Abstract
Transcription is a noisy and stochastic process that produces sibling‐to‐sibling variations in physiology across a population of genetically identical cells. This pattern of diversity reflects, in part, the burst‐like nature of transcription. Transcription bursting has many causes and a failure to remove the supercoils that accumulate in DNA during transcription elongation is an important contributor. Positive supercoiling of the DNA ahead of the transcription elongation complex can result in RNA polymerase stalling if this DNA topological roadblock is not removed. The relaxation of these positive supercoils is performed by the ATP‐dependent type II topoisomerases DNA gyrase and topoisomerase IV. Interference with the action of these topoisomerases involving, inter alia, topoisomerase poisons, fluctuations in the [ATP]/[ADP] ratio, and/or the intervention of nucleoid‐associated proteins with GapR‐like or YejK‐like activities, may have consequences for the smooth operation of the transcriptional machinery. Antibiotic‐tolerant (but not resistant) persister cells are among the phenotypic outliers that may emerge. However, interference with type II topoisomerase activity can have much broader consequences, making it an important epigenetic driver of physiological diversity in the bacterial population.
Keywords: DNA gyrase, DNA supercoiling, persisters, topoisomerases, transcription bursting
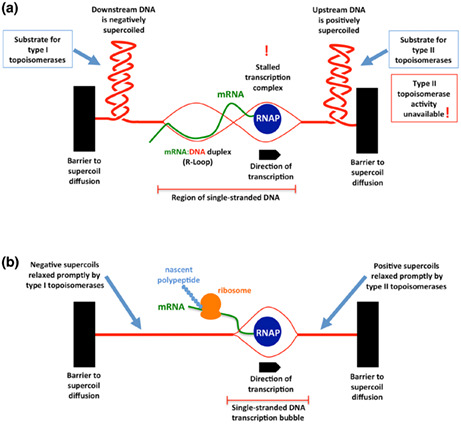
Positive supercoils in DNA impede transcription: (a) failure to relax positive supercoils halts transcription elongation; (b) removal of the supercoils facilitates transcription.
1. INTRODUCTION
In 1902, Almroth Wright, an Irish‐Swedish bacteriologist who had trained in medicine at Trinity College Dublin (TCD), founded the Bacteriology Department at St. Mary's Hospital in London. In 1912, Wright's department provided training in bacteriology for Adrian Stokes, another medical student from TCD, who would become that university's first Professor of Bacteriology and Preventive Medicine in 1919 (Coakley, 1992). Stokes' successor in the TCD chair was Joseph Warwick Bigger, the person who described the phenomenon of “persisters” in bacterial populations (Bigger, 1944). It is believed that Alexander Fleming, working in Wright's department at St. Mary's, was attempting to repeat Bigger's experiments on small colony variants of Staphylococcus aureus (Bigger et al., 1927), when he made the breakthrough in 1928 that led to the discovery of penicillin (Greenwood, 2008). These events illustrate the interconnectedness of institutions, individuals, and ideas in the development of scientific thought.
Persisters are bacterial cells that have become non‐susceptible to antimicrobial treatment without becoming resistant (Balaban et al., 2019; Bigger, 1944). They are phenotypic outliers with an environment‐specific enhanced survival trait, and they arise stochastically in a population of otherwise antimicrobial‐susceptible siblings (Hobby et al., 1942; Lewis, 2010; Wainwright et al., 2021). Many cellular pathways can lead to the production of persister cells, and one of these involves stochastic interference with the machinery that manages DNA topology. A key component of this machinery is DNA gyrase, a target for many antimicrobial agents, and one that is inhibited in some persister‐generating pathways (Harms et al., 2015; Kato et al., 2022). However, the contributions of bacterial topoisomerases to the production of cell‐to‐cell physiological diversity, in populations of genetically identical siblings, extend beyond the phenomenon of persistence.
2. DNA TOPOISOMERASES AND TRANSCRIPTION
Topoisomerases are enzymes that manage the topology of DNA through strand breakage and religation, with the enzyme forming a transient covalent bond with the DNA phosphate backbone at the site of cleavage (Bates & Maxwell, 2005). The model bacterium Escherichia coli has four DNA topoisomerases: topo I, topo II (gyrase), topo III, and topo IV (McKie et al., 2021). Topo I (encoded by the topA gene) and topo III (encoded by topB) are type IA topoisomerases that alter the linking number of DNA in steps of 1. They break one DNA strand, allowing the DNA to swivel around the intact strand before religating the nick. This swivelase activity alters the number of times the DNA strands cross one another in the double helix. The energy to drive the process comes from the pent‐up torsional stress in the under‐wound, negatively supercoiled DNA molecule. Topo I relaxes negatively supercoiled DNA, restarts stalled transcription complexes, and suppresses aberrant DnaA‐independent initiation of chromosome replication away from oriC, the natural origin of bacterial chromosome replication (Drolet et al., 1994; Kuzminov, 2018; Lang & Merrikh, 2018; Leela et al., 2021; Leng et al., 2004; Usongo et al., 2008). Topo III is a decatenase and is concerned with the smooth progression of chromosome segregation at cell division (DiGate & Marians, 1989; Nurse et al., 2003; Perez‐Cheeks et al., 2012). By removing negative supercoils, type I enzymes also play an essential role in preventing the formation of RNA–DNA hybrids, known as R‐loops, in the hyper‐negatively supercoiled DNA that accumulates behind stalled transcription elongation complexes (Brochu et al., 2018; Sutormin et al., 2022) (Figure 1).
FIGURE 1.
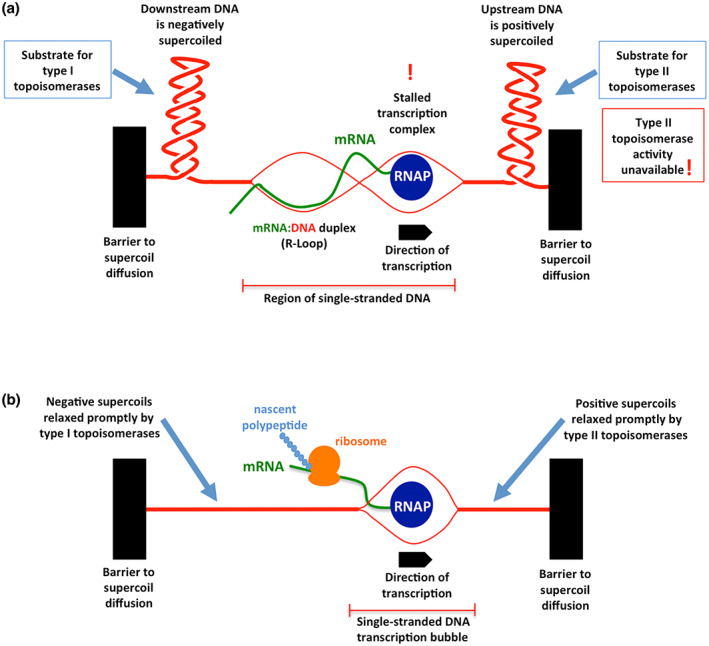
Topoisomerases ensure the smooth operation of RNA polymerase during transcription. Identical copies of a gene are being transcribed in two genetically identical bacterial siblings. (a) Transcription of the gene has stalled because further progress by RNA polymerase (RNAP) is blocked as a result of a failure by type II topoisomerases to relax the positive supercoils that have accumulated in its path. The transcriptionally induced supercoils cannot diffuse due to the presence of topological barriers (black solid rectangles) and viscous drag prevents RNAP from rotating around the DNA to relieve the torsional strain. Type II topoisomerases may be physically absent or present‐but‐inactive, preventing them from adjusting the topology of the DNA. If type I enzymes are unavailable, hypernegative supercoiling can allow the mRNA to hybridize with its complementary DNA template, forming an R‐loop that prevents its translation into a polypeptide chain by ribosomes. (b) an identical copy of the same gene in another cell is undergoing smooth transcription due to the timely relaxation of the transcriptionally induced supercoiling domains by topoisomerases. Messenger RNA transcribed by RNAP is available for translation by ribosomes.
Type II topoisomerases change the linking number of the DNA duplex in steps of 2, using a double‐strand‐breakage, intact‐duplex‐segment‐passage, and religation mechanism (Deweese & Osheroff, 2009; Dong & Berger, 2007). Type II enzymes bind and hydrolyse ATP in order to complete each reaction cycle (Gellert et al., 1976; Higgins et al., 1978; Mueller‐Planitz & Herschlag, 2008; Wang, 1998). In bacteria, DNA gyrase, a type II topoisomerase, introduces negative supercoils into DNA and eliminates positive supercoils, processes that are vital for the smooth operation of both the chromosomal replication and the transcriptional machineries (Rovinskiy et al., 2012; Stracy et al., 2019; Zechiedrich et al., 2000). A second type II enzyme, topoisomerase IV, removes DNA catenanes that are generated during chromosome replication and which, if not removed, are inhibitory to chromosome segregation at cell division (Hirsch & Klostermeier, 2021; Lopez et al., 2012; Nolivos et al., 2016; Zechiedrich & Cozzarelli, 1995). Topo IV also relaxes both positive and negative supercoils (Crisona et al., 2000; Kato et al., 1992; Khodursky et al., 2000).
Early work in Salmonella linked transcription and topoisomerase activity. Here, supX mutations in the topA gene (encoding DNA topoisomerase I) were found to suppress the Leu− phenotype caused by a point mutation in the promoter of the l‐leucine biosynthetic operon, leuABCD (Margolin et al., 1985; Pruss & Drlica, 1985; Trucksis & Depew, 1981; Trucksis et al., 1981). The mutation made the promoter's Pribnow box more G+C‐rich and thus more difficult to melt. The elimination of the DNA relaxing activity provided by topo I allowed negative supercoils generated in a neighboring promoter relay to accumulate, melting the recalcitrant leuABCD promoter and restoring leucine prototrophy (Chen et al., 1992; Dorman & Dorman, 2016; El Hanafi & Bossi, 2000; Richardson et al., 1988). The intellectual challenge of reconciling the genotypic and phenotypic properties of the leu + topA − mutant led to the breakthrough in understanding provided by the twin‐DNA‐supercoiling‐domain model (Liu & Wang, 1987) (Figure 1). Application of the model is not restricted to transcription; it also applies to DNA replication, another process that generates local domains of differentially supercoiled DNA. The model has been validated experimentally in prokaryotes and eukaryotes (Ma & Wang, 2014; Rahmouni & Wells, 1992; Tsao et al., 1989; Wu et al., 1988) and is proving to be very useful in advancing understanding of an important source of gene‐copy‐to‐gene‐copy transcriptional variation.
3. TRANSCRIPTION IS NOT ALWAYS A SMOOTH PROCESS
Identical gene copies are not transcribed in a synchronized manner across a microbial population, even if the required activation signal is evenly distributed throughout the environment that the bacteria share. Transcriptional “burstiness” means that, at any given time, some gene copies remain silent while others are firing, and those genes that are firing do so in bursts. This phenomenon is not restricted to bacteria: it occurs in all cell types, including those of humans (Larsson et al., 2021; Naik et al., 2021). Bursting reflects the stochastic operation of numerous processes that facilitate or inhibit RNA polymerase activity in both simple and complex organisms (Bahar Halpern et al., 2015; Chubb et al., 2006; Corrigan et al., 2016; Dar et al., 2012; Golding et al., 2005; Levsky et al., 2002; Raj et al., 2006; Raj & van Oudenaarden, 2008, 2009; Suter et al., 2011). For example, the availability and activities of transcription factors influence the recruitment of RNA polymerases to promoters, affecting the initiation of transcription (Mazzocca et al., 2021; Mejia‐Almonte et al., 2020). The interactions between RNA polymerases queuing along the DNA template also contribute to “bursty” transcription (Fujita et al., 2016; Tripathi et al., 2022). In eukaryotes, the number of cis‐acting regulatory elements that a gene possesses, and their affinity for the factors that they bind, influences burst size, while trans‐acting regulatory factor availability is a determinant of burst frequency; both aspects of bursting are modulated by histone modification (Chen et al., 2019; Nicolas et al., 2017; Wang et al., 2018). In prokaryotes, the nature of the sigma factor subunit of RNA polymerase influences the pattern of the transcription burst that a gene may exhibit, with the more eukaryotic‐like promoters that depend on sigma‐54 displaying bursting with a eukaryotic character that emphasizes burst frequency over burst size (Engl et al., 2020).
4. TRANSCRIPTION BURSTS
In bacteria, interference with RNA polymerase translocation by supercoil accumulation contributes to what has been termed transcription “bursts” (Figure 1). Bursting refers to the stochastic production of RNA by transcription. Single‐molecule methods have allowed the transcription of individual genes to be studied in vitro and in real time. In one experimental set up, linear DNA templates containing a bacterial promoter are tethered to a surface in a flow cell, RNA polymerase and nucleoside triphosphates are introduced, and RNA production is measured fluorescently (Chong et al., 2014). Hundreds of identical DNA templates are viewed simultaneously. When the DNA molecules are free to rotate at their ends, transcription proceeds smoothly along each individual template. When the DNA molecules are fixed at multiple points to the substrate, free rotation is blocked and transcript elongation is inhibited stochastically across the field of transcription complexes. The introduction of DNA gyrase overcomes the interference with transcription elongation. The data indicate that over‐twisting of the DNA in front of an elongation complex inhibits further transcription of the affected gene until either DNA gyrase removes the obstruction or the supercoils diffuse away from the site by DNA rotation (Ancona et al., 2019; Chong et al., 2014; Geng et al., 2022; Klindziuk & Kolomeisky, 2021). Supercoil diffusion will not be possible if the DNA is not free to rotate, so the intervention of gyrase is essential. This proposal represents a practical application of the twin‐DNA‐supercoiling‐domain hypothesis (Liu & Wang, 1987). In the case of genes that encode proteins, the inhibition of transcription complex rotation around the DNA template by viscous drag in the cytoplasm is likely to be exacerbated by the coupling of transcription and translation in bacteria (Chen et al., 1992) (Figure 1b). If DNA gyrase is absent from the affected gene, or is present‐but‐inactive due to an unfavorable [ATP]/[ADP] ratio, or has been inhibited by a gyrase poison, then the expression of that particular gene copy will remain interrupted. Single‐molecule studies in flow cells, with tethered circular DNA molecules that contain a bacterial promoter, have shown that the accumulated positive supercoils feed back to inhibit transcription initiation, silencing the affected transcription unit by inhibiting the formation of an open transcription complex (Chong et al., 2014; Revyakin et al., 2004). Extrapolating to the in vivo context, if one copy of a gene is being inhibited by positive supercoil accumulation in one bacterial cell, and an identical copy of the same gene in a sibling bacterium is undergoing efficient transcription, the result will be a physiological difference between the two cells. Amplified across the population, stochastic interference with transcription has the potential to generate considerable diversity at the levels of the transcriptome and proteome, and at the level of whole‐cell physiology.
5. NUCLEOID‐ASSOCIATED PROTEINS AND RELAXATION OF POSITIVE SUPERCOILS
Given the importance of positive supercoiling in current models of transcription bursting in bacteria, it will be interesting to discover if the GapR family of nucleoid‐associated proteins, that targets positively supercoiled DNA, contributes to transcription stochasticity. This tetrameric DNA binding protein from Caulobacter crescentus targets over‐wound DNA and stimulates type II to remove the positive supercoils, facilitating DNA replication (Ahmed & Dröge, 2020; Guo et al., 2018, 2021; Huang et al., 2021; Lourenço et al., 2020). GapR also targets positive supercoils that are generated by RNA polymerase activity (Guo et al., 2021). The finding that GapR contains structural motifs that are distantly related to those of the H‐NS nucleoid‐associated protein makes the possibility that GapR affects transcription output stochasticity all the more compelling: H‐NS has a well‐established role in transcription silencing in A+T‐rich DNA sequences (Lourenço et al., 2020; Turner & Dorman, 2007) and GapR also has a preference for binding to A+T‐rich DNA (Huang et al., 2021). The YejK nucleoid‐associated protein from E. coli interacts with type II topoisomerases, stimulating the relaxation of positively supercoiled DNA by topoisomerase IV (Lee & Marians, 2013). These findings indicate that the relaxation of positive supercoils by type II topoisomerases is modulated by nucleoid‐associated proteins such as GapR and YejK, introducing yet another variable into the generation of randomness in the transcription process.
6. SPECIES DIFFERENCES IN STOCHASTIC TRANSCRIPTION
Comprehensive surveys that make direct comparisons of DNA supercoiling set points across bacterial species are lacking, but some data indicate that species‐to‐species variation does occur. For example, S. enterica has average DNA superhelical densities that are more relaxed than those of E. coli, despite the very high level of conservation of genome structure and gene content seen in these bacteria (Cameron et al., 2011; Champion & Higgins, 2007; Rovinskiy et al., 2019). These differences in DNA supercoiling may reflect a need on the part of S. enterica to manage carefully the expression of the relatively large complement of energetically “expensive” genes that it has acquired by horizontal gene transfer (Pozdeev et al., 2021, 2022; Srinivasan et al., 2013) and which are silenced by H‐NS (Dorman, 2007; Lucchini et al., 2006; Navarre et al., 2006). Inter‐species differences in DNA supercoiling may result in identical copies of a gene exhibiting distinct patterns of stochastic transcription in different host backgrounds, even if the gene is placed in identical genomic locations.
Even within a single bacterium, variations in supercoiling values around the chromosome (Guo et al., 2021; Lal et al., 2016) may result in location‐dependent variations in transcriptional stochasticity. Differences in the distribution of topoisomerases around the chromosome (Ahmed et al., 2017; Hsu et al., 2006; Stracy et al., 2019; Sutormin et al., 2019) could affect the efficiency of positive supercoil extinction at different chromosomal sites, and hence the probability that a given gene will be transcribed. Increasing, or decreasing, the unpredictability of expression of a given gene may, in turn, have unpredictable consequences for the competitive fitness of the individual bacterium and for the wider population of its siblings. The base composition of a gene, especially one that has been acquired by horizontal gene transfer, seems to influence its sensitivity to DNA supercoiling changes (Dorman & Dorman, 2022), and this is likely to influence the stochasticity of its transcription.
7. GYRASE POISONS AND PERSISTERS
Toxin/antitoxin (TA) systems play prominent roles in the production of persister cells. Typically, these systems consist of a chemically stable toxin and a labile antitoxin. As long as de novo antitoxin production is sustained, the inhibitory effect of the toxin is neutralized. Stochastic removal of the antitoxin allows the toxin to exert its negative influence on some aspect of bacterial cell physiology. The resulting growth inhibition renders the bacterium tolerant to the presence of antimicrobial agents that affect only active cells.
The FicTA protein partnership is a useful example of a TA system that targets topoisomerase function (Figure 2). The FicA protein is an antitoxin, while the FicT toxin is an inhibitor of bacterial ATP‐dependent type II topoisomerase activities. Removal of FicA from a cell allows FicT to inhibit DNA gyrase and topoisomerase IV by adenylylation at their ATP binding sites, preventing the topoisomerases from binding ATP (Harms et al., 2015). The resulting (reversible) interruption of type II topoisomerase activity causes a growth arrest that allows the affected cell to exhibit a persister phenotype (Figure 2). In the context of type II topoisomerase modulation and the promotion of physiological diversity in bacterial populations, it is perhaps significant that fluctuation in the cellular concentration of ATP is itself a factor in the emergence of persister cells (Conlon et al., 2016).
FIGURE 2.
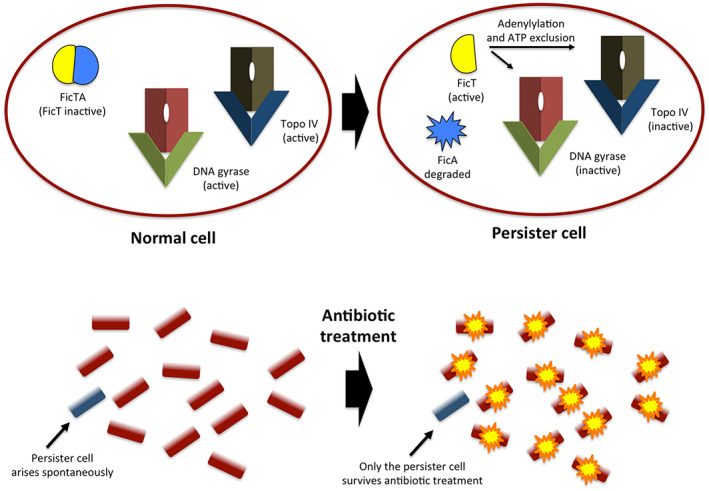
The role of the FicTA toxin/antitoxin system in the stochastic emergence of a persister cell in a bacterial population. The normal cell (top, left) has produced the FicT toxin and the FicA antitoxin. These have formed a complex in which the toxin is maintained in an inactive state. The targets of FicT, the type II topoisomerases DNA gyrase and topoisomerase IV, are active and participate in cellular metabolism. Following the loss of the ficA and ficT genes, the unstable FicA antitoxin breaks down and FicT can now inhibit the type II topoisomerases by an adenylylation mechanism that prevents them from binding and hydrolyzing ATP, an essential part of their enzymatic function (top right). The resulting loss of type II topoisomerase activity brings about a cessation of normal metabolic function in the affected cell. This cell (shown at bottom left) is a physiological outlier in the population. Its metabolic quiescence protects it from an antibiotic challenge that inhibits, or kills, its siblings (bottom right). Consequently, it persists and may emerge to colonize the local environment once the antibiotic challenge passes. The persister cell displays tolerance, but not resistance, to the antibiotic. Its ability to survive is contingent on the stochastic appearance of unrestrained FicT toxic activity following the random loss of the gene that supplies the FicA antitoxin.
The FicTA system, which deprives the type II topoisomerases of their ATP supply, represents a physiologically gentle mechanism for causing cell growth arrest. Other TA systems target DNA gyrase via a poisoning mechanism that stabilizes the gyrase‐DNA cleavage complex, leading to cell death. The F‐plasmid‐encoded CcdA/CcdB TA partnership is one of these more aggressive systems. It uses post‐segregational killing to eliminate any daughter cell that fails to inherit a copy of the F plasmid and its ccdA antitoxin gene (Bahassi et al., 1999). However, modulation of CcdB toxin production, by changes to the culture's growth conditions, can rescue some of the bacteria from the cell death pathway, leading to the emergence of persisters (Tripathi et al., 2012).
8. FLUCTUATING [ATP]/[ADP] RATIOS AND GYRASE ACTIVITY AS GENERATORS OF DIVERSITY
The intersection between ATP synthesis, DNA gyrase activity, and the stochastic nature of transcription has implications for bacterial physiological diversity that extend beyond the special case of persister cell generation. Variations in metabolic flux throughout the growth cycle and the vagaries of environmental stress impose changes in [ATP]/[ADP] ratios that influence DNA supercoiling (Hsieh, Burger, et al., 1991; Hsieh, Rouvière‐Yaniv, et al., 1991; van Workum et al., 1996). In crude terms, cells in the lag and stationary phases of the growth cycle have low levels of metabolic flux, and hence low [ATP]/[ADP] ratios, whereas rapidly growing bacteria undergoing exponential expansion of the population have a high energy charge. These fluctuations in the [ATP]/[ADP] ratio affect the ATP‐dependent activities of type II topoisomerases (van Workum et al., 1996).
Each phase of the bacterial growth cycle has a characteristic DNA topological profile (Bordes et al., 2003; Conter et al., 1997; Gomez‐Gomez et al., 1996; Kusano et al., 1996; Muskhelishvili et al., 2022; Sobetzko et al., 2012). Cells emerging from lag phase and entering exponential growth experience a growth spurt that is accompanied by increased negative supercoiling of DNA (Gomez‐Gomez et al., 1996). A delay in making the transition from lag to exponential growth, perhaps due to an environmental stress, is associated with the stochastic appearance of small colony variants and persister cells within the population (Vulin et al., 2018). The transition from exponential growth to stationary phase is another significant life event for the bacterium and it is marked by a loss of negative supercoils (Bordes et al., 2003; Conter et al., 1997; Dorman et al., 1988; Kusano et al., 1996).
These DNA topological changes are both the causes of, and the consequences of, shifts in the expression of the transcriptome (Dorman, 2019). They are a manifestation of the combined actions of the topoisomerases that manage the DNA topological shifts and of the altered activities of RNA polymerase at hundreds of transcription units (Cameron et al., 2017; Geng et al., 2022; Kim et al., 2019; Stracy et al., 2019). In addition to the task of expressing genetic information, transcription also sets the 3D architecture of the nucleoid (Badrinarayanan et al., 2015; Cagliero et al., 2013; Jeong et al., 2004; Le & Laub, 2016; Shen & Landick, 2019). Precise synchronization of these transcription patterns across every cell in the population is impossible, resulting in cell‐to‐cell variations in the outputs from copies of the same genes in different cells. Transcriptional activity is modulated not only by the genome‐wide distribution of RNA polymerase and the topoisomerases, but also by the actions of, inter alia, transcription factors, nucleoid‐associated proteins, sigma factors, intrinsic and extrinsic transcription terminators, and by the stalling and backtracking of RNA polymerase (Figueroa‐Bossi et al., 2022; Janissen et al., 2022; Schwall et al., 2021). This produces a population of cells exhibiting transcription patterns that are noisy and randomized (Urchueguía et al., 2021). Shocks to the cell arising from environmental stress introduce additional sources of transcriptional fluctuation, including the premature onset of stationary phase. It is probably for this reason that the stationary‐phase sigma factor, RpoS, is also closely associated with stress responses during exponential growth (Battesti et al., 2011; Klauck et al., 2007; Schellhorn, 2020), and has a preference for DNA that is relaxed rather than negatively supercoiled (Bordes et al., 2003; Kusano et al., 1996; Typas et al., 2007).
9. CONCLUSIONS
The influences on gene expression that were summarized above produce a population of bacterial siblings with a transcriptomic profile distributed around a statistical mean. Due to the many stochastic inputs that displace the profile from the mean in individual cells, the population contains physiological outliers. These outliers allow the population to hedge its bets by increasing the probability that suitably prepared, “stress‐pre‐adapted” members will be present if the environment changes. The same strategy drives the emergence of antibiotic‐tolerant persister cells (Balaban et al., 2004). It is also useful during host infection by pathogens, where factors that influence DNA topology and nucleoid architecture play prominent roles in producing transcriptional noise (Figueroa‐Bossi et al., 2022). On the other hand, it may sometimes be advantageous to override stochasticity by imposing deterministic transcriptional outcomes that better fit a majority of the population to the environment. In the case of phase‐variable expression of the operon encoding type 1 fimbriae in E. coli, this is achieved through negative modulation of DNA gyrase activity such that a population entering stationary phase has more fimbriate than afimbriate members (Corcoran & Dorman, 2009; Dorman, 2022; Dove & Dorman, 1994; Muller et al., 2009). The result is a shift from a planktonic lifestyle to an attached one within a biofilm (Anderson et al., 2003; Justice et al., 2004; Wright et al., 2007). The enlisting of variable DNA topology to achieve stochastic outcomes under some circumstances and deterministic ones under others is an example of microbial molecular pragmatism at work.
Future research could usefully expand the investigation of variable DNA topology in the generation of transcriptional stochasticity and physiological diversity beyond the usual model organisms such as E. coli. Direct comparisons of transcriptional outputs from identical gene copies, placed in different bacterial species and/or at different genomic locations in the same species, would make valuable contributions to knowledge. Probing for the presence of positive supercoils, using methods such as GapR‐seq (Guo et al., 2021) would permit correlations between transcription bursting, topoisomerase activity, and positive supercoiling of the DNA template to be studied at a whole‐genome level. What other nucleoid‐associated proteins, other than GapR and YejK, might modulate the relaxation of positive supercoils by type II topoisomerases, and thus the burst‐like nature of transcription? What factors influence the production/activity of these nucleoid‐associated proteins? Do they affect differently the expression of genes from the core genome and genes that have been acquired by horizontal transfer? Addressing these issues will both deepen our understanding of naturally occurring bacteria (including pathogens) and improve our ability to fashion useful microbial tools in synthetic biology. It is also likely to cast light on the contributions of stochastic transcription on the production of immunodiversity and neurodiversity in higher organisms, including humans (Dorman, 2022).
AUTHOR CONTRIBUTIONS
Charles J. Dorman: Conceptualization; Investigation; Writing—original draft; Writing—review & editing.
ACKNOWLEDGMENT
Open access funding provided by IReL.
CONFLICT OF INTEREST
The author declares that he has no conflicts of interest.
10. ETHICS STATEMENT
The work presented here did not include human or animal subjects nor human or animal material or data. Thus, no formal consent or approval was necessary.
Dorman, C.J. (2023) Variable DNA topology is an epigenetic generator of physiological heterogeneity in bacterial populations. Molecular Microbiology, 119, 19–28. 10.1111/mmi.15014
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were generated.
REFERENCES
- Ahmed, S.M. & Dröge, P. (2020) Chromatin architectural factors as safeguards against excessive supercoiling during DNA replication. International Journal of Molecular Sciences, 21, 4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, W. , Sala, C. , Hegde, S.R. , Jha, R.K. , Cole, S.T. & Nagaraja, V. (2017) Transcription facilitated genome‐wide recruitment of topoisomerase I and DNA gyrase. PLoS Genetics, 13, e1006754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancona, M. , Bentivoglio, A. , Brackley, C.A. , Gonnella, G. & Marenduzzo, D. (2019) Transcriptional bursts in a nonequilibrium model for gene regulation by supercoiling. Biophysical Journal, 117, 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, G.G. , Palermo, J.J. , Schilling, J.D. , Roth, R. , Heuser, J. & Hultgren, S.J. (2003) Intracellular bacterial biofilm‐like pods in urinary tract infections. Science, 301, 105–107. [DOI] [PubMed] [Google Scholar]
- Badrinarayanan, A. , Le, T.B. & Laub, M.T. (2015) Bacterial chromosome organization and segregation. Annual Review of Cell and Developmental Biology, 31, 171–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahar Halpern, K. , Tanami, S. , Landen, S. , Chapal, M. , Hutzler, A. , Nizberg, A. et al. (2015) Bursty gene expression in the intact mammalian liver. Molecular Cell, 58, 147–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahassi, E.M. , O'Dea, M.H. , Allali, N. , Messens, J. , Gellert, M. & Couturier, M. (1999) Interactions of CcdB with DNA gyrase. Inactivation of GyrA, poisoning of the gyrase‐DNA complex, and the antidote action of CcdA. Journal of Biological Chemistry, 274, 10936–10944. [DOI] [PubMed] [Google Scholar]
- Balaban, N.Q. , Helaine, S. , Lewis, K. , Ackermann, M. , Aldridge, B. , Andersson, D.I. et al. (2019) Definitions and guidelines for research on antibiotic persistence. Nature Reviews Microbiology, 17, 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban, N.Q. , Merrin, J. , Chait, R. , Kowalik, L. & Leibler, S. (2004) Bacterial persistence as a phenotypic switch. Science, 305, 1622–1625. [DOI] [PubMed] [Google Scholar]
- Bates, A.D. & Maxwell, A. (2005) DNA topology. Oxford: Oxford University Press. [Google Scholar]
- Battesti, A. , Majdalani, N. & Gottesman, S. (2011) The RpoS‐mediated general stress response in Escherichia coli . Annual Review of Microbiology, 65, 189–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger, J.W. (1944) Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet, 244, 497–500. [Google Scholar]
- Bigger, J.W. , Boland, C.R. & O'Meara, R.A.Q. (1927) Variant colonies of Staphylococcus aureus . The Journal of Pathology and Bacteriology, 30, 261–269. [Google Scholar]
- Bordes, P. , Conter, A. , Morales, V. , Bouvier, J. , Kolb, A. & Gutierrez, C. (2003) DNA supercoiling contributes to disconnect σS accumulation from σS‐dependent transcription in Escherichia coli . Molecular Microbiology, 48, 561–571. [DOI] [PubMed] [Google Scholar]
- Brochu, J. , Vlachos‐Breton, E. , Sutherland, S. , Martel, M. & Drolet, M. (2018) Topoisomerases I and III inhibit R‐loop formation to prevent unregulated replication in the chromosomal Ter region of Escherichia coli . PLoS Genetics, 14, 1007668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliero, C. , Grand, R.S. , Jones, M.B. , Jin, D.J. & O'Sullivan, J.M. (2013) Genome conformation capture reveals that the Escherichia coli chromosome is organized by replication and transcription. Nucleic Acids Research, 41, 6058–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, A.D.S. , Dillon, S.C. , Kröger, C. , Beran, L. & Dorman, C.J. (2017) Broad scale redistribution of mRNA abundance and transcriptional machinery in response to growth rate in Salmonella enterica serovar Typhimurium. Microbial Genomics, 3, 00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, A.D.S. , Stoebel, D.M. & Dorman, C.J. (2011) DNA supercoiling is differentially regulated by environmental factors and FIS in Escherichia coli and Salmonella enterica . Molecular Microbiology, 80, 85–101. [DOI] [PubMed] [Google Scholar]
- Champion, K. & Higgins, N.P. (2007) Growth rate toxicity phenotypes and homeostatic supercoil control differentiate Escherichia coli from Salmonella enterica serovar Typhimurium. Journal of Bacteriology, 189, 5839–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D. , Bowater, R. , Dorman, C.J. & Lilley, D.M.J. (1992) Activity of a plasmid‐borne leu‐500 promoter depends on the transcription and translation of an adjacent gene. Proceedings of the National Academy of Sciences of the United States of America, 89, 8784–8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.‐F. , Lin, Y.T. , Gallegos, D.A. , Hazlett, M.F. , Gomez‐Schiavon, M. , Yang, M.G. et al. (2019) Enhancer histone acetylation modulates transcriptional bursting dynamics of neuronal activity‐inducible genes. Cell Reports, 26, 1174–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, S.S. , Chen, C.Y. , Ge, H. & Xie, X.S. (2014) Mechanism of transcriptional bursting in bacteria. Cell, 158, 314–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb, J.R. , Trcek, T. , Shenoy, S.M. & Singer, R.H. (2006) Transcriptional pulsing of a developmental gene. Current Biology, 16, 1018–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coakley, D. (1992) Masters in medicine. Dublin: Town House. [Google Scholar]
- Conlon, B.P. , Rowe, S.E. , Gandt, A.B. , Nuxoll, A.S. , Donegan, N.P. , Zalis, E.A. et al. (2016) Persister formation in Staphylococcus aureus is associated with ATP depletion. Nature Microbiology, 1, 16051. [DOI] [PubMed] [Google Scholar]
- Conter, A. , Menchon, C. & Gutierrez, C. (1997) Role of DNA supercoiling and rpoS sigma factor in the osmotic and growth phase‐dependent induction of the gene osmE of Escherichia coli K12. Journal of Molecular Biology, 273, 75–83. [DOI] [PubMed] [Google Scholar]
- Corcoran, C.P. & Dorman, C.J. (2009) DNA relaxation dependent phase biasing of the fim genetic switch in Escherichia coli depends on the interplay of H‐NS, IHF and LRP. Molecular Microbiology, 74, 1071–1082. [DOI] [PubMed] [Google Scholar]
- Corrigan, A.M. , Tunnacliffe, E. , Cannon, D. & Chubb, J.R. (2016) A continuum model of transcriptional bursting. eLife, 5, e13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisona, N.J. , Strick, T.R. , Bensimon, D. , Croquette, V. & Cozzarelli, N.R. (2000) Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single‐molecule and ensemble measurements. Genes and Development, 14, 2881–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar, R.D. , Razooky, B.S. , Singh, A. , Trimeloni, T.V. , McCollum, J.M. , Cox, C.D. et al. (2012) Transcriptional burst frequency and burst size are equally modulated across the human genome. Proceedings of the National Academy of Sciences of the United States of America, 109, 17454–17459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deweese, J.E. & Osheroff, N. (2009) The DNA cleavage reaction of topoisomerase II: wolf in sheep's clothing. Nucleic Acids Research, 37, 738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGate, R.J. & Marians, K.J. (1989) Molecular cloning and DNA sequence analysis of Escherichia coli topB, the gene encoding topoisomerase III. Journal of Biological Chemistry, 264, 17924–17930. [PubMed] [Google Scholar]
- Dong, K.C. & Berger, J.M. (2007) Structural basis for gate‐DNA recognition and bending by type IIa topoisomerases. Nature, 450, 1201–1205. [DOI] [PubMed] [Google Scholar]
- Dorman, C.J. (2007) H‐NS, the genome sentinel. Nature Reviews Microbiology, 5, 157–161. [DOI] [PubMed] [Google Scholar]
- Dorman, C.J. (2019) DNA supercoiling and transcription: a two‐way street. BMC Molecular Cell Biology, 20, 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman, C.J. (2022) Mechanistic insights from molecular microbiology into the production of immunological and neuronal diversity. Molecular Microbiology. Available from: 10.1111/mmi.14997 [DOI] [PubMed] [Google Scholar]
- Dorman, C.J. , Barr, G.C. , Ní Bhriain, N. & Higgins, C.F. (1988) DNA supercoiling and the anaerobic and growth‐phase regulation of tonB gene expression. Journal of Bacteriology, 170, 2816–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman, C.J. & Dorman, M.J. (2016) DNA supercoiling is a fundamental regulatory principle in the control of bacterial gene expression. Biophysical Reviews, 8, 209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman, C.J. & Dorman, M.J. (2022) Physiological robustness of model Gram‐negative bacteria in response to genome rewiring. Microbial Physiology, 32, 158–176. Available from: 10.1159/000526651 [DOI] [PubMed] [Google Scholar]
- Dove, S.L. & Dorman, C.J. (1994) The site‐specific recombination system regulating expression of the type 1 fimbrial subunit gene of Escherichia coli is sensitive to changes in DNA supercoiling. Molecular Microbiology, 14, 975–988. [DOI] [PubMed] [Google Scholar]
- Drolet, M. , Bi, X. & Liu, L.F. (1994) Hypernegative supercoiling of the DNA template during transcription elongation in vitro . Journal of Biological Chemistry, 269, 2068–2074. [PubMed] [Google Scholar]
- El Hanafi, D. & Bossi, L. (2000) Activation and silencing of leu‐500 promoter by transcription‐induced DNA supercoiling in the Salmonella chromosome. Molecular Microbiology, 37, 583–594. [DOI] [PubMed] [Google Scholar]
- Engl, C. , Jovanovic, G. , Brackston, R.D. , Kotta‐Loizou, I. & Buck, M. (2020) The route to transcription initiation determines the mode of transcriptional bursting in E. coli . Nature Communications, 11, 2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa‐Bossi, N. , Sánchez‐Romero, M.A. , Kerboriou, P. , Naquin, D. , Mendes, C. , Bouloc, P. et al. (2022) Pervasive transcription enhances the accessibility of H‐NS‐silenced promoters and generates bistability in Salmonella virulence gene expression. Proceedings of the National Academy of Sciences of the United States of America, 119, e2203011119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, K. , Iwaki, M. & Yanagida, T. (2016) Transcriptional bursting is intrinsically caused by interplay between RNA polymerases on DNA. Nature Communications, 7, 13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert, M. , Mizuuchi, K. , O'Dea, M.H. & Nash, H.A. (1976) DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proceedings of the National Academy of Sciences of the United States of America, 73, 3872–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng, Y. , Bohrer, C.H. , Yehya, N. , Hendrix, H. , Shachaf, L. , Liu, J. et al. (2022) A spatially resolved stochastic model reveals the role of supercoiling in transcription regulation. PLoS Computational Biology, 18, e1009788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding, I. , Paulsson, J. , Zawilski, S.M. & Cox, E.C. (2005) Real‐time kinetics of gene activity in individual bacteria. Cell, 123, 1025–1036. [DOI] [PubMed] [Google Scholar]
- Gomez‐Gomez, J.M. , Baquero, F. & Blazquez, J. (1996) Cyclic AMP receptor protein positively controls gyrA transcription and alters DNA topology after nutritional upshift in Escherichia coli . Journal of Bacteriology, 178, 3331–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood, D. (2008) Antimicrobial drugs: a Chronicle of a Twentieth Century Medical Triumph. Oxford: Oxford University Press. [Google Scholar]
- Guo, M.S. , Haakonsen, D.L. , Zeng, W. , Schumacher, M.A. & Laub, M.T. (2018) A bacterial chromosome structuring protein binds overtwisted DNA to stimulate type II topoisomerases and enable DNA replication. Cell, 175, 583–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, M.S. , Kawamura, R. , Littlehale, M.L. , Marko, J.F. & Laub, M.T. (2021) High‐resolution, genome‐wide mapping of positive supercoiling in chromosomes. eLife, 10, e67236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms, A. , Stanger, F.V. , Scheu, P.D. , de Jong, I.G. , Goepfert, A. , Glatter, T. et al. (2015) Adenylylation of gyrase and topo IV by FicT toxins disrupts bacterial DNA topology. Cell Reports, 12, 1497–1507. [DOI] [PubMed] [Google Scholar]
- Higgins, N.P. , Peebles, C.L. , Sugino, A. & Cozzarelli, N.R. (1978) Purification of subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic activity. Proceedings of the National Academy of Sciences of the United States of America, 75, 1773–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, J. & Klostermeier, D. (2021) What makes a type IIA topoisomerase a gyrase or a topo IV? Nucleic Acids Research, 49, 6027–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobby, G.L. , Meyer, K. & Chaffee, E. (1942) Observations on the mechanism of action of penicillin. Proceedings of the Society of Experimental Biology (New York), 50, 281–285. [Google Scholar]
- Hsieh, L.S. , Burger, R.M. & Drlica, K. (1991) Bacterial DNA supercoiling and [ATP]/[ADP]. Changes associated with a transition to anaerobic growth. Journal of Molecular Biology, 219, 443–450. [DOI] [PubMed] [Google Scholar]
- Hsieh, L.S. , Rouvière‐Yaniv, J. & Drlica, K. (1991) Bacterial DNA supercoiling and [ATP]/[ADP] ratio: changes associated with salt shock. Journal of Bacteriology, 173, 3914–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, Y.H. , Chung, M.W. & Li, T.K. (2006) Distribution of gyrase and topoisomerase IV on bacterial nucleoid: implications for nucleoid organization. Nucleic Acids Research, 34, 3128–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Q. , Duan, B. , Qu, Z. , Fan, S. & Xia, B. (2021) The DNA recognition motif of GapR has an intrinsic DNA binding preference towards AT‐rich DNA. Molecules, 26, 5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janissen, R. , Eslami‐Mossallam, B. , Artsimovitch, I. , Depken, M. & Dekker, N.H. (2022) High‐throughput single‐molecule experiments reveal heterogeneity, state switching, and three interconnected pause states in transcription. Cell Reports, 39, 110749. [DOI] [PubMed] [Google Scholar]
- Jeong, K.S. , Ahn, J. & Khodursky, A.B. (2004) Spatial patterns of transcriptional activity in the chromosome of Escherichia coli . Genome Biology, 5, R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice, S.S. , Hung, C. , Theriot, J.A. , Fletcher, D.A. , Anderson, G.G. , Footer, M.J. et al. (2004) Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proceedings of the National Academy of Sciences of the United States of America, 101, 1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, J. , Suzuki, H. & Ikeda, H. (1992) Purification and characterization of DNA topoisomerase IV in Escherichia coli . Journal of Biological Chemistry, 267, 25676–25684. [PubMed] [Google Scholar]
- Kato, F. , Yamaguchi, Y. , Inouye, K. , Matsuo, K. , Ishida, Y. & Inouye, M. (2022) A novel gyrase inhibitor from toxin‐antitoxin system expressed by Staphylococcus aureus . FEBS Journal. Available from: 10.1111/febs.16634 [DOI] [PubMed] [Google Scholar]
- Khodursky, A.B. , Peter, B.J. , Schmid, M.B. , DeRisi, J. , Botstein, D. , Brown, P.O. et al. (2000) Analysis of topoisomerase function in bacterial replication fork movement: use of microarrays. Proceedings of the National Academy of Sciences of the United States of America, 97, 9419–9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. , Beltran, B. , Irnov, I. & Jacobs‐Wagner, C. (2019) Long‐distance cooperative and antagonistic RNA polymerase dynamics via DNA supercoiling. Cell, 179, 106–119. [DOI] [PubMed] [Google Scholar]
- Klauck, E. , Typas, A. & Hengge, R. (2007) The sigmaS subunit of RNA polymerase as a signal integrator and network master regulator in the general stress response in Escherichia coli . Science Progress, 90, 103–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindziuk, A. & Kolomeisky, A.B. (2021) Long‐range supercoiling‐mediated RNA polymerase cooperation in transcription. The Journal of Physical Chemistry B, 125, 4692–4700. [DOI] [PubMed] [Google Scholar]
- Kusano, S. , Ding, Q. , Fujita, N. & Ishihama, A. (1996) Promoter selectivity of Escherichia coli RNA polymerase Eσ70 and Eσ38 holoenzymes. Effect of DNA supercoiling. Journal of Biological Chemistry, 271, 1998–2004. [DOI] [PubMed] [Google Scholar]
- Kuzminov, A. (2018) When DNA topology turns deadly—RNA polymerases dig in their R‐loops to stand their ground: new positive and negative (super)twists in the replication‐transcription conflict. Trends in Genetics, 34, 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal, A. , Dhar, A. , Trostel, A. , Kouzine, F. , Seshasayee, A.S.N. & Adhya, A. (2016) Genome scale patterns of supercoiling in a bacterial chromosome. Nature Communications, 7, 11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, K.S. & Merrikh, H. (2018) The clash of macromolecular titans: replication transcription conflicts in bacteria. Annual Review of Microbiology, 8, 71–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, A.J.M. , Ziegenhain, C. , Hagemann‐Jensen, M. , Reinus, B. , Jacob, T. , Dalessandri, T. et al. (2021) Transcriptional bursts explain autosomal random monoallelic expression and affect allelic imbalance. PLoS Computational Biology, 17, e1008772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, T.B. & Laub, M.T. (2016) Transcription rate and transcript length drive formation of chromosomal interaction domain boundaries. EMBO Journal, 35, 1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. & Marians, K.J. (2013) Characterization of the nucleoid‐associated protein YejK. Journal of Biological Chemistry, 288, 31503–31516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leela, J.K. , Raghunathan, N. & Gowrishankar, J. (2021) Topoisomerase I essentiality, DnaA‐independent chromosomal replication, and transcription‐replication conflict in Escherichia coli . Journal of Bacteriology, 203, 0019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng, F. , Amado, L. & McMacken, R. (2004) Coupling DNA supercoiling to transcription in defined protein systems. Journal of Biological Chemistry, 279, 47564–47571. [DOI] [PubMed] [Google Scholar]
- Levsky, J.M. , Shenoy, S.M. , Pezo, R.C. & Singer, R.H. (2002) Single‐cell gene expression profiling. Science, 297, 836–840. [DOI] [PubMed] [Google Scholar]
- Lewis, K. (2010) Persister cells. Annual Review of Microbiology, 64, 357–372. [DOI] [PubMed] [Google Scholar]
- Liu, L.F. & Wang, J.C. (1987) Supercoiling of the DNA template during transcription. Proceedings of the National Academy of Sciences, 84, 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, V. , Martinez‐Robles, M.L. , Hernandez, P. , Krimer, D.B. & Schvartzman, J.B. (2012) Topo IV is the topoisomerase that knots and unknots sister duplexes during DNA replication. Nucleic Acids Research, 40, 3563–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenço, R.F. , Saurabh, S. , Herrmann, J. , Wakatsuki, S. & Shapiro, L. (2020) The nucleoid‐associated protein GapR uses conserved structural elements to oligomerize and bind DNA. MBio, 11, e00448–e00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini, S. , Rowley, G. , Goldberg, M.D. , Hurd, D. , Harrison, M. & Hinton, J.C. (2006) H‐NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathogens, 2, e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J. & Wang, M.D. (2014) RNA polymerase is a powerful torsional motor. Cell Cycle, 13, 337–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin, P. , Zumstein, L. , Sternglanz, R. & Wang, J.C. (1985) The Escherichia coli supX locus is topA, the structural gene for DNA topoisomerase I. Proceedings of the National Academy of Sciences of the United States of America, 82, 5437–5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzocca, M. , Colombo, E. , Callegari, A. & Mazza, D. (2021) Transcription factor binding kinetics and transcriptional bursting: what do we really know? Current Opinion in Structural Biology, 71, 239–248. [DOI] [PubMed] [Google Scholar]
- McKie, S.J. , Neuman, K.C. & Maxwell, A. (2021) DNA topoisomerases: advances in understanding of cellular roles and multi‐protein complexes via structure‐function analysis. BioEssays, 43, 2000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia‐Almonte, C. , Busby, S.J.W. , Wade, J.T. , van Helden, J. , Arkin, A.P. , Stormo, G.D. et al. (2020) Redefining fundamental concepts of transcription initiation in bacteria. Nature Reviews Genetics, 21, 699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller‐Planitz, F. & Herschlag, D. (2008) Coupling between ATP binding and DNA cleavage by DNA topoisomerase II: a unifying kinetic and structural mechanism. Journal of Biological Chemistry, 283, 17463–17476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, C.M. , Aberg, A. , Straseviciene, J. , Uhlin, B.E. & Balsalobre, C. (2009) Type 1 fimbriae, a colonization factor of uropathogenic Escherichia coli, are controlled by the metabolic sensor CRP‐cAMP. PLoS Pathogens, 5, e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskhelishvili, G. , Sobetzko, P. & Travers, A. (2022) Spatiotemporal coupling of DNA supercoiling and genomic sequence organization—a timing chain for the bacterial growth cycle? Biomolecules, 12, 831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik, H.C. , Hari, K. , Chandel, D. , Mandal, S. , Jolly, M.K. & Gayen, S. (2021) Semi‐coordinated allelic‐bursting shape dynamic random monoalleleic expression in pregastrulation embryos. iScience, 24, 102954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre, W.W. , Porwollik, S. , Wang, Y. , McClelland, M. , Rosen, H. , Libby, S.J. et al. (2006) Selective silencing of foreign DNA with low GC content by the H‐NS protein in Salmonella . Science, 313, 236–238. [DOI] [PubMed] [Google Scholar]
- Nicolas, D. , Philips, N.E. & Naef, F. (2017) What shapes eukaryotic transcriptional bursting? Molecular BioSystems, 13, 1280–1290. [DOI] [PubMed] [Google Scholar]
- Nolivos, S. , Upton, A.L. , Badrinarayanan, A. , Muller, J. , Zawadzka, K. , Wiktor, J. et al. (2016) MatP regulates the coordinated action of topoisomerase IV and MukBEF in chromosome segregation. Nature Communications, 7, 10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse, P. , Levine, C. , Hassing, H. & Marians, K.J. (2003) Topoisomerase III can serve as a cellular decatenase in Escherichia coli . Journal of Biological Chemistry, 278, 8653–8660. [DOI] [PubMed] [Google Scholar]
- Perez‐Cheeks, B.A. , Lee, C. , Hayama, R. & Marians, K.J. (2012) A role for topoisomerase III in Escherichia coli chromosome segregation. Molecular Microbiology, 86, 1007–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozdeev, G. , Beckett, M.C. , Mogre, A. , Thomson, N.R. & Dorman, C.J. (2022) Reciprocally rewiring and repositioning the integration host factor (IHF) subunit genes in Salmonella enterica serovar Typhimurium: impacts on physiology and virulence. Microbial Genomics, 8, 00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozdeev, G. , Mogre, A. & Dorman, C.J. (2021) Consequences of producing DNA gyrase from a synthetic gyrBA operon in Salmonella enterica serovar Typhimurium. Molecular Microbiology, 155, 1410–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss, G.J. & Drlica, K. (1985) DNA supercoiling and suppression of the leu‐500 promoter mutation. Journal of Bacteriology, 164, 947–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmouni, A.R. & Wells, R.D. (1992) Direct evidence for the effect of transcription on local DNA supercoiling in vivo . Journal of Molecular Biology, 223, 131–144. [DOI] [PubMed] [Google Scholar]
- Raj, A. , Peskin, C.S. , Tranchina, D. , Vargas, D.Y. & Tyagi, S. (2006) Stochastic mRNA synthesis in mammalian cells. PLoS Biology, 4, e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj, A. & van Oudenaarden, A. (2008) Nature, nurture, or chance: stochastic gene expression and its consequences. Cell, 135, 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj, A. & van Oudenaarden, A. (2009) Single‐molecular approaches to stochastic gene expression. Annual Review of Biophysics, 38, 255–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revyakin, A. , Ebright, R.H. & Strick, T.R. (2004) Promoter unwinding and promoter clearance by RNA polymerase: detection by single‐molecule DNA nanomanipulation. Proceedings of the National Academy of Sciences of the United States of America, 101, 4776–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, S.M. , Higgins, C.F. & Lilley, D.M.J. (1988) DNA supercoiling and the leu‐500 promoter mutation of Salmonella typhimurium . EMBO Journal, 7, 1863–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovinskiy, N.S. , Agbleke, A.A. , Chesnokova, O.N. & Higgins, N.P. (2019) Supercoil levels in E. coli and Salmonella chromosomes are regulated by the C‐terminal 35–38 amino acids of GyrA. Microorganisms, 7, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovinskiy, N.S. , Agbleke, A.A. , Chesnokova, O. , Pang, Z. & Higgins, N.P. (2012) Rates of gyrase supercoiling and transcription elongation control supercoil density in a bacterial chromosome. PLoS Genetics, 8, e1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellhorn, H.E. (2020) Function, evolution, and composition of the RpoS regulon in Escherichia coli . Frontiers in Microbiology, 11, 560099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwall, C.P. , Loman, T.E. , Martins, B.M.C. , Cortijo, S. , Villava, C. , Kusmartsev, V. et al. (2021) Tunable phenotypic variability through an autoregulatory alternative sigma factor circuit. Molecular Systems Biology, 17, e9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, B.A. & Landick, R. (2019) Transcription of bacterial chromatin. Journal of Molecular Biology, 431, 4040–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobetzko, P. , Travers, A. & Muskhelishvili, G. (2012) Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proceedings of the National Academy of Sciences of the United States of America, 109, E42–E50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, R. , Chandraprakash, D. , Krishnamurthi, R. , Singh, P. , Scolari, V.F. , Krishna, S. et al. (2013) Genomic analysis reveals epistatic silencing of "expensive" genes in Escherichia coli K‐12. Molecular BioSystems, 9, 2021–2033. [DOI] [PubMed] [Google Scholar]
- Stracy, M. , Wollman, A.J.M. , Kaja, E. , Gapinski, J. , Lee, J.‐E. , Leek, V.A. et al. (2019) Single‐molecule imaging of DNA gyrase activity in living Escherichia coli . Nucleic Acids Research, 47, 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter, D.M. , Molina, N. , Gatfield, D. , Schneider, K. , Schibler, U. & Naef, F. (2011) Mammalian genes are transcribed with widely different bursting kinetics. Science, 332, 472–474. [DOI] [PubMed] [Google Scholar]
- Sutormin, D. , Galivondzhyan, A. , Musharova, O. , Travin, D. , Rusanova, A. , Obraztsova, K. et al. (2022) Interaction between transcribing RNA polymerase and topoisomerase I prevents R‐loop formation in E. coli. Nature . Communications, 13, 4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutormin, D. , Rubanova, N. , Logacheva, M. , Ghilarov, D. & Severinov, K. (2019) Single‐nucleotide‐resolution mapping of DNA gyrase cleavage sites across the Escherichia coli genome. Nucleic Acids Research, 47, 1373–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi, S. , Brahmachari, S. , Onuchic, J.N. & Levine, H. (2022) DNA supercoiling‐mediated collective behaviour of co‐transcribing RNA polymerases. Nucleic Acids Research, 50, 1269–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi, A. , Dewan, P.C. , Barua, B. & Varadarajan, R. (2012) Additional role for the ccd operon of F‐plasmid as a transmissible persistence factor. Proceedings of the National Academy of Sciences of the United States of America, 109, 12497–12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucksis, M. & Depew, R.E. (1981) Identification and localization of a gene that specifies production of Escherichia coli DNA topoisomerase I. Proceedings of the National Academy of Sciences of the United States of America, 78, 2164–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucksis, M. , Golub, E.I. , Zabel, D.J. & Depew, R.E. (1981) Escherichia coli and Salmonella typhimurium supX genes specify deoxyribonucleic acid topoisomerase I. Journal of Bacteriology, 147, 679–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao, Y.P. , Wu, H.Y. & Liu, L.F. (1989) Transcription‐driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell, 56, 111–118. [DOI] [PubMed] [Google Scholar]
- Turner, E.C. & Dorman, C.J. (2007) H‐NS antagonism in Shigella flexneri by VirB, a virulence gene transcription regulator that is closely related to plasmid partition factors. Journal of Bacteriology, 189, 3403–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas, A. , Becker, G. & Hengge, R. (2007) The molecular basis of selective promoter activation by the sigmaS subunit of RNA polymerase. Molecular Microbiology, 63, 1296–1306. [DOI] [PubMed] [Google Scholar]
- Urchueguía, A. , Galbusera, L. , Chauvin, D. , Bellement, G. , Julou, T. & van Nimwegen, E. (2021) Genome‐wide gene expression noise in Escherichia coli is condition‐dependent and determined by propagation of noise through the regulatory network. PLoS Biology, 19, e3001491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usongo, V. , Nolent, F. , Sanscartier, P. , Tanguay, C. , Broccoli, S. , Baaklini, I. et al. (2008) Depletion of RNase HI activity in Escherichia coli lacking DNA topoisomerase I leads to defects in DNA supercoiling and segregation. Molecular Microbiology, 69, 968–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulin, C. , Leimer, N. , Huerner, M. , Ackermann, M. & Zinkernagel, A.S. (2018) Prolonged bacterial lag time results in small colony variants that represent a sub‐population of persisters. Nature Communications, 9, 4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainwright, J. , Hobbs, G. & Nakouti, I. (2021) Persister cells: formation, resuscitation and combative therapies. Archives of Microbiology, 203, 5899–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.C. (1998) Moving one DNA double helix through another by a type II topoisomerase: the story of a simple molecular machine. Quarterly Reviews of Biophysics, 31, 107–114. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Ni, T. , Wang, W. & Liu, F. (2018) Gene transcription in bursting: a unified mode for realizing accuracy and stochasticity. Biological Reviews of the Cambridge Philosophical Society, 94, 248–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Workum, M. , van Dooren, S.J. , Oldenburg, N. , Molenaar, D. , Jensen, P.R. , Snoep, J.L. et al. (1996) DNA supercoiling depends on the phosphorylation potential in Escherichia coli . Molecular Microbiology, 20, 351–360. [DOI] [PubMed] [Google Scholar]
- Wright, K.J. , Seed, P.C. & Hultgren, S.J. (2007) Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cellular Microbiology, 9, 2230–2241. [DOI] [PubMed] [Google Scholar]
- Wu, H.Y. , Shyy, S. , Wang, J.C. & Liu, L.F. (1988) Transcription generates positively and negatively supercoiled domains in the template. Cell, 53, 433–440. [DOI] [PubMed] [Google Scholar]
- Zechiedrich, E.L. & Cozzarelli, N.R. (1995) Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli . Genes and Development, 9, 2859–2869. [DOI] [PubMed] [Google Scholar]
- Zechiedrich, E.L. , Khodursky, A.B. , Bachellier, S. , Schneider, R. , Chen, D. , Lilley, D.M.J. et al. (2000) Roles of topoisomerases in maintaining steady‐state DNA supercoiling in Escherichia coli . Journal of Biological Chemistry, 275, 8103–8113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were generated.


