Summary
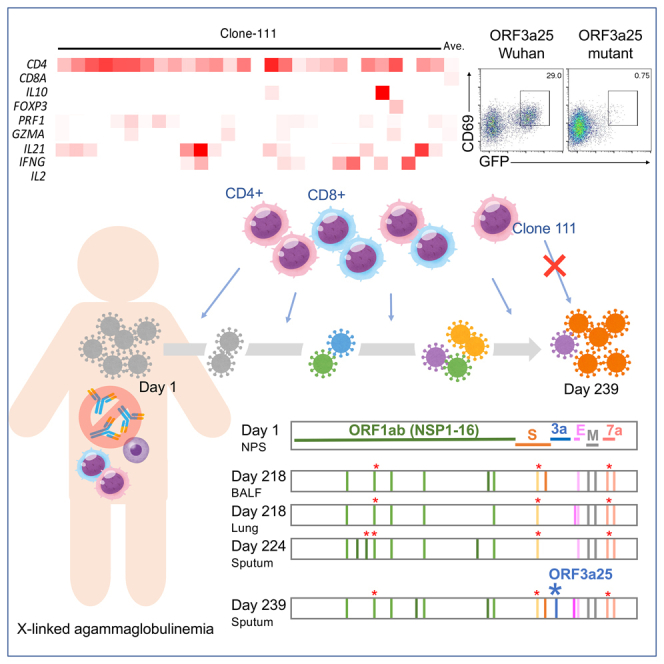
A 25-year-old patient with a primary immunodeficiency lacking immunoglobulin production experienced a relapse after a 239-day period of persistent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Viral genetic sequencing demonstrated that SARS-CoV-2 had evolved during the infection period, with at least five mutations associated with host cellular immune recognition. Among them, the T32I mutation in ORF3a was found to evade recognition by CD4+ T cells. The virus found after relapse showed an increased proliferative capacity in vitro. SARS-CoV-2 may have evolved to evade recognition by CD4+ T cells and increased in its proliferative capacity during the persistent infection, likely leading to relapse. These mutations may further affect viral clearance in hosts with similar types of human leukocyte antigens. The early elimination of SARS-CoV-2 in immunocompromised patients is therefore important not only to improve the condition of patients but also to prevent the emergence of mutants that threaten public health.
Subject areas: Immunology
Graphical abstract
Highlights
-
•
SARS-CoV-2 persistently infected a man with humoral immunodeficiency for 239 days
-
•
COVID-19 relapsed and SARS-CoV-2 evolved during the infection period
-
•
The virus isolated after relapse had an increased proliferative capacity in vitro
-
•
SARS-CoV-2 may have evolved to evade recognition by CD4+ T-cells
Immunology
Introduction
X-linked agammaglobulinemia (XLA) is a rare primary humoral immunodeficiency that was first reported by Bruton in 1952. In XLA, abnormalities in Bruton’s tyrosine kinase prevent normal B-cell maturation, resulting in agammaglobulinemia.1 The prognosis is generally good if globulin preparations are administered regularly.1
Coronavirus disease 2019 (COVID-19) can become chronic and relapse in patients with primary2 and secondary3,4,5,6,7 antibody deficiencies. Previous reports have shown that convalescent plasma,7,8 remdesivir plus convalescent plasma,5,9,10 and remdesivir plus a monoclonal antibody cocktail11 have therapeutic effects in COVID-19 cases with XLA. Polyfunctional CD8+ T-cell responses have also been observed during the course of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in patients with XLA.6 A recent analysis of SARS-CoV-2 infection in an immunocompromised patient demonstrated viral evolution and reduced sensitivity to neutralizing antibodies, which was either dependent on12 or independent of convalescent plasma therapy.13
Here, we present an XLA case in which SARS-CoV-2 infection persisted for more than 239 days without specific humoral immunity to SARS-CoV-2. The patient experienced a relapse as SARS-CoV-2 evolved during the infection period. The clinical course of the patient, intra-host evolution of SARS-CoV-2 during persistent and relapsing infection for more than 239 days, and SARS-CoV-2-specific T-cell responses in the absence of humoral immunity have been described.
Results
Clinical presentation of an agammaglobulinemia patient who experienced relapse following persistent SARS-CoV-2 infection
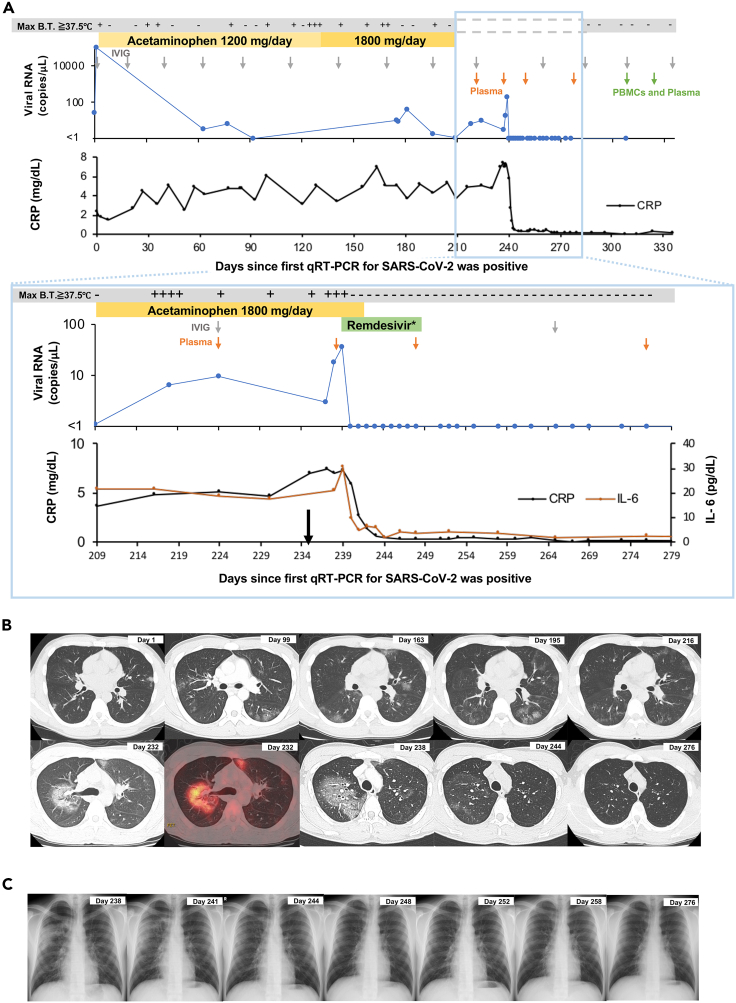
A 25-year-old male patient developed fever at the end of July 2020. He had been genetically diagnosed with XLA due to an exon 6 skip mutation (splicing site mutation, c.392-2A>G) at 11 months of age. The CD19 lymphocyte subset in this patient was undetectable (Table S1). The same exon 6 skip mutation has been reported to notably reduce the levels of BTK expression in monocytes.14 The neutrophil sterilizing function and natural killer cell activity in the patient were within normal limits, but there was a decrease in his neutrophil phagocytosis function (Table S1). He was diagnosed with COVID-19 based on a positive result from a quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) test for SARS-CoV-2 in early August. One day after the COVID-19 diagnosis (day 1), the patient was admitted to Osaka City General Hospital as this was a requirement from the Japanese government at the time. Because the early prognosis of the patient was thought to be good and his general condition remained stable after he had been admitted, no antiviral drugs, convalescent plasma, or antibody cocktails were administered, and he was discharged on day 9. After being discharged, the patient occasionally experienced malaise and fever. Blood tests showed slightly but persistently elevated levels of C-reactive protein (CRP) (range, 2.51–7.49 mg/dL; median, 4.85 mg/dL) (Figure 1A). Computed tomography (CT) performed on days 99, 163, 195, 216, and 232 showed pneumonia with migrating ground glass opacities, and [18F]-2-fluoro-2-deoxy-D-glucose (FDG)-positron emission tomography (PET)/CT on day 232 showed an accumulation of FDG only at the site of pneumonia (maximum standardized uptake value, 5.6) (Figure 1B). Considering the possibility of organizing pneumonia, a bronchoscopy with bronchoalveolarlavage was performed on day 218, which confirmed non-specific inflammation in the biopsy tissue of the pneumonia-affected region. The qRT-PCR testing for SARS-CoV-2 in sputum and saliva samples taken on days 63, 77, 175, 176, and 181 detected SARS-CoV-2 RNA with high cycle threshold (Ct) values (33.82–36.5; median, 35.3) (Figure 1A). Because the Ct values were high, COVID-19 was initially considered unlikely to be the main cause of the pneumonia and fever.
Figure 1.
Clinical and laboratory data from the patient during the observation period
(A) Results of qRT-PCR for SARS-CoV-2; days in which there was a maximum body temperature above 37.5°C; and blood tests during the observation period. The black arrow indicates the time of symptom exacerbation. The gray arrow indicates the administration time of intravenous immunoglobulin (IVIG). The green and orange arrows indicate the collection time of PBMCs for the T-cell analysis and/or plasma for the measurement of the anti-SARS-CoV-2 spike antibody, respectively. qRT-PCR, real-time quantitative reverse transcriptase polymerase chain reaction; Max B.T., maximum body temperature of the day. ∗ Remdesivir was administered at 200 mg/day on the first day and 100 mg/day for the following 9 days.
(B) Computed tomography (black and white images) and positron emission tomography/computed tomography (colored image) results during the observation period.
(C) Chest X-ray results during the observation period, particularly after starting the administration of remdesivir. See also Tables S1–S3 and S5.
On approximately day 235, the general condition of the patient worsened significantly. He presented chills, shivering, malaise, and uncontrollably high fever (approximately 40 °C, treated with 1,800 mg of acetaminophen per day). Blood testing showed elevated levels of CRP and interleukin (IL)-6, and CT showed worsening pneumonia. The qRT-PCR test for SARS-CoV-2 on day 239 showed a decreased Ct value. The multiplex PCR testing of sputum samples for 19 respiratory tract infections (BioFire FilmArray Respiratory Panel 2.1) on the same day detected SARS-CoV-2 but no other viruses or bacteria causing respiratory tract infection (Table S2). The patient was readmitted to Osaka City General Hospital and treated with remdesivir from day 239. Remdesivir was administered at 200 mg/day on the first day and 100 mg/day from the second day onwards for 10 days in total, based on methods described in previous studies.3,10 On the second day of remdesivir administration (day 240), the fever subsided, and the chills, shivering, and malaise disappeared. Blood testing showed rapid decreases in the levels of CRP and IL-6 (Figure 1A, Table S3), and imaging tests showed the resolution of pneumonia (Figure 1B). The qRT-PCR testing of sputum samples for SARS-CoV-2 showed elevated Ct values, and the patient tested negative for the virus on day 243 (Figure 1A, Table S4). No SARS-CoV-2 was isolated from the sputum samples after the administration of remdesivir. The patient was discharged on day 248 after all of his symptoms had been resolved and the blood and imaging examination results had improved. No flare-ups have occurred for 9 months since his discharge, indicating that the treatment was successful. The patient was given intravenous immunoglobulin (IVIG) injections every month (Figure 1A, Table S5) and was not vaccinated against SARS-CoV-2 during the entire course of this study. Tests for the anti-SARS-CoV-2 spike (S) protein in the plasma and IVIG lots administered to the patient during the study period yielded negative results (Figure S1).
Viral genome and proliferative capacity analysis
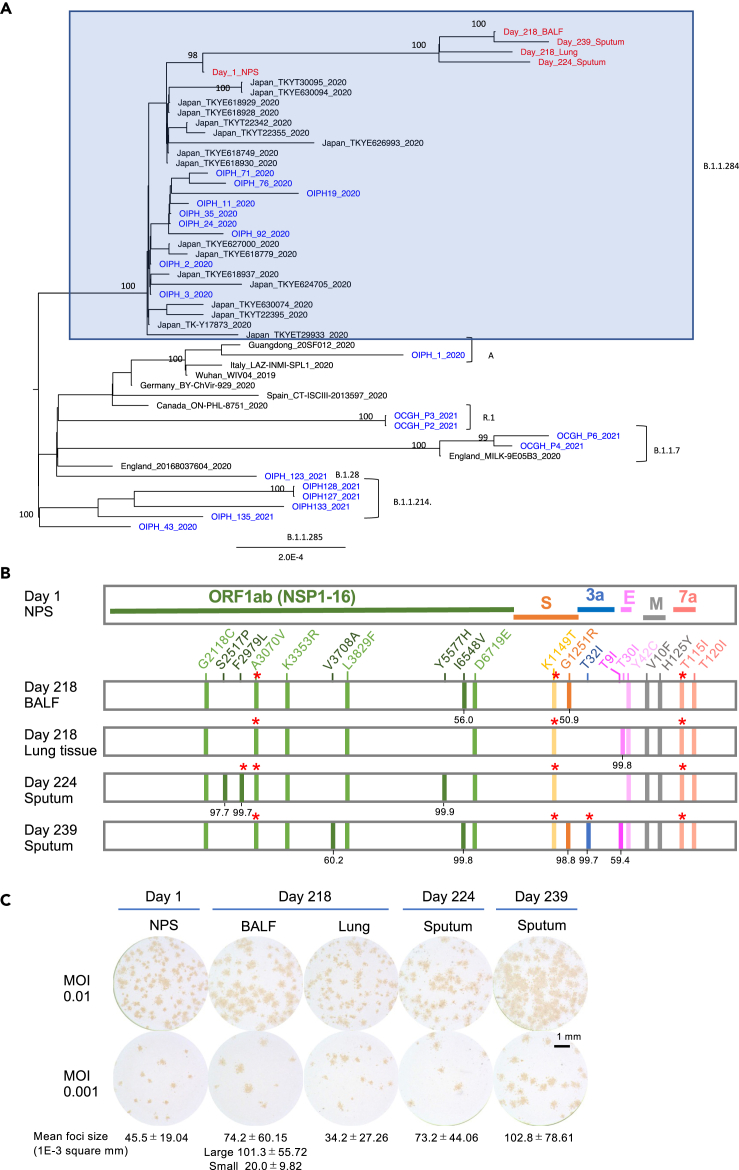
Infectious viruses were isolated from cryopreserved specimens from a nasopharyngeal swab (NPS) on day 1, bronchoalveolar lavage fluid (BALF) and lung tissue on day 218, sputum on day 224, and fresh sputum on day 239 (Table S4). Whole-genome sequences were obtained from the five isolates and day-1 NPS specimen. A phylogenetic analysis showed that all of the viruses belonged to the clade B.1.1.284, which was a prevalent clade in Japan during the period in which the patient was infected. The isolated day-1 virus showed the same consensus sequence as the day-1 NPS specimen, and the other viruses formed a distinct cluster with the day-1 viruses (Figure 2A, Table S6). It should be noted that most of the viruses in Japan between days 218–239, during which the mutated viruses in the patient were isolated, belonged to clade B.1.1.7. In contrast, the viruses isolated from the patient belonged to B.1.1.284, which nearly disappeared between days 218–239 in Japan.15,16,17 These results demonstrate that the worsening of the condition of the patient after day 235 was due to a relapse of COVID-19, ruling out the possibility of reinfection with another virus. Nearly two-thirds of the mutations were non-synonymous (Table S7), suggesting the presence of positive selection pressure. Compared with the day-1 viruses, 11 amino acid changes and two synonymous nucleotide substitutions were commonly observed in all of the other isolates (Figure 2B). An additional two, one, three, and five amino acid changes were found in the isolates from the BALF and lung tissue from day 218 and those from days 224 and 239, respectively. Apart from the K3353R mutation in ORF1ab, these mutations have rarely been found in the CoV-GLUE database18 (Table S7), indicating a unique evolution of SARS-CoV-2 in this patient. These mutations were unevenly distributed in the viral genome, and the average numbers of mutations per 1,000 nucleotides in these viruses exceeded 1 in the regions of E (6.67), M (3.00), and ORF7a (6.89), whereas those in ORF1a (0.454), ORF1b (0.278), S (0.720), and ORF3a (0.303) were less than 1. There were no mutations in the ORF6, ORF8, and N regions. The evolution rate of SARS-CoV-2 in this patient was higher (9.37 × 10−4/site/year) than that reported in general populations (3.95–6.67 × 10−4/site/year) (Table 1).19,20,21,22,23 We calculated the SARS-CoV-2 evolution rate based on published sequence data from other immunocompromised patients2,4,10,12,13,23,24 and confirmed that SARS-CoV-2 evolves faster in immunocompromised hosts than in the general population (Table 1).
Figure 2.
Whole-genome sequencing, phylogenetic relationships, and proliferative capacity of SARS-CoV-2 isolated from the patient
(A) Phylogenetic tree of the viruses isolated from the patient on days 1, 218, 224, and 239 (red), along with representative global and local (Osaka, Japan) sequences. Viruses isolated from the patient and other local patients are shown in red and blue, respectively.
(B) Locations of amino acid changes in the viruses isolated on days 218, 224, and 239 in the patient compared with those in the day-1 virus. The numbers below the mutations indicate the percentage of the mutation. ORF, open reading frame; NSP, non-structural protein; S, spike; 3a, ORF3a; E, envelope; M, membrane; 7a, ORF7a. ∗ Amino acid changes involved in T-cell recognition.
(C) Focus phenotypes of the isolated viruses (passage #1) were characterized in VeroE6/TMPRSS2 cells. MOI, multiplicity of infection. Scale bar depicted in (C) is 1 mm. See also Tables S4, S6, and S7.
Table 1.
Evolution rate of SARS-CoV-2 in the general population and immunocompromised hosts
| General population | ||
|---|---|---|
| Population | Reference | Evolution rate/site/year (95% CI)a |
| Africa | Motayo et al.19 | 4.13 × 10−4 |
| USA | Wang et al.20 | 6.67 × 10−4 |
| UK | Hill et al.21 | 4.6 × 10−4 |
| GISAID till March 2020 | Shen et al.22 | 3.95 × 10−4 |
| GISAID | Chaguzaet al.23 | 5.83 × 10−4 (5.56–6.11 × 10−4) |
| Immunocompromised patient | |||
|---|---|---|---|
| Deficiency | Reference | Disease | Evolution rate/site/year (95% CI)a |
| Primary | Present study | X-linked agammaglobulinemia | 9.37 × 10−4 (9.10–9.65 × 10−4)b |
| Avanzato et al.2 | Chronic lymphocytic leukemia | 12.1 × 10−4 (5.88–18.2 × 10−4)b | |
| Buckland et al.10 | X-linked agammaglobulinemia | 27.3 × 10−4 (22.5–32.0 × 10−4)b | |
| Secondary | Choi et al.4 | Severe antiphospholipid syndrome | 17.0 × 10−4 (14.8–19.2 × 10−4)b |
| Borges et al.24 | Non-Hodgkin lymphoma | 11.3 × 10−4b | |
| Kemp et al.12 | Marginal B cell lymphoma | 11.4 × 10−4 (8.19–14.6 × 10−4)b | |
| Williamson et al.13 | Chronic lymphocytic leukemia | 12.0 × 10−4 (8.61–15.4 × 10−4)b | |
| Chaguzaet al.23 | Advanced lymphocytic leukemia and B-cell lymphoma | 12.1 × 10−4 (10.7–13.4 × 10−4) | |
95% confidential intervals.
Calculated in the present study.
The proliferative capacities of the isolated viruses were evaluated using a focus-forming assay. Passage #1 viruses isolated from the NPS specimen on day 1, BALF and lung tissue on day 218, and sputum on days 224 and 239 were inoculated in VeroE6/TMPRSS2 cells, which were seeded in 24-well plates, at infection multiplicities of 0.01 and 0.001. The focus sizes of the viruses were compared 1 day after infection (Figure 2C). The viruses isolated from the day-1 NPS specimen had small foci (mean size, 45.5 ± 19.04 × 10−3 mm2), whereas those isolated from the sputum on day 239 had large foci (mean size, 102.8 ± 78.61 × 10−3 mm2). Although the viruses isolated from the lung tissue on day 218 formed small foci (mean size, 34.2 ± 27.26 × 10−3 mm2), those isolated from the BALF on day 218 formed both large (mean size, 101.3 ± 55.72 × 10−3 mm2) and small foci (mean size, 20.0 ± 9.82 × 10−3 mm2), suggesting that these isolates comprised a mixture of more than one viral clone. The viruses isolated from the sputum on day 224 formed medium-sized foci (mean size, 73.2 ± 44.06 × 10−3 mm2). Although several factors, including the binding and destruction of the receptor of the viruses, can affect focus or plaque sizes,25,26 it is generally believed that the sizes of foci are correlated with the proliferative capacities of viruses. These results suggest that SARS-CoV-2 evolved with an increased proliferative capacity in this patient.
The T32I mutation in ORF3a evades T-cell recognition
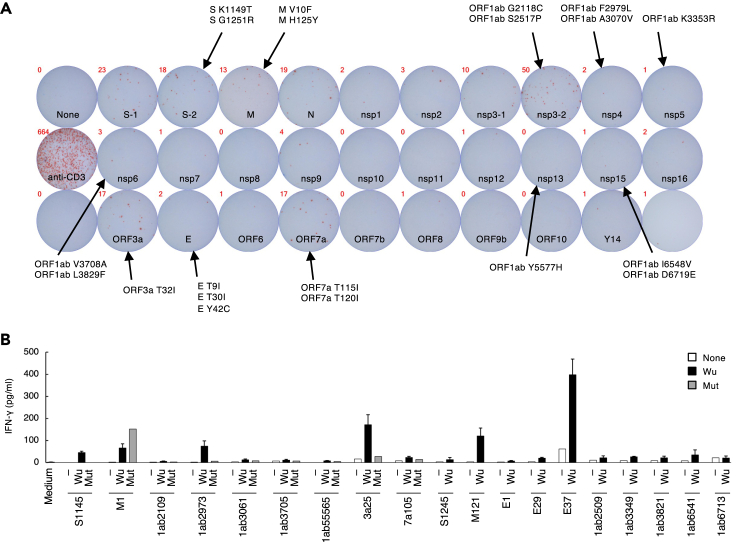
To profile the immune response against SARS-CoV-2 in the patient, the T-cell response against SARS-CoV-2 antigens was first examined. Peripheral blood mononuclear cells (PBMCs) were stimulated with peptide pools derived from proteins of the SARS-CoV-2 Wuhan-Hu-1 strain (Wu) in an ELISPOT assay (Figure 3A). Interferon (IFN)-γ-expressing cells were mainly observed when the PBMCs were stimulated with structural spike (S), membrane (M), nucleocapsid (N) proteins, accessory factors (ORF3a and ORF7a), and a non-structural protein (NSP3), suggesting that T-cell epitopes were mostly located in these proteins. These epitope hotspots included almost half of the mutations detected in the patient, implying that they were selected under pressure from the host T-cell response.
Figure 3.
SARS-CoV-2 epitope recognition by peripheral blood mononuclear cells (PBMCs) from the patient
(A) Reaction of PBMCs to the peptide pools derived from proteins of the SARS-CoV-2 Wuhan strain. PBMCs collected on day 308 were stimulated with peptide pools for 48 h, and IFN-γ expression was determined using an ELISPOT assay. SARS-CoV-2 proteins with mutations detected in the patient are indicated.
(B) Concentrations of IFN-γ in the supernatant of PBMCs stimulated with SARS-CoV-2 peptides. PBMCs collected from the patient on day 324 were stimulated with the indicated individual peptides from the SARS-CoV-2 Wuhan-Hu-1 strain (Wu) for 12 days before being washed and re-stimulated with nothing (−), the same Wu peptides, or corresponding mutant peptides (Mut) for 20 h. Error bars indicate mean ± SD of duplicates. Data are representative of three independent experiments. See also Figure S2 and Table S8.
To further investigate the pivotal mutations that aggravated the infection, the T-cell response to single epitopes covering the mutated regions was examined. PBMCs from the patient were expanded with peptides from Wu that covered the mutated regions (Figure 3A) before they were re-stimulated with the same or corresponding mutant (mut) peptides (Table S8). The S1145, ORF1ab2973, and ORF3a25 Wu peptides induced greater IFN-γ expression than their corresponding mutant peptides (Figure 3B). In addition, a decreased expression of IFN-γ induced by ORF1ab3061 mut and ORF7a105 T115I mut peptides was repeatedly observed in three experiments, indicating that mutations in these five regions were likely involved in the immune escape of the viral variants.
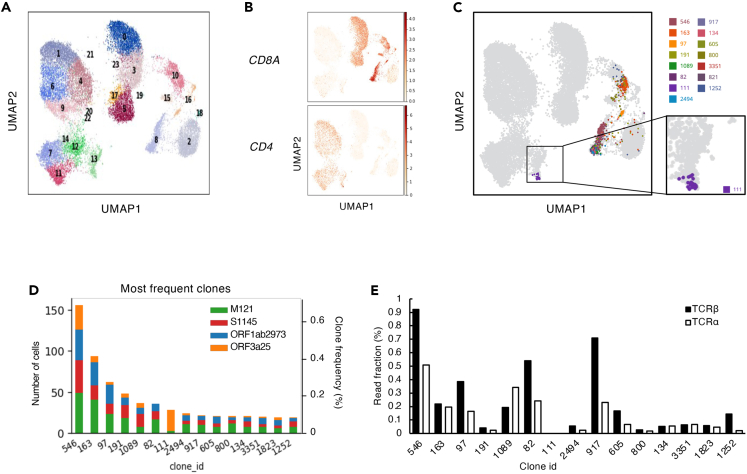
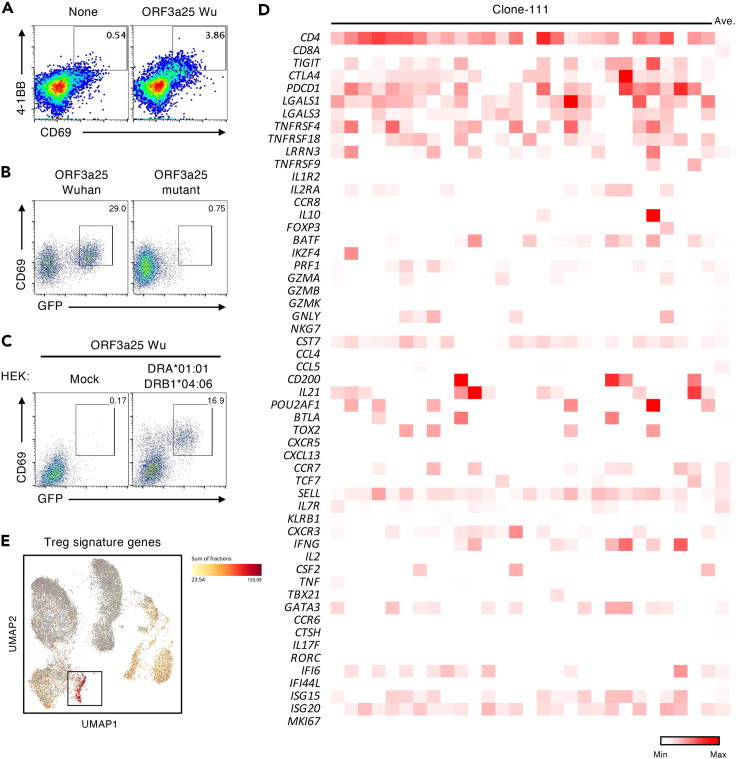
To further elucidate the characteristics of the T cells targeting these critical regions, T cells were sorted after being stimulated with S1145, ORF1ab2973, ORF3a25, or M121 Wu peptides, pooled, and analyzed using single-cell T-cell receptor (TCR)- and RNA-sequencing analyses (Figure 4A). The TCR sequencing results showed that some of the clonotypes had higher frequencies after the in vitro stimulations within both the CD4+ and CD8+ clusters (Figure 4B). Within the top-15 frequent T-cell clones identified in the single-cell analyses, 14 clones were located in clusters reflecting CD8+ CTL-like cells (Figure 4C). The remaining clone-111 comprised CD4+ cells (Figure 4C) and was predominantly from ORF3a25-stimulated cells (Figure 4D), suggesting that this clone was preferentially expanded by the stimulation of ORF3a25. All of the top 15 frequent clones, except clone-111, were detected in the PBMCs of the patient before the in vitro stimulation (Figure 4E), suggesting that clone-111 had a low frequency before the stimulation and became dominant afterward. Consistent with this, the ORF3a25 Wu peptide was found to activate the in vitro-expanded CD4+ T cells upon re-stimulation (Figure 5A). To confirm the epitope of clone-111, TCR-α and-β of clone-111 were reconstituted in a TCR-deficient reporter cell line. The reporter cells were activated by the ORF3a25 Wu peptide in the presence of autologous antigen-presenting cells, but the T32I mutant peptide lost antigenicity (Figure 5B). Of the human leukocyte antigen class II genes in the patient, DRB1∗04:06 was identified as a restricting allele for ORF3a epitope recognition (Figure 5C). These results indicate that the T32I mutation in ORF3a resulted in the evasion of T-cell recognition.
Figure 4.
CD4 T-cell clone-111 was specifically expanded following ORF3a25 stimulation
(A) Results of single-cell TCR- and RNA-seq analyses. PBMCs collected from the patient on day 324 were stimulated with S1145, ORF1ab2973, ORF3a25, or M121 Wu peptides in individual wells for 15 days before the CD3+T cells were sorted, hashtagged, mixed, and analyzed by single-cell TCR- and RNA-seq. Each dot in the UMAP plot corresponds to a single cell and is colored according to the Leiden cluster.
(B) mRNA expression of CD4 and CD8A in the single cells from the single-cell analyses described in (A).
(C) The top-15 most frequent T-cell clones detected in the single-cell analyses described in (A) are shown in the UMAP plot.
(D) Clone size (left yaxis), clone fraction (right yaxis), and origin of the top-15 frequent clones detected in the single-cell analyses shown in (A).
(E) Frequencies of the 15 clones shown in (D) before stimulation were determined by bulk TCR-seq using unstimulated PBMCs. The read fractions of the TCR-α and-β sequences are shown. See also Tables S8 and S9.
Figure 5.
The T32I mutation in ORF3a evades recognition by a CD4+ T-cell clone
(A) Percentages of CD69+4-1BB+ populations in CD4+ T-cell gates of PBMCs stimulated with the ORF3a25 Wu peptide for 12 days before being washed and re-stimulated with nothing (None) or the same peptide (ORF3a25 Wu) for 24 h.
(B) Reporter cells expressing the TCRs of clone-111 were stimulated with ORF3a25 Wu or Mut peptides in the presence of autologous PBMCs and analyzed for GFP and CD69 expression.
(C) The restricting human leukocyte antigen of clone-111 was DRB1∗04:06. Reporter cells expressing the TCRs of clone-111 were stimulated with the ORF3a25 Wu peptide in the presence of HEK293T cells transfected with nothing or DRA∗01:01-DRB1∗04:06-expressing plasmids and were analyzed for GFP and CD69 expression.
(D) mRNA expression of the characteristic genes of CD4+ T-cell subsets in clone-111 cells.
(E) Expression of Treg signature genes (FOXP3, CTLA4, DUSP4, IL2RB, STAM, DOK2, and TNFRSF1B) in the single cells in Figure 4A. The black box indicates the cluster containing clone-111. The sum of fractions of the Treg signature genes was calculated using BBrowser and is shown as a heatmap. Data are representative of three independent experiments (A–C). See also Tables S1 and S8.
Clone-111, which was detected as 28 different barcoded cells in a CD4+ cluster (Figure 4C), showed the gene signatures of highly activated cells (Figure 5D). These cells also expressed genes related to heterogeneous functions, such as IFN-γ, IL-21, PRF1, and IL-10 (Figure 5D). Notably, clone-111 was located in the cluster with a high expression of T regulatory (Treg) signature genes (Figure 5E),27 although most of the clone-111 cells did not highly express FOXP3. These observations suggest the helper function, cytotoxicity, and regulatory activity of this clonotype during infection.20,27,28,29 The finding that SARS-CoV-2 escaped the CD4+ clonotype with multiple potentials in the absence of humoral immunity further suggests a previously unclear role of CD4+ cells in the immune protection against COVID-19.
Discussion
We evaluated a patient with XLA in whom SARS-CoV-2 evolved uniquely for 239 days. During the whole infection period, 20 non-synonymous mutations were acquired by the virus, and many of these mutations were previously unreported (GISAID, https://www.gisaid.org). Among them, the ORF3a T32I mutation evaded recognition by CD4+ T cells with multi-functional characteristics, such as cytotoxic and regulatory function. Indeed, infection was controlled for 8 months when the original ORF3a epitope (ORF3a T32) was preserved, suggesting that T cells recognizing this epitope have a potent protective function even in the absence of humoral immunity. Consistently, when the mutation was acquired on day 239, the patient suffered severe deterioration with drastically increased levels of IL-6 and CRP and the sputum viral load. Thus, this variant is considered to acquire escape capacity from protective T cells, which results in viral expansion and the subsequent exacerbation of pneumonia in the patient.
The patient was given immunoglobulin every month because no normal mature B cells were present in his peripheral blood. The globulin products administered during the observation period were manufactured from blood collected before the emergence of COVID-19 (Table S5). Therefore, no SARS-CoV-2-specific antibodies were present in the serum of the patient (Figure S1). This case illustrates the pathophysiology of SARS-CoV-2 infection in the absence of specific humoral immunity.
Several precedents for SARS-CoV-2 evolution in immunocompromised patients with partially attenuated humoral responses have been described.4,12,24,30,31,32,33 SARS-CoV-2 evolved faster in these patients (Table 1), and its mutations showed a certain degree of similarity to those in several variants of concern, such as B.1.17, B.1.135, and P1, because these viruses carry deletions in the N-terminal domain and E484K or N501Y substitutions in the spike protein. The observed SARS-CoV-2 mutations in our patient, however, have rarely been reported, except for the K3353R mutation in ORF1ab, possibly due to a near-complete lack of humoral immune responses.
In the absence of humoral immunity, cellular immunity should exert protective effects against pathogens. The 28 CD4+ T cells from clone-111 had the same TCRs but showed the characteristics of different cell types, such as T follicular helper (Tfh), CD4 CTL, and Treg cells (Figures 5D and 5E).20,27,28,29 These results suggest that this CD4+ T-cell clonotype may exhibit functional plasticity and balance the T-cell responses in the protective immunity against SARS-CoV-2 infection. It has been reported that epitope affinity may affect the fate of T-cell clonotypes.34,35 However, in the case evaluated in our study, the development of heterogeneous T-cell types did not seem to be restricted by the epitope.
In our patient, CD4-mediated responses were likely to be dominant, because, in addition to ORF3a, four more mutations (ORF1ab F2979L in NSP4, ORF1ab L3829F in NSP6, K1149T in S, and T1155I in ORF7a) escaped from CD4+ T-cell recognition, whereas no mutations lost antigenicity to CD8+ T cells (Figure S2). Several mutant peptides, such as M1, ORF1ab2509, and ORF1ab3061, activated more CD4+ T cells than the corresponding Wu peptides did. These mutations may have occurred due to viral fitness, which warrants further investigation. The reason for the dominance of CD4-mediated responses is unclear at present. Nevertheless, it is known that MHC class II deficiency, but not MHC class I deficiency, incapacitates the immune system during viral infection,36which suggests an indispensable role of CD4+ T cells in antiviral immunity. Therefore, the identification of de novo mutations that avoid T-cell recognition in the absence of protective antibodies may help to predict future “T-escaping” variants when broad neutralization antibodies prevail in developing vaccines.37,38
The ORF3a protein is unique for the coronavirus subgenus Sarbecovirus, which includes SARS-CoV and SARS-CoV-2. ORF3a works as a viroporin, assisting the entry and release of the virus.39 Because ORF3a has multiple roles in the life cycle of SARS-CoV-2 and the induction of host immune responses, it is hard to predict the influence of the T32I mutation in ORF3a. Nevertheless, given that we observed a deteriorated infection after the occurrence of this mutation, we suspect that this mutation does not disrupt or even benefit ORF3a function. The replicative speed of this mutant virus isolated on day 239 was higher than that of the day-1 virus without the ORF3a mutation.
As shown in the results in Figure 2C, the day-1 virus formed small foci, whereas the day-239 virus formed large foci. Like the day-1 virus, the day-218 lung tissue virus formed small foci, but the day-218-BALF virus formed both small and large foci. The day-239 and day-218 BALF viruses shared ORF1ab I6548V and S G1251R mutations, both of which were absent in the other viruses. These mutations were detected in approximately half of the day-218-BALF virus population and nearly all of the day-239 virus population (Figure 2B). These results suggest that ORF1ab I6548V and/or S G1251R mutations may have been involved in the increased proliferative ability of the virus. This is because the S protein plays an important role in viral entry into cells,40 and ORF1ab 6548 is located at NSP15, which is an endoribonuclease important for viral replication.41
The administration of remdesivir dramatically improved the symptoms, inflammatory markers, and imaging test results of the patient, and the shedding of infectious viral particles stopped immediately. Remdesivir induced strong antiviral activity in the patient, which was consistent with results from a previous report. SARS-CoV-2 may therefore be eliminated by remdesivir alone without SARS-CoV-2-specific antibodies.42
In conclusion, SARS-CoV-2 evolved to evade recognition by CD4+ T cells and increased in its proliferative capacity during the persistent infection in our reported patient, which raises the possibility of the emergence of such variants in future even in immunocompetent patients, particularly when the escape from neutralizing antibodies is not sufficient for immune evasion. Thus, the control of chronic SARS-CoV-2 infection would also be beneficial for the prevention of virulent variants that evade such multi-functional T cells. In particular, immunocompromised patients should be treated with available antiviral drugs to prevent the emergence of public health-threatening mutants. We recently reported that non-spike epitopes recognized by CD8+ T cells contribute to the protection.43 Given the importance of multi-functional CD4+ T cells for protection in this study, the epitopes of such CD4+ T cells can be promising candidate antigens for the next-generation vaccines, because these epitopes are conserved in currently-reported VOCs. Extensive investigation of variants and T cell responses is therefore a crucial measure for the sustainable control of SARS-CoV-2 worldwide.
Limitations of the study
Despite the importance of our findings, there were several limitations in this study. These included the number of cases, because we could not investigate other patients with XLA who were also infected with SARS-CoV-2 as such cases are rare. Nevertheless, we were able to show the accelerated evolution of SARS-CoV-2 in the case evaluated herein as well as in other reported immunocompromised cases. In addition, the clinical specimens, especially the PBMCs that were used for the T-cell analysis, were mainly collected after day 210, and we failed to obtain evidence for the immune evasion of some of the mutations that were identified. We were, however, able to identify ade novomutation that evaded the recognition of dominant T cells in the patient with XLA.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti Coronavirus (SARS-CoV-2 and SARS-CoV NP) | EastCoast Bio | Cat#HM1054 |
| Rabbit polyclonal anti-mouse IgG (whole molecule) | MP Biomedicals | Cat#55436 |
| Goat polyclonal anti-rabbit IgG (whole molecule) | MP Biomedicals | Cat#55602 |
| Rabbit peroxidase-anti-peroxidase | Jackson Immuno Research | Cat#323-005-024; RRID: AB_2315781 |
| Peroxidase-conjugated AffiniPure Alpaca Anti-Human IgG (H+L) | Jackson Immuno Research | Cat#609-035-213; RRID: AB_2721849 |
| Anti-human CD3e FITC | BioLegend | Cat#300306; RRID: AB_314042 |
| Anti-human CD8a PE | BioLegend | Cat#301008; RRID: AB_314126 |
| Anti-human CD4 AF-700 | BioLegend | Cat#344621; RRID: AB_2563149 |
| Anti-human CD69 PE | BioLegend | Cat#310906; RRID: AB_314841 |
| Anti-human CD69 PECy7 | BioLegend | Cat#310911; RRID: AB_314846 |
| Anti-human CD137 APC | BioLegend | Cat#309810; RRID: AB_830672 |
| Anti-human Hashtags 1 | BioLegend | Cat#394661; RRID: AB_2801031 |
| Anti-human Hashtags 2 | BioLegend | Cat#394663; RRID: AB_2801032 |
| Anti-human Hashtags 3 | BioLegend | Cat#394665; RRID: AB_2801033 |
| Anti-human Hashtags 4 | BioLegend | Cat#394667; RRID: AB_2801034 |
| Anti-mouse CD69 APC | BioLegend | Cat#104514; RRID: AB_492843 |
| Bacterial and viral strains | ||
| hCoV–19/Japan/RIMD–DVI–C10B/2021 | This paper | GISAD: EPI_ISL_4935777 |
| hCoV–19/Japan/RIMD–DVI–C10L/2021 | This paper | GISAD: EPI_ISL_4935949 |
| hCoV–19/Japan/RIMD–DVI–C16/2021 | This paper | GISAD: EPI_ISL_4936095 |
| hCoV–19/Japan/RIMD–DVI–H06/2020 | This paper | GISAD: EPI_ISL_4936243 |
| hCoV–19/Japan/RIMD–DVI–C31/2021 | This paper | GISAD: EPI_ISL_4936533 |
| Biological samples | ||
| Patient nasopharyngeal swabs | This paper | Osaka City General Hospital |
| Patient bronchoalveolar lavage fluid | This paper | Osaka City General Hospital |
| Patient sputum | This paper | Osaka City General Hospital |
| Patient saliva | This paper | Osaka City General Hospital |
| Patient lung tissue obtained by biopsy | This paper | Osaka City General Hospital |
| Patient suction phlegm | This paper | Osaka City General Hospital |
| Patient blood | This paper | Osaka City General Hospital |
| Patient urine | This paper | Osaka City General Hospital |
| Human intravenous immunoglobulin (Pirivigen 10% I.V. Drip Infusion 10 g/100 mL) | CSL Behring | 23100AMX00288 |
| Healthy donor sera | This paper | Osaka City General Hospital |
| First WHO International Standard for anti-SARS-CoV-2 immunoglobulin (human) | NIBSC | Code: 20/136 |
| Chemicals, peptides, and recombinant proteins | ||
| 3,3′-Diaminobenzidine tetrahydrochloride | Sigma-Aldrich | Cat#D5905 |
| Hydrogen peroxide | FUJIFILM Wako Pure Chemical Corporation | Cat#084-07441 |
| SARS-CoV-2 Spike Protein (S1+S2 ECD, His tag) | Sino Biological | Cat#40589-V08B1 |
| TMB Substrate Kit | Thermo Scientific | Cat#34021 |
| Non-Essential Amino Acid Solution | Gibco-BRL | Cat#11140-050 |
| Sodium pyruvate | Gibco-BRL | Cat#06977-34 |
| Synthesized peptides | Genscript | Customized |
| PepMix™ SARS-CoV-2 (Spike Glycoprotein) | JPT | PM-WCPV-S-2 |
| PepMix™ SARS-CoV-2 (VME1) | JPT | PM-WCPV-VME |
| PepMix™ SARS-CoV-2 (NCAP) | JPT | PM-WCPV-NCAP |
| PepMix™ SARS-CoV-2 (Nsp1) | JPT | PM-WCPV-NSP01-1 |
| PepMix™ SARS-CoV-2 (Nsp2) | JPT | PM-WCPV-NSP02-1 |
| PepMix™ SARS-CoV-2 (Nsp3) | JPT | PM-WCPV-NSP03-1 |
| PepMix™ SARS-CoV-2 (Nsp4) | JPT | PM-WCPV-NSP04-1 |
| PepMix™ SARS-CoV-2 (Nsp5) | JPT | PM-WCPV-NSP5-1 |
| PepMix™ SARS-CoV-2 (Nsp6) | JPT | PM-WCPV-NSP06-1 |
| PepMix™ SARS-CoV-2 (Nsp7) | JPT | PM-WCPV-NSP07-1 |
| PepMix™ SARS-CoV-2 (Nsp8) | JPT | PM-WCPV-NSP08-1 |
| PepMix™ SARS-CoV-2 (Nsp9) | JPT | PM-WCPV-NSP09-1 |
| PepMix™ SARS-CoV-2 (Nsp10) | JPT | PM-WCPV-NSP10-1 |
| PepMix™ SARS-CoV-2 (C+Nsp11) | JPT | PM-WCPV-CLEAVE+Nsp11 |
| PepMix™ SARS-CoV-2 (Nsp12) | JPT | PM-WCPV-NSP12-1 |
| PepMix™ SARS-CoV-2 (Nsp13) | JPT | PM-WCPV-NSP13-1 |
| PepMix™ SARS-CoV-2 (Nsp15) | JPT | PM-WCPV-NSP15-1 |
| PepMix™ SARS-CoV-2 (Nsp16) | JPT | PM-WCPV-NSP16-1 |
| PepMix™ SARS-CoV-2 (AP3A) | JPT | PM-WCPV-AP3A |
| PepMix™ SARS-CoV-2 (VEMP) | JPT | PM-WCPV-VEMP |
| PepMix™ SARS-CoV-2 (NS6) | JPT | PM-WCPV-NS6 |
| PepMix™ SARS-CoV-2 (NS7a) | JPT | PM-WCPV-NS7A |
| PepMix™ SARS-CoV-2 (NS7B) | JPT | PM-WCPV-NS7B |
| PepMix™ SARS-CoV-2 (NS8) | JPT | PM-WCPV-NS8 |
| PepMix™ SARS-CoV-2 (ORF9B) | JPT | PM-WCPV-ORF9B |
| PepMix™ SARS-CoV-2 (ORF10) | JPT | PM-WCPV-ORF10 |
| PepMix™ SARS-CoV-2 (Y14) | JPT | PM-WCPV-Y14 |
| Recombinant human IL-2 | Peprotech | Cat#212-12 |
| Recombinant human IL-15 | Peprotech | Cat#200-15 |
| Recombinant human granulocyte–macrophage colony–stimulating factor (GM-CSF) | Peprotech | Cat#300-03 |
| Recombinant human IL-4 | Peprotech | Cat#200-04 |
| Recombinant human IL-7 | BioLegend | Cat#581902 |
| Critical commercial assays | ||
| QIAamp Viral RNA Mini kit | QIAGEN | Cat#52906 |
| One Step PrimeScript™ RT-PCR Kit | Takara | Cat# RR064A |
| GenNext RamDA-seq Single Cell Kit | TOYOBO | Cat#RMD-101 |
| Nextera XT DNA Library Prep Kit | Illumina | Cat#FC-131-1096 |
| Nextera XT Index Kit v2 | Illumina | Cat#FC-131-2001 |
| LEGENDplex Human CD8/NK Panel (13-plex) | BioLegend | Cat#740267 |
| Human interferon-γ/IL-4 Double-Color ELISPOT | ImmunoSpot | Cat#hIFNgIL4-1M/10 |
| Human CD14 MicroBeads | Miltenyi Biotec | Cat#130-050-201 |
| QIAzol | Qiagen | Cat#79306 |
| Deposited data | ||
| Single-cell TCR- and RNA-sequencing data | This paper | GEO datasets: GSE190895 |
| hCoV-19/Japan/RIMD-DVI-C26H/2021 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4929525 |
| hCoV-19/Japan/RIMD-DVI-C26W/2021 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4930839 |
| hCoV-19/Japan/RIMD-DVI-D09/2021 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4931056 |
| hCoV-19/Japan/RIMD-DVI-D21/2021 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4931846 |
| hCoV-19/Japan/OIPH14/2020 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4932136 |
| hCoV-19/Japan/OIPH16/2020 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4932550 |
| hCoV-19/Japan/OIPH21/2020 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4932712 |
| hCoV-19Japan/OIPH29/2020 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4932860 |
| hCoV-19/Japan/OIPH34/2020 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4933015 |
| hCoV-19/Japan/OIPH6/2020 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4933271 |
| hCoV-19/Japan/OIPH51/2020 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4933542 |
| hCoV-19/Japan/OIPH54/2020 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4933741 |
| hCoV-19Japan/OIPH61/2020 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4933859 |
| hCoV-19/Japan/OIPH1/2021 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4933990 |
| hCoV-19/Japan/OIPH25/2020 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4934247 |
| hCoV-19/Japan/OIPH28/2020 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4934751 |
| hCoV-19/Japan/OIPH5/2021 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4934873 |
| hCoV-19/Japan/OIPH6/2021 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4934996 |
| hCoV-19/Japan/OIPH10/2021 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4935185 |
| hCoV-19/Japan/OIPH12/2021 (SARS-CoV-2 sequence) | This paper | GSAID: EPI_ISL_4935429 |
| Experimental models: Cell lines | ||
| VeroE6/TMPRSS2 | JCRB cell bank | Cat#JCRB1819 |
| NFAT-GFP reporter mouse T cell hybridoma | Osaka University | Matsumoto et al.44 |
| Oligonucleotides | ||
| NIID_2019-nCOV_N_F2 AAATTTTGGGGACCAGGAAC |
This paper | https://www.niid.go.jp/niid/images/epi/corona/2019-nCoVmanual20200217-en.pdf |
| NIID_2019-nCOV_N_R2 TGGCAGCTGTGTAGGTCAAC∗ |
This paper | https://www.niid.go.jp/niid/images/epi/corona/2019-nCoVmanual20200217-en.pdf |
| NIID_2019-nCOV_N_P2 FAM-ATGTCGCGCATTGGCATGGA-BHQ |
This paper | https://www.niid.go.jp/niid/images/epi/corona/2019-nCoVmanual20200217-en.pdf |
| Recombinant DNA | ||
| TCR sequences of clone-111 | IDT | Customized |
| Software and algorithms | ||
| Cutadapt | Martin45 | https://cutadapt.readthedocs.io/en/stable/index.html |
| bwa | Li and Durbin46 | http://bio-bwa.sourceforge.net/ |
| Mutect2 (GATK 4.1.2.0) | Cibulskis et al.47 | https://gatk.broadinstitute.org/hc/en-us/articles/360037593851-Mutect2 |
| snpEff | Cingolani et al.48 | http://pcingola.github.io/SnpEff/ |
| Haplotype Caller (GATK 4.1.2.0) | Poplin et al.49 | https://gatk.broadinstitute.org/hc/en-us/articles/360037225632-HaplotypeCaller |
| bcftools ver1.9 | Li50 | https://samtools.github.io/bcftools/bcftools.html |
| W-IQ-TREE | Trifinopoulos et al.51 | http://iqtree.cibiv.univie.ac.at |
| ModelFinder | Kalyaanamoorthy et al.52 | http://iqtree.org/ |
| Ultrafast bootstrap | Hoang et al.53 | http://iqtree.org/ |
| Other | ||
| 2019-nCoV one-step RT-qPCR Positive control RNA for N2 set | Nihon Gene Research Laboratories, Inc. | Cat#JP-NN2-PC |
| Human AB serum (off the clot) | Gemini Bio | Cat#100-318 |
Resource availability
Lead contact
Requests for further information, resources, and reagents should be directed to and will be fulfilled by the Lead Contact, Tatsuo Shioda (shioda@biken.osaka-u.ac.jp).
Materials availability
This study did not generate new reagents.
Experimental model and subject details
Ethical considerations
Written informed consent for the present study was obtained from the patient. The protocols of the present study were approved by the Local Incorporated Administrative Agency Osaka City Hospital Organization and Osaka City General Hospital Research Ethics Review Board (approval numbers: 2105017 and 2107048) and the institutional review board of the Research Institute for Microbial Diseases, Osaka University (approval number: 2021–4). The authors vouch for the accuracy and completeness of the data in this report.
COVID-19 patient
The patient, a 25-year-old man, experienced repeated bacterial infections from approximately 8 months of age and was genetically diagnosed with X-linked agammaglobulinemia (XLA) due to an exon 6 skip mutation at 11 months. He has since been given immunoglobulin every month (Table S5) but no prophylactic antibiotics. He developed fever at the end of July 2020 and was diagnosed with COVID-19 based on a positive result from a quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) test for SARS-CoV-2 in early August.
Cell lines
VeroE6/TMPRSS2 cells were obtained from the JCRB cell bank (cat. JCRB1819). The cells were maintained at 37 °C and with 5% CO2 in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum, 1 mM L-glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin. Mouse T-cell hybridoma with a nuclear factor of activated T cells (NFAT)-green fluorescent protein (GFP) reporter gene44 were maintained at 37 °C and with 5% CO2 in RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin, streptomycin, and 2-mercaptoethanol.
Method details
Sample preparation for PCR testing and viral infectivity studies
Fresh samples (obtained within 24 h of sampling) and those that had been cryopreserved at 80 °C were obtained from a nasopharyngeal swab (NPS), bronchoalveolar lavage fluid, sputum and saliva, lung tissue obtained by a biopsy, and suction phlegm and used for PCR tests and viral infectivity studies. Before extracting the RNA, the sputum and saliva samples were diluted two- or four-fold using phosphate-buffered saline (PBS). A lung tissue sample was homogenized by adding a four-fold volume of PBS.
PCR test
A qRT-PCR was performed as previously described,54 with minor modifications. In brief, the samples were centrifuged at 10,000 rpm for 30 s to remove insoluble material. Viral RNA was extracted from 140 μL of the supernatant using the QIAamp Viral RNA Mini Kit (QIAGEN) according to the manufacturer’s instructions. Assays for the qRT-PCR were performed using the OneStep PrimeScript™ RT-PCR Kit (Takara Bio) with the N2 set of primers and a probe designed according to the methods published by the National Institute of Infectious Diseases, Japan. The conditions for the qRT-PCR were as follows: 52 °C for 5 min and 95 °C for 10 s, followed by 40 cycles at 95 °C for 5 s and 60 °C for 30 s using the QuantStudio™ 3 Real-Time PCR System (Life Technologies).
In-house ELISA of the anti-SARS-CoV-2 spike (S) protein antibody
A 96-well flat-bottom microplate was coated with 0.1 μg/well of the S protein (Sino Biological) in 50 μL/well of carbonate-bicarbonate buffer (Sigma-Aldrich) and incubated at 4 °C overnight. The plate was washed with 0.05% Tween 20 in PBS (PBS-T) and blocked with 300 μL/well of 25% BlockAce for 2hat 37 °C. After washing the plate with PBS-T, 50 μL/well of diluted patient plasma (1:100), control sera, and diluted 10% intravenous immunoglobulin (1:1,000) were added and incubated for 1hat 37 °C. After washing the sample three times with PBS-T, 50 μL/well of diluted peroxidase-conjugated alpaca anti-human IgG (H+L; 1:50,000) (Jackson ImmunoResearch) was added and incubated for 1hat 37 °C. After washing the sample three times, 50 μL/well of 3,3',5,5'-tetramethylbenzidine solution (TMB substrate kit; Thermo Scientific) was added and incubated for 5minat 25 ± 2 °C. The peroxidase reaction was stopped with 50 μL/well of 1 M sulfuric acid. The optical density at 450 nm was measured using a Multigrading Microplate Reader (SH-9500Lab; Corona). Antibody titers were calculated in units of binding antibody unit (BAU)/mL with calibrators assigned to the first WHO international standard for anti-SARS-CoV-2 immunoglobulin from the National Institute for Biological Standards and Control (code 20/136) as 1,000 BAU/mL.55,56
Focus-forming assay
The level of viral infectivity in the clinical specimens was measured using a focus-forming assay as previously described.54 In brief, VeroE6/TMPRSS2 cells were seeded to 24-well plates (2 x105 cells/well) 1 day before the experiment. The medium was removed, and the cell monolayers in each well were inoculated with 100 μL of samples that had been serially diluted 10-fold. After 30 min, the samples were removed and washed three times with serum-free Minimum Essential Medium (MEM). The cells were covered with 0.5 mL of DMEM containing 1% carboxymethyl cellulose, 2% fetal bovine serum, and antibiotics (200 units/mL penicillin, 200 μg/mL streptomycin, and 0.25 μg/mL amphotericin B). Eighteen hours after being inoculated, the cells were fixed with 10% formalin for 30minat room temperature. They were then washed three times with PBS and fixed in absolute ethanol for 5minat room temperature. To visualize the virus-infected cells, the peroxidase-anti-peroxidase (PAP) technique was applied. Each well was treated for 40minat room temperature with 400 μL of each antibody in the following order: anti-SARS/SARS-2 NP mouse monoclonal antibody (EastCoast Bio) diluted to 1 μg/mL; anti-mouse rabbit immunoglobulin (Ig) G antibody (MP Biomedicals, Irvine, CA) diluted to 1:1,000; anti-rabbit goat IgG antibody (MP Biomedicals) diluted to 1:500; and rabbit PAP (Jackson Immuno Research) diluted to 1:200. After each reaction, the cells were washed three times with PBS. In the final step, the peroxidase reaction was developed for approximately 5 min using 0.01% H2O2 and 0.3 mg/mL of 3-3'-diaminobenzidine tetrahydrochloride in PBS. The cells were then washed with tap water and dried, and the stained foci were macroscopically counted. Viral infectivity was expressed as focus-forming units (FFUs). Focus sizes were measured using an ELISPOT analyzer (ImmunoSpot S6 VERSA Analyzer; M&S TechnoSystems, Inc., Japan).
Viral isolation
Viral isolation from the clinical specimens was performed as previously described.54 In brief, VeroE6/TMPRSS2 cells were seeded to 24-well plates (2 × 105 cells/well) 1 day before the experiment. The medium was removed, and 100 μL of the samples were added to each well. After 30 min, the samples were removed, and the cells were washed three times with serum-free MEM. The cells were then cultured in 1 mL of DMEM containing 2% fetal bovine serum, 200 units/mL of penicillin, 200 μg/mL of streptomycin, and 0.25 μg/mL of amphotericin B in an atmosphere with 5% CO2 and at 37 °C for up to 5 days. Cytopathic effects (CPEs) were macroscopically observed each day. When CPEs appeared, the culture supernatant was collected (passage #0). The isolated viruses were passaged once (passage #1) and titrated using focus-forming assays.
Viral sequencing and phylogenetic analysis
For viral sequencing, RNA was extracted from the culture supernatant of the isolated viruses (passage #0) using the Qiagen Viral RNA Mini kit (QIAGEN). cDNA synthesis was performed using the GenNext RamDA-seq Single Cell Kit (TOYOBO). An Illumina library was constructed using the Nextera XT DNA Library Prep Kit (Illumina) and Nextera XT Index Kit v2 (Illumina). The resulting DNA libraries were sequenced using the NovaSeq6000 instrument (Illumina). The obtained reads were analyzed using Cutadapt to trim adapters; bwa to map the reads to the reference genome (NC_045512.2); Mutect2 to call mutations; snpEff to make annotations; and HaplotypeCaller and bcftools to make consensus sequences.
A phylogenetic tree was inferred from the alignment using the maximum likelihood approach in W-IQ-TREE.51 The best-fit model was selected by ModelFinder,52 and the ultrafast bootstrap53 with 1,000 replicates was calculated. The viruses used for the phylogenetic analyses have been listed in Table S3. We used BLAST to obtain similar sequences to the virus from the patient using the NCBI database. We picked 17 sequences based on the highest identity score. As references, we added 8 sequences from various lineages. We then added 4 sequences from the same hospital and 16 sequences from the same prefecture of Japan, all of which were isolated and analyzed in-house during the period in which the patient was infected and recovered after 240 days of being infected.
In vitroT-cell expansion and stimulation
Cryopreserved peripheral blood mononuclear cells (PBMCs) were thawed and washed with RPMI 1640 medium (Sigma-Aldrich) supplemented with 5% human AB serum (Gemini Bio), penicillin (Sigma-Aldrich), streptomycin (MP Biomedicals), and 2-mercaptoethanol (Nacalai Tesque). The cells were expanded with 4 μg/mL of single peptides derived from the SARS-CoV-2 Wuhan-Hu-1 strain in the same medium for 12 to 15 days. We used 15-mer peptides since they are widely used for activating both CD4+ T cells and CD8+ T cells,57 presumably because the exo-degradation of peptides during culture creates the appropriate length of peptides for MHC class I binding. Recombinant human IL-2 (10 IU/mL; Peprotech), human IL-7 (5 ng/mL; BioLegend), and human IL-15 (5 ng/mL; Peprotech) were supplied every 3 days from day 2 onwards. For the re-stimulation assay, the PBMCs that had been expanded in vitro were harvested at the indicated time points and washed before being re-stimulated with 4 μg/mL of the Wuhan or mutant peptides for the indicated durations. The resultant cells were used for subsequent assays. Cytokines were analyzed using the LEGENDplex Human CD8/NK Panel (13-plex) (BioLegend). An ELISPOT assay was performed using a human interferon-γ/IL-4 Double-Color ELISPOT (ImmunoSpot). The PBMCs were stimulated with peptide pools derived from proteins of the SARS-CoV-2 Wuhan-Hu-1 strain (1 μg/mL per peptide; JPT) for 48 h. The expression of the cytokines was determined based on the manufacturer’s protocol. The single peptides used for the stimulation of the T cells have been listed in Table S6.
Reporter cell establishment and stimulation
TCR-α and-β chain cDNA sequences were introduced into a mouse T-cell hybridoma with an NFAT-GFP reporter gene44 using retroviral vectors. TCR-reconstituted cells were co-cultured with 1 μg/mL of single peptides in the presence of PBMCs from the patient as antigen-presenting cells. After 20 h, cell activation was assessed based on the expression of GFP and CD69.
Single-cell-based transcriptome and TCR repertoire analyses
Single-cell libraries were prepared using reagents from 10× Genomics following the manufacturer’s instructions. RNA from the cells was reverse-transcribed; cDNA was amplified for 14 cycles; and up to 50 ng of cDNA were used for the gene expression and TCR libraries. Clustering was performed based on the top-4,000 highly variable genes, and the cell types were annotated as previously described.58
The single-cell RNA-seq fastq files were processed using 10× Genomics Cell Ranger version 6.059 with default settings to yield the gene expression profiles of the hashtag-attached cell barcodes. The transcriptomes of the 13,777 and 13,078 barcodes from two different libraries were merged and subjected to downstream analyses using Scanpy 1.8.1.60 Information on the T-cell clonotypes was integrated with the gene expression profiles of the single-cell RNA-seq data using Scirpy.61 The read counts of the hashtags of the cell barcodes were extracted and scaled from 0 to 10. The samples from which the cell barcodes originated were then estimated based on the hashtag read counts of each barcode. To avoid the potential bias of clustered results, ribosomal genes, TRAV, TRAJ, TRBJ, TRBV, IGHV, IGKV, and IGLV were excluded from the downstream analyses. Scrublet62 was used to predict and filter potential doublets. The distributions of the read counts, gene counts per barcode, ribosomal gene concentration, mitochondrial gene concentration, and hemoglobin gene concentration within the samples were each fitted with a mixture of two Gaussian distributions.63 Filtration thresholds for the quality control of the cell barcodes were then determined based on the distance of the fitted mean values of the Gaussian distributions. The resulting 25,961 cells, which consisted of 9,708 cells from the M121 sample, 7,669 cells from the S1145 sample, 7,082 cells from the 1ab2973 sample, and 1,502 cells from the 3a25 sample, were used in the final analysis (see below for each sample name). The top-4,000 highly variable genes were selected using Cellranger flavor integrated with Scanpy.59 Batch effect correction was conducted using BBKNN.64 Leiden clustering65 and PAGA graphs66 with Reingold–Tilford layouts67 were integrated with UMAP.68 The cell typing of the T cells was conducted by mapping the transcriptomic data onto the reference PBMC dataset published by Hao et al.,58which was downloaded from the UCSC Cell Browser Homepage.69
Bulk TCR sequencing and analysis
A total of 3 x 105 fresh PBMCs were lysed in 200 μL of QIAzol (QIAGEN). Full-length cDNA was synthesized using SMARTer (Takara Bio) before variable regions of the TCR-α and-β genes were amplified. Each pair of reads was assigned a clonotype (defined as TR(A/B)V and TR(A/B)J genes and the complementarity-determining region 3) using MiXCR software.70
Monocyte-derived dendritic cells
CD14+ cells were isolated from PBMCs using human CD14 MicroBeads (Miltenyi Biotec) and cultured in RPMI 1640 medium supplemented with 5% human AB serum, 0.1 mM Non-Essential Amino Acid Solution (Gibco-BRL), 1 mM sodium pyruvate (Gibco-BRL), 10 ng/mL of human granulocyte–macrophage colony-stimulating factor (GM-CSF) (Peprotech), and 10 ng/mL of IL-4 (Peprotech) for 13 days.
Quantification and statistical analysis
The cytokines were measured in duplicate. The data are shown as mean ± standard deviation.
Acknowledgments
We are grateful to Dr. Ko Iida and Dr. Hidenori Nakagawa for their helpful discussions and comments. We are grateful to Dr. Charles Schutt for helpful discussion and editing this manuscript. We are grateful to Dr. Naoki Takahashi for providing technical assistance. We are profoundly grateful to the patient and his family for their kindness and generosity. This work was supported by funding from the Japan Agency for Medical Research and Development (JP20nf0101623 and JP20nk0101602). We gratefully acknowledge the contributions of the nurses and all of those who participated in caring for the patient. We also thank Editage (www.editage.jp) for editing this manuscript.
Author contributions
R.M., Y.I., K.A., T.G., J.H., and M.S. provided clinical care for the patient; R.M. collected clinical data and performed data analyses; Y.F. carried out and supervised the laboratory tests; R.K.K. performed viral infectivity studies and data analyses; X.L. performed T-cell analyses; Tadahiro S. and E.E.N. performed viral genetic analyses; Y.L., D.O., J.B.W., and D.M. performed single-cell analyses; S.Y., M.S., and Tatsuo S. supervised and designed the experiments; R.M., R.K.K., and X.L. wrote the original manuscript, with input from all the other authors. All authors contributed to the analyses of the present results.
Declaration of interests
M.S. received honoraria for lectures from Gilead Sciences Inc. All the other authors declare no competing interests.
Published: April 20, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106685.
Contributor Information
Michinori Shirano, Email: shirano@wonder.ocn.ne.jp.
Sho Yamasaki, Email: yamasaki@biken.osaka-u.ac.jp.
Tatsuo Shioda, Email: shioda@biken.osaka-u.ac.jp.
Supplemental information
Data and code availability
-
•
Single-cell TCR- and RNA-sequencing data have been deposited in Gene Expression Omnibus (GEO) datasets (accession number: GSE190895). The newly obtained viral sequence data may be downloaded from GISAID (https://www.gisaid.org) as accession numbers listed in key resources table. Patient-related data not included in the paper were generated as part of the clinical examinations and may be subjected to patient confidentiality.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Winkelstein J.A., Marino M.C., Lederman H.M., Jones S.M., Sullivan K., Burks A.W., Conley M.E., Cunningham-Rundles C., Ochs H.D. X-linked agammaglobulinemia: report on a United States registry of 201 patients. Medicine. 2006;85:193–202. doi: 10.1097/01.md.0000229482.27398.ad. [DOI] [PubMed] [Google Scholar]
- 2.Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., Barbian K., Judson S.D., Fischer E.R., Martens C., et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with Cancer. Cell. 2020;183:1901–1912.e9. doi: 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., et al. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.H., Boucau J., Bowman K., et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin H., Reed J.C., Liu S.T.H., Ho H.E., Lopes J.P., Ramsey N.B., Waqar O., Rahman F., Aberg J.A., Bouvier N.M., et al. Three patients with X-linked agammaglobulinemia hospitalized for COVID-19 improved with convalescent plasma. J. Allergy Clin. Immunol. Pract. 2020;8:3594–3596.e3. doi: 10.1016/j.jaip.2020.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.London J., Boutboul D., Lacombe K., Pirenne F., Heym B., Zeller V., Baudet A., Ouedrani A., Bérezné A. Severe COVID-19 in patients with B cell alymphocytosis and response to convalescent plasma therapy. J. Clin. Immunol. 2021;41:356–361. doi: 10.1007/s10875-020-00904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mira E., Yarce O.A., Ortega C., Fernández S., Pascual N.M., Gómez C., Alvarez M.A., Molina I.J., Lama R., Santamaria M. Rapid recovery of a SARS-CoV-2-infected X-linked agammaglobulinemia patient after infusion of COVID-19 convalescent plasma. J. Allergy Clin. Immunol. Pract. 2020;8:2793–2795. doi: 10.1016/j.jaip.2020.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hovey J.G., Tolbert D., Howell D. Burton's agammaglobulinemia and COVID-19. Cureus. 2020;12:e11701. doi: 10.7759/cureus.11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iaboni A., Wong N., Betschel S.D. A patient with X-linked agammaglobulinemia and COVID-19 infection treated with remdesivir and convalescent plasma. J. Clin. Immunol. 2021;41:923–925. doi: 10.1007/s10875-021-00983-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buckland M.S., Galloway J.B., Fhogartaigh C.N., Meredith L., Provine N.M., Bloor S., Ogbe A., Zelek W.M., Smielewska A., Yakovleva A., et al. Treatment of COVID-19 with remdesivir in the absence of humoral immunity: a case report. Nat. Commun. 2020;11:6385. doi: 10.1038/s41467-020-19761-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palomba E., Carrabba M., Zuglian G., Alagna L., Saltini P., Fortina V., Hu C., Bandera A., Fabio G., Gori A., Muscatello A. Treatment of SARS-CoV-2 relapse with remdesivir and neutralizing antibodies cocktail in a patient with X-linked agammaglobulinaemia. Int. J. Infect. Dis. 2021;110:338–340. doi: 10.1016/j.ijid.2021.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kemp S.A., Collier D.A., Datir R.P., Ferreira I.A.T.M., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U., et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson M.K., Hamilton F., Hutchings S., Pymont H.M., Hackett M., Arnold D., Maskell N.A., MacGowan A., Albur M., Jenkins M., et al. Chronic SARS-CoV-2 infection and viral evolution in a hypogammaglobulinaemic individual. medRxiv. 2021 doi: 10.1101/2021.05.31.21257591. Preprint at. [DOI] [Google Scholar]
- 14.Kanegane H., Tsukada S., Iwata T., Futatani T., Nomura K., Yamamoto J., Yoshida T., Agematsu K., Komiyama A., Miyawaki T. Detection of Bruton's tyrosine kinase mutations in hypogammaglobulinaemic males registered as common variable immunodeficiency (CVID) in the Japanese Immunodeficiency Registry. Clin. Exp. Immunol. 2000;120:512–517. doi: 10.1046/j.1365-2249.2000.01244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes L., Gangavarapu K., Latif A.A., Mullen J., Alkuzweny M., Hufbauer E., Tsueng G., Haag E., Zeller M., Aceves C., et al. Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. Res. Sq. 2022 doi: 10.21203/rs.3.rs-1723829/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes L., Wu C., Su A., Andersen K. Outbreak.info. https://outbreak.info/
- 17.Tsueng G., Mullen J.L., Alkuzweny M., Cano M., Rush B., Haag E., Zhou X., Lin J., Welzel D.J., Qian Z., et al. Outbreak.info research library: a standardized, searchable platform to discover and explore COVID-19 resources. bioRxiv. 2022 doi: 10.1101/2022.01.20.477133. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singer J., Gifford R., Cotten M., Robertson D. CoV-GLUE: a web application for tracking SARS-CoV-2 genomic variation. Preprints. 2020 doi: 10.20944/preprints202006.0225.v1. Preprint at. [DOI] [Google Scholar]
- 19.Motayo B.O., Oluwasemowo O.O., Olusola B.A., Akinduti P.A., Arege O.T., Obafemi Y.D., Faneye A.O., Isibor P.O., Aworunse O.S., Oranusi S.U. Evolution and genetic diversity of SARS-CoV-2 in Africa using whole genome sequences. Int. J. Infect. Dis. 2021;103:282–287. doi: 10.1016/j.ijid.2020.11.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., Chen Z., Wang T., Guo H., Liu Y., Dang N., Hu S., Wu L., Zhang C., Ye K., Shi B. A novel CD4+ CTL subtype characterized by chemotaxis and inflammation is involved in the pathogenesis of Graves' orbitopathy. Cell. Mol. Immunol. 2021;18:735–745. doi: 10.1038/s41423-020-00615-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill V., Du Plessis L., Peacock T.P., Aggarwal D., Colquhoun R., Carabelli A.M., Ellaby N., Gallagher E., Groves N., Jackson B., et al. The origins and molecular evolution of SARS-CoV-2 lineage B.1.1.7 in the UK. bioRxiv. 2022 doi: 10.1101/2022.03.08.481609. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen S., Zhang Z., He F. The phylogenetic relationship within SARS-CoV-2s: an expanding basal clade. Mol. Phylogenet. Evol. 2021;157:107017. doi: 10.1016/j.ympev.2020.107017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaguza C., Hahn A.M., Petrone M.E., Zhou S., Ferguson D., Breban M.I., Pham K., Peña-Hernández M.A., Castaldi C., Hill V., et al. Accelerated SARS-CoV-2 intrahost evolution leading to distinct genotypes during chronic infection. medRxiv. 2022 doi: 10.1101/2022.06.29.22276868. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borges V., Isidro J., Cunha M., Cochicho D., Martins L., Banha L., Figueiredo M., Rebelo L., Trindade M.C., Duarte S., et al. Long-term evolution of SARS-CoV-2 in an immunocompromised patient with non-hodgkin lymphoma. mSphere. 2021;6:e0024421. doi: 10.1128/mSphere.00244-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shioda T., Wakao S., Suzu S., Shibuta H. Differences in bovine parainfluenza 3 virus variants studied by sequencing of the genes of viral envelope proteins. Virology. 1988;162:388–396. doi: 10.1016/0042-6822(88)90479-5. [DOI] [PubMed] [Google Scholar]
- 26.McKimm-Breschkin J.L., Williams J., Barrett S., Jachno K., McDonald M., Mohr P.G., Saito T., Tashiro M. Reduced susceptibility to all neuraminidase inhibitors of influenza H1N1 viruses with haemagglutinin mutations and mutations in non-conserved residues of the neuraminidase. J. Antimicrob. Chemother. 2013;68:2210–2221. doi: 10.1093/jac/dkt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmiedel B.J., Singh D., Madrigal A., Valdovino-Gonzalez A.G., White B.M., Zapardiel-Gonzalo J., Ha B., Altay G., Greenbaum J.A., McVicker G., et al. Impact of genetic polymorphisms on human immune cell gene expression. Cell. 2018;175:1701–1715.e16. doi: 10.1016/j.cell.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meckiff B.J., Ramírez-Suástegui C., Fajardo V., Chee S.J., Kusnadi A., Simon H., Eschweiler S., Grifoni A., Pelosi E., Weiskopf D., et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4(+) T cells in COVID-19. Cell. 2020;183:1340–1353.e16. doi: 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Appay V., Zaunders J.J., Papagno L., Sutton J., Jaramillo A., Waters A., Easterbrook P., Grey P., Smith D., McMichael A.J., et al. Characterization of CD4(+) CTLs ex vivo. J. Immunol. 2002;168:5954–5958. doi: 10.4049/jimmunol.168.11.5954. [DOI] [PubMed] [Google Scholar]
- 30.Hensley M.K., Bain W.G., Jacobs J., Nambulli S., Parikh U., Cillo A., Staines B., Heaps A., Sobolewski M.D., Rennick L.J., et al. Intractable coronavirus disease 2019 (COVID-19) and prolonged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replication in a Chimeric antigen receptor-modified T-cell therapy recipient: a case study. Clin. Infect. Dis. 2021;73:e815–e821. doi: 10.1093/cid/ciab072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Duprex W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371:1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark S.A., Clark L.E., Pan J., Coscia A., McKay L.G.A., Shankar S., Johnson R.I., Brusic V., Choudhary M.C., Regan J., et al. SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell. 2021;184:2605–2617.e18. doi: 10.1016/j.cell.2021.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen B., Luebke N., Feldt T., Keitel V., Brandenburger T., Kindgen-Milles D., Lutterbeck M., Freise N.F., Schoeler D., Haas R., et al. Emergence of the E484K mutation in SARS-COV-2-infected immunocompromised patients treated with bamlanivimab in Germany. Lancet Reg. Health. Eur. 2021;8:100164. doi: 10.1016/j.lanepe.2021.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotov D.I., Mitchell J.S., Pengo T., Ruedl C., Way S.S., Langlois R.A., Fife B.T., Jenkins M.K. TCR Affinity biases Th cell differentiation by regulating CD25, Eef1e1, and Gbp2. J. Immunol. 2019;202:2535–2545. doi: 10.4049/jimmunol.1801609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khatun A., Kasmani M.Y., Zander R., Schauder D.M., Snook J.P., Shen J., Wu X., Burns R., Chen Y.G., Lin C.W., et al. Single-cell lineage mapping of a diverse virus-specific naive CD4 T cell repertoire. J. Exp. Med. 2021;218:e20200650. doi: 10.1084/jem.20200650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geha R., Notarangelo L. Garland Science; 2016. Case Studies in Immunology: A Clinical Companion. [Google Scholar]
- 37.Ota M.O., Ndhlovu Z., Oh S., Piyasirisilp S., Berzofsky J.A., Moss W.J., Griffin D.E. Hemagglutinin protein is a primary target of the measles virus-specific HLA-A2-restricted CD8+ T cell response during measles and after vaccination. J. Infect. Dis. 2007;195:1799–1807. doi: 10.1086/518006. [DOI] [PubMed] [Google Scholar]
- 38.Kaaijk P., Emmelot M.E., Meiring H.D., van Els C.A.C.M., de Wit J. Novel mumps virus epitopes reveal robust cytotoxic T cell responses after natural infection but not after vaccination. Sci. Rep. 2021;11:13664. doi: 10.1038/s41598-021-92926-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J., Ejikemeuwa A., Gerzanich V., Nasr M., Tang Q., Simard J.M., Zhao R.Y. Understanding the role of SARS-CoV-2 ORF3a in viral pathogenesis and COVID-19. Front. Microbiol. 2022;13:854567. doi: 10.3389/fmicb.2022.854567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gottlieb R.L., Vaca C.E., Paredes R., Mera J., Webb B.J., Perez G., Oguchi G., Ryan P., Nielsen B.U., Brown M., et al. Early remdesivir to prevent progression to severe Covid-19 in outpatients. N. Engl. J. Med. 2022;386:305–315. doi: 10.1056/NEJMoa2116846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogura H., Gohda J., Lu X., Yamamoto M., Takesue Y., Son A., Doi S., Matsushita K., Isobe F., Fukuda Y., et al. Dysfunctional Sars-CoV-2-M protein-specific cytotoxic T lymphocytes in patients reco- vering from severe COVID-19. Nat. Commun. 2022;13:7063. doi: 10.1038/s41467-022-34655-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsumoto Y., Kishida K., Matsumoto M., Matsuoka S., Kohyama M., Suenaga T., Arase H. A TCR-like antibody against a proinsulin-containing fusion peptide ameliorates type 1 diabetes in NOD mice. Biochem. Biophys. Res. Commun. 2021;534:680–686. doi: 10.1016/j.bbrc.2020.11.019. [DOI] [PubMed] [Google Scholar]
- 45.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011;17 https://journal.embnet.org/index.php/embnetjournal/article/view/200 [Google Scholar]
- 46.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, Kling DE, Gauthier LD, Levy-Moonshine A, Roazen D, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv. 2017:201178. doi: 10.1101/201178. Preprint at. [DOI] [Google Scholar]
- 50.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hiroi S., Kubota-Koketsu R., Sasaki T., Morikawa S., Motomura K., Nakayama E.E., Okuno Y., Shioda T. Infectivity assay for detection of SARS-CoV-2 in samples from patients with COVID-19. J. Med. Virol. 2021;93:5917–5923. doi: 10.1002/jmv.27145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kristiansen P.A., Page M., Bernasconi V., Mattiuzzo G., Dull P., Makar K., Plotkin S., Knezevic I. WHO International Standard for anti-SARS-CoV-2 immunoglobulin. Lancet. 2021;397:1347–1348. doi: 10.1016/S0140-6736(21)00527-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mattiuzzo G., Bentley E., Hassall M., Routley S., Richardson S., Bernasconi V., Kristiansen P. Expert Committee On Biological Standardization; 2020. WHO/BS.2020.2403 Establishment of the WHO International Standard and Reference Panel for Anti-SARS-CoV-2 Antibody. [Google Scholar]
- 57.Kiecker F., Streitz M., Ay B., Cherepnev G., Volk H.D., Volkmer-Engert R., Kern F. Analysis of antigen-specific T-cell responses with synthetic peptides–what kind of peptide for which purpose? Hum. Immunol. 2004;65:523–536. doi: 10.1016/j.humimm.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 58.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sturm G., Szabo T., Fotakis G., Haider M., Rieder D., Trajanoski Z., Finotello F. Scirpy: a Scanpy extension for analyzing single-cell T-cell receptor-sequencing data. Bioinformatics. 2020;36:4817–4818. doi: 10.1093/bioinformatics/btaa611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolock S.L., Lopez R., Klein A.M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8:281–291.e9. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pedregosa F., Varoquaux G., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J., et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 64.Polański K., Young M.D., Miao Z., Meyer K.B., Teichmann S.A., Park J.E. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics. 2020;36:964–965. doi: 10.1093/bioinformatics/btz625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Traag V.A., Waltman L., van Eck N.J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 2019;9:5233. doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolf F.A., Hamey F.K., Plass M., Solana J., Dahlin J.S., Göttgens B., Rajewsky N., Simon L., Theis F.J. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019;20:59. doi: 10.1186/s13059-019-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buchheim C., Jünger M., Leipert S. Drawing rooted trees in linear time. Softw:. Pract. Exper. 2006;36:651–665. [Google Scholar]
- 68.Mclnnes L., Healy J., Saul N., Großberger L. UMAP: uniform manifold approximation and projection. J. Open Source Softw. 2018;3:861–862. [Google Scholar]
- 69.Speir M.L., Bhaduri A., Markov N.S., Moreno P., Nowakowski T.J., Papatheodorou I., Pollen A.A., Raney B.J., Seninge L., Kent W.J., Haeussler M. UCSC cell browser: visualize your single-cell data. Bioinformatics. 2021;37:4578–4580. doi: 10.1093/bioinformatics/btab503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bolotin D.A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I.Z., Putintseva E.V., Chudakov D.M. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Single-cell TCR- and RNA-sequencing data have been deposited in Gene Expression Omnibus (GEO) datasets (accession number: GSE190895). The newly obtained viral sequence data may be downloaded from GISAID (https://www.gisaid.org) as accession numbers listed in key resources table. Patient-related data not included in the paper were generated as part of the clinical examinations and may be subjected to patient confidentiality.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.