Significance
L-amino acids are crucial for protein synthesis and energy production in all organisms. Mammals synthesize most amino acids selectively in L-configurations, but mammals and their symbiotic microbes also convert L-amino acids into their D-configurations. While such chiral conversion of amino acids is linked to immune responses to symbiotic microbes, how mammals cope with D-amino acids is poorly understood. Here, we report that mammals sustain systemic predominance of L-amino acids against microbial D-amino acids by stereo-selective catabolism and urinary excretion of D-enantiomers. This study illuminates how mammals ensure an environment dominated by L-amino acids despite symbiosis with microbes.
Keywords: chirality, amino acid, D-amino acid oxidase, symbiosis
Abstract
Mammals exhibit systemic homochirality of amino acids in L-configurations. While ribosomal protein synthesis requires rigorous chiral selection for L-amino acids, both endogenous and microbial enzymes convert diverse L-amino acids to D-configurations in mammals. However, it is not clear how mammals manage such diverse D-enantiomers. Here, we show that mammals sustain systemic stereo dominance of L-amino acids through both enzymatic degradation and excretion of D-amino acids. Multidimensional high performance liquidchromatography analyses revealed that in blood, humans and mice maintain D-amino acids at less than several percent of the corresponding L-enantiomers, while D-amino acids comprise ten to fifty percent of the L-enantiomers in urine and feces. Germ-free experiments showed that vast majority of D-amino acids, except for D-serine, detected in mice are of microbial origin. Experiments involving mice that lack enzymatic activity to catabolize D-amino acids showed that catabolism is central to the elimination of diverse microbial D-amino acids, whereas excretion into urine is of minor importance under physiological conditions. Such active regulation of amino acid homochirality depends on maternal catabolism during the prenatal period, which switches developmentally to juvenile catabolism along with the growth of symbiotic microbes after birth. Thus, microbial symbiosis largely disturbs homochirality of amino acids in mice, whereas active host catabolism of microbial D-amino acids maintains systemic predominance of L-amino acids. Our findings provide fundamental insight into how the chiral balance of amino acids is governed in mammals and further expand the understanding of interdomain molecular homeostasis in host-microbial symbiosis.
Creating a homochiral milieu of amino acids is a signature of life. While D- and L-enantiomers of amino acids have equivalent chemical and physical properties, ribosomal protein synthesis employs only L-amino acids as building blocks to define a unique structure from each codon. Accordingly, organisms have evolved metabolic pathways and transport systems to establish predominantly L-amino acid environments. Synthesis and uptake selective for L-enantiomers were thought to maintain amino acid homochirality, but technological progress has revealed that D-amino acids are also present, albeit at low levels, in all domains of life (1, 2). Therefore, regulatory mechanisms for both enantiomers must exist to sustain amino acid homochirality.
Systemic concentrations of amino acids are tightly regulated by transport, metabolism, and protein biosynthesis/degradation in mammals. Organs work in concert to achieve systemic amino acid homeostasis: i) intestines absorb amino acids and peptides, ii) kidneys reabsorb amino acids to recycle, iii) all organs, especially the liver, metabolize amino acids, iv) muscles participate in protein biosynthesis and degradation, and v) the nervous and endocrine systems modulate such transport and metabolism (3). However, such basic understanding of amino acid homeostasis is based mostly on evidence that lacks chiral resolution. On the other hand, growing evidence shows that both endogenous and exogenous D-amino acids exist in mammals (4, 5), just as there are nonessential and essential L-enantiomers. Of note, mammalian amino acid racemases convert serine and aspartate from L- to D-configurations to modulate neurotransmission (6, 7), neurogenesis (8, 9), and/or hormone synthesis in the central nervous and endocrine systems (10, 11). Symbiotic bacteria have several metabolic pathways to synthesize various D-amino acids, such as D-alanine and D-proline, which initiate intestinal immune responses to control symbiosis with mammals (5, 12). D-amino acids such as D-serine, D-alanine, and D-aspartate are also included in common foods such as seafood and fermented foods (13). Despite our knowledge of physiological functions and occurrence of D-amino acids in mammals, how they handle such diverse D-enantiomers to sustain systemic dominance of L-amino acids is little known.
Under physiological conditions in mammals, most amino acids in plasma have L-configurations, while urine contains similar or larger amounts of D-amino acids than L-counterparts (14). Our recent study in humans shows that plasma D-amino acids are maintained within narrow ranges (low micromolar levels) and have different associations with organ functions than L-amino acids (15). Therefore, mammals appear to have totally distinct transport systems and metabolism for D- and L-enantiomers to sustain a specific chiral equilibrium in each organ. While transport systems of D-amino acids are largely unknown, key metabolic pathways for D-amino acids have been studied. Mammals express two FAD-dependent enzymes, D-amino acid oxidase (DAO) and D-aspartate oxidase (DDO), to degrade D-amino acids (16, 17). Since mammalian DDO degrades only D-aspartate (18), DAO catalyzes oxidation of nearly all other D-amino acids detected in mammals. Indeed, systemic loss of DAO activity increases some D-amino acids in blood, urine, the central nervous system, and peripheral organs (14, 19, 20), suggesting that DAO is crucial to regulate amino acid homochirality in mammals.
In this study, we sought to understand chiral homeostasis of amino acids in mammals and to clarify its regulation from the fetal period to adulthood in mice. Our results reveal that DAO is essential for interdomain homeostasis of amino acid chirality between mammals and microbes.
Results
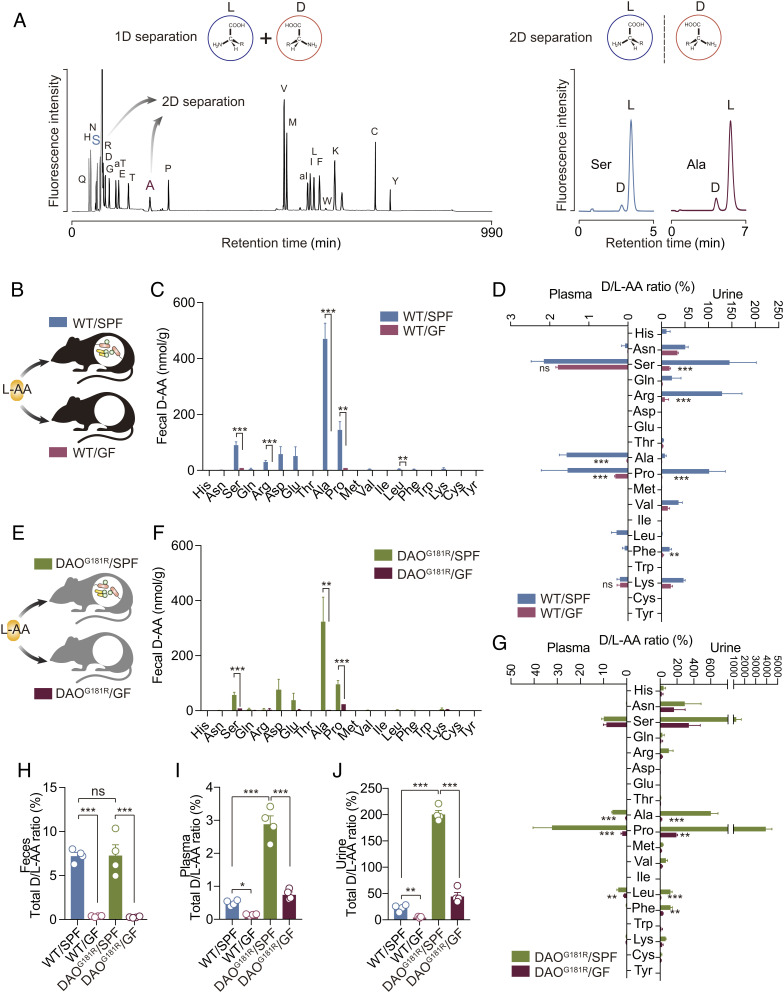
Unlike other organisms, bacteria produce a variety of D-amino acids as essential components of their cell walls and as regulators of diverse cellular processes (21, 22). Therefore, we hypothesized that symbiotic bacteria may disrupt chiral homeostasis of amino acids in mammals. In this study, to determine amino acid enantiomers in biological samples, we used a two-dimensional high-performance liquid chromatography (2D-HPLC) system (23), in which the first dimension separated respective amino acids, followed by chiral separation into D- and L-enantiomers in the second dimension (Fig. 1A). Samples that included small amount of D-amino acids were further confirmed in combination with tandem mass spectrometry (MS) (23). First, to test whether symbiotic bacteria synthesize D-amino acids in mammals, we fed adult C57BL/6 mice with an L-amino acid diet (L-AA) (Fig. 1B), which contained 16.7% (w/w) amino acids with >99.9% selectivity for L-enantiomers (SI Appendix, Fig. S1) and included no protein as a nitrogen source. In a specific pathogen-free (SPF) condition, D-serine, D-arginine, D-aspartate, D-glutamate, D-alanine, and D-proline were found in the feces at concentrations ranging from 30 nmol/g to 500 nmol/g (Fig. 1C). D-glutamine, D-valine, D-leucine, D-phenylalanine, and D-lysine were detected at trace levels (Fig. 1C). Among D-amino acids, D-alanine was most abundant (Fig. 1C), reflecting the fact that most bacteria require D-alanine as a component of their cell walls. In contrast, no D-amino acids, except for trace levels of D-serine and D-proline, were detected in feces when mice were raised in germ-free (GF) conditions (Fig. 1C). On the other hand, fecal L-amino acid levels in GF mice were generally higher than those in SPF mice with a pattern distinct from that of fecal D-amino acids (SI Appendix, Fig. S2A). These findings suggest that microbes actively and almost exclusively produce many kinds of D-amino acids in the gut, whereas several L-amino acids including L-aspartate, L-glutamate, L-alanine, L-valine, L-leucine, and L-lysine, are metabolized by gut microbes.
Fig. 1.
Mammalian catabolism/excretion of D-amino acids maintains amino acid homochirality disturbed by symbiotic microbes. (A) Chromatograms show representative separation of amino acids in the first dimension (Left) followed by separations of enantiomers in the second dimension (Right) of two-dimensional HPLC (2D-HPLC). (B–J) Wild-type (WT, B–D) or DAOG181R mice (E–G) fed with the L-amino acid diet (L-AA) were raised in SPF or GF conditions. Amino acid enantiomers were quantified in feces, plasma, and urine of WT or DAOG181R mice using 2D-HPLC. D-Enantiomer levels of feces are shown in C and F. D/L-Enantiomer ratios (D/L-AA ratios) for each amino acid in plasma and urine are indicated in D and G, and those for total amino acids are in H (feces), I (plasma), and J (urine). N = 4, each group (exception: n = 3 for urine from WT/GF). Error bars, mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, analyzed with multiple t test corrected for multiple comparisons using Bonferroni–Dunn method (C, D, F, and G) or one-way ANOVA followed by Tukey’s test (H–J). ‘ns’, not significant.
In the plasma, some D-amino acids, such as D-serine, D-alanine, D-proline, D-leucine, D-phenylalanine, and D-lysine, detected in the feces, were also found at low micromolar levels under SPF conditions, whereas most D-amino acids were significantly reduced under GF conditions (SI Appendix, Fig. S2B). Importantly, plasma D-serine and D-lysine levels were not affected by the absence of microbes (SI Appendix, Fig. S2B). Those results indicate that microbial D-amino acids are absorbed to some extent and many of them circulate in mice but that plasma D-serine and D-lysine may have murine origins. On the other hand, the absence of microbes did not influence plasma levels of L-amino acids (SI Appendix, Fig. S2C), possibly because the diet met the L-amino acid requirement and half of the microbe-metabolizing L-amino acids, such as L-aspartate, L-glutamate, and L-alanine, can be synthesized de novo by mice as well. Consequently, plasma D-/L-amino acid ratios were up to 2% in SPF conditions, while those in GF conditions were less than 0.5%, except for serine, reflecting the reduction in D-enantiomers (Fig. 1D). Such reductions of D-/L-amino acid ratios in GF conditions were more evident in urine (Fig. 1D). D-/L-Amino acid ratios were generally much higher in urine than in plasma and those for serine, arginine, and proline showed a ratio >50% in urine under SPF conditions (Fig. 1D). In GF conditions, most amino acids including serine showed significant declines in D-/L-enantiomer ratios (Fig. 1D). These results show that levels of D-amino acids in mammalian body fluids are influenced by the presence of microbes, while L-amino acid levels are independent of microbes. Furthermore, low D-/L-amino acid ratios in the plasma compared with those in urine indicate the presence of active regulatory mechanisms for chiral balance of amino acid enantiomers.
Next, we tested whether mammalian metabolism also influences levels of D-amino acids originating from symbiotic microbes. DAO is an unusual enzyme that catabolizes D-amino acids in mammals and has broad substrate specificity for D-amino acids with strict chiral selectivity (16). DAO degrades D-serine in the central nervous system (24), whereas microbial D-amino acids are catabolized by peripheral DAO (5, 12). To understand the extent to which DAO degrades microbial D-amino acids in mammals, we fed an L-AA to mice lacking DAO activity, due to homozygous G181R point mutations in the DAO gene (DAOG181R) raised in SPF or GF conditions (Fig. 1E). DAOG181R mice showed similar levels of fecal D- and L-amino acids to wild-type (WT) C57BL6 mice in SPF conditions and trace levels of D-amino acids in GF conditions (Fig. 1 C and F and SI Appendix, Fig. S2A and S3A), showing that fecal microbes produce D-amino acids independent of host DAO activity. On the other hand, loss of DAO activity significantly increased plasma levels of D-amino acids, including D-serine, D-alanine, D-proline, and D-leucine (SI Appendix, Fig. S3B), but not those of L-counterparts (SI Appendix, Fig. S3C), which resulted in D-/L-amino acid ratios elevated to 5 to 35% in the plasma under SPF conditions (Fig. 1G). An absence of microbes reduced D-alanine, D-proline, and D-leucine, but did not affect D-serine or L-amino acids in plasma (SI Appendix, Fig. S3 B and C), as reflected in D-/L-amino acid ratios (Fig. 1G). Therefore, DAO degrades both endogenous and microbial D-amino acids but impacts predominantly microbial D-amino acids in amount and variety (Fig. 1G and SI Appendix, Fig. S3B). This view is further supported by urinary D-/L-amino acid ratios in DAOG181R mice. In the presence of microbes, several D-amino acids, such as D-asparagine, D-serine, D-alanine, D-proline, D-leucine, and D-phenylalanine, were even more abundant compared to L-enantiomers in urine of DAOG181R mice (Fig. 1G). On the other hand, lack of microbes resulted in significant reduction of urinary D-/L-amino acid ratios including serine, alanine, proline, leucine, and phenylalanine (Fig. 1G). These results indicate that urinary excretion of microbial D-amino acids compensates in part for lack of DAO activity to sustain chiral homeostasis of amino acids in plasma.
To evaluate overall chiral homeostasis in WT and DAOG181R mice raised under SPF or GF conditions, levels of all proteinogenic L-amino acids or their D-counterparts, with the exception of glycine, which is achiral, were summed (SI Appendix, Fig. S4 A–D). The total fecal D-/L-amino acid ratio was constant at about 7% regardless of DAO activity in SPF conditions and low at around 0.3% in GF conditions (Fig. 1H). On the other hand, loss of DAO activity increased ratios of total D-/L-amino acids in plasma from 0.50 to 2.9% in SPF conditions, whereas, in GF conditions, ratios were only minimally elevated from 0.14 to 0.76% (Fig. 1I). A similar tendency was apparent in urine. Urinary total D-/L-amino acid ratios were 23% in WT animals and 200% in DAOG181R mice under SPF conditions, whereas they were 5.1% in WT mice and 45% in DAOG181R mice in GF conditions (Fig. 1J). These results suggest that symbiotic microbes disrupt the homochirality of amino acids and that mammals sustain chiral homeostasis by selective degradation and excretion of D-enantiomers.
We wondered when such degradation and excretion of D-enantiomers occurs during development. To understand the regulation of chiral homeostasis of amino acids in the prenatal stage, we transplanted WT or DAOG181R embryos to either WT or DAOG181R pseudopregnant females and measured amino acid enantiomers in dams and fetuses on embryonic day 16 (Fig. 2A). No gross abnormality or stunting was observed in fetuses in the presence or absence of maternal or fetal DAO activity. We selected alanine to evaluate a microbial D-amino acid and serine as a mammalian D-amino acid. DAOG181R dams had higher D-/L-alanine and D-/L-serine ratios in the plasma than WT dams (Fig. 2B and SI Appendix, Fig. S5 A and B), as observed in nonpregnant animals (Fig. 1). Lack of DAO activity in the dams significantly increased D-/L-alanine and D-/L-serine ratios in fetal plasma, but fetal genotype did not alter D-/L-amino acid ratios in fetal plasma (Fig. 2C and SI Appendix, Fig. S6 A and B). Together with the fact that DAO is not expressed in fetal kidneys (SI Appendix, Fig. S6C), these results indicated that fetal DAO is absent or inactive throughout the fetal body. Of note, amniotic fluid, which contains fetal urine, had similar ratios of D-/L-amino acids compared to plasma (Fig. 2D and SI Appendix, Fig. S6 D and E), suggesting that D-amino acids are not actively synthesized due to lack of symbiotic microbes nor excreted into the amniotic cavity in the fetus. Again, loss of DAO activity in dams but not fetuses enhanced D-/L-alanine ratios in amniotic fluid (Fig. 2D and SI Appendix, Fig. S6 D and E). A similar trend was observed in the placenta (Fig. 2E and SI Appendix, Fig. S6 F and G). Those findings suggest that maternal D-amino acids with both endogenous and microbial origins influence the chiral balance of amino acids in the fetal circulation but that fetuses are unable to regulate homeostasis in utero. Therefore, chiral homeostasis appears to occur after birth.
Fig. 2.
Maternal catabolism of D-enantiomers regulates fetal amino acid homochirality. (A) WT or DAOG181R embryos were transplanted into WT or DAOG181R surrogate mothers. (B–E) Enantiomers for alanine and serine quantified in dam plasma (B), fetal plasma (C), amniotic fluid (D), and placenta (E) on E16 using 2D-HPLC and D/L-AA ratios are shown. N = 3, each group. Error bars, mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, analyzed with Student’s t test (B) or one-way ANOVA followed by Tukey’s test (C–E). ‘ns’, not significant.
Mammals are colonized by microbes immediately after birth, and levels and diversity of microbes increase thereafter. Therefore, we further monitored the transition of D- or L-amino acid levels in C57BL6 mice after birth. To understand how the production of D-amino acids by microbes changes in the intestine, we compared the levels of D-alanine and D-serine to bacterial abundance during the postnatal period. Relative amounts of total intestinal bacteria, evaluated by quantitative PCR for the 16s rRNA gene, gradually increased after birth and showed a striking surge of 100 to 350-fold after postnatal day 14 (P14), coinciding with weaning (Fig. 3A). D-alanine and D-serine levels in the intestines were both elevated and positively correlated with bacterial colonization density, to a greater extent for D-alanine (Fig. 3 A and B), suggesting that both D-amino acids originate from microbes in the intestine. Plasma D-alanine was slightly but similarly increased after P14 (SI Appendix, Fig. S7A), whereas plasma D-serine decreased after birth (SI Appendix, Fig. S7A). In contrast, D-/L-serine and D-/L-alanine ratios were both constant during development (Fig. 3C and SI Appendix, Fig. S7 A and B), suggesting that the equilibrium of enantiomers is regulated in terms of D-/L-ratios in blood. Such equilibrium can be maintained in part by urinary excretion of amino acid enantiomers, especially during the neonatal period. Urinary levels of amino acid enantiomers, standardized by urinary creatinine concentration, showed a consistent decline after birth until P14 (SI Appendix, Fig. S7 C and D), which may coincide with postnatal development of tubular function. In fact, fractional excretion (FE) of amino acid enantiomers, percentages of enantiomers in urine among those filtered by the kidneys, decreased dramatically after P14 (Fig. 3D). These results indicate that both D- and L-enantiomers of serine and alanine are actively reabsorbed in mature kidney tubules. However, since FEs of D-amino acids were much higher than those of L-enantiomers (Fig. 3D), D-amino acids appear to be less efficiently reabsorbed by the tubules compared to L-enantiomers. Indeed, D-/L-amino acid ratios in urine were significantly larger than those in plasma (SI Appendix, Fig. S7E). Another factor to control the equilibrium of D- and L-amino acids in the blood is degradation of D-amino acids by DAO (Fig. 1), expressed primarily in kidney tubules (16, 25). Renal DAO expression gradually increased after birth and reached adult levels around P14 (Fig. 3E). Considering that intestinal D-alanine levels increased dramatically after P14 (Fig. 3A), we speculated that enhanced DAO expression compensates for the disruption of chiral homeostasis by microbes during development.
Fig. 3.
Developmental alterations in systemic chiral balance of amino acids. (A) D-amino acids (Left vertical axis) and relative abundance of bacterial 16s rRNA gene (Right) in whole intestinal contents in C57BL6 mice were quantified along with development. (B) Correlations between D-amino acids and 16s rRNA gene abundance are shown. (C and D) Plasma D/L-amino acid ratios (C) and fractional excretion of amino acid enantiomers (D) during development were calculated. (E) Western blotting for DAO and GAPDH in developing kidneys is shown. N = 3, each timepoint. Error bars, mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001, analyzed with one-way ANOVA followed by Dunnet’s test (comparisons to P0) (A, C, and D) or Spearman’s correlation coefficient (B). ‘ns’, not significant.
Finally, we studied whether humans also have such chiral homeostasis. Enantiomers of 19 amino acids were quantified in the feces, plasma, and urine from 24 healthy adult volunteers (12 males and 12 females). As observed in mice, human feces contain a variety of D-amino acids, including D-serine, D-arginine, D-aspartate, D-glutamate, D-alanine, D-proline, D-leucine, and D-lysine, ranging from 10 to 800 nmol/g (SI Appendix, Fig. S8A). Notably, despite the lack of dietary uniformity, D-/L-amino acid ratios in feces were quite similar between individuals (Fig. 4A and SI Appendix, Fig. S8 A and B). Some D-/L-amino acid ratios such as for aspartate and valine had positive correlations (SI Appendix, Fig. S9A). On the other hand, in plasma, we detected D-serine, D-alanine, and D-proline at trace levels in all subjects, but no other D-amino acids (SI Appendix, Fig. S8C), whereas all tested L-amino acids, except L-histidine, were detectable at 5 to 500 nmol/mL (SI Appendix, Fig. S8D). Resultant plasma D-/L-amino acid ratios were <2% with small variations (serine, 1.75 ± 0.08%; alanine, 0.21 ± 0.02%; and proline, 0.26 ± 0.03%) (Fig. 4B) and with mild positive associations with each other (SI Appendix, Fig. S9B), suggesting that chiral homeostasis is strictly maintained in humans as well. Urinary D-/L-amino acid ratios were much higher than those of plasma in humans, with serine presenting the highest ratio, as was observed in mice (Figs. 1D and 4C). Ratios for asparagine, serine, glutamine, threonine, and lysine, or those for valine and leucine had positive correlations with each other (SI Appendix, Fig. S9C). Exceptionally, unlike mice, humans had low urinary ratios of D-/L-arginine and D-/L-proline (Fig. 4C). FEs for L-amino acids were <1% (SI Appendix, Fig. S8E), while those for D-amino acids were much higher than for L-counterparts (D-serine, 30.4% ± 2.1%; L-serine, 0.72 ± 0.13 %; D-alanine, 23.2 ± 3.0 %; and L-alanine, 0.18 ± 0.03 %) (SI Appendix, Fig. S8F). Therefore, although reabsorption patterns of enantiomers in humans may differ somewhat from those in mice, our results suggest that D-amino acids are less efficiently reabsorbed in the kidney than L-enantiomers in humans and mice. To further assess overall chiral homeostasis of amino acids in humans, all proteinogenic L-amino acids except glycine or their D-counterparts were summed. Chiral equilibrium evaluated by total D-/L-amino acid ratios was maintained within narrow ranges in plasma (0.13 ± 0.01%), urine (17.04 ± 1.39%), and feces (17.89 ± 1.56%) (Fig. 4D) with no associations with each other (SI Appendix, Fig. S9D). There were no sex differences in chiral equilibrium in plasma or feces, while urinary D-/L-amino acid ratios in females were slightly higher than those in males (Fig. 4E). Therefore, given our findings in mice (Fig. 1), those results suggest that humans also strictly maintain plasma amino acid homochirality against chiral conversion by microbes, in part through urinary excretion of D-enantiomers. We further examined whether specific microbial taxa in feces are associated with total D-/L-amino acid ratios in feces, plasma, or urine. Alpha or beta diversity for fecal microbiota, analyzed using 16s rRNA gene sequencing varied among individuals with no sex differences (SI Appendix, Fig. S10 A and B). Therefore, we tested correlations between relative abundance of microbial taxa (SI Appendix, Fig. S10 C and D) and D-/L-amino acid ratios, without categorizing subjects. There were no correlations between abundances of detectable bacterial classes and total D-/L-amino acid ratios in feces, plasma, or urine (Fig. 4F), but the relative abundance of Clostridia, one of the most abundant classes of human gut microbiota, had positive correlations with D-/L-ratios for plasma proline, urinary tyrosine, and fecal aspartate (SI Appendix, Fig. S11 A and B). In contrast, D-/L-ratios for fecal alanine and proline, which were significantly high among those of fecal amino acids (Fig. 4A), did not show correlations with abundances of bacterial classes. This may indicate that their chiral conversion is conducted by gut microbiota in general. In fact, relative fecal bacterial level, estimated by qPCR for the 16s rRNA gene, was positively correlated with fecal total D-/L-amino acid ratio (Fig. 4G). On the other hand, fecal bacterial level showed no correlations with plasma or urinary D-/L-amino acid ratios (Fig. 4G), which was likely because plasma or urinary parameters mediate additional physiological regulatory processes. Thus, those results suggested that humans also have chiral equilibrium of amino acids in body fluids, despite chiral conversion by symbiotic bacteria.
Fig. 4.
Chiral homeostasis of amino acids in humans. (A–C) Amino acid enantiomers were quantified using 2D-HPLC and ratios of D/L-amino acids in feces (A), plasma (B), and urine (C) are shown in healthy individuals (n = 24). (D and E) Ratios of total D/L-amino acids (D) and their sex differences (E, n =12 each) are shown. (F) Heat maps show correlation coefficients between ratios of total D/L-amino acids in feces, urine, or plasma and relative abundances of fecal bacterial phyla (Left) and classes (Right). (G) Correlations between total D/L-amino acid ratios and relative bacterial amounts estimated by 16s rRNA gene qPCR are shown. Error bars, mean ± SEM. *P < 0.05 and ***P < 0.001, analyzed with Kruskal–Wallis followed by Dunn’s multiple comparison test (D), Mann–Whitney U test (E), Spearman’s correlation coefficient (F), or linear regression (G). ‘ns’, not significant.
Discussion
We found that symbiotic microbes synthesize diverse D-amino acids and disturb the homochiral environment of amino acids in mammals. In the prenatal period, the maternal homochiral environment of amino acids preserves fetal homeostasis of enantiomers. After birth, kidney DAO and urinary excretion remove D-amino acids synthesized by symbiotic microbes to maintain amino acid homochirality. Catabolism of D-amino acids seems central to achieve homochirality since urinary excretion of D-amino acids declines after birth, in contrast to increased DAO in the kidneys with growth of symbiotic microbes. Thus, mammals adapt to the heterochiral amino acid environment generated by symbionts with support from their mothers during the prenatal period and via activity of the kidneys after birth.
Symbiotic microbes produce a specific, abundant set of D-amino acids, including D-serine, D-aspartate, D-glutamate, D-alanine, and D-proline in mammals (Figs. 1 C and F and 4A). Among symbionts, bacteria have a unique capacity to synthesize diverse D-amino acids and to incorporate them into peptidoglycans (26), a major component of bacterial cell walls. Peptidoglycans, net-like heteropolymers, are composed of glycan strands that are cross-linked by peptide stems containing essentially D-amino acids (22), such as D-alanine and D-glutamate in general, as observed in mouse and human feces (Figs. 1 C and F and 4A). Since most mammalian proteases cleave peptide bonds only between L-amino acids, D-amino acids in peptidoglycans make bacteria resistant to digestive enzymes and govern bacterial fitness in complex gut ecosystems (27). Intriguingly, different types of D-amino acids in the peptide stem may show distinct cell wall chemistry in the gut. For instance, the presence of D-serine or D-aspartate at the terminal position of peptide stems affords resistance to certain bactericidal agents such as vancomycin (28, 29). Moreover, other noncanonical D-amino acids, such as D-methionine, D-leucine, D-tyrosine, and D-phenylalanine, control robustness of peptidoglycan to changing osmolarity or stresses such as starvation (26, 30). In addition, noncanonical D-amino acids also regulate growth (31), spore germination (32), and biofilm dispersal (33) in certain species of bacteria. Therefore, we assume that intestinal microbes chiral-convert amino acids to adapt to environmental changes, enabling them to reside in the gut. Although we detected similar sets and abundance of D-amino acids among healthy subjects, including rodents and humans (Figs. 1 and 4), emergence of different microbes due to antibiotic resistance or inflammation may result in D-amino acids other than those detected in this study.
The absence of symbiosis with microbes strikingly reduces D-amino acids in the intestine and body fluids, allowing mice to maintain systemic homochirality without metabolic burdens (Fig. 1). Therefore, our findings support the classical view that de novo synthesis of most amino acids is selective to L-enantiomers in mammals. However, regardless of microbial symbiosis, serine and lysine have low, but constant D/L-enantiomer ratios in blood (Fig. 1 C and F), suggesting endogenous production of their D-enantiomers. While lysine remains uncharacterized, serine is an exceptional amino acid that can be chiral-converted by a mammalian enzyme, serine racemase (SR) (6). SR accounts for 90% of D-serine synthesis in the central nervous system (34, 35), where D-serine binds to N-methyl-D-aspartate receptors to modulate excitatory neurotransmission (36, 37). In contrast to our understanding of D-serine as a neuromodulator in the central nervous system, the significance of D-serine production in the periphery remains largely unknown. SR-knockout in mice does not influence blood D-serine levels (38) and neither do microbes (Fig. 1 C and F), implying the existence of a previously unidentified mammalian enzyme to synthesize D-serine (35), which should be characterized in future studies. On the other hand, peripheral D-amino acids originating from symbiotic microbes are involved in murine immune responses to microbes. Since catabolism of D-amino acids by DAO generates hydrogen peroxide, DAO has been linked to innate defense against bacteria by leukocytes (39, 40). More recent reports indicate immunomodulatory roles of microbial D-amino acids in both innate and acquired responses in the mucosa. Intestinal epithelium induces DAO in response to microbes, which helps to limit microbial growth, as well as to catabolize microbial D-amino acids (5). Induction of DAO is linked to inhibition, survival, and differentiation of intestinal B lymphocytes and it reduces excessive production of immunoglobulin A against symbiotic microbes (12). As we found that DAO regulates microbial D-amino acids (Fig. 1), DAO appears to maintain symbiosis with gut microbes. Also, in the upper airway, microbial D-amino acids, such as D-leucine and D-phenylalanine, directly inhibit release of antimicrobial peptides by binding to sweet taste receptors (T1R2/R3) (41). Furthermore, microbial D-tryptophan inhibits the growth of enteric pathogens (42) and also modulates the gut microbial community to attenuate allergic responses by influencing T lymphocytes in the lower airway (43). Therefore, microbes appear to release D-amino acids to attenuate host immune responses, which benefits their symbiotic relationships with mammalian hosts. Notably, since whole-exome sequencing in humans has revealed an association between DAO and energy expenditure (44), microbial D-amino acids may also influence symbiosis with regard to energy metabolism, although the underlying mechanism remains elusive. Thus, those studies suggest that loss of homochirality triggers aberrant immune responses and/or energy metabolism in mammals.
Selective synthesis and absorption/reabsorption of L-enantiomers were thought to account for the homochirality of amino acids in mammals. Contrary to this idea, the ability of bacteria to metabolize D-alanine has led some researchers to propose the presence of bacterial D-amino acids in mammals. In 1965, Hoeprich first suggested that the gut microbiota might be the source of serum D-alanine in rodents (45). Konno showed in his seminal works using DAO-null mice that the majority of urinary D-alanine is of microbial origin (46, 47). Consistent with their observations, our findings clearly show that microbes actively chiral-convert diverse L-amino acids to their D-counterparts, whereas mice maintain the predominance of L-enantiomers through D-enantiomer-specific catabolism by DAO and urinary excretion (Figs. 1–3). Along with exponential growth of gut microbiota after birth, accelerated chiral conversion by microbes is countered by an increase of kidney DAO expression, while the FE of D-amino acids declines in an opposite manner (Fig. 3). Therefore, catabolism, but not urinary excretion of D-amino acids appears central to the maintenance of homochirality in mature individuals. In fact, loss of DAO markedly elevates urinary excretion of D-amino acids in adult mice, and the resultant increase of blood D-amino acids indicates that renal excretion capacity for D-enantiomers cannot compensate for microbial chiral conversion (Fig. 1). Since Hans Krebs first documented in 1935 that kidneys have much greater catabolic activity for D-amino acids than L-enantiomers (48), the physiological significance of kidney DAO has remained obscure for more than 80 y. As DAO catalyzes oxidation of a wide variety of D-amino acids synthesized by symbiotic bacteria (Fig. 1), we propose that mammalian DAO in the kidneys serves as a critical regulator to sustain amino acid homochirality. While the ultimate understanding of the basis for amino acid homochirality in mammals requires additional studies that will illuminate transport systems for D-amino acids especially in the intestines and kidneys, this study provides fundamental insight into the mechanism by which the mammals ensure an environment dominated by L-amino acids.
Materials and Methods
Animals.
All animal experiments were approved by the institutional Animal Experiment Committee and conducted in accordance with Institutional Guidelines on Animal Experimentation at Keio University. DAOG181R/G181R mice were on the C57BL6 background (49). DAOWT/WT mice used in this study were generated by in vitro fertilization of DAOG181R/WT heterozygote breeders and transplantation to surrogate mothers.
DAOG181R/G181R and DAOWT/WT male mice were raised in GF or SPF conditions in our institution or CLEA Japan, and after weaning for 3 wk until sampling, they were fed L-AAs (Dyets, # 510025), which contained only purified L-amino acids as a nitrogen source. The purity of L-amino acids in the diet was confirmed using 2D HPLC (SI Appendix, Fig. S1).
For embryonic amino acids, embryos generated by in vitro fertilization of DAOG181R/WT heterozygote breeders were transplanted into DAOWT/WT and DAOG181R/G181R surrogate mothers in vitro. Mutant DAO embryos and their surrogate mothers were sacrificed on embryonic day 16. For the detection of amino acids during development, pregnant C57BL6 mice were obtained from CLEA Japan and neonates were killed on postnatal days 0, 3, 6, 14, 21, and 28.
Human Samples.
The study protocol was approved by the Ethics Committee of Keio University School of Medicine (Approval No. 20190258). Human samples were collected from healthy adult volunteers recruited at the Shinanomachi Campus of Keio University. Subjects were males or females between the ages of 20 and 45. Consent was obtained from 12 males (age, 30.2 ± 1.8; BMI, 22.9 ± 0.6) and 12 females (age, 30.0 ± 1.1; BMI, 20.9 ± 0.7). Recruits had no antibiotics within 3 wk prior to the day of sampling and had only drinking water with no food on the day of sampling.
Blood was drawn from antecubital vein to microtubes containing EDTA-2Na. After more than 30 min on ice, blood was centrifuged at 1,200 × g for 10 min. Plasma samples were kept at −80 °C until use. Urine samples were taken from midstream of the first urination in the morning on the sampling day. Stool samples were obtained in a stool collection container (TOYO KIZAI, Japan) within 24 h before or after blood sampling. Urine and stool samples were stored at 4 °C as soon as they were collected and transferred to −80 °C within 12 h after collection.
Quantification of Amino Acid Enantiomers with Two-Dimensional HPLC.
Feces were homogenized at 3,500 rpm for 10 s in 20-fold v/w of H2O at 4 °C using a micro-homogenizing system (Micro Smash MS-100R; TOMY), incubated on ice for 15 min, and centrifuged at 4 °C at 12,100 × g for 5 min. Fetal blood was collected from the carotid artery exposed by decapitation with a micro hematocrit tube containing K2-EDTA and centrifuged at 3,500 × g for 5 min to obtain plasma. Amniotic fluid was collected from the fetal sac by puncture aspiration. Those samples were homogenized in 10 or 20-fold v/w of H2O at 3,500 rpm for 2 min at 4 °C using the microhomogenizing system and centrifuged at 12,100 × g for 10 min. Placentas were removed from E16 embryos and homogenized in 10 or 20-fold volume/weight of H2O with metal beads (ø3.2 mm) at 3,500 rpm for 2 min at 4 °C using the microhomogenizing system and centrifuged at 12,100 × g for 10 min at 4 °C. Supernatants were stored at −80 °C until use.
Amino acid enantiomers were quantified using a 2D-HPLC system (NANOSPACE SI-2 series, Shiseido) in part in combination with the MS/MS system, as previously described (23). Briefly, protein in the samples homogenized in H2O was removed by addition of 9 volumes of methanol. The mixture was vortexed vigorously and centrifuged at 12,100 × g for 5 min at 4 °C. The supernatant was evaporated to dryness, suspended in 200 mM sodium borate, and then derivatized with 4-fluoro-7-nitro-2,1,3-benzoxadiazole (NBD-F). NBD-conjugated amino acids were separated on an octadecylsilyl column (Singularity RP18, 1.0 mm inner diameter (ID) × 250 mm) (designed by Kyushu University and KAGAMI Co. Ltd., Osaka, Japan) for first dimensional (1D) separation. Enantiomers of amino acids were separated using a Pirkle-type enantioselective column (Singularity CSP-001S, 1.5 mm ID × 250 mm) for 2D separation (designed by Kyushu University and KAGAMI). Fluorescence of NBD-amino acids was detected at 530 nm with excitation at 470 nm. The isolated NBD-amino acids after 2D separations were used for the MS/MS analysis.
FE.
FEs of D- and L-amino acids (FED-aa and FEL-aa) was calculated by clearance of substrate divided by that of creatinine: FE = substrate clearance/creatinine clearance = US × PCr/UCr × PS (US and PS, levels of urinary and plasma substrate; UCr and PCr, levels of urinary and plasma Cr).
16S rRNA Gene Sequencing and Analysis.
Stools collected from healthy adult volunteers were homogenized in 20-fold v/w PBS with metal beads (ø3.2 mm, TOMY) using a Micro Smash at 3,500 rpm for 20 s. A mixture of 300 μL homogenate, 400 μL of an extraction buffer (200 mM Tris-HCl pH 8.0, 200 mM NaCl, and 20 mM EDTA), a 300-μL slurry of glass beads (ø0.1 mm, TOMY), 50 µl of 10% SDS solution, and 500 μL of PCI solution (phenol : chloroform : isoamyl-alcohol, 25 : 24 : 1; Invitrogen) was further homogenized using the Micro Smash at 3,500 rpm for 3 min and centrifuged at 14,000 × g for 5 min. Subsequently, 500 μL of supernatant was mixed with the same volume of chloroform, vortexed for 10 s, and centrifuged at 14,000 × g for 5 min. DNA was precipitated by adding 300 μL of isopropanol to 300 μL of the aqueous phase, pelleted by centrifugation at 14,000 × g for 5 min and resuspended with 100 μL of 10 mM Tris-HCl (pH 8.5). Then, the DNA extract was purified with a Spin Smart column (CM 400-50, Denville, South Plainfield, NJ, USA) and bacterial genomic DNA was eluted with 10 mM Tris-HCl (pH 8.5). Then, the V1–V2 region of 16S ribosomal RNA gene was PCR amplified (12.5 ng purified DNA per reaction; Phusion polymerase, New England Biolab, Ipswich, MA, USA) (25 cycles: 95 °C for 30 s, 50 °C for 30 s, and 72 °C for 30 s) (primer pair: 27Fmod/338R with overhang adapters) (adapter-27Fmod: 5′-tcg tcg gca gcg tca gat gtg tat aag aga cag AGR GTT TGA TYM TGG CTG AG-3′; adapter-338R: 5′-gtc tcg tgg gct cgg aga tgt gta taa gag aca gTG CTG CCT CCC GTA GGA GT-3′). PCR products were purified (MinElute, QIAGEN) and resuspended in 25 µL of 10 mM Tris-HCL pH 8.5. V1–V2 PCR products were indexed with a Nextera XT Index kit (Illumina, San Diego, CA, USA) by PCR (2.5 μL PCR product; Nextera XT Index primers; Phusion polymerase) (8 cycles: 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s). 16S rRNA amplicons with indices were purified (MinElute, QIAGEN), resuspended in 25 μL of 10 mM Tris-HCl (pH 8.5), quantified with a Qubit 2.0 Fluorometer (Life Technologies), pooled at a concentration of 4 nM, denatured, diluted to a final concentration of 8 pM, and sequenced using the MiSeq Reagent Kit v2 (500-cycle, paired-end, Illumina) on a MiSeq sequencer (Illumina). 16S rRNA sequencing analysis with Qiime was previously described (5).
Western Blotting.
Tissues removed from mice were homogenized in a lysis buffer [15 mM sodium chloride, 20 mM EDTA, 50 mM Tris-HCl (pH 7.4), 1% Triton X-100 and a protease inhibitor cocktail, Complete EDTA-free (Roche, Basel, Switzerland)] and centrifuged at 12,000 × g at 4 °C for 10 min. Cerebellar lysate was used as a positive control, and the cerebral cortex was used as a negative control. Supernatants were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and protein was transferred to PVDF membranes. Blots were blocked in 5% skim milk in PBS with 0.1% Tween-20 (PBST) with constant shaking for 60 min. Membranes were rinsed in PBST and immersed in PBST with appropriate primary antibodies [a rabbit polyclonal antibody to mouse DAO (5); a rabbit monoclonal antibody to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (14C10), Cell Signaling Tech (Danvers, MA, USA)] at 4 °C overnight. Membranes were subsequently rinsed in PBST and then incubated with an appropriate secondary antibody conjugated with horseradish peroxidase for 30 min. Membranes were rinsed in PBST, and bound antibodies were detected with the Pierce ECL Plus Western Blotting Substrate (Thermo Scientific).
qPCR Analysis.
The amount of genomic DNA extracted from intestinal luminal contents was normalized to the mass of the contents. Real-time quantitative PCR analysis was performed in a QuantStudio 1 Real-Time PCR System (Applied Biosystems). The following PCR primers were used: 16S rRNA gene (forward (1048 to 1067: 5′-GTGSTGCAYGGYTGTCGTCA-3′; reverse: 5′-ACGTCRTCCMCACCTTCCTC-3′).
Statistical Analysis.
No statistical methods were used to predetermine sample size. Blinding was not possible for animal experiments. No randomization was used. Prism 9 (GraphPad Software) was used for data plotting and statistical analyses. Statistical significance was determined with two-sided unpaired t tests to compare two groups, or one-way analysis of variance (ANOVA) for multiple comparisons when data were normally distributed and had equal variance. If variances of the data were not equal, then nonparametric tests were performed. Simple correlation was analyzed using Spearman’s correlation coefficient by R studio and visualized using corrplot packages. D-/L-amino acid ratios of His, Ile, Cys, and Tyr in the feces, those of His, Asp, Glu, Trp, and Cys in the urine, and those of His, Asn, Gln, Arg, Asp, Glu, Thr, Met, Val, Ile, Leu, Phe, Trp, Lys, Cys, and Tyr in the plasma were below the detection limit and did not contribute to the correlation matrix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Maiko Nakane, Hiroshi Imoto, Tatsuhiko Ikeda, Eiichi Negishi, and Shoto Ishigo for the technical support on chiral amino acid analysis and Steven D. Aird for editing the manuscript. This work was funded by Japan Agency for Medical Research and Development (19gm6010017h0002) (J.S.), JSPS KAKENHI Grant number 21H02982 and 22K19408 (J.S.), Keio Gijuku Fukuzawa Memorial Fund for the Advancement of Education and Research (J.S.), and Keio Program for the Promotion of Next Generation Research Projects Type A (J.S.).
Author contributions
K.H. and J.S. designed research; Y.G., A.M., K.A., C.I., M.S., A.O., and M.M. performed research; C.I., M.M., and K.H. contributed new reagents/analytic tools; Y.G., N.N., Y.O., T.S., M.Y., K.H., and J.S. analyzed data; Y.O., T.S., and M.Y. supervised research; and Y.G. and J.S. wrote the paper.
Competing interests
M.M. is a founder and CEO of KAGAMI INC., a company working on analysis of chiral amino acids.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Fujii N., Saito T., Homochirality and life. Chem. Record 4, 267–278 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Sasabe J., Suzuki M., Distinctive roles of D-Amino acids in the homochiral world: Chirality of amino acids modulates mammalian physiology and pathology. Keio J. Med. 68, 1–16 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Broer S., Gauthier-Coles G., Amino acid homeostasis in mammalian cells with a focus on amino acid transport. J. Nutr. 152, 16–28 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashimoto A., et al. , The presence of free D-serine in rat brain. FEBS Lett. 296, 33–36 (1992). [DOI] [PubMed] [Google Scholar]
- 5.Sasabe J., et al. , Interplay between microbial d-amino acids and host d-amino acid oxidase modifies murine mucosal defence and gut microbiota. Nat. Microbiol. 1, 16125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolosker H., et al. , Purification of serine racemase: Biosynthesis of the neuromodulator D-serine. Proc. Natl. Acad. Sci. U.S.A. 96, 721–725 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mothet J. P., et al. , D-serine is an endogenous ligand for the glycine site of the N-methyl-D-aspartate receptor. Proc. Natl. Acad. Sci. U.S.A. 97, 4926–4931 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakai K., et al. , Emergence of D-aspartic acid in the differentiating neurons of the rat central nervous system. Brain Res. 808, 65–71 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Wolosker H., D’Aniello A., Snyder S. H., D-aspartate disposition in neuronal and endocrine tissues: Ontogeny, biosynthesis and release. Neuroscience 100, 183–189 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Schell M. J., Cooper O. B., Snyder S. H., D-aspartate localizations imply neuronal and neuroendocrine roles. Proc. Natl. Acad. Sci. U.S.A. 94, 2013–2018 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakai K., et al. , Localization of D-aspartic acid in elongate spermatids in rat testis. Arch. Biochem. Biophys. 351, 96–105 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Suzuki M., et al. , Host-microbe cross-talk governs amino acid chirality to regulate survival and differentiation of B cells. Sci. Adv. 7, eabd6480 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genchi G., An overview on D-amino acids. Amino acids 49, 1521–1533 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Miyoshi Y., et al. , Determination of D-serine and D-alanine in the tissues and physiological fluids of mice with various D-amino-acid oxidase activities using two-dimensional high-performance liquid chromatography with fluorescence detection. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 877, 2506–2512 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Suzuki M., Shimizu-Hirota R., Mita M., Hamase K., Sasabe J., Chiral resolution of plasma amino acids reveals enantiomer-selective associations with organ functions. Amino Acids 54, 421–432 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Pollegioni L., Piubelli L., Sacchi S., Pilone M. S., Molla G., Physiological functions of D-amino acid oxidases: From yeast to humans. Cell. Mol. Life Sci. 64, 1373–1394 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katane M., Homma H., D-aspartate oxidase: The sole catabolic enzyme acting on free D-aspartate in mammals. Chem. Biodiversity 7, 1435–1449 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Han H., et al. , Changes in D-aspartic acid and D-glutamic acid levels in the tissues and physiological fluids of mice with various D-aspartate oxidase activities. J. Pharm. Biomed. Analysis 116, 47–52 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Nagata Y., Konno R., Yasumura Y., Akino T., Involvement of D-amino acid oxidase in elimination of free D-amino acids in mice. Biochem. J. 257, 291–292 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyoshi Y., et al. , Simultaneous two-dimensional HPLC determination of free D-serine and D-alanine in the brain and periphery of mutant rats lacking D-amino-acid oxidase. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 879, 3184–3189 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Cava F., Lam H., de Pedro M. A., Waldor M. K., Emerging knowledge of regulatory roles of D-amino acids in bacteria. Cell Mol. Life Sci. 68, 817–831 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radkov A. D., Moe L. A., Bacterial synthesis of D-amino acids. Appl. Microbiol. Biotechnol. 98, 5363–5374 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Ishii C., Akita T., Mita M., Ide T., Hamase K., Development of an online two-dimensional high-performance liquid chromatographic system in combination with tandem mass spectrometric detection for enantiomeric analysis of free amino acids in human physiological fluid. J. Chromatogr. A 1570, 91–98 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Gonda Y., et al. , Astrocytic d-amino acid oxidase degrades d-serine in the hindbrain. FEBS Lett. 596, 2889–2897 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Perotti M. E., Gavazzi E., Trussardo L., Malgaretti N., Curti B., Immunoelectron microscopic localization of D-amino acid oxidase in rat kidney and liver. Histochem. J. 19, 157–169 (1987). [DOI] [PubMed] [Google Scholar]
- 26.Lam H., et al. , D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325, 1552–1555 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aliashkevich A., Alvarez L., Cava F., New insights into the mechanisms and biological roles of D-amino acids in complex eco-systems. Front. Microbiol. 9, 683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds P. E., Courvalin P., Vancomycin resistance in enterococci due to synthesis of precursors terminating in D-alanyl-D-serine. Antimicrobial Agents Chemother. 49, 21–25 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellais S., et al. , Aslfm, the D-aspartate ligase responsible for the addition of D-aspartic acid onto the peptidoglycan precursor of Enterococcus faecium. J. Biol. Chem. 281, 11586–11594 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Cava F., de Pedro M. A., Lam H., Davis B. M., Waldor M. K., Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J. 30, 3442–3453 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alvarez L., Aliashkevich A., de Pedro M. A., Cava F., Bacterial secretion of D-arginine controls environmental microbial biodiversity. ISME J. 12, 438–450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hills G. M., Chemical factors in the germination of spore-bearing aerobes; the effect of yeast extract on the germination of Bacillus anthracis and its replacement by adenosine. Biochem. J. 45, 353–362 (1949). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bucher T., Oppenheimer-Shaanan Y., Savidor A., Bloom-Ackermann Z., Kolodkin-Gal I., Disturbance of the bacterial cell wall specifically interferes with biofilm formation. Environ. Microbiol. Rep. 7, 990–1004 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Official J., R. Inoue, K. Hashimoto, T. Harai, H. Mori, NMDA- and beta-amyloid1-42-induced neurotoxicity is attenuated in serine racemase knock-out mice. J. Neurosci. Soc. Neurosci. 28, 14486–14491 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osaki A., et al. , Endogenous d-serine exists in the mammalian brain independent of synthesis by serine racemase. Biochem. Biophys. Res. Commun. 641, 186–191 (2022). [DOI] [PubMed] [Google Scholar]
- 36.Basu A. C., et al. , Targeted disruption of serine racemase affects glutamatergic neurotransmission and behavior. Mol. Psychiatry 14, 719–727 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papouin T., et al. , Synaptic and extrasynaptic NMDA receptors are gated by different endogenous coagonists. Cell 150, 633–646 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Miyoshi Y., et al. , Alteration of intrinsic amounts of D-serine in the mice lacking serine racemase and D-amino acid oxidase. Amino Acids 43, 1919–1931 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Cline M. J., Lehrer R. I., D-amino acid oxidase in leukocytes: A possible D-amino-acid-linked antimicrobial system. Proc. Natl. Acad. Sci. U.S.A. 62, 756–763 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuinema B. R., Reid-Yu S. A., Coombes B. K., Salmonella evades D-amino acid oxidase to promote infection in neutrophils. mBio 5, e01886 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee R. J., et al. , Bacterial d-amino acids suppress sinonasal innate immunity through sweet taste receptors in solitary chemosensory cells. Sci. Signaling 10, eaam7703 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seki N., et al. , D-Tryptophan suppresses enteric pathogen and pathobionts and prevents colitis by modulating microbial tryptophan metabolism. iScience 25, 104838. (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kepert I., et al. , D-tryptophan from probiotic bacteria influences the gut microbiome and allergic airway disease. J. Allergy Clin. Immunol. 139, 1525–1535 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Piaggi P., et al. , Exome sequencing identifies a nonsense variant in DAO associated with reduced energy expenditure in American Indians. J. Clin. Endocrinol. Metab. 105, e3989–4000 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoeprich P. D., Alanine: Cycloserine antagonism. Vi., Demonstration of D-alanine in the serum of guinea pigs and mice. J. Biol. Chem. 240, 1654–1660 (1965). [PubMed] [Google Scholar]
- 46.Konno R., Niwa A., Yasumura Y., Intestinal bacterial origin of D-alanine in urine of mutant mice lacking D-amino-acid oxidase. Biochem. J. 268, 263–265 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konno R., et al. , Origin of D-alanine present in urine of mutant mice lacking D-amino-acid oxidase activity. Am. J. Physiol. 265, G699–G703 (1993). [DOI] [PubMed] [Google Scholar]
- 48.Krebs H. A., Metabolism of amino-acids: Deamination of amino-acids. Biochem. J. 29, 1620–1644 (1935). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sasabe J., et al. , D-amino acid oxidase controls motoneuron degeneration through D-serine. Proc. Natl. Acad. Sci. U.S.A. 109, 627–632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.