In this Resource/Methodology, Scheepbouwer et al. developed ALL-tRNAseq, which combines the highly processive MarathonRT and RNA demethylation for the robust assessment of tRNA expression, together with a randomized adapter ligation strategy prior to reverse transcription to assess tRNA fragmentation levels in both cell lines and tissues. Their profiling strategy effectively improves classification of oncogenic signatures in glioblastoma and diffuse large B-cell lymphoma tissues, particularly for samples presenting higher levels of RNA fragmentation, further highlighting the utility of ALL-tRNAseq for translational research
Keywords: cancer, high-throughput sequencing, tissue samples, transfer RNA
Abstract
Transfer RNAs (tRNAs) are small adaptor RNAs essential for mRNA translation. Alterations in the cellular tRNA population can directly affect mRNA decoding rates and translational efficiency during cancer development and progression. To evaluate changes in the composition of the tRNA pool, multiple sequencing approaches have been developed to overcome reverse transcription blocks caused by the stable structures of these molecules and their numerous base modifications. However, it remains unclear whether current sequencing protocols faithfully capture tRNAs existing in cells or tissues. This is specifically challenging for clinical tissue samples that often present variable RNA qualities. For this reason, we developed ALL-tRNAseq, which combines the highly processive MarathonRT and RNA demethylation for the robust assessment of tRNA expression, together with a randomized adapter ligation strategy prior to reverse transcription to assess tRNA fragmentation levels in both cell lines and tissues. Incorporation of tRNA fragments not only informed on sample integrity but also significantly improved tRNA profiling of tissue samples. Our data showed that our profiling strategy effectively improves classification of oncogenic signatures in glioblastoma and diffuse large B-cell lymphoma tissues, particularly for samples presenting higher levels of RNA fragmentation, further highlighting the utility of ALL-tRNAseq for translational research.
Transfer RNAs (tRNAs) are the most abundant class of RNAs in the cell and play a crucial function in protein synthesis by bridging the genetic code to the amino acid sequence of proteins (Rak et al. 2018). Their central role in translation demands a high level of processing as part of a strict quality control mechanism that ensures their stability and functionality (Pan 2018). This involves removal of leader and trailer (as well as intronic) sequences, addition of the trinucleotide CCA to the 3′ mature end of each tRNA, and the addition of an average of 13 nucleobase modifications per molecule (Pan 2018). The dynamic regulation of tRNA abundance and their aminoacylation further increases the complexity of their regulatory control on translation rates, mRNA stability, and protein folding and has been shown to facilitate oncogene translation (Pavon-Eternod et al. 2009; Evans et al. 2017; Hernandez-Alias et al. 2020; Yang et al. 2020; Rak et al. 2021). Multiple studies have now established that tRNA expression levels are differentially modulated in tissues and cell types and are selectively altered in tumor tissues (Dittmar et al. 2006; Gingold et al. 2014; Schmitt et al. 2014; Sagi et al. 2016; Hernandez-Alias et al. 2020; Pinkard et al. 2020). Additionally, tRNAs can also be processed into tRNA-derived small RNAs (tsRNAs) in response to different cellular and environmental cues (Kumar et al. 2016; Shen et al. 2018; Drino et al. 2020; Shi et al. 2021). These tRNA fragments have been described to play additional regulatory roles in RNA silencing and epigenetic regulation but also in cancer development and progression (Haussecker et al. 2010; Maute et al. 2013; Kumar et al. 2014; Goodarzi et al. 2015; Schorn et al. 2017; Martinez 2018; Boskovic et al. 2020). However, tissue collection and processing procedures were shown to impact RNA integrity and modification analysis (’t Hoen et al. 2013; Richter et al. 2022), which could be particularly challenging for robust tRNA profiling in patient-derived tumor samples.
tRNA expression levels assessed by conventional sequencing-based methods suffer from various technical biases and cannot always be replicated by hybridization-based detection approaches (Motorin et al. 2007; Drino et al. 2020). During RNA-seq library preparation, the rigid tertiary structure and presence of highly modified nucleotides in the Watson–Crick face of tRNA bases lead to RT arrest or result in nucleotide misincorporations (Wittig and Wittig 1978; Motorin et al. 2007). Although tRNAs are the most abundant molecules in the cell (Westermann et al. 2012; Palazzo and Lee 2015), the combination of their specific biological properties and the usage of inadequate sequencing approaches often results in strong underestimation of tRNA expression in cells and tissue samples. Specifically, a large number of otherwise full-length tRNAs may appear as truncated species or fragments in sequencing data because of prematurely terminated cDNAs that complicate reliable quantitative assessment of tRNAs via high-throughput sequencing. Monitoring the dynamic changes in the composition of cellular tRNA repertoires and/or assessing the potential use of tRNA expression signatures as cancer-specific biomarkers require more robust approaches to determine the relative abundance of tRNAs.
Multiple high-resolution approaches have been developed with the aim to advance tRNA detection and analysis. The first major improvement was the removal of the base modifications 1-methyladenosine (m1A), 3-methylcytosine (m3C), and 1-methylguanosine (m1G) with the demethylating enzyme AlkB from Escherichia coli, which has been used in library preparation workflows for the quantification of mature tRNA transcripts (Zheng et al. 2015; Hu et al. 2021; Warren et al. 2021) or tsRNA products (Cozen et al. 2015; Shi et al. 2021). In addition to removing known RT-blocking modifications, the detection of mature full-length tRNAs has been further improved by incorporating thermostable group II intron reverse transcriptase (TGIRT) into the library construction workflow of both DM-tRNA-seq (Zheng et al. 2015) and mim-tRNAseq (Behrens et al. 2021). In contrast to retroviral RTs, which suffer from low processivity and higher misincorporation rates (Wittig and Wittig 1978; Zhao et al. 2018; Minshall and Git 2020; Werner et al. 2020), TGIRT was shown to efficiently reverse-transcribe highly structured tRNAs into complementary DNA (cDNA) (Mohr et al. 2013). Alternatively, both RNA fragmentation strategies (Gogakos et al. 2017) and improved adapter ligation steps (Pang et al. 2014; Shigematsu et al. 2017; Erber et al. 2020; Pinkard et al. 2020; Hu et al. 2021) were implemented into different library preparation protocols aimed at obtaining mature full-length tRNA libraries. All of these recently developed methodologies, however, may potentially also inherently introduce composition bias due to their size fractionation steps, adapter ligation strategies, or single-strand cDNA circularization (Zheng et al. 2015; Erber et al. 2020; Pinkard et al. 2020; Behrens et al. 2021; Hu et al. 2021).
In this study, we describe the development of adapter-ligated libraries of tRNA-derived sequences (ALL-tRNAseq), a protocol complementary to the existing arsenal of tRNA profiling approaches that is specifically designed for the detection of full-length tRNA molecules and tRNA-derived products in clinical tissue samples. Whereas DM-tRNA-seq and mim-tRNAseq improved tRNA sequencing using TGIRT, we incorporated the highly processive group II intron maturase MarathonRT that was shown to outperform TGIRT (and retroviral RTs) for the sequencing of long and structured RNAs (Zhao et al. 2018; Guo et al. 2020, 2022). To further improve tRNA profiling, we removed the most interfering methyl marks with the RNA demethylating enzyme AlkB, together with a randomized adapter ligation strategy facilitated by molecular crowding (Kim et al. 2019), to capture both full-length species and RNA fragments already present in the assessed samples (or potentially generated prior reverse transcription). To the best of our knowledge, ALL-tRNAseq the first tRNA profiling approach that can capture the entire repertoire of full-length tRNA molecules and provide compensation for possible variability in RNA integrity by inclusion of all tRNA mapping reads. This approach provided a more precise and reliable quantification of the total tRNA repertoire, which is required for a detailed assessment of steady-state and dynamic changes in tRNA levels in both cultured cells and clinical tissue samples. Our data revealed that the cell type-specific regulation of tRNA repertoires was not as pronounced as previously thought across different cell lines or during chemically induced differentiation. Finally, we showed that ALL-tRNAseq can effectively improve classification of tumor-associated tRNA signatures in clinical tissue samples.
Results
The ultraprocessive group II intron maturase MarathonRT combined with RNA demethylation overcomes tRNA length bias
Studies that rely on tissue sample collection often face differences in sample handling, storage time, and transport conditions that could ultimately influence RNA stability and its subsequent RNA analyses (Micke et al. 2006; Gallego Romero et al. 2014). For this reason, we initiated the development of ALL-tRNAseq to enable robust assessment of tRNA expression signatures in human tissue samples that may suffer from a higher degree of variability in RNA quality. Hence, in contrast to most recent tRNA sequencing methods that apply reverse transcription prior to adapter ligation (Pinkard et al. 2020; Behrens et al. 2021; Hu et al. 2021; Watkins et al. 2022), our approach relies on performing the reverse transcription step with the ultraprocessive group II intron maturase MarathonRT after adapter ligation to allow analysis of both full-length tRNA molecules and their derived fragments (Fig. 1A). ALL-tRNAseq included a demethylation step (Zheng et al. 2015; Hrabeta-Robinson et al. 2017; Zhou et al. 2019) for removal of potentially hindering modifications at the Watson–Crick face prior to reverse transcription. Additionally, a sequential adapter ligation strategy was performed using two adapters, ending with four fully degenerated positions to reduce ligation bias. The presence of a high concentration of polyethylene glycol (PEG) increased the effective concentration of target RNAs and adapters via macromolecular crowding (Kim et al. 2019). Last, a size selection step (via polyacrylamide gel electrophoresis) between both adapter ligation steps was included to further purify small RNAs and simultaneously exclude free adapter contaminants.
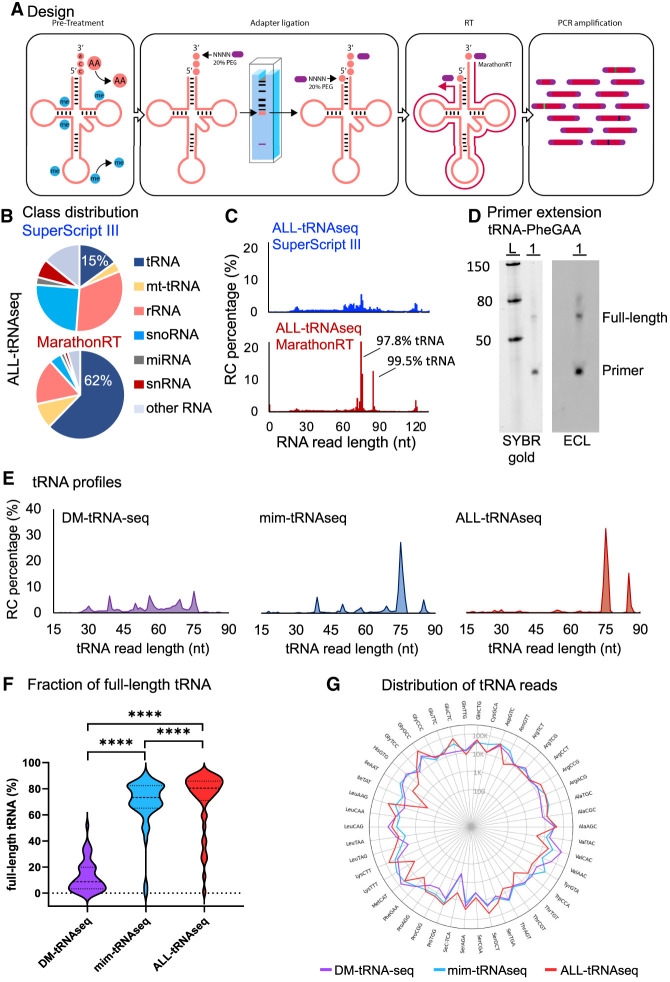
Figure 1.
The ultraprocessive group II intron maturase MarathonRT combined with RNA demethylation overcomes tRNA length bias. (A) Workflow of ALL-tRNAseq depicting steps for obtaining the full tRNA repertoire (including type I and type II tRNAs), consisting of tRNA deacylation and demethylation, sequential adapter ligation (adapters indicated in purple) with a gel size selection step, reverse transcription with incorporation of the highly processive MarathonRT, and PCR amplification. (B) RNA class distribution in percentage of total normalized reads for the ALL-tRNAseq library preparation protocol using SuperScript III and MarathonRT in SNB-75 cells. (C) Full RNA read length analysis in percentage after adapter trimming in SuperScript III and MarathonRT prepared libraries from SNB-75 cell line RNA. The increased proportion of reads mapping to tRNA sequences at the most dominant peaks of 75 and 85 nt are indicated. (D) Primer extension analysis of yW37 in tRNA-PheGAA using 2 μg of demethylated total RNA from HEK293T cells. (Left panel) SYBR Gold staining of small RNA ladder (L; 50, 80, and 150 nt) and cDNA yield (1) separated on a 10% denaturing polyacrylamide gel. (Right panel) Chemiluminescent detection of biotin-labeled cDNA products of tRNA-PheGAA. (E) tRNA read length distribution comparison between DM-tRNA-seq, mim-tRNAseq, and ALL-tRNAseq, shown as percentage of all reads mapping to cytoplasmic tRNA from HEK293T cells. (F) Violin plot of the full-length tRNA read fraction with the 3′-terminal CCA triplet detected in DM-tRNA-seq (n = 2), mim-tRNAseq (n = 2), and ALL-tRNAseq (n = 2) in HEK293T cells. Statistics were performed using Wilcoxon rank sum test. (****) P < 0.001. (G) Radar plot of tRNA anticodon reads per million mapping to cytoplasmic tRNA showing the distribution of reads per tRNA anticodon for DM-tRNA-seq (purple line), mim-tRNAseq (blue line), and ALL-tRNAseq (red line). Data are represented as log10 values on the radius.
To assess the performance of our approach, we first profiled tRNA expression in a glioblastoma (GBM) model cell line (sample with RNA integrity number [RIN] 8.7) (Supplemental Fig. S1A) to evaluate whether incorporation of MarathonRT in place of the commonly used retroviral reverse transcriptase (RT) SuperScript III could improve detection of tRNA molecules. MarathonRT was shown to reverse-transcribe highly structured RNAs at low temperatures with higher efficiency and processivity than both SuperScript and the thermostable group II intron maturase TGIRT (Zhao et al. 2018; Guo et al. 2020, 2022). Indeed, an analysis of all sequencing reads corresponding to small RNAs up to 120 nt showed that inclusion of MarathonRT enabled a dramatic increase in detection of tRNA reads from 15% to 62% of total reads when compared with SuperScript III (Fig. 1B). Moreover, read length analysis of the complete RNA repertoire further emphasized tRNA enrichment by a pronounced and specific increase of 75-, 76-, and 85-nt-long RNA sequences that almost entirely consisted of mature full-length (including the terminal CCA addition) type I and type II tRNAs (Fig. 1C). Although the processivity of MarathonRT has already been extensively validated by primer extension assays using long RNA molecules (Zhao et al. 2018; Guo et al. 2020, 2022), the ability of MarathonRT to produce full-length cDNAs for stably folded and heavily modified tRNAs has not been demonstrated yet. Therefore, we assessed MarathonRT performance in a primer extension assay for tRNA-PheGAA using total RNA of HEK293T cells. This tRNA contains a structurally complex modification, wybutosine (yW), at position 37 (Perche-Letuvée et al. 2014) that can block Watson–Crick base pairing (Werner et al. 2020) and was one of the first modifications known to lead to premature termination in reverse transcription reactions (Wittig and Wittig 1978). Only full-length cDNA products could be detected in this assay (Fig. 1D), suggesting that MarathonRT is not hindered by this bulky RNA modification. Similar results were observed for tRNA-GlyGCC and a lysine tRNA (Supplemental Fig. S1B), indicating that truncated cDNA products are not generated by this enzyme at detectable rates for the three types of tRNA tested. Last, overall reproducibility of ALL-tRNAseq library preparation was evaluated by comparing expression of all tRNA anticodon families between two technical replicates from a matching RNA source, which revealed high correlation between the two samples (Pearson correlation r:0.98) (Supplemental Fig. S1C).
To benchmark our method to established tRNA sequencing methods, we retrieved publicly available data sets using another group II intron-encoded RT, TGIRT, in combination with circularization-based ligation approaches. Both methods used HEK293T cell line-derived RNA for tRNA analysis. Complete preprocessing, data normalization, and analysis of all raw sequencing data were performed with our recently updated computational pipeline, sRNAbench (Aparicio-Puerta et al. 2019, 2022). DM-tRNA-seq (Zheng et al. 2015) displayed a wide read length distribution of tRNA reads in the absence of a type II tRNA peak (Fig. 1E, left). On the other hand, mim-tRNAseq (Behrens et al. 2021) and ALL-tRNAseq showed a nearly identical read length distribution, with a notable exception for type II tRNAs (Fig. 1E, middle and right). Next, we extended read length comparisons to mitochondrial tRNAs (mt-tRNAs), which revealed highly similar profile lengths with a distinct peak at 72 nt for ALL-tRNAseq, DM-tRNA-seq, and mim-tRNAseq, with additional peaks at 36 nt for the latter two methods (Supplemental Fig. S1D). In contrast to ALL-tRNAseq, and for both cytoplasmic and mitochondrial tRNAs, DM-tRNA-seq and mim-tRNAseq showed a higher relative abundance of truncated tRNA reads (Fig. 1E, left and middle; Supplemental Fig. S1D, left and middle) as a result of single-adapter or circularization protocols that cannot differentiate between 5′ degradation of full-length tRNA and truncated reads generated during reverse transcription. This is particularly surprising in the case of the mim-tRNAseq procedure, which relies on multiple size fractionation steps of full-length species. This implies that combining RNA demethylation and the use of MarathonRT with a two-adapter ligation strategy prior to RT may be useful in eliminating some of the technical hurdles present in currently available sequencing methodologies. To further evaluate whether the ALL-tRNAseq approach reduced bias caused by premature cDNA termination at modified bases, an additional analysis on the relationship between full-length tRNA reads and total tRNA reads per tRNA isodecoder was performed. This analysis revealed higher correlation for ALL-tRNAseq (Pearson correlation r: 0.99) (Supplemental Fig. S1E) compared with mim-tRNAseq (Pearson correlation r: 0.7) (Supplemental Fig. S1F) and DM-tRNA-seq (Pearson correlation r: 0.85) (Supplemental Fig. S1G). The near absence of full-length tRNA-LysTTT and tRNA-tRNA-TyrGTA mainly contributed to the relatively low correlation between full-length tRNA and all tRNA reads for mim-tRNAseq despite the high fraction of full-length tRNA reads for most tRNA anticodon families (Supplemental Fig. S1F). In contrast, the full-length tRNA anticodons that got relatively more read counts compared with other tRNA anticodons in DM-tRNA-seq appeared proportionate to the number of all tRNA reads (Supplemental Fig. S1G). We extended this analysis by directly comparing the total fraction of full-length tRNA molecules containing CCA ending for each method by determining the percentage of full-length tRNA reads out of the total reads assigned to tRNAs for each amino acid. This analysis showed a median of 80% full-length tRNA in ALL-tRNAseq compared with a median of 73% and 9% full-length tRNA reads for mim-tRNAseq and DM-tRNA-seq (Wilcoxon rank-sum P-value < 0.001) (Fig. 1F), respectively. Last, we assessed global tRNA expression in the HEK293T cell line using all three methods. In general, our analysis showed divergent tRNA expression profiles between the three protocols (Fig. 1G). Whereas the majority of reads for DM-tRNA-seq were represented by tRNA-Val (19% of tRNA reads), as similarly observed by Behrens et al. (2021), tRNA coverage appeared to be more evenly distributed for mim-tRNAseq in comparison with ALL-tRNAseq, which showed distinct tRNA expression patterns including higher abundance of type II tRNAs (tRNA-Leu and tRNA-Ser) (Fig. 1G).
Altogether, these data indicate that the combination of AlkB treatment with the high modification readthrough efficiency of MarathonRT in ALL-tRNAseq can overcome potential roadblocks generating biases in tRNA length, particularly for the recovery of type II tRNA molecules.
Inclusion of all tRNA reads detected by ALL-tRNAseq improves robustness of tRNA profiling
To evaluate whether ALL-tRNAseq can accurately monitor differences as well as dynamic change in cellular tRNA repertoires, we next examined tRNA read length distribution in different cell lines (SU-DHL-5, SNB-75, and HEK293T) and during hESC proliferation and differentiation by retinoic acid (Gingold et al. 2014). The differentiation process of the latter was monitored by bright-field microscopy at different time points (Supplemental Fig. S2A) and confirmed by RT-qPCR of relevant differentiation markers (Supplemental Fig. S2B). The tRNA read length profiles of all cell lines displayed a high degree of similarity with comparable levels of type I tRNA, albeit differences were observed in the levels of type II tRNA between all cell lines (Fig. 2A). Next, we examined differences in full-length tRNA expression between cell lines using principal component analysis (PCA). Here, we observed high resemblance across the different cell lines, including an unexpected clustering of the hESC-derived samples from both proliferative and differentiated cells. In contrast, PCA showed wider variability among samples within the same cell line, particularly between SU-DHL-5 samples (Fig. 2B). Thus, we decided to explore the origin of the full-length tRNA variability among SU-DHL-5 replicates in more detail. First, we confirmed that RNA integrity in the three SU-DHL-5 replicates was comparable by investigating their bioanalyzer profiles and RNA integrity number (RIN) values (Supplemental Fig. S3A) as well as the percentage of full-length tRNAs within the total pool of a given tRNA isodecoder. These analyses revealed that although bioanalyzer profiles and RIN values were nearly identical for the three replicates, a striking decrease in the fraction of full-length tRNAs could be observed in replicate 1 compared with replicate 2 and 3 (Fig. 2C). This suggested that tRNAs in SU-DHL-5 cells of replicate 1 were subjected to a higher degree of small RNA processing or degradation into fragments, consistent with the PCA (Fig. 2B). We next decided to confirm this analysis by evaluating the fragmentation pattern of a specific tRNA by Northern blot. For this purpose, we focused on the well-characterized tRNA-GlyGCC-derived fragments (Goodarzi et al. 2015; Drino et al. 2020) and probed for 5′-tsRNA GlyCCC/GCC. Similar to the sequencing data, both tRNA-GlyCCC/GCC tsRNAs and full-length tRNAs were detected in SU-DHL-5 replicate 1 (Supplemental Fig. S3B), while full-length tRNAs were predominantly observed in an additional SU-DHL-5 sample (Supplemental Fig. S3C). To ensure that the designed probe could detect fragments of different sizes reliably, we included a serial dilution of a DNA oligonucleotides pool of 35 and 74 nt corresponding to tRNA GlyCCC/GCC full-length (74 nt) and fragments (35 nt) into our analysis. Northern blotting confirmed the possible detection of both length variants reliably up to 40 fmol (Supplemental Fig. S3C). Since Northern blotting was performed directly after RNA isolation, these results further confirmed that tRNA fragments detected in sequencing of SU-DHL-5 sample replicate 1 were not generated during library preparation.
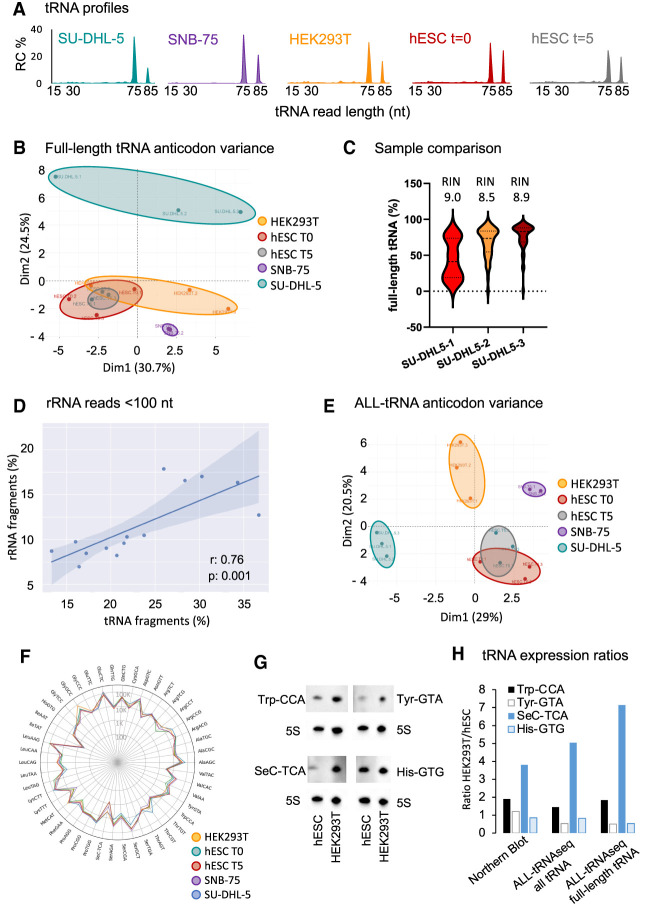
Figure 2.
Inclusion of all tRNA reads detected by ALL-tRNAseq improves robustness of tRNA profiling. (A) tRNA read length distribution in percentage in cell lines SU-DHL-5, SNB-75, and HEK293T as well as proliferating and differentiated hESCs. (B) Principal component analysis (PCA) of full-length tRNA anticodon expression in HEK293T (n = 3), SNB-75 (n = 2), SU-DHL-5 (n = 3), hESC (n = 3), and differentiated hESCs (n = 3). (C) Violin plot of the full-length tRNA read fraction with the 3′-terminal CCA triplet in three replicates of the SU-DHL-5 cell line. (D) Pearson correlation of normalized tRNA fragments reads per million and normalized rRNA reads per million <100 nt. (E) PCA of tRNA anticodon expression including full-length and short tRNA-derived reads in HEK293T (n = 3), SNB-75 (n = 2), SU-DHL-5 (n = 3), proliferating hESCs (n = 3), and differentiated hESCs (n = 3). (F) Radar plot of all tRNA anticodon reads per million mapping to cytoplasmic tRNA, showing the distribution of reads per tRNA anticodon for SU-DHL-5 (blue line), HEK293T (orange line), proliferating hESCs (green line), differentiated hESCs (red line), and SNB-75 (purple line). Data are represented as log10 values on the radius. (G) Detection of tRNA-Trp-CCA, tRNA-Tyr-GTA, tRNA-His-GTG, and tRNA-SeC-TCA in 1 µg of total RNA isolated from proliferating hESCs (T = day 0) and HEK293T cell lines by Northern blot. 5S rRNA was used as a loading control. (H) Ratio in tRNA abundance between hESCs and HEK293T by quantification of Northern blot signal (left) compared with relative abundance detected by ALL-tRNAseq including all tRNA reads (middle) and full-length tRNA reads only (right). Band intensities were quantified by ImageJ, background-subtracted, and normalized to 5S rRNA signal.
To further exclude possible variation originating from our RNA isolation procedures, we next re-evaluated RNA treatments included in our study. First, we examined our tRNA deacylation treatment for either 30 min at 37°C, 1 h at 37°C, or 2 h at 50°C. Next, RNA integrity was assessed using urea–polyacrylamide gel (Supplemental Fig. S4A, left panel). Northern blotting analysis of tRNA-GlyGCC/CCC showed signal for the control oligos at 18, 35, and 74 nt (Supplemental Fig. S4A, middle panel), and all treated RNA samples revealed full-length tRNA-GlyGCC/CCC exclusively (Supplemental Fig. S4A, right panel). We then re-evaluated the performance of the demethylation step (Zhou et al. 2019) as a possible source of RNA decay. Whereas total RNA incubations with the AlkB enzyme mix (AlkB WT and AlkB mutant D135S) resulted in a high fraction of full-length tRNA reads (Fig. 2A; Supplemental Fig. S4B), a replicate RNA sample incubated with the reaction buffer only (buffer-treated sample) showed a high degree of small RNA fragmentation with <2% mature type I tRNA and a nearly complete absence of full-length type II tRNA molecules (Supplemental Fig. S4C, gray profile). In striking contrast, sequencing libraries prepared with untreated RNA provided a majority of mature full-length tRNA (Supplemental Fig. S4C, yellow profile), albeit with a reduced level of type II tRNA compared with AlkB-treated samples, additionally illustrating the importance of the demethylation treatment to fully transcribe type II tRNAs. This is also consistent with the differences we previously observed between ALL-tRNAseq and mim-tRNAseq (Fig. 1E), which does not include a demethylation step. These results suggested the possibility of a buffer-induced RNA fragmentation rather than enzyme-induced RNA fragmentation (Shi et al. 2021). The absence of RNA degradation in our untreated and enzyme-treated samples suggested that this phenomenon may be caused by increased Fe(II) availability in the absence of the dioxygenase AlkB, leading to nucleic acid cleavage via a Fenton-type chemical reaction (Dallas et al. 2004). To confirm our hypothesis, RNA integrity was further assessed using urea–polyacrylamide gels. Whereas RNA integrity profiles for untreated and DM-treated total RNA are similar, the buffer-treated sample (no enzymes but iron-containing buffer) showed increased RNA degradation (Supplemental Fig. S4D). To validate these results, RNA samples were incubated with different components of the AlkB reaction, further confirming that RNA degradation occurs only in the presence of iron (Supplemental Fig. S4E). In agreement with this observation, the Pandora-seq method also reported the issue of AlkB treatment-induced RNA fragmentation that was circumvented rather than resolved by applying size selection prior to demethylation (Shi et al. 2021). Thus, based on these results, Fe(II) availability in the reaction buffer should be treated with caution during the demethylation step. Furthermore, despite the enrichment of type I mature tRNA in the untreated condition, we also observed higher levels of specific tRNA fragments around 32 and 54 nt (Supplemental Fig. S4C). Although our ligation strategy prior to reverse transcription prevented detection of incomplete cDNA synthesis, the increased presence of these tRNA fragments in the untreated samples was compatible with a less efficient reverse transcription that artificially increased the contribution of tRNA fragments in these sequencing data. Altogether, these results indicate that the detected tRNA fragments in SU-DHL-5 replicate 1 were already present in the total RNA isolate but that the conditions of the demethylation steps are also critical to avoid further treatment-induced fragmentation.
Since RINs did not explain the observed variation in the tRNA population of the RNA isolates, we aimed to assess whether similar levels of RNA fragments could also be observed for other highly abundant RNAs of similar sequence length in our cell line samples. Therefore, we categorized fragmented 5S, 5.8S, 18S, and 28S rRNA reads up to 100 nt to exclude potential contaminating full-length 5S rRNA of ∼120 nt long (which is just above our size selection during library preparation) and study rRNA fragmentation patterns in the same sequence length as tRNA molecules. All tRNA molecules that were not identified as full-length tRNA were considered as tRNA fragments to allow all possible fragment lengths into the analysis. Similar to a recent study that identified characteristic rRNA and tRNA modification markers in the small RNA fraction, indicative of the degradation status (Richter et al. 2022), our analysis in SU-DHL-5 sample (replicate 1) revealed a high correlation (Pearson correlation r: 0.76) between the amount of tRNA fragmentation and the occurrence of rRNA fragmentation (Fig. 2D). Therefore, we incorporated all reads in the analysis to evaluate whether intersample variability was still present and to further account for any unexpected technical variation/fragmentation that may be introduced during sample collection and/or RNA isolation. PCA of tRNA anticodon expression demonstrated clear separation between cell lines of different origins and high resemblance among replicates within the same cell line (Fig. 2, cf. B and E). The added benefit of including all tRNA reads was further evident from improved separation between the proliferative and differentiated hESCs. We further compared and visualized relative tRNA anticodon expression in our cell lines using the ALL-tRNAseq approach. The overall relative tRNA anticodon abundance exhibited comparable distribution across all cell lines, as similarly observed in other studies (Fig. 2F; Dittmar et al. 2006; Aharon-Hefetz et al. 2020). However, our analysis also indicated cell line-specific differences, with the most dynamic change observed for tRNA-SeC-TCA (Fig. 2F). Despite the overall similarity in tRNA expression profiles, the heat map derived from hierarchical cluster analysis also clearly separated all cell lines based on tRNA anticodon expression using the ALL-tRNAseq approach (Supplemental Fig. S5A). Interestingly, ALL-tRNAseq generally detected relatively modest changes in the tRNA pool between proliferating and differentiating cells compared with what has been previously reported (Gingold et al. 2014; Aharon-Hefetz et al. 2020). The relative changes in tRNA anticodon expression were further validated by hybridization-based Northern blotting of four tRNAs (tRNA-Tyr-GTA, tRNA-SeC-TCA, tRNA-Trp-CCA, and tRNA-His-GTG) in two cell lines: hESCs and HEK293T (Fig. 2G). We next compared Northern blot results with ALL-tRNAseq full-length tRNA expression profiles (Supplemental Fig. S4F) and tRNA profiles including all tRNA-derived reads (Fig. 2F). Quantification of Northern blot signals showed an overall high similarity to ALL-tRNAseq results, with inclusion of all tRNA reads providing a better evaluation of full-length ratios in the two cell lines compared with hybridization-based methods (Fig. 2H). It should be noted that a direct comparison of the relative levels of a specific tRNA (as assessed by high-throughput sequencing of size-selected RNA samples) with their corresponding absolute levels in total RNA samples (as measured by Northern blotting) may be affected by potential differences in total tRNA levels between the cell lines compared. Nevertheless, these results further confirmed the added benefit of our profiling strategy including all reads for the relative tRNA abundance by ALL-tRNAseq is highly concordant with hybridization-based Northern blotting results.
tRNA expression profiling distinguishes normal from hematological cancer tissues
To assess the utility of ALL-tRNAseq for the discovery of tumor-associated tRNA signatures, we first examined in more detail whether RNA quality in tissue samples had an impact on full-length tRNA analysis. We obtained freshly frozen tissue samples from patients diagnosed with the hematological malignancy diffuse large B-cell lymphoma (DLBCL; n = 15) versus reactive lymph nodes (n = 3) that all had RIN values ≥7. An unsupervised hierarchical cluster analysis showed that the expression of full-length tRNAs alone is not sufficient to fully differentiate between reactive lymph nodes and DLBCL tumor biopsy tissues. However, the full-length tRNA matrix showed relative changes in expression of tRNA anticodon families in DLBCL samples compared with normal tissues (Fig. 3A). Specifically, tRNA-TyrGTA expression in most DLBCL samples appeared strongly down-regulated, as previously observed by Gingold et al. (2014). Next, we also evaluated the contribution of tRNA fragmentation patterns to the tRNA expression profiles by exclusively analyzing tRNA reads <60 nt. Our analysis again showed insufficient clustering of reactive lymph nodes, which was not fully separated from DLBCL samples (Fig. 3B). Therefore, similar to our approach of tRNA read analysis in cell lines, we next examined whether the relationship between RNA quality (using RINs) and the proportion of full-length tRNAs between the tumor biopsies and healthy lymph node samples can be compared. This analysis indicated that, also for tissue samples, RIN values were not correlated or were poorly correlated with the degree of tRNA fragmentation (Pearson correlation r: −0.14) (Fig. 3C). Alternatively, an additional analysis showed that the levels of rRNA fragments were clearly inversely correlated with the levels of full-length species (Pearson correlation r: −85) (Fig. 3D), as expected based on the cell line results.
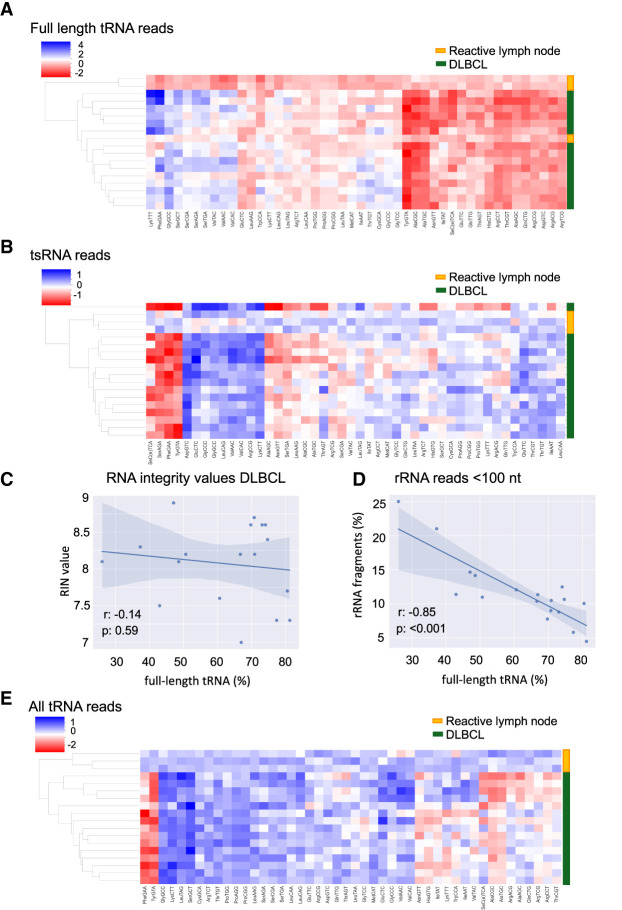
Figure 3.
tRNA expression profiling distinguishes normal from hematological cancer tissues. (A) Heat map visualization showing clustering of full-length tRNA reads in DLBCL samples. Each horizontal line represents a biological sample (15 DLBCL and three healthy, reactive lymph node samples). Each row represents a tRNA type, grouped by anticodon, displaying 46 tRNA anticodons in total. The color code depicts the log2 fold change of each tRNA isodecoder group in every sample relative to the average of the three healthy samples. (B) Unsupervised hierarchical clustering analysis of normalized tRNA reads <60 nt for 15 DLBCL samples and three reactive lymph nodes. Each row represents a tRNA type, grouped by anticodon, displaying 46 tRNA anticodons in total. (C) Pearson correlation (r: −0.14) between RIN values and normalized full-length tRNA reads in 15 DLBCL samples and three healthy, reactive lymph node samples of good RNA quality (RIN values between 7 and 9). (D) Pearson correlation (r: −0.85) between normalized rRNA reads <100 nt and normalized full-length tRNA reads in 15 DLBCL samples and three healthy, reactive lymph node samples. (E) Heat map visualization showing improved clustering of DLBCL (n = 15) and reactive lymph node samples (n = 3) after inclusion of all tRNA reads.
Since the contribution of shorter tRNA sequences to the tRNA expression profiles compensated for possible intersample variations in our cell lines (Fig. 2E), inclusion of all tRNA-derived reads into our ALL-tRNAseq workflow could also further improve clustering analysis in tissue samples. In line with previous studies that validated the accuracy of their tRNA profiling methods with the detection of brain-specific tRNA-Arg-TCT-4-1 (Ishimura et al. 2014; Pinkard et al. 2020; Behrens et al. 2021), we first explored whether we could observe similar tissue-restricted expression using ALL-tRNAseq with inclusion of all tRNA reads. Comparison of tRNA expression profiles between histologically normal brain tissue (n = 3) and reactive lymph nodes (n = 3) indeed revealed a strong expression of tRNA ArgTCT-4-1 (Supplemental Fig. S5B). Next, in a subsequent analysis of all tRNA reads in our DLBCL set, we showed that implementation of all tRNA read counts for unsupervised hierarchical cluster analyses to detect tumor-associated tRNA signatures also strongly differentiated between DLBCL and reactive lymph nodes (Fig. 3E). In addition, the obtained signature from our previous full-length tRNA analysis was further enriched with multiple relative changes to the tRNA pool, including the down-regulated tRNA-Tyr-GTA expression that was observed in the full-length tRNA analysis (Fig. 3E).
ALL-tRNAseq allows tRNA expression profiling in brain tumor tissues of highly variable RNA integrity
Meaningful studies of tRNA repertoire composition during cancer development and its clinical consequences can be challenging due to difficulties in obtaining high-quality RNA from clinical tissue samples and the limited availability of such specimens. Although lower RNA quality in samples is often a reason for exclusion, collection and processing procedures of clinical tissue samples often influence RNA preservation (Gallego Romero et al. 2014). Based on our results with DLBCL analyses of samples with good RNA integrity, we questioned whether similar full-length tRNA expression profiles could be observed in GBM samples (n = 18) that showed higher variation in RNA quality (RIN values in range 4–9) (Supplemental Fig. S6). Hierarchical cluster analysis again indicated full-length tRNA could not fully separate GBM from normal brain tissue (n = 3), with the latter samples branching out from a subgroup of GBMs (Fig. 4A). In contrast, the expression of tsRNAs alone enabled strong differentiation between GBM tissues of highly variable RNA quality (RIN 4–9) and histologically normal brain tissue (RIN >7) that may have resulted from the higher degree of variation in RNA integrity and thus RNA fragmentation in GBM samples (Fig. 4B). Therefore, we again evaluated how the six low-quality tissue samples affected the correlation between the RINs and the proportion of full-length tRNA in histologically normal brain samples (n = 3) and GBM samples (n = 18) (Supplemental Fig. S6). Despite inclusion of low-quality samples, RIN values were still poorly correlated with the proportion of full-length tRNA (Pearson correlation r: 0.35) (Supplemental Fig. S5C), as a wide range between 4% and 41% of full-length tRNA proportion could be observed for the low-quality samples with RIN values <5. Nevertheless, our approach of correlating the proportion of rRNA reads <100 nt with the proportion of full-length tRNA proved to be beneficial for the determination of tRNA transcript integrity in GBM and histologically normal brain samples (r = −0.78) (Supplemental Fig. S5D). When we applied hierarchical cluster analysis to these samples, similar to our observations in DLBCL tissues, GBM samples still strongly differentiated from histologically normal brain samples once we incorporated all tRNA reads into the analysis (Fig. 4C).
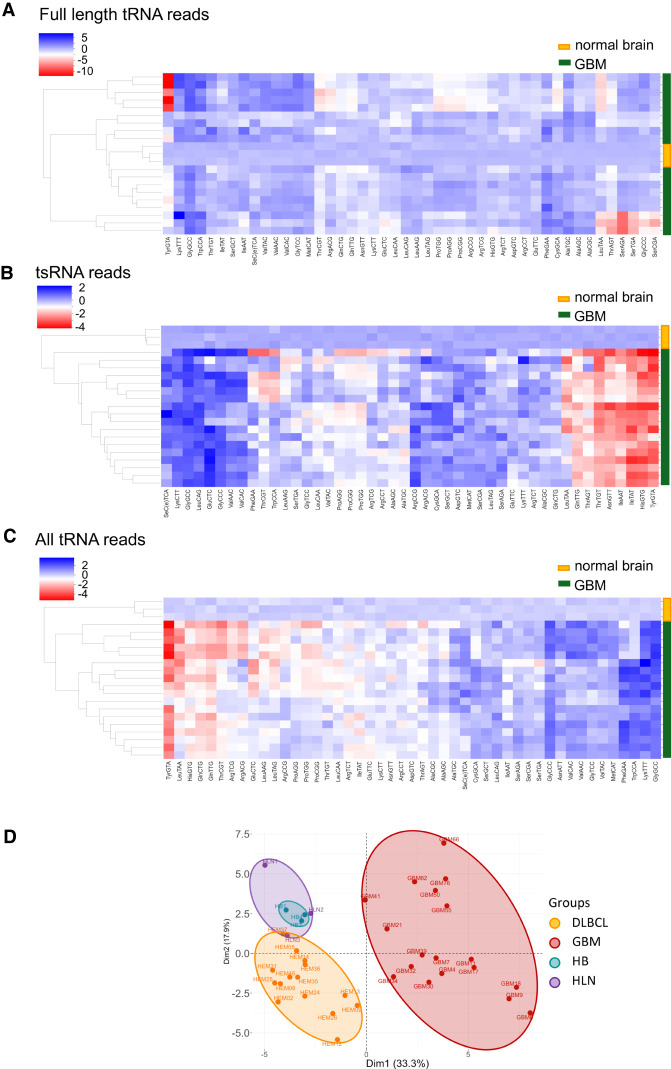
Figure 4.
ALL-tRNAseq allows tRNA expression profiling in brain tumor tissues of highly variable RNA integrity. (A) Heat map visualization showing clustering of full-length tRNA reads in glioblastoma (GBM) samples. Each horizontal line represents a biological sample; 18 GBM samples, and three normal brain samples. (B) Hierarchical clustering of GBM (n = 18) and normal brain samples (n = 3). tRNA reads <60 nt are normalized per million. Each row represents a tRNA type, grouped by anticodon, displaying 46 tRNA anticodons in total. (C) Heat map visualization showing clustering changes of GBM and normal brain samples after inclusion of all tRNA reads. (D) PCA of tRNA anticodon expression using all normalized reads per million mapping to cytoplasmic tRNA in reactive lymph nodes (n = 3), normal brain tissue (n = 3), DLBCL (n = 15), and GBM (n = 18).
Finally, we used PCA on tRNA-derived reads from all tissue samples to understand whether the difference in RNA quality is affecting variability in tRNA expression. Despite a significant variability in RNA integrity, healthy tissue samples clustered together, while both cancer types were fully separated (Fig. 4D). A higher variability in tRNA expression was observed in GBM samples, which could also reflect a higher degree of tumor heterogeneity in GBMs (Sottoriva et al. 2013). Notably, these PCA results recapitulated the unsupervised hierarchical cluster analyses (Fig. 4C). Altogether, these results confirm that tRNA read coverage by ALL-tRNAseq-based profiling is useful to assess sample integrity and enhances the identification and classification of tumor-associated tRNA signatures.
Discussion
Recent studies have revealed that differential mRNA translation rates were controlled, at least in part, by alteration of the expressed cellular tRNA repertoire (Gingold et al. 2014; Aharon-Hefetz et al. 2020; Rak et al. 2021). Increasing evidence suggests that mature tRNAs are also involved in cancer development and progression (Pavon-Eternod et al. 2009; Gingold et al. 2014; Goodarzi et al. 2015; Hernandez-Alias et al. 2020), prompting the development of optimal techniques enabling the reliable quantification of tRNA repertoires in human cells and tissues. However, the presence of RT-blocking modifications on the highly structured tRNAs fundamentally hinders accurate quantification via high-throughput sequencing technologies (Motorin et al. 2007), which limits in-depth investigation of tRNAs’ contribution during cancer development. Our results revealed that ALL-tRNAseq offers a robust high-throughput sequencing technology that is useful to explore relative mature tRNA abundance and to simultaneously assess tRNA integrity in cell lines as well as clinical tissue samples. We showed that ALL-tRNAseq can detect cell type-specific regulation of tRNA expression, providing an additional layer of information that improves classification of cancer-specific tRNA signatures in tumor tissues.
Combined analysis of full-length tRNA molecules and tRNA fragmentation patterning requires classical two-adapter-based ligation strategies, which may introduce a source of ligation bias for some RNA sequences and structures (Fuchs et al. 2015). To overcome this problem, ALL-tRNAseq applies an adapter ligation approach that has been previously shown to minimize ligation bias in miRNA sequencing methods (Giraldez et al. 2018; Kim et al. 2019). In addition, introduction of RNA-3′ adapter size selection simultaneously allowed for removal of excess free 3′ adapters, which enabled efficient ligation to the full RNA repertoire by using high concentrations of 3′ adapter. Incorporation of a demethylation step (Zhou et al. 2019) with a combination of wild-type and D135S mutant AlkB enzymes from Escherichia coli increased recovery of mature full-length tRNA sequences, dramatically shifting relative anticodon abundance in demethylated samples. One of the crucial modification roadblocks to overcome is m22G26 in type II tRNAs, which is sandwiched between the coaxially stacked D-loop and the tRNA anticodon and forms a tight pair with nucleotide 44, making it less accessible for reverse transcription (Clark et al. 2016). In line with the previously reported high processivity of MarathonRT in reverse-transcribing long and structured RNAs, we further showed that the ALL-tRNAseq protocol allowed for the bypass of RT roadblocks present on highly structured tRNA molecules, including the bulky yW modification in tRNA-Phe GAA. In addition, our protocol was able to dramatically increase cDNA yields of type II tRNAs even under relatively low temperatures compared with other RTs.
The main goal of ALL-tRNAseq development was to generate a sequencing approach that was suitable for profiling clinical tissue samples that often suffer from higher variability in RNA integrity. We showed that incorporation of all tRNA sequencing reads compensated for compromised RNA quality during sample processing. Although the Agilent 2100 bioanalyzer platform is an automated, highly reproducible approach for the evaluation of RNA integrity (Schroeder et al. 2006), we showed in this study that this RNA quality measurement is not sufficient to explain the extent of rRNA and tRNA fragmentation in both cell line-derived and tissue-derived samples. With the application of ALL-tRNAseq, we can provide this layer of information on tRNA integrity and, more importantly, compensate for technical tRNA fragmentation during sample collection and processing procedures.
Several additional enzymatic and/or chemical treatments can be incorporated into our sequencing approach according to the goal or application of the study. In the current study, analysis of biological tRNA-derived fragments was not included as a result of library preparation choices to make the workflow optimal for tRNA analysis in clinical tissue samples. The 3′ phosphate (3′-P) and 2′3′ cyclic phosphate (2′3′-cP) that are enzymatically generated at the ends of tRNA fragments during biogenesis require a treatment with T4 polynucleotide kinase (T4PNK) to convert the 3′-P or 2′3′-cP into ligation-competent ends (Hu et al. 2021; Shi et al. 2021). Although T4 PNK treatment is not incorporated into the current protocol, we foresee implementation of this additional step in the future to allow simultaneous assessment of all fragments, including tsRNAs.
A potential limitation of the ALL-tRNAseq procedure is that this protocol is incompatible with reverse transcription-induced modification calling that has been addressed by several other high-throughput tRNA sequencing methods. Even though this provides an essential additional layer of information on tRNA stability and processing, tRNA-seq-based modification calling needs to be implemented with care. Differences in RT processivity may alter RT-specific mismatch signatures that need to be considered during analysis of mismatch sites. Furthermore, we showed that inclusion of the demethylation step significantly affected the relative assessment of type II tRNA expression. Hence, assessment of tRNA methylation status by a reverse transcription-based approach may come with the disadvantage of less accurate profiling of tRNA expression. In addition, incorporation of MarathonRT did not allow us to compare reverse transcription-induced tRNA modification signatures due to its high processivity compared with other RTs. Nevertheless, MarathonRT has previously been reported to generate 2′O methylation and pseudouridylation signatures in long RNAs (Guo et al. 2020). Together with incorporation of mass spectrometry data and combining specific RT signatures, the utility of these misincorporation-induced modification signatures can be implemented into the ALL-tRNAseq workflow in the future. We showed that ALL-tRNAseq can accommodate detection of mature full-length tRNAs by alleviation of known RT roadblocks without losing information on possible tRNA fragmentation patterns. However, similar to all other sequencing-based protocols published so far, changes in relative abundance of a few tRNA isodecoders can possibly still be linked to persistent bulky RT blocks present on these tRNAs rather than their abundance. For instance, modification levels of 2-methylthio-6-threonylcarbamoyl-A (ms2t6A) in lysine tRNAs or yW in tRNA-PheGAA may be reduced in some conditions, leading to better accessibility of the RT and thus artificially increasing their abundance in sequencing-based quantification (Rak et al. 2021), as also described by others (Behrens et al. 2021). Whether the abundance of such tRNAs appears affected in relation to a change in their modification status or their expression, sequencing-based protocols remain powerful tools to identify attractive targets for further investigation.
Altogether, with ALL-tRNAseq, we introduced a method that incorporates an optimized strategy for mature tRNA quantification while making the workflow amenable for samples that may suffer from compromised RNA quality. Applying ALL-tRNAseq to profile human cell lines of different tissue origins as well as clinical tissue samples, we identified cell lineage- and tumor-specific tRNA profiles that effectively improve classification of oncogenic signatures in samples presenting higher levels of RNA fragmentation. This illustrates the high potential of implementing ALL-tRNAseq for the discovery of oncogenic tRNA signatures in human malignancies.
Materials and methods
Cell culture
SNB-75 and HEK293T were cultured at 37°C in a humidified atmosphere containing 5% CO2 in complete Dulbecco's modified Eagle medium (DMEM; Sigma Aldrich D5796). All passaging was performed with trypsin ethylenediaminetetraacetic acid (EDTA) solution (Sigma Aldrich T3924) according to the manufacturer's instructions. Media were supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) and 1% penicillin/streptomycin. SU-DHL-5 cells were cultured in suspension at 37°C with 5% CO2 in Roswell Park Memorial Institute (RPMI)-1640 medium (Sigma Aldrich R8758). Medium was supplemented with 10% heat-inactivated fetal bovine serum (FBS; Thermo Fisher Scientific) and 1% penicillin/streptomycin. hESC line WA09 from WiCell was maintained in complete TESR-E8 medium (Stem Cell Technologies 05990) supplemented with 1% penicillin/streptomycin on Vitronectin XF-coated (Stem Cell Technologies 07180) plates. Passaging was performed with gentle cell dissociation reagent (Stem Cell Technologies 100-0485). Media were supplemented with 10 µM ROCK inhibitor (RI; SelleckChem S1049) after passaging. All cell lines were subjected to mycoplasma testing and only used for experiments when confirmed negative.
Differentiation assay
hESCs were exposed to all-trans retinoic acid (ATRA in DMSO; MP Biomedicals 02190269.6) at a final concentration of 1 µM in plain TESR-E8 medium supplemented with 1% penicillin/streptomycin (Thermo Fisher Scientific 15140122). Medium was changed daily. Cells were harvested in mirVana miRNA isolation kit lysis buffer (Ambion AM1560) after 0 d (no ATRA) or 1, 3, and 5 d following ATRA induction. Real-time qPCR was used to quantify levels of mRNA expression of four selected genes as previously described in Gingold et al. (2014) and Sagi et al. (2016). First strand cDNA was synthesized from 100 ng of total RNA using SuperScript III RT (Thermo Fisher Scientific 18080093) and random hexamers according to the manufacturer's instructions. qPCR was performed using SYBR Green PCR master mix (Applied Biosystems 10187094) on an Applied Biosystems 7500 real-time PCR system (primer sequences are listed in Supplemental Table S1).
Clinical samples
Surgical tissue samples of diffuse large B-cell lymphoma (DLBCL; n = 15) and glioblastoma (GBM; n = 18) were obtained from the Department of Pathology of Amsterdam University Medical Center, Vrije Universiteit, in Amsterdam, and HemoBase Registry (Kibbelaar et al. 2017) in Leeuwarden, the Netherlands. Complete data on clinical features at presentation, treatment, and outcome are available for all samples. As control tissues, reactive lymph node tissue samples (n = 3) and histologically normal brain tissue samples (n = 3) were included. All material and clinical data have been fully anonymized, and the study was performed according to the guidelines of the Helsinki Declaration. Approval of the Medical Ethical Committee at Amsterdam University Medical Center has been obtained for the use of all included tissues and related clinical data.
RNA isolation
Total RNA of cell lines and tissue samples was extracted using mirVana isolation kit (Ambion AM1560). All tissue samples were kept on dry ice during the initial processing steps. Approximately 30–80 mg of freshly frozen tissue was cut on a cryostat at −20°C and directly disrupted by TissueRuptor II (Qiagen 9002756) with TissueRuptor disposable probes (Qiagen 990890) in mirVana miRNA isolation kit lysis buffer (Ambion AM1560). First, total RNA isolation was performed in an RNase-free environment, followed by on-column RNase-free DNase treatment (Qiagen 79254). Deacylation was then performed by incubating total RNA in 0.1 M Tris-HCL (pH 9.0) and 1 mM EDTA for 30 min at 37°C (Blanco et al. 2014). After ethanol precipitation, total RNA was resuspended in RNase-free H2O. Quality and integrity of all RNA samples were measured on a NanodropOne (Thermo Fisher Scientific) and Agilent 2100 bioanalyzer (Agilent Technologies) using an RNA 6000 Nano kit (Agilent Technologies 5067-1511).
Expression and purification of AlkB WT and mutant enzymes
Expression and purification methods were adapted from previously published methods (Cozen et al. 2015; Zheng et al. 2015; Hrabeta-Robinson et al. 2017). pET30a-AlkB-WT and pET30a-AlkB-D135S, a truncated AlkB with deletion of the N-terminal 11 amino acids, were a gift from Tao Pan (Addgene plasmid #79050 [http://n2t.net/addgene:79050; RRID: Addgene_79050] and #79051 [http://n2t.net/addgene:79051; RRID: Addgene_79051], respectively). AlkB in pET30a vector was overexpressed in E. coli BL21 (DE3) pLysS, and cells were grown in 12 L of LB medium at 37°C in the presence of kanamycin until an OD600 of 0.5 was reached. Cells were then incubated for 4 h with 1 mM IPTG (Sigma Aldrich I1312) at 30°C to express the His-3C-AlkB fusion protein. Cells were collected and pelleted at 4600g for 20 min and resuspended in sonication buffer (20 mM HEPES-KOH at pH 7.5, 5% glycerol, 1 M NaCl, 2 mM β-mercaptoethanol, 2 µg/mL leupeptin, 1 µg/mL pepstatin, 1 mM Pefabloc). Cells were lysed by sonication eight times for 10 sec with 1 min rest and then centrifuged at 17,420g for 20 min. The soluble proteins were affinity-purified on Talon metal affinity resin (Takara CL635502), concentrated to 0.5 mL, and then further purified by size exclusion chromatography over a HiLoad 16/60 Superdex 200 prepgrade column (GE Healthcare Life Sciences 28-9893-35) using an Ultimate 3000 high-performance liquid chromatography (HPLC) system (Dionex). The system was composed of an LPG-3400SD pump, a WPS-3000 autosampler, a VWD-3100 UV detector, and an AFC-3000 fraction collector. The column was equilibrated with five column volumes (CVs) of elution buffer with an isocratic flow rate of 1 mL/min. Protein aggregates were removed by centrifugation on a centrifugal filter unit with 0.22 µM Durapore PVDF membrane (Millipore Sigma UFC30GV25), and 0.5 mL of sample was injected. Detection was carried out at 280 nm, and collected fraction size was 3 mL. Collected fractions were pooled, concentrated, and then stored in buffer containing 20 mM Tris-HCl (pH 8.0), 50% glycerol, 0.2 M NaCl, and 2 mM dithiothreitol at −80°C. All protein purification steps were performed at 4°C. Protein concentration was measured by Pierce BCA protein assay kit (Thermo Fisher 23227). Yield and protein quality were further assessed by SDS-PAGE and Coomassie staining.
Demethylation reaction
Demethylation reaction was adapted from a previously published method (Hrabeta-Robinson et al. 2017) with some additional modifications (Zhou et al. 2019) and was performed on total RNAs. Briefly, 2 µg of total RNA was treated with 19 µM AlkB consisting of 8.5 µM AlkB WT and 10.5 µM AlkB-D135S mutant (ratio adapted from a previously published method) (Zheng et al. 2015), 10 mM KCl, 2 mM MgCl2, 283 µM freshly prepared (NH4)2Fe(SO4)2, 6H2O, 0.3 mM 2-ketoglutarate, 2 mM freshly made L-ascorbic acid, 40 U of RNase inhibitor, and 50 mM MES buffer (pH 5.0) (Fisher Scientific 15474529) in a total volume of 40 µL. Reactions were incubated for 2 h at 25°C and quenched with 5 mM EDTA (Ambion AM9260). RNA was recovered by an oligo Clean&Concentrator-5 kit (Zymo Research D4060) and eluted in 6 µL of nuclease-free water (Ambion AM9937). RNA quality and integrity were assessed by Agilent 2100 bioanalyzer (Agilent Technologies) using an RNA 6000 Nano kit (Agilent Technologies 5067-1511).
Small RNA-seq library preparation
Demethylated total RNA was used for each cell line and tissue sample. For sequencing libraries of ALL-tRNAseq, a 5′ phosphorylated 3′ end adapter with four randomized nucleotides at the 5′ end and a 3′ blocking group (3C Spacer; IDT 3SpC3) was adenylated by Mth RNA ligase (NEB M2611A) and then ligated to the RNA template using a truncated KQ T4 RNA ligase 2 (NEB M0373L) for 1 h at 25°C in the presence of 20% PEG8000 (NEB). Ligated RNA was gel-purified and size-fractionated on a Novex TBE–urea 10% denaturing polyacrylamide gel (Invitrogen EC68752BOX) to enrich small RNA molecules using two FAM markers of 25 and 39 nt (from a previously published protocol) (Kim et al. 2019) and the low-range ssRNA ladder (NEB N0364S). Gels were stained using 1× SYBR Gold nucleic acid stain (Thermo Fisher Scientific S11494) in 1× TBE for 10 min, and 3′adapter-ligated RNA in size range of 39–150 nt was excised on a blue-light transilluminator. RNA was recovered from excised polyacrylamide gel pieces by crushing the gel fragments and soaking them in 0.3 M NaCl overnight at 4°C, followed by ethanol precipitation. Subsequently, 5′ adapters with four terminal randomized nucleotides at the 3′ end were ligated using T4 RNA ligase (NEB M0204S). Ligation was performed for 1 h at 25°C in the presence of 20% PEG8000. Thereafter, reverse transcription was performed for 1 h at 50°C or 42°C using SuperScript III (Invitrogen 18080093) or MarathonRT (Kerafast EYU007), respectively. Reactions were terminated by incubation with 0.25 M NaOH for 3 min at 95°C and then neutralized with 0.25 M HCl. Next, cDNA was purified using MinElute reaction cleanup kit (Qiagen 28006), and PCR was amplified using Phusion high-fidelity PCR master mix (Fisher Scientific F531L) for 12 cycles of 5 sec at 98°C, 10 sec at 62°C, and 10 sec at 72°C. Primers, randomized adapters, Illumina Multiplex, and barcode primers were obtained from IDT and are listed in Supplemental Table S1. PCR products were equimolarly pooled for cluster generation without additional size selection. The quality of the sequence libraries was validated using Agilent high-sensitivity DNA kit (Agilent Technologies), and a fragment analyzer system (Agilent Technologies) was used to check size, purity, and concentration. Sequencing of ALL-tRNAseq was performed as single-end reads for 150 cycles on a NovaSeq 6000 at GenomeScan.
Sequencing analysis
Raw Illumina reads were first preprocessed using sRNAbench (Aparicio-Puerta et al. 2019, 2022) for adapter removal and quality control. The standard sRNAbench mapping step using bowtie was extended with a second round of Smith–Waterman alignment implemented using BioJava, which further allows gaps and/or mismatches, to recover additional tRNA-derived reads. Reference libraries were obtained from GtRNAdb 2.0 (Homo sapiens GRCh38/hg38) for nuclear tRNAs and mitotRNAdb for mitochondrial tRNAs. tRNA expression was profiled at different levels: tRNA gene level, anticodon level, and amino acid level. Reads containing the 5′ end of the complete sequence with CCA ending (allowing 1-nt truncation) were classified as mature full-length transcripts.
To compare ALL-tRNAseq libraries with previously published methods, we downloaded publicly available data sets from the NCBI Gene Expression Omnibus repository for DM-tRNA-seq (GSE66550) (Zheng et al. 2015) and mim-tRNAseq (GSE152621) (Behrens et al. 2021) that we processed using the same pipeline described above. All raw sequencing files generated for this study are available at GEO accession GSE186736.
Northern blotting
Northern blots were largely performed as described by Gerber et al. (2020) with the following modifications. Total RNA was resolved on a 10% urea–polyacrylamide gel. After electrophoresis, RNAs were transferred onto a Hybond N+ membrane (Thermo Fisher RPN2020B) for 1 h at 30 V using a wet electrophoretic transfer system. RNAs were UV cross-linked (0.24 J on the RNA side and 0.12 J on the other side of the membrane). Prehybridization was performed in ULTRAhyb oligo hybridization buffer (Thermo Fisher AM8663) for 1 h at 42°C with gentle agitation. Hybridization was then performed with 50 nM biotinylated oligonucleotide probes overnight at 37°C. Membranes were subsequently washed twice for 10 min with 5× SSC and 0.5% SDS at room temperature and then blocked for 15 min with 2× SSC, 0.5% SDS, and 3% BSA at room temperature. Membranes were then incubated with streptavidin-HRP (1:40,000; Thermo Fisher N100) in 2× SSC with 3% BSA and 0.5% SDS for 30 min. Membranes were subsequently washed with ABS buffer (10% BSA, 1% Triton X-100 in 2× SSC) and washed twice for 5 min with 2× SSC. Membranes were rinsed briefly with PBS prior to exposure using SuperSignal WestPico Plus ECL-based detection (Thermo Fisher 34580). Finally, membranes were exposed on a Uvitec chemiluminescence imaging system. Band intensity was quantified using ImageJ. Oligonucleotide sequences are listed in Supplemental Table S1.
Primer extension analysis
Demethylated total RNA (2 µg) was used for each primer extension condition. Three biotin-labeled primers were heated to 80°C together with demethylated RNA and dNTPs in RT buffer and slowly ramped down to 42°C. At 42°C, MarathonRT was added and incubated for 1 h to allow primer extension. Reactions were terminated by incubation with 0.25 M NaOH for 3 min at 95°C and then neutralized with 0.25 M HCl. Next, cDNA was purified using MinElute reaction cleanup kit (Qiagen 28006) and resolved on a 10% urea–polyacrylamide gel. After electrophoresis, cDNAs were visualized using SYBR Gold staining and subsequently transferred onto a Hybond N+ membrane (Thermo Fisher RPN2020B) for 1 h at 30 V using a wet electrophoretic transfer system. cDNAs were UV cross-linked (0.24 J on the RNA side, and 0.12 J on the other side of the membrane). Membranes were blocked for 1 h with 2× SSC, 0.5% SDS, and 3% BSA at room temperature. Membranes were then incubated with streptavidin-HRP (1:40,000; Thermofisher N100) in 2× SSC with 3% BSA and 0.5% SDS for 30 min. Membranes were subsequently washed with ABS buffer (10% BSA, 1% Triton X-100 in 2× SSC) and washed twice for 5 min with 2× SSC. Membranes were rinsed briefly with PBS prior to exposure using SuperSignal WestPico Plus ECL-based detection (Thermo Fisher 34580). Finally, membranes were exposed on a Uvitec chemiluminescence imaging system.
Statistical analysis
RT-qPCR data were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH), cycle threshold (Ct) values were obtained, and ΔΔCt values were calculated to determine relative abundance and are reported as mean ± SD. All expression values derived from sequencing data presented throughout this work are expressed as normalized RPM (reads per million) per reference library or total mapped reads, as indicated in the figure legends. We performed statistical analysis (Wilcoxon rank sum test for significance) using GraphPad Prism9 software. Pearson correlations were calculated using the function pearsonr from the scipy package (v1.8.1). PCA was carried out by means of the prcomp function in R software (http://www.r-project.org), and visualization was performed using the factoextra R package. Heat maps were generated using the clustermap function from the package seaborn. Samples were clustered using single-linkage clustering implemented in the cluster.hierarchy.linkage method from scipy. We applied hierarchical clustering using average linkage based on Euclidean distance.
Data and code availability
The sequencing data reported here have been deposited in NCBI's Gene Expression Omnibus under accession number GSE186736. The ALL-tRNAseq computational pipeline is available at https://github.com/bioinfoUGR/sRNAtoolbox. All custom scripts for data visualization are available at https://github.com/sert23/ALL-tRNAseq. Original blot scans were deposited as a Mendeley data set and are accessible at http://dx.doi.org/10.17632/ctzn8mf42n.1.
Supplementary Material
Acknowledgments
This work was supported by the Dutch Cancer Society (KWF 2016-10476 to D.K.-L.), the Cancer Center Amsterdam Foundation (CCA2017-2-16 to D.K.-L.), and “Stichting MRD Hodgkin Lymphoma” (to D.M.P.). C.S was supported by a Cancer Center Amsterdam Foundation grant (CCA2021-5-26 to A.G.). A.G. was supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) Talent Programme Vidi grant (VI.Vidi.193.107). S.G., M.H., D.K.-L., and T.W. received funding from the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement number 765492. We thank Disa Tehler for stimulating discussions, and we are grateful to Hakan Kalay for assistance with the purification of AlkB enzymes. We thank Bram van den Broek for his contribution to the graphical images.
Author contributions: C.S., D.d.J., D.M.P., M.H., A.G., and D.K.-L. conceived the study. C.S., E.A.-P., H.V., V.M.H., M.H., T.W., A.G., and D.K.-L. performed the methodology. E.A.-P., C.G.-M., and M.H. ran the software. C.S., E.A.-P., C.G.-M., and A.G performed the formal analysis. C.S., E.A.-P., C.G.-M., H.V., M.v.E., L.E.W., S.G., and L.G. performed the investigation. N.H., H.T.v.d.G., R.S.v.R., E.A., R.K., V.M.H., P.W., D.P.N., W.P.V., D.d.J, D.M.P., M.H., and D.K.-L. acquired the resources. C.S, D.M.P., and A.G. wrote the original draft of the manuscript. All of the authors reviewed and edited the manuscript. C.S., E.A.-P., C.G.-M., and A.G. visualized the study. D.d.J., D.M.P., T.W., A.G., and D.K.-L. supervised the study. L.E.W., D.d.J., and D.K.-L. were the project administrators. D.M.P., A.G., and D.K.-L. acquired the funding.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.350233.122.
Competing interest statement
The authors declare no competing interests.
References
- Aharon-Hefetz N, Frumkin I, Mayshar Y, Dahan O, Pilpel Y, Rak R. 2020. Manipulation of the human tRNA pool reveals distinct tRNA sets that act in cellular proliferation or cell cycle arrest. Elife 9: e58461. 10.7554/eLife.58461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio-Puerta E, Lebrón R, Rueda A, Gómez-Martín C, Giannoukakos S, Jaspez D, Medina JM, Zubkovic A, Jurak I, Fromm B, et al. 2019. sRNAbench and sRNAtoolbox 2019: intuitive fast small RNA profiling and differential expression. Nucleic Acids Res 47: W530–W535. 10.1093/nar/gkz415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio-Puerta E, Gómez-Martín C, Giannoukakos S, Medina JM, Scheepbouwer C, García-Moreno A, Carmona-Saez P, Fromm B, Pegtel M, Keller A, et al. 2022. sRNAbench and sRNAtoolbox 2022 update: accurate miRNA and sncRNA profiling for model and non-model organisms. Nucleic Acids Res 50: W710–W717. 10.1093/nar/gkac363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens A, Rodschinka G, Nedialkova DD. 2021. High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol Cell 81: 1802–1815.e7. 10.1016/j.molcel.2021.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al. 2014. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 33: 2020–2039. 10.15252/embj.201489282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskovic A, Bing XY, Kaymak E, Rando OJ. 2020. Control of noncoding RNA production and histone levels by a 5′ tRNA fragment. Genes Dev 34: 118–131. 10.1101/gad.332783.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark WC, Evans ME, Dominissini D, Zheng G, Pan T. 2016. tRNA base methylation identification and quantification via high-throughput sequencing. RNA 22: 1771–1784. 10.1261/rna.056531.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozen AE, Quartley E, Holmes AD, Robinson EH, Phizicky EM, Lowe TM. 2015. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods 12: 879–884. 10.1038/nmeth.3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas A, Vlassov AV, Kazakov SA. 2004. Principles of nucleic acid cleavage by metal ions. In Artificial nucleases. Nucleic acids and molecular biology, vol 13 (ed. Zenkova MA), pp. 61–88. Springer, Berlin. 10.1007/978-3-642-18510-6_6 [DOI] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T. 2006. Tissue-specific differences in human transfer RNA expression. PLoS Genet 2: e221. 10.1371/journal.pgen.0020221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drino A, Oberbauer V, Troger C, Janisiw E, Anrather D, Hartl M, Kaiser S, Kellner S, Schaefer MR. 2020. Production and purification of endogenously modified tRNA-derived small RNAs. RNA Biol 17: 1104–1115. 10.1080/15476286.2020.1733798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erber L, Hoffmann A, Fallmann J, Betat H, Stadler PF, Mörl M. 2020. LOTTE-seq (Long hairpin oligonucleotide based tRNA high-throughput sequencing): specific selection of tRNAs with 3′-CCA end for high-throughput sequencing. RNA Biol 17: 23–32. 10.1080/15476286.2019.1664250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans ME, Clark WC, Zheng G, Pan T. 2017. Determination of tRNA aminoacylation levels by high-throughput sequencing. Nucleic Acids Res 45: e133. 10.1093/nar/gkx514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs RT, Sun Z, Zhuang F, Robb GB. 2015. Bias in ligation-based small RNA sequencing library construction is determined by adaptor and RNA structure. PLoS One 10: e0126049. 10.1371/journal.pone.0126049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego Romero I, Pai AA, Tung J, Gilad Y. 2014. RNA-seq: impact of RNA degradation on transcript quantification. BMC Biol 12: 42. 10.1186/1741-7007-12-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A, Ito K, Chu CS, Roeder RG. 2020. Gene-specific control of tRNA expression by RNA polymerase II. Mol Cell 78: 765–778.e7. 10.1016/j.molcel.2020.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sørensen KD, et al. 2014. A dual program for translation regulation in cellular proliferation and differentiation. Cell 158: 1281–1292. 10.1016/j.cell.2014.08.011 [DOI] [PubMed] [Google Scholar]
- Giraldez M, Spengler R, Etheridge A, Godoy P, Barczak A, Srinivasan S, De Hoff P, Tanriverdi K, Courtright A, Lu S, et al. 2018. Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat Biotechnol 36: 746–757. 10.1038/nbt.4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogakos T, Brown M, Garzia A, Meyer C, Hafner M, Tuschl T. 2017. Characterizing expression and processing of precursor and mature human tRNAs by hydro-tRNAseq and PAR-CLIP. Cell Rep 20: 1463–1475. 10.1016/j.celrep.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Liu X, Nguyen HCB, Zhang S, Fish L, Tavazoie SF. 2015. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161: 790–802. 10.1016/j.cell.2015.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L-T, Adams RL, Wan H, Huston NC, Potapova O, Olson S, Gallardo CM, Graveley BR, Torbett BE, Pyle AM. 2020. Sequencing and structure probing of long RNAs using MarathonRT: a next-generation reverse transcriptase. J Mol Biol 432: 3338–3352. 10.1016/j.jmb.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L-T, Olson S, Patel S, Graveley BR, Pyle AM. 2022. Direct tracking of reverse-transcriptase speed and template sensitivity: implications for sequencing and analysis of long RNA molecules. Nucleic Acids Res 50: 6980–6989. 10.1093/nar/gkac518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. 2010. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16: 673–695. 10.1261/rna.2000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Alias X, Benisty H, Schaefer MH, Serrano L. 2020. Translational efficiency across healthy and tumor tissues is proliferation-related. Mol Syst Biol 16: e9275. 10.15252/msb.20199275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabeta-Robinson E, Marcus E, Cozen AE, Phizicky EM, Lowe TM. 2017. High-throughput small RNA sequencing enhanced by AlkB-facilitated RNA de-methylation (ARM-seq). Methods Mol Biol 1562: 231–243. 10.1007/978-1-4939-6807-7_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JF, Yim D, Ma D, Huber SM, Davis N, Bacusmo JM, Vermeulen S, Zhou J, Begley TJ, DeMott MS, et al. 2021. Quantitative mapping of the cellular small RNA landscape with AQRNA-seq. Nat Biotechnol 39: 978–988. 10.1038/s41587-021-00874-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Zhou H, Yang X-L, Schimmel P, Senju S, Nishimura Y, Chuang JH, Ackerman SL. 2014. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345: 455–459. 10.1126/science.1249749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibbelaar RE, Oortgiesen BE, van der Wal-Oost AM, Boslooper K, Coebergh JW, Veeger NJGM, Joosten P, Storm H, van Roon EN, Hoogendoorn M. 2017. Bridging the gap between the randomised clinical trial world and the real world by combination of population-based registry and electronic health record data: a case study in haemato-oncology. Eur J Cancer 86: 178–185. 10.1016/j.ejca.2017.09.007 [DOI] [PubMed] [Google Scholar]
- Kim H, Kim J, Kim K, Chang H, You K, Kim VN. 2019. Bias-minimized quantification of microRNA reveals widespread alternative processing and 3′ end modification. Nucleic Acids Res 47: 2630–2640. 10.1093/nar/gky1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Anaya J, Mudunuri SB, Dutta A. 2014. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol 12: 78. 10.1186/s12915-014-0078-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Kuscu C, Dutta A. 2016. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem Sci 41: 679–689. 10.1016/j.tibs.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G. 2018. tRNA-derived small RNAs: new players in genome protection against retrotransposons. RNA Biol 15: 170–175. 10.1080/15476286.2017.1403000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. 2013. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci 110: 1404–1409. 10.1073/pnas.1206761110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micke P, Ohshima M, Tahmasebpoor S, Ren Z-P, Östman A, Pontén F, Botling J. 2006. Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Lab Invest 86: 202–211. 10.1038/labinvest.3700372 [DOI] [PubMed] [Google Scholar]
- Minshall N, Git A. 2020. Enzyme- and gene-specific biases in reverse transcription of RNA raise concerns for evaluating gene expression. Sci Rep 10. 10.1038/s41598-020-65005-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S, Ghanem E, Smith W, Sheeter D, Qin Y, King O, Polioudakis D, Iyer VR, Hunicke-Smith S, Swamy S, et al. 2013. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA 19: 958–970. 10.1261/rna.039743.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Muller S, Behm-Ansmant I, Branlant C. 2007. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol 425: 21–53. 10.1016/S0076-6879(07)25002-5 [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Lee ES. 2015. Non-coding RNA: what is functional and what is junk? Front Genet 6: 2. 10.3389/fgene.2015.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T. 2018. Modifications and functional genomics of human transfer RNA. Cell Res 28: 395–404. 10.1038/s41422-018-0013-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang YLJ, Abo R, Levine SS, Dedon PC. 2014. Diverse cell stresses induce unique patterns of tRNA up- and down-regulation: tRNA-seq for quantifying changes in tRNA copy number. Nucleic Acids Res 42: e170. 10.1093/nar/gku945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. 2009. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res 37: 7268–7280. 10.1093/nar/gkp787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perche-Letuvée P, Molle T, Forouhar F, Mulliez E, Atta M. 2014. Wybutosine biosynthesis: structural and mechanistic overview. RNA Biol 11: 1508–1518. 10.4161/15476286.2014.992271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkard O, McFarland S, Sweet T, Coller J. 2020. Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. Nat Commun 11: 4104. 10.1038/s41467-020-17879-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak R, Dahan O, Pilpel Y. 2018. Repertoires of tRNAs: the couplers of genomics and proteomics. Annu Rev Cell Dev Biol 34: 239–264. 10.1146/annurev-cellbio-100617-062754 [DOI] [PubMed] [Google Scholar]
- Rak R, Polonsky M, Eizenberg-Magar I, Mo Y, Sakaguchi Y, Mizrahi O, Nachshon A, Reich-Zeliger S, Stern-Ginossar N, Dahan O, et al. 2021. Dynamic changes in tRNA modifications and abundance during T cell activation. Proc Natl Acad Sci 118: e2106556118. 10.1073/pnas.2106556118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter F, Plehn JE, Bessler L, Hertler J, Jörg M, Cirzi C, Tuorto F, Friedland K, Helm M. 2022. RNA marker modifications reveal the necessity for rigorous preparation protocols to avoid artifacts in epitranscriptomic analysis. Nucleic Acids Res 50: 4201–4215. 10.1093/nar/gkab1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi D, Rak R, Gingold H, Adir I, Maayan G, Dahan O, Broday L, Pilpel Y, Rechavi O. 2016. Tissue- and time-specific expression of otherwise identical tRNA genes. PLoS Genet 12: e1006264. 10.1371/journal.pgen.1006264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt BM, Rudolph KLM, Karagianni P, Fonseca NA, White RJ, Talianidis I, Odom DT, Marioni JC, Kutter C. 2014. High-resolution mapping of transcriptional dynamics across tissue development reveals a stable mRNA–tRNA interface. Genome Res 24: 1797–1807. 10.1101/gr.176784.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. 2017. LTR-retrotransposon control by tRNA-derived small RNAs. Cell 170: 61–71.e11. 10.1016/j.cell.2017.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. 2006. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol 7: 3. 10.1186/1471-2199-7-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yu X, Zhu L, Li T, Yan Z, Guo J. 2018. Transfer RNA-derived fragments and tRNA halves: biogenesis, biological functions and their roles in diseases. J Mol Med (Berl) 96: 1167–1176. 10.1007/s00109-018-1693-y [DOI] [PubMed] [Google Scholar]
- Shi J, Zhang Y, Tan D, Zhang X, Yan M, Zhang Y, Franklin R, Shahbazi M, Mackinlay K, Liu S, et al. 2021. Pandora-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat Cell Biol 23: 424–436. 10.1038/s41556-021-00652-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu M, Honda S, Loher P, Telonis AG, Rigoutsos I, Kirino Y. 2017. YAMAT-seq: an efficient method for high-throughput sequencing of mature transfer RNAs. Nucleic Acids Res 45: e70. 10.1093/nar/gkx005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Piccirillo SGM, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavaré S. 2013. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci 110: 4009–4014. 10.1073/pnas.1219747110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ‘t Hoen PAC, Friedländer MR, Almlöf J, Sammeth M, Pulyakhina I, Anvar SY, Laros JFJ, Buermans HPJ, Karlberg O, Brännvall M, et al. 2013. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat Biotechnol 31: 1015–1022. 10.1038/nbt.2702 [DOI] [PubMed] [Google Scholar]
- Warren JM, Salinas-Giegé T, Hummel G, Coots NL, Svendsen JM, Brown KC, Drouard L, Sloan DB. 2021. Combining tRNA sequencing methods to characterize plant tRNA expression and post-transcriptional modification. RNA Biol 18: 64–78. 10.1080/15476286.2020.1792089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins CP, Zhang W, Wylder AC, Katanski CD, Pan T. 2022. A multiplex platform for small RNA sequencing elucidates multifaceted tRNA stress response and translational regulation. Nat Commun 13: 2491. 10.1038/s41467-022-30261-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Schmidt L, Marchand V, Kemmer T, Falschlunger C, Sednev MV, Bec G, Ennifar E, Höbartner C, Micura R, et al. 2020. Machine learning of reverse transcription signatures of variegated polymerases allows mapping and discrimination of methylated purines in limited transcriptomes. Nucleic Acids Res 48: 3734–3746. 10.1093/nar/gkaa113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann AJ, Gorski SA, Vogel J. 2012. Dual RNA-seq of pathogen and host. Nat Rev Microbiol 10: 618–630. 10.1038/nrmicro2852 [DOI] [PubMed] [Google Scholar]
- Wittig B, Wittig S. 1978. Reverse transcription of tRNA. Nucleic Acids Res 5: 1165–1178. 10.1093/nar/5.4.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Smith DK, Ni H, Wu K, Huang D, Pan S, Sathe AA, Tang Y, Liu M-L, Xing C, et al. 2020. SOX4-mediated repression of specific tRNAs inhibits proliferation of human glioblastoma cells. Proc Natl Acad Sci 117: 5782–5790. 10.1073/pnas.1920200117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Liu F, Pyle AM. 2018. An ultraprocessive, accurate reverse transcriptase encoded by a metazoan group II intron. RNA 24: 183–195. 10.1261/rna.063479.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. 2015. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods 12: 835–837. 10.1038/nmeth.3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Rauch S, Dai Q, Cui X, Zhang Z, Nachtergaele S, Sepich C, He C, Dickinson BC. 2019. Evolution of a reverse transcriptase to map N1-methyladenosine in human messenger RNA. Nat Methods 16: 1281–1288. 10.1038/s41592-019-0550-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data reported here have been deposited in NCBI's Gene Expression Omnibus under accession number GSE186736. The ALL-tRNAseq computational pipeline is available at https://github.com/bioinfoUGR/sRNAtoolbox. All custom scripts for data visualization are available at https://github.com/sert23/ALL-tRNAseq. Original blot scans were deposited as a Mendeley data set and are accessible at http://dx.doi.org/10.17632/ctzn8mf42n.1.