SUMMARY
Disruption of the nuclear pore complex (NPC) and nucleocytoplasmic transport (NCT) have been implicated in the pathogenesis of neurodegenerative diseases. A GGGGCC hexanucleotide repeat expansion (HRE) in an intron of the C9orf72 gene is the most common genetic cause of amyotrophic lateral sclerosis and frontotemporal dementia, but the mechanism by which the HRE disrupts NCT is incompletely understood. We find that expression of GGGGCC repeats in Drosophila neurons induces proteasome-mediated degradation of select nucleoporins of the NPC. This process requires the Vps4 ATPase and the endosomal-sorting complex required for transport complex-III (ESCRT-III), as knockdown of ESCRT-III/Vps4 genes rescues nucleoporin levels, normalizes NCT, and suppresses GGGGCC-mediated neurodegeneration. GGGGCC expression upregulates nuclear ESCRT-III/Vps4 expression, and expansion microscopy demonstrates that the nucleoporins are translocated into the cytoplasm before undergoing proteasome-mediated degradation. These findings demonstrate a mechanism for nucleoporin degradation and NPC dysfunction in neurodegenerative disease.
Graphical Abstract
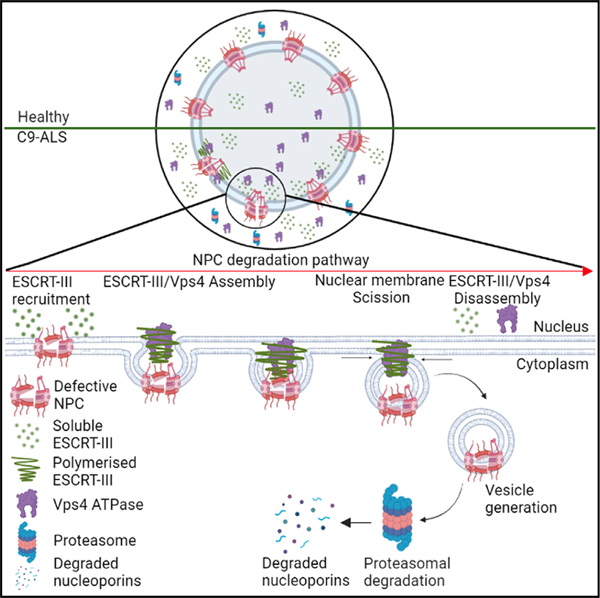
In brief
Dubey et al. show that increased nuclear levels of the ESCRT-III/Vps4 complex in neurons causes degradation of nucleoporins that make up the nuclear pore complex through the proteasome pathway in fly models of C9-ALS/FTD. Knockdown of ESCRT-III/Vps4 complex proteins restores nucleoporin levels and nucleocytoplasmic transport and suppresses neurodegeneration.
INTRODUCTION
The nuclear pore complex (NPC) is one of the largest and longest-lived protein complexes in eukaryotic cells and consists of hundreds of copies of approximately 30 different types of nuclear pore proteins called nucleoporins (Nups). A key function of the ~50–100 MDa NPC is to mediate the selective transport of macromolecules across the nuclear envelope, where phenylalanine-glycine (FG)-repeat-containing segments in nucleoporins form a diffusion barrier (Mohr et al., 2009; Ribbeck and Görlich, 2001; Rout et al., 2000). Defects in NCT have been associated with aging, traumatic brain injury, and neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), Alzheimer’s disease (AD), and Huntington disease (Zhang et al., 2015; Freibaum et al., 2015; Jovičić et al., 2015; Chou et al., 2018; Eftekharzadeh et al., 2019; Gasset-Rosa et al., 2017; Grima et al., 2017; Anderson et al., 2021). The mechanisms underlying NCT defects in neurodegenerative diseases remain incompletely understood.
As a whole, NPCs are extremely long lived in non-dividing cells and show age-dependent deterioration in postmitotic neurons. However, individual nucleoporin constituents of the NPC have variably long half-lives (D’Angelo et al., 2009; Hetzer, 2010; Savas et al., 2012; Toyama et al., 2013). Recently, nucleoporins in yeast were initially shown to form sub-complexes that assemble in a hierarchical way from the center to the periphery, with individual nucleoporins maturing at different rates (minutes to ~1 h) (Onischenko et al., 2020). However, very little is known about NPC maturation and turnover in neurons.
In yeast, defective NPC assemblies are degraded by the ESCRT-III/Vps4 complex (Webster et al., 2014). The endosomal-sorting complexes required for transport (ESCRT) pathway are composed of five core complexes: ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III, and the AAA-ATPase Vps4, which mediate cellular membrane budding and fission (McCullough et al., 2013). ESCRT-III also plays a key role in HIV budding, plasma membrane wound repair, cytokinesis, centrosome duplication, and exosome secretion (Baietti et al., 2012; Carlton and Martin-Serrano, 2007; Garrus et al., 2001; Jimenez et al., 2014; Morita et al., 2007, 2010; Votteler and Sundquist, 2013). Polymerization of ESCRT-III subunits at membranes is driven by the Vps4 ATPase, which induces membrane deformation and fission (Pfitzner et al., 2020).
We and others previously reported that the C9orf72 hexanucleotide repeat expansion (HRE) disrupts NCT in multiple models of C9-ALS, including mouse and fly models, induced pluripotent stem cell (iPSC)-derived neurons, and postmortem brain (Zhang et al., 2015; Freibaum et al., 2015; Jovičić et al., 2015). A GGGGCC HRE in the first intron of C9orf72 is the most common genetic cause of ALS and FTD (DeJesus-Hernandez et al., 2011; Renton et al., 2011). A common pathologic hallmark of both ALS and FTD is the mislocalization of TAR-DNA binding protein 43 (TDP-43), which is normally nuclear but mislocalizes to the cytoplasm and forms aggregates in neurons of ALS and FTD patients. Healthy individuals typically carry two to five GGGGCC repeats in the first intron of the C9orf72 gene, whereas ALS/FTD patients may carry several hundred to thousands of repeats (Balendra and Isaacs, 2018). The HRE impairs C9orf72 transcription, and haploinsufficiency has been suggested to contribute to ALS/FTD pathogenesis (van Blitterswijk et al., 2015; Shi et al., 2018; Zhu et al., 2020). However, the preponderance of evidence suggests that ALS/FTD is caused by a gain-of-function mechanism downstream of bidirectional HRE transcription (Donnelly et al., 2013; Lee et al., 2013; Zu et al., 2011). Sense (GGGGCC) and antisense (GGCCCC) RNA species accumulate predominantly in nuclear RNA foci in patient cells and tissues (Cooper-Knock et al., 2015; Donnelly et al., 2013; Gomez-Deza et al., 2015; Mackenzie et al., 2015; Zu et al., 2013). Furthermore, the C9orf72 HRE-RNA is translated by a non-canonical AUG-independent mechanism to produce five different dipeptide repeat proteins (DPRs; poly-GP, poly-GA, poly-GR, poly-PA, and poly-PR) (Ash et al., 2013; Mori et al., 2013; Zu et al., 2011). These DPRs form predominantly cytoplasmic inclusions in neurons from C9-ALS/FTD postmortem brains (Ash et al., 2013; Mori et al., 2013; Balendra and Isaacs, 2018). Both the HRE-RNA and DPRs have been suggested to cause toxicity through a variety of mechanisms; however, the primary cause of toxicity remains unclear.
Arginine-rich DPRs may bind and sequester low-complexity domain-containing proteins through a liquid-liquid phase separation mechanism into membrane-less organelles, including RNA granules, nucleoli, and the NPC (Lee et al., 2016; Lin et al., 2016). Additionally, a series of genetic, imaging, and biochemical studies in C9-ALS/FTD models identified different components of the nuclear transport machinery including nucleoporins and nuclear transport receptors as strong modifiers of GGGGCC repeat toxicity in both fly and iPS neuron models of C9-ALS (Boeynaems et al., 2016; Coyne et al., 2020; Cunningham et al., 2020; Freibaum et al., 2015; Kramer et al., 2018; Lee et al., 2016; Zhang et al., 2015). Both RanGAP1, a master regulator of NCT, and nucleoporins have been found to be mislocalized in fly and mouse models of C9-ALS as well as in postmortem human tissue (Freibaum et al., 2015; Chew et al., 2019; Zhang et al., 2015, 2018). A recent study in iPS neurons suggests that the GGGGCC RNA expansion disrupts NPC composition via nuclear accumulation of the ESCRT-III protein CHMP7 (Coyne and Rothstein, 2021). A subsequent study suggests that CHMP7-mediated recruitment of VPS4 may contribute to NPC injury in ALS iPS neurons (Coyne and Rothstein, 2021; Coyne et al., 2021), though the molecular mechanisms inducing nucleoporin degradation in neurons remain incompletely understood.
In this study, we found that a subset of nucleoporins is reduced in neurons expressing expanded GGGGCC repeats in the adult Drosophila brain. We used super-resolution expansion microscopy (ExM) to find that the NPC is disrupted and nucleoporins accumulate in the cytoplasm of neurons in fly C9-ALS/FTD models. A nuclear ESCRT-III/Vps4 complex mechanistically contributes to the reduction of select nucleoporins through the proteasomal pathway. Importantly, downregulation of ESCRT-III/Vps4 complex components prevents nucleoporin loss, rescues NCT, and suppresses neurodegeneration in flies expressing GGGGCC repeats.
RESULTS
Select nucleoporins are reduced in GGGGCC-repeat-expressing neurons of the adult fly brain
Since we previously showed that expression of 30 GGGGCC repeats (30R) in flies inhibits NCT (Zhang et al., 2015), we asked whether 30R expression causes loss of nucleoporin proteins in vivo. We expressed 30R in adult fly neurons using ELAV-Gene Switch (E-GS-GAL4, an RU486-inducible GAL4) (Osterwalder et al., 2001), and we examined nucleoporin expression in the brain after feeding newly eclosed adult flies RU486. The nucleoporin Nup214 normally localizes to the cytoplasmic ring and filaments of NPCs, whereas we observe a marked reduction of Nup214 in 30R-expressing neurons (labeled with anti-Elav) in 15-day-old fly brains, but not in adjacent glia (labeled with ‘‘*’’, Figures 1A and 1B). Similarly, Nup98 normally localizes to the central channel of NPCs but is also significantly downregulated in 30R-expressing neurons of the adult fly brain (Figures 1C and 1D). Since ~90% of cells of the adult fly brain are neurons, we confirmed downregulation of Nup214 and Nup98 in 30R-expressing neurons by performing western blot analysis of fly brains (Figures 1E and 1F). Similarly, we also performed western blot analysis of additional nucleoporins including Megator, the Drosophila homolog of Tpr (Lince-Faria et al., 2009; Vaquerizas et al., 2010), Nup50, a nuclear basket nucleoporin, Nup93, a scaffolding nucleoporin, and LaminB, a nuclear envelope protein. While Nup214, Nup98, Megator, and Nup50 are markedly downregulated in 30R brains, Nup93 and LaminB are not significantly affected (Figures 1E and 1F). We next examined nucleoporins in other fly models of C9-ALS/FTD that express GGGGCC repeats and found that overexpression of 36 repeats (36R) or 44 repeats (44R) also significantly reduces Megator levels (Figures S1A and S1B). In experiments examining the kinetics of nucleoporin downregulation, we observe significant downregulation of Nup98 and Nup50 by 10 days of RU486 feeding (Figures S1C–S1E). Next, we measured mRNA levels of these nucleoporins using qRT-PCR and found that transcript levels are not changed in 30R-expressing adult brains of 10- and 15-day-old flies (Figures S1F and S1G). These data demonstrate that in response to 30R expression, both cytoplasmic- and nuclear-facing nucleoporin proteins are downregulated, whereas the scaffolding nucleoporin Nup93 and nuclear envelope protein Lamin are expressed at normal levels, similar to what has been observed in iPS neurons and postmortem tissue from C9-ALS patients (Coyne et al., 2020).
Figure 1. Specific nucleoporins are reduced in G4C2-repeat-expressing adult brain.
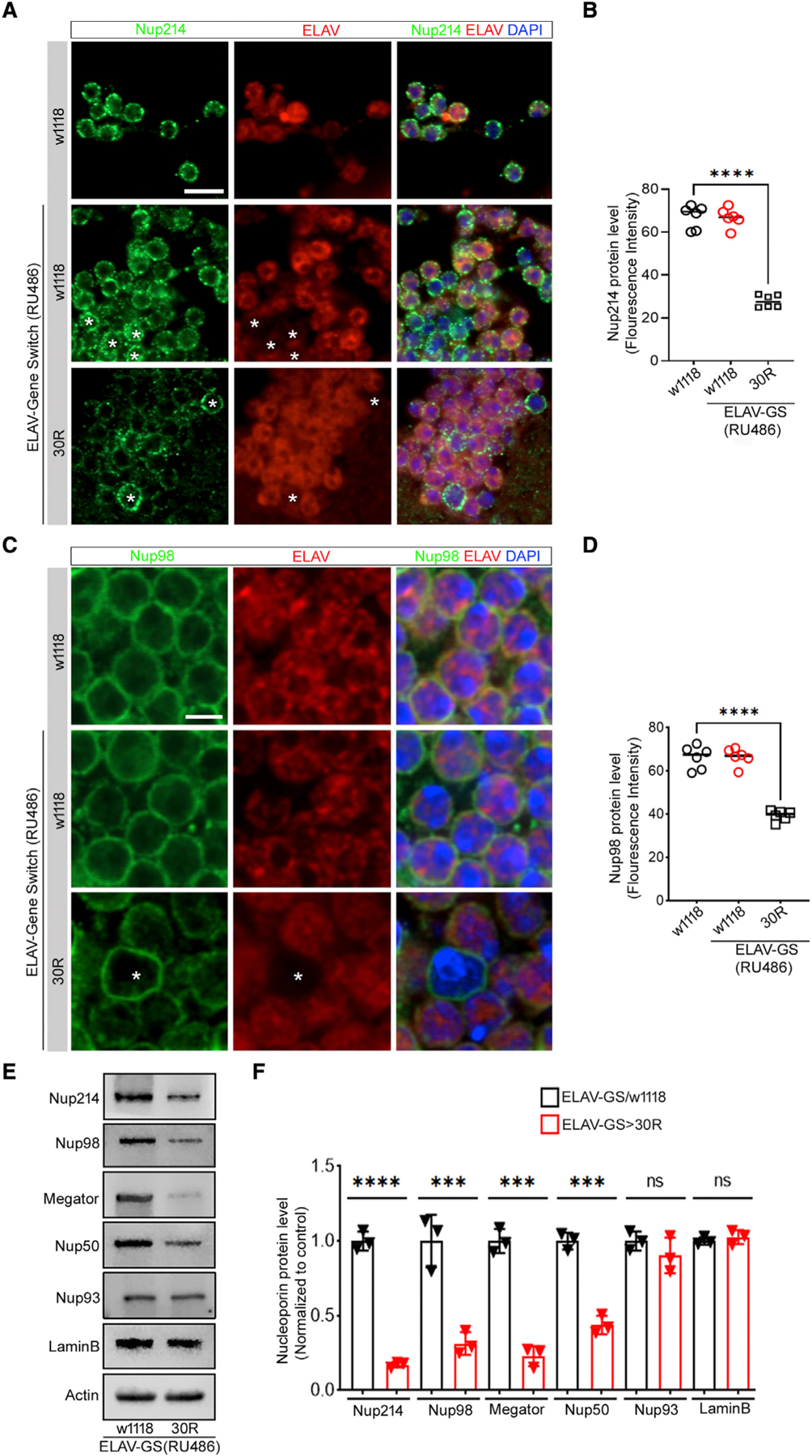
(A) 15-day-old Drosophila adult brain expressing (G4C2)30 repeats (30R) under the control of ELAV-Gene Switch GAL4 (ELAV-GS) induced with 100 mM RU486, stained with Nup214 (green), neuronal marker ELAV (red), and DAPI (blue), (scale bar represents 5 mm, n = 15 brains per genotype). ‘‘*’’ represents non-neuronal cells.
(B) Quantification of neuronal Nup214 immunofluorescence is reported as mean ± SEM (one-way ANOVA, followed by Tukey’s test, ****p < 0.0001, n > 60 cells per genotype).
(C and D) Nup98 (green), ELAV (red), and DAPI (scale bar represents 2 mm, n = 15 brains per genotype). ‘‘*’’ represents non-neuronal cells. Quantification of neuronal Nup98 fluorescence intensity, data are reported as mean ± SEM (one-way ANOVA, followed by Tukey’s test, ****p < 0.0001, n > 60 cells per genotype).
(E) Western blot of adult brains of ELAV-GS/+ and ELAV-GS>30R flies fed RU486.
(F) Quantification of western blot in (E) normalized with actin. One-way ANOVA, followed by Sidak’s multiple comparisons (n = 3, ***p < 0.001, ****p < 0.0001, ns = not significant). Error bars indicate ± SEM.
Since disruption of NCT and NPCs has been observed in other neurodegenerative disease models, including FUS-mediated ALS and Huntington’s disease (Grima et al., 2017; Gasseet-Rosa et al., 2017; Lin et al., 2021), we next examined nucleoporin levels in other fly models of neurodegenerative diseases. Interestingly, the level of these nucleoporins (Nup214, Nup98, Megator, and Nup50) is not affected in fly models of FUS-ALS (R518K or R521C) or Huntington’s disease (Htt128Q) (Figures S1H–S1L), suggesting that loss of this subset of nucleoporins may be specific for C9-ALS/FTD. Furthermore, expression of 30R specifically in glial cells using Repo-Gene Switch does not alter Nup98 levels in 15-day-old adult flies, suggesting that 30R-mediated nucleoporin downregulation is neuron-specific (Figures S2A and S2B), consistent with prior reports in iPS models and postmortem patient tissues (Coyne et al., 2020). Together, these data suggest that select nucleoporin proteins are specifically lost in neurons in fly models of C9-ALS.
Inhibition of Rpn10 proteasomal subunit rescues nucleoporin levels and suppresses toxicity in GGGGCC-repeat-expressing flies
Both the proteasome and autophagy have been implicated in quality control of nucleoporins and the NPC in yeast (Lee et al., 2020; Tomioka et al., 2020; Webster et al., 2014). Autophagic degradation of NPCs has been suggested to occur via direct binding of Atg8 (yeast LC3) with Nup159 (yeast ortholog of Nup214) in the cytoplasm (Lee et al., 2020). Therefore, we first tested whether macroautophagy is required for 30R-mediated nucleoporin degradation by RNAi-mediated knockdown of Atg8, known to be critical for selective autophagy in Drosophila (Mauvezin et al., 2014). However, Atg8 knockdown does not affect 30R-mediated loss of Nup214 immunofluorescence in the adult fly brain (Figures S3A and S3B). Similarly, the marked loss of Nup214 and Nup98 caused by 30R expression detected with western blot analysis is unaffected by Atg8 RNAi (Figures S3C–S3E). p62 is increased and forms ubiquitinated aggregates in fly models of C9-ALS due to autophagolysosomal dysfunction in neurons (Cunningham et al., 2020), and it is even further increased with Atg8-RNAi, confirming that autophagy is inhibited in these experiments (Figures S3C and S3F). These data suggest that the downregulation of select nucleoporins in fly models of C9-ALS does not occur via Atg8-mediated autophagy.
In yeast, many nucleoporins have been shown to be degraded via the proteasome (Webster et al., 2014; Lee et al., 2020). To determine whether loss of nucleoporins induced by 30R expression occurs via proteasomal degradation, we first inhibited the proteasome by feeding flies the proteasome inhibitor epoxomicin. As shown in Figure 2, feeding flies epoxomicin at the same time as RU468 to induce 30R expression fully rescues Nup98 levels in the adult brain (Figures 2A–2D) in a dose-dependent manner (Figures S3G and S3H).
Figure 2. Inhibition of proteasome restores nucleoporins in 30R-expressing adult brain.
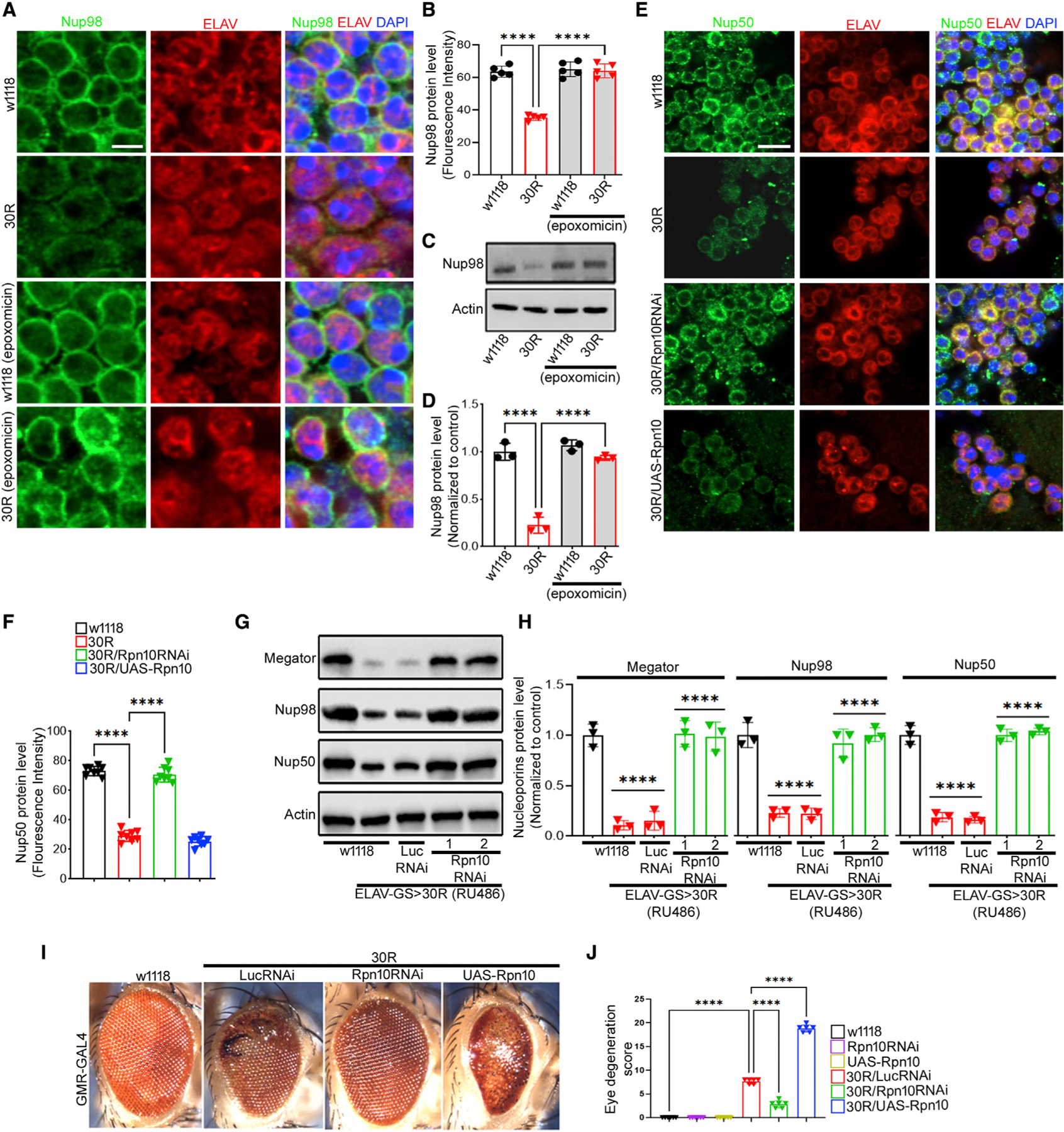
(A) Adult brain from 15-day-old fly labeled with indicated antibodies and genotypes treated with proteasome inhibitor epoxomicin or control (scale bar represents 2 mm, n = 10 brains per genotype).
(B) Quantification of neuronal Nup98 fluorescence intensity in (A). Data are reported as mean ± SEM (one-way ANOVA, followed by Tukey’s test, ****p < 0.0001, n > 50 cells per genotype).
(C) Western blot of adult brain from flies fed epoxomicin or control.
(D) Quantification of Nup98 from (C) normalized with actin. One-way ANOVA, p < 0.0001, followed by Sidak’s multiple comparisons (n = 3, ***p < 0.001, ****p < 0.0001). Error bars indicate ± SEM.
(E) Immunofluorescence staining of 15-day-old adult brains with Nup50 (green), ELAV (red), and DAPI (scale bar represents 5 mm, n = 10 brains per genotype).
(F) Quantification of (E). Data are reported as mean ± SEM (one-way ANOVA, followed by Tukey’s test, ****p < 0.0001, n > 50 cells per genotype).
(G) Western blot of 15-day-old adult brains.
(H) Quantification of blots in (G). One-way ANOVA, p < 0.0001, followed by Sidak’s multiple comparisons (n = 3, ***p < 0.001, ****p < 0.0001). Error bars indicate ± SEM.
(I)15-day-old fly eyes expressing 30R using GMR-GAL4 alone or with indicated transgenes.
(J) Quantification of eye degeneration. Data are reported as mean ± SEM. Kruskal-Wallis test, p < 0.0001, followed by Dunn’s multiple comparisons, n > 15 adults.
Next, since the proteasomal ubiquitin receptor Rpn10 has been implicated in nucleoporin degradation in yeast (Lee et al., 2020), we tested whether fly Rpn10 is required for 30R-mediated nucleoporin degradation in neurons. Rpn10 RNAi expression reduced Rpn10 protein levels by ~50%, and overexpression increased protein level ~1.5-fold compared with control (Figures S3I and S3J). Indeed, RNAi-mediated knockdown of Rpn10 rescues the loss of nucleoporins Megator, Nup98, and Nup50 in 30R-expressing fly brains (Figures 2E–2H), whereas overexpression of Rpn10 does not significantly enhance the effects of 30R expression (Figures 2E and 2F). Importantly, nucleoporin protein levels are not altered with overexpression or knockdown of Rpn10 in wild-type flies, suggesting that manipulation of Rpn10 levels alone is not sufficient to regulate nucleoporin levels (Figures S4A–S4E). Interestingly, cytoplasmic puncta of Nup98 colocalize with the proteasome regulatory subunit Rpt5 in neurons expressing 30R for 10 days in the adult fly brain (Figure S4F), suggesting that nucleoporins may become localized to the cytoplasm for proteasomal degradation.
Given the striking rescue of nucleoporin expression with proteasome inhibition, we next asked whether Rpn10 knockdown also rescues 30R-mediated eye degeneration. While expression of 30R in the eye using GMR-Gal4 induces photoreceptor degeneration, ommatidial disruption, necrotic patches, and an irregular bristle lattice by day 15 (Figure 2I) (Xu et al., 2013; Zhang et al., 2015), knockdown of Rpn10 in 30R-expressing flies suppresses these phenotypes, whereas overexpression of Rpn10 in 30R-expressing flies strongly enhances them (Figures 2I and 2J). Knockdown or overexpression of Rpn10 itself does not show an eye degeneration phenotype (Figures 2J and S4G). These data demonstrate that 30R-mediated neurodegeneration requires Rpn10 function, perhaps at least in part via proteasome-mediated degradation of nucleoporins.
Vps4 is necessary and sufficient for nucleoporin downregulation and neurodegeneration
Since the ESCRT-III/Vps4 complex has been implicated in nucleoporin degradation upstream of Rpn10 in yeast (Webster et al., 2014; Lee et al., 2020), we next examined the role of the ATPase Vps4 in nucleoporin degradation in 30R-expressing neurons. Vps4 transgenic RNAi lines reduced Vps4 protein expression by ~75%, and overexpression increased protein level ~2-fold compared with control (Figures S5A and S5B). Interestingly, immunofluorescence analysis of adult brains reveals that knockdown of Vps4 using three independent RNAi lines fully rescues levels of Nup214, Nup50, Nup98, and Megator in 30R-expressing neurons (Figures 3A, 3B, and S6A–S6F), similar to what is observed with proteasome inhibition. Similarly, western blot analysis of these same nucleoporins confirms that levels are rescued to control levels in 30R-expressing fly brains (Figures 3C–3E). In contrast, overexpression of Vps4 in 30R-expressing animals does not further lower nucleoporin levels. Since these data reveal that Vps4 is required for 30R-mediated nucleoporin loss, we next asked whether it is sufficient. Indeed, overexpression of Vps4 in wild-type flies significantly reduces Nup98 and Nup50 protein levels by western blot (Figures 3F and 3G). Together, these data demonstrate that Vps4 is both necessary and sufficient for nucleoporin degradation in fly neurons expressing 30R.
Figure 3. Depletion of Vps4 restores nucleoporins in 30R-expressing adult brain.
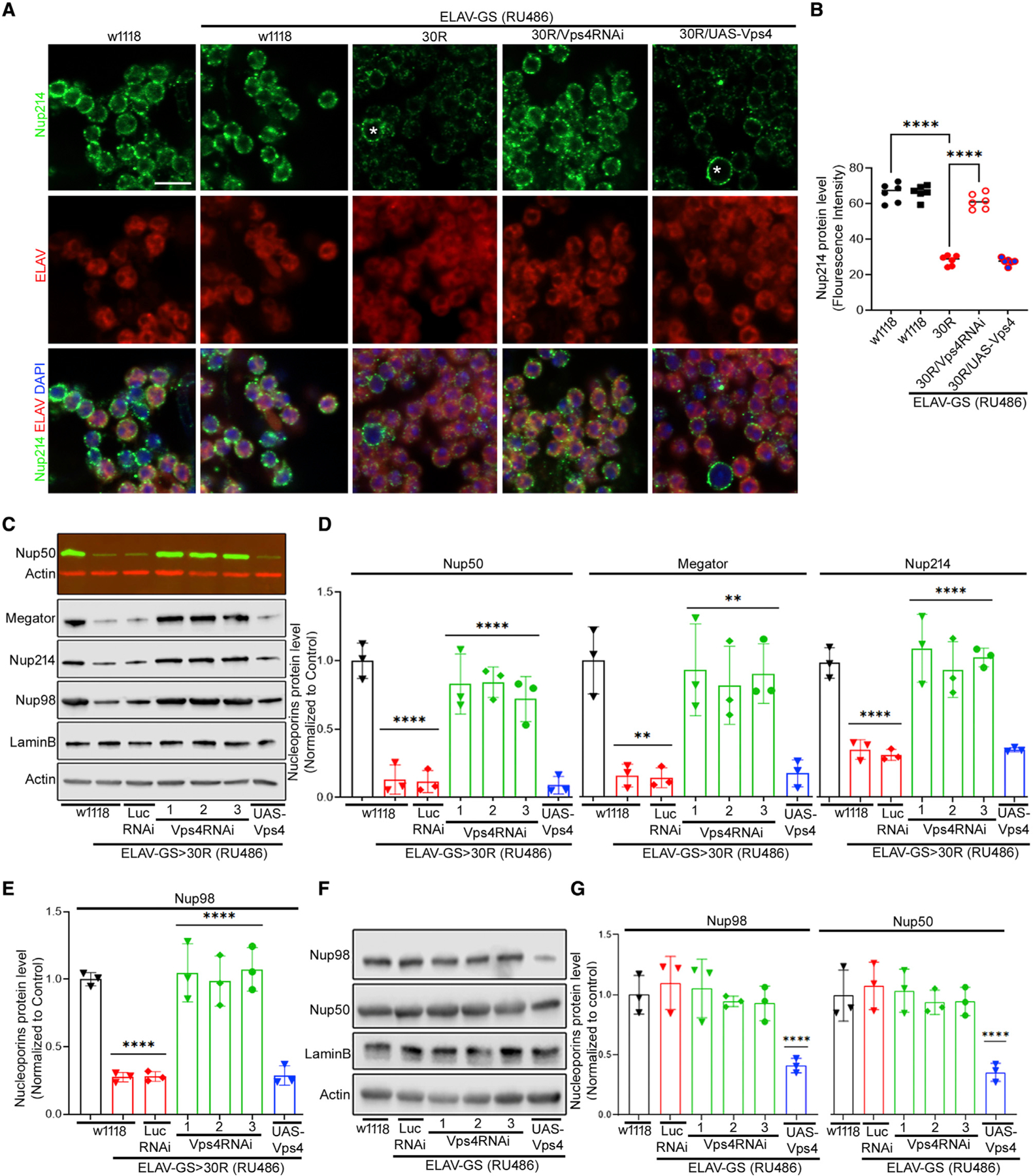
(A) 15-day-old fly brains labeled with Nup214 (green), ELAV (red), and DAPI (blue) (scale bar represents 5 mm, n = 10 brains per genotype).
(B) Quantification of (A). Data are reported as mean ± SEM (one-way ANOVA, followed by Tukey’s test, ****p < 0.0001, n > 60 cells per genotype).
(C) Western blot of 15-day-old brains. All genotypes were raised on RU486, except w1118.
(D and E) Quantification of (C). One-way ANOVA, p < 0.0001, followed by Sidak’s multiple comparisons (n = 3, **p < 0.01, ***p < 0.001, ****p < 0.0001). Error bars indicate ± SEM.
(F) Western blot of adult brains from indicated genotypes.
(G) Quantification of (F). One-way ANOVA, p < 0.0001, followed by Sidak’s multiple comparisons (n = 3, ***p < 0.001, ****p < 0.0001). Error bars indicate ±SEM.
Given the critical role of Vps4 in regulation of nucleoporin levels in fly neurons, we next determined the effect of Vps4 levels on 30R-mediated neurodegeneration. Interestingly, overexpression of Vps4 in the fly eye disrupts ommatidial organization and induces necrotic patches, similar to what is seen with 30R expression alone, whereas knockdown of Vps4 does not cause an obvious eye phenotype (Figures 4A and 4B). Furthermore, in fly eyes expressing 30R, overexpression of Vps4 enhances the rough eye phenotype, whereas knockdown of Vps4 strongly suppresses it. Since the C9orf72 HRE predominantly affects motor neurons in ALS patients, we next determined the effect of Vps4 on 30R toxicity in fly motor neurons. Overexpression of 30R in motor neurons using OK371-Gal4 causes paralysis and lethality during pupal development (Figure 4C) (Zhang et al., 2015; Cunningham et al., 2020). Importantly, this phenotype is enhanced with Vps4 overexpression and suppressed with Vps4 knockdown (Figure 4C). Since Vps4 overexpression does not significantly reduce Nup levels further in 30R expressing brains (Figure 3), these data suggest that Vps4 overexpression has additional toxic effects in addition to reducing Nup levels. Together, these data indicate that Vps4 is necessary and sufficient for neurodegeneration downstream of 30R expression.
Figure 4. Downregulation of Vps4 suppresses G4C2 toxicity.
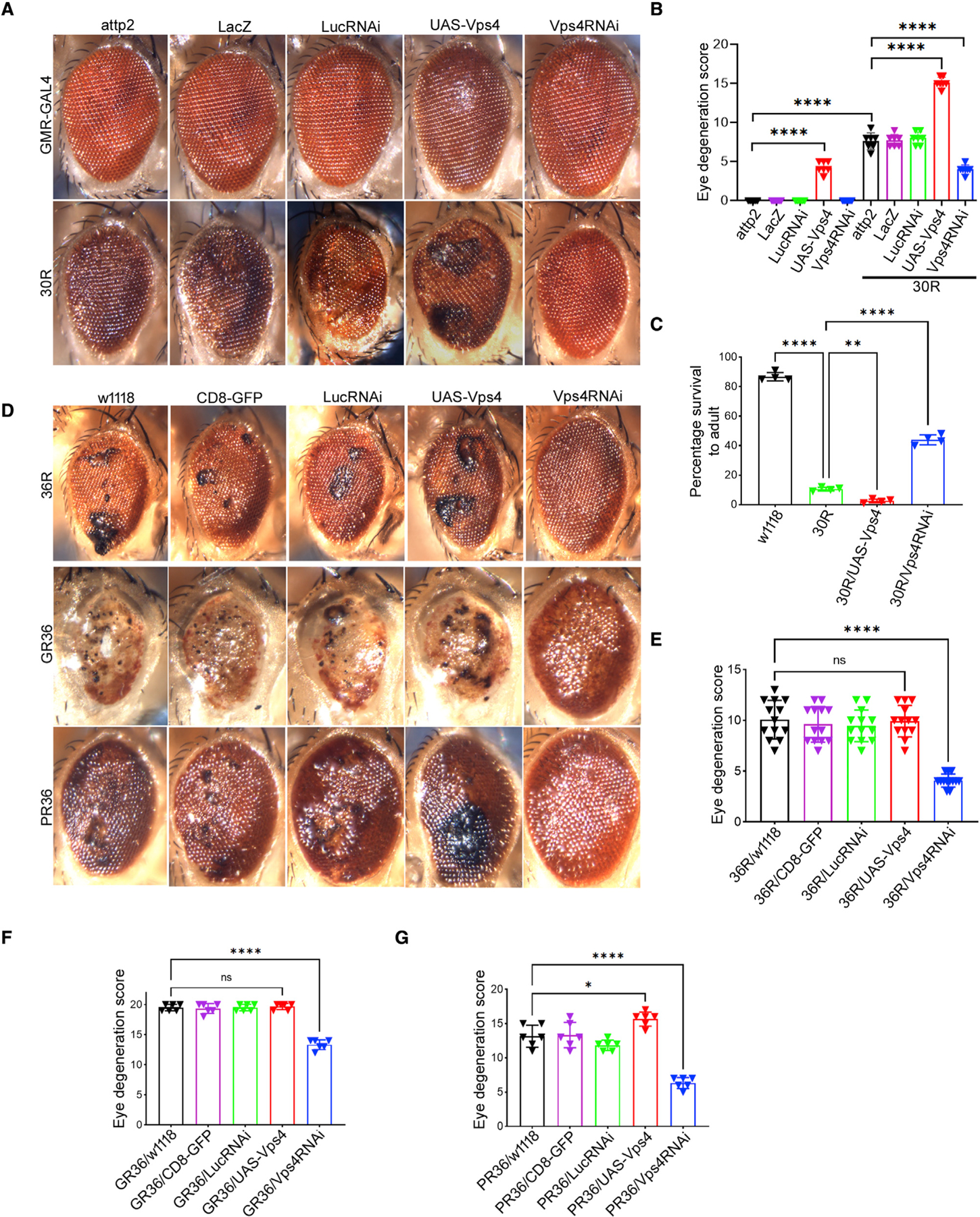
(A) External eye of 15-day-old flies.
(B) Quantification of eye degeneration in (A). Data are presented as mean ± SEM. One-way ANOVA, Sidak’s multiple comparisons (n ≥ 25 adults, ****p < 0.0001).
(C) Percent of pupal eclosion of flies expressing UAS-30R under the control of vGlut-GAL4 with UAS-Vps4 or UAS-Vps4RNAi. Sidak’s multiple comparisons (n ≥ 100 pupa, **p < 0.01, ****p < 0.0001).
(D) External eye of 15-day-old flies expressing 36R, poly(GR)36, or poly(PR)36 under control of GMR-GAL4 in control (w1118, CD8-GFP, LucRNAi), Vps4RNAi, or Vps4 overexpression background.
(E–G) Quantification of eye degeneration in (D). Data are presented as mean ± SEM, one-way ANOVA, Sidak’s multiple comparisons, (n ≥ 25 adults, *p < 0.05, ****p < 0.0001, ns = not signifiicant).
Increasing evidence from multiple model systems suggests that both the HRE and arginine-containing DPRs generated via repeat-associated non-AUG (RAN) translation may contribute to neurodegeneration (Mackenzie et al., 2015; Jovičić et al., 2015; Lee et al., 2016). Thus, we next tested whether Vps4 is required for neurodegeneration in additional fly models of C9-ALS/FTD. Importantly, Vps4 knockdown suppresses eye degeneration in flies expressing 36 GGGGCC repeats (36R) (Figures 4D and 4E), demonstrating that this effect is also seen in an independent fly model of GGGGCC repeat toxicity. Furthermore, Vps4 knockdown also rescues the rough eye phenotype induced by poly-GR (GR36) and poly-PR (PR36) expression, and Vps4 overexpression slightly enhances the effect of PR36 expression (Figures 4D, 4F, and 4G). These data indicate that Vps4 is necessary for neurodegeneration mediated by both G4C2 repeats and by arginine-containing DPR proteins in C9-ALS/FTD.
Downregulation of ESCRT-III rescues nucleoporins and suppresses GGGGCC repeat toxicity
Vps4 primarily functions with the ESCRT-III complex to mediate its membrane trafficking functions in yeast (McCullough et al., 2013; Votteler and Sundquist, 2013; Pfitzner et al., 2020). Therefore, we next investigated whether knockdown of ESCRT-III genes would rescue nucleoporin levels to a similar extent as Vps4. Humans express 12 ESCRT-III proteins, grouped into eight different families called charged multivesicular body proteins (CHMP1-8) (Christ et al., 2017; Schöneberg et al., 2017). In Drosophila, there are eight known genes in the ESCRT-III complex, including CHMP1, Vps2 (Drosophila homolog of CHMP2A), CHMP2B, Vps24 (Drosophila homolog of CHMP3), Shrub (Drosophila homolog of snf7/CHMP4), Vps60 (Drosophila homolog of CHMP5), Vps20 (Drosophila homolog of CHMP6), and CG5498 (Drosophila homolog of CHMP7) (Bertin et al., 2020; McCullough et al., 2013; Teis et al., 2008). Efficacy of transgenic RNAi lines targeting CHMP1, CHMP2B, Vps20, and Shrub was confirmed to reduce protein expression by ~50% (Figures S5C–S5F). Since antibodies are not available for Vps2, Vps24, and CG5498, efficacy of RNAi-mediated knockdown was confirmed by qRT-PCR (Figures S5G–S5I). We tested the effects of RNAi-mediated knockdown of each of these genes on nucleoporin protein expression and found that downregulation of CHMP1, CHMP2B, Vps24, CG5498, and Shrub significantly rescued nucleoporin levels in 30R-expressing neurons, whereas knockdown of Vps2 and Vps20 did not show a significant effect (Figures 5A–5E). Of note, Vps2 (CHMP2A) may be partially compensated for by CHMP2B, and Vps20 acts in ESCRT-III nucleation but is not required for ESCRT-III filament assembly or for NPC surveillance in yeast (Webster et al., 2014). These data suggest that specific ESCRT-III family members including CHMP1, CHMP2B, Vps24, CG5498, and Shrub are required to degrade nucleoporins in 30R-expressing neurons.
Figure 5. Downregulation of ESCRT-III components restores nucleoporins and suppresses toxicity in 30R flies.
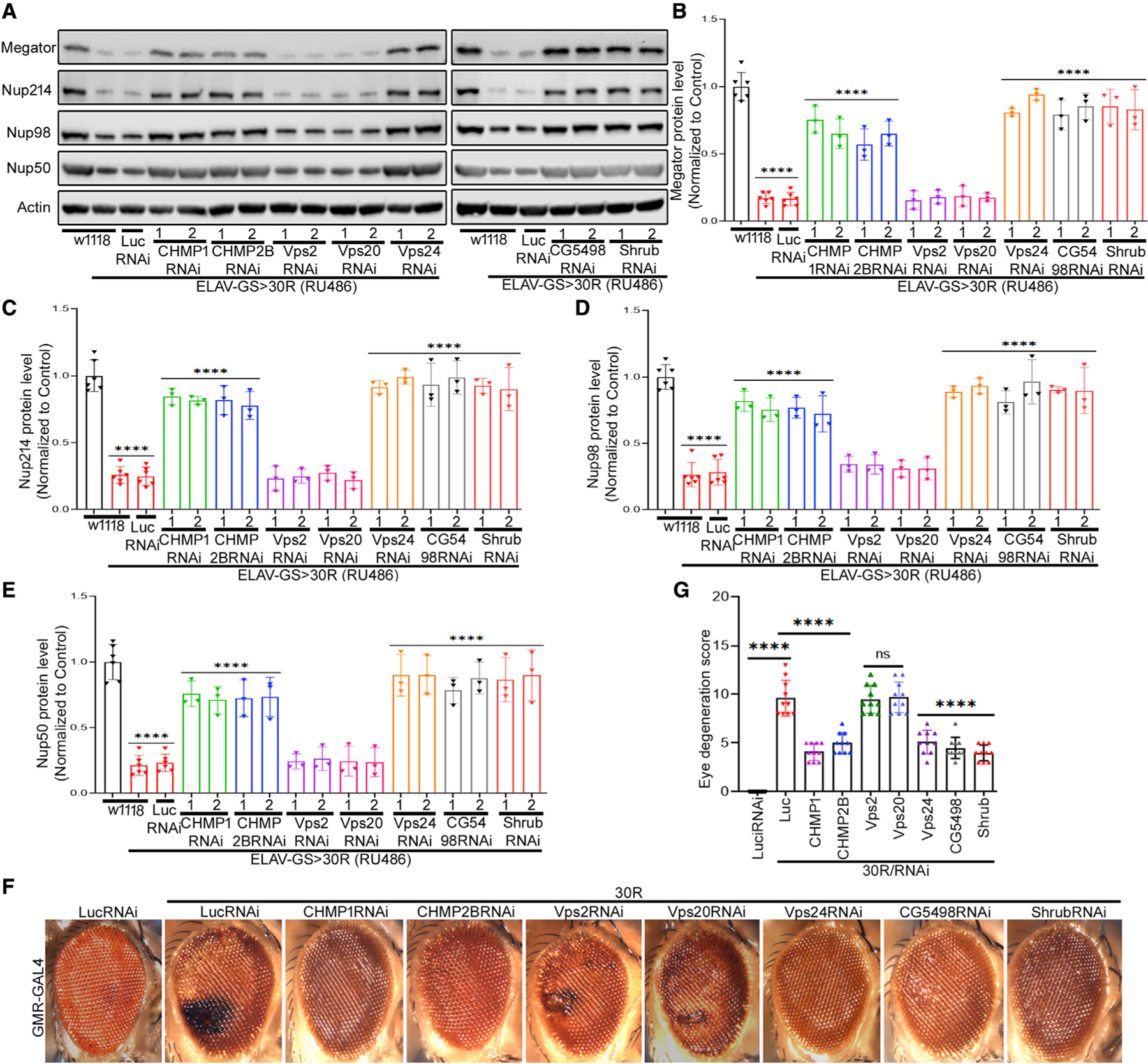
(A) Western blot of nucleoporins of adult brains from 15-day-old flies raised on RU486-containing food.
(B–E) Quantification of (A). One-way ANOVA, followed by Dunnett’s multiple comparisons test (n = 3, ****p < 0.0001). Error bars indicate ± SEM.
(F) External eye of 15-day-old flies of indicated genotype.
(G) Quantification of eye degeneration from (F). Data are presented as mean ± SEM. One-way ANOVA, followed by Tukey’s test, (n ≥ 25 adults, ****p < 0.0001, ns = not significant).
Given that ESCRT-III downregulation rescues nucleoporin levels in 30R-expressing brains, we next tested whether ESCRT-III downregulation also suppresses 30R-mediated neurodegeneration. Interestingly, knockdown of ESCRT-III genes that rescued nucleoporin protein expression, including CHMP1, CHMP2B, Vps24, CG5498, and Shrub, suppressed 30R-mediated eye degeneration, whereas knockdown of Vps2 and Vps20 had no effect (Figures 5F and 5G). These data indicate that 30R-mediated nucleoporin degradation and neurodegeneration require core components of the ESCRT-III complex.
ESCRT-III/Vps4 expression is increased in GGGGCC-repeat-expressing neurons
Given the striking requirement for Vps4 and the ESCRT-III complex in 30R-mediated downregulation of nucleoporins, we next examined the localization and expression of the Vps4/ESCRT-III complex in 30R-expressing neurons. Although this complex is best characterized to function in the cytoplasm to mediate membrane scission, Vps4/ESCRT-III also localizes to the inner nuclear membrane to allow viral budding and nuclear envelope sealing after mitosis (Arii et al., 2018; Olmos and Carlton, 2016; Stoten and Carlton, 2018; Vietri et al., 2020). To determine if Vps4 localization is altered in 30R-expressing neurons, we first examined Vps4 protein expression in 30R-expressing neurons using immunofluorescence microscopy of the adult brain. Interestingly, Vps4 protein expression is increased ~2-fold in 30R-expressing neurons, where it appears to be enriched in the nucleus (Figures 6A and 6B). Similarly, the expression of Shrub, the fly ortholog of Vps32/Snf7/CHMP4, a filament-forming subunit of the ESCRT-III complex, is also increased ~2-fold in 30R-expressing neurons (Figures 6C and 6D). This increase in protein expression is confirmed by western blot that also shows significant upregulation of both Vps4 and Shrub protein in 30R-expressing adult brains compared with wild-type (Figures 6E and 6F). The increased Vps4 protein can be attributed to an ~2-fold increase in transcript expression in 30R-expressing larva, as measured by qRT-PCR (Figure 6G). Together, these data suggest that 30R expression leads to increased Vps4 mRNA and protein levels, which then induce degradation of select nucleoporins.
Figure 6. Increased nuclear Vps4/ESCRT-III complex in G4C2-repeat-expressing adult brain.
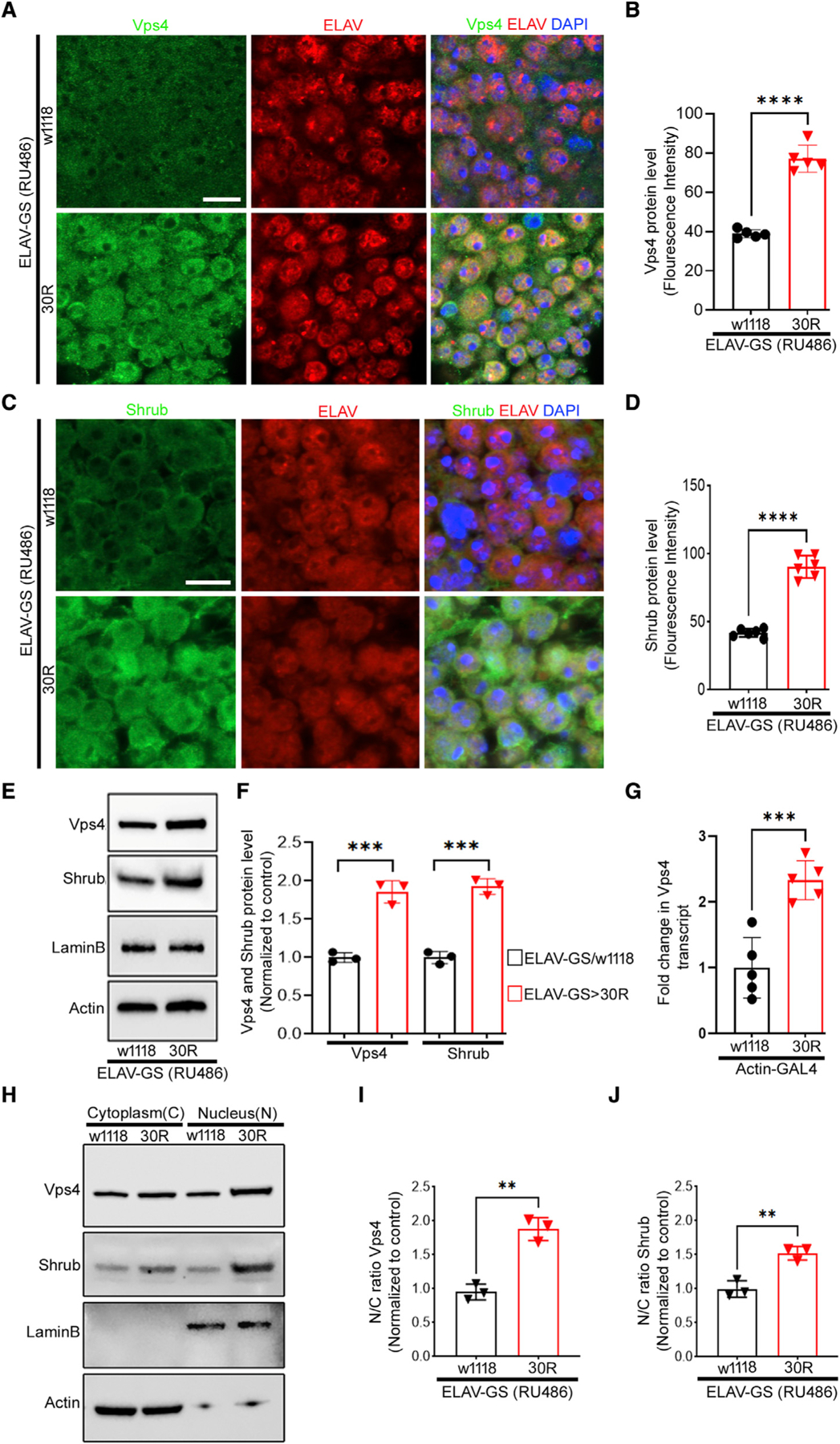
(A) Immunofluorescence imaging of 15-day-old adult fly brains (scale bar represents 5 mm, n = 10 brains per genotype).
(B) Quantification of (A), data are reported as mean ± SEM (unpaired t test, ****p < 0.0001, n > 60 cells per genotype).
(C) Immunostaining of 15-day-old adult brains (scale bar represents 5 mm, n = 10 brains per genotype).
(D) Quantification of (C), data are reported as mean ± SEM (unpaired t test, ****p < 0.0001, n > 60 cells per genotype).
(E) Western blot of 15-day-old adult brains.
(F) Quantification of (E). Data are presented as mean ± SEM. Unpaired t test (n = 3, ***p < 0.001).
(G) qRT-PCR of Vps4 transcript in larvae. Data are presented as mean ± SEM. Unpaired t test (n = 5, ***p < 0.001).
(H) Western blot of cytoplasmic and nuclear fractions from 15-day-old fly brains.
(I and J) Quantification of nucleocytoplasmic ratio from (H). Actin was used to normalize cytoplasmic fraction and LaminB for nuclear fraction. (n = 3, unpaired t test, **p < 0.01).
Whereas Vps4 and Shrub are localized in both the cytoplasm and nucleus in control neurons, immunofluorescence analysis suggests that Vps4 and Shrub localization are increased in the nucleus of 30R-expressing neurons. To confirm the enrichment of Vps4 and Shrub in the nucleus, we performed nuclear/cytoplasmic fractionation in control and 30R-expressing adult brains and probed fractions for both proteins that confirm that both Vps4 and Shrub are markedly enriched in the nucleus compared with the cytoplasm (Figures 6H–6J). These data suggest that GGGGCC repeats induce nucleoporin degradation by increasing nuclear ESCRT-III/Vps4 localization in fly brains.
Downregulation of Vps4 restores nuclear nucleoporins and NCT in GGGGCC-repeat-expressing neurons
To further investigate the mechanism of nucleoporin degradation caused by 30R expression, we examined nucleoporin localization after 10 days of 30R expression, before it is fully degraded (Figures S1C–S1E). To determine the subcellular localization of nucleoporins, we performed super-resolution expansion microscopy using probes for both the nuclear envelope marker LaminB and for the nucleoporin Nup98. As shown in Figure 7A, Nup98 is at the nuclear envelope in control neurons. However, in 30R-expressing neurons, there are numerous cytoplasmic puncta of Nup98, suggesting that cytoplasmic mislocalization of nucleoporins precedes their degradation (Figures 7A and 7B). Double labeling of the cytoplasmic nucleoporin Nup214 and the nuclear nucleoporin Nup98 suggests that they remain closely opposed in the cytoplasm, suggesting that they may remain in a complex (Figure S7A). Interestingly, we also observed Nup98 at the neuromuscular junction in larvae expressing 30R in motor neurons (Figures S7B and S7C), consistent with our finding that nucleoporins become mislocalized to the cytoplasm. These data suggest that prior to degradation, Nups become mislocalized to the cytoplasm in 30R-expressing neurons.
Figure 7. Knockdown of Vps4 restores nuclear nucleoporins and rescues NCT in G4C2-expressing motor neurons.
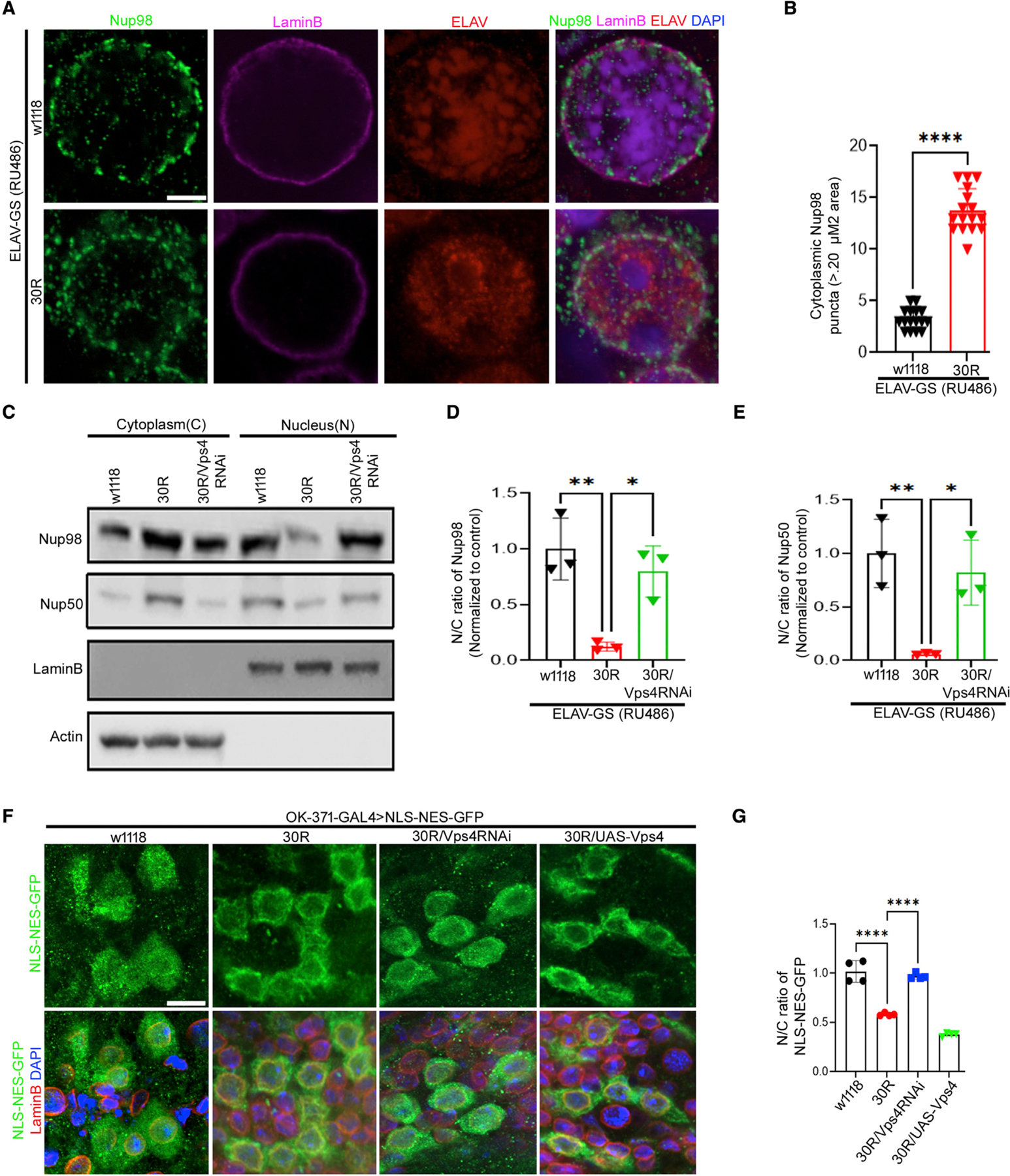
(A) Expansion microscopy (ExM) shows Nup98 (green), LaminB (magenta), ELAV (red), and DAPI (blue) of 10-day-old adult brains (scale bar represents 5 mm, n = 10 brain per genotype).
(B) Quantification of number of cytoplasmic Nup98 puncta (>0.20 μM area) in (A). Data are reported as mean ± SEM (unpaired t test, ****p < 0.0001, n > 30 cells per genotype).
(C) Western blot of cytoplasmic and nuclear fractions from 10-day-old adult brains raised on RU486-containing food.
(D and E) Quantification of nucleocytoplasmic ratio from (C). Actin was used to normalize cytoplasmic fraction and LaminB for nuclear fraction (n = 3, Tukey’s multiple comparisons test, *p < 0.05, **p < 0.01).
(F) Images showing localization of NLS-NES-GFP (green), LaminB (red), and DAPI in larval motor neurons (scale bar represents 10 mm, n = 10 brains per genotype).
(G) Quantification of nucleocytoplasmic (N/C) ratio of NLS-NES-GFP in motor neurons in F (n > 40 cells per genotype; Tukey’s multiple comparisons test, ****p < 0.0001).
To further test the hypothesis that increased nuclear ESCRT-III/Vps4 complex causes translocation of nucleoporins into the cytoplasm, we analyzed nucleoporins in nuclear and cytoplasmic fractions of fly brains after 10 days RU486 feeding. As predicted, nucleoporins Nup98 and Nup50 are strongly enriched in the nucleus of control adult brains, whereas they are strongly reduced in the nucleus and increased in the cytoplasm in 30R-expressing brains (Figures 7C–7E). Furthermore, knockdown of Vps4 in 30R-expressing adult brains rescues localization of nucleoporins in the nucleus to a level indistinguishable from the control (Figures 7C–7E). Next, Nup93 protein was probed in nuclear and cytoplasmic fractions to examine whether select nucleoporins are removed from the nuclear envelope and transported to cytoplasm since expression of Nup93 is not reduced in the 30R-expressing brain. Like Nup98 and Nup50, Nup93 is also increased in the cytoplasm and reduced in the nucleus in 30R-expressing brains, and Vps4 knockdown in 30R-expressing adult brains rescues this mislocalization (Figures S7D and S7E). These data suggest that ESCRT-III/Vps4 complex mediates the mislocalization of Nups from the nuclear envelope to the cytoplasm in 30R-expressing brains.
Impairment of NCT has been previously observed in both fly and iPS models of C9-ALS/FTD (Freibaum et al., 2015; Zhang et al., 2015). Here, we expressed an NCT reporter, a GFP protein tagged with an NLS and NES (NLS-NES-GFP) that shuttles between the nucleus and cytoplasm of neurons. In control larval motor neurons, NLS-NES-GFP localizes to both the nucleus and cytoplasm (Figures 7F and 7G), whereas in cells expressing 30R, it localizes primarily to the cytoplasm, leading to a significantly reduced nucleocytoplasmic ratio (N/C ratio) (Figures 7F and 7G), confirming our prior data employing this NCT reporter construct (Zhang et al., 2015). Vps4 knockdown in 30R-expressing motor neurons rescues this impairment in NCT, whereas overexpression of Vps4 enhances it (Figures 7F and 7G). To determine whether Vps4 is sufficient to alter NCT in motor neurons, we overexpressed or knocked down Vps4 in wild-type flies. While Vps4 knockdown does not significantly alter NCT, Vps4 overexpression significantly reduces the N/C ratio in motor neurons (Figures S7F and S7G). These data demonstrate that altered Vps4 levels can contribute to impaired nuclear import of NLS-containing proteins in addition to the observed NPC deficits in 30R-expressing flies.
DISCUSSION
Increasing evidence suggests that NCT disruption is a common mechanism underlying neurodegenerative diseases including ALS/FTD, Huntington’s disease, and AD (Eftekharzadeh et al., 2019; Freibaum et al., 2015; Grima et al., 2017; Zhang et al., 2015). This evidence for altered NCT is primarily based on mislocalization of proteins, such as NLS-NES reporter proteins (Boeynaems et al., 2016; Chou et al., 2018; Freibaum et al., 2015; Giampetruzzi et al., 2019; Li and Lagier-Tourenne, 2018; Zhang et al., 2015). However, much less is known about the mechanisms that lead to impaired nuclear pore function. Earlier studies of DPR proteins in C9-ALS/FTD implicated arginine-containing DPRs in ‘‘plugging’’ the central channel of the NPC (Shi et al., 2017); however, more recent evidence suggests that DPRs indirectly alter NCT through interactions with karyopherins (Hayes et al., 2020; Solomon et al., 2018). Recent evidence in iPS neurons derived from C9-ALS patients suggests that select nucleoporins are downregulated, and that this alteration in the NPC significantly contributes to the disruption in NCT (Coyne et al., 2020). Indeed, our findings in Drosophila models of C9-ALS/FTD support this model whereby NPC alterations can impact functional NCT. Importantly, inhibiting nucleoporin degradation restores NCT and rescues neurodegenerative phenotypes, suggesting that the NPCs remain functional if they are stabilized, and also that the loss of nucleoporins is a significant contributor to neurodegeneration rather than merely an indicator of an unhealthy neuron.
How does the C9orf72 HRE trigger the loss of nucleoporins? Data from iPS neurons suggests that post-translational downregulation of the transmembrane nucleoporin POM121 leads to loss of seven other nucleoporins, as restoring POM121 protein expression after NPC injury has occurred rescues nucleoporin levels and NCT (Coyne et al., 2020). However, Drosophila lack POM121, and yet expression of expanded GGGGCC repeats also results in a severe loss of multiple nucleoporins, primarily those more frequently turned over (Savas et al., 2012). Since expression of arginine-containing DPRs causes severe neurotoxicity (Figure 4D), it is tempting to speculate that interactions between GR/PR and low-complexity domains of FG-repeat containing Nups (Lee et al., 2016) may trigger their removal from the NPC. Alternatively, injury of the nuclear envelope may signal nuclear localization and increased expression of ESCRT-III, leading to enhanced nucleoporin turnover.
Recent work has shown that aberrant nuclear accumulation of the ESCRT-III protein CHMP7 can initiate Nup reduction in ALS iPS neurons via a mechanism that may at least in part involve recruitment and function of VPS4 (Coyne and Rothstein, 2021; Coyne et al., 2021), though the initiating signal for nucleoporin removal remains unknown. Our data strongly implicate the Vps4/ESCRT-III machinery as necessary and sufficient for nucleoporin degradation in fly models of C9-ALS/FTD. Of note, the Vps20 subunit of ESCRT-III does not appear to be required for this process, similar to what has been observed in the yeast NPC surveillance pathway (Webster et al., 2014), suggesting that the mechanisms of ESCRT-III function in NPC maintenance are distinct from those of endosomal sorting. The C9orf72 HRE induces expression of Vps4, and both Vps4 and ESCRT-III subsequently become enriched in the nucleus (Figures 6A–6J). Since knockdown of Vps4 and ESCRT-III genes rescue nucleoporin levels and NCT abnormalities in our fly C9-ALS models, our data indicate that nuclear Vps4/ESCRT-III is required for nucleoporin downregulation downstream of the HRE.
Since the Vps4/ESCRT-III complex is involved in surveillance and clearing of defective NPC assembly intermediates in yeast (Webster et al., 2014), we hypothesize that nuclear Vps4/ESCRT-III mediates scission of NPCs from the inner nuclear membrane, and then vesicles containing NPCs are transported into the cytoplasm. This model is consistent with the known mechanism of the Vps4/ESCRT-III complex in viral budding from the nuclear membrane (Arii et al., 2018). Alternatively, Vps4/ESCRT-III could selectively remove specific nucleoporin complexes from the NPC. Arguing against this possibility, though, is our super-resolution imaging data that shows what appear to be relatively intact NPCs (with nuclear and cytoplasmic Nups appropriately juxtaposed) in the cytoplasm of neurons expressing GGGGCC repeats. Furthermore, even though total protein levels of the scaffolding Nup93 remain unaltered in our fly models of C9-ALS/FTD (Figures 1E and 1F), Nup93 does become mislocalized to the cytoplasm (Figures S7D and S7E). Given that the ESCRT-III/Vps4 complex is best known as a membrane scission complex, we favor the possibility that entire NPCs are removed from the nuclear membrane and exported into the cytoplasm where select nucleoporins are degraded via the proteasome.
Our results are consistent with two very recent reports showing that Vps4 and CHMP7 are increased in nuclei of iPSC-derived motor neurons and postmortem human motor cortex from patients with familial and sporadic ALS (Coyne and Rothstein, 2021; Coyne et al., 2021). Knockdown of the ESCRT-III component CHMP7 following NPC alterations alleviates the nucleoporin reduction and TDP-43 dysfunction observed in iPS neurons derived from patients with both C9orf72 and sporadic ALS. These findings suggest that nuclear expression of CHMP7 in vertebrate neurons is sufficient for nucleoporin downregulation, similar to our findings in Drosophila that ESCRT-III/Vps4 is necessary and sufficient to drive nucleoporin mislocalization and degradation.
Once nucleoporin complexes (or intact NPCs) enter the cytoplasm, they associate with proteasomal subunits (Figure S4F), consistent with our observation that they undergo proteasome-mediated degradation (Figure 2). Since either Rpn10 knockdown or a chemical inhibitor of the proteasome (epoxomicin) fully rescues nucleoporin reduction, our data suggest that nucleoporins are primarily degraded via the proteasome in neurons in C9-ALS, rather than via the autophagy machinery as has been shown with other stressors such as nitrogen starvation or TORC1 inhibition (Lee et al., 2016, 2020; Tomioka et al., 2020). Intriguingly, even though proteasome dysfunction has been implicated in the pathogenesis of ALS, genetic knockdown of the Rpn10 regulatory subunit strongly rescues GGGGCC-repeat-mediated neurode-generation in the fly eye (Figures 2I and 2J). Rpn10 is one of several ubiquitin-binding proteasome regulatory proteins and is not essential for most proteasome functions, so it could potentially be considered as a therapeutic target if validated in other ALS models. While we cannot exclude the possibility that Rpn10 knockdown suppresses degradation of additional neuroprotective factors, our findings suggest that preventing Rpn10-mediated nucleoporin degradation is neuroprotective.
In conclusion, our findings suggest a model whereby transcriptional upregulation and increased neuronal and nuclear expression of the Vps4/ESCRT-III complex contribute to NPC dysfunction in C9-ALS. Based on the known function of the ESCRT-III complex and Vps4 ATPase in membrane scission, we hypothesize that the NPC-containing inner nuclear membrane buds into the cytoplasm where it undergoes scission and transport for proteasome-mediated degradation of specific Nups. Since primarily peripheral Nups are degraded (and the central scaffolding Nup93 is not altered), it is possible that Nups that are not targeted for proteasomal degradation may be recycled or remain in the cytoplasm. Restoring NPC function via inhibition of the nuclear ESCRT-III/Vps4 complex could represent a feasible therapeutic strategy in C9-ALS/FTD as has previously been suggested through use of antisense oligonucleotides targeting the ESCRT-III protein CHMP7 in an iPS neuron model of C9-ALS/FTD (Coyne et al., 2021).
Limitations of the study
There are several important differences between Drosophila NPCs and vertebrate NPCs that may limit translation of these findings to human disease. For example, loss of POM121 seems to be the critical initiating factor of nucleoporin loss in iPS neuron models of C9-ALS/FTD (Coyne et al., 2020), and yet, Drosophila lack a clear POM121 ortholog. Nonetheless, since our fly C9-ALS/FTD models show a similar ESCRT-III-dependent loss of specific Nups, our findings suggest an evolutionarily conserved nucleoporin degradation pathway in neurons that involves nuclear accumulation of the ESCRT-III/Vps4 complex. While our study does not identify the initiating factors that mediate this nuclear localization in C9-ALS/FTD, future efforts to understand this mechanism may shed light into upstream triggers of neurodegeneration in ALS/FTD.
An additional limitation is that the genetic and pharmacologic rescue experiments were performed at the same time the C9 HRE expression was initiated. While the proteasome inhibitor epoxomicin restores nucleoporin levels in the fly C9-ALS/FTD model, it is unlikely that broad proteasome inhibition will suppress neurodegeneration in these models. Future studies will investigate whether Rpn10 or Vps4/ESCRT-III inhibition initiated after induction of C9 HRE expression and NPC degradation will also rescue neurodegenerative phenotypes to determine if these are valid therapeutic strategies.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Thomas E. Lloyd (tlloyd4@jhmi.edu).
Materials availability
This study did not generate new unique reagents. All materials are publicly available. Please contact Thomas E. Lloyd for additional information.
Data and code availability
All data generated and reported in this paper are available from the lead contact upon request.
This paper does not report original program code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All flies were raised on standard cornmeal-molasses food at 25°C. All stocks were obtained from Bloomington Drosophila Stock Center (BDSC) or The Vienna Drosophila Resource Center (VDRC). Stock genotypes and availability is listed in the Key Resources Table. Male flies were used for all experiments. Bloomington stock: w1118 was used as wild-type control. The attp2, LacZ, CD8-GFP and LucRNAi are used with GAL4 drivers as a control for experiments. Eye degeneration phenotypes were examined for the presence of supernumerary interommatidial bristles, abnormal orientation of interommatidial bristles, necrotic patches, a decrease in size, retinal collapse, fusion of ommatidia, disorganization of ommatidial array, and loss of pigmentation, and given points for abnormalities on a 20-point scale as described (Ritson et al., 2010). GMR-Gal4; UAS-(G4C2)30/CyO were crossed to UAS-modifier lines or background controls and GMR-GAL4; UAS-30R/UAS-modifier lines were selected and aged at 25°C for 15 days. In eye degeneration scoring, points were added if there was complete loss of interommatidial bristles, necrotic patches, retinal collapse, loss of ommatidial structure, and/or depigmentation of the eye. Eye images were obtained using a Nikon SMZ 1500 Microscope and Infinity 3 Luminera Camera with Image Pro Insight 9.1 software. For pupal survival assays, male flies of the genotype vGlut-Gal4; UAS-30R/TM6-Gal80(Tb) were crossed to female flies containing UAS-modifier lines or background controls. Overexpression of 30R in motor neurons causes lethality due to paralysis, preventing eclosion of the adult from the pupal case. Parental flies were transferred to fresh new vials every 2–3 days. After 15 days, non-tubby pupae were scored as eclosed (alive) or non-enclosed (dead).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Chicken polyclonal anti-GFP | Abcam | Cat# ab13970, RRID:AB_300798 |
| Mouse monoclonal anti- Beta actin (clone C4) | EMD Millipore | Cat# MAB1501, RRID:AB_2223041 |
| Mouse monoclonal anti- ELAV | Developmental Studies Hybridoma Bank | Elav-9F8A9; RRID:AB_528217 |
| Mouse monoclonal anti- LaminB | Developmental Studies Hybridoma Bank | ADL46; RRID:AB_528335 |
| Mouse monoclonal anti-Repo | Developmental Studies Hybridoma Bank | 8D12; RRID:AB_528448 |
| Rabbit polyclonal anti-Nup98 | Pascual-Garcia et al., 2014 | Maya Capelson Lab (Upenn) |
| Rabbit polyclonal anti-Nup98 | Cell Signaling | #2288; RRID:AB_561204 |
| Rabbit anti-Nup50 | Brandt et al., 2006 | Jörg Grosshans Lab |
| Guinea pig anti-Nup214 | Roth et al., 2003 | Christos Samakovlis Lab |
| Guinea pig anti-Nup93 | Gozalo et al., 2020 | Maya Capelson Lab (Upenn) |
| Mouse monoclonal anti-Megator | Developmental Studies Hybridoma Bank | 12F10-F11; RRID:AB_2721935 |
| Rabbit anti-Shrub | Sweeney et al., 2006 | Fen-Biao Gao Lab |
| Rabbit polyclonal anti-ref(2)P | Gabor Juhasz laboratory | N/A |
| Rabbit ant-Vps4 | Millipore Sigma | SAB4200025; RRID:AB_10603251 |
| Mouse Anti-26S Proteasome p50 (Rpt5) | Santa Cruz | sc-65745; RRID:AB_1118470 |
| Mouse Anti-Rpn10 | Lipinszki et al., 2009 | Zoltan Lipinszki Lab |
| Rabbit Anti-CHMP1B | Thermo Fischer Scientific | PA5-100437; RRID:AB_2849948 |
| Rabbit Anti-CHMP2B | Cell Signaling | 76173; RRID:AB_2799880 |
| Rabbit Anti-CHMP6 (Vps20) | Thermo Fischer Scientific | PA5-113238; RRID:AB_2867972 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| SuperScript III First-Strand Synthesis System | Thermo Fischer Scientific | Cat #18080051 |
| SYBR Select Master Mix | Thermo Fischer Scientific | Cat #4472908 |
| TRIzol | Thermo Fischer Scientific | Cat #15596018 |
| TRIzol | Millipore Sigma | Cat #M8046 |
| Epoxomicin | UBPBio | Cat# F1400; CAS: 134,381–21-8 |
| Protease Inhibitor Cocktail | Roche | Cat#11873580001 |
| Chemical compound, drug Blotting Grade Blocker (nonfat dry milk) | Bio-Rad | Cat #1706404 |
| Commercial assay or kit NE-PER Nuclear and Cytoplasmic Extraction Kit | Thermo Fischer Scientific | Cat #78833 |
| Expansion Microscopy Reagent Acryloyl-X SE (AcX) | Thermo Fisher Scientific | Cat #A20770 |
|
| ||
| Oligonucleotides | ||
|
| ||
| qRT-PCR Primer Vps4 F: 5′ACTGCGTCTGTATGAGCACG-3′ Vps4 R: 5′CTCCTTTAGCTTCTCGGCCC-3′ |
Integrated DNA Technologies IDT | N/A |
| qRT-PCR Primer Actin F: 5′-GCGCGGT TACTCTTTCACCA-3′ Actin R: 5′-ATGTCACGGACGATTTCACG-3′ |
Integrated DNA Technologies IDT | N/A |
| qRT-PCR Primer Vps2 F: 5’-CGATGAGATGCTGCGTA AGA −3′′Vps2 R: 5′-CTCCTTGGCCATCTTCTTGATA-3′ |
Integrated DNA Technologies IDT | N/A |
| qRT-PCR Primer Vps24 F: 5’-CAGTTAGTGCCATGGGCTTAT-3′ ps24 R: 5’-GTTGCCCTCTTTCCGTATCTT-3′ |
Integrated DNA Technologies IDT | N/A |
| qRT-PCR Primer CG5498 F: 5’-GCCTGTTGGCCTTAAATGTTC-3′ CG5498 R: 5’-GCCTGGTCTTCTTGGCTTAT-3′ |
Integrated DNA Technologies IDT | N/A |
| qRT-PCR Primer Nup98 F: 5′-TCCTGCTTTCGGACAGACTAAC-3′ Nup98 R: 5′-TGGAGCTCCAAATCCAAAACCT-3′ |
Integrated DNA Technologies IDT | N/A |
| qRT-PCR Primer Nup214 F: 5′-ACCAGCAAGCCTACGAATGTAA-3′ Nup214 R: 5′-CTCCTCCAAAAATGTTGGCTGG-3′ |
Integrated DNA Technologies IDT | N/A |
| qRT-PCR Primer Nup93 F: 5′-ACACCGTCCGCGAAATAC −3′ ′Nup93 R: 5′-ACTCAACCGCCACCTTAAC-3′ |
Integrated DNA Technologies IDT | N/A |
| qRT-PCR Primer Nup50F:5′-GTCGAGTTTAAACAGGTTGTGG AGG-3′ Nup50F:5′-GCGCGGACTAACAGTTGGATC-3′ |
Integrated DNA Technologies IDT | N/A |
| qRT-PCR Primer Megator F:5′GAACAACCAGAGTCAGGCCA-3′ Megator F: 5′-CTCCACAGAGCTTTCGTTGC-3′ |
Integrated DNA Technologies IDT | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| (D. melanogaster) GMR-Gal4; w*; P{GAL4-ninaE.GMR}12 | Bloomington Drosophila Stock Center | BDSC:1104 |
| (D. melanogaster) 30R; w[1118]; UAS-(G4C2)30 | Peng Jin (Xu et al., 2013) | FlyBase:FBal0294759 |
| (D. melanogaster) vGlut-Gal4;w[1118]; P{w[+mW.hs] = GawB}VGlut[OK371] | Bloomington Drosophila Stock Center | BDSC: 26160 |
| (D. melanogaster) w1118; w[1118] | Bloomington Drosophila Stock Center | BDSC: 3605 |
| (D. melanogaster) ELAV-GS;w[*];P{elavSwich.O} GSG301 | Adrian Isaacs (Mizielinska et al., 2014) | Flybase: FBtp0015149 |
| (D. melanogaster) UAS-poly(GR)36; w[1118]; P{{y[+t7.7] w[+mC] = UAS poly-GR.P O −36} attP40 | Adrian Isaacs (Mizielinska et al., 2014) | Flybase: FBtp0097185 |
| (D. melanogaster) UAS-poly(PR)36 | Adrian Isaacs (Mizielinska et al., 2014) | Flybase: FBti0166229 |
| (D. melanogaster) Act-Gal4; y[1] w[*]; P{Act5C-GAL4}17bF O 1/TM6B, Tb1 | Bloomington Drosophila Stock Center | BDSC: 3954 |
| (D. melanogaster) UAS-36R; UAS-(G4C2)36; w[1118]; P{y[+t7.7] w[+mC] = UAS GGGGCC.36}attP40 | Adrian Isaacs (Mizielinska et al., 2014) | Flybase: FBtp0097184 |
| (D. melanogaster) UAS-LacZ; w[1118]; P{w[+mC] = UAS-lacZ.NZ}J312 | Bloomington Drosophila Stock Center | BDSC: 3956 |
| (D. melanogaster) UAS-luciferaseRNAi; y[1] v[1]; P{y[+t7.7] v[+t1.8] = TRiP.JF01355}attP2 | Bloomington Drosophila Stock Center | BDSC: 31,603 |
| (D. melanogaster) UAS-S-GFP; w[1118]; P{w[+mC] = UAS-NLS-NES[+]-GFP}5A | Bloomington Drosophila Stock Center | BDSC: 7032 |
| (D. melanogaster) UAS-CD8:GFP; y[1] w[*]; P{w[+mC] = UAS-mCD8::GFP.L}LL5, P{UAS-mCD8::GFP.L}2 | Bloomington Drosophila Stock Center | BDSC:5137 |
| (D. melanogaster) Repo-GS; P{repo-GAL4::GS} | Amita Sehgal lab | Flybase: FBtp0143340 |
| (D. melanogaster) UAS-Atg8aRNAi; y[1] v[1]; P{y[+t7.7] v[+t1.8] = TRiP.JF02895}attP2 e[*]/TM3, Sb[1] | Bloomington Drosophila Stock Center | BDSC: 28,989 |
| (D. melanogaster) UAS-Vps4; w*; P{UAS-HA-Vps4}2 | Bloomington Drosophila Stock Center | BDSC:63799 |
| (D. melanogaster) UAS-Vps4RNAi#1; y1 v1; P{TRiP.HM04061}attP2 | Bloomington Drosophila Stock Center | BDSC:31751 |
| (D. melanogaster) UAS-Vps4RNAi#2; w1118; P{GD12054}v35125 | VDRC | 35125 |
| (D. melanogaster) UAS-Vps4RNAi#3; w1118; P{GD12054}v35126 | VDRC | 35126 |
| (D. melanogaster) UAS-Vps4RNAi#4; P{KK101722}VIE-260B | VDRC | 105977 |
| (D. melanogaster) UAS- ShrubRNAi#1; y1v1; P{TRiP.HMS01767}attP40 | Bloomington Drosophila Stock Center | BDSC:38305 |
| (D. melanogaster) UAS- ShrubRNAi#2; P{KK108557}VIE-260B | VDRC | VDRC: 106823 |
| (D. melanogaster) UAS- Vps24RNAi#1; y1 sc* v1 sev21; P{TRiP.HMS01733}attP40 | Bloomington Drosophila Stock Center | BDSC:38281 |
| (D. melanogaster) UAS- Vps24RNAi#2 w1118; P{GD14676}v29275 | VDRC | VDRC: 29275 |
| (D. melanogaster) UAS-Chmp1RNAi#1;y1v1; P{TRiP.HM05117}attP2 | Bloomington Drosophila Stock Center | BDSC:28906 |
| (D. melanogaster) UAS- Chmp1RNAi#2; y1 sc* v1 sev21; P{TRiP.HMS00877}attP2 | Bloomington Drosophila Stock Center | BDSC:33928 |
| (D. melanogaster) UAS- Chmp2BRNAi#1; y1 v1; P{TRiP.HM05017}attP2 | Bloomington Drosophila Stock Center | BDSC: 28531 |
| (D. melanogaster) UAS- Chmp2BRNAi#2; y1 v1; P{TRiP.HMS01844}attP40 | Bloomington Drosophila Stock Center | BDSC:41993 |
| (D. melanogaster) UAS- CG5498RNAi#1; w1118; P{GD11674}v27439 | VDRC | 27439 |
| (D. melanogaster) UAS- CG5498RNAi#2; P{KK100759}VIE-260B | VDRC | 103711 |
| (D. melanogaster) UAS- Vps20RNA#1; y1 v1; P{TRiP.HMS02142}attP40 | Bloomington Drosophila Stock Center | BDSC: 40894 |
| (D. melanogaster) UAS- Vps20RNA#2; w1118; P{GD11211}v26387 | VDRC | 26387 |
| (D. melanogaster) UAS- Vps2RNAi#1; y1 sc* v1 sev21; P{TRiP.HMS01911}attP40 | Bloomington Drosophila Stock Center | BDSC: 38995 |
| (D. melanogaster) UAS- Vps2RNAi#2; w1118; P{GD8363}v24869/TM3 | VDRC | 24869 |
| (D. melanogaster) Rpn10-EP; y1 w*; P{EP}Rpn10G6601/TM3, Sb1 Ser1 | Bloomington Drosophila Stock Center | BDSC: 27212 |
| (D. melanogaster) UAS- Rpn10RNAi#1; y1 sc* v1 sev21; P{TRiP.HMS01039}attP2 | Bloomington Drosophila Stock Center | BDSC: 34566 |
| (D. melanogaster) UAS- Rpn10RNAi#2; P{KK100347}VIE-260B | VDRC | 110659 |
| (D. melanogaster) Fus-WT | Udai Bhan Pandey Lab | N/A |
| (D. melanogaster) Fus-R518K | Udai Bhan Pandey Lab | N/A |
| (D. melanogaster) Fus-R521C | Udai Bhan Pandey Lab | N/A |
| (D. melanogaster) Htt128Q; P{UAS-htt.128Q.FL}f27b | Bloomington Drosophila Stock Center | BDSC:33808 |
|
| ||
| Software and algorithm | ||
|
| ||
| GraphPad Prism 8 | https://www.graphpad.com/scientific-software/prism/ | N/A |
| Image Studio™ Lite | https://www.licor.com/bio/image-studio-lite/ | N/A |
| Adobe Photoshop | https://www.adobe.com/products/photoshop.html | N/A |
| Biorender | www.Biorender.com | N/A |
| Zen zeiss software | https://www.zeiss.com/microscopy/us/products/microscope-software/zen-lite.html | N/A |
METHOD DETAILS
Drosophila drug feeding
Cornmeal-molasses food was melted and cooled for 3–5 min before being mixed with concentration of mifepristone (RU486:100uM) and epoxomicin (0.25, 0.50, 0.75 and 1uM) and cooled to room temperature (RT). Parental flies were crossed on normal food and 1-day-old male adult progenies were transferred to vials containing drug food or ethanol vehicle alone and transferred to new vials every 2–3 days. Adult flies were aged on the drug containing food for 10 and 15 days.
Immunofluorescence staining and imaging
For Drosophila adults and larvae, brains from desired genotypes were dissected in 1× PBS and fixed in 4% paraformaldehyde for 20 min. Tissues were rinsed in PBST (1X PBS, 0.1% Triton X-100). The tissues were blocked for 2h (PBS with 5% normal goat serum (NGS) and 0.1% PBX) for 2h at RT followed by incubation with primary antibodies at 4°C overnight. Post-antibody incubation, tissues were washed in 0.1% PBST (3 × 20 min each) and were incubated in 2° antibody at 4°C overnight. They were then washed in 0.1% PBST and counterstained by DAPI (DAPI was added to the prep at a final concentration of 1 mg/mL). They were again rinsed in 0.1% PBST (3 × 20 min each) and mounted in antifadant, Fluoromount-G (Invitrogen).
Imaging was done using LSM800 confocal microscope (Carl Zeiss). Images were processed with Adobe Photoshop7. To quantify fluorescence intensities, Zen Zeiss software was used to quantify the intensities of certain areas/bands. The presented values are the mean fluorescence intensity of the NPC/protein of individual ELAV-marked neuron cell bodies.
Expansion microscopy
Samples were processed for expansion microscopy after immunohistochemistry according to a protocol from the Boyden Lab (Chen et al., 2015; Tillberg et al., 2016). Antibody-labeled brains were treated with 0.1 mg/mL Acryloyl-X SE (AcX) in PBS for 12 h (ThermoFisher Scientific, Catalog #A20770, Waltham, MA) overnight at room temperature. The brains were washed in PBS three times before the gelation. The polyelectrolyte gel solution was added and incubated on ice for 45 min at room temperature before pipetting the brains onto the Gel Chamber composed of a glass slide, spacers, and a coverglass. After incubation, assembled Gel Chamber with the brains were incubated at 37°C for about 2 h. Solidified brain samples were excised and trimmed away from the Gel Chamber and processed for digestion with fresh Proteinase K in six-well plates (1:100) for 24 h. The brains were washed with water (ddH2O) at least three times, 20 min each. Finally, expanded brains (~4 times original size) were nearly transparent. The expanded brains were placed on a slide with dH2O for imaging using a Zeiss LSM800 confocal.
Western blotting
Adult brains from desired genotype were dissected in 13 PBS, and protein samples were homogenized and lysed in RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, and 1% Triton X-100) supplemented with protease inhibitor cocktail, using microcentrifuge pestles, and then incubated in RIPA buffer on ice for 20 min. Protein samples were mixed with 4x Laemmli buffer (Bio-Rad) and 2-Mercaptoethanol and heated at 98°C for 10 min. Sample was centrifuged at 12,000 rpm for 10 min and supernatant was collected. Protein sample was resolved on 4–15% TGX gels (Bio-Rad). Protein samples were transferred onto PVDF membrane using wet electrotransfer apparatus at 90 V for 2h at 4°C. PVDF membranes were incubated in blocking solution (TBST (50 mM Tris-HCl pH 7.4, 1% Triton X-100) with 5% non-fat milk (Bio-Rad) was used for blocking), followed by primary antibody incubation for overnight at 4°C. Membranes were washed in TBST (3 × 15 min each) and were incubated in 2° antibody for 2h at RT followed by three PBST washes. Signals were detected by Enhanced chemiluminescence (ECL) method on Odyssey Fc Imaging System - LI-COR Biosciences. For nucleocytoplasmic fractionation of adult brain, fractionation was performed with the NE-PER Nuclear and Cytoplasmic Extraction Kit according to the manufacturer protocol (Thermo Fisher Scientific).
RNA isolation and q-RT-PCR
Total cellular RNA was isolated from 5 larvae of each genotype using TRIzol reagent (Invitrogen) following the manufacturer protocol. Reverse transcription was performed using 1 microgram of RNA using SuperScript III First-Strand synthesis kit (manufacturer protocol). q-RT-PCR was performed using the SYBR Green PCR system on a 7900HT fast Real-Time PCR system (Applied Biosystem). Data was extracted and analyzed using excel. Primers used are detailed in key resources table.
QUANTIFICATION AND STATISTICAL ANALYSIS
All quantitative data were derived from independent experiments. All statistical significance tests were performed with Prism version 8.3.1. All datasets were assembled in Microsoft Excel 365. Each n value in data representing biological replicates is indicated in the figure legends. Adobe photoshop was used to make all the figures. Each value is from at least 3 different experiments and reported as mean ± SEM.
Supplementary Material
Highlights.
Neuronal GGGGCC repeat expression induces degradation of nucleoporins in vivo
Rpn10 and the proteasome are required for GGGGCC-mediated nucleoporin degradation
ESCRT-III/Vps4 is increased in the nucleus of GGGGCC-repeat-expressing neurons
ESCRT-III/Vps4 knockdown restores nucleoporins and suppresses repeat toxicity
ACKNOWLEDGMENTS
This work was supported by NINDS R01NS094239 and NIA R01AG068043, ALSA, the Robert Packard Center for ALS Research, and Target ALS. We thank Udai Pandey, Peng Jin, Adrian Isaacs, Nancy Bonini, the Bloomington Drosophila Stock Center (NIH P40ODO18537), and Vienna Drosophila Research Center for Drosophila lines, and the Multiphoton Imaging Core (NS050274). We thank Maya Capelson, Jörg Grosshans, Christos Samakovlis, Zoltan Lipinszki, and Fen-Biao Gao for antibodies and Alyssa Coyne for feedback on this manuscript.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111379.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Anderson EN, Morera AA, Kour S, Cherry JD, Ramesh N, Gleixner A, Schwartz JC, Ebmeier C, Old W, Donnelly CJ, et al. (2021). Traumatic injury compromises nucleocytoplasmic transport and leads to TDP-43 pathology. Elife 10, e67587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arii J, Watanabe M, Maeda F, Tokai-Nishizumi N, Chihara T, Miura M, Maruzuru Y, Koyanagi N, Kato A, and Kawaguchi Y (2018). ESCRT-III mediates budding across the inner nuclear membrane and regulates its integrity. Nat. Commun 9, 3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin W-L, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW 3rd, Rademakers R, et al. (2013). Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al. (2012). Syndecan–syntenin–ALIX regulates the biogenesis of exosomes. Nat. Cell Biol 14, 677–685. [DOI] [PubMed] [Google Scholar]
- Balendra R, and Isaacs AM (2018). C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat. Rev. Neurol 14, 544–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, de Franceschi N, de la Mora E, Maity S, Alqabandi M, Miguet N, di Cicco A, Roos WH, Mangenot S, Weissenhorn W, et al. (2020). Author Correction: human ESCRT-III polymers assemble on positively curved membranes and induce helical membrane tube formation. Nat. Commun 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blitterswijk M, Gendron TF, Baker MC, DeJesus-Hernandez M, Finch NA, Brown PH, Daughrity LM, Murray ME, Heckman MG, Jiang J, et al. (2015). Novel clinical associations with specific C9ORF72 transcripts in patients with repeat expansions in C9ORF72. Acta Neuropathol 130, 863–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Michiels E, Gijselinck I, Sieben A, Jovičić A, De Baets G, Scheveneels W, Steyaert J, Cuijt I, et al. (2016). Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci. Rep 6, 20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt A, Papagiannouli F, Wagner N, Wilsch-Bräuninger M., Braun M., Furlong EE., Loserth S, Wenzl C, Pilot F., Vogt N., et al. (2006). Developmental control of nuclear size. Lea 2, 3–74. [DOI] [PubMed] [Google Scholar]
- Carlton JG, and Martin-Serrano J (2007). Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316, 1908–1912. [DOI] [PubMed] [Google Scholar]
- Chen F, Tillberg PW, and Boyden ES (2015). Expansion microscopy. Science 347, 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew J, Cook C, Gendron TF, Jansen-West K, del Rosso G, Daughrity LM, Castanedes-Casey M, Kurti A, Stankowski JN, Disney MD, et al. (2019). Aberrant deposition of stress granule-resident proteins linked to C9orf72-associated TDP-43 proteinopathy. Mol. Neurodegener 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C-C, Zhang Y, Umoh ME, Vaughan SW, Lorenzini I, Liu F, Sayegh M, Donlin-Asp PG, Chen YH, Duong DM, et al. (2018). TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci 21, 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ L, Raiborg C, Wenzel EM, Campsteijn C, and Stenmark H (2017). Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem. Sci 42, 42–56. [DOI] [PubMed] [Google Scholar]
- Cooper-Knock J, Higginbottom A, Stopford MJ, Highley JR, Ince PG, Wharton SB, Pickering-Brown S, Kirby J, Hautbergue GM, and Shaw PJ (2015). Antisense RNA foci in the motor neurons of C9ORF72-ALS patients are associated with TDP-43 proteinopathy. Acta Neuropathol 130, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne AN, Zaepfel BL, Hayes L, Fitchman B, Salzberg Y, Luo E-C, Bowen K, Trost H, Aigner S, Rigo F, et al. (2020). G4C2 repeat RNA initiates a POM121-mediated reduction in specific nucleoporins in C9orf72 ALS/FTD. Neuron 107, 1124–1140.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne AN, and Rothstein JD (2021). The ESCRT-III protein VPS4, but not CHMP4B or CHMP2B, is pathologically increased in familial and sporadic ALS neuronal nuclei. Acta Neuropathol. Commun 9, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne AN, Baskerville V, Zaepfel BL, Dickson DW, Rigo F, Bennett F, Lusk CP, and Rothstein JD (2021). Nuclear accumulation of CHMP7 initiates nuclear pore complex injury and subsequent TDP-43 dysfunction in sporadic and familial ALS. Sci. Transl. Med 13, eabe1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham KM, Maulding K, Ruan K, Senturk M, Grima JC, Sung H, Zuo Z, Song H, Gao J, Dubey S, et al. (2020). TFEB/Mitf links impaired nuclear import to autophagolysosomal dysfunction in C9-ALS. eLife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, and Hetzer MW (2009). Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136, 284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. (2011). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang P-W, Pham JT, Haeusler AR, Mistry NA, Viden -sky S, Daley EL, Poth EM, Hoover B, Fines DM, et al. (2013). RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 80, 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftekharzadeh B, Daigle JG, Kapinos LE, Coyne A, Schiantarelli J, Carlomagno Y, Cook C, Miller SJ, Dujardin S, Amaral AS, et al. (2019). Tau protein disrupts nucleocytoplasmic transport in Alzheimer’s disease. Neuron 101, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee K-H, Badders N, Valentine M, Miller BL, Wong PC, et al. (2015). GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525, 129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Côté M, Rich RL., et al. (2001). Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107, 55–65. [DOI] [PubMed] [Google Scholar]
- Gasset-Rosa F, Chillon-Marinas C, Goginashvili A, Atwal RS, Artates JW, Tabet R, Wheeler VC, Bang AG, Cleveland DW, and Lagier-Tourenne C (2017). Polyglutamine-expanded huntingtin exacerbates age-related disruption of nuclear integrity and nucleocytoplasmic transport. Neuron 94, 48–57.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giampetruzzi A, Danielson EW, Jeon M, Gumina V, Boopathy S, Brown RH, Landers JE, and Fallini C (2019). Modulation of actin polymerization affects nucleocytoplasmic transport in multiple forms of Amyotrophic Lateral Sclerosis. Nat. Comm 10, 3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Deza J, Lee Y-B, Troakes C, Nolan M, Al-Sarraj S, Gallo J-M, and Shaw CE (2015). Dipeptide repeat protein inclusions are rare in the spinal cord and almost absent from motor neurons in C9ORF72 mutant amyotrophic lateral sclerosis and are unlikely to cause their degeneration. Acta Neuropathol. Commun 3, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozalo A, Duke A, Lan Y, Pascual-Garcia P, Talamas JA, Nguyen SC, Shah PP, Jain R, Joyce EF, and Capelson M (2020). Core components of the nuclear pore bind distinct states of chromatin and contribute to polycomb repression. Mol. Cell 77, 67–81.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima JC, Daigle JG, Arbez N, Cunningham KC, Zhang K, Ochaba J, Geater C, Morozko E, Stocksdale J, Glatzer JC, et al. (2017). Mutant huntingtin disrupts the nuclear pore complex. Neuron 94, 93–107.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes LR, Duan L, Bowen K, Kalab P, and Rothstein JD (2020). C9orf72 arginine-rich dipeptide repeat proteins disrupt karyopherin-mediated nuclear import. Elife 9, e51685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW (2010). The nuclear envelope. Cold Spring Harb. Perspect. Biol 2, a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez AJ, Maiuri P, Lafaurie-Janvore J, Divoux S, Piel M, and Perez F (2014). ESCRT machinery is required for plasma membrane repair. Science 343, 1247136. [DOI] [PubMed] [Google Scholar]
- Jovičić A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW 3rd, Sun S, Herdy JR, Bieri G, et al. (2015). Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci 18, 1226–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer NJ, Haney MS, Morgens DW, Jovičić A, Couthouis J, Li A, Ousey J, Ma R, Bieri G, Tsui CK, et al. (2018). CRISPR-Cas9 screens in human cells and primary neurons identify modifiers of C9ORF72 dipeptide-repeat-protein toxicity. Nat. Genet 50, 603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-W, Wilfling F, Ronchi P, Allegretti M, Mosalaganti S, Jentsch S, Beck M, and Pfander B (2020). Selective autophagy degrades nuclear pore complexes. Nat. Cell Biol 22, 159–166. [DOI] [PubMed] [Google Scholar]
- Lee K-H, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, et al. (2016). C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-B, Chen H-J, Peres JN, Gomez-Deza J, Attig J, Štalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C, et al. (2013). Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep 5, 1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, and Lagier-Tourenne C (2018). Nuclear pores: the gate to neurodegeneration. Nat. Neurosci 21, 156–158. [DOI] [PubMed] [Google Scholar]
- Lin Y, Mori E, Kato M, Xiang S, Wu L, Kwon I, and McKnight SL (2016). Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167, 789–802.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Kumar MS, Ramesh N, Anderson EN, Nguyen AT, Kim B, Cheung S, McDonough JA, Skarnes WC, Lopez-Gonzalez R, et al. (2021). Lin Interactions between ALS-linked FUS and nucleoporins are associated with defects in the nucleocytoplasmic transport pathway. Nature Neurosci 24, 1077–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lince-Faria M, Maffini S, Orr B, Ding Y, Florindo C, Sunkel CE, Tavares Á ., Johansen J, Johansen KM, and Maiato H. (2009). Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J. Cell Biol 184, 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinszki Z, Kiss P, Pál M, Deák P., Szabó A., Hunyadi-Gulyas E., Klement E, Medzihradszky KF., and Udvardy A (2009). Developmental-stage-specific regulation of the polyubiquitin receptors in Drosophila melanogaster. J. Cell Sci 122, 3083–3092. [DOI] [PubMed] [Google Scholar]
- Mackenzie IRA, Frick P, Grässer FA., Gendron TF, Petrucelli L., Cashman NR., Edbauer D, Kremmer E., Prudlo J., Troost D., et al. (2015). Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol 130, 845–861. [DOI] [PubMed] [Google Scholar]
- Mauvezin C, Ayala C, Braden CR, Kim J, and Neufeld TP (2014). Assays to monitor autophagy in Drosophila. Methods 68, 134–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Colf LA, and Sundquist WI (2013). Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem 82, 663–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Grönke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IOC, Pietrzyk J, et al. (2014). C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345, 1192–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr D, Frey S, Fischer T, Güttler T, and Görlich D. (2009). Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J 28, 2541–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng S-M, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. (2013). The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338. [DOI] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung H-Y, Morham SG, Gygi SP, Rodesch CK, and Sundquist WI (2007). Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J 26, 4215–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, and Sundquist WI (2010). Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc. Natl. Acad. Sci. USA 107, 12889–12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y, and Carlton JG (2016). The ESCRT machinery: new roles at new holes. Curr. Opin. Cell Biol 38, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E, Noor E, Fischer JS, Gillet L, Wojtynek M, Vallotton P, and Weis K (2020). Maturation kinetics of a multiprotein complex revealed by metabolic labeling. Cell 183, 1785–1800.e26. [DOI] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, and Keshishian H (2001). A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. USA 98, 12596–12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Garcia P, Jeong J, and Capelson M (2014). Nucleoporin Nup98 associates with trx/MLL and NSL histone-modifying complexes and regulates hox gene expression. Cell Rep 9, 1981. [DOI] [PubMed] [Google Scholar]
- Pfitzner A-K, Mercier V, Jiang X, von Filseck JM, Baum B, Šarić A., and Roux A. (2020). An ESCRT-III polymerization sequence drives membrane deformation and fission. Cell 182, 1140–1155.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J., Rollinson S, Gibbs JR, Schymick JC., Laaksovirta H., Swieten JC., Myllykangas L, et al. (2011). A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, and Görlich D (2001). Kinetic analysis of translocation through nuclear pore complexes. EMBO J 20, 1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritson GP, Custer SK, Freibaum BD, Guinto JB, Geffel D, Moore J, Tang W, Winton MJ, Neumann M, Trojanowski JQ, et al. (2010). TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J. Neurosci 30, 7729–7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth P, Xylourgidis N, Sabri N, Uv A, Fornerod M, and Samakovlis C (2003). The Drosophila nucleoporin DNup88 localizes DNup214 and CRM1 on the nuclear envelope and attenuates NES-mediated nuclear export. J. Cell Biol 163, 701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, and Chait BT (2000). The yeast nuclear pore complex. J. Cell Biol 148, 635–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas JN, Toyama BH, Xu T, Yates JR, and Hetzer MW (2012). Extremely long-lived nuclear pore proteins in the rat brain. Science 335, 942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöneberg J, Lee I-H, Iwasa JH, and Hurley JH (2017). Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol 18, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi KY, Mori E, Nizami ZF, Lin Y, Kato M, Xiang S, Wu LC, Ding M, Yu Y, Gall JG, et al. (2017). Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc. Natl. Acad. Sci. USA 114, E1111–E1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lin S, Staats KA, Li Y, Chang W-H, Hung S-T, Hendricks E, Linares GR, Wang Y, Son EY, et al. (2018). Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat. Med 24, 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA, Stepto A, Au WH, Adachi Y, Diaper DC, Hall R, Rekhi A, Boudi A, Tziortzouda P, Lee Y-B, et al. (2018). A feedback loop between dipeptide-repeat protein, TDP-43 and karyopherin-a mediates C9orf72-related neurodegeneration. Brain 141, 2908–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoten CL, and Carlton JG (2018). ESCRT-dependent control of membrane remodelling during cell division. Semin. Cell Dev. Biol 74, 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney NT, Brenman JE, Jan YN, and Gao F-B (2006). The coiledcoil protein shrub controls neuronal morphogenesis in Drosophila. Curr. Biol 16, 1006–1011. [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, and Emr SD (2008). Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell 15, 578–589. [DOI] [PubMed] [Google Scholar]
- Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu C-CJ, English BP, Gao L, Martorell A, Suk H-J, Yoshida F, et al. (2016). Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol 34, 987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka Y, Kotani T, Kirisako H, Oikawa Y, Kimura Y, Hirano H, Ohsumi Y, and Nakatogawa H (2020). TORC1 inactivation stimulates autophagy of nucleoporin and nuclear pore complexes. J. Cell Biol 219. e201910063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR, and Hetzer MW (2013). Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 154, 971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, and Akhtar A (2010). Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet 6, e1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri M, Radulovic M, and Stenmark H (2020). The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol 21, 25–42. [DOI] [PubMed] [Google Scholar]
- Votteler J, and Sundquist WI (2013). Virus budding and the ESCRT pathway. Cell Host Microbe 14, 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BM, Colombi P, Jäger J, and Patrick Lusk C. (2014). Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell 159, 388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS, et al. (2013). Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc. Natl. Acad. Sci. USA 110, 7778–7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. (2015). The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Daigle JG, Cunningham KM, Coyne AN, Ruan K, Grima JC, Bowen KE, Wadhwa H, Yang P, Rigo F, et al. (2018). Stress granule assembly disrupts nucleocytoplasmic transport. Cell 173, 958–971.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Jiang J, Gendron TF, McAlonis-Downes M, Jiang L, Taylor A, Diaz Garcia S, Ghosh Dastidar S, Rodriguez MJ, King P, et al. (2020). Reduced C9ORF72 function exacerbates gain of toxicity from ALS/FTD-causing repeat expansion in C9orf72. Nat. Neurosci 23, 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MAC, et al. (2011). Non-ATG–initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. USA 108, 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. (2013). RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. USA 110, E4968–E4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and reported in this paper are available from the lead contact upon request.
This paper does not report original program code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


