Abstract
The fibroblast growth factor (FGF) family is composed of 18 secreted signaling proteins consisting of canonical FGFs and endocrine FGFs that activate four receptor tyrosine kinases (FGFRs 1–4) and four intracellular proteins (intracellular FGFs or iFGFs) that primarily function to regulate the activity of voltage-gated sodium channels and other molecules. The canonical FGFs, endocrine FGFs, and iFGFs have been reviewed extensively by us and others. In this review, we briefly summarize past reviews and then focus on new developments in the FGF field since our last review in 2015. Some of the highlights in the past 6 years include the use of optogenetic tools, viral vectors, and inducible transgenes to experimentally modulate FGF signaling, the clinical use of small molecule FGFR inhibitors, an expanded understanding of endocrine FGF signaling, functions for FGF signaling in stem cell pluripotency and differentiation, roles for FGF signaling in tissue homeostasis and regeneration, a continuing elaboration of mechanisms of FGF signaling in development, and an expanding appreciation of roles for FGF signaling in neuropsychiatric diseases.
Keywords: fibroblast growth factors, organogenesis, receptor tyrosine kinase, regeneration, tyrosine kinase inhibitors
This article is categorized under: Cardiovascular Diseases > Molecular and Cellular Physiology, Neurological Diseases > Molecular and Cellular Physiology, Congenital Diseases > Stem Cells and Development, Cancer > Stem Cells and Development
1 |. INTRODUCTION
A proteinaceous “fibroblast growth factor” (FGF) activity was first identified nearly 50 years ago by Armelin (1973) and Gospodarowicz (1975). The purification of different FGF proteins, the identification of FGF receptors (FGFRs), and the cloning and identification of the whole mammalian FGF–FGFR family has been reviewed by us and others (Gong, 2014; Itoh et al., 2015; X. Li, Wang, Xiao, et al., 2016; Maddaluno et al., 2017; Ornitz & Itoh, 2015; Pablo & Pitt, 2016; Turner & Grose, 2010; Xie, Su, et al., 2020). Four mammalian FGFRs have been identified and key alternative splicing events result in seven functionally distinct receptors, FGFRs 1b, 1c, 2b, 2c, 3b, 3c, and 4, with distinct ligand binding properties (Figure 1a,b). Tissue-specific splicing of b and c exons and tissue-specific expression of FGF ligands allows for specific epithelial to mesenchymal and mesenchymal to epithelial signaling in many organs. The alternative use of b versus c exons is regulated by the epithelial splicing-regulatory protein 1 (ESRP1), which is required for expression of FGFR2b (Bowler & Oltean, 2019; Gong, 2014; Ishiwata, 2018; Ornitz & Itoh, 2015). Additionally, there are several less common splicing events, such as exclusion of the linker region between Ig-like domain I and II, which contains a cluster of acidic residues, or complete elimination of Ig-like domain I and the I–II linker region (Bowler & Oltean, 2019). These splicing events are thought to affect the autoregulatory properties of the FGF receptor (Gong, 2014).
FIGURE 1.
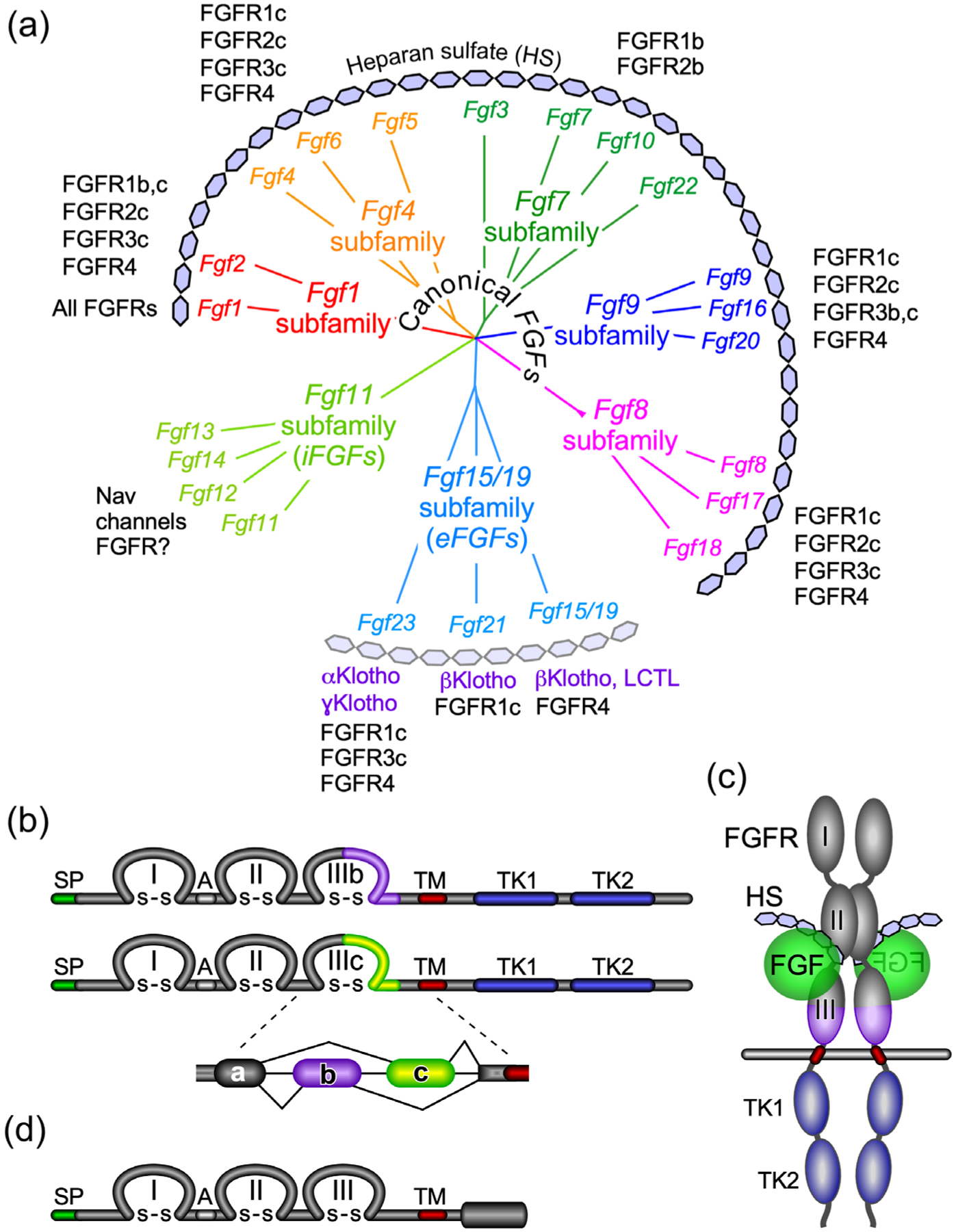
The FGF and FGFR family. (a) Phylogenetic analysis shows the organization of the 22 Fgf genes into seven subfamilies. Branch lengths represent the evolutionary distance between each gene. Canonical FGFs include the Fgf1, Fgf4, Fgf7, Fgf8, and Fgf9 subfamilies that interact with the cofactor heparin/heparan sulfate (HS) for binding and activation of different splice variants (b and c) of the four FGFRs. The Fgf15/19 subfamily members encode endocrine FGFs (eFGFs), which use αKlotho (KL) or βKlotho (KLB) as the primary cofactors for binding and activation of FGFRs. FGF15/19 can also interact with Lactase-like Klotho (LCTL) also called Klotho-LPH related protein (KLPH) or γKlotho. eFGFs have lower affinity for HS but still required HS for optimal receptor binding. The Fgf11 subfamily genes encode intracellular FGFs (iFGFs), which are thought to be non-signaling proteins serving as regulators of voltage-gated sodium (Nav) channels and other molecules. Recent data open the possibility that iFGFs can activate FGFRs under certain conditions. (b) Diagram showing FGFR domain structures. The FGFR extracellular domain includes three immunoglobulin-like domains (I, II, and III) linked with disulfide (S–S) bonds and an acidic region (A), a transmembrane domain (TM), and two intracellular tyrosine kinase domains (TK1 and TK2). SP indicates a cleavable secreted signal sequence. Of the four Fgfr genes, Fgfr1–Fgfr3 generate two major splice variants in immunoglobulin-like domain III, referred to as IIIb and IIIc, which are essential determinants of ligand-binding specificity. (c) Schematic showing the relative orientation of an active signaling complex with a canonical FGF ligand, HS, and an FGFR forming a 2:2:2 dimer. (d) Diagram of FGFRL1/FGFR5 protein structure. FGFRL1 has a similar extracellular domain structural to other FGFRs with three extracellular immunoglobulin-like domains (I, II, and III) and a transmembrane domain (TM). Unlike other FGFRs, FGFRL1 has a short intracellular tail with no tyrosine kinase domain. SP indicates a cleavable secreted signal sequence
Sequence homology and biological activity classify the 22 members of the FGF family into three functional groups and seven subfamilies (Figure 1a). The biochemical characteristics of the FGF subfamilies, their specificity for different FGFRs and other molecules, and their co-factor dependence are diagramed in Figure 1a and have been reviewed extensively (Brewer et al., 2016; Edmonston & Wolf, 2020; Gong, 2014; Ho & Bergwitz, 2021; Itoh et al., 2015; Kliewer & Mangelsdorf, 2019; X. Li, Wang, Xiao, et al., 2016; Ornitz & Itoh, 2015; Pablo & Pitt, 2016; Takashi & Fukumoto, 2020b; Xie, Su, et al., 2020).
The canonical or locally acting secreted FGFs include the FGF1, 4, 7, 8, and 9 subfamilies. These FGFs use heparan sulfate (HS) as a co-factor for binding to FGFRs. The endocrine FGFs are in the FGF15/19 subfamily, they have a reduced affinity for HS, and they require either αKlotho (KL) or βKlotho (KLB) as a cofactor for receptor binding (Kuro-o, 2019). The intracellular FGFs (iFGFs) are in the FGF11 subfamily and interact with and regulate voltage-gated sodium channels and other proteins. New evidence suggests that under some conditions iFGFs may also be able to activate FGFRs (H. Lin et al., 2019; Sochacka et al., 2020).
Here, we focus on several areas of FGF research that have seen a significant advance since 2015 and several emerging areas of FGF research and emerging clinical applications of FGF research in genetic and metabolic disease, wound healing and tissue repair, cancer, and neurological diseases.
2 |. REGULATION OF FGFR SIGNAL TRANSDUCTION BY EXTRACELLULAR AND INTEGRAL MEMBRANE FACTORS
Canonical FGFs bind to the extracellular domain of FGFRs (Figure 1c) leading to conformational changes in receptor dimers that activate the receptor’s intracellular tyrosine kinase domain (ICD). The specificity of binding is determined by conserved and unique structures of the ligands and their interaction with alternatively spliced variants of FGFRs. The specificity and affinity of FGFRs for FGF ligands are further enhanced by heparan sulfate polysaccharides which directly interact with both ligands and receptors and by other integral membrane, cell surface, and extracellular proteins and other extracellular molecules. Intracellular signaling requires transphosphorylation of FGFR tyrosine kinase domains, phosphorylation/dephosphorylation of the FGFR by other kinases/phosphatases, and regulated activation/inactivation of adaptor proteins that interact with and are phosphorylated by the FGFR tyrosine kinase (Ornitz & Itoh, 2015; Turner & Grose, 2010; Xie, Su, et al., 2020).
2.1 |. Regulation of FGF1 and FGF2 secretion
FGF1 and FGF2 are potent mitogenic and angiogenic factors but they lack secretory signal peptides and are secreted using noncanonical mechanisms. Despite the abundance of extracellular FGF1 and FGF2, most adult tissues are not actively growing, suggesting that the reason for noncanonical secretion may involve sequestration of these FGFs in a specialized and protected extracellular environment that allows for rapid mobilization and activation in response to stress or injury. Experimentally, if a secretory signal peptide is added to the N-terminus of FGF1 or FGF2 to circumvent noncanonical secretion, they are transformed from stress and injury response proteins to angiogenic and oncogenic proteins (Forough et al., 1993; Rogelj et al., 1988).
FGF2 is released from cells through an unconventional mechanism involving direct translocation across the plasma membrane as a fully folded protein (Figure 2a). The FGF2 secretory process involves localization to the inner leaflet of the plasma membrane through interaction with the catalytic α1 subunit of the Na,K-ATPase (ATP1A1) and with phosphoinositide PI(4,5)P2 (Legrand et al., 2020; Steringer et al., 2017; Zacherl et al., 2015). The cardenolides, ouabain and reevesioside, bind ATP1A1 and increase the secretion of FGF2 (G. H. Zhao, Qiu, et al., 2020). Two surface cysteines in FGF2 form disulfide bridges that drive oligomerization and PI(4,5)P2-mediated membrane insertion of the tethered protein on the inner leaflet of the plasma membrane (Muller et al., 2015). FGF2 oligomers forms pores in the plasma membrane which are facilitated by phosphorylation of FGF2 by TEC protein tyrosine kinase. Released FGF2 is then trapped by cell surface membrane-proximal heparan sulfate proteoglycans (La Venuta et al., 2015; Steringer et al., 2015).
FIGURE 2.

FGF secretion, dimerization, and receptor shedding regulate the availability and activity of FGFs. (a) Unconventional secretion of FGF2 (adapted from Zacherl et al., 2015). ATP1A1, the α1-chain of Na/K-ATPase, interacts with FGF2 and recruits it to the inner plasma membrane, and promotes oligomerization. FGF2 oligomerization requires phosphatidylinositol 4,5-bisphosphate and FGF2 surface cysteines. Phosphorylation of FGF2 by Tec Kinase further promotes membrane insertion of FGF2 oligomers to form a membrane pore. FGF2, which is released into the extracellular space is bound and sequestered by cell surface HSPGs. (b) Dimerization of the FGF9 subfamily increases affinity for HS and prevents receptor binding by blocking the receptor-binding interface (blue). Mutations in FGF9 prevent homodimerization, decrease affinity for HS, and increase ligand diffusion. Only FGF9 monomers are able to interact with FGFRs. (c) Proteolytic cleavage of FGFRs by ADAM proteases 10 and 17 can release a soluble extracellular domain of the receptor that has the ability to bind and sequester FGF ligands
2.2 |. Regulation by ligand oligomerization
FGF9 subfamily members (FGF9, FGF16, and FGF20) undergo a reversible homodimerization which regulates their affinity for HS, diffusion, and activity (Figure 2b) (Goetz & Mohammadi, 2013; Harada et al., 2009; Kalinina et al., 2009). A mutation in Fgf9 in mice causing Elbow knee synostosis (Eks, FGF9 p.N143T) prevents homodimerization and lowers the affinity of monomers for HS (Harada et al., 2009). When this mutation is bred to homozygosity, increased width of long bones is observed at birth (Harada & Akita, 2020). In human, the FGF9 p.S99N mutation causes multiple synostosis syndrome 3 (SYNS3). This mutation does not affect dimerization but does decrease affinity for HS and impair receptor binding and activation. A mouse model containing the p.S99N mutation has similar phenotypes to SYNS3 patients (Tang et al., 2017; Wu et al., 2009). A second mutation in humans, FGF9 p.R62G causes both craniosynostosis and multiple synostosis. This mutation impaired dimerization and prevented binding and activation of FGFR3 (Rodriguez-Zabala et al., 2017). A third mutation in FGF9 p.P189R impairs FGF9 homodimerization, but not FGFR3c binding (Thuresson et al., 2021).
Structural studies show that deletion of the FGF9 N-terminus or mutation in the C-terminus disrupts FGF9 dimerization and allows binding to inappropriate “b” splice variants of FGFR2 (Liu et al., 2017). Engineered mutants in FGF9 or FGF20 with reduced dimerization exhibited reduced heparin binding, an increased radius of heparan sulfate-dependent diffusion, and a net increased biologic activity (Kalinina et al., 2009).
Engineered covalently linked FGF2 dimers showed increased stability and biological activity and reduced dependence on heparin (Nawrocka et al., 2020). Multivalent conjugates of FGF2 to hyaluronic acid polymer chains showed enhanced bioactivity in assays of cell proliferation and migration (Zbinden et al., 2018).
2.3 |. Regulation by proteolytic processing
FGFRs 1–4 are proteolytically processed by the ADAM (a disintegrin and metalloproteinase) metalloproteases, ADAM10 and ADAM17, to release an extracellular soluble receptor (Figure 2c). Soluble FGFR extracellular domains can function as ligand traps that bind and sequester FGFs and inhibit FGF signaling. Constitutive shedding, phorbolester-induced (activates protein kinase C), and ionomycin-induced (stimulates calcium influx) processing of FGFRs 1, 3, and 4 are mediated by ADAM17. FGFR2 processing is mediated by ADAM10 when activated by calcium influx (Dixit et al., 2021; Hanneken et al., 2021).
The endocrine FGF ligands, FGF21 and FGF23, are proteolytically processed as a means of rapid inactivation. FGF21 and FGF23 are cleaved by the enzyme FAP (fibroblast activation protein) and proprotein convertases, respectively (discussed later) (Al Rifai et al., 2021; Dunshee et al., 2016).
2.4 |. Regulation of FGFR internalization
FGFR internalization (endocytosis) is critical for proper signaling and is mediated by clathrin-dependent and clathrin-independent mechanisms, the spatial distribution of the receptor in the plasma membrane, and ligand binding, dimerization, and trans-autophosphorylation of the receptors (Szybowska et al., 2021).
Engineered anti-FGFR1 antibodies of different valency, were used to show that dimerization of FGFR1 with a bivalent antibody triggers clathrin-mediated endocytosis and that clustering of FGFR1 into larger oligomers with a tetravalent antibody increased efficient FGFR1 uptake through both clathrin-mediated and dynamin-dependent clathrin-independent endocytosis. Furthermore, FGFR1 internalization did not require receptor activation (Pozniak et al., 2020). Engineered anti-FGFR1 antibodies with high affinity to a specific epitope show faster internalization kinetics compared to antibodies with lower affinity to the same epitope (Opalinski et al., 2018).
Lectin family members, Galectin-1 (LGALS1) and Galectin-3 (LGALS3), were found to directly bind to the sugar chains of N-glycosylated FGFR1. Galectin-1 mimics FGF1 and efficiently activates FGFR1 and downstream signaling pathways. In contrast, galectin-3 binds with higher affinity and induces clustering of FGFR1 on the cell surface that inhibits constitutive receptor internalization (Kucinska et al., 2019).
Desulfation at the 2-O position of HS led to increased FGFR1 signaling and premature cell senescence (Jung et al., 2016). Similarly, depletion of the HSPG syndecan-1 (SDC1), or its level of sulfation, decreased FGFR1 internalization, and increased responsiveness to prolonged activation by FGF2, also leading to increased cellular senescence (Kang et al., 2020). Thus, the level of HS sulfation may prevent cell senescence through the regulation of FGFR1 endocytosis.
Biophysical analysis of the FGFR3 mutation (p.G380R) that causes Achondroplasia shows that a primary effect of this activating mutation is to increase receptor dimerization in the absence of ligand (Sarabipour & Hristova, 2016a). However, FGFR3 (p.G380R) also shows an enhanced response to FGF ligands (Ornitz & Legeai-Mallet, 2017).
FGFR1 and FGFR4 were shown to use clathrin-mediated endocytosis for internalization, while FGFR3 was internalized by both clathrin-dependent and clathrin-independent mechanisms. Depletion of clathrin heavy chain led to accumulation of FGFR1 and FGFR4 at the cell surface. For FGFR1, this led to increased levels of phospho-FGFR1, phospho-FRS2, and phospho-MAPK. For FGFR4, this led to increased levels of phospho-FGFR4 and PLCγ, but decreased phospho-AKT and phospho-ERK1/2 (Haugsten et al., 2011, 2016). These studies suggest that internalization and signaling are receptor-specific but also might be affected by the host cell type.
2.5 |. Regulation by heparan sulfate proteoglycans
Heparan sulfate (HS), in the form of heparan sulfate proteoglycans (HSPG), regulates the diffusion of FGFs through the extracellular matrix (ECM) and the binding of FGFs to FGFRs (Bernfield et al., 1999; Hassan et al., 2021; Lian et al., 2020; Xie & Li, 2019; Xu & Esko, 2014). HS was also the first identified co-factor to regulate FGF–FGFR binding (Rapraeger et al., 1991; Yayon et al., 1991). HS interacts with both FGFs and FGFRs to cooperatively increase the affinity of an FGF–FGFR dimer by binding to a cleft formed between the HS binding sites on FGFs and the N-terminal region of immunoglobulin-like domain 2 in FGFRs (Figure 1c, 3). This interaction leads to a conformational change, stabilization, and activation of an FGFR dimer (Goetz & Mohammadi, 2013; Ornitz & Itoh, 2015). Perlecan (Hspg2) is a large HSPG that is a key component of basement membranes and connective tissue pericellular matrix. Mice containing mutant Hspg2 that lacks exon 3 containing the HS attachment sites (Hspg2Δ3/Δ3) show reduced pulmonary artery smooth muscle cell proliferation, pulmonary vascular muscularization, and right ventricular pressure in response to hypoxia (Chang et al., 2015). Hspg2Δ3/Δ3 mice are also protected from induced osteoarthritis (discussed later) (C. C. Shu, Jackson, et al., 2016). These phenotypes may be a consequence of reduced FGF2–FGFR1 signaling.
FIGURE 3.

Cell surface and extracellular molecules that interact with FGF ligands and receptors. FGFR showing immunoglobulin-like domains I, II, and III and intracellular tyrosine kinase domains, TK1 and TK2. FGF ligands bind to the heparan sulfate (HS) chains that decorate heparan sulfate proteoglycans (HSPGs). FGF ligands also bind to long pentraxin 3 (PTX3), FGF binding proteins (FGFBP), and latent TGFβ binding protein 2 (LTBP2). FGFR–FGF interactions are stabilized by HS, α, β, and ɣ Klotho, Anosmin 1 (ANOS1), and fibronectin leucine-rich transmembrane proteins (FLRTs). ANOS1 can form a complex with FGFR and L1 cell adhesion molecule (L1CAM). Cell adhesion molecules, including neural cell adhesion molecule (NCAM) and N-cadherin (CDH2) can activate low-level FGFR signaling independent of FGF ligands. Inorganic phosphate (Pi) can directly activate FGFR signaling independent of FGF ligands. FGF receptor-like 1 (FGFRL1) lacks an intracellular tyrosine kinase domain and is involved in cell adhesion and may act as a decoy receptor through binding FGF ligands. Relative strength of signaling is indicated below each receptor
Some HSPGs may have additional FGF-related activities that are independent of mediating FGF–FGFR binding. An example is Syndecan-4 (SDC4), which has an intracellular domain that interacts with various cytoplasmic adaptor proteins and the signaling molecule protein kinase Cα (PRKCA). Deletion of the SDC4 cytoplasmic tail abolishes FGF signaling even though extracellular HS levels are not altered (Murakami et al., 2008).
Exostosins (EXT1, EXT2, EXTL1, EXTL2, and EXTL3) are glycosyltransferase genes that control HS biosynthesis. The expression levels and activity of exostosin regulate the amount and chain length of HS (Busse-Wicher et al., 2014). A role for HS in promoting FGF1 signaling was demonstrated in mice that conditionally lack one copy of Ext1 in adipocytes. Adipocytes in these mice had reduced expression of PPARɣ and reduced bone morphogenetic protein 4 (BMP4) and FGF1 signaling, smaller lipid droplets, and reduced levels of enzymes involved in lipid synthesis (Matsuzawa et al., 2021). The requirement for HS for cellular functions was demonstrated in zebrafish by mutations in ext2 and extl3 which resulted in loss of cell polarity and a failure of collective cell migration of the lateral line primordium. This defect was attributed to enhanced FGF diffusion and decreased FGF signaling leading to ectopic activation of Wingless/Int1 (Wnt)/β-catenin signaling (Venero Galanternik et al., 2015). EXTL2 differs from other EXTs in that it functions to limit the number of HS chains. In cultured cells, loss of EXTL2 increased the number of HS chains, increased FGF2 endocytosis, and reduced FGF2 signaling (Nadanaka & Kitagawa, 2018).
The affinity of HS for FGFs is specified by the pattern and level of sulfation of the HS chains (Barbosa et al., 2021; Hassan et al., 2021; Xu & Esko, 2014). Several recent studies have probed the mechanism of HS regulation of FGF signaling by manipulating HS modifying enzymes or identifying functional domains of the HS chain. The N-acetylglucosamine N-deacetylase/N-sulfotransferases (NDSTs) add a sulfate group at position 2 of N-acetylglucosamine (2-N HS sulfation) in the HS chain. This is the first sulfation step during the synthesis of HS. Developmental heart defects were observed in mice bearing a targeted disruption of NDST1. These defects closely resembled cardiac anomalies observed in mice with hypomorphic mutations in the cardiogenic regulator FGF8 (Abu-Issa et al., 2002; Frank et al., 2002; Pan et al., 2014). The HS sulfotransferases, HS2ST and HS6ST1, which add a sulfate group at position 2 of uronic acid (2-O HS sulfation) or position 6 of N-sulfated glucosamine (6-O HS sulfation), respectively, were shown to differentially modulate the properties of emerging FGF8 protein concentration gradients and FGF–FGFR-regulated MAPK signaling (Chan et al., 2017). Overexpression of HS6ST2 enhanced the effects of FGF2 on chondrocyte growth, while knockdown of HS6ST2 reduced FGF2 activity (W. Wang, Ju, et al., 2015). The sulfation level of HS was shown to be dynamically regulated during muscle differentiation and to regulate the response of muscle satellite cells (muscle stem cells) to FGF2 (Ghadiali et al., 2017). Synthetic HS oligosaccharides with defined structures demonstrated differential regulation of the specificity and activity of FGF1–FGFR1c and FGF2–FGFR1c ligand-receptor pairs (Schultz et al., 2017). Similarly, a library of HS tetrasaccharides with different levels and patterns of sulfation identified molecules with different binding properties for FGF2 (Zong et al., 2017). Interestingly, the binding preferences of FGFs for a library of HS-like molecules showed that the selectivity for binding structures in sulfated polysaccharides and the pattern of secondary binding sites on the surface of FGFs follows the phylogenetic relationship of the FGFs, suggesting that evolutionary selective pressures lead to expansion of the FGF family (Y. Li, Sun, Yates, et al., 2016).
The surface of vascular endothelial cells (ECs) is covered by a HS-rich layer of glycosaminoglycans and associated proteoglycans called the endothelial glycocalyx. The endothelial glycocalyx is essential for vascular homeostasis and is degraded in vascular disease. After vascular injury, levels of Ext1 and Fgfr1 in the lung were increased. Mice lacking EC Fgfr1/2 had reduced expression of Ext1 and impaired recovery of the endothelial glycocalyx in pulmonary ECs (Y. Yang, Haeger, et al., 2017). In ECs, it was found that FGF2 binding kinetics were altered in flow adapted cells due to changes in the quantity, availability, and binding kinetics, to cell surface HSPGs (Garcia et al., 2019). HS fragments released from mouse lungs treated with heparinase-III bind FGF2 and increase its biological activity (Y. Yang, Haeger, et al., 2017).
2.6 |. Regulation by FGF and FGFR interacting peptides and proteins
Classical mechanisms by which canonical and endocrine FGFs activate FGFRs use HS and α/β Klotho, respectively, as co-factors for receptor binding. In addition to HS and Klotho proteins, an increasing number of other molecules have been identified that interact with and exert biological effects on and through FGFs and FGFRs (Figure 3). These molecules have been referred to as noncanonical ligands or co-receptors, and include FGF binding proteins (FGFBPs), integrins, thrombospondin, neural cell adhesion molecule (NCAM1), L1 cell adhesion molecule (L1CAM), N-cadherin (CDH2), anosmin1 (ANOS1), fibronectin leucine-rich transmembrane proteins (FLRT1–3), G-protein-coupled receptors (GPCRs), Latent TGFβ binding protein 2 (LTBP2), and other receptor tyrosine kinases (RTKs) (Di Liberto et al., 2017; Latko et al., 2019; Margosio et al., 2008; Menz et al., 2015; Polanska et al., 2009; Shin et al., 2019; Sideek et al., 2016; Taraboletti et al., 1997). Inorganic phosphate (Pi) has also been shown to directly activate FGFR1 signaling (Takashi et al., 2019).
2.6.1 |. Klotho proteins (KL, KLB, and LCTL)
The Klotho proteins, αKlotho (KL), βKlotho (KLB), and ɣKlotho (Lactase-like, LCTL or Klotho-LPH related, KLPH, KLG) are essential cofactors for high-affinity binding of endocrine FGFs to FGFRs (Figure 1a) (Potthoff et al., 2012). Klotho proteins may additionally suppress canonical FGF signaling through interference with their receptor binding (Goetz et al., 2012). αKlotho and βKlotho have been shown to function as a tumor suppressor through interference with FGF, IGF1, and Wnt/β-Catenin signaling (Wolf et al., 2008).
αKlotho is a single-pass transmembrane protein that is primarily expressed in distal tubule epithelial cells of the kidney. αKlotho specifically interacts with FGF23 to form a signaling complex with FGFR1c or FGFR4 (Figure 3). In addition to the full-length form of αKlotho, two short forms of αKlotho have been detected in mice and humans (Xu & Sun, 2015). Proteolytic cleavage of full-length αKlotho leads to shedding of the extracellular domain, releasing a soluble form of αKlotho into the circulation (Chen et al., 2007). A secreted form of αKlotho can be produced by alternative splicing and is the major form found in the circulation (Matsumura et al., 1998). Tissue-specific inactivation of αKlotho or inhibition of ADAM metalloproteinases shows that the kidney is the principal contributor of circulating αKlotho. The soluble ectodomain of αKlotho can also function as a cofactor for FGF23 signaling, although with much weaker activity compared to full-length membrane αKlotho (G. Chen, Liu, et al., 2018; Erben & Andrukhova, 2017; Hu et al., 2016; Lindberg et al., 2014; Quarles, 2019; van Loon et al., 2015). Interestingly, the FGF23/αKlotho/FGFR signaling complex still requires HS for full activity (G. Chen, Liu, et al., 2018).
βKlotho specifically interacts with FGF15/19 to form a signaling complex with FGFR1c or FGFR4, and with FGF21 to form a signaling complex with FGFR1c. βKlotho biology has been recently reviewed (Kilkenny & Rocheleau, 2016; Kuro-o, 2019; Owen et al., 2015; Tan et al., 2014). Activation of βKlotho/FGFR1c by FGF21 induces sustained energy expenditure in brown adipose tissue, browning of white adipose tissue, weight loss, and improvements in obesity-associated metabolic derangements. To mimic this activity, a monoclonal antibody has been identified that can interact with and activate βKlotho/FGFR1c without interfering with FGF21 binding to the complex (Kolumam et al., 2015; Min et al., 2018). In the liver, interleukin 1β (IL-1β) strongly inhibits βKlotho expression and FGF15/19-induced ERK1/2 phosphorylation and cell proliferation (Y. Zhao, Meng, et al., 2016).
ɣKlotho has been shown to enhance signaling of FGF15/19 in human embryonic kidney 293 (HEK 293) cells and to directly interact with FGFs and FGFRs (Fon Tacer et al., 2010). ɣKlotho expression is increased in several types of cancer and is necessary for cancer cell survival (Hori et al., 2018; Onishi et al., 2020; Trost et al., 2016). Depletion of ɣKlotho in triple-negative breast cancer cells, bladder carcinoma cells, or prostate cancer cells led to increased oxidative stress, cell cycle arrest, and apoptosis (Hori et al., 2018; Onishi et al., 2020; Trost et al., 2016). Interestingly, depletion of ɣKlotho resulted in constitutive activation of ERK and a reduced induction of ERK in response to canonical FGF2 (Trost et al., 2016). ɣKlotho is highly expressed in gliomas and is associated with high tumor aggressiveness. Protein interaction analysis suggests that ɣKlotho may directly interact with FGF23, FGFR1, FGFR4, and FGFRL1 and may have an immunosuppressive function mediated by FGFR signaling (Fon Tacer et al., 2010; Trost et al., 2016). Further discussion of the actions of the endocrine FGFs can be found below.
2.6.2 |. FGF binding proteins
FGF binding proteins (FGFBPs) are secreted proteins that release locally stored FGFs from the extracellular HSPG-rich matrix to facilitate activation of cells that express FGFRs (Figure 3) (Taetzsch et al., 2018; Tassi et al., 2011). FGFBP1 is expressed in skeletal muscle and enriched in the neuromuscular junction (NMJ). FGFBP1 expression is reduced in a mouse model of amyotrophic lateral sclerosis (ALS) and in aging muscles. Mechanisms leading to reduced expression of Fgfbp1 are thought to be downstream of transforming growth factor beta (TGFβ). In mice lacking Fgfbp1, there is an accelerated progression of ALS pathology and age-related NMJ degeneration (Taetzsch et al., 2017), epidermal thickening, decreased epidermal papilloma formation, and delayed healing of skin wounds (M. O. Schmidt et al., 2018). FGFBP1 is expressed in ECs in the central nervous system (CNS) during blood-brain barrier formation. Conditional inactivation of Fgfbp1 in ECs leads to transient hypervascularization and delayed maturation of the blood-brain barrier (Cottarelli et al., 2020). Induced expression of Fgfbp1 in mice was shown to activate FGF signaling, increase angiogenesis, and increase blood pressure by sensitizing resistance vessels to angiotensin II (AngII, Agtr1b; Tassi et al., 2011; Tassi, Lai, et al., 2018).
Patients with a rare IgG4-related disease (IgG4-RD) have a frameshift mutation in FGFBP2. The expression of FGFBP2 in cytotoxic T cells suggests a role for FGFBP2 in the pathophysiology of this disease (Newman et al., 2019). Loss of function mutations in FGFBP2 are associated with congenital heart disease (McKean et al., 2016).
FGFBP3 modulates fat and glucose metabolism in mouse models of metabolic syndrome. FGFBP3 interacts with endocrine FGFs through its C-terminus and enhances their signaling. In obese mice, expression of exogenous FGFBP3 reduced hyperglycemia, hepatosteatosis, and weight gain, blunted de novo lipogenesis in liver and adipose tissues, increased circulating adiponectin, and decreased nonesterified fatty acids (Tassi, Garman, et al., 2018).
2.6.3 |. FGF receptor-like 1
FGF receptor-like 1 (FGFRL1; also referred to as FGFR5) is a single-span transmembrane non-tyrosine kinase FGFR that binds to heparin and a subset of FGF ligands (Figures 1d and 3). No function has been attributed to the short intracellular histidine-rich domain of FGFRL1, as mice in which this domain is replaced with green fluorescent protein (GFP) are phenotypically normal (Bluteau et al., 2014). The critical molecular functions of FGFRL1 are not well defined; however, proposed functions include binding FGF ligands as a decoy receptor, dimerization-induced inhibition of tyrosine kinase FGFRs, modulation of receptor turnover or signaling, or FGF-independent regulation of cell adhesion or cell fusion (Trueb, 2011; Zhuang et al., 2016). Other studies show that FGFRL1 may activate FGF signaling pathways and promote resistance to chemotherapy in small cell lung cancer cell lines (R. Chen, Li, Zheng, et al., 2020).
FGFRL1 binds FGFs 2, 3, 4, 8, 10, and 22, but not other ligands tested, including FGF9 or FGF20 (Steinberg et al., 2010). High-affinity binding of FGF8 required the presence of Ig-like domains 2 and 3 (Gerber et al., 2020; Zhuang et al., 2020). Overexpression of FGFRL1 did not affect cell proliferation or ERK1/2 phosphorylation in response to FGF2 but did promote cell adhesion, suggesting that its primary function may not involve FGF signaling (X. Yang, Steinberg, et al., 2016). In vertebrates, the third immunoglobulin-like domain of FGFRL1 is required for cell fusion activity (Zhuang & Trueb, 2017).
When FGFRL1 is overexpressed in cultured cells, it forms a homodimer on the cell surface and promotes cell adhesion through binding to HS (Rieckmann et al., 2008). Additionally, FGFRL1 was shown to induce cell clustering, cell–cell fusion, and formation of large multinucleated syncytia (Rieckmann et al., 2008; Steinberg et al., 2010; X. Yang, Steinberg, et al., 2016; Zhuang et al., 2015; Zhuang & Trueb, 2017).
Mice lacking FGFRL1 have agenesis of slow muscle fibers leading to defects in the diaphragm, and bilateral agenesis of the metanephric kidneys. It is not clear whether these phenotypes are caused by effects on FGF signaling or effects on cell adhesion; however, Fgf8 is expressed in the developing kidneys and Fgf8−/− mice have a similar kidney agenesis phenotype to Fgfrl1−/− mice suggesting that FGFRL1 may be a physiological receptor for FGF8 in the kidney (Amann et al., 2014; Gerber et al., 2020; Zhuang et al., 2020).
2.6.4 |. Thrombospondin 1
Thrombospondin 1 (THBS1) is a serum glycoprotein that can associate with the cell surface through binding to HS. THBS1 inhibits the angiogenic properties of FGF2 through direct binding to FGF2 through calcium-binding, type III repeats in its carboxy-terminal domain (Margosio et al., 2008; Taraboletti et al., 1997). A 60 kDa recombinant fragment derived from the THBS1 type III repeats inhibits FGF2 binding to ECs and FGF2-induced angiogenesis in vivo (Margosio et al., 2008). Additionally, a 15 amino acid peptide derived from the type III repeat sequences and a small molecule mimetic (SM27) of this peptide was also able to inhibit FGF2 EC binding and angiogenic activity by directly interfering with the heparin-binding site of FGF2 and inducing allosteric changes along with the FGF2/FGFR1 interface (Colombo et al., 2010; Pagano et al., 2012). Second-generation molecules based on SM27 have improved affinities and abilities to inhibit FGF2/HS/FGFR binding (Foglieni et al., 2016).
2.6.5 |. Long pentraxin-3
Pentraxin-3 (PTX3) is in the superfamily of cyclic multimeric soluble pattern recognition receptors (sPRRs). PTX3 is induced locally in ECs and immune cells at sites of inflammation or cell damage and is a positive regulator of the inflammatory response, conferring resistance to viral and bacterial infection (Smole et al., 2020). PTX3 binds FGFs 2, 6, 8b, 10, and 17 and specifically inhibits FGF2-induced EC and vascular smooth muscle (VSMC) cell proliferation (Figure 3) (Camozzi et al., 2005; Giacomini et al., 2015; Presta et al., 2018; Rusnati et al., 2004). Transgenic expression of PTX3 in ECs blocks FGFR1 activation in response to FGF2 and inhibits tumor growth and angiogenesis (Rezzola et al., 2019; Ronca et al., 2017; Ronca, Giacomini, Di Salle, et al., 2015). A similar activity was elicited by FGF-binding pentapeptide (aa100–104, ARPCA) derived from the N-terminus of PTX3 (Giacomini et al., 2015; Ronca, Giacomini, Di Salle, et al., 2015).
Based on the structure of the ARPCA pentapeptide, a synthetic small molecule, NSC12, was synthesized and shown to block FGF2 binding to FGFR1 but did not interfere with FGF2 binding to heparin. NSC12 binds to FGF2 with a Kd of 51 μM and to FGF3, FGF4, FGF6, FGF8, FGF16, FGF18, FGF20, and FGF22 with Kd values ranging from 16 to 120 μM (Ronca, Giacomini, Di Salle, et al., 2015). In functional assays, NSC12 and its derivatives were shown to inhibit angiogenesis and tumor growth in vivo (Castelli et al., 2021; Presta et al., 2018; Ronca et al., 2017; Ronca, Giacomini, Di Salle, et al., 2015).
In developing and injured bone, PTX3 is produced by osteoblasts (Parente et al., 2019). Mice lacking Ptx3 have reduced osteoblast function resulting in decreased bone mass and bone mineralization, but no change in active osteoclasts populating the bone surface. Ptx3−/− mice also have impaired callus mineralization during fracture healing. In vitro, FGF2 inhibits osteoblast differentiation, and this can be blocked by PTX3 (Grčevi c et al., 2018). These data suggest that PTX3 may modulate direct effects of FGF2 signaling to osteoblasts; however, effects of FGF signaling to the skeletal vasculature may also affect bone formation, homeostasis, and response to injury.
In zebrafish, inactivation of ptx3b increases FGF ligands availability, causing sustained activation of FGF signaling. Sustained FGFR signaling resulted in shortened primary cilia leading to defects in the left–right asymmetry determination (Guerra et al., 2020). Increased expression of PTX3 or treatment with NSC12 increased the length of the primary cilium in several FGF-dependent tumor cell lines (Guerra et al., 2020).
2.6.6 |. Anosmin 1 and Kallmann syndrome
Anosmin 1 (ANOS1; formerly called KAL1) is the gene responsible for the X-linked form of Kallmann syndrome, a type of congenital hypogonadotropic hypogonadism (CHH) (de Castro, Seal, Maggi, & Group of HGNC Consultants for KAL1 Nomenclature, 2017; Korsensky & Ron, 2016). Mutations in FGF8 and FGFR1 cause an autosomal form of Kallmann syndrome, accounting for 12% of cases (Falardeau et al., 2008; Hardelin & Dode, 2008; Trarbach et al., 2010). Mutations in FGF17, IL17RD, DUSP6, SPRY4, and FLRT3 have also been identified in people with CHH (Miraoui et al., 2013). ANOS1 is a secreted heparin-binding glycoprotein and has been shown to function as a co-factor that promotes FGFR signaling in developing gonadotropin-releasing hormone (GnRH) neurons (Gonzalez-Martinez et al., 2004). Mechanistically, ANOS1 potentiates FGFR signaling in a FGF ligand-dependent manner through direct interactions with the FGFR IG-like domain II–III linker region and with FGF2 (Figure 3) (Hu et al., 2009; Korsensky & Ron, 2016). More recently, ANOS1 was shown to form a complex with L1 cell adhesion molecule (L1CAM) and FGFR, which is required for neurite branching in Caenorhabditis elegans (Diaz-Balzac et al., 2015).
Agenesis of the corpus callosum occurs in Kallmann syndrome patients with hypomorphic mutations in FGF8 (McCabe et al., 2011; Moldrich et al., 2010). Perinatal FGF8 signaling regulates the timing of the onset of anterior–dorsal Glial fibrillary acidic protein (Gfap) expression, identifying a role in midline glial cell development. Fgf8 hypomorphic mice have impaired midline glial development leading to agenesis of the corpus callosum (Stewart et al., 2016).
In the zebrafish lateral line primordium, ANOS1 enhances FGF signaling by directly binding and facilitating the diffusion of FGF10a through the ECM to increase the FGF10a signaling range (J. Wang, Yin, et al., 2018). Of note, mice and rats lack an identifiable ANOS1 orthologue (de Castro et al., 2014; Rugarli et al., 1993); however, overexpression of human ANOS1 in mice led to activation of FGFR1/ERK1/2 signaling and resulted in enhanced oligodendrocyte precursor cell (OPC) proliferation, migration, and myelination (Murcia-Belmonte et al., 2016).
2.6.7 |. Fibronectin leucine-rich transmembrane proteins
Fibronectin leucine-rich transmembrane proteins (FLRTs) 1–3 are single transmembrane domain glycoproteins that regulate cell adhesion and synaptogenesis. FLRT1, 2, and 3 interact with FGFR1 and FGFR2 and increase their expression and signaling in a positive feedback loop (Figure 3) (Korsensky & Ron, 2016; Latko et al., 2019). FLRT interactions with FGFRs involve the membrane-proximal fibronectin type III domain and the short cytoplasmic tail (Wei et al., 2011; L. Yang, Hansen Falkesgaard, et al., 2017). Formation of FLRT1–FGFR1 complexes enhance FGFR signaling in the presence of FGF ligand (Wheldon et al., 2010). Rare variants in FLRT3, FGF8, FGF17, and SPRY4 have been identified in patients with Kallmann syndrome or CHH further supporting a role for FLRT3 in the anosmin–FGFR1 signaling axis (Amato et al., 2019; Falardeau et al., 2008; Korsensky & Ron, 2016; Miraoui et al., 2013). In X. laevis anterior–posterior patterning, BMP signaling induces the transcription factor TBX2, which suppresses Flrt3 expression, resulting in decreased FGF8–FGFR1 signaling and anterior tissue formation (Cho et al., 2017).
2.6.8 |. Cubilin
Cubilin (CUBN) is a highly conserved membrane protein that interacts with and facilitates the endocytosis of multiple proteins, sugars, and phospholipids. During mouse development, CUBN is expressed in embryonic and extra-embryonic tissues, and mouse knockouts of Cubn result in early embryonic lethality (Kozyraki & Cases, 2020). CUBN binds FGF8 and most likely FGF17 and FGF18 with high affinity, but not members of other FGF subfamilies. Conditional knockouts of Cubn showed that it acts synergistically with FGF8 for cephalic neural crest cell survival, migration, proliferation, and telencephalic patterning. Mechanistically, CUBN is necessary for optimal FGF8 mediated MAPK/ERK1/2 signaling, although it does not form a ternary complex with FGF8 and FGFRs (Cases et al., 2013).
2.6.9 |. Interaction with non-FGFR tyrosine kinases
FGFRs interact with several other RTKs and cytosolic tyrosine kinases, including platelet-derived growth factor receptors (PDGFRs) and erythropoietin-producing hepatocellular (Eph) receptors. The formation of complexes with other kinases can result in a conformational change and/or phosphorylation of the FGFR leading to activation of downstream signaling pathways, which in some cases is independent of FGF ligand. The literature on these interactions has been extensively reviewed (Latko et al., 2019; Turner et al., 2017). Using fluorescence resonance energy transfer (FRET)-based techniques, it was demonstrated that the strength of RTK heterodimers was similar to that of homodimers, strongly suggesting that RTK-hetero-interactions may be biologically important (Paul et al., 2020). Eph receptors are a large class of RTKs that are activated by membrane-anchored ephrin ligands. Several Eph receptors have been shown to interact with FGFRs through their cytoplasmic domains (Paul & Hristova, 2019; Yokote et al., 2005). In an analysis of mouse embryonic neural stem/progenitor cell differentiation, ephrin-A1 was found to regulate neuronal differentiation through a RAS–MAPK pathway that was dependent on FGFR signaling. These data suggest that FGFs activate a ternary complex of EphA, FGFR, and the fibroblast growth factor receptor substrate 2 (FRS2) adaptor protein, to regulate self-renewal and differentiation of mouse embryonic neural stem/progenitor cells (Sawada et al., 2015). In the developing lens, mouse Disks Large 1 (DLG1) functions as a scaffolding protein to regulate macromolecular complexes between EphA2 and FGFRs 1, 2, and 3. Inactivation of Dlg1 in the lens disrupts lens fiber differentiation and correlates with decreased FGFR2 signaling (Lee & Griep, 2014; S. Lee, Shatadal, & Griep, 2016).
In a recent study, protein tyrosine kinase 7 (PTK7), a catalytically defective receptor protein tyrosine kinase, was found to bind FGFR1 and FGFR3 through extracellular domain interactions. These interactions were not affected by FGF1 ligand and were found to activate FGFR1 independent of the ligand to promote oncogenesis (Figure 3) (Shin et al., 2019).
2.6.10 |. Interaction with cell adhesion molecules
Cell adhesion molecules (CAMs) that stimulate neurite outgrowth (NCAM, CDH2, and L1-CAM) were the first identified noncanonical FGFR1 interacting proteins (Doherty & Walsh, 1996; Latko et al., 2019; Nguyen & Mege, 2016; Williams et al., 1994).
The affinity of FGFRs for NCAM is 106-fold lower than for FGFs; however, the functional interaction is driven by the high cell-surface concentration of NCAM (Kiselyov et al., 2005). The interaction between NCAM and FGFRs requires the FGFR Ig-like domains II and III (Figure 1b, 3) and this interaction can be inhibited by extracellular ATP binding to NCAM (Kiselyov et al., 2003; Rasmussen et al., 2018). FGF2 and NCAM promote different FGFR1 assembly and dynamics at the plasma membrane. NCAM stimulation elicits long-lasting cycles of unstable FGFR1 homomultimers causing rapid FGFR1 internalization and recycling resulting in sustained low-level signaling that is sufficient to induce cell migration. Low concentrations of FGF2 have similar effects. In contrast, high concentrations of FGF2 induced stable dimerization, robust signaling, and cell proliferation (Zamai et al., 2019).
A short heptapeptide, AKTVKFK (Enreptin), derived from the N-terminus of FGFR Ig-like domain II binds NCAM and partly overlaps with the FGFR self-dimerization site. This peptide acts as a dual agonist for NCAM and FGFR and enhances neurite outgrowth (Enevoldsen et al., 2012; Ilieva et al., 2019). Similarly, peptides derived from NCAM1 and NCAM2 can also promote neurite outgrowth in vitro and improve cognitive function in vivo through activation of FGFR1 (Hansen et al., 2010; Rasmussen et al., 2018).
CDH2 (N-cadherin) and FGFR1 have been shown to function together to regulate cell survival and migration in the setting of embryonic development and cancer (Suyama et al., 2002). FGFR1 and CDH2 support each other in a feed-forward loop where FGF signaling regulates the expression of CDH2 and CDH2 regulates sustained activation of FGFR1 (Nguyen & Mege, 2016; Qian et al., 2014; Suyama et al., 2002). Consistent with this, treatment of kidney proximal tubule epithelial cells with TGFβ rapidly and transiently induced both FGFR1 and CDH2, and inhibition of FGFR1 signaling blocked these cells from an epithelial–mesenchymal transition (EMT) (Zivotic et al., 2018). In early embryonic development, CDH2 is highly expressed in mouse epiblast stem cells where it functions to stabilize FGFR1 expression. Knockdown of either Cdh2 or Fgfr1 reduces pluripotency of epiblast stem cells. However, in mouse embryonic stem cells, CDH2 increases FGF2-mediated cell differentiation (Takehara et al., 2015). In neuronal differentiation, FGF signaling must be decreased to allow the acquisition of a neuronal cell fate. In a model of embryonic stem cell differentiation into neurons, the expression of CDH2 promotes neuronal differentiation by dampening FGF signaling (Francavilla et al., 2009; Punovuori et al., 2019).
In migrating cortical neurons different effects of CAMs were observed. CDH2 was shown to bind FGFRs cell autonomously and inhibit FGFR polyubiquitination and lysosomal degradation, leading to increased FGFR protein, prolonged ERK1/2 phosphorylation, and increased neuronal migration (Kon et al., 2019). A similar effect on neuronal migration was observed with the cell adhesion molecule, neuronal growth regulator 1 (NEGR1), which also directly interacts with FGFR2. Loss of NEGR1 increased FGFR2 degradation and impaired FGFR2-dependent ERK1/2 and AKT signaling and neuronal migration (Szczurkowska et al., 2018). In an in vitro model in which Fgfr1- and Cdh2-transfected human embryonic kidney (HEK) cells were allowed to migrate on a CDH2 coated surface, FGFR1 and CDH2 were co-localized at cell–cell contact sites, cell–cell adhesion was strengthened, and cell migration was reduced. These effects were attributed to a positive feedback loop between CDH2 and FGFR1 at adhesion sites (Nguyen et al., 2019). In Ciona, heart progenitor cells adhere to the ECM through integrin receptors. Cell adhesion was shown to inhibit mitotic FGF receptor internalization, leading to receptor enrichment along adherent membranes leading to signal polarization (Cota & Davidson, 2015).
In non small cell lung cancer cells, an FGFR4 variant (p.G388R) induced an EMT phenotype (expression of Cdh2, Vim, Snai1, and Twist1). Expression of CDH2 was necessary for the pro-tumorigenic effects of this FGFR variant (Quintanal-Villalonga et al., 2018). Additionally, co-expression of FGFR1 and/or FGFR4 with CDH2 predicted a poorer outcome for a variety of cancer types (Quintanal-Villalonga et al., 2020). However, in this model, it is not known if CDH2 affects FGFR expression of activity.
The question of whether canonical FGFR signaling pathways are required to mediate cell adhesion was addressed by comparing FGFR null mutants, FGFR mutants that uncouple activation of all major downstream signaling pathways, and mutants in which the tyrosine kinase domain is inactivated. Interestingly, the effects of FGF-induced cell–matrix or cell–cell interactions were lost in the FGFR null mutants and in the kinase-dead mutants, but were retained in mutants that lacked connections to the canonical downstream signaling pathways (Ray et al., 2020). These data suggest that FGF–FGFR signaling regulates cell adhesion through kinase-dependent mechanisms that are distinct from the classical downstream signaling pathways. Direct interaction of FGFRs with cadherins is also supported by these data, as FGFRs with the most severe signaling mutations still formed β-catenin positive cell–cell contacts, which were not formed in the receptor null mutants.
2.6.11 |. Latent TGFβ binding protein 2
Latent TGFβ binding protein 2 (LTBP2) is a highly conserved member of the LTBP family. Most LTBP family members bind Fibrillin 1 (FBN1) in the ECM to retain pro-forms of TGFβ (Hirani et al., 2007). However, unlike other family members, LTBP2 does not bind TGFβ, but does bind FGF2 and inhibits FGF2-stimulated cell proliferation (Menz et al., 2015). Furthermore, LTBP2 and FGF2 co-localize in hypertrophic scar tissue suggesting a functional relationship between LTBP2 and FGF2 (Sideek et al., 2016).
2.6.12 |. Family with sequence similarity 3 member B (FAM3B)/PANDER (pancreatic-derived factor)
Family with sequence similarity 3 member B (FAM3B) is a secreted factor involved in glucose metabolism, type 2 diabetes (T2D), and cancer. FAM3B was shown to act as a FGFR ligand in Xenopus embryos. Overexpression of Xenopus fam3b mRNA or injection of recombinant FAM3B protein into the blastocoel cavity inhibited cephalic structures and induced ectopic tail-like structures, indicating FGF-like activity. In vitro, FAM3B protein bound to FGFR extracellular domain-Fc fusion proteins for FGFRs 1–4 and activated the downstream ERK signaling in an FGFR-dependent manner (F. Zhang, Zhu, et al., 2021).
2.7 |. Regulation by inorganic phosphate
High dietary levels of inorganic phosphate (Pi) were found to increase the expression of Galnt3 (polypeptide N-acetylgalactosaminyltransferase 3) in bone tissue and increase circulating levels of FGF23 (Takashi et al., 2019). Proteomic analysis identified unliganded FGFR1 as the sensor for Pi. In response to high extracellular Pi, osteoblastic UMR106 cells increased phosphorylation of FGFR1c, increased pERK, and increased GALNT3 expression. These effects could be blocked with the FGFR inhibitor PD173074 (Takashi et al., 2019; Takashi & Fukumoto, 2020a, 2020b). As discussed below, Pi sensing by FGFR and downstream regulation of FGF23 activation forms a feedback loop to maintain phosphate homeostasis.
2.8 |. Quantitative effects of FGFR activation on the cellular response
Sequence differences in FGFs and FGFRs and mutations in these proteins affect the specificity and affinity of FGF–FGFR interactions. Quantitative differences in FGF–FGFR interactions affect the stability of the active FGFR dimer and regulate downstream signaling.
The strength of an FGF–FGFR dimer is one determinant of downstream signaling and cellular responses. High-affinity FGF–FGFR interactions elicit a stronger intracellular response (MAPK phosphorylation and cell proliferation) compared to weaker FGF–FGFR interactions. This principle was demonstrated by showing that suboptimal FGFR activation by a mutant FGF1 that forms a weak FGF1–FGFR dimer evokes a poor proliferative response in NIH3T3 fibroblasts but a full metabolic response in 3T3L1 preadipocytes (Z. Huang, Tan, et al., 2017; Suh et al., 2014).
Similarly, comparing NCAM to FGF activation of FGFRs, stimulation with a soluble NCAM-Fc molecule elicited long-lasting cycles of short-lived FGFR1 monomers and multimers, possibly promoting rapid FGFR1 internalization and recycling. In contrast, at a dose that stimulates cell proliferation, FGF2 induced stable dimerization, and degradation of endocytosed receptors (Francavilla et al., 2009; Zamai et al., 2019).
Synovial sarcoma (SS) is a soft tissue malignancy. SS cell lines express multiple FGF ligands and receptors and inhibition of FGFR signaling slows tumor growth (Ishibe et al., 2005). The SS-associated protein, SYT–SSX2, directly upregulated the expression of FGFR2 (Garcia et al., 2012). A mouse model for SS identified expression of FGFRs 1, 2, and 3 in SS tumors, with FGFR2 and FGFR3 levels induced relative to surrounding tissue. All three receptors were required for tumor growth through MAPK–ERK activation of ETV4 and ETV5 expression (DeSalvo et al., 2021).
Data showing differential activation of FGFRs and corresponding distinct cellular responses support a threshold model for FGFR signaling specificity. In this model, quantitative differences in the strength and longevity of ligand-stabilized receptor dimers on the cell surface correlate with quantitative differences in the phosphorylation of the activation loop in the tyrosine kinase domain leading to the recruitment and activation of distinct substrates and intracellular signaling pathways (Zinkle & Mohammadi, 2018).
3 |. INTRACELLULAR FGFR SIGNAL TRANSDUCTION AND FEEDBACK REGULATION
Several RTKs, including FGFRs, exist as unliganded dimers that are then stabilized and undergo conformational changes upon ligand binding (Belov & Mohammadi, 2012; Livnah et al., 1999; Sarabipour & Hristova, 2015, 2016b). Ligand-induced conformational changes in FGFR dimers lead to activation of the FGFR tyrosine kinase and the sequential transphosphorylation of five tyrosine residues in the catalytic core of the FGFR (Lew et al., 2009; Ornitz & Itoh, 2015). Transphosphorylation of tyrosine residues in the activation loop (A-loop) is a prerequisite for subsequent tyrosine phosphorylation (Furdui et al., 2006). In the absence of ligand, the A-loop asymmetric dimer is electrostatically destabilized. Ligand binding stabilizes the transphosphorylating dimer. Differences in the stability of ligand-induced extracellular dimerization promote the formation of this dimeric complex to varying extents, to modulate intracellular kinase activity and signaling intensity (L. Chen, Marsiglia, Chen, et al., 2020). The intracellular juxtamembrane domain may also contribute to maintaining the FGFR dimer in the absence of ligand (Sarabipour & Hristova, 2015).
After ligand binding and A-loop phosphorylation, subsequent tyrosine phosphorylation enhances and stabilizes tyrosine kinase domain activity and forms specific binding sites for some adaptor proteins. Phosphorylation of adaptor proteins activates four classical intracellular signal transduction pathways (RAS–MAPK, PI3K–AKT, PLCɣ, and STAT). Classical intracellular FGFR signaling has been extensively reviewed (Brewer et al., 2016; Ornitz & Itoh, 2015; Xie, Su, et al., 2020; Zinkle & Mohammadi, 2018). FGFR signaling through FRS2 is the primary pathway that activates downstream RAS–MAPK and PI3K–AKT intracellular signaling cascades. Complexities of downstream signal transduction are illustrated by conditional knockouts of Frs2 and Shp2 in the developing lens, where inactivation of both was required to prevent lens development. Genetic interaction experiments showed that direct binding of SHP2 to FRS2 is necessary for the activation of ERK signaling (H. Li, Tao, et al., 2014).
The requirement for cooperativity of obligate receptor dimers acting in trans is illustrated by PLCɣ signaling, where the PLCɣ SH2 domain is bound by one receptor and phosphorylated by the second receptor (Huang et al., 2016). Activation of multiple downstream signaling pathways by a single FGFR is illustrated by FGFR3 in chondrocytes where MAPK and STAT1 pathways are activated and in the inner ear where FGFR1 activates MAPK and PI3K to regulate sensory cell differentiation (Ornitz & Legeai-Mallet, 2017; Su et al., 2021).
In addition to the activation of intracellular signaling pathways, there are several feedback regulators of FGFR signaling, some of which are induced or activated by FGFR signaling. These include four members of the Sprouty family (SPRYs 1–4), similar expression to FGF (SEF, IL17RD), Sprouty-related EVH1 domain-containing (SPREDs 1 and 2), several dual-specificity phosphatases (DUSPs 1, 4, and 6), MAPK phosphatase 3 (MKP3), direct phosphorylation of serine residues in the FGFR, and tyrosine dephosphorylation by tyrosine phosphatase receptor type G (PTPRG) (Azami et al., 2019; Huh et al., 2020; J. Y. Kim, Lee, Kim, et al., 2019; Kostas, Haugsten, et al., 2018; Missinato et al., 2018; Szybowska et al., 2021; Umair et al., 2020; van Boxtel et al., 2018; Wakioka et al., 2001; Zakrzewska et al., 2013, 2019). Others signaling pathways such as G-protein-coupled receptors and natriuretic peptide receptor B (NPR2) can directly or indirectly regulate FGFR signaling.
3.1 |. Regulation by interleukin17 receptor D (IL17RD, SEF)
Sef (Similar Expression to Fgf) encodes a type I single transmembrane domain protein with an extracellular region containing an immunoglobulin-like motif and a type III-like fibronectin repeat, and an intracellular domain with similarity to the interleukin 17 receptor (Grothe et al., 2008; Ron et al., 2008). Sef expression is induced by FGF signaling in different cell types, and it functions as a feedback inhibitor of FGF signaling primarily by blocking the activation of MAPK, but in some cell-types also by blocking the activation of AKT (Korsensky & Ron, 2016; Latko et al., 2019; Ornitz & Itoh, 2015). A splice variant of Sef, Sef-b, is a cytosolic protein that inhibits ERK/MAPK (Korsensky & Ron, 2016).
In vivo, Sef is expressed in developing bone (periosteum and the chondro-osseous junction), and mice lacking Sef have increased cortical bone mass and enhanced osteoclastogenesis (He et al., 2014). In a breast cancer cell line, SEF functions to inhibit EMT phenotypes that promote cell migration and invasion (He et al., 2016). Expression of SEF is decreased in some aggressive tumors. Targeted overexpression of SEF in vivo in tumors reduced proliferation and blood vessel density and the local expression of FGF2 and matrix metalloproteinase 9 (MMP9), suggesting that SEF may have both cell autonomous and cell non-autonomous effects (Mishel et al., 2017).
3.2 |. Regulation by C-type natriuretic peptide signaling
FGFR3 is expressed in growth plate chondrocytes and FGFR3 signaling inhibits endochondral bone growth. Activating mutations in FGFR3 result in Achondroplasia and other related forms of skeletal dwarfism (discussed later) (Hogler & Ward, 2020; Legeai-Mallet & Savarirayan, 2020; Ornitz & Legeai-Mallet, 2017; Unger et al., 2017). C-Type natriuretic peptide (CNP) is expressed in multiple tissues, including chondrocytes, and its primary function is to increase bone growth through signaling through the natriuretic peptide receptor B (NPR2) in chondrocytes. Mice or humans with loss of function mutations in CNP or NPR2 develop severe dwarfism (acromesomelic dysplasia, type Maroteaux) (Potter et al., 2006). Conversely, overexpression of CNP or activating mutations in NPR2 lead to skeletal overgrowth (Bocciardi & Ravazzolo, 2009; Hannema et al., 2013; Miura et al., 2014; Wagner et al., 2021).
CNP signaling through NPR2 functions to inhibit FGFR3 induced ERK phosphorylation in growth plate chondrocytes (Ozasa et al., 2005). In chondrosarcoma cells and in growth plate chondrocytes, activation of FGFR3 signaling results in dephosphorylation of NPR2, which is required to inactivate the receptor (Robinson et al., 2017; Shuhaibar et al., 2017). A mutation in NPR2 that prevents dephosphorylation mitigated the short limb phenotype of mice homozygous for the p.G380R Achondroplasia mutation in FGFR3 (Wagner et al., 2021). Suppression of CNP signaling by activating mutations in FGFR3 is an additional mechanism for inhibition of bone growth. CNP analogs with an increased serum half-life (TransCon CNP; Vosoritide) are in clinical trials for treating patients with Achondroplasia (Breinholt et al., 2019; Klag & Horton, 2016; Legeai-Mallet & Savarirayan, 2020; Ornitz & Legeai-Mallet, 2017; Savarirayan et al., 2020, 2021; Wendt et al., 2015). Vosoritide (Voxzogo) has recently been approved for the treatment of Achondroplasia in the EU and by the FDA (Duggan, 2021) (https://www.fda.gov/news-events/press-announcements/fda-approves-first-drug-improve-growth-children-most-common-form-dwarfism).
NPR2 expression has also been found in cranial sutures. However, a mouse model of craniosynostosis that results from activating mutations in FGFR2 (Apert syndrome) did not show a response to CNP analogs (Holmes, Zhang, et al., 2018).
3.3 |. Regulation by G-protein-coupled receptors
G-protein-coupled receptors (GPCRs) that interact with FGFRs have been identified using yeast two-hybrid screens, co-immunoprecipitation, proximity ligation assays, or direct activation assays. These include the adenosine receptor (Adora2a), cannabinoid receptor 1 (Cnr1), muscarinic acetylcholine receptor (mAChR, Chrm1), 5-hydroxytryptamine (serotonin) receptor (Htr1a, Htr2a), and the Mu-opioid receptor (Oprm1) (Borroto-Escuela et al., 2015, 2016; Di Liberto et al., 2017, 2019; Latko et al., 2019; Narvaez et al., 2020). Disruption of interactions between FGFR1 and serotonin receptors may contribute to the development of major depression and anxiety disorders (Borroto-Escuela et al., 2021).
Bradykinin (BK; a 9 amino acid peptide cleaved from kininogen) and FGF2 have been implicated in the pathogenesis of inflammatory and angiogenic disorders by inducing a pro-inflammatory signature in ECs. The GPCRs, BK receptors 1 and 2 (Bdkrb1, Bdkrb2), increased FGF2 expression and FGFR1 signaling in human umbilical vein endothelial cells (HUVECs) and in retinal capillary endothelial cells (HRECs). FGFR1 phosphorylation triggered by BK was mediated by cSrc and was independent FGF2 upregulation (Terzuoli et al., 2018).
In Drosophila, Folded gastrulation (Fog) is a secreted ligand that signals through the G-protein-coupled receptors Mist and Smog where they activate downstream effectors to elicit cell-shape change during gastrulation, axon guidance in the developing nervous system, and glial morphogenesis. The FGFR, Heartless (Htl), is expressed in glia and regulates glial morphogenesis (Stork et al., 2014). Htl was found to interact with Smog in a synergistic manner to suppress Fog signaling (Shweta et al., 2021).
3.4 |. Feedback regulation of intracellular signaling pathways
Feedback inhibition of FGFR signaling is necessary for the precise control of cellular function and for the prevention of pathological responses to injury and cancer. The MAPK/ERK pathway is one of the major downstream pathways activated by FGFRs and other RTKs. Direct feedback regulation of FGFR1 by ERK1/2 occurs through phosphorylation of a serine residue in the carboxyl terminus of FGFR1 (Ser777 in FGFR1) which directly reduces tyrosine phosphorylation and activity of the tyrosine kinase domain (Zakrzewska et al., 2013). Direct inactivation of FGFR1 occurs through tyrosine dephosphorylation by protein tyrosine phosphatase receptor type G (PTPRG) (Kostas, Haugsten, et al., 2018).
Less direct feedback mechanisms occur through phosphorylation of threonine residues in the adaptor protein FRS2 by ERK1/2 and P38, which decreases FRS2 tyrosine phosphorylation and attenuates binding to Grb2 and coupling to downstream MAPK signaling (Lax et al., 2002; Zakrzewska et al., 2019). Further downstream, dual-specificity phosphatase (DUSP) family phosphatases are responsible for dephosphorylation and inactivation of the MAPKs (Huang & Tan, 2012). FGF activated ERK1/2 signaling directly induces DUSP6 expression, and, through its downstream target p90RSK, phosphorylates the transcriptional repressor CIC leading to its nuclear export, resulting in derepression of DUSP6 (Ren et al., 2020).
In zebrafish, the specification of germ layers is regulated by a feedforward interaction between Nodal and FGF signaling to pattern endoderm and mesoderm (Pinheiro & Heisenberg, 2020). Nodal induces long-range FGF signaling while simultaneously inducing proximal cell autonomous expression of DUSP4 which specifically dephosphorylates phospho-ERK1/2. Attenuation of proximal FGF signaling allows specification of endoderm progenitors by Nodal, while more distal cells receive Nodal and FGF signaling and are specified as mesoderm (van Boxtel et al., 2018).
In the mouse blastocyst, FGF4 activates ERK signaling in inner cell mass precursor cells and in differentiated primitive endoderm and definitive epiblast cells. In the primitive endoderm, DUSP4 expression is induced by ERK signaling and in epiblast cells, ETV5 is induced by Nanog, independent of ERK. DUSP4 reports FGF signaling in primitive endoderm; however, its functional role in this cell type is not yet known (Azami et al., 2019).
Spred and Sprouty (Spry) genes encode intracellular negative regulators of RTK signaling that target the RAS/MAPK pathway. SPRED sequesters RAF and a RAS/RAF complex to the plasma membrane, preventing RAF activation by phosphorylation. SPRY, activated by phosphorylation, disrupts the GRB2/SOS complex to block RAS/RAF signaling (Neben et al., 2019). Furthermore, the expression of Spry2 and Spry4 is induced by ERK activation and are effective reporters of FGF signaling in many cell types (Morgani, Saiz, et al., 2018). Spry genes are involved in many developmental processes and in tumorigenesis (Kawazoe & Taniguchi, 2019). For example, in kidney development, SPRY is expressed in nephron progenitors and counters FGF9 and FGF20 signaling to maintain progenitor cell stemness (Huh et al., 2020). In optic vesicle development, SPRY2 regulates the movement of retinal progenitors to properly position progenitors within the optic vesicle (Sun, Yoon, et al., 2020). In lens fiber development, SPRY and SPRED proteins are expressed in the lens epithelium and redundantly suppress FGF-induced ERK1/2-signaling in lens epithelial cells. Overexpression of Spry2 in lens cells in vitro suppressed FGF-induced ERK1/2 phosphorylation and expression of the fiber-specific marker, Prox1, but not the accumulation of β-crystallins (G. Zhao, Bailey, et al., 2018). Overexpression of Spred1 or Spred2 in the lens in vivo resulted in a small lens phenotype caused by reduced epithelial cell proliferation and fiber differentiation (Susanto et al., 2019).
FGF signaling is required for salivary gland development. Gain-of-function mutation in Fgfr2 in Apert syndrome mice increases epithelial branching of the submandibular glands (Yamaji et al., 2018). FGFs also suppress Wnt signals that are required for parasympathetic ganglia that are necessary for salivary gland innervation. SPRY1/2 function to suppress FGF signals allowing parasympathetic gangliogenesis and innervation of the salivary gland (Knosp et al., 2015).
In primary fetal lung epithelial cells, hypoxia favored nuclear localization of SPRY2 which interacts with regions of the rat and human VEGF-A promoter spanning the hypoxia response element (HRE) to repress VEGF-A expression. Treatment with FGF10 reduced the interaction of Spry2 with the HRE to promote VEGF-A expression and coordinate epithelial and vascular morphogenesis (Walker & Land, 2018).
3.5 |. New downstream targets of FGFR signaling
The repertoire of proteins that interact with FGFRs has been explored through immunoprecipitation coupled with mass spectrometry. Using this approach, 66 proteins were found to interact with the FGFR3 intracellular domain. Validated examples include the adapter protein STAM1, the transcriptional regulator SHOX2, the translation elongation factor eEF1A1, serine/threonine kinases ICK, MAK, and CCRK, and the inositol phosphatase SHIP2 (Balek et al., 2018). In another example, a yeast 2-hybrid screen for proteins that interact with FGFR3 (K650E), an activating mutation found in multiple myeloma and bladder cancer, identified TGFβ-activated kinase 1 (TAK1). Follow-up experiments showed that wild-type FGFRs 1, 2, 3, and 4 interact with TAK1 and that FGFR3 phosphorylates and activates TAK1. TAK1 is an important regulator of NFkβ activity and may be a potential therapeutic target downstream of FGFR3 dependent cancers (Salazar et al., 2014). An overexpression screen of RTKs identified FGFR1 and FGFR2 and several other RTKs as activators of the Hippo pathway. FGFR1 and FGFR2 were found to directly interact with and phosphorylate multiple tyrosine residues, and thereby activate YAP/TAZ independent of upstream Hippo signaling (Azad et al., 2020).
3.6 |. Cell and receptor-specific properties of FGFR downstream signaling pathways
RTKs share common intracellular signaling pathways yet have distinct cell-type specific effects. Comparison of the transcriptional response to platelet-derived growth factor (PDGF) and FGF in embryonic palatal mesenchyme cells showed that the FGF response was MEK dependent, while the PDGF response was PI3K dependent, resulting in the promotion of proliferation and differentiation, respectively (Vasudevan et al., 2015). Similarly, during the development of the blastocyst, FGFR1 regulates primitive endoderm (PrE) specification through ERK1/2 signaling while PDGFRα (and likely FGFR2) regulates PrE survival through PI3K signaling (Molotkov & Soriano, 2018).
Within the FGFR family, different cell types can exhibit diverse responses to FGFR signaling where an FGFR transmits receptor-specific downstream signals that are cell-type specific. For example, growth plate chondrocytes decrease proliferation and differentiation in response to FGFR3 whereas in malignant melanoma FGFR3 signaling promotes growth, metastasis, and EMT phenotypes (L. Li, Zhang, et al., 2019; Ornitz & Legeai-Mallet, 2017). In a classic study, FGFR1 and FGFR2 were found to have unique functions in cranial bone formation. The formation of a FGF ligand gradient, from high levels in the differentiated region to low levels in the environment of the osteogenic stem cells, modulates differential expression of Fgfr1 and Fgfr2. Signaling through FGFR2 was found to regulate stem cell proliferation whereas signaling through FGFR1 regulated osteogenic differentiation (Iseki et al., 1999).
In lung epithelial cells, FGF9 signaled through epithelial FGFR3 to directly promote distal epithelial fate specification and inhibit epithelial differentiation, while FGF10 signaled through epithelial FGFR2b to promote epithelial proliferation and differentiation. Analysis of downstream signaling pathways revealed that FGFR3 preferentially activated phosphoinositide 3-kinase (PI3K) pathways, whereas FGFR2b preferentially activated mitogen-activated protein kinase (MAPK) pathways. These data show that in lung epithelial cells, different FGFRs function independently to direct distinct developmental functions through the activation of distinct downstream signaling pathways (Yin & Ornitz, 2020).
3.7 |. Functions of cytoplasmic and nuclear FGF/FGFRs
FGF ligands and receptors have been observed in the nucleus and several studies have shown biological activity that is independent of FGFR tyrosine kinase activity (Decker et al., 2020; Forthmann et al., 2015; Ornitz & Itoh, 2015; Stachowiak et al., 2015; Stachowiak & Stachowiak, 2016; Tuzon et al., 2019).
Under stress conditions, exogenous FGF1 and FGF2 can be translocated into the cell via the endosomal membrane in a receptor-dependent manner. Intracellular FGF1 and FGF2 show anti-apoptotic activity, independent of receptor activation, and downstream signaling (Kostas, Lampart, et al., 2018). FGF1 and FGF2 can also be internalized in complex with FGFR1 or FGFR4 (not FGFR2 or FGFR3) and translocated from the endosome to the cytosol and then to the nucleus (Sluzalska et al., 2021). Nucleolin is an abundant nuclear phosphoprotein that directly binds FGF1 (Bober et al., 2016). Nucleolin mediates the intranuclear phosphorylation of FGF1 by protein kinase C δ (PKCδ) resulting in its export from the nucleus (Sletten et al., 2014).
FGF10 has two putative nuclear localization sequences (NLSs). In lacrimo-auriculo-dento-digital (LADD) syndrome, the p.G138E FGF10 mutation in NLS1 blocks nuclear translocation of FGF10 and reduces its secretion from cells. This study suggests that in addition to its paracrine roles, FGF10 may also have intracrine functions in FGF10-producing cells (Mikolajczak et al., 2016).
The diverse roles of nuclear FGFR1 have been termed Integrative Nuclear FGFR1 Signaling (INFS) (Decker et al., 2020; Stachowiak et al., 2015). FGFR1 has a unique transmembrane domain structure, which in the presence of FGF2 and β-Importin allows translocation to the nucleus (Stachowiak & Stachowiak, 2016). Nuclear FGFR1 has been shown to associate with topological domains in chromatin to regulate gene expression (Decker et al., 2020).
Under hypoxic conditions, FGFR2 and HIF1α co-localize and associate in the nucleus. Functionally, this interaction blocks the recruitment of coactivator p300, resulting in the repression of HIF target genes (Lee et al., 2019). Dominant mutations in the transmembrane domain of FGFR2 that cause Bent Bone Dysplasia (BBD) reduced canonical FGFR2 signaling mediated by extracellular FGFs, but increased FGFR2 nucleolar localization and epigenetically activated transcriptionally silent ribosomal DNA. This led to nucleolar disorganization, altered ribosome biogenesis, and activated the Rpl11-Mdm2-p53 nucleolar stress response pathways (Merrill et al., 2012; Neben et al., 2017; Stichelbout et al., 2016). Overexpression of BBD mutations in FGFR2 or wild-type FGFR2 with added nuclear and nucleolar localization signals, in chick lateral plate mesoderm, induced angulated hindlimbs associated with defects in skeletal muscle patterning and tendon-to-bone attachment (Salva et al., 2019; Tuzon et al., 2019). These studies link nuclear localization of FGFR2 with the etiology of BBD disease phenotypes.
4 |. TOOLS TO MANIPULATE AND MONITOR FGF SIGNALING IN VITRO AND IN VIVO
4.1 |. Optogenetics
Optogenetics commonly refers to a biological technique by which the activity of cellular proteins is controlled by light. Using natural or engineered photosensory proteins, this technique can control basic cellular functions, protein localization, and protein–protein interactions (Kim et al., 2016; Pathak et al., 2013). Optically controlled FGFR1 (opto-FGFR1) was engineered by generating a chimeric protein containing a membrane tethering myristoylation domain, the intracellular kinase domain of FGFR1, and the light-oxygen-voltage (LOV)-sensing domain of aureochrome1 (from the yellow-green alga, Vaucheria frigida). Exposure to blue light can rapidly and reversibly control intracellular FGFR1 signaling and mimic complex mitogenic and morphogenic cell behavior that would normally be induced by FGFs (Grusch et al., 2014). A similar optically controlled FGFR1 was engineered using the Arabidopsis photosensory protein cryptochrome 2 (CRY2). This construct conferred spatiotemporal control of cell polarity and induced directed cell migration (Kim et al., 2014, 2016).
A third approach was designed to cluster and activate endogenous RTKs using non-covalent interactions with engineered CRY2, in which mCherry-CRY2 was fused to the SH2 domain from PLCɣ (SH2-N). This technology, termed Clustering Indirectly using Cryptochrome 2 (CLICR), conferred light-inducible activation of endogenous RTKs. In the presence of blue light, SH2-N and FGFR-GFP showed light-dependent membrane translocation and formation of puncta with GFP and mCherry that could be blocked with an FGFR kinase inhibitor. Although this technique lacks receptor specificity it has the advantage of activating endogenous receptors (Bugaj et al., 2015).
Optogenetic tools were used to assess the relative importance of membrane-tethered FGFR1 versus cytosolic FGFR1 using mem-opto-FGFR1 containing an N-terminal myristoylation domain (Grusch et al., 2014) and cyto-opto-FGFR1 that lacks the membrane tethering signal. In response to light, mem-opto-FGFR1 induced neurite outgrowth in PC12 cells whereas cyto-opto-FGFR1 did not. Both constructs stimulated pERK in HEK293 cells. Cyto-opto-FGFR1 did not stimulate pERK in PC12 cells. These data suggest that membrane-associated FGFR1 is necessary for PC12 cell differentiation (Csanaky et al., 2019). One limitation of membrane-targeted opto-FGFR1 is its relatively high background activity even in the absence of light. In vivo, mem-opto-FGFR1 resulted in embryonic death in Xenopus laevis. To overcome this, a binary system was developed using the FGFR intracellular domain fused to CRY2 and a membrane-tethered cryptochrome-interacting basic helix–loop–helix N-terminal domain (CIBN). Together, these constructs conferred light-inducible FGFR ICD translocation to the plasma membrane (Krishnamurthy et al., 2020). Optical activation of this binary system allowed for light-mediated induction of ectopic tail-like structures in X. laevis embryos (Krishnamurthy et al., 2020). An opto-FGFR2 gene anchored the intracellular domain and LOV domain to the membrane with a myristoylation signal. Activation of opto-FGFR2 in keratinocytes in vivo turned on classical FGFR signaling pathways and target genes. However, opto-FGFR2 signaling was transient and opto-FGFR2 expression was rapidly downregulated with loss of response to light (Rauschendorfer et al., 2021).
4.2 |. Acoustically responsive scaffolds
Delivery of growth factors in a controlled and spatially regulated manner is an ongoing challenge in regenerative medicine. Acoustically responsive scaffolds (ARS) are composite fibrin hydrogels containing payload-carrying monodispersed perfluorocarbon emulsions that can be disrupted by ultrasound. Focused ultrasound results in vaporization of the perfluorocarbon emulsion (acoustic droplet vaporization). The use of ARSs and focused ultrasound provides a mean for temporal and spatial control of growth factor release. Spatially patterning suprathreshold ultrasound within FGF2 loaded ARS can be used to elicit a spatially directed response from the host (Huang et al., 2021; Lu et al., 2020). Subcutaneous implantation of an ARS loaded with FGF2 increased vascular perfusion and blood vessel density in response to suprathreshold ultrasound (Moncion et al., 2017). Sequential release of multiple payloads can be achieved with bi-layer ARSs containing different payloads within each layer that can be released by different frequencies in an ultrasound standing wave field. This technology was demonstrated by the sequential release of FGF2 and PDGFB, two growth factors that induce different angiogenic responses (Aliabouzar et al., 2020). In a hindlimb ischemia model, implantation of an ARS loaded with FGF2 improved angiogenesis and reduced fibrosis in response to focused ultrasound (Jin et al., 2021).
4.3 |. Genetic and viral vector-mediated regulation of FGF signaling
Genetic approaches to manipulate FGF signaling in vivo use mice in which ligands (FGF2, FGF7, FGF9, FGF10, and FGF18) or a constitutively activated FGFR1 can be induced to activate cell non-autonomous or cell autonomous FGF signaling, respectively (Cilvik et al., 2013; Clark et al., 2001; Koo et al., 2018; Tichelaar et al., 2000; Volckaert et al., 2017; White et al., 2006; Whitsett et al., 2002). Similarly, induced expression of dominant-negative FGFR1 or FGFR2b or the FGFR downstream pathway inhibitor SPRY4, allow cell non-autonomous or cell autonomous, respectively, suppression of FGF signaling (Eckenstein et al., 2006; Parsa et al., 2008; Perl et al., 2003; Urness et al., 2018). In zebrafish, a transgenic allele encoding a dominant-negative Fgfr1a with a fluorescent tag (fgfr1-dn-cargo) placed under combined Cre/lox and heat shock control allows spatiotemporal perturbation of FGF signaling (Kirchgeorg et al., 2018).
Lentiviral, adenoviral, and adeno-associated viral vectors have also been used to deliver FGFs, small hairpin RNAs (shRNAs), and inhibitors of FGF signaling, to locally activate or inhibit the FGF signaling pathway. Lentiviruses have been used to express Fgf2, Fgf13, Fgf23, Spry1, shFgf9, and shSpry1 (Allodi et al., 2014; Li, Wang, Wang, et al., 2018; R. Li, Tao, Huang, et al., 2021; Mao et al., 2020; Qiao et al., 2018; Tu et al., 2019; Zhao et al., 2015).
Adenoviral (Ad) vectors have been used to express Fgf1, Fgf2, Fgf4, Fgf9, and Fgf21 (Jerebtsova et al., 2015; Justet et al., 2017; Y. Li, Wong, et al., 2014; Ren et al., 2019; Rissanen et al., 2004).
Adeno-associated virus (AAV) vectors have been used to express Fgf1, Fgf2, Fgf4, Fgf10, Fgf16, Fgf15/19, Fgf21, Fgf22, shFgf22, Fgfr1, and shFgfr2 (Frisch et al., 2016; Gadaleta et al., 2020; Haenzi et al., 2016; Jazwa et al., 2010; V. Jimenez, Jambrina, et al., 2018; Li, Wang, Cai, et al., 2018; S. Li, He, et al., 2020; Luo et al., 2018; Madry et al., 2013; Sponton & Kajimura, 2018; Tang et al., 2016; Yu et al., 2016; J. Zhang, Gupte, et al., 2017). AAV-FGF10 has been shown to protect against hepatic ischemia–reperfusion (IR) injury in mice (S. Li, Zhu, Xue, et al., 2021). Mice injected with AAV-FGF15/19 (non-mitogenic variant) showed reduced bile acid synthesis and bile acid pool size and protection from intestinal inflammation (Gadaleta et al., 2020).
4.4 |. Reporter alleles for the FGF signaling pathway
A transcriptional readout of FGF/ERK activity in mice was developed by targeting a histone H2B-linked Venus fluorophore to the endogenous Spry4 locus. The Spry4H2B-Venus reporter recapitulated the expression pattern of Spry4 and localized it to sites of known FGF/ERK activity including the inner cell mass of the pre-implantation embryo, limb buds, somites, and highly localized patterns in adult organs (Morgani, Saiz, et al., 2018). However, this reporter was not sensitive enough to visualize short-term changes in signaling activity in single cells.
To develop a more sensitive reporter for FGFR and other RTK signaling, a ratiometric ERK-kinase translocation reporter was expressed in all cells of a mouse. When the ERK signaling pathway is active, activated phospho-ERK phosphorylates the mClover sensor and increases its nuclear export, resulting in cytoplasmic enrichment. When the ERK signaling pathway is inactive, the unphosphorylated sensor is imported into the nucleus, resulting in nuclear enrichment of mClover fluorescence. Therefore, the cytoplasmic to nuclear ratio of the ERK kinase translocation reporter indicates ERK activity (Regot et al., 2014; Simon et al., 2020). Imaging the mouse blastocyst revealed spatial and temporal lineage-specific dynamics in ERK signaling (Simon et al., 2020).
Etv1 (Er81), Etv4 (Pea3), and Etv5 (Erm) are direct transcriptional effectors of FGF signaling (Brent & Tabin, 2004; DeSalvo et al., 2021; di Martino et al., 2019; Ebeid & Huh, 2020; Herriges et al., 2015; Jones et al., 2019; Peluso et al., 2017; Willardsen et al., 2014; Yamamoto-Shiraishi et al., 2014; L. M. Yang, Stout, et al., 2020). Transgenic reporter mice have been developed in which a green fluorescent protein (GFP) reporter gene was inserted into the Etv4 locus in a large bacterial artificial chromosome (Lamballe et al., 2011) or in which a CreER-RFP gene was inserted into the endogenous Etv5 gene locus (Jones et al., 2019). In the mouse embryo, ETV4-GFP was detected in sites consistent with active FGF signaling (nasal placode, mandibular and maxillary processes, midbrain/hindbrain junction, anterior limb bud, mammary line, otic placode, and tailbud). In the embryonic lung, GFP was expressed in the epithelial tips and surrounding mesenchyme, sites consistent with FGF10 and FGF9 signaling, respectively (Jones et al., 2019; Yin & Ornitz, 2020). ETV5-RFP was expressed in the lung epithelium, a target of FGF10 signaling, and was decreased when FGF10 signaling was inhibited by the expression of a soluble FGFR2b extracellular domain which functions as a ligand trap (Jones et al., 2019). However, Etvs 1, 4, and 5 are not exclusively regulated by FGFR signaling. For example, in primitive endoderm, FGF signaling regulates DUSP4, but ETV5 production does not depend on direct activation by ERK but instead requires NANOG activity (Azami et al., 2019).
In zebrafish, the Dusp6 promoter driving GFP or a Dusp6 enhancer trap line expressing GFP has been used to monitor patterns of FGF signaling (Cavanah et al., 2021; Molina et al., 2007).
5 |. THERAPEUTIC ACTIVATION OF FGF SIGNALING PATHWAYS
Activation of the FGF signaling pathway can be achieved by the delivery of FGF ligands, by molecules that mimic FGF ligands, or by molecules that increase the stability of FGF ligands or receptors (Table 1).
TABLE 1.
Therapeutic activation of FGF signaling pathways
| Mechanism, molecular target | Clinical, animal, or cellular target | Alternative or trade name | FDA approved/clinical trial | References | |
|---|---|---|---|---|---|
| (A) Paracrine FGF signaling | |||||
| Recombinant FGF proteins | |||||
| FGF1 (Mutant, reduced HS binding) | Pan FGFR | Decrease in myocardial infract size (rat) | C. Huang, Liu, et al., 2017 | ||
| FGF1 (Stabilized derivative) | Pan FGFR | Protection in a chemical injury corneal epithelium | TTHX1114 | Clinical trial | Eveleth et al., 2018, 2020 |
| FGF2 | FGFR (c splice variant) | Wound healing | Barrientos et al., 2014; Koike et al., 2020 | ||
| LET-bFGF (FGF2) | FGFR (c splice variant) | Lung endothelial targeted FGF2. Protection in radiation-induced lung injury | LET-bFGF | Giordano et al., 2008; Guan et al., 2018 | |
| FGF2 (Lacks N-terminal 26 amino acid residues) | FGFR (c splice variant) | FGFR1-mediated oligodendrocyte mitogenicity | F2 V2 | Thummler et al., 2019 | |
| FGF7 | FGFR (b splice variant) | Protection in oral mucositis | Palifermin | FDA approved | Raber-Durlacher et al., 2013 |
| FGF9 (Oleosin-FGF9 fusion protein) | FGFR (c splice variant) | Promotes hair growth and wound healing | Oleosin-rhFGF9 | Cai et al., 2018 | |
| FGF10 | FGFR (b splice variant) | Corneal wound healing | KGF-2 | Cai et al., 2019 | |
| FGF10 | FGFR (b splice variant) | Protection in middle cerebral artery occlusion (mouse) | Y. H. Li, Fu, Tian, et al., 2016 | ||
| FGF10 | FGFR (b splice variant) | Protection in lung injury (rat) | KGF-2 | Tong et al., 2016; Bi et al., 2014; Fang et al., 2014; Feng et al., 2016; Liu, Song, et al., 2020; L. Liu, Xia, et al., 2019; Tenghao et al., 2019, 2020 | |
| FGF10 | FGFR (b splice variant) | Protection in myocardial infract (mouse) | Z. Wang, Huang, et al., 2021 | ||
| FGF18 | FGFR (c splice variant) | Improvement in Parkinson's disease outcomes (rat) | Guo et al., 2017 | ||
| FGF18 | FGFR (c splice variant) | Treatment of osteoarthritis | Sprifermin | Clinical trial | McClurg et al., 2021; Xie, Zinkle, et al., 2020 |
| Antibody agonists | |||||
| FGFR1-specific monovalent antibody fragments | FGFR1 N-terminal D1 domain | Activates FGFR1 in the absence of ligand | Opalinski et al., 2017 | ||
| FGFR1/βKlotho-bispecific antibody | FGF21 mimetic | Sustained stimulation of thermogenesis BAT (mouse) | Kolumam et al., 2015; Sonoda et al., 2017 | ||
| Oligonucleotide aptamer agonists | |||||
| DNA aptamer (76-mer ssDNA tandem dimer) | FGF2 mimetic | Self-renewal and pluripotency of iPSCs | TD0 | Ueki et al., 2019 | |
| RNA aptamer dimer | FGFR3 | BaF3 cell line expression FGFR3 | Kamatkar et al., 2019 | ||
| B. Endocrine FGF signaling | |||||
| Recombinant FGF proteins | |||||
| FGF15/19 (Nontumorigenic variant biased to FGFR4) | FGFR4/βKlotho | Primary sclerosing cholangitis. Nonalcoholic steatohepatitis |
NGM282 | Clinical trial | Luo et al., 2014; Hirschfield et al., 2019; Harrison et al., 2018 |
| FGF21-PEG (extended half-life) | FGFR1/βKlotho | Normalized plasma glucose in mouse models for diabetes | Diener et al., 2021 | ||
| FGF21-PEG (extended half-life) | FGFR1/βKlotho | Nonalcoholic steatohepatitis (mice, monkey) | B1344 | Cui et al., 2020; Sanyal et al., 2019 | |
| FGF21 antibody conjugate (extended half-life) | FGFR1/βKlotho | Decreased food intake and body weight (monkey) | PF-05231023 | Huang et al., 2013; Talukdar, Zhou, et al., 2016 | |
| Fc-FGF21 (extended halflife) | FGFR1/βKlotho | Improvement in insulin sensitivity and lipid metabolism | AKR-001 | Clinical trial | Kaufman et al., 2020 |
| Antibody agonist | |||||
| Anti-FGFR1/βKlotho antibody | FGFR1/αKlotho FGFR1/βKlotho |
Improvement of obesity-related metabolic derangements; sweet and alcohol preference; improvement of motor performance; anti-inflammatory effects in ALS (mouse) | R1Mab1, 39F7, BFKB8488A, bFKB1 | Wu et al., 2011, 2013; Kolumam et al., 2015; M. Z. Chen, Chang, et al., 2017; Lan et al., 2017; Sonoda et al., 2017; Han, Du, et al., 2018; Min et al., 2018; S. Y. Shi, Lu, et al., 2018; Baruch et al., 2020; Delaye et al., 2021 | |
| Small molecular inhibitor | |||||
| FAP (fibroblast activation protein) inhibitor | FAP | Inhibits protease responsible for Cterminal cleavage of FGF21; increases serum FGF21 levels (mouse) | Talabostat, BR103354 | Cho et al., 2020; Sanchez-Garrido et al., 2016 | |
5.1 |. Recombinant proteins
The simplest way to activate the FGF pathway is to deliver recombinant FGF ligand to a target tissue. Recombinant FGFs are being used or evaluated for tissue repair and regeneration or prevention of tissue damage. FGFs that are in the therapeutic pipeline or being used in the clinic include FGF1, FGF2, FGF7, FGF10, FGF18, FGF15/19, and FGF21 (Hui et al., 2018; Xie, Zinkle, et al., 2020).
In a rat model for myocardial infarction, treatment with an FGF1 mutant with reduced HS binding decreased infarct size compared treatment with wild-type FGF1 (C. Huang, Liu, et al., 2017).
Chemical and physical agents can damage the corneal epithelium, leading to pain, and impaired vision. During development, FGFR2 is important for corneal epithelial proliferation and differentiation (Zhang et al., 2015). Corneal endothelial dystrophies, such as Fuchs endothelial corneal dystrophy, have a progressive loss of corneal ECs leading to edema, corneal opacity, and loss of vision (Vedana et al., 2016). A stabilized derivative of FGF1 (TTHX1114) was found to be protective in a chemical injury corneal epithelial explant assay and to stimulate proliferation in cultured human corneal ECs and is currently in clinical trials (Eveleth et al., 2018, 2020). In vivo, in cryo-lesioned cornea, TTHX1001 (a similar mutant FGF1), enhanced corneal EC regeneration and showed a significant clearing of opacity relative to untreated controls (Weant et al., 2021). Comparison of the efficacy of FGF2 and FGF10 in healing a corneal wound (alkali burn model) showed that FGF10 (KGF2) was more effective than FGF2, reflecting differences in either the relative expression levels or signaling properties of FGFR2b versus FGFR2c splice variants (Cai et al., 2019). Notably, FGF1 can activate all FGFR splice variants including FGFR2b and FGFR2c (Ornitz et al., 1996).
FGF2, either wild type or a stabilized mutant, is in clinical use for diabetic ulcer and burn wound healing (Barrientos et al., 2014). FGF2 treatment of wounds promotes keratinocyte EMT to accelerate wound closure (Koike et al., 2020). However, the stimulation of EMT and fibrosis by FGFs is context-dependent and in a mouse model of pulmonary fibrosis, intratracheal administration of recombinant FGF2 decreased fibrosis in part through decreased collagen expression and decreased fibroblast to myofibroblast differentiation (Dolivo et al., 2017; Koo et al., 2018). In FGFR1-amplified lung cancer, FGF2 promoted cell proliferation, EMT, and metastasis through FGFR1-ERK1/2-SOX2 signaling (K. Wang, Ji, et al., 2018).
FGF2 is protective for radiation-induced pulmonary EC apoptosis and subsequent radiation-induced lung injury. To improve the delivery of FGF2 to pulmonary ECs, the lung EC targeting peptide (LET) was fused to the FGF2 coding sequence (LET-bFGF) (Giordano et al., 2008; Guan et al., 2018). Following intravenous injection, recombinant LET-bFGF compared to native FGF2 preferentially accumulated in lung tissue and showed improved protection against radiation-induced EC apoptosis (Guan et al., 2018).
Deletion of the N-terminal 26 amino acids of FGF2 resulted in a protein (F2 V2) that retained the ability of the native protein to activate FGFR1, but significantly reduced its ability to activate FGFR2, FGFR3, and FGFR4. In oligodendrocyte regeneration, F2 V2 retained FGFR1-mediated mitogenicity without FGFR2 or FGFR3 mediated inhibition of myelin production (Thummler et al., 2019).
Oleosin is a structural oil body membrane protein in safflower seed oil bodies. A fusion protein of oleosin and FGF9 (Oleosin–rhFGF9) allowed production of oil body-bound FGF9. Oil body-bound oleosin–rhFGF9 was shown to promote both hair growth and wound healing when applied topically (Cai et al., 2018).
Oral mucositis is a severe complication during chemo- and/or radiation therapy. Oral mucositis can be prevented by treatment with recombinant FGF7 (Palifermin) (Raber-Durlacher et al., 2013; Sadeghi et al., 2021).
In a mouse middle cerebral artery occlusion model, administration of FGF10 into the lateral ventricle decreased brain infarct volume and neurological deficits and reduced neuronal cell death and expression of inflammatory factors (TNF-α and IL-6) (Y. H. Li, Fu, Tian, et al., 2016). In the lung, intratracheal delivery of FGF10 was shown to activate lung-resident mesenchymal stem cells that are protective in a rat model of LPS-induced lung injury (Tong et al., 2016). FGF10 was also protective in models of lung injury induced by particulate matter, oleic acid, bacterial infection, ischemia–reperfusion injury, and ventilator-induced lung injury (Bi et al., 2014; Fang et al., 2014; Feng et al., 2016; L. Liu, Song, et al., 2020; L. Liu, Xia, et al., 2019; Tenghao et al., 2019, 2020). FGF10, stabilized as a biodegradable coacervate containing heparin was protective in a myocardial infarct model when injected directly into the heart just after infarction. Free FGF10 with heparin showed no effect in this model (Z. Wang, Huang, et al., 2021).
Striatal infusion of FGF18 improved outcomes in a rat model of Parkinson’s disease (Guo et al., 2017). Infusion of FGF18 into joints has shown promising results for the treatment of osteoarthritis (OA; McClurg et al., 2021; Xie, Zinkle, et al., 2020). This is discussed in more detail as follows.
The therapeutic uses of endocrine FGFs (FGF15/19, 21, and 23) are discussed as follows.
5.2 |. Antibody agonists
The N-terminal D1 domain of FGFR1 negatively regulates ligand binding to the receptor. A D1-specific monovalent antibody fragment can activate FGFR1 and its downstream signaling cascades in the absence of ligand (Opalinski et al., 2017).
A monoclonal anti-FGFR1 antibody (R1MAb1) activates FGFR1 in the kidney distal tubules to normalized blood pressure and attenuate left ventricular hypertrophy in the Hyp mouse model of excess FGF23 (X. Han, Ross, et al., 2018; Wu et al., 2013). The R1MAb1 antibody may also activate FGF21 signaling pathways (Wu et al., 2011). Additional recombinant bispecific antibodies have been developed that activate the FGFR1/βKlotho complex to mimic the activity of FGF21 (discussed later) (Kolumam et al., 2015; Sonoda et al., 2017).
5.3 |. Oligonucleotide aptamer agonists
Aptamers are short single-stranded DNA or RNA oligonucleotides that fold into a vast set of tertiary structures depending on their unique primary structures. Aptamers can function as molecular mimics of proteins including antibodies and can be used to enhance or suppress a variety of molecular interactions (Adachi & Nakamura, 2019; Nakamura, 2018).
A DNA aptamer synthesized as a tandem dimer (TD0) was developed that binds FGFR1 and functionally mimics FGF2 receptor activation. The TD0 aptamer could fully replace FGF2 to support the self-renewal and pluripotency of iPSCs in short term, but not long term, cultures (Ueki et al., 2019). Similarly, an RNA aptamer was developed that binds FGFR3 but not FGFR1 or FGFR2. As a monomer, this aptamer inhibits FGFR3 signaling; however, when dimerized this aptamer is converted into a functional activator of FGFR3 signaling (Kamatkar et al., 2019).
6 |. DEVELOPMENT OF FGF PATHWAY INHIBITORS AS PHARMACEUTICALS
Monoclonal antibodies targeting FGFs and FGFRs, ligand traps (soluble FGFR ligand binding domains), small-molecule FGFR inhibitors, and aptamers are being developed for the treatment of a variety of genetic, metabolic, and oncologic human diseases (Table 2) (Chioni & Grose, 2021; Grabner et al., 2015; Herbert et al., 2014; Ho et al., 2009; Liu et al., 2016; Ornitz & Itoh, 2015; Ornitz & Legeai-Mallet, 2017; Presta et al., 2017; Szymczyk et al., 2021; Weaver & Bossaer, 2021). Among them, some medications including Burosumab, Erdafitnib, and Pemigatinib have been approved for therapeutic use by the US Food and Drug Administration (FDA).
TABLE 2.
Therapeutic inhibition of FGF signaling pathways
| Mechanism, molecular target | Clinical, animal, or cellular target | Alternative or trade name | FDA approved/clinical trial | References | |
|---|---|---|---|---|---|
| A. Paracrine FGF signaling | |||||
| Small molecule inhibitors | |||||
| FGFR inhibitor | FGFR2, FGFR3 | Metastatic urothelial carcinoma | Erafitibnib | FDA approved | Hanna, 2019 |
| FGFR inhibitor | FGFR2, FGFR3 | Cholangiocarcinoma | Pemiganib, Infigratinib | FDA approved | Hoy, 2020; Kang, 2021; Rizzo et al., 2021 |
| FGFR inhibitor | FGFR2, FGFR3 | Achondroplasia (mouse), urothelial, hepatocellular, squamous cell lung cancer (human) | ASP5878 | Clinical trial | Futami et al., 2017; Kikuchi et al., 2017; Ozaki et al., 2020; Yamamoto et al., 2020 |
| FGFR inhibitor | FGFR2, FGFR3 | Pulmonary fibrosis; gastrointestinal stromal tumor; hepatocellular carcinoma; urothelial carcinoma; tumor-induced osteomalacia; Achondroplasia(mouse) | Infigratinib (BGJ398) | Komla-Ebri et al., 2016; Morizumi et al., 2020; Botrus et al., 2021; Florenzano et al., 2021; Hartley et al., 2020; Kelly et al., 2019; Pal et al., 2018; Prawira et al., 2021 | |
| FGFR inhibitor | FGFRs 1–4 | Urothelial, hepatocellular, squamous cell lung carcinoma | ASP5878 | Clinical trial | Yamamoto et al., 2020 |
| FGFR inhibitor | FGFRs 1–3 | Intrahepatic carcinoma | Derazantinib (ARQ087) | Clinical trial | Papadopoulos et al., 2017; Mazzaferro et al., 2019 |
| FGFR inhibitor (covalent) | FGFRs 1–4 | TAS-120 | Clinical trial | Kalyukina et al., 2019 | |
| FGFR inhibitor (covalent) | FGFRs 1–4 | Cancer cells, xenograft | PRN1371 | Clinical trial | Venetsanakos et al., 2017; Brameld et al., 2017 |
| FGFR inhibitor (covalent) | FGFRs 1–4 | Cholangiocarcinoma | Futibatinib | Clinical trial | Meric-Bernstam et al., 2021 |
| FGFR inhibitor (covalent) | FGFR4 selective | Hepatocellular carcinoma | H3B-6527 | Clinical trial | Joshi et al., 2017 |
| FGFR inhibitor (covalent) | FGFR4 selective | Hepatocellular carcinoma | Fisogatinib (BLU-554) | Clinical trial | Hatlen et al., 2019; Kim, Joe, Choi, et al., 2019 |
| FGFR inhibitor (covalent, reversible) | FGFR4 selective | Hepatocellular carcinoma | Roblitinib | Clinical trial | Fairhurst et al., 2020 |
| THBS1 or PTX3 based compounds | FGF2 | Antiangiogenic drugs (see NSC12 below) | Foglieni et al., 2016; Pagano et al., 2012; Presta et al., 2018; Ronca, Giacomini, Di Salle, et al., 2015; Ronca et al., 2017 | ||
| NSC12 (steroid derivative) | Pan-FGF | Extracellular pan-FGF trap derived from PTX3 structure | NSC12 | Castelli et al., 2016, 2021; Ronca, Giacomini, Di Salle, et al., 2015 | |
| MEK inhibitor and FGFR inhibitor | MEK and FGFR1 | Synergistical inhibition of cell proliferation in KRAS-mutant lung cancer | Trametinib and Ponatinib | Manchado et al., 2016 | |
| Peptide inhibitors | |||||
| FGF8b (13 amino acid peptide) | FGF8b–FGFR | Inhibition of prostate cancer cell line and HUVEC proliferation | 8b-13 | Li et al., 2013; Lin et al., 2017 | |
| 12 amino acid peptide | FGF2–FGFR1 | Attenuation of cartilage loss in osteoarthritis; attenuation of tumor growth and angiogenesis in xenograft model (mouse) | R1-P2, R1-P1 | Tan, Chen, et al., 2018; Tan, Wang, et al., 2018 | |
| Cyclic 10 amino acid peptide | FGF1–FGFR1 | Inhibition of FGF1 induced proliferation of BaF3 cells that express FGFR1c | F8 | Lipok et al., 2019; Ornitz et al., 1996 | |
| 12 amino acid peptide | FGFR3 | Rescues of viability of mice with Thanatophoric dysplasia type II mutation; inhibition of 9-cisRAinduced tracheal lymphangiogenesis (mouse) | P3 | Jin et al., 2012; Perrault et al., 2019 | |
| C-type natriuretic peptide (CNP) | NPR2 | Achondroplasia; Inhibition of FGFR3 induced ERK phosphorylation in growth plate chondrocytes | TransCon CNP; Vosoritide | Clinical trial | Breinholt et al., 2019; Savarirayan et al., 2020 |
| Protein or antibody inhibitors | |||||
| Soluble Fc-FGFR1c decoy receptor | FGF ligands | Decrease of tumor-associated angiogenesis in vivo; mesothelioma; non-small cell lung cancer | GSK3052230, FP-1039 | Clinical trial | Harding et al., 2013; Blackwell et al., 2016; Morgensztern et al., 2019; van Brummelen et al., 2020 |
| Anti-FGFR2b antibody | FGFR2b | Inhibition of advanced gastroesophageal cancer | Bemarituzumab | Clinical trial | Catenacci et al., 2019, 2020 |
| Anti-FGFR2-auristatin | FGFR2/microtubuledisrupting agent | Adverse effects in clinical trials | Aprutumab ixadotin | Clinical trial | S. B. Kim, Meric-Bernstam, Kalyan, et al., 2019 |
| Anti-FGFR3 antibody | FGFR3 | Inhibition of lung adenocarcinoma (mouse) | D11 | Yin et al., 2016 | |
| Soluble FGFR3 decoy receptor | FGF ligands | Increase of skeletal growth and reduction of visceral obesity in Achondroplasia (mouse) | Recifercept | Garcia et al., 2013; Goncalves et al., 2020; Saint-Laurent et al., 2018; Unger et al., 2017 | |
| Oligonucleotide aptamer inhibitors | |||||
| RNA aptamer to FGF2 | FGF2 | Improvement of arthritis, osteoporosis, and Achondroplasia, inhibition of neoangiogenesis in models of agerelated macular degeneration (mouse, rat); strong analgesic effect for bone cancer pain (mouse) | RBM-007 | Clinical trial | Jin et al., 2016; Hamamoto et al., 2018; Matsuda et al., 2019; Kimura et al., 2021; Nakamura, 2018, 2021 |
| RNA aptamer to FGF5 | FGF5 | Inhibition of FGF5-induced cell proliferation | F5f1–56 | Amano et al., 2021 | |
| DNA aptamer to FGFR1 | FGFR1 | Killing of FGFR-1 expressing human osteosarcoma cells | A11-G18T | Jurek et al., 2017 | |
| B. Endocrine FGF signaling | |||||
| Protein or antibody inhibitors | |||||
| Anti-FGF15/19 N-terminus monoclonal antibody | FGF15/19 | Inhibition hepatocellular carcinoma growth without bile-acid related side effects (mouse, monkey) | H. Liu, Zheng, et al., 2020 | ||
| FGF23 C-tail-Fc fusion | FGFR/αKlotho | Increase of serum phosphate in X-linked | FGF23 c-tail Fc | Johnson et al., 2017 protein | hypophosphatemia (mouse) |
| Anti-FGF23 monoclonal antibody | FGF23 | Treatment of chronic kidney disease; Xlinked hypophosphatemia | Burosumab (KRN23) | FDA approved | Carpenter et al., 2014, 2018; Fukumoto, 2014, 2021; X. Zhang, Ibrahimi, et al., 2006; Kutilek, 2017; Insogna et al., 2018; Lamb, 2018; Imel et al., 2019; Imel, 2021; Michigami, 2019; Dahir et al., 2020; Schindeler et al., 2020; Balani & Perwad, 2020; Schindeler et al., 2020; Gladding et al., 2021; Verbueken & Moe, 2021; Jan de Beur et al., 2021 |
| Anti-FGFR4 antibody | FGFR4 | Inhibition of FGF23-induced left ventricular hypertrophy (rat) | Grabner et al., 2015, 2017 | ||
| Peptide inhibitor | |||||
| FGF23 C-terminal 72 amino acid peptide | FGFR1/αKlotho | Prevention of FGF23-mediated anemia and iron deficiency (hypoferremia) in chronic kidney disease (mouse) | Agoro et al., 2018, 2021 |
6.1 |. Inhibitors with FDA approval
6.1.1 |. Burosumab (anti-FGF23 antibody)
Increased expression of FGF23 is a pathogenic feature of X-linked hypophosphatemia (XLH) and other conditions that cause hypophosphatemia, including tumor-induced osteomalacia (TIO), fibrous dysplasia of the bone, and cutaneous skeletal hypophosphatemia syndrome (Dahir et al., 2020; Imel et al., 2019). To treat these and related diseases, a neutralizing antibody for FGF23 (Burosumab, previously KRN23) has been developed and is now FDA approved for treating XLH (Carpenter et al., 2014, 2018; Insogna et al., 2018; Kutilek, 2017; Lamb, 2018; X. Zhang, Imel, et al., 2016). Treatment with Burosumab improves biochemical abnormalities (serum phosphate, alkaline phosphatase, and 1,25 (OH)2D levels), radiographic signs of rickets (delayed growth, bow legs, weakness, and bone pain due to hypophosphatemia), growth, fracture healing, and impaired mineralization in patients with XLH (Dahir et al., 2020; Fukumoto, 2014, 2021; Gladding et al., 2021; Imel, 2021; Michigami, 2019; Schindeler et al., 2020; Verbueken & Moe, 2021). In adults with TIO, Burosumab was associated with improvements in phosphate metabolism and osteomalacia (Imel et al., 2015; Jan de Beur et al., 2021; Whyte, 2021).
Increased FGF23 levels are a pathogenic feature of chronic kidney disease (CKD) (Czaya & Faul, 2019). High serum levels of FGF23 are also a direct cause of left ventricular hypertrophy (mediated by cardiomyocyte FGFR4) and are associated with increased cardiovascular mortality (Faul, 2018; Grabner et al., 2015). In a rat model of CKD, blockade of FGFR4 attenuated cardiac hypertrophy, suggesting a therapeutic approach to cardiac effects of CKD (Grabner et al., 2017). FGF23 blockade with Burosumab is being evaluated as a therapy for cardiovascular complications of CKD (Balani & Perwad, 2020).
6.1.2 |. Erdafitnib, Pemigatinib, and Infigratinib (small molecule FGFR tyrosine kinase inhibitors)
Deregulation of FGF signaling pathways has been implicated in many types of human cancers. Deregulation can occur at the level of gene/protein expression of ligands or receptors, which can result from changes in transcriptional activity or gene amplification. Deregulation can also result from mutations in FGF ligands, receptors, or downstream signaling pathways (Katoh, 2016; Krook et al., 2021; Ornitz & Itoh, 2015). Erdafitnib (JNJ-42756493), Pemigatinib (INCB054828), and Infigratinib (BGJ398) are small molecule drugs with distinct molecular structures that block FGFR signaling by inhibiting their intracellular tyrosine kinase activities. Erdafitinib was approved for the treatment of locally advanced or metastatic urothelial carcinoma with FGFR2 and FGFR3 alterations (Al-Obaidy & Cheng, 2021; Bansal et al., 2021; D’Angelo et al., 2020; Hanna, 2019; Loriot et al., 2019; Nauseef et al., 2020; Perera et al., 2017; Roubal et al., 2020). Pemigatinib and Infigratinib were approved for the treatment of unresectable or metastatic cholangiocarcinoma with FGFR2 alterations (Hoy, 2020; Kang, 2021; Rizzo et al., 2021).
6.1.3 |. Potential adverse effects of FGFR inhibition
FGF23 is an endocrine hormone that is primarily produced by osteocytes and signals to the kidney proximal tubules through a heterodimeric receptor complex, comprising predominantly FGFR1 and its co-receptor αKlotho. FGF23 signaling suppresses urinary phosphate resorption and suppresses circulating concentrations of 1,25-dihydroxyvitamin D. These and other activities of FGF23 lead to decreased serum phosphate (Bacchetta et al., 2020; Erben, 2018; Erben & Andrukhova, 2017; Ho & Bergwitz, 2021). In contrast, in αKlotho or Fgf23 deficient mice, levels of 1,25-dihydroxyvitamin D are increased causing hypercalcemia, hyperphosphatemia, ectopic calcifications, impaired bone mineralization, and early lethality. Similarly, mice that inactivate Fgfr1 in proximal renal tubules showed increased sodium-dependent phosphate co-transporter expression and hyperphosphatemia consistent with a direct role of FGFR1 in mediating the proximal tubular phosphate responses to FGF23 (Han et al., 2016). Treatment of rats with a small molecule FGFR inhibitor (PD-0176067) for 4 days resulted in increased levels of serum FGF23 and phosphate (Yanochko et al., 2013). These and many other data suggest that inhibition of the FGF23–FGFR1 signaling axis in the kidney proximal tubule would perturb phosphate homeostasis. Consistent with this, the major reported adverse effects of treatment with Erdafitinib (median 5.3 months) or other FGFR inhibitors were hyperphosphatemia, musculoskeletal pain, increased alkaline phosphatase, and metastatic calcinosis cutis (a dermatological condition caused by widespread deposition of calcium phosphate in the skin) (Arudra et al., 2018; Carr et al., 2019; Janssen_Pharmaceuticals, 2019; Markham, 2019). These and other adverse effects of FGFR inhibition could limit their long-term use as cancer therapeutics.
In a recent study, Fgfr1 (or Fgfr1 and Fgfr2) was inactivated in mature osteoblasts and osteocytes. Histopathological analyses show a striking loss of osteocytes and secondarily increased bone characterized by increased periosteal apposition, increased and disorganized endocortical bone with increased porosity, and biomechanical properties that reflect increased bone mass but impaired material properties. These data identify a homeostatic role for FGFR1 signaling in mature osteoblasts/osteocytes that is directly or indirectly required for osteocyte survival (McKenzie et al., 2019). It is not known whether therapeutic inhibition of FGFR signaling could adversely affect the homeostatic properties of bone.
6.2 |. The FGF/FGFR inhibitor pipeline
6.2.1 |. Small molecule inhibitors
A relatively large number of additional small-molecule FGFR inhibitors have been developed and are being evaluated for genetic and physiological diseases and cancer (Al-Obaidy & Cheng, 2021; Chioni & Grose, 2021; Weaver & Bossaer, 2021). For example, in preclinical models, ASP5878 and Infigratinib (BGJ398) show efficacy in a mouse model for Achondroplasia, which is caused by activating mutations in FGFR3 (Komla-Ebri et al., 2016; Ornitz & Legeai-Mallet, 2017; Ozaki et al., 2020). However, in a mouse model of pulmonary fibrosis, Infigratinib reduced fibrosis, but also inhibited epithelial homeostasis and repair, leading to increased mortality (Morizumi et al., 2020), demonstrating the need for more selective FGFR inhibitors or cell-type-specific targeting.
In human clinical trials, Infigratinib is being evaluated as a therapy for cholangiocarcinoma (now FDA approved), advanced gastrointestinal stromal tumor, hepatocellular carcinoma, and urothelial carcinoma with FGFR3 alterations, and TIO (Botrus et al., 2021; Hartley et al., 2020; Kardoust Parizi et al., 2021; Kang, 2021; Kelly et al., 2019; Pal et al., 2018; Prawira et al., 2021). ASP5878 is being evaluated for treatment of patients with urothelial carcinoma, hepatocellular carcinoma, or squamous cell lung carcinoma (Futami et al., 2017; Kikuchi et al., 2017; Yamamoto et al., 2020). ARQ087 (Derazantinib) is being evaluated for intrahepatic cholangiocarcinomas with FGFR2 gene fusions (Mazzaferro et al., 2019; Papadopoulos et al., 2017). In the case of hepatocellular carcinoma, resistance to Infigratinib has been attributed to increased expression of lp-ErbB2 and p-ErbB3 (Prawira et al., 2021). In gastric cancer cell lines, combination treatment with an FGFR inhibitor and the multikinase inhibitor, regorafenib, delayed MAPK/ERK reactivation and synergistically inhibited proliferation, circumventing acquired resistance to FGFR only (Infigratinib, Erdafitinib, or TAS-120) inhibition (Lau et al., 2021).
To achieve long-term FGFR inhibition for cancer, irreversible covalent FGFR inhibitors have the advantage of a longer effective half-life and possibly reduced adverse effects. Futibatinib, Roblitinib, H3B-6527, Fisogatinib (BLU-554), TAS-120, and PRN1371, were identified (Brameld et al., 2017; Fairhurst et al., 2020; Hatlen et al., 2019; Joshi et al., 2017; Kalyukina et al., 2019; R. D. Kim, Sarker, Meyer, et al., 2019; Kommalapati et al., 2021; Lee et al., 2021; Meric-Bernstam et al., 2021; Roskoski Jr., 2020; Venetsanakos et al., 2017). These inhibitors target FGFRs by interacting with a cysteine residue within the kinase domain active site.
Small molecule inhibitors derived from FGF2 binding peptides in thrombospondin-1 (THBS1) and long pentraxin-3 (PTX3; discussed earlier) show promise as antiangiogenic drugs (Castelli et al., 2021; Foglieni et al., 2016; Pagano et al., 2012; Presta et al., 2018; Ronca et al., 2017; Ronca, Giacomini, Di Salle, et al., 2015). Thrombospondin-1 based molecules are 7 bi-naphthalenic compounds and PTX3 based molecules are steroidal derivatives with IC50s in the low μM range (Castelli et al., 2021; Foglieni et al., 2016).
In KRAS-mutant lung cancer, treatment with the MEK inhibitor, Trametinib, leads to a compensatory (adaptive resistance) increased FGFR1 signaling. Combining treatment with Trametinib and the multikinase FGFR1 inhibitor Ponatinib synergistically inhibited cell proliferation and FRS2 phosphorylation (Manchado et al., 2016).
6.2.2 |. Peptide inhibitors
As discussed above, long PTX3 is an effective inhibitor of tumor growth and angiogenesis (Ronca, Giacomini, Di Salle, et al., 2015). To generate a potential pharmacological inhibitor, an N-terminal pentapeptide derived from PTX3 was identified that binds FGF2 and FGF8b and inhibits their ability to form a ternary complex with FGFR1 and HS and their ability to induce EC angiogenic activity (Giacomini et al., 2015; Ronca, Giacomini, Di Salle, et al., 2015). Based on the binding properties of the PTX3-derived FGF-binding pentapeptide with FGF2, a small-molecule steroidal derivative (NSC12) was identified that functions as an extracellular FGF trap that blocks FGF2 receptor binding without interfering with heparin binding (Castelli et al., 2016; Ronca, Giacomini, Di Salle, et al., 2015). Recently, NSC12 was shown to trigger mitochondrial oxidative stress, DNA damage, and apoptotic cell death via the proteasomal degradation of c-Myc in multiple myeloma cells or xenografts (Ronca et al., 2020).
Inhibitory peptides can also be derived directly from ligand sequences. A 13 amino acid peptide that mimics part of FGF8b inhibited the proliferation of prostate cancer cells and the proliferation and migration of human umbilical vein endothelial cells (HUVECs) (Li et al., 2013; Lin et al., 2017). When injected intracerebroventricularly (third ventricle), this peptide inhibited endocrine FGF signaling (FGFs 15/19 and 21) and improved glucose tolerance (S. Liu, Marcelin, et al., 2018).
A 72 amino acid peptide derived from the C-terminus of FGF23 was shown to block its binding to the FGFR1-αKlotho complex (Goetz et al., 2010). This peptide was effective at preventing FGF23 mediated anemia, iron deficiency (hypoferremia), and diabetic nephropathy that are complications of CKD or inflammation (Agoro et al., 2018, 2021; X. Zhang, Guo, et al., 2018). A FGF23 C-tail-Fc fusion protein similarly inhibits FGF23 binding and increases serum phosphate in a mouse model of hypophosphatemic rickets (Hyp mouse) (Johnson et al., 2017). However, it has also been suggested that endogenous FGF23 C-terminal peptide is unlikely to inhibit FGF23 binding, as it is present in high levels in humans with iron deficiency and FGF23 deficiency phenotypes (tumoral calcinosis) are not seen in these patients (Johnson et al., 2017).
Two different 12 amino acid peptides that bind to and inhibit FGFR1 were identified and shown to functionally attenuate cartilage loss in a mouse model for osteoarthritis (OA) and in a xenograft model for tumor growth and angiogenesis (Tan, Chen, et al., 2018; Tan, Wang, et al., 2018). A cyclic 10 amino acid peptide that binds the FGF1–FGFR1 interface was identified by phage display and shown to inhibit FGF1 induced proliferation of BaF3 cells that express FGFR1c (Lipok et al., 2019; Ornitz et al., 1996).
A 12 amino acid peptide (P3) that binds and inhibits FGFR3 was identified through screening a phage library. The P3 peptide binds the extracellular domain of FGFR3 and inhibits downstream signaling. In vivo, the P3 peptide was able to rescue the viability of mice with the strong activating Thanatophoric dysplasia type II (TDII) mutation (Jin et al., 2012). The P3 peptide was able to inhibit FGFR3-mediated 9-cis retinoic acid (9-cisRA)-stimulated lymphatic EC proliferation in vitro and 9-cisRA-induced tracheal lymphangiogenesis in vivo (Perrault et al., 2019).
6.2.3 |. Protein and antibody inhibitors
Activation of FGFRs by congenital mutation, acquired mutation, or FGF ligand expression, results in genetic diseases (e.g., Achondroplasia, craniofacial malformations, and craniosynostosis), metabolic disease, and a variety of cancers (Acquaviva et al., 2014; J. Chen, Liu, et al., 2017; Desai & Adjei, 2016; Moosa & Wollnik, 2016; Narayana & Horton, 2015; Ornitz & Legeai-Mallet, 2017; Shinmura et al., 2014). Monoclonal antibodies have been developed that target the FGFR extracellular domain to inhibit ligand binding or receptor dimerization. Soluble FGFR extracellular ligand binding domains function as ligand traps that sponge FGF ligands and block their signaling to membrane FGFRs. The use of receptor-specific monoclonal antibodies to FGFRs 2, 3, or 4 and soluble FGFR ligand traps have advantages over pan-FGFR tyrosine kinase inhibitors in that hyperphosphatemia, the main adverse effect of FGFR1 inhibition in the kidney, is avoided. Additionally, antibody–drug conjugates are being developed to target cancers that have increased expression of specific FGFRs.
Soluble extracellular domains of FGFR1c, FGFR2b, FGFR2c, and FGFR3c have been developed as ligand trap inhibitors and have been evaluated in vivo (Blackwell et al., 2016; Garcia et al., 2013; Goncalves et al., 2020; Liu et al., 2016; Saint-Laurent et al., 2018; Tolcher et al., 2016). As an example, mesothelioma and other lung cancers express high levels of FGF2 and FGFR1 (Marek et al., 2014; Wynes et al., 2014). A soluble Fc-FGFR1c decoy receptor (GSK3052230) inhibited proliferation of mesothelioma cell lines in vitro and decreased tumor-associated angiogenesis in vivo. Notably, the FGFR1 decoy receptor does not bind endocrine FGFs and therefore should not affect phosphate homeostasis and other endocrine FGF activities (Blackwell et al., 2016; Harding et al., 2013). In several Phase I clinical trials with cancer patients, GSK3052230 was well tolerated in combination with chemotherapy and did not show adverse effects such as hyperphosphatemia (Morgensztern et al., 2019; Tolcher et al., 2016; van Brummelen et al., 2020).
Amplification of FGFR2b occurs in ~4%–5% of gastroesophageal cancers and triple-negative breast cancers (Lei & Deng, 2017; Matsumoto et al., 2012; D. Wang, Yang, et al., 2019). Several antibodies that block FGFR2 activation are in various stages of preclinical to clinical development (Catenacci et al., 2019; S. T. Kim, Lee, Rom, et al., 2019). Bemarituzumab (anti-FGFR2b monoclonal antibody) is in phase III clinical trials for FGFR2b-overexpressing advanced gastroesophageal cancer (Catenacci et al., 2019, 2020). Aprutumab ixadotin uses an anti-FGFR2 antibody coupled to auristatin (microtubule-disrupting agent). Though promising in patient-derived xenograft models, phase I clinical trials in humans showed a variety of adverse effects, preventing further use (S. B. Kim, Meric-Bernstam, Kalyan, et al., 2019).
Activating mutations in FGFR3 cause Achondroplasia, the most common form of dwarfism in humans. Somatic activating mutations or amplification of FGFR3 are pathogenic for several cancers including urothelial, CNS, multiple myeloma, and hepatocellular carcinoma (Ardizzone et al., 2020; Chesi & Bergsagel, 2015; Ertl et al., 2020; Katoh & Nakagama, 2014; Paur et al., 2015). Several inhibitory FGFR3-specific antibodies have been developed for use in cancers with amplified or activated FGFR3 (Casadei et al., 2019; Kamath et al., 2012; Yin et al., 2016). In a mouse model for lung adenocarcinoma, the D11 anti-FGFR3 antibody was shown to inhibit tumor growth and tumor-induced mortality (Yin et al., 2016). Another approach being used to block FGFR3 uses a decoy receptor consisting of a soluble extracellular ligand-binding domain. In a mouse model for Achondroplasia injection of soluble FGFR3 (Recifercept) was shown to increase skeletal growth and reduce visceral obesity (Garcia et al., 2013; Goncalves et al., 2020; Saint-Laurent et al., 2018; Unger et al., 2017).
Aberrant FGF15/19 signaling through FGFR4 has been identified as an oncogenic driver for a subset of patients with hepatocellular carcinoma (Lu et al., 2019; Repana & Ross, 2015). In a xenograft model, monoclonal antibodies to FGF15/19 that bock its interaction with FGFR4 prevented tumor growth (Desnoyers et al., 2008). However, high dose antibody caused gastrointestinal side effects due to increased bile acid synthesis and reduced uptake (Pai et al., 2012). A monoclonal antibody that targets the N-terminus of FGF15/19 was recently developed and found effective at inhibiting tumor growth without bile-acid-related side effects (H. Liu, Zheng, et al., 2020). A monoclonal antibody that targets FGFR4 blocks FGF23 induced left ventricular hypertrophy (Grabner et al., 2015, 2017).
A human anti-NCAM single-chain antibody fragment was shown to specifically bind NCAM-expressing cells, interfere with NCAM-FGFR1 binding, and partially decrease migration of a cancer cell line (Flego et al., 2021).
6.2.4 |. Oligonucleotide aptamer inhibitors
Oligonucleotide aptamers, introduced above, are generally designed as inhibitors of protein–protein interactions (Nakamura, 2018, 2021). An RNA aptamer that binds FGF2 (RBM-007, APT-F2) has been shown to block the interaction of FGF2 with all FGFRs and inhibit FGF2-induced downstream signaling. A polyethylene glycol-conjugated (PEGylated) form of RBM-007 was shown to improve the histopathological phenotype and clinical score in mouse and rat models of arthritis, osteoporosis, and Achondroplasia (Jin et al., 2016; Kimura et al., 2021). RBM-007 showed a strong analgesic effect (equivalent to morphine) in a mouse model of bone cancer pain (Jin et al., 2016). RBM-007 also inhibited proliferation and induced apoptosis of an FGF2-dependent lung cancer cell line in vitro (Hamamoto et al., 2018). In a mouse model for lung adenocar-cinoma, cancer-associated fibroblasts (CAFs) were shown to express FGF2 and support tumor growth. Aptamer inhibition of FGF2 in a co-culture of CAFs and tumor cells resulted in decreased colony number and size, demonstrating the importance of CAF-derived FGF2 (Hegab et al., 2019). In vivo, RBM-007 inhibited angiogenesis in an FGF2-induced matrigel plug assay and in a laser-induced choroidal neovascularization (CNV) assay. In addition, RBM-007 was effective in a CNV plus fibrosis model, making it a potentially useful therapy for neovascular age-related macular degeneration (nAMD, often referred to as “wet” AMD), which is currently treated with anti-VEGF therapies (Matsuda et al., 2019). An RNA aptamer that binds FGF5 with high affinity was shown to inhibit FGF5-induced cell proliferation, but not FGF2-induced cell proliferation. As mutations or knockout of Fgf5 results in abnormally long hair, this aptamer may be useful to enhance hair growth (Amano et al., 2021; Hébert et al., 1994).
A DNA aptamer that binds to the extracellular domain of FGFR1 was combined with iron oxide nanoparticles to produce aptamer superparamagnetic conjugates that selectively kill FGFR1-expressing human osteosarcoma cells in the presence of magnetic field irradiation (Jurek et al., 2017). Aptamers that bind to FGFR1 or FGFR3, that when dimerized activate the respective receptors are described above.
7 |. INTRACELLULAR FGFS
Intracellular FGFs (iFGFs, also known as FGF homologous factors or FHFs; Figure 1a) are essential regulators of neuronal and myocardial excitability. iFGFs lack a signal peptide and are thought to function as intracellular proteins. The primary intracellular binding partners of iFGFs are voltage-gated sodium (Nav) channels and the best characterized activity of iFGFs is to regulate the activity of Nav channels. FGF14 or peptides derived from FGF14 directly interact with and regulate the activity of Nav channels (Ali et al., 2016; Bosch et al., 2016; Laezza et al., 2007; Lou et al., 2005; Singh et al., 2021). Though still controversial, other interacting proteins have been identified in various studies including; IB2 (MAPK8IP2, Mitogen-activated protein kinase 8-interacting protein 2), β-tubulin, NEMO (NF-κB essential modulator), hypoxia-inducible factor-1a (HIF-1a), and casein kinase 2 (CK2) (Dib-Hajj, 2016; Hoxha et al., 2019; Hsu et al., 2016; K. W. Lee, Yim, et al., 2017; Ornitz & Itoh, 2015; Pablo & Pitt, 2016; Sun, Niu, et al., 2020; Q. Wang, Yang, et al., 2021). In cardiac hypertrophy, expression and nuclear localization of FGF13 were increased. FGF13 was found to interact with and activate the p65 subunit of NF-κβ, which has been linked to the development of pathological cardiac hypertrophy (Sun, Niu, et al., 2020). Conditional inactivation of Fgf13 in cardiomyocytes resulted in a reduction in peak Na+ channel current density and increased arrhythmia susceptibility in the setting of Na+ channel blockade (X. Wang, Tang, et al., 2017). In a recent study, FGF13 was shown to modulate sodium channel function in dorsal root ganglion (DRG) neurons through stabilization of microtubules (Q. Wang, Yang, et al., 2021). Following peripheral nerve injury, lentiviral delivery of FGF13 was shown to improve motor and sensory functional recovery and enhance axon elongation and remyelination (Li, Tao, Huang, et al., 2021). Although the primary function of iFGFs involves modulation of Na+ channels, interactions with other proteins are uncovering novel activities of this protein family.
iFGFs have a high degree of homology with canonical FGFs. Structural studies suggested that differences in two conserved amino acid residues would greatly reduce the ability of iFGFs to interact with FGFRs (Olsen et al., 2003). However, a recent study demonstrated that recombinant FGF13 (FHF2) can induce proliferation of NIH3T3 cells in a heparin and FGFR-dependent manner (H. Lin, Lu, et al., 2019). Another study showed that FGF12 (FHF1) internalization is protective against radiation injury, but independent of FGFRs (Nakayama et al., 2014). A third study shows that FGF12 can bind to and activate all four FGFRs (Sochacka et al., 2020). However, unlike with canonical FGFs, FGF12 was not capable of stimulating cell proliferation in NIH3T3 cells or glucose uptake in L1–3T3 cells but was able to support cell survival of staurosporine-induced apoptosis in U2OS-R1 cells. FGF12 contains a cell membrane penetrating peptide sequence and therefore could be released from cells (Nakayama et al., 2011). Collectively, these studies suggest that FGF12 has a weaker but sustained ability to activate FGFRs.
8 |. MICRORNA REGULATION OF FGF SIGNALING
MicroRNAs (miRNAs) are small noncoding RNAs with approximately 21–24 nucleotides, which function as posttranscriptional regulators of gene expression. miRNAs regulate diverse biological processes including development, differentiation, cell proliferation, and metabolism, as well as function in human diseases including metabolic diseases and cancers. Many aspects of FGF signaling pathways are regulated by miRNAs (Ornitz & Itoh, 2015). Recently, new functions for miRNA as modulators of FGF signaling have been reported.
8.1 |. Embryonic development
miR-214 attenuates osteogenesis by inhibiting FGF signaling through FGFR1 indicating that targeting miR-214 could be used as a potential therapy for the treatment of postmenopausal osteoporosis (L. Yang, Ge, et al., 2016). Decreased expression of miR-195 promoted chondrocyte proliferation and maintained chondrogenic phenotypes by increasing the production of FGF18, indicating that the miR-195/FGF18 axis could be a potential target for the treatment of cartilage lesions (Y. Wang, Yang, et al., 2017). miR-339, which is highly expressed in the cardiovascular system, inhibits FGF2-induced proliferation of pulmonary artery smooth muscle cells (PASMCs). FRS2 is a potential direct target of miR-339. The biological role of miR-339 in regulating proliferation of PASMCs by targeting FGF signaling provides new mechanistic insights into PASMC proliferation and pathogenesis of pulmonary artery hypertension (J. Chen, Cui, et al., 2017).
miR-133a and miR-135a are involved in the inhibition of osteogenic factors including runt-related transcription factor 2 (Runx2) and Smad5. Recombinant FGF18, released from collagen membranes, promoted osteoblastic activity by decreasing the expression of miR-133a and miR-135a to allow increased expression of osteogenic factors (Imamura et al., 2018). Cyclic mechanical loading-induced osteogenic differentiation of human periodontal ligament cells and decreased expression of endogenous miR-195–5p. In response to cyclic tension strain, production of the miR-195–5p targets, WNT3A, FGF2, and BMPR1A was increased, leading to osteogenic differentiation of periodontal ligament cells (Chang et al., 2017).
The endoribonuclease DICER (Dicer1) promotes the maturation of miRNAs. DICER expression was elevated in neural crest progenitors where it promoted the maturation of a set of cell-type-specific miRNAs that target components of the FGF signaling pathway that are involved in neural induction. These studies showed that posttranscriptional attenuation of FGF signaling by miRNAs is essential for neural crest induction (Copeland & Simoes-Costa, 2020). Pleuropulmonary Blastoma (PPB) can lead to rare mesenchymal pediatric lung cancers. In PPB, loss of expression of DICER in lung epithelium and subsequent loss of miR-140 led to increased expression of FGF9 which promoted mesenchymal hyperplasia which could predispose to mesenchymal oncogenic transformation (Yin et al., 2015).
8.2 |. Neuropathic pain
FGFR1 is expressed in dorsal root ganglia (DRG) neurons. FGF2 was shown to induce Erk1/2 phosphorylation in nociceptive neurons and increased the current density of NaV1.8 channels leading to increased pain (Andres et al., 2013). Gabapentin is a γ-aminobutyric acid analog that is used as an anticonvulsant and is also an effective analgesic agent in neuropathic and inflammatory pain (Cheng & Chiou, 2006). In a rat model for arthritis, gabapentin was found to relieve arthritis pain (reduce the paw withdrawal reflex) by increasing the expression of microRNA-15a in dorsal root ganglia. MicroRNA-15a was shown to bind and suppress the 3′ UTR of FGF2 and FGFR1 (D. Sun, Yang, et al., 2017).
8.3 |. Metabolic diseases
Nonalcoholic fatty liver disease (NAFLD) includes pathologies ranging from uncomplicated hepatic fat accumulation (steatosis) to a state of lobular inflammation and hepatocyte ballooning, known as nonalcoholic steatohepatitis (NASH). Obesity and insulin resistance are primary etiologies of NAFLD. Mice given a high-fat diet have increased expression of miR-212 and miR-149, which were reduced by exercise. HepG2 cells (human hepatocellular carcinoma cells) grown in the presence of long-chain fatty acids increased expression of miR-212 and miR-149 and decreased expression of FGF21. miR-212 and miR-149 functioned to promote lipogenesis and suppress translation of FGF21 in HepG2 cells. Inhibition of miR-212 or miR-149 reduced lipogenesis, and this effect was blocked by knockdown of Fgf21. These data link the benefit of exercise and miR-212/149 downregulation to FGF21 production in preventing NAFLD (Xiao, Bei, et al., 2016; Xiao, Lv, et al., 2016).
Diabetic retinopathy is a leading cause of acquired blindness. Protection of retinal ganglion cells (RGCs) from high glucose-induced injury is a promising strategy for delaying diabetic retinopathy. miR-145–5p is significantly increased in RGC-5 cells grown in the presence of high glucose levels. Inhibition of miR-145–5p decreased expression of proinflammatory cytokines and cell apoptosis and increased cell viability and proliferation and expression of FGF5, a direct target of miR-145–5p. Knockdown of Fgf5 partially reversed the protective effects of miR-145–5p inhibition on RGC-5 cells (J. Zhang, Cui, & Xu, 2019).
Exposure of microvascular ECs to high glucose increased cell proliferation and decreased expression of miR-144–3p. FGF16 is a direct target of miR-144–3p. miR-144–3p mimics significantly inhibited high glucose-induced EC proliferation and MAPK activation by suppressing FGF16 production (C. Chen, Zhao, Gu, Cui, & Wu, 2019). In a mouse model for traumatic brain injury, FGF2 reduces neurofunctional deficits and preserves blood–brain barrier integrity (Z. G. Wang, Cheng, et al., 2016). This results in part from FGF2 protection against oxygen–glucose deprivation and reoxygenation-induced EC permeability (Lin et al., 2018).
8.4 |. Tumorigenesis
Malignant gliomas are the most common tumor in the CNS. Glioblastoma multiforme (GBM) is a highly invasive and aggressive type of malignant glioma and aberrant activation of RTK signaling is one of the most frequent molecular alterations in glioblastoma (Jimenez-Pascual & Siebzehnrubl, 2019). Invasive and migratory abilities of GBM cells and growth of xenograft tumors were significantly inhibited by miR-186–5p, which directly targets and suppresses expression of FGF2 and RelA. The miR-186–5p/FGF2/RelA pathway may be a potential molecular target for the clinical treatment of GBM (F. Wang, Jiang, et al., 2017). The level of miR-144 was decreased in higher grade glioma tissue and in glioma cell lines. miR-144 suppressed glioma cell proliferation, sensitized gliomas to chemotherapeutics, and inhibited metastasis through direct suppression of FGF7 and Caveolin 2 (CAV2). The anti-tumor function of miR-144 was reversed by overexpression of FGF7 and CAV2 (Z. Q. Liu, Ren, et al., 2020).
Malignant pleural mesothelioma (MPM) is an aggressive malignancy. Expression of miR-15/16 is decreased in MPM, and activation of miR-15/16 inhibited the growth of MPM cell lines. Several FGFs (1, 2, and 18) and FGFRs (1 and 4) are direct targets of miR-15/16. Activation of miR-15/16 increased sensitivity to FGFR inhibition, suggesting a potential therapeutic combination strategy to treat MPM (Schelch et al., 2018).
FGF18 is overexpressed in primary gastric cancer (GC) samples and cell lines. FGF18 is a direct target of the tumor suppressor, miR-590–5p. Overexpression of FGF18 partly abolished the tumor-suppressive effect of miR-590–5p (J. Zhang, Zhou, et al., 2019). miR-5582–5p and miR-187 are significantly decreased in nonsmall cell lung cancer (NSCLC) tissues and cell lines. Overexpression of miR-5582–5p and miR-187 inhibited the proliferation and migration of NSCLC cells (Han et al., 2019; Liang et al., 2020). miR-5582–5p binds to the 3′-untranslated region of FGF10 and decreased the expression of FGF10 in NSCLC cells. FGF10 expression is inversely correlated with the level of miR-5582–5p and overexpression of FGF10 significantly reversed the inhibitory effect of miR-5582–5p on the proliferation of NSCLC cells (Han et al., 2019). Similarly, miR-187 expression is lower in NSCLC and cancer cells. FGF9 is a target of miR-187 and miR-187 overexpression reduced the expression of FGF9, cyclin D1, CDK4, and CDK6 (Liang et al., 2020).
Fgfbp1 is a direct target of miR-6887–5p in squamous cell carcinoma (SCC) cells. Overexpression of miR-6887–5p in SCC cells inhibited cell proliferation and colony formation in vitro, and inhibited tumor growth in vivo (Higaki et al., 2020).
9 |. LONG NONCODING RNA REGULATION OF FGF SIGNALING
Long noncoding RNAs (lncRNAs) comprising approximately 200 nucleotides or more can function as posttranscriptional regulators of gene expression. lncRNAs are involved in diverse biological processes including development and disease (Chen & Shan, 2020). New evidence for roles of lncRNAs in diverse FGF signaling has been reported.
9.1 |. Development
FGF2 is implicated in muscle satellite cell self-renewal and differentiation by repressing MyoD. Overexpression of the lncRNA Linc-RAM (long intergenic noncoding-RNA activator of myogenesis) rescues FGF2-induced inhibition of mouse myoblast cell differentiation indicating that inhibition of Linc-RAM is required for FGF2-mediated suppression of myogenic differentiation (Y. Zhao, Cao, et al., 2018). The expression of lncRNA-H19 is significantly increased in human dental pulp stem cells (hDPSCs) undergoing odontoblastic differentiation and forced expression of H19 further promotes differentiation. H19 binds and sequesters miR-140–5p and miR-140–5p to directly suppress translation of Bmp2 and Fgf9 mRNAs. Thus, H19 promotes odontoblastic differentiation of hDPSCs by regulating the miR-140–5p/BMP2/FGF9 axis (Zhong et al., 2020).
lncRNA-TUG1 (taurine upregulated gene 1) functions to promote osteogenic differentiation of several cell types including tendon stem/progenitor cells. TUG1 was shown to interact with FGF2 and promote the ubiquitination and degradation of FGF2 protein, which is necessary for osteogenic differentiation (Yu et al., 2020). During the differentiation of adipose-derived stem cells (ADSCs) to ECs, TUG1, and FGF1 levels were increased. TUG1 was shown to interact with and suppress the expression of miR-143. Decreased miR-143 led to increased expression of its direct target, Fgf1, and increased markers of EC differentiation (Xue et al., 2019).
The E3 ubiquitin ligase TRIM71/Lin41 promotes FGF/ERK signaling by binding and enhancing the stability of Shc SH2-binding protein 1 (SHCBP1) in neural progenitor cells (J. Chen, Lai, & Niswander, 2012). The Trincr1 (TRIM71 interacting lncRNA 1) binds and represses TRIM71/LIN-41. Knocking out Trincr1 led to increased TRIM71, phosphorylated ERK1/2, and ERK pathway target genes, and decreased embryonic stem cell self-renewal. Ectopic expression of Trincr1 repressed FGF/ERK signaling and the self-renewal of neural progenitor cells (Y. P. Li, Duan, et al., 2019).
9.2 |. Cardiovascular diseases
Vascular calcification (VC) is a pathological process commonly found in patients with atherosclerosis, hypertension, and diabetes. lncRNA-SNHG29 inhibits vascular smooth muscle cell (VSMC) calcification by downregulating miR-200b-3p to activate the αKlotho/FGFR1/FGF23 axis (Huang et al., 2020). Vascular remodeling is a characteristic pathological feature of hypertension. lncRNA-TUG1 is highly expressed in the aorta of spontaneously hypertensive rats. TUG1 was shown to bind and sequester miR-145–5p to allow increased production of the miR-145–5p target, Fgf10. TUG1 and FGF10 were shown to promote the expression of β-catenin and activation of Wnt signaling, which promoted the proliferation and migration of VMSCs in spontaneously hypertensive rats (L. Shi, Tian, et al., 2018; Watson & Francavilla, 2018). Atherosclerotic lesion formation results from vascular wall inflammation, damage to ECs, and dysregulated proliferation of VSMCs. TUG1 expression was increased in the aorta in a mouse model for atherosclerosis. TUG1 interacts with and suppresses miR-133a. In macrophage and VSMC cell lines, overexpression of TUG1 decreased miR-133a, which led to increased expression of its direct target, Fgf1, and increased cell proliferation. Inhibiting TUG1 may therefore protect against the formation of atherosclerotic lesions (L. Zhang, Cheng, et al., 2018).
A correlation was observed between cardiac remodeling and lncRNA-FAF (FGF9-associated factor) expression in the heart. Expression of FAF and FGF9 was decreased in hypoxic cardiomyocytes and heart tissue after acute myocardial infarction. Overexpression of FAF increased the expression of FGF9 and inhibited cardiomyocyte apoptosis induced by ischemia and hypoxia (Shi et al., 2019). FAF was enriched in neonatal rat cardiac fibroblasts. Increased FAF suppressed angiotensin II-induced increased cell proliferation, differentiation, and collagen accumulation in cardiac fibroblasts. FAF functioned to increase TGFβ1-P-Smad2/3 and FGF9 signaling in cardiac fibroblasts (J. Sun, Wang, et al., 2020).
TUG1 expression was increased after acute myocardial infarction and knockdown of TUG1 reduced cardiac fibrosis following acute myocardial infarction. Inhibition of TUG1 decreased the proliferation, migration, and collagen I expression of TGFβ1 treated cardiac fibroblasts. TUG1 functioned to suppress miR-590–5p, which suppresses FGF1 production. Increased TUG1 and resulting increased FGF1 were shown to promote cardiac fibrosis following acute myocardial infarction (Q. Sun, Luo, et al., 2021).
9.3 |. Ischemia
Ischemia exerts a negative impact on mitochondrial function resulting in neuronal damage. In a mouse model of ischemia-reperfusion (IR)-mediated hippocampal injury expression of lncRNA-AK005401 was increased and expression of FGF21 was reduced. The resulting decreased PI3K/Akt signaling promoted reactive oxygen species (ROS) generation, mitochondria dysfunction, and cell apoptosis. This pathology was attenuated by knockdown of AK005401 (Wan et al., 2019).
lncRNA-GUSBP5-AS is upregulated in endothelial progenitor cells (EPCs) of deep vein thrombosis patients. GUSBP5-AS increased angiogenesis, proliferation, and homing ability of EPCs to promote resolution and recanalization of thrombi in a mouse model of deep vein thrombosis. GUSBP5-AS binds and sequesters miR-223–3p, which results in increased expression of the FOXO1 transcription factor. FOXO1 expression in EPCs increased AKT signaling and FGF2 and matrix metalloproteinase 2/9 expression (L. L. Sun, Lei, et al., 2020).
9.4 |. Tumorigenesis
lncRNA-MALAT1 and FGF2 were expressed in thyroid cancer tissues and in thyroid cancer cell lines and tumor-associated macrophages (TAMs). MALAT1 was shown to promote FGF2 secretion from TAMs. FGF2 inhibited inflammatory cytokine release, promoted proliferation, migration, and invasion of follicular thyroid carcinoma (FTC133) cells, and induced tumor angiogenesis (J. K. Huang, Ma, et al., 2017). lncRNA-RHPN1-AS1 expression was increased in cervical cancer tissues. RHPN1-AS1 promoted cervical cancer progression by targeting the miR-299–3p/FGF2 axis. RHPN1-AS1 overexpression effects on cell proliferation, growth, invasion, and migration in human cervical cancer (SiHa) cells were attenuated by overexpression of miR-299–3p or inhibition of FGF2 (Duan et al., 2019). lncRNA-HOXD-AS1 is increased in cervical cancer tissues and higher levels of HOXD-AS1 predict worse prognosis of cervical cancer patients. HOXD-AS1 functions as a pro-oncogenic molecule by suppressing miR-877–3p and inducing FGF2 production (Chen & Li, 2020). lncRNA-MT1JP levels were significantly decreased in osteosarcoma tissues. MT1JP and miR-646 overexpression inhibited the migration and invasion of osteosarcoma cells and overexpression of MT1JP increased levels of miR-646, which suppressed production of FGF2. FGF2 overexpression suppressed the effects of MT1JP and miR-646 overexpression on osteosarcoma cell migration and invasion (L. Yang, Liu, et al., 2020). Silencing of lncRNA-PCGEM1 repressed cell proliferation and migration and promoted apoptosis in renal cell carcinoma. PCGEM1 binds and sequesters miR-433–3p. Inhibition of PCGEM1 led to increased miR-433–3p, which suppressed production of FGF2 (Cai et al., 2020).
lncRNA-Linc00460 expression was increased in breast cancer tissue and correlated with increased lymphatic metastasis and poor survival. Linc00460 binds and sequesters miR-489–5p, which further suppresses Fgf7 translation. Linc00460 promotes breast cancer progression partly through the miR-489–5p/FGF7/AKT axis to increase the expression of FGF7 (Zhu et al., 2019). FGF9 levels were increased in muscle-invasive bladder cancer (MIBC) tissues. lncRNA-AFAP1-AS1 promoted gastric cancer cell proliferation, migration, and invasion by sequestering miR-155–5p and increasing levels of the miR-155–5p target, FGF7 (Ma et al., 2020).
lncRNA-LINC01140 was positively correlated with FGF9 levels and was significantly increased in muscle-invasive bladder cancer tissues. LINC01140 binds and sequesters miR-140–5p, which suppresses FGF9 production. Increased FGF9 was shown to increase the severity of the bladder cancer phenotype (Wu et al., 2020). lncRNA-H19 expression was increased in small cell lung cancer (SCLC) tissue. Through inhibition of miR-140–5p, H19 increased FGF9 levels to promote SCLC progression (X. Li, Lv, et al., 2020). FGF9 may additionally function to promote the transdifferentiation of lung adenocarcinoma to a SCLC phenotype (Ishioka et al., 2021).
lncRNA-SNHG16 expression was increased and associated with poor prognosis in hepatocellular carcinoma. SNHG16 binds and sequesters miR-302a-3p. miR-302a-3p suppresses the production of FGF19 in liver cancer cells. SNHG16 promoted liver cancer cell proliferation by suppressing miR-302a-3p and increasing FGF19 (W. Li, Xu, et al., 2019).
10 |. SELECTED TOPICS IN GENETICS, DEVELOPMENT, REGENERATION, AND DISEASE
FGF signaling in genetic disease and roles in organogenesis have been reviewed recently (Itoh & Ornitz, 2011; X. Li, Wang, Xiao, et al., 2016; Mossahebi-Mohammadi et al., 2020; Ornitz & Itoh, 2015; Ornitz & Itoh, 2017; Turner & Grose, 2010; Xie, Su, et al., 2020). The role of FGF signaling in tissue regeneration and repair has recently been reviewed (El Agha et al., 2016; Farooq et al., 2021; X. Li, Wang, Xiao, et al., 2016; Maddaluno et al., 2017). In the following sections, we focus on more recent advances in genetics, development, and regeneration.
10.1 |. FGF regulation of stem cells pluripotency and differentiation
Pluripotent stem cells (PSCs) are self-renewing stem cells that, through communication with their microenvironment, can differentiate into a variety of specialized cell types that contribute to the development, maintenance, and regeneration of tissues and organs. FGF signaling has important roles in maintaining PSC pluripotency and regulating PSC differentiation (Lanner & Rossant, 2010). There are at least two states of embryonic PSC pluripotency, naïve, and primed (Nichols & Smith, 2009). Mouse embryonic stem cells (mESCs) are naïve pluripotent cells; they resemble the pre-implantation epiblast and have unbiased developmental potential. Human ESCs, human-induced PSCs (iPSCs), and mouse post-implantation derived epiblast stem cells (mEpiSCs) are referred to as primed PSCs; they resemble the post-implantation epiblast and show signs of lineage bias during differentiation (C. Dong, Fischer, & Theunissen, 2019; Theunissen & Jaenisch, 2017).
Several FGFs, including FGF2, FGF4, FGF6, FGF7, FGF8, and FGF9 have the capacity to maintain the stemness of primed PSCs but induce differentiation of less mature naïve PSCs (Li & Belmonte, 2017; Mossahebi-Mohammadi et al., 2020). Spred1 and Spred2, genes that encode inhibitors of ERK/MAPK, are expressed in mESCs where they promote self-renewal and inhibit mesodermal differentiation (Azami et al., 2018; Muhl et al., 2015). In mEpiSCs, FGFR1 is stabilized by high levels of CDH2, and both proteins are required to maintain pluripotency (Takehara et al., 2015). In vitro, FGF2 is commonly used to maintain primed pluripotency in culture and withdrawal of FGF2 leads to ectodermal differentiation (Haghighi et al., 2018). In vivo, FGF4 appears to be the endogenous key regulator of stem cell pluripotency and differentiation (Lanner & Rossant, 2010).
Complete blockage of autocrine FGF signaling led to a homogeneous population of naive PSCs (An et al., 2020). Conversion of human primed PSCs into less mature naive PSCs is accompanied by genome-wide loss of methylation that includes loss of imprinting. High levels of FGF signaling suppressed the extent of loss of imprinting during this conversion, whereas low levels of FGF signaling promoted loss of imprinting (Keshet & Benvenisty, 2021).
10.1.1 |. FGF signaling in preimplantation development
In the blastocyst, the inner cell mass (ICM) will differentiate into the epiblast (EPI), which gives rise to most somatic and germ cells, and the primitive endoderm (PrE), which will form the yolk sac (Kang et al., 2017). In the late-stage mouse blastocyst, Fgf4 is expressed in PrE-biased cells. Fgf4 gene inactivation shows that FGF4 is required for ICM proliferation and for the formation of the PrE (Kang et al., 2013, 2017; Krawchuk et al., 2013; Molotkov et al., 2017; Ohnishi et al., 2014). Modeling and experimental perturbation of lineage-restricted cell composition in the blastocyst identified a gene regulatory network involving the transcription factors Nanog and Gata6 and FGF4/Erk signaling to regulate the fate of progenitors to robustly generate cell types in the correct proportions (De Caluwe et al., 2019; De Mot et al., 2016). Direct analysis of ICM lineage specification at the single-cell level shows that a consistent ratio of EPI and PrE lineages is achieved through incremental allocation of cells from a common progenitor pool and that the lineage composition of the ICM is conserved regardless of its size (Saiz et al., 2016). Mechanistically, changes in the local concentration of FGF4 provide a basis for the adaptive abilities of the blastocyst to coordinate fate decisions at the population level (Saiz et al., 2020). Using Fgf4 mutant cells and an FGF signaling reporter (Spry4H2B-Venus), short-range FGF4 signaling was found to constitute the minimal molecular mechanism underlying EPI and PrE lineage allocation (Morgani, Saiz, et al., 2018; Raina et al., 2021).
FGFR1 and FGFR2 are potential receptors for FGF4. Fgfr1 is expressed in all cell populations of the blastocyst, while Fgfr2 expression becomes restricted to extraembryonic lineages. Loss of Fgfr1 and Fgfr2, or only Fgfr1, prevents the development of the PrE. However, the activity of both receptors is required for lineage establishment within the ICM. Thus, unique and additive activities of FGFR1 and FGFR2 within the ICM coordinate establishment of two distinct embryonic lineages (Brewer et al., 2015; Kang et al., 2017; Molotkov et al., 2017). Cells outside of the ICM give rise to the trophectoderm, and FGFR1 is required for differentiation of trophectoderm cells and maintenance of their polarity (Kurowski et al., 2019). PDGFRA signaling also interacts with FGFR1 and FGFR2 in PrE development. FGFR2 and PDGFRA regulate PrE cell survival while FGFR1 controls PrE cell specification (Molotkov & Soriano, 2018).
Activation of the MAPK/ERK signaling pathway is essential for the differentiation of the ICM during mouse preimplantation development. ERK1/2 phosphorylation occurs in ICM precursor cells, in differentiated PrE cells, as well as in the mature, formative state epiblast. DUSP4 and ETV5 are often involved in negative-feedback regulation of the FGF pathway. Whereas the presence of DUSP4 clearly depends on ERK phosphorylation in PrE cells, ETV5 localizes mainly to EPI cells. ETV5 accumulation does not depend on direct activation by ERK but requires NANOG activity. In pluripotent early EPI cells, NANOG induces the expression of both FGF4 and ETV5 to enable the differentiation of neighboring cells into the PrE while protecting EPI identity from autocrine signaling (Azami et al., 2019).
Human preimplantation development can be modeled in vitro with 2D micropatterned and 3D gastruloids (Morgani & Hadjantonakis, 2020; Morgani, Metzger, et al., 2018). BMP4 treatment of hESCs cultured on ECM microdiscs produced microcolonies termed “gastruloids” with three prospective germ layers. Single-cell RNA sequencing identified mesodermal cell populations that express FGFR1 and its downstream target SNAI1 (Minn et al., 2020). At the early time points of gastruloid development (0–12 h), FGF2, FGFR1, and FGFR signaling antagonists, SPRY1, DUSP6, and CBL were expressed at high levels. During differentiation (44 h) these FGF signaling components decreased, and FGF17 increased in mesoderm and endoderm along with expression of BMP, WNT, NODAL, and HIPPO signaling components (Minn et al., 2021). Comparison of mouse gastrulation and human gastruloids showed that mouse expresses high levels of Fgf8, which is required for cell migration, while human gastruloid mesenchyme expressed high FGF17 and very low FGF8 (Minn et al., 2020). Thus, although evolutionarily conserved pathways are used to drive EMT and cell migration during gastrulation, specific components differ between mouse and human.
10.1.2 |. FGF signaling in adult stem cells
Adult stem cells are tissue-resident stem cells with properties of self-renewal, maintenance of tissue homeostasis, and regeneration of damaged tissue. FGFs are common components of the adult stem cell niche and function to maintain quiescence and regulate self-renewal and differentiation (Coutu & Galipeau, 2011; Keyes & Fuchs, 2018; Otsuka et al., 2021).
Comparison of the effects of different growth factors on the differentiation of neuronal stem cells that were isolated from rat adult dorsal root ganglia showed that FGF2 was a much stronger inducer of glial differentiation compared to nerve growth factor (NGF) and neuregulin1-β (NRG1-β) (Gu et al., 2014).
Human primary synovium-derived stem cells showed increased glycosaminoglycan deposition, pellet size, and chondrogenic gene expression when treated with FGF2 and grown in chondrogenic differentiation medium and increased calcium deposition and alkaline phosphatase activity when treated with FGF2 and grown in osteogenic differentiation medium (Pizzute et al., 2016).
Muscle stem cells (satellite cells) are lost with aging, leading to impaired muscle regeneration. Increased FGF signaling in the satellite cell niche led to loss of quiescence, satellite cell depletion, and diminished regenerative capacity; and inhibition of FGFR1 in satellite cells prevented their depletion (Chakkalakal et al., 2012).
In the lung, FGF10 is expressed in several epithelial stem cell niches, and through signaling to FGFR2b, FGF10 maintains stem cell populations and activates stem cells in response to injury (Volckaert & De Langhe, 2014; Watson & Francavilla, 2018). For example, type II alveolar epithelial cells (AEC2s) are stem cells that can regenerate type I alveolar epithelial cells (AEC1s) in response to alveolar injury. Inactivation of FGFR2 in adult AEC2s led to fewer AEC2s, decreased surfactant protein C (Sftpc) gene expression, increased alveolar diameter, increased collagen deposition, and, in response to injury, increased mortality (Dorry et al., 2020).
Bone marrow-derived mesenchymal stem cells (BMSCs) are discussed as follows.
10.2 |. Regulation of body axis elongation and segmentation
The establishment of the vertebrate body plan requires body axis elongation from a posterior growth zone. This growth zone generates the presomitic mesoderm (PSM). The anterior region of the PSM periodically segments into somites that flank the neural tube. Early studies demonstrated that FGF activity was required for this process and suggested that FGF8 was the principal signaling ligand (Dubrulle et al., 2001; Sawada et al., 2001). However, PSM-specific Fgf8 inactivation in mice revealed no axis defect (Perantoni et al., 2005). Furthermore, inactivation of the Fgf8 subfamily (Fgf8, Fgf17, and Fgf18) resulted in only a subtle effect on somite size, but, surprisingly, a significant effect on closing the ventral body wall (Boylan et al., 2020). Concurrent inactivation of both Fgf4 and Fgf8 revealed that both ligands act redundantly to maintain the posterior growth zone (Benazeraf & Pourquie, 2013; Boulet & Capecchi, 2012; Naiche et al., 2011). At early somite stages (E8.5), Wnt3a is expressed in the ectodermal layer of the primitive streak and is also required to maintain the posterior growth zone (Benazeraf & Pourquie, 2013; Boulet & Capecchi, 2012; Naiche et al., 2011). At this stage, Wnt8a is expressed throughout the epiblast and primitive streak (Cunningham et al., 2015). Although in mammals Wnt3a is most important, mice lacking Wnt3a and Wnt8a demonstrate that these Wnts cooperate to maintain Fgf8 expression and prevent premature Sox2 upregulation in the axial stem cell niche (adjacent to the tailbud) (Cunningham et al., 2015). Thus, redundancy in FGF and WNT signaling components maintains the posterior growth zone.
In mouse, FGF8 and FGFR1 are required for posterior mesoderm patterning during gastrulation (Deng et al., 1994; Sun et al., 1999; Yamaguchi et al., 1994). However, in zebrafish, fgf8a and fgf24 are both required for posterior mesodermal development and their loss results in shortening of the embryonic axis (Draper et al., 2003). Additional redundancy was also found for FGFRs where posterior mesoderm development requires zygotically expressed fgfr1a and fgfr1b, and maternally expressed fgfr1a (Leerberg et al., 2019).
As cells progress to the PSM they no longer synthesize Fgf8 mRNA and the decay of Fgf8 mRNA (and potentially Fgf4) establishes a decreasing posterior to anterior gradient of FGF signaling (Dubrulle & Pourquie, 2004). This FGF gradient is established in part by direct suppression of Fgf8 transcription by retinoic acid (RA) (Kumar et al., 2016; Kumar & Duester, 2014). This FGF gradient controls elongation by maintaining a posterior-to-anterior random cell motility gradient in the PSM (Benazeraf & Pourquie, 2013). Interestingly, increased cell mobility is dependent on increased glycolytic activity which is regulated by the level of FGF signaling (Oginuma et al., 2017). MBTPS1/SKI-1/S1P (membrane-bound transcription factor protease, subtilisin kexin isozyme-1, or site 1 protease) is an auto-catalytically activatable member of the proprotein convertase family of serine proteases that activates a small family of bZIP transcription factors. Conditional inactivation of Mbtps1 in PSM leads to complete loss of Fgf8 expression in the PSM and axial truncation. Loss of Fgf8 expression is consistent with elevated apoptosis in the PSM and axis elongation phenotypes (Achilleos et al., 2015).
Progressing anteriorly, segmentation of the PSM forms the somites, which will form the vertebral column and associated body segments containing muscle, connective tissue, and dermis. Although both Fgf4 and Fgf8 are required for PSM elongation, only Fgf4 is required for actual somitogenesis, where it maintains Notch oscillatory activity through maintaining Hes7 levels. Fgf4 may be important for some human segmentation defects associated with impaired Notch oscillations (Anderson et al., 2020). Vertebral defects due to gestational hypoxia are thought to be caused by a sensitivity of FGF signaling to a drop in oxygen during mid-gestation (8 h at E9.5), and FGF4 may be a component of this oxygen-sensing machinery (Anderson et al., 2020; Sparrow et al., 2012).
The vertebrate spinal cord is also patterned in all three embryonic axes. FGF3 activity from the PSM regulates caudal neural tube BMP signaling that in turn regulates neural crest formation, neural pore closure, and cessation of axis extension (Anderson, Schimmang, & Lewandoski, 2016). FGF and RA signaling have been shown to regulate Hox gene expression in the developing neural tube (Leung & Shimeld, 2019).
In the hindbrain, FGF3, FGF8, and RA signaling from hindbrain primordia are essential for setting up the initial identities of hindbrain segments (rhombomeres) by regulating the expression of transcription factors such as Krox20 that control segment identity (Frank & Sela-Donenfeld, 2019; Parker et al., 2016). In recent studies, rhombomeres 2 and 4 were found to function as temporally dynamic signaling centers that use FGF to regulate the expression of Ephrin type-A receptor 4 (EphA4) independent of Krox20 in adjacent rhombomeres 3 and 5 (Cambronero et al., 2020).
10.3 |. Invertebrate morphogenesis and regeneration
10.3.1 |. Cnidarian (Hydra)
The freshwater cnidarian polyp, Hydra, contains evolutionarily conserved signaling pathways including FGFs (Ghaskadbi, 2020). At least five Fgf genes have been identified in Hydra, and expression studies show localization to boundary regions and tentacle tips (Lange et al., 2014).
Hydra reproduces by budding (an extreme case of morphogenesis). Bud formation can occur in the absence of FGFR signaling; however, separation of adjacent epithelial cells and bud detachment requires FGFR signaling (Hasse et al., 2014). Mechanistically, FGFR signaling controls a Rho-ROCK-myosin II pathway that regulates cell shape changes required for bud detachment (Holz et al., 2017). Hydra has a remarkable ability to regenerate. Phylogenetic analysis identified Hydra sequences with significant similarity to vertebrate FGF1 and FGFR1. Pharmacological inhibition of FGFR1 or VEGFR2 delayed head regeneration and reduced expression of head- and tentacle-specific marker genes (Turwankar & Ghaskadbi, 2019).
10.3.2 |. Platyhelminthes (Planaria)
Planaria contain two FGFRs and a homolog of FGFRL1 (ndl-3). Ndl-3 is expressed in the trunk region and functions with wntP-2 and ptk7 to suppress posterior trunk expansion (Lander & Petersen, 2016). Single-cell RNA sequencing revealed two distinct FGFRL-Wnt circuits with juxtaposed anterior FGFRL and posterior Wnt expression domains that control planarian head and trunk patterning (Scimone et al., 2016). Djfgf, a Fgf gene in Dugesia japonica, encodes a putative secreted protein with a core FGF domain. Djfgf expression was highly induced during head regeneration compared to tail regeneration (Auwal et al., 2020).
10.3.3 |. Arthropod (Drosophila)
Drosophila contains three FGF ligands, branchless (Bnl), pyramus (Pyr), and thisbe (Ths) and two FGF receptors, breathless (Btl), and heartless (Htl) (Huang & Stern, 2005; Shilo, 2014). Mutations in the Drosophila FGF signaling pathway generally disrupt mesoderm cell migration and differentiation by controlling cell shape and adhesion. During gastrulation, FGFR signaling (Htl) regulates multiple properties of mesodermal cells to facilitate collective migration. These include mesoderm tube collapse, cell protrusions, suppression of adherens junction number, and promotion of mesoderm cell division (J. Sun, Macabenta, et al., 2020). Fog elicits cell-shape change during gastrulation through the G-protein-coupled receptors Mist and Smog. FGFR (Htl) interacts synergistically with Smog to suppress Fog signaling (Shweta et al., 2021).
The FGF ligand branchless (Bnl) signals to breathless (Btl) to provide an essential developmental cue for tracheal cells (Shilo, 2016). In tracheal cells, FGF signaling can act as a chemoattractant to promote collective cell migration (Lebreton & Casanova, 2016). At the tracheal tip, two cell fates respond differentially to FGF signals. Tip cells are attracted by FGF while fusion cells are repelled (Miao & Hayashi, 2015, 2016). FGF signaling also induces the expression of the spectraplakin Short-stop (Shot) in cells undergoing the initial steps of subcellular branching. Shot promotes interaction between microtubules and actin, which is required for the extension and guidance of the subcellular lumen within the tracheal tip cell cytoplasm (Ricolo & Araujo, 2020).
In flight muscle, regulated subcellular FGF (Bnl) trafficking controls tracheal cell invasion into T-tubules that are required to meet the high oxygen demands of this tissue (Peterson & Krasnow, 2015). Drosophila mechanosensory neurons express the breathless (Btl) FGFR which regulates axonal branch number and length (Dos Santos et al., 2019). In the Drosophila hematopoietic lymph gland, FGF (bnl), produced by vascular cells, activates FGF signaling in hematopoietic progenitors to maintain the progenitor pool and prevent blood cell differentiation (Destalminil-Letourneau et al., 2021). Damage to the intestinal epithelium or tumors increases FGF (Bnl) expression in the intestinal epithelium and reactive oxygen species that stabilize hypoxia-inducible factor-1α (HIF-1α) in gut-associated terminal tracheal cells (TTC). HIF-1α upregulates FGFR (Btl) expression in TCC, which, in response to FGF (Bnl), leads to vascular remodeling and indirectly to intestinal stem cell proliferation (Perochon et al., 2021; Tamamouna et al., 2021).
10.3.4 |. Ascidian (Ciona intestinalis)
In Ciona, “mesendoderm” precursors give rise to part of the endoderm and mesoderm. Foxa.a, Foxd, and Fgf9/16/20 are transcriptional targets of β-catenin and are required for the correct initiation of both the mesoderm and endoderm gene regulatory networks. The combinatorial activity of these three factors is sufficient to produce a mesendoderm ground state that can be further programmed into mesoderm or endoderm lineages (Hudson et al., 2016).
FGF and Nodal signals specify lateral and ventral neural progenitors and control cellular behaviors underlying morphogenesis of the neural tube (Navarrete & Levine, 2016). FGF-responsive Ets family transcription factors differentially pattern neural plate medial and lateral lineages with partially redundant function (Gainous et al., 2015). At later stages of nervous system development, FGF signaling directs pigment cell identity at the expense of anterior neural fates (Racioppi et al., 2014).
In the presumptive notochord, Ets family transcription factors regulate the expression of Brachyury through two distinct enhancers. These enhancers drive qualitatively similar expression patterns but show quantitatively distinct responses to FGF. Together these enhancers drive high levels of Brachyury expression with a characteristic input/output relationship (Harder et al., 2021). Vegetal FGF signaling was shown to regulate Foxa.a expression in the notochord, which directly activates Brachyury expression (Reeves et al., 2021). Single-cell RNA sequencing of Ciona embryos treated with the MAPK inhibitor U0126 showed that most FGF-dependent cell bifurcations were translated to the sibling cell type revealing that most cell fate decisions were dependent on MAPK signaling through the Ets family transcription factor Elk1/3/4 (Winkley et al., 2021).
In the development of bipolar tail neurons, early FGF signaling is required for Neurog expression and bipolar tail neuron specification. At later stages, Fgf8/17/18 expression in tail tip cells suppresses sustained Neurog expression in the adjacent anterior bipolar tail neuron allowing the more distal cell to maintain Neurog expression and become a neuron (K. Kim, Gibboney, et al., 2020).
In cardiopharyngeal development, differential FGF–MAPK signaling distinguishes between heart and pharyngeal muscle precursors (Razy-Krajka et al., 2018).
10.4 |. Cardiovascular system
10.4.1 |. Heart development
Contracting muscular tubes and primitive hearts are found throughout the animal kingdom, and FGF signaling has been identified as a key pathway that drives the cardiac fate (Ahmad, 2017; Dobrzycki et al., 2020; Poelmann & Gittenberger-de Groot, 2019). In mammalian development, multiple studies have identified essential roles for FGF signaling in early specification, differentiation, and morphogenesis of the heart (Itoh, Ohta, et al., 2016; Khosravi et al., 2021). FGFs that function as paracrine growth factors during embryonic heart development include FGFs 3, 8, 9, 10, 15/19, and 16 (Dong et al., 2021; Hubert et al., 2018; Itoh, Ohta, et al., 2016; Meganathan et al., 2015; S. Wang, Li, et al., 2018). In mammals, Notch and FGF8 signaling are required for second heart field (SHF) development, which is required for the formation of the right ventricle and outflow tract. One of the Notch ligands, Delta-like ligand 4 (Dll4), is expressed in the SHF and was shown to regulate Fgf8 expression. Mice heterozygous for Fgf8 and Dll4 showed defects in outflow tract alignment (De Zoysa et al., 2020). Transient gestational hypoxia decreases FGF signaling (Sparrow et al., 2012). Reporters of FGF signaling, Spry1, Spry2, and Spry4, were reduced in the SHF after 8 h exposure to hypoxia at E9.5, resulting in congenital heart defects (Shi et al., 2016).
In humans, damaging mutations in FGF8 and FGF10 are associated with conotruncal defects, consistent with the expression of FGF8 and FGF10 in the outflow tract during human embryonic development (Zhou et al., 2020). During late embryonic development in the mouse, Fgf10 mRNA and protein were preferentially expressed in the right ventricle compared to left ventricle. Analysis of sorted cells showed expression of Fgf10 and its receptor, Fgfr2b, in cardiomyocytes, but not in fibroblasts. Fgf10−/− embryos showed reduced thickness, proliferation, and increased expression of p27kip1 in the right ventricular wall compared to the left ventricular wall suggesting autocrine FGF10 signaling (Rochais et al., 2014). In zebrafish, inhibition of FGFR signaling after the onset of atrial and ventricular differentiation led to ectopic expression of atrial genes in the ventricle. FGF signaling was shown to function upstream of Nkx transcription factors which are important for specifying ventricular identity (Pradhan et al., 2017).
In chicken embryos, inducing left ventricular pressure overload resulted in increased Fgf2 mRNA expression in the heart, increased levels of FGF2 in the serum, and increased myocyte proliferation. Pharmacological inhibition of FGFR signaling decreased myocyte proliferation and resulted in epicardial hemorrhages (Krejci et al., 2016).
In zebrafish, inactivation of the endocardial transcription factors, Kruppel-like factor 2a and 2b (Klf2a, Klf2b), results in abluminal extrusion of cardiomyocytes. Klf2 mutants had reduced expression of FGF ligands and receptors, and pharmacological inhibition of FGFR signaling resulted in a similar cardiomyocyte extrusion phenotype. These data suggest that FGF signaling is required downstream of KLF2 to maintain myocardial wall integrity (Rasouli et al., 2018).
10.4.2 |. Blood and lymphatic vasculature
The blood and lymphatic vasculatures are involved in the maintenance of tissue oxygenation and fluid homeostasis. Assembly of these vascular networks involves sprouting, migration, and proliferation of ECs. ECs closely associated with pericytes in the capillaries and VSMCs in larger vessels (Lilly, 2014; Sweeney & Foldes, 2018). Complex interactions between ECs and VSMCs regulate vascular physiology and response to injury.
In addition to well-established regulation by vascular endothelial growth factor (VEGF), ECs are also highly responsive to FGF signaling. ECs express high levels of the c splice variants of Fgfr1 and Fgfr3, lower levels of Fgfr2, and little or no Fgfr4 (Antoine et al., 2005; De Smet et al., 2014; Oladipupo et al., 2014; X. Yang, Liaw, et al., 2015; Y. Yang, Haeger, et al., 2017; P. Yu, Wilhelm, et al., 2017). ECs express canonical FGFs (FGFs 1, 2, 5, 7, 8, 16, and 18) and iFGFs (iFGFs 11 and 12) (Antoine et al., 2005; De Smet et al., 2014; Domouzoglou et al., 2015; Oladipupo et al., 2014; Seo et al., 2016; X. Yang, Liaw, et al., 2015; P. Yu, Wilhelm, et al., 2017). The expression of canonical FGFs in ECs is a potential source of autocrine signaling. In addition to FGFRs 1–3, canonical FGFs, and iFGFs, ECs express βKlotho, enabling endocrine FGF21–FGFR1/βKlotho signaling (J. Chen, Hu, et al., 2018; Domouzoglou et al., 2015; W. Huang, Shao, et al., 2019; Yaqoob et al., 2014).
FGF11, a member of the iFGF subfamily (Figure 1a) is induced by hypoxia. Overexpression of FGF11 in human umbilical vein endothelial cells (HUVECs) stimulated capillary-like tube formation and expression of tight junction proteins but did not affect cell migration (J. Yang, Kim, et al., 2015). Interestingly, FGF12, another member of the iFGF family, is required for maintaining the quiescent and contractile phenotypes of VSMCs. FGF12 inhibited cell proliferation through the p53 pathway and induced VSMC lineage differentiation through the p38 MAPK pathway (Song et al., 2016). BMP signaling suppresses pathological VSMC proliferation and promotes differentiation. FGF12 expression is induced by BMP signaling and is necessary for the BMP-regulated maintenance of the quiescent and differentiated phenotype of human pulmonary arterial VSMCs. FGF12 was found to regulate the effects of BMP signaling on VSMCs by inducing MEF2a (myocyte enhancer factor 2a) phosphorylation via p38MAPK signaling (Yeo et al., 2020). It is not known whether FGF11 and FGF12 are signaling through FGFRs or through other mechanisms to mediate these effects on ECs and VSMCs.
VSMCs express FGFs 1, 2, 5, 8, 16, and 18, high levels of Fgfr1, low levels of Fgfr2 and Fgfr3, and high levels of Fgfrl1 (Antoine et al., 2005; P. Y. Chen, Qin, et al., 2016a). In response to FGF2, VSMC TGFβ signaling was decreased leading to a switch from a contractile to a proliferative phenotypic (P. Y. Chen, Qin, et al., 2016b). FGFs and TGFβ thus contribute to developmental and pathophysiological interactions between ECs and VSMCs. FGF12 was expressed at high levels in contractile VSMCs. FGF12 induces a quiescent and contractile VSMC phenotype and directly promoted VSMC lineage differentiation. These effects required p38 MAPK signaling, but it is not known if FGFR activation is involved (Song et al., 2016).
Angiogenic FGFR signaling
In zebrafish development, inhibition of FGFR signaling with a highly specific multi-FGFR allosteric inhibitor impaired vascular outgrowth and branching and caused a loss of structural integrity of existing vessels (De Smet et al., 2014).
In mice, inactivation of Fgfr1 and Fgfr2 in ECs did not affect vascular development but did impair injury-induced angiogenesis (Oladipupo et al., 2014). In contrast, mice lacking endothelial Fgfr1 and Fgfr3 showed defects in blood and lymphatic vasculature. These defects were similar to those seen with endothelial loss of the glycolytic enzyme hexokinase 2 (Hk2), and overexpression of Hk2 partly rescued the defects caused by loss of EC FGFR signaling. These data indicate that FGF-dependent regulation of endothelial glycolysis is critical for developmental and adult vascular growth (P. Yu, Wilhelm, et al., 2017).
Bone morphogenetic protein EC precursor–derived regulator (BMPER) is a secreted glycoprotein that enhances BMP signaling. BMPER can enhance the angiogenic response of ECs and is required for coronary vascular remodeling (Esser et al., 2015). The effects of BMPER on ECs are mediated by increased expression of FGF2 and activation of the FGF signaling pathway and decreased expression of thrombospondin-1, which has antiangiogenic properties (Esser et al., 2015).
Endothelial to mesenchymal transition
Developmental, reparative, and pathological angiogenesis often involve vascular remodeling, a process that involves proliferation and migration of ECs and VSMCs and conversion of ECs into a multipotent mesenchymal progenitor that can then form VSMC or other mesenchymal cell types, a process referred to as endothelial to mesenchymal transition (EndMT). In addition to physiological effectors, such as hypoxia and shear stress, TGFβ and FGFs are signaling molecules that regulate EndMT (Akatsu et al., 2019; Chen et al., 2014; Good et al., 2015; Leopold & Maron, 2016; Sanchez-Duffhues et al., 2018; Simons, 2021; Woo et al., 2021; Xiao & Dudley, 2017). TGFβ2 promotes EndMT by activating canonical SMAD2/3 and noncanonical pathways (Leopold & Maron, 2016). TGFβ mediated EndMT is inhibited by FGFR signaling. Knocking down the adapter protein FRS2 in human umbilical artery endothelial cells (HUVECs) caused an increase in the production of the smooth muscle cell markers, αSMA (ACTA2), and vimentin (VIM) (P. Y. Chen, Qin, et al., 2012). EndMT associated with atherosclerosis is enhanced by EC-specific knockout of Fgfr1 or Frs2 (P. Y. Chen, Qin, et al., 2015; Chen et al., 2014). EndMT associated with hypoxia-induced pulmonary hypertension is increased by EC-specific knockout of Fgfr1/2 and reduced by EC-specific expression of a constitutively active FGFR1 (Woo et al., 2021). These studies show that EC FGF signaling limits EndMT through inhibition of EC TGFβ signaling. In support of this mechanism, in tumor-associated ECs, FGF signaling inhibits TGFβ-induced endothelial-to-myofibroblast transition, a form of EndMT (Akatsu et al., 2019).
The endogenous tetrapeptide, N-acetyl-seryl-aspartyl-lysyl-proline (AcSDKP) generated from thymosin β4 inhibits EndMT through induction of FGFR1 and KLB in ECs (R. Gao, Kanasaki, et al., 2019). AcSDKP and FGF21 synergize to suppress EndMT (R. Gao, Kanasaki, et al., 2019; J. Li, Shi, et al., 2017). Inactivation of EC FGFR1 or KLB blocked the ability of AcSDKP to inhibit EndMT (R. Gao, Kanasaki, et al., 2019; J. Li, Liu, et al., 2020).
Atherosclerosis
Atherosclerosis is a chronic metabolic disease of the arteries that leads to multifocal plaque development. In high-fat diet (HFD)-fed apolipoprotein E deficient (ApoE−/−) mice, an in vivo model of atherosclerosis, the parenteral administration of FGF1 facilitates the progression of atherosclerosis, with increased expression of PPARα and inflammatory cytokines (J. Han, Du, et al., 2018). In HFD-fed ApoE−/− mice, inactivation of Fgf2 (18 kDa isoform) attenuated the progression of atherosclerosis by reducing aortic plaques, macrophage infiltration, and oxidative stress. These data suggest that FGF2 worsens inflammatory reactions that promote atherosclerosis (W. Liang, Wang, et al., 2018). In contrast, in HFD-fed ApoE−/− mice, phosphorylation of hepatic farnesoid X receptor by FGF15/19 signaling maintained serum cholesterol levels and protected against atherosclerosis (Byun et al., 2019). In patients with type 2 diabetes (T2D), serum FGF15/19 levels are independently and inversely associated with arteriosclerosis parameters, supporting a protective role for serum FGF15/19 in atherosclerosis progression (W. S. Liu, Tang, et al., 2020). FGF21 also mitigates atherosclerosis in HFD-fed ApoE−/− mice (Maeng et al., 2020). FGF21 also reduced Fas-mediated apoptosis induced by oxidized LDL in HUVECs and in ApoE−/− mice (Yan et al., 2018). However, in patients with T2D, serum FGF21 levels correlate with carotid intima-media thickness and were predictive of subclinical atherosclerosis (Yafei et al., 2019), and serum FGF23 levels were independently and positively correlated with lower extremity atherosclerotic disease (He et al., 2017). In patients with gestational diabetes, serum FGF23 levels positively correlated with subclinical atherosclerosis as assessed by carotid artery intima-media thickness (Tuzun et al., 2018).
Tumor angiogenesis
Angiogenesis is essential for the growth of solid tumors (Folkman, 1971). FGFs were originally identified as angiogenic factors and can synergize and potentially replace VEGF to support EC growth (Giacomini et al., 2016; Haibe et al., 2020; Ornitz & Itoh, 2015; Presta et al., 2005, 2017; Turner & Grose, 2010; Zahra et al., 2021; Zhao & Adjei, 2015). Coupled with the observations that many tumors produce FGF ligands, inhibiting the FGF pathway has long been considered as a viable adjunct antitumor therapeutic strategy (Ronca, Giacomini, Rusnati, & Presta, 2015; Touat et al., 2015; Turner & Grose, 2010; Zhao & Adjei, 2015).
Hypoxia is a characteristic feature of solid tumors. Hypoxia-inducible factor-2α (HIF-2α) was shown to promote cMyc-mediated repression of miR15a and miR16, which normally posttranscriptionally inhibits FGF2. Thus, hypoxia can lead to increased expression of FGF2 as a pathway to promote tumor angiogenesis (Xue et al., 2015). In a breast cancer xenograft model, antibody inhibition of FGF2 reduced tumor cell proliferation and microvascular density (Cai et al., 2016). An in vivo model in which either VEGF-A (121 aa) or FGF4 drives tumor angiogenesis was used to determine differential effects on tumor blood vessels. Vessel density was increased by both factors, but vascular permeability was greater with VEGF-A compared to FGF4. FGF4-stimulated vessels had increased numbers of pericytes, suggesting that the pericyte may also directly respond to FGF4 signaling (Hori et al., 2017).
Using a mouse genetic model and skin carcinogenesis or tumor transplantation, haploinsufficiency of VEGFR2 was shown to decrease tumor growth, but surprisingly, knockout of EC FGFR1/2 did not affect tumor growth (Oladipupo et al., 2018). Given the established roles of FGFR signaling in tumor angiogenesis, this suggests that FGFR3, alone or in combination with FGFR1/2, should be further evaluated for synergy with VEGFR2 in tumor angiogenesis.
Consistent with potential redundancies with angiogenic RTKs, inhibitors that target multiple RTKs (VEGFRs, FGFRs, PDGFRs) have shown efficacy in xenograft models using several different tumor types (Caglevic et al., 2015; D. Li, Xie, Zhang, et al., 2016, Li, Fu, Zhao, et al., 2016; Yamamoto et al., 2014).
10.4.3 |. Regeneration and response to injury
Canonical FGF signaling
FGF signaling is cardioprotective in various models of myocardial infarction (MI) (Kardami et al., 2007; Khosravi et al., 2021; Tanajak et al., 2015; J. Wang, Sontag, & Cattini, 2015; S. Wang, Li, et al., 2018). Mature cardiomyocytes express high levels of Fgfr1 and Fgfr3, lower levels of Fgfr2 and Fgfr4, and the endocrine FGF cofactor, βKlotho (Grabner et al., 2015; Liu et al., 2013; Planavila et al., 2013). Cardiomyocytes do not express detectable levels of αKlotho (Faul et al., 2011). The adult heart expresses high levels of FGF2 and FGF16 and lower levels of FGF9 and FGF10 (Hotta et al., 2008; House et al., 2015; Kardami et al., 2007; Miyake et al., 1998; Rochais et al., 2014).
In mouse modes of neonatal heart injury, inactivation of the GATA4 transcription factor in cardiomyocytes worsened outcomes. Fgf16 expression was decreased in hearts lacking GATA4, and GATA4 was found to directly regulate Fgf16 expression. Cardiac-specific overexpression of FGF16 in GATA4 mutant hearts partially rescued the injury-induced cardiac hypertrophy (Yu et al., 2016).
In zebrafish amputation-induced heart injury, FGF signaling is important for neovascularization of the regenerating myocardium through ERK activation in ECs (Lowe et al., 2021). Additionally, following heart injury, the number of cardiomyocytes with activated FGFR-induced AKT signaling was increased. Inhibition of AKT led to increased cardiomyocyte death, suggesting that FGFR-AKT signaling functions as a survival factor that is necessary for subsequent heart regeneration (Tahara et al., 2021).
In a model of closed-chest ischemia–reperfusion (IR) injury in mice, inactivation of EC Fgfr1 and Fgfr2 resulted in increased infarct size, demonstrating that EC FGFR1/2 signaling is necessary for the angiogenic response to IR injury (House et al., 2016). In a rat IR injury model, intravenous treatment with a FGF1 mutant with reduced heparin-binding affinity reduced infarct size and improved cardiac function (C. Huang, Liu, et al., 2017). In a mouse MI model, injection of recombinant FGF9 post-MI into the peri-infarct region reduced vessel apoptosis and increased left ventricular output (Singla & Wang, 2016). Intraperitoneal injection of FGF21 reduced the occurrence of ventricular tachycardia, improved epicardial conduction velocity, and shorted action potential duration following MI (J. Li, Xu, et al., 2020). In a rat MI model, intraperitoneal administration of FGF21 before and after MI resulted in reduced infarct size and improved ejection fraction and reduced myocardial inflammation and fibrosis (J. Li, Gong, Zhang, et al., 2021). In vitro studies showed that FGF21 induced expression of early growth response protein 1 (EGR1) by suppressing miR-143. EGR1 transcriptionally induced the expression of sodium and potassium channels (SCN5A and KCNJ2) and suppressed the expression of inflammatory cytokines (J. Li, Gong, Zhang, et al., 2021; J. Li, Xu, et al., 2020).
FGF16 pretreatment was found to protect against doxorubicin-induced cardiotoxicity (Sontag et al., 2013). In response to doxorubicin, endogenous levels of Fgf16 mRNA were reduced secondary to reduced binding of the transcription factor Nkx2–5 to the Fgf16 promoter (J. Wang, Jin, & Cattini, 2017). Rat neonatal cardiomyocyte cultures treated with a mutant non-mitogenic form of FGF2 (S117A-FGF2) protected against doxorubicin-induced cardiotoxicity (Koleini et al., 2018).
In a mouse model of pressure-overload-induced systolic dysfunction, mice lacking the high molecular weight isoform of FGF2 showed improved systolic function. These mice had increased levels of FGFR1, increased FGFR-regulated expression of mTOR, and increased expression of the cardioprotective molecule, NR1D1 (nuclear receptor subfamily 1 group D member 1) (Koleini et al., 2021).
Regeneration of damaged myocardium with cell-based therapy is a goal of regenerative medicine. Adult bone marrow-derived mesenchymal stem cells (BMSCs) are a heterogeneous population of cells with stem cell-like properties (discussed later). BMSCs were shown to differentiate into a noncontractile cardiomyocyte-like cell when treated with FGF2 and hydrocortisone. This method was found to be more efficient and potentially safer than the traditional use of 5-azacytidine to induce cardiomyocyte differentiation (Hafez et al., 2016). A combination of FGF2, FGF10, and VEGF was able to promote cardiac reprogramming of fibroblasts into beating cardiomyocyte-like cells under defined serum-free conditions through activation of the p38 MAPK and AKT pathways (Yamakawa et al., 2015).
Pathologic neovascularization occurs in many diseases. In the eye, in advanced age-related macular degeneration, pathologic neovascularization can cause blindness. In a mouse model of choroidal neovascularization, FGF2 was identified as the pathogenic ligand that regulates pathogenic angiogenesis via STAT3 activation through signaling to endothelial FGFR1/2 (Z. Dong, Santeford, et al., 2019; Oladipupo et al., 2014).
In a hindlimb ischemia model, Fgf2−/− mice showed delayed recovery of limb function. However, the vascular growth response to ischemia was similar between wild type and Fgf2−/− hindlimbs. The delayed recovery was attributed to increased expression of inflammatory proteins and increased neutrophil or monocyte recruitment/infiltration in Fgf2−/− ischemic muscle (Adeyemo et al., 2020).
Endocrine FGF signaling
In the heart, hypertension, cardiomyopathy, heart failure, and MI induce cardiac FGF21 and KLB expression (Planavila et al., 2015). FGF21 induced in the heart protects against hypertensive heart disease and alcohol-induced cardiomyopathy, indicating that FGF21 may function as a paracrine factor in the heart (Ferrer-Curriu et al., 2019, 2021; Khosravi et al., 2021). In a mouse model of traumatic brain injury, administration of recombinant FGF21 reduced neurofunctional behavior deficits, cerebral edema, brain tissue loss, and neuronal apoptosis by preserving the integrity of the blood–brain barrier (J. Chen, Hu, et al., 2018). Recombinant FGF21 also promoted angiogenesis and migration of human brain microvascular ECs through activation of PPARɣ and increased expression of endothelial nitric oxide synthase (eNOS) (W. Huang, Shao, et al., 2019).
In diabetic mice (db/db), FGF21 was found to alleviate aortic EC disfunction by reducing hyperglycemia and insulin resistance (Ying et al., 2019). Ectopic mineral depositions in blood vessel walls (vascular calcification) leads to increased vascular stiffness and decreased vascular compliance and can cause serious adverse cardiovascular events. Vascular calcification involves the differentiation of VSMCs to an osteoblast-like phenotype. VSMCs express FGFR1, FGFR2, and KLB, co-receptors for endocrine FGF21. In an in vitro model system, FGF21 suppressed VSMC calcification through activation of p38 MAPK and the pre-osteoblast transcription factor, RUNX2 (Cao et al., 2017).
Circulating levels of FGF23 are greatly increased in CKD. FGF23 was found to signal to cardiomyocytes in an αKlotho-independent mechanism to stimulate left ventricular hypertrophy (Faul et al., 2011). This effect was abolished in mice lacking FGFR4 or in mice treated with blocking antibodies to FGFR4 (Grabner et al., 2015). Importantly, blockade of FGFR4 also reversed established left ventricular hypertrophy (Grabner et al., 2017).
In the heart, FGF23 is expressed in cardiomyocytes, fibroblasts, VSMCs, coronary artery ECs, and inflammatory macrophages. FGF23 directly induces pro-hypertrophic genes and promotes the progression of left ventricular hypertrophy through both autocrine and paracrine signaling, and can stimulate secretion of pro-fibrotic factors from cardiomyocytes to promote cardiac fibrosis (Leifheit-Nestler & Haffner, 2018). Elevated levels of FGF23 can contribute to the development of heart failure with preserved ejection fraction (HFpEF) by suppressing coronary microvascular function and by promoting angiotensin II-induced vascular and myocardial fibrosis (van de Wouw et al., 2019). HFpEF patients, compared with controls of similar age and sex, had higher levels of FGF23, and the level of FGF23 was a strong predictor of poor outcomes (Roy et al., 2020).
10.5 |. Respiratory system
10.5.1 |. Lung development
FGF signaling has essential roles in all stages of lung development (Chao et al., 2015, 2016; Danopoulos, Shiosaki, & Al Alam, 2019; El Agha, Seeger, & Bellusci, 2017; Herriges & Morrisey, 2014; Kiyokawa & Morimoto, 2021; Ornitz & Yin, 2012; Rankin et al., 2015; Vila Ellis & Chen, 2021; Volckaert & De Langhe, 2015; Wu et al., 2018; Xiao & Dudley, 2017).
Embryonic-pseudoglandular stage lung development
At the earliest stages of lung development, Fgf10 is expressed in mesenchyme adjacent to the sites of lung bud formation where it signals to FGFR2b in foregut endoderm to induce expression of Nkx2–1, a transcription factor that defines the foregut lung field. Mice that lack Fgf10 fail to form primary lung buds. Conditional inactivation of Fgf10 or its receptor, Fgfr2b, after formation of the primary lung buds results in decreased epithelial branching. Similarly, expression of a Fgfr2b ligand trap in pig lung epithelium inhibited normal branching (Q. Chen, Fang, et al., 2018). In an experimental model of doxorubicin-induced lung hypoplasia, decreased expression of Fgf10 and its downstream effectors, Bmp4 and Cathepsin H (Ctsh) were associated with impaired airway branching and epithelial cell development (J. Wang, Liu, et al., 2018). Timed inhibition of FGF10 through the induced expression of a soluble FGFR2b ligand trap showed that FGF10 regulates lobular septation of the right lung at mouse embryonic day 9 (E9) and regulates branching morphogenesis after E11 (Taghizadeh et al., 2020). This supports the existence of an early embryonic stage that is temporally and functionally distinct from the subsequent pseudoglandular stage.
In lung development, a bipotential progenitor gives rise to alveolar type 1 (AEC1) and alveolar type 2 (AEC2) cells (Frank et al., 2019). AEC1 cells are very thin cells that allow gas exchange with the underlying capillary network. AEC2 cells produce surfactant and in lung regeneration function as progenitors that give rise to AEC1 cells. Induced expression of a soluble FGFR2b ligand trap or conditional inactivation of Fgfr2 during the pseudoglandular stage of lung development (E12.5) decreased expression of markers of differentiated epithelium (Jones et al., 2018; Yin & Ornitz, 2020). Induction of the FGFR2b ligand trap during midgestation (E14.5) development resulted in reduced AEC2 cell proliferation (M. R. Jones, Lingampally, et al., 2020). FGF10 was also shown to induce protrusion of a subset of alveolar epithelial cells toward the mesenchyme to maintain their AEC2 cell fate and counter mechanical stretching forces that favor AEC1 cell differentiation (Li, Wang, Chu, et al., 2018). Culture of embryonic (E13.5) lung-tip epithelium in matrigel in the presence of insulin, transferrin, selenium, and FGF7 induced the formation of an alveolus-like organoid that express markers of AEC1 and AEC2 cells (Seiji et al., 2019). This is consistent with FGF7 and FGF10 having similar receptor specificities (Zhang, Ibrahimi, et al., 2006).
In mouse embryonic lung explants, treatment with FGF10 induces branching which can be blocked by inhibiting MAPK (Yin & Ornitz, 2020). In contrast, treatment of human embryonic lung explants with FGF10 induced epithelial cyst formation (Danopoulos, Thornton, et al., 2019). These discordant responses to FGF10 could be due to different relative stages of development or species-specific differences. Etv4 and Etv5 are transcriptional targets of FGF signaling (Brent & Tabin, 2004). Inactivation of Etv4/5 in lung epithelium directly led to reduced SHH expression and indirectly to increased FGF10 production, resulting in increased epithelial outgrowth and delayed formation of new branches (Herriges et al., 2015).
Fgf9 is expressed in the mesothelium and epithelium of the developing lung at early stages of branching morphogenesis (Danopoulos, Thornton, et al., 2019; White et al., 2006; Yin et al., 2011). Mice lacking Fgf9 have severely hypoplastic lung development, characterized by reduced distal mesenchyme and decreased epithelial branching (Ornitz & Yin, 2012). FGF9 derived from the mesothelium signals to FGFR1 and FGFR2 in lung mesenchyme and epithelial-derived FGF9 is important for branching (White et al., 2006; Yin et al., 2011). Deregulated expression of FGF9 in embryonic lung epithelium, as occurs in pleuropulmonary blastoma, contributes to the mesenchymal hyperplasia characteristic of this disease (Yin et al., 2015). Treatment of mouse embryonic lung explant cultures with FGF9 induced expansion of FGF10 lineage positive cells and decreased numbers of ACTA2+ airway smooth muscle cells (El Agha, Kheirollahi, et al., 2017; White et al., 2006). Treatment of either human or mouse explants with FGF9 resulted in expansion of SOX9-expressing distal epithelium (Danopoulos, Thornton, et al., 2019; Yin & Ornitz, 2020). FGF9 treatment of mouse embryonic explants led to epithelial dilation, a response that could be partially blocked with inhibition of PI3K but not MAPK (Yin & Ornitz, 2020). Collectively, these studies show that within lung epithelial cells, different FGFRs function independently by binding receptor-specific ligands and activating distinct downstream signaling pathways to direct distinct developmental functions.
Saccular–alveolar stage lung development
Alveologenesis is the final stage of lung development where the gas exchange surface area of the lung is increased by the formation of secondary septae in alveolar saccules, followed by thinning of the septal walls. Impaired alveologenesis is a major cause of morbidity in extremely preterm infants and often results in bronchopulmonary dysplasia (BPD) (Chao et al., 2016; Lignelli et al., 2019; Vila Ellis & Chen, 2021). Alveologenesis can be divided into three stages: pre-alveologenesis in which AEC1 cells flatten in the saccular stage lung to expand lung surface area and permit gas exchange; primary alveologenesis where formation of secondary septae that contain contractile myofibroblasts increase the number and surface area of alveoli; and secondary alveologenesis characterized by septal remodeling and alveolar expansion in the absence of alveolar myofibroblasts (Vila Ellis & Chen, 2021).
Alveolar septae have matrix fibroblasts that contain lipid droplets and elastin-producing myofibroblasts. Matrix fibroblasts express low levels of the Pdgfra-GFP reporter and high levels of Fgfr3, Fgfr4, and Fgf10, and myofibroblasts express high levels of the Pdgfra-GFP reporter and Fgf18 (McGowan & McCoy, 2015; Ruiz-Camp & Morty, 2015). During late embryonic development, FGF10 signaling to FGFR1b and FGFR2b is important for generation of lipofibroblasts (a type of matrix fibroblast), a cell type that is retained in adult lung and is important for maintaining epithelial stem cells (Al Alam, El Agha, et al., 2015; Chao et al., 2016).
Inactivation of Fgfr3 and Fgfr4, globally or broadly within embryonic lung mesenchyme, resulted in impaired alveolar septation demonstrating that FGFR3 and FGFR4 are necessary for alveologenesis (R. Li, Herriges, et al., 2017; Srisuma et al., 2010; Weinstein et al., 1998). Mice lacking Fgfr3 and Fgfr4 showed a disorganization of the elastin-rich ECM in developing alveolar septae. Tissue-specific gene inactivation showed that loss of mesenchymal FGFR3 was critical for recapitulating the phenotype of the whole organism knockouts (R. Li, Herriges, et al., 2017).
Ligand specificity studies show that FGFR3 and FGFR4 are potential receptors for FGF18 (Zhang, Ibrahimi, et al., 2006). Fgf18 is expressed at low levels in the saccular stage lung and increases dramatically during primary alveologenesis, suggesting that FGF18 may be a critical FGF ligand that activates FGFR3 and FGFR4 (Boucherat et al., 2007; Chailley-Heu et al., 2005; Franco-Montoya et al., 2011). In neonatal mouse lung, RA is produced by pulmonary ECs and regulates pulmonary angiogenesis and elastin synthesis by induction of Vegfa and Fgf18, respectively. Treatment of neonatal lung fibroblasts with RA increased expression of Fgf18 (Yun et al., 2016).
Expression analysis of neonatal lung mesenchymal cells sorted based on expression of the Pdgfra-GFP reporter shows that Fgf18 is primarily expressed in alveolar myofibroblasts (AMFs) and Fgf10 is expressed in lipofibroblasts (McGowan & McCoy, 2015). Lineage tracing with an Fgf18CreER knockin allele showed that AEC1 and AMFs are the major cell populations that express Fgf18 during pre- and primary stages of alveologenesis and that the majority of AMFs are cleared from the lung by the end of the primary stage of alveologenesis (Hagan et al., 2019, 2020; Vila Ellis & Chen, 2021). In adult mice, most of the AEC1 cells that were lineage-labeled during alveologenesis are retained (Hagan et al., 2020). During secondary alveologenesis, AEC1 cells continue to express Fgf18 suggesting a distinct role for FGF18 in septal remodeling (Hagan et al., 2020). Single cell RNA seq data confirm these expression patterns (Du et al., 2015).
10.5.2 |. Trachea development
The tracheal primordium arises from the ventral anterior foregut. Multiple signaling pathways, including Wnt, Bmp, RA, Shh, and Fgf, coordinate interactions between the epithelium and mesenchyme to specify tracheal identity and regulate separation from the esophagus (Kiyokawa & Morimoto, 2021). The tracheal epithelium is established and maintained by basal progenitor cells. Inactivation of mesenchymal WNT signaling or epithelial FGFR2 signaling results in reduced numbers of basal progenitor cells (Hou et al., 2019).
Cartilaginous C-shaped tracheal rings are important to maintain distension of the trachea. In patients with syndromic craniosynostosis syndromes (e.g., Pfeiffer, Crouzon, Apert, Beare-Stevenson, and Saethre-Chotzen) failure of segmentation of the C-shaped tracheal rings can occur, increasing the risk of mortality in these patients (Mahmud et al., 2021; Pickrell et al., 2017; Seki et al., 2020; Wenger et al., 2017). Mice heterozygous or homozygous for activating mutations in Fgfr2 (p.C342Y) that cause Crouzon syndrome fail to segment tracheal cartilage resulting in a rigid tracheal sleeve. Prior to segmentation, mutant mice show increased chondrocyte proliferation (Hines et al., 2019; Lam et al., 2021; Peskett et al., 2017).
Submucosal glands that line the airway produce mucus in the tracheal and nasal airways. Conditional knockout studies show that Fgf10 is required for submucosal gland development, regulating the number of glands and extent of epithelial branching (May et al., 2019).
10.5.3 |. Lung homeostasis and response to injury
Bronchopulmonary dysplasia
Exposure of premature infants to hyperoxia is essential for survival but can lead to bronchopulmonary dysplasia, characterized by alveolar simplification, pulmonary vascular rarefaction, and increased vascular muscularization. Heterozygosity of Fgf10 or inhibition of FGF10 signaling with expression of a soluble FGFR2b ligand trap further decreased the density of the pulmonary vasculature and increased muscularization of remaining vessels, a phenotype that can cause pulmonary hypertension, a severe and often fatal outcome of bronchopulmonary dysplasia (Chao et al., 2019). Genetic analysis of patients that died of neonatal lung diseases identified rare coding variants and hypomorphic noncoding single-nucleotide variants in the genes encoding TBX4 and FGF10, suggesting a complex compound inheritance predisposing to pulmonary hypoplasia (Karolak et al., 2019). In a neonatal mouse model of hyperoxia-induced lung injury, expression levels of FGF18 were reduced. Treatment with FGF18 increased viability of AEC2 cells and reduced oxidative stress and inflammation (X. G. Li, Song, Wang, et al., 2021).
Pulmonary fibrosis
Idiopathic pulmonary fibrosis (IPF) is a progressive interstitial lung disease in which alveolar tissue remodeling and fibrosis leads to respiratory failure (Goodwin & Jenkins, 2016). In whole lung homogenates from IPF patients, FGF1 expression and downstream targets of FGF-signaling, phospho-ERK1/2, and phospho-AKT, were increased (MacKenzie, Korfei, et al., 2015). Remodeled alveolar epithelium close to the fibroblast foci in IPF lungs expressed FGF9, FGF18, and all FGFRs. In vitro analysis of human lung fibroblasts showed that FGF9, and to a lesser extent FGF18, act as antiapoptotic and promigratory growth factors to maintain fibroblasts in an undifferentiated state contributing to the fibrotic process (Joannes et al., 2016). In patients with progressive compared to stable IPF, mesenchymal stromal cells (MSCs) isolated from bronchoalveolar lavage fluid showed activation of TGFβ and SHH signaling pathways and decreased expression of FGF10. TGFβ and SHH signaling were shown to suppress FGF10 expression in MSCs isolated from patients with progressive IPF (Chanda et al., 2016).
In primary lung fibroblasts from human IPF patients and mouse bleomycin-induced lung fibrosis, levels of αKlotho (KL) were reduced. In a mouse bleomycin model, co-administration of KL and FGF23 were protective, with the reduction in TGFβ-induced fibrosis and inflammation (Barnes et al., 2019).
Overexpression of FGF7 or FGF10, ligands for FGFR2b, were protective to bleomycin-induced injury; however, endogenous FGF7 or FGF10 did not significantly protect against bleomycin-injury (MacKenzie, Henneke, et al., 2015). In a rat model of TGFβ overexpression, adenoviral-mediated expression of FGF1 attenuated TGFβ-induced myofibroblast differentiation and pulmonary fibrosis. In vitro, FGF1 attenuated the TGFβ signaling pathway in alveolar epithelial cells (AECs) and primary human lung fibroblasts (Shimbori et al., 2016). In a mouse bleomycin-induced injury model, Wnt/β-catenin signaling was activated and TGFβ and FGF2 expression were increased. Inhibition of Wnt/β-catenin signaling led to decreased pulmonary fibrosis and decreased production of TGFβ and FGF2 in bleomycin-treated mice and in AEC2 cells cultured with bleomycin (X. Chen, Shi, et al., 2016). In mice, overexpression of FGF2 protected against bleomycin-induced pulmonary fibrosis in vivo and reversed TGFβ-induced fibroblast to myofibroblast differentiation in vitro. FGF2 expression did not affect inflammation or epithelial gene expression (Koo et al., 2018).
In adult lung, Fgfr2 was expressed at high levels in AEC2 and club cells, at low levels in AEC1 cells, and was not detected in ciliated or neuroendocrine cells. Following bleomycin-induced injury, Fgfr2 expression was increased in distal airway club cells (Yuan et al., 2020). Inactivation of Fgfrs 1, 2, and 3 in AEC2 cells in adult mice resulted in depletion of targeted AEC2 cells, decreased Sftpc (surfactant protein C) gene expression, increased alveolar diameter, increased collagen deposition, and an impaired response to bleomycin-induced injury. Induced inactivation of Fgfr2 or Fgfr3 individually showed that Fgfr2, but not Fgfr3, was responsible for the increased mortality and lung injury after bleomycin administration (Dorry et al., 2020). Similarly, inactivation of Fgfr2b in mature AEC2 cells of adult mice resulted in a nearly complete loss of targeted AEC2 cells (Yuan et al., 2019). A third study showed that Fgfr2-deficient AEC2 cells had decreased proliferation and increased differentiation into AEC1 cells in areas of active alveolar remodeling but no depletion of AEC2 cells under homeostatic conditions (Liberti et al., 2021). Consistent with a role for FGF signaling in both epithelial cells and fibroblasts, blockade of FGFR signaling after bleomycin injury with the inhibitor BGJ398 (Infigratinib) reduced lung fibrosis but increased mortality due to suppression of AEC regeneration (Morizumi et al., 2020).
Spred2 is a negative regulator of the MAPK–ERK pathway and is predominantly expressed in bronchial epithelial cells. In response to bleomycin, mice lacking Spred2 show increased proliferation of bronchial epithelial cells which may contribute to the observed milder fibrosis phenotype (Kawara et al., 2020).
Epithelial injury
Mice lacking Fgfr4 undergo normal embryonic and neonatal development; however, as adults Fgfr4−/− mice show airway inflammation and an emphysema-like phenotype with widened airway spaces, increased airway inflammation, bronchial obstruction, and right ventricular hypertrophy. Whole lungs from Fgfr4−/− mice had a complete loss of phosphorylated p38 but no significant change in phospho-ERK1/2 (Easter et al., 2020).
Injury to distal conducting airways with naphthalene, which kills club cells, leads to the emergence of a distinct population of PDGFRA expressing cells that are distinct from airway smooth muscle cells. This repair-supportive mesenchymal cell expresses high levels of FGF10 which supports the regeneration of club cells (Moiseenko et al., 2020).
Basal cells (BCs) are a stem cell population in the trachea that maintain self-renewing luminal secretory cells and terminally differentiated ciliated cells. Inactivation of Fgfr1 or Spry2 in BCs of the adult mouse trachea caused an increase in steady-state BC proliferation. In response to FGFR1 signaling, SPRY2 is posttranslationally modified allowing it to inhibit intracellular signaling downstream of other RTKs and restrain BC proliferation. Consistent with these in vivo data, treatment of primary BC cultures with FGF2 led to smaller colony formation (Balasooriya et al., 2016). Inactivation of one copy of Fgfr2 in basal cells leads to gradual loss of mutant basal and luminal cells, demonstrating that the FGFR2 signaling is important for homeostatic maintenance of BCs and their progeny. Treatment of primary BCs with FGF7, a ligand for FGFR2b, increased colony size (Balasooriya et al., 2017). These studies show that FGFR1 and FGFR2 have independent functions in airway BCs.
Asthma
Asthma is a chronicinflammatory disease of the airways of the lungs. The proliferation and migration of airway smooth muscle cells (ASMCs) play an important role in asthma. In a rat model for asthma induced by ovalbumin injection, the long noncoding RNA, TUG1, promoted ASMC proliferation and migration, and reduced apoptosis. As discussed above, TUG1 increased FGF1 expression through suppression of miR-590–5p, resulting in increased proliferation and migration of ASMCs (J. Lin, Feng, et al., 2019). Patients with asthma have lower levels of miR-192–5p, which functions to suppress airway smooth muscle cell proliferation and expression of matrix metalloproteinase 16 (MMP16) and autophagy-related 7 (ATG7). In a mouse model for asthma, treatment with a miR-192–5p agomir attenuated airway remodeling, possibly mediated by reduction of FGF23 and MMPs 2, 9, and 16 (Lou et al., 2020).
FGF2 has been implicated as an immunomodulatory factor in asthma and COPD (Tan et al., 2020). In a house dust mite antigen-induced mouse models of asthma, IL17 produced by infiltrating eosinophils and neutrophils led to increased FGF2 expression, which was associated with smooth muscle hypertrophy/hyperplasia (Ogawa et al., 2018).
Chronic obstructive pulmonary disease
Chronic obstructive pulmonary disease (COPD) is a chronic inflammatory airway disease. COPD and asthma have different etiologies but share pathological features including chronic inflammation, airflow limitation, and airway wall remodeling. FGF2 was decreased in the lungs of mice exposed to cigarette smoke or with elastase-induced COPD, and in serum from human patients with COPD. Administration of FGF2 (intranasally) reduced inflammation and alveolar destruction in smoke-exposed mice and improved regeneration in mice with elastase-induced COPD (Y. S. Kim, Hong, et al., 2018).
Genome-wide association studies of COPD patients indicate that variants in the branching structure of the lung is associated with intronic single nucleotide polymorphisms (SNPs) in FGF10 and an increased risk for COPD (Smith et al., 2018) Another study identified variants in FGF7 that are associated with COPD (X. Zhang, Guo, et al., 2019). Serum FGF23 levels were significantly elevated and KL levels were reduced in patients with mild to moderate COPD but not severe COPD. Exposure of airway epithelial cells to cigarette smoke and FGF23 activated FGFR4/PLCɣ independent of KL to increase the release of interleukin-1β (Krick et al., 2018). Consistent with this study, serum levels of FGF23 were positively correlated with COPD exacerbations (Gulati et al., 2019). Inhibition of FGF23 signaling might therefore serve as a novel anti-inflammatory strategy in COPD. However, another study showed reduced serum levels of full-length FGF23 in COPD patients but did not suggest a causative role for FGF23 (Stroda et al., 2018).
10.6 |. Gastrointestinal tract
10.6.1 |. Intestine
FGF9 and FGF10 have essential roles in the development of the gastrointestinal (GI) tract. Fgf10 is expressed in GI tract mesenchyme and signals to epithelium. Mice lacking Fgf10 or Fgfr2b have impaired development of the stomach, duodenum, cecum, and colon (Danopoulos et al., 2017; Kowalkowski et al., 2020; Lv et al., 2019). For example, genetic knockout of Fgf10 or Fgfr2b results in duodenal or colonic atresia in mice (M. L. M. Jones, Sarila, et al., 2020; Kowalkowski et al., 2020; Teague et al., 2018). Examination of the colonic epithelium in Fgfr2b−/− mice showed increased apoptosis but normal levels of proliferation. Epithelial histology showed lack of a pseudostratified columnar architecture and discontinuity of the adjacent basal lamina with individual E-cadherin (Cdh1) expressing cells found in the colonic mesenchyme (Kowalkowski et al., 2020).
Fgf9 is expressed in GI tract epithelium and signals to mesenchyme. Mice lacking Fgf9 have a shortened small intestine and lack a cecal bud (Al Alam et al., 2012; Danopoulos et al., 2017; Geske et al., 2008; Zhang, Stappenbeck, et al., 2006).
FGF10 may also have a role in intestinal homeostasis. Overexpression of FGF10 in the adult leads to an increase in crypt depth and villus height in the small intestine, and it induces goblet cell differentiation at the expense of Paneth cells (Al Alam, Danopoulos, et al., 2015). FGFR3 may antagonize FGF10 signaling, as mice lacking FGFR3 have reduced numbers of intestinal crypts and reduced Paneth cell specification and differentiation (Vidrich et al., 2009).
10.6.2 |. Liver
Canonical and endocrine FGF signaling have essential roles in liver development, response to injury, and tumorigenesis (Z. Chen, Jiang, et al., 2021; Itoh, Nakayama, & Konishi, 2016; Seitz & Hellerbrand, 2021; Y. Wang, Liu, et al., 2021).
Liver development
Fgf8 is expressed in the prospective hepatic endoderm and the adjacent splanchnic mesoderm in the early embryonic stages of mouse development (J. Wang, Rhee, et al., 2015). FGF8 is expected to be required for liver development. However, as Fgf8−/− mice are lethal at the gastrulation stage, its roles in liver development remain to be elucidated (Sun et al., 1999). Fgf10 is also expressed in the mesoderm proximal to the hepatic endoderm (J. Wang, Rhee, et al., 2015). Fgf10−/− mouse embryos with smaller livers exhibit reduced proliferation and survival of hepatoblasts, indicating that FGF10 is essential for liver growth during embryogenesis and for hepatoblast growth (Berg et al., 2007). Human induced PSCs (iPSCs) can be differentiated into hepatocyte-like cells through definitive endoderm cells and hepatoblast-like cells. During hepatoblast differentiation from human iPSCs, the levels of hepatoblast markers are upregulated by the removal of FGFs. Hepatoblast differentiation is also promoted by an FGFR inhibitor. These results indicate FGF signals are not necessary for hepatoblast differentiation in vitro (Toba et al., 2019).
Liver injury
Hepatic injury is associated with the activation of hepatic stellate cells. Fgf9 expression is increased in hepatic stellate cells in an acute liver slice injury model in vitro. FGF9 significantly stimulates the incorporation of thymidine by hepatocytes. FGF9 is a paracrine mitogenic signal to hepatocytes during acute liver injury (Antoine et al., 2007). In mice lacking Fgf7, following toxin-induced hepatic injury liver progenitor cell expansion is decreased, and mice have increased mortality. In addition, in transgenic mice that overexpress FGF7, liver progenitor cells are increased and hepatic dysfunction is ameliorated. These results indicate that FGF7 is a regulator of liver progenitor cells in response to liver injury (Takase et al., 2013). Recombinant FGF7 protein also exerted a protective effect on mice with CCl4-induced acute liver injury (M. Liu, Chen, et al., 2018).
Inducible loss (mediated by siRNA) of Fgfr4 in the liver severely impaired liver regeneration following partial hepatectomy and combined loss of Fgfr1, Fgfr2, and Fgfr4 in hepatocytes caused liver failure following partial hepatectomy. Serum levels of FGF15/19 were increased following partial hepatectomy and further increased in mice lacking hepatocyte Fgfrs 1, 2, and 4 (Padrissa-Altes et al., 2015). These data demonstrate partial redundancy of FGFRs in the liver and an essential role for FGFR signaling in liver regeneration. In iron overload-induced liver diseases, FGF21 administration or Fgf21 overexpression protected against iron overload-induced hepatocyte mitochondria damage, liver injury, and fibrosis, by inhibiting ferroptosis (Wu et al., 2021).
In zebrafish liver, Fgf signaling from surrounding mesenchymal cells maintains an extrahepatic niche by directly preventing premature differentiation and allocation of extrahepatic duct progenitors to the liver. Intrahepatic duct cells regenerate after activation of Jag/Notch signaling from multipotent progenitors originating from this Fgf-dependent extrahepatic stem cell niche (Zhao et al., 2021).
Hepatocellular carcinoma
Hepatocellular carcinoma (HCC) is closely associated with liver fibrosis (Alvarez-Sola et al., 2017). In a mouse model for HCC, mice lacking Fgf15/19 showed fewer and smaller tumors, decreased hepatocellular proliferation, and decreased expression of alpha-fetoprotein. Mechanistically, FGF15/19 induced the expression of connective tissue growth factor (CTGF) in hepatocytes, which promoted liver fibrosis, an established enhancer of HCC (Uriarte et al., 2015). Selective inhibition of FGFR4 with Fisogatinib (BLU-554) showed clinical benefit and tumor regression in patients with HCC with aberrant FGF19 expression (Hatlen et al., 2019).
High FGF9 expression levels in human HCC significantly correlated with poor patient survival. FGF9 is expressed in hepatic stellate cells but not HCC cells and promotes tumorigenicity. FGF9 appears as a potential prognostic marker and novel therapeutic target in HCC (Seitz et al., 2020).
Mice maintained on a high-fat high sucrose diet develop NAFLD which, in 6% of wild-type mice, can progress to HCC after 52 weeks. Mice lacking Fgf21 have a greatly accelerated development of hepatosteatosis and worse fibrosis, and 78% of mice progressed to HCC (Singhal et al., 2018). Thus, FGF21 limits the progression from NAFLD to HCC in response to prolonged exposure to an obesogenic diet.
10.6.3 |. Pancreas
The pancreas is a glandular organ that participates in the regulation of food digestion and systemic glucose metabolism by its exocrine and endocrine functions, respectively.
Pancreas development
Development of the early pancreas relies on active crosstalk between the pancreatic epithelium and the surrounding mesenchyme (Bastidas-Ponce et al., 2017). FGF10 and its receptor FGFR2b are expressed in the epithelium and mesenchyme, respectively. During mouse embryonic development, FGF10 signaling regulates epithelial cell proliferation, maintenance of progenitor cell fate, and branching morphogenesis in the pancreas (Ndlovu et al., 2018).
In human PSCs, FGF2 and FGF17 function as a potent anti-pancreatic factors. FGF2 and FGF17 inhibited early pancreatic lineage specification during differentiation of human embryonic stem cells through activation of FGFR1c or FGFR3c and downstream MEK/ERK signaling, whereas FGF7 and FGF10 (through activation of FGFR2b), and EGF increased the cell mass and retained pancreatic identity (PDX1 expression) (Dettmer et al., 2020). FGF10 signaling from the mesenchyme has been proposed to expand pancreas progenitors in mice. However, a single-cell transcriptome atlas of the human fetal pancreas shows that FGF7 and FGF10 are expressed at much lower levels than FGF2, FGF9, and FGF13, indicating these activities in the pancreas would be interesting to study (Goncalves et al., 2021).
Pancreas physiology
FGF21 is highly expressed in the exocrine pancreas. FGF21 expression was decreased with fasting and elevated with obesity. Mice lacking Fgf21 develop islet hyperplasia and periductal lymphocytic inflammation when fed a high-fat diet (Singhal et al., 2016). Furthermore, mice lacking Fgf21 or with acinar-specific deletion of Klb accumulate zymogen granules and are susceptible to pancreatic ER stress. FGF21 signaling in acinar cells triggers intracellular calcium release via PLC-IP3R signaling to stimulate digestive enzyme secretion. Pancreatic FGF21 functions as a digestive enzyme secretagogue, whose physiologic function is to maintain acinar cell proteostasis (Coate et al., 2017).
Pancreas injury
Acute pancreatitis (AP) is an acute inflammatory disease that injures pancreatic exocrine cells. In humans with AP and in a rat model of AP, serum levels of FGF1 and FGF2, and amylase and lipase activities were increased but the amylase protein concentration in serum was decreased. Injection of recombinant FGF1 or FGF2 in rats with AP was shown to inhibit inflammation and the production of inflammatory cytokines and normalize amylase and lipase activities. In contrast, the injection of FGF1 and FGF2 inhibitory antibodies worsened the AP phenotype (Tu et al., 2020).
In both patients and cerulein-induced mice with AP, serum FGF21 levels were significantly increased. However, in cerulein-induced AP in mice, FGF21 levels in the pancreas though initially elevated (at 4 h) were then reduced to nearly undetectable levels by 18 h. Pharmacologic replacement of FGF21 was shown to mitigate AP in this mouse model (Hernandez et al., 2020). Mechanistically, the administration of recombinant FGF21 reversed cerulein-induced AP by increasing Sirtuin-1 (Sirt1) expression and Sirt1 restoration of autophagy (Q. Chen, Li, Ma, et al., 2020).
Chronic pancreatitis (CP) is a progressive, irreversible inflammatory and fibrotic disease. l-arginine-induced CP in mice leads to pathological changes including acinar atrophy, loss of pancreas morphology, inflammatory cells infiltration, and elevated serum amylase activity. Treatment with FGF21 improved pancreatic fibrogenesis in CP via activation of the mTOR pathway (N. Wang, Zhao, et al., 2019).
10.7 |. Urogenital tract
10.7.1 |. Kidney development
FGF signaling is required at multiple stages of kidney development from Wolffian (nephric) duct (WD) development through nephrogenesis (Krause et al., 2015; McMahon, 2016; Okazawa et al., 2015; Trivedi & Kumar, 2021; Walker et al., 2016). In the chicken, developing WD tubular elongation and lumen formation (epithelialization) proceed simultaneously in a spatiotemporally coordinated manner. FGF8, produced in the caudal region of the embryo, acts as a chemoattractant on the leader cells of the elongating WD and prevents them from epithelialization, thus coordinating this developmental process (Atsuta & Takahashi, 2015). However, in the mouse, FGF signaling is necessary for nephric duct migration, but does not regulate directionality (Attia et al., 2015). Fgfr2 is expressed throughout the WD. Inactivation of Fgfr2 in the mouse WD epithelia resulted in the regression of the caudal part of the WD and abnormal male reproductive tract development. Cranial (rostral) WD formation and ureteric budding were not affected (Okazawa et al., 2015). To determine if FGF signaling contributes to formation of the ureteric bud, chimeric embryos containing wild-type cells and either Spry1−/− or Fgfr2 UB−/− (conditional knockout of Fgfr2 in the ureteric bud) were generated. Analysis of cellular contributions to the ureteric bud showed that FGFR2 signaling promotes ureteric bud cell rearrangements that form the tip domain, similar to GDNF/Ret signaling (Leclerc & Costantini, 2016).
FGF9 and FGF20 are required to maintain the stemness of nephron progenitor cells (Barak et al., 2012). FGF8 also contributes to the maintenance of nephron progenitors and is important for differentiation of nephron progenitors to an epithelial renal vesicle (Grieshammer et al., 2005; Perantoni et al., 2005). Sprouty 1 (Spry1), a feedback inhibitor of RTKs, antagonizes FGF9/20 and FGF8 signaling to balance progenitor cell survival, proliferation, and differentiation (Huh et al., 2020).
Inactivation of Esrp1 in mice results in loss of epithelial Fgfr2b and ectopic expression of the mesenchymal receptor, Fgfr2c, reduced kidney size with fewer ureteric tips, and reduced nephron numbers. This study indicates that signaling through ectopic FGFR2c is not sufficient to comensate for loss of FGFR2B in the developing kidney (Bebee et al., 2016; Ishiwata, 2018).
The corpuscles of Stannius (CS) comprise a unique endocrine organ in teleost fish that maintains calcium homeostasis through secretion of stanniocalcin-1 (Stc1). In the distal zebrafish pronephros, FGF signaling is required to commit tubular epithelial cells to differentiate into stanniocalcin-1-expressing corpuscles of Stannius cells (Klingbeil et al., 2021).
10.7.2 |. Bladder development
Bladder development is regulated by signaling interactions involving sonic hedgehog (SHH), TGFβ, BMP4, and FGFR2 that control epithelial–mesenchymal interactions between endoderm of the urogenital sinus and mesodermal mesenchyme (Liaw et al., 2018). Inactivation of Fgfr2 in bladder mesenchyme resulted in thin muscle layers with reduced α-smooth muscle actin (Acta2) levels at E16.5 and progressive muscle loss during the first month of life. Fgfr2 conditional knockout embryos had increased hedgehog (HH) signaling, likely due to increased expression of the HH coreceptors, Cdo (cell adhesion molecule, downregulated by oncogenes), and Boc (brother of Cdo) (Ikeda et al., 2017).
10.7.3 |. External genitalia development
Development of the external genitalia involves key interactions between FGF, HH, Wnt, and BMP signaling pathways in both epithelial and mesenchymal tissues (Cohn, 2011; Tarulli et al., 2021). Developmental gene expression patterns and mechanisms are conserved across species that diverge in external genitalia structure. For example, in both mouse and Asian house musk shrew (Suncus murinus) expression patterns of Fgf8 and Shh are similar in the genital tubercle epithelium (Miyado et al., 2017). Consistent with FGF8 signaling to mesenchymal FGFRs, tissue-specific inactivation of Fgfr2 shows that mesenchymal FGF signaling in the early-stage genital tubercle is required for its outgrowth (Gredler et al., 2015; Harada et al., 2015).
In genital tubercle mesenchyme, the transcription factor ISL1 was shown to directly regulate Bmp4, Fgf10, and Wnt5a, factors that are essential for genital tubercle formation (Ching et al., 2018). Later in development, FGFR2 regulates epithelial maturation and cell cycle progression in the urethral endoderm and surface ectoderm. Consistent with FGF10 signaling to epithelial FGFRs, ectodermal deletion of Fgfr2 resulted in hypospadias showing that FGFR2 signaling is required for epithelial maturation and maintenance of a closed urethral tube (Gredler et al., 2015; Harada et al., 2015).
10.7.4 |. Homeostasis and response to injury
Ischemia–reperfusion (IR) injury is a major cause of acute kidney injury (AKI). FGFs have been proposed as biomarkers for AKI and as potential therapeutic targets (Deng et al., 2020; Trivedi & Kumar, 2021). In AKI, circulating levels of FGF23 are dramatically increased (discussed below) (Bar et al., 2019; Christov et al., 2019). In zebrafish, FGF signaling is rapidly induced after kidney injury and inhibitor experiments show that FGF signaling is required for recruitment of renal progenitor cells to sites of new nephron formation (Gallegos et al., 2019). FGFBP1 and FGF2 are induced and released following AKI (L. Zhao, Cao, et al., 2020). In mouse, FGFBP1 expression sensitizes peripheral resistance vessels to angiotensin II (AngII) constriction. This effect is absent in Fgf2−/− mice and can be fully blocked with FGFR kinase inhibitors (Tassi, Lai, et al., 2018).
In a mouse model of cyclophosphamide-induced bladder injury, pretreatment (subcutaneous administration) with FGF7 was protective. At early time points, FGF7 suppressed intermediate and basal cell apoptosis likely through AKT signaling. FGF7 also induced proliferation of KRT5+/KRT14− epithelial cells through ERK1/2 signaling (Narla et al., 2020). Mice lacking urothelial Fgfr2 had impaired epithelial regeneration, larger basal cells and nuclei, more persistent basal and ectopic luminal KRT14+ cells, and signs of metaplasia (Narla et al., 2021).
10.8 |. Skeletal development, homeostasis, and regeneration
FGF signaling is required at all stages of skeletal development, as well as in skeletal remodeling, homeostasis, and response to injury. In the embryonic mammalian and avian limb bud, an FGF8–FGF10 feedback loop operating between the apical ectodermal ridge (FGF8) and underlying mesenchyme (FGF10) is required for limb bud outgrowth (L. Jin, Wu, et al., 2018; Mariani et al., 2017; Ornitz & Marie, 2015; Tickle, 2015; Young et al., 2019). FGF signaling in limb bud mesenchyme regulates Shh expression which regulates anterior–posterior patterning of the limb bud (Peluso et al., 2017; Tickle, 2015). FGF signaling has also been shown to convey positional information along with the limb bud proximal–distal axis by regulating the level of MEIS (myeloid ectopic viral integration site) transcription factors (Delgado et al., 2020). Apical ectoderm ridge-derived FGFs were found to regulate planar cell polarity (PCP) along the proximal–distal axis in a Wnt5a-dependent manner. A Wnt5a gradient was shown to be instructive for establishing PCP in the limb mesenchyme and permissive for FGF signaling to orient PCP (Gao et al., 2018). FGF8 and SHH were found to act synergistically to regulate expression of Hoxd13 in limb bud mesenchymal cells in vitro (Rodrigues et al., 2017). Later in development, FGFs regulate formation of the mesenchymal condensations that give rise to the skeleton (Ornitz & Marie, 2015).
Limb development in the axolotl differs from mammals and birds in that axolotls do not have an FGF producing apical ectodermal ridge but do have mesenchymal FGFs. Inhibition of FGF-signaling in axolotl demonstrated that FGFs regulate cell proliferation across all three limb axes, in contrast to mammals and birds where FGF-signaling regulates cell survival and proximodistal patterning (Purushothaman et al., 2019).
FGF signaling also regulates growth plate and articular chondrocytes, osteogenesis, and tendon. FGF signaling in bone and joint development and response to injury has been extensively reviewed (Marie et al., 2019; Ornitz & Marie, 2015, 2019; Su et al., 2014; Tickle, 2015; Tuzon et al., 2019; Xie et al., 2014). Mutations in FGFRs and in genes that regulate the FGF signaling pathway have been linked to the etiology of skeletal dysplasia syndromes. Most mutations in FGFRs are dominant and activate FGFR signaling in diseases such as Achondroplasia (FGFR3) and in several craniosynostosis syndromes (FGFR1 and FGFR2) including Apert, Crouzon, and Pfeifer syndromes. These have been extensively reviewed (Albino et al., 2015; J. Chen, Liu, et al., 2017; Klag & Horton, 2016; Legeai-Mallet, 2016; Moosa & Wollnik, 2016; Ornitz & Legeai-Mallet, 2017; Ornitz & Marie, 2015, 2019).
Dominant mutations in the transmembrane domain of FGFR2 that cause Bent Bone Dysplasia (BBD) affect receptor trafficking to the cell surface and to the nucleus (discussed below) (Merrill et al., 2012; Neben et al., 2017; Salva et al., 2019; Stichelbout et al., 2016). A recessive loss of function mutation in FGFR2 was identified in a patient with lethal ectrodactyly/split hand-foot malformation and pulmonary acinar dysplasia (Barnett et al., 2016). Recessive loss of function mutations in FGFR3 cause Camptodactyly, Tall Stature, and Hearing Loss, referred to as CATSHL syndrome in humans or spider lamb syndrome in sheep, which mimics the phenotype of mice that lack Fgfr3 (Beever et al., 2006; Colvin et al., 1996; Deng et al., 1996; Escobar et al., 2016; Makrythanasis et al., 2014; Smith et al., 2006; X. Sun, Zhang, et al., 2020; Toydemir et al., 2006).
Ellis–van Creveld syndrome (EvC) patients have short limbs, postaxial polydactyly, and dental abnormalities. EvC is caused by mutations in EVC (EVC ciliary complex subunit 1) and EVC2 (EVC ciliary complex subunit 2) which are transmembrane proteins localized at the base of the primary cilium. Evc2 is expressed in growth plate chondrocytes and in the perichondrium. Evc2 mutant mice showed a dwarfism phenotype with reduced growth plate HH signaling and increased FGF signaling. Increased expression of Fgf18 in the perichondrium of Evc2 mutant mice was shown to be a critical contributor to the dwarfism phenotype (H. Zhang, Kamiya, et al., 2016).
10.8.1 |. Embryonic skeletal development
Appendicular skeletal development
During the development of the appendicular skeleton, Fgf2, Fgf9, and Fgf18 are expressed in mesenchyme surrounding the chondrogenic condensations that form the skeletal anlage. In growing bone, Fgf9 and Fgf18 are expressed in the perichondrium and periosteum and Fgf2 is expressed in chondrocytes (Coffin et al., 2018; Hagan et al., 2019; Hung et al., 2007; Karuppaiah et al., 2016; Lazarus et al., 2007; Ornitz & Marie, 2015). These and possibly other FGF ligands signal to FGFRs in the mesenchymal components of bone and the skeletal vasculature. Fgfr2 is expressed in the chondrogenic condensations, Fgfr1 and Fgfr2 are expressed in the perichondrium and periosteum, Fgfr3 and Fgfr4 are expressed in proliferating chondrocytes, Fgfr1 is expressed in hypertrophic chondrocytes, and Fgfr1 and Fgfr3 are expressed in articular chondrocytes (Lazarus et al., 2007; Ornitz & Marie, 2002, 2015; Z. Wang, Huang, et al., 2018; Zhou et al., 2016).
During embryonic development, Fgf9 and Fgf18 were shown to function redundantly to regulate chondrogenesis in the proximal limb with differential potency along the proximodistal limb axis (Hung et al., 2016). Mesenchymal inactivation of Msh homeobox 1 (Msx1) and Msx2, transcription factors that directly regulate Fgf9 and Fgf18, results in decreased expression of Fgf9 and Fgf18 and decreased ERK1/2 phosphorylation, which results in defects in limb development (Y. Yang, Zhu, et al., 2020). However, these defects are not as severe as in mice with germline inactivation of both Fgf9 and Fgf18. Knockout of Fgf2 alone did not show a bone developmental phenotype; however, aging Fgf2−/− mice developed osteoporosis (Coffin et al., 2018). The possibility of synergism between Fgf2 and Fgf9 or Fgf18 during skeletal development has not been explored.
Several breeds of dogs have a chondrodystrophy phenotype that resembles Achondroplasia. A multibreed association analysis identified a FGF4 retrogene on Canis familiaris chromosome 18 (CFA18), about 30 Mb from the endogenous FGF4, that is strongly associated with a short-legged chondrodysplasia phenotype of at least 19 dog breeds (Parker et al., 2009). A second mapping study of 76 distinct breeds of dogs identified another FGF4 retrogene insertion on CFA12 that was associated with short limb skeletal dysplasia and intervertebral disc disease (IVDD) (Brown et al., 2017). Whole-genome sequencing further identified five additional and distinct FGF4 retrocopies in canids with IVDD (Batcher et al., 2020). These studies demonstrate that FGF4 retrotransposition is a frequent event that can account for phenotypic diversity in canids.
Synovial joints are fluid-filled cavities that separate the articulating surfaces of the bones. The formation of the inter-zone, a dense population of prechondrogenic mesenchymal cells that lie between adjacent cartilaginous anlagen, is the first morphological change in a developing synovial joint. The interzone is marked by the expression of the BMP family member, Gdf5, which is necessary but not sufficient for joint formation. In addition to GDF5, HH, WNT, and FGF signaling is also required for joint formation. Several studies show that opposing Wnt/β-catenin-FGF and Indian hedgehog (IHH)-BMP signals regulate cell fate in the interzone (Rockel et al., 2016; Salva & Merrill, 2017).
Craniofacial development
Analysis of mice with Fgfr2 mutations found in Apert syndrome, showed that activation of FGFR2 differentially affects cartilage formation and intramembranous ossification of dermal bone leading to defects in mandibular morphogenesis (Motch Perrine et al., 2019). Apert syndrome mice also showed increased proliferation of nasal septal chondrocytes at E14.5, increased nasal septal chondrocyte hypertrophy, and abnormal thickening of the nasal septum which likely contributes to midface deformities and upper airway anomalies seen in Apert syndrome (Holmes, O’Rourke, et al., 2018; Kim et al., 2021). The coronal sutures of Apert syndrome mice show increased expression of Lrp5 and Lrp6, indicating activation of WNT signaling and potential involvement of WNT signaling in the pathogenesis of Apert syndrome (Min Swe et al., 2021).
Mice homozygous for gain of function mutations in Fgfr2 that cause Crouzon syndrome show enhanced osteoblast function and maturation along with increased ERK–MAPK activation (Pfaff et al., 2016). Additionally, these mice show increased chondrogenesis in parts of the endochondral and cartilaginous skeleton that correlates with expansion of the Sox9 expression domain in mesenchymal condensations (Peskett et al., 2017). In calvarial bone development, moderate Fgf8 overexpression resulted in delayed ossification and craniosynostosis of the coronal suture whereas higher levels of Fgf8 overexpression suppressed ossification and promoted cartilage formation throughout the skull. Thus, there is plasticity in mesenchymal cells of the skull that are normally fated to form intramembranous bone (L. Schmidt, Taiyab, et al., 2018).
The skeletogenic mesenchyme of the maxillary facial prominence (MxP) and mandibular facial prominence gives rise to the maxilla and mandible and requires tissue separation at the maxillomandibular junction (MMJ). Syngnathia is a congenital craniofacial disorder characterized by bony or soft tissue fusion of the upper and lower jaws. Fgf8 and Bmp4 are expressed in partially overlapping regions of the MMJ epithelium and are required for normal separation of the maxilla and mandible (Tak et al., 2017).
10.8.2 |. Regulation of the bone–tendon interface
During axial tendon development in chicken embryos, induction of Scleraxis (Scx)-expressing tendon progenitors requires FGF signaling from the neighboring myotome (Havis et al., 2016; Huang et al., 2015). In mice lacking Fgf9, the size of the deltoid tuberosity was enlarged in newborn mice (Hung et al., 2007). Functional analysis of the deltoid tuberosity showed increased chondrocyte hypertrophy but reduced cell proliferation at the deltoid tuberosity attachment site (Leek et al., submitted). Fgf9 is expressed in the perichondrium and muscle (Hung et al., 2007).
Mice with conditional inactivation of Fgf9 in muscle are viable. These mice had an enlarged deltoid tuberosity and lateral supracondylar ridge of the humerus (Leek et al., 2021), similar to mice with mutations that induce muscle hypertrophy (e.g., Myostatin−/−) (Elkasrawy & Hamrick, 2010). Analysis of differentially expressed genes in muscle of control and global knockout mice suggests autocrine FGF9 signaling in muscle with direct or indirect effects on the adjacent developing tuberosities which may be mediated by muscle fiber type switching and metabolic changes in mitochondrial bioenergetics and glycolytic processes (Leek et al., 2021).
Using a chicken flexor tendon injury model, adeno-associated virus 2 (AAV2) mediated delivery of FGF2 (or VEGF) increased tenocyte proliferation, production of type I collagen, and tendon strength (Tang et al., 2016). In a rat model for tendon repair, treatment with FGF2 (in a gelatin hydrogel) improved mechanical strength. Histological analysis showed increased proliferation of Sox9-, Scx-, and Tenomodulin (Tnmd)-positive progenitor cells (Tokunaga et al., 2015; Yonemitsu et al., 2019; Zhang et al., 2020).
Inactivation of Fgfr2 in the mouse tendon–bone interface reduced Scx expression in Scx+/Sox9+ progenitors and promoted their differentiation into Sox9+ chondrocytes (Bobzin et al., 2021; Roberts et al., 2019). FGF signaling thus helps to pattern the boundary between tendon and bone by regulating cell fate decisions of progenitor cells.
Lacrimo-auriculo-dento-digital (LADD) syndrome
Lacrimo-auriculo-dento-digital (LADD) syndrome is caused by loss-of-function mutations in FGFR2, FGFR3, or FGF10 (Garg & Zhang, 2017; Mikolajczak et al., 2016; Milunsky et al., 2006; Rigueur et al., 2019; Rohmann et al., 2006; Ryu et al., 2020; Talebi et al., 2017). The multiple congenital anomalies in LADD syndrome affect the lacrimal and salivary glands and ducts, ears, teeth, and distal limb segments. Hearing loss associated with LADD syndrome is usually conductive and involves the external and middle ear. A mouse model of conditional inactivation of Fgfr2 in the neural crest lineage, showed decreased auditory function that correlated with hypoplasia of the auditory bulla and ectopic bone growth at sites of tendon/ligament attachment, providing an explanation for conductive hearing loss in LADD syndrome patients (Rigueur et al., 2019).
Mutations in human FGF10 can lead to congenital lacrimal gland defects (Entesarian et al., 2005; Rohmann et al., 2006). Analysis of Fgf10 gene regulation in mice showed that Shp2 is required for the expression of the homeodomain transcription factor ALX4, which directly controls Fgf10 expression in the periocular mesenchyme through interacting with an intronic evolutionary conserved element in terrestrial animals (Garg et al., 2017). The Etv4 (Pea3) transcription factor functions downstream of FGF signaling to regulate lacrimal gland duct elongation and branching (Garg et al., 2018).
10.8.3 |. Development of the hard and soft palate
Cleft lip and palate are common craniofacial birth defects in humans. Genetic analysis has implicated multiple FGF ligands and receptors in both syndromic and non-syndromic cleft lip and palate (Bush & Jiang, 2012; Riley et al., 2007; Weng et al., 2018). Gain- and loss-of-function studies in mice have identified roles for Fgfr1, Fgfr2, Fgf7, Fgf8, Fgf9, Fgf10, and Fgf18 in palatogenesis.
Single nucleotide polymorphisms in FGF10 are potential risk factors for cleft lip and palate (Itoh, 2016). Fgf10 is expressed in the anterior palatal mesenchyme and induces Shh expression through its receptor Fgfr2b in the palatal epithelium. Inactivation of either Fgf10 or Fgfr2b results in cleft palate (Rice et al., 2004). Inactivation of Fgf10 in the neural crest lineage resulted in cleft palate caused by a failure in the development of the palatal shelves (Prochazkova et al., 2018; Teshima et al., 2016).
Epithelial Shh regulates mesenchymal proliferation and expression of forkhead-box transcription factors (Foxf1a and Foxf2) and Fgf10 (Bush & Jiang, 2012; Rice et al., 2004). Inactivation of Foxf2 in neural crest-derived palatal mesenchyme resulted in ectopic activation of Fgf18 expression throughout the palatal mesenchyme and loss of Shh expression throughout the overlying palatal epithelium resulting in cleft palate (J. Xu, Liu, et al., 2016). These and other data establish an epithelial–mesenchymal feedback loop involving Shh, Fgf10, and Fgf18 that regulates the growth of the palatal shelves. Although null alleles for Fgf18 resulted in cleft palate, inactivation of Fgf18 in the neural crest mesenchyme resulted in partially penetrant cleft palate and inactivation of Fgf18 in the Twist2 (Dermo1) lineage did not result in cleft palate (Hagan et al., 2019; Liu et al., 2002; Ohbayashi et al., 2002; Yue et al., 2021). This suggests that epithelial sources of Fgf18 and/or redundancy with other Fgfs may also be required for palate development. It is likely that FGF18 signals through mesenchymal FGFR1 and FGFR2 (K. Yu, Karuppaiah, & Ornitz, 2015). Whether FGF18 directly regulates epithelial Shh is not known, but this could involve epithelial FGFR3 or another FGFR. MutaFlorenzanotions in FGFR3 that cause Muenke syndrome have been associated with a high arch palate, submucosal cleft palate, or cleft palate (Agochukwu et al., 2012; Anderson et al., 2013; Owall et al., 2020).
Analysis of genome-wide association studies (GWAS) data for nonsyndromic orofacial clefts identified a noncoding mutation that disrupted the activity of a neural crest enhancer downstream of FGFR2 (Leslie et al., 2015). Conditional inactivation of Fgfr2 in epithelium reduced epithelial proliferation and resulted in cleft palate (Hosokawa et al., 2009).
Hartsfield syndrome is characterized by holoprosencephaly (failure of complete separation of the two cerebral hemispheres), ectrodactyly (split hand and foot anomaly), and variable cleft lip/palate and is caused by heterozygous mutations in the tyrosine kinase domain of FGFR1, homozygous mutations in the extracellular domain in FGFR1, or in one case, a splicing mutation in FGFR1 (Courage et al., 2019; Hong et al., 2016; Palumbo et al., 2019; Prasad et al., 2016; Takagi et al., 2016). Analysis of FGFR kinase domain mutants in zebrafish or in cell culture showed that these alleles are likely loss of function or dominant-negative mutations (Hong et al., 2016; Palumbo et al., 2019).
Conditional inactivation of Fgfr1 in the neural crest lineage in mice resulted in severe midfacial clefting including cleft lip and palate and caused heterotopic chondrogenesis and osteogenesis of the anterior frontal bones (Kawai et al., 2019; Wang et al., 2013). Conditional inactivation of Fgfr1 and Fgfr2 in palate mesenchyme, at later stages of development, impaired palatal shelf elevation and resulted in cleft palate. Additionally, decreased growth of the mandible prevented tongue displacement from the oral–nasal cavity which obstructed palatal shelf elevation (K. Yu, Karuppaiah, & Ornitz, 2015).
Collectively, these studies link loss of function or dominant-negative mutations in mesenchymal FGFR1 and FGFR2 with the formation of cleft palate.
10.8.4 |. Postnatal regulation of the growth plate
During late embryonic and postnatal bone development, FGF signaling through FGFR3 suppresses chondrocyte proliferation and differentiation. Loss of function mutations in FGFR3 leads to a tall stature syndrome, CATSHL, and associated enchondromas, and activating mutations in FGFR3 cause several dwarfism syndromes including, Hypochondroplasia, Achondroplasia, and Thanatophoric dysplasia. Several mouse models have been developed that express Achondroplasia mutations in FGFR3 using a transgenic or knock-in approach (Komla-Ebri et al., 2016; Y. C. Lee, Song, et al., 2017; Martin et al., 2018; Shazeeb et al., 2018). Loss of function mutations in Fgfr3 in mice or zebrafish lead to skeletal overgrowth mimicking CATSHL syndrome (Colvin et al., 1996; Deng et al., 1996; X. Sun, Zhang, et al., 2020).
Established primary signaling pathways downstream of FGFR3 include STAT1, which suppresses chondrocyte proliferation, and MAPK, which suppresses chondrocyte differentiation (Escobar et al., 2016; Hogler & Ward, 2020; Y. C. Lee, Song, et al., 2017; Legeai-Mallet & Savarirayan, 2020; Merker, Neumeyer, Hertel, Grigelioniene, Makitie, et al., 2018; Merker, Neumeyer, Hertel, Grigelioniene, Mohnike, & Hagenas, 2018; Ornitz & Legeai-Mallet, 2017; Ornitz & Marie, 2019; Shazeeb et al., 2018; X. Sun, Zhang, et al., 2020; Unger et al., 2017). Ligands that are likely to activate FGFR3 in the growth plate include FGF2, FGF9, and FGF18 (Ornitz & Legeai-Mallet, 2017). Inhibition of FGF2 restored defective skeletal growth in a mouse model of Achondroplasia (Kimura et al., 2021). Overexpression of Fgf9 or Fgf18 in perichondrial tissue suppressed growth plate chondrocytes (Karuppaiah et al., 2016; H. Zhang, Kamiya, et al., 2016).
Further exploration of signaling pathways found that FGF18 induced the expression of autophagy genes in growth plate chondrocytes through signaling to FGFR4. The autophagy pathway was shown to be important for the secretion of type II collagen (COL2), a major component of the cartilaginous ECM (Cinque et al., 2015). In another study, FGFR3 activation was also shown to inhibit autophagic activity in chondrocytes in vivo through direct interactions with autophagy protein 5 (ATG5) to suppress chondrogenesis (X. Wang, Qi, et al., 2015).
To identify additional mechanisms by which FGFR3 signaling suppresses chondrogenesis, the response of rat chondrosarcoma cells to FGF1 was analyzed using phosphoproteomics (Chapman et al., 2017). This analysis showed strong expression of phospho-ERK1/2 but not phospho-STAT1, suggesting possible phosphorylation independent STAT1 function in chondrocytes (Krejci et al., 2008). Inhibition of FGF9 promotes the growth and differentiation of ATDC5 chondroprogenitor cells through inhibition of phosphorylation of AKT and glycogen synthase kinase-3β (GSK3β) (Zhang, Weng, et al., 2021). Inhibition of GSK3β in cultured metatarsal bones increased FGF18 expression and suppressed chondrogenesis through regulation of β-catenin levels. This phenotype was suppressed in tissue lacking FGFR3 (Kapadia et al., 2005). These studies identify feedback mechanisms between FGF and β-catenin that regulate chondrogenesis.
Analysis of the function of suppressor of cytokine signaling 3 (SOCS3), a negative regulator of cytokine signaling, showed that mice lacking SOCS3 in chondrocytes had decreased longitudinal bone growth, attributed to increased FGFR3–MAPK signaling (X. Liu, D’Cruz, et al., 2019). Mice lacking all four tissue inhibitor of metalloprotienase (TIMP) genes showed reduced bone growth and premature closure of the growth plates. This was attributed to increased FGF2 expression and signaling in chondrocytes. Mechanistically, TIMPs regulate metalloproteinase-mediated release of FGF2 from aggrecan in cartilage matrix (Saw et al., 2019).
Another consequence of elevated constitutive FGFR3 signaling in chondrocytes was shortening of the primary cilia, while transient FGFR3 activation resulted in elongated primary cilia. Constitutively active FGFR3 signaling was found to suppress intraflagellar transport velocity and cilia-mediated HH signaling (Kunova Bosakova et al., 2018; Martin et al., 2018). Consistent with this, in a zebrafish model for CATSHL syndrome, deficiency of fgfr3 led to enhanced IHH and canonical Wnt/β-catenin signaling (X. Sun, Zhang, et al., 2020).
10.8.5 |. Postnatal regulation of cortical and membranous bone
Genetic inactivation of Fgfr1 and Fgfr2 in osteoprogenitor cells identified a cell autonomous role in osteogenesis and a cell non-autonomous effect on the adjacent growth plate that suppressed chondrogenesis impairing longitudinal bone growth by 50% (Karuppaiah et al., 2016).
Inactivation of FGFR3 in chondrocytes led to increased chondrocyte hypertrophy and to a cell non-autonomous increase in bone mass resulting from increased expression of osteogenic factors, Bmps, Ihh, Wnt4, and Tgfβ, in Fgfr3-deficient chondrocytes (Wen et al., 2016). This is in contrast to adult mice that lack Fgfr3, which show bone overgrowth and osteopenia (Colvin et al., 1996; Eswarakumar & Schlessinger, 2007; Valverde-Franco et al., 2004). These phenotypes show that FGFR3 has distinct cell autonomous and cell non-autonomous functions in multiple bone cell types. Activation of FGFR3 in immature osteoblasts led to osteopenia and reduced cortical thickness in long bones of adult mice and activation of FGFR3 in hypertrophic chondrocytes led to decreased bone growth (Biosse Duplan et al., 2021).
Inactivation of FGFR3 in osteoclasts impaired their ability to resorb bone, resulting in increased bone mass (Su et al., 2016).
10.8.6 |. Skeletal homeostasis
Multiple canonical Fgfs (including Fgfs 1, 3, 4, 5, 7, 8, 10, and 18) were expressed in total bone tissue (including bone marrow) from 4- to 6-week-old mice. In an osteocyte cell line, Fgfs 7 and 10 were expressed at higher levels compared to other Fgfs. In vitro, FGF7 promoted the elongation of osteocyte processes and connexin 43 (Gja1) expression (X. Y. Liu, Li, et al., 2018).
Mice lacking Fgf2 develop normally but become osteoporotic as they age, indicating that Fgf2 is required to maintain bone mass (Coffin et al., 2018). Mechanistically, in the absence of FGF2, the anabolic effects of parathyroid hormone (PTH) were decreased, in part through uncoupling to Wnt signaling (Xiao et al., 2009, 2018). Three-month-old male (but not female) mice that conditionally inactivated Fgf9 in osteoblasts, showed decreased trabecular and cortical bone thickness, demonstrating an autocrine role for FGF9 in regulating bone mass in males (L. Wang, Roth, et al., 2017).
Mice lacking the negative regulator of FGF signaling, SEF, show no embryonic skeletal phenotype, but develop a high bone mass phenotype by 2 months of age (He et al., 2014). Bone marrow cells from Sef−/− mice showed increased response to FGF2 and primary periosteal cells from Sef−/− mice showed increased osteoblast differentiation.
Fgfr1 is expressed in mature articular chondrocytes. To determine if FGFFR1 signaling was required to maintain articular cartilage, the Fgfr1 gene was conditionally inactivated in chondrocytes of 8-week-old mice. After aging to 18 months, wild-type controls mice showed spontaneous degeneration of the temporomandibular joint surface, while conditional knockout mice were protected (Z. Wang, Huang, et al., 2018). These data are consistent with proposed catabolic roles for FGFR1 and anabolic roles for FGFR3 signaling in articular cartilage (Xie, Zinkle, et al., 2020).
In postnatal growing bone, Fgfr1 expression in mature osteoblasts and osteocytes is required for bone homeostasis and maintaining the viability of osteocytes. Mice lacking Fgfr1 or both Fgfr1 and Fgfr2 are phenotypically normal at birth; however, these mice show a delayed (>6 weeks of age) high bone mass phenotype with increased periosteal apposition and increased and disorganized endocortical bone. The increased bone mass was thought to be secondary to osteocyte death and activation of Wnt signaling (McKenzie et al., 2019).
10.8.7 |. Skeletal regeneration
FGF signaling in appendage regeneration in nonmammalian vertebrates
The ability to regenerate amputated or injured appendage tissues, with the exception of the very tip of the digit, was lost in mammals during vertebrate evolution. However, appendage regeneration readily occurs in nonmammalian vertebrates including amphibians, lizards, and zebrafish. Understanding the mechanism of appendage regeneration in non-mammalian vertebrates is expected to provide a pathway for the development of effective regenerative strategies in mammals (Daponte et al., 2021).
In axolotls and other urodele amphibians, limb regeneration can be induced by grafting a blastema onto a limb stump. In blastema tissue that expresses FGF8 (anterior), SHH expression maintains FGF8 expression and is sufficient to induce complete limb regeneration. In blastema tissue that lacks FGF8 expression (posterior), a combination of SHH and FGF8 can induce complete limb regeneration (Nacu et al., 2016). In an ectopic limb formation model in axolotls, it was found that limb wounds that are first treated with a combination of FGF2, FGF8, and BMP2, followed by RA treatment of the resultant mid-bud stage blastema, resulted in the generation of limbs with a complete proximal/distal and anterior/posterior limb axes (Vieira et al., 2019).
In urodele amphibians (salamanders, including newts and axolotls), limb regeneration requires neural input. Experimental innervation of wounded skin leads to formation of an accessory limb. In place of experimental innervation, application of BMP2 or BMP7 in combination with FGF2 and FGF8 could also induce accessory limb formation in wounded skin (Makanae et al., 2014). Using gain- and loss-of function experiments, FGF8 and BMP7 were identified as the necessary factors, made in dorsal root ganglion (DRG) neural cells, and delivered to regenerating limb regions through DRG axons (Satoh et al., 2016). FGF2, FGF8, and BMP7 also show similar inductive effects in tail regeneration in the axolotl, Ambystoma mexicanum (Makanae et al., 2016). Similarly, gill regeneration in the axolotl requires FGF2, FGF8, BMP2, and BMP7 input from the trigeminal ganglia (Saito et al., 2019). Cyclooxygenase-2 (COX-2) converts arachidonic acid to prostaglandins such as PGE2, which affects cell proliferation, apoptosis, angiogenesis, inflammation, and immune surveillance. Inhibition of COX-2 showed an interaction of prostaglandin signaling with matrix metalloproteinases (MMPs) and FGF2 that was essential for the initiation of tail regeneration in the Gecko, Hemidactylus flaviviridis (Buch et al., 2018). Xenopus laevis (anuran amphibian) can only partially regenerate severed limbs. Hyperinnervation of the blastema could improve regeneration leading to a branched structure in the regenerating limb that could be partially recapitulated with electroporation of Bmp7, Fgf2, and Fgf8 (Mitogawa et al., 2018).
In zebrafish fin regeneration, Laminin beta 1a (Lamb1a) is a key component of the regeneration epidermis, a specialized structure required for appendage regeneration. FGF20a, a paralog of FGF20, is required for fin regeneration and was found to directly regulate the expression of Lamb1a, providing a function for FGF20a in formation of a signaling-competent regeneration epidermis (C. H. Chen, Merriman, et al., 2015). Gene expression analyses in zebrafish identified HSPGS (glypicans and syndecans) in the distal and lateral basal layers of the wound epidermis, the distal most blastema, and more proximal blastema regions, as potential regulators of growth factor signaling (Keil et al., 2021). FGF20a, and FGF3 and FGF10a, were the major FGF ligands expressed in the wound epidermis and blastema, respectively. Early epidermal FGF20a and the later blastemal FGF3 and FGF10a are directly required for fin ray mesenchyme to form the blastema at the early pre-blastema stage and to activate regenerative cell proliferation at a later post-blastema stage, respectively (Shibata et al., 2016). The skeleton of adult zebrafish fins comprises lepidotrichia, which are dermal bones of the rays, and actinotrichia, which are non-mineralized spicules at the distal margin of the fin. Inhibition of FGF signaling shows that FGF signaling is necessary for maintenance of the actinotrichia-specific structural protein, actinodin, and its de novo deposition during fin regeneration (Konig et al., 2018).
FGF signaling in mammalian digital tip regeneration
The capacity for regeneration of most tissues was lost in mammalian evolution. However, digit tip regeneration is one of the few examples of true multi-tissue regeneration that occurs in adult mammals. The blastema is a transient proliferating cell mass that forms at the tip of an amputated digit. The regenerative environment promotes various local cells to acquire a blastema state which can then generate the different cell types required for digit regeneration. Understanding blastema formation and function should provide insights into therapeutic strategies to promote mammalian tissue regeneration and repair (Qu et al., 2020; Storer et al., 2020).
Amputation in the middle of the terminal phalanx, distal to the nail matrix, results in the formation of a blastema and complete skeletal regeneration, whereas amputation at more proximal locations fail to form a blastema and results in scar formation (Muller et al., 1999). The homeobox-containing gene, Msx1, is expressed in the blastema of amphibians and mammals and is required for digit tip regeneration (Han et al., 2005). In mouse bone marrow-derived mesenchymal stem cells (mBMSCs), Msx1/2 gene overexpression increased the expression of specific blastemal markers and enhanced the proliferation rate and osteogenesis of blastema-like cells via induced expression of Fgf8 and Bmp4. When Msx1/2-transduced mBMSCs were injected into a non-regenerative proximal amputation, the full digit tip was regenerated (Taghiyar et al., 2017). In another transplantation model, in which a non-regenerating amputation was made in the mouse middle phalange (P2), transplanted iPSC-derived limb progenitor-like cells were also able to stimulate phalange regrowth (Y. Chen, Xu, & Lin, 2017).
Another example of blastema formation occurs in wound-induced hair follicle neogenesis (WIHN). A process in which fully functional hair follicles regenerate de novo in the center of large excisional wounds. WIHN largely duplicates the morphological and signaling features of normal embryonic hair development. Similar to hair development, WIHN critically depends on the activation of canonical WNT signaling. However, unlike hair development, WNT activation in WIHN is dependent on FGF9 signaling generated by the immune system’s gamma delta T cells (Gay et al., 2013; X. Wang, Hsi, et al., 2015).
Stem cells in skeletal repair
Dental pulp may provide a relevant and accessible source of adult mesenchymal stem cells for the repair and regeneration of craniofacial tissues, as dental pulp stem cells display strong osteogenic properties and are efficient at bone formation and repair. Treatment of dental pulp stem cells with FGF2 and hypoxia increased proliferation and osteogenic differentiation leading to enhanced intramembranous bone formation in xenograft models with calvarial defects (Novais et al., 2019). Adipose-derived mesenchymal stem cells (ADSCs) engineered to overexpress FGF2 showed increased vascular endothelial growth factor (VEGF) expression. When incorporated into newly formed fracture callus these cells stimulated angiogenesis and resident stem cell differentiation into osteoblasts, to accelerate fracture repair (H. Zhang, Kot, et al., 2017). Neural crest-derived osteogenic progenitors (OPs) were found to express high levels of FGF1. Knockdown of Fgf1 in these cells resulted in decreased ERK1/2 levels and RUNX2 expression, suggesting an autocrine role for FGF signaling (Kidwai et al., 2020). Analysis of an FGF9 (p.S99N) loss-of-function mutation in mice was used to show that FGF9 functions to inhibit BMSC osteogenesis and mineralization and increase osteoclastogenesis suggesting that endogenous FGF9 is a negative regulator of bone homeostasis (Tang et al., 2021). In vitro, FGF9 suppressed osteogenic differentiation of BMSCs (Lu et al., 2015). Another readily available source of human mesenchymal stem cells (MSCs) is the palatine tonsils. Tonsil-derived mesenchymal stem cells (T-MSCs) were found to express high levels of FGF5. Knockdown of Fgf5 inhibited T-MSC proliferation and differentiation (G. C. Park, Song, et al., 2016). Synovium-derived stem cells (SDSCs) may be useful for cartilage regenerative therapies. FGF2, but not FGF10, was shown to increase glycosaminoglycan deposition, pellet size, and chondrogenic gene expression, following chondrogenic induction and osteogenic genes following osteogenic differentiation (Pizzute et al., 2016). SDSCs that were cultured with FGF2 were able to repair osteochondral defects induced in the femoral trochlea (Okamura et al., 2020). FGF2 and FGF18 were shown to promote chondrogenesis of BMSCs grown in micromass culture under chondrogenic conditions (C. Shu, Smith, et al., 2016). Differentiation of human MSCs to an articular chondrogenic fate was enhanced by sequential treatment with FGF2, which increased cell proliferation and expression of SOX9. Subsequently, in combination with TGFβ, FGF9 and FGF18 promoted an anabolic effect and inhibited terminal hypertrophic differentiation (Correa et al., 2015).
FGF signaling in cartilage repair
Articular cartilage expresses FGFR1 and FGFR3 (Ornitz & Marie, 2015; Z. Wang, Huang, et al., 2018; Weng et al., 2012; Yan et al., 2011; Zhou et al., 2016). In models for osteoarthritis (OA), FGFR1 signaling is catabolic (worsening outcomes), while FGFR3 is anabolic (protecting against OA) (T. M. Chen, Chen, Sun, & Tsai, 2019; Ellman et al., 2013; Xie, Zinkle, et al., 2020). Consistent with this model, expression of FGFR3 p.G380R (activating mutation in Achondroplasia) in chondrocytes was found to prevent sclerotic changes of the subchondral bone and subsequent cartilage degeneration (Okura et al., 2018). Additionally, patients with Achondroplasia have lower incidence of OA despite having other risk factors (obesity, bowed legs) (Klag & Horton, 2016; Valverde-Franco et al., 2006). Mice lacking the N-terminal glycosaminoglycan attachment sites in perlecan (Hspg2), a heparan sulfate proteoglycan that is localized to the basement membrane, maintain FGFR3 expression and have less severe pathology after induction of OA. This study suggests that perlecan HS may inhibit FGFR3 signaling in articular chondrocytes (C. C. Shu, Jackson, et al., 2016). In the DMM (destabilization of the medial meniscus) model for OA, treatment with FGF21 reduced OA phenotypes. In vitro, FGF21 protected chondrocytes from apoptosis, senescence, and ECM catabolism in response to oxidative stress (Lu et al., 2021).
In a temporomandibular joint (TMJ) model for OA, inactivation of Fgfr1 in chondrocytes of adult mice was protective. In this model, Fgfr1 deficiency decreased expression of matrix metallopeptidase 13 (MMP13), ADAM metallopeptidase with thrombospondin type 1 motif 5 (ADAMTS5), and collagen type X alpha 1 chain (COL10A1) but increased aggrecan (ACAN) expression and autophagy pathway components (Z. Wang, Huang, et al., 2018). Similarly, a small molecule inhibitor that is selective for FGFR1 improved outcomes in a mouse model for OA and reduced catabolic activity in human articular chondrocytes in vitro (W. Xu, Xie, et al., 2016).
FGF1 and FGF2 are expressed in human articular chondrocytes (Abd El Kader et al., 2014; Vincent et al., 2007). FGF1 had catabolic effects on cultured articular chondrocytes from OA patients (El-Seoudi et al., 2017). Mice that overexpress high molecular weight FGF2, which preferentially activates FGFR1, develop an OA phenotype after 2 months of age (Meo Burt et al., 2016). This phenotype was mitigated by treatment with the FGFR tyrosine kinase inhibitor NVP-BGJ398 (Xiao et al., 2020).
Articular chondrocytes also express FGF18 (Hagan et al., 2019; Mori et al., 2014). FGF18 selectively activates FGFR3 and is protective for articular chondrocytes (Barr et al., 2014; Howard et al., 2015; Mori et al., 2014; Power et al., 2014; Zhang, Ibrahimi, et al., 2006). In primary articular chondrocytes, recombinant FGF18 increased ECM production and cell proliferation but strongly suppressed type I collagen expression and did not induce markers of hypertrophy (Gigout et al., 2017; Muller et al., 2020). In a model in which articular chondrocytes were seeded in a Fibrin-Hyaluronan based hydrogel, mechanical loading in combination with FGF18 induced expression of articular chondrocyte matrix genes, ACAN, cartilage oligomeric matrix protein (COMP), COL2A1, and proteoglycan 4 (PRG4/Lubricin), and suppressed expression of MMP9 and MMP13 (Antunes et al., 2020). In human knee OA articular cartilage explants, FGF18 induced biphasic cartilage remodeling, with early-phase increased aggrecanase activity and late-phase increased type II collagen formation (Reker et al., 2020). These studies suggest that FGF18 may be a useful therapeutic for OA. Several clinical trials have used recombinant FGF18 (Sprifermin) in patients with OA resulting in a statistically significant increased total femorotibial joint cartilage thickness after 2 years. However, it is not yet known whether FGF18 treatment improves clinical outcomes (Dahlberg et al., 2016; Eckstein et al., 2020; Hochberg et al., 2019; J. Li, Wang, Ruan, et al., 2021; McClurg et al., 2021; Xie, Zinkle, et al., 2020).
FGF signaling in bone repair
The expression of FGF ligands and receptors have been studied in several models of bone repair, including long bone fracture healing, cortical and calvarial bone defect healing, and distraction osteogenesis (a bone lengthening procedure used in Achondroplasia patients) (Charoenlarp et al., 2017; Osawa et al., 2017). Skeletal fracture healing involves both endochondral and intramembranous programs (Einhorn & Gerstenfeld, 2015; Ko & Sumner, 2021).
In a skeletal fracture model in mice, Fgfs 1, 2, 5, 6, 9, 16, 17, and 18 and Fgfrs 1, 2, and 3 were expressed soon after injury, in the fracture callus, or in mineralizing bone (Charoenlarp et al., 2017; Schmid et al., 2009). In a stabilized fracture model, mice lacking Fgfr3 had increased cartilaginous callus, accelerated endochondral ossification, and callus mineralization (Xie et al., 2017). In contrast, mice expressing activating mutations in Fgfr3 in periosteal cells had impaired bone healing attributed to a failure of cartilage-to-bone transformation that is required for remodeling of the fracture callus (Julien et al., 2020). Fgfr3Ach mice express activated FGFR3 (p.G380R) in chondrocytes driven by the Col2A1 promoter (Naski et al., 1998). In a mouse model of distraction osteogenesis, activated FGFR3 signaling (Fgfr3Ach mice) led to enhanced new bone regeneration in the consolidation phase by accelerating endochondral ossification (Osawa et al., 2017).
In a calvarial bone repair model, FGF2 (18 kDa) enhanced healing of the bone defect and improved the efficacy of low concentrations of BMP2 (L. Xiao, Ueno, et al., 2014). FGF2 and BMP2 synergized to promote osteoblastic differentiation of C2C12 myoblasts in vitro and local sustained administration of FGF2 and BMP2 promoted ectopic bone formation in a rat model (Song et al., 2017).
FGF signaling in rheumatoid arthritis
Rheumatoid arthritis (RhA) is a chronic systemic autoimmune disease that primarily affects synovial joints. Continuous joint inflammation promotes cartilage and bone damage and eventually systemic complications (Lopez-Pedrera et al., 2020). Serum and synovial fluid FGF21 levels are elevated in patients with RhA (Hulejova et al., 2012). In a mouse model of collagen-induced arthritis (CIA), treatment with FGF21 improved clinical symptoms and joint histology, reduced levels of pro-inflammatory cytokines, and reduced obesity-mediated inflammation (S. M. Li, Yu, Li, et al., 2016; W. F. Wang, Li, et al., 2015; Y. Yu, Li, et al., 2015). Treatment of CIA mice with FGF21 showed similar efficacy to treatment with the FDA-approved drug, Adalimumab (D. Yu, Ye, et al., 2017). FGF21 therapy, in combination with dexamethasone, improved the treatment of arthritis in CIA mice, possibly by mitigating some of the adverse effects associated with dexamethasone (X. Sun, Xie, et al., 2021). Metformin (an AMP-activated protein kinase activator), a drug used to treat T2D and obesity, has shown efficacy in experimental autoimmune arthritis. In obese CIA mice treated with metformin, FGF21-expressing cells were increased in the spleen, liver, and brown adipose tissue (BAT) (E. K. Kim, Lee, et al., 2018). These studies suggest that FGF21 may be a promising therapeutic agent for RhA patients. In patients with RhA, those with the highest levels of serum FGF21 were associated with worsening in physical functioning, indicating that FGF21 might serve as a biomarker to identify patients at risk for functional decline (Gould et al., 2020). Similarly, in patients with RhA, serum FGF23 levels were correlated with inflammation, disease severity, and bone absorption markers, indicating that FGF23 may be associated with abnormal bone absorption related to inflammation in RhA (Sato et al., 2016).
Ankylosing spondylitis (AS) is an inflammatory disease that affects the axial skeleton and joints. An important pathological feature of AS is the replacement of articular cartilage with vascularized granulation tissue. Activating transcription factor 6 (ATF6) was identified as an important regulator of pathological angiogenesis in AS and ATF6 was shown to directly promote FGF2 transcription in chondrocytes (Ma et al., 2021). Increased FGF2 may both promote pathological angiogenesis and catabolic effects on chondrocytes through FGFR1 activation in ECs and articular chondrocytes, respectively (Presta et al., 2009; Weng et al., 2012).
10.9 |. Skeletal muscle
10.9.1 |. Skeletal muscle development and aging
During Drosophila metamorphosis, skeletal muscles of the adult appendages are generated from pools of quiescent adult muscle progenitors (AMPs) that were specified during embryogenesis. FGF ligands, thisbe (Ths) and pyramus (Pyr), are expressed in wing imaginal disc epithelium. The proliferation of AMPs in the wing disk that gives rise to the indirect flight muscles requires signaling through the heartless (Htl) FGFR (Everetts et al., 2021;Vishal et al., 2020). Mechanistically, Htl enhances Wingless (Wg)/Wnt-associated Armadillo (Arm)/β-Catenin signaling to promote AMP proliferation, suggesting that combinatorial FGF/Wnt signals regulate adult myoblast cell numbers (Vishal et al., 2020). FGF signaling in the wing imaginal disk is also required to target migrating myoblasts to domains of adult myogenesis (Everetts et al., 2021). In developing skeletal muscle in the Drosophila embryo, elongating myotubes make specific attachments to tendons. In the absence of the FGFR Htl or its ligand FGF Pyr, myotube guidance was disrupted. This phenotype was attributed to effects on the cytoskeleton by FGF-regulated Rho/Rac GTPases in nascent myotubes (S. Yang, Weske, et al., 2020).
FGF signaling is a critical pathway for myogenesis in vertebates. The expression of FGFR4 and Spry1 are direct targets of Pax3, a transcription factor expressed in paraxial mesoderm that is required for myogenesis (Lagha et al., 2008). The embryonic form of myosin heavy chain 3 (Myh3) is expressed in myofibers but not in myogenic progenitors (myoblasts). Loss of Myh3 led to a cell non-autonomous acceleration of myoblast differentiation leading to depletion of progenitors. This was accompanied by decreased levels of phospho-FGFR4, phospho-AKT, phospho-STAT3, the feedback regulator Spry2, and expression of FGF1 and FGF2, suggesting reduced FGF signaling. This differentiation phenotype could be rescued with exogenous FGF (Agarwal et al., 2020). Fgf9 is expressed in muscle and periosteum. In vitro studies show that FGF9 inhibits myogenic differentiation of C2C12 myoblasts and human skeletal muscle cells (J. Huang, Wang, et al., 2019).
In zebrafish during the late stages of gastrulation, FGF signaling is required for the initiation of myogenesis and acts in cooperation with Tbx16/Tbxta to induce slow and fast trunk muscle precursors at distinct dorsoventral positions. Tbx16 directly activates the myf5 and myod genes, while Tbxta only regulates Myod (Osborn et al., 2020). During somitogenesis, FGF signals directly pattern the fast muscle progenitors along the somite anterior-posterior axis. FGF also acts indirectly to determine the precise temporal window of exposure of slow muscle progenitors to Shh and BMP signaling through its direct effect on fast muscle differentiation (Yin et al., 2018).
Studies in mice showed that the expression of FGF2 was increased with age in injured myofibers that were in close proximity to satellite cells. FGF2 was not present in satellite cells, which robustly express the FGFR signaling antagonist, Spry1 (Chakkalakal et al., 2012). Consistent with this, high levels of FGF2 in aged muscle do not result in increased pERK1/2 in satellite cells (Li et al., 2015). However, another study showed that the level of Fgf2 mRNA was decreased in old compared to young mouse skeletal muscle (Homer-Bouthiette et al., 2021). Mice lacking Fgf2 showed increased fibrosis at 5 months of age and looked histologically similar to wild-type mice at 20 months of age. Compared to old wild-type mice, old Fgf2−/− mice had decreased muscle strength and increased skeletal muscle inflammation (Homer-Bouthiette et al., 2021). Lack of FGF2 thus exacerbates a sarcopenia-like phenotype in aging mice. Together, these studies suggest that in old mice there may be a non-muscle cell source of FGF2.
Aged skeletal muscle is characterized by intramuscular fatty infiltration. In human skeletal muscle, FGF2 expression positively correlates with aging. Although FGF2 stimulates muscle growth, it also promotes intramuscular adipogenesis by suppression of the secreted adipogenic inhibitor SPARC (Mathes et al., 2021). Skeletal muscle-associated mitochondrial disease can result in hyperlactatemia. In response to lactate, skeletal muscle cell lines were shown to release high levels of FGF21. FGF21 could mediate some of the systemic metabolic changes seen in hyperlactatemic patients (Villarroya et al., 2018).
10.9.2 |. Skeletal muscle regeneration
FGFs are essential for activation and proliferation of skeletal muscle stem cells (satellite cells) and are required for maintenance and repair of skeletal muscle. FGF2 and FGF6 are the only FGFs known to regulate satellite cell function in vivo. The role of FGF in the regulation of satellite cells in vertebrates shows some similarities to FGF regulation of adult muscle progenitors (AMPs) in Drosophila.
During aging, reduced responsiveness to FGF diminished satellite cell self-renewal, leading to impaired skeletal muscle regeneration and depletion of satellite cells (Pawlikowski et al., 2017). This decline in self-renewal with age and impaired FGF signaling is accompanied by altered β1-integrin activity (Bernet et al., 2014; Rozo et al., 2016). β1-integrin was shown to cooperate with FGF2 to maintain satellite cell homeostasis and to sustain their expansion and self-renewal during regeneration (Rozo et al., 2016).
Cdon (also called Cdo), is a cell surface protein that interacts with N-cadherin at contact sites between skeletal myoblasts. Inactivation of Cdon specifically in satellite cells impaired muscle regeneration, increased fibrosis, and increased cellular senescence in satellite cells. Analysis of cell signaling showed reduced phospho-ERK1/2 and impaired β1-integrin activation. Cdon forms complexes with FGFR1 and FGFR4, and Cdon inactivation reduces cell surface levels of FGFRs (Bae et al., 2020).
Adult zebrafish extraocular muscles (EOM) regenerate through a process of myocyte dedifferentiation, which involves a muscle-to-mesenchyme transition, followed by cell cycle reentry of differentiated myocytes. Inhibition of FGFR signaling impaired muscle regeneration by reducing cell proliferation and active caspase 3 levels (Saera-Vila et al., 2016). In zebrafish, caspase 3 has a non-apoptotic role in promoting skeletal muscle differentiation, thus linking FGF signaling with the early steps of myocyte dedifferentiation.
A functional role for endocrine FGFs, FGF15/19, FGF21, and FGF23, was identified in skeletal muscle in vivo. FGF15/19 caused skeletal muscle hypertrophy in mice. Moreover, mice lacking the obligate co-receptor for FGF15/19, βKlotho (KLB), in skeletal muscle were unresponsive to the hypertrophic effect of FGF15/19. FGF15/19 reduced the amount of skeletal muscle atrophy induced by glucocorticoid treatment or obesity and age-related loss of muscle mass (sarcopenia). The ability of FGF15/19 to increase skeletal muscle mass suggests that it may have therapeutic potential for the treatment of muscle wasting (Benoit et al., 2017).
FGF21 is expressed at very low levels in normal healthy skeletal muscle. However, FGF21 expression and secretion are induced in skeletal muscle in both mice and humans in response to physiological (exercise) and pathological conditions, including fasting, ER stress, mitochondrial myopathies, inflammation, and metabolic disorders (Tezze et al., 2019). In response to mitochondrial uncoupling, increased circulating levels of FGF21 caused browning of white adipose tissue. However, there are conflicting data as to whether FGF21 functions as the key metabolic mediator of the mitochondrial stress response in muscle (Keipert et al., 2014; Ost et al., 2016; Pereira et al., 2017; Rodriguez-Nuevo et al., 2018; Tezze et al., 2017; Vandanmagsar et al., 2016).
In addition to endocrine effects, skeletal muscle-derived FGF21 also has local paracrine activity. Although FGF21 does not contribute to muscle homeostasis under basal conditions, muscle-specific Fgf21 knockout mice indicate that FGF21 is required for fasting-induced muscle atrophy and weakness. The contribution of FGF21 to the atrophy program was also supported by in vivo FGF21 overexpression in muscles, which was found to induce autophagy and muscle loss. FGF21 is therefore proposed to be a novel regulator of muscle mass (Oost et al., 2019). In skeletal muscle fibers in vitro, low levels of FGF21 induced glucose uptake mediated by the glucose transporter, GLUT4. These data suggest that muscle contractile activity, known to induce FGF21 production, may improve glucose uptake into muscle via FGF21 signaling (Rosales-Soto et al., 2020; Tezze et al., 2019).
Skeletal muscle inflammation induced by obesity contributes to skeletal muscle loss/atrophy and is implicated in metabolic complications such as insulin resistance. In FGF21-deficient obese mice, there was increased expression of factors associated with muscle atrophy, increased levels of inflammatory cytokines, and reduced AMP-activated protein kinase (AMPK) phosphorylation. These studies suggest that in obese mice, FGF21 is protective for inflammation-mediated atrophy through AMPK signaling (C. S. Kim, Joe, Choi, et al., 2019).
Sustained exercise also increased FGF23 mRNA and protein expression in skeletal muscle but not in other tissues. Exogenous FGF23 treatment improved exercise performance by extending the time to exhaustion and reducing exercise-induced reactive oxygen species (ROS) and H2O2 production. Exercise-stimulated FGF23 may thus enhance mitochondrial function in skeletal muscle to improve performance (D. J. Li, Fu, Zhao, et al., 2016).
10.10 |. Skin and wound healing
10.10.1 |. Skin development
Epidermal development begins at around E8.5 in mice at which time the embryonic ectoderm begins to express the transcription factor p63, which is expressed in basal cells (ectodermal stem cells). p63 is essential for maintaining basal cell stemness, proliferation, and capacity for asymmetric cell division. By E14.5, basal cells initiate asymmetric division to form an intermediate layer. By P0, the intermediate layer forms the postmitotic suprabasal stratified layers of keratinocytes (spinous, granular, and stratum corneum layers), which are required to form a functional barrier (Flora & Ezhkova, 2020). Epidermal production of Wnt ligands is essential for the formation of the spinous layer. Mechanistically, Wnt, produced by proliferating basal cells activates a BMP-FGF signaling cascade in the underlying dermis. BMP4 cell non-autonomously activates dermal expression of FGF7 and FGF10, which promotes basal cell proliferation (Zhu et al., 2014).
FGFR2b signaling regulates all stages of keratinocyte differentiation (Rosato et al., 2018). FGF7 signaling to FGFR2b induced autophagy in keratinocytes through activation of PLCɣ and Jun N-terminal protein kinase 1 (JNK1) signaling. Inhibitors of autophagy blocked FGF7-induced differentiation of keratinocytes (Belleudi et al., 2014; Nanni, Ranieri, Rosato, et al., 2018). The level of expression of FGFR2b also regulated the rate of phagocytosis and autophagy in keratinocytes, suggesting that FGFR2b signaling could affect skin pigmentation by regulating the number of melanosomes in keratinocytes (Nanni, Ranieri, Raffa, et al., 2018).
10.10.2 |. Hair follicle development
Several intercellular signaling pathways are critical for initiating and patterning hair follicle development (Biggs & Mikkola, 2014; de Groot et al., 2021; Saxena et al., 2019). Hair follicle primordia contain high levels of WNT activity and are a source of FGFs, BMPs, BMP inhibitors, and TGFβ (de Groot et al., 2021). Fgf20 is expressed in primary ectodermal hair placodes prior to detectable markers of the dermal condensation (Huh et al., 2013). However, patterning the location of hair follicles involves an inherent capacity for the dermis to self-organize. Mesenchymal self-organization depends on restricted TGFβ signaling, which facilitates cell movement to locally pre-patterned sources of FGF (Glover et al., 2017). The hair follicle dermal condensation is the precursor to the dermal papilla, which regulates hair cycling throughout life. Formation of the dermal condensation requires FGF20, which regulates cell cycle exit, cell shape changes, and directional cell migration (Biggs et al., 2018).
Hair follicles go through continuous cycles of growth (anagen), regression (catagen), and rest (telogen) throughout their lifetime (Alonso & Fuchs, 2006; Houschyar et al., 2020). The hair follicle cycle is regulated in part by Wnt and FGF signaling interactions within the dermal papilla, which regulates the production of Wnt agonists and antagonists which regulate epithelial Wnt signaling and the hair follicle cycle. Inactivation of Fgfr1 and Fgfr2 in the dermal papilla resulted in an extended anagen phase and hair overgrowth (Harshuk-Shabso et al., 2020).
Mice with a mutation in Fgf5 have abnormally long hair (Angora mouse) (Hébert et al., 1994). Similarly, disruption of Fgf5 in Syrian hamsters, goats, rabbits, or longhair cats causes hair overgrowth (Hébert et al., 1994; Shaffer et al., 2021; X. Wang, Cai, et al., 2016; Y. Xu, Liu, et al., 2020; Yoshizawa et al., 2015). Consistent with these animal models, familial trichomegaly (extreme eyelash growth) in humans is caused by pathogenic loss of function mutations in FGF5 (Higgins et al., 2014). In hair follicle organ cultures, FGF5 induced regression of hair follicles (Higgins et al., 2014).
Merkel cells are innervated mechanosensory cells responsible for light-touch sensations. SOX9-positive cells in hair follicles give rise to hair follicle stem cells and Merkel Cells. SOX9 is critical for hair follicle stem cell specification, whereas FGFR2 is required for Merkel cell formation (Nguyen et al., 2018).
10.10.3 |. Wound healing
Cutaneous wounding causes an immediate inflammatory response, epithelial, endothelial, and fibroblast proliferation and migration, and contraction and remodeling of the ECM. At the signaling level, the production of TGFβ1 is rapidly increased by wounding. As the wound is re-vascularized, fibroblasts acquire an activated, highly contractile myofibroblast phenotype, which though necessary to close the wound, can also form dense collagen-I containing scar tissue (Grella et al., 2016). The FGF7 family of FGF ligands (FGFs 7, 10, and 22) are also increased in response to wounding. These FGFs signal to FGFRs 1b and 2b in keratinocytes (Maddaluno et al., 2017).
Mice lacking FGFRs 1 and 2 in keratinocytes develop a chronic dermatitis phenotype due to an impaired epidermal barrier resulting from loss of tight junction components (Yang et al., 2010). Epithelial barrier defects were made worse with the additional inactivation of FGFR3 in keratinocytes (Meyer et al., 2020). Furthermore, wound repair is severely delayed, with impaired wound contraction and delayed re-epithelialization caused by reduced expression of components of focal adhesions including focal adhesion kinase and paxillin (Meyer et al., 2012).
In the dermis, TGFβ1-SMAD2/3 signaling regulates transcription of Integrin a11 (ITGA11), a major fibrillar collagen receptor in fibroblasts, to promote the formation of scar tissue (Grella et al., 2016). FGF2 has been shown to reduce scarring during wound healing by slowing the differentiation of epidermal progenitor cells into myofibroblasts (Xie et al., 2008). Treatment of wounded skin in mice with FGF2 showed that wound edge keratinocytes took on mesenchymal character (decreased E-cadherin and increased vimentin) allowing enhanced migration toward the wound center. In vitro, FGF2 alone was insufficient to promote EMT; however, FGF2 enhanced TGFβ-induced EMT (Koike et al., 2020). Depletion of ESRP1, the splicing factor required for FGFR2b, resulted in increased expression of FGFR2c, activation of PKCε signaling, and expression of the EMT-related transcription factors STAT3, Snail1, and FRA1 (Ranieri et al., 2020). FGF2 and physiological low oxygen levels altered fibroblast participation in wound healing, leading to a more pro-regenerative phenotype (Kashpur et al., 2013; Page et al., 2011). Mechanistically, FGF2 suppressed TGFβ1 induction of ITGA11 expression at least in part through activation of ERK1/2 (Grella et al., 2016).
Adult mammalian skin has the capacity to regenerate fully functional hair follicles de novo in the center of large excisional wounds (wound-induced hair follicle neogenesis, WIHN) (Wier & Garza, 2020). Unlike hair development, WNT activation in WIHN is dependent on FGF9 signaling generated by γδ T cells (Gay et al., 2013; X. Wang, Hsi, et al., 2015). Additionally, M2 macrophages produce IGF1 and FGF2, which could also promote WIHN (Kasuya et al., 2018).
In a tissue-expansion model of skin regeneration, depletion of macrophages resulted in decreased skin thickness and collagen density and reduced vascularization. After 5 weeks of tissue expansion, levels of Fgf2 were decreased in skin but other growth factors (Egf, Tgfβ, and Vegf) were not changed, suggesting a requirement for FGF2 in skin regeneration (Ding et al., 2019).
Systemic sclerosis (SSc) is a prototypical fibrosing connective tissue disease. In SSc, TGFβ selectively upregulates FGFR3 and its ligand FGF9. FGFR3 activated multiple downstream signaling pathways to promote a fibroblast-to-myofibroblast transition in a mouse model (bleomycin-induced skin fibrosis) of SSc (Chakraborty et al., 2020). Psoriasis is a chronic inflammatory disease characterized by epidermal hyperplasia. Inhibition of FGF10 using monoclonal antibodies decreased keratinocyte proliferation and reduced the inflammatory response and dermal neovascularization in a guinea pig model of psoriasis (Xia et al., 2014).
Physiologic aging of the skin is associated with loss of fibrous and elastic tissue, reduced cell turnover, reduction of the vascular and glandular network, and impaired epidermal barrier function. Activation of FGF1 or FGF2 has been shown to improve skin elasticity and induce the synthesis of collagen and elastin through signaling to dermal fibroblasts (de Araujo et al., 2019).
10.10.4 |. Skin cancer
Seborrheic keratoses are common, benign, and genetically stable epidermal lesions. FGFR3 and the transcription factor forkhead box N1 (FOXN1) were highly expressed in seborrheic keratoses and were nearly undetectable in squamous cell carcinomas. A positive regulatory loop between FGFR3 and FOXN1 was shown to promote a benign versus malignant skin tumor phenotype (Mandinova et al., 2009). Activating mutations in FGFR3 were identified in nearly half of seborrheic keratosis lesions examined, supporting a role for FGFR3 in maintaining a differentiated phenotype and suppressing malignant progression (Heidenreich et al., 2017). Whole exome sequencing of skin tags (acrochordons, polypoid skin lesions) identified epidermal somatic mutations in FGFR3, HRAS, or KRAS, which are commonly seen in seborrheic keratoses (Aoki et al., 2021). Consistent with these findings, activating FGFR3 mutations were found to cause mild hyperplasia in human skin, but were insufficient to drive benign or malignant skin tumors (Duperret et al., 2014).
Elevated PI3K signaling in Pten (Phosphatase and tensin homolog)-deficient epidermis led to an mTOR-regulated increase in Fgf10 translation which promoted skin tumorigenesis (Hertzler-Schaefer et al., 2014). Analysis of downstream FGF signaling identified a requirement for both Frs2 and Shp2 for tumorigenesis. Furthermore, RAF–MEK–ERK signaling was found to cooperate with Pten deletion by enhancing PI3K signaling, as well as directly promoting epidermal hyperplasia (Mathew et al., 2016).
Mice lacking FGFBP1 showed thickening of the epidermis associated with a decreased transepidermal water loss and increased proinflammatory gene expression. These mice showed delayed wound healing and when challenged with a two-hit model for skin carcinogenesis using DMBA/TPA (7,12-dimethylbenz(a)anthracene/12-O-tetradecanoyl-phorbol-13-acetate), they showed delayed and reduced papillomatosis. Mechanistically, it was shown that both tissue-resident and circulating Fgfbp1-expressing cells modulate skin carcinogenesis and inflammation (M. O. Schmidt et al., 2018).
Melanoma is the most aggressive form of skin cancer. Most melanoma cell lines overexpress FGF2, FGF5, or FGF18, and different splice variants of FGFRs 1–4 (Czyz, 2019; Herraiz et al., 2018; Metzner et al., 2011). FGF5 protein expression was found in over half of the samples of melanoma and benign nevi, and human melanoma xenografts overexpressing FGF5 showed enhanced tumor growth (Ghassemi et al., 2017). FGFR3 expression was significantly increased in melanoma tissue and its expression was correlated with thickness and lymph node metastasis (L. Li, Zhang, et al., 2019).
11 |. FGF SIGNALING AND METABOLIC DISEASE
11.1 |. Endocrine FGF signaling
FGF15/19, FGF21, and FGF23 are the members of the FGF15/19 subfamily of endocrine FGFs (Figure 1). Endocrine FGFs have relatively low affinity for HS and are easily released from tissues to enter the circulation and function as endocrine hormones. Endocrine FGFs activate FGFRs but require αKlotho (for FGF23) and βKlotho (for FGF15/19 and FGF21) for high-affinity binding and receptor activation. Accumulating data provide evidence that endocrine FGFs are involved in human metabolic diseases and that activating or inhibiting endocrine FGFs may have therapeutic utility (Degirolamo et al., 2016; Gadaleta & Moschetta, 2019; Geng et al., 2020; Li, 2019; Markan & Potthoff, 2016; Nies et al., 2015; Noonan & White, 2019; Phan et al., 2021).
11.1.1 |. Fibroblast growth factor 15/19
In adult humans, FGF19 (and in mice, FGF15) is selectively expressed in the ileum and signals to FGFR4/βKotho in hepatocytes to regulate bile acid synthesis. A key function of FGF15/19 is to reduce bile acid synthesis after eating (Somm & Jornayvaz, 2018). FGF15/19 also regulates hepatic lipid storage, whole-body energy, glucose homeostasis, and skeletal muscle growth (Benoit et al., 2017; Gadaleta & Moschetta, 2019; Y. C. Kim, Seok, et al., 2020; Li, 2019). However, during development, FGF15/19 is expressed in multiple tissues and functions in the development of the ear, eye, brain, and heart, probably through paracrine signaling (Somm & Jornayvaz, 2018). In human, FGF15/19 is thought to function in metabolic diseases including obesity, type 2 diabetes, hepatic steatosis, biliary disorders, chronic diarrhea, inflammatory bowel disease, renal diseases, and cardiovascular disorders (Gadaleta & Moschetta, 2019; Li, 2019). FGF15/19 also regulates skeletal muscle mass (hypertrophy) and is protective against muscle atrophy (Benoit et al., 2017). Although FGF15/19 is expressed in growth plate chondrocytes, a direct role for FGF15/19 signaling in bone growth is not known (H. Chen, Li, et al., 2021).
A primary function of FGF15/19 is to prevent bile acid-induced liver damage. However, because FGF15/19 also stimulates the proliferation of hepatocytes, its therapeutic use comes with a potential risk of promoting hepatocellular carcinogenesis (Gadaleta & Moschetta, 2019; Niu et al., 2020; Raja et al., 2019; Somm & Jornayvaz, 2018; Uriarte et al., 2015; Zhou et al., 2017). A nontumorigenic FGF15/19 variant, M70 (also called NGM282), fully retains its ability to suppress bile acid production, but lacks proliferative and tumorigenic activity (Zhou et al., 2014). Administration of the NGM282 protein to healthy humans potently reduced serum levels of 7α-hydroxy-4-cholesten-3-one, establishing a potential therapeutic for the prevention and treatment of cholestatic liver disease and other disorders associated with bile acid dysregulation (Luo et al., 2014). Primary sclerosing cholangitis (PSC) is an inflammatory, cholestatic, and progressively fibrotic liver disease that has no effective medical intervention. In patients with PSC, NGM282 potently inhibited bile acid synthesis and decreased fibrosis markers without significantly affecting alkaline phosphatase levels (Hirschfield et al., 2019). NASH is a chronic liver disease characterized by hepatic steatosis, inflammation, and hepatocellular injury. Treatment with NGM282 in a phase 2 clinical trial produced significant reductions in liver fat content with an acceptable safety profile (Harrison et al., 2018). Structure-based mutagenesis of FGF15/19 identified three variants with reduced binding to FGFRs, HS, or βKlotho. All three variants were non-mitogenic but retained the full glucose-lowering and bile acid regulatory activities compared to wild-type FGF15/19 (Niu et al., 2020).
11.1.2 |. Fibroblast growth factor 21
FGF21 is expressed at the highest levels in the liver and exocrine pancreas, and in mice its expression and serum levels increase maximally with a low-protein high carbohydrate diet, but it is also increased with fasting, a ketogenic diet, alcohol, or protein and amino acid restriction (Morrison & Laeger, 2015; Solon-Biet et al., 2016). In humans and mice, sucrose, fructose, and alcohol rapidly increase serum FGF21 levels (Fisher & Maratos-Flier, 2016; Hernandez et al., 2020; Song et al., 2018; von Holstein-Rathlou et al., 2016; von Holstein-Rathlou & Gillum, 2019). Paradoxically, in mice and humans, FGF21 levels in the liver and serum also increase in response to high fat diet (Fisher et al., 2010).
Interleukin-4 (IL4) and IL13 are the major T helper 2 (Th2) cytokines that regulate adipose tissue metabolism. IL4 and IL13 were shown to increase the secretion of FGF21 in the liver. The IL4/IL13-STAT6 signaling axis thus regulates metabolic homeostasis through the induction of hepatic FGF21 (Kang et al., 2021). Under various stress conditions, FGF21 expression is increased in several tissues including brown adipose tissue (BAT), white adipose tissue (WAT), muscle, exocrine pancreas, and heart (Fisher & Maratos-Flier, 2016). In the exocrine pancreas, FGF21 is induced by feeding and is thought to signal as a paracrine factor to FGFR1 and βKlotho (KLB) to stimulate zymogen release (Coate et al., 2017).
The cJun NH2-terminal kinase (JNK) signaling pathway is activated in multiple organs by metabolic stress and promotes the development of metabolic syndrome (hyperglycemia, hyperlipidemia, and insulin resistance). Hepatic JNK signaling suppresses Fgf21 expression and circulating FGF21 (Vernia et al., 2014). FGF21 produced by hepatocytes mitigates systemic insulin resistance and the development of metabolic syndrome (Vernia et al., 2016). JNK signaling in adipocytes was also found to regulate metabolism by indirectly regulating hepatic Fgf21 expression. Inactivation of JNK signaling in adipocytes reduced hepatic steatosis and prevented whole body insulin resistance in response to a high fat diet. Mechanistically, JNK-regulated FGF21 autocrine signaling in adipocytes promoted increased expression of circulating adiponectin, which induced hepatic expression of FGF21 (Han et al., 2021).
FGF21 exerts various physiological and pharmacological effects on glucose and lipid metabolism through its endocrine actions in both the CNS and peripheral tissues. In humans, FGF21 is thought to be involved in metabolic diseases including nonalcoholic fatty liver disease (NAFLD), type 2 diabetes (T2D), CKD, and cardiovascular disorders (Geng et al., 2020; Li, 2019; Tucker et al., 2020; X. Zhou, Zhang, & Wang, 2021).
FGF21 signaling to adipose tissue is required for the acute insulin-sensitizing effects of FGF21 but not for its chronic effects to increase energy expenditure and lower body weight (BonDurant et al., 2017). FGF21 promotes fatty acid oxidation and suppresses lipolysis (Berglund et al., 2009; J. G. Park, Xu, et al., 2016; Xu et al., 2009). Fasting-induced FGF21 or pharmacological administration of FGF21 induced VEGF in WAT and increased WAT angiogenesis (Hua et al., 2021). Pharmacological administration of FGF21 also improved hypercholesterolemia by accelerating triglyceride-rich lipoprotein turnover by activating BAT and by inducing browning of WAT, thereby reducing atherosclerotic lesion severity (C. Liu, Schonke, et al., 2021). Peroxisome proliferator-activated receptor-α (PPARα) is a master regulator of fatty acid oxidation. In WAT of wild type mice, but not of Fgf21−/− mice, the PPARα agonist, fenofibrate, enhanced the expression of genes related to brown adipocyte functions, including Ucp1, Pgc1a, and Cpt1b, to increase energy expenditure and attenuate obesity (Goto et al., 2017).
Long-term metabolic effects of FGF21 on bodyweight appear to be mediated through signaling to neurons and not hepatocytes or adipocytes (Lan et al., 2017). FGF21 was shown to suppress intake of simple sugars and alcohol in mice and consumption of the artificial sweetener saccharin in primates and humans (Frayling et al., 2018; Owen et al., 2015; Talukdar, Owen, et al., 2016; von Holstein-Rathlou et al., 2016). In zebrafish, FGF21 stimulates food intake and the expression of FGF21, and its receptors were increased preprandially and decreased post-feeding in the foregut and/or liver (Blanco et al., 2020). Many of the effects of liver-derived FGF21 are mediated through signaling in the CNS as discussed as follows.
For effective clinical use, long-acting FGF21 analogs have been developed (Geng et al., 2020). In streptozotocin-treated mice, a model of type 1 diabetes (T1D), treatment with a polyethylene glycol-conjugated (PEG) and half-life extended form of FGF21 (FGF21–PEG) normalized plasma glucose, without restoring pancreatic β-cell function. FGF21–PEG also normalized plasma glucose levels and improved glucose tolerance in mice chronically treated with a competitive insulin receptor antagonist, a model of autoimmune/Type-B insulin resistance (Diener et al., 2021). In obese cynomolgus monkeys and overweight/obese humans with T2D, administration of a long-acting FGF21 analog, PF-05231023, decreased food intake resulting in a significant decrease in body weight and improved plasma lipoprotein profile, without significant effects on glycemic control (Huang et al., 2013; Talukdar, Zhou, et al., 2016). In humans with T2D, another long-acting FGF21 analog, Fc-FGF21 fusion protein, demonstrated a significant improvement in insulin sensitivity and an improved lipoprotein profile, including triglycerides, non-high-density lipoprotein (non-HDL) cholesterol, HDL-C, and apolipoproteins B and C3 (Kaufman et al., 2020).
NAFLD is the most common liver disease and is strongly associated with obesity and insulin resistance. NAFLD increases the risk of developing more severe forms of liver disease such as NASH, progressive fibrosis, cirrhosis, and hepatocellular carcinoma (Verzijl et al., 2020). FGF21 has been shown to protect against NASH (Liu et al., 2015; Maratos-Flier, 2017). Mice lacking PPARβ/δ have elevated very low-density lipoprotein receptor (VLDLR) and develop hepatosteatosis. Administration of recombinant FGF21 protected against hepatosteatosis by reducing endoplasmic reticulum (ER) stress-induced VLDLR upregulation (Zarei et al., 2018). A PEGylated FGF21 analog, with increased stability significantly reduced hepatosteatosis in humans with NAFLD (Cui et al., 2020; Sanyal et al., 2019).
FGF21 is proteolytically cleaved in the circulation rending it inactive (Kharitonenkov et al., 2005, 2007; Tezze et al., 2019; Zhen et al., 2016). Fibroblast activation protein (FAP) was implicated as the protease responsible for C-terminal cleavage of FGF21 after Pro171 and its resulting inactivation (Dunshee et al., 2016; Zhen et al., 2016). Talabostat (TB) and BR103354 are small molecule inhibitors of FAP (Cho et al., 2020; Sanchez-Garrido et al., 2016). Pharmacological inhibition of FAP with TB increased serum levels of FGF21 in diet-induced obese mice and led to decreased bodyweight, reduced food consumption and adiposity, increased energy expenditure, improved glucose tolerance, and insulin sensitivity, and lowered cholesterol levels (Sanchez-Garrido et al., 2016). In ob/ob mice (leptin deficiency), single-dose administration of BR103354 with recombinant hFGF21 reduced nonfasting blood glucose concentrations, and chronic treatment with BR103354 reduced nonfasting blood glucose concentrations, improved glucose tolerance, and reduced hepatic triglyceride content. In a mouse model for NASH, BR103354 reduced hepatic steatosis, and fibrosis (Cho et al., 2020).
Another way to increase FGF21 signaling is through the use of recombinant bispecific antibodies that directly activate the FGFR1/βKlotho complex to mimic the activity of FGF21 (Kolumam et al., 2015; Sonoda et al., 2017; Wu et al., 2011). In vivo use of these antibodies show that they likely activate FGFR1/βKlotho in the CNS to indirectly stimulate brown fat thermogenesis, promote weight loss, improve obesity-related metabolic derangements, and suppress sweet and alcohol preference (Baruch et al., 2020; M. Z. Chen, Chang, et al., 2017; Kolumam et al., 2015; Lan et al., 2017; Min et al., 2018; S. Y. Shi, Lu, et al., 2018; Sonoda et al., 2017). In a mouse model for ALS, treatment with an FGFR1 agonist antibody (R1MAb1) that mimics FGF21 activity resulted in a mild improvement in motor performance and reduced inflammation (Delaye et al., 2021).
11.1.3 |. FGF23
FGF23 is mainly expressed in osteoblasts and osteocytes and primarily signals to FGFR1 in the kidney to suppress phosphate resorption in the proximal tubule and vitamin D synthesis (resulting in reduced intestinal phosphate uptake), and to FGFR1 in the parathyroid gland to suppress parathyroid hormone secretion (to reduce phosphate levels) (Balani & Perwad, 2019; Bar et al., 2019; Erben & Andrukhova, 2017; Ho & Bergwitz, 2021; Kuro-o, 2019; Li, 2019; Musgrove & Wolf, 2020; Robling & Bonewald, 2020; Shimada et al., 2004). Interestingly, expression of the FGF23 receptor complex (FGFR1 and αKLOTHO) is also found in osteocytes and osteoblasts, suggesting that autocrine or paracrine signaling could occur in bone (Rhee et al., 2011).
FGF23 binding to FGFR1 requires the cofactor, αKlotho (KL), which functions as a molecular scaffold linking the third Ig domain of FGFR1 to the C-terminal tail of FGF23 (G. Chen, Liu, et al., 2018; Goetz et al., 2010). Within the C-terminal tail of FGF23, there are two tandem repeats that interact with KL with high affinity and can promote KL dimerization on the cell surface (Suzuki et al., 2020). However, only the first repeat region is necessary for physiological activity (Goetz et al., 2010).
Circulating levels of FGF23 are increased dramatically in response to acute kidney injury (AKI) and CKD (Christov et al., 2019; Leifheit-Nestler & Haffner, 2021). Inflammation and iron deficiency associated with CKD may directly and via iron-related mechanisms induce Fgf23 production in bone (David et al., 2016; Francis et al., 2019; Francis & David, 2016). The increased production of Fgf23 in response to inflammation and CKD was blocked by inactivation of neutrophil gelatinase-associated lipocalin (Lcn2), a pro-inflammatory and iron-shuttling molecule that is secreted in response to kidney injury (Courbon et al., 2021). In a mouse model of AKI, Fgf23 mRNA levels were increased 2-fold in bone and 5- to 15-fold in thymus, spleen, and heart (Egli-Spichtig et al., 2018). Thus, multiple organs could respond to AKI with increased FGF23 production. In AKI, increased expression of Fgf23 mRNA in bone and circulating FGF23 protein was blocked by pharmacological inhibition of FGFR1 signaling (Hassan et al., 2016). Consistent with a role for osteoblast and osteocyte Fgfr1 regulation of FGF23 production, conditional inactivation of Fgfr1 in osteoblasts and osteocytes resulted in a 50% decrease in FGF23 protein levels in bones of 6-week-old mice (Z. Xiao, Huang, et al., 2014). However, similar conditional knockout mice showed increased Fgf23 mRNA levels in cortical bone at 3- and 12-weeks-of-age (McKenzie et al., 2019). Activation of Fgfr1 in osteoblasts and osteocytes may be regulated by locally produced FGF ligands, circulating FGF23, and noncanonical mechanisms. The massive increased expression in response to kidney disease may be also controlled by kidney-derived glycerol-3-phosphate signaling to bone and bone marrow stromal cells (Simic et al., 2020; W. Zhou, Simic, & Rhee, 2021).
In human, FGF23 is thought to be involved in metabolic diseases including CKD and cardiovascular disorders (Bar et al., 2019; Faul, 2018; Ho & Bergwitz, 2021; Kuro-o, 2019; Li, 2019). FGF23 is expressed at low levels in the heart and its expression is increased during cardiac remodeling or heart failure. FGF23 at high concentrations (as occurs in CKD) signals to cardiomyocytes through FGFR4, but independent of αKlotho, to directly promote cardiac hypertrophy and indirectly promote fibrosis (Faul, 2018; Leifheit-Nestler et al., 2016; Leifheit-Nestler & Haffner, 2018). Notably, FGF23-induced cardiac hypertrophy is reversible upon removal of the hypertrophic stimulus using an FGFR4-blocking antibody (Grabner et al., 2015, 2017).
Humans with CKD have a greatly enhanced risk of cardiovascular morbidity and mortality. Disturbed calcium phosphate metabolism with elevated serum FGF23 levels is partly accountable for this enhanced risk. Lowering the level of serum FGF23 with a human anti-FGF23 antibody is being investigated as a therapeutic approach (Bouma-de Krijger & Vervloet, 2020). Increased (supraphysiological) serum levels of FGF23, as occurs in CKD and end-stage renal disease patients, have been associated with the onset of diabetes. These pathological responses to FGF23 may involve direct signaling to pancreatic β cells through non-αKlotho-dependent binding of FGF23 to FGFRs and activation of PLCγ/calcineurin (CN)/nuclear factor of activated T cells (NFAT) signaling (Donate-Correa et al., 2021).
The intestine includes a diverse microbial community (gut microbiota), which affects host physiology. In germ-free mice, serum FGF23 levels are high and vitamin D levels are low. Reintroduction of gut microbes lowered FGF23 levels and normalized vitamin D levels. Mechanistically, reintroduction of gut microbiota induced inflammation, which inhibited FGF23, indicating that the gut microbiota, through FGF23, regulates vitamin D metabolism (Bora et al., 2018).
Secreted active FGF23 is regulated by proteolytic cleavage at the 176RXXR179 motif which separates the FGF core homology domain from its 72-amino acid C-terminal tail rendering it inactive (Ratsma et al., 2021; Shimada et al., 2001; White et al., 2001). Furthermore, the FGF23 C-terminal tail functions as a competitive inhibitor of FGF23 binding to the FGFR1–αKlotho signaling complex (Goetz et al., 2010). In vitro, two proprotein convertases, furin, and PC5, were shown to cleave FGF23 at RHTR179. Inactivation of furin in vivo in osteoblasts and osteocytes increased circulating FGF23 by 25%. Thus, the in vivo processing of FGF23 likely involves the redundant action of multiple proprotein convertases or of other peptidases (Al Rifai et al., 2021). Mutations in FGF23 that cause autosomal-dominant hypophosphatemic rickets (ADHR) disrupt this cleavage site resulting in higher circulating levels of FGF23 (White et al., 2001; Wolf & White, 2014). Iron deficiency has been identified as an environmental trigger that increases FGF23 expression and disease severity in ADHR (Wolf & White, 2014). In a prospective clinical trial, oral iron administration normalized FGF23 and phosphorus in symptomatic, iron-deficient ADHR subjects (Imel et al., 2020).
The primary function of FGF23 is to maintain phosphate homeostasis. Thus, circulating levels of FGF23 should be regulated by circulating levels of inorganic phosphorus. As discussed above, direct regulation of FGFR1 signaling by inorganic phosphate in osteoblasts and osteocytes regulates the expression of Galnt3. GALNT3, polypeptide N-acetylgalactosaminyltransferase 3, is the enzyme that initiates O-linked glycosylation of FGF23 at Thr178. Glycosylation at this site prevents proteolytic cleavage and increases the levels of full-length active FGF23. Phosphorylation of FGF23 at Ser180 blocks glycosylation and leads to proteolytic cleavage of FGF23 (Edmonston & Wolf, 2020; Ho & Bergwitz, 2021; Ratsma et al., 2021; Takashi & Fukumoto, 2020b).
Expression of FGF23 is controlled by multiple mechanisms. Parathyroid-hormone-type 1 receptor (PTH1R) and FGFR1 signaling both induce expression of Fgf23 through downstream MAPK and PKC signaling pathways (Fan et al., 2016; Han et al., 2015; He et al., 2019). Sclerostin, the product of the Sost gene, induced Galnt3 and Fgf23 expression in an osteocyte cell line. Thus, Sclerostin directly regulates Fgf23 expression and, through GALNT3, indirectly promotes production of full-length FGF23 protein (Ito et al., 2021). Consistent with a role for Fgfr1 regulation of FGF23 production, conditional inactivation of Fgfr1 in mature osteoblasts and osteocytes decreased FGF23 protein levels in bones and in serum of 6-week-old mice (Z. Xiao, Huang, et al., 2014). Similar conditional knockout mice did not show increased serum FGF23 or increased expression of Galnt3 in response to a high phosphate diet in 8-week-old mice (Takashi et al., 2021). Thus, Pi sensing by FGFR1 and downstream regulation of FGF23 activation forms a feedback loop to maintain phosphate homeostasis.
11.2 |. Adipose tissue and thermogenesis
11.2.1 |. FGF regulation of thermogenesis
In mammals, endothermy requires a source of endogenous heat production, which is mostly achieved through non-shivering thermogenesis. Increased thermogenesis in response to cold or β-adrenergic agonists involves beiging (or browning) of WAT, in which small pockets of WAT begin to express uncoupling protein 1 (UCP1). UCP1 allows these beige adipocytes to consume large amounts of glucose and fatty acids to drive non-shivering thermogenesis (Cohen & Spiegelman, 2015; Reinisch et al., 2020).
Several FGFs are involved in the formation of beige adipocytes. FGF 21, an endocrine FGF, is well known to play essential roles in thermogenesis and has been reviewed extensively (BonDurant & Potthoff, 2018; Cuevas-Ramos et al., 2019; Klein Hazebroek & Keipert, 2020). β-Adrenergic stimulation of beiging requires autocrine FGF21 signaling in adipocytes where it promotes phosphorylation of phospholipase C-ɣ and mobilization of intracellular calcium which is required for thermogenic gene expression (Abu-Odeh et al., 2021). FGF21 signaling in the brain may also induce beiging of WAT. Administration of FGF21 into the lateral ventricle increased norepinephrine turnover in target tissues including WAT and BAT and stimulated beiging of WAT in a β-adrenergic receptor-dependent manner (Douris et al., 2015).
Several canonical (paracrine) FGFs including FGF1, FGF6, FGF9, and FGF10, are also involved in thermogenesis. β-Adrenergic stimulation increases FGF10 levels to promote preadipocyte differentiation into beige adipocytes. Gain- and loss-of-function experiments in mice reveal that miR-327 suppresses FGF10 to prevent beige adipogenesis. These studies identify an autocrine/paracrine signaling loop involving the miR-327/FGF10/FGFR2b signaling axis that regulates beige adipocyte formation (Fischer et al., 2017; Lv et al., 2021). Fgf9 is highly expressed in WAT in obese humans and mice and was decreased by cold stress. Furthermore, Fgf9 overexpression inhibited thermogenic genes involved in beige adipocyte differentiation in vitro. Adipose-derived FGF9 thus functions as an inhibitor of the browning of white adipocytes (Sun et al., 2019). In another study, FGF6 and FGF9 were identified as potent inducers of UCP1 expression in BAT and preadipocytes independent of adipogenesis. Fgf6 and Fgf9 expression was increased by exercise and cold stress, respectively. BAT-specific loss of Fgf9 impaired cold tolerance and BAT thermogenesis. In vivo, administration of FGF9 increased Ucp1 expression and thermogenic capacity. FGF6 and FGF9 are thus adipokines that can regulate UCP1 through a transcriptional network that is distinct from brown adipogenesis (Shamsi et al., 2020).
11.2.2 |. FGF signaling in diet-induced obesity
In response to high-fat diet-induced obesity or genetic deletion of leptin (ob/ob), mRNA levels of Fgfr1, and phosphorylation of FGFR1 were increased in epididymal fat pads (Choi et al., 2016). High-fat diet also led to activation of the mechanosensitive ion channel, PIEZO1, in hypertrophic adipocytes, possibly in response to increased membrane tension. Activated PIEZO1 led to the release of FGF1 through nonclassical secretory mechanisms and activation of FGFR1 on preadipocytes to promote adipocyte precursor differentiation. By this mechanism, mature adipocytes control adipogenesis during the development of obesity (S. Wang, Cao, et al., 2020). Obesity can lead to chronic inflammation in WAT, which is considered as a primary etiology of insulin resistance and T2D. This can be counteracted by treatment with recombinant FGF1, which leads to insulin sensitization by increasing insulin-dependent glucose uptake in skeletal muscle and suppressing the hepatic production of glucose (Suh et al., 2014). Mechanistically, this is achieved by inhibition of macrophage recruitment and inflammatory responses in adipose tissue by activation of adipocyte mTORC2/Rictor signaling and inhibition of C-C chemokine ligand 2 (CCL2) production (L. Zhao, Fan, et al., 2020).
Insulin-mediated pseudoacromegaly (IMPA) is a rare disease characterized by acanthosis nigricans, hirsutism, acromegalic features, normal growth hormone secretion, and extremely elevated insulin levels. Whole exome sequencing identified monoallelic, predicted deleterious variants in FGFR1 and KLB. Functional analysis in vitro showed an attenuated response to FGF21, but not FGF2, suggesting that these variants act synergistically to inhibit endocrine FGF21 signaling but not canonical FGF2 signaling (Stone et al., 2020).
11.2.3 |. Adipose-derived mesenchymal stem cells
Adipose tissue is an abundant source of adipose-derived stem cells (ASCs). However, endogenously, ASCs are important for the function of adipose tissue and could also be involved in the dysfunction of adipose tissue associated with obesity and in the onset of disorders associated with metabolic syndrome. FGF2 has been shown to increase the rate of proliferation and the clonogenic potential of ASCs in vitro mediated by FGFR1c (Cheng et al., 2020; Ma et al., 2019; Villageois et al., 2012; Zaragosi et al., 2006). Regulation of ASCs may be involved in the variations in adipose cellularity that occur in response to metabolic state. Interestingly, a worsening metabolic profile in patients was accompanied by a decrease in the proliferative rate, clonogenic potential, and export of FGF2 to the cell surface of ASCs. In obese patients without metabolic syndrome, ASCs may contribute to the protective effects of subcutaneous adipose-tissue expansion through autocrine FGF2 signaling (Oliva-Olivera et al., 2017).
11.3 |. Diabetes
Type 2, or insulin resistant, diabetes (T2D) is the most common form of diabetes, with the ensuing hyperglycemia having detrimental effects on many organ systems. Circulating concentrations of FGF1 are significantly increased in T2D patients (S. Wang, Yang, et al., 2016). Several studies have shown that activation of FGF signaling, either systemically or in the CNS, can mitigate some of the pathologies associated with diabetes (Deng et al., 2021; Gasser et al., 2017; Izaguirre et al., 2017; Nies et al., 2015). A single dose of recombinant FGF1 was shown to lower glucose levels in the leptin-deficient (ob/ob) or leptin receptor-deficient (db/db) mouse model of T2D. Chronic treatment with recombinant FGF1 sensitized the response to insulin by increasing insulin-dependent glucose uptake in skeletal muscle and suppressing the hepatic production of glucose (Suh et al., 2014). Administration of FGF1 was also shown to mitigate many of the complications of diabetes, including diabetic nephropathy, hepatocyte apoptosis, and inflammation (Fan et al., 2019; G. Liang, Song, et al., 2018; Z. Xu, Wu, et al., 2020; Zheng et al., 2021). In a high-fat diet-induced model of T2D, systemic administration of FGF1 reduced sciatic nerve demyelination possibly by reducing hyperglycemia-induced oxidative stress in Schwann cells (R. Li, Wang, Wu, et al., 2021). In zebrafish, overfeeding induced compensatory β-cell differentiation through a paracrine FGF1 signal from persistently activated β-cells. Activation of cultured mammalian β-cells (INS-1832/13 cells) that overexpress Fgf1 with the insulin secretagogue, glibenclamide, led to increased FGF1 release. Mechanistically, it was shown that ER stress was sufficient to trigger FGF1 release and necessary for persistent activation-induced FGF1 release (M. Li, Page-McCaw, & Chen, 2016). Expression of the long noncoding RNA, TUG1, was decreased in islets from high-fat diet-fed mice, and in mouse islet β cells (Min6) cultured in high glucose. Further knockdown of TUG1 decreased proliferation and increased apoptosis of Min6 cells. TUG1 was shown to bind and sequester miR-188–3p. Decreased TUG1 led to increased miR-188–3p and decreased translation of the miR-188–3p target, Fgf5 (P. Zhang, Li, et al., 2021). Of note, Fgf1 is also a target of miR-188–3p (Mi et al., 2020). In the OVE26 transgenic mouse model of Type I diabetes (T1D, insulin-dependent) nephropathy, the renal abnormalities were significantly attenuated after 3-month treatment with FGF21 (Weng et al., 2020).
A significant part of the protective properties of endocrine and canonical FGFs is likely mediated centrally, through the hypothalamic–pituitary–adrenal (HPA) axis. In streptozotocin-induced rodent models of T1D, intracerebroventricular injections of FGF1 or FGF15/19 led to a 60% reduction in hepatic glucose production, hepatic acetyl CoA content, and whole-body lipolysis (Perry et al., 2015). Similarly, in the ob/ob mouse, intracerebroventricular injection of FGF1 led to a ~25% reduction in fasting blood glucose (Scarlett et al., 2016). Patch-clamp recordings following 4th ventricle injection of FGF15/19 showed altered synaptic and intrinsic membrane properties of the dorsal motor nucleus of the vagus nerve, which could contribute to the normalization of blood glucose levels (Wean & Smith, 2021).
Serum FGF21 levels were significantly higher in obesity and metabolic syndrome populations and in patients with T2D (Chen et al., 2008; R. Y. Gao, Hsu, et al., 2019; Zhang et al., 2008). In New Zealand obese (NZO) mice, a model for polygenetic obesity and T2D, treatment with recombinant FGF21 prevented islet destruction and the onset of hyperglycemia, improved glucose clearance, and increased energy expenditure by inducing browning in subcutaneous WAT (Laeger et al., 2017).
12 |. FGF SIGNALING IN THE BRAIN AND SENSORY ORGANS
12.1 |. Sensory organ development and disease
12.1.1 |. Eye development
During vertebrate eye morphogenesis, the eye field gives rise to the optic vesicle and subsequently the optic cup. Transient inhibition of FGF signaling as the eye field first emerges disrupts patterning along the dorsoventral axis of the developing eye and the periocular mesenchyme resulting in coloboma (a ventral gap in the eye) (Atkinson-Leadbeater et al., 2014). In zebrafish, the evaginating optic vesicles become partitioned into prospective nasal and temporal domains by the opposing actions of Fgfs and Shh emanating from dorsal and ventral domains, respectively, of the forebrain primordium. Shh activity is required to activate forkhead box D1 (Foxd1) expression to initiate temporal retinal identity. However, this function of Shh requires FGF signaling (Hernandez-Bejarano et al., 2015). The LIM-homeodomain transcription factor, Lhx2, is essential for eye development. Inactivation of Lhx2 in the neuroretina led to decreased expression of Fgf3, Fgf9, and Fgf15/19 in the neuroretina, reduced FGF signaling (Etv1, Etv5, Spry2) in the lens, and an arrest in lens fiber development along with severe microphthalmia. This phenotype could be partially rescued by forced overexpression of FGF10 (Thein et al., 2016). Inhibitors of FGF signaling, Spry and Spred, are also important for lens development (discussed above) (Susanto et al., 2019; G. Zhao, Bailey, et al., 2018).
The GATA3 transcription factor is expressed in the posterior lens vesicle. BMP and FGF signaling were shown to increase expression of a lens-specific Gata3 enhancer element in differentiating lens fiber cells (Martynova et al., 2018).
12.1.2 |. Eye disease
In mouse models of diabetic retinopathy, administration of a long-acting FGF21 analog (PF-05231023) improved photoreceptor function and morphology and reduced retinal inflammation (Fu et al., 2018). Retinitis pigmentosa is a neurodegenerative disease that affects photoreceptors. Treatment of neonatal mice with genetically induced retinitis pigmentosa (heterozygous P23H mutation of rod opsin) with FGF21 preserved photoreceptor function and normalized Müller glial cell morphology. FGF21 predominantly affected genes in Müller glia/astrocytes (Fu et al., 2021).
Pathological neovascularization is a leading cause of blindness and occurs in retinopathy of prematurity, diabetic retinopathy, age-related macular degeneration, and idiopathic choroidal neovascularization. Serum levels of FGF2, VEGF, and other cytokines were elevated in idiopathic choroidal neovascularization patients compared to controls (Guo et al., 2019). Pathological neovascularization can be modeled using a mouse model of hypoxia-driven retinal neovascularization (oxygen-induced retinopathy) or choroidal neovascularization. FGF21 administration was shown to suppress pathological retinal neovascularization in an adiponectin-dependent manner (Fu et al., 2017). In contrast, FGF2 expression promoted pathological neovascularization and inactivation of Fgf2, but not Fgf8 or Fgf9, reduced pathological neovascularization (Z. Dong, Santeford, et al., 2019).
Herpes simplex virus type 1 (HSV-1) infection of the cornea can lead to pathological lymphangiogenesis that continues to develop after resolution of the infection. Several pro-angiogenic factors are expressed in the cornea following virus clearance. In a mouse model, antibody neutralization of FGF2, but not other angiogenic factors, suppressed pathological neovascularization that occurred after resolution of the infection (Gurung et al., 2018). Treatment with a stabilized FGF1 (TTHX1114) concurrent with the primary infection was beneficial and showed reduction in recurrent stromal keratitis and blepharitis, without affecting viral replication (Dhanushkodi et al., 2021). Thus, FGF signaling has time sensitive effects on the pathogenesis of corneal HSV-1 infection.
12.1.3 |. Inner ear
During vertebrate inner ear morphogenesis, the otic placode gives rise to the otocyst, which gives rise to the complex structures of the cochlear and vestibular domains of the inner ear. FGF signals are required to induce early otic placode markers from competent pre-placodal ectoderm. Fgf3 and Fgf4 are expressed in the developing hindbrain adjacent to the otic placode. Mice lacking Fgf3 (Int-2) have defects in inner ear development that include failure of endolymphatic duct and common crus formation and reduced cochlear coiling. However, these phenotypes exhibited reduced penetrance and variable expressivity (Hatch et al., 2007; Mansour et al., 1993). This was partially explained by redundancy with Fgf8 and Fgf10, which are expressed in the neighboring endoderm and mesoderm; however, it may also be explained by reduced expression of Fgf4 (Alvarez et al., 2003; Schimmang, 2007; Zelarayan et al., 2021). Fgf3 and Fgf4 are tightly linked on mouse chromosome 7 (Itoh & Ornitz, 2008). To address redundancy between these Fgfs, mice were generated that are null for Fgf3 and conditional null for Fgf4 (Fgf3Δ–Fgf4 flox-cis mice; Anderson, Southon, et al., 2016). Inactivation of Fgf3Δ–Fgf4 flox-cis mice in the otic placode or in the ectoderm and endoderm of pharyngeal arches resulted in the absence of otic tissue at the otic vesicle stage (Zelarayan et al., 2021).
In response to FGF signaling, expression of negative regulators, Spry1 and Spry2, was increased and limited placode size. FGF activates Wnt signaling upstream of TCF/LEF transcription factors. FGF signaling is also involved in patterning multiple axes of the otocyst (Anwar et al., 2017; Basch et al., 2016; Ebeid & Huh, 2017; Groves & Fekete, 2012; Riley, 2021; Sai & Ladher, 2015; Wright et al., 2015). Neuronal lineages in the inner ear arise from the anterior–ventral region of the otocyst to form the statoacoustic ganglion, which will become the spiral and vestibular ganglia. The transcription factor TBX2 represses Fgf8 expression and is required to restrict neurogenesis to the anterior-ventral region of the otocyst (Kaiser et al., 2021).
Mice heterozygous for Fgf10 have a small or absent posterior semicircular canal. Mice lacking Fgf10 have a shortened and narrower cochlear duct, as well as defects in non-sensory regions of the inner ear (Munnamalai & Fekete, 2020; Urness et al., 2015). Inhibition of FGF3 and FGF10 at the otic placode stage with a soluble FGFR2b transgene show that both ligands are required after otocyst formation for maintenance of otic neuroblasts and for patterning and proliferation of the epithelium of both the cochlear and vestibular domains (Urness et al., 2018).
Inactivation of Fgfr1 or its ligand, Fgf20, leads to the loss of hair cells and supporting cells from the organ of Corti (Ebeid & Huh, 2017; Hayashi et al., 2008; Huh et al., 2012, 2015; Munnamalai & Fekete, 2020; Ono et al., 2014; Pirvola et al., 2002). FGF9 and FGF20 are expressed in the non-sensory and sensory epithelium, respectively, during otic development. FGF9 and FGF20 signal to mesenchymal FGFRs and activate ETV4 and ETV5 to indirectly regulate the size of the sensory progenitor population and overall cochlear length. These FGFs also signal to epithelial FGFR1 to regulate the differentiation of outer hair cells and supporting cells (Ebeid & Huh, 2020; Huh et al., 2015; L. M. Yang, Stout, et al., 2020). In mouse explant cultures, FGF20–FGFR1 signaling activates MAPK and PI3K signaling to regulate hair cell and support cell differentiation (Su et al., 2021). FGFR1 has an FGF20-independent, Sox2-dependent role in specification, and Sox2 and Fgf20 interact to ensure that specification occurs before differentiation toward the cochlear base (Yang et al., 2019). Genes regulated by FGF20 in the developing cochlea include Etv4, Etv5, Etv1, Dusp6, Hey1, Hey2, Heyl, Tectb, Fat3, Cpxm2, Sall1, Sall3, and the cell cycle regulator Cdc20 (L. M. Yang, Stout, et al., 2020).
Fgf12 is expressed in spiral and vestibular ganglion neurons. Mice lacking Fgf12 have morphologically normal inner ears but have functional deficits in hearing and balance (Hanada et al., 2018). Fgf13 is expressed in the organ of Corti, spiral ganglion neurons, stria vascularis, and supporting cells. Conditional knockout of Fgf13 in hair cells and supporting cells led to sensorineural deafness without changes in the number or morphology of hair cells. A secondary consequence of conditional knockout of Fgf13 was loss of or aberrant development of spiral ganglion neurons (Yu et al., 2021).
Whole exome sequencing in patients with sensorineural hearing loss identified pathogenic mutations in the alternative splice regulator, ESRP1. Esrp1−/− mouse embryos had defects in cochlear morphogenesis, auditory hair cell differentiation, and cell fate specification resulting from aberrant splicing of Fgfr2 from the b (epithelial) isoform to the c (mesenchymal) isoform. The resulting FGF9 activation of ectopic FGFR2c caused an altered cell identity along the cochlear lateral wall (Rohacek et al., 2017).
In zebrafish, FGFR-PI3K/AKT signaling regulates statoacoustic ganglia development, while FGFR-ERK1/2 signaling regulates utricular hair cell development (J. Wang, Wu, et al., 2015). Zebrafish neuromasts contain hair cells with similar functions to mammalian hair cells. In hair cell regeneration, c-MYC and FGF are required for proliferation and differentiation, respectively, of fgfr1a-expressing neuromast support cells that will replace damaged hair cells (S. G. Lee, Huang, et al., 2016). Wnt and FGF pathways interact to modulate the proliferation of neuromast progenitor cells during early development and to regulate regenerative cell proliferation (Tang et al., 2019).
Inner hair cell ribbon synapses in the cochlea transmit sound signals to the auditory nerve. FGF22 is expressed in inner hair cells. In an ototoxic injury model, FGF22 infusion preserved ribbon synapses and restored hearing (S. Li, Hang, & Ma, 2016).
12.1.4 |. Olfactory development
The olfactory placode gives rise to the olfactory epithelium, a neurosensory organ required for the sense of smell, and other non-neuronal structures such as respiratory epithelium. The olfactory placode forms as a focal thickening of the surface ectoderm in the ventrolateral part of the frontonasal prominence. Signaling by BMPs, FGFs, and RA regulates olfactory placode formation. Subsequent asymmetric growth leads to formation of the nasal pit and then nasal cavity (Suzuki & Osumi, 2015). Ectodermal inactivation of Wls (Wntless, Gpr177), which blocks secretion of Wnt ligands, results in severe facial deformities due to a combined loss of WNT, FGF (FGFs 3, 4, 7, 8, 9, and 10), and BMP4 signaling. In the invaginating nasal pit, inactivation of Wls disrupts migration of olfactory epithelial cells into adjacent mesenchyme (Zhu et al., 2016). Interestingly, GnRH neurons originate in the olfactory placode under the control of FGF8 and migrate to the hypothalamus (Chung et al., 2016). Mutations in FGF8 and FGFR1 disrupt GnRH neuron development and can lead to autosomal Kallmann syndrome (Akkus et al., 2017; Choi et al., 2021; Falardeau et al., 2008; Hardelin & Dode, 2008; Hero et al., 2015; Hong et al., 2016; B. F. Jin, Ji, et al., 2018; Luo et al., 2017; Scavone et al., 2019; Trarbach et al., 2010; Villanueva et al., 2015).
Turbinates are the bony projections from the nasal cavity wall that function to increase the nasal cavity surface area that is lined by the olfactory epithelium. Wnt signaling suppresses differentiation of Fgf20-expressing epithelium spanning progenitor cells. FGF20 signals to the underlying mesenchyme to regulate the growth of turbinates, thus allowing olfactory epithelial progenitor cells to regulate the olfactory epithelial surface area (Yang et al., 2018).
The Drosophila olfactory circuit contains olfactory receptor neurons and projection neurons that form synaptic connections in glomerular compartments in the antennal lobe. Each circuit is separated by ensheathing glia. The FGF ligand, Thisbe, is released from olfactory neurons and instructs the FGFR Heartless-expressing ensheathing glia to wrap each glomerulus (Wu et al., 2017).
12.1.5 |. Taste bud development
Taste buds are clusters of neuroepithelial receptor cells in the oral cavity. The FGF pathway has been shown to regulate circumvallate papilla (large posterior taste buds) development in mice. FGF10 functions as an inductive, mesenchyme-derived factor for taste papillae. Inactivation of Sprouty (Spry1 and 2) alleles progressively increases the number of circumvallate papilla (Barlow & Klein, 2015; Petersen et al., 2011). In embryonic tongue explant cultures, pharmacological activation of Wnt signaling reduced mesenchymal Fgf10 expression and epithelial Lgr5 expression, identifying a feedback loop between epithelial Wnt and mesenchymal FGF10 signaling (S. Zhang, Choi, et al., 2018). Mesenchyme-derived FGF10 signaling is also required for the development of fungiform papillae, the taste buds on the anterior tongue. FGF10 inhibits the size but not the number of the fungiform papillae, possibly by regulating the ECM and Wnt ligand diffusion (Prochazkova et al., 2017).
12.2 |. Oligodendrocyte development
Oligodendrocytes are specialized glial cells in the central nervous system (CNS) that produce myelin. FGF signaling through FGFR1 and FGFR2, but not FGFR3, is required for the initial generation of oligodendrocyte progenitors in the mouse ventral forebrain (Furusho et al., 2011). Interestingly, inactivation of Fgfr1 and Fgfr2 in oligodendrocyte-lineage cells did not affect their proliferation or differentiation. However, beginning in 2-week-old mice, the growth of CNS myelin was significantly reduced, a phenotype that continued in adult mice (Furusho et al., 2012). Conditional inactivation of Frs2 (encoding the FGFR adaptor protein FRS2) showed that it is required for specification of oligodendrocyte progenitors in the embryonic telencephalon downstream of Fgfr1 (Furusho et al., 2020).
Stimulating oligodendrocyte production from endogenous progenitor cells may be useful to treat demyelinating diseases. Increasing FGFR3 activity in adult subventricular zone stem cells redirected their differentiation from the neuronal to the oligodendroglial lineage after pathological demyelination, leading to improved oligodendrocyte regeneration and myelin repair (Kang et al., 2019). Further studies showed that growth of the myelin sheath was dependent on FGFR2, but not FGFR1, and that increased ERK1/2 activation converges with the PI3K/Akt/mTOR pathway at the level of mTORC1 to regulate myelin gene expression (Furusho et al., 2017). Interestingly, FGFR2 signaling did not require the adaptor protein, FRS2, for the growth of the myelin sheath (Furusho et al., 2020).
In spinal cord injury, co-grafting of Schwann cells (myelin forming cells in the peripheral nervous system) and OPCs promoted better survival, proliferation, and migration of the grafted OPCs. Schwann cells were shown to promote proliferation and migration of OPCs through secretion of PDGFA and FGF2 (Y. J. Chen, Zhang, et al., 2015).
12.3 |. Synaptogenesis
Members of the FGF7 subfamily, FGFs 7, 10, and 22, function as synaptic organizing molecules (Umemori et al., 2004). Distinct sets of overlapping FGFRs, FGFR2b, and FGFR1b, mediate excitatory or inhibitory presynaptic differentiation. FGFR2b and FGFR1b are required for an excitatory presynaptic response to FGF22, and FGFR2b alone is required for an inhibitory presynaptic response to FGF7 (Dabrowski et al., 2015; Dabrowski & Umemori, 2016). In the hippocampus, Fgf22 is produced by CA3 pyramidal neurons. During development, FGF22 organizes the differentiation of excitatory nerve terminals from dentate granule cells (DGCs) that innervate CA3 dendrites. In DGCs, FGF22 signaling induces the expression of insulin-like growth factor 2 (IGF2), which then functions to stabilize the DGC presynaptic terminals (Terauchi et al., 2016). Conditional knockout of Fgf22 in CA3 neurons led to reduced excitatory synapse formation and depressive-like behavioral phenotypes similar to those observed in FGF22 null mice (Terauchi et al., 2017; Williams et al., 2016). Quantification of the distal region of CA3 showed that in Fgf22−/− mice, the number of synaptic vesicles, the bouton volume, and the number of vesicles in axonal regions were reduced, indicating a smaller presynaptic size (Pasaoglu & Schikorski, 2016).
12.4 |. Nervous system development and morphogenesis
Neural crest cells delaminate from the neural tube and migrate ventrally to generate the trunk peripheral nervous system. Inhibition of FGFR signaling showed that FGFR1 signaling is required for the ventral migration of newly delaminated neural crest cells. In the absence of FGF signaling, the dorsally stalled neural crest cells lose their dorsal/ventral oriented polarity and instead adopt rounded morphology (Dunkel et al., 2020).
In rat brain development, FGF2 expression is localized to neuroepithelial precursors close to the neuroepithelial–cerebrospinal fluid interface. This suggests that FGF2 present in embryonic cerebrospinal fluid may originate from apical secretion from the neuroepithelium (Lamus et al., 2020).
The evolution of cortical folds allows for an increase in the number of neurons in the cerebral cortex. The expression of FGFRs in the developing cerebral cortex of ferrets showed regional differences before cortical folds were formed and inhibition of FGF signaling impaired cortical folding. FGF signaling was shown to regulate progenitors in the outer subventricular zone and the growth of the upper layers of the cerebral cortex (Matsumoto et al., 2017).
12.5 |. Neuronal homeostasis
Neuronal homeostasis requires the production of mature neurons from progenitor cells in the adult brain, a process called adult neurogenesis. Failures in adult neurogenesis are linked to the etiology of neurodegenerative and psychiatric disorders. FGF14, an iFGF that regulates neuronal excitability and synaptic plasticity, was also found to regulate adult neurogenesis in the dentate gyrus (DG). Mice lacking Fgf14 have reduced numbers of proliferating cells and impaired maturation of newly born adult granule cell neurons (Alshammari et al., 2016). It is not known if these effects are mediated through direct activation of FGFR signaling.
Adult mice lacking Fgf22 are resistant to induced seizures. In response to pentylenetetrazol (PTZ), Fgf22−/− mice show reduced neurogenesis in the DG, reduced ectopic migration of hilar DGCs, and reduced hilar cell death after seizures (Lee & Umemori, 2013). It is proposed that FGF22 that is secreted from CA3 neurons indirectly controls adult neurogenesis by inducing genes for secreted proteins in DGCs that affect neuronal stem cells in the neighboring sub-granular zone to regulate adult neurogenesis in the maturing hippocampus (Dabrowski & Umemori, 2016).
Adult neurogenesis in the subventricular zone (SVZ) of the lateral ventricle decreases with age. Specialized ECM structures in the SVZ contact neural stem cells (NSCs) and regulate neurogenesis. This matrix captures and stores FGF2 via heparan sulfate (HS) binding and may contribute to the NSC niche. In aging, critical HS 6-O-sulfation is reduced, and this may contribute to reduced neurogenic activity in aging brains (Yamada et al., 2017).
12.6 |. Astrocytes and neuronal regeneration
FGF1 or FGF2 can promote the maturation of human and mouse stem cell-derived astrocytes (Roybon et al., 2013). FGF2 strongly increased glutamate transporter 1 (GLT1) expression and enhanced astrocyte proliferation, while FGF16 and FGF18 promoted maturation (GLT1 expression) but not proliferation (Savchenko et al., 2019). During neocortical development, the cell fate switch from neurons to astrocytes determines the proper numbers of neurons and astrocytes. FGF signaling is activated in ventricular zone radial glial cells at the time that this fate choice is made and promotes neuronal fate through activation of MAPK signaling (Dinh Duong et al., 2019).
Astrocytes become activated in response to neuronal injury and disease and can lead to astrogliosis (glial scar formation). During this process, some astrocytes dedifferentiate and express markers found in NSCs. This property of astrocytes may be exploited as an approach to treat neurodegenerative disease and injuries. FGF2 signaling was found to convert mouse glial fibrillary acidic protein (GFAP)-positive stem cell-derived astrocytes into proliferating nestin-positive NSCs. This de-differentiation to NSCs was suppressed by inflammatory mediators. This antagonistic relationship between FGF and inflammatory signals may explain why astrocytes do not readily form NSCs in most neurodegenerative diseases and brain injuries (Kleiderman et al., 2016).
12.7 |. FGF signaling in the CNS regulates peripheral metabolism
The endocrine FGFs, FGF15/19, and FGF21, are mainly produced by the liver and small intestine, respectively. They primarily target peripheral tissues to regulate metabolic homeostasis (Itoh, Ohta, et al., 2016; Ornitz & Itoh, 2015). However, many recent studies identified direct actions of endocrine FGFs on CNS neurons in the hypothalamus.
FGF15/19 is released postprandially from the intestine. FGF15/19 administered in the periphery in mice has hypoglycemic actions. Intracerebroventricular infusions of FGF15/19 also improve glycemic status in the periphery indicating that the CNS responds to FGF15/19, which have been shown to activate FGFRs in hypothalamic agouti-related protein (AGRP)/neuropeptide Y (NPY) neurons. The activation of FGFR signaling subsequently silences AGRP/NPY neurons, leading to improved glucose tolerance (S. Liu, Marcelin, et al., 2018; Marcelin et al., 2014).
FGF15/19 and FGF21 have different physiologic functions as hormonal regulators; however, their pharmacologic administration causes similar increases in energy expenditure, weight loss, and enhanced insulin sensitivity in obese mice. Both FGF15/19 action in the liver and FGF21 action in adipose tissue are not required for their longer-term weight loss and glycemic effects. In contrast, βKlotho in CNS neurons is essential for both FGF15/19 and FGF21 to cause weight loss and lower glucose and insulin levels, indicating the importance of the CNS in mediating the beneficial effects of endocrine FGFs (Lan et al., 2017).
In humans, a statistically significant association was identified with a variant in FGF21 and sweet consumption (Soberg et al., 2017). Systemic administration of FGF21 reduced sweet and alcohol preference in mice and sweet preference in monkeys (Talukdar, Owen, et al., 2016). FGF21 was shown to signal directly to the hypothalamic neurons to suppress simple sugar and alcohol intake and stimulate water intake (Kliewer & Mangelsdorf, 2019; Owen et al., 2015; Song et al., 2018; von Holstein-Rathlou et al., 2016; von Holstein-Rathlou & Gillum, 2019). In the hypothalamus, FGF21 signals to glutamatergic neurons to mediate FGF21-induced sugar suppression and sweet-taste preference (von Holstein-Rathlou et al., 2016).
Alcohol increases water consumption in mice. Serum FGF21 levels are increased by alcohol consumption in mice. Pharmacologic administration of FGF21 stimulates water drinking behavior in mice. In contrast, mice lacking FGF21 fail to increase water intake in response to alcohol. The effect of FGF21 on drinking is mediated in part by neurons of the hypothalamus and is inhibited by β-adrenergic receptor antagonists. FGF21 is expected to be a neurotropic hormone that governs water balance in response to specific nutrient stresses (Song et al., 2018). Hepatic FGF21 is increased in response to macronutrient imbalance and signals to the brain to suppress sugar intake and sweet-taste preference. Hepatic FGF21 signaling to glutamatergic neurons in the ventromedial hypothalamus (VNM) mediates sugar suppression and sweet-taste preference. FGF21 affects neuronal activity by increasing activation and excitability of neurons in the VMH. FGF21 signaling in the VMH functions to regulate sucrose intake (Jensen-Cody et al., 2020).
Reduced dietary protein intake induces adaptive physiological changes in macronutrient preference, energy expenditure, growth, and glucose homeostasis. Brain-specific deletion of βKlotho in mice blocks the ability to mount a physiological response to protein restriction. The effect is replicated by whole-body deletion of FGF21. FGF21 is suggested to be an endocrine signal linking the liver and brain, which regulates adaptive and homeostatic changes in metabolism and feeding behavior during protein restriction (Hill et al., 2019). βKlotho expression in glutamatergic neurons in the brain is required for protection against weight gain associated with dietary protein restriction, indicating that FGF21 signaling in glutamatergic neurons is required for weight loss associated with dietary protein dilution (Flippo et al., 2020).
Glucagon promotes weight loss by regulating glucose and lipid metabolism. Mice deficient for liver FGF21 are partially resistant to glucagon-receptor-mediated weight loss, indicating that FGF21 is a regulator of glucagon’s weight-loss effects. Mice deficient for neuronal βKlotho in the brain exhibit a partial reduction in body weight with chronic glucagon-receptor-agonism, indicating a role for central FGF21 signaling in glucagon-receptor-mediated weight loss (Nason et al., 2021).
12.8 |. FGF signaling in the CNS in neurological, neurodegenerative, and psychiatric disorders
Brain diseases include a variety of neurological, neurodegenerative, and psychiatric disorders. FGF signaling in the brain is important for neuronal homeostasis, injury response, and is involved in several neurological diseases (Dremencov et al., 2021; Terwisscha van Scheltinga et al., 2013; Turner et al., 2016).
12.8.1 |. Multiple sclerosis
Multiple sclerosis (MS) is a demyelinating disease resulting in brain and spinal cord damage (Huang & Dreyfus, 2016). Remyelination is essential for the regenerative response to demyelinating diseases such as MS. Injection of myelin oligodendrocyte glycoprotein peptide (MOG35–55) results in experimental autoimmune encephalomyelitis (EAE), a model for MS. Oligodendrocyte-specific Fgfr1 deficient mice show a milder disease course, with reduced myelin and axonal loss and reduced lymphocyte and macrophage/microglia infiltration in spinal cord white matter (Rajendran et al., 2018). In the cerebellum, these mice showed reduced myelin and axonal degeneration and reduced inflammatory infiltrates compared to controls (Rajendran, Rajendran, et al., 2021). Thus, cell-specific inactivation of FGFR1 in oligodendrocytes has anti-inflammatory and neuroprotective effects, and pharmacological inhibition of FGFRs could have therapeutic benefit (Rajendran, Bottiger, et al., 2021).
Similarly, oligodendrocyte-specific conditional inactivation of Fgfr2 resulted in a milder disease course, less myelin damage, and enhanced axonal density, without change in the number of oligodendrocytes in demyelinated areas, reduced inflammatory infiltrates, or reduced production of inflammatory cytokines (Kamali et al., 2021). Inactivation of both Fgfr1 and Fgfr2 in oligodendrocytes and their precursors showed normal differentiation of oligodendrocytes and recovery of myelin in the corpus callosum of mice with acute cuprizone-induced demyelination. In contrast, with chronic cuprizone-induced demyelination, there were fewer differentiated oligodendrocytes and less efficient myelin recovery, showing that FGF signaling through FGFR1 and FGFR2 may promote regeneration in chronic demyelinating diseases (Furusho et al., 2015).
FGF2 is elevated in the cerebrospinal fluid and serum of MS patients with recurrent or chronic disease. However, the role of FGF2 is complicated, with some studies showing therapeutic efficacy of administered FGF2 for demyelinating models and other studies showing improved outcomes in Fgf2−/− mice (Huang & Dreyfus, 2016). Inflammatory activity in MS lesions correlates with increased FGF2 expression in astrocytes. FGF2 was shown to inhibit myelination through FGFR2-mediated activation of Wnt-signaling. However, preferential activation of FGFR1 was found to be protective, promoting oligodendrocyte progenitor mitogenesis and expression of neuroprotective and anti-inflammatory factors (Thummler et al., 2019). Although remyelination of the CNS, which is isolated from the peripheral circulation by the blood–brain barrier, is thought to be controlled by its microenvironment, circulating factors have also been found to affect the CNS regenerative response. FGF21 derived from peripheral tissue was shown to leak into the CNS after injury and promote remyelination in toxin-induced demyelinated mice (Kuroda et al., 2017).
12.8.2 |. Parkinson’s and Huntington’s disease
Parkinson’s disease (PD) is a neurodegenerative disorder that predominately affects dopaminergic neurons in the substantia nigra of the brain (Y. Liu, Deng, et al., 2021). Genetic association analysis indicates that polymorphisms in FGF20 may be significantly associated with an increased risk of PD in Chinese and Indian populations (Jing et al., 2015; Rudakou et al., 2021; Sadhukhan et al., 2018; X. Y. Sun, Wang, et al., 2017; X. Wang, Sun, et al., 2017; X. Zhao, Wu, et al., 2016). Dopaminergic neurons co-express tyrosine-hydroxylase (TH) and FGFRs 1, 3, and 4, while GFAP-positive astrocytes express FGF20.
PD can be modeled in rats by treatment with 6-hydrodopamine, which induces nigrostriatal cell loss. In the PD rat model, treatment with FGF2 (P. H. Yang, Zhu, et al., 2016; Zheng et al., 2016), FGF18 (Guo et al., 2017), or FGF20 (Boshoff et al., 2018; Niu et al., 2018) protected against loss of TH-positive neurons. In PD, pathological proteins such as alpha-synuclein (α-Syn) are transmitted in extracellular vesicles between neurons. Ras-associated binding proteins (Rabs) can modulate α-Syn induced toxicity, and FGF2 was shown to enhance the release of extracellular vesicles that are enriched for Rab8b and Rab31, suggesting that these Rabs may limit the toxicity of α-Syn and other exosomal proteins (Kumar et al., 2020). FGF21 also protects against neurodegeneration in mouse models of PD (Y. Chen, Shen, Qi, et al., 2020; Fang et al., 2020). FGF21 is neuroprotective for several neurodegenerative diseases possibly by regulating the amount of autophagy (Y. Chen, Shen, Qi, et al., 2020; Kakoty et al., 2020).
In Huntington’s disease, FGF9 was shown to protect striatal cells in vitro by increasing proliferation and reducing cell death in response to starvation stress. FGF9 strongly induced expression of glial cell line-derived neurotrophic factor (GDNF), the anti-apoptotic marker, Bcl-xL, the anti-oxidative stress gene, nuclear factor erythroid 2-like 2 (Nrf2), and nuclear factor kappa B (NF-kB) signaling, through ERK signaling pathways (Yusuf et al., 2018, 2019, 2021).
12.8.3 |. Alzheimer’s disease
Alzheimer’s disease (AD) is a neurogenerative disorder associated with β-amyloid (Aβ) plaques, tau neurofibrillary tangles, and neuroinflammation (Long & Holtzman, 2019). Genetic association analysis identified a variant in FGF1 that may interact with a variant in apolipoprotein E (APOE)-ε4 to increase the risk of episodic memory deficits, an early symptom of AD (Chang et al., 2019; Tao et al., 2014). Serum levels of FGF21 were elevated in healthy aging and were lower in patients with AD (Conte et al., 2021). FGF21 was shown to reduce neurodegeneration in rat and cellular models of AD (S. Chen, Chen, Sun, Xu, et al., 2019). FGF21 may function by attenuating Aβ generation, inflammation, oxidative stress, and insulin resistance (Taliyan et al., 2019). The FGF21 analog LY2405319 was shown to activate Aβ degrading genes in vitro but did not affect Aβ plaque size in vivo. However, LY2405319 may be useful to treat AD-related neuroinflammation and increase neuronal activity (Ruhlmann et al., 2021). FGF7 signaling through the FGFR2/PI3K/AKT pathway was also found to reduce damage in a cellular model of AD (W. Chen, Wu, Hu, et al., 2020).
12.8.4 |. Depressive disorder
Depressive disorder is the most common psychiatric disorder characterized by depressed mood, diminished interests, impaired cognitive function, and vegetative symptoms (Otte et al., 2016). Recent studies identify a role for FGFs in depressive disorder (Amidfar et al., 2021; Y. H. Xu, Zhu, et al., 2021). Genetic association analysis identified a functional polymorphism in FGF20 that is associated with an increased risk of depressive symptoms (K. M. Jimenez, Pereira-Morales, et al., 2018).
Serum FGF2 levels were lower in children with anxiety and depressive symptoms (Lebowitz et al., 2021). FGF2 potentially acts as an anti-depressive agent in rodents. Fluoxetine, an antidepressant drug, is effective for depressive and anxiety behaviors in wild-type mice, but not in Fgf2 knockout mice, indicating that FGF2 is necessary for the anti-depressant effects of fluoxetine (Simard et al., 2018).
Chronic unpredictable stress (CUS) induced depression-like behaviors in rats and was associated with increased hippocampal expression of miR-497. Knockdown of miR-497 reduced CUS-induced depressive-like behavior. FGF2 is a direct target of miR-497 in microglia and overexpression of FGF2 inhibited miR-497-induced proinflammatory cytokines. These studies suggest that miR-497 enhanced hippocampal microglial activation in CUS-induced depression in part by suppressing production of FGF2 (Zhai et al., 2020).
FGF9 expression was increased and FGF2 was decreased in the hippocampus of humans with major depressive disorder (MDD). Rats subjected to chronic social stress recapitulate some aspects of MDD and showed a significant increase in hippocampal FGF9 levels. Chronic intracerebroventricular administration of FGF9 increased anxiety- and depression-like behavior in rats and selective knockdown of Fgf9 in the dentate gyrus decreased anxiety-like behavior but did not affect depression-like behavior. Of particular interest is the observation that the effects of FGF9 were opposite to those of FGF2. These studies suggest that inhibition of hippocampal FGF9 may be a therapeutic approach for the treatment of anxiety and depression (Aurbach et al., 2015). Adiponectin (ADPN) is a protein hormone that modulates a number of metabolic processes. ADPN and FGF9 have anti-depressive and pro-depressive effects, respectively. In mice, ADPN may be a key negative regulator of FGF9/FGFR3 signaling in depressive disorder, and dysfunction of ADPN–FGF9 signaling may promote stress-induced depression (X. Q. Wang, Li, et al., 2020). In a model of poststroke depression (PSD) in rats, Fgf9 mRNA and protein was increased in the dentate gyrus (A. Li, Tian, Yang, et al., 2021).
Serum FGF21 levels are higher in patients with bipolar disorder and are increased in patients treated with valproic acid (Chang et al., 2018; Hu et al., 2019; Omileke et al., 2020). Administration of FGF21 was shown to have antidepressant effects on lipopolysaccharide (LPS)-induced depressive-like behaviors in mice. Mechanistically, FGF21 decreased microglial expression of proinflammatory cytokines by suppressing the nuclear factor-kB (NF-kB) signing pathway (X. Wang, Zhu, et al., 2020). In male rodents, social defeat stress increased plasma FGF21 concentrations. Mice lacking FGF21 showed increased social avoidance and augmented immobility behavior in a forced swimming test administered after social defeat stress (Usui et al., 2021).
Fgf22 knockout mice show a depressive-like phenotype in the forced swim test and increased immobility in the tail suspension test, indicating a role for FGF22 in affective behaviors (Williams et al., 2016). These phenotypes require FGF22 function in neonatal, but not adult, neurons, indicating a role for FGF22 in synaptogenesis and/or neurogenesis may underlie depression in adults (discussed above) (Terauchi et al., 2020). Analysis of serum from patients with depression showed a negative correlation between FGF22 levels and interleukin-1β (IL-1β) levels. Experiments in rats and primary hippocampal neurons showed that treatment with FGF22 reduced IL-1β expression and reduced depression-like phenotypes in response to chronic stress (Xu et al., 2017).
FGFR1 forms a heterodimer with the serotonin 1A receptor (5-HT1A; discussed above). Combined treatment with FGF2 and a 5-HT1A agonist increased the formation of these heterocomplexes, enhanced FGFR1 signaling, and was associated with antidepressant effects (Borroto-Escuela et al., 2015, 2016, 2018, 2021).
12.8.5 |. Schizophrenia
Schizophrenia is a psychiatric disorder characterized by continuous or relapsing episodes of psychosis. The FGF family has been proposed as a potential drug target for the treatment of schizophrenia (Talaei et al., 2020). Serum FGF21 levels are significantly higher in patients with schizophrenia suggesting that FGF21 might serve as a biomarker for schizophrenia (Qing et al., 2015). Serum FGF10 levels were significantly decreased in patients with schizophrenia. Both first-episode, drug-free patients and chronically medicated patients also had lower levels of FGF10. The association between FGF10 and schizophrenia supports a pathogenic role for neurotrophic factor in schizophrenia (Yu et al., 2019).
12.8.6 |. Autism spectrum disorder
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by impaired social, emotional, and communication skills. Analysis of serum FGF2 in children with ASD and healthy controls found lower levels of FGF2 in the ASD group (Esnafoglu & Ayyildiz, 2017). Suppression of the cell adhesion molecule NEGR1 or FGFR2 in the lateral ventricles of E15.5 mouse embryos resulted in behavioral abnormalities consistent with ASD. Overexpression of FGFR2 could rescue behavioral phenotypes resulting from knockdown of NEGR1 (Szczurkowska et al., 2018).
12.8.7 |. Alcohol use disorder
Alcohol use disorder (AUD) is defined by uncontrolled excessive alcohol intake that results in significant mental or physical health problems (Abrahao et al., 2017). Long-term alcohol intake leads to neuroadaptations in the corticostriatal dopamine systems, resulting in addiction phenotypes (Koob & Volkow, 2016).
In rats, alcohol-induced Fgf2 expression in the dorsal hippocampus, nucleus accumbens, and dorsal striatum. Systemic administration or infusion of recombinant FGF2 into the dorsal striatum (or dorsomedial striatum) increased alcohol consumption and preference, without affecting sweet preference. Fgf2 is thus an alcohol-responsive gene constituting a positive regulatory feedback loop that regulates alcohol consumption (Even-Chen et al., 2017). In addition, long-term voluntary alcohol intake increased the expression of Fgfr1 selectively in the dorsomedial striatum in rats. Inhibition of FGFR1 activity by a selective receptor antagonist reduced consumption and preference, without affecting other reward behaviors. The effects of FGF2–FGFR1 signaling on excessive alcohol consumption are mediated by activation of the PI3K signaling pathway (Even-Chen & Barak, 2019a).
A genome-wide association meta-analysis in Europeans identified βKlotho (KLB) as a locus associated with alcohol consumption. SNP analysis of Chinese patients with alcohol dependence identified a genetic interaction of specific SNPs in FGF19 and FGF21 that were predictive of high-level alcohol dependence. The combination of these SNPs in FGF19 and FGF21 with a SNP in FGF23 was predictive of high-level aggression and alcohol dependence (J. Xu, Wu, et al., 2021).
Brain-specific Klb knockout mice showed an increased alcohol preference, and systemic administration of FGF21 inhibited alcohol consumption in wild-type mice but not in brain-specific Klb conditional knockout mice (Schumann et al., 2016). Serum FGF21 levels were markedly increased by both acute and sub-chronic alcohol intake in humans, and systemic FGF21 administration reduced alcohol intake in mice. FGF21 is thus considered an endocrine inhibitor of alcohol preference (Soberg et al., 2018; Song et al., 2018; Talukdar, Owen, et al., 2016; von Holstein-Rathlou & Gillum, 2019; Wagner-Skacel et al., 2021). FGF21 was shown to signal to neurons in the paraventricular nucleus to stimulate water consumption in response to alcohol (Song et al., 2018). These studies and others suggest that RTKs and FGFRs, in particular, may be viable drug targets to modify alcohol abuse behaviors (Hamada & Lasek, 2020).
12.8.8 |. Drug addiction
Drug addiction is a chronic and relapsing psychiatric disorder that is characterized by excessive compulsive drug seeking and consumption. Cocaine directly affects mesolimbic dopamine neurotransmission in the reward pathway by increasing extracellular dopamine concentrations through the inhibition of dopamine reuptake in the ventral tegmental area (Smith & Laiks, 2018).
Following cocaine exposure in rats, FGF2 is increased in addiction-related brain regions including the infralimbic medial prefrontal cortex (IL-mPFC). Blocking FGF2 by administration of a neutralizing antibody directly into the IL-mPFC before extinction facilitated the extinction of cocaine-seeking behavior. These results indicate that cocaine-induced overexpression of FGF2 inhibits extinction and blocking FGF2 enhances extinction (Hafenbreidel et al., 2015). Fgf2 expression is altered in a region-specific manner following cocaine-seeking and extinction in rats, indicating that FGF2 uniquely mediates drug seeking and extinction in specific brain regions (Doncheck et al., 2018). These and other results indicate that cocaine increases FGF2 levels in several mesocorticolimbic brain regions, and that FGF2 levels positively correlate with cocaine consumption and sensitization. These studies suggest that reducing FGF2 activity may provide a novel therapeutic approach for treating substance use disorders (Even-Chen & Barak, 2019b).
13 |. FGF REGULATION OF HOST–PATHOGEN INTERACTIONS
13.1 |. SARS-CoV-2 virus
Disease severity of patients infected with SARS-CoV-2, the causative agent of COVID-19, is linked to physiological and pathogenic factors including mitochondrial dysfunction and metabolic alterations with increased glycolysis. FGF21, which regulates metabolic function, has been proposed as a biomarker for mitochondrial dysfunction (Tezze et al., 2019). Increased FGF21 levels in plasma of SARS-CoV-2-infected patients were found to correlate with COVID-19 disease severity and mortality. Targeting factors that regulate mitochondrial metabolic pathways may be useful to treat patients with COVID-19 (Ajaz et al., 2021).
Increased plasma levels of FGF2 in COVID-19 patients were strongly associated with severe disease, ICU admission, and in-hospital mortality (Petrey et al., 2021; Smadja et al., 2021). Increased new vessel growth through intussusceptive angiogenesis has been reported in lungs of patients who died from COVID-19. Increased levels of FGF2 may contribute to this increased angiogenesis which may be further promoted by inflammation and hypoxia. Suppression of FGF2 signaling may be useful in limiting the pathological response to COVID-19 (Meini et al., 2020).
13.2 |. Influenza virus
Influenza virus pneumonia causes severe damage to the lung epithelium and can lead to respiratory failure. Organoid cultures and in vivo infection models in mice showed that the regenerative response of the lung epithelium to influenza virus-induced injury required FGFR2b signaling. Intratracheal application of recombinant FGF10 recruited noninfected epithelial stem/progenitor cells that express high levels of FGFR2b to restore epithelial barrier function and promote epithelial regeneration (Quantius et al., 2016). Following influenza virus infection, AEC2 cells that conditionally lack Fgfr2 showed decreased proliferation and increased differentiation to AEC1 cells in lung regions undergoing active regeneration (Liberti et al., 2021). However, mitogenic stimulation with FGF7 along with influenza virus infection markedly accelerated the spread of viral infection from the airways to the alveoli and increased disease severity and mortality in mice (Nikolaidis et al., 2017).
Serum levels of FGF2 were markedly increased in humans and mice infected with influenza virus. Depletion or knockout of Fgf2 increased influenza-associated disease pathology by impairing neutrophil recruitment and activation. Intravenous administration of FGF2 reduced the severity of influenza virus-induced lung injury and promoted the survival of influenza virus-infected mice. These studies show that FGF2 may be an effective therapy for influenza virus infection (K. Wang, Lai, et al., 2018). In volunteer patients infected with influenza virus, serum levels of FGF9 were increased 1 day after infections specifically in asymptomatic patients suggesting that FGF9 may affect the early response to influenza infection (Linel et al., 2014).
13.3 |. Zika virus
Zika virus can cause microcephaly and other neurological defects in developing human fetuses. Zika virus infection of human fetal astrocytes results in elevated expression and secretion of FGF2, which was shown to enhance the replication and spread of the Zika virus. FGF2 suppressed the interferon response, which may contribute to its proviral effects. Therapeutic inhibition of FGF2 signaling may provide a strategy to limit the replication of Zika virus (Limonta et al., 2019).
13.4 |. Human immunodeficiency virus
Human immunodeficiency virus (HIV) causes acquired immunodeficiency syndrome (AIDS), which results in progressive failure of the immune system allowing for opportunistic infections and cancer. HIV-infected patients have higher bone turnover and bone loss. Elevated serum FGF21 levels were shown to be associated with poor bone homeostasis in HIV-infected patients. Increased FGF21 levels may serve as a biomarker of altered bone homeostasis in HIV-infected patients (Gallego-Escuredo et al., 2017). Elevated serum FGF21 levels were shown to serve as a stronger predictor of severe hepatic steatosis in HIV-infected patients (Praktiknjo et al., 2019). In contrast, the decrease of serum FGF21 levels due to antiretroviral therapy is associated with the alteration in lipid profile and an increased risk for cardiovascular diseases. FGF21 levels are predictors of inflammatory status in HIV patients using antiretroviral therapy (Ruiz-Padilla et al., 2020). Serum FGF23 levels were also found to be predictive of increased risk for cardiovascular and kidney diseases and were associated with greater risk of frailty in HIV-infected patients (Atta et al., 2016; R. Wang, Shlipak, et al., 2019).
High serum FGF2 levels were predictive of children at risk of developing HIV-associated nephropathy (HIVAN). In mice, FGF2 activated phospho-ERK in renal epithelial cells and induced transient and reversible HIVAN-like lesions, including proteinuria and glomerular enlargement (Das et al., 2021).
13.5 |. Virus-induced interferon response
Interferons are proteins made by host cells in response to the presence of viruses. A screen of 756 human secreted proteins identified FGF16 as an inhibitor of vesicular stomatitis virus (VSV) replication. FGF16 (and other FGFs, including FGF1, 8b, 9, and 20) prevented an antiviral interferon signature in multiple cell types and was also effective in preventing infection by coxsackievirus (van Asten et al., 2018). Inhibition of FGFR signaling in keratinocytes promoted the expression of interferon-stimulated genes (ISGs) and proteins. FGF7 or FGF10 treatment of keratinocytes suppressed ISG gene expression independent of signaling through the interferon-α/β receptor. FGFs promoted the replication of herpes simplex virus-1, lymphocytic choriomeningitis virus, and Zika virus. Inhibition of FGFR signaling blocked herpes simplex virus-1 replication in cultured human keratinocytes and in mice. These studies suggest that FGFR kinase inhibitors may be useful for the treatment of viral infections (Maddaluno et al., 2020).
13.6 |. Tapeworm
Infection by the tapeworm Echinococcus multilocularis causes tumor-like growth of the larval stage worm within the host liver, leading to extensive fibrosis and organ-failure. Larval stage E. multilocularis expresses three FGFRs with homology to human FGFRs. E. multilocularis FGFRs can be activated by human FGF1 and FGF2 and inhibited with a FGFR kinase inhibitor drug (BIBF 1120). These studies show that FGFs, which are present in the liver and upregulated during fibrosis, support the establishment of E. metacestode during larval stage infection by acting on an evolutionarily conserved parasite FGF signaling system (Forster et al., 2019).
14 |. FGF SIGNALING IN EVOLUTIONARY ADAPTATION
14.1 |. Avian adaptation to flight
The avian genome is characterized by its constrained size with the loss of repetitive elements, segmental deletions, and gene loss, driven in part by the metabolic intensity of powered flight (Wright et al., 2014; Zhang et al., 2014). FGF11, FGF17, and FGF21 were lost in the avian lineage (Oulion et al., 2012). FGF21 plays essential roles in energy metabolism including glucose and lipid metabolism in humans and mice (Itoh et al., 2015). The loss of FGF21 may therefore be an adaptation for the high metabolic demands of powered flight in birds.
Birds can be classified into altricial and precocial birds. Altricial bird hatchlings are almost naked, whereas precocial birds are covered with natal down. Zebra finch and chicken are altricial and precocial birds, respectively. Ectopic expression of FGF16 in embryonic chicken leg skin suppressed feather bud elongation by upregulating expression of FGF10, which suppresses feather growth. This is similar to embryonic zebra finch skin. FGF16-related signals are suggested to suppress natal down elongation and cause the naked skin phenotype in the zebra finch (C. K. Chen, Ng, et al., 2016). Emus are flightless birds. Regulatory changes in the Fgf10 gene result in lower Fgf10 expression leading to failure to express genes required for limb proliferation in the emu wing bud (Young et al., 2019).
14.2 |. Mammalian adaptation to flight
Bats are the only mammal capable of powered flight. This adaptation was enabled by the acquisition of a unique set of wing muscles. The wing membrane, which is an extension of the skin that contains epidermis and dermis, may regulate the patterning of wing muscles. Fgf10 expression is uniquely activated in the lateral plate mesoderm primordia of wing membranes and at later stages in wing membrane connective tissue in the proximity of wing muscles, indicating that FGF10 is likely to be involved in the development and evolution of bat flight muscles (Tokita et al., 2012).
14.3 |. Environmental adaptation
Sheep have successfully adapted to extreme high-altitude regions of the Himalayas by increasing lung function. Comparison of high- and low-altitude adapted sheep identified variants in Fgf7 gene regulatory regions that may account for increased Fgf7 expression in the high-altitude adapted sheep (Gorkhali et al., 2016). Cetaceans (whale, dolphin, porpoise) have adapted their body structure and physiology to their aquatic environment. Several changes in Fgf genes may account for cetacean-specific phenotypes such as loss of body hair, rigid flippers, ear and tooth development, and tolerance for hypoxia. Loss of a functional Fgf22 may account for hair loss. A variant in Fgf9 may affect FGF9 homodimerization and contribute to the acquisition of rigid flippers. Increased expression of Fgf23 in the cetacean liver under hypoxic conditions and may contribute to reduced bone density which is a critical component of the cetacean buoyancy control mechanism (Nam et al., 2017). Mammalian dental morphology is under strong evolutionary pressure controlled by the available diet. Changes in evolutionarily conserved regions that function as enhancers in Fgf9 and Fgf10 were shown to control dental morphology (molar cusp shape and crown height) in rodents (Tapaltsyan et al., 2016).
14.4 |. Mechanical regulation of morphogenesis
A relationship between FGF signaling and species-specific jaw architecture may involve regulation of gene expression by mechanical forces. In the duck jaw, but not in chicken or quail, mechanical load was shown to regulate secondary cartilage at the tendon insertion of the adductor muscle through the differential regulation of FGF and TGFβ signaling (Woronowicz et al., 2018).
14.5 |. Sex determination
Fgf9 is a testis determining gene expressed by somatic cells in the seminiferous tubules (Capel, 2017; Colvin et al., 2001). In embryonic gonadal somatic cells, FGF9 signaling to FGFR2 is required to maintain SOX9 expression which is required for Sertoli cell fate. Mice with a hypomorphic mutation in Fgfr2 (hobbyhorse) or knockout of the 2c splice variant for FGFR2 have complete XY sex reversal (Bagheri-Fam et al., 2017; Siggers et al., 2014). Gene expression studies showed that FGFR2c functions to repress both the WNT4- and FOXL2-driven ovarian-determining pathways (Bagheri-Fam et al., 2017). Overexpression of FGF9 in vivo (lentivirus-mediated) resulted in arrested spermatogenesis and accumulation of undifferentiated spermatogonia. Functionally, FGF9 regulated spermatogonial stem cell proliferation through p38 MAPK phosphorylation and upregulation of Etv5 and Bcl6b (Yang et al., 2021).
In zebrafish, Fgf24 signaling is required for the early growth phase of the bipotential gonad where it is required for proliferation, differentiation, and morphogenesis of the somatic tissues that are in direct contact with germ cells. The role of zebrafish Fgf24 is functionally similar to mammalian FGF9 in early gonad development (Leerberg et al., 2017).
In moles, males have normal testes, but genotypic females develop ovotestes instead of ovaries. Genomic analysis identified an intrachromosomal inversion involving FGF9 gene regulatory elements that are specific to moles and not present in other mammals. In mole gonads, FGF9 expression is maintained after sex determination in both sexes and becomes restricted to the ovarian part of the ovotestes at later stages (Real et al., 2020).
15 |. FGF SIGNALING IN EPIDEMIOLOGY (MENDELIAN RANDOMIZATION)
The integration of genetic information into population-based epidemiological studies presents translational opportunities. Mendelian randomization (MR) is a method to generate more reliable evidence regarding interventions that should produce health benefits by using genetic variants that are robustly associated with modifiable factors that potentially influence disease risk (Davey Smith & Hemani, 2014). Efficiently transforming genetic associations into drug targets requires evidence that a particular gene and its protein contribute causally to a disease.
Recently, MR studies identified FGF5, FGF21, and FGF23 as likely to have a causal role in human disease pathogenesis. MR analysis of 154 quantitative trait loci and 846 traits identified 38 proteins that significantly predicted variation in traits and diseases. Further analysis identified FGF5 and other factors as likely to have a causal role in cardiovascular disease (Bretherick et al., 2020).
MR analysis of a common variant in FGF21 demonstrated an association with lower intakes of total sugars and alcohol, higher intakes of protein and fat, as well as favorable lipid levels, blood pressure traits, waist-to-hip ratio, systemic inflammation, cardiovascular outcomes, Alzheimer’s disease risk, and lifespan. These findings anticipate the effects of pharmacologically increasing FGF21 signaling (Larsson & Gill, 2021).
Several MR studies have focused on FGF23. Two studies looked at serum FGF23 levels and bone mineral density and identified a causal relationship between FGF23 and heel and femoral neck bone mineral density in adults (Y. Wang, Wang, & Chen, 2020; Yokomoto-Umakoshi et al., 2021). FGF23 also regulates cardiovascular functions. No association was found between FGF23 and cardiometabolic traits or ischemic stroke; however, there was a significant causal association between genetically predicted FGF23 levels and the risk of large-artery atherosclerotic stroke (Yokomoto-Umakoshi et al., 2021; Zheng et al., 2020).
16 |. CONCLUSIONS
The FGF family consists of secreted canonical ligands, endocrine ligands, and intracellular proteins. Four of the five dedicated FGFRs activate intracellular tyrosine kinase domains that regulate multiple signaling pathways in cell and tissue-specific patterns. FGFs and FGFRs also regulate cellular functions independent of RTK activity through actions in the nucleus and regulation of other intracellular signaling pathways. During the last 6 years, the repertoire of factors that interact with and regulate both ligands and receptors has grown substantially. The functions of FGFs and FGFRs encompass many aspects of biology from the earliest stages of embryonic development to roles in regulating tissue homeostasis, tissue regeneration, and an increasing importance in cancer biology. Biotechnology and pharmaceutical efforts have focused on the manipulation of the FGF signaling pathway for promoting cell and tissue growth and differentiation for uses in regenerative medicine. Many FGFR selective tyrosine kinase inhibitors have been developed for use in treating genetic diseases caused by activating mutations in FGFRs and in cancers that acquire activating mutations in FGFRs or increased expression of FGF ligands.
Future directions will continue to explore the biological functions of FGFs and FGFRs, as well as to develop more specific agonists and antagonists that can be tailored to treat disease with minimal off-target activities.
ACKNOWLEDGMENT
We thank A. Bowman, B. Hiller, A. Johnson, M. Lewandoski, T. Theunissen, and Y. Yin for critically reading the manuscript.
Funding information
This work was supported by the Childrens Discovery Institute, St. Louis Childrens hospital; Washington University Hope Center for Neurological Disorders; Musculoskeletal Research Center P30 AR057235; Washington University Institute of Clinical and Translational Sciences grant UL1TR002345; and NIH grants, HL111190, HL154747, HD09887202.
Footnotes
TOC is available in online supplemental data.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- Abd El Kader T, Kubota S, Anno K, Tanaka S, Nishida T, Furumatsu T, Aoyama E, Kuboki T, & Takigawa M (2014). Direct interaction between CCN family protein 2 and fibroblast growth factor 1. Journal of Cell Communication and Signaling, 8(2), 157–163. 10.1007/s12079-014-0232-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahao KP, Salinas AG, & Lovinger DM (2017). Alcohol and the brain: Neuronal molecular targets, synapses, and circuits. Neuron, 96(6), 1223–1238. 10.1016/j.neuron.2017.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Issa R, Smyth G, Smoak I, Yamamura K, & Meyers EN (2002). Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development, 129(19), 4613–4625. [DOI] [PubMed] [Google Scholar]
- Abu-Odeh M, Zhang Y, Reilly SM, Ebadat N, Keinan O, Valentine JM, Hafezi-Bakhtiari M, Ashayer H, Mamoun L, Zhou X, Zhang J, Yu RT, Dai Y, Liddle C, Downes M, Evans RM, Kliewer SA, Mangelsdorf DJ, & Saltiel AR (2021). FGF21 promotes thermogenic gene expression as an autocrine factor in adipocytes. Cell Reports, 35(13), 109331. 10.1016/j.celrep.2021.109331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achilleos A, Huffman NT, Marcinkiewicyz E, Seidah NG, Chen Q, Dallas SL, Trainor PA, & Gorski JP (2015). MBTPS1/SKI-1/S1P proprotein convertase is required for ECM signaling and axial elongation during somitogenesis and vertebral developmentdagger. Human Molecular Genetics, 24(10), 2884–2898. 10.1093/hmg/ddv050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acquaviva J, He S, Zhang C, Jimenez JP, Nagai M, Sang J, Sequeira M, Smith DL, Ogawa LS, Inoue T, Tatsuta N, Knowles MA, Bates RC, & Proia DA (2014). FGFR3 translocations in bladder cancer: Differential sensitivity to HSP90 inhibition based on drug metabolism. Molecular Cancer Research, 12(7), 1042–1054. 10.1158/1541-7786.MCR-14-0004 [DOI] [PubMed] [Google Scholar]
- Adachi T, & Nakamura Y (2019). Aptamers: A review of their chemical properties and modifications for therapeutic application. Molecules, 24(23), 4229. 10.3390/molecules24234229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeyemo A, Johnson C, Stiene A, LaSance K, Qi Z, Lemen L, & Schultz JEJ (2020). Limb functional recovery is impaired in fibroblast growth factor-2 (FGF2) deficient mice despite chronic ischaemia-induced vascular growth. Growth Factors, 38(2), 75–93. 10.1080/08977194.2020.1767612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal M, Sharma A, Kumar P, Kumar A, Bharadwaj A, Saini M, Kardon G, & Mathew SJ (2020). Myosin heavy chain-embryonic regulates skeletal muscle differentiation during mammalian development. Development, 147(7), dev184507. 10.1242/dev.184507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agochukwu NB, Solomon BD, Doherty ES, & Muenke M (2012). Palatal and oral manifestations of Muenke syndrome (FGFR3-related craniosynostosis). The Journal of Craniofacial Surgery, 23(3), 664–668. 10.1097/SCS.0b013e31824db8bb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agoro R, Montagna A, Goetz R, Aligbe O, Singh G, Coe LM, Mohammadi M, Rivella S, & Sitara D (2018). Inhibition of fibroblast growth factor 23 (FGF23) signaling rescues renal anemia. The FASEB Journal, 32(7), 3752–3764. 10.1096/fj.201700667R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agoro R, Park MY, Le Henaff C, Jankauskas S, Gaias A, Chen G, Mohammadi M, & Sitara D (2021). C-FGF23 peptide alleviates hypoferremia during acute inflammation. Haematologica, 106(2), 391–403. 10.3324/haematol.2019.237040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad SM (2017). Conserved signaling mechanisms in Drosophila heart development. Developmental Dynamics, 246(9), 641–656. 10.1002/dvdy.24530 [DOI] [PubMed] [Google Scholar]
- Ajaz S, McPhail MJ, Singh KK, Mujib S, Trovato FM, Napoli S, & Agarwal K (2021). Mitochondrial metabolic manipulation by SARS-CoV-2 in peripheral blood mononuclear cells of patients with COVID-19. American Journal of Physiology. Cell Physiology, 320(1), C57–C65. 10.1152/ajpcell.00426.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akatsu Y, Takahashi N, Yoshimatsu Y, Kimuro S, Muramatsu T, Katsura A, Maishi N, Suzuki HI, Inazawa J, Hida K, Miyazono K, & Watabe T (2019). Fibroblast growth factor signals regulate transforming growth factor-beta-induced endothelial-to-myofibroblast transition of tumor endothelial cells via Elk1. Molecular Oncology, 13(8), 1706–1724. 10.1002/1878-0261.12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkus G, Kotan LD, Durmaz E, Mengen E, Turan I, Ulubay A, Gurbuz F, Yuksel B, Tetiker T, & Topaloglu AK (2017). Hypogonadotropic hypogonadism due to novel FGFR1 mutations. Journal of Clinical Research in Pediatric Endocrinology, 9(2), 95–100. 10.4274/jcrpe.3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Alam D, Danopoulos S, Schall K, Sala FG, Almohazey D, Fernandez GE, Georgia S, Frey MR, Ford HR, Grikscheit T, & Bellusci S (2015). Fibroblast growth factor 10 alters the balance between goblet and Paneth cells in the adult mouse small intestine. American Journal of Physiology. Gastrointestinal and Liver Physiology, 308(8), G678–G690. 10.1152/ajpgi.00158.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Alam D, El Agha E, Sakurai R, Kheirollahi V, Moiseenko A, Danopoulos S, Shrestha A, Schmoldt C, Quantius J, Herold S, Chao CM, Tiozzo C, De Langhe S, Plikus MV, Thornton M, Grubbs B, Minoo P, Rehan VK, & Bellusci S (2015). Evidence for the involvement of fibroblast growth factor 10 in lipofibroblast formation during embryonic lung development. Development, 142(23), 4139–4150. 10.1242/dev.109173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Alam D, Sala FG, Baptista S, Galzote R, Danopoulos S, Tiozzo C, Gage P, Grikscheit T, Warburton D, Frey MR, & Bellusci S (2012). FGF9-Pitx2-FGF10 signaling controls cecal formation in mice. Developmental Biology, 369(2), 340–348. 10.1016/j.ydbio.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Rifai O, Susan-Resiga D, Essalmani R, Creemers JWM, Seidah NG, & Ferron M (2021). In vivo analysis of the contribution of proprotein convertases to the processing of FGF23. Frontiers in Endocrinology, 12, 690681. 10.3389/fendo.2021.690681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albino FP, Wood BC, Oluigbo CO, Lee AC, Oh AK, & Rogers GF (2015). Achondroplasia and multiple-suture craniosynostosis. The Journal of Craniofacial Surgery, 26(1), 222–225. 10.1097/SCS.0000000000001267 [DOI] [PubMed] [Google Scholar]
- Ali SR, Singh AK, & Laezza F (2016). Identification of amino acid residues in fibroblast growth factor 14 (FGF14) required for structure-function interactions with voltage-gated sodium channel Nav1.6. The Journal of Biological Chemistry, 291(21), 11268–11284. 10.1074/jbc.M115.703868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliabouzar M, Jivani A, Lu X, Kripfgans OD, Fowlkes JB, & Fabiilli ML (2020). Standing wave-assisted acoustic droplet vaporization for single and dual payload release in acoustically-responsive scaffolds. Ultrasonics Sonochemistry, 66, 105109. 10.1016/j.ultsonch.2020.105109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allodi I, Mecollari V, Gonzalez-Perez F, Eggers R, Hoyng S, Verhaagen J, Navarro X, & Udina E (2014). Schwann cells transduced with a lentiviral vector encoding Fgf-2 promote motor neuron regeneration following sciatic nerve injury. Glia, 62(10), 1736–1746. 10.1002/glia.22712 [DOI] [PubMed] [Google Scholar]
- Al-Obaidy KI, & Cheng L (2021). Fibroblast growth factor receptor (FGFR) gene: Pathogenesis and treatment implications in urothelial carcinoma of the bladder. Journal of Clinical Pathology, 74(8), 491–495. 10.1136/jclinpath-2020-207115 [DOI] [PubMed] [Google Scholar]
- Alonso L, & Fuchs E (2006). The hair cycle. Journal of Cell Science, 119(Pt 3), 391–393. 10.1242/jcs02793 [DOI] [PubMed] [Google Scholar]
- Alshammari MA, Alshammari TK, Nenov MN, Scala F, & Laezza F (2016). Fibroblast growth factor 14 modulates the neurogenesis of granule neurons in the adult dentate gyrus. Molecular Neurobiology, 53(10), 7254–7270. 10.1007/s12035-015-9568-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez Y, Alonso MT, Vendrell V, Zelarayan LC, Chamero P, Theil T, Bosl MR, Kato S, Maconochie M, Riethmacher D, & Schimmang T (2003). Requirements for FGF3 and FGF10 during inner ear formation. Development, 130(25), 6329–6338. 10.1242/dev.00881 [DOI] [PubMed] [Google Scholar]
- Alvarez-Sola G, Uriarte I, Latasa MU, Urtasun R, Barcena-Varela M, Elizalde M, Jimenez M, Rodriguez-Ortigosa CM, Corrales FJ, Fernandez-Barrena MG, Berasain C, & Avila MA (2017). Fibroblast growth factor 15/19 in hepatocarcinogenesis. Digestive Diseases, 35(3), 158–165. 10.1159/000450905 [DOI] [PubMed] [Google Scholar]
- Amann R, Wyder S, Slavotinek AM, & Trueb B (2014). The FgfrL1 receptor is required for development of slow muscle fibers. Developmental Biology, 394(2), 228–241. 10.1016/j.ydbio.2014.08.016 [DOI] [PubMed] [Google Scholar]
- Amano R, Namekata M, Horiuchi M, Saso M, Yanagisawa T, Tanaka Y, Ghani FI, Yamamoto M, & Sakamoto T (2021). Specific inhibition of FGF5-induced cell proliferation by RNA aptamers. Scientific Reports, 11(1), 2976. 10.1038/s41598-021-82350-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato LGL, Montenegro LR, Lerario AM, Jorge AAL, Guerra Junior G, Schnoll C, Renck AC, Trarbach EB, Costa EMF, Mendonca BB, Latronico AC, & Silveira LFG (2019). New genetic findings in a large cohort of congenital hypogonadotropic hypogonadism. European Journal of Endocrinology, 181(2), 103–119. 10.1530/EJE-18-0764 [DOI] [PubMed] [Google Scholar]
- Amidfar M, Reus GZ, de Moura AB, Quevedo J, & Kim YK (2021). The role of neurotrophic factors in pathophysiology of major depressive disorder. Advances in Experimental Medicine and Biology, 1305, 257–272. 10.1007/978-981-33-6044-0_14 [DOI] [PubMed] [Google Scholar]
- An C, Feng G, Zhang J, Cao S, Wang Y, Wang N, Lu F, Zhou Q, & Wang H (2020). Overcoming autocrine FGF signaling-induced heterogeneity in naive human ESCs enables modeling of random X chromosome inactivation. Cell Stem Cell, 27(3), 482–497.e484. 10.1016/j.stem.2020.06.002 [DOI] [PubMed] [Google Scholar]
- Anderson MJ, Magidson V, Kageyama R, & Lewandoski M (2020). Fgf4 maintains Hes7 levels critical for normal somite segmentation clock function. eLife, 9, e55608. 10.7554/eLife.55608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ, Schimmang T, & Lewandoski M (2016). An FGF3-BMP signaling axis regulates caudal neural tube closure, neural crest specification and anterior-posterior axis extension. PLoS Genetics, 12(5), e1006018. 10.1371/journal.pgen.1006018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MJ, Southon E, Tessarollo L, & Lewandoski M (2016). Fgf3-Fgf4-cis: A new mouse line for studying Fgf functions during mouse development. Genesis, 54(2), 91–98. 10.1002/dvg.22913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson PJ, Snell B, & Moore MH (2013). Muencke syndrome with cleft lip and palate. The Journal of Craniofacial Surgery, 24(4), 1484–1485. 10.1097/SCS.0b013e31829035c3 [DOI] [PubMed] [Google Scholar]
- Andres C, Hasenauer J, Ahn HS, Joseph EK, Isensee J, Theis FJ, Allgower F, Levine JD, Dib-Hajj SD, Waxman SG, & Hucho T (2013). Wound-healing growth factor, basic FGF, induces Erk1/2-dependent mechanical hyperalgesia. Pain, 154(10), 2216–2226. 10.1016/j.pain.2013.07.005 [DOI] [PubMed] [Google Scholar]
- Antoine M, Wirz W, Tag CG, Gressner AM, Marvituna M, Wycislo M, Hellerbrand C, & Kiefer P (2007). Expression and function of fibroblast growth factor (FGF) 9 in hepatic stellate cells and its role in toxic liver injury. Biochemical and Biophysical Research Communications, 361(2), 335–341. 10.1016/j.bbrc.2007.06.189 [DOI] [PubMed] [Google Scholar]
- Antoine M, Wirz W, Tag CG, Mavituna M, Emans N, Korff T, Stoldt V, Gressner AM, & Kiefer P (2005). Expression pattern of fibroblast growth factors (FGFs), their receptors and antagonists in primary endothelial cells and vascular smooth muscle cells. Growth Factors, 23(2), 87–95. 10.1080/08977190500096004 [DOI] [PubMed] [Google Scholar]
- Antunes BP, Vainieri ML, Alini M, Monsonego-Ornan E, Grad S, & Yayon A (2020). Enhanced chondrogenic phenotype of primary bovine articular chondrocytes in Fibrin-Hyaluronan hydrogel by multi-axial mechanical loading and FGF18. Acta Biomaterialia, 105, 170–179. 10.1016/j.actbio.2020.01.032 [DOI] [PubMed] [Google Scholar]
- Anwar M, Tambalo M, Ranganathan R, Grocott T, & Streit A (2017). A gene network regulated by FGF signalling during ear development. Scientific Reports, 7(1), 6162. 10.1038/s41598-017-05472-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki S, Hirata Y, Kawai T, Nakabayashi K, Hata K, Suzuki H, Kosaki K, Amagai M, & Kubo A (2021). Frequent FGFR3 and Ras gene mutations in skin tags or acrochordons. The Journal of Investigative Dermatology, 141(11), 2756–2760.e2758. 10.1016/j.jid.2021.03.028 [DOI] [PubMed] [Google Scholar]
- Ardizzone A, Scuderi SA, Giuffrida D, Colarossi C, Puglisi C, Campolo M, Cuzzocrea S, Esposito E, & Paterniti I (2020). Role of fibroblast growth factors receptors (FGFRs) in brain tumors, focus on astrocytoma and glioblastoma. Cancers (Basel), 12(12), 3825. 10.3390/cancers12123825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armelin HA (1973). Pituitary extracts and steroid hormones in the control of 3T3 cell growth. Proceedings of the National Academy of Sciences of the United States of America, 70(9), 2702–2706. 10.1073/pnas.70.9.2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arudra K, Patel R, Tetzlaff MT, Hymes S, Subbiah V, Meric-Bernstam F, Torres-Cabala C, Aung PP, Nagarajan P, Diab A, Prieto VG, Nelson K, & Curry JL (2018). Calcinosis cutis dermatologic toxicity associated with fibroblast growth factor receptor inhibitor for the treatment of Wilms tumor. Journal of Cutaneous Pathology, 45(10), 786–790. 10.1111/cup.13319 [DOI] [PubMed] [Google Scholar]
- Atkinson-Leadbeater K, Hehr CL, & McFarlane S (2014). Fgfr signaling is required as the early eye field forms to promote later patterning and morphogenesis of the eye. Developmental Dynamics, 243(5), 663–675. 10.1002/dvdy.24113 [DOI] [PubMed] [Google Scholar]
- Atsuta Y, & Takahashi Y (2015). FGF8 coordinates tissue elongation and cell epithelialization during early kidney tubulogenesis. Development, 142(13), 2329–2337. 10.1242/dev.122408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atta MG, Estrella MM, Fine DM, Zook K, Monroy Trujillo JM, Stein JH, & Lucas GM (2016). Correlates and longitudinal renal and cardiovascular implications of FGF23 levels in HIV-positive individuals. PLoS One, 11(5), e0155312. 10.1371/journal.pone.0155312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attia L, Schneider J, Yelin R, & Schultheiss TM (2015). Collective cell migration of the nephric duct requires FGF signaling. Developmental Dynamics, 244(2), 157–167. 10.1002/dvdy.24241 [DOI] [PubMed] [Google Scholar]
- Aurbach EL, Inui EG, Turner CA, Hagenauer MH, Prater KE, Li JZ, Absher D, Shah N, Blandino P Jr., Bunney WE, Myers RM, Barchas JD, Schatzberg AF, Watson SJ Jr., & Akil H (2015). Fibroblast growth factor 9 is a novel modulator of negative affect. Proceedings of the National Academy of Sciences of the United States of America, 112(38), 11953–11958. 10.1073/pnas.1510456112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auwal MA, Kashima M, Nishimura O, Hosoda K, Motoishi M, Kamimura A, Okumura A, Agata K, & Umesono Y (2020). Identification and characterization of a fibroblast growth factor gene in the planarian Dugesia japonica. Development, Growth & Differentiation, 62(9), 527–539. 10.1111/dgd.12696 [DOI] [PubMed] [Google Scholar]
- Azad T, Nouri K, Janse van Rensburg HJ, Maritan SM, Wu L, Hao Y, Montminy T, Yu J, Khanal P, Mulligan LM, & Yang X (2020). A gain-of-functional screen identifies the Hippo pathway as a central mediator of receptor tyrosine kinases during tumorigenesis. Oncogene, 39(2), 334–355. 10.1038/s41388-019-0988-y [DOI] [PubMed] [Google Scholar]
- Azami T, Bassalert C, Allegre N, Valverde Estrella L, Pouchin P, Ema M, & Chazaud C (2019). Regulation of the ERK signalling pathway in the developing mouse blastocyst. Development, 146(14), dev177139. 10.1242/dev.177139 [DOI] [PubMed] [Google Scholar]
- Azami T, Matsumoto K, Jeon H, Waku T, Muratani M, Niwa H, Takahashi S, & Ema M (2018). Klf5 suppresses ERK signaling in mouse pluripotent stem cells. PLoS One, 13(11), e0207321. 10.1371/journal.pone.0207321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchetta J, Bardet C, & Prie D (2020). Physiology of FGF23 and overview of genetic diseases associated with renal phosphate wasting. Metabolism, 103S, 153865. 10.1016/j.metabol.2019.01.006 [DOI] [PubMed] [Google Scholar]
- Bae JH, Hong M, Jeong HJ, Kim H, Lee SJ, Ryu D, Bae GU, Cho SC, Lee YS, Krauss RS, & Kang JS (2020). Satellite cell-specific ablation of Cdon impairs integrin activation, FGF signalling, and muscle regeneration. Journal of Cachexia, Sarcopenia and Muscle, 11(4), 1089–1103. 10.1002/jcsm.12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagheri-Fam S, Bird AD, Zhao L, Ryan JM, Yong M, Wilhelm D, Koopman P, Eswarakumar VP, & Harley VR (2017). Testis determination requires a specific FGFR2 isoform to repress FOXL2. Endocrinology, 158(11), 3832–3843. 10.1210/en.2017-00674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balani S, & Perwad F (2019). Fibroblast growth factor 23 and phosphate homeostasis. Current Opinion in Nephrology and Hypertension, 28(5), 465–473. 10.1097/MNH.0000000000000526 [DOI] [PubMed] [Google Scholar]
- Balani S, & Perwad F (2020). Burosumab in X-linked hypophosphatemia and perspective for chronic kidney disease. Current Opinion in Nephrology and Hypertension, 29(5), 531–536. 10.1097/MNH.0000000000000631 [DOI] [PubMed] [Google Scholar]
- Balasooriya GI, Goschorska M, Piddini E, & Rawlins EL (2017). FGFR2 is required for airway basal cell self-renewal and terminal differentiation. Development, 144(9), 1600–1606. 10.1242/dev.135681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasooriya GI, Johnson JA, Basson MA, & Rawlins EL (2016). An FGFR1-SPRY2 signaling axis limits basal cell proliferation in the steady-state airway epithelium. Developmental Cell, 37(1), 85–97. 10.1016/j.devcel.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balek L, Nemec P, Konik P, Kunova Bosakova M, Varecha M, Gudernova I, Medalova J, Krakow D, & Krejci P (2018). Proteomic analyses of signalling complexes associated with receptor tyrosine kinase identify novel members of fibroblast growth factor receptor 3 interactome. Cellular Signalling, 42, 144–154. 10.1016/j.cellsig.2017.10.003 [DOI] [PubMed] [Google Scholar]
- Bansal P, Dwivedi DK, Hatwal D, Sharma P, Gupta V, Goyal S, & Maithani M (2021). Erdafitinib as a novel and advanced treatment strategy of metastatic urothelial carcinoma. Anti-Cancer Agents in Medicinal Chemistry, 21(18), 2478–2486. 10.2174/1871520621666210121093852 [DOI] [PubMed] [Google Scholar]
- Bar L, Stournaras C, Lang F, & Foller M (2019). Regulation of fibroblast growth factor 23 (FGF23) in health and disease. FEBS Letters, 593(15), 1879–1900. 10.1002/1873-3468.13494 [DOI] [PubMed] [Google Scholar]
- Barak H, Huh SH, Chen S, Jeanpierre C, Martinovic J, Parisot M, Bole-Feysot C, Nitschke P, Salomon R, Antignac C, Ornitz DM, & Kopan R (2012). FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Developmental Cell, 22(6), 1191–1207. 10.1016/j.devcel.2012.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa GO, Biancardi MF, & Carvalho HF (2021). Heparan sulfate fine-tunes stromal-epithelial communication in the prostate gland. Developmental Dynamics, 250(5), 618–628. 10.1002/dvdy.281 [DOI] [PubMed] [Google Scholar]
- Barlow LA, & Klein OD (2015). Developing and regenerating a sense of taste. Current Topics in Developmental Biology, 111, 401–419. 10.1016/bs.ctdb.2014.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes JW, Duncan D, Helton S, Hutcheson S, Kurundkar D, Logsdon NJ, Locy M, Garth J, Denson R, Farver C, Vo HT, King G, Kentrup D, Faul C, Kulkarni T, De Andrade JA, Yu Z, Matalon S, Thannickal VJ, & Krick S (2019). Role of fibroblast growth factor 23 and klotho cross talk in idiopathic pulmonary fibrosis. American Journal of Physiology. Lung Cellular and Molecular Physiology, 317(1), L141–L154. 10.1152/ajplung.00246.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett CP, Nataren NJ, Klingler-Hoffmann M, Schwarz Q, Chong CE, Lee YK, Bruno DL, Lipsett J, McPhee AJ, Schreiber AW, Feng J, Hahn CN, & Scott HS (2016). Ectrodactyly and lethal pulmonary acinar dysplasia associated with homozygous FGFR2 mutations identified by exome sequencing. Human Mutation, 37(9), 955–963. 10.1002/humu.23032 [DOI] [PubMed] [Google Scholar]
- Barr L, Getgood A, Guehring H, Rushton N, & Henson FM (2014). The effect of recombinant human fibroblast growth factor-18 on articular cartilage following single impact load. Journal of Orthopaedic Research, 32(7), 923–927. 10.1002/jor.22622 [DOI] [PubMed] [Google Scholar]
- Barrientos S, Brem H, Stojadinovic O, & Tomic-Canic M (2014). Clinical application of growth factors and cytokines in wound healing. Wound Repair and Regeneration, 22(5), 569–578. 10.1111/wrr.12205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch A, Wong C, Chinn LW, Vaze A, Sonoda J, Gelzleichter T, Chen S, Lewin-Koh N, Morrow L, Dheerendra S, Boismenu R, Gutierrez J, Wakshull E, Wilson ME, & Arora PS (2020). Antibody-mediated activation of the FGFR1/Klothobeta complex corrects metabolic dysfunction and alters food preference in obese humans. Proceedings of the National Academy of Sciences of the United States of America, 117(46), 28992–29000. 10.1073/pnas.2012073117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch ML, Brown RM 2nd, Jen HI, & Groves AK (2016). Where hearing starts: The development of the mammalian cochlea. Journal of Anatomy, 228(2), 233–254. 10.1111/joa.12314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas-Ponce A, Scheibner K, Lickert H, & Bakhti M (2017). Cellular and molecular mechanisms coordinating pancreas development. Development, 144(16), 2873–2888. 10.1242/dev.140756 [DOI] [PubMed] [Google Scholar]
- Batcher K, Dickinson P, Maciejczyk K, Brzeski K, Rasouliha SH, Letko A, Drögemüller C, Leeb T, & Bannasch D (2020). Multiple FGF4 retrocopies recently derived within canids. Genes (Basel), 11(8), 839. 10.3390/genes11080839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebee TW, Sims-Lucas S, Park JW, Bushnell D, Cieply B, Xing Y, Bates CM, & Carstens RP (2016). Ablation of the epithelial-specific splicing factor Esrp1 results in ureteric branching defects and reduced nephron number. Developmental Dynamics, 245(10), 991–1000. 10.1002/dvdy.24431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beever JE, Smit MA, Meyers SN, Hadfield TS, Bottema C, Albretsen J, & Cockett NE (2006). A single-base change in the tyrosine kinase II domain of ovine FGFR3 causes hereditary chondrodysplasia in sheep. Animal Genetics, 37(1), 66–71. 10.1111/j.1365-2052.2005.01398.x [DOI] [PubMed] [Google Scholar]
- Belleudi F, Purpura V, Caputo S, & Torrisi MR (2014). FGF7/KGF regulates autophagy in keratinocytes: A novel dual role in the induction of both assembly and turnover of autophagosomes. Autophagy, 10(5), 803–821. 10.4161/auto.28145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov AA, & Mohammadi M (2012). Grb2, a double-edged sword of receptor tyrosine kinase signaling. Science Signaling, 5(249), pe49. 10.1126/scisignal.2003576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benazeraf B, & Pourquie O (2013). Formation and segmentation of the vertebrate body axis. Annual Review of Cell and Developmental Biology, 29, 1–26. 10.1146/annurev-cellbio-101011-155703 [DOI] [PubMed] [Google Scholar]
- Benoit B, Meugnier E, Castelli M, Chanon S, Vieille-Marchiset A, Durand C, Bendridi N, Pesenti S, Monternier PA, Durieux AC, Freyssenet D, Rieusset J, Lefai E, Vidal H, & Ruzzin J (2017). Fibroblast growth factor 19 regulates skeletal muscle mass and ameliorates muscle wasting in mice. Nature Medicine, 23(8), 990–996. 10.1038/nm.4363 [DOI] [PubMed] [Google Scholar]
- Berg T, Rountree CB, Lee L, Estrada J, Sala FG, Choe A, Veltmaat JM, De Langhe S, Lee R, Tsukamoto H, Crooks GM, Bellusci S, & Wang KS (2007). Fibroblast growth factor 10 is critical for liver growth during embryogenesis and controls hepatoblast survival via beta-catenin activation. Hepatology, 46(4), 1187–1197. 10.1002/hep.21814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund ED, Li CY, Bina HA, Lynes SE, Michael MD, Shanafelt AB, Kharitonenkov A, & Wasserman DH (2009). Fibroblast growth factor 21 controls glycemia via regulation of hepatic glucose flux and insulin sensitivity. Endocrinology, 150(9), 4084–4093. 10.1210/en.2009-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernet JD, Doles JD, Hall JK, Kelly Tanaka K, Carter TA, & Olwin BB (2014). p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nature Medicine, 20(3), 265–271. 10.1038/nm.3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, & Zako M (1999). Functions of cell surface heparan sulfate proteoglycans. Annual Review of Biochemistry, 68, 729–777. 10.1146/annurev.biochem.68.1.729 [DOI] [PubMed] [Google Scholar]
- Bi J, Tong L, Zhu X, Yang D, Bai C, Song Y, & She J (2014). Keratinocyte growth factor-2 intratracheal instillation significantly attenuates ventilator-induced lung injury in rats. Journal of Cellular and Molecular Medicine, 18(6), 1226–1235. 10.1111/jcmm.12269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs LC, Makela OJ, Myllymaki SM, Das Roy R, Narhi K, Pispa J, Mustonen T, & Mikkola ML (2018). Hair follicle dermal condensation forms via Fgf20 primed cell cycle exit, cell motility, and aggregation. eLife, 7, e36468. 10.7554/eLife.36468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs LC, & Mikkola ML (2014). Early inductive events in ectodermal appendage morphogenesis. Seminars in Cell & Developmental Biology, 25–26, 11–21. 10.1016/j.semcdb.2014.01.007 [DOI] [PubMed] [Google Scholar]
- Biosse Duplan M, Dambroise E, Estibals V, Veziers J, Guicheux J, & Legeai-Mallet L (2021). An Fgfr3-activating mutation in immature murine osteoblasts affects the appendicular and craniofacial skeleton. Disease Models & Mechanisms, 14(4), dmm048272. 10.1242/dmm.048272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell C, Sherk C, Fricko M, Ganji G, Barnette M, Hoang B, Tunstead J, Skedzielewski T, Alsaid H, Jucker BM, Minthorn E, Kumar R, & DeYoung MP (2016). Inhibition of FGF/FGFR autocrine signaling in mesothelioma with the FGF ligand trap, FP-1039/GSK3052230. Oncotarget, 7(26), 39861–39871. 10.18632/oncotarget.9515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco AM, Bertucci JI, & Unniappan S (2020). FGF21 Mimics a Fasting-Induced Metabolic State and Increases Appetite in Zebrafish. Scientific Reports, 10(1), 6993. 10.1038/s41598-020-63726-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluteau G, Zhuang L, Amann R, & Trueb B (2014). Targeted disruption of the intracellular domain of receptor FgfrL1 in mice. PLoS One, 9(8), e105210. 10.1371/journal.pone.0105210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bober J, Olsnes S, Kostas M, Bogacz M, Zakrzewska M, & Otlewski J (2016). Identification of new FGF1 binding partners-Implications for its intracellular function. IUBMB Life, 68(3), 242–251. 10.1002/iub.1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobzin L, Roberts RR, Chen HJ, Crump JG, & Merrill AE (2021). Development and maintenance of tendons and ligaments. Development, 148(8), dev186916. 10.1242/dev.186916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocciardi R, & Ravazzolo R (2009). C-type natriuretic peptide and overgrowth. Endocrine Development, 14, 61–66. 10.1159/000207477 [DOI] [PubMed] [Google Scholar]
- BonDurant LD, Ameka M, Naber MC, Markan KR, Idiga SO, Acevedo MR, Walsh SA, Ornitz DM, & Potthoff MJ (2017). FGF21 regulates metabolism through adipose-dependent and -independent mechanisms. Cell Metabolism, 25(4), 935–944.e934. 10.1016/j.cmet.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BonDurant LD, & Potthoff MJ (2018). Fibroblast growth factor 21: A versatile regulator of metabolic homeostasis. Annual Review of Nutrition, 38, 173–196. 10.1146/annurev-nutr-071816-064800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bora SA, Kennett MJ, Smith PB, Patterson AD, & Cantorna MT (2018). The gut microbiota regulates endocrine vitamin D metabolism through fibroblast growth factor 23. Frontiers in Immunology, 9, 408. 10.3389/fimmu.2018.00408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroto-Escuela DO, Ambrogini P, Chruscicka B, Lindskog M, Crespo-Ramirez M, Hernandez-Mondragon JC, Perez de la Mora M, Schellekens H, & Fuxe K (2021). The role of central serotonin neurons and 5-HT heteroreceptor complexes in the pathophysiology of depression: A historical perspective and future prospects. International Journal of Molecular Sciences, 22(4), 1927. 10.3390/ijms22041927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroto-Escuela DO, Narvaez M, Ambrogini P, Ferraro L, Brito I, Romero-Fernandez W, Andrade-Talavera Y, Flores-Burgess A, Millon C, Gago B, Narvaez JA, Odagaki Y, Palkovits M, Diaz-Cabiale Z, & Fuxe K (2018). Receptor(−)receptor interactions in multiple 5-HT1A heteroreceptor complexes in raphe-hippocampal 5-HT transmission and their relevance for depression and its treatment. Molecules, 23(6), 1341. 10.3390/molecules23061341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroto-Escuela DO, Perez-Alea M, Narvaez M, Tarakanov AO, Mudo G, Jimenez-Beristain A, Agnati LF, Ciruela F, Belluardo N, & Fuxe K (2015). Enhancement of the FGFR1 signaling in the FGFR1–5-HT1A heteroreceptor complex in midbrain raphe 5-HT neuron systems. Relevance for neuroplasticity and depression. Biochemical and Biophysical Research Communications, 463(3), 180–186. 10.1016/j.bbrc.2015.04.133 [DOI] [PubMed] [Google Scholar]
- Borroto-Escuela DO, Tarakanov AO, & Fuxe K (2016). FGFR1–5-HT1A heteroreceptor complexes: Implications for understanding and treating major depression. Trends in Neurosciences, 39(1), 5–15. 10.1016/j.tins.2015.11.003 [DOI] [PubMed] [Google Scholar]
- Bosch MK, Nerbonne JM, Townsend RR, Miyazaki H, Nukina N, Ornitz DM, & Marionneau C (2016). Proteomic analysis of native cerebellar iFGF14 complexes. Channels (Austin, TX), 10(4), 297–312. 10.1080/19336950.2016.1153203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshoff EL, Fletcher EJR, & Duty S (2018). Fibroblast growth factor 20 is protective towards dopaminergic neurons in vivo in a paracrine manner. Neuropharmacology, 137, 156–163. 10.1016/j.neuropharm.2018.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botrus G, Raman P, Oliver T, & Bekaii-Saab T (2021). Infigratinib (BGJ398): An investigational agent for the treatment of FGFR-altered intrahepatic cholangiocarcinoma. Expert Opinion on Investigational Drugs, 30(4), 309–316. 10.1080/13543784.2021.1864320 [DOI] [PubMed] [Google Scholar]
- Boucherat O, Benachi A, Barlier-Mur AM, Franco-Montoya ML, Martinovic J, Thebaud B, Chailley-Heu B, & Bourbon JR (2007). Decreased lung fibroblast growth factor 18 and elastin in human congenital diaphragmatic hernia and animal models. American Journal of Respiratory and Critical Care Medicine, 175(10), 1066–1077. 10.1164/rccm.200601-050OC [DOI] [PubMed] [Google Scholar]
- Boulet AM, & Capecchi MR (2012). Signaling by FGF4 and FGF8 is required for axial elongation of the mouse embryo. Developmental Biology, 371(2), 235–245. 10.1016/j.ydbio.2012.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma-de Krijger A, & Vervloet MG (2020). Fibroblast growth factor 23: Are we ready to use it in clinical practice? Journal of Nephrology, 33(3), 509–527. 10.1007/s40620-020-00715-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler E, & Oltean S (2019). Alternative splicing in angiogenesis. International Journal of Molecular Sciences, 20(9), 2067. 10.3390/ijms20092067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan M, Anderson MJ, Ornitz DM, & Lewandoski M (2020). The Fgf8 subfamily (Fgf8, Fgf17 and Fgf18) is required for closure of the embryonic ventral body wall. Development, 147(21), dev189506. 10.1242/dev.189506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brameld KA, Owens TD, Verner E, Venetsanakos E, Bradshaw JM, Phan VT, Tam D, Leung K, Shu J, LaStant J, Loughhead DG, Ton T, Karr DE, Gerritsen ME, Goldstein DM, & Funk JO (2017). Discovery of the irreversible covalent FGFR inhibitor 8-(3-(4-acryloylpiperazin-1-yl)propyl)-6-(2,6-dichloro-3,5-dimethoxyphenyl)-2-(me thylamino)pyrido[2,3-d]pyrimidin-7 (8H)-one (PRN1371) for the treatment of solid tumors. Journal of Medicinal Chemistry, 60(15), 6516–6527. 10.1021/acs.jmedchem.7b00360 [DOI] [PubMed] [Google Scholar]
- Breinholt VM, Rasmussen CE, Mygind PH, Kjelgaard-Hansen M, Faltinger F, Bernhard A, Zettler J, & Hersel U (2019). TransCon CNP, a sustained-release c-type natriuretic peptide prodrug, a potentially safe and efficacious new therapeutic modality for the treatment of comorbidities associated with fibroblast growth factor receptor 3-related skeletal dysplasias. The Journal of Pharmacology and Experimental Therapeutics, 370(3), 459–471. 10.1124/jpet.119.258251 [DOI] [PubMed] [Google Scholar]
- Brent AE, & Tabin CJ (2004). FGF acts directly on the somitic tendon progenitors through the Ets transcription factors Pea3 and Erm to regulate scleraxis expression. Development, 131(16), 3885–3896. 10.1242/dev.01275 [DOI] [PubMed] [Google Scholar]
- Bretherick AD, Canela-Xandri O, Joshi PK, Clark DW, Rawlik K, Boutin TS, Zeng Y, Amador C, Navarro P, Rudan I, Wright AF, Campbell H, Vitart V, Hayward C, Wilson JF, Tenesa A, Ponting CP, Baillie JK, & Haley C (2020). Linking protein to phenotype with Mendelian randomization detects 38 proteins with causal roles in human diseases and traits. PLoS Genetics, 16(7), e1008785. 10.1371/journal.pgen.1008785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer JR, Mazot P, & Soriano P (2016). Genetic insights into the mechanisms of Fgf signaling. Genes & Development, 30(7), 751–771. 10.1101/gad.277137.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer JR, Molotkov A, Mazot P, Hoch RV, & Soriano P (2015). Fgfr1 regulates development through the combinatorial use of signaling proteins. Genes & Development, 29(17), 1863–1874. 10.1101/gad.264994.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EA, Dickinson PJ, Mansour T, Sturges BK, Aguilar M, Young AE, Korff C, Lind J, Ettinger CL, Varon S, Pollard R, Brown CT, Raudsepp T, & Bannasch DL (2017). FGF4 retrogene on CFA12 is responsible for chondrodystrophy and intervertebral disc disease in dogs. Proceedings of the National Academy of Sciences of the United States of America, 114(43), 11476–11481. 10.1073/pnas.1709082114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch PR, Ranadive I, Desai I, & Balarakrishnan S (2018). Cyclooxygenase-2 interacts with MMP and FGF pathways to promote epimorphic regeneration in lizard Hemidactylus flaviviridis. Growth Factors, 36(1–2), 69–77. 10.1080/08977194.2018.1497021 [DOI] [PubMed] [Google Scholar]
- Bugaj LJ, Spelke DP, Mesuda CK, Varedi M, Kane RS, & Schaffer DV (2015). Regulation of endogenous transmembrane receptors through optogenetic Cry2 clustering. Nature Communications, 6, 6898. 10.1038/ncomms7898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JO, & Jiang R (2012). Palatogenesis: Morphogenetic and molecular mechanisms of secondary palate development. Development, 139(2), 231–243. 10.1242/dev.067082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse-Wicher M, Wicher KB, & Kusche-Gullberg M (2014). The exostosin family: Proteins with many functions. Matrix Biology, 35, 25–33. 10.1016/j.matbio.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Byun S, Jung H, Chen J, Kim YC, Kim DH, Kong B, Guo G, Kemper B, & Kemper JK (2019). Phosphorylation of hepatic farnesoid X receptor by FGF19 signaling-activated Src maintains cholesterol levels and protects from atherosclerosis. The Journal of Biological Chemistry, 294(22), 8732–8744. 10.1074/jbc.RA119.008360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caglevic C, Grassi M, Raez L, Listi A, Giallombardo M, Bustamante E, Gil-Bazo I, & Rolfo C (2015). Nintedanib in non-small cell lung cancer: From preclinical to approval. Therapeutic Advances in Respiratory Disease, 9(4), 164–172. 10.1177/1753465815579608 [DOI] [PubMed] [Google Scholar]
- Cai J, Wen R, Li W, Wang X, Tian H, Yi S, Zhang L, Li X, Jiang C, & Li H (2018). Oil body bound oleosin-rhFGF9 fusion protein expressed in safflower (Carthamus tinctorius L.) stimulates hair growth and wound healing in mice. BMC Biotechnology, 18(1), 51. 10.1186/s12896-018-0433-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhou Q, Wang Z, Guo R, Yang R, Yang X, Li W, Ahmad N, Chen Q, Hui Q, & Wang X (2019). Comparative analysis of kgf-2 and bfgf in prevention of excessive wound healing and scar formation in a corneal alkali burn model. Cornea, 38(11), 1430–1437. 10.1097/ICO.0000000000002134 [DOI] [PubMed] [Google Scholar]
- Cai X, Zhang X, Mo L, Zhu J, & Yu H (2020). LncRNA PCGEM1 promotes renal carcinoma progression by targeting miR-433–3p to regulate FGF2 expression. Cancer Biomarkers, 27(4), 493–504. 10.3233/CBM-190669 [DOI] [PubMed] [Google Scholar]
- Cai Y, Zhang J, Lao X, Jiang H, Yu Y, Deng Y, Zhong J, Liang Y, Xiong L, & Deng N (2016). Construction of a disulfide-stabilized diabody against fibroblast growth factor-2 and the inhibition activity in targeting breast cancer. Cancer Science, 107(8), 1141–1150. 10.1111/cas.12981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronero F, Ariza-McNaughton L, Wiedemann LM, & Krumlauf R (2020). Inter-rhombomeric interactions reveal roles for fibroblast growth factors signaling in segmental regulation of EphA4 expression. Developmental Dynamics, 249(3), 354–368. 10.1002/dvdy.101 [DOI] [PubMed] [Google Scholar]
- Camozzi M, Zacchigna S, Rusnati M, Coltrini D, Ramirez-Correa G, Bottazzi B, Mantovani A, Giacca M, & Presta M (2005). Pentraxin 3 inhibits fibroblast growth factor 2-dependent activation of smooth muscle cells in vitro and neointima formation in vivo. Arteriosclerosis, Thrombosis, and Vascular Biology, 25(9), 1837–1842. 10.1161/01.ATV.0000177807.54959.7d [DOI] [PubMed] [Google Scholar]
- Cao F, Wang S, Cao X, Liu X, Fu K, Hao P, & Liu J (2017). Fibroblast growth factor 21 attenuates calcification of vascular smooth muscle cells in vitro. The Journal of Pharmacy and Pharmacology, 69(12), 1802–1816. 10.1111/jphp.12826 [DOI] [PubMed] [Google Scholar]
- Capel B (2017). Vertebrate sex determination: Evolutionary plasticity of a fundamental switch. Nature Reviews. Genetics, 18(11), 675–689. 10.1038/nrg.2017.60 [DOI] [PubMed] [Google Scholar]
- Carpenter TO, Imel EA, Ruppe MD, Weber TJ, Klausner MA, Wooddell MM, Kawakami T, Ito T, Zhang X, Humphrey J, Insogna KL, & Peacock M (2014). Randomized trial of the anti-FGF23 antibody KRN23 in X-linked hypophosphatemia. Journal of Clinical Investigation, 124(4), 1587–1597. 10.1172/JCI72829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter TO, Whyte MP, Imel EA, Boot AM, Hogler W, Linglart A, Padidela R, Van’t Hoff W, Mao M, Chen CY, Skrinar A, Kakkis E, San Martin J, & Portale AA (2018). Burosumab therapy in children with X-linked hypophosphatemia. The New England Journal of Medicine, 378(21), 1987–1998. 10.1056/NEJMoa1714641 [DOI] [PubMed] [Google Scholar]
- Carr DR, Pootrakul L, Chen HZ, & Chung CG (2019). Metastatic calcinosis cutis associated with a selective FGFR inhibitor. JAMA Dermatology, 155(1), 122–123. 10.1001/jamadermatol.2018.4070 [DOI] [PubMed] [Google Scholar]
- Casadei C, Dizman N, Schepisi G, Cursano MC, Basso U, Santini D, Pal SK, & De Giorgi U (2019). Targeted therapies for advanced bladder cancer: New strategies with FGFR inhibitors. Therapeutic Advances in Medical Oncology, 11, 1758835919890285. 10.1177/1758835919890285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases O, Perea-Gomez A, Aguiar DP, Nykjaer A, Amsellem S, Chandellier J, Umbhauer M, Cereghini S, Madsen M, Collignon J, Verroust P, Riou JF, Creuzet SE, & Kozyraki R (2013). Cubilin, a high affinity receptor for fibroblast growth factor 8, is required for cell survival in the developing vertebrate head. The Journal of Biological Chemistry, 288(23), 16655–16670. 10.1074/jbc.M113.451070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli R, Giacomini A, Anselmi M, Bozza N, Vacondio F, Rivara S, Matarazzo S, Presta M, Mor M, & Ronca R (2016). Synthesis, structural elucidation, and biological evaluation of NSC12, an orally available fibroblast growth factor (FGF) ligand trap for the treatment of FGF-dependent lung tumors. Journal of Medicinal Chemistry, 59(10), 4651–4663. 10.1021/acs.jmedchem.5b02021 [DOI] [PubMed] [Google Scholar]
- Castelli R, Taranto S, Furiassi L, Bozza N, Marseglia G, Ferlenghi F, Rivara S, Retini M, Bedini A, Spadoni G, Matarazzo S, Ronca R, Presta M, Mor M, & Giacomini A (2021). Chemical modification of NSC12 leads to a specific FGF-trap with antitumor activity in multiple myeloma. European Journal of Medicinal Chemistry, 221, 113529. 10.1016/j.ejmech.2021.113529 [DOI] [PubMed] [Google Scholar]
- Catenacci DV, Tesfaye A, Tejani M, Cheung E, Eisenberg P, Scott AJ, Eng C, Hnatyszyn J, Marina N, Powers J, & Wainberg Z (2019). Bemarituzumab with modified FOLFOX6 for advanced FGFR2-positive gastroesophageal cancer: FIGHT Phase III study design. Future Oncology, 15(18), 2073–2082. 10.2217/fon-2019-0141 [DOI] [PubMed] [Google Scholar]
- Catenacci DVT, Rasco D, Lee J, Rha SY, Lee KW, Bang YJ, Bendell J, Enzinger P, Marina N, Xiang H, Deng W, Powers J, & Wainberg ZA (2020). Phase I escalation and expansion study of bemarituzumab (FPA144) in patients with advanced solid tumors and FGFR2b-selected gastroesophageal adenocarcinoma. Journal of Clinical Oncology, 38(21), 2418–2426. 10.1200/JCO.19.01834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanah P, Itou J, Rusman Y, Tahara N, Williams JM, Salomon CE, & Kawakami Y (2021). A nontoxic fungal natural product modulates fin regeneration in zebrafish larvae upstream of FGF-WNT developmental signaling. Developmental Dynamics, 250(2), 160–174. 10.1002/dvdy.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chailley-Heu B, Boucherat O, Barlier-Mur AM, & Bourbon JR (2005). FGF-18 is upregulated in the postnatal rat lung and enhances elastogenesis in myofibroblasts. American Journal of Physiology. Lung Cellular and Molecular Physiology, 288(1), L43–L51. 10.1152/ajplung.00096.2004 [DOI] [PubMed] [Google Scholar]
- Chakkalakal JV, Jones KM, Basson MA, & Brack AS (2012). The aged niche disrupts muscle stem cell quiescence. Nature, 490(7420), 355–360. 10.1038/nature11438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty D, Zhu H, Jungel A, Summa L, Li YN, Matei AE, Zhou X, Huang J, Trinh-Minh T, Chen CW, Lafyatis R, Dees C, Bergmann C, Soare A, Luo H, Ramming A, Schett G, Distler O, & Distler JHW (2020). Fibroblast growth factor receptor 3 activates a network of profibrotic signaling pathways to promote fibrosis in systemic sclerosis. Science Translational Medicine, 12(563), eaaz5506. 10.1126/scitranslmed.aaz5506 [DOI] [PubMed] [Google Scholar]
- Chan WK, Price DJ, & Pratt T (2017). FGF8 morphogen gradients are differentially regulated by heparan sulphotransferases Hs2st and Hs6st1 in the developing brain. Biology Open, 6(12), 1933–1942. 10.1242/bio.028605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda D, Kurundkar A, Rangarajan S, Locy M, Bernard K, Sharma NS, Logsdon NJ, Liu H, Crossman DK, Horowitz JC, De Langhe S, & Thannickal VJ (2016). Developmental reprogramming in mesenchymal stromal cells of human subjects with idiopathic pulmonary fibrosis. Scientific Reports, 6, 37445. 10.1038/srep37445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HH, Chen PS, Cheng YW, Wang TY, Yang YK, & Lu RB (2018). FGF21 is associated with metabolic effects and treatment response in depressed bipolar ii disorder patients treated with valproate. The International Journal of Neuropsychopharmacology, 21(4), 319–324. 10.1093/ijnp/pyx093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Lin H, Fu H, Wang B, Han G, & Fan M (2017). MicroRNA-195–5p regulates osteogenic differentiation of periodontal ligament cells under mechanical loading. Journal of Cellular Physiology, 232(12), 3762–3774. 10.1002/jcp.25856 [DOI] [PubMed] [Google Scholar]
- Chang YT, Kazui H, Ikeda M, Huang CW, Huang SH, Hsu SW, Chang WN, & Chang CC (2019). Genetic interaction of APOE and FGF1 is associated with memory impairment and hippocampal atrophy in Alzheimer’s disease. Aging and Disease, 10(3), 510–519 10.14336/AD.2018.0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YT, Tseng CN, Tannenberg P, Eriksson L, Yuan K, de Jesus Perez VA, Lundberg J, Lengquist M, Botusan IR, Catrina SB, Tran PK, Hedin U, & Tran-Lundmark K (2015). Perlecan heparan sulfate deficiency impairs pulmonary vascular development and attenuates hypoxic pulmonary hypertension. Cardiovascular Research, 107(1), 20–31. 10.1093/cvr/cvv143 [DOI] [PubMed] [Google Scholar]
- Chao CM, El Agha E, Tiozzo C, Minoo P, & Bellusci S (2015). A breath of fresh air on the mesenchyme: Impact of impaired mesenchymal development on the pathogenesis of bronchopulmonary dysplasia. Frontiers in Medicine, 2, 27. 10.3389/fmed.2015.00027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao CM, Moiseenko A, Kosanovic D, Rivetti S, El Agha E, Wilhelm J, Kampschulte M, Yahya F, Ehrhardt H, Zimmer KP, Barreto G, Rizvanov AA, Schermuly RT, Reiss I, Morty RE, Rottier RJ, Bellusci S, & Zhang JS (2019). Impact of Fgf10 deficiency on pulmonary vasculature formation in a mouse model of bronchopulmonary dysplasia. Human Molecular Genetics, 28(9), 1429–1444. 10.1093/hmg/ddy439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao CM, Moiseenko A, Zimmer KP, & Bellusci S (2016). Alveologenesis: Key cellular players and fibroblast growth factor 10 signaling. Molecular and Cellular Pediatrics, 3(1), 17. 10.1186/s40348-016-0045-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Katsara O, Ruoff R, Morgenstern D, Nayak S, Basilico C, Ueberheide B, & Kolupaeva V (2017). Phosphoproteomics of fibroblast growth factor 1 (FGF1) signaling in chondrocytes: Identifying the signature of inhibitory response. Molecular & Cellular Proteomics, 16(6), 1126–1137. 10.1074/mcp.M116.064980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charoenlarp P, Rajendran AK, & Iseki S (2017). Role of fibroblast growth factors in bone regeneration. Inflammation and Regeneration, 37, 10. 10.1186/s41232-017-0043-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhao C, Gu C, Cui X, & Wu J (2019). MiRNA-144–3p inhibits high glucose induced cell proliferation through suppressing FGF16. Bioscience Reports, 39(7), BSR20181788. 10.1042/BSR20181788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CD, Podvin S, Gillespie E, Leeman SE, & Abraham CR (2007). Insulin stimulates the cleavage and release of the extracellular domain of Klotho by ADAM10 and ADAM17. Proceedings of the National Academy of Sciences of the United States of America, 104(50), 19796–19801. 10.1073/pnas.0709805104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Merriman AF, Savage J, Willer J, Wahlig T, Katsanis N, Yin VP, & Poss KD (2015). Transient laminin beta 1a induction defines the wound epidermis during zebrafish fin regeneration. PLoS Genetics, 11(8), e1005437. 10.1371/journal.pgen.1005437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Ng CS, Wu SM, Chen JJ, Cheng PL, Wu P, Lu MY, Chen DR, Chuong CM, Cheng HC, Ting CT, & Li WH (2016). Regulatory differences in natal down development between altricial zebra finch and precocial chicken. Molecular Biology and Evolution, 33(8), 2030–2043. 10.1093/molbev/msw085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Liu Y, Goetz R, Fu L, Jayaraman S, Hu MC, Moe OW, Liang G, Li X, & Mohammadi M (2018). alpha-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling. Nature, 553(7689), 461–466. 10.1038/nature25451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Li J, Zhang D, Zhou X, & Xie J (2021). Role of the fibroblast growth factor 19 in the skeletal system. Life Sciences, 265, 118804. 10.1016/j.lfs.2020.118804 [DOI] [PubMed] [Google Scholar]
- Chen H, & Shan G (2020). The physiological function of long-noncoding RNAs. Noncoding RNA Research, 5(4), 178–184. 10.1016/j.ncrna.2020.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Cui X, Li L, Qu J, Raj JU, & Gou D (2017). MiR-339 inhibits proliferation of pulmonary artery smooth muscle cell by targeting FGF signaling. Physiological Reports, 5(18), e13441 10.14814/phy2.13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Hu J, Liu H, Xiong Y, Zou Y, Huang W, Shao M, Wu J, Yu L, Wang X, Wang X, & Lin L (2018). FGF21 protects the blood-brain barrier by upregulating PPARgamma via FGFR1/beta-klotho after traumatic brain injury. Journal of Neurotrauma, 35(17), 2091–2103. 10.1089/neu.2017.5271 [DOI] [PubMed] [Google Scholar]
- Chen J, Lai F, & Niswander L (2012). The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes & Development, 26(8), 803–815. 10.1101/gad.187641.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Liu J, Zhou Y, Liu S, Liu G, Zuo Y, Wu Z, Wu N, & Qiu G (2017). Molecular therapeutic strategies for FGFR3 gene-related skeletal dysplasia. Journal of Molecular Medicine (Berlin, Germany), 95(12), 1303–1313. 10.1007/s00109-017-1602-9 [DOI] [PubMed] [Google Scholar]
- Chen L, Marsiglia WM, Chen H, Katigbak J, Erdjument-Bromage H, Kemble DJ, Fu L, Ma J, Sun G, Zhang Y, Liang G, Neubert TA, Li X, Traaseth NJ, & Mohammadi M (2020). Molecular basis for receptor tyrosine kinase A-loop tyrosine transphosphorylation. Nature Chemical Biology, 16(3), 267–277. 10.1038/s41589-019-0455-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MZ, Chang JC, Zavala-Solorio J, Kates L, Thai M, Ogasawara A, Bai X, Flanagan S, Nunez V, Phamluong K, Ziai J, Newman R, Warming S, Kolumam G, & Sonoda J (2017). FGF21 mimetic antibody stimulates UCP1-independent brown fat thermogenesis via FGFR1/betaKlotho complex in non-adipocytes. Mol Metab, 6(11), 1454–1467. 10.1016/j.molmet.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Qin L, Baeyens N, Li G, Afolabi T, Budatha M, Tellides G, Schwartz MA, & Simons M (2015). Endothelial-to-mesenchymal transition drives atherosclerosis progression. Journal of Clinical Investigation, 125(12), 4514–4528. 10.1172/JCI82719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Qin L, Barnes C, Charisse K, Yi T, Zhang X, Ali R, Medina PP, Yu J, Slack FJ, Anderson DG, Kotelianski V, Wang F, Tellides G, & Simons M (2012). FGF regulates TGF-beta signaling and endothelial-to-mesenchymal transition via control of let-7 miRNA expression. Cell Reports, 2(6), 1684–1696. 10.1016/j.celrep.2012.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Qin L, Li G, Tellides G, & Simons M (2016a). Fibroblast growth factor (FGF) signaling regulates transforming growth factor beta (TGFbeta)-dependent smooth muscle cell phenotype modulation. Scientific Reports, 6, 33407. 10.1038/srep33407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Qin L, Li G, Tellides G, & Simons M (2016b). Smooth muscle FGF/TGFbeta cross talk regulates atherosclerosis progression. EMBO Molecular Medicine, 8(7), 712–728. 10.15252/emmm.201506181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Qin L, Tellides G, & Simons M (2014). Fibroblast growth factor receptor 1 is a key inhibitor of TGFbeta signaling in the endothelium. Science Signaling, 7(344), ra90. 10.1126/scisignal.2005504 [DOI] [PubMed] [Google Scholar]
- Chen Q, Fang B, Wang Y, Li C, Li X, Wang R, Xiong Q, Zhang L, Jin Y, Zhang M, Liu X, Li L, Mou L, Li R, Yang H, & Dai Y (2018). Overexpressing dominant-negative FGFR2-IIIb impedes lung branching morphogenesis in pigs. Journal of Genetics and Genomics, 45(3), 147–154. 10.1016/j.jgg.2018.02.002 [DOI] [PubMed] [Google Scholar]
- Chen Q, Li J, Ma J, Yang X, Ni M, Zhang Y, Li X, Lin Z, & Gong F (2020). Fibroblast growth factor 21 alleviates acute pancreatitis via activation of the Sirt1-autophagy signalling pathway. Journal of Cellular and Molecular Medicine, 24(9), 5341–5351. 10.1111/jcmm.15190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Li D, Zheng M, Chen B, Wei T, Wang Y, Li M, Huang W, Tong Q, Wang Q, Zhu Y, Fang W, Guo L, & Fang S (2020). FGFRL1 affects chemoresistance of small-cell lung cancer by modulating the PI3K/Akt pathway via ENO1. Journal of Cellular and Molecular Medicine, 24(3), 2123–2134. 10.1111/jcmm.14763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Chen ST, Sun Y, Xu Z, Wang Y, Yao SY, Yao WB, & Gao XD (2019). Fibroblast growth factor 21 ameliorates neurodegeneration in rat and cellular models of Alzheimer’s disease. Redox Biology, 22, 101133. 10.1016/j.redox.2019.101133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, & Li K (2020). HOXD-AS1 facilitates cell migration and invasion as an oncogenic lncRNA by competitively binding to miR-877–3p and upregulating FGF2 in human cervical cancer. BMC Cancer, 20(1), 924. 10.1186/s12885-020-07441-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TM, Chen YH, Sun HS, & Tsai SJ (2019). Fibroblast growth factors: Potential novel targets for regenerative therapy of osteoarthritis. The Chinese Journal of Physiology, 62(1), 2–10. 10.4103/CJP.CJP_11_19 [DOI] [PubMed] [Google Scholar]
- Chen W, Wu L, Hu Y, Jiang L, Liang N, Chen J, Qin H, & Tang N (2020). MicroRNA-107 ameliorates damage in a cell model of Alzheimer’s disease by mediating the FGF7/FGFR2/PI3K/Akt pathway. Journal of Molecular Neuroscience, 70(10), 1589–1597. 10.1007/s12031-020-01600-0 [DOI] [PubMed] [Google Scholar]
- Chen WW, Li L, Yang GY, Li K, Qi XY, Zhu W, Tang Y, Liu H, & Boden G (2008). Circulating FGF-21 levels in normal subjects and in newly diagnose patients with Type 2 diabetes mellitus. Experimental and Clinical Endocrinology & Diabetes, 116(1), 65–68. 10.1055/s-2007-985148 [DOI] [PubMed] [Google Scholar]
- Chen X, Shi C, Meng X, Zhang K, Li X, Wang C, Xiang Z, Hu K, & Han X (2016). Inhibition of Wnt/beta-catenin signaling suppresses bleomycin-induced pulmonary fibrosis by attenuating the expression of TGF-beta1 and FGF-2. Experimental and Molecular Pathology, 101(1), 22–30. 10.1016/j.yexmp.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Shen J, Qi G, Zha Q, Zhang C, Yao W, Gao X, & Chen S (2020). Potential therapeutic role of fibroblast growth factor 21 in neurodegeneration: Evidence for ameliorating parkinsonism via silent information regulator 2 homolog 1 and implication for gene therapy. Neuropharmacology, 181, 108335. 10.1016/j.neuropharm.2020.108335 [DOI] [PubMed] [Google Scholar]
- Chen Y, Xu H, & Lin G (2017). Generation of iPSC-derived limb progenitor-like cells for stimulating phalange regeneration in the adult mouse. Cell Discovery, 3, 17046. 10.1038/celldisc.2017.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Zhang JX, Shen L, Qi Q, Cheng XX, Zhong ZR, Jiang ZQ, Wang R, Lu HZ, & Hu JG (2015). Schwann cells induce proliferation and migration of oligodendrocyte precursor cells through secretion of PDGF-AA and FGF-2. Journal of Molecular Neuroscience, 56(4), 999–1008. 10.1007/s12031-015-0570-1 [DOI] [PubMed] [Google Scholar]
- Chen Z, Jiang L, Liang L, Koral K, Zhang Q, Zhao L, Lu S, & Tao J (2021). The role of fibroblast growth factor 19 in hepatocellular carcinoma. The American Journal of Pathology, 191(7), 1180–1192. 10.1016/j.ajpath.2021.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JK, & Chiou LC (2006). Mechanisms of the antinociceptive action of gabapentin. Journal of Pharmacological Sciences, 100(5), 471–486. 10.1254/jphs.cr0050020 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Lin KH, Young TH, & Cheng NC (2020). The influence of fibroblast growth factor 2 on the senescence of human adipose-derived mesenchymal stem cells during long-term culture. Stem Cells Translational Medicine, 9(4), 518–530. 10.1002/sctm.19-0234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesi M, & Bergsagel PL (2015). Advances in the pathogenesis and diagnosis of multiple myeloma. International Journal of Laboratory Hematology, 37(Suppl 1), 108–114. 10.1111/ijlh.12360 [DOI] [PubMed] [Google Scholar]
- Ching ST, Infante CR, Du W, Sharir A, Park S, Menke DB, & Klein OD (2018). Isl1 mediates mesenchymal expansion in the developing external genitalia via regulation of Bmp4, Fgf10 and Wnt5a. Human Molecular Genetics, 27(1), 107–119. 10.1093/hmg/ddx388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chioni AM, & Grose RP (2021). Biological significance and targeting of the FGFR axis in cancer. Cancers (Basel), 13(22), 1–27. 10.3390/cancers13225681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho GS, Park DS, Choi SC, & Han JK (2017). Tbx2 regulates anterior neural specification by repressing FGF signaling pathway. Developmental Biology, 421(2), 183–193. 10.1016/j.ydbio.2016.11.020 [DOI] [PubMed] [Google Scholar]
- Cho JM, Yang EH, Quan W, Nam EH, & Cheon HG (2020). Discovery of a novel fibroblast activation protein (FAP) inhibitor, BR103354, with anti-diabetic and anti-steatotic effects. Scientific Reports, 10(1), 21280. 10.1038/s41598-020-77978-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Oh A, Lee Y, Kim GH, & Yoo HW (2021). Functional characteristics of novel FGFR1 mutations in patients with isolated gonadotropin-releasing hormone deficiency. Experimental and Clinical Endocrinology & Diabetes, 129(6), 457–463. 10.1055/a-1151-4800 [DOI] [PubMed] [Google Scholar]
- Choi Y, Jang S, Choi MS, Ryoo ZY, & Park T (2016). Increased expression of FGF1-mediated signaling molecules in adipose tissue of obese mice. Journal of Physiology and Biochemistry, 72(2), 157–167. 10.1007/s13105-016-0468-6 [DOI] [PubMed] [Google Scholar]
- Christov M, Neyra JA, Gupta S, & Leaf DE (2019). Fibroblast growth factor 23 and Klotho in AKI. Seminars in Nephrology, 39(1), 57–75. 10.1016/j.semnephrol.2018.10.005 [DOI] [PubMed] [Google Scholar]
- Chung WC, Linscott ML, Rodriguez KM, & Stewart CE (2016). The regulation and function of fibroblast growth factor 8 and its function during gonadotropin-releasing hormone neuron development. Frontiers in Endocrinology, 7, 114. 10.3389/fendo.2016.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilvik SN, Wang JI, Lavine KJ, Uchida K, Castro A, Gierasch CM, Weinheimer CJ, House SL, Kovacs A, Nichols CG, & Ornitz DM (2013). Fibroblast growth factor receptor 1 signaling in adult cardiomyocytes increases contractility and results in a hypertrophic cardiomyopathy. PLoS One, 8(12), e82979. 10.1371/journal.pone.0082979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinque L, Forrester A, Bartolomeo R, Svelto M, Venditti R, Montefusco S, Polishchuk E, Nusco E, Rossi A, Medina DL, Polishchuk R, De Matteis MA, & Settembre C (2015). FGF signalling regulates bone growth through autophagy. Nature, 528(7581), 272–275. 10.1038/nature16063 [DOI] [PubMed] [Google Scholar]
- Clark JC, Tichelaar JW, Wert SE, Itoh N, Perl AK, Stahlman MT, & Whitsett JA (2001). FGF-10 disrupts lung morphogenesis and causes pulmonary adenomas in vivo. American Journal of Physiology. Lung Cellular and Molecular Physiology, 280(4), L705–L715. 10.1152/ajplung.2001.280.4.L705 [DOI] [PubMed] [Google Scholar]
- Coate KC, Hernandez G, Thorne CA, Sun S, Le TDV, Vale K, Kliewer SA, & Mangelsdorf DJ (2017). FGF21 is an exocrine pancreas secretagogue. Cell Metabolism, 25(2), 472–480. 10.1016/j.cmet.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JD, Homer-Bouthiette C, & Hurley MM (2018). Fibroblast growth factor 2 and its receptors in bone biology and disease. Journal of the Endocrine Society, 2(7), 657–671. 10.1210/js.2018-00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, & Spiegelman BM (2015). Brown and beige fat: Molecular parts of a thermogenic machine. Diabetes, 64(7), 2346–2351. 10.2337/db15-0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn MJ (2011). Development of the external genitalia: Conserved and divergent mechanisms of appendage patterning. Developmental Dynamics, 240(5), 1108–1115. 10.1002/dvdy.22631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo G, Margosio B, Ragona L, Neves M, Bonifacio S, Annis DS, Stravalaci M, Tomaselli S, Giavazzi R, Rusnati M, Presta M, Zetta L, Mosher DF, Ribatti D, Gobbi M, & Taraboletti G (2010). Non-peptidic thrombospondin-1 mimics as fibroblast growth factor-2 inhibitors: An integrated strategy for the development of new antiangiogenic compounds. The Journal of Biological Chemistry, 285(12), 8733–8742. 10.1074/jbc.M109.085605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin JS, Bohne BA, Harding GW, McEwen DG, & Ornitz DM (1996). Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nature Genetics, 12(4), 390–397. 10.1038/ng0496-390 [DOI] [PubMed] [Google Scholar]
- Colvin JS, Green RP, Schmahl J, Capel B, & Ornitz DM (2001). Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cells, 104(6), 875–889. 10.1016/s0092-8674(01)00284-7 [DOI] [PubMed] [Google Scholar]
- Conte M, Sabbatinelli J, Chiariello A, Martucci M, Santoro A, Monti D, Arcaro M, Galimberti D, Scarpini E, Bonfigli AR, Giuliani A, Olivieri F, Franceschi C, & Salvioli S (2021). Disease-specific plasma levels of mitokines FGF21, GDF15, and Humanin in type II diabetes and Alzheimer’s disease in comparison with healthy aging. Geroscience, 43(2), 985–1001. 10.1007/s11357-020-00287-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland J, & Simoes-Costa M (2020). Post-transcriptional tuning of FGF signaling mediates neural crest induction. Proceedings of the National Academy of Sciences of the United States of America, 117(52), 33305–33316. 10.1073/pnas.2009997117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa D, Somoza RA, Lin P, Greenberg S, Rom E, Duesler L, Welter JF, Yayon A, & Caplan AI (2015). Sequential exposure to fibroblast growth factors (FGF) 2, 9 and 18 enhances hMSC chondrogenic differentiation. Osteoarthritis and Cartilage, 23(3), 443–453. 10.1016/j.joca.2014.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cota CD, & Davidson B (2015). Mitotic membrane turnover coordinates differential induction of the heart progenitor lineage. Developmental Cell, 34(5), 505–519. 10.1016/j.devcel.2015.07.001 [DOI] [PubMed] [Google Scholar]
- Cottarelli A, Corada M, Beznoussenko GV, Mironov AA, Globisch MA, Biswas S, Huang H, Dimberg A, Magnusson PU, Agalliu D, Lampugnani MG, & Dejana E (2020). Fgfbp1 promotes blood-brain barrier development by regulating collagen IV deposition and maintaining Wnt/beta-catenin signaling. Development, 147(16), 1–16. 10.1242/dev.185140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courage C, Jackson CB, Owczarek-Lipska M, Jamsheer A, Sowinska-Seidler A, Piotrowicz M, Jakubowski L, Dalleves F, Riesch E, Neidhardt J, & Lemke JR (2019). Novel synonymous and missense variants in FGFR1 causing Hartsfield syndrome. American Journal of Medical Genetics. Part A, 179(12), 2447–2453. 10.1002/ajmg.a.61354 [DOI] [PubMed] [Google Scholar]
- Courbon G, Francis C, Gerber C, Neuburg S, Wang X, Lynch E, Isakova T, Babitt JL, Wolf M, Martin A, & David V (2021). Lipocalin 2 stimulates bone fibroblast growth factor 23 production in chronic kidney disease. Bone Research, 9(1), 35. 10.1038/s41413-021-00154-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutu DL, & Galipeau J (2011). Roles of FGF signaling in stem cell self-renewal, senescence and aging. Aging (Albany NY), 3(10), 920–933. 10.18632/aging.100369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csanaky K, Hess MW, & Klimaschewski L (2019). Membrane-associated, not cytoplasmic or nuclear, FGFR1 induces neuronal differentiation. Cells, 8(3), 1–15. 10.3390/cells8030243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Ramos D, Mehta R, & Aguilar-Salinas CA (2019). Fibroblast growth factor 21 and browning of white adipose tissue. Frontiers in Physiology, 10, 37. 10.3389/fphys.2019.00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui A, Li J, Ji S, Ma F, Wang G, Xue Y, Liu Z, Gao J, Han J, Tai P, Wang T, Chen J, Ma X, & Li Y (2020). The effects of B1344, a novel fibroblast growth factor 21 analog, on nonalcoholic steatohepatitis in nonhuman primates. Diabetes, 69(8), 1611–1623. 10.2337/db20-0209 [DOI] [PubMed] [Google Scholar]
- Cunningham TJ, Kumar S, Yamaguchi TP, & Duester G (2015). Wnt8a and Wnt3a cooperate in the axial stem cell niche to promote mammalian body axis extension. Developmental Dynamics, 244(6), 797–807. 10.1002/dvdy.24275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaya B, & Faul C (2019). The role of fibroblast growth factor 23 in inflammation and anemia. International Journal of Molecular Sciences, 20(17), 4195. 10.3390/ijms20174195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyz M (2019). Fibroblast growth factor receptor signaling in skin cancers. Cells, 8(6), 540. 10.3390/cells8060540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrowski A, Terauchi A, Strong C, & Umemori H (2015). Distinct sets of FGF receptors sculpt excitatory and inhibitory synaptogenesis. Development, 142(10), 1818–1830. 10.1242/dev.115568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabrowski A, & Umemori H (2016). Buttressing a balanced brain: Target-derived FGF signaling regulates excitatory/inhibitory tone and adult neurogenesis within the maturating hippocampal network. Neurogenesis (Austin), 3(1), e1168504. 10.1080/23262133.2016.1168504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahir K, Roberts MS, Krolczyk S, & Simmons JH (2020). X-linked hypophosphatemia: A new era in management. Journal of the Endocrine Society, 4(12), bvaa151. 10.1210/jendso/bvaa151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg LE, Aydemir A, Muurahainen N, Guhring H, Fredberg Edebo H, Krarup-Jensen N, Ladel CH, & Jurvelin JS (2016). A first-in-human, double-blind, randomised, placebo-controlled, dose ascending study of intra-articular rhFGF18 (sprifermin) in patients with advanced knee osteoarthritis. Clinical and Experimental Rheumatology, 34(3), 445–450. [PubMed] [Google Scholar]
- D’Angelo A, Bagby S, Galli IC, Bortoletti C, & Roviello G (2020). Overview of the clinical use of erdafitinib as a treatment option for the metastatic urothelial carcinoma: Where do we stand. Expert Review of Clinical Pharmacology, 13(10), 1139–1146. 10.1080/17512433.2020.1823830 [DOI] [PubMed] [Google Scholar]
- Danopoulos S, Schlieve CR, Grikscheit TC, & Al Alam D (2017). Fibroblast growth factors in the gastrointestinal tract: Twists and turns. Developmental Dynamics, 246(4), 344–352. 10.1002/dvdy.24491 [DOI] [PubMed] [Google Scholar]
- Danopoulos S, Shiosaki J, & Al Alam D (2019). FGF signaling in lung development and disease: Human versus mouse. Frontiers in Genetics, 10, 170. 10.3389/fgene.2019.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danopoulos S, Thornton ME, Grubbs BH, Frey MR, Warburton D, Bellusci S, & Al Alam D (2019). Discordant roles for FGF ligands in lung branching morphogenesis between human and mouse. The Journal of Pathology, 247(2), 254–265. 10.1002/path.5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daponte V, Tylzanowski P, & Forlino A (2021). Appendage regeneration in vertebrates: What makes this possible? Cells, 10(2), 242. 10.3390/cells10020242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das JR, Jerebtsova M, Tang P, Li J, Yu J, & Ray PE (2021). Circulating fibroblast growth factor-2 precipitates HIV nephropathy in mice. Disease Models & Mechanisms, 14(7), dmm048980. 10.1242/dmm.048980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey Smith G, & Hemani G (2014). Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Human Molecular Genetics, 23(R1), R89–R98. 10.1093/hmg/ddu328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David V, Martin A, Isakova T, Spaulding C, Qi L, Ramirez V, Zumbrennen-Bullough KB, Sun CC, Lin HY, Babitt JL, & Wolf M (2016). Inflammation and functional iron deficiency regulate fibroblast growth factor 23 production. Kidney International, 89(1), 135–146. 10.1038/ki.2015.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Araujo R, Lobo M, Trindade K, Silva DF, & Pereira N (2019). Fibroblast growth factors: A controlling mechanism of skin aging. Skin Pharmacology and Physiology, 32(5), 275–282. 10.1159/000501145 [DOI] [PubMed] [Google Scholar]
- De Caluwe J, Tosenberger A, Gonze D, & Dupont G (2019). Signalling-modulated gene regulatory networks in early mammalian development. Journal of Theoretical Biology, 463, 56–66. 10.1016/j.jtbi.2018.12.008 [DOI] [PubMed] [Google Scholar]
- de Castro F, Esteban PF, Bribian A, Murcia-Belmonte V, Garcia-Gonzalez D, & Clemente D (2014). The adhesion molecule anosmin-1 in neurology: Kallmann syndrome and beyond. Advances in Neurobiology, 8, 273–292. 10.1007/978-1-4614-8090-7_12 [DOI] [PubMed] [Google Scholar]
- de Castro F, Seal R, Maggi R, & Group of HGNC Consultants for KAL1 Nomenclature. (2017). ANOS1: A unified nomenclature for Kallmann syndrome 1 gene (KAL1) and anosmin-1. Briefings in Functional Genomics, 16(4), 205–210. 10.1093/bfgp/elw037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot SC, Ulrich MMW, Gho CG, & Huisman MA (2021). Back to the future: From appendage development toward future human hair follicle neogenesis. Frontiers in Cell and Development Biology, 9, 661787. 10.3389/fcell.2021.661787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mot L, Gonze D, Bessonnard S, Chazaud C, Goldbeter A, & Dupont G (2016). Cell fate specification based on tristability in the inner cell mass of mouse blastocysts. Biophysical Journal, 110(3), 710–722. 10.1016/j.bpj.2015.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet F, Tembuyser B, Lenard A, Claes F, Zhang J, Michielsen C, Van Schepdael A, Herbert JM, Bono F, Affolter M, Dewerchin M, & Carmeliet P (2014). Fibroblast growth factor signaling affects vascular outgrowth and is required for the maintenance of blood vessel integrity. Chemistry & Biology, 21(10), 1310–1317. 10.1016/j.chembiol.2014.07.018 [DOI] [PubMed] [Google Scholar]
- De Zoysa P, Liu J, Toubat O, Choi J, Moon A, Gill PS, Duarte A, Sucov HM, & Kumar SR (2020). Delta-like ligand 4-mediated Notch signaling controls proliferation of second heart field progenitor cells by regulating Fgf8 expression. Development, 147(17), dev185249. 10.1242/dev.185249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker B, Liput M, Abdellatif H, Yergeau D, Bae Y, Jornet JM, Stachowiak EK, & Stachowiak MK (2020). Global genome conformational programming during neuronal development is associated with CTCF and nuclear FGFR1-the genome archipelago model. International Journal of Molecular Sciences, 22(1), 347. 10.3390/ijms22010347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degirolamo C, Sabba C, & Moschetta A (2016). Therapeutic potential of the endocrine fibroblast growth factors FGF19, FGF21 and FGF23. Nature Reviews. Drug Discovery, 15(1), 51–69. 10.1038/nrd.2015.9 [DOI] [PubMed] [Google Scholar]
- Delaye JB, Lanznaster D, Veyrat-Durebex C, Fontaine A, Bacle G, Lefevre A, Hergesheimer R, Lecron JC, Vourc’h P, Andres CR, Maillot F, Corcia P, Emond P, & Blasco H (2021). Behavioral, hormonal, inflammatory, and metabolic effects associated with FGF21-pathway activation in an ALS mouse model. Neurotherapeutics, 18(1), 297–308. 10.1007/s13311-020-00933-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado I, Lopez-Delgado AC, Rosello-Diez A, Giovinazzo G, Cadenas V, Fernandez-de-Manuel L, Sanchez-Cabo F, Anderson MJ, Lewandoski M, & Torres M (2020). Proximo-distal positional information encoded by an Fgf-regulated gradient of homeodomain transcription factors in the vertebrate limb. Science Advances, 6(23), eaaz0742. 10.1126/sciadv.aaz0742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng CX, Wynshaw-Boris A, Shen MM, Daugherty C, Ornitz DM, & Leder P (1994). Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes & Development, 8(24), 3045–3057. 10.1101/gad.8.24.3045 [DOI] [PubMed] [Google Scholar]
- Deng CX, WynshawBoris A, Zhou F, Kuo A, & Leder P (1996). Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cells, 84(6), 911–921. 10.1016/S0092-8674(00)81069-7 [DOI] [PubMed] [Google Scholar]
- Deng J, Liu Y, Liu Y, Li W, & Nie X (2021). The multiple roles of fibroblast growth factor in diabetic nephropathy. Journal of Inflammation Research, 14, 5273–5290. 10.2147/JIR.S334996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng LC, Alinejad T, Bellusci S, & Zhang JS (2020). Fibroblast growth factors in the management of acute kidney injury following ischemia-reperfusion. Frontiers in Pharmacology, 11, 426. 10.3389/fphar.2020.00426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, & Adjei AA (2016). FGFR signaling as a target for lung cancer therapy. Journal of Thoracic Oncology, 11(1), 9–20. 10.1016/j.jtho.2015.08.003 [DOI] [PubMed] [Google Scholar]
- DeSalvo J, Ban Y, Li L, Sun X, Jiang Z, Kerr DA, Khanlari M, Boulina M, Capecchi MR, Partanen JM, Chen L, Kondo T, Ornitz DM, Trent JC, & Eid JE (2021). ETV4 and ETV5 drive synovial sarcoma through cell cycle and DUX4 embryonic pathway control. Journal of Clinical Investigation, 131(13), e141908. 10.1172/JCI141908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers LR, Pai R, Ferrando RE, Hotzel K, Le T, Ross J, Carano R, D’Souza A, Qing J, Mohtashemi I, Ashkenazi A, & French DM (2008). Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene, 27(1), 85–97. 10.1038/sj.onc.1210623 [DOI] [PubMed] [Google Scholar]
- Destalminil-Letourneau M, Morin-Poulard I, Tian Y, Vanzo N, & Crozatier M (2021). The vascular niche controls Drosophila hematopoiesis via fibroblast growth factor signaling. eLife, 10, e64672. 10.7554/eLife.64672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer R, Cirksena K, Munchhoff J, Kresse J, Diekmann U, Niwolik I, Buettner FFR, & Naujok O (2020). FGF2 inhibits early pancreatic lineage specification during differentiation of human embryonic stem cells. Cells, 9(9), 1927. 10.3390/cells9091927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanushkodi NR, Srivastava R, Coulon PA, Prakash S, Roy S, Bagnol D, David ED, & BenMohamed L (2021). Healing of ocular herpetic disease following treatment with an engineered FGF-1 is associated with increased corneal anti-inflammatory M2 macrophages. Frontiers in Immunology, 12, 673763. 10.3389/fimmu.2021.673763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Liberto V, Borroto-Escuela DO, Frinchi M, Verdi V, Fuxe K, Belluardo N, & Mudo G (2017). Existence of muscarinic acetylcholine receptor (mAChR) and fibroblast growth factor receptor (FGFR) heteroreceptor complexes and their enhancement of neurite outgrowth in neural hippocampal cultures. Biochimica et Biophysica Acta - General Subjects, 1861(2), 235–245. 10.1016/j.bbagen.2016.10.026 [DOI] [PubMed] [Google Scholar]
- Di Liberto V, Mudo G, & Belluardo N (2019). Crosstalk between receptor tyrosine kinases (RTKs) and G protein-coupled receptors (GPCR) in the brain: Focus on heteroreceptor complexes and related functional neurotrophic effects. Neuropharmacology, 152, 67–77. 10.1016/j.neuropharm.2018.11.018 [DOI] [PubMed] [Google Scholar]
- di Martino E, Alder O, Hurst CD, & Knowles MA (2019). ETV5 links the FGFR3 and Hippo signalling pathways in bladder cancer. Scientific Reports, 9(1), 5740. 10.1038/s41598-018-36456-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Balzac CA, Lazaro-Pena MI, Ramos-Ortiz GA, & Bulow HE (2015). The adhesion molecule KAL-1/anosmin-1 regulates neurite branching through a SAX-7/L1CAM-EGL-15/FGFR receptor complex. Cell Reports, 11(9), 1377–1384. 10.1016/j.celrep.2015.04.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib-Hajj SD (2016). iFGF14-Navs: A monogamous partnership? Channels (Austin, TX), 10(6), 435–436. 10.1080/19336950.2016.1204829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener JL, Mowbray S, Huang WJ, Yowe D, Xu J, Caplan S, Misra A, Kapur A, Shapiro J, Ke X, Wu X, Bose A, Panza D, Chen M, Beaulieu V, & Gao J (2021). FGF21 normalizes plasma glucose in mouse models of type 1 diabetes and insulin receptor dysfunction. Endocrinology, 162(9), bqab092. 10.1210/endocr/bqab092 [DOI] [PubMed] [Google Scholar]
- Ding J, Lei L, Liu S, Zhang Y, Yu Z, Su Y, & Ma X (2019). Macrophages are necessary for skin regeneration during tissue expansion. Journal of Translational Medicine, 17(1), 36. 10.1186/s12967-019-1780-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh Duong TA, Hoshiba Y, Saito K, Kawasaki K, Ichikawa Y, Matsumoto N, Shinmyo Y, & Kawasaki H (2019). FGF signaling directs the cell fate switch from neurons to astrocytes in the developing mouse cerebral cortex. The Journal of Neuroscience, 39(31), 6081–6094. 10.1523/JNEUROSCI.2195-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit G, Schanz W, Pappas BA, & Maretzky T (2021). Members of the fibroblast growth factor receptor superfamily are proteolytically cleaved by two differently activated metalloproteases. International Journal of Molecular Sciences, 22(6), 3165. 10.3390/ijms22063165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrzycki T, Lalwani M, Telfer C, Monteiro R, & Patient R (2020). The roles and controls of GATA factors in blood and cardiac development. IUBMB Life, 72(1), 39–44. 10.1002/iub.2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty P, & Walsh FS (1996). CAM-FGF receptor interactions: A model for axonal growth. Molecular and Cellular Neuroscience, 8(2–3), 99–111. 10.1006/mcne.1996.0049 [DOI] [PubMed] [Google Scholar]
- Dolivo DM, Larson SA, & Dominko T (2017). Fibroblast growth factor 2 as an antifibrotic: Antagonism of myofibroblast differentiation and suppression of pro-fibrotic gene expression. Cytokine & Growth Factor Reviews, 38, 49–58. 10.1016/j.cytogfr.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domouzoglou EM, Naka KK, Vlahos AP, Papafaklis MI, Michalis LK, Tsatsoulis A, & Maratos-Flier E (2015). Fibroblast growth factors in cardiovascular disease: The emerging role of FGF21. American Journal of Physiology. Heart and Circulatory Physiology, 309(6), H1029–H1038. 10.1152/ajpheart.00527.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donate-Correa J, Martin-Nunez E, Gonzalez-Luis A, Ferri CM, Luis-Rodriguez D, Tagua VG, Mora-Fernandez C, & Navarro-Gonzalez JF (2021). Pathophysiological implications of imbalances in fibroblast growth factor 23 in the development of diabetes. Journal of Clinical Medicine, 10(12), 2583. 10.3390/jcm10122583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncheck EM, Hafenbreidel M, Ruder SA, Fitzgerald MK, Torres L, & Mueller D (2018). bFGF expression is differentially regulated by cocaine seeking versus extinction in learning-related brain regions. Learning & Memory, 25(8), 361–368. 10.1101/lm.047530.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Fischer LA, & Theunissen TW (2019). Recent insights into the naive state of human pluripotency and its applications. Experimental Cell Research, 385(1), 111645. 10.1016/j.yexcr.2019.111645 [DOI] [PubMed] [Google Scholar]
- Dong Y, Qian L, & Liu J (2021). Molecular and cellular basis of embryonic cardiac chamber maturation. Seminars in Cell & Developmental Biology, 118, 144–149. 10.1016/j.semcdb.2021.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Santeford A, Ban N, Lee TJ, Smith C, Ornitz DM, & Apte RS (2019). FGF2-induced STAT3 activation regulates pathologic neovascularization. Experimental Eye Research, 187, 107775. 10.1016/j.exer.2019.107775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorry SJ, Ansbro BO, Ornitz DM, Mutlu GM, & Guzy RD (2020). FGFR2 is required for AEC2 homeostasis and survival after bleomycin-induced lung injury. American Journal of Respiratory Cell and Molecular Biology, 62(5), 608–621. 10.1165/rcmb.2019-0079OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos JV, Yu RY, Terceros A, & Chen BE (2019). FGF receptors are required for proper axonal branch targeting in Drosophila. Molecular Brain, 12(1), 84. 10.1186/s13041-019-0503-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douris N, Stevanovic DM, Fisher FM, Cisu TI, Chee MJ, Nguyen NL, Zarebidaki E, Adams AC, Kharitonenkov A, Flier JS, Bartness TJ, & Maratos-Flier E (2015). Central fibroblast growth factor 21 browns white fat via sympathetic action in male mice. Endocrinology, 156(7), 2470–2481. 10.1210/en.2014-2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Stock DW, & Kimmel CB (2003). Zebrafish fgf24 functions with fgf8 to promote posterior mesodermal development. Development, 130(19), 4639–4654. [DOI] [PubMed] [Google Scholar]
- Dremencov E, Jezova D, Barak S, Gaburjakova J, Gaburjakova M, Kutna V, & Ovsepian SV (2021). Trophic factors as potential therapies for treatment of major mental disorders. Neuroscience Letters, 764, 136194. 10.1016/j.neulet.2021.136194 [DOI] [PubMed] [Google Scholar]
- Du Y, Guo M, Whitsett JA, & Xu Y (2015). ‘LungGENS’: A web-based tool for mapping single-cell gene expression in the developing lung. Thorax, 70(11), 1092–1094. 10.1136/thoraxjnl-2015-207035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Li X, Chen Y, Wang Y, & Li Z (2019). LncRNA RHPN1-AS1 promoted cell proliferation, invasion and migration in cervical cancer via the modulation of miR-299–3p/FGF2 axis. Life Sciences, 239, 116856. 10.1016/j.lfs.2019.116856 [DOI] [PubMed] [Google Scholar]
- Dubrulle J, McGrew MJ, & Pourquie O (2001). FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal Hox gene activation. Cells, 106(2), 219–232. 10.1016/s0092-8674(01)00437-8 [DOI] [PubMed] [Google Scholar]
- Dubrulle J, & Pourquie O (2004). fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature, 427(6973), 419–422. 10.1038/nature02216 [DOI] [PubMed] [Google Scholar]
- Duggan S (2021). Vosoritide: First approval. Drugs, 81 (17), 2057–2062. 10.1007/s40265-021-01623-w [DOI] [PubMed] [Google Scholar]
- Dunkel H, Chaverra M, Bradley R, & Lefcort F (2020). FGF signaling is required for chemokinesis and ventral migration of trunk neural crest cells. Developmental Dynamics, 249(9), 1077–1097. 10.1002/dvdy.190 [DOI] [PubMed] [Google Scholar]
- Dunshee DR, Bainbridge TW, Kljavin NM, Zavala-Solorio J, Schroeder AC, Chan R, Corpuz R, Wong M, Zhou W, Deshmukh G, Ly J, Sutherlin DP, Ernst JA, & Sonoda J (2016). Fibroblast activation protein cleaves and inactivates fibroblast growth factor 21. The Journal of Biological Chemistry, 291(11), 5986–5996. 10.1074/jbc.M115.710582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duperret EK, Oh SJ, McNeal A, Prouty SM, & Ridky TW (2014). Activating FGFR3 mutations cause mild hyperplasia in human skin, but are insufficient to drive benign or malignant skin tumors. Cell Cycle, 13(10), 1551–1559. 10.4161/cc.28492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easter M, Garth J, Harris ES, Shei RJ, Helton ES, Wei Y, Denson R, Zaharias R, Rowe SM, Geraghty P, Faul C, Barnes JW, & Krick S (2020). Fibroblast growth factor receptor 4 deficiency mediates airway inflammation in the adult healthy lung? Frontiers in Medicine, 7, 317. 10.3389/fmed.2020.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebeid M, & Huh SH (2017). FGF signaling: Diverse roles during cochlear development. BMB Reports, 50(10), 487–495. 10.5483/bmbrep.2017.50.10.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebeid M, & Huh SH (2020). Mesenchymal ETV transcription factors regulate cochlear length. Hearing Research, 396, 108039. 10.1016/j.heares.2020.108039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckenstein FP, McGovern T, Kern D, & Deignan J (2006). Neuronal vulnerability in transgenic mice expressing an inducible dominant-negative FGF receptor. Experimental Neurology, 198(2), 338–349. 10.1016/j.expneurol.2005.12.020 [DOI] [PubMed] [Google Scholar]
- Eckstein F, Kraines JL, Aydemir A, Wirth W, Maschek S, & Hochberg MC (2020). Intra-articular sprifermin reduces cartilage loss in addition to increasing cartilage gain independent of location in the femorotibial joint: Post-hoc analysis of a randomised, placebo-controlled phase II clinical trial. Annals of the Rheumatic Diseases, 79(4), 525–528. 10.1136/annrheumdis-2019-216453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonston D, & Wolf M (2020). FGF23 at the crossroads of phosphate, iron economy and erythropoiesis. Nature Reviews. Nephrology, 16(1), 7–19. 10.1038/s41581-019-0189-5 [DOI] [PubMed] [Google Scholar]
- Egli-Spichtig D, Zhang MYH, & Perwad F (2018). Fibroblast growth factor 23 expression is increased in multiple organs in mice with folic acid-induced acute kidney injury. Frontiers in Physiology, 9, 1494. 10.3389/fphys.2018.01494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einhorn TA, & Gerstenfeld LC (2015). Fracture healing: Mechanisms and interventions. Nature Reviews Rheumatology, 11(1), 45–54. 10.1038/nrrheum.2014.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Agha E, Kheirollahi V, Moiseenko A, Seeger W, & Bellusci S (2017). Ex vivo analysis of the contribution of FGF10(+) cells to airway smooth muscle cell formation during early lung development. Developmental Dynamics, 246(7), 531–538. 10.1002/dvdy.24504 [DOI] [PubMed] [Google Scholar]
- El Agha E, Kosanovic D, Schermuly RT, & Bellusci S (2016). Role of fibroblast growth factors in organ regeneration and repair. Seminars in Cell & Developmental Biology, 53, 76–84. 10.1016/j.semcdb.2015.10.009 [DOI] [PubMed] [Google Scholar]
- El Agha E, Seeger W, & Bellusci S (2017). Therapeutic and pathological roles of fibroblast growth factors in pulmonary diseases [Review]. Developmental Dynamics, 246(4), 235–244. 10.1002/dvdy.24468 [DOI] [PubMed] [Google Scholar]
- Elkasrawy MN, & Hamrick MW (2010). Myostatin (GDF-8) as a key factor linking muscle mass and bone structure. Journal of Musculoskeletal & Neuronal Interactions, 10(1), 56–63. [PMC free article] [PubMed] [Google Scholar]
- Ellman MB, Yan D, Ahmadinia K, Chen D, An HS, & Im HJ (2013). Fibroblast growth factor control of cartilage homeostasis. Journal of Cellular Biochemistry, 114(4), 735–742. 10.1002/jcb.24418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Seoudi A, Abd El Kader T, Nishida T, Eguchi T, Aoyama E, Takigawa M, & Kubota S (2017). Catabolic effects of FGF-1 on chondrocytes and its possible role in osteoarthritis. Journal of Cell Communication and Signaling, 11(3), 255–263. 10.1007/s12079-017-0384-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enevoldsen MN, Kochoyan A, Jurgenson M, Jaako K, Dmytriyeva O, Walmod PS, Nielsen JD, Nielsen J, Li S, Korshunova I, Klementiev B, Novikova T, Zharkovsky A, Berezin V, & Bock E (2012). Neuroprotective and memory enhancing properties of a dual agonist of the FGF receptor and NCAM. Neurobiology of Disease, 48(3), 533–545. 10.1016/j.nbd.2012.07.016 [DOI] [PubMed] [Google Scholar]
- Entesarian M, Matsson H, Klar J, Bergendal B, Olson L, Arakaki R, Hayashi Y, Ohuchi H, Falahat B, Bolstad AI, Jonsson R, Wahren-Herlenius M, & Dahl N (2005). Mutations in the gene encoding fibroblast growth factor 10 are associated with aplasia of lacrimal and salivary glands. Nature Genetics, 37(2), 125–127. 10.1038/ng1507 [DOI] [PubMed] [Google Scholar]
- Erben RG (2018). Physiological actions of fibroblast growth factor-23. Frontiers in Endocrinology, 9, 267. 10.3389/fendo.2018.00267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erben RG, & Andrukhova O (2017). FGF23-Klotho signaling axis in the kidney. Bone, 100, 62–68. 10.1016/j.bone.2016.09.010 [DOI] [PubMed] [Google Scholar]
- Ertl IE, Shariat SF, Mostafaei H, Ilijazi D, & Loriot Y (2020). Fibroblast growth factor receptors across urothelial carcinoma landscape. Current Opinion in Urology, 30(4), 557–565. 10.1097/MOU.0000000000000782 [DOI] [PubMed] [Google Scholar]
- Escobar LF, Tucker M, & Bamshad M (2016). A second family with CATSHL syndrome: Confirmatory report of another unique FGFR3 syndrome. American Journal of Medical Genetics. Part A, 170(7), 1908–1911. 10.1002/ajmg.a.37676 [DOI] [PubMed] [Google Scholar]
- Esnafoglu E, & Ayyildiz SN (2017). Decreased levels of serum fibroblast growth factor-2 in children with autism spectrum disorder. Psychiatry Research, 257, 79–83. 10.1016/j.psychres.2017.07.028 [DOI] [PubMed] [Google Scholar]
- Esser JS, Rahner S, Deckler M, Bode C, Patterson C, & Moser M (2015). Fibroblast growth factor signaling pathway in endothelial cells is activated by BMPER to promote angiogenesis. Arteriosclerosis, Thrombosis, and Vascular Biology, 35(2), 358–367. 10.1161/ATVBAHA.114.304345 [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, & Schlessinger J (2007). Skeletal overgrowth is mediated by deficiency in a specific isoform of fibroblast growth factor receptor 3. Proceedings of the National Academy of Sciences of the United States of America, 104(10), 3937–3942. 10.1073/pnas.0700012104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveleth D, Pizzuto S, Weant J, Jenkins-Eveleth J, & Bradshaw RA (2020). Proliferation of human corneal endothelia in organ culture stimulated by wounding and the engineered human fibroblast growth factor 1 derivative TTHX1114. Journal of Ocular Pharmacology and Therapeutics, 36(9), 686–696. 10.1089/jop.2019.0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eveleth DD, Eveleth JJ, Subramaniam A, Hahn R, Zhou P, Gordon MK, & Bradshaw RA (2018). An engineered human fibroblast growth factor-1 derivative, TTHX1114, ameliorates short-term corneal nitrogen mustard injury in rabbit organ cultures. Investigative Ophthalmology & Visual Science, 59(11), 4720–4730. 10.1167/iovs.18-24568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Chen O, & Barak S (2019a). Inhibition of FGF receptor-1 suppresses alcohol consumption: Role of PI3 kinase signaling in dorsomedial striatum. The Journal of Neuroscience, 39(40), 7947–7957. 10.1523/JNEUROSCI.0805-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Chen O, & Barak S (2019b). The role of fibroblast growth factor 2 in drug addiction. The European Journal of Neuroscience, 50(3), 2552–2561. 10.1111/ejn.14133 [DOI] [PubMed] [Google Scholar]
- Even-Chen O, Sadot-Sogrin Y, Shaham O, & Barak S (2017). Fibroblast growth factor 2 in the dorsomedial striatum is a novel positive regulator of alcohol consumption. The Journal of Neuroscience, 37(36), 8742–8754. 10.1523/JNEUROSCI.0890-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everetts NJ, Worley MI, Yasutomi R, Yosef N, & Hariharan IK (2021). Single-cell transcriptomics of the Drosophila wing disc reveals instructive epithelium-to-myoblast interactions. eLife, 10, e61276. 10.7554/eLife.61276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhurst RA, Knoepfel T, Buschmann N, Leblanc C, Mah R, Todorov M, Nimsgern P, Ripoche S, Niklaus M, Warin N, Luu VH, Madoerin M, Wirth J, Graus-Porta D, Weiss A, Kiffe M, Wartmann M, Kinyamu-Akunda J, Sterker D, … Furet P (2020). Discovery of roblitinib (FGF401) as a reversible-covalent inhibitor of the kinase activity of fibroblast growth factor receptor 4. Journal of Medicinal Chemistry, 63(21), 12542–12573. 10.1021/acs.jmedchem.0c01019 [DOI] [PubMed] [Google Scholar]
- Falardeau J, Chung WC, Beenken A, Raivio T, Plummer L, Sidis Y, Jacobson-Dickman EE, Eliseenkova AV, Ma J, Dwyer A, Quinton R, Na S, Hall JE, Huot C, Alois N, Pearce SH, Cole LW, Hughes V, Mohammadi M, … Pitteloud N (2008). Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. Journal of Clinical Investigation, 118(8), 2822–2831. 10.1172/JCI34538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Ding L, Lan J, Niu J, He Y, & Song L (2019). Fibroblast growth factor-1 improves insulin resistance via repression of JNK-mediated inflammation. Frontiers in Pharmacology, 10, 1478. 10.3389/fphar.2019.01478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Bi R, Densmore MJ, Sato T, Kobayashi T, Yuan Q, Zhou X, Erben RG, & Lanske B (2016). Parathyroid hormone 1 receptor is essential to induce FGF23 production and maintain systemic mineral ion homeostasis. The FASEB Journal, 30(1), 428–440. 10.1096/fj.15-278184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Ma J, Mu D, Li B, Lian B, & Sun C (2020). FGF21 protects dopaminergic neurons in Parkinson’s disease models via repression of neuroinflammation. Neurotoxicity Research, 37(3), 616–627. 10.1007/s12640-019-00151-6 [DOI] [PubMed] [Google Scholar]
- Fang X, Wang L, Shi L, Chen C, Wang Q, Bai C, & Wang X (2014). Protective effects of keratinocyte growth factor-2 on ischemia-reperfusion-induced lung injury in rats. American Journal of Respiratory Cell and Molecular Biology, 50(6), 1156–1165. 10.1165/rcmb.2013-0268OC [DOI] [PubMed] [Google Scholar]
- Farooq M, Khan AW, Kim MS, & Choi S (2021). The role of fibroblast growth factor (FGF) signaling in tissue repair and regeneration. Cells, 10(11), 3242. 10.3390/cells10113242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faul C (2018). FGF23 effects on the heart-levels, time, source, and context matter. Kidney International, 94(1), 7–11. 10.1016/j.kint.2018.03.024 [DOI] [PubMed] [Google Scholar]
- Faul C, Amaral AP, Oskouei B, Hu MC, Sloan A, Isakova T, Gutierrez OM, Aguillon-Prada R, Lincoln J, Hare JM, Mundel P, Morales A, Scialla J, Fischer M, Soliman EZ, Chen J, Go AS, Rosas SE, Nessel L, … Wolf M (2011). FGF23 induces left ventricular hypertrophy. Journal of Clinical Investigation, 121(11), 4393–4408. 10.1172/JCI46122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng N, Wang Q, Zhou J, Li J, Wen X, Chen S, Zhu Z, Bai C, Song Y, & Li H (2016). Keratinocyte growth factor-2 inhibits bacterial infection with Pseudomonas aeruginosa pneumonia in a mouse model. Journal of Infection and Chemotherapy, 22(1), 44–52. 10.1016/j.jiac.2015.10.005 [DOI] [PubMed] [Google Scholar]
- Ferrer-Curriu G, Guitart-Mampel M, Ruperez C, Zamora M, Crispi F, Villarroya F, Fernandez-Sola J, Garrabou G, & Planavila A (2021). The protective effect of fibroblast growth factor-21 in alcoholic cardiomyopathy: A role in protecting cardiac mitochondrial function. The Journal of Pathology, 253(2), 198–208. 10.1002/path.5573 [DOI] [PubMed] [Google Scholar]
- Ferrer-Curriu G, Redondo-Angulo I, Guitart-Mampel M, Ruperez C, Mas-Stachurska A, Sitges M, Garrabou G, Villarroya F, Fernandez-Sola J, & Planavila A (2019). Fibroblast growth factor-21 protects against fibrosis in hypertensive heart disease. The Journal of Pathology, 248(1), 30–40. 10.1002/path.5226 [DOI] [PubMed] [Google Scholar]
- Fischer C, Seki T, Lim S, Nakamura M, Andersson P, Yang Y, Honek J, Wang Y, Gao Y, Chen F, Samani NJ, Zhang J, Miyake M, Oyadomari S, Yasue A, Li X, Zhang Y, Liu Y, & Cao Y (2017). A miR-327-FGF10-FGFR2-mediated autocrine signaling mechanism controls white fat browning. Nature Communications, 8(1), 2079. 10.1038/s41467-017-02158-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher FM, Chui PC, Antonellis PJ, Bina HA, Kharitonenkov A, Flier JS, & Maratos-Flier E (2010). Obesity is a fibroblast growth factor 21 (FGF21)-resistant state. Diabetes, 59(11), 2781–2789. 10.2337/db10-0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher FM, & Maratos-Flier E (2016). Understanding the physiology of FGF21. Annual Review of Physiology, 78, 223–241. 10.1146/annurev-physiol-021115-105339 [DOI] [PubMed] [Google Scholar]
- Flego M, Colotti G, Ascione A, Dupuis ML, Petrucci E, Riccioni R, Andreotti M, Raggi C, Boe A, Barca S, Gellini M, Vella S, & Mallano A (2021). Isolation and preliminary characterization of a human ‘phage display’-derived antibody against neural adhesion molecule-1 antigen interfering with fibroblast growth factor receptor-1 binding. Human Antibodies, 29(1), 63–84. 10.3233/HAB-200431 [DOI] [PubMed] [Google Scholar]
- Flippo KH, Jensen-Cody SO, Claflin KE, & Potthoff MJ (2020). FGF21 signaling in glutamatergic neurons is required for weight loss associated with dietary protein dilution. Scientific Reports, 10(1), 19521. 10.1038/s41598-020-76593-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora P, & Ezhkova E (2020). Regulatory mechanisms governing epidermal stem cell function during development and homeostasis. Development, 147(22), dev194100. 10.1242/dev.194100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florenzano P, Hartley IR, Jimenez M, Roszko K, Gafni RI, & Collins MT (2021). Tumor-induced osteomalacia. Calcified Tissue International, 108(1), 128–142. 10.1007/s00223-020-00691-6 [DOI] [PubMed] [Google Scholar]
- Foglieni C, Pagano K, Lessi M, Bugatti A, Moroni E, Pinessi D, Resovi A, Ribatti D, Bertini S, Ragona L, Bellina F, Rusnati M, Colombo G, & Taraboletti G (2016). Integrating computational and chemical biology tools in the discovery of antiangiogenic small molecule ligands of FGF2 derived from endogenous inhibitors. Scientific Reports, 6, 23432. 10.1038/srep23432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkman J (1971). Tumor angiogenesis: Therapeutic implications. The New England Journal of Medicine, 285(21), 1182–1186. 10.1056/NEJM197111182852108 [DOI] [PubMed] [Google Scholar]
- Fon Tacer K, Bookout AL, Ding X, Kurosu H, John GB, Wang L, Goetz R, Mohammadi M, Kuro-o M, Mangelsdorf DJ, & Kliewer SA (2010). Research resource: Comprehensive expression atlas of the fibroblast growth factor system in adult mouse. Molecular Endocrinology, 24(10), 2050–2064. 10.1210/me.2010-0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forough R, Xi Z, MacPhee M, Friedman S, Engleka KA, Sayers T, Wiltrout RH, & Maciag T (1993). Differential transforming abilities of non-secreted and secreted forms of human fibroblast growth factor-1. The Journal of Biological Chemistry, 268(4), 2960–2968. [PubMed] [Google Scholar]
- Forster S, Koziol U, Schafer T, Duvoisin R, Cailliau K, Vanderstraete M, Dissous C, & Brehm K (2019). The role of fibroblast growth factor signalling in Echinococcus multilocularis development and host-parasite interaction. PLoS Neglected Tropical Diseases, 13(3), e0006959. 10.1371/journal.pntd.0006959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forthmann B, Grothe C, & Claus P (2015). A nuclear odyssey: Fibroblast growth factor-2 (FGF-2) as a regulator of nuclear homeostasis in the nervous system. Cellular and Molecular Life Sciences, 72(9), 1651–1662. 10.1007/s00018-014-1818-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francavilla C, Cattaneo P, Berezin V, Bock E, Ami D, de Marco A, Christofori G, & Cavallaro U (2009). The binding of NCAM to FGFR1 induces a specific cellular response mediated by receptor trafficking. The Journal of Cell Biology, 187(7), 1101–1116. 10.1083/jcb.200903030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis C, Courbon G, Gerber C, Neuburg S, Wang X, Dussold C, Capella M, Qi L, Isakova T, Mehta R, Martin A, Wolf M, & David V (2019). Ferric citrate reduces fibroblast growth factor 23 levels and improves renal and cardiac function in a mouse model of chronic kidney disease. Kidney International, 96(6), 1346–1358. 10.1016/j.kint.2019.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis C, & David V (2016). Inflammation regulates fibroblast growth factor 23 production. Current Opinion in Nephrology and Hypertension, 25(4), 325–332. 10.1097/MNH.0000000000000232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Montoya ML, Boucherat O, Thibault C, Chailley-Heu B, Incitti R, Delacourt C, & Bourbon JR (2011). Profiling target genes of FGF18 in the postnatal mouse lung: Possible relevance for alveolar development. Physiological Genomics, 43(21), 1226–1240. 10.1152/physiolgenomics.00034.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D, & Sela-Donenfeld D (2019). Hindbrain induction and patterning during early vertebrate development. Cellular and Molecular Life Sciences, 76(5), 941–960. 10.1007/s00018-018-2974-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DB, Penkala IJ, Zepp JA, Sivakumar A, Linares-Saldana R, Zacharias WJ, Stolz KG, Pankin J, Lu M, Wang Q, Babu A, Li L, Zhou S, Morley MP, Jain R, Morrisey EE (2019). Early lineage specification defines alveolar epithelial ontogeny in the murine lung. Proceedings of the National Academy of Sciences, 116(10), 4362–4371. 10.1073/pnas.1813952116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DU, Fotheringham LK, Brewer JA, Muglia LJ, Tristani-Firouzi M, Capecchi MR, & Moon AM (2002). An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development, 129(19), 4591–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling TM, Beaumont RN, Jones SE, Yaghootkar H, Tuke MA, Ruth KS, Casanova F, West B, Locke J, Sharp S, Ji Y, Thompson W, Harrison J, Etheridge AS, Gallins PJ, Jima D, Wright F, Zhou Y, Innocenti F, … Wood AR (2018). A common allele in FGF21 associated with sugar intake is associated with body shape, lower total body-fat percentage, and higher blood pressure. Cell Reports, 23(2), 327–336. 10.1016/j.celrep.2018.03.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch J, Venkatesan JK, Rey-Rico A, Zawada AM, Schmitt G, Madry H, & Cucchiarini M (2016). Effects of rAAV-mediated FGF-2 gene transfer and overexpression upon the chondrogenic differentiation processes in human bone marrow aspirates. Journal of Experimental Orthopaedics, 3(1), 16. 10.1186/s40634-016-0052-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Gong Y, Liegl R, Wang Z, Liu CH, Meng SS, Burnim SB, Saba NJ, Fredrick TW, Morss PC, Hellstrom A, Talukdar S, & Smith LE (2017). FGF21 administration suppresses retinal and choroidal neovascularization in mice. Cell Reports, 18(7), 1606–1613. 10.1016/j.celrep.2017.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Qiu C, Cagnone G, Tomita Y, Huang S, Cakir B, Kotoda Y, Allen W, Bull E, Akula JD, Joyal JS, Hellstrom A, Talukdar S, & Smith LEH (2021). Retinal glial remodeling by FGF21 preserves retinal function during photoreceptor degeneration. iScience, 24(4), 102376. 10.1016/j.isci.2021.102376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Wang Z, Liu CH, Gong Y, Cakir B, Liegl R, Sun Y, Meng SS, Burnim SB, Arellano I, Moran E, Duran R, Poblete A, Cho SS, Talukdar S, Akula JD, Hellstrom A, & Smith LEH (2018). Fibroblast growth factor 21 protects photoreceptor function in type 1 diabetic mice. Diabetes, 67(5), 974–985. 10.2337/db17-0830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto S (2014). Anti-fibroblast growth factor 23 antibody therapy. Current Opinion in Nephrology and Hypertension, 23(4), 346–351. 10.1097/01.mnh.0000447012.98357.da [DOI] [PubMed] [Google Scholar]
- Fukumoto S (2021). FGF23-related hypophosphatemic rickets/osteomalacia: Diagnosis and new treatment. Journal of Molecular Endocrinology, 66(2), R57–R65. 10.1530/JME-20-0089 [DOI] [PubMed] [Google Scholar]
- Furdui CM, Lew ED, Schlessinger J, & Anderson KS (2006). Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Molecular Cell, 21(5), 711–717. 10.1016/j.molcel.2006.01.022 [DOI] [PubMed] [Google Scholar]
- Furusho M, Dupree JL, Nave KA, & Bansal R (2012). Fibroblast growth factor receptor signaling in oligodendrocytes regulates myelin sheath thickness. The Journal of Neuroscience, 32(19), 6631–6641. 10.1523/JNEUROSCI.6005-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusho M, Ishii A, & Bansal R (2017). Signaling by FGF receptor 2, not FGF receptor 1, regulates myelin thickness through activation of ERK1/2-MAPK, which promotes mTORC1 activity in an Akt-independent manner. The Journal of Neuroscience, 37(11), 2931–2946. 10.1523/JNEUROSCI.3316-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusho M, Ishii A, Hebert JM, & Bansal R (2020). Developmental stage-specific role of Frs adapters as mediators of FGF receptor signaling in the oligodendrocyte lineage cells. Glia, 68(3), 617–630. 10.1002/glia.23743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusho M, Kaga Y, Ishii A, Hebert JM, & Bansal R (2011). Fibroblast growth factor signaling is required for the generation of oligodendrocyte progenitors from the embryonic forebrain. The Journal of Neuroscience, 31(13), 5055–5066. 10.1523/JNEUROSCI.4800-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusho M, Roulois AJ, Franklin RJ, & Bansal R (2015). Fibroblast growth factor signaling in oligodendrocyte-lineage cells facilitates recovery of chronically demyelinated lesions but is redundant in acute lesions. Glia, 63(10), 1714–1728. 10.1002/glia.22838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futami T, Okada H, Kihara R, Kawase T, Nakayama A, Suzuki T, Kameda M, Shindoh N, Terasaka T, Hirano M, & Kuromitsu S (2017). ASP5878, a novel inhibitor of FGFR1, 2, 3, and 4, inhibits the growth of FGF19-expressing hepatocellular carcinoma. Molecular Cancer Therapeutics, 16(1), 68–75. 10.1158/1535-7163.MCT-16-0188 [DOI] [PubMed] [Google Scholar]
- Gadaleta RM, Garcia-Irigoyen O, Cariello M, Scialpi N, Peres C, Vetrano S, Fiorino G, Danese S, Ko B, Luo J, Porru E, Roda A, Sabba C, & Moschetta A (2020). Fibroblast growth factor 19 modulates intestinal microbiota and inflammation in presence of farnesoid X receptor. eBioMedicine, 54, 102719. 10.1016/j.ebiom.2020.102719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadaleta RM, & Moschetta A (2019). Metabolic messengers: Fibroblast growth factor 15/19. Nature Metabolism, 1(6), 588–594. 10.1038/s42255-019-0074-3 [DOI] [PubMed] [Google Scholar]
- Gainous TB, Wagner E, & Levine M (2015). Diverse ETS transcription factors mediate FGF signaling in the Ciona anterior neural plate. Developmental Biology, 399(2), 218–225. 10.1016/j.ydbio.2014.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Escuredo JM, Lamarca MK, Villarroya J, Domingo JC, Mateo MG, Gutierrez MDM, Vidal F, Villarroya F, Domingo P, & Giralt M (2017). High FGF21 levels are associated with altered bone homeostasis in HIV-1-infected patients. Metabolism, 71, 163–170. 10.1016/j.metabol.2017.03.014 [DOI] [PubMed] [Google Scholar]
- Gallegos TF, Kamei CN, Rohly M, & Drummond IA (2019). Fibroblast growth factor signaling mediates progenitor cell aggregation and nephron regeneration in the adult zebrafish kidney. Developmental Biology, 454(1), 44–51. 10.1016/j.ydbio.2019.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Ajima R, Yang W, Li C, Song H, Anderson MJ, Liu RR, Lewandoski MB, Yamaguchi TP, & Yang Y (2018). Coordinated directional outgrowth and pattern formation by integration of Wnt5a and Fgf signaling in planar cell polarity. Development, 145(8), dev163824. 10.1242/dev.163824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Kanasaki K, Li J, Kitada M, Okazaki T, & Koya D (2019). betaklotho is essential for the anti-endothelial mesenchymal transition effects of N-acetyl-seryl-aspartyl-lysyl-proline. FEBS Open Bio, 9(5), 1029–1038. 10.1002/2211-5463.12638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao RY, Hsu BG, Wu DA, Hou JS, & Chen MC (2019). Serum fibroblast growth factor 21 levels are positively associated with metabolic syndrome in patients with type 2 diabetes. International Journal of Endocrinology, 2019, 5163245. 10.1155/2019/5163245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia CB, Shaffer CM, Alfaro MP, Smith AL, Sun J, Zhao Z, Young PP, VanSaun MN, & Eid JE (2012). Reprogramming of mesenchymal stem cells by the synovial sarcoma-associated oncogene SYT-SSX2. Oncogene, 31(18), 2323–2334. 10.1038/onc.2011.418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J, Patel N, Basehore S, & Clyne AM (2019). Fibroblast growth factor-2 binding to heparan sulfate proteoglycans varies with shear stress in flow-adapted cells. Annals of Biomedical Engineering, 47(4), 1078–1093. 10.1007/s10439-019-02202-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia S, Dirat B, Tognacci T, Rochet N, Mouska X, Bonnafous S, Patouraux S, Tran A, Gual P, Le Marchand-Brustel Y, Gennero I, & Gouze E (2013). Postnatal soluble FGFR3 therapy rescues achondroplasia symptoms and restores bone growth in mice. Science Translational Medicine, 5(203), 203ra124. 10.1126/scitranslmed.3006247 [DOI] [PubMed] [Google Scholar]
- Garg A, Bansal M, Gotoh N, Feng GS, Zhong J, Wang F, Kariminejad A, Brooks S, & Zhang X (2017). Alx4 relays sequential FGF signaling to induce lacrimal gland morphogenesis. PLoS Genetics, 13(10), e1007047. 10.1371/journal.pgen.1007047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A, Hannan A, Wang Q, Collins T, Teng S, Bansal M, Zhong J, Xu K, & Zhang X (2018). FGF-induced Pea3 transcription factors program the genetic landscape for cell fate determination. PLoS Genetics, 14(9), e1007660. 10.1371/journal.pgen.1007660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A, & Zhang X (2017). Lacrimal gland development: From signaling interactions to regenerative medicine. Developmental Dynamics, 246(12), 970–980. 10.1002/dvdy.24551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser E, Moutos CP, Downes M, & Evans RM (2017). FGF1—A new weapon to control type 2 diabetes mellitus. Nature Reviews. Endocrinology, 13(10), 599–609. 10.1038/nrendo.2017.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay D, Kwon O, Zhang Z, Spata M, Plikus MV, Holler PD, Ito M, Yang Z, Treffeisen E, Kim CD, Nace A, Zhang X, Baratono S, Wang F, Ornitz DM, Millar SE, & Cotsarelis G (2013). Fgf9 from dermal gammadelta T cells induces hair follicle neogenesis after wounding. Nature Medicine, 19(7), 916–923. 10.1038/nm.3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L, Lam KSL, & Xu A (2020). The therapeutic potential of FGF21 in metabolic diseases: From bench to clinic. Nature Reviews. Endocrinology, 16(11), 654–667. 10.1038/s41574-020-0386-0 [DOI] [PubMed] [Google Scholar]
- Gerber SD, Beauchamp P, Zhuang L, Villiger PM, & Trueb B (2020). Functional domains of the FgfrL1 receptor. Developmental Biology, 461(1), 43–54. 10.1016/j.ydbio.2020.01.003 [DOI] [PubMed] [Google Scholar]
- Geske MJ, Zhang X, Patel KK, Ornitz DM, & Stappenbeck TS (2008). Fgf9 signaling regulates small intestinal elongation and mesenchymal development. Development, 135(17), 2959–2968. 10.1242/dev.020453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadiali RS, Guimond SE, Turnbull JE, & Pisconti A (2017). Dynamic changes in heparan sulfate during muscle differentiation and ageing regulate myoblast cell fate and FGF2 signalling. Matrix Biology, 59, 54–68. 10.1016/j.matbio.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaskadbi S (2020). Cell signaling molecules in hydra: Insights into evolutionarily ancient functions of signaling pathways. The International Journal of Developmental Biology, 64(1–2–3), 141–149. 10.1387/ijdb.190243sg [DOI] [PubMed] [Google Scholar]
- Ghassemi S, Vejdovszky K, Sahin E, Ratzinger L, Schelch K, Mohr T, Peter-Vorosmarty B, Brankovic J, Lackner A, Leopoldi A, Meindl D, Pirker C, Hegedus B, Marian B, Holzmann K, Grasl-Kraupp B, Heffeter P, Berger W, & Grusch M (2017). FGF5 is expressed in melanoma and enhances malignancy in vitro and in vivo. Oncotarget, 8(50), 87750, 10.18632/oncotarget.21184-87762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomini A, Chiodelli P, Matarazzo S, Rusnati M, Presta M, & Ronca R (2016). Blocking the FGF/FGFR system as a “two-compartment” antiangiogenic/antitumor approach in cancer therapy. Pharmacological Research, 107, 172–185. 10.1016/j.phrs.2016.03.024 [DOI] [PubMed] [Google Scholar]
- Giacomini A, Matarazzo S, Pagano K, Ragona L, Rezzola S, Corsini M, Di Salle E, Presta M, & Ronca R (2015). A long pentraxin-3-derived pentapeptide for the therapy of FGF8b-driven steroid hormone-regulated cancers. Oncotarget, 6(15), 13790–13802. 10.18632/oncotarget.3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigout A, Guehring H, Froemel D, Meurer A, Ladel C, Reker D, Bay-Jensen AC, Karsdal MA, & Lindemann S (2017). Sprifermin (rhFGF18) enables proliferation of chondrocytes producing a hyaline cartilage matrix. Osteoarthritis and Cartilage, 25(11), 1858–1867. 10.1016/j.joca.2017.08.004 [DOI] [PubMed] [Google Scholar]
- Giordano RJ, Lahdenranta J, Zhen L, Chukwueke U, Petrache I, Langley RR, Fidler IJ, Pasqualini R, Tuder RM, & Arap W (2008). Targeted induction of lung endothelial cell apoptosis causes emphysema-like changes in the mouse. The Journal of Biological Chemistry, 283(43), 29447–29460. 10.1074/jbc.M804595200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladding A, Szymczuk V, Auble BA, & Boyce AM (2021). Burosumab treatment for fibrous dysplasia. Bone, 150, 116004. 10.1016/j.bone.2021.116004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JD, Wells KL, Matthaus F, Painter KJ, Ho W, Riddell J, Johansson JA, Ford MJ, Jahoda CAB, Klika V, Mort RL, & Headon DJ (2017). Hierarchical patterning modes orchestrate hair follicle morphogenesis. PLoS Biology, 15(7), e2002117. 10.1371/journal.pbio.2002117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz R, & Mohammadi M (2013). Exploring mechanisms of FGF signalling through the lens of structural biology. Nature Reviews. Molecular Cell Biology, 14(3), 166–180. 10.1038/nrm3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz R, Nakada Y, Hu MC, Kurosu H, Wang L, Nakatani T, Shi M, Eliseenkova AV, Razzaque MS, Moe OW, Kuroo M, & Mohammadi M (2010). Isolated C-terminal tail of FGF23 alleviates hypophosphatemia by inhibiting FGF23-FGFR-Klotho complex formation. Proceedings of the National Academy of Sciences of the United States of America, 107(1), 407–412. 10.1073/pnas.0902006107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz R, Ohnishi M, Ding X, Kurosu H, Wang L, Akiyoshi J, Ma J, Gai W, Sidis Y, Pitteloud N, Kuro OM, Razzaque MS, & Mohammadi M (2012). Klotho coreceptors inhibit signaling by paracrine fibroblast growth factor 8 subfamily ligands. Molecular and Cellular Biology, 32(10), 1944–1954. 10.1128/mcb.06603-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves CA, Larsen M, Jung S, Stratmann J, Nakamura A, Leuschner M, Hersemann L, Keshara R, Perlman S, Lundvall L, Thuesen LL, Hare KJ, Amit I, Jorgensen A, Kim YH, Del Sol A, & Grapin-Botton A (2021). A 3D system to model human pancreas development and its reference single-cell transcriptome atlas identify signaling pathways required for progenitor expansion. Nature Communications, 12(1), 3144. 10.1038/s41467-021-23295-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves D, Rignol G, Dellugat P, Hartmann G, Sarrazy Garcia S, Stavenhagen J, Santarelli L, Gouze E, & Czech C (2020). In vitro and in vivo characterization of Recifercept, a soluble fibroblast growth factor receptor 3, as treatment for achondroplasia. PLoS One, 15(12), e0244368. 10.1371/journal.pone.0244368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong SG (2014). Isoforms of receptors of fibroblast growth factors. Journal of Cellular Physiology, 229(12), 1887–1895. 10.1002/jcp.24649 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Martinez D, Kim SH, Hu Y, Guimond S, Schofield J, Winyard P, Vannelli GB, Turnbull J, & Bouloux PM (2004). Anosmin-1 modulates fibroblast growth factor receptor 1 signaling in human gonadotropin-releasing hormone olfactory neuroblasts through a heparan sulfate-dependent mechanism. The Journal of Neuroscience, 24(46), 10384–10392. 10.1523/JNEUROSCI.3400-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good RB, Gilbane AJ, Trinder SL, Denton CP, Coghlan G, Abraham DJ, & Holmes AM (2015). Endothelial to mesenchymal transition contributes to endothelial dysfunction in pulmonary arterial hypertension. The American Journal of Pathology, 185(7), 1850–1858. 10.1016/j.ajpath.2015.03.019 [DOI] [PubMed] [Google Scholar]
- Goodwin AT, & Jenkins G (2016). Molecular endotyping of pulmonary fibrosis. Chest, 149(1), 228–237. 10.1378/chest.15-1511 [DOI] [PubMed] [Google Scholar]
- Gorkhali NA, Dong K, Yang M, Song S, Kader A, Shrestha BS, He X, Zhao Q, Pu Y, Li X, Kijas J, Guan W, Han J, Jiang L, & Ma Y (2016). Genomic analysis identified a potential novel molecular mechanism for high-altitude adaptation in sheep at the Himalayas. Scientific Reports, 6, 29963. 10.1038/srep29963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gospodarowicz D (1975). Purification of a fibroblast growth factor from bovine pituitary. The Journal of Biological Chemistry, 250(7), 2515–2520. [PubMed] [Google Scholar]
- Goto T, Hirata M, Aoki Y, Iwase M, Takahashi H, Kim M, Li Y, Jheng HF, Nomura W, Takahashi N, Kim CS, Yu R, Seno S, Matsuda H, Aizawa-Abe M, Ebihara K, Itoh N, & Kawada T (2017). The hepatokine FGF21 is crucial for peroxisome proliferator-activated receptor-alpha agonist-induced amelioration of metabolic disorders in obese mice. The Journal of Biological Chemistry, 292(22), 9175–9190. 10.1074/jbc.M116.767590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould PW, Zemel BS, Taratuta EG, & Baker JF (2020). Circulating fibroblast growth factor-21 levels in rheumatoid arthritis: Associations with disease characteristics, body composition, and physical functioning. The Journal of Rheumatology, 48(4), 504–512. 10.3899/jrheum.200673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabner A, Amaral AP, Schramm K, Singh S, Sloan A, Yanucil C, Li J, Shehadeh LA, Hare JM, David V, Martin A, Fornoni A, Di Marco GS, Kentrup D, Reuter S, Mayer AB, Pavenstadt H, Stypmann J, Kuhn C, … Faul C (2015). Activation of cardiac fibroblast growth factor receptor 4 causes left ventricular hypertrophy. Cell Metabolism, 22(6), 1020–1032. 10.1016/j.cmet.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabner A, Schramm K, Silswal N, Hendrix M, Yanucil C, Czaya B, Singh S, Wolf M, Hermann S, Stypmann J, Di Marco GS, Brand M, Wacker MJ, & Faul C (2017). FGF23/FGFR4-mediated left ventricular hypertrophy is reversible. Scientific Reports, 7(1), 1993. 10.1038/s41598-017-02068-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grčevi c D, Sironi M, Valentino S, Deban L, Cvija H, Inforzato A, Kovači c N, Katavi c, V., Kelava T, Kalajzi c, I., Mantovani A, & Bottazzi B (2018). The long pentraxin 3 plays a role in bone turnover and repair. Frontiers in Immunology, 9, 417. 10.3389/fimmu.2018.00417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gredler ML, Seifert AW, & Cohn MJ (2015). Tissue-specific roles of Fgfr2 in development of the external genitalia. Development, 142(12), 2203–2212. 10.1242/dev.119891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grella A, Kole D, Holmes W, & Dominko T (2016). FGF2 overrides TGFbeta1-driven integrin ITGA11 expression in human dermal fibroblasts. Journal of Cellular Biochemistry, 117(4), 1000–1008. 10.1002/jcb.25386 [DOI] [PubMed] [Google Scholar]
- Grieshammer U, Cebrian C, Ilagan R, Meyers E, Herzlinger D, & Martin GR (2005). FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development, 132(17), 3847–3857. 10.1242/dev.01944 [DOI] [PubMed] [Google Scholar]
- Grothe C, Claus P, Haastert K, Lutwak E, & Ron D (2008). Expression and regulation of Sef, a novel signaling inhibitor of receptor tyrosine kinases-mediated signaling in the nervous system. Acta Histochemica, 110(2), 155–162. 10.1016/j.acthis.2007.08.006 [DOI] [PubMed] [Google Scholar]
- Groves AK, & Fekete DM (2012). Shaping sound in space: The regulation of inner ear patterning. Development, 139(2), 245–257. 10.1242/dev.067074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusch M, Schelch K, Riedler R, Reichhart E, Differ C, Berger W, Ingles-Prieto A, & Janovjak H (2014). Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. The EMBO Journal, 33(15), 1713–1726. 10.15252/embj.201387695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Xue C, Zhu J, Sun H, Ding F, Cao Z, & Gu X (2014). Basic fibroblast growth factor (bFGF) facilitates differentiation of adult dorsal root ganglia-derived neural stem cells toward Schwann cells by binding to FGFR-1 through MAPK/ERK activation. Journal of Molecular Neuroscience, 52(4), 538–551. 10.1007/s12031-013-0109-2 [DOI] [PubMed] [Google Scholar]
- Guan D, Mi J, Chen X, Wu Y, Yao Y, Wang L, Xiao Z, Zhao Y, Chen B, & Dai J (2018). Lung endothelial cell-targeted peptide-guided bFGF promotes the regeneration after radiation induced lung injury. Biomaterials, 184, 10–19. 10.1016/j.biomaterials.2018.08.061 [DOI] [PubMed] [Google Scholar]
- Guerra J, Chiodelli P, Tobia C, Gerri C, & Presta M (2020). Long-pentraxin 3 affects primary cilium in zebrafish embryo and cancer cells via the FGF system. Cancers (Basel), 12(7), 1756. 10.3390/cancers12071756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati S, Wells JM, Urdaneta GP, Balestrini K, Vital I, Tovar K, Barnes JW, Bhatt SP, Campos M, & Krick S (2019). Fibroblast growth factor 23 is associated with a frequent exacerbator phenotype in COPD: A cross-sectional pilot study. International Journal of Molecular Sciences, 20(9), 2292. 10.3390/ijms20092292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Yin H, Zheng M, Tang Y, Lu B, Chen X, Fu Q, Qin Z, Lyu D, Tang Q, Zhang L, Ma J, Zhang L, & Fang X (2019). Cytokine profiling reveals increased serum inflammatory cytokines in idiopathic choroidal neovascularization. BMC Ophthalmology, 19(1), 94. 10.1186/s12886-019-1101-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Liu T, Zhao D, Wang X, Liu D, He Y, Shan C, Kong Y, Hu W, Tao B, Sun L, Zhao H, Li S, & Liu J (2017). FGF18 protects against 6-hydroxydopamine-induced nigrostriatal damage in a rat model of Parkinson’s disease. Neuroscience, 356, 229–241. 10.1016/j.neuroscience.2017.05.007 [DOI] [PubMed] [Google Scholar]
- Gurung HR, Carr MM, Bryant K, Chucair-Elliott AJ, & Carr DJ (2018). Fibroblast growth factor-2 drives and maintains progressive corneal neovascularization following HSV-1 infection. Mucosal Immunology, 11(1), 172–185. 10.1038/mi.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenzi B, Gers-Barlag K, Akhoundzadeh H, Hutson TH, Menezes SC, Bunge MB, & Moon LD (2016). Overexpression of the fibroblast growth factor receptor 1 (FGFR1) in a model of spinal cord injury in rats. PLoS One, 11(3), e0150541. 10.1371/journal.pone.0150541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafenbreidel M, Twining RC, Rafa Todd C, & Mueller D (2015). Blocking infralimbic basic fibroblast growth factor (bFGF or FGF2) facilitates extinction of drug seeking after cocaine self-administration. Neuropsychopharmacology, 40(13), 2907–2915. 10.1038/npp.2015.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafez P, Jose S, Chowdhury SR, Ng MH, Ruszymah BH, & Abdul Rahman Mohd R (2016). Cardiomyogenic differentiation of human sternal bone marrow mesenchymal stem cells using a combination of basic fibroblast growth factor and hydrocortisone. Cell Biology International, 40(1), 55–64. 10.1002/cbin.10536 [DOI] [PubMed] [Google Scholar]
- Hagan AS, Boylan M, Smith C, Perez-Santamarina E, Kowalska K, Hung IH, Lewis RM, Hajihosseini MK, Lewandoski M, & Ornitz DM (2019). Generation and validation of novel conditional flox and inducible Cre alleles targeting fibroblast growth factor 18 (Fgf18). Developmental Dynamics, 248(9), 882–893. 10.1002/dvdy.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan AS, Zhang B, & Ornitz DM (2020). Identification of a FGF18-expressing alveolar myofibroblast that is developmentally cleared during alveologenesis. Development, 147(2), dev181032. 10.1242/dev.181032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighi F, Dahlmann J, Nakhaei-Rad S, Lang A, Kutschka I, Zenker M, Kensah G, Piekorz RP, & Ahmadian MR (2018). bFGF-mediated pluripotency maintenance in human induced pluripotent stem cells is associated with NRAS-MAPK signaling. Cell Communication and Signaling, 16(1), 96. 10.1186/s12964-018-0307-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haibe Y, Kreidieh M, El Hajj H, Khalifeh I, Mukherji D, Temraz S, & Shamseddine A (2020). Resistance mechanisms to anti-angiogenic therapies in cancer. Frontiers in Oncology, 10, 221. 10.3389/fonc.2020.00221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada K, & Lasek AW (2020). Receptor tyrosine kinases as therapeutic targets for alcohol use disorder. Neurotherapeutics, 17(1), 4–16. 10.1007/s13311-019-00795-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto J, Yasuda H, Nonaka Y, Fujiwara M, Nakamura Y, Soejima K, & Betsuyaku T (2018). The FGF2 aptamer inhibits the growth of FGF2-FGFR pathway driven lung cancer cells. Biochemical and Biophysical Research Communications, 503(3), 1330–1334. 10.1016/j.bbrc.2018.07.044 [DOI] [PubMed] [Google Scholar]
- Han J, Du Y, Wang L, Chen X, Jiang L, & Xu J (2018). Acid fibroblast growth factor facilitates the progression of atherosclerotic plaques regardless of alterations in serum lipid expression levels in HFDfed ApoE/mice. Molecular Medicine Reports, 18(1), 1025–1030. 10.3892/mmr.2018.9060 [DOI] [PubMed] [Google Scholar]
- Han M, Yang X, Taylor G, Burdsal CA, Anderson RA, & Muneoka K (2005). Limb regeneration in higher vertebrates: Developing a roadmap. Anatomical Record. Part B, New Anatomist, 287(1), 14–24. 10.1002/ar.b.20082 [DOI] [PubMed] [Google Scholar]
- Han MS, Perry RJ, Camporez JP, Scherer PE, Shulman GI, Gao G, & Davis RJ (2021). A feed-forward regulatory loop in adipose tissue promotes signaling by the hepatokine FGF21. Genes & Development, 35(1–2), 133–146. 10.1101/gad.344556.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Mei Y, Wang Y, & Feng Z (2019). miR-5582–5p inhibits cell proliferation of non-small cell lung cancer through targeting FGF-10. International Journal of Clinical and Experimental Pathology, 12(3), 1087–1094. [PMC free article] [PubMed] [Google Scholar]
- Han X, Ross J, Kolumam G, Pi M, Sonoda J, King G, & Quarles LD (2018). Cardiovascular effects of renal distal tubule deletion of the FGF receptor 1 gene. Journal of the American Society of Nephrology, 29(1), 69–80. 10.1681/ASN.2017040412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Xiao Z, & Quarles LD (2015). Membrane and integrative nuclear fibroblastic growth factor receptor (FGFR) regulation of FGF-23. The Journal of Biological Chemistry, 290(16), 10447–10459. 10.1074/jbc.M114.609230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yang J, Li L, Huang J, King G, & Quarles LD (2016). Conditional deletion of Fgfr1 in the proximal and distal tubule identifies distinct roles in phosphate and calcium transport. PLoS One, 11(2), e0147845. 10.1371/journal.pone.0147845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada Y, Nakamura Y, Ozono Y, Ishida Y, Takimoto Y, Taniguchi M, Ohata K, Koyama Y, Imai T, Morihana T, Kondo M, Sato T, Inohara H, & Shimada S (2018). Fibroblast growth factor 12 is expressed in spiral and vestibular ganglia and necessary for auditory and equilibrium function. Scientific Reports, 8(1), 11491. 10.1038/s41598-018-28618-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna KS (2019). Erdafitinib to treat urothelial carcinoma. Drugs of Today (Barcelona, Spain: 1998), 55(8), 495–501. 10.1358/dot.2019.55.8.3010573 [DOI] [PubMed] [Google Scholar]
- Hanneken A, Mercado M, & Maher P (2021). Constitutive and regulated shedding of soluble FGF receptors releases biologically active inhibitors of FGF-2. International Journal of Molecular Sciences, 22(5), 2712. 10.3390/ijms22052712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannema SE, van Duyvenvoorde HA, Premsler T, Yang RB, Mueller TD, Gassner B, Oberwinkler H, Roelfsema F, Santen GW, Prickett T, Kant SG, Verkerk AJ, Uitterlinden AG, Espiner E, Ruivenkamp CA, Oostdijk W, Pereira AM, Losekoot M, Kuhn M, & Wit JM (2013). An activating mutation in the kinase homology domain of the natriuretic peptide receptor-2 causes extremely tall stature without skeletal deformities. The Journal of Clinical Endocrinology and Metabolism, 98(12), E1988–E1998. 10.1210/jc.2013-2358 [DOI] [PubMed] [Google Scholar]
- Hansen SM, Li S, Bock E, & Berezin V (2010). Synthetic NCAM-derived ligands of the fibroblast growth factor receptor. Advances in Experimental Medicine and Biology, 663, 355–372. 10.1007/978-1-4419-1170-4_22 [DOI] [PubMed] [Google Scholar]
- Harada M, & Akita K (2020). Mouse fibroblast growth factor 9 N143T mutation leads to wide chondrogenic condensation of long bones. Histochemistry and Cell Biology, 153(4), 215–223. 10.1007/s00418-020-01844-2 [DOI] [PubMed] [Google Scholar]
- Harada M, Murakami H, Okawa A, Okimoto N, Hiraoka S, Nakahara T, Akasaka R, Shiraishi Y, Futatsugi N, Mizutani-Koseki Y, Kuroiwa A, Shirouzu M, Yokoyama S, Taiji M, Iseki S, Ornitz DM, & Koseki H (2009). FGF9 monomer-dimer equilibrium regulates extracellular matrix affinity and tissue diffusion. Nature Genetics, 41(3), 289–298. 10.1038/ng.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M, Omori A, Nakahara C, Nakagata N, Akita K, & Yamada G (2015). Tissue-specific roles of FGF signaling in external genitalia development. Developmental Dynamics, 244(6), 759–773. 10.1002/dvdy.24277 [DOI] [PubMed] [Google Scholar]
- Hardelin JP, & Dode C (2008). The complex genetics of Kallmann syndrome: KAL1, FGFR1, FGF8, PROKR2, PROK2, et al. Sexual Development, 2(4–5), 181–193. 10.1159/000152034 [DOI] [PubMed] [Google Scholar]
- Harder MJ, Hix J, Reeves WM, & Veeman MT (2021). Ciona Brachyury proximal and distal enhancers have different FGF dose-response relationships. PLoS Genetics, 17(1), e1009305. 10.1371/journal.pgen.1009305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding TC, Long L, Palencia S, Zhang H, Sadra A, Hestir K, Patil N, Levin A, Hsu AW, Charych D, Brennan T, Zanghi J, Halenbeck R, Marshall SA, Qin M, Doberstein SK, Hollenbaugh D, Kavanaugh WM, Williams LT, & Baker KP (2013). Blockade of nonhormonal fibroblast growth factors by FP-1039 inhibits growth of multiple types of cancer. Science Translational Medicine, 5(178), 178ra139. 10.1126/scitranslmed.3005414 [DOI] [PubMed] [Google Scholar]
- Harrison SA, Rinella ME, Abdelmalek MF, Trotter JF, Paredes AH, Arnold HL, Kugelmas M, Bashir MR, Jaros MJ, Ling L, Rossi SJ, DePaoli AM, & Loomba R (2018). NGM282 for treatment of non-alcoholic steatohepatitis: A multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet, 391(10126), 1174–1185. 10.1016/S0140-6736(18)30474-4 [DOI] [PubMed] [Google Scholar]
- Harshuk-Shabso S, Dressler H, Niehrs C, Aamar E, & Enshell-Seijffers D (2020). Fgf and Wnt signaling interaction in the mesenchymal niche regulates the murine hair cycle clock. Nature Communications, 11(1), 5114. 10.1038/s41467-020-18643-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley IR, Miller CB, Papadakis GZ, Bergwitz C, Del Rivero J, Blau JE, Florenzano P, Berglund JA, Tassone J, Roszko KL, Moran S, Gafni RI, Isaacs R, & Collins MT (2020). Targeted FGFR blockade for the treatment of tumor-induced osteomalacia. The New England Journal of Medicine, 383(14), 1387–1389. 10.1056/NEJMc2020399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A, Durlacher K, Silver J, Naveh-Many T, & Levi R (2016). The fibroblast growth factor receptor mediates the increased FGF23 expression in acute and chronic uremia. American Journal of Physiology. Renal Physiology, 310(3), F217–F221. 10.1152/ajprenal.00332.2015 [DOI] [PubMed] [Google Scholar]
- Hassan N, Greve B, Espinoza-Sanchez NA, & Gotte M (2021). Cell-surface heparan sulfate proteoglycans as multifunctional integrators of signaling in cancer. Cellular Signalling, 77, 109822. 10.1016/j.cellsig.2020.109822 [DOI] [PubMed] [Google Scholar]
- Hasse C, Holz O, Lange E, Pisowodzki L, Rebscher N, Christin Eder M, Hobmayer B, & Hassel M (2014). FGFR-ERK signaling is an essential component of tissue separation. Developmental Biology, 395(1), 154–166. 10.1016/j.ydbio.2014.08.010 [DOI] [PubMed] [Google Scholar]
- Hatch EP, Noyes CA, Wang X, Wright TJ, & Mansour SL (2007). Fgf3 is required for dorsal patterning and morphogenesis of the inner ear epithelium. Development, 134(20), 3615–3625. 10.1242/dev.006627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatlen MA, Schmidt-Kittler O, Sherwin CA, Rozsahegyi E, Rubin N, Sheets MP, Kim JL, Miduturu C, Bifulco N, Brooijmans N, Shi H, Guzi T, Boral A, Lengauer C, Dorsch M, Kim RD, Kang YK, Wolf BB, & Hoeflich KP (2019). Acquired on-target clinical resistance validates FGFR4 as a driver of hepatocellular carcinoma. Cancer Discovery, 9(12), 1686–1695. 10.1158/2159-8290.CD-19-0367 [DOI] [PubMed] [Google Scholar]
- Haugsten EM, Sorensen V, Kunova Bosakova M, de Souza GA, Krejci P, Wiedlocha A, & Wesche J (2016). Proximity labeling reveals molecular determinants of FGFR4 endosomal transport. Journal of Proteome Research, 15(10), 3841–3855. 10.1021/acs.jproteome.6b00652 [DOI] [PubMed] [Google Scholar]
- Haugsten EM, Zakrzewska M, Brech A, Pust S, Olsnes S, Sandvig K, & Wesche J (2011). Clathrin- and dynamin-independent endocytosis of FGFR3—Implications for signalling. PLoS One, 6(7), e21708. 10.1371/journal.pone.0021708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havis E, Bonnin MA, Esteves de Lima J, Charvet B, Milet C, & Duprez D (2016). TGFbeta and FGF promote tendon progenitor fate and act downstream of muscle contraction to regulate tendon differentiation during chick limb development. Development, 143(20), 3839–3851. 10.1242/dev.136242 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Ray CA, & Bermingham-McDonogh O (2008). Fgf20 is required for sensory epithelial specification in the developing cochlea. The Journal of Neuroscience, 28(23), 5991–5999. 10.1523/JNEUROSCI.1690-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Gong Y, Gower L, Yang X, & Friesel RE (2016). Sef regulates epithelial-mesenchymal transition in breast cancer cells. Journal of Cellular Biochemistry, 117(10), 2346–2356. 10.1002/jcb.25532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Shumate LT, Matthias J, Aydin C, Wein MN, Spatz JM, Goetz R, Mohammadi M, Plagge A, Divieti Pajevic P, & Bastepe M (2019). A G protein-coupled, IP3/protein kinase C pathway controlling the synthesis of phosphaturic hormone FGF23. JCI Insight, 4(17), e125007. 10.1172/jci.insight.125007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Yang X, Gong Y, Kovalenko D, Canalis E, Rosen CJ, & Friesel RE (2014). Deficiency of Sef is associated with increased post-natal cortical bone mass by regulating Runx2 activity. Journal of Bone and Mineral Research, 29(5), 1217–1231. 10.1002/jbmr.2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Hu X, Ma X, Su H, Ying L, Peng J, Pan X, Bao Y, Zhou J, & Jia W (2017). Elevated serum fibroblast growth factor 23 levels as an indicator of lower extremity atherosclerotic disease in Chinese patients with type 2 diabetes mellitus. Cardiovascular Diabetology, 16(1), 77. 10.1186/s12933-017-0559-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert JM, Rosenquist T, Götz J, & Martin GR (1994). FGF5 as a regulator of the hair growth cycle: Evidence from targeted and spontaneous mutations. Cells, 78(6), 1017–1025. 10.1016/0092-8674(94)90276-3 [DOI] [PubMed] [Google Scholar]
- Hegab AE, Ozaki M, Kameyama N, Gao J, Kagawa S, Yasuda H, Soejima K, Yin Y, Guzy RD, Nakamura Y, Ornitz DM, & Betsuyaku T (2019). Effect of FGF/FGFR pathway blocking on lung adenocarcinoma and its cancer-associated fibroblasts. The Journal of Pathology, 249(2), 193–205. 10.1002/path.5290 [DOI] [PubMed] [Google Scholar]
- Heidenreich B, Denisova E, Rachakonda S, Sanmartin O, Dereani T, Hosen I, Nagore E, & Kumar R (2017). Genetic alterations in seborrheic keratoses. Oncotarget, 8(22), 36639–36649. 10.18632/oncotarget.16698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert C, Lassalle G, Alcouffe C, & Bono F (2014). Approaches targeting the FGF-FGFR system: A review of the recent patent literature and associated advanced therapeutic agents. Pharmaceutical Patent Analyst, 3(6), 585–612. 10.4155/ppa.14.45 [DOI] [PubMed] [Google Scholar]
- Hernandez G, Luo T, Javed TA, Wen L, Kalwat MA, Vale K, Ammouri F, Husain SZ, Kliewer SA, & Mangelsdorf DJ (2020). Pancreatitis is an FGF21-deficient state that is corrected by replacement therapy. Science Translational Medicine, 12(525), eaay5186. 10.1126/scitranslmed.aay5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Bejarano M, Gestri G, Spawls L, Nieto-Lopez F, Picker A, Tada M, Brand M, Bovolenta P, Wilson SW, & Cavodeassi F (2015). Opposing Shh and Fgf signals initiate nasotemporal patterning of the zebrafish retina. Development, 142(22), 3933–3942. 10.1242/dev.125120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hero M, Laitinen EM, Varimo T, Vaaralahti K, Tommiska J, & Raivio T (2015). Childhood growth of females with Kallmann syndrome and FGFR1 mutations. Clinical Endocrinology, 82(1), 122–126. 10.1111/cen.12504 [DOI] [PubMed] [Google Scholar]
- Herraiz C, Jimenez-Cervantes C, Sanchez-Laorden B, & Garcia-Borron JC (2018). Functional interplay between secreted ligands and receptors in melanoma. Seminars in Cell & Developmental Biology, 78, 73–84. 10.1016/j.semcdb.2017.06.021 [DOI] [PubMed] [Google Scholar]
- Herriges JC, Verheyden JM, Zhang Z, Sui P, Zhang Y, Anderson MJ, Swing DA, Zhang Y, Lewandoski M, & Sun X (2015). FGF-regulated ETV transcription factors control FGF-SHH feedback loop in lung branching. Developmental Cell, 35(3), 322–332. 10.1016/j.devcel.2015.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriges M, & Morrisey EE (2014). Lung development: Orchestrating the generation and regeneration of a complex organ. Development, 141(3), 502–513. 10.1242/dev.098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzler-Schaefer K, Mathew G, Somani AK, Tholpady S, Kadakia MP, Chen Y, Spandau DF, & Zhang X (2014). Pten loss induces autocrine FGF signaling to promote skin tumorigenesis. Cell Reports, 6(5), 818–826. 10.1016/j.celrep.2014.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki M, Shintani T, Hamada A, Rosli SNZ, & Okamoto T (2020). Eldecalcitol (ED-71)-induced exosomal miR-6887–5p suppresses squamous cell carcinoma cell growth by targeting heparin-binding protein 17/fibroblast growth factor-binding protein-1 (HBp17/FGFBP-1). In Vitro Cellular & Developmental Biology. Animal, 56(3), 222–233. 10.1007/s11626-020-00440-x [DOI] [PubMed] [Google Scholar]
- Higgins CA, Petukhova L, Harel S, Ho YY, Drill E, Shapiro L, Wajid M, & Christiano AM (2014). FGF5 is a crucial regulator of hair length in humans. Proceedings of the National Academy of Sciences of the United States of America, 111(29), 10648–10653. 10.1073/pnas.1402862111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CM, Laeger T, Dehner M, Albarado DC, Clarke B, Wanders D, Burke SJ, Collier JJ, Qualls-Creekmore E, Solon-Biet SM, Simpson SJ, Berthoud HR, Munzberg H, & Morrison CD (2019). FGF21 signals protein status to the brain and adaptively regulates food choice and metabolism. Cell Reports, 27(10), 2934–2947.e2933. 10.1016/j.celrep.2019.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines EA, Jones MN, Harvey JF, Perlyn C, Ornitz DM, Sun X, & Verheyden JM (2019). Crouzon syndrome mouse model exhibits cartilage hyperproliferation and defective segmentation in the developing trachea. Science China. Life Sciences, 62(10), 1375–1380. 10.1007/s11427-019-9568-x [DOI] [PubMed] [Google Scholar]
- Hirani R, Hanssen E, & Gibson MA (2007). LTBP-2 specifically interacts with the amino-terminal region of fibrillin-1 and competes with LTBP-1 for binding to this microfibrillar protein. Matrix Biology, 26(4), 213–223. 10.1016/j.matbio.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Hirschfield GM, Chazouilleres O, Drenth JP, Thorburn D, Harrison SA, Landis CS, Mayo MJ, Muir AJ, Trotter JF, Leeming DJ, Karsdal MA, Jaros MJ, Ling L, Kim KH, Rossi SJ, Somaratne RM, DePaoli AM, & Beuers U (2019). Effect of NGM282, an FGF19 analogue, in primary sclerosing cholangitis: A multicenter, randomized, double-blind, placebo-controlled phase II trial. Journal of Hepatology, 70(3), 483–493. 10.1016/j.jhep.2018.10.035 [DOI] [PubMed] [Google Scholar]
- Ho BB, & Bergwitz C (2021). FGF23 signalling and physiology. Journal of Molecular Endocrinology, 66(2), R23–R32. 10.1530/JME-20-0178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho HK, Pok S, Streit S, Ruhe JE, Hart S, Lim KS, Loo HL, Aung MO, Lim SG, & Ullrich A (2009). Fibroblast growth factor receptor 4 regulates proliferation, anti-apoptosis and alpha-fetoprotein secretion during hepatocellular carcinoma progression and represents a potential target for therapeutic intervention. Journal of Hepatology, 50(1), 118–127. 10.1016/j.jhep.2008.08.015 [DOI] [PubMed] [Google Scholar]
- Hochberg MC, Guermazi A, Guehring H, Aydemir A, Wax S, Fleuranceau-Morel P, Reinstrup Bihlet A, Byrjalsen I, Ragnar Andersen J, & Eckstein F (2019). Effect of intra-articular sprifermin vs placebo on femorotibial joint cartilage thickness in patients with osteoarthritis: The FORWARD randomized clinical trial. JAMA, 322(14), 1360–1370. 10.1001/jama.2019.14735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogler W, & Ward LM (2020). New developments in the management of achondroplasia. Wiener Medizinische Wochenschrift (1946), 170(5–6), 104–111. 10.1007/s10354-020-00741-6 (Neue Entwicklungen im Management der Achondroplasie.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes G, O’Rourke C, Motch Perrine SM, Lu N, van Bakel H, Richtsmeier JT, & Jabs EW (2018). Midface and upper airway dysgenesis in FGFR2-related craniosynostosis involves multiple tissue-specific and cell cycle effects. Development, 145(19), dev166488. 10.1242/dev.166488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes G, Zhang L, Rivera J, Murphy R, Assouline C, Sullivan L, Oppeneer T, & Jabs EW (2018). C-type natriuretic peptide analog treatment of craniosynostosis in a Crouzon syndrome mouse model. PLoS One, 13(7), e0201492. 10.1371/journal.pone.0201492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz O, Apel D, Steinmetz P, Lange E, Hopfenmuller S, Ohler K, Sudhop S, & Hassel M (2017). Bud detachment in hydra requires activation of fibroblast growth factor receptor and a Rho-ROCK-myosin II signaling pathway to ensure formation of a basal constriction. Developmental Dynamics, 246(7), 502–516. 10.1002/dvdy.24508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer-Bouthiette C, Xiao L, & Hurley MM (2021). Gait disturbances and muscle dysfunction in fibroblast growth factor 2 knockout mice. Scientific Reports, 11(1), 11005. 10.1038/s41598-021-90565-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Hu P, Marino J, Hufnagel SB, Hopkin RJ, Toromanovic A, Richieri-Costa A, Ribeiro-Bicudo LA, Kruszka P, Roessler E, & Muenke M (2016). Dominant-negative kinase domain mutations in FGFR1 can explain the clinical severity of Hartsfield syndrome. Human Molecular Genetics, 25(10), 1912–1922. 10.1093/hmg/ddw064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori S, Miyake M, Tatsumi Y, Morizawa Y, Nakai Y, Onishi S, Onishi K, Iida K, Gotoh D, Tanaka N, & Fujimoto K (2018). Gamma-Klotho exhibits multiple roles in tumor growth of human bladder cancer. Oncotarget, 9(28), 19508–19524. 10.18632/oncotarget.24628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori Y, Ito K, Hamamichi S, Ozawa Y, Matsui J, Umeda IO, & Fujii H (2017). Functional characterization of VEGF- and FGF-induced tumor blood vessel models in human cancer xenografts. Anticancer Research, 37(12), 6629–6638. 10.21873/anticanres.12120 [DOI] [PubMed] [Google Scholar]
- Hosokawa R, Deng X, Takamori K, Xu X, Urata M, Bringas P Jr., & Chai Y (2009). Epithelial-specific requirement of FGFR2 signaling during tooth and palate development. Journal of Experimental Zoology. Part B, Molecular and Developmental Evolution, 312B(4), 343–350. 10.1002/jez.b.21274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta Y, Sasaki S, Konishi M, Kinoshita H, Kuwahara K, Nakao K, & Itoh N (2008). Fgf16 is required for cardiomyocyte proliferation in the mouse embryonic heart. Developmental Dynamics, 237(10), 2947–2954. 10.1002/dvdy.21726 [DOI] [PubMed] [Google Scholar]
- Hou Z, Wu Q, Sun X, Chen H, Li Y, Zhang Y, Mori M, Yang Y, Que J, & Jiang M (2019). Wnt/Fgf crosstalk is required for the specification of basal cells in the mouse trachea. Development, 146(3), dev171496. 10.1242/dev.171496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houschyar KS, Borrelli MR, Tapking C, Popp D, Puladi B, Ooms M, Chelliah MP, Rein S, Pforringer D, Thor D, Reumuth G, Wallner C, Branski LK, Siemers F, Grieb G, Lehnhardt M, Yazdi AS, Maan ZN, & Duscher D (2020). Molecular mechanisms of hair growth and regeneration: Current understanding and novel paradigms. Dermatology, 236(4), 271–280. 10.1159/000506155 [DOI] [PubMed] [Google Scholar]
- House SL, Castro AM, Lupu TS, Weinheimer C, Smith C, Kovacs A, & Ornitz DM (2016). Endothelial fibroblast growth factor receptor signaling is required for vascular remodeling following cardiac ischemia-reperfusion injury. American Journal of Physiology. Heart and Circulatory Physiology, 310(5), H559–H571. 10.1152/ajpheart.00758.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- House SL, Wang J, Castro AM, Weinheimer C, Kovacs A, & Ornitz DM (2015). Fibroblast growth factor 2 is an essential cardioprotective factor in a closed-chest model of cardiac ischemia-reperfusion injury. Physiological Reports, 3(1), e12278. 10.14814/phy2.12278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard D, Wardale J, Guehring H, & Henson F (2015). Delivering rhFGF-18 via a bilayer collagen membrane to enhance microfracture treatment of chondral defects in a large animal model. Journal of Orthopaedic Research, 33(8), 1120–1127. 10.1002/jor.22882 [DOI] [PubMed] [Google Scholar]
- Hoxha E, Marcinno A, Montarolo F, Masante L, Balbo I, Ravera F, Laezza F, & Tempia F (2019). Emerging roles of Fgf14 in behavioral control. Behavioural Brain Research, 356, 257–265. 10.1016/j.bbr.2018.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy SM (2020). Pemigatinib: First approval. Drugs, 80(9), 923–929. 10.1007/s40265-020-01330-y [DOI] [PubMed] [Google Scholar]
- Hsu WC, Scala F, Nenov MN, Wildburger NC, Elferink H, Singh AK, Chesson CB, Buzhdygan T, Sohail M, Shavkunov AS, Panova NI, Nilsson CL, Rudra JS, Lichti CF, & Laezza F (2016). CK2 activity is required for the interaction of FGF14 with voltage-gated sodium channels and neuronal excitability. The FASEB Journal, 30(6), 2171–2186. 10.1096/fj.201500161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MC, Shi M, Zhang J, Addo T, Cho HJ, Barker SL, Ravikumar P, Gillings N, Bian A, Sidhu SS, Kuro-o M, & Moe OW (2016). Renal production, uptake, and handling of circulating alphaKlotho. Journal of the American Society of Nephrology, 27(1), 79–90. 10.1681/ASN.2014101030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Wang C, Liu F, He J, Wang F, Wang W, & You P (2019). High serum levels of FGF21 are decreased in bipolar mania patients during psychotropic medication treatment and are associated with increased metabolism disturbance. Psychiatry Research, 272, 643–648. 10.1016/j.psychres.2018.12.159 [DOI] [PubMed] [Google Scholar]
- Hu Y, Guimond SE, Travers P, Cadman S, Hohenester E, Turnbull JE, Kim SH, & Bouloux PM (2009). Novel mechanisms of fibroblast growth factor receptor 1 regulation by extracellular matrix protein anosmin-1. The Journal of Biological Chemistry, 284(43), 29905–29920. 10.1074/jbc.M109.049155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua L, Li J, Feng B, Jiang D, Jiang X, Luo T, Che L, Xu S, Lin Y, Fang Z, Wu D, & Zhuo Y, (2021). Dietary intake regulates white adipose tissues angiogenesis via liver fibroblast growth factor 21 in male mice. Endocrinology, 162(3), bqaa244. 10.1210/endocr/bqaa244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang AH, Lu HH, & Schweitzer R (2015). Molecular regulation of tendon cell fate during development. Journal of Orthopaedic Research, 33(6), 800–812. 10.1002/jor.22834 [DOI] [PubMed] [Google Scholar]
- Huang C, Liu Y, Beenken A, Jiang L, Gao X, Huang Z, Hsu A, Gross GJ, Wang YG, Mohammadi M, & Schultz JEJ (2017). A novel fibroblast growth factor-1 ligand with reduced heparin binding protects the heart against ischemia-reperfusion injury in the presence of heparin co-administration. Cardiovascular Research, 113(13), 1585–1602. 10.1093/cvr/cvx165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Zhan JF, Chen YX, Xu CY, & Chen Y (2020). LncRNA-SNHG29 inhibits vascular smooth muscle cell calcification by downregulating miR-200b-3p to activate the alpha-Klotho/FGFR1/FGF23 axis. Cytokine, 136, 155243. 10.1016/j.cyto.2020.155243 [DOI] [PubMed] [Google Scholar]
- Huang CY, & Tan TH (2012). DUSPs, to MAP kinases and beyond. Cell & Bioscience, 2(1), 24. 10.1186/2045-3701-2-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ishino T, Chen G, Rolzin P, Osothprarop TF, Retting K, Li L, Jin P, Matin MJ, Huyghe B, Talukdar S, Bradshaw CW, Palanki M, Violand BN, Woodnutt G, Lappe RW, Ogilvie K, & Levin N (2013). Development of a novel long-acting antidiabetic FGF21 mimetic by targeted conjugation to a scaffold antibody. The Journal of Pharmacology and Experimental Therapeutics, 346(2), 270–280. 10.1124/jpet.113.204420 [DOI] [PubMed] [Google Scholar]
- Huang J, Wang K, Shiflett LA, Brotto L, Bonewald LF, Wacker MJ, Dallas SL, & Brotto M (2019). Fibroblast growth factor 9 (FGF9) inhibits myogenic differentiation of C2C12 and human muscle cells. Cell Cycle, 18(24), 3562–3580. 10.1080/15384101.2019.1691796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JK, Ma L, Song WH, Lu BY, Huang YB, Dong HM, Ma XK, Zhu ZZ, & Zhou R (2017). LncRNA-MALAT1 Promotes Angiogenesis of Thyroid Cancer by Modulating Tumor-Associated Macrophage FGF2 Protein Secretion. Journal of Cellular Biochemistry, 118(12), 4821–4830. 10.1002/jcb.26153 [DOI] [PubMed] [Google Scholar]
- Huang L, Quesada C, Aliabouzar M, Fowlkes JB, Franceschi RT, Liu Z, Putnam AJ, & Fabiilli ML (2021). Spatially-directed angiogenesis using ultrasound-controlled release of basic fibroblast growth factor from acoustically-responsive scaffolds. Acta Biomaterialia, 129, 73–83. 10.1016/j.actbio.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, & Stern MJ (2005). FGF signaling in flies and worms: More and more relevant to vertebrate biology. Cytokine & Growth Factor Reviews, 16(2), 151–158. 10.1016/j.cytogfr.2005.03.002 [DOI] [PubMed] [Google Scholar]
- Huang W, Shao M, Liu H, Chen J, Hu J, Zhu L, Liu F, Wang D, Zou Y, Xiong Y, & Wang X (2019). Fibroblast growth factor 21 enhances angiogenesis and wound healing of human brain microvascular endothelial cells by activating PPARgamma. Journal of Pharmacological Sciences, 140(2), 120–127. 10.1016/j.jphs.2019.03.010 [DOI] [PubMed] [Google Scholar]
- Huang Y, & Dreyfus CF (2016). The role of growth factors as a therapeutic approach to demyelinating disease. Experimental Neurology, 283(Pt B), 531–540. 10.1016/j.expneurol.2016.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Marsiglia WM, Basu Roy U, Rahimi N, Ilghari D, Wang H, Chen H, Gai W, Blais S, Neubert TA, Mansukhani A, Traaseth NJ, Li X, & Mohammadi M (2016). Two FGF receptor kinase molecules act in concert to recruit and transphosphorylate phospholipase Cgamma. Molecular Cell, 61(1), 98–110. 10.1016/j.molcel.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Tan Y, Gu J, Liu Y, Song L, Niu J, Zhao L, Srinivasan L, Lin Q, Deng J, Li Y, Conklin DJ, Neubert TA, Cai L, Li X, & Mohammadi M (2017). Uncoupling the mitogenic and metabolic functions of FGF1 by tuning FGF1-FGF receptor dimer stability. Cell Reports, 20(7), 1717–1728. 10.1016/j.celrep.2017.06.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert F, Payan SM, & Rochais F (2018). FGF10 signaling in heart development, homeostasis, disease and repair. Frontiers in Genetics, 9, 599. 10.3389/fgene.2018.00599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson C, Sirour C, & Yasuo H (2016). Co-expression of Foxa.a, Foxd and Fgf9/16/20 defines a transient mesendoderm regulatory state in ascidian embryos. eLife, 5, e14692. 10.7554/eLife.14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SH, Ha L, & Jang HS (2020). Nephron progenitor maintenance is controlled through fibroblast growth factors and Sprouty1 interaction. Journal of the American Society of Nephrology, 31(11), 2559–2572. 10.1681/ASN.2020040401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SH, Jones J, Warchol ME, & Ornitz DM (2012). Differentiation of the lateral compartment of the cochlea requires a temporally restricted FGF20 signal. PLoS Biology, 10(1), e1001231. 10.1371/journal.pbio.1001231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SH, Narhi K, Lindfors PH, Haara O, Yang L, Ornitz DM, & Mikkola ML (2013). Fgf20 governs formation of primary and secondary dermal condensations in developing hair follicles. Genes & Development, 27(4), 450–458. 10.1101/gad.198945.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh SH, Warchol ME, & Ornitz DM (2015). Cochlear progenitor number is controlled through mesenchymal FGF receptor signaling. eLife, 4, 1–17. 10.7554/eLife.05921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui Q, Jin Z, Li X, Liu C, & Wang X (2018). FGF family: From drug development to clinical application. International Journal of Molecular Sciences, 19(7), 1875. 10.3390/ijms19071875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulejova H, Andres Cerezo L, Kuklova M, Pecha O, Vondracek T, Pavelka K, Vencovsky J, Haluzik M, & Senolt L (2012). Novel adipokine fibroblast growth factor 21 is increased in rheumatoid arthritis. Physiological Research, 61(5), 489–494. 10.33549/physiolres.932324 [DOI] [PubMed] [Google Scholar]
- Hung IH, Schoenwolf GC, Lewandoski M, & Ornitz DM (2016). A combined series of Fgf9 and Fgf18 mutant alleles identifies unique and redundant roles in skeletal development. Developmental Biology, 411(1), 72–84. 10.1016/j.ydbio.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung IH, Yu K, Lavine KJ, & Ornitz DM (2007). FGF9 regulates early hypertrophic chondrocyte differentiation and skeletal vascularization in the developing stylopod. Developmental Biology, 307(2), 300–313. 10.1016/j.ydbio.2007.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Zabbarova I, Schaefer CM, Bushnell D, De Groat WC, Kanai A, & Bates CM (2017). Fgfr2 is integral for bladder mesenchyme patterning and function. American Journal of Physiology. Renal Physiology, 312(4), F607–F618. 10.1152/ajprenal.00463.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilieva M, Nielsen TT, Michel T, & Pankratova S (2019). FGF2 and dual agonist of NCAM and FGF receptor 1, Enreptin, rescue neurite outgrowth loss in hippocampal neurons expressing mutated huntingtin proteins. Journal of Neural Transmission (Vienna), 126(11), 1493–1500. 10.1007/s00702-019-02073-1 [DOI] [PubMed] [Google Scholar]
- Imamura K, Tachi K, Takayama T, Shohara R, Kasai H, Dai J, & Yamano S (2018). Released fibroblast growth factor18 from a collagen membrane induces osteoblastic activity involved with downregulation of miR-133a and miR-135a. Journal of Biomaterials Applications, 32(10), 1382–1391. 10.1177/0885328218763318 [DOI] [PubMed] [Google Scholar]
- Imel EA (2021). Burosumab for pediatric X-linked hypophosphatemia. Current Osteoporosis Reports, 19(3), 271–277. 10.1007/s11914-021-00669-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imel EA, Biggin A, Schindeler A, & Munns CF (2019). FGF23, hypophosphatemia, and emerging treatments. JBMR Plus, 3(8), e10190. 10.1002/jbm4.10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imel EA, Liu Z, Coffman M, Acton D, Mehta R, & Econs MJ (2020). Oral iron replacement normalizes fibroblast growth factor 23 in iron-deficient patients with autosomal dominant hypophosphatemic rickets. Journal of Bone and Mineral Research, 35(2), 231–238. 10.1002/jbmr.3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imel EA, Zhang X, Ruppe MD, Weber TJ, Klausner MA, Ito T, Vergeire M, Humphrey JS, Glorieux FH, Portale AA, Insogna K, Peacock M, & Carpenter TO (2015). Prolonged correction of serum phosphorus in adults with X-linked hypophosphatemia using monthly doses of KRN23. The Journal of Clinical Endocrinology and Metabolism, 100(7), 2565–2573. 10.1210/jc.2015-1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insogna KL, Briot K, Imel EA, Kamenicky P, Ruppe MD, Portale AA, Weber T, Pitukcheewanont P, Cheong HI, Jan de Beur S, Imanishi Y, Ito N, Lachmann RH, Tanaka H, Perwad F, Zhang L, Chen CY, Theodore-Oklota C, Mealiffe M, … AXLES 1 Investigators. (2018). A randomized, double-blind, placebo-controlled, phase 3 trial evaluating the efficacy of burosumab, an anti-FGF23 antibody, in adults with X-linked hypophosphatemia: Week 24 primary analysis. Journal of Bone and Mineral Research, 33(8), 1383–1393. 10.1002/jbmr.3475 [DOI] [PubMed] [Google Scholar]
- Iseki S, Wilkie AOM, & Morriss-Kay GM (1999). Fgfr1 and Fgfr2 have distinct differentiation- and proliferation-related roles in the developing mouse skull vault. Development, 126(24), 5611–5620. [DOI] [PubMed] [Google Scholar]
- Ishibe T, Nakayama T, Okamoto T, Aoyama T, Nishijo K, Shibata KR, Shima Y, Nagayama S, Katagiri T, Nakamura Y, Nakamura T, & Toguchida J (2005). Disruption of fibroblast growth factor signal pathway inhibits the growth of synovial sarcomas: Potential application of signal inhibitors to molecular target therapy. Clinical Cancer Research, 11(7), 2702–2712. 10.1158/1078-0432.CCR-04-2057 [DOI] [PubMed] [Google Scholar]
- Ishioka K, Yasuda H, Hamamoto J, Terai H, Emoto K, Kim TJ, Hirose S, Kamatani T, Mimaki S, Arai D, Ohgino K, Tani T, Masuzawa K, Manabe T, Shinozaki T, Mitsuishi A, Ebisudani T, Fukushima T, Ozaki M, … Fukunaga K (2021). Upregulation of FGF9 in lung adenocarcinoma transdifferentiation to small cell lung cancer. Cancer Research, 81(14), 3916–3929. 10.1158/0008-5472.CAN-20-4048 [DOI] [PubMed] [Google Scholar]
- Ishiwata T (2018). Role of fibroblast growth factor receptor-2 splicing in normal and cancer cells. Frontiers in Bioscience-Landmark, 23, 626–639. 10.2741/4609 [DOI] [PubMed] [Google Scholar]
- Ito N, Prideaux M, Wijenayaka AR, Yang D, Ormsby RT, Bonewald LF, & Atkins GJ (2021). Sclerostin directly stimulates osteocyte synthesis of fibroblast growth factor-23. Calcified Tissue International, 109(1), 66–76. 10.1007/s00223-021-00823-6 [DOI] [PubMed] [Google Scholar]
- Itoh N (2016). FGF10: A multifunctional mesenchymal-epithelial signaling growth factor in development, health, and disease. Cytokine & Growth Factor Reviews, 28, 63–69. 10.1016/j.cytogfr.2015.10.001 [DOI] [PubMed] [Google Scholar]
- Itoh N, Nakayama Y, & Konishi M (2016). Roles of FGFs as paracrine or endocrine signals in liver development, health, and disease. Frontiers in Cell and Development Biology, 4, 30. 10.3389/fcell.2016.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Ohta H, & Konishi M (2015). Endocrine FGFs: Evolution, physiology, pathophysiology, and pharmacotherapy. Frontiers in Endocrinology, 6, 154. 10.3389/fendo.2015.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Ohta H, Nakayama Y, & Konishi M (2016). Roles of FGF signals in heart development, health, and disease. Frontiers in Cell and Development Biology, 4, 110. 10.3389/fcell.2016.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, & Ornitz DM (2008). Functional evolutionary history of the mouse Fgf gene family. Developmental Dynamics, 237(1), 18–27. 10.1002/dvdy.21388 [DOI] [PubMed] [Google Scholar]
- Itoh N, & Ornitz DM (2011). Fibroblast growth factors: From molecular evolution to roles in development, metabolism and disease. Journal of Biochemistry, 149(2), 121–130. 10.1093/jb/mvq121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaguirre M, Gil MJ, Monreal I, Montecucco F, Fruhbeck G, & Catalan V (2017). The role and potential therapeutic implications of the fibroblast growth factors in energy balance and type 2 diabetes. Current Diabetes Reports, 17(6), 43. 10.1007/s11892-017-0866-3 [DOI] [PubMed] [Google Scholar]
- Jan de Beur SM, Miller PD, Weber TJ, Peacock M, Insogna K, Kumar R, Rauch F, Luca D, Cimms T, Roberts MS, San Martin J, & Carpenter TO (2021). Burosumab for the treatment of tumor-induced osteomalacia. Journal of Bone and Mineral Research, 36(4), 627–635. 10.1002/jbmr.4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen_Pharmaceuticals. (2019). BALVERSATM (erdafitinib). (Reference ID: 4418725). Retrieved from http://www.accessdata.fda.gov/drugsatfda_docs/label/2019/212018s000lbl.pdf.
- Jazwa A, Kucharzewska P, Leja J, Zagorska A, Sierpniowska A, Stepniewski J, Kozakowska M, Taha H, Ochiya T, Derlacz R, Vahakangas E, Yla-Herttuala S, Jozkowicz A, & Dulak J (2010). Combined vascular endothelial growth factor-A and fibroblast growth factor 4 gene transfer improves wound healing in diabetic mice. Genetic Vaccines and Therapy, 8, 6. 10.1186/1479-0556-8-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen-Cody SO, Flippo KH, Claflin KE, Yavuz Y, Sapouckey SA, Walters GC, Usachev YM, Atasoy D, Gillum MP, & Potthoff MJ (2020). FGF21 signals to glutamatergic neurons in the ventromedial hypothalamus to suppress carbohydrate intake. Cell Metabolism, 32(2), 273–286.e276. 10.1016/j.cmet.2020.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerebtsova M, Das JR, Tang P, Wong E, & Ray PE (2015). Angiopoietin-1 prevents severe bleeding complications induced by heparin-like drugs and fibroblast growth factor-2 in mice. American Journal of Physiology. Heart and Circulatory Physiology, 309(8), H1314–H1325. 10.1152/ajpheart.00373.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez KM, Pereira-Morales AJ, Adan A, Lopez-Leon S, & Forero DA (2018). Depressive symptoms are associated with a functional polymorphism in a miR-433 binding site in the FGF20 gene. Molecular Brain, 11(1), 53. 10.1186/s13041-018-0397-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez V, Jambrina C, Casana E, Sacristan V, Munoz S, Darriba S, Rodo J, Mallol C, Garcia M, Leon X, Marco S, Ribera A, Elias I, Casellas A, Grass I, Elias G, Ferre T, Motas S, Franckhauser S, … Bosch F (2018). FGF21 gene therapy as treatment for obesity and insulin resistance. EMBO Molecular Medicine, 10(8), e8791. 10.15252/emmm.201708791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Pascual A, & Siebzehnrubl FA (2019). Fibroblast growth factor receptor functions in glioblastoma. Cells, 8(7), 715. 10.3390/cells8070715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin BF, Ji ZY, Su ZY, Mei LB, Huang XJ, Lin SB, Li P, & Sha YW (2018). Identification of a novel mutation in FGFR1 gene in patients with Kallmann syndrome by high throughput sequencing. Systems Biology in Reproductive Medicine, 64(3), 202–206. 10.1080/19396368.2018.1458919 [DOI] [PubMed] [Google Scholar]
- Jin H, Quesada C, Aliabouzar M, Kripfgans OD, Franceschi RT, Liu J, Putnam AJ, & Fabiilli ML (2021). Release of basic fibroblast growth factor from acoustically-responsive scaffolds promotes therapeutic angiogenesis in the hind limb ischemia model. Journal of Controlled Release, 338, 773–783. 10.1016/j.jconrel.2021.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Nonaka Y, Miyakawa S, Fujiwara M, & Nakamura Y (2016). Dual therapeutic action of a neutralizing anti-FGF2 aptamer in bone disease and bone cancer pain. Molecular Therapy, 24(11), 1974–1986. 10.1038/mt.2016.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Wu J, Bellusci S, & Zhang JS (2018). Fibroblast growth factor 10 and vertebrate limb development. Frontiers in Genetics, 9, 705. 10.3389/fgene.2018.00705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Yu Y, Qi H, Xie Y, Su N, Wang X, Tan Q, Luo F, Zhu Y, Wang Q, Du X, Xian CJ, Liu P, Huang H, Shen Y, Deng CX, Chen D, & Chen L (2012). A novel FGFR3-binding peptide inhibits FGFR3 signaling and reverses the lethal phenotype of mice mimicking human thanatophoric dysplasia. Human Molecular Genetics, 21(26), 5443–5455. 10.1093/hmg/dds390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing CC, Luo XG, Cui HG, Li FR, Li P, Jiang EZ, Ren Y, & Pang H (2015). Screening of polymorphisms located in the FGF20 and TMEM175 genes in North Chinese Parkinson’s disease patients. Genetics and Molecular Research, 14(4), 13679–13687. 10.4238/2015.October.28.30 [DOI] [PubMed] [Google Scholar]
- Joannes A, Brayer S, Besnard V, Marchal-Somme J, Jaillet M, Mordant P, Mal H, Borie R, Crestani B, & Mailleux AA (2016). FGF9 and FGF18 in idiopathic pulmonary fibrosis promote survival and migration and inhibit myofibroblast differentiation of human lung fibroblasts in vitro. American Journal of Physiology. Lung Cellular and Molecular Physiology, 310(7), L615–L629. 10.1152/ajplung.00185.2015 [DOI] [PubMed] [Google Scholar]
- Johnson K, Levine K, Sergi J, Chamoun J, Roach R, Vekich J, Favis M, Horn M, Cao X, Miller B, Snyder W, Aivazian D, Reagan W, Berryman E, Colangelo J, Markiewicz V, Bagi CM, Brown TP, Coyle A, … Magram J (2017). Therapeutic effects of FGF23 c-tail Fc in a murine preclinical model of X-linked hypophosphatemia via the selective modulation of phosphate reabsorption. Journal of Bone and Mineral Research, 32(10), 2062–2073. 10.1002/jbmr.3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MLM, Sarila G, Chapuis P, Hutson JM, King SK, & Teague WJ (2020). The role of fibroblast growth factor 10 signaling in duodenal atresia. Frontiers in Pharmacology, 11, 250. 10.3389/fphar.2020.00250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Dilai S, Lingampally A, Chao CM, Danopoulos S, Carraro G, Mukhametshina R, Wilhelm J, Baumgart-Vogt E, Al Alam D, Chen C, Minoo P, Zhang JS, & Bellusci S (2018). A comprehensive analysis of fibroblast growth factor receptor 2b signaling on epithelial tip progenitor cells during early mouse lung branching morphogenesis. Frontiers in Genetics, 9, 746. 10.3389/fgene.2018.00746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Lingampally A, Dilai S, Shrestha A, Stripp B, Helmbacher F, Chen C, Chao CM, & Bellusci S (2019). Characterization of Tg(Etv4-GFP) and Etv5 (RFP) reporter lines in the context of fibroblast growth factor 10 signaling during mouse embryonic lung development. Frontiers in Genetics, 10, 178. 10.3389/fgene.2019.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MR, Lingampally A, Wu J, Sedighi J, Ahmadvand N, Wilhelm J, Vazquez-Armendariz AI, Herold S, Chen C, Zhang JS, Bellusci S, & Chao CM (2020). Evidence for overlapping and distinct biological activities and transcriptional targets triggered by fibroblast growth factor receptor 2b signaling between mid- and early pseudoglandular stages of mouse lung development. Cells, 9(5), 1274. 10.3390/cells9051274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi JJ, Coffey H, Corcoran E, Tsai J, Huang CL, Ichikawa K, Prajapati S, Hao MH, Bailey S, Wu J, Rimkunas V, Karr C, Subramanian V, Kumar P, MacKenzie C, Hurley R, Satoh T, Yu K, Park E, … Selvaraj A (2017). H3B-6527 is a potent and selective inhibitor of FGFR4 in FGF19-driven hepatocellular carcinoma. Cancer Research, 77(24), 6999–7013. 10.1158/0008-5472.CAN-17-1865 [DOI] [PubMed] [Google Scholar]
- Julien A, Perrin S, Duchamp de Lageneste O, Carvalho C, Bensidhoum M, Legeai-Mallet L, & Colnot C (2020). FGFR3 in periosteal cells drives cartilage-to-bone transformation in bone repair. Stem Cell Reports, 15(4), 955–967. 10.1016/j.stemcr.2020.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SH, Lee HC, Yu DM, Kim BC, Park SM, Lee YS, Park HJ, Ko YG, & Lee JS (2016). Heparan sulfation is essential for the prevention of cellular senescence. Cell Death and Differentiation, 23(3), 417–429. 10.1038/cdd.2015.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurek PM, Zablocki K, Wasko U, Mazurek MP, Otlewski J, & Jelen F (2017). Anti-FGFR1 aptamer-tagged superparamagnetic conjugates for anticancer hyperthermia therapy. International Journal of Nanomedicine, 12, 2941–2950. 10.2147/IJN.S125231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justet A, Joannes A, Besnard V, Marchal-Somme J, Jaillet M, Bonniaud P, Sallenave JM, Solhonne B, Castier Y, Mordant P, Mal H, Cazes A, Borie R, Mailleux AA, & Crestani B (2017). FGF9 prevents pleural fibrosis induced by intrapleural adenovirus injection in mice. American Journal of Physiology. Lung Cellular and Molecular Physiology, 313(5), L781–L795. 10.1152/ajplung.00508.2016 [DOI] [PubMed] [Google Scholar]
- Kaiser M, Wojahn I, Rudat C, Ludtke TH, Christoffels VM, Moon A, Kispert A, & Trowe MO (2021). Regulation of otocyst patterning by Tbx2 and Tbx3 is required for inner ear morphogenesis in the mouse. Development, 148(8), dev195651. 10.1242/dev.195651 [DOI] [PubMed] [Google Scholar]
- Kakoty V, Sarathlal KC, Tang RD, Yang CH, Dubey SK, & Taliyan R (2020). Fibroblast growth factor 21 and autophagy: A complex interplay in Parkinson disease. Biomedicine & Pharmacotherapy, 127, 110145. 10.1016/j.biopha.2020.110145 [DOI] [PubMed] [Google Scholar]
- Kalinina J, Byron SA, Makarenkova HP, Olsen SK, Eliseenkova AV, Larochelle WJ, Dhanabal M, Blais S, Ornitz DM, Day LA, Neubert TA, Pollock PM, & Mohammadi M (2009). Homodimerization controls the fibroblast growth factor 9 subfamily’s receptor binding and heparan sulfate-dependent diffusion in the extracellular matrix. Molecular and Cellular Biology, 29(17), 4663–4678. 10.1128/MCB.01780-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyukina M, Yosaatmadja Y, Middleditch MJ, Patterson AV, Smaill JB, & Squire CJ (2019). TAS-120 cancer target binding: Defining reactivity and revealing the first fibroblast growth factor receptor 1 (FGFR1) irreversible structure. ChemMedChem, 14(4), 494–500. 10.1002/cmdc.201800719 [DOI] [PubMed] [Google Scholar]
- Kamali S, Rajendran R, Stadelmann C, Karnati S, Rajendran V, Giraldo-Velasquez M, & Berghoff M (2021). Oligodendrocyte-specific deletion of FGFR2 ameliorates MOG35–55 -induced EAE through ERK and Akt signalling. Brain Pathology, 31(2), 297–311. 10.1111/bpa.12916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath AV, Lu D, Gupta P, Jin D, Xin Y, Brady A, Stephan JP, Li H, Tien J, Qing J, & Damico-Beyer LA (2012). Preclinical pharmacokinetics of MFGR1877A, a human monoclonal antibody to FGFR3, and prediction of its efficacious clinical dose for the treatment of t(4;14)-positive multiple myeloma. Cancer Chemotherapy and Pharmacology, 69(4), 1071–1078. 10.1007/s00280-011-1807-5 [DOI] [PubMed] [Google Scholar]
- Kamatkar N, Levy M, & Hebert JM (2019). Development of a monomeric inhibitory RNA aptamer specific for FGFR3 that acts as an activator when dimerized. Molecular Therapy. Nucleic Acids, 17, 530–539. 10.1016/j.omtn.2019.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C (2021). Infigratinib: First approval. Drugs, 81(11), 1355–1360. 10.1007/s40265-021-01567-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Jung SH, Lee GH, Lee S, Park HJ, Ko YG, Kim YN, & Lee JS (2020). Sulfated syndecan 1 is critical to preventing cellular senescence by modulating fibroblast growth factor receptor endocytosis. The FASEB Journal, 34(8), 10316–10328. 10.1096/fj.201902714R [DOI] [PubMed] [Google Scholar]
- Kang M, Garg V, & Hadjantonakis AK (2017). Lineage establishment and progression within the inner cell mass of the mouse blastocyst requires FGFR1 and FGFR2. Developmental Cell, 41(5), 496–510.e495. 10.1016/j.devcel.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M, Piliszek A, Artus J, & Hadjantonakis AK (2013). FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development, 140(2), 267–279. 10.1242/dev.084996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Lee SE, Choi MJ, Chang JY, Kim JT, Zhang BY, Kang YE, Lee JH, Yi HS, & Shong M (2021). Th2 cytokines increase the expression of fibroblast growth factor 21 in the liver. Cells, 10(6), 1298. 10.3390/cells10061298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W, Nguyen KCQ, & Hebert JM (2019). Transient redirection of SVZ stem cells to oligodendrogenesis by FGFR3 activation promotes remyelination. Stem Cell Reports, 12(6), 1223–1231. 10.1016/j.stemcr.2019.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia RM, Guntur AR, Reinhold MI, & Naski MC (2005). Glycogen synthase kinase 3 controls endochondral bone development: Contribution of fibroblast growth factor 18. Developmental Biology, 285(2), 496–507. 10.1016/j.ydbio.2005.07.029 [DOI] [PubMed] [Google Scholar]
- Kardami E, Detillieux K, Ma X, Jiang Z, Santiago JJ, Jimenez SK, & Cattini PA (2007). Fibroblast growth factor-2 and cardioprotection. Heart Failure Reviews, 12(3–4), 267–277. 10.1007/s10741-007-9027-0 [DOI] [PubMed] [Google Scholar]
- Kardoust Parizi M, Margulis V, Lotan Y, Mori K, & Shariat SF (2021). Fibroblast growth factor receptor: A systematic review and meta-analysis of prognostic value and therapeutic options in patients with urothelial bladder carcinoma. Urologic Oncology, 39(7), 409–421. 10.1016/j.urolonc.2021.01.025 [DOI] [PubMed] [Google Scholar]
- Karolak JA, Vincent M, Deutsch G, Gambin T, Cogne B, Pichon O, Vetrini F, Mefford HC, Dines JN, Golden-Grant K, Dipple K, Freed AS, Leppig KA, Dishop M, Mowat D, Bennetts B, Gifford AJ, Weber MA, Lee AF, … Stankiewicz P (2019). Complex compound inheritance of lethal lung developmental disorders due to disruption of the TBX-FGF pathway. American Journal of Human Genetics, 104(2), 213–228. 10.1016/j.ajhg.2018.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karuppaiah K, Yu K, Lim J, Chen J, Smith C, Long F, & Ornitz DM (2016). FGF signaling in the osteoprogenitor lineage non-autonomously regulates postnatal chondrocyte proliferation and skeletal growth. Development, 143(10), 1811–1822. 10.1242/dev.131722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashpur O, LaPointe D, Ambady S, Ryder EF, & Dominko T (2013). FGF2-induced effects on transcriptome associated with regeneration competence in adult human fibroblasts. BMC Genomics, 14, 656. 10.1186/1471-2164-14-656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuya A, Ito T, & Tokura Y (2018). M2 macrophages promote wound-induced hair neogenesis. Journal of Dermatological Science, 91(3), 250–255. 10.1016/j.jdermsci.2018.05.004 [DOI] [PubMed] [Google Scholar]
- Katoh M (2016). FGFR inhibitors: Effects on cancer cells, tumor microenvironment and whole-body homeostasis (Review). International Journal of Molecular Medicine, 38(1), 3–15. 10.3892/ijmm.2016.2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M, & Nakagama H (2014). FGF receptors: Cancer biology and therapeutics. Medicinal Research Reviews, 34(2), 280–300. 10.1002/med.21288 [DOI] [PubMed] [Google Scholar]
- Kaufman A, Abuqayyas L, Denney WS, Tillman EJ, & Rolph T (2020). AKR-001, an Fc-FGF21 analog, showed sustained pharmaco-dynamic effects on insulin sensitivity and lipid metabolism in type 2 diabetes patients. Cell Reports Medicine, 1(4), 100057. 10.1016/j.xcrm.2020.100057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Herrmann D, Fuchs A, Cheng S, Ferrer-Vaquer A, Gotz R, Driller K, Neubuser A, & Ohura K (2019). Fgfr1 conditional-knockout in neural crest cells induces heterotopic chondrogenesis and osteogenesis in mouse frontal bones. Medical Molecular Morphology, 52(3), 156–163. 10.1007/s00795-018-0213-z [DOI] [PubMed] [Google Scholar]
- Kawara A, Mizuta R, Fujisawa M, Ito T, Li C, Nakamura K, Sun C, Kuwabara M, Kitabatake M, Yoshimura T, & Matsukawa A (2020). Spred2-deficiency enhances the proliferation of lung epithelial cells and alleviates pulmonary fibrosis induced by bleomycin. Scientific Reports, 10(1), 16490. 10.1038/s41598-020-73752-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazoe T, & Taniguchi K (2019). The sprouty/spred family as tumor suppressors: Coming of age. Cancer Science, 110(5), 1525–1535. 10.1111/cas.13999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil S, Gupta M, Brand M, & Knopf F (2021). Heparan sulfate proteoglycan expression in the regenerating zebrafish fin. Developmental Dynamics, 250(9), 1368–1380. 10.1002/dvdy.321 [DOI] [PubMed] [Google Scholar]
- Keipert S, Ost M, Johann K, Imber F, Jastroch M, van Schothorst EM, Keijer J, & Klaus S (2014). Skeletal muscle mitochondrial uncoupling drives endocrine cross-talk through the induction of FGF21 as a myokine. American Journal of Physiology. Endocrinology and Metabolism, 306(5), E469–E482. 10.1152/ajpendo.00330.2013 [DOI] [PubMed] [Google Scholar]
- Kelly CM, Shoushtari AN, Qin LX, D’Angelo SP, Dickson MA, Gounder MM, Keohan ML, McFadyen C, Sjoberg A, Singer S, DeMatteo RP, Hwang S, Heinemann MH, Francis JH, Antonescu CR, Chi P, & Tap WD (2019). A phase Ib study of BGJ398, a pan-FGFR kinase inhibitor in combination with imatinib in patients with advanced gastrointestinal stromal tumor. Investigational New Drugs, 37(2), 282–290. 10.1007/s10637-018-0648-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet G, & Benvenisty N (2021). Large-scale analysis of imprinting in naive human pluripotent stem cells reveals recurrent aberrations and a potential link to FGF signaling. Stem Cell Reports, 16(10), 2520–2533. 10.1016/j.stemcr.2021.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes BE, & Fuchs E (2018). Stem cells: Aging and transcriptional fingerprints. The Journal of Cell Biology, 217(1), 79–92. 10.1083/jcb.201708099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitonenkov A, Shiyanova TL, Koester A, Ford AM, Micanovic R, Galbreath EJ, Sandusky GE, Hammond LJ, Moyers JS, Owens RA, Gromada J, Brozinick JT, Hawkins ED, Wroblewski VJ, Li DS, Mehrbod F, Jaskunas SR, & Shanafelt AB (2005). FGF-21 as a novel metabolic regulator. Journal of Clinical Investigation, 115(6), 1627–1635. 10.1172/JCI23606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitonenkov A, Wroblewski VJ, Koester A, Chen YF, Clutinger CK, Tigno XT, Hansen BC, Shanafelt AB, & Etgen GJ (2007). The metabolic state of diabetic monkeys is regulated by fibroblast growth factor-21. Endocrinology, 148(2), 774–781. 10.1210/en.2006-1168 [DOI] [PubMed] [Google Scholar]
- Khosravi F, Ahmadvand N, Bellusci S, & Sauer H (2021). The multifunctional contribution of FGF signaling to cardiac development, homeostasis, disease and repair. Frontiers in Cell and Development Biology, 9, 672935. 10.3389/fcell.2021.672935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwai F, Mui BWH, Arora D, Iqbal K, Hockaday M, de Castro Diaz LF, Cherman N, Martin D, Myneni VD, Ahmad M, Futrega K, Ali S, Merling RK, Kaufman DS, Lee J, & Robey PG (2020). Lineage-specific differentiation of osteogenic progenitors from pluripotent stem cells reveals the FGF1-RUNX2 association in neural crest-derived osteoprogenitors. Stem Cells, 38(9), 1107–1123. 10.1002/stem.3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A, Suzuki T, Nakazawa T, Iizuka M, Nakayama A, Ozawa T, Kameda M, Shindoh N, Terasaka T, Hirano M, & Kuromitsu S (2017). ASP5878, a selective FGFR inhibitor, to treat FGFR3-dependent urothelial cancer with or without chemoresistance. Cancer Science, 108(2), 236–242. 10.1111/cas.13124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny DM, & Rocheleau JV (2016). The FGF21 receptor signaling complex: Klothobeta, FGFR1c, and other regulatory interactions. Vitamins and Hormones, 101, 17–58. 10.1016/bs.vh.2016.02.008 [DOI] [PubMed] [Google Scholar]
- Kim BS, Shin HR, Kim HJ, Yoon H, Cho YD, Choi KY, Choi JY, Kim WJ, & Ryoo HM (2021). Septal chondrocyte hypertrophy contributes to midface deformity in a mouse model of Apert syndrome. Scientific Reports, 11(1), 7979. 10.1038/s41598-021-87260-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CS, Joe Y, Choi HS, Back SH, Park JW, Chung HT, Roh E, Kim MS, Ha TY, & Yu R (2019). Deficiency of fibroblast growth factor 21 aggravates obesity-induced atrophic responses in skeletal muscle. Journal of Inflammation, 16, 17. 10.1186/s12950-019-0221-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EK, Lee SH, Lee SY, Kim JK, Jhun JY, Na HS, Kim SY, Choi JY, Yang CW, Park SH, & Cho ML (2018). Met-formin ameliorates experimental-obesity-associated autoimmune arthritis by inducing FGF21 expression and brown adipocyte differentiation. Experimental & Molecular Medicine, 50(1), e432. 10.1038/emm.2017.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Lee DH, Kim JK, Choi HS, Dwivedi B, Rupji M, Kowalski J, Green SJ, Song H, Park WJ, Chang JY, Kim TM, & Park C (2019). ETV2/ER71 regulates the generation of FLK1(+) cells from mouse embryonic stem cells through miR-126-MAPK signaling. Stem Cell Research & Therapy, 10(1), 328. 10.1186/s13287-019-1466-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Gibboney S, Razy-Krajka F, Lowe EK, Wang W, & Stolfi A (2020). Regulation of neurogenesis by FGF signaling and neurogenin in the invertebrate chordate ciona. Frontiers in Cell and Development Biology, 8, 477. 10.3389/fcell.2020.00477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Kim JM, & Heo WD (2016). Optogenetic control of fibroblast growth factor receptor signaling. Methods in Molecular Biology, 1408, 345–362. 10.1007/978-1-4939-3512-3_24 [DOI] [PubMed] [Google Scholar]
- Kim N, Kim JM, Lee M, Kim CY, Chang KY, & Heo WD (2014). Spatiotemporal control of fibroblast growth factor receptor signals by blue light. Chemistry & Biology, 21(7), 903–912. 10.1016/j.chembiol.2014.05.013 [DOI] [PubMed] [Google Scholar]
- Kim RD, Sarker D, Meyer T, Yau T, Macarulla T, Park JW, Choo SP, Hollebecque A, Sung MW, Lim HY, Mazzaferro V, Trojan J, Zhu AX, Yoon JH, Sharma S, Lin ZZ, Chan SL, Faivre S, Feun LG, … Kang YK (2019). First-in-human phase I study of fisogatinib (BLU-554) validates aberrant FGF19 signaling as a driver event in hepatocellular carcinoma. Cancer Discovery, 9(12), 1696–1707. 10.1158/2159-8290.CD-19-0555 [DOI] [PubMed] [Google Scholar]
- Kim SB, Meric-Bernstam F, Kalyan A, Babich A, Liu R, Tanigawa T, Sommer A, Osada M, Reetz F, Laurent D, Wittemer-Rump S, & Berlin J (2019). First-in-human phase I study of aprutumab ixadotin, a fibroblast growth factor receptor 2 antibody-drug conjugate (BAY 1187982) in patients with advanced cancer. Targeted Oncology, 14(5), 591–601. 10.1007/s11523-019-00670-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Lee IK, Rom E, Sirkis R, Park SH, Park JO, Park YS, Lim HY, Kang WK, Kim KM, Yayon A, & Lee J (2019). Neutralizing antibody to FGFR2 can act as a selective biomarker and potential therapeutic agent for gastric cancer with FGFR2 amplification. American Journal of Translational Research, 11(7), 4508–4515. [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Seok S, Zhang Y, Ma J, Kong B, Guo G, Kemper B, & Kemper JK (2020). Intestinal FGF15/19 physiologically repress hepatic lipogenesis in the late fed-state by activating SHP and DNMT3A. Nature Communications, 11(1), 5969. 10.1038/s41467-020-19803-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Hong G, Kim DH, Kim YM, Kim YK, Oh YM, & Jee YK (2018). The role of FGF-2 in smoke-induced emphysema and the therapeutic potential of recombinant FGF-2 in patients with COPD. Experimental & Molecular Medicine, 50(11), 1–10. 10.1038/s12276-018-0178-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Bosakova M, Nonaka Y, Hruba E, Yasuda K, Futakawa S, Kubota T, Fafilek B, Gregor T, Abraham SP, Gomolkova R, Belaskova S, Pesl M, Csukasi F, Duran I, Fujiwara M, Kavkova M, Zikmund T, Kaiser J, … Krejci P (2021). An RNA aptamer restores defective bone growth in FGFR3-related skeletal dysplasia in mice. Science Translational Medicine, 13(592), eaba4226. 10.1126/scitranslmed.aba4226 [DOI] [PubMed] [Google Scholar]
- Kirchgeorg L, Felker A, van Oostrom M, Chiavacci E, & Mosimann C (2018). Cre/lox-controlled spatiotemporal perturbation of FGF signaling in zebrafish. Developmental Dynamics, 247(10), 1146–1159. 10.1002/dvdy.24668 [DOI] [PubMed] [Google Scholar]
- Kiselyov VV, Skladchikova G, Hinsby AM, Jensen PH, Kulahin N, Soroka V, Pedersen N, Tsetlin V, Poulsen FM, Berezin V, & Bock E (2003). Structural basis for a direct interaction between FGFR1 and NCAM and evidence for a regulatory role of ATP. Structure, 11(6), 691–701. 10.1016/s0969-2126(03)00096-0 [DOI] [PubMed] [Google Scholar]
- Kiselyov VV, Soroka V, Berezin V, & Bock E (2005). Structural biology of NCAM homophilic binding and activation of FGFR. Journal of Neurochemistry, 94(5), 1169–1179. 10.1111/j.1471-4159.2005.03284.x [DOI] [PubMed] [Google Scholar]
- Kiyokawa H, & Morimoto M (2021). Molecular crosstalk in tracheal development and its recurrence in adult tissue regeneration. Developmental Dynamics, 250(11), 1552–1567. 10.1002/dvdy.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klag KA, & Horton WA (2016). Advances in treatment of achondroplasia and osteoarthritis. Human Molecular Genetics, 25(R1), R2–R8. 10.1093/hmg/ddv419 [DOI] [PubMed] [Google Scholar]
- Kleiderman S, Gutbier S, Ugur Tufekci K, Ortega F, Sa JV, Teixeira AP, Brito C, Glaab E, Berninger B, Alves PM, & Leist M (2016). Conversion of nonproliferating astrocytes into neurogenic neural stem cells: Control by FGF2 and interferon-gamma. Stem Cells, 34(12), 2861–2874. 10.1002/stem.2483 [DOI] [PubMed] [Google Scholar]
- Klein Hazebroek M, & Keipert S (2020). Adapting to the cold: A role for endogenous fibroblast growth factor 21 in thermoregulation? Frontiers in Endocrinology, 11, 389. 10.3389/fendo.2020.00389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, & Mangelsdorf DJ (2019). A dozen years of discovery: Insights into the physiology and pharmacology of FGF21. Cell Metabolism, 29(2), 246–253. 10.1016/j.cmet.2019.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingbeil K, Nguyen TQ, Fahrner A, Guthmann C, Wang H, Schoels M, Lilienkamp M, Franz H, Eckert P, Walz G, & Yakulov TA (2021). Corpuscles of Stannius development requires FGF signaling. Developmental Biology, 481, 160–171. 10.1016/j.ydbio.2021.10.005 [DOI] [PubMed] [Google Scholar]
- Knosp WM, Knox SM, Lombaert IM, Haddox CL, Patel VN, & Hoffman MP (2015). Submandibular parasympathetic gangliogenesis requires sprouty-dependent Wnt signals from epithelial progenitors. Developmental Cell, 32(6), 667–677. 10.1016/j.devcel.2015.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko FC, & Sumner DR (2021). How faithfully does intramembranous bone regeneration recapitulate embryonic skeletal development? Developmental Dynamics, 250(3), 377–392. 10.1002/dvdy.240 [DOI] [PubMed] [Google Scholar]
- Koike Y, Yozaki M, Utani A, & Murota H (2020). Fibroblast growth factor 2 accelerates the epithelial-mesenchymal transition in keratinocytes during wound healing process. Scientific Reports, 10(1), 18545. 10.1038/s41598-020-75584-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleini N, Nickel BE, Edel AL, Fandrich RR, Ravandi A, & Kardami E (2018). Non-mitogenic FGF2 protects cardiomyocytes from acute doxorubicin-induced toxicity independently of the protein kinase CK2/heme oxygenase-1 pathway. Cell and Tissue Research, 374(3), 607–617. 10.1007/s00441-018-2905-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleini N, Nickel BE, Nagalingam RS, Landry NM, Fandrich RR, Cheung DYC, Dixon IM, Czubryt MP, Jassal DS, Cattini PA, & Kardami E (2021). Elimination of endogenous high molecular weight FGF2 prevents pressure-overload-induced systolic dysfunction, linked to increased FGFR1 activity and NR1D1 expression. Cell and Tissue Research, 385(3), 753–768. 10.1007/s00441-021-03465-0 [DOI] [PubMed] [Google Scholar]
- Kolumam G, Chen MZ, Tong R, Zavala-Solorio J, Kates L, van Bruggen N, Ross J, Wyatt SK, Gandham VD, Carano RA, Dunshee DR, Wu AL, Haley B, Anderson K, Warming S, Rairdan XY, Lewin-Koh N, Zhang Y, Gutierrez J, … Sonoda J (2015). Sustained brown fat stimulation and insulin sensitization by a humanized bispecific antibody agonist for fibroblast growth factor receptor 1/betaKlotho complex. eBioMedicine, 2(7), 730–743. 10.1016/j.ebiom.2015.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komla-Ebri D, Dambroise E, Kramer I, Benoist-Lasselin C, Kaci N, Le Gall C, Martin L, Busca P, Barbault F, Graus-Porta D, Munnich A, Kneissel M, Di Rocco F, Biosse-Duplan M, & Legeai-Mallet L (2016). Tyrosine kinase inhibitor NVP-BGJ398 functionally improves FGFR3-related dwarfism in mouse model. Journal of Clinical Investigation, 126(5), 1871–1884. 10.1172/JCI83926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kommalapati A, Tella SH, Borad M, Javle M, & Mahipal A (2021). FGFR inhibitors in oncology: Insight on the management of toxicities in clinical practice. Cancers (Basel), 13(12), 2968. 10.3390/cancers13122968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon E, Calvo-Jimenez E, Cossard A, Na Y, Cooper JA, & Jossin Y (2019). N-cadherin-regulated FGFR ubiquitination and degradation control mammalian neocortical projection neuron migration. eLife, 8, e47673. 10.7554/eLife.47673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig D, Page L, Chassot B, & Jazwinska A (2018). Dynamics of actinotrichia regeneration in the adult zebrafish fin. Developmental Biology, 433(2), 416–432. 10.1016/j.ydbio.2017.07.024 [DOI] [PubMed] [Google Scholar]
- Koo HY, El-Baz LM, House S, Cilvik SN, Dorry SJ, Shoukry NM, Salem ML, Hafez HS, Dulin NO, Ornitz DM, & Guzy RD (2018). Fibroblast growth factor 2 decreases bleomycin-induced pulmonary fibrosis and inhibits fibroblast collagen production and myofibroblast differentiation. The Journal of Pathology, 246(1), 54–66. 10.1002/path.5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, & Volkow ND (2016). Neurobiology of addiction: A neurocircuitry analysis. Lancet Psychiatry, 3(8), 760–773. 10.1016/S2215-0366(16)00104-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsensky L, & Ron D (2016). Regulation of FGF signaling: Recent insights from studying positive and negative modulators. Seminars in Cell & Developmental Biology, 53, 101–114. 10.1016/j.semcdb.2016.01.023 [DOI] [PubMed] [Google Scholar]
- Kostas M, Haugsten EM, Zhen Y, Sorensen V, Szybowska P, Fiorito E, Lorenz S, Jones N, de Souza GA, Wiedlocha A, & Wesche J (2018). Protein tyrosine phosphatase receptor type G (PTPRG) controls fibroblast growth factor receptor (FGFR) 1 activity and influences sensitivity to FGFR kinase inhibitors. Molecular & Cellular Proteomics, 17(5), 850–870. 10.1074/mcp.RA117.000538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostas M, Lampart A, Bober J, Wiedlocha A, Tomala J, Krowarsch D, Otlewski J, & Zakrzewska M (2018). Translocation of exogenous FGF1 and FGF2 protects the cell against apoptosis independently of receptor activation. Journal of Molecular Biology, 430(21), 4087–4101. 10.1016/j.jmb.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Kowalkowski A, Zaremba KM, Rogers AP, Hoffman OR, Turco AE, & Nichol PF (2020). Lack of discreet colocalization of epithelial apoptosis to the atretic precursor in the colon of the Fibroblast growth factor receptor 2IIIb mouse and staining consistent with cellular movement suggest a revised model of atresia formation. Developmental Dynamics, 249(6), 741–753. 10.1002/dvdy.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozyraki R, & Cases O (2020). Cubilin, the intrinsic factor-vitamin B12 receptor in development and disease. Current Medicinal Chemistry, 27(19), 3123–3150. 10.2174/0929867325666181008143945 [DOI] [PubMed] [Google Scholar]
- Krause M, Rak-Raszewska A, Pietila I, Quaggin SE, & Vainio S (2015). Signaling during kidney development. Cells, 4(2), 112–132. 10.3390/cells4020112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawchuk D, Honma-Yamanaka N, Anani S, & Yamanaka Y (2013). FGF4 is a limiting factor controlling the proportions of primitive endoderm and epiblast in the ICM of the mouse blastocyst. Developmental Biology, 384(1), 65–71. 10.1016/j.ydbio.2013.09.023 [DOI] [PubMed] [Google Scholar]
- Krejci E, Pesevski Z, Nanka O, & Sedmera D (2016). Physiological role of FGF signaling in growth and remodeling of developing cardiovascular system. Physiological Research, 65(3), 425–435. 10.33549/physiolres.933216 [DOI] [PubMed] [Google Scholar]
- Krejci P, Salazar L, Goodridge HS, Kashiwada TA, Schibler MJ, Jelinkova P, Thompson LM, & Wilcox WR (2008). STAT1 and STAT3 do not participate in FGF-mediated growth arrest in chondrocytes. Journal of Cell Science, 121(Pt 3), 272–281. 10.1242/jcs.017160 [DOI] [PubMed] [Google Scholar]
- Krick S, Grabner A, Baumlin N, Yanucil C, Helton S, Grosche A, Sailland J, Geraghty P, Viera L, Russell DW, Wells JM, Xu X, Gaggar A, Barnes J, King GD, Campos M, Faul C, & Salathe M (2018). Fibroblast growth factor 23 and Klotho contribute to airway inflammation. The European Respiratory Journal, 52(1), 1–13. 10.1183/13993003.00236-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy VV, Fu J, Oh TJ, Khamo J, Yang J, & Zhang K (2020). A generalizable optogenetic strategy to regulate receptor tyrosine kinases during vertebrate embryonic development. Journal of Molecular Biology, 432(10), 3149–3158. 10.1016/j.jmb.2020.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krook MA, Reeser JW, Ernst G, Barker H, Wilberding M, Li G, Chen HZ, & Roychowdhury S (2021). Fibroblast growth factor receptors in cancer: Genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance. British Journal of Cancer, 124(5), 880–892. 10.1038/s41416-020-01157-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucinska M, Porebska N, Lampart A, Latko M, Knapik A, Zakrzewska M, Otlewski J, & Opalinski L (2019). Differential regulation of fibroblast growth factor receptor 1 trafficking and function by extracellular galectins. Cell Communication and Signaling, 17(1), 65. 10.1186/s12964-019-0371-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Donakonda S, Muller SA, Botzel K, Hoglinger GU, & Koeglsperger T (2020). FGF2 affects Parkinson’s disease-associated molecular networks through exosomal Rab8b/Rab31. Frontiers in Genetics, 11, 572058. 10.3389/fgene.2020.572058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Cunningham TJ, & Duester G (2016). Nuclear receptor corepressors Ncor1 and Ncor2 (Smrt) are required for retinoic acid-dependent repression of Fgf8 during somitogenesis. Developmental Biology, 418(1), 204–215. 10.1016/j.ydbio.2016.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, & Duester G (2014). Retinoic acid controls body axis extension by directly repressing Fgf8 transcription. Development, 141(15), 2972–2977. 10.1242/dev.112367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunova Bosakova M, Varecha M, Hampl M, Duran I, Nita A, Buchtova M, Dosedelova H, Machat R, Xie Y, Ni Z, Martin JH, Chen L, Jansen G, Krakow D, & Krejci P (2018). Regulation of ciliary function by fibroblast growth factor signaling identifies FGFR3-related disorders achondroplasia and thanatophoric dysplasia as ciliopathies. Human Molecular Genetics, 27(6), 1093–1105. 10.1093/hmg/ddy031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuro-o M (2019). The Klotho proteins in health and disease. Nature Reviews. Nephrology, 15(1), 27–44. 10.1038/s41581-018-0078-3 [DOI] [PubMed] [Google Scholar]
- Kuroda M, Muramatsu R, Maedera N, Koyama Y, Hamaguchi M, Fujimura H, Yoshida M, Konishi M, Itoh N, Mochizuki H, & Yamashita T (2017). Peripherally derived FGF21 promotes remyelination in the central nervous system. Journal of Clinical Investigation, 127(9), 3496–3509. 10.1172/JCI94337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowski A, Molotkov A, & Soriano P (2019). FGFR1 regulates trophectoderm development and facilitates blastocyst implantation. Developmental Biology, 446(1), 94–101. 10.1016/j.ydbio.2018.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutilek S (2017). Burosumab: A new drug to treat hypophosphatemic rickets. Sudanese Journal of Paediatrics, 17(2), 71–73. 10.24911/SJP.2017.2.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Venuta G, Zeitler M, Steringer JP, Muller HM, & Nickel W (2015). The startling properties of fibroblast growth factor 2: How to exit mammalian cells without a signal peptide at hand. The Journal of Biological Chemistry, 290(45), 27015–27020. 10.1074/jbc.R115.689257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeger T, Baumeier C, Wilhelmi I, Wurfel J, Kamitz A, & Schurmann A (2017). FGF21 improves glucose homeostasis in an obese diabetes-prone mouse model independent of body fat changes. Diabetologia, 60(11), 2274–2284. 10.1007/s00125-017-4389-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laezza F, Gerber BR, Lou JY, Kozel MA, Hartman H, Craig AM, Ornitz DM, & Nerbonne JM (2007). The FGF14(F145S) mutation disrupts the interaction of FGF14 with voltage-gated Na+ channels and impairs neuronal excitability. The Journal of Neuroscience, 27(44), 12033–12044. 10.1523/JNEUROSCI.2282-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagha M, Sato T, Bajard L, Daubas P, Esner M, Montarras D, Relaix F, & Buckingham M (2008). Regulation of skeletal muscle stem cell behavior by Pax3 and Pax7. Cold Spring Harbor Symposia on Quantitative Biology, 73, 307–315. 10.1101/sqb.2008.73.006 [DOI] [PubMed] [Google Scholar]
- Lam AS, Liu CC, Deutsch GH, Rivera J, Perkins JA, Holmes G, Jabs EW, Cunningham ML, & Dahl JP (2021). Genotype-phenotype correlation of tracheal cartilaginous sleeves and Fgfr2 mutations in mice. Laryngoscope, 131(4), E1349–E1356. 10.1002/lary.29060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb YN (2018). Burosumab: First global approval. Drugs, 78(6), 707–714. 10.1007/s40265-018-0905-7 [DOI] [PubMed] [Google Scholar]
- Lamballe F, Genestine M, Caruso N, Arce V, Richelme S, Helmbacher F, & Maina F (2011). Pool-specific regulation of motor neuron survival by neurotrophic support. The Journal of Neuroscience, 31(31), 11144–11158. 10.1523/JNEUROSCI.2198-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamus F, Martin C, Carnicero E, Moro JA, Fernandez JMF, Mano A, Gato A, & Alonso MI (2020). FGF2/EGF contributes to brain neuroepithelial precursor proliferation and neurogenesis in rat embryos: The involvement of embryonic cerebrospinal fluid. Developmental Dynamics, 249(1), 141–153. 10.1002/dvdy.135 [DOI] [PubMed] [Google Scholar]
- Lan T, Morgan DA, Rahmouni K, Sonoda J, Fu X, Burgess SC, Holland WL, Kliewer SA, & Mangelsdorf DJ (2017). FGF19, FGF21, and an FGFR1/beta-Klotho-activating antibody act on the nervous system to regulate body weight and glycemia. Cell Metabolism, 26(5), 709–718.e703. 10.1016/j.cmet.2017.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander R, & Petersen CP (2016). Wnt, Ptk7, and FGFRL expression gradients control trunk positional identity in planarian regeneration. eLife, 5, e12850. 10.7554/eLife.12850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange E, Bertrand S, Holz O, Rebscher N, & Hassel M (2014). Dynamic expression of a Hydra FGF at boundaries and termini. Development Genes and Evolution, 224(4–6), 235–244. 10.1007/s00427-014-0480-1 [DOI] [PubMed] [Google Scholar]
- Lanner F, & Rossant J (2010). The role of FGF/Erk signaling in pluripotent cells. Development, 137(20), 3351–3360. 10.1242/dev.050146 [DOI] [PubMed] [Google Scholar]
- Larsson SC, & Gill D (2021). Genetic evidence supporting fibroblast growth factor 21 signalling as a pharmacological target for cardiometabolic outcomes and Alzheimer’s disease. Nutrients, 13(5), 1504. 10.3390/nu13051504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latko M, Czyrek A, Porebska N, Kucinska M, Otlewski J, Zakrzewska M, & Opalinski L (2019). Cross-talk between fibroblast growth factor receptors and other cell surface proteins. Cells, 8(5), 455. 10.3390/cells8050455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau DK, Luk IY, Jenkins LJ, Martin A, Williams DS, Schoffer KL, Chionh F, Buchert M, Sjoquist K, Boussioutas A, Hayes SA, Ernst M, Weickhardt AJ, Pavlakis N, Tebbutt NC, & Mariadason JM (2021). Rapid resistance of FGFR-driven gastric cancers to regorafenib and targeted FGFR inhibitors can be overcome by parallel inhibition of MEK. Molecular Cancer Therapeutics, 20(4), 704–715. 10.1158/1535-7163.MCT-20-0836 [DOI] [PubMed] [Google Scholar]
- Lax I, Wong A, Lamothe B, Lee A, Frost A, Hawes J, & Schlessinger J (2002). The docking protein FRS2alpha controls a MAP kinase-mediated negative feedback mechanism for signaling by FGF receptors. Molecular Cell, 10(4), 709–719. 10.1016/s1097-2765(02)00689-5 [DOI] [PubMed] [Google Scholar]
- Lazarus JE, Hegde A, Andrade AC, Nilsson O, & Baron J (2007). Fibroblast growth factor expression in the postnatal growth plate. Bone, 40(3), 577–586. 10.1016/j.bone.2006.10.013 [DOI] [PubMed] [Google Scholar]
- Lebowitz ER, Orbach M, Marin CE, Salmaso N, Vaccarino FM, & Silverman WK (2021). Fibroblast growth factor 2 implicated in childhood anxiety and depression symptoms. Journal of Affective Disorders, 282, 611–616. 10.1016/j.jad.2020.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton G, & Casanova J (2016). Ligand-binding and constitutive FGF receptors in single Drosophila tracheal cells: Implications for the role of FGF in collective migration. Developmental Dynamics, 245(3), 372–378. 10.1002/dvdy.24345 [DOI] [PubMed] [Google Scholar]
- Leclerc K, & Costantini F (2016). Mosaic analysis of cell rearrangements during ureteric bud branching in dissociated/reaggregated kidney cultures and in vivo. Developmental Dynamics, 245(4), 483–496. 10.1002/dvdy.24387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, & Umemori H (2013). Suppression of epileptogenesis-associated changes in response to seizures in FGF22-deficient mice. Frontiers in Cellular Neuroscience, 7, 43. 10.3389/fncel.2013.00043 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lee SG, Huang M, Obholzer ND, Sun S, Li W, Petrillo M, Dai P, Zhou Y, Cotanche DA, Megason SG, Li H, & Chen ZY (2016). Myc and Fgf are required for zebrafish neuromast hair cell regeneration. PLoS One, 11(6), e0157768. 10.1371/journal.pone.0157768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Shin SH, Shin HW, Chun YS, & Park JW (2019). Nuclear FGFR2 negatively regulates hypoxia-induced cell invasion in prostate cancer by interacting with HIF-1 and HIF-2. Scientific Reports, 9(1), 3480. 10.1038/s41598-019-39843-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Yim HS, Shin J, Lee C, Lee JH, & Jeong JY (2017). FGF11 induced by hypoxia interacts with HIF-1alpha and enhances its stability. FEBS Letters, 591(2), 348–357. 10.1002/1873-3468.12547 [DOI] [PubMed] [Google Scholar]
- Lee PC, Hendifar A, Osipov A, Cho M, Li D, & Gong J (2021). Targeting the fibroblast growth factor receptor (FGFR) in advanced cholangiocarcinoma: Clinical trial progress and future considerations. Cancers (Basel), 13(7), 1706. 10.3390/cancers13071706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, & Griep AE (2014). Loss of Dlg-1 in the mouse lens impairs fibroblast growth factor receptor signaling. PLoS One, 9(5), e97470. 10.1371/journal.pone.0097470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Shatadal S, & Griep AE (2016). Dlg-1 interacts with and regulates the activities of fibroblast growth factor receptors and EphA2 in the mouse lens. Investigative Ophthalmology & Visual Science, 57(2), 707–718. 10.1167/iovs.15-17727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Song IW, Pai YJ, Chen SD, & Chen YT (2017). Knock-in human FGFR3 achondroplasia mutation as a mouse model for human skeletal dysplasia. Scientific Reports, 7, 43220. 10.1038/srep43220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek CC, Soulas JM, Bhattacharya I, Ganji E, Locke RC, Smith MC, Bhavsar JD, Polson SW, Ornitz DM, & Killian ML (2021). Deletion of Fibroblast growth factor 9 globally and in skeletal muscle results in enlarged tuberosities at sites of deltoid tendon attachments. Developmental Dynamics, 250(12), 1778–1795. 10.1002/dvdy.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leerberg DM, Hopton RE, & Draper BW (2019). Fibroblast growth factor receptors function redundantly during zebrafish embryonic development. Genetics, 212(4), 1301–1319. 10.1534/genetics.119.302345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leerberg DM, Sano K, & Draper BW (2017). Fibroblast growth factor signaling is required for early somatic gonad development in zebrafish. PLoS Genetics, 13(9), e1006993. 10.1371/journal.pgen.1006993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legeai-Mallet L (2016). C-Type natriuretic peptide analog as therapy for achondroplasia. Endocrine Development, 30, 98–105. 10.1159/000439334 [DOI] [PubMed] [Google Scholar]
- Legeai-Mallet L, & Savarirayan R (2020). Novel therapeutic approaches for the treatment of achondroplasia. Bone, 141, 115579. 10.1016/j.bone.2020.115579 [DOI] [PubMed] [Google Scholar]
- Legrand C, Saleppico R, Sticht J, Lolicato F, Muller HM, Wegehingel S, Dimou E, Steringer JP, Ewers H, Vattulainen I, Freund C, & Nickel W (2020). The Na,K-ATPase acts upstream of phosphoinositide PI(4,5)P2 facilitating unconventional secretion of fibroblast growth factor 2. Communications Biology, 3(1), 141. 10.1038/s42003-020-0871-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, & Deng CX (2017). Fibroblast growth factor receptor 2 signaling in breast cancer. International Journal of Biological Sciences, 13(9), 1163–1171. 10.7150/ijbs.20792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifheit-Nestler M, Grosse Siemer R, Flasbart K, Richter B, Kirchhoff F, Ziegler WH, Klintschar M, Becker JU, Erbersdobler A, Aufricht C, Seeman T, Fischer DC, Faul C, & Haffner D (2016). Induction of cardiac FGF23/FGFR4 expression is associated with left ventricular hypertrophy in patients with chronic kidney disease. Nephrology, Dialysis, Transplantation, 31(7), 1088–1099. 10.1093/ndt/gfv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifheit-Nestler M, & Haffner D (2018). Paracrine effects of FGF23 on the heart. Frontiers in Endocrinology, 9, 278. 10.3389/fendo.2018.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifheit-Nestler M, & Haffner D (2021). How FGF23 shapes multiple organs in chronic kidney disease. Molecular and Cellular Pediatrics, 8(1), 12. 10.1186/s40348-021-00123-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold JA, & Maron BA (2016). Molecular mechanisms of pulmonary vascular remodeling in pulmonary arterial hypertension. International Journal of Molecular Sciences, 17(5), 761. 10.3390/ijms17050761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Taub MA, Liu H, Steinberg KM, Koboldt DC, Zhang Q, Carlson JC, Hetmanski JB, Wang H, Larson DE, Fulton RS, Kousa YA, Fakhouri WD, Naji A, Ruczinski I, Begum F, Parker MM, Busch T, Standley J, … Murray JC (2015). Identification of functional variants for cleft lip with or without cleft palate in or near PAX7, FGFR2, and NOG by targeted sequencing of GWAS loci. American Journal of Human Genetics, 96(3), 397–411. 10.1016/j.ajhg.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung B, & Shimeld SM (2019). Evolution of vertebrate spinal cord patterning. Developmental Dynamics, 248(11), 1028–1043. 10.1002/dvdy.77 [DOI] [PubMed] [Google Scholar]
- Lew ED, Furdui CM, Anderson KS, & Schlessinger J (2009). The precise sequence of FGF receptor autophosphorylation is kinetically driven and is disrupted by oncogenic mutations. Science Signaling, 2(58), ra6. 10.1126/scisignal.2000021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Tian J, Yang J, Ren X, Zhou Z, & Zhou W (2021). Expression of fibroblast growth factor 9 and its receptors in the dentate gyrus of hippocampus in poststroke depression rats. Neuroreport, 32(4), 321–325. 10.1097/WNR.0000000000001591 [DOI] [PubMed] [Google Scholar]
- Li D, Xie K, Zhang L, Yao X, Li H, Xu Q, Wang X, Jiang J, & Fang J (2016). Dual blockade of vascular endothelial growth factor (VEGF) and basic fibroblast growth factor (FGF-2) exhibits potent anti-angiogenic effects. Cancer Letters, 377(2), 164–173. 10.1016/j.canlet.2016.04.036 [DOI] [PubMed] [Google Scholar]
- Li DJ, Fu H, Zhao T, Ni M, & Shen FM (2016). Exercise-stimulated FGF23 promotes exercise performance via controlling the excess reactive oxygen species production and enhancing mitochondrial function in skeletal muscle. Metabolism, 65(5), 747–756. 10.1016/j.metabol.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Li H, Tao C, Cai Z, Hertzler-Schaefer K, Collins TN, Wang F, Feng GS, Gotoh N, & Zhang X (2014). Frs2alpha and Shp2 signal independently of Gab to mediate FGF signaling in lens development. Journal of Cell Science, 127(Pt 3), 571–582. 10.1242/jcs.134478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Gong L, Zhang R, Li S, Yu H, Liu Y, Xue Y, Huang D, Xu N, Wang Y, Xu Y, Zhao Y, Li Q, Li M, Li P, Liu M, Zhang Z, Li X, Du W, & Wang N (2021). Fibroblast growth factor 21 inhibited inflammation and fibrosis after myocardial infarction via EGR1. European Journal of Pharmacology, 910, 174470. 10.1016/j.ejphar.2021.174470 [DOI] [PubMed] [Google Scholar]
- Li J, Han S, Cousin W, & Conboy IM (2015). Age-specific functional epigenetic changes in p21 and p16 in injury-activated satellite cells. Stem Cells, 33(3), 951–961. 10.1002/stem.1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu H, Srivastava SP, Hu Q, Gao R, Li S, Kitada M, Wu G, Koya D, & Kanasaki K (2020). Endothelial FGFR1 (fibroblast growth factor receptor 1) deficiency contributes differential fibrogenic effects in kidney and heart of diabetic mice. Hypertension, 76(6), 1935–1944. 10.1161/HYPERTENSIONAHA.120.15587 [DOI] [PubMed] [Google Scholar]
- Li J, Shi S, Srivastava SP, Kitada M, Nagai T, Nitta K, Kohno M, Kanasaki K, & Koya D (2017). FGFR1 is critical for the anti-endothelial mesenchymal transition effect of N-acetyl-seryl-aspartyl-lysyl-proline via induction of the MAP4K4 pathway. Cell Death & Disease, 8(8), e2965. 10.1038/cddis.2017.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang Q, Cai H, He Z, Wang H, Chen J, Zheng Z, Yin J, Liao Z, Xu H, Xiao J, & Gong F (2018). FGF1 improves functional recovery through inducing PRDX1 to regulate autophagy and anti-ROS after spinal cord injury. Journal of Cellular and Molecular Medicine, 22(5), 2727–2738. 10.1111/jcmm.13566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang Q, Wang H, Wu Y, Yin J, Chen J, Zheng Z, Jiang T, Xie L, Wu F, Zhang H, Li X, Xu H, & Xiao J (2018). Lentivirus mediating FGF13 enhances axon regeneration after spinal cord injury by stabilizing microtubule and improving mitochondrial function. Journal of Neurotrauma, 35(3), 548–559. 10.1089/neu.2017.5205 [DOI] [PubMed] [Google Scholar]
- Li J, Wang X, Ruan G, Zhu Z, & Ding C (2021). Sprifermin: A recombinant human fibroblast growth factor 18 for the treatment of knee osteoarthritis. Expert Opinion on Investigational Drugs, 30(9), 923–930. 10.1080/13543784.2021.1972970 [DOI] [PubMed] [Google Scholar]
- Li J, Wang Z, Chu Q, Jiang K, Li J, & Tang N (2018). The strength of mechanical forces determines the differentiation of alveolar epithelial cells. Developmental Cell, 44(3), 297–312 e295. 10.1016/j.devcel.2018.01.008 [DOI] [PubMed] [Google Scholar]
- Li J, Xu C, Liu Y, Li Y, Du S, Zhang R, Sun Y, Zhang R, Wang Y, Xue H, Ni S, Asiya M, Xue G, Li Y, Shi L, Li D, Pan Z, Zhang Y, Wang Z, … Yang B (2020). Fibroblast growth factor 21 inhibited ischemic arrhythmias via targeting miR-143/EGR1 axis. Basic Research in Cardiology, 115(2), 9. 10.1007/s00395-019-0768-4 [DOI] [PubMed] [Google Scholar]
- Li L, Zhang S, Li H, & Chou H (2019). FGFR3 promotes the growth and malignancy of melanoma by influencing EMT and the phosphorylation of ERK, AKT, and EGFR. BMC Cancer, 19(1), 963. 10.1186/s12885-019-6161-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, & Belmonte JC (2017). Ground rules of the pluripotency gene regulatory network. Nature Reviews. Genetics, 18(3), 180–191. 10.1038/nrg.2016.156 [DOI] [PubMed] [Google Scholar]
- Li M, Page-McCaw P, & Chen W (2016). FGF1 mediates overnutrition-induced compensatory beta-cell differentiation. Diabetes, 65(1), 96–109. 10.2337/db15-0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Herriges JC, Chen L, Mecham RP, & Sun X (2017). FGF receptors control alveolar elastogenesis. Development, 144(24), 4563–4572. 10.1242/dev.149443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Tao X, Huang M, Peng Y, Liang J, Wu Y, & Jiang Y (2021). Fibroblast growth factor 13 facilitates peripheral nerve regeneration through maintaining microtubule stability. Oxidative Medicine and Cellular Longevity, 2021, 5481228. 10.1155/2021/5481228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Wang B, Wu C, Li D, Wu Y, Ye L, Ye L, Chen X, Li P, Yuan Y, Zhang H, Xie L, Li X, Xiao J, & Wang J (2021). Acidic fibroblast growth factor attenuates type 2 diabetes-induced demyelination via suppressing oxidative stress damage. Cell Death & Disease, 12(1), 107. 10.1038/s41419-021-03407-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hang L, & Ma Y (2016). FGF22 protects hearing function from gentamycin ototoxicity by maintaining ribbon synapse number. Hearing Research, 332, 39–45. 10.1016/j.heares.2015.11.011 [DOI] [PubMed] [Google Scholar]
- Li S, He J, Liu Y, & Yang J (2020). FGF22 promotes generation of ribbon synapses through downregulating MEF2D. Aging (Albany NY), 12(7), 6456–6466. 10.18632/aging.103042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhu Z, Xue M, Pan X, Tong G, Yi X, Fan J, Li Y, Li W, Dong Y, Shen E, Gong W, Wang X, Yu Y, Maeng YJ, Li X, Lee KY, Jin L, & Cong W (2021). The protective effects of fibroblast growth factor 10 against hepatic ischemia-reperfusion injury in mice. Redox Biology, 40, 101859. 10.1016/j.redox.2021.101859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SM, Yu YH, Li L, Wang WF, & Li DS (2016). Treatment of CIA Mice with FGF21 Down-regulates TH17-IL-17 Axis. Inflammation, 39(1), 309–319. 10.1007/s10753-015-0251-9 [DOI] [PubMed] [Google Scholar]
- Li T, Luo W, He D, Wang R, Huang Y, Zeng X, Wang W, Chen X, Gao S, Yu Y, Li X, & Wu X (2013). A short peptide derived from the gN helix domain of FGF8b suppresses the growth of human prostate cancer cells. Cancer Letters, 339(2), 226–236. 10.1016/j.canlet.2013.06.001 [DOI] [PubMed] [Google Scholar]
- Li W, Xu W, Song JS, Wu T, & Wang WX (2019). LncRNA SNHG16 promotes cell proliferation through miR-302a-3p/FGF19 axis in hepatocellular carcinoma. Neoplasma, 66(3), 397–404. 10.4149/neo_2018_180720N504 [DOI] [PubMed] [Google Scholar]
- Li X (2019). The FGF metabolic axis. Frontiers in Medicine, 13(5), 511–530. 10.1007/s11684-019-0711-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lv F, Li F, Du M, Liang Y, Ju S, Liu Z, Wang B, & Gao Y (2020). Long noncoding RNA H19 facilitates small cell lung cancer tumorigenesis through miR-140–5p/FGF9 axis. OncoTargets and Therapy, 13, 3525–3534. 10.2147/OTT.S245710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang C, Xiao J, McKeehan WL, & Wang F (2016). Fibroblast growth factors, old kids on the new block. Seminars in Cell & Developmental Biology, 53, 155–167. 10.1016/j.semcdb.2015.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XG, Song X, Wang JY, Sun CH, Li ZQ, Meng LL, & Chi SH (2021). Fibroblast growth factor 18 alleviates hyperoxia-induced lung injury in mice by adjusting oxidative stress and inflammation. European Review for Medical and Pharmacological Sciences, 25(3), 1485–1494. 10.26355/eurrev_202102_24856 [DOI] [PubMed] [Google Scholar]
- Li Y, Sun C, Yates EA, Jiang C, Wilkinson MC, & Fernig DG (2016). Heparin binding preference and structures in the fibroblast growth factor family parallel their evolutionary diversification. Open Biology, 6(3), e150275. 10.1098/rsob.150275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wong K, Giles A, Jiang J, Lee JW, Adams AC, Kharitonenkov A, Yang Q, Gao B, Guarente L, & Zang M (2014). Hepatic SIRT1 attenuates hepatic steatosis and controls energy balance in mice by inducing fibroblast growth factor 21. Gastroenterology, 146(2), 539–549.e537. 10.1053/j.gastro.2013.10.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Fu HL, Tian ML, Wang YQ, Chen W, Cai LL, Zhou XH, & Yuan HB (2016). Neuron-derived FGF10 ameliorates cerebral ischemia injury via inhibiting NF-kappaB-dependent neuroinflammation and activating PI3K/Akt survival signaling pathway in mice. Scientific Reports, 6, 19869. 10.1038/srep19869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YP, Duan FF, Zhao YT, Gu KL, Liao LQ, Su HB, Hao J, Zhang K, Yang N, & Wang Y (2019). A TRIM71 binding long noncoding RNA Trincr1 represses FGF/ERK signaling in embryonic stem cells. Nature Communications, 10(1), 1368. 10.1038/s41467-019-08911-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Z, Du W, Zhang Y, Fu Y, Liu T, Wang A, Cai T, Zhu J, Zeng Y, Liu Z, & Huang JA (2020). Anlotinib can overcome acquired resistance to EGFR-TKIs via FGFR1 signaling in non-small cell lung cancer without harboring EGFR T790M mutation. Thoracic Cancer, 11(7), 1934–1943. 10.1111/1759-7714.13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Song L, Chen Z, Qian Y, Xie J, Zhao L, Lin Q, Zhu G, Tan Y, Li X, Mohammadi M, & Huang Z (2018). Fibroblast growth factor 1 ameliorates diabetic nephropathy by an anti-inflammatory mechanism. Kidney International, 93(1), 95–109. 10.1016/j.kint.2017.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Wang Q, Ma H, Yan W, & Yang J (2018). Knockout of low molecular weight FGF2 attenuates atherosclerosis by reducing macrophage infiltration and oxidative stress in mice. Cellular Physiology and Biochemistry, 45(4), 1434–1443. 10.1159/000487569 [DOI] [PubMed] [Google Scholar]
- Liang Z, Xu J, Ma Z, Li G, & Zhu W (2020). MiR-187 suppresses non-small-cell lung cancer cell proliferation by targeting FGF9. Bioengineered, 11(1), 70–80. 10.1080/21655979.2019.1706287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A, Cunha GR, Shen J, Cao M, Liu G, Sinclair A, & Baskin L (2018). Development of the human bladder and ureterovesical junction. Differentiation, 103, 66–73. 10.1016/j.diff.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Liberti DC, Kremp MM, Liberti WA 3rd, Penkala IJ, Li S, Zhou S, & Morrisey EE (2021). Alveolar epithelial cell fate is maintained in a spatially restricted manner to promote lung regeneration after acute injury. Cell Reports, 35(6), 109092. 10.1016/j.celrep.2021.109092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lignelli E, Palumbo F, Myti D, & Morty RE (2019). Recent advances in our understanding of the mechanisms of lung alveolarization and bronchopulmonary dysplasia. American Journal of Physiology. Lung Cellular and Molecular Physiology, 317(6), L832–L887. 10.1152/ajplung.00369.2019 [DOI] [PubMed] [Google Scholar]
- Lilly B (2014). We have contact: Endothelial cell-smooth muscle cell interactions. Physiology (Bethesda), 29(4), 234–241. 10.1152/physiol.00047.2013 [DOI] [PubMed] [Google Scholar]
- Limonta D, Jovel J, Kumar A, Lu J, Hou S, Airo AM, Lopez-Orozco J, Wong CP, Saito L, Branton W, Wong GK, Mason A, Power C, & Hobman TC (2019). Fibroblast growth factor 2 enhances zika virus infection in human fetal brain. The Journal of Infectious Diseases, 220(8), 1377–1387. 10.1093/infdis/jiz073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Lu P, Zhou M, Wu F, Weng L, Meng K, Yang D, Li S, Jiang C, & Tian H (2019). Purification of recombinant human fibroblast growth factor 13 in E. coli and its molecular mechanism of mitogenesis. Applied Microbiology and Biotechnology, 103(17), 7017–7027. 10.1007/s00253-019-09973-y [DOI] [PubMed] [Google Scholar]
- Lin J, Feng X, Zhang J, & Tong Z (2019). Long noncoding RNA TUG1 promotes airway smooth muscle cells proliferation and migration via sponging miR-590–5p/FGF1 in asthma. American Journal of Translational Research, 11(5), 3159–3166. [PMC free article] [PubMed] [Google Scholar]
- Lin L, Wang Q, Qian K, Cao Z, Xiao J, Wang X, Li X, & Yu Z (2018). bFGF protects against oxygen glucose deprivation/reoxygenation-induced endothelial monolayer permeability via S1PR1-dependent mechanisms. Molecular Neurobiology, 55(4), 3131–3142. 10.1007/s12035-017-0544-0 [DOI] [PubMed] [Google Scholar]
- Lin X, Song L, He D, Zeng X, Wu J, Luo W, Yang Q, Wang J, Wang T, Cai J, Lin Y, Lai F, Peng W, & Wu X (2017). An FGF8b-mimicking peptide with potent antiangiogenic activity. Molecular Medicine Reports, 16(1), 894–900. 10.3892/mmr.2017.6651 [DOI] [PubMed] [Google Scholar]
- Lindberg K, Amin R, Moe OW, Hu MC, Erben RG, Ostman Wernerson A, Lanske B, Olauson H, & Larsson TE (2014). The kidney is the principal organ mediating klotho effects. Journal of the American Society of Nephrology, 25(10), 2169–2175. 10.1681/ASN.2013111209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linel P, Wu S, Deng N, & Wu H (2014). Dynamic transcriptional signatures and network responses for clinical symptoms in influenza-infected human subjects using systems biology approaches. Journal of Pharmacokinetics and Pharmacodynamics, 41(5), 509–521. 10.1007/s10928-014-9365-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipok M, Szlachcic A, Kindela K, Czyrek A, & Otlewski J (2019). Identification of a peptide antagonist of the FGF1-FGFR1 signaling axis by phage display selection. FEBS Open Bio, 9(5), 914–924. 10.1002/2211-5463.12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Schonke M, Zhou E, Li Z, Kooijman S, Boon MR, Larsson M, Wallenius K, Dekker N, Barlind L, Peng XR, Wang Y, & Rensen PCN (2021). Pharmacological treatment with FGF21 strongly improves plasma cholesterol metabolism to reduce atherosclerosis. Cardiovascular Research, 1–14. 10.1093/cvr/cvab076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zheng S, Hou X, Liu X, Du K, Lv X, Li Y, Yang F, Li W, & Sui J (2020). Novel Abs targeting the N-terminus of fibroblast growth factor 19 inhibit hepatocellular carcinoma growth without bile-acid-related side-effects. Cancer Science, 111(5), 1750–1760. 10.1111/cas.14353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xu Y, Hu Y, & Wang G (2015). The role of fibroblast growth factor 21 in the pathogenesis of non-alcoholic fatty liver disease and implications for therapy. Metabolism, 64(3), 380–390. 10.1016/j.metabol.2014.11.009 [DOI] [PubMed] [Google Scholar]
- Liu L, Song C, Li J, Wang Q, Zhu M, Hu Y, Chen J, Chen C, Zhang JS, Dong N, & Chen C (2020). Fibroblast growth factor 10 alleviates particulate matter-induced lung injury by inhibiting the HMGB1-TLR4 pathway. Aging (Albany NY), 12(2), 1186–1200. 10.18632/aging.102676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Xia Z, Li J, Hu Y, Wang Q, Chen J, Fan S, Wu J, Dong N, & Chen C (2019). Fibroblast growth factor 10 protects against particulate matter-induced airway inflammatory response through regulating inflammatory signaling and apoptosis. American Journal of Translational Research, 11(11), 6977–6988. [PMC free article] [PubMed] [Google Scholar]
- Liu M, Chen Z, Huang S, Chu S, Issaro N, Tian H, Hu H, & Jiang C (2018). Effective protection on acute liver injury by halo tag-flanked recombinant fibroblast growth factor 7. Biotechnology Journal, 13(7), e1700411. 10.1002/biot.201700411 [DOI] [PubMed] [Google Scholar]
- Liu S, Marcelin G, Blouet C, Jeong JH, Jo YH, Schwartz GJ, & Chua S Jr. (2018). A gut-brain axis regulating glucose metabolism mediated by bile acids and competitive fibroblast growth factor actions at the hypothalamus. Molecular Metabolism, 8, 37–50. 10.1016/j.molmet.2017.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SQ, Roberts D, Kharitonenkov A, Zhang B, Hanson SM, Li YC, Zhang LQ, & Wu YH (2013). Endocrine protection of ischemic myocardium by FGF21 from the liver and adipose tissue. Scientific Reports, 3, 2767. 10.1038/srep02767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WS, Tang MJ, Xu TL, Su JB, Wang XQ, Xu F, Zhang DM, Zhu Q, Cao J, & Wang H (2020). Association of serum fibroblast growth factor 19 levels with arteriosclerosis parameters assessed by arterial stiffness and atherogenic index of plasma in patients with type 2 diabetes. Diabetology and Metabolic Syndrome, 12, 44. 10.1186/s13098-020-00552-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, D’Cruz AA, Hansen J, Croker BA, Lawlor KE, Sims NA, & Wicks IP (2019). Deleting suppressor of cytokine signaling-3 in chondrocytes reduces bone growth by disrupting mitogen-activated protein kinase signaling. Osteoarthritis and Cartilage, 27(10), 1557–1563. 10.1016/j.joca.2019.05.018 [DOI] [PubMed] [Google Scholar]
- Liu XY, Li X, Bai MR, Chen X, Wang CL, Xie J, & Ye L (2018). FGF-7 dictates osteocyte cell processes through beta-catenin transduction. Scientific Reports, 8(1), 14792. 10.1038/s41598-018-33247-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Deng J, Liu Y, Li W, & Nie X (2021). FGF, mechanism of action, role in Parkinson’s disease, and therapeutics. Frontiers in Pharmacology, 12, 675725. 10.3389/fphar.2021.675725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ma J, Beenken A, Srinivasan L, Eliseenkova AV, & Mohammadi M (2017). Regulation of receptor binding specificity of FGF9 by an autoinhibitory homodimerization. Structure, 25(9), 1325–1336.e1323. 10.1016/j.str.2017.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Liu G, Zhang GL, Li J, He YQ, Zhang SS, Wang Y, He WY, Cheng GH, Yang X, Xu J, & Wang J (2016). Binding of human recombinant mutant soluble ectodomain of FGFR2IIIc to c subtype of FGFRs: Implications for anticancer activity. Oncotarget, 7(42), 68473–68488. 10.18632/oncotarget.12067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Xu J, Colvin JS, & Ornitz DM (2002). Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes & Development, 16(7), 859–869. 10.1101/gad.965602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZQ, Ren JJ, Zhao JL, Zang J, Long QF, Du JJ, Jia XT, Gu NB, Di ZL, Qian YH, & Li SZ (2020). MicroRNA-144 represses gliomas progression and elevates susceptibility to Temozolomide by targeting CAV2 and FGF7. Scientific Reports, 10(1), 4155. 10.1038/s41598-020-60218-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livnah O, Stura EA, Middleton SA, Johnson DL, Jolliffe LK, & Wilson IA (1999). Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science, 283(5404), 987–990. 10.1126/science.283.5404.987 [DOI] [PubMed] [Google Scholar]
- Long JM, & Holtzman DM (2019). Alzheimer disease: An update on pathobiology and treatment strategies. Cells, 179(2), 312–339. 10.1016/j.cell.2019.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Pedrera C, Barbarroja N, Patino-Trives AM, Luque-Tevar M, Collantes-Estevez E, Escudero-Contreras A, & Perez-Sanchez C (2020). Effects of biological therapies on molecular features of rheumatoid arthritis. International Journal of Molecular Sciences, 21(23), 9067. 10.3390/ijms21239067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loriot Y, Necchi A, Park SH, Garcia-Donas J, Huddart R, Burgess E, Fleming M, Rezazadeh A, Mellado B, Varlamov S, Joshi M, Duran I, Tagawa ST, Zakharia Y, Zhong B, Stuyckens K, Santiago-Walker A, De Porre P, O’Hagan A, … BLC2001 Study Group. (2019). Erdafitinib in locally advanced or metastatic urothelial carcinoma. The New England Journal of Medicine, 381(4), 338–348. 10.1056/NEJMoa1817323 [DOI] [PubMed] [Google Scholar]
- Lou JY, Laezza F, Gerber BR, Xiao M, Yamada KA, Hartmann H, Craig AM, Nerbonne JM, & Ornitz DM (2005). Fibroblast growth factor 14 is an intracellular modulator of voltage-gated sodium channels. The Journal of Physiology, 569(Pt 1), 179–193. 10.1113/jphysiol.2005.097220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou L, Tian M, Chang J, Li F, & Zhang G (2020). MiRNA-192–5p attenuates airway remodeling and autophagy in asthma by targeting MMP-16 and ATG7. Biomedicine & Pharmacotherapy, 122, 109692. 10.1016/j.biopha.2019.109692 [DOI] [PubMed] [Google Scholar]
- Lowe V, Wisniewski L, & Pellet-Many C (2021). The zebrafish cardiac endothelial cell-roles in development and regeneration. Journal of Cardiovascular Development and Disease, 8(5), 49. 10.3390/jcdd8050049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Jia C, Wu D, Jin H, Lin Z, Pan J, Li X, & Wang W (2021). Fibroblast growth factor 21 (FGF21) alleviates senescence, apoptosis, and extracellular matrix degradation in osteoarthritis via the SIRT1-mTOR signaling pathway. Cell Death & Disease, 12(10), 865. 10.1038/s41419-021-04157-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Dai J, Wang X, Zhang M, Zhang P, Sun H, Zhang X, Yu H, Zhang W, Zhang L, Jiang X, & Shen SG (2015). Effect of fibroblast growth factor 9 on the osteogenic differentiation of bone marrow stromal stem cells and dental pulp stem cells. Molecular Medicine Reports, 11(3), 1661–1668. 10.3892/mmr.2014.2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Chen H, Patterson AV, Smaill JB, & Ding K (2019). Fibroblast growth factor receptor 4 (FGFR4) selective inhibitors as hepatocellular carcinoma therapy: Advances and prospects. Journal of Medicinal Chemistry, 62(6), 2905–2915. 10.1021/acs.jmedchem.8b01531 [DOI] [PubMed] [Google Scholar]
- Lu X, Jin H, Quesada C, Farrell EC, Huang L, Aliabouzar M, Kripfgans OD, Fowlkes JB, Franceschi RT, Putnam AJ, & Fabiilli ML (2020). Spatially-directed cell migration in acoustically-responsive scaffolds through the controlled delivery of basic fibroblast growth factor. Acta Biomaterialia, 113, 217–227. 10.1016/j.actbio.2020.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo F, Xie Y, Wang Z, Huang J, Tan Q, Sun X, Li F, Li C, Liu M, Zhang D, Xu M, Su N, Ni Z, Jiang W, Chang J, Chen H, Chen S, Xu X, Deng C, … Chen L (2018). Adeno-associated virus-mediated RNAi against mutant alleles attenuates abnormal calvarial phenotypes in an Apert syndrome mouse model. Molecular Therapy. Nucleic Acids, 13, 291–302. 10.1016/j.omtn.2018.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Zheng R, Zhao Y, Wu J, Li J, Jiang F, Chen DN, Zhou XT, & Li JD (2017). A dominant negative FGFR1 mutation identified in a Kallmann syndrome patient. Gene, 621, 1–4. 10.1016/j.gene.2017.04.017 [DOI] [PubMed] [Google Scholar]
- Luo J, Ko B, Elliott M, Zhou M, Lindhout DA, Phung V, To C, Learned RM, Tian H, DePaoli AM, & Ling L (2014). A nontumorigenic variant of FGF19 treats cholestatic liver diseases. Science Translational Medicine, 6(247), 247ra100. 10.1126/scitranslmed.3009098 [DOI] [PubMed] [Google Scholar]
- Lv YQ, Dhlamini Q, Chen C, Li X, Bellusci S, & Zhang JS (2021). FGF10 and lipofibroblasts in lung homeostasis and disease: Insights gained from the adipocytes. Frontiers in Cell and Development Biology, 9, 645400. 10.3389/fcell.2021.645400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv YQ, Wu J, Li XK, Zhang JS, & Bellusci S (2019). Role of FGF10/FGFR2b signaling in mouse digestive tract development, repair and regeneration following injury. Frontiers in Cell and Development Biology, 7, 326. 10.3389/fcell.2019.00326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HW, Xi DY, Ma JZ, Guo M, Ma L, Ma DH, Li PW, & Guo CA (2020). Long noncoding RNA AFAP1-AS1 promotes cell proliferation and metastasis via the miR-155–5p/FGF7 axis and predicts poor prognosis in gastric cancer. Disease Markers, 2020, 8140989. 10.1155/2020/8140989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Li H, Wang P, Yang W, Mi R, Zhuang J, Jiang Y, Lu Y, Shen X, Wu Y, & Shen H (2021). ATF6 aggravates angiogenesis-osteogenesis coupling during ankylosing spondylitis by mediating FGF2 expression in chondrocytes. iScience, 24(7), 102791. 10.1016/j.isci.2021.102791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Kakudo N, Morimoto N, Lai F, Taketani S, & Kusumoto K (2019). Fibroblast growth factor-2 stimulates proliferation of human adipose-derived stem cells via Src activation. Stem Cell Research & Therapy, 10(1), 350. 10.1186/s13287-019-1462-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie B, Henneke I, Hezel S, Al Alam D, El Agha E, Chao CM, Quantius J, Wilhelm J, Jones M, Goth K, Li X, Seeger W, Konigshoff M, Herold S, Rizvanov AA, Gunther A, & Bellusci S (2015). Attenuating endogenous Fgfr2b ligands during bleomycin-induced lung fibrosis does not compromise murine lung repair. American Journal of Physiology. Lung Cellular and Molecular Physiology, 308(10), L1014–L1024. 10.1152/ajplung.00291.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie B, Korfei M, Henneke I, Sibinska Z, Tian X, Hezel S, Dilai S, Wasnick R, Schneider B, Wilhelm J, El Agha E, Klepetko W, Seeger W, Schermuly R, Gunther A, & Bellusci S (2015). Increased FGF1-FGFRc expression in idiopathic pulmonary fibrosis. Respiratory Research, 16, 83. 10.1186/s12931-015-0242-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddaluno L, Urwyler C, Rauschendorfer T, Meyer M, Stefanova D, Sporri R, Wietecha M, Ferrarese L, Stoycheva D, Bender D, Li N, Strittmatter G, Nasirujjaman K, Beer HD, Staeheli P, Hildt E, Oxenius A, & Werner S (2020). Antagonism of interferon signaling by fibroblast growth factors promotes viral replication. EMBO Molecular Medicine, 12(9), e11793. 10.15252/emmm.201911793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddaluno L, Urwyler C, & Werner S (2017). Fibroblast growth factors: Key players in regeneration and tissue repair. Development, 144(22), 4047–4060. 10.1242/dev.152587 [DOI] [PubMed] [Google Scholar]
- Madry H, Kohn D, & Cucchiarini M (2013). Direct FGF-2 gene transfer via recombinant adeno-associated virus vectors stimulates cell proliferation, collagen production, and the repair of experimental lesions in the human ACL. The American Journal of Sports Medicine, 41(1), 194–202. 10.1177/0363546512465840 [DOI] [PubMed] [Google Scholar]
- Maeng HJ, Lee GY, Bae JH, & Lim S (2020). Effect of fibroblast growth factor 21 on the development of atheromatous plaque and lipid metabolic profiles in an atherosclerosis-prone mouse model. International Journal of Molecular Sciences, 21(18), 6836. 10.3390/ijms21186836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud N, Abdul Latif H, Mohd Zaki F, & Goh BS (2021). Tracheal cartilaginous sleeve in Pfeiffer syndrome: Lesson learnt from its rarity. BML Case Reports, 14(4), e236888. 10.1136/bcr-2020-236888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makanae A, Mitogawa K, & Satoh A (2014). Co-operative Bmp- and Fgf-signaling inputs convert skin wound healing to limb formation in urodele amphibians. Developmental Biology, 396(1), 57–66. 10.1016/j.ydbio.2014.09.021 [DOI] [PubMed] [Google Scholar]
- Makanae A, Mitogawa K, & Satoh A (2016). Cooperative inputs of Bmp and Fgf signaling induce tail regeneration in urodele amphibians. Developmental Biology, 410(1), 45–55. 10.1016/j.ydbio.2015.12.012 [DOI] [PubMed] [Google Scholar]
- Makrythanasis P, Temtamy S, Aglan MS, Otaify GA, Hamamy H, & Antonarakis SE (2014). A novel homozygous mutation in FGFR3 causes tall stature, severe lateral tibial deviation, scoliosis, hearing impairment, camptodactyly, and arachnodactyly. Human Mutation, 35(8), 959–963. 10.1002/humu.22597 [DOI] [PubMed] [Google Scholar]
- Manchado E, Weissmueller S, Morris JP 4th, Chen CC, Wullenkord R, Lujambio A, de Stanchina E, Poirier JT, Gainor JF, Corcoran RB, Engelman JA, Rudin CM, Rosen N, & Lowe SW (2016). A combinatorial strategy for treating KRAS-mutant lung cancer. Nature, 534(7609), 647–651. 10.1038/nature18600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandinova A, Kolev V, Neel V, Hu B, Stonely W, Lieb J, Wu X, Colli C, Han R, Pazin MJ, Ostano P, Dummer R, Brissette JL, & Dotto GP (2009). A positive FGFR3/FOXN1 feedback loop underlies benign skin keratosis versus squamous cell carcinoma formation in humans. Journal of Clinical Investigation, 119(10), 3127–3137. 10.1172/JCI38543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SL, Goddard JM, & Capecchi MR (1993). Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development, 117(1), 13–28. [DOI] [PubMed] [Google Scholar]
- Mao Y, Liu XQ, Song Y, Zhai CG, Xu XL, Zhang L, & Zhang Y (2020). Fibroblast growth factor-2/platelet-derived growth factor enhances atherosclerotic plaque stability. Journal of Cellular and Molecular Medicine, 24(1), 1128–1140. 10.1111/jcmm.14850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maratos-Flier E (2017). Fatty liver and FGF21 physiology. Experimental Cell Research, 360(1), 2–5. 10.1016/j.yexcr.2017.05.006 [DOI] [PubMed] [Google Scholar]
- Marcelin G, Jo YH, Li X, Schwartz GJ, Zhang Y, Dun NJ, Lyu RM, Blouet C, Chang JK, & Chua S Jr. (2014). Central action of FGF19 reduces hypothalamic AGRP/NPY neuron activity and improves glucose metabolism. Mol Metab, 3(1), 19–28. 10.1016/j.molmet.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek LA, Hinz TK, von Massenhausen A, Olszewski KA, Kleczko EK, Boehm D, Weiser-Evans MC, Nemenoff RA, Hoffmann H, Warth A, Gozgit JM, Perner S, & Heasley LE (2014). Nonamplified FGFR1 is a growth driver in malignant pleural mesothelioma. Molecular Cancer Research, 12(10), 1460–1469. 10.1158/1541-7786.MCR-14-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margosio B, Rusnati M, Bonezzi K, Cordes BL, Annis DS, Urbinati C, Giavazzi R, Presta M, Ribatti D, Mosher DF, & Taraboletti G (2008). Fibroblast growth factor-2 binding to the thrombospondin-1 type III repeats, a novel antiangiogenic domain. The International Journal of Biochemistry & Cell Biology, 40(4), 700–709. 10.1016/j.biocel.2007.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani FV, Fernandez-Teran M, & Ros MA (2017). Ectoderm-mesoderm crosstalk in the embryonic limb: The role of fibroblast growth factor signaling. Developmental Dynamics, 246(4), 208–216. 10.1002/dvdy.24480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie PJ, Hurley M, & Ornitz DM (2019). Fibroblast growth factor (FGF) and FGF receptor families in bone. In Bilezikian J, Martin TJ, Clemens T, & Rosen C (Eds.), Principles of Bone Biology (Vol. 1–2, p. 2024). Academic Press. 10.1016/B978-0-12-814841-9.00045-2 [DOI] [Google Scholar]
- Markan KR, & Potthoff MJ (2016). Metabolic fibroblast growth factors (FGFs): Mediators of energy homeostasis. Seminars in Cell & Developmental Biology, 53, 85–93. 10.1016/j.semcdb.2015.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham A (2019). Erdafitinib: First global approval. Drugs, 79(9), 1017–1021. 10.1007/s40265-019-01142-9 [DOI] [PubMed] [Google Scholar]
- Martin L, Kaci N, Estibals V, Goudin N, Garfa-Traore M, Benoist-Lasselin C, Dambroise E, & Legeai-Mallet L (2018). Constitutively-active FGFR3 disrupts primary cilium length and IFT20 trafficking in various chondrocyte models of achondroplasia. Human Molecular Genetics, 27(1), 1–13. 10.1093/hmg/ddx374 [DOI] [PubMed] [Google Scholar]
- Martynova E, Bouchard M, Musil LS, & Cvekl A (2018). Identification of novel Gata3 distal enhancers active in mouse embryonic lens. Developmental Dynamics, 247(11), 1186–1198. 10.1002/dvdy.24677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathes S, Fahrner A, Ghoshdastider U, Rudiger HA, Leunig M, Wolfrum C, & Krutzfeldt J (2021). FGF-2-dependent signaling activated in aged human skeletal muscle promotes intramuscular adipogenesis. Proceedings of the National Academy of Sciences of the United States of America, 118(37), e2021013118. 10.1073/pnas.2021013118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew G, Hannan A, Hertzler-Schaefer K, Wang F, Feng GS, Zhong J, Zhao JJ, Downward J, & Zhang X (2016). Targeting of Ras-mediated FGF signaling suppresses Pten-deficient skin tumor. Proceedings of the National Academy of Sciences of the United States of America, 113(46), 13156–13161. 10.1073/pnas.1604450113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Nonaka Y, Futakawa S, Imai H, Akita K, Nishihata T, Fujiwara M, Ali Y, Bhisitkul RB, & Nakamura Y (2019). Anti-angiogenic and anti-scarring dual action of an anti-fibroblast growth factor 2 aptamer in animal models of retinal disease. Molecular Therapy. Nucleic Acids, 17, 819–828. 10.1016/j.omtn.2019.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Arao T, Hamaguchi T, Shimada Y, Kato K, Oda I, Taniguchi H, Koizumi F, Yanagihara K, Sasaki H, Nishio K, & Yamada Y (2012). FGFR2 gene amplification and clinicopathological features in gastric cancer. British Journal of Cancer, 106(4), 727–732. 10.1038/bjc.2011.603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto N, Shinmyo Y, Ichikawa Y, & Kawasaki H (2017). Gyrification of the cerebral cortex requires FGF signaling in the mammalian brain. eLife, 6, e29285. 10.7554/eLife.29285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y, Aizawa H, Shiraki-Iida T, Nagai R, Kuro-o M, & Nabeshima Y (1998). Identification of the human klotho gene and its two transcripts encoding membrane and secreted klotho protein. Biochemical and Biophysical Research Communications, 242(3), 626–630. 10.1006/bbrc.1997.8019 [DOI] [PubMed] [Google Scholar]
- Matsuzawa T, Morita M, Shimane A, Otsuka R, Mei Y, Irie F, Yamaguchi Y, Yanai K, & Yoshikawa T (2021). Heparan sulfate promotes differentiation of white adipocytes to maintain insulin sensitivity and glucose homeostasis. The Journal of Biological Chemistry, 297(3), 101006. 10.1016/j.jbc.2021.101006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May AJ, Teshima THN, Noble A, & Tucker AS (2019). FGF10 is an essential regulator of tracheal submucosal gland morphogenesis. Developmental Biology, 451(2), 158–166. 10.1016/j.ydbio.2019.03.017 [DOI] [PubMed] [Google Scholar]
- Mazzaferro V, El-Rayes BF, Droz Dit Busset M, Cotsoglou C, Harris WP, Damjanov N, Masi G, Rimassa L, Personeni N, Braiteh F, Zagonel V, Papadopoulos KP, Hall T, Wang Y, Schwartz B, Kazakin J, Bhoori S, de Braud F, & Shaib WL (2019). Derazantinib (ARQ 087) in advanced or inoperable FGFR2 gene fusion-positive intrahepatic cholangiocarcinoma. British Journal of Cancer, 120(2), 165–171. 10.1038/s41416-018-0334-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MJ, Gaston-Massuet C, Tziaferi V, Gregory LC, Alatzoglou KS, Signore M, Puelles E, Gerrelli D, Farooqi IS, Raza J, Walker J, Kavanaugh SI, Tsai PS, Pitteloud N, Martinez-Barbera JP, & Dattani MT (2011). Novel FGF8 mutations associated with recessive holoprosencephaly, craniofacial defects, and hypothalamo-pituitary dysfunction. The Journal of Clinical Endocrinology and Metabolism, 96(10), E1709–E1718. 10.1210/jc.2011-0454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClurg O, Tinson R, & Troeberg L (2021). Targeting cartilage degradation in osteoarthritis. Pharmaceuticals (Basel), 14(2), 126. 10.3390/ph14020126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan SE, & McCoy DM (2015). Fibroblast growth factor signaling in myofibroblasts differs from lipofibroblasts during alveolar septation in mice. American Journal of Physiology. Lung Cellular and Molecular Physiology, 309(5), L463–L474. 10.1152/ajplung.00013.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean DM, Homsy J, Wakimoto H, Patel N, Gorham J, DePalma SR, Ware JS, Zaidi S, Ma W, Patel N, Lifton RP, Chung WK, Kim R, Shen Y, Brueckner M, Goldmuntz E, Sharp AJ, Seidman CE, Gelb BD, & Seidman JG (2016). Loss of RNA expression and allele-specific expression associated with congenital heart disease. Nature Communications, 7, 12824. 10.1038/ncomms12824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie J, Smith C, Karuppaiah K, Langberg J, Silva MJ, & Ornitz DM (2019). Osteocyte death and bone overgrowth in mice lacking fibroblast growth factor receptors 1 and 2 in mature osteoblasts and osteocytes. Journal of Bone and Mineral Research, 34(9), 1660–1675. 10.1002/jbmr.3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AP (2016). Development of the mammalian kidney. Current Topics in Developmental Biology, 117, 31–64. 10.1016/bs.ctdb.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meganathan K, Sotiriadou I, Natarajan K, Hescheler J, & Sachinidis A (2015). Signaling molecules, transcription growth factors and other regulators revealed from in-vivo and in-vitro models for the regulation of cardiac development. International Journal of Cardiology, 183, 117–128. 10.1016/j.ijcard.2015.01.049 [DOI] [PubMed] [Google Scholar]
- Meini S, Giani T, & Tascini C (2020). Intussusceptive angiogenesis in Covid-19: Hypothesis on the significance and focus on the possible role of FGF2. Molecular Biology Reports, 47(10), 8301–8304. 10.1007/s11033-020-05831-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz C, Parsi MK, Adams JR, Sideek MA, Kopecki Z, Cowin AJ, & Gibson MA (2015). LTBP-2 has a single high-affinity binding site for FGF-2 and blocks FGF-2-induced cell proliferation. PLoS One, 10(8), e0135577. 10.1371/journal.pone.0135577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meo Burt P, Xiao L, Dealy C, Fisher MC, & Hurley MM (2016). FGF2 high molecular weight isoforms contribute to osteoarthropathy in male mice. Endocrinology, 157(12), 4602–4614. 10.1210/en.2016-1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric-Bernstam F, Bahleda R, Hierro C, Sanson M, Bridgewater J, Arkenau HT, Tran B, Kelley RK, Park JO, Javle M, He Y, Benhadji KA, & Goyal L (2021). Futibatinib, an irreversible FGFR1–4 inhibitor, in patients with advanced solid tumors harboring FGF/FGFR aberrations: A phase I dose-expansion study. Cancer Discovery, 1–14. 10.1158/2159-8290.CD-21-0697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker A, Neumeyer L, Hertel NT, Grigelioniene G, Makitie O, Mohnike K, & Hagenas L (2018). Growth in achondroplasia: Development of height, weight, head circumference, and body mass index in a European cohort. American Journal of Medical Genetics. Part A, 176(8), 1723–1734. 10.1002/ajmg.a.38853 [DOI] [PubMed] [Google Scholar]
- Merker A, Neumeyer L, Hertel NT, Grigelioniene G, Mohnike K, & Hagenas L (2018). Development of body proportions in achondroplasia: Sitting height, leg length, arm span, and foot length. American Journal of Medical Genetics. Part A, 176(9), 1819–1829. 10.1002/ajmg.a.40356 [DOI] [PubMed] [Google Scholar]
- Merrill AE, Sarukhanov A, Krejci P, Idoni B, Camacho N, Estrada KD, Lyons KM, Deixler H, Robinson H, Chitayat D, Curry CJ, Lachman RS, Wilcox WR, & Krakow D (2012). Bent bone dysplasia-FGFR2 type, a distinct skeletal disorder, has deficient canonical FGF signaling. American Journal of Human Genetics, 90(3), 550–557. 10.1016/j.ajhg.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzner T, Bedeir A, Held G, Peter-Vorosmarty B, Ghassemi S, Heinzle C, Spiegl-Kreinecker S, Marian B, Holzmann K, Grasl-Kraupp B, Pirker C, Micksche M, Berger W, Heffeter P, & Grusch M (2011). Fibroblast growth factor receptors as therapeutic targets in human melanoma: Synergism with BRAF inhibition. The Journal of Investigative Dermatology, 131(10), 2087–2095. 10.1038/jid.2011.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Ben-Yehuda Greenwald M, Rauschendorfer T, Sanger C, Jukic M, Iizuka H, Kubo F, Chen L, Ornitz DM, & Werner S (2020). Mouse genetics identifies unique and overlapping functions of fibroblast growth factor receptors in keratinocytes. Journal of Cellular and Molecular Medicine, 24(2), 1774–1785. 10.1111/jcmm.14871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Muller AK, Yang J, Moik D, Ponzio G, Ornitz DM, Grose R, & Werner S (2012). FGF receptors 1 and 2 are key regulators of keratinocyte migration in vitro and in wounded skin. Journal of Cell Science, 125(Pt 23), 5690–5701. 10.1242/jcs.108167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Wang P, & Lin L (2020). miR-188–3p inhibits vascular smooth muscle cell proliferation and migration by targeting fibroblast growth factor 1 (FGF1). Medical Science Monitor, 26, e924394. 10.12659/MSM.924394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao G, & Hayashi S (2015). Manipulation of gene expression by infrared laser heat shock and its application to the study of tracheal development in Drosophila. Developmental Dynamics, 244(3), 479–487. 10.1002/dvdy.24192 [DOI] [PubMed] [Google Scholar]
- Miao G, & Hayashi S (2016). Escargot controls the sequential specification of two tracheal tip cell types by suppressing FGF signaling in Drosophila. Development, 143(22), 4261–4271. 10.1242/dev.133322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michigami T (2019). Skeletal mineralization: Mechanisms and diseases. Annals of Pediatric Endocrinology & Metabolism, 24(4), 213–219. 10.6065/apem.2019.24.4.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolajczak M, Goodman T, & Hajihosseini MK (2016). Interrogation of a lacrimo-auriculo-dento-digital syndrome protein reveals novel modes of fibroblast growth factor 10 (FGF10) function. The Biochemical Journal, 473(24), 4593–4607. 10.1042/BCJ20160441 [DOI] [PubMed] [Google Scholar]
- Milunsky JM, Zhao G, Maher TA, Colby R, & Everman DB (2006). LADD syndrome is caused by FGF10 mutations. Clinical Genetics, 69(4), 349–354. 10.1111/j.1399-0004.2006.00597.x [DOI] [PubMed] [Google Scholar]
- Min Swe NM, Kobayashi Y, Kamimoto H, & Moriyama K (2021). Aberrantly activated Wnt/beta-catenin pathway co-receptors LRP5 and LRP6 regulate osteoblast differentiation in the developing coronal sutures of an Apert syndrome (Fgfr2(S252W) (/+)) mouse model. Developmental Dynamics, 250(3), 465–476. 10.1002/dvdy.239 [DOI] [PubMed] [Google Scholar]
- Min X, Weiszmann J, Johnstone S, Wang W, Yu X, Romanow W, Thibault S, Li Y, & Wang Z (2018). Agonistic beta-Klotho antibody mimics fibroblast growth factor 21 (FGF21) functions. The Journal of Biological Chemistry, 293(38), 14678–14688. 10.1074/jbc.RA118.004343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn KT, Dietmann S, Waye SE, Morris SA, & Solnica-Krezel L (2021). Gene expression dynamics underlying cell fate emergence in 2D micropatterned human embryonic stem cell gastruloids. Stem Cell Reports, 16(5), 1210–1227. 10.1016/j.stemcr.2021.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn KT, Fu YC, He S, Dietmann S, George SC, Anastasio MA, Morris SA, & Solnica-Krezel L (2020). High-resolution transcriptional and morphogenetic profiling of cells from micropatterned human ESC gastruloid cultures. eLife, 9, e59445. 10.7554/eLife.59445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miraoui H, Dwyer AA, Sykiotis GP, Plummer L, Chung W, Feng B, Beenken A, Clarke J, Pers TH, Dworzynski P, Keefe K, Niedziela M, Raivio T, Crowley WF Jr., Seminara SB, Quinton R, Hughes VA, Kumanov P, Young J, … Pitteloud N (2013). Mutations in FGF17, IL17RD, DUSP6, SPRY4, and FLRT3 are identified in individuals with congenital hypogonadotropic hypogonadism. American Journal of Human Genetics, 92(5), 725–743. 10.1016/j.ajhg.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishel S, Shneyer B, Korsensky L, Goldshmidt-Tran O, Haber T, Machluf M, & Ron D (2017). Delivery of the gene encoding the tumor suppressor Sef into prostate tumors by therapeutic-ultrasound inhibits both tumor angiogenesis and growth. Scientific Reports, 7(1), 15060. 10.1038/s41598-017-12408-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missinato MA, Saydmohammed M, Zuppo DA, Rao KS, Opie GW, Kuhn B, & Tsang M (2018). Dusp6 attenuates Ras/MAPK signaling to limit zebrafish heart regeneration. Development, 145(5), dev157206. 10.1242/dev.157206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitogawa K, Makanae A, & Satoh A (2018). Hyperinnervation improves Xenopus laevis limb regeneration. Developmental Biology, 433(2), 276–286. 10.1016/j.ydbio.2017.10.007 [DOI] [PubMed] [Google Scholar]
- Miura K, Kim OH, Lee HR, Namba N, Michigami T, Yoo WJ, Choi IH, Ozono K, & Cho TJ (2014). Overgrowth syndrome associated with a gain-of-function mutation of the natriuretic peptide receptor 2 (NPR2) gene. American Journal of Medical Genetics. Part A, 164A(1), 156–163. 10.1002/ajmg.a.36218 [DOI] [PubMed] [Google Scholar]
- Miyado M, Miyado K, Nakamura A, Fukami M, Yamada G, & Oda SI (2017). Expression patterns of Fgf8 and Shh in the developing external genitalia of Suncus murinus. Reproduction, 153(2), 187–195. 10.1530/REP-16-0231 [DOI] [PubMed] [Google Scholar]
- Miyake A, Konishi M, Martin FH, Hernday NA, Ozaki K, Yamamoto S, Mikami T, Arakawa T, & Itoh N (1998). Structure and expression of a novel member, FGF-16, on the fibroblast growth factor family. Biochemical and Biophysical Research Communications, 243(1), 148–152. 10.1006/bbrc.1998.8073 [DOI] [PubMed] [Google Scholar]
- Moiseenko A, Vazquez-Armendariz AI, Kheirollahi V, Chu X, Tata A, Rivetti S, Gunther S, Lebrigand K, Herold S, Braun T, Mari B, De Langhe S, Kwapiszewska G, Gunther A, Chen C, Seeger W, Tata PR, Zhang JS, Bellusci S, & El Agha E (2020). Identification of a repair-supportive mesenchymal cell population during airway epithelial regeneration. Cell Reports, 33(12), 108549. 10.1016/j.celrep.2020.108549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldrich RX, Gobius I, Pollak T, Zhang J, Ren T, Brown L, Mori S, De Juan Romero C, Britanova O, Tarabykin V, & Richards LJ (2010). Molecular regulation of the developing commissural plate. The Journal of Comparative Neurology, 518(18), 3645–3661. 10.1002/cne.22445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina GA, Watkins SC, & Tsang M (2007). Generation of FGF reporter transgenic zebrafish and their utility in chemical screens. BMC Developmental Biology, 7, 62. 10.1186/1471-213X-7-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotkov A, Mazot P, Brewer JR, Cinalli RM, & Soriano P (2017). Distinct requirements for FGFR1 and FGFR2 in primitive endoderm development and exit from pluripotency. Developmental Cell, 41(5), 511–526.e514. 10.1016/j.devcel.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotkov A, & Soriano P (2018). Distinct mechanisms for PDGF and FGF signaling in primitive endoderm development. Developmental Biology, 442(1), 155–161. 10.1016/j.ydbio.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncion A, Lin M, O’Neill EG, Franceschi RT, Kripfgans OD, Putnam AJ, & Fabiilli ML (2017). Controlled release of basic fibroblast growth factor for angiogenesis using acoustically-responsive scaffolds. Biomaterials, 140, 26–36. 10.1016/j.biomaterials.2017.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosa S, & Wollnik B (2016). Altered FGF signalling in congenital craniofacial and skeletal disorders. Seminars in Cell & Developmental Biology, 53, 115–125. 10.1016/j.semcdb.2015.12.005 [DOI] [PubMed] [Google Scholar]
- Morgani SM, & Hadjantonakis AK (2020). Signaling regulation during gastrulation: Insights from mouse embryos and in vitro systems. Current Topics in Developmental Biology, 137, 391–431. 10.1016/bs.ctdb.2019.11.011 [DOI] [PubMed] [Google Scholar]
- Morgani SM, Metzger JJ, Nichols J, Siggia ED, & Hadjantonakis AK (2018). Micropattern differentiation of mouse pluripotent stem cells recapitulates embryo regionalized cell fate patterning. eLife, 7, e32839. 10.7554/eLife.32839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgani SM, Saiz N, Garg V, Raina D, Simon CS, Kang M, Arias AM, Nichols J, Schroter C, & Hadjantonakis AK (2018). A Sprouty4 reporter to monitor FGF/ERK signaling activity in ESCs and mice. Developmental Biology, 441(1), 104–126. 10.1016/j.ydbio.2018.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgensztern D, Karaseva N, Felip E, Delgado I, Burdaeva O, Domine M, Lara P, Paik PK, Lassen U, Orlov S, Trigo J, Shomova M, Baker-Neblett K, Vasquez J, Wang X, Yan L, Mitrica I, DeYoung MP, & Garrido P (2019). An open-label phase IB study to evaluate GSK3052230 in combination with paclitaxel and carboplatin, or docetaxel, in FGFR1-amplified non-small cell lung cancer. Lung cancer (Amsterdam, Netherlands), 136, 74–79. 10.1016/j.lungcan.2019.08.011 [DOI] [PubMed] [Google Scholar]
- Mori Y, Saito T, Chang SH, Kobayashi H, Ladel CH, Guehring H, Chung UI, & Kawaguchi H (2014). Identification of fibroblast growth factor-18 as a molecule to protect adult articular cartilage by gene expression profiling. The Journal of Biological Chemistry, 289(14), 10192–10200. 10.1074/jbc.M113.524090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizumi S, Sato S, Koyama K, Okazaki H, Chen Y, Goto H, Kagawa K, Ogawa H, Nishimura H, Kawano H, Toyoda Y, Uehara H, & Nishioka Y (2020). Blockade of pan-fibroblast growth factor receptors mediates bidirectional effects in lung fibrosis. American Journal of Respiratory Cell and Molecular Biology, 63(3), 317–326. 10.1165/rcmb.2019-0090OC [DOI] [PubMed] [Google Scholar]
- Morrison CD, & Laeger T (2015). Protein-dependent regulation of feeding and metabolism. Trends in Endocrinology and Metabolism, 26(5), 256–262. 10.1016/j.tem.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossahebi-Mohammadi M, Quan M, Zhang JS, & Li X (2020). FGF signaling pathway: A key regulator of stem cell pluripotency. Frontiers in Cell and Development Biology, 8, 79. 10.3389/fcell.2020.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motch Perrine SM, Wu M, Stephens NB, Kriti D, van Bakel H, Jabs EW, & Richtsmeier JT (2019). Mandibular dysmorphology due to abnormal embryonic osteogenesis in FGFR2-related craniosynostosis mice. Disease Models & Mechanisms, 12(5), dmm038513. 10.1242/dmm.038513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhl B, Hagele J, Tasdogan A, Loula P, Schuh K, & Bundschu K (2015). SPREDs (Sprouty related proteins with EVH1 domain) promote self-renewal and inhibit mesodermal differentiation in murine embryonic stem cells. Developmental Dynamics, 244(4), 591–606. 10.1002/dvdy.24261 [DOI] [PubMed] [Google Scholar]
- Muller HM, Steringer JP, Wegehingel S, Bleicken S, Munster M, Dimou E, Unger S, Weidmann G, Andreas H, Garcia-Saez AJ, Wild K, Sinning I, & Nickel W (2015). Formation of disulfide bridges drives oligomerization, membrane pore formation, and translocation of fibroblast growth factor 2 to cell surfaces. The Journal of Biological Chemistry, 290(14), 8925–8937. 10.1074/jbc.M114.622456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Lindemann S, & Gigout A (2020). Effects of sprifermin, IGF1, IGF2, BMP7, or CNP on bovine chondrocytes in monolayer and 3D culture. Journal of Orthopaedic Research, 38(3), 653–662. 10.1002/jor.24491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller TL, Ngo-Muller V, Reginelli A, Taylor G, Anderson R, & Muneoka K (1999). Regeneration in higher vertebrates: Limb buds and digit tips. Seminars in Cell & Developmental Biology, 10(4), 405–413. 10.1006/scdb.1999.0327 [DOI] [PubMed] [Google Scholar]
- Munnamalai V, & Fekete DM (2020). The acquisition of positional information across the radial axis of the cochlea. Developmental Dynamics, 249(3), 281–297. 10.1002/dvdy.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Elfenbein A, & Simons M (2008). Non-canonical fibroblast growth factor signalling in angiogenesis. Cardiovascular Research, 78(2), 223–231. 10.1093/cvr/cvm086 [DOI] [PubMed] [Google Scholar]
- Murcia-Belmonte V, Esteban PF, Martinez-Hernandez J, Gruart A, Lujan R, Delgado-Garcia JM, & de Castro F (2016). Anosmin-1 over-expression regulates oligodendrocyte precursor cell proliferation, migration and myelin sheath thickness. Brain Structure & Function, 221(3), 1365–1385. 10.1007/s00429-014-0977-4 [DOI] [PubMed] [Google Scholar]
- Musgrove J, & Wolf M (2020). Regulation and effects of FGF23 in chronic kidney disease. Annual Review of Physiology, 82, 365–390. 10.1146/annurev-physiol-021119-034650 [DOI] [PubMed] [Google Scholar]
- Nacu E, Gromberg E, Oliveira CR, Drechsel D, & Tanaka EM (2016). FGF8 and SHH substitute for anterior-posterior tissue interactions to induce limb regeneration. Nature, 533(7603), 407–410. 10.1038/nature17972 [DOI] [PubMed] [Google Scholar]
- Nadanaka S, & Kitagawa H (2018). Exostosin-like 2 regulates FGF2 signaling by controlling the endocytosis of FGF2. Biochimica et Biophysica Acta - General Subjects, 1862(4), 791–799. 10.1016/j.bbagen.2018.01.002 [DOI] [PubMed] [Google Scholar]
- Naiche LA, Holder N, & Lewandoski M (2011). FGF4 and FGF8 comprise the wavefront activity that controls somitogenesis. Proceedings of the National Academy of Sciences of the United States of America, 108(10), 4018–4023. 10.1073/pnas.1007417108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y (2018). Aptamers as therapeutic middle molecules. Biochimie, 145, 22–33. 10.1016/j.biochi.2017.10.006 [DOI] [PubMed] [Google Scholar]
- Nakamura Y (2021). Multiple therapeutic applications of RBM-007, an anti-FGF2 aptamer. Cells, 10(7), 1617. 10.3390/cells10071617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama F, Umeda S, Yasuda T, Fujita M, Asada M, Meineke V, Imamura T, & Imai T (2014). Cellular internalization of fibroblast growth factor-12 exerts radioprotective effects on intestinal radiation damage independently of FGFR signaling. International Journal of Radiation Oncology, Biology, Physics, 88(2), 377–384. 10.1016/j.ijrobp.2013.10.035 [DOI] [PubMed] [Google Scholar]
- Nakayama F, Yasuda T, Umeda S, Asada M, Imamura T, Meineke V, & Akashi M (2011). Fibroblast growth factor-12 (FGF12) translocation into intestinal epithelial cells is dependent on a novel cell-penetrating peptide domain: Involvement of internalization in the in vivo role of exogenous FGF12. The Journal of Biological Chemistry, 286(29), 25823–25834. 10.1074/jbc.M110.198267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K, Lee KW, Chung O, Yim HS, Cha SS, Lee SW, Jun J, Cho YS, Bhak J, Magalhaes JP, Lee JH, & Jeong JY (2017). Analysis of the FGF gene family provides insights into aquatic adaptation in cetaceans. Scientific Reports, 7, 40233. 10.1038/srep40233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanni M, Ranieri D, Raffa S, Torrisi MR, & Belleudi F (2018). Interplay between FGFR2b-induced autophagy and phagocytosis: Role of PLCgamma-mediated signalling. Journal of Cellular and Molecular Medicine, 22(1), 668–683. 10.1111/jcmm.13352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanni M, Ranieri D, Rosato B, Torrisi MR, & Belleudi F (2018). Role of fibroblast growth factor receptor 2b in the cross talk between autophagy and differentiation: Involvement of Jun N-terminal protein kinase signaling. Molecular and Cellular Biology, 38(13), e00119–18. 10.1128/MCB.00119-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayana J, & Horton WA (2015). FGFR3 biology and skeletal disease. Connective Tissue Research, 56(6), 427–433. 10.3109/03008207.2015.1051224 [DOI] [PubMed] [Google Scholar]
- Narla ST, Bushnell DS, Schaefer CM, Nouraie M, & Bates CM (2020). Keratinocyte growth factor reduces injury and leads to early recovery from cyclophosphamide bladder injury. The American Journal of Pathology, 190(1), 108–124. 10.1016/j.ajpath.2019.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla ST, Bushnell DS, Schaefer CM, Nouraie M, Tometich JT, Hand TW, & Bates CM (2021). Loss of fibroblast growth factor receptor 2 (FGFR2) leads to defective bladder urothelial regeneration after cyclophosphamide injury. The American Journal of Pathology, 191(4), 631–651. 10.1016/j.ajpath.2020.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narvaez M, Andrade-Talavera Y, Valladolid-Acebes I, Fredriksson M, Siegele P, Hernandez-Sosa A, Fisahn A, Fuxe K, & Borroto-Escuela DO (2020). Existence of FGFR1–5-HT1AR heteroreceptor complexes in hippocampal astrocytes. Putative link to 5-HT and FGF2 modulation of hippocampal gamma oscillations. Neuropharmacology, 170, 108070. 10.1016/j.neuropharm.2020.108070 [DOI] [PubMed] [Google Scholar]
- Naski MC, Colvin JS, Coffin JD, & Ornitz DM (1998). Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by fibroblast growth factor receptor 3. Development, 125(24), 4977–4988. [DOI] [PubMed] [Google Scholar]
- Nason SR, Antipenko J, Presedo N, Cunningham SE, Pierre TH, Kim T, Paul JR, Holleman C, Young ME, Gamble KL, Finan B, DiMarchi R, Hunter CS, Kharitonenkov A, & Habegger KM (2021). Glucagon receptor signaling regulates weight loss via central KLB receptor complexes. JCI Insight, 6(4), e141323. 10.1172/jci.insight.141323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauseef JT, Villamar DM, Lebenthal J, Vlachostergios PJ, & Tagawa ST (2020). An evaluation of the efficacy and safety of erdafitinib for the treatment of bladder cancer. Expert Opinion on Pharmacotherapy, 21(8), 863–870. 10.1080/14656566.2020.1736036 [DOI] [PubMed] [Google Scholar]
- Navarrete IA, & Levine M (2016). Nodal and FGF coordinate ascidian neural tube morphogenesis. Development, 143(24), 4665–4675. 10.1242/dev.144733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocka D, Krzyscik MA, Opalinski L, Zakrzewska M, & Otlewski J (2020). Stable fibroblast growth factor 2 dimers with high prosurvival and mitogenic potential. International Journal of Molecular Sciences, 21(11), 4108. 10.3390/ijms21114108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndlovu R, Deng LC, Wu J, Li XK, & Zhang JS (2018). Fibroblast growth factor 10 in pancreas development and pancreatic cancer. Frontiers in Genetics, 9, 482. 10.3389/fgene.2018.00482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neben CL, Lo M, Jura N, & Klein OD (2019). Feedback regulation of RTK signaling in development. Developmental Biology, 447(1), 71–89. 10.1016/j.ydbio.2017.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neben CL, Tuzon CT, Mao X, Lay FD, & Merrill AE (2017). FGFR2 mutations in bent bone dysplasia syndrome activate nucleolar stress and perturb cell fate determination. Human Molecular Genetics, 26(17), 3253–3270. 10.1093/hmg/ddx209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JH, Shaver A, Sheehan JH, Mallal S, Stone JH, Pillai S, Bastarache L, Riebau D, Allard-Chamard H, Stone WM, Perugino C, Pilkinton M, Smith SA, McDonnell WJ, Capra JA, Meiler J, Cogan J, Xing K, Mahajan VS, … Undiagnosed Disease N (2019). IgG4-related disease: Association with a rare gene variant expressed in cytotoxic T cells. Molecular Genetics & Genomic Medicine, 7(6), e686. 10.1002/mgg3.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MB, Cohen I, Kumar V, Xu Z, Bar C, Dauber-Decker KL, Tsai PC, Marangoni P, Klein OD, Hsu YC, Chen T, Mikkola ML, & Ezhkova E (2018). FGF signalling controls the specification of hair placode-derived SOX9 positive progenitors to Merkel cells. Nature Communications, 9(1), 1–10. 10.1038/s41467-018-04399-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Duchesne L, Sankara Narayana GHN, Boggetto N, Fernig DD, Uttamrao Murade C, Ladoux B, & Mege RM (2019). Enhanced cell-cell contact stability and decreased N-cadherin-mediated migration upon fibroblast growth factor receptor-N-cadherin cross talk. Oncogene, 38(35), 6283–6300. 10.1038/s41388-019-0875-6 [DOI] [PubMed] [Google Scholar]
- Nguyen T, & Mege RM (2016). N-cadherin and fibroblast growth factor receptors crosstalk in the control of developmental and cancer cell migrations. European Journal of Cell Biology, 95(11), 415–426. 10.1016/j.ejcb.2016.05.002 [DOI] [PubMed] [Google Scholar]
- Nichols J, & Smith A (2009). Naive and primed pluripotent states. Cell Stem Cell, 4(6), 487–492. 10.1016/j.stem.2009.05.015 [DOI] [PubMed] [Google Scholar]
- Nies VJ, Sancar G, Liu W, van Zutphen T, Struik D, Yu RT, Atkins AR, Evans RM, Jonker JW, & Downes MR (2015). Fibroblast growth factor signaling in metabolic regulation. Frontiers in Endocrinology, 6, 193. 10.3389/fendo.2015.00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaidis NM, Noel JG, Pitstick LB, Gardner JC, Uehara Y, Wu H, Saito A, Lewnard KE, Liu H, White MR, Hartshorn KL, & McCormack FX (2017). Mitogenic stimulation accelerates influenza-induced mortality by increasing susceptibility of alveolar type II cells to infection. Proceedings of the National Academy of Sciences of the United States of America, 114(32), E6613–E6622. 10.1073/pnas.1621172114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J, Xie J, Guo K, Zhang X, Xia F, Zhao X, Song L, Zhuge D, Li X, Zhao Y, & Huang Z (2018). Efficient treatment of Parkinson’s disease using ultrasonography-guided rhFGF20 proteoliposomes. Drug Delivery, 25(1), 1560–1569. 10.1080/10717544.2018.1482972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J, Zhao J, Wu J, Qiao G, Gu J, Zhou C, Li Q, Ying L, Wang D, Lin H, Li X, Mohammadi M, & Huang Z (2020). Curtailing FGF19’s mitogenicity by suppressing its receptor dimerization ability. Proceedings of the National Academy of Sciences of the United States of America, 117(46), 29025–29034. 10.1073/pnas.2010984117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan ML, & White KE (2019). FGF23 Synthesis and Activity. Current Issues in Molecular Biology, 5(1), 18–25. 10.1007/s40610-019-0111-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais A, Lesieur J, Sadoine J, Slimani L, Baroukh B, Saubamea B, Schmitt A, Vital S, Poliard A, Helary C, Rochefort GY, Chaussain C, & Gorin C (2019). Priming dental pulp stem cells from human exfoliated deciduous teeth with fibroblast growth factor-2 enhances mineralization within tissue-engineered constructs implanted in craniofacial bone defects. Stem Cells Translational Medicine, 8(8), 844–857. 10.1002/sctm.18-0182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Azuma M, Tsunematsu T, Morimoto Y, Kondo M, Tezuka T, Nishioka Y, & Tsuneyama K (2018). Neutrophils induce smooth muscle hyperplasia via neutrophil elastase-induced FGF-2 in a mouse model of asthma with mixed inflammation. Clinical and Experimental Allergy, 48(12), 1715–1725. 10.1111/cea.13263 [DOI] [PubMed] [Google Scholar]
- Oginuma M, Moncuquet P, Xiong F, Karoly E, Chal J, Guevorkian K, & Pourquie O (2017). A gradient of glycolytic activity coordinates FGF and Wnt signaling during elongation of the body axis in amniote embryos. Developmental Cell, 40(4), 342–353 e310. 10.1016/j.devcel.2017.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi N, Shibayama M, Kurotaki Y, Imanishi M, Fujimori T, Itoh N, & Takada S (2002). FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes & Development, 16(7), 870–879. 10.1101/gad.965702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y, Huber W, Tsumura A, Kang M, Xenopoulos P, Kurimoto K, Oles AK, Arauzo-Bravo MJ, Saitou M, Hadjantonakis AK, & Hiiragi T (2014). Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nature Cell Biology, 16(1), 27–37. 10.1038/ncb2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura G, Ebina K, Hirao M, Chijimatsu R, Yonetani Y, Etani Y, Miyama A, Takami K, Goshima A, Yoshikawa H, Ishimoto T, Nakano T, Hamada M, Kanamoto T, & Nakata K (2020). Promoting effect of basic fibroblast growth factor in synovial mesenchymal stem cell-based cartilage regeneration. International Journal of Molecular Sciences, 22(1), 300. 10.3390/ijms22010300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazawa M, Murashima A, Harada M, Nakagata N, Noguchi M, Morimoto M, Kimura T, Ornitz DM, & Yamada G (2015). Region-specific regulation of cell proliferation by FGF receptor signaling during the Wolffian duct development. Developmental Biology, 400(1), 139–147. 10.1016/j.ydbio.2015.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura T, Matsushita M, Mishima K, Esaki R, Seki T, Ishiguro N, & Kitoh H (2018). Activated FGFR3 prevents subchondral bone sclerosis during the development of osteoarthritis in transgenic mice with achondroplasia. Journal of Orthopaedic Research, 36(1), 300–308. 10.1002/jor.23608 [DOI] [PubMed] [Google Scholar]
- Oladipupo SS, Kabir AU, Smith C, Choi K, & Ornitz DM (2018). Impaired tumor growth and angiogenesis in mice heterozygous for Vegfr2 (Flk1). Scientific Reports, 8(1), 14724. 10.1038/s41598-018-33037-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladipupo SS, Smith C, Santeford A, Park C, Sene A, Wiley LA, Osei-Owusu P, Hsu J, Zapata N, Liu F, Nakamura R, Lavine KJ, Blumer KJ, Choi K, Apte RS, & Ornitz DM (2014). Endothelial cell FGF signaling is required for injury response but not for vascular homeostasis. Proceedings of the National Academy of Sciences of the United States of America, 111(37), 13379–13384. 10.1073/pnas.1324235111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva-Olivera W, Coin-Araguez L, Lhamyani S, Clemente-Postigo M, Torres JA, Bernal-Lopez MR, El Bekay R, & Tinahones FJ (2017). Adipogenic impairment of adipose tissue-derived mesenchymal stem cells in subjects with metabolic syndrome: Possible protective role of FGF2. The Journal of Clinical Endocrinology and Metabolism, 102(2), 478–487. 10.1210/jc.2016-2256 [DOI] [PubMed] [Google Scholar]
- Olsen SK, Garbi M, Zampieri N, Eliseenkova AV, Ornitz DM, Goldfarb M, & Mohammadi M (2003). Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. The Journal of Biological Chemistry, 278(36), 34226–34236. 10.1074/jbc.M303183200 [DOI] [PubMed] [Google Scholar]
- Omileke F, Ishiwata S, Matsuo J, Yoshida F, Hidese S, Hattori K, & Kunugi H (2020). Possible associations between plasma fibroblast growth factor 21 levels and cognition in bipolar disorder. Neuropsychopharmacology Reports, 40(2), 175–181. 10.1002/npr2.12102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi K, Miyake M, Hori S, Onishi S, Iida K, Morizawa Y, Tatsumi Y, Nakai Y, Tanaka N, & Fujimoto K (2020). gamma-Klotho is correlated with resistance to docetaxel in castration-resistant prostate cancer. Oncology Letters, 19(3), 2306–2316. 10.3892/ol.2020.11308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Kita T, Sato S, O’Neill P, Mak SS, Paschaki M, Ito M, Gotoh N, Kawakami K, Sasai Y, & Ladher RK (2014). FGFR1-Frs2/3 signalling maintains sensory progenitors during inner ear hair cell formation. PLoS Genetics, 10(1), e1004118. 10.1371/journal.pgen.1004118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oost LJ, Kustermann M, Armani A, Blaauw B, & Romanello V (2019). Fibroblast growth factor 21 controls mitophagy and muscle mass. Journal of Cachexia, Sarcopenia and Muscle, 10(3), 630–642. 10.1002/jcsm.12409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalinski L, Szczepara M, Sokolowska-Wedzina A, Zakrzewska M, & Otlewski J (2017). The autoinhibitory function of D1 domain of FGFR1 goes beyond the inhibition of ligand binding. The International Journal of Biochemistry & Cell Biology, 89, 193–198. 10.1016/j.biocel.2017.06.015 [DOI] [PubMed] [Google Scholar]
- Opalinski L, Szymczyk J, Szczepara M, Kucinska M, Krowarsch D, Zakrzewska M, & Otlewski J (2018). High affinity promotes internalization of engineered antibodies targeting FGFR1. International Journal of Molecular Sciences, 19(5), 1435. 10.3390/ijms19051435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, & Itoh N (2015). The fibroblast growth factor signaling pathway. WIREs: Developmental Biology, 4(3), 215–266. 10.1002/wdev.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, & Itoh N (2017). An Introduction to the Fibroblast Growth Factors. In Simons M (Ed.), Fibroblast Growth Factors: Biology and Clinical Application: FGF Biology and Therapeutics (pp. 1–39). World Scientific. https://www.worldscientific.com/doi/10.1142/9789813143371_0001 [Google Scholar]
- Ornitz DM, & Legeai-Mallet L (2017). Achondroplasia: Development, pathogenesis, and therapy. Developmental Dynamics, 246(4), 291–309. 10.1002/dvdy.24479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, & Marie PJ (2002). FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes & Development, 16(12), 1446–1465. 10.1101/gad.990702 [DOI] [PubMed] [Google Scholar]
- Ornitz DM, & Marie PJ (2015). Fibroblast growth factor signaling in skeletal development and disease. Genes & Development, 29(14), 1463–1486. 10.1101/gad.266551.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, & Marie PJ (2019). Fibroblast growth factors in skeletal development. Current Topics in Developmental Biology, 133, 195–234. 10.1016/bs.ctdb.2018.11.020 [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, & Goldfarb M (1996). Receptor specificity of the fibroblast growth factor family. The Journal of Biological Chemistry, 271(25), 15292–15297. 10.1074/jbc.271.25.15292 [DOI] [PubMed] [Google Scholar]
- Ornitz DM, & Yin Y (2012). Signaling networks regulating development of the lower respiratory tract. Cold Spring Harbor Perspectives in Biology, 4(5), 1–19. 10.1101/cshperspect.a008318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa Y, Matsushita M, Hasegawa S, Esaki R, Fujio M, Ohkawara B, Ishiguro N, Ohno K, & Kitoh H (2017). Activated FGFR3 promotes bone formation via accelerating endochondral ossification in mouse model of distraction osteogenesis. Bone, 105, 42–49. 10.1016/j.bone.2017.05.016 [DOI] [PubMed] [Google Scholar]
- Osborn DPS, Li K, Cutty SJ, Nelson AC, Wardle FC, Hinits Y, & Hughes SM (2020). Fgf-driven Tbx protein activities directly induce myf5 and myod to initiate zebrafish myogenesis. Development, 147(8), dev184689. 10.1242/dev.184689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ost M, Coleman V, Voigt A, van Schothorst EM, Keipert S, van der Stelt I, Ringel S, Graja A, Ambrosi T, Kipp AP, Jastroch M, Schulz TJ, Keijer J, & Klaus S (2016). Muscle mitochondrial stress adaptation operates independently of endogenous FGF21 action. Molecular Metabolism, 5(2), 79–90. 10.1016/j.molmet.2015.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka T, Mengsteab PY, & Laurencin CT (2021). Control of mesenchymal cell fate via application of FGF-8b in vitro. Stem Cell Research, 51, 102155. 10.1016/j.scr.2021.102155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte C, Gold SM, Penninx BW, Pariante CM, Etkin A, Fava M, Mohr DC, & Schatzberg AF (2016). Major depressive disorder. Nature Reviews. Disease Primers, 2, 16065. 10.1038/nrdp.2016.65 [DOI] [PubMed] [Google Scholar]
- Oulion S, Bertrand S, & Escriva H (2012). Evolution of the FGF gene family. International Journal of Evolutionary Biology, 2012, 298147. 10.1155/2012/298147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owall L, Kreiborg S, Duno M, Hermann NV, Darvann TA, & Hove H (2020). Phenotypic variability in Muenke syndrome-observations from five Danish families. Clinical Dysmorphology, 29(1), 1–9. 10.1097/MCD.0000000000000300 [DOI] [PubMed] [Google Scholar]
- Owen BM, Mangelsdorf DJ, & Kliewer SA (2015). Tissue-specific actions of the metabolic hormones FGF15/19 and FGF21. Trends in Endocrinology and Metabolism, 26(1), 22–29. 10.1016/j.tem.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki T, Kawamoto T, Iimori Y, Takeshita N, Yamagishi Y, Nakamura H, Kamohara M, Fujita K, Tanahashi M, & Tsumaki N (2020). Evaluation of FGFR inhibitor ASP5878 as a drug candidate for achondroplasia. Scientific Reports, 10(1), 20915. 10.1038/s41598-020-77345-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozasa A, Komatsu Y, Yasoda A, Miura M, Sakuma Y, Nakatsuru Y, Arai H, Itoh N, & Nakao K (2005). Complementary antagonistic actions between C-type natriuretic peptide and the MAPK pathway through FGFR-3 in ATDC5 cells. Bone, 36(6), 1056–1064. 10.1016/j.bone.2005.03.006 [DOI] [PubMed] [Google Scholar]
- Pablo JL, & Pitt GS (2016). Fibroblast growth factor homologous factors: New roles in neuronal health and disease [Review]. The Neuroscientist, 22(1), 19–25. 10.1177/1073858414562217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrissa-Altes S, Bachofner M, Bogorad RL, Pohlmeier L, Rossolini T, Bohm F, Liebisch G, Hellerbrand C, Koteliansky V, Speicher T, & Werner S (2015). Control of hepatocyte proliferation and survival by Fgf receptors is essential for liver regeneration in mice. Gut, 64(9), 1444–1453. 10.1136/gutjnl-2014-307874 [DOI] [PubMed] [Google Scholar]
- Pagano K, Torella R, Foglieni C, Bugatti A, Tomaselli S, Zetta L, Presta M, Rusnati M, Taraboletti G, Colombo G, & Ragona L (2012). Direct and allosteric inhibition of the FGF2/HSPGs/FGFR1 ternary complex formation by an antiangiogenic, thrombospondin-1-mimic small molecule. PLoS One, 7(5), e36990. 10.1371/journal.pone.0036990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RL, Malcuit C, Vilner L, Vojtic I, Shaw S, Hedblom E, Hu J, Pins GD, Rolle MW, & Dominko T (2011). Restoration of skeletal muscle defects with adult human cells delivered on fibrin microthreads. Tissue Engineering. Part A, 17(21–22), 2629–2640. 10.1089/ten.TEA.2011.0024 [DOI] [PubMed] [Google Scholar]
- Pai R, French D, Ma N, Hotzel K, Plise E, Salphati L, Setchell KD, Ware J, Lauriault V, Schutt L, Hartley D, & Dambach D (2012). Antibody-mediated inhibition of fibroblast growth factor 19 results in increased bile acids synthesis and ileal malabsorption of bile acids in cynomolgus monkeys. Toxicological Sciences, 126(2), 446–456. 10.1093/toxsci/kfs011 [DOI] [PubMed] [Google Scholar]
- Pal SK, Rosenberg JE, Hoffman-Censits JH, Berger R, Quinn DI, Galsky MD, Wolf J, Dittrich C, Keam B, Delord JP, Schellens JHM, Gravis G, Medioni J, Maroto P, Sriuranpong V, Charoentum C, Burris HA, Grunwald V, Petrylak D, … Bajorin DF (2018). Efficacy of BGJ398, a fibroblast growth factor receptor 1–3 inhibitor, in patients with previously treated advanced urothelial carcinoma with FGFR3 alterations. Cancer Discovery, 8(7), 812–821. 10.1158/2159-8290.CD-18-0229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo P, Petracca A, Maggi R, Biagini T, Nardella G, Sacco MC, Di Schiavi E, Carella M, Micale L, & Castori M (2019). A novel dominant-negative FGFR1 variant causes Hartsfield syndrome by deregulating RAS/ERK1/2 pathway. European Journal of Human Genetics, 27(7), 1113–1120. 10.1038/s41431-019-0350-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Carbe C, Kupich S, Pickhinke U, Ohlig S, Frye M, Seelige R, Pallerla SR, Moon AM, Lawrence R, Esko JD, Zhang X, & Grobe K (2014). Heparan sulfate expression in the neural crest is essential for mouse cardiogenesis. Matrix Biology, 35, 253–265. 10.1016/j.matbio.2013.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos KP, El-Rayes BF, Tolcher AW, Patnaik A, Rasco DW, Harvey RD, LoRusso PM, Sachdev JC, Abbadessa G, Savage RE, Hall T, Schwartz B, Wang Y, Kazakin J, & Shaib WL (2017). A Phase 1 study of ARQ 087, an oral pan-FGFR inhibitor in patients with advanced solid tumours. British Journal of Cancer, 117(11), 1592–1599. 10.1038/bjc.2017.330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parente R, Sobacchi C, Bottazzi B, Mantovani A, Grcevic D, & Inforzato A (2019). The long pentraxin PTX3 in bone homeostasis and pathology. Frontiers in Immunology, 10, 2628. 10.3389/fimmu.2019.02628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park GC, Song JS, Park HY, Shin SC, Jang JY, Lee JC, Wang SG, Lee BJ, & Jung JS (2016). Role of fibroblast growth factor-5 on the proliferation of human tonsil-derived mesenchymal stem cells. Stem Cells and Development, 25(15), 1149–1160. 10.1089/scd.2016.0061 [DOI] [PubMed] [Google Scholar]
- Park JG, Xu X, Cho S, Hur KY, Lee MS, Kersten S, & Lee AH (2016). CREBH-FGF21 axis improves hepatic steatosis by suppressing adipose tissue lipolysis. Scientific Reports, 6, 27938. 10.1038/srep27938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, VonHoldt BM, Quignon P, Margulies EH, Shao S, Mosher DS, Spady TC, Elkahloun A, Cargill M, Jones PG, Maslen CL, Acland GM, Sutter NB, Kuroki K, Bustamante CD, Wayne RK, & Ostrander EA (2009). An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science, 325(5943), 995–998. 10.1126/science.1173275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HJ, Bronner ME, & Krumlauf R (2016). The vertebrate Hox gene regulatory network for hindbrain segmentation: Evolution and diversification: Coupling of a Hox gene regulatory network to hindbrain segmentation is an ancient trait originating at the base of vertebrates. BioEssays, 38(6), 526–538. 10.1002/bies.201600010 [DOI] [PubMed] [Google Scholar]
- Parsa S, Ramasamy SK, De Langhe S, Gupte VV, Haigh JJ, Medina D, & Bellusci S (2008). Terminal end bud maintenance in mammary gland is dependent upon FGFR2b signaling. Developmental Biology, 317(1), 121–131. 10.1016/j.ydbio.2008.02.014 [DOI] [PubMed] [Google Scholar]
- Pasaoglu T, & Schikorski T (2016). Presynaptic size of associational/commissural CA3 synapses is controlled by fibroblast growth factor 22 in adult mice. Hippocampus, 26(2), 151–160. 10.1002/hipo.22499 [DOI] [PubMed] [Google Scholar]
- Pathak GP, Vrana JD, & Tucker CL (2013). Optogenetic control of cell function using engineered photoreceptors. Biology of the Cell, 105(2), 59–72. 10.1111/boc.201200056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MD, Grubb HN, & Hristova K (2020). Quantifying the strength of heterointeractions among receptor tyrosine kinases from different subfamilies: Implications for cell signaling. The Journal of Biological Chemistry, 295(29), 9917–9933. 10.1074/jbc.RA120.013639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MD, & Hristova K (2019). The RTK interactome: Overview and perspective on RTK heterointeractions. Chemical Reviews, 119(9), 5881–5921. 10.1021/acs.chemrev.8b00467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paur J, Nika L, Maier C, Moscu-Gregor A, Kostka J, Huber D, Mohr T, Heffeter P, Schrottmaier WC, Kappel S, Kandioler D, Holzmann K, Marian B, Berger W, Grusch M, & Grasl-Kraupp B (2015). Fibroblast growth factor receptor 3 isoforms: Novel therapeutic targets for hepatocellular carcinoma? Hepatology, 62(6), 1767–1778. 10.1002/hep.28023 [DOI] [PubMed] [Google Scholar]
- Pawlikowski B, Vogler TO, Gadek K, & Olwin BB (2017). Regulation of skeletal muscle stem cells by fibroblast growth factors. Developmental Dynamics, 246(5), 359–367. 10.1002/dvdy.24495 [DOI] [PubMed] [Google Scholar]
- Peluso S, Douglas A, Hill A, De Angelis C, Moore BL, Grimes G, Petrovich G, Essafi A, & Hill RE (2017). Fibroblast growth factors (FGFs) prime the limb specific Shh enhancer for chromatin changes that balance histone acetylation mediated by E26 transformation-specific (ETS) factors. eLife, 6, e28590. 10.7554/eLife.28590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perantoni AO, Timofeeva O, Naillat F, Richman C, Pajni-Underwood S, Wilson C, Vainio S, Dove LF, & Lewandoski M (2005). Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development, 132(17), 3859–3871. 10.1242/dev.01945 [DOI] [PubMed] [Google Scholar]
- Pereira RO, Tadinada SM, Zasadny FM, Oliveira KJ, Pires KMP, Olvera A, Jeffers J, Souvenir R, McGlauflin R, Seei A, Funari T, Sesaki H, Potthoff MJ, Adams CM, Anderson EJ, & Abel ED (2017). OPA1 deficiency promotes secretion of FGF21 from muscle that prevents obesity and insulin resistance. The EMBO Journal, 36(14), 2126–2145. 10.15252/embj.201696179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera TPS, Jovcheva E, Mevellec L, Vialard J, De Lange D, Verhulst T, Paulussen C, Van De Ven K, King P, Freyne E, Rees DC, Squires M, Saxty G, Page M, Murray CW, Gilissen R, Ward G, Thompson NT, Newell DR, … Lorenzi MV (2017). Discovery and pharmacological characterization of JNJ-42756493 (Erdafitinib), a functionally selective small-molecule FGFR family inhibitor. Molecular Cancer Therapeutics, 16(6), 1010–1020. 10.1158/1535-7163.MCT-16-0589 [DOI] [PubMed] [Google Scholar]
- Perl AK, Hokuto I, Impagnatiello MA, Christofori G, & Whitsett JA (2003). Temporal effects of Sprouty on lung morphogenesis. Developmental Biology, 258(1), 154–168. 10.1016/s0012-1606(03)00106-4 [DOI] [PubMed] [Google Scholar]
- Perochon J, Yu Y, Aughey GN, Medina AB, Southall TD, & Cordero JB (2021). Dynamic adult tracheal plasticity drives stem cell adaptation to changes in intestinal homeostasis in Drosophila. Nature Cell Biology, 23(5), 485–496. 10.1038/s41556-021-00676-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault DP, Lee GK, Park SY, Lee S, Choi D, Jung E, Seong YJ, Park EK, Sung C, Yu R, Bouz A, Pourmoussa A, Kim SJ, Hong YK, & Wong AK (2019). Small peptide modulation of fibroblast growth factor receptor 3-dependent postnatal lymphangiogenesis. Lymphatic Research and Biology, 17(1), 19–29. 10.1089/lrb.2018.0035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RJ, Lee S, Ma L, Zhang D, Schlessinger J, & Shulman GI (2015). FGF1 and FGF19 reverse diabetes by suppression of the hypothalamic-pituitary-adrenal axis. Nature Communications, 6, 6980. 10.1038/ncomms7980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskett E, Kumar S, Baird W, Jaiswal J, Li M, Patel P, Britto JA, & Pauws E (2017). Analysis of the Fgfr2(C342Y) mouse model shows condensation defects due to misregulation of Sox9 expression in prechondrocytic mesenchyme. Biology Open, 6(2), 223–231. 10.1242/bio.022178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CI, Jheon AH, Mostowfi P, Charles C, Ching S, Thirumangalathu S, Barlow LA, & Klein OD (2011). FGF signaling regulates the number of posterior taste papillae by controlling progenitor field size. PLoS Genetics, 7(6), e1002098. 10.1371/journal.pgen.1002098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SJ, & Krasnow MA (2015). Subcellular trafficking of FGF controls tracheal invasion of Drosophila flight muscle. Cells, 160(1–2), 313–323. 10.1016/j.cell.2014.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrey AC, Qeadan F, Middleton EA, Pinchuk IV, Campbell RA, & Beswick EJ (2021). Cytokine release syndrome in COVID-19: Innate immune, vascular, and platelet pathogenic factors differ in severity of disease and sex. Journal of Leukocyte Biology, 109(1), 55–66. 10.1002/JLB.3COVA0820-410RRR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff MJ, Xue K, Li L, Horowitz MC, Steinbacher DM, & Eswarakumar JVP (2016). FGFR2c-mediated ERK-MAPK activity regulates coronal suture development. Developmental Biology, 415(2), 242–250. 10.1016/j.ydbio.2016.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan P, Saikia BB, Sonnaila S, Agrawal S, Alraawi Z, Kumar TKS, & Iyer S (2021). The saga of endocrine FGFs. Cells, 10(9), 2418. 10.3390/cells10092418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell BB, Meaike JD, Canadas KT, Chandy BM, & Buchanan EP (2017). Tracheal cartilaginous sleeve in syndromic craniosynostosis: An underrecognized source of significant morbidity and mortality. The Journal of Craniofacial Surgery, 28(3), 696–699. 10.1097/SCS.0000000000003489 [DOI] [PubMed] [Google Scholar]
- Pinheiro D, & Heisenberg CP (2020). Zebrafish gastrulation: Putting fate in motion. Current Topics in Developmental Biology, 136, 343–375. 10.1016/bs.ctdb.2019.10.009 [DOI] [PubMed] [Google Scholar]
- Pirvola U, Ylikoski J, Trokovic R, Hebert JM, McConnell SK, & Partanen J (2002). FGFR1 is required for the development of the auditory sensory epithelium. Neuron, 35(4), 671–680. 10.1016/s0896-6273(02)00824-3 [DOI] [PubMed] [Google Scholar]
- Pizzute T, Li J, Zhang Y, Davis ME, & Pei M (2016). Fibroblast growth factor ligand dependent proliferation and chondrogenic differentiation of synovium-derived stem cells and concomitant adaptation of Wnt/mitogen-activated protein kinase signals. Tissue Engineering. Part A, 22(15–16), 1036–1046. 10.1089/ten.TEA.2016.0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planavila A, Redondo I, Hondares E, Vinciguerra M, Munts C, Iglesias R, Gabrielli LA, Sitges M, Giralt M, van Bilsen M, & Villarroya F (2013). Fibroblast growth factor 21 protects against cardiac hypertrophy in mice. Nature Communications, 4, 2019. 10.1038/ncomms3019 [DOI] [PubMed] [Google Scholar]
- Planavila A, Redondo-Angulo I, & Villarroya F (2015). FGF21 and cardiac physiopathology. Frontiers in Endocrinology, 6, 133. 10.3389/fendo.2015.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelmann RE, & Gittenberger-de Groot AC (2019). Development and evolution of the metazoan heart. Developmental Dynamics, 248(8), 634–656. 10.1002/dvdy.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanska UM, Fernig DG, & Kinnunen T (2009). Extracellular interactome of the FGF receptor-ligand system: Complexities and the relative simplicity of the worm. Developmental Dynamics, 238(2), 277–293. 10.1002/dvdy.21757 [DOI] [PubMed] [Google Scholar]
- Potter LR, Abbey-Hosch S, & Dickey DM (2006). Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocrine Reviews, 27(1), 47–72. 10.1210/er.2005-0014 [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Kliewer SA, & Mangelsdorf DJ (2012). Endocrine fibroblast growth factors 15/19 and 21: From feast to famine. Genes & Development, 26(4), 312–324. 10.1101/gad.184788.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J, Hernandez P, Guehring H, Getgood A, & Henson F (2014). Intra-articular injection of rhFGF-18 improves the healing in microfracture treated chondral defects in an ovine model. Journal of Orthopaedic Research, 32(5), 669–676. 10.1002/jor.22580 [DOI] [PubMed] [Google Scholar]
- Pozniak M, Sokolowska-Wedzina A, Jastrzebski K, Szymczyk J, Porebska N, Krzyscik MA, Zakrzewska M, Miaczynska M, Otlewski J, & Opalinski L (2020). FGFR1 clustering with engineered tetravalent antibody improves the efficiency and modifies the mechanism of receptor internalization. Molecular Oncology, 14(9), 1998–2021. 10.1002/1878-0261.12740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan A, Zeng XI, Sidhwani P, Marques SR, George V, Targoff KL, Chi NC, & Yelon D (2017). FGF signaling enforces cardiac chamber identity in the developing ventricle. Development, 144(7), 1328–1338. 10.1242/dev.143719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praktiknjo M, Djayadi N, Mohr R, Schierwagen R, Bischoff J, Dold L, Pohlmann A, Schwarze-Zander C, Wasmuth JC, Boesecke C, Rockstroh JK, & Trebicka J (2019). Fibroblast growth factor 21 is independently associated with severe hepatic steatosis in non-obese HIV-infected patients. Liver International, 39(8), 1514–1520. 10.1111/liv.14107 [DOI] [PubMed] [Google Scholar]
- Prasad R, Brewer C, & Burren CP (2016). Hartsfield syndrome associated with a novel heterozygous missense mutation in FGFR1 and incorporating tumoral calcinosis. American Journal of Medical Genetics. Part A, 170(8), 2222–2225. 10.1002/ajmg.a.37731 [DOI] [PubMed] [Google Scholar]
- Prawira A, Le TBU, Ho RZW, & Huynh H (2021). Upregulation of the ErbB family by EZH2 in hepatocellular carcinoma confers resistance to FGFR inhibitor. Journal of Cancer Research and Clinical Oncology, 147(10), 2955–2968. 10.1007/s00432-021-03703-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presta M, Andres G, Leali D, Dell’Era P, & Ronca R (2009). Inflammatory cells and chemokines sustain FGF2-induced angiogenesis. European Cytokine Network, 20(2), 39–50. 10.1684/ecn.2009.0155 [DOI] [PubMed] [Google Scholar]
- Presta M, Chiodelli P, Giacomini A, Rusnati M, & Ronca R (2017). Fibroblast growth factors (FGFs) in cancer: FGF traps as a new therapeutic approach. Pharmacology & Therapeutics, 179, 171–187. 10.1016/j.pharmthera.2017.05.013 [DOI] [PubMed] [Google Scholar]
- Presta M, Dell’Era P, Mitola S, Moroni E, Ronca R, & Rusnati M (2005). Fibroblast growth factor/fibroblast growth factor receptor system in angiogenesis. Cytokine & Growth Factor Reviews, 16(2), 159–178. 10.1016/j.cytogfr.2005.01.004 [DOI] [PubMed] [Google Scholar]
- Presta M, Foglio E, Churruca Schuind A, & Ronca R (2018). Long pentraxin-3 modulates the angiogenic activity of fibroblast growth factor-2. Frontiers in Immunology, 9, 2327. 10.3389/fimmu.2018.02327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochazkova M, Hakkinen TJ, Prochazka J, Spoutil F, Jheon AH, Ahn Y, Krumlauf R, Jernvall J, & Klein OD (2017). FGF signaling refines Wnt gradients to regulate the patterning of taste papillae. Development, 144(12), 2212–2221. 10.1242/dev.148080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochazkova M, Prochazka J, Marangoni P, & Klein OD (2018). Bones, glands, ears and more: The multiple roles of FGF10 in craniofacial development. Frontiers in Genetics, 9, 542. 10.3389/fgene.2018.00542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punovuori K, Migueles RP, Malaguti M, Blin G, Macleod KG, Carragher NO, Pieters T, van Roy F, Stemmler MP, & Lowell S (2019). N-cadherin stabilises neural identity by dampening anti-neural signals. Development, 146(21), dev183269. 10.1242/dev.183269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushothaman S, Elewa A, & Seifert AW (2019). Fgf-signaling is compartmentalized within the mesenchyme and controls proliferation during salamander limb development. eLife, 8, e48507. 10.7554/eLife.48507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Anzovino A, Kim S, Suyama K, Yao J, Hulit J, Agiostratidou G, Chandiramani N, McDaid HM, Nagi C, Cohen HW, Phillips GR, Norton L, & Hazan RB (2014). N-cadherin/FGFR promotes metastasis through epithelial-to-mesenchymal transition and stem/progenitor cell-like properties. Oncogene, 33(26), 3411–3421. 10.1038/onc.2013.310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao G, Xia D, Cheng Z, & Zhang G (2018). Role of Sprouty1 (Spry1) in the pathogenesis of atrial fibrosis. Pathology, Research and Practice, 214(2), 308–313. 10.1016/j.prp.2017.04.021 [DOI] [PubMed] [Google Scholar]
- Qing Y, Yang J, & Wan C (2015). Increased serum fibroblast growth factor 21 levels in patients with schizophrenia. The Australian and New Zealand Journal of Psychiatry, 49(9), 849–850. 10.1177/0004867415575380 [DOI] [PubMed] [Google Scholar]
- Qu F, Palte IC, Gontarz PM, Zhang B, & Guilak F (2020). Transcriptomic analysis of bone and fibrous tissue morphogenesis during digit tip regeneration in the adult mouse. The FASEB Journal, 34(7), 9740–9754. 10.1096/fj.202000330R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quantius J, Schmoldt C, Vazquez-Armendariz AI, Becker C, El Agha E, Wilhelm J, Morty RE, Vadasz I, Mayer K, Gattenloehner S, Fink L, Matrosovich M, Li X, Seeger W, Lohmeyer J, Bellusci S, & Herold S (2016). Influenza virus infects epithelial stem/progenitor cells of the distal lung: Impact on Fgfr2b-driven epithelial repair. PLoS Pathogens, 12(6), e1005544. 10.1371/journal.ppat.1005544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarles LD (2019). Fibroblast growth factor 23 and alpha-Klotho co-dependent and independent functions. Current Opinion in Nephrology and Hypertension, 28(1), 16–25. 10.1097/MNH.0000000000000467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanal-Villalonga A, Ferrer I, Guruceaga E, Cirauqui C, Marrugal A, Ojeda L, Garcia S, Zugazagoitia J, Munoz-Galvan S, Lopez-Rios F, Montuenga L, Vicent S, Molina-Pinelo S, Carnero A, & Paz-Ares L (2020). FGFR1 and FGFR4 oncogenicity depends on n-cadherin and their co-expression may predict FGFR-targeted therapy efficacy. eBioMedicine, 53, 102683. 10.1016/j.ebiom.2020.102683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanal-Villalonga A, Ojeda-Marquez L, Marrugal A, Yague P, Ponce-Aix S, Salinas A, Carnero A, Ferrer I, Molina-Pinelo S, & Paz-Ares L (2018). The FGFR4–388arg variant promotes lung cancer progression by N-cadherin induction. Scientific Reports, 8(1), 2394. 10.1038/s41598-018-20570-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raber-Durlacher JE, von Bultzingslowen I, Logan RM, Bowen J, Al-Azri AR, Everaus H, Gerber E, Gomez JG, Pettersson BG, Soga Y, Spijkervet FK, Tissing WJ, Epstein JB, Elad S, Lalla RV, & Mucositis Study Group of the Multinational Association of Supportive Care in Cancer/International Society of Oral Oncology (MASCC/ISOO). (2013). Systematic review of cytokines and growth factors for the management of oral mucositis in cancer patients. Support Care Cancer, 21(1), 343–355. 10.1007/s00520-012-1594-5 [DOI] [PubMed] [Google Scholar]
- Racioppi C, Kamal AK, Razy-Krajka F, Gambardella G, Zanetti L, di Bernardo D, Sanges R, Christiaen LA, & Ristoratore F (2014). Fibroblast growth factor signalling controls nervous system patterning and pigment cell formation in Ciona intestinalis. Nature Communications, 5, 4830. 10.1038/ncomms5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina D, Bahadori A, Stanoev A, Protzek M, Koseska A, & Schroter C (2021). Cell-cell communication through FGF4 generates and maintains robust proportions of differentiated cell types in embryonic stem cells. Development, 148(21), dev199926. 10.1242/dev.199926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja A, Park I, Haq F, & Ahn SM (2019). FGF19-FGFR4 signaling in hepatocellular carcinoma. Cells, 8(6), 1–16. 10.3390/cells8060536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran R, Bottiger G, Stadelmann C, Karnati S, & Berghoff M (2021). FGF/FGFR pathways in multiple sclerosis and in its disease models. Cells, 10(4), 884. 10.3390/cells10040884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran R, Giraldo-Velasquez M, Stadelmann C, & Berghoff M (2018). Oligodendroglial fibroblast growth factor receptor 1 gene targeting protects mice from experimental autoimmune encephalomyelitis through ERK/AKT phosphorylation. Brain Pathology, 28(2), 212–224. 10.1111/bpa.12487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran R, Rajendran V, Giraldo-Velasquez M, Megalofonou FF, Gurski F, Stadelmann C, Karnati S, & Berghoff M (2021). Oligodendrocyte-specific deletion of FGFR1 reduces cerebellar inflammation and neurodegeneration in MOG35–55-induced EAE. International Journal of Molecular Sciences, 22(17), 9495. 10.3390/ijms22179495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranieri D, Nanni M, Persechino F, Torrisi MR, & Belleudi F (2020). Role of PKCepsilon in the epithelial-mesenchymal transition induced by FGFR2 isoform switch. Cell Communication and Signaling, 18(1), 76. 10.1186/s12964-020-00582-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, Thi Tran H, Wlizla M, Mancini P, Shifley ET, Bloor SD, Han L, Vleminckx K, Wert SE, & Zorn AM (2015). A molecular atlas of Xenopus respiratory system development. Developmental Dynamics, 244(1), 69–85. 10.1002/dvdy.24180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapraeger AC, Krufka A, & Olwin BB (1991). Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science, 252(5013), 1705–1708. 10.1126/science.1646484 [DOI] [PubMed] [Google Scholar]
- Rasmussen KK, Falkesgaard MH, Winther M, Roed NK, Quistgaard CL, Teisen MN, Edslev SM, Petersen DL, Aljubouri A, Christensen C, Thulstrup PW, Lo Leggio L, Teilum K, & Walmod PS (2018). NCAM2 fibronectin type-III domains form a rigid structure that binds and activates the fibroblast growth factor receptor. Scientific Reports, 8(1), 8957. 10.1038/s41598-018-27089-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasouli SJ, El-Brolosy M, Tsedeke AT, Bensimon-Brito A, Ghanbari P, Maischein HM, Kuenne C, & Stainier DY (2018). The flow responsive transcription factor Klf2 is required for myocardial wall integrity by modulating Fgf signaling. eLife, 7, e38889. 10.7554/eLife.38889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratsma DMA, Zillikens MC, & van der Eerden BCJ (2021). Upstream regulators of fibroblast growth factor 23. Frontiers in Endocrinology, 12, 588096. 10.3389/fendo.2021.588096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauschendorfer T, Gurri S, Heggli I, Maddaluno L, Meyer M, Ingles-Prieto A, Janovjak H, & Werner S (2021). Acute and chronic effects of a light-activated FGF receptor in keratinocytes in vitro and in mice. Life Science Alliance, 4(11), e202101100. 10.26508/lsa.202101100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray AT, Mazot P, Brewer JR, Catela C, Dinsmore CJ, & Soriano P (2020). FGF signaling regulates development by processes beyond canonical pathways. Genes & Development, 34(23–24), 1735–1752. 10.1101/gad.342956.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razy-Krajka F, Gravez B, Kaplan N, Racioppi C, Wang W, & Christiaen L (2018). An FGF-driven feed-forward circuit patterns the cardiopharyngeal mesoderm in space and time. eLife, 7, e29656. 10.7554/eLife.29656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real FM, Haas SA, Franchini P, Xiong P, Simakov O, Kuhl H, Schöpflin R, Heller D, Moeinzadeh MH, Heinrich V, Krannich T, Bressin A, Hartmann MF, Wudy SA, Dechmann D, Hurtado A, Barrionuevo FJ, Schindler M, Harabula I, … Lupi añez DG (2020). The mole genome reveals regulatory rearrangements associated with adaptive intersexuality. Science, 370(6513), 208–214. 10.1126/science.aaz2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves WM, Shimai K, Winkley KM, & Veeman MT (2021). Brachyury controls Ciona notochord fate as part of a feed-forward network. Development, 148(3), dev195230. 10.1242/dev.195230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regot S, Hughey JJ, Bajar BT, Carrasco S, & Covert MW (2014). High-sensitivity measurements of multiple kinase activities in live single cells. Cells, 157(7), 1724–1734. 10.1016/j.cell.2014.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch I, Schreiber R, & Prokesch A (2020). Regulation of thermogenic adipocytes during fasting and cold. Molecular and Cellular Endocrinology, 512, 110869. 10.1016/j.mce.2020.110869 [DOI] [PubMed] [Google Scholar]
- Reker D, Siebuhr AS, Thudium CS, Gantzel T, Ladel C, Michaelis M, Aspberg A, Berchtold MW, Karsdal MA, Gigout A, & Bay-Jensen AC (2020). Sprifermin (rhFGF18) versus vehicle induces a biphasic process of extracellular matrix remodeling in human knee OA articular cartilage ex vivo. Scientific Reports, 10(1), 6011. 10.1038/s41598-020-63216-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Ouyang Z, Hou Z, Yan Y, Zhi Z, Shi M, Du M, Liu H, Wen Y, & Shao Y (2020). CIC is a mediator of the ERK1/2-DUSP6 negative feedback loop. iScience, 23(11), 101635. 10.1016/j.isci.2020.101635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Xiao W, Zeng Y, Liu MH, Li GH, Tang ZH, Qu SL, Hao YM, Yuan HQ, & Jiang ZS (2019). Fibroblast growth factor-21 alleviates hypoxia/reoxygenation injury in H9c2 cardiomyocytes by promoting autophagic flux. International Journal of Molecular Medicine, 43(3), 1321–1330. 10.3892/ijmm.2019.4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repana D, & Ross P (2015). Targeting FGF19/FGFR4 pathway: A novel therapeutic strategy for hepatocellular carcinoma. Diseases, 3(4), 294–305. 10.3390/diseases3040294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezzola S, Ronca R, Loda A, Nawaz MI, Tobia C, Paganini G, Maccarinelli F, Giacomini A, Semeraro F, Mor M, & Presta M (2019). The autocrine FGF/FGFR system in both skin and uveal melanoma: FGF trapping as a possible therapeutic approach. Cancers (Basel), 11(9), 1305. 10.3390/cancers11091305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee Y, Bivi N, Farrow E, Lezcano V, Plotkin LI, White KE, & Bellido T (2011). Parathyroid hormone receptor signaling in osteocytes increases the expression of fibroblast growth factor-23 in vitro and in vivo. Bone, 49(4), 636–643. 10.1016/j.bone.2011.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, Thesleff I, & Rice DP (2004). Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. Journal of Clinical Investigation, 113(12), 1692–1700. 10.1172/JCI20384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricolo D, & Araujo SJ (2020). Coordinated crosstalk between microtubules and actin by a spectraplakin regulates lumen formation and branching. eLife, 9, e61111. 10.7554/eLife.61111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckmann T, Kotevic I, & Trueb B (2008). The cell surface receptor FGFRL1 forms constitutive dimers that promote cell adhesion. Experimental Cell Research, 314(5), 1071–1081. 10.1016/j.yexcr.2007.10.029 [DOI] [PubMed] [Google Scholar]
- Rigueur D, Roberts RR, Bobzin L, & Merrill AE (2019). A requirement for Fgfr2 in middle ear development. Genesis, 57(1), e23252. 10.1002/dvg.23252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BB (2021). Comparative assessment of Fgf’s diverse roles in inner ear development: A zebrafish perspective. Developmental Dynamics, 250(11), 1524–1551. 10.1002/dvdy.343 [DOI] [PubMed] [Google Scholar]
- Riley BM, Mansilla MA, Ma J, Daack-Hirsch S, Maher BS, Raffensperger LM, Russo ET, Vieira AR, Dode C, Mohammadi M, Marazita ML, & Murray JC (2007). Impaired FGF signaling contributes to cleft lip and palate. Proceedings of the National Academy of Sciences of the United States of America, 104(11), 4512–4517. 10.1073/pnas.0607956104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissanen TT, Rutanen J, & Yla-Herttuala S (2004). Gene transfer for therapeutic vascular growth in myocardial and peripheral ischemia. Advances in Genetics, 52, 117–164. 10.1016/S0065-2660(04)52004-7 [DOI] [PubMed] [Google Scholar]
- Rizzo A, Ricci AD, Frega G, Di Federico A, & Brandi G (2021). FGFR inhibitors in elderly patients with advanced biliary tract cancer: An unsolved issue. Expert Review of Gastroenterology & Hepatology, 15(5), 567–574. 10.1080/17474124.2021.1911646 [DOI] [PubMed] [Google Scholar]
- Roberts RR, Bobzin L, Teng CS, Pal D, Tuzon CT, Schweitzer R, & Merrill AE (2019). FGF signaling patterns cell fate at the interface between tendon and bone. Development, 146(15), dev170241. 10.1242/dev.170241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JW, Egbert JR, Davydova J, Schmidt H, Jaffe LA, & Potter LR (2017). Dephosphorylation is the mechanism of fibroblast growth factor inhibition of guanylyl cyclase-B. Cellular Signalling, 40, 222–229. 10.1016/j.cellsig.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robling AG, & Bonewald LF (2020). The osteocyte: New insights. Annual Review of Physiology, 82, 485–506. 10.1146/annurev-physiol-021119-034332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochais F, Sturny R, Chao CM, Mesbah K, Bennett M, Mohun TJ, Bellusci S, & Kelly RG (2014). FGF10 promotes regional foetal cardiomyocyte proliferation and adult cardiomyocyte cell-cycle re-entry. Cardiovascular Research, 104(3), 432–442. 10.1093/cvr/cvu232 [DOI] [PubMed] [Google Scholar]
- Rockel JS, Yu C, Whetstone H, Craft AM, Reilly K, Ma H, Tsushima H, Puviindran V, Al-Jazrawe M, Keller GM, & Alman BA (2016). Hedgehog inhibits beta-catenin activity in synovial joint development and osteoarthritis. Journal of Clinical Investigation, 126(5), 1649–1663. 10.1172/JCI80205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues AR, Yakushiji-Kaminatsui N, Atsuta Y, Andrey G, Schorderet P, Duboule D, & Tabin CJ (2017). Integration of Shh and Fgf signaling in controlling Hox gene expression in cultured limb cells. Proceedings of the National Academy of Sciences of the United States of America, 114(12), 3139–3144. 10.1073/pnas.1620767114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Nuevo A, Diaz-Ramos A, Noguera E, Diaz-Saez F, Duran X, Munoz JP, Romero M, Plana N, Sebastian D, Tezze C, Romanello V, Ribas F, Seco J, Planet E, Doctrow SR, Gonzalez J, Borras M, Liesa M, Palacin M, … Zorzano A (2018). Mitochondrial DNA and TLR9 drive muscle inflammation upon Opa1 deficiency. The EMBO Journal, 37(10), e96553. 10.15252/embj.201796553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Zabala M, Aza-Carmona M, Rivera-Pedroza CI, Belinchon A, Guerrero-Zapata I, Barraza-Garcia J, Vallespin E, Lu M, Del Pozo A, Glucksman MJ, Santos-Simarro F, & Heath KE (2017). FGF9 mutation causes craniosynostosis along with multiple synostoses. Human Mutation, 38(11), 1471–1476. 10.1002/humu.23292 [DOI] [PubMed] [Google Scholar]
- Rogelj S, Weinberg RA, Fanning P, & Klagsbrun M (1988). Basic fibroblast growth factor fused to a signal peptide transforms cells. Nature, 331(6152), 173–175. 10.1038/331173a0 [DOI] [PubMed] [Google Scholar]
- Rohacek AM, Bebee TW, Tilton RK, Radens CM, McDermott-Roe C, Peart N, Kaur M, Zaykaner M, Cieply B, Musunuru K, Barash Y, Germiller JA, Krantz ID, Carstens RP, & Epstein DJ (2017). ESRP1 mutations cause hearing loss due to defects in alternative splicing that disrupt cochlear development. Developmental Cell, 43(3), 318–331.e315. 10.1016/j.devcel.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmann E, Brunner HG, Kayserili H, Uyguner O, Nurnberg G, Lew ED, Dobbie A, Eswarakumar VP, Uzumcu A, Ulubil-Emeroglu M, Leroy JG, Li Y, Becker C, Lehnerdt K, Cremers CW, Yuksel-Apak M, Nurnberg P, Kubisch C, Schlessinger J, … Wollnik B (2006). Mutations in different components of FGF signaling in LADD syndrome. Nature Genetics, 38(4), 414–417. 10.1038/ng1757 [DOI] [PubMed] [Google Scholar]
- Ron D, Fuchs Y, & Chorev DS (2008). Know thy Sef: A novel class of feedback antagonists of receptor tyrosine kinase signaling. The International Journal of Biochemistry & Cell Biology, 40(10), 2040–2052. 10.1016/j.biocel.2008.03.013 [DOI] [PubMed] [Google Scholar]
- Ronca R, Ghedini GC, Maccarinelli F, Sacco A, Locatelli SL, Foglio E, Taranto S, Grillo E, Matarazzo S, Castelli R, Paganini G, Desantis V, Cattane N, Cattaneo A, Mor M, Carlo-Stella C, Belotti A, Roccaro AM, Presta M, & Giacomini A (2020). FGF trapping inhibits multiple myeloma growth through c-Myc degradation-induced mitochondrial oxidative stress. Cancer Research, 80(11), 2340–2354. 10.1158/0008-5472.CAN-19-2714 [DOI] [PubMed] [Google Scholar]
- Ronca R, Giacomini A, Di Salle E, Coltrini D, Pagano K, Ragona L, Matarazzo S, Rezzola S, Maiolo D, Torrella R, Moroni E, Mazzieri R, Escobar G, Mor M, Colombo G, & Presta M (2015). Long-pentraxin 3 derivative as a small-molecule FGF trap for cancer therapy. Cancer Cell, 28(2), 225–239. 10.1016/j.ccell.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Ronca R, Giacomini A, Rusnati M, & Presta M (2015). The potential of fibroblast growth factor/fibroblast growth factor receptor signaling as a therapeutic target in tumor angiogenesis. Expert Opinion on Therapeutic Targets, 19(10), 1361–1377. 10.1517/14728222.2015.1062475 [DOI] [PubMed] [Google Scholar]
- Ronca R, Tamma R, Coltrini D, Ruggieri S, Presta M, & Ribatti D (2017). Fibroblast growth factor modulates mast cell recruitment in a murine model of prostate cancer. Oncotarget, 8(47), 82583–82592. 10.18632/oncotarget.19773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales-Soto G, Diaz-Vegas A, Casas M, Contreras-Ferrat A, & Jaimovich E (2020). Fibroblast growth factor-21 potentiates glucose transport in skeletal muscle fibers. Journal of Molecular Endocrinology, 65(3), 85–95. 10.1530/JME-19-0210 [DOI] [PubMed] [Google Scholar]
- Rosato B, Ranieri D, Nanni M, Torrisi MR, & Belleudi F (2018). Role of FGFR2b expression and signaling in keratinocyte differentiation: Sequential involvement of PKCdelta and PKCalpha. Cell Death & Disease, 9(5), 565. 10.1038/s41419-018-0509-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R Jr. (2020). The role of fibroblast growth factor receptor (FGFR) protein-tyrosine kinase inhibitors in the treatment of cancers including those of the urinary bladder. Pharmacological Research, 151, 104567. 10.1016/j.phrs.2019.104567 [DOI] [PubMed] [Google Scholar]
- Roubal K, Myint ZW, & Kolesar JM (2020). Erdafitinib: A novel therapy for FGFR-mutated urothelial cancer. American Journal of Health-System Pharmacy, 77(5), 346–351. 10.1093/ajhp/zxz329 [DOI] [PubMed] [Google Scholar]
- Roy C, Lejeune S, Slimani A, de Meester C, Ahn As SA, Rousseau MF, Mihaela A, Ginion A, Ferracin B, Pasquet A, Vancraeynest D, Beauloye C, Vanoverschelde JL, Horman S, Gruson D, Gerber BL, & Pouleur AC (2020). Fibroblast growth factor 23: A biomarker of fibrosis and prognosis in heart failure with preserved ejection fraction. ESC Heart Failure, 7(5), 2494–2507. 10.1002/ehf2.12816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybon L, Lamas NJ, Garcia AD, Yang EJ, Sattler R, Lewis VJ, Kim YA, Kachel CA, Rothstein JD, Przedborski S, Wichterle H, & Henderson CE (2013). Human stem cell-derived spinal cord astrocytes with defined mature or reactive phenotypes. Cell Reports, 4(5), 1035–1048. 10.1016/j.celrep.2013.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozo M, Li L, & Fan CM (2016). Targeting beta1-integrin signaling enhances regeneration in aged and dystrophic muscle in mice. Nature Medicine, 22(8), 889–896. 10.1038/nm.4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudakou U, Yu E, Krohn L, Ruskey JA, Asayesh F, Dauvilliers Y, Spiegelman D, Greenbaum L, Fahn S, Waters CH, Dupre N, Rouleau GA, Hassin-Baer S, Fon EA, Alcalay RN, & Gan-Or Z (2021). Targeted sequencing of Parkinson’s disease loci genes highlights SYT11, FGF20 and other associations. Brain, 144(2), 462–472. 10.1093/brain/awaa401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugarli EI, Lutz B, Kuratani SC, Wawersik S, Borsani G, Ballabio A, & Eichele G (1993). Expression pattern of the Kallmann syndrome gene in the olfactory system suggests a role in neuronal targeting. Nature Genetics, 4(1), 19–26. 10.1038/ng0593-19 [DOI] [PubMed] [Google Scholar]
- Ruhlmann C, Dannehl D, Brodtruck M, Adams AC, Stenzel J, Lindner T, Krause BJ, Vollmar B, & Kuhla A (2021). Neuroprotective effects of the FGF21 analogue LY2405319. Journal of Alzheimer’s Disease, 80(1), 357–369. 10.3233/JAD-200837 [DOI] [PubMed] [Google Scholar]
- Ruiz-Camp J, & Morty RE (2015). Divergent fibroblast growth factor signaling pathways in lung fibroblast subsets: Where do we go from here? American Journal of Physiology. Lung Cellular and Molecular Physiology, 309(8), L751–L755. 10.1152/ajplung.00298.2015 [DOI] [PubMed] [Google Scholar]
- Ruiz-Padilla AJ, Ruiz-Noa Y, Del Rocio Ibarra-Reynoso L, Lazo-de-la-Vega-Monroy ML, Alonso-Castro AJ, Sanchez-Barajas M, Alvarez-Alvarez RM, & Del Carmen Preciado-Puga M (2020). FGF21 and its relationship with inflammatory and metabolic parameters in HIV patients after antiretroviral treatment. Current HIV Research, 18(5), 308–314. 10.2174/1570162X18666200719235625 [DOI] [PubMed] [Google Scholar]
- Rusnati M, Camozzi M, Moroni E, Bottazzi B, Peri G, Indraccolo S, Amadori A, Mantovani A, & Presta M (2004). Selective recognition of fibroblast growth factor-2 by the long pentraxin PTX3 inhibits angiogenesis. Blood, 104(1), 92–99. 10.1182/blood-2003-10-3433 [DOI] [PubMed] [Google Scholar]
- Ryu YH, Kyun Chae J, Kim JW, & Lee S (2020). Lacrimo-auriculo-dento-digital syndrome: A novel mutation in a Korean family and review of literature. Molecular Genetics & Genomic Medicine, 8(10), e1412. 10.1002/mgg3.1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi S, Kalhor H, Panahi M, Abolhasani H, Rahimi B, Kalhor R, Vahdatinia M, & Rahimi H (2021). Keratinocyte growth factor in focus: A comprehensive review from structural and functional aspects to therapeutic applications of palifermin. International Journal of Biological Macromolecules, 191, 1175–1190. 10.1016/j.ijbiomac.2021.09.151 [DOI] [PubMed] [Google Scholar]
- Sadhukhan D, Das G, Biswas A, Ghosh S, Das SK, Ray K, & Ray J (2018). Evaluation of FGF 20 variants for susceptibility to Parkinson’s disease in Eastern Indians. Neuroscience Letters, 675, 68–73. 10.1016/j.neulet.2018.03.059 [DOI] [PubMed] [Google Scholar]
- Saera-Vila A, Kish PE, & Kahana A (2016). Fgf regulates dedifferentiation during skeletal muscle regeneration in adult zebrafish. Cellular Signalling, 28(9), 1196–1204. 10.1016/j.cellsig.2016.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai X, & Ladher RK (2015). Early steps in inner ear development: Induction and morphogenesis of the otic placode. Frontiers in Pharmacology, 6, 19. 10.3389/fphar.2015.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Laurent C, Garcia S, Sarrazy V, Dumas K, Authier F, Sore S, Tran A, Gual P, Gennero I, Salles JP, & Gouze E (2018). Early postnatal soluble FGFR3 therapy prevents the atypical development of obesity in achondroplasia. PLoS One, 13(4), e0195876. 10.1371/journal.pone.0195876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito N, Nishimura K, Makanae A, & Satoh A (2019). Fgf- and Bmp-signaling regulate gill regeneration in Ambystoma mexicanum. Developmental Biology, 452(2), 104–113. 10.1016/j.ydbio.2019.04.011 [DOI] [PubMed] [Google Scholar]
- Saiz N, Mora-Bitria L, Rahman S, George H, Herder JP, Garcia-Ojalvo J, & Hadjantonakis AK (2020). Growth-factor-mediated coupling between lineage size and cell fate choice underlies robustness of mammalian development. eLife, 9, e56079. 10.7554/eLife.56079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiz N, Williams KM, Seshan VE, & Hadjantonakis AK (2016). Asynchronous fate decisions by single cells collectively ensure consistent lineage composition in the mouse blastocyst. Nature Communications, 7, 13463. 10.1038/ncomms13463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar L, Kashiwada T, Krejci P, Meyer AN, Casale M, Hallowell M, Wilcox WR, Donoghue DJ, & Thompson LM (2014). Fibroblast growth factor receptor 3 interacts with and activates TGFbeta-activated kinase 1 tyrosine phosphorylation and NFkappaB signaling in multiple myeloma and bladder cancer. PLoS One, 9(1), e86470. 10.1371/journal.pone.0086470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salva JE, & Merrill AE (2017). Signaling networks in joint development. Developmental Dynamics, 246(4), 262–274. 10.1002/dvdy.24472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salva JE, Roberts RR, Stucky TS, & Merrill AE (2019). Nuclear FGFR2 regulates musculoskeletal integration within the developing limb. Developmental Dynamics, 248(3), 233–246. 10.1002/dvdy.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Duffhues G, Garcia de Vinuesa A, & Ten Dijke P (2018). Endothelial-to-mesenchymal transition in cardiovascular diseases: Developmental signaling pathways gone awry. Developmental Dynamics, 247(3), 492–508. 10.1002/dvdy.24589 [DOI] [PubMed] [Google Scholar]
- Sanchez-Garrido MA, Habegger KM, Clemmensen C, Holleman C, Muller TD, Perez-Tilve D, Li P, Agrawal AS, Finan B, Drucker DJ, Tschop MH, DiMarchi RD, & Kharitonenkov A (2016). Fibroblast activation protein (FAP) as a novel metabolic target. Molecular Metabolism, 5(10), 1015–1024. 10.1016/j.molmet.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Charles ED, Neuschwander-Tetri BA, Loomba R, Harrison SA, Abdelmalek MF, Lawitz EJ, Halegoua-DeMarzio D, Kundu S, Noviello S, Luo Y, & Christian R (2019). Pegbelfermin (BMS-986036), a PEGylated fibroblast growth factor 21 analogue, in patients with non-alcoholic steatohepatitis: A randomised, double-blind, placebo-controlled, phase 2a trial. Lancet, 392(10165), 2705–2717. 10.1016/S0140-6736(18)31785-9 [DOI] [PubMed] [Google Scholar]
- Sarabipour S, & Hristova K (2015). FGFR3 unliganded dimer stabilization by the juxtamembrane domain. Journal of Molecular Biology, 427(8), 1705–1714. 10.1016/j.jmb.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarabipour S, & Hristova K (2016a). Effect of the achondroplasia mutation on FGFR3 dimerization and FGFR3 structural response to fgf1 and fgf2: A quantitative FRET study in osmotically derived plasma membrane vesicles. Biochimica et Biophysica Acta, 1858(7 Pt A), 1436–1442. 10.1016/j.bbamem.2016.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarabipour S, & Hristova K (2016b). Mechanism of FGF receptor dimerization and activation. Nature Communications, 7, 10262. 10.1038/ncomms10262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Kazama JJ, Murasawa A, Otani H, Abe A, Ito S, Ishikawa H, Nakazono K, Kuroda T, Nakano M, & Narita I (2016). Serum fibroblast growth factor 23 (FGF23) in patients with rheumatoid arthritis. Internal Medicine, 55(2), 121–126. 10.2169/internalmedicine.55.5507 [DOI] [PubMed] [Google Scholar]
- Satoh A, Makanae A, Nishimoto Y, & Mitogawa K (2016). FGF and BMP derived from dorsal root ganglia regulate blastema induction in limb regeneration in Ambystoma mexicanum. Developmental Biology, 417(1), 114–125. 10.1016/j.ydbio.2016.07.005 [DOI] [PubMed] [Google Scholar]
- Savarirayan R, Tofts L, Irving M, Wilcox WR, Bacino CA, Hoover-Fong J, Font RU, Harmatz P, Rutsch F, Bober MB, Polgreen LE, Ginebreda I, Mohnike K, Charrow J, Hoernschemeyer D, Ozono K, Alanay Y, Arundel P, Kotani Y, … Day JRS (2021). Safe and persistent growth-promoting effects of vosoritide in children with achondroplasia: 2-year results from an open-label, phase 3 extension study. Genetics in Medicine, 23(12), 2443–2447. 10.1038/s41436-021-01287-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarirayan R, Tofts L, Irving M, Wilcox W, Bacino CA, Hoover-Fong J, Ullot Font R, Harmatz P, Rutsch F, Bober MB, Polgreen LE, Ginebreda I, Mohnike K, Charrow J, Hoernschemeyer D, Ozono K, Alanay Y, Arundel P, Kagami S, … Day J (2020). Once-daily, subcutaneous vosoritide therapy in children with achondroplasia: A randomised, double-blind, phase 3, placebo-controlled, multicentre trial. Lancet, 396(10252), 684–692. 10.1016/S0140-6736(20)31541-5 [DOI] [PubMed] [Google Scholar]
- Savchenko E, Teku GN, Boza-Serrano A, Russ K, Berns M, Deierborg T, Lamas NJ, Wichterle H, Rothstein J, Henderson CE, Vihinen M, & Roybon L (2019). FGF family members differentially regulate maturation and proliferation of stem cell-derived astrocytes. Scientific Reports, 9(1), 9610. 10.1038/s41598-019-46110-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saw S, Aiken A, Fang H, McKee TD, Bregant S, Sanchez O, Chen Y, Weiss A, Dickson BC, Czarny B, Sinha A, Fosang A, Dive V, Waterhouse PD, Kislinger T, & Khokha R (2019). Metalloprotease inhibitor TIMP proteins control FGF-2 bioavailability and regulate skeletal growth. The Journal of Cell Biology, 218(9), 3134–3152. 10.1083/jcb.201906059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada A, Shinya M, Jiang YJ, Kawakami A, Kuroiwa A, & Takeda H (2001). Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development, 128(23), 4873–4880. [DOI] [PubMed] [Google Scholar]
- Sawada T, Arai D, Jing X, Furushima K, Chen Q, Kawakami K, Yokote H, Miyajima M, & Sakaguchi K (2015). Trans-activation between EphA and FGFR regulates self-renewal and differentiation of mouse embryonic neural stem/progenitor cells via differential activation of FRS2alpha. PLoS One, 10(5), e0128826. 10.1371/journal.pone.0128826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena N, Mok KW, & Rendl M (2019). An updated classification of hair follicle morphogenesis. Experimental Dermatology, 28(4), 332–344. 10.1111/exd.13913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlett JM, Rojas JM, Matsen ME, Kaiyala KJ, Stefanovski D, Bergman RN, Nguyen HT, Dorfman MD, Lantier L, Wasserman DH, Mirzadeh Z, Unterman TG, Morton GJ, & Schwartz MW (2016). Central injection of fibroblast growth factor 1 induces sustained remission of diabetic hyperglycemia in rodents. Nature Medicine, 22(7), 800–806. 10.1038/nm.4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavone M, Chiarello P, Talarico V, Mascaro I, Caglioti C, Galati MC, & Raiola G (2019). A case of Kallmann syndrome associated to a novel missense mutation of the FGFR1 gene. Acta Biomed, 90(4), 577–579. 10.23750/abm.v90i4.7170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelch K, Kirschner MB, Williams M, Cheng YY, van Zandwijk N, Grusch M, & Reid G (2018). A link between the fibroblast growth factor axis and the miR-16 family reveals potential new treatment combinations in mesothelioma. Molecular Oncology, 12(1), 58–73. 10.1002/1878-0261.12150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmang T (2007). Expression and functions of FGF ligands during early otic development. The International Journal of Developmental Biology, 51(6–7), 473–481. 10.1387/ijdb.072334ts [DOI] [PubMed] [Google Scholar]
- Schindeler A, Biggin A, & Munns CF (2020). Clinical evidence for the benefits of burosumab therapy for X-linked hypophosphatemia (XLH) and other conditions in adults and children. Frontiers in Endocrinology, 11, 338. 10.3389/fendo.2020.00338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid GJ, Kobayashi C, Sandell LJ, & Ornitz DM (2009). Fibroblast growth factor expression during skeletal fracture healing in mice. Developmental Dynamics, 238(3), 766–774. 10.1002/dvdy.21882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt L, Taiyab A, Melvin VS, Jones KL, & Williams T (2018). Increased FGF8 signaling promotes chondrogenic rather than osteogenic development in the embryonic skull. Disease Models & Mechanisms, 11(6), dmm031526. 10.1242/dmm.031526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MO, Garman KA, Lee YG, Zuo C, Beck PJ, Tan M, Aguilar-Pimentel JA, Ollert M, Schmidt-Weber C, Fuchs H, Gailus-Durner V, Hrabe de Angelis M, German Mouse Clinic Consortium, Tassi E, Riegel AT, & Wellstein A (2018). The role of fibroblast growth factor-binding protein 1 in skin carcinogenesis and inflammation. The Journal of Investigative Dermatology, 138(1), 179–188. 10.1016/j.jid.2017.07.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz V, Suflita M, Liu X, Zhang X, Yu Y, Li L, Green DE, Xu Y, Zhang F, DeAngelis PL, Liu J, & Linhardt RJ (2017). Heparan sulfate domains required for fibroblast growth factor 1 and 2 signaling through fibroblast growth factor receptor 1c. The Journal of Biological Chemistry, 292(6), 2495–2509. 10.1074/jbc.M116.761585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Liu C, O’Reilly P, Gao H, Song P, Xu B, Ruggeri B, Amin N, Jia T, Preis S, Segura Lepe M, Akira S, Barbieri C, Baumeister S, Cauchi S, Clarke TK, Enroth S, Fischer K, Hallfors J, … Elliott P (2016). KLB is associated with alcohol drinking, and its gene product beta-Klotho is necessary for FGF21 regulation of alcohol preference. Proceedings of the National Academy of Sciences of the United States of America, 113(50), 14372–14377. 10.1073/pnas.1611243113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Cote LE, Rogers T, & Reddien PW (2016). Two FGFRL-Wnt circuits organize the planarian anteroposterior axis. eLife, 5, e12845. 10.7554/eLife.12845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiji Y, Ito T, Nakamura Y, Nakaishi-Fukuchi Y, Matsuo A, Sato N, & Nogawa H (2019). Alveolus-like organoid from isolated tip epithelium of embryonic mouse lung. Human Cell, 32(2), 103–113. 10.1007/s13577-019-00236-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz T, Freese K, Dietrich P, Thasler WE, Bosserhoff A, & Hellerbrand C (2020). Fibroblast growth factor 9 is expressed by activated hepatic stellate cells and promotes progression of hepatocellular carcinoma. Scientific Reports, 10(1), 4546. 10.1038/s41598-020-61510-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz T, & Hellerbrand C (2021). Role of fibroblast growth factor signalling in hepatic fibrosis. Liver International, 41(6), 1201–1215. 10.1111/liv.14863 [DOI] [PubMed] [Google Scholar]
- Seki E, Enomoto K, Tanoue K, Tanaka M, & Kurosawa K (2020). Tracheal cartilaginous sleeve in patients with Beare-Stevenson syndrome. Congenital Anomalies, 60(3), 97–99. 10.1111/cga.12352 [DOI] [PubMed] [Google Scholar]
- Seo HR, Jeong HE, Joo HJ, Choi SC, Park CY, Kim JH, Choi JH, Cui LH, Hong SJ, Chung S, & Lim DS (2016). Intrinsic FGF2 and FGF5 promotes angiogenesis of human aortic endothelial cells in 3D microfluidic angiogenesis system. Scientific Reports, 6, 28832. 10.1038/srep28832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer GD, Ballif BC, Meurs K, Shaffer LG, & Flores-Smith H (2021). Identification of a novel missense mutation in the fibroblast growth factor 5 gene associated with longhair in the Maine Coon Cat. Human Genetics, 140(11), 1517–1523. 10.1007/s00439-021-02373-1 [DOI] [PubMed] [Google Scholar]
- Shamsi F, Xue R, Huang TL, Lundh M, Liu Y, Leiria LO, Lynes MD, Kempf E, Wang CH, Sugimoto S, Nigro P, Landgraf K, Schulz T, Li Y, Emanuelli B, Kothakota S, Williams LT, Jessen N, Pedersen SB, … Tseng YH (2020). FGF6 and FGF9 regulate UCP1 expression independent of brown adipogenesis. Nature Communications, 11(1), 1421. 10.1038/s41467-020-15055-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shazeeb MS, Cox MK, Gupta A, Tang W, Singh K, Pryce CT, Fogle R, Mu Y, Weber WD, Bangari DS, Ying X, & Sabbagh Y (2018). Skeletal characterization of the Fgfr3 mouse model of achondroplasia using micro-CT and MRI volumetric imaging. Scientific Reports, 8(1), 469. 10.1038/s41598-017-18801-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, O’Reilly VC, Moreau JL, Bewes TR, Yam MX, Chapman BE, Grieve SM, Stocker R, Graham RM, Chapman G, Sparrow DB, & Dunwoodie SL (2016). Gestational stress induces the unfolded protein response, resulting in heart defects. Development, 143(14), 2561–2572. 10.1242/dev.136820 [DOI] [PubMed] [Google Scholar]
- Shi HJ, Wang MW, Sun JT, Wang H, Li YF, Chen BR, Fan Y, Wang SB, Wang ZM, Wang QM, & Wang LS (2019). A novel long noncoding RNA FAF inhibits apoptosis via upregulating FGF9 through PI3K/AKT signaling pathway in ischemia-hypoxia cardiomyocytes. Journal of Cellular Physiology, 234(12), 21973–21987. 10.1002/jcp.28760 [DOI] [PubMed] [Google Scholar]
- Shi L, Tian C, Sun L, Cao F, & Meng Z (2018). The lncRNA TUG1/miR-145–5p/FGF10 regulates proliferation and migration in VSMCs of hypertension. Biochemical and Biophysical Research Communications, 501(3), 688–695. 10.1016/j.bbrc.2018.05.049 [DOI] [PubMed] [Google Scholar]
- Shi SY, Lu YW, Liu Z, Stevens J, Murawsky CM, Wilson V, Hu Z, Richards WG, Michaels ML, Zhang J, Yan W, & Li Y (2018). A biparatopic agonistic antibody that mimics fibroblast growth factor 21 ligand activity. The Journal of Biological Chemistry, 293(16), 5909–5919. 10.1074/jbc.RA118.001752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata E, Yokota Y, Horita N, Kudo A, Abe G, Kawakami K, & Kawakami A (2016). Fgf signalling controls diverse aspects of fin regeneration. Development, 143(16), 2920–2929. 10.1242/dev.140699 [DOI] [PubMed] [Google Scholar]
- Shilo BZ (2014). The regulation and functions of MAPK pathways in Drosophila. Methods, 68(1), 151–159. 10.1016/j.ymeth.2014.01.020 [DOI] [PubMed] [Google Scholar]
- Shilo BZ (2016). New twists in Drosophila cell signaling. The Journal of Biological Chemistry, 291(15), 7805–7808. 10.1074/jbc.R115.711473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Hasegawa H, Yamazaki Y, Muto T, Hino R, Takeuchi Y, Fujita T, Nakahara K, Fukumoto S, & Yamashita T (2004). FGF-23 is a potent regulator of vitamin D metabolism and phosphate homeostasis. Journal of Bone and Mineral Research, 19(3), 429–435. 10.1359/JBMR.0301264 [DOI] [PubMed] [Google Scholar]
- Shimada T, Mizutani S, Muto T, Yoneya T, Hino R, Takeda S, Takeuchi Y, Fujita T, Fukumoto S, & Yamashita T (2001). Cloning and characterization of FGF23 as a causative factor of tumor-induced osteomalacia. Proceedings of the National Academy of Sciences of the United States of America, 98(11), 6500–6505. 10.1073/pnas.101545198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimbori C, Bellaye PS, Xia J, Gauldie J, Ask K, Ramos C, Becerril C, Pardo A, Selman M, & Kolb M (2016). Fibroblast growth factor-1 attenuates TGF-beta1-induced lung fibrosis. The Journal of Pathology, 240(2), 197–210. 10.1002/path.4768 [DOI] [PubMed] [Google Scholar]
- Shin WS, Lee HW, & Lee ST (2019). Catalytically inactive receptor tyrosine kinase PTK7 activates FGFR1 independent of FGF. The FASEB Journal, 33(11), 12960–12971. 10.1096/fj.201900932R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinmura K, Kato H, Matsuura S, Inoue Y, Igarashi H, Nagura K, Nakamura S, Maruyama K, Tajima M, Funai K, Ogawa H, Tanahashi M, Niwa H, & Sugimura H (2014). A novel somatic FGFR3 mutation in primary lung cancer. Oncology Reports, 31(3), 1219–1224. 10.3892/or.2014.2984 [DOI] [PubMed] [Google Scholar]
- Shu C, Smith SM, Little CB, & Melrose J (2016). Use of FGF-2 and FGF-18 to direct bone marrow stromal stem cells to chondrogenic and osteogenic lineages. Future Science OA, 2(4), FSO142. 10.4155/fsoa-2016-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu CC, Jackson MT, Smith MM, Smith SM, Penm S, Lord MS, Whitelock JM, Little CB, & Melrose J (2016). Ablation of perlecan domain 1 heparan sulfate reduces progressive cartilage degradation, synovitis, and osteophyte size in a preclinical model of posttraumatic osteoarthritis. Arthritis & Rhematology, 68(4), 868–879. 10.1002/art.39529 [DOI] [PubMed] [Google Scholar]
- Shuhaibar LC, Robinson JW, Vigone G, Shuhaibar NP, Egbert JR, Baena V, Uliasz TF, Kaback D, Yee SP, Feil R, Fisher MC, Dealy CN, Potter LR, & Jaffe LA (2017). Dephosphorylation of the NPR2 guanylyl cyclase contributes to inhibition of bone growth by fibroblast growth factor. eLife, 6, e31343. 10.7554/eLife.31343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shweta K, Basargekar A, & Ratnaparkhi A (2021). FGFR/Heartless and Smog interact synergistically to negatively regulate Fog mediated G-protein coupled receptor signaling in the Drosophila nervous system. G3 (Bethesda), 11(3), kaa029. 10.1093/g3journal/jkaa029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideek MA, Teia A, Kopecki Z, Cowin AJ, & Gibson MA (2016). Co-localization of LTBP-2 with FGF-2 in fibrotic human keloid and hypertrophic scar. Journal of Molecular Histology, 47(1), 35–45. 10.1007/s10735-015-9645-0 [DOI] [PubMed] [Google Scholar]
- Siggers P, Carre GA, Bogani D, Warr N, Wells S, Hilton H, Esapa C, Hajihosseini MK, & Greenfield A (2014). A novel mouse Fgfr2 mutant, hobbyhorse (hob), exhibits complete XY gonadal sex reversal. PLoS One, 9(6), e100447. 10.1371/journal.pone.0100447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard S, Shail P, MacGregor J, El Sayed M, Duman RS, Vaccarino FM, & Salmaso N (2018). Fibroblast growth factor 2 is necessary for the antidepressant effects of fluoxetine. PLoS One, 13(10), e0204980. 10.1371/journal.pone.0204980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic P, Kim W, Zhou W, Pierce KA, Chang W, Sykes DB, Aziz NB, Elmariah S, Ngo D, Pajevic PD, Govea N, Kestenbaum BR, de Boer IH, Cheng Z, Christov M, Chun J, Leaf DE, Waikar SS, Tager AM, … Rhee EP (2020). Glycerol-3-phosphate is an FGF23 regulator derived from the injured kidney. Journal of Clinical Investigation, 130(3), 1513–1526. 10.1172/JCI131190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon CS, Rahman S, Raina D, Schroter C, & Hadjantonakis AK (2020). Live visualization of ERK activity in the mouse blastocyst reveals lineage-specific signaling dynamics. Developmental Cell, 55(3), 341–353 e345. 10.1016/j.devcel.2020.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M (2021). Fibroblast growth factors: The keepers of endothelial normalcy. Journal of Clinical Investigation, 131(17), e152716. 10.1172/JCI152716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, Dvorak NM, Tapia CM, Mosebarger A, Ali SR, Bullock Z, Chen H, Zhou J, & Laezza F (2021). Differential modulation of the voltage-gated Na(+) channel 1.6 by peptides derived from fibroblast growth factor 14. Frontiers in Molecular Biosciences, 8, 742903. 10.3389/fmolb.2021.742903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal G, Fisher FM, Chee MJ, Tan TG, El Ouaamari A, Adams AC, Najarian R, Kulkarni RN, Benoist C, Flier JS, & Maratos-Flier E (2016). Fibroblast growth factor 21 (FGF21) protects against high fat diet induced inflammation and islet hyperplasia in pancreas. PLoS One, 11(2), e0148252. 10.1371/journal.pone.0148252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal G, Kumar G, Chan S, Fisher FM, Ma Y, Vardeh HG, Nasser IA, Flier JS, & Maratos-Flier E (2018). Deficiency of fibroblast growth factor 21 (FGF21) promotes hepatocellular carcinoma (HCC) in mice on a long term obesogenic diet. Molecular Metabolism, 13, 56–66. 10.1016/j.molmet.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla D, & Wang J (2016). Fibroblast growth factor-9 activates c-kit progenitor cells and enhances angiogenesis in the infarcted diabetic heart. Oxidative Medicine and Cellular Longevity, 2016, 5810908. 10.1155/2016/5810908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletten T, Kostas M, Bober J, Sorensen V, Yadollahi M, Olsnes S, Tomala J, Otlewski J, Zakrzewska M, & Wiedlocha A (2014). Nucleolin regulates phosphorylation and nuclear export of fibroblast growth factor 1 (FGF1). PLoS One, 9(3), e90687. 10.1371/journal.pone.0090687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluzalska KD, Slawski J, Sochacka M, Lampart A, Otlewski J, & Zakrzewska M (2021). Intracellular partners of fibroblast growth factors 1 and 2 - implications for functions. Cytokine & Growth Factor Reviews, 57, 93–111. 10.1016/j.cytogfr.2020.05.004 [DOI] [PubMed] [Google Scholar]
- Smadja DM, Philippe A, Bory O, Gendron N, Beauvais A, Gruest M, Peron N, Khider L, Guerin CL, Goudot G, Levavasseur F, Duchemin J, Pene F, Cheurfa C, Szwebel TA, Sourdeau E, Planquette B, Hauw-Berlemont C, Hermann B, … Chocron R (2021). Placental growth factor level in plasma predicts COVID-19 severity and in-hospital mortality. Journal of Thrombosis and Haemostasis, 19(7), 1823–1830. 10.1111/jth.15339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BM, Traboulsi H, Austin JHM, Manichaikul A, Hoffman EA, Bleecker ER, Cardoso WV, Cooper C, Couper DJ, Dashnaw SM, Guo J, Han MK, Hansel NN, Hughes EW, Jacobs DR Jr., Kanner RE, Kaufman JD, Kleerup E, Lin CL, … MESA Lung and SPIROMICS Investigators. (2018). Human airway branch variation and chronic obstructive pulmonary disease. Proceedings of the National Academy of Sciences of the United States of America, 115(5), E974–E981. 10.1073/pnas.1715564115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LB, Dally MR, Sainz RD, Rodrigue KL, & Oberbauer AM (2006). Enhanced skeletal growth of sheep heterozygous for an inactivated fibroblast growth factor receptor 3. Journal of Animal Science, 84(11), 2942–2949. 10.2527/jas.2006-255 [DOI] [PubMed] [Google Scholar]
- Smith RJ, & Laiks LS (2018). Behavioral and neural mechanisms underlying habitual and compulsive drug seeking. Progress in Neuro-Psychopharmacology & Biological Psychiatry, 87(Pt A), 11–21. 10.1016/j.pnpbp.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smole U, Kratzer B, & Pickl WF (2020). Soluble pattern recognition molecules: Guardians and regulators of homeostasis at airway mucosal surfaces. European Journal of Immunology, 50(5), 624–642. 10.1002/eji.201847811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberg S, Andersen ES, Dalsgaard NB, Jarlhelt I, Hansen NL, Hoffmann N, Vilsboll T, Chenchar A, Jensen M, Grevengoed TJ, Trammell SAJ, Knop FK, & Gillum MP (2018). FGF21, a liver hormone that inhibits alcohol intake in mice, increases in human circulation after acute alcohol ingestion and sustained binge drinking at Oktoberfest. Molecular Metabolism, 11, 96–103. 10.1016/j.molmet.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberg S, Sandholt CH, Jespersen NZ, Toft U, Madsen AL, von Holstein-Rathlou S, Grevengoed TJ, Christensen KB, Bredie WLP, Potthoff MJ, Solomon TPJ, Scheele C, Linneberg A, Jorgensen T, Pedersen O, Hansen T, Gillum MP, & Grarup N (2017). FGF21 is a sugar-induced hormone associated with sweet intake and preference in humans. Cell Metabolism, 25(5), 1045–1053.e1046. 10.1016/j.cmet.2017.04.009 [DOI] [PubMed] [Google Scholar]
- Sochacka M, Opalinski L, Szymczyk J, Zimoch MB, Czyrek A, Krowarsch D, Otlewski J, & Zakrzewska M (2020). FHF1 is a bona fide fibroblast growth factor that activates cellular signaling in FGFR-dependent manner. Cell Communication and Signaling, 18(1), 69. 10.1186/s12964-020-00573-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon-Biet SM, Cogger VC, Pulpitel T, Heblinski M, Wahl D, McMahon AC, Warren A, Durrant-Whyte J, Walters KA, Krycer JR, Ponton F, Gokarn R, Wali JA, Ruohonen K, Conigrave AD, James DE, Raubenheimer D, Morrison CD, Le Couteur DG, & Simpson SJ (2016). Defining the nutritional and metabolic context of FGF21 using the geometric framework. Cell Metabolism, 24(4), 555–565. 10.1016/j.cmet.2016.09.001 [DOI] [PubMed] [Google Scholar]
- Somm E, & Jornayvaz FR (2018). Fibroblast growth factor 15/19: From basic functions to therapeutic perspectives. Endocrine Reviews, 39(6), 960–989. 10.1210/er.2018-00134 [DOI] [PubMed] [Google Scholar]
- Song P, Zechner C, Hernandez G, Canovas J, Xie Y, Sondhi V, Wagner M, Stadlbauer V, Horvath A, Leber B, Hu MC, Moe OW, Mangelsdorf DJ, & Kliewer SA (2018). The hormone FGF21 stimulates water drinking in response to ketogenic diet and alcohol. Cell Metabolism, 27(6), 1338–1347.e1334. 10.1016/j.cmet.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Wang D, Zeng R, & Wang J (2017). Synergistic effects of fibroblast growth factor-2 and bone morphogenetic protein-2 on bone induction. Molecular Medicine Reports, 16(4), 4483–4492. 10.3892/mmr.2017.7183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SH, Kim K, Jo EK, Kim YW, Kwon JS, Bae SS, Sung JH, Park SG, Kim JT, & Suh W (2016). Fibroblast growth factor 12 is a novel regulator of vascular smooth muscle cell plasticity and fate. Arteriosclerosis, Thrombosis, and Vascular Biology, 36(9), 1928–1936. 10.1161/ATVBAHA.116.308017 [DOI] [PubMed] [Google Scholar]
- Sonoda J, Chen MZ, & Baruch A (2017). FGF21-receptor agonists: An emerging therapeutic class for obesity-related diseases. Hormone Molecular Biology and Clinical Investigation, 30(2), 1–13. 10.1515/hmbci-2017-0002 [DOI] [PubMed] [Google Scholar]
- Sontag DP, Wang J, Kardami E, & Cattini PA (2013). FGF-2 and FGF-16 protect isolated perfused mouse hearts from acute doxorubicin-induced contractile dysfunction. Cardiovascular Toxicology, 13(3), 244–253. 10.1007/s12012-013-9203-5 [DOI] [PubMed] [Google Scholar]
- Sparrow DB, Chapman G, Smith AJ, Mattar MZ, Major JA, O’Reilly VC, Saga Y, Zackai EH, Dormans JP, Alman BA, McGregor L, Kageyama R, Kusumi K, & Dunwoodie SL (2012). A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cells, 149(2), 295–306. 10.1016/j.cell.2012.02.054 [DOI] [PubMed] [Google Scholar]
- Sponton CH, & Kajimura S (2018). AAV-mediated gene therapy as a strategy to fight obesity and metabolic diseases. EMBO Molecular Medicine, 10(8), e9431. 10.15252/emmm.201809431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisuma S, Bhattacharya S, Simon DM, Solleti SK, Tyagi S, Starcher B, & Mariani TJ (2010). Fibroblast growth factor receptors control epithelial-mesenchymal interactions necessary for alveolar elastogenesis. American Journal of Respiratory and Critical Care Medicine, 181(8), 838–850. 10.1164/rccm.200904-0544OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak MK, Birkaya B, Aletta JM, Narla ST, Benson CA, Decker B, & Stachowiak EK (2015). Nuclear FGF receptor-1 and CREB binding protein: An integrative signaling module. Journal of Cellular Physiology, 230(5), 989–1002. 10.1002/jcp.24879 [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, & Stachowiak EK (2016). Evidence-based theory for integrated genome regulation of ontogeny—An unprecedented role of nuclear FGFR1 signaling. Journal of Cellular Physiology, 231(6), 1199–1218. 10.1002/jcp.25298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg F, Zhuang L, Beyeler M, Kalin RE, Mullis PE, Brandli AW, & Trueb B (2010). The FGFRL1 receptor is shed from cell membranes, binds fibroblast growth factors (FGFs), and antagonizes FGF signaling in Xenopus embryos. The Journal of Biological Chemistry, 285(3), 2193–2202. 10.1074/jbc.M109.058248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steringer JP, Lange S, Cujova S, Sachl R, Poojari C, Lolicato F, Beutel O, Muller HM, Unger S, Coskun U, Honigmann A, Vattulainen I, Hof M, Freund C, & Nickel W (2017). Key steps in unconventional secretion of fibroblast growth factor 2 reconstituted with purified components. eLife, 6, e28985. 10.7554/eLife.28985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steringer JP, Muller HM, & Nickel W (2015). Unconventional secretion of fibroblast growth factor 2—A novel type of protein translocation across membranes? Journal of Molecular Biology, 427(6 Pt A), 1202–1210. 10.1016/j.jmb.2014.07.012 [DOI] [PubMed] [Google Scholar]
- Stewart CE, Corella KM, Samberg BD, Jones PT, Linscott ML, & Chung WCJ (2016). Perinatal midline astrocyte development is impaired in fibroblast growth factor 8 hypomorphic mice. Brain Research, 1646, 287–296. 10.1016/j.brainres.2016.06.015 [DOI] [PubMed] [Google Scholar]
- Stichelbout M, Dieux-Coeslier A, Clouqueur E, Collet C, & Petit F (2016). A new case of bent bone dysplasia—FGFR2 type and review of the literature. American Journal of Medical Genetics. Part A, 170(3), 785–789. 10.1002/ajmg.a.37473 [DOI] [PubMed] [Google Scholar]
- Stone SI, Wegner DJ, Wambach JA, Cole FS, Urano F, & Ornitz DM (2020). Digenic variants in the FGF21 signaling pathway associated with severe insulin resistance and pseudoacromegaly. Journal of the Endocrine Society, 4(12), bvaa138. 10.1210/jendso/bvaa138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storer MA, Mahmud N, Karamboulas K, Borrett MJ, Yuzwa SA, Gont A, Androschuk A, Sefton MV, Kaplan DR, & Miller FD (2020). Acquisition of a unique mesenchymal precursor-like blastema state underlies successful adult mammalian digit tip regeneration. Developmental Cell, 52(4), 509–524 e509. 10.1016/j.devcel.2019.12.004 [DOI] [PubMed] [Google Scholar]
- Stork T, Sheehan A, Tasdemir-Yilmaz OE, & Freeman MR (2014). Neuron-glia interactions through the heartless FGF receptor signaling pathway mediate morphogenesis of Drosophila astrocytes. Neuron, 83(2), 388–403. 10.1016/j.neuron.2014.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroda A, Brandenburg V, Daher A, Cornelissen C, Goettsch C, Keszei A, & Dreher M (2018). Serum phosphate and phosphate-regulatory hormones in COPD patients. Respiratory Research, 19(1), 183. 10.1186/s12931-018-0889-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su N, Jin M, & Chen L (2014). Role of FGF/FGFR signaling in skeletal development and homeostasis: Learning from mouse models 580525613[Review Article]. Bone Research, 2, 1–24. 10.1038/boneres.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su N, Li X, Tang Y, Yang J, Wen X, Guo J, Tang J, Du X, & Chen L (2016). Deletion of FGFR3 in osteoclast lineage cells results in increased bone mass in mice by inhibiting osteoclastic bone resorption. Journal of Bone and Mineral Research, 31(9), 1676–1687. 10.1002/jbmr.2839 [DOI] [PubMed] [Google Scholar]
- Su Y, Yang LM, & Ornitz DM (2021). FGF20-FGFR1 signaling through MAPK and PI3K controls sensory progenitor differentiation in the organ of Corti. Developmental Dynamics, 250(2), 134–144. 10.1002/dvdy.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh JM, Jonker JW, Ahmadian M, Goetz R, Lackey D, Osborn O, Huang Z, Liu W, Yoshihara E, van Dijk TH, Havinga R, Fan W, Yin YQ, Yu RT, Liddle C, Atkins AR, Olefsky JM, Mohammadi M, Downes M, & Evans RM (2014). Endocrinization of FGF1 produces a neomorphic and potent insulin sensitizer. Nature, 513(7518), 436–439. 10.1038/nature13540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Yang J, Wang D, Li X, Wang L, Yu Q, Qin S, & Tang R (2017). Gabapentin regulates expression of FGF2 and FGFR1 in dorsal root ganglia via microRNA-15a in the arthritis rat model. Journal of Orthopaedic Science, 22(6), 1112–1119. 10.1016/j.jos.2017.08.017 [DOI] [PubMed] [Google Scholar]
- Sun J, Macabenta F, Akos Z, & Stathopoulos A (2020). Collective migrations of drosophila embryonic trunk and caudal mesoderm-derived muscle precursor cells. Genetics, 215(2), 297–322. 10.1534/genetics.120.303258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Niu C, Ye W, An N, Chen G, Huang X, Wang J, Chen X, Shen Y, Huang S, Wang Y, Wang X, Wang Y, Jin L, Cong W, & Li X (2020). FGF13 is a novel regulator of NF-kappaB and potentiates pathological cardiac hypertrophy. iScience, 23(10), 101627. 10.1016/j.isci.2020.101627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Wang Z, Shi H, Gu L, Wang S, Wang H, Li Y, Wei T, Wang Q, & Wang L (2020). LncRNA FAF inhibits fibrosis induced by angiotensinogen II via the TGFbeta1-P-Smad2/3 signalling by targeting FGF9 in cardiac fibroblasts. Biochemical and Biophysical Research Communications, 521(3), 814–820. 10.1016/j.bbrc.2019.10.175 [DOI] [PubMed] [Google Scholar]
- Sun J, Yoon J, Lee M, Hwang YS, & Daar IO (2020). Sprouty2 regulates positioning of retinal progenitors through suppressing the Ras/Raf/MAPK pathway. Scientific Reports, 10(1), 13752. 10.1038/s41598-020-70670-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LL, Lei FR, Jiang XD, Du XL, Xiao L, Li WD, & Li XQ (2020). LncRNA GUSBP5-AS promotes EPC migration and angiogenesis and deep vein thrombosis resolution by regulating FGF2 and MMP2/9 through the miR-223–3p/FOXO1/Akt pathway. Aging (Albany NY), 12(5), 4506–4526. 10.18632/aging.102904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Luo M, Gao Z, Han X, Yan Z, Xie S, Zhao H, & Sun H (2021). TUG1 knockdown suppresses cardiac fibrosis after myocardial infarction. Mammalian Genome, 32(6), 435–442. 10.1007/s00335-021-09895-z [DOI] [PubMed] [Google Scholar]
- Sun X, Meyers EN, Lewandoski M, & Martin GR (1999). Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes & Development, 13(14), 1834–1846. 10.1101/gad.13.14.1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Xie YZ, Jiang YY, Wang GY, Wang YJ, Mei Y, Gao RH, Li YH, Xiao W, Wang WF, & Li DS (2021). FGF21 enhances therapeutic efficacy and reduces side effects of dexamethasone in treatment of rheumatoid arthritis. Inflammation, 44(1), 249–260. 10.1007/s10753-020-01327-5 [DOI] [PubMed] [Google Scholar]
- Sun X, Zhang R, Chen H, Du X, Chen S, Huang J, Liu M, Xu M, Luo F, Jin M, Su N, Qi H, Yang J, Tan Q, Zhang D, Ni Z, Liang S, Zhang B, Chen D, … Xie Y (2020). Fgfr3 mutation disrupts chondrogenesis and bone ossification in zebrafish model mimicking CATSHL syndrome partially via enhanced Wnt/beta-catenin signaling. Theranostics, 10(16), 7111–7130. 10.7150/thno.45286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XY, Wang L, Cheng L, Li NN, Lu ZJ, Li JY, & Peng R (2017). Genetic analysis of FGF20 in Chinese patients with Parkinson’s disease. Neurological Sciences, 38(5), 887–891. 10.1007/s10072-017-2868-y [DOI] [PubMed] [Google Scholar]
- Sun Y, Wang R, Zhao S, Li W, Liu W, Tang L, Wang Z, Wang W, Liu R, Ning G, Wang J, & Hong J (2019). FGF9 inhibits browning program of white adipocytes and associates with human obesity. Journal of Molecular Endocrinology, 62(2), 79–90. 10.1530/JME-18-0151 [DOI] [PubMed] [Google Scholar]
- Susanto A, Zhao G, Wazin F, Feng Y, Rasko JEJ, Bailey CG, & Lovicu FJ (2019). Spred negatively regulates lens growth by modulating epithelial cell proliferation and fiber differentiation. Experimental Eye Research, 178, 160–175. 10.1016/j.exer.2018.09.019 [DOI] [PubMed] [Google Scholar]
- Suyama K, Shapiro I, Guttman M, & Hazan RB (2002). A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell, 2(4), 301–314. 10.1016/s1535-6108(02)00150-2 [DOI] [PubMed] [Google Scholar]
- Suzuki J, & Osumi N (2015). Neural crest and placode contributions to olfactory development. Current Topics in Developmental Biology, 111, 351–374. 10.1016/bs.ctdb.2014.11.010 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Kuzina E, An SJ, Tome F, Mohanty J, Li W, Lee S, Liu Y, Lax I, & Schlessinger J (2020). FGF23 contains two distinct high-affinity binding sites enabling bivalent interactions with alpha-Klotho. Proceedings of the National Academy of Sciences of the United States of America, 117(50), 31800–31807. 10.1073/pnas.2018554117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M, & Foldes G (2018). It takes two: Endothelial-perivascular cell cross-talk in vascular development and disease. Frontiers in Cardiovascular Medicine, 5, 154. 10.3389/fcvm.2018.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczurkowska J, Pischedda F, Pinto B, Manago F, Haas CA, Summa M, Bertorelli R, Papaleo F, Schafer MK, Piccoli G, & Cancedda L (2018). NEGR1 and FGFR2 cooperatively regulate cortical development and core behaviours related to autism disorders in mice. Brain, 141(9), 2772–2794. 10.1093/brain/awy190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szybowska P, Kostas M, Wesche J, Haugsten EM, & Wiedlocha A (2021). Negative regulation of FGFR (fibroblast growth factor receptor) signaling. Cells, 10(6), 1342. 10.3390/cells10061342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczyk J, Sluzalska KD, Materla I, Opalinski L, Otlewski J, & Zakrzewska M (2021). FGF/FGFR-dependent molecular mechanisms underlying anti-cancer drug resistance. Cancers (Basel), 13(22), 5796. 10.3390/cancers13225796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taetzsch T, Brayman VL, & Valdez G (2018). FGF binding proteins (FGFBPs): Modulators of FGF signaling in the developing, adult, and stressed nervous system. Biochimica et Biophysica Acta. Molecular Basis of Disease, 1864(9 Pt B), 2983–2991. 10.1016/j.bbadis.2018.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taetzsch T, Tenga MJ, & Valdez G (2017). Muscle fibers secrete FGFBP1 to slow degeneration of neuromuscular synapses during aging and progression of ALS. The Journal of Neuroscience, 37(1), 70–82. 10.1523/JNEUROSCI.2992-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghiyar L, Hesaraki M, Sayahpour FA, Satarian L, Hosseini S, Aghdami N, & Baghaban Eslaminejad M (2017). Msh homeobox 1 (Msx1)- and Msx2-overexpressing bone marrow-derived mesenchymal stem cells resemble blastema cells and enhance regeneration in mice. The Journal of Biological Chemistry, 292(25), 10520–10533. 10.1074/jbc.M116.774265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghizadeh S, Jones MR, Olmer R, Ulrich S, Danopoulos S, Shen C, Chen C, Wilhelm J, Martin U, Chen C, Al Alam D, & Bellusci S (2020). Fgf10 signaling-based evidence for the existence of an embryonic stage distinct from the pseudoglandular stage during mouse lung development. Frontiers in Cell and Development Biology, 8, 576604. 10.3389/fcell.2020.576604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara N, Akiyama R, Wang J, Kawakami H, Bessho Y, & Kawakami Y (2021). The FGF-AKT pathway is necessary for cardiomyocyte survival for heart regeneration in zebrafish. Developmental Biology, 472, 30–37. 10.1016/j.ydbio.2020.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak HJ, Park TJ, Piao Z, & Lee SH (2017). Separate development of the maxilla and mandible is controlled by regional signaling of the maxillomandibular junction during avian development. Developmental Dynamics, 246(1), 28–40. 10.1002/dvdy.24465 [DOI] [PubMed] [Google Scholar]
- Takagi M, Miyoshi T, Nagashima Y, Shibata N, Yagi H, Fukuzawa R, & Hasegawa T (2016). Novel heterozygous mutation in the extracellular domain of FGFR1 associated with Hartsfield syndrome. Human Genome Variation, 3, 16034. 10.1038/hgv.2016.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase HM, Itoh T, Ino S, Wang T, Koji T, Akira S, Takikawa Y, & Miyajima A (2013). FGF7 is a functional niche signal required for stimulation of adult liver progenitor cells that support liver regeneration. Genes & Development, 27(2), 169–181. 10.1101/gad.204776.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashi Y, & Fukumoto S (2020a). Fibroblast growth factor receptor as a potential candidate for phosphate sensing. Current Opinion in Nephrology and Hypertension, 29(4), 446–452. 10.1097/MNH.0000000000000618 [DOI] [PubMed] [Google Scholar]
- Takashi Y, & Fukumoto S (2020b). Phosphate-sensing and regulatory mechanism of FGF23 production. Journal of Endocrinological Investigation, 43(7), 877–883. 10.1007/s40618-020-01205-9 [DOI] [PubMed] [Google Scholar]
- Takashi Y, Kosako H, Sawatsubashi S, Kinoshita Y, Ito N, Tsoumpra MK, Nangaku M, Abe M, Matsuhisa M, Kato S, Matsumoto T, & Fukumoto S (2019). Activation of unliganded FGF receptor by extracellular phosphate potentiates proteolytic protection of FGF23 by its O-glycosylation. Proceedings of the National Academy of Sciences of the United States of America, 116(23), 11418–11427. 10.1073/pnas.1815166116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashi Y, Sawatsubashi S, Endo I, Ohnishi Y, Abe M, Matsuhisa M, Kawanami D, Matsumoto T, & Fukumoto S (2021). Skeletal FGFR1 signaling is necessary for regulation of serum phosphate level by FGF23 and normal life span. Biochemistry and Biophysics Reports, 27, 101107. 10.1016/j.bbrep.2021.101107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehara T, Teramura T, Onodera Y, Frampton J, & Fukuda K (2015). Cdh2 stabilizes FGFR1 and contributes to primed-state pluripotency in mouse epiblast stem cells. Scientific Reports, 5, 14722. 10.1038/srep14722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaei A, Farkhondeh T, & Forouzanfar F (2020). Fibroblast growth factor: Promising target for schizophrenia. Current Drug Targets, 21(13), 1344–1353. 10.2174/1389450121666200628114843 [DOI] [PubMed] [Google Scholar]
- Talebi F, Ghanbari Mardasi F, Mohammadi Asl J, Bavarsad AH, & Tizno S (2017). Identification of a novel missence mutation in FGFR3 gene in an Iranian family with LADD syndrome by next-generation sequencing. International Journal of Pediatric Otorhinolaryngology, 97, 192–196. 10.1016/j.ijporl.2017.04.016 [DOI] [PubMed] [Google Scholar]
- Taliyan R, Chandran SK, & Kakoty V (2019). Therapeutic approaches to Alzheimer’s type of dementia: A focus on FGF21 mediated neuroprotection. Current Pharmaceutical Design, 25(23), 2555–2568. 10.2174/1381612825666190716101411 [DOI] [PubMed] [Google Scholar]
- Talukdar S, Owen BM, Song P, Hernandez G, Zhang Y, Zhou Y, Scott WT, Paratala B, Turner T, Smith A, Bernardo B, Muller CP, Tang H, Mangelsdorf DJ, Goodwin B, & Kliewer SA (2016). FGF21 regulates sweet and alcohol preference. Cell Metabolism, 23(2), 344–349. 10.1016/j.cmet.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukdar S, Zhou Y, Li D, Rossulek M, Dong J, Somayaji V, Weng Y, Clark R, Lanba A, Owen BM, Brenner MB, Trimmer JK, Gropp KE, Chabot JR, Erion DM, Rolph TP, Goodwin B, & Calle RA (2016). A long-acting FGF21 molecule, PF-05231023, decreases body weight and improves lipid profile in non-human primates and type 2 diabetic subjects. Cell Metabolism, 23(3), 427–440. 10.1016/j.cmet.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Tamamouna V, Rahman MM, Petersson M, Charalambous I, Kux K, Mainor H, Bolender V, Isbilir B, Edgar BA, & Pitsouli C (2021). Remodelling of oxygen-transporting tracheoles drives intestinal regeneration and tumorigenesis in Drosophila. Nature Cell Biology, 23(5), 497–510. 10.1038/s41556-021-00674-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Chen B, Wang Q, Xu W, Wang Y, Lin Z, Luo F, Huang S, Zhu Y, Su N, Jin M, Li C, Kuang L, Qi H, Ni Z, Wang Z, Luo X, Jiang W, Chen H, … Chen L (2018). A novel FGFR1-binding peptide attenuates the degeneration of articular cartilage in adult mice. Osteoarthritis and Cartilage, 26(12), 1733–1743. 10.1016/j.joca.2018.08.012 [DOI] [PubMed] [Google Scholar]
- Tan Q, Wang Z, Wang Q, Wang Y, Huang Z, Su N, Jin M, Kuang L, Qi H, Ni Z, Li C, Zhu Y, Jiang W, Chen H, Deng C, Du X, Xie Y, & Chen L (2018). A novel FGFR1-binding peptide exhibits anti-tumor effect on lung cancer by inhibiting proliferation and angiogenesis. International Journal of Biological Sciences, 14(10), 1389–1398. 10.7150/ijbs.24739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SJ, Smith ER, Hewitson TD, Holt SG, & Toussaint ND (2014). The importance of klotho in phosphate metabolism and kidney disease. Nephrology (Carlton), 19(8), 439–449. 10.1111/nep.12268 [DOI] [PubMed] [Google Scholar]
- Tan Y, Qiao Y, Chen Z, Liu J, Guo Y, Tran T, Tan KS, Wang DY, & Yan Y (2020). FGF2, an immunomodulatory factor in asthma and chronic obstructive pulmonary disease (COPD). Frontiers in Cell and Development Biology, 8, 223. 10.3389/fcell.2020.00223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanajak P, Chattipakorn SC, & Chattipakorn N (2015). Effects of fibroblast growth factor 21 on the heart. The Journal of Endocrinology, 227(2), R13–R30. 10.1530/JOE-15-0289 [DOI] [PubMed] [Google Scholar]
- Tang D, He Y, Li W, & Li H (2019). Wnt/beta-catenin interacts with the FGF pathway to promote proliferation and regenerative cell proliferation in the zebrafish lateral line neuromast. Experimental & Molecular Medicine, 51(5), 1–16. 10.1038/s12276-019-0247-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JB, Wu YF, Cao Y, Chen CH, Zhou YL, Avanessian B, Shimada M, Wang XT, & Liu PY (2016). Basic FGF or VEGF gene therapy corrects insufficiency in the intrinsic healing capacity of tendons. Scientific Reports, 6, 20643. 10.1038/srep20643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Wu M, Lu S, Zhang H, Shen Y, Shen C, Liang H, Ge H, Ding X, & Wang Z (2021). Fgf9 negatively regulates bone mass by inhibiting osteogenesis and promoting osteoclastogenesis via MAPK and PI3K/AKT signaling. Journal of Bone and Mineral Research, 36(4), 779–791. 10.1002/jbmr.4230 [DOI] [PubMed] [Google Scholar]
- Tang L, Wu X, Zhang H, Lu S, Wu M, Shen C, Chen X, Wang Y, Wang W, Shen Y, Gu M, Ding X, Jin X, Fei J, & Wang Z (2017). A point mutation in Fgf9 impedes joint interzone formation leading to multiple synostoses syndrome. Human Molecular Genetics, 26(7), 1280–1293. 10.1093/hmg/ddx029 [DOI] [PubMed] [Google Scholar]
- Tao QQ, Sun YM, Liu ZJ, Ni W, Yang P, Li HL, Lu SJ, & Wu ZY (2014). A variant within FGF1 is associated with Alzheimer’s disease in the Han Chinese population. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics, 165B(2), 131–136. 10.1002/ajmg.b.32205 [DOI] [PubMed] [Google Scholar]
- Tapaltsyan V, Charles C, Hu J, Mindell D, Ahituv N, Wilson GM, Black BL, Viriot L, & Klein OD (2016). Identification of novel Fgf enhancers and their role in dental evolution. Evolution & Development, 18(1), 31–40. 10.1111/ede.12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboletti G, Belotti D, Borsotti P, Vergani V, Rusnati M, Presta M, & Giavazzi R (1997). The 140-kilodalton antiangiogenic fragment of thrombospondin-1 binds to basic fibroblast growth factor. Cell Growth & Differentiation, 8(4), 471–479. [PubMed] [Google Scholar]
- Tarulli GA, Cripps SM, Pask AJ, & Renfree MB (2021). Spatiotemporal map of key signalling factors during early penis development. Developmental Dynamics, 1–16. 10.1002/dvdy.433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassi E, Garman KA, Schmidt MO, Ma X, Kabbara KW, Uren A, Tomita Y, Goetz R, Mohammadi M, Wilcox CS, Riegel AT, Carlstrom M, & Wellstein A (2018). Fibroblast growth factor binding protein 3 (FGFBP3) impacts carbohydrate and lipid metabolism. Scientific Reports, 8(1), 15973. 10.1038/s41598-018-34238-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassi E, Lai EY, Li L, Solis G, Chen Y, Kietzman WE, Ray PE, Riegel AT, Welch WJ, Wilcox CS, & Wellstein A (2018). Blood pressure control by a secreted FGFBP1 (fibroblast growth factor-binding protein). Hypertension, 71(1), 160–167. 10.1161/HYPERTENSIONAHA.117.10268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassi E, McDonnell K, Gibby KA, Tilan JU, Kim SE, Kodack DP, Schmidt MO, Sharif GM, Wilcox CS, Welch WJ, Gallicano GI, Johnson MD, Riegel AT, & Wellstein A (2011). Impact of fibroblast growth factor-binding protein-1 expression on angiogenesis and wound healing. The American Journal of Pathology, 179(5), 2220–2232. 10.1016/j.ajpath.2011.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teague WJ, Jones MLM, Hawkey L, Smyth IM, Catubig A, King SK, Sarila G, Li R, & Hutson JM (2018). FGF10 and the mystery of duodenal atresia in humans. Frontiers in Genetics, 9, 530. 10.3389/fgene.2018.00530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenghao S, Ning C, Shenghai W, Qinlong S, Jiaqian W, Kuo W, Zhanbiao Y, & Xigang M (2020). Keratinocyte growth factor-2 reduces inflammatory response to acute lung injury induced by oleic acid in rats by regulating key proteins of the Wnt/beta-catenin signaling pathway. Evidence-based Complementary and Alternative Medicine, 2020, 8350579. 10.1155/2020/8350579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenghao S, Shenmao M, Zhaojun W, Jijia B, Wenjie Z, Wenyan Z, & Xigang M (2019). Keratinocyte growth factor-2 is protective in oleic acid-induced acute lung injury in rats. Evidence-based Complementary and Alternative Medicine, 2019, 9406580. 10.1155/2019/9406580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi A, Durlacher E, Pitino J, & Umemori H (2020). Neuronal fibroblast growth factor 22 signaling during development, but not in adults, is involved in anhedonia. Neuroreport, 31(2), 125–130. 10.1097/WNR.0000000000001399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi A, Gavin E, Wilson J, & Umemori H (2017). Selective inactivation of fibroblast growth factor 22 (FGF22) in CA3 pyramidal neurons impairs local synaptogenesis and affective behavior without affecting dentate neurogenesis. Frontiers in Synaptic Neuroscience, 9, 17. 10.3389/fnsyn.2017.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi A, Johnson-Venkatesh EM, Bullock B, Lehtinen MK, & Umemori H (2016). Retrograde fibroblast growth factor 22 (FGF22) signaling regulates insulin-like growth factor 2 (IGF2) expression for activity-dependent synapse stabilization in the mammalian brain. eLife, 5, e12151. 10.7554/eLife.12151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwisscha van Scheltinga AF, Bakker SC, Kahn RS, & Kas MJ (2013). Fibroblast growth factors in neurodevelopment and psychopathology. The Neuroscientist, 19(5), 479–494. 10.1177/1073858412472399 [DOI] [PubMed] [Google Scholar]
- Terzuoli E, Corti F, Nannelli G, Giachetti A, Donnini S, & Ziche M (2018). Bradykinin B2 receptor contributes to inflammatory responses in human endothelial cells by the transactivation of the fibroblast growth factor receptor FGFR-1. International Journal of Molecular Sciences, 19(9), 2638. 10.3390/ijms19092638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima TH, Lourenco SV, & Tucker AS (2016). Multiple cranial organ defects after conditionally knocking out Fgf10 in the neural crest. Frontiers in Physiology, 7, 488. 10.3389/fphys.2016.00488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezze C, Romanello V, Desbats MA, Fadini GP, Albiero M, Favaro G, Ciciliot S, Soriano ME, Morbidoni V, Cerqua C, Loefler S, Kern H, Franceschi C, Salvioli S, Conte M, Blaauw B, Zampieri S, Salviati L, Scorrano L, & Sandri M (2017). Age-associated loss of OPA1 in muscle impacts muscle mass, metabolic homeostasis, systemic inflammation, and epithelial senescence. Cell Metabolism, 25(6), 1374–1389 e1376. 10.1016/j.cmet.2017.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezze C, Romanello V, & Sandri M (2019). FGF21 as modulator of metabolism in health and disease. Frontiers in Physiology, 10, 419. 10.3389/fphys.2019.00419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein T, de Melo J, Zibetti C, Clark BS, Juarez F, & Blackshaw S (2016). Control of lens development by Lhx2-regulated neuroretinal FGFs. Development, 143(21), 3994–4002. 10.1242/dev.137760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, & Jaenisch R (2017). Mechanisms of gene regulation in human embryos and pluripotent stem cells. Development, 144(24), 4496–4509. 10.1242/dev.157404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummler K, Rom E, Zeis T, Lindner M, Brunner S, Cole JJ, Arseni D, Mucklisch S, Edgar JM, Schaeren-Wiemers N, Yayon A, & Linington C (2019). Polarizing receptor activation dissociates fibroblast growth factor 2 mediated inhibition of myelination from its neuroprotective potential. Acta Neuropathologica Communications, 7(1), 212. 10.1186/s40478-019-0864-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuresson AC, Croft B, Hailer YD, Liminga G, Arvidsson CG, Harley VR, & Stattin EL (2021). A novel heterozygous variant in FGF9 associated with previously unreported features of multiple synostosis syndrome 3. Clinical Genetics, 99(2), 325–329. 10.1111/cge.13880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichelaar JW, Lu W, & Whitsett JA (2000). Conditional expression of fibroblast growth factor-7 in the developing and mature lung. The Journal of Biological Chemistry, 275(16), 11858–11864. 10.1074/jbc.275.16.11858 [DOI] [PubMed] [Google Scholar]
- Tickle C (2015). How the embryo makes a limb: Determination, polarity and identity. Journal of Anatomy, 227(4), 418–430. 10.1111/joa.12361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba Y, Kiso A, Nakamae S, Sakurai F, Takayama K, & Mizuguchi H (2019). FGF signal is not required for hepatoblast differentiation of human iPS cells. Scientific Reports, 9(1), 3713. 10.1038/s41598-019-40305-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokita M, Abe T, & Suzuki K (2012). The developmental basis of bat wing muscle. Nature Communications, 3, 1302. 10.1038/ncomms2298 [DOI] [PubMed] [Google Scholar]
- Tokunaga T, Shukunami C, Okamoto N, Taniwaki T, Oka K, Sakamoto H, Ide J, Mizuta H, & Hiraki Y (2015). FGF-2 stimulates the growth of tenogenic progenitor cells to facilitate the generation of tenomodulin-positive tenocytes in a rat rotator cuff healing model. The American Journal of Sports Medicine, 43(10), 2411–2422. 10.1177/0363546515597488 [DOI] [PubMed] [Google Scholar]
- Tolcher AW, Papadopoulos KP, Patnaik A, Wilson K, Thayer S, Zanghi J, Gemo AT, Kavanaugh WM, Keer HN, & LoRusso PM (2016). A phase I, first in human study of FP-1039 (GSK3052230), a novel FGF ligand trap, in patients with advanced solid tumors. Annals of Oncology, 27(3), 526–532. 10.1093/annonc/mdv591 [DOI] [PubMed] [Google Scholar]
- Tong L, Zhou J, Rong L, Seeley EJ, Pan J, Zhu X, Liu J, Wang Q, Tang X, Qu J, Bai C, & Song Y (2016). Fibroblast growth factor-10 (FGF-10) mobilizes lung-resident mesenchymal stem cells and protects against acute lung injury. Scientific Reports, 6, 21642. 10.1038/srep21642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touat M, Ileana E, Postel-Vinay S, Andre F, & Soria JC (2015). Targeting FGFR signaling in cancer. Clinical Cancer Research, 21(12), 2684–2694. 10.1158/1078-0432.CCR-14-2329 [DOI] [PubMed] [Google Scholar]
- Toydemir RM, Brassington AE, Bayrak-Toydemir P, Krakowiak PA, Jorde LB, Whitby FG, Longo N, Viskochil DH, Carey JC, & Bamshad MJ (2006). A novel mutation in FGFR3 causes camptodactyly, tall stature, and hearing loss (CATSHL) syndrome. American Journal of Human Genetics, 79(5), 935–941. 10.1086/508433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trarbach EB, Abreu AP, Silveira LF, Garmes HM, Baptista MT, Teles MG, Costa EM, Mohammadi M, Pitteloud N, Mendonca BB, & Latronico AC (2010). Nonsense mutations in FGF8 gene causing different degrees of human gonadotropin-releasing deficiency. The Journal of Clinical Endocrinology and Metabolism, 95(7), 3491–3496. 10.1210/jc.2010-0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi N, & Kumar D (2021). Fibroblast growth factor and kidney disease: Updates for emerging novel therapeutics. Journal of Cellular Physiology, 236(12), 7909–7925. 10.1002/jcp.30497 [DOI] [PubMed] [Google Scholar]
- Trost N, Pena-Llopis S, Koirala S, Stojan J, Potts PR, Fon Tacer K, & Martinez ED (2016). gammaKlotho is a novel marker and cell survival factor in a subset of triple negative breast cancers. Oncotarget, 7(3), 2611–2628. 10.18632/oncotarget.6006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueb B (2011). Biology of FGFRL1, the fifth fibroblast growth factor receptor. Cellular and Molecular Life Sciences, 68(6), 951–964. 10.1007/s00018-010-0576-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu HJ, Zhao CF, Chen ZW, Lin W, & Jiang YC (2020). Fibroblast growth factor (FGF) signaling protects against acute pancreatitis-induced damage by modulating inflammatory responses. Medical Science Monitor, 26, e920684. 10.12659/MSM.920684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y, Qu T, & Chen F (2019). Mutant hFGF23(A12D) stimulates osteoblast differentiation through FGFR3. Journal of Cellular and Molecular Medicine, 23(4), 2933–2942. 10.1111/jcmm.14201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker B, McClelland RL, Allison MA, Budoff MJ, Wu BJ, Barter PJ, Rye KA, & Ong KL (2020). Relationship of fibroblast growth factor 21 levels with inflammation, lipoproteins and non-alcoholic fatty liver disease. Atherosclerosis, 299, 38–44. 10.1016/j.atherosclerosis.2020.03.009 [DOI] [PubMed] [Google Scholar]
- Turner CA, Eren-Kocak E, Inui EG, Watson SJ, & Akil H (2016). Dysregulated fibroblast growth factor (FGF) signaling in neurological and psychiatric disorders. Seminars in Cell & Developmental Biology, 53, 136–143. 10.1016/j.semcdb.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CA, Flagel SB, Blandino P Jr., Watson SJ Jr., & Akil H (2017). Utilizing a unique animal model to better understand human temperament. Current Opinion in Behavioral Sciences, 14, 108–114. 10.1016/j.cobeha.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner N, & Grose R (2010). Fibroblast growth factor signalling: From development to cancer [Review]. Nature Reviews. Cancer, 10(2), 116–129. 10.1038/nrc2780 [DOI] [PubMed] [Google Scholar]
- Turwankar A, & Ghaskadbi S (2019). VEGF and FGF signaling during head regeneration in hydra. Gene, 717, 144047. 10.1016/j.gene.2019.144047 [DOI] [PubMed] [Google Scholar]
- Tuzon CT, Rigueur D, & Merrill AE (2019). Nuclear fibroblast growth factor receptor signaling in skeletal development and disease. Current Osteoporosis Reports, 17(3), 138–146. 10.1007/s11914-019-00512-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun D, Oguz A, Aydin MN, Kurutas EB, Ercan O, Sahin M, Unsal V, Ceren I, Akcay A, & Gul K (2018). Is FGF-23 an early indicator of atherosclerosis and cardiac dysfunction in patients with gestational diabetes? Archives of Endocrinology and Metabolism, 62(5), 506–513. 10.20945/2359-3997000000070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki R, Atsuta S, Ueki A, Hoshiyama J, Li J, Hayashi Y, & Sando S (2019). DNA aptamer assemblies as fibroblast growth factor mimics and their application in stem cell culture. Chemical Communication, 55(18), 2672–2675. 10.1039/c8cc08080a [DOI] [PubMed] [Google Scholar]
- Umair Z, Kumar S, Rafiq K, Kumar V, Reman ZU, Lee SH, Kim S, Lee JY, Lee U, & Kim J (2020). Dusp1 modulates activin/smad2 mediated germ layer specification via FGF signal inhibition in Xenopus embryos. Animal Cells and Systems (Seoul), 24(6), 359–370. 10.1080/19768354.2020.1847732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemori H, Linhoff MW, Ornitz DM, & Sanes JR (2004). FGF22 and its close relatives are presynaptic organizing molecules in the mammalian brain. Cells, 118(2), 257–270. 10.1016/j.cell.2004.06.025 [DOI] [PubMed] [Google Scholar]
- Unger S, Bonafe L, & Gouze E (2017). Current care and investigational therapies in achondroplasia. Current Osteoporosis Reports, 15(2), 53–60. 10.1007/s11914-017-0347-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uriarte I, Latasa MU, Carotti S, Fernandez-Barrena MG, Garcia-Irigoyen O, Elizalde M, Urtasun R, Vespasiani-Gentilucci U, Morini S, de Mingo A, Mari M, Corrales FJ, Prieto J, Berasain C, & Avila MA (2015). Ileal FGF15 contributes to fibrosis-associated hepatocellular carcinoma development. International Journal of Cancer, 136(10), 2469–2475. 10.1002/ijc.29287 [DOI] [PubMed] [Google Scholar]
- Urness LD, Wang X, Doan H, Shumway N, Noyes CA, Gutierrez-Magana E, Lu R, & Mansour SL (2018). Spatial and temporal inhibition of FGFR2b ligands reveals continuous requirements and novel targets in mouse inner ear morphogenesis. Development, 145(24), dev170142. 10.1242/dev.170142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urness LD, Wang X, Shibata S, Ohyama T, & Mansour SL (2015). Fgf10 is required for specification of non-sensory regions of the cochlear epithelium. Developmental Biology, 400(1), 59–71. 10.1016/j.ydbio.2015.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui N, Yoshida M, Takayanagi Y, Nasanbuyan N, Inutsuka A, Kurosu H, Mizukami H, Mori Y, Kuro OM, & Onaka T (2021). Roles of fibroblast growth factor 21 in the control of depression-like behaviours after social defeat stress in male rodents. Journal of Neuroendocrinology, 33(10), e13026. 10.1111/jne.13026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde-Franco G, Binette JS, Li W, Wang H, Chai S, Laflamme F, Tran-Khanh N, Quenneville E, Meijers T, Poole AR, Mort JS, Buschmann MD, & Henderson JE (2006). Defects in articular cartilage metabolism and early arthritis in fibroblast growth factor receptor 3 deficient mice. Human Molecular Genetics, 15(11), 1783–1792. 10.1093/hmg/ddl100 [DOI] [PubMed] [Google Scholar]
- Valverde-Franco G, Liu H, Davidson D, Chai S, Valderrama-Carvajal H, Goltzman D, Ornitz DM, & Henderson JE (2004). Defective bone mineralization and osteopenia in young adult FGFR3−/− mice. Human Molecular Genetics, 13(3), 271–284. 10.1093/hmg/ddh034 [DOI] [PubMed] [Google Scholar]
- van Asten SD, Raaben M, Nota B, & Spaapen RM (2018). Secretome screening reveals fibroblast growth factors as novel inhibitors of viral replication. Journal of Virology, 92(16), e00260–18. 10.1128/JVI.00260-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boxtel AL, Economou AD, Heliot C, & Hill CS (2018). Long-range signaling activation and local inhibition separate the mesoderm and endoderm lineages. Developmental Cell, 44(2), 179–191.e175. 10.1016/j.devcel.2017.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brummelen EMJ, Levchenko E, Domine M, Fennell DA, Kindler HL, Viteri S, Gadgeel S, Lopez PG, Kostorov V, Morgensztern D, Orlov S, Zauderer MG, Vansteenkiste JF, Baker-Neblett K, Vasquez J, Wang X, Bellovin DI, Schellens JHM, Yan L, … Trigo J (2020). A phase Ib study of GSK3052230, an FGF ligand trap in combination with pemetrexed and cisplatin in patients with malignant pleural mesothelioma. Investigational New Drugs, 38(2), 457–467. 10.1007/s10637-019-00783-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wouw J, Broekhuizen M, Sorop O, Joles JA, Verhaar MC, Duncker DJ, Danser AHJ, & Merkus D (2019). Chronic kidney disease as a risk factor for heart failure with preserved ejection fraction: A focus on microcirculatory factors and therapeutic targets. Frontiers in Physiology, 10, 1108. 10.3389/fphys.2019.01108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon EP, Pulskens WP, van der Hagen EA, Lavrijsen M, Vervloet MG, van Goor H, Bindels RJ, & Hoenderop JG (2015). Shedding of klotho by ADAMs in the kidney. American Journal of Physiology. Renal Physiology, 309(4), F359–F368. 10.1152/ajprenal.00240.2014 [DOI] [PubMed] [Google Scholar]
- Vandanmagsar B, Warfel JD, Wicks SE, Ghosh S, Salbaum JM, Burk D, Dubuisson OS, Mendoza TM, Zhang J, Noland RC, & Mynatt RL (2016). Impaired Mitochondrial Fat Oxidation Induces FGF21 in Muscle. Cell Reports, 15(8), 1686–1699. 10.1016/j.celrep.2016.04.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan HN, Mazot P, He F, & Soriano P (2015). Receptor tyrosine kinases modulate distinct transcriptional programs by differential usage of intracellular pathways. eLife, 4, e07186. 10.7554/eLife.07186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedana G, Villarreal G Jr., & Jun AS (2016). Fuchs endothelial corneal dystrophy: Current perspectives. Clinical Ophthalmology, 10, 321–330. 10.2147/OPTH.S83467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venero Galanternik M, Kramer KL, & Piotrowski T (2015). Heparan sulfate proteoglycans regulate Fgf signaling and cell polarity during collective cell migration. Cell Reports, 10(3), 414–428. 10.1016/j.celrep.2014.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venetsanakos E, Brameld KA, Phan VT, Verner E, Owens TD, Xing Y, Tam D, LaStant J, Leung K, Karr DE, Hill RJ, Gerritsen ME, Goldstein DM, Funk JO, & Bradshaw JM (2017). The irreversible covalent fibroblast growth factor receptor inhibitor PRN1371 exhibits sustained inhibition of FGFR after drug clearance. Molecular Cancer Therapeutics, 16(12), 2668–2676. 10.1158/1535-7163.MCT-17-0309 [DOI] [PubMed] [Google Scholar]
- Verbueken D, & Moe OW (2021). Strategies to lower fibroblast growth factor-23 bioactivity. Nephrology, Dialysis, Transplantation, 1–8. 10.1093/ndt/gfab012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernia S, Cavanagh-Kyros J, Barrett T, Tournier C, & Davis RJ (2016). Fibroblast growth factor 21 mediates glycemic regulation by hepatic JNK. Cell Reports, 14(10), 2273–2280. 10.1016/j.celrep.2016.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernia S, Cavanagh-Kyros J, Garcia-Haro L, Sabio G, Barrett T, Jung DY, Kim JK, Xu J, Shulha HP, Garber M, Gao G, & Davis RJ (2014). The PPARalpha-FGF21 hormone axis contributes to metabolic regulation by the hepatic JNK signaling pathway. Cell Metabolism, 20(3), 512–525. 10.1016/j.cmet.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verzijl CRC, Van De Peppel IP, Struik D, & Jonker JW (2020). Pegbelfermin (BMS-986036): An investigational PEGylated fibroblast growth factor 21 analogue for the treatment of nonalcoholic steatohepatitis. Expert Opinion on Investigational Drugs, 29(2), 125–133. 10.1080/13543784.2020.1708898 [DOI] [PubMed] [Google Scholar]
- Vidrich A, Buzan JM, Brodrick B, Ilo C, Bradley L, Fendig KS, Sturgill T, & Cohn SM (2009). Fibroblast growth factor receptor-3 regulates Paneth cell lineage allocation and accrual of epithelial stem cells during murine intestinal development. American Journal of Physiology. Gastrointestinal and Liver Physiology, 297(1), G168–G178. 10.1152/ajpgi.90589.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira WA, Wells KM, Raymond MJ, De Souza L, Garcia E, & McCusker CD (2019). FGF, BMP, and RA signaling are sufficient for the induction of complete limb regeneration from non-regenerating wounds on Ambystoma mexicanum limbs. Developmental Biology, 451(2), 146–157. 10.1016/j.ydbio.2019.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila Ellis L, & Chen J (2021). A cell-centric view of lung alveologenesis. Developmental Dynamics, 250(4), 482–496. 10.1002/dvdy.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villageois P, Wdziekonski B, Zaragosi LE, Plaisant M, Mohsen-Kanson T, Lay N, Ladoux A, Peraldi P, & Dani C (2012). Regulators of human adipose-derived stem cell self-renewal. American Journal of Stem Cells, 1(1), 42–47. [PMC free article] [PubMed] [Google Scholar]
- Villanueva C, Jacobson-Dickman E, Xu C, Manouvrier S, Dwyer AA, Sykiotis GP, Beenken A, Liu Y, Tommiska J, Hu Y, Tiosano D, Gerard M, Leger J, Drouin-Garraud V, Lefebvre H, Polak M, Carel JC, Phan-Hug F, Hauschild M, … Pitteloud N (2015). Congenital hypogonadotropic hypogonadism with split hand/foot malformation: A clinical entity with a high frequency of FGFR1 mutations. Genetics in Medicine, 17(8), 651–659. 10.1038/gim.2014.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya J, Campderros L, Ribas-Aulinas F, Carriere A, Casteilla L, Giralt M, & Villarroya F (2018). Lactate induces expression and secretion of fibroblast growth factor-21 by muscle cells. Endocrine, 61(1), 165–168. 10.1007/s12020-018-1612-6 [DOI] [PubMed] [Google Scholar]
- Vincent TL, McLean CJ, Full LE, Peston D, & Saklatvala J (2007). FGF-2 is bound to perlecan in the pericellular matrix of articular cartilage, where it acts as a chondrocyte mechanotransducer. Osteoarthritis and Cartilage, 15(7), 752–763. 10.1016/j.joca.2007.01.021 [DOI] [PubMed] [Google Scholar]
- Vishal K, Lovato TL, Bragg C, Chechenova MB, & Cripps RM (2020). FGF signaling promotes myoblast proliferation through activation of wingless signaling. Developmental Biology, 464(1), 1–10. 10.1016/j.ydbio.2020.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T, & De Langhe S (2014). Lung epithelial stem cells and their niches: Fgf10 takes center stage. Fibrogenesis & Tissue Repair, 7, 8. 10.1186/1755-1536-7-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T, & De Langhe SP (2015). Wnt and FGF mediated epithelial-mesenchymal crosstalk during lung development. Developmental Dynamics, 244(3), 342–366. 10.1002/dvdy.24234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T, Yuan T, Chao CM, Bell H, Sitaula A, Szimmtenings L, El Agha E, Chanda D, Majka S, Bellusci S, Thannickal VJ, Fassler R, & De Langhe SP (2017). Fgf10-hippo epithelial-mesenchymal crosstalk maintains and recruits lung basal stem cells. Developmental Cell, 43(1), 48–59 e45. 10.1016/j.devcel.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Holstein-Rathlou S, BonDurant LD, Peltekian L, Naber MC, Yin TC, Claflin KE, Urizar AI, Madsen AN, Ratner C, Holst B, Karstoft K, Vandenbeuch A, Anderson CB, Cassell MD, Thompson AP, Solomon TP, Rahmouni K, Kinnamon SC, Pieper AA, … Potthoff MJ (2016). FGF21 mediates endocrine control of simple sugar intake and sweet taste preference by the liver. Cell Metabolism, 23(2), 335–343. 10.1016/j.cmet.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Holstein-Rathlou S, & Gillum MP (2019). Fibroblast growth factor 21: An endocrine inhibitor of sugar and alcohol appetite. The Journal of Physiology, 597(14), 3539–3548. 10.1113/JP277117 [DOI] [PubMed] [Google Scholar]
- Wagner BM, Robinson JW, Lin YW, Lee YC, Kaci N, Legeai-Mallet L, & Potter LR (2021). Prevention of guanylyl cyclase-B dephosphorylation rescues achondroplastic dwarfism. JCI Insight, 6(9), e147832. 10.1172/jci.insight.147832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Skacel J, Horvath A, Grande P, Wenninger J, Matzer F, Fazekas C, Morkl S, Meinitzer A, & Stadlbauer V (2021). Association of fibroblast growth factor 21 with alcohol consumption and alcohol liver cirrhosis. Neuropsychiatrie, 35(3), 140–146. 10.1007/s40211-020-00380-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakioka T, Sasaki A, Kato R, Shouda T, Matsumoto A, Miyoshi K, Tsuneoka M, Komiya S, Baron R, & Yoshimura A (2001). Spred is a Sprouty-related suppressor of Ras signalling. Nature, 412(6847), 647–651. 10.1038/35088082 [DOI] [PubMed] [Google Scholar]
- Walker DJ, & Land SC (2018). Regulation of vascular signalling by nuclear Sprouty2 in fetal lung epithelial cells: Implications for coordinated airway and vascular branching in lung development. Comparative Biochemistry and Physiology. Part B, Biochemistry & Molecular Biology, 224, 105–114. 10.1016/j.cbpb.2018.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker KA, Sims-Lucas S, & Bates CM (2016). Fibroblast growth factor receptor signaling in kidney and lower urinary tract development. Pediatric Nephrology, 31(6), 885–895. 10.1007/s00467-015-3151-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Yang Y, Li M, Liu X, Sun Y, Wang K, Zhang C, Zheng Q, & Wang C (2019). Activation of AK005401 aggravates acute ischemia/reperfusion mediated hippocampal injury by directly targeting YY1/FGF21. Aging (Albany, NY), 11(14), 5108–5123. 10.18632/aging.102106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Chang JY, Yang C, Huang Y, Liu J, You P, McKeehan WL, Wang F, & Li X (2013). Type 1 fibroblast growth factor receptor in cranial neural crest cell-derived mesenchyme is required for palatogenesis. The Journal of Biological Chemistry, 288(30), 22174–22183. 10.1074/jbc.M113.463620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Yang L, Yu W, & Zhang Y (2019). Investigational fibroblast growth factor receptor 2 antagonists in early phase clinical trials to treat solid tumors. Expert Opinion on Investigational Drugs, 28(10), 903–916. 10.1080/13543784.2019.1672655 [DOI] [PubMed] [Google Scholar]
- Wang F, Jiang H, Wang S, & Chen B (2017). Dual functional microRNA-186–5p targets both FGF2 and RelA to suppress tumorigenesis of glioblastoma multiforme. Cellular and Molecular Neurobiology, 37(8), 1433–1442. 10.1007/s10571-017-0474-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Jin Y, & Cattini PA (2017). Expression of the cardiac maintenance and survival factor FGF-16 gene is regulated by Csx/Nkx2.5 and is an early target of doxorubicin cardiotoxicity. DNA and Cell Biology, 36(2), 117–126. 10.1089/dna.2016.3507 [DOI] [PubMed] [Google Scholar]
- Wang J, Liu H, Gao L, & Liu X (2018). Impaired FGF10 signaling and epithelial development in experimental lung hypoplasia with esophageal atresia. Frontiers in Pediatrics, 6, 109. 10.3389/fped.2018.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rhee S, Palaria A, & Tremblay KD (2015). FGF signaling is required for anterior but not posterior specification of the murine liver bud. Developmental Dynamics, 244(3), 431–443. 10.1002/dvdy.24215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sontag D, & Cattini PA (2015). Heart-specific expression of FGF-16 and a potential role in postnatal cardioprotection. Cytokine & Growth Factor Reviews, 26(1), 59–66. 10.1016/j.cytogfr.2014.07.007 [DOI] [PubMed] [Google Scholar]
- Wang J, Wu Y, Zhao F, Wu Y, Dong W, Zhao J, Zhu Z, & Liu D (2015). Fgf-signaling-dependent Sox9a and Atoh1a regulate otic neural development in zebrafish. The Journal of Neuroscience, 35(1), 234–244. 10.1523/JNEUROSCI.3353-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yin Y, Lau S, Sankaran J, Rothenberg E, Wohland T, Meier-Schellersheim M, & Knaut H (2018). Anosmin1 shuttles Fgf to facilitate its diffusion, increase its local concentration, and induce sensory organs. Developmental Cell, 46(6), 751–766.e712. 10.1016/j.devcel.2018.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Ji W, Yu Y, Li Z, Niu X, Xia W, & Lu S (2018). FGFR1-ERK1/2-SOX2 axis promotes cell proliferation, epithelial-mesenchymal transition, and metastasis in FGFR1-amplified lung cancer. Oncogene, 37(39), 5340–5354. 10.1038/s41388-018-0311-3 [DOI] [PubMed] [Google Scholar]
- Wang K, Lai C, Li T, Wang C, Wang W, Ni B, Bai C, Zhang S, Han L, Gu H, Zhao Z, Duan Y, Yang X, Xing L, Zhao L, Zhou S, Xia M, Jiang C, Wang X, & Yang P (2018). Basic fibroblast growth factor protects against influenza A virus-induced acute lung injury by recruiting neutrophils. Journal of Molecular Cell Biology, 10(6), 573–585. 10.1093/jmcb/mjx047 [DOI] [PubMed] [Google Scholar]
- Wang L, Roth T, Abbott M, Ho L, Wattanachanya L, & Nissenson RA (2017). Osteoblast-derived FGF9 regulates skeletal homeostasis. Bone, 98, 18–25. 10.1016/j.bone.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Zhao TT, Li SM, Li YH, Wang YJ, Li DS, & Wang WF (2019). Fibroblast growth factor 21 ameliorates pancreatic fibrogenesis via regulating polarization of macrophages. Experimental Cell Research, 382(1), 111457. 10.1016/j.yexcr.2019.06.002 [DOI] [PubMed] [Google Scholar]
- Wang Q, Yang J, Wang H, Shan B, Yin C, Yu H, Zhang X, Dong Z, Yu Y, Zhao R, Liu B, Zhang H, & Wang C (2021). Fibroblast growth factor 13 stabilizes microtubules to promote Na(+) channel function in nociceptive DRG neurons and modulates inflammatory pain. Journal of Advanced Research, 31, 97–111. 10.1016/j.jare.2020.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Shlipak MG, Ix JH, Brown TT, Jacobson LP, Palella FJ Jr., Lake JE, Koletar SL, Semba RD, & Estrella MM (2019). Association of fibroblast growth factor-23 (FGF-23) with incident frailty in HIV-infected and HIV-uninfected individuals. Journal of Acquired Immune Deficiency Syndromes, 80(1), 118–125. 10.1097/QAI.0000000000001868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Cao S, Arhatte M, Li D, Shi Y, Kurz S, Hu J, Wang L, Shao J, Atzberger A, Wang Z, Wang C, Zang W, Fleming I, Wettschureck N, Honore E, & Offermanns S (2020). Adipocyte Piezo1 mediates obesogenic adipogenesis through the FGF1/FGFR1 signaling pathway in mice. Nature Communications, 11(1), 2303. 10.1038/s41467-020-16026-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Li Y, Jiang C, & Tian H (2018). Fibroblast growth factor 9 subfamily and the heart. Applied Microbiology and Biotechnology, 102(2), 605–613. 10.1007/s00253-017-8652-3 [DOI] [PubMed] [Google Scholar]
- Wang S, Yang Q, Yu S, Pan R, Jiang D, Liu Y, Hu H, Sun W, Hong X, Xue H, Qian W, Wang D, Zhou L, Mao C, & Yuan G (2016). Fibroblast growth factor 1 levels are elevated in newly diagnosed type 2 diabetes compared to normal glucose tolerance controls. Endocrine Journal, 63(4), 359–365. 10.1507/endocrj.EJ15-0627 [DOI] [PubMed] [Google Scholar]
- Wang W, Ju X, Sun Z, Hou W, Yang L, & Zhang R (2015). Overexpression of heparan sulfate 6-O-sulfotransferase-2 enhances fibroblast growth factor-mediated chondrocyte growth and differentiation. International Journal of Molecular Medicine, 36(3), 825–832. 10.3892/ijmm.2015.2272 [DOI] [PubMed] [Google Scholar]
- Wang WF, Li SM, Ren GP, Zheng W, Lu YJ, Yu YH, Xu WJ, Li TH, Zhou LH, Liu Y, & Li DS (2015). Recombinant murine fibroblast growth factor 21 ameliorates obesity-related inflammation in monosodium glutamate-induced obesity rats. Endocrine, 49(1), 119–129. 10.1007/s12020-014-0433-5 [DOI] [PubMed] [Google Scholar]
- Wang X, Cai B, Zhou J, Zhu H, Niu Y, Ma B, Yu H, Lei A, Yan H, Shen Q, Shi L, Zhao X, Hua J, Huang X, Qu L, & Chen Y (2016). Disruption of FGF5 in cashmere goats using CRISPR/Cas9 results in more secondary hair follicles and longer fibers. PLoS One, 11(10), e0164640. 10.1371/journal.pone.0164640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hsi TC, Guerrero-Juarez CF, Pham K, Cho K, McCusker CD, Monuki ES, Cho KW, Gay DL, & Plikus MV (2015). Principles and mechanisms of regeneration in the mouse model for wound-induced hair follicle neogenesis. Regeneration (Oxford), 2(4), 169–181. 10.1002/reg2.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Qi H, Wang Q, Zhu Y, Wang X, Jin M, Tan Q, Huang Q, Xu W, Li X, Kuang L, Tang Y, Du X, Chen D, & Chen L (2015). FGFR3/fibroblast growth factor receptor 3 inhibits autophagy through decreasing the ATG12-ATG5 conjugate, leading to the delay of cartilage development in achondroplasia. Autophagy, 11(11), 1998–2013. 10.1080/15548627.2015.1091551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sun X, Zhang X, Li H, & Xie A (2017). Quantitative assessment of the effect of FGF20 rs12720208 variant on the risk of Parkinson’s disease: A meta-analysis. Neurological Research, 39(4), 374–380. 10.1080/01616412.2017.1286542 [DOI] [PubMed] [Google Scholar]
- Wang X, Tang H, Wei EQ, Wang Z, Yang J, Yang R, Wang S, Zhang Y, Pitt GS, Zhang H, & Wang C (2017). Conditional knockout of Fgf13 in murine hearts increases arrhythmia susceptibility and reveals novel ion channel modulatory roles. Journal of Molecular and Cellular Cardiology, 104, 63–74. 10.1016/j.yjmcc.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhu L, Hu J, Guo R, Ye S, Liu F, Wang D, Zhao Y, Hu A, Wang X, Guo K, & Lin L (2020). FGF21 attenuated LPS-induced depressive-like behavior via inhibiting the inflammatory pathway. Frontiers in Pharmacology, 11, 154. 10.3389/fphar.2020.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XQ, Li WH, Tang YH, Wu LF, Zeng GR, Wang YH, Cheng ZN, & Jiang DJ (2020). The correlation between adiponectin and FGF9 in depression disorder. Brain Research, 1729, 146596. 10.1016/j.brainres.2019.146596 [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu D, Zhang T, & Xia L (2021). FGF/FGFR signaling in hepatocellular carcinoma: From carcinogenesis to recent therapeutic intervention. Cancers (Basel), 13(6), 1360. 10.3390/cancers13061360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang H, & Chen P (2020). Higher fibroblast growth factor 23 levels are causally associated with lower bone mineral density of heel and femoral neck: Evidence from two-sample Mendelian randomization analysis. Frontiers in Public Health, 8, 467. 10.3389/fpubh.2020.00467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang T, Liu Y, Zhao W, Zhang Z, Lu M, & Zhang W (2017). Decrease of miR-195 promotes chondrocytes proliferation and maintenance of chondrogenic phenotype via targeting FGF-18 pathway. International Journal of Molecular Sciences, 18(5), 975. 10.3390/ijms18050975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Huang J, Zhou S, Luo F, Tan Q, Sun X, Ni Z, Chen H, Du X, Xie Y, & Chen L (2018). Loss of Fgfr1 in chondrocytes inhibits osteoarthritis by promoting autophagic activity in temporomandibular joint. The Journal of Biological Chemistry, 293(23), 8761–8774. 10.1074/jbc.RA118.002293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Huang Y, He Y, Khor S, Zhong X, Xiao J, Ye Q, & Li X (2021). Myocardial protection by heparin-based coacervate of FGF10. Bioactive Materials, 6(7), 1867–1877. 10.1016/j.bioactmat.2020.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZG, Cheng Y, Yu XC, Ye LB, Xia QH, Johnson NR, Wei X, Chen DQ, Cao G, Fu XB, Li XK, Zhang HY, & Xiao J (2016). bFGF protects against blood-brain barrier damage through junction protein regulation via PI3K-Akt-Rac1 pathway following traumatic brain injury. Molecular Neurobiology, 53(10), 7298–7311. 10.1007/s12035-015-9583-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J, & Francavilla C (2018). Regulation of FGF10 signaling in development and disease. Frontiers in Genetics, 9, 500. 10.3389/fgene.2018.00500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wean JB, & Smith BN (2021). FGF19 in the hindbrain lowers blood glucose and alters excitability of vagal motor neurons in hyperglycemic mice. Endocrinology, 162(4), bqab021. 10.1210/endocr/bqab021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weant J, Eveleth DD, Subramaniam A, Jenkins-Eveleth J, Blaber M, Li L, Ornitz DM, Alimardanov A, Broadt T, Dong H, Vyas V, Yang X, & Bradshaw RA (2021). Regenerative responses of rabbit corneal endothelial cells to stimulation by fibroblast growth factor 1 (FGF1) derivatives, TTHX1001 and TTHX1114. Growth Factors, 1–14. 10.1080/08977194.2021.2012468 [DOI] [PubMed] [Google Scholar]
- Weaver A, & Bossaer JB (2021). Fibroblast growth factor receptor (FGFR) inhibitors: A review of a novel therapeutic class. Journal of Oncology Pharmacy Practice, 27(3), 702–710. 10.1177/1078155220983425 [DOI] [PubMed] [Google Scholar]
- Wei K, Xu Y, Tse H, Manolson MF, & Gong SG (2011). Mouse FLRT2 interacts with the extracellular and intracellular regions of FGFR2. Journal of Dental Research, 90(10), 1234–1239. 10.1177/0022034511415272 [DOI] [PubMed] [Google Scholar]
- Weinstein M, Xu X, Ohyama K, & Deng CX (1998). FGFR-3 and FGFR-4 function cooperatively to direct alveogenesis in the murine lung. Development, 125(18), 3615–3623. [DOI] [PubMed] [Google Scholar]
- Wen X, Li X, Tang Y, Tang J, Zhou S, Xie Y, Guo J, Yang J, Du X, Su N, & Chen L (2016). Chondrocyte FGFR3 regulates bone mass by inhibiting osteogenesis. The Journal of Biological Chemistry, 291(48), 24912–24921. 10.1074/jbc.M116.730093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt DJ, Dvorak-Ewell M, Bullens S, Lorget F, Bell SM, Peng J, Castillo S, Aoyagi-Scharber M, O’Neill CA, Krejci P, Wilcox WR, Rimoin DL, & Bunting S (2015). Neutral endopeptidase-resistant C-type natriuretic peptide variant represents a new therapeutic approach for treatment of fibroblast growth factor receptor 3-related dwarfism. The Journal of Pharmacology and Experimental Therapeutics, 353(1), 132–149. 10.1124/jpet.114.218560 [DOI] [PubMed] [Google Scholar]
- Weng M, Chen Z, Xiao Q, Li R, & Chen Z (2018). A review of FGF signaling in palate development. Biomedicine & Pharmacotherapy, 103, 240–247. 10.1016/j.biopha.2018.04.026 [DOI] [PubMed] [Google Scholar]
- Weng T, Yi L, Huang J, Luo F, Wen X, Du X, Chen Q, Deng C, Chen D, & Chen L (2012). Genetic inhibition of fibroblast growth factor receptor 1 in knee cartilage attenuates the degeneration of articular cartilage in adult mice. Arthritis and Rheumatism, 64(12), 3982–3992. 10.1002/art.34645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng W, Ge T, Wang Y, He L, Liu T, Wang W, Zheng Z, Yu L, Zhang C, & Lu X (2020). Therapeutic effects of fibroblast growth factor-21 on diabetic nephropathy and the possible mechanism in type 1 diabetes mellitus mice. Diabetes and Metabolism Journal, 44(4), 566–580. 10.4093/dmj.2019.0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger TL, Dahl J, Bhoj EJ, Rosen A, McDonald-McGinn D, Zackai E, Jacobs I, Heike CL, Hing A, Santani A, Inglis AF, Sie KC, Cunningham M, & Perkins J (2017). Tracheal cartilaginous sleeves in children with syndromic craniosynostosis. Genetics in Medicine, 19(1), 62–68. 10.1038/gim.2016.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheldon LM, Haines BP, Rajappa R, Mason I, Rigby PW, & Heath JK (2010). Critical role of FLRT1 phosphorylation in the interdependent regulation of FLRT1 function and FGF receptor signalling. PLoS One, 5(4), e10264. 10.1371/journal.pone.0010264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AC, Xu J, Yin Y, Smith C, Schmid G, & Ornitz DM (2006). FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development, 133(8), 1507–1517. 10.1242/dev.02313 [DOI] [PubMed] [Google Scholar]
- White KE, Carn G, Lorenz-Depiereux B, Benet-Pages A, Strom TM, & Econs MJ (2001). Autosomal-dominant hypophosphatemic rickets (ADHR) mutations stabilize FGF-23. Kidney International, 60(6), 2079–2086. 10.1046/j.1523-1755.2001.00064.x [DOI] [PubMed] [Google Scholar]
- Whitsett JA, Clark JC, Picard L, Tichelaar JW, Wert SE, Itoh N, Perl AK, & Stahlman MT (2002). Fibroblast growth factor 18 influences proximal programming during lung morphogenesis. The Journal of Biological Chemistry, 277(25), 22743–22749. 10.1074/jbc.M202253200 [DOI] [PubMed] [Google Scholar]
- Whyte MP (2021). Tumor-induced osteomalacia: Treatment progress using burosumab, an anti-FGF23 monoclonal antibody. Journal of Bone and Mineral Research, 36(4), 625–626. 10.1002/jbmr.4280 [DOI] [PubMed] [Google Scholar]
- Wier EM, & Garza LA (2020). Through the lens of hair follicle neogenesis, a new focus on mechanisms of skin regeneration after wounding. Seminars in Cell & Developmental Biology, 100, 122–129. 10.1016/j.semcdb.2019.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willardsen M, Hutcheson DA, Moore KB, & Vetter ML (2014). The ETS transcription factor Etv1 mediates FGF signaling to initiate proneural gene expression during Xenopus laevis retinal development. Mechanisms of Development, 131, 57–67. 10.1016/j.mod.2013.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AJ, Yee P, Smith MC, Murphy GG, & Umemori H (2016). Deletion of fibroblast growth factor 22 (FGF22) causes a depression-like phenotype in adult mice. Behavioural Brain Research, 307, 11–17. 10.1016/j.bbr.2016.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EJ, Furness J, Walsh FS, & Doherty P (1994). Activation of the FGF receptor underlies neurite outgrowth stimulated by L1, N-CAM, and N-cadherin. Neuron, 13(3), 583–594. 10.1016/0896-6273(94)90027-2 [DOI] [PubMed] [Google Scholar]
- Winkley KM, Reeves WM, & Veeman MT (2021). Single-cell analysis of cell fate bifurcation in the chordate Ciona. BMC Biology, 19(1), 180. 10.1186/s12915-021-01122-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf I, Levanon-Cohen S, Bose S, Ligumsky H, Sredni B, Kanety H, Kuro-o M, Karlan B, Kaufman B, Koeffler HP, & Rubinek T (2008). Klotho: A tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene, 27(56), 7094–7105. 10.1038/onc.2008.292 [DOI] [PubMed] [Google Scholar]
- Wolf M, & White KE (2014). Coupling fibroblast growth factor 23 production and cleavage: Iron deficiency, rickets, and kidney disease. Current Opinion in Nephrology and Hypertension, 23(4), 411–419. 10.1097/01.mnh.0000447020.74593.6f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo KV, Shen IY, Weinheimer CJ, Kovacs A, Nigro J, Lin CY, Chakinala M, Byers DE, & Ornitz DM (2021). Endothelial FGF signaling is protective in hypoxia-induced pulmonary hypertension. Journal of Clinical Investigation, 131(17), e141467. 10.1172/JCI141467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woronowicz KC, Gline SE, Herfat ST, Fields AJ, & Schneider RA (2018). FGF and TGFbeta signaling link form and function during jaw development and evolution. Developmental Biology, 444(Suppl 1), S219–S236. 10.1016/j.ydbio.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KD, Mahoney Rogers AA, Zhang J, & Shim K (2015). Cooperative and independent functions of FGF and Wnt signaling during early inner ear development. BMC Developmental Biology, 15(1), 33. 10.1186/s12861-015-0083-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright NA, Gregory TR, & Witt CC (2014). Metabolic ‘engines’ of flight drive genome size reduction in birds. Proceedings of the Biological Sciences, 281(1779), 20132780. 10.1098/rspb.2013.2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Feng B, Yu J, Yan L, Che L, Zhuo Y, Luo Y, Yu B, Wu D, & Chen D (2021). Fibroblast growth factor 21 attenuates iron overload-induced liver injury and fibrosis by inhibiting ferroptosis. Redox Biology, 46, 102131. 10.1016/j.redox.2021.102131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AL, Feng B, Chen MZ, Kolumam G, Zavala-Solorio J, Wyatt SK, Gandham VD, Carano RA, & Sonoda J (2013). Antibody-mediated activation of FGFR1 induces FGF23 production and hypophosphatemia. PLoS One, 8(2), e57322. 10.1371/journal.pone.0057322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AL, Kolumam G, Stawicki S, Chen Y, Li J, Zavala-Solorio J, Phamluong K, Feng B, Li L, Marsters S, Kates L, van Bruggen N, Leabman M, Wong A, West D, Stern H, Luis E, Kim HS, Yansura D, … Sonoda J (2011). Amelioration of type 2 diabetes by antibody-mediated activation of fibroblast growth factor receptor 1. Science Translational Medicine, 3(113), 113ra126. 10.1126/scitranslmed.3002669 [DOI] [PubMed] [Google Scholar]
- Wu B, Li J, Chou YH, Luginbuhl D, & Luo L (2017). Fibroblast growth factor signaling instructs ensheathing glia wrapping of Drosophila olfactory glomeruli. Proceedings of the National Academy of Sciences of the United States of America, 114(29), 7505–7512. 10.1073/pnas.1706533114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Chu X, Chen C, & Bellusci S (2018). Role of fibroblast growth factor 10 in mesenchymal cell differentiation during lung development and disease. Frontiers in Genetics, 9, 545. 10.3389/fgene.2018.00545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Xu R, Zhu X, He H, Zhang J, Zeng Q, Wang Y, & Zhao X (2020). The long noncoding RNA LINC01140/miR-140–5p/FGF9 axis modulates bladder cancer cell aggressiveness and macrophage M2 polarization. Aging (Albany NY), 12(24), 25845–25864. 10.18632/aging.202147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XL, Gu MM, Huang L, Liu XS, Zhang HX, Ding XY, Xu JQ, Cui B, Wang L, Lu SY, Chen XY, Zhang HG, Huang W, Yuan WT, Yang JM, Gu Q, Fei J, Chen Z, Yuan ZM, & Wang ZG (2009). Multiple synostoses syndrome is due to a missense mutation in exon 2 of FGF9 gene. American Journal of Human Genetics, 85(1), 53–63. 10.1016/j.ajhg.2009.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynes MW, Hinz TK, Gao D, Martini M, Marek LA, Ware KE, Edwards MG, Bohm D, Perner S, Helfrich BA, Dziadziuszko R, Jassem J, Wojtylak S, Sejda A, Gozgit JM, Bunn PA Jr., Camidge DR, Tan AC, Hirsch FR, & Heasley LE (2014). FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clinical Cancer Research, 20(12), 3299–3309. 10.1158/1078-0432.CCR-13-3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia JX, Mei XL, Zhu WJ, Li X, Jin XH, Mou Y, Yu K, Wang YY, & Li FQ (2014). Effect of FGF10 monoclonal antibody on psoriasis-like model in guinea pigs. International Journal of Clinical and Experimental Pathology, 7(5), 2219–2228. [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Bei Y, Liu J, Dimitrova-Shumkovska J, Kuang D, Zhou Q, Li J, Yang Y, Xiang Y, Wang F, Yang C, & Yang W (2016). miR-212 downregulation contributes to the protective effect of exercise against non-alcoholic fatty liver via targeting FGF-21. Journal of Cellular and Molecular Medicine, 20(2), 204–216. 10.1111/jcmm.12733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Lv D, Zhao Y, Chen X, Song M, Liu J, Bei Y, Wang F, Yang W, & Yang C (2016). miR-149 controls non-alcoholic fatty liver by targeting FGF-21. Journal of Cellular and Molecular Medicine, 20(8), 1603–1608. 10.1111/jcmm.12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, & Dudley AC (2017). Fine-tuning vascular fate during endothelial-mesenchymal transition. The Journal of Pathology, 241(1), 25–35. 10.1002/path.4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Fei Y, & Hurley MM (2018). FGF2 crosstalk with Wnt signaling in mediating the anabolic action of PTH on bone formation. Bone Rep, 9, 136–144. 10.1016/j.bonr.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Liu P, Li X, Doetschman T, Coffin JD, Drissi H, & Hurley MM (2009). Exported 18-kDa isoform of fibroblast growth factor-2 is a critical determinant of bone mass in mice. The Journal of Biological Chemistry, 284(5), 3170–3182. 10.1074/jbc.M804900200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Ueno D, Catros S, Homer-Bouthiette C, Charles L, Kuhn L, & Hurley MM (2014). Fibroblast growth factor-2 isoform (low molecular weight/18 kDa) overexpression in preosteoblast cells promotes bone regeneration in critical size calvarial defects in male mice. Endocrinology, 155(3), 965–974. 10.1210/en.2013-1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Williams D, & Hurley MM (2020). Inhibition of FGFR signaling partially rescues osteoarthritis in mice overexpressing high molecular weight FGF2 isoforms. Endocrinology, 161(1), bqz016. 10.1210/endocr/bqz016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Huang J, Cao L, Liang Y, Han X, & Quarles LD (2014). Osteocyte-specific deletion of Fgfr1 suppresses FGF23. PLoS One, 9(8), e104154. 10.1371/journal.pone.0104154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie JL, Bian HN, Qi SH, Chen HD, Li HD, Xu YB, Li TZ, Liu XS, Liang HZ, Xin BR, & Huan Y (2008). Basic fibroblast growth factor (bFGF) alleviates the scar of the rabbit ear model in wound healing. Wound Repair and Regeneration, 16(4), 576–581. 10.1111/j.1524-475X.2008.00405.x [DOI] [PubMed] [Google Scholar]
- Xie M, & Li JP (2019). Heparan sulfate proteoglycan—A common receptor for diverse cytokines. Cellular Signalling, 54, 115–121. 10.1016/j.cellsig.2018.11.022 [DOI] [PubMed] [Google Scholar]
- Xie Y, Luo F, Xu W, Wang Z, Sun X, Xu M, Huang J, Zhang D, Tan Q, Chen B, Jiang W, Du X, & Chen L (2017). FGFR3 deficient mice have accelerated fracture repair. International Journal of Biological Sciences, 13(8), 1029–1037. 10.7150/ijbs.19309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Su N, Yang J, Tan Q, Huang S, Jin M, Ni Z, Zhang B, Zhang D, Luo F, Chen H, Sun X, Feng JQ, Qi H, & Chen L (2020). FGF/FGFR signaling in health and disease. Signal Transduction and Targeted Therapy, 5(1), 181. 10.1038/s41392-020-00222-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Zhou S, Chen H, Du X, & Chen L (2014). Recent research on the growth plate: Advances in fibroblast growth factor signaling in growth plate development and disorders. Journal of Molecular Endocrinology, 53(1), T11–T34. 10.1530/JME-14-0012 [DOI] [PubMed] [Google Scholar]
- Xie Y, Zinkle A, Chen L, & Mohammadi M (2020). Fibroblast growth factor signalling in osteoarthritis and cartilage repair. Nature Reviews Rheumatology, 16(10), 547–564. 10.1038/s41584-020-0469-2 [DOI] [PubMed] [Google Scholar]
- Xu D, & Esko JD (2014). Demystifying heparan sulfate-protein interactions. Annual Review of Biochemistry, 83, 129–157. 10.1146/annurev-biochem-060713-035314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Liu H, Lan Y, Aronow BJ, Kalinichenko VV, & Jiang R (2016). A Shh-Foxf-Fgf18-Shh Molecular Circuit Regulating Palate Development. PLoS Genetics, 12(1), e1005769. 10.1371/journal.pgen.1005769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lloyd DJ, Hale C, Stanislaus S, Chen M, Sivits G, Vonderfecht S, Hecht R, Li YS, Lindberg RA, Chen JL, Jung DY, Zhang Z, Ko HJ, Kim JK, & Veniant MM (2009). Fibroblast growth factor 21 reverses hepatic steatosis, increases energy expenditure, and improves insulin sensitivity in diet-induced obese mice. Diabetes, 58(1), 250–259. 10.2337/db08-0392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wu F, Wang F, Yang F, Liu M, Lou M, Wu L, Li H, Lin W, Fan Y, Chen L, Liu Y, Xu H, & He J (2021). The interaction of single nucleotide polymorphisms on fibroblast growth factor 19 superfamily genes is associated with alcohol dependence-related aggression. Frontiers in Genetics, 12, 695835. 10.3389/fgene.2021.695835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Xie Y, Wang Q, Wang X, Luo F, Zhou S, Wang Z, Huang J, Tan Q, Jin M, Qi H, Tang J, Chen L, Du X, Zhao C, Liang G, & Chen L (2016). A novel fibroblast growth factor receptor 1 inhibitor protects against cartilage degradation in a murine model of osteoarthritis. Scientific Reports, 6, 24042. 10.1038/srep24042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Liu H, Pan H, Wang X, Zhang Y, Yao B, Li N, Lai L, & Li Z (2020). CRISPR/Cas9-mediated disruption of fibroblast growth factor 5 in rabbits results in a systemic long hair phenotype by prolonging anagen. Genes (Basel), 11(3), 297. 10.3390/genes11030297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, & Sun Z (2015). Molecular basis of Klotho: From gene to function in aging. Endocrine Reviews, 36(2), 174–193. 10.1210/er.2013-1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YH, Yu M, Wei H, Yao S, Chen SY, Zhu XL, & Li YF (2017). Fibroblast growth factor 22 is a novel modulator of depression through interleukin-1beta. CNS Neuroscience & Therapeutics, 23(11), 907–916. 10.1111/cns.12760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YH, Zhu Y, Zhu YY, Wei H, Zhang NN, Qin JS, Zhu XL, Yu M, & Li YF (2021). Abnormalities in FGF family members and their roles in modulating depression-related molecules. The European Journal of Neuroscience, 53(1), 140–150. 10.1111/ejn.14570 [DOI] [PubMed] [Google Scholar]
- Xu Z, Wu Y, Wang F, Li X, Wang P, Li Y, Wu J, Li Y, Jiang T, Pan X, Zhang X, Xie L, Xiao J, & Liu Y (2020). Fibroblast growth factor 1 ameliorates diabetes-induced liver injury by reducing cellular stress and restoring autophagy. Frontiers in Pharmacology, 11, 52. 10.3389/fphar.2020.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G, Yan HL, Zhang Y, Hao LQ, Zhu XT, Mei Q, & Sun SH (2015). c-Myc-mediated repression of miR-15–16 in hypoxia is induced by increased HIF-2alpha and promotes tumor angiogenesis and metastasis by upregulating FGF2. Oncogene, 34(11), 1393–1406. 10.1038/onc.2014.82 [DOI] [PubMed] [Google Scholar]
- Xue YN, Yan Y, Chen ZZ, Chen J, Tang FJ, Xie HQ, Tang SJ, Cao K, Zhou X, Wang AJ, & Zhou JD (2019). LncRNA TUG1 regulates FGF1 to enhance endothelial differentiation of adipose-derived stem cells by sponging miR-143. Journal of Cellular Biochemistry, 120(11), 19087–19097. 10.1002/jcb.29232 [DOI] [PubMed] [Google Scholar]
- Yafei S, Elsewy F, Youssef E, Ayman M, & El-Shafei M (2019). Fibroblast growth factor 21 association with subclinical atherosclerosis and arterial stiffness in type 2 diabetes. Diabetes and Metabolic Syndrome: Clinical Research and Reviews, 13(1), 882–888. 10.1016/j.dsx.2018.12.007 [DOI] [PubMed] [Google Scholar]
- Yamada T, Kerever A, Yoshimura Y, Suzuki Y, Nonaka R, Higashi K, Toida T, Mercier F, & Arikawa-Hirasawa E (2017). Heparan sulfate alterations in extracellular matrix structures and fibroblast growth factor-2 signaling impairment in the aged neurogenic niche. Journal of Neurochemistry, 142(4), 534–544. 10.1111/jnc.14081 [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Harpal K, Henkemeyer M, & Rossant J (1994). FGFR-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes & Development, 8, 3032–3044. [DOI] [PubMed] [Google Scholar]
- Yamaji K, Morita J, Watanabe T, Gunjigake K, Nakatomi M, Shiga M, Ono K, Moriyama K, & Kawamoto T (2018). Mal-development of the submandibular gland in a mouse model of apert syndrome. Developmental Dynamics, 247(11), 1175–1185. 10.1002/dvdy.24673 [DOI] [PubMed] [Google Scholar]
- Yamakawa H, Muraoka N, Miyamoto K, Sadahiro T, Isomi M, Haginiwa S, Kojima H, Umei T, Akiyama M, Kuishi Y, Kurokawa J, Furukawa T, Fukuda K, & Ieda M (2015). Fibroblast growth factors and vascular endothelial growth factor promote cardiac reprogramming under defined conditions. Stem Cell Reports, 5(6), 1128–1142. 10.1016/j.stemcr.2015.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Ryoo BY, Keam B, Kudo M, Lin CC, Kunieda F, Ball HA, Moran D, Komatsu K, Takeda K, Fukuda M, Furuse J, Morita S, & Doi T (2020). A phase 1 study of oral ASP5878, a selective small-molecule inhibitor of fibroblast growth factor receptors 1–4, as a single dose and multiple doses in patients with solid malignancies. Investigational New Drugs, 38(2), 445–456. 10.1007/s10637-019-00780-w [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Matsui J, Matsushima T, Obaishi H, Miyazaki K, Nakamura K, Tohyama O, Semba T, Yamaguchi A, Hoshi SS, Mimura F, Haneda T, Fukuda Y, Kamata JI, Takahashi K, Matsukura M, Wakabayashi T, Asada M, Nomoto KI, … Tsuruoka A (2014). Lenvatinib, an angiogenesis inhibitor targeting VEGFR/FGFR, shows broad antitumor activity in human tumor xenograft models associated with microvessel density and pericyte coverage. Vascular Cell, 6, 18. 10.1186/2045-824X-6-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto-Shiraishi Y, Higuchi H, Yamamoto S, Hirano M, & Kuroiwa A (2014). Etv1 and Ewsr1 cooperatively regulate limb mesenchymal Fgf10 expression in response to apical ectodermal ridge-derived fibroblast growth factor signal. Developmental Biology, 394(1), 181–190. 10.1016/j.ydbio.2014.07.022 [DOI] [PubMed] [Google Scholar]
- Yan D, Chen D, Cool SM, van Wijnen AJ, Mikecz K, Murphy G, & Im HJ (2011). Fibroblast growth factor receptor 1 is principally responsible for fibroblast growth factor 2-induced catabolic activities in human articular chondrocytes. Arthritis Research & Therapy, 13(4), R130. 10.1186/ar3441 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yan X, Gou Z, Li Y, Wang Y, Zhu J, Xu G, & Zhang Q (2018). Fibroblast growth factor 21 inhibits atherosclerosis in apoE−/− mice by ameliorating Fas-mediated apoptosis. Lipids in Health and Disease, 17(1), 203. 10.1186/s12944-018-0846-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Whelan EC, Guan X, Deng B, Wang S, Sun J, Avarbock MR, Wu X, & Brinster RL (2021). FGF9 promotes mouse spermatogonial stem cell proliferation mediated by p38 MAPK signalling. Cell Proliferation, 54(1), e12933. 10.1111/cpr.12933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Kim WJ, Jun HO, Lee EJ, Lee KW, Jeong JY, & Lee SW (2015). Hypoxia-induced fibroblast growth factor 11 stimulates capillary-like endothelial tube formation. Oncology Reports, 34(5), 2745–2751. 10.3892/or.2015.4223 [DOI] [PubMed] [Google Scholar]
- Yang J, Meyer M, Muller AK, Bohm F, Grose R, Dauwalder T, Verrey F, Kopf M, Partanen J, Bloch W, Ornitz DM, & Werner S (2010). Fibroblast growth factor receptors 1 and 2 in keratinocytes control the epidermal barrier and cutaneous homeostasis. The Journal of Cell Biology, 188(6), 935–952. 10.1083/jcb.200910126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ge D, Cao X, Ge Y, Chen H, Wang W, & Zhang H (2016). MiR-214 attenuates osteogenic differentiation of mesenchymal stem cells via targeting FGFR1. Cellular Physiology and Biochemistry, 38(2), 809–820. 10.1159/000443036 [DOI] [PubMed] [Google Scholar]
- Yang L, Hansen Falkesgaard M, Thulstrup PW, Walmod PS, Lo Leggio L, & Krighaar Rasmussen K (2017). Expression, refolding and spectroscopic characterization of fibronectin type III (FnIII)-homology domains derived from human fibronectin leucine rich transmembrane protein (FLRT)-1, −2, and −3. PeerJ, 5, e3550. 10.7717/peerj.3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu G, Xiao S, Wang L, Liu X, Tan Q, & Li Z (2020). Long noncoding MT1JP enhanced the inhibitory effects of miR-646 on FGF2 in osteosarcoma. Cancer Biotherapy & Radiopharmaceuticals, 35(5), 371–376. 10.1089/cbr.2019.3328 [DOI] [PubMed] [Google Scholar]
- Yang LM, Cheah KSE, Huh SH, & Ornitz DM (2019). Sox2 and FGF20 interact to regulate organ of Corti hair cell and supporting cell development in a spatially-graded manner. PLoS Genetics, 15(7), e1008254. 10.1371/journal.pgen.1008254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LM, Huh SH, & Ornitz DM (2018). FGF20-expressing, Wnt-responsive olfactory epithelial progenitors regulate underlying turbinate growth to optimize surface area. Developmental Cell, 46(5), 564–580.e565. 10.1016/j.devcel.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LM, Stout L, Rauchman M, & Ornitz DM (2020). Analysis of FGF20-regulated genes in organ of Corti progenitors by translating ribosome affinity purification. Developmental Dynamics, 249(10), 1217–1242. 10.1002/dvdy.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PH, Zhu JX, Huang YD, Zhang XY, Lei P, Bush AI, Xiang Q, Su ZJ, & Zhang QH (2016). Human basic fibroblast growth factor inhibits tau phosphorylation via the PI3K/Akt-GSK3beta signaling pathway in a 6-hydroxydopamine-induced model of Parkinson’s disease. Neurodegenerative Diseases, 16(5–6), 357–369. 10.1159/000445871 [DOI] [PubMed] [Google Scholar]
- Yang S, Weske A, Du Y, Valera JM, Jones KL, & Johnson AN (2020). FGF signaling directs myotube guidance by regulating Rac activity. Development, 147(3), dev183624. 10.1242/dev.183624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Liaw L, Prudovsky I, Brooks PC, Vary C, Oxburgh L, & Friesel R (2015). Fibroblast growth factor signaling in the vasculature. Current Atherosclerosis Reports, 17(6), 509. 10.1007/s11883-015-0509-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Steinberg F, Zhuang L, Bessey R, & Trueb B (2016). Receptor FGFRL1 does not promote cell proliferation but induces cell adhesion. International Journal of Molecular Medicine, 38(1), 30–38. 10.3892/ijmm.2016.2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Haeger SM, Suflita MA, Zhang F, Dailey KL, Colbert JF, Ford JA, Picon MA, Stearman RS, Lin L, Liu X, Han X, Linhardt RJ, & Schmidt EP (2017). Fibroblast growth factor signaling mediates pulmonary endothelial glycocalyx reconstitution. American Journal of Respiratory Cell and Molecular Biology, 56(6), 727–737. 10.1165/rcmb.2016-0338OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhu X, Jia X, Hou W, Zhou G, Ma Z, Yu B, Pi Y, Zhang X, Wang J, & Wang G (2020). Phosphorylation of Msx1 promotes cell proliferation through the Fgf9/18-MAPK signaling pathway during embryonic limb development. Nucleic Acids Research, 48(20), 11452–11467. 10.1093/nar/gkaa905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanochko GM, Vitsky A, Heyen JR, Hirakawa B, Lam JL, May J, Nichols T, Sace F, Trajkovic D, & Blasi E (2013). Pan-FGFR inhibition leads to blockade of FGF23 signaling, soft tissue mineralization, and cardiovascular dysfunction. Toxicological Sciences, 135(2), 451–464. 10.1093/toxsci/kft161 [DOI] [PubMed] [Google Scholar]
- Yaqoob U, Jagavelu K, Shergill U, de Assuncao T, Cao S, & Shah VH (2014). FGF21 promotes endothelial cell angiogenesis through a dynamin-2 and Rab5 dependent pathway. PLoS One, 9(5), e98130. 10.1371/journal.pone.0098130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yayon A, Klagsbrun M, Esko JD, Leder P, & Ornitz DM (1991). Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cells, 64(4), 841–848. 10.1016/0092-8674(91)90512-w [DOI] [PubMed] [Google Scholar]
- Yeo Y, Yi ES, Kim JM, Jo EK, Seo S, Kim RI, Kim KL, Sung JH, Park SG, & Suh W (2020). FGF12 (fibroblast growth factor 12) inhibits vascular smooth muscle cell remodeling in pulmonary arterial hypertension. Hypertension, 76(6), 1778–1786. 10.1161/HYPERTENSIONAHA.120.15068 [DOI] [PubMed] [Google Scholar]
- Yin J, Lee R, Ono Y, Ingham PW, & Saunders TE (2018). Spatiotemporal coordination of FGF and Shh signaling underlies the specification of myoblasts in the zebrafish embryo. Developmental Cell, 46(6), 735–750.e734. 10.1016/j.devcel.2018.08.024 [DOI] [PubMed] [Google Scholar]
- Yin Y, Castro AM, Hoekstra M, Yan TJ, Kanakamedala AC, Dehner LP, Hill DA, & Ornitz DM (2015). Fibroblast growth factor 9 regulation by microRNAs controls lung development and links DICER1 loss to the pathogenesis of pleuropulmonary blastoma. PLoS Genetics, 11(5), e1005242. 10.1371/journal.pgen.1005242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, & Ornitz DM (2020). FGF9 and FGF10 activate distinct signaling pathways to direct lung epithelial specification and branching. Science Signaling, 13(621), eaay4353. 10.1126/scisignal.aay4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Ren X, Smith C, Guo Q, Malabunga M, Guernah I, Zhang Y, Shen J, Sun H, Chehab N, Loizos N, Ludwig DL, & Ornitz DM (2016). Inhibition of fibroblast growth factor receptor 3-dependent lung adenocarcinoma with a human monoclonal antibody. Disease Models & Mechanisms, 9(5), 563–571. 10.1242/dmm.024760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang F, & Ornitz DM (2011). Mesothelial- and epithelial-derived FGF9 have distinct functions in the regulation of lung development. Development, 138(15), 3169–3177. 10.1242/dev.065110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying L, Li N, He Z, Zeng X, Nan Y, Chen J, Miao P, Ying Y, Lin W, Zhao X, Lu L, Chen M, Cen W, Guo T, Li X, Huang Z, & Wang Y (2019). Fibroblast growth factor 21 Ameliorates diabetes-induced endothelial dysfunction in mouse aorta via activation of the CaMKK2/AMPKalpha signaling pathway. Cell Death & Disease, 10(9), 665. 10.1038/s41419-019-1893-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomoto-Umakoshi M, Umakoshi H, Miyazawa T, Ogata M, Sakamoto R, & Ogawa Y (2021). Investigating the causal effect of fibroblast growth factor 23 on osteoporosis and cardiometabolic disorders: A Mendelian randomization study. Bone, 143, 115777. 10.1016/j.bone.2020.115777 [DOI] [PubMed] [Google Scholar]
- Yokote H, Fujita K, Jing X, Sawada T, Liang S, Yao L, Yan X, Zhang Y, Schlessinger J, & Sakaguchi K (2005). Trans-activation of EphA4 and FGF receptors mediated by direct interactions between their cytoplasmic domains. Proceedings of the National Academy of Sciences of the United States of America, 102(52), 18866–18871. 10.1073/pnas.0509741102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemitsu R, Tokunaga T, Shukunami C, Ideo K, Arimura H, Karasugi T, Nakamura E, Ide J, Hiraki Y, & Mizuta H (2019). Fibroblast growth factor 2 enhances tendon-to-bone healing in a rat rotator cuff repair of chronic tears. The American Journal of Sports Medicine, 47(7), 1701–1712. 10.1177/0363546519836959 [DOI] [PubMed] [Google Scholar]
- Yoshizawa Y, Wada K, Shimoi G, Kameyama Y, Wakabayashi Y, Fukuta K, & Hashizume R (2015). A 1-bp deletion in Fgf5 causes male-dominant long hair in the Syrian hamster. Mammalian Genome, 26(11–12), 630–637. 10.1007/s00335-015-9608-5 [DOI] [PubMed] [Google Scholar]
- Young JJ, Grayson P, Edwards SV, & Tabin CJ (2019). Attenuated Fgf signaling underlies the forelimb heterochrony in the emu Dromaius novaehollandiae. Current Biology, 29(21), 3681–3691.e3685. 10.1016/j.cub.2019.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ye X, Che R, Wu Q, Qi J, Song L, Guo X, Zhang S, Wu H, Ren G, & Li D (2017). FGF21 exerts comparable pharmacological efficacy with Adalimumab in ameliorating collagen-induced rheumatoid arthritis by regulating systematic inflammatory response. Biomedicine & Pharmacotherapy, 89, 751–760. 10.1016/j.biopha.2017.02.059 [DOI] [PubMed] [Google Scholar]
- Yu K, Karuppaiah K, & Ornitz DM (2015). Mesenchymal fibroblast growth factor receptor signaling regulates palatal shelf elevation during secondary palate formation. Developmental Dynamics, 244(11), 1427–1438. 10.1002/dvdy.24319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Wilhelm K, Dubrac A, Tung JK, Alves TC, Fang JS, Xie Y, Zhu J, Chen Z, De Smet F, Zhang J, Jin SW, Sun L, Sun H, Kibbey RG, Hirschi KK, Hay N, Carmeliet P, Chittenden TW, … Simons M (2017). FGF-dependent metabolic control of vascular development. Nature, 545(7653), 224–228. 10.1038/nature22322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Huang X, Tian X, Zhang H, He L, Wang Y, Nie Y, Hu S, Lin Z, Zhou B, Pu W, Lui KO, & Zhou B (2016). GATA4 regulates Fgf16 to promote heart repair after injury. Development, 143(6), 936–949. 10.1242/dev.130971 [DOI] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Zheng YJ, Weng QH, Zhu SP, & Zhou DS (2020). LncRNA TUG1 promoted osteogenic differentiation through promoting bFGF ubiquitination. In Vitro Cellular & Developmental Biology. Animal, 56(1), 42–48. 10.1007/s11626-019-00410-y [DOI] [PubMed] [Google Scholar]
- Yu Y, Li S, Liu Y, Tian G, Yuan Q, Bai F, Wang W, Zhang Z, Ren G, Zhang Y, & Li D (2015). Fibroblast growth factor 21 (FGF21) ameliorates collagen-induced arthritis through modulating oxidative stress and suppressing nuclear factor-kappa B pathway. International Immunopharmacology, 25(1), 74–82. 10.1016/j.intimp.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Yu Y, Xie GJ, Hu Y, Li XS, Chen GY, Zheng GE, Chen X, & Cheng Y (2019). Dysregulation of fibroblast growth factor 10 in the peripheral blood of patients with schizophrenia. Journal of Molecular Neuroscience, 69(1), 69–74. 10.1007/s12031-019-01331-x [DOI] [PubMed] [Google Scholar]
- Yu Y, Yang J, Luan F, Gu G, Zhao R, Wang Q, Dong Z, Tang J, Wang W, Sun J, Lv P, Zhang H, & Wang C (2021). Sensorineural hearing loss and mitochondrial apoptosis of cochlear spiral ganglion neurons in fibroblast growth factor 13 knockout mice. Frontiers in Cellular Neuroscience, 15, 658586. 10.3389/fncel.2021.658586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T, Klinkhammer K, Lyu H, Gao S, Yuan J, Hopkins S, Zhang JS, & De Langhe SP (2020). Temporospatial expression of Fgfr1 and 2 during lung development, homeostasis, and regeneration. Frontiers in Pharmacology, 11, 120. 10.3389/fphar.2020.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T, Volckaert T, Redente EF, Hopkins S, Klinkhammer K, Wasnick R, Chao CM, Yuan J, Zhang JS, Yao C, Majka S, Stripp BR, Gunther A, Riches DWH, Bellusci S, Thannickal VJ, & De Langhe SP (2019). FGF10-FGFR2B signaling generates basal cells and drives alveolar epithelial regeneration by bronchial epithelial stem cells after lung injury. Stem Cell Reports, 12(5), 1041–1055. 10.1016/j.stemcr.2019.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue M, Lan Y, Liu H, Wu Z, Imamura T, & Jiang R (2021). Tissue-specific analysis of Fgf18 gene function in palate development. Developmental Dynamics, 250(4), 562–573. 10.1002/dvdy.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun EJ, Lorizio W, Seedorf G, Abman SH, & Vu TH (2016). VEGF and endothelium-derived retinoic acid regulate lung vascular and alveolar development. American Journal of Physiology. Lung Cellular and Molecular Physiology, 310(4), L287–L298. 10.1152/ajplung.00229.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf IO, Chen HM, Cheng PH, Chang CY, Tsai SJ, Chuang JI, Wu CC, Huang BM, Sun HS, Chen CM, & Yang SH (2021). Fibroblast growth factor 9 stimulates neuronal length through NF-kB signaling in striatal cell Huntington’s disease models. Molecular Neurobiology, 58(5), 2396–2406. 10.1007/s12035-020-02220-w [DOI] [PubMed] [Google Scholar]
- Yusuf IO, Chen HM, Cheng PH, Chang CY, Tsai SJ, Chuang JI, Wu CC, Huang BM, Sun HS, & Yang SH (2019). Fibroblast growth factor 9 activates anti-oxidative functions of Nrf2 through ERK signalling in striatal cell models of Huntington’s disease. Free Radical Biology & Medicine, 130, 256–266. 10.1016/j.freeradbiomed.2018.10.455 [DOI] [PubMed] [Google Scholar]
- Yusuf IO, Cheng PH, Chen HM, Chang YF, Chang CY, Yang HI, Lin CW, Tsai SJ, Chuang JI, Wu CC, Huang BM, Sun HS, & Yang SH (2018). Fibroblast growth factor 9 suppresses striatal cell death dominantly through ERK signaling in Huntington’s disease. Cellular Physiology and Biochemistry, 48(2), 605–617. 10.1159/000491889 [DOI] [PubMed] [Google Scholar]
- Zacherl S, La Venuta G, Muller HM, Wegehingel S, Dimou E, Sehr P, Lewis JD, Erfle H, Pepperkok R, & Nickel W (2015). A direct role for ATP1A1 in unconventional secretion of fibroblast growth factor 2. The Journal of Biological Chemistry, 290(6), 3654–3665. 10.1074/jbc.M114.590067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahra FT, Sajib MS, & Mikelis CM (2021). Role of bFGF in acquired resistance upon anti-VEGF therapy in cancer. Cancers (Basel), 13(6), 1422. 10.3390/cancers13061422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska M, Haugsten EM, Nadratowska-Wesolowska B, Oppelt A, Hausott B, Jin Y, Otlewski J, Wesche J, & Wiedlocha A (2013). ERK-mediated phosphorylation of fibroblast growth factor receptor 1 on Ser777 inhibits signaling. Science Signaling, 6(262), ra11. 10.1126/scisignal.2003087 [DOI] [PubMed] [Google Scholar]
- Zakrzewska M, Opalinski L, Haugsten EM, Otlewski J, & Wiedlocha A (2019). Crosstalk between p38 and Erk 1/2 in downregulation of FGF1-induced signaling. International Journal of Molecular Sciences, 20(8), 1826. 10.3390/ijms20081826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamai M, Trullo A, Giordano M, Corti V, Arza Cuesta E, Francavilla C, Cavallaro U, & Caiolfa VR (2019). Number and brightness analysis reveals that NCAM and FGF2 elicit different assembly and dynamics of FGFR1 in live cells. Journal of Cell Science, 132(1), jcs220624. 10.1242/jcs.220624 [DOI] [PubMed] [Google Scholar]
- Zaragosi LE, Ailhaud G, & Dani C (2006). Autocrine fibroblast growth factor 2 signaling is critical for self-renewal of human multipotent adipose-derived stem cells. Stem Cells, 24(11), 2412–2419. 10.1634/stemcells.2006-0006 [DOI] [PubMed] [Google Scholar]
- Zarei M, Barroso E, Palomer X, Dai J, Rada P, Quesada-Lopez T, Escola-Gil JC, Cedo L, Zali MR, Molaei M, Dabiri R, Vazquez S, Pujol E, Valverde AM, Villarroya F, Liu Y, Wahli W, & Vazquez-Carrera M (2018). Hepatic regulation of VLDL receptor by PPARbeta/delta and FGF21 modulates non-alcoholic fatty liver disease. Molecular Metabolism, 8, 117–131. 10.1016/j.molmet.2017.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zbinden A, Browne S, Altiok EI, Svedlund FL, Jackson WM, & Healy KE (2018). Multivalent conjugates of basic fibroblast growth factor enhance in vitro proliferation and migration of endothelial cells. Biomaterials Science, 6(5), 1076–1083. 10.1039/c7bm01052d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelarayan L, Vendrell V, Dominguez-Frutos E, Lopez-Hernandez I, Schimmang-Alonso K, Alonso MT, Alvarez Y, Maier H, Anderson MJ, Lewandoski M, & Schimmang T (2021). Inactivation of Fgf3 and Fgf4 within the Fgf3/Fgf4/Fgf15 gene cluster reveals their redundant requirement for mouse inner ear induction and embryonic survival. Developmental Dynamics, 1–8. 10.1002/dvdy.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai X, Liu J, Ni A, & Ye J (2020). MiR-497 promotes microglia activation and proinflammatory cytokines production in chronic unpredictable stress-induced depression via targeting FGF2. Journal of Chemical Neuroanatomy, 110, 101872. 10.1016/j.jchemneu.2020.101872 [DOI] [PubMed] [Google Scholar]
- Zhang F, Zhu X, Wang P, He Q, Huang H, Zheng T, Li Y, Jia H, Xu L, Zhao H, Colozza G, Tao Q, De Robertis EM, & Ding Y (2021). The cytokine FAM3B/PANDER is an FGFR ligand that promotes posterior development in Xenopus. Proceedings of the National Academy of Sciences of the United States of America, 118(20), e2100342118. 10.1073/pnas.2100342118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li C, Li Q, Li B, Larkin DM, Lee C, Storz JF, Antunes A, Greenwold MJ, Meredith RW, Odeen A, Cui J, Zhou Q, Xu L, Pan H, Wang Z, Jin L, Zhang P, Hu H, … Wang J (2014). Comparative genomics reveals insights into avian genome evolution and adaptation. Science, 346(6215), 1311–1320. 10.1126/science.1251385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kamiya N, Tsuji T, Takeda H, Scott G, Rajderkar S, Ray MK, Mochida Y, Allen B, Lefebvre V, Hung IH, Ornitz DM, Kunieda T, & Mishina Y (2016). Elevated fibroblast growth factor signaling is critical for the pathogenesis of the Dwarfism in Evc2/Limbin mutant mice. PLoS Genetics, 12(12), e1006510. 10.1371/journal.pgen.1006510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kot A, Lay YE, Fierro FA, Chen H, Lane NE, & Yao W (2017). Acceleration of fracture healing by overexpression of basic fibroblast growth factor in the mesenchymal stromal cells. Stem Cells Translational Medicine, 6(10), 1880–1893. 10.1002/sctm.17-0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cui C, & Xu H (2019). Downregulation of miR-145–5p elevates retinal ganglion cell survival to delay diabetic retinopathy progress by targeting FGF5. Bioscience, Biotechnology, and Biochemistry, 83(9), 1655–1662. 10.1080/09168451.2019.1630251 [DOI] [PubMed] [Google Scholar]
- Zhang J, Gupte J, Gong Y, Weiszmann J, Zhang Y, Lee KJ, Richards WG, & Li Y (2017). Chronic over-expression of fibroblast growth factor 21 increases bile acid biosynthesis by opposing FGF15/19 action. eBioMedicine, 15, 173–183. 10.1016/j.ebiom.2016.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu Z, Li Y, You Q, Yang J, Jin Y, Zou G, Tang J, Ge Z, & Liu Y (2020). FGF2: A key regulator augmenting tendon-to-bone healing and cartilage repair. Regenerative Medicine, 15(9), 2129–2142. 10.2217/rme-2019-0080 [DOI] [PubMed] [Google Scholar]
- Zhang J, Upadhya D, Lu L, & Reneker LW (2015). Fibroblast growth factor receptor 2 (FGFR2) is required for corneal epithelial cell proliferation and differentiation during embryonic development. PLoS One, 10(1), e0117089. 10.1371/journal.pone.0117089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhou Y, Huang T, Wu F, Pan Y, Dong Y, Wang Y, Chan AKY, Liu L, Kwan JSH, Cheung AHK, Wong CC, Lo AKF, Cheng ASL, Yu J, Lo KW, Kang W, & To KF (2019). FGF18, a prominent player in FGF signaling, promotes gastric tumorigenesis through autocrine manner and is negatively regulated by miR-590–5p. Oncogene, 38(1), 33–46. 10.1038/s41388-018-0430-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Cheng H, Yue Y, Li S, Zhang D, & He R (2018). TUG1 knockdown ameliorates atherosclerosis via up-regulating the expression of miR-133a target gene FGF1. Cardiovascular Pathology, 33, 6–15. 10.1016/j.carpath.2017.11.004 [DOI] [PubMed] [Google Scholar]
- Zhang P, Li YN, Tu S, & Cheng XB (2021). SP1-induced lncRNA TUG1 regulates proliferation and apoptosis in islet cells of type 2 diabetes mellitus via the miR-188–3p/FGF5 axis. European Review for Medical and Pharmacological Sciences, 25(4), 1959–1966. 10.26355/eurrev_202102_25096 [DOI] [PubMed] [Google Scholar]
- Zhang S, Choi HS, Jung HS, & Lee JM (2018). FGF10 Is Required for Circumvallate Papilla Morphogenesis by Maintaining Lgr5 Activity. Frontiers in Physiology, 9, 1192. 10.3389/fphys.2018.01192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo K, Xia F, Zhao X, Huang Z, & Niu J (2018). FGF23(C-tail) improves diabetic nephropathy by attenuating renal fibrosis and inflammation. BMC Biotechnology, 18(1), 33. 10.1186/s12896-018-0449-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo Y, Yang J, Niu J, Du L, Li H, & Li X (2019). A functional variant alters binding of activating protein 1 regulating expression of FGF7 gene associated with chronic obstructive pulmonary disease. BMC Medical Genetics, 20(1), 33. 10.1186/s12881-019-0761-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, & Ornitz DM (2006). Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. The Journal of Biological Chemistry, 281(23), 15694–15700. 10.1074/jbc.M601252200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Imel EA, Ruppe MD, Weber TJ, Klausner MA, Ito T, Vergeire M, Humphrey J, Glorieux FH, Portale AA, Insogna K, Carpenter TO, & Peacock M (2016). Pharmacokinetics and pharmacodynamics of a human monoclonal anti-FGF23 antibody (KRN23) in the first multiple ascending-dose trial treating adults with X-linked hypophosphatemia. Journal of Clinical Pharmacology, 56(2), 176–185. 10.1002/jcph.570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Stappenbeck TS, White AC, Lavine KJ, Gordon JI, & Ornitz DM (2006). Reciprocal epithelial-mesenchymal FGF signaling is required for cecal development. Development, 133(1), 173–180. 10.1242/dev.02175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Weng M, & Chen Z (2021). Fibroblast Growth Factor 9 (FGF9) negatively regulates the early stage of chondrogenic differentiation. Plos One, 16(2), e0241281. 10.1371/journal.pone.0241281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yeung DC, Karpisek M, Stejskal D, Zhou ZG, Liu F, Wong RL, Chow WS, Tso AW, Lam KS, & Xu A (2008). Serum FGF21 levels are increased in obesity and are independently associated with the metabolic syndrome in humans. Diabetes, 57(5), 1246–1253. 10.2337/db07-1476 [DOI] [PubMed] [Google Scholar]
- Zhao C, Lancman JJ, Yang Y, Gates KP, Cao D, Barske L, Matalonga J, Pan X, He J, Graves A, Huisken J, Chen C, & Dong PDS (2021). Intrahepatic cholangiocyte regeneration from an Fgf-dependent extrahepatic progenitor niche in a zebrafish model of Alagille Syndrome. Hepatology, 1–17. 10.1002/hep.32173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Bailey CG, Feng Y, Rasko J, & Lovicu FJ (2018). Negative regulation of lens fiber cell differentiation by RTK antagonists Spry and Spred. Experimental Eye Research, 170, 148–159. 10.1016/j.exer.2018.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao GH, Qiu YQ, Yang CW, Chen IS, Chen CY, & Lee SJ (2020). The cardenolides ouabain and reevesioside A promote FGF2 secretion and subsequent FGFR1 phosphorylation via converged ERK1/2 activation. Biochemical Pharmacology, 172, 113741. 10.1016/j.bcp.2019.113741 [DOI] [PubMed] [Google Scholar]
- Zhao L, Cao X, Li L, Wang X, Wang Q, Jiang S, Tang C, Zhou S, Xu N, Cui Y, Hu W, Fei L, Zheng Z, Chen L, Schmidt MO, Wei Q, Zhao J, Labes R, Patzak A, … Lai EY (2020). Acute kidney injury sensitizes the brain vasculature to Ang II (Angiotensin II) constriction via FGFBP1 (fibroblast growth factor binding protein 1). Hypertension, 76(6), 1924–1934. 10.1161/HYPERTENSIONAHA.120.15582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Fan M, Zhao L, Yun H, Yang Y, Wang C, & Qin D (2020). Fibroblast growth factor 1 ameliorates adipose tissue inflammation and systemic insulin resistance via enhancing adipocyte mTORC2/Rictor signal. Journal of Cellular and Molecular Medicine, 24(21), 12813–12825. 10.1111/jcmm.15872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Wu Y, Zhao C, & Feng M (2016). Association between FGF20 rs12720208 gene polymorphism and Parkinson’s disease: A meta-analysis. Neurological Sciences, 37(7), 1119–1126. 10.1007/s10072-016-2559-0 [DOI] [PubMed] [Google Scholar]
- Zhao Y, & Adjei AA (2015). Targeting angiogenesis in cancer therapy: Moving beyond vascular endothelial growth factor. The Oncologist, 20(6), 660–673. 10.1634/theoncologist.2014-0465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Cao F, Yu X, Chen C, Meng J, Zhong R, Zhang Y, & Zhu D (2018). Linc-RAM is required for FGF2 function in regulating myogenic cell differentiation. RNA Biology, 15(3), 404–412. 10.1080/15476286.2018.1431494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Meng C, Wang Y, Huang H, Liu W, Zhang JF, Zhao H, Feng B, Leung PS, & Xia Y (2016). IL-1beta inhibits beta-Klotho expression and FGF19 signaling in hepatocytes. American Journal of Physiology. Endocrinology and Metabolism, 310(4), E289–E300. 10.1152/ajpendo.00356.2015 [DOI] [PubMed] [Google Scholar]
- Zhao YF, Luo YM, Xiong W, Ding W, Li YR, Zhao W, Zeng HZ, Gao HC, & Wu XL (2015). Mesenchymal stem cell-based FGF2 gene therapy for acute lung injury induced by lipopolysaccharide in mice. European Review for Medical and Pharmacological Sciences, 19(5), 857–865. [PubMed] [Google Scholar]
- Zhen EY, Jin Z, Ackermann BL, Thomas MK, & Gutierrez JA (2016). Circulating FGF21 proteolytic processing mediated by fibroblast activation protein. The Biochemical Journal, 473(5), 605–614. 10.1042/BJ20151085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Zhao L, Xia H, Xu C, Wang D, Liu K, Lin L, Li X, Yan Z, & Gao H (2016). NMR-based metabolomics reveal a recovery from metabolic changes in the striatum of 6-OHDA-induced rats treated with basic fibroblast growth factor. Molecular Neurobiology, 53(10), 6690–6697. 10.1007/s12035-015-9579-2 [DOI] [PubMed] [Google Scholar]
- Zheng K, Lin L, Cui P, Liu T, Chen L, Yang C, & Jiang W (2020). Association of fibroblast growth factor 23 with ischemic stroke and its subtypes: A Mendelian randomization study. Frontiers in Genetics, 11, 608517. 10.3389/fgene.2020.608517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Tang Z, Xiong J, Wang B, Xu J, Chen L, Cai S, Wu C, Ye L, Xu K, Chen Z, Wu Y, & Xiao J (2021). RAGE: A potential therapeutic target during FGF1 treatment of diabetes-mediated liver injury. Journal of Cellular and Molecular Medicine, 25(10), 4776–4785. 10.1111/jcmm.16446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Tu X, Kong Y, Guo L, Li B, Zhong W, Cheng Y, Jiang Y, & Jiang Q (2020). LncRNA H19 promotes odontoblastic differentiation of human dental pulp stem cells by regulating miR-140–5p and BMP-2/FGF9. Stem Cell Research & Therapy, 11(1), 202. 10.1186/s13287-020-01698-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Chen Y, Cao C, Chen X, Gao W, & Zhang L (2015). Inactivation of glycogen synthase kinase-3beta up-regulates beta-catenin and promotes chondrogenesis. Cell and Tissue Banking, 16(1), 135–142. 10.1007/s10561-014-9449-6 [DOI] [PubMed] [Google Scholar]
- Zhou M, Wang X, Phung V, Lindhout DA, Mondal K, Hsu JY, Yang H, Humphrey M, Ding X, Arora T, Learned RM, DePaoli AM, Tian H, & Ling L (2014). Separating tumorigenicity from bile acid regulatory activity for endocrine hormone FGF19. Cancer Research, 74(12), 3306–3316. 10.1158/0008-5472.CAN-14-0208 [DOI] [PubMed] [Google Scholar]
- Zhou M, Yang H, Learned RM, Tian H, & Ling L (2017). Non-cell-autonomous activation of IL-6/STAT3 signaling mediates FGF19-driven hepatocarcinogenesis. Nature Communications, 8, 15433. 10.1038/ncomms15433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Wang Q, Meng Z, Peng J, Zhou Y, Song W, Wang J, Chen S, & Sun K (2020). Mutations in fibroblast growth factor (FGF8) and FGF10 identified in patients with conotruncal defects. Journal of Translational Medicine, 18(1), 283. 10.1186/s12967-020-02445-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Xie Y, Li W, Huang J, Wang Z, Tang J, Xu W, Sun X, Tan Q, Huang S, Luo F, Xu M, Wang J, Wu T, Chen L, Chen H, Su N, Du X, Shen Y, & Chen L (2016). Conditional deletion of Fgfr3 in chondrocytes leads to osteoarthritis-like defects in temporomandibular joint of adult mice. Scientific Reports, 6, 24039. 10.1038/srep24039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Simic P, & Rhee EP (2021). Fibroblast growth factor 23 regulation and acute kidney injury. Nephron, 1–4. 10.1159/000517734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhang Y, & Wang N (2021). Regulation and potential biological role of fibroblast growth factor 21 in chronic kidney disease. Frontiers in Physiology, 12, 764503. 10.3389/fphys.2021.764503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XJ, Liu Y, Dai ZM, Zhang X, Yang X, Li Y, Qiu M, Fu J, Hsu W, Chen Y, & Zhang Z (2014). BMP-FGF signaling axis mediates Wnt-induced epidermal stratification in developing mammalian skin. PLoS Genetics, 10(10), e1004687. 10.1371/journal.pgen.1004687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XJ, Liu Y, Yuan X, Wang M, Zhao W, Yang X, Zhang X, Hsu W, Qiu M, Zhang Z, & Zhang Z (2016). Ectodermal Wnt controls nasal pit morphogenesis through modulation of the BMP/FGF/JNK signaling axis. Developmental Dynamics, 245(3), 414–426. 10.1002/dvdy.24376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Yang L, Chong QY, Yan H, Zhang W, Qian W, Tan S, Wu Z, Lobie PE, & Zhu T (2019). Long noncoding RNA Linc00460 promotes breast cancer progression by regulating the miR-489–5p/FGF7/AKT axis. Cancer Management and Research, 11, 5983–6001. 10.2147/CMAR.S207084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang L, Pandey AV, Villiger PM, & Trueb B (2015). Cell-cell fusion induced by the Ig3 domain of receptor FGFRL1 in CHO cells. Biochimica et Biophysica Acta, 1853(10 Pt A), 2273–2285. 10.1016/j.bbamcr.2015.05.027 [DOI] [PubMed] [Google Scholar]
- Zhuang L, Steinberg F, & Trueb B (2016). Receptor FGFRL1 acts as a tumor suppressor in nude mice when overexpressed in HEK 293 Tet-On cells. Oncology Letters, 12(6), 4524–4530. 10.3892/ol.2016.5245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang L, & Trueb B (2017). Evolution of the fusogenic activity of the receptor FGFRL1. Archives of Biochemistry and Biophysics, 625–626, 54–64. 10.1016/j.abb.2017.06.002 [DOI] [PubMed] [Google Scholar]
- Zhuang L, Vogel M, Villiger PM, & Trueb B (2020). Dissecting the interaction of FGF8 with receptor FGFRL1. Biomolecules, 10(10), 1399. 10.3390/biom10101399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkle A, & Mohammadi M (2018). A threshold model for receptor tyrosine kinase signaling specificity and cell fate determination. F1000Research, 7, F1000 Faculty Rev-872. 10.12688/f1000research.14143.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivotic M, Tampe B, Muller G, Muller C, Lipkovski A, Xu X, Nyamsuren G, Zeisberg M, & Markovic-Lipkovski J (2018). Modulation of NCAM/FGFR1 signaling suppresses EMT program in human proximal tubular epithelial cells. PLoS One, 13(11), e0206786. 10.1371/journal.pone.0206786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong C, Venot A, Li X, Lu W, Xiao W, Wilkes JL, Salanga CL, Handel TM, Wang L, Wolfert MA, & Boons GJ (2017). Heparan sulfate microarray reveals that heparan sulfate-protein binding exhibits different ligand requirements. Journal of the American Chemical Society, 139(28), 9534–9543. 10.1021/jacs.7b01399 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


