstream splicing predictions was carried out using SPLINTER69.
Cytokine and autoantibody quantification in mice
Mice were anesthetized by isoflurane inhalation and then bled retroorbitally using heparinized micro-hematocrit capillary tubes (Fisherbrand). 20–50 μl of blood were collected in 1 ml Eppendorf tubes and centrifuged for 10 min at 9,500 g to separate the serum.
Concentration of cytokines was measured using Bio-Plex Pro Mouse Cytokine 8-plex Assay (Bio-Rad), according to the manufacturer’s protocol. Data was acquired using a Luminex 100/200 plate reader.
For autoantibody measurement, 10 μl of serum were analyzed using an Autoantigen Microarray Super Panel (128 antigen panel) by the Genomic and Microarray Core Facility of University of Texas Southwestern Medical Center, Dallas, TX, USA. Data were analyzed using R (version 4.0.5) and the package ComplexHeatMap (version 2.6.2).
Mass cytometry staining and acquisition
Fresh mouse blood cells were first incubated in a 37ºC water bath for 20 minutes with Cell-ID Rh103 Intercalator (Fluidigm,) to label non-viable cells. Samples were then blocked with Fc receptor blocking solution, Fc anti-mouse CD16/32 (Biolegend), and stained with a cocktail of surface antibodies for 30 minutes on ice. All antibodies were either conjugated in-house using Fluidigm’s ×8 polymer conjugation kits or purchased commercially from Fluidigm. Next, samples were fixed with freshly diluted 2.4% formaldehyde in PBS containing 0.02% saponin and iridium intercalator to label nucleated cells and barcoded and pooled using Fluidigm’s 20-Plex Pd barcoding kit (Fluidigm). The pooled sample was then then stored as pellets in PBS until acquisition.
Immediately prior to acquisition, the pooled sample was washed with Cell Staining Buffer and Cell Acquisition Solution (Fluidigm) and resuspended in Cell Acquisition Solution at a concentration of 106 cells/ml containing a 1:20 dilution of EQ normalization beads. The sample was acquired on the Fluidigm Helios mass cytometer using the wide bore injector configuration at an acquisition speed of < 400 cells per second.
The resulting FCS files were normalized and concatenated using Fluidigm’s CyTOF software 7.0.8493 and then de-multiplexed using the Zunder lab single-cell debarocder (https://github.com/zunderlab/single-cell-debarcoder). The FCS files were further cleaned on Cytobank 7.2.1 by removing EQ beads, low DNA debris and gaussian multiplets. Barcoding multiplets were also removed based on the mahalanobis distance and barcode separation distance parameters provided by the Zunder lab debarcoder.
The following isotope conjugated anti-mouse antibodies were used at 1:100 dilution:
113 In CD45.1 (clone A20, eBioscience, Catalog # 14–0453-85), 115 In CD45.2 (clone 104, eBioscience, Catalog # 14–0454-85), 141 Pr Ly6G (clone 1A8, Fluidigm, Catalog # 3141008B), 142 Nd CD11c (clone N418, Biolegend, Catalog # 117302), 143 Nd TCRbeta (clone H57–597, Fluidigm, Catalog # 3143010B), 144 Nd CD24 (clone M1/69, Biolegend, Catalog # 101802),145 Nd CD69 (clone H1.2F3, Fluidigm, Catalog # 3145005B), 146 Nd F4/80 (clone BM8, Fluidigm), 147 Sm CD357/GITR (clone DTA-1, Biolegend), 148 Nd CD11b (clone M1/70, Biolegend, Catalog #), 149 Sm CD19 (clone 6D5, Fluidigm, Catalog #), 150 Nd IgD (clone 11–26c.2a, Biolegend, Catalog #), 151 Eu CD25 (clone 3C7, Fluidigm, Catalog # 3146008B), 152 Sm SiglecF (clone S17007L, Biolegend, Catalog # 155502), 153 Eu NKp46 (clone 29A1.4, Fluidigm, Catalog # 3153006B), 155 Gd CD73 (clone TY/11.8, Biolegend, Catalog # 127202), 156 Gd CD64 (clone X54–5/7.1, Biolegend, Catalog # 139302), 158 Gd CD117 (clone 2B8, Biolegend, Catalog # 105802), 159 Tb PD-1 (clone RMP1, Biolegend, Catalog #), 160 Gd CD62L (clone MEL-14, Fluidigm, Catalog # 109113),162 Dy Ly6C (clone HK1.4, Biolegend, Catalog # 128002),163 Dy anti-APC (Biolegend, Catalog # 408002),164 Dy Sca-1 (clone D7, Fluidigm, Catalog # 3164005B), 165 Ho anti-PE (Biolegend, Catalog # 408102), 166 Er KLRG1 (clone 2F1/KLRG1, Biolegend, Catalog # 93135),167 Er CXCR3 (clone CXCR3–173, Biolegend, Catalog # 126526), 168 Er CD8alpha (clone 53–6.7, Biolegend, Catalog # 100702), 169 Tm CD206 (clone C068C2, Fluidigm, Catalog # 3169021B), 170 Er NK1.1 (clone PK136, Fluidigm, Catalog # 3170002B), 171 Yb CD44 (clone IM7, Biolegend, Catalog # 103002), 172 Yb CD4 (clone RM4–5, Fluidigm, Catalog # 3172003B), 174 Yb MHCII (I-A/I-E) (clone M5/114.15.2, Biolegend, Catalog # 107602), 175 Lu CD127 (clone A7R34, Biolegend, Catalog # 135002), 176 Yb B220 (RA3–6B2, clone Biolegend, Catalog # 103202). Anti-mouse CD68-PE (clone FA-11, Catalog # 137014) and CD88-APC (clone C5aR, Catalog # 135808) were purchased from Biolegend.
Single cell CITE-sequencing
PBMCs were incubated on ice for 20 min with 100 μl of the hash mixture in PBS 2% BSA, and then washed three time before CD8 T cell negative selection using a Dynabeads Untouched Human CD8 T Cells Kit (Invitrogen), according to the manufacturer’s protocol.
Hashed cells were counted on a Nexcelom Autocounter 2000 using AO/PI dye and pooled based on live cell counts. Pooled samples were centrifuged and subsequently resuspended in 100 μl of BioLegend’s Total Seq C CITE-Seq antibody panel and incubated on ice for 30 minutes. Antibodies were used at a concentration of 1:100 and diluted in wash buffer (PBS, 0.5% BSA, 1mM EDTA). After 3 washes, cells were resuspended in loading buffer (PBS, 0.5% BSA) and counted. Cells were loaded on one lane of the 10× 5’v1.1 assay with a targeted cell recovery of 20,000 cells.
For scRNA seq and scTCR seq, base call files from a NovaSeq6000 Sequencer (Illumina) and a NextSeq550 Sequencer (Illumina), respectively, were generated by the Human Immune Monitoring Center and the Genomic CoRE at the ISMMS.
BCL files were base-called and demultiplexed using cellranger mkfastq v3.1.0. Alignment of gene expression libraries to human reference GRCh38 as well as the counting of unique features of CITESeq libraries were performed using cellranger count v3.1.0.
The following antibodies were used for CITE-seq, purchased from Biolegend and used at 1:100 dilution:
TotalSeq-C0046 anti-human CD8 (clone SK1, Catalog # 344753); TotalSeq-C0049 anti-human CD3 (clone SK7, Catalog # 344849);TotalSeq-C0063 anti-human CD45RA (clone HI100, Catalog # 304163); TotalSeq-C0148 anti-human CD197 (CCR7) (clone G043H7, Catalog # 353251); TotalSeq-C0390 anti-human CD127 (IL-7Ralpha) (clone A019D5, Catalog # 351356);TotalSeq-C0088 anti-human CD279 (PD-1) (clone EH12.2H7, Catalog # 329963); TotalSeq-C0154 anti-human CD27 (clone O323, Catalog # 302853); TotalSeq-C0386 anti-human CD28 (clone CD28.2, Catalog # 302963 ); TotalSeq-C0140 anti-human CD183 (CXCR3) (clone G025H7, Catalog # 353747); TotalSeq-C0071 anti-human CD194 (CCR4) (clone L291H4, Catalog # 359425); TotalSeq-C0141 anti-human CD195 (CCR5) (clone J418F1, Catalog # 359137); TotalSeq-C0143 anti-human CD196 (CCR6) (clone G034E3, Catalog # 353440); TotalSeq-C0160 anti-human CD1c (clone L161, Catalog # 331547 ); TotalSeq-C0085 anti-human CD25 (clone BC96, Catalog # 302649); TotalSeq-C0171 anti-human/mouse/rat CD278 (ICOS) (clone C398.4A, Catalog # 313553 ); TotalSeq-C0410 anti-human CD38 (clone HB-7, Catalog # 356637); TotalSeq-C0087 anti-human CD45RO (clone UCHL1, Catalog # 304259 ); TotalSeq-C0168 anti-human CD57 (clone QA17A04, Catalog # 393321 ); TotalSeq-C0159 anti-human HLA-DR (clone L243, Catalog # 307663 ); TotalSeq-C0089 anti-human TIGIT (VSTM3) (clone A15153G, Catalog # 372729); TotalSeq-C0250 anti-mouse/human KLRG1 (MAFA) (clone 2F1/KLRG1, Catalog # 138433).
Analysis of single cell CITE-sequencing
The Cell Ranger filtered feature matrices were used to generate a UMI count matrix with the R package Seurat (v4.0.1)70. A Seurat Object was created where each feature was detected in at least 3 cells with cells having at least 200 features. Independent assays for RNA, Antibody-capture, and Cell Hashing were added to the Seurat Object. Cells having between 200 and 2500 unique features containing less than 10% mitochondrial reads were retained and TCR features were excluded. Read counts were normalized using the NormalizeData Seurat function with default parameters. Antibody-capture and Cell Hashing counts were normalized with a centered log-ratio transformation. Cells were demultiplexed using the HTODemux Seurat function and cells exhibiting a single unique identity were retained for downstream analysis. Following all filtering steps, 16,163 genes remained across 10,112 cells. Uniform Manifold Approximation and Projection clustering analysis was performed after PCA on the scaled RNA expression data using the first 25 principal components as input features and a 0.8 cluster resolution. Contaminant clusters 3, 10, 11, 12, i.e. non CD8 T cells, were subsequently filtered out using the Seurat subset function, leaving 16,163 genes across 8,609 cells. Uniform Manifold Approximation and Projection clustering analysis was repeated on the remaining cells using the first 20 principal components as input features and a 0.8 cluster resolution. Differentially expressed genes between all clusters were identified with the FindAllMarkers function using the Wilcoxon Rank Sum test where genes were detected in at least 30% of cells. A TEMRA-only Seurat Object was created using clusters 1 and 3 as the subset. Differentially expressed genes between ALS4 patients and healthy controls within the TEMRA subset were identified with the FindMarkers function incorporating the MAST (v1.16.0) model. The differentially expressed genes were visualized using ggplot2 (v3.3.3) and ggrepel (0.9.1) to plot the average log2 fold change and negative log10 transformed P value from the MAST test. Points were highlighted in red if they had a log2 fold change greater than 0.6 and a Bonferroni-corrected P value less than 0.05. Points were highlighted in blue if they had a log2 fold change less than −0.6 and a Bonferroni-corrected P value less than 0.05.
The filtered_contig_annotations output from Cell Ranger was used to analyze TCR clonality with the scRepertoire (v1.1.4) R package71. The combineTCR function was used to create a list object of TCR genes and CDR3 sequences by cell barcodes. The combineExpression function was used to integrate the clonotype information with the filtered Seurat Object. The frequency of clonotypes were binned by patient with the following parameters: Single=1, Small=5, Medium=20, Large=100, Hyperexpanded=500. Clonotypes were determined using use the VDJC genes comprising the TCR. Distribution of clonotype bins were visualized using the Seurat DimPlot function.
To visualize TCR clonality in the TEMRA compartment of individual donors, clonotypes were first binned as described above. Cells that did not have both their TCRα and TCRβ chains sequenced were removed from the analysis. Thereafter, the frequency of cells corresponding to each clonotype group per patient and only within clusters 1 and 3 (TEMRA only cluster) were visualized in donut plots.
Our list of TCR genes and CDR3 sequences was corroborated by intersecting our dataset with reported sequence datasets of TCRs specific to EBV and CMV44–46 (https://doi.org/10.21417/B7001Z; http://friedmanlab.weizmann.ac.il/McPAS-TCR/; EMBL-GenBank-DDBJ under accession numbers AJ010874–877, AJ010878–883, A010884–886, AJ010887–895, and AJ010896–900).
Accelerating rotarod
Mice were trained on an elevated accelerating rod (3 cm in diameter, Omnitech Electronics Inc) for 3 days as following: on day 0 mice were allowed to run free on the rotarod without acceleration (4 rpm/min) for 5 minutes; on day 1 they underwent 3 trials/each mouse with an acceleration of 4–40 rpm in 5 min; on day 2 they underwent 3 trials/each mouse with an acceleration of 4–40 rpm in 3 min. On day 3, mice were placed on a rotarod for comparative testing with an acceleration of 4–40 rpm in 3 min and the time of falling from the rotarod were recorded for 3 trails. Each trial lasted for a maximum of 3 min, during which the rotarod underwent a linear acceleration from 4 to 40 rpm. A 20-min interval was used between trials to avoid fatigue. Animals were scored for their latency to fall.
Tumor models
Tumor-derived neural stem cells (NSCs) overexpressing PDGFB (Platelet Derived Growth Factor Subunit B) and deficient for p53 (tumor protein P53) and Nf1 (Neurofibromin 1), a kind gift from M. Squatrito, were generated as described39. NSCs and tumorspheres were grown in Mouse NeuroCult proliferation kit (Stem Cell Technologies), supplemented with 10 ng/ml recombinant human EGF (Gibco), 20 ng/ml basic-FGF (Sigma), and 0.2 μg/ml Heparin (Stem Cell Technologies). Spheres were periodically dissociated with Accumax (Accutase) following the manufacturer’s instructions. After dissociation, 5×105 cells were resuspended in 100 μl of Mouse NeuroCult proliferation kit and Matrigel (Corning) in 1:1 ratio, and subcutaneously injected in the mouse flank.
The B16-F10 (ATCC CRL-6475) cell line was grown in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% FBS, 100 μg/ml penicillin and 100 μg/ml streptomycin. After dissociation with Trypsin-EDTA (0.25%) (Gibco), 5×105 cells were resuspended in 100 μl of DMEM and subcutaneously injected in the mouse flank.
B16-F10 were purchased with validation statements from ATCC.
NSCs and B16-F10 cells were further authenticate by PCR and flow cytometry (NSCs, GFP), by morphology, and in vivo and in vitro growth (B16-F10 and NSCs). NSCs and B16-F10 cells resulted negative from mycoplasma contamination.
Tumor measurement was started from the first day of tumor detection in at least one animal, and determined using a digital caliper. Tumor volume was calculated using the formula: Tumor volume (V) = L × W2, where L, or tumor length, is the larger diameter, and W, or tumor width, is the smallest diameter72. Animals showing solid and visible tumors larger than 1 cm sphere or multiple tumors cumulatively >2 cm were evaluated for euthanasia upon consultation with the Attending Veterinarian.
Statistical analysis
ANOVA and t-test, with the corresponding post-tests, were performed as indicated in the figure legends using Prism 5 (GraphPad).
Data availability
CITE-seq, SMART-seq and RNAseq datasets have been deposited online in the Gene Expression Omnibus (GEO), accession codes: GSE180410, GSE199663, GSE199664, GSE199666.
Extended Data
Extended Data Fig. 1. Characterization of KI mice.
a, Average latency to fall (sec) from an accelerating rotarod of WT and KI mice tested at 6 and 8 months of age (2 independent experiments, n=11 (WT 6 months and KI 8 months) and n= 13 (WT 8 months and KI 6 months) individual mice per group; mean +/− SEM). p-value calculated using the one-way Repeated Measures ANOVA (**p=0.0012) and Bonferroni’s post-test. (b,c) Bone marrow derived dendritic cells (BMDCs) from Setx WT and KI (b,c), and HET and KO (b) mice were infected with Citrobacter rodentium (multiplicity of infection=10, b) or stimulated with 5 μg/ml of cGAMP (c). Bar graphs show the relative induction of the indicated transcripts by RT-qPCR analysis at 2 hours post-infection or cGAMP stimulation in KO and KI cells normalized to their controls (2 independent experiments, n=4 mice/group, mean +/− SEM). No expression of the indicated genes was detected in uninfected cells. d, Concentration of the indicated cytokines measured by Luminex Assay in the serum of KI and WT (8–9 months old) animals (1 out of 2 independent experiments, n=6 (WT) and n=5 (KI) mice/group, mean +/− SEM). e, RT-qPCR analysis for the indicated inflammatory genes in the spinal cord of 10 months old WT and KI mice (Log2 relative expression against Actb, 2 independent experiments, n=5 mice/group, mean +/− SEM). f, Absolute numbers of indicated immune cell subsets in the spleen of 8–9 month old KI and WT mice. pDC: plasmacytoid dendritic cells; cDC: conventional dendritic cells. g, Expression of the activation marker CD86 in cDC and CD11b+ cells from the spleen of 8–9 months old WT and KI mice. (f,g) 2 independent experiments, n=4 mice/group, mean +/− SEM.
Extended Data Fig. 2. Autoantibody secretion in KI and WT mice.
Heat maps showing an unsupervised hierarchical clustering based on IgM (top) and IgG (bottom) reactivity against 128 self-antigens in the serum of 10–12 month old KI and WT mice (n=5 mice/group).
Extended Data Fig. 3. Related to Figure 1.
a, b, Peripheral blood cells from 8 month old WT and KI mice were analyzed by mass cytometry. a, t-SNE plots after gating on live CD45high (top) and CD19−NK1.1−CD11b−CD11c−TCRβ+ (bottom). b, Frequency of PD-1+ expressing cells after gating on CD44+ CD8+ T cells. Three mice/group were pooled for mass cytometry analysis. c, Gating strategy for flow cytometry analysis of CD8 T cells in the CNS. d, Frequency of PD-1+ CD8 T cells in the spleen and lungs of WT and KI mice at the indicated ages (2 independent experiments, 4 mice/group; mean +/− SEM). d, Frequency of PD-1+ (left) and Foxp3+ (right) CD4 T cells in the peripheral blood of WT and KI mice at the indicated ages (3 independent experiments, n=6 (left) and 2 independent experiments, n=4 (8 months) and n=6 (right) individual mice/group. SB=super bright.
Extended Data Fig. 4. PD-1+ CD8 T cells are effector memory T cells activated in an antigen fashion.
. a, Expression of indicated surface markers in CD8 T cells from the spinal cord (top), brain (middle) and peripheral blood (bottom) of 10–12 month old KI animals after gating on PD-1+ and PD-1− CD8 T cells. Histograms are representative of 3 independent experiment (n=6 mice). b, Median fluorescence intensity (MFI) of the indicated markers in CD44+CD62L− PD-1+ and PD-1− CD8 T cells isolated from the spinal cord (top) and blood (bottom) of 10–12 month old KI mice (2 independent experiments, n=6 (blood TIGIT and TOX), n=5 (spinal cord TIGIT and TOX), n=4 (TCF-1) individual mice/group, mean +/− SEM). p-value calculated using a two-tailed unpaired t test with Welch’s correction (spinal cord: ***p=0.0007; *p=0.045; **p=0.0066; blood: ***p=0.0004 (TOX); ***p=0.0002 (TIGIT and TCF-1). c, KI mice were crossed to OT-I mice and the offspring analyzed at 10 months of age. Dot plots from spinal cords, SC (left), and bar graphs (right) show the frequency of PD-1+ among OVA specific (gated on Vα2+Vβ5+) and non-TCR transgenic (gated on Vα2−) CD8 T cells (3 independent experiments, n=6, brain and spinal cord, and n=5, blood, individual mice; mean+/− SEM). p-value calculated using a two-tailed unpaired t test with Welch’s correction. **p=0.0031 (brain) ; ***p=0.0004 (SC); ***p=0.0003 (blood). eF=eFluor.
Extended Data Fig.5. Anti-tumor immunity and alternative splicing in KI mice.
a, Kinetics of tumor growth in 10–12 month old KI and WT mice subcutaneously injected with B16-F10 cells. Two independent experiments, n=10 individual mice/group; mean+/− SEM. ns = not significant, p-value calculated using a two-tailed Wilcoxon matched-pairs signed rank test. b, Scans of CD8 staining inside glioma isolated from 3 individual WT and KI from 2 independent experiments at day 27 post-tumor detection. c, Spinal cords from 10 to 12 month old WT (n= 4) and KI (n=3) mice (top) and in vitro activated WT CD8 T cells (n=3) (bottom) were analyzed by RNA-seq. WT CD8 T cells were used as positive control since increased AS upon T cell activation has been reported73. Bar graphs show transcript counts of significantly changed (| deltaPSI | > 0.15, p<0.05, FDR < 0.05) AS events using mm10 annotation. n= individual mice per group. SE, exon skipping; RI, intron retention; MXE, mutually exclusive exons; A5SS, alternative 5′ splice site; A3SS, alternative 3′ splice site.
Extended Data Fig. 6. Related to Figure 3.
a, Schematic of bone marrow cell transplantation experiments. b, Representative dot plots of the frequency of CD45.2 (recipient) and CD45.1 (donor) cells in the peripheral blood of chimeric mice 8 weeks after irradiation and reconstitution. c, Average of motor neuron area calculated in the lumbar region of spinal cord sections. n=5 mice/group, mean +/− SD. p-value calculated using one-way ANOVA Kruskal-Wallis test and Dunn’s post-hoc test (*p=0.0002). d, Axon areas (left) and number of large caliber axons (right) in the sciatic nerve. n=6 (WT/WT, KI/KI, KI/WT) and 7 (WT/KI) animals per group; mean +/− SEM. p-value calculated using one-way ANOVA Kruskal-Wallis test and Dunn’s post-hoc test (*p=0.026, ns=not statistically significant). Chimeric mice in (c,d) were analyzed at 12–13 months after reconstitution. e, Peripheral blood cells from two mice of the indicated group were pooled and analyzed by mass cytometry. Left, Details of markers and cell subsets identified by SPADE (spanning-tree progression analysis for density-normalized events) analysis (WT/WT group is shown). Right, SPADE analysis showing the distribution of immune cells into 4 main populations. Size and color of circles are dependent on the number of cells and the median intensity of expression for each marker, respectively. Red circles highlight PD-1+ CD8 T cells.
Extended Data Fig. 7. Autoantibody secretion in KI/KI and WT/WT chimeric mice.
Heat maps showing an unsupervised hierarchical clustering based on IgM (top) and IgG (bottom) reactivity against 128 self-antigens in the serum of chimeric mice at 12 months after reconstitution (n=3 WT/WT and n=2 KI/KI chimera).
Extended Data Fig. 8. Absence of T cell signature in FusP517L/WT and SOD1-G93A mice.
(a,d) Cell suspensions from brain, spinal cord, spleen, and peripheral blood of 22–24 month old FusP517L/WT and FusWT/WT littermates were analyzed by spectral flow cytometry. (a,b) t-SNE of total live CD45+ cells (a) and frequency of TCRβ+ CD8+ and CD4+ T cells expressing CD62L, CD44, PD-1+ and Foxp3 (b,c) from the spinal cord of FusP517L/WT and FusWT/WT mice after concatenation of all of the 5 samples/group. d, Frequency of CD8 T cell subsets in the indicated organs from the 2 genotypes (2 independent experiments, 5 individual mice/group; mean +/− SEM). e, Frequency of CD8 T cell subsets in the indicated organs from 3–4 month old non carrier and SOD1-G93A mice by spectral flow cytometry analysis (2 independent experiments, n=5 (spleen), n=6 (brain, WT blood) and n=7 individual mice/group; mean +/− SEM).
N= naïve, CD62L+CD44−; CM= central memory, CD62L+CD44+; EM= effector memory CD62L−CD44+. T cells are gated on live CD45high CD19−NK1.1−CD11b−CD11c−TCRγδ−TCRβ+ CD8+ or CD4+ cells.
Extended Data Fig. 9. Related to Fig. 4.
a, PBMCs from age-matched controls and patients were analyzed by spectral flow cytometry. Frequencies of naïve (CD45RA+ CD45RO− CD27+CD28+) and central memory (TCM, CD45RO+ CD45RA− CD28+) were determined after gating on CCR7+ CD8 (left) or CD4 (right) T cells. Frequencies of effector (TEff, CD45RA+/− CD45RO+/− CD27+CD28+), effector memory (TEM, CD45RA− CD45RO+ CD27+CD28+) and terminally differentiated effector memory (TEMRA, CD45RO− CD45RA+ CD28−) were determined after gating on CCR7− CD8 or CD4 T cells. T cells were gated on live CD19−CD16− CD56−CD3+ cells. Each dot represents one individual donor and graph show results from 2 independent experiments (n=5 ALS4 and n=10 controls). mean +/− SEM. p-value calculated for each T cell subset using Unpaired two-tailed t test with Welch’s correction (Teff: ***p=0.0002; TEMRA:***p=0.0067). ns= not significant. b, Levels of expression of TOX, TIGIT and TCF-1 in the indicated CD8 T cell subsets from the peripheral blood of ALS4 patients (n=4) and age-matched controls (n=8). Two independent experiments, mean +/− SEM. p-value calculated by comparing TEMRA CD8 T cells from patients and controls pool together to the other subsets using one-way ANOVA and Tukey post-hoc test. c,d, UMAP projections of 20,000 single cells from controls (n=3) and ALS4 patients (n=3) colored by cluster (c) or by cell subsets (d) are shown on the left and Z-score normalized mean expression of differentially expressed genes in the indicated clusters (c) or CD8 T cell subsets (d) are shown on the right.
Extended Data Fig. 10. Phenotype of CD8 T cells from ALS4 patients and controls.
a,b, UMAP projections of identified clusters showing differential expression of surface markers (by oligo-conjugated antibodies) (a) and of the top genes (b) in ALS4 versus control donors. c, Spectral flow cytometry analysis of the 6 samples used for CITE-seq. Histograms show the frequency of CCR7+ CD8 T cells and dot plots the proportion of the indicated subsets after gating on CCR7pos or neg CD8 T cells as indicated.
Extended Data Fig. 11. Clonal expansion and antigen specificity of TEMRA CD8 T cells from ALS4 patients.
a-c, CD8 T cells from controls and ALS4 patients were analyzed by CITE-seq. a, Donut plots depicting T cell clonality per CD8 T cell subset and per group. b, Donut plots indicating the number of TEMRA CD8 T cells for each indicated clonal category. Each patient and the corresponding age-matched controls are individually shown. c, Z-score normalized mean expression of differentially expressed surface markers (by oligo-conjugated antibodies) in “High” (clonality >20) versus “Low” expanded TEMRA CD8 T cells from ALS4 patients. “High” and “Low” clones have been subsampled for equal representation on the heatmap. d, PBMCs from 2 ALS4 patients and 4 age-matched controls were analyzed by spectral flow cytometry using a mixture of fluorophore-conjugated antibodies against 24 human TCR Vβ. Graph bars show the frequencies of the top 5 TCR VB expressed by TEMRA CD8 T cells in each donor. Top 5 VB chains are indicated in the table below the graph. e, Concentration of IFN-γ in the supernatant of PBMCs from controls (n=10) and ALS4 (n=4) patients stimulated for 6 days with 15mers overlapping TDP-43 (106 peptides), Senataxin (703 peptides divided in 2 sub-pools), or with a pool of 80 pathogen-derived peptides. Two independent experiments, mean +/− SEM.
Extended Data Fig. 12. CD8 T cells are detected in the ventral horn of lumbar spinal cords from ALS4 patients.
CD8 T cell staining in the indicated post-mortem paraffin-embedded tissues from ALS4 patients and controls. For spinal cord sections, scans show the ventral horn. Narrows indicate CD8 T cells. n=2.
Supplementary Material
Acknowledgments.
We thank Nan Zhao and Simin Zheng from the Marazzi’s lab, all the teams at the Icahn School of Medicine at Mount Sinai, Elodie Drapeau for teaching the use of the rotarod, Thomas Kraus for use of the Luminex 100/200 plate reader, Sara Cuadrado-Castano and Aryana Javaher for help with the B16 melanoma model, the Flow Cytometry and Mouse facilities, the NGS Platform of the Center for Advanced Genomics Technology, Travis Dawson, Darwin D’souza, Bhaskar Upadhyaya and all the team of the Human Immune Monitoring Center, Alan Soto, Monica Garcia-Barros and the Biorepository and Pathology Dean’s CoRE. We are grateful to Lara Manganaro, Julien Moretti and Andrew Chan for their help in the preliminary phase of the project, Matthew J. Miller (McMaster University, Hamilton, Ontario, Canada) and Martin F. Lavin (University of Queensland, Brisbane, Australia) for Setx deficient mice, The Genomic and Microarray Core Facility of University of Texas Southwestern Medical Center, Dallas, TX, The Duke Electron Microscopy Service of the Duke University Pathology Core, Durham, NC, The Johns Hopkins ALS Postmortem Core, Kathleen Wilsbach, and Kathryn Gallo at the Johns Hopkins School of Medicine, Baltimore, MD, and the New York Blood Center. We are indebted to ALS4.org for help and support.
This work was supported by The Burroughs Wellcome Fund 1017892 (I.M.), the Chan Zuckerberg Initiative Neurodegeneration Challenge Network Ben Barres Early Career Acceleration Awards (to I.M., 2018-191895, and N.J., 2018-191856 (5022)), the Hirschl Young Investigator fellowship (I.M.), the National Institutes of Health grants U01AI150748, R01AI143840 and R01AI168130 (I.M.), R33CA225539 and R01AG067581 (N.J.), R35 NS122140 (A.R.L.), T32 AG00096 (F.A.), 5R01NS106236 (N.A.S.), R01HL153974 and U01AI150748 (M.B.), the startup fund of the School of Engineering and Applied Science at University of Pennsylvania (N.J.), ARISLA Italy, ALS Association ALSA, USA, and the Italian Ministry of Health CCM2011 (C.L.). M.S. received support from the Seve Ballesteros Foundation. M.G. is a recipient of a post-doctoral fellowship from the Fundación Alfonso Martín Escudero (Spain).
Footnotes
Competing interests. C.L. is the Scientific Director of the company NEMO LAB, and he served on a scientific advisory board for Mitsubishi Tanabe Pharma Europe, Cytokinetics, Neuraltus and Italfarmaco. The other authors declare no competing interests.
Supplementary Information is available for this paper.
References
- 1.Brown RH & Al-Chalabi A. Amyotrophic Lateral Sclerosis. N Engl J Med 377, 162–172, doi: 10.1056/NEJMra1603471 (2017). [DOI] [PubMed] [Google Scholar]
- 2.McCauley ME & Baloh RH Inflammation in ALS/FTD pathogenesis. Acta Neuropathol 137, 715–730, doi: 10.1007/s00401-018-1933-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grunseich C. et al. Clinical and Molecular Aspects of Senataxin Mutations in Amyotrophic Lateral Sclerosis 4. Ann Neurol 87, 547–555, doi: 10.1002/ana.25681 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boillee S. et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science 312, 1389–1392, doi: 10.1126/science.1123511 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Yamanaka K. et al. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci 11, 251–253, doi: 10.1038/nn2047 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butovsky O. et al. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J Clin Invest 122, 3063–3087, doi: 10.1172/JCI62636 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCauley ME et al. C9orf72 in myeloid cells suppresses STING-induced inflammation. Nature 585, 96–101, doi: 10.1038/s41586-020-2625-x (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Rourke JG et al. C9orf72 is required for proper macrophage and microglial function in mice. Science 351, 1324–1329, doi: 10.1126/science.aaf1064 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atanasio A. et al. C9orf72 ablation causes immune dysregulation characterized by leukocyte expansion, autoantibody production, and glomerulonephropathy in mice. Sci Rep 6, 23204, doi: 10.1038/srep23204 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burberry A. et al. Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Sci Transl Med 8, 347ra393, doi: 10.1126/scitranslmed.aaf6038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oakes JA, Davies MC & Collins MO TBK1: a new player in ALS linking autophagy and neuroinflammation. Mol Brain 10, 5, doi: 10.1186/s13041-017-0287-x (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao Y. et al. The kinase TBK1 functions in dendritic cells to regulate T cell homeostasis, autoimmunity, and antitumor immunity. J Exp Med 214, 1493–1507, doi: 10.1084/jem.20161524 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becherel OJ et al. Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet 9, e1003435, doi: 10.1371/journal.pgen.1003435 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller MS et al. Senataxin suppresses the antiviral transcriptional response and controls viral biogenesis. Nat Immunol 16, 485–494, doi: 10.1038/ni.3132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett CL et al. Senataxin mutations elicit motor neuron degeneration phenotypes and yield TDP-43 mislocalization in ALS4 mice and human patients. Acta Neuropathol 136, 425–443, doi: 10.1007/s00401-018-1852-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rathinam VA et al. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell 150, 606–619, doi: 10.1016/j.cell.2012.07.007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu CH et al. TDP-43 Triggers Mitochondrial DNA Release via mPTP to Activate cGAS/STING in ALS. Cell 183, 636–649 e618, doi: 10.1016/j.cell.2020.09.020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gate D. et al. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature 577, 399–404, doi: 10.1038/s41586-019-1895-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gate D. et al. CD4(+) T cells contribute to neurodegeneration in Lewy body dementia. Science 374, 868–874, doi: 10.1126/science.abf7266 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindestam Arlehamn CS et al. alpha-Synuclein-specific T cell reactivity is associated with preclinical and early Parkinson’s disease. Nat Commun 11, 1875, doi: 10.1038/s41467-020-15626-w (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sulzer D. et al. T cells from patients with Parkinson’s disease recognize alpha-synuclein peptides. Nature 546, 656–661, doi: 10.1038/nature22815 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimoto M. et al. CD8 T Cell Exhaustion in Chronic Infection and Cancer: Opportunities for Interventions. Annu Rev Med 69, 301–318, doi: 10.1146/annurev-med-012017-043208 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Baruch K. et al. PD-1 immune checkpoint blockade reduces pathology and improves memory in mouse models of Alzheimer’s disease. Nat Med 22, 135–137, doi: 10.1038/nm.4022 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Schenkel JM, Fraser KA & Masopust D. Cutting edge: resident memory CD8 T cells occupy frontline niches in secondary lymphoid organs. J Immunol 192, 2961–2964, doi: 10.4049/jimmunol.1400003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baron JL, Madri JA, Ruddle NH, Hashim G. & Janeway CA Jr. Surface expression of alpha 4 integrin by CD4 T cells is required for their entry into brain parenchyma. J Exp Med 177, 57–68, doi: 10.1084/jem.177.1.57 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young KG, Maclean S, Dudani R, Krishnan L. & Sad S. CD8+ T cells primed in the periphery provide time-bound immune-surveillance to the central nervous system. J Immunol 187, 1192–1200, doi: 10.4049/jimmunol.1100695 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campisi L. et al. Apoptosis in response to microbial infection induces autoreactive TH17 cells. Nat Immunol 17, 1084–1092, doi: 10.1038/ni.3512 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma A, Koka R. & Burkett P. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu Rev Immunol 24, 657–679, doi: 10.1146/annurev.immunol.24.021605.090727 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Sekine T. et al. TOX is expressed by exhausted and polyfunctional human effector memory CD8(+) T cells. Sci Immunol 5, doi: 10.1126/sciimmunol.aba7918 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Markovic-Plese S, Cortese I, Wandinger KP, McFarland HF & Martin R. CD4+CD28- costimulation-independent T cells in multiple sclerosis. J Clin Invest 108, 1185–1194, doi: 10.1172/JCI12516 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt D, Goronzy JJ & Weyand CM CD4+ CD7- CD28- T cells are expanded in rheumatoid arthritis and are characterized by autoreactivity. J Clin Invest 97, 2027–2037, doi: 10.1172/JCI118638 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gearty SV et al. An autoimmune stem-like CD8 T cell population drives type 1 diabetes. Nature, doi: 10.1038/s41586-021-04248-x (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Page N. et al. Persistence of self-reactive CD8+ T cells in the CNS requires TOX-dependent chromatin remodeling. Nat Commun 12, 1009, doi: 10.1038/s41467-021-21109-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dulken BW et al. Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature 571, 205–210, doi: 10.1038/s41586-019-1362-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derecki NC et al. Regulation of learning and memory by meningeal immunity: a key role for IL-4. J Exp Med 207, 1067–1080, doi: 10.1084/jem.20091419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filiano AJ, Gadani SP & Kipnis J. How and why do T cells and their derived cytokines affect the injured and healthy brain? Nat Rev Neurosci 18, 375–384, doi: 10.1038/nrn.2017.39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kipnis J. Multifaceted interactions between adaptive immunity and the central nervous system. Science 353, 766–771, doi: 10.1126/science.aag2638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiffelle J. et al. T-cell repertoire analysis and metrics of diversity and clonality. Curr Opin Biotechnol 65, 284–295, doi: 10.1016/j.copbio.2020.07.010 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Oldrini B. et al. Somatic genome editing with the RCAS-TVA-CRISPR-Cas9 system for precision tumor modeling. Nat Commun 9, 1466, doi: 10.1038/s41467-018-03731-w (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kahles A. et al. Comprehensive Analysis of Alternative Splicing Across Tumors from 8,705 Patients. Cancer Cell 34, 211–224 e216, doi: 10.1016/j.ccell.2018.07.001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren P. et al. Alternative Splicing: A New Cause and Potential Therapeutic Target in Autoimmune Disease. Front Immunol 12, 713540, doi: 10.3389/fimmu.2021.713540 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurepa Z, Su J. & Forman J. Memory phenotype of CD8+ T cells in MHC class Ia-deficient mice. J Immunol 170, 5414–5420, doi: 10.4049/jimmunol.170.11.5414 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Korobeynikov VA, Lyashchenko AK, Blanco-Redondo B, Jafar-Nejad P. & Shneider NA Antisense oligonucleotide silencing of FUS expression as a therapeutic approach in amyotrophic lateral sclerosis. Nat Med 28, 104–116, doi: 10.1038/s41591-021-01615-z (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gurney ME et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264, 1772–1775, doi: 10.1126/science.8209258 (1994). [DOI] [PubMed] [Google Scholar]
- 45.Tu PH et al. Transgenic mice carrying a human mutant superoxide dismutase transgene develop neuronal cytoskeletal pathology resembling human amyotrophic lateral sclerosis lesions. Proc Natl Acad Sci U S A 93, 3155–3160, doi: 10.1073/pnas.93.7.3155 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sallusto F, Lenig D, Forster R, Lipp M. & Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712, doi: 10.1038/44385 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Patil VS et al. Precursors of human CD4(+) cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci Immunol 3, doi: 10.1126/sciimmunol.aan8664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emerson RO et al. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat Genet 49, 659–665, doi: 10.1038/ng.3822 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Tickotsky N, Sagiv T, Prilusky J, Shifrut E. & Friedman N. McPAS-TCR: a manually curated catalogue of pathology-associated T cell receptor sequences. Bioinformatics 33, 2924–2929, doi: 10.1093/bioinformatics/btx286 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Weekes MP, Wills MR, Mynard K, Carmichael AJ & Sissons JG The memory cytotoxic T-lymphocyte (CTL) response to human cytomegalovirus infection contains individual peptide-specific CTL clones that have undergone extensive expansion in vivo. J Virol 73, 2099–2108, doi: 10.1128/JVI.73.3.2099-2108.1999 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Latorre D. et al. T cells in patients with narcolepsy target self-antigens of hypocretin neurons. Nature 562, 63–68, doi: 10.1038/s41586-018-0540-1 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Campisi L, Cummings RJ & Blander JM Death-defining immune responses after apoptosis. Am J Transplant 14, 1488–1498, doi: 10.1111/ajt.12736 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weyand CM & Goronzy JJ The immunology of rheumatoid arthritis. Nat Immunol 22, 10–18, doi: 10.1038/s41590-020-00816-x (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garretti F, Agalliu D, Lindestam Arlehamn CS, Sette A. & Sulzer D. Autoimmunity in Parkinson’s Disease: The Role of alpha-Synuclein-Specific T Cells. Front Immunol 10, 303, doi: 10.3389/fimmu.2019.00303 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rabin BA et al. Autosomal dominant juvenile amyotrophic lateral sclerosis. Brain 122 ( Pt 8), 1539–1550, doi: 10.1093/brain/122.8.1539 (1999). [DOI] [PubMed] [Google Scholar]
- 56.Lamoreaux L, Roederer M. & Koup R. Intracellular cytokine optimization and standard operating procedure. Nat Protoc 1, 1507–1516, doi: 10.1038/nprot.2006.268 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Campisi L. In vitro Antigen-presentation Assay for Self- and Microbial-derived Antigens. Bio-protocol 7, e2307, doi: 10.21769/BioProtoc.2307 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korin B, Dubovik T. & Rolls A. Mass cytometry analysis of immune cells in the brain. Nat Protoc 13, 377–391, doi: 10.1038/nprot.2017.155 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Tan YS & Lei YL Isolation of Tumor-Infiltrating Lymphocytes by Ficoll-Paque Density Gradient Centrifugation. Methods Mol Biol 1960, 93–99, doi: 10.1007/978-1-4939-9167-9_8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liao Y, Smyth GK & Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41, e108, doi: 10.1093/nar/gkt214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, doi: 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47, doi: 10.1093/nar/gkv007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 17, 3, doi: 10.14806/ej.17.1.200 (2011). [DOI] [Google Scholar]
- 65.Liao Y, Smyth GK & Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930, doi: 10.1093/bioinformatics/btt656 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Law CW, Chen Y, Shi W. & Smyth GK voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29, doi: 10.1186/gb-2014-15-2-r29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu G, Wang LG, Han Y. & He QY clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287, doi: 10.1089/omi.2011.0118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen S. et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A 111, E5593–5601, doi: 10.1073/pnas.1419161111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.SPLINTER: Splice Interpreter Of Transcripts. version 1.4.0. (2017. ). [Google Scholar]
- 70.Hao Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529, doi: 10.1016/j.cell.2021.04.048 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Borcherding N, Bormann NL & Kraus G. scRepertoire: An R-based toolkit for single-cell immune receptor analysis. F1000Res 9, 47, doi: 10.12688/f1000research.22139.2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cuadrado-Castano S. et al. Enhancement of the proapoptotic properties of newcastle disease virus promotes tumor remission in syngeneic murine cancer models. Mol Cancer Ther 14, 1247–1258, doi: 10.1158/1535-7163.MCT-14-0913 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martinez NM & Lynch KW Control of alternative splicing in immune responses: many regulators, many predictions, much still to learn. Immunol Rev 253, 216–236, doi: 10.1111/imr.12047 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CITE-seq, SMART-seq and RNAseq datasets have been deposited online in the Gene Expression Omnibus (GEO), accession codes: GSE180410, GSE199663, GSE199664, GSE199666.