Significance
The flowering time and architecture of soybean is extremely sensitive to photoperiod, which limits the suitable planting area of a certain soybean cultivar to a small latitudinal span. It is known that light signaling through phytochrome A (E3/E4) modulates the activity of J, a key component of circadian Evening Complex (EC), to control photoperiodic flowering. However, the molecular mechanism remains unclear. Here, we revealed that GmEID1 acts as a bridge to link the light signals perceived by E3/E4 to the activity of EC. In addition to this unique insight into flowering time mechanisms, our results suggest that GmEID1 is a potential target for adjustment of soybean flowering time to improve adaptation and yield, via gene editing and classical breeding.
Keywords: phytochrome A, flowering, GmEID1, circadian Evening Complex, yield
Abstract
Soybean (Glycine max) morphogenesis and flowering time are accurately regulated by photoperiod, which determine the yield potential and limit soybean cultivars to a narrow latitudinal range. The E3 and E4 genes, which encode phytochrome A photoreceptors in soybean, promote the expression of the legume-specific flowering repressor E1 to delay floral transition under long-day (LD) conditions. However, the underlying molecular mechanism remains unclear. Here, we show that the diurnal expression pattern of GmEID1 is opposite to that of E1 and targeted mutations in the GmEID1 gene delay soybean flowering regardless of daylength. GmEID1 interacts with J, a key component of circadian Evening Complex (EC), to inhibit E1 transcription. Photoactivated E3/E4 interacts with GmEID1 to inhibit GmEID1–J interaction, promoting J degradation resulting in a negative correlation between daylength and the level of J protein. Notably, targeted mutations in GmEID1 improved soybean adaptability by enhancing yield per plant up to 55.3% compared to WT in field trials performed in a broad latitudinal span of more than 24°. Together, this study reveals a unique mechanism in which E3/E4-GmEID1-EC module controls flowering time and provides an effective strategy to improve soybean adaptability and production for molecular breeding.
Soybean (Glycine max L.) is one of the most economically important legume crops that provides plant oil and protein to humans and livestock around the world. As a facultative short-day (SD) plant, soybean flowers earlier under SD conditions than under long-day (LD) conditions (1). This photoperiodic response significantly limits the yield potential of soybean cultivars at different latitudes (2). Molecular breeding by modulating flowering time and reducing sensitivity to daylength are effective ways to improve the adaptability and production of soybean (3).
Garner and Allard discovered photoperiodism by comparing the flowering time of plants (including soybean) grown under SD or LD conditions in 1920 (4). To date, more than a dozen genes/loci related to flowering time and maturity have been identified in soybean, including E1 to E11, J, TOF5, TOF11, TOF12, TOF16, and TOF18. Among them, E1 encodes a legume-specific flowering repressor that inhibits flowering under LD conditions (5–7); E2 is homologous to Arabidopsis GIGANTEA (8); E3 and E4 encode phytochrome A homologs, named GmPHYA3 and GmPHYA2, respectively (9, 10); E6 and J, renowned for “long-juvenile” gene, encode a homolog of EARLY FLOWERING 3, named GmELF3a (11–13), a key component of the circadian Evening Complex (EC) consisting of ELF3, ELF4, and LUX ARRHYTHMO (LUX) (14); E9 encodes a homolog of FLOWERING LOCUS T, named GmFT2a (15, 16); and TOF5 encodes a homolog of Arabidopsis FRUITFULL (FUL) (17, 18). TOF11 and TOF12 encode homologs of Arabidopsis PSEUDO-RESPONSE REGULATOR 3, named GmPRR3a and GmPRR3b, respectively (19–21). TOF16 encodes a homolog of LATE ELONGATED HYPOCOTYL (LHY) (22). TOF18 encodes a homolog of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1/AGL20) (23). It should be noted that E1, as the central flowering repressor in the photoperiodic flowering pathway in soybean, expresses at a high level under LD conditions to inhibit GmFT2a and GmFT5a transcription (5, 24, 25). J, as one of the EC components, represses E1 transcription by directly binding the E1 promoter (11–13). In cultivars with loss-of-function j alleles, E1 expression is released from the inhibition of EC, allowing E1 to delay flowering and maturation and increase yield by 30 to 50% in low latitudinal regions (12). Light signals are perceived by E3 and E4 that indirectly up-regulate E1 expression under LD conditions (12, 26). However, the factors that link the perception of light or photoperiod by E3 and E4 to the transcriptional upregulation of E1 remain largely unknown.
The EID1 gene (EMPFINDLICHER IM DUNNKELROTEN LICHT 1) encodes an F-box protein that functions as a negative regulator in the light signaling cascade downstream of the photoreceptor PHYA in Arabidopsis (27–29). Meanwhile, a domesticated gene (Solyc09g075080) homologous to EID1 is responsible for the deceleration of the circadian clock in cultivated tomato (30–32). In this study, we identified an orthologous gene for EID1 in soybean (named GmEID1) that regulates flowering time by physically interacting with and stabilizing J protein. Furthermore, E3 and E4 interact with GmEID1 in a light-dependent manner to interfere with the GmEID1–J interaction, which reduces J protein abundance under LD conditions. The CRISPR-Cas9 engineered mutations in the GmEID1 gene dramatically increased yield in field trials, providing a promising approach to substantially improve soybean adaptability and production.
Results
Identification of GmEID1 as a Flowering Regulator.
The E1 gene expression pattern is characterized by a robust rhythmic fluctuation with a low level during the day except for a slight hump at ZT4, peaking at dusk and subsequently decreasing during the night under LD conditions (6, 12, 25). We surmised that the gene upstream of E1 may be characterized with an opposite or consistent expression pattern compared to E1. Therefore, we screened flowering candidate genes exhibiting such expression patterns by transcriptome sequencing (RNA-seq) analysis within an LD photoperiodic cycle (Datasets S1 and S2) (33). Bioinformatic analysis identified four opposite-pattern genes and ten consistent-pattern genes compared to E1 (SI Appendix, Fig. S1). We noticed that one of the opposite-pattern genes, named GmEID1 hereafter, is orthologous to Arabidopsis EID1 (AtEID1) that encodes an F-box protein involved in the PHYA-mediated light signaling pathway (27) (Fig. 1A and SI Appendix, Fig. S1). We found five homologous proteins for EID1 in soybean, among which GmEID1 is mostly conserved with AtEID1 (SI Appendix, Figs. S2 and S3). Tissue-specific expression analysis showed that the GmEID1 gene is highly expressed in above-ground tissues, including leaves and shoot tips (SI Appendix, Fig. S4A), which is similar to the J gene (18). The subcellular localization experiment demonstrated that GmEID1 is localized in the nucleus (SI Appendix, Fig. S4B), which is reminiscent to that of AtEID1 (29), suggesting that GmEID1 may also be involved in the GmPHYA-mediated light signaling pathway in soybean.
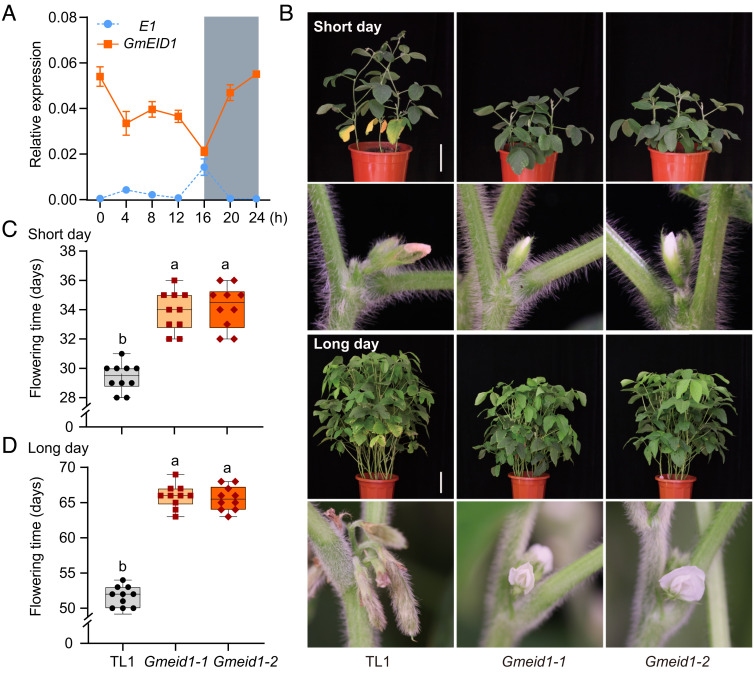
Fig. 1.
GmEID1 is a flowering enhancer. (A) Dynamic mRNA levels of E1 and GmEID1 in wild-type TL1 plants grown under LD conditions (16 h light/8 h dark). New fully expanded unifoliate leaves were collected for qRT-PCR analysis. Data are mean ± SD of three biological replicates. The relative expression level of each indicated gene was calculated using the GmActin gene as an internal control. (B) Photos of wild-type TL1 and Gmeid1 mutant lines grown under SD conditions (12 h light/12 h dark, upper two panels) or LD conditions (lower two panels) in phytotrons. (Scale bar, 10 cm.) (C and D) Flowering time of indicated lines grown under SD conditions (C) or LD conditions (D) as in B. Mean values ± SD (n > 8) are shown. The lowercase letters above the dots indicate significant differences (P < 0.01, ANOVA with Tukey’s post-test).
To test this possibility, we generated mutants engineered with CRISPR/Cas9 in two genetic backgrounds (Gmeid1-1 and Gmeid1-2 in the Tianlong1 (TL1) background, Gmeid1-3 and Gmeid1-4 in the Williams82 (W82) background) (SI Appendix, Fig. S5 A and B). Meanwhile, we made the 35S::YFP-GmEID1 and 35S::GmEID1-YFP overexpression constructs and obtained multiple transgenic lines in the TL1 background (SI Appendix, Fig. S5 C–E). Phenotypic analysis demonstrated that knockout or overexpression of the GmEID1 gene resulted in significantly later or early flowering phenotype, respectively, under both LD and SD conditions (Fig. 1 B–D and SI Appendix, Fig. S6 A–F). The above results demonstrate that the GmEID1 gene functions as a photoperiod-independent flowering regulator in soybean.
GmEID1 Inhibits E1 Transcription.
To gain insight into how GmEID1 accelerates flowering, we compared the diurnal transcript levels of key flowering genes in wild-type TL1 and Gmeid1 mutants grown under LD or SD conditions. The qRT-PCR results demonstrated that the mRNA levels of GmFT2a and GmFT5a were much lower in the Gmeid1 mutants (Fig. 2 A–D) but increased in the GmEID1 overexpression lines compared to TL1 (SI Appendix, Fig. S7 A and B). Consistently, the E1 mRNA level was significantly up-regulated in the Gmeid1 mutants (Fig. 2 E and F) but down-regulated in the GmEID1 overexpression lines (SI Appendix, Fig. S7C). Interestingly, the expression levels of J, GmCCA1a, and GmPRR3b, which are circadian clock component genes upstream of E1 (19), did not show significant changes in the Gmeid1 mutants (SI Appendix, Fig. S8 A and B) and GmEID1 overexpression lines at most of the time points tested under diurnal conditions (SI Appendix, Fig. S7 D–F). Taken together, the above results suggest that the GmEID1 gene promotes flowering through inhibiting E1 expression in soybean.
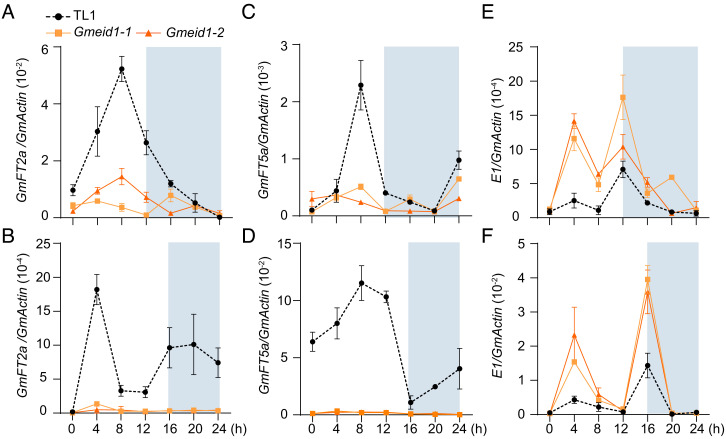
Fig. 2.
Temporal expressions of flowering time-associated genes in the indicated lines. (A–F) Diurnal variation in the transcript levels of GmFT2aGmFT5a, and E1 in the wild-type TL1 and Gmeid1 mutants under SD conditions (A, C, and E, respectively) and LD conditions (B, D, and F, respectively). Second trifoliate leaves of 20-d-old plants were collected for qRT-PCR analysis. Mean values ± SD (n = 3) are shown. GmActin was used as an internal control.
GmEID1 Interacts with J to Promote Flowering.
Given that GmEID1 and J behave similarly in the aspects of repressing E1 expression and promoting flowering, together with the fact that J transcription level does not change significantly in both Gmeid1 mutants and GmEID1 overexpression lines (SI Appendix, Figs. S7 D–F and S8 A and B), we hypothesized that GmEID1 may function by physically interacting with J. Consistent with this hypothesis, the β-galactosidase activity assay in yeast demonstrated that GmEID1 interacted not only with J/GmELF3a (Fig. 3A), but also with the other two ELF3 coorthologs, GmELF3b-1 and GmELF3b-2 (12). Furthermore, other potential EC components including GmELF4a and GmELF4b, but not GmLUX1 and GmLUX2, were also shown detectable interaction with GmEID1 (Fig. 3A). Physical associations between GmEID1 and GmELF3s were further confirmed by coimmunoprecipitation (Co-IP) assays and dual-luciferase assays in tobacco leaves (Fig. 3B and SI Appendix, Fig. S9). Taken together, these results suggested that GmEID1 may affect the activity of the EC by interacting with GmELF3s and GmELF4s to modulate flowering time.
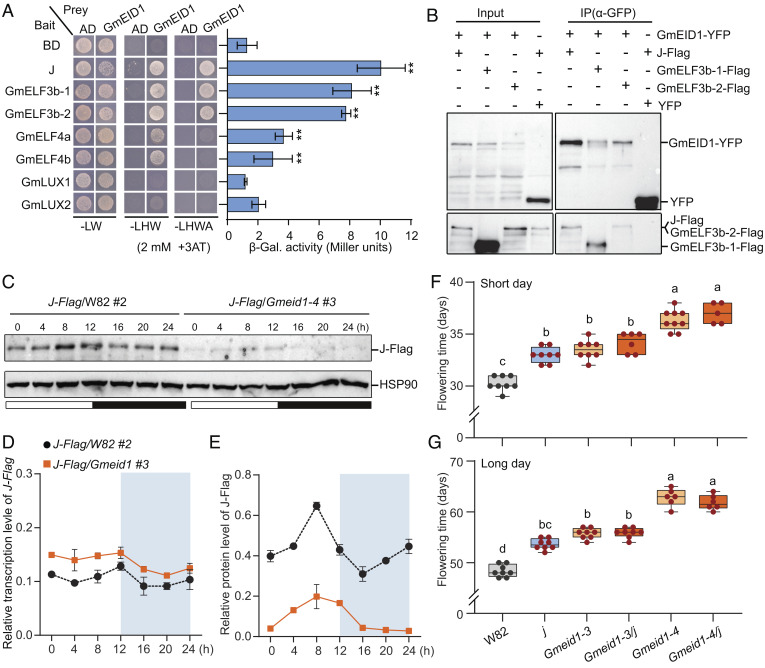
Fig. 3.
GmEID1 interacts with EC and influences the abundance of J protein to regulate flowering time. (A) The interaction between GmEID1 and each EC component (J, GmELF3b-1, GmELF3b-2, GmELF4a, GmELF4b, GmLUX1, or GmLUX2) in yeast. For auxotrophic analysis, yeast cells transformed with the indicated genes were selected on -LW (lacking Leu and Trp), -LWH (lacking Leu, Trp, and His), and -LWHA (lacking Leu, Trp, His, and Ade) media. β-galactosidase assays show the interaction strength of GmEID1 with each EC component. Data are means ± SD (n = 3) with significant differences determined by two-tailed Student’s t test (*P < 0.05, **P < 0.01). (B) Coimmunoprecipitation (Co-IP) showing the interaction of GmEID1 with J, GmELF3b-1, or GmELF3b-2 in tobacco leaves. The YFP protein was used as a negative control. (C) Immunoblot showing the fluctuation of J-Flag protein levels in the wild-type W82 background or the Gmeid1-4 mutant background under diurnal conditions. The membrane was probed by the anti-Flag antibody, stripped, and then probed by the anti-HSP90 antibody. (D and E) Quantitative assays of J-Flag transcript levels relative to GmActin (D) and J-Flag protein levels relative to HSP90 (E) in samples as in C. (F and G) Flowering time of the indicated lines grown under SD conditions (F) and LD conditions (G). Data are means ± SD (n ≥ 5). The lowercase letters above the dots indicate significant differences (P < 0.01, ANOVA with Tukey’s post-test).
GmEID1 Enhances the Abundance of J Protein.
Considering that GmEID1 is an F-box protein that is supposed to function as an E3 ligase destabilizing target proteins through the ubiquitin pathway, we tested whether GmEID1 affects the stability of J protein through the root-induced callus expression (RICE) system in soybean (19, 34, 35). Briefly, the 35S::J-3xFlag construct was transformed into hairy root which was consequently induced into uniform callus by tissue culture. Multiple independent transgenic callus lines in the wild-type W82 background or in the Gmeid1-4 mutant background were used to compare the abundance of J protein. Interestingly, the western blot results showed that the levels of the J-Flag protein were prone to be higher in the W82 background than those in the Gmeid1-4 mutant background (SI Appendix, Fig. S10A), suggesting that GmEID1 is positively correlated with the abundance of J protein. In particular, the abundance of the J-Flag protein was positively correlated with its transgenic mRNA level in the W82 background, but not in the Gmeid1-4 mutant background (SI Appendix, Fig. S10B). Consistent with this, the overall expression levels of E1 were higher in the Gmeid1-4 mutant background than those in the wild-type W82 background (SI Appendix, Fig. S10C). Next, we tested how GmEID1 affects J protein abundance in a time-course manner under diurnal conditions. We selected two representative callus lines expressing similar levels of transgenic J-Flag mRNA in the wild-type W82 background (J-Flag/W82 #2) and in the Gmeid1 mutant background (J-Flag/Gmeid1 #3) (Fig. 3 C and D). The immunoblot results demonstrated that the levels of the J-Flag protein were constitutively lower in the Gmeid1 mutant than those in the wild-type W82 (Fig. 3 C and E). To exclude the influence of different genetic backgrounds, we tested the abundance of J protein in the callus of the Gmeid1-1 mutant in the TL1 cultivar background, and the result further supported that GmEID1 enhances the accumulation of J protein (SI Appendix, Fig. S11 A and B). Consistent with the above observations, E1 transcriptional levels were significantly higher in the Gmeid1 mutant callus than those in wild-type callus (SI Appendix, Fig. S11C). Meanwhile, E1 protein levels increased significantly in the Gmeid1 mutant compared to wild type, especially after dusk (SI Appendix, Fig. S11 D and E).
To test the genetic relationship between GmEID1 and J, we used the Gmeid1-3 and Gmeid1-4 mutants to cross with the j mutant, which are all in the W82 background (SI Appendix, Fig. S12 A and B). Molecular analysis showed that Gmeid1-4 is likely a null mutant due to a frameshift mutation, while Gmeid1-3 is possibly a weak mutant expressing an incomplete GmEID1 protein missing 17 amino acids (SI Appendix, Fig. S5 A and B). Consistently, the Gmeid1-4 mutant showed a more severely late flowering phenotype than that of the Gmeid1-3 mutant (SI Appendix, Fig. S6 E and F). The flowering time of the j mutant was similar to that of the Gmeid1-3 mutant, but earlier than that of the Gmeid1-4 mutant under both SD and LD conditions (Fig. 3 F and G), suggesting that GmEID1 stabilizes not only J but also other J-like proteins, including GmELF3b-1 and GmELF3b-2 (Fig. 3 A and B and SI Appendix, Fig. S9). Furthermore, the flowering time of Gmeid1-3/j and Gmeid1-4/j was similar to that of Gmeid1-3 and Gmeid1-4, respectively (Fig. 3 F and G), supporting that GmEID1 and J function in a same genetic pathway.
Light-Dependent Interaction between E3/E4 and GmEID1 Interferes with the GmEID1–J Interaction.
It has been documented that J mediates the regulation of E1 expression through a GmPHYA (E3 and E4) signaling pathway (12). Moreover, we were unable to detect the direct interaction between E3/4 and J (SI Appendix, Fig. S13) (36). In this context, we tested whether E3 and E4 may interact directly with GmEID1. Both the β-galactosidase activity assay and the auxotrophic assay showed that the light receptors E3 and E4 were able to interact with GmEID1 in a red or far-red light-dependent manner in yeast cells (Fig. 4A and SI Appendix, Fig. S14A). The E3/E4–GmEID1 interactions were further confirmed by dual-luciferase and Co-IP assays by transient expression in tobacco leaves (SI Appendix, Fig. S14 B and C). Next, we crossed the CRISPR-Cas9-engineered e3 mutant which has an early flowering phenotype (37), with the Gmeid1-1 mutant which has a later flowering phenotype, to obtain the Gmeid1/e3 double mutant (SI Appendix, Fig. S15A). Phenotypic analysis showed that the Gmeid1/e3 mutant flowered as late as the Gmeid1 mutant under natural LD conditions (Fig. 4B), demonstrating that the Gmeid1 mutant could completely suppress the early flowering phenotype of the e3 mutant. Consistent with this, the expression levels of E1, GmFT2a, and GmFT5a in the Gmeid1/e3 mutant were similar to that in the Gmeid1-1 mutant (SI Appendix, Fig. S15B), confirming that the GmEID1 gene is epistatic to the E3 gene in terms of genetic relationship.
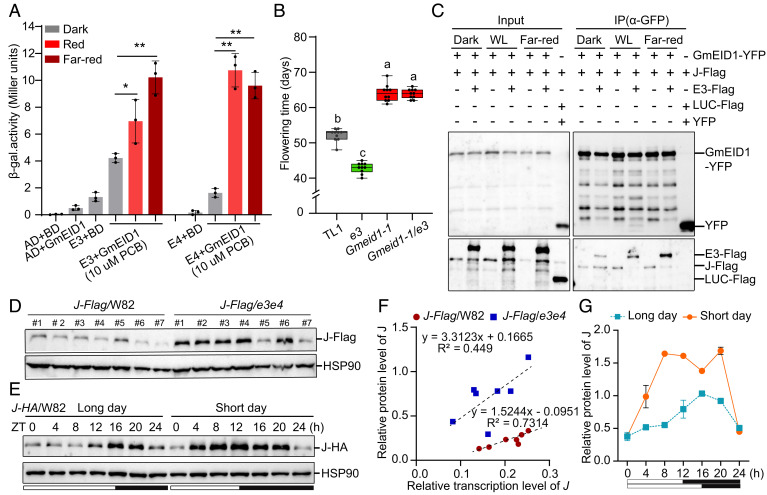
Fig. 4.
Photoactivated E3/E4 interacts with GmEID1 to inhibit the GmEID1–J interaction and promotes the degradation of J protein. (A) β-galactosidase assays to compare the interacting strength between GmEID1 and E3/E4 in yeast cells treated with red light (30 μmol m–2 s–1), far-red light (30 μmol m–2 s–1), or darkness. AD, activation domain; BD, binding domain. PCB, phycocyanobilin, which is the natural precursor of the phytochrome chromophore. Data are means ± SD (n = 3) with significant differences determined by two-tailed Student’s t test (*P < 0.05, **P < 0.01). (B) Flowering time of indicated lines. Data are means ± SD (n = 10). The lowercase letters above the dots indicate significant differences (P < 0.01, ANOVA with Tukey’s post-test). (C) Co-IP assay showing the light-dependent attenuation of the GmEID1–J interaction by E3 in tobacco leaves. The 35S::J-Flag35S::YFP-GmEID135S::E3-Flag, 35S::YFP, and 35S::LUC-Flag constructs were combined into tobacco leaves as indicated and incubated at 25 °C for 12 h in the dark and then transferred to white light (WL, 80 μmol m–2 s–1), far-red light (30 μmol m–2 s–1) or kept in the dark for 36 h, then the leaves were collected for the Co-IP experiment. (D) Immunoblot to compare the J-Flag protein levels in the indicated root hair callus lines cultured under LD conditions. The 35S::J-Flag construct was transformed into the hair root of e3e4 mutant in W82 background (J-Flag/e3e4). Multiple transgenic callus lines were harvested at ZT0 for immunoblot using anti-Flag antibody. HSP90 was used as a loading control. (E) Comparison of transgenic J-HA protein levels under LD and SD conditions by immunoblot using HA antibody. The first trifoliate leaves of J-HA overexpression line in W82 background were collected at the indicated time points. (F) Scatter plot to compare the correlation between transcript levels and protein levels of transgenic J-Flag in the e3e4 mutant and wild-type W82 as in D. (G) Quantitative assay of J-HA protein levels relative to HSP90 in samples as in E.
Next, we investigated whether E3 may affect the interaction between GmEID1 and J by Co-IP (Fig. 4C and SI Appendix, Fig. S16A). Briefly, the GmEID1-YFP and J-Flag proteins were coexpressed with or without the E3-Flag protein in tobacco leaves. The GmEID1–J interaction was compared in the presence or absence of E3-Flag in response to white light (WL) or far-red light treatment. We found that the J-Flag was equally coprecipitated by GmEID1-YFP in the absence of E3-Flag regardless of light or dark treatment. However, in the presence of E3-Flag, a much less amount of J-Flag was coprecipitated by GmEID1-YFP upon WL or far-red light treatment compared to that of dark-adapted control (Fig. 4C and SI Appendix, Fig. S16A). This mechanism was further confirmed by yeast three-hybrid assay and dual-luciferase assays in plant cells (SI Appendix, Fig. S16 B–D). For yeast three-hybrid assay, the interaction between the bait (BD-GmEID1) and the prey (AD-J) was tested in the absence or presence of the third protein bait mate (BM-E3) in response to far-red light treatment. As expected, GmEID1–J interaction was not affected by far-red light in the absence of PCB or E3. However, the extent of GmEID1–J interaction was significantly reduced in the presence of both E3 and PCB when yeast cells were exposed to far-red light (SI Appendix, Fig. S16B), demonstrating that E3 could inhibit the GmEID1–J interaction in a far-red light-dependent manner. The above results demonstrate that photoactivated E3/E4 can disrupt the GmEID1–J interaction and suggest that E3/E4 may promote the degradation of J protein.
To test this possibility, we compared the abundance of J-Flag protein in the presence or absence of E3/E4 by RICE system. The western blot results showed that the levels of the J-Flag protein were prone to be higher in the e3 or e3e4 mutant background than those in the wild-type TL1 or W82 background, respectively (Fig. 4D and SI Appendix, Figs. S17A and S18). Correlation analysis between J-Flag mRNA and J-Flag protein levels confirmed that J-Flag proteins were more efficiently accumulated in the e3e4 or e3 mutant than those in the wild-type W82 or TL1, respectively (Fig. 4F and SI Appendix, Fig. S17B). Given that E3 and E4 mediate photoperiod signals to regulate flowering time, we tested if daylength affects the abundance of J protein using a stable transgenic soybean line overexpressing J-HA protein. The immunoblot results demonstrated that the J-HA protein levels were higher under SD conditions than those under LD conditions (Fig. 4 E and G). Intriguingly, the J-HA protein levels gradually increased during the light period, which is likely associated with the gradual decline of E3/E4 transcripts and proteins during the day (Fig. 4G and SI Appendix, Fig. S17 C–F). Taken together, it is conceivable that photoactivated E3 and E4 act as a competitive inhibitor of the GmEID1–J interaction and consequently promote E1 expression to inhibit flowering in soybean (SI Appendix, Fig. S19).
Deactivation of GmEID1 Improved Adaptability and Yield Performance.
Given that the natural variations of the J gene have been successfully utilized for soybean breeding (11, 12, 38) together with the fact that GmEID1 affects the abundance of J protein, we tested the performance of Gmeid1 mutants and GmEID1-OX lines in field trials in Beijing for two consecutive years in 2020 and 2021. Attractively, besides the later flowering phenotype, the Gmeid1 mutants exhibited multiple preferential agronomic traits, including more branch number, thicker main stem with more nodes, shorter internode length, and greater biomass and yield per plant compared to those of wild-type TL1 (Fig. 5A and SI Appendix, Fig. S20 A–E). Consistently, overexpression of GmEID1 resulted in opposite phenotypic changes including less branch and node number and lower biomass and yield per plant compared to the absence of GmEID1 (SI Appendix, Fig. S20).
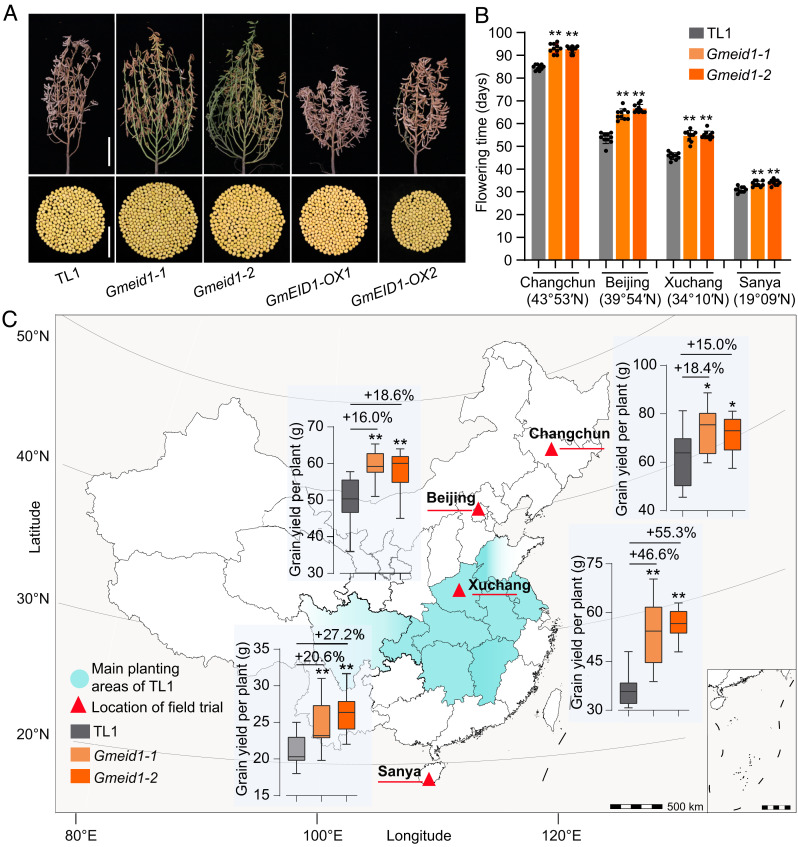
Fig. 5.
Disruption of GmEID1 enhances soybean yield in a broad latitudinal region. (A, Upper) Photographs of wild-type TL1, Gmeid1 mutants, and GmEID1 overexpression lines at the R8 stage grown under Beijing field conditions in 2021. (Scale bar, 20 cm.) (Lower) Whole seeds produced by the respective plants in the Upper panel. (Scale bar, 5 cm.) (B) Comparison of flowering time of wild-type TL1 and Gmeid1 mutants grown in four locations (Changchun, Beijing, Xuchang, and Sanya) in 2021. (C) The geographical distribution of field trials in 2021. The indicated lines were cultivated under a planting density of 67,000 plants/ha in the fields of different latitudes. Wathet blue indicates the main planting areas of the TL1 cultivar. The yield per plant of each line is shown in the bar graph. Data are means ± SD (n = 10) with significant differences determined by two-tailed Student’s t test (*P < 0.05, **P < 0.01).
Next, we tested whether target mutations in GmEID1 could improve soybean performance at different latitudes by field trials in Changchun (44°N), Beijing (39°N), Xuchang (34°N), and Sanya (18°N). As expected, the Gmeid1 mutants displayed consistent beneficial traits, including delayed flowering time and increased number of nodes, branches, and pods per plant at all planting locations (Fig. 5B and SI Appendix, Fig. S21 A–D). Consequently, the yield per plant of the mutant lines increased by at least 15.0% compared to that of wild-type TL1. To be noted, the yield per plant of Gmeid1 increased by 55.3% in Xuchang, which is the origin and main planting area (Huang-Huaihai region of China) for the TL1 cultivar, and by 20.6% in Sanya under typical SD conditions of tropic region (Fig. 5C). The yield per plant of TL1 increased in Changchun and Beijing relative to its main planting area, likely due to increased vegetative growth under LD conditions. The above results demonstrated that targeted mutagenesis of the GmEID1 gene can improve the adaptability of soybean by enhancing yield potential at different latitudinal regions.
Discussion
In summary, we identified a unique flowering-time regulator GmEID1 that could conditionally interact with either J or E3/E4 in soybean in response to light variations. The GmEID1–J interaction enhances the accumulation of the J protein, which directly inhibits the E1 transcript to accelerate flowering. The light-dependent GmEID1–E3/E4 interaction interferes with the GmEID1–J interaction to reduce J protein level and modulate flowering time. Although the regulatory mechanism of how GmEID1-stabilizing J protein remains to be studied in the future, our findings bridge the gap between light signal perception by E3 and E4 to flowering time regulation in soybean. Importantly, the CRISPR/Cas9-engineered Gmeid1 mutants generated in this study provide valuable genetic resources and an approach to breed high-yield soybean cultivars characterized with wide adaptability to different latitudes.
Previous investigations have proposed a possible genetic pathway that controls photoperiodic flowering in soybean: Red/far-red light receptors E3 (GmPHYA3) and E4 (GmPHYA2) act upstream of J, and J directly inhibits E1 expression to regulate flowering time (12). Consistent with this hypothesis, E1 transcript levels were significantly reduced by the mutations in E3 and E4 (5, 6, 26), but up-regulated in the absence of J (12), while the transcript of J was not obviously affected by the dysfunction of E1 (12). However, in contrast to the striking downregulation of E1, the transcript levels of J only slightly increased in the absence of E3 and E4 (12). Our findings that the photoactivated E3 and E4 suppress the accumulation of the J protein by competitive interaction with GmEID1, together with the fact that the J protein tends to accumulate to a higher level under SD conditions than under LD conditions (Fig. 4 E and G), at least partially explain how E3 and E4 dynamically perceive the photoperiod signals to modulate soybean flowering time. EID1 was first discovered as a negatively acting component in the phyA-dependent light pathway, and the expression of the eid1 phenotype requires the presence of functional phyA in Arabidopsis (32). However, the Gmeid1/e3 mutant showed the same phenotype as Gmeid1 (Fig. 4B), indicating a legume-specific function of GmEID1 in soybean.
Gmeid1 mutants exhibit a range of traits, including delayed flowering time, a thick main stem, short internode length, and increased node number and branch number, under natural field conditions (Fig. 5A and SI Appendix, Fig. S20 A–E). These traits are in line with the breeding goal of the “Soybean Green Revolution” (39) and have been shown to increase yield in four different locations compared to that of the wild-type TL1 cultivar (Fig. 5C). The TL1 cultivar is susceptible to lodging under LD conditions, which limits its ability to be grown commercially in northern latitudes. Mutations in GmEID1 result in a stout main stem and short internodes (SI Appendix, Fig. S20 D and E), which can improve the lodging resistant ability and thus expand the adaptive range of TL1 to northern latitudes. On the contrary, short-day conditions lead to early flowering, less vegetative growth, and reduced yield at low latitudes, while dysfunction of GmEID1 delays flowering time, increases the number of internodes and branches, and enhances yield, thus expanding the adaptability of TL1 to southern regions.
To be noted, the field trials in this study were conducted at a lower planting density (67,000 plants/hectare) than normal. This is due to TL1 being prone to lodging at normal densities (200,000 to 250,000 plants/hectare) at high latitudes (Beijing and Changchun), which often leads to problems with growth uniformity. As such, we investigated the agronomic traits of the Gmeid1 mutant and TL1 under lower planting densities to avoid any serious morphological differences among individual plants. All tested lines were planted under the same low planting density in different field locations, in order to assess the effect of different latitude/photoperiods on soybean morphology and yield. Indeed, this low planting density led to a much higher increase in the yield of Gmeid1 mutant compared to that of TL1 (reaching up to 55.3% in yield per plant in Xuchang). Subsequently, we evaluated the yield performance further under its normal planting density (200,000 plants/hectare) in Xuchang. The results showed that the yield per plant of Gmeid1 mutant (16.8 g/plant) was around 16% higher than that of TL1 (14.5 g/plant) (SI Appendix, Fig. S20 E). According to the plot yield level (up to 2,850 kg/hectare) and planting density (200,000 to 250,000 plants/hectare) of TL1 in the main producing area, about 14 g/plant is estimated for the locally grown TL1, which is in agreement with our result for TL1 under similar planting density in this study.
The GmEID1 gene has been neither recovered in flowering time Quantitative Trait Locus (QTLs) (33) (https://www.soybase.org/) and nor identified in previous screens for flowering-time mutants in soybean, which is likely due to the existence of few allelic variations associated with flowering time in natural populations. Therefore, saturation mutagenesis at GmEID1 by target genome editing is worth to explore in future to precisely manipulating flowering time and other agronomic traits in soybean. In conclusion, this study not only outlines a fundamental difference in EID1 function between model plant Arabidopsis and soybean, but also provides an important perspective on photoperiodic flowering, latitudinal adaptation, and high yield breeding of soybean which is the key crop for sustainable consumption of plant protein and oil products in the world.
Materials and Methods
Plant Materials, Growth Conditions, and Phenotyping.
The soybean [Glycine max (L.) Merr.] Tianlong 1 (TL1) or Williams 82 (W82) cultivar was used as wild-type control. The CRISPR-Cas9-engineered mutants and transgenic lines were generated in the TL1 and W82 background. For the analysis of flowering time, the indicated lines were grown under LD conditions (16 h light/8 h dark, 26 °C) or SD conditions (12 h light/12 h dark, 26 °C) in controlled growth chambers. TL1 is a commercial cultivar raised by the Oil Crops Research Institute of Chinese Academy of Agricultural Sciences in 2008 (approval number: State-approved bean 2008023). TL1 is a relatively high-yield variety (up to 2,850 kg/hectare) suitable for spring planting in the Yangtze River Basin in China, which has been used as the control of field trials in the Yangtze River Basin for national soybean variety certification since 2017. For field trials, the indicated lines were grown under natural conditions on the farmlands of Beijing, Changchun, and Xuchang in the summer and Sanya in the winter with a plant spacing of 30 cm and a row spacing of 50 cm in a 3 × 2.5-m plot (50 plants/plot, about 67,000 plants/hectare). To compare with the locally grown TL1 under normal planting density (200,000 to 250,000 plants/hectare) in the main producing area, the seeds of each line were sown with a plant spacing of 10 cm and a row spacing of 50 cm in a 3 × 2.5-m plot (150 plants/plot, about 200,000 plants/hectare). At least ten plants inside the plot were randomly selected for phenotypic analysis. All field experiments were performed in three independent plots. The flowering time was recorded at the R1 stage (days from emergence to the first open flower appeared at any node on the main stem). Other agronomic traits were recorded at harvest.
Yeast Three-Hybrid Experiments.
The pBridge vector expressing both the bait-BD fusion protein and the bait mate protein and the pGADT7 vector expressing the prey-AD fusion protein were constructed for yeast three-hybrid assay. The CDS of GmEID1 was fused with BD to construct the pBridge-GmEID1 vector. The CDS of E3 was inserted into the pBridge-GmEID1 vector to generate the pBridge-GmEID1-E3 vector. The CDS of J was fused with AD to make the pGADT7-J vector. The yeast strain AH109 was transformed with the indicated vector combinations (SI Appendix, Fig. S16B). The individual colony was selected and cultured in a 10-mL centrifuge tube containing 4 mL SD medium (−Leu/-Met/−Trp/+Asp) at 28 °C, 180 rpm in the dark until OD600 = 0.1. An aliquot of 2 mL yeast culture was divided into 8 mL YPDA culture solution with or without 10 mM PCB in a 50-mL centrifuge tube and cultured at 25 °C, 180 rpm under far-red light (30 μmol m−2 s−1) or dark conditions until OD600 = 0.5 to 0.8 prior to the β-galactosidase assay.
Coimmunoprecipitation Assays (Co-IP).
After infiltration with the indicated constructs, the tobacco plants were incubated in the dark at 25 °C for 12 h, and then transferred to light growth conditions for an additional 36 h. Samples were harvested and ground in lysate buffer (1 mM MgCl2, 10 mM EDTA [pH 8.0], 1 mM PMSF, 5 mM DTT, Roche protease inhibitor cocktail). The extracts were centrifuged at 14,000 rpm at 4 °C for 30 min. The supernatant was incubated with 20 μL anti-GFP Trap Agarose (Chromotek, catalog number gta-20) at 4 °C for 2 h and then washed 3 times with lysate buffer. The samples were boiled in SDS-PAGE sample buffer, and the supernatant was analyzed by immunoblot probed with anti-GFP or anti-Flag antibody. To test the effect of E3 on the GmEID1–J interaction, 35S::J-Flag, 35S::GmEID1-YFP, and 35S::E3-Flag constructs were cotransferred into tobacco leaves and incubated at 25 °C for 12 h in the dark and then transferred to white light (80 μmol m−2 s−1), far-red light (30 μmol m−2 s−1) or kept in the dark for an additional 36 h prior to Co-IP analysis.
Details of RNA sequencing and data analysis, gene expression analysis, plasmid construct, plant transformation, subcellular localization in protoplasts, yeast two-hybrid experiments, dual-luciferase assay, in-vitro pull-down assay, RICE system to investigate J protein levels, immunoblot assay, statistical analysis, and primers and accession numbers are provided in SI Appendix, Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Acknowledgments
This work was supported by the National Key Research and Development Plan (2021YFF1001201), the National Natural Science Foundation of China (31422041, 31871705), the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences, and the Central Public-Interest Scientific Institution Basal Research Fund. A Chinese patent has been applied for the GmEID1 gene (application no. ZL202110555960.4).
Author contributions
B.L., F.K., and C.Q. designed research; C.Q. and Haiyang Li performed research; Xiaoya Lin, Z.J., F.Z., X.W., Y.J., Z.L., Z.N., Y.Z., Xiaojiao Li, Hongyu Li, T.Z., J.L., Haiyan Li, and Y.L. contributed new reagents/analytic tools; C.Q. and S.Z. analyzed data; and B.L., F.K., and C.Q. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Although PNAS asks authors to adhere to United Nations naming conventions for maps (https://www.un.org/geospatial/mapsgeo), our policy is to publish maps as provided by the authors.
Contributor Information
Fanjiang Kong, Email: kongfj@gzhu.edu.cn.
Bin Liu, Email: liubin05@caas.cn.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix. Gene sequences were obtained from the Phytozome database (https://phytozome-next.jgi.doe.gov/) by selecting the reference genome Glycine max Wm82.a2.v1 with accession number ACUP01000000 (40). The genes used in this study and their respective identifiers are as follows: GmEID1 (Glyma.03G214300), E3 (Glyma.19G224200), E4 (Glyma.20G090000), J (Glyma.04G050200), GmELF3b-1 (Glyma.14G091900), GmELF3b-2 (Glyma.17G231600), GmELF4a (Glyma.11G229700), GmELF4b (Glyma.18G027500), GmLUX1 (Glyma.12G060200), GmLUX2 (Glyma.11G136600), E1 (Glyma.06G207800), GmFT2a (Glyma.16G150700), GmFT5a (Glyma.16G044100), GmCCA1a (Glyma.07G048500), GmPRR3b (Glyma.12G073900), and GmActin (Glyma.18G290800).
Supporting Information
References
- 1.Watanabe S., Harada K., Abe J., Genetic and molecular bases of photoperiod responses of flowering in soybean. Breed Sci. 61, 531–543 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Destro D., Carpentieri-Pipolo V., Kiihl R. A. D. S., Almeida L. A. D., Photoperiodism and genetic control of the long juvenile period in soybean: A review. Crop Breed. Appl. Biot. 1, 72–92 (2001). [Google Scholar]
- 3.Lin X., Liu B., Weller J. L., Abe J., Kong F., Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J. Integr. Plant Biol. 63, 981–994 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Garner W. W., Allard H. A., Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. Crop Breed. Appl. Biot. 18, 157–158 (1920). [Google Scholar]
- 5.Xia Z., et al. , Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. U.S.A. 109, E2155–2164 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu M., et al. , The soybean-specific maturity gene E1 family of floral repressors controls night-break responses through down-regulation of FLOWERING LOCUS T orthologs. Plant Physiol. 168, 1735–1746 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cober E. R., Voldeng H. D., A new soybean maturity and photoperiod-sensitivity locus linked to E1 and T. Crop Sci. 41, 698–701 (2001). [Google Scholar]
- 8.Watanabe S., et al. , A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188, 395–407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe S., et al. , Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182, 1251–1262 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu B., et al. , Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180, 995–1007 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue Y., et al. , A single nucleotide deletion in J encoding GmELF3 confers long juvenility and is associated with adaption of tropic soybean. Mol. Plant 10, 656–658 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Lu S., et al. , Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49, 773–779 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Fang C., et al. , A recent retrotransposon insertion of J caused E6 locus facilitating soybean adaptation into low latitude. J. Integr. Plant Biol. 63, 995–1003 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Nusinow D. A., et al. , The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao C., et al. , A recessive allele for delayed flowering at the soybean maturity locus E9 is a leaky allele of FT2a, a FLOWERING LOCUS T ortholog. BMC Plant Biol. 16, 20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong F., et al. , A new dominant gene E9 conditions early flowering and maturity in soybean. Crop Sci. 54, 2529–2535 (2014). [Google Scholar]
- 17.Dong L., et al. , Parallel selection of distinct Tof5 alleles drove the adaptation of cultivated and wild soybean to high latitudes. Mol. Plant 15, 308–321 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Bu T., et al. , A critical role of the soybean evening complex in the control of photoperiod sensitivity and adaptation. Proc. Natl. Acad. Sci. U.S.A. 118, e2010241118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C., et al. , A domestication-associated gene GmPRR3b regulates the circadian clock and flowering time in soybean. Mol. Plant 13, 745–759 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Wang L., et al. , Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol. J. 18, 1869–1881 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu S., et al. , Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 52, 428–436 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Dong L., et al. , Genetic basis and adaptation trajectory of soybean from its temperate origin to tropics. Nat. Commun. 12, 5445 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kou K., et al. , A functionally divergent SOC1 homolog improves soybean yield and latitudinal adaptation. Curr. Biol. 32, 1728–1742.e6 (2022). [DOI] [PubMed] [Google Scholar]
- 24.Zhang X., et al. , Functional conservation and diversification of the soybean maturity gene E1 and its homologs in legumes. Sci. Rep. 6, 29548 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhai H., et al. , GmMDE genes bridge the maturity gene E1 and florigens in photoperiodic regulation of flowering in soybean. Plant Physiol. 189, 1021–1036 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J., et al. , Loss of function of the E1-Like-b gene associates with early flowering under long-day conditions in soybean. Front. Plant Sci. 9, 1867 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dieterle M., Zhou Y. C., Schafer E., Funk M., Kretsch T., EID1, an F-box protein involved in phytochrome A-specific light signaling. Genes Dev. 15, 939–944 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y. C., Dieterle M., Buche C., Kretsch T., The negatively acting factors EID1 and SPA1 have distinct functions in phytochrome A-specific light signaling. Plant Physiol. 128, 1098–1108 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marrocco K., et al. , Functional analysis of EID1, an F-box protein involved in phytochrome A-dependent light signal transduction. Plant J. 45, 423–438 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Muller N. A., et al. , Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat. Genet. 48, 89–93 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Muller N. A., Zhang L., Koornneef M., Jimenez-Gomez J. M., Mutations in EID1 and LNK2 caused light-conditional clock deceleration during tomato domestication. Proc. Natl. Acad. Sci. U.S.A. 115, 7135–7140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Büche C., Poppe C., Schäfer E., Kretsch T., eid1: A new Arabidopsis mutant hypersensitive in phytochrome A-dependent high-irradiance responses. Plant Cell 12, 547–558 (2000). [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang S. R., et al. , Photoperiodism dynamics during the domestication and improvement of soybean. Sci. China Life Sci. 60, 1416–1427 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Kereszt A., et al. , Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2, 948–952 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Chen L., et al. , Soybean hairy roots produced in vitro by Agrobacterium rhizogenes-mediated transformation. Crop J. 6, 162–171 (2018). [Google Scholar]
- 36.Lin X., et al. , Novel and multifaceted regulations of photoperiodic flowering by phytochrome A in soybean. Proc. Natl. Acad. Sci. U.S.A. 119, e2208708119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao F., et al. , CRISPR/Cas9-engineered mutation to identify the roles of phytochromes in regulating photomorphogenesis and flowering time in soybean. Crop J. 10, 2214–5141 (2022). [Google Scholar]
- 38.Fang X., et al. , Modulation of evening complex activity enables north-to-south adaptation of soybean. Sci. China Life Sci. 64, 179–195 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Liu S., Zhang M., Feng F., Tian Z., Toward a "Green Revolution" for soybean. Mol. Plant 13, 688–697 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Schmutz J., et al. , Glycine max cultivar Williams 82, whole genome shotgun sequencing project. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/nuccore/ACUP01000000. Deposited 4 August 2014.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Data Availability Statement
All study data are included in the article and/or SI Appendix. Gene sequences were obtained from the Phytozome database (https://phytozome-next.jgi.doe.gov/) by selecting the reference genome Glycine max Wm82.a2.v1 with accession number ACUP01000000 (40). The genes used in this study and their respective identifiers are as follows: GmEID1 (Glyma.03G214300), E3 (Glyma.19G224200), E4 (Glyma.20G090000), J (Glyma.04G050200), GmELF3b-1 (Glyma.14G091900), GmELF3b-2 (Glyma.17G231600), GmELF4a (Glyma.11G229700), GmELF4b (Glyma.18G027500), GmLUX1 (Glyma.12G060200), GmLUX2 (Glyma.11G136600), E1 (Glyma.06G207800), GmFT2a (Glyma.16G150700), GmFT5a (Glyma.16G044100), GmCCA1a (Glyma.07G048500), GmPRR3b (Glyma.12G073900), and GmActin (Glyma.18G290800).