Abstract
Acute exercise elicits dynamic transcriptional changes that, when repeated, form the fundamental basis of health, resilience, and performance adaptations. While moderate-intensity endurance training combined with conventional resistance training (traditional, TRAD) is often prescribed and recommended by public health guidance, high-intensity training combining maximal-effort intervals with intensive, limited-rest resistance training is a time-efficient alternative that may be used tactically (HITT) to confer similar benefits. Mechanisms of action of these distinct stimuli are incompletely characterized and have not been directly compared. We assessed transcriptome-wide responses in skeletal muscle and circulating extracellular vesicles (EVs) to a single exercise bout in young adults randomized to TRAD (n = 21, 12 M/9 F, 22 ± 3 yr) or HITT (n = 19, 11 M/8 F, 22 ± 2 yr). Next-generation sequencing captured small, long, and circular RNA in muscle and EVs. Analysis identified differentially expressed transcripts (|log2FC|>1, FDR ≤ 0.05) immediately (h0, EVs only), h3, and h24 postexercise within and between exercise protocols. In aaddition, all apparently responsive transcripts (FDR < 0.2) underwent singular value decomposition to summarize data structures into latent variables (LVs) to deconvolve molecular expression circuits and interregulatory relationships. LVs were compared across time and exercise protocol. TRAD, a longer but less intense stimulus, generally elicited a stronger transcriptional response than HITT, but considerable overlap and key differences existed. Findings reveal shared and unique molecular responses to the exercise stimuli and lay groundwork toward establishing relationships between protein-coding genes and lesser-understood transcripts that serve regulatory roles following exercise. Future work should advance the understanding of these circuits and whether they repeat in other populations or following other types of exercise/stress.
NEW & NOTEWORTHY We examined small and long transcriptomics in skeletal muscle and serum-derived extracellular vesicles before and after a single exposure to traditional combined exercise (TRAD) and high-intensity tactical training (HITT). Across 40 young adults, we found more consistent protein-coding gene responses to TRAD, whereas HITT elicited differential expression of microRNA enriched in brain regions. Follow-up analysis revealed relationships and temporal dynamics across transcript networks, highlighting potential avenues for research into mechanisms of exercise response and adaptation.
Keywords: exercise physiology, extracellular vesicles, gene expression, skeletal muscle, transcriptomics
INTRODUCTION
Exercise is a powerful and effective master regulator of metabolic health, physical performance, and overall wellness throughout the lifespan. As our team has recently reviewed (1), regular exposure to exercise improves cardiorespiratory fitness, neuromuscular function, cognitive function, metabolic efficiency, body composition, disease resilience, and healthspan, among a host of other benefits that continue to be identified. However, despite generally well-defined health and performance benefits, the optimal exercise “dose” continues to be an elusive research pursuit and likely varies based on age, fitness level, presence of comorbidities, other demographic confounders, and goal outcomes for training and/or overall health (2).
Exercise intensity and duration are the primary determinants of overall workload via their combined demands on neuromuscular activation patterns, cardiorespiratory demand, energy metabolism, and underlying molecular signaling required to support continued activity (1). Public health guidelines (e.g., American College of Sports Medicine, World Health Organization) encourage a combination of regular resistance and endurance training performed at a moderate-to-high intensity. In contrast to this more traditional (TRAD) approach, an increasingly popular modality both in the public (e.g., CrossFit, Tabata) and among military personnel [high intensity tactical training, or HITT (3)] exists in the form of a circuit of movements performed at maximum velocity for brief time intervals with some movements performed explosively (i.e., maximum power) against bodyweight and/or added resistance. A range of functional and time-saving benefits are associated with HITT training, and some evidence indicates that the long-term physiological benefits are similar to TRAD training with regard to aerobic fitness, energy metabolism (4), and even longevity (5). However, whether these benefits arise through similar molecular mechanisms is not known. A better understanding of the so-called molecular circuits linked to TRAD versus HITT responses may help guide exercise prescription for individuals, including identification of those whose intrinsic molecular phenotype suggests benefiting more from one stimulus than the other (2).
Insight into the intensity-dependent molecular response to exercise is likely to provide valuable guidance toward optimizing and personalizing exercise dosing. The purest comparison may be best achieved in the exercise-naïve state following a single exposure to a relatively unaccustomed bout of exercise (6). Following acute exercise exposure, transient changes in gene expression form the basis of long-term adaptations at the protein level (7, 8), and therefore are most likely to lead to the myriad structural and functional changes in skeletal muscle and other organs and tissues. Accordingly, the current understanding of exercise transcriptomics is mostly limited to the subset of transcripts that encode protein (i.e., mRNA) and their ontological relationships to biological pathways via curated resources for annotation (9, 10). However, the vast majority of transcripts do not encode protein and are instead classified across a wide range of other “biotypes” that may serve a variety of signaling, regulatory, or otherwise poorly understood cellular roles. For example, regulatory transcripts such as microRNAs (miRNAs) are a class of posttranscriptional modifiers that have been recognized to play important roles in exercise response and adaptation (11, 12). Although produced in most tissues, miRNAs are commonly packaged into extracellular vesicles (EVs) and circulated to peripheral tissues (13), where they may affect a wide range of potential target genes. In addition, a range of other noncoding biotypes of both small (e.g., piRNA, snRNA) and long (e.g., lncRNA, circRNA) nucleotide length is likely to exert regulatory effects on cellular processes including but not limited to transcription and translation (14, 15).
Bolstered by extensive research from our group (16–18) and many others (19–21), the “protein-coding gene” expression response to acute and chronic exercise provides a solid basis yet a still-incomplete picture of coordinated changes in the molecular environment. As such, this study examined the exercise-dependent, coordinated transcriptomic responses to two combined exercise stimuli in a sex-balanced cohort to provide valuable insight across multiple knowledge gaps, including 1) clarity regarding acute molecular responses to HITT versus TRAD, 2) the relative coordinated temporal dynamics and potential regulatory response circuitry at the transcriptomic level, and 3) associations with established biological pathways linked to exercise adaptation. Profiling this response in a mixed cohort of healthy, young males and females not only strengthens the existing knowledge base but also provides a foundation for more complex future studies investigating comparator populations (e.g., older adults, highly trained athletes, individuals with comorbidities, etc.), different exercise stimuli, and/or the effects of combined wellness strategies (e.g., exercise plus behavioral, nutritional, or pharmaceutical interventions).
METHODS
Participant Recruitment, Screening, and Randomization
For this investigation into the transcriptomic responses to acute exercise, 40 young adults (23 M/17 F, 22 ± 3 yr) were studied, representing a subset of participants enrolled in the parent randomized clinical efficacy trial (Department of Defense, Office of Naval Research grant: N000141613159, ClinicalTrials.gov Identifier: NCT03380923). Participants were recruited from the Greater Birmingham, Alabama area and surrounding communities. Inclusion/exclusion criteria and screening procedures are reported in NCT03380923. Briefly, all participants were aged 18–27 yr and generally healthy, as assessed by questionnaires detailing health history and medical history, including screening for mental health disorders. Participants reported no history of regular exercise training in the past 12 mo, had a body mass index (BMI) < 30, and were nonsmokers. The trial was approved and monitored by The University of Alabama at Birmingham (UAB) Institutional Review Board for Human Use (IRB) and conducted in accordance with the Declaration of Helsinki. All participants provided written informed consent to participate and allowed their biospecimens to be used for future research. A schematic of the general study design is presented in Fig. 1. Following consent and screening, participants were randomized to either traditional combined exercise training (TRAD, n = 21, 12 M/9 F, 22 ± 3 yr) or high-intensity tactical training (HITT, n = 19, 11 M/8 F, 22 ± 2 yr). Randomization was stratified only by sex to ensure similar distributions of females and males in TRAD and HITT.
Figure 1.
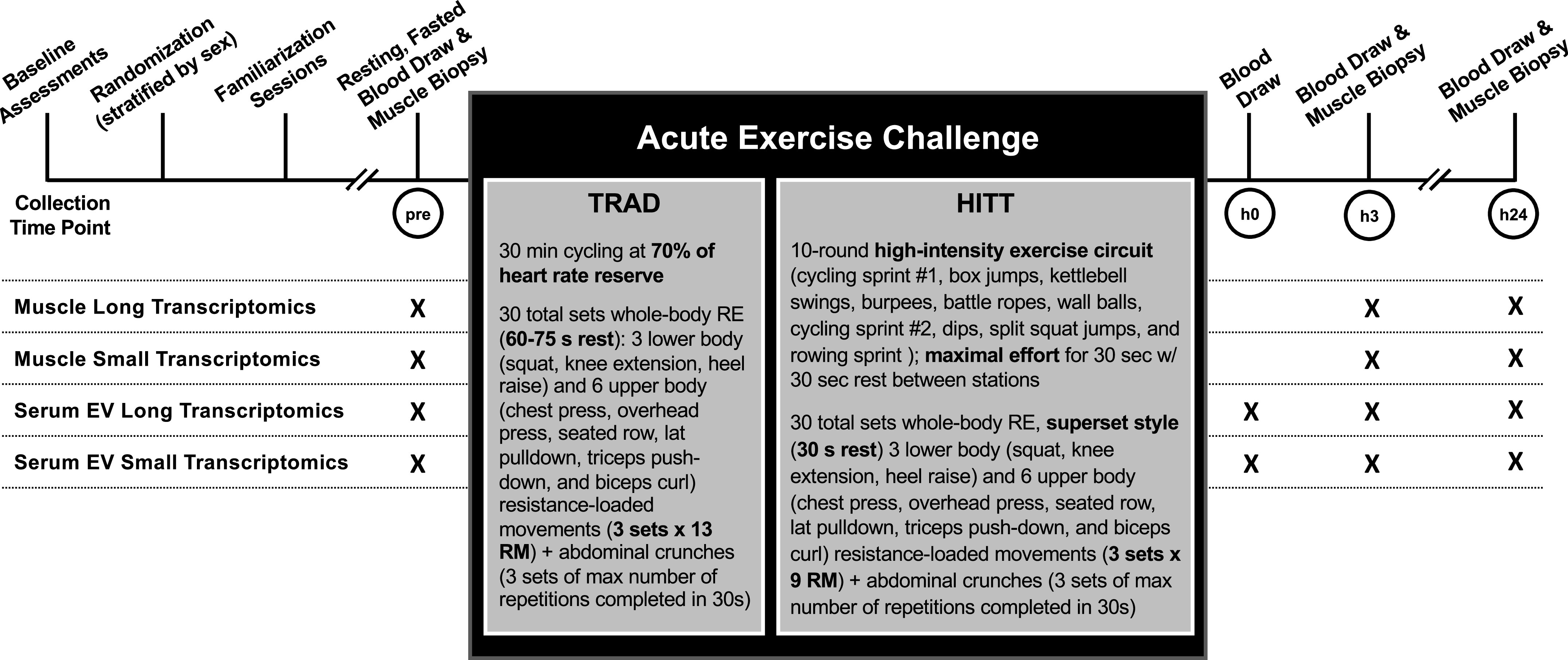
Schematic showing general study processes conducted around the acute exercise challenge and details for each bout. At the indicated time points, participant underwent a muscle biopsy (preexercise, h3, and h24 postexercise) and blood draw (preexercise, h0, h3, and h24 postexercise) to be processed for extraction of serum extracellular vesicles (EVs). Small and long transcriptomics were performed on RNA isolated from biospecimens collected at each time point.
Phenotyping
Participants underwent several relevant phenotyping assessments to provide a detailed depiction of the cohort yielding the biospecimens and ultimately molecular response circuits (Table 1). Baseline phenotyping included standardized and well-validated tests to evaluate body composition (dual-energy X-ray absorptiometry, DXA), cardiorespiratory fitness or aerobic power (V̇o2peak on a cycle ergometer), anaerobic power (30-s Wingate test), one-repetition maximum (1RM) upper and lower body dynamic strength on five movements, unilateral maximal voluntary isometric knee extension strength, MVC), and unilateral knee extension peak power. Methods for each of these tests is provided in detail elsewhere (22–25).
Table 1.
Demographic and phenotyping results
| TRAD | HITT | |
|---|---|---|
| n | n = 21 (12 M, 9 F) | n = 19 (11 M, 8 F) |
| Age, yr | 22 ± 3 | 22 ± 2 |
| Height, cm | 174 ± 10 | 173 ± 6 |
| Weight, kg | 74 ± 12 | 73 ± 11 |
| BMI, kg/m2 | 24 ± 3 | 24 ± 3 |
| Body composition | n = 20 | n = 19 |
| Total lean mass, kg | 50.5 ± 9.4 | 49.7 ± 9.9 |
| Total fat mass, kg | 20.0 ± 8.0 | 20.0 ± 6.1 |
| Body fat, % | 27.0 ± 8.7 | 27.6 ± 7.6 |
| One repetition maximum strength | n = 20 | n = 18 |
| Overhead press, kg | 42 ± 14 | 40 ± 19 |
| Chest press, kg | 61 ± 25 | 61 ± 37 |
| Lat pull down, kg | 46 ± 15 | 47 ± 18 |
| Squat, kg | 85 ± 30 | 83 ± 37 |
| Knee extension, kg | 95 ± 32 | 88 ± 36 |
| Unilateral maximal knee extension | n = 20 | n = 18 |
| Isometric MVC peak torque, Nm | 228 ± 70 | 218 ± 80 |
| Isotonic average power, W | 183.2 ± 81.3 | 210.5 ± 130.0 |
| Isotonic peak work, J | 98.3 ± 26.7 | 100.9 ± 38.1 |
| Isotonic total work, J | 449.7 ± 121.6 | 460.7 ± 169.7 |
| Aerobic power | n = 21 | n = 19 |
| Absolute V̇o2max, L/min | 2.6 ± 0.7 | 2.4 ± 0.6 |
| Relative V̇o2max, mL/kg/min | 35.3 ± 7.1 | 32.7 ± 5.0 |
| RER at V̇o2max | 1.21 ± 0.06 | 1.24 ± 0.09 |
| Anaerobic power | n = 19 | n = 18 |
| Absolute peak power, W | 732 ± 235 | 680 ± 235 |
| Relative peak power, W/kg | 9.68 ± 2.25 | 9.29 ± 2.02 |
Values are means ± SD. BMI, body mass index; HITT, high-intensity tactical training; MVC, maximum voluntary contraction; RER, respiratory exchange ratio; TRAD, traditional combined exercise; V̇o2max, maximal oxygen consumption.
Familiarization
Participants completed four progressive familiarization sessions before the acute exercise challenge. These familiarization sessions were designed to 1) ensure that all exercises were performed safely and correctly, 2) dial in the target intensity and volume for the acute bout, and 3) ensure that the acute bout indeed yielded a molecular exercise response in the untrained state as opposed to an overt tissue damage response in the truly naïve state. The four familiarizations followed a standardized progression in intensity and volume such that each participant’s first exposure to the full stimulus was the acute bout itself.
Acute Exercise Challenge with Biospecimen Collection
All acute exercise bouts were performed between the hours of 0600 and 0900. Following an overnight fast (∼10 h), individuals rested in a supine position for ∼30 min. A rested, fasting blood draw was obtained from an antecubital vein followed by a percutaneous muscle biopsy of the vastus lateralis using established methods previously described in detail (16, 18). Muscle tissue was cleared of all visible connective tissue and fat, and ∼25–30-mg portions were immediately snap-frozen in liquid nitrogen for RNA isolation and other assays and stored at −80°C until analysis. For histological analysis, a separate portion ∼3–4 mm in cross-sectional diameter was mounted on cork using Tissue-Tek Optimal Cutting Temperature (O.C.T.) compound mixed with tragacanth gum and frozen in liquid nitrogen-cooled isopentane. Blood was allowed to clot for 30 min and then spun at 2,000 g for 10 min at 4°C to separate the serum fraction. Serum aliquots were stored at −80°C until analysis.
Following the muscle biopsy, each participant performed the acute exercise challenge of the study arm to which they had been randomized (TRAD or HITT).
TRAD prescription.
TRAD participants first performed 30 min of steady-state cycle ergometry at an intensity targeting 70% of heart rate reserve (HRR), as calculated from resting HR and peak HR attained during the cycle ergometer-based maximal aerobic power (V̇o2max) test. This aerobic exercise was followed by 30 total sets of whole body resistance exercise (RE) consisting of three lower body (squat, knee extension, and heel raise) and six upper body (chest press, overhead press, seated row, lat pulldown, triceps push-down, and biceps curl) resistance loaded movements, along with abdominal crunches. For the nine loaded RE movements, three sets of each movement were completed with loads targeting an intensity of 13 repetition-maximum (13RM). Crunches were performed as three sets × maximum number of repetitions completed in 30 s per set. The total of 30 RE sets were performed with 60–75 s rest between sets. Given the focus on muscle biopsies of the vastus lateralis, the squat and knee extension were performed last in the sequence.
HITT prescription.
In lieu of steady-state aerobic exercise, HITT participants first performed a 10-round circuit of high-intensity, explosive exercises in the following order: cycling sprint #1, box jumps, kettlebell swings, burpees, battle ropes, wall balls, cycling sprint #2, dips, split squat jumps, and rowing sprint each at maximal effort for 30 s with 30 s rest between stations. This was followed by whole body RE with the same 10 movements × 3 sets each prescribed in TRAD. However, major differences in intensity and work-rest ratio were prescribed to HITT. Specifically, intensity of the loaded RE movements targeted 9RM resistance loads, supersets were performed consisting of two paired movements that activated antagonistic or independent muscle groups, and supersets were separated by only 30 s rest intervals. These differences were prescribed to contrast TRAD with a substantially more intense RE exposure in HITT. The prescriptions are summarized in Fig. 1. The entire protocol duration was ∼90 min for TRAD and ∼45 min for HITT.
No structured physical activity was allowed postexercise through the h24 biospecimen collections. Immediately after the acute exercise bout, blood was collected (within ∼10 min, h0). Participants were then provided with a standardized protein shake (Ensure Plus, Abbott Laboratories, Abbott Park, IL; 350 kcal, 13 g protein, 51 g carbohydrate, and 11 g fat) and permitted to leave the exercise facility (with the activity restriction monitored by a wearable device) and returned for a postexercise muscle biopsy and blood collection at h3. Subjects repeated an evening meal as similar as possible to the preceding night to reduce the influence of dietary variability within an individual, followed by an overnight fast (∼10 h). The following morning, 24 ± 2 h from the cessation of the acute exercise bout, participants returned in the fasted state and underwent a final blood draw and muscle biopsy.
RNA Isolation
RNA was isolated from both skeletal muscle and serum extracellular vesicles (EVs) for each study time point. Isolations were performed at UAB by two independent processors for each biospecimen type in batches balanced for sex and exercise group (TRAD/HITT), and all samples from a participant were always included in the same batch. For skeletal muscle, RNA was isolated from snap-frozen tissue (27.8 ± 5.4 mg) using the Qiagen miRNeasy kit (Cat. No. 217004, Qiagen, Germantown, MD). The miRNeasy kit enables collection of all RNAs (including those <200 nt) and is therefore ideal for transcriptome-wide sequencing operations, capturing both long and small RNA. Muscle-derived RNA was confirmed to be of excellent quality via Agilent Bioanalyzer (Agilent, Santa Clara, CA) [average ± SD RNA integrity number (RIN) 8.5 ± 0.4, and 28S/18S ratio 1.7 ± 0.4].
EVs were isolated from ∼1 mL of serum. Any cryoprecipitates accumulated during freezing were cleared via centrifugation, and RNA was subsequently liberated from EVs using the exoRNeasy midi kit (Cat. No. 77144, Qiagen). The exoRNeasy kit is used in the only Clinical Laboratory Improvement Amendments (CLIA)-validated EV-based liquid biopsy tests and has been shown to have the highest reproducibility in replicates across multiple laboratories (26, 27). The kit has an affinity membrane that binds and isolates all EVs at once and eliminates contaminating RNA-binding proteins, lipoproteins, and other RNA carriers by allowing them to pass though the membrane. Thus, RNA samples extracted from EVs were highly likely to be of sufficient quality for next-generation sequencing. Quant-iT Ribogreen RNA Assay (Cat. No. R11490, ThermoFisher, Waltham, MA) using the low-range concentration protocol confirmed good concentration across total EV-derived RNA samples (0.78 ± 0.41 ng/µL); however, due to the quantity of RNA necessary to determine RIN, bioanalysis was not performed. When total sample RNA exceeded 10 ng, 5 ng RNA was put into whole transcriptome library prep, and 5 ng RNA was put into small RNA library prep. For samples with less than 10 ng of total RNA, half of the total RNA was put into whole transcriptome library prep and half was put into small RNA library prep.
Next-Generation Sequencing
Preparation and quality control.
RNA was prepared for sequencing and subsequently sequenced at Translational Genomics Institute (TGen, Phoenix, AZ). Due to differing optimal downstream sequencing approaches for detection and quantification of long and small transcriptomics, separate laboratory workflows were used to prepare fractions for long and small transcriptomic sequencing from the same original RNA aliquot. Total RNA from skeletal muscle was DNase-treated with TURBO DNA-free (Cat. No. AM1907, ThermoFisher), then purified with RNeasy MinElute Cleanup (Cat. No. 74204, Qiagen) using a modified MinElute Cleanup protocol to capture both small and long RNA species; namely, in step 2 of the standard MinElute protocol, 950 µL 100% EtOH was added to each sample instead of 250 µL. RNA was measured for quantity with Quant-iT Ribogreen RNA Assay (Cat. No. R11490, ThermoFisher) and quality with High Sensitivity RNA ScreenTape and buffer (Cat. Nos. 5067–5579 and 5067–5580, Agilent).
Long RNA library preparation and next-generation sequencing.
For each RNA sample, a uniquely dual-indexed, Illumina-compatible, double-stranded cDNA whole transcriptome library was synthesized from 10 ng (for skeletal muscle) or up to 5 ng (for serum EVs) total RNA with the SMARTer Stranded Total RNA-Seq kit v2 Pico Input Mammalian (Cat. No. 634418, Takara Bio, San Jose, CA) and SMARTer RNA Unique Dual Index Kit (Cat. No. 634452, Takara Bio). Briefly, this library preparation included RNA fragmentation, a 5-cycle Indexing PCR, ribosomal cDNA depletion, and enrichment PCR (12 cycles for skeletal muscle, 16 for serum EVs). RNA fragmentation incubation time and temperature were dictated by each sample’s RIN (when available) and %DV200 according to the library prep protocol. Each library was measured for size with Agilent High Sensitivity D1000 ScreenTape and buffer (Cat. Nos. 5067–5584 and 5067–5603, Agilent). Next, 1 µL of each library was combined into a nonequimolar pool, which was then measured for size via TapeStation and concentration via the KAPA SYBR FAST Universal qPCR Kit (Cat. No. KK4824, Roche, Wilmington, MA), diluted to 70 pM, then loaded into an iSeq flowcell cartridge (Cat. No. 20031371, Illumina, San Diego, CA) with a 1% vol/vol PhiX Control v3 spike-in (Cat. No. FC-110–3001, Illumina), and sequenced at 101 × 8 × 8 × 101 cycles. Passing filter cluster counts per library were generated from this data and used to make a rebalanced pool, which was subsequently measured for size and concentration, diluted to 2 nM with a 1% vol/vol PhiX Control v3 spike-in, denatured and further diluted, loaded into a NovaSeq 6000 flow cell cartridge (Cat. No. 20028313, Illumina), and sequenced at 101 × 9 × 9 × 101 cycles with standard workflow and a final flow cell concentration of 400 pM. Muscle cDNA libraries were sequenced to at least 40 M read pairs (or 80 M paired-end reads), and serum EV cDNA libraries were sequenced to at least 50 M read pairs (or 100 M paired-end reads).
Small RNA library preparation and next-generation sequencing.
Small RNA libraries were generated from 125 ng of skeletal muscle RNA or 5 ng of serum EV RNA using NEXTflex Small RNA Library Prep Kit v3 (Cat. No. NOVA-5132-23, Perkin Elmer, Waltham, MA). The manufacturer’s instructions were followed with the following modifications 1) 3′ and 5′ adapters were diluted by half, 2) adapter inactivation (protocol step C) was not performed, 3) 2-µL adapter inactivation buffer was added after protocol step B, and 4) an additional 0.5 µL of water was added to each well. Eighteen cycles of PCR amplification were used for serum EVs and 16 cycles for skeletal muscle. Libraries between 150 and 200 bp in size were excised and purified using 6% Tris/Borate/EDTA gels, followed by DNA cleaning, concentrating (Zymo, Irvine, CA), and eluting in 30 µL of water. The quality and quantity of gel-purified libraries was assessed using Agilent 2100 Bioanalyzer with High-Sensitivity DNA chips. Equimolar amounts of libraries were pooled and quantified. Pooled libraries were then normalized and denatured at a working concentration range of 15 pM with 5% PhiX spike-in for flow cell cluster generation. Libraries were sequenced on the HiSeq 2500 with cluster generation on the cBot Flow Cell (Illumina). For sequencing small RNA, kits with specialized adaptors that have four randomized nucleotides on the end for ligation were used to reduce bias in small RNASeq (28–30).
Alignment.
The Extracellular RNA Communications Consortium (exceRpt) pipeline was used for alignment of small RNA (31). This pipeline is specifically optimized and validated for extracellular RNAs with alignments in parallel for miRNA (miRBase, piRNABank, tRNA, etc.). Data were normalized to read depth. Long RNA was aligned using STAR v2.6+ (32) to GRCh38 and to additional lncRNAs from the highly curated LNCpedia (33) and GENCODE datasets. Circular RNA (circRNA) was identified from reads in the long transcriptomics dataset using CIRI2 following mapping with Burrows-Wheeler Alignment (bwa), as described (34). Briefly, a 19-nucleotide overlap between the read and both sides of a backspliced junction was required to identify a circRNA. Single nucleotide polymorphism (SNP) calls available for the Arizona Study of Aging and Neurodegenerative Disease (AZSAND) Brain and Body Donation Program (BBDP) were leveraged to provide additional insight into genetics and exRNA detection. Count data were created using featureCounts (35). All FASTQ files and raw count matrices are available in the Gene Expression Omnibus (GEO) database under accession number GSE209880.
Computational Analysis
Data processing.
The general workflow, including the gene × sample matrix dimensions for each data stream throughout processing, is presented in Table 2. All downstream data processing and analysis was performed using R v4.1.2. Each transcriptomics data stream was analyzed independently and in parallel according to the same general computational pipeline, with minor adjustments made where necessary to optimize the workflow for small, long, and circular RNA in both skeletal muscle and serum EVs. Each data stream was initially cleaned to remove all corresponding postexercise samples from individuals whose preexercise samples failed sequencing QC. The rationale for this approach was that a missing preexercise sample would preclude interpretation of an exercise-induced change at any other time point; however, if the outlier was a sample collected at any postexercise time point, all corresponding samples were retained to maximize the statistical power of the dataset.
Table 2.
Computational workflow and dimensionality for analyzed data streams
| Skeletal Muscle |
Serum EVs |
|||||
|---|---|---|---|---|---|---|
| Transcript-Level Differential Expression Analysis | Long RNA | Small RNA | circRNA | Long RNA | Small RNA | circRNA |
| Raw counts (transcripts × samples) | 94,946 × 120 | 85,542 × 120 | 39,577 × 120 | 94,946 × 148 | 85,542 × 154 | 197,605 × 148 |
| Filtering Steps: | ||||||
| 1. to remove participants missing pre-exercise sample | 94,946 × 120 | 85,542 × 120 | 39,577 × 120 | 94,946 × 140 | 85,542 × 154 | 197,605 × 140 |
| 2. by biotype (based on length at transcript maturity) | 86,636 × 120 | 5,398 × 120 | 39,577 × 120 | 86,636 × 140 | 5,398 × 154 | 197,605 × 140 |
| 3. for expression (cpm >0) in ≥25% of samples | 32,295 × 120 | 2,673 × 120 | 1,812 × 120 | 62,772 × 140 | 1,313 × 154 | 1,120 × 140 |
| Batch effect identification (adjusted with ComBat_seq if needed) | Adjusted | |||||
| Outlier (±6 MAD from median) identification | 32,295 × 120 | 2,673 × 120 | 1,812 × 120 | 62,772 × 133 | 1,313 × 146 | 1,120 × 140 |
| Filtering by expression (long RNA only) | 23,329 × 121 | 57,025 × 133 | ||||
| limma differential expression analysis with voom correction | ||||||
| Exploratory Transcriptomic Network Discovery | ||||||
| FDR < 0.2 at any time point | 19,444 | 1,728 | 148 | 55,325 | 772 | 310 |
| All biotype counts collapsed within tissue & filtered for shared samples | 21,320 × 120 | 56,407 × 128 | ||||
| PLIER singular value decomposition: | ||||||
| n LVs generated (n showing main effect of exercise) | 40 (23) | 32 (4) | ||||
See text for complete workflow, including steps that do not affect data dimensionality. cpm, counts per million; EVs, extracellular vesicles; FDR, false discovery rate; LVs, latent variables; MAD, mean absolute deviation; PLIER, Pathway-Level information extractor.
RNA biotypes were analyzed as part of the data stream corresponding with transcript length in mature form [e.g., we discarded fragments that mapped to protein-coding reads in the small dataset, or incompletely processed pri-miRNA in the long RNA dataset (36)]. Based on an available predefined threshold (37), biotypes with a mean nucleotide (nt) length >200 were treated as long RNA (e.g., protein-coding, lncRNA, antisense, and multiple subclasses of pseudogenes), and those <200 nt were considered small RNA (e.g., miRNA, rRNA, tRNA, snRNA, snoRNA, and vaultRNA). A total of 20 long RNA biotypes and 9 small RNA biotypes were retained following this filtering step.
Count data were filtered by expression based on non-zero expression in at least 25% of samples. Next, principal components analysis (PCA) was used to identify the influence of any technical confounders, such as processing and/or sequencing batch. Where identified, batch effects were adjusted using ComBat_seq via sva (38). After applying a log2cpm normalization corrected for within-subject variance, statistical outliers were identified using PCA. Samples with scores more than six median absolute deviations from the median for PCs 1 and 2 (together, contributing to ∼35–40% of between-sample variance) were flagged as statistical outliers. An approach similar to that described above was used to clean the data for data points detected as statistical outliers and related samples. As a result, there was imperfect overlap across the six data streams, which we allowed to maximize the statistical power of each independent analysis.
Transcript-level differential expression analysis.
While all biotypes were retained in the transcript-level differential expression analysis, interpretation and visualization were limited to protein-coding transcripts and microRNAs, based on resources currently available for annotation of biological roles. The limma package (39) was used to perform linear mixed models to compare transcript expression in each group over time, as well as any potential interaction of group and time, as described (40). To calculate predicted variance-based weights for each feature, voom normalization was performed across all transcripts (41), and all steps within the pipeline were blocked using participant ID as a random effect to control for between-subject variability. Resultant differentially expressed genes of interest were identified based on dual criteria for fold-change [absolute log2fold-change(FC) >1, corresponding to a doubling of halving of expression] and Benjamini-Hochberg adjusted false discovery rate (FDR < 0.05). These thresholds were uniformly applied to all gene sets to isolate salient and consistent patterns in differential transcript expression.
Differences between groups were qualitatively visualized for all protein-coding transcripts and miRNAs surpassing fold-change and significance thresholds. Protein-coding transcripts (from long RNA) and miRNA (from small RNA) were grouped into contrasts based on directional changes per time point per group. Differentially expressed circRNA were not pursued due to the low number of differentially expressed transcripts and insufficient availability of data for biological annotation, leaving a total of four data streams (protein coding and miRNA for both muscle and serum EV). Within each, “intersections” (i.e., overlaps between transcript sets) were identified for all contrasts using upsetR (42), then ranked according to the number of genes in each intersection. For the eight largest intersections, downstream gene annotation was performed on protein-coding genes to identify biological pathways using overrepresentation analysis in both Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Pathway annotations were filtered for unadjusted P < 0.001.
Exploratory transcriptomic network discovery.
To identify potentially important regulatory relationships across all biotypes, any transcript with an FDR < 0.2 versus preexercise at any postexercise time point was considered responsive to acute exercise and included in the discovery analysis (an example for h3 vs. preexercise in skeletal muscle and serum EVs of the TRAD group is shown in Supplemental Fig. S1). This threshold was applied to reduce noise from unresponsive transcripts while allowing for inclusion of transcripts strongly trending toward significance, so as not to limit the discovery potential of the analysis. Separately but in parallel, all responsive transcripts within muscle (n = 21,320) and serum EVs (n = 56,407) were pooled and prepared for downstream analysis using pathway-level information extractor (PLIER) (43). Tissues were handled separately due to the differential impact of exercise on body compartments [e.g., only ∼33% (18,586 of 56,407) of genes meeting criteria for exercise responsiveness in serum EVs met the criteria in muscle], as well as the incompatible number of subjects between the two data streams as a result of strict upstream filtering (e.g., 96 of 120 samples in muscle were represented in serum EVs).
The PLIER algorithm is built around a singular value decomposition (SVD) anchored to biological pathway annotations for genes, as previously described (43) and used by our research team (16, 17). Presently, however, prior knowledge information was not provided to allow all unannotated transcripts (and therefore all biotypes) to remain in the algorithm; thus, PLIER essentially performed unsupervised SVD clustering across all transcripts to summarize patterns in the data independently of transcript biotype and/or known biological roles. The resulting data structures are known as latent variables, or LVs (43) and represent the variation across all samples. Each transcript is given a value within the dimension of the newly defined LV [matrix Z (43)], whereas each sample is given a score representing the LV [matrix B (43)]. Based on the structure of each data stream, PLIER identified 40 LVs from muscle and 32 LVs from serum EV responsive transcripts.
Using the generated scores, LVs were tested across samples using a group × time analysis of variance (ANOVA) design approximately analogous to that applied during the transcript-level analysis, and a correction factor for the number of LVs was applied to all ANOVA results to identify LVs of interest (P < 0.05/number of LVs). Where an LV reached this strict threshold (i.e., P < 0.00125 in skeletal muscle and P < 0.00156 in serum EVs), it was followed up with a post hoc test to identify the contrast for which a main or interaction effect was detected. All post hoc tests were adjusted internally using a Benjamini-Hochberg adjustment for multiple comparisons; no additional adjustment was performed for the total number of LVs. All LVs surviving post hoc testing were pursued for downstream interpretation. To strategically reduce the number of features pertinent to each LV, top transcripts were defined as any with a loading value greater than half the loading value for the top transcript in the LV (e.g., if the top-loaded transcript had a value of 0.70, all transcripts with values <0.35 were excluded). Transcripts were grouped by biotype and visualized to represent the biological makeup of the LV, and the protein-coding fraction of the LV was annotated using an approach identical to that described above (Gene Ontology and KEGG overrepresentation analysis). When possible, pathway annotations were filtered for P < 0.001; where this approach yielded nothing, nominally significantly related pathways (P < 0.05) were presented. Finally, LVs were classified based on general temporal pattern independently of direction into “early response” (i.e., different at h0 and/or h3 post-exercise), “late response” (i.e., different at h24 post-exercise), or “sustained response” (i.e., different at h0 and/or h3 point and not returned to preexercise by h24).
RESULTS
Transcript-Level Differential Expression Analysis
Skeletal muscle long RNA.
Differential skeletal muscle long transcript expression patterns following the acute exercise challenge are shown in Table 3. Based on the imposed thresholds, a total of 811 long transcripts (54% protein-coding) were upregulated at h3 postexercise versus preexercise in the TRAD group, whereas 425 long transcripts (also 54% protein-coding) were upregulated in HITT; of these, 352 (206 protein-coding) were shared. Conversely, 2,316 long transcripts (24% protein-coding) were downregulated in TRAD and 726 (22% protein-coding) in HITT at h3 postexercise, with 584 (108 protein-coding) in common. At h24 postexercise, a total of 932 (77% protein-coding) transcripts were upregulated versus preexercise in TRAD, whereas 783 (73% protein-coding) transcripts were upregulated in HITT; of these, 481 (396 protein-coding) were shared. TRAD also showed downregulation of 1,058 long transcripts (31% protein-coding), while 479 (37% protein-coding) were downregulated in HITT, and 341 (131 protein-coding) transcripts were shared. Figure 2 shows the patterns in protein-coding genes at h3 (Fig. 2A) and h24 (Fig. 2B) postexercise with respect to the FDR and log2fold-change versus preexercise in TRAD only, given the overall greater magnitude of the acute skeletal muscle long transcript expression response in TRAD. Green circles represent transcripts where a significant change in expression was also detected in HITT.
Table 3.
Number of differentially expressed transcripts in skeletal muscle
| Exercise Stimulus | All Long RNA | Protein-Coding | All Small RNA | Micro RNA | Circular RNA | |
|---|---|---|---|---|---|---|
| h3 vs. pre-exercise | ||||||
| Up | TRAD | 811 | 434 | 52 | 37 | 4 |
| HITT | 425 | 230 | 2 | 2 | 0 | |
| Down | TRAD | 2,316 | 546 | 16 | 15 | 7 |
| HITT | 726 | 160 | 0 | 0 | 0 | |
| h24 vs. pre-exercise | ||||||
| Up | TRAD | 932 | 721 | 100 | 41 | 1 |
| HITT | 783 | 575 | 77 | 21 | 0 | |
| Down | TRAD | 1,058 | 324 | 8 | 5 | 2 |
| HITT | 479 | 176 | 3 | 3 | 0 | |
All transcripts satisfied both |log2FC| >1 and false discovery rate < 0.05. Down, downregulated; Up, upregulated.
Figure 2.
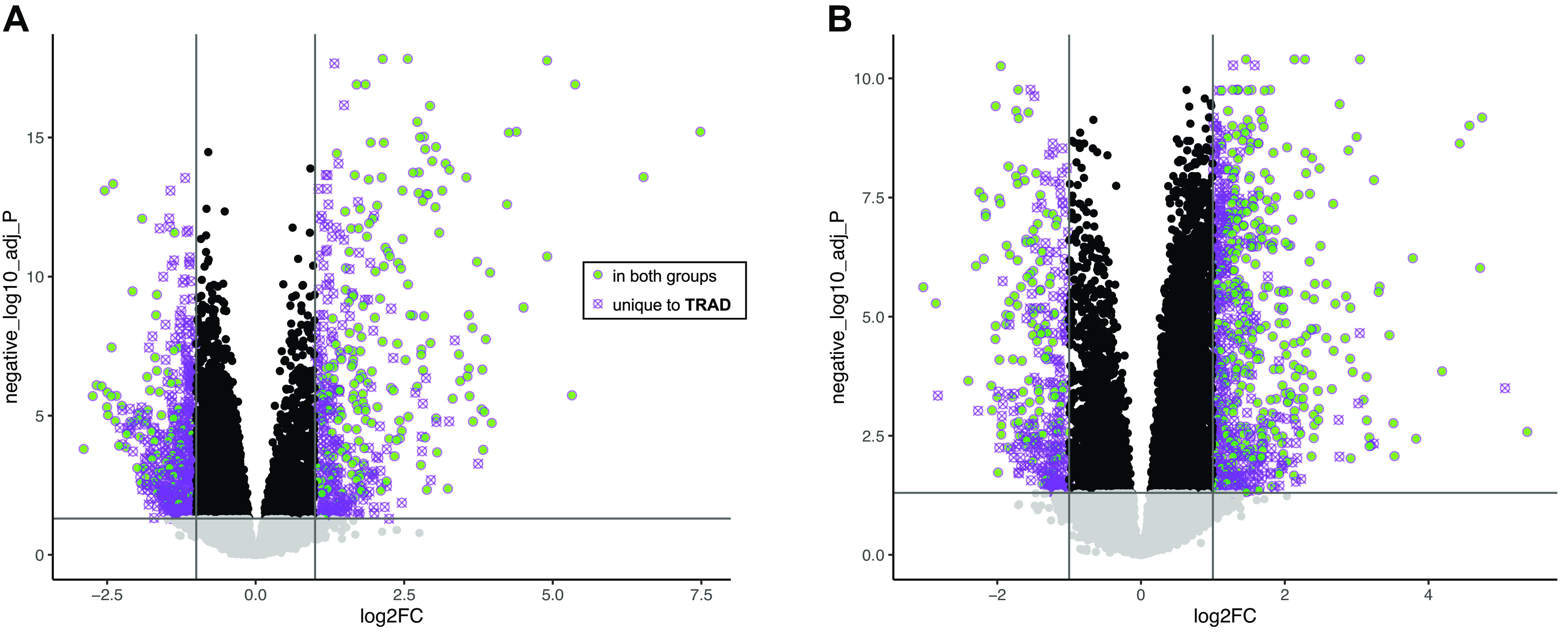
Volcano plot showing changes in protein-coding transcript expression h3 (A) and h24 (B) postexercise in skeletal muscle of individuals performing TRAD exercise (|log2FC| >1, FDR < 0.05). Transcripts meeting identical thresholds in HITT are represented as green circles, whereas those shown as crossed-out purple circles were only significant in TRAD (exact values plotted for log2FC and FDR correspond only to TRAD). FDR, false discovery rate; HITT, high-intensity tactical training; TRAD, traditional exercise.
To establish insight into coordinated patterns in gene expression across time for both exercise protocols, upsetR plots were constructed from the lists of up- and downregulated protein-coding transcripts at each time point versus preexercise (Fig. 3A). The largest intersection contained 356 genes uniquely downregulated in TRAD at h3. Follow-up annotation revealed associations to Gene Ontology (GO) pathways including cell adhesion (GO:0007155, P = 1.5 × 10−7), inflammatory response (GO:0006954, P = 5.2 × 10−7), and cell activation (GO:0001775, P = 8.5 × 10−7), among others. A common set of 282 genes upregulated in both groups at h24 postexercise annotated primarily to metabolic processes related to glycerol-3-phosphate (GO:0006072; P = 1.1 × 10−5), alditol phosphate (GO:0052646, P = 2.3e−5), and carbohydrate (GO:0005975, P = 7.3 × 10−5), among other pathways. These and further annotations for the top eight intersections (Fig. 3A), where available, are shown in Supplemental Table S1.
Figure 3.
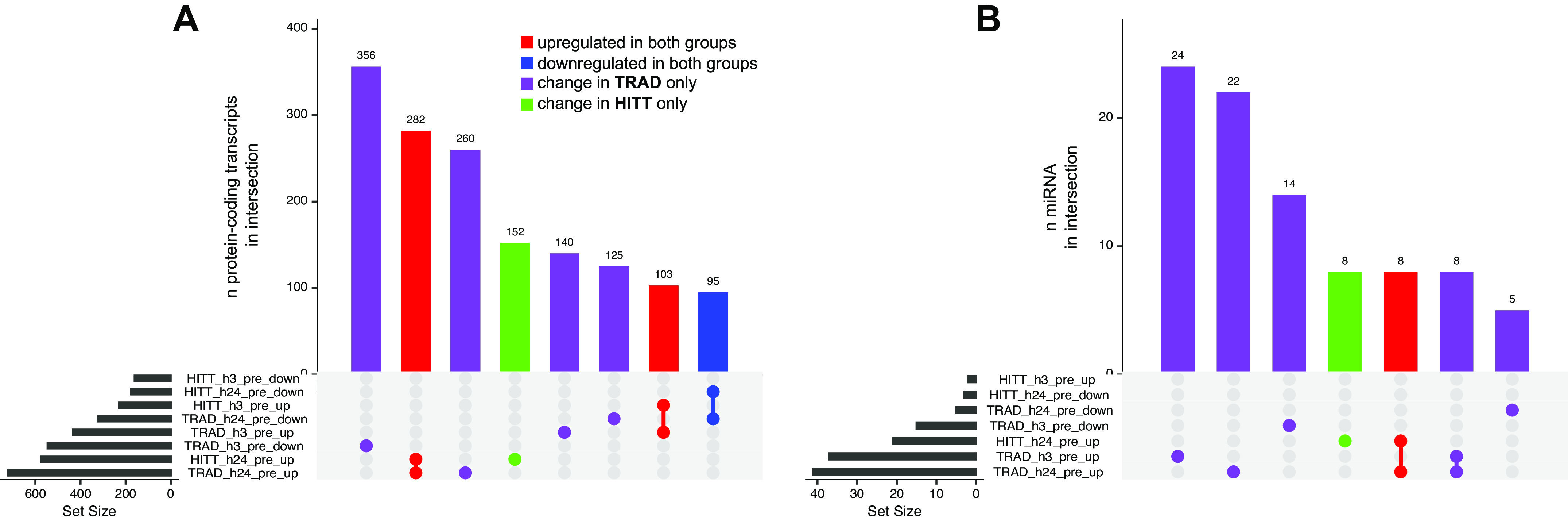
UpsetR plots indicating shared overlap (intersection) of directional transcript expression changes vs. preexercise in protein-coding transcripts (A) and miRNA (B) in skeletal muscle. Bar height represents the number of genes in each intersection, whereas dots represent comparison(s) in which transcripts were significantly up- or downregulated vs. preexercise (|log2FC| >1 and false discovery rate < 0.05). Gene intersections are shown in red where upregulation was detected at any time point independently of group, in blue where downregulation was detected independently of group, in purple where a change was detected in traditional exercise (TRAD) only, and in green where a change was detected in high-intensity tactical training (HITT) only. Gene Ontology analysis for each intersection in A is available in Supplemental Table S1.
Skeletal muscle small RNA.
At h3 postexercise versus preexercise, 52 small transcripts (71% miRNA) were upregulated in TRAD, whereas 2 of these (both miRNAs) were upregulated in HITT. Conversely, 16 transcripts (94% miRNA) were downregulated in TRAD at h3, although none was significantly downregulated in HITT. At h24 postexercise versus preexercise, a total of 100 small transcripts (41% miRNA) were upregulated in TRAD, whereas 77 small transcripts (27% miRNA) were upregulated in HITT; of these, 40 (11 miRNA) were shared. TRAD also showed downregulated expression of eight small transcripts (5 miRNA), while three (all miRNA) were downregulated in HITT, though there was no overlap between groups. Supplemental Fig. S2 shows the patterns in miRNA at h3 (Supplemental Fig. S2A) and h24 (Supplemental Fig. S2B) postexercise with respect to the FDR and log2fold-change versus preexercise in TRAD only, with filled shapes representing transcripts where a significant change in expression was also detected in HITT. As shown in Fig. 3B, the small transcript response in TRAD was largely unique to this cohort, as the largest three intersections contained miRNA with unique responses in TRAD.
Skeletal muscle circular RNA.
At h3 postexercise versus preexercise in TRAD, four circRNAs were upregulated and seven were downregulated. At h24 versus preexercise in TRAD, one circRNA was upregulated and two were downregulated. No circRNAs were significantly different from preexercise at any point in HITT. Given that the response in skeletal muscle circular RNA was overall more muted compared with the other data streams, these acute changes are not visually presented.
Serum EV long RNA.
Differential serum EV long transcript expression patterns following the acute exercise challenge are shown in Table 4. A total of 33,029 long transcripts (13% protein-coding) were upregulated at h0 postexercise versus preexercise in TRAD, whereas 15,664 (perhaps notably, also 13% protein coding) were upregulated in HITT. Of these, 14,141 (1,682 protein-coding) were common to both groups. Conversely, 259 long transcripts (81% protein-coding) were downregulated in TRAD and 223 (82% protein-coding) in HITT at h0 pos-exercise, with 59 (55 protein-coding) in common. At h3 postexercise versus preexercise, a total of 29,766 long transcripts (13% protein-coding) were upregulated in TRAD, whereas 20,629 (14% protein-coding) transcripts were upregulated in HITT; of these, 17,762 (2,370 protein-coding) were shared. TRAD also exhibited downregulation of 198 long transcripts (75% protein-coding), whereas 374 (83% protein-coding) were downregulated in HITT, and 156 (128 protein-coding) transcripts were shared. Finally, at h24 postexercise versus preexercise, a total of 26,895 (12% protein-coding) transcripts were upregulated in TRAD, whereas 16,018 (14% protein-coding) transcripts were upregulated in HITT; of these, 13,310 (1,716 protein-coding) were shared. TRAD also led to downregulation of 78 long transcripts (71% protein-coding), while 177 (78% protein-coding) were downregulated in HITT, and 59 (47 protein-coding) transcripts were common to both groups. Supplemental Fig. S3 shows the patterns in protein-coding transcripts at h0 (Supplemental Fig. S3A), h3 (Supplemental Fig. S3B), and h24 (Supplemental Fig. S3C) postexercise with respect to the FDR and log2FC versus preexercise in TRAD only, with filled shapes representing where a significant change in expression was also detected in HITT.
Table 4.
Number of differentially expressed transcripts in serum extracellular vesicles
| Exercise Stimulus | All Long RNA | Protein-Coding | All Small RNA | Micro RNA | Circular RNA | |
|---|---|---|---|---|---|---|
| h0 vs. pre-exercise | ||||||
| Up | TRAD | 33,029 | 4,197 | 2 | 2 | 1 |
| HITT | 15,664 | 2,056 | 24 | 9 | 30 | |
| Down | TRAD | 259 | 211 | 0 | 0 | 0 |
| HITT | 223 | 183 | 0 | 0 | 6 | |
| h3 vs. pre-exercise | ||||||
| Up | TRAD | 29,766 | 3,967 | 4 | 0 | 0 |
| HITT | 20,629 | 2,975 | 8 | 2 | 2 | |
| Down | TRAD | 198 | 149 | 52 | 47 | 0 |
| HITT | 374 | 309 | 148 | 139 | 3 | |
| h24 vs. pre-exercise | ||||||
| Up | TRAD | 26,895 | 3,272 | 7 | 0 | 0 |
| HITT | 16,018 | 2,285 | 0 | 0 | 3 | |
| Down | TRAD | 78 | 55 | 50 | 49 | 0 |
| HITT | 177 | 138 | 126 | 121 | 1 | |
All transcripts satisfied both |log2FC| >1 and false discovery rate < 0.05. HITT, high-intensity tactical training; down, downregulated; TRAD, traditional exercise; Up, upregulated.
As shown in Fig. 4A, the largest intersection contained 1,199 genes commonly upregulated at all postexercise time points in both groups. GO analysis revealed these genes were primarily associated with protein-containing complex organization (GO:0043933, P = 9.3 × 10−10), organelle organization (GO:0006996, P = 7.8 × 10−9), and cellular component assembly (GO:0022607, P = 1.1 × 10−8), among others. The next largest intersection contained 839 genes upregulated at every postexercise time point in TRAD only. These genes are primarily annotated to biological processes such as plasma membrane-bounded cell projection organization (GO:0120036, P = 3.6 × 10−6) and neuron development (GO: 0048666, P = 1.1 × 10−5). Other annotations, where available, are presented in Supplemental Table S2.
Figure 4.
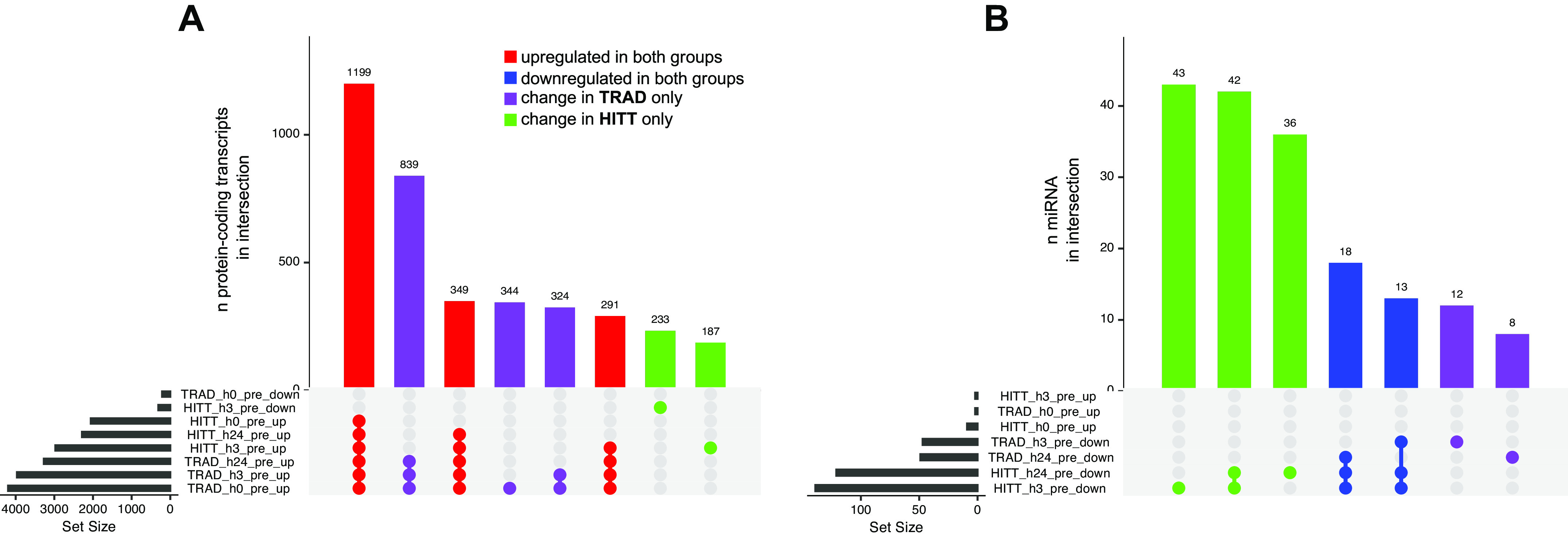
UpsetR plots indicating shared overlap (intersection) of directional transcript expression changes vs. preexercise in protein-coding transcripts (A) and miRNA (B) in serum extracellular vesicles (EVs). Bar height represents the number of genes in each intersection, whereas dots represent comparison(s) in which transcripts were significantly up- or downregulated vs. preexercise (|log2FC| >1 and false discovery rate < 0.05). Gene intersections are shown in red where upregulation was detected at any time point independently of group, in blue where downregulation was detected independently of group, in purple where a change was detected in traditional exercise (TRAD) only, and in green where a change was detected in high-intensity tactical training (HITT) only. Gene Ontology analysis for each intersection in A is available in Supplemental Table S2.
Serum EV small RNA.
Following acute exposure to TRAD, 2 small transcripts were upregulated at h0 postexercise versus preexercise (both miRNA), compared with 24 (38% miRNA) in HITT, including the 2 seen in TRAD. No small transcripts in serum EVs were significantly downregulated from preexercise in either group at this time point. At h3 postexercise in TRAD, four small transcripts (0% miRNA) were upregulated, and eight small transcripts (25% miRNAs) were upregulated in HITT, including the four significantly changed in TRAD. Conversely, 52 transcripts (90% miRNA) were downregulated in TRAD at h3 versus preexercise, and 148 (94% miRNA) were downregulated in HITT. Of these, 32 (30 miRNA) were shared between groups. Finally, at h24 postexercise versus preexercise, a total of seven small transcripts (no miRNA) were upregulated in TRAD, and none were significantly changed in HITT. A total of 50 small transcripts (98% miRNA) were downregulated in TRAD compared with 126 (96% miRNA) in HITT. Of these, 29 (all miRNA) were common to both groups. Because this data stream exhibited a contrary pattern wherein HITT elicited a larger gene expression response, Supplemental Fig. S4 shows the patterns in miRNA at h0 (Supplemental Fig. S4A), h3 (Supplemental Fig. S4B), and h24 (Supplemental Fig. S4C) postexercise with respect to the FDR and log2FC versus preexercise in HITT only, with filled shapes representing where a significant change in expression was also detected in TRAD. Figure 4B shows that the three largest intersections in small RNA from serum EVs were unique to HITT only.
Serum EV circular RNA.
At h0 postexercise versus preexercise, 1 circRNA was upregulated and 0 downregulated in TRAD versus 30 upregulated and 6 downregulated in HITT. No significant changes were seen in TRAD circRNA expression at h3 or h24 postexercise versus preexercise. However, at h3 postexercise versus preexercise, two circRNA were upregulated and three downregulated in HITT. At h24 versus preexercise, three were upregulated and one downregulated in HITT. Given that the response in serum EV circular RNA was overall more muted compared with the other data streams, these acute changes are not visually presented.
Exploratory Transcriptomic Network Discovery
Regulatory networks in skeletal muscle.
Skeletal muscle transcript expression networks were constructed from a total of 21,320 “responsive” transcripts that were different from preexercise at any time point based on FDR < 0.20. PLIER-guided SVD generated 40 LVs from skeletal muscle transcripts, of which 23 showed a significant (P < 0.00125) main effect of exercise (11 early, 6 late, 6 sustained). Table 5 shows the manually selected biological annotations associated with each of the 23 LVs, derived from detailed results of GO/KEGG enrichment analysis available in Supplemental Table S3. LVs demonstrating a group × time interaction effect at the post hoc level of significance are presented in Fig. 5, A–E, along with the proportion of other RNA biotypes associated with the LV that passed 50% of the maximum loading threshold. A full list of all transcripts above this threshold is available for each of the 23 LVs in Supplemental Table S4. In particular, skeletal muscle transcript LVs 1 (intracellular signaling) and 34 (lipid metabolism) were significantly different at h3 postexercise in both groups and also different at h3 between HITT versus TRAD. LVs 4 (kinase activity, muscle tissue development) and 14 (negative regulation of muscle tissue development) were significantly different at h3 postexercise in TRAD and different from HITT at the same time point. LV20 (skin and cardiac muscle development, hormone response) was different at h3 postexercise in HITT and different from TRAD at h3; within TRAD, LV20 was different at h3 versus h24.
Table 5.
Values for LVs of interest in the muscle transcriptomics network analysis for each exercise dose
| TRAD |
HITT |
||||||
|---|---|---|---|---|---|---|---|
| LV | Relevant Biological Annotation | Pre | h3 | h24 | Pre | h3 | h24 |
| Early response§ | |||||||
| LV1 | intracellular signaling (STAT, MAPK) | −0.337 ± 0.438 | 0.757 ± 1.077*‡ | −0.193 ± 0.391 | −0.327 ± 0.276 | 0.257 ± 0.533* | −0.181 ± 0.303 |
| LV4 | kinase activity, muscle tissue development | −0.126 ± 0.265 | 0.583 ± 0.698*‡ | −0.213 ± 0.22 | −0.161 ± 0.187 | 0.053 ± 0.351 | −0.162 ± 0.177 |
| LV5 | protein metabolism, post-translational modification | 0.567 ± 1.075 | −0.736 ± 0.705† | 0.004 ± 1.315 | 0.167 ± 1.084 | −0.202 ± 1 | 0.216 ± 1.318 |
| LV9 | AMPK signaling, mitochondrial function | −0.16 ± 0.232 | 0.505 ± 0.975* | −0.291 ± 0.373 | 0.061 ± 1.076 | 0.195 ± 0.327 | −0.317 ± 0.218 |
| LV13 | cell respiration, aerobic metabolism | −0.596 ± 0.798 | 0.664 ± 1.178* | −0.088 ± 1.012 | −0.216 ± 1.142 | 0.381 ± 0.935 | −0.143 ± 0.659 |
| LV14 | negative regulation of muscle tissue development | −0.115 ± 0.273 | 0.554 ± 1.144*‡ | −0.332 ± 0.344 | −0.036 ± 0.207 | 0.114 ± 0.416 | −0.197 ± 0.373 |
| LV15 | protein metabolism | 0.119 ± 0.835 | −0.312 ± 0.622ϕ | 0.411 ± 1.378 | 0.13 ± 0.625 | −0.665 ± 0.559* | 0.294 ± 1.244 |
| LV20 | skin and cardiac muscle development, hormone response | −0.221 ± 0.172 | 0.159 ± 0.285ϕ | −0.267 ± 0.251 | −0.039 ± 0.486 | 0.609 ± 1.263*‡ | −0.207 ± 0.15 |
| LV21 | MAPK, Wnt signaling | −0.231 ± 0.206 | 0.283 ± 0.216 | −0.153 ± 0.287 | −0.097 ± 0.258 | 0.386 ± 1.567 | −0.176 ± 0.285 |
| LV32 | histamine metabolism | −0.047 ± 0.137 | 0.101 ± 0.124 | −0.194 ± 0.158 | −0.087 ± 0.174 | 0.38 ± 1.143* | −0.138 ± 0.128 |
| LV34 | lipid metabolism | −0.235 ± 0.321 | 0.636 ± 0.907*‡ | −0.206 ± 0.3 | −0.153 ± 0.382 | 0.264 ± 0.445* | −0.326 ± 0.214 |
| Late response | |||||||
| LV8 | muscle contraction and development | −0.361 ± 0.185 | −0.06 ± 0.69 | 0.464 ± 0.806* | −0.385 ± 0.206 | −0.101 ± 0.517 | 0.439 ± 0.71* |
| LV12 | ribosome activity, RNA processing | −0.652 ± 0.438 | −0.513 ± 0.344 | 1.094 ± 1.815* | −0.509 ± 0.434 | −0.426 ± 0.568 | 1.015 ± 1.707* |
| LV17 | IL-8 signaling, synaptic transmission | −0.275 ± 0.244 | −0.354 ± 0.175 | 0.804 ± 1.587* | −0.345 ± 0.24 | −0.288 ± 0.315 | 0.44 ± 0.54* |
| LV19 | axon guidance, chemotaxis | 0.023 ± 0.244 | −0.209 ± 0.209ϕ | 0.305 ± 0.982 | −0.088 ± 0.16 | −0.143 ± 0.333 | 0.098 ± 0.257 |
| LV22 | RNA metabolism | −0.425 ± 0.219 | 0.006 ± 0.418 | 0.579 ± 1.629† | −0.262 ± 0.258 | −0.233 ± 0.458 | 0.319 ± 0.685 |
| LV38 | protein metabolism and assembly | −0.162 ± 0.477 | −0.438 ± 0.32ϕ | 0.229 ± 0.492 | −0.066 ± 0.62 | −0.196 ± 0.623 | 0.673 ± 1.114* |
| Sustained response | |||||||
| LV16 | response to pain, meiosis | 0.329 ± 0.932* | −0.114 ± 0.37 | −0.11 ± 0.301 | 0.034 ± 0.21 | −0.019 ± 0.188 | −0.131 ± 0.175 |
| LV18 | membrane fusion, focal adhesion | 0.474 ± 1.007* | −0.09 ± 0.295 | −0.272 ± 0.221 | 0.23 ± 0.389* | −0.163 ± 0.136 | −0.19 ± 0.256 |
| LV29 | inflammatory signaling (NFκB, JNK) | 0.158 ± 0.197 | −0.185 ± 0.159 | −0.202 ± 0.216 | 0.254 ± 1.016 | 0.036 ± 0.517 | −0.036 ± 0.55 |
| LV30 | energy metabolism | 0.647 ± 0.774* | −0.546 ± 0.313 | −0.398 ± 0.584 | 0.542 ± 0.748 | −0.447 ± 0.301† | 0.234 ± 2.041 |
| LV33 | calcium signaling | 0.481 ± 0.699* | −0.172 ± 0.23 | −0.329 ± 0.287 | 0.381 ± 0.59* | −0.194 ± 0.253 | −0.165 ± 0.526 |
| LV36 | leukocyte chemotaxis | 0.298 ± 0.924* | −0.058 ± 0.276 | −0.057 ± 0.346 | 0.068 ± 0.225 | −0.076 ± 0.202 | −0.194 ± 0.148 |
Values are means ± SD. Emergent biology annotation based on predominant results in GO/KEGG parallel annotation as shown in Supplemental Table S3. LV, latent variable. §LV temporal response patterns based on significant differences across time in pooled groups. Early, different at h3 vs. all other time points; late, different at h24 vs. all other time points; sustained, different at h3 and h24 vs. preexercise. Differences are not based on directionality. *Difference vs. other time points within group. ‡Difference between groups. †Difference vs. preexercise time point within group. ϕDifference vs. h24 time point within group.
Figure 5.
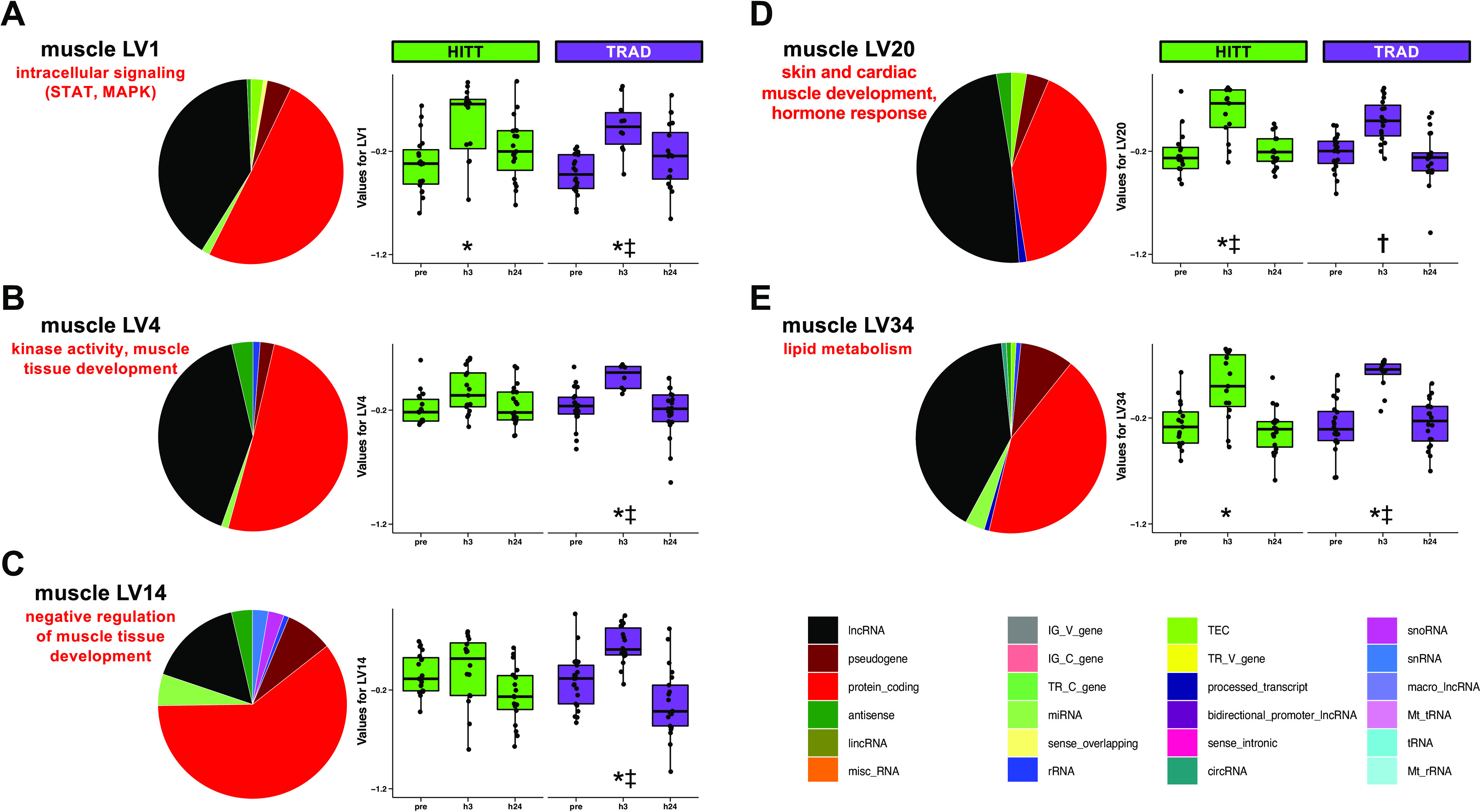
Latent variables (LVs) in skeletal muscle where a group × time difference in LV was detected via post hoc testing of Pathway-Level Information Extractor (PLIER)-generated LVs. For each LV (A–E), the fraction of transcripts most highly representative of the LV is summarized for biotype, and then the protein-coding fraction of each LV was annotated using a combination of Gene Ontology and Kyoto Encyclopedia of Genes and Genomes, as summarized on the right of each pie chart. Additional LVs exhibited a main effect of exercise across both groups and are shown in Table 5. *Post hoc P < 0.05 vs. preexercise within group; ‡post hoc P < 0.05 vs. other group at the same time point; †post hoc P < 0.05 vs. h24 time point within group. TRAD, traditional exercise; HITT, high-intensity tactical training.
Networks in serum extracellular vesicles.
Serum EV transcript expression networks were constructed from a total of 56,407 “responsive” transcripts that were different from preexercise at any time point based on FDR < 0.20. PLIER-guided SVD generated 32 LVs from serum EV transcripts, of which 4 showed a significant (P < 0.00156) main effect of exercise. Table 6 shows the general biological annotations associated with each of the four LVs, derived from detailed results of GO/KEGG analysis available in Supplemental Table S5. These four LVs are presented in Fig. 6, A–D, along with the proportion of other RNA biotypes associated with the LV that passed 50% of the maximum loading threshold. A list of all transcripts above this threshold is available for each of the four LVs in Supplemental Table S6. Briefly, serum EV transcript LVs 10 (protein metabolism and biosynthesis), 17 (leukocyte activation and immune response), and 19 (blood coagulation and platelet biology) were significantly different at every postexercise time point versus preexercise in TRAD only. Serum EV LV2 (sensory perception and cell mobility) was different postexercise overall but did not survive post hoc tests when the groups were separated.
Table 6.
Values for LVs of interest in the serum EV transcriptomics network analysis for each exercise dose
| TRAD |
HITT |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| LV | Relevant Biological Annotation | Pre | h0 | h3 | h24 | Pre | h0 | h3 | h24 |
| Sustained response§ | |||||||||
| LV2 | Sensory perception and cell mobility | −0.853 ± 0.773 | −0.224 ± 1.138 | 0.109 ± 1.192 | 0.061 ± 0.954 | 0.06 ± 2.201 | −0.186 ± 0.751 | 0.658 ± 1.183 | 0.793 ± 1.058 |
| LV10 | Protein metabolism and biosynthesis | 1.169 ± 2.963* | −0.312 ± 0.369 | −0.229 ± 0.536 | 0.011 ± 0.497 | 0.159 ± 0.868 | −0.348 ± 0.294 | −0.431 ± 0.242 | −0.326 ± 0.309 |
| LV17 | Leukocyte activation and immune response | 0.93 ± 2.296* | −0.248 ± 0.376 | −0.165 ± 0.634 | −0.001 ± 0.414 | 0.134 ± 0.704 | −0.265 ± 0.355 | −0.37 ± 0.19 | −0.262 ± 0.326 |
| LV19 | Blood coagulation and platelet biology | 0.842 ± 1.137* | −0.261 ± 0.409 | −0.336 ± 0.378 | −0.074 ± 0.467 | 0.935 ± 2.618 | −0.329 ± 0.288 | −0.532 ± 0.2 | −0.402 ± 0.297 |
Values are means ± SD. LV, latent variable. Emergent biology annotation based on predominant results in GO/KEGG parallel annotation as shown in Supplemental Table S5. §LV temporal response patterns based on significant differences across time in pooled groups. Sustained, different at h0, h3, and h24 vs. preexercise (LV2 follows general pattern but was not statistically different at h0 vs. pre). Differences are not based on directionality. HITT, high-intensity tactical training; TRAD, traditional exercise. *Significant difference vs. other time points within group.
Figure 6.
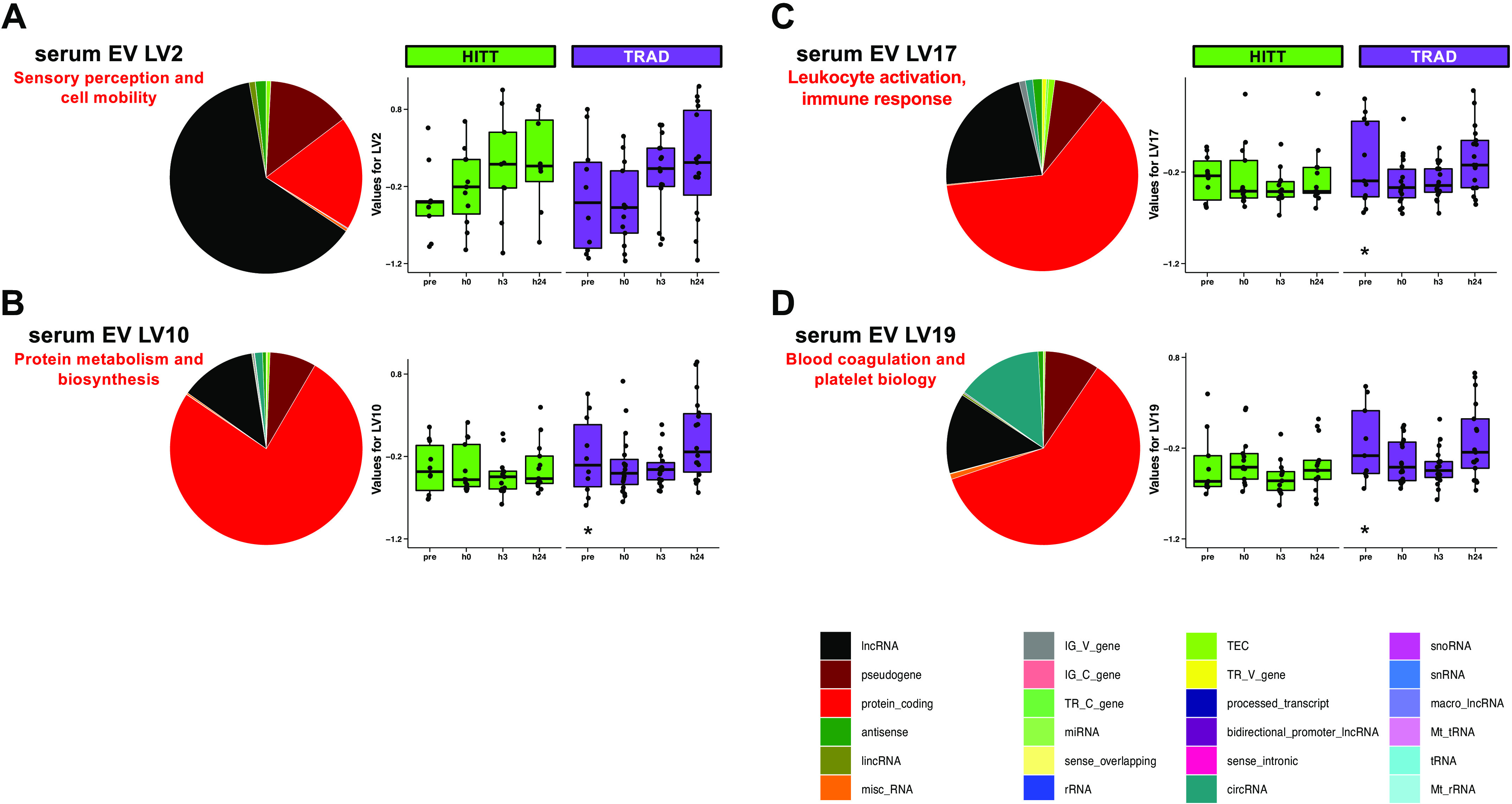
Latent variables (LVs) in serum extracellular vesicles (EVs) where a difference in LV was detected via post hoc testing of Pathway-Level Information Extractor (PLIER)-generated LVs. For each LV (A–D), the fraction of transcripts most highly representative of the LV is summarized for biotype, and then the protein-coding fraction of each LV was annotated using a combination of Gene Ontology and Kyoto Encyclopedia of Genes and Genomes, as summarized on the right of each pie chart. *Post hoc P < 0.05 vs. pre-exercise within group. TRAD, traditional exercise; HITT, high-intensity tactical training.
DISCUSSION
This study represents a keystone for exercise ‘omics in presenting a highly practical and more widely applicable dataset of mechanistic gene expression profiling in response to two relatively understudied exercise stimuli in both male and female young adults. This is particularly valuable, as acute exercise provides insight into molecular patterns that may form the basis of adaptations conferred through repeated exposures over the long term (i.e., consistent exercise training). This could aid in establishing direction toward personalized exercise training prescriptions that leverage interindividual variation in molecular phenotype and associated whole body phenotypes. In the present study, we compared the acute transcriptomic responses to two innovative and potent yet distinct, combined exercise prescriptions and examined the interrelationships across transcript biotypes. Our findings provide insight into the temporal dynamics of the transcriptome circuitry and patterns that may suggest regulatory or other molecular interactions among transcripts. To our knowledge, this is the largest molecular mapping study of its kind to date in terms of sample size, comparisons across two randomized combined exercise stimuli, and depth and breadth of skeletal muscle and serum EV transcriptomic profiling.
Impact of Exercise Stimulus
Concurrent aerobic and resistance exercise is highly recommended by physical activity guidelines for the maintenance of cardiovascular fitness and strength (1, 44), but the molecular map of integrated effects of these modalities is often not a focus of research (20, 45). Considering this, the present study addresses a knowledge gap in examining the acute response to traditional combined exercise and further builds upon this innovation by comparing it to HITT, an emerging popular and effective training stimulus. In both the transcript-level and network-level analyses, the overall transcriptomic response was larger and/or more consistent across individuals in TRAD versus HITT, although the composition of differentially expressed transcripts in both serum EV and skeletal muscle (e.g., % protein-coding fraction) appeared generally similar between groups. While it may be argued that similar overall directional patterns were evident in HITT and declaration of significant difference was restricted by the rigorous yet still arbitrary significance and fold-change cutoffs (Fig. 2), this effect would likely be seen regardless of the specific thresholds imposed. Thus, it is potentially important that in well-balanced, similarly sized groups with no apparent different physical characteristics at baseline, a larger response consistently emerged in TRAD. A potential explanation for these findings is that the TRAD stimulus lasted approximately twice as long in duration as HITT, and this might account for lower variability across individuals by the time the biopsies and blood draws were collected. However, the more likely explanation is that the longer exercise regimen presented a sustained, unique metabolic challenge not experienced by participants undergoing HITT. Presently, we are unable to disentangle these effects, and this represents a limitation of this study. Detection of truly divergent responses might require a larger sample size to account for interindividual response heterogeneity in the exercise-related response ability to detect changes. Nevertheless, heterogeneity itself is important and may present opportunities for follow-up research, particularly as it relates to prescription of and adaptation to HITT across individuals.
On the contrary, miRNA in serum EVs exhibited a greater response following HITT exercise, with most of these downregulated (Fig. 4B). As shown in Supplemental Fig. S4, these differences do not tend to cluster below a particular fold-change cutoff, as was apparent in the other data streams. While we did not assess protein-level changes in response to acute exercise, it would be informative to establish whether chronic exposure to HITT might differentially impact the abundance of critical downstream proteins that modulate some adaptations.
A comparison to a tissue enrichment atlas developed by members of our team (46) showed a key miRNA downregulated in HITT only at h24 (hsa-mir-9-3) is exclusively and significantly enriched in central nervous system regions including the cerebellum, cerebral cortex, spinal cord, and brain white matter. This suggests a potential cross talk originating in the nervous system that is unique to the high-intensity stimulus, possibly necessitated by the nature of the bout (a timed sequence of varied functional exercises) and its demand on cognition, concentration, memory, neuromuscular control (balance, proprioception, coordination), maximal motor unit activation, and other neural processes. In support, other neuroactive molecules have been reported to exhibit intensity-dependent effects (47, 48), while accustomed moderate-intensity cycling facilitates reduced cognitive burden during exercise (49). It could be highly informative to establish whether this effect of HITT is sustained over time and has long-term benefits for cognition over TRAD, but this would likely be best illustrated in a population other than young, healthy adults with normal cognitive function.
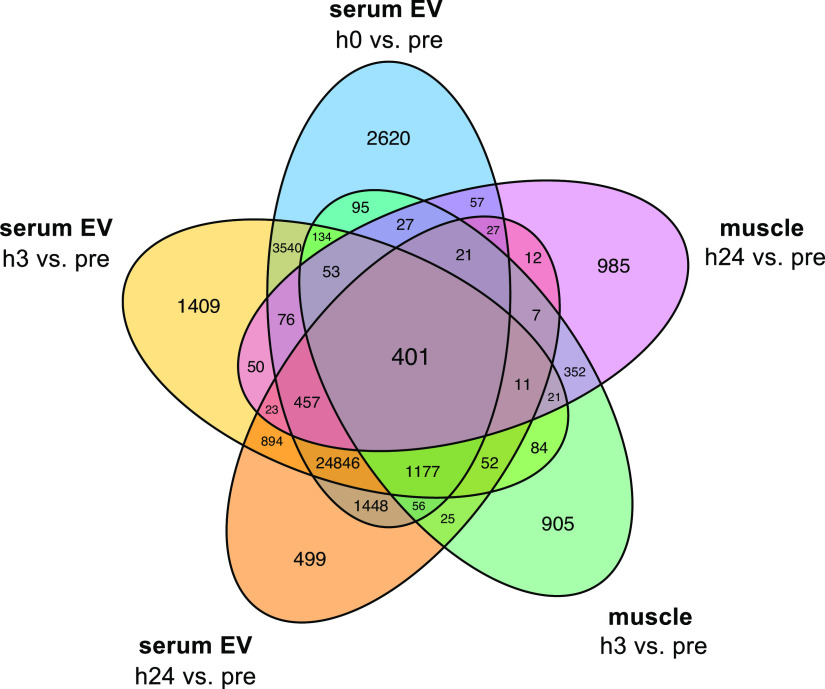
Insight from Serum EVs
Circulating EVs are an integrated and accessible reflection of the exercise “secretome” (50). While a large portion of differentially expressed transcripts was unique to serum EVs, 401 transcripts across all RNA biotypes were significantly impacted at all measured time points within both body compartments (Fig. 7). Despite this, protein-coding transcripts and validated functional targets of differentially expressed miRNA covered a wide and nonoverlapping range of biological processes (Fig. 8), including cytokine signaling, kinase activity, and growth factor activity for the targets of differentially expressed miRNA, and G-protein coupled signaling, hormone activity, and ion channel regulation for differentially expressed protein-coding genes. This is perhaps not entirely surprising, given that miRNAs act posttranscriptionally and would not necessarily be expected to correlate with changes in expression of their potential target genes. Additional interregulatory dynamics, such as tissue cross talk and temporal regulation (e.g., miRNA changing in serum at postexercise h0 impacting translation of mRNA in muscle at postexercise h3 or h24) must be taken into account when attempting to integrate such multidimensional data. For these reasons, the agnostic SVD approach in PLIER provides a solid mathematical basis by which relatedness across transcript biotypes is uncovered. Continued mechanistic experiments to understand their physical interactions are certainly necessary to inform design of future multiomics studies.
Figure 7.
Venn diagram showing shared overlap of differentially expressed transcripts in both muscle and serum extracellular vesicles (EVs) at all postexercise time points (h0, h3, and h24) vs. preexercise (pre). All transcripts satisfying both |log2FC| >1 and false discovery rate < 0.05 were collapsed regardless of directional change or RNA biotype.
Figure 8.
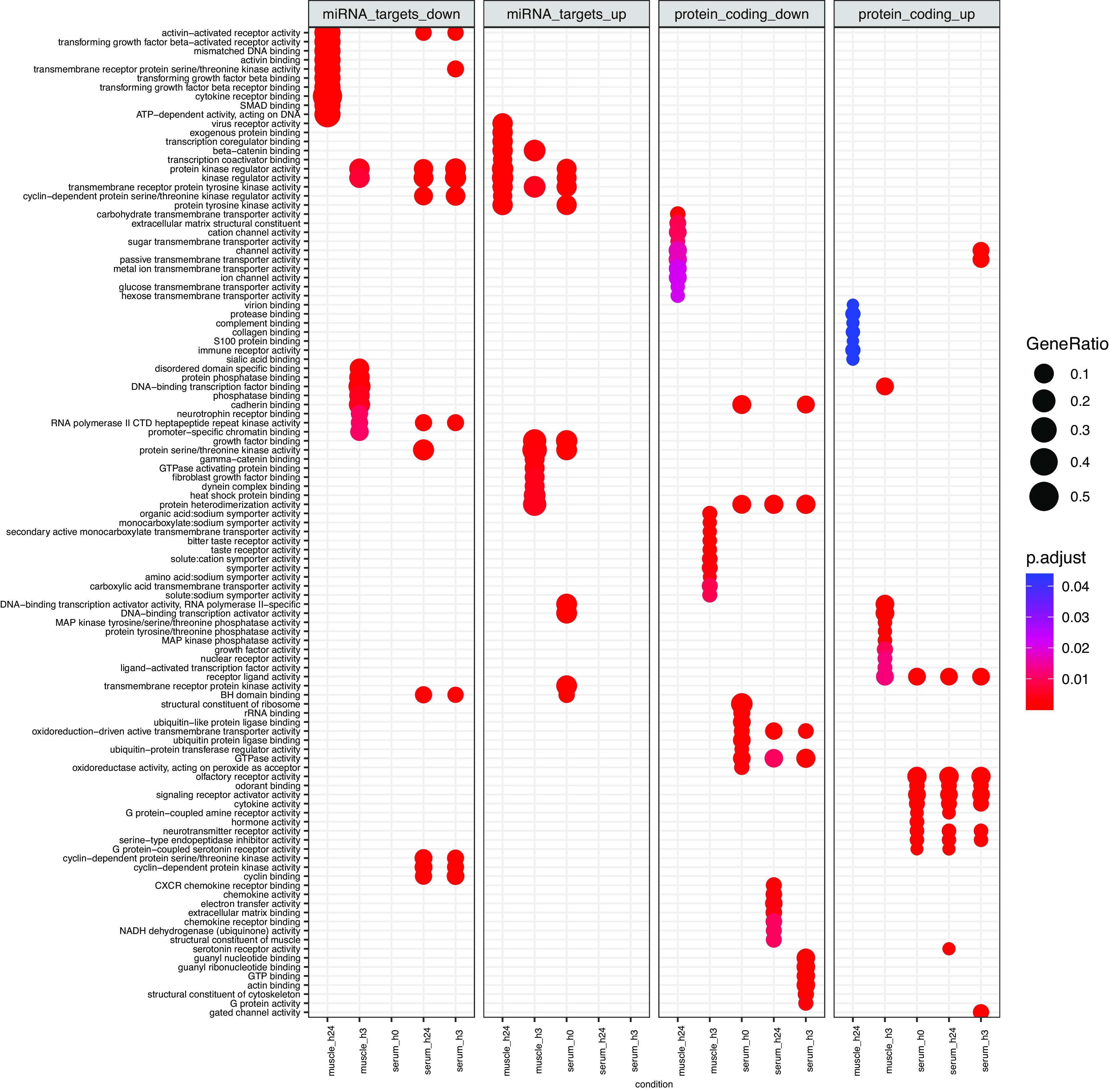
Biological processes associated with differentially expressed transcripts in skeletal muscle and serum extracellular vesicles at all postexercise time points (h0, h3, and h24) with false discovery rate < 0.05 and absolute log2fold-change >1 vs. preexercise, as curated by Gene Ontology. For miRNA targets (left two panels), multiMiR (v1.16) was used to identify the top 5% of predicted protein-coding mRNAs that functionally interact with miRNA differentially expressed following acute exercise. Targets were combined for all miRNA upregulated (up) or downregulated (down) at a given time point within muscle and serum extracellular vesicles and entered into Gene Ontology analysis. Differentially expressed protein-coding transcripts (right two panels) were directly input into Gene Ontology. GeneRatio (circle size) represents the proportion of genes shared between gene query and biological pathway, whereas likelihood of overlap based on the reference genome is represented by p.adjust (circle color).
As an exploratory approach to interpret patterns in the protein-coding transcriptome after acute HITT and TRAD exercise, we used the Human Protein Atlas (51) to map transcripts to protein expression patterns enriched at or above a medium level of confidence and supported reliability or better in bodily tissues of interest (Supplemental Fig. S5). This revealed an integrative whole body response to the acute exercise stimulus via shared genes upregulated at all timepoints in both exercise protocols. Skeletal muscle-enriched genes formed the basis of the largest intersection (n = 18) and were mostly associated with processes of contractile function and muscle development. It could be meaningful to establish whether these EVs may be originating from or targeting skeletal muscle. Tissue-specific EV patterns such as surface markers and/or cargo may be measured for this purpose in future investigations.
The dramatically asymmetrical response in serum EV long RNA (strongly positive) was critically evaluated during quality control and determined not to be the result of technical artifact (e.g., laboratory technician or sample batching). While the asymmetry persisted, it may be of interest that the imbalanced ratio of up- to downregulated transcripts in serum EVs after exercise is notably diminished when considering only the proportion of protein-coding genes, which was remarkably consistent at each postexercise time point (∼12–14% of all differentially expressed transcripts). While the biological roles of the larger, noncoding fraction of differentially expressed transcripts are unclear, this imbalance may suggest an overall transcriptional stress response to acute exercise throughout the body. It would be informative to establish whether this signature persists in the trained state.
PLIER provided key insight into the potential relationship of upregulated transcripts to biological pathways associated with circulating blood cell types. For instance, serum EV LV17 was related to leukocyte activation and immunity, whereas serum EV LV19 was related to clotting and platelet biology. Supporting this as a likely source of variation, a recent study examining the multiomic patterns in a specialized subset of blood cells (e.g., peripheral blood mononuclear cells) found that the transcriptomic response following acute exercise is relatively well-balanced between the number of up- and downregulated genes (21). While insight into more direct contributions of different blood cell types might have been gained by leveraging a prior knowledge database (43), we elected to perform PLIER agnostically to retain unannotated transcripts, which would otherwise be automatically removed by the algorithm (43). As such, a follow-up analysis would be necessary to establish whether the observed changes reflect the expected exercise-induced shifts in circulating cell composition (52), under the potentially tenuous assumption that resting gene expression signature is representative of the postexercise state in these blood cells.
As opposed to other LVs of interest associated with mostly protein-coding mRNA (>60% protein-coding in serum EVs, ∼46% in skeletal muscle), serum EV LV2 was unique in that it was associated with a large component of lncRNAs (63% in top 50th percentile). This population of transcripts has been recently studied in the context of exercise adaptation (14, 53), although its biological roles are incompletely understood. The 10 lncRNAs loaded into serum EV LV2 are poorly annotated, hinging interpretation of this LV completely on the relatively smaller protein-coding component (19%). However, future transcriptome-wide studies employing PLIER may establish whether these lncRNAs and other underannotated biotypes consistently associate with the same functional categories of protein-coding genes. Thus, it may be possible computationally and iteratively to build a resource of likely biological roles for these biotypes to be experimentally validated via standard biochemical assays.
Temporal Dynamics of the Transcriptomic Response
Of 23 LVs in muscle that showed a main effect of exercise, 11 were responsive at the early time point (i.e., significantly different from both other time points at h3 only). These played roles including activation of multiple cellular signaling cascades implicated in energy metabolism (AMPK), growth (Wnt), and inflammation (e.g., MAPK, STAT). Considerable information could be gained by exploring the nonprotein-coding fraction of these LVs and potential regulatory roles of transcripts in a cell/tissue culture model or other laboratory-based assays.
Previous studies have established 3–4 h postexercise as a highly transcriptionally active time point (54). While the present findings substantiate this, the temporal gene expression response within skeletal muscle tissue generally suggests that the molecular environment is still perturbed from rest at h24 after exposure to the exercise bout. In support, when PLIER was used to collapse transcripts into LVs, 12 of 23 that exhibited a significant time effect were different at h24 (i.e., either late or sustained). The “late” LVs (different at h24 only) in skeletal muscle were primarily associated with protein-coding genes linked to processes related to protein synthesis and metabolism, chemokine signaling, and neuromuscular communication. This suggests a role for these transcripts in recovery and mechanistic changes potentially underlying gains in strength, power, and muscle mass.
In serum EVs, all four LVs that showed a main effect of exercise were sustained at h24. This may be due to the unaccustomed nature of the exercise bout itself in untrained individuals. While all participants underwent four progressive familiarization sessions to reduce undue muscular pain and soreness, they remained relatively exercise-naïve. We suspect some of the molecular responses sustained at h24 may not be as protracted in exercise-trained individuals. It is well-established that a single exposure to an unaccustomed bout can reduce the magnitude of the effect of subsequent exposures, particularly in terms of a classical inflammatory response (6). Fittingly, transcripts associated with serum EV LV17 were linked to processes including leukocyte activation and immune response. Although acute inflammation is considered important for exercise-induced muscle adaptation (54–56), chronic elevation has negative consequences. As a comparator, testing for these sustained response muscle LVs in trained participants would perhaps be a valuable means of filtering LVs (and identifying novel member genes) most likely to promote positive exercise adaptations. Studying the response of transcripts associated with LV17 to a chronic training regimen might provide such insight. In addition, while typically restricted to skeletal muscle, prolonged proinflammation often seen before (18) and after (55) training in older adults is thought to be a barrier to adaptation. It could be informative to establish whether similar patterns in LV17 are upheld after unaccustomed exercise in older populations.
Future Directions
Our research team is highly cognizant of the imbalance in mechanistic exercise data from male-only versus mixed-sex populations (20, 57). The decision not to covary transcript expression patterns by sex was based on several considerations: first, both males and females were included in relative balance by design (randomization stratified by sex only). As validation of the stratified randomization approach and confirmation that mixed-sex cohorts randomized to TRAD versus HITT (the primary comparison of interest) were balanced at baseline, expression of transcripts at baseline was not significantly different between TRAD and HITT, with the exception of a single muscle circRNA (genomic coordinates chr4:109922159|109945275). As described in sex-cognizant research literature (57), using sex as a categorical covariate uniformly across all genes is inappropriate because sex is a nonrandom factor. While some transcripts are likely influenced by sex, others are not; thus, the relationship between gene expression and sex, if one exists, is likely not similar across all transcripts (58). Furthermore, covarying for sex as a categorical variable forces transcript expression to a representation of an average, “sexless” phenotype, which is inappropriate for the present dataset and difficult to extrapolate to binary populations. Best practices for managing sex as a biological variable in future research are described by our group (57) and others (58) but include stratified randomization, adequate sample size, and possibly collection and statistical normalization based on typically sex-specific outcomes such as lean body mass, testosterone levels, and estrogen levels (which, of course, also vary based on menstrual cycle phase). Despite this, we concede that this study is limited by lack of menstrual cycle control and is underpowered to test true sex differences, at least in the context of a stimulus-dependent exercise response. Nevertheless, that biologically meaningful differences emerge despite this and other aspects not being explicitly controlled is encouraging and may provide direction for the field in designing studies with a higher degree of ecological validity.
Additional investigations in populations of different age, health, or training status will also likely benefit from this description of short-term transcriptomic changes following exposure to an acute exercise stimulus. By continuing to build an extensive repository of rigorous datasets across molecular levels and by enriching the computational tools available to integrate and interpret them, a clearer understanding of the normal, healthy transcriptome map of exercise may eventually be built. By using this resource as a comparator dataset, it may become possible to identify and target biological pathways dysregulated by disease to maximize exercise responsiveness for the greatest number of individuals (16) or augment the impact of combinatorial therapies for disease management.
Given the complex and integrated signaling responses to an acute exercise stress, examining exercise-induced changes at multiple molecular levels (i.e., via multiomics) is likely to provide even further insight into coordination and control of molecular processes underlying adaptation to exercise. This goal has led to the recent establishment of noteworthy efforts such as the Molecular Transducers of Physical Activity Consortium (MoTrPAC) (45), the Athlome (59), and numerous smaller-scale studies (20) designed to characterize molecular responses to exercise. While the larger studies remain years away from interpretation and distribution of meaningful findings, they are likely to provide valuable multiomic data that may be leveraged as a healthy comparator dataset for independent examinations of molecular phenotypes across levels. The present findings would be similarly expanded by integrating other ‘omics (e.g., metabolomics, proteomics, methylomics), assessing alternative posttranscriptional splicing, correlating patterns with phenotypes (e.g., any measures shown in Table 1) across individuals to predict or explain phenotypic or response heterogeneity (16), and characterizing the molecular responses to chronic HITT and TRAD exercise training.
Conclusions
While both traditional and high-intensity exercises confer numerous health- and performance-related benefits when completed chronically, the underlying mechanisms driving these adaptations have not been mechanistically elucidated nor directly compared. Our findings suggest that, at the selected time points, traditional combined exercise elicits a stronger or more consistent transcriptome-wide response in skeletal muscle and serum EVs of young, healthy adults. In addition, our findings point to patterns across transcripts that may aid in laying the foundation for annotation of transcripts based on parameters other than the cellular behavior of functional proteins. Continued attention should be directed toward these understudied biotypes, especially since they exhibit considerable plasticity following a bout of exercise and are therefore likely to play roles in molecular regulation and long-term adaptation. When integrated in a multiomic fashion with other molecular responses to acute and chronic exercise, the transcriptome may provide guidance toward optimizing exercise prescription and maximizing the benefits while increasing enjoyment and reducing time burden for exercise participants. This is of utmost importance if exercise is to become a public health focus and a habitual component of a lifestyle change for individuals across the lifespan.
DATA AVAILABILITY
All FASTQ files and raw count matrices are openly available in the Gene Expression Omnibus (GEO) database under accession number GSE209880.
SUPPLEMENTAL DATA
Supplemental Figs. S1–S5 and Supplemental Tables S1–S6: https://doi.org/10.6084/m9.figshare.21094795.
GRANTS
Parent randomized clinical trial was supported by Department of Defense Office of Naval Research (DoD ONR) Grant N000141613159. Z.A.G. is supported by the Department of Veterans Affairs Rehabilitation Research and Development Service (IK2RX002781). Research reported in this publication included work performed in the Integrated Mass Spectrometry Shared Resource supported by the National Cancer Institute of the National Institutes of Health under Grant P30CA033572.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
K.M.L., K.V.K.-J., M.J.H., P.P., T.J.B., and M.M.B. conceived and designed research; K.M.L., J.S.M., S.M.O., D.D., M.B.B., C.J.K., B.P., S.C.T., R.S.S., R.R., E.A., E.H., J.A., A.B., B.M., J.P., J.S.T., A.S., I.A., and K.P. performed experiments; K.M.L., J.S.M., M.E.L., E.A., E.H., J.A., and J.P. analyzed data; K.M.L., J.S.M., S.M.O., D.D., M.E.L., K.V.K.-J., M.J.H., P.P., Z.A.G., T.J.B., and M.M.B. interpreted results of experiments; K.M.L. prepared figures; K.M.L. drafted manuscript; K.M.L., J.S.M., S.M.O., D.D., M.E.L., K.V.K.-J., P.P., E.A., E.H., Z.A.G., T.J.B., and M.M.B. edited and revised manuscript; K.M.L., Z.A.G., J.S.M., S.M.O., D.D., M.B.B., C.J.K., M.E.L. B.P., S.C.T., R.S.S., K.V.K.-J., M.J.H., P.P., R.R., E.A., E.H., J.A., A.B., B.M., J.P., J.S.T., A.S., I.A., K.P., T.J.B., and M.M.B. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank all of the participants in this trial, in addition to the study coordinators, interventionists, all involved with physical performance testing, biospecimen collection, and inventorying, and the UAB Center for Exercise Medicine Core Muscle Research Lab and Exercise Clinical Training Facility personnel.
Preprint is available at https://doi.org/10.1101/2022.09.14.507939.
REFERENCES
- 1. Lavin KM, Coen PM, Baptista LC, Bell MB, Drummer D, Harper SA, Lixandrao ME, McAdam JS, O'Bryan SM, Ramos S, Roberts LM, Vega RB, Goodpaster BH, Bamman MM, Buford TW. State of knowledge on molecular adaptations to exercise in humans: historical perspectives and future directions. Compr Physiol 12: 3193–3279, 2022. doi: 10.1002/cphy.c200033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lavin KM, Roberts BM, Fry CS, Moro T, Rasmussen BB, Bamman MM. The importance of resistance exercise training to combat neuromuscular aging. Physiology (Bethesda) 34: 112–122, 2019. doi: 10.1152/physiol.00044.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Haddock CK, Poston WSC, Heinrich KM, Jahnke SA, Jitnarin N. The benefits of high-intensity functional training fitness programs for military personnel. Mil Med 181: e1508–e1514, 2016. doi: 10.7205/MILMED-D-15-00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. MacInnis MJ, Gibala MJ. Physiological adaptations to interval training and the role of exercise intensity. J Physiol 595: 2915–2930, 2017. doi: 10.1113/JP273196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stensvold D, Viken H, Steinshamn SL, Dalen H, Stoylen A, Loennechen JP, Reitlo LS, Zisko N, Baekkerud FH, Tari AR, Sandbakk SB, Carlsen T, Ingebrigtsen JE, Lydersen S, Mattsson E, Anderssen SA, Fiatarone Singh MA, Coombes JS, Skogvoll E, Vatten LJ, Helbostad JL, Rognmo O, Wisloff U. Effect of exercise training for five years on all cause mortality in older adults-the Generation 100 study: randomised controlled trial. BMJ 371: m3485, 2020. doi: 10.1136/bmj.m3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hyldahl RD, Chen TC, Nosaka K. Mechanisms and mediators of the skeletal muscle repeated bout effect. Exerc Sport Sci Rev 45: 24–33, 2017. doi: 10.1249/JES.0000000000000095. [DOI] [PubMed] [Google Scholar]
- 7. Neufer PD, Dohm GL. Exercise induces a transient increase in transcription of the GLUT-4 gene in skeletal muscle. Am J Physiol Cell Physiol 265: C1597–C1603, 1993. doi: 10.1152/ajpcell.1993.265.6.C1597. [DOI] [PubMed] [Google Scholar]
- 8. Perry CG, Lally J, Holloway GP, Heigenhauser GJ, Bonen A, Spriet LL. Repeated transient mRNA bursts precede increases in transcriptional and mitochondrial proteins during training in human skeletal muscle. J Physiol 588: 4795–4810, 2010. doi: 10.1113/jphysiol.2010.199448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47: D330–D338, 2019. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44: D457–D462, 2016. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garner RT, Solfest JS, Nie Y, Kuang S, Stout J, Gavin TP. Multivesicular body and exosome pathway responses to acute exercise. Exp Physiol 105: 511–521, 2020. doi: 10.1113/EP088017. [DOI] [PubMed] [Google Scholar]
- 12. Nair VD, Ge Y, Li S, Pincas H, Jain N, Seenarine N, Amper MAS, Goodpaster BH, Walsh MJ, Coen PM, Sealfon SC. Sedentary and trained older men have distinct circulating exosomal microRNA profiles at baseline and in response to acute exercise. Front Physiol 11: 605, 2020. doi: 10.3389/fphys.2020.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Estrada AL, Valenti ZJ, Hehn G, Amorese AJ, Williams NS, Balestrieri NP, Deighan C, Allen CP, Spangenburg EE, Kruh-Garcia NA, Lark DS. Extracellular vesicle secretion is tissue-dependent ex vivo and skeletal muscle myofiber extracellular vesicles reach the circulation in vivo. Am J Physiol Cell Physiol 322: C246–C259, 2022. doi: 10.1152/ajpcell.00580.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bonilauri B, Dallagiovanna B. Long non-coding RNAs are differentially expressed after different exercise training programs. Front Physiol 11: 567614, 2020. doi: 10.3389/fphys.2020.567614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greco S, Cardinali B, Falcone G, Martelli F. Circular RNAs in muscle function and disease. Int J Mol Sci 19: 3454, 2018. doi: 10.3390/ijms19113454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lavin KM, Bell MB, McAdam JS, Peck BD, Walton RG, Windham ST, Tuggle SC, Long DE, Kern PA, Peterson CA, Bamman MM. Muscle transcriptional networks linked to resistance exercise training hypertrophic response heterogeneity. Physiol Genomics 53: 206–221, 2021. doi: 10.1152/physiolgenomics.00154.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lavin KM, Ge Y, Sealfon SC, Nair VD, Wilk K, McAdam JS, Windham ST, Kumar PL, McDonald M-LN, Bamman MM. Rehabilitative impact of exercise training on human skeletal muscle transcriptional programs in Parkinson’s disease. Front Physiol 11: 653, 2020. doi: 10.3389/fphys.2020.00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thalacker-Mercer A, Stec M, Cui X, Cross J, Windham S, Bamman M. Cluster analysis reveals differential transcript profiles associated with resistance training-induced human skeletal muscle hypertrophy. Physiol Genomics 45: 499–507, 2013. doi: 10.1152/physiolgenomics.00167.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rubenstein AB, Hinkley JM, Nair VD, Nudelman G, Standley RA, Yi F, Yu G, Trappe TA, Bamman MM, Trappe SW, Sparks LM, Goodpaster BH, Vega RB, Sealfon SC, Zaslavsky E, Coen PM. Skeletal muscle transcriptome response to a bout of endurance exercise in physically active and sedentary older adults. Am J Physiol Endocrinol Metab 322: E260–E277, 2022. doi: 10.1152/ajpendo.00378.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pillon NJ, Gabriel BM, Dollet L, Smith JAB, Sardon Puig L, Botella J, Bishop DJ, Krook A, Zierath JR. Transcriptomic profiling of skeletal muscle adaptations to exercise and inactivity. Nat Commun 11: 470, 2020. doi: 10.1038/s41467-019-13869-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Contrepois K, Wu S, Moneghetti KJ, Hornburg D, Ahadi S, Tsai MS, Metwally AA, Wei E, Lee-McMullen B, Quijada JV, Chen S, Christle JW, Ellenberger M, Balliu B, Taylor S, Durrant MG, Knowles DA, Choudhry H, Ashland A, Bahmani A, Enslen B, Amsallem A, Kobayashi Y, Avina M, Perelman D, Schüssler-Fiorenza Rose SM, Zhou W, Ashley EA, Montgomery SB, Chaib H, Haddad F, Snyder MP. Molecular choreography of acute exercise. Cell 181: 1112–1130.e16, 2020. doi: 10.1016/j.cell.2020.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bar-Or O. The Wingate anaerobic test. An update on methodology, reliability and validity. Sports Med 4: 381–394, 1987. doi: 10.2165/00007256-198704060-00001. [DOI] [PubMed] [Google Scholar]
- 23. Roberts BM, Lavin KM, Many GM, Thalacker-Mercer A, Merritt EK, Bickel CS, Mayhew DL, Tuggle SC, Cross JM, Kosek DJ, Petrella JK, Brown CJ, Hunter GR, Windham ST, Allman RM, Bamman MM. Human neuromuscular aging: sex differences revealed at the myocellular level. Exp Gerontol 106: 116–124, 2018. doi: 10.1016/j.exger.2018.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stec MJ, Thalacker-Mercer A, Mayhew DL, Kelly NA, Tuggle SC, Merritt EK, Brown CJ, Windham ST, Dell'Italia LJ, Bickel CS, Roberts BM, Vaughn KM, Isakova-Donahue I, Many GM, Bamman MM. Randomized, four-arm, dose-response clinical trial to optimize resistance exercise training for older adults with age-related muscle atrophy. Exp Gerontol 99: 98–109, 2017. doi: 10.1016/j.exger.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kelly NA, Ford MP, Standaert DG, Watts RL, Bickel CS, Moellering DR, Tuggle SC, Williams JY, Lieb L, Windham ST, Bamman MM. Novel, high-intensity exercise prescription improves muscle mass, mitochondrial function, and physical capacity in individuals with Parkinson's disease. J Appl Physiol (1985) 116: 582–592, 2014. doi: 10.1152/japplphysiol.01277.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saugstad JA, Lusardi TA, Van Keuren-Jensen KR, Phillips JI, Lind B, Harrington CA, McFarland TJ, Courtright AL, Reiman RA, Yeri AS, Kalani MYS, Adelson PD, Arango J, Nolan JP, Duggan E, Messer K, Akers JC, Galasko DR, Quinn JF, Carter BS, Hochberg FH. Analysis of extracellular RNA in cerebrospinal fluid. J Extracell Vesicles 6: 1317577, 2017. doi: 10.1080/20013078.2017.1317577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Srinivasan S, Yeri A, Cheah PS, Chung A, Danielson K, De Hoff P, Filant J, Laurent CD, Laurent LD, Magee R, Moeller C, Murthy VL, Nejad P, Paul A, Rigoutsos I, Rodosthenous R, Shah RV, Simonson B, To C, Wong D, Yan IK, Zhang X, Balaj L, Breakefield XO, Daaboul G, Gandhi R, Lapidus J, Londin E, Patel T, Raffai RL, Sood AK, Alexander RP, Das S, Laurent LC. Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell 177: 446–462.e16, 2019. doi: 10.1016/j.cell.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pine PS, Lund SP, Parsons JR, Vang LK, Mahabal AA, Cinquini L, Kelly SC, Kincaid H, Crichton DJ, Spira A, Liu G, Gower AC, Pass HI, Goparaju C, Dubinett SM, Krysan K, Stass SA, Kukuruga D, Van Keuren-Jensen K, Courtright-Lim A, Thompson KL, Rosenzweig BA, Sorbara L, Srivastava S, Salit ML. Summarizing performance for genome scale measurement of miRNA: reference samples and metrics. BMC Genomics 19: 180, 2018. doi: 10.1186/s12864-018-4496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yeri A, Courtright A, Danielson K, Hutchins E, Alsop E, Carlson E, Hsieh M, Ziegler O, Das A, Shah RV, Rozowsky J, Das S, Van Keuren-Jensen K. Evaluation of commercially available small RNASeq library preparation kits using low input RNA. BMC Genomics 19: 331, 2018. doi: 10.1186/s12864-018-4726-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Giraldez MD, Spengler RM, Etheridge A, Godoy PM, Barczak AJ, Srinivasan S, De Hoff PL, Tanriverdi K, Courtright A, Lu S, Khoory J, Rubio R, Baxter D, Driedonks TAP, Buermans HPJ, Nolte-'t Hoen ENM, Jiang H, Wang K, Ghiran I, Wang YE, Van Keuren-Jensen K, Freedman JE, Woodruff PG, Laurent LC, Erle DJ, Galas DJ, Tewari M. Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat Biotechnol 36: 746–757, 2018. doi: 10.1038/nbt.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rozowsky J, Kitchen RR, Park JJ, Galeev TR, Diao J, Warrell J, Thistlethwaite W, Subramanian SL, Milosavljevic A, Gerstein M. exceRpt: a comprehensive analytic platform for extracellular RNA profiling. Cell Syst 8: 352–357.e3, 2019. doi: 10.1016/j.cels.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21, 2013. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Volders PJ, Helsens K, Wang X, Menten B, Martens L, Gevaert K, Vandesompele J, Mestdagh P. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res 41: D246–D251, 2013. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hutchins E, Reiman R, Winarta J, Beecroft T, Richholt R, De Both M, Shahbander K, Carlson E, Janss A, Siniard A, Balak C, Bruhns R, Whitsett TG, McCoy R, Anastasi M, Allen A, Churas B, Huentelman M, Van Keuren-Jensen K. Extracellular circular RNA profiles in plasma and urine of healthy, male college athletes. Sci Data 8: 276, 2021. doi: 10.1038/s41597-021-01056-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419, 2017. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11: 228–234, 2009. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 37. Rodosthenous RS, Hutchins E, Reiman R, Yeri AS, Srinivasan S, Whitsett TG, Ghiran I, Silverman MG, Laurent LC, Van Keuren-Jensen K, Das S. Profiling extracellular long RNA transcriptome in human plasma and extracellular vesicles for biomarker discovery. iScience 23: 101182, 2020. doi: 10.1016/j.isci.2020.101182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y, Parmigiani G, Johnson WE. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom Bioinform 2: lqaa078, 2020. doi: 10.1093/nargab/lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Law CW, Alhamdoosh M, Su S, Dong X, Tian L, Smyth GK, Ritchie ME. RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR. F1000Res 5: ISCB Comm J-1408, 2016. doi: 10.12688/f1000research.9005.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Law CW, Zeglinski K, Dong X, Alhamdoosh M, Smyth GK, Ritchie ME. A guide to creating design matrices for gene expression experiments. F1000Res 9: 1444, 2020. doi: 10.12688/f1000research.27893.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15: R29, 2014. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33: 2938–2940, 2017. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mao W, Zaslavsky E, Hartmann BM, Sealfon SC, Chikina M. Pathway-level information extractor (PLIER) for gene expression data. Nat Methods 16: 607–610, 2019. doi: 10.1038/s41592-019-0456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bull FC, Al-Ansari SS, Biddle S, Borodulin K, Buman MP, Cardon G, Carty C, Chaput JP, Chastin S, Chou R, Dempsey PC, DiPietro L, Ekelund U, Firth J, Friedenreich CM, Garcia L, Gichu M, Jago R, Katzmarzyk PT, Lambert E, Leitzmann M, Milton K, Ortega FB, Ranasinghe C, Stamatakis E, Tiedemann A, Troiano RP, van der Ploeg HP, Wari V, Willumsen JF. World Health Organization 2020 guidelines on physical activity and sedentary behaviour. Br J Sports Med 54: 1451–1462, 2020. doi: 10.1136/bjsports-2020-102955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sanford JA, Nogiec CD, Lindholm ME, Adkins JN, Amar D, Dasari S, Drugan JK, Fernandez FM, Radom-Aizik S, Schenk S, Snyder MP, Tracy RP, Vanderboom P, Trappe S, Walsh MJ; Molecular Transducers of Physical Activity Consortium. Molecular transducers of physical activity consortium (MoTrPAC): mapping the dynamic responses to exercise. Cell 181: 1464–1474, 2020. doi: 10.1016/j.cell.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alsop E, Meechoovet B, Kitchen R, Sweeney T, Beach TG, Serrano GE, Hutchins E, Ghiran I, Reiman R, Syring M, Hsieh M, Courtright-Lim A, Valkov N, Whitsett TG, Rakela J, Pockros P, Rozowsky J, Gallego J, Huentelman MJ, Shah R, Nakaji P, Kalani MYS, Laurent L, Das S, Van Keuren-Jensen K. A novel tissue atlas and online tool for the interrogation of small RNA expression in human tissues and biofluids. Front Cell Dev Biol 10: 804164, 2022. doi: 10.3389/fcell.2022.804164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Eaton M, Granata C, Barry J, Safdar A, Bishop D, Little JP. Impact of a single bout of high-intensity interval exercise and short-term interval training on interleukin-6, FNDC5, and METRNL mRNA expression in human skeletal muscle. J Sport Health Sci 7: 191–196, 2018. doi: 10.1016/j.jshs.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McKay BR, Nederveen JP, Fortino SA, Snijders T, Joanisse S, Kumbhare DA, Parise G. Brain-derived neurotrophic factor is associated with human muscle satellite cell differentiation in response to muscle-damaging exercise. Appl Physiol Nutr Metab 45: 581–590, 2020. doi: 10.1139/apnm-2019-0501. [DOI] [PubMed] [Google Scholar]
- 49. Ludyga S, Gronwald T, Hottenrott K. The athlete's brain: cross-sectional evidence for neural efficiency during cycling exercise. Neural Plast 2016: 4583674, 2016. doi: 10.1155/2016/4583674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Whitham M, Febbraio MA. Redefining tissue crosstalk via shotgun proteomic analyses of plasma extracellular vesicles. Proteomics 19: e1800154, 2019. doi: 10.1002/pmic.201800154. [DOI] [PubMed] [Google Scholar]
- 51. Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science 347: 1260419, 2015. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 52. Shinkai S, Shore S, Shek PN, Shephard RJ. Acute exercise and immune function. Relationship between lymphocyte activity and changes in subset counts. Int J Sports Med 13: 452–461, 1992. doi: 10.1055/s-2007-1021297. [DOI] [PubMed] [Google Scholar]
- 53. De Sanctis P, Filardo G, Abruzzo PM, Astolfi A, Bolotta A, Indio V, Di Martino A, Hofer C, Kern H, Lofler S, Marcacci M, Marini M, Zampieri S, Zucchini C. Non-coding RNAs in the transcriptional network that differentiates skeletal muscles of sedentary from long-term endurance- and resistance-trained elderly. Int J Mol Sci 22: 1539, 2021. doi: 10.3390/ijms22041539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Louis E, Raue U, Yang Y, Jemiolo B, Trappe S. Time course of proteolytic, cytokine, and myostatin gene expression after acute exercise in human skeletal muscle. J Appl Physiol (1985) 103: 1744–1751, 2007. doi: 10.1152/japplphysiol.00679.2007. [DOI] [PubMed] [Google Scholar]
- 55. Lavin KM, Perkins RK, Jemiolo B, Raue U, Trappe SW, Trappe TA. Effects of aging and lifelong aerobic exercise on basal and exercise-induced inflammation. J Appl Physiol (1985) 128: 87–99, 2020. doi: 10.1152/japplphysiol.00495.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Raue U, Jemiolo B, Yang Y, Trappe S. TWEAK-Fn14 pathway activation after exercise in human skeletal muscle: insights from two exercise modes and a time course investigation. J Appl Physiol (1985) 118: 569–578, 2015. doi: 10.1152/japplphysiol.00759.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. O’Bryan SM, Connor KR, Drummer D, Lavin KM, Bamman MM. Considerations for sex-cognizant research in exercise biology and medicine. Front Sports Act Living 4: 903992, 2022. doi: 10.3389/fspor.2022.903992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Beltz AM, Beery AK, Becker JB. Analysis of sex differences in pre-clinical and clinical data sets. Neuropsychopharmacology 44: 2155–2158, 2019. doi: 10.1038/s41386-019-0524-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sellami M, Elrayess MA, Puce L, Bragazzi NL. Molecular big data in sports sciences: state-of-art and future prospects of OMICS-based sports sciences. Front Mol Biosci 8: 815410, 2021. doi: 10.3389/fmolb.2021.815410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figs. S1–S5 and Supplemental Tables S1–S6: https://doi.org/10.6084/m9.figshare.21094795.
Data Availability Statement
All FASTQ files and raw count matrices are openly available in the Gene Expression Omnibus (GEO) database under accession number GSE209880.