Abstract
The Gram positive bacterium Bacillus subtilis and its relatives are capable of forming a durable dormant long-lasting spore. Although spores can remain dormant for years, they possess the remarkable capacity to rapidly resume life and convert into actively growing cells. This cellular transition initiates with a most enigmatic irreversible event, termed germination, lasting only for a few minutes. Germination is typified by a morphological conversion that culminates in loss of spore resilient properties. Yet, the molecular events occurring during this brief critical phase are largely unknown. The current widely accepted view considers germination to occur without the need for any macromolecule synthesis; however, accumulating data from our laboratory and others, highlighted here, provide evidence that both transcription and translation occur during germination and are required for its execution. We further underline numerous overlooked studies, conducted mainly during the 1960s–1970s, reinforcing this notion. We propose to revisit the fascinating process of spore germination and redefine it as a pathway involving macromolecule synthesis. We expect our perspective to shed new light on the awakening process of a variety of spore-forming environmental, commensal, and pathogenic bacteria and possibly be applicable to additional organisms displaying a quiescent life form.
Keywords: Bacillus subtilis, sporulation, spore germination, spore dormancy, spore revival
In this review, we synthesize old and current data to evoke that, in contrast to current dogma, bacterial spore germination demands macromolecule synthesis.
Introduction: Entering and exiting dormancy
Members of the Gram-positive Bacillus and Clostridium genera respond to starvation by initiating the developmental process of sporulation, resulting in the formation of a highly durable spore, among the most resilient life forms known (Stragier and Losick 1996, Driks 2002). Spores are long-lived dormant cells, exhibiting exceptional resistance to extremes of heat, desiccation, radiation, and toxic chemicals. Due to their remarkable robustness, spore-forming pathogenic bacteria, such as C. difficile and B. anthracis, are highly resistant to antibacterial treatments and are difficult to eradicate (Driks 2002, Henriques and Moran 2007, Hutchison et al. 2014, Paredes-Sabja et al. 2014, Wells-Bennik et al. 2016). The non-pathogenic ubiquitous soil bacterium B. subtilis evolved throughout the years to become a paradigm for the study of spore biology [e.g. (Stragier and Losick 1996, Higgins and Dworkin 2012)], and as such will be the focus of this review.
The process of sporulation is typified by the formation of an asymmetric septum, dividing the progenitor cell into a small forespore compartment, which designates the future spore, and a larger mother-cell portion, which nurtures the maturing spore. Subsequently, the forespore is engulfed by the mother-cell, and a thick layer of peptidoglycan, called the cortex, inner and outer shells of proteinaceous coats, and a glycoprotein crust are deposited around the spore. These shielding layers protect the spore from its surroundings, providing resilience properties (Stragier and Losick 1996, Henriques and Moran 2007, McKenney et al. 2010, Higgins and Dworkin 2012). In parallel, the spore chromosome is bounded by small acid soluble proteins, mainly SspA and SspB, defending it from damage during the dormant phase (Setlow 2007). Such tight packaging could potentially modify the DNA to become inaccessible for transcription. Molecular dormancy is further achieved by dehydration of the spore core, mainly through the accumulation of pyridine-2, 6-dicarboxylic acid (DPA) in complex with Ca2+ (Moir 2006). Entering dormancy appears to proceed several days, during which the spore is still responsive to extracellular cues. Throughout this time, the spore RNA content is modified, according to spore age and temperature of incubation (Segev et al. 2012, Camilleri et al. 2019). Along with the RNA pool, the levels of additional cytoplasmic molecules, such as 3-phosphoglyceric acid (3PGA) and ribonucleotides, are altered even 30 days post-sporulation (Ghosh et al. 2015).
Spores can remain dormant for years, but still retain the striking potential to promptly resume the vegetative life form once nutrients become reachable (Stragier and Losick 1996, Setlow 2003, Higgins and Dworkin 2012). The process of spore revival consists of three major consecutive phases: (i) Germination, persisting only for a few minutes, is characterized by a transition from a phase-bright to a phase grey spore, which is then converted to a phase-dark cell (Fig. 1A). During this progression, the spore undergoes rehydration, release of DPA, cortex hydrolysis, and coat disassembly (Setlow 2003, Moir 2006, 2013, Setlow et al. 2017); (ii) Ripening period, a lag phase that is dedicated to molecular reorganization that includes massive synthesis of translational and transcriptional components, along with essential metabolic enzymes (Fig. 1A) (Segev et al. 2013, Sinai et al. 2015); (iii) Outgrowth, in which the spore emerges from the disintegrating shells, and activates the synthesis of macromolecule pathways needed for cell growth and division (Fig. 1A) (Santo and Doi 1974, Sloma and Smith 1979, Setlow 2003, Moir 2006, 2013).
Figure 1.
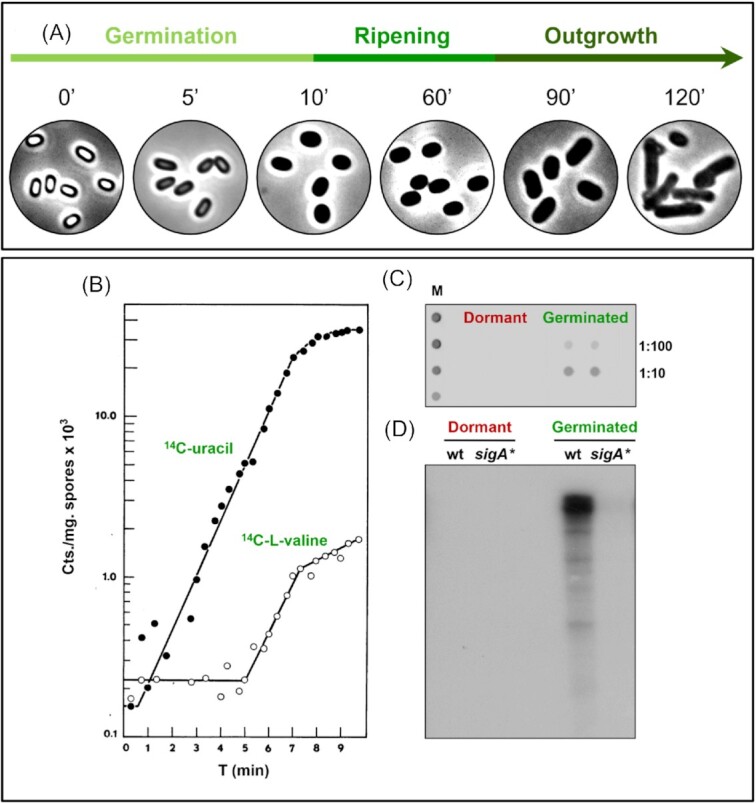
Evidence for the occurrence of transcription and translation during spore germination. (A) The morphological sequence of events occurring during spore revival is shown as captured by phase contrast images at the indicated time points [min]. Germination is visualized by a switch from a bright spore to a grey spore and subsequently to a fully germinated dark cell. The ripening period is not associated with an evident morphological change, and outgrowth is characterized by increasing cell length. (B) Incorporation of 14C-uracil and 14C-L-valine during Bacilli spore germination provides indication for transcription and translation during germination. Labels were added for clarity. Adopted from (Torriani and Levinthal 1967). Reprinted with permission from the ASM. (C) Spores were induced to germinate with the non-nutrient germinant Ca-DPA in the presence of methionine analogue (azidohomoalaine, AHA). Shown is a dot blot analysis of protein samples (in duplicates) that were collected from dormant and germinated spores. Samples were diluted (1:100 and 1:10) and spotted on a membrane that was subsequently probed with anti-biotin antibodies, indicating the presence of AHA. The marker (M) represents known amounts of biotinylated BSA. Adopted from (Sinai et al. 2015). (D)In vitro transcription reaction was carried out in whole extracts from dormant and germinated spores of WT and sigA* phospho-mutant strains, in transcription buffer supplemented with NTPs (ATP, CTP, GTP, UTP, [α-32P]-UTP). After 40 min of incubation the reaction was stopped, RNA was purified and radioactively labeled RNAs were analyzed in polyacrylamide gel. Adopted from (Zhou et al. 2019).
The process of germination, the earliest revival event, is considered most puzzling, and holds the secret to understanding the rapid transition from cellular silencing to cellular awakening. Germination is triggered by the binding of nutrients, termed germinants, to multiple germination receptors (Ger-receptors) located in the spore membrane (Paredes-Sabja et al. 2011). The Ger-receptors are assembled into a large germinosome complex, and serve as “aerials” to survey for external germinant factors that include amino acids, sugars, purine nucleosides, DPA, and cell wall derived muropeptides (Setlow 2003, Shah et al. 2008, Griffiths et al. 2011, Christie and Setlow 2020). In B. subtilis, the GerA receptor binds L-alanine, while the GerB and GerK receptors respond jointly to a germinant mixture consisting of asparagine, glucose, fructose, and potassium ions (AGFK) (Setlow 2003, Moir 2006, 2013), Artzi et al. 2021. These ligand-receptor interactions activate scarcely explored downstream signaling events, guiding the spore to revive (Setlow 2003, Moir 2006, Dworkin and Shah 2010, Amon et al. 2021). The bacterial post-translational modification, arginine (Arg) dephosphorylation, guided by the Arg-phosphatase YwlE, was found to facilitate germination by dephosphorylation of downstream target proteins. Consequently, the absence of the cognate Arg-kinase, McsB, prompts the process (Fuhrmann et al. 2009, Elsholz et al. 2012, Schmidt et al. 2014, Zhou et al. 2019, Huang et al. 2021, Zhang et al. 2021). As we shall discuss here, the Arg-phosphoproteome provides a major clue to elucidate molecular pathways succeeding the activation of the Ger-receptors.
Germination is broadly considered to be completed without the need for macromolecule synthesis, while the subsequent stages of revival are characterized by the massive production of RNA and proteins (Steinberg et al. 1965, Vinter 1970, Setlow 2003, 2013, Christie and Setlow 2020). This assessment heavily relies on previous studies, mostly conducted during the 1960s–1970s, showing that Bacilli spores can undergo germination in the presence of RNA and protein synthesis inhibitors [e.g. (Steinberg and Halvorson 1968, Vinter 1970)]. Still, several studies performed back then implied that macromolecule synthesis occurs very early in reviving spores, at a time coinciding with germination, and even in media lacking any growth factors [e.g. (Balassa and Contesse 1965, Torriani and Levinthal 1967, Armstrong and Sueoka 1968)]. Nevertheless, the dogma affirming that germination does not require RNA or protein synthesis was almost unanimously accepted [e.g. (Setlow 2003, Moir 2006, 2013)]. In recent years, our laboratory provided evidence indicating that both transcription and translation are activated during germination and required for its completion. We would like to employ this platform to portray our revised view in conjunction with supporting past and current experiments and discuss this conceptual change and its implications.
Evidence for the occurrence of translation during germination
The fundamental question of whether protein synthesis is a prerequisite for spore germination has been the subject of numerous earlier studies, executed mainly during the 1960s–1970s. A key strategy of those studies was to supplement spores with an array of translation inhibitors and assess germination capacity. Such assays failed to detect an effect on germination, thus leading to the conclusion that germination is independent of protein synthesis [e.g. (Steinberg et al. 1965, Vinter 1970)]. Other researchers challenged these interpretations at that time, claiming that the tested compounds could not cross the impermeable spore shells to reach the targeted ribosomes (Tisdale and DeBusk 1972, Sussman and Douthit 1973). Furthermore, these results were inconsistent with several studies demonstrating the incorporation of radioactive amino acids into newly synthesized proteins within a few minutes following germination induction. For example, Torriani and Levinthal could detect 14C-L-valine incorporation into proteins 3 to 4 minutes post-stimulation of germination (Torriani and Levinthal 1967) (Fig. 1B). Similarly, Shaw and Armstrong assessed protein synthesis in germinating spores by following 3H-phenylalanine incorporation specifically into components composing the 50S ribosomal subunit. Surprisingly, they found that synthesis of the majority of the 50S ribosomal proteins is detectable as early as 5–10 minutes post-germination triggering (Shaw and Armstrong 1972), signifying that translation is likely to initiate even earlier than the detected time, and hence overlaps with the process of germination. In keeping with these findings, polysomes were shown to exist in dormant spores (Chambon et al. 1968, Feinsod and Douthit 1970), hinting that they could be rapidly tuned to translate the associated transcripts upon germination. Coupled with this, biochemical studies showed the existence of different-size ribosomal particles from dormant and germinating spores, inferring prompt modulation of ribosomal complexes during the transition from dormancy to germination [e.g. (Chambon et al. 1968, Idriss and Halvorson 1969, Feinsod and Douthit 1970)].
In the current years, our lab provided evidence reinforcing the view that translation occurs during germination and is essential for its execution (Sinai et al. 2015, Zhou et al. 2019). These unexpected findings came as a surprise while employing the BONCAT (BioOrthogonal Non-Canonical Amino-acid Tagging) protein tagging technique to study the temporal landscape of newly synthesized proteins during spore revival (Dieterich et al. 2007, Sinai et al. 2015). Using this approach, the generation of labeled proteins was monitored when spores were triggered to germinate with L-alanine or with the “non-nutrient” germinant Ca-DPA, in the absence of any other nutrients that are required to support successive revival events (Fig. 1C) (Sinai et al. 2015). By treating dormant spores with ethanol to increase permeability (Tanimoto et al. 1996), we could introduce into the spore core ribosome-targeting antibiotics that subsequently halted labeled-protein production. Furthermore, the antibiotic-treated spores could initiate germination, as indicated by DPA release, but were paused prior to cortex hydrolysis and thus remained phase bright (Sinai et al. 2015). The DPA release suggests that at least partial core rehydration occurred, allowing resumption of protein synthesis among other enzymatic activities. Based on the BONCAT methodology, a set of 30 proteins synthesized during this early stage was defined as the basic germination proteome. The proteome contained proteins required for glycolysis, malate utilization and translation, along with enzymes composing the vital pyruvate dehydrogenase complex (Sinai et al. 2015). Notably, included in the germination proteome were two bona fide translational factors, RpmE, a ribosomal component constituting the 50S subunit (Akanuma et al. 2012), and Tig, a ribosome-associated chaperone (Wegrzyn and Deuerling 2005). Spores lacking these factors were halted in germination due to deficiency in protein synthesis, directly linking these factors with the process of germination-induced translation (Sinai et al. 2015). In line with these findings, dephosphorylation of Tig, phosphorylated on the Arg-residue (R45) by YwlE, was found to propel germination, enabling the association of Tig with the ribosomes to reactivate protein synthesis (Zhou et al. 2019).
The conceptual shift that translation occurs during germination and is required for its occurrence was received with skepticism by some researchers, opposing this prospect (Korza et al. 2016, Christie and Setlow 2020, Swarge et al. 2020b). We carefully assessed these studies, and, to our understanding, the illustrated experiments seem indecisive and lack appropriate controls [e.g. (Sinai and Ben-Yehuda 2016)]. For example, the use of antibiotics or the employment of rpmE or tig translational mutants were not examined by these researchers to sustain or rule out our findings (Boone and Driks 2016). Besides, as we mentioned above, our results are in agreement with numerous earlier deserted studies, reporting that translation can be monitored within a few minutes post-germination triggering (Fig. 1B) [e.g. (Torriani and Levinthal 1967, Rodenberg et al. 1968, Steinberg and Halvorson 1968, Shaw and Armstrong 1972)].
Activation of and the requirement for transcription during germination
The detection of germination-induced translation raises the immediate question as to whether de novo transcription is entailed in the process. In fact, numerous early observations showed the occurrence of RNA synthesis in spores almost immediately after prompting germination [e.g. (Torriani and Levinthal 1967, Armstrong and Sueoka 1968, Rodenberg et al. 1968, Cohen and Keynan 1970, Matsuda and Kameyama 1986)]. Torriani and Levinthal, for instance, reported that incorporation of 14C-uracil into germinating spore RNA occurs as fast as 30–60 sec following germination induction, whereas Amstrong and Sueoka monitored uridine incorporation within two minutes after germinant supplementation (Torriani and Levinthal 1967; Fig. 1B). Armstrong and Sueoka 1968 Co-examination of both RNA and protein synthesis in germinating spores by various studies steadily showed that RNA production precedes protein manufacturing [e.g. (Torriani and Levinthal 1967, Armstrong and Sueoka 1968, Rodenberg et al. 1968)], hinting that there is a firm hierarchy in reactivation of these processes. In a recent transcriptomic analysis of reviving spores, Swarge et al (Swarge et al. 2020a) concluded that there is no significant change in the spore transcriptome during germination. However, they do detect that at the time coinciding with germination completion, transcripts associated with purine and pyrimidine biosynthesis (14 transcripts), amino acid production (9 transcripts), and translational components (31 transcripts) were upregulated, which seems consistent with our view.
Evidence for the necessity for transcription during germination was unexpectedly gained while exploring the spore Arg-phosphoproteome, through which the occurrence of spore-unique Arg-phosphorylation of the housekeeping sigma factor σA, was uncovered (Haldenwang 1995, Zhou et al. 2019). The construction of a mutant strain, mimicking a constitutive phosphorylation state, unveiled the corresponding spores to have a profound deficiency in germination, without any apparent influence on vegetative growth (Zhou et al. 2019). Further examination revealed that the mutant spores were stalled before DPA release and remained heat resistant, indicating a deficiency at an early germination stage, prior to protein synthesis activation. Subsequent analysis monitored an increase in transcript levels of genes included in the germination proteome (i.e., tig and rpmE) in wild type, but not in spores harboring a constitutively phosphorylated sigA mutation. An in vitro system devised for monitoring the transcriptional activity of whole extracts failed to detect transcription by dormant spore extracts. However, triggering germination was sufficient to reactivate the transcriptional machinery in extracts from wild type spores but was undetectable in extracts of sigA phospho-mutant germinating spores (Fig. 1D) (Zhou et al. 2019). These findings indicate that the transcription machinery ceases at the course of dormancy, reinitiates early during germination, and is required to advance the process. This rapid molecular awakening is mediated, at least in part, by Arg-dephosphorylation of SigA by YwlE. The massive RNA degradation observed in spores entering dormancy (Segev et al. 2012, Segev et al. 2013, Camilleri et al. 2019) alludes that the resultant ribonucleotides are used as a reservoir to be consumed through germination. We assume that the subsequent awakening of the translational machinery is fueled by the germination-synthesized transcripts that couple with the ribosomes. In the future, examination of these early-produced transcripts might assist in illuminating the process.
Germination is propelled by phosphorylation and dephosphorylation events
Among the most puzzling germination events is the delivery of a signal from the stimulated germinant receptor into the spore core, conveying the fast resumption of multiple cellular processes including transcription and translation. This primary event is still enigmatic, yet accumulating evidence implies that the co-initiation of various processes is facilitated, at least partially, by protein phosphorylation, a ubiquitous means for mediating rapid cellular responses to external stimuli (Kobir et al. 2011). In fact, an assortment of protein kinase inhibitors was found to interfere with spore awakening in the bacterium Streptomyces coelicolor (Paleckova et al. 2007), whereas muropeptides were shown to trigger spore germination in B. subtilis by activating the eukaryotic-like membrane serine/threonine kinase PrkC (Shah et al. 2008). In line with these findings, phosphoproteomic analysis, defining the Ser/Thr/Tyr phosphoproteome of dormant and reviving spores, supports the notion that phospho-modifications play a central role in rapidly modulating protein activity throughout revival. However, evidence for the direct impact of these phosphorylation events on the actual germination process has not been assigned (Rosenberg et al. 2015). By conducting a genetic unbiased screen to isolate mutants, deficient in germination, we recently identified the arginine phosphatase YwlE as a facilitator of the process (Zhou et al. 2019). Arg- phosphorylation, unique to Gram positive bacteria (Elsholz et al. 2012, Schmidt et al. 2014, Trentini et al. 2016), seems to acquire an exclusive role in spore-forming bacteria, controlling spore dormancy and awakening. Besides the role of YwlE in resuming translation and transcription by dephosphorylating Tig and SigA respectively, the spore Arg-phosphoproteome hints that it could also stimulate the awakening of key metabolic pathways. For instance, GapA, glyceraldehyde-3-phosphate dehydrogenase and Pgi, glucose-6-phosphate isomerase, harbor Arg-phosphorylation sites unique to spores (Zhou et al. 2019), suggesting that YwlE instantly reactivates a plethora of pathways, propelling the exit from dormancy. Though, how the phosphatase activity is unleashed during germination, how it receives the signal from the germination receptors, and whether arginine phosphorylation is only part of an array of multiple phosphorylation-dephosphorylation events that act to resume life remains unrevealed.
Spores are not all made equal
Besides obvious genetic and environmental factors that affect spore germination properties, it becomes evident that phenotypic variation in the spore's temporary molecular cargo impacts its ability to germinate. A prominent observation was that a poorly germinating subpopulation exists in a genetically identical spore population. The germination deficiency of these superdormant spores was found to originate from low levels of germination receptor components due to phenotypic variation (Ghosh and Setlow 2009, Ghosh et al. 2012, Chen et al. 2014). In complementary findings, Sturm and Dworkin showed that spores germinate stochastically due to fluctuating expression of the sporulation transcription factor GerE, involved in the last steps of spore assembly, with spores having the lowest expression of gerE showed the highest frequency of spontaneous germination (Sturm and Dworkin 2015). A more recent study that followed the life cycle of individual cells using time-lapse microscopy exposed that sporulation timing controls spore revival, most probably due to variations in the levels of the metabolic enzyme alanine dehydrogenase (Mutlu et al. 2018, Mutlu et al. 2020). This is consistent with the observations that varying sporulation conditions interferes with germination [reviewed in (Bressuire-Isoard et al. 2018)]. For instance, spores formed in nutrient-poor sporulation medium germinated more slowly than spores formed in nutrient-rich medium (Ramirez-Peralta et al. 2012). Taken together, these findings reinforce the view that phenotypic diversity during the pre-spore period affects spore germination and revival capabilities. Furthermore, while following the RNA content of mature spores over time, we found that a few days after sporulation, the RNA profile is dynamically modified, depending on the spore age and the temperature of incubation. Consequently, the ability to germinate and propagate varies, implying that molecular changes occurring after the conclusion of sporulation continue to shape the spore molecular pool and influence the emergence from quiescence (Segev et al. 2012). As B. subtilis and its relatives reside in diverse ecological niches, different characteristics of each habitat are likely to produce spores harboring dissimilar molecular cargo, impacting germination kinetics.
Perspective
Dormant bacterial spores can survive prolonged periods and withstand extreme conditions but preserve the extraordinary capacity to switch within minutes into actively growing cells, with germination being the initial and most critical event of this transformable process. As we attempted to illuminate throughout this perspective, we suggest that the notion of germination being independent of macromolecule synthesis needs to be reviewed, as accumulating data concomitantly with previous observations, indicate that both transcription and translation are restored during germination and required for its culmination. Our current viewpoint is that the intricate process of spore germination initiates by activation of Ger-receptors that, in turn, activate downstream signal transduction pathways, with the key path comprising the arginine phosphatase YwlE, which dephosphorylates target proteins implicated in metabolism, transcription, and translation. Dephosphorylation of SigA activates transcription (before DPA release), whereas subsequent dephosphorylation of Tig activates translation (after DPA release) (Fig. 2A). Intriguingly, our model greatly resembles that of Hansen, Spiegelman, and Halvorson proposed based on their data obtained approximately 50 years ago (Fig. 2B; Hansen et al. 1970). Our model is also supported by previous studies showing that the spore reservoir contains all elements needed for initial macromolecule synthesis, including nucleotides, and amino acids [e.g. (Kieras et al. 1978, Setlow and Kornberg, 1970, Setlow and Primus, 1975)]. Moreover, the spore holds high concentrations of potential energy sources such as PGA and malate that could be employed for raising the ATP levels during germination (Nelson and Kornberg 1970, Ghosh et al. 2015, Sinai et al. 2015). The revised view we present here might pave the way for developing innovative strategies to interfere with spore germination of dangerous pathogens and those causing food spoilage. It could further enlighten the awakening process of various bacteria exhibiting a latent phase, and be applicable to diverse organisms displaying a dormant form, including fungi spores and plant seeds.
Figure 2.
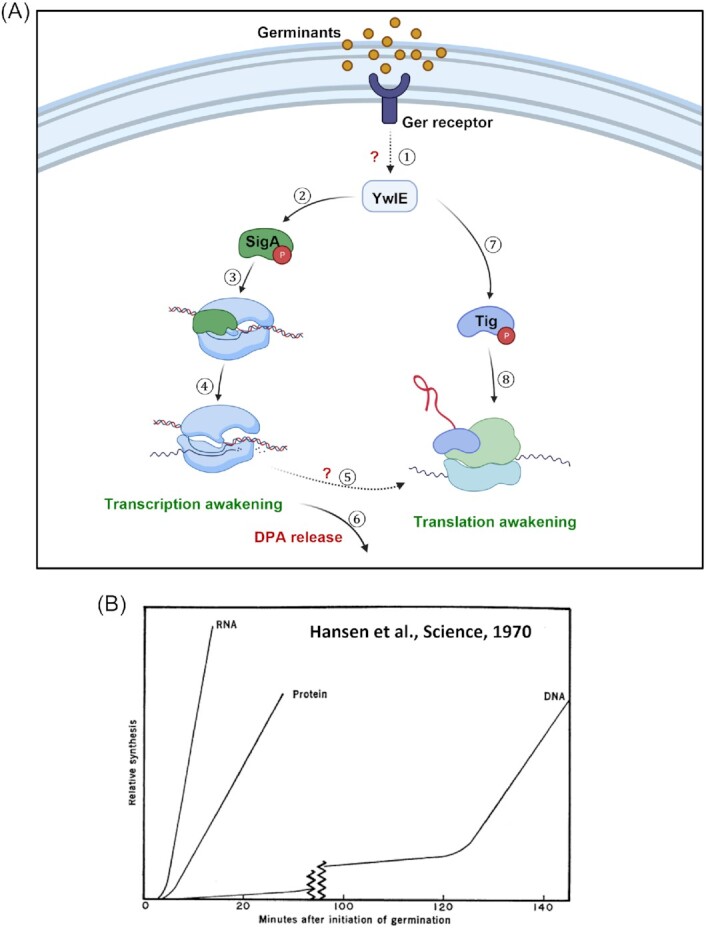
A revised model for bacterial spore germination. (A) A revised view for bacterial spore germination. Following nutrient sensing, the germination receptors activate the Arg phosphatase YwlE (①). Consequently, SigA is dephosphorylated by active YwlE to reactivate the transcription machinery (②③④). Next, translation is reestablished by the YwlE-mediated dephosphorylation of Tig after DPA release (⑥⑦⑧). The source of the transcripts for protein synthesis during germination could be derived from the newly transcribed RNA (⑤). (B) Our revised model very much resembles that of Hansen et al. (Hansen et al. 1970) based on their results. Reprinted with permission from the AAAS.
Funding
This work was supported by the Israel Science Foundation (grant no. 774/16, 308/21) awarded to S. B-Y.
ACKNOWLEDGEMENTS
The authors declare no conflict of interest. We are emendable to members of the Ben-Yehuda laboratory and Ilan Rosenshine (Hebrew University, IL) for valuable discussions and comments. Artwork in Fig. 2 was prepared using BioRender (Agreement Number CQ23NTEHO5).
Contributor Information
Bing Zhou, Department of Microbiology and Molecular Genetics, Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, POB 12272, The Hebrew University of Jerusalem, 91120 Jerusalem, Israel.
Sima Alon, Department of Microbiology and Molecular Genetics, Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, POB 12272, The Hebrew University of Jerusalem, 91120 Jerusalem, Israel.
Lei Rao, Department of Microbiology and Molecular Genetics, Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, POB 12272, The Hebrew University of Jerusalem, 91120 Jerusalem, Israel.
Lior Sinai, Department of Microbiology and Molecular Genetics, Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, POB 12272, The Hebrew University of Jerusalem, 91120 Jerusalem, Israel.
Sigal Ben-Yehuda, Department of Microbiology and Molecular Genetics, Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, POB 12272, The Hebrew University of Jerusalem, 91120 Jerusalem, Israel.
Conflict of interest statement
None declared.
References
- Akanuma G, Nanamiya H, Natori Yet al. Inactivation of ribosomal protein genes in Bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation. J Bacteriol. 2012;194:6282–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon JD, Artzi L, Rudner DZ. Genetic evidence for signal transduction within the Bacillus subtilis GerA germinant receptor. J Bacteriol. 2022;204:e0047021. DOI: 10.1128/JB.00470-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong RL, Sueoka N. Phase transitions in ribonucleic acid synthesis during germination of Bacillus subtilis spores. Proc Natl Acad Sci. 1968;59:153–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artzi L, Alon A, Brock KPet al. Dormant spores sense amino acids through the B subunits of their germination receptors. Nat Commun. 2021;12:6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balassa G, Contesse G. [Macromolecular synthesis during the germination of spores of B. subtilis. I. Kinetics]. Ann Inst Pasteur (Paris). 1965;109:683–705. [PubMed] [Google Scholar]
- Boone T, Driks A. Protein synthesis during germination: shedding new light on a classical question. J Bacteriol. 2016;198:3251–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressuire-Isoard C, Broussolle V, Carlin F. Sporulation environment influences spore properties in Bacillus: evidence and insights on underlying molecular and physiological mechanisms. FEMS Microbiol Rev. 2018;42:614–26. [DOI] [PubMed] [Google Scholar]
- Camilleri E, Korza G, Green Jet al. Properties of Aged Spores of Bacillus subtilis. J Bacteriol. 2019;201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon P, Deutscher MP, Kornberg A. Biochemical studies of bacterial sporulation and germination. X. Ribosomes and nucleic acids of vegetative cells and spores of Bacillus megaterium. J Biol Chem. 1968;243:5110–6. [PubMed] [Google Scholar]
- Chen Y, Ray WK, Helm RFet al. Levels of germination proteins in Bacillus subtilis dormant, superdormant, and germinating spores. PLoS One. 2014;9:e95781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie G, Setlow P. Bacillus spore germination: knowns, unknowns and what we need to learn. Cell Signal. 2020;74:109729. [DOI] [PubMed] [Google Scholar]
- Cohen A, Keynan A. Synthesis of a factor stimulating transcription in outgrowing Bacillus cereus spores. Biochem Biophys Res Commun. 1970;38:744–9. [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Lee JJ, Link AJet al. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc. 2007;2:532–40. [DOI] [PubMed] [Google Scholar]
- Driks A. Maximum shields: the assembly and function of the bacterial spore coat. Trends Microbiol. 2002;10:251–4. [DOI] [PubMed] [Google Scholar]
- Dworkin J, Shah IM. Exit from dormancy in microbial organisms. Nat Rev Microbiol. 2010l;8:890–6. [DOI] [PubMed] [Google Scholar]
- Elsholz AK, Turgay K, Michalik Set al. Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis. Proc Natl Acad Sci. 2012;109:7451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinsod FM, Douthit HA. Ribosomes from spores of Bacillus cereus T. Science. 1970;168:991. [DOI] [PubMed] [Google Scholar]
- Fuhrmann J, Schmidt A, Spiess Set al. McsB is a protein arginine kinase that phosphorylates and inhibits the heat-shock regulator CtsR. Science. 2009;324:1323–7. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Korza G, Maciejewski Met al. Analysis of metabolism in dormant spores of Bacillus species by 31P nuclear magnetic resonance analysis of low-molecular-weight compounds. J Bacteriol. 2015;197:992–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Scotland M, Setlow P. Levels of germination proteins in dormant and superdormant spores of Bacillus subtilis. J Bacteriol. 2012;194:2221–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Setlow P. Isolation and characterization of superdormant spores of Bacillus species. J Bacteriol. 2009; 191:1787–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths KK, Zhang J, Cowan AEet al. Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Mol Microbiol. 2011;81:1061–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldenwang WG. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JN, Spiegelman G, Halvorson HO. Bacterial spore outgrowth: its regulation. Science. 1970;168:1291–8. [DOI] [PubMed] [Google Scholar]
- Henriques AO, Moran CP Jr. Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol. 2007;61:555–88. [DOI] [PubMed] [Google Scholar]
- Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev. 2012;36:131–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Zhao Z, Zhao Yet al. Protein arginine phosphorylation in organisms. Int J Biol Macromol. 2021;171:414–22. [DOI] [PubMed] [Google Scholar]
- Hutchison EA, Miller DA, Angert ER. Sporulation in Bacteria: beyond the Standard Model. Microbiol Spect. 2014;2. [DOI] [PubMed] [Google Scholar]
- Idriss JM, Halvorson HO. The nature of ribosomes of spores of Bacillus cereus T. and Bacillus megaterium. Arch Biochem Biophys. 1969;133:442–53. [DOI] [PubMed] [Google Scholar]
- Kieras RM, Preston RA, Douthit HA. Isolation of stable ribosomal subunits from spores of Bacillus cereus. J Bacteriol. 1978;136:209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobir A, Shi L, Boskovic Aet al. Protein phosphorylation in bacterial signal transduction. Biochim Biophys Acta. 2011;1810:989–94. [DOI] [PubMed] [Google Scholar]
- Korza G, Setlow B, Rao Let al. Changes in Bacillus spore small molecules, rRNA, germination and outgrowth after extended sub-lethal exposure to various temperatures: evidence that protein synthesis is not essential for spore germination. J Bacteriol. 2016;198:3254–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Kameyama T. Fractionation of ribonucleic acid transcripts synthesized during spore germination in Bacillus subtilis. J Basic Microbiol. 1986;26:91–9. [DOI] [PubMed] [Google Scholar]
- McKenney PT, Driks A, Eskandarian HAet al. A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat. Curr Biol. 2010;20:934–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir A. How do spores germinate?. J Appl Microbiol. 2006;101:526–30. [DOI] [PubMed] [Google Scholar]
- Mutlu A, Kaspar C, Becker Net al. A spore quality-quantity tradeoff favors diverse sporulation strategies in Bacillus subtilis. ISME J. 2020;14:2703–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu A, Trauth S, Ziesack Met al. Phenotypic memory in Bacillus subtilis links dormancy entry and exit by a spore quantity-quality tradeoff. Nat Commun. 2018;9:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DL, Kornberg A. Biochemical studies of bacterial sporulation and germination. 18. Free amino acids in spores. J Biol Chem. 1970;245:1128–36. [PubMed] [Google Scholar]
- Paleckova P, Kontrova F, Kofronova Oet al. Effect of protein kinase inhibitors on protein phosphorylation and germination of aerial spores from Streptomyces coelicolor. Folia Microbiol (Praha). 2007;52:215–22. [DOI] [PubMed] [Google Scholar]
- Paredes-Sabja D, Setlow P, Sarker MR. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 2011;19:85–94. [DOI] [PubMed] [Google Scholar]
- Paredes-Sabja D, Shen A, Sorg JA. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol. 2014;22:406–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Peralta A, Zhang P, Li YQet al. Effects of sporulation conditions on the germination and germination protein levels of Bacillus subtilis spores. Appl Environ Microbiol. 2012;78:2689–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenberg S, Steinberg W, Piper Jet al. Relationship between protein and ribonucleic acid synthesis during outgrowth of spores of Bacillus cereus. J Bacteriol. 1968;96:492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg A, Soufi B, Ravikumar Vet al. Phosphoproteome dynamics mediate revival of bacterial spores. BMC Biol. 2015;13:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo LY, Doi RH. Ultrastructural analysis during germination and outgrowth of Bacillus subtilis spores. J Bacteriol. 1974;120:475–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Trentini DB, Spiess Set al. Quantitative phosphoproteomics reveals the role of protein arginine phosphorylation in the bacterial stress response. Mol Cell Proteomics. 2014;13:537–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev E, Rosenberg A, Mamou Get al. Molecular kinetics of reviving bacterial spores. J Bacteriol. 2013;195:1875–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev E, Smith Y, Ben-Yehuda S. RNA dynamics in aging bacterial spores. Cell. 2012;148:139–49. [DOI] [PubMed] [Google Scholar]
- Setlow P, Kornberg A. Biochemical studies of bacterial sporulation and germination .22. Energy metabolism in early stages of germination of Bacillus megaterium spores. J Biol Chem. 1970;245:3637–44. [PubMed] [Google Scholar]
- Setlow P, Primus G. Protein metabolism during germination of Bacillus megaterium spores. I. Protein synthesis and amino acid metabolism. J Biol Chem. 1975;250:623–30. [PubMed] [Google Scholar]
- Setlow P, Wang S, Li YQ. Germination of spores of the orders Bacillales and Clostridiales. Annu Rev Microbiol. 2017;71:459–77. [DOI] [PubMed] [Google Scholar]
- Setlow P. I will survive: DNA protection in bacterial spores. Trends Microbiol. 2007;15:172–80. [DOI] [PubMed] [Google Scholar]
- Setlow P. Spore germination. Curr Opin Microbiol. 2003;6:550–6. [DOI] [PubMed] [Google Scholar]
- Setlow P. Summer meeting 201–when the sleepers wake: the germination of spores of Bacillus species. J Appl Microbiol. 2013;115:1251–68. [DOI] [PubMed] [Google Scholar]
- Shah IM, Laaberki MH, Popham DLet al. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw MV, Armstrong RL. Biosynthesis of 50S ribosomal proteins during the outgrowth of Bacillus subtilis spores. J Bacteriol. 1972;109:276–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinai L, Ben-Yehuda S. Commentary: changes in Bacillus spore small molecules, rRNA, germination, and outgrowth after extended sublethal exposure to various temperatures: evidence that protein synthesis Is not essential for spore germination. Front Microbiol. 2016;7:2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinai L, Rosenberg A, Smith Yet al. The molecular timeline of a reviving bacterial spore. Mol Cell. 2015;57:695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloma A, Smith I. RNA synthesis during spore germination in Bacillus subtilis. Mol Gen Genet. 1979;175:113–20. [DOI] [PubMed] [Google Scholar]
- Steinberg W, Halvorso H, Keynan Aet al. Timing of protein synthesis during germination and outgrowth of spores of Bacillus cereus strain T. Nature. 1965;208:710–1. [Google Scholar]
- Steinberg W, Halvorson HO. Timing of enzyme synthesis during outgrowth of spores of Bacillus cereus. I. Ordered enzyme synthesis. J Bacteriol. 1968;95:469–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. [DOI] [PubMed] [Google Scholar]
- Sturm A, Dworkin J. Phenotypicdiversity as a mechanism to exit cellular dormancy. Curr Biol. 2015;25:2272–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman AS, Douthit HA. Dormancy in microbial spores. Annu Rev Plant Phys. 1973;24:311–52. [Google Scholar]
- Swarge B, Abhyankar W, Jonker Met al. Integrative Analysis of Proteome and Transcriptome Dynamics during Bacillus subtilis Spore Revival. mSphere. 2020a;5:e00463–20. DOI: 10.1128/mSphere.00463-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarge B, Nafid C, Vischer Net al. Investigating synthesis of the MalS malic enzyme during Bacillus subtilis spore germination and outgrowth and the Influence of spore maturation and sporulation conditions. mSphere. 2020b;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto Y, Ichikawa Y, Yasuda Yet al. Permeability of dormant spores of Bacillus subtilis to gramicidin S. FEMS Microbiol Lett. 1996;136:151–6. [DOI] [PubMed] [Google Scholar]
- Tisdale JH, DeBusk AG. Permeability problems encountered when treating conidia of neurospora crassa with RNA synthesis inhibitors. Biochem Biophys Res Commun. 1972;48:816–22. [DOI] [PubMed] [Google Scholar]
- Torriani A, Levinthal C. Ordered synthesis of proteins during outgrowth of spores of Bacillus cereus. J Bacteriol. 1967;94:176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentini DB, Suskiewicz MJ, Heuck Aet al. Arginine phosphorylation marks proteins for degradation by a Clp protease. Nature. 2016;539:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinter V. Symposium on bacterial spores: V. Germination and outgrowth: effect of inhibitors. J Appl Bacteriol. 1970;33:50–9. [DOI] [PubMed] [Google Scholar]
- Wegrzyn RD, Deuerling E. Molecular guardians for newborn proteins: ribosome-associated chaperones and their role in protein folding. Cell Mol Life Sci. 2005;62:2727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells-Bennik MH, Eijlander RT, den Besten HMet al. Bacterial Spores in Food: survival, Emergence, and Outgrowth. Annu Rev Food Sci Technol. 2016;7:457–82. [DOI] [PubMed] [Google Scholar]
- Zhang A, Pompeo F, Galinier A. Overview of protein phosphorylation in bacteria with a main focus on unusual protein kinases in Bacillus subtilis. Res Microbiol. 2021;172:103871. [DOI] [PubMed] [Google Scholar]
- Zhou B, Semanjski M, Orlovetskie Net al. Arginine dephosphorylation propels spore germination in bacteria. Proc Natl Acad Sci. 2019;116:14228–37. [DOI] [PMC free article] [PubMed] [Google Scholar]


