ABSTRACT
Individuals often experience ailments such as allergies, asthma and respiratory tract infections throughout the year. Weather reports often include estimations of common allergens that can affect these individuals. To describe the local ‘atmospheric microbiome’ in Lubbock, Texas, USA, we examined the culturable fungal and bacterial microbiome present in the air on calm and dust storm days using internal transcribed spacer (ITS)-1 and 16S rRNA amplicon sequencing, respectively. While some types of airborne fungi were frequently present throughout the year, distinct differences were also observed between calm and dust storm days. We also observed the influence of the origin of air parcels and wind elevation of the air trajectory. The most abundant genera of fungi identified during the study period were Cryptococcus, Aureobasidium, Alternaria, Cladosporium and Filobasidium. This observation was not surprising considering the agricultural intensive environment of West Texas. Interestingly, Cladosporium, a common allergenic mold, was increased during days with dust storm events. The predominant bacterial genera observed were Bacillus, Pseudomonas, Psychrobacter, Massilia and Exiguobacterium. The relative abundance of the psychrophiles, Psychrobacter and Exiguobacterium, was surprising, given the semi-aridity of West Texas. Coupling our observations with back trajectories of the wind (Hybrid Single-Particle Lagrangian Integrated Trajectory models) demonstrated that dust storms, regional anthropogenic activity and origin of air parcels are important influences on the diversity and temporal presence of the atmospheric microbiome.
Keywords: fungi, bacteria, atmospheric microbiome, air parcels, dust storms, West Texas, allergens
Dust storms impact the atmospheric microbiome; for example they increase the abundance of Cladosporium, an allergenic fungus that may affect the health of susceptible individuals.
INTRODUCTION
The total mass of the atmosphere is ∼5.14 × 1018 kg (Trenberth and Guillemot 1994). The atmosphere is mainly constituted of gases and particulate matter (PM) of different sizes. A constant part of our life as humans is the direct exposure to air. Although airborne microorganisms represent a significant part of our surrounding fungal and bacterial microbiome, they are greatly underestimated and understudied (Meadow et al. 2015; Fujiyoshi, Tanaka and Maruyama 2017). This continuous exposure may have beneficial and detrimental effects, which need to be investigated carefully (Prussin and Marr 2015).
Recent work has focused on characterizing the upper troposphere microbiome and its potential role in cloud formation (DeLeon-Rodriguez et al. 2013). Other studies have attempted to investigate the built environment ‘indoor-air’ microbiome, including homes (Dunn et al. 2013), university buildings (Meadow et al. 2014), urban subway systems (Leung et al. 2014) and hospitals (Tong et al. 2017). The effect of dust storms on the atmospheric microbiome was examined by Mazar et al. (2016). It was found that dust storms influence the airborne microbiome to become dominated by the soil-associated microbiome instead of the anthropogenic-associated microbiome (Mazar et al. 2016; Marone et al. 2020). Several reports have suggested that thunderstorms are linked to an increase in respiratory problems (e.g. asthma), which were linked to allergens (i.e. fungal spores or airborne pollens) transported by thunderstorms (Nasser and Pulimood 2009; Yair et al. 2019).
Discussions have been raised regarding the potential role of the atmospheric microbiome on global health (Hanson et al. 2016; Gat et al. 2017). The average human breathes in ∼20 000 L of air every day. For individuals with chronic respiratory disorders, including asthma, cystic fibrosis and other lung infections, microbes and allergens suspended as PM in the air can negatively impact their health (Douwes et al. 2003). Particulate matter is a mixture of solid particles of different sizes (2.5, 10 and >10 μm designated PM2.5, PM10, respectively), and minute liquid droplets known as aerosols, some of which like dust and smoke are visible to the naked eye when present at high concentrations. Bioaerosols, however, are microscopic fragments suspended in the air and are present in particles like pollen, and microbes (bacteria, fungi and viruses), which may be pathogenic and/or allergenic (Fischer and Dott 2003; Haas et al. 2013). Suspended PM can remain in the atmosphere for extended periods of time, with smaller and lighter particles (PM2.5) transported by the wind for extended time periods and over a long distances (thousands of kilometers) (Kim, Kabir and Kabir 2015). Thus, it is important to leverage biometeorological prediction models to identify the origin of air parcels as well as the wind direction and particle concentration. Information on the atmospheric microbiome is also important due to their potential impact on cloud formation (Spracklen and Heald 2014) and human health (Griffin 2007).
The City of Lubbock, Texas is located in a semi-arid region, and thus has conditions conducive to dust storms due to wind erosion of dry surface soils. Our study site at Texas Tech University is surrounded by millions of acres of cattle, dairy farms, cotton fields and vineyards. Dust storms are responsible for the majority of PM in the local environment (Neff et al. 2013; Kelley et al. 2020). High levels of PM influence bioaerosol generation and dispersal, and increased concentrations of PM and bioaerosols generally track together (Adhikari et al. 2006). Bacteria tend to be associated with coarser particles, while fungal spores are associated with smaller particles (Haas et al. 2013). Inhalation of bioaerosols can lead to lung irritation, respiratory illnesses and susceptibility to viral, fungal and bacterial pathogens (Douwes et al. 2003). Several airborne fungi such as Cladosporium, Penicillium, Aspergillus and Alternaria have been implicated in allergies (Levetin et al. 2016). For example, Alternaria alternata can exacerbate asthma and allergy flare-ups (Denning et al. 2006; Levetin et al. 2016). Moreover, the abundance of airborne Cladosporium and Alternaria was correlated with asthma exacerbation episodes (Fukutomi and Taniguchi 2015). Numerous health studies in the last two decades have linked high concentrations of particulate air pollution with an increase in emergency room visits, hospital admissions and even premature death (Anderson, Thundiyil and Stolbach 2012; Karanasiou et al. 2012). Epidemiological studies have linked PM prevalence to the number of daily deaths and hospitalizations, most likely as a result of respiratory and cardiovascular diseases (Karanasiou et al. 2012). According to Cohen et al. (2004), PM2.5 (particulate matter with aerodynamic diameter ≤2.5 µm) are responsible for ∼1.4% of deaths worldwide (Cohen et al. 2004). In order for healthcare workers to adequately address their patients’ needs, they need to know which types of microbes may be present as a result of changes in weather patterns. The physicians at the Texas Tech University Health Sciences Center note that the majority of their patients with asthma are seen following high-wind episodes, and information on airborne microbes could provide valuable information when treating these patients (Tarbox, pers. comm. 2016).
In order to describe the local ‘atmospheric microbiome’ in Lubbock, TX, a total of 47 air samples were collected on 22 days from September 2015 through November 2016. Meteorological information during the sampled days was collected from the local airport meteorological station, while the origins of the air parcels were tracked using Hybrid Single-Particle Lagrangian Integrated Trajectory (HYSPLIT) back trajectory analyses. To examine the atmospheric microbiome, we used a modified approach by culturing the bacteria and fungi prior to amplicon [internal transcribed spacer (ITS)-1] or 16S rRNA gene amplicon next-generation sequencing to identify fungal and bacterial genera that comprised these communities. We also examined the influence of dust storm events on the diversity and composition of the atmospheric microbiome.
MATERIALS AND METHODS
Sample collection
Forty-seven samples across 22 days were collected at Texas Tech University campus (33° 35' 5.4456'' N, 101° 52' 41.178'' W) at ground level and ∼60 m (Figure S1, Supporting Information)] over a period from September 2015 to November 2016 in Lubbock, TX (Table S1, Supporting Information). All samples were obtained between 4:00 and 6:00 p.m. local standard time (LST), which is when soil moisture and relative humidity are known to be the lowest in the air, and there is sufficient time for the air to mix with the high concentrations of microbes and dust that can impact humans (Tong 1999). Samples were collected at least once a month during calm days (control), and experimental samples were taken when dust storms (wind speeds > 48km/h) occurred.
Fungal and bacterial atmospheric microbiome sample collection
Previous studies investigating the atmospheric microbiome have used culturing techniques or direct next-generation sequencing of air samples to analyze the atmospheric microbiome. Limitations of culturing techniques or direct next-generation sequencing are well-known (Stephens et al. 2015). We chose to investigate the culturable microorganisms only in order to eliminate the possibility of sequencing DNA artifacts carried by the air. Sampling was conducted using Petri plates containing potato dextrose agar (PDA) (Thermo-Fisher Scientific, Waltham, MA, USA) or tryptic soy agar (TSA) (Thermo-Fisher Scientific, Waltham, MA, USA). The plates were held open into the incoming air for 2 min to collect airborne fungi and bacteria. The plates were then incubated at room temperature for 48 h and stored at 4°C until sequencing. Originally, 47 samples for 22 days were collected using both PDA and TSA media. The number of replicates per day varied from 2 to 4. However, some samples were discarded because of their low-read quality after sequencing and that they would not represent the true microbial diversity in the environment. Therefore, we analyzed the culturable fungal microbiome using 47 replicates collected from 22 days, while for the culturable bacterial microbiome, 36 replicates collected from 17 days were used.
ITS and 16S rRNA DNA sequencing and analysis
DNA extraction and amplicon-specific sequencing were performed using the previously collected plates by Molecular Research LP (MR DNA), Shallowater, TX, USA as described at MR DNA Laboratory (www.mrdnalab.com). The ITS-1 rRNA region was targeted to characterize the fungal community, while the 16S rRNA gene V4 variable region was used to identify the bacterial community. Sequencing was performed on Illumina MiSeq platform following the manufacturer's guidelines. Raw sequencing data were deposited under BioProject accession number PRJNA637810 in the National Center for Biotechnology Information BioProject database. ITS-1 and 16S rRNA sequence data were analyzed using QIIME 2 (Bolyen et al. 2019). In summary, sequences were filtered, denoised and joined. Then, DADA2 plugin was used within QIIME 2 to determine exact amplicon sequence variants (ASVs). For taxonomy assignment of ITS and 16S rRNA ASVs, SILVA 132 (Pruesse, Peplies and Glöckner 2012) and UNITE 7.2 (Community 2017) databases were used, respectively.
Back trajectory analysis and meteorological information
In order to determine the link between the microbial species present within the local environment and the origin of the air mass moving through the area, back trajectories and surface meteorological variables were used to determine the atmospheric conditions that result in high levels of specific fungi and bacteria. The HYSPLIT model (https://www.arl.noaa.gov/) was used to create backward trajectories of air parcels moving through the air over 72-hour periods from the coordinates of the collection site. Using a Lagrangian framework to time-integrate air parcel advection backward through the domain, HYSPLIT outputs 3D paths of atmospheric transport based on observational and model data (Draxler and Hess 1997). The Eta Data Assimilation System 40 km (EDAS40) dataset was used to force the HYSPLIT runs. Back trajectories at 500 m above ground level were used to capture the conditions that could be expected to be representative of a well-mixed daytime boundary layer.
Meteorological information, such as hourly ambient temperature, relative humidity, wind speed, wind direction, wind gust, visibility, station pressure and precipitation, was retrieved from the local National Weather Service (NWS 2019) meteorological station, which is located at the Lubbock Preston Smith International Airport (33° 39' 48.96'' N, 101° 49' 22.8'' W), ∼9.8 km north from the sample collection. Dust storm days were identified using the station Present Weather Code. Information of main meteorological conditions of each sample can be found in Table S1 (Supporting Information).
RESULTS
The culturable atmospheric fungal microbiome
Across the analyzed fungal microbiome samples, 695 ASVs were identified, with a mean frequency of ∼ 112,753 (range: 29 676–326 859). Ascomycota and Basidiomycota were the top two prevalent phyla among all samples with an average relative abundance 57% and 40%, respectively (Fig. 1A). Other phyla (e.g. Mucoromycota and Mortierellomycota) were found with an average relative abundance below 5% (Fig. 1A). Among the most resolved ASVs (at the species and genus levels), four genera and six species had the most abundant average relative abundance ≥1% (Fig. 1B). The most abundant taxa were Aureobasidium pullulans (with an average relative abundance of ∼18%), Cryptococcus consortionis (∼17%), Alternaria (∼16%), Cladosporium flabelliforme (10%) and Filobasidium (∼8%) (Fig. 1B).
Figure 1.
The culturable atmospheric fungal microbiome. (A) A stacked bar plot shows phyla belong to fungi. (B)Most abundant genera and species of the culturable atmospheric fungal microbiome. Relative abundance of the top taxa with an average relative abundance ≥ 1% shown as boxplot.
The culturable atmospheric bacterial microbiome
Across the analyzed bacterial microbiome samples, 1,153 ASVs were identified, with a mean frequency of ∼39,158 (range: 10 012–22 754). The most abundant bacterial phyla were Firmicutes, Proteobacteria, Actinobacteria and Bacteroidetes with average relative abundance of 40%, 35%, 19% and 1%, respectively (Fig. 2A). Among the most resolved ASVs (at the genus level), 10 genera had the highest average relative abundance ≥1% (Fig. 2B). The most abundant taxa were Bacillus (with an average relative abundance of ∼26%), Pseudomonas (∼9%), Psychrobacter (∼8%) and Massilia (∼5%) (Fig. 2B).
Figure 2.
The culturable atmospheric bacterial microbiome. (A) A stacked bar plot shows phyla belong to bacteria. (B) Most abundant genera and species of the culturable atmospheric bacterial microbiome. Relative abundance of the top taxa with an average relative abundance ≥ 1% shown as boxplot.
The impact of dust storm events and the origin of the air on the local atmospheric microbiome
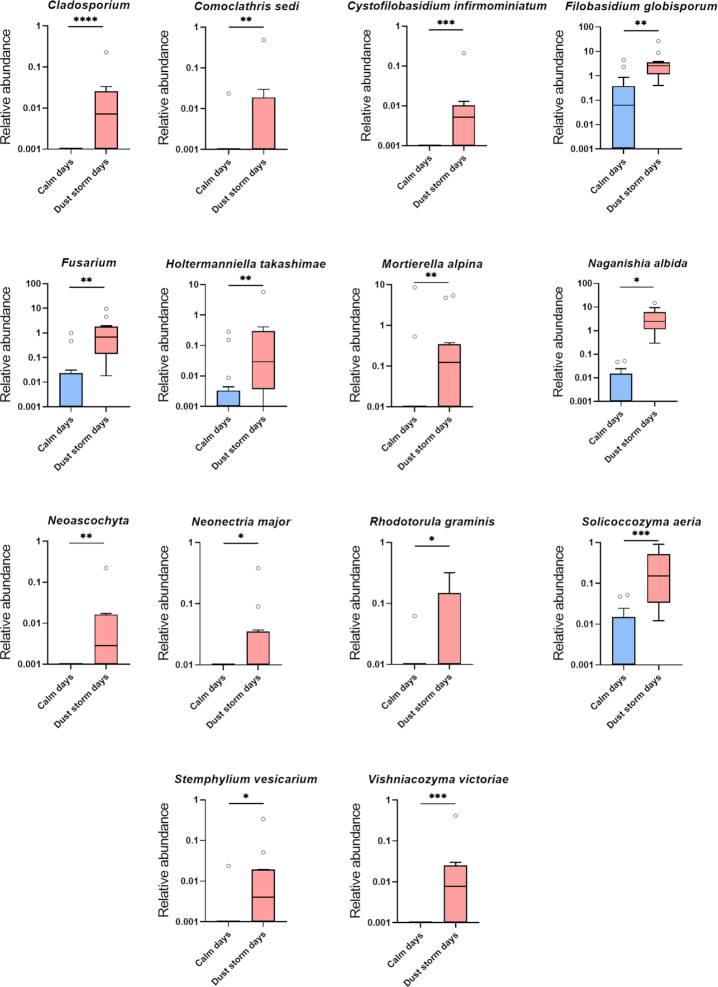
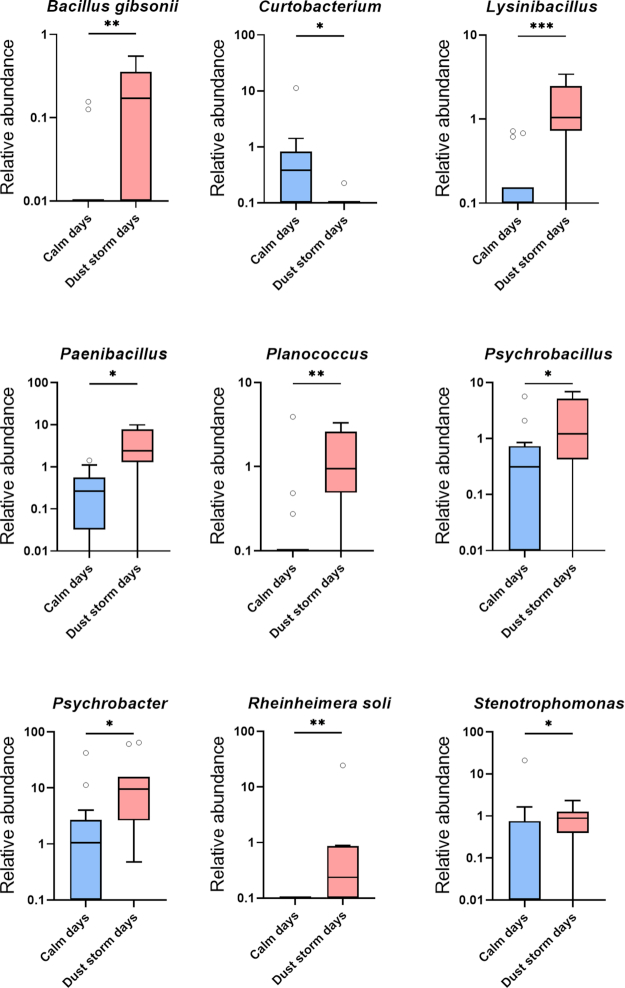
One of the goals of the current study was to examine the effect of dust storm events on the atmospheric microbiome. To do so, we used a subset of the samples collected during days of blowing dust storms, identified by the NWS station, in comparison to calm days. Six high-wind days with dust storms with a median wind speed of ∼48.3 km/h and seven calm (control) days with a median speed of 22.5 km/h were identified. We detected 14 fungal genera and species (Fig. 3) and 9 bacterial genera and species (Fig. 4) whose relative abundance was significantly increased by dust storms (except for Curtobacterium that was decreased). This analysis was performed using Wilcoxon rank-sum test with Benjamini–Hochberg false discovery rate correction. Fungal taxa included Neonectria major, Rhodotorula graminis, Cystofilobasidium infirmominiatum, Filobasidium globisporum, Naganishia albida, Solicoccozyma aeria, Holtermanniella takashimae, Vishniacozyma victoriae, Mortierella alpine, Fusarium, Comoclathris sedi, Stemphylium vesicarium, Cladosporium and Neoascochyta (Fig. 3). Bacterial taxa included Curtobacterium, Bacillus gibsonii, Paenibacillus, Lysinibacillus, Planococcus, Psychrobacillus, Rheinheimera soli, Psychrobacter and Stenotrophomonas (Fig. 4). We also observed that the richness of the fungal microbiome using Faith's phylogenetic diversity was significantly increased in dust storm days (Fig. 5), but no change was observed in the richness of the bacterial microbiome between calm and dust storm days.
Figure 3.
The impact of dust storms on the relative abundance of the culturable atmospheric fungal microbiome. Genera and species of fungal microbiome with statistical significance in abundance between calm and dust storm days are shown (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). The Wilcoxon Rank-Sum test was used to determine statistical significance, followed by the Benjamini–Hochberg procedure to correct the P values. Data are represented as boxplots, which extend from the 25th to 75th percentiles and the line in the middle of the box represents the median.
Figure 4.
The impact of dust storms on the relative abundance of the culturable atmospheric bacterial microbiome. Genera and species of bacterial microbiome with statistical significance in abundance between calm and dust storm daysare shown (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). The Wilcoxon Rank-Sum test was used to determine statistical significance, followed by the Benjamini–Hochberg procedure to correct the P values. Data are represented as boxplots, which extend from the 25th to 75th percentiles and the line in the middle of the box represents the median.
Figure 5.
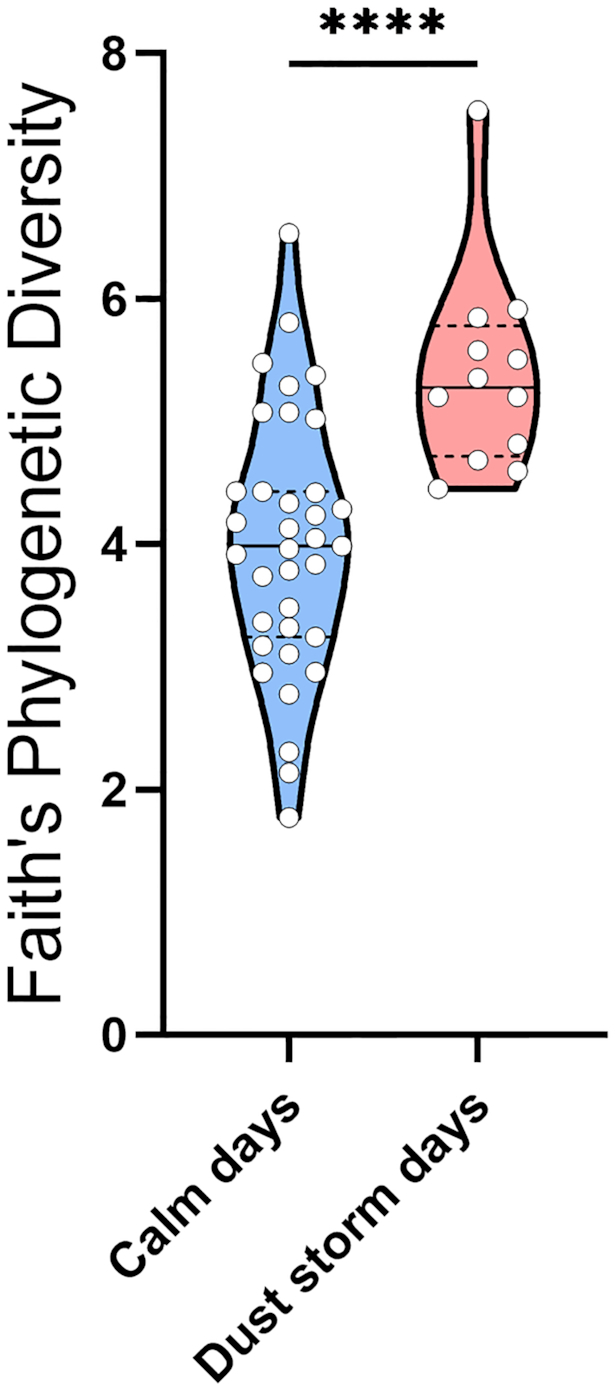
The impact of dust storms on the alpha diversity (richness) of the culturable atmospheric fungal microbiome. The non-parametric Mann–Whitney U test was used to determine statistical significance. Genera and species of fungal microbiome with statistical significance in abundance between calm and dust storm days (****P < 0.0001). Data are represented as violin plot. The line in the middle of the box represents the median
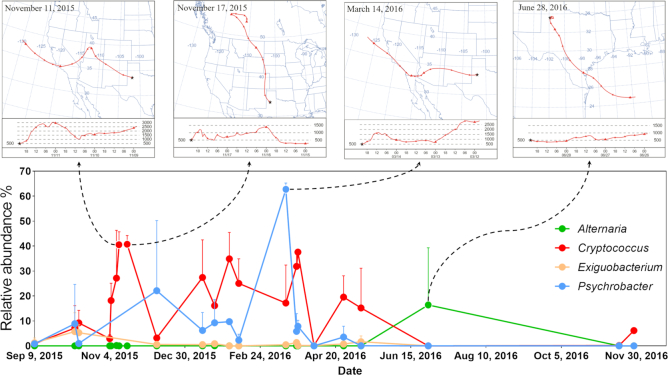
We unexpectedly observed psychrophiles belonging to the genera Psychrobacter and Exiguobacterium in a number of our samples. Examination of the back trajectories (Fig. 6) revealed that these genera were most abundant in air parcels that originated in the North Pacific or Northern Canada and when the elevation of the parcel was above 2000 m (Figure S2, Supporting Information). Psychrobacter was among the three most abundant bacterial genera observed over the entire collection period (Fig. 2B).
Figure 6.
Correlations between microbiome composition temporal spike and origin of air parcels.
DISCUSSION
This study emanated from an earlier qualitative observation of a large diversity of bacterial and fungal species in the atmosphere of the Texas Tech University campus during a very windy day in the spring of 2014. This area is among those with frequent dust storms in the United States due to the persistent winds (Orgill and Sehmel 1976; Deane and Gutmann 2003). As a region used for cotton, sorghum and grape growing, as well as livestock rearing, large areas of land may be left fallow for significant periods of time. Approximately 3.7 million acres of land is devoted to cotton cultivation annually in the Lubbock area high plains. These large tracts of land can contain dust particles that may be lifted up from the surface by winds. Particulates in the wind have been associated with respiratory ailments such as allergies and asthma (Hanson et al. 2016; Gat et al. 2017). In this study, we sampled air on the Texas Tech University campus over the 2015 and 2016 calendar years to assess the composition and relative abundance of bacteria and fungi based on calm and dust storm days.
Airborne particulates have increasingly been observed to spike during dust storms and act as carriers of bacteria and fungi, resulting in an increase in the microbial burden as well (Nourmoradi et al. 2015). Correlations between dust storms and the incidence of respiratory diseases have been long recognized (Hefflin et al. 1994; Kim, Kabir and Kabir 2015). A link between exposure to PM and hospital visits due to upper respiratory ailments has been observed by others (Tian et al. 2019; Ma et al. 2020), in some cases, a link was established between the long-term exposure and mortality rate in elderly (Wu et al. 2020). A recent study suggests that severe air pollution (enhanced PM2.5 levels as well as nitric oxide) may potentiate SARS-CoV-2 infections (Frontera et al. 2020). While the ubiquity of bacteria and fungi in the atmosphere have been assessed in different environments, the primary focus of previous studies has been on bacteria and not the fungi. Studies on atmospheric fungal microbiomes have largely focused on agricultural regions and practices or built environments (Dietzel et al. 2019). Abdel-Hafez et al. (1990) studied the fungal microbiome of wheat and sorghum harvester dusts and reported the common occurrence of Alternaria, Aspergillus, Fusarium and Penicillium species. Among these, many thermophilic taxa were also detected (Abdel-Hafez et al. 1990). In one study that assessed aerial transport of bacteria from cattle feed yards on PM in the area surrounding Lubbock, Texas, the authors identified antibiotic resistance genes present in these bacteria. Genes encoding resistance to tetracycline were more abundant in particulates downwind of the feed yards compared with collections made upwind (McEachran et al. 2015). Seasonal variations were correlated with changes in the airborne microbiome in agreement with atmospheric circulation (Cáliz et al. 2018).
Our study focused on airborne fungi and bacteria in an urban environment. We investigated the longitudinal abundance and diversity of fungal and bacterial genera and related these observations to the origin of the wind using back trajectories. Fungal genera and species present in our sampling with high relative abundance included Aureobasidium pullulans, Cryptococcus consortionis, Alternaria, Cladosporium flabelliforme and Filobasidium. These taxa belong to two phyla, Basidiomycota and Ascomycota. A. pullulans is a ubiquitous fungus that can cause several forms of infections in humans (Oliveira et al. 2013). Species of Aureobasidium have been observed in dust events in Africa (Griffin et al. 2003), sulfur-contaminated soils (Wainwright 1978), vineyards (Bozoudi and Tsaltas 2018) and were occasionally associated with Cryptococcus in vineyards (Comitini and Ciani 2008). Several Cryptococcus spp. were identified in this study. This genus contains species that are commonly associated with feces of birds (Kwon-Chung et al. 2014), particularly pigeons and canaries. The great abundance of pigeons on the Texas Tech University campus was very likely an influence on the Cryptococcus sampling. Species of Cryptococcus have been found to be sensitive to high temperatures but can tolerate alkaline soils, similar to those found in the Lubbock area. Species of Cryptococcus species, e.g. C. neoformans and C. laurentii, have been associated with human diseases (Byrnes et al. 2011; Molina-Leyva et al. 2013). Other spp., e.g. C. infirmo-miniatus, have also been used in the control of plant disease as part of integrated pest management (Spotts, Cervantes and Facteau 2002). Cladosporium flabelliforme is among the Cladosporium species that have been associated with clinical samples (Sandoval-Denis et al. 2015). The most interesting species of Cryptococcus we identified was C. consortionis first described in 1985, isolated from the Ross Desert of Antarctica (Vishniac 1985). Indeed C. consortionis was the second most abundant species of all the fungal samples identified in this study (Figs 1B and 6).
Mortierellomycota and Mucoromycota were the least represented fungi in our dataset. They were only identified in small number of days and at much lower abundance than other phyla. In the Mucorales order (belongs to Mucoromycota) several species are pathogenic and can cause skin and lung infections (Tomecki et al. 1989). Intriguingly, Mucorales-associated infections were found to be prevalent after natural disasters such as hurricane Katrina and the 2004 Indian Ocean tsunami (Kontoyiannis 2012). The reason behind the underrepresentation of these fungi in our dataset is that Lubbock area is not impacted by any hurricane air flow. Furthermore, the wind currents for most of the samples in our study originated in the higher latitudes (greater than 35°N latitude) and the northern Pacific Ocean.
Coccidioides immitis, the cause of the valley fever, is endemic in the southwestern parts of the United States, including Texas (Thompson et al. 2019). Interestingly, we did not detect C. immitis at any abundance in any day of our study. But we cannot definitively exclude the possibility of its presence as it could be a limitation of our culturing step.
The most predominant bacterial genera were Bacillus, Pseudomonas, Psychrobacter, Massilia and Exiguobacterium; of these, only members of the genus Bacillus form endospores. Endospores provide resistance to desiccation, temperature changes, chemical stress and have been known to survive for hundreds of years (Nicholson et al. 2000). Together with their ubiquity, it is not surprising that this genus is the most abundant, representing ∼50% of all genera. Pseudomonas, a bacterium with a large genome is ubiquitous and is well known for inhabiting a wide variety of habitats from soils and plants to hospital environments and is an opportunistic pathogen (Moradali, Ghods and Rehm 2017). Members of this genus are also known for their resistance to a broad variety of environmental chemicals and antibiotics and being excellent competitors by producing antimicrobial chemicals (Moradali, Ghods and Rehm 2017). Pseudomonas is also known to be an ice nucleator in clouds (Maki et al. 1974; Du et al. 2017). The abundance of both Bacillus and Pseudomonas species in our analyses is, therefore, not surprising. Massilia species have been isolated from different environmental samples (e.g. soil, air and water) and some of them have been associated with human infections (Peta, Raths and Bücking 2019). The occurrence of the two abundant genera, Psychrobacter and Exiguobacterium, was unexpected. Members of the Psychrobacter are known to be cold-loving and are often found in glaciers (Rodrigues et al. 2009). A few species of this genus are found associated with wound infections (Caspar et al. 2013). Psychrobacter was also in high abundance when wind elevations exceeded 2500 m or originated from the North Pacific Ocean (9 December 2015 and 14 March 2016). Species in the genus Exiguobacterium are known to have broad temperature tolerances (−12 to 55°C), and are also found in Himalayan soils, permafrost in Siberia and hot springs (White et al. 2018). The relative abundance of Exiguobacterium was highest on 12 October 2015 (9%), in contrast with an average of <1% across the rest of the days. When we observed the back trajectory maps (Figure S2, Supporting Information) we noticed that on this day, the air mass originated from the North Pacific Ocean.
The impact of dust storms on the atmospheric microbiome is of interest due to possible implications in human health and agriculture. The increase in the richness of the fungal atmospheric microbiome was notable as it resulted in the increase of several fungal genera. Among these was Stemphylium vesicarium, which is a plant pathogen and the cause of Welsh onion leaf blight (Aveling and Snyman 1993; Misawa and Yasuoka 2012). Solicoccozyma aeria (formerly Cryptococcus aerius) was isolated from soil samples and utilized in lipid production (Ghanavati, Nahvi and Karimi 2015). Mortierella alpine was another oleaginous fungus that we observed increased in dust storms days. Naganishia albida (formerly Cryptococcus albidus) has been associated with human infections (Lee et al. 2004). Other species of Naganishia have been collected in high elevation soils of the Atacama, Chile and Bolivia. These fungi also demonstrate high ultraviolet light resistance (Schmidt et al. 2017). Rhodotorula graminis, an endophytic yeast, is a carotenoid producer and plant growth-promoting fungus (Firrincieli et al. 2015; Lyman et al. 2019). Vishniacozyma victoriae (formerly Cryptococcus victoriae) was recently found to be prevalent in homes, especially with dogs present (Rush et al. 2020). Other species belonging to the genus Vishniacozyma have been identified from the Canadian High Arctic (Tsuji et al. 2019). The relative abundance of C. consortionis was also very high on dust storm days.
Cladosporium, Alternaria, Aspergillus and Penicillium species are notorious allergens (Kurup 2003). Interestingly, only Cladosporium and Alternaria were detected at high relative abundance (>5% on average) and in all collection times. In contrast, Aspergillus and Penicillium were observed in a few days with a relatively low abundance (<0.5% on average). This is because Aspergillus and Penicillium are more common indoors, while Cladosporium and Alternaria are ubiquitous both indoors and outdoors (Bozek and Pyrkosz 2017). The prevalence of these airborne allergens has an important implication in the health of asthmatic patients. For example, patients who are allergic and exposed to Alternaria have frequent exacerbations of their asthma that require hospitalization (Kołodziejczyk et al. 2016). Moreover, because dust storms are associated with increased asthma hospitalizations (Kanatani et al. 2010), we were interested to test the hypothesis that allergenic molds are increased in the air during dust storms and may contribute to this. Our results revealed that Cladosporium was the only mold (among the four allergens: Cladosporium, Alternaria, Aspergillus and Penicillium) that was increased during dust storms. This is an exciting result that requires further investigation, to find out whether the patients admitted to hospitals due to asthmatic complication (after a dust storm event) are allergic to Cladosporium. This knowledge can help susceptible patients and the healthcare system to avoid overburden after dust storms.
Among the bacterial taxa, Stenotrophomonas was increased in dust storm days and its species S. maltophilia is an emerging respiratory pathogen (Brooke 2012). Paenibacillus is one of the endospore-forming bacteria and its species is relevant to human health and agriculture (Grady et al. 2016). Rheinheimera soli findings are of interest in that it was significantly increased in dust storm days (i.e. it was only detected in dust storm days). Rheinheimera soli was first isolated from a soil sample in South Korea (Ryu et al. 2008). Other related Rheinheimera species were isolated from aquatic or marine habitats (Brettar, Christen and Höfle 2002; Romanenko et al. 2003). Linking the abundance of this fungus to back trajectory data showed that it had an abundance of 13% on 23 March 2016, when the wind originated from the North Pacific Ocean and reached an elevation of over 4000 m.
This research contributes to our broader understanding and efforts to appreciate the immense diversity of the Earth's microbiome (deciphering bacterial and fungal distribution), and efforts to harness and manage it (Alivisatos et al. 2015). Our study reveals that broad microbial diversity of fungi and bacteria in local environments is influenced by dust storm events, regional anthropogenic practices such as farming and the origins of air parcels. Interdisciplinary research linking microbiome, wind trajectory and environmental information with medical and public health data will enable researchers to establish a comprehensive epidemiological analysis of outbreaks to better understand their etiology and broad impacts on human, plant and wildlife health.
ACKNOWLEDGEMENTS
We would like to thank the Departments of Biological Sciences and Geosciences for their support, and the Graduate School for support of MME.
Supplementary Material
Contributor Information
Moamen M Elmassry, Department of Biological Sciences, Texas Tech University, Lubbock, TX 79409, USA.
Nandini Ray, Department of Biological Sciences, Texas Tech University, Lubbock, TX 79409, USA.
Sara Sorge, Department of Biological Sciences, Texas Tech University, Lubbock, TX 79409, USA.
Jennifer Webster, Department of Biological Sciences, Texas Tech University, Lubbock, TX 79409, USA.
Kyle Merry, Department of Biological Sciences, Texas Tech University, Lubbock, TX 79409, USA.
Angelica Caserio, Department of Biological Sciences, Texas Tech University, Lubbock, TX 79409, USA.
Daniel J Vecellio, Department of Geography, Texas A&M University, College Station, TX 77843, USA.
Cassandra Kruczek, Department of Medical Education, Texas Tech University Health Sciences Center, Lubbock, TX 79430, USA.
Scot Dowd, Molecular Research LP, Clovis Road, Shallowater, TX 79363, USA.
Karin Ardon-Dryer, Department of Geosciences, Atmospheric Science Group, Texas Tech University, Lubbock, TX 79409, USA.
Jennifer Vanos, School of Sustainability, Arizona State University, Tempe, AZ 85281, USA.
Michael J San Francisco, Department of Biological Sciences, Texas Tech University, Lubbock, TX 79409, USA; Honors College, Texas Tech University, Lubbock, TX 79410, USA.
AUTHOR CONTRIBUTIONS
MME: data analysis and writing; NR, SS, JW, KM, AC and DJV: data collection and writing; CK: data analysis and writing; SD: DNA sequencing and analysis; KA-D and JV: wind back trajectories, data analysis and writing; MJSF: conception, data analysis and writing.
Conflict of Interest
None declared.
REFERENCES
- Abdel-Hafez SI, Moubasher AH, Shoreit AAet al. . Fungal flora associated with combine harvester wheat and sorghum dusts from Egypt. J Basic Microbiol. 1990;30:467–79. [DOI] [PubMed] [Google Scholar]
- Adhikari A, Reponen T, Grinshpun SAet al. . Correlation of ambient inhalable bioaerosols with particulate matter and ozone: a two-year study. Environ Pollut. 2006;140:16–28. [DOI] [PubMed] [Google Scholar]
- Alivisatos AP, et al. MICROBIOME. A unified initiative to harness Earth's microbiomes. Science. 2015;350:507–8. [DOI] [PubMed] [Google Scholar]
- Anderson JO, Thundiyil JG, Stolbach A. Clearing the air: a review of the effects of particulate matter air pollution on human health. J Med Toxicol. 2012;8:166–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aveling TAS, Snyman HG. Infection studies of Stemphylium vesicarium on onion leaves. Mycol Res. 1993;97:984–8. [Google Scholar]
- Bolyen E, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozek A, Pyrkosz K. Immunotherapy of mold allergy: a review. Hum Vaccin Immunother. 2017;13:2397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozoudi D, Tsaltas D. The multiple and versatile roles of Aureobasidium pullulans in the vitivinicultural sector. Fermentation. 2018;4:85. [Google Scholar]
- Brettar I, Christen R, Höfle MG. Rheinheimera baltica gen. nov., sp. nov., a blue-coloured bacterium isolated from the central Baltic Sea. Int J Syst Evol Microbiol. 2002;52:1851–7. [DOI] [PubMed] [Google Scholar]
- Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev. 2012;25:2–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes EJ, Bartlett KH, Perfect JRet al. . Cryptococcus gattii: an emerging fungal pathogen infecting humans and animals. Microbes Infect. 2011;13:895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar Y, Recule C, Pouzol Pet al. . Psychrobacter arenosus bacteremia after blood transfusion, France. Emerging Infect Dis. 2013;19:1118–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AJ, et al. Mortality impacts of urban air pollution. In: Majid E, Lopez AD, Rodgers AA, Murray CJL (eds). Comparative Quantification of Health Risks: Global and Regional Burden of Disease Attributable to Selected Major Risk Factors, Vol. 2. Geneva: World Health Organization, 2004, 1353–433. [Google Scholar]
- Comitini F, Ciani M. Influence of fungicide treatments on the occurrence of yeast flora associated with wine grapes. Ann Microbiol. 2008;58:489–93. [Google Scholar]
- Community U. UNITE QIIME release. UNITE Community, 2017. 10.15156/BIO/587481; https://unite.ut.ee/repository.php (14 January 2021, date last accessed). [DOI] [Google Scholar]
- Cáliz J, Triadó-Margarit X, Camarero Let al. . A long-term survey unveils strong seasonal patterns in the airborne microbiome coupled to general and regional atmospheric circulations. Proc Natl Acad Sci USA. 2018;115:12229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane G, Gutmann MP. Blowin’ down the road: investigating bilateral causality between dust storms and population in the Great Plains. Population Res Policy Rev. 2003;22:297–331. [Google Scholar]
- DeLeon-Rodriguez N, et al. Microbiome of the upper troposphere: species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc Natl Acad Sci USA. 2013;110:2575–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning DW, O'Driscoll BR, Hogaboam CMet al. . The link between fungi and severe asthma: a summary of the evidence. Eur Respir J. 2006;27:615–26. [DOI] [PubMed] [Google Scholar]
- Dietzel K, Valle D, Fierer Net al. . Geographical distribution of fungal plant pathogens in dust across the United States. Front Ecol Evol. 2019;7:304. [Google Scholar]
- Douwes J, Thorne P, Pearce Net al. . Bioaerosol health effects and exposure assessment: progress and prospects. Ann Occup Hyg. 2003;47:187–200. [DOI] [PubMed] [Google Scholar]
- Draxler RR, Hess GD. Description of the HYSPLIT_4 modeling system, Maryland: Air Resources Laboratory, Silver Spring, 1997. [Google Scholar]
- Dunn RR, Fierer N, Henley JBet al. . Home life: factors structuring the bacterial diversity found within and between homes. PLoS One. 2013;8:e64133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du R, Du P, Lu Zet al. . Evidence for a missing source of efficient ice nuclei. Sci Rep. 2017;7:39673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firrincieli A, et al. Genome sequence of the plant growth promoting endophytic yeast Rhodotorula graminis WP1. Front Microbiol. 2015;6:978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G, Dott W. Relevance of airborne fungi and their secondary metabolites for environmental, occupational and indoor hygiene. Arch Microbiol. 2003;179:75–82. [DOI] [PubMed] [Google Scholar]
- Frontera A, Cianfanelli L, Vlachos Ket al. . Severe air pollution links to higher mortality in COVID-19 patients: the “double-hit” hypothesis. J Infect. 2020;81:255–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyoshi S, Tanaka D, Maruyama F. Transmission of airborne bacteria across built environments and its measurement standards: a review. Front Microbiol. 2017;8:2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukutomi Y, Taniguchi M. Sensitization to fungal allergens: resolved and unresolved issues. Allergol Int. 2015;64:321–31. [DOI] [PubMed] [Google Scholar]
- Gat D, Mazar Y, Cytryn Eet al. . Origin-dependent variations in the atmospheric microbiome community in Eastern Mediterranean dust storms. Environ Sci Technol. 2017;51:6709–18. [DOI] [PubMed] [Google Scholar]
- Ghanavati H, Nahvi I, Karimi K. Organic fraction of municipal solid waste as a suitable feedstock for the production of lipid by oleaginous yeast Cryptococcus aerius. Waste Manag. 2015;38:141–8. [DOI] [PubMed] [Google Scholar]
- Grady EN, MacDonald J, Liu Let al. . Current knowledge and perspectives of Paenibacillus: a review. Microb Cell Fact. 2016;15:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DW, Kellogg CA, Garrison VHet al. . Atmospheric microbiology in the northern Caribbean during African dust events. Aerobiologia (Bologna). 2003;19:143–57. [Google Scholar]
- Griffin DW. Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin Microbiol Rev. 2007;20:459–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas D, Galler H, Luxner Jet al. . The concentrations of culturable microorganisms in relation to particulate matter in urban air. Atmos Environ. 2013;65:215–22. [Google Scholar]
- Hanson B, et al. Characterization of the bacterial and fungal microbiome in indoor dust and outdoor air samples: a pilot study. Environ Sci Process Impacts. 2016;18:713–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefflin BJ, Jalaludin B, McClure Eet al. . Surveillance for dust storms and respiratory diseases in Washington State, 1991. Arch Environ Health. 1994;49:170–74. [DOI] [PubMed] [Google Scholar]
- Kanatani KT, Ito I, Al-Delaimy WKet al. . Desert dust exposure is associated with increased risk of asthma hospitalization in children. Am J Respir Crit Care Med. 2010;182:1475–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanasiou A, Moreno N, Moreno Tet al. . Health effects from Sahara dust episodes in Europe: literature review and research gaps. Environ Int. 2012;47:107–14. [DOI] [PubMed] [Google Scholar]
- Kelley MC, Brown MM, Fedler CBet al. . Long-term measurements of PM2.5 concentrations in Lubbock, Texas. Aerosol Air Qual Res. 2020;20:1306–18. [Google Scholar]
- Kim K-H, Kabir E, Kabir S. A review on the human health impact of airborne particulate matter. Environ Int. 2015;74:136–43. [DOI] [PubMed] [Google Scholar]
- Kontoyiannis DP. Mucormycosis. In: Goldman's Cecil Medicine, Philadelphia, PA: Elsevier, 2012, 1994–7. [Google Scholar]
- Kołodziejczyk K, Bożek A, Jarząb Jet al. . The clinical differences of asthma in patients with molds allergy. Pneumonol Alergol Pol. 2016;84:81–6. [DOI] [PubMed] [Google Scholar]
- Kurup VP. Fungal allergens. Curr Allergy Asthma Rep. 2003;3:416–23. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Fraser JA, Doering TLet al. . Cryptococcus neoformans and Cryptococcus gattii, the etiologic agents of cryptococcosis. Cold Spring Harb Perspect Med. 2014;4:a019760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YA, Kim HJ, Lee TWet al. . First report of Cryptococcus albidus-induced disseminated cryptococcosis in a renal transplant recipient. Korean J Intern Med. 2004;19:53–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung MHY, Wilkins D, Li EKTet al. . Indoor-air microbiome in an urban subway network: diversity and dynamics. Appl Environ Microbiol. 2014;80:6760–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levetin E, Horner WE, Scott JAet al. . Taxonomy of allergenic fungi. J Allergy Clin Immunol Pract. 2016;4:375–85.e1. [DOI] [PubMed] [Google Scholar]
- Lyman M, Urbin S, Strout Cet al. . The oleaginous red yeast rhodotorula/rhodosporidium: a factory for industrial bioproducts. In: Yeasts in Biotechnology. IntechOpen, 2019. [Google Scholar]
- Maki LR, Galyan EL, Chang-Chien MMet al. . Ice nucleation induced by Pseudomonas syringae. Appl Microbiol. 1974;28:456–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marone A, Kane CT, Mbengue Met al. . Characterization of bacteria on aerosols from dust events in Dakar, Senegal, West Africa. GeoHealth. 2020;4:e2019GH000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Yue L, Liu Jet al. . Association of air pollution with outpatient visits for respiratory diseases of children in an ex-heavily polluted Northwestern city, China. BMC Public Health. 2020;20:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazar Y, Cytryn E, Erel Yet al. . Effect of dust storms on the atmospheric microbiome in the Eastern Mediterranean. Environ Sci Technol. 2016;50:4194–202. [DOI] [PubMed] [Google Scholar]
- McEachran AD, Blackwell BR, Hanson JDet al. . Antibiotics, bacteria, and antibiotic resistance genes: aerial transport from cattle feed yards via particulate matter. Environ Health Perspect. 2015;123:337–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadow JF, Altrichter AE, Bateman ACet al. . Humans differ in their personal microbial cloud. PeerJ. 2015;3:e1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadow JF, et al. Indoor airborne bacterial communities are influenced by ventilation, occupancy, and outdoor air source. Indoor Air. 2014;24:41–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa T, Yasuoka S. The life cycle of Stemphylium vesicarium, the causal agent of Welsh onion leaf blight. J Gen Plant Pathol. 2012;78:18–29. [Google Scholar]
- Molina-Leyva A, Ruiz-Carrascosa JC, Leyva-Garcia Aet al. . Cutaneous Cryptococcus laurentii infection in an immunocompetent child. Int J Infect Dis. 2013;17:e1232–3. [DOI] [PubMed] [Google Scholar]
- Moradali MF, Ghods S, Rehm BHA. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol. 2017;7:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser SM, Pulimood TB. Allergens and thunderstorm asthma. Curr Allergy Asthma Rep. 2009;9:384–90. [DOI] [PubMed] [Google Scholar]
- Neff JC, Reynolds RL, Munson SMet al. . The role of dust storms in total atmospheric particle concentrations at two sites in the western U.S. J Geophys Res Atmos. 2013;118:1–12. [Google Scholar]
- Nicholson WL, Munakata N, Horneck Get al. . Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev. 2000;64:548–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourmoradi H, Moradnejadi K, Moghadam FMet al. . The effect of dust storm on the microbial quality of ambient air in Sanandaj: a city located in the west of Iran. Glob J Health Sci. 2015;7:114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NWS . National Weather Service: Lubbock Preston Smith International Airport meteorological information. 2019. https://mesonet.agron.iastate.edu/request/download.phtml (29 May 2020, date last accessed).
- Oliveira de LR, Moraes-Souza H, Maltos ALet al. . Aureobasidium pullulans infection in a patient with chronic lymphocytic leukemia. Rev Soc Bras Med Trop. 2013;46:660–2. [DOI] [PubMed] [Google Scholar]
- Orgill MM, Sehmel GA. Frequency and diurnal variation of dust storms in the contiguous U.S.A. Atmos Environ. 1976;10:813–25. [Google Scholar]
- Peta V, Raths R, Bücking H. Draft genome sequence of Massilia sp. strain ONC3, a novel bacterial species of the Oxalobacteraceae family isolated from garden soil. Microbiol Resour Announc. 2019;8, e00377–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prussin AJ, Marr LC. Sources of airborne microorganisms in the built environment. Microbiome. 2015;3:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues DF, da C Jesus E, Ayala-Del-Río HLet al. . Biogeography of two cold-adapted genera: Psychrobacter and Exiguobacterium. ISME J. 2009;3:658–65. [DOI] [PubMed] [Google Scholar]
- Romanenko LA, Uchino M, Falsen Eet al. . Rheinheimera pacifica sp. nov., a novel halotolerant bacterium isolated from deep sea water of the Pacific. Int J Syst Evol Microbiol. 2003;53:1973–7. [DOI] [PubMed] [Google Scholar]
- Rush R, et al. Detection of environmentally ubiquitous Vishniacozyma victoriae (syn. Cryptococcus victoriae) in the Homes of asthmatic and non-asthmatic children in New York City. J Allergy Clin Immunol. 2020;145:AB164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu SH, Chung BS, Park Met al. . Rheinheimera soli sp. nov., a gammaproteobacterium isolated from soil in Korea. Int J Syst Evol Microbiol. 2008;58:2271–4. [DOI] [PubMed] [Google Scholar]
- Sandoval-Denis M, Sutton DA, Martin-Vicente Aet al. . Cladosporium species recovered from clinical samples in the United States. J Clin Microbiol. 2015;53:2990–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt SK, Vimercati L, Darcy JLet al. . A Naganishia in high places: functioning populations or dormant cells from the atmosphere? Mycology. 2017;8:153–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spotts RA, Cervantes LA, Facteau TJ. Integrated control of brown rot of sweet cherry fruit with a preharvest fungicide, a postharvest yeast, modified atmosphere packaging, and cold storage temperature. Postharvest Biol Technol. 2002;24:251–7. [Google Scholar]
- Spracklen DV, Heald CL. The contribution of fungal spores and bacteria to regional and global aerosol number and ice nucleation immersion freezing rates. Atmos Chem Phys. 2014;14:9051–9. [Google Scholar]
- Stephens B, Adams RI, Bhangar Set al. . From commensalism to mutualism: integrating the microbial ecology, building science, and indoor air communities to advance research on the indoor microbiome. Indoor Air. 2015;25:1–3. [DOI] [PubMed] [Google Scholar]
- Tarbox J. Personal communication. 2016.
- Thompson GR, Lewis JS, Nix DEet al. . Current concepts and future directions in the pharmacology and treatment of coccidioidomycosis. Med Mycol. 2019;57:S76–84. [DOI] [PubMed] [Google Scholar]
- Tian Y, et al. Ambient particulate matter pollution and adult hospital admissions for pneumonia in urban China: a national time series analysis for 2014 through 2017. PLoS Med. 2019;16:e1003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki KJ, Steck WD, Hall GSet al. . Subcutaneous mycoses. J Am Acad Dermatol. 1989;21:785–90. [DOI] [PubMed] [Google Scholar]
- Tong X, Xu H, Zou Let al. . High diversity of airborne fungi in the hospital environment as revealed by meta-sequencing-based microbiome analysis. Sci Rep. 2017;7:39606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y. Diurnal distribution of total and culturable atmospheric bacteria at a rural site. Aerosol Sci Technol. 1999;30:246–54. [Google Scholar]
- Trenberth KE, Guillemot CJ. The total mass of the atmosphere. J Geophys Res. 1994;99:23079. [Google Scholar]
- Tsuji M, Tanabe Y, Vincent WFet al. . Vishniacozyma ellesmerensis sp. nov., a psychrophilic yeast isolated from a retreating glacier in the Canadian High Arctic. Int J Syst Evol Microbiol. 2019;69:696–700. [DOI] [PubMed] [Google Scholar]
- Vishniac HS. Cryptococcus socialis sp. nov. and Cryptococcus consortionis sp. nov., Antarctic basidioblastomycetes. Int J Syst Bacteriol. 1985;35:119–22. [DOI] [PubMed] [Google Scholar]
- Wainwright M. Sulphur-oxidising micro-organisms on vegetation and in soils exposed to atmospheric pollution. Environ Pollut. 1978;17:167–74. [Google Scholar]
- White RA, Soles SA, Gavelis Get al. . The complete genome and physiological analysis of the eurythermal Firmicute Exiguobacterium chiriqhucha strain RW2 isolated from a freshwater microbialite, widely adaptable to broad thermal, pH, and salinity ranges. Front Microbiol. 2018;9:3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Braun D, Schwartz Jet al. . Evaluating the impact of long-term exposure to fine particulate matter on mortality among the elderly. Sci Adv. 2020;6:eaba5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yair Yoav, Yair Yifat, Rubin Bet al. . First reported case of thunderstorm asthma in Israel. Nat Hazards Earth Syst Sci. 2019;19:2715–25. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.