Significance
Sensing gram-negative bacterial lipopolysaccharide by human caspase-4 is critical for host defense to intracellular gram-negative bacterial pathogens. Human guanylate-binding proteins (GBPs) facilitate caspase-4 activation in response to gram-negative infections by a poorly understood mechanism. The prevailing model suggests that GBP1 binding to bacteria and consequential recruitment of caspase-4 to the bacterial surface are essential for triggering this host response. Here, we show that GBP1 binding to bacteria is dispensable for caspase-4 activation and identify GBP2 as an additional lipopolysaccharide-binding protein that can functionally replace GBP1. We demonstrate that GBP1 and GBP2 share the ability to form lipopolysaccharide-protein complexes, which promote caspase-4 activation. Our study provides a revised mechanistic framework for cytosolic LPS sensing and noncanonical inflammasome activation.
Keywords: cell-autonomous immunity, interferon-stimulated genes, guanylate-binding proteins, lipopolysaccharide, inflammasome
Abstract
The gamma-interferon (IFNγ)-inducible guanylate-binding proteins (GBPs) promote host defense against gram-negative cytosolic bacteria in part through the induction of an inflammatory cell death pathway called pyroptosis. To activate pyroptosis, GBPs facilitate sensing of the gram-negative bacterial outer membrane component lipopolysaccharide (LPS) by the noncanonical caspase-4 inflammasome. There are seven human GBP paralogs, and it is unclear how each GBP contributes to LPS sensing and pyroptosis induction. GBP1 forms a multimeric microcapsule on the surface of cytosolic bacteria through direct interactions with LPS. The GBP1 microcapsule recruits caspase-4 to bacteria, a process deemed essential for caspase-4 activation. In contrast to GBP1, closely related paralog GBP2 is unable to bind bacteria on its own but requires GBP1 for direct bacterial binding. Unexpectedly, we find that GBP2 overexpression can restore gram-negative-induced pyroptosis in GBP1KO cells, without GBP2 binding to the bacterial surface. A mutant of GBP1 that lacks the triple arginine motif required for microcapsule formation also rescues pyroptosis in GBP1KO cells, showing that binding to bacteria is dispensable for GBPs to promote pyroptosis. Instead, we find that GBP2, like GBP1, directly binds and aggregates “free” LPS through protein polymerization. We demonstrate that supplementation of either recombinant polymerized GBP1 or GBP2 to an in vitro reaction is sufficient to enhance LPS-induced caspase-4 activation. This provides a revised mechanistic framework for noncanonical inflammasome activation where GBP1 or GBP2 assembles cytosol-contaminating LPS into a protein-LPS interface for caspase-4 activation as part of a coordinated host response to gram-negative bacterial infections.
Cell-autonomous immune responses to intracellular pathogens are a powerful defense against infection (1). Within a single cell, there are highly orchestrated pathways that sense and respond to invading microbes. During infection with gram-negative bacteria, immune sensors detect the presence of lipopolysaccharide (LPS), an abundant component of the gram-negative cell wall. Extracellular LPS is detected through TLR4, causing proinflammatory transcriptional changes within the cell, and cytosolic LPS is sensed through the caspase-4 noncanonical inflammasome, triggering inflammatory cell death through pyroptosis (2–8). Activation of caspase-4 not only occurs in professional immune sentinel cells but also in nonimmune cells such as colonic epithelial cells, where caspase-4 plays an essential role in host defense against enteric bacterial pathogens (9–11).
Pyroptosis exerts host defense through the destruction of the replicative niche of intracellular bacteria. Additionally, pyroptotic cell death is proinflammatory and thereby promotes the recruitment and activation of professional immune cells to further restrict infection (12). Pyroptosis can be triggered by diverse stimuli which include cytosolic LPS. Cytosolic LPS binds and activates caspase-4, which then cleaves gasdermin D (GSDMD). The cleaved N terminus of GSDMD forms a pore in the plasma membrane (13, 14), disrupting the ionic balance of the cell and leading to cell swelling and eventual rupture of the plasma membrane through a programmed cell lysis pathway (15). The GSDMD pore also allows secretion of proinflammatory alarmins and cytokines such as IL-18. Due to this inflammatory nature of pyroptosis, professional immune cells are recruited to and activated at the site of infection, where they provide critical immune effector functions. The recruited immune cells release additional cytokines to enhance local responses to pathogens. For example, innate and adaptive lymphocytes activated during a type 1 immune response secrete the potent antibacterial cytokine gamma-interferon (IFNγ). IFNγ induces expression of hundreds of proteins encoded by interferon-stimulated genes, including the dynamin-related guanylate-binding proteins (GBPs). In both mouse and human cells, GBPs have been shown to promote pyroptosis in response to LPS introduced into the cytosol, either through LPS transfection, uptake of outer membrane vesicles containing LPS, or during gram-negative bacterial infection (16–22).
Humans have seven GBPs, of which GBP1, GBP2, GBP3, GBP4, and GBP5 are widely expressed in many cell types, and are some of the most highly up-regulated genes in response to IFNγ (23). GBP1 is the most well-characterized paralog and can target gram-negative bacteria in the cytosol due to GBP1’s ability to bind LPS (24–26). Following initial docking on the bacterial surface, GBP1 intercalates into the bacterial outer membrane and forms a stable coat called the GBP1 microcapsule (24). GBP1 recruits GBP2, GBP3, and GBP4 to the bacterial surface through unknown mechanisms (27–29). It was proposed that this protein coat consisting of multiple GBP paralogs forms a signaling platform on the surface of bacteria, allowing recruitment and activation of caspase-4 and consequential pyroptosis in human epithelial cells (25, 26). Whether and how each GBP paralog contributes to pyroptosis or restriction of bacterial growth during gram-negative infection of epithelial cells has not been clearly defined. Similarly, it has remained unclear how the model of a GBP signaling platform on the surface of bacteria relates to GBP-mediated caspase-4 activation following outer membrane vesicle uptake or LPS transfection.
GBP2, the protein with the highest degree of homology to GBP1, is unable to encapsulate bacteria in cells lacking its interaction partner and bacterial encapsulation factor GBP1. Although GBP2 cannot bind to bacteria in the absence of GBP1, we show here that GBP2 overexpression restores pyroptotic cell death in response to gram-negative infections in GBP1 knockout cells. We demonstrate that GBP2 binds to LPS and forms LPS aggregates, thus sharing critical biochemical properties with GBP1. A GBP1 mutant lacking its unique C-terminal polybasic motif essential for GBP1 microcapsule formation maintains the ability to aggregate LPS and to activate pyroptosis in response to gram-negative infections or LPS transfections. Recombinant GBP1 and GBP2 also promote LPS-dependent caspase-4 activation in a cell-free in vitro system. Our studies therefore demonstrate that GBP1 or GBP2 binding to the bacterial surface is dispensable for caspase-4 activation in response to infections with cytosolic gram-negative bacteria and instead support a model in which GBP1 and GBP2 facilitate pyroptosis independent of each other through the formation of protein-LPS aggregates.
Results
GBP1 Targeting of S. flexneri Does Not Correlate with Pyroptosis.
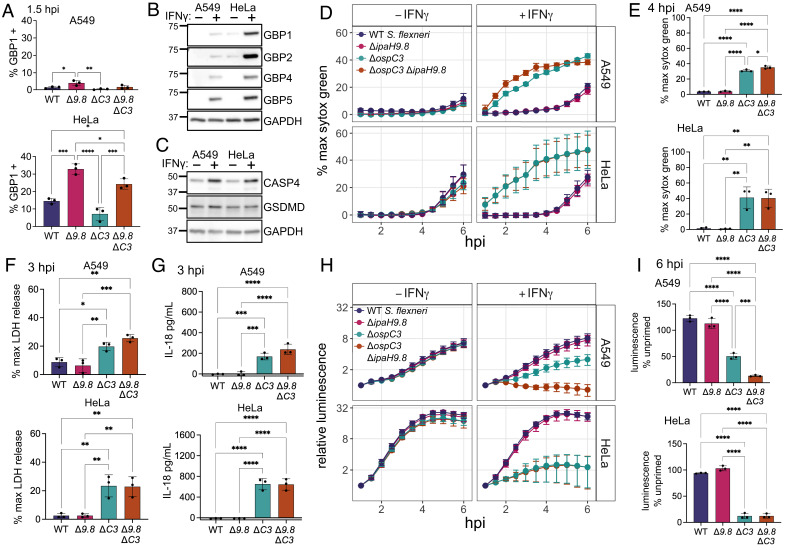
The prevailing model of caspase-4 activation during infections with cytosolic gram-negative bacteria postulates that GBP1 binding to the bacterial outer membrane recruits GBP2, 3, and 4, and that this complex assembled on the bacterial surface forms an essential platform for caspase-4 activation (25, 26). Following caspase-4 activation, cells undergo pyroptosis and invading bacteria are killed. While GBP1, 2, 3, and 4 are all present on the bacterial surface, it is unclear how each GBP contributes to caspase-4 activation. Shigella flexneri is a useful model pathogen for studying GBP regulation of the caspase-4 inflammasome, because it encodes specific effectors that modulate this process; OspC3 blocks caspase-4 activation and IpaH9.8 reduces GBP bacterial targeting through degradation of GBP1, 2, and 4 (10, 27, 28, 30, 31). S. flexneri ΔospC3 induces pyroptosis that is dependent on caspase-4. However, it is unclear whether IpaH9.8 affects cell death by reducing the targeting of GBPs to the S. flexneri surface (10). We previously showed that there is significantly less binding of GBP1 to S. flexneri in A549 cells compared to HeLa cells, making these two cell lines useful for testing how different levels of GBP targeting to the S. flexneri surface affect cell death or bacterial growth restriction (27). To investigate the role of GBP binding in the activation of pyroptosis or bacterial killing, we infected A549 and HeLa cells with four S. flexneri strains: wild-type, ΔipaH9.8, ΔospC3, and ΔospC3ΔipaH9.8. Since GBP1 binding to S. flexneri is required for subsequent recruitment of other GBP family members, we used GBP1 targeting as a proxy for all GBP recruitment (27, 28). Using immunofluorescence microscopy, we again saw that GBP1 targeting of bacteria in A549 cells was significantly lower than that in HeLa cells, with 3.9% of S. flexneri ΔipaH9.8 targeted by GBP1 in A549 cells compared to 32.9% of S. flexneri ΔipaH9.8 targeted in HeLa cells at 1.5 h post infection (Fig. 1A and SI Appendix, Fig. S1 A and B). This may be due to lower levels of GBP expression in A549 cells (Fig. 1B). While GBPs exhibit expression differences between A549 and HeLa cells, other known proteins in the pyroptosis pathway, caspase-4 and GSDMD, were expressed at similar levels between the two cell lines (Fig. 1C). Strains lacking OspC3 had slightly lower levels of targeting than isogenic controls, likely due to cell death or bacterial killing, but knockout (KO) of IpaH9.8 in S. flexneri ΔospC3 still substantially increased the levels of GBP1 targeting (Fig. 1A).
Fig. 1.
GBP1 targeting of S. flexneri is reduced in A549 cells compared to HeLa cells, but pyroptosis levels are similar. (A) A549 and HeLa cells were infected with the indicated strains, fixed at 1.5 h post infection, and immunostained for GBP1. S. flexneri with GBP1 surrounding >50% of the bacterial surface were counted as GBP1 positive. (B and C) A549 and HeLa cells were grown with or without 100 U/mL IFNγ overnight, then lysed for western blotting. Membranes were probed with antibodies against the indicated GBPs and GAPDH (B), or CASP4, GSDMD, and GAPDH (C). (D–I) A549 and HeLa cells unprimed or primed with 100 U/mL IFNγ overnight were infected with the indicated S. flexneri strains expressing a bioluminescent reporter plasmid. Cell death was measured over time using sytox green fluorescence (D). 4 h timepoint of IFNγ primed cells was used for statistical analysis (E). Supernatant from IFNγ-primed cells infected with S. flexneri was removed at 3 hpi, and LDH levels (F) or IL-18 secretion (G) was measured. Bacterial luminescence was measured over time (H). Luminescence measurements from the 6 h timepoint were used to calculate the growth of each strain in primed cells relative to unprimed cells (I). Caspase-4 – CASP4. Graphs are averages from three independent experiments and are represented by mean ± SD. One-way ANOVA with Tukey’s multiple comparisons test was used, all statistically significant comparisons are shown. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
If GBP binding to the bacterial surface is required for cell death, we hypothesized that A549 cells would exhibit lower levels of pyroptosis during S. flexneri infection, and that bacterial killing would be similarly reduced. We also hypothesized that S. flexneri ΔospC3ΔipaH9.8 would induce higher levels of cell death than S. flexneri ΔospC3. To study pyroptosis and bacterial killing concurrently over time, we infected cells with S. flexneri containing a bioluminescent reporter plasmid to monitor bacterial growth. Pyroptosis was measured using the DNA dye sytox green, which fluoresces in cells with compromised plasma membrane integrity, such as pyroptotic cells with GSDMD pores (32). When we infected A549 and HeLa cells, there was significant IFNγ-dependent pyroptosis triggered by both S. flexneri ΔospC3 and S. flexneri ΔospC3ΔipaH9.8, while there was no detectable cell death for wild-type or S. flexneri ΔipaH9.8 (Fig. 1 D and E). We saw similar results using lactate dehydrogenase (LDH) release as a measure of the cellular rupture that occurs during pyroptosis (Fig. 1F). Interestingly, in IFNγ primed cells, there was a similar amount of cell death in both cell lines, despite HeLa cells having significantly more GBP1 targeting of S. flexneri. There was also very little difference in the amount of cell death induced by infection with S. flexneri ΔospC3 or S. flexneri ΔospC3ΔipaH9.8, suggesting that IpaH9.8 plays a limited role in reducing cell death through GBP degradation. As an independent measure of inflammasome activation, we measured IL-18 secretion. IFNγ-dependent IL-18 processing and release only occurred after infection with S. flexneri ΔospC3 or S. flexneri ΔospC3ΔipaH9.8, and there was no significant difference in the amount of IL-18 secretion induced by these two strains (Fig. 1G). For bacterial restriction, there was no difference in growth of the different strains in unprimed cells; however, IFNγ priming elicited significant growth restriction of both S. flexneri ΔospC3 and S. flexneri ΔospC3ΔipaH9.8 (Fig. 1 H and I). While in HeLa cells there was no significant difference in restriction between S. flexneri ΔospC3 and S. flexneri ΔospC3ΔipaH9.8, in A549 cells, S. flexneri ΔospC3 was restricted significantly less than S. flexneri ΔospC3ΔipaH9.8 (Fig. 1I). The differences between HeLa and A549 cells regarding the effect of IpaH9.8 on bacterial burden are not readily explained and may reflect cell line–specific variation in any activity modulated by IpaH9.8, which not only targets GBPs but also NEMO, a protein that regulates NF-κB-dependent signaling (33). Because S. flexneri ΔospC3 and S. flexneri ΔospC3ΔipaH9.8 displayed comparable phenotypes across all assays in both cell lines, we focused our studies on the comparison of wild-type and S. flexneri ΔospC3ΔipaH9.8 from here on forward.
Endogenous GBP1 Is the Only GBP Required for Cell Death or Bacterial Killing.
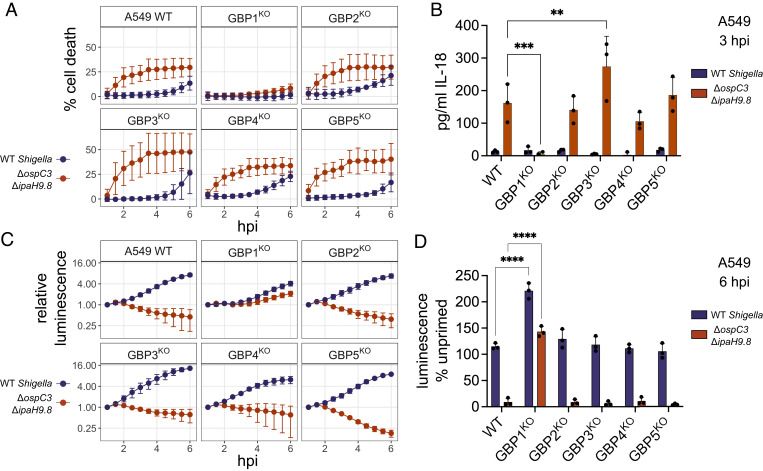
Human GBP1, GBP2, GBP3, and GBP4 have been suggested to form a complex on the bacterial membrane that is essential for pyroptosis and bacterial killing. However, in epithelial cells, only knockout cells of GBP1 have been tested, whereas other GBPs were targeted for reduced expression using siRNAs (25, 26). To avoid possible off-target effects prevalent with siRNA technology and to test the role of each GBP independently, we generated CRISPR knockout lines of GBP1, 2, 3, 4, or 5 in A549 and HeLa cells (SI Appendix, Fig. S2 A–C). In the A549 knockout cells, only knockout of GBP1 led to a significant reduction in cell death. GBP2, 3, 4, and 5KO cells had similar levels of cell death to wild-type cells in response to infection with S. flexneri ΔospC3ΔipaH9.8 (Fig. 2A). IL-18 secretion was completely lost in GBP1KO cells, while secretion from GBP2-5KO cells was comparable to levels observed with wild-type cells (Fig. 2B). For bacterial restriction, there were similar results; S. flexneri ΔospC3ΔipaH9.8 growth was only restored in the GBP1KO cells, while there was still IFNγ-dependent growth restriction in GBP2-5KO cells (Fig. 2 C and D). In HeLa cells, we saw identical results to A549 cells, where only GBP1 was required for pyroptosis, IL-18 secretion, or restriction of bacterial growth (SI Appendix, Fig. S3 A–D). Since we observed very similar results between HeLa and A549 cells for all assays, and GBP1 is the only GBP required in both A549 and HeLa cells, we focused on a single cell line, A549 GBP1KO cells, from here on to investigate why GBP1, but not the other GBPs, is required for pyroptosis during S. flexneri infection. We confirmed that cell death induced by S. flexneri ΔospC3ΔipaH9.8 in A549 cells is caspase-4 dependent (SI Appendix, Fig. S4 A–F), and that restriction of bacterial growth also requires caspase-4 (SI Appendix, Fig. S4 G and H).
Fig. 2.
A549 cells require endogenous GBP1 for pyroptosis and restriction of S. flexneri growth. A549 cells were primed overnight with 100 U/mL IFNγ, then infected with wild-type S. flexneri or S. flexneri ΔipaH9.8ΔospC3 expressing a bioluminescent reporter plasmid. Cell death was measured using sytox green (A). Supernatant was taken at 3 hpi to measure IL-18 secretion (B). Bacterial growth was monitored by luminescence (C). Luminescence measurements from the 6 h timepoint were used to calculate the growth of each strain in primed cells relative to unprimed cells (D). Data are averages from three independent experiments and are represented by mean ± SD. Two-way ANOVA with Dunnett’s multiple comparisons test was used; for each bacterial strain, values for each knockout cell line were compared to the wild-type cells. All statistically significant comparisons are shown. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Overexpression of GBP2 Promotes Pyroptosis in the Absence of GBP1.
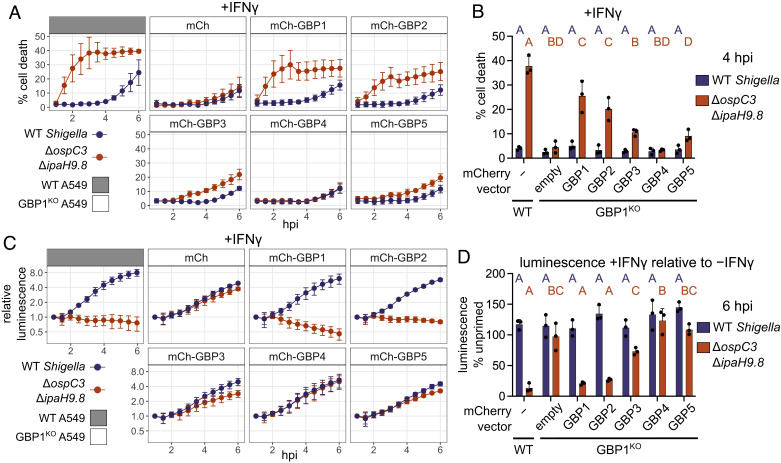
Although endogenous GBP2, 3, 4, and 5 are not essential for pyroptosis or bacterial restriction, they share many structural similarities with GBP1 and could have some functional redundancy with GBP1. Individual GBP paralogs differ from each other in their respective expression levels, and it is therefore unclear whether GBP1 is the only GBP capable of promoting pyroptosis, or alternatively whether endogenous GBP1 is the only GBP expressed at high enough levels to promote pyroptosis in the cells we tested. We hypothesized that overexpression of each GBP individually would reveal which paralogs can promote pyroptosis or restrict S. flexneri growth. We overexpressed GBP1, 2, 3, 4, and 5 in GBP1KO cells and measured bacterial binding, cell death, and bacterial restriction during S. flexneri infection. We first confirmed previous studies (24–27, 29) showing that overexpressed GBP2, 3, 4, and 5 are not recruited to bacteria in the absence of GBP1 (SI Appendix, Fig. S5 A–C). In unprimed cells, there was minimal cell death with any construct, although GBP1 overexpression did promote some cell death (SI Appendix, Fig. S6 A and B). In IFNγ-primed cells, overexpression of either GBP1 or GBP2 rescued cell death in GBP1KO cells, while overexpression of GBP3, 4, and 5 did not (Fig. 3 A and B). For bacterial growth, there was minimal restriction in unprimed cells, with GBP1 overexpression reducing the S. flexneri ΔospC3ΔipaH9.8 bioluminescence signal by approximately half (SI Appendix, Fig. S6 C). In IFNγ-primed cells, both GBP1 and GBP2 rescued S. flexneri restriction in GBP1KO cells (Fig. 3 C and D). It is unclear whether the requirement for IFNγ is due to the upregulation of GBP3, GBP4, and GBP5, or if there are other ISGs that are required for efficient execution of pyroptosis. In primed cells, there was no significant difference between the efficiency of GBP1 or GBP2 at executing pyroptosis or restricting bacterial growth. In these overexpression experiments, GBP2 was expressed at a higher level than the other GBPs (SI Appendix, Fig. S5B). Therefore, to ensure that the phenotype observed was not due to differences in the levels of ectopic GBP expression, we repeated this experiment across a range of GBP1 or GBP2 expression levels. We titrated the concentration of anhydrotetracycline (aTc) used to induce GBP1 and GBP2 expression (SI Appendix, Fig. S7A) in GBP1KO cells prior to infection with S. flexneri ΔospC3ΔipaH9.8 and found that even at low levels of GBP1 or GBP2 expression, cell death was significantly elevated compared to cells expressing GBP3, GBP4, or GBP5 at their maximum levels (SI Appendix, Fig. S7 B–E). At four hours post infection, GBP3 overexpression led to significantly more death than the mCherry control, although it was still lower than the level of cell death observed for GBP1 or GBP2 (SI Appendix, Fig. S7 E). This observation suggests that GBP3 may have some capacity to respond to LPS, but much less efficiently so than GBP1 or GBP2. For bacterial growth, both GBP1 and GBP2 overexpression at several concentrations of aTc significantly reduced bacterial replication, while mCherry, GBP3, GBP4, or GBP5 overexpression did not reduce bacterial growth (SI Appendix, Fig. S7F). Collectively, these data show that GBP1 is not unique among GBPs as an antibacterial inducer of pyroptosis but that GBP2 can also promote pyroptosis and bacterial restriction.
Fig. 3.
Overexpression of GBP1 or GBP2 can rescue pyroptosis and bacterial restriction in GBP1KO cells. A549 cells were stably transduced to express mCherry or mCherry-GBPs. Cells were then infected with bioluminescent S. flexneri and cell death was measured over time using sytox green fluorescence in cells primed with 100 U/mL IFNγ overnight before infection (A). The sytox green signal at 4 h was used to determine statistical significance (B). Bacterial luminescence was measured over time (C). Luminescence measurements from the 6 h timepoint were used to calculate the growth of each strain in primed cells relative to unprimed cells (D). Data are averages from three independent experiments and are represented by mean ± SD. Two-way ANOVA with Tukey’s multiple comparisons test was used. Statistical comparisons are shown by letters, with bars sharing no matching letters being significantly different. Purple letters correspond to statistical comparisons for wild-type S. flexneri, and orange letters correspond to S. flexneri ΔipaH9.8ΔospC3.
Binding of GBP2 to the Bacterial Surface Requires the Formation of Mixed Polymers with GBP1.
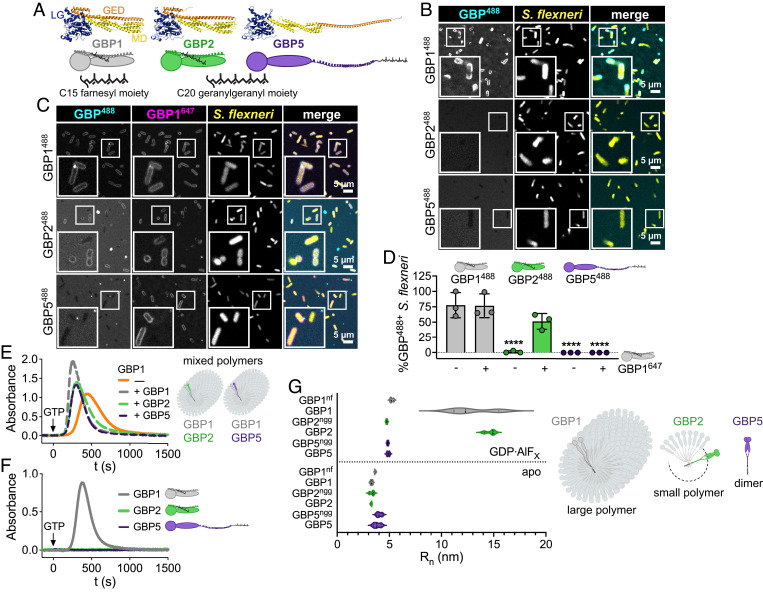
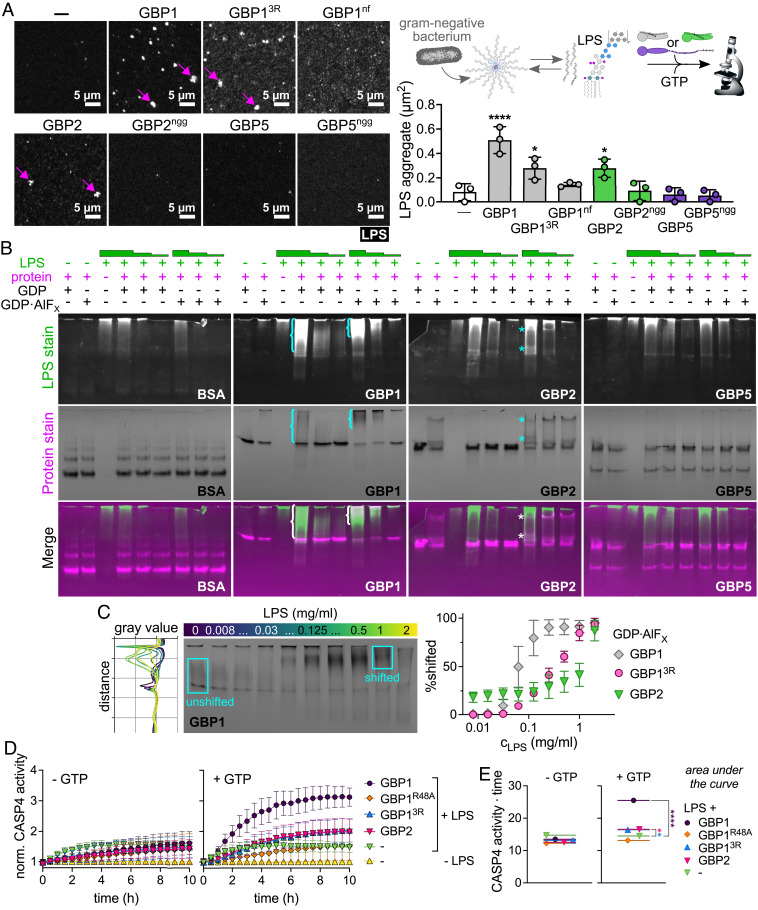
Our finding that GBP2 overexpression can restore host cell pyroptosis in response to S. flexneri invasion of GBP1-deficient epithelial cells led us to hypothesize that GBP2 shares specific biochemical characteristics with GBP1 that are required for pyroptosis induction. Previous work showed that following GTP hydrolysis, GBP1 exits its closed monomeric state, adopts a dimeric outstretched state, and forms dimers (SI Appendix, Fig. S8A) (34). GBP1 dimers can assemble into large polymers holding over 1,000 molecules (35, 36). These GBP1 polymers attach to the bacterial surface and transition into a GBP1 microcapsule encasing the entire bacteria (24). Because GBP1 polymerization and microcapsule formation are dependent on the posttranslational attachment of farnesyl lipid moieties to GBP1 molecules, we asked whether the other two lipidated members of the GBP family, geranylgeranylated GBP2 and GBP5 (37) (Fig. 4A), could similarly form polymers and attach directly to bacteria. Although GBP2 and other GBP paralogs are unable to dock on cytosolic bacteria in GBP1-deficient epithelial cells (SI Appendix, Fig. S5 A–C), we hypothesized that lipidated GBP2 or GBP5 may bind bacteria transiently or at low levels that are difficult to detect in cells but could be detectable in a highly manipulable and more sensitive in vitro GBP-bacteria binding assay. We recently developed such an in vitro binding assay for GBP1 (24) and here leveraged an analogous system to test the ability of GBP2 and GBP5 to bind to bacteria. For these experiments, we added recombinant Alexa Fluor 488 (488)-labeled GBP1, GBP2, or GBP5 supplemented with GTP to broth cultured RFP-expressing S. flexneri and observed that only GBP1 but not GBP2 or GBP5 formed a microcapsule surrounding S. flexneri in vitro (Fig. 4B, and Movie S1). These data further supported the concept that GBP1 but not GBP2 or GBP5 can directly and autonomously bind bacteria.
Fig. 4.
GBP2 fails to bind bacteria on its own but forms small polymers. (A) Structures and schematics for GBP1, GBP2, and GBP5. All GBP paralogs consist of a large GTPase domain (LG), a middle domain (MD), and a GTPase effector domain (GED), whereby C-terminal isoprenyl moieties vary with GBP1 becoming post transcriptionally farnesylated, and GBP2 and GBP5 geranylgeranylated. (B–D) Recombinant Alexa Fluor 488-labeled GBP1, GBP2, and GBP5 alone (B) or mixed with Alexa Fluor 647-labeled GBP1 (C) were supplemented with GTP and added to formaldehyde-fixed RFP-expressing S. flexneri. Confocal microscopy time-lapses were recorded, and targeted bacteria were quantified after 1 h (D). (E and F) GBP polymerization was monitored in UV-absorbance-based light scattering experiments. Equal molar ratios of recombinant GBP1, GBP2, or GBP5 were mixed with GBP1, and polymerization was induced with GTP (E). GTP-induced polymerization of single GBPs was monitored over time (F). (G) Number-weighted mean radius (Rn) of nucleotide-free (apo) or GDP·AlFX-bound nonisoprenylated (nonfarnesylated - nf, nongeranylgeranylated - ngg) and isoprenylated GBPs was determined in DLS experiments. (A) PDB entry 1F5N and AlphaFold models AF-P32456, AF-Q96PP8. (B and C) Representative images of three independent experiments. (D and G) Graphs are averages from three independent experiments and are represented by mean ± SD. (D) One-way ANOVA with Dunnett’s multiple comparisons test comparing to GBP1488+ S. flexneri was used, all statistically significant comparisons are shown. ****P < 0.0001. (E and F) Representative graphs from two independent experiments.
We next investigated the mechanism by which GBP1 facilitates GBP2 binding to cytosolic bacteria in infected cells, which was not previously determined (25–28). We found that upon addition of GTP, Alexa Fluor 647 (647)-labeled GBP1 formed mixed polymers with 488-labeled GBP1, GBP2, and GBP5, which associated with the bacterial surface (SI Appendix, Fig. S8B). Over time, the surface-attached GBP1-GBP1 and GBP1-GBP2 polymers formed a microcapsule surrounding the bacterial cell, whereas GBP5 failed to be incorporated in the GBP1 microcapsule (Fig. 4 C and D and Movie S2). Incorporation of GBP2 and GBP5 into GBP1 polymers was confirmed in UV-absorption-based light scattering experiments, where we observed that the addition of equimolar amounts of GBP1, GBP2, or GBP5 to GBP1 accelerated the initiation of polymerization and increased the absorbance signal (Fig. 4E). Together, these experiments showed that although GBP1 recruits both GBP2 and GBP5 to the bacterial surface as mixed polymers, only the GBP1-GBP2 mixed polymer transforms into a microcapsule. This explains the reported observation that GBP5 is not part of the GBP coat surrounding cytosolic bacteria in infected cells (25–28). Our present observations further implied that GBP2 only interacts with the bacterial outer membrane when stabilized in its active conformation by GBP1 through mixed polymer formation. Because GBP2-driven pyroptosis did not require GBP1 (Fig. 3), we can conclude that mixed GBP1-GBP2 polymer formation and the resulting GBP1-dependent binding of GBP2 to the bacterial surface are dispensable for GBP2-dependent cell death.
GBP2 Self-Assembles into Small Polymers Independently of GBP1.
We previously reported that reversible GBP1 polymerization precedes encapsulation of gram-negative bacteria. We also showed that GBP1 polymerization promotes clustering of LPS (24). To determine whether, like farnesylated GBP1, geranylgeranylated GBP2 and GBP5 self-assemble into polymers, we monitored GTP-induced polymerization of GBPs using the UV-absorption-based turbidity assay in which light scattering by large protein polymers leads to an increase in absorbance (35, 36). We observed that only GBP1 but not GBP2 or GBP5 formed large polymers during GTP hydrolysis which dissociated with GTP depletion (Fig. 4F). To stabilize smaller, short-lived GBP complexes, we utilized the GTP transition state analog GDP·AlFX in dynamic light scattering (DLS) experiments. We determined the number-weighted mean radius (Rn) as a measure of size for nonisoprenylated and isoprenylated GBPs in their nucleotide-free, inactive (apo), and GDP·AlFX-bound, activated states (Fig. 4G and SI Appendix, Fig. S8C). As expected, all GBPs were monomeric in their nucleotide-free resting state with nonisoprenylated and isoprenylated GBP5 appearing slightly larger in size due to their outstretched conformations (Fig. 4A). In the presence of GDP·AlFX, the Rn of nonisoprenylated GBP1, GBP2, and GBP5 increased to approximately 5 nm indicating that dimers formed by different GBP paralogs have a uniform size (Fig. 4G and SI Appendix, Fig. S8C). While GDP·AlFX-bound geranylgeranylated GBP5 remained dimeric, farnesylated GBP1 and geranylgeranylated GBP2 self-assembled further into complexes with Rn of approximately 13 to 15 nm, corresponding to small polymers. Together, these light scattering experiments demonstrate that, although GBP1 is unique in forming large polymers, GBP2, like GBP1, can self-assemble into small polymers. Since GBP2 can form polymers on its own, but not bind bacteria, this suggested that polymerization rather than bacterial binding is an important characteristic that allows GBP2 to promote pyroptosis during infection.
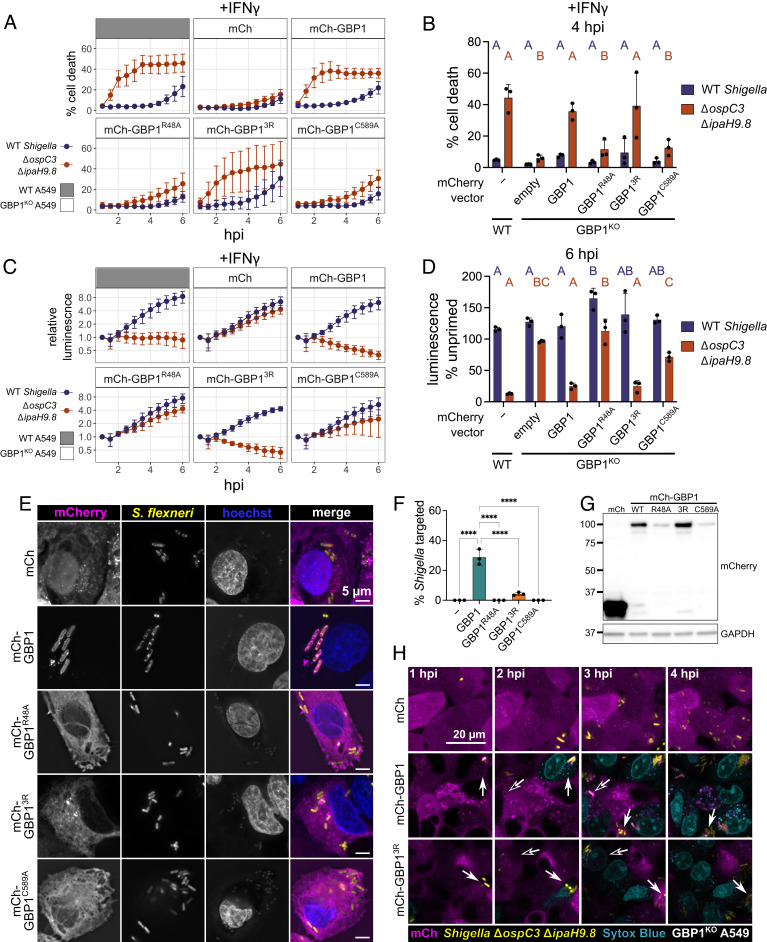
GBP1 Binding to Bacterial Surface Is Dispensable for Cell Death and Bacterial Killing.
Because GBP1 binding to the bacterial surface precedes cell death during Salmonella enterica Typhimurium or S. flexneri infection, it has been suggested that GBP1 binding to bacteria is required for caspase-4 activation (25, 26). We showed that GBP2 promotes pyroptosis without binding to bacteria and therefore hypothesized that a GBP1 mutant that can bind free LPS and polymerize, but not bind the bacterial outer membrane, would also be able to promote pyroptosis. To test this hypothesis, we monitored pyroptosis or bacterial killing of infected cells that overexpress wild-type and mutant GBP1 variants in GBP1KO cells. As expected, wild-type GBP1 completely restored pyroptosis and bacterial killing in GBP1KO cells, whereas there was no rescue with the empty mCherry vector (Fig. 5 A and B). We previously showed that a polybasic motif of three arginines (3R) at the C terminus of GBP1 is important for stable binding of GBP1 to the bacterial surface (24, 27, 29). Importantly, like wild-type GBP1, GBP13R is still able to polymerize, as well as binds and clusters free LPS (24). In GBP1KO cells, overexpression of GBP13R restored cell death and bacterial killing to similar levels as wild-type GBP1 (Fig. 5 A–D). This shows that GBP1 binding to the S. flexneri surface is dispensable for both pyroptosis and restriction of bacterial growth. GBP1R48A, which lacks GTPase activity and cannot polymerize, was unable to promote cell death or bacterial killing (Fig. 5 A–D). Similarly, GBP1C589A, which lacks the farnesyl tail and is unable to form polymers, cluster free LPS, or bind the bacterial surface, did not restore cell death or bacterial restriction.
Fig. 5.
GBP1-dependent pyroptosis and restriction of S. flexneri growth is independent of GBP1 binding to bacteria. (A–D) Wild-type A549 or GBP1KO cells expressing mCherry or mCherry-GBP1 mutants were unprimed or primed with 100 U/mL IFNγ overnight, where indicated. Cells were infected with S. flexneri expressing a bioluminescent reporter and cell death was measured over time using sytox green fluorescence (A). The sytox green signal at 4 h was used to determine statistical significance (B). Bacterial luminescence was measured over time (C). Luminescence measurements from the 6 h timepoint were used to calculate the growth of each strain in primed cells relative to unprimed cells (D). (E and F) Wild-type A549 or GBP1KO cells expressing mCherry or mCherry-GBP1 mutants were primed overnight with IFNγ, then infected with GFP expressing S. flexneri ΔipaH9.8, and fixed at 2 h post infection. (E) Coverslips were imaged at 100× magnification using widefield microscopy. Images were deconvolved and z-projections are shown, scale bar are 5 μm. (F) Coverslips were imaged at 63× magnification, with images taken from five independent fields. Targeting of S. flexneri by each overexpressed protein was quantified using ImageJ. S. flexneri with indicated protein around at least 50% of the bacterial membrane were counted as targeted. (G) Expression levels of indicated overexpressed proteins determined by western blot. (H) Frames from timelapse microscopy at indicated time points for GBP1KO cells expressing mCherry, mCherry-GBP1, or mCherry-GBP13R infected with GFP expressing S. flexneri ΔospC3ΔipaH9.8. Dying cells are shown in blue (sytox blue). All graphs show averages from three independent experiments and are represented by mean ± SD. (B and D) Significance was determined using two-way ANOVA with Tukey’s multiple comparisons test. Statistical comparisons are shown by letters, with bars sharing no matching letters being significantly different. Purple letters correspond to statistical comparisons for wild-type S. flexneri, and orange letters correspond to S. flexneri ΔipaH9.8ΔospC3. (F) One-way ANOVA with Tukey’s multiple comparisons test was used. All significant comparisons are shown. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
We next confirmed that GBP1, but not GBP13R, binds to the surface of S. flexneri ΔipaH9.8 (Fig. 5 E–G) within infected cells. To check whether there was transient binding of GBP13R immediately preceding cell death, we used timelapse microscopy to view cells expressing mCherry-GBP1 or GBP13R and infected with GFP expressing S. flexneri ΔospC3ΔipaH9.8. Using the cell-impermeant dye sytox blue to label cells undergoing pyroptosis, we confirmed prior studies showing that GBP1 targeting of S. flexneri ΔospC3ΔipaH9.8 precedes cell death (Fig. 5H and Movie S3). Importantly, in cells expressing mCherry-GBP13R, we observed pyroptosis in cells without appreciable binding of GBP13R to the bacterial surface (Fig. 5H and Movie S3). This supports a revised model where GBP1 or GBP2 can promote caspase-4 activation without binding the bacterial surface. Demonstrating that this result was not specific to S. flexneri, we found that GBP1, GBP13R, and GBP2 were also able to rescue pyroptosis in GBP1KO cells during S. Typhimurium infection (SI Appendix, Fig. S9 A and B). Because GBP2 and GBP13R cannot bind S. Typhimurium in the absence of wild-type GBP1 (29), we conclude that GBP binding to the surface of S. flexneri or S. Typhimurium is dispensable for GBP-dependent pyroptosis.
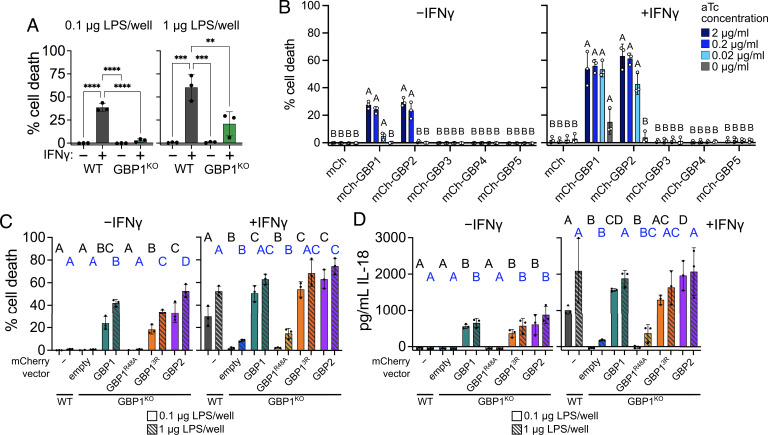
GBP1, GBP13R, or GBP2 Can Mediate Cell Death in Response to LPS Transfection.
Since we observed rescue of pyroptosis by both GBP13R and GBP2 during S. flexneri and S. Typhimurium infection, we wondered whether these proteins could mediate pyroptosis in response to direct delivery of LPS to the cytosol. First, we tested two methods of LPS delivery to the cytosol: electroporation and transfection. For electroporation, although there was lower cell death in GBP1KO cells at early timepoints with lower concentrations of LPS, GBP1 was largely dispensable for pyroptosis (SI Appendix, Fig. S10 A and B). This suggests that electroporation is not a good model for GBP1-dependent pyroptosis, likely due to the high efficiency of cytosolic LPS delivery by electroporation and consequential direct caspase-4 activation. Conversely, there was a significant reduction in pyroptosis in GBP1KO cells in response to LPS delivered through transfection (Fig. 6A). We therefore used LPS transfection to probe for the role of individual GBPs in LPS-triggered pyroptosis. To do so, we transduced GBP1KO A549 cells with tetracycline-inducible mCherry or mCherry-GBP expression constructs. In IFNγ-primed cells, GBP1 or GBP2 was able to restore pyroptosis in GBP1KO cells across a 2-log range of aTc concentrations (Fig. 6B). Higher aTc concentrations were needed to induce GBP1 or GBP2 expression sufficiently for the induction of pyroptosis in unprimed cells. GBP3 expression did not lead to any detectable pyroptosis, a notable difference from results obtained in S. flexneri infection experiments. Even at 24 h post transfection, only GBP1- and GBP2-expressing cells had higher levels of cell death compared to the mCherry control (SI Appendix, Fig. S10C). To test if GBP13R, like GBP1 and GBP2, could promote pyroptosis in response to cytosolic-free LPS, we overexpressed GBP1, GBP13R, or GBP2 in GBP1KO cells and measured cell death following LPS transfection. With two concentrations of LPS, expression of GBP1, GBP13R, or GBP2 promoted similar levels of cell death in GBP1KO cells (Fig. 6C). There was also no significant difference in IL-18 secretion between cells expressing GBP1, GBP13R, or GBP2 (Fig. 6D).
Fig. 6.
Ectopically expressed GBP1, GBP13R, and GBP2 can rescue pyroptosis in GBP1KO cells transfected with LPS. (A) Wild-type and GBP1KO A549 cells in 96-well plates were unprimed or primed with 100 U/mL IFNγ overnight, then transfected with 0.1 μg or 1 μg per well E. coli O55:B5 LPS. Cell death was measured using sytox green fluorescence at 6 h post transfection. (B) GBP1KO A549 cells were transduced with tet-inducible expression vectors, and expression of each construct was titrated with different concentrations of anhydrotetracycline (aTc). Cells were unprimed or primed with 100 U/mL IFNγ overnight, then transfected with 1 μg E. coli O55:B5 LPS per well. Cell death was measured using sytox green fluorescence at 6 h post transfection. (C and D) Wild-type A549 or GBP1KO A549 cells overexpressing mCherry or the indicated mCherry-GBPs in 96-well plates were unprimed or primed with 100 U/mL IFNγ overnight, then transfected with 0.1 μg (solid bars) or 1 μg (striped bars) E. coli O55:B5 LPS per well. Cell death was measured using sytox green fluorescence at 6 h post transfection (B). IL-18 secretion was measured in supernatants taken at 6 h post transfection (C). All graphs show averages from three independent experiments and are represented by mean ± SD. (A) One-way ANOVA with Tukey’s multiple comparisons test was used. All significant comparisons are shown. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (B–D) Significance was determined using two-way ANOVA with Tukey’s multiple comparisons test. Statistical comparisons are shown by letters, with bars sharing no matching letters being significantly different. (C and D) Black letters correspond to statistical comparisons for 0.1 μg per well LPS, and blue letters correspond to 1 μg per well LPS.
Interestingly, even in the absence of IFNγ priming, there was significant cell death and IL-18 secretion in cells overexpressing GBP1, GBP13R, or GBP2, indicating that expression of additional GBPs or other interferon-stimulated genes is not required for cell death in response to cytosolic LPS (Fig. 6 B and C). It was previously suggested that even with concurrent GBP1, 3, and 4 overexpression in unprimed cells, there were significantly lower levels of cell death following LPS transfection compared to IFNγ-primed cells (25). Our results suggest that overexpression of GBP1 or GBP2 alone is sufficient to promote pyroptosis, although addition of IFNγ does enhance the response.
GBP2 Acts as a Surfactant and Aggregates LPS.
Because GBP2 induces pyroptosis in response to “free” LPS in the cytosol, we hypothesized that it can directly recognize LPS similar to GBP1. We reported previously that GBP1 is an LPS-binding surfactant which clusters soluble LPS into larger LPS aggregates independent of its 3R motif (24). To test whether GBP2, like GBP1, can induce LPS clustering, we mixed fluorescently labeled LPS with recombinant GBPs and GTP and assessed the LPS particle number and size with fluorescence microscopy (Fig. 7A and SI Appendix, Fig. S11A). We found that GBP2, like GBP1 and GBP13R, increased the LPS aggregate area during GTP hydrolysis, although fewer aggregates were formed by GBP2. Nonisoprenylated GBPs and geranylgeranylated GBP5, on the other hand, failed to induce clustering of LPS to larger aggregates. We further observed that GDP·AlFX-induced GBP1 and GBP2 polymers shifted to larger and smaller Rn when supplemented with LPS (SI Appendix, Fig. S11B), which suggest changes in polymer size and/or polymer conformation due to LPS incorporation. However, these DLS experiments were inconclusive, because sizes of LPS aggregates comprised a broad range and could not be definitively distinguished from GBP monomers and complexes. We therefore employed native polyacrylamide gel electrophoresis (NPAGE) to separate GBP, LPS, and GBP-LPS aggregates not only by size but also by charge, then stained successively for LPS and protein to detect changes in LPS and GBP mobility. To stabilize either the resting state or the active state of the respective GBP in these experiments, we supplemented protein-LPS mixtures with GDP or GDP·AlFX. The migration of bovine serum albumin (BSA, negative control) and GBP5 was unchanged in the presence of LPS (Fig. 7B). However, GBP1 and GBP2 showed shifts in mobility with increasing LPS concentrations (Fig. 7B). This confirms that GBP1 and GBP2 interact with LPS directly and form mixed aggregates. To estimate the affinities of GBP-LPS complexes, we titrated GBP1, GBP13R, and GBP2 with LPS in the presence of GDP·AlFX and analyzed the gray values of the NPAGE gels after protein staining. We then plotted the percentage of protein signal with altered mobility against the LPS concentration (Fig. 7C and SI Appendix, Fig. S11C). GBP13R required roughly 10 times and GBP2 roughly 100 times higher LPS concentrations than GBP1 to generate similar shifts in mobility, suggesting that GBP2 binds LPS and forms LPS-protein aggregates with lower affinity than GBP1. Together, these data identify GBP2 as an LPS clustering surfactant, a property that GBP2 shares with GBP1. We hypothesized that their LPS clustering activities account for the role GBP1 and GBP2 play in caspase-4 activation during gram-negative bacterial infections.
Fig. 7.
GBP1 and GBP2 bind and aggregate LPS and promote caspase-4 activation in vitro. (A) Recombinant isoprenylated and nonisoprenylated GBPs were supplemented with GTP and added to Alexa Fluor 568-labeled E. coli O55:B5 LPS. LPS particles were analyzed from different fields of view taken after 20 min with Fiji, and the average aggregate area was plotted. (B and C) Following mixing of recombinant proteins with E. coli O55:B5 LPS in the presence of different nucleotides, protein-LPS complexes were resolved with NPAGE, and gels were stained successively for LPS and proteins. (B) NPAGEs of BSA, GBP1, GBP2, and GBP5 supplemented with GDP or GDP·AlFX in the absence or presence of LPS (final concentrations 1 mg/mL, 0.1 mg/L, or 0.01 mg/mL). (C) NPAGE of GBP1 titrated with LPS (final concentrations 2 mg/mL to 0.008 mg/mL) supplemented with GDP·AlFX. Gray values for each gel lane were plotted with Fiji and areas were defined representing unshifted and shifted protein fractions. Percentage of shifted protein was plotted against LPS concentrations. (D and E) Following mixing of recombinant proteins with E. coli O55:B5 LPS in the absence or presence of GTP, protein-LPS complexes were added to recombinant caspase-4, and caspase-4 activity was determined by monitoring release of free 7-Amino-4-methylcoumarin (AMC) upon cleavage of fluorogenic caspase substrate Z-VAD-AMC over time. (D) Fluorescence intensities were normalized to basal caspase-4 activities. (E) The area under the curve was used to determine statistical significance. Caspase-4 – CASP4. All graphs show averages from three independent experiments and are represented by mean ± SD. (A and E) One-way ANOVA with Dunnett’s multiple comparisons test comparing to control (no GBP addition) was used. All significant comparisons are shown. *P < 0.05, ****P < 0.0001. (B and C) Representative NPAGEs from two (B) or three (C) independent experiments are shown.
GBP1-LPS and GBP2-LPS Complexes Enhance Caspase-4 Activity In Vitro.
To test whether GBP-LPS complexes directly activate caspase-4, we reconstituted caspase-4 activation in vitro. We incubated LPS with recombinant GBP1, GBP1R48A (negative control), GBP13R, and GBP2 in the presence and absence of GTP, added these reactions to recombinant caspase-4, and monitored cleavage of the fluorogenic caspase substrate Z-VAD-AMC over time (Fig. 7D). LPS addition alone increased caspase-4 activity as previously reported (4)(here: 1.5-fold), and this activity was not enhanced by addition of nucleotide-free GBPs (Fig. 7 D and E). Importantly, when incubated with LPS in the presence of GTP, GBP1, GBP13R, and GBP2, but not GBP1R48A, increased caspase-4 activity significantly (threefold with GBP1 and twofold with GBP13R, GBP2) (Fig. 7 D and E). Of note, GTP-induced GBP1 polymers in the absence of LPS did not increase caspase-4 activity (SI Appendix, Fig. S11D). Together, these data show that GBP1-LPS and GBP2-LPS complexes are sufficient to significantly enhance caspase-4 activity.
Discussion
The cytokine IFNγ enhances noncanonical inflammasome activation in response to cytosolic LPS through upregulation of the GBP protein family. GBPs promote pyroptosis in different cell types and species; however, the exact role of each human GBP paralog and the mechanism by which GBPs promote pyroptosis have not been clear (38–42). It was previously thought that GBP1 binding to the surface of gram-negative bacteria and the subsequent recruitment of GBP2, GBP3, and GBP4 were essential steps in initiating LPS-triggered and caspase-4-dependent pyroptosis during infection. While GBP2 can only bind gram-negative bacteria when GBP1 is present, in this study, we showed that GBP2 promotes pyroptosis in the absence of GBP1, indicating that bacterial targeting is dispensable for GBP-dependent pyroptosis. We confirmed this by showing that a mutant of GBP1 that binds free LPS but does not form a bacteria-encasing microcapsule can induce pyroptosis during infection. We also found that, like GBP1, GBP2 directly binds and aggregates LPS, and we show that this LPS clustering by GBPs facilitates caspase-4 activation and pyroptosis.
A major question remaining is why GBP1 and GBP2 have seemingly redundant roles in response to cytosolic LPS. One potential explanation is that GBP1 and GBP2 have unique specificities for different LPS species, allowing detection of a broader range of bacteria. Gram-negative bacteria contain a diverse repertoire of LPS structures, with many variants existing in the O-antigen, core oligosaccharide, and lipid A portions of LPS molecules (43). LPS variants have different abilities to activate sensors TLR4 or caspase-4 (43, 44). Supporting the hypothesis that GBP2 could preferentially drive inflammasome responses to a subset of LPS structures, it was shown that GBP2 was required for efficient activation of caspase-4 in response to tetra-acylated LPS from Francisella novicida, while it was dispensable for pyroptosis in response to hexa-acylated E. coli LPS (20).
The functions of GBP1 and GBP2 may also be defined by the route of entry of LPS into the cytosol, similar to the differential role for human caspase-4 and caspase-5 in LPS sensing. Caspase-4 and caspase-5 are structurally similar, and both are able to directly bind LPS and induce pyroptosis (4), yet in many assays of LPS-induced pyroptosis, caspase-4 is essential and caspase-5 is dispensable (9, 11, 26). Recently, it was shown that caspase-5, but not caspase-4, responds to LPS present in OMVs trafficked through endosomes of intestinal epithelial cells (45). This suggests that the cell may have different ways of responding depending on the delivery route by which LPS reaches the cytosol.
Another reason for GBP1 and GBP2 redundancy may be to allow either cell type-specific or tissue-specific responses to gram-negative infection, as these GBPs are each expressed at different levels in cell lines and tissues (46, 47). We found that GBP2 has a lower affinity for LPS, so it may be preferentially used to detect cytosolic LPS in tissues where a less robust response is warranted, for example, in the gut where there is abundant LPS released by commensal bacteria. Further studies investigating the differences between GBP1 and GBP2 function may provide an answer for the importance of each protein.
Our study highlights the ability of GBPs to promote immune defense even in the absence of direct binding to pathogens. While many studies have investigated the pathogen-targeting-dependent functions of GBPs in immunity, there are now several examples of GBPs restricting pathogen growth without translocation to pathogens or pathogen-containing vacuoles. GBPs restrict growth of protozoan parasites Leishmania donovani and Toxoplasma gondii without detectable binding to the parasitophorous vacuoles (48–50). GBPs also promote pyroptosis in response to avirulent E. coli in macrophages, without significant GBP targeting of these bacteria in cells (18). We show here that GBP1 and GBP2 promote pyroptosis without binding the surface of bacteria, likely through aggregation of LPS released during infection. This could be a more universal strategy for GBPs to act as pattern recognition receptors, where pathogen-associated molecular patterns (PAMPs), such as LPS but also others, are released into the host cytosol and are detected by GBPs. PAMPs bound by GBPs could then facilitate activation of other defense signaling pathways. Future studies could test this hypothesis and determine which additional PAMPs are detected by GBPs, and if these are similarly being released from pathogen-containing vacuoles and directly bound by GBPs. For pathogens where GBP targeting is observed, it will also be informative for future studies to use GBP mutants to separate targeting-dependent and -independent functions.
Our study identifies GBP2 as an immune sensor that aggregates LPS and allows caspase-4 activation. While GBPs were known to be important for activation of the noncanonical inflammasome in response to “free” LPS, LPS contained in outer membrane vesicles, or in response to gram-negative bacterial infection, it was unclear how GBPs function to detect these different forms of LPS. During infection with cytosolic gram-negative bacteria, one model proposes that GBP binding to the surface of invading bacteria provides a necessary platform for caspase-4 activation. The recent finding that GBP1 directly binds and clusters “free” LPS provides another model for caspase-4 activation, where GBP1 acts as a surfactant to aggregate LPS into larger structures that are a preferred substrate for caspase-4 (24, 51) (model illustration in SI Appendix, Fig. S12). Since gram-negative bacteria constitutively shed “free” LPS (52), aggregation of LPS in the cytosol explains both responses to invading bacteria and “free” LPS. Our current study supports a model where GBP1 or GBP2 promotes caspase-4 activation through aggregation of LPS released into the cytosol during infection.
Materials and Methods
Cell Lines, Cell Culture, Cell Transduction, and Cell Transfection.
For IFNγ priming, cells were treated overnight with 100 U/mL IFNγ. LPS transfection was done essentially as previously described (25) using E. coli O55:B5 LPS at the indicated concentrations per well. A detailed description of all CRISPR-generated mutant and parental cell lines and their culture conditions and a description of all mammalian expression constructs, lentiviral transduction procedures, LPS transfection, and electroporation protocols are provided in SI Appendix, Materials and Methods.
Bacterial Strains and Infections.
Parental Shigella flexneri strain 2457T was used. S. flexneri ΔipaH9.8, ΔospC3, and ΔipaH9.8, ΔospC3 were previously described (24, 27, 53). For Salmonella enterica Typhimurium, strain 14028s was used. Bacterial plasmids, bacterial culture conditions, and infection methods are detailed in SI Appendix, Materials and Methods.
Cell Death Assays, Bacterial Luminescence, IL-18 ELISA, and Western Blotting.
Cells were infected or transfected/electroporated with LPS and cell death was measured at indicated timepoints using sytox green fluorescence, while bacterial growth was assessed using luminescence over time. Supernatants were removed at indicated timepoints for measuring LDH release (cell death) or performing IL-18 ELISA. Detailed procedures are provided in SI Appendix, Materials and Methods. For western blotting, cells were lysed in RIPA buffer and processed as described in SI Appendix, Materials and Methods. All antibodies used to detect proteins are listed in SI Appendix, Materials and Methods.
Immunofluorescence and Time-Lapse Microscopy.
For microscopy, cells were plated on glass coverslips and infected the next day with S. flexneri at an MOI of 5, then processed for microscopy at the indicated time points. Detailed description of the applied cell staining and time-lapse microscopy protocols are provided in SI Appendix, Materials and Methods.
Expression, Purification, Prenylation, and Labeling of Recombinant Protein.
Protein purification and prenylation was essentially performed as described previously (34) and detailed in SI Materials and Methods. Briefly, His-tagged GBP1, GBP2, GBP5, FTase, and GGTase were expressed in E. coli. Recombinant proteins were purified by immobilized metal affinity chromatography (IMAC) followed by size-exclusion chromatography (SEC) (36). Fractions with monomeric GBPs, FTase, or GGTase were pooled and concentrated. Monomeric GBP1, GBP2, and GBP5 were incubated with farnesyl pyrophosphate (FPP) or geranylgeranyl pyrophosphate (GGPP), and FTase or GGTase. Prenylated GBPs were further purified by hydrophobic interaction chromatography (HIC) and SEC. Recombinant GBPs were labeled with Alexa Fluor 488 C5 maleimide dye or Alexa Fluor 647 C2 maleimide dye.
GBP Bacterial Binding, LPS Aggregation Assays, Light Scattering, and Native Polyacrylamide Gel Electrophoresis.
Bacterial binding and LPS aggregation assays were performed essentially as described previously (24). Particle sizes were measured by dynamic light scattering. Absorbance-based light scattering experiments were performed to detect the formation of large GBP polymers. Native GBP-LPS complexes were analyzed with native polyacrylamide gel electrophoresis (NPAGE). Detailed information for all of these methods are provided in SI Appendix, Materials and Methods.
Caspase-4 Activation.
Caspase-4 activity was determined essentially as described previously (4) with some modifications detailed in SI Appendix, Materials and Methods. Recombinant GBP1, GBP1R48A, GBP13R, and GBP2 were mixed with E. coli O55:B5 LPS and GTP before incubation with GST-tagged full-length caspase-4 (purified from LPS-free wheat germ) and the caspase-4 substrate Z-VAD-AMC. The colorimetric signal resulting from substrate cleavage was measured.
Supplementary Material
Appendix 01 (PDF)
GBP2 and GBP5 fail to encapsulate bacteria on their own. Recombinant Alexa Fluor 488-labeled GBP1, GBP2, or GBP5 were supplemented with GTP and added to formaldehyde-fixed RFP-expressing S. flexneri.
Both GBP2 and GBP5 associate with bacteria GBP1-dependently but only GBP2 incorporates in the microcapsule. Recombinant Alexa Fluor 488-labeled GBP1, GBP2, or GBP5 mixed with Alexa-647-labeled GBP1 were supplemented with GTP and added to formaldehyde-fixed RFP-expressing S. flexneri.
Pyroptosis of S. flexneri does not require GBP1 binding to bacteria. GBP1KO cells primed with 100 U/ml IFNγ overnight expressing mCherry (control), mCherry-GBP1, or mCherry-GBP13R (magenta) infected with GFP-expressing S. flexneri ΔospC3 ΔipaH9.8 (yellow). Dying cells are shown in blue (sytox blue).
Acknowledgments
We thank Dr. Edward Miao for sharing Salmonella strains with us and Dr. Gerrit Praefcke for sharing the pRSF-Duet1-FTase plasmid. This work was supported by NIH grants AI139425 (to J.C.), AI064285 (C.F.L.), and AI28360 (C.F.L.); by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) Research Fellowship 427472513 (to M.K.) and by research grant HE2679/6-1 (to C.H.). J.C. holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. This work was performed in part at the Duke University Light Microscopy Core Facility (LMCF) and Duke Functional Genomics. We thank LMCF members Drs. Benjamin Carlsen, Yasheng Gao, and Lisa Cameron for providing training and Dr. So Young Kim for generating knockout cell lines.
Author contributions
M.S.D., M.K., and J.C. designed research; M.S.D., M.K., and L.S. performed research; L.S., D.H., A.S.P., D.N., C.F.L., and C.H. contributed new reagents/analytic tools; M.S.D., M.K., L.S., D.N., C.H., and J.C. analyzed data; M.K., C.F.L, C.H., and J.C. obtained funding; and M.S.D., M.K., and J.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Randow F., MacMicking J. D., James L. C., Cellular self-defense: How cell-autonomous immunity protects against pathogens. Science 340, 701–706 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poltorak A., et al. , Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science 282, 2085–2088 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Rosadini C. V., Kagan J. C., Early innate immune responses to bacterial LPS. Curr. Opin. Immunol. 44, 14–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi J., et al. , Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Aachoui Y., et al. , Caspase-11 protects against bacteria that escape the vacuole. Science 339, 975–978 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagar J. A., Powell D. A., Aachoui Y., Ernst R. K., Miao E. A., Cytoplasmic LPS activates caspase-11: Implications in TLR4-independent endotoxic shock. Science 341, 1250–1253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kayagaki N., et al. , Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341, 1246–1249 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald K. A., Kagan J. C., Toll-like receptors and the control of immunity. Cell 180, 1044–1066 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knodler L. A., et al. , Noncanonical inflammasome activation of caspase-4/caspase-11 mediates epithelial defenses against enteric bacterial pathogens. Cell Host Microbe 16, 249–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi T., et al. , The Shigella OspC3 effector inhibits caspase-4, antagonizes inflammatory cell death, and promotes epithelial infection. Cell Host Microbe 13, 570–583 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Naseer N., et al. , Salmonella enterica serovar typhimurium induces NAIP/NLRC4- and NLRP3/ASC-independent, caspase-4-dependent inflammasome activation in human intestinal epithelial cells. Infect. Immun. 90, e0066321 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan A. H., Schroder K., Inflammasome signaling and regulation of interleukin-1 family cytokines. J. Exp. Med. 217, e20190314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi J., et al. , Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Liu X., et al. , Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kayagaki N., et al. , NINJ1 mediates plasma membrane rupture during lytic cell death. Nature 591, 131–136 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Pilla D. M., et al. , Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc. Natl. Acad. Sci. U.S.A. 111, 6046–6051 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meunier E., et al. , Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature 509, 366–370 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Finethy R., et al. , Inflammasome activation by bacterial outer membrane vesicles requires guanylate binding proteins. mBio 8, e01188–01117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisch D., et al. , Human GBP1 is a microbe-specific gatekeeper of macrophage apoptosis and pyroptosis. EMBO J. 38, e100926 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagrange B., et al. , Human caspase-4 detects tetra-acylated LPS and cytosolic Francisella and functions differently from murine caspase-11. Nat. Commun. 9, 242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santos J. C., et al. , LPS targets host guanylate-binding proteins to the bacterial outer membrane for non-canonical inflammasome activation. EMBO J. 37, e98089 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xavier A., Al-Zeer M. A., Meyer T. F., Daumke O., hGBP1 coordinates chlamydia restriction and inflammasome activation through sequential GTP hydrolysis. Cell Rep. 31, 107667 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Kutsch M., Coers J., Human guanylate binding proteins: Nanomachines orchestrating host defense. FEBS J. 288, 5826–5849 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kutsch M., et al. , Direct binding of polymeric GBP1 to LPS disrupts bacterial cell envelope functions. EMBO J. 39, e104926 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santos J. C., et al. , Human GBP1 binds LPS to initiate assembly of a caspase-4 activating platform on cytosolic bacteria. Nat. Commun. 11, 3276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wandel M. P., et al. , Guanylate-binding proteins convert cytosolic bacteria into caspase-4 signaling platforms. Nat. Immunol. 21, 880–891 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piro A. S., et al. , Detection of cytosolic Shigella flexneri via a C-terminal triple-arginine motif of GBP1 inhibits actin-based motility. mBio 8, e01979–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wandel M. P., et al. , GBPs inhibit motility of shigella flexneri but are targeted for degradation by the bacterial ubiquitin ligase IpaH9.8. Cell Host Microbe 22, 507–518.e505 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohler K. M., et al. , A rapidly evolving polybasic motif modulates bacterial detection by guanylate binding proteins. mBio 11, e00340–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li P., et al. , Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature 551, 378–383 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Oh C., Verma A., Hafeez M., Hogland B., Aachoui Y., Shigella OspC3 suppresses murine cytosolic LPS sensing. iScience 24, 102910 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gregor C., Gwosch K. C., Sahl S. J., Hell S. W., Strongly enhanced bacterial bioluminescence with the ilux operon for single-cell imaging. Proc. Natl. Acad. Sci. U.S.A. 115, 962–967 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashida H., et al. , A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kappaB-mediated inflammatory response. Nat. Cell Biol. 12, 66–73; sup pp 61–69 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ince S., Kutsch M., Shydlovskyi S., Herrmann C., The human guanylate-binding proteins hGBP-1 and hGBP-5 cycle between monomers and dimers only. FEBS J. 284, 2284–2301 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Shydlovskyi S., et al. , Nucleotide-dependent farnesyl switch orchestrates polymerization and membrane binding of human guanylate-binding protein 1. Proc. Natl. Acad. Sci. U.S.A. 114, E5559–E5568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sistemich L., et al. , The molecular mechanism of polymer formation of farnesylated human Guanylate-binding Protein 1. J. Mol. Biol. 432, 2164–2185 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Britzen-Laurent N., et al. , Intracellular trafficking of guanylate-binding proteins is regulated by heterodimerization in a hierarchical manner. PLoS One 5, e14246 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meunier E., Broz P., Interferon-inducible GTPases in cell autonomous and innate immunity. Cell Microbiol. 18, 168–180 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Man S. M., Place D. E., Kuriakose T., Kanneganti T. D., Interferon-inducible guanylate-binding proteins at the interface of cell-autonomous immunity and inflammasome activation. J. Leukoc. Biol. 101, 143–150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ingram J. P., Brodsky I. E., Balachandran S., Interferon-gamma in Salmonella pathogenesis: New tricks for an old dog. Cytokine 98, 27–32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomes M. T. R., Cerqueira D. M., Guimarães E. S., Campos P. C., Oliveira S. C., Guanylate-binding proteins at the crossroad of noncanonical inflammasome activation during bacterial infections. J. Leukoc Biol. 106, 553–562 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oh C., Verma A., Aachoui Y., Caspase-11 non-canonical inflammasomes in the lung. Front Immunol. 11, 1895 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simpson B. W., Trent M. S., Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol. 17, 403–416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alexander-Floyd J., et al. , Lipid A variants activate human TLR4 and the noncanonical inflammasome differently and require the core oligosaccharide for inflammasome activation. Infect. Immun. 90, e0020822 (2022), 10.1128/iai.00208-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X., et al. , SNX10-mediated LPS sensing causes intestinal barrier dysfunction via a caspase-5-dependent signaling cascade. EMBO J. 40, e108080 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uhlen M., et al. , Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Human Protein Atlas, proteinatlas.org (Accessed August 20, 2022).
- 48.Haldar A. K., Nigam U., Yamamoto M., Coers J., Goyal N., Guanylate binding proteins restrict Leishmania donovani growth in nonphagocytic cells independent of parasitophorous vacuolar targeting. mBio 11, e01464–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnston A. C., et al. , Human GBP1 does not localize to pathogen vacuoles but restricts Toxoplasma gondii. Cell Microbiol. 18, 1056–1064 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fisch D., Clough B., Khan R., Healy L., Frickel E. M., Toxoplasma-proximal and distal control by GBPs in human macrophages. Pathog Dis. 79, ftab058 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wacker M. A., Teghanemt A., Weiss J. P., Barker J. H., High-affinity caspase-4 binding to LPS presented as high molecular mass aggregates or in outer membrane vesicles. Innate Immun. 23, 336–344 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prins J. M., van Deventer S. J., Kuijper E. J., Speelman P., Clinical relevance of antibiotic-induced endotoxin release. Antimicrob Agents Chemother. 38, 1211–1218 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mou X., Souter S., Du J., Reeves A. Z., Lesser C. F., Synthetic bottom-up approach reveals the complex interplay of Shigella effectors in regulation of epithelial cell death. Proc. Natl. Acad. Sci. U.S.A. 115, 6452–6457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
GBP2 and GBP5 fail to encapsulate bacteria on their own. Recombinant Alexa Fluor 488-labeled GBP1, GBP2, or GBP5 were supplemented with GTP and added to formaldehyde-fixed RFP-expressing S. flexneri.
Both GBP2 and GBP5 associate with bacteria GBP1-dependently but only GBP2 incorporates in the microcapsule. Recombinant Alexa Fluor 488-labeled GBP1, GBP2, or GBP5 mixed with Alexa-647-labeled GBP1 were supplemented with GTP and added to formaldehyde-fixed RFP-expressing S. flexneri.
Pyroptosis of S. flexneri does not require GBP1 binding to bacteria. GBP1KO cells primed with 100 U/ml IFNγ overnight expressing mCherry (control), mCherry-GBP1, or mCherry-GBP13R (magenta) infected with GFP-expressing S. flexneri ΔospC3 ΔipaH9.8 (yellow). Dying cells are shown in blue (sytox blue).
Data Availability Statement
All study data are included in the article and/or SI Appendix.