Abstract
Efficient and regulated nucleocytoplasmic trafficking of macromolecules to the correct subcellular compartment is critical for proper functions of the eukaryotic cell. The majority of the macromolecular traffic across the nuclear pores is mediated by the Karyopherin-β (or Kap) family of nuclear transport receptors. Work over more than two decades has shed considerable light on how the different Kap family members bring their respective cargoes into the nucleus or the cytoplasm in efficient and highly regulated manners. In this Review, we overview the main features and established functions of Kap family members, describe how Kaps recognize their cargoes and discuss the different ways in which these Kap–cargo interactions can be regulated, highlighting new findings and open questions. We also describe current knowledge of the import and export of the components of three large gene expression machines — the core replisome, RNA polymerase II and the ribosome — pointing out the questions that persist about how such large macromolecular complexes are trafficked to serve their function in a designated subcellular location.
The nucleus is bound by the double-membraned nuclear envelope, which separates the genome from the rest of the eukaryotic cell. Nuclear pore complexes (NPCs) span the nuclear envelope, serving both as the main conduit for molecules between the nucleus and cytoplasm and as a permeability barrier to limit the passage of macromolecules (Supplementary Box 1) and to ensure the maintenance of nuclear composition. The conserved Karyopherin-β (Kap) family of nuclear transport receptors mediates the majority of transport of macromolecules, especially of proteins, across the NPC into the nucleus (importins), out of the nucleus (exportins) or in both directions (biportins)1–3. The exceptions of a few Kap-independent transport pathways for a few specific classes of macromolecules are described in Supplementary Box 2. Although the exact numbers of Kap family members vary in different organisms, most are identifiable and conserved across eukaryotes, suggesting a conservation of function throughout the eukaryotic kingdom4 (Supplementary Box 3). Of the 20 Kaps identified in human cells, 10 are importins, 5 are exportins, 3 are biportins and the functions of 2 Kaps remain unknown (TABLE 1). Kaps are critically involved in many eukaryotic cellular processes that include response to the environment, signal transduction, regulation and maintenance of the cell cycle. It is therefore not surprising that disruptions of Kap functions contribute to diseases such as neurodegeneration, cancer, inflammation and viral infections, making these transport receptors promising therapeutic targets (BOX 1).
Table 1 |.
Members of the Karyopherin-β (Kap) family
| Kap | Other names | Isoformsa | Yeast homologue | Modes of cargo bindingb — consensus sequence (binding affinity/dissociation constants) | Functions of putative cargoesc | 59 ptRAN–GTP binding affinity |
|---|---|---|---|---|---|---|
| Importins | ||||||
| Importin-β (IMPβ) | p97, Kapβ1, KPNB1, IPO1, IPOB, IMB1, NTF97 | 2 | KAP95 | cNLS via IMPα, with monopartite (K-K/R-X-K/R) or bipartite (K/R-K/R-X10–12-K/R3/5) sequences (single-digit nanomolar to micromolar21,27,221,222) | Broad functions in gene expression and cell cycle regulation9, inner nuclear membrane proteins, spliceosomal small nuclear ribonucleoproteins via SNUPN | Hundreds of picomolars95 |
| IBB of adaptors (~2 nM1) | ||||||
| Diverse folded domains (~2–200 nM1) | ||||||
| Transportin/Karyopherin-β 2 (KAPβ2)d | Impβ2, KPNB2, IPO2, TNPO1, TRN1 | 3 | KAP104 | PY-NLS — hydrophobic/basic and R/K/H-X2–5-P-Y (single-digit to tens of nanomolars30–32,223) | mRNA processing9 | Hundreds of picomolars95 |
| RGG regions (hundreds of nanomolars49,51) | ||||||
| Transportin 2/Karyopherin-β2b (KAPβ2b)d | Impβ2b, KPNB2B, IPO3, TNPO2, TRN2 | 2 | NA | PY-NLS | NA | Unknown |
| Transportin 3/Transportin-SR (TNPO3) | TrnSR, TrnSR2, Trn3, Imp12, IPO12 | 4 | MTR10 | RS-NLS — phospho-RS or RD/RE dipeptide repeats (hundreds of nanomolars to low micromolar36,51) | mRNA splicing and export9 | Hundreds of nanomolars95 |
| Importin 4 (IPO4) | RanBP4, Imp4, Imp4b | 2 | YRB4/KAP123 | Linear segments | Chromatin organization9, vesicular transport and cytoskeleton organization8, ribosome biogenesis (yeast)197 | Tens of nanomolars95 |
| Diverse folded domainsb | ||||||
| Importin 5 (IPO5)d | Impβ3, Kapβ3, RanBP5, KPNB3, Imp5, IMB3 | 3 | PSE1/KAP121 | IK-NLS — K-V/I-X-K-X1–2-K/H/R (tens of nanomolars33) | Chromatin organization, ribosome biogenesis, cytokinesis9, vesicular transport and cytoskeleton organization8 | Hundreds of picomolars95 |
| Diverse folded domainsb | ||||||
| Importin 7 (IPO7)d | RanBP7, IPO7 | 1 | NMD5/KAP119 | Diverse folded domainsb | Cell cycle regulation, ribosome biogenesis9 | Single-digit to tens of nanomolars95 |
| Importin 8 (IPO8)d | RanBP8, Imp8 | 2 | SXM1/KAP108 | Diverse folded domainsb | Ribosome biogenesis8,9 | Single-digit to tens of nanomolars95 |
| Importin 9 (IPO9) | RanBP9, Imp9 | 1 | KAP114 | Diverse folded domains (low nanomolar59) | Nucleosome organization, ribosome biogenesis9 | Tens of nanomolars95 |
| Importin 11 (IPO11) | RanBP11, Imp11 | 2 | KAP120 | Diverse folded domainsb | Developmental processes9 | Hundreds of nanomolars95 |
| Exportins | ||||||
| Chromatin Region Maintenance 1/Exportin 1 (CRM1/XPO1) | Exp1, emb | 1 | CRM1 | NES, class 1a–1d: Φ-X2–3-Φ-X2–3-Φ-X-Φ, class 2: Φ-X-Φ-X2-Φ-X-Φ, class 3: Φ-X2-Φ-X3-Φ-X2-Φ, class 4: Φ-X3-Φ-X2-Φ-X3-Φ, class 1a-R–1d-R: Φ-X-Φ-X2–3-Φ-X2–3-Φ (tens of nanomolars to tens of micromolars112) |
Broadly functioning protein cargoes11, and various RNA via adaptors | Undetectable95 |
| Cellular Apoptosis Susceptibility/Exportin 2 (CAS/XPO2) | Exp2, CSE1L, Impα re-exporter | 4 | CSE1 | Autoinhibited and folded Impα (~1 nM224) | IMPα and, possibly, more proteins8 | Undetectable95 |
| Exportin-t (XPOT) | Exp-T, XPO3 | 1 | LOS1 | RNA structures (~3 nM225) | tRNA and other RNAs77 | Undetectable95 |
| Exportin 5 (XPO5) | Exp5, RanBP21 | 1 | MSN5 | RNA structures | Pre-miRNA and other RNAs77,156 | Tens of nanomolars95 |
| Undefined phospho-NESb | ||||||
| Exportin 6 (XPO6) | Exp6, RanBP20 | 2 | NA | Undefined folded domainsb | Actin–profilin226 | Unknown |
| Biportins | ||||||
| Importin 13 (IPO13) | Kap13, RanBP13, IPO13 | 1 | PDR6/KAP122 | Diverse folded domains (import cargo: hundreds of nanomolars; export cargo: single-digit micromolar68) | Transcription factors and RNA binding proteins9 | Tens to hundreds of nanomolars68,95 |
| Exportin 4 (XPO4) | Exp4 | 1 | NA | Diverse folded domains (export cargo: ~2 nM71) | RNA processing, ribosome biogenesis9 | ~40 nM71 |
| Exportin 7 (XPO7)d | Exp7, RanBP16 | 1 | NA | Diverse folded domainsb | Broadly functioning protein cargoes11 | Unknown |
| Unknown function | ||||||
| RanBP6d | NA | 1 | NA | NA | NA | NA |
| RanBP17d | NA | 2 | NA | NA | NA | NA |
cNLS, classical nuclear localization signal; IBB, amino-terminal IMPβ-binding; IK-NLS, isoleucine-lysine nuclear localization signal; NA, not available; NES, nuclear export signal; PY-NLS, proline-tyrosine nuclear localization signal; RS-NLS, arginine-serine repeat nuclear localization signal; Φ, hydrophobic residue; SNUPN, Snurportin-1; X, any amino acid.
These are described isoforms from alternative splicing reported in UNIPROT.
Unknown modes of interactions.
These pairs of Kaps are similar with high sequence identities: 85.3% for KAPβ2/KAPβ2b, 79.2% for IPO5/RanBP6, 64.4% for IPO7/IPO8 and 66.9% for XPO7/RanBP17.
Box 1 |. Karyopherin-specific inhibitors.
Specific inhibitors for several Karyopherins (Kaps) are currently available. Many of them are routinely used for studying import/export mechanisms of macromolecules and are critical for the verification of Kap-specific cargoes and their embedded signals. Development of specific inhibitors for other lesser known Kaps can greatly facilitate the elucidation of their functional importance and mechanisms. Some inhibitors are also developed with pharmacological properties suitable for drugs, targeting various human diseases such as viral infections and cancer, as import and export pathways are critical for environmental response and cell cycle progression.
IMPα, IMPβ or IMPα/β inhibitors
Peptide inhibitors
In vitro measured affinities of classical nuclear localization signals (cNLSs) for ImPα–ImPβ are linearly correlated with nuclear import activities within a functional dissociation constant (KD) in the range of one to hundreds of nanomolars27. More recently, micromolar affinity NLSs were also shown to be active in cells21,221,222. Peptides that bind ImPα–ImPβ tighter than the high-affinity limit cannot be released efficiently by RAN–GTP in the nucleus, and thus act as import inhibitors. Bimax1 and Bimax2 peptides bind ImPα with picomolar affinities and are effective inhibitors of ImPα–ImPβ223,228.
Small-molecule inhibitors
Inhibitors that target ImPα (ivermectin and gossypol229) or ImPβ (58H5–6 (REF.230), INI-43 (REF.231), INI-60 (REF.232), Karyostatin 1a (REF.233), Importazole234) were reviewed previously235, but little is known about their mechanisms of inhibition. INI-34, Karyostatin 1A and Importazole were suggested to interfere with ImPβ–RAN interaction231,233,234, whereas 58H5–6 is predicted to disrupt the interactions between ImPβ and the phenylalanine-glycine (FG) nucleoporins in the nuclear pore complex (NPC)230 (see Supplementary Box 1). Ivermectin is approved for use as an antiparasitic agent236. It was also shown to act as a broad-spectrum antiviral (completed phase II trials for treatment of dengue virus) by binding to the ImPα armadillo (ARM) domain and decreasing ImPα thermostability237. Import inhibitors that target viral proteins include compounds that bind to the NLS of HIV-1 integrase (mifepristone229, budesonide and flunisolide238) and the NLS of flavivirus non-structural protein NS5 (4-HPR/fenretinide239).
KAPβ2 inhibitors
The M9M peptide inhibitor effectively blocks KAPβ2-mediated import in mammalian cells but not in yeast. m9m is a chimeric peptide that contains the KaPβ2-binding hotspot motifs of the proline-tyrosine NLSs (PY-NLSs) from heterogeneous nuclear RNP (hnRNP) A1 (also known as the M9 sequence) and hnRNP M. M9M binds KAPβ2 with picomolar affinity, outcompeting both natural PY-NLSs and RAN–GTP223,228.
IPO5 inhibitor
A small-molecule natural product inhibitor named goyazensolide was recently found to inhibit IPo5 by covalently binding to an unidentified cysteine residue240. Goyazensolide treatment prevented binding of IPO5 to NLS peptides of Rev protein from HIV-1 and the RNA polymerase catalytic subunit PB1 from influenza A virus (interestingly, neither of the NLSs matches the isoleucine-lysine NLS (IK-NLS) consensus). Goyazensolide also decreased nuclear localization of the IPO5 cargo Ras GTPase-activating protein RaSal2 (REF.240).
CRM1 inhibitors
Peptide inhibitors
Affinities of nuclear export signals (NESs) measured in vitro for CRM1 are linearly correlated with nuclear export activities within a functional KD range of 10 nm–30 μm112. Tight binding NESs with picomolar affinities include several synthetic NESs: S1, Super PKI-NS2 and CDC7-NS2 (REFS112,241). These peptides inhibit nuclear export of CRM1 cargoes and S1 also sequesters CRM1 at the nuclear pore.
Small-molecule inhibitors
Most known small-molecule inhibitors of CRM1 bind the NES-binding groove to block binding of NES-containing cargoes to CRM1. These inhibitors covalently conjugate to a cysteine side chain in the groove (reviewed elsewhere242), with one exception, compound NCI-1, which is reported to bind non-covalently to the groove243. Covalent inhibitors include: natural product inhibitors such as antifungal agent Leptomycin B (lmB) and its analogues from bacteria244, acetoxychavicol acetate (ACA)245 and other diverse compounds extracted from various plants and fungi; human prostaglandin 15d-PGJ(2) (REF.246); and different synthetic pharmacological compounds such as KPT-SINEs247–250, CBS9106 (REF.251), S109 (REF.252) and lFS-829 (REF.253). The most commonly used inhibitor, LMB, is hydrolysed by CRM1, causing extremely low reversibility of the conjugating bond, and is therefore highly toxic to cells244; on the contrary, synthetic compounds are slowly reversible and cause CRM1 degradation, and are better tolerated in cells247,250–252,254,255. The KPT-SINE (marketed as Selinexor, Xpovio or KPT-330) is approved for treatment of refractory/relapsed multiple myeloma256 and diffuse large B cell lymphoma257, and is in preclinical and clinical trials for various cancers and other diseases such as traumatic brain injury and viral infections.
Nanobodies as Kap inhibitors
Plasmid-encoded nanobodies represent a novel type of nuclear transport inhibitor. anti-XPo7 nanobodies were first used in proteomics analysis to study XPo7 cargoes11. Intracellular nanobodies are highly specific to their target Kap and can be generated for any Kap. They are a valuable research tool that can assist with cargo discovery, cargo validation, and characterization of nuclear import and export pathways.
Nanobodies.
Small, single-domain antibodies derived from camelids (camels, alpacas and llamas) that lack light chains.
For a Kap to transport its cargoes between the nucleus and the cytoplasm in the correct direction, it must first bind the cargo in the originating compartment (FIG. 1a). Cargoes use linear elements known as nuclear localization or export signals (NLSs or NESs) and/or folded domains to bind specific Kaps. The Kap–cargo complex then passes through the NPC by binding the phenylalanine-glycine (FG) repeats of nucleoporins located in the central pore of the NPC (so-called FG Nups) (Supplementary Box 1 and Supplementary Box 3). Finally, the cargo must be released from the Kap in the destination compartment.
Fig. 1 |. Kap-mediated nucleocytoplasmic transport and principles of Kap–cargo interactions and RAN–GTP-dependent loading/unloading.
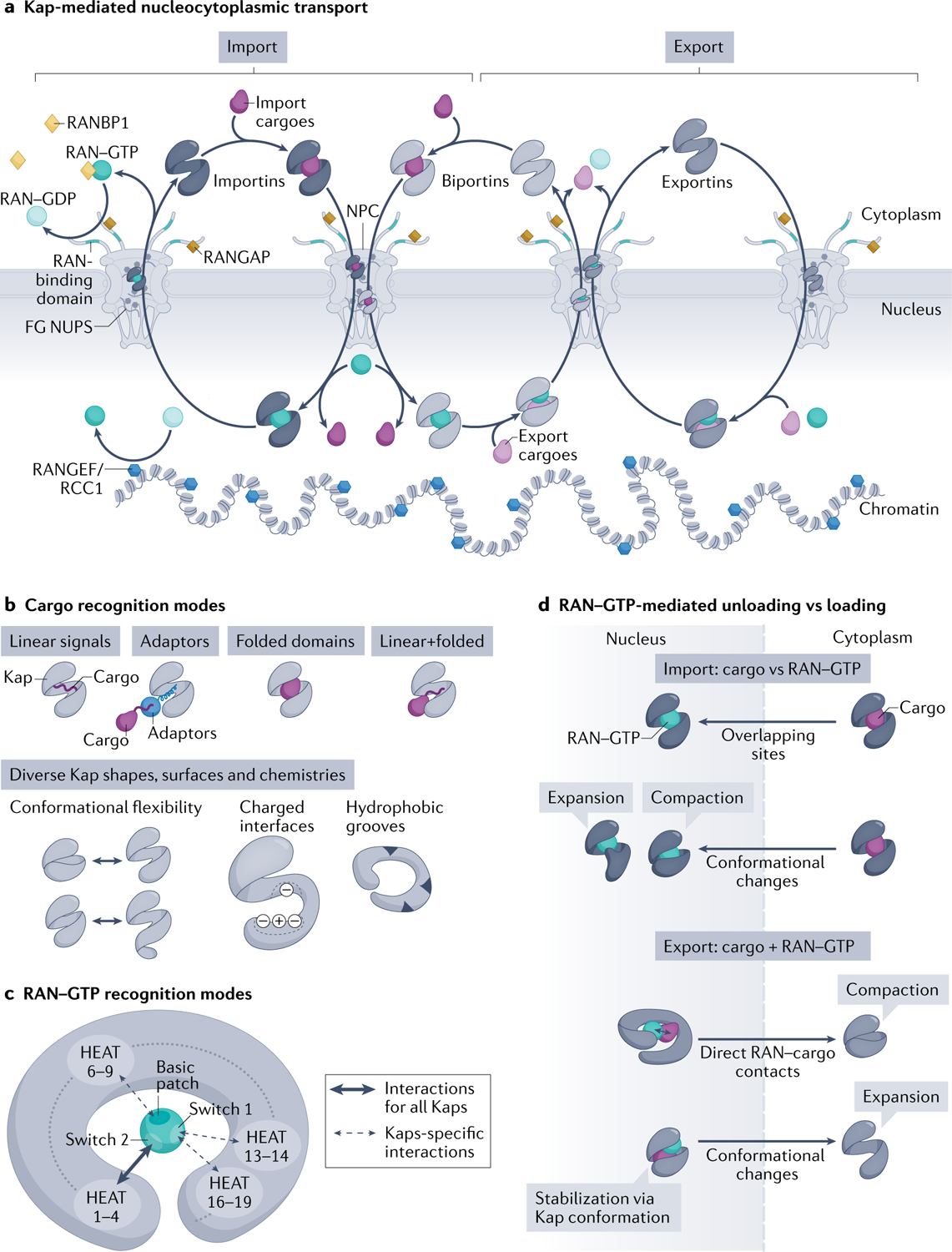
a | Karyopherins (Kaps) (comprising importins, biportins and exportins; shown in grey) and the RAN–GTP gradient control directional transport of cargoes across nuclear pore complexes (NPCs). RAN–GTP (dark teal circles) enriched in the nucleus by the action of chromatin-bound nucleotide exchange factor RANGEF/RCC1 (blue hexagons). Cytoplasmic RAN–GDP (light teal circles) enriched by action of cytoplasmic RAN GTPase-activating protein (RANGAP) (gold squares), RAN-binding domains in nucleoporin RANBP2 (teal fibrils) and cytosolic RANBP1 (yellow diamonds), which accelerate GTP hydrolysis. Importins (left) and biportins (middle) bind cytosolic import cargoes (dark purple), translocate across NPCs by interacting with phenylalanine-glycine (FG) Nups and bind RAN–GTP in the nucleus to release cargoes. Importin–RAN–GTP complexes then exit the nucleus; RAN–GTP hydrolysis and release of RAN–GDP in the cytoplasm then frees importins for the next round of import. Biportins and exportins (right) cooperatively bind export cargoes (light purple) and RAN–GTP in the nucleus. GTP hydrolysis of RAN in the cytosol releases export cargoes and RAN–GDP. Biportins are free for import, whereas unliganded exportins return to the nucleus. b | Kaps use their HEAT repeats to bind cargoes. Kap-binding elements can be specific linear signals (nuclear localization signals and nuclear export signals) within the cargoes, linear signals within adaptor proteins (which then bind cargoes with linear signals), folded domains or the combination of both linear and folded domains. Kaps display various charged and hydrophobic surfaces and their HEAT repeats are also arranged into diverse shapes that undergo different conformational changes, all allowing engagement of many diverse cargoes. c | RAN–GTP recognition is achieved by contacts made between RAN–GTP-specific switch 1, switch 2 and the basic patch with various HEAT repeats in the concave surface of all Kaps. HEAT repeats 1–4 are used by all Kaps, whereas other HEAT repeats are additionally used only by specific Kaps. Some Kaps also use long loops between or within HEAT repeats to contact switch 1 and the basic patch of RAN–GTP (not shown). d | Import: Kap binding to RAN–GTP and import cargoes is mutually exclusive, as a result of overlapping binding sites and steric hindrance and/or conformational changes in the superhelix of Kap that favour binding to RAN–GTP or cargo. Export: Kaps bind cargo and RAN–GTP cooperatively; RAN–GTP–cargo contacts and Kap conformational changes stabilize the ternary export complexes.
The directionality of Kap-mediated transport is controlled by the RAN GTPase3 (FIG. 1a). RAN–GTP and RAN–GDP are asymmetrically localized in the nucleus and cytoplasm, respectively. Nuclear RAN is mostly bound to GTP because its guanine nucleotide exchange factor RCC1 is bound to chromatin, whereas cytoplasmic RAN GTPase-activating protein (RANGAP) and RAN-binding protein RANBP1 activate GTP hydrolysis to keep cytosolic RAN in the GDP-bound state. Kaps bind import cargoes and RAN–GTP in a mutually exclusive manner but bind export cargoes and RAN–GTP cooperatively. Thus, importins and biportins can bind import cargoes in the cytoplasm and translocate across NPCs to the nucleus where they bind RAN–GTP and release the cargoes. Conversely, exportins and biportins form ternary complexes with export cargoes and RAN–GTP in the nucleus, which translocate across NPCs to the cytosol where they disassemble upon GTP hydrolysis.
This Review describes the many ways Kaps recognize cargoes and how Kap–cargo interactions are modulated. We also cover knowledge about the trafficking of three gene expression machineries, one each involved in replication, transcription and translation, as case studies to showcase our current, albeit still limited, understanding of how they enter/exit the nucleus and how these processes are coordinated with their assembly/disassembly and functions.
Traffic in and out of nucleus
The organization of nucleocytoplasmic traffic mediated by different members of the Kap family and the links to biological function for individual Kaps are evolving topics of study and discussion. Dozens to hundreds of cargoes were biochemically identified to be transported by well-studied Kaps such as IMPβ (via cargo adaptor IMPα), KAPβ2 and CRM1, but such lists for other Kaps were much smaller1. Fortunately, several recent proteomics studies expanded the lists of candidate cargoes for many importins, a few exportins and a few biportins; it is important to note, however, that most of the interactions of these cargoes with the Kaps have not been individually verified as yet5–12. Similar numbers of candidate import cargoes were identified for each Kap; one study found 49–66 import cargoes for each of the 10 human importins and 2 biportins, whereas another listed ~400–800 interaction partners (direct and indirect) for each of the 6 importins and 2 biportins studied8,9. This relatively even distribution of import traffic amongst Kaps is different from previous assumptions that the most-studied IMPα–IMPβ pathway imports more cargoes than other importin systems13. Some importins seem to import functionally related cargoes (TABLE 1); for instance, IMPβ imports many cargoes linked to gene expression, KAPβ2 cargoes include many mRNA processing factors and TNPO3 imports many cargoes linked to transcription elongation, mRNA splicing and export9. However, current understanding of the traffic mediated by many importins remains limited and it is difficult to define the functional preferences or significance of individual Kaps.
In studies of nuclear export traffic, a proteomics study of CRM1 identified ~1,000 potential export cargoes. Only ~10% of these proteins were previously reported as CRM1 cargoes. This suggested that CRM1 carries a far larger traffic volume than previously thought, covering broadly functioning cargoes, and that CRM1 may have a role in generally keeping cytoplasmic proteins out of the nucleus11. Additionally, proximity-dependent biotin identification (BioID) found hundreds of proteins that interact with the exportin CAS, which is generally thought to export only IMPα8. However, further analysis showed that many of these proteins are IMPα cargoes that could have been identified due to the close proximity of CAS with IMPα–IMPβ–cargo complexes during the disassembly of import complexes in the nucleus8. More comprehensive traffic analyses for other exportins have not been reported. Proteomics studies also reported new candidate export and import cargoes for the biportins XPO7 and IPO13 (REFS8,10,11). Gene Ontology analysis of candidate XPO7 cargoes indicates broad cellular functions, suggesting that XPO7 may be a more general exporter. Gene Ontology analyses were not reported for candidate CAS and IPO13 cargoes.
Proteomics analysis also revealed that a subset of import cargoes are transported by multiple importins, especially by a group consisting of IPO4, IPO5 and XPO7 (REF.8) and by highly conserved pairs of Kaps: KAPβ2 and KAPβ2b, as well as IPO7 and IPO8 (REF.9). Considerably fewer cargoes seem to be exported by multiple exportins8. An important caveat of proteomics studies is that individual identified cargoes have not been validated, and thus cannot be considered true cargoes. Experiments showing direct and RAN–GTP-sensitive Kap–cargo interactions (for import cargoes) or RAN–GTP-cooperative Kap–cargo interactions (for export cargoes) are necessary to eliminate false positives. Overall, the understanding of Kap traffic flow and overlap are just beginning to emerge, and will be improved by careful use of Kap-specific inhibitors (BOX 1).
Recognition of cargoes by Kaps
Kaps are proteins of molecular weight around 95–140 kDa and are composed of 19–24 HEAT repeats arranged in a superhelical or solenoid structure. The convex and concave surfaces of the superhelical structure as well as hydrophobic grooves between HEAT repeat helices and loops of various length that connect the HEAT repeats are utilized in different ways to recognize cargo, RAN–GTP and FG Nups. Kaps use diverse modes to bind cargoes, binding linear sequence elements (NLSs or NESs) and/or folded domains within the cargoes (FIG. 1b–d). The mechanisms of cargo recognition are known for some Kaps but remain undefined for many other Kaps.
HEAT repeats.
Structural motifs composed of two antiparallel α-helices that are usually connected by a loop. HEAT repeats occur in tandem to form solenoid or superhelical structures.
Recognition of linear targeting signals
Protein cargoes commonly use NLSs and NESs to bind their cognate Kaps for import and export, respectively. Only four classes of NLSs and one class of NESs have been defined, each for a specific Kap, and targeting signals for many Kaps remain undefined14,15. Each NLS/NES class has distinct sequence patterns and binds to different sites on their cognate Kap. NLSs/NESs do share some common features: they are often found in intrinsically disordered regions (IDRs) or at termini of cargoes; they become ordered, adopting either extended or helical conformations, upon binding Kaps; many NLSs are enriched in basic residues as Kaps are acidic; and the peptides in an NLS/NES class can be highly diverse in sequence. These common features and the exceptions are discussed below.
Intrinsically disordered regions.
(IDRs). Protein regions that do not have persistent tertiary structures.
NLS and NES classes.
The oldest class of NLS is the highly basic classical NLS (cNLS) that binds IMPα proteins (seven subtypes in humans; see Supplementary Table 1)16, which are adaptors for IMPβ. The two subclasses of cNLS are the monopartite cNLS that is 5–7 residues long (consensus: K-K/R-X-K/R) and the bipartite cNLS with 2 linker-connected basic clusters (consensus: K/R-K/R-X10–12-K/R3/5, where X10–12 is a linker of 10–12 residues and K/R3/5 refers to three basic residues within five consecutive residues)17,18. Recent findings suggest that the linkers can be much longer than the classical ones of 10–12 residues and may contain α-helical elements or even entire folded domains19,20. cNLSs bind two sites, major and minor, on the concave surface of the IMPα armadillo (ARM) repeat domain; most monopartite cNLSs bind to the major site and a few bind to the minor site, whereas bipartite cNLSs occupy both sites21. The residues that form the concave surface of IMPα, including the major and minor cNLS binding sites, are virtually identical between IMPα subtypes16. The amino-terminal IMPβ-binding (IBB) region of IMPα contains an NLS-like49KRRNV53 segment that binds the major cNLS-binding site of unliganded IMPα and is thus autoinhibitory22,23. IMPβ binding to the IBB frees up the IMPα major site to bind cNLSs on cargoes, increasing cargo-binding affinity by >100-fold24–27 (BOX 1; TABLE 1). However, some cNLSs bind to IMPα with such high affinity as to displace the IBB in the absence of IMPβ; examples include cNLSs of several inner nuclear membrane proteins (Supplementary Box 2). The extent of IBB autoinhibition varies for different IMPα subtypes, conferring subtype specificity for some cargoes that can bind IMPα in the absence of IMPβ28,29. The invariant cNLS binding surface shared by all IMPα subtypes suggests that subtype specificity is determined by interactions outside the cNLS-binding sites16,29. Additional examples of IMPα subtype specificity are discussed below (also reviewed elsewhere16).
Armadillo (ARM) repeat domain.
A protein domain typically 40 residues long that shares homology with repeating units in the ARM protein family. it contains two or three helices per repeat, which stack into a solenoid arrangement.
The second type of NLS defined is the proline-tyrosine NLS (PY-NLS) that binds KAPβ2 or KAPβ2b. PY-NLSs are 15 to >100 residues long, have overall positive charge and bind in mostly extended conformations to a negatively charged site of KAPβ2 (REFS1,30–32) (FIG. 2). PY-NLSs are very diverse in sequence and best defined by a set of physical, chemical and sequence rules. Sequence motifs include an N-terminal hydrophobic or basic motif and a carboxy-terminal R/K/H-X2–5-P-Y or R/K/H-X2–5-P-ϕ motif (ϕ, hydrophobic; H, histidine; P, proline; Y, tyrosine). The N-terminal, R/K/H and PY motifs quasi-independently contribute to overall binding energy and can be used in combination such that two of the three motifs can sometimes confer import activity. Hence, some PY-NLSs do not have a clearly recognizable PY motif30.
Fig. 2 |. Comparison of unliganded, cargo-bound and RAN-bound importins.
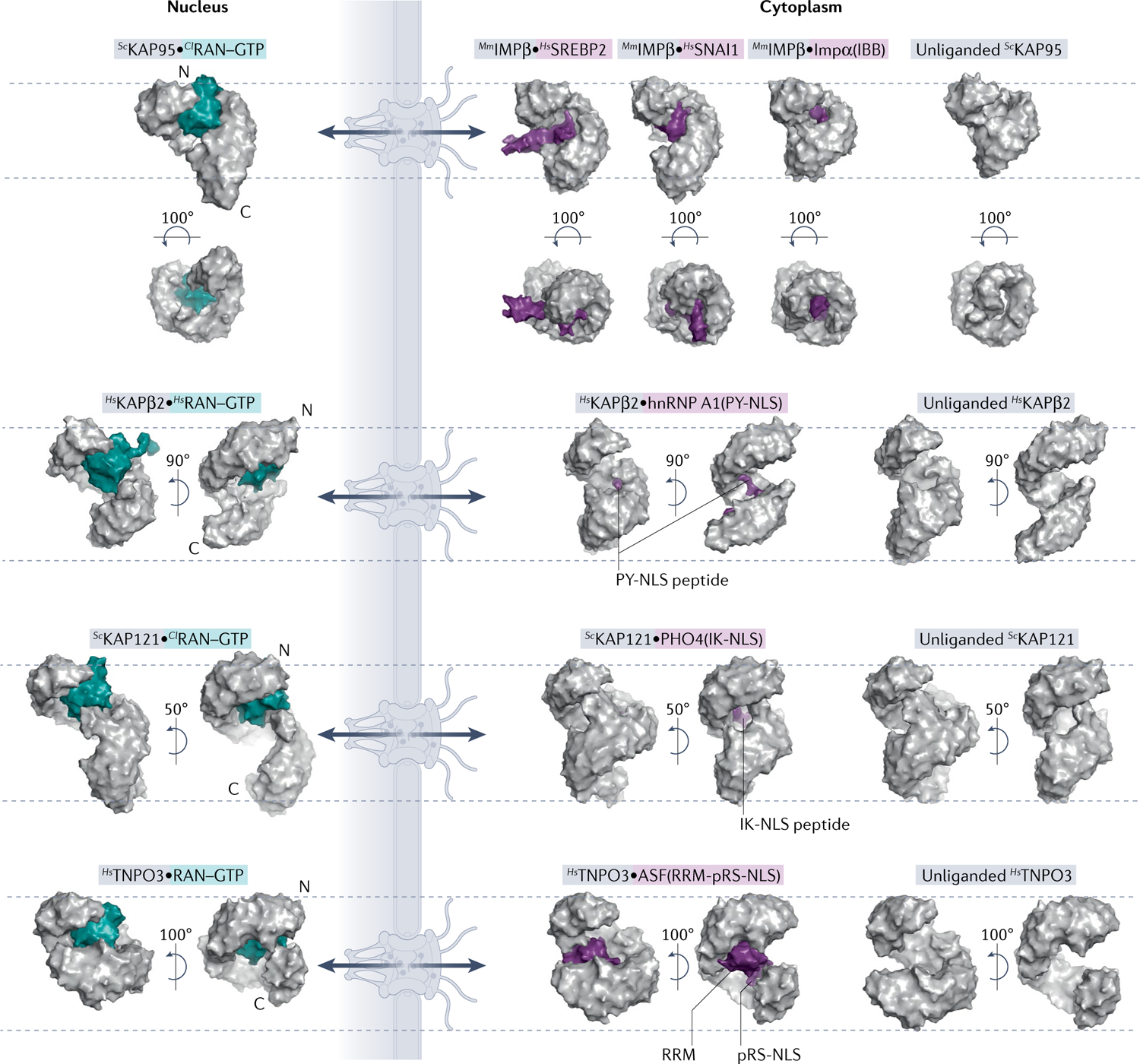
Surface representations of nuclear and cytoplasmic complexes of importins (grey) with RAN–GTP (teal) or cargo (purple). All importins are structurally aligned at residues 1–200 and displayed in the same orientation. Dashed lines mark top of first HEAT repeat and bottom of last HEAT repeat of each unliganded importin to help visualize importin conformational changes and differences in superhelical compactness. Cargo release by RAN–GTP results from either steric clash between RAN–GTP and cargo or conformational changes to the importin superhelix. PDB IDs (left to right): IMPβ [PDB:2BKU, PDB:1UKL, PDB:3W5K, PDB:1QGK, PDB:3ND2]; KAPβ2 [PDB:1QBK, PDB:2H4M, PDB:2QMR]; KAP121 [PDB:3W3Z, PDB:3W3X, PDB:3S3T]; and TNPO3 [PDB:4C0Q, PDB:4C0O, PDB:4C0P]. ASF, splicing factor 1; Cl, Canis lupus; hnRNP, heterogeneous nuclear RNP; Hs, Homo sapien; IBB, amino-terminal IMPβ-binding; IK-NLS, isoleucine-lysine nuclear localization signal; Kaps, Karyopherins; Mm, Mus musculus; PY-NLS, proline-tyrosine nuclear localization signal; RRM, RNA-recognition motif; RS-NLS, arginine-serine repeat nuclear localization signal; Sc, Saccharomyces cerevisiae; SREBP2, sterol regulatory element-binding protein 2.
The third NLS class is the isoleucine-lysine NLS (IK-NLS) that binds yeast KAP121 (FIG. 2). IK-NLSs have the consensus sequence K-V/I-X-K-X1–2-K/H/R, where the valine (V)/isoleucine (I) and the lysine (K) in the fourth NLS position are Kap121-binding hotspots and Kap–cargo specificity determinants33,34. Little is known about sequences that bind the homologous human IPO5, but KAP121 residues that contact IK-NLSs are conserved in IPO5, suggesting that it likely also binds IK-NLS-containing cargoes33. Both the IK-NLS and monopartite cNLS contain multiple lysine residues and are compact, but with different sequence patterns (K-V/I-X-K-X1–2-K/H/R versus K-K/R-X-K/R). Little is known about whether they bind KAP121/IPO5 and IMPα interchangeably. One study showed that overexpression of IK-NLS of Ubiquitin-like-specific protease 1 (ULP1) inhibited the KAP121 but not the IMPα pathway35, but potential cross-interactions need further investigation.
The fourth class of NLS is the TNPO3-binding arginine-serine repeat NLS (RS-NLS) found in RS domains (>50 residues long, >40% RS dipeptide) of many serine-arginine (SR) splicing factors. The RS-NLS is often immediately C-terminal to an RNA-recognition motif (RRM) domain. Many RS-NLSs must be phosphorylated on serine residues to bind TNPO3 (REFS36–38); the alternating arginine and phospho-serine residues bind complementary acidic and basic patches on TNPO3 (FIG. 2). Some cargoes, such as pre-mRNA cleavage factor CPSF6, have arginine-glutamate/aspartate instead of phospho-RS dipeptides36.
At this time, the only known export signal class is the one that binds CRM1, hence it is simply called the NES. NESs are usually 8–15 residues long and contain 4–5 hydrophobic anchor residues that interact with CRM1, arranged with various spacings14. The first NES sequences discovered had many leucines, but it is now obvious that NESs are very diverse in sequence and structure14,39,40. NESs can bind in both polypeptide directions and occupy different extents of a structurally invariant hydrophobic groove on the convex/outside surface of the ring-shaped CRM1 (REFS14,41–44) (FIG. 3a). NES peptides adopt various helix, β-strand, loop or turn structures that position hydrophobic side chains into hydrophobic pockets in the groove of CRM1. The only conserved structural element of the NES is a single helical turn that forms hydrogen bonds with a conserved CRM1 lysine that acts as a selectivity filter14. An active NES has to meet only this one structural requirement of three or four residues and the rest of it can be structurally diverse, resulting in the large sequence variability of NESs. At this time, there are 11 NES consensus patterns (TABLE 1), but there may be more to be uncovered. The very relaxed sequence requirements of an NES allow CRM1 to recognize thousands of diverse NESs in broadly functioning protein cargoes.
Fig. 3 |. Comparison of unliganded, cargo-bound and RAN-bound exportins and biportins.
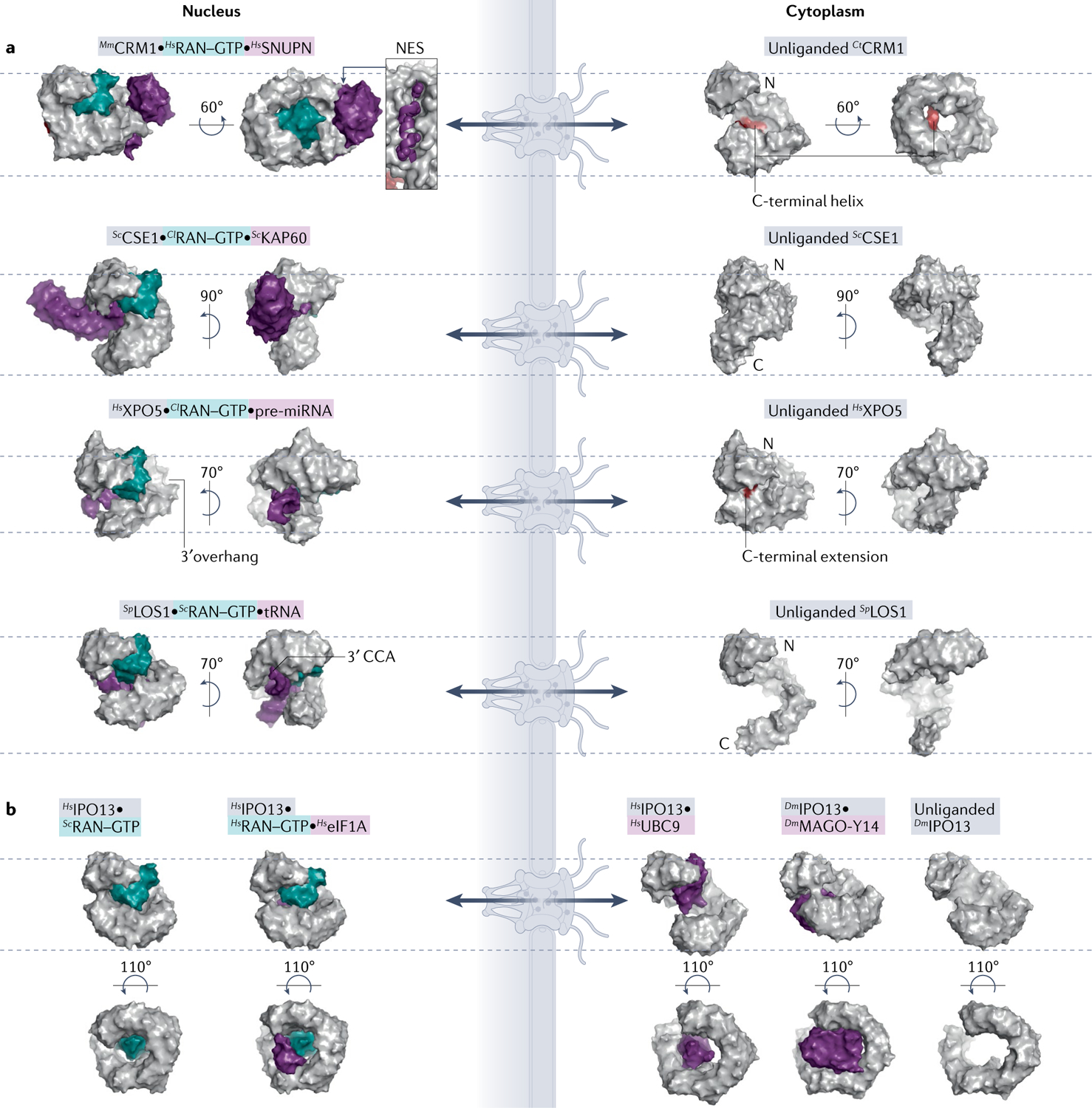
Surface representations of Karyopherins (Kaps; grey), Kap–RAN–GTP (teal)–cargo (purple) or Kap–RAN–GTP complexes. All Kaps are aligned, displayed in same orientation and marked with dashed lines as in FIG. 2. a | Exportin structures. Autoinhibitory carboxy-terminal helix/loop (red) of CRM1 and XPO5 are shown in the unliganded exportins. Locations of 3′ overhangs of pre-microRNA (pre-miRNA) and tRNA, recognized by different regions of XPO5 and LOS1, respectively, are also indicated. RAN–GTP binding to XPO1 and XPOT cause ring compaction for cargo binding, whereas RAN–GTP binding to CSE1 and XPO5 opens them up to accommodate cargoes. b | Biportin IPO13 structures. Cargo release from IPO13 is induced by RAN–GTP using steric clash mechanism and conformational changes to the Kap superhelix, to bind specific import and export cargoes. PDB IDs (left to right): CRM1 [PDB:3GJX, PDB:4FGV]; CSE1 [PDB:1WA5, PDB:1Z3H]; XPO5 [PDB:3A6P, PDB:5YU7]; LOS1 [PDB:3ICQ, PDB:3IBV]; and IPO13 [PDB:2X19, PDB:3ZJY, PDB:2XWU, PDB:2X1G, PDB:3ZKV]. Cl, Canis lupus; Ct, Chaetomium thermophilum; Dm, Drosophila melanogaster; eIF1A, eukaryotic translation initiation factor 1A; Mm, Mus musculus; NES, nuclear export signal; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe.
Potential NLSs and NESs.
Other Kap-binding segments within IDRs of cargoes have been identified, but their Kap-binding elements have not been determined and/or they have not been found in multiple different cargoes that are recognized by the same Kap. Therefore, they are yet to be classified as NLSs or NESs.
IMPβ binds directly to linear segments of several cargoes. The IBB in IMPα and other IMPβ adaptors, such as the U small nuclear RNP-specific nuclear import adapter Snurportin 1 (SNUPN)45 and Replication Protein A (RPA) complex import adaptor RPA-interacting protein (RIP)46, is not usually thought of as an NLS but it is much more similar to a linear signal than a folded domain. This ~40-residue region or motif, which is enriched in basic and hydrophobic residues, is intrinsically disordered but becomes a long helix that is wrapped around by the IMPβ superhelix upon IMPβ binding1,47 (FIG. 2). IMPβ also binds directly, without using IMPα, to at least 29 different cargo proteins that share no obvious common elements that bind IMPβ (TABLE 1). Of these, a few use disordered and basic linear cargo segments to bind IMPβ; they are often simply called ‘non-classical NLSs’. In particular, the neuroendocrine peptide parathyroid hormone-related protein (PTHrP) uses a 28-residue IDR segment to bind in an extended conformation to the N-terminal half of IMPβ, in a site that partially overlaps with the IBB-binding site48. PTHrP sequence motifs that are important for IBB have not been mapped and it is unclear whether PTHrP-like peptides exist in other IMPβ cargoes. Further studies of IBB-like and PTHrP-like cargo segments are needed before they can be classified as NLSs.
KAPβ2 also binds at least one other type of linear targeting element. The importin binds arginineglycine-glycine or RGG IDRs in many RNA-binding proteins, including the translational repressor cold-inducible RNA-binding protein (CIRBP) and multifunctional RNA/DNA binding protein fused in sarcoma (FUS) (Supplementary BOX 4). RGG regions bind dynamically to KAPβ2 sites that overlap with the PY-NLS binding site49–51. They bind weaker than PY-NLSs (TABLE 1) and are outcompeted by neighbouring PY-NLSs. For example, wild-type FUS uses its high-affinity PY-NLS to bind Kapβ2, but the amyotrophic lateral sclerosis-associated variant FUS(R495X) that is missing the PY-NLS uses RGG regions to bind KAPβ2 (REF.49). The prevalence of cargoes that use RGGs to bind KAPβ2 is undetermined and it is currently unknown whether there are additional sequence motifs unrelated to RGGs and the PY-NLS that also bind KAPβ2.
Amyotrophic lateral sclerosis.
A fatal neurodegenerative disease, with progressive loss of motor neuron control that leads to paralysis. amyotrophic lateral sclerosis onset and progression may be a consequence of protein misfolding/aggregation.
The remaining importins (IPO4, IPO5, IPO7, IPO8, IPO9 and IPO11) bind linear sequence elements in some cargoes, but the NLSs have not been defined due to lack of biochemical and structural information about how they bind their importins1. IPO5 is predicted to recognize IK-NLS-containing cargoes, but it is unclear whether it also recognizes other types of NLSs. Recent studies showed several importins (IPO4, IPO5, IPO7 and IPO9 along with IMPα, IMPβ and KAPβ2) binding the basic disordered tails of histones H3 and H4, but it is unclear how important these interactions are for nuclear import52. Slightly more is known for IPO7 as it was shown to mediate stimulated import of a few kinases (SYK, ERK2 and MEK1) and transcription factors SMAD3 and EGR1 upon phosphorylation of their serine-proline-serine motifs. It is not known whether the motifs bind IPO7 directly or whether phosphorylation exposes a separate NLS or an importin-binding surface. Comprehensive biochemical and structural analyses of candidate cargoes obtained from proteomics studies will likely reveal many new NLS classes.
No additional NES class, other than the one for CRM1, has been defined for other exportins. No other linear segments found in multiple cargoes are known to direct CRM1-mediated export. This is true for other exportins and biportins except for the yeast exportin MSN5, which exports many phosphorylated proteins by binding to a still undefined phospho-NES that is located in the IDRs of its cargoes. For example, transcription factor PHO4 must be phosphorylated at two specific sites within a 150-residue-long IDR inter-domain linker53. Sixteen other MSN5 cargoes, all involved in either environmental response or cell cycle regulation, contain IDR segments that drive export only when phosphorylated. No common sequence/structural features have been identified for this potential phospho-NES (Supplementary Table 2). Although the human homologue of MSN5 (XPO5) is known to export RNAs, its one putative protein cargo is the phosphorylated RNA editing enzyme ADAR1, suggesting that the recognition of a phospho-NES may be evolutionarily conserved54,55. Further studies are needed to understand how this phospho-NES binds its exportin.
Altogether, although five NES/NLS classes have been defined, cargoes for many Kaps need to be studied to reveal new NLS and NES classes. Detailed knowledge of the sequence and structural requirements for interactions between Kaps and linear signals within their cargoes will improve existing NLS/NES predictors (BOX 2) and enable development of new predictors.
Box 2 |. Prediction tools for nuclear localization and export signals.
Various tools using different approaches such as sequence similarity and machine learning are now available to predict nuclear localization signals (NLSs) and the CRM1-dependent nuclear export signals (NESs). All tools utilize sequence information from known NLSs/NESs to look for putative signals, whereas some include additional known features of NLSs/NESs such as higher disorder propensity. Performance of these tools are often evaluated by precision (percentage of true signals amongst predicted hits) and recall (percentage of true signals retrieved by prediction). Prediction precision is moderate for NLSs and poor for the NES. Therefore, predicted signals must be experimentally verified both in isolation (as peptides) and in the context of the full-length proteins.
Machine learning.
A branch of artificial intelligence that automates iterative analytical model building based solely on training data, with minimal human intervention.
NLS predictors
PSoRT (or PSoRT II)258,259 and cNlS mapper260 are focused on the prediction of classical NLSs (cNLSs). PSORT uses machine learning with a training set of eukaryotic proteins with known subcellular localization to predict the localization of an input query and look for motifs in the query that match defining features of the monopartite and bipartite cNLS consensus sequences. It has the best recall of all methods, but its precision is among the lowest. cNLS Mapper uses scoring of individual residues based on a nuclear import activity profile generated by extensive mutagenesis screening of several classes of cNLSs in a yeast import assay to predict whether any motifs are putative NLSs. PredictNlS matches motifs within the query to a database of expanded putative NLS sequences that include variants of known cNLSs and the M9 sequence (the proline-tyrosine NLS (PY-NLS) of heterogeneous nuclear RNP (hnRNP) A1)261. Several other tools that implement machine leaning approaches are also available, using different algorithms to mine NLS defining features from different training data sets. NucPred uses nuclear and non-nuclear proteins as a training set for their algorithm262. NucImport also uses proteomes as their training set similar to NucPred but further incorporates information from NLSdb263 (a database of NLSs that are not exclusively cNLSs) and protein interaction networks264. NlStradamus uses a data set of experimentally verified yeast NLSs (not limited to cNLSs)265. SeqNlS uses verified NLSs from NLSdb as a training set and included bipartite NLS motifs, disorder prediction and conservation with the NLS sequence266. INSP uses a slightly bigger data set from the more updated version of NLSdb267 and also integrates information such as the physical chemistry of amino acids, disorder propensity and evolutionary conservation268. SeqNLS has the highest precision (~60–70%) out of all methods for multi-organism data sets, whereas NLStradamus and cNLS Mapper perform better (~80%) in yeast266. INSP is the newest tool and has shown improvement in recall (~70% versus 40–50% of SeqNLS) but precision is reduced to ~50%, which is not ideal. All of the predictors above use the cNLS consensus or a database of known NLSs (mostly cNLSs) as a training set, and therefore cannot effectively predict other NLSs such as PY-NLS, isoleucine-lysine NLS (IK-NLS) or arginine-serine repeat NLS (RS-NLS). Although PredictNLS included M9 sequences in its data set, due to the low sequence conservation of PY-NLSs, performance for looking for PY-NLSs in particular is expected to be poor.
NES predictors
All NES predictors uses machine learning methods with known NES sequences as training data sets. NetNeS is trained on NESbase269 (the first NES database of 75 entries) using sequence information only270. NetNES and NESbase were developed in 2004 prior to knowledge of what NESs look like when bound to CRM1. NESsential (no longer available) was developed to use a larger training data set derived from NESbase with a different algorithm, applying two NES consensus patterns, as well as secondary structure, disorder and solvent accessibility predictions271. Wregrex is trained on the same set of sequences as NESsential with a different algorithm272. NeSmapper uses a larger data set (including newly discovered NESs and synthetic sequences) and incorporates activity-based scoring (similar to cNLS Mapper) of five NES consensus patterns along with properties of flanking sequences, which improved prediction performance over NESsential and Wregex273. locNeS uses a similarly sized data set to NESmapper and integrates algorithms used in NESsential and Wregrex with an expanded NES consensus (six patterns) and disorder scoring, and has the best performance to date with ~50% precision at 20% recall274. Nologo used a different algorithm on yeast-only NESs without limiting the NES consensus pattern and length; however, it did not improve performance over LocNES275. Recently, a structure-based method using the binding energy of putative NES sequences modelled into the groove of diverse template structures of NES-bound CRM1 groove was found to reduce false positives in NES predictions, but a larger data set of validated NESs with binding affinities is needed to improve the method276.
Recognition of folded domains
Some protein cargoes use folded domains rather than linear NLSs/NESs to bind their Kaps. Although IMPα predominantly binds to linear cNLSs, a few cargoes exclusively use folded domains to bind a distinct non-cNLS-binding site on IMPα, thus conferring IMPα subtype specificity (for example, Ebola virus secondary matrix protein VP24 binding to IMPα6 (REF.56)). Several IMPβ−cargo interactions also involve globular cargo domains. Examples include the cholesterol regulator sterol regulatory element-binding protein 2 (SREBP2) and the transcription factor zinc finger protein SNAI1, which bind IMPβ through their dimeric helix–loop–helix leucine zipper domain and zinc finger domains, respectively. Both IMPβ-binding domains are enriched in basic residues, and the flexible IMPβ superhelix adopts different helical pitches and uses different sites to bind these very differently sized and shaped domains57,58 (FIG. 2). Due to sterically clashing binding sites and/or allosteric conformation changes of IMPβ, the binding of these domains and the IMPα IBB to IMPβ is mutually exclusive, with the IBB binding with higher affinity. It is likely that more cargoes will be found to bind IMPβ through folded globular domains.
Other importins also bind folded domains of their import cargoes. IPO9 was recently shown to bind the highly basic histone H2A–H2B dimer by wrapping around its globular histone-fold domain59. IPO9 was also reported to bind very differently folded cargo regions such as the C-terminal HEAT repeats of serine-threonine-protein phosphatase PP2A and the homeobox domain of developmental regulator Aristaless60,61. Similar to IMPβ, IPO9 probably uses different sites and binding modes to bind structurally diverse cargo domains. IPO4, IPO5, IPO7, IPO8 and IPO11 all bind diverse folded cargo domains but little information is available about these interactions1,11. IPO5 binds to a helical region of the high-density lipoprotein component apolipoprotein A-I, IPO8 binds the globular single-domain eIF4E and IPO11 binds the compact five-helix matrix domain of the Rous sarcoma virus Gag polyprotein1,62.
Biportins IPO13, XPO4 and XPO7 also bind folded domains of their import cargoes. Recent structures showed IPO13 using different sites to bind the diverse domains of two different import cargoes: the exon junction complex MAGO-Y14 (REF.63) and SUMO-conjugating enzyme UBC9 (REF.64) (FIG. 3b). The yeast homologue of IPO13, PDR6, also imports yeast UBC9 but uses a binding mode different from that of IPO13 (REFS12,64,65). IPO13 and PDR6 bind different regions of UBC9 using their N-terminal and C-terminal HEAT repeats, respectively. XPO4 binds the three-helix HMG box domains of import cargoes Sex-determining Region Y protein (SRY) and SRY-box transcription factor 2 (SOX2)66. A recent proteomics study identified structurally diverse candidate import cargoes for XPO7 but it is not yet known whether they bind using linear elements or folded domains11.
Exon junction complex.
A multiprotein complex that binds to the junction between exons in nuclear precursor mrNas and remains bound during their export to the cytoplasm.
Many export protein cargoes use folded domains to bind exportins (other than CRM1 and MSN5) and biportins. The exportin CSE1 (yeast homologue of CAS) wraps around both the ARM domain of autoinhibited KAP60 (yeast IMPα homologue) and RAN–GTP (FIG. 3a). XPO6 exports the profilin–actin complex, which has only folded globular domains and no IDRs; however, XPO6 binding to profilin–actin has not been biochemically demonstrated and it is unclear whether additional factors, potentially with linear motifs, are involved. Notably, the nuclear tumour suppressor protein RASSF1A appears necessary for RAN–GTP interaction with XPO6 and for nuclear export of profilin–actin67. Biportins IPO13, XPO4 and XPO7 also recognize diverse folded domains, such as histone fold heterodimers and helical homeodomains, in their export cargoes (reviewed elsewhere1). IPO13 binds the folded domain of export cargo eukaryotic translation initiation factor 1A (eIF1A) that binds to a site distinct from those for import cargoes68 (FIG. 3b). XPO4 binds the MH2 domain of SMAD3 (REF.69), as well as the SH3-like domain and OB domain of eIF5A (REFS70,71).
MH2 domain.
A protein domain found in the carboxy-terminal portion of sMaD proteins. it comprises a β-sandwich fold with a three-helix bundle on one end and a loop–helix region on the other.
SH3-like domain.
A small globular domain that binds poly-proline motifs, comprising five or six β-strands tightly packed into antiparallel β-sheets.
OB domain.
A small globular domain composed of two three-stranded antiparallel β-strands packed into a flattened β-barrel.
In summary, Kaps use many different binding sites to bind diverse folded cargo domains. The flexible superhelical architecture also enables Kaps to adopt various conformations to accommodate diverse cargo domains.
The combined use of motifs
A Kap may also bind to both a linear element and folded domain in a cargo. For example, folded domains immediately adjacent to linear cNLSs (such as in RCC1) require certain IMPα subtypes with more flexible ARM domains such as IMPα3 to accommodate the bulky fold29,72,73. The unusual IMPβ–IPO7 heterodimer binds both the globular histone-fold domain of histone H1 and its C-terminal disordered tail74. TNPO3 binds both the RS-NLS and the RRM domain of splicing factor 1 ASF1 (also known as SF2) (FIG. 2); the RS-NLS is the primary TNPO3-binding determinant and the RRM a secondary element1,36,75,76. Many TNPO3 cargoes have RRM domains but many candidate TNPO3 cargoes identified by proteomics do not75; it is unclear how prevalent TNPO3–RRM interactions are. Lastly, SNUPN binds CRM1 in a multipartite manner using an N-terminal NES, a folded globular m3g cap-binding domain and a disordered C-terminal tail, all binding to the outer convex surface of CRM1 (REFS41,42) (FIG. 3a). It is not known whether other cargoes also use folded domains, with or without an NES, to bind CRM1.
m 3G cap.
A 2,2,7-trimethylated guanosine cap structure of uridylate-rich small nuclear rNas (U small nuclear rNas), which are the rNa components of spliceosomal ribonucleoproteins (small nuclear RNP).
RNA cargoes
The exportins XPO5 and XPOT are known to export RNA rather than protein cargoes, being primary exporters of pre-microRNAs (pre-miRNAs) and tRNAs, respectively. Both XPO5 and LOS1 (Schizosaccharomyces pombe homologue of XPOT) wrap around their double-stranded RNA cargoes, making electrostatic interactions with the negatively charged RNA backbones and the pre-miRNA 3′ overhang or the tRNA 3′ CCA motif (FIG. 3a; reviewed in REF.2). Notably however, these RNA species can be exported by multiple exportins77. For instance, XPO5 also exports tRNAs but likely binds them differently to pre-miRNAs78,79.
Some RNA cargoes do not bind directly to their Kaps but bind via adaptor proteins. SNUPN is an import adaptor that binds m3G-cap-containing spliceosomal U small nuclear RNPs and IMPβ45. Many different RNAs including mRNA80, tRNA77, small nuclear RNA and small nucleolar RNA81, miRNA82,83, telomerase RNA84 and viral RNA85 are exported by CRM1, all by binding to different adaptor proteins, such as nuclear RNA export factor NXF3 (REF.86), RNA helicase DBP5 (REF.87), phosphorylated adaptor for RNA export (PHAX)88 and HIV transactivating protein Rev89, which drive export via their NESs. It is unclear whether the RNA cargoes make direct contact with CRM1 in addition to binding the adaptors.
In general, shape and charge complementarity govern all Kap–cargo (including protein and RNA) interactions. The flexible HEAT repeat architecture allows recognition of diverse signals and/or structural features/shapes of the cargoes. The use of adaptors further diversifies cargo recognition modes and expands Kap cargo repertoires.
Cargo loading and unloading via RAN–GTP
RAN–GTP, but not RAN–GDP, binds all Kaps to regulate cargo binding and allow cargo loading/unloading in both transport directions. All Kaps use their N-terminal portion comprising ~100 residues (HEAT repeats 1–4) to bind the switch 2 feature of RAN–GTP. Kaps also use additional HEAT repeats in the N termini and, sometimes, C termini to bind switch 1 and the basic patch of RAN–GTP33,42,63,65,68,70,75,90–94 (FIG. 1c). Additionally, IMPβ, KAPβ2, TNPO3, KAP121 and CSE1 use long acidic loops between or within HEAT repeats to bind switch 1 or the basic patch of RAN–GTP33,75,90,91,94. RAN–GTP binds tightly to all importins, has high-to-moderate affinity for biportins and shows weak to no binding to most unliganded exportins68,71,95 (TABLE 1). Here, we review the different ways Kaps use their flexible HEAT repeat architecture to achieve mutually exclusive binding of RAN–GTP versus import cargo, or the RAN–GTP-dependent binding of export cargo (FIG. 1d).
RAN–GTP-regulated cargo import
For nuclear import, the binding of RAN–GTP versus cargo is mutually exclusive. This selection is driven by the existence of a steric clash between import cargo and RAN–GTP when binding importin (steric mechanism), by the diverse conformational changes of the importin upon binding RAN–GTP that abolish cargo binding sites (allosteric mechanism) or a combination of the two mechanisms (FIGS 1b–d and 2). TNPO3 uses a steric clash mechanism, with overlapping RAN–GTP and RS-NLS binding sites75, whereas IMPβ primarily rearranges HEAT repeat packing into a conformation that is incompatible with cargo binding96,97. Other importins use combinations of steric and allosteric mechanisms. A long acidic loop between the helices of HEAT repeat 8 in KAPβ2, which is disordered in the KAPβ2–cargo complex, undergoes a drastic conformational change when it interacts with RAN–GTP, allowing it to engage the PY-NLS binding site and displace the cargo94. For KAP121, RAN–GTP binds at a site that partially overlaps with the IK-NLS binding site, and also causes the KAP121 superhelix to elongate, closing the IK-NLS binding pockets33.
Although, in most cases, binding of RAN–GTP to the importin is sufficient to release the cargo, there are a few reports of importin–cargo complexes where RAN–GTP alone is not able to mediate this release. The most clearly demonstrated example is the RAN–GTP inability to release histones H2A–H2B from IPO9 (REF.59) or from its yeast homologue KAP114 (REFS98,99). RAN–GTP is also unable to release another KAP114 cargo, the TATA-binding protein100, or the mRNA-binding protein NPL3 from its importin, MTR10 (REF.101). Other examples with conflicting data also exist, such as the release of RNA-binding proteins NAB2 and NAB4/HRP1 from yeast KAP104 (KAPβ2 homologue)31,102; further studies are needed to confirm the RAN–GTP insensitivity of this release. In all of these cases, nucleic acid targets (chromatin, RNA) of the cargoes are required along with RAN–GTP for efficient cargo release, suggesting that locations of cargo release in the nucleus may be specified by downstream functions of the cargoes. The mechanisms of this cargo release mediated by the combined effect of the nuclear target and RAN–GTP are unknown at this time.
RAN–GTP-regulated cargo export
To enable nuclear export, exportins show little or no binding to cargo or RAN–GTP alone, and instead bind both cooperatively. XPO5/MSN5 is an exception as it binds stably to RAN–GTP alone and to some primary miRNA precursors95,103,104, perhaps suggesting additional miRNA processing roles for this exportin104. Coupled cargo and RAN–GTP binding involves various large conformational changes in most exportins and direct contacts between RAN–GTP and cargo that stabilize the ternary complex (FIGS 1b–d and 3a; CRM1 is an exception, see below). Unliganded CSE1 and XPO5 form closed-ring structures that open up when binding both cargo and RAN–GTP to accommodate both molecules inside the exportin solenoids91,93,103,105. Conversely, unliganded CRM1 and LOS1 adopt open conformations, but undergo compaction when binding cargo and RAN–GTP to interact with both molecules favourably; compaction of CRM1 is one of the many conformational changes that leads to the opening of the NES binding groove (reviewed elsewhere106 and more below) whereas the LOS1 superhelix clamps onto both RAN–GTP and cargo41,42,92,107. Additionally, binding of RAN–GTP displaces C-terminal autoinhibitory tails/helices of both CRM1 and XPO5, to facilitate the opening or closing of the exportin superhelix, respectively103,108,109.
As mentioned earlier, CRM1 is the only exportin where the RAN–GTP and cargo binding sites are distant, with no direct RAN–GTP–cargo contacts. This is because CRM1 uses its outer/convex surface to bind cargoes, whereas Ran–GTP interacts with the inner/concave surface. Coupling of RAN–GTP and cargo binding is thus driven by conformational changes to open or close the NES-binding groove in CRM1. The precise mechanisms for CRM1 export cargo dissociation vary in yeast and human even though both involve RanBP1. Yeast RanBP1 binds the inner CRM1 surface in the RAN–GTP–CRM1–cargo complex, which induces a conformational change that closes the NES-binding groove to release cargo110. By contrast, human RanBP1 strips RAN–GTP from the RAN–GTP–CRM1–cargo complex to cause subsequent cargo release111.
Cargo loading/unloading in biportins
Biportins use the same mechanisms as importins and exportins to achieve RAN–GTP selective loading/unloading of cargoes (FIG. 3b). RAN–GTP binds to the cargo (UBC9) binding sites of IPO13 and PDR6 to sterically displace the cargo; RAN–GTP binding also causes a slight compaction of the biportin superhelices64,65. However, RAN binding and slight compaction of IPO13 do not release import cargo MAGO-Y14, which requires export cargo eIF1A for efficient dissociation in vitro63,64, suggesting that import and export of some cargoes by may be coupled. For export cargoes, RAN–GTP binding to IPO13, PDR6 and XPO4 causes their superhelices to twist and tighten to achieve more contacts with export cargoes, and RAN–GTP also contacts cargoes to stabilize the export complexes65,68,70.
In summary, Kaps adapt to RAN–GTP binding quite differently to enable either nuclear import or export. Kap interactions with RAN–GTP either abolish binding sites for the import cargo to release cargo in the nucleus, or they help establish favourable binding sites for export cargo to increase its binding in the nucleus. RAN–GTP itself may also inhibit the binding of import cargo through steric hindrance or may facilitate the binding of export cargo by contacting both cargo and Kap.
Regulation of Kap-mediated transport
The nuclear versus cytoplasmic distributions of many proteins remain steady over the long term, but the distributions of many other proteins are dynamic, changing according to the progression of cell cycle, according to environmental cues or to generate specific cellular responses (FIG. 4a). This can be achieved by modifications of the Kap system via several mechanisms involving the cargo itself and/or the Kaps, which impact the transport rates of some or all cargoes in a particular direction.
Fig. 4 |. Regulation of Kap–cargo interactions.
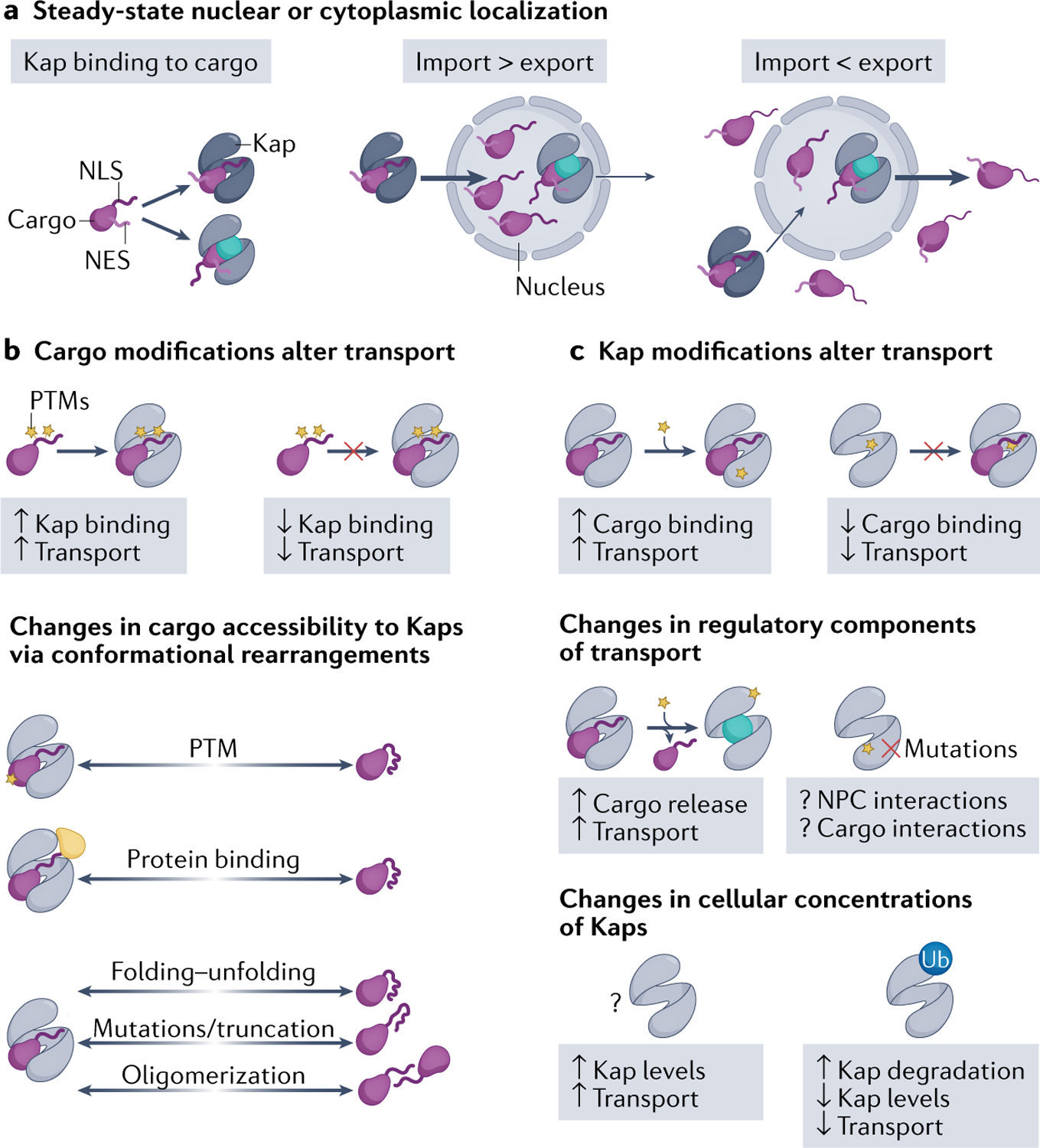
a | At steady state, cellular localization of cargo depends on whether the nuclear import or export flux is stronger. Overall transport flux can change due to various modifications on either particular cargoes (part b) or the Karyopherins (Kaps) that may affect transport of some or all cargoes (part c). b | Cargo modifications include post-translational modifications (PTMs), binding to a protein partner, mutations/truncations and changes to protein folding–unfolding and to cargo oligomerization state. PTMs can either activate or inactivate nuclear localization/export signals and directly affect Kap–cargo affinity to increase or decrease transport. Cargo modifications can also indirectly affect Kap–cargo affinity, by inducing conformational changes in the cargoes that then alter accessibility of the cargo elements that bind Kaps. c | Changes to Kaps that control transport flux include PTMs of Kaps and changes in Kap expression levels. PTMs of Kap can directly affect Kap–cargo binding affinities. PTMs can also alter Kap interactions with other regulatory components of transport, such as RAN–GTP, nucleoporins and other Kaps. Overall traffic can also be regulated by changes in Kap expression levels, whether physiological regulation by PTMs or aberrant accumulation in cancers and disease. NES, nuclear export signal; NLS, nuclear localization signal; NPC, nuclear pore complex; Ub, ubiquitylation.
Changes to cargoes
Studies of the IMPα–IMPβ and CRM1 systems show that higher cNLS/NES binding affinities correlate with greater nuclear or cytoplasmic localization within an optimal range, respectively27,112 (TABLE 1). Post-translational modifications (PTMs) in or adjacent to signals in the cargo that bind to Kaps can change Kap–cargo affinities and thus cause a change in their cellular localization (FIG. 4b). The effects of PTMs are usually highly dependent on their position within the full-length cargo proteins. This topic was previously reviewed113,114 and we highlight recent examples here. PTMs of NLSs/NESs, such as phosphorylation, acetylation or methylation, are known to affect Kap affinities. In some instances, the effects are predictable; for example, phosphorylation of the MSN5-binding cargo segments and the TNPO3-binding RS-NLS is critical for Kap binding, whereas arginine methylation of the FUS RGG regions impairs FUS interaction with KAPβ2 (REF.115). In other cases, the effects are difficult to predict; for example, phosphorylation of cNLSs inhibits import of some cargoes but enhances import of others. Recent examples of cNLS phosphorylation include serine phosphorylation of the bipartite cNLS linkers of tumour suppressor protein 53BP1 (REF.116) and peptide hormone GLP1 (REF.117) that decreased importin binding and nuclear import. Acetylation of lysines in the cNLSs of the glucose metabolic enzyme PFKFB3 (REF.118) and the antiviral protein IFIX119 also inhibited import, but lysine acetylation of the tyrosyl-tRNA synthetase (TyrRS) cNLS promoted import; in the latter case, the unacetylated cNLS is helical and acetylation is suspected to destabilize the helix to generate an extended and active cNLS120. Effects of NES modifications are similarly difficult to predict. Phosphorylation near the NES of transcription factor TFEB121 increased CRM1 binding but phosphorylation of the hypoxia-inducible factor HIF2α NES is inhibitory122. Methylation of a lysine adjacent to the NES of the transcriptional regulator YAP1 also inhibited CRM1 binding123.
Modifications that change the accessibility of NLSs/NESs to Kaps can also affect Kap–cargo binding (FIG. 4b). Conformational changes of the cargo or changes in cargo oligomerization may expose or sequester targeting signals. Both types of changes may be induced by PTMs or by binding to additional macromolecule partners. Classic examples include tetramerization of the tumour suppressor protein p53 that masks its NES, and the masking of the NLS in the transcription factor NF-κB/RELA by its binding to a regulatory partner protein IκB (reviewed in REF.124). More recently, dephosphorylation of serine residues distant from the putative NLS of transcription factor YB1 was suggested to increase NLS accessibility and nuclear entry during the late G2/M phase of the cell cycle, mediated by a conformational change125. By contrast, phosphorylation of the apoptotic regulator caspase 2 causes binding of scaffolding protein 14-3-3 that masks the caspase 2 cNLS, inhibiting its nuclear import and activation126,127. Similar control of NES accessibility has also been observed. Acetylation of the N-terminal IDR region in the fly homeobox protein Ubx, which is far from its NES, seems to mask the NES via an unknown mechanism, to inhibit export128.
Mutations of NLSs and NESs, such as those that occur in neurodegenerative diseases and cancers, change cargo localization by disrupting functional targeting signals or by generating new ones. Many mutations in and/or adjacent to the PY-NLSs of mRNA binding proteins FUS and heterogeneous nuclear RNPs (hnRNPs) A1 and A2B1 were found in patients with familial amyotrophic lateral sclerosis and multisystem proteinopathy129–132. Mutations in the PY-NLSs of hnRNPs H1 and H2 are also thought to drive the mental retardation, X-linked, syndromic, Bain type (MRXSB) disease133,134. All these mutations likely decrease KAPβ2 binding and disrupt nuclear import. Conversely, frameshift mutations in nucleophosmin create potent NESs that lead to aberrant CRM1-mediated nuclear export in a subtype of acute myeloid leukaemia135. The W1038X truncation mutation in tumour suppressor PALB2 also exposes an otherwise occluded NES, mislocalizing the protein in breast/ovarian cancers136. Alternatively, amyotrophic lateral sclerosis-linked mutations in superoxide dismutase 1 (SOD1) cause SOD1 misfolding that exposes an NES for CRM1 export137.
Multisystem proteinopathy.
A group of inherited disorders that cause neurodegeneration, myopathy and bone disease, and can manifest as amyotrophic lateral sclerosis, frontotemporal dementia, inclusion body myopathy, Paget’s disease of bone or their combination.
In summary, modifications to cargoes, both within and outside Kap-binding regions, can change Kap–cargo binding affinities in many different ways. These changes to cargoes ultimately result in their altered subcellular localization.
Changes to Kaps
Multiple examples of PTMs of Kaps have been reported. These modifications can change Kap–cargo affinities, either by direct chemical changes to cargo binding sites or, indirectly, by allosterically changing the conformations of cargo binding sites (FIG. 4c). An example of the former includes S-nitrosylation of cysteines at the NES-binding groove of CRM1 that inhibits NES binding138. Examples of allosteric mechanisms include phosphorylation of the C-terminal autoinhibitory tail of CRM1 that activates export139, and XPO5 phosphorylation by kinase ERK that enables binding to the peptidyl-prolyl cis/trans isomerase PIN1, which stabilizes a XPO5 conformation that disfavours pre-miRNA binding140. More unusual examples of Kap modifications include disulfide linkage of KAPβ2 to the transcription factor FOXO4 in response to reactive oxygen species, which leads to FOXO4 nuclear entry141.
PTMs of Kaps can also modulate interactions with other proteins to affect nuclear transport. sumoylation of KAP114 stimulates import by facilitating RAN-induced cargo release, but the mechanism is still unclear142. Phosphorylation of IPO13 by kinase PKA upon hormonal stimulation reduced nuclear entry of the Kap and increased binding of cargo transcription factor PAX6 but reduced its nuclear import, suggesting that factors other than Kap–cargo affinity decreased PAX6 import143. PTMs of Kaps also lead to changes in their cellular concentrations and thus change their global import/export profiles. CRM1 phosphorylation by kinase AKT3 induces CRM1 degradation, causing nuclear accumulation of cargo transcriptional coactivator PGC1α, which is critical for regulating mitochondrial biogenesis and driving prostate cancer metastasis144,145.
Many PTMs and mutations of Kaps as well as changes in their cellular concentrations are associated with diseases. CRM1 has emerged as a viable therapeutic target146,147 as it is overexpressed in many cancers, and mutations in its NES-binding groove are associated with tumorigenesis and with poor prognosis in blood cancers148–151. The oncogenic, charge-reversal E571K mutation in CRM1 was reported to affect binding of a subset of NESs/cargoes152–154. A SILAC-based mass spectrometry study identified some cargoes with altered cellular localization in B cells carrying the heterozygous E571K CRM1 mutation153. A separate study showed that export cargo P-body protein eIF4E transporter was mislocalized to the nucleus in cultured cells and in leukaemia cells carrying the E571K mutation in CRM1. This was accompanied by decreased binding of mutated CRM1 to eIF4E transporter, suggesting that E571K mutation impairs cargo–CRM1 interaction, resulting in defective export154. However, a recent study showed that this mutation does not affect CRM1-mediated export, but that mutated CRM1 is instead enriched at the nuclear envelope via an unknown IMPβ-dependent mechanism with an unknown effect155. It remains unclear how CRM1 mutations drive tumorigenesis. XPO5 overexpression, mutations and modifications are also found in various cancers156. Phosphorylation and sumoylation of XPO5 suppress pre-miRNA export, which leads to aberrant expression of oncogenic proteins140,157.
P-body.
A type of cytoplasmic granule containing mrNas and proteins that is involved in rNa metabolism with liquid droplet properties.
Similar to cargo modifications, Kap modifications affect cellular localization of their cargoes via many different mechanisms. However, the modification of a Kap likely has a larger effect as it affects at least a subset if not all of its cargoes. Diverse changes to cargoes and Kaps thus allow complex regulation of the nucleocytoplasmic transport and the subcellular distribution of cargoes that is integrated into a wider cell biology, allowing appropriate responses to various cues. However, it is also apparent that aberrant changes to Kaps and/or their cargo can perturb the physiological distribution of cargo, which may contribute to disease.
Kaps in biogenesis of large assemblies
Many large (hundreds to thousands of kilodaltons) macromolecular machineries/assemblies are either assembled in the nucleus and/or function in the nucleus, raising questions of whether they are assembled before or after nuclear import and how assembly is coordinated with import/export. We review these questions for three assemblies that each function in major gene expression processes: replication, transcription and translation.
Import of the replisome core
Assembly of the multiprotein DNA replication machinery, or replisome, on DNA has been reviewed extensively, but its trafficking to the nucleus is rarely discussed. Components of the eukaryotic replisome core include the CMG helicase (named for its components — cell division control protein 45 (CDC45), minichromosome maintenance complex (MCM2–MCM7) and the GINS complex (SLD5–PSF1–PSF2–PSF3)), three DNA polymerases (Polε, Polδ and Polα-primase), processivity clamp proliferating cell nuclear antigen (PCNA) and clamp loader replication factor C (RFC), clamp unloader RFC-like complexes (RLC) and the single-stranded DNA-binding protein RPA (Fig. 5a). The components of the CMG helicase seem to be imported separately prior to their assembly in the nucleus. Yeast IMPα–IMPβ is responsible for importing the bipartite cNLS-containing CDC45 (REFS158,159) and the MCM2–MCM7 complex (where MCM2 and MCM3 have cNLSs)160–162. The latter is assembled in the cytoplasm by loading factor CDT1, which is imported with the MCM complex to load it onto DNA163,164. It is not known how the GINS complex is trafficked. Mechanisms of trafficking for the remaining replisome components are largely unclear, with some conflicting reports. The p66 and p125 subunits of human Polδ have putative cNLSs165,166. However, the C-terminal α-helical putative cNLS of p125 is atypical and it is unclear whether it is functional for import167. No information is found on nuclear import of Polε. The mouse Polα-primase is imported as a heterotetrameric complex; putative cNLSs in its p180 and p54 subunits direct import of smaller assemblies but are dispensable for import of the larger complex168,169. A bipartite cNLS in p180 is buried in a folded domain, unlikely to drive import170,171. The homotrimeric PCNA binds IMPβ but not IMPα, and the IMPβ-binding region (conserved from human to yeast) is partially located at the trimer interface172,173, suggesting that PCNA is trafficked as a monomer either by binding directly to an importin or by binding the cNLS-containing cyclin-dependent kinase inhibitor p21 (REF.174). The large subunit of the penta meric human RFC clamp loader, replication factor C subunit 1 (RFC1), and that of an RLC clamp unloader, ATPase family AAA domain-containing protein 5 (ATAD5), have putative N-terminal NLSs that have not been verified experimentally175,176. Nuclear import of RPA requires its C-terminal tetramerization domain, but there are conflicting reports of different importers including KAP95 and MSN5 in yeast, and the involvement of RIP as an IBB-containing IMPβ adaptor for RPA in the frog46,177–179.
Fig. 5 |. Nuclear trafficking of three different gene ▶expression machineries.
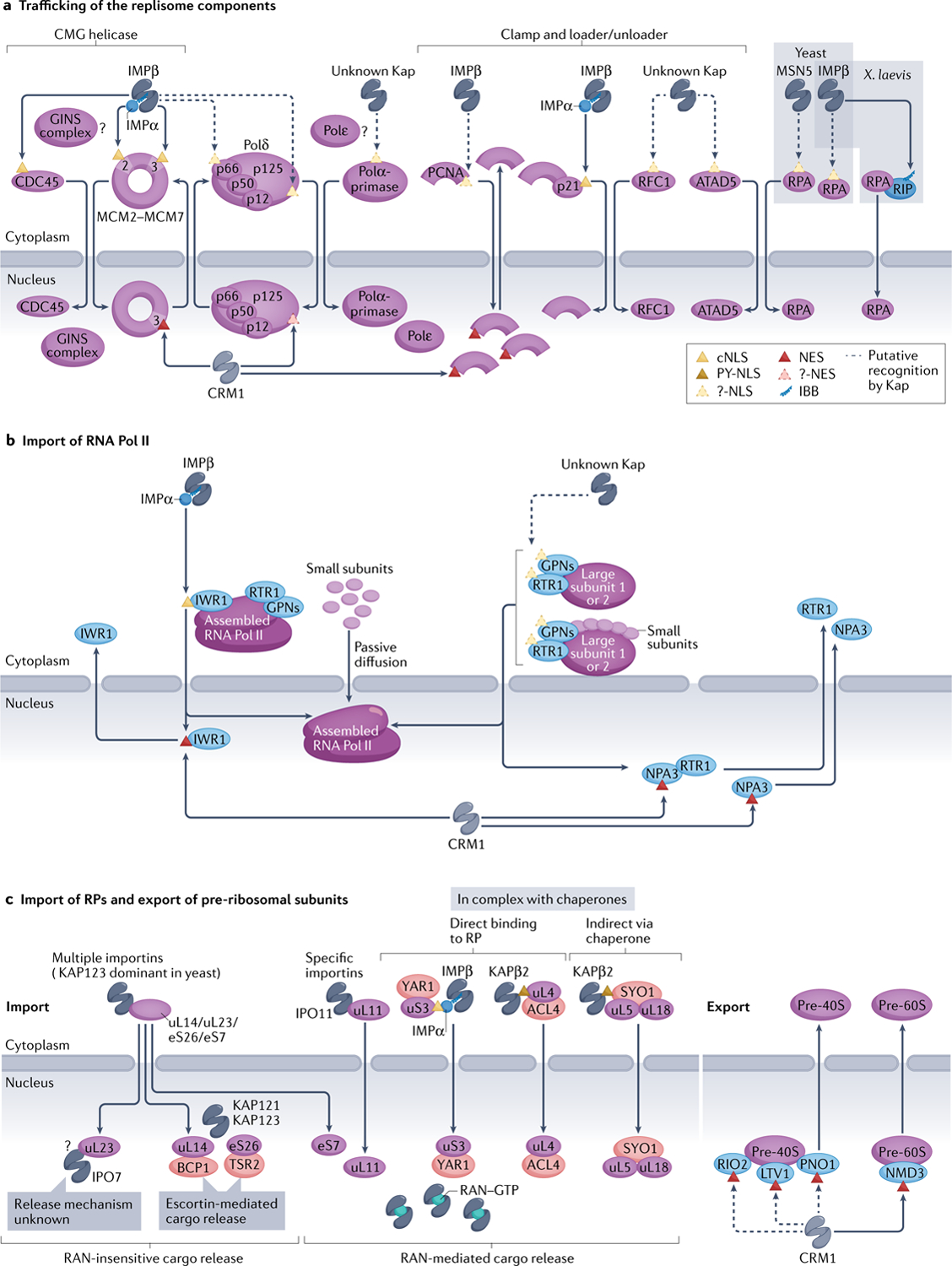
a | Most replisome components are believed to be imported by importin IMPα–IMPβ, but only two proteins — cell division control protein 45 (CDC45) and minichrosome maintenance (MCM) complex — have validated classical nuclear localization signals (cNLSs; yellow triangles), whereas various others have only unverified NLSs or their NLSs have not be identified (?-NLS; dashed, yellow triangles). Xenopus laevis replication protein A (RPA) may be imported by IMPβ via adaptor RPA-interacting protein (RIP), which contains an IMPβ-binding (IBB) region. A few components of the replisome may be imported by other Karyopherins (Kaps), and trafficking of others, such as GINS complex and polymerase ε (Polε), are unstudied (?).Similarly, only a few components seem to have nuclear export signals (NESs; red triangles) and are exported by CRM1 — it is unclear whether others are recycled back to the cytosol. b | Import of the yeast RNA polymerase II (Pol II) complex. Interacting with RNA polymerase II protein 1 (IWR1) acts as an adaptor and binds IMPα–IMPβ to import fully assembled RNA Pol II. IWR1 is recycled back to the cytosol by CRM1 exportin. Alternative import pathways of partially assembled RNA Pol II were proposed, such as via passive diffusion of small components or additional factors such as Regulator of transcription 1 (RTR1), as well as Glycine-proline-asparagine (GPN)-loop GTPase proteins. These factors are exported by CRM1 possibly via the NES within GPN protein NPA3. c | Several ribosomal proteins (RPs), such as uL14, uL23, eS7 and eS26, are imported by multiple importins. Others, such as uL11, are imported by specific importins, and yet others are imported bound to their chaperones (red ovals). Nuclear RAN–GTP dissociates most Kap–RP complexes, releasing the RPs into the nucleoplasm for complex assembly; exceptions include those that require dedicated chaperones, called escortins, for release or other, unexplored mechanisms (?). Assembled and mature pre-40S and pre-60S subunits are exported either by CRM1 or alternate pathways that involve mRNA export receptor MEX67-MTR2 (not pictured). ACL4, assembly chaperone of RPL4; ATAD5, ATPase family AAA domain-containing protein 5; BCP1, Bacterioferritin co-migratory protein 1; GINS, ‘go-ichi-ni-san’ in Japanese meaning ‘5-3-2-1’ (SLD5-PSF1-PSF2-PSF3); LTV1, low-temperature viability protein 1; NMD3, nonsense-mediated mRNA decay protein 3; PCNA, proliferating cell nuclear antigen; PNO1, partner of NOB1 (nuclear integrity protein 1 (NIN1) binding protein 1); PY-NLS, proline-tyrosine nuclear localization signal; RFC1, replication factor C subunit 1; RIO2, RIO (right open reading frame)-type kinase 2; SYO, symportin 1; TSR2, 20S ribosomal RNA accumulation protein 2; YAR1, yeast ankyrin repeat-containing protein 1.
Some replisome components are found to also exit the nucleus. The MCM complex is exported after replication in subsequent S, G2 and M phases — probably by CRM1 (the NES was identified in yeast MCM3) — which is regulated by cyclin-dependent kinases162,180. Human p125 is also exported by CRM1 but no NES has been reported166. CRM1 exports human PCNA by binding an NES accessible only in the monomer181, consistent with export of monomeric PCNA and its assembly and disassembly on DNA.
In summary, a few replisome components have been found to carry cNLSs and some others may ‘piggy-back’ on cNLS-containing proteins, such as the PCNA binding to p21, for import. The use of cNLSs for the import of the replisome components is accompanied by the involvement of KAP60 in regulating G1/S and G2/M transitions in yeast, suggesting that key DNA replication and cell cycle checkpoint proteins are imported by IMPα–IMPβ to coordinate cell cycle progression with genome replication182,183. However, import mechanisms for the majority of replisome components remain unknown. Similarly, CRM1-mediated export of three replisome components was reported, but it is unclear whether more components are actively exported out of the nucleus and how this export impacts the activity of the replisome.
Import of RNA polymerase II
We focus on RNA polymerase II (RNA Pol II) to describe nuclear import of a large transcription machinery (FIG. 5b). The 12-subunit RNA Pol II is fully assembled in the cytoplasm before import (reviewed in REF.184). Yeast RNA Pol II nuclear localization protein IWR1 contains a cNLS conserved from yeast to human, which binds IMPα–IMPβ, and is believed to be the major import adaptor for assembled RNA Pol II185; the effect of the human homologue of IWR1 has not been verified. Subunit sub-assemblies and small individual subunits can also enter the nucleus in yeast, prior to assembly, by active transport and passive diffusion, respectively186. Regulator of transcription 1 (RTR1; RPAP2 in human) may be the import factor that mediates the entry of individual large subunits of the RNA Pol II and subunit assemblies186,187; however, no NLS has been identified for RTR1. RPAP2 is also implicated in the import of human RNA Pol II187. Glycine-proline-asparagine (GPN)-loop GTPases were also reported as putative import factors in both human and yeast (reviewed in REF.184). Pull-down mass spectrometry studies revealed possible interactions of human GPN1/2/3 with IMPα2 (although no cNLSs were identified)188, but no such interaction was shown for yeast NPA3 (GPN1 homologue)189. However, some studies suggested that GPNs act upstream of IWR1 as cytosolic chaperones for RNA Pol II assembly, rather than as import adaptors190,191.
Several RNA Pol II import adaptors are exported from the nucleus via CRM1, but export of the polymerase has not been reported. The RNA Pol II machinery may be passively retained in the nucleus due to its large size and association with DNA, consistent with studies showing that the nuclear proteome is primarily maintained by complex assembly and passive retention rather than active export6. IWR1 is displaced from RNA Pol II by transcription initiation factors and then exported via a putative NES192. RTR1/RPAP2 is exported in a CRM1-dependent manner (but no NES has been identified)187,193, possibly through binding NES-containing GPNs (the NES was so far identified in GPN1/NPA3 (REF.194); reviewed elsewhere184).
In summary, generally, RNA Pol II complex assembles in the cytoplasm and uses import adaptors for nuclear import which are recycled back to the cytoplasm by CRM1. However, other mechanisms for RNA Pol II nuclear import, involving individual subunits and their smaller assemblies, were also reported. The mechanism of function of these import adaptors and their evolutionary conservation remain mostly unclear. The contribution of the multiple import mechanisms to RNA Pol II biogenesis and function also remains to be addressed.
Import/export of ribosome components
Ribosomes are complex machineries composed of ~80 individual ribosomal proteins (RPs) together with ribosomal RNAs. The assembly of pre-ribosomal particles occurs in the nucleoplasm, where ribosomal RNAs are transcribed, and hence necessitates import of the RPs into the nucleus. Despite the importance of the process, nuclear import mechanisms are so far known for only a few RPs (FIG. 5c). Early studies suggested that some RPs (eS1, eS7, eS26, eL6, eL20, uL4, uL14, uL18 and uL23) could be imported by multiple importins. Yeast cells primarily use KAP123 for RP import; the roles of other Kaps are much less pronounced, and they mostly function as backup pathways to reduce competition for import by KAP123 or when KAP123 is absent195–201. Interestingly, yeast cells show a particularly large import flux for RPs, and KAP123 is correspondingly the most abundant yeast Kap197, underscoring the observation that cellular concentrations of Kaps are linked with their respective cargo loads202. Although the role of KAP123 in RP import is dominant, not all RPs use this route for import. For example, uL11 is exclusively imported by IPO11 (REF.203). Furthermore, cNLS-containing uS3 and PY-NLS-containing uL4 are imported by IMPα–IMPβ and KAPβ2, respectively; this transport is orchestrated by uS3 and uL4 while bound to their cognate chaperones, yeast ankyrin repeat-containing protein 1 (YAR1)204,205 and assembly chaperone of RPL4 (ACL4)206,207, which may prevent these RPs from aggregation and/or ensure proper ribosome assembly. Transport of uL5 and uL18 in yeast is also supported by a chaperone, Symportin 1 (SYO1), which binds both RPs as well as KAP104 via its N-terminal basic PY-NLS for import208. RAN–GTP is presumed to release most imported RPs in the nucleus (see above discussion in Cargo loading and unloading via RAN–GTP). Nevertheless, several exceptions were reported. For example, RAN–GTP is inefficient in releasing uL23 from IPO7, which suggests the involvement of other factors196. Release of RPs eS26 and uL14 from KAP123 and KAP121/KAP123 also occurs in a RAN–GTP-insensitive manner and requires the action of dedicated chaperones/escortins 20S ribosomal RNA accumulation protein 2 (TSR2) (for eS26)200,209 and Bacterioferritin co-migratory protein 1 (BCP1) (for uL14)201. It is unclear what prevents these escortins from prematurely dissociating cytoplasmic importin–RP complexes.
Following import, RPs are assembled and matured into the pre-60S and pre-40S ribosomal subunits before nuclear export for further cytoplasmic processing and the assembly of the mature ribosome. This export occurs via two routes, involving CRM1 and mRNA export receptor Mex67-MTr2 (reviewed elsewhere210). The pre-60S subunit is exported by CRM1 via NES-containing adaptor protein nonsense-mediated mRNA decay protein 3 (NMD3)211, whereas ribosome biogenesis factors low-temperature viability protein 1 (LTV1), partner of NOB1 (PNO1) and RIO (right open reading frame)-type kinase 2 (RIO2) were proposed to function as redundant export adaptors for pre-40S export212–214.
MEX67-MTR2.
A heterodimeric mrNa export receptor that also functions in ribosomal export and is conserved in eukaryotes.
Overall, similar to the replisome, import mechanisms are known for only a small fraction of RPs. Unlike replisome components and RNA Pol II, which seem to use almost exclusively the IMPα–IMPβ pathway, RPs seem to use various importins. Because eukaryotic cells require hundreds of thousands to millions of ribosomes215,216, the availability of multiple import pathways for RPs may reduce the import burden for individual Kaps, as well as provide multiple backup pathways, to ensure efficient delivery of RPs into the nucleus. Assembled ribosomal subunits also use different pathways and adaptors for nuclear exit.
Conclusions and perspectives
Importins, exportins and biportins all share a common spiral-shaped or ring-shaped architecture that is conformationally adaptable with large surface areas to bind many different partners (FIGS 1b–d and 4). Unusual surface chemistries of Kaps include hydrophobic residues as well as arginines, histidines and cysteines that confer solubility, allowing them to permeate the FG-repeat populated NPC barrier (Supplementary Boxes 1 and 3) and act as anti-aggregation chaperones for their cargo (Supplementary Box 4). Kaps mediate the majority of nucleocytoplasmic protein traffic with the import load relatively evenly divided among the ten importins and biportins; it is still unclear how traffic out of the nucleus is parsed. Kaps bind to NLS/NES linear motifs and/or folded domain(s) of cargoes (TABLE 1), and the interactions are regulated in many different ways to control import versus export versus nuclear/cytoplasmic retention to, ultimately, control localization and function of a plethora of macromolecules (FIG. 4). Finally, the biogenesis of many gene expression machineries is coordinated with the nucleocytoplasmic traffic of their components, but the current knowledge of the Kaps and signals responsible for these trafficking events is far from complete (FIG. 5).
Our knowledge of how macromolecules are trafficked between the nucleus and the cytoplasm has advanced tremendously since discovery of the Kap family, the RAN GTPase and its regulators in the 1990s. For some Kaps, namely IMPα–IMPβ, KAPβ2, TNPO3 and CRM1, we know many of the cargoes and how these Kaps recognize these cargoes, but much of the knowledge for the other Kaps is missing. More generally, it is still unclear why nucleocytoplasmic traffic is mediated by 20 different Kaps. Is this diversity in Kaps related to different activities of individual Kaps in regulating cell function? Or, perhaps, could the presence of multiple Kaps be linked to the robustness of nucleocytoplasmic transport? What are the mechanistic differences between importins, exportins and biportins, and how is the directionality of transport orchestrated by individual Kap properties? Will we learn in the future that importins/exportins are all biportins?
An emerging theme in Kap–cargo recognition, observed in the IMPα–IMPβ, KAPβ2 and CRM1 systems, involves Kap-binding elements that are very diverse in both sequence and structure, often used in multipartite and combinatorial manners. Such promiscuity in cargo recognition allows each Kap to function as a transport receptor for hundreds to thousands of different protein cargoes. Cargo selection by Kaps extends beyond very diverse NLS/NES sequences to the binding of diverse folded domains. It is unclear whether any common patterns exist for interactions of Kaps with folded domains of cargoes. Specifically, it remains to be addressed whether Kaps are capable of recognizing particular domain types or whether the recognition of folded domains is specific to individual domains, and therefore not general. Missing knowledge of how Kaps bind cargoes contributes to the current inability to robustly predict cargoes for the different Kaps. NLS/NES/cargo prediction for even the best studied IMPα–IMPβ and CRM1 systems is still poor (BOX 2). Expanding the knowledge of how Kaps bind cargoes will also lead to design/development of Kap-specific inhibitors that currently do not exist for most Kaps. Nuclear import and export inhibitors are not just critical as laboratory reagents to control nucleocytoplasmic localization of macromolecules of interest but also as therapeutic agents for the many diseases (cancer, neurodegeneration, inflammation and viral infections) that arise from aberrant nucleocytoplasmic trafficking (BOX 1). Lastly, it is clear that Kap–cargo interactions are regulated to control protein localization that affect many cellular processes such as cell cycle regulation and environmental sensing. Are the regulatory mechanisms used for nucleocytoplasmic transport also used in non-transport functions of Kaps, such as during mitosis? Some examples in spindle assembly and cytokinesis seem to suggest so217–220, but more studies are needed to focus on Kaps in non-transport roles.
Supplementary Material
Acknowledgements
This work was funded by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health (NIH) under Awards R01GM069909 and R35GM144137 (Y.M.C.), the Welch Foundation Grant I-1532 (Y.M.C.), Cancer Prevention Research Institute of Texas (CPRIT) Grant RP180410 (Y.M.C.), support from the Alfred and Mabel Gilman Chair in Molecular Pharmacology, Eugene McDermott Scholar in Biomedical Research (Y.M.C.), the Gilman Special Opportunities Award (H.Y.J.F.) and NIGMS Molecular Biophysics Training Program T32GM131963 (C.E.W.).
RelATed liNKs
cNLs Mapper: http://nls-mapper.iab.keio.ac.jp
iNsP: http://www.csbio.sjtu.edu.cn/bioinf/INSP/
LocNes: http://prodata.swmed.edu/LocNES/LocNES.php
Nesmapper: https://sourceforge.net/projects/nesmapper/
NetNes: https://services.healthtech.dtu.dk/service.php?NetNES-1.1
NLstradamus: http://www.moseslab.csb.utoronto.ca/NLStradamus/
NoLogo: https://github.com/mppl1/NoLogo
NucPred: https://nucpred.bioinfo.se/nucpred/
Nucimport: http://bioinf.scmb.uq.edu.au:8080/NucImport/
PsORT: https://www.genscript.com/psort.html
PsORT ii: https://psort.hgc.jp/form2.html
PredictNLs: https://rostlab.org/owiki/index.php/PredictNLS
seqNLs: http://mleg.cse.sc.edu/seqNLS/
Footnotes
Competing interests
Y.M.C. is a consultant for Faze Medicines. The remaining authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at https://doi.org/10.1038/s41580-021-00446-7.
References
- 1.Chook YM & Suel KE Nuclear import by karyopherin-βs: recognition and inhibition. Biochim. Biophys. Acta 1813, 1593–1606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuura Y Mechanistic insights from structural analyses of Ran-GTPase-driven nuclear export of proteins and RNAs. J. Mol. Biol 428, 2025–2039 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Kalita J, Kapinos LE & Lim RYH On the asymmetric partitioning of nucleocytoplasmic transport — recent insights and open questions. J. Cell Sci 134, jcs240382 (2021). [DOI] [PubMed] [Google Scholar]
- 4.O’Reilly AJ, Dacks JB & Field MC Evolution of the karyopherin-β family of nucleocytoplasmic transport factors; ancient origins and continued specialization. PLoS ONE 6, e19308 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thakar K, Karaca S, Port SA, Urlaub H & Kehlenbach RH Identification of CRM1-dependent nuclear export cargos using quantitative mass spectrometry. Mol. Cell Proteom 12, 664–678 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wuhr M et al. The nuclear proteome of a vertebrate. Curr. Biol 25, 2663–2671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kırlı K et al. A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning. eLife 4, e11466 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mackmull MT et al. Landscape of nuclear transport receptor cargo specificity. Mol. Syst. Biol 13, 962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This proteomics study identifies interaction partners for several Kaps and IMPαs using proximity ligation, which can differentiate direct versus ‘piggyback’ transport.
- 9.Kimura M et al. Extensive cargo identification reveals distinct biological roles of the 12 importin pathways. eLife 6, e21184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This proteomics study identifies import cargoes for the 12 mammalian importins and biportins.
- 10.Baade I, Spillner C, Schmitt K, Valerius O & Kehlenbach RH Extensive identification and in-depth validation of importin 13 cargoes. Mol. Cell Proteom 17, 1337–1353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This proteomics study is the first to employ an experimental approach that can identify and differentiate both import and export cargoes of a biportin.
- 11.Aksu M et al. Xpo7 is a broad-spectrum exportin and a nuclear import receptor. J. Cell Biol 217, 2329–2340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article shows the first use of nanobodies as Kap-specific inhibitors and demonstrates that XPO7 functions as a biportin.
- 12.Vera Rodriguez A, Frey S & Gorlich D Engineered SUMO/protease system identifies Pdr6 as a bidirectional nuclear transport receptor. J. Cell Biol 218, 2006–2020 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article shows that PDR6 functions as a biportin and identifies novel import and export cargoes.
- 13.Lange A et al. Classical nuclear localization signals: definition, function, and interaction with importin α. J. Biol. Chem 282, 5101–5105 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fung HY, Fu SC & Chook YM Nuclear export receptor CRM1 recognizes diverse conformations in nuclear export signals. eLife 6, e23961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article shows how diverse NESs bind CRM1 and further expands the number of NES consensus sequences.
- 15.Soniat M & Chook YM Nuclear localization signals for four distinct karyopherin-β nuclear import systems. Biochem. J 468, 353–362 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Pumroy RA & Cingolani G Diversification of importin-α isoforms in cellular trafficking and disease states. Biochem. J 466, 13–28 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbins J, Dilworth SM, Laskey RA & Dingwall C Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64, 615–623 (1991). [DOI] [PubMed] [Google Scholar]
- 18.Kalderon D, Richardson WD, Markham AF & Smith AE Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311, 33–38 (1984). [DOI] [PubMed] [Google Scholar]
- 19.Chang CC et al. Ran pathway-independent regulation of mitotic Golgi disassembly by Importin-α. Nat. Commun 10, 4307 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jagga B et al. Structural basis for nuclear import selectivity of pioneer transcription factor SOX2. Nat. Commun 12, 28 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article shows how IMPα3 recognizes SOX2 using two NLSs linked by a folded domain.
- 21.Lott K, Bhardwaj A, Sims PJ & Cingolani G A minimal nuclear localization signal (NLS) in human phospholipid scramblase 4 that binds only the minor NLS-binding site of importin α1. J. Biol. Chem 286, 28160–28169 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conti E, Uy M, Leighton L, Blobel G & Kuriyan J Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell 94, 193–204 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Kobe B Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nat. Struct. Biol 6, 388–397 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Catimel B et al. Biophysical characterization of interactions involving importin-α during nuclear import. J. Biol. Chem 276, 34189–34198 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Fanara P, Hodel MR, Corbett AH & Hodel AE Quantitative analysis of nuclear localization signal (NLS)-importin α interaction through fluorescence depolarization. Evidence for auto-inhibitory regulation of NLS binding. J. Biol. Chem 275, 21218–21223 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Hodel MR, Corbett AH & Hodel AE Dissection of a nuclear localization signal. J. Biol. Chem 276, 1317–1325 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Hodel AE et al. Nuclear localization signal receptor affinity correlates with in vivo localization in Saccharomyces cerevisiae. J. Biol. Chem 281, 23545–23556 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Nardozzi J, Wenta N, Yasuhara N, Vinkemeier U & Cingolani G Molecular basis for the recognition of phosphorylated STAT1 by importin α5. J. Mol. Biol 402, 83–100 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pumroy RA, Ke S, Hart DJ, Zachariae U & Cingolani G Molecular determinants for nuclear import of influenza A PB2 by importin α isoforms 3 and 7. Structure 23, 374–384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soniat M & Chook YM Karyopherin-β2 recognition of a PY-NLS variant that lacks the proline-tyrosine motif. Structure 24, 1802–1809 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies the first PY-NLS that lacks the PY motif, illustrating the sequence diversity of PY-NLSs.
- 31.Suel KE, Gu H & Chook YM Modular organization and combinatorial energetics of proline-tyrosine nuclear localization signals. PLoS Biol 6, e137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee BJ et al. Rules for nuclear localization sequence recognition by karyopherin β 2. Cell 126, 543–558 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi J & Matsuura Y Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p. J. Mol. Biol 425, 1852–1868 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Kobayashi J, Hirano H & Matsuura Y Crystal structure of the karyopherin Kap121p bound to the extreme C-terminus of the protein phosphatase Cdc14p. Biochem. Biophys. Res. Commun 463, 309–314 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Makhnevych T, Ptak C, Lusk CP, Aitchison JD & Wozniak RW The role of karyopherins in the regulated sumoylation of septins. J. Cell Biol 177, 39–49 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jang S et al. Differential role for phosphorylation in alternative polyadenylation function versus nuclear import of SR-like protein CPSF6. Nucleic Acids Res. 47, 4663–468 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows how TNPO3 recognizes non-phosphorylated RS-like domains.
- 37.Yun CY, Velazquez-Dones AL, Lyman SK & Fu XD Phosphorylation-dependent and -independent nuclear import of RS domain-containing splicing factors and regulators. J. Biol. Chem 278, 18050–18055 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Lai MC, Lin RI & Tarn WY Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl Acad. Sci. USA 98, 10154–10159 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wen W, Meinkoth JL, Tsien RY & Taylor SS Identification of a signal for rapid export of proteins from the nucleus. Cell 82, 463–473 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Fischer U, Huber J, Boelens WC, Mattaj IW & Luhrmann R The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82, 475–483 (1995). [DOI] [PubMed] [Google Scholar]
- 41.Dong X et al. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature 458, 1136–1141 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monecke T et al. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science 324, 1087–1091 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Guttler T et al. NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat. Struct. Mol. Biol 17, 1367–1376 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Fung HY, Fu SC, Brautigam CA & Chook YM Structural determinants of nuclear export signal orientation in binding to exportin CRM1. eLife 4, e10034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huber J et al. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 17, 4114–4126 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jullien D, Gorlich D, Laemmli UK & Adachi Y Nuclear import of RPA in Xenopus egg extracts requires a novel protein XRIPα but not importin α. EMBO J. 18, 4348–4358 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Görlich D, Henklein P, Laskey RA & Hartmann EA 41 amino acid motif in importin-α confers binding to importin-β and hence transit into the nucleus. EMBO J. 15, 1810–1817 (1996). [PMC free article] [PubMed] [Google Scholar]
- 48.Cingolani G, Bednenko J, Gillespie MT & Gerace L Molecular basis for the recognition of a nonclassical nuclear localization signal by importin β. Mol. Cell 10, 1345–1353 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Gonzalez A et al. Mechanism of karyopherin-β2 binding and nuclear import of ALS variants FUS(P525L) and FUS(R495X). Sci. Rep 11, 3754 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baade I et al. The RNA-binding protein FUS is chaperoned and imported into the nucleus by a network of import receptors. J. Biol. Chem 296, 100659 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bourgeois B et al. Nonclassical nuclear localization signals mediate nuclear import of CIRBP. Proc. Natl Acad. Sci. USA 117, 8503–8514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies the novel RGG motif recognized by KAPβ2.
- 52.Soniat M, Cagatay T & Chook YM Recognition elements in the histone H3 and H4 tails for seven different importins. J. Biol. Chem 291, 21171–21183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Komeili A & O’Shea EK Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284, 977–980 (1999). [DOI] [PubMed] [Google Scholar]
- 54.Fritz J et al. RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1. Mol. Cell Biol 29, 1487–1497 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakurai M et al. ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay. Nat. Struct. Mol. Biol 24, 534–543 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu W et al. Ebola virus VP24 targets a unique NLS binding site on karyopherin α 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe 16, 187–200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi S et al. Structural basis for the selective nuclear import of the C2H2 zinc-finger protein Snail by importin β. Acta Crystallogr. D. Biol. Crystallogr 70, 1050–1060 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Lee SJ et al. The structure of importin-β bound to SREBP-2: nuclear import of a transcription factor. Science 302, 1571–1575 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Padavannil A et al. Importin-9 wraps around the H2A–H2B core to act as nuclear importer and histone chaperone. eLife 8, e43630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article shows the first structure of IPO9 bound to histone dimer H2A–H2B in a chaperone-like manner.
- 60.Lin W et al. The roles of multiple importins for nuclear import of murine aristaless-related homeobox protein. J. Biol. Chem 284, 20428–20439 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lubert EJ & Sarge KD Interaction between protein phosphatase 2A and members of the importin β superfamily. Biochem. Biophys. Res. Commun 303, 908–913 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Volpon L et al. Importin 8 mediates m7G cap-sensitive nuclear import of the eukaryotic translation initiation factor eIF4E. Proc. Natl Acad. Sci. USA 113, 5263–5268 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bono F, Cook AG, Grunwald M, Ebert J & Conti E Nuclear import mechanism of the EJC component Mago-Y14 revealed by structural studies of importin 13. Mol. Cell 37, 211–222 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Grunwald M & Bono F Structure of Importin13–Ubc9 complex: nuclear import and release of a key regulator of sumoylation. EMBO J. 30, 427–438 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows how biportin PDR6 recognizes its import cargo UBC9 and export cargo eIF5A using a mechanism distinct from that of mammalian IPO13 and XPO4, respectively.
- 65.Aksu M, Trakhanov S, Vera Rodriguez A & Gorlich D Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122. J. Cell Biol 218, 1839–1852 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gontan C et al. Exportin 4 mediates a novel nuclear import pathway for Sox family transcription factors. J. Cell Biol 185, 27–34 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chatzifrangkeskou M et al. RASSF1A is required for the maintenance of nuclear actin levels. EMBO J. 38, e101168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grunwald M, Lazzaretti D & Bono F Structural basis for the nuclear export activity of Importin13. EMBO J. 32, 899–913 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kurisaki A et al. The mechanism of nuclear export of Smad3 involves exportin 4 and Ran. Mol. Cell Biol 26, 1318–1332 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aksu M, Trakhanov S & Gorlich D Structure of the exportin Xpo4 in complex with RanGTP and the hypusine-containing translation factor eIF5A. Nat. Commun 7, 11952 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lipowsky G et al. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J. 19, 4362–4371 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith KM et al. Structural basis for importin α 3 specificity of W proteins in Hendra and Nipah viruses. Nat. Commun 9, 3703 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sankhala RS et al. Three-dimensional context rather than NLS amino acid sequence determines importin α subtype specificity for RCC1. Nat. Commun 8, 979 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ivic N et al. Fuzzy interactions form and shape the histone transport complex. Mol. Cell 73, 1191–1203.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses cryo electron microscopy to demonstrate how the IMPβ–IPO7 heterodimer binds and imports H1.
- 75.Maertens GN et al. Structural basis for nuclear import of splicing factors by human Transportin 3. Proc. Natl Acad. Sci. USA 111, 2728–2733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Caceres JF, Misteli T, Screaton GR, Spector DL & Krainer AR Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol 138, 225–238 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hopper AK & Nostramo RT tRNA processing and subcellular trafficking proteins multitask in pathways for other RNAs. Front. Genet 10, 96 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Calado A, Treichel N, Muller EC, Otto A & Kutay U Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 21, 6216–6224 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bohnsack MT et al. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 21, 6205–6215 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okamura M, Inose H & Masuda S RNA export through the NPC in eukaryotes. Genes 6, 124–149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Verheggen C & Bertrand E CRM1 plays a nuclear role in transporting snoRNPs to nucleoli in higher eukaryotes. Nucleus 3, 132–137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martinez I et al. An Exportin-1-dependent microRNA biogenesis pathway during human cell quiescence. Proc. Natl Acad. Sci. USA 114, E4961–E4970 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sheng P et al. Dicer cleaves 5′-extended microRNA precursors originating from RNA polymerase II transcription start sites. Nucleic Acids Res 46, 5737–5752 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu H, Becker D & Krebber H Telomerase RNA TLC1 shuttling to the cytoplasm requires mRNA export factors and is important for telomere maintenance. Cell Rep. 8, 1630–1638 (2014). [DOI] [PubMed] [Google Scholar]
- 85.Gales JP, Kubina J, Geldreich A & Dimitrova M Strength in diversity: nuclear export of viral RNAs. Viruses 12, 1014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang J, Bogerd HP, Wang PJ, Page DC & Cullen BR Two closely related human nuclear export factors utilize entirely distinct export pathways. Mol. Cell 8, 397–406 (2001). [DOI] [PubMed] [Google Scholar]
- 87.Lari A et al. A nuclear role for the DEAD-box protein Dbp5 in tRNA export. eLife 8, e48410 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ohno M, Segref A, Bachi A, Wilm M & Mattaj IW PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101, 187–198 (2000). [DOI] [PubMed] [Google Scholar]
- 89.Malim MH, Hauber J, Le SY, Maizel JV & Cullen BR The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338, 254–257 (1989). [DOI] [PubMed] [Google Scholar]
- 90.Lee SJ, Matsuura Y, Liu SM & Stewart M Structural basis for nuclear import complex dissociation by RanGTP. Nature 435, 693–696 (2005). [DOI] [PubMed] [Google Scholar]
- 91.Matsuura Y & Stewart M Structural basis for the assembly of a nuclear export complex. Nature 432, 872–877 (2004). [DOI] [PubMed] [Google Scholar]
- 92.Cook AG, Fukuhara N, Jinek M & Conti E Structures of the tRNA export factor in the nuclear and cytosolic states. Nature 461, 60–65 (2009). [DOI] [PubMed] [Google Scholar]
- 93.Okada C et al. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 326, 1275–1279 (2009). [DOI] [PubMed] [Google Scholar]
- 94.Chook YM & Blobel G Structure of the nuclear transport complex karyopherin-β2-Ran × GppNHp. Nature 399, 230–237 (1999). [DOI] [PubMed] [Google Scholar]
- 95.Hahn S & Schlenstedt G Importin β-type nuclear transport receptors have distinct binding affinities for Ran-GTP. Biochem. Biophys. Res. Commun 406, 383–388 (2011). [DOI] [PubMed] [Google Scholar]
- 96.Cingolani G, Petosa C, Weis K & Muller CW Structure of importin-β bound to the IBB domain of importin-α. Nature 399, 221–229 (1999). [DOI] [PubMed] [Google Scholar]
- 97.Mitrousis G, Olia AS, Walker-Kopp N & Cingolani G Molecular basis for the recognition of snurportin 1 by importin β. J. Biol. Chem 283, 7877–7884 (2008). [DOI] [PubMed] [Google Scholar]
- 98.Mosammaparast N, Ewart CS & Pemberton LF A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 21, 6527–6538 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mosammaparast N, Del Rosario BC & Pemberton LF Modulation of histone deposition by the karyopherin kap114. Mol. Cell Biol 25, 1764–1778 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pemberton LF, Rosenblum JS & Blobel G Nuclear Import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J. Cell Biol 145, 1407–1417 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Senger B et al. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J. 17, 2196–2207 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee DC & Aitchison JD Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and RNA. J. Biol. Chem 274, 29031–29037 (1999). [DOI] [PubMed] [Google Scholar]
- 103.Yamazawa R et al. Structural basis for selective binding of export cargoes by Exportin-5. Structure 26, 1393–1398.e2 (2018). [DOI] [PubMed] [Google Scholar]; This study shows how XPO5 recognizes mature tRNAs.
- 104.Wang J et al. XPO5 promotes primary miRNA processing independently of RanGTP. Nat. Commun 11, 1845 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cook A et al. The structure of the nuclear export receptor Cse1 in its cytosolic state reveals a closed conformation incompatible with cargo binding. Mol. Cell 18, 355–367 (2005). [DOI] [PubMed] [Google Scholar]
- 106.Fung HY & Chook YM Atomic basis of CRM1–cargo recognition, release and inhibition. Semin. Cancer Biol 27, 52–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Monecke T et al. Structural basis for cooperativity of CRM1 export complex formation. Proc. Natl Acad. Sci. USA 110, 960–965 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dong X, Biswas A & Chook YM Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nat. Struct. Mol. Biol 16, 558–560 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dian C et al. Structure of a truncation mutant of the nuclear export factor CRM1 provides insights into the auto-inhibitory role of its C-terminal helix. Structure 21, 1338–1349 (2013). [DOI] [PubMed] [Google Scholar]
- 110.Koyama M & Matsuura Y An allosteric mechanism to displace nuclear export cargo from CRM1 and RanGTP by RanBP1. EMBO J. 29, 2002–201 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li Y et al. Distinct RanBP1 nuclear export and cargo dissociation mechanisms between fungi and animals. eLife 8, e41331 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fu SC, Fung HYJ, Cagatay T, Baumhardt J & Chook YM Correlation of CRM1–NES affinity with nuclear export activity. Mol. Biol. Cell 29, 2037–2044 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Christie M et al. Structural biology and regulation of protein import into the nucleus. J. Mol. Biol 428, 2060–2090 (2016). [DOI] [PubMed] [Google Scholar]
- 114.Cautain B, Hill R, de Pedro N & Link W Components and regulation of nuclear transport processes. FEBS J. 282, 445–462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Suarez-Calvet M et al. Monomethylated and unmethylated FUS exhibit increased binding to Transportin and distinguish FTLD-FUS from ALS-FUS. Acta Neuropathol 131, 587–604 (2016). [DOI] [PubMed] [Google Scholar]
- 116.von Morgen P, Lidak T, Horejsi Z & Macurek L Nuclear localisation of 53BP1 is regulated by phosphorylation of the nuclear localisation signal. Biol. Cell 110, 137–146 (2018). [DOI] [PubMed] [Google Scholar]
- 117.Zhang X, Fan S, Zhang L & Shi Y Glucagon-like peptide-1 receptor undergoes importin-α-dependent nuclear localization in rat aortic smooth muscle cells. FEBS Lett. 594, 1506–1516 (2020). [DOI] [PubMed] [Google Scholar]
- 118.Li FL et al. Acetylation accumulates PFKFB3 in cytoplasm to promote glycolysis and protects cells from cisplatin-induced apoptosis. Nat. Commun 9, 508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Howard TR et al. The DNA sensor IFIX drives proteome alterations to mobilize nuclear and cytoplasmic antiviral responses, with its acetylation acting as a localization Toggle. mSystems 6, e0039721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cao X et al. Acetylation promotes TyrRS nuclear translocation to prevent oxidative damage. Proc. Natl Acad. Sci. USA 114, 687–692 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Napolitano G et al. mTOR-dependent phosphorylation controls TFEB nuclear export. Nat. Commun 9, 3312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gkotinakou IM, Befani C, Simos G & Liakos P ERK1/2 phosphorylates HIF-2α and regulates its activity by controlling its CRM1-dependent nuclear shuttling. J. Cell Sci 132, jcs225698 (2019). [DOI] [PubMed] [Google Scholar]
- 123.Fang L et al. SET1A-mediated mono-methylation at K342 regulates YAP activation by blocking its nuclear export and promotes tumorigenesis. Cancer Cell 34, 103–118.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 124.Poon IK & Jans DA Regulation of nuclear transport: central role in development and transformation? Traffic 6, 173–186 (2005). [DOI] [PubMed] [Google Scholar]
- 125.Mehta S et al. Dephosphorylation of YB-1 is required for nuclear localisation during G2 phase of the cell cycle. Cancers (Basel) 12, 315 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Smidova A et al. 14-3-3 protein masks the nuclear localization sequence of caspase-2. FEBS J. 285, 4196–4213 (2018). [DOI] [PubMed] [Google Scholar]
- 127.Kalabova D et al. 14-3-3 protein binding blocks the dimerization interface of caspase-2. FEBS J. 287, 3494–3510 (2020). [DOI] [PubMed] [Google Scholar]
- 128.Duffraisse M et al. Role of a versatile peptide motif controlling Hox nuclear export and autophagy in the Drosophila fat body. J. Cell Sci 133, jcs241943 (2020). [DOI] [PubMed] [Google Scholar]
- 129.Purice MD & Taylor JP Linking hnRNP function to ALS and FTD pathology. Front. Neurosci 12, 326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Beijer D et al. Characterization of HNRNPA1 mutations defines diversity in pathogenic mechanisms and clinical presentation. JCI Insight 6, e148363 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Naruse H et al. Molecular epidemiological study of familial amyotrophic lateral sclerosis in Japanese population by whole-exome sequencing and identification of novel HNRNPA1 mutation. Neurobiol. Aging 61, 255.e9–255.e16 (2018). [DOI] [PubMed] [Google Scholar]
- 132.Liu Q et al. Whole-exome sequencing identifies a missense mutation in hnRNPA1 in a family with flail arm ALS. Neurology 87, 1763–1769 (2016). [DOI] [PubMed] [Google Scholar]
- 133.Pilch J et al. Evidence for HNRNPH1 being another gene for Bain type syndromic mental retardation. Clin. Genet 94, 381–385 (2018). [DOI] [PubMed] [Google Scholar]
- 134.Bain JM et al. Variants in HNRNPH2 on the X chromosome are associated with a neurodevelopmental disorder in females. Am. J. Hum. Genet 99, 728–734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Falini B et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc + AML. Blood 107, 4514–4523 (2006). [DOI] [PubMed] [Google Scholar]
- 136.Pauty J et al. Cancer-causing mutations in the tumor suppressor PALB2 reveal a novel cancer mechanism using a hidden nuclear export signal in the WD40 repeat motif. Nucleic Acids Res. 45, 2644–2657 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhong Y et al. Nuclear export of misfolded SOD1 mediated by a normally buried NES-like sequence reduces proteotoxicity in the nucleus. eLife 6, e23759 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows how a mutation in SOD1 exposes a novel NES recognized by CRM1.
- 138.Wang P et al. Repression of classical nuclear export by S-nitrosylation of CRM1. J. Cell Sci 122, 3772–3779 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Martin AP et al. STK38 kinase acts as XPO1 gatekeeper regulating the nuclear export of autophagy proteins and other cargoes. EMBO Rep. 20, e48150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sun HL et al. ERK activation globally downregulates miRNAs through phosphorylating Exportin-5. Cancer Cell 30, 723–736 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows how phosphorylation of XPO5 enables recruitment of a protein that causes subsequent conformation change in XPO5, leading to aberrant transport and disease.
- 141.Putker M et al. Redox-dependent control of FOXO/DAF-16 by transportin-1. Mol. Cell 49, 730–742 (2013). [DOI] [PubMed] [Google Scholar]
- 142.Rothenbusch U, Sawatzki M, Chang Y, Caesar S & Schlenstedt G Sumoylation regulates Kap114-mediated nuclear transport. EMBO J. 31, 2461–2472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu X et al. PKA-site phosphorylation of importin13 regulates its subcellular localization and nuclear transport function. Biochem. J 475, 2699–2712 (2018). [DOI] [PubMed] [Google Scholar]
- 144.Corum DG, Tsichlis PN & Muise-Helmericks RC AKT3 controls mitochondrial biogenesis and autophagy via regulation of the major nuclear export protein CRM-1. FASEB J 28, 395–407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Galbraith LCA et al. PPAR-γ induced AKT3 expression increases levels of mitochondrial biogenesis driving prostate cancer. Oncogene 40, 2355–2366 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Azizian NG & Li Y XPO1-dependent nuclear export as a target for cancer therapy. J. Hematol. Oncol 13, 61 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Azmi AS, Uddin MH & Mohammad RM The nuclear export protein XPO1 — from biology to targeted therapy. Nat. Rev. Clin. Oncol 18, 152–169 (2021). [DOI] [PubMed] [Google Scholar]
- 148.Jain P et al. Clinical and molecular characteristics of XPO1 mutations in patients with chronic lymphocytic leukemia. Am. J. Hematol 91, E478–E479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Navarro-Bailon A et al. Exportin-1 E571K mutation is a common finding in patients with classical Hodgkin lymphoma. Hematol. Oncol 37, 215–218 (2019). [DOI] [PubMed] [Google Scholar]
- 150.Hing ZA et al. Exploring the role of the recurrent exportin 1 (XPO1/CRM1) mutations E571G and E571K in chronic lymphocytic leukemia. Blood 128, 972–972 (2016). [Google Scholar]
- 151.Walker JS et al. Recurrent XPO1 mutations alter pathogenesis of chronic lymphocytic leukemia. J. Hematol. Oncol 14, 17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Garcia-Santisteban I et al. A cellular reporter to evaluate CRM1 nuclear export activity: functional analysis of the cancer-related mutant E571K. Cell Mol. Life Sci 73, 4685–4699 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Taylor J et al. Altered nuclear export signal recognition as a driver of oncogenesis. Cancer Discov. 9, 1452–1467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Baumhardt JM et al. Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation. Mol. Biol. Cell 31, 1879 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Miloudi H et al. XPO1(E571K) mutation modifies Exportin 1 localisation and interactome in B-cell lymphoma. Cancers (Basel) 12, 2829 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wu K, He J, Pu W & Peng Y The role of Exportin-5 in microRNA biogenesis and cancer. Genomics Proteom. Bioinforma 16, 120–126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lin D et al. Exportin-5 SUMOylation promotes hepatocellular carcinoma progression. Exp. Cell Res 395, 112219 (2020). [DOI] [PubMed] [Google Scholar]
- 158.Hahn S, Maurer P, Caesar S & Schlenstedt G Classical NLS proteins from Saccharomyces cerevisiae. J. Mol. Biol 379, 678–694 (2008). [DOI] [PubMed] [Google Scholar]
- 159.Hopwood B & Dalton S Cdc45p assembles into a complex with Cdc46p/Mcm5p, is required for minichromosome maintenance, and is essential for chromosomal DNA replication. Proc. Natl Acad. Sci. USA 93, 12309–12314 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Young MR, Suzuki K, Yan H, Gibson S & Tye BK Nuclear accumulation of Saccharomyces cerevisiae Mcm3 is dependent on its nuclear localization sequence. Genes Cell 2, 631–643 (1997). [DOI] [PubMed] [Google Scholar]
- 161.Pasion SG & Forsburg SL Nuclear localization of Schizosaccharomyces pombe Mcm2/Cdc19p requires MCM complex assembly. Mol. Biol. Cell 10, 4043–4057 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liku ME, Nguyen VQ, Rosales AW, Irie K & Li JJ CDK phosphorylation of a novel NLS–NES module distributed between two subunits of the Mcm2–7 complex prevents chromosomal rereplication. Mol. Biol. Cell 16, 5026–5039 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wu R, Wang J & Liang C Cdt1p, through its interaction with Mcm6p, is required for the formation, nuclear accumulation and chromatin loading of the MCM complex. J. Cell Sci 125, 209–219 (2012). [DOI] [PubMed] [Google Scholar]
- 164.Frigola J et al. Cdt1 stabilizes an open MCM ring for helicase loading. Nat. Commun 8, 15720 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pohler JR, Otterlei M & Warbrick E An in vivo analysis of the localisation and interactions of human p66 DNA polymerase δ subunit. BMC Mol. Biol 6, 17 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Shen Y, Wang K & Qi RZ The catalytic subunit of DNA polymerase δ is a nucleocytoplasmic shuttling protein. Exp. Cell Res 375, 36–40 (2019). [DOI] [PubMed] [Google Scholar]
- 167.Lancey C et al. Structure of the processive human Pol δ holoenzyme. Nat. Commun 11, 1109 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kelich JM, Papaioannou H & Skordalakes E Pol α-primase dependent nuclear localization of the mammalian CST complex. Commun. Biol 4, 349 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mizuno T, Yamagishi K, Miyazawa H & Hanaoka F Molecular architecture of the mouse DNA polymerase α-primase complex. Mol. Cell Biol 19, 7886–7896 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mizuno T et al. The second-largest subunit of the mouse DNA polymerase α-primase complex facilitates both production and nuclear translocation of the catalytic subunit of DNA polymerase α. Mol. Cell Biol 18, 3552–3562 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Baranovskiy AG et al. Mechanism of concerted RNA–DNA primer synthesis by the human primosome. J. Biol. Chem 291, 10006–10020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kim BJ & Lee H Lys-110 is essential for targeting PCNA to replication and repair foci, and the K110A mutant activates apoptosis. Biol. Cell 100, 675–686 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M & Kuriyan J Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87, 297–306 (1996). [DOI] [PubMed] [Google Scholar]
- 174.Rodriguez-Vilarrupla A et al. Identification of the nuclear localization signal of p21(cip1) and consequences of its mutation on cell proliferation. FEBS Lett. 531, 319–323 (2002). [DOI] [PubMed] [Google Scholar]
- 175.Merkle CJ, Karnitz LM, Henry-Sanchez JT & Chen J Cloning and characterization of hCTF18, hCTF8, and hDCC1. Human homologs of a Saccharomyces cerevisiae complex involved in sister chromatid cohesion establishment. J. Biol. Chem 278, 30051–30056 (2003). [DOI] [PubMed] [Google Scholar]
- 176.Kang MS et al. Regulation of PCNA cycling on replicating DNA by RFC and RFC-like complexes. Nat. Commun 10, 2420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Belanger KD et al. The karyopherin Msn5/Kap142 requires Nup82 for nuclear export and performs a function distinct from translocation in RPA protein import. J. Biol. Chem 279, 43530–43539 (2004). [DOI] [PubMed] [Google Scholar]
- 178.Yoshida K & Blobel G The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J. Cell Biol 152, 729–740 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Belanger KD et al. The karyopherin Kap95 and the C-termini of Rfa1, Rfa2, and Rfa3 are necessary for efficient nuclear import of functional RPA complex proteins in Saccharomyces cerevisiae. DNA Cell Biol. 30, 641–651 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Nguyen VQ, Co C, Irie K & Li JJ Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2–7. Curr. Biol 10, 195–205 (2000). [DOI] [PubMed] [Google Scholar]
- 181.Bouayad D et al. Nuclear-to-cytoplasmic relocalization of the proliferating cell nuclear antigen (PCNA) during differentiation involves a chromosome region maintenance 1 (CRM1)-dependent export and is a prerequisite for PCNA antiapoptotic activity in mature neutrophils. J. Biol. Chem 287, 33812–33825 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Loeb JD et al. The yeast nuclear import receptor is required for mitosis. Proc. Natl Acad. Sci. USA 92, 7647–7651 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Pulliam KF, Fasken MB, McLane LM, Pulliam JV & Corbett AH The classical nuclear localization signal receptor, importin-α, is required for efficient transition through the G1/S stage of the cell cycle in Saccharomyces cerevisiae. Genetics 181, 105–118 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Garrido-Godino AI, Gutierrez-Santiago F & Navarro F Biogenesis of RNA polymerases in yeast. Front. Mol. Biosci 8, 669300 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Czeko E, Seizl M, Augsberger C, Mielke T & Cramer P Iwr1 directs RNA polymerase II nuclear import. Mol. Cell 42, 261–266 (2011). [DOI] [PubMed] [Google Scholar]
- 186.Gomez-Navarro N & Estruch F Different pathways for the nuclear import of yeast RNA polymerase II. Biochim. Biophys. Acta 1849, 1354–1362 (2015). [DOI] [PubMed] [Google Scholar]
- 187.Forget D et al. Nuclear import of RNA polymerase II is coupled with nucleocytoplasmic shuttling of the RNA polymerase II-associated protein 2. Nucleic Acids Res. 41, 6881–6891 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Forget D et al. The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol. Cell Proteom 9, 2827–2839 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Staresincic L, Walker J, Dirac-Svejstrup AB, Mitter R & Svejstrup JQ GTP-dependent binding and nuclear transport of RNA polymerase II by Npa3 protein. J. Biol. Chem 286, 35553–35561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Minaker SW, Filiatrault MC, Ben-Aroya S, Hieter P & Stirling PC Biogenesis of RNA polymerases II and III requires the conserved GPN small GTPases in Saccharomyces cerevisiae. Genetics 193, 853–864 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Niesser J, Wagner FR, Kostrewa D, Muhlbacher W & Cramer P Structure of GPN-loop GTPase Npa3 and implications for RNA polymerase II assembly. Mol. Cell Biol 36, 820–831 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Peiro-Chova L & Estruch F The yeast RNA polymerase II-associated factor Iwr1p is involved in the basal and regulated transcription of specific genes. J. Biol. Chem 284, 28958–28967 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gibney PA, Fries T, Bailer SM & Morano KA Rtr1 is the Saccharomyces cerevisiae homolog of a novel family of RNA polymerase II-binding proteins. Eukaryot. Cell 7, 938–948 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Reyes-Pardo H et al. A nuclear export sequence in GPN-loop GTPase 1, an essential protein for nuclear targeting of RNA polymerase II, is necessary and sufficient for nuclear export. Biochim. Biophys. Acta 1823, 1756–1766 (2012). [DOI] [PubMed] [Google Scholar]
- 195.Jakel S, Mingot JM, Schwarzmaier P, Hartmann E & Gorlich D Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 21, 377–386 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jakel S & Gorlich D Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17, 4491–4502 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Rout MP, Blobel G & Aitchison JD A distinct nuclear import pathway used by ribosomal proteins. Cell 89, 715–725 (1997). [DOI] [PubMed] [Google Scholar]
- 198.Sydorskyy Y et al. Intersection of the Kap123p-mediated nuclear import and ribosome export pathways. Mol. Cell Biol 23, 2042–2054 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Rosenblum JS, Pemberton LF & Blobel G A nuclear import pathway for a protein involved in tRNA maturation. J. Cell Biol 139, 1655–1661 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Schutz S et al. A RanGTP-independent mechanism allows ribosomal protein nuclear import for ribosome assembly. eLife 3, e03473 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ting YH et al. Bcp1 is the nuclear chaperone of Rpl23 in Saccharomyces cerevisiae. J. Biol. Chem 292, 585–596 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Timney BL et al. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J. Cell Biol 175, 579–593 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Plafker SM & Macara IG Ribosomal protein L12 uses a distinct nuclear import pathway mediated by importin 11. Mol. Cell. Biol 22, 1266–1275 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mitterer V et al. Sequential domain assembly of ribosomal protein S3 drives 40S subunit maturation. Nat. Commun 7, 10336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mitterer V et al. Nuclear import of dimerized ribosomal protein Rps3 in complex with its chaperone Yar1. Sci. Rep 6, 36714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Pillet B et al. The dedicated chaperone Acl4 escorts ribosomal protein Rpl4 to its nuclear pre-60S assembly site. PLoS Genet. 11, e1005565 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Stelter P et al. Coordinated ribosomal L4 protein assembly into the pre-ribosome is regulated by its eukaryote-specific extension. Mol. Cell 58, 854–862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kressler D et al. Synchronizing nuclear import of ribosomal proteins with ribosome assembly. Science 338, 666–671 (2012). [DOI] [PubMed] [Google Scholar]
- 209.Schutz S et al. Molecular basis for disassembly of an importin:ribosomal protein complex by the escortin Tsr2. Nat. Commun 9, 3669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Ting et al. (2017), this paper identifies how nuclear chaperones mediate the RAN–GTP-independent release of RPs from Kaps, coordinating nuclear import and ribosome biogenesis.
- 210.Nerurkar P et al. Eukaryotic ribosome assembly and nuclear export. Int. Rev. Cell Mol. Biol 319, 107–140 (2015). [DOI] [PubMed] [Google Scholar]
- 211.Thomas F & Kutay U Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci 116, 2409–2419 (2003). [DOI] [PubMed] [Google Scholar]
- 212.Zemp I et al. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J. Cell Biol 185, 1167–1180 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Seiser RM et al. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics 174, 679–691 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Vanrobays E et al. TOR regulates the subcellular distribution of DIM2, a KH domain protein required for cotranscriptional ribosome assembly and pre-40S ribosome export. RNA 14, 2061–2073 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Schnapp A, Schnapp G, Erny B & Grummt I Function of the growth-regulated transcription initiation factor TIF-IA in initiation complex formation at the murine ribosomal gene promoter. Mol. Cell Biol 13, 6723–6732 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Warner JR The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci 24, 437–440 (1999). [DOI] [PubMed] [Google Scholar]
- 217.Wu Z, Jiang Q, Clarke PR & Zhang C Phosphorylation of Crm1 by CDK1–cyclin-B promotes Ran-dependent mitotic spindle assembly. J. Cell Sci 126, 3417–3428 (2013). [DOI] [PubMed] [Google Scholar]
- 218.Guo H, Wei JH, Zhang Y & Seemann J Importin α phosphorylation promotes TPX2 activation by GM130 to control astral microtubules and spindle orientation. J. Cell Sci 134, jcs258356 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Beaudet D, Pham N, Skaik N & Piekny A Importin binding mediates the intramolecular regulation of anillin during cytokinesis. Mol. Biol. Cell 31, 1124–1139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Guo L et al. Phosphorylation of importin-α1 by CDK1–cyclin B1 controls mitotic spindle assembly. J. Cell Sci 132, jcs232314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Nakada R, Hirano H & Matsuura Y Structure of importin-α bound to a non-classical nuclear localization signal of the influenza A virus nucleoprotein. Sci. Rep 5, 15055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wu W et al. Synergy of two low-affinity NLSs determines the high avidity of influenza A virus nucleoprotein NP for human importin α isoforms. Sci. Rep 7, 11381 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Cansizoglu AE, Lee BJ, Zhang ZC, Fontoura BM & Chook YM Structure-based design of a pathway-specific nuclear import inhibitor. Nat. Struct. Mol. Biol 14, 452–454 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kutay U, Bischoff FR, Kostka S, Kraft R & Gorlich D Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell 90, 1061–1071 (1997). [DOI] [PubMed] [Google Scholar]
- 225.Lipowsky G et al. Coordination of tRNA nuclear export with processing of tRNA. RNA 5, 539–549 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Stuven T, Hartmann E & Gorlich D Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J. 22, 5928–5940 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Çağatay T & Chook YM Karyopherins in cancer. Curr. Opin. Cell Biol 52, 30–42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kosugi S et al. Design of peptide inhibitors for the importin α/β nuclear import pathway by activity-based profiling. Chem. Biol 15, 940–949 (2008). [DOI] [PubMed] [Google Scholar]
- 229.Wagstaff KM, Rawlinson SM, Hearps AC & Jans DA An AlphaScreen®-based assay for high-throughput screening for specific inhibitors of nuclear import. J. Biomol. Screen 16, 192–200 (2011). [DOI] [PubMed] [Google Scholar]
- 230.Ambrus G et al. Small molecule peptidomimetic inhibitors of importin α/β mediated nuclear transport. Bioorg. Med. Chem 18, 7611–7620 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.van der Watt PJ et al. Targeting the nuclear import receptor kpnβ1 as an anticancer therapeutic. Mol. Cancer Ther 15, 560–573 (2016). [DOI] [PubMed] [Google Scholar]
- 232.Ajayi-Smith A et al. Novel small molecule inhibitor of Kpnβ1 induces cell cycle arrest and apoptosis in cancer cells. Exp. Cell Res 404, 112637 (2021). [DOI] [PubMed] [Google Scholar]
- 233.Hintersteiner M et al. Identification of a small molecule inhibitor of importin β mediated nuclear import by confocal on-bead screening of tagged one-bead one-compound libraries. ACS Chem. Biol 5, 967–979 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Soderholm JF et al. Importazole, a small molecule inhibitor of the transport receptor importin-β. ACS Chem. Biol 6, 700–708 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Jans DA, Martin AJ & Wagstaff KM Inhibitors of nuclear transport. Curr. Opin. Cell Biol 58, 50–60 (2019). [DOI] [PubMed] [Google Scholar]
- 236.Gonzalez Canga A et al. The pharmacokinetics and interactions of ivermectin in humans — a mini-review. AAPS J. 10, 42–46 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yang SNY et al. The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antivir. Res 177, 104760 (2020). [DOI] [PubMed] [Google Scholar]
- 238.Wagstaff KM et al. Molecular dissection of an inhibitor targeting the HIV integrase dependent preintegration complex nuclear import. Cell Microbiol. 21, e12953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Fraser JE et al. A nuclear transport inhibitor that modulates the unfolded protein response and provides in vivo protection against lethal dengue virus infection. J. Infect. Dis 210, 1780–1791 (2014). [DOI] [PubMed] [Google Scholar]
- 240.Liu W et al. Identification of a covalent importin-5 inhibitor, goyazensolide, from a collective synthesis of furanoheliangolides. ACS Cent. Sci 7, 954–962 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This article identifies the first IPO5 inhibitor.
- 241.Engelsma D, Bernad R, Calafat J & Fornerod M Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 23, 3643–3652 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ferreira BI, Cautain B, Grenho I & Link W Small molecule inhibitors of CRM1. Front. Pharmacol 11, 625 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Lei Y et al. Structure-guided design of the first noncovalent small-molecule inhibitor of CRM1. J. Med. Chem 64, 6596–6607 (2021). [DOI] [PubMed] [Google Scholar]
- 244.Sun Q et al. Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1. Proc. Natl Acad. Sci. USA 110, 1303–1308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ye Y & Li B 1′S-1′-acetoxychavicol acetate isolated from Alpinia galanga inhibits human immunodeficiency virus type 1 replication by blocking Rev transport. J. Gen. Virol 87, 2047–2053 (2006). [DOI] [PubMed] [Google Scholar]
- 246.Hilliard M et al. The anti-inflammatory prostaglandin 15-deoxy-Δ(12,14)-PGJ2 inhibits CRM1-dependent nuclear protein export. J. Biol. Chem 285, 22202–22210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Hing ZA et al. Next-generation XPO1 inhibitor shows improved efficacy and in vivo tolerability in hematological malignancies. Leukemia 30, 2364–2372 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Etchin J et al. Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells. Leukemia 27, 66–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Lapalombella R et al. Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia. Blood 120, 4621–4634 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Haines JD et al. Nuclear export inhibitors avert progression in preclinical models of inflammatory demyelination. Nat. Neurosci 18, 511–520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Sakakibara K et al. CBS9106 is a novel reversible oral CRM1 inhibitor with CRM1 degrading activity. Blood 118, 3922–3931 (2011). [DOI] [PubMed] [Google Scholar]
- 252.Niu M, Chong Y, Han Y & Liu X Novel reversible selective inhibitor of nuclear export shows that CRM1 is a target in colorectal cancer cells. Cancer Biol. Ther 16, 1110–1118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tian X et al. Small-molecule antagonist targeting exportin-1 via rational structure-based discovery. J. Med. Chem 63, 3881–3895 (2020). [DOI] [PubMed] [Google Scholar]
- 254.Xu HW et al. A low toxic CRM1 degrader: synthesis and anti-proliferation on MGC803 and HGC27. Eur. J. Med. Chem 206, 112708 (2020). [DOI] [PubMed] [Google Scholar]
- 255.Saito N et al. CBS9106-induced CRM1 degradation is mediated by cullin ring ligase activity and the neddylation pathway. Mol. Cancer Ther 13, 3013–3023 (2014). [DOI] [PubMed] [Google Scholar]
- 256.Peterson TJ, Orozco J & Buege M Selinexor: a first-in-class nuclear export inhibitor for management of multiply relapsed multiple myeloma. Ann. Pharmacother 54, 577–582 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Ben-Barouch S & Kuruvilla J Selinexor (KTP-330) — a selective inhibitor of nuclear export (SINE): anti-tumor activity in diffuse large B-cell lymphoma (DLBCL). Expert Opin. Investig. Drugs 29, 15–21 (2020). [DOI] [PubMed] [Google Scholar]
- 258.Nakai K & Horton P PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci 24, 34–36 (1999). [DOI] [PubMed] [Google Scholar]
- 259.Nakai K & Kanehisa M A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14, 897–911 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kosugi S, Hasebe M, Tomita M & Yanagawa H Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl Acad. Sci. USA 106, 10171–10176 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Cokol M, Nair R & Rost B Finding nuclear localization signals. EMBO Rep. 1, 411–415 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Brameier M, Krings A & MacCallum RM NucPred — predicting nuclear localization of proteins. Bioinformatics 23, 1159–1160 (2007). [DOI] [PubMed] [Google Scholar]
- 263.Nair R, Carter P & Rost B NLSdb: database of nuclear localization signals. Nucleic Acids Res. 31, 397–399 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Mehdi AM, Sehgal MS, Kobe B, Bailey TL & Boden M A probabilistic model of nuclear import of proteins. Bioinformatics 27, 1239–1246 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Nguyen Ba AN, Pogoutse A, Provart N & Moses AM NLStradamus: a simple hidden Markov model for nuclear localization signal prediction. BMC Bioinforma 10, 202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Lin JR & Hu J SeqNLS: nuclear localization signal prediction based on frequent pattern mining and linear motif scoring. PLoS ONE 8, e76864 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper introduces SeqNLS, which is the current NLS predictor with the best balance in performance.
- 267.Bernhofer M et al. NLSdb — major update for database of nuclear localization signals and nuclear export signals. Nucleic Acids Res. 46, D503–D508 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Guo Y, Yang Y, Huang Y & Shen HB Discovering nuclear targeting signal sequence through protein language learning and multivariate analysis. Anal. Biochem 591, 113565 (2020). [DOI] [PubMed] [Google Scholar]
- 269.la Cour T et al. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 31, 393–396 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.la Cour T et al. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel 17, 527–536 (2004). [DOI] [PubMed] [Google Scholar]
- 271.Fu SC, Imai K & Horton P Prediction of leucine-rich nuclear export signal containing proteins with NESsential. Nucleic Acids Res. 39, e111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Prieto G, Fullaondo A & Rodriguez JA Prediction of nuclear export signals using weighted regular expressions (Wregex). Bioinformatics 30, 1220–1227 (2014). [DOI] [PubMed] [Google Scholar]
- 273.Kosugi S, Yanagawa H, Terauchi R & Tabata S NESmapper: accurate prediction of leucine-rich nuclear export signals using activity-based profiles. PLoS Comput. Biol 10, e1003841 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Xu D et al. LocNES: a computational tool for locating classical NESs in CRM1 cargo proteins. Bioinformatics 31, 1357–1365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper introduces LocNES, which is the top-performing NES predictor available currently.
- 275.Liku ME, Legere EA & Moses AM NoLogo: a new statistical model highlights the diversity and suggests new classes of Crm1-dependent nuclear export signals. BMC Bioinforma. 19, 65 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Lee Y, Pei J, Baumhardt JM, Chook YM & Grishin NV Structural prerequisites for CRM1-dependent nuclear export signaling peptides: accessibility, adapting conformation, and the stability at the binding site. Sci. Rep 9, 6627 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


