SUMMARY
EGFR-RAS-ERK signaling promotes growth and proliferation in many cell types, and genetic hyperactivation of RAS-ERK signaling drives many cancers. Yet despite intensive study of upstream components in EGFR signal transduction, the identities and functions of downstream effectors in the pathway are poorly understood. In Drosophila intestinal stem cells (ISC) the transcriptional repressor Capicua (cic) and its targets, the ETS-type transcriptional activators Pointed (pnt) and Ets21C, are essential downstream effectors of mitogenic EGFR signaling. Here we show that these factors promote EGFR-dependent metabolic changes that increase ISC mass, mitochondrial growth, and mitochondrial activity. Gene target analysis using RNA- and DamID-sequencing revealed that Pnt and Ets21C directly up-regulate not only DNA replication and cell cycle genes, but genes for oxidative phosphorylation, the TCA cycle, and fatty acid beta-oxidation. Metabolite analysis substantiated these metabolic functions. The mitochondrial transcription factor B2 (mtTFB2), a direct target of Pnt, was required and partially sufficient for EGFR-driven ISC growth, mitochondrial biogenesis and proliferation. MEK-dependent EGF signaling stimulated mitochondrial biogenesis in human RPE-1 cells, indicating conservation of these metabolic effects. This work illustrates how EGFR signaling alters metabolism to coordinately activate cell growth and cell division.
Keywords: Intestinal stem cell (ISC), proliferation, Pointed, Ets21C, mtTFB2, mitochondrial biogenesis
In Brief
Zhang et al. show that through its downstream transcriptional effectors Cic, Pnt, and Est21C, EGFR signaling upregulates mtTFB2, and β-oxidation, OXPHOS, and TCA cycle genes. These transcriptomic effects drive mitochondrial biogenesis and increase mitochondria activity to facilitate intestinal stem cell growth and proliferation.
INTRODUCTION
The EGFR signaling pathway can promote cell proliferation, differentiation, survival, and/or migration in many animal cell types. Although the receptor-proximal elements of this highly conserved signaling pathway are well characterized, the downstream effectors that mediate EGFR’s effects on cell growth and proliferation are less well understood. An extensive literature confirms that the downstream kinases, RAF (a MAP3K), MEK (a MAP2K) and ERK (a MAPK) are essential signaling transducers, but ERK is a broad-spectrum serine-threonine kinase with many nuclear and cytoplasmic targets. Understanding what these targets are and how they direct growth-associated cellular functions can advance our understanding of development, tissue maintenance and regeneration, and reveal potential targets for cancer therapy.
Genetic studies in Drosophila show that EGFR/Ras/Raf/Erk signaling robustly induces intestinal stem cell (ISC) proliferation in the adult midgut (intestine)1–4. The pathway is required for ISC proliferation and maintenance in the fly’s midgut, but has little if any role in regulating cell differentiation in this context3,4. In these respects the functions of EGFR signaling in Drosophila and mammalian ISCs are nearly identical5–9. Upon damage or stress, visceral muscle, progenitor cells, and enterocytes (EC) in the gut produce the neuregulin-type EGFR ligands Vn, Spi, and Krn, respectively. These signals activate EGFR signaling in ISCs and post-mitotic enteroblasts (EBs), promoting the division of ISCs and the growth and DNA endoreplication of EBs, thus driving gut epithelial regeneration2,3,10. In this study, we used over-expression of a secreted form of Spi (sSpi)11, or Krn, or enteric infection with Pseudomonas entomophila (P.e.), to trigger EGFR signaling in vivo in the fly’s midgut3.
ETS family transcription factors are known downstream effectors of EGFR signaling, and some are directly phosphorylated by ERK. All ETS transcription factors share a conserved ETS DNA binding domain, and ~30% have a PNT domain, which mediates Erk signaling regulation via a MAPK phosphorylation site12. Pointed (Pnt), the Drosophila homolog of human ETS1 and ETS2, regulates stem cell proliferation in adult tissues including Drosophila ISCs13, renal and nephric stem cells14, female germline stem cells15, and neural stem cells16,17. Ets21C, the closest paralog of Pnt and a homolog of human ERG, is also important for Drosophila ISC proliferation13,18. Ets21C and Pnt share a predicted DNA binding site seed sequence (ACCGGAAAT), suggesting that their gene targets overlap. Previous studies demonstrated that Ets21C can be activated by the JNK pathway during infections19,20,21,22, and promotes epithelial renewal in the Drosophila midgut downstream of JNK signaling.
In an earlier study we reported that Pnt and Ets21C were up-regulated by the ERK-dependent inactivation of the transcriptional repressor Capicua (Cic). Cic, Pnt, and Ets21C are each required for and sufficient to regulate ISC proliferation13. Consistent with these findings in Drosophila, studies in mammals found CIC and several ETS factors to be transcriptional regulators of EGFR/Ras/MAPK pathway genes23–25 and showed that CIC downregulation and ETS factor upregulation can confer resistance to RAF and MEK inhibitors in cancer26,27. Yet despite being critical nuclear mediators of EGFR/Ras/Erk signaling in cell proliferation and tumor progression, the transcriptional regulatory network comprising Cic, Pnt, and Ets21C has not been thoroughly investigated, and the identities of their downstream target genes remain unclear. Moreover, the mechanisms via which EGFR/Ras/Raf/Erk signaling promotes cell growth are poorly understood in any cell type. In this study we investigate the functions and gene targets of Pnt and Ets21C in adult Drosophila ISCs, in order to understand how they promote cell growth and proliferation.
RESULTS
EGFR signaling regulates ISC growth and division via Pnt and Ets21C
To activate EGFR/Erk signaling specifically in Drosophila ISCs, we used a temperature-dependent ISC-specific Gal4 driver (esg-Gal4 UAS-2xEYFP; Su(H)GBE-Gal80 tub-Gal80ts; henceforth referred to as esgts; Su(H)-Gal80) to induce UAS-linked target genes28. Activating EGFR signaling in ISCs by inducing the activated EGFR ligand, sSpi, strongly promoted ISC divisions after as few as 8 hours (Figures 1A and 1B). We also tested the downstream ETS-type transcription factors Pointed (Pnt) and Ets21C13. Pnt has two splice isoforms, PntP1 and PntP229, which are equivalent to the Pnt-PC and Pnt-PB isoforms, respectively. PntP1 and P2 have the same DNA binding domain, but the shorter PntP1 isoform lacks the PNT domain, which contains a regulatory MAPK phosphorylation site30,31. A sequential activation model from Drosophila eye discs proposed that Erk signaling activates pre-existing PntP2 by phosphorylation, and that PntP2 in turn activates PntP1 transcriptionally. Auto-feedback of PntP1 can then provide lasting Pnt transcription factor activity in response to transient Erk activation32. In our experiments, overexpressed PntP1 or PntP2 both caused significant ISC proliferation within 8 hours of induction in ISCs, with more robust increases observed after 24h (Figures 1A and 1B). Consistent with the sequential activation model above, PntP1 could promote ISC divisions when co-expressed with MEKRNAi, whereas PntP2 could not (Figures S1A-S1C). Of the two Ets21C protein isoforms, only the longer Ets21C-PC isoform promoted ISC division when over-expressed (Figures S1D-S1E). We performed further experiments only with Ets21C-PC, referred to hereafter simply as Ets21C.
Figure 1. EGFR signaling regulates ISC growth and division.
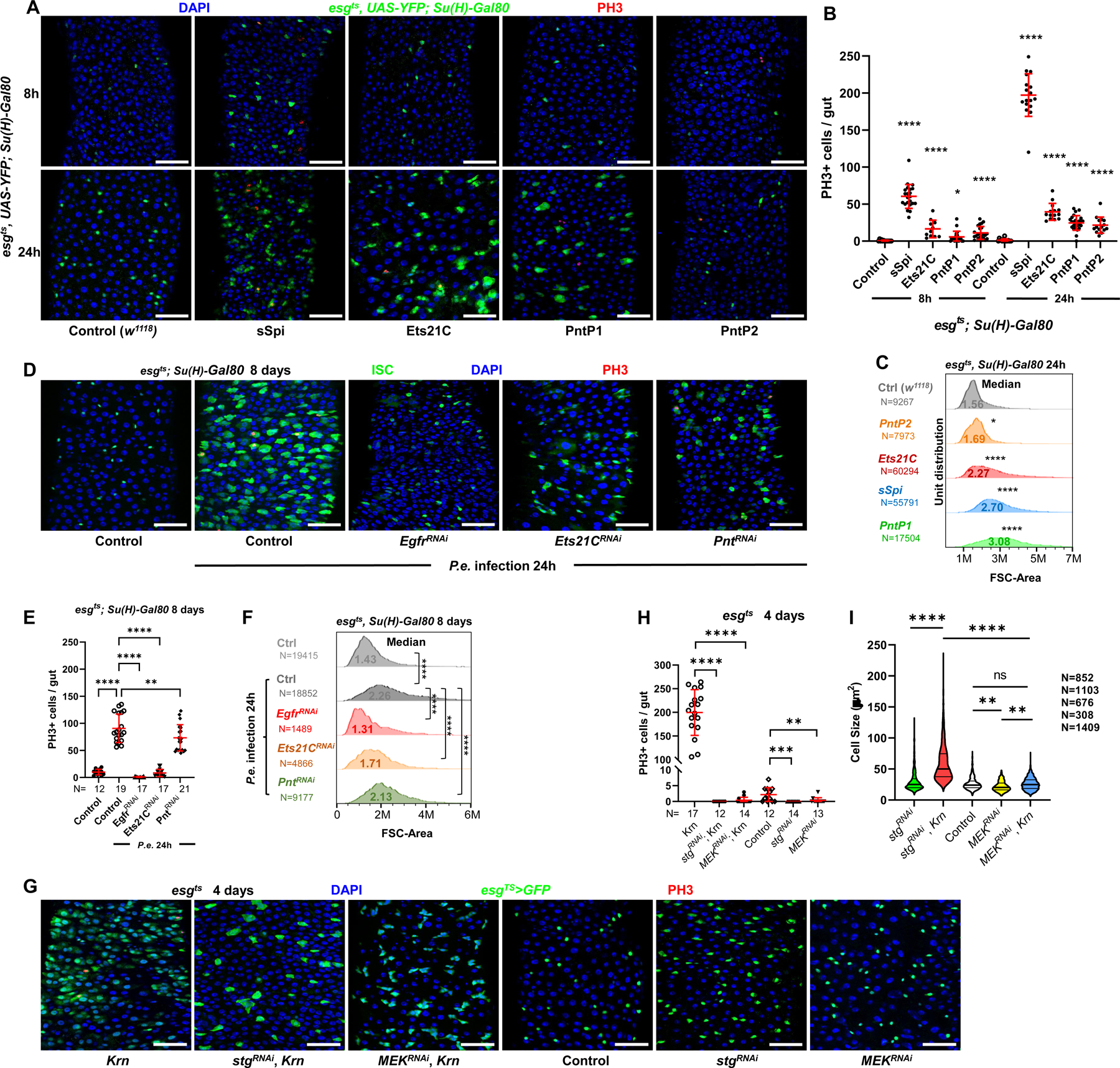
(A) Confocal images of posterior midguts, showing the induction of ISC proliferation and growth by EGFR signaling. Guts were stained with Phosphorylated Histone H3 (PH3) and DAPI. Top panels: 8h transgene induction in ISCs, showing a very low level of YFP fluorescent protein accumulation. Bottom panels: 24h induction. From left to right: control (w1118), sSpi OE, Ets21C-PC OE, PntP1 OE, and PntP2 OE. Scale bar is 50μM. (B) Quantification of PH3+ mitotic cells in whole guts. Both 8h and 24h activation of EGFR signaling led to significantly increased mitotic cell numbers, with 24h activation having a more robust proliferative effect. (C) Flow cytometry unit distribution of Forward Scatter (FSC) Area of YFP+ ISCs upon 24h activation of EGFR signaling by EGF ligand sSpi or downstream transcription factors Pnt and Ets21C. ISC cell size was increased by EGFR activation. (D-F) The requirements of EGFR signaling and downstream transcription factors Pnt and Ets21C for ISC proliferation and growth during tissue repair. (D) Confocal images of posterior midguts infected with Pseudomonas entomophila (P.e.). Guts were stained for PH3 and with DAPI. (E) The increase of mitotic cell numbers upon P.e. infection is dependent on EGFR signaling. (F) Flow cytometry unit distribution of FSC-Area of YFP+ ISCs in response to P.e. infection. EGFR signaling and transcription factors Pnt and Ets21C are required for ISC growth. (G-I) The growth effect of EGFR signaling is independent of proliferation, and depend on MEK-ERK cascade. (G) Confocal images of posterior midguts. Scale bar is 50μM. (H) Both Stg and MEK knockdown blocked ISC proliferation, (I) but only MEK knockdown blocked cellular growth. Violin plot showing ISC cell size. Thick line represents the median and thin lines represent quartiles. See also Figure S1. (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; ns, not significant)
In addition to driving ISC divisions, activation of EGFR signaling by sSpi, Pnt, or Ets21C also increased ISC size (Figures 1A, 1C, S1F). RNAi-mediated knockdown of Cic in ISCs also increased ISC size (Figure S1G). Conversely, disrupting EGFR signaling by expressing RNAi’s against Egfr, Pnt, or Ets21C reduced ISC size (Figure S1G). When midguts were infected with Pseudomonas entomophila (P.e.), which activates EGFR signaling and ISC proliferation3, ISC size also increased significantly (Figures 1D–1F). Knockdown of Egfr completely abolished the P.e.-dependent activation of ISC division and cell growth, while knockdown of Ets21C or Pnt partially suppressed these effects (Figures 1D–1F). Thus, ISC growth caused by P.e. infection requires EGFR signaling and its downstream transcriptional effectors, Pnt and Ets21C.
To determine whether cellular growth driven by EGFR signaling is dependent on cell division, an RNAi against string (stg), a Cdc25C homolog, was used to specifically block ISC mitoses. Progenitor cells expressing Krn along with stgRNAi for 4 days were significantly larger than control cells (Figures 1G, 1I). In addition, RNAi targeting MEK (Dsor1) completely blocked ISC proliferation caused by Krn (Figures 1G–1H). Progenitor cells co-expressing Krn and MEKRNAi were smaller than those expressing Krn and stgRNAi (Figures 1G, 1I). Furthermore, growth and endoreplication in polyploid enteroblasts (EB) could be blocked by MEKRNAi (Figures S6G-S6H). Thus, ISC and EB growth driven by EGFR signaling can occur without cell division, and is dependent on MEK activity.
Gene targets of EGFR signaling
To understand how EGFR signaling promotes ISC growth and proliferation, we performed mRNA sequencing (mRNA-Seq) on FACS-sorted ISCs after overexpressing PntP1, PntP2, Ets21C, or sSpi in ISCs for 8 or 24hr, using the esgts; Su(H)-Gal80 driver (see33,34 for method). To determine the DNA binding profiles of Pnt and Ets21C, we also performed ISC-targeted DNA adenine methylase Identification-DNA Sequencing (DamID-Seq) by expressing Dam-Pnt or Dam-Ets21C fusion proteins in ISCs (see13,35 for methods). “Direct targets” of Pnt and Ets21C were defined as genes that were significantly up or down regulated (adjusted p < 0.05, absolute log2 fold change > 0.5) as determined by mRNA-Seq, and which also had one or more significant DamID-Seq peaks assigned to the gene body or known cis regulatory modules (Tables S1, Data S1-S2).
Approximately 20% of DamID binding sites were associated with genes showing altered mRNA levels after PntP1, PntP2, or Ets21C overexpression, indicating significant overlap (Figures S2A-S2C). DamID binding peaks aligned strongly with Transcription Start Sites (TSS) and Cis-Regulatory Modules (CRM) as defined by the Red Fly Database (Figures S2D-S2I). For both Pnt and Ets21C, ~half of the direct targets were down-regulated, suggesting that these ETS factors might act as both transcriptional activators and repressors (Figures 2A, 2B, see also36,37). Many genes that were up-regulated by Ets21C, PntP1, or PntP2 had DamID binding sites for these factors (41.36%, 35.58%, 37.43%, respectively; Figures 2A, 2B). PntP1 and PntP2 had very similar transcriptional regulatory effects (Figures S2J-S2N), showing a ~60% overlap in DamID sites (Figures S2N), with PntP1 affecting more genes than PntP2 (Figures S2J-S2K). Indeed, the majority of PntP2 targets were also PntP1 targets.
Figure 2. Pnt and Ets21C directly upregulate DNA replication and cell cycle genes.
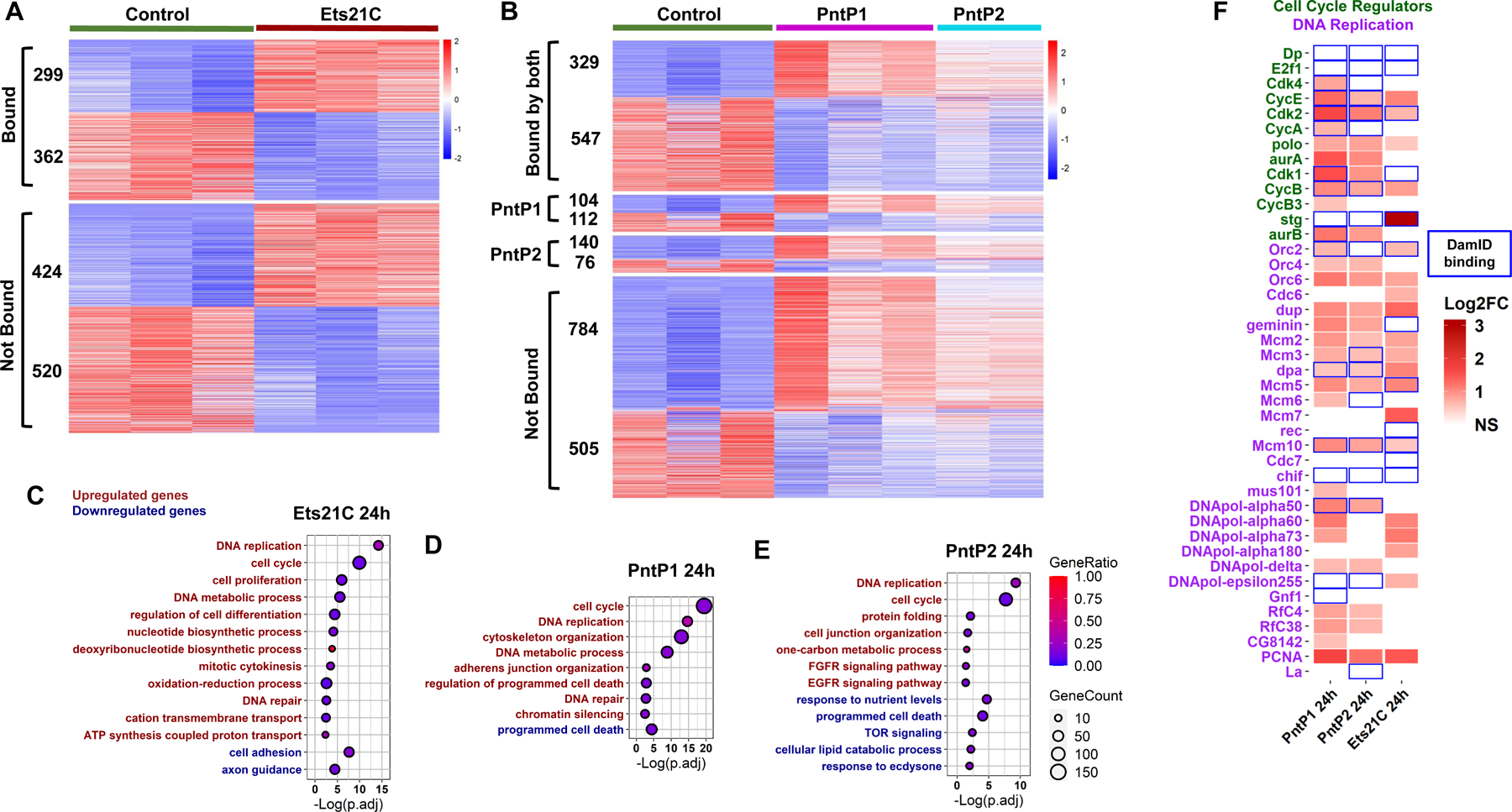
(A-B) Heatmap showing the genes differentially expressed by 24h of (A) Ets21C-OE, (B) PntP1-OE or PntP2-OE in ISCs. Genes marked as “bound” (~40% of differentially expressed genes) had DamID peaks falling in the gene body (from transcription start site to termination site) or known regulatory regions (experimentally defined in vivo and collected in the RedFly database) of that gene. Transcripts per million (TPM) values were z-score normalized along rows before plotting. Genes were manually grouped by category. Within each category, genes were ordered based on decreasing average expression across samples. See also Table S1, Data S1 and Data S2. (C-E) Dot plots of selected Gene Ontology Biological Process Terms (GO-terms) enriched in response to Ets21C, PntP1, or PntP2 OE in ISC. For the full list of enriched GO-terms, see Data S3. Red labels indicate terms for significantly up-regulated genes, while green ones indicate terms for significantly down-regulated genes. The size of the dot indicates the gene count for the GO-terms, and the color indicates the proportion of genes in the GO term that are significantly differentially expressed. (F) Heatmap of Log2FC of cell cycle and DNA replication genes regulated by PntP1, PntP2, and Ets21C. Blue boxes indicate the presence of a direct DamID binding site or sites on the gene. See also Figure S2.
Cross-comparisons of our mRNA-Seq data showed that genes which responded to Ets21C and Pnt strongly overlapped with sSpi responding genes (Figures 3G, S3A-S3C), although sSpi modulated the expression of a larger set of genes and its transcriptional output was dynamic in time (Figures S2O and S2P). For instance, Pnt- and Ets21C-regulated genes made up 56% of genes affected by sSpi at 8h, of which 51% were Pnt and/or Ets21C direct targets based on DamID. Together with epistasis tests (Figures S1A-S1C), this strong overlap of targets confirms that Pnt and Ets21C are important effectors of EGFR signaling in ISCs. The DamID binding gene targets of Pnt and Ets21C also strongly overlapped with those of the ERK-dependent repressor Capicua (Cic; p≈0)(Figure S3D), suggesting that ERK may have a dual action on its gene targets, simultaneously relieving Cic-mediated repression and promoting the binding of ETS transcriptional activators.
Figure 3. EGFR signaling up-regulates oxidative phosphorylation and TCA cycle genes in ISCs.
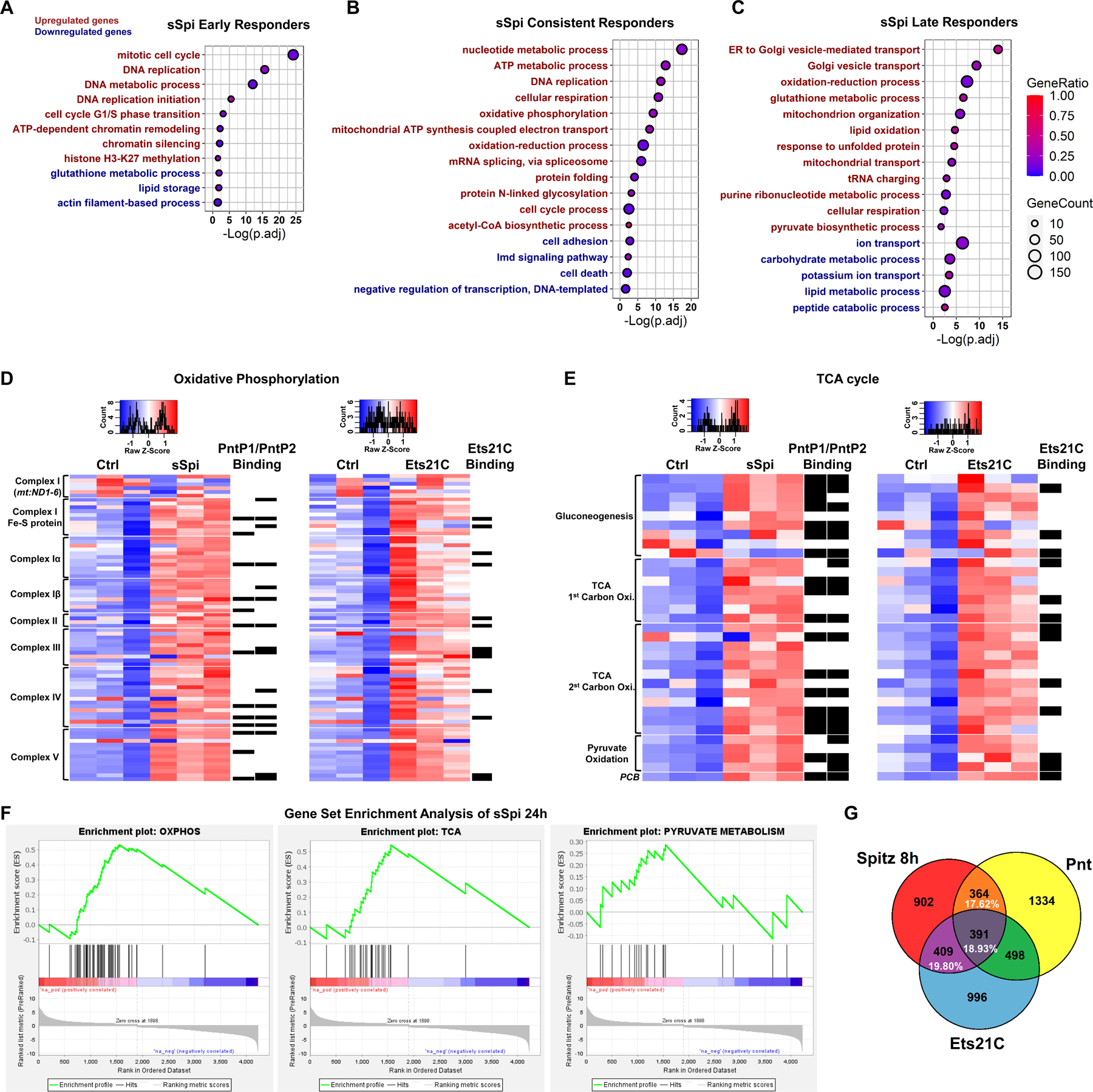
(A-C) Dot plot of selected enriched GO-terms of early, consistent, and late responder genes to sSpi over-expression, which are the genes significantly changed only at 8h, only at 24h, or at both 8h and 24h (with the same trend), respectively. For the full list of enriched GO-terms, see Data S4. Red indicates significantly enriched terms for the up-regulated gene set, while green indicates significantly enriched terms for the down-regulated gene set. The size of the dot indicates the gene count for the GO-term, and the color indicates the proportion of genes in the GO term that are significantly differentially expressed. (D-E) Heatmap of (D) OXPHOS and (E) TCA cycle genes after sSpi and Ets21C 24h over-expression. All genes with known function involved in metabolic pathways (KEGG dme00190 and dme00020) and expressed in ISCs were included. Black boxes on the right of the heatmaps indicate DamID binding for the specified TF. Transcripts per million (TPM) values were z-score normalized along rows before plotting. Genes were manually ordered. (F) Gene Set Enrichment Analysis of sSpi 24h differentially expressed genes showing OXPHOS, TCA, and Pyruvate Metabolism were significantly up-regulated. (G) Venn diagram shows that Pnt and Ets21C differentially expressed genes are essential for EGFR signaling early activation, making up to 56.35% of sSpi 8h transcription output. The overlap is significant, P=1.24e-276. See also Figure S3.
EGFR signaling regulates genes in DNA replication, cell cycle, cell death, cell adhesion, and chromatin remodeling
From Gene Ontology (GO) analysis “DNA replication” and “cell cycle” or “mitotic cell cycle” were the most significantly enriched GO biological processes amongst Pnt, Ets21C and sSpi up-regulated genes. Many genes in these sets were direct Pnt and/or Ets21C targets by DamID-Seq (Figures 2C–2F); CycB, CycE, and cdk2 were direct targets of PntP1 and PntP2; Cdk1, cdk4, CycA, and aurB were direct targets of PntP1; stg was a direct target of Ets21C (Figure 2F). These genes were also upregulated by sSpi (Data S1). These highly significant effects help explain the activation of the ISC cell cycle by EGFR signaling. “Programmed cell death” or “cell death” was one of the most significant GO processes amongst PntP1, PntP2, and sSpi downregulated genes (Figure 2E, 3B, Data S3-S4), many of which were DamID targets (Data S2). This suggests a role for EGFR signaling in ISC survival. Cell adhesion and axon guidance were enriched biological processes amongst Ets21C and sSpi downregulated genes and most of these had Dam-Ets21C binding sites. Such targets may help explain ETS-TF functions during tumor progression and metastasis.
We also noted evidence of signaling crosstalk. PntP1 and PntP2 both up-regulated three ligands in FGF signaling (bnl, ths, pyr). Of these bnl is known to be pro-proliferative for ISCs38,39. sSpi, Pnt, and Ets21C all up-regulated the cytokine upd3, indicating crosstalk between EGFR signaling and Jak-STAT signaling. Additionally, sSpi, Pnt, and Ets21C each altered the expression of many components (Figure S3E) and feedback regulators in the EGFR signaling pathway itself, and Pnt and Ets21C bound many of these targets by DamID (Figure S3F). This is likely an indication of positive and negative feedback interactions within the pathway, as previously characterized in other contexts40.
Of the early responders to sSpi, “chromatin remodeling” was an enriched biological process (Figure 3A). Several genes in the Brahma complex, nucleosome remodeling factors, and histone modifiers were up-regulated (Data S4). This rapid, transient expression of chromatin remodelers in response to EGFR activation may prepare the genome for transcriptional changes that facilitate ISC proliferation or daughter cell fate determination. Consistently, brm is required for ISC proliferation and damage-induced midgut regeneration41.
EGFR signaling up-regulates oxidative phosphorylation and TCA cycle genes
Tellingly, GO term enrichment analysis showed that EGFR signaling upregulated numerous metabolic processes related to mitochondrial function (Figures 2C–2E, 3D-3F, Data S4). For instance, “nucleotide metabolism”, “oxidative phosphorylation” (OXPHOS), “mitochondrial ATP biogenesis”, “acetyl-CoA biogenesis”, and “oxidation-reduction” were among the most significant biological processes consistently up-regulated by sSpi at both 8h and 24h (Figure 3B). A large fraction of genes used in OXPHOS (mitochondrial electron transport chain) and the TCA cycle were upregulated by sSpi, Pnt and/or Ets21C, and many of these bound Pnt and/or Ets21C by DamID (Figures 3D and 3E). Late positive responders to sSpi included genes falling under “oxidation-reduction” and “glutathione metabolic process” GO terms, suggesting that the up-regulation of OXPHOS might increase ROS levels. The up-regulation of “lipid oxidation” genes at 24h suggested that fatty acid β-oxidation may be commandeered to fuel OXPHOS and ATP synthesis after prolonged sSpi stimulation (Figure 3C). Consistent with our GO analysis, gene set enrichment analysis (GSEA) showed that OXPHOS, TCA, and pyruvate metabolism genes were significantly up-regulated by sSpi (Figure 3F). sSpi also upregulated a number of genes encoding mitochondrial factors including the mitochondrial pyruvate transporter Mpc1, mitochondrial TOM complex elements, and clueless (Figure S4C).
EGFR signaling reconfigures midgut metabolism
To determine whether the expression of genes for mitochondrial functions actually altered ISC metabolism, we performed metabolite profiling on whole midguts. sSpi was over-expressed in esg+ progenitor cells for 4 days prior to analysis. Compared to controls, sSpi-expressing midguts had decreased levels of glucose, mannose, fructose, glucose-6P, fructose-6P, and pyruvate (Figure 4A). We also detected depletions of fatty acids and fatty acid esters, the TCA cycle intermediate α-ketoglutarate, and the pentose phosphate pathway intermediate sedoheptulose-7P. These decreases in glycolytic intermediates, fatty acids and their derivatives are indicative of increased glycolysis and fatty acid oxidation, and could be due to the high demand for substrates like nucleotides and amino acids, and for energy (ATP), during cell proliferation. Interestingly, we did not observe decreases in ATP/ADP, NADH/NAD, or NADPH/NADP ratios (Figure 4F), indicating that cellular energy levels were maintained. However, process enrichment analysis indicated that fatty acid biogenesis, β-oxidation, fructose and mannose degradation, and glycolysis were all significantly altered by EGFR signaling (Figure 4B).
Figure 4. EGFR signaling reprograms ISC metabolism.
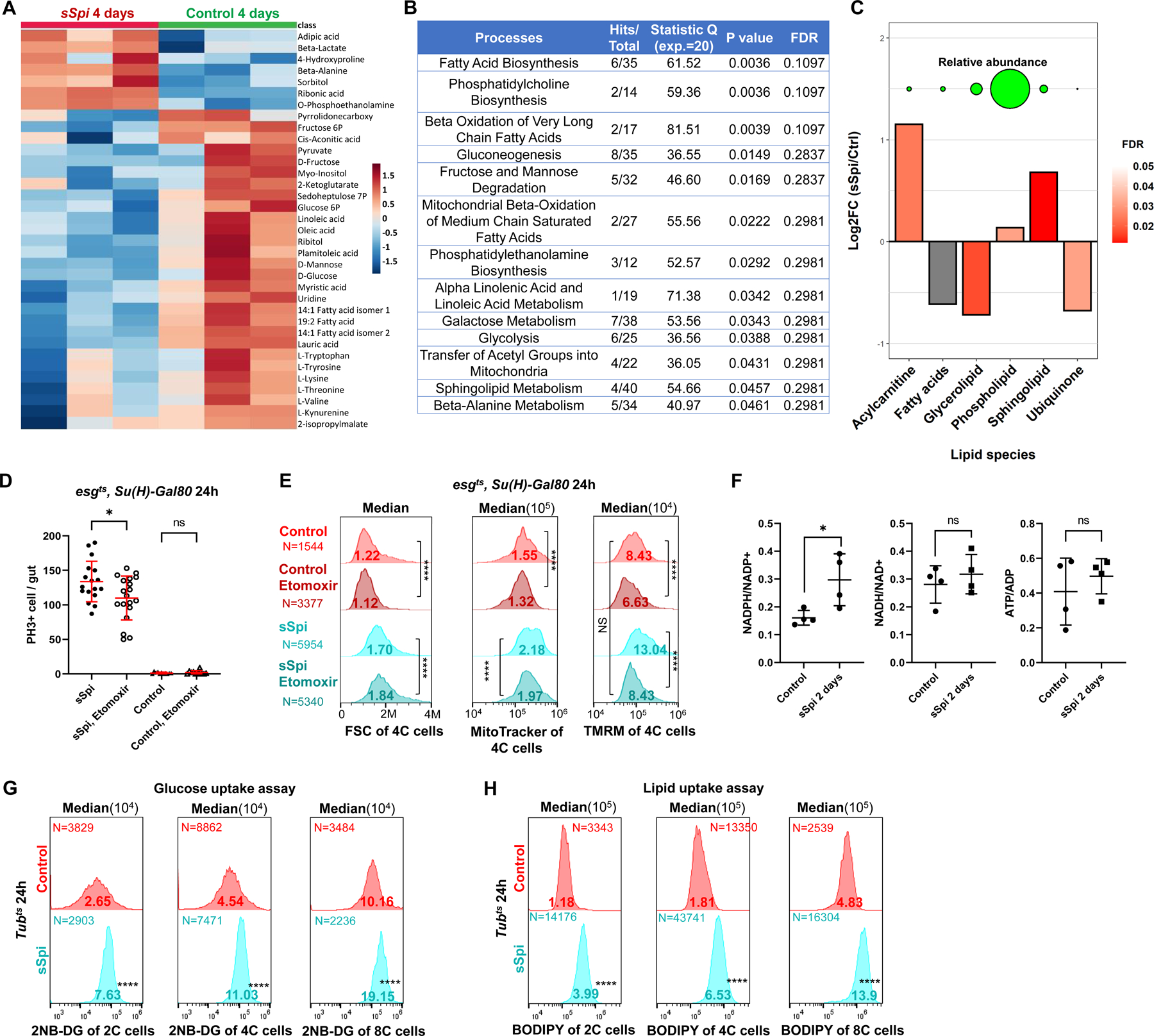
(A) GC-MS metabolomics profiling from 3×100 control midguts or midguts overexpressing sSpi in esg+ cells for 4 days. Heatmap of top 35 altered metabolites (Top 35 by P value). Raw data was log transformed and normalized by Pareto scaling, and then Z-scores were calculated across all samples. Each row represents one metabolite. For complete data see Data S5. (B) Enrichment analysis results of metabolomics following sSpi overexpression, from MetaboAnalystR. (C) Differences in abundance of major lipid classes after sSpi overexpression for 4 days. Y-axis represents log2 fold-changes between sSpi and control samples. For each class, mass-spectrometry peaks belonging to lipids in the class were summed. Values from control and sSpi-overexpression biological replicates were then compared via t-test. Significance is depicted by the bars’ color, with grey bars indicating that differences are not significant (p>0.05). Circles on top of the bar-plot depict the relative abundance of each lipid class in sSpi overexpression samples. For the full list of lipids, see Data S5. (D) EGFR signaling-induced ISC proliferation was repressed by CPT1 inhibition. Etomoxir (25μM), a CPT1 inhibitor, was mixed in fly food and fed to flies 36h prior to dissection. Etomoxir had no effect on control guts, but reduced mitotic cell number in sSpi-overexpressing guts. (E) Flow cytogram showing that EGFR signaling increased ISC mitochondrial membrane potential (TMRM staining), which was repressed by CPT1 inhibition. (F) The Ratios of redox and energy upon 2 days EGFR signaling activation by targeted metabolomics, NADPH/NADP+ was found to be increased. For complete data see Data S5. (G-H) Flow cytogram showing that EGFR signaling increased 2NE-DG (G) and BODIPY uptake; (H) fluorescent dodecanoic acid uptake in 2C, 4C, and 8C cells of midgut epithelial. See also Figure S4.
Lipidomics data revealed a significant increase of total acylcarnitines. (Figure 4C). Synthesized by the carnitine palmitoyltransferase, CPT1, at the outer mitochondrial membrane, acylcarnitines are an intermediate in the carnitine shuttle system used to transport long-chain fatty acids into mitochondria for β-oxidation. Increased acylcarnitine therefore indicates an increase in β-oxidation, as also suggested by the depletion of various fatty acids (Figures 4A–4C). Consistent with this, our mRNA-Seq data showed that the mRNA encoding fatty acid binding protein (fabp), used to transport fatty acids, was up-regulated by sSpi, Ets21C, and Pnt (Figure S4A), and that expression of β-oxidation enzymes including Acsl, Mtpalpha, Mtpbeta, yip2, ACOX1, Fdh and Aldh was up-regulated by sSpi (Figure S4B).
To test whether EGFR-driven ISC proliferation required fatty acid β-oxidation, we treated the flies with Etomoxir, an inhibitor of CPT1. Etomoxir significantly reduced ISC divisions in response to sSpi OE (Figure 4D), and effectively blocked the increase in mitochondrial membrane potential induced by sSpi (Figure 4E). Thus, sSpi-induced increases in mitochondrial activity and ISC proliferation require β-oxidation. The observed depletions of sugars, glycolytic intermediates, and fatty acids could result from increased consumption, or decreased uptake. To distinguish these possibilities, we used flow cytometry to measure the uptake of 2NB-DG (a fluorescent glucose analog) or BODIPY (a fluorescent dodecanoic acid analog) in cells dissociated from midguts. Midgut cells with 2C, 4C, and 8C DNA content all showed large, highly significant increases in 2NB-DG and BODIPY uptake following in vivo exposure to sSpi for 24h (Figures 4G–4H). This suggests that the observed decreases in sugars, glycolytic intermediates, and fatty acids were consequences of increased consumption.
EGFR signaling promotes mitochondrial biogenesis in Drosophila ISCs
Finding that EGFR signaling up-regulated a broad array of genes involved in mitochondrial function prompted us to investigate mitochondrial biogenesis as a potential effector of EGFR-driven cell growth. Using the mitochondrial dye MitoTracker and live imaging, we determined that mitochondria/cell area ratios increased significantly after sSpi, Ets21C, PntP1 or PntP2 OE, and decreased after expressing EgfrRNAi (Figures 5A–5B). These phenotypes were confirmed by transmission electron microscope (TEM; Figures S5A-S5C). Thus, mitochondrial biogenesis rates exceed cell growth rates after EGFR signaling activation. By expressing a mito-GFP marker in esg+ cells, we determined that sSpi expression or P.e. infection increased mitochondrial volume threefold (Figures 5C–5D). We also assayed ISC mitochondrial volumes and membrane potential (as a metric of mitochondrial OXPHOS activity) using flow cytometry and the MitoTracker and Tetramethylrhodamine (TMRM) dyes, respectively. EGFR signaling increased not only mitochondrial volumes, but also mitochondrial activity as assayed by TMRM accumulation, on a per-cell basis (Figure 5E). Conversely, EgfrRNAi decreased mitochondrial volumes and activity (Figure 5E). Similarly, P.e. infection promoted ISC mitochondrial biogenesis and activity, and this effect required Egfr, Ets21C, and Pnt (Figure 5G). To test whether mitochondrial biogenesis required cell division, stgRNAi was co-expressed along with the Egfr ligand Krn, to block ISC mitosis. Flow cytometry of 4C esg+ cells showed that the Krn-dependent increases in cell size and mitochondrial mass were not affected by blocking mitosis with stgRNAi (Figure 5F, see also Figure 1I). However, MEKRNAi did suppress Krn-dependent increases in cell size, mitochondrial mass and activity (Figures 5F, S6I-S6K), showing that these EGFR-dependent effects are mediated by MEK.
Figure 5. EGFR signaling regulates ISC growth by controlling mitochondrial biogenesis.
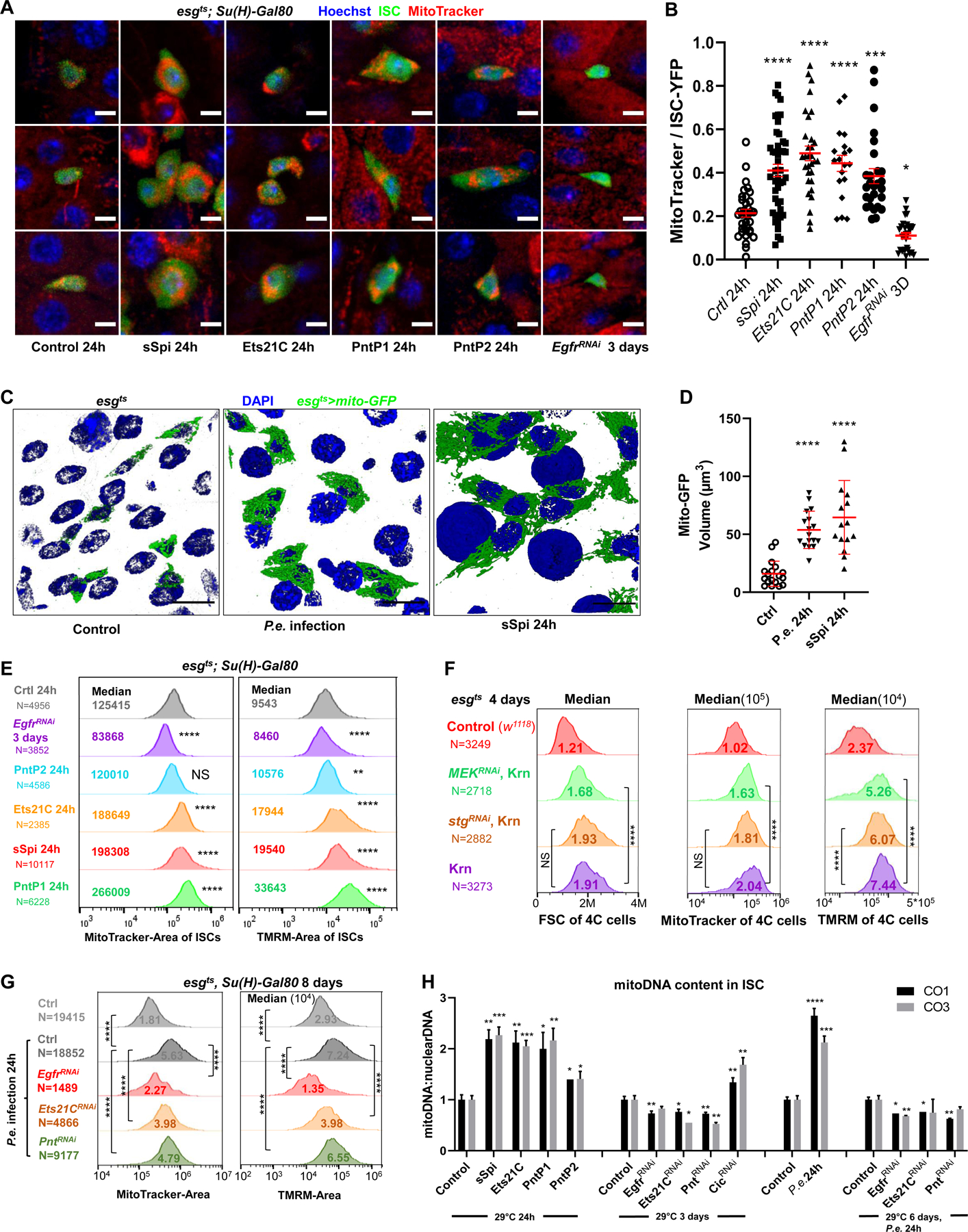
(A-B) Live-imaging of ISCts >YFP ISC cells (green), stained with Hoechst dye (blue), and MitoTracker dye (red). Scale bar is 5μM. The ratio of MitoTracker area to ISC-YFP area across all Z-slices of an ISC cell was calculated. Compared to controls, ISCs expressing sSpi, Ets21C, PntP1, or PntP2 for 24h had more MitoTracker stained areas relative to their cell size, while the ISCs expressing EgfrRNAi had less. (C-D) Mitochondria of progenitor cells marked by esgts >mito-GFP. The 3D re-construction of gut epithelia showed that after 24h P.e. infection or sSpi OE, the mitochondrial volume in the progenitor cells was significantly increased. Scale bar is 10μM. (E) Flow cytometry unit distribution of MitoTracker-Area and TMRM-Area of YFP positive ISCs upon activation or repression of EGFR signaling. In ISCs, sSpi, PntP1, PntP2, and Ets21C over-expression increased mitochondrial area and activity; EGFR knockdown decreased mitochondria area and activity. (F) Flow cytometry unit distribution of FSC-Area, MitoTracker-Area, and TMRM-Area of 4C state esg>GFP cells. The mitochondrial biogenesis effect of EGFR signaling is independent of proliferation, and dependent on MEK-ERK cascade. (G) Flow cytometry unit distribution of MitoTracker-Area and TMRM-Area of YFP positive ISCs. P.e. infection promoted mitochondria growth and activity, and required EGFR, Ets21C, and Pnt. (H) EGFR signaling affects Mitochondria DNA content in ISCs. Relative mitoDNA content was calculated using the DNA level of two mitochondria genes (CO1 and CO3) relative to two chromosomal genes (Ets21C and β-tub56D). qPCR was performed on total DNA samples extracted from sorted ISCs. Error bar represents SEM. See also Figure S5.
Consistent with these results, the mitochondrial DNA content of ISCs more than doubled after 24h induction of sSpi, Ets21C, or PntP1 (Figure 5H). PntP2 over-expression for 24h or Cic knockdown for 3 days also increased mitochondrial DNA copy number. Consistently, Pnt and Ets21C knockdown significantly reduced mitochondrial DNA content. P.e. infection also strongly increased ISC mitochondrial DNA content, and depletion of Egfr by RNAi suppressed this increase. These data show that EGFR signaling via Cic, Pnt, and Ets21C is required for and sufficient to promote mitochondrial biogenesis.
EGFR signaling regulates mitochondrial biogenesis in human cells
To test whether the metabolic effects of EGFR signaling are conserved, we investigated hTERT immortalized human Retinal Pigment Epithelial-1 (RPE-1) cells. EGFR signaling promotes RPE-1 cell proliferation, survival, and migration through the activation of ERK and AKT42–45, and RPE-1 cells do not carry any known mutations in the EGFR pathway. In our study, RPE-1 cells were synchronized by 20h serum starvation, forcing 85–90% of cells into G1 arrest. Human EGF (50ng/ml), MEK inhibitor (Binimetinib 500nM), or both were then added to basal media for 24h. After 24h of EGF stimulation, mitochondrial mass and activity were significantly increased as compared to controls (Figures S5D-S5F). Interestingly, MEK inhibition blocked the mitochondrial biogenesis effect of EGF (Figures S5G-S5H), but failed to suppress the EGF-dependent increase in mitochondrial membrane potential (Figures S5G and S5I). These data indicate that, as in Drosophila ISCs, mitochondrial biogenesis is regulated by the EGFR/MEK/ERK cascade. However, in contrast to what we found in Drosophila ISCs, MEK was not required for EGF to stimulate mitochondrial activity in RPE-1 cells. Thus, other effectors of EGFR signaling, for instance PI3K/AKT, may be more potent in regulating mitochondria activity, at least in human cells.
Pnt promotes mitochondrial biogenesis via mtTFB2
Despite these profound effects on mitochondrial biogenesis, the mRNA expression of known master transcriptional regulators of mitochondrial biogenesis like Spargel (PGC-1α), ewg (NRF-1), Ets97D (NRF-2), and TFAM (mitochondrial transcription factor A/mtTFA) was not significantly changed by EGFR signaling activation (Figure S4C). PGC-1α and NRF-1 might be regulated postranslationally46–49, and this could not be ruled out. We noted, however, that many of the OXPHOS and TCA cycle genes up-regulated by EGFR signaling were detected as Pnt and/or Ets21C binding targets by DamID (Figure 3D), suggesting direct regulation.
A second mechanism was suggested by sequence analysis showing that Pnt and Ets21C are homologs of the GABPα subunit of human Nuclear Respiratory Factor 2 (NRF-2). The closest Drosophila homolog of GABPα is Ets97D, but Pnt and Ets21C are very similar, having highly conserved ETS DNA binding and PNT domains, and nearly identical predicted DNA binding seed sequences (Figure 6A). Drosophila Ets97D, PntP2, and Ets21C have a percent coverage (aligned amino acids) of 95.6%, 87.0%, and 75.8% of human GABPα, and percent identity (amino acids) of 38%, 21%, and 27%, respectively (EMBL-EBI Multiple Sequence Alignment). Ets97D is dispensable in the gut, where Pnt and/or Ets21C might fulfill its function50. Indeed, Ets97D is poorly expressed in the midgut, whereas Pnt and Ets21C are highly expressed (Data S1)33.
Figure 6. mtTFB2, a transcriptional target of Pnt, is required for ISC proliferation and cellular growth.
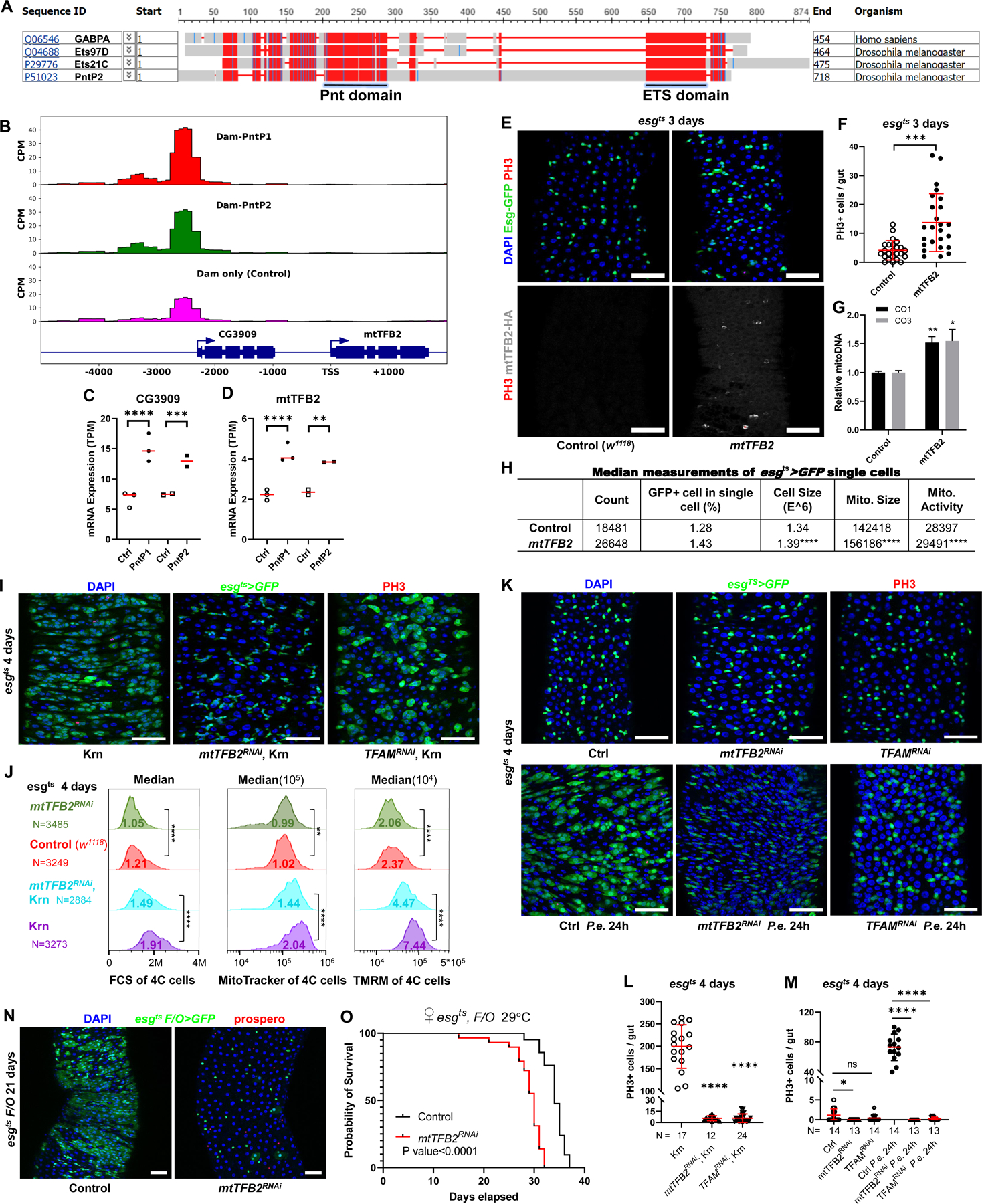
(A) COBALT Constraint-based multiple alignment (https://www.ncbi.nlm.nih.gov/tools/cobalt/cobalt.cgi) of human NRF2, Drosophila Ets97D, PntP2, and Ets21C protein showing the conserved Pnt and ETS domains. This is a column-based method that highlights highly conserved and less conserved columns based on amino acids’ relative entropy threshold. Alignment columns with no gaps are colored in red (highly conserved) or blue (less conserved). (B) DamID binding peaks (Significant peaks when compared to Dam alone control) of PntP1 and PntP2 upstream of mtTFB2 gene. (C-D) PntP1 and PntP2 up-regulated the expression of CG3909 and mtTFB2 by RNASeq. (E-F) mtTFB2 is sufficient to mildly induce ISC proliferation. Confocal images of posterior midgut. Guts were stained for GFP (green), PH3 (red), mtTFB2-HA (white), and with DAPI (blue). mtTFB2-HA protein localized distinctly in mitochondria. Scale bar is 50μM. (G) Over-expression of mtTFB2 increased mitochondria DNA content in ISCs. Relative mitoDNA content was calculated using the DNA level of two mitochondria genes (CO1 and CO3) relative to two chromosomal genes (Ets21C and β-tub56D). qPCR was performed on total DNA samples extracted from sorted ISCs. Error bar represents SEM. (H) FACS showed mtTFB2 OE increased the percentage of esgts>GFP cell in total single live cell. The FCS-Area, MitoTracker-Area, and TMRM-Area of esg+ cell over-expressing mtTFB2 were increased. (I) Knockdown of mtTFB2 or TFAM was sufficient to block proliferation induced by Krn. Confocal images of posterior midguts. Guts were stained for PH3 (red), GFP (green), and with DAPI (blue). Scale bar is 50μM. (J) Flow cytometry unit distribution of FSC-Area, MitoTracker-Area, and TMRM-Area of 4C state esgts>GFP cells. mtTFB2 knockdown impacted cell size, mitochondria size and activity in both physiological conditions and upon EGFR signaling activation. (K) mtTFB2 and TFAM are necessary for ISC proliferation induced by P.e. infection. Confocal images of posterior midguts, with and without P.e. infection. Scale bar is 50μM. (L-M) Quantification of PH3 positive cells in whole guts. (N) Image of esgts F/O driving mtTFB2 knockdown for 21 days, showing that clone formation requires mtTFB2. Scale bar is 50μM. (O) Survival assay showing that mtTFB2 knockdown in the esg lineage significantly reduced the life span of flies. See also Figure S6.
Human NRF-2 regulates mitochondrial biogenesis by controlling the mitochondria transcription factors B1 and B2 (mtTFB1 and mtTFB2)51. Drosophila mtTFB2 is required for larval development, and mtTFB2RNAi severely impairs transcription of the mtDNA, suppressing OXPHOS and promoting anaerobic glycolysis in larvae52. mtTFB1 shows very low expression in all cell types of the gut epithelium33, and was not induced by EGFR signaling (Data S1). mtTFB2, however, was significantly up-regulated by PntP1 and PntP2 (Figure 6D; Data S1). sSpi stimulation also up-regulated mtTFB2 although not as significantly (adjusted P = 0.064). PntP1 and PntP2 had DamID binding sites 3kb upstream of the mtTFB2 gene transcript (Figure 6B). Between these binding sites and the mtTFB2 coding sequence, there is another gene (CG3909) on the same strand, which was also up-regulated by PntP1 and PntP2 (Figures 6B and 6C). These observations suggest that Pnt activates an enhancer up-stream of mtTFB2.
Overexpressed mtTFB2 was sufficient to mildly induce ISC proliferation and increase the number of esg+ progenitor cells (Figures 6E and 6F). Flow cytometry confirmed the increase in progenitors, and revealed that mtTFB2 increased esg+ cell size, mitochondrial size, and mitochondrial activity (Figure 6H). Consistently, mtTFB2 overexpression increased mitochondrial DNA content (Figure 6G). However, mtTFB2 overexpression failed to promote proliferation when EGFR signaling was blocked, indicating that EGFR has other targets required for proliferation (Figure S6A). Thus, Pnt promotes mitochondrial biogenesis and ISC growth and proliferation in part by up-regulating mtTFB2.
mtTFB2, TFAM and mitochondrial biogenesis are required for ISC proliferation
We next tested the requirement of mtTFB2. Expression of mtTFB2RNAi in esg+ cells completely blocked their ability to proliferate in response to Krn or P.e. infection (Figures 6I–6M). Progenitor cells lacking mtTFB2 also showed reduced size, mitochondrial mass, and mitochondrial activity despite the activation of EGFR signaling (Figure 6J). Flow cytometry of 8C and 16C esg+ enteroblasts showed the mtTFB2RNAi also suppressed the EGFR-dependent growth and endoreplication of these postmitotic progenitors (Figures S6G-S6H). Thus, mtTFB2 is required for cell growth and mitochondrial biogenesis driven by EGFR signaling.
To test the long-term effect of mtTFB2 knockdown, we used the esgts F/O driver53. ISC clones expressing mtTFB2RNAi arrested as single cells, whereas control clones grew profusely and populated most of the epithelium in the R4 region within 21 days (Figure 6N). In some guts mtTFB2RNAi caused progenitor cell loss, probably due to the failure of stem cell self-renewal. Midguts expressing mtTFB2RNAi in esg+ progenitor cells were shrunken in aged flies, and these animals died earlier than controls (Figure 6O).
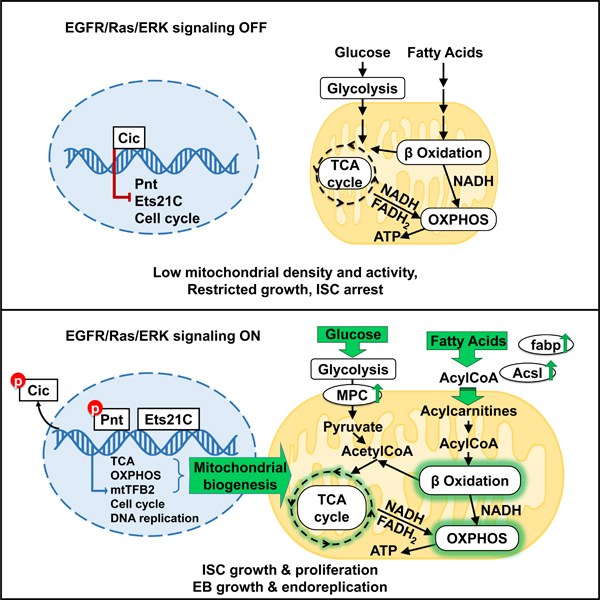
Previous investigations found that TFAM (mtTFA) and mtTFB2 are the only DNA binding proteins required for transcription of the mitochondrial genome54, and are both indispensable for mitochondrial biogenesis. Hence, we also investigated TFAM. As with mtTFB2, esg+ cells expressing TFAMRNAi failed to proliferate in response to P.e. infection or Krn expression (Figures 6I, 6K-6M, S6D-S6F). However, esg+ cells expressing TFAMRNAi showed only modest decreases in size under normal conditions, and were able to grow and increase their mitochondrial mass in response to EGFR activation by Krn (Figures 6I, 6K, S6B-S6C). However, TFAMRNAi decreased the mitochondrial membrane potential both at homeostasis and after EGFR activation, as assayed by TMRM staining (Figures S6B-S6C). These results indicate that TFAM and mtTFB2 have distinct functions in mitochondrial biogenesis, but are both required for mitochondrial activity and ISC proliferation. To summarize, we report that EGFR signaling via Cic, Pnt, Ets21C, and mtTFB2 alters stem cell metabolism to facilitate cell growth and proliferation, and that this works through promoting mitochondrial biogenesis and activity by increasing fatty acid β-oxidation, the TCA cycle and OXPHOS (Figure 7).
Figure 7. Effects of EGFR signaling on ISC gene expression and metabolism.
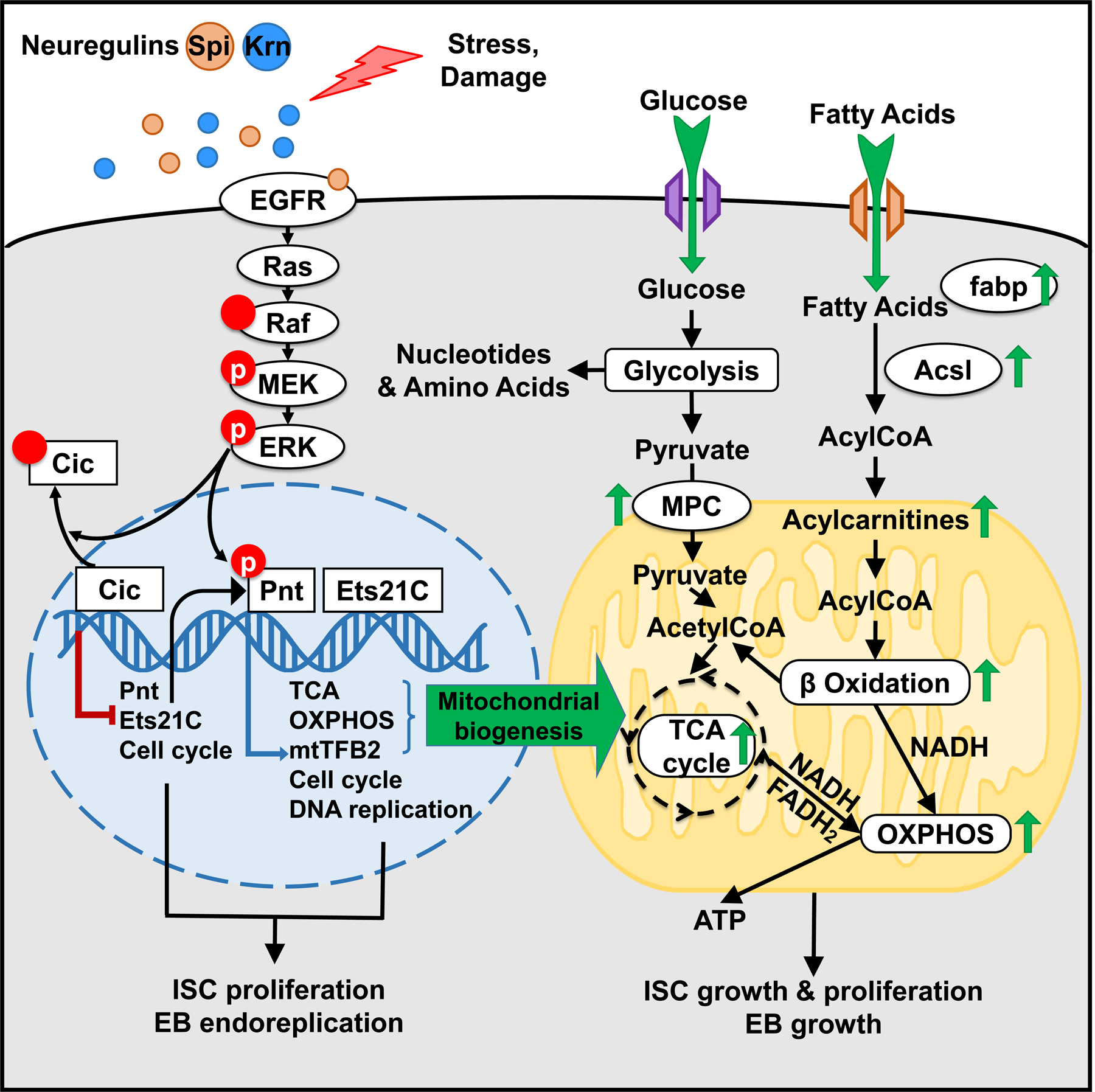
We show the effects of upregulating EGFR signaling in the ISC nucleus (blue), mitochondrion (yellow), and cytoplasm (grey). Green arrows indicate example processes, gene products, and metabolites that are up-regulated by EGFR signaling in response to the EGFR-dependent transcription factors Cic, Pnt and Ets21C.
DISCUSSION
EGFR signaling activates Drosophila intestinal stem cells (ISC) for growth and division, and enteroblasts (EB) for growth and DNA endoreplication. Together, these effects drive gut epithelial regeneration1–4,10. In this study we report that the EGFR ligand Spitz (Spi) and the downstream transcription factors Pnt and Ets21C not only up-regulate cell cycle genes, but also a large set of genes used for mitochondrial biogenesis, the TCA cycle, oxidative phosphorylation, and fatty acid oxidation. Many of these genes are bound by Pnt and/or Ets21C, indicating direct regulation. EGFR signaling-dependent gene expression is sufficient to increase both uptake and usage of sugars and fatty acids, a combination that can in principle enhance the synthesis of nucleotides and amino acids without throttling energy supplies (ATP, NADH), thus potentiating anabolic cell growth. Our findings align well with results reported by Mundorf et al., who also assessed Ets21C target genes in Drosophila midgut18, as well as studies of ErbB signaling in the mouse intestine6.
Early studies of proliferation in cultured cells show that growth factors trigger a stepwise temporal program of gene expression that culminates in DNA replication and, later, mitosis55. Transcriptional activation of primary responder (“immediate early”) genes relies on pre-existing receptors and transcriptional regulators and can occur without de novo protein synthesis, whereas secondary responder (“delayed early”) genes require new expression of additional transcription factors55–58. To assess this growth factor stimulation process in a physiological context, we assayed ISC transcriptomes after 8h and 24h of sSpi induction. We found that many more genes were affected after 24h, indicating that the transcriptional output of EGFR signaling is dynamic. In Drosophila ISCs, Cic appears to act as an central pre-existing primary transcription regulator controlled directly by ERK phosphorylation13. PntP2, which can be activated by phosphorylation, may share this function. PntP1 and Ets21C appear to function as required “immediate early” responders that activate downstream target genes. Both the Pnt- and Ets21C-regulated transcriptomes shared a high degree of overlap with the transcriptional profile triggered by sSpi, confirming the centrality of Pnt and Ets21C to EGFR signaling transcriptional output.
EGFR signaling is essential for ISC maintenance and proliferation in Drosophila, mice, and humans. In the mammalian gut, ligands for the EGFR/ErbB-type receptors are secreted by cells in the crypt-localized stem cell niche, namely Paneth cells and the subepithelial mesenchyme7,8. EGFR is expressed in ISCs and transit amplifying cells (TA)8,9, whereas ErbB2, and ErbB3 are expressed throughout the crypt-villus axis6. These receptors are required for crypt basal cell proliferation5,6. Consistent with our results, mammalian ISCs deprived of EGFR/ErbB signaling downregulate metabolic processes such as glycolysis, oxidative phosphorylation, and cholesterol metabolism5. NRG1, a neuregulin type ligand that activates ErbB2/ErbB3, is the principal driver of proliferation in mouse intestinal crypts6. Mammalian ISCs maintain a mitochondrial state distinct from their differentiated progeny. They express high levels of the mitochondrial biogenesis genes TFAM and NRF-1, and have higher mitochondria content, membrane potential, and PDK1 expression than differentiated intestinal epithelial cells59. This aligns with our proposal that proliferative ISCs and TAs require mitochondrial biogenesis and high levels of mitochondrial activity to maintain their functions in gut epithelial regeneration. It is also noteworthy that RAS signaling promotes mitochondrial biogenesis in Schwann cells and that, consistent with our results, this requires ERK signaling60.
We found that EGFR signaling promoted mitochondrial biogenesis, fatty acid oxidation, the TCA cycle, and oxidative phosphorylation by up-regulating ETS factor target genes, activating Drosophila ISCs from quiescence to cycling. Glucose and fatty acid uptake were also increased by EGFR activation, and ATP and NADH levels were maintained even as stored lipids and sugars were depleted. A similar increase in OXPHOS gene expression, mitochondrial content and activity has been observed in muscle stem cells transitioning from quiescence to an adaptive state for rapid re-entry into the cell cycle61. In Drosophila ISCs this up-regulation is evidently required, since impairing mitochondrial biogenesis by knockdown of either mtTFB2 or TFAM caused ISC arrest. We find it notable that mtTFB2, a target of EGFR signaling and Pnt, was not only required, but also sufficient to stimulate a modest amount of ISC proliferation when overexpressed. This suggests that the metabolic changes resulting from EGFR activation may be instructive for ISC activation.
Our metabolomics data from EGFR-activated intestines showed depletions of sugars, glycolysis intermediates and fatty acids, despite increased uptake of glucose and fatty acid, suggesting that glycolysis and fatty acid β-oxidation were both activated by EGFR signaling. Hence we suggest that, following EGFR activation, glycolysis may be more heavily utilized for generating metabolic intermediates required for rapid cell proliferation, namely nucleotides for nucleic acid synthesis and amino acids for growth, whereas fatty acid β-oxidation may be preferentially used to provide acetyl-CoA to the TCA cycle to provide the energy intermediates NADH and ATP. A recent study using genetic tools to visualize ATP/ADP ratios in vivo in ISCs reported that ATP levels decreased transiently during rapid ISC proliferation, but rapidly rebounded when ISCs returned to quiescence62. Another study highlighted an ISC-specific requirement for lipolysis63. These results support the notion that rapid ISC proliferation increases the demand not only for biosynthesis, but also for energy, and therefore requires a re-structuring of metabolism that favors fatty acid catabolism. Future studies of how mitochondrial fuel selection and metabolite flux change as cells get activated for proliferation should prove interesting.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Bruce A. Edgar (bruce.edgar@hci.utah.edu).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
RNA-seq data and Dam-seq data have been deposited at GEO and are publicly available, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181583.
All original code used for Dam-seq analysis and image processing is available from the lead contact upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
We maintained Drosophila melanogaster stocks on standard cornmeal food at 18°C. Fly stocks used in this study are listed in the key resources table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rat anti-HA | Roche | 3F10 |
| rabbit anti-PH3 | Millipore Sigma | 06–570 |
| chicken anti-GFP | Invitrogen | A10262 |
| mouse anti-Pnt | Christian Klambt lab | N/A |
| Bacterial and virus strains | ||
| Pseudomonas entomophila | Edgar lab | N/A |
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Phusion DNA polymerases | Thermo Scientific | F530 |
| Collagenase I | Gibco | 17018029 |
| Propidium Iodide | Invitrogen | P1304MP |
| Hoechst 33342 | Thermo Scientific | 62249 |
| MitoTracker Deep Red | Invitrogen | M22426 |
| Tetramethylrhodamine | Invitrogen | T668 |
| 2NB-DG | Invitrogen | N13195 |
| Etomoxir | Cayman Chemicals | 11969 |
| human EGF | PeproTech | AF-100–15 |
| Binimetinib | LC Laboratories | N/A |
| MitoView™ 650 | Biotium | 70075 |
| JC-1 | Invitrogen | T3168 |
| Critical commercial assays | ||
| Arcturus picoPure RNA Isolation Kit | Applied Biosystems | Cat#12204–01 |
| QBT™ Fatty Acid Uptake Assay Kit | Molecular Devices | PN:R8132 |
| QIAamp DNA Micro Kit | Qiagen | Cat#56304 |
| Illumina TruSeq Stranded mRNA Kit | Illumina | 20020594 |
| TruSeq RNA Single Indexes Set A | Illumina | 20020492 |
| TruSeq DNA PCR-Free Sample Preparation kit | Illumina | FC-121–3001 |
| Deposited data | ||
| mRNA Sequence, DamID Sequence, and analyzed sequencing data | This paper | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181583 |
| Experimental models: Cell lines | ||
| Human RPE-1 cell | ATCC | CRL-4000 |
| Experimental models: Organisms/strains | ||
| D. melanogaster: esgts: esg-Gal4/Cyo; tubGal80ts UAS-GFP/TM6B | Bruce Edgar Lab | BAE418 |
| D. melanogaster: esgts F/O: esg-Gal4 tubGal80ts UAS-GFP/Cyo; UAS-flp>CD2>Gal4/TM6B | Bruce Edgar Lab | BAE419 |
| D. melanogaster: esg ts ; Su(H)-Gal80: esg-Gal4 UAS-2XEYFP; Su(H)GBE-Gal80 tub-Gal80 ts | Bruce Edgar Lab | CZ16 |
| D. melanogaster: W 1118 | Bloomington DSC | BL3605 |
| D. melanogaster: UAS-sSpi | Bloomington DSC | BL63134 |
| D. melanogaster: UAS-PntP1 | Bloomington DSC | BL869 |
| D. melanogaster: UAS-PntP2 | Bloomington DSC | BL399 |
| D. melanogaster: UAS-Ets21C-PC | FlyORF | F000624 |
| D. melanogaster: UAS-Krn | FlyORF | F002754 |
| D. melanogaster: UAS-Egfr-RNAi | Bloomington DSC | BL31525 |
| D. melanogaster: UAS-Pnt-RNAi | Bloomington DSC | BL31936 |
| D. melanogaster: UAS-Pnt-RNAi | Bloomington DSC | BL35808 |
| D. melanogaster: UAS-Ets21C-RNAi | VDRC | KK103211 |
| D. melanogaster: UAS-MEK-RNAi | VDRC | KK107276 |
| D. melanogaster: UAS-stg-RNAi (II) | Kyoto Stock Center | 1395R-1 |
| D. melanogaster: UAS-mtTFB2-RNAi | Bloomington DSC | BL27055 |
| D. melanogaster: UAS-TFAM-RNAi 1 64 | Kyoto Stock Center | 4217R-2 |
| D. melanogaster: UAS-TFAM-RNAi 2 | VDRC | 37819 |
| D. melanogaster: UAS-TFAM-RNAi 3 | VDRC | 10719 |
| D. melanogaster: UAS-mito-HA-GFP (II) | Bloomington DSC | BL8442 |
| D. melanogaster: UAS-mito-HA-GFP (III) | Bloomington DSC | BL8443 |
| D. melanogaster: UAS-Dam-Ets21C-PC | This paper | CZ39 |
| D. melanogaster: UAS-Dam-PntP1 | This paper | JYH182 |
| D. melanogaster: UAS-Dam-PntP2 | This paper | JYH183 |
| D. melanogaster: UAS-Ets21C-PB (II) | This paper | CZ51 |
| D. melanogaster: UAS-Ets21C-PC (II) | This paper | CZ22 |
| D. melanogaster: UAS-UAS-mtTFB2-Flag-HA | This paper | CZ229 |
| Oligonucleotides | ||
| For Dam-PntP1 vector, PntP1 start: CCTC GAG ATGCCGCCCTCTGCGTTTTT. Templet cDNA from Drosophila midgut | This paper | N/A |
| For Dam-PntP2 vector, PntP2 start: CCTC GAG ATGGAATTGGCGATTTGTAAAAC. Templet: cDNA from Drosophila midgut | This paper | N/A |
| For Dam-PntP1 and Dam-PntP2 vectors, Pnt stop: CTCT AGA CTAATCCACATCTTTTTTCTCAA. Templet: cDNA from Drosophila midgut | This paper | N/A |
| For Dam-Ets21C-PC vector, Ets21C-PC start: CCTC GAG ATGGCCATTCTACAGAATAGCC. Templet: cDNA from Drosophila midgut | This paper | N/A |
| For Dam-Ets21C-PC vector, Ets21C stop: CTCT AGA TCAGTTGAATGCATTTGTGGTG. Templet: cDNA from Drosophila midgut | This paper | N/A |
| For UAS-Ets21C-PB vector, Ets21C PB start: ATGAGCGTCAGCGTGGACGTG. Templet: cDNA from Drosophila midgut | This paper | N/A |
| For UAS-Ets21C-PC vector, Ets21C PC start: ATGGCCATTCTACAGAATAGCC. Templet: cDNA from Drosophila midgut | This paper | N/A |
| For UAS-Ets21C-PB and Dam-Ets21C-PC vectors, Ets21C rev (without stop codon): GTTGAATGCATTTGTGGTGG. Templet: cDNA from Drosophila midgut | This paper | N/A |
| For Mito-DNA content assay, Ets21C Forward: CCCTGACTATCTCGGGTGAA | This paper | N/A |
| For Mito-DNA content assay, Ets21C Reverse: CACTTCACTTTGGCCCTGTT | This paper | N/A |
| For Mito-DNA content assay, β-tub56D Forward: ACATCCCGCCCCGTGGTC | This paper | N/A |
| For Mito-DNA content assay, β-tub56D Reverse: AGAAAGCCTTGCGCCTGAACATAG | This paper | N/A |
| For Mito-DNA content assay, CO1 Forward: TGACTTCTACCTCCTGCTCT | This paper | N/A |
| For Mito-DNA content assay, CO1 Reverse: GCAATTCCAGCGGATAGAGG | This paper | N/A |
| For Mito-DNA content assay, CO3 Forward: TCACAGAAGTTTATCACCCGC | This paper | N/A |
| For Mito-DNA content assay, CO3 Reverse: TGGTGGGCTCAAGTTACAGT | This paper | N/A |
| Recombinant DNA | ||
| The UAS controlled overexpression vector for mtTFB2: UAS-mtTFB2-Flag-HA | Drosophila Genetics Resource Center | UFO03038 |
| Software and algorithms | ||
| Prism | GraphPad | SCR_002798 |
| Fiji | https://imagej.net | SCR_002285 |
| Bowtie 2 | http://bowtie-bio.sourceforge.net/ | SCR_016368 |
| STAR | https://github.com/alexdobin/STAR | SCR_004463 |
| HTSeq | https://pypi.org/project/HTSeq/ | SCR_005514 |
| Bedtools | https://github.com/arq5x/bedtools2 | SCR_006646 |
| R | www.r-project.org/ | SCR_001905 |
| Python | www.python.org/ | SCR_008394 |
| GSEA | www.gsea-msigdb.org/ | SCR_003199 |
| MetaboAnalyst | www.metaboanalyst.ca/ | SCR_015539 |
| Other | ||
METHOD DETAILS
Generation of transgenic flies
UAS-Dam-PntP1, UAS-Dam-PntP2, and UAS-Dam-Ets21C-PC were generated in our lab. Total RNA was extracted from adult fly midgut and converted to cDNA. The cDNA of each TF was cloned and inserted into the pUAST attB-LT3-NDam vector (gift from Andrea Brand lab)35 and then integrated into the attB site on the Drosophila third chromosome.
Primers for UAS-Dam-PntP1, UAS-Dam-PntP2, and UAS-Dam-Ets21C-PC cloning are in KEY RESOURCES TABLES.
UAS-Ets21C-PB and UAS-Ets21C-PC also have versions generated in our lab. Ets21C-PC and Ets21C-PB coding sequence without stop codon was amplified from cDNA library prepared from midgut by using Phusion DNA polymerases (Thermo Fisher Scientific). Poly-A tails were added to the purified PCR products via Tag polymerase (Thermo Fisher Scientific) at 72°C for 10min. Then the A-tailed PCR products were sub-cloned to pCRTM8/GW/TOPO vector (Invitrogen). Through LR recombination reaction, they were inserted into pUASg-attB-HA vector65. For the insertion, the constructs were prepared with Qiagen Maxi prep kit, sent to Genetic Services, and injected into attp2 line to generate transgenic flies.
Primers for UAS-Ets21C-PB and UAS-Ets21C-PC cloning are in KEY RESOURCES TABLES.
UAS-mtTFB2-Flag-HA was generated using UFO03038 vector from Drosophila Genetics Resource Center BDGP Tagged ORF Collection. This vector has the mtTFB2 cDNA followed by one Flag and one HA tag on the C terminal and was inserted in the attP2 site of third chromosome. The injection and selection were done by BestGene Inc.
Cell-type specific mRNA Seq
Cell type specificity was achieved by using an ISC-specific temperature-sensitive driver (esgts; Su(H)-Gal80). The over-expression of ectopic Pnt, Ets21C, or sSpi was spatio-temporally controlled by esgts; Su(H)-Gal80 system. With this system, UAS driven transgene only expresses in esg positive and Su(H) negative ISCs and only at temperatures above 29°C. For this experiment young female flies were selected three days post-eclosion and shifted to 29°C for 8 or 24 hours, allowing ectopic protein expression in ISCs. In the control group, for each replicate, 120 guts were dissected right after induction. Due to EGFR signaling activated guts having more ISCs than controls, around 90 guts were dissected in each experimental group to get an equal number of ISC. Experimental groups had the same genetic background, sex, and treatment with control groups. After 1h collagenase digestion, ISCs were sorted by flow cytometry and collected into lysis buffer with RNAse inhibitor, as previously described34. YFP positive and Propidium Iodide (PI) negative single small cells were collected. Total RNA from ISC was extracted using Arcturus picoPure RNA Isolation Kit (Cat.No. 12204–01) in an RNAse free environment. All of the total RNA samples had an RNA Integrity Number (RIN) greater than 8.5. Library preparation had been carried out by Illumina TruSeq Stranded mRNA Kit according to its protocol. This kit used oligo-dT beads to capture and enrich RNA species containing a polyA tail. Sequencing was performed by Illumina High Seq 2500, with a single-end (1 × 50 cycle) high output run mode.
Cell-type specific DamID Seq
DamID-Seq (DNA Adenine Methyltransferase Identification via Sequencing) was used to identify the binding profiles of Ets21C and Pnt on the ISC genome. Dam protein from E.coli can methylate the N6-position of adenine in the sequence GATC of the eukaryote genome. Adenine methylation is common across bacterial phyla, but is mostly absent in eukaryotic cells. In the Drosophila genome GATC sites occur on average every 200–300bp66. In this experiment, Dam protein was fused with PntP1, PntP2, and Ets21C-PC isoforms at the N-terminus. The cDNA of each TF was inserted into the pUAST attB-LT3-NDam vector (from Andrea Brand lab) and then integrated into the attB site on the Drosophila third chromosome35. This transgene is expressed at a very low level when Gal4 is present due to a non-coding primary ORF introduced in front of Dam-TF fusion ORF, which causes the ejection of ribosomes from the mRNA35. In our experiment, Dam-TF fusion proteins or Dam control protein were only expressed in ISC at a very low dose for 24h to achieve specific DamID without toxicity, thus avoiding damage-dependent EGFR activation. For all of the TFs, the binding profiles were revealed under physiological condition. Dam or Dam-TF fused proteins were expressed for 24 hours in ISC of newly enclosed females. Around 100 midguts were dissected right after Dam or Dam-TF induction for DNA extraction. For each experiment, there were at least three biological replicates for Dam control and Dam-TF fusion proteins. The previously described DamID protocol was used to extract genomic DNA, process and amplify methylated DNA fragments67. Libraries for DNA sequencing were prepared by TruSeq DNA PCR-Free Sample Preparation kit (Ilumina FC-121–3001), following its protocol.
Flow cytometry
Midguts were dissected in nuclease free cold PBS. While dissecting, hindgut and malpighian tubules were carefully removed from the midgut. Midguts were transferred to 1.5 ml Eppendorf tube with 500 µl nuclease/RNAse free PBS on ice. Collagenase I (1:1000 of 100U/µl stock solution, Gibco 17018029) and EDTA (final concentration of 2mM) were then added and samples were incubated for 1 hour at 29°C in thermomixer with 450 rpm. Samples were vigorously pipetted 30 times using a cut tip every 15 mins to help dissociation. After dissociation, samples were centrifuged at 600 g for 10 mins. Supernatant was discarded and 500 µl of cold fresh nuclease free PBS with Propidium Iodide (PI) (Invitrogen™ P1304MP) were added. Cells were re-suspended and pipetted onto a 40um strainer cap FACS tube, tapping gently to help flowing through. BD FACSAria cell sorter was used to sort and collect YFP positive and PI negative single ISC.
For cell cycle analysis, Hoechst dye 10μM (Thermo Scientific™ 62249) added to the dissociation buffer was used to quantify DNA content. Since ISC did not easily incorporate Hoechst, a high concentration was necessary. For mitochondria volume and activity, MitoTracker Deep Red FM 100nM (Invitrogen™ M22426) and Tetramethylrhodamine 40nM (Invitrogen™ T668) were used to stain mitochondria. These dyes were added in the dissociation buffer and incubated with the cells for 60 mins.
For glucose uptake assay, 2NB-DG (Invitrogen™, catalog number: N13195) at working concentration 250μM was added into the dissociation buffer and incubated with the cells for 30mins and Hoechst 10μM for 60mins. For fatty acid uptake assay, cells were incubated with Hoechst 10μM for 60mins while dissociating, then with BODIPY (QBT™ Fatty Acid Uptake Assay Kit from Molecular Devices) 1:10 of the loading buffer suggested by its protocol for 5 mins after dissociation. Cells were centrifuged to remove BODIPY buffer, then re-suspended for FACS. We reduced the loading concentration and incubation time of BODIPY, since the accumulation was quick and the fluorescent intensity was high in the cells. CytoFLEX analyzer was used to perform the cell cycle, mitochondria activity, glucose and fatty acid uptake assays.
Immunofluorescence staining
Female adult flies were dissected in cold PBS. Midguts were fixed in PBS with 4% paraformaldehyde (PFA) at room temperature for 30 minutes or 4°C overnight. Midguts were then washed in PBS with 0.1% Triton x-100 (PBST) for 3×10 minutes each. Samples were blocked in PBST with 3% BSA and 10% Normal Goat Serum (NGS) for 30 min at room temperature. All samples were incubated with primary antibodies overnight at the following dilutions: rat anti-HA (1:500; Roche 3F10), rabbit anti-PH3 (1:1000, Millipore 06–570), chicken anti-GFP (1:1000, Invitrogen™ A10262), and mouse anti-Pnt (1:500, a gift from Christian Klambt lab)30. After washing 3×10 minutes in PBST, samples were incubated with secondary antibodies for at least 2 hours at room temperature at a dilution of 1:1000. DNA was stained with DAPI (0.1mg/ml, Sigma), diluted 1:200.
Mito-DNA content assays
Real Time quantitative PCR (RT-qPCR) was used to determine the ratio between Mito-DNA and Chromatin-DNA. This ratio reflects mitochondrial DNA copy number in a cell. YFP positive ISCs were dissociated and isolated from freshly dissected female midguts by FACS. Total DNA, including mitochondrial DNA, was extracted from ISC using QIAamp DNA Micro Kit, according to its protocol. The total DNA sample was diluted to desired concentration for RT-qPCR. Two pairs of primers targeting genomic DNA and two pairs of primers targeting mitochondrial DNA were used. Primers are listed below:
Ets21C Forward: CCCTGACTATCTCGGGTGAA
Ets21C Reverse: CACTTCACTTTGGCCCTGTT
PRODUCT SIZE: 143bp. This pair of primers targeting 5’UTR region of Ets21C and will not recognize UAS:Ets21C-HA (FlyORF F000624) in the Ets21C OE flies.
β-tub56D Forward: ACATCCCGCCCCGTGGTC
β-tub56D Reverse: AGAAAGCCTTGCGCCTGAACATAG
PRODUCT SIZE: 120bp. Last exon of β-Tub56D.
CO1 Forward: TGACTTCTACCTCCTGCTCT
CO1 Reverse: GCAATTCCAGCGGATAGAGG
PRODUCT SIZE: 104bp
CO3 Forward: TCACAGAAGTTTATCACCCGC
CO3 Reverse: TGGTGGGCTCAAGTTACAGT
PRODUCT SIZE: 141bp
For data analysis, the two genomic DNA genes were considered as controls, since the copy number of genomic genes should be consistent in all ISC except those in S, G2, and early M phase of cell cycle. Delta delta Ct analysis was used to calculate the relative content of mitoDNA in comparison to the W1118 control.
Metabolomics and lipidomics
Flies were dissected in cold PBS and their explanted intestines flash frozen in liquid nitrogen. For each replicate, 100 or 40 intestines were collected for metabolomics and lipidomics analyses, respectively.
For metabolite extraction, each sample was transferred to 2.0 mL ceramic bead mill tubes (Qiagen Catalog Number 13116–50). 450 μL of cold 90% methanol (MeOH) solution containing the internal standard d4-succinic acid (Sigma 293075) was added to each sample. The samples were then homogenized in an OMNI Bead Ruptor 24. Homogenized samples were then incubated at −20 ˚C for 1 hr. After incubation the samples were centrifuged at 20,000 x g for 10 minutes at 4 °C. 400 μl of supernatant was then transferred from each bead mill tube into a labeled, fresh microcentrifuge tubes. Another internal standard, d27-myristic acid, was then added to each sample. Pooled quality control samples were made by removing a fraction of collected supernatant from each sample. Process blanks were made using only extraction solvent and went through the same process steps as actual samples. Everything was then dried en vacuo.
All GC-MS analysis was performed with an Agilent 7200 GC-QTOF fit with an Agilent 7693A automatic liquid sampler. Dried samples were suspended in 40 µL of a 40 mg/mL O-methoxylamine hydrochloride (MOX) (MP Bio #155405) in dry pyridine (EMD Millipore #PX2012–7) and incubated for one hour at 37 °C in a sand bath. 25 µL of this solution was added to auto sampler vials. 60 µL of N-methyl-N-trimethylsilyltrifluoracetamide (MSTFA with 1%TMCS, Thermo #TS48913) was added automatically via the auto sampler and incubated for 30 minutes at 37 °C. After incubation, samples were vortexed and 1 µL of the prepared sample was injected into the gas chromatograph inlet in the split mode with the inlet temperature held at 250 °C. A 10:1 split ratio was used for analysis for the majority of metabolites. Any metabolites that saturated the instrument at the 10:1 split were analyzed at a 50:1 split ratio. The gas chromatograph had an initial temperature of 60 °C for one minute followed by a 10 °C/min ramp to 325 °C and a hold time of 10 minutes. A 30-meter Agilent Zorbax DB-5MS with 10 m Duraguard capillary column was employed for chromatographic separation. Helium was used as the carrier gas at a rate of 1 mL/min. Below is a description of the two-step derivatization process used to convert non-volatile metabolites to a volatile form amenable to GC-MS.
Sample extraction protocol for lipidomics based on Matyash et al. J Lipid Res 49(5) (2008) 1137–1146.
Lipid extracts were separated on an Acquity UPLC CSH C18 column (2.1 × 100 mm; 1.7 µm) coupled to an Acquity UPLC CSH C18 VanGuard precolumn (5 × 2.1 mm; 1.7 µm) (Waters, Milford, MA) maintained at 65 °C connected to an Agilent HiP 1290 Sampler, Agilent 1290 Infinity pump, and Agilent 6545 Accurate Mass Q-TOF dual AJS-ESI mass spectrometer (Agilent Technologies, Santa Clara, CA). Samples were analyzed in a randomized order in both positive and negative ionization modes in separate experiments acquiring with the scan range m/z 100 – 1700. For positive mode, the source gas temperature was set to 225 °C, with a drying gas flow of 11 L/min, nebulizer pressure of 40 psig, sheath gas temp of 350 °C and sheath gas flow of 11 L/min. VCap voltage is set at 3500 V, nozzle voltage 500V, fragmentor at 110 V, skimmer at 85 V and octopole RF peak at 750 V. For negative mode, the source gas temperature was set to 300 °C, with a drying gas flow of 11 L/min, a nebulizer pressure of 30 psig, sheath gas temp of 350 °C and sheath gas flow 11 L/min. VCap voltage was set at 3500 V, nozzle voltage 75 V, fragmentor at 175 V, skimmer at 75 V and octopole RF peak at 750 V. Mobile phase A consisted of ACN:H2O (60:40 v/v) in 10 mM ammonium formate and 0.1% formic acid, and mobile phase B consisted of IPA:ACN:H2O (90:9:1 v/v/v) in 10 mM ammonium formate and 0.1% formic acid. For negative mode analysis the modifiers were changed to 10 mM ammonium acetate. The chromatography gradient for both positive and negative modes started at 15% mobile phase B then increased to 30% B over 2.4 min, it then increased to 48% B from 2.4 – 3.0 min, then increased to 82% B from 3 – 13.2 min, then increased to 99% B from 13.2 – 13.8 min where it’s held until 16.7 min and then returned to the initial conditions and equilibrated for 5 min. Flow was 0.4 mL/min throughout, with injection volumes of 1 µL for positive and 10 µL negative mode. Tandem mass spectrometry was conducted using iterative exclusion, the same LC gradient at collision energies of 20 V and 27.5 V in positive and negative modes, respectively.
Etomoxir treatment
Etomoxir was mixed with freshly made fly food (Bloomington Formulation) at a final concentration of 25μM. Flies were fed with Etomoxir 36h prior to dissection, and were flipped into flesh Etomoxir food every 12 hours.
Transmission electron microscopy
Posterior midguts were dissected and fixed in the fixative solution [2.5% Glutaraldehyde, 1.25% Paraformaldehyde, and 0.03% picric acid in 0.1 M sodium cacodylate buffer (pH 7.4)] at 4°C overnight, washed in 0.1M cacodylate buffer and postfixed with 1% osmiumtetroxide (OsO4) in 1.5% potassium ferrocyanide (KFeCN6) for 1 hour, washed 2 times in water, 1 time in Maleate buffer (MB) and incubated in 1% uranyl acetate in MB for 1 hour followed by 2 washes in water and subsequent dehydration in grades of alcohol (10min each; 50%, 70%, 90%, 2×100%).
The samples were then incubated in propylene oxide for 30 minutes and infiltrated in a 1:1 mixture of propylene oxide and TAAB Epon (Marivac Canada Inc. St. Laurent, Canada). The following day the samples were embedded in TAAB Epon and polymerized at 60°C for 48 hr. Ultrathin epoxy resin sections (∼60nm) were cut on a Reichert Ultracut-S microtome, placed on copper grids, and contrasted with 0.3% lead citrate. Grids with the resin sections were imaged using a JEOL 1200-EX transmission electron microscope operating at 80 kV with an AMT 2k CCD camera.
RPE-1 live cell imaging
Human RPE-1 cells were seeded on µ-Slide 8 Well (iBiDi 80826), 1.2×105 cells / well. RPE-1 cells were grown in complete medium (DMEM F12, 10% FBS, 50U/mL Penicillin-Streptomycin, and 0.01 mg/ml hygromycin B) for 6h until fully adherent. Medium was then changed to starvation medium (DMEM F12, 50U/mL Penicillin-Streptomycin, and 0.01 mg/ml hygromycin B) for 20h to synchronize the cells in G1 phase. Following arrest, human EGF (50ng/ml), MEK inhibitor (Binimetinib 500nM), or both were added to basal medium for 24h. For Fig. 7A-7C, 2μM Hoechst and 100nM MitoView™ 650 (Biotium Catalog no. 70075), 2μg/mL JC-1 Dye (Invitrogen™ T3168) were used to stain nuclei and mitochondria for 30mins prior to live-imaging. For Fig. 7D-7F, 2μM Hoechst and 100nM MitoTracker Deep Red FM (Invitrogen™ M22426) and 100nM Tetramethylrhodamine (Invitrogen™ T668) were used to stain nuclei and mitochondria for 30 mins prior to live-imaging.
QUANTIFICATION AND STATISTICAL ANALYSIS
mRNA Seq
Samples were aligned to recent genome assembly (BDGP 6.94) with STAR. Aligned reads were then assigned to genes using HTSeq with mode “Union”. Raw counts were then used for a Differential Expression Analysis using the DESeq2 algorithm. Genes having absolute Log2 Fold Change > 0.5 and adjusted p-value < 0.05 were then selected as significantly differently expressed.
Gene ontology enrichment analysis
Gene Ontology biological process terms enrichment analyses were conducted using the R package topGO68 and the Drosophila melanogaster annotation file released on 2019–07-01 from the Gene Ontology Consortium website.
DamID-Seq
For each sequencing sample, using Bowtie 2, raw reads were aligned to the Drosophila genome. Aligned reads with a quality score ≥ 30 were extended 5’ to 3’ to a length of 350bp and mapped to the genome subdivided into 75bp contiguous fragments. The number of reads mapping to each fragment were quantified and statistically significant differential binding was computed with DESeq2, using default settings and a paired design between DAM-fusion and DAM-control samples, which reduced the noise from random DAM binding and PCR bias. Bound fragments with an adjusted p-value ≤ 0.01 and log2 Fold-Change between DAM-fusion and DAM-control samples > 0 were selected as significant. Adjacent significant fragments were then joined together into peaks, and their score recalculated as the log2 of the mean of each original log2 score elevated to the power of 2. Peaks were annotated using Bedtools Intersect command (“-wao” option) to Drosophila annotated genome (BDGP6.94) and to known cis regulatory modules experimentally defined in vivo (CRM) (RedFly 5.5.1), independently. Peaks falling outside of gene regions and associated CRM were extended 500bp on both 5’ and 3’ ends and re-annotated. Peaks annotated to the Drosophila annotated genome or the RedFly database were marked as “gene” and “regulatory region”, respectively, in the “Element Type” field of Data S2.
Venn diagrams
In-scale Venn diagrams were generated using the R package VennDiagram69. Significance of overlaps was calculated by hypergeometric test.
Dot-plots
Significantly enriched Gene Ontology terms were plotted using the R package ggplot2 (https://ggplot2.tidyverse.org/)70. Dot plots were based on the design from the R package clusterProfiler71.
Heatmaps
Heatmaps were generated using the heatmap.2 function from the R package gplots (https://cran.r-project.org/package=gplots). Raw reads were converted to Transcripts Per Million (TPM), z-score normalized, manually grouped and ordered, and plotted without unsupervised clustering.
GSEA
Gene Set Enrichment Analyses were performed using the algorithm from the official website (http://www.gsea-msigdb.org/)72. Genes from RNASeq data were ranked according to their log2 fold-change and tested for enrichment of specific metabolic gene sets downloaded from the KEGG database.
LC-MS Data Processing
For data processing, Agilent MassHunter (MH) Workstation and software packages MH Qualitiative and MH Quantitative were used. The pooled QC (n=8) and process blank (n=4) were injected throughout the sample queue to ensure the reliability of acquired lipidomics data. For lipid annotation, accurate mass and MS/MS matching was used with the Agilent Lipid Annotator library and LipidMatch (See Ref below). Results from the positive and negative ionization modes from Lipid Annotator were merged based on the class of lipid identified. Data exported from MH Quantitative was evaluated using Excel where initial lipid targets are parsed based on the following criteria. Only lipids with relative standard deviations (RSD) less than 30% in QC samples are used for data analysis. Additionally, only lipids with background AUC counts in process blanks that are less than 30% of QC are used for data analysis. The parsed excel data tables are normalized based on the ratio to class-specific internal standards, then to tissue mass and sum prior to statistical analysis.
Metabolomics and lipidomics data analysis
Peaks data was analyzed using the MetaboAnalyst software. Log2 fold-changes were calculated on raw peaks values. Prior to statistical analysis, raw peaks were log10-transformed, then, for each metabolite, peaks were mean-centered and divided by the square root of their standard deviation (Pareto scaling).
Immunofluorescence staining image analysis
All images were processed using ImageJ. Cell sizes were measured using a freehand selection tool to outline individual cells. Similarly, pixel intensity was measured as the average within the selection.
PH3 counts statistical analysis
PH3+ cells were manually counted. Each dot indicates the PH3 count for a single intestines. All analyses were performed using Prism 9 (GraphPad Software, San Diego, CA). Statistical significance was assessed using t-test, with appropriate corrections and modifications for multiple testing between all samples (Tukey’s method) or between samples and a shared control (Dunnett’s test).
Supplementary Material
Data S1: mRNA Seq summary tables, including mRNA Seq results of PntP1 24h, PntP2 24h, Ets21C-PC 8h, Ets21C-PC 24h, sSpi 8h, and sSpi 24h. Related to Figure 2, Figure 3, Figure S2, Figure S3, and Figure S4.
Data S2: DamID binding peaks summary tables, including PntP1, PntP2, and Ets21C-PC DamID Seq results. Related to Figure 2, Figure 3, Figure 6B, Figure S2, and Figure S3.
Data S3: Full list of enriched Gene Ontology Biological Processes terms of PntP1, PntP2 and, Ets21C 24h up- or down-regulated genes. Related to Figure 2.
Data S4: Full list of enriched Gene Ontology Biological Processes terms of sSpi early/consistent/late, up- or down-regulated genes. Related to Figure 3.
Data S5: metabolomics, lipidomics, and targeted metabolomics data of sSpi overexpression vs control. Related to Figure 4.
Supplementary Figures S1-S6 and Supplementary references
Table S1: Lists of direct gene targets of PntP1, PntP2, and Ets21C-PC. Related to Figure 2, Figure S2, and Figure S3.
Highlights.
EGFR signaling promotes intestinal stem cell (ISC) growth, division, and mitochondrial biogenesis
Mitochondrial biogenesis is required for EGFR-dependent ISC proliferation
Uptake of glucose and fatty acids by ISCs are increased upon EGFR activation
EGFR signaling upregulates β-oxidation to increase mitochondrial membrane potential
ACKNOWLEDGEMENTS
This work was supported by the Huntsman Cancer Foundation and National Institute of Health Grant R01 GM124434, R35 GM140900 (to B.A.E.) and P30 CA042014. We thank C. Klambt, and A. Brand for reagents and the Kyoto, FlyORF and Bloomington Drosophila Stock Centers for Drosophila. We thank the Huntsman Cancer Institute High-Throughput Genomics and Bioinformatic Analysis Shared Resources, and the University of Utah Metabolomics, Flow Cytometry, Cell Imaging and Electron Microscopy Cores for assistance. Our Metabolomics Core was supported by NCRR Shared Instrumentation Grants 1S10OD016232–01, 1S10OD018210–01A1 and 1S10OD021505–01. Our Flow Cytometry Core was supported by NIH director’s office Award S10OD026959 and NCI Award 5P30CA042014–24.
Inclusion and diversity
One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTEREST
The authors declare no competing interests.
SUPPLEMENTARY INFORMATION
REFERENCES
- 1.Biteau B, and Jasper H (2011). EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development 138, 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchon N, Broderick NA, Kuraishi T, and Lemaitre B (2010). DrosophilaEGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biology 8, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang H, Grenley MO, Bravo M-J, Blumhagen RZ, and Edgar BA (2011). EGFR/Ras/MAPK Signaling Mediates Adult Midgut Epithelial Homeostasis and Regeneration in Drosophila. Cell Stem Cell 8, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu N, Wang SQ, Tan D, Gao Y, Lin G, and Xi R (2011). EGFR, Wingless and JAK/STAT signaling cooperatively maintain Drosophila intestinal stem cells. Developmental Biology 354, 31–43. [DOI] [PubMed] [Google Scholar]
- 5.Basak O, Beumer J, Wiebrands K, Seno H, van Oudenaarden A, and Clevers H (2017). Induced Quiescence of Lgr5+ Stem Cells in Intestinal Organoids Enables Differentiation of Hormone-Producing Enteroendocrine Cells. Cell Stem Cell 20, 177–190.e4. [DOI] [PubMed] [Google Scholar]
- 6.Jardé T, Chan WH, Rossello FJ, Kaur Kahlon T, Theocharous M, Kurian Arackal T, Flores T, Giraud M, Richards E, Chan E, et al. (2020). Mesenchymal Niche-Derived Neuregulin-1 Drives Intestinal Stem Cell Proliferation and Regeneration of Damaged Epithelium. Cell Stem Cell 27, 646–662.e7. [DOI] [PubMed] [Google Scholar]
- 7.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, and Clevers H (2011). Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki A, Sekiya S, Gunshima E, Fujii S, and Taniguchi H (2010). EGF signaling activates proliferation and blocks apoptosis of mouse and human intestinal stem/progenitor cells in long-term monolayer cell culture. Lab Invest 90, 1425–1436. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y-P, Ma H, Starchenko A, Huh WJ, Li W, Hickman FE, Zhang Q, Franklin JL, Mortlock DP, Fuhrmann S, et al. (2017). A Chimeric Egfr Protein Reporter Mouse Reveals Egfr Localization and Trafficking In Vivo. Cell Rep 19, 1257–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiang J, Bandura J, Zhang P, Jin Y, Reuter H, and Edgar BA (2017). EGFR-dependent TOR-independent endocycles support Drosophila gut epithelial regeneration. Nature Communications 8, 15125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schweitzer R, Shaharabany M, Seger R, and Shilo BZ (1995). Secreted Spitz triggers the DER signaling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes Dev 9, 1518–1529. [DOI] [PubMed] [Google Scholar]
- 12.Slupsky CM, Gentile LN, Donaldson LW, Mackereth CD, Seidel JJ, Graves BJ, and McIntosh LP (1998). Structure of the Ets-1 pointed domain and mitogen-activated protein kinase phosphorylation site. Proc Natl Acad Sci U S A 95, 12129–12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin Y, Ha N, Forés M, Xiang J, Gläßer C, Maldera J, Jiménez G, and Edgar BA (2015). EGFR/Ras Signaling Controls Drosophila Intestinal Stem Cell Proliferation via Capicua-Regulated Genes. PLoS Genet 11, e1005634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Liu S, and Cai Y (2015). EGFR/MAPK Signaling Regulates the Proliferation of Drosophila Renal and Nephric Stem Cells. Journal of Genetics and Genomics 42, 9–20. [DOI] [PubMed] [Google Scholar]
- 15.Liu M, Lim TM, and Cai Y (2010). The Drosophila female germline stem cell lineage acts to spatially restrict DPP function within the niche. Sci Signal 3, ra57. [DOI] [PubMed] [Google Scholar]
- 16.Xie Y, Li X, Zhang X, Mei S, Li H, Urso A, and Zhu S (2014). The Drosophila Sp8 transcription factor Buttonhead prevents premature differentiation of intermediate neural progenitors. eLife 3, e03596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie Y, Li X, Deng X, Hou Y, O’Hara K, Urso A, Peng Y, Chen L, and Zhu S (2016). The Ets protein Pointed prevents both premature differentiation and dedifferentiation of Drosophila intermediate neural progenitors. Development 143, 3109–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mundorf J, Donohoe CD, McClure CD, Southall TD, and Uhlirova M (2019). Ets21c Governs Tissue Renewal, Stress Tolerance, and Aging in the Drosophila Intestine. Cell Reports 27, 3019–3033.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park JM, Brady H, Ruocco MG, Sun H, Williams D, Lee SJ, Kato T, Richards N, Chan K, Mercurio F, et al. (2004). Targeting of TAK1 by the NF-κB protein Relish regulates the JNK-mediated immune response in Drosophila. Genes Dev 18, 584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boutros M, Agaisse H, and Perrimon N (2002). Sequential Activation of Signaling Pathways during Innate Immune Responses in Drosophila. Developmental Cell 3, 711–722. [DOI] [PubMed] [Google Scholar]
- 21.Toggweiler J, Willecke M, and Basler K (2016). The transcription factor Ets21C drives tumor growth by cooperating with AP-1. Scientific Reports 6, 34725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Külshammer E, Mundorf J, Kilinc M, Frommolt P, Wagle P, and Uhlirova M (2015). Interplay among Drosophila transcription factors Ets21c, Fos and Ftz-F1 drives JNK-mediated tumor malignancy. Disease Models & Mechanisms 8, 1279–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dissanayake K, Toth R, Blakey J, Olsson O, Campbell DG, Prescott AR, and MacKintosh C (2011). ERK/p90RSK/14–3-3 signalling has an impact on expression of PEA3 Ets transcription factors via the transcriptional repressor capicúa. Biochemical Journal 433, 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad ST, Rogers AD, Chen MJ, Dixit R, Adnani L, Frankiw LS, Lawn SO, Blough MD, Alshehri M, Wu W, et al. (2019). Capicua regulates neural stem cell proliferation and lineage specification through control of Ets factors. Nat Commun 10, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weissmann S, Cloos PA, Sidoli S, Jensen ON, Pollard S, and Helin K (2018). The Tumor Suppressor CIC Directly Regulates MAPK Pathway Genes via Histone Deacetylation. Cancer Res 78, 4114–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B, Krall EB, Aguirre AJ, Kim M, Widlund HR, Doshi MB, Sicinska E, Sulahian R, Goodale A, Cowley GS, et al. (2017). ATXN1L, CIC, and ETS Transcription Factors Modulate Sensitivity to MAPK Pathway Inhibition. Cell Reports 18, 1543–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simón-Carrasco L, Jiménez G, Barbacid M, and Drosten M (2018). The Capicua tumor suppressor: a gatekeeper of Ras signaling in development and cancer. Cell Cycle 17, 702–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Zeng X, Ryoo HD, and Jasper H (2014). Integration of UPRER and Oxidative Stress Signaling in the Control of Intestinal Stem Cell Proliferation. PLOS Genetics 10, e1004568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klambt C (1993). The Drosophila gene pointed encodes two ETS-like proteins which are involved in the development of the midline glial cells. Development 117, 163–176. [DOI] [PubMed] [Google Scholar]
- 30.Brunner D, Dücker K, Oellers N, Hafen E, Scholzi H, and Klambt C (1994). The ETS domain protein Pointed-P2 is a target of MAP kinase in the Sevenless signal transduction pathway. Nature 370, 386–389. [DOI] [PubMed] [Google Scholar]
- 31.O’Neill EM, Rebay I, Tjian R, and Rubin GM (1994). The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78, 137–147. [DOI] [PubMed] [Google Scholar]
- 32.Shwartz A, Yogev S, Schejter ED, and Shilo B-Z (2013). Sequential activation of ETS proteins provides a sustained transcriptional response to EGFR signaling. Development 140, 2746–2754. [DOI] [PubMed] [Google Scholar]
- 33.Dutta D, Dobson AJ, Houtz PL, Gläßer C, Revah J, Korzelius J, Patel PH, Edgar BA, and Buchon N (2015). Regional Cell-Specific Transcriptome Mapping Reveals Regulatory Complexity in the Adult Drosophila Midgut. Cell Reports 12, 346–358. [DOI] [PubMed] [Google Scholar]
- 34.Dutta D, Xiang J, and Edgar BA (2013). RNA expression profiling from FACS-isolated cells of the Drosophila intestine. Curr Protoc Stem Cell Biol 27, Unit 2F.2. [DOI] [PubMed] [Google Scholar]
- 35.Southall TD, Gold KS, Egger B, Davidson CM, Caygill EE, Marshall OJ, and Brand AH (2013). Cell-Type-Specific Profiling of Gene Expression and Chromatin Binding without Cell Isolation: Assaying RNA Pol II Occupancy in Neural Stem Cells. Developmental Cell 26, 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plotnik JP, Budka JA, Ferris MW, and Hollenhorst PC (2014). ETS1 is a genome-wide effector of RAS/ERK signaling in epithelial cells. Nucleic Acids Res 42, 11928–11940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Webber JL, Zhang J, Massey A, Sanchez-Luege N, and Rebay I (2018). Collaborative repressive action of the antagonistic ETS transcription factors Pointed and Yan fine-tunes gene expression to confer robustness in Drosophila. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamamouna V, Rahman MM, Petersson M, Charalambous I, Kux K, Mainor H, Bolender V, Isbilir B, Edgar BA, and Pitsouli C (2021). Remodelling of oxygen-transporting tracheoles drives intestinal regeneration and tumorigenesis in Drosophila. Nat Cell Biol 23, 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perochon J, Yu Y, Aughey GN, Medina AB, Southall TD, and Cordero JB (2021). Dynamic adult tracheal plasticity drives stem cell adaptation to changes in intestinal homeostasis in Drosophila. Nat Cell Biol 23, 485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harden N (2017). New Insights from Drosophila into the Regulation of EGFR Signaling. Methods Mol Biol 1652, 37–42. [DOI] [PubMed] [Google Scholar]
- 41.Jin Y, Xu J, Yin M-X, Lu Y, Hu L, Li P, Zhang P, Yuan Z, Ho MS, Ji H, et al. (2013). Brahma is essential for Drosophila intestinal stem cell proliferation and regulated by Hippo signaling. eLife 2, e00999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anchan RM, Reh TA, Angello J, Balliet A, and Walker M (1991). EGF and TGF-alpha stimulate retinal neuroepithelial cell proliferation in vitro. Neuron 6, 923–936. [DOI] [PubMed] [Google Scholar]
- 43.Leschey KH, Hackett SF, Singer JH, and Campochiaro PA (1990). Growth factor responsiveness of human retinal pigment epithelial cells. Invest. Ophthalmol. Vis. Sci 31, 839–846. [PubMed] [Google Scholar]
- 44.Defoe DM, and Grindstaff RD (2004). Epidermal growth factor stimulation of RPE cell survival: contribution of phosphatidylinositol 3-kinase and mitogen-activated protein kinase pathways. Experimental Eye Research 79, 51–59. [DOI] [PubMed] [Google Scholar]
- 45.Jiang Q, Zhou C, Bi Z, and Wan Y (2006). EGF-Induced Cell Migration Is Mediated by ERK and PI3K/AKT Pathways in Cultured Human Lens Epithelial Cells. Journal of Ocular Pharmacology and Therapeutics 22, 93–102. [DOI] [PubMed] [Google Scholar]
- 46.Jäger S, Handschin C, St-Pierre J, and Spiegelman BM (2007). AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci U S A 104, 12017–12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson RM, Barger JL, Edwards MG, Braun KH, O’Connor CE, Prolla TA, and Weindruch R (2008). Dynamic regulation of PGC-1alpha localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell 7, 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandez-Marcos PJ, and Auwerx J (2011). Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr 93, 884S–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C, Li Z, Lu Y, Du R, Katiyar S, Yang J, Fu M, Leader JE, Quong A, Novikoff PM, et al. (2006). Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. PNAS 103, 11567–11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baltzer C, Tiefenböck SK, Marti M, and Frei C (2009). Nutrition Controls Mitochondrial Biogenesis in the Drosophila Adipose Tissue through Delg and Cyclin D/Cdk4. PLOS ONE 4, e6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Z-F, Drumea K, Mott S, Wang J, and Rosmarin AG (2014). GABP Transcription Factor (Nuclear Respiratory Factor 2) Is Required for Mitochondrial Biogenesis. Molecular and Cellular Biology 34, 3194–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adán C, Matsushima Y, Hernández-Sierra R, Marco-Ferreres R, Fernández-Moreno MÁ, González-Vioque E, Calleja M, Aragón JJ, Kaguni LS, and Garesse R (2008). Mitochondrial Transcription Factor B2 Is Essential for Metabolic Function in Drosophila melanogaster Development. J. Biol. Chem 283, 12333–12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, and Edgar BA (2009). Cytokine/Jak/Stat Signaling Mediates Regeneration and Homeostasis in the Drosophila Midgut. Cell 137, 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Litonin D, Sologub M, Shi Y, Savkina M, Anikin M, Falkenberg M, Gustafsson C, and Temiakov D (2010). Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro. J. Biol. Chem, jbc.C110.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pardee AB (1989). G1 events and regulation of cell proliferation. Science 246, 603–608. [DOI] [PubMed] [Google Scholar]
- 56.Cochran BH, Reffel AC, and Stiles CD (1983). Molecular cloning of gene sequences regulated by platelet-derived growth factor. Cell 33, 939–947. [DOI] [PubMed] [Google Scholar]
- 57.Hendrickson SL, and Scher CD (1983). Platelet-derived growth factor-modulated translatable mRNAs. Mol Cell Biol 3, 1478–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelly K, Cochran BH, Stiles CD, and Leder P (1983). Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell 35, 603–610. [DOI] [PubMed] [Google Scholar]
- 59.Schell JC, Wisidagama DR, Bensard C, Zhao H, Wei P, Tanner J, Flores A, Mohlman J, Sorensen LK, Earl CS, et al. (2017). Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nat. Cell Biol 19, 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Echave P, Machado-da-Silva G, Arkell RS, Duchen MR, Jacobson J, Mitter R, and Lloyd AC (2009). Extracellular growth factors and mitogens cooperate to drive mitochondrial biogenesis. Journal of Cell Science 122, 4516–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodgers JT, King KY, Brett JO, Cromie MJ, Charville GW, Maguire KK, Brunson C, Mastey N, Liu L, Tsai C-R, et al. (2014). mTORC1 controls the adaptive transition of quiescent stem cells from G 0 to G Alert. Nature 510, 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morris O, Deng H, Tam C, and Jasper H (2020). Warburg-like Metabolic Reprogramming in Aging Intestinal Stem Cells Contributes to Tissue Hyperplasia. Cell Reports 33, 108423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh SR, Zeng X, Zhao J, Liu Y, Hou G, Liu H, and Hou SX (2016). The lipolysis pathway sustains normal and transformed stem cells in adult Drosophila. Nature 538, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Icreverzi A, de la Cruz AF, Van Voorhies WA, and Edgar BA (2012). Drosophila cyclin D/Cdk4 regulates mitochondrial biogenesis and aging and sensitizes animals to hypoxic stress. Cell Cycle 11, 554–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bischof J, Björklund M, Furger E, Schertel C, Taipale J, and Basler K (2013). A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development 140, 2434–2442. [DOI] [PubMed] [Google Scholar]
- 66.Steensel B. van, and Henikoff S (2000). Identification of in vivo DNA targets of chromatin proteins using tethered Dam methyltransferase. Nature Biotechnology 18, 424. [DOI] [PubMed] [Google Scholar]
- 67.Papagiannouli F, Schardt L, Grajcarek J, Ha N, and Lohmann I (2014). The Hox gene Abd-B controls stem cell niche function in the Drosophila testis. Dev Cell 28, 189–202. [DOI] [PubMed] [Google Scholar]
- 68.Alexa A, and Rahnenfuhrer J (2021). topGO: Enrichment Analysis for Gene Ontology (Bioconductor version: Release (3.12)) [Google Scholar]
- 69.Chen H, and Boutros PC (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis 2nd ed. (Springer International Publishing) [Google Scholar]
- 71.Yu G, Wang L-G, Han Y, and He Q-Y (2012). clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS: A Journal of Integrative Biology 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. PNAS 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1: mRNA Seq summary tables, including mRNA Seq results of PntP1 24h, PntP2 24h, Ets21C-PC 8h, Ets21C-PC 24h, sSpi 8h, and sSpi 24h. Related to Figure 2, Figure 3, Figure S2, Figure S3, and Figure S4.
Data S2: DamID binding peaks summary tables, including PntP1, PntP2, and Ets21C-PC DamID Seq results. Related to Figure 2, Figure 3, Figure 6B, Figure S2, and Figure S3.
Data S3: Full list of enriched Gene Ontology Biological Processes terms of PntP1, PntP2 and, Ets21C 24h up- or down-regulated genes. Related to Figure 2.
Data S4: Full list of enriched Gene Ontology Biological Processes terms of sSpi early/consistent/late, up- or down-regulated genes. Related to Figure 3.
Data S5: metabolomics, lipidomics, and targeted metabolomics data of sSpi overexpression vs control. Related to Figure 4.
Supplementary Figures S1-S6 and Supplementary references
Table S1: Lists of direct gene targets of PntP1, PntP2, and Ets21C-PC. Related to Figure 2, Figure S2, and Figure S3.
Data Availability Statement
RNA-seq data and Dam-seq data have been deposited at GEO and are publicly available, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181583.
All original code used for Dam-seq analysis and image processing is available from the lead contact upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


