Abstract
The present study reviewed the occurrence of SARS-CoV-2 RNA and the evaluation of virus infectivity in feces and environmental matrices. The detection of SARS-CoV-2 RNA in feces and wastewater samples, reported in several studies, has generated interest and concern regarding the possible fecal–oral route of SARS-CoV-2 transmission. To date, the presence of viable SARS-CoV-2 in feces of COVID-19 infected people is not clearly confirmed although its isolation from feces of six different patients. Further, there is no documented evidence on the infectivity of SARS-CoV-2 in wastewater, sludge and environmental water samples, although the viral genome has been detected in these matrices. Decay data revealed that SARS-CoV-2 RNA persisted longer than infectious particle in all aquatic environment, indicating that genome quantification of SARS-CoV-2 does not imply the presence of infective viral particles. In addition, this review also outlined the fate of SARS-CoV-2 RNA during the different steps in the wastewater treatment plant and focusing on the virus elimination along the sludge treatment line. Studies showed complete removal of SARS-CoV-2 during the tertiary treatment. Moreover, thermophilic sludge treatments present high efficiency in SARS-CoV-2 inactivation. Further studies are required to provide more evidence with respect to the inactivation behavior of infectious SARS-CoV-2 in different environmental matrices and to examine factors affecting SARS-CoV-2 persistence.
Keywords: SARS-CoV-2, Detection methods, Wastewater, Wastewater treatment plants, Decay, Fate
Graphical abstract
1. Introduction
SARS-CoV-2, a virus belonging to the subgenus Sarbecovirus of the genus Betacoronavirus in the Coronaviridae family, is the causative agent of the pandemic coronavirus disease 2019 (COVID-19) which is the most global health crisis since the era of the influenza pandemic of 1918 (Cascella et al., 2022). It has a positive sense single- stranded RNA genome enclosed within a protein capsid coated with a bilayer lipid envelope. The genome of ∼30,000 nucleotides contains four structural proteins, that include envelope (E), nucleocapsid (N), membrane (M), and spike (S), and 25 nonstructural proteins (Feng et al., 2020). According to the World Health Organization data, as of 28 December 2022, over 649 million confirmed cases and 6.6 million deaths have been reported worldwide (WHO, 2022).
Although SARS-CoV-2 is predominantly a respiratory virus, it can cause gastrointestinal symptoms such as nausea, abdominal pain, vomiting and diarrhea, with the latter being the most frequent (Moura et al., 2022; Lin et al., 2020). It has been demonstrated that people infected with SARS-CoV-2 shed the virus in feces in addition to saliva, nasopharyngeal secretions, sputum (Ahmed et al., 2020c; Cevik et al., 2021) and to lesser extent in urine (Jeong et al., 2020), which are collected in sewerage. Accordingly, numerous studies have reported the presence of SARS-CoV-2 genome in human feces (Wölfel et al., 2020; Wang, et al. 2020a; Xiao et al., 2020; Yong et al., 2020), where approximately 50 % of COVID-19 patients shed fecal RNA in the week after infection and 4 % of patients shed fecal viral RNA up to 10 months after diagnosis (Natarajan et al., 2022; Zhou et al., 2022; Zhang et al., 2021). Further, SARS-CoV-2 genome has been detected throughout different processes of WWTPs, where the viral RNA was found not only in WWTP effluent, primary sludge and secondary sludge, but also in some treated sludge (biosolids) (Foladori et al., 2022). Besides, the occurrence of SARS-CoV-2 genome has also been reported in surface water, sediments and aquatic biota (Rimoldi et al., 2020; Guerrero-Latorre et al., 2020; Kolarević et al., 2021; Tandukar et al., 2022; Polo et al., 2021; Mancusi et al., 2022; Yang et al., 2022).
Following the detection of SARS-CoV-2 RNA in water bodies, the risk of virus transmission to human via the water route was discussed and the key question is whether the detected SARS-CoV-2 is infectious or not?
In fact, the positive detection of SARS-CoV-2 RNA does not provide evidence on the infectivity of the virus in these environmental samples and the possible oral-fecal transmission, because the presence of fragments of viral genome in environmental sample does not necessarily imply that the virus is structurally intact and viable (Bivins et al., 2020; Foladori et al., 2022; de Oliveira et al., 2021; Ahmed et al., 2020a). Cell culture assay remains the only method used to assess the infectivity of such samples as it provide the more reliable information to evaluate the risk of transmission of SARS-CoV-2 in the environment.
To date, viable cases of SARS-CoV-2 have been reported in the feces of only six different patients (Xiao et al., 2020; Zhang et al., 2020; Dergham et al., 2021; Wang et al. 2020b). Moreover, no cases of infection through contact with fecally contaminated samples have been reported and the few published studies revealed the absence of infectious SARS-CoV-2 in environmental samples.
The small number of studies addressing the infectious potential of SARS-CoV-2 in environmental samples may be due to the limited access to biosafety level 3 laboratories to work with SARS-CoV-2 in cell culture assays (CDC, 2021) and to the presence of toxic compounds and micro-organisms, which would hinder cell culture assay (Monteiro et al., 2022).
Persistence and decay data are necessary to evaluate the infectivity risk of the detected virus in wastewater, sludge, biosolids, and other environmental matrices. Actually, there is still little knowledge about the infectivity of this virus in these matrices. However, inactivation and decay studies demonstrated that SARS-CoV-2 RNA persisted longer than infectious viruses when seeded in wastewater, surface water and seawater.
The objective of this review was to (i) provide an overview on the environmental contamination with SARS-CoV-2; (ii) discuss on the applicability and limitations of the relevant methods used for its detection in the environmental samples; (iii) collect the available decay data of infectious SARS-CoV-2 and its genome in different aquatic environments and discuss the parameters influencing its persistence; and (iv) present the efficiency of different treatments in WWTPs on the removal of SARS-CoV-2.
2. SARS-CoV-2 in the environment and potential contamination modes
SARS-CoV-2 is an airborne virus whose transmission routes involve human-to-human that occurs mainly by aerosol droplets released from the infected person's mouth and nose (Patel et al., 2021). High viral loads have been found in the respiratory tract of infected individuals whose can shed the virus and its genetic material via their sputum, nasopharyngeal secretions and saliva (Patel et al., 2021; Cevik et al., 2021).
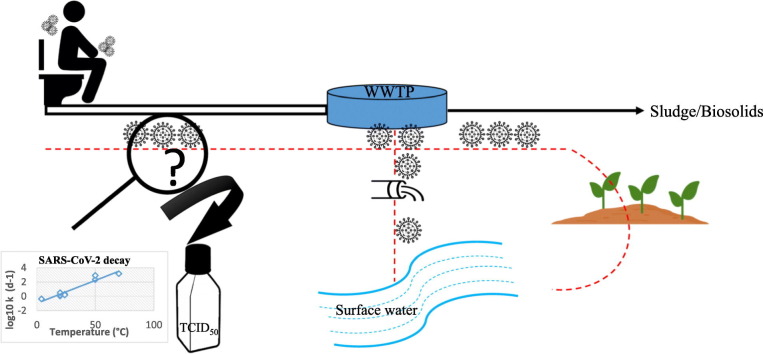
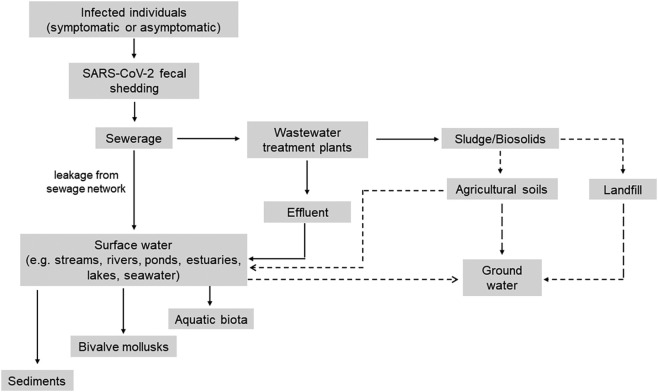
It has been demonstrated that SARS-CoV-2 RNA can be shed from people infected with COVID-19 (Wang et al., 2020b; Wölfel et al., 2020; Xiao et al., 2020; Yong et al., 2020; Kim et al., 2020). Fecal shedding of SARS-CoV-2 RNA was also observed in asymptomatic individuals and it was reported that the duration of fecal RNA shedding persisted from 1 to 50 days following the complete resolution of symptoms (Foladori et al., 2020; Park et al., 2021; van Doorn et al., 2020; Gupta et al., 2020). Since SARS-CoV-2 RNA can be shed in the feces of individuals with symptomatic or asymptomatic infection, it can be discharged into the sewerage to the central WWTP (Fig. 1 ) and hence could be transmitted to the environment by several routes (Wölfel et al., 2020; Foladori et al., 2020).
Fig. 1.
Potential modes of environmental contamination by SARS-CoV-2. Dashed arrow indicates suspected contamination routes where no data has been collected.
Treated wastewater, such as discharged secondary effluents, may release SARS-CoV-2 RNA into the aquatic ecosystems (Fig. 1) and, in particular surface water systems (e.g. rivers, lakes, seawater, ponds and estuaries) and then to sediments, bivalve shellfish and aquatic biota (Bosch et al., 2006; Polo et al., 2021; Le Guernic et al., 2022).
Moreover, sewer overflows caused by heavy rainfalls, leakage from sewage network systems like sewers and septic tanks can act as the source of viral contamination to the surface water (Fig. 1) (Bernard et al., 2022). Additionally, poor treatment at WWTPs, or lack of complete infrastructure in some countries are factors that can result in SARS-CoV-2 environmental contamination (Bogler et al., 2020).
In WWTPs, a partial accumulation of SARS-CoV-2 may take place in the separated solids due to its hydrophobic properties. Sewage sludge (solids) generated in the WWTPs is usually treated before disposal or recycling. Once treated, biosolids can be recycled or disposed of using three main routes: recycling to agriculture via landspreading, incineration or landfilling (Li et al., 2021). In this regards, surface water may also be contaminated through stormwater runoff from agricultural soil (Bernard et al., 2022). Further, it has also been proposed that groundwater may become contaminated with SARS-CoV-2 from a sewage sludge landfill (Anand et al., 2022), through agricultural soils, or from fecally contaminated surface water (Fig. 1).
Despite the detection of SARS-CoV-2 RNA in various environmental matrices in different studies (as shown by bold arrow in Fig. 1), infectivity of SARS-CoV-2 was not detected (Rimoldi et al., 2020; Westhaus et al., 2021) or not investigated in these matrices. However, based on the this evidence and the efficacy of most WWTPs in virus reduction, Ahmed et al. (2021); Albert et al. (2021); Sobsey (2022); Cerrada-Romero et al. (2022) asserted that fecal-oral transmission of SARS-CoV-2 associated with wastewater is likely to be low compared to well-documented person-to-person transmission via respiratory droplets/aerosols.
3. Detection methods of SARS-CoV-2 in wastewater
Most environmental monitoring of SARS-CoV-2 to date uses quantitative PCR-based methods to detect viral RNA. Given the low concentrations of SARS-CoV-2 RNA in wastewater (concentrations ranged between 20 GC/L and 3 × 106 GC/L), several concentration/enrichment protocols, such as ultracentrifugation, ultrafiltration, electronegative membrane filtration and precipitation with polyethylene glycol (PEG), have been developed and applied before viral RNA extraction (Ahmed et al., 2020b; Kumblathan et al., 2021; Ahmed et al., 2022). SARS-CoV-2 RNA detection and quantification is then performed using reverse transcription quantitative real-time PCR (RT-qPCR) based on a calibration curve, or reverse transcription digital PCR (RT-dPCR) without the necessity for a calibration curve (Kumblathan et al., 2021; Ahmed et al., 2022). Several gene targets specific to the SARS-CoV-2 have been used in molecular assays including a combination of structural (i.e., envelope (E), nucleocapsid (N) and spike (S)) and/or non-structural (i.e., ORF1ab, RdRp) genes in simplex or multiplex formats (Corman et al., 2020). However, data in the literature underline the absence of information regarding the SARS-CoV-2 recovery efficiency of different concentration methods and the lack of method standardization. This, is turn, highlights the challenge that need to be addressed to obtain accurate quantification, especially when low viral RNA quantities are present in the environmental samples. Another limiting factor for SARS-CoV-2 RNA detection is that wastewater contains a wide range of PCR-inhibitors such as proteins, fats, carbohydrates, polyphenols, metal ions, and RNAses (Ahmed et al., 2022) that can affect the proper PCR amplification and can also give false negative results (Foladori et al., 2021).
Nonetheless, quantitative PCR-based methods do not provide information on the presence of the infective viable virus in wastewater because viral genome detection does not necessarily indicate the presence of infective viable virus (Foladori et al., 2022). A new method referred as viability RT-qPCR or capsid integrity was employed to assess SARS-CoV-2 infectivity in environmental samples (Desdouits et al., 2021; Polo et al., 2021; Monteiro et al., 2022; Cuevas-Ferrando et al., 2021). This technique combines the use of photoactivatable dye pretreatment, such as ethidium monoazide (EMA), propidium monoazide (PMA) or platinum chloride (PtCl4), with qPCR. These molecules penetrate only damaged or destroyed capsids where they intercalate covalently into viral genome RNA, interfering with PCR amplification (Elizaquível et al., 2014). However, the efficacy of such strategy is limited to many factors such as dye concentration, type of the light source, and incubation conditions (Leifels et al., 2021). Actually, robustness of these methods should be evaluated considering the diversity of wastewater characteristics and composition that could widely vary according to location and weather.
Cell culture has long been considered the gold standard approach for isolating infectious virus particles. Nevertheless, several factors make it difficult to use this method to determine the possible presence of SARS-CoV-2 particles in wastewater:
-
i)
the requirement for a biosafety level 3 (BSL-3) laboratory for SARS-CoV-2 manipulation (CDC, 2021).
-
ii)
the considerable costs and time needed to establish cell culture tests.
-
iii)
the diversity of toxic compounds and micro-organisms resulting from wastewater concentration that are difficult to eliminate before sample inoculation, which would hinder cell culture assay (Monteiro et al., 2022).
-
iv)
the variation in cell line behavior in response to infection by the SARS-CoV-2 virus. In this regard, Decimo et al. (2022) examined the behavior of Vero E6 / kidney cell line originating from monkeys using 4 sub clones from 4 different laboratories. In light microscopy, Vero E6 cells were grouped under 2 morphological phenotypes, the fibroblastic phenotype and the epithelial one. Both phenotypes varied in response to infection by the SARS-CoV-2. For instance, cells of fibroblast phenotype were detached between 48 and 72 h after infection and continuously produced virus at high titers (> 106 PFU/mL) without the cell layer being damaged and this type of cells could be used for cell line production. In contrast, the cells of the epithelial phenotype were partially or totally destroyed within 48 and 72 h and this type of cells could be used for TCID50 or phage lysis assays. Transcriptomic analyzes carried out 24 h after infection confirmed these results (Decimo et al., 2022).
-
v)
It is well known that Vero E6 kidney cell line is widely used in coronavirus research for virus stock propagation and antiviral assays. Although it does express the ACE2 receptor for SARS-CoV-2 attachment, it lacks the TMPRSS2 protease required for entry into human cells. Instead, viral entry into Vero E6 is likely cathepsin-mediated and may not accurately mimic the infection event in human cells (Mautner et al., 2022). Besides, Caco-2 cells, an intestinal epithelium cell line originating from humans, and Calu-3 cells, a pulmonary epithelium cell line also originating from humans seem to be preferential modeling cell lines. Different viral isolates replicate similarly in Caco-2 cells, but show very different replicative capacities in Calu-3 cells (de Souza et al., 2021).
4. Occurrence and infectivity evaluation of SARS-CoV-2 in feces and environmental matrices
4.1. Feces
SARS-CoV-2 RNA shedding by infected patients has been detected in feces at concentrations between 106 and 1010 genome copies per L of feces (GC/L). The detected levels varied according to the day of sampling post infection initiation (Wölfel et al., 2020; Foladori et al., 2020), where the highest levels were recorded during the first week of symptoms. Even though the detection of SARS-CoV-2 genetic signal in feces samples, the presence of infectious particles in these samples is not confirmed. In fact, studies have examined the presence of infectious SARS-CoV-2 in feces from infected individuals with contradictory results. Until date, viable SARS-CoV-2 has been reported in the feces of only six different patients (Table 1 ), but with no data on quantities of infectious SARS-CoV-2. Using the Vero E6 cell line, Xiao et al. (2020) were able to find infectious SARS-CoV-2 in 2 fece samples from one infected patient, while Wölfel et al. (2020) were unable to detect infectious SARS-CoV-2 in two separate laboratories by using the same cell line, despite the positive RT-qPCR tests with high RNA load. In another study, Jeong et al. (2020) failed to demonstrate the presence of viable SARS-CoV-2 in fecal samples using Vero cells, but viable SARS-CoV-2 was isolated from the nasal washes of ferret inoculated with one COVID-19 patient's stool. Additionally, Zhang et al. (2020) were able to observe viral particles with typical morphology of a coronavirus using electron microscopy after inoculating stool suspension into Vero cells (Table 1).
Table 1.
Infectious SARS-CoV-2 evaluation in stool and wastewater samples.
| Reference | Country | Type of samples | Cell culture assay | Results |
|---|---|---|---|---|
| (Xiao et al., 2020) | China | Stool | 3 Positive stool samples for SARS-CoV-2 RNA obtained from 2 patients were tested for infectivity using the Vero E6 cell line | 2 out of 3 were positive for infectious viral particles |
| (Wölfel et al., 2020) | Germany | Stool | Experiment conducted using the same cell line used by Xiao et al. (2020), in two separate laboratories | No infectious SARS-CoV-2 particles was detected despite the high viral RNA load detected by RT-qPCR |
| (Zhang et al., 2020) | China | Stool | Vero cells were used for viral isolation from stool samples of unreported number of COVID-19 patients The presence of SARS-CoV-2 was confirmed by electron microscopic observation |
A virus particle with typical morphology of coronavirus was observed in one sample |
| (Jeong et al., 2020) | Republic of Korea | Stool | 3 Positive qPCR samples were subjected to SARS-CoV-2 isolation in Vero cells. One fecal specimen was selected to experimentally infect ferret and then viable virus titres in nasal washes were checked on 2, 4, 6 and 8 days post infection | No cultures were positive, however viable SARS-CoV-2 was isolated from the nasal washes of the stool treated ferret |
| (Kim et al., 2020) | Republic of Korea | Stool | 129 stool samples were tested for infectivity test using CaCo-2 cell line | No cultures were positive |
| (Wang et al., 2020a) | China | Stool | Four SARS-CoV-2 positive fecal specimens with high copy numbers were cultured to detect live virus. No details on cultivation method was reported | Viable SARS-CoV-2 was observed in the stool sample from 2 patients |
| (Albert et al., 2021) | Spain | Stool and sewage | Fecal sewage samples with highest RNA concentrations were used to inoculate Vero E6 cells | No cytopathic effect on Vero E6 cells was observed in any of the analyzed samples |
| (Dergham et al., 2021) | France | Stool | Vero cells were used for viral isolation from 106 stool samples of 46 COVID-19 patients | Viable SARS-CoV-2 was detected in 2 stool samples from 1 patient undergone a kidney transplant 21 years ago |
| (Cerrada-Romero et al., 2022) | Spain | Stool | Vero cells were used for viral isolation from 79 stools sample collected from 62 adult COVID-19 patients | No cultures were positive |
| (Rimoldi et al., 2020) | Italy | Raw and tertiary treated wastewater | SARS-CoV-2 infectivity in positive and negative samples for SARS-CoV-2 RNA was evaluated using Vero E6 cells | No infectious SARS-CoV-2 particles was detected in all samples |
| (Westhaus et al., 2021) | Germany | Influent raw wastewater, effluent and effluent after tertiary treatment | Infectivity of purified and concentrated influent and effluent samples (in both liquid and solid phases) were evaluated using differentiated Caco-2 cells | No infectious SARS-CoV-2 particle was detected in all samples |
| (Robinson et al., 2022) | USA | Raw wastewater | 10 positive raw wastewater samples for SARS-CoV-2 RNA were tested for infectivity using Vero E6 cells | No cytopathic effects were obtained |
| (Monteiro et al., 2022) | Portugal | Secondary treated wastewater (effluents) | Positive secondary-treated wastewater samples for SARS-CoV-2 RNA were tested for infectivity using Vero E6 cells | No infectious SARS-CoV-2 viral particles was obtained despite the high viral RNA load detected by RT-qPCR |
It is well known that other human coronaviruses, such as SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV), are excreted in the stools of infected patients and remain viable under conditions that could facilitate fecal–oral transmission (Cuicchi et al., 2021), whereas, there is no current evidence showing that SARS-CoV-2 could also be transmitted via this route. Potential SARS-CoV-2 infection in the gastrointestinal tract has been discussed in regard to the intestine expression of ACE2 receptors required for SARS-CoV-2 infection and to the prolonged viral shedding (Giacobbo et al., 2021; Cerrada-Romero et al., 2022). Recently, Cerrada-Romero et al. (2022) evaluated the viability of the SARS-CoV-2 viral particles excreted in 79 stools sample collected from 62 adult COVID-19 patients. They showed that SARS-CoV-2 RNA was detected in stools samples from 27 (43.5 %) out of the 62 patients. However, SARS-CoV-2 replication, assessed by the generation of cytopathic effects followed by viral load quantification by RT-PCR assay, was not revealed in any of stool samples, suggesting that SARS-CoV-2 replicative capacity is null or very limited in stool samples, and thus, they suggested that the fecal–oral transmission of SARS-CoV-2 as an alternative infection route is highly unlikely. Furthermore, Zang et al. (2020) reported that SARS-CoV-2 entered the intestinal lumen was inactivated by simulated human colonic fluid, and hence the virus is likely to be inactivated before it is expelled.
4.2. Wastewater
The viral RNA concentrations measured in raw wastewater were at least 4 log10 unit lower than those detected in feces and varied between 20 GC/L and 3 × 106 GC/L (Ahmed et al., 2020a; Foladori et al., 2020). It was reported that this viral dilution is due to many factors such as the large domestic daily water consumption per person, the presence of rainwater and parasitic inflow in the sewerage, industrial discharges, and the limited percentage of COVID-19 positive persons in the community served by the WWTP (Foladori et al., 2022).
Due to the strict biosafety requirements for SARS-CoV-2 and the fast evolving COVID-19 pandemic, research has been concerned primarily on the detection of SARS-CoV-2 RNA in wastewater and only a few investigations have examined the potential presence of infectious SARS-CoV-2 in raw and treated wastewaters with negative results (Table 1). Rimoldi et al. (2020) used the RNA positive and negative raw and treated wastewater samples to inoculate Vero E6 cells. Infectivity was assessed daily by screening cells for cytopathic effects under reverse-phase light microscope. No infectious SARS-CoV-2 particles was detected in the analyzed samples.
Using the CaCo-2 cell line, the absence of infective viable SARS-CoV-2 was also obtained by Westhaus et al. (2021) in influent raw wastewater and in both effluent and tertiary treated effluent wastewater samples despite the presence of SARS-CoV-2 genome in all the analyzed samples.
Recently, Robinson et al. (2022) used 10 raw wastewater samples with SARS-CoV-2 RNA levels ranging from 16.9 × 104 to 3.255 × 106 GC/L to inoculate Vero E6 cells within one week from collection. The authors did not find infectious SARS-CoV-2 in the analyzed samples. Moreover, according to Monteiro et al. (2022), the detected SARS-CoV-2 RNA in secondary treated wastewater at concentrations up to 104 GC/L was found to be non-infectious in cell culture using Vero E6 for 5 days.
These results align with recent evidence suggesting that wastewater does not appear to be a be a route of SARS-CoV-2 transmission (Albert et al., 2021; Cerrada-Romero et al., 2022; Ahmed et al., 2021; Sobsey, 2022).
4.3. River water
There are only few studies that have examined the occurrence of SARS-CoV-2 in the receiving water systems. Rimoldi et al. (2020) surveyed three rivers (near Milano, Italy) during the epidemic peak outbreak in April 2020 (Table 2 ). They found SARS-CoV-2 RNA in all investigated river samples. However, the viral load was not reported by the authors. Because SARS-CoV-2 detection was correlated with caffeine detection in the river samples, they related viral presence to untreated or ineffectively treated wastewater discharged into surface waters. Infectivity of positive RNA samples was evaluated by screening cells for cytopathic effects under reverse-phase light microscope. No infectious SARS-CoV-2 was detected in all positive samples.
Table 2.
Detection of SARS-CoV-2 RNA in river water and marine environment.
| Reference | Country | Type of samples | Location | Target regions used for RT-qPCR detection | Concentration range (GC/L) |
|---|---|---|---|---|---|
| (Rimoldi et al., 2020) | Italy | River water | Lambro River, Vettabbia Canal, and Lambro Meridionale River in the two provinces of Milan and Monza and Brianza | Nucleocapsid (N), ORF1ab and E | Not reported |
| (Guerrero-Latorre et al., 2020) | Ecuador | River water | Quito's river | Nucleocapsid N1 and N2 | 2.91 × 105 to 3.19 × 106 GC/L (N1) 2.07 × 105 to 2.22 × 106 GC/L (N2) |
| (Kolarević et al., 2021) | Serbia | River water | Danube River | Nucleocapsid (N1 and N2) and Envelope (E) | 5.96 × 103 to 1.30 × 104 GC/L |
| (Tandukar et al., 2022) | Nepal | River water | Bagmati River | CDC-N1, CDC-N2, NIID_2019-nCOV_N, and N_Sarbeco | 4–5.1 log10 GC/L |
| (Yang et al., 2022) | China | River water | Beijing, China | E_Sarbeco, CDC_N1 | 9.7 × 101 and 9.52 × 102 GC/L |
| (Polo et al., 2021) | Spain | Bivalve mollusks: Ruditapes philippinarum and Ruditapes decussatus Marine sediments |
Estuaries and river catchment in Galicia (NW of Spain) | IP4, E and N1 | Below limit of quantification to 4.48 log10 GC/g of digestive tissue Below limit of quantification to 3.60 log10 GC/g of sediment |
| (Mancusi et al., 2022) | Italy | Bivalve mollusks (Mytilus galloprovincialis) | Coastal sites from Gulf of Naples (Campania region, Italy). | Orf1b nsp14, RdRp and E | 7.8 × 101 to 2.6 × 103 GC/g (Orf1b nsp14) 7.2 × 101 to 4.9 × 103 GC/g (RdRp) 1.3 × 102 to 5.0 × 102 GC/g (E) |
| (Yamazaki et al., 2022) | Japan | Cultivated oysters (Crassostrea gigas) | Kyoto Hiroshima, Okayama, Hyogo and Yamaguchi prefectures, Japan | Nucleocapsid N2 | SARS-CoV-2 was not detected in the analyzed oysters samples |
| (Ransome et al., 2023) | UK & Serbia | Cultivated oysters (Crassostrea gigas), river water and sediments | The River Thames, UK Sava and Danube rivers, Serbia |
Nucleocapsid N1 and E genes | None of the collected samples were positive for the N1 or E gene, and no infectious SARS-CoV-2 was recovered from any of these samples |
Guerrero-Latorre et al. (2020) examined the presence of SARS-CoV-2 RNA in rivers from urban streams in Quito, Ecuador, where wastewater is discharged directly into receiving waters (Table 2). SARS-CoV-2 RNA detected in the analyzed samples ranged from 2.91 × 105 to 3.19 × 106 GC/L using N1 assay and from 2.07 × 105 to 2.22 × 106 GC/L using N2 assay. The higher SARS-CoV-2 RNA concentration was recorded during COVID-19 peak. A study conducted in Serbia to detect the SARS-CoV-2 RNA in Danube River showed that RNA viral load in the analyzed sampling sites ranged from 5.96 × 103 up to 1.30 × 104 GC/L (Table 2). SARS-CoV-2 genome detection was associated with the discharge of untreated wastewaters (Kolarević et al., 2021) and no infectious virus was recovered in any environmental samples. Tandukar et al. (2022) detected SARS-CoV-2 RNA in 9/13 river water samples collected from the Bagmati River in Nepal (Table 2). The mean concentration of the viral RNA ranged from 4 to 5.1 log10 GC/L according to the RT-qPCR. In another study, 9 river samples were taken from 3 locations from upstream to downstream of a river in Beijing, China (Yang et al., 2022). Samples were collected 17 days before to 19 days after the end of the second wave of the COVID-19 epidemic. Results showed that SARS-CoV-2 RNA was detected in 9 river samples and concentrations ranged between 9.7 × 101 and 9.52 × 102 GC/L (Table 2). In Argentina, La Caldera, Mojotoro, and Arenales Rivers were monitored for the presence of SARS-CoV-2 (Maidana-Kulesza et al., 2022). SARS-CoV-2 RNA was found in about half of samples in low concentrations in La Caldera and Mojotoro Rivers, while it was high in Arenales River (concentrations between 106 and 107 GC/L).
Recently, river water samples spiked with infectious SARS-CoV-2 showed that infectious SARS-CoV-2 inoculum is stable in water and sediment for <3 days, while SARS-CoV-2 RNA is detectable for at least seven days (Ransome et al., 2023).
4.4. Marine environment
Polo et al. (2021) detected SARS-CoV-2 genome in 9/12 bivalve mollusks and 3/12 estuarine sediments (Table 2). For bivalve mollusks samples, the quantification values ranged from below limit of quantification to 4.48 log10 GC/g of digestive tissue. Concerning the marine sediment samples, the detected SARS-CoV-2 load ranged from below limit of quantification to 3.60 log10 GC/g of sediment. However, using viability RT-qPCR assay, they showed that the detected SARS-CoV-2 RNA did not correspond to intact capsids and, therefore, to infectious viral particles.
Recently, Mancusi et al. (2022) reported the presence of SARS-CoV-2, using RT-ddPCR, in 27/179 (15.1 %) of bivalve mollusks (Mytilus galloprovincialis) harvested from Southern Italy area (Table 2). Viral concentration range was 7.8 × 101 to 2.6 × 103 GC/g using Orf1b nsp14 region, 7.2 × 101 to 4.9 × 103 GC/g using RdRp gene and 1.3 × 102 to 5.0 × 102 GC/g using E gene.
Yamazaki et al. (2022) examined cultivated oysters sold in Japan for the presence of SARS-CoV-2 between October 2021 and April 2022 to clarify the extent of viral contamination and evaluate the risk of food-borne transmission of SARS-CoV-2. Despite a marked increase in infections caused by the Omicron variant from January to April 2022 in Japan, SARS-CoV-2 was not detected in any of the 145 raw oyster samples surveyed from Kyoto Hiroshima, Okayama, Hyogo and Yamaguchi prefectures (Yamazaki et al., 2022).
5. Persistence of infectious SARS-CoV-2 and its genome in wastewater
Human enveloped viruses, like coronaviruses, are considered to have a rapid decay rate in the water environment (Kampf et al., 2020; Ye et al., 2016). After the SARS epidemic of 2003–2004, an experimental study showed that SARS-CoV-1 stability under an infectious form was only 2 days at 20 °C, but 14 days at 4 °C (Wang et al., 2005) in urban and hospital wastewater. Survival of SARS-CoV-2 and other viruses in wastewater could be influenced by several factors. These factors include viral structure, the composition of the wastewater, pH, temperature (Amoah et al., 2020) or even the microbial composition (Wurtzer et al., 2021). Because of the limited reports about the presence and behavior of infective SARS-CoV-2 in wastewater and the absence of suggested evidence that water and wastewater play a role in SARS-CoV-2 transmission, inactivation and persistence data may allow us to evaluate the infectivity risk of the virus in water and wastewater.
In this section, we present the persistence data of infectious SARS-CoV-2 and SARS-CoV-2 RNA in wastewater and discuss the different factors, mainly temperature, influencing its persistence. In addition, the persistence of SARS-CoV-2 in wastewater is compared to other water matrices.
5.1. Persistence of infectious SARS-CoV-2 in wastewater
Bivins et al. (2020) studied the persistence of infectious SARS-CoV-2 in wastewater spiked with high (105 TCID50 /mL) and low (103 TCID50/mL) titers of infective SARS-CoV-2 at 20 °C. They reported that the virus decay at both titers was not significally different and observed that it takes 1.6–2.1 days at high and low titers respectively for 90 % inactivation (Table 3 ). Further, the authors reported that infective SARS-CoV-2 could be detected for the entire 7 days at 20 °C during the higher titer experiment (105 TCID50 /mL) and for 3 days during the lower titer experiment (103 TCID50 /mL).
Table 3.
Persistence of SARS-CoV-2 particles and RNA in different water matrices.
| Reference | Matrix | Test conditions | Method of measurement | Temperature | T90 |
|---|---|---|---|---|---|
| (Bivins et al., 2020) | Wastewater influent | Samples were inoculated with low titer (103 TCID50/mL) of SARS-CoV-2. The experiment lasted for 7 days | TCID50/mL RNA quantification |
20 °C | 2.1 days 26.2 days |
| Bivins et al., 2020) | Wastewater influent | Samples were inoculated with high titer (105 TCID50/mL) of SARS-CoV-2. The experiment lasted for 7 days | TCID50/mL RNA quantification |
20 °C | 1.6 days 3.3 days |
| Bivins et al., 2020) | Wastewater influent | Samples were inoculated with high titer (105 TCID50/mL) of SARS-CoV-2. The experiment lasted for 7 days | TCID50/mL | 50 °C 70 °C |
15 min 2.2 min |
| Bivins et al., 2020) | Tap water | Samples were inoculated with high titer (105 TCID50/mL) of SARS-CoV-2 | TCID50/mL RNA quantification |
20 °C | 2 days 33.2 days |
| (Ahmed et al., 2020a) | Wastewater influent | Samples were spiked with gamma irradiated SARS-CoV-2: 7.03 ± 0.19 log10 GC in 15 mL. The experiment lasted for 33 days | RNA quantification | 4 °C 15 °C 25 °C 37 °C |
27.8 days 20.4 days 12.6 days 8.04 days |
| (Ahmed et al., 2020a) | Wastewater influent | Samples were autoclaved and spiked with gamma irradiated SARS-CoV-2: 7.03 ± 0.19 log10 GC in 15 mL. The experiment lasted for 33 days | RNA quantification | 4 °C 15 °C 25 °C 37 °C |
43.2 days 29.9 days 13.5 days 5.7 days |
| (Ahmed et al., 2020a) | Tap water | Samples were spiked with gamma irradiated SARS-CoV-2: 7.03 ± 0.19 log10 GC in 15 mL. The experiment lasted for 33 days | RNA quantification | 4 °C 15 °C 25 °C 37 °C |
58.6 days 51.2 days 15.2 days 9.4 days |
| (Varbanov et al., 2021) | Wastewater influent | Samples were spiked with SARS-CoV-2 (105–106 TCID50/mL). The experiment lasted for 7 days | TCID50/mL | 20 °C 50 °C |
18 h 4 min |
| (Fukuta et al., 2021) | Mineral water | Samples were spiked with SARS-CoV-2 (105 PFU/mL). The experiment lasted for 11 weeks | PFU/mL | 4 °C | 175.43 days |
| (Fukuta et al., 2021) | Tap water | Samples were spiked with SARS-CoV-2 (105 PFU/mL). The experiment lasted for 11 weeks | PFU/mL | 4 °C | 50.25 days |
| (Fukuta et al., 2021) | Distilled water | Samples were spiked with SARS-CoV-2 (105 PFU/mL). The experiment lasted for 11 weeks | PFU/mL | 4 °C | 85.47 days |
| (Sala-Comorera et al., 2021) | Sterilized filtered River water | Samples were spiked with SARS-CoV-2 (3.16 × 104 TCID50/mL). The experiment lasted for 20 days | TCID50/mL | 4 °C 20 °C |
3.77 days 2.27 days |
| (Sala-Comorera et al., 2021) | Sterilized filtered sea water | Samples were spiked with SARS-CoV-2 (3.16 × 104 TCID50/mL). The experiment lasted for 20 days | TCID50/mL | 4 °C 20 °C |
2.15 days 1.13 days |
| (de Oliveira et al., 2021) | Autoclaved River water | Samples were spiked with SARS-CoV-2 (2 × 104 PFU/mL). The experiment lasted for 15 days | PFU/mL | 4 °C 24 °C |
7.7 days 1.9 days |
| (de Oliveira et al., 2021) | Autoclaved filtered River water | Samples were spiked with SARS-CoV-2 (2 × 104 PFU/mL). The experiment lasted for 15 days | PFU/mL | 24 °C | 3.3 days |
| (de Oliveira et al., 2021) | Autoclaved wastewater | Samples were spiked with SARS-CoV-2 (2 × 104 PFU/mL). The experiment lasted for 15 days | PFU/mL | 4 °C 24 °C |
5.5 days 1.2 days |
| (de Oliveira et al., 2021) | Autoclaved filtered wastewater | Samples were spiked with SARS-CoV-2 (2 × 104 PFU/mL). The experiment lasted for 15 days | PFU/mL | 24 °C | 1.5 days |
| (Hokajärvi et al., 2021) | Wastewater influent | Samples were spiked with SARS-CoV-2 (titer was not specified). The experiment lasted for 25 days | RNA quantification | 4 °C | 52 days for E-Sarbeco 36 days for N2 |
| (Roldan-Hernandez et al., 2022) | Wastewater primary settled solids from 2 wastewater treatment plant (A and B) | Study performed with endogenous SARS-CoV-2 at concentrations determined using N1 and N2: 4.48 ± 3.93 log10 GC/g (A) and 4.60 ± 3.69 log10 GC/g (B) for N1 4.45 ± 3.89 log10 GC/g (A) and 4.55 ± 3.56 log10 GC/g (B) for N2 The experiment lasted for 10 days |
RNA quantification | 4 °C 22 °C 37 °C |
95 (A), 64.6 (B) 85.9 (A), 36.5 (B) 36.6 (A), 25.3 (B) for N1 gene |
| RNA quantification | 4 °C 22 °C 37 °C |
214.7 (A), 75.4 (B) 107.3(A), 26(B) 49.4 (A), 23.5 (B) for N2 gene |
|||
| (Yang et al., 2022) | Raw wastewater | Study performed with endogenous SARS-CoV-2 (5 × 103 GC/L). The experiment lasted for 4 days | RNA quantification | 4 °C 26 °C |
17.17 days 8.4 days |
5.2. Persistence of SARS-CoV-2 RNA in wastewater
SARS-CoV-2 RNA was found to be significantly more persistent than infectious particles where T90 for SARS-CoV-2 RNA at 20 °C was 3.3 and 26.2 days in wastewater spiked at high (105 TCID50 /mL) and low (103 TCID50/mL) titers, respectively (Bivins et al., 2020) compared to 1.6 and 2.1 days for infectious SARS-CoV-2, respectively (Table 3).
Hokajärvi et al. (2021) also observed high SARS-CoV-2 genome persistence (Table 3). They found that T90 of SARS-CoV-2 RNA in wastewater at 4 °C was in the range 36–52 days compared to 5.5 days for infectious SARS-CoV-2 at 4 °C (de Oliveira et al., 2021).
Recently, Yang et al. (2022) studied the persistence of endogenous SARS-CoV-2 in wastewater sample where its initial load was ~5 × 103 GC/L. They showed that the T90 values of SARS-CoV-2 RNA were 17.17 and 7.68 days, respectively, at 4 °C and 26 °C smaller than that obtained by Ahmed et al. (2020b) 27.8 and 12.6, respectively at 4 °C and 25 °C (Table 3). This was interpreted by the authors as being related to the fact that their decay experiment was performed with endogenous SARS-CoV-2 and not with spiked one as achieved by Ahmed et al., 2020b and others studies (Hokajärvi et al., 2021; Bivins et al., 2020), where incomplete viral structure may present in the wastewater making viral RNA more prone to degradation. In this context, Wurtzer et al. (2021) indicated that SARS-CoV-2 viral genome could persist under several forms in wastewaters: RNA protected within an infectious particles, RNA protected in a non-infectious particles and free total or partial RNA. Moreover, they showed that <10 % of the total viral RNA was under a protected form in raw wastewater samples collected in Greater Paris area, suggesting the presence of minor part of intact particles in the analyzed samples (Wurtzer et al., 2021).
5.3. Effect of temperature on the persistence
Infectivity of SARS-CoV-2 in wastewater was shown to be affected by temperature. The persistence was significantly decreased with increasing temperature (Table 3), where T90 values for infective SARS-CoV-2, seeded at 105 TCID50 /mL, reduced to 15 and 2 min at 50 °C and 70 °C respectively compared to 1.6 days at 20 °C (Bivins et al., 2020). Similar finding was reported by Varbanov et al. (2021) where they showed that T90 for infective SARS-CoV-2 in wastewater, spiked with 105–106 TCID50 /mL titer was 18 h and 4 min at 20 °C and 50 °C, respectively (Table 3).
It was reported that the decrease in virus survival with increasing temperature could be associated to the denaturation of proteins and nucleic acids (Gundy et al., 2009; Ye et al., 2016; Ahmed et al., 2020b; Hokajärvi et al., 2021).
On the other hand, studies have shown that low temperature favors viral persistence. de Oliveira et al. (2021) studied the persistence of infectious SARS-CoV-2 in autoclaved wastewater and found that it takes 5.5 days at 4 °C for 90 % inactivation while T90 at 24 °C was 1.2 days (Table 3). These findings suggest that SARS-CoV-2 may persist longer in wastewater in temperate or colder regions than in tropical regions.
As for infectious SARS-CoV-2, the persistence of viral RNA is also negatively affected by increasing the temperature. Observations by Ahmed et al. (2020b) showed that times for 1-log10 reduction (90 % reduction) of SARS-CoV-2 RNA in untreated wastewater under 4, 15, 25 and 37 °C were 27.8, 20.4, 12.6, 8.04 days, respectively (Table 3). As the increase of temperature could have an impact on the SARS-CoV-2 RNA persistence, this parameter should be recorded when the genome is used to follow virus circulation in the population using a wastewater surveillance between summer and winter. Hence, the interest of integrating the collection temperature in the metadata associated with wastewater.
5.4. Effect of wastewater characteristics/composition on the persistence
Besides temperature, another factor affecting the persistence of SARS-CoV-2 is the wastewater characteristics/composition. In the study of Wurtzer et al. (2021), the persistence of infectious SARS-CoV-2 was studied by spiking five negative wastewater samples. After 24 h-incubation, they observed that SARS-CoV-2 infectivity was reduced by about 1-log10 in all samples at 4 °C while a reduction of >3 log10 and <1-log10 was observed in 3 out of 5 wastewater samples and 2 over 5 samples respectively at 20 °C. This variation in SARS-CoV-2 infectivity at 20 °C may be explained by the difference in chemical and/or microbial composition of wastewater samples. In another study, de Oliveira et al. (2021) reported that decay rate of infectious SARS-CoV-2 was lower in autoclaved filtered wastewater (T90 = 1.5 days) than in autoclaved wastewater (T90 = 1.2 days) at 24 °C (Table 3).
With respect to SARS-CoV-2 RNA, Roldan-Hernandez et al. (2022) reported a high persistence of SARS-CoV-2 RNA in primary settled solids compared to persistence in raw wastewater. Persistence study was conducted with endogenous SARS-CoV-2 RNA in two WWTPs (POTW A, 14.07 % of solids and POTW B, 16.57 % of solids) at 4 °C, 22 °C and 37 °C. T90 values ranged from 24 to 214 days, according to temperature conditions, target gene and the degree of sorption to solids where faster RNA decay rates were obtained at POTW B compared to POTW A (Table 3).
As for infectious SARS-CoV-2, genome persistence in wastewater was also shown to be influenced by microbial activities in raw wastewater. In autoclaved wastewater, T90 values of SARS-CoV-2 RNA were 5.71 to 43.2 days with decreasing temperature from 37 to 4 °C, and were higher than the T90 for untreated wastewater (8.04 to 27.8 days) at all temperatures except 37 °C (Ahmed et al., 2020b).
5.5. Effect of pH on the persistence
There is limited information on the impact of pH on the survival of infectious SARS-CoV-2 in wastewater matrix. However, Varbanov et al. (2021) investigated the effect of a pH range of 9–12 on the inactivation of infectious SARS-CoV-2 in wastewater for 10 min at room temperature. They observed a slight decrease (<1-log10 unit) in infective SARS-CoV-2 at pH 9 or 10, while an approximately 5.5 log10 units reduction was observed at pH 11. They also reported that the inactivation of infectious SARS-CoV-2 was not affected when the exposure time was increased at pH 9 or 10 from 10 to 60 min.
This considerable reduction of infective SARS-CoV-2 at pH >11 may provide useful information about the stability of the virus during lime treatment of sludge. In fact, lime applied to sludge leads to an enormous increase in pH, where it usually elevates the pH to higher than 11 and even 12 (Parmar et al., 2001) and may have a rigorous effect on pathogen reduction and the associated risk of exposure.
Infectious SARS-CoV-2 was shown to be stable over a wide range of pH (3−10) in suspension at room temperature (Chin et al., 2020) in contrast to SARS-CoV-1 where its survival was shown to be affected by the pH of feces (Lai et al., 2005). The survival time of SARS-CoV-1 ranged from three hours in slightly acidic feces of a newborn to four days in diarrheal feces of an adult with a pH of up to 9 (Lai et al., 2005). Xie et al. (2022) showed that the complex structures of hACE2 and the S proteins of SARS-CoV/SARS-CoV-2 are stable at pH values ranging from 7.5 to 9. Moreover, the presence of a polybasic cleavage site in SARS-CoV-2 spike may be the factor in the increased survival of SARS-CoV-2 over wide range of pH compared to SARS-CoV-1 (Winstone et al., 2021).
5.6. SARS-CoV-2 persistence in other water matrices
SARS-CoV-2 can persist longer in different aquatic matrices (Bivins et al., 2020; Ahmed et al., 2020b) than in wastewater. Bivins et al. (2020) showed that T90 of infectious virus particles in tap water at room temperature was 2 days compared to 1.6 days in sewage (Table 3). In another study, starting with a spiked concentration of 105 PFU/mL, the half-life value of infectious SARS-CoV-2 were 52.79, 25.77 and 15.123 days in mineral water, distilled water and tap water, at 4 °C respectively (Fukuta et al., 2021), the values which represented a calculated T90 values of 175.4, 50.25 and 85.4 days, respectively (Table 3). Further, de Oliveira et al. (2021) demonstrated that SARS-CoV-2 persisted more in river water than in wastewater with T90 values were 7.7–1.9 days for river water and 5.5–1.2 days for wastewater at 4–24 °C, respectively. Sala-Comorera et al. (2021) showed there was rapid inactivation of infectious SARS-CoV-2 in sterilized filtered seawater (T90 = 2.15 and 1.14 at 4 °C and 20 °C, respectively) compared to sterilized filtered river water (T90 = 3.77 and 2.28 at 4 °C and 20 °C, respectively) (Table 3).
With respect to RNA, Bivins et al. (2020) showed that the SARS-CoV-2 RNA could persist in tap water at room temperature for up to 33.2 days, while it persisted for 3.3 days in wastewater. Ahmed et al. (2020b) observed 90 % RNA reduction of SARS-CoV-2 at 4–37 °C after 9.4–58.6 days in tap water compared to 8.04–27.8 days in raw wastewater (Table 3).
5.7. Temperature sensitivity of SARS-CoV-2 decay in aquatic environment
First-order decay rate constants (k) of both infectious SARS-CoV-2 and SARS-CoV-2 RNA in wastewater and other water matrices (tap water, mineral water and river water) were collected from previous works (Table 3) and presented in units of inverse days (d−1). In case a study only reported T90 values, they were converted to first-order decay rate constants according to Chick's law:
Data compiled from this analysis are presented in Table S1.
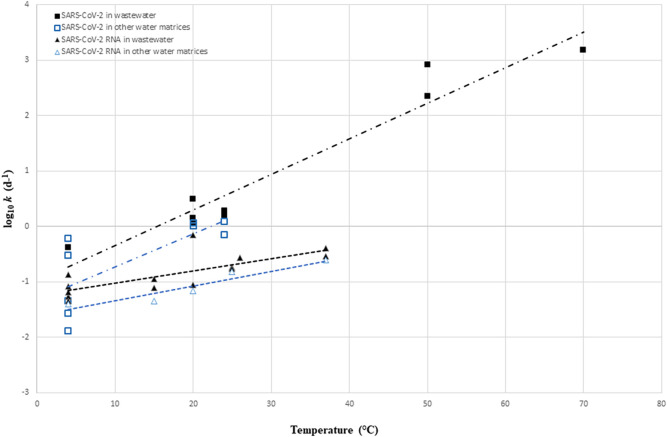
To evaluate the change in the decay rate constant with temperature, k at different temperatures of infectious SARS-CoV-2 and SARS-CoV-2 RNA in wastewater and other water matrices were log10 transformed (log10 k) and a linear regression was performed (Fig. 2 ).Values of the linear regression for each target are summarized in Table 4 .
Fig. 2.
log10 k of infections SARS-CoV-2 and SARS-CoV-2 RNA in wastewater and other water matrices at different temperatures. Dashed lines represent the linear regression fits for each target. Individual k values are obtained from previous studies and presented in Table S1.
log10 k of infections SARS-CoV-2 and SARS-CoV-2 RNA in wastewater and other water matrices at different temperatures. Dashed lines represent the linear regression fits for each target. Individual k values are obtained from previous studies and presented in Table S1.
Table 4.
Values of slopes, y-intercept and regression coefficients (R2) of linear regression of the log10-transformed first-order decay rate constants (in units of inverse days) of infectious SARS-CoV-2 and SARS-CoV-2 RNA as a function of temperature in wastewater and other water matrices.
| Virus | Wastewater |
Other water matrices |
||||
|---|---|---|---|---|---|---|
| Slope | y-intercept | R2 | Slope | y-intercept | R2 | |
| Infectious SARS-CoV-2 | 0.064 | −0.989 | 0.93 | 0.059 | −1.3307 | 0.55 |
| SARS-CoV-2 RNA | 0.022 | −1.239 | 0.56 | 0.026 | −1.6054 | 0.89 |
Fig. 2 showed that k of infectious SARS-CoV-2 and SARS-CoV-2 RNA increase with temperature regardless of the experimental water matrix. As shown in Fig. 2, the first-order decay rate constant for infectious SARS-CoV-2 showed slightly higher sensitivity to temperature in wastewater (slope = 0.064) than in water matrices (slope = 0.059) (Table 4). Moreover, infectious SARS-CoV-2 is more persistent in water matrices than in wastewater at a similar range of temperature (lower y-intercept in water matrices) (Fig. 2, Table 4). In comparison to other studies, similar slopes were obtained from the linear regression performed for infectious coronaviruses in wastewater (slope = 0.065) by Silverman and Boehm (2020) and for infectious SARS-CoV-2 in wastewater (slope = 0.07) performed by Bivins et al. (2020).
As for infectious SARS-CoV-2, comparable trend was observed for SARS-CoV-2 RNA in both wastewater and other water matrices in relation to temperature (Fig. 2, Table 4). Notably, within the range of environmentally relevant temperatures (i.e., 4–37 °C) infectious SARS-CoV-2 decayed at a faster rate than SARS-CoV-2 RNA in wastewater and other water matrices. For instance, in wastewater, when temperature increase 1 °C, log10 k increase by 0.064 (i.e k increase by 1.15) for infectious SARS-CoV-2 and increase by 0.022 (i.e k increase by 1.05) for SARS-CoV-2 RNA. It is important to note that SARS-CoV-2 RNA is less affected by temperature increase regardless the type of matrix than the infectious particle.
Additionally, SARS-CoV-2 RNA showed higher persistence than infectious SARS-CoV-2 in other water matrices than in wastewater at a similar range of temperature (lower y-intercept in other water matrices).
In conclusion, k is higher in wastewater and lowest in water matrices for both infectious SARS-CoV-2 and SARS-CoV-2 RNA. Several studies have indicated the low capacity of SARS-CoV-2 to persist in wastewater because of the presence of organic matter, pollutants and microbes that may increase the inactivation rates of the viruses (Foladori et al., 2022; Pinon and Vialette, 2018).
6. Elimination of SARS-CoV-2 in wastewater treatment plants
Traditional wastewater treatment techniques are intended to efficiently remove organic matter, suspended solids and bacteria (Adelodun et al., 2020). However, it has been shown to present an efficiency in removing viruses when disinfection is applied (Foladori et al., 2021). Wastewater treatment line comprises a series of steps: pre-treatments, primary treatment, secondary treatment, and tertiary treatment. It was reported from the literature that the efficiency to eliminate SARS-CoV-2, from the influent to the effluent wastewater, depends on the processes involved in the WWTP.
6.1. Pre-treatments stage
Pre-treatments involve the separation of coarse materials by mechanical screening (removal of particles over 5 mm in size), sieving (removal of particles over 0.25 mm in size), grit removal, and oil or grease removal (Zhou et al., 2015; Foladori et al., 2022). It was reported that these treatments might not have significant effects in reducing the viral load in wastewater (Zhou et al., 2015).
6.2. Primary treatment
Primary treatment aims to remove the settleable solids by sedimentation and producing primary sludge, which has 1–2 % total solids content, higher than 0.01–0.05 % in raw wastewater (Peccia et al., 2020). The effluent from primary sedimentation units is referred to as primary effluent. The retention time in the primary sedimentation tank or clarifier is between 2 and 3 h (Foladori et al., 2022). Because virus particles are small and having similar density to water, they cannot settle spontaneously and efficiently during this treatment. However, SARS-CoV-2, as in the case of others enveloped viruses, has a hydrophobic envelope, which decreases virus solubility in water and increases its ability to adsorb on solids (Gundy et al., 2009). Thus, this makes the virus to concentrate in sludge when it adsorbs on settled suspended solids. Moreover, the capacity of viral separation may increase during the flocculation process, where viral particles aggregate in the suspension and form larger particles with higher density (Bhatt et al., 2020). About 1-log10 removal of SARS-CoV-2 RNA was obtained after primary treatment by Abu Ali et al. (2021), indicating that most of viral RNA was adsorbed to settled solids while a 0.48 ± 1.17 log10 removal of SARS-CoV-2 RNA was obtained by Serra-Compte et al. (2021).
6.3. Secondary treatment
Secondary treatment involves biological treatments aiming at the removal of biodegradable compounds and the separation of inert particulate solids. The commonly adopted method in most WWTPs is the activated sludge process. It mainly consists of two steps. The first one involves aeration or anoxic tank, in which wastewater is treated with the aid of active microorganisms (i.e., activated sludge). During the second step, the secondary treated effluent and the activated sludge are separated in the sedimentation tank, or secondary clarifier (Amoah et al., 2020). Due to the hydrophobicity of the viral envelope, SARS-CoV-2 removal may be due to the adsorption on suspended solids (biological flocs) and subsequent sedimentation in the secondary clarifier (Mohapatra et al., 2021) where an additional 1-log removal was obtained after secondary settling by Abu Ali et al. (2021). Moreover, the retention time of the activated sludge process may contribute to viral removal through a spontaneous decaying process (Amoah et al., 2020). Randazzo et al. (2020) published a preliminary study on SARS-CoV-2 presence in wastewater after secondary treatment by activated sludge process and showed that 11 % of samples were positive to SARS-CoV-2 RNA. Balboa et al. (2021) confirmed these results when studying the presence of SARS-CoV-2 in several point of a real WWTP in Spain. Recently, no infectious SARS-CoV-2 viral particles was detected by Monteiro et al. (2022) in secondary treated wastewater samples despite their high viral RNA load.
When using a membrane bioreactor (MBR) system as secondary treatment, the biological process is combined with membrane filtration without the need for a secondary settler (Foladori et al., 2022). The degradation of biomass is performed inside the bioreactor tank, while the solid–liquid separation is achieved in a membrane module (Al-Asheh et al., 2021). Microfiltration and ultrafiltration, with size ranges of 0.1–0.2 μm and 0.005–10 μm, respectively, are the most common types of filtration utilized in MBR procedures (Foladori et al., 2022). A higher viral removal efficiency was achieved in MBR compared to conventional activated sludge process (Simmons and Xagoraraki, 2011). For instance, a 1.96 log10 removal of SARS-CoV-2 was obtained after MBR treatment compared to 1.03 log10 removal after activated sludge treatment (Serra-Compte et al., 2021). These results indicate that sludge obtained after MBR treatment may contain higher SARS-Cov-2 load than the sludge obtained after activated sludge process.
6.4. Tertiary treatment
Tertiary treatment involves the disinfection of the effluents with physical or chemical processes such as chlorination, UV irradiation, or ozonation. These treatments showed high efficiency in complete SARS-CoV-2 inactivation (Patel et al., 2021). Moreover, SARS-CoV-2 RNA was not detected in the tertiary effluent in most of the conducted studies. In the preliminary study of Randazzo et al. (2020), all samples were negative to SARS-CoV-2 RNA after tertiary treatment (using disinfection with NaClO and in some cases coupled with UV). This finding was also confirmed by the study of Balboa et al. (2021). Additionally, cell culture assays showed the absence of infectious SARS-CoV-2 particle in tertiary treated effluent wastewater samples (Rimoldi et al., 2020; Westhaus et al., 2021).
7. Partitioning and fate of SARS-CoV-2 RNA along the water and sludge treatment lines
Several studies have evaluated the partitioning and fate of SARS-CoV-2 during wastewater treatment processes (Table 5 ). Kocamemi et al. (2020) investigated the presence of SARS-CoV-2 in two primary sludge samples and seven secondary sludge samples collected from two WWTPs in Istanbul. The copy number values of SARS-CoV-2 in sludge samples ranged between 11.7 and 40.2 GC/mL and were higher than that observed in influent wastewater, indicating a partial accumulation of SARS-CoV-2 in the sludge. This finding was also confirmed by Westhaus et al. (2021) who studied SARS-CoV-2 partitioning in influent wastewater by comparing the aqueous and solid phases of the samples after centrifugation. They observed that SARS-CoV-2 RNA copy number was higher in the solid fraction (25 GC/mL) by approximately one order of magnitude than in the aqueous fraction (1.8 GC/mL).
Table 5.
Data on the detection of SARS-CoV-2 RNA in various types of samples along the wastewater and sludge treatment lines in WWTPs.
| Reference | Country | Sample type and/or treatment conditions | No of samples | number of positive samples or % of detection by RT-qPCR | Concentration range (GC/L) |
|---|---|---|---|---|---|
| Peccia et al., 2020 | USA | Primary sludge | 44 | 44 | 1.7 × 103–4.6 × 103 |
| Kocamemi et al., 2020 | Turkey | Primary sludge Secondary sludge |
2 7 |
2 7 |
12.5 × 103 –23.3 × 103 11.7 × 103–40.2 × 103 |
| Westhaus et al., 2021 | Germany | Influent raw wastewater Effluent Effluent after tertiary treatment Effluent after ozonation and filtration |
9 2 1 1 |
9 2 1 1 |
3 × 103–2 × 104 2.7 × 103–37 × 103 for all untreated and treated effluent samples |
| Balboa et al., 2021 | Spain | Influent raw wastewater Treated effluent Primary sludge Biological sludge Thickened sludge Digested sludge (Thermal hydrolysis and anaerobic digestion) |
5 5 5 10 10 10 |
5 0 4 1 9 0 |
2.15 × 103–9.8 × 103 NDa 1.3 × 103–24.5 × 103 1.9 × 103 1.9 × 103–18.8 × 103 ND |
| Serra-Compte et al., 2021 | France and Spain | Primary treated effluent Secondary treated effluent Effluent after activated sludge plus nutrient removal Tertiary treated effluent Membrane bioreactor treatment Primary sludge Activated sludge Thickened sludge Anaerobic digested sludge Anaerobic digested sludge plus thermal hydrolysis |
5 30 11 2 11 6 14 13 7 5 |
40 % 23.3 % 18.2 % 0 % 0 % 83.3 % 57.1 % 69.2 % 71.4 % 0 % |
Not reported |
| D'Aoust et al., 2021 | Canada | Post grit sludge Primary clarified sludge |
24 24 |
79.2 % (N1) and 82.3 % (N2) 92.7 % (N1) and 90.6 %(N2) |
1.7 × 103–78 × 103 for all positive samples |
| Pourakbar et al., 2022 | Iran/East Azerbaijan | Raw wastewater Final chlorination effluents Primary sludge Activated sludge Anaerobically digested sludge |
4 8 4 4 8 |
4 0 2 3 0 |
3.8 × 103–28 × 103 ND 3.2 × 103–13 × 103 7.1 × 103–31 × 103 ND |
| Carraturo et al., 2022 | Italy | Mature digestate collected during 13 months from the storage tank of a full-scale anaerobic digestion plant | 11 | 0 | ND |
| Yang et al., 2022 | China | Influent samples from 4 municipal WWTPs in Beijing Hospitals influent Secondary treated effluents Tertiary treated effluent water Sewer sediment |
15 6 5 12 2 |
6 3 3 8 2 |
8.5 × 101–8.8 × 103 1.60 × 102–9.61 × 102 3.7 × 102–2.34 × 103 4.64 × 102–8.62 × 102 4 × 103 |
ND: not detected.
In a collaborative study performed by six laboratories across the USA, Kim et al. (2022) compared SARS-CoV-2 RNA in primary settled solids obtained from primary clarifiers and raw wastewater influent samples collected from five publicly owned treatment works (POTWs). They showed that SARS-CoV-2 RNA concentrations, on a mass equivalent basis, were approximately 3 log10 unit higher in primary settled solids than in influent samples (Table 5).
In another study, D'Aoust et al. (2021), quantified SARS-CoV-2 RNA in 24 wastewater influent solids or post grit solids (PGS), and 24 primary clarified sludge (PCS) samples in two water resource recovery facilities in Canada, using CDC N1 and N2 assays. SARS-CoV-2 RNA was detected in high frequency in PCS (92.7 and 90.6 % for N1 and N2, respectively) as compared with PGS samples (79.2 and 82.3 % for N1 and N2, respectively) (Table 5).
Furthermore, Balboa et al. (2021) studied the prevalence of SARS-CoV-2 in various samples in a WWTP located in north-western Spain (Table 5). The analyzed samples were 5 influent raw wastewater, 5 treated effluent, 5 primary sludge, 10 biological sludge, 10 thickened sludge and 10 anaerobically digested sludge. SARS-CoV-2 RNA was detected in 100 % of influent samples at a generally low concentration, at most up to 9.8 × 103 GC/L but reached >2 × 104 GC/L in some sludge samples. No SARS-CoV-2 RNA was detected in the effluent samples as they were adsorb to solids. Hence, SARS-CoV-2 RNA was found in the majority of the primary (5/4) and thickened sludge (9/10) samples. Interestingly, no RNA signal was detected in the digested sludge samples, which is likely due to the high temperature applied during anaerobic digestion (Balboa et al., 2021). The authors hypothesized that the primary settler and the sludge thickeners could act as “concentrators” of SARS-CoV-2 RNA. Similarly, Serra-Compte et al. (2021) detected SARS-CoV-2 RNA with high frequency in primary, activated and thickened sludge samples (Table 5). It was also detected in anaerobically digested sludge samples but was eliminated in sludge samples when thermal hydrolysis was applied during anaerobic digestion.
Pourakbar et al. (2022) also studied the fate of SARS-CoV-2 RNA in two different WWTPs in Iran using sequencing batch reactor (SBR) and conventional activated sludge (CAS) (Table 5). They showed that SARS-CoV-2 RNA was detected in all raw wastewater samples and was absent in the final chlorination effluents. They observed that the viral RNA in the WWTPs has higher affinity to biosolids rather than liquid phase, with higher concentrations in the secondary sludge (0.71 × 104– 3.1 × 104 GC/L) than the primary sludge (0.32 × 104– 1.3 × 104 GC/L) of the conventional activated sludge process. This fact could be due to the higher solids retention time in the secondary treatment units, which was about 16–20 days. However, SARS-CoV-2 RNA was shown to be completely destroyed during anaerobic digestion with solids retention time value of about 30 days (Pourakbar et al., 2022).
Recently, Carraturo et al. (2022) studied the effectiveness of a full-scale thermophilic (55 °C) anaerobic digestion process by monitoring the hygienic characteristics of mature digestate samples collected during 13 months. They showed the absence of SARS-CoV-2 RNA in all samples despite it has been detected in the inlet flux of organic solids (Table 5).
According to the Italian National Institute of Health (COVID-19, 2020) and U.S. National Research Council (Council, 2002), the presence of viruses in sludge is not directly indicative of a potential hazard of the matrix as an effective transmission capacity of the pathogen is not proven.
8. Concluding remarks
The occurrence of SARS-CoV-2 RNA and the evaluation of virus infectivity were reviewed in different compartments presented in Fig. 1. Viral RNA of SARS-CoV-2 have been detected in wastewater, sludge, effluent, river water, sediments and bivalve mollusks. However, data are lacking for certain compartments, for example in groundwater, that allows not to have a global overview of the spreading of SARS-CoV-2 in the environment.
Based on the information reviewed in this study, there is no clear evidence for the presence of infectious SARS-CoV-2 in feces although its isolation from stool samples of six different patients. Additionally, infectivity of virus was not revealed in wastewater and sludge samples when SARS CoV-2 RNA were detected. However, cell culture method, which actually considered the gold standard technique to confirm the presence of infectious particle, remain complex to use especially when concentrated wastewater is tested. Furthermore, samples containing high level of microorganisms and the diversity of chemical compound could provide interference, which complicate the interpretation. The use of new methods combining different strategy such as PMA, PtCl4, EMA-RT-PCR should allow, in the future, obtaining more data on this aspect.
Data on the partitioning and removal of SARS-CoV-2 RNA during the wastewater and sludge treatments revealed that
-
(a)
tertiary treatments of wastewater present high efficiency in complete removal of SARS-CoV-2
-
(b)
SARS-CoV-2 RNA copy number was higher in the solid fraction than in the aqueous fraction and genome concentrations increase during the sludge thickening process
-
(c)
Sludge digestion treatments, mainly in thermophilic condition show an efficiency in SARS-CoV-2 inactivation.
Regarding SARS-CoV-2 persistence, SARS-CoV-2 RNA was found to be more persistent than infectious particles in aquatic environment and SARS-CoV-2 decay occurs at a higher rate in wastewater than in others water matrices at a similar range of temperature. Besides temperature, SARS-CoV-2 persistence in wastewater was shown to be also influenced by pH and wastewater characteristics and composition. The high persistence of SARS-CoV-2 RNA makes its detection and quantification a useful indicator to monitor the disease among the population. More studies are needed to understand the different mechanisms of SARS-CoV-2 particles and RNA inactivation in wastewater. Finally, it is also important to study in depth the fate of SARS-CoV-2 and the mechanism of its decay through the stage of wastewater and sludge treatment lines.
The following is the supplementary data related to this article.
Decay Kinetics of infectious SARS-CoV-2 and SARS-CoV-2 RNA in wastewater and other water matrices at different temperatures
CRediT authorship contribution statement
Ali Atoui: Writing - Original draft preparation, Conceptualization, Writing - Reviewing and Editing.
Christophe Cordevant: Writing - Reviewing and Editing.
Thierry Chesnot: Writing - Reviewing and Editing.
Benoit Gassilloud: Conceptualization- Writing - Reviewing and Editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are grateful to Gilles Salvat, the General Director for Research and Reference at Anses, for his critical reviewing and support. We also thank Damien Mouly and Frédéric Jourdain from Santé publique France for reading the manuscript and for providing criticism and helpful suggestions. This work was supported by One Health European Joint Programme under grant agreement No 773830: COVRIN One Health research integration on SARS-CoV-2 emergence, risk assessment and preparedness.
Editor: Damia Barcelo
Data availability
Data will be made available on request.
References
- Abu Ali H., Yaniv K., Bar-Zeev E., Chaudhury S., Shagan M., Lakkakula S., Ronen Z., Kushmaro A., Nir O. 2021. Tracking SARS-CoV-2 RNA Through the Wastewater Treatment Process. [DOI] [PubMed] [Google Scholar]
- Adelodun Bashir, Ogunshina Matthew Segun, Ajibade Fidelis Odedishemi, Abdulkadir Taofeeq Sholagberu, Bakare Hashim Olalekan, Choi Kyung Sook. Kinetic and prediction modeling studies of organic pollutants removal from municipal wastewater using Moringa oleifera biomass as a coagulant. Water. 2020;12:2052. [Google Scholar]
- Ahmed Warish, Angel Nicola, Edson Janette, Bibby Kyle, Bivins Aaron, O’Brien Jake W., Choi Phil M., Kitajima Masaaki, Simpson Stuart L., Li Jiaying, Tscharke Ben, Verhagen Rory, Smith Wendy J.M., Zaugg Julian, Dierens Leanne, Hugenholtz Philip, Thomas Kevin V., Mueller Jochen F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Warish, Bibby Kyle, D'Aoust Patrick M., Delatolla Robert, Gerba Charles P., Haas Charles N., Hamilton Kerry A., Hewitt Joanne, Julian Timothy R., Kaya Devrim, Monis Paul, Moulin Laurent, Naughton Colleen, Noble Rachel T., Shrestha Abhilasha, Tiwari Ananda, Simpson Stuart L., Wurtzer Sebastien, Bivins Aaron. Differentiating between the possibility and probability of SARS-CoV-2 transmission associated with wastewater: empirical evidence is needed to substantiate risk. FEMS Microbes. 2021;2 doi: 10.1093/femsmc/xtab007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Warish, Bivins Aaron, Bertsch Paul M., Bibby Kyle, Choi Phil M., Farkas Kata, Gyawali Pradip, Hamilton Kerry A., Haramoto Eiji, Kitajima Masaaki, Simpson Stuart L., Tandukar Sarmila, Thomas Kevin, Mueller Jochen F. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimisation and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020;17 doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Warish, Simpson Stuart L., Bertsch Paul M., Bibby Kyle, Bivins Aaron, Blackall Linda L., Bofill-Mas Sílvia, Bosch Albert, Brandão João, Choi Phil M., Ciesielski Mark, Donner Erica, D'Souza Nishita, Farnleitner Andreas H., Gerrity Daniel, Gonzalez Raul, Griffith John F., Gyawali Pradip, Haas Charles N., Hamilton Kerry A., Hapuarachchi Hapuarachchige Chanditha, Harwood Valerie J., Haque Rehnuma, Jackson Greg, Khan Stuart J., Khan Wesaal, Kitajima Masaaki, Korajkic Asja, La Rosa Giuseppina, Layton Blythe A., Lipp Erin, McLellan Sandra L., McMinn Brian, Medema Gertjan, Metcalfe Suzanne, Meijer Wim G., Mueller Jochen F., Murphy Heather, Naughton Coleen C., Noble Rachel T., Payyappat Sudhi, Petterson Susan, Pitkänen Tarja, Rajal Veronica B., Reyneke Brandon, Roman Fernando A., Rose Joan B., Rusiñol Marta, Sadowsky Michael J., Sala-Comorera Laura, Setoh Yin Xiang, Sherchan Samendra P., Sirikanchana Kwanrawee, Smith Wendy, Steele Joshua A., Sabburg Rosalie, Symonds Erin M., Thai Phong, Thomas Kevin V., Tynan Josh, Toze Simon, Thompson Janelle, Whiteley Andy S., Wong Judith Chui Ching, Sano Daisuke, Wuertz Stefan, Xagoraraki Irene, Zhang Qian, Zimmer-Faust Amity G., Shanks Orin C. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.149877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Asheh Sameer, Bagheri Marzieh, Aidan Ahmed. Membrane bioreactor for wastewater treatment: a review. Case Stud. Chem. Environ. Eng. 2021;4 [Google Scholar]
- Albert S., Ruíz A., Pemán J., Salavert M., Domingo-Calap P. Lack of evidence for infectious SARS-CoV-2 in feces and sewage. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:2665–2667. doi: 10.1007/s10096-021-04304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoah Dennis, Isaac Sheena Kumari, Bux Faizal. Coronaviruses in wastewater processes: source, fate and potential risks. Environ. Int. 2020;143 doi: 10.1016/j.envint.2020.105962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand U., Li X., Sunita K., Lokhandwala S., Gautam P., Suresh S., Sarma H., Vellingiri B., Dey A., Bontempi E., Jiang G. SARS-CoV-2 and other pathogens in municipal wastewater, landfill leachate, and solid waste: a review about virus surveillance, infectivity, and inactivation. Environ. Res. 2022;203 doi: 10.1016/j.envres.2021.111839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodriguez S., Martinez-Lamas L., Vasallo F.J., Regueiro B., Lema J.M. The fate of SARS-COV-2 in WWTPS points out the sludge line as a suitable spot for detection of COVID-19. Sci. Total Environ. 2021;772 doi: 10.1016/j.scitotenv.2021.145268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard K., Davis A., Simpson I.M., Hale V.L., Lee J., Winston R.J. Detection of SARS-CoV-2 in urban stormwater: an environmental reservoir and potential interface between human and animal sources. Sci. Total Environ. 2022;807 doi: 10.1016/j.scitotenv.2021.151046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt A., Arora P., Prajapati S.K. Occurrence, fates and potential treatment approaches for removal of viruses from wastewater: a review with emphasis on SARS-CoV-2. J. Environ. Chem. Eng. 2020;8 doi: 10.1016/j.jece.2020.104429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins Aaron, Greaves Justin, Fischer Robert, Yinda Kwe Claude, Ahmed Warish, Kitajima Masaaki, Munster Vincent J., Bibby Kyle. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7(12):937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogler Anne, Packman Aaron, Furman Alex, Gross Amit, Kushmaro Ariel, Ronen Avner, Dagot Christophe, Hill Colin, Vaizel-Ohayon Dalit, Morgenroth Eberhard, Bertuzzo Enrico, Wells George, Kiperwas Hadas Raanan, Horn Harald, Negev Ido, Zucker Ines, Bar-Or Itay, Moran-Gilad Jacob, Balcazar Jose Luis, Bibby Kyle, Elimelech Menachem, Weisbrod Noam, Nir Oded, Sued Oded, Gillor Osnat, Alvarez Pedro J., Crameri Sandra, Arnon Shai, Walker Sharon, Yaron Sima, Nguyen Thanh H., Berchenko Yakir, Hu Yunxia, Ronen Zeev, Bar-Zeev Edo. Rethinking wastewater risks and monitoring in light of the COVID-19 pandemic. Nat. Sustain. 2020;3:981–990. [Google Scholar]
- Bosch A., Pintó R.M., Abad F.X. Survival and transport of enteric viruses in the environment. Viruses Foods. 2006:151–187. [Google Scholar]
- Carraturo F., Panico A., Giordano A., Libralato G., Aliberti F., Galdiero E., Guida M. Hygienic assessment of digestate from a high solids anaerobic co-digestion of sewage sludge with biowaste by testing salmonella typhimurium, Escherichia coli and SARS-CoV-2. Environ. Res. 2022;206 doi: 10.1016/j.envres.2021.112585. [DOI] [PubMed] [Google Scholar]
- Cascella M., Rajnik M., Aleem A., Dulebohn S.C., Napoli R.Di. StatPearls. StatPearls Publishing Copyright © 2022, StatPearls Publishing LLC.; Treasure Island (FL): 2022. Features, evaluation, and treatment of Coronavirus (COVID-19) [PubMed] [Google Scholar]
- CDC Interim laboratory biosafety guidelines for handling and processing specimens associated with Coronavirus Disease 2019 (COVID-19) 2021. https://www.cdc.gov/coronavirus/2019-ncov/lab/lab-biosafety-guidelines.html
- Cerrada-Romero C., Berastegui-Cabrera J., Camacho-Martínez P., Goikoetxea-Aguirre J., Pérez-Palacios P., Santibáñez S., José Blanco-Vidal M., Valiente A., Alba J., Rodríguez-Álvarez R., Pascual Á., Oteo J.A., Miguel Cisneros J., Pachón J., Casas-Flecha I., Cordero E., Pozo F., Sánchez-Céspedes J. Excretion and viability of SARS-CoV-2 in feces and its association with the clinical outcome of COVID-19. Sci. Rep. 2022;12:7397. doi: 10.1038/s41598-022-11439-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik Muge, Tate Matthew, Lloyd Ollie, Maraolo Alberto Enrico, Schafers Jenna, Ho Antonia. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A.W.H., Chu J.T.S., Perera M.R.A., Hui K.P.Y., Yen H.L., Chan M.C.W., Peiris M., Poon L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1 doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Council, National Research . The National Academies Press; Washington, DC: 2002. Biosolids Applied to Land: Advancing Standards and Practices. [Google Scholar]
- COVID-19, Gruppo di Lavoro ISS Ambiente-Rifiuti . Istituto Superiore di Sanità; Roma: 2020. Indicazioni ad interim sulla gestione dei fanghi di depurazione per la prevenzione della diffusione del virus SARS-CoV-2. [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020:25. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Ferrando E., Randazzo W., Pérez-Cataluña A., Falcó I., Navarro D., Martin-Latil S., Díaz-Reolid A., Girón-Guzmán I., Allende A., Sánchez G. Platinum chloride-based viability RT-qPCR for SARS-CoV-2 detection in complex samples. Sci. Rep. 2021;11:18120. doi: 10.1038/s41598-021-97700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuicchi D., Lazzarotto T., Poggioli G. Fecal-oral transmission of SARS-CoV-2: review of laboratory-confirmed virus in gastrointestinal system. Int. J. Color. Dis. 2021;36:437–444. doi: 10.1007/s00384-020-03785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust Patrick M., Mercier Elisabeth, Montpetit Danika, Jia Jian-Jun, Alexandrov Ilya, Neault Nafisa, Baig Aiman Tariq, Janice Mayne Xu., Zhang Tommy Alain, Langlois Marc-André, Servos Mark R., MacKenzie Malcolm, Figeys Daniel, MacKenzie Alex E., Graber Tyson E., Delatolla Robert. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188 doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decimo Didier, Labaronne Emmanuel, Ricci Emiliano, Mathieu Cyrille, Goujon Caroline, Terrier Olivier. XXIVème Journées Francophones de Virologie. Université de Strasbourg, Strasbourg-France: Société Française de Virologie. 2022. Avez-vous les bonnes Vero-E6? [Google Scholar]
- Dergham J., Delerce J., Bedotto M., La Scola B., Moal V. Isolation of viable SARS-CoV-2 virus from feces of an immunocompromised patient suggesting a possible fecal mode of transmission. J. Clin. Med. 2021;10 doi: 10.3390/jcm10122696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desdouits Marion, Piquet Jean-Côme, Wacrenier Candice, Le Mennec Cécile, Parnaudeau Sylvain, Jousse Sarah, Rocq Sophie, Bigault Lionel, Contrant Ma.ud., Garry Pascal, Chavanon Fabienne, Gabellec Raoul, Lamort Laure, Lebrun Luc, Le Gall Patrik, Meteigner Claire, Schmitt Anne, Seugnet Jean Luc, Serais Ophélie, Peltier Cécile, Bressolette-Bodin Céline, Blanchard Yannick, Le Guyader Françoise S. Can shellfish be used to monitor SARS-CoV-2 in the coastal environment? Sci. Total Environ. 2021;778 doi: 10.1016/j.scitotenv.2021.146270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn A.S., Meijer B., Frampton C.M.A., Barclay M.L., de Boer N.K.H. Systematic review with meta-analysis: SARS-CoV-2 stool testing and the potential for faecal-oral transmission. Aliment. Pharmacol. Ther. 2020;52:1276–1288. doi: 10.1111/apt.16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elizaquível P., Aznar R., Sánchez G. Recent developments in the use of viability dyes and quantitative PCR in the food microbiology field. J. Appl. Microbiol. 2014;116:1–13. doi: 10.1111/jam.12365. [DOI] [PubMed] [Google Scholar]
- Feng W., Newbigging A.M., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D.B., Cui M., Tao J., Tyrrell D.L., Zhang X.E., Zhang H., Le X.C. Molecular diagnosis of COVID-19: challenges and research needs. Anal. Chem. 2020;92:10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Cadonna M., Manara S., Maestrini F. Environmental and Health Management of Novel Coronavirus Disease (COVID-19 ) 2021. Route of SARS-CoV-2 in sewerage and wastewater treatment plants: dilution, decay, removal, and environmental transmission; pp. 145–176. [Google Scholar]
- Foladori P., Cutrupi F., Cadonna M., Manara S. Coronaviruses and SARS-CoV-2 in sewerage and their removal: step by step in wastewater treatment plants. Environ. Res. 2022;207 doi: 10.1016/j.envres.2021.112204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuta Mizuki, Mao Zhan Qiu, Morita Kouichi, Moi Meng Ling. Stability and infectivity of SARS-CoV-2 and viral RNA in water, commercial beverages, and bodily fluids. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.667956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacobbo Alexandre, Rodrigues Marco Antônio Siqueira, Ferreira Jane Zoppas, Bernardes Andréa Moura, de Pinho Maria Norberta. A critical review on SARS-CoV-2 infectivity in water and wastewater. What do we know? Sci. Total Environ. 2021;774 doi: 10.1016/j.scitotenv.2021.145721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Latorre Laura, Ballesteros Isabel, Irina Villacrés-Granda M., Granda Genoveva, Freire-Paspuel Byron, Ríos-Touma Blanca. SARS-CoV-2 in river water: implications in low sanitation countries. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundy Patricia M., Gerba Charles P., Pepper Ian L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2009;1:10. [Google Scholar]
- Gupta S., Parker J., Smits S., Underwood J., Dolwani S. Persistent viral shedding of SARS-CoV-2 in faeces - a rapid review. Color. Dis. 2020;22:611–620. doi: 10.1111/codi.15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokajärvi Anna-Maria, Rytkönen Annastiina, Tiwari Ananda, Kauppinen Ari, Oikarinen Sami, Lehto Kirsi-Maarit, Kankaanpää Aino, Gunnar Teemu, Al-Hello Haider, Blomqvist Soile, Miettinen Ilkka T., Savolainen-Kopra Carita, Pitkänen Tarja. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H.W., Kim S.M., Kim H.S., Kim Y.I., Kim J.H., Cho J.Y., Kim S.H., Kang H., Kim S.G., Park S.J., Kim E.H., Choi Y.K. Viable SARS-CoV-2 in various specimens from COVID-19 patients. Clin. Microbiol. Infect. 2020;26:1520–1524. doi: 10.1016/j.cmi.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf G., Voss A., Scheithauer S. Inactivation of coronaviruses by heat. J. Hosp. Infect. 2020;105:348–349. doi: 10.1016/j.jhin.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Kim H.M., Lee E.J., Jo H.J., Yoon Y., Lee N.J., Son J., Lee Y.J., Kim M.S., Lee Y.P., Chae S.J., Park K.R., Cho S.R., Park S., Kim S.J., Wang E., Woo S., Lim A., Park S.J., Jang J., Chung Y.S., Chin B.S., Lee J.S., Lim D., Han M.G., Yoo C.K. Detection and isolation of SARS-CoV-2 in serum, urine, and stool specimens of COVID-19 patients from the Republic of Korea. Osong. Public Health Res. Perspect. 2020;11:112–117. doi: 10.24171/j.phrp.2020.11.3.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Sooyeol, Kennedy Lauren C., Wolfe Marlene K., Criddle Craig S., Duong Dorothea H., Topol Aaron, White Bradley J., Kantor Rose S., Nelson Kara L., Steele Joshua A., Langlois Kylie, Griffith John F., Zimmer-Faust Amity G., McLellan Sandra L., Schussman Melissa K., Ammerman Michelle, Wigginton Krista R., Bakker Kevin M., Boehm Alexandria B. SARS-CoV-2 RNA is enriched by orders of magnitude in primary settled solids relative to liquid wastewater at publicly owned treatment works. Environ. Sci.: Water Res. Technol. 2022;8:757–770. doi: 10.1039/d1ew00826a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocamemi Bilge Alpaslan, Kurt Halil, Sait Ahmet, Sarac Fahriye, Saatci Ahmet Mete, Pakdemirli Bekir. medRxiv: 2020.05.12.20099358; 2020. SARS-CoV-2 Detection in Istanbul Wastewater Treatment Plant Sludges. [Google Scholar]
- Kolarević Stoimir, Micsinai Adrienn, Szántó-Egész Réka, Lukács Alena, Kračun-Kolarević Margareta, Lundy Lian, Kirschner Alexander K.T., Farnleitner Andreas H., Djukic Aleksandar, Čolić Jasna, Nenin Tanja, Sunjog Karolina, Paunović Momir. Detection of SARS-CoV-2 RNA in the Danube River in Serbia associated with the discharge of untreated wastewaters. Sci. Total Environ. 2021;783 doi: 10.1016/j.scitotenv.2021.146967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumblathan Teresa, Liu Yanming, Uppal Gursharan K., Hrudey Steve E., Li Xing-Fang. Wastewater-based epidemiology for community monitoring of SARS-CoV-2: Progress and challenges. ACS Environ. Au. 2021;1:18–31. doi: 10.1021/acsenvironau.1c00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Mary Y.Y., Cheng Peter K.C., Lim Wilina W.L. Survival of severe acute respiratory syndrome coronavirus. Clin. Infect. Dis. 2005;41:e67–e71. doi: 10.1086/433186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guernic A., Palos Ladeiro M., Boudaud N., Do Nascimento J., Gantzer C., Inglard J.C., Mouchel J.M., Pochet C., Moulin L., Rocher V., Waldman P., Wurtzer S., Geffard A. First evidence of SARS-CoV-2 genome detection in zebra mussel (Dreissena polymorpha) J. Environ. Manag. 2022;301 doi: 10.1016/j.jenvman.2021.113866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifels Mats, Cheng Dan, Sozzi Emanuele, Shoults David C., Wuertz Stefan, Mongkolsuk Skorn, Sirikanchana Kwanrawee. Capsid integrity quantitative PCR to determine virus infectivity in environmental and food applications – a systematic review. Water Res. X. 2021;11 doi: 10.1016/j.wroa.2020.100080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Mengtian, Song Ge, Liu Ruiping, Huang Xia, Liu Huijuan. Inactivation and risk control of pathogenic microorganisms in municipal sludge treatment: a review. Front. Environ. Sci. Eng. 2021;16:70. doi: 10.1007/s11783-021-1504-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Jiang X., Zhang Z., Huang S., Zhang Z., Fang Z., Gu Z., Gao L., Shi H., Mai L., Liu Y., Lin X., Lai R., Yan Z., Li X., Shan H. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- Maidana-Kulesza M.N., Poma H.R., Sanguino-Jorquera D.G., Reyes S.I., Del Milagro Said-Adamo M., Mainardi-Remis J.M., Gutiérrez-Cacciabue D., Cristóbal H.A., Cruz M.C., Aparicio González M., Rajal V.B. Tracking SARS-CoV-2 in rivers as a tool for epidemiological surveillance. Sci. Total Environ. 2022;848 doi: 10.1016/j.scitotenv.2022.157707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancusi A., Capuano F., Girardi S., Di Maro O., Suffredini E., Di Concilio D., Vassallo L., Cuomo M.C., Tafuro M., Signorelli D., Pierri A., Pizzolante A., Cerino P., La Rosa G., Proroga Y.T.R., Pierri B. Detection of SARS-CoV-2 RNA in bivalve mollusks by droplet digital RT-PCR (dd RT-PCR) Int. J. Environ. Res. Public Health. 2022;19 doi: 10.3390/ijerph19020943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mautner L., Hoyos M., Dangel A., Berger C., Ehrhardt A., Baiker A. Replication kinetics and infectivity of SARS-CoV-2 variants of concern in common cell culture models. Virol. J. 2022;19:76. doi: 10.1186/s12985-022-01802-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra S., Menon N.G., Mohapatra G., Pisharody L., Pattnaik A., Menon N.G., Bhukya P.L., Srivastava M., Singh M., Barman M.K., Gin K.Y., Mukherji S. The novel SARS-CoV-2 pandemic: possible environmental transmission, detection, persistence and fate during wastewater and water treatment. Sci. Total Environ. 2021;765 doi: 10.1016/j.scitotenv.2020.142746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro S., Rente D., Cunha M.V., Marques T.A., Cardoso E., Vilaça J., Coelho N., Brôco N., Carvalho M., Santos R. Discrimination and surveillance of infectious severe acute respiratory syndrome coronavirus 2 in wastewater using cell culture and RT-qPCR. Sci. Total Environ. 2022;815 doi: 10.1016/j.scitotenv.2022.152914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura I.B., Buckley A.M., Wilcox M.H. Can SARS-CoV-2 be transmitted via faeces? Curr. Opin. Gastroenterol. 2022;38:26–29. doi: 10.1097/MOG.0000000000000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan A., Zlitni S., Brooks E.F., Vance S.E., Dahlen A., Hedlin H., Park R.M., Han A., Schmidtke D.T., Verma R., Jacobson K.B., Parsonnet J., Bonilla H.F., Singh U., Pinsky B.A., Andrews J.R., Jagannathan P., Bhatt A.S. Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection. Med (N Y) 2022;3:371–387. doi: 10.1016/j.medj.2022.04.001. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.K., Lee C.W., Park D.I., Woo H.Y., Cheong H.S., Shin H.C., Ahn K., Kwon M.J., Joo E.J. Detection of SARS-CoV-2 in fecal samples from patients with asymptomatic and mild COVID-19 in Korea. Clin. Gastroenterol. Hepatol. 2021;19:1387–1394. doi: 10.1016/j.cgh.2020.06.005. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar Nagina, Singh Ajay, Ward Owen P. Characterization of the combined effects of enzyme, pH and temperature treatments for removal of pathogens from sewage sludge. World J. Microbiol. Biotechnol. 2001;17:169–172. [Google Scholar]
- Patel Manvendra, Chaubey Abhishek Kumar, Pittman Charles U., Mlsna Todd, Mohan Dinesh. Coronavirus (SARS-CoV-2) in the environment: occurrence, persistence, analysis in aquatic systems and possible management. Sci. Total Environ. 2021;765 doi: 10.1016/j.scitotenv.2020.142698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia Jordan, Zulli Alessandro, Brackney Doug E., Grubaugh Nathan D., Kaplan Edward H., Casanovas-Massana Arnau, Ko Albert I., Malik Amyn A., Wang Dennis, Wang Mike, Warren Joshua L., Weinberger Daniel M., Arnold Wyatt, Omer Sa.ad.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon A., Vialette M. Survival of viruses in water. Intervirology. 2018;61:214–222. doi: 10.1159/000484899. [DOI] [PubMed] [Google Scholar]
- Polo D., Lois M., Fernández-Núñez M.T., Romalde J.L. Detection of SARS-CoV-2 RNA in bivalve mollusks and marine sediments. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourakbar M., Abdolahnejad A., Raeghi S., Ghayourdoost F., Yousefi R., Behnami A. Comprehensive investigation of SARS-CoV-2 fate in wastewater and finding the virus transfer and destruction route through conventional activated sludge and sequencing batch reactor. Sci. Total Environ. 2022;806 doi: 10.1016/j.scitotenv.2021.151391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransome E., Hobbs F., Jones S., Coleman C.M., Harris N.D., Woodward G., Bell T., Trew J., Kolarević S., Kračun-Kolarević M., Savolainen V. Evaluating the transmission risk of SARS-CoV-2 from sewage pollution. Sci. Total Environ. 2023;858 doi: 10.1016/j.scitotenv.2022.159161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi Sara Giordana, Stefani Fabrizio, Gigantiello Anna, Polesello Stefano, Comandatore Francesco, Mileto Davide, Maresca Mafalda, Longobardi Concetta, Mancon Alessandro, Romeri Francesca, Pagani Cristina, Cappelli Francesca, Roscioli Claudio, Moja Lorenzo, Gismondo Maria Rita, Salerno Franco. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson Carolyn A., Hsieh Hsin-Yeh, Hsu Sh.u.-Yu., Wang Yang, Salcedo Braxton T., Belenchia Anthony, Klutts Jessica, Zemmer Sally, Reynolds Melissa, Semkiw Elizabeth, Foley Trevor, Wan XiuFeng, Wieberg Chris G., Wenzel Jeff, Lin Chung-Ho, Johnson Marc C. Defining biological and biophysical properties of SARS-CoV-2 genetic material in wastewater. Sci. Total Environ. 2022;807 doi: 10.1016/j.scitotenv.2021.150786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan-Hernandez L., Graham K.E., Duong D., Boehm A.B. Persistence of endogenous SARS-CoV-2 and pepper mild mottle virus RNA in wastewater-settled solids. ACS ES T Water. 2022;2(11):1944–1952. doi: 10.1021/acsestwater.2c00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Comorera Laura, Reynolds Liam J., Martin Niamh A., O'Sullivan John J., Meijer Wim G., Fletcher Nicola F. Decay of infectious SARS-CoV-2 and surrogates in aquatic environments. Water Res. 2021;201 doi: 10.1016/j.watres.2021.117090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Compte Albert, González Susana, Arnaldos Marina, Berlendis Sabrina, Courtois Sophie, Loret Jean Francois, Schlosser Olivier, Yáñez Adela M., Soria-Soria Elena, Fittipaldi Mariana, Saucedo Gemma, Pinar-Méndez Anna, Paraira Miquel, Galofré Belén, Lema Juan M., Balboa Sabela, Mauricio-Iglesias Miguel, Bosch Albert, Pintó Rosa M., Bertrand Isabelle, Gantzer Christophe, Montero Carlos, Litrico Xavier. Elimination of SARS-CoV-2 along wastewater and sludge treatment processes. Water Res. 2021;202 doi: 10.1016/j.watres.2021.117435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman Andrea I., Boehm Alexandria B. Systematic review and meta-analysis of the persistence and disinfection of human coronaviruses and their viral surrogates in water and wastewater. Environ. Sci. Technol. Lett. 2020;7(8):544–553. doi: 10.1021/acs.estlett.0c00313. [DOI] [PubMed] [Google Scholar]
- Simmons F.J., Xagoraraki I. Release of infectious human enteric viruses by full-scale wastewater utilities. Water Res. 2011;45:3590–3598. doi: 10.1016/j.watres.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Sobsey Mark D. Absence of virological and epidemiological evidence that SARS-CoV-2 poses COVID-19 risks from environmental fecal waste, wastewater and water exposures. J. Water Health. 2022;20:126–138. doi: 10.2166/wh.2021.182. [DOI] [PubMed] [Google Scholar]
- de Oliveira Leonardo, Camilo Andrés Felipe, Torres-Franco Bruna Coelho, Lopes Beatriz Senra, da Silva Álvares, Santos Erica Azevedo, Costa Michelle S., Costa Marcus Tulius, Reis P., Melo Marília C., Polizzi Rodrigo Bicalho, Teixeira Mauro Martins, Mota César R. Viability of SARS-CoV-2 in river water and wastewater at different temperatures and solids content. Water Res. 2021;195 doi: 10.1016/j.watres.2021.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza G.A.P., Le Bideau M., Boschi C., Ferreira L., Wurtz N., Devaux C., Colson P., La Scola B. Emerging SARS-CoV-2 genotypes show different replication patterns in human pulmonary and intestinal epithelial cells. Viruses. 2021;14 doi: 10.3390/v14010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandukar Sarmila, Sthapit Niva, Thakali Ocean, Malla Bikash, Sherchan Samendra P., Shakya Bijay Man, Shrestha Laxman P., Sherchand Jeevan B., Joshi Dev Raj, Lama Bhupendra, Haramoto Eiji. Detection of SARS-CoV-2 RNA in wastewater, river water, and hospital wastewater of Nepal. Sci. Total Environ. 2022;824 doi: 10.1016/j.scitotenv.2022.153816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varbanov M., Bertrand I., Philippot S., Retourney C., Gardette M., Hartard C., Jeulin H., Duval R.E., Loret J.F., Schvoerer E., Gantzer C. Somatic coliphages are conservative indicators of SARS-CoV-2 inactivation during heat and alkaline pH treatments. Sci. Total Environ. 2021;797 doi: 10.1016/j.scitotenv.2021.149112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Li J.S., Guo T.K., Zhen B., Kong Q.X., Yi B., Li Z., Song N., Jin M., Xiao W.J., Zhu X.M., Gu C.Q., Yin J., Wei W., Yao W., Liu C., Li J.F., Ou G.R., Wang M.N., Fang T.Y., Wang G.J., Qiu Y.H., Wu H.H., Chao F.H., Li J.W. Concentration and detection of SARS coronavirus in sewage from xiao tang Shan hospital and the 309th hospital. J. Virol. Methods. 2005;128:156–161. doi: 10.1016/j.jviromet.2005.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Xiaoming, Zheng Jingwei, Guo Lei, Yao Hao, Wang Lingya, XiaoDong Xia, Zhang Weixi. Fecal viral shedding in COVID-19 patients: clinical significance, viral load dynamics and survival analysis. Virus Res. 2020;289 doi: 10.1016/j.virusres.2020.198147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus Sandra, Weber Frank-Andreas, Schiwy Sabrina, Linnemann Volker, Brinkmann Markus, Widera Marek, Greve Carola, Janke Axel, Hollert Henner, Wintgens Thomas, Ciesek Sandra. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Coronavirus disease (COVID-19) weekly epidemiological update and weekly operational update. 2022. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- Winstone H., Lista M.J., Reid A.C., Bouton C., Pickering S., Galao R.P., Kerridge C., Doores K.J., Swanson C.M., Neil S.J.D. The polybasic cleavage site in SARS-CoV-2 spike modulates viral sensitivity to type I interferon and IFITM2. J. Virol. 2021;95 doi: 10.1128/JVI.02422-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel Roman, Corman Victor M., Guggemos Wolfgang, Seilmaier Michael, Zange Sabine, Müller Marcel A., Niemeyer Daniela, Jones Terry C., Vollmar Patrick, Rothe Camilla, Hoelscher Michael, Bleicker Tobias, Brünink Sebastian, Schneider Julia, Ehmann Rosina, Zwirglmaier Katrin, Drosten Christian, Wendtner Clemens. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wurtzer S., Waldman P., Ferrier-Rembert A., Frenois-Veyrat G., Mouchel J.M., Boni M., Maday Y., Marechal V., Moulin L. Several forms of SARS-CoV-2 RNA can be detected in wastewaters: implication for wastewater-based epidemiology and risk assessment. Water Res. 2021;198 doi: 10.1016/j.watres.2021.117183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao J., Huang J., Zhao J. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Guo W., Lopez-Hernadez A., Teng S., Li L. The pH effects on SARS-CoV and SARS-CoV-2 spike proteins in the process of binding to hACE2. Pathogens. 2022;11 doi: 10.3390/pathogens11020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y., Thongchankaew-Seo U., Yamazaki W. Very low likelihood that cultivated oysters are a vehicle for SARS-CoV-2: 2021–2022 seasonal survey at supermarkets in Kyoto, Japan. Heliyon. 2022;8 doi: 10.1016/j.heliyon.2022.e10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Shaolin, Dong Qian, Li Siqi, Cheng Zhao, Kang Xiaofeng, Ren Daheng, Chenyang Xu., Zhou Xiaohong, Liang Peng, Sun Lingli, Zhao Jianhong, Jiao Yang, Han Taoli, Liu Yanchen, Qian Yi, Liu Yi, Huang Xia, Jiuhui Qu. Persistence of SARS-CoV-2 RNA in wastewater after the end of the COVID-19 epidemics. J. Hazard. Mater. 2022;429 doi: 10.1016/j.jhazmat.2022.128358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Yinyin, Ellenberg Robert M., Graham Katherine E., Wigginton Krista R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Yong Zhang, Cao Chen, Shuangli Zh.u., Chang Sh.u., Dongyan Wang, Jingdong Song, Yang Song, Wei Zhen, Feng Zijian Wu., Jun Guizhen, Xu, Wenbo Xu. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the coronavirus disease 2019 (COVID-19) China CDC Weekly. 2020;2:123–124. [PMC free article] [PubMed] [Google Scholar]
- Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., Diamond M.S., Ciorba M.A., Whelan S.P.J., Ding S. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen C., Zhu S., Shu C., Wang D., Song J., Song Y., Zhen W., Feng Z., Wu G., Xu J., Xu W. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the coronavirus disease 2019 (COVID-19) China CDC Wkly. 2020;2:123–124. [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cen M., Hu M., Du L., Hu W., Kim J.J., Dai N. Prevalence and persistent shedding of fecal SARS-CoV-2 RNA in patients with COVID-19 infection: a systematic review and meta-analysis. Clin. Transl. Gastroenterol. 2021;12 doi: 10.14309/ctg.0000000000000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang X.C., Ji Z., Xu L., Yu Z. Source identification of bacterial and viral pathogens and their survival/fading in the process of wastewater treatment, reclamation, and environmental reuse. World J. Microbiol. Biotechnol. 2015;31:109–120. doi: 10.1007/s11274-014-1770-5. [DOI] [PubMed] [Google Scholar]
- Zhou J.Q., Liu G.X., Huang X.L., Gan H.T. The importance of fecal nucleic acid detection in patients with coronavirus disease (COVID-19): a systematic review and meta-analysis. J. Med. Virol. 2022;94:2317–2330. doi: 10.1002/jmv.27652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Decay Kinetics of infectious SARS-CoV-2 and SARS-CoV-2 RNA in wastewater and other water matrices at different temperatures
Data Availability Statement
Data will be made available on request.