Abstract
Prenatal opioid exposure is associated with significantly adverse medical, developmental, and behavioral outcomes in offspring, though the underlying mechanisms driving these impairments are still unclear. Accumulating evidence implicates gut microbial dysbiosis as a potential modulator of these adverse effects. However, how opioid exposure during pregnancy alters the maternal and neonatal microbiome remain to be elucidated. Here, we utilize a murine model of brief hydromorphone exposure during pregnancy (gestation day 11-13; i.p.; 10mg/kg) to examine its impact on the maternal and neonatal microbiome. Fecal samples were collected at various timepoints in dams (4 days post hydromorphone exposure, birth, and weaning) and offspring (2, 3, and 5 weeks) to interrogate longitudinal changes in the microbiome. Stomach contents at 2 weeks were also collected as a surrogate for breastmilk and microbial analysis was performed using 16S rRNA sequencing. Alongside alterations in the maternal gut microbial composition, offspring gut microbiota exhibited distinct communities at 2 and 3 weeks. Furthermore, functional profiling of microbial communities revealed significant differences in microbial community-level phenotypes gram-negative, gram-positive, and potentially pathogenic in maternal and/or neonatal hydromorphone exposed groups compared with controls. We also observed differences in stomach microbiota in opioid-exposed vs non-exposed offspring, which suggests breast milk may also play a role in shaping the development of the neonatal gut microbiota. Together, we provide evidence of maternal and neonatal microbial dysbiosis provoked even with brief hydromorphone exposure during pregnancy.
Keywords: prenatal drug, pregnancy, opioids, gut microbiome; breastmilk microbiome; stomach microbiome; hydromorphone; high-throughput sequencing
Introduction
Paralleling the opioid epidemic, there has been a concomitant rise in neonates prenatally exposed to opioids. As of 2019, estimates stand at up to 6.6% of women reporting prescription opioid use during pregnancy (Ko et al. 2020). Opioids (such as hydromorphone, morphine, and oxycodone) and medications used to manage opioid use disorder (such as buprenorphine and methadone) cross the placenta, subjecting offspring to the adverse effects of prenatal opioid exposure (POE). For instance, neonatal opioid withdrawal syndrome (NOWS), which affects 21-94% of neonates exposed to opioids, is characterized by profound neonatal neurobehavioral dysregulation (Ross et al. 2015; McQueen and Murphy-Oikonen 2016). Additionally, growing evidence suggests that POE has detrimental effects on offspring that extend past the neonatal period with some evidence of behavioral disorders and motor and cognitive deficits in adolescence and early adulthood, though research in this field is lacking (Ornoy et al. 2001; Hunt et al. 2008; McQueen and Murphy-Oikonen 2016; Nygaard et al. 2017; Abu and Roy 2021). In particular, there is an urgent need to investigate potential mechanisms driving adverse effects of POE, alongside developmental consequences.
The microbiome has emerged as a key player in modulating health and disease, particularly in pregnancy. Maternal exposures to antibiotics (Gonzalez-Perez et al. 2016), high fat diet (Xie et al. 2018), infection (Nyangahu and Jaspan 2019), metabolic and gastrointestinal diseases (Wang et al. 2018b; Torres et al. 2020), and various environmental exposures (Zhang et al. 2020) are associated with intestinal dysbiosis during pregnancy. Accumulating evidence has implicated maternal microbial dysbiosis in promoting atopic, gastrointestinal, immunological and neuropsychiatric dysfunction in offspring, potentially through the development of neonatal intestinal dysbiosis (McKeever et al. 2002; Bruce-Keller et al. 2017; Schulfer et al. 2018; Xie et al. 2018; Jašarević and Bale 2019).
There is compelling evidence that maternal exposure to opioids during pregnancy may cause maternal and neonatal intestinal dysbiosis. We have previously shown that opioid exposure causes microbial dysbiosis in adult mice (Meng et al. 2013; Banerjee et al. 2016; Wang et al. 2018a; Zhang et al. 2019). Importantly, this highlights the fact that women of reproductive age taking opioids may have dysbiotic gut microbiomes, which they may maintain throughout pregnancy, though this has never been shown. Here we explore how maternal opioid exposure during pregnancy impacts the maternal and neonatal gut microbiome using a model of brief hydromorphone exposure.
Methods
Experimental design:
All experimental procedures were performed according to the guidelines of the Institutional Animal Care and Use Committee at University of Miami, Miller School of Medicine. Female 10–12-week C57 BL/6 mice were mated by housing two females and an adult male per cage. Following confirmation of a copulation plug (gestation day 0 (G0) of pregnancy), males were removed. Dams received an intraperitoneal injection of either saline or hydromorphone (10mg/kg, once daily) from G11-G13. This period overlaps with the beginning of gastrointestinal development in the growing fetus and supports investigations of brief opioid exposure during sensitive periods of time in pregnancy. The dose was chosen to balance viability of pups with an abuse model of brief opioid exposure. To determine longitudinal changes in the maternal gut microbiome following brief opioid exposure, large intestine (LI) samples were collected from dams at G17 (4 days post last hydromorphone/saline injection), birth and weaning. To assess alterations in the neonatal microbiome, pups were sacrificed 2 (pre-weaning), 3 (weaning), or 5 (post-weaning) weeks after birth. Large intestinal samples (collected at all time points) and stomach contents (collected in offspring at 2 weeks) as a proxy for breastmilk were subjected to microbiome analysis. Male and female offspring were included in analysis, but this study was not powered to detect sex-based differences.
16S rRNA gene sequencing:
DNA was isolated using DNeasy 96 PowerSoil Pro QIAcube HT Kit with QIAcube HT liquid-handling machine (Qiagen, Maryland, USA). Sequencing was performed by the University of Minnesota Genomics Center. The hypervariable regions V4 region of 16S rRNA gene was PCR amplified using the forward primer 515F (GTGCCAFCMGCCGCGGTAA), reverse primer 806R (GGACTACHVGGGTWTCTAAT). The amplicons were sequenced with the Illumina MiSeq v.3 platform, generating 300-bp paired-end reads.
Bioinformatic analysis:
Demultiplexed raw sequence reads were filtered and clustered into amplicon sequence variants (ASVs) with the DADA2 package (Callahan et al. 2016). QIIME2 (Bolyen et al. 2019) pipeline was used for diversity analyses and taxonomic assignment. LDA Effect Size (LEfSe) (Segata et al. 2011) was used to detect differentially enriched taxa across groups. The threshold on the logarithmic LDA score for discriminative features was set to 2. BugBase (Ward et al. 2017) software was used to predict high-level microbial phenotypes. MicrobiomeAnalyst (Chong et al. 2020) was used for graphic visualization. SourceTracker (Knights et al. 2011) was used to predict the source of microbial communities of the large intestine of 2-week-old offspring from known (offspring stomach contents or maternal gut samples at weaning) and unknown environments.
Statistical analysis:
Kruskal-Wallis test was used to detect if α diversity differed across treatments. Permutational multivariate analysis of variance (PERMANOVA) was used to detect if β diversity differed across treatments.
Results
Impact of POE on gut and stomach microbiota diversity
After quality control of raw sequencing, 1252 unique ASVs were identified among all samples (Supplemental Table S1). Prenatal opioid exposure (POE) did not change α diversity in terms of richness (measured by Faith’s Phylogenetic Diversity) or evenness (measured by Pielou’s Evenness) during the first 3 weeks of life in offspring LI samples (Fig 1A). However, POE offspring displayed higher richness than controls at 5 weeks, q = 0.038 (Fig 1A). No effect on α diversity was observed in dams 4 days post (4dp) last hydromorphone/saline treatment (G17), at birth, or at weaning (Fig 1A). Weighted UniFrac was used to calculate distance for β diversity and principal-coordinate analysis (PCoA) plots was used for visualization of principal-coordinate analysis (PCoA) plots (Fig 1B–D). Brief hydromorphone treatment during pregnancy had a significant impact on β diversity in both the maternal and offspring microbiome. Microbial composition of fecal samples from POE mothers was significantly different from control mothers 4dp last hydromorphone/saline treatment (q < 0.01) and at parturition (q = 0.014) (Fig 1D). However, there was no significant difference between POE and control dams at the time of weaning (q = 0.2) (Fig 1D). Among the fecal samples from offspring, the microbial composition in POE offspring was significantly different from controls at 2 weeks (q = 0.01) and 3 weeks (q< 0.001) (Fig 1B). However, at 5 weeks, the difference was no longer significant (q= 0.059), though to some extent, distinct clustering was observed (Fig 1B). Stomach samples were also collected from 2-week-old offspring as a surrogate for the maternal breast milk microbiome. Similar to fecal samples, there was no change in α diversity between the POE and control stomach microbiomes (Fig 1A). Notably, in terms of β diversity, stomach samples from POE mice were significantly different from control mice (q = 0.02) (Fig 1C).
Figure 1.
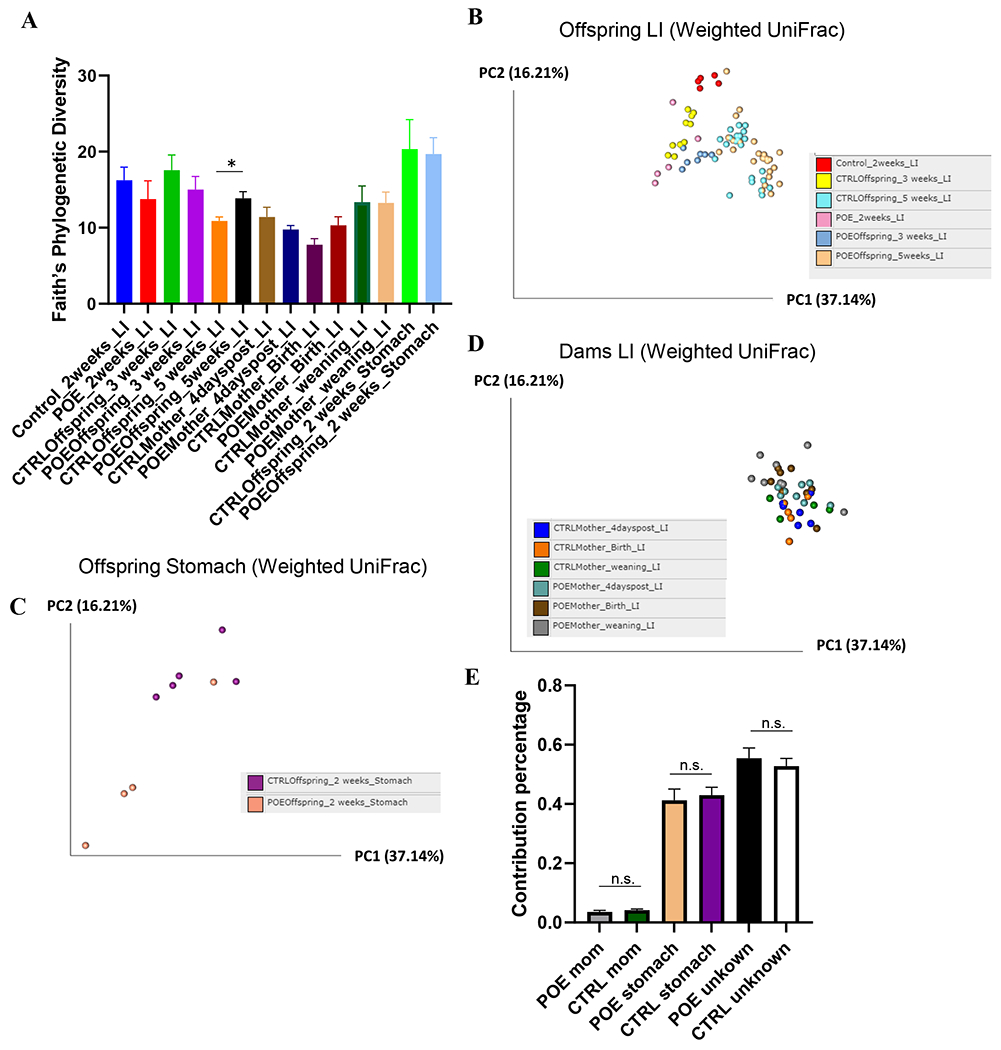
Analysis of α and β-diversity of samples. Maternal fecal samples include CTRLMom_4dp (n=6), POEMom_4dp (n=8), CTRLMom_Bir (n=5), POEMom_Bir (n=8), CTRLMom_wean (n=4), and POEMom_wean (n=8). Offspring fecal samples include CTRL_2wk (n=5), POE_2wk (n=5), CTRLOff_3wk (n=10), POEOff_3wk (n=7), CTRLOff_5wk (n=22), POEOff_5wk (n=25). Offspring stomach samples include CTRLOff_2wk (n=5) and POEOff_2wk (n=5). (A) Faith Phylogenic Diversity (metric of α-diversity) at sequencing depth 20000. q > 0.05 among all groups, except in offspring at 5 weeks (q = 0.038). Principal coordinates analysis (PCoA) plot of weighted UniFrac distances (metric of β -diversity) in (B) offspring LI, (C) offspring stomach, and (D) maternal LI samples. (E) Sourcetracker estimates of the proportion of microbes in offspring stool identical to maternal stool or offspring stomach (surrogate for breastmilk). Error bars represent standard error of the mean (SEM). P>0.05 for all groups. CTRL, control; 4dp, 4 days post last hydromorphone or saline exposure (gestation day 17); Bir, birth; wean, weaning; Off, offspring; Sto, stomach; LI, large intestine; POE, prenatal opioid (hydromorphone) exposure.
Impact of POE on gut and stomach microbiota composition
LEfSe was used to identify differentially enriched microbial taxa from the phylum to the genus level in hydromorphone-exposed and unexposed mice (Fig 2).
Figure 2.
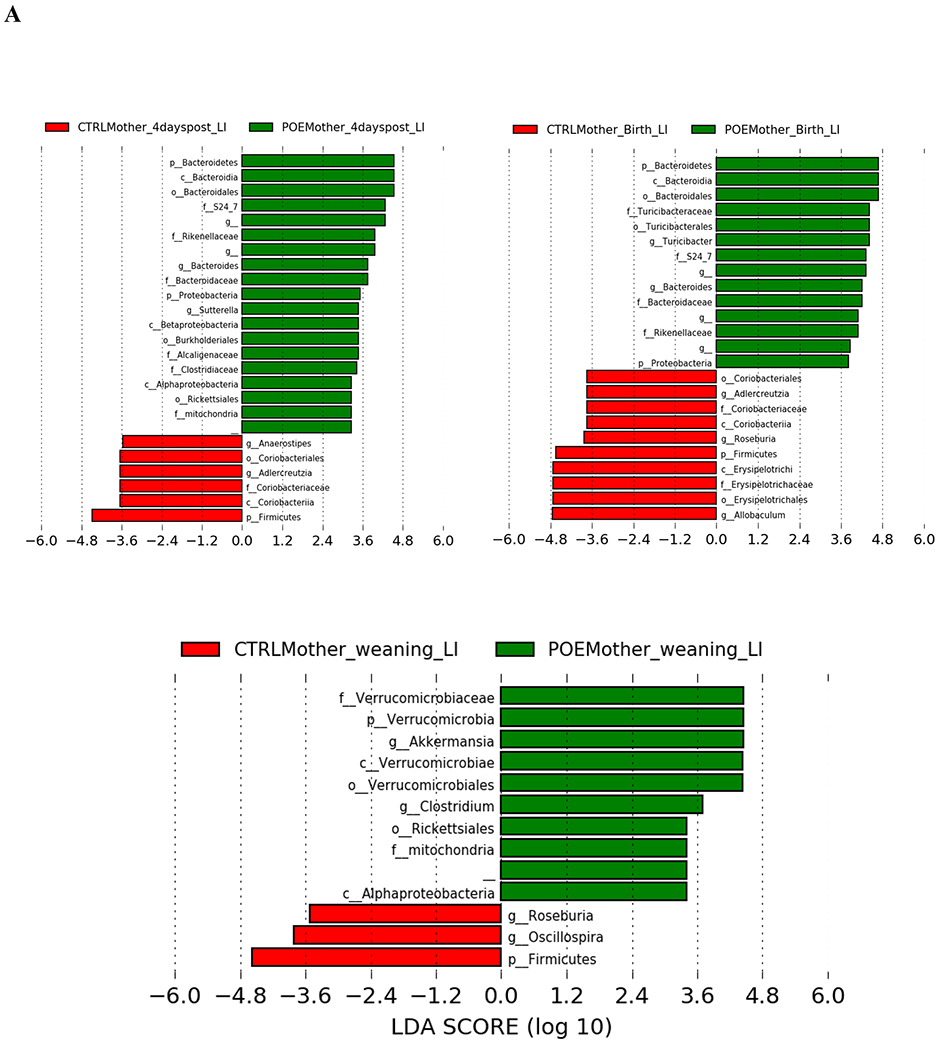
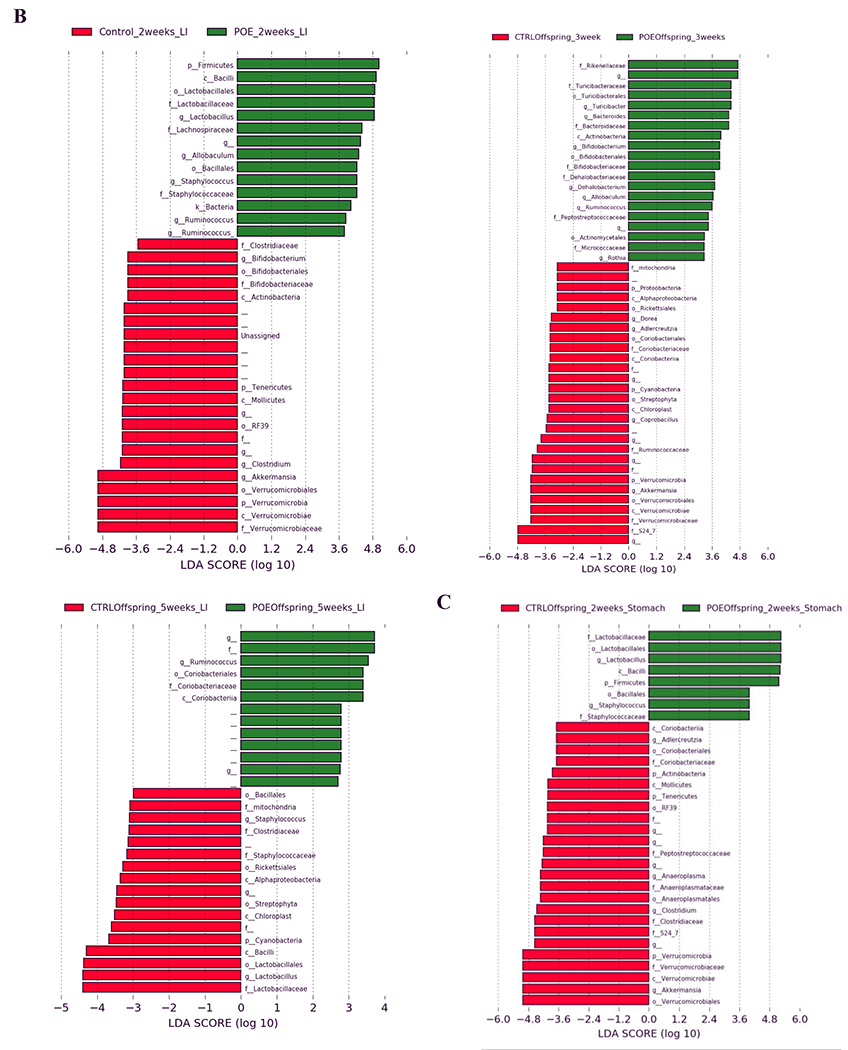
LEfSe (Linear Discriminant Analysis Effect Size) analysis of bacterial taxa among samples. Empty label names indicate unidentified taxa. (A) Top discriminative bacteria taxa between stool sample from hydromorphone exposed or control dams at the following timepoints: 4 days post last hydromorphone/saline exposure (gestation day 17), birth or weaning. (B) Top discriminative bacterial taxa between stool sample from prenatal hydromorphone exposed or control offspring at the following timepoints: 2, 3, 5 weeks. (C) Top discriminative bacterial taxa between stomach sample from prenatal hydromorphone exposed or control offspring at 2 weeks. CTRL, control; 4dp, 4 days post last hydromorphone or saline exposure (gestation day 17); Bir, birth; wean, weaning; Off, offspring; Sto, stomach; LI, large intestine; POE, prenatal opioid (hydromorphone) exposure.
Four days post last hydromorphone (HYD)/saline treatment, bacteria from phyla Bacteoidetes and Proteobacteria, and genera Bacteroides (from phylum Bacteoidetes) and Sutterella (from phylum Proteobacteria), were more abundant in fecal samples from HYD-treated dams (Fig 2A). Bacteria from phylum Firmicutes, genera Adlercreutzia (from phylum Actinobacteria), and genera Anaerostipes (from phylum Firmicutes) were more abundant from control dams (Fig 2A).
At the time of birth, bacteria from phyla Bacteoidetes and Proteobacteria, and genera Bacteroides (from phylum Bacteoidetes) and Turicibacter (from phylum Firmicutes), were more enriched in fecal samples from HYD-treated mothers (Fig 2A). Bacteria from phylum Firmicutes and genera Allobaculum (from phylum Firmicutes) and Roseburia (from phylum Firmicutes), were more enriched from control mothers (Fig 2A).
At the time of weaning, bacteria from phylum Verrucomicrobia, and genera Clostridium (from phylum Firmicutes) and Akkermansia (from phylum Verrucomicrobia) were more prevalent in fecal samples from HYD-treated mothers (Fig 2A). Bacteria from phylum Firmicutes and genera Oscillospira (from phylum Firmicutes) and Roseburia (from phylum Firmicutes), were more prevalent from control mothers (Fig 2A).
At two weeks, bacteria from phylum Firmicutes and genera Lactobacillus, Ruminococcus and Allobaculum (all genera from phylum Firmicutes), were more prevalent in fecal samples from POE offspring (Fig 2B). Bacteria from phyla Verrucomicrobia and Tenericutes, and genera Akkermansia (from phylum Verrucomicrobia), Clostridium (from phylum Firmicutes), and Bifidobacterium (from phylum Actinobacteria), were more prevalent from control offspring (Fig 2B).
At three weeks, bacteria from genera Turicibacter (from phylum Firmicutes), Bacteroides (from phylum Bacteoidetes), Bifidobacterium (from phylum Actinobacteria), Allobaculum (from phylum Firmicutes) and Dehalobacterium (from phylum Firmicutes) were more abundant in fecal samples from POE offspring (Fig 2B). Bacteria from phyla Verrucomicrobia and Proteobacteria; and genera Akkermansia (from phylum Verrucomicrobia), Coprobacillus (from phylum Firmicutes), Dorea (from phylum Firmicutes) and Adlercreutzia (from phylum Actinobacteria), were more abundant from control offspring (Fig 2B).
At five weeks, bacteria from genera Ruminococcus (from phylum Firmicutes) were more abundant in fecal samples from POE offspring (Fig 2B). Bacteria from genera Lactobacillus (from phylum Firmicutes) and Staphylococcus (from phylum Firmicutes) were more enriched from control offspring (Fig 2B).
Bacteria from genera Staphylococcus (from phylum Firmicutes) and Lactobacillus (from phylum Firmicutes) were more prevalent in stomach samples from POE offspring (Fig 2C). Genera Akkermansia (from phylum Verrucomicrobia), Clostridium (from phylum Firmicutes) and an unknown genus from family S24-7 (from phylum Bacteoidetes) were more prevalent in stomach samples from control offspring (Fig 2C).
To study the contribution of the maternal gut and breast milk microbiome on the gut microbiome in offspring, we utilized Sourcetracker to estimate the proportion of microbes contributed from known sources (maternal LI samples at weaning, offspring stomach contents at 2 weeks) and unknown sources (Fig 1E). The analysis of the gut microbiome from two-week old control pups showed that the maternal microbiome contributed 4.2 ± 0.8% to the pup gut microbiome, while stomach samples contributed 43 ± 5.9% (Fig 1E). The percentage contribution of the POE maternal microbiome to pups was 3.6 ± 1.1% while the contribution of stomach samples to pups is 41.2 ± 8.6 % (Fig 1E).
Impact of POE on gut and stomach microbiota function
Using BugBase, high level potential phenotypes (gram-negative, gram-positive, and potentially pathogenic) were predicted among opioid exposed and unexposed groups (Fig 3).
Figure 3.
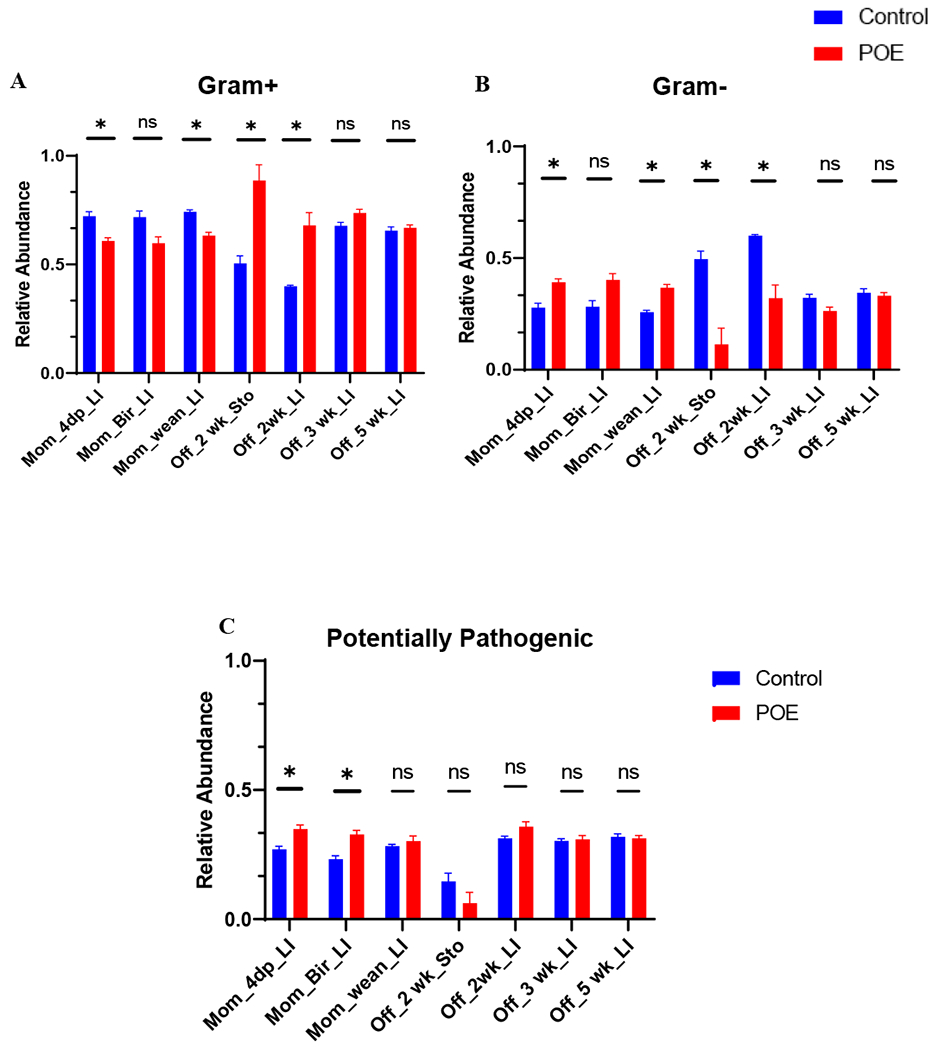
High-level phenotypic analysis predicted by BugBase software. Statistical analysis of predicted phenotypes displayed in Supplemental Figures 2–4. (A) predicted relative abundance of Gram-positive bacteria. P < 0.05 between POEMom_4dp_LI vs control, POEMom_wean_LI vs control, and POEOff_2wk_Sto vs. control, POEOff_2wk_LI vs control. (B) predicted relative abundance of Gram-negative bacteria. P < 0.05 between POEMom_4dp_LI vs control, POEMom_wean_LI vs control, and POEOff_2wk_Sto vs. control, POEOff_2wk_LI vs control. (C) predicted relative abundance of potentially pathogenic bacteria. P < 0.05 between POEMom_4dp_LI vs control, POEMom_bir_LI vs control. CTRL, control; 4dp, 4 days post last hydromorphone or saline exposure (gestation day 17); Bir, birth; wean, weaning; Off, offspring; Sto, stomach; LI, large intestine; POE, prenatal opioid (hydromorphone) exposure.
Four days post last HYD/saline treatment, gram-negative bacteria were significantly higher (p < 0.01) and gram-positive bacteria were significantly lower (p < 0.01) in samples from POE dams compared with control dams sampled (Fig 3A/B). Notably, there were more potentially pathogenic bacteria in POE dams at this timepoint (Fig 3C).
At the time of birth, there was no significant difference in gram-positive or gram-negative bacterial abundance (Fig 3A/B). However, there were more potentially pathogenic bacteria in the POE group (p < 0.01) (Fig 3C).
At the time of weaning, similar to that observed 4dp HYD treatment, gram-negative bacteria in POE maternal samples were significantly higher (p < 0.01), while gram-positive bacteria were significantly lower (p < 0.01) (Fig 3A/B). However, at this timepoint, there was no significance in pathogenic bacteria (p=0.2) (Fig 3C).
In 2-week-old offspring, gram-negative bacteria were significantly lower in LI and stomach samples from POE offspring than in control (p < 0.01) (Fig 3B). On the contrary, gram-positive bacteria were significantly higher (p < 0.01) in this group (Fig 3A). However, there was no difference (p > 0.1) in potentially pathogenic bacteria between control and POE 2-week-old offspring samples (Fig 3C). Additionally, there was no difference (p > 0.3) in gram-positive, gram-negative, or potentially pathogenic bacteria in 3-week or 5-week control and POE offspring sampled (Fig 3).
Discussion
Here, we show that brief hydromorphone exposure during pregnancy is sufficient to induce longitudinal changes in both the maternal and neonatal gut microbiota, with perturbations also observed in the stomach contents (as a proxy for breastmilk). We further show that the proportion of gram negative, gram positive, and potentially pathogenic phenotypes was significantly altered in hydromorphone-exposed dams, with gram positive species significantly increased and gram-negative species significantly decreased in 2-week-old offspring.
Our results further show that opioid exposure in dams does not have an impact on α diversity in terms of richness or evenness. Previous studies have shown no change or decreased α diversity with opioid treatment (Banerjee et al. 2016; Lee et al. 2018; Wang et al. 2018a); however, these studies were not conducted during pregnancy, and also utilized different paradigms of acute and chronic opioid exposure. Recently, using a ~60-day chronic methadone treatment, it was shown that prenatal and early postnatal opioid exposure resulted in increased α diversity in dams and offspring at weaning (Grecco et al. 2021). However, in our studies of brief opioid exposure, in offspring, POE did not affect α diversity during the first 3 weeks of life but did result in higher richness than controls at 5 weeks (q = 0.038).
The β and LEfSe analysis in our study did reveal significant differences in bacterial clades from phylum to species level between groups. Bacteria from phylum Bacteroidetes and Proteobacteria, genus Bacteroides were consistently enriched in maternal LI samples post HYD treatment to birth. The microbiome continued to change during weaning, whereby bacteria from phylum Verrucomicrobia, genera Clostridium (from phylum Firmicutes), and Akkermansia (from phylum Verrucomicrobia) were more prevalent. Consistently, one study found that Akkermansia was enriched in methadone-exposed dams at weaning (Grecco et al. 2021); however, in this study Lachnospiracea NK4A136 was more profoundly enriched in both dams and offspring at weaning, which we did not find at this timepoint (Grecco et al. 2021). Rather, while we show differential abundance of Lachnospiracea in 2-week-old POE offspring, this was not statistically significant at 3 or 5 weeks, per our LEfSe analyses. Indeed, multiple reports have shown that morphine has a direct impact on the gut microbiome composition at various levels in mice models (Banerjee et al. 2016; Lee et al. 2018; Sindberg et al. 2019; Zhang et al. 2019). For instance, consistent with our study, Sharma et al., 2020 reported Proteobacteria and Verrucomicrobia were more abundant in hydromorphone treated adulted male mice. However, we also discovered Firmicutes were less abundant in HYD treated mothers, whereas Sharma et al., 2020 did not find significant differences. Differences in reported gut microbial composition post hydromorphone exposure may be due to variation in sampling times, length/duration of opioid exposure, the use of males/females and the presence/absence of pregnancy. Furthermore, our sampling periods in mothers (e.g., 4dp last hydromorphone injection at G17) most likely resembles a gut microbiome during hydromorphone withdrawal, as dysbiosis induced by morphine treatment may occur as rapidly as 24 hours (Wang et al. 2018a). Additionally, our experimental regimen of hydromorphone injections may have subjected dams to withdrawal between injections, which may reflect in their microbiomes. Future studies may limit opioid withdrawal during pregnancy or probe earlier sampling timepoints. Nonetheless, to date, there have been no reported studies documenting how long dysbiosis can persist after discontinuation of opioids, which we have examined here with our longitudinal analyses.
In our studies, we also found significant differences in gut and stomach microbial composition in POE offspring. Bacteria from genera Ruminococcus (from phylum Firmicutes) and Allobaculum (from phylum Firmicutes) were overrepresented in POE pup gut microbiota at 2 and 3 weeks. At five weeks, Ruminococcus was still enriched even post-weaning. Interestingly, Staphylococcus and Lactobacillus were also enriched in both stomach and gut microbiota samples at 2 weeks. We further utilized the SourceTracker algorithm to probe contributions from 2-week-old stomach contents (as a surrogate of breastmilk) to offspring’s LI gut microbiota and found up to a 50% contribution. This is in agreement with LEfSe analysis, which found that Staphylococcus and Lactobacillus (both from phylum Firmicutes) were enriched in both stomach and gut microbiota samples at 2 weeks. Besides the contribution from breastmilk, around 45% of the taxa present in the offspring gut microbiota were from unknown sources, which indicate that other microbial sources such as the vagina, skin, oral or environmental microbiota may contribute to the development of the neonatal gut microbiota. For instance, Nyangahu et al., 2018 reported less than a 5% contribution from breastmilk and maternal gut combined compared with a 95% contribution from unknown sources (Nyangahu et al. 2018). These observed differences may be due to variations in animal strains, housing, or weaning conditions between studies. While mechanisms whereby maternal hydromorphone exposure contribute to changes in the neonatal microbiome are unknown, several theories have been posited and remain to be investigated. For instance, opioid exposure may alter the maternal gut metabolome, and these metabolites may indirectly influence the neonatal microbiome in utero at the maternal-fetal interface or postnatally through sources such as breastmilk via the enteromammary pathway (Nyangahu and Jaspan 2019). In line, in our model of brief hydromorphone exposure, changes in microbial composition of stomach contents likely represent an indirect effect of maternal hydromorphone exposure on breastmilk microbial composition; given the 2.3-hour termination elimination half-life of hydromorphone, hydromorphone and its metabolites (e.g., hydromorphone-3-glucuronide) are unlikely to be found in maternal circulation at birth and transmitted to offspring via breastmilk. Still, direct mechanisms by which in utero opioid exposure affect the fetus or maternal gut translocation to the placenta remain to be examined. Future studies that include cross-fostering or litter swapping to discern postnatal from prenatal influences will also be required to provide mechanistic insights.
At the microbial community level, the proportion of gram-positive, gram-negative, and potentially pathogenic bacteria were altered with hydromorphone exposure. A significant increase in gram-negative bacteria and a significant decrease in gram-positive bacteria were observed in dams 4dp last hydromorphone treatment and at weaning. Recent reports have correlated enrichment of gram-negative and potentially pathogenic to the increased abundance of Bacteroidetes, which we also observed (Han et al. 2019). Notably, this trend was inverted in the fecal and stomach samples of two-week-old POE offspring, which had significantly decreased gram-negative bacteria and increased gram-positive bacteria. Potentially pathogenic bacteria were elevated 4dp last hydromorphone treatment and birth in HYD-treated dams but were not significantly different at weaning or in offspring at any timepoint. Even though there was numerical decrease in the percentage of potential pathogens in POE-offspring compared to control-POE, the pairwise comparison failed to yield a significant result (p = 0.155) (Sup. Fig. 4). Why opposite trends in abundance of gram-negative and gram-positive bacteria exist in HYD treated dams and 2-week-old POE offspring remains to be investigated.
This work is the first to look at longitudinal changes in microbial diversity, composition, and function in dams and offspring after hydromorphone exposure and opens avenues for much needed work in this field. While there is no evidence for a causal relationship between brief hydromorphone exposure and intestinal flora imbalance in dams and neonates, our work provides a compelling association and is in line with several studies showing that maternal dysbiosis during pregnancy is associated with dysbiosis in offspring. This investigation did not look at microbial contributions from the genital tract and the breastmilk microbiome directly, which will be necessary to elucidate important contributions to the development of the neonatal gut microbiome. Sex-specific effects also need to be clarified. Future studies can also determine how the metabolome is impacted by brief hydromorphone exposure and the consequences of these changes in mothers and offspring. Importantly, further work will be required to determine how gut microbial dysbiosis contributes to known neurodevelopmental effects of prenatal opioid exposure, and how the gut microbiome in pre-exposed offspring responds to subsequent opioid challenges.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Grants (R01 DA043252, R01 DA037843, R01 DA044582, R01 DA047089 and R01 DA050542). Additionally, we thank Dr. Valerie Gramling from the University of Miami Writing Center for help with reading and revising this manuscript. Graphical abstract created with BioRender.com.
Footnotes
Conflict of Interest
All authors declare that they have no conflict of interest in this manuscript.
References
- Abu Y, Roy S (2021) Prenatal opioid exposure and vulnerability to future substance use disorders in offspring. Exp Neurol 339:113621. 10.1016/j.expneurol.2021.113621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Sindberg G, Wang F, et al. (2016) Opioid-induced gut microbial disruption and bile dysregulation leads to gut barrier compromise and sustained systemic inflammation. Mucosal Immunol 9:1418–1428. 10.1038/mi.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, et al. (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce-Keller AJ, Fernandez-Kim SO, Townsend RL, et al. (2017) Maternal obese-Type gut microbiota differentially impact cognition, anxiety and compulsive behavior in male and female offspring in mice. PLoS One 12:1–20. 10.1371/journal.pone.0175577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, et al. (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Liu P, Zhou G, Xia J (2020) Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc 15:799–821. 10.1038/s41596-019-0264-1 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez G, Hicks AL, Tekieli TM, et al. (2016) Maternal Antibiotic Treatment Impacts Development of the Neonatal Intestinal Microbiome and Antiviral Immunity. J Immunol 196:3768–3779. 10.4049/jimmunol.1502322 [DOI] [PubMed] [Google Scholar]
- Grecco GG, Gao Y, Gao H, et al. (2021) Prenatal opioid administration induces shared alterations to the maternal and offspring gut microbiome: A preliminary analysis. Drug Alcohol Depend 227:108914. 10.1016/j.drugalcdep.2021.108914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M-M, Zhu X-Y, Peng Y-F, et al. (2019) The alterations of gut microbiota in mice with chronic pancreatitis. Ann Transl Med 7:464–464. 10.21037/atm.2019.08.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt RW, Tzioumi D, Collins E, Jeffery HE (2008) Adverse neurodevelopmental outcome of infants exposed to opiate in-utero. Early Hum Dev 84:29–35. 10.1016/j.earlhumdev.2007.01.013 [DOI] [PubMed] [Google Scholar]
- Jašarević E, Bale TL (2019) Prenatal and postnatal contributions of the maternal microbiome on offspring programming. Front Neuroendocrinol 55:. 10.1016/j.yfrne.2019.100797 [DOI] [PubMed] [Google Scholar]
- Knights D, Kuczynski J, Charlson ES, et al. (2011) Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8:761–765. 10.1038/nmeth.1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JY, D’Angelo DV, Haight SC, et al. (2020) Vital Signs: Prescription Opioid Pain Reliever Use During Pregnancy — 34 U.S. Jurisdictions, 2019 . MMWR Morb Mortal Wkly Rep 69:897–903. 10.15585/mmwr.mm6928a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Vuong HE, Nusbaum DJ, et al. (2018) The gut microbiota mediates reward and sensory responses associated with regimen-selective morphine dependence. Neuropsychopharmacology 43:2606–2614. 10.1038/s41386-018-0211-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeever TM, Lewis SA, Smith C, Hubbard R (2002) The importance of prenatal exposures on the development of allergic disease: A birth cohort study using the West Midlands General Practice Database. Am J Respir Crit Care Med 166:827–832. 10.1164/rccm.200202-158OC [DOI] [PubMed] [Google Scholar]
- McQueen K, Murphy-Oikonen J (2016) Neonatal abstinence syndrome. N Engl J Med 375:2468–2479. 10.1056/NEJMra1600879 [DOI] [PubMed] [Google Scholar]
- Meng J, Yu H, Ma J, et al. (2013) Morphine Induces Bacterial Translocation in Mice by Compromising Intestinal Barrier Function in a TLR-Dependent Manner. PLoS One 8:. 10.1371/journal.pone.0054040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyangahu DD, Jaspan HB (2019) Influence of maternal microbiota during pregnancy on infant immunity. Clin Exp Immunol 198:47–56. 10.1111/cei.13331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyangahu DD, Lennard KS, Brown BP, et al. (2018) Disruption of maternal gut microbiota during gestation alters offspring microbiota and immunity. Microbiome 6:1–10. 10.1186/s40168-018-0511-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard E, Slinning K, Moe V, Walhovd KB (2017) Cognitive function of youths born to mothers with opioid and poly-substance abuse problems during pregnancy. Child Neuropsychol 23:159–187. 10.1080/09297049.2015.1092509 [DOI] [PubMed] [Google Scholar]
- Ornoy A, Segal J, Bar-Hamburger R, Greenbaum C (2001) Developmental outcome of school-age children born to mothers with heroin dependency: Importance of environmental factors. Dev Med Child Neurol 43:668–675. 10.1111/j.1469-8749.2001.tb00140.x [DOI] [PubMed] [Google Scholar]
- Ross EJ, Graham DL, Money KM, Stanwood GD (2015) Developmental consequences of fetal exposure to drugs: What we know and what we still must learn. Neuropsychopharmacology 40:61–87. 10.1038/npp.2014.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulfer AF, Battaglia T, Alvarez Y, et al. (2018) Intergenerational transfer of antibiotic-perturbed microbiota enhances colitis in susceptible mice. Nat Microbiol 3:234–242. 10.1038/s41564-017-0075-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, et al. (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J, Hu J, Seki A, et al. (2020) Infants born to mothers with IBD present with altered gut microbiome that transfers abnormalities of the adaptive immune system to germ-free mice. Gut 69:42–51. 10.1136/gutjnl-2018-317855 [DOI] [PubMed] [Google Scholar]
- Wang F, Meng J, Zhang L, et al. (2018a) Morphine induces changes in the gut microbiome and metabolome in a morphine dependence model. Sci Rep 8:1–15. 10.1038/s41598-018-21915-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zheng J, Shi W, et al. (2018b) Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut 67:1614–1625. 10.1136/gutjnl-2018-315988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T, Larson J, Meulemans J, et al. (2017) BugBase predicts organism-level microbiome phenotypes. bioRxiv [Google Scholar]
- Xie R, Sun Y, Wu J, et al. (2018) Maternal high fat diet alters gut microbiota of offspring and exacerbates dss-induced colitis in adulthood. Front Immunol 9:1–17. 10.3389/fimmu.2018.02608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Ren Q, Ma S, et al. (2020) Intergenerational transfer of Dechlorane Plus and the associated long-term effects on the structure and function of gut microbiota in offspring. Environ Int 141:105770. 10.1016/j.envint.2020.105770 [DOI] [PubMed] [Google Scholar]
- Zhang L, Meng J, Ban Y, et al. (2019) Morphine tolerance is attenuated in germfree mice and reversed by probiotics, implicating the role of gut microbiome. Proc Natl Acad Sci U S A 116:13523–13532. 10.1073/pnas.1901182116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


