Abstract
Acinetobacter baumannii is one of the most problematic nosocomial pathogens that can efficiently thrive within hospital settings, mainly due to resistances toward antibiotics, desiccation, disinfectants, human serum and oxidative stress. Recently, increased resistance against last-resort antibiotics earns this bacterium the highest priority concern classified by the Centers for Disease Control and Prevention and the World Health Organization. An obvious hallmark of this bacterium is the high heterogeneity observed among A. baumannii isolates, with a limited core genome. This feature complexifies the study of A. baumannii bacteria as an entity, subsequently reflected in a diversity of phenotypes of not only antimicrobial and environmental resistance but also virulence. A high degree of genome plasticity, along with the use of a limited subset of established strains, can lead to strain-specific observations, decreasing the global understanding of this pathogenic agent. Phenotypic variability of A. baumannii strains is easily observable such as with the macrocolony morphologies, in vitro and in vivo virulence, natural competence level, production of different capsular polysaccharide structures and cellular densities. Some strains encode several virulence factors, while others, including the established strains, lack key ones. The lack/excess of genes or specific physiological processes might interfere with in vivo and in vitro experiments, thus providing a limited impact on the global understanding of Acinetobacter bacteria. As an answer to the high heterogeneity among A. baumannii strains, we propose a first comprehensive database that includes the bacterial strains and the associated phenotypic and genetic data. This new repository, freely accessible to the entire scientific community, allows selecting the best bacterial isolate(s) related to any biological question, using an efficient and fast exchange platform.
Database URL: https://acinetobase.vib.be/
Introduction
Acinetobacter baumannii is an opportunistic nosocomial pathogen thriving in hospital environment and endangering, especially critically ill patients (1). This Gram-negative bacterium is part of the most problematic ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa and Enterobacter species) group of human pathogens, against which therapeutic approaches become limited (2). Increased isolations of antibiotic-resistant strains, especially to last-resort antibiotics, earn this bacterium the consideration as an ‘urgent threat to public health’ by the Centers for Disease Control and Prevention (3) and as ‘a bacterium for which research and development of new antibiotics is critically needed’ by the World Health Organization (4). An obvious hallmark of A. baumannii bacteria is the high heterogeneity observed among the isolates (5). Acinetobacter baumannii bacteria have a dynamic genome, with an estimated conserved core genome of only 16.5%, while 25% of the genome is unique to each strain, meaning that there is no counterpart in any other A. baumannii genome (5). Our recent study (6) on 47 strains of A. baumannii shows even lower core genome of 2009 genes from the pangenome of 13 611 genes, accounting for 14.76%. Acinetobacter baumannii isolates are impressively variable, at both genetic and phenotypic levels. For example, the capsular polysaccharide, which impacts antibiotic and environmental resistances, host response and virulence, is encoded by at least 237 various capsule locus types, while the outer core of lipooligosaccharide is encoded by at least 13 locus types (7, 8). This genetic variability translates into phenotypic heterogeneity, observable using electron microscopy where the thickness and rigidity of the capsule differ, along with different cellular densities (9). The established strains such as ATCC19606, ATCC17978, DSM30011 or AB5075 are widely used type strains from which significant and validated observations were and are still generated (10–13). Altogether, these established strains greatly contributed to the state of the art of the current and growing A. baumannii field. The established strains AB5075, ATCC17978 and ATCC19606 are multidrug-resistant (MDR) strains of clinical origin (14, 15), while DSM30011 is a non-MDR environmental strain obtained from plant microbiota (16). The observations gained about these established strains shed a strong base of the knowledge about A. baumannii. However, on the other hand, type strains also showed the limitations of only using these strains when current clinical isolates of A. baumannii show extreme genetic and phenotypic variabilities (6). In order to fill this modern gap of knowledge, grasp the complexity of A. baumannii and widen the possibilities of the community to use and efficiently share genotypically and phenotypically characterized strains, we propose the database and strain repository Acinetobase as a new community tool (Figure 1). This new data and strains sharing platform is especially important in the context of novel therapeutic discoveries, for which conserved targets and antimicrobial modes of actions among the highest proportion of problematic and clinically relevant A. baumannii isolates are required.
Figure 1.
‘About’ page of Acinetobase.
Results
Construction and content
This first version of Acinetobase consists of 43 clinical isolates, 4 widely used strains (AB5075-VUB, ATCC19606-VUB, ATCC17978-VUB and DSM30011-VUB) and 1 naturally occurring mutant of A. baumannii AB5075-VUB, disrupted in the itrA gene designated AB5075-VUB-itrA::ISAba13 (9). We generated whole-genome sequences and de novo assembled the complete chromosomal sequences with high quality of all these strains, rendering them publicly available via GenBank accession number. In addition, the Acinetobase includes a variety of phenotypical characteristics associated with a strain:
-
(i)
Quantification of the capsular polysaccharide using the gradient density method for each strain (9)
-
(ii)
Type of macrocolony produced for each strain (designated with letters A to F) with a corresponding photograph (6)
-
(iii)
Virulence in Galleria mellonella infection model for nine strains (9)
-
(iv)
Transmission electron microscopy (TEM) photograph depicting the amount and density of the capsule for 11 strains (6)
-
(v)
Antimicrobial susceptibility testing with minimum inhibitory concentrations for 43 clinical isolates (20)
The database also describes the basic genotypical characteristics of each strain:
-
(i)
Multilocus sequence type (ST) according to Pasteur (17) and Oxford (18, 19) typing scheme
- (ii)
- (iii)
-
(iv)
Antibiotic resistance genes (ARGs) obtained using the ResFinder 4.0 (24) database as described earlier (20)
Furthermore, the metadata for each strain are provided along with a reference to the publication providing a more profound description:
-
(i)
Origin of the strain (e.g. clinical, environmental and animal)
-
(ii)
Source of isolation (e.g. wound, sputum and feces)
-
(iii)
Year of isolation
The above-mentioned genotypical and phenotypical traits along with metadata can be used to search the database and filter the strains for specific criteria. This way users can efficiently find the strain of desired characteristics and request it. The bacterial strains of A. baumannii were deposited and are available in lyophilized form as per order from the Belgian Coordinated Collection of Microorganisms/Lab of Microbiology (https://bccm.belspo.be/). The example of an entry of AB5075-VUB is depicted in Figure 2.
Figure 2.
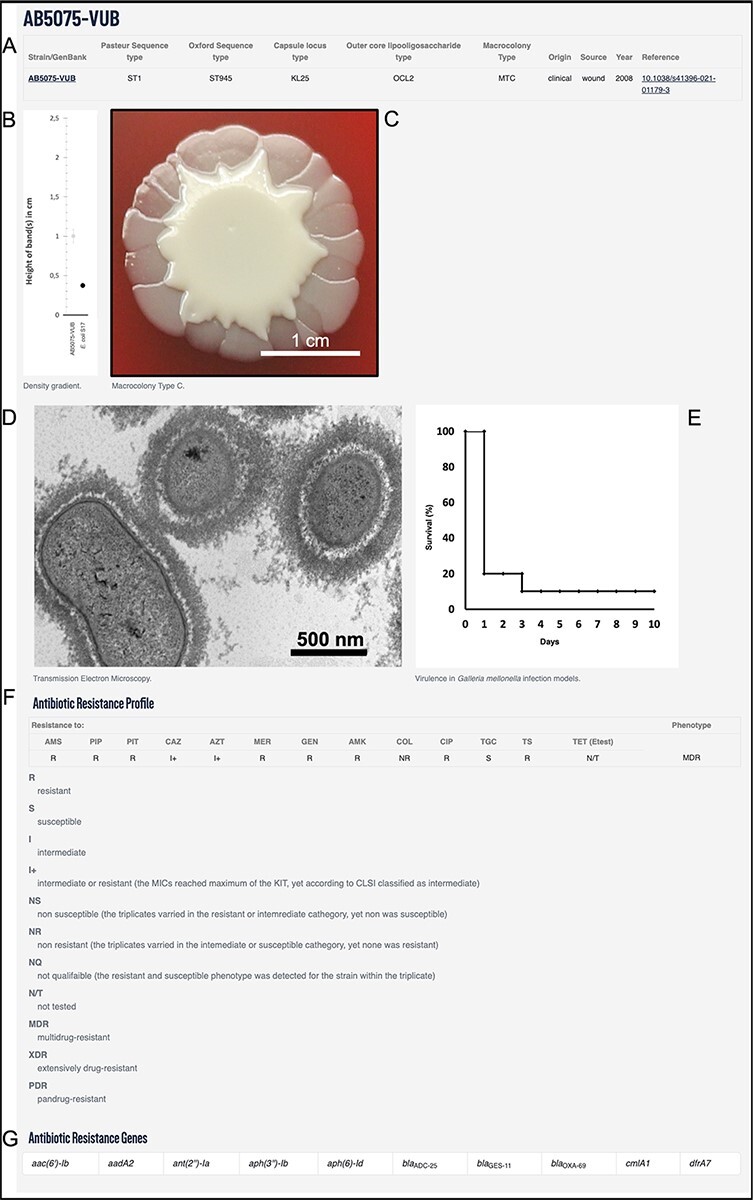
Example using AB5075-VUB entry depicting: (A) description of the genotype (KL, STOx/Pas and OCL), origin, source and year of isolation with a hyperlink for the complete chromosomal sequence and a reference publication; (B) quantification of the capsule; (C) macrocolony type; (D) TEM micrograph of the capsule; (E) virulence in G. mellonella infection model; (F) antibiotic resistance profile and (G) ARGs.
Discussion
Existing databases
The decreasing costs of the whole-genome sequencing allowed genomic databases, filled with high-quality data, to thrive. There are well-established databases such as EnteroBase (22), which do not include data on Acinetobacter. In contrast, existing databases that provide whole-genome sequencing data on A. baumannii, such as BIGSdb (23), include metadata and genotype; however, BIGSdb does not include phenotypic data unrelated to the genotype nor the strains themselves. Moreover, centralization of the laboratory providing strains with the genotype and phenotype characteristics will help to decrease the chance of working with subcultured variants with altered genotype and phenotype, a pitfall that becomes more and more apparent in the A. baumannii field (9, 25, 26).
Browsing
The users can browse the ‘Collection’ page for the strains (Figure 3) and pick according to their genomic description. By clicking on the name of the selected strain, the full entry with genomic and phenotypic details appears (Figure 2).
Figure 3.
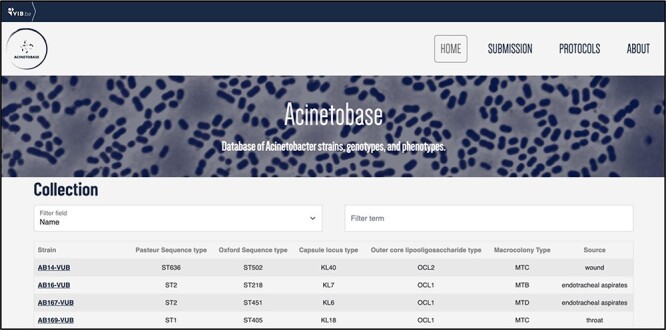
‘Collection’ page depicting the name, Pasteur and Oxford STs, KL, OCL, macrocolony types and source of each isolate.
Searching
The users can filter the entries (Figure 4) according to each genetic- and phenotypic-based criterium depicted in the ‘Collection’ page. This feature helps to select specific strains of A. baumannii according to criteria demanded by the user, appropriate to their biological question.
Figure 4.
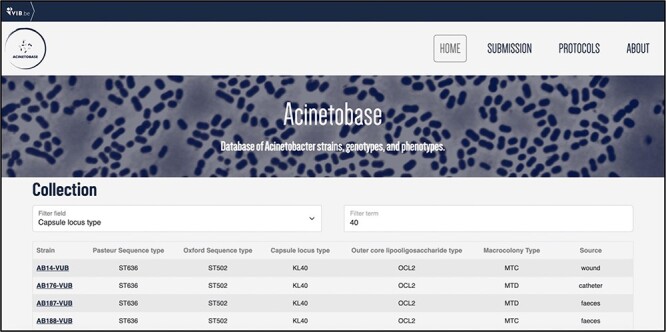
Example of entries filtering based on the KL type 40.
Download
To avoid unnecessary redundancy, downloading the complete chromosomal sequences and the whole-genome assemblies is facilitated via GenBank (https://www.ncbi.nlm.nih.gov/). The source information on the strains can be found in the corresponding research articles found in the ‘Reference’ column using hyperlinks (using a digital object identifier). However, the main information such as metadata for each strain and protocols for each methodology is displayed in Acinetobase. In the corresponding peer-reviewed study, authors of the study provide additional or further information on how the strains were obtained their origin, original figures and the protocols for assessing the phenotypical traits. The authors of the study will be directly responsible for the information and data they provided and submitted to Acinetobase.
Submissions
The Acinetobase is open for submissions of the genomic and phenotypic data of Acinetobacter strains by other research teams and laboratories. However, there are requirements that must be met prior to contacting the curators of the Acinetobase with an inquiry of submission. We also propose ‘Optional’ information to be provided by submitters. The requirements and optional information are discussed in the next subsection.
Requirements
-
(i)
You are willing to share the strain upon demand.
-
(ii)
You can provide information on the strain such as the source of isolation and year (no Protected Health Information).
Optional
-
(i)
The maximum genetic and phenotypic characterization.
-
(ii)
The hyperlink for the whole-genome sequences within a repository (e.g. GenBank)—optimally complete genomes (closed circular genomes).
-
(iii)
Reference
Future perspectives
Acinetobase has a significant potential to influence the field of microbiology, especially the A. baumannii community, and other research fields working with similar pathogens and biological questions might be inspired. Further implementation potential of Acinetobase resides in new strain acquisition and the accumulation of additional experimental analyses and data. This will provide valuable information to researchers on whether one (or a subset of available strains) is an appropriate isolate to work with, depending on the biological question aimed. The proposed project is so far aimed to A. baumannii, yet not limited to a single species of Acinetobacter. With an increasing clinical relevance of other Acinetobacter species such as A. pittii, A. nosocomialis, A. seifertti and A. dijkshoorniae (27), the Acinetobase initiative is open to all Acinetobacter species. Acinetobase accepts submission of the phenotypic and genotypic data including strains from other laboratories and research groups, and the authors (curators) of the Acinetobase do intend to provide further genomic and phenotypic data in form of strain submissions. This initiative will, over time, result in a growing database and a platform for sharing knowledge and Acinetobacter spp. strains and might include further molecular tools such as fluorescently labeled strains or characterized deletion mutant strains.
Conclusion
In conclusion, Acinetobase is the first database dedicated to Acinetobacter genotypes and associated phenotypes that also serves as a repository of characterized strains. Acinetobase not only will be the proof of concept of high heterogeneity among A. baumannii strains but also will transform this pitfall into a new strength, benefiting the entire community by helping to select an appropriate strain related to specific conditions, experiments and biological questions. The subculturing steps of the current strain-sharing strategy will therefore decrease, helping to keep the genetic background of these dynamic bacteria as much as possible.
Acknowledgements
We would like to thank to Belgian Coordinated Collections of Microorganisms of Ghent University, Faculty of Sciences, Belgium, and the VIB Bioinformatics Core for their support in developing the website, especially to Christof De Bo who helped with graphics. We would also like to thank the National Reference Center Laboratory for Antibiotic-Resistant Gram-Negative Bacilli (CHU UCL Namur, Yvoir, Belgium) for sharing the modern clinical isolates of A. baumannii and related information.
Contributor Information
Adam Valcek, Microbial Resistance and Drug Discovery, VIB-VUB Center for Structural Biology, VIB, Flanders Institute for Biotechnology, Brussels, Belgium; Structural Biology Brussels, Vrije Universiteit Brussel (VUB), Brussels, Belgium.
James Collier, VIB Bioinformatics Core, Rijvischestraat 126 3R, Zwijnaarde-Ghent 9052, Belgium.
Alexander Botzki, VIB Bioinformatics Core, Rijvischestraat 126 3R, Zwijnaarde-Ghent 9052, Belgium.
Charles Van der Henst, Microbial Resistance and Drug Discovery, VIB-VUB Center for Structural Biology, VIB, Flanders Institute for Biotechnology, Brussels, Belgium; Structural Biology Brussels, Vrije Universiteit Brussel (VUB), Brussels, Belgium.
Data availability
The datasets generated and analyzed during the current study are linked from the Acinetobase website (http://acinetobase.vib.be/).
Author contributions
A.V. and C.V.d.H. wrote the manuscript. C.V.d.H. conceptualized the Acinetobase. J.C. wrote and designed the website, data storage and update mechanisms. A.B. assisted with the design of the website, is the project sponsor and reviewed the manuscript.
Ethics approval and consent to participate
Not applicable.
Funding
This project was supported by the Flanders Institute for Biotechnology (VIB). AV is a recipient of a Junior postdoctoral fellowship of the Research Foundation – Flanders (FWO, File number: File number: 1287223N).
Conflict of interest
The authors declare that they have no competing interests.
References
- 1. Whiteway C., Breine A., Philippe C.. et al. (2022) Acinetobacter baumannii. Trends Microbiol., 30, 199–200. [DOI] [PubMed] [Google Scholar]
- 2. Pendleton J.N., Gorman S.P. and Gilmore B.F. (2014) Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther., 11, 297–308. [DOI] [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention (U.S.), National Center for Emerging Zoonotic and Infectious Diseases (U.S.), Division of Healthcare Quality Promotion, Antibiotic Resistance Coordination and Strategy Unit . (2019) Antibiotic Resistance Threats in the United States, 2019. Atlanta and U.S. Department of Health and Human Services. https://stacks.cdc.gov/view/cdc/82532.
- 4. Tacconelli E., Carrara E., Savoldi A.. et al. (2018) Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis., 18, 318–327. [DOI] [PubMed] [Google Scholar]
- 5. Imperi F., Antunes L.C.S., Blom J.. et al. (2011) The genomics of Acinetobacter baumannii: insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB Life, 63, 1068–1074. [DOI] [PubMed] [Google Scholar]
- 6. Philippe C., Valcek A., Whiteway C.. et al. (2022) What are the reference strains of Acinetobacter baumannii referring to? bioRxiv.doi: 10.1101/2022.02.27.482139. [DOI]
- 7. Wyres K.L., Cahill S.M., Holt K.E.. et al. (2020) Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microb. Genom., 6, e000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kenyon J.J. and Hall R.M. (2022) Updated analysis of the surface carbohydrate gene clusters in a diverse panel of Acinetobacter baumannii isolates. Antimicrob. Agents Chemother., 66, e0180721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Whiteway C., Valcek A., Philippe C.. et al. (2021) Scarless excision of an insertion sequence restores capsule production and virulence in Acinetobacter baumannii. ISME J, 2022, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roussin M., Rabarioelina S., Cluzeau L.. et al. (2019) Identification of a contact-dependent growth inhibition (CDI) system that reduces biofilm formation and host cell adhesion of Acinetobacter baumannii DSM30011 strain. Front Microbiol, 10, 2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bravo Z., Orruño M., Parada C.. et al. (2016) The long-term survival of Acinetobacter baumannii ATCC 19606T under nutrient-deprived conditions does not require the entry into the viable but non-culturable state. Arch. Microbiol., 198, 399–407. [DOI] [PubMed] [Google Scholar]
- 12. Jacobs A.C., Thompson M.G., Black C.C.. et al. (2014) AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio, 5, e01076–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Silva P.M., Chong P., Fernando D.M.. et al. (2017) Effect of incubation temperature on antibiotic resistance and virulence factors of Acinetobacter baumannii ATCC 17978. Antimicrob. Agents Chemother., 62, e01514–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gallagher L.A., Ramage E., Weiss E.J.. et al. (2015) Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J. Bacteriol., 197, 2027–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harding C.M., Hennon S.W. and Feldman M.F. (2017) Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol., 16, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Repizo G.D., Viale A.M., Borges V.. et al. (2017) The environmental Acinetobacter baumannii isolate DSM30011 reveals clues into the preantibiotic era genome diversity, virulence potential, and niche range of a predominant nosocomial pathogen. Genome Biol Evol, 9, 2292–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Diancourt L., Passet V., Nemec A.. et al. (2010) The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One, 5, e10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bartual S.G., Seifert H., Hippler C.. et al. (2005) Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol., 43, 4382–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wisplinghoff H., Hippler C., Bartual S.G.. et al. (2008) Molecular epidemiology of clinical Acinetobacter baumannii and Acinetobacter genomic species 13TU isolates using a multilocus sequencing typing scheme. Clin. Microbiol. Infect., 14, 708–715. [DOI] [PubMed] [Google Scholar]
- 20. Valcek A., Nesporova K., Whiteway C.. et al. (2022) Genomic analysis of a strain collection containing multidrug-, extensively drug-, pandrug-, and carbapenem-resistant modern clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother., 15, e0089222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wick R.R., Heinz E., Holt K.E.. et al. (2018) Kaptive web: user-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J. Clin. Microbiol., 56, 197–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou Z., Alikhan N.-F., Mohamed K.. et al. (2020) The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res., 30, 138–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jolley K.A., Bray J.E. and Maiden M.C.J. (2018) Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res., 3, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bortolaia V., Kaas R.S. and Ruppe E. (2020) ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother., 75, 3491–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Artuso I., Lucidi M., Visaggio D.. et al. (2022) Genome diversity of domesticated Acinetobacter baumannii ATCC 19606T strains. Microb. Genom., 8, 000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wijers C.D.M., Pham L., Menon S.. et al. (2021) Identification of two variants of Acinetobacter baumannii strain ATCC 17978 with distinct genotypes and phenotypes. Infect. Immun., 89, e0045421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vijayakumar S., Biswas I. and Veeraraghavan B. (2019) Accurate identification of clinically important Acinetobacter spp.: an update. Future Sci. OA, 5, 395–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study are linked from the Acinetobase website (http://acinetobase.vib.be/).


