Abstract
Knowledge of locations and activities of cis-regulatory elements (CREs) is needed to decipher basic mechanisms of gene regulation and to understand the impact of genetic variants on complex traits. Previous studies identified candidate CREs (cCREs) using epigenetic features in one species, making comparisons difficult across species. In contrast, we conducted a cross-species study defining epigenetic states and identifying cCREs in blood cell types to generate regulatory maps that are comparable across species. This study used integrative modeling of eight epigenetic features jointly in human and mouse in our Validated Systematic Integration (VISION) Project. The contribution of each epigenetic state in cCREs to gene regulation was estimated from a multivariate regression against gene expression across cell types. We used these values to estimate epigenetic state Regulatory Potential (esRP) scores for each cCRE in each cell type, which are useful for visualizing and categorizing dynamic changes in cCREs. Groups of cCREs displaying similar patterns of regulatory activity in human and mouse cell types, obtained by joint clustering on esRP scores, harbored distinctive transcription factor binding motifs that were similar across species. Genetic variants associated with blood cell phenotypes were highly and specifically enriched in the catalog of human VISION cCREs, supporting its utility for understanding impacts of noncoding genetic variants on blood cell-related traits. A cross-species comparison of cCREs, based on the joint modeling, revealed both conserved and lineage-specific patterns of epigenetic evolution, even in the absence of genomic sequence alignment. We provide these resources through tools and browsers at http://usevision.org.
Keywords: epigenetics, gene regulatory elements, dimensional reduction, regulatory potential
INTRODUCTION
The morphology and function of different cell types are determined by the expression of distinctive sets of genes in each cell type. This differential gene expression is regulated by the interplay of transcription factors (TFs) binding to cis-regulatory elements (CREs) in the genomic DNA, such as promoters and enhancers, forging interactions among the CREs and components of transcriptional apparatus and ultimately leading to patterns of gene activation and repression characteristic of each cell type (Maston et al. 2006; Hamamoto and Fukaya 2022). Epigenetic features such as accessibility of DNA and modifications of histone tails in chromatin have pronounced impacts on the ability of TFs to bind to CREs, and furthermore, they serve as a molecular memory of transcription and repression (Strahl and Allis 2000; Ringrose and Paro 2004). Frequently occurring sets of chromatin features define epigenetic states, which are associated with gene regulation and expression (Ernst and Kellis 2010; Hoffman et al. 2013; Zhang et al. 2016). Genome-wide assignment of DNA intervals to epigenetic states (annotation) provides a view of the regulatory landscape that can be compared across cell types, which in turn leads to insights into the processes regulating gene expression (Libbrecht et al. 2021).
Comprehensive mapping of CREs within the context of the regulatory landscape in different cell types is needed to achieve a broad understanding of differential gene expression. Maps of candidate CREs (cCREs) provide guidance in understanding how changes in cCREs, including single nucleotide variants and indels, can lead to altered expression (Hardison 2012), and they can inform approaches for activation or repression of specific genes in potential strategies for therapies (Bauer et al. 2013). Indeed, most human genetic variants associated with common traits and diseases are localized in or near cCREs (Hindorff et al. 2009; Maurano et al. 2012; The_ENCODE_Project_Consortium 2012). Thus, knowledge of the activity and epigenetic state of cCREs in each cell type can facilitate understanding the impact of trait-associated genetic variants on specific phenotypes. Furthermore, genome editing approaches in somatic cells have recently been demonstrated to have promise as therapeutic modalities (Frangoul et al. 2021), and a full set of cCREs annotated by activity and state can help advance similar applications.
The different types of blood cells in humans and mice are particularly tractable systems for studying many aspects of gene regulation during differentiation. The striking differences among mature cell types result from progressive differentiation starting from a common hematopoietic stem cell (HSC) (Kondo et al. 2003). While single cell analyses reveal a pattern of ostensibly continuous expression change along each hematopoietic lineage (Laurenti and Göttgens 2018), intermediate populations of multi-lineage progenitor cells with decreasing differentiation potential have been defined, which provide an overall summary and nomenclature for major stages in differentiation. These stem, progenitor, and mature cell populations can be isolated using characteristic cell surface markers (Spangrude et al. 1988; Payne and Crooks 2002), albeit with dramatically fewer cells in progenitor populations. Many lineage-restricted transcription factors exert critical roles in gene regulation during hematopoiesis (Orkin 1995; Blobel and Weiss 2009). In addition to the primary blood cells, several immortalized cell lines provide amenable systems for intensive study of various aspects of gene regulation during differentiation and maturation of blood cells (Weiss et al. 1997).
The VISION project aims to produce a Validated Systematic Integration of hematopoietic epigenomes, harvesting the extensive epigenetic and transcriptomic datasets from previous work from many investigators and large consortia into concise, systematically integrated summaries of regulatory landscapes and cCREs (Hardison et al. 2020). We previously published the results of these analyses for progenitor and mature blood cell types from mouse (Xiang et al. 2020b). In the current study, we generated additional epigenetic datasets and expanded the integrative analyses to include data across both human and mouse cell types to facilitate cross-species analyses for insights into both evolution and function of the cCREs. Here we describe (a) our systematic integrative analyses of epigenetic features across progenitor and mature blood cell types jointly for human and mouse to produce genome-wide views of the epigenetic states (the regulatory landscapes) that are comparable across species, (b) catalogs of cCREs, annotated by accessibility and epigenetic state in each cell type and each species along with overlaps with orthogonal sets of genomic elements, (c) estimates of regulatory output from each cCRE in each cell type, based on the epigenetic state assignments, (d) discovery of distinctive and discriminatory motifs for many categories of cCREs, (e) a demonstration of the utility of the cCREs for gaining inferences into the way that noncoding genetic variants may be impacting complex blood cell-related traits, and (f) a study of the evolution of DNA sequences and inferred function of cCREs between human and mouse. Together, this work provides valuable community resources that enable researchers to leverage the extensive existing epigenomic data into further mechanistic regulatory studies of both individual loci and genome-wide trends in human and mouse blood cells.
RESULTS
Data collation, preprocessing, normalization, and denoising
The input for our joint integrative analysis of human and mouse regulatory landscapes across progenitor and mature blood cell types was a large number of data sets of epigenetic features related to gene regulation and expression (404 data sets, 216 in human and 188 in mouse) generated by both consortia and individual laboratories, including several that were generated for this work (Figure 1 and Supplementary Table S1). Most data were from primary cell populations, and data from commonly used cell lines were also included. Chromatin accessibility is a general feature of almost all regulatory elements, and it was measured by the Assay for Transposase Accessible Chromatin with high throughput sequencing (ATAC-seq, Buenrostro et al. 2013; Corces et al. 2016) or by DNase-seq (Thurman et al. 2012) for almost all cell types in both species. Available ChIP-seq data for up to six histone modifications provided information related to different elements or processes in gene expression, specifically H3K4me3 for promoters and H3K4me1 for enhancers (Birney et al. 2007; Heintzman et al. 2007), H3K27ac for activation (Roh et al. 2005; Smith and Shilatifard 2014), H3K36me3 for transcriptional elongation (Li et al. 2002), H3K27me3 for repression by the Polycomb repressor complex (Muller et al. 2002; Schwartz et al. 2006), and H3K9me3 for heterochromatin (Padeken et al. 2022). ChIP-seq data on occupancy by the structural protein CTCF associated with insulation (West et al. 2002) were available in many cell types. Bulk RNA-seq data were collected for all cell types.
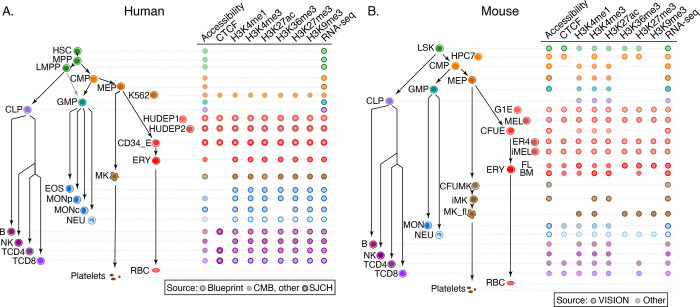
Figure 1. Cell types and datasets used for systematic integration of epigenetic features of blood cells.
(A) The tree on the left shows the populations of stem, progenitor, and mature blood cells in human. The diagram on the right indicates the epigenetic features and transcriptomes for which genome-wide datasets were collected, with distinctive icons for the major sources of data, specifically the Blueprint project (Martens and Stunnenberg 2013; Stunnenberg et al. 2016), Corces et al. (2016), abbreviated CMB, and St. Jude Children’s Research Hospital (SJCRH, Cheng et al. 2021; Qi et al. 2021). (B) Cell types and epigenetic data sets in mouse, diagrammed as for panel A. Sources were described in Xiang et al. (2020b) and Supplementary Table S1. Abbreviations for blood cells and lines are: HSC = hematopoietic stem cell, MPP = multipotent progenitor cell, LMPP = lymphoid-myeloid primed progenitor cell, CMP = common myeloid progenitor cell, MEP = megakaryocyte-erythrocyte progenitor cell, K562 = a human cancer cell line with some features of early megakaryocytic and erythroid cells, HUDEP = immortalized human umbilical cord blood-derived erythroid progenitor cell lines expressing fetal globin genes (HUDEP1) or adult globin genes (HUDEP2), CD34_E = human erythroid cells generated by differentiation from CD34+ blood cells, ERY = erythroblast, RBC = mature red blood cell, MK = megakaryocyte, GMP = granulocyte monocyte progenitor cell, EOS = eosinophil, MON = monocyte, MONp = primary monocyte, MONc = classical monocyte, NEU = neutrophil, CLP = common lymphoid progenitor cell, B = B cell, NK = natural killer cell, TCD4 = CD4+ T cell, TCD8 = CD8+ T cell, LSK = Lin-Sca1+Kit+ cells from mouse bone marrow containing hematopoietic stem and progenitor cells, HPC7 = immortalized mouse cell line capable of differentiation in vitro into more mature myeloid cells, G1E = immortalized mouse cell line blocked in erythroid maturation by a knockout of the Gata1 gene and its subline ER4 that will further differentiate after restoration of Gata1 function in an estrogen inducible manner (Weiss et al. 1997), MEL = murine erythroleukemia cell line that can undergo further maturation upon induction (designated iMEL), CFUE = colony forming unit erythroid, FL = designates ERY derived from fetal liver, BM = designates ERY derived from adult bone marrow, CFUMK = colony forming unit megakaryocyte, iMK = immature megakaryocyte, MK_fl = megakaryocyte derived from fetal liver.
These epigenomic data from multiple sources differed in many properties, including sequencing depth, fraction of reads on target, and signal-to-noise ratio (Xiang et al. 2020a). To reduce the impact of these technical differences, we used an improved version of the S3norm method, called S3V2, to normalize and denoise all data sets. S3V2 (Xiang et al. 2021) was designed to match the ranges of both peak and background signal intensities and their variances across epigenetic datasets (see Methods). This adjustment produced a stronger and more consistent correlation by feature across cell types, indicating that the denoising and normalization were effective (Supplementary Figure S1).
Extracting epigenetic states by modeling epigenomic information jointly in human and mouse
A powerful class of methods for integrative analysis of epigenomes involves statistical modeling to discover frequently occurring combinations of epigenetic features, comprising epigenetic states, and then assigning DNA intervals (often of 200 bp) to those states to produce regulatory annotations across the genome. These segmentation and genome annotation (SAGA) methods (Libbrecht et al. 2021) include ChromHMM (Ernst and Kellis 2012), SegWay (Hoffman et al. 2012), and IDEAS (Zhang et al. 2016; Zhang and Hardison 2017). We employed IDEAS because its simultaneous two-dimensional modeling along chromosomes and across cell types provides a consistent and well-resolved annotation while leveraging epigenetic information from related cell types when assigning states in cell types with missing data (Zhang and Mahony 2019). Moreover, its Bayesian statistical framework allows the incorporation of epigenetic models from different studies and even from different species.
We conducted an iterative, joint training on the epigenomic data of both human and mouse blood cells to ensure that the same set of epigenetic states was learned and applied for both species. Previous studies showed that the epigenetic states uncovered by SAGA methods such as ChromHMM (Ernst and Kellis 2012) were similar in both mouse and human (Yue et al. 2014; Roadmap_Epigenomics_Consortium et al. 2015; Gorkin et al. 2020). Indeed, when the epigenomic data from mouse or human were used separately as input to IDEAS, most of the resulting states were shared between the species (Supplementary Figure S2). The states specific to human or mouse were often similar to the shared states but with small variations in one or more epigenetic features; no clear evidence for a state specific to either species was found. The joint modeling began with a search for epigenetic states that exhibit similar combinatorial patterns across different epigenetic features in both human and mouse (see Methods), which we defined as reproducible epigenetic states. This search led to the retention of 27 reproducible states (Figure 2A, steps 1 and 2). Then, to analyze the full epigenomic information in each species, we used these 27 states as prior information to sequentially run the IDEAS genome segmentation on the human and mouse data sets, updating the signal compositions of the epigenetic states after each run (Figure 2A: steps 3a and 3b). Two heterogenous states, identified by their coefficient of variance (Supplementary Figure S3), were removed, because such states previously had been observed to be composites of low frequency states (Xiang et al. 2020b). This process produced a model with 25 epigenetic states (Figure 2B). As observed in previous IDEAS modeling studies (Xiang et al. 2020b), the states capture combinations of epigenetic features characteristic of regulatory elements such as promoters and enhancers, transcribed regions, repressed regions marked by either Polycomb (H3K27me3) or heterochromatin (H3K9me3), including states that differ quantitatively in the contribution of particular features to each state. For example, H3K4me1 is the predominant component of states E1 and E, but E1 has a lower contribution of that histone modification. The proportions of the genomes covered by each state were similar in human and mouse (Figure 2B). The largest portions of the genomes were in the quiescent state 0, characterized by no significant detectable contribution from any feature.
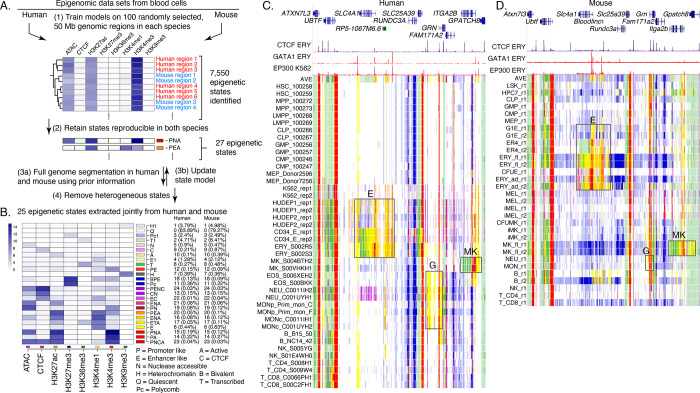
Figure 2. Genome segmentation and annotation jointly between human and mouse using IDEAS.
(A) Workflow for joint modeling. (1) Initial epigenetic states from 100 randomly selected regions separately in human and mouse hematopoietic cell types were identified in IDEAS runs. (2) States that were reproducible and shared in both species (see Methods) were retained. (3a and 3b) The profile of epigenetic feature contribution to each of the reproducible states was sequentially refined by applying IDEAS across the full genomes of human and of mouse, updating the state model after each IDEAS run. (4) Two heterogeneous states were removed to generate the final joint epigenetic states in the two species. (B) The 25 joint epigenetic states for human and mouse hematopoietic cell types. The average signal of the epigenetic features for each state are shown in the heatmap. The corresponding state colors, the state names based on the function, and the average proportions of the genome covered by each state across cell types are listed on the right-side of the heatmap. (C) Annotation of epigenetic states in a large genomic interval containing SLC4A1 and surrounding genes across human blood cell types. The genomic interval is 210kb, GRCh38 chr17:44,192,001–44,402,000, with gene annotations from GENCODE V38. Binding patterns for selected transcription factors are from the VISION project ChIP-seq tracks (CTCF and GATA1 in adult erythroblasts, signal tracks from MACS, track heights 100 and 80, respectively) or from the ENCODE data portal (EP300 in K562 cells, experiment ENCSR000EGE, signal track is fold change over background, track height is 50). The epigenetic state assigned to each genomic bin in the different cell types is designated by the color coding shown in panel (B). The replicates in each cell type examined in Blueprint are labeled by the id for the donor of biosamples. Genes and regulatory regions active specifically in erythroid (E), granulocytes (G), and megakaryocytes (MK) are marked by gray rectangles. (D) Annotation of epigenetic states in a large genomic interval containing Slc4a1 and surrounding genes across mouse blood cell types. The genomic interval is 198kb, mm10 chr11:102,290,001–102,488,000, with gene annotations from GENCODE VM23. Binding patterns for selected transcription factors are from the VISION project ChIP-seq tracks (CTCF in adult erythroblasts, GATA1 and EP300 from the highly erythroid fetal liver, signal tracks from MACS, track heights 200, 200, and 150, respectively; the EP300 track was made by re-mapping reads from ENCODE experiment ENCSR982LJQ). The tracks of epigenetic states and highlighted regions are indicated as in panel (C).
All genomic regions of each hematopoietic cell type in both human and mouse were then assigned to one of the 25 states from the IDEAS joint modeling. The resulting annotation of blood cell types provides a concise view of the epigenetic landscape and how it changes across cell types, using labels and color conventions consistently for human and mouse. The value of this concise view can be illustrated in the orthologous genomic intervals containing genes expressed preferentially in different cell lineages as well as genes that are uniformly expressed (Figure 2C, D). The gene SLC4A1/Slc4a1 encodes the anion transporter in the erythrocyte plasma membrane, and it is expressed in the later stages of erythroid maturation (Dore and Crispino 2011). This gene, its flanking regions, and a non-coding gene located upstream (to its right, Bloodlinc in mouse), were assigned to epigenetic states indicative of enhancers (yellow and orange), promoters (red), and transcribed regions (green) in erythroid cell types in both human and mouse, with indications of stronger activation in the more mature erythroblasts (region boxed and labeled E in Figure 2 C, D). Several of the elements assigned as enhancer-like were also occupied by the transcription factor GATA1 (Xu et al. 2012; Pimkin et al. 2014) and co-activator EP300 (ENCODE datasets ENCSR000EGE and ENCSR982LJQ), which are associated with erythroid enhancers. The GRN/Grn gene, encoding the granulin precursor protein, is more broadly expressed but with high levels in granulocytes and monocytes (Jian et al. 2013). This gene and upstream regions (to its left) were assigned to epigenetic states indicative of enhancers and promoters in those cell types, with a larger region in enhancer-like states in human cells (region boxed and labeled G). The ITGA2B/Itga2b gene, encoding the alpha 2b subunit of integrin, is highly expressed in mature megakaryocytes (van Pampus et al. 1992; Pimkin et al. 2014). Again, this gene and upstream regions (to its right in Figure 2) were assigned to epigenetic states indicative of enhancers and promoters in mature megakaryocytes, along with a transcribed state in the gene body (region boxed and labeled MK). Genes expressed in all the blood cell types, such as UBTF/Ubtf, were assigned to active promoter states and transcribed states across the cell types. These concise summaries of the epigenetic landscapes across cell types showed patterns of activity in chromatin for both differentially and uniformly expressed genes in blood cells, along with indications of potential regulatory regions. Furthermore, the consistent state assignments from the joint modeling revealed similar epigenetic landscapes in human and mouse.
While these resources are useful, some limitations should be kept in mind. For example, IDEAS uses data from similar cell types to improve state assignments in cell types with missing data, but the effectiveness of this approach may be impacted by the pattern of missing data. In particular, the epigenetic data on human stem and progenitor cell types was largely limited to ATAC-seq data, whereas histone modification data and CTCF occupancy was available for the analogous cell types in mouse (Figure 1). Thus, the state assignments for epigenomes in human stem and progenitor cells may not be as robust as those for similar cell types in mouse. Another limitation is the broad range of quality in the data sets that cannot be completely adjusted by normalization, which leads to over- or under-representation of some epigenetic signals in some cell types. For example, state 5 (light mauve) for low ATAC-seq signal was prominent in human HUDEP cells, neutrophils, and K562 cells (Figure 2C). The abundance of this state assignment may result from stronger ATAC-seq signals overall in the data sets from those cell types, leading to some regions with low ATAC-seq signal outside the peak regions that were assigned to state 5. Despite the limitations in the input data, the annotation of blood cell epigenomes after normalization and joint modeling of epigenetic states produces a highly informative painting of the activity and regulatory landscapes across the genomes of human and mouse blood cells.
Identification of candidate cis-regulatory elements in human and mouse
We define a candidate cis-regulatory element, or cCRE, as a DNA interval with a high signal for chromatin accessibility in any cell type (Xiang et al. 2020b). When peaks of accessibility are called independently on different cell types and then combined across cell types, the genomic intervals inferred as peaks can enlarge excessively unless special procedures are employed to prevent expansion (Meuleman et al. 2020; The_ENCODE_Project_Consortium et al. 2020). We reasoned that this expansion could be avoided by using both a combination of all the chromatin accessibility signals and the original data for each cell type as input for modeling across all these datasets to call peaks. We utilized a version of the IDEAS methodology for this purpose, running it in the signal intensity state (IS) mode on ATAC-seq and DNase-seq signals only (Xiang et al. 2021), in contrast to the previously described epigenetic state mode used for integrating data on multiple epigenetic features. The chromatin accessibility signals for each replicate of each cell type plus a track of combined average signal were modeled to define discrete signal intensity states and assign them to all the epigenomes. Genomic intervals in the higher intensity states were called as peaks of chromatin accessibility, following a hierarchical process to ensure that the collection included both peaks present in many cell types as well as those in a single cell type (Figure 3A, also see Methods). The preference given to the peaks in the average signal track helped prevent excessive lengthening of the peak calls after combining them.
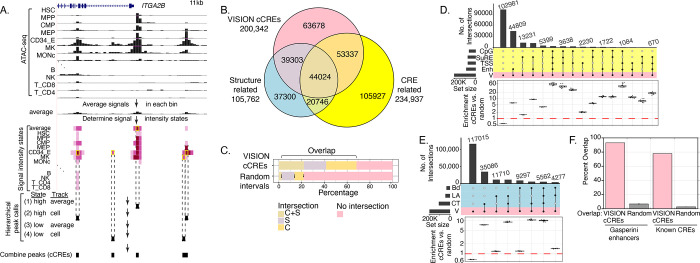
Figure 3. Predicting cCREs in the VISION project and comparisons with other catalogs.
(A) Method for calling cCREs using S3V2-IDEAS in the IS mode. The normalized ATAC-seq signals (expressed as the negative log10 p-value for fitting a negative binomial distribution, signal range 0–10, 200bp bins) are shown for a selected subset of the 39 human biosamples plus the average signal track in an 11kb genomic interval around the transcription start site (TSS) of the ITGB2B gene is shown (GRCh38 chr17:44,384,001–44,395,000). The signal intensity states learned by IDEAS in the IS mode are shown as shades of violet (state 0 is white, darker shades represent higher signal states). Genomic intervals in high signal states were called as peaks (yellow rectangles) in a four-step hierarchical process designed to limit the peak calls to local maxima while also finding cell type-specific peaks (see Methods). Peaks in this genomic region illustrate calls at steps 1, 2, and 4 of the hierarchical process. Panels B-F present intersections of human VISION cCREs with genome-wide datasets of structural and CRE-related elements. (B) The Venn diagram displays the result of intersecting VISION cCREs with a combined superset of elements associated with nuclear structure (CTCF OSs, loop anchors, and boundaries) and with a combined superset of DNA intervals associated with cis-regulatory elements (CREs). (C) The proportions of cCREs and randomly selected, matched sets of intervals in the overlap categories are compared in the bar graph (right). For the random sets, the bar shows the mean, and the dots show the values for each of ten random sets. (D) A higher resolution view (an UpSet plot) of the intersections of VISION cCREs with the four groups of CRE-related elements, specifically enhancer-related (Enh), transcription start sites (TSS), Survey of Regulatory Elements (SuRE), and CpG islands (CpG). The enrichment for the cCRE overlaps compared to those in randomly selected, matched sets of intervals are shown in the boxplots below each overlap subset, with dots for the enrichment relative to individual random sets. (E) Overlaps and enrichments of VISION cCREs for three sets of structure-related elements, specifically CTCF OSs (CT), loop anchors (LA), and TAD boundary elements. (F) Overlaps of VISION cCREs with two sets of experimentally determined blood cell cCREs.
We employed the same peak-calling procedure for the blood cell epigenomes of human as well as mouse, resulting in 200,342 peaks of chromatin accessibility for human blood cell types and 96,084 peaks for mouse blood cell types. For comparison, we also called peaks on the human ATAC-seq data using MACS3 (Zhang et al. 2008), which generated a larger number of peaks compared to IDEAS-IS (277,064 versus 200,342), but those additional peaks tended to have low signal, and they had less enrichment for overlap with other function-related genomic datasets (Supplementary Figure S4). Unlike the set of accessibility peaks used in earlier work (Xiang et al. 2020b), which were called using the HOMER program (Heinz et al. 2010), all of the IDEAS-IS peaks were in a non-quiescent state in at least one cell type. Thus, the sets of IDEAS-IS peaks comprised the sets of VISION cCREs. The larger number of cCREs called in human than in mouse results at least in part from the very high signal in chromatin accessibility data from some human cell lines (HUDEP1, HUDEP2, and K562) and cell types (e.g., monocytes).
The ENCODE Project has released regulatory element predictions in a broad spectrum of cell types in the Index of DHSs (Meuleman et al. 2020) and the SCREEN cCRE catalog (The_ENCODE_Project_Consortium et al. 2020), using data that were largely orthogonal to those utilized for the VISION analyses. Almost all the VISION cCRE calls in human blood cells were included in the regulatory element predictions from ENCODE (Supplementary Figure S5A), supporting the quality of the VISION cCRE calls. Furthermore, as expected from its focus on blood cell types, the VISION cCRE catalog shows stronger enrichment for CREs and other indicators of regulatory function in blood cells (Supplementary Figure S5B).
cCRE co-localization with orthogonal features related to structure or function
Having generated catalogs of cCREs along with an assignment of their epigenetic states in each cell type, we sought to gain additional information by connecting the VISION cCREs to other, orthogonal (not included in VISION predictions) datasets of DNA elements implicated in gene regulation or in chromatin structure and architecture (termed structure-related) (Figure 3B, Supplementary Figure S6, Methods). About two-thirds (136,664 or 68%) of the VISION human cCREs overlapped with elements in the broad groups of CRE-related and structure-related elements (Figure 3B, C). Specifically, 97,361 cCREs overlapped with CRE-related elements and 83,327 cCREs overlapped with structure-related elements, with 44,024 cCREs overlapping elements in both categories. In contrast, ten sets of randomly chosen DNA intervals, matched in length and GC-content with the human cCRE list, showed much less overlap (about 22%) with the orthogonal sets of elements (Figure 3C). Of the CRE-related superset, the enhancer-related group of datasets contributed the most overlap with VISION cCREs, followed by SuRE peaks, which measure promoter activity in a massively parallel reporter assay (van Arensbergen et al. 2017), and CpG islands (Figure 3D). The extent of these overlaps was much higher (most ranged from 4- to 60-fold) than that observed for overlaps with the random matched intervals, with particularly high enrichments for cCREs overlapping with multiple features (Figure 3D). Of the structure-related superset, the set of CTCF occupied segments (OSs) contributed the most overlap, followed by chromatin loop anchors, again with high enrichment relative to overlaps with random matched sets (Figure 3E). Considering the VISION cCREs that intersect with both structure- and CRE-related elements, the largest group are those that overlap with enhancers and CTCF OSs, followed by enhancers and loop anchors, and then promoter-like elements and CTCF OSs (Supplementary Figure S7). Furthermore, the VISION cCREs capture known blood cell CREs (Supplementary Table S3) and CREs demonstrated to impact a specific target gene in a high throughput analysis (Gasperini et al. 2019) (Figure 3F).
The intersections with orthogonal, function- or structure-related elements lend strong support for the biological significance of the VISION cCRE calls and add to the annotation of potential functions for each cCRE. Those annotations for both human and mouse cCREs are recorded in our cCRE database with a web-based query interface, available at our website (http://usevision.org; cCRE_db).
Actuation, epigenetic states, and regulatory impact of cCREs during differentiation
Actuation and state assignments of cCREs during differentiation
We examined the epigenetic states assigned to cCREs to determine the major trends in changes for all cCREs across cell types and to map the cCREs that show changes in activity during differentiation. For clarity, we emphasize that while a cCRE is defined by epigenetic features in specific cell types, it is a DNA element present in all cell types. Inferences about the activity of a cCRE in a given cell type are based on whether the cCRE was actuated and which epigenetic state was assigned to the actuated cCRE. The cCREs in peaks of chromatin accessibility were considered to be actuated, but they can be in states associated with activation (e.g., enhancer-like or promoter-like) or repression (associated with polycomb or heterochromatin). For both activation and repression studied here, the epigenetic states result from dynamic histone modification processes. Most, but not all, actuated cCREs were in states associated with activation.
Changes in predicted activity of epigenetic states of individual cCREs during differentiation
The epigenetic state assignments represent a systematic integration of all the available epigenetic features for a genomic element, and therefore, we reasoned that they should reflect potential biological functions more comprehensively compared to individual epigenetic features. While previous work has used signals from single or multiple individual features such as chromatin accessibility or histone modifications in regression modeling to explain gene expression (e.g., Karlić et al. 2010; Dong et al. 2012), we used the more comprehensive integration of epigenetic features in state assignments in a multivariate regression model (see Methods) to estimate the impact of each state on the expression of local genes. That impact was captured as β coefficients showing the expected strong positive impact for promoter and enhancer associated states and a negative impact from heterochromatin and polycomb states (Figure 4A). The β coefficients provide a quantitative estimate of the regulatory impact of each state. In contrast to using the state assignments as categorical annotations, which is commonly done in SAGA methods, our regression modeling has mapped this set of categorical states into a continuous variable. The differences in the values for β coefficients between states provides an estimate of the change in regulatory impact as a cCRE shifts between states during differentiation (difference matrix to the left of the β coefficient values in Figure 4A).
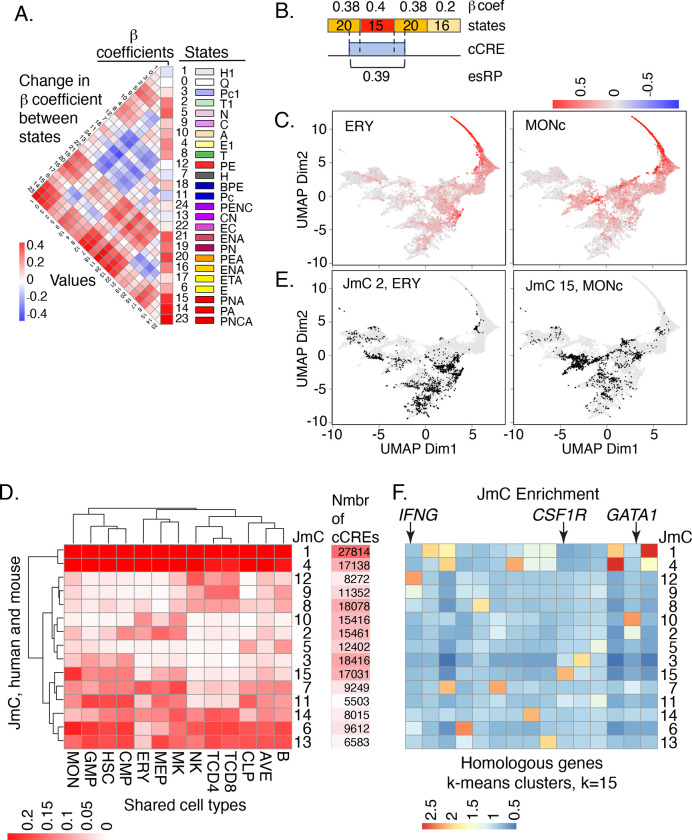
Figure 4. Beta coefficients of states, esRP scores of cCREs, and joint HM metaclusters of cCREs based on esRP scores.
(A) Beta coefficients and the difference of beta coefficients of the 25 epigenetic states. The vertical columns on the right show the beta coefficients, the state ID, state color, and the state names based on the function of all 25 joint epigenetic states. The triangular heatmap shows the difference of the beta coefficients between two states in the right columns. Each value in the triangle heatmap shows the beta coefficient of state on top minus the beta coefficient of the state below based on the order of state in the right columns. (B) An example of calculating esRP score for a cCRE in a cell type based on the beta coefficients of states. For a cCRE covering more than one 200bp bin, the esRP equals the weighted sum of beta coefficients of states that covers the cCRE, where the weights are the region covered by different states. (C) UMAP of cCREs based on their esRP scores across all cell-types. The points are colored by the esRP scores in the designated cell type. (D) The average esRP score of all cCREs in JmC across all cell-types shared by both human and mouse. The right column shows the number of human cCREs in each JmC. (E) UMAP of cCREs based on their esRP scores across all cell-types, with the points colored by the binary label indicating whether a cCRE belongs to the specified JmC. (F) The average enrichment of JmCs in 15 homologous gene clusters. The genes are clustered based on the JmCs’ enrichments by K-means.
The β coefficient values were used to generate an epigenetic state Regulatory Potential (esRP) score for each cCRE in each cell type, calculated as the β coefficient values for the epigenetic states assigned to the cCRE weighted by the coverage of the cCRE by each state (Figure 4B). These esRP scores were the basis for visualizing the collection of cCREs and how their regulatory impact changed across differentiation. Starting with the large matrix of about 200,000 human cCREs with esRP scores for each cell type and replicate, we employed the dimensional reduction visualization method UMAP to project all cCREs onto a plane that keeps together the cCREs with similar esRP scores across cell types (Figure 4C). The resulting image showed multiple clusters populated with cCREs (dots) that are colored to visualize their activity in individual cell types, with the color determined by the esRP score in that cell type. The darker red dots indicate cCREs more strongly implicated in gene activation in that cell type, as illustrated for erythroblasts (ERY) and monocytes (MON) in Figure 4C. UMAPs annotated by esRP scores for all cell types in both human and mouse along with movies showing the changes in estimated impact of cCREs across human hematopoietic differentiation are provided in the Supplementary Materials and on our VISION website (http://usevision.org). These annotated UMAP projections revealed both cCREs active in all cell types, such as the long arc of red cCREs in the upper right of the graphs, as well as shifts in cCRE activation as cells differentiate.
The esRP scores across cell types and replicates were also used to cluster the cCREs. In previous work, cCREs have been clustered based on levels of chromatin accessibility or histone modifications across cell types (e.g., Heintzman et al. 2009; Meuleman et al. 2020), whereas in our approach the clustering is based on estimated regulatory impact of the cCREs. Focusing on the esRP scores in 12 cell types shared between human and mouse along with the average across cell types, we identified clusters jointly in both species. We conducted a series of clustering steps to find reproducible K-means clusters for the combined human and mouse cCREs, identify the clusters shared by cCREs in both species, and then further group those shared K-means clusters hierarchically to define fifteen joint metaclusters (JmCs) (see Methods and Supplementary Figure S8). Each cCRE in both mouse and human was assigned to one of the fifteen JmCs, and each JmC was populated with cCREs from both mouse and human.
These JmCs establish discrete categories for the cCREs based on the cell type distribution of their regulatory impact (Figure 4D). Shading the cCRE UMAPs by metacluster assignment revealed well-defined clusters of cCREs within the zones of active cCREs visualized from the esRP scores (Figure 4E). The clusters of cCREs with high esRP scores across cell types were highly enriched for promoter elements (Supplementary Figure S9A). The cell type-restricted clusters of cCREs showed enrichment both for selected enhancer catalogs and for functional terms associated with those cell types (Supplementary Figures S9A and S9B, respectively). Furthermore, clustering of genes by the JmC assignments of cCREs in a 100kb interval centered on their TSS revealed a strong enrichment for JmCs with high activity in the cell type(s) in which the genes are expressed. Examples include IFNG showing enrichment for JmC 12, which has high esRP scores in T and NK cells, CSF1R showing enrichment for JmC 15, which has high scores in monocytes, and GATA1 showing enrichment for JmC 10, which has high scores in erythroid cells and megakaryocytes (Figure 4F).
In summary, we show that the β coefficients and esRP scores provide valuable estimates of regulatory impacts of states and cCREs, respectively. The UMAP visualization portrays the activation and repression of cCREs during differentiation accompanied by other cCREs with an invariant epigenetic state. The esRP-driven joint metaclusters provide refined subsets of cCREs that should be highly informative for investigating cell type-specific and general functions of cCREs. The cCRE annotations are organized into a database for further investigation at our website (http://usevision.org; cCRE_db). As a complementary approach to systematic integration of epigenetic features and RNA data across cell types, we also built self-organizing maps (Supplementary Figure S10, Jansen et al. 2019) and provided an interactive viewing tool for them on our website (http://usevision.org; SOM).
Motif enrichment in human and mouse cCREs
We examined the sets of cCREs in each JmC to ascertain enrichment for transcription factor binding site (TFBS) motifs because these enriched motifs suggest the families of transcription factors that play a major role in regulation by each category of cCREs. Furthermore, having sets of cCREs determined and clustered for comparable blood cell types in human and mouse provided the opportunity to discover which TFBS motifs were shared between species and whether any were predominant in only one species. Our examination of cCREs grouped by their similar profiles of esRP scores across cell types differed from previous analyses in which all cCREs active in a specific cell type or those that are distinctive for a cell type were examined (e.g., Neph et al. 2012; Vierstra et al. 2020). The JmCs bring out not only sets of cCREs that are distinctive for a lineage, but also sets of cCREs with a broader distribution of activity.
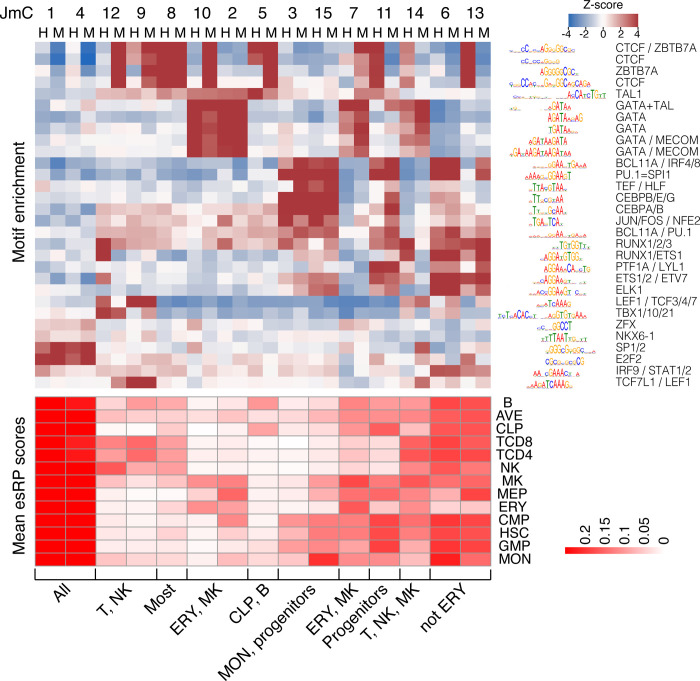
To find TFBS motifs associated with each JmC, we calculated enrichment for all non-redundant motifs in the Cis-BP database (Weirauch et al. 2014) using Maelstrom from GimmeMotifs (Bruse and van Heeringen 2018). The results confirmed previously established roles of specific TFs in cell lineages and showed little evidence for novel motifs (Figure 5). JmCs 2 and 10, which have high esRP scores in progenitor and mature cells in the erythroid and megakaryocytic lineages, were enriched in TFBS motifs for the GATA family of transcription factors, as expected for the known roles of GATA1 and GATA2 in this lineage (Blobel and Weiss 2009; Fujiwara et al. 2009). This pattern was also observed for JmC 7, which has high esRP scores in a broader range of cell types including erythroid and megakaryocytic cells. JmC 14 was also enriched for the GATA motif, as expected for the role of GATA3 in natural killer (NK) and T cells (Rothenberg and Taghon 2005). The cCREs in JmCs 9 and 12, which also have high esRP scores in NK and T cells, were enriched in motifs for the known lymphoid transcription factors TBX21, TCF7L1, and LEF1 (Chi et al. 2009). The cCREs in JmCs 3 and 15 were active in progenitor cells and monocytes, and they were enriched in the binding site motifs for myeloid-determining transcription factors CEBPA and CEBPB (Graf and Enver 2009) and the myeloid transcription factor PU.1 (Tenen et al. 1997). Broadly active cCREs in JmCs 1 and 4 were enriched in TFBS motifs for promoter-associated transcription factors such as E2F2 and SP1 (Dynan and Tjian 1983; Kaczynski et al. 2003). These patterns of motif enrichments in the JmCs fit well with the expectations from the previous studies of activities of transcription factors in the various lineages of blood cells, and thus, they lend further credence to the value of the cCRE calls and the JmC groupings for further studies of regulation in the blood cell types.
Figure 5. Motifs enriched in joint metaclusters.
The top heatmap shows the enrichment of motifs in the cCREs in each JmC in human (H) and mouse (M) as a Z-score. The logo for each motif is given to the right of the heat map, labeled by the family of transcription factors that recognize that motif. The heatmap below is aligned with the motif enrichment heatmap, showing the mean esRP score for the cCREs in each JmC for all the common cell types examined between human and mouse. A summary description of the cell types in which the cCREs in each JmC are more active is given at the bottom.
The genome-wide collection of cCREs across many blood cell types in human and mouse provided an opportunity for an unbiased and large-scale search for indications of transcription factors that may be active in one species for a shared cell type. Prior studies of transcription factors have shown homologous transcription factors used in analogous cell types across species (e.g., Carroll 2008; Noyes et al. 2008; Schmidt et al. 2010; Cheng et al. 2014; Villar et al. 2014), but it is not clear if there are significant exceptions. In our study, we found that for the most part, the motif enrichments were quite similar between the human and mouse cCREs in each JmC. Note that these similarities were not forced by requiring sequence matches between species; the cCREs were grouped into JmCs based on their pattern of activity, as reflected in the esRP scores, across cell types, not by requiring homologous sequences. This similarity between species indicates that the same transcription factors tend to be active in similar groups of cell types in both mouse and human.
The enrichment of TFBS motifs for CTCF and ZBTB7A presented some potential exceptions to the sharing of motifs across species. The cCREs in JmC 8 showed the expected strong enrichment for these motifs in both human and mouse, with little enrichment for binding site motifs of other TFs (Figure 5). These cCREs had modest regulatory impact, as estimated by esRP scores, across most cell types, suggesting the hypothesis that cCREs in JmC 8 may consist of CTCF-bound sites that are not involved in gene activation, such as insulators. Indeed, examination of ChIP-seq results showed that the cCREs in JmC 8 were enriched for CTCF occupancy and overlap with loop anchors (Supplementary Table S4). In contrast, the cCREs in several JmCs were enriched for CTCF and ZBTB7A motifs only in mouse (JmCs 12, 9, and 10) or only in human (JmCs 13 and 14). In these cases, the cCREs were also enriched for binding sites for other TFs, with those other motifs enriched in the cCREs from both species. The frequency of occupancy by CTCF in the cCREs in these latter JmCs corresponded well with the enrichments for the motifs (Supplementary Table S4). A parallel analysis of the cCREs in human and mouse JmCs by the multi-label discriminative motif-finder SeqUnwinder (Kakumanu et al. 2017) did uncover enrichment for some apparently species-specific motifs, but the magnitude of enrichment was limited (Supplementary Figure S11).
In summary, after grouping the cCREs in both human and mouse by their inferred regulatory impact across blood cell in a manner agnostic to DNA sequence or occupancy by TFs, the enrichment for TFBS motifs within those groups recapitulated known activities of TFs both broadly and in specific cell lineages. The results also showed considerable sharing of inferred TF activity in both human and mouse, as expected. Indications of preferential usage of CTCF and/or ZBTB7A in human or mouse in a context with other TFs were observed, which could serve as the basis for more detailed studies in the future.
Enrichment of genetic variants for blood cell related traits in the human cCRE collection
Most genetic variants associated with complex phenotypes occur in noncoding regions of the genome (Hindorff et al. 2009), and candidate regulatory elements are strongly enriched for such variants (Maurano et al. 2012; The_ENCODE_Project_Consortium 2012). Thus, collections of high quality predictions of CREs could provide insights into the functional mechanisms by which noncoding variants mediate phenotypic variation (Hardison 2012). We reasoned that our collection of cCREs would be particularly informative for interpreting non-coding variants influencing blood cell traits and blood diseases, and therefore, we examined the overlap between the human VISION cCREs and several databases of phenotype-associated variants. Our initial examination of variants from the NHGRI-EBI GWAS Catalog (Buniello et al. 2019) associated with blood cell traits showed that they overlapped with the VISION cCREs much more frequently than with random genomic intervals, and more variants associated with traits of specific cell types overlapped with the cCREs actuated in those cell types (Supplementary Figure S12).
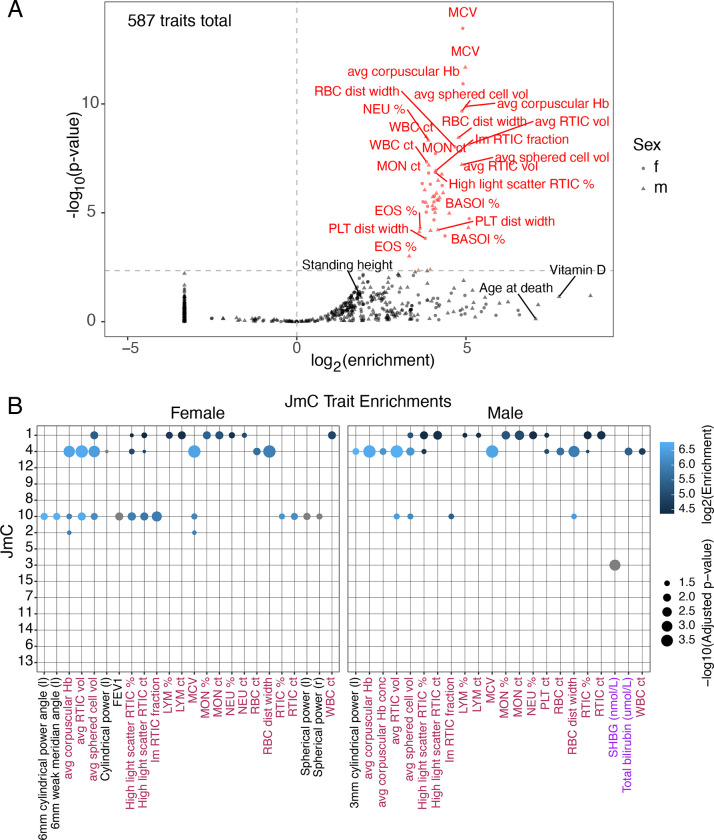
When measuring such enrichments from GWAS data, it is important to consider the haplotype structure of human genomes, whereby association signals measured at assayed genetic markers likely reflect an indirect effect driven by linkage disequilibrium (LD) with a causal variant (that may or may not have been genotyped). Stratified linkage disequilibrium score regression (sLDSC, Finucane et al. 2015) offers one principled approach to account for LD structure and estimate the proportion of heritability of each trait explained by a given genomic annotation. We applied sLDSC to quantify the enrichment of heritability in 587 traits from the UK Biobank (UKBB) GWAS (Ge et al. 2017 and http://www.nealelab.is/uk-biobank/) within the VISION cCREs relative to the rest of the genome. GWAS were conducted separately in males and females and as such, the data include 292 unique traits with GWAS results from both males and females and 3 traits with results only from males. Importantly, the traits included in our analysis encompassed 114 blood-related traits and 473 non-blood-related traits, allowing us to assess the specific relevance of the cCREs to regulation of blood-related versus other phenotypes. Of the 114 blood-related traits, 54 were “blood count” traits that measure properties including size and counts of specific blood cell-types, and 60 were “blood biochemistry” traits that measure lipid, enzyme, and other molecular concentrations within whole blood samples.
At a 5% FDR threshold, we discovered 53 traits for which cCREs were significantly enriched in heritability (Figure 6A). Strikingly, 52 (98%) of these traits were blood-related, and only a single trait was not related to blood (male pulse wave arterial stiffness index; 0.2% of non-blood related traits). Of the 52 significant blood-related traits, 50 were blood count traits, representing 93% of all UKBB blood count traits included in our analysis. The remaining 2 significant traits were blood biochemistry traits, specifically, the male and female glycated hemoglobin concentrations. The 58 non-enriched blood biochemistry traits were from screenings of metabolites, proteins, and enzymes that were not produced in blood cells, but rather by the liver (e.g., albumin, alkaline phosphatase, alanine aminotransferase, apolipoproteins, aspartate aminotransferase, bilirubin, urea, cholesterol), kidney (e.g., creatinine), or other organs. While these were labeled as blood-related traits by the UKBB, they are largely controlled by organs, tissues, and cell types that we did not assay when developing the VISION CRE annotation. These metrics and observations together lend support to the VISION CRE annotation being composed of informative genomic regions associated with regulation of genes involved in development of blood cell traits.
Figure 6. Enrichment of SNPs associated with blood cell traits from UK Biobank in VISION cCREs.
(A) Human cCREs are enriched in the heritability of blood-related traits. Results of the initial sLDSC analysis of all cCREs considered as a single annotation. The plot shows enrichment of the cCRE annotation in heritability of each trait on the x-axis, and the significance of the enrichment on the y-axis. The vertical dotted line indicates an enrichment of 1, and the horizontal dotted line delineates the 5% FDR significance threshold. Points and labels in red represent the traits for which there was significant enrichment of SNPs associated with the VISION cCREs. Traits with a negative enrichment were assigned an arbitrary enrichment of 0.1 for plotting and appear as the column of points at the bottom left of the plot. The shape of the point delineates the sex in which the GWAS analysis was performed for each trait. (B) Results of the JmC sLDSC analysis where each set of cCREs within a JmC was considered as a separate annotation. The plot lists a trait on the x-axis if any JmC had a significant enrichment for it. The labels for these traits are maroon for blood count traits, purple for blood biochemistry traits, and black for non-blood related traits. The plot lists the JmC on the y-axis. For a given JmC and trait combination, a dot is plotted if and only if there was an observed significant enrichment for that combination. Size of the dot reflects the significance of the enrichment, and the color of the dot reflects the size of the enrichment itself. Negative enrichments are colored gray. Panels separate the sex in which the GWAS analysis was performed for each trait.
We next sought to determine the enrichment of trait-associated SNPs in the subsets of VISION cCREs based on activity within groups of cell types, i.e., the joint metaclusters (JmCs). We re-analyzed the blood trait-associated variants by running sLDSC with fifteen separate annotations, each annotation defined by a JmC, and found five JmCs with significant results at a 5% FDR (Figure 6B). The cCREs more active in erythroid and megakaryocytic cells, i.e., those in JmCs 10 and 2, were significantly enriched for heritability of several blood traits, including many related to erythroid cells. Several of the enrichments were for cCREs in JmCs 1 and 4, which are active across all cell types examined (Figure 4E) and are themselves highly enriched for proximal regulatory regions such as promoters (Supplementary Figure S9A). While this result may suggest that many blood-trait associated variants were in proximal regulatory regions of genes with active epigenetic marks broadly present across blood cells, more study is needed to establish such a relationship. One caveat is that the large number of cCREs in JmCs 1 and 4 makes it more likely for them to overlap with any feature, and thus the large overlap with proximal regulatory regions could be separable, at least in part, from the overlap with trait-associated variants. Many of the JmCs showed no significant enrichment, perhaps reflecting a reduced power for JmCs comprising fewer cCREs.
We conclude that the VISION cCREs offer a valuable resource for further studies of genetic variants associated with complex phenotypes involving blood cells, especially those that may impact the phenotypes by altering gene regulation.
Evolution of sequence and inferred function of cCREs
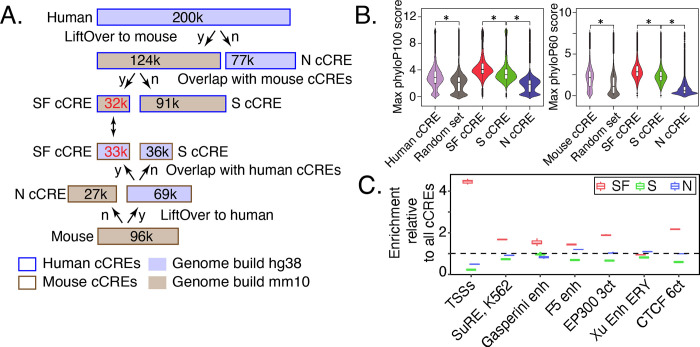
The human and mouse cCREs from blood cells were assigned to three distinct lines of evolution (Figure 7A). About one-third of the cCREs were present only in the reference species (39% for human, 28% for mouse), as inferred by the failure to find a matching orthologous sequence in whole-genome alignments with the other species. We refer to these as nonconserved (N) cCREs. Of the two-thirds of cCREs with an orthologous sequence in the second species (124,000 in human and 69,000 in mouse), slightly over 30,000 were also identified as cCREs in the second species. The latter cCREs comprise the set of cCREs conserved in both sequence and inferred function, which we call SF conserved (SF) cCREs. A similar number of cCREs in both species fall into the SF category. The degree of chromatin accessibility in orthologous SF cCREs was positively correlated between the two species (Supplementary Figure S13). The remaining cCREs (91,000 in human and 36,000 in mouse) were conserved in sequence but not in the inferred function as a regulatory element, and we call them S conserved (S) cCREs. The latter group could result from turnover of regulatory motifs or acquisition of different functions in the second species.
Figure 7. Evolutionary and epigenetic comparisons of cCREs.
(A) Workflow of generating three evolutionary categories for blood cell cCREs in human and mouse. N=nonconserved, S=conserved in sequence but not inferred function, SF=conserved in both sequence and inferred function as a cCRE, y=yes, n=no. (B) PhyloP scores for three evolutionary categories of cCREs in human and mouse. The maximum phyloP score for each genomic interval was used to represent the score for each cCRE. The distribution of phyloP scores for each group are displayed as a violin plot. The asterisk (*) over brackets indicates comparison for which the P values for Welch’s t-test is less than 2.2e-16. (C) Enrichment of SF-conserved human cCREs for TSSs. The number of elements in seven sets of function-related DNA intervals that overlap with the 32,422 SF human VISION cCREs was determined (bedtools), along with the number that overlap with three randomly selected subsets (32,422 each) from the full set of 200,342 human cCREs. The ratio of the number of function-related elements overlapping SF-cCREs to the number overlapping a randomly chosen subset of all cCREs gave the estimate of enrichment plotted in the graph. The mean for the three determinations of enrichment is indicated by the horizontal line for each set. Results are also shown for a similar analysis for the S and N cCREs.
The levels of cCRE sequence conservation across mammalian genomes differed among the evolutionary categories. We used the maximum phyloP score (Pollard et al. 2010) in each cCRE interval as an estimate of the level of sequence similarity across genome sequence alignments of 100 species with human as the reference (phyloP100) and across genome sequence alignments of 60 species with mouse as the reference (phyloP60). For both human and mouse, the distribution of phyloP scores for all cCREs was higher than those for ten sets of randomly chosen genomic intervals matched to the cCRE intervals in length and G+C content (Figure 7B; all ten random sets had distributions similar to the one shown; p-value < 2.2e-16, Welch two sample t-test). In both species, the distribution of phyloP scores for SF cCREs was significantly higher than the distribution for S cCREs (Figure 7B), which indicates that the preservation of inferred function was associated with more stringent evolutionary constraint on the SF cCREs. The N cCREs had a lower distribution of phyloP scores than other groups, as expected given the absence of orthologous DNA sequence in some clades of mammals.
The distributions of epigenetic states assigned to the blood cell cCREs in each of the three evolutionary categories were similar between human and mouse, but those distributions differed dramatically between evolutionary categories, with significantly more SF cCREs assigned to promoter-like states than were S or N cCREs (Supplementary Figure S14). Indeed, the SF cCREs tended to be close to or encompass the TSSs of genes, showing a substantial enrichment in overlap with TSSs compared to the overlap observed for all cCREs (Figure 7C). Many of the S and N cCREs were assigned to enhancer-like states (Supplementary Figure S14D), giving a level of enrichment for overlap with enhancer datasets comparable to that observed for the full set of cCREs (Figure 7C). These results indicated that the stringency of conservation of cCREs was related to their inferred function, and specifically they suggest stronger selection has been exerted on promoter-associated genomic intervals to conserve DNA sequences and preserve biological function.
Comparison of epigenetic states in homologous regions of human and mouse
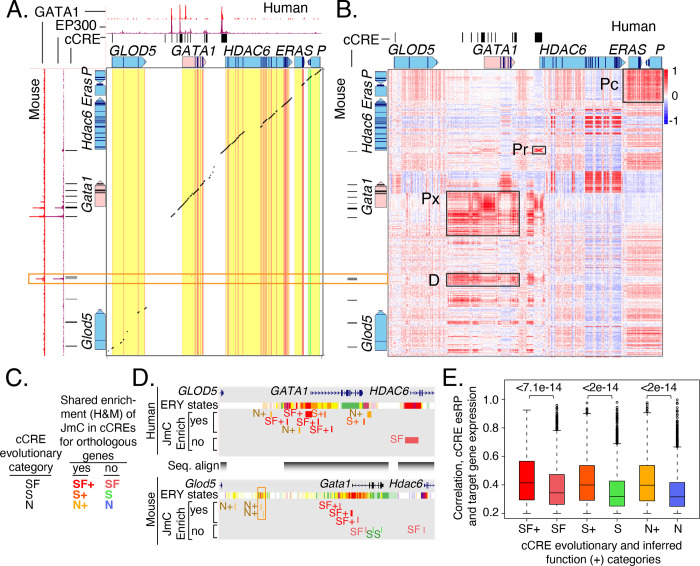
The consistent state assignments from the joint modeling facilitated comparisons between species. Such comparisons are particularly interesting for orthologous genes with similar expression patterns but some differences in their regulatory landscapes. For example, the orthologous genes GATA1 in human and Gata1 in mouse each encode a transcription factor with a major role in regulating gene expression in erythroid cells, megakaryocytes, and eosinophils (Ferreira et al. 2005), and they showed a strong correlation in expression levels across blood cell types in both species (Supplementary Figure S15). The genomic DNA sequences aligned between the orthologous human GATA1 and mouse Gata1 genes, including their promoters and proximal enhancers; the alignments continued through the genes downstream of GATA1/Gata1 (Figure 8A). An additional, distal regulatory element is located upstream of the mouse Gata1 gene, corresponding to the binding by GATA1 and EP300. However, this element was found only in mouse (Valverde-Garduno et al. 2004), and the DNA sequences of this upstream interval harboring the mouse regulatory element did not align between mouse and human except in portions of the GLOD5/Glod5 genes (Figure 8A). Thus, the interspecies sequence alignments provide limited information about this distal regulatory element, which led us to explore whether comparisons of epigenetic information would be more informative.
Figure 8. Epigenetic comparisons of regulatory landscapes and cCREs.
(A and B) DNA sequence alignments and correlations of epigenetic states in human GATA1 and mouse Gata1 genes and flanking genes. (A) Dot-plot view of chained blastZ alignments by PipMaker (Schwartz et al. 2000) between genomic intervals encompassing and surrounding the human GATA1 (GRCh38 chrX:48,760,001–48,836,000; 76kb) and mouse Gata1 (mm10 chrX:7,919,401–8,020,800; 101.4kb, reverse complement of reference genome) genes. The axes are annotated with gene locations (GENCODE), predicted cis-regulatory elements (cCREs), and binding patterns for GATA1 and EP300 in erythroid cells. (B) Matrix of Pearson correlation values between epigenetic states (quantitative contributions of each epigenetic feature to the assigned state) across 15 cell types analogous for human and mouse. The correlation is shown for each 200bp bin in one species with all the bins in the other species, using a red-blue heat map to indicate the value of the correlation. Axes are annotated with genes and cCREs in each species. (C and D) JmC enrichment tracks of cCREs at the human GATA1 and mouse Gata1 gene loci. A “+” sign assigned to a cCRE indicates that the JmC to which it belongs was enriched at the GATA1/Gata1 gene locus. (E) The correlation between the cCRE’s esRP score and the target gene expression level. The results for each set of cCREs are shown as box plots summarizing the distribution of correlations observed for all loci with orthologous protein-coding genes. The cCREs in each evolutionary category were separated into those that are members of the JmCs enriched for a gene locus (indicated by a +) or those that are not (labeled SF, S and N).
We compared the chromatin activity landscapes across species by utilizing the consistent assignment of epigenetic states in both human and mouse, without relying on DNA sequence alignment. In the large genomic regions (76kb and 101kb in the two species) encompassing the orthologous human GATA1 and mouse Gata1 genes and surrounding genes, we computed the correlation between the epigenetic state assignments of each bin across cell types in one species and that in the other species for all the bins (Supplementary Figure S16 and Methods). This all-versus-all comparison yielded a matrix of correlation values showing similarities and differences in profiles of epigenetic states in the two species (Figure 8B). The conserved promoter and proximal enhancers of the GATA1/Gata1 genes were highly correlated in epigenetic states across cell types between the two species, clustering largely along a diagonal through the center of the matrix that encompassed the aligning DNA sequences (labeled Px in Figure 8B). In contrast, whereas the mouse-specific distal regulatory element gave no signal in the DNA sequence alignment, the epigenetic states annotating it presented high correlations with active epigenetic states in the human GATA1 locus (labeled D in Figure 8B).
This comparison of epigenetic state profiles across cell types provided a means to categorize cCREs across species that did not require a match in the underlying genomic DNA sequence. We used that information to identify cCREs that may be playing a similar role in regulation in both species despite their lack of conservation in DNA sequence. Specifically, we hypothesized that most cCREs regulating expression of a gene would show a similar epigenetic profile across cell types in both species, regardless of whether the element was conserved in sequence. We leveraged the membership of each cCRE in the joint metaclusters (JmCs) determined in human and mouse because those JmCs reflect the inferred activity (deduced from epigenetic states) of the cCREs across cell types, reasoning that most cCREs regulating a given gene would be in one of the JmCs found frequently in the locus. Orthologous loci in mouse and human were defined as 100 kb genomic DNA intervals centered on the TSS of a gene with an identical name in the two species. Within these orthologous loci, we calculated the enrichment of each joint metacluster (JmC) in the collection of cCREs and assessed whether each individual cCRE was a member of the enriched JmC (Supplementary Figure S17). Thus, each cCRE was assessed both for its evolutionary history, which relied on DNA sequence alignments, and its regulatory potential deduced from epigenetic state profiles, which did not rely on DNA sequence alignments. The cCREs in these orthologous loci were assigned to a subdivision of the conservation categories; the cCREs in JmCs enriched for a specific orthologous locus were labeled SF+, S+, and N+, whereas those not in enriched JmCs were labeled SF, S, and N cCREs (Figure 8C and Supplementary Figure S17). Using this approach, one can deduce that even cCREs in non-aligning genomic regions, such as the one upstream of Gata1, have epigenetic state profiles suggestive of a role in regulation of the orthologous gene (Figure 8D). Inclusion of the JmC enrichment along with the evolutionary categories increased the correlation between the esRP scores of cCREs and the expression of their inferred target genes (Figure 8E). The increase in correlation was observed for cCREs in all three evolutionary categories, including the species-specific N category, consistent with our hypothesis of common epigenetic profiles across cell types for relevant regulatory elements regardless of evolutionary category. These JmC enrichments provided an opportunity to delve into assessment of potential functions across species even in regions of the genome that no longer align between species.
In summary, the IDEAS joint modeling on the input data compiled here and consistent state assignments in both mouse and human confirmed and extended previous observations on known regulatory elements, and they revealed both shared and distinctive candidate regulatory elements and states across species. Correlations of state profiles across species provided a comparison of chromatin landscapes even in regions with DNA sequences that were not conserved between species.
DISCUSSION
In this paper, the VISION consortium introduces a set of resources describing the regulatory landscapes of both human and mouse blood cell epigenomes. One major resource is the annotation of the epigenetic states across the epigenomes of progenitor and mature blood cells. These state maps show the epigenetic landscape in a compact form, capturing information from the input data on multiple histone modifications, CTCF occupancy, and chromatin accessibility. The state assignments reveal the patterns of epigenetic activity associated with gene expression and regulation, and they enable comparisons across cell types and species.
A second major resource is a catalog of cCREs (candidate cis-regulatory elements) actuated in one or more of the blood cell types in each species. The cCREs are predictions of discrete DNA segments likely involved in gene regulation, based on the patterns of chromatin accessibility across cell types. The epigenetic state annotations in each cell type suggest the type of activity for each cCRE in that cell type, such as serving as a promoter or enhancer, participating in repression, or inactivity. A key, novel aspect of our work is that the systematic integrative modeling that generated these resources was conducted jointly across the data from both species, which enables robust comparisons between species. Thus, our novel approach enables comparison of epigenetic landscape between human and mouse blood cells without being limited by sequence alignments, allowing comparisons in non-conserved and lineage-specific genomic regions.
A third major resource is a quantitative estimate of the regulatory impact of human and mouse cCREs on gene expression in each cell type, i.e., an esRP score, derived from multivariate regression modeling of the epigenetic states in cCREs as predictors of gene expression. The esRP scores are a continuous variable capturing not only the integration of the input epigenetic data, but also the inferred impacts on gene expression. These scores build on the foundation of categorical assignment of epigenetic states in SAGA methods to provide a rich and flexible prediction of regulatory impact. Currently, the state annotations are used to assign labels about inferred function to genomic intervals, such as strong enhancer, promoter-like, or bivalent element. The esRP scores complement and extend those annotations by providing quantitative predictions of impact, without regard to the label on the cCRE. They are useful for many downstream analyses, which we illustrated with visualization across differentiation (using dimensional reduction methods) and determining informative groups of cCREs by clustering analysis. These resources along with browsers for visualization and tools for analysis are provided at our project website, http://usevision.org. Among these tools is cCRE_db, which records the several dimensions of annotation of the cCREs and provides a query interface to support custom queries from users.
Our human blood cell cCRE catalog should be valuable for mechanistic interpretations of trait-related human genetic variants. The documented strong enrichment of trait-associated, non-protein-coding variants in candidate regulatory elements active in relevant cell types (Maurano et al. 2012; The_ENCODE_Project_Consortium 2012) has served as a basis for many studies that strive to establish a mechanistic connection between a specific set of variants (the genotype) and a trait of interest (the phenotype) (e.g., Bauer et al. 2013; Claussnitzer et al. 2015; Joslin et al. 2021). We showed that human genetic variants associated with traits intrinsic to blood cells were significantly enriched in the VISION cCRE catalog, whereas variants associated with a broad diversity of other traits were not enriched. We expect that the extensive annotations in our cCRE catalog combined with information about TFBS motifs and TF occupancy should lead to specific, refined hypotheses for mechanisms by which a variant impacts expression, such as alterations in TF binding, which can be tested experimentally in further work.
The jointly learned state maps and cCRE predictions allowed us to extend previous work on the evolution of regulatory elements between mouse and human. Several previous studies focused on transcription factor (TF) occupancy, e.g. examining key TFs in one tissue across multiple species (Schmidt et al. 2010; Ballester et al. 2014; Villar et al. 2014), or a diverse set of TFs in multiple cell types and in mouse and human (Cheng et al. 2014; Yue et al. 2014; Denas et al. 2015). Other studies focused on discrete regions of high chromatin accessibility, i.e., DNase hypersensitive sites (DHSs), in multiple cell types and tissues between mouse and human (Stergachis et al. 2014; Vierstra et al. 2014). These previous studies revealed that the fraction of elements that were conserved both in genomic sequence and in inferred function (TF occupancy or DHS) was small and varied considerably, especially for TF occupancy. A notable fraction of elements showed evidence of considerable change during mammalian diversification, including turnover of TF binding site motifs and repurposing of elements (Schmidt et al. 2010; Cheng et al. 2014; Stergachis et al. 2014; Denas et al. 2015). These prior studies were primarily limited to the regions of the genome with sequences that aligned between human and mouse. The non-aligning regions were used to infer that some elements were lineage-specific and that many were derived from transposable elements and endogenous retroviruses (Bourque 2009; Rebollo et al. 2012; Jacques et al. 2013; Sundaram et al. 2014).
Our evolutionary analyses confirmed the previous observations, e.g., finding about one-third of cCREs are conserved in both sequence and inferred function between human and mouse, and further showing that this evolutionary category was highly enriched for proximal regulatory elements. Going beyond the prior studies, our jointly learned epigenetic state maps provided a representation of multiple epigenetic features, not just TF occupancy or DHSs, and they are continuous in bins across genomes of both species. Thus, they provide a basis for comparisons of the epigenetic profiles between species. We showed that these epigenetic comparisons were a strong complement to genomic sequence alignments, allowing us to find elements with similar epigenetic profiles even in genomic regions that do not align between species. In the current work, we used both a correlation between profiles of epigenetic states and joint clusterings of cCREs across species by esRP scores as initial explorations of these epigenetic comparisons. The results of these initial studies suggest that it would be productive to pursue additional work developing methods and exploring the utility of comparisons of epigenetic states between species.
Other work compares epigenetic profiles across species, such as the phylo-HMGP method to find different evolutionary states in multi-species epigenomic data (Yang et al. 2018) and the LECIF scores to find evidence of conservation from functional genomic data (Kwon and Ernst 2021). These approaches are powerful but limited to the genomic regions with DNA sequences that align between the species. Importantly, the approach of correlating epigenetic states introduced here is agnostic to the underlying DNA sequence alignments (or absence of them), and thus it complements other approaches and expands the scope of the analysis, potentially to the whole genome.
Several innovations were developed to produce the resources introduced here. A major innovation was to extend the IDEAS framework (Zhang et al. 2016) to jointly learn epigenetic states and assign them to annotate the epigenomes in human and mouse blood cells. The IDEAS method employs a Bayesian approach to the modeling to learn the states, which we utilized to bring in states learned from the data in one species as priors for learning states in the data from the second species. Iterative applications of this approach led to the final joint model. Another extension of the IDEAS framework was to learn states based on one feature, specifically ATAC-seq data, defining discrete signal intensity states. This approach was used for calling cCREs, implemented as the IDEAS-IS method (Xiang et al. 2021). The approach is relatively simple and benefits from joint modeling across the input datasets. Other methods for predicting cCREs based on chromatin accessibility across many cell types prevented excessive expansion of the summary calls for overlapping peaks by employing a centroid determination for the DNase hypersensitive sites (DHS) index (Meuleman et al. 2020) or by choosing the highest signal peak for the ENCODE cCRE catalog (The_ENCODE_Project_Consortium et al. 2020). The ENCODE cCRE catalog paired DHS peaks with individual chromatin modifications or CTCF occupancy, which led to complications when data on diagnostic features was missing from some cell types. The VISION cCRE sets were generated by state modeling jointly across cell types in the IDEAS framework both for calling peaks (in the IS mode) and for identifying peaks in regions of dynamic chromatin modification (in epigenetic state mode), leveraging data in related cell types to ameliorate the impact of missing data.
While the resources introduced here are valuable for many applications, it is prudent to acknowledge their limitations. First, the quality of the products of integrated analyses are limited by the quality and completeness of the input, raw data. We endeavored to reduce the impact of variances in the input data by normalization. The S3V2 procedure (Xiang et al. 2021) systematically normalized the input data to adjust for differences in signal-to-noise and variance in signal across the datasets. Some epigenetic features were not determined in some cell types, and we used the IDEAS method in part because it is able to assign an epigenetic state even in the context of missing data by learning patterns from local similarities in cell types for which the data are present (Zhang and Mahony 2019). However, these approaches cannot completely overcome all issues with variance in input data. Second, the resolution of both the epigenetic state assignments and the cCRE inference is limited to 200 bp, which is the window size we utilized in the IDEAS analyses. Other resources, such as DHS calls (Meuleman et al. 2020), DNase footprints (Vierstra et al. 2020), and motif instances (Weirauch et al. 2014), achieve a higher resolution. Indeed, one can use these higher resolution datasets to derive further information about cCREs, such as TFs that are likely to be binding to them. With regard to the esRP scores, a third limitation is that we do not make explicit assignments for target genes of cCREs. Predictions of a large number of target gene-cCRE pairs were made in our prior work (Xiang et al. 2020b); these assignments cover large genomic intervals around each gene and are most useful when used with further filtering, such as restricting cCREs and target genes to the same topologically associated domains. On-going work is examining other models and approaches for assigning likely target genes to cCREs. A fourth limitation is that our inference of repression-related cCREs apply only to those with stable histone modifications. Elements that had been involved in initiation of repression but eventually were packaged into quiescent chromatin, e.g., via a hit-and-run mechanism (Shah et al. 2019), would not be detected.
In conclusion, we present several important new resources to enable further and more detailed studies of gene regulation in human and mouse blood cells both during normal differentiation and in pathological contexts. The patterns of epigenetic states in cCREs across cell types show value in developing an understanding of how genetic variants impact blood cell traits and diseases. Furthermore, the joint modeling across species opens avenues for further exploration of comparisons of epigenetic landscapes in addition to sequence alignments for insights into evolution and function of regulatory elements across species.
METHODS
Collection and initial processing of epigenetic and transcriptomic data from human and mouse blood cells
Sources for all 404 data sets used as input for the systematic integrative analysis are listed in Supplementary Table S1. Much of the epigenetic data for mature blood cell types in humans was collected as mapped reads (human genome build GRCh38) in bigWig format from the data portal for the BLUEPRINT Project (Martens and Stunnenberg 2013; Stunnenberg et al. 2016). Additional data, including ATAC-seq for human progenitor cells (Corces et al. 2016), and the full set of features in HUDEP1 (generated for this paper), HUDEP2 (Cheng et al. 2021; Qi et al. 2021), and K562 (The_ENCODE_Project_Consortium et al. 2020) cell lines were collected as sequencing reads and processed through the mapping pipelines described in a previous VISION paper (Xiang et al. 2020b), mapping the reads to human genome build GRCh38. Replicate data were obtained for most but not all features across the cell types, especially for the human blood cell types (Supplementary Table S1), and integrative analysis was conducted keeping the replicate sets separate for each cell type. The data for mouse hematopoietic cells were described previously, with reads mapped to mouse genome build mm10 (Xiang et al. 2020b).
Normalization
The normalization method S3V2 was designed to match the ranges of both signal intensities and variances across epigenetic datasets (Xiang et al. 2021). In this procedure, we first generated a reference signal track for each epigenetic feature by computing the mean signal of all data sets for that feature at each genomic location (200 bp bins). Then, the peak means and the background non-zero means of the reference signal tracks for the different epigenetic features were equalized by the S3norm method (Xiang et al. 2020a). We then used these mean-adjusted references as the new reference signal tracks for each epigenetic feature. For all datasets of the same epigenetic feature, we normalized their signal against the reference signal track using the recently developed S3V2 method (Xiang et al. 2021). The S3V2 version of the method was designed to adjust both the non-zero means and the standard deviations of the background regions, so that it can better reduce background noise in some data sets with higher variance at the background regions.
Joint systematic integration of human and mouse blood cell epigenomes by IDEAS
The joint training in IDEAS to identify epigenetic states was done iteratively, as illustrated in Figure 2. Initially, 200 sets of epigenetic states were identified by IDEAS at 100 randomly selected, 50 MB regions from each species, using the S3V2-IDEAS pipeline (Xiang et al. 2021) on both species. The reproducible IDEAS states were selected by an internal combineState function in the IDEAS pipeline. We wanted to include as reproducible states not only those found at high frequency in all the runs on data from both species but also those that were either highly reproducible in one species and also found in the other species or those that were moderately reproducible in both species. By requiring that a state appear in at least 52% of the 200 runs, we ensured that the collected states included the latter two categories. The set of 27 reproducible IDEAS states were used as the priors for the distribution parameters in the two rounds of IDEAS runs in both species. The two rounds of IDEAS runs for the two species were performed sequentially, alternating between human and mouse. After each round of IDEAS run, the frequency, mean and variance parameters for each epigenetic state were updated, so that the information of the species at the current round was then integrated into the next IDEAS runs. The heterogeneous states (Xiang et al. 2020b) were also removed after each round of IDEAS run (Supplementary Figure S3). After the two rounds of IDEAS runs for the two species, a set of IDEAS states were used as the final joint epigenetic states for both species. To assign these final joint epigenetic states to each genomic location in each cell type in both species, another two rounds of IDEAS runs for the two species were performed in parallel.
Peak calling for ATAC-seq data across cell types
We adopted an extension of the IDEAS methodology, specifically the S3V2-IDEAS pipeline (Xiang et al. 2021) in the signal intensity state (IS) mode, to call peaks of chromatin accessibility across the blood cell types. The input data were the ATAC-seq signals for each replicate of each cell type plus a track of combined average ATAC-seq signal. This combined average was computed by averaging the normalized ATAC-seq signal in 200 bp bins for each cell type, and then averaging these values per bin for all cell types (Fig. 3A). IDEAS in the IS mode learned four signal intensity states, with state 0 being no detectable signal and state 3 being the highest signal state, which were then used to annotate the genomes of all cell types plus the combined cell data. The peaks were called using a hierarchical process designed to find genomic DNA intervals in the high signal intensity states, compared to the local background, both in many cell types and in restricted sets of cell types (Fig. 3A). Specifically, in the first step (1), the DNA bins in the higher signal states, compared to the local background, in the average track were collected as peaks. If a contiguous series of bins were in higher signal states, indicating an accessible region, only the bin(s) in the highest signal state were called as peaks (Fig. 3A). In the second step (2) bins in a high signal state in individual cell types were included in the set of peaks. The next two steps added bins in a lower signal state, but still above the local background, as peaks, with step (3) adding such bins from the average signal track and step (4) adding such bins from signal tracks from individual cells. Juxtaposed peak calls were combined into a single peak. If replicate determinations were available for chromatin accessibility in a given cell type, the peak call had to be replicated.
Annotation of VISION cCREs utilizing orthogonal datasets of elements related to regulation and chromatin structure
The VISION human blood cell cCREs were found to be a sub-set of two large collections of cCREs predicted from ENCODE data on a much larger number of cell types. These large collections were the 926,535 ENCODE cCREs provided in Supplementary Table 10 of reference (The_ENCODE_Project_Consortium et al. 2020) and the 3,591,898 elements in the Index of DNase hypersensitive sites from Meuleman et al. (2020).
Additional datasets, orthogonal to those included in the prediction and epigenetic state annotation of VISION cCREs, that annotate potential roles of human genomic intervals in transcriptional regulation (CREs) or in chromatin structure (chromatin architecture) were curated from the literature and associated databases (Figure S6). The orthogonal sets of CREs included TSSs from the GENCODE basic gene set (Frankish et al. 2021), peaks from the Survey of Regulatory Elements (SuRE), which is a massively parallel reporter assay that reveals both promoter and enhancer activity, in K562 cells (van Arensbergen et al. 2017), unmasked CpG islands downloaded from UCSC genome browser (Nassar et al. 2023), and a group of enhancer-related elements. The latter group of enhancer-related elements were a combination of three sets: (1) enhancers predicted from eRNAs in hundreds of human cell types (Andersson et al. 2014; https://fantom.gsc.riken.jp/5/datafiles/reprocessed/hg38_latest/extra/enhancer/), (2) a combined set of EP300 ChIP-seq peaks from K562 and GM12878 cell lines (The_ENCODE_Project_Consortium et al. 2020) and erythroblasts (Su et al. 2013), and (3) the erythroid Enhancer Repertoire deduced from histone modification data (Huang et al. 2016). The chromatin structure category included CTCF-occupied DNA segments (CTCF OSs), chromatin loop anchors, and TAD boundaries in primary blood cells and related cell lines. The set of CTCF OSs was generated by combining peaks from ChIP-seq experiments in human fetal and adult erythroblasts (Huang et al. 2017), and from K562, MM1S, Delta47, and GM12878 cell lines (Sánchez-Castillo et al. 2015; The_ENCODE_Project_Consortium et al. 2020). Loop anchors determined by HICCUPS from Hi-C data in K562 and GM12878 cell lines (Rao et al. 2014) were downloaded from GEO [accession GSE63525] and combined. TAD boundaries were called by OnTAD (An et al. 2019) using Hi-C data at 10kb resolution in K562 and GM12878 cell lines (Rao et al. 2014).
The human VISION human cCREs were also compared to two sets of known CREs. One was a compilation of 109 demonstrated regulatory elements in blood cells from the literature (Supplementary Table S3). The other set was 664 enhancers with target genes, determined by a high throughput mutagenesis and eQTL analysis in K562 cells (Gasperini et al. 2019).
Pairwise overlaps between these orthogonal sets of elements and the VISION cCREs were computed using the bedtool intersect tool (-u option) (Quinlan and Hall 2010). Intersections among multiple datasets were computed using the intervene tool, version 0.6.5 (Khan and Mathelier 2017), and displayed in UpSet plots using the UpSetR package (Conway et al. 2017) and 3-way Venn diagrams using the eulerr package (https://CRAN.R-project.org/package=eulerr).
Ten different randomly chosen sets of human genomic DNA intervals, matched with the cCREs in length and G+C content, were generated by the script in the following link https://github.com/YenLab/Tn5InsertPrefer/blob/main/StandaloneScripts/Negetive_sequence_matched_length. Enrichments of overlaps between the VISION cCREs relative to those in the random set of intervals were determined by computing overlaps (using tools described above) between the sets of function- and structure-related elements and each of the ten sets of random intervals, dividing the number of overlapped cCREs by the number of overlapped random intervals (ten times), and using the average of the ten quotients as the enrichment. All ten results were shown in boxplots.
Inferring epigenetic state effects on gene expression regulation
Calculating β coefficients for epigenetic states and esRPs for cCREs across all cell types
In order to use the categorical state assignments to estimate the impact of each cCRE in each cell type on gene expression, we applied a modified version of the iterative multivariable regression model developed previously (Xiang et al. 2020b) to quantify the biological functions of epigenetic state in terms of regulating gene expression. In this model, we introduced two measurements: β coefficients for each epigenetic state and an esRP (epigenetic state Regulatory Potential) score for each cCRE in each cell type or sample. The biological interpretation of the two measurements are as follows. The β coefficients measure the contribution of each epigenetic state to the expression of local genes; they are calculated in a multivariate regression evaluating how changes in the coverage of cCREs and promoters by each epigenetic state across cell types impact expression levels. The esRP score measures the contribution of individual cCREs on regulating its target gene’s expression level; it is calculated from the overall epigenetic state coverage of the cCRE in each cell type (Figure 4B). In contrast to our previous modeling (Xiang et al. 2020b), our current model does not aim to identify the likely target gene(s) for each cCRE; that will be the subject of a subsequent report. In brief, for the current regression model, the epigenetic state coverage was computed on all cCREs and promoter regions within 50 kb on both sides of the TSS of each gene (an interval of 100kb). We first calculated the β coefficients of the promoter intervals and distal cCREs as separate terms in the regression model. For further analyses and visualization, including computation of the esRP scores, the β coefficients of each state were merged into a single value that was the average of the β coefficients for promoters and for distal cCREs. A more detailed presentation on the calculations of the β coefficients and the esRPs is in the Supplemental Methods.
Visualizing the esRP matrix by UMAP
We used the UMAP method (McInnes et al. 2018) to visualize the esRP matrix of all cCREs across all available cell types. This method for visualizing high dimensional data in lower dimensional space has been widely used in single cell data analysis where each cell’s transcriptomic or epigenomic profile is projected onto a 2-dimensional UMAP space. Similarly, we projected each cCRE’s esRP scores onto a 2-dimensional UMAP space. Thus, each point on our UMAP represents a cCRE rather than a cell, and cCREs with similar vectors of esRP scores across cell types are placed in similar locations on the UMAP plane. We used the umap library’s umap function in R with default settings to transform the data and generate the UMAP images.
Clustering of cCRE based on esRP scores
To infer the potential biological functions of the cCREs, we clustered them based on their esRP scores across different cell types. The traditional methods for clustering cCREs are based on a pairwise distance matrix of signals measured in different epigenomic datasets across different cell-types. However, these approaches cannot capture the information of the potential biological effects of the epigenetic modifications on gene expression regulation. For example, transitions from an initial state with only H3K4me3 to a second state with only H3K9me3 or to a second state with only H3K27ac are very different with regard to their biological associations (repression versus activation, respectively), but the pairwise distances determined from the vectors of epigenetic signals would be very similar. In contrast to the signals measured directly by epigenomic datasets, the esRP score has already integrated all available epigenetic features’ information as a quantitative score that can reflect the overall potential of each cCRE for regulating gene expression. Thus, we hypothesized that clustering the cCREs using a distance based on esRP score would produce groups that better capture the changes of biological effects of the cCRE across different cell types.
In this study, we conducted a series of clustering steps to generate robust clusters representing the prevalent patterns of inferred regulatory potential (esRP scores) across cell types in both human and mouse species (Supplementary Figure S8). We first created a combined matrix of esRP scores for all human and mouse cCREs across all shared cell types in both species. This ensured that the identified clusters were based on esRP patterns across the same set of cell types in both species. To mitigate the potential issues of cross-cell-type collinearity and overfitting, we performed a principal component analysis and selected the top 10 principal components (PCs), which account for 99% of the variance. This generated a matrix containing 10 PC values corresponding to each cCRE. Although we had data for neutrophils from both species, we excluded the esRP scores for cCREs in neutrophils due to the significant noise in the ATAC-seq, which affected the overall quality of our results.
To identify robust clusters, we employed an iterative K-means clustering strategy, finding clusters with high consensus across repeated clustering rounds and then re-assigning all cCREs to these robust clusters. The initial clustering round (Step 1, Supplementary Figure S8) used K-means clustering (K=100) based on the pairwise Euclidean distance of PCs of esRP scores across all cell types. This was performed 100 times, generating 10,000 clusters that could contain both human and mouse cCREs or cCREs from only one species. To identify consensus clusters across the 100 K-means runs, we combined the vectors of mean PC values for each of the 10,000 clusters from Step 1 into a matrix and clustered them again using K-means (K=100, Step 2, Supplementary Figure S8). This step partitions the Step 1 clusters with similar characteristics into groups of clusters, where a large group size implies high consensus.
We considered groups with more than 70 Step 1 clusters as high consensus clusters, determined by calculating the Z score for the group size and finding the group size corresponding to the upper tail probability of 0.05. We identified 61 such groups, which we called robust clusters. However, these 61 clusters did not contain all cCREs, so we reassigned all cCREs in both human and mouse to one of the 61 robust clusters based on the cosine distance between each cCRE’s esRP score profile and each cluster’s average esRP score profile, which was computed by averaging the mean-esRP-signal-vectors of the Step 1 clusters within each robust cluster.
Since our goal was to find clusters with cCREs in both species, we counted the number of cCREs from each species in each robust cluster and calculated the proportion of cCREs in each species for each robust cluster. We identified 44 clusters in which the proportion of cCREs did not significantly differ between the two species (i.e., the absolute log2 fold changes of cCRE proportion between the species were <= 2.18 (p-value = 0.05)), which we designated as robust clusters shared across species. Using the cosine distance, we reassigned each cCRE in each species to one of these 44 shared robust clusters.
Lastly, we observed that the average esRP score profiles across cell types for some subsets of the 44 clusters were similar, indicating they were not distinct clusters. To better differentiate groups of cCREs shared between species, we created joint metaclusters (JmCs) by clustering the clusters (Step 3, Supplementary Figure S8). We combined the 44 clusters to generate 15 JmCs using hierarchical clustering (Hclust) and dynamic trimming with dynamicTreeCut (Murtagh and Legendre 2014). We ran Hclust followed by dynamicTreeCut 100 times, adding different noise (uniformly distributed within a range of −0.001 to 0.001) for each run and using the cosine distance matrix. We then created a count matrix to record the frequency of each cluster being found in the same JmC. We used Hclust to cluster the count matrix and applied dynamicTreeCut to cut the Hclust tree into 15 JmCs. As a result, each cCRE in mouse and human was assigned to one of the 15 JmCs. These JmCs provide discrete categories for cCREs based on the cell type distribution of their estimated regulatory impact.
Here, we also calculated the enrichment of JmCs at gene loci (TSS +/−50kb) in both species. For each , the enrichment of is defined as follows:
where the is the number cCRE assigned to at the locus in human or mouse, the is the expected number of cCRE assigned to at the locus in human or mouse by random chance.
Enrichment for transcription factor binding site motifs in joint metaclusters of cCREs
We used the Maelstrom tool in the GimmeMotifs suite (v0.17.1) to identify motifs that are differentially enriched across JmCs (Bruse and van Heeringen 2018). We first labeled all cCREs according to their JmC membership. We then ran separate Maelstrom analyses on human and mouse cCRE sets to find enrichment of motifs in GimmeMotif’s default “gimme.vertebrate.v5.0.pfm” collection of non-redundant clustered vertebrate motifs derived from the Cis-BP database (Weirauch et al. 2014). Maelstrom’s --filter-cutoff parameter was set to the default value of 0.8, which has the effect of filtering redundant motif enrichment results based on scores across the input sets. We filtered out motifs that did not achieve a Maelstrom z-score of at least 4 in any JmC. We then combined results across human and mouse (which required running Maelstrom again for each species using the --no-filter option to fill in z-scores for motifs that were found in one species but not the other). Heatmaps were constructed using the Python seaborn package (Waksom 2021) and motif logos were plotted using WebLogo3 (Crooks et al. 2004). Putative TF names were associated with each motif by examining the identities of Cis-BP motifs that were clustered into the relevant non-redundant motifs within the GimmeMotifs non-redundant set, and by matching enriched motifs against mouse and human motifs from Cis-BP (v.2.0) using STAMP (v.1.0) (Mahony and Benos 2007) with arguments “-cc PCC -align SWU”.
Separately, we used the SeqUnwinder multi-label discriminative motif finder to discover de novo motifs associated with each JmC and to search for motifs that were potentially constitutively differentially enriched across species. We first gave every cCRE two labels: their JmC membership and their species of origin. We then filtered out sequences that were larger than 1kbp and randomly selected 50,000 sequences from the remaining set. We then provided these doubly-labeled sequences to SeqUnwinder (v.0.1.5) (Kakumanu et al. 2017) and ran analysis using the following options: --threads 8 --win 200 --mink 4 --maxk 5 --r 10 --x 3 --a 200 --hillsthresh 0.1 --memesearchwin 16 --minsubclass 150. Heatmaps were again constructed using the Python seaborn package (Waksom 2021) and motif logos were plotted using WebLogo3 (Crooks et al. 2004). Putative TF names were associated with each SeqUnwinder-discovered motif by matching against mouse and human motifs from Cis-BP (v.2.0) using STAMP (v.1.0) (Mahony and Benos 2007) with arguments “-cc PCC -align SWU”.
Enrichment of genetic variants for blood cell related traits in the human cCRE collection
To assess whether the cCREs were associated with regulation of genes involved in development of blood cell traits, we examined overlaps between phenotype-associated genetic variants and the human cCRE catalog. The initial analysis examined SNPs associated with blood cell traits that were obtained from the GWAS catalog (Buniello et al. 2019). The red blood cell traits included the following: red blood cell distribution width, erythrocyte measurement, hematocrit, mean corpuscular volume, mean corpuscular hemoglobin, erythrocyte count, hemoglobin measurement, mean corpuscular hemoglobin concentration, and erythrocyte indices.
We then proceeded to examine overlaps of VISION human cCREs with genetic variants associated with a large number of traits from the United Kingdom BioBank (UKBB) in a manner that takes into account linkage disequilibrium. The UKBB (Ge et al. 2017 and http://www.nealelab.is/uk-biobank/) is a database comprising genotypic data as well as data for several medical traits from over 400,000 individuals, and GWAS summary statistic results are publicly available for a number of these traits, stratified by sex. We used all 587 sex-stratified traits for which inverse rank-normalized data was available (representing 295 unique traits) in our analysis, including 54 traits labeled “blood count”-related traits by UKBB, 60 traits labeled “blood biochemistry”-related, and 473 traits that are not blood-related. Blood count-related traits reflect cell morphology and number while blood biochemistry-related traits reflect the concentrations of certain proteins and metabolic products. (Note: hemoglobin concentration is an exception and considered a blood count trait).
For each of these 587 traits, we used sLDSC (Finucane et al. 2015) to quantify the extent to which our cCRE annotation is enriched in the heritability of the trait. Using SNPs within some window of the annotation, this approach regresses the GWAS chi square summary statistic (for the focal trait) of these SNPs onto the LD scores of the SNPs with respect to the annotation. The LD score of a SNP reflects the extent to which that SNP is in linkage disequilibrium with the annotation. If the annotation is associated with the focal trait, we expect a linear relationship between the LD scores of the tested SNPs with the annotation and the chi square values of those SNPs. The slope of the regression line is an estimate of the SNP heritability of the trait with respect to the annotation. By dividing this estimate by the overall SNP heritability of the trait, we obtain an estimate of the proportion of heritability explained by the annotation. Finally, dividing this by the portion of SNPs falling within the annotation provides an estimate of the enrichment of that annotation in heritability of the focal trait.
The sLDSC tool recommends using a set of SNPs from HapMap 3 for analysis, and because these SNPs are reported on GRCh37, we first lifted over (Hinrichs et al. 2006) our cCRE annotations from GRCh38 to GRCh37. A total of 826 cCREs (0.4% of all cCREs) failed to liftover and were excluded from analysis.
Using LDSC v1.0.1, and with these lifted over annotations, we first computed LD scores for HapMap 3 SNPs within 1 cM of cCRE annotations. Then, for each of the 587 UKBB traits, we performed sLDSC, regressing the trait summary statistics onto the LD scores. From this, we obtain an estimate of the enrichment of the cCRE annotation in the heritability of each trait.
We repeated this analysis, using the 15 Joint metaclusters as 15 separate annotations. Running these annotations through the same pipeline described above, we obtained estimates of the enrichment of each JmC in the heritability of each of 587 traits.
Apportioning cCREs to evolutionary categories based on DNA sequence alignments and cCRE calls across species
The full list of human cCREs was mapped to mm10 using the liftOver tool at the UCSC Genome Browser (Hinrichs et al. 2006). Human cCREs that failed to map to mm10 were grouped as N cCREs. Then, those human cCREs that could be mapped by liftOver to genome build mm10 were compared with mouse cCREs, using bedtools intersect. Human cCREs that overlapped with mouse cCREs were labeled as SF cCREs. Human cCREs that mapped to mm10 but did not have any matched mouse cCREs were labeled as S cCREs. A similar process was performed on the full list of mouse cCREs (using liftOver to map to genome build GRCh38/hg38) as well. By this procedure, we generated three evolutionary categories of cCREs for human and mouse.
Comparing integrated epigenetic features between human and mouse blood cells
Calculating gene expression correlation coefficients between human and mouse
We identified 14 cell types matched between human and mouse with RNA-seq datasets in the VISION project. For each gene, the correlation coefficient was calculated between the two vectors of 14 values for log2(TPM+1), generating one correlation coefficient value per cell type per gene. When calculating the correlation coefficients, we added random noise (mean=0, sd=1) to the raw values to avoid high correlation coefficients created between vectors with low signals.
Calculating bin-to-bin pairwise correlation coefficients between human and mouse
We introduced a bin-to-bin pairwise correlation coefficient to quantify the similarity of cross-cell-type epigenetic landscape between two DNA regions in human and mouse. For each 200bp bin in one cell type in one species, the assigned epigenetic state was replaced by a vector of mean signals of 8 epigenetic features in the IDEAS state model. After replacing the states in all 15 matched cell types in the two species, the original two categorical state vectors with 15 elements can be converted into two numeric vectors with 120 numbers (Supplementary Figure S16). The similarity of cross-cell-type epigenetic landscape between two bins in the two species was defined as the correlation coefficient between each pair of numeric vectors with 120 numbers. When calculating the correlation coefficients, we added a random noise (mean=0, sd=0.2) to the raw values to avoid high correlation coefficients created between regions with states that have low signals.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health: R24DK106766 to RCH, GAB, MJW, YZ, FY, JT, MS, DB, DH, JRH, BG; R01DK054937 to GAB; R01GM121613 to YZ and SM; R01GM109453 to QL; R35GM133747 to RCM; F31HG012900 to DJT; R01HG011139; and intramural funds from the National Human Genome Research Institute.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing interests.
DATA ACCESS
Most of the data used in the integrative analysis are already publicly available; accession numbers for each file are listed in Supplementary Table S1. Regarding the new experiments, all raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE_______.
Resources developed in the VISION project are available at the website https://usevision.org. The epigenetic state annotations can be visualized in the BX VISION browser (http://main.genome-browser.bx.psu.edu). The database cCRE_db (https://globin.bx.psu.edu/cgi-bin/hlab/ccre_query) supports flexible user queries on extensive annotation of the cCREs, including epigenetic states and esRP scores across cell types, chromatin accessibility scores across cell types, membership in JmCs, and evolutionary categories. Intersections with other cCRE collections and epigenetic features are supported. The website section “Human and mouse cCREs and visualization” (https://usevision.org/data/ccre/) contains downloadable files of the cCREs along with annotation such as ATAC-seq signals and esRP scores, the joint metaclusters, the matched random regions, the UMAP visualizations of cCREs colored by esRP scores in each cell type, and movies of the changes in esRP scores across selected lineages of differentiation. The SOMs can be examined and analyzed at (https://usevision.org/som/DNA_V5/). The Downloads section contains data from both human and mouse for the input raw epigenetic data, gene expression levels from RNA-seq data, and epigenetic state assignments from IDEAS.
Code used in this study are in the following repositories:
| Process | URL |
|---|---|
| Joint human-mouse IDEAS pipeline | https://github.com/guanjue/Joint_Human_Mouse_IDEAS_State |
| S3V2 pipeline | https://github.com/guanjue/S3V2_IDEAS_ESMP |
| Build self-organizing maps | https://github.com/csjansen/SOMatic |
| sLDSC analysis | https://github.com/usevision/cre_heritability |
REFERENCES
- An L, Yang T, Yang J, Nuebler J, Xiang G, Hardison RC, Li Q, Zhang Y. 2019. Hierarchical Domain Structure Reveals the Divergence of Activity among TADs and Boundaries. Genome Biology 20: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature 507: 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester B, Medina-Rivera A, Schmidt D, Gonzalez-Porta M, Carlucci M, Chen X, Chessman K, Faure AJ, Funnell AP, Goncalves A et al. 2014. Multi-species, multi-transcription factor binding highlights conserved control of tissue-specific biological pathways. eLife 3: e02626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, Shao Z, Canver MC, Smith EC, Pinello L et al. 2013. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342: 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA, Weiss MJ. 2009. Nuclear Factors that Regulate Erythropoiesis. In Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management, (ed. Steinberg MH, et al. ), pp. 62–85. Cambridge University Press, Cambridge. [Google Scholar]
- Bourque G. 2009. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr Opin Genet Dev 19: 607–612. [DOI] [PubMed] [Google Scholar]
- Bruse N, van Heeringen SJ. 2018. GimmeMotifs: an analysis framework for transcription factor motif analysis. BioRxiv doi: 10.1101/474403. [DOI] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E et al. 2019. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47: D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36. [DOI] [PubMed] [Google Scholar]
- Cheng L, Li Y, Qi Q, Xu P, Feng R, Palmer L, Chen J, Wu R, Yee T, Zhang J et al. 2021. Single-nucleotide-level mapping of DNA regulatory elements that control fetal hemoglobin expression. Nat Genet 53: 869–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Ma Z, Kim BH, Wu W, Cayting P, Boyle AP, Sundaram V, Xing X, Dogan N, Li J et al. 2014. Principles of regulatory information conservation between mouse and human. Nature 515: 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi AW, Bell JJ, Zlotoff DA, Bhandoola A. 2009. Untangling the T branch of the hematopoiesis tree. Curr Opin Immunol 21: 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V et al. 2015. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. The New England journal of medicine 373: 895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JR, Lex A, Gehlenborg N. 2017. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33: 2938–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ et al. 2016. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 48: 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denas O, Sandstrom R, Cheng Y, Beal K, Herrero J, Hardison RC, Taylor J. 2015. Genomewide comparative analysis reveals human-mouse regulatory landscape and evolution. BMC Genomics 16: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Greven MC, Kundaje A, Djebali S, Brown JB, Cheng C, Gingeras TR, Gerstein M, Guigo R, Birney E et al. 2012. Modeling gene expression using chromatin features in various cellular contexts. Genome Biol 13: R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore LC, Crispino JD. 2011. Transcription factor networks in erythroid cell and megakaryocyte development. Blood 118: 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan WS, Tjian R. 1983. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell 35: 79–87. [DOI] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2010. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 28: 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2012. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9: 215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R, Ohneda K, Yamamoto M, Philipsen S. 2005. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol 25: 1215–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, Anttila V, Xu H, Zang C, Farh K et al. 2015. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47: 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangoul H, Altshuler D, Cappellini MD, Chen YS, Domm J, Eustace BK, Foell J, de la Fuente J, Grupp S, Handgretinger R et al. 2021. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and beta-Thalassemia. The New England journal of medicine 384: 252–260. [DOI] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Jungreis I, Lagarde J, Loveland JE, Mudge JM, Sisu C, Wright JC, Armstrong J, Barnes I et al. 2021. Gencode 2021. Nucleic Acids Res 49: D916–D923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, O’Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. 2009. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell 36: 667–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini M, Hill AJ, McFaline-Figueroa JL, Martin B, Kim S, Zhang MD, Jackson D, Leith A, Schreiber J, Noble WS et al. 2019. A Genome-wide Framework for Mapping Gene Regulation via Cellular Genetic Screens. Cell 176: 377–390 e319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Chen CY, Neale BM, Sabuncu MR, Smoller JW. 2017. Phenome-wide heritability analysis of the UK Biobank. PLoS Genet 13: e1006711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorkin DU, Barozzi I, Zhao Y, Zhang Y, Huang H, Lee AY, Li B, Chiou J, Wildberg A, Ding B et al. 2020. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature 583: 744–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, Enver T. 2009. Forcing cells to change lineages. Nature 462: 587–594. [DOI] [PubMed] [Google Scholar]
- Hamamoto K, Fukaya T. 2022. Molecular architecture of enhancer-promoter interaction. Curr Opin Cell Biol 74: 62–70. [DOI] [PubMed] [Google Scholar]
- Hardison RC. 2012. Genome-wide epigenetic data facilitate understanding of disease susceptibility association studies. J Biol Chem 287: 30932–30940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison RC, Zhang Y, Keller CA, Xiang G, Heuston EF, An L, Lichtenberg J, Giardine BM, Bodine D, Mahony S et al. 2020. Systematic integration of GATA transcription factors and epigenomes via IDEAS paints the regulatory landscape of hematopoietic cells. IUBMB Life 72: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 106: 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F et al. 2006. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res 34: D590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MM, Buske OJ, Wang J, Weng Z, Bilmes JA, Noble WS. 2012. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods 9: 473–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman MM, Ernst J, Wilder SP, Kundaje A, Harris RS, Libbrecht M, Giardine B, Ellenbogen PM, Bilmes JA, Birney E et al. 2013. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res 41: 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liu X, Li D, Shao Z, Cao H, Zhang Y, Trompouki E, Bowman TV, Zon LI, Yuan GC et al. 2016. Dynamic Control of Enhancer Repertoires Drives Lineage and Stage-Specific Transcription during Hematopoiesis. Dev Cell 36: 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Keller CA, Giardine B, Grevet JD, Davies JOJ, Hughes JR, Kurita R, Nakamura Y, Hardison RC, Blobel GA. 2017. Comparative analysis of three-dimensional chromosomal architecture identifies a novel fetal hemoglobin regulatory element. Genes Dev 31: 1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques PE, Jeyakani J, Bourque G. 2013. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet 9: e1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen C, Ramirez RN, El-Ali NC, Gomez-Cabrero D, Tegner J, Merkenschlager M, Conesa A, Mortazavi A. 2019. Building gene regulatory networks from scATAC-seq and scRNA-seq using Linked Self Organizing Maps. PLoS Comput Biol 15: e1006555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian J, Konopka J, Liu C. 2013. Insights into the role of progranulin in immunity, infection, and inflammation. J Leukoc Biol 93: 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joslin AC, Sobreira DR, Hansen GT, Sakabe NJ, Aneas I, Montefiori LE, Farris KM, Gu J, Lehman DM, Ober C et al. 2021. A functional genomics pipeline identifies pleiotropy and cross-tissue effects within obesity-associated GWAS loci. Nature communications 12: 5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R. 2003. Sp1- and Kruppel-like transcription factors. Genome Biol 4: 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakumanu A, Velasco S, Mazzoni E, Mahony S. 2017. Deconvolving sequence features that discriminate between overlapping regulatory annotations. PLoS Comput Biol 13: e1005795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlić R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. 2010. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci U S A 107: 2926–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Mathelier A. 2017. Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinformatics 18: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, Shizuru JA, Weissman IL. 2003. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol 21: 759–806. [DOI] [PubMed] [Google Scholar]
- Kwon SB, Ernst J. 2021. Learning a genome-wide score of human-mouse conservation at the functional genomics level. Nature communications 12: 2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E, Göttgens B. 2018. From haematopoietic stem cells to complex differentiation landscapes. Nature 553: 418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Moazed D, Gygi SP. 2002. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem 277: 49383–49388. [DOI] [PubMed] [Google Scholar]
- Libbrecht MW, Chan RCW, Hoffman MM. 2021. Segmentation and genome annotation algorithms for identifying chromatin state and other genomic patterns. PLoS Comput Biol 17: e1009423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony S, Benos PV. 2007. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res 35: W253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JH, Stunnenberg HG. 2013. BLUEPRINT: mapping human blood cell epigenomes. Haematologica 98: 1487–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maston GA, Evans SK, Green MR. 2006. Transcriptional Regulatory Elements in the Human Genome. Annu Rev Genomics Hum Genet 7: 29–59. [DOI] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J et al. 2012. Systematic localization of common disease-associated variation in regulatory DNA. Science 337: 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L, Healy J, Saul N, Grossberger L. 2018. UMAP: Uniform Manifold Approximation and Projection. Journal of Open Source Software 3: 861. [Google Scholar]
- Meuleman W, Muratov A, Rynes E, Halow J, Lee K, Bates D, Diegel M, Dunn D, Neri F, Teodosiadis A et al. 2020. Index and biological spectrum of human DNase I hypersensitive sites. Nature 584: 244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208. [DOI] [PubMed] [Google Scholar]
- Murtagh F, Legendre P. 2014. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? Journal of Classification 31: 274–295. [Google Scholar]
- Nassar LR, Barber GP, Benet-Pages A, Casper J, Clawson H, Diekhans M, Fischer C, Gonzalez JN, Hinrichs AS, Lee BT et al. 2023. The UCSC Genome Browser database: 2023 update. Nucleic Acids Res 51: D1188–D1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK et al. 2012. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. 2008. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133: 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH. 1995. Transcription factors and hematopoietic development. J Biol Chem 270: 4955–4958. [DOI] [PubMed] [Google Scholar]
- Padeken J, Methot SP, Gasser SM. 2022. Establishment of H3K9-methylated heterochromatin and its functions in tissue differentiation and maintenance. Nat Rev Mol Cell Biol 23: 623–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne KJ, Crooks GM. 2002. Human hematopoietic lineage commitment. Immunol Rev 187: 48–64. [DOI] [PubMed] [Google Scholar]
- Pimkin M, Kossenkov AV, Mishra T, Morrissey CS, Wu W, Keller CA, Blobel GA, Lee D, Beer MA, Hardison RC et al. 2014. Divergent functions of hematopoietic transcription factors in lineage priming and differentiation during erythro-megakaryopoiesis. Genome Res 24: 1932–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q, Cheng L, Tang X, He Y, Li Y, Yee T, Shrestha D, Feng R, Xu P, Zhou X et al. 2021. Dynamic CTCF binding directly mediates interactions among cis-regulatory elements essential for hematopoiesis. Blood 137: 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. 2012. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet 46: 21–42. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. 2004. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet 38: 413–443. [DOI] [PubMed] [Google Scholar]
- Roadmap_Epigenomics_Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Zhao K. 2005. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev 19: 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Taghon T. 2005. Molecular genetics of T cell development. Annu Rev Immunol 23: 601–649. [DOI] [PubMed] [Google Scholar]
- Sánchez-Castillo M, Ruau D, Wilkinson AC, Ng FS, Hannah R, Diamanti E, Lombard P, Wilson NK, Göttgens B. 2015. CODEX: a next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic Acids Res 43: D1117–D1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S et al. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. 2000. PipMaker-A web server for aligning two genomic DNA sequences. Genome Res 10: 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38: 700–705. [DOI] [PubMed] [Google Scholar]
- Shah M, Funnell APW, Quinlan KGR, Crossley M. 2019. Hit and Run Transcriptional Repressors Are Difficult to Catch in the Act. Bioessays 41: e1900041. [DOI] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. 2014. Enhancer biology and enhanceropathies. Nat Struct Mol Biol 21: 210–219. [DOI] [PubMed] [Google Scholar]
- Spangrude GJ, Heimfeld S, Weissman IL. 1988. Purification and characterization of mouse hematopoietic stem cells. Science 241: 58–62. [DOI] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Sandstrom R, Haugen E, Reynolds AP, Zhang M, Byron R, Canfield T, Stelhing-Sun S, Lee K et al. 2014. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature 515: 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403: 41–45. [DOI] [PubMed] [Google Scholar]
- Stunnenberg HG, International Human Epigenome C, Hirst M. 2016. The International Human Epigenome Consortium: A Blueprint for Scientific Collaboration and Discovery. Cell 167: 1145–1149. [DOI] [PubMed] [Google Scholar]
- Su MY, Steiner LA, Bogardus H, Mishra T, Schulz VP, Hardison RC, Gallagher PG. 2013. Identification of biologically relevant enhancers in human erythroid cells. J Biol Chem 288: 8433–8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T. 2014. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res 24: 1963–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenen DG, Hromas R, Licht JD, Zhang DE. 1997. Transcription factors, normal myeloid development, and leukemia. Blood 90: 489–519. [PubMed] [Google Scholar]
- The_ENCODE_Project_Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The_ENCODE_Project_Consortium, Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, Kawli T, Davis CA, Dobin A et al. 2020. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583: 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde-Garduno V, Guyot B, Anguita E, Hamlett I, Porcher C, Vyas P. 2004. Differences in the chromatin structure and cis-element organization of the human and mouse GATA1 loci: implications for cis-element identification. Blood 104: 3106–3116. [DOI] [PubMed] [Google Scholar]
- van Arensbergen J, FitzPatrick VD, de Haas M, Pagie L, Sluimer J, Bussemaker HJ, van Steensel B. 2017. Genome-wide mapping of autonomous promoter activity in human cells. Nat Biotechnol 35: 145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Pampus EC, Denkers IA, van Geel BJ, Huijgens PC, Zevenbergen A, Ossenkoppele GJ, Langenhuijsen MM. 1992. Expression of adhesion antigens of human bone marrow megakaryocytes, circulating megakaryocytes and blood platelets. Eur J Haematol 49: 122–127. [DOI] [PubMed] [Google Scholar]
- Vierstra J, Lazar J, Sandstrom R, Halow J, Lee K, Bates D, Diegel M, Dunn D, Neri F, Haugen E et al. 2020. Global reference mapping of human transcription factor footprints. Nature 583: 729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, Rynes E, Sandstrom R, Zhang M, Canfield T, Hansen RS, Stehling-Sun S, Sabo PJ, Byron R, Humbert R et al. 2014. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346: 1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Flicek P, Odom DT. 2014. Evolution of transcription factor binding in metazoans - mechanisms and functional implications. Nat Rev Genet 15: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waksom ML. 2021. seaborn: statistical data visualization. Journal of Open Source Software 6: 3021. [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K et al. 2014. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH. 1997. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol 17: 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AG, Gaszner M, Felsenfeld G. 2002. Insulators: many functions, many mechanisms. Genes Dev 16: 271–288. [DOI] [PubMed] [Google Scholar]
- Xiang G, Giardine BM, Mahony S, Zhang Y, Hardison RC. 2021. S3V2-IDEAS: a package for normalizing, denoising and integrating epigenomic datasets across different cell types. Bioinformatics 37: 3011–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang G, Keller CA, Giardine B, An L, Li Q, Zhang Y, Hardison RC. 2020a. S3norm: simultaneous normalization of sequencing depth and signal-to-noise ratio in epigenomic data. Nucleic Acids Res 48: e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang G, Keller CA, Heuston E, Giardine BM, An L, Wixom AQ, Miller A, Cockburn A, Sauria MEG, Weaver K et al. 2020b. An integrative view of the regulatory and transcriptional landscapes in mouse hematopoiesis. Genome Res 30: 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Shao Z, Glass K, Bauer DE, Pinello L, Van Handel B, Hou S, Stamatoyannopoulos JA, Mikkola HK, Yuan GC et al. 2012. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev Cell 23: 796–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gu Q, Zhang Y, Sasaki T, Crivello J, O’Neill RJ, Gilbert DM, Ma J. 2018. Continuous-Trait Probabilistic Model for Comparing Multi-species Functional Genomic Data. Cell Syst 7: 208–218 e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD et al. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, An L, Yue F, Hardison RC. 2016. Jointly characterizing epigenetic dynamics across multiple human cell types. Nucleic Acids Res 44: 6721–6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hardison RC. 2017. Accurate and reproducible functional maps in 127 human cell types via 2D genome segmentation. Nucleic Acids Res 45: 9823–9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mahony S. 2019. Direct prediction of regulatory elements from partial data without imputation. PLoS Comput Biol 15: e1007399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Most of the data used in the integrative analysis are already publicly available; accession numbers for each file are listed in Supplementary Table S1. Regarding the new experiments, all raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE_______.
Resources developed in the VISION project are available at the website https://usevision.org. The epigenetic state annotations can be visualized in the BX VISION browser (http://main.genome-browser.bx.psu.edu). The database cCRE_db (https://globin.bx.psu.edu/cgi-bin/hlab/ccre_query) supports flexible user queries on extensive annotation of the cCREs, including epigenetic states and esRP scores across cell types, chromatin accessibility scores across cell types, membership in JmCs, and evolutionary categories. Intersections with other cCRE collections and epigenetic features are supported. The website section “Human and mouse cCREs and visualization” (https://usevision.org/data/ccre/) contains downloadable files of the cCREs along with annotation such as ATAC-seq signals and esRP scores, the joint metaclusters, the matched random regions, the UMAP visualizations of cCREs colored by esRP scores in each cell type, and movies of the changes in esRP scores across selected lineages of differentiation. The SOMs can be examined and analyzed at (https://usevision.org/som/DNA_V5/). The Downloads section contains data from both human and mouse for the input raw epigenetic data, gene expression levels from RNA-seq data, and epigenetic state assignments from IDEAS.
Code used in this study are in the following repositories:
| Process | URL |
|---|---|
| Joint human-mouse IDEAS pipeline | https://github.com/guanjue/Joint_Human_Mouse_IDEAS_State |
| S3V2 pipeline | https://github.com/guanjue/S3V2_IDEAS_ESMP |
| Build self-organizing maps | https://github.com/csjansen/SOMatic |
| sLDSC analysis | https://github.com/usevision/cre_heritability |