Abstract
The prion protein (PrP) is a broadly expressed glycoprotein linked with a multitude of (suggested) biological and pathological implications. Some of these roles seem to be due to constitutively generated proteolytic fragments of the protein. Among them is a soluble PrP form, which is released from the surface of neurons and other cell types by action of the metalloprotease ADAM10 in a process termed ‘shedding’. The latter aspect is the focus of this review, which aims to provide a comprehensive overview on (i) the relevance of proteolytic processing in regulating cellular PrP functions, (ii) currently described involvement of shed PrP in neurodegenerative diseases (including prion diseases and Alzheimer’s disease), (iii) shed PrP’s expected roles in intercellular communication in many more (patho)physiological conditions (such as stroke, cancer or immune responses), (iv) and the need for improved research tools in respective (future) studies. Deeper mechanistic insight into roles played by PrP shedding and its resulting fragment may pave the way for improved diagnostics and future therapeutic approaches in diseases of the brain and beyond.
Keywords: Biomarker, Immune responses, Intercellular communication, Neurodegeneration, Proteolytic cleavage
Introducing the proteolytic processing of the prion protein: a brief overview on a protein’s enzymatic fragmentation
Since its discovery in the context of unravelling a mysterious group of fatal and transmissible neurodegenerative disease in humans and animals (now collectively termed prion diseases) (Colby and Prusiner 2011; Prusiner 1993), a variety of biological functions and diverse (patho)physiological implications (reviewed in (Aguzzi et al. 2008; Hirsch et al. 2017; Manni et al. 2020; Watts et al. 2018; Wulf et al. 2017)) have been attributed to the evolutionary conserved cellular prion protein (PrP) (Basler et al. 1986; Oesch et al. 1985; Westaway and Prusiner 1986). In prion diseases (including Creutzfeldt-Jakob disease [CJD] in humans or bovine spongiform encephalopathy [BSE] in cattle), PrP undergoes progressive, templated three-dimensional misfolding (into its pathological ‘scrapie’ isoform PrPSc) and aggregation, and its expression is thus prerequisite and driving force of these ultimately fatal neurodegenerative conditions (Bockman et al. 1985; Prusiner 1982). Another pathological implication was found roughly a decade ago, when it was first shown (Gimbel et al. 2010; Laurén et al. 2009) and subsequently firmly established (Beraldo et al. 2016; Chen et al. 2010; Chung et al. 2010; Dohler et al. 2014; Freir et al. 2011; Gomes et al. 2019; Hu et al. 2014; Klyubin et al. 2014; Larson et al. 2012; Nicoll et al. 2013; Resenberger et al. 2011; Salazar et al. 2017; Um et al. 2013, 2012) that harmful protein conformers associated with more common neurodegenerative proteinopathies bind to PrP at the neuronal cell surface and thereby initiate neurotoxic signalling cascades. To date, this detrimental interaction with PrP has been shown for oligomers of Amyloid-β (Aβ), tau, and α-synuclein, which are critically associated with Alzheimer’s disease (AD), frontotemporal dementia and other tauopathies, or Parkinson’s disease, respectively (Corbett et al. 2020; Ferreira et al. 2017; Hu et al. 2018; Ondrejcak et al. 2018).
But what about the multitude of suggested physiological functions? A rather small glycoprotein at the cell surface acting like a ‘Jack-of-all-trades’? Even though some suggested roles are clearly controversial, have been challenged, or did not withstand experimental verification (e.g., upon developing improved knockout mice lacking genetic confounding effects (Nuvolone et al. 2016)), PrPC most certainly can be regarded as a ‘multifunctional protein’. However, this multifunctional character might not solely be restricted and immanent to the — so far — primarily studied mature, membrane-anchored and full-length form of PrP (fl-PrP). Likewise, it might not be limited to the nervous system, the area where most prion research of the past has focused on. Increasing evidence reveals that both, transient functional interactions with diverse binding partners (Aguzzi et al. 2008; Béland and Roucou 2012; Linden 2017) and endogenously produced forms or fragments of PrP holding intrinsic biological properties (Collins et al. 2018; Guillot-Sestier and Checler 2012; Linsenmeier et al. 2017), critically contribute to the protein’s apparent versatility. The latter aspect of enzymatically generated fragments constitutes the focus of this review. In particular, the release of nearly full-length PrP upon membrane-proximate cleavage by a metalloprotease, the so-called ‘shedding’ event, and current knowledge and perspectives in that regard will be discussed in detail. For the sake of completeness, however, we will start with a brief introduction of additional cleavage events occurring on PrP and their relevance in physiological and/or pathological conditions. For a more comprehensive view of these cleavages, we refer to earlier review articles (Altmeppen et al. 2013, 2012; Dexter and Kong 2021a, b; Liang and Kong 2012; Linsenmeier et al. 2017).
A process termed α-cleavage in the middle of the protein sequence separates the two structurally different parts of PrP (Chen et al. 1995; Haigh et al. 2009b; Harris et al. 1993; Linsenmeier et al. 2017): The intrinsically disordered N-terminal half or ‘flexible tail’, an important hub for interactions with diverse physiological and pathological ligands within fl-PrP (Béland and Roucou 2012; Carulla et al. 2015; Resenberger et al. 2011; Trevitt et al. 2014; Turnbaugh et al. 2012, 2011), is released into the extracellular space as a soluble (and rather instable (Mohammadi et al. 2020)) N1 fragment. N1 is a ligand linked with (neuro)protective and apparently myelin-maintaining activities as well as regulatory roles in diverse cellular processes and cell-to-cell communication (Carroll et al. 2020; Collins et al. 2018; Guillot-Sestier et al. 2012, 2009; Küffer et al. 2016; Mohammadi et al. 2021, 2020). The counterpart of N1, a globularly structured, N-glycosylated and stable C1 fragment, remains attached to the cell surface via PrP’s C-terminal GPI-anchor (Chen et al. 1995; Harris et al. 1993; Shyng et al. 1993). Upon α-cleavage, PrP’s central hydrophobic domain gets exposed as C1’s new N-terminus, which may have functional consequences, for instance, in cell-to-cell interactions or binding of certain ligands (Altmeppen et al. 2012; Bremer et al. 2010; Brenna et al. 2020; Harris et al. 1993; Linsenmeier et al. 2017). Since the N-terminal tail is critical for binding toxic protein assemblies mentioned above, α-cleavage can be regarded as a protective event rendering PrP unresponsive to these conformers. Moreover, the C1 fragment is resistant to misfolding in prion diseases and can even impair this process (Lewis et al. 2009; Westergard et al. 2011). Although α-cleavage represents the major physiological cleavage event of PrP in many cell types and its resulting fragments may hold relevant functions, there is enduring controversy on the responsible protease(s) (Altmeppen et al. 2011; Béland et al. 2012; Haigh et al. 2009b; Laffont-Proust et al. 2005; Liang et al. 2012; Mays et al. 2014; McDonald et al. 2014; Oliveira-Martins et al. 2010; Pietri et al. 2013; Taylor et al. 2009; Vincent et al. 2001; Wik et al. 2012), and it may well be that more than just one proteolytic entity ensures this important cleavage. Independent of the identification of the relevant protease(s), recent reports have shown that dimerization of PrP (Béland et al. 2012) or binding of PrP-directed peptide aptamers (Corda et al. 2018) causes increased α-cleavage, which may hold therapeutic relevance.
Around amino acid 90 and, thus, slightly N-terminally shifted from the α-cleavage site, the so-called β-cleavage by proteases or reactive oxygen species (with the latter causing a Fenton reaction) may occur, which is increased under pathological conditions and/or in response to oxidative stress (Castle et al. 2019; Chen et al. 1995; Mangé et al. 2004; Mays et al. 2014; McMahon et al. 2001). In consequence of this cleavage, a shorter N2 fragment is released while a longer C2 fragment stays attached to the membrane. Both fragments have been suggested to hold intrinsic functions and pathological roles (Haigh et al. 2015, 2009a, b; Lau et al. 2015; Sunyach et al. 2007).
The γ-cleavage represents the most recently described cleavage event (Lewis et al. 2016), preferentially occurs on unglycosylated PrP and separates a long N-terminal (N3 of ~ 20 kDa) from a short C-terminal fragment (C3 of ~ 6 kDa). Knowledge of its (patho)physiological implications is limited, yet it appears to be upregulated in prion disease.
Additional proteolytic fragmentation of synthetic PrP peptides by certain metalloproteases has been shown in vitro (Kojima et al. 2014), yet whether all cleavages found with recombinant protease-substrate mixtures in the test tube also hold in vivo-relevance is currently unsolved (Linsenmeier et al. 2017; McDonald et al. 2014).
Following this overview, we will now concentrate on another relevant cleavage event, termed shedding, occurring in the far C-terminal part of PrP and increasingly raising scientific attention.
Difficulties detecting shed PrP: new research tools enabling novel insights
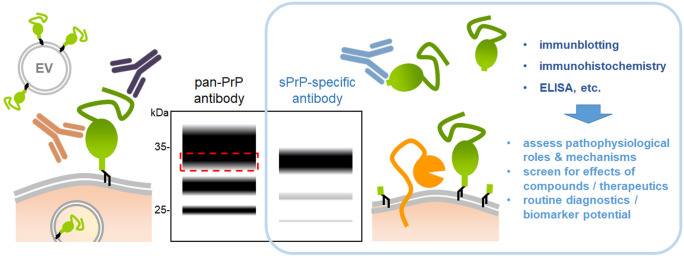
The presence of extracellular PrP molecules with nearly full-length protein sequence has been described roughly three decades ago (Borchelt et al. 1993; Harris et al. 1993; Tagliavini et al. 1992), and although their physiological production by the endogenous metalloproteinase ADAM10 (in a process referred to as ‘proteolytic shedding’) is known for more than a decade by now (Altmeppen et al. 2011; Taylor et al. 2009), only little insight into the biological relevance of this shed PrP (sPrP) has been gained until recently. Difficulties in reliable identification of sPrP in experimental models and complex biological tissues certainly represent the main reason for this lack of knowledge. In contrast to truncated released or cell-associated fragments resulting from the α- or β-cleavage, which differ remarkably from fl-PrP in size/molecular weight and may be easily discriminated by western blot analyses of conditioned media/body fluids or cell/tissue lysates, respectively, sPrP is usually masked by levels of fl-PrP present in vast excess and, thus, simply ‘overlooked’ when using common (pan-) PrP antibodies for detection (Fig. 1). To solve this problem, we have recently generated a cleavage-site specific antibody for sensitive and reliable detection of sPrP (Linsenmeier et al. 2018), based on sequence and cleavage-site information for murine PrP published earlier (Taylor et al. 2009). The glycosylation state of PrP critically influences PrP’s biology and pathophysiological roles (DeArmond et al. 1997; Makarava et al. 2020; Nishina et al. 2006; Priola and Lawson 2001; Puig et al. 2011). Our new sPrP antibody revealed that PrP is mostly shed in a diglycosylated state, which likely represents the physiological status-quo at the cell surface, where shedding by ADAM10 is thought to occur. Moreover, we have subsequently identified the shedding process as a relevant part of a posttranslationally active regulatory network controlling cellular PrP homeostasis (Linsenmeier et al. 2018). This compensatory network also involves cellular uptake and degradation, as well as release of PrP via extracellular vesicles (Brenna et al. 2020; D’Arrigo et al. 2021; Falker et al. 2016; Fevrier et al. 2004; Guo et al. 2016; Ritchie et al. 2013; Wik et al. 2012). Hence, PrP release via EVs (Heisler et al. 2018) or the ADAM10-mediated shedding are increased upon lysosomal inhibition (Linsenmeier et al. 2018) and in mice lacking intracellular regulators of retrograde sorting and vesicular transport to lysosomes (Heisler et al. 2018; Linsenmeier et al. 2018; Uchiyama et al. 2017). Vice versa, pharmacological inhibition of proteolytic PrP shedding is compensated by an elevated release of EVs with increased PrP load (Linsenmeier et al. 2018). We also found evidence for ‘cleavage cascades’ occurring on PrP, as the truncated C1 fragment resulting from the α-cleavage can subsequently be shed by ADAM10 and thus be detected with a sPrP-specific antibody, for instance, in conditioned media (Linsenmeier et al. 2018). The same seems plausible for the β-cleavage product C2 (Perini et al. 1996). This further complicates the variety of released PrP fragments and investigations on their particular functions or pathological relevance. Therefore, improved assays, such as capillary western analysis (Castle et al. 2019), in combination with powerful site-specific antibodies will lead to better discrimination, reliable quantifications, and eventually biological insight (Fig. 1). For sPrP, this has already been achieved in parts and will certainly continue to reveal novel (patho)physiological implications, as discussed in the following paragraphs.
Fig. 1.
Challenging assessment of sPrP and the advantage of site-specific antibodies. Besides membrane-attached forms of PrP (green) in/on cells or on extracellular vesicles (EV), plenty of different cleaved fragments (not all depicted here) are present in biological samples. Due to the similar size of sPrP (released by ADAM10; orange) and fl-PrP and the usually vast excess of the latter, pan-PrP antibodies do not discriminate between these forms (e.g., in immunoblots), and sPrP is therefore masked (as indicated by the schematic immunoblot in the middle (red striped box). The three bands typical for PrP are caused by its glycosylation state (with di-, mono- , and unglycosylated forms; N-glycans are not depicted here to simplify matters). Generation of cleavage site-specific antibodies (blue) allows for reliable detection of sPrP (right lane in the blot; note the slightly lower molecular weight due to the lack of the GPI-anchor, and the strong predominance of diglycosylated sPrP (Linsenmeier et al. 2018)). Such fragment-specific antibodies allow for a reliable assessment of specific PrP derivatives (in this case sPrP) in standard research and routine diagnostic methods, such as western blot, histological approaches, or ELISA. This enables studies on the (patho)physiological relevance as well as on the therapeutic and/or diagnostic potential of certain PrP fragments
Biological roles and assessment of sPrP in neurodegenerative proteinopathies
Prion diseases
In stark contrast to the aforementioned harmful roles in neurodegenerative diseases played by fl-PrP at the cell surface, diffusible extracellular forms or derivatives of PrP have been shown to protect from prion misfolding and act against toxic protein assemblies. Expression of a soluble dimerized PrP in prion-infected mice interfered with PrPSc formation and disease progression (Meier et al. 2003). Similar ‘anti-prion’ effects were also observed for recombinant PrP (Priola et al. 1994; Yuan et al. 2013) and for PrP being released upon treatment with lipid-raft disturbing drugs (Bate et al. 2009; Marella et al. 2002; Taraboulos et al. 1995) or overexpression of a sorting factor (SNX33) in prion-infected cell cultures (Heiseke et al. 2008). In the latter study, release of PrP was likely accomplished by phospholipases cleaving within the GPI-anchor structure (Caughey et al. 1989; Harris et al. 1993; Stahl et al. 1987). Soon after that study, the zinc-dependent metalloprotease ADAM10 was shown to mediate the proteolytic shedding of PrP just a few amino acids away from the GPI-anchor in vitro (Taylor et al. 2009). Subsequent studies in transgenic mice not only confirmed ADAM10 as the major PrP sheddase in vivo (Altmeppen et al. 2011), but also revealed its protective and disease-modifying effects upon prion infection (Altmeppen et al. 2015; Endres et al. 2009) (Fig. 2a). Until now, ADAM10 even seems to be the only relevant sheddase of PrP (Altmeppen et al. 2011; Linsenmeier et al. 2018; McDonald et al. 2014; Taylor et al. 2009). Shed PrP, the physiological correlate of soluble PrP forms mentioned above, seems to bind and block critical PrPSc assemblies (‘seeds’) in the extracellular space and thereby interfere with the conversion process, as indicated by an inverse correlation of sPrP and PrPSc levels shown in a recent study (Linsenmeier et al. 2021) and reflected in Fig. 2b, c. In addition to this blocking effect, sPrP may also act as a ligand (similar to PrP’s N1 fragment) inducing neuroprotective signalling cascades or rescuing PrP functions in transgenic mice expressing toxic PrP mutants (Race et al. 2009). Accordingly, lack of protective sPrP in transgenic mice expressing PrP with a C-terminal deletion (Δ214-229 (Puig et al. 2016)) and reduced PrP shedding in cells and mice expressing PrP with an altered GPI-anchor and, hence, shifted membrane localization (Puig et al. 2019, 2011) may contribute to the respective pathological phenotypes observed in these models.
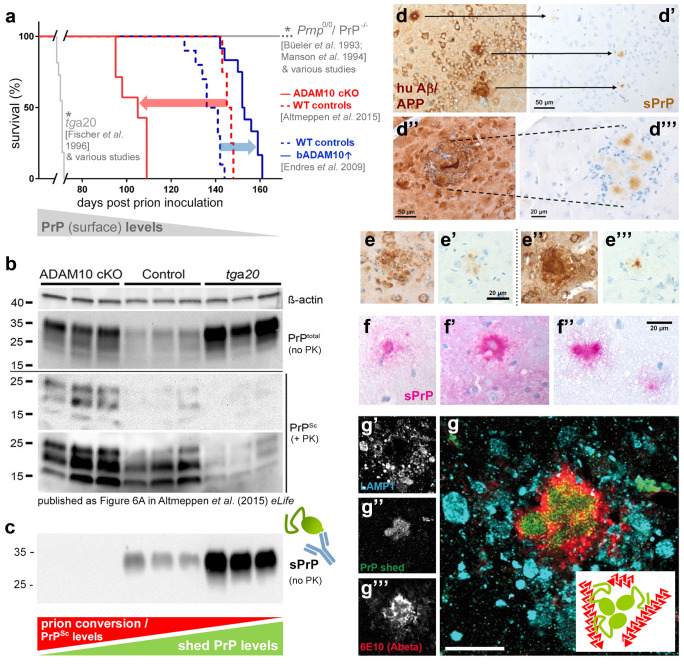
Fig. 2.
Consequences of the ADAM10-mediated shedding of PrP in neurodegenerative diseases. (a) Kaplan–Meier survival curves summarizing two in vivo studies that assessed the role of ADAM10 in prion diseases (with both studies using the Rocky Mountain Laboratory (RML) prion strain). While moderate overexpression of bovine ADAM10 in mice (bADAM10↑; blue line) in the study of Endres et al. (2009) resulted in prolonged survival (blue arrow; wild-type controls represented by dotted blue line), lack of ADAM10 in forebrain neurons (in ADAM10 cKO mice; red line) caused shortened incubation times (red arrow) compared to controls (dotted red line) (Altmeppen et al. 2015). For comparison, the diagram also schematically presents prion protein knockout mice (e.g., Prnp0/0 [(Büeler et al. 1993)] or PrP−/− [(Manson et al. 1994)]), which are resistant to prion infection, as well as PrP-overexpressing mice (e.g., tga20 [(Fischer et al. 1996)]), which succumb to disease very early. *Note that the curves for these models reflect typical study outcome rather than exact datasets. Taken together, all models/genotypes depicted here fit the view that levels of (cell-associated) PrPC critically determine survival times in prion diseases (Manson et al. 1994; Sandberg et al. 2011) (see grey ‘correlation bar’ below). (b) Western blot data reproduced from Fig. 6A in Altmeppen et al. (2015) eLife ((Altmeppen et al. 2015); https://elifesciences.org/articles/04260) published under a CC BY 4.0 license (https://creativecommons.org/licenses/by/4.0/). In non-PK digested samples, highest levels in total PrP (i.e., PrPC and PrPSc) were found in terminally diseased tga20 mice (at 65 days post-inoculation, dpi), followed by ADAM10 cKO and wild-type control mice (both at 95 dpi). In contrast, prion conversion (judged by PrPSc amounts detectable after PK digestion) was highest in ADAM10 cKO while barely detectable in tga20 mouse brains. (c) Re-analysis of aforementioned samples in a replica blot probed with a new sPrP-specific antibody demonstrates lack of detectable sPrP in ADAM10 cKO and efficient sPrP production in tga20 mice. In connection with (b) this may indicate an inverse correlation (see red/green ‘correlation bars’) between PrP shedding and pathogenic prion conversion (though deeper insight is clearly required). (d, d″) Amyloid plaques composed of human Aβ (left) in brain sections of 8 months-old APP23 mice heterozygous for PrP (Prnp + / −). Note that endogenous sPrP (stained for in serial sections shown on the right; d′, d‴) is enriched in the plaques (showing multiple foci in the lower panel; d‴). (e, e′, e″, e‴) In APP23 mice with normal PrP expression (Prnp + / +) colocalization of sPrP with diffuse (left panel) and dense amyloid plaques (right panel) is already detectable at 5 months of age. (f, f′, f″) Plaque-like appearance of sPrP (detected with alkaline phosphatase, hence the pink signal) in brain sections of another AD mouse model (5xFAD mouse, 5 months old). (g) Immunofluorescence analysis showing a brain section of a 5xFAD mouse stained for Aβ plaques (red), sPrP (green), and endosomal/lysosomal marker LAMP1 (bright blue; mostly representing dystrophic neurites around plaques). Note that sPrP is enriched in the centre of this amyloid plaque (merged channels; g). Scale bar in the magnified merge picture is 30 µm. Inlay shows a model of the conceivable Aβ (red) sequestrating and plaque-promoting action of sPrP (green). Respective non-coloured single channels are shown on the left (g′, g″, g‴)
A (seemingly) opposing finding, however, challenges the view of soluble PrP forms being protective: Anchorless PrP expressed in transgenic mice is a potent substrate for prion conversion and aggregate formation (Chesebro et al. 2010, 2005; Rangel et al. 2013; Rogers et al. 1993; Stöhr et al. 2011). Anchorless forms of PrP are also associated with some genetic forms of prion diseases in humans (Choi et al. 2016; Jansen et al. 2010; Zanusso et al. 2014). However, a key difference that may resolve this contradiction is the different N-glycosylation pattern between transgenically expressed or mutation-derived anchorless PrP on the one hand, and physiological sPrP on the other hand (Fig. 3): while the latter is predominantly diglycosylated (due to its transport through the secretory pathway as a GPI-anchored protein and subsequent cleavage after reaching the cell surface (Linsenmeier et al. 2018)), anchorless PrP is secreted in an underglycosylated state. This may have a profound impact on how these PrP forms encounter and affect extracellular PrPSc assemblies. In fact, an influence of the N-glycans on PrP’s susceptibility to prion conversion (depending on the respective prion strains) has been firmly established (DeArmond et al. 1997; Makarava et al. 2020; Nishina et al. 2006; Priola and Lawson 2001; Sevillano et al. 2020). In many experimental paradigms, underglycosylated PrP was efficiently converted, whereas diglycosylated PrP was a rather poor substrate for the templated misfolding (Camacho et al. 2019; Cheng et al. 2017; DeMarco and Daggett 2009; Kang et al. 2020; Priola and Lawson 2001; Xiao et al. 2013). This may well translate into sPrP’s blocking activity against the amplification of certain prion strains.
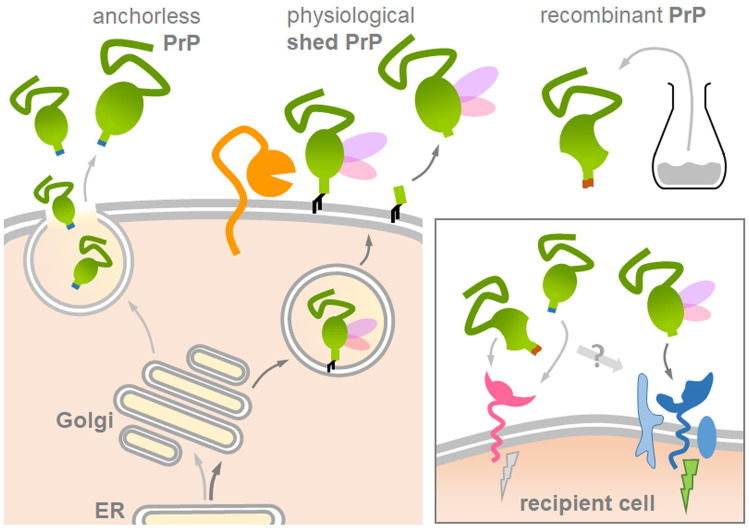
Fig. 3.
Structural differences between experimentally employed ‘anchorless’ PrP or recombinant PrP and physiological sPrP may affect biological functions and study outcome. Several studies on diverse topics covered in this review used transgenically expressed anchorless PrP (on the left) or recombinant PrP (on the upper right) as soluble PrP forms in assumed analogy to physiologically generated sPrP (middle). However, sPrP likely differs from these forms: recPrP is unglycosylated and anchorless PrP typically underglycosylated (no or one N-glycan), whereas sPrP is mainly diglycosylated (pink/purple structures). Moreover, the C-terminus is altered and the overall structure may be different. In sum, these features could feasibly affect critical ligand-to-receptor interactions and downstream effects (box on the lower right) and other biological implications. This should be considered and controlled for in experimental paradigms
The situation, however, might be more complicated, as one in vitro study showed that ADAM10, in principal, is also able to shed misfolded PrPSc (Taylor et al. 2009). This could -to some degree- contribute to the spread of anchorless PrPSc assemblies and pathology within the brain (Altmeppen et al. 2015). Surprisingly, in contrast to the peptide bond hydrolysis by ADAM10, experimentally applied phospholipase C is incapable of cleaving the GPI-anchor of PrPSc (Caughey et al. 1990; Stahl et al. 1990). It remains to be studied whether ‘proteolytic shedding’ by ADAM10 and release of putative shed PrPSc seeds into body fluids also plays a role in the ‘environmental shedding’ of prions resulting in the high contagiosity observed in chronic wasting disease (CWD), a prion disease affecting deer, moose, and elk (Bessen et al. 2010; Denkers et al. 2020; Moore et al. 2016; Moreno and Telling 2018; Tennant et al. 2020). Expression of PrP with a single amino acid variation (found in cervid PrP) in mice affected prion strain selection upon infection with CWD prions (Bian et al. 2021). Though not investigated in that study, it would be interesting to assess whether this variation in close proximity to the cleavage site and GPI-anchor affects the shedding, with possible consequences for strain and disease features. Notably, a recent study investigating the role of the extracellular matrix component heparan sulfate as a cofactor in prion diseases revealed that prion deposits in brains of transgenic mice infected with CWD prions largely consisted of heparan sulfate-associated ADAM10-cleaved PrPSc (Aguilar-Calvo et al. 2020). Subsequent studies, however, provided evidence that ADAM10-cleaved PrPSc was mostly associated with large perivascular and possibly inert plaques, whereas diffusible oligomeric or sub-fibrillar (and presumably more neurotoxic) PrPSc assemblies dominating in many prion diseases mostly result from the conversion of GPI-anchored PrP (Aguilar-Calvo et al. 2020; Callender et al. 2020; Sevillano et al. 2020). In conclusion, the ADAM10-mediated shedding might play a dual role in prion diseases, and whether it is protective or rather disease-supporting might depend on critical molecular stoichiometries, cofactors, and currently unknown cellular modalities as well as species and prion strains. Regarding PrPSc formation and survival times of prion-infected mice, recent studies suggest that protective effects may dominate (Altmeppen et al. 2015; Endres et al. 2009; Linsenmeier et al. 2021).
Alzheimer’s disease and other neurodegenerative proteinopathies
Considering cell surface PrP’s role as a receptor and toxicity mediator of harmful protein assemblies (introduced in paragraph 1), it is not surprising that released forms of PrP harbouring the relevant binding sites (Chen et al. 2010; Laurén et al. 2009) instead have the ability to bind and sequester oligomeric assemblies in the extracellular space and interfere with their neurotoxicity in respective model systems. This has most convincingly been shown for Aβ oligomers and their ‘neutralization’ by the N1 fragment resulting from the α-cleavage (Béland et al. 2014; Fluharty et al. 2013; Guillot-Sestier et al. 2012; Nieznanski et al. 2012; Resenberger et al. 2011; Scott-McKean et al. 2016). Several studies also employed anchorless or recombinant PrP (mimicking physiological sPrP) and found similar binding affinities for and protective effects against Aβ oligomers (Calella et al. 2010; Fluharty et al. 2013; König et al. 2021; Nieznanski et al. 2012; Scott-McKean et al. 2016). In fact, an additional binding site targeting the ends of Aβ fibrils has recently been located in PrP’s C-terminal half, which is therefore preserved in sPrP yet absent in N1 (Amin and Harris 2021; Bove-Fenderson et al. 2017). Some reports also indicated increased α-cleavage and shedding of PrP in AD models and brains, possibly reflecting a protective feedback loop (Béland et al. 2014; Ostapchenko et al. 2013). Similar to prion diseases discussed above, existing data for Aβ (possibly holding true for other harmful conformers alike) supports the view of a two-level protection conferred by the ADAM10-mediated shedding: First, this process reduces amounts of PrP as the toxicity receptor at the neuronal surface (Jarosz-Griffiths et al. 2019). Second, sPrP blocks toxic conformers and may support their sequestration into less toxic plaques. In fact, a plaque-promoting effect and presence of PrP in amyloid deposits have been demonstrated earlier (Boon et al. 2020; Ferrer et al. 2001; Schwarze-Eicker et al. 2005; Takahashi et al. 2021). These studies, however, used pan-PrP antibodies for detection. Most recently, using our site-specific antibody, we provided further insight that sPrP is enriched in the centre of amyloid plaques in mouse models for AD-associated amyloidosis (Linsenmeier et al. 2021) (Fig. 2d–g). Although doubtlessly many proteins are found (enriched) in plaques (among them rather specific interactors of Aβ or its precursor protein (APP), but also others just being trapped by these ‘sticky’ extracellular structures), many of the aforementioned studies speak in favour for sPrP playing an Aβ-sequestrating and plaque-promoting role. Given that large deposits, such as plaques, are currently considered less harmful than diffusible, toxic oligomeric species of neurodegeneration-associated misfolded proteins, this would indicate a protective role of sPrP. However, deeper mechanistic insight is certainly required.
The ability of PrP to bind toxic conformers has also been linked with a role in mediating their cellular uptake (De Cecco et al. 2020; Foley et al. 2020; Legname and Scialò 2020). Though not studied to date, in that scenario, ADAM10-mediated shedding of surface PrP could regulate these phagocytic activities. Moreover, sPrP bound to harmful extracellular oligomers could represent a signal triggering the binding to a given cell surface receptor (with homophilic interactions with membrane-anchored PrP being one conceivable possibility) and subsequent uptake.
Therapeutic and biomarker potential of sPrP in neurodegeneration
Based on the above, stimulation of the ADAM10-mediated PrP shedding could represent a therapeutic option in prion and other neurodegenerative diseases (Jarosz-Griffiths et al. 2019). ADAM10 is already pharmacologically targeted in certain skin disease treatments and, in a recent trial in AD patients, its activity has already been stimulated using the vitamin A analog acitretin (Endres et al. 2014). However, in view of the multitude of critical roles and substrates of this protease in the brain and throughout the body (mentioned in part in the following sections), unwanted side effects to be expected with this rather systematic treatment may pose major challenges (Kuhn et al. 2016; Saftig and Lichtenthaler 2015; Wetzel et al. 2017). We may now have overcome this hurdle by identifying a substrate-specific approach (Linsenmeier et al. 2021), in which binding of certain ligands (e.g., antibodies) to PrP causes increased sPrP levels in the absence of overt toxicity. This may — at least in part — explain the protective effects of PrP-directed antibodies found in diverse cellular and animal models of prion diseases and AD (Chung et al. 2010; Enari et al. 2001; Féraudet et al. 2005; Freir et al. 2011; Gilch et al. 2003; Heppner et al. 2001; Laurén et al. 2009; Peretz et al. 2001).
A promising therapeutic approach currently pursued against prion diseases (that could likewise be beneficial in other neurodegenerative diseases) is to reduce the overall expression of PrP via antisense oligonucleotides (Minikel et al. 2020; Raymond et al. 2019; Vallabh et al. 2019). A possible combination therapy, i.e., reducing total PrP expression and stimulating the release of remaining PrP molecules, may even increase the benefit while preserving putative physiological functions of soluble extracellular PrP fragments.
Both, verification and implementation of any PrP-modifying therapy in pre-clinical and clinical trials (Minikel et al. 2019) as well as urgently required improved (and earlier) diagnosis of specific neurodegenerative diseases, will critically depend on the assessment of reliable biomarkers. Detection of alterations in a defined subset of PrP molecules will most certainly be superior to detection of bulk PrP with its diverse forms and fragments found in biological samples (Vallabh et al. 2019). This highlights the relevance of fragment-specific antibodies and warrants the need for future studies on the potential of sPrP as a meaningful biomarker (Fig. 1) (Linsenmeier et al. 2018). A recent report of increased levels of ADAM10 in the brains of patients with CJD further supports this notion (Diaz-Lucena et al. 2021).
Although not directly assessed to date and therefore rather hypothesized below, it is conceivable that sPrP is mechanistically involved or could at least represent a relevant biomarker in several other pathological processes, as will be discussed in the next paragraph.
Potential relevance of PrP shedding in other pathological processes
Both, the prion protein (Rubenstein et al. 2017; Sekar et al. 2019) and its sheddase ADAM10 (Appel et al. 2021; Sun et al. 2017; Warren et al. 2012; Zohar et al. 2011) have — so far independently — been implicated in pathological and recovery-associated processes following traumatic brain injury (TBI). Moreover, levels of soluble PrP were found elevated in blood plasma where they may serve as a diagnostic marker for TBI and sport-related concussion (Persad et al. 2021; Pham et al. 2015a, b). Also, ADAM10 levels correlated with clinical grade (Persad et al. 2021). Although it seems likely that soluble PrP assessed in these studies correlates with sPrP, final proof needs to be obtained in systematic studies using specific antibodies to discriminate from other PrP fragments or from PrP released via extracellular vesicles. This will also help to investigate potential protective or regenerative processes conferred by sPrP. However, as inhibition of ADAM10 in a mouse model for TBI reduced tissue injury and inflammatory responses, it appears questionable if sPrP would act beneficially in this context (Appel et al. 2021).
PrP and its released fragments also seem to play beneficial roles in hypoxic conditions affecting the CNS, such as stroke (Doeppner et al. 2015; Guillot-Sestier et al. 2009; McLennan et al. 2004; Shyu et al. 2005; Spudich et al. 2005; Weise et al. 2006, 2004). While signalling and downstream effects mediated via cell surface PrP most certainly underlies some of the protective roles (reviewed in (Puig et al. 2020)), release of PrP fragments, such as sPrP and PrP, on extracellular vesicles may be relevant for intercellular communication with nearby or distant brain regions, neuron-glia interactions, or recruitment/activation of cell types required for the induction of regenerative processes, such as angiogenesis (Brenna et al. 2020; D’Arrigo et al. 2021; Guitart et al. 2016; Mitsios et al. 2007; Turu et al. 2008). Again, clarification of specific roles of sPrP in these aspects seems reasonable, as they could be employed therapeutically.
While all previously discussed (potential) roles of sPrP were exclusively focussed on the CNS, sPrP may reveal itself as a relevant molecule beyond this organ system. Moreover, in contrast to the aforementioned beneficial implications, sPrP may also carry out negative roles as discussed first for the aspect of tumorigenesis and cancer.
Why cancer? Increased PrP expression has been found in various types of malignant tumours ranging from brain tumours to breast, gastric, skin, and colorectal cancer. In these and other tumour entities, PrP was shown to support tumorigenesis and metastasis by engaging in a variety of pathogenic processes, including anti-apoptotic signalling cascades, cancer stem cell survival, angiogenesis, and even resistance towards chemotherapy and radiation (Atkinson et al. 2019; Barbieri et al. 2011; Bernardino-Sgherri et al. 2021; Corsaro et al. 2016; de Lacerda et al. 2016; Du et al. 2013; Ghazi et al. 2021; Le Corre et al. 2019; Li et al. 2009; Liang et al. 2007; Lopes et al. 2015; Luo et al. 2017; Pan et al. 2006; Roucou et al. 2005; Thellung et al. 2019; Wang et al. 2016; Yap and Say 2012). As such, elevated PrP levels may correlate with malignancy and are considered a sign for poor prognosis.
Strikingly, a huge amount of published evidence also implicates ADAM10 in various aspects of cancer development and progression. Previous studies mostly focussed on ADAM10’s role in extracellular matrix degradation for angiogenesis and metastasis or on its processing of cellular substrates regulating differentiation and cancer cell survival, yet did not consider or reveal any link to PrP (Crawford et al. 2009; Dempsey 2017; Ostalecki et al. 2017; Smith et al. 2020). However, this may now change given that a recent report found both, PrP and its sheddase, to be associated in the pathogenic process of breast cancer progression (Cheng et al. 2021). From the combination of elevated PrP levels and increased ADAM10 expression/activity found in various cancer types, one can anticipate that sPrP, the product likely generated by this molecular encounter, may be mechanistically involved in certain oncogenic processes. Considering the well-established role of PrP in signalling (Chiarini et al. 2002; Mattei et al. 2020; Mouillet-Richard et al. 2000) and the growing evidence for sPrP acting as a ligand or trophic factor in intercellular communication (discussed herein), further studies are warranted to check for a mechanistic relevance in cancer biology. But even if sPrP turns out to be not much more than a bystander, the combination of upregulated ADAM10 and PrP independently described in various cancer types and models could well point towards a diagnostic biomarker potential of sPrP assessable in body fluids.
Notably, two recent reports specifically linked released PrP with development of certain CNS tumours (Provenzano et al. 2017) and with chemotherapy resistance in breast cancer (Wiegmans et al. 2019). However, these studies did not strictly discriminate between PrP on extracellular vesicles or sPrP, which supports the need for further studies using differentiating antibodies and protocols.
In recent years, protective functions have also been attributed to PrP in the kidney (Han et al. 2020; Yoon et al. 2021; Zhang et al. 2015), and PrP is ‘secreted’ into the urine in response to chronic kidney disease and ER stress in kidney injury (Bignon et al. 2020). However, before assessment of PrP in plasma can be used as a reliable biomarker, differentiation between sPrP and other PrP forms again seems appropriate.
Lastly, shedding of PrP might be relevant in immune regulation, the immune privilege of certain organs, and inflammatory responses during (viral) infections. In fact, PrP is highly expressed in lymphoid tissues and has long been discussed to be important in the communication between immune cells (Bakkebø et al. 2015). It is tempting to speculate that sPrP (and other released PrP fragments) act similar to chemokines or interleukins and mediate intercellular crosstalk between diverse cell types as well as between different tissues and organ systems, such as neuro-immune interactions (Salvesen et al. 2019). These processes might be diverted and hence become harmful in tumorigenesis or CNS infections as mentioned earlier. Platelets (Perini et al. 1996), primary lymphoid cells (Parizek et al. 2001), and mast cells were shown to release PrP (Haddon et al. 2009), the latter especially upon activation, thus indicating functional relevance. It seems likely that this release is mediated by ADAM10 at the cell surface. After finding increased levels of soluble PrP in the CSF (Roberts et al. 2010) of patients with HIV-associated neurological impairment (Price et al. 1988) and suggesting soluble PrP as a respective biomarker (Megra et al. 2013), one group recently proposed a disease-accelerating role for shed PrP in HIV neuropathogenesis (Megra et al. 2017). In that scenario, active ADAM10 and PrP shedding are upregulated on astrocytes in response to certain inflammatory mediators. This, in turn, triggers a cascade of events eventually leading to increased monocyte recruitment to the brain and worsening of brain damage. Again, since pan-PrP antibodies were used in that study and no ultracentrifugation was performed (to exclude extracellular vesicles), a clear demonstration that bona-fide sPrP caused this effect is still pending.
Further biological roles influenced by shedding or played by ‘sPrP’: a few facts and some fiction
An outstanding aspect common to many of the aforementioned putative implications of sPrP is its supposed role as a diffusible ligand in intercellular communication. In this regard, it appears likely that sPrP and/or the N1 fragment are the physiologically relevant ligands for a G-protein coupled receptor on Schwann cells, ensuring myelin maintenance in the peripheral nervous system (Henzi et al. 2020; Küffer et al. 2016).
Further support for the view that sPrP acts as a ligand in various processes also comes from several studies using recombinant PrP. Treatment of neurons with the latter, for instance, causes neuronal polarization, increased axon length and dendritic differentiation as well as synapse formation. Notably, C- or N-terminal PrP fragments were not sufficient to elicit this effect (Kanaani et al. 2005). Moreover, axons grow towards a source of recPrP, and cell surface PrP itself seems to act as ‘its own’ neuronal receptor in this process (Amin et al. 2016). This suggests that physiological sPrP may hold properties as both, a growth factor-like and chemoattractant molecule in neuronal differentiation, neuritogenesis, and synaptic homeostasis. Fittingly, ADAM10 holds key roles in brain development, axon targeting and functioning of synapses (Jorissen et al. 2010; Kuhn et al. 2016; Malinverno et al. 2010; Prox et al. 2013), although its substrate PrP was not specifically assessed in these studies.
One way by which PrP modulates signalling cascades is via interaction with different transmembrane partners. An inhibitory effect on excitotoxicity is mediated by binding of PrP to the NMDA receptor (Huang et al. 2021; Khosravani et al. 2008; Meneghetti et al. 2019; Petit-Paitel et al. 2012), whereas its interaction with this receptor and metabotropic glutamate receptor mGluR5 mediates toxicity upon binding of harmful protein oligomers (Hamilton et al. 2015; Hu et al. 2014; Resenberger et al. 2011; Um et al. 2013; You et al. 2012). Binding of PrP to the 37 kDa/67 kDa laminin receptor precursor (LRP/LR) (Gauczynski et al. 2001; Hundt et al. 2001; Simoneau et al. 2003) or the low-density lipoprotein receptor-related protein 1 (LRP1) (Parkyn et al. 2008; Taylor and Hooper 2007) (note the similar nomenclature!) regulates PrP’s cellular trafficking and homeostasis and may affect PrP-related signalling and prion conversion (Leucht et al. 2003; Mattei et al. 2020; Pinnock et al. 2016; Rushworth et al. 2013). Interaction of PrP with the neural cell adhesion protein (NCAM) at the cell surface is involved in cell adhesion and morphogenesis, neuronal differentiation, and neurite branching (Brethour et al. 2017; Prodromidou et al. 2014; Santuccione et al. 2005; Schmitt-Ulms et al. 2001; Slapšak et al. 2016). Like PrP and ADAM10 (discussed in Sect. 4), many of these PrP interactors are also associated with different processes during cancerogenesis (reviewed in (Colombo and Meldolesi 2015; Gonias and Campana 2014; Vania et al. 2019)). Since all relevant binding sites for interactions with these and other receptors are preserved in sPrP, this calls for detailed studies investigating whether sPrP may act as an antagonistic, agonistic or regulatory ligand of the diverse processes mentioned above. In fact, functional interaction with NCAM in trans was shown for recPrP (Chen et al. 2003; Santuccione et al. 2005). Intriguingly, a recent report demonstrated that experimentally administered recPrP induced MAP kinase Erk1/2 signalling via engagement of both, LRP1 and NMDA receptor (Mantuano et al. 2020). This caused neurite outgrowth in a neuronal cell line and migration of Schwann cells, thus indicating relevance in the central and peripheral nervous system, respectively. Another report showed that recPrP induced phosphorylation of Erk1/2 and additional signalling causing neuronal differentiation of stem cells (Martellucci et al. 2019a, b). As in other conditions mentioned earlier (Amin et al. 2016), this required expression of PrP at the cell surface suggesting an underlying homophilic encounter between sPrP as ligand and PrP as (co-)receptor. In sum, interaction of PrP with diverse partners is key to its established roles in regulating (stem) cell proliferation, maintenance, and viability as well as morphological and functional differentiation (such as epithelial-to-mesenchymal transition) in the central nervous system and beyond (Brethour et al. 2017; Lee and Baskakov 2013; Mehrabian et al. 2015; Prodromidou et al. 2014; Steele et al. 2006; Zhang et al. 2006). These fine-tuned processes are relevant during development and for regenerative processes yet may be corrupted in pathogenic conditions such as cancer. The role played by sPrP in these regards is out for debate and investigation.
Although it seems at least possible, if not likely, that sPrP is the physiological correlate in many of the pathophysiological processes covered in this review, detailed studies on this are lacking to date. All of the aforementioned studies using recPrP or transgenically expressed anchorless PrP as a ‘proxy’ for physiological sPrP are -without doubt- very interesting and may help to unravel sPrP’s real functions. However, pending further direct proof and considering the possibly relevant structural differences of these forms compared to physiologically shed PrP (Fig. 3), one should, at least at the present state, be careful with generalizing experimental findings. In this consideration, we would rather disagree with recent statements that these PrP versions are ‘basically the same as’ shed PrP (Dexter and Kong 2021b).
Apart from the focus on released PrP fragments, proteolytic processing may also have implications from the perspective of membrane-bound PrP. In many of the biological implications discussed herein, a gradual engagement of proteolytic cleavages is conceivable: by releasing the N-terminal half of PrP, α-cleavage may inhibit certain interactions, while simultaneous production of the C1 fragment with its exposed hydrophobic sequence likely enables others. In this scenario, the PrP shedding by ADAM10 might be a mechanism to regulate or terminate all PrP interactions and downstream effects. This assumption could be particularly important for two partially connected aspects, namely the established involvement of cell surface PrP in both, diverse cellular signalling cascades (Chiarini et al. 2002; Mattei et al. 2020; Mouillet-Richard et al. 2000) and cell adhesion (Kaiser et al. 2012; Málaga-Trillo et al. 2009; Mangé et al. 2002; Petit et al. 2013; Solis et al. 2013). Moreover, PrP and its C1 fragment are highly enriched on EVs and may serve important regulatory functions regarding the fate of EVs upon interaction with recipient cells and delivery of cargo and information (Brenna et al. 2020; D’Arrigo et al. 2021; Falker et al. 2016; Guo et al. 2015; Linsenmeier et al. 2018; Vella et al. 2008). EVs are considered as rather stable extracellular structures able to cross tissue borders, including the blood–brain barrier, whereas half-life of sPrP as a soluble factor in tissue environment or body fluids might be rather short. Strikingly, EVs also carry the active form of ADAM10 and, though not reported yet, it is conceivable that PrP shedding continues on EVs, as has been shown for other ADAM10 substrates (Folkesson et al. 2015; Padro et al. 2013; Pérez-González et al. 2020; Stoeck et al. 2006). It appears tempting to speculate (and investigate) whether EVs could function as a ‘carrier rocket’ enabling transport to distant target organs, followed by subsequent local release of sPrP to exhibit its functions.
Conclusion and outlook
As it stands now, clear conclusions on whether PrP shedding and sPrP play harmful (e.g., in cancer) or beneficial roles (e.g., in neurodegeneration) cannot be easily drawn — it all rather seems to be a matter of perspective (and pathophysiological context). Plenty of biological implications may arise for the ADAM10-mediated shedding and its product sPrP (summarized in Fig. 4). And even though shedding may not be of mechanistic relevance in certain processes mentioned herein, considering the broad expression pattern of both, PrP and its sheddase ADAM10, alterations in sPrP levels in body fluids may qualify as diagnostic tool in some pathological conditions. Also, PrP shedding may well affect processes in organs/tissues not covered in this review. However, all of this requires detailed analyses and new research tools able to reliably differentiate between sPrP and other PrP derivatives. While sPrP-specific antibodies for murine models are available, identification of the shedding site and generation of respective antibodies for the human system and other species will fuel further studies and provide insight into both, biological effects and biomarker potential of sPrP. Moreover, given that PrP appears to be exclusively shed by ADAM10 (with no contribution by other proteases, such as the closely related and often redundantly working ADAM17), detection of sPrP as a surrogate read-out could become a convenient way to investigate the efficacy of any pharmacological approaches aiming to manipulate ADAM10 activity in general. With regard to PrP, we recently presented a substrate-specific approach that, depending on the applied PrP-directed ligand, enables stimulated shedding as well as (transient) downregulation of total PrP (Linsenmeier et al. 2021). Future studies into this direction, but also on endogenous regulators (e.g., certain tetraspanin molecules (Matthews et al. 2017; Saint-Pol et al. 2017; Seipold et al. 2018)) or pharmacological modulators of ADAM10 trafficking, maturation and activity, could pave the way for future therapeutic avenues in neurodegeneration and beyond.
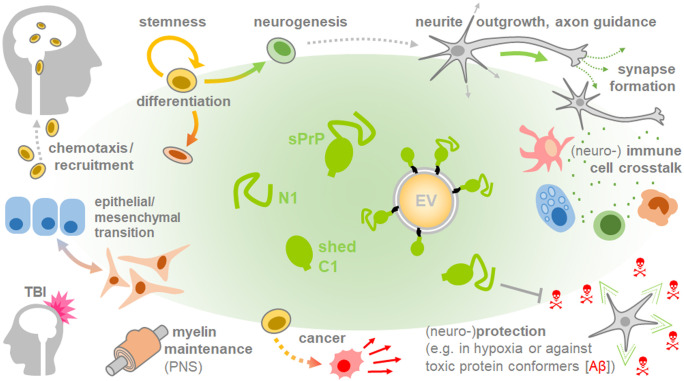
Fig. 4.
Overview of potential (patho)physiological implications of sPrP. A plethora of studies suggest that released fragments of PrP are linked with intrinsic functions, which may partially explain the multitude of roles attributed to this evolutionary conserved protein. Putative roles of sPrP addressed in this review may be beneficial (e.g., in neurodegeneration, development/differentiation) or detrimental (e.g., in cancer progression or immune response aggravation). It should be noted, however, that many of the roles discussed herein are, thus far, rather speculative and based on the combination of described PrP participation on the one hand, and documented involvement of (increased) ADAM10 expression/activity on the other hand. Clear experimental proof in these and other conditions is mostly pending and will eventually require use of research tools able to discriminate between different PrP derivatives
Acknowledgements
The authors apologize to all colleagues whose important scientific contributions to aspects covered herein could not be cited due to space and format limitations.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the Alzheimer Forschung Initiative e.V. (AFI; to HCA), the Creutzfeldt-Jakob Disease Foundation, Inc. (to HCA), the Werner-Otto-Stiftung (to BP and HCA), and the Deutsche Forschungsgemeinschaft (DFG; GRK1459 to BM and MG; CRC877 project A12 to MG).
Declarations
Ethical approval
No animal experiments or research on human subjects/samples/cells have been performed for this review article.
Informed consent
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aguilar-Calvo P, Sevillano AM, Bapat J, Soldau K, Sandoval DR, Altmeppen HC, Linsenmeier L, Pizzo DP, Geschwind MD, Sanchez H, Appleby BS, Cohen ML, Safar JG, Edland SD, Glatzel M, Nilsson KPR, Esko JD, Sigurdson CJ. Shortening heparan sulfate chains prolongs survival and reduces parenchymal plaques in prion disease caused by mobile, ADAM10-cleaved prions. Acta Neuropathol. 2020;139:527–546. doi: 10.1007/s00401-019-02085-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguzzi A, Baumann F, Bremer J. The prion’s elusive reason for being. Annu Rev Neurosci. 2008;31:439–477. doi: 10.1146/annurev.neuro.31.060407.125620. [DOI] [PubMed] [Google Scholar]
- Altmeppen HC, Prox J, Krasemann S, Puig B, Kruszewski K, Dohler F, Bernreuther C, Hoxha A, Linsenmeier L, Sikorska B, Liberski PP, Bartsch U, Saftig P, Glatzel M (2015) The sheddase ADAM10 is a potent modulator of prion disease. Elife 4 [DOI] [PMC free article] [PubMed]
- Altmeppen HC, Prox J, Puig B, Dohler F, Falker C, Krasemann S, Glatzel M. Roles of endoproteolytic α-cleavage and shedding of the prion protein in neurodegeneration. FEBS J. 2013;280:4338–4347. doi: 10.1111/febs.12196. [DOI] [PubMed] [Google Scholar]
- Altmeppen HC, Prox J, Puig B, Kluth MA, Bernreuther C, Thurm D, Jorissen E, Petrowitz B, Bartsch U, De Strooper B, Saftig P, Glatzel M. Lack of a-disintegrin-and-metalloproteinase ADAM10 leads to intracellular accumulation and loss of shedding of the cellular prion protein in vivo. Mol Neurodegener. 2011;6:36. doi: 10.1186/1750-1326-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeppen HC, Puig B, Dohler F, Thurm DK, Falker C, Krasemann S, Glatzel M. Proteolytic processing of the prion protein in health and disease. Am J Neurodegener Dis. 2012;1:15–31. [PMC free article] [PubMed] [Google Scholar]
- Amin L, Harris DA. Aβ receptors specifically recognize molecular features displayed by fibril ends and neurotoxic oligomers. Nat Commun. 2021;12:3451. doi: 10.1038/s41467-021-23507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin L, Nguyen XTA, Rolle IG, D’Este E, Giachin G, Tran TH, Šerbec VČ, Cojoc D, Legname G. Characterization of prion protein function by focal neurite stimulation. J Cell Sci. 2016;129:3878–3891. doi: 10.1242/jcs.183137. [DOI] [PubMed] [Google Scholar]
- Appel D, Hummel R, Weidemeier M, Endres K, Gölz C, Schäfer MKE. Pharmacologic inhibition of ADAM10 attenuates brain tissue loss, axonal injury and pro-inflammatory gene expression following traumatic brain injury in mice. Front Cell Dev Biol. 2021;9:661462. doi: 10.3389/fcell.2021.661462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson CJ, Kawamata F, Liu C, Ham S, Győrffy B, Munn AL, Wei MQ, Möller A, Whitehall V, Wiegmans AP. EGFR and Prion protein promote signaling via FOXO3a-KLF5 resulting in clinical resistance to platinum agents in colorectal cancer. Mol Oncol. 2019;13:725–737. doi: 10.1002/1878-0261.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkebø MK, Mouillet-Richard S, Espenes A, Goldmann W, Tatzelt J, Tranulis MA. The Cellular Prion Protein: A Player in Immunological Quiescence. Front Immunol. 2015;6:450. doi: 10.3389/fimmu.2015.00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri G, Palumbo S, Gabrusiewicz K, Azzalin A, Marchesi N, Spedito A, Biggiogera M, Sbalchiero E, Mazzini G, Miracco C, Pirtoli L, Kaminska B, Comincini S. Silencing of cellular prion protein (PrPC) expression by DNA-antisense oligonucleotides induces autophagy-dependent cell death in glioma cells. Autophagy. 2011;7:840–853. doi: 10.4161/auto.7.8.15615. [DOI] [PubMed] [Google Scholar]
- Basler K, Oesch B, Scott M, Westaway D, Wälchli M, Groth DF, McKinley MP, Prusiner SB, Weissmann C. Scrapie and cellular PrP isoforms are encoded by the same chromosomal gene. Cell. 1986;46:417–428. doi: 10.1016/0092-8674(86)90662-8. [DOI] [PubMed] [Google Scholar]
- Bate C, Tayebi M, Diomede L, Salmona M, Williams A. Glimepiride reduces the expression of PrPc, prevents PrPSc formation and protects against prion mediated neurotoxicity in cell lines. PLoS ONE. 2009;4:e8221. doi: 10.1371/journal.pone.0008221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béland M, Bédard M, Tremblay G, Lavigne P, Roucou X. Aβ induces its own prion protein N-terminal fragment (PrPN1)-mediated neutralization in amorphous aggregates. Neurobiol Aging. 2014;35:1537–1548. doi: 10.1016/j.neurobiolaging.2014.02.001. [DOI] [PubMed] [Google Scholar]
- Béland M, Motard J, Barbarin A, Roucou X. PrP(C) homodimerization stimulates the production of PrPC cleaved fragments PrPN1 and PrPC1. J Neurosci. 2012;32:13255–13263. doi: 10.1523/JNEUROSCI.2236-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béland M, Roucou X. The prion protein unstructured N-terminal region is a broad-spectrum molecular sensor with diverse and contrasting potential functions. J Neurochem. 2012;120:853–868. doi: 10.1111/j.1471-4159.2011.07613.x. [DOI] [PubMed] [Google Scholar]
- Beraldo FH, Ostapchenko VG, Caetano FA, Guimaraes ALS, Ferretti GDS, Daude N, Bertram L, Nogueira KOPC, Silva JL, Westaway D, Cashman NR, Martins VR, Prado VF, Prado MAM. Regulation of Amyloid β Oligomer Binding to Neurons and Neurotoxicity by the Prion Protein-mGluR5 Complex. J Biol Chem. 2016;291:21945–21955. doi: 10.1074/jbc.M116.738286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardino-Sgherri J, Siberchicot C, Auvré F, Busso D, Brocas C, El Masri G, Lioutsko A, Ferri F, Radicella JP, Romeo P-H, Bravard A. Tumor resistance to radiotherapy is triggered by an ATM/TAK1-dependent-increased expression of the cellular prion protein. Oncogene. 2021;40:3460–3469. doi: 10.1038/s41388-021-01746-0. [DOI] [PubMed] [Google Scholar]
- Bessen RA, Shearin H, Martinka S, Boharski R, Lowe D, Wilham JM, Caughey B, Wiley JA. Prion shedding from olfactory neurons into nasal secretions. PLoS Pathog. 2010;6:e1000837. doi: 10.1371/journal.ppat.1000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian J, Kim S, Kane SJ, Crowell J, Sun JL, Christiansen J, Saijo E, Moreno JA, DiLisio J, Burnett E, Pritzkow S, Gorski D, Soto C, Kreeger TJ, Balachandran A, Mitchell G, Miller MW, Nonno R, Vikøren T, Våge J, Madslien K, Tran L, Vuong TT, Benestad SL, Telling GC. Adaptive selection of a prion strain conformer corresponding to established North American CWD during propagation of novel emergent Norwegian strains in mice expressing elk or deer prion protein. PLoS Pathog. 2021;17:e1009748. doi: 10.1371/journal.ppat.1009748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignon Y, Poindessous V, Lazareth H, Passet B, Vilotte J-L, Djouadi F, Mouillet-Richard S, Pallet N. The cellular prion protein is a stress protein secreted by renal tubular cells and a urinary marker of kidney injury. Cell Death Dis. 2020;11:243. doi: 10.1038/s41419-020-2430-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockman JM, Kingsbury DT, McKinley MP, Bendheim PE, Prusiner SB. Creutzfeldt-Jakob disease prion proteins in human brains. N Engl J Med. 1985;312:73–78. doi: 10.1056/NEJM198501103120202. [DOI] [PubMed] [Google Scholar]
- Boon BDC, Bulk M, Jonker AJ, Morrema THJ, van den Berg E, Popovic M, Walter J, Kumar S, van der Lee SJ, Holstege H, Zhu X, Van Nostrand WE, Natté R, van der Weerd L, Bouwman FH, van de Berg WDJ, Rozemuller AJM, Hoozemans JJM. The coarse-grained plaque: a divergent Aβ plaque-type in early-onset Alzheimer’s disease. Acta Neuropathol. 2020;140:811–830. doi: 10.1007/s00401-020-02198-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchelt DR, Rogers M, Stahl N, Telling G, Prusiner SB. Release of the cellular prion protein from cultured cells after loss of its glycoinositol phospholipid anchor. Glycobiology. 1993;3:319–329. doi: 10.1093/glycob/3.4.319. [DOI] [PubMed] [Google Scholar]
- Bove-Fenderson E, Urano R, Straub JE, Harris DA. Cellular prion protein targets amyloid-β fibril ends via its C-terminal domain to prevent elongation. J Biol Chem. 2017;292:16858–16871. doi: 10.1074/jbc.M117.789990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer J, Baumann F, Tiberi C, Wessig C, Fischer H, Schwarz P, Steele AD, Toyka KV, Nave K-A, Weis J, Aguzzi A. Axonal prion protein is required for peripheral myelin maintenance. Nat Neurosci. 2010;13:310–318. doi: 10.1038/nn.2483. [DOI] [PubMed] [Google Scholar]
- Brenna S, Altmeppen HC, Mohammadi B, Rissiek B, Schlink F, Ludewig P, Krisp C, Schlüter H, Failla AV, Schneider C, Glatzel M, Puig B, Magnus T. Characterization of brain-derived extracellular vesicles reveals changes in cellular origin after stroke and enrichment of the prion protein with a potential role in cellular uptake. J Extracell Vesicles. 2020;9:1809065. doi: 10.1080/20013078.2020.1809065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brethour D, Mehrabian M, Williams D, Wang X, Ghodrati F, Ehsani S, Rubie EA, Woodgett JR, Sevalle J, Xi Z, Rogaeva E, Schmitt-Ulms G. A ZIP6-ZIP10 heteromer controls NCAM1 phosphorylation and integration into focal adhesion complexes during epithelial-to-mesenchymal transition. Sci Rep. 2017;7:40313. doi: 10.1038/srep40313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büeler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- Calella AM, Farinelli M, Nuvolone M, Mirante O, Moos R, Falsig J, Mansuy IM, Aguzzi A. Prion protein and Abeta-related synaptic toxicity impairment. EMBO Mol Med. 2010;2:306–314. doi: 10.1002/emmm.201000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callender JA, Sevillano AM, Soldau K, Kurt TD, Schumann T, Pizzo DP, Altmeppen H, Glatzel M, Esko JD, Sigurdson CJ. Prion protein post-translational modifications modulate heparan sulfate binding and limit aggregate size in prion disease. Neurobiol Dis. 2020;142:104955. doi: 10.1016/j.nbd.2020.104955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho MV, Telling G, Kong Q, Gambetti P, Notari S. Role of prion protein glycosylation in replication of human prions by protein misfolding cyclic amplification. Lab Invest. 2019;99:1741–1748. doi: 10.1038/s41374-019-0282-1. [DOI] [PubMed] [Google Scholar]
- Carroll JA, Groveman BR, Williams K, Moore R, Race B, Haigh CL. Prion protein N1 cleavage peptides stimulate microglial interaction with surrounding cells. Sci Rep. 2020;10:6654. doi: 10.1038/s41598-020-63472-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carulla P, Llorens F, Matamoros-Angles A, Aguilar-Calvo P, Espinosa JC, Gavín R, Ferrer I, Legname G, Torres JM, del Río JA. Involvement of PrP(C) in kainate-induced excitotoxicity in several mouse strains. Sci Rep. 2015;5:11971. doi: 10.1038/srep11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle AR, Daude N, Gilch S, Westaway D. Application of high-throughput, capillary-based Western analysis to modulated cleavage of the cellular prion protein. J Biol Chem. 2019;294:2642–2650. doi: 10.1074/jbc.RA118.006367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B, Neary K, Buller R, Ernst D, Perry LL, Chesebro B, Race RE. Normal and scrapie-associated forms of prion protein differ in their sensitivities to phospholipase and proteases in intact neuroblastoma cells. J Virol. 1990;64:1093–1101. doi: 10.1128/jvi.64.3.1093-1101.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B, Race RE, Ernst D, Buchmeier MJ, Chesebro B. Prion protein biosynthesis in scrapie-infected and uninfected neuroblastoma cells. J Virol. 1989;63:175–181. doi: 10.1128/jvi.63.1.175-181.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Mangé A, Dong L, Lehmann S, Schachner M. Prion protein as trans-interacting partner for neurons is involved in neurite outgrowth and neuronal survival. Mol Cell Neurosci. 2003;22:227–233. doi: 10.1016/S1044-7431(02)00014-3. [DOI] [PubMed] [Google Scholar]
- Chen S, Yadav SP, Surewicz WK. Interaction between human prion protein and amyloid-beta (Abeta) oligomers: role OF N-terminal residues. J Biol Chem. 2010;285:26377–26383. doi: 10.1074/jbc.M110.145516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SG, Teplow DB, Parchi P, Teller JK, Gambetti P, Autilio-Gambetti L. Truncated forms of the human prion protein in normal brain and in prion diseases. J Biol Chem. 1995;270:19173–19180. doi: 10.1074/jbc.270.32.19173. [DOI] [PubMed] [Google Scholar]
- Cheng CJ, Koldsø H, Van der Kamp MW, Schiøtt B, Daggett V. Simulations of membrane-bound diglycosylated human prion protein reveal potential protective mechanisms against misfolding. J Neurochem. 2017;142:171–182. doi: 10.1111/jnc.14044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Lin L, Li X, Lu A, Hou C, Wu Q, Hu X, Zhou Z, Chen Z, Tang F. ADAM10 is involved in the oncogenic process and chemo-resistance of triple-negative breast cancer via regulating Notch1 signaling pathway, CD44 and PrPc. Cancer Cell Int. 2021;21:32. doi: 10.1186/s12935-020-01727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Race B, Meade-White K, Lacasse R, Race R, Klingeborn M, Striebel J, Dorward D, McGovern G, Jeffrey M. Fatal transmissible amyloid encephalopathy: a new type of prion disease associated with lack of prion protein membrane anchoring. PLoS Pathog. 2010;6:e1000800. doi: 10.1371/journal.ppat.1000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B, Trifilo M, Race R, Meade-White K, Teng C, LaCasse R, Raymond L, Favara C, Baron G, Priola S, Caughey B, Masliah E, Oldstone M. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science. 2005;308:1435–1439. doi: 10.1126/science.1110837. [DOI] [PubMed] [Google Scholar]
- Chiarini LB, Freitas ARO, Zanata SM, Brentani RR, Martins VR, Linden R. Cellular prion protein transduces neuroprotective signals. EMBO J. 2002;21:3317–3326. doi: 10.1093/emboj/cdf324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J-K, Cali I, Surewicz K, Kong Q, Gambetti P, Surewicz WK. Amyloid fibrils from the N-terminal prion protein fragment are infectious. Proc Natl Acad Sci U S A. 2016;113:13851–13856. doi: 10.1073/pnas.1610716113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E, Ji Y, Sun Y, Kascsak RJ, Kascsak RB, Mehta PD, Strittmatter SM, Wisniewski T. Anti-PrPC monoclonal antibody infusion as a novel treatment for cognitive deficits in an Alzheimer’s disease model mouse. BMC Neurosci. 2010;11:130. doi: 10.1186/1471-2202-11-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby DW, Prusiner SB. Prions. Cold Spring Harb Perspect Biol. 2011;3:a006833. doi: 10.1101/cshperspect.a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SJ, Tumpach C, Groveman BR, Drew SC, Haigh CL. Prion protein cleavage fragments regulate adult neural stem cell quiescence through redox modulation of mitochondrial fission and SOD2 expression. Cell Mol Life Sci. 2018;75:3231–3249. doi: 10.1007/s00018-018-2790-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo F, Meldolesi J. L1-CAM and N-CAM: from adhesion proteins to pharmacological targets. Trends Pharmacol Sci. 2015;36:769–781. doi: 10.1016/j.tips.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Corbett GT, Wang Z, Hong W, Colom-Cadena M, Rose J, Liao M, Asfaw A, Hall TC, Ding L, DeSousa A, Frosch MP, Collinge J, Harris DA, Perkinton MS, Spires-Jones TL, Young-Pearse TL, Billinton A, Walsh DM. PrP is a central player in toxicity mediated by soluble aggregates of neurodegeneration-causing proteins. Acta Neuropathol. 2020;139:503–526. doi: 10.1007/s00401-019-02114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corda E, Du X, Shim SY, Klein AN, Siltberg-Liberles J, Gilch S. Interaction of peptide aptamers with prion protein central domain promotes α-cleavage of PrPC. Mol Neurobiol. 2018;55:7758–7774. doi: 10.1007/s12035-018-0944-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsaro A, Bajetto A, Thellung S, Begani G, Villa V, Nizzari M, Pattarozzi A, Solari A, Gatti M, Pagano A, Würth R, Daga A, Barbieri F, Florio T. Cellular prion protein controls stem cell-like properties of human glioblastoma tumor-initiating cells. Oncotarget. 2016;7:38638–38657. doi: 10.18632/oncotarget.9575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford HC, Dempsey PJ, Brown G, Adam L, Moss ML. ADAM10 as a therapeutic target for cancer and inflammation. Curr Pharm Des. 2009;15:2288–2299. doi: 10.2174/138161209788682442. [DOI] [PubMed] [Google Scholar]
- D’Arrigo G, Gabrielli M, Scaroni F, Swuec P, Amin L, Pegoraro A, Adinolfi E, Di Virgilio F, Cojoc D, Legname G, Verderio C. Astrocytes-derived extracellular vesicles in motion at the neuron surface: Involvement of the prion protein. J Extracell Vesicles. 2021;10:e12114. doi: 10.1002/jev2.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco E, Celauro L, Vanni S, Grandolfo M, Bistaffa E, Moda F, Aguzzi A, Legname G. The uptake of tau amyloid fibrils is facilitated by the cellular prion protein and hampers prion propagation in cultured cells. J Neurochem. 2020;155:577–591. doi: 10.1111/jnc.15040. [DOI] [PubMed] [Google Scholar]
- de Lacerda TCS, Costa-Silva B, Giudice FS, Dias MVS, de Oliveira GP, Teixeira BL, Dos Santos TG, Martins VR. Prion protein binding to HOP modulates the migration and invasion of colorectal cancer cells. Clin Exp Metastasis. 2016;33:441–451. doi: 10.1007/s10585-016-9788-8. [DOI] [PubMed] [Google Scholar]
- DeArmond SJ, Sánchez H, Yehiely F, Qiu Y, Ninchak-Casey A, Daggett V, Camerino AP, Cayetano J, Rogers M, Groth D, Torchia M, Tremblay P, Scott MR, Cohen FE, Prusiner SB. Selective neuronal targeting in prion disease. Neuron. 1997;19:1337–1348. doi: 10.1016/S0896-6273(00)80424-9. [DOI] [PubMed] [Google Scholar]
- DeMarco ML, Daggett V. Characterization of cell-surface prion protein relative to its recombinant analogue: insights from molecular dynamics simulations of diglycosylated, membrane-bound human prion protein. J Neurochem. 2009;109:60–73. doi: 10.1111/j.1471-4159.2009.05892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey PJ. Role of ADAM10 in intestinal crypt homeostasis and tumorigenesis. Biochim Biophys Acta Mol Cell Res. 2017;1864:2228–2239. doi: 10.1016/j.bbamcr.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denkers ND, Hoover CE, Davenport KA, Henderson DM, McNulty EE, Nalls AV, Mathiason CK, Hoover EA. Very low oral exposure to prions of brain or saliva origin can transmit chronic wasting disease. PLoS ONE. 2020;15:e0237410. doi: 10.1371/journal.pone.0237410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter E, Kong Q. Neuroprotective effect and potential of cellular prion protein and its cleavage products for treatment of neurodegenerative disorders part I. a literature review. Expert Rev Neurother. 2021;21:969–982. doi: 10.1080/14737175.2021.1965881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter E, Kong Q. Neuroprotective effect and potential of cellular prion protein and its cleavage products for treatment of neurodegenerative disorders part II: strategies for therapeutics development. Expert Rev Neurother. 2021;21:983–991. doi: 10.1080/14737175.2021.1965882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Lucena D, Kruse N, Thüne K, Schmitz M, Villar-Piqué A, da Cunha JEG, Hermann P, López-Pérez Ó, Andrés-Benito P, Ladogana A, Calero M, Vidal E, Riggert J, Pineau H, Sim V, Zetterberg H, Blennow K, Del Río JA, Marín-Moreno A, Espinosa JC, Torres JM, Sánchez-Valle R, Mollenhauer B, Ferrer I, Zerr I, Llorens F. TREM2 expression in the brain and biological fluids in prion diseases. Acta Neuropathol. 2021;141:841–859. doi: 10.1007/s00401-021-02296-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doeppner TR, Kaltwasser B, Schlechter J, Jaschke J, Kilic E, Bähr M, Hermann DM, Weise J. Cellular prion protein promotes post-ischemic neuronal survival, angioneurogenesis and enhances neural progenitor cell homing via proteasome inhibition. Cell Death Dis. 2015;6:e2024. doi: 10.1038/cddis.2015.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohler F, Sepulveda-Falla D, Krasemann S, Altmeppen H, Schlüter H, Hildebrand D, Zerr I, Matschke J, Glatzel M. High molecular mass assemblies of amyloid-β oligomers bind prion protein in patients with Alzheimer’s disease. Brain. 2014;137:873–886. doi: 10.1093/brain/awt375. [DOI] [PubMed] [Google Scholar]
- Du L, Rao G, Wang H, Li B, Tian W, Cui J, He L, Laffin B, Tian X, Hao C, Liu H, Sun X, Zhu Y, Tang DG, Mehrpour M, Lu Y, Chen Q. CD44-positive cancer stem cells expressing cellular prion protein contribute to metastatic capacity in colorectal cancer. Cancer Res. 2013;73:2682–2694. doi: 10.1158/0008-5472.CAN-12-3759. [DOI] [PubMed] [Google Scholar]
- Enari M, Flechsig E, Weissmann C. Scrapie prion protein accumulation by scrapie-infected neuroblastoma cells abrogated by exposure to a prion protein antibody. Proc Natl Acad Sci U S A. 2001;98:9295–9299. doi: 10.1073/pnas.151242598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres K, Fahrenholz F, Lotz J, Hiemke C, Teipel S, Lieb K, Tüscher O, Fellgiebel A. Increased CSF APPs-α levels in patients with Alzheimer disease treated with acitretin. Neurology. 2014;83:1930–1935. doi: 10.1212/WNL.0000000000001017. [DOI] [PubMed] [Google Scholar]
- Endres K, Mitteregger G, Kojro E, Kretzschmar H, Fahrenholz F. Influence of ADAM10 on prion protein processing and scrapie infectiosity in vivo. Neurobiol Dis. 2009;36:233–241. doi: 10.1016/j.nbd.2009.07.015. [DOI] [PubMed] [Google Scholar]
- Falker C, Hartmann A, Guett I, Dohler F, Altmeppen H, Betzel C, Schubert R, Thurm D, Wegwitz F, Joshi P, Verderio C, Krasemann S, Glatzel M. Exosomal cellular prion protein drives fibrillization of amyloid beta and counteracts amyloid beta-mediated neurotoxicity. J Neurochem. 2016;137:88–100. doi: 10.1111/jnc.13514. [DOI] [PubMed] [Google Scholar]
- Féraudet C, Morel N, Simon S, Volland H, Frobert Y, Créminon C, Vilette D, Lehmann S, Grassi J. Screening of 145 anti-PrP monoclonal antibodies for their capacity to inhibit PrPSc replication in infected cells. J Biol Chem. 2005;280:11247–11258. doi: 10.1074/jbc.M407006200. [DOI] [PubMed] [Google Scholar]
- Ferreira DG, Temido-Ferreira M, Vicente Miranda H, Batalha VL, Coelho JE, Szegö ÉM, Marques-Morgado I, Vaz SH, Rhee JS, Schmitz M, Zerr I, Lopes LV, Outeiro TF. α-synuclein interacts with PrPC to induce cognitive impairment through mGluR5 and NMDAR2B. Nat Neurosci. 2017;20:1569–1579. doi: 10.1038/nn.4648. [DOI] [PubMed] [Google Scholar]
- Ferrer I, Blanco R, Carmona M, Puig B, Ribera R, Rey MJ, Ribalta T. Prion protein expression in senile plaques in Alzheimer’s disease. Acta Neuropathol. 2001;101:49–56. doi: 10.1007/s004010000271. [DOI] [PubMed] [Google Scholar]
- Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, Laude H, Raposo G. Cells release prions in association with exosomes. Proc Natl Acad Sci U S A. 2004;101:9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Rülicke T, Raeber A, Sailer A, Moser M, Oesch B, Brandner S, Aguzzi A, Weissmann C. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 1996;15:1255–1264. doi: 10.1002/j.1460-2075.1996.tb00467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluharty BR, Biasini E, Stravalaci M, Sclip A, Diomede L, Balducci C, La Vitola P, Messa M, Colombo L, Forloni G, Borsello T, Gobbi M, Harris DA. An N-terminal fragment of the prion protein binds to amyloid-β oligomers and inhibits their neurotoxicity in vivo. J Biol Chem. 2013;288:7857–7866. doi: 10.1074/jbc.M112.423954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley AR, Roseman GP, Chan K, Smart A, Finn TS, Yang K, Lokey RS, Millhauser GL, Raskatov JA. Evidence for aggregation-independent, PrPC-mediated Aβ cellular internalization. Proc Natl Acad Sci U S A. 2020;117:28625–28631. doi: 10.1073/pnas.2009238117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkesson M, Li C, Frebelius S, Swedenborg J, Wågsäter D, Williams KJ, Eriksson P, Roy J, Liu M-L. Proteolytically active ADAM10 and ADAM17 carried on membrane microvesicles in human abdominal aortic aneurysms. Thromb Haemost. 2015;114:1165–1174. doi: 10.1160/TH14-10-0899. [DOI] [PubMed] [Google Scholar]
- Freir DB, Nicoll AJ, Klyubin I, Panico S, Mc Donald JM, Risse E, Asante EA, Farrow MA, Sessions RB, Saibil HR, Clarke AR, Rowan MJ, Walsh DM, Collinge J. Interaction between prion protein and toxic amyloid β assemblies can be therapeutically targeted at multiple sites. Nat Commun. 2011;2:336. doi: 10.1038/ncomms1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauczynski S, Peyrin JM, Haïk S, Leucht C, Hundt C, Rieger R, Krasemann S, Deslys JP, Dormont D, Lasmézas CI, Weiss S. The 37-kDa/67-kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein. EMBO J. 2001;20:5863–5875. doi: 10.1093/emboj/20.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazi A, Le Corre D, Pilati C, Taieb J, Aparicio T, Didelot A, Dedhar S, Mulot C, Le Malicot K, Djouadi F, de Reynies A, Launay J-M, Laurent-Puig P, Mouillet-Richard S. Prognostic value of the PrPC-ILK-IDO1 axis in the mesenchymal colorectal cancer subtype. Oncoimmunology. 2021;10:1940674. doi: 10.1080/2162402X.2021.1940674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilch S, Wopfner F, Renner-Müller I, Kremmer E, Bauer C, Wolf E, Brem G, Groschup MH, Schätzl HM. Polyclonal anti-PrP auto-antibodies induced with dimeric PrP interfere efficiently with PrPSc propagation in prion-infected cells. J Biol Chem. 2003;278:18524–18531. doi: 10.1074/jbc.M210723200. [DOI] [PubMed] [Google Scholar]
- Gimbel DA, Nygaard HB, Coffey EE, Gunther EC, Laurén J, Gimbel ZA, Strittmatter SM. Memory impairment in transgenic Alzheimer mice requires cellular prion protein. J Neurosci. 2010;30:6367–6374. doi: 10.1523/JNEUROSCI.0395-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LA, Hipp SA, Rijal Upadhaya A, Balakrishnan K, Ospitalieri S, Koper MJ, Largo-Barrientos P, Uytterhoeven V, Reichwald J, Rabe S, Vandenberghe R, von Arnim CAF, Tousseyn T, Feederle R, Giudici C, Willem M, Staufenbiel M, Thal DR. Aβ-induced acceleration of Alzheimer-related τ-pathology spreading and its association with prion protein. Acta Neuropathol. 2019;138:913–941. doi: 10.1007/s00401-019-02053-5. [DOI] [PubMed] [Google Scholar]
- Gonias SL, Campana WM. LDL receptor-related protein-1: a regulator of inflammation in atherosclerosis, cancer, and injury to the nervous system. Am J Pathol. 2014;184:18–27. doi: 10.1016/j.ajpath.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot-Sestier M-V, Checler F. Cellular prion and its catabolites in the brain: production and function. Curr Mol Med. 2012;12:304–315. doi: 10.2174/156652412799218912. [DOI] [PubMed] [Google Scholar]
- Guillot-Sestier M-V, Sunyach C, Druon C, Scarzello S, Checler F. The alpha-secretase-derived N-terminal product of cellular prion, N1, displays neuroprotective function in vitro and in vivo. J Biol Chem. 2009;284:35973–35986. doi: 10.1074/jbc.M109.051086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot-Sestier M-V, Sunyach C, Ferreira ST, Marzolo M-P, Bauer C, Thevenet A, Checler F. α-Secretase-derived fragment of cellular prion, N1, protects against monomeric and oligomeric amyloid β (Aβ)-associated cell death. J Biol Chem. 2012;287:5021–5032. doi: 10.1074/jbc.M111.323626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guitart K, Loers G, Buck F, Bork U, Schachner M, Kleene R. Improvement of neuronal cell survival by astrocyte-derived exosomes under hypoxic and ischemic conditions depends on prion protein. Glia. 2016;64:896–910. doi: 10.1002/glia.22963. [DOI] [PubMed] [Google Scholar]
- Guo BB, Bellingham SA, Hill AF. Stimulating the Release of Exosomes Increases the Intercellular Transfer of Prions. J Biol Chem. 2016;291:5128–5137. doi: 10.1074/jbc.M115.684258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo BB, Bellingham SA, Hill AF. The neutral sphingomyelinase pathway regulates packaging of the prion protein into exosomes. J Biol Chem. 2015;290:3455–3467. doi: 10.1074/jbc.M114.605253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddon DJ, Hughes MR, Antignano F, Westaway D, Cashman NR, McNagny KM. Prion protein expression and release by mast cells after activation. J Infect Dis. 2009;200:827–831. doi: 10.1086/605022. [DOI] [PubMed] [Google Scholar]
- Haigh CL, Drew SC, Boland MP, Masters CL, Barnham KJ, Lawson VA, Collins SJ. Dominant roles of the polybasic proline motif and copper in the PrP23-89-mediated stress protection response. J Cell Sci. 2009;122:1518–1528. doi: 10.1242/jcs.043604. [DOI] [PubMed] [Google Scholar]
- Haigh CL, Lewis VA, Vella LJ, Masters CL, Hill AF, Lawson VA, Collins SJ. PrPC-related signal transduction is influenced by copper, membrane integrity and the alpha cleavage site. Cell Res. 2009;19:1062–1078. doi: 10.1038/cr.2009.86. [DOI] [PubMed] [Google Scholar]
- Haigh CL, McGlade AR, Collins SJ. MEK1 transduces the prion protein N2 fragment antioxidant effects. Cell Mol Life Sci. 2015;72:1613–1629. doi: 10.1007/s00018-014-1777-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A, Zamponi GW, Ferguson SSG. Glutamate receptors function as scaffolds for the regulation of β-amyloid and cellular prion protein signaling complexes. Mol Brain. 2015;8:18. doi: 10.1186/s13041-015-0107-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y-S, Yoon YM, Go G, Lee JH, Lee SH. Melatonin protects human renal proximal tubule epithelial cells against high glucose-mediated fibrosis via the cellular prion protein-TGF-β-smad signaling axis. Int J Med Sci. 2020;17:1235–1245. doi: 10.7150/ijms.42603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris DA, Huber MT, van Dijken P, Shyng SL, Chait BT, Wang R. Processing of a cellular prion protein: identification of N- and C-terminal cleavage sites. Biochemistry. 1993;32:1009–1016. doi: 10.1021/bi00055a003. [DOI] [PubMed] [Google Scholar]
- Heiseke A, Schöbel S, Lichtenthaler SF, Vorberg I, Groschup MH, Kretzschmar H, Schätzl HM, Nunziante M. The novel sorting nexin SNX33 interferes with cellular PrP formation by modulation of PrP shedding. Traffic. 2008;9:1116–1129. doi: 10.1111/j.1600-0854.2008.00750.x. [DOI] [PubMed] [Google Scholar]
- Heisler FF, Pechmann Y, Wieser I, Altmeppen HC, Veenendaal L, Muhia M, Schweizer M, Glatzel M, Krasemann S, Kneussel M. Muskelin coordinates PrPC lysosome versus exosome targeting and impacts prion disease progression. Neuron. 2018;99:1155–1169.e9. doi: 10.1016/j.neuron.2018.08.010. [DOI] [PubMed] [Google Scholar]
- Henzi A, Senatore A, Lakkaraju AKK, Scheckel C, Mühle J, Reimann R, Sorce S, Schertler G, Toyka KV, Aguzzi A (2020) Soluble dimeric prion protein ligand activates Adgrg6 receptor but does not rescue early signs of demyelination in PrP-deficient mice. PLoS One 15:e0242137 [DOI] [PMC free article] [PubMed]
- Heppner FL, Musahl C, Arrighi I, Klein MA, Rülicke T, Oesch B, Zinkernagel RM, Kalinke U, Aguzzi A. Prevention of scrapie pathogenesis by transgenic expression of anti-prion protein antibodies. Science. 2001;294:178–182. doi: 10.1126/science.1063093. [DOI] [PubMed] [Google Scholar]
- Hirsch TZ, Martin-Lannerée S, Mouillet-Richard S. Functions of the prion protein. Prog Mol Biol Transl Sci. 2017;150:1–34. doi: 10.1016/bs.pmbts.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Hu N-W, Corbett GT, Moore S, Klyubin I, O’Malley TT, Walsh DM, Livesey FJ, Rowan MJ. Extracellular forms of Aβ and tau from iPSC models of Alzheimer’s disease disrupt synaptic plasticity. Cell Rep. 2018;23:1932–1938. doi: 10.1016/j.celrep.2018.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N-W, Nicoll AJ, Zhang D, Mably AJ, O’Malley T, Purro SA, Terry C, Collinge J, Walsh DM, Rowan MJ. mGlu5 receptors and cellular prion protein mediate amyloid-β-facilitated synaptic long-term depression in vivo. Nat Commun. 2014;5:3374. doi: 10.1038/ncomms4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Black SA, Huang J, Stys PK, Zamponi GW. Mutation of copper binding sites on cellular prion protein abolishes its inhibitory action on NMDA receptors in mouse hippocampal neurons. Mol Brain. 2021;14:117. doi: 10.1186/s13041-021-00828-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundt C, Peyrin JM, Haïk S, Gauczynski S, Leucht C, Rieger R, Riley ML, Deslys JP, Dormont D, Lasmézas CI, Weiss S. Identification of interaction domains of the prion protein with its 37-kDa/67-kDa laminin receptor. EMBO J. 2001;20:5876–5886. doi: 10.1093/emboj/20.21.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen C, Parchi P, Capellari S, Vermeij AJ, Corrado P, Baas F, Strammiello R, van Gool WA, van Swieten JC, Rozemuller AJM. Prion protein amyloidosis with divergent phenotype associated with two novel nonsense mutations in PRNP. Acta Neuropathol. 2010;119:189–197. doi: 10.1007/s00401-009-0609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz-Griffiths HH, Corbett NJ, Rowland HA, Fisher K, Jones AC, Baron J, Howell GJ, Cowley SA, Chintawar S, Cader MZ, Kellett KAB, Hooper NM. Proteolytic shedding of the prion protein via activation of metallopeptidase ADAM10 reduces cellular binding and toxicity of amyloid-β oligomers. J Biol Chem. 2019;294:7085–7097. doi: 10.1074/jbc.RA118.005364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorissen E, Prox J, Bernreuther C, Weber S, Schwanbeck R, Serneels L, Snellinx A, Craessaerts K, Thathiah A, Tesseur I, Bartsch U, Weskamp G, Blobel CP, Glatzel M, De Strooper B, Saftig P. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J Neurosci. 2010;30:4833–4844. doi: 10.1523/JNEUROSCI.5221-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser DM, Acharya M, Leighton PLA, Wang H, Daude N, Wohlgemuth S, Shi B, Allison WT. Amyloid beta precursor protein and prion protein have a conserved interaction affecting cell adhesion and CNS development. PLoS ONE. 2012;7:e51305. doi: 10.1371/journal.pone.0051305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaani J, Prusiner SB, Diacovo J, Baekkeskov S, Legname G. Recombinant prion protein induces rapid polarization and development of synapses in embryonic rat hippocampal neurons in vitro. J Neurochem. 2005;95:1373–1386. doi: 10.1111/j.1471-4159.2005.03469.x. [DOI] [PubMed] [Google Scholar]
- Kang H-E, Bian J, Kane SJ, Kim S, Selwyn V, Crowell J, Bartz JC, Telling GC. Incomplete glycosylation during prion infection unmasks a prion protein epitope that facilitates prion detection and strain discrimination. J Biol Chem. 2020;295:10420–10433. doi: 10.1074/jbc.RA120.012796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravani H, Zhang Y, Tsutsui S, Hameed S, Altier C, Hamid J, Chen L, Villemaire M, Ali Z, Jirik FR, Zamponi GW. Prion protein attenuates excitotoxicity by inhibiting NMDA receptors. J Cell Biol. 2008;181:551–565. doi: 10.1083/jcb.200711002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klyubin I, Nicoll AJ, Khalili-Shirazi A, Farmer M, Canning S, Mably A, Linehan J, Brown A, Wakeling M, Brandner S, Walsh DM, Rowan MJ, Collinge J. Peripheral administration of a humanized anti-PrP antibody blocks Alzheimer’s disease Aβ synaptotoxicity. J Neurosci. 2014;34:6140–6145. doi: 10.1523/JNEUROSCI.3526-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima A, Konishi M, Akizawa T. Prion fragment peptides are digested with membrane type matrix metalloproteinases and acquire enzyme resistance through Cu2+-binding. Biomolecules. 2014;4:510–526. doi: 10.3390/biom4020510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König AS, Rösener NS, Gremer L, Tusche M, Flender D, Reinartz E, Hoyer W, Neudecker P, Willbold D, Heise H. Structural details of amyloid β oligomers in complex with human prion protein as revealed by solid-state MAS NMR spectroscopy. J Biol Chem. 2021;296:100499. doi: 10.1016/j.jbc.2021.100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küffer A, Lakkaraju AKK, Mogha A, Petersen SC, Airich K, Doucerain C, Marpakwar R, Bakirci P, Senatore A, Monnard A, Schiavi C, Nuvolone M, Grosshans B, Hornemann S, Bassilana F, Monk KR, Aguzzi A. The prion protein is an agonistic ligand of the G protein-coupled receptor Adgrg6. Nature. 2016;536:464–468. doi: 10.1038/nature19312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn P-H, Colombo AV, Schusser B, Dreymueller D, Wetzel S, Schepers U, Herber J, Ludwig A, Kremmer E, Montag D, Müller U, Schweizer M, Saftig P, Bräse S, Lichtenthaler SF. Systematic substrate identification indicates a central role for the metalloprotease ADAM10 in axon targeting and synapse function. Elife. 2016;5:e12748. doi: 10.7554/eLife.12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffont-Proust I, Faucheux BA, Hässig R, Sazdovitch V, Simon S, Grassi J, Hauw J-J, Moya KL, Haïk S. The N-terminal cleavage of cellular prion protein in the human brain. FEBS Lett. 2005;579:6333–6337. doi: 10.1016/j.febslet.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Larson M, Sherman MA, Amar F, Nuvolone M, Schneider JA, Bennett DA, Aguzzi A, Lesné SE. The complex PrP(c)-Fyn couples human oligomeric Aβ with pathological tau changes in Alzheimer’s disease. J Neurosci. 2012;32:16857–16871a. doi: 10.1523/JNEUROSCI.1858-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A, McDonald A, Daude N, Mays CE, Walter ED, Aglietti R, Mercer RCC, Wohlgemuth S, van der Merwe J, Yang J, Gapeshina H, Kim C, Grams J, Shi B, Wille H, Balachandran A, Schmitt-Ulms G, Safar JG, Millhauser GL, Westaway D. Octarepeat region flexibility impacts prion function, endoproteolysis and disease manifestation. EMBO Mol Med. 2015;7:339–356. doi: 10.15252/emmm.201404588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurén J, Gimbel DA, Nygaard HB, Gilbert JW, Strittmatter SM. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers. Nature. 2009;457:1128–1132. doi: 10.1038/nature07761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Corre D, Ghazi A, Balogoun R, Pilati C, Aparicio T, Martin-Lannerée S, Marisa L, Djouadi F, Poindessous V, Crozet C, Emile J-F, Mulot C, Le Malicot K, Boige V, Blons H, de Reynies A, Taieb J, Ghiringhelli F, Bennouna J, Launay J-M, Laurent-Puig P, Mouillet-Richard S. The cellular prion protein controls the mesenchymal-like molecular subtype and predicts disease outcome in colorectal cancer. EBioMedicine. 2019;46:94–104. doi: 10.1016/j.ebiom.2019.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Baskakov IV. The cellular form of the prion protein is involved in controlling cell cycle dynamics, self-renewal, and the fate of human embryonic stem cell differentiation. J Neurochem. 2013;124:310–322. doi: 10.1111/j.1471-4159.2012.07913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legname G, Scialò C. On the role of the cellular prion protein in the uptake and signaling of pathological aggregates in neurodegenerative diseases. Prion. 2020;14:257–270. doi: 10.1080/19336896.2020.1854034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucht C, Simoneau S, Rey C, Vana K, Rieger R, Lasmézas CI, Weiss S. The 37 kDa/67 kDa laminin receptor is required for PrP(Sc) propagation in scrapie-infected neuronal cells. EMBO Rep. 2003;4:290–295. doi: 10.1038/sj.embor.embor768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis V, Hill AF, Haigh CL, Klug GM, Masters CL, Lawson VA, Collins SJ. Increased proportions of C1 truncated prion protein protect against cellular M1000 prion infection. J Neuropathol Exp Neurol. 2009;68:1125–1135. doi: 10.1097/NEN.0b013e3181b96981. [DOI] [PubMed] [Google Scholar]
- Lewis V, Johanssen VA, Crouch PJ, Klug GM, Hooper NM, Collins SJ. Prion protein “gamma-cleavage”: characterizing a novel endoproteolytic processing event. Cell Mol Life Sci. 2016;73:667–683. doi: 10.1007/s00018-015-2022-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q-Q, Cao X-X, Xu J-D, Chen Q, Wang W-J, Tang F, Chen Z-Q, Liu X-P, Xu Z-D. The role of P-glycoprotein/cellular prion protein interaction in multidrug-resistant breast cancer cells treated with paclitaxel. Cell Mol Life Sci. 2009;66:504–515. doi: 10.1007/s00018-008-8548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Kong Q. α-Cleavage of cellular prion protein. Prion. 2012;6:453–460. doi: 10.4161/pri.22511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Pan Y, Zhang D, Guo C, Shi Y, Wang J, Chen Y, Wang X, Liu J, Guo X, Chen Z, Qiao T, Fan D. Cellular prion protein promotes proliferation and G1/S transition of human gastric cancer cells SGC7901 and AGS. FASEB J. 2007;21:2247–2256. doi: 10.1096/fj.06-7799com. [DOI] [PubMed] [Google Scholar]
- Liang J, Wang W, Sorensen D, Medina S, Ilchenko S, Kiselar J, Surewicz WK, Booth SA, Kong Q. Cellular prion protein regulates its own α-cleavage through ADAM8 in skeletal muscle. J Biol Chem. 2012;287:16510–16520. doi: 10.1074/jbc.M112.360891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden R. The biological function of the prion protein: a cell surface scaffold of signaling modules. Front Mol Neurosci. 2017;10:77. doi: 10.3389/fnmol.2017.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsenmeier L, Altmeppen HC, Wetzel S, Mohammadi B, Saftig P, Glatzel M. Diverse functions of the prion protein — does proteolytic processing hold the key? Biochim Biophys Acta Mol Cell Res. 2017;1864:2128–2137. doi: 10.1016/j.bbamcr.2017.06.022. [DOI] [PubMed] [Google Scholar]
- Linsenmeier L, Mohammadi B, Shafiq M, Frontzek K, Bär J, Shrivastava AN, Damme M, Song F, Schwarz A, Da Vela S, Massignan T, Jung S, Correia A, Schmitz M, Puig B, Hornemann S, Zerr I, Tatzelt J, Biasini E, Saftig P, Schweizer M, Svergun D, Amin L, Mazzola F, Varani L, Thapa S, Gilch S, Schätzl H, Harris DA, Triller A, Mikhaylova M, Aguzzi A, Altmeppen HC, Glatzel M. Ligands binding to the prion protein induce its proteolytic release with therapeutic potential in neurodegenerative proteinopathies. Sci Adv. 2021;7:eabj1826. doi: 10.1126/sciadv.abj1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsenmeier L, Mohammadi B, Wetzel S, Puig B, Jackson WS, Hartmann A, Uchiyama K, Sakaguchi S, Endres K, Tatzelt J, Saftig P, Glatzel M, Altmeppen HC. Structural and mechanistic aspects influencing the ADAM10-mediated shedding of the prion protein. Mol Neurodegener. 2018;13:18. doi: 10.1186/s13024-018-0248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes MH, Santos TG, Rodrigues BR, Queiroz-Hazarbassanov N, Cunha IW, Wasilewska-Sampaio AP, Costa-Silva B, Marchi FA, Bleggi-Torres LF, Sanematsu PI, Suzuki SH, Oba-Shinjo SM, Marie SKN, Toulmin E, Hill AF, Martins VR. Disruption of prion protein-HOP engagement impairs glioblastoma growth and cognitive decline and improves overall survival. Oncogene. 2015;34:3305–3314. doi: 10.1038/onc.2014.261. [DOI] [PubMed] [Google Scholar]
- Luo G, Wang W, Wu Q, Lu Y, Su T, Gu N, Li K, Wang J, Du R, Zhao X, Li X, Fan R, Zhang H, Nie Y, Zhou X, Shi Y, Liang J, Wang X, Fan D. MGr1-Antigen/37 kDa laminin receptor precursor promotes cellular prion protein induced multi-drug-resistance of gastric cancer. Oncotarget. 2017;8:71630–71641. doi: 10.18632/oncotarget.17795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarava N, Chang JC-Y, Molesworth K, Baskakov IV. Posttranslational modifications define course of prion strain adaptation and disease phenotype. J Clin Invest. 2020;130:4382–4395. doi: 10.1172/JCI138677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Málaga-Trillo E, Solis GP, Schrock Y, Geiss C, Luncz L, Thomanetz V, Stuermer CAO. Regulation of embryonic cell adhesion by the prion protein. PLoS Biol. 2009;7:e55. doi: 10.1371/journal.pbio.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinverno M, Carta M, Epis R, Marcello E, Verpelli C, Cattabeni F, Sala C, Mulle C, Di Luca M, Gardoni F. Synaptic localization and activity of ADAM10 regulate excitatory synapses through N-cadherin cleavage. J Neurosci. 2010;30:16343–16355. doi: 10.1523/JNEUROSCI.1984-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangé A, Béranger F, Peoc’h K, Onodera T, Frobert Y, Lehmann S. Alpha- and beta- cleavages of the amino-terminus of the cellular prion protein. Biol Cell. 2004;96:125–132. doi: 10.1016/j.biolcel.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Mangé A, Milhavet O, Umlauf D, Harris D, Lehmann S. PrP-dependent cell adhesion in N2a neuroblastoma cells. FEBS Lett. 2002;514:159–162. doi: 10.1016/S0014-5793(02)02338-4. [DOI] [PubMed] [Google Scholar]
- Manni G, Lewis V, Senesi M, Spagnolli G, Fallarino F, Collins SJ, Mouillet-Richard S, Biasini E. The cellular prion protein beyond prion diseases. Swiss Med Wkly. 2020;150:w20222. doi: 10.4414/smw.2020.20222. [DOI] [PubMed] [Google Scholar]
- Manson JC, Clarke AR, McBride PA, McConnell I, Hope J. PrP gene dosage determines the timing but not the final intensity or distribution of lesions in scrapie pathology. Neurodegeneration. 1994;3:331–340. [PubMed] [Google Scholar]
- Mantuano E, Azmoon P, Banki MA, Lam MS, Sigurdson CJ, Gonias SL. A soluble derivative of PrPC activates cell-signaling and regulates cell physiology through LRP1 and the NMDA receptor. J Biol Chem. 2020;295:14178–14188. doi: 10.1074/jbc.RA120.013779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marella M, Lehmann S, Grassi J, Chabry J. Filipin prevents pathological prion protein accumulation by reducing endocytosis and inducing cellular PrP release. J Biol Chem. 2002;277:25457–25464. doi: 10.1074/jbc.M203248200. [DOI] [PubMed] [Google Scholar]
- Martellucci S, Santacroce C, Manganelli V, Santilli F, Piccoli L, Cassetta M, Misasi R, Sorice M, Mattei V. Isolation, propagation, and prion protein expression during neuronal differentiation of human dental pulp stem cells. J vis Exp. 2019 doi: 10.3791/59282. [DOI] [PubMed] [Google Scholar]
- Martellucci S, Santacroce C, Santilli F, Piccoli L, Delle Monache S, Angelucci A, Misasi R, Sorice M, Mattei V. Cellular and Molecular Mechanisms Mediated by recPrPC Involved in the Neuronal Differentiation Process of Mesenchymal Stem Cells. Int J Mol Sci. 2019;20:E345. doi: 10.3390/ijms20020345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei V, Manganelli V, Martellucci S, Capozzi A, Mantuano E, Longo A, Ferri A, Garofalo T, Sorice M, Misasi R. A multimolecular signaling complex including PrPC and LRP1 is strictly dependent on lipid rafts and is essential for the function of tissue plasminogen activator. J Neurochem. 2020;152:468–481. doi: 10.1111/jnc.14891. [DOI] [PubMed] [Google Scholar]
- Matthews AL, Szyroka J, Collier R, Noy PJ, Tomlinson MG. Scissor sisters: regulation of ADAM10 by the TspanC8 tetraspanins. Biochem Soc Trans. 2017;45:719–730. doi: 10.1042/BST20160290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays CE, Coomaraswamy J, Watts JC, Yang J, Ko KWS, Strome B, Mercer RCC, Wohlgemuth SL, Schmitt-Ulms G, Westaway D. Endoproteolytic processing of the mammalian prion glycoprotein family. FEBS J. 2014;281:862–876. doi: 10.1111/febs.12654. [DOI] [PubMed] [Google Scholar]
- McDonald AJ, Dibble JP, Evans EGB, Millhauser GL. A new paradigm for enzymatic control of α-cleavage and β-cleavage of the prion protein. J Biol Chem. 2014;289:803–813. doi: 10.1074/jbc.M113.502351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan NF, Brennan PM, McNeill A, Davies I, Fotheringham A, Rennison KA, Ritchie D, Brannan F, Head MW, Ironside JW, Williams A, Bell JE. Prion protein accumulation and neuroprotection in hypoxic brain damage. Am J Pathol. 2004;165:227–235. doi: 10.1016/S0002-9440(10)63291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HE, Mangé A, Nishida N, Créminon C, Casanova D, Lehmann S. Cleavage of the amino terminus of the prion protein by reactive oxygen species. J Biol Chem. 2001;276:2286–2291. doi: 10.1074/jbc.M007243200. [DOI] [PubMed] [Google Scholar]
- Megra B, Eugenin E, Roberts T, Morgello S, Berman JW. Protease resistant protein cellular isoform (PrP(c)) as a biomarker: clues into the pathogenesis of HAND. J Neuroimmune Pharmacol. 2013;8:1159–1166. doi: 10.1007/s11481-013-9458-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megra BW, Eugenin EA, Berman JW. The role of Shed PrPc in the neuropathogenesis of HIV infection. J Immunol. 2017;199:224–232. doi: 10.4049/jimmunol.1601041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabian M, Brethour D, Wang H, Xi Z, Rogaeva E, Schmitt-Ulms G. The prion protein controls polysialylation of neural cell adhesion molecule 1 during cellular morphogenesis. PLoS ONE. 2015;10:e0133741. doi: 10.1371/journal.pone.0133741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier P, Genoud N, Prinz M, Maissen M, Rülicke T, Zurbriggen A, Raeber AJ, Aguzzi A. Soluble dimeric prion protein binds PrP(Sc) in vivo and antagonizes prion disease. Cell. 2003;113:49–60. doi: 10.1016/S0092-8674(03)00201-0. [DOI] [PubMed] [Google Scholar]
- Meneghetti E, Gasperini L, Virgilio T, Moda F, Tagliavini F, Benetti F, Legname G. Prions strongly reduce NMDA receptor S-nitrosylation levels at pre-symptomatic and terminal stages of prion diseases. Mol Neurobiol. 2019;56:6035–6045. doi: 10.1007/s12035-019-1505-6. [DOI] [PubMed] [Google Scholar]
- Minikel EV, Kuhn E, Cocco AR, Vallabh SM, Hartigan CR, Reidenbach AG, Safar JG, Raymond GJ, McCarthy MD, O’Keefe R, Llorens F, Zerr I, Capellari S, Parchi P, Schreiber SL, Carr SA. Domain-specific quantification of prion protein in cerebrospinal fluid by targeted mass spectrometry. Mol Cell Proteomics. 2019;18:2388–2400. doi: 10.1074/mcp.RA119.001702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minikel EV, Zhao HT, Le J, O’Moore J, Pitstick R, Graffam S, Carlson GA, Kavanaugh MP, Kriz J, Kim JB, Ma J, Wille H, Aiken J, McKenzie D, Doh-Ura K, Beck M, O’Keefe R, Stathopoulos J, Caron T, Schreiber SL, Carroll JB, Kordasiewicz HB, Cabin DE, Vallabh SM. Prion protein lowering is a disease-modifying therapy across prion disease stages, strains and endpoints. Nucleic Acids Res. 2020;48:10615–10631. doi: 10.1093/nar/gkaa616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsios N, Saka M, Krupinski J, Pennucci R, Sanfeliu C, Miguel Turu M, Gaffney J, Kumar P, Kumar S, Sullivan M, Slevin M. Cellular prion protein is increased in the plasma and peri-infarcted brain tissue after acute stroke. J Neurosci Res. 2007;85:602–611. doi: 10.1002/jnr.21142. [DOI] [PubMed] [Google Scholar]
- Mohammadi B, Glatzel M, Altmeppen HC. Disordered structure and flexible roles: using the prion protein N1 fragment for neuroprotective and regenerative therapy. Neural Regen Res. 2021;16:1431–1432. doi: 10.4103/1673-5374.301008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi B, Linsenmeier L, Shafiq M, Puig B, Galliciotti G, Giudici C, Willem M, Eden T, Koch-Nolte F, Lin Y-H, Tatzelt J, Glatzel M, Altmeppen HC. Transgenic overexpression of the disordered prion protein N1 fragment in mice does not protect against neurodegenerative diseases due to impaired ER translocation. Mol Neurobiol. 2020;57:2812–2829. doi: 10.1007/s12035-020-01917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SJ, Kunkle R, Greenlee MHW, Nicholson E, Richt J, Hamir A, Waters WR, Greenlee J. Horizontal transmission of chronic wasting disease in reindeer. Emerg Infect Dis. 2016;22:2142–2145. doi: 10.3201/eid2212.160635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JA, Telling GC. Molecular mechanisms of chronic wasting disease prion propagation. Cold Spring Harb Perspect Med. 2018;8:a024448. doi: 10.1101/cshperspect.a024448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillet-Richard S, Ermonval M, Chebassier C, Laplanche JL, Lehmann S, Launay JM, Kellermann O. Signal transduction through prion protein. Science. 2000;289:1925–1928. doi: 10.1126/science.289.5486.1925. [DOI] [PubMed] [Google Scholar]
- Nicoll AJ, Panico S, Freir DB, Wright D, Terry C, Risse E, Herron CE, O’Malley T, Wadsworth JDF, Farrow MA, Walsh DM, Saibil HR, Collinge J. Amyloid-β nanotubes are associated with prion protein-dependent synaptotoxicity. Nat Commun. 2013;4:2416. doi: 10.1038/ncomms3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieznanski K, Choi J-K, Chen S, Surewicz K, Surewicz WK. Soluble prion protein inhibits amyloid-β (Aβ) fibrillization and toxicity. J Biol Chem. 2012;287:33104–33108. doi: 10.1074/jbc.C112.400614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishina KA, Deleault NR, Mahal SP, Baskakov I, Luhrs T, Riek R, Supattapone S. The stoichiometry of host PrPC glycoforms modulates the efficiency of PrPSc formation in vitro. Biochemistry. 2006;45:14129–14139. doi: 10.1021/bi061526k. [DOI] [PubMed] [Google Scholar]
- Nuvolone M, Hermann M, Sorce S, Russo G, Tiberi C, Schwarz P, Minikel E, Sanoudou D, Pelczar P, Aguzzi A. Strictly co-isogenic C57BL/6J-Prnp-/- mice: a rigorous resource for prion science. J Exp Med. 2016;213:313–327. doi: 10.1084/jem.20151610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesch B, Westaway D, Wälchli M, McKinley MP, Kent SB, Aebersold R, Barry RA, Tempst P, Teplow DB, Hood LE. A cellular gene encodes scrapie PrP 27–30 protein. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- Oliveira-Martins JB, Yusa S, Calella AM, Bridel C, Baumann F, Dametto P, Aguzzi A. Unexpected tolerance of alpha-cleavage of the prion protein to sequence variations. PLoS ONE. 2010;5:e9107. doi: 10.1371/journal.pone.0009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondrejcak T, Klyubin I, Corbett GT, Fraser G, Hong W, Mably AJ, Gardener M, Hammersley J, Perkinton MS, Billinton A, Walsh DM, Rowan MJ. Cellular prion protein mediates the disruption of hippocampal synaptic plasticity by soluble tau in vivo. J Neurosci. 2018;38:10595–10606. doi: 10.1523/JNEUROSCI.1700-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostalecki C, Lee J-H, Dindorf J, Collenburg L, Schierer S, Simon B, Schliep S, Kremmer E, Schuler G, Baur AS. Multiepitope tissue analysis reveals SPPL3-mediated ADAM10 activation as a key step in the transformation of melanocytes. Sci Signal. 2017;10:eaai8288. doi: 10.1126/scisignal.aai8288. [DOI] [PubMed] [Google Scholar]
- Ostapchenko VG, Beraldo FH, Guimarães ALS, Mishra S, Guzman M, Fan J, Martins VR, Prado VF, Prado MAM. Increased prion protein processing and expression of metabotropic glutamate receptor 1 in a mouse model of Alzheimer’s disease. J Neurochem. 2013;127:415–425. doi: 10.1111/jnc.12296. [DOI] [PubMed] [Google Scholar]
- Padro CJ, Shawler TM, Gormley MG, Sanders VM. Adrenergic regulation of IgE involves modulation of CD23 and ADAM10 expression on exosomes. J Immunol. 2013;191:5383–5397. doi: 10.4049/jimmunol.1301019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Zhao L, Liang J, Liu J, Shi Y, Liu N, Zhang G, Jin H, Gao J, Xie H, Wang J, Liu Z, Fan D. Cellular prion protein promotes invasion and metastasis of gastric cancer. FASEB J. 2006;20:1886–1888. doi: 10.1096/fj.06-6138fje. [DOI] [PubMed] [Google Scholar]
- Parizek P, Roeckl C, Weber J, Flechsig E, Aguzzi A, Raeber AJ. Similar turnover and shedding of the cellular prion protein in primary lymphoid and neuronal cells. J Biol Chem. 2001;276:44627–44632. doi: 10.1074/jbc.M107458200. [DOI] [PubMed] [Google Scholar]
- Parkyn CJ, Vermeulen EGM, Mootoosamy RC, Sunyach C, Jacobsen C, Oxvig C, Moestrup S, Liu Q, Bu G, Jen A, Morris RJ. LRP1 controls biosynthetic and endocytic trafficking of neuronal prion protein. J Cell Sci. 2008;121:773–783. doi: 10.1242/jcs.021816. [DOI] [PubMed] [Google Scholar]
- Peretz D, Williamson RA, Kaneko K, Vergara J, Leclerc E, Schmitt-Ulms G, Mehlhorn IR, Legname G, Wormald MR, Rudd PM, Dwek RA, Burton DR, Prusiner SB. Antibodies inhibit prion propagation and clear cell cultures of prion infectivity. Nature. 2001;412:739–743. doi: 10.1038/35089090. [DOI] [PubMed] [Google Scholar]
- Pérez-González R, Kim Y, Miller C, Pacheco-Quinto J, Eckman EA, Levy E. Extracellular vesicles: where the amyloid precursor protein carboxyl-terminal fragments accumulate and amyloid-β oligomerizes. FASEB J. 2020;34:12922–12931. doi: 10.1096/fj.202000823R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perini F, Vidal R, Ghetti B, Tagliavini F, Frangione B, Prelli F. PrP27-30 is a normal soluble prion protein fragment released by human platelets. Biochem Biophys Res Commun. 1996;223:572–577. doi: 10.1006/bbrc.1996.0936. [DOI] [PubMed] [Google Scholar]
- Persad A, Pham N, Moien-Afshari F, Gormley W, Yan S, Mannix R, Taghibiglou C. Plasma PrPC and ADAM-10 as novel biomarkers for traumatic brain injury and concussion: a pilot study. Brain Inj. 2021;35:734–741. doi: 10.1080/02699052.2021.1900602. [DOI] [PubMed] [Google Scholar]
- Petit CSV, Besnier L, Morel E, Rousset M, Thenet S. Roles of the cellular prion protein in the regulation of cell-cell junctions and barrier function. Tissue Barriers. 2013;1:e24377. doi: 10.4161/tisb.24377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit-Paitel A, Ménard B, Guyon A, Béringue V, Nahon J-L, Zsürger N, Chabry J. Prion protein is a key determinant of alcohol sensitivity through the modulation of N-methyl-D-aspartate receptor (NMDAR) activity. PLoS ONE. 2012;7:e34691. doi: 10.1371/journal.pone.0034691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham N, Akonasu H, Shishkin R, Taghibiglou C. Plasma soluble prion protein, a potential biomarker for sport-related concussions: a pilot study. PLoS ONE. 2015;10:e0117286. doi: 10.1371/journal.pone.0117286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham N, Sawyer TW, Wang Y, Jazii FR, Vair C, Taghibiglou C. Primary blast-induced traumatic brain injury in rats leads to increased prion protein in plasma: a potential biomarker for blast-induced traumatic brain injury. J Neurotrauma. 2015;32:58–65. doi: 10.1089/neu.2014.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietri M, Dakowski C, Hannaoui S, Alleaume-Butaux A, Hernandez-Rapp J, Ragagnin A, Mouillet-Richard S, Haik S, Bailly Y, Peyrin J-M, Launay J-M, Kellermann O, Schneider B. PDK1 decreases TACE-mediated α-secretase activity and promotes disease progression in prion and Alzheimer’s diseases. Nat Med. 2013;19:1124–1131. doi: 10.1038/nm.3302. [DOI] [PubMed] [Google Scholar]
- Pinnock EC, Jovanovic K, Pinto MG, Ferreira E, Dias BDC, Penny C, Knackmuss S, Reusch U, Little M, Schatzl HM, Weiss SFT. LRP/LR antibody mediated rescuing of amyloid-β-induced cytotoxicity is dependent on PrPc in Alzheimer’s disease. J Alzheimers Dis. 2016;49:645–657. doi: 10.3233/JAD-150482. [DOI] [PubMed] [Google Scholar]
- Price RW, Brew B, Sidtis J, Rosenblum M, Scheck AC, Cleary P. The brain in AIDS: central nervous system HIV-1 infection and AIDS dementia complex. Science. 1988;239:586–592. doi: 10.1126/science.3277272. [DOI] [PubMed] [Google Scholar]
- Priola SA, Caughey B, Race RE, Chesebro B. Heterologous PrP molecules interfere with accumulation of protease-resistant PrP in scrapie-infected murine neuroblastoma cells. J Virol. 1994;68:4873–4878. doi: 10.1128/jvi.68.8.4873-4878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priola SA, Lawson VA. Glycosylation influences cross-species formation of protease-resistant prion protein. EMBO J. 2001;20:6692–6699. doi: 10.1093/emboj/20.23.6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromidou K, Papastefanaki F, Sklaviadis T, Matsas R. Functional cross-talk between the cellular prion protein and the neural cell adhesion molecule is critical for neuronal differentiation of neural stem/precursor cells. Stem Cells. 2014;32:1674–1687. doi: 10.1002/stem.1663. [DOI] [PubMed] [Google Scholar]
- Provenzano L, Ryan Y, Hilton DA, Lyons-Rimmer J, Dave F, Maze EA, Adams CL, Rigby-Jones R, Ammoun S, Hanemann CO. Cellular prion protein (PrPC) in the development of Merlin-deficient tumours. Oncogene. 2017;36:6132–6142. doi: 10.1038/onc.2017.200. [DOI] [PubMed] [Google Scholar]
- Prox J, Bernreuther C, Altmeppen H, Grendel J, Glatzel M, D’Hooge R, Stroobants S, Ahmed T, Balschun D, Willem M, Lammich S, Isbrandt D, Schweizer M, Horré K, De Strooper B, Saftig P. Postnatal disruption of the disintegrin/metalloproteinase ADAM10 in brain causes epileptic seizures, learning deficits, altered spine morphology, and defective synaptic functions. J Neurosci. 2013;33:12915–12928. doi: 10.1523/JNEUROSCI.5910-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Prion encephalopathies of animals and humans. Dev Biol Stand. 1993;80:31–44. [PubMed] [Google Scholar]
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- Puig B, Altmeppen HC, Linsenmeier L, Chakroun K, Wegwitz F, Piontek UK, Tatzelt J, Bate C, Magnus T, Glatzel M. GPI-anchor signal sequence influences PrPC sorting, shedding and signalling, and impacts on different pathomechanistic aspects of prion disease in mice. PLoS Pathog. 2019;15:e1007520. doi: 10.1371/journal.ppat.1007520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig B, Altmeppen HC, Thurm D, Geissen M, Conrad C, Braulke T, Glatzel M. N-glycans and glycosylphosphatidylinositol-anchor act on polarized sorting of mouse PrP(C) in Madin-Darby canine kidney cells. PLoS ONE. 2011;6:e24624. doi: 10.1371/journal.pone.0024624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig B, Altmeppen HC, Ulbrich S, Linsenmeier L, Krasemann S, Chakroun K, Acevedo-Morantes CY, Wille H, Tatzelt J, Glatzel M. Secretory pathway retention of mutant prion protein induces p38-MAPK activation and lethal disease in mice. Sci Rep. 2016;6:24970. doi: 10.1038/srep24970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig B, Yang D, Brenna S, Altmeppen HC, Magnus T. Show me your friends and I tell you who you are: The many facets of prion protein in stroke. Cells. 2020;9:E1609. doi: 10.3390/cells9071609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race B, Meade-White K, Race R, Baumann F, Aguzzi A, Chesebro B. Prion protein on astrocytes or in extracellular fluid impedes neurodegeneration induced by truncated prion protein. Exp Neurol. 2009;217:347–352. doi: 10.1016/j.expneurol.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel A, Race B, Klingeborn M, Striebel J, Chesebro B. Unusual cerebral vascular prion protein amyloid distribution in scrapie-infected transgenic mice expressing anchorless prion protein. Acta Neuropathol Commun. 2013;1:25. doi: 10.1186/2051-5960-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond GJ, Zhao HT, Race B, Raymond LD, Williams K, Swayze EE, Graffam S, Le J, Caron T, Stathopoulos J, O’Keefe R, Lubke LL, Reidenbach AG, Kraus A, Schreiber SL, Mazur C, Cabin DE, Carroll JB, Minikel EV, Kordasiewicz H, Caughey B, Vallabh SM. Antisense oligonucleotides extend survival of prion-infected mice. JCI Insight. 2019;5:131175. doi: 10.1172/jci.insight.131175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resenberger UK, Harmeier A, Woerner AC, Goodman JL, Müller V, Krishnan R, Vabulas RM, Kretzschmar HA, Lindquist S, Hartl FU, Multhaup G, Winklhofer KF, Tatzelt J. The cellular prion protein mediates neurotoxic signalling of β-sheet-rich conformers independent of prion replication. EMBO J. 2011;30:2057–2070. doi: 10.1038/emboj.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie AJ, Crawford DM, Ferguson DJP, Burthem J, Roberts DJ. Normal prion protein is expressed on exosomes isolated from human plasma. Br J Haematol. 2013;163:678–680. doi: 10.1111/bjh.12543. [DOI] [PubMed] [Google Scholar]
- Roberts TK, Eugenin EA, Morgello S, Clements JE, Zink MC, Berman JW. PrPC, the cellular isoform of the human prion protein, is a novel biomarker of HIV-associated neurocognitive impairment and mediates neuroinflammation. Am J Pathol. 2010;177:1848–1860. doi: 10.2353/ajpath.2010.091006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M, Yehiely F, Scott M, Prusiner SB. Conversion of truncated and elongated prion proteins into the scrapie isoform in cultured cells. Proc Natl Acad Sci U S A. 1993;90:3182–3186. doi: 10.1073/pnas.90.8.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roucou X, Giannopoulos PN, Zhang Y, Jodoin J, Goodyer CG, LeBlanc A. Cellular prion protein inhibits proapoptotic Bax conformational change in human neurons and in breast carcinoma MCF-7 cells. Cell Death Differ. 2005;12:783–795. doi: 10.1038/sj.cdd.4401629. [DOI] [PubMed] [Google Scholar]
- Rubenstein R, Chang B, Grinkina N, Drummond E, Davies P, Ruditzky M, Sharma D, Wang K, Wisniewski T. Tau phosphorylation induced by severe closed head traumatic brain injury is linked to the cellular prion protein. Acta Neuropathol Commun. 2017;5:30. doi: 10.1186/s40478-017-0435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushworth JV, Griffiths HH, Watt NT, Hooper NM. Prion protein-mediated toxicity of amyloid-β oligomers requires lipid rafts and the transmembrane LRP1. J Biol Chem. 2013;288:8935–8951. doi: 10.1074/jbc.M112.400358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftig P, Lichtenthaler SF. The alpha secretase ADAM10: A metalloprotease with multiple functions in the brain. Prog Neurobiol. 2015;135:1–20. doi: 10.1016/j.pneurobio.2015.10.003. [DOI] [PubMed] [Google Scholar]
- Saint-Pol J, Eschenbrenner E, Dornier E, Boucheix C, Charrin S, Rubinstein E. Regulation of the trafficking and the function of the metalloprotease ADAM10 by tetraspanins. Biochem Soc Trans. 2017;45:937–944. doi: 10.1042/BST20160296. [DOI] [PubMed] [Google Scholar]
- Salazar SV, Gallardo C, Kaufman AC, Herber CS, Haas LT, Robinson S, Manson JC, Lee MK, Strittmatter SM. Conditional deletion of Prnp rescues behavioral and synaptic deficits after disease onset in transgenic Alzheimer’s disease. J Neurosci. 2017;37:9207–9221. doi: 10.1523/JNEUROSCI.0722-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen Ø, Tatzelt J, Tranulis MA. The prion protein in neuroimmune crosstalk. Neurochem Int. 2019;130:104335. doi: 10.1016/j.neuint.2018.11.010. [DOI] [PubMed] [Google Scholar]
- Sandberg MK, Al-Doujaily H, Sharps B, Clarke AR, Collinge J. Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature. 2011;470:540–542. doi: 10.1038/nature09768. [DOI] [PubMed] [Google Scholar]
- Santuccione A, Sytnyk V, Leshchyns’ka I, Schachner M. Prion protein recruits its neuronal receptor NCAM to lipid rafts to activate p59fyn and to enhance neurite outgrowth. J Cell Biol. 2005;169:341–354. doi: 10.1083/jcb.200409127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt-Ulms G, Legname G, Baldwin MA, Ball HL, Bradon N, Bosque PJ, Crossin KL, Edelman GM, DeArmond SJ, Cohen FE, Prusiner SB. Binding of neural cell adhesion molecules (N-CAMs) to the cellular prion protein. J Mol Biol. 2001;314:1209–1225. doi: 10.1006/jmbi.2000.5183. [DOI] [PubMed] [Google Scholar]
- Schwarze-Eicker K, Keyvani K, Görtz N, Westaway D, Sachser N, Paulus W. Prion protein (PrPc) promotes beta-amyloid plaque formation. Neurobiol Aging. 2005;26:1177–1182. doi: 10.1016/j.neurobiolaging.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Scott-McKean JJ, Surewicz K, Choi J-K, Ruffin VA, Salameh AI, Nieznanski K, Costa ACS, Surewicz WK. Soluble prion protein and its N-terminal fragment prevent impairment of synaptic plasticity by Aβ oligomers: Implications for novel therapeutic strategy in Alzheimer’s disease. Neurobiol Dis. 2016;91:124–131. doi: 10.1016/j.nbd.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipold L, Altmeppen H, Koudelka T, Tholey A, Kasparek P, Sedlacek R, Schweizer M, Bär J, Mikhaylova M, Glatzel M, Saftig P. In vivo regulation of the A disintegrin and metalloproteinase 10 (ADAM10) by the tetraspanin 15. Cell Mol Life Sci. 2018;75:3251–3267. doi: 10.1007/s00018-018-2791-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar S, Zhang Y, Miranzadeh Mahabadi H, Parvizi A, Taghibiglou C. Low-field magnetic stimulation restores cognitive and motor functions in the mouse model of repeated traumatic brain injury: role of cellular prion protein. J Neurotrauma. 2019;36:3103–3114. doi: 10.1089/neu.2018.5918. [DOI] [PubMed] [Google Scholar]
- Sevillano AM, Aguilar-Calvo P, Kurt TD, Lawrence JA, Soldau K, Nam TH, Schumann T, Pizzo DP, Nyström S, Choudhury B, Altmeppen H, Esko JD, Glatzel M, Nilsson KPR, Sigurdson CJ. Prion protein glycans reduce intracerebral fibril formation and spongiosis in prion disease. J Clin Invest. 2020;130:1350–1362. doi: 10.1172/JCI131564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyng SL, Huber MT, Harris DA. A prion protein cycles between the cell surface and an endocytic compartment in cultured neuroblastoma cells. J Biol Chem. 1993;268:15922–15928. doi: 10.1016/S0021-9258(18)82340-7. [DOI] [PubMed] [Google Scholar]
- Shyu W-C, Lin S-Z, Chiang M-F, Ding D-C, Li K-W, Chen S-F, Yang H-I, Li H. Overexpression of PrPC by adenovirus-mediated gene targeting reduces ischemic injury in a stroke rat model. J Neurosci. 2005;25:8967–8977. doi: 10.1523/JNEUROSCI.1115-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoneau S, Haïk S, Leucht C, Dormont D, Deslys J-P, Weiss S, Lasmézas C. Different isoforms of the non-integrin laminin receptor are present in mouse brain and bind PrP. Biol Chem. 2003;384:243–246. doi: 10.1515/BC.2003.027. [DOI] [PubMed] [Google Scholar]
- Slapšak U, Salzano G, Amin L, Abskharon RNN, Ilc G, Zupančič B, Biljan I, Plavec J, Giachin G, Legname G. The N terminus of the prion protein mediates functional interactions with the neuronal cell adhesion molecule (NCAM) fibronectin domain. J Biol Chem. 2016;291:21857–21868. doi: 10.1074/jbc.M116.743435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TM, Tharakan A, Martin RK. Targeting ADAM10 in cancer and autoimmunity. Front Immunol. 2020;11:499. doi: 10.3389/fimmu.2020.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis GP, Radon Y, Sempou E, Jechow K, Stuermer CAO, Málaga-Trillo E. Conserved roles of the prion protein domains on subcellular localization and cell-cell adhesion. PLoS ONE. 2013;8:e70327. doi: 10.1371/journal.pone.0070327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich A, Frigg R, Kilic E, Kilic U, Oesch B, Raeber A, Bassetti CL, Hermann DM. Aggravation of ischemic brain injury by prion protein deficiency: role of ERK-1/-2 and STAT-1. Neurobiol Dis. 2005;20:442–449. doi: 10.1016/j.nbd.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Stahl N, Borchelt DR, Hsiao K, Prusiner SB. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51:229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- Stahl N, Borchelt DR, Prusiner SB. Differential release of cellular and scrapie prion proteins from cellular membranes by phosphatidylinositol-specific phospholipase C. Biochemistry. 1990;29:5405–5412. doi: 10.1021/bi00474a028. [DOI] [PubMed] [Google Scholar]
- Steele AD, Emsley JG, Ozdinler PH, Lindquist S, Macklis JD. Prion protein (PrPc) positively regulates neural precursor proliferation during developmental and adult mammalian neurogenesis. Proc Natl Acad Sci U S A. 2006;103:3416–3421. doi: 10.1073/pnas.0511290103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck A, Keller S, Riedle S, Sanderson MP, Runz S, Le Naour F, Gutwein P, Ludwig A, Rubinstein E, Altevogt P. A role for exosomes in the constitutive and stimulus-induced ectodomain cleavage of L1 and CD44. Biochem J. 2006;393:609–618. doi: 10.1042/BJ20051013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöhr J, Watts JC, Legname G, Oehler A, Lemus A, Nguyen H-OB, Sussman J, Wille H, DeArmond SJ, Prusiner SB, Giles K. Spontaneous generation of anchorless prions in transgenic mice. Proc Natl Acad Sci U S A. 2011;108:21223–21228. doi: 10.1073/pnas.1117827108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhao M, Zhang J, Liu A, Ji W, Li Y, Yang X, Wu Z. MiR-144 promotes β-amyloid accumulation-induced cognitive impairments by targeting ADAM10 following traumatic brain injury. Oncotarget. 2017;8:59181–59203. doi: 10.18632/oncotarget.19469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyach C, Cisse MA, da Costa CA, Vincent B, Checler F. The C-terminal products of cellular prion protein processing, C1 and C2, exert distinct influence on p53-dependent staurosporine-induced caspase-3 activation. J Biol Chem. 2007;282:1956–1963. doi: 10.1074/jbc.M609663200. [DOI] [PubMed] [Google Scholar]
- Tagliavini F, Prelli F, Porro M, Salmona M, Bugiani O, Frangione B. A soluble form of prion protein in human cerebrospinal fluid: implications for prion-related encephalopathies. Biochem Biophys Res Commun. 1992;184:1398–1404. doi: 10.1016/S0006-291X(05)80038-5. [DOI] [PubMed] [Google Scholar]
- Takahashi RH, Yokotsuka M, Tobiume M, Sato Y, Hasegawa H, Nagao T, Gouras GK. Accumulation of cellular prion protein within β-amyloid oligomer plaques in aged human brains. Brain Pathol. 2021;31:e12941. doi: 10.1111/bpa.12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboulos A, Scott M, Semenov A, Avrahami D, Laszlo L, Prusiner SB, Avraham D. Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J Cell Biol. 1995;129:121–132. doi: 10.1083/jcb.129.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR, Hooper NM. The low-density lipoprotein receptor-related protein 1 (LRP1) mediates the endocytosis of the cellular prion protein. Biochem J. 2007;402:17–23. doi: 10.1042/BJ20061736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR, Parkin ET, Cocklin SL, Ault JR, Ashcroft AE, Turner AJ, Hooper NM. Role of ADAMs in the ectodomain shedding and conformational conversion of the prion protein. J Biol Chem. 2009;284:22590–22600. doi: 10.1074/jbc.M109.032599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant JM, Li M, Henderson DM, Tyer ML, Denkers ND, Haley NJ, Mathiason CK, Hoover EA. Shedding and stability of CWD prion seeding activity in cervid feces. PLoS ONE. 2020;15:e0227094. doi: 10.1371/journal.pone.0227094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellung S, Corsaro A, Bosio AG, Zambito M, Barbieri F, Mazzanti M, Florio T. Emerging role of cellular prion protein in the maintenance and expansion of glioma stem cells. Cells. 2019;8:E1458. doi: 10.3390/cells8111458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevitt CR, Hosszu LLP, Batchelor M, Panico S, Terry C, Nicoll AJ, Risse E, Taylor WA, Sandberg MK, Al-Doujaily H, Linehan JM, Saibil HR, Scott DJ, Collinge J, Waltho JP, Clarke AR. N-terminal domain of prion protein directs its oligomeric association. J Biol Chem. 2014;289:25497–25508. doi: 10.1074/jbc.M114.566588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh JA, Unterberger U, Saá P, Massignan T, Fluharty BR, Bowman FP, Miller MB, Supattapone S, Biasini E, Harris DA. The N-terminal, polybasic region of PrP(C) dictates the efficiency of prion propagation by binding to PrP(Sc) J Neurosci. 2012;32:8817–8830. doi: 10.1523/JNEUROSCI.1103-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh JA, Westergard L, Unterberger U, Biasini E, Harris DA. The N-terminal, polybasic region is critical for prion protein neuroprotective activity. PLoS ONE. 2011;6:e25675. doi: 10.1371/journal.pone.0025675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turu M, Slevin M, Ethirajan P, Luque A, Elasbali A, Font A, Gaffney J, Cairols M, Kumar P, Kumar S, Krupinski J. The normal cellular prion protein and its possible role in angiogenesis. Front Biosci. 2008;13:6491–6500. doi: 10.2741/3169. [DOI] [PubMed] [Google Scholar]
- Uchiyama K, Tomita M, Yano M, Chida J, Hara H, Das NR, Nykjaer A, Sakaguchi S. Prions amplify through degradation of the VPS10P sorting receptor sortilin. PLoS Pathog. 2017;13:e1006470. doi: 10.1371/journal.ppat.1006470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JW, Kaufman AC, Kostylev M, Heiss JK, Stagi M, Takahashi H, Kerrisk ME, Vortmeyer A, Wisniewski T, Koleske AJ, Gunther EC, Nygaard HB, Strittmatter SM. Metabotropic glutamate receptor 5 is a coreceptor for Alzheimer aβ oligomer bound to cellular prion protein. Neuron. 2013;79:887–902. doi: 10.1016/j.neuron.2013.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JW, Nygaard HB, Heiss JK, Kostylev MA, Stagi M, Vortmeyer A, Wisniewski T, Gunther EC, Strittmatter SM. Alzheimer amyloid-β oligomer bound to postsynaptic prion protein activates Fyn to impair neurons. Nat Neurosci. 2012;15:1227–1235. doi: 10.1038/nn.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabh SM, Nobuhara CK, Llorens F, Zerr I, Parchi P, Capellari S, Kuhn E, Klickstein J, Safar JG, Nery FC, Swoboda KJ, Geschwind MD, Zetterberg H, Arnold SE, Minikel EV, Schreiber SL. Prion protein quantification in human cerebrospinal fluid as a tool for prion disease drug development. Proc Natl Acad Sci U S A. 2019;116:7793–7798. doi: 10.1073/pnas.1901947116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vania L, Morris G, Otgaar TC, Bignoux MJ, Bernert M, Burns J, Gabathuse A, Singh E, Ferreira E, Weiss SFT. Patented therapeutic approaches targeting LRP/LR for cancer treatment. Expert Opin Ther Pat. 2019;29:987–1009. doi: 10.1080/13543776.2019.1693543. [DOI] [PubMed] [Google Scholar]
- Vella LJ, Greenwood DLV, Cappai R, Scheerlinck J-PY, Hill AF. Enrichment of prion protein in exosomes derived from ovine cerebral spinal fluid. Vet Immunol Immunopathol. 2008;124:385–393. doi: 10.1016/j.vetimm.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Vincent B, Paitel E, Saftig P, Frobert Y, Hartmann D, De Strooper B, Grassi J, Lopez-Perez E, Checler F. The disintegrins ADAM10 and TACE contribute to the constitutive and phorbol ester-regulated normal cleavage of the cellular prion protein. J Biol Chem. 2001;276:37743–37746. doi: 10.1074/jbc.M105677200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yu S, Huang D, Cui M, Hu H, Zhang L, Wang W, Parameswaran N, Jackson M, Osborne B, Bedogni B, Li C, Sy M-S, Xin W, Zhou L. Cellular prion protein mediates pancreatic cancer cell survival and invasion through association with and enhanced signaling of Notch1. Am J Pathol. 2016;186:2945–2956. doi: 10.1016/j.ajpath.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren KM, Reeves TM, Phillips LL. MT5-MMP, ADAM-10, and N-cadherin act in concert to facilitate synapse reorganization after traumatic brain injury. J Neurotrauma. 2012;29:1922–1940. doi: 10.1089/neu.2012.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JC, Bourkas MEC, Arshad H. The function of the cellular prion protein in health and disease. Acta Neuropathol. 2018;135:159–178. doi: 10.1007/s00401-017-1790-y. [DOI] [PubMed] [Google Scholar]
- Weise J, Crome O, Sandau R, Schulz-Schaeffer W, Bähr M, Zerr I. Upregulation of cellular prion protein (PrPc) after focal cerebral ischemia and influence of lesion severity. Neurosci Lett. 2004;372:146–150. doi: 10.1016/j.neulet.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Weise J, Sandau R, Schwarting S, Crome O, Wrede A, Schulz-Schaeffer W, Zerr I, Bähr M. Deletion of cellular prion protein results in reduced Akt activation, enhanced postischemic caspase-3 activation, and exacerbation of ischemic brain injury. Stroke. 2006;37:1296–1300. doi: 10.1161/01.STR.0000217262.03192.d4. [DOI] [PubMed] [Google Scholar]
- Westaway D, Prusiner SB. Conservation of the cellular gene encoding the scrapie prion protein. Nucleic Acids Res. 1986;14:2035–2044. doi: 10.1093/nar/14.5.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergard L, Turnbaugh JA, Harris DA. A naturally occurring C-terminal fragment of the prion protein (PrP) delays disease and acts as a dominant-negative inhibitor of PrPSc formation. J Biol Chem. 2011;286:44234–44242. doi: 10.1074/jbc.M111.286195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel S, Seipold L, Saftig P. The metalloproteinase ADAM10: A useful therapeutic target? Biochim Biophys Acta Mol Cell Res. 2017;1864:2071–2081. doi: 10.1016/j.bbamcr.2017.06.005. [DOI] [PubMed] [Google Scholar]
- Wiegmans AP, Saunus JM, Ham S, Lobb R, Kutasovic JR, Dalley AJ, Miranda M, Atkinson C, Foliaki ST, Ferguson K, Niland C, Johnstone CN, Lewis V, Collins SJ, Lakhani SR, Al-Ejeh F, Möller A. Secreted cellular prion protein binds doxorubicin and correlates with anthracycline resistance in breast cancer. JCI Insight. 2019;5:124092. doi: 10.1172/jci.insight.124092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wik L, Klingeborn M, Willander H, Linne T. Separate mechanisms act concurrently to shed and release the prion protein from the cell. Prion. 2012;6:498–509. doi: 10.4161/pri.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf M-A, Senatore A, Aguzzi A. The biological function of the cellular prion protein: an update. BMC Biol. 2017;15:34. doi: 10.1186/s12915-017-0375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Yuan J, Haïk S, Cali I, Zhan Y, Moudjou M, Li B, Laplanche J-L, Laude H, Langeveld J, Gambetti P, Kitamoto T, Kong Q, Brandel J-P, Cobb BA, Petersen RB, Zou W-Q. Glycoform-selective prion formation in sporadic and familial forms of prion disease. PLoS ONE. 2013;8:e58786. doi: 10.1371/journal.pone.0058786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap YH-Y, Say Y-H. Resistance against tumour necrosis factor α apoptosis by the cellular prion protein is cell-specific for oral, colon and kidney cancer cell lines. Cell Biol Int. 2012;36:273–277. doi: 10.1042/CBI20110088. [DOI] [PubMed] [Google Scholar]
- Yoon S, Go G, Yoon Y, Lim J, Lee G, Lee S. Harnessing the physiological functions of cellular prion protein in the kidneys: applications for treating renal diseases. Biomolecules. 2021;11:784. doi: 10.3390/biom11060784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You H, Tsutsui S, Hameed S, Kannanayakal TJ, Chen L, Xia P, Engbers JDT, Lipton SA, Stys PK, Zamponi GW. Aβ neurotoxicity depends on interactions between copper ions, prion protein, and N-methyl-D-aspartate receptors. Proc Natl Acad Sci U S A. 2012;109:1737–1742. doi: 10.1073/pnas.1110789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Zhan Y-A, Abskharon R, Xiao X, Martinez MC, Zhou X, Kneale G, Mikol J, Lehmann S, Surewicz WK, Castilla J, Steyaert J, Zhang S, Kong Q, Petersen RB, Wohlkonig A, Zou W-Q. Recombinant human prion protein inhibits prion propagation in vitro. Sci Rep. 2013;3:2911. doi: 10.1038/srep02911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanusso G, Fiorini M, Ferrari S, Meade-White K, Barbieri I, Brocchi E, Ghetti B, Monaco S. Gerstmann-Sträussler-Scheinker disease and “anchorless prion protein” mice share prion conformational properties diverging from sporadic Creutzfeldt-Jakob disease. J Biol Chem. 2014;289:4870–4881. doi: 10.1074/jbc.M113.531335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Cowden D, Zhang F, Yuan J, Siedlak S, Abouelsaad M, Zeng L, Zhou X, O’Toole J, Das AS, Kofskey D, Warren M, Bian Z, Cui Y, Tan T, Kresak A, Wyza RE, Petersen RB, Wang G-X, Kong Q, Wang X, Sedor J, Zhu X, Zhu H, Zou W-Q. Prion protein protects against renal ischemia/reperfusion injury. PLoS ONE. 2015;10:e0136923. doi: 10.1371/journal.pone.0136923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CC, Steele AD, Lindquist S, Lodish HF. Prion protein is expressed on long-term repopulating hematopoietic stem cells and is important for their self-renewal. Proc Natl Acad Sci U S A. 2006;103:2184–2189. doi: 10.1073/pnas.0510577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar O, Lavy R, Zi X, Nelson TJ, Hongpaisan J, Pick CG, Alkon DL. PKC activator therapeutic for mild traumatic brain injury in mice. Neurobiol Dis. 2011;41:329–337. doi: 10.1016/j.nbd.2010.10.001. [DOI] [PubMed] [Google Scholar]