Abstract
Inflammation is a common feature in neurodegenerative diseases that contributes to neuronal loss. Previously, we demonstrated that the basal inflammatory tone differed between brain regions and, consequently, the reaction generated to a pro‐inflammatory stimulus was different. In this study, we assessed the innate immune reaction in the midbrain and in the striatum using an experimental model of Parkinson's disease. An adeno‐associated virus serotype 9 expressing the α‐synuclein and mCherry genes or the mCherry gene was administered into the substantia nigra. Myeloid cells (CD11b+) and astrocytes (ACSA2+) were purified from the midbrain and striatum for bulk RNA sequencing. In the parkinsonian midbrain, CD11b+ cells presented a unique anti‐inflammatory transcriptomic profile that differed from degenerative microglia signatures described in experimental models for other neurodegenerative conditions. By contrast, striatal CD11b+ cells showed a pro‐inflammatory state and were similar to disease‐associated microglia. In the midbrain, a prominent increase of infiltrated monocytes/macrophages was observed and, together with microglia, participated actively in the phagocytosis of dopaminergic neuronal bodies. Although striatal microglia presented a phagocytic transcriptomic profile, morphology and cell density was preserved and no active phagocytosis was detected. Interestingly, astrocytes presented a pro‐inflammatory fingerprint in the midbrain and a low number of differentially displayed transcripts in the striatum. During α‐synuclein‐dependent degeneration, microglia and astrocytes experience context‐dependent activation states with a different contribution to the inflammatory reaction. Our results point towards the relevance of selecting appropriate cell targets to design neuroprotective strategies aimed to modulate the innate immune system during the active phase of dopaminergic degeneration.
Keywords: astrocyte, inflammation, microglia, myeloid, neurodegeneration, Parkinson's disease, synuclein
Main Points
Myeloid cells adopt an anti‐inflammatory phenotype in the midbrain and a pro‐inflammatory profile in the striatum of α‐synuclein‐overexpressing mice.
Astrocytes in the midbrain of α‐synuclein mice are involved in the pro‐inflammatory reaction.
Abbreviations
- AAV
adeno‐associated virus
- AAV9
adeno‐associated virus serotype 9
- AD
Alzheimer's disease
- ALS
amyotrophic lateral sclerosis
- BBB
blood–brain barrier
- CB
cytometry buffer
- DAM
disease‐associated microglia
- DAMP
damage‐associated molecular patterns
- GSEA
gene set enrichment analysis
- IFNγ
interferon gamma
- IL1β
interleukin 1 beta
- IL6
interleukin 6
- LPS
lipopolysaccharide
- LXR
liver X receptor; MFI, median flouorescence intensity
- MGnD
neurodegenerative microglia
- MHC‐II
major histocompatibility complex class II
- MS
multiple sclerosis
- NLRP3
NOD‐, LRR‐, and pyrin domain‐containing protein 3
- PD
Parkinson's disease
- PFA
paraformaldehyde
- RNA‐seq
RNA sequencing
- RXR
retinoid X receptor
- SN
substantia nigra
- SNpc
substantia nigra pars compacta
- TH
tyrosine hydroxylase
- TLR
toll‐like receptor
- TNFα
tumor necrosis factor alpha
- αSyn
alpha‐synuclein
1. INTRODUCTION
Neuroinflammation is a common feature in neurodegenerative diseases. The inflammatory reaction is generated by the release of immune modulators, mainly by glial and immune cells, in response to neuronal damage (Becher et al., 2017). The course and degree of neuroinflammation will depend on the context, duration, and type of stimulus. In this regard, little is known about the specific neuroinflammatory responses associated to each neurodegenerative process. Evidence for a neuroinflammatory response in Parkinson's disease (PD) was first addressed by McGeer and colleagues as they described the presence of large numbers of human leukocyte antigen‐DR isotype (HLA‐DR)‐positive microglia/macrophages infiltrated in the substantia nigra (SN) (McGeer et al., 1988). Subsequent studies reported elevated levels of pro‐inflammatory cytokines and proteins including interleukin 1 beta (IL1β), IL6, tumor necrosis factor alpha (TNFα) (Knott et al., 2000; Mogi, Harada, Kondo, et al., 1994; Mogi, Harada, Riederer, et al., 1994), complement activation (Loeffler et al., 2006) and T lymphocyte infiltration (Brochard et al., 2009) in the SN of postmortem tissue. An increasing body of evidence suggests that early changes in innate and adaptive immune cells are key drivers in PD pathogenesis (Harms et al., 2021). Furthermore, the associations between single‐nucleotide polymorphisms in the MHC‐II regions and the risk for PD (Ahmed et al., 2012; Hamza et al., 2010; International Parkinson Disease Genomics Consortium et al., 2011; Wissemann et al., 2013), may influence CD4 T cell activation towards a pro‐inflammatory response explaining the influence of the environment in the predisposition of individuals to PD (Kannarkat et al., 2015). In this regard, a higher incidence of PD is observed in patients with inflammatory bowel disease, which can be reduced with early anti‐TNF therapy (Peter et al., 2018). These data support a role of inflammation in the pathogenesis of PD.
In the absence of infection, a sterile inflammatory response is triggered by self‐derived danger signals called damage‐associated molecular patterns (DAMPs), which are primarily released from injured tissue. They are sensed through specialized pattern‐recognition receptors promoting the release of pro‐inflammatory mediators with the subsequent activation of peripheral immune cells (Banjara & Ghosh, 2017). Microglia and astrocytes are the major brain cells that express innate immune receptors and release pro‐inflammatory cytokines and chemokines when stimulated by DAMPs. Alpha‐synuclein (αSyn) is a key protein involved in PD (Ibáñez et al., 2004; Nalls et al., 2014; Polymeropoulos et al., 1997; Zarranz et al., 2004) that could act as a DAMP through the activation of toll‐like receptors (TLR) leading to chronic inflammation and exacerbating neuronal dysfunction and loss (Béraud et al., 2011; Harms et al., 2021; Kouli et al., 2019; Zhang et al., 2005). The same process that initiates a pro‐inflammatory response activates microglial phagocytic activity, which is necessary to promote αSyn clearance (Fellner et al., 2013), suggesting that it may be neuroprotective. Thus, microglia reactivity in the context of neurodegeneration may have different effects on neuronal survival.
The presence of specialized subsets of microglia to serve a broad range of biological roles has been known for many years. The development of single‐cell analysis has revealed the spatial, temporal, and functional diversity of microglia during development, homeostasis and disease (Masuda et al., 2020). Although with some controversy, microglia in adulthood is considered quite homogeneous showing a transcriptional continuum across the CNS regions rather than different subclasses (Grabert et al., 2016; Hammond et al., 2019; Keren‐Shaul et al., 2017; Li et al., 2019; Masuda et al., 2019). The homogeneity of homeostatic microglia is disrupted under pathological conditions, switching gene expression to react to the surrounding alterations (Keren‐Shaul et al., 2017; Masuda et al., 2019; Sousa et al., 2018). From studies using different experimental models of disease it is possible to conclude that there are disease/condition‐specific microglial activation states (Masuda et al., 2020). The functional diversity of astrocytes beyond their morphological classification, fibrous or protoplasmic, is being addressed with studies reporting heterogeneity between and within brain regions (Batiuk et al., 2020; Ben Haim & Rowitch, 2017; Chai et al., 2017; Khakh & Deneen, 2019; Khakh & Sofroniew, 2015). Astrocyte reactivity in response to inflammation or damage shows a pronounced context‐dependent heterogeneity across central nervous system. Modulation of this reactivity alters disorder outcome (Burda et al., 2022).
In this regard, we have previously shown that the inflammatory tone in homeostasis differs within the midbrain and the striatum. The reaction of microglia and immune cell infiltration to peripheral lipopolysaccharide administration shows a clear immunosuppressive response in the midbrain compared to the striatum (Abellanas et al., 2019). This prompted us to explore the specific inflammatory reaction at the level of innate immune cells that takes place in the midbrain and in the striatum in the context of αSyn‐dependent dopaminergic degeneration. We hypothesized that the neurodegenerative environment, the loss of neuronal cell bodies in the midbrain versus dopaminergic terminals in the striatum, would trigger a differential inflammatory reaction. To generate the PD experimental mouse model, we prepared an adeno‐associated virus serotype 9 (AAV9) that overexpressed αSyn and mCherry and an AAV9 overexpressing only mCherry for control animals. In this study, we describe a switch in the activation of microglia and astrocytes in the midbrain and in the striatum of animals overexpressing αSyn. Interestingly, the activation pattern differs between regions suggesting that the design of immunomodulatory therapies aimed to prevent dopaminergic degeneration should consider this factor.
2. MATERIAL AND METHODS
2.1. Animals
Adult male 3‐month‐old C57BL6JRccHsd (20–30 g) were obtained from Envigo (Barcelona, Spain). Mice were housed at 21°C in a humidity‐controlled environment on a 12 h light/dark cycle, fed ad libitum with standard rodent pellet diet (Envigo) and free access to water. All procedures involving animals were carried out in accordance with the Spanish National Research Council's guide for the care and use of laboratory animals following the protocols approved by the Ethical Committee for Animal Testing at the University of Navarra (ref. 064‐19).
2.2. Virus generation
To generate AAV9‐Syn‐IRES‐mCherry and AAV9‐IRES‐mCherry viral expression plasmids, pCAAGs‐IRES‐Ch and pCAAGs‐Syn‐IRES‐Ch plasmid versions were first generated. mCherry was amplified by PCR from pCAGGs‐mCherry with the oligonucleotides FSmaI: GCTTACCCGGGATCATTTTGGCAAAGAATTCCATGGTGAGC and RSacI: AGTGAATTGAGCTCGATATCGGTACC. The PCR product was digested with SmaI/SacI and subcloned into pCAGGs‐IRES‐GFP, pCAGGs‐Syn‐IRES‐GFP (Íñigo‐Marco et al., 2017) replacing GFP. Next, pCAAGs‐IRES‐mCherry and pCAAGs‐Syn‐IRES‐mCherry plasmids were digested with SpeI, treated with Klewow and further digested with BglII. The inserts containing the CAG promoter (the ubiquitous cytomegalovirus major immediate‐early enhancer combined with the chicken beta‐actin promoter) and Syn‐IRES‐mCherry or IRES‐mCherry were ligated into the pAAV‐MCS plasmid previously digested with MluI, Klenow treated and secondly digested with BglII. All plasmids were verified by DNA sequencing.
AAV9 vectors were produced by polyethylenimine (PEI) (Polysciences, Warrington, PA) mediated co‐transfection of a mixture of 20 μg of specific‐pAAV plasmid and 55 μg of pDP9 (Kindly provided by Dr Mueller, DKFZ, Germany) in HEK293 cells. The supernatant was collected after 72 h and treated with polyethylene glycol solution (PEG8000, 8% vol/vol final concentration) for 48–72 h at 4°C and centrifuged at 3000 rpm for 15 min. PEG‐pellet and harvested cells were resuspended in lysis buffer (50 mM Tris–HCl, 150 mM NaCl, 2 mM MgCl2, 0.1% Triton X‐100) and subjected to three cycles of freeze and thaw by using a dry ice/ethanol bath. After centrifugation, purification was performed by ultracentrifugation in iodixanol gradients as described (Zolotukhin et al., 1999). Viral batches were further concentrated by using centricon tubes (YM‐100; Millipore, Burlington, MA) and stocks were kept at –80°C until use. Viral titers (viral genomes [vg]/ml) were determined by quantitative‐PCR for viral genome copies extracted from DNase‐treated viral particles (High Pure Viral Nucleic Acid Kit, Roche, Basel, Switzerland). Quantitative PCR was performed in triplicate with primers for the ITR region of AAV (D'Costa et al., 2016).
2.3. Stereotaxic surgery
Mice were deeply anesthetized with ketamine (75 mg/kg) and xylacine (10 mg/kg) and placed in a stereotaxic frame (Kopf Instruments, Tujunga, CA). Animals were bilaterally injected with 1 μl of AAV9‐Control and AAV9‐Syn (5.5 × 1012 vg/ml) at a rate of 0.2 μl/min into the substantia nigra pars compacta (SNpc) using a 10 μl Hamilton Neuros syringe (model 1701 RN; Hamilton, Reno, NV) and a pump (Stoelting Co, Wood Lane, IL). The coordinates of the SNpc were calculated with respect to bregma using the atlas of Paxinos and Watson (Paxinos & Franklin, 2001): anteroposterior −3.5 mm, mediolateral +/− 1.3 mm and dorsoventral −4 mm. Before and after administration of the AAV, the needle was left for 3 min at the injection site.
2.4. Motor behavior
Behavioral tests were performed 24 h prior sacrifice under low light conditions. For the pole test, animals were placed heading up on the top of a vertical wooden pole of 50 cm height and 1 cm in diameter covered with a bandage. Animals were pre‐trained before the surgery until they were able to turn the head down and to descend from the pole in less than 5 s. The average time to turn the head down and to completely descend the pole was measured in three trials with a resting time of 15 min between them. For the catalepsy bar test, mice were placed with their forepaws on a bar oriented parallel to the ground at 4 cm height. The average time to correct the posture was measured in three trials.
2.5. Histological techniques
For immunochemistry on free‐floating sections, animals were anesthetized with ketamine (75 mg/kg) and xylacine (10 mg/kg) and transcardially perfused during 5 min with Ringer's solution (145.4 mM NaCl, 3.4 mM KCl, 2.4 mM NaHCO₃, pH 7.4) at a rate of 9.5 ml/min, followed by 4% paraformaldehyde (PFA; Panreac, Barcelona, Spain) in 0.125 M phosphate buffered saline (PBS, pH 7.4) for 10 min at the same rate and three additional minutes at 16 ml/min. The brain was removed, post‐fixed overnight in 4% PFA, and stored in 30% sucrose/PBS. Coronal 40 μm thick sections were obtained using a Leica SM2000R sliding microtome (Leica, Wetzlar, Germany). Free‐floating sections were washed with PBS and endogenous peroxidase activity was inactivated by incubation in 0.03% H2O2 (Sigma‐Aldrich)/methanol (Panreac) for 30 min. After washing three times with PBS, the tissue was incubated, first with the blocking solution (4% normal goat serum and/or normal donkey serum, 0.05% Triton X‐100 [Sigma‐Aldrich] and 4% BSA [Merck, Darmstadt, Germany] in PBS) for 40 min, and then with primary antibodies diluted in blocking solution at room temperature (RT) overnight. The primary antibodies used were: rabbit anti‐tyrosine hydroxylase (TH; 1:1000; Merck Millipore), sheep anti‐TH (1:1000; Abcam, Cambridge), mouse anti‐mCherry (1:500; Abcam), mouse anti‐αSyn (1:500; Life Technologies, Carlsbad, CA), guinea pig anti‐Iba1 (1:1000; Synaptic Systems, Göttingen, Germany), rabbit anti‐Pu.1 (1:200; Cell Signaling, Technology, Danvers, MA), rabbit anti‐Ki67 (1:200; Thermo Fisher Scientific, Waltham, MA), rabbit anti‐TMEM119 (1:400; Cell Signaling) and mouse anti‐GFAP (1:500; Cell Signaling, Technology, Danvers, MA). For colorimetric immunostaining, sections were incubated with the biotinylated secondary antibodies goat anti‐rabbit (1:500; Jackson ImmunoResearch, Ely) and goat‐anti mouse (1:500; Jackson ImmunoResearch) in blocking solution for 2 h at RT, followed by incubation with peroxidase‐conjugated avidin in PBS (1:5000; Sigma‐Aldrich) during 90 min at RT. After washing with PBS, the tissue was incubated in 0.05% diaminobenzidine (Sigma‐Aldrich), 0.03% H2O2 and Trizma‐HCl buffer (pH 7.6). For immunofluorescence staining, sections were incubated for 2 h at RT with the following secondary antibodies diluted in blocking solution: Alexa Fluor 488 donkey anti‐sheep (1:500; Jackson ImmunoResearch), Alexa Fluor 568 donkey anti‐mouse (1:500; Thermo Fisher Scientific), Alexa Fluor 594 goat anti‐guinea pig (1:500; Invitrogen, Carlsbad, CA), Alexa Fluor 647 goat anti‐rabbit (1:250; Invitrogen), Alexa Fluor 488 donkey anti‐rabbit (1:250; Invitrogen) and Alexa Fluor 546 donkey anti‐mouse (1:250; Invitrogen) and finally stained with DAPI (1:50,000; Sigma‐Aldrich). Sections were mounted on glass slides in a 0.2% solution of gelatin in 0.05 M Tris–HCl buffer (pH 7.6) (Sigma‐Aldrich), dried and dehydrated in toluene (Panreac) for 12 min before coverslipping with DPX (BDH Chemicals, Poole).
2.6. Image analysis
The number of TH+ neurons present in the SNpc was determined by unbiased design‐based stereology using a Bx61 microscope (Olympus, Hicksville, NY) equipped with a camera DP71 (Olympus,), a stage connected to a xyz stepper (H101BX, PRIOR) and using the Stereo Investigator software (version 2021.1.1; MBF Bioscience, Williston, VT). Stereological counting was performed in 6 or 7 coronal SNpc sections (40 μm thick) taken at uniform intervals (160 μm) that covered the entire rostrocaudal extent of the nucleus between −2.92 and −3.64 mm relative to bregma (Paxinos & Franklin, 2001). The reference volume (Vr) of the SNpc was calculated from images obtained with the 2× objective using a point count array according to Cavalieri principles (Gundersen & Jensen, 1987). The cross‐sectional area of the nucleus was measured and the Vr for the entire SNpc was estimated using the following equation:
where T is the section thickness, a/p is the area of each point, and Pi is the number of points falling within the SNpc. The SNpc was outlined with the 10× objective to estimate the area. The number of labeled neurons was obtained at 100× magnification under oil immersion, using randomized meander sampling and the optical dissector methods. The optical dissector height was 11 μm, keeping an upper guard zone of 2 μm, to count 100–150 cells per animal using a sampling frame of 4900 μm2 and sampling steps of 140 μm × 140 μm (dx, dy). Unbiased counting was performed blindly and the total number of TH‐positive neurons (N) was calculated using the following formula:
where ∑Q− is the total number of particles counted, t is the mean section thickness, h is the height of the optical dissector, asf is the area sampling fraction, and ssf is the section sampling fraction. Neuronal density (D) was determined using the following formula: D = N/Vr. Gunderson's coefficients of error were <0.1 for all stereological quantifications.
Images for immunostained sections of TH and αSyn were acquired on an Aperio CS2 Digital Pathology Slide Scanner (Leica) at a 20× magnification. Optical density values of striatal TH and αSyn immunoreactivity were obtained using ImageJ (National Institutes of Health, MD). For αSyn staining analysis, a random region of the cortex was used as blank, and its value was subtracted to the average intensity of both hemispheres. For the analysis of TH, a random region of the cortex was used as blank and its value was subtracted to the intensity of the most degenerated hemisphere. Confocal images of double/triple staining TH/mCherry, TH/Iba1/Pu.1, TH/GFAP, Iba1/Ki67, and TMEM119/Iba1 immunofluorescences were acquired on an LSM510 confocal microscope (Zeiss, Jena, Germany) using the 20× and 63× oil objectives. A projection stack with the same number of images per slice was analyzed to calculate the number of Iba1+/Pu.1+, Iba1+/Ki67+, and TMEM119+/Iba1+/DAPI+ cells. For the morphological analysis of Iba1+ cells 4 types of morphologies previously described were selected (Wyatt‐Johnson et al., 2017): ramified, hypertrophic, bushy and amoeboid. Ramified microglia are characterized by long, thin processes and a small cell body. Hypertrophic morphology is characterized by long, thicker processes and a bigger cell body. Bushy microglia have a big cell body and short processes. Amoeboid microglia have a macrophage‐like morphology with few or no processes. This classification was made manually from two stacks obtained from each region in each hemisphere. An ImageJ plugin was used to calculate the volume of TH+ signal comprised in Iba1+ signal and in GFAP+ signal.
2.7. Cell suspensions
Mice were anesthetized with ketamine/xylazine and perfused transcardially with ice‐cold PBS. The striatum and midbrain were dissected on ice and incubated with 2 mg/ml of papain (Worthington, Lakewood, NJ) or 400 units/ml of collagenase D (Roche, Mannheim, Germany), containing 50 μg/ml of DNase I (Sigma‐Aldrich) in Dulbecco's PBS (Lonza, Basel, Switzerland) for 30 or 15 min, respectively, at 37°C in rotation. After enzymatic digestion, tissue was mechanically processed with a glass Pasteur pipette, filtered through a 70 μm nylon cell strainer and centrifuged at 300 g for 15 min. A 25% Percoll gradient was used to remove cell debris and myelin by centrifugation at 1000 g for 10 min at RT. The cell pellet was resuspended in the appropriate buffer for flow cytometry or RNA sequencing (RNA‐seq).
2.8. Flow cytometry
For the analysis of cell surface markers, cell pellets obtained from the brain were resuspended in 100 μl of cytometry buffer (CB): 5 mM EDTA (Thermo Fisher Scientific), 0.5% FBS (Gibco, Paisley), 100 U/ml penicillin G (Gibco), 100 μg/ml streptomycin (Gibco) in PBS and were incubated with Zombie NIR Dye (1:2000; BioLegend) in PBS to assess their viability for 5 min at RT. The Zombie NIR Dye was quenched with CB and cells were centrifuged at 2000 rpm for 1 min. Then, samples were incubated with different panels of fluorescent antibodies (Table 1) and the FcR Blocking Reagent (1:50; Miltenyi Biotec, Bergisch Gladbach, Germany) during 15 min at 4°C. Once labeled, samples were washed with CB, acquired on a CytoFLEX LX flow cytometer (Beckman Coulter, Brea, CA) and analyzed using the CytExpert 2.3 (Beckman Coulter) and FlowJo 10.0.7r2 (BD Biosciences, Franklin Lakes, NJ) softwares.
TABLE 1.
Fluorescent primary antibodies for flow cytometry staining
| Antigen | Fluorophore | Dilution | Clone | Trademark |
|---|---|---|---|---|
| CD11b | BV510 | 1:500 | M1/70 | BioLegend |
| CD45 | BV421 | 1:1000 | 30F11 | BioLegend |
| TLR4 | PE‐Cy7 | 1:500 | SA15‐21 | BioLegend |
| MHC‐II | PE | 1:1500 | AF6‐120.1 | BioLegend |
| CD80 | APC | 1:100 | 16‐10A1 | BioLegend |
2.9. RNA sequencing and analysis
Cells obtained from the striatum and from the midbrain were resuspended in 100 μl of CB and incubated with FcR Blocking Reagent (1:10; Miltenyi Biotec) and CD11b MicroBeads (1:10; Miltenyi Biotec) in CB. Microglial CD11b+ cells were separated in an autoMACS Pro Separator (Miltenyi Biotec). The CD11b− fraction was collected and incubated with FcR Blocking Reagent (1:10; Miltenyi Biotec) and ACSA2 MicroBeads (1:10; Miltenyi Biotec) in CB and separated in an autoMACS. CD11b+ and ACSA2+ cells were pelleted and resuspended in the lysis/binding buffer from the Dynabeads mRNA Direct Kit (Ambion, Foster City, CA), and stored at −80°C for further processing. An aliquot of the separated cells was stained with CD11b‐PE (1:100; Miltenyi Biotec), CD45‐FITC (1:50; Miltenyi Biotec), and ACSA2‐APC (1:50; Miltenyi Biotec). RNA sequencing (RNA‐seq) was performed using MARS‐seq adapted for bulk RNA‐seq (Jaitin et al., 2014; Lavin et al., 2017) with minor modifications. Briefly, poly‐A RNA was extracted with Dynabeads Oligo (dT) (Thermo Fisher Scientific) and reverse transcribed with AffinityScript Multiple Temperature Reverse Transcriptase (Agilent Technologies, Santa Clara, CA) using poly‐dT oligos carrying a 7‐base pair (bp) index. Upon indexing, samples were pooled and subjected to linear amplification using HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs, Ipswich, MA). The resulting antisense RNA was fragmented into 250–350 bp fragments using RNA Fragmentation Reagents (Thermo Fisher Scientific) and dephosphorylated for 15 min at 37°C with 1 U FastAP (Thermo Fisher Scientific). Partial Illumina adaptor sequences (Jaitin et al., 2014) were ligated to the fragments with T4 RNA Ligase 1 (New England Biolabs) and reverse transcription was repeated. Full Illumina adaptor sequences were added during library amplification with KAPA HiFi DNA Polymerase (Kapa Biosystems, Wilmington, MA). The libraries were then quantified using a Qubit 3.0 Fluorometer (Life Technologies), and their size profiles were examined in an Agilent 4200 TapeStation System. Libraries were sequenced in an Illumina NextSeq 500 instrument at a sequence depth of 10 million reads per sample. The Kallisto pseudoaligner (version 0.46) was applied for Mus musculus transcriptome quantification. First, the fasta file of the GRCm38.p6 assembly was used to create the index file. Then, measurement of the expression was carried out with quant mode. Applied parameters were: single‐end, 100 bootstrap, length = 68 and standard deviation = 20. The identification of differentially expressed transcripts was carried out in sleuth. Transcripts with less than 50 counts in 50% of the samples were excluded for the analysis. A cutoff p‐value <.01 was applied to select the most relevant transcripts. Enrichment analysis was carried out using the Ingenuity Pathway Analysis expression analysis and gene set enrichment analysis (GSEA). The data are publicly available in NCBI's Gene Expression Omnibus (GEO) (Edgar et al., 2002), and are accessible through GEO Series accession number GSE191131.
2.10. Polymerase chain reaction
CD11b−ACSA2− cells obtained after magnetic bead separation were analyzed for the expression of mCherry to confirm correct AAV administration. Poly‐A RNA was selected with Dynabeads Oligo (dT) (Ambion) and reverse‐transcribed with AffinityScript Multiple Temperature Reverse Transcriptase (Agilent) using poly‐dT oligos. The mRNA expression was studied by semi‐quantitative real‐time PCR using iQ SYBR Green Supermix (Bio‐Rad, Hercules, CA) in a CFX96 Touch real‐time detection system (Bio‐Rad). The following primers were used: mCherry forward‐ CCCCGTAATGCAGAAGAAGA, mCherry reverse‐TTGACCTCAGCGTCGTAGTCG; β‐actin forward‐CCTGACAGACTACCTCATG, β‐actin reverse‐CCATCTCTTGCTCGAAGTCT. Each PCR was carried out in duplicates to obtain an average Ct value. The results were normalized to β‐actin, and the amount of each transcript was expressed as 2ΔCt (ΔCt = Ct [β‐actin] − Ct [gene]).
2.11. Statistics
GraphPad Prism version 7.0 was used to create the graphs. All data are represented as mean with 95% confidence intervals (CI). The normal distribution of data was analyzed with a Shapiro–Wilk test. Pairwise comparisons of data following a normal distribution were analyzed with a Student's t‐test (two‐tailed) for equal variances. If variances were significantly different, the Welch's correction was applied. Data not following a normal distribution were analyzed with Mann–Whitney U test. For multiple comparisons the two‐way ANOVA followed by Bonferroni's test was used.
3. RESULTS
3.1. Differential activation of microglia and astrocytes in the striatum and in the midbrain of mice overexpressing αSyn
To study the neuroinflammatory response triggered by the dopaminergic degeneration dependent of αSyn, we generated an AAV9 that expressed simultaneously αSyn, under the control of the CAG promoter, and mCherry, under the control of an internal ribosome entry site (IRES) (AAV9‐Syn). The control AAV9 lacked αSyn but maintained the IRES and the mCherry sequence (AAV9‐Control) (Figure 1a). In this way, infected neurons would overexpress mCherry, a non‐toxic protein (Íñigo‐Marco et al., 2017). The AAV9s were administered bilaterally in the SNpc by stereotaxic surgery and animals were sacrificed at 2 and 4 weeks post‐transduction (Figure 1b). Mice presented evident motor deficits in the pole and in the catalepsy test, at 2 (Figure 1c) and at 4 weeks (Figure 1d) indicating that overexpression of αSyn but not mCherry was responsible for the motor impairment. The mCherry protein was predominantly expressed in dopaminergic cell bodies positive for tyrosine hydroxylase (TH+) located in the SNpc of mice that received the AAV9‐Control, but not in the dopaminergic terminals in the striatum (Figure S1a). Due to the degeneration of the dopaminergic neurons, mCherry expression was lower in the animals injected with the AAV9‐Syn (Figure S1a). Furthermore, the immunohistochemistry to detect the human αSyn showed that the protein expressed by dopaminergic neurons was successfully delivered to the presynaptic terminals in the striatum (Figure S1b). The integrity of the neurons constituting the nigrostriatal pathway was analyzed by tyrosine hydroxylase (TH) immunohistochemistry. A progressive loss of TH+ terminals in the striatum was observed from 2 to 4 weeks (Figure 1e), which was accompanied by a significant death of TH+ neurons in the SNpc, very prominent at 2 weeks and maintained at 4 weeks (Figure 1f). Altogether, these results show that αSyn is efficiently expressed by the AAV9 in neurons, inducing a progressive loss of the nigrostriatal pathway.
FIGURE 1.
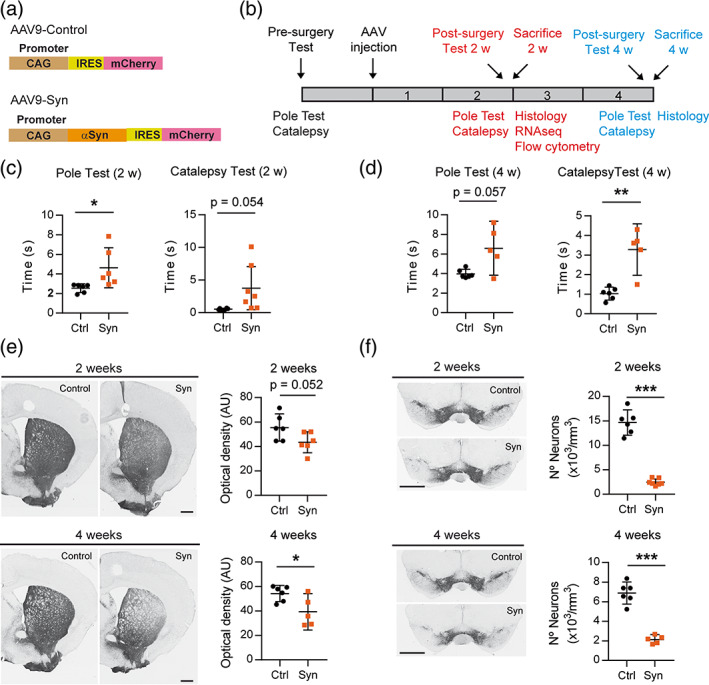
Overexpression of αSyn by the injection of AAV9 in the SN induces motor deficits and nigrostriatal degeneration in mice. (a) Two AAV9s were prepared, the AAV9‐Control that overexpresses the protein mCherry and the AAV9‐Syn virus that co‐expresses independently αSyn and mCherry in the infected neurons. The expression of both proteins was regulated by an ubiquitous promoter. (b) Schedule of AAV9 administration to mice, motor tests and procedures performed at 2 and 4 weeks (2 and 4 weeks, respectively). (c) Motor behavior evaluated at 2 weeks after AAV9 injection. Motor coordination was analyzed in the pole test quantifying the time required by the mice to turn down and descend the pole. Cataleptic behavior was evaluated in the bar test by measuring the time that the mice took to recover the position of the upper paws. (d) Motor behavior evaluated in the pole and in the bar test at 4 weeks. (e) Representative photomicrographs showing TH‐immunoreactivity in the striatum and photomicrographs showing TH+ dopaminergic terminals at 2 and 4 weeks. (f) Representative photomicrographs showing TH+ dopaminergic cell bodies in the midbrain and quantification of the positive cells at 2 and 4 weeks. The data represent the mean ± 95% CI from 5 to 6 animals per group. Statistical analysis: (c and d) Welch's test, (e and f) t‐test. *p < .05, **p < .01, ***p < .001. Magnification bar: (e and f) 1 mm
To determine the type of glial activation generated by αSyn‐dependent neuronal loss, mice were sacrificed at 2 weeks post‐transduction, in the phase of the active neurodegenerative process. The midbrain and the striatum were dissected out from animals that showed a motor deficit in the pole and in the catalepsy test (Figure S2a). A cell suspension for each region was prepared and myeloid cells and astrocytes were subsequently separated based on the CD11b and ACSA2 cell surface expression, respectively (Figure S2b). The correct transduction of the AAV9 was checked by analyzing the expression of mCherry by PCR in the CD11b−ACSA2− negative fraction, which contains the neuronal cell bodies. The control protein was detected in the midbrain but not in the striatum, indicating a correct viral infection (Figure S2c). The midbrain and striatal transcriptomes of each cell type were examined independently by RNA‐seq. As a result of the nigrostriatal degeneration, both regions showed a different microglial activation profile with an overlap of 5 common differentially expressed genes (Hspb3, Rps9, Tmem147, Pik3ip1, and H2‐Aa) (Figure 2a). The network generated with Ingenuity Pathway Analysis for the midbrain showed a clear anti‐inflammatory response and an upregulation of the liver X receptor (LXR)/retinoid X receptor (RXR) pathway which is involved in the phagocytosis of apoptotic cells (Blander, 2017; Savage et al., 2015) (Figure 2b), while the striatal network showed a clear pro‐inflammatory profile predicting a central role for interferon gamma (IFNγ)‐derived responses (Figure 2c). We further investigated microglial phenotypes by gene set enrichment analysis (GSEA). To determine whether CD11b+ cells presented a neurodegenerative phenotype, two molecular signatures were selected, one for neurodegenerative microglia (MGnD) which was established from different mouse models of neurological diseases, including amyotrophic lateral sclerosis (ALS), Alzheimers disease (AD), and multiple sclerosis (MS) (Deczkowska et al., 2018), and another one for disease‐associated microglia (DAM) obtained from an AD transgenic mouse model (Keren‐Shaul et al., 2017). Midbrain CD11b+ cells in parkinsonian mice showed a significant enrichment in genes downregulated in the MGnD and DAM signatures (Figure 2d) which are associated with the loss of homeostatic microglial genes (Deczkowska et al., 2018; Keren‐Shaul et al., 2017). In addition, an overlap with transcripts downregulated in the phagocytic MGnD signature but not with a gene ontology biological process (GOBP) annotation for phagocytosis was observed (Figure 2d). By contrast, striatal microglia in parkinsonian mice exhibited a DAM but not a MGnD phenotype and presented a clear overlap with phagocytic genes in the GOBP but not in the MGnD signature (Figure 2e). The analysis performed with the M1/M2 fingerprint (Coates et al., 2008) indicated an anti‐inflammatory profile for midbrain microglia/macrophages (Figure 2f), and no significant enrichment in M1 or M2 genes for striatal cells under αSyn‐dependent degeneration (Figure 2g). The analysis of the astrocytic (ACSA2+ cells) transcriptome showed that most of the differentially expressed genes corresponded to midbrain cells while degeneration of dopaminergic terminals seemed to have a low impact on striatal transcripts (Figure 3a). The network generated in the midbrain of parkinsonian mice foresees the involvement in pro‐inflammatory responses mediated by TNFα, IFNγ, and IL6 (Figure 3b). This observation was confirmed by the significant enrichment in lipopolysaccharide (LPS)‐induced astrogliosis genes (Figure 3c). These results suggest that the death of neuronal bodies in the midbrain induces an inflammatory tone that is cell‐specific, anti‐inflammatory in microglia, and pro‐inflammatory in astrocytes. Microglia from both regions show a neurodegenerative and phagocytic phenotype, midbrain microglia are closer to the MGnD signature, probably because the signature was generated from experimental models that have neuronal death. By contrast, striatal microglia, which are exposed to degenerating terminals but not to neuronal cell bodies, show a different phagocytic profile and a neurodegenerative fingerprint closer to an AD mouse model with minimal neuronal degeneration.
FIGURE 2.
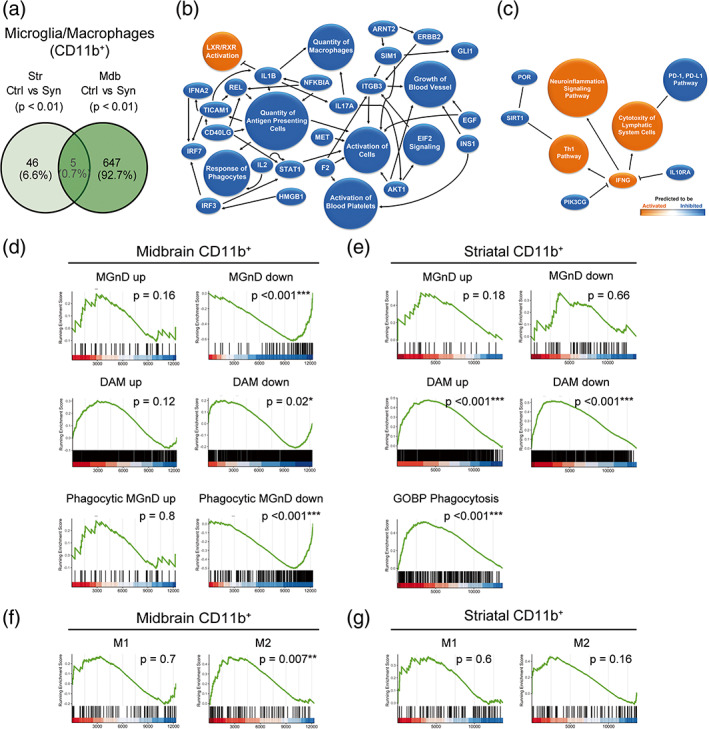
Transcriptomic analysis of myeloid cells purified from the striatum and the midbrain. Animals were sacrificed at 2 weeks after AAV9 administration in the SNpc. The striatum and the midbrain were dissected out and a cell suspension was prepared from these regions. Myeloid cells were separated based on the CD11b expression for RNA sequencing. (a) Venn diagram showing overlap of differentially expressed genes (p < .01) between CD11b+ cells from control and αSyn mice of the two regions. (n = 3 animals/group). (b) Graphical summary of the pathways, upstream regulators, and biological functions predicted to be altered in CD11b+ cells from the midbrain of αSyn mice. (c) Graphical summary of the pathways, upstream regulators, and biological functions predicted to be altered in CD11b+ cells from the striatum of αSyn mice. (d) Gene set enrichment analysis (GSEA) plots of the signatures enriched in midbrain myeloid cells of αSyn mice: Neurodegenerative microglia (MGnD), disease‐associated microglia (DAM) and phagocytic MGnD. GSEA was performed with the upregulated (“signature” up) and downregulated (“signature” down) genes of the signatures separately. (e) GSEA plots of the MGnD, DAM, and GOBP phagocytosis signatures enriched in striatal myeloid cells of αSyn overexpressing mice. GSEA was performed with the upregulated (“signature” up) and downregulated (“signature” down) genes of the MGnD and DAM signatures separately. (f) GSEA plots of the M1/M2 enrichment analysis in midbrain and (g) striatal myeloid cells of αSyn mice.
FIGURE 3.
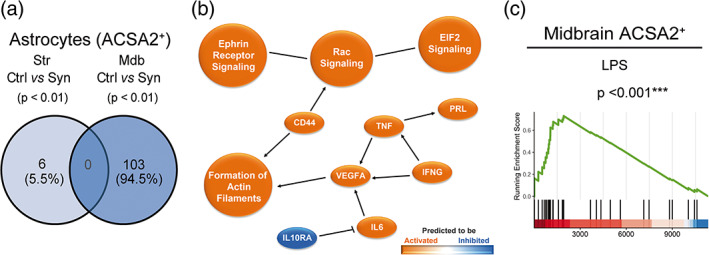
Transcriptomic analysis of astrocytes purified from the striatum and the midbrain of αSyn overexpressing mice. Animals were sacrificed at 2 weeks after AAV9 administration in the SNpc. The striatum and the midbrain were dissected out and a cell suspension was prepared from these regions. Astrocytes were separated based on ACSA2 expression for RNA sequencing. (a) Venn diagram showing overlap of differentially expressed genes (p < .01) between ACSA2+ cells from control and αSyn mice of the two regions(n = 3 animals/group). (b) Graphical summary of the pathways, upstream regulators and biological functions predicted to be altered in ACSA2+ cells from the midbrain of αSyn mice. (c) GSEA plot of the LPS reactive astrocyte signature enriched in midbrain astrocytes of αSyn mice.
3.2. Alpha‐synuclein‐dependent neurodegeneration promotes the appearance of cells with a CD11b + CD45high phenotype in the brain parenchyma
Next, we examined myeloid cell activation in the midbrain and in the striatum by flow cytometry. Cell populations were analyzed independently based on the cell surface expression of CD45 (Figure 4a). The CD11b+CD45low gate was ascribed to resident microglia and the CD11b+CD45high to infiltrated myeloid cells and resident microglia that under pathological conditions are known to upregulate CD45 expression (Gate et al., 2010; Honarpisheh et al., 2020). The proportion of CD11b+CD45low cells among total viable cells decreased significantly in the midbrain of parkinsonian animals compared to animals receiving the AAV9‐Control (Figure 4b). The decreased ratio could be due to both, to the increase in the number of cells caused by the prominent immune cell infiltration, and/or to an upregulation of CD45 expression. No variation in the proportion of CD11b+CD45low cells was detected in the striatum. The CD11b+CD45high subset was much more abundant in the midbrain (≈10%) than in the striatum (≈0.5%). In both regions, its proportion increased in αSyn overexpressing animals (Figure 4c). The absence of changes in the proportion of Iba1+ cells that express Ki67 suggests a peripheral myeloid infiltration (Figure S3). The cell surface expression of the activation marker TLR4 decreased significantly in CD11b+CD45low cells (Figure 4d) and in midbrain CD11b+CD45high cells (Figure 4e). The striatal CD11b+CD45high subpopulation presented lower expression levels of TLR4 than midbrain cells, which were preserved under parkinsonian conditions (Figure 4e). These observations are consistent with the downregulation of the pro‐inflammatory‐related transcripts found in the RNA‐seq analysis of midbrain CD11b+ cells (e.g., Tlr3, Tlr4, Casp1, C3, and Il1a). An upregulation of the MHC‐II expression was detected in CD11b+CD45low cells (Figure 4f) and in striatal CD11b+CD45high cells (Figure 4g), but not in the midbrain CD11b+CD45high subpopulation (Figure 4g). The differential regulation of median fluorescence intensity (MFI) MHC‐II in midbrain CD11b+CD45high cells could be due to the high basal values presented by this subpopulation. In the midbrain, the co‐stimulatory molecule CD80 increased in the cell surface of CD11b+CD45low (Figure 4h) and in CD11b+CD45high cells (Figure 4i), while in the striatum, this increase was restricted to the CD11b+CD45high subpopulation (Figure 4i). In summary, our data indicate that the expression of αSyn in the midbrain shifts the balance from CD11b+CD45low to CD11b+CD45high. In the striatum of αSyn mice, an increase in the CD11b+CD45high cell population is also observed. Both populations from both regions experience similar changes in the expression of inflammation‐related molecules, such as TLR4, MHC‐II, and CD80.
FIGURE 4.
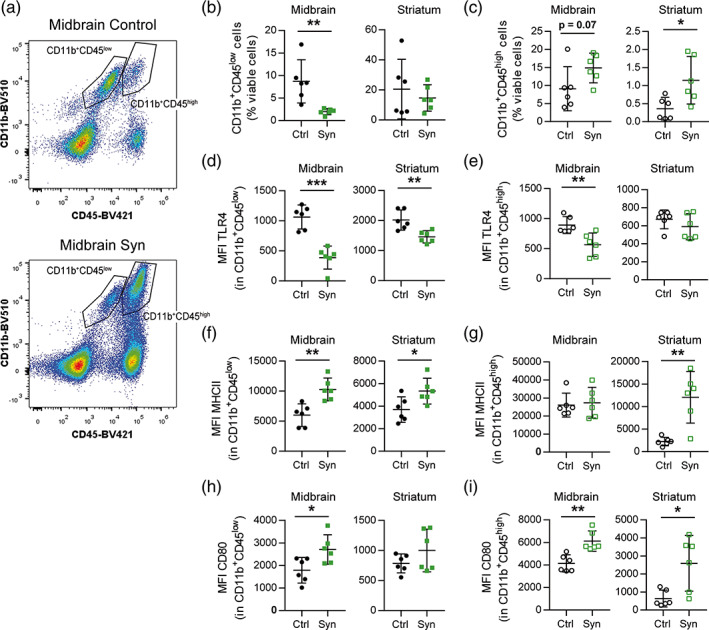
Recruitment of myeloid cells and modulation of cell surface marker expression in the midbrain. Animals were sacrificed at 2 weeks after AAV9 administration in the SNpc. The striatum and the midbrain were dissected out and a cell suspension was prepared from these regions for flow cytometry analysis. (a) Gating strategy for CD11b+CD45low and CD11b+CD45high myeloid cells in control and αSyn mice. (b) Frequency of CD11b+CD45low and (c) CD11b+CD45high cells out of viable cells in the midbrain and striatum of control and αSyn mice. (d) MFI of TLR4 in CD11b+CD45low and (e) CD11b+CD45high cells. (f) MFI of MHCII in CD11b+CD45low and (g) CD11b+CD45high cells. (h) MFI of CD80 in CD11b+CD45low and (i) CD11b+CD45high cells. The data represent the mean ± 95% CI from 6 animals per group. Statistical analysis: (b, d, e, f, and h) t‐test, (c) midbrain and (i) Mann–Whitney test and (c) striatum and (g) Welch's t‐test. *p < .05, **p < .01, ***p < .001
3.3. Phagocytic capacity of myeloid cells
Next, we explored the morphology and the phagocytic state of microglia/macrophage cells. Using Iba1 immunostaining, it was possible to detect myeloid cells with different morphologies. In the SNpc, the ramified phenotype was the predominant form in control animals together with a few hypertrophic and bushy cells (Figure 5a,b), probably due to the transduction of the AAV9‐Control in this region. By contrast, the density of Iba1+ cells increased significantly in the SNpc injected with the AAV9‐Syn, mainly caused by the appearance of a new small sized amoeboid subset of cells not detected in control animals (Figure 5a,b). These were the most abundant Iba1+ cells in the degenerating SNpc and may correspond to the CD11b+CD45high population detected by flow cytometry. The amoeboid Iba1+ cells were located at the site of neuronal death, participating actively in the phagocytosis of TH+ cells (Figure 5b, Syn field 1). In the same mice, the bushy and hypertrophic Iba1+ cells predominated in regions where TH+ cells presented an apparently healthy morphology, in some cases surrounding TH+ neurons. They also contained TH+ phagocytic vesicles in their cytoplasm (Figure 5b, Syn field 2). In the striatum, the density and the morphology of Iba1+ cells was maintained in regions with a clear depletion of TH+ terminals (Figure 5a,b). These observations suggest that the increase in CD45 expression in striatal CD11b+ cells detected by flow cytometry might be due to the increased expression in microglia rather than to the infiltration of monocyte/macrophages from the periphery. To assess phagocytosis, we quantitated the fraction of TH+ signal found inside Iba1+ cells (Figure 5c). In the midbrain, Iba1+ cells with different morphologies contributed to remove dying TH+ neurons while in the striatum no active phagocytic cells were detected (Figure 5c). The transmembrane protein 119 (TMEM119) is a specific microglia marker proposed as a tool to differentiate microglia from monocytes/macrophages (Bennett et al., 2016). Thus, TMEM119 immunofluorescence was assessed in Iba1+ cells to determine the presence of infiltrated myeloid cells, Figure 5d shows representative images of the double immunostaining in the midbrain. In animals that received the AAV9‐Control vector, the Iba1 and TMEM119 signal co‐localized in most cells (Figure 5d). By contrast, two Iba1+ cell subsets were clearly differentiated in the midbrain of parkinsonian mice, one co‐expressing TMEM119 and exhibiting microglial morphology and a second one lacking TMEM119 in cells with amoeboid shape compatible with infiltrated monocytes/macrophages (Figure 5d). GFAP+ astrocytes were not in contact with the degenerating neurons and lacked TH+ fragments (Figure 5e), indicating that they were not participating in the removal of cellular debris. Our results show that myeloid cells with different morphologies contribute to the removal of dying TH+ neurons by phagocytosis, and suggest that the amoeboid phenotype observed in parkinsonian animals might correspond to monocytes/macrophages infiltrated from the periphery.
FIGURE 5.
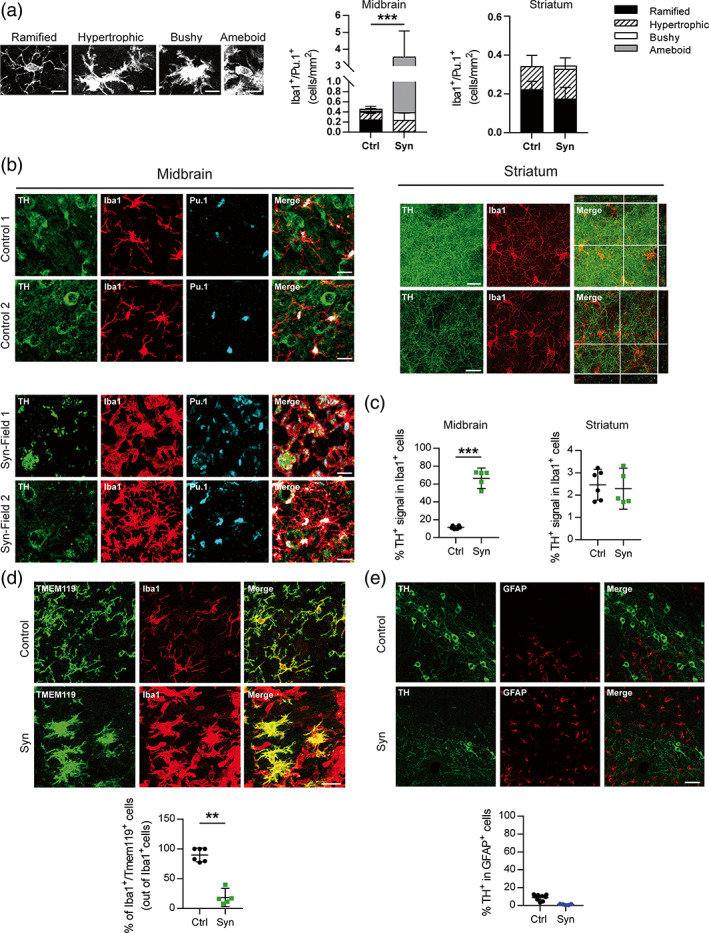
Morphological and phagocytic analysis of glial cells from the striatum and the midbrain. Animals were perfused at 2 weeks after AAV9 administration by stereotaxic surgery in the SNpc (a) Four types of morphology were identified and quantified for Iba1+/Pu.1+ cells in the midbrain and striatum of control and αSyn mice: Ramified, hypertrophic, bushy, and amoeboid. Ramified microglia are characterized by long, thin processes and a small cell body. Hypertrophic morphology is characterized by long, thicker processes and a bigger cell body. Bushy microglia have a big cell body and short processes. Amoeboid microglia have a macrophage‐like morphology with few or no processes. (b) Representative images of TH/Iba1/Pu.1 immunofluorescence staining in the midbrain and striatum of control and αSyn mice. (c) Percentage of TH+ signal in Iba1+ cells. (d) Representative images of Tmem119/Iba1 immunofluorescence staining in the midbrain of control and αSyn overexpressing mice and percentage of Iba1+/Tmem119+ cells out of total Iba1+ cells. (e) Representative images of TH/GFAP immunofluorescence staining in the midbrain of control and αSyn mice and percentage of TH+ signal in GFAP+ cells. The data represent the mean ± 95% CI from 5 to 6 animals per group. Statistical analysis: (a) t‐test, (c) Welch's t‐test (d) t‐test. **p < .01, ***p < .001. Magnification bars: (a) 10 μm, (b and d) 20 μm, (e) 50 μm
4. DISCUSSION
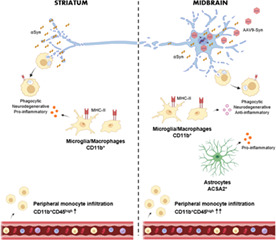
In this study, we explored the neuroinflammatory reaction generated in the striatum and in the midbrain by the αSyn‐dependent degeneration. The loss of the nigrostriatal pathway shifted myeloid cell activation from a homeostatic to a neurodegenerative and phagocytic phenotype in both regions. However, these cells exhibited anti‐inflammatory properties in the midbrain, while in the striatum a pro‐inflammatory profile was detected. An extravasation of peripheral CD11b+CD45high cells into the brain parenchyma that was compatible with the Iba1+TMEM119− amoeboid subpopulation was observed in the midbrain. In this region, all Iba1+ cells participated actively in the elimination of dopaminergic cell debris while no active phagocytic cells were detected in the striatum. Only in the midbrain, astrocytes acquired LPS‐induced pro‐inflammatory properties not observed in their striatal counterparts.
During sterile inflammatory reactions, the recruitment of peripheral immune cells is preceded by a local cell response that senses the damage and produces alarm signals and cytokines. In the experimental model used in this study, degeneration of the nigrostriatal pathway was induced by the stereotaxic administration of AAV9‐Syn vector; control animals were also injected with an AAV9‐Control to overexpress the mCherry protein. After the AAV9 injection, αSyn was translated causing a neuronal death that became evident at 2 weeks post‐transduction. The AAV9 used in this study exhibits primarily neuronal tropism in vivo which is in line with other studies that administer AAV9s intraparenchymally (Castle et al., 2016; Cearley & Wolfe, 2006). In the phases of molecular and cellular inflammation after neuronal damage, the inflammasome activation is an early and transient event, while monocytes and lymphocytes begin to arrive days later and can remain during weeks (Celorrio et al., 2021; Gadani et al., 2015). DAMPs such as ATP, or in this case αSyn, are produced by damaged neurons and trigger the activation of the NLRP3 inflammasome resulting in the production of the inflammatory cytokines IL1β and IL18, which exert broad pro‐inflammatory effects (Lukens et al., 2012; Swanson et al., 2019). In experimental models of PD, activation of the CNS innate immune system precedes neuronal loss (Cicchetti et al., 2002; Depino et al., 2003; Marinova‐Mutafchieva et al., 2009; Purisai et al., 2007). Different strategies directed to prevent priming of myeloid cells, including inflammasome inhibition, are neuroprotective (Broom et al., 2011; Du et al., 2001; Gordon et al., 2018; Harms et al., 2011; Pabon et al., 2011; Teismann & Ferger, 2001; Wu et al., 2002), suggesting a neurotoxic role for the innate immune system. Studies investigating the underlying molecular mechanisms that regulate the transition from homeostatic to disease‐activated microglia have established the MGnD phenotype obtained from experimental mouse models of ALS, AD, and MS, but did not include models of PD (Krasemann et al., 2017). The MGnD activation pattern was induced by apoptotic neurons and contained two clusters of genes, one cluster of downregulated transcripts associated with the loss of homeostatic microglial genes and a second cluster of upregulated inflammatory molecules (Krasemann et al., 2017). A subtype of disease‐associate microglia (DAM) actively involved in the elimination of β‐amyloid plaques was defined in a transgenic mouse model of AD (Keren‐Shaul et al., 2017), in which upregulated genes were related to lipid metabolism and phagocytosis, and downregulated genes with the loss of homeostatic functions (Keren‐Shaul et al., 2017). In our experimental model of PD, the transcriptome of midbrain CD11b+ cells was similar to the downregulated transcripts observed in both signatures, indicating that they all shared a decrease in homeostatic genes. Trem2 (triggering receptor expressed on myeloid cells 2) and Apoe are involved in the regulation of MGnD and DAM activation profiles (Keren‐Shaul et al., 2017; Krasemann et al., 2017). Apoe was upregulated in midbrain CD11b+ cells, but not Trem2, and no enrichment of transcripts was found in the upregulated list of genes from these microglial signatures. By contrast, striatal myeloid cells showed a clear DAM profile, but no similarities to the MGnD signature. Different factors could be behind the different activation states of midbrain versus striatal microglia. We have demonstrated that the basal inflammatory tone of the midbrain with respect to the striatum is pro‐inflammatory (Abellanas et al., 2019), thus the specific environment in each region could determine microglial state and activation under neurodegenerative conditions. It is also necessary to consider that the triggering stimulus in these regions is different, the loss of dopaminergic terminals in the striatum would switch microglial state towards a DAM phenotype, while the loss of neuronal cell bodies would induce an alternative neurodegenerative profile not included in DAM or MGnD signatures. Another possibility could be that our results reflect different activation dynamics of a similar activation pattern. In the striatum the dopaminergic terminals have been effectively eliminated and a phagocytic profile is only detected in the differentially displayed transcripts. On the contrary, an active phagocytic state is observed in the midbrain. The different phagocytic dynamics might be related to the ability of myeloid cells to deal with the elimination of cell bodies versus neuronal terminals and could be associated to the different inflammatory state of these cells. One of the best‐known consequences of apoptotic cell clearance is immunosuppression, a process that is mediated by a set of receptors that include LXR and RXR (A‐Gonzalez et al., 2009; Blander, 2017; Roszer et al., 2011). This pathway was upregulated in midbrain but not in striatal CD11b+ cells and may explain the different inflammatory profile detected in these cells. Midbrain CD11b+ cells showed a M2‐like anti‐inflammatory profile, not present in MGnD, DAM or striatal CD11b+ cells. This anti‐inflammatory phenotype may be a consequence of an active phagocytic process to remove neuronal cell debris in the midbrain. Another possibility is that it could simply reflect a delayed response to control an early inflammatory event (Celorrio et al., 2021; Gadani et al., 2015) or the acquisition of a wound healing profile by the infiltrated monocytes/macrophages (Kubota & Frangogiannis, 2022; Zhang et al., 2019). Thus, at the same experimental point striatal and midbrain myeloid cells experience a different transition from homeostatic to neurodegenerative in the αSyn experimental model of PD. The striatal CD11b+ cells exhibit a DAM profile, while midbrain CD11b+ cells present a unique phenotype characterized by the decreased expression of homeostatic microglial genes and by the acquisition of anti‐inflammatory properties.
To delve into the inflammatory reaction, we also sequenced the transcriptome of astrocytes. Differentially displayed genes in midbrain astrocytes showed a pro‐inflammatory profile similar to LPS reactive astrocytes (Zamanian et al., 2012). Although it has been proposed that microglial activation is required to promote astrogliosis (Liddelow et al., 2017; Tejera et al., 2019), the astrocytic reactivity observed in this study might be either a consequence of an initial microglial activation or due to the direct pro‐inflammatory effect of αSyn‐dependent neurodegeneration (Kim et al., 2018). We discarded the possibility of an AAV9 transduction in astrocytes because no αSyn transcripts were detected in these cells. The lack of phagocytic activity in the astrocytes is consistent with the impairment of this function in LPS‐induced A1 astrocytes (Liddelow et al., 2017). The uptake of myelin by astrocytes in MS has been proposed as an early event that occurs before myelin has been cleared by myeloid cells (Ponath et al., 2017). In vitro experiments indicate that, while astrocytes do not contribute significantly to myelin removal compared to professional phagocytes, they initiate a pro‐inflammatory response that may lead to the recruitment of immune cells to the lesion site (Ponath et al., 2017). Pro‐inflammatory (A1) astrocytes are present in post‐mortem tissue in most neurodegenerative diseases suggesting that they may contribute to neuronal death (Liddelow et al., 2017). Our results suggest that reactive astrocytes play an active role in the generation of a pro‐inflammatory environment when exposed to αSyn‐dependent neuronal death. This type of astrocytic activation may cause damage by either the release of pro‐inflammatory molecules and/or the loss of support functions for the neurons (Kam et al., 2020). Therefore, reactive astrocytes have been proposed as a therapeutic target for PD, by switching this pathological phenotype to a neuroprotective one (L'Episcopo et al., 2018; Serapide et al., 2020; Yun et al., 2018). However, this astrocytic phenotype was not detected in the striatum, indicating that myeloid cells would be the main cell type involved in the removal of αSyn‐overexpressing dopaminergic terminals and contributing to the pro‐inflammatory response in this region. Previously, we demonstrated that the systemic administration of LPS increased the expression of CD80, and TLR4 in striatal microglia that resulted in an immunosuppressive environment in the midbrain that prevented the entrance of CD4 T cells (Abellanas et al., 2019). The αSyn‐dependent degeneration of the nigrostriatal pathway used in this study led us to identify specific inflammatory responses in the midbrain and in the striatum that converged in an increased infiltration of CD11b+CD45high and differed in the magnitude of the response. The increased expression of the antigen‐presenting molecules MHC‐II and CD80 in myeloid cells suggests that both, CD11b+CD45low and CD11b+CD45high subsets, participate in the modulation of CD4+ T cell responses. In fact, CD4+ but not CD8+ T cells are associated with dopaminergic degeneration in experimental models of PD (Brochard et al., 2009; Schonhoff et al., 2020; Williams et al., 2021). The contribution of microglia and other myeloid cells to neuronal degeneration is not clear. In experimental models of AD, a positive role is attributed to monocytes/macrophages for their capacity to remove β‐amyloid (Prinz & Priller, 2014). Boosting the immune system in APP/PS1 transgenic mice promoted the infiltration of Iba1+/CD45high macrophages that engulfed Aβ and produced anti‐inflammatory cytokines, ameliorating the pathology (Koronyo‐Hamaoui et al., 2009). A similar beneficial effect was obtained by blocking the PD‐1/PD‐L1 checkpoint (Rosenzweig et al., 2019). In addition, microglial transition to DAM has been proposed as a protective mechanism aiming to contain/remove neuronal damage (Deczkowska et al., 2018). By contrast, activated microglia may contribute to amyloid‐β‐induced neurotoxicity by generating reactive oxygen species and peroxynitrite (Prinz & Priller, 2014). In demyelinating diseases microglia engulf myelin more efficiently than peripheral monocytes/macrophages, and the inflammatory phenotype is related to the phagocytic capacity being the M2‐like myeloid cells the ones with the greatest capacity (Durafourt et al., 2012; Mosley & Cuzner, 1996; Smith, 1993). In PD, genetic deletion of the C‐C chemokine receptor type 2 prevents the infiltration of pro‐inflammatory monocytes into the SNpc and is neuroprotective in an αSyn overexpression mouse model (Harms et al., 2018). Still, it is not clear whether infiltrated monocyte/macrophages, microglia, or both contribute to the degenerative process. If their specific activation is context‐dependent, they may play different roles in different diseases.
5. CONCLUSION
In summary, we have demonstrated that the αSyn‐dependent neurodegeneration of the nigrostriatal pathway promotes a specific glial activation. The loss of neuronal bodies in the midbrain promotes the infiltration of monocytes/macrophages from the periphery, with a great phagocytic capacity. Under these conditions, myeloid cells show an anti‐inflammatory profile while astrocytes exhibit a pro‐inflammatory phenotype. By contrast, the loss of dopaminergic terminals in the striatum activates a pro‐inflammatory profile in myeloid cells similar to the DAM signature. These data provide valuable information for the design of neuroprotective strategies for the treatment of Parkinson's disease aimed to modulate the inflammatory response.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Figure S1 Characterization of the αSyn‐overexpressing mouse model of dopaminergic neuron degeneration. Mice injected with the AAV9‐Control or the AAV9‐Syn were sacrificed at 2 and 4 weeks after virus administration in the SNpc. (a) Dopaminergic neurons were immunostained to detect the expression of the dopaminergic marker TH and the fluorescent protein mCherry. Representative photomicrographs corresponding to the immunofluorescence staining of dopaminergic terminals in the striatum and to neuronal cell bodies in the SNpc from Control and αSyn‐overexpressing animals are shown. (b) Representative photomicrographs of the immunohistochemistry to detect the correct localization of the human αSyn in the striatum and the midbrain and their corresponding quantifications at 2 and 4 weeks. The data represent the mean ± 95% CI from 5 to 6 animals per group. Statistical analysis: t‐test. ***p < .001. Magnification bars: (a) 20 μm, (b) 1 mm.
Figure S2: RNA sequencing analysis of myeloid cells and astrocytes from the striatum and the midbrain of αSyn mice. Mice injected with the AAV9‐Control or the AAV9‐Syn were sacrificed at 2 weeks after virus administration in the SNpc. (a) Motor behavioral assessment at 2 weeks of αSyn overexpression using the pole test and the catalepsy test. (b) Schematic representation of the purification of CD11b+ and ACSA2+ cells from the striatum and the midbrain of Control and αSyn mice. Cell suspensions were labeled with beads anti‐CD11b and separated on a magnetic cell sorter. CD11b+ cells were collected for RNA sequencing and the CD11b− fraction was labeled with beads anti‐ACSA2 to purify ACSA2+ cells and sequence the RNA. (c) After magnetic bead separation, the negative fraction (CD11b−ACSA2− cells) was analyzed to confirm correct AAV administration in the substantia nigra pars compacta by measuring the expression of mCherry. The data represent the mean from 3 animals/group. Statistica analysis: p < 0.05.
Figure S3: Myeloid cell division analysis in the SNpc of αSyn‐overexpressing mice. (a) Representative photomicrographs for Iba1 and Ki67 immunofluorescence staining in Control and αSyn mice. (b) Percentage of Ki67+ cells out of Iba1+ cells. The data represent the mean ± 95% CI from 5 animals/group. Magnification bars: 20 μm.
ACKNOWLEDGMENTS
We acknowledge the support of Diego Alignani from the flow cytometry facility for his guidance with the FACS analysis. We acknowledge the technical support of Beatriz Paternain and Marta Zamarbide. This work was supported by the Spanish Government (ISCIII‐FEDER) PI20/01063 and PI18/00556 and by Navarra Government (PC 060‐061 and PC 192‐193) and FEDER/Ministerio de Ciencia, Innovación y Universidades‐Agencia Estatal de Investigación/Project BFU2017‐90043‐P and Fundación Gangoiti. Leyre Basurco and Leyre Ayerra were funded by the Ministerio de Universidades, FPU018/02244 and FPU19/0325, respectively.
Basurco, L. , Abellanas, M. A. , Ayerra, L. , Conde, E. , Vinueza‐Gavilanes, R. , Luquin, E. , Vales, A. , Vilas, A. , Martin‐Uriz, P. S. , Tamayo, I. , Alonso, M. M. , Hernaez, M. , Gonzalez‐Aseguinolaza, G. , Clavero, P. , Mengual, E. , Arrasate, M. , Hervás‐Stubbs, S. , & Aymerich, M. S. (2023). Microglia and astrocyte activation is region‐dependent in the α‐synuclein mouse model of Parkinson's disease. Glia, 71(3), 571–587. 10.1002/glia.24295
Sandra Hervás‐Stubbs and Maria S. Aymerich are senior authors of this manuscript.
Funding information Agencia Estatal de Investigación, Grant/Award Number: BFU2017‐90043‐P; Dirección General de Industria y Energia, Gobierno de Navarra. Proyectos Colaborativos, Grant/Award Numbers: PC 060‐061, PC 192‐193; Fundacion Gangoiti; Instituto de Salud Carlos III‐FEDER, Grant/Award Numbers: PI20/01063, PI18/00556; LB was funded by the Ministerio de Universidades; LA was funded by the Ministerio de Universidades
DATA AVAILABILITY STATEMENT
The transcriptome data are publicly available at NCBI's Gene Expression Omnibus (GEO) (Edgar et al., 2002) and are accessible through GEO Series accession number GSE191131. All other data that support the findings of this study are available upon reasonable request to the corresponding author.
REFERENCES
- Abellanas, M. A. , Zamarbide, M. , Basurco, L. , Luquin, E. , Garcia‐Granero, M. , Clavero, P. , San Martin‐Uriz, P. , Vilas, A. , Mengua, E. , Hervas‐Stubbs, S. , & Aymerich, M. S. (2019). Midbrain microglia mediate a specific immunosuppressive response under inflammatory conditions. Journal of Neuroinflammation, 16, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A‐Gonzalez, N. , Bensinger, S. J. , Hong, C. , Beceiro, S. , Bradley, M. N. , Zelcer, N. , Deniz, J. , Ramirez, C. , Díaz, M. , Gallardo, G. , de Galarreta, C. R. , Salazar, J. , Lopez, F. , Edwards, P. , Parks, J. , Andujar, M. , Tontonoz, P. , & Castrillo, A. (2009). Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity, 31, 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, I. , Tamouza, R. , Delord, M. , Krishnamoorthy, R. , Tzourio, C. , Mulot, C. , Nacfer, M. , Lambert, J. C. , Beaune, P. , Laurent‐Puig, P. , Loriot, M. A. , Charron, D. , & Elbaz, A. (2012). Association between Parkinson's disease and the HLA‐DRB1 locus. Movement Disorders, 27, 1104–1110. [DOI] [PubMed] [Google Scholar]
- Banjara, M. , & Ghosh, C. (2017). Sterile neuroinflammation and strategies for therapeutic intervention. Journal of Inflammation, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batiuk, M. Y. , Martirosyan, A. , Wahis, J. , de Vin, F. , Marneffe, C. , Kusserow, C. , Koeppen, J. , Viana, J. F. , Oliveira, J. F. , Voet, T. , Ponting, C. P. , Belgard, T. G. , & Holt, M. G. (2020). Identification of region‐specific astrocyte subtypes at single cell resolution. Nature Communications, 11, 1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher, B. , Spath, S. , & Goverman, J. (2017). Cytokine networks in neuroinflammation. Nature Reviews. Immunology, 17, 49–59. [DOI] [PubMed] [Google Scholar]
- Ben Haim, L. , & Rowitch, D. H. (2017). Functional diversity of astrocytes in neural circuit regulation. Nature Reviews. Neuroscience, 18, 31–41. [DOI] [PubMed] [Google Scholar]
- Bennett, M. L. , Bennett, F. C. , Liddelow, S. A. , Ajami, B. , Zamanian, J. L. , Fernhoff, N. B. , Mulinyawe, S. B. , Bohlen, C. J. , Adil, A. , Tucker, A. , Weissman, I. L. , Chang, E. F. , Li, G. , Grant, G. A. , Hayden Gephart, M. G. , & Barres, B. A. (2016). New tools for studying microglia in the mouse and human CNS. Proceedings of the National Academy of Sciences of the United States of America, 113, E1738–E1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béraud, D. , Twomey, M. , Bloom, B. , Mittereder, A. , Ton, V. , Neitzke, K. , Chasovskikh, S. , Mhyre, T. R. , & Maguire‐Zeiss, K. A. (2011). α‐Synuclein alters toll‐like receptor expression. Frontiers in Neuroscience, 5, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander, J. M. (2017). The many ways tissue phagocytes respond to dying cells. Immunological Reviews, 277, 158–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochard, V. , Combadière, B. , Prigent, A. , Laouar, Y. , Perrin, A. , Beray‐Berthat, V. , Bonduelle, O. , Alvarez‐Fischer, D. , Callebert, J. , Launay, J.‐M. , Duyckaerts, C. , Flavell, R. A. , Hirsch, E. C. , & Hunot, S. (2009). Infiltration of CD4+ lymphocytes into the brain contributes to neurodegeneration in a mouse model of Parkinson disease. The Journal of Clinical Investigation, 119, 182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broom, L. , Marinova‐Mutafchieva, L. , Sadeghian, M. , Davis, J. B. , Medhurst, A. D. , & Dexter, D. T. (2011). Neuroprotection by the selective iNOS inhibitor GW274150 in a model of Parkinson disease. Free Radical Biology & Medicine, 50, 633–640. [DOI] [PubMed] [Google Scholar]
- Burda, J. E. , O'Shea, T. M. , Ao, Y. , Suresh, K. B. , Wang, S. , Bernstein, A. M. , Chandra, A. , Deverasetty, S. , Kawaguchi, R. , Kim, J. H. , McCallum, S. , Rogers, A. , Wahane, S. , & Sofroniew, M. V. (2022). Divergent transcriptional regulation of astrocyte reactivity across disorders. Nature, 606, 557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle, M. J. , Turunen, H. T. , Vandenberghe, L. H. , & Wolfe, J. H. (2016). Controlling AAV tropism in the nervous system with natural and engineered capsids. Methods in Molecular Biology, 1382, 133–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cearley, C. N. , & Wolfe, J. H. (2006). Transduction characteristics of adeno‐associated virus vectors expressing cap serotypes 7, 8, 9, and Rh10 in the mouse brain. Molecular Therapy, 13, 528–537. [DOI] [PubMed] [Google Scholar]
- Celorrio, M. , Abellanas, M. A. , Rhodes, J. , Goodwin, V. , Moritz, J. , Vadivelu, S. , Wang, L. , Rodgers, R. , Xiao, S. , Anabayan, I. , Payne, C. , Perry, A. M. , Baldridge, M. T. , Aymerich, M. S. , Steed, A. , & Friess, S. H. (2021). Gut microbial dysbiosis after traumatic brain injury modulates the immune response and impairs neurogenesis. Acta Neuropathologica Communications, 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, H. , Diaz‐Castro, B. , Shigetomi, E. , Monte, E. , Octeau, J. C. , Yu, X. , Cohn, W. , Rajendran, P. S. , Vondriska, T. M. , Whitelegge, J. P. , Coppola, G. , & Khakh, B. S. (2017). Neural circuit‐specialized astrocytes: Transcriptomic, proteomic, morphological, and functional evidence. Neuron, 95, 531–549.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchetti, F. , Brownell, A. L. , Williams, K. , Chen, Y. I. , Livni, E. , & Isacson, O. (2002). Neuroinflammation of the nigrostriatal pathway during progressive 6‐OHDA dopamine degeneration in rats monitored by immunohistochemistry and PET imaging. The European Journal of Neuroscience, 15, 991–998. [DOI] [PubMed] [Google Scholar]
- Coates, P. J. , Rundle, J. K. , Lorimore, S. A. , & Wright, E. G. (2008). Indirect macrophage responses to ionizing radiation: Implications for genotype‐dependent bystander signaling. Cancer Research, 68, 450–456. [DOI] [PubMed] [Google Scholar]
- D'Costa, S. , Blouin, V. , Broucque, F. , Penaud‐Budloo, M. , François, A. , Perez, I. C. , le Bec, C. , Moullier, P. , Snyder, R. O. , & Ayuso, E. (2016). Practical utilization of recombinant AAV vector reference standards: Focus on vector genomes titration by free ITR qPCR. Molecular Therapy ‐ Methods & Clinical Development, 5, 16019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deczkowska, A. , Keren‐Shaul, H. , Weiner, A. , Colonna, M. , Schwartz, M. , & Amit, I. (2018). Disease‐associated microglia: A universal immune sensor of neurodegeneration. Cell, 173, 1073–1081. [DOI] [PubMed] [Google Scholar]
- Depino, A. M. , Earl, C. , Kaczmarczyk, E. , Ferrari, C. , Besedovsky, H. , del Rey, A. , Pitossi, F. J. , & Oertel, W. H. (2003). Microglial activation with atypical proinflammatory cytokine expression in a rat model of Parkinson's disease. The European Journal of Neuroscience, 18, 2731–2742. [DOI] [PubMed] [Google Scholar]
- Du, Y. , Ma, Z. , Lin, S. , Dodel, R. C. , Gao, F. , Bales, K. R. , Triarhou, L. C. , Chernet, E. , Perry, K. W. , Nelson, D. L. , Luecke, S. , Phebus, L. A. , Bymaster, F. P. , & Paul, S. M. (2001). Minocycline prevents nigrostriatal dopaminergic neurodegeneration in the MPTP model of Parkinson's disease. Proceedings of the National Academy of Sciences of the United States of America, 98, 14669–14674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durafourt, B. A. , Moore, C. S. , Zammit, D. A. , Johnson, T. A. , Zaguia, F. , Guiot, M.‐C. , Bar‐Or, A. , & Antel, J. P. (2012). Comparison of polarization properties of human adult microglia and blood‐derived macrophages. Glia, 60, 717–727. [DOI] [PubMed] [Google Scholar]
- Edgar, R. , Domrachev, M. , & Lash, A. E. (2002). Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research, 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellner, L. , Irschick, R. , Schanda, K. , Reindl, M. , Klimaschewski, L. , Poewe, W. , Wenning, G. K. , & Stefanova, N. (2013). Toll‐like receptor 4 is required for α‐synuclein dependent activation of microglia and astroglia. Glia, 61, 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadani, S. P. , Walsh, J. T. , Lukens, J. R. , & Kipnis, J. (2015). Dealing with danger in the CNS: The response of the immune system to injury. Neuron, 87, 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gate, D. , Rezai‐Zadeh, K. , Jodry, D. , Rentsendorj, A. , & Town, T. (2010). Macrophages in Alzheimer's disease: The blood‐borne identity. Journal of Neural Transmission, 117, 961–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, R. , Albornoz, E. A. , Christie, D. C. , Langley, M. R. , Kumar, V. , Mantovani, S. , Robertson, A. A. B. , Butler, M. S. , Rowe, D. B. , O'Neill, L. A. , Kanthasamy, A. G. , Schroder, K. , Cooper, M. A. , & Woodruff, T. M. (2018). Inflammasome inhibition prevents α‐synuclein pathology and dopaminergic neurodegeneration in mice. Science Translational Medicine, 10, eaah4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabert, K. , Michoel, T. , Karavolos, M. H. , Clohisey, S. , Baillie, J. K. , Stevens, M. P. , Freeman, T. C. , Summers, K. M. , & McColl, B. W. (2016). Microglial brain region‐dependent diversity and selective regional sensitivities to aging. Nature Neuroscience, 19, 504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen, H. J. , & Jensen, E. B. (1987). The efficiency of systematic sampling in stereology and its prediction. Journal of Microscopy, 147, 229–263. [DOI] [PubMed] [Google Scholar]
- Hammond, T. R. , Dufort, C. , Dissing‐Olesen, L. , Giera, S. , Young, A. , Wysoker, A. , Walker, A. J. , Gergits, F. , Segel, M. , Nemesh, J. , Marsh, S. E. , Saunders, A. , Macosko, E. , Ginhoux, F. , Chen, J. , Franklin, R. J. M. , Piao, X. , McCarroll, S. A. , & Stevens, B. (2019). Single‐cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell‐state changes. Immunity, 50, 253–271.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza, T. H. , Zabetian, C. P. , Tenesa, A. , Laederach, A. , Montimurro, J. , Yearout, D. , Kay, D. M. , Doheny, K. F. , Paschall, J. , Pugh, E. , Kusel, V. I. , Collura, R. , Roberts, J. , Griffith, A. , Samii, A. , Scott, W. K. , Nutt, J. , Factor, S. A. , & Payami, H. (2010). Common genetic variation in the HLA region is associated with late‐onset sporadic Parkinson's disease. Nature Genetics, 42, 781–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms, A. S. , Barnum, C. J. , Ruhn, K. A. , Varghese, S. , Treviño, I. , Blesch, A. , & Tansey, M. G. (2011). Delayed dominant‐negative TNF gene therapy halts progressive loss of nigral dopaminergic neurons in a rat model of Parkinson's disease. Molecular Therapy, 19, 46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms, A. S. , Ferreira, S. A. , & Romero‐Ramos, M. (2021). Periphery and brain, innate and adaptive immunity in Parkinson's disease. Acta Neuropathologica, 141, 527–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms, A. S. , Thome, A. D. , Yan, Z. , Schonhoff, A. M. , Williams, G. P. , Li, X. , Liu, Y. , Qin, H. , Benveniste, E. N. , & Standaert, D. G. (2018). Peripheral monocyte entry is required for alpha‐Synuclein induced inflammation and neurodegeneration in a model of Parkinson disease. Experimental Neurology, 300, 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honarpisheh, P. , Lee, J. , Banerjee, A. , Blasco‐Conesa, M. P. , Honarpisheh, P. , d'Aigle, J. , Mamun, A. A. , Ritzel, R. M. , Chauhan, A. , Ganesh, B. P. , & McCullough, L. D. (2020). Potential caveats of putative microglia‐specific markers for assessment of age‐related cerebrovascular neuroinflammation. Journal of Neuroinflammation, 17, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez, P. , Bonnet, A.‐M. , Débarges, B. , Lohmann, E. , Tison, F. , Pollak, P. , Agid, Y. , Dürr, A. , & Brice, A. (2004). Causal relation between alpha‐synuclein gene duplication and familial Parkinson's disease. Lancet, 364, 1169–1171. [DOI] [PubMed] [Google Scholar]
- Íñigo‐Marco, I. , Valencia, M. , Larrea, L. , Bugallo, R. , Martínez‐Goikoetxea, M. , Zuriguel, I. , & Arrasate, M. (2017). E46K α‐synuclein pathological mutation causes cell‐autonomous toxicity without altering protein turnover or aggregation. Proceedings of the National Academy of Sciences, 114, E8274–E8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Parkinson Disease Genomics Consortium , Nalls, M. A. , Plagnol, V. , Hernandez, D. G. , Sharma, M. , Sheerin, U.‐M. , Saad, M. , Simón‐Sánchez, J. , Schulte, C. , Lesage, S. , Sveinbjörnsdóttir, S. , Stefánsson, K. , Martinez, M. , Hardy, J. , Heutink, P. , Brice, A. , Gasser, T. , Singleton, A. B. , & Wood, N. W. (2011). Imputation of sequence variants for identification of genetic risks for Parkinson's disease: A meta‐analysis of genome‐wide association studies. Lancet (London, England), 377, 641–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin, D. A. , Kenigsberg, E. , Keren‐Shaul, H. , Elefant, N. , Paul, F. , Zaretsky, I. , Mildner, A. , Cohen, N. , Jung, S. , Tanay, A. , & Amit, I. (2014). Massively parallel single‐cell RNA‐Seq for marker‐free decomposition of tissues into cell types. Science, 343, 776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam, T.‐I. , Hinkle, J. T. , Dawson, T. M. , & Dawson, V. L. (2020). Microglia and astrocyte dysfunction in parkinson's disease. Neurobiology of Disease, 144, 105028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannarkat, G. , Cook, D. , Lee, J. , Chung, J. , Sandy, E. , Paul, K. , Ritz, B. , Bronstein, J. , Factor, S. A. , Boss, J. , & Tansey, M. (2015). Common genetic variant association with altered HLA expression, synergy with pyrethroid exposure, and risk for Parkinson's disease: An observational and case‐control study. NPJ Parkinson's Disease, 1, 15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren‐Shaul, H. , Spinrad, A. , Weiner, A. , Matcovitch‐Natan, O. , Dvir‐Szternfeld, R. , Ulland, T. K. , David, E. , Baruch, K. , Lara‐Astaiso, D. , Toth, B. , Itzkovitz, S. , Colonna, M. , Schwartz, M. , & Amit, I. (2017). A unique microglia type associated with restricting development of Alzheimer's disease. Cell, 169, 1276–1290. [DOI] [PubMed] [Google Scholar]
- Khakh, B. S. , & Deneen, B. (2019). The emerging nature of astrocyte diversity. Annual Review of Neuroscience, 42, 187–207. [DOI] [PubMed] [Google Scholar]
- Khakh, B. S. , & Sofroniew, M. V. (2015). Diversity of astrocyte functions and phenotypes in neural circuits. Nature Neuroscience, 18, 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C. , Spencer, B. , Rockenstein, E. , Yamakado, H. , Mante, M. , Adame, A. , Fields, J. A. , Masliah, D. , Iba, M. , Lee, H.‐J. , Rissman, R. A. , Lee, S.‐J. , & Masliah, E. (2018). Immunotherapy targeting toll‐like receptor 2 alleviates neurodegeneration in models of synucleinopathy by modulating α‐synuclein transmission and neuroinflammation. Molecular Neurodegeneration, 13, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott, C. , Stern, G. , & Wilkin, G. P. (2000). Inflammatory regulators in Parkinson's disease: iNOS, lipocortin‐1, and cyclooxygenases‐1 and ‐2. Molecular and Cellular Neurosciences, 16, 724–739. [DOI] [PubMed] [Google Scholar]
- Koronyo‐Hamaoui, M. , Ko, M. K. , Koronyo, Y. , Azoulay, D. , Seksenyan, A. , Kunis, G. , Pham, M. , Bakhsheshian, J. , Rogeri, P. , Black, K. L. , Farkas, D. L. , & Schwartz, M. (2009). Attenuation of AD‐like neuropathology by harnessing peripheral immune cells: Local elevation of IL‐10 and MMP‐9. Journal of Neurochemistry, 111, 1409–1424. [DOI] [PubMed] [Google Scholar]
- Kouli, A. , Horne, C. B. , & Williams‐Gray, C. H. (2019). Toll‐like receptors and their therapeutic potential in Parkinson's disease and α‐synucleinopathies. Brain, Behavior, and Immunity, 81, 41–51. [DOI] [PubMed] [Google Scholar]
- Krasemann, S. , Madore, C. , Cialic, R. , Baufeld, C. , Calcagno, N. , El Fatimy, R. , Beckers, L. , O'Loughlin, E. , Xu, Y. , Fanek, Z. , Greco, D. J. , Smith, S. T. , Tweet, G. , Humulock, Z. , Zrzavy, T. , Conde‐Sanroman, P. , Gacias, M. , Weng, Z. , Chen, H. , … Butovsky, O. (2017). The TREM2‐APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity, 47, 566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota, A. , & Frangogiannis, N. G. (2022). Macrophages in myocardial infarction. American Journal of Physiology. Cell Physiology, 323, C1304–C1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin, Y. , Kobayashi, S. , Leader, A. , Amir, E. D. , Elefant, N. , Bigenwald, C. , Remark, R. , Sweeney, R. , Becker, C. D. , Levine, J. H. , Meinhof, K. , Chow, A. , Kim‐Shulze, S. , Wolf, A. , Medaglia, C. , Li, H. , Rytlewski, J. A. , Emerson, R. O. , Solovyov, A. , … Merad, M. (2017). Innate immune landscape in early lung adenocarcinoma by paired single‐cell analyses. Cell, 169, 750–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Episcopo, F. , Tirolo, C. , Peruzzotti‐Jametti, L. , Serapide, M. F. , Testa, N. , Caniglia, S. , Balzarotti, B. , Pluchino, S. , & Marchetti, B. (2018). Neural stem cell grafts promote astroglia‐driven neurorestoration in the aged parkinsonian brain via Wnt/β‐catenin signaling. Stem Cells, 36, 1179–1197. [DOI] [PubMed] [Google Scholar]
- Li, Q. , Cheng, Z. , Zhou, L. , Darmanis, S. , Neff, N. F. , Okamoto, J. , Gulati, G. , Bennett, M. L. , Sun, L. O. , Clarke, L. E. , Marschallinger, J. , Yu, G. , Quake, S. R. , Wyss‐Coray, T. , & Barres, B. A. (2019). Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single‐cell RNA sequencing. Neuron, 101, 207–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow, S. A. , Guttenplan, K. A. , Clarke, L. E. , Bennett, F. C. , Bohlen, C. J. , Schirmer, L. , Bennett, M. L. , Münch, A. E. , Chung, W.‐S. , Peterson, T. C. , Wilton, D. K. , Frouin, A. , Napier, B. A. , Panicker, N. , Kumar, M. , Buckwalter, M. S. , Rowitch, D. H. , Dawson, V. L. , Dawson, T. M. , … Barres, B. A. (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature, 541, 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeffler, D. A. , Camp, D. M. , & Conant, S. B. (2006). Complement activation in the Parkinson's disease substantia nigra: An immunocytochemical study. Journal of Neuroinflammation, 3, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens, J. R. , Gross, J. M. , & Kanneganti, T.‐D. (2012). IL‐1 family cytokines trigger sterile inflammatory disease. Frontiers in Immunology, 3, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinova‐Mutafchieva, L. , Sadeghian, M. , Broom, L. , Davis, J. B. , Medhurst, A. D. , & Dexter, D. T. (2009). Relationship between microglial activation and dopaminergic neuronal loss in the substantia nigra: A time course study in a 6‐hydroxydopamine model of Parkinson's disease. Journal of Neurochemistry, 110, 966–975. [DOI] [PubMed] [Google Scholar]
- Masuda, T. , Sankowski, R. , Staszewski, O. , Böttcher, C. , Amann, L. , Sagar, S. C. , Nessler, S. , Kunz, P. , van Loo, G. , Coenen, V. A. , Reinacher, P. C. , Michel, A. , Sure, U. , Gold, R. , Grün, D. , Priller, J. , Stadelmann, C. , & Prinz, M. (2019). Spatial and temporal heterogeneity of mouse and human microglia at single‐cell resolution. Nature, 566, 388–392. [DOI] [PubMed] [Google Scholar]
- Masuda, T. , Sankowski, R. , Staszewski, O. , & Prinz, M. (2020). Microglia heterogeneity in the single‐cell era. Cell Reports, 30, 1271–1281. [DOI] [PubMed] [Google Scholar]
- McGeer, P. L. , Itagaki, S. , Boyes, B. E. , & McGeer, E. G. (1988). Reactive microglia are positive for HLA‐DR in the substantia nigra of Parkinson's and Alzheimer's disease brains. Neurology, 38, 1285–1291. [DOI] [PubMed] [Google Scholar]
- Mogi, M. , Harada, M. , Kondo, T. , Riederer, P. , Inagaki, H. , Minami, M. , & Nagatsu, T. (1994). Interleukin‐1β, interleukin‐6, epidermal growth factor and transforming growth factor‐α are elevated in the brain from parkinsonian patients. Neuroscience Letters, 180, 147–150. [DOI] [PubMed] [Google Scholar]
- Mogi, M. , Harada, M. , Riederer, P. , Narabayashi, H. , Fujita, K. , & Nagatsu, T. (1994). Tumor necrosis factor‐alpha (TNF‐alpha) increases both in the brain and in the cerebrospinal fluid from parkinsonian patients. Neuroscience Letters, 165, 208–210. [DOI] [PubMed] [Google Scholar]
- Mosley, K. , & Cuzner, M. L. (1996). Receptor‐mediated phagocytosis of myelin by macrophages and microglia: Effect of opsonization and receptor blocking agents. Neurochemical Research, 21, 481–487. [DOI] [PubMed] [Google Scholar]
- Nalls, M. A. , Pankratz, N. , Lill, C. M. , Do CB, H. D. G. , Saad, M. , AL, D. S. , Kara, E. , Bras, J. , Sharma, M. , Schulte, C. , Keller, M. F. , Arepalli, S. , Letson, C. , Edsall, C. , Stefansson, H. , Liu, X. , Pliner, H. , Lee, J. H. , Cheng, R. , … Singleton, A. B. (2014). Large‐scale meta‐analysis of genome‐wide association data identifies six new risk loci for Parkinson's disease. Nature Genetics, 46, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabon, M. M. , Bachstetter, A. D. , Hudson, C. E. , Gemma, C. , & Bickford, P. C. (2011). CX3CL1 reduces neurotoxicity and microglial activation in a rat model of Parkinson's disease. Journal of Neuroinflammation, 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. 2001. The mouse brain in sterotaxic coordinates. Academic Press. [Google Scholar]
- Peter, I. , Dubinsky, M. , Bressman, S. , Park, A. , Lu, C. , Chen, N. , & Wang, A. (2018). Anti‐tumor necrosis Factor therapy and incidence of Parkinson disease among patients with inflammatory bowel disease. JAMA Neurology, 75, 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos, M. H. , Lavedan, C. , Leroy, E. , Ide, S. E. , Dehejia, A. , Dutra, A. , Pike, B. , Root, H. , Rubenstein, J. , Boyer, R. , Stenroos, E. S. , Chandrasekharappa, S. , Athanassiadou, A. , Papapetropoulos, T. , Johnson, W. G. , Lazzarini, A. M. , Duvoisin, R. C. , Di Iorio, G. , Golbe, L. I. , & Nussbaum, R. L. (1997). Mutation in the alpha‐synuclein gene identified in families with Parkinson's disease. Science, 276, 2045–2047. [DOI] [PubMed] [Google Scholar]
- Ponath, G. , Ramanan, S. , Mubarak, M. , Housley, W. , Lee, S. , Sahinkaya, F. R. , Vortmeyer, A. , Raine, C. S. , & Pitt, D. (2017). Myelin phagocytosis by astrocytes after myelin damage promotes lesion pathology. Brain, 140, 399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz, M. , & Priller, J. (2014). Microglia and brain macrophages in the molecular age: From origin to neuropsychiatric disease. Nature Reviews Neuroscience, 155, 300–312. [DOI] [PubMed] [Google Scholar]
- Purisai, M. G. , McCormack, A. L. , Cumine, S. , Li, J. , Isla, M. Z. , & Di Monte, D. A. (2007). Microglial activation as a priming event leading to paraquat‐induced dopaminergic cell degeneration. Neurobiology of Disease, 25, 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig, N. , Dvir‐Szternfeld, R. , Tsitsou‐Kampeli, A. , Keren‐Shaul, H. , Ben‐Yehuda, H. , Weill‐Raynal, P. , Cahalon, L. , Kertser, A. , Baruch, K. , Amit, I. , Weiner, A. , & Schwartz, M. (2019). PD‐1/PD‐L1 checkpoint blockade harnesses monocyte‐derived macrophages to combat cognitive impairment in a tauopathy mouse model. Nature Communications, 10, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszer, T. , Menéndez‐Gutiérrez, M. P. , Lefterova, M. I. , Alameda, D. , Núñez, V. , Lazar, M. A. , Fischer, T. , & Ricote, M. (2011). Autoimmune kidney disease and impaired engulfment of apoptotic cells in mice with macrophage peroxisome proliferator‐activated receptor gamma or retinoid X receptor alpha deficiency. Journal of Immunology, 186, 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage, J. C. , Jay, T. , Goduni, E. , Quigley, C. , Mariani, M. M. , Malm, T. , Ransohoff, R. M. , Lamb, B. T. , & Landreth, G. E. (2015). Nuclear receptors license phagocytosis by trem2+ myeloid cells in mouse models of Alzheimer's disease. The Journal of Neuroscience, 35, 6532–6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonhoff, A. M. , Williams, G. P. , Wallen, Z. D. , Standaert, D. G. , & Harms, A. S. (2020). Innate and adaptive immune responses in Parkinson's disease. Progress in Brain Research, 252, 169–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serapide, M. F. , L'Episcopo, F. , Tirolo, C. , Testa, N. , Caniglia, S. , Giachino, C. , & Marchetti, B. (2020). Boosting antioxidant self‐defenses by grafting astrocytes rejuvenates the aged microenvironment and mitigates nigrostriatal toxicity in parkinsonian brain via an Nrf2‐driven Wnt/β‐catenin prosurvival axis. Frontiers in Aging Neuroscience, 12, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M. E. (1993). Phagocytosis of myelin by microglia in vitro. Journal of Neuroscience Research, 35, 480–487. [DOI] [PubMed] [Google Scholar]
- Sousa, C. , Golebiewska, A. , Poovathingal, S. K. , Kaoma, T. , Pires‐Afonso, Y. , Martina, S. , Coowar, D. , Azuaje, F. , Skupin, A. , Balling, R. , Biber, K. , Niclou, S. P. , & Michelucci, A. (2018). Single‐cell transcriptomics reveals distinct inflammation‐induced microglia signatures. EMBO Reports, 19, E46171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, K. V. , Deng, M. , & Ting, J. P.‐Y. (2019). The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nature Reviews. Immunology, 19, 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teismann, P. , & Ferger, B. (2001). Inhibition of the cyclooxygenase isoenzymes COX‐1 and COX‐2 provide neuroprotection in the MPTP‐mouse model of Parkinson's disease. Synapse, 39, 167–174. [DOI] [PubMed] [Google Scholar]
- Tejera, D. , Mercan, D. , Sanchez‐Caro, J. M. , Hanan, M. , Greenberg, D. , Soreq, H. , Latz, E. , Golenbock, D. , & Heneka, M. T. (2019). Systemic inflammation impairs microglial Aβ clearance through NLRP3 inflammasome. The EMBO Journal, 38, e101064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, G. P. , Schonhoff, A. M. , Jurkuvenaite, A. , Gallups, N. J. , Standaert, D. G. , & Harms, A. S. (2021). CD4 T cells mediate brain inflammation and neurodegeneration in a mouse model of Parkinson's disease. Brain, 144, 2047–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissemann, W. T. , Hill‐Burns, E. M. , Zabetian, C. P. , Factor, S. A. , Patsopoulos, N. , Hoglund, B. , Holcomb, C. , Donahue, R. J. , Thomson, G. , Erlich, H. , & Payami, H. (2013). Association of Parkinson disease with structural and regulatory variants in the HLA region. American Journal of Human Genetics, 93, 984–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, D. C. , Jackson‐Lewis, V. , Vila, M. , Tieu, K. , Teismann, P. , Vadseth, C. , Choi, D.‐K. , Ischiropoulos, H. , & Przedborski, S. (2002). Blockade of microglial activation is neuroprotective in the 1‐methyl‐4‐phenyl‐1,2,3,6‐tetrahydropyridine mouse model of Parkinson disease. The Journal of Neuroscience, 22, 1763–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt‐Johnson, S. K. , Herr, S. A. , & Brewster, A. L. (2017). Status epilepticus triggers time‐dependent alterations in microglia abundance and morphological phenotypes in the hippocampus. Frontiers in Neurology, 8, 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, S. P. , Kam, T.‐I. , Panicker, N. , Kim, S. , Oh, Y. , Park, J.‐S. , Kwon, S.‐H. , Park, Y. J. , Karuppagounder, S. S. , Park, H. , Kim, S. , Oh, N. , Kim, N. A. , Lee, S. , Brahmachari, S. , Mao, X. , Lee, J. H. , Kumar, M. , An, D. , … Ko, H. S. (2018). Block of A1 astrocyte conversion by microglia is neuroprotective in models of Parkinson's disease. Nature Medicine, 24, 931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian, J. L. , Xu, L. , Foo, L. C. , Nouri, N. , Zhou, L. , Giffard, R. G. , & Barres, B. A. (2012). Genomic analysis of reactive astrogliosis. The Journal of Neuroscience, 32, 6391–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarranz, J. J. , Alegre, J. , Gómez‐Esteban, J. C. , Lezcano, E. , Ros, R. , Ampuero, I. , Vidal, L. , Hoenicka, J. , Rodriguez, O. , Atarés, B. , Llorens, V. , Tortosa, E. G. , del Ser, T. , Muñoz, D. G. , & de Yebenes, J. G. (2004). The new mutation, E46K, of α‐synuclein causes Parkinson and Lewy body dementia. Annals of Neurology, 55, 164–173. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Qu, C. , Li, T. , Cui, W. , Wang, X. , & Du, J. (2019). Phagocytosis mediated by scavenger receptor class BI promotes macrophage transition during skeletal muscle regeneration. The Journal of Biological Chemistry, 294, 15672–15685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W. , Wang, T. , Pei, Z. , Miller, D. S. , Wu, X. , Block, M. L. , Wilson, B. , Zhang, W. , Zhou, Y. , Hong, J.‐S. , & Zhang, J. (2005). Aggregated alpha‐synuclein activates microglia: A process leading to disease progression in Parkinson's disease. The FASEB Journal, 19, 533–542. [DOI] [PubMed] [Google Scholar]
- Zolotukhin, S. , Byrne, B. J. , Mason, E. , Zolotukhin, I. , Potter, M. , Chesnut, K. , Summerford, C. , Samulski, R. J. , & Muzyczka, N. (1999). Recombinant adeno‐associated virus purification using novel methods improves infectious titer and yield. Gene Therapy, 6, 973–985. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Characterization of the αSyn‐overexpressing mouse model of dopaminergic neuron degeneration. Mice injected with the AAV9‐Control or the AAV9‐Syn were sacrificed at 2 and 4 weeks after virus administration in the SNpc. (a) Dopaminergic neurons were immunostained to detect the expression of the dopaminergic marker TH and the fluorescent protein mCherry. Representative photomicrographs corresponding to the immunofluorescence staining of dopaminergic terminals in the striatum and to neuronal cell bodies in the SNpc from Control and αSyn‐overexpressing animals are shown. (b) Representative photomicrographs of the immunohistochemistry to detect the correct localization of the human αSyn in the striatum and the midbrain and their corresponding quantifications at 2 and 4 weeks. The data represent the mean ± 95% CI from 5 to 6 animals per group. Statistical analysis: t‐test. ***p < .001. Magnification bars: (a) 20 μm, (b) 1 mm.
Figure S2: RNA sequencing analysis of myeloid cells and astrocytes from the striatum and the midbrain of αSyn mice. Mice injected with the AAV9‐Control or the AAV9‐Syn were sacrificed at 2 weeks after virus administration in the SNpc. (a) Motor behavioral assessment at 2 weeks of αSyn overexpression using the pole test and the catalepsy test. (b) Schematic representation of the purification of CD11b+ and ACSA2+ cells from the striatum and the midbrain of Control and αSyn mice. Cell suspensions were labeled with beads anti‐CD11b and separated on a magnetic cell sorter. CD11b+ cells were collected for RNA sequencing and the CD11b− fraction was labeled with beads anti‐ACSA2 to purify ACSA2+ cells and sequence the RNA. (c) After magnetic bead separation, the negative fraction (CD11b−ACSA2− cells) was analyzed to confirm correct AAV administration in the substantia nigra pars compacta by measuring the expression of mCherry. The data represent the mean from 3 animals/group. Statistica analysis: p < 0.05.
Figure S3: Myeloid cell division analysis in the SNpc of αSyn‐overexpressing mice. (a) Representative photomicrographs for Iba1 and Ki67 immunofluorescence staining in Control and αSyn mice. (b) Percentage of Ki67+ cells out of Iba1+ cells. The data represent the mean ± 95% CI from 5 animals/group. Magnification bars: 20 μm.
Data Availability Statement
The transcriptome data are publicly available at NCBI's Gene Expression Omnibus (GEO) (Edgar et al., 2002) and are accessible through GEO Series accession number GSE191131. All other data that support the findings of this study are available upon reasonable request to the corresponding author.


