Summary
Diet profoundly influences physiology. Whereas over-nutrition elevates risk for disease via its influence on immunity and metabolism, caloric restriction and fasting appear to be salutogenic. Despite multiple correlations observed between diet and health, the underlying biology remains unclear. Here, we identified a fasting-induced switch in leukocyte migration that prolongs monocyte lifespan and alters disease susceptibility in mice. We show that fasting during the active phase induced rapid return of monocytes from blood to the bone marrow. Monocyte re-entry was orchestrated by hypothalamic-pituitary-adrenal (HPA)-axis-dependent release of corticosterone, which augmented the CXCR4 chemokine receptor. Though the marrow is a safe haven for monocytes during nutrient scarcity, re-feeding prompted mobilization culminating in monocytosis of chronologically older and transcriptionally distinct monocytes. These shifts altered response to infection. Our study shows that diet— in particular a diet’s temporal dynamic balance—modulates monocyte lifespan, with consequences on adaptation to external stressors.
Keywords: monocyte, fasting, hematopoiesis, bone marrow, infection
eTOC blurb
Fasting influences the distribution of leukocytes throughout the body, but mechanisms and implications are only partly understood. Here, Janssen et al. reveal how fasting leads to homing of monocytes into the bone marrow. Re-feeding gives rise to a surge of circulating monocytes with deleterious effects on bacterial host response.
Graphical Abstract
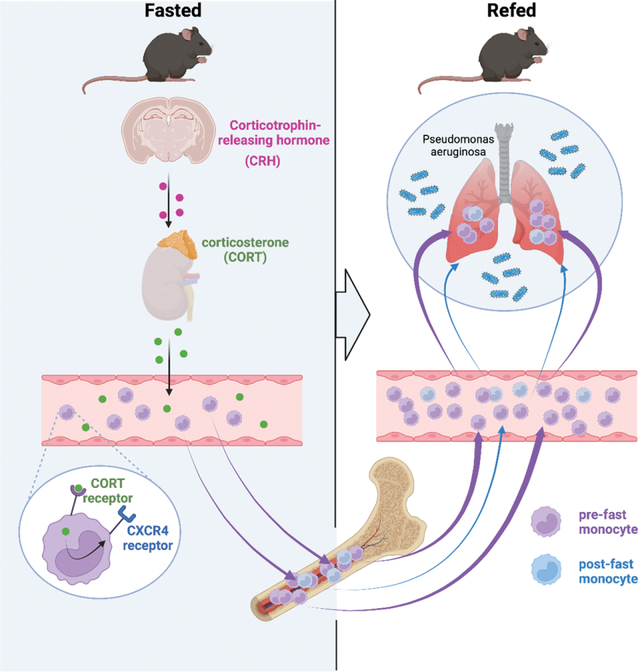
Introduction
Caloric excess reduces organismal lifespan1–3, increases systemic inflammation4,5, elevates risk for cardiometabolic diseases4,6, and exacerbates bacterial infection3,7. Conversely, caloric restriction and fasting have been shown to inversely correlate with hypertension, atherosclerosis, diabetes, obesity, colitis, asthma, and psoriasis 8–12. As ingested energy sources engage with multiple metabolic processes that affect physiology, the precise mechanisms linking diet with tissue, cellular, and molecular function remain poorly understood.
Recent studies have indicated that diet influences the distribution of leukocytes throughout the body. For example, Belkaid and colleagues showed that T lymphocytes relocate from secondary lymphoid organs to the bone marrow during caloric restriction13, Hase and colleagues discovered that B cells leave Peyer’s patches14, while Merad and colleagues have demonstrated that fasting reduces the number of circulating monocytes in mice and humans by preventing their mobilization from the bone marrow15. All studies reveal that diet-dependent shifts in leukocyte distribution can have major effects on disease outcome, and in particular to the response during infection, oral tolerance, tumor growth, and experimental autoimmune encephalomyelitis. These insights raise a number of fundamental questions concerning diet’s control of leukocyte biology. As dietary restriction leads to hunger, it remains unclear how the brain participates in these phenomena, particularly in light of recent studies on the central nervous system’s regulation of peripheral leukocytes16,17. Moreover, the fate of leukocytes during fasting is unclear: monocytes, for example, are typically short-lived, raising the question as to whether fasting influences their lifespan, production, and death. Unless fasting continues to starvation, it is followed by re-feeding, the consequences of which are unknown. In this study we focused on the mechanism and effects of fasting and re-feeding on monocyte dynamics and homeostasis.
Results
Fasting reorganizes the leukocyte distribution landscape
We started our study by enumerating major leukocyte classes (B and T lymphocytes, monocytes, and neutrophils) across 16 organs and tissues in response to a 24h fast. Our data, tabulated (Table 1 and S1) or represented graphically (Fig. 1A and B), illustrate major and rapid shifts for each leukocyte type. Whereas some tissues experienced an increase, others saw a decrease in leukocyte number after fasting. Notably, every tissue except the bone marrow had a reduced number of Ly-6Chi monocytes compared to the corresponding tissue of non-fasted animals, which is consistent with recent work in both mice and humans15. The relative increase of monocytes in the bone marrow after a 24h fast is likewise consistent with the idea that fasting prevents monocyte mobilization from the bone marrow to the blood15.
Table 1.
Absolute counts of major leukocyte populations in selected organs in mice continuously fed or fasted for 24h.
| Monocytes | Neutrophils | |||
| Fed | 24h Fast | Fed | 24h Fast | |
| Blood (×103 cells/ml) | 724±212 | 37±14 | 1539±291 | 424±38 |
| n=5 | n=5, p<0.05 | n=4 | n=4, p<0.05 | |
| Spleen (×103 cells/organ) | 185±7.7 | 14±2.0 | 191±20 | 82±26 |
| n=3 | n=3, p<0.001 | n=3 | n=3, p<0.05 | |
| Liver (×104 cells/organ) | 7.0±0.4 | 0.6±0.2 | 8.5±0.7 | 4.9±3.0 |
| n=3 | n=3, p<0.001 | n=3 | n=3, p=n.s. | |
| Lung (×103 cells/organ) | 148±32 | 31±3.7 | 150±20 | 67±11 |
| n=3 | n=3, p<0.05 | n=3 | n=3, p<0.05 | |
| WAT (Cells/organ) | 626±247 | 61±14 | 2104±997 | 379±79 |
| n=3 | n=3, p=n.s. | n=4 | n=4, p=n.s. | |
| BAT (×103 cells/organ) | 2.1±0.4 | 0.6±0.3 | 6.8±2.4 | 3.3±0.9 |
| n=4 | n=4, p<0.05 | n=4 | n=4, p=n.s. | |
| Heart (×103 cells/organ) | 2.0±0.2 | 1.4±0.4 | 2.4±0.6 | 3.1±0.4 |
| n=3 | n=3, p=n.s. | n=3 | n=3, p=n.s. | |
| Muscle (Cells/mg) | 1.7±0.3 | 0.9±0.3 | 1.8±0.3 | 4.7±2.3 |
| n=5 | n=4, p=n.s. | n=5 | n=5, p=n.s. | |
| Pancreas (×103 cells/organ) | 1.8±0.6 | 0.9±0.2 | 1.2±0.2 | 0.4±0.1 |
| n=5 | n=5, p=n.s. | n=5 | n=5, p<0.05 | |
| SI lamina propria (×103 cells/organ) | 2.9±0.5 | 1.0±0.2 | 0.9±0.3 | 1.7±0.6 |
| n=3 | n=3, p<0.05 | n=4 | n=5, p=n.s. | |
| LI lamina propria (×103 cells/organ) | 0.7±0.2 | 0.6±0.3 | 0.8±0.2 | 0.6±0.2 |
| n=4 | n=4, p=n.s. | n=4 | n=4, p=n.s. | |
| MLN (×103 cells/LN) | 0.27±0.03 | 0.06±0.01 | 0.26±0.01 | 0.13±0.03 |
| n=4 | n=4, p<0.001 | n=4 | n=4, p<0.01 | |
| Peyer Patches (×103 cells/PP) | 0.028±0.017 | 0.006±0.002 | 0.004±0.001 | 0.001±0.0002 |
| n=5 | n=5, p=n.s. | n=5 | n=5, p<0.01 | |
| Peritoneum (×103 cells/organ) | 1.9±0.4 | 0.8±0.1 | 1.0±0.7 | 0.6±0.3 |
| n=3 | n=4, p<0.05 | n=3 | n=4, p=n.s. | |
| Mesenteric AT (×103 cells/organ) | 8.8±1.6 | 0.8±0.1 | 3.1±1.4 | 1.2±0.2 |
| n=3 | n=3, p<0.01 | n=3 | n=3, p=n.s. | |
| Bone marrow (×104 cells/femur) | 13±1.1 | 18±0.8 | 105±8.8 | 107±6.5 |
| n=3 | n=3, p<0.05 | n=3 | n=3, p=n.s. | |
| T cells | B cells | |||
| Fed | 24h Fast | Fed | 24h Fast | |
| Blood (×103 cells/ml) | 1409±181 | 760±106 | 4619±396 | 2138±253 |
| n=4 | n=4, p<0.05 | n=4 | n=4, p<0.01 | |
| Spleen (×103 cells/organ) | 8498±1317 | 6790±337 | 18010±1703 | 12686±1391 |
| n=3 | n=3, p=n.s. | n=3 | n=3, p=n.s. | |
| Liver (×104 cells/organ) | 57±9.6 | 36±12 | 191±30 | 96±46 |
| n=3 | n=3, p=n.s. | n=3 | n=3, p=n.s. | |
| Lung (×103 cells/organ) | 598±77 | 280±61 | 1007±85 | 459±61 |
| n=3 | n=3, p<0.05 | n=3 | n=3, p<0.01 | |
| WAT (Cells/organ) | 16967±5009 | 4099±1278 | 2437±551 | 621±175 |
| n=4 | n=4, p=n.s. | n=4 | n=4, p<0.05 | |
| BAT (×103 cells/organ) | 9.9±2.0 | 4.3±0.6 | 14±3.8 | 5.4±1.6 |
| n=4 | n=4, p<0.05 | n=4 | n=4, p=n.s. | |
| Heart (×103 cells/organ) | 3.8±0.8 | 2.8±0.3 | 15±2.4 | 6.9±0.9 |
| n=3 | n=3, p=n.s. | n=3 | n=3, p<0.05 | |
| Muscle (Cells/mg) | 3.5±0.6 | 4.4±1.9 | 4.7±0.9 | 11±6.6 |
| n=5 | n=5, p=n.s. | n=5 | n=5, p=n.s. | |
| Pancreas (×103 cells/organ) | 12±3.2 | 59±25 | 11±4.3 | 59±27 |
| n=5 | n=5, p=n.s. | n=5 | n=5, p=n.s. | |
| SI lamina propria (×103 cells/organ) | 17±5.3 | 103±17 | 105±52 | 460±123 |
| n=4 | n=5, p<0.01 | n=4 | n=5, p=n.s. | |
| LI lamina propria (×103 cells/organ) | 22±4.1 | 21±3.6 | 114±37 | 52±11 |
| n=4 | n=4, p=n.s. | n=4 | n=4, p=n.s. | |
| MLN (×103 cells/LN) | 239±25 | 130±4.4 | 211±27 | 96±8.2 |
| n=4 | n=4, p<0.01 | n=4 | n=4, p<0.01 | |
| Peyer Patches (×103 cells/PP) | 8.9±0.7 | 4.2±0.4 | 51±14 | 30±4.5 |
| n=5 | n=5, p<0.001 | n=5 | n=5, p=n.s. | |
| Peritoneum (×103 cells/organ) | 26±3.4 | 54±11 | 246±40 | 353±63 |
| n=3 | n=4, p=n.s. | n=3 | n=4, p=n.s. | |
| Mesenteric AT (×103 cells/organ) | 25±7.2 | 17±0.9 | 9.1±1.4 | 6.3±1.3 |
| n=3 | n=3, p=n.s. | n=3 | n=3, p=n.s. | |
| Bone marrow (×104 cells/femur) | 2.5±0.2 | 9.2±1.4 | 173±4.6 | 166±9.4 |
| n=3 | n=3, p<0.05 | n=3 | n=3, p=n.s. | |
Unpaired t test. Data presented as mean ±SEM
p < 0.05
p<0.01
p < 0.001.
(WAT: White adipose tissue, BAT: Brown adipose tissue, SI: Small intestine, LI: Large intestine, MLN: Mesenteric Lymph nodes, AT: Adipose tissue). Please also see Figure 1.
Figure 1: Fasting reorganizes the leukocyte distribution landscape.
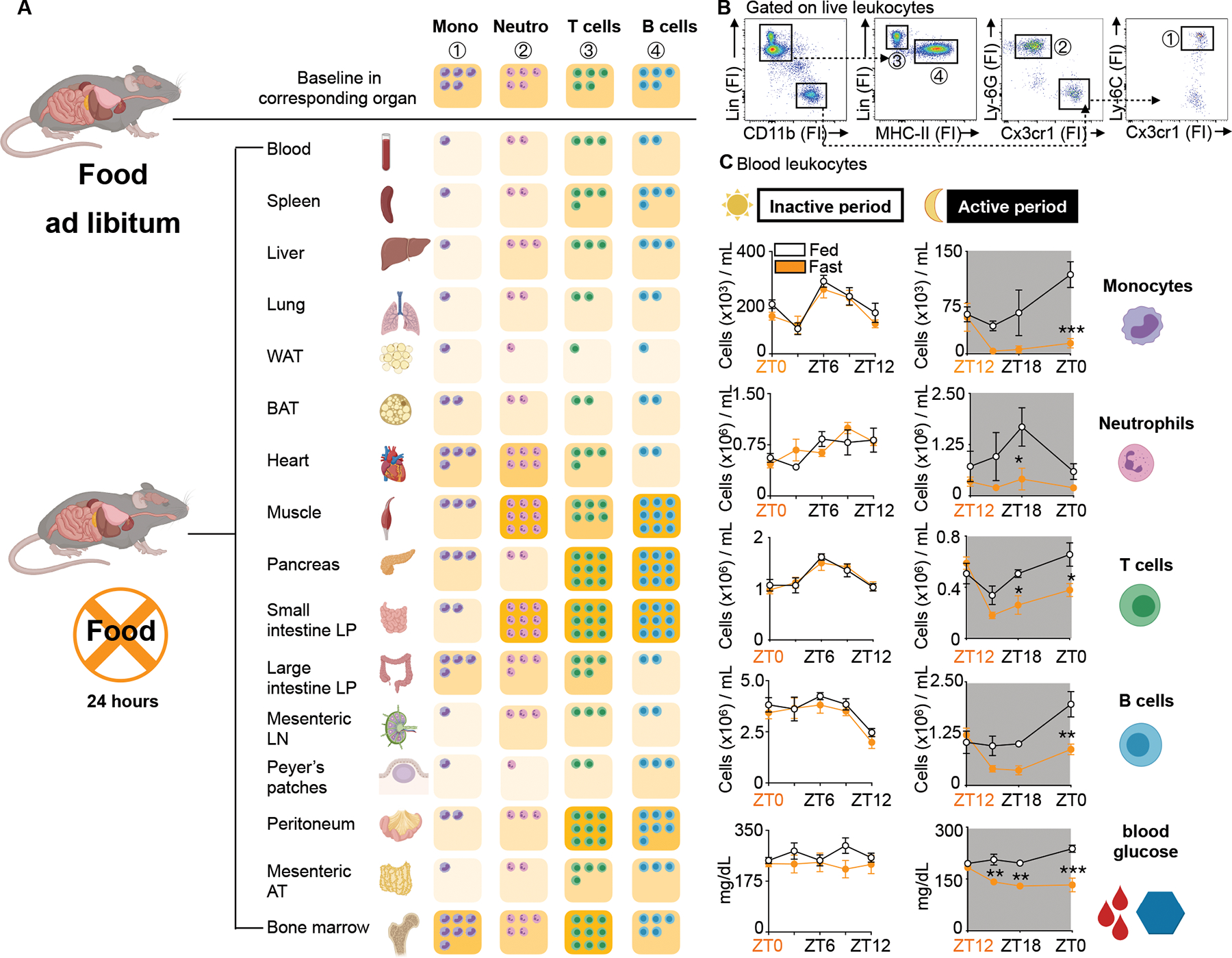
A. Relative shifts of major leukocyte populations in depicted organs compared between mice fed ad libitum and after a 24h fast. Baseline cell content in each organ is defined as “5 cells”. Fasted results are calculated in relative relation to fed. Ly-6Chi monocytes are defined as CD45+ CD11b+ LIN1(CD19, CD3, NK 1.1, CD90−) Ly-6G− CX3CR1+ Ly-6Chi, neutrophils as CD45+ CD11b+ LIN1− Ly-6G+, T cells as CD45+ CD11b− LIN1+ MHCII−, B cells as CD45+ CD11b− LIN1+ MHCII+. Absolute numbers for each organ are depicted in Table 1. (n=3–5 mice per group). Unpaired t test.
B. Representative gating strategy of all major leukocyte populations in the blood.
C. Absolute blood leukocyte count and blood glucose concentration divided by inactive (light) and active (dark) period compared between mice fed ad libitum or fasted for up to 12h. Orange zeitgeber (ZT) indicates start of the fast (n=3–10 per group). Two-way ANOVA. Data presented as mean ±SEM, *p < 0.05, **p<0.01, ***p < 0.001. Mono: Ly-6Chi monocytes, Neutro: Neutrophils, WAT: White adipose tissue, BAT: Brown adipose tissue, LP: Lamina propria, LN: Lymph node, AT: Adipose tissue, FI: Fluorescence intensity, ZT: Zeitgeber. Please also see Table 1 and Supplement Table 1.
Monocyte numbers in the blood fluctuate according to circadian rhythms18. In mice, monocyte numbers peak in the blood during the day shortly after mice go to sleep (~ZT6) and reach their nadir at night shortly after mice wake up (~ZT16). The mechanisms that control this circadian rhythm are incompletely understood19,20. We performed a time course analysis, asking whether the fasting effect on Ly-6Chi monocytes depended on the time of day. We found that food restriction during the active period (ZT12-ZT24/0) but not during the inactive period (ZT0-ZT12) rapidly reduced blood glucose concentration and circulating Ly-6Chi monocytes, suggesting that meals early after waking prevent drops in leukocyte numbers (Fig. 1C). We found similar patterns for neutrophils, T cells, and B cells.
Fasting returns Ly-6Chi monocytes back from circulation to the bone marrow
Having documented a decrease of monocytes in the periphery, including the blood, which we verified with intravital microscopy (Fig. S1A and Video S1 and S2), along with a corresponding increase in the bone marrow, we next sought to identify the relevant mechanism. Earlier work suggested that a fasting-mediated mobilization shutdown from the bone marrow drives the phenomenon15. However, we speculated there may be additional mechanisms. First, Ly-6Chi monocytes disappeared from the blood 4h after food deprivation in the active period (Fig. 1C). This appeared to be much faster than expected. Under the assumptions that fasting leads to a complete shutdown of bone marrow Ly-6Chi monocyte egress and a typical half-life of circulating Ly-6Chi monocytes of approximately 20h21, the expected reduction of blood monocytes after a 20h fast, in the absence of any additional mechanisms, would be ~50% (Fig. S1B). However, we observed a ~90% reduction of circulating Ly-6Chi monocytes in mice after only 4h, suggesting the contribution of additional mechanisms beyond bone marrow monocyte egress (Fig. 1C). Second, mice lacking Ccr2, the chemokine receptor responsible for mobilization from the bone marrow, also experienced a fasting monocyte phenotype (Fig. S1C). The caveat that these mice already have reduced monocyte numbers in the blood at steady state notwithstanding, the data nevertheless point to a mobilization-independent mechanism. Third, we excluded several candidate mechanisms, noting that the disappearance of Ly-6Chi monocytes in response to fasting was independent of CD18/LFA1 and CX3CR1-mediated patrolling behavior (Fig. S1C), and independent of myeloid-derived AMPK (Prkaa1), the clock gene Arntl, and Glut-1 (Slc2a1) (Fig. S1D). Moreover, the disappearance of Ly-6Chi monocytes in response to fasting was not caused by monocyte-platelet aggregation (Fig. S1E), was independent of T or B cells (Fig. S1F), and it occurred in aged mice (Fig. S1G), both sexes (Fig. S1G), and different genetic backgrounds (Fig. S1H). When mice were fasted for 24h every other day for a period of 2 weeks, the effect persisted (Fig. S1I), and was therefore not subjected to adaptation.
We elected to conduct a pulse-chase experiment involving the adoptive transfer of GFP+ monocytes to either fed or fasted mice (Fig. 2A). 24h after transfer (and after fasting) a GFP+ monocyte population was present in the blood of fed mice, but absent in the blood of fasted mice (Fig. 2B and D). As hypothesized, it was the bone marrow of fasted mice that not only contained the transferred GFP+ cells, but contained more of them compared to the fed ad libitum group (Fig. 2C and D). Because GFP+ monocytes were transferred intravenously, their appearance in the bone marrow necessarily required migration from blood to the bone marrow. The adoptive transfer experiments suggested that reverse mobilization (i.e., the migration of monocytes from the blood to the bone marrow) is an important mechanism contributing to the fasting-induced phenomenon.
Figure 2: Fasting returns Ly-6Chi monocytes back from circulation to the bone marrow.
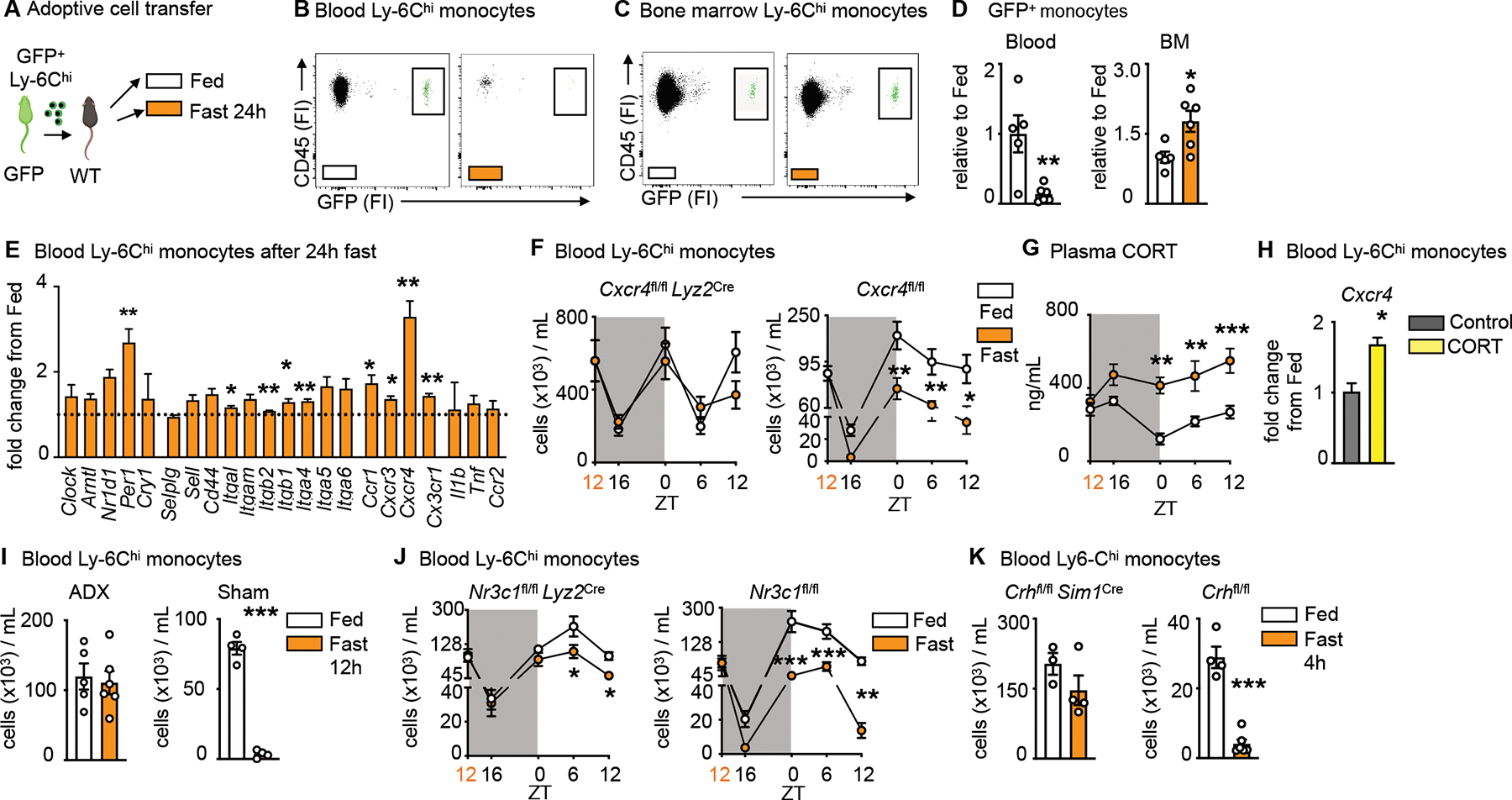
A. Ly-6Chi monocytes isolated from GFP positive mice were injected intravenously into wild type mice directly prior to initiation of a 24h fast or feeding ad libitum.
B. Representative flow cytometry plots of GFP positive monocytes after feeding ad libitum and fasting in the blood. Adoptively transferred monocytes are defined as CD45+ GFP+. (n=5–6 per group, two independent experiments combined)
C. Representative flow cytometry plots of GFP positive monocytes after feeding ad libitum and fasting in the bone marrow (BM). Adoptively transferred monocytes are defined as CD45+ GFP+. (n=5–6 per group, two independent experiments combined)
D. Relative decrease of GFP positive monocytes in the blood and relative increase of GFP positive monocytes in the bone marrow after 24h fasting compared to fed control (n=5–6 per group, two independent experiments combined). Unpaired t test.
E. Relative expression of target genes of sorted blood Ly-6Chi monocytes after 24h fasting normalized to fed control (n=3 mice per group). Unpaired t test.
F. Circulating Ly-6Chi monocytes measured at indicated timepoints in Cxcr4fl/fl Lyz2Cre and Cxcr4fl/fl under fed and fasted conditions (n=4–10, timepoints are independent experiments). Start of the fast is indicated by orange zeitgeber (ZT). Two-way ANOVA.
G. Plasma corticosterone (CORT) in Cxcr4fl/fl under fed and fasted conditions at indicated timepoints (n=4–8, timepoints are independent experiments). Start of the fast is indicated by orange zeitgeber (ZT). Two-way ANOVA.
H. Relative expression of Cxcr4 on sorted blood Ly-6Chi monocytes after incubation with CORT (n=10 mice pooled for sort, n=3 technical replicates per group). Unpaired t test.
I. Mice underwent bilateral adrenalectomy (ADX) or sham surgery and were allowed to recover for 2 weeks, prior to submission to feeding ad libitum or a 12h fast. Blood Ly-6Chi monocytes in mice 2 weeks post ADX or sham surgery after fasting compared to fed ad libitum (n=4–6 per group). Unpaired t test.
J. Blood Ly-6Chi monocytes measured at indicated times in Nr3c1fl/fl Lyz2Cre and Nr3c1fl/fl under fed and fasted conditions (n=4–11, timepoints areindependent experiments). Start of the fast is indicated by orange ZT. Two-way ANOVA.
K. Blood Ly-6Chi monocytes in Crhfl/fl Sim1Cre and Crhfl/fl after 4h of fasting compared to fed ad libitum (n=3–6 per group). Unpaired t test. Data presented as mean ±SEM, *p < 0.05, **p<0.01, ***p < 0.001. FI: Fluorescence intensity, BM: Bone marrow, CORT:
Corticosterone, ADX: Adrenalectomy. Please also see Figure S1.
To explore the reverse mobilization idea mechanistically, we measured a cassette of potentially relevant genes, such as clock genes and genes important to leukocyte adhesion and migration, on monocytes retrieved from the blood of fasted and fed mice (Fig. 2E). Among transcripts measured, Cxcr4 was particularly notable because it was expressed nearly four-fold above fed conditions and because it is known to facilitate medullary cell retention, as the loss of Cxcr4 expression mobilizes leukocytes to the blood22. We therefore generated mice lacking Cxcr4 on myeloid cells. The rapid and lasting fasting-induced monocytopenia in controls was abolished in the absence of Cxcr4 (Fig. 2F, S1J), without affecting the typical circadian fluctuations that are known to be controlled by the circadian clocks22. The data provide further evidence for reverse migration of monocytes from the blood to the bone marrow.
Previous studies from us and others have shown that corticosterone augments Cxcr4 expression17,23. We therefore wondered whether corticosterone was responsible for the phenomenon. We first observed that fasting rapidly and consistently increased corticosterone in the blood (Fig. 2G), which is in keeping with the idea that fasting induces a corticosterone-mediated stress-like response. The addition of corticosterone to sorted monocytes in vitro increased Cxcr4 expression (Fig. 2H), and adrenalectomy, which prevented a corticosterone spike in response to fasting (Fig. S1K), also prevented monocytopenia (Fig 2I) and an increase of Ly-6Chi monocytes in the bone marrow (Fig. S1L). Fasting is known to increase plasma free fatty acids via lipolysis of adipose tissue24. This partly depends on the melanocortin-3 receptor and thereby on the activation of the hypothalamic-pituitary-adrenal (HPA) axis25. We found that fasted and adrenolectomized mice developed severe hypoglycemia with no weight loss and no change in plasma fatty acids (Fig. 2M–O). These observations led us to ask whether corticosterone was directly engineering reverse mobilization, or if this was a phenomenon induced by another mechanism downstream of the adrenal gland. We deleted the glucocorticoid receptor (Nr3c1) from myeloid cells and indeed discovered that fasting no longer induced the disappearance of monocytes from the blood in those mice (Fig. 2J, S1P).
Corticosterone, produced by the adrenal glands, is an end product of the HPA-axis, which originates in the paraventricular hypothalamus (PVH) containing a small cluster of corticotropin releasing hormone (CRH) neurons. To determine whether mass migration of monocytes from the blood to the bone marrow was controlled by this cluster — as opposed to other possible mechanisms that converge on corticosterone production — we generated mice with cell-specific deletion of Crh in neurons of the hypothalamus (Crhfl/fl Sim1Cre). Compared to controls, we failed to detect monocytopenia in fasted mice lacking CRH expression (Fig. 2K). To confirm that the lack of fasting-induced monocytopenia was indeed a result of an impaired HPA-axis response, we injected Crhfl/fl Sim1Cre and littermate controls with corticosterone (Fig. S1Q). Hyper-corticosteronemia increased CXCR4 and reduced circulating monocytes in Crhfl/fl Sim1Cre and Crhfl/fl mice (Fig. S1R, S). Thus, monocyte reverse medullary migration during fasting is controlled by a hypothalamic brain axis that induces monocytic CXCR4 expression via direct corticosterone engagement.
Re-feeding leads to a surge of monocytes into the bloodstream
Seclusion of monocytes in the bone marrow during fasting raises questions as to their fate. Rather than exploring how even more severe fasting, which would lead to starvation and eventual death, affects monocytes, we wondered how circulating monocyte numbers would change upon re-feeding. We anticipated that monocytes would re-enter the blood, but could not surmise how quickly this would happen and to what extent. We therefore enumerated circulating monocytes in mice that were fed ad libitum, fasted, or fasted and re-fed (Fig. 3A). Next to the characteristic monocytopenia in fasted conditions, mice that were re-fed developed monocytosis 4h after re-feeding, dwarfing the typical monocyte numbers found in ad libitum-fed mice at this stage of the circadian clock (Fig. 3B and C). We observed a similar, if slightly muted, surge among neutrophils, but not among lymphocytes (Fig. S2A). The monocytic surge correlated with blood glucose (Fig. 3D) and inversely correlated with plasma corticosterone (Fig. 3E). The data point to a robust liberation of sequestered monocytes into the blood shortly after food reintroduction.
Figure 3: Re-feeding leads to a surge of monocytes into the bloodstream.
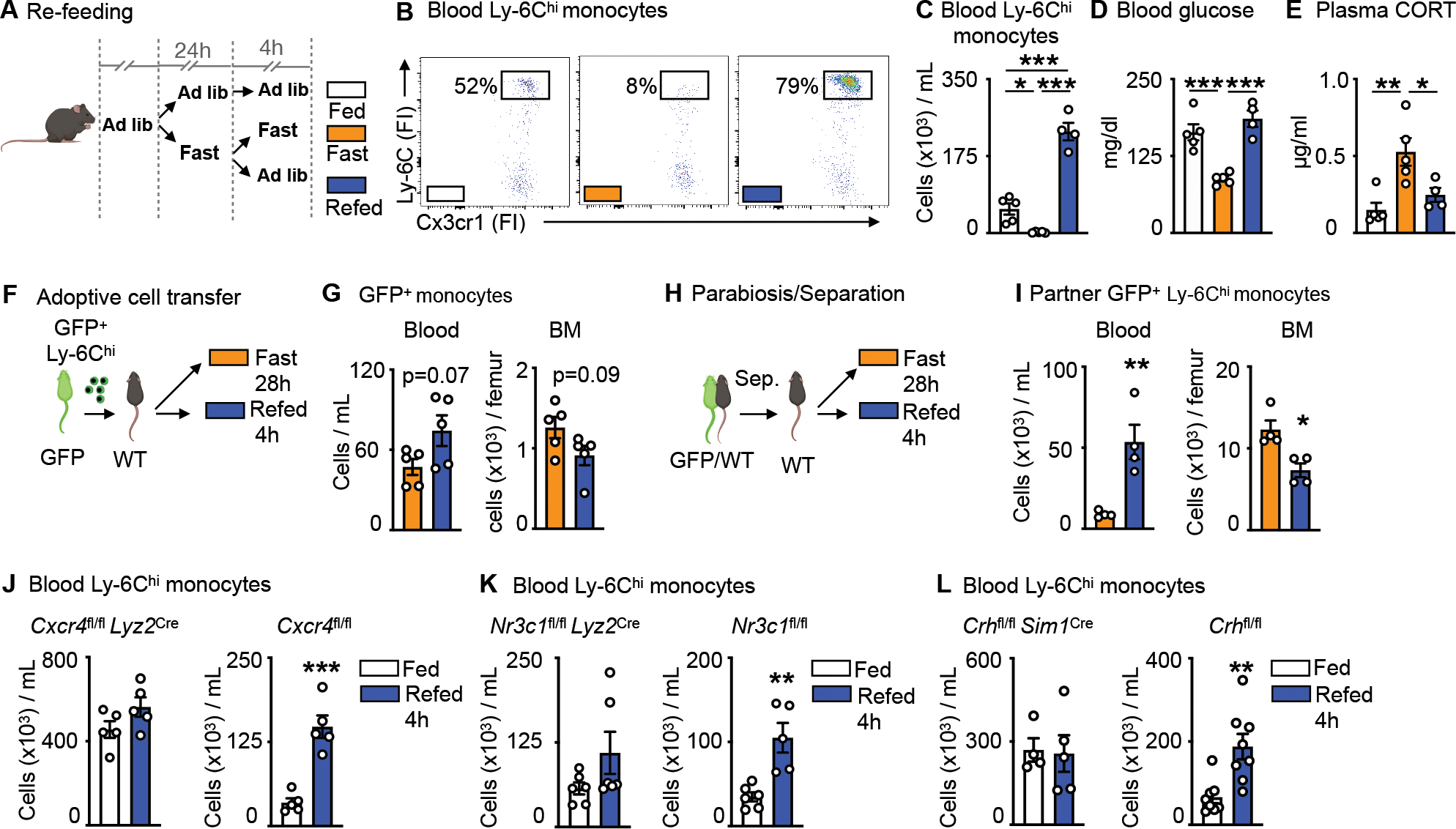
A. Mice were fasted for 24h and divided into further fasting or re-feeding for 4h. Control mice were fed ad libitum.
B. Representative flow cytometry plots of blood Ly-6Chi monocytes after feeding ad libitum, fasting for 28h or fasting for 24h followed by 4h of re-feeding. Ly-6Chi monocytes are defined as CD45+ CD11b+ LIN1− Ly-6G− CX3CR1+ Ly-6Chi (n=4–5 per group).
C. Blood Ly-6Chi monocytes after feeding ad libitum, fasting or fasting+re-feeding (n=4–5 per group). One-way ANOVA.
D. Blood glucose concentration after feeding ad libitum, fasting or fasting+re-feeding (n=4–5 per group). One-way ANOVA.
E. Plasma corticosterone (CORT) concentration after feeding ad libitum, fasting or fasting+re-feeding (n=4–5 per group). One-way ANOVA
F. Ly-6Chi monocytes sorted from GFP positive mice were injected intravenously into wild type mice directly prior to submission to 28h of fasting or 24h of fasting and 4h of re-feeding.
G. Adoptively transferred monocytes in the blood and bone marrow (BM) of wild type recipients. Adoptively transferred monocytes are defined as CD45+ GFP+. (n=5 per group). Unpaired t test.
H. Parabiosis of GFP positive and wild type mice. After two weeks, mice were separated and wild type mice were either fasted for 28h or fasted for 24h followed by 4h of re-feeding.
I. GFP positive Ly-6Chi monocytes in the blood and BM of wild type parabionts after fasting or fasting+re-feeding. (n=4 per group). Unpaired t test.
J. Blood Ly-6Chi monocytes in Cxcr4fl/fl Lyz2Cre and Cxcr4fl/fl after feeding ad libitum or fasting+re-feeding (n=5 per group). Unpaired t test.
K. Blood Ly-6Chi monocytes in Nr3c1fl/fl Lyz2Cre and Nr3c1fl/fl after feeding ad libitum or fasting+re-feeding (n=5–6 per group). Unpaired t test.
L. Blood Ly-6Chi monocytes in Crhfl/fl Sim1Cre and Crhfl/fl after feeding ad libitum or fasting+re-feeding (n=4–8 per group). Unpaired t test. Data presented as mean ±SEM, *p < 0.05, **p<0.01, ***p < 0.001. FI: Fluorescence intensity CORT: Corticosterone, BM: Bone marrow. Please also see Figure S2.
To chart mobilization more directly, we performed two types of cell tracking experiments. First, we adoptively transferred GFP+ monocytes to mice that were subsequently fasted or fasted and re-fed (Fig. 3F). Similar to our prior experiment, we noted slightly more monocytes in blood and fewer monocytes in bone marrow of re-fed mice, suggesting mobilization after re-feeding (Fig. 3G). These re-appearing GFP+Ly-6Chi monocytes upon re-feeding were alive and not eaten by endogenous GFP−Ly-6Chi monocytes (Fig. S2B). Second, we supplemented this experiment with the use of parabiosis, partnering GFP+ mice with wild type mice for 2 weeks to establish chimeric equilibrium (Fig. 3H and Fig. S2C), and then separating the parabionts before subjecting them to either fasting or re-feeding. We tracked the GFP+Ly-6Chi monocytes in partner mice. Despite the invasive surgical interventions, we detected notable differences in the location of GFP+Ly-6Chi monocytes, finding considerably more monocytes in the blood and, conversely, fewer monocytes in the bone marrow of re-fed mice (Fig. 3I), yet again supporting the idea that re-feeding led to massive mobilization of sequestered cells.
The re-feeding data revealed a surge of rapidly mobilized monocytes back to the blood. We reasoned that the surge likely consisted of both newly-generated monocytes that were trapped during fasting and of so-called returners — that is, mature circulating monocytes that returned to the bone marrow via CXCR4 during fasting and that now were again released to the blood. We tested for hematopoiesis by measuring BrdU incorporation to hematopoietic stem and progenitor cells (HSPC) and found a notable decrease in hematopoiesis during fasting that remained low 4h after re-feeding (Fig. S2D). We tested for the returners by enumerating monocytes under re-feeding conditions in mice with impaired re-mobilization: mice lacking CXCR4 (Fig. 3J), the glucocorticoid receptor on monocytes (Fig. 3K), and CRH in neurons of the PVH (Fig. 3L). In all three cases, we did not see a surge of monocytes in blood after re-feeding, strongly suggesting that a major component of the surge consisted of “returners”.
The monocyte surge is dominated by aged Ly-6Chi monocytes
The observation that the surge consisted of formerly-circulating monocytes that had re-mobilized to the bone marrow during fasting and then returned to the blood upon re-feeding raised a crucial question: Were monocytes sequestered in the bone marrow surviving longer than their circulating counterparts? By extension, was the re-feeding surge characterized by a chronologically older monocyte population? To begin testing these ideas, we implemented a double pulse-chase strategy involving sequential injection of the nucleotide analogues EdU and BrdU (Fig. 4A). Analysis of EdU and BrdU incorporation into medullary Ly-6Chi monocytes revealed 4 populations (Fig. 4B): the double negatives (BrdU−EdU−), representing cells that never had the opportunity to proliferate with either nucleotide analogue present, the double positives (BrdU+EdU+), representing cells that proliferated once or more with both nucleotide analogue present, and the single positives. Among the single positives, the EdU+BrdU− monocytes (EdU monocytes henceforth) would have arisen prior to fasting, as this was when EdU was available before BrdU was injected. Conversely, the single positive EdU−BrdU+ monocytes (BrdU monocytes henceforth) would have arisen later, only after BrdU became available. Thus, we hypothesized that EdU monocytes were likely “old” (arisen pre-fast) whereas BrdU monocytes were “young” (arisen during fast) monocytes.
Figure 4: The monocyte surge is dominated by aged Ly-6Chi monocytes.
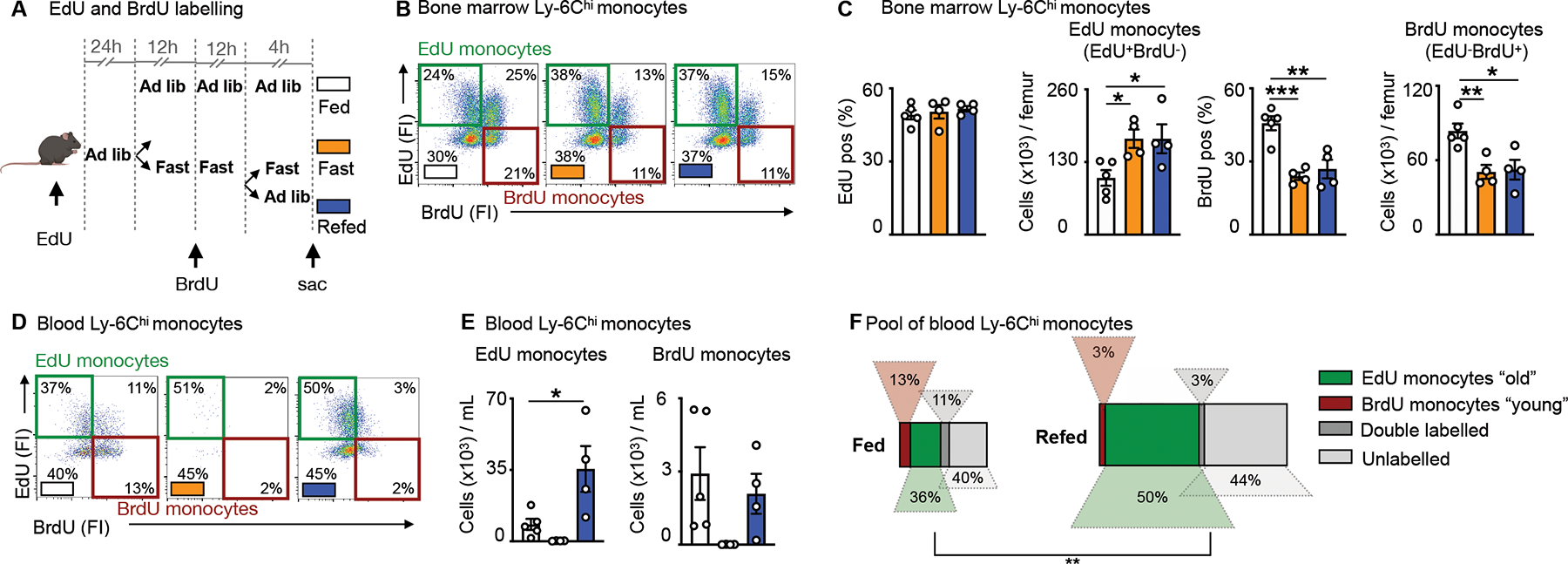
A. EdU and BrdU were sequentially injected for labelling of Ly-6Chi monocytes under feeding ad libitum, fasting and fasting+re-feeding conditions. EdU was injected 24h prior to fasting. BrdU was injected half-way through the fast. After a 24h fast, fasted mice were divided into groups submitted to a further 4h fast or 4h of re-feeding.
B. Representative flow cytometry plots for Ly-6Chi monocytes in the bone marrow under conditions of feeding ad libitum, fasting and fasting+re-feeding. Ly-6Chi monocytes are defined as CD45+ CD11b+ LIN1− Ly-6G− CX3CR1+ Ly-6Chi. “EdU monocytes” are Ly-6Chi monocytes with EdU-only label (labelled prior to fasting). “BrdU monocytes” are Ly-6Chi monocytes with BrdU-only label (labelled under fasting). (n=4–5 per group)
C. Relative EdU positivity of Ly-6Chi monocytes and absolute EdU Ly-6Chi monocyte count under feeding ad libitum, fasting and fasting+re-feeding conditions in the bone marrow. Relative BrdU positivity of Ly-6Chi monocytes and absolute BrdU Ly-6Chi monocyte count under feeding ad libitum, fasting and fasting+re-feeding conditions in the bone marrow (n=4–5 per group). One-way ANOVA.
D. Representative flow cytometry plots for Ly-6Chi monocytes in the blood under conditions of feeding ad libitum, fasting and fasting+re-feeding. Ly-6Chi monocytes are defined as CD45+ CD11b+ LIN1− Ly-6G− CX3CR1+ Ly-6Chi. “EdU monocytes” are Ly-6Chi monocytes with EdU-only label (labelled prior to fasting). “BrdU monocytes” are Ly-6Chi monocytes with BrdU-only label (labelled under fasting). (n=4–5 per group)
E. Absolute count of EdU monocytes and BrdU monocytes in the blood under feeding, fasting or fasting+re-feeding conditions (n=4–5 per group). One-way ANOVA.
F. Relative quantification of Ly-6Chi monocyte subpopulations, based on EdU and BrdU label in the blood after feeding or fasting+re-feeding conditions (n=4–5 per group). The surface of the rectangle resembles the size of the whole pool of Ly-6Chi monocytes in the blood. EdU monocytes can be considered chronologically older (“old”) than BrdU monocytes (“young”) (n=4–5 per group). Chi-square test. Data presented as mean ±SEM, *p < 0.05, **p<0.01, ***p < 0.001. Please also see Figure S3.
With this in mind, we made several observations in the bone marrow: First, EdU was similarly incorporated across the three groups (fed, fasted, fasted+re-fed) in HSPC, indicating no major effects on progenitor proliferation, and thus no major effects on upstream hematopoiesis. BrdU incorporation was slightly reduced in certain HSPCs under fasting and fasting+re-feeding, in-line with our prior finding of fasting suppressing hematopoiesis (Fig. S3A, B and S2D). Second, while we found a similar rate of relative EdU incorporation in monocytes in the bone marrow, we found numerically more of these cells overall in the fasted and re-fed groups, compared to the ad libitum-fed mice (Fig. 4C). Third, relative BrdU incorporation was lower in monocytes under fasting and fasting+re-feeding conditions, in line with previous findings, resulting in fewer BrdU monocytes in the bone marrow of fasted and re-fed mice compared to fed ad libitum mice (Fig. 4C). These bone marrow data suggested the persistence of chronological “old” EdU monocytes in mice subjected to fasting along with reduced generation of chronological “new” BrdU monocytes in those groups. In the blood, we likewise detected 4 monocyte populations (Fig. 4D). Enumerating single EdU and single BrdU populations in the fed, fasted, and fasted+re-fed groups, we found that the surge during re-feeding was dominated by EdU, rather than BrdU monocytes (Fig. 4E), thus indicating that the surge was dominated by old returners rather than newly-produced cells. Altogether, these data provide evidence that the enlarged monocyte pool shortly after re-feeding consisted predominantly of older monocytes that had been sequestered in the bone marrow during the fast (Fig. 4F).
Fasting and re-feeding generates a chronologically older and transcriptionally distinct monocyte population
EdU and BrdU labelling revealed that the Ly-6Chi monocyte surge into the bloodstream occurring 4h after re-feeding largely consisted of cells produced prior to fasting. We sought to investigate if this pool was qualitatively different from ad libitum-fed conditions. We started by performing bulk RNAseq of sorted blood Ly-6Chi monocytes retrieved after fasting+re-feeding and feeding ad libitum (Fig. 5A and S4A). As expected, RNA expression differences overall were small, but 5 genes stood out as significantly and robustly differentially expressed after p-value adjustment on genes pre-filtered by absolute logFC > 1 (Fig. 5B): Ly-6i, a member of the Ly-6 superfamily, expressed on immature monocytes26 and Tppp3 were decreased under fasting+re-feeding conditions, whereas Lars2, Plppr3 and Chi3l3 (also known as Ym1), the latter of which is a known myeloid cell product and modulator27,28 were increased.
Figure 5: Fasting and re-feeding generates a chronologically older and transcriptionally distinct monocyte population.
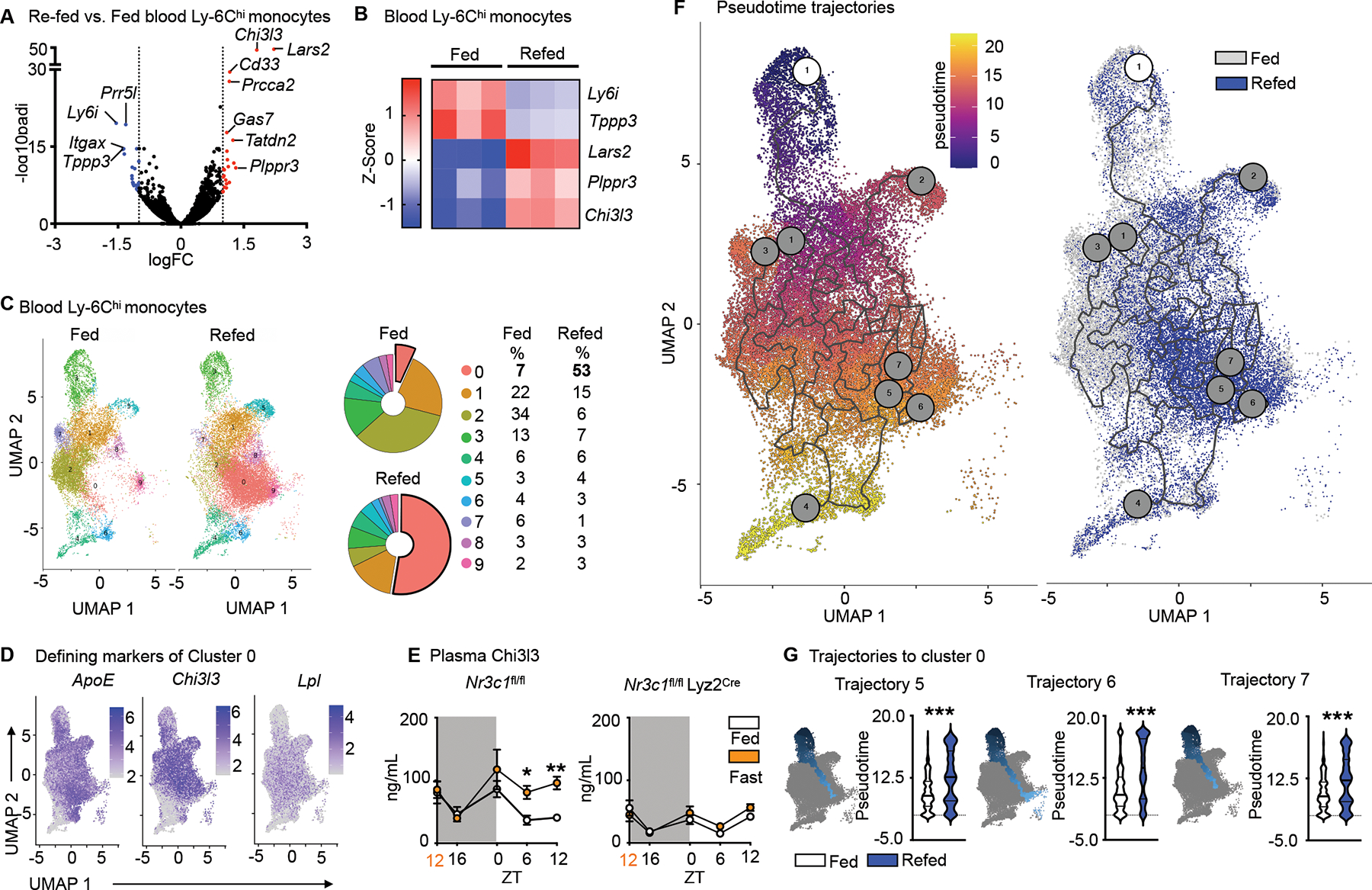
A. Volcano plot indicating differentially regulated genes (FC>1.0, FDR<0.05, p<0.05) of sorted blood Ly-6Chi monocytes after feeding ad libitum or fasting for 24h followed by 4h re-feeding. (n=3 samples per group, 4 mice pooled per sample).
B. Heatmap of differentially expressed genes after conservative hierarchical clustering of sorted blood Ly-6Chi monocytes after feeding ad libitum or fasting+re-feeding. (n=3 samples per group, 4 mice pooled per sample)
C. UMAP representations, colored by clusters computed by FindClusters function of Seurat and relative representation of the clusters of sorted blood Ly-6Chi monocytes under fed and fasting+re-fed conditions. (n=1 sample per group, 5 mice pooled per sample)
D. Expression of Top 3 defining marker genes of cluster 0 visualized by feature plots.
E. Plasma Chi3l3 in Nr3c1fl/fl and Nr3c1fl/fl Lyz2Cre under feeding ad libitum or fasting conditions. Orange zeitgeber (ZT) indicates start of the fast (n=4–12, timepoints are independent experiments). Two-way ANOVA.
F. Pseudotime computed with Monocle 3. The white circled 1 indicates the root cell from where pseudotime analaysis begins. Gray circles correspond to the different outcomes in the trajectory. UMAP representation overlaying conditions of feeding ad libitum and fasting+re-feeding.
G. Pseudotime of trajectories 5, 6 and 7 projecting into cluster 0 in mice under feeding ad libitum and fasting+re-feeding. Kolmogorov-Smirnov test. Data presented as mean ±SEM, *p < 0.05, **p<0.01, ***p < 0.001. ZT: zeitgeber. Please also see Figure S4.
For more in-depth analysis, we performed single-cell RNA sequencing on sorted blood Ly-6Chi monocytes. We identified nine clusters, among which cluster 0, defined by ApoE, Chi3l3 and Lpl was considerably larger in the re-fed group (Fig. 5C and D). Because bulk RNAseq also revealed increased Chi3l3 expression (Fig. 5A and B), we measured Chi3l3 in plasma, detecting higher protein concentration during fasting (Fig. 5E). As Nr3c1fl/fl Lyz2Cre mice did not augment Chi3l3 during fasting (Fig. 5E), we concluded that Chi3l3 secretion under these conditions depended on glucocorticoid control of myeloid cells.
To acquire a more nuanced understanding of the temporal relationships between the monocyte clusters, we computed a trajectory graph from the UMAP dimensional space and ordered the cells in pseudotime. According to the principal graph, clusters 3 and 4 were the most viable trajectory graph starting points. Cluster 3 was chosen as the more likely starting point because cluster 4 was enriched for Ly-6C intermediate marker genes (Cd74, H2-Aa, H2-Eb1 and H2-Ab1); Ly-6Chi monocytes lose Ly-6C as they age and can convert to Ly-6C intermediate monocytes29 (Fig. 5F). Out of eight trajectories, three projected through or terminated at cluster 0, with trajectories falling later in pseudotime under fasting+re-feeding conditions (Fig. 5G and S5B). Pseudotime analysis, therefore, supported our hypothesis that under fasting and subsequent re-feeding, older monocytes are released into the blood stream.
Fasting and re-feeding alters host response to infection
Having demonstrated that Ly-6Chi monocytes released from the bone marrow upon re-feeding are chronologically older and transcriptionally different, as well as larger in numbers, we wondered if the older pool would dominate a site of acute inflammation. To keep observations chronologically in line with our previous findings with the EdU and BrdU double pulse-chase method, we tracked monocytes of fed and fasted mice in response to LPS administered upon re-feeding (Fig. 6A). After excluding Ly-6Chi monocytes in the vasculature by intravenous CD45 labelling (Fig. 6B), we found the expected four populations in the lung parenchyma, including “old” EdU positive and “young” BrdU positive monocytes. Mice that had been fasted and re-fed along with LPS administration, had a more pronounced monocyte recruitment to the lung, involving enhanced TNFα, and a larger contribution of “old” EdU monocytes compared to mice that were fed ad libitum (Fig. 6C).
Figure 6. Fasting and re-feeding alters host response to infection.
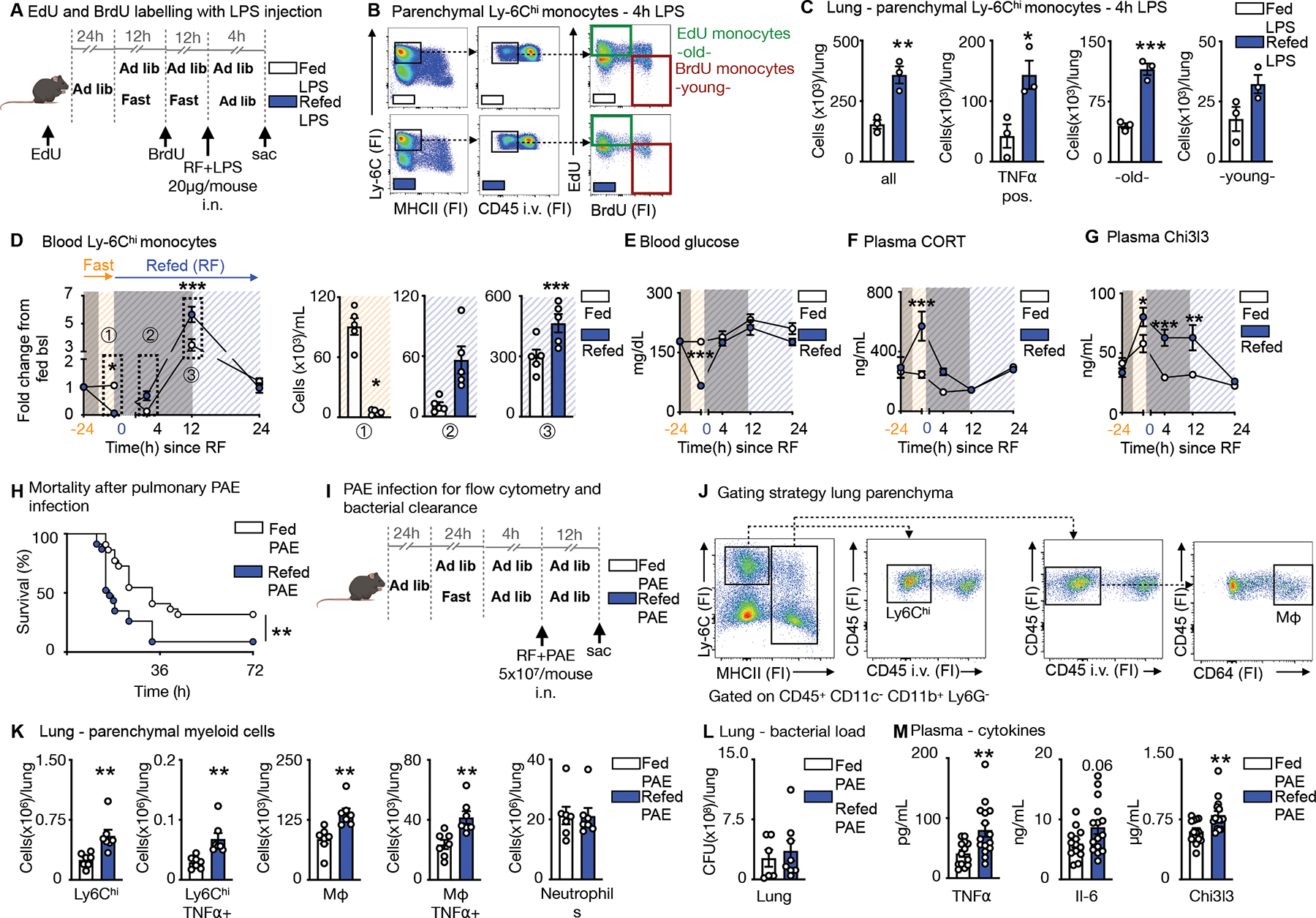
A. EdU and BrdU were sequentially injected for labelling of Ly-6Chi monocytes in mice under feeding ad libitum and fasting+re-feeding conditions. EdU was injected 24h prior to fasting. BrdU was injected half-way through the fast. After a 24h fast, fasted mice were refed for 4h. LPS (20μg/mouse) was administered intranasally at the timepoint of re-feeding and mice sacrificed 4h later.
B. Representative flow cytometry plots for double labelling of Ly-6Chi monocytes in the lung parenchyma. Intravascular Ly-6Chi monocytes were excluded by intravenous CD45 staining prior to sacrifice. Ly-6Chi monocytes are defined as CD45+ CD11b+ LIN1− Ly-6G− CX3CR1+ MHCII− Ly-6Chi. EdU monocytes are Ly-6Chi monocytes with EdU-only label (labelled prior to fasting, old). BrdU monocytes are Ly-6Chi monocytes with BrdU-only label (labelled under fasting, young). (n=3 per group)
C. Absolute count of Ly-6Chi monocytes and TNFα-positive Ly-6Chi monocytes in the parenchyma after intranasal LPS challenge in mice undergoing feeding ad libitum or fasting+re-feeding. Absolute counts of “old” and “young” Ly-6Chi monocytes in the parenychma. “Old” are Ly-6Chi monocytes with EdU-only label. “Young” are Ly-6Chi monocytes with BrdU-only label (n=3 per group). Unpaired t test.
D. Blood Ly-6Chi monocytes under feeding or fasting+re-feeding conditions followed for 24h after re-feeding normalized to group’s baseline before fasting. Orange −24h indicates baseline before start of a 24h fast. Blue 0 indicates start of re-feeding. Bar graphs represent absolute Ly-6Chi monocytes counts of indicated timepoints (n=4–5 per group, timepoints are independent experiments). Two-way ANOVA.
E. Blood glucose under feeding or fasting+re-feeding conditions followed for 24h after re-feeding. Orange −24h indicates baseline before start of a 24h fast. Blue 0 indicates start of re-feeding (n=4–5 per group, timepoints are independent experiments). Two-way ANOVA.
F. Plasma corticosterone (CORT) under under feeding or fasting+re-feeding conditions followed for 24h after re-feeding. Orange −24h indicates baseline before start of a 24h fast. Blue 0 indicates start of re-feeding (n=4–5 per group, timepoints are independent experiments). Two-way ANOVA.
G. Plasma Chi3l3 concentration under feeding or fasting+re-feeding conditions followed for 24h after re-feeding. Orange −24h indicates baseline before start of a 24h fast. Blue 0 indicates start of re-feeding (n=4–5 per group, timepoints are independent experiments). Two-way ANOVA.
H. Mice were infected intranasally with Pseudomonas aeruginosa (PAE) at a dose of 5×107/mouse. Refed mice were infected after a 24h fast, followed by a 4h re-feeding period. Control mice were fed ad libitum and both groups followed for survival (n= 22–23 per group, two independent experiments combined). Log-rank (Mantel-Cox) test.
I. Fed ad libitum mice and mice after 24h fast followed by 4h of re-feeding were intranasally infected with PAE at a dose of 5×107/mouse and sacrificed 12h after.
J. To distinguish between circulating and parenchymal leukocytes, CD45 was injected i.v. before sacrifice. Representative gating strategy for flow cytometery on parenchymal Ly-6Chi monocytes defined as CD45+CD11c−CD11b+Ly6G−Ly-6Chi MHCII− and macrophages (Mφ) defined as CD45+CD11c−CD11b+Ly6G−MHCII+CD64+ in the lung. (n=7 per group)
K. Parenchymal myeloid cell content in the lung after infection with PAE as described in panel I (n=7 per group). Unpaired t test.
L. Bacterial load after infection as described in panel I from homogenized lungs comparing fed and fasted+re-fed mice (n=6–8 per group). Unpaired t test.
M. Plasma concentration of cytokines after infection as described in panel I (n=13–15 per group, two experiments combined). Unpaired t test. Data presented as mean ±SEM, *p < 0.05, **p<0.01, ***p < 0.001. FI: Fluorescence intensity, RF: Re-feeding, PAE: Pseudomonas aeruginosa, Mφ: Macrophages. Please also see Figure S5 and S6.
Because a 4h re-feeding window gave mice enough time to normalize blood glucose and corticosterone concentrations (Fig. 3D, E), we next asked how long fasting+re-feeding monocytosis would last. We noticed that the post re-feeding surge persisted for at least 12h before the system normalized itself by 24h (Fig. 6D). This return to monocytic homeostasis, at least numerically, lagged behind normalization of blood glucose concentration (Fig. 6E) and plasma corticosterone (Fig. 6F) but correlated well with Chi3l3 plasma concentrations (Fig. 6G), consistent with the idea that Chi3I3 is a fasting-induced myeloid product. We therefore surmised that, even though repetitive fasting may have long-term consequences, acute effects on monocyte function within a day of fasting and re-feeding are also potentially meaningful. Pathway analysis from bulkRNA seq revealed decreased activity of class-II antigen presentation, IFNβ, and innate immune signatures in monocytes of fasted+re-fed mice, suggesting that fasting+re-feeding may affect Ly-6Chi monocytes ‘potential to respond to external challenges (Fig. S5A, B and S6).
Gram-negative bacteria, such as Pseudomonas aeruginosa (PAE) are a major cause of nosocomial pneumonia30. We infected mice intranasally with PAE after a 24h fast followed by re-feeding for 4h and tracked survival. Mice that underwent fasting+re-feeding died earlier and in larger numbers compared to ad libitum-fed mice (Fig. 6H). To correlate higher mortality with monocyte recruitment into the lung, we assessed myeloid cell content in the parenchyma 12h after infection (Fig. 6I, J). Fasting+re-feeding prior to infection with PAE led to higher content of Ly-6Chi monocytes, macrophages, as well as enhanced TNFα positivity, with no difference in neutrophil numbers (Fig. 6K). Although we did not detect a difference in bacterial load in the lung (Fig. 6L) or bronchoalveolar lavage (Fig. S5C), we did observe more inflammation in the plasma, as assessed by higher circulating cytokine concentrations (Fig. 6M). The monocytes were equally phagocytic (Fig. S5D, E). These data suggest that the ensuing monocytic disequilibrium that occurs during prolonged fasting followed by re-feeding increases inflammation, altering host response, leading to higher mortality compared to ad libitum feeding.
Discussion
A key theme throughout this study is, arguably, the body’s ability to limit energy expenditure during nutrient scarcity. Among leukocytes, monocytes (and neutrophils) are energetically costly if only because of their massive daily production in the bone marrow. Unlike B and T cells, these circulating myeloid cells have a short half-life and thus rely on myelopoiesis for their replenishment. While the benefits of this phenomenon may be the availability of freshly-made innate immune responders with finely-tuned molecular machinery to meet the challenges of infection and injury, the process does seem to rely, this study suggests, on an ad libitum supply of dietary energy. In the absence of such supply, the body slows down metabolic expenditure31–33, limiting production and preserving—and thus extending—the lifespan of these already-made, short-lived cells.
The bone marrow as a hibernating tissue of choice for leukocytes has been explored before. Several recent studies have documented migration of leukocytes back to the marrow in response to environmental stimuli13,14,17, although the consequence of such migration is poorly understood. As the birthplace of both red and white blood cells, and the cradle of hematopoietic stem cells, the bone marrow is, almost by definition, a privileged tissue tasked with ensuring that the blood pool remains refreshed throughout life. The reverse migration of monocytes during fasting described here raises a series of additional questions. First, it is not yet clear how monocytes survive in the bone marrow during fasting. Perhaps the marrow produces its own energy to provide and nourish its resident cells during nutrient scarcity34. Or it simply shields sequestered monocytes from the harsh extra-medullary environments that they would otherwise encounter in the circulation. Second, it remains to be seen whether a mechanistic link exists between re-mobilization of circulating monocytes and the reduction of hematopoiesis during fasting. Are re-mobilized monocytes calibrating hematopoiesis, as aged neutrophils do35, or are these phenomena independent? Third, the mechanism orchestrating the return of monocytes to the circulation remains unclear and will need to be further studied. While it is likely that return of monocytes to the circulation coincides with lower CXCR4, it remains to be determined whether this is a passive consequence of less corticosterone or whether other, more active mechanisms contribute.
The role of the central nervous system in orchestrating large-scale leukocyte shifts documented here and elsewhere16,17,36 underscores the importance of focusing on brain-body communication in physiology and disease. While previous work indicated that hepatocyte-derived AMPK is responsible for shutting down monocyte mobilization15, our study establishes the brain as the most upstream and consequential contributor. The series of signaling events that originate in the PVH and terminate at corticosterone release are known37,38, but in the context of this study offer a perspective that merges fasting with the perception of hunger, stress and complex changes in the leukocyte distribution map. Heightened corticosterone itself, whether induced by extrinsic stressors or intrinsically by fasting, offers a potential explanation for altered host response via immunosuppressive mechanisms, but is quickly normalized upon the removal of the stressor. Indeed, it may be monocytes staying “hangry” even after stress resolution—that are key to the leukocytic reorganization and aggravated response in the periphery. Further inquiry will need to look deeper and more upstream, using synaptic retrograde tracing techniques, to better understand the links between metabolic sensing, the neurobiology of stress, and efficiency of energetically-costly hematopoiesis. Further work is also needed to better understand the implications of this work to human physiology. The concentration of plasma cortisol is expected to rise only after severe fasting in humans39. Thus, our data may be more relevant to conditions of more severe food scarcity or eating disorders where cortisol concentration is known to rise40.
One of the interpretations of our data based on dual pulse-chase tracking and pseudotime analyses of sequenced cells is that hibernating monocytes returning back to the blood upon re-feeding are chronologically older and transcriptionally distinct. To what extent these transcriptional differences offer a glimpse of biological aging, as opposed to chronological aging, which is simply marked by the passage of time, is unclear, although we do note that the transcriptional differences are rather small. Nevertheless, the correlation between the preponderance of older monocytes, heightened inflammation, and altered functionality in response to infection, does suggest a disequilibrium, with an altered response of the monocyte population, perhaps evoking a few key features of biological aging. Regardless of how these cells are defined, there is a potential benefit to reducing the rate of hematopoiesis, whether by prolonged fasting41, exercising42, better sleep hygiene16,43,44 or better diet45. Recent data suggest that accelerating the rate of leukocyte production precipitates clonal hematopoiesis, thus reducing hematopoietic diversity and conferring heightened risk of cardiovascular disease43,44. It follows that measures aimed at reducing hematopoiesis may provide long-term benefit by virtue of preserving a diverse, non-clonal hematopoietic pool. Future studies will need to investigate these relationships in more mechanistic depth.
In aggregate, our study provides a sequence of events occurring during fasting and re-feeding that link the HPA-axis with monocytes and bone marrow. Although fasting can protect against disease in numerous instances, our work argues there are limits, or at least a cost, to habits involving prolonged fasting and re-feeding. It is well documented that during starvation various bodily systems shut down or greatly diminish in a predictable sequence46, likely reflecting a hierarchy related to the survival imperative. In this context, continuous replenishment of circulating monocytes through myelopoiesis may be a type of luxury of plenty that is sacrificed relatively early even at a physiological cost.
Limitations of Study
Our study has limitations for translation into human physiology regarding the length of the fast and the related stress response. A 24h fast in mice is different in humans, which makes our findings potentially more translatable to situations of severe food scarcity or eating disorders. We do not investigate mechanistically how monocytes are released back into circulation upon re-feeding but hypothesize that it is the result of reestablished normalized plasma corticosterone concentration. Fasting+re-feeding led to detrimental outcomes in mice infected with Pseudomonas aeruginosa, which was chosen as it is a common strain for nosocomial pneumonia. We cannot exclude that other bacterial strains would have caused a different outcome.
STAR methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Filip K. Swirski (filip.swirski@mssm.edu).
Materials availability
This study did not generate unique reagents.
Data and code availability
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Brilliant Violet 711™ anti-mouse CD45 clone 30-F11 | BioLegend | Cat# 103147 |
| PE anti-mouse CD45.1 clone A20 | BioLegend | Cat# 110707 |
| PE/Cyanine7 anti-mouse CD45.2 clone 104 | BioLegend | Cat# 109829 |
| PE anti-mouse CD3 clone 17A2 | BioLegend | Cat# 100206 |
| PE anti-mouse CD90.2 clone 3O-H12 | BioLegend | Cat# 105308 |
| PE anti-mouse CD19 clone 6D5 | BioLegend | Cat# 115508 |
| PE anti-mouse B220 clone RA3-6B2 | BD Biosciences | Cat# 553089 |
| PE anti-mouse NK-1.1 clone PK136 | BioLegend | Cat# 108708 |
| APC anti-mouse Ly-6G clone 1A8 | BioLegend | Cat# 127614 |
| FITC anti-mouse Ly-6C clone HK1.4 | BioLegend | Cat# 128006 |
| Alexa Fluor® 700 anti-mouse I-A/I-E clone M5/114.15.2 | BioLegend | Cat# 107602 |
| APC/Fire™ 810 anti-mouse F4/80 clone BM8 | BioLegend | Cat# 123166 |
| APC/Cyanine7 anti-mouse/human CD11b clone M1/70 | BioLegend | Cat# 101226 |
| Brilliant Violet 605™ anti-mouse CD115 clone AFS98 | BioLegend | Cat# 135517 |
| PE anti-mouse TER-119 clone TER-119 | BioLegend | Cat# 116208 |
| FITC anti-mouse CD34 clone RAM34 | ThermoFisher | Cat# 11-0341-85 |
| APC anti-mouse CD11c clone N418 | BioLegend | Cat# 117310 |
| PE anti-mouse CD127 clone SB/199 | BioLegend | Cat# 121112 |
| PerCP/Cyanine5.5 anti-mouse CD16/32 clone 93 | BioLegend | Cat# 101324 |
| PerCP/Cyanine5.5 anti-mouse CD150 | BioLegend | Cat# 115922 |
| APC anti-mouse CD135 clone A2F10 | BioLegend | Cat# 135310 |
| Alexa Fluor® 700 anti-mouse CD48 clone HM48-1 | BioLegend | Cat# 103426 |
| PE/Cyanine7 anti-mouse CD117 (c-Kit) clone 2B8 | BioLegend | Cat# 105814 |
| APC/Cyanine7 anti-mouse Ly-6A/E (Sca-1) clone D7 | BioLegend | Cat# 108126 |
| APC anti-mouse CD8a clone 53-6.7 | BD Biosciences | Cat# 553035 |
| Pacific Blue™ anti-mouse CD4 clone GK1.5 | BioLegend | Cat# 100428 |
| PE anti-mouse Siglec-F clone E50-2440 | BD Biosciences | Cat# 562068 |
| PE anti-mouse CXCR4 clone 2B11 | ThermoFisher | Cat# 12-9991-82 |
| PE/Cyanine7 anti-mouse CX3CR1 clone SA011F11 | BioLegend | Cat# 149016 |
| PE/Cyanine7 anti-mouse TNF-α clone MP6-XT22 | BioLegend | Cat# 506324 |
| PE/Dazzle™ 594 anti-mouse CD64 clone X54-5/7.1 | BioLegend | Cat# 139320 |
| AF647™ anti-BrdU clone MoBU-1 | ThermoFisher | Cat# B35133 |
| Brilliant Violet 650™ anti-mouse CD192 (CCR2) | BioLegend | Cat# 150613 |
| Bacterial and virus strains | ||
| Pseudomonas aeruginosa, Strain Boston 41501 | ATCC | ATCC# 27853 |
| Chemicals, peptides, and recombinant proteins | ||
| Corticosterone: HBC complex | Sigma-Aldrich | Cat# C174 |
| pHrodo™ Red Phagocytosis Particle Labeling Kit for Flow Cytometry | Thermo Fisher | Cat# A10026 |
| pHrodo™ Red E. coli BioParticles™ Conjugate for Phagocytosis | Thermo Fisher | Cat# P35361 |
| Critical commercial assays | ||
| RNeasy Mini Kit | Qiagen | Cat# 74104 |
| High-Capacity cDNA Reverse Transcription Kit | Thermo Fisher | Cat# 4368814 |
| Corticosterone ELISA kit | Abcam | Cat# ab108821 |
| Mouse YM1/Chitinase 3-like 3 DuoSet ELISA | R&D Systems | Cat# DY2446 |
| Mouse IL-6 Quantikine ELISA Kit | R&D Systems | Cat# M6000B |
| Mouse TNF-alpha Quantikine ELISA Kit | R&D Systems | Cat# MTA00B |
| Free Fatty Acid Quantitation Kit | Sigma-Aldrich | Cat# MAK044 |
| Click-iT™ Plus EdU Pacific Blue™ Flow Cytometry Assay Kit | Thermo Fisher | Cat# C10636 |
| Deposited data | ||
| Single cell RNA sequencing data | Gene Expression Omnibus | |
| Bulk RNA sequencing data | Gene Expression Omnibus | |
| Experimental models: Organisms/strains | ||
| C57BL/6J | The Jackson Laboratory | Strain 000664 |
| C57BL/6-Tg(UBC-GFP)30Scha/J | The Jackson Laboratory | Strain 004353 |
| B6.129P2-Lyz2tm1(cre)Ifo/J | The Jackson Laboratory | Strain 004781 |
| B6.129P2-Cxcr4tm2Yzo/J | The Jackson Laboratory | Strain 008767 |
| B6.Cg-Nr3c1tm1.1Jda/J | The Jackson Laboratory | Strain 021021 |
| STOCK Prkaa1tm1.1Sjm/J | The Jackson Laboratory | Strain 014141 |
| B6.129S4(Cg)-Arntltm1Weit/J | The Jackson Laboratory | Strain 007668 |
| STOCK Slc2a1tm1.1Stma/AbelJ | The Jackson Laboratory | Strain 031871 |
| B6.FVB(129X1)-Tg(Sim1Cre)1Lowl/J | The Jackson Laboratory | Strain 006451 |
| B6.129S4(Cg)-Crhtm2.1 Maj/J | The Jackson Laboratory | Strain 030110 |
| B6.129S4-Ccr2tm1Ifc/J | The Jackson Laboratory | Strain 004999 |
| C.129S7(B6)-Itgb2tm2Bay/AbhmJ | The Jackson Laboratory | Strain 027003 |
| B6.129P2(Cg)-Cx3cr1tm1Litt/J | The Jackson Laboratory | Strain 005582 |
| B6.129S7-Rag1tm1Mom/J | The Jackson Laboratory | Strain 002216 |
| BALB/cJ | The Jackson Laboratory | Strain 000651 |
| Oligonucleotides | ||
| Mouse GAPD Mm99999915_g1 | Thermo Fisher | Cat# 4331182 |
| Mouse ACTB Mm00607939_s1 | Thermo Fisher | Cat# 4331182 |
| Mouse Clock Mm00455950_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Arntl Mm00500226_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Nr1d2 Mm01310356_g1 | Thermo Fisher | Cat# 4331182 |
| Mouse Per1 Mm00501813_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Cry1 Mm00514392_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Selplg Mm01204601_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Cd44 Mm01277161_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Itgal Mm00801807_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Itgam Mm00434455_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Itgb1 Mm01253230_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Itgb2 Mm00434513_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Itga4 Mm01277951_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Itga5 Mm00439797_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Itga6 Mm00434375_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Ccr1 Mm00438260_s1 | Thermo Fisher | Cat# 4331182 |
| Mouse Cxcr3 Mm9999054_s1 | Thermo Fisher | Cat# 4331182 |
| Mouse Cxcr4 Mm01996749_s1 | Thermo Fisher | Cat# 4331182 |
| Mouse Cx3cr1 Mm02620111_s1 | Thermo Fisher | Cat# 4331182 |
| Mouse Ilb1 Mm00434228_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Tnf Mm00443258_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Ccr2 Mm99999051_gH | Thermo Fisher | Cat# 4331182 |
| Software and algorithms | ||
| FlowJo v10 | FlowJo | https://www.flowjo.com/ |
| GraphPad Prism v9 | GraphPad Software | https://www.graphpad.com/ |
| BioRender | BioRender | https://biorender.com |
| R Differential gene expression analysis: | ||
| R version 4.2.1, Platform: aarch64-apple-darwin20 (64-bit); OS: macOS Monterey 12.6.1 | aarch64-apple-darwin20 (64-bit) | N/A |
| scales_1.2.1 | CRAN/BiocManager | N/A |
| clusterProfiler_4.6.0 | CRAN/BiocManager | N/A |
| tidyr_1.2.1 | CRAN/BiocManager | N/A |
| dplyr_1.0.10 | CRAN/BiocManager | N/A |
| ggplot2_3.3.6 | CRAN/BiocManager | N/A |
| GenomicRanges_1.50.0 | CRAN/BiocManager | N/A |
| MatrixGenerics_1.10.0 | CRAN/BiocManager | N/A |
| ggthemes_4.2.4 | CRAN/BiocManager | N/A |
| org.Hs.eg.db_3.16.0 | CRAN/BiocManager | N/A |
| immunarch_0.8.0 | CRAN/BiocManager | N/A |
| EnhancedVolcano_1.16.0 | CRAN/BiocManager | N/A |
| genefilter_1.80.0 | CRAN/BiocManager | N/A |
| GenomeInfoDb_1.34.0 | CRAN/BiocManager | N/A |
| matrixStats_0.62.0 | CRAN/BiocManager | N/A |
| msigdbr_7.5.1 | CRAN/BiocManager | N/A |
| org.Mm.eg.db_3.16.0 | CRAN/BiocManager | N/A |
| patchwork_1.1.2 | CRAN/BiocManager | N/A |
| ggrepel_0.9.1 | CRAN/BiocManager | N/A |
| DESeq2_1.38.0 | CRAN/BiocManager | N/A |
| IRanges_2.32.0 | CRAN/BiocManager | N/A |
| glmpca_0.2.0 | CRAN/BiocManager | N/A |
| pathview_1.38.0 | CRAN/BiocManager | N/A |
| AnnotationDbi_1.60.0 | CRAN/BiocManager | N/A |
| data.table_1.14.4 | CRAN/BiocManager | N/A |
| glmGamPoi_1.10.0 | CRAN/BiocManager | N/A |
| SummarizedExperiment_1.28.0 | CRAN/BiocManager | N/A |
| S4Vectors_0.36.0 | CRAN/BiocManager | N/A |
| RColorBrewer_1.1-3 | CRAN/BiocManager | N/A |
| enrichplot_1.18.0 | CRAN/BiocManager | N/A |
| gridExtra_2.3 | CRAN/BiocManager | N/A |
| dtplyr_1.2.2 | CRAN/BiocManager | N/A |
| ggbeeswarm_0.6.0 | CRAN/BiocManager | N/A |
| Biobase_2.58.0 | CRAN/BiocManager | N/A |
| BiocGenerics_0.44.0 | CRAN/BiocManager | N/A |
| pheatmap_1.0.12 | CRAN/BiocManager | N/A |
| R scRNA analysis: | ||
| monocle3_1.2.9 | GitHub | N/A |
| IRanges_2.32.0 | CRAN/BiocManager | N/A |
| BiocGenerics_0.44.0 | CRAN/BiocManager | N/A |
| SeuratObject_4.1.2 | GitHub | N/A |
| SingleCellExperiment_1.20.0 | CRAN/BiocManager | N/A |
| S4Vectors_0.36.0 | CRAN/BiocManager | N/A |
| SeuratDisk_0.0.0.9020 | GitHub | N/A |
| Seurat_4.2.0 | GitHub | N/A |
| SummarizedExperiment_1.28.0 | CRAN/BiocManager | N/A |
| MatrixGenerics_1.10.0 | GitHub | N/A |
| SeuratWrappers_0.3.1 | GitHub | N/A |
| magrittr_2.0.3 | CRAN/BiocManager | N/A |
| GenomicRanges_1.50.0 | CRAN/BiocManager | N/A |
| matrixStats_0.62.0 | CRAN/BiocManager | N/A |
| SeuratData_0.2.2 | GitHub | N/A |
| patchwork_1.1.2 | CRAN/BiocManager | N/A |
| GenomeInfoDb_1.34.0 | CRAN/BiocManager | N/A |
| Biobase_2.58.0 | CRAN/BiocManager | N/A |
| sp_1.5-0 | CRAN/BiocManager | N/A |
| ggplot2_3.3.6 | CRAN/BiocManager | N/A |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6J, C57BL/6-Tg(UBC-GFP)30Scha/J (GFP-UBI), B6.129P2-Lyz2tm1(cre)Ifo/J (Lyz2Cre), B6.129P2-Cxcr4tm2Yzo/J (Cxcr4fl/fl), B6.Cg-Nr3c1tm1.1Jda/J (Nr3c1fl/fl), STOCK Prkaa1tm1.1Sjm/J (Prkaa1fl/fl), B6.129S4(Cg)-Arntltm1Weit/J (Arntlfl/fl), STOCK Slc2a1tm1.1Stma/AbelJ (Slc2a1fl/fl), B6.FVB(129X1)-Tg(Sim1Cre)1Lowl/J (Sim1Cre), B6.129S4(Cg)-Crhtm2.1Maj/J (Crhfl/fl), B6.129S4-Ccr2tm1Ifc/J (Ccr2−/−), C.129S7(B6)-Itgb2tm2Bay/AbhmJ (Cd18−/−), B6.129P2(Cg)-Cx3cr1tm1Litt/J (Cx3cr1ΔGFP/GFP), B6.129S7-Rag1tm1Mom/J (Rag1−/−), and BALB/cJ were purchased from The Jackson Laboratory. Genotyping for each strain was performed as recommended on The Jackson Laboratory website. Relevant mice were crossed to generate Cxcr4fl/fl Lyz2Cre, Nr3c1fl/fl Lyz2Cre, Crhfl/fl Sim1Cre, Prkaa1fl/flLyz2Cre, Arntlfl/flLyz2Cre, Slc2a1fl/flLyz2Cre and respective litter mate controls. All experiments were performed on male and female 7 week- to 1 year-old mice with age- and sex-matched controls. All mice were housed on a 12h/12h light/dark cycle at 22°C with unrestricted access to food and water. Where appropriate, mice were randomly assigned to groups and experiments were performed blinded. All protocols were approved by the Animal Review Committee at the Massachusetts General Hospital (protocols 2011N000035 and 2015N000044) and/or the Animal Review Committee at the Mount Sinai Hospital (PROTO202000262 and PROTO202200000111) and complied with all relevant ethical regulations.
Bacteria
The ATCC 27853 strain Pseudomonas aeruginosa was grown overnight on tryptic soy agar plates at 37°C. Single colonies were picked and grown overnight in tryptic soy broth at 37°C with constant shaking.
METHOD DETAILS
Feeding and Fasting
Mice were single- or group-housed on a 12:12-h light-dark cycle at 22°C with free access to chow diet and water. Under fasting conditions, mice were fasted for periods up to 28 hours starting at ZT12 with unlimited access to water. Fed controls had food available ad libitum.
Re-feeding
After a 24h fast, chow diet was put back into the cage, mice were allowed to eat ad libitum.
Platelet depletion
36h prior to fasting, mice were i.p. injected with anti-GP1BA/ CD42b antibodies or isotope control to deplete platelets.
Cell sorting
Blood, spleen or/and bone marrow cell suspensions were stained to identify the indicated cell populations and cells were sorted on a FACS Aria II cell sorter (BD Biosciences) directly into either RLT buffer for subsequent RNA isolation or collecting medium.
Adoptive transfer
An equal amount of sorted Ly-6Chi monocytes from GFP-UBI mice was injected into the retrobulbar plexus directly before initiation of fasting.
Parabiosis
The procedure, adapted from was conducted as previously described47. Briefly, after shaving the corresponding lateral aspects of a GFP-UBI and a wild type mouse, matching skin incisions were made from behind the ear to the tail of each mouse, and the subcutaneous fascia was bluntly dissected to create about 0.5 cm of free skin. The scapulas were sutured using a mono-nylon 5.0 (Ethicon, Albuquerque, NM), and the dorsal and ventral skins were approximated by continuous suture. Mice were joined for 2 weeks. After, mice in parabiosis were surgically separated by a reversal of the procedure. Percent chimerism in the blood was defined for gated Ly-6Chi monocytes as %GFP+ (%GFP+ & %GFP−)−1 in wild type mice. Mice in the fasting group were fasted directly after separation for 28 hrs and mice in the refed group refed after 24h for 4h.
Intravital microscopy
Fed mice versus fasted mice were anesthetized with 1–3% isoflurane and 2 l/min oxygen anesthesia for monocyte imaging. Mice were kept on a 37°C heating plate during the whole procedure. Fluorescent agents, FITC dextran (2 million Da) for labelling the vasculature and PE-labeled anti-CD115 antibodies to visualize Ly-6Chi monocytes (5 μl antibody stock in 50 μl PBS), were injected i.v. The femoral vein was exposed on a 37°C heated plate and the vessel lumen was imaged confocally. The entire surgical and imaging procedure was kept to a maximum of 1h. Imaging was done with an Olympus XLUMPLFLN 20X W NA:1.00 water immersion objective on an Olympus custom made confocal multi-photon microscope using 473 nm and 559 nm diode-lasers with a DM405/473/559 dichroic mirror, a SDM560 beam splitter, and BA490–540 and BA575–675 emission filters (Olympus America Inc.).
Adrenalectomy
Both adrenal glands were surgically removed via bilateral dorso-lateral subcostal access routes. Sham-operated mice served as controls. Post surgery, adrenalectomized mice received saline as drinking water. Mice were allowed to recover for >7 d before further manipulation.
Corticosterone injections
Mice were intraperitoneally injected with either vehicle (PBS) or corticosterone (5 mg/kg bodyweight, Corticosterone: HBC complex, Sigma-Aldrich, C174) under short isoflurane anaesthesia. Blood was withdrawn and analyzed 1 h after injection.
EdU and BrdU incorporation
To assess cell proliferation, 1 mg BrdU was injected intraperitoneally 2 h before euthanasia. For cell labelling assays, 1 mg BrdU and 250 μg EdU were injected intraperitoneally at indicated time points. BrdU-postive cells were stained with a BrdU flow kit (BD Biosciences) and EdU-positive cells with a Click-iT™ Plus EdU Flow Cytometry assay kit (Thermo Fisher)
pHrodo™ labeling of Pseudomonas aeruginosa particles
Pseudomonas aeruginosa particles were labeled based on the manufacturers ‘instructions of the pHrodo™ Red Phagocytosis Particle Labeling Kit for Flow Cytometry (Thermo Fisher, A10026).
LPS inoculation
Mice were anesthetized with isoflurane and intranasally infected with 20μg LPS in a volume of 25 μl of sterile PBS.
pHrodo™-labeled Pseudomonas aeruginosa particle inoculation
Mice were anesthetized with isoflurane and intranasally infected with 4×107 particles in a volume of 50 μl of sterile PBS and sacrificed 4h after.
Pseudomonas aeruginosa infection
Pseudomonas aeruginosa was subcultured in tryptic soy broth for 3 hours at 37°C before injection, washed with sterile PBS, OD600 recorded and resuspended in PBS. Injected colony forming units (CFU) were retrospectively confirmed in counts of serial dilutions on blood agar plates. Mice were anesthetized with isoflurane and intranasally infected with stated concentration in a total volume of 50 μl (2×25μl).
Bronchoalveolar lavage (BAL)
Mice were anesthetized with isoflurane. The trachea was exposed and cannulated with 20G catheter. 1 ml of PBS (4°C) was gently infused followed by aspiration. This was repeated 5 times and total volume was recovered.
Bacterial load assay
CFU were determined by plating titrated amounts of lung homogenate or BAL on blood agar plates. Briefly, lung and BAL were harvested at indicated times post-infection. Tissue homogenates were generated by pushing the tissue through a 70 mm cell strainer using the plunger of a 5 ml syringe. Titrated 10-fold dilutions of tissue homogenate and BAL were serially diluted in PBS, plated on blood agar plates, and grown overnight at 37°C.
In vitro pHrodo™ assay
100 μl whole blood was incubated with 5 μl of pHrodo™ Red E. coli BioParticles™ (Thermo Fisher Scientific, P35361) for 30 minutes at 37°C.
Cell isolation and flow cytometry
Blood was collected and lysed with RBC buffer (Biolegend) twice. After transcardiac perfusion with PBS (Thermo Fisher Scientific), solid organs were harvested, minced, and digested in a mixture of 450 U ml™1 collagenase I, 125 U ml™1 collagenase XI, 60 U ml™1 DNase, and 60 U ml™1 hyaluronidase (Sigma-Aldrich) in PBS on a shaker (800rpm) at 37°C for 20min (liver and pancreas), or 1 h (white/brown/mesenteric adipose tissue, skeletal muscle, heart and lung). Afterwards, the digested organ was flushed through a 40-μm cell strainer. Spleens were pressed through a 40-μm cell strainer and lysed with RBC. Peyer’s patches and lymph nodes were pressed through a 40-μm cell strainer. Bone marrow cells were flushed from the bone marrow cavities and brought into single-cell suspension by pipetting up and down, before lysis with RBC. Total viable cell numbers were counted using counting beads (Thermo Fisher Scientific). Small and large intestinal lamina propria was isolated as follows: after excision of the intestine, the Peyer’s patches were removed under a microscope and the gut was cut open longitudinally to wash off the lumen contents in HBSS buffer. The gut was then cut into 1–2-cm pieces and subjected to 3× dissociation in EDTA-containing buffer (7.5 mM HEPES, 2% FCS, 2 mM EDTA, 10,000 U ml−1 penicillin–streptomycin, 50 μg ml−1 gentamycin in HBSS; all Thermo Fisher Scientific) in a shaker at 37 °C for 15 min. After dissociation the epithelial layer was isolated (by filtering through a mesh) and discarded and the lamina propria digested in a mixture of 1M CaCl2 (Sigma), 60 U/ml Dnase I, 5mg/ml Liberase TM (Roche), RPMI with L-Glutamin (Corning), 10% FCS, 5 mM Sodiumpyruvat (Corning cellgro), 5 mM HEPES (1M), 1% Pen/Strep. at 37°C for 40min. Single-cell suspensions were stained in FACS buffer (0.5% BSA and 2mM EDTA in PBS) containing fluorophore-coupled antibodies at a concentration of 1:700 at 4 °C for 20 min, unless otherwise indicated. To differentiate between live and dead cells, the cell suspensions were stained with Zombie Aqua (BioLegend) at a concentration of 1:1,000 in PBS at 4 °C for 15 min or by adding propidium iodide directly before flow cytometry analyses. Cells were identified as
Ly-6Chi monocytes (CD45+Lin1−CD11b+CD115+CX3CR1+F4/80−MHCII−Ly-6Chi),
Ly-6Clo monocytes (CD45+Lin1−CD11b+CD115+CX3CR1+F4/80−MHCII−Ly-6Clow)
neutrophils (CD45+Lin1−CD11b+CX3CR1−Ly-6G+),
B cells(CD45+Lin1+CD11b−MHCII+),
T cells (CD45+Lin1+CD11b−MHCII−)
LSK cells (CD45+Lin2−Kit+Sca1+),
common myeloid progenitor (CD45+Lin2−Kit+Sca1−CD34+CD16/32mid),
granulocyte–macrophage progenitor (CD45+Lin2−Kit+Sca1−CD34+CD16/32highCD115−),
monocyte–dendritic cell progenitor (CD45+Lin2−Kit+Sca1−CD34+CD16/32highCD115+); for neutrophil populations
Macrophages (CD45+CD11c−CD11b+Ly6G−MHCII+CD64+)
Lineages were defined as Lin1: CD3, CD90.2, CD19, NK1.1 and Lin2: B220, CD19, CD49b, Ter119, CD90.2, CD11b, CD11c, Ly-6G, IL-7Rα. Data were acquired on a LSRII (BD Biosciences) and Cytek Aurora (Cytek). Data was analyzed with FlowJo (Tree Star)
RNA extraction and cDNA
PCR: Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Venlo, Netherlands) according to the manufacturer’s instructions. RNA quality and quantity were determined using NanoDrop (Thermo Fisher Scientific). cDNA was generated from a max. of 1 μg of total RNA per sample using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) was used to generate cDNA from up to 1 μg of total RNA per sample.
Corticosterone ELISA
Plasma corticosterone concentrations were measured using a commercial corticosterone ELISA kit (Abcam, ab108821) following the manufacturer’s instructions.
Chitinase 3-like 3 ELISA
Chi3l3 was measured in blood plasma and BAL supernatant using a self-coating ELISA kit (R&D Systems, DY2446).
Interleukin-6 ELISA
Mouse IL-6 Quantikine ELISA Kit (R&D Systems, M6000B) was used, following the manufacturer’s instructions.
TNF-alpha ELISA
Mouse TNF-alpha Quantikine ELISA Kit (R&D Systems, MTA00B) was used, following the manufacturer’s instructions.
Free Fatty Acid Quantitation Kit
Free fatty acids in plasma were measured, following the manufacturer’s instructions (Sigma-Aldrich, MAK044).
Bulk RNA-seq
For bulk RNA sequencing, blood Ly-6Chi monocytes (each sample pooled from n=5 mice) were FACS sorted (CD45+; Lin−; CD11b−; Ly-6G−CXCR1+Ly-6C+) into empty 1.5 mL tubes (4 mice pooled per sample, n=3 samples per group (fed vs. fasted+re-fed), total of n=24 mice). After centrifugation, the supernatant was removed and the cell pellet was lysed in RLT lysis buffer, from the RNeasy Micro Kit (Qiagen). The RNeasy Micro Kit (Qiagen) was subsequently used to extract the RNA following the manufacturer’s instructions. RNA-seq libraries were constructed from total RNA using Clontech SMARTer v4 kit (Takara), followed by sequencing on an Illumina HiSeq 2500 instrument, resulting in 20–30 million 50 bp reads per sample. FASTQs files were processed by nf-core rnaseq pipeline with star_salmon configuration48, including multi QC49, alignment to the mm10 genome using STAR aligner50 and counted using unstranded count function per gene_biotype feature by Salmon51 using reference mouse gene models in standard GFF formal. Downstream processing, including count normalization and differential gene expression analysis is conducted using the DESeq252 applying a generalized linear model with the Wald statistical test under the assumption that underlying gene expression count data are distributed as a negative binomial distribution. Pathway enrichment analysis is done using clusterProlifer53 following the standard analytical procedures as described for fastGSEA algorithm54). RNA sequencing data and unnormalized counts will be deposited for public access in the NCBI GEO database prior to publication of this manuscript.
Single-cell RNA-seq
For single-cell RNA sequencing, blood Ly-6Chi monocytes were FACS sorted (CD45+; Lin−; CD11b−; Ly-6G−CXCR1+Ly-6C+) into 1.5 mL tubes (5 mice pooled per sample, n=1 sample per group (fed vs. fasted+re-fed), total of n=10 mice,). After cell counting and viability control, 6,601 (fed) and 7,560 (re-fed) cells were subjected to a single-cell RNA-seq. The library was constructed on the Chromium 10x instrument using Chromium single cell 3 ‘reagent v3.0 kits, followed by sequencing on Illumina HiSeq 2500 instruments, which resulted in approximately 205–209 million reads per sample. FASTQs files were aligned to the mm10 genome provided from 10x (refdata-gex-mm10–2020-A. Alignment, filtering, barcode counting, UMI counting and identification of cells was performed with cellranger version 6.1.2. Count matrices were done aggregated using cellranger aggr with no normalization applied. Low quality cells identified by having less than 200 genes and dying cells with greater than 20% mitochondrial content were filtered out. Seurat was used for subsequent downstream analysis55. Each sample was independently normalized by dividing the gene count for each cell by the total counts of the cell, multiplied by a factor 10,000 and natural log transformed. Variable genes that were common across all samples was used as anchors for integration. Principal Component Analysis (PCA) was performed and the first 30 components were used for calculating the Uniform Manifold Approximation and Projection (UMAP) coordinates and k-nearest neighbors. The Louvain algorithm was used to construct the Shared nearest neighbor (SNN) graph with a resolution of 0.5. Marker genes were identified for each cluster Wilcoxon Rank Sum test. Trajectory and pseudotime inference were performed by Monocle356. Clusters enriched in red blood cells as well as those enriched in mitochondrial and housekeeping genes were excluded first, before running clustering again using the leiden algorithm. The clustering algorithm from monocle3 also outputs 2 “partitions” of the data, and we keep the partition with the largest amount of cells to build one trajectory graph using reversed graph embedding. Cluster 7 from monocle’s clustering was chosen as the root node to order the cells according to pseudotime.
QUANTIFICATION AND STATISTICAL ANALYSIS
Results are shown as mean ±SEM. Statistical tests included unpaired, 2-tailed nonparametric Mann-Whitney tests (when Gaussian distribution was not assumed). For multiple comparisons, nonparametric multiple comparisons test comparing mean rank of each group (when Gaussian distribution was not assumed) or 1-way ANOVA followed by Tukey’s test were performed. Survival was assessed by Log-rank (Mantel-Cox) test. P values of 0.05 or less were considered to denote statistical significance.
Supplementary Material
Video S1. Intravital microscopy in mice with food ad libitum. Related to Figure 2 and Figure S1A
Video S2: Intravital microscopy in mice fasted for 24 h. Related to Figure 2 and Figure S1A
Highlights.
Circulating monocytes migrate to the bone marrow upon fasting
Monocytes augment CXCR4 via a fasting-induced hormonal stress response
Re-feeding after prolonged fasting results in a surge of monocytes into circulation
Prolonged fasting and re-feeding alters the immune response to bacterial infection
Acknowledgements
This work was funded by the National Institutes of Health R35 HL135752, P01 HL131478, P01 HL142494 (to FKS). HJ was supported by the Deutsche Forschungsgemeinschaft (DFG, JA 2545/2-1. FK was supported by scholarships of the Deutsche Forschungsgemeinschaft (DFG, KA 4639/1-1) and the Deutsche Herzstiftung (S/03/19). LH was supported by Boehringer-Ingelheim-Fonds MD fellowship. CSM. was supported by NIH K99/R00 HL151750, R01 HL158534, and the Cure Alzheimer’s Fund.
Footnotes
Declaration of Interest
MN has received funds or material research support from Alnylam, Biotronik, CSL Behring, GlycoMimetics, GSK, Medtronic, Novartis and Pfizer, as well as consulting fees from Lilly, Biogen, Gimv, IFM Therapeutics, Molecular Imaging, Sigilon and Verseau Therapeutics.
FKS has received funds or material research support from Novartis, Partner Therapeutics, Pfizer, Verseu Therapeutics
We support inclusive, diverse, and equitable conduct of research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Dehghan M et al. (2017). Associations of fats and carbohydrate intake with cardiovascular disease and mortality in 18 countries from five continents (PURE): a prospective cohort study. Lancet 390, 2050–2062. [DOI] [PubMed] [Google Scholar]
- 2.Flegal KM, Kit BK, Orpana H, and Graubard BI (2013). Association of all-cause mortality with overweight and obesity using standard body mass index categories: a systematic review and meta-analysis. JAMA 309, 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Q, Zhang Z, Gregg EW, Flanders WD, Merritt R, and Hu FB (2014). Added sugar intake and cardiovascular diseases mortality among US adults. JAMA Intern Med 174, 516–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lumeng CN, and Saltiel AR (2011). Inflammatory links between obesity and metabolic disease. J Clin Invest 121, 2111–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma T et al. (2011). Sucrose counteracts the anti-inflammatory effect of fish oil in adipose tissue and increases obesity development in mice. PLoS One 6, e21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilmot EG et al. (2012). Sedentary time in adults and the association with diabetes, cardiovascular disease and death: systematic review and meta-analysis. Diabetologia 55, 2895–2905. [DOI] [PubMed] [Google Scholar]
- 7.Papadimitriou-Olivgeris M et al. (2016). The Role of Obesity in Sepsis Outcome among Critically Ill Patients: A Retrospective Cohort Analysis. Biomed Res Int 2016, 5941279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandhorst S, and Longo VD (2019). Dietary Restrictions and Nutrition in the Prevention and Treatment of Cardiovascular Disease. Circ Res 124, 952–965. [DOI] [PubMed] [Google Scholar]
- 9.Di Francesco A, Di Germanio C, Bernier M, and de Cabo R (2018). A time to fast. Science 362, 770–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen P et al. (2016). Long-term effects of weight reduction on the severity of psoriasis in a cohort derived from a randomized trial: a prospective observational follow-up study. Am J Clin Nutr 104, 259–265. [DOI] [PubMed] [Google Scholar]
- 11.Johnson JB et al. (2007). Alternate day calorie restriction improves clinical findings and reduces markers of oxidative stress and inflammation in overweight adults with moderate asthma. Free Radic Biol Med 42, 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shibolet O, Alper R, Avraham Y, Berry EM, and Ilan Y (2002). Immunomodulation of experimental colitis via caloric restriction: role of Nk1.1+ T cells. Clin Immunol 105, 48–56. [DOI] [PubMed] [Google Scholar]
- 13.Collins N et al. (2019). The Bone Marrow Protects and Optimizes Immunological Memory during Dietary Restriction. Cell 178, 1088–1101.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagai M et al. (2019). Fasting-Refeeding Impacts Immune Cell Dynamics and Mucosal Immune Responses. Cell 178, 1072–1087.e14. [DOI] [PubMed] [Google Scholar]
- 15.Jordan S et al. (2019). Dietary Intake Regulates the Circulating Inflammatory Monocyte Pool. Cell 178, 1102–1114.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McAlpine CS et al. (2019). Sleep modulates haematopoiesis and protects against atherosclerosis. Nature 566, 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poller WC et al. (2022). Brain motor and fear circuits regulate leukocytes during acute stress. Nature 607, 578–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pick R, He W, Chen CS, and Scheiermann C (2019). Time-of-Day-Dependent Trafficking and Function of Leukocyte Subsets. Trends Immunol 40, 524–537. [DOI] [PubMed] [Google Scholar]
- 19.Scheiermann C et al. (2012). Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity 37, 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Lutes LK, Barnoud C, and Scheiermann C (2022). The circadian immune system. Sci Immunol 7, eabm2465. [DOI] [PubMed] [Google Scholar]
- 21.Yona S et al. (2013). Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pick R, He W, Chen CS, and Scheiermann C (2019). Time-of-Day-Dependent Trafficking and Function of Leukocyte Subsets. Trends Immunol 40, 524–537. [DOI] [PubMed] [Google Scholar]
- 23.Cain DW, Bortner CD, Diaz-Jimenez D, Petrillo MG, Gruver-Yates A, and Cidlowski JA (2020). Murine Glucocorticoid Receptors Orchestrate B Cell Migration Selectively between Bone Marrow and Blood. J Immunol 205, 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barradas M et al. (2022). Fatty acids homeostasis during fasting predicts protection from chemotherapy toxicity. Nat Commun 13, 5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renquist BJ, Murphy JG, Larson EA, Olsen D, Klein RF, Ellacott KL, and Cone RD (2012). Melanocortin-3 receptor regulates the normal fasting response. Proc Natl Acad Sci U S A 109, E1489–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pflugh DL, Maher SE, and Bothwell AL (2000). Ly-6I, a new member of the murine Ly-6 superfamily with a distinct pattern of expression. J Immunol 165, 313–321. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda N et al. (2018). Emergence of immunoregulatory Ym1(+)Ly6C(hi) monocytes during recovery phase of tissue injury. Sci Immunol 3, [DOI] [PubMed] [Google Scholar]
- 28.Shibuya T et al. (2021). Immunoregulatory Monocyte Subset Promotes Metastasis Associated With Therapeutic Intervention for Primary Tumor. Front Immunol 12, 663115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mildner A et al. (2017). Genomic Characterization of Murine Monocytes Reveals C/EBPbeta Transcription Factor Dependence of Ly6C(−) Cells. Immunity 46, 849–862.e7. [DOI] [PubMed] [Google Scholar]
- 30.Magill SS et al. (2014). Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370, 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buono R, and Longo VD (2018). Starvation, Stress Resistance, and Cancer. Trends Endocrinol Metab 29, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longo VD, and Mattson MP (2014). Fasting: molecular mechanisms and clinical applications. Cell Metab 19, 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura K, Nakamura Y, and Kataoka N (2022). A hypothalamomedullary network for physiological responses to environmental stresses. Nat Rev Neurosci 23, 35–52. [DOI] [PubMed] [Google Scholar]
- 34.Devlin MJ (2011). Why does starvation make bones fat. Am J Hum Biol 23, 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casanova-Acebes M et al. (2013). Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell 153, 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Courties G et al. (2019). Glucocorticoids Regulate Bone Marrow B Lymphopoiesis After Stroke. Circ Res 124, 1372–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers MGJ, and Olson DP (2012). Central nervous system control of metabolism. Nature 491, 357–363. [DOI] [PubMed] [Google Scholar]
- 38.Perry RJ et al. (2019). Leptin’s hunger-suppressing effects are mediated by the hypothalamic-pituitary-adrenocortical axis in rodents. Proc Natl Acad Sci U S A 116, 13670–13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnstone AM, Faber P, Andrew R, Gibney ER, Elia M, Lobley G, Stubbs RJ, and Walker BR (2004). Influence of short-term dietary weight loss on cortisol secretion and metabolism in obese men. Eur J Endocrinol 150, 185–194. [DOI] [PubMed] [Google Scholar]
- 40.Culbert KM, Racine SE, and Klump KL (2016). Hormonal Factors and Disturbances in Eating Disorders. Curr Psychiatry Rep 18, 65. [DOI] [PubMed] [Google Scholar]
- 41.Cheng CW et al. (2014). Prolonged fasting reduces IGF-1/PKA to promote hematopoietic-stem-cell-based regeneration and reverse immunosuppression. Cell Stem Cell 14, 810–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frodermann V et al. (2019). Exercise reduces inflammatory cell production and cardiovascular inflammation via instruction of hematopoietic progenitor cells. Nat Med 25, 1761–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heyde A et al. (2021). Increased stem cell proliferation in atherosclerosis accelerates clonal hematopoiesis. Cell 184, 1348–1361.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McAlpine CS et al. (2022). Sleep exerts lasting effects on hematopoietic stem cell function and diversity. J Exp Med 219, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhattacharya R et al. (2021). Association of Diet Quality With Prevalence of Clonal Hematopoiesis and Adverse Cardiovascular Events. JAMA Cardiol 6, 1069–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cahill GFJ (1970). Starvation in man. N Engl J Med 282, 668–675. [DOI] [PubMed] [Google Scholar]
- 47.Robbins CS et al. (2013). Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat Med 19, 1166–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ewels PA et al. (2020). The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol 38, 276–278. [DOI] [PubMed] [Google Scholar]
- 49.Ewels P, Magnusson M, Lundin S, and Kaller M (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu T et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2, 100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subramanian A et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hao Y et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, 979–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Intravital microscopy in mice with food ad libitum. Related to Figure 2 and Figure S1A
Video S2: Intravital microscopy in mice fasted for 24 h. Related to Figure 2 and Figure S1A
Data Availability Statement
Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Brilliant Violet 711™ anti-mouse CD45 clone 30-F11 | BioLegend | Cat# 103147 |
| PE anti-mouse CD45.1 clone A20 | BioLegend | Cat# 110707 |
| PE/Cyanine7 anti-mouse CD45.2 clone 104 | BioLegend | Cat# 109829 |
| PE anti-mouse CD3 clone 17A2 | BioLegend | Cat# 100206 |
| PE anti-mouse CD90.2 clone 3O-H12 | BioLegend | Cat# 105308 |
| PE anti-mouse CD19 clone 6D5 | BioLegend | Cat# 115508 |
| PE anti-mouse B220 clone RA3-6B2 | BD Biosciences | Cat# 553089 |
| PE anti-mouse NK-1.1 clone PK136 | BioLegend | Cat# 108708 |
| APC anti-mouse Ly-6G clone 1A8 | BioLegend | Cat# 127614 |
| FITC anti-mouse Ly-6C clone HK1.4 | BioLegend | Cat# 128006 |
| Alexa Fluor® 700 anti-mouse I-A/I-E clone M5/114.15.2 | BioLegend | Cat# 107602 |
| APC/Fire™ 810 anti-mouse F4/80 clone BM8 | BioLegend | Cat# 123166 |
| APC/Cyanine7 anti-mouse/human CD11b clone M1/70 | BioLegend | Cat# 101226 |
| Brilliant Violet 605™ anti-mouse CD115 clone AFS98 | BioLegend | Cat# 135517 |
| PE anti-mouse TER-119 clone TER-119 | BioLegend | Cat# 116208 |
| FITC anti-mouse CD34 clone RAM34 | ThermoFisher | Cat# 11-0341-85 |
| APC anti-mouse CD11c clone N418 | BioLegend | Cat# 117310 |
| PE anti-mouse CD127 clone SB/199 | BioLegend | Cat# 121112 |
| PerCP/Cyanine5.5 anti-mouse CD16/32 clone 93 | BioLegend | Cat# 101324 |
| PerCP/Cyanine5.5 anti-mouse CD150 | BioLegend | Cat# 115922 |
| APC anti-mouse CD135 clone A2F10 | BioLegend | Cat# 135310 |
| Alexa Fluor® 700 anti-mouse CD48 clone HM48-1 | BioLegend | Cat# 103426 |
| PE/Cyanine7 anti-mouse CD117 (c-Kit) clone 2B8 | BioLegend | Cat# 105814 |
| APC/Cyanine7 anti-mouse Ly-6A/E (Sca-1) clone D7 | BioLegend | Cat# 108126 |
| APC anti-mouse CD8a clone 53-6.7 | BD Biosciences | Cat# 553035 |
| Pacific Blue™ anti-mouse CD4 clone GK1.5 | BioLegend | Cat# 100428 |
| PE anti-mouse Siglec-F clone E50-2440 | BD Biosciences | Cat# 562068 |
| PE anti-mouse CXCR4 clone 2B11 | ThermoFisher | Cat# 12-9991-82 |
| PE/Cyanine7 anti-mouse CX3CR1 clone SA011F11 | BioLegend | Cat# 149016 |
| PE/Cyanine7 anti-mouse TNF-α clone MP6-XT22 | BioLegend | Cat# 506324 |
| PE/Dazzle™ 594 anti-mouse CD64 clone X54-5/7.1 | BioLegend | Cat# 139320 |
| AF647™ anti-BrdU clone MoBU-1 | ThermoFisher | Cat# B35133 |
| Brilliant Violet 650™ anti-mouse CD192 (CCR2) | BioLegend | Cat# 150613 |
| Bacterial and virus strains | ||
| Pseudomonas aeruginosa, Strain Boston 41501 | ATCC | ATCC# 27853 |
| Chemicals, peptides, and recombinant proteins | ||
| Corticosterone: HBC complex | Sigma-Aldrich | Cat# C174 |
| pHrodo™ Red Phagocytosis Particle Labeling Kit for Flow Cytometry | Thermo Fisher | Cat# A10026 |
| pHrodo™ Red E. coli BioParticles™ Conjugate for Phagocytosis | Thermo Fisher | Cat# P35361 |
| Critical commercial assays | ||
| RNeasy Mini Kit | Qiagen | Cat# 74104 |
| High-Capacity cDNA Reverse Transcription Kit | Thermo Fisher | Cat# 4368814 |
| Corticosterone ELISA kit | Abcam | Cat# ab108821 |
| Mouse YM1/Chitinase 3-like 3 DuoSet ELISA | R&D Systems | Cat# DY2446 |
| Mouse IL-6 Quantikine ELISA Kit | R&D Systems | Cat# M6000B |
| Mouse TNF-alpha Quantikine ELISA Kit | R&D Systems | Cat# MTA00B |
| Free Fatty Acid Quantitation Kit | Sigma-Aldrich | Cat# MAK044 |
| Click-iT™ Plus EdU Pacific Blue™ Flow Cytometry Assay Kit | Thermo Fisher | Cat# C10636 |
| Deposited data | ||
| Single cell RNA sequencing data | Gene Expression Omnibus | |
| Bulk RNA sequencing data | Gene Expression Omnibus | |
| Experimental models: Organisms/strains | ||
| C57BL/6J | The Jackson Laboratory | Strain 000664 |
| C57BL/6-Tg(UBC-GFP)30Scha/J | The Jackson Laboratory | Strain 004353 |
| B6.129P2-Lyz2tm1(cre)Ifo/J | The Jackson Laboratory | Strain 004781 |
| B6.129P2-Cxcr4tm2Yzo/J | The Jackson Laboratory | Strain 008767 |
| B6.Cg-Nr3c1tm1.1Jda/J | The Jackson Laboratory | Strain 021021 |
| STOCK Prkaa1tm1.1Sjm/J | The Jackson Laboratory | Strain 014141 |
| B6.129S4(Cg)-Arntltm1Weit/J | The Jackson Laboratory | Strain 007668 |
| STOCK Slc2a1tm1.1Stma/AbelJ | The Jackson Laboratory | Strain 031871 |
| B6.FVB(129X1)-Tg(Sim1Cre)1Lowl/J | The Jackson Laboratory | Strain 006451 |
| B6.129S4(Cg)-Crhtm2.1 Maj/J | The Jackson Laboratory | Strain 030110 |
| B6.129S4-Ccr2tm1Ifc/J | The Jackson Laboratory | Strain 004999 |
| C.129S7(B6)-Itgb2tm2Bay/AbhmJ | The Jackson Laboratory | Strain 027003 |
| B6.129P2(Cg)-Cx3cr1tm1Litt/J | The Jackson Laboratory | Strain 005582 |
| B6.129S7-Rag1tm1Mom/J | The Jackson Laboratory | Strain 002216 |
| BALB/cJ | The Jackson Laboratory | Strain 000651 |
| Oligonucleotides | ||
| Mouse GAPD Mm99999915_g1 | Thermo Fisher | Cat# 4331182 |
| Mouse ACTB Mm00607939_s1 | Thermo Fisher | Cat# 4331182 |
| Mouse Clock Mm00455950_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Arntl Mm00500226_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Nr1d2 Mm01310356_g1 | Thermo Fisher | Cat# 4331182 |
| Mouse Per1 Mm00501813_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Cry1 Mm00514392_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Selplg Mm01204601_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Cd44 Mm01277161_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Itgal Mm00801807_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Itgam Mm00434455_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Itgb1 Mm01253230_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Itgb2 Mm00434513_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Itga4 Mm01277951_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Itga5 Mm00439797_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Itga6 Mm00434375_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Ccr1 Mm00438260_s1 | Thermo Fisher | Cat# 4331182 |
| Mouse Cxcr3 Mm9999054_s1 | Thermo Fisher | Cat# 4331182 |
| Mouse Cxcr4 Mm01996749_s1 | Thermo Fisher | Cat# 4331182 |
| Mouse Cx3cr1 Mm02620111_s1 | Thermo Fisher | Cat# 4331182 |
| Mouse Ilb1 Mm00434228_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Tnf Mm00443258_m1 | Thermo Fisher | Cat# 4331182 |
| Mouse Ccr2 Mm99999051_gH | Thermo Fisher | Cat# 4331182 |
| Software and algorithms | ||
| FlowJo v10 | FlowJo | https://www.flowjo.com/ |
| GraphPad Prism v9 | GraphPad Software | https://www.graphpad.com/ |
| BioRender | BioRender | https://biorender.com |
| R Differential gene expression analysis: | ||
| R version 4.2.1, Platform: aarch64-apple-darwin20 (64-bit); OS: macOS Monterey 12.6.1 | aarch64-apple-darwin20 (64-bit) | N/A |
| scales_1.2.1 | CRAN/BiocManager | N/A |
| clusterProfiler_4.6.0 | CRAN/BiocManager | N/A |
| tidyr_1.2.1 | CRAN/BiocManager | N/A |
| dplyr_1.0.10 | CRAN/BiocManager | N/A |
| ggplot2_3.3.6 | CRAN/BiocManager | N/A |
| GenomicRanges_1.50.0 | CRAN/BiocManager | N/A |
| MatrixGenerics_1.10.0 | CRAN/BiocManager | N/A |
| ggthemes_4.2.4 | CRAN/BiocManager | N/A |
| org.Hs.eg.db_3.16.0 | CRAN/BiocManager | N/A |
| immunarch_0.8.0 | CRAN/BiocManager | N/A |
| EnhancedVolcano_1.16.0 | CRAN/BiocManager | N/A |
| genefilter_1.80.0 | CRAN/BiocManager | N/A |
| GenomeInfoDb_1.34.0 | CRAN/BiocManager | N/A |
| matrixStats_0.62.0 | CRAN/BiocManager | N/A |
| msigdbr_7.5.1 | CRAN/BiocManager | N/A |
| org.Mm.eg.db_3.16.0 | CRAN/BiocManager | N/A |
| patchwork_1.1.2 | CRAN/BiocManager | N/A |
| ggrepel_0.9.1 | CRAN/BiocManager | N/A |
| DESeq2_1.38.0 | CRAN/BiocManager | N/A |
| IRanges_2.32.0 | CRAN/BiocManager | N/A |
| glmpca_0.2.0 | CRAN/BiocManager | N/A |
| pathview_1.38.0 | CRAN/BiocManager | N/A |
| AnnotationDbi_1.60.0 | CRAN/BiocManager | N/A |
| data.table_1.14.4 | CRAN/BiocManager | N/A |
| glmGamPoi_1.10.0 | CRAN/BiocManager | N/A |
| SummarizedExperiment_1.28.0 | CRAN/BiocManager | N/A |
| S4Vectors_0.36.0 | CRAN/BiocManager | N/A |
| RColorBrewer_1.1-3 | CRAN/BiocManager | N/A |
| enrichplot_1.18.0 | CRAN/BiocManager | N/A |
| gridExtra_2.3 | CRAN/BiocManager | N/A |
| dtplyr_1.2.2 | CRAN/BiocManager | N/A |
| ggbeeswarm_0.6.0 | CRAN/BiocManager | N/A |
| Biobase_2.58.0 | CRAN/BiocManager | N/A |
| BiocGenerics_0.44.0 | CRAN/BiocManager | N/A |
| pheatmap_1.0.12 | CRAN/BiocManager | N/A |
| R scRNA analysis: | ||
| monocle3_1.2.9 | GitHub | N/A |
| IRanges_2.32.0 | CRAN/BiocManager | N/A |
| BiocGenerics_0.44.0 | CRAN/BiocManager | N/A |
| SeuratObject_4.1.2 | GitHub | N/A |
| SingleCellExperiment_1.20.0 | CRAN/BiocManager | N/A |
| S4Vectors_0.36.0 | CRAN/BiocManager | N/A |
| SeuratDisk_0.0.0.9020 | GitHub | N/A |
| Seurat_4.2.0 | GitHub | N/A |
| SummarizedExperiment_1.28.0 | CRAN/BiocManager | N/A |
| MatrixGenerics_1.10.0 | GitHub | N/A |
| SeuratWrappers_0.3.1 | GitHub | N/A |
| magrittr_2.0.3 | CRAN/BiocManager | N/A |
| GenomicRanges_1.50.0 | CRAN/BiocManager | N/A |
| matrixStats_0.62.0 | CRAN/BiocManager | N/A |
| SeuratData_0.2.2 | GitHub | N/A |
| patchwork_1.1.2 | CRAN/BiocManager | N/A |
| GenomeInfoDb_1.34.0 | CRAN/BiocManager | N/A |
| Biobase_2.58.0 | CRAN/BiocManager | N/A |
| sp_1.5-0 | CRAN/BiocManager | N/A |
| ggplot2_3.3.6 | CRAN/BiocManager | N/A |


