Abstract
Objective
Mitophagy removes damaged mitochondria to maintain cellular homeostasis. Aryl hydrocarbon receptor (AhR) expression in the liver plays a crucial role in supporting normal liver functions, but its impact on mitochondrial function is unclear. Here, we identified a new role of AhR in the regulation of mitophagy to control hepatic energy homeostasis.
Methods
In this study, we utilized primary hepatocytes from AhR knockout (KO) mice and AhR knockdown AML12 hepatocytes. An endogenous AhR ligand, kynurenine (Kyn), was used to activate AhR in AML12 hepatocytes. Mitochondrial function and mitophagy process were comprehensively assessed by MitoSOX and mt-Keima fluorescence imaging, Seahorse XF-based oxygen consumption rate measurement, and Mitoplate S-1 mitochondrial substrate utilization analysis.
Results
Transcriptomic analysis indicated that mitochondria-related gene sets were dysregulated in AhR KO liver. In both primary mouse hepatocytes and AML12 hepatocyte cell lines, AhR inhibition strongly suppressed mitochondrial respiration rate and substrate utilization. AhR inhibition also blunted the fasting response of several essential autophagy genes and the mitophagy process. We further identified BCL2 interacting protein 3 (BNIP3), a mitophagy receptor that senses nutrient stress, as an AhR target gene. AhR is directly recruited to the Bnip3 genomic locus, and Bnip3 transcription was enhanced by AhR endogenous ligand treatment in wild-type liver and abolished entirely in AhR KO liver. Mechanistically, overexpression of Bnip3 in AhR knockdown cells mitigated the production of mitochondrial reactive oxygen species (ROS) and restored functional mitophagy.
Conclusions
AhR regulation of the mitophagy receptor BNIP3 coordinates hepatic mitochondrial function. Loss of AhR induces mitochondrial ROS production and impairs mitochondrial respiration. These findings provide new insight into how endogenous AhR governs hepatic mitochondrial homeostasis.
Keywords: Mitophagy, Autophagy, Reactive oxygen species, BNIP3, Kynurenine
Graphical abstract
Highlights
-
•
AhR has diverse roles in normal liver physiology and pathology.
-
•
AhR inhibition suppresses the mitophagy process in the fasted liver, which leads to mitochondrial dysfunction.
-
•
A mitophagy receptor BNIP3 is a new direct target of AhR to modulate mitochondria homeostasis.
1. Introduction
The mitochondrion is an essential organelle for metabolism and energy homeostasis. Nutrient demands cause dynamic morphology changes and functional responses through mitochondrial fusion, fission, and mitophagy processes [1]. For example, an energy-deprived condition such as calorie restriction and prolonged fasting induces mitochondrial fusion and stimulates mitophagy, which effectively removes damaged mitochondria and restores mitochondrial function [2,3].
Autophagic degradation of mitochondria, also known as mitophagy, regulates mitochondrial quality by selectively removing damaged mitochondria [4]. Mitophagy can be initiated by ubiquitin-autophagy adaptors or direct mitophagy receptors to recruit autophagy machinery to mitochondria [5]. The Pink1-Parkin signaling pathway is the best known ubiquitin-dependent mitophagy mechanism. The protein kinase PINK1 activates the E3 ubiquitin ligase Parkin on the outer membrane of damaged mitochondria, which collectively induces subsequent catabolism of mitochondria by autophagosome formation [6]. The alternate mitophagy receptor-mediated pathway is promoted by the interaction between LC3 and key mitophagy receptors, which directly recruits autophagosomes to mitochondria [7]. Several mitophagy receptors have been reported, including Bcl2/adenovirus E1B 19 kDa protein-interacting protein 3 (BNIP3), BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 like (BNIP3L, also called NIX), and Fun14 Domain containing 1 [8]. Although several stress-activated transcription factors regulate BNIP3 and NIX mitophagy receptor expressions under specific disease contexts [9,10], little is known about the regulatory mechanisms of mitophagy receptor-mediated autophagic processes in the liver, particularly during normal physiology.
The aryl hydrocarbon receptor (AhR) is a ligand-activated basic helix-loop-helix transcription factor and the inactive form is predominantly located in the cytosol [11]. Binding of an AhR ligand dissociates chaperone proteins from AhR to promote translocation into the nucleus and enable target gene regulation. AhR is expressed in various organs, such as the liver, lung, and immune cells for the sensing of environmental signals and activation of multiple biological processes [12]. The most well-characterized AhR function is its role as a xenobiotic sensor to accelerate toxicant and xenobiotic metabolism in various organs [13]. In parallel, additional studies using AhR knockout (AhR KO) mice and AhR endogenous ligands such as kynurenine (Kyn) highlight endogenous AhR functions in the cardiovascular, gastrointestinal, nervous, and immune systems [14]. Initial studies identified that AhR mediates harmful effects of toxic environmental chemicals like dioxin, partly through increased mitochondrial reactive oxygen species (ROS) production and impaired mitochondrial function [15,16]. However, detrimental effects of AhR ablation have been documented in organ development and are closely associated with mitochondrial dysfunction [17,18]. These discrepant results suggest diverse AhR effects in tissue-, age-, and context-dependent manner.
In the current study, we uncovered a novel impact of AhR function on hepatic mitochondrial homeostasis. Our data indicate that AhR inhibition impairs mitochondrial function, which is linked to induced mitochondrial ROS and defective mitophagy. Mechanistically, we identified BNIP3 as a direct target of AhR to regulate AhR-dependent mitophagy. These results present significant new insight into AhR function in hepatic mitochondrial homeostasis.
2. Materials and methods
2.1. Materials
Kynurenine (K8625) and bafilomycin A1 (baf1, 196000) were purchased from Sigma Aldrich (St.Louis, MO). Anti β-actin (4967s), anti-phospho-Drp1 Ser616 (3455), anti-p62 (5144), anti-LC3A (4599), anti-Tom20 (42406S), and secondary antibodies, horseradish peroxidase-conjugated anti-rabbit (G-21234) and anti-mouse (G-21040) IgGs were purchased from Cell Signaling Technology (Beverley, MA). Antibodies against BNIP3 (nbp1-77683s), LC3A (NB100-2331), and LC3B (NB600-1384) were obtained from Novus Biologicals (Littleton, CO). Anti-COX IV (ab14744) was purchased from Abcam (Cambridge, MA). Fluorescence secondary antibodies anti-mouse IgG Alexa Fluor 488 (A11029) and anti-rabbit IgG Alexa Fluor 568 (A11036) were obtained from Thermo Scientific (Waltham, MA).
2.2. Gene Expression Omnibus data
Transcriptome data from AhR KO mice with AhR ligand TCDD (2,3,7,8-tetrachlorodibenzo-p-dioxin) treatment and primary hepatocytes treated with TCDD were downloaded from Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). For Gene ontology analysis of GSE15858, DAVID 6.7 (http://david.avcc.ncifcrf.gov/ref) online bioinformatics was used. GSE15858, GSE10082, and GSE17925 were analyzed using gene set enrichment analysis (GSEA) 4.2.3 software. ‘Gene ontology (GO)’ and ‘canonical pathways’ from Molecular Signature Database (MSigDB, http://software.broadinstitute.org/gsea/msigdb) v6.2 was employed. False discovery rate (FDR) was used for the statistical significance assessment of the normalized enrichment score (NES). Liver AhR ChIP-seq data (GSE97634, GSM2573807) were processed at the Cistrome DataBase analysis tool (http://cistrome.org) and transferred to UCSC Genome Browser for visualization.
2.3. Animal studies
All animal studies and procedures were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine. All mice used were male and had C57BL/6 background. The 8-weeks old AhR KO mice and littermates were fed ad libitum or fasted for 24 h and euthanized for tissue collection. Harvested liver tissues were immediately frozen in liquid nitrogen for further studies.
2.4. Oxygen consumption rate measurement
Primary hepatocytes were plated on XF96 cell culture microplate (101085-004; Agilent) and cultured in William's E medium overnight. The rest of the process was performed according to the manufacturer's protocol. For SeaHorse assay, oligomycin at 2 μM, FCCP at 2 μM, and rotenone/antimycin at 0.5 μM (103015-100; Agilent) were administered to the cells in SeaHorse XF assay medium (102365-100; Agilent) supplemented with glucose and pyruvate.
2.5. Mitochondrial fractionation
Mitochondria were isolated by Mitochondria/Cytosol Fractionation Kit (ab65320, Abcam). Briefly, the cells were suspended in Cytosolic Extraction Buffer Mix and incubated on ice for 10 min, and then homogenized. Homogenates were centrifuged at 700 g for 10 min, and the supernatant was collected. Cellular debris was separated by centrifugation at 10,000 g for 30 min at 4 °C. The pellets containing the crude mitochondria were lysed in the Mitochondrial Extraction Buffer Mix, and mitochondrial protein was prepared for the immunoblotting.
2.6. Mitochondrial ROS measurement
Mitochondrial ROS was measured using MitoSOX Red Mitochondrial Superoxide Indicator (Life Technologies) for 20 min as per the manufacturer's instructions. AML12 cells were seeded in a black 96 well plate with clear bottom, and MitoSOX dye in HBSS buffer was added. Fluorescence was measured using Cytation 5 microplate reader (BioTek) at 510 nm excitation and 580 nm emission wavelengths and normalized to the total protein levels.
2.7. Primary hepatocytes and cell lines
Primary hepatocytes were isolated from C57BL/6 mice. Under deep anesthesia with Isofluorane, livers were perfused with Earle's balanced salt solution containing 5 mM EGTA, followed by continuous collagenase in Hank's balanced salt solution. The liver was removed and massaged to obtain dissociated cells in the hepatocytes wash medium (17704-024; Invitrogen). The cell suspension was filtered through the cell strainer (70 μm) and purified with 40% Percoll gradient. Then the cells were cultured in William's E medium (12551; Invitrogen). Kyn was treated at 4–6 h after the initial plating. AML12 cells were maintained in DMEM/F12 high glucose (Invitrogen, # 11330-057) supplemented with 10% FBS, 1% ITS, 1% penicillin/streptomycin antibiotics, and 40 ng/mL dexamethasone. HEK293 cells were maintained in high glucose DMEM (Genedepot, # CM002-050) supplemented with 10% FBS, 1% penicillin/streptomycin antibiotics. For starvation, an HBSS medium with Ca2+ and Mg2+ supplemented with 10 mM HEPES (Invitrogen, # 15630) was used.
2.8. Real-time PCR quantification
Total RNA was isolated using TRIzol reagent (15596; Thermo Fisher Scientific) and cDNA was synthesized by qScript cDNA Synthesis Kit (95047; Quanta Biosciences). Gene expression level was determined by real-time PCR using LightCycler 480 Real-Time PCR System (Roche) with KAPA SYBR FAST Universal qPCR Master Mix (KK 4618; Kapa Biosystems). Relative mRNA level was calculated with the delta–delta Ct method and normalized by Tbp or 36b4 genes. Primer information can be found in Supplementary Table S1.
2.9. Transient transfection
The plasmid encoding for Bnip3 was purchased from Addgene (http://www.addgene.org). For the siRNA-mediated knockdown experiment, scrambled siRNA control (siCon) and siRNAs targeting AhR (siAhR, sc-29655) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Cells were transfected with the indicated plasmid (1 μg) and each siRNA (50 pmol) using lipofectamine 2000 (Invitrogen) or RNAiMAX (Invitrogen) according to the manufacturer's instructions.
2.10. Mitoplate S-1 mitochondrial function assay
Mitochondrial substrate utilization was measured by Mitoplate S-1 assay following the manufacturer's instructions. Briefly, the assay mix containing 2× Biolog MAS, 6× Redox Dye MC, and saponin was dispensed into the plate and incubated at 37 °C for 1 h. Cells transfected with siAhR or siCon for 48 h were harvested and resuspended in 1× Biolog MAS and dispensed into each well of a MitoPlate (final concentration of saponin 30 μg/mL). Load the MicroPlate into an OmniLog® for kinetic reading of the rate of purple color formation at OD590.
2.11. Immunohistochemistry
Formalin-fixed and paraffin-embedded liver specimens were prepared for histology. Liver sections (4 μm) deparaffinized and rehydrated were then incubated with primary antibodies against LC3A and BNIP3 overnight. Subsequently, the sections were incubated in biotinylated rabbit-specific secondary antibody (Abcam, ab64621) for 1 h at room temperature and streptavidin for 10 min. The reaction was revealed by using 3,3′-diaminobenzidine. The intensity was quantified by Image J software.
2.12. Immunoblot analysis
Cells and liver tissues were solubilized in RIPA buffer (25 mM Tris–HCL [pH 7.6], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, and 1 mM EDTA) supplemented with protease and phosphatase inhibitors. Protein concentration was determined using Pierce™ BCA Protein Assay Kit (Thermo Scientific, Rockford, IL). Proteins were resolved through gel electrophoresis and transferred onto a nitrocellulose membrane (Millipore, Bedford, MA). The membrane was blocked with 5% non-fat dried milk in a TBST buffer (20 mM Tris–HCl, 150 mM NaCl, and 0.1% Tween 20, pH 7.4) for 1 h and incubated overnight with each primary antibody at 4 °C. After washing, the membranes were incubated with secondary antibodies for 1 h at room temperature. Equal loading of samples was verified by immunoblotting for β-actin. The band intensities were quantified using Photoshop CS5 (Adobe Systems, San Jose, CA, USA).
2.13. Mitophagy analysis
AML12 cells treated with siCon and siAhR were stained with MitoTracker (Thermo Fisher Scientific, M7512) and LysoTracker (Thermo Fisher Scientific, L7526) and then observed under a confocal microscope. For mitophagy quantification, pMitophagy Keima-Red mPark2 was used according to the manufacturer's protocols (MBL International Corporation, WOBURN, MA). Briefly, cells were transfected with mixed pMitophagy-mt-Keima-Red mPark2 and siCon, siAhR, or siAhR plus Bnip3 plasmids, respectively. The mt-Keima signals were excited at 488 or 561 nm light, indicating mitochondria in the cytoplasm (green) or the lysosomes (red).
2.14. Immunocytochemistry
AML12 cells were grown on a coverslip and fixed in a 4% paraformaldehyde solution, followed by permeabilization with 0.3% Triton X-100 (Sigma Aldrich). After washing with PBS, cells were immunostained with antibodies against LC3A. Alexa Fluor 488–conjugated anti-mouse IgG (1:500, Invitrogen, Carlsbad, CA, USA) was used as the secondary antibody. Then, samples were mounted with media containing Hoechst 33342 NucBlue® Live ReadyProbes TM Reagent (Invitrogen, Carlsbad, CA, USA) and visualized by Leica TCS SPE confocal microscope.
2.15. Chromatin immunoprecipitation (ChIP) assays
AML12 cells were transfected with pCMX and pCMX-Ahr using Lipofectamine 2000. After 18 h, Kyn (100 μM) was treated for 4 h. The cells were fixed with 1% formaldehyde and ChIP assay was performed according to the protocol we previously described [19] with few modifications. Chromatin was sheared by BioRuptor (Diagnenode) for 10 cycles (30 s on and 30 s off). Each DNA was incubated with IgG (Santacruz, sc-2025) or anti-AHR antibody (Thermo Fisher, MA1513). PCR was done using the primers flanking the putative AhR binding sites located in the enhancer/promoter/intron regions of mouse Cyp1a1 and Bnip3 loci.
2.16. Luciferase reporter assay
The 3 copies of −10 kb Bnip3 enhancer region containing putative ARE were cloned into the pGL3 basic vector (Promega). Mutation of vectors were done by replacing the sequence of AhR binding element from 5′-TGGCGTGGCA-3′ (WT) to 5′-CGACGTGGAC-3′ (Mut1) or 5′-TGATAGGGCA-3′ (Mut2) (bolds indicate mutated bases). HEK293 cells were transfected with pCMX and pCMX-Ahr using Lipofectamine 2000. After 12 h, Kyn was treated for 24 h and luciferase activity was measured according to the manufacturer's instructions (Promega, Madison, WI, USA).
2.17. Data analysis
Statistically significant differences were assessed by the Student's t-test or one-way analysis of variance tests followed by the Tukey or Newman–Keuls methods for multiple comparisons. The data were expressed as the mean ± SEM. The coefficient of correlation was determined by the Pearson correlation. The criterion for statistical significance was set at P < 0.05 or P < 0.01.
3. Results
3.1. Dysregulated hepatic mitochondrial function by AhR inhibition
Previous results showed that genetic ablation of AhR results in smaller liver and hepatocyte sizes, potentially due to increased proliferation and impaired polyploidy [20]. AhR deletion also showed accelerated hepatic injury and ROS production in response to high-fat diet and alcohol treatment [21,22]. These findings suggest endogenous AhR function supports liver development and function. To identify AhR-modulated metabolic pathways, we analyzed public datasets of AhR KO livers [23,24] and found that mitochondrial gene signature was highly attenuated in AhR KO liver in Gene Ontology and Gene Set Enrichment Analysis (Figure 1A and B). In addition, AhR activation by TCDD was strongly associated with mitochondrial function and ROS production in primary hepatocytes and the liver (Supplemental Fig. S1A and B), suggesting a regulatory role of AhR on mitochondrial function.
Figure 1.
Decreased mitochondrial gene signatures in AhR KO. (A) Gene ontology analyses of AhR KO liver treated with TCDD (GSE15858). Down-regulated genes (fold change >1.5, p-value < 0.05) in AhR KO livers were further analyzed for the enrichment of biological process (upper), cellular component (middle), and molecular function (lower) using David bioinformatics software. Top 5 terms in each category were shown. (B) Gene set enrichment analysis of AhR KO liver with TCDD treatment (GSE10082). Enrichment plots are shown enriched signaling pathways in WT (left) and AhR KO (right) livers with TCDD treatment.
To validate the AhR effect on mitochondrial function, we monitored oxygen consumption rates (OCR) in AhR KO primary hepatocytes. Both basal and maximal OCR levels were significantly suppressed in AhR KO primary hepatocytes (Figure 2A). Decreased mitochondrial function in response to AhR inhibition was associated with increased mitochondrial ROS stress, as quantified by MitoSox (Figure 2B). Furthermore, MitoPlate S-1 analysis showed that overall utilization of tricarboxylic acid cycle-specific substrates (e.g., citrate and α-ketoglutarate) was broadly suppressed by AhR inhibition (Figure 2C), suggesting that endogenous AhR function supports basal mitochondrial function in the liver.
Figure 2.
Mitochondrial dysfunction by AhR inhibition. (A) OCRs in mouse primary hepatocytes from WT or AhR KO mice. Real-time readings (upper) and indicated rates of oxygen consumption (lower) are shown (n = 4 each, four to six replicates per group for each experiment). (B) Mitochondrial superoxide level was quantified by MitoSOX™ in siCon or siAhR transfected AML12 cells. (C) Mitoplate S-1 assay to measure substrate-specific mitochondrial function. AML12 cells were transfected with siCon or siAhR for 48h. The change in metabolic rate for each substrate/imtermediate of mitochondrial/glycolytic pathways is shown as a heatmap (upper) and significantly changed substrates for the TCA cycle were shown as a bar graph (lower). Data represented the mean ± SEM, ∗p < 0.05, ∗∗p < 0.01. PPP; pentose phosphate pathway.
3.2. Defective mitophagy by AhR inhibition
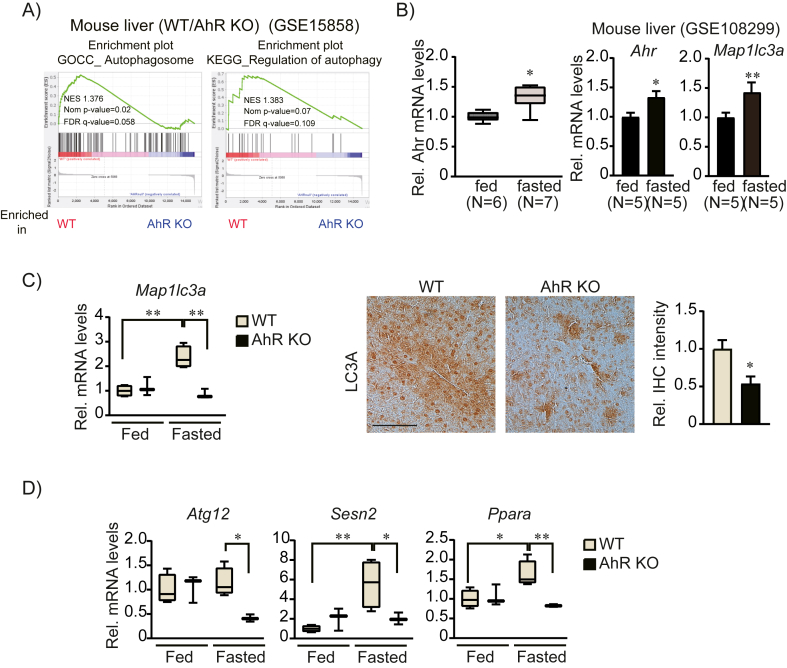
Regulation of mitochondrial dynamics is essential for the maintenance of mitochondrial homeostasis [25]. Prolonged starvation changes mitochondrial morphology and activates autophagy/mitophagy processes [26]. Interestingly, GO analysis of AhR KO indicated that AhR deletion reduced the expression of a group of genes involved in autophagy and autophagosome formation (Figure 3A), suggesting a role for AhR in the regulation of the autophagic process. Fasting increased Ahr expression (Figure 3B), which is consistent with the public dataset from fed/fasted liver (GSE108499). To gain insight into how AhR affects autophagy-related gene expression, we further investigated expression of key autophagy genes in AhR KO liver. Fasting-induced expression of the autophagy marker Map1lc3a mRNA was dysregulated (Figure 3C) and other autophagy-related gene expressions [19] were disrupted in the AhR KO liver (Figure 3D). At the protein level, LC3A expression was also attenuated in response to AhR inhibition, whereas AhR activation by the endogenous ligand kynurenine (Kyn) enhanced LC3A expression (Supplemental Fig. S2). These findings suggest that AhR functions as a crucial regulator of the hepatic autophagy process.
Figure 3.
Attenuated autophagy gene expressions by AhR inhibition. (A) Gene set enrichment analysis of autophagosome and regulation of autophagy gene sets in AhR KO livers. (B) The mRNA level of Ahr and autophagy marker Map1lc3a in the GEO dataset (GSE108299) and our real-time PCR (n = 6 or 7). Data represented the mean ± SEM, ∗p < 0.05, ∗∗p < 0.01. (C) The mRNA levels of Map1lc3a in WT and AhR KO livers during feeding and fasting (left, n = 3 or 4). Immunohistochemical analysis of LC3a (middle) in the fasted liver and quantification of LC3a expression (right) by Image J. Representative liver sections were shown (n = 3 each). Scale bar, 100 μm. (D) Gene expression analysis of autophagy-related genes in WT and AhR KO livers during feeding and fasting. (B)–(D), data were shown as box and whisker plots. Box, interquartile range (IQR); whiskers, min to the max; and horizontal line within box, median, ∗p < 0.05, ∗∗p < 0.01.
Since AhR inhibition largely attenuated mitochondrial function, we questioned whether AhR-regulated autophagy directly targets mitochondria. We discovered that the autophagy marker LC3A was highly enriched in the mitochondrial fraction, which was strongly suppressed by AhR inhibition (Figure 4A). The mitochondria-targeting fluorescent Keima protein (called mt-Keima) emits red fluorescence in the acidic lysosome (pH 4.5–5) and green fluorescence in the cytoplasm, which allows detection of mitophagy inside cells [27]. As shown in Figure 4B, the control cells exhibited both green and red puncta, indicating normal mitophagy, whereas AhR inhibition selectively decreased red mitophagy signals. This observation was further confirmed by the dramatic reduction in the overlay of mitochondria (MitoTracker) and lysosome (LysoTracker) markers in response to AhR inhibition (Figure 4C). These results indicate that AhR suppression results in defective mitophagy, which is closely associated with mitochondrial ROS accumulation as shown in Figure 2B.
Figure 4.
Inhibition of mitophagy by AhR inhibition. (A) Reduced autophagy marker LC3A in the mitochondrial fraction. AML12 cells were transfected with siCon or siAhR for 48 h. Then, the cytosolic and mitochondrial fraction was separately isolated for LC3A immunoblotting. Tom20 was used as a mitochondrial marker. (B) Direct assessment of mitophagy using mt-Keima plasmid. AML12 cells were transfected with siCon or siAhR along with mt-Keima for 48h. Green fluorescence represented normal mitochondria (pH 8.0), whereas red fluorescence displayed mitochondria in an acidic condition (pH 4.5) in the lysosome compartment. Representative images are presented. Scale bar, 50 μm. (C) Co-staining of mitochondria (Mitotracker) and lysosome (Lysotracker) markers in AML12 cells transfected with siCon or siAhR. Representative images are presented. Scale bar, 50 μm. (B) and (C), fluorescence intensities were quantified by ImageJ.
3.3. BNIP3 as a direct transcriptional target of AhR
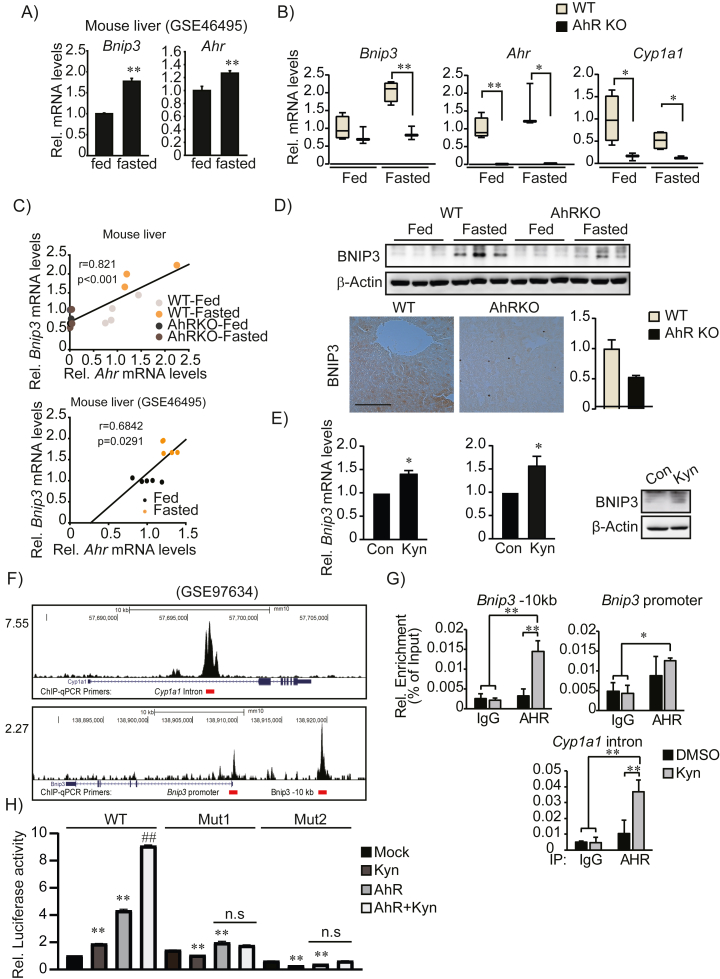
Stress conditions like starvation or hypoxia activate receptor-mediated mitophagy mechanisms [28]. Since our data suggested that AhR might regulate mitophagy, we assessed the expression of the well-described mitophagy receptors BNIP3 and BNIP3L in fed or fasted wild-type and AhR KO livers. Interestingly, Bnip3 expression was strongly upregulated by fasting in wild-type livers (Figure 5A) and this was completely prevented in AhR KO liver (Figure 5B). Moreover, we detected a strong positive correlation between Ahr and Bnip3 transcription among the samples (Figure 5C). Immunoblotting and IHC images confirmed that AhR suppressed BNIP3 expression (Figure 5D). Activation of AhR by the endogenous agonist Kyn significantly increased Bnip3 transcription and protein levels in primary hepatocytes and AML12 cells (Figure 5E). In contrast, Bnip3l level was not correlated with AhR activity and its expression was unchanged during fasting and by Kyn treatment (Supplemental Figs. S3A-C).
Figure 5.
Direct regulation of Bnip3 transcription by AhR. (A) The mRNA level of Bnip3 and Ahr in the GEO dataset (GSE46495) during fed and fasted conditions. (n = 5, each). (B) Expression levels of Bnip3, Ahr, and Cyp1a1 in fed and fasted livers of WT or AhR KO mice (n = 3 or 4). Data were shown as box and whisker plots. Box, interquartile range (IQR); whiskers, min to the max; and horizontal line within box, median, ∗p < 0.05, ∗∗p < 0.01. (C) Pearson's correlation between Ahr and Bnip3 mRNA levels. (D) BNIP3 protein levels in fed and fasted livers of WT and AhR KO mice (upper) and IHC for BNIP3 in fasted livers (lower, left). Representative images were shown (n = 3 each). Scale bar, 100 μm. The quantification of BNIP3 expression (lower, right) was measured by Image J. (E) Increased BNIP3 expression by endogenous AhR ligand, kynurenine (Kyn) treatment (100 μM, 24 h) in mouse primary hepatocyte (left) and AML12 cells (middle and right). (F) AhR recruitment to the Bnip3 genomic locus in TCDD-treated liver (GSE97634). (G) ChIP-PCR analysis of AhR binding to Bnip3 genomic locus. (H) Reporter assays using the pGL3-basic vector containing WT ARE or Mutated ARE. HEK293 cells transfected with mock or AhR overexpression vector together with the reporter vector and treated Kyn for 24h. Results was normalized to WT control. (D), (E), (G) and (H), Data represented the mean ± SEM, ∗p < 0.05, ∗∗p < 0.01. (H), ## was compared to AhR overexpressed group.
We further examined whether AhR directly serves as a direct transcriptional regulator of the Bnip3 gene. Cistrome analysis of AhR ChIP seq data (GSE97634) identified AhR recruitment to the −10 kb enhancer and promoter regions of Bnip3 gene locus (Figure 5F), which have multiple AhR-binding motifs (TGnnTG; data not shown). Our ChIP-PCR also confirmed that AhR recruitment was significantly increased by Kyn treatment (Figure 5G). The functional interaction of AhR with AhR binding motifs at the Bnip3 enhancer region (−10 kb) was further validated in luciferase reporter assays (Figure 5H). These data collectively suggest that AhR is a direct upstream regulator of Bnip3 transcription.
3.4. Restored mitochondrial function by BNIP3 activation
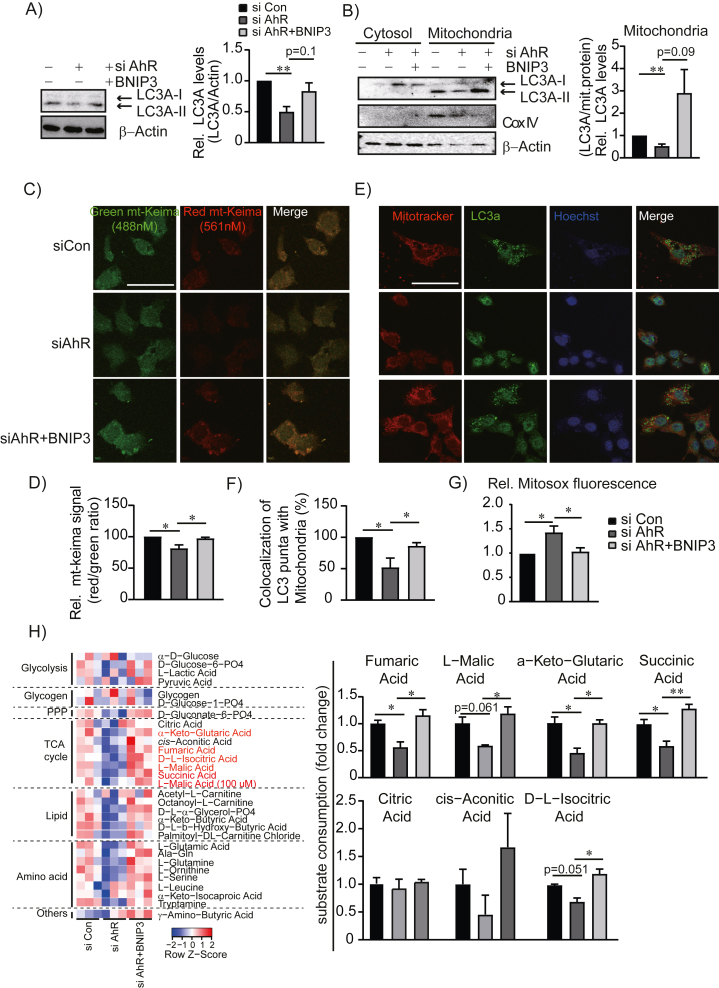
To assess whether AhR regulation of mitophagy is dependent on BNIP3, we transfected AML12 cells with siAhR or siAhR plus BNIP3-overexpressing plasmid (Supplementary Fig. S4). As expected, AhR inhibition suppressed LC3A expression, whereas BNIP3 overexpression restored LC3A levels (Figure 6A). Mitochondria-associated LC3A-II level was more sensitively regulated by AhR inhibition and BNIP3 overexpression (Figure 6B). Similarly, mitophagy-specific quantification using mt-Keima identified that reduced mitophagy by AhR knockdown was recovered upon BNIP3 overexpression (Figure 6C and D), which was confirmed by colocalization of the mitochondria marker (MitoTracker) and autophagy marker (LC3A) (Figure 6E and F). Moreover, increased mitochondrial ROS by AhR inhibition was entirely reversed by BNIP3 overexpression (Figure 6G) and restored the mitochondrial functionality to utilize tricarboxylic acid cycle-specific substrates in MitoPlate S-1 assay (Figure 6H). These results collectively suggest a functional significance of AhR-BNIP3 axis on mitophagy process and mitochondrial homeostasis.
Figure 6.
Restored mitophagy by BNIP3 overexpression. (A) Changes of LC3A protein levels by Bnip3 overexpression in AhR knockdown cells. AML12 cells were co-transfected with siCon, siAhR, or siAhR with Bnip3-overexpressing plasmids. Band intensities represent values relative to siCon. (B) Changes of mitochondria-associated LC3A levels. Cytosolic and mitochondrial fractions were subjected to Western blotting. Cox IV was used for the control of mitochondrial fraction. (C) and (D) Visualization of mitophagy using mt-Keima assay and quantification. (E) Dual staining of mitochondria (Mitotracker) and autophagy marker (LC3A). Representative images are presented. Scale bar, 50 μm. (F) Quantification of colocalization of LC3A with mitotracker. (G) Mitochondrial superoxide levels measured by MitoSOX™ in AML12 cells transfected with siCon, siAhR, or siAhR with Bnip3 overexpression plasmids. (H) Mitoplate S-1 assay to measure mitochondrial substrate utilization. AML12 cells were transfected with siCon, siAhR, or siAhR with Bnip3-overexpressing plasmids for 48 h. The change in metabolic rate for each substrate/intermediate of mitochondrial/glycolytic pathways was shown in a heatmap (left) and change of substrate utilization in TCA cycle were shown as a bar graph (right). (A),(B), (D–H), Data represented the mean ± SEM, ∗p < 0.05, ∗∗p < 0.01. PPP; pentose phosphate pathway.
4. Discussion
AhR is an environmental sensor and a transcription factor with various physiological and homeostatic functions. AhR function is still controversial in liver pathology. Deletion of AhR induced liver fibrosis and liver injury [29], but another study reported that activation of AhR accelerated nonalcoholic steatohepatitis [30]. Generally, toxin-induced AhR activation is detrimental since it has been shown that AhR activation by environmental toxicants such as dioxin induces cellular damage [31,32]. However, beneficial roles of AhR in several tissues, such as the liver [33] and intestine [34], have been documented. In the current study, we focused on the endogenous and physiologic function of AhR in mitochondrial homeostasis. We discovered that AhR inhibition decreases mitochondrial respiration and induces mitochondrial stress, which is in line with previous reports that showed AhR inhibition suppressed NAD(P)H quinone dehydrogenase 1 level and increased oxidative stress in alcohol-challenged mice [22]. It has also been reported that AhR inhibition induces oxidative stress and mitochondrial dysfunction [35,36]. These reports strongly support our observation that AhR is an essential player in the maintenance of mitochondrial function.
Mitochondrial homeostasis is tightly regulated by mitochondrial fission/fusion, mitophagy, and biogenesis pathways under various physiological and pathological conditions [37]. We first hypothesized that morphological dynamics might be affected by AhR, but our analysis of fission and fusion gene expression showed that both fusion (Opa1 and Mfn1) and fission genes (Drp1 and Mff) were decreased (Supplemental Fig. S5). Other genes such as Mfn2 and Fis1 were unchanged. Instead, we found that AhR regulates hepatic mitophagy. Consistent to our demonstration using an endogenous hepatic AhR activation, environmental toxicants such as TCDD directly affect autophagy and mitophagy in other contexts [38,39]. For example, TCDD-activated AhR enhances autophagy to ameliorate ROS-triggered cytotoxicity in neuronal cells [40]. In human keratinocytes, benzo[a]pyrene induces mitophagy in an AhR-dependent manner [41] and AhR knockdown attenuated autophagy genes [42]. However, an opposing result also has been documented [43], possibly due to the distinct biological properties of xenobiotic and endobiotic AhR ligands in tissue- and context-specific manner. Together, these results imply diverse effects of AhR on autophagy in tissue-, ligand- or disease model-dependent manner.
Accumulation of damaged or dysfunctional mitochondria induces oxidative stress, inflammation, and cell death in various liver diseases. Elimination of damaged mitochondria is vital for normal liver function and autophagy/mitophagy is essential for the maintenance of mitochondrial function. We first asked whether AhR directly modulates key autophagy gene expression (Fig. 3). However, we did not observe AhR binding to autophagy gene loci in our cistrome analysis (data not shown). Instead, we identified a direct regulatory mechanism of AhR on mitophagy processes through the key mitophagy receptor BNIP3. BNIP3 is generally increased in stressful conditions and activates mitochondrial surveillance programs [44]. One of the BNIP3-activating signals is nutrient deprivation, as previously reported that glucagon induces BNIP3-mediated mitophagy in hepatocytes [45]. Others also showed that BNIP3 activation is generally protective against high fat-induced liver damage [46] and hypoxia-reoxygenation apoptosis [47]. These observations were compatible with our data indicating that the AhR-BNIP3 axis mediates fasting-induced mitophagy and ultimately reduces mitochondrial stress. However, further studies are needed to define molecular mechanisms underlying the selective regulation of autophagy genes by AhR.
Overall, we conclude that AhR regulation of BNIP3 activates mitophagy in the liver. Inhibition of AhR impairs mitochondrial energetics and increases mitochondrial ROS. These findings expand our knowledge of how AhR signaling contributes to mitochondrial homeostasis in response to nutrient-related cellular and/or organ stress. This study suggests the utility of AhR targeting therapeutics for aberrant mitochondrial function in liver diseases.
Author contributions
MJH, KHK, and DDM conceptualized the study, designed experiments, and wrote the manuscript. MJH, JHS, and SHL conducted experiments. KLP, YAA, BM, and SMH revised the manuscript. BM provided AhR KO mice. KHK, JHS, and YAA reviewed raw data. KHK and DDM supervised the study.
Financial support
This work was supported by the NIH R01DK126656 (to KKH), R00AA026648 (to KLP), K01DK125447 (to YAA), P30DK056338 (to YAA), R01DK114356 (to SMH), P42ES027725, R01ES029382, and R01HL129794 to BM, and P01DK113954 (to DDM) as well as the American Heart Association Grant 19CDA34660196 (to KKH).
Acknowledgment
We thanked to In Hyuk Bang for technical assistance on immunohistochemistry.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2023.101717.
Contributor Information
David D. Moore, Email: daviddmoore@berkeley.edu.
Kang Ho Kim, Email: Kangho.Kim@uth.tmc.edu.
Conflict of interest
Nothing to report.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Published datasets (GEO Dataset) were used for the study.
References
- 1.Ma K., Chen G., Li W., Kepp O., Zhu Y., Chen Q. Mitophagy, mitochondrial homeostasis, and cell fate. Front Cell Dev Biol. 2020;8:467. doi: 10.3389/fcell.2020.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao A.W., Canto C., Houtkooper R.H. Mitochondrial response to nutrient availability and its role in metabolic disease. EMBO Mol Med. 2014;6:580–589. doi: 10.1002/emmm.201303782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehrabani S., Bagherniya M., Askari G., Read M.I., Sahebkar A. The effect of fasting or calorie restriction on mitophagy induction: a literature review. J Cachexia Sarcopenia Muscle. 2020;11:1447–1458. doi: 10.1002/jcsm.12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding W.X., Yin X.M. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem. 2012;393:547–564. doi: 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onishi M., Yamano K., Sato M., Matsuda N., Okamoto K. Molecular mechanisms and physiological functions of mitophagy. EMBO J. 2021;40 doi: 10.15252/embj.2020104705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vives-Bauza C., Zhou C., Huang Y., Cui M., De Vries R.L., Kim J., et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu L., Sakakibara K., Chen Q., Okamoto K. Receptor-mediated mitophagy in yeast and mammalian systems. Cell Res. 2014;24:787–795. doi: 10.1038/cr.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamacher-Brady A., Brady N.R. Mitophagy programs: mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell Mol Life Sci. 2016;73:775–795. doi: 10.1007/s00018-015-2087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin A., Yao J., Zhuang L., Wang D., Han J., Lam E.W., et al. The FoxO-BNIP3 axis exerts a unique regulation of mTORC1 and cell survival under energy stress. Oncogene. 2014;33:3183–3194. doi: 10.1038/onc.2013.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sowter H.M., Ratcliffe P.J., Watson P., Greenberg A.H., Harris A.L. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001;61:6669–6673. [PubMed] [Google Scholar]
- 11.Mimura J., Ema M., Sogawa K., Fujii-Kuriyama Y. Identification of a novel mechanism of regulation of Ah (dioxin) receptor function. Gene Dev. 1999;13:20–25. doi: 10.1101/gad.13.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marszalek-Grabska M., Walczak K., Gawel K., Wicha-Komsta K., Wnorowska S., Wnorowski A., et al. Kynurenine emerges from the shadows - current knowledge on its fate and function. Pharmacol Ther. 2021;225 doi: 10.1016/j.pharmthera.2021.107845. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt J.V., Bradfield C.A. Ah receptor signaling pathways. Annu Rev Cell Dev Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- 14.Kou Z., Dai W. Aryl hydrocarbon receptor: its roles in physiology. Biochem Pharmacol. 2021;185 doi: 10.1016/j.bcp.2021.114428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poland A., Clover E., Kende A.S., DeCamp M., Giandomenico C.M. 3,4,3′,4′-Tetrachloro azoxybenzene and azobenzene: potent inducers of aryl hydrocarbon hydroxylase. Science. 1976;194:627–630. doi: 10.1126/science.136041. [DOI] [PubMed] [Google Scholar]
- 16.Senft A.P., Dalton T.P., Nebert D.W., Genter M.B., Puga A., Hutchinson R.J., et al. Mitochondrial reactive oxygen production is dependent on the aromatic hydrocarbon receptor. Free Radic Biol Med. 2002;33:1268–1278. doi: 10.1016/s0891-5849(02)01014-6. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Salguero P., Pineau T., Hilbert D.M., McPhail T., Lee S.S., Kimura S., et al. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science. 1995;268:722–726. doi: 10.1126/science.7732381. [DOI] [PubMed] [Google Scholar]
- 18.Carreira V.S., Fan Y., Wang Q., Zhang X., Kurita H., Ko C.I., et al. Ah receptor signaling controls the expression of cardiac development and homeostasis genes. Toxicol Sci. 2015;147:425–435. doi: 10.1093/toxsci/kfv138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J.M., Wagner M., Xiao R., Kim K.H., Feng D., Lazar M.A., et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreno-Marin N., Merino J.M., Alvarez-Barrientos A., Patel D.P., Takahashi S., Gonzalez-Sancho J.M., et al. Aryl hydrocarbon receptor promotes liver polyploidization and inhibits PI3K, ERK, and Wnt/beta-catenin signaling. iScience. 2018;4:44–63. doi: 10.1016/j.isci.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wada T., Sunaga H., Miyata K., Shirasaki H., Uchiyama Y., Shimba S. Aryl hydrocarbon receptor plays protective roles against high fat diet (HFD)-induced hepatic steatosis and the subsequent lipotoxicity via direct transcriptional regulation of Socs3 gene expression. J Biol Chem. 2016;291:7004–7016. doi: 10.1074/jbc.M115.693655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong H., Hao L., Zhang W., Zhong W., Guo W., Yue R., et al. Activation of AhR-NQO1 signaling pathway protects against alcohol-induced liver injury by improving redox balance. Cell Mol Gastroenterol Hepatol. 2021;12:793–811. doi: 10.1016/j.jcmgh.2021.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boutros P.C., Bielefeld K.A., Pohjanvirta R., Harper P.A. Dioxin-dependent and dioxin-independent gene batteries: comparison of liver and kidney in AHR-null mice. Toxicol Sci. 2009;112:245–256. doi: 10.1093/toxsci/kfp191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tijet N., Boutros P.C., Moffat I.D., Okey A.B., Tuomisto J., Pohjanvirta R. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol Pharmacol. 2006;69:140–153. doi: 10.1124/mol.105.018705. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Liu H.H., Cao Y.T., Zhang L.L., Huang F., Yi C. The role of mitochondrial dynamics and mitophagy in carcinogenesis, metastasis and therapy. Front Cell Dev Biol. 2020;8:413. doi: 10.3389/fcell.2020.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank M., Duvezin-Caubet S., Koob S., Occhipinti A., Jagasia R., Petcherski A., et al. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim Biophys Acta. 2012;1823:2297–2310. doi: 10.1016/j.bbamcr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Katayama H., Kogure T., Mizushima N., Yoshimori T., Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol. 2011;18:1042–1052. doi: 10.1016/j.chembiol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 28.Palikaras K., Lionaki E., Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20:1013–1022. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt J.V., Su G.H., Reddy J.K., Simon M.C., Bradfield C.A. Characterization of a murine Ahr null allele: involvement of the Ah receptor in hepatic growth and development. Proc Natl Acad Sci U S A. 1996;93:6731–6736. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J.H., Wada T., Febbraio M., He J., Matsubara T., Lee M.J., et al. A novel role for the dioxin receptor in fatty acid metabolism and hepatic steatosis. Gastroenterology. 2010;139:653–663. doi: 10.1053/j.gastro.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forgacs A.L., Burgoon L.D., Lynn S.G., LaPres J.J., Zacharewski T. Effects of TCDD on the expression of nuclear encoded mitochondrial genes. Toxicol Appl Pharmacol. 2010;246:58–65. doi: 10.1016/j.taap.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elbekai R.H., Korashy H.M., Wills K., Gharavi N., El-Kadi A.O. Benzo[a]pyrene, 3-methylcholanthrene and beta-naphthoflavone induce oxidative stress in hepatoma hepa 1c1c7 Cells by an AHR-dependent pathway. Free Radic Res. 2004;38:1191–1200. doi: 10.1080/10715760400017319. [DOI] [PubMed] [Google Scholar]
- 33.Yan J., Tung H.C., Li S., Niu Y., Garbacz W.G., Lu P., et al. Aryl hydrocarbon receptor signaling prevents activation of hepatic stellate cells and liver fibrogenesis in mice. Gastroenterology. 2019;157:793–806 e714. doi: 10.1053/j.gastro.2019.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metidji A., Omenetti S., Crotta S., Li Y., Nye E., Ross E., et al. The environmental sensor AHR protects from inflammatory damage by maintaining intestinal stem cell homeostasis and barrier integrity. Immunity. 2018;49:353–362 e355. doi: 10.1016/j.immuni.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rico de Souza A., Zago M., Pollock S.J., Sime P.J., Phipps R.P., Baglole C.J. Genetic ablation of the aryl hydrocarbon receptor causes cigarette smoke-induced mitochondrial dysfunction and apoptosis. J Biol Chem. 2011;286:43214–43228. doi: 10.1074/jbc.M111.258764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X., Li S., Liu L., Jian Z., Cui T., Yang Y., et al. Role of the aryl hydrocarbon receptor signaling pathway in promoting mitochondrial biogenesis against oxidative damage in human melanocytes. J Dermatol Sci. 2019;96:33–41. doi: 10.1016/j.jdermsci.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 37.Wu N.N., Zhang Y., Ren J. Mitophagy, mitochondrial dynamics, and homeostasis in cardiovascular aging. Oxid Med Cell Longev. 2019;2019 doi: 10.1155/2019/9825061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van de Veerdonk F.L., Renga G., Pariano M., Bellet M.M., Servillo G., Fallarino F., et al. Anakinra restores cellular proteostasis by coupling mitochondrial redox balance to autophagy. J Clin Invest. 2022:132. doi: 10.1172/JCI144983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jang H.S., Lee J.E., Myung C.H., Park J.I., Jo C.S., Hwang J.S. Particulate matter-induced aryl hydrocarbon receptor regulates autophagy in keratinocytes. Biomol Ther. 2019:570–576. doi: 10.4062/biomolther.2019.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J., Tang C., Nie X., Xi H., Jiang S., Jiang J., et al. Autophagy potentially protects against 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin induced apoptosis in SH-SY5Y cells. Environ Toxicol. 2016;31:1068–1079. doi: 10.1002/tox.22116. [DOI] [PubMed] [Google Scholar]
- 41.Das D.N., Naik P.P., Mukhopadhyay S., Panda P.K., Sinha N., Meher B.R., et al. Elimination of dysfunctional mitochondria through mitophagy suppresses benzo [a] pyrene-induced apoptosis. Free Radic Biol Med. 2017;112:452–463. doi: 10.1016/j.freeradbiomed.2017.08.020. [DOI] [PubMed] [Google Scholar]
- 42.Kim H.R., Kang S.Y., Kim H.O., Park C.W., Chung B.Y. Role of aryl hydrocarbon receptor activation and autophagy in psoriasis-related inflammation. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21062195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim H.R., Kim J.C., Kang S.Y., Kim H.O., Park C.W., Chung B.Y. Rapamycin alleviates 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin-induced aggravated dermatitis in mice with imiquimod-induced psoriasis-like dermatitis by inducing autophagy. Int J Mol Sci. 2021;22:3968. doi: 10.3390/ijms22083968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao A., Jiang J., Xie F., Chen L. Bnip3 in mitophagy: novel insights and potential therapeutic target for diseases of secondary mitochondrial dysfunction. Clin Chim Acta. 2020;506:72–83. doi: 10.1016/j.cca.2020.02.024. [DOI] [PubMed] [Google Scholar]
- 45.Springer M.Z., Poole L.P., Drake L.E., Bock-Hughes A., Boland M.L., Smith A.G., et al. BNIP3-dependent mitophagy promotes cytosolic localization of LC3B and metabolic homeostasis in the liver. Autophagy. 2021;17:3530–3546. doi: 10.1080/15548627.2021.1877469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li R., Xin T., Li D., Wang C., Zhu H., Zhou H. Therapeutic effect of Sirtuin 3 on ameliorating nonalcoholic fatty liver disease: the role of the ERK-CREB pathway and Bnip3-mediated mitophagy. Redox Biol. 2018;18:229–243. doi: 10.1016/j.redox.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu Z.J., Wang Z.Y., Xu L., Chen X.H., Li X.X., Liao W.T., et al. HIF-1alpha-BNIP3-mediated mitophagy in tubular cells protects against renal ischemia/reperfusion injury. Redox Biol. 2020;36 doi: 10.1016/j.redox.2020.101671. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Published datasets (GEO Dataset) were used for the study.