ABSTRACT
Methyltransferase (MTases) enzymes transfer methyl groups particularly on proteins and nucleotides, thereby participating in controlling the epigenetic information in both prokaryotes and eukaryotes. The concept of epigenetic regulation by DNA methylation has been extensively described for eukaryotes. However, recent studies have extended this concept to bacteria showing that DNA methylation can also exert epigenetic control on bacterial phenotypes. Indeed, the addition of epigenetic information to nucleotide sequences confers adaptive traits including virulence-related characteristics to bacterial cells. In eukaryotes, an additional layer of epigenetic regulation is obtained by post-translational modifications of histone proteins. Interestingly, in the last decades it was shown that bacterial MTases, besides playing an important role in epigenetic regulations at the microbe level by exerting an epigenetic control on their own gene expression, are also important players in host–microbe interactions. Indeed, secreted nucleomodulins, bacterial effectors that target the nucleus of infected cells, have been shown to directly modify the epigenetic landscape of the host. A subclass of nucleomodulins encodes MTase activities, targeting both host DNA and histone proteins, leading to important transcriptional changes in the host cell. In this review, we will focus on lysine and arginine MTases of bacteria and their hosts. The identification and characterization of these enzymes will help to fight bacterial pathogens as they may emerge as promising targets for the development of novel epigenetic inhibitors in both bacteria and the host cells they infect.
Keywords: methyltransferase, epigenetics, bacterial pathogens, Legionella
Bacterial methyltransferases play a role in epigenetic regulations in bacteria as well as in controlling the host epigenetic landscape during infection
Introduction
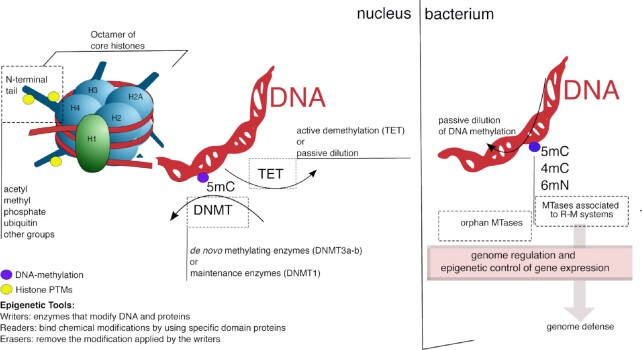
The epigenome consists of a network of modifications on nucleotides or, in the case of eukaryotes, on histone proteins, that lead to the alteration of the biochemical landscape of DNA, without altering the DNA sequence, but directly impacting its structural conformation and, therefore, affecting transcriptional regulation. In eukaryotes, epigenetic regulation involves DNA methylation and histone post-translational modifications, whereas in bacteria, which lack histone proteins, epigenetic control relies on DNA methylation only (Fig. 1).
Figure 1.
The epigenomes of eukaryotes and bacteria. In eukaryotes, epigenetic modifications involve DNA methylation (purple tag) and histone modifications [yellow tag; for a complete list of possible chemical modifications on histone proteins please refer to Huang et al. (2014) and Zhao and Garcia (2015)]. The DNA is packaged in a structure called chromatin, which regulates its activity and inheritance and is organized in fundamental structures called nucleosomes. Each nucleosome consists of a segment of 147 bp of DNA wrapped around an octamer of proteins containing two copies each of four different histones: H2A, H2B, H3, and H4. This arrangement of 11 nm of DNA and its associated proteins forms a fiber, which plays a major role in the cell. In fact, the regulatory proteins that interact with target subunits of the fiber can increase or decrease the compactness of the chromatin structure, leading to enhancing or reducing gene expression. Separate enzymes are responsible for de novo methylation and the maintenance of DNA methylation. DNA demethylation can occur by an active process driven by dedicated proteins (TET enzymes), or by a passive one, where DNA methylation is diluted upon DNA replication. Typically, the methylated base in eukaryotes is C5m, whereas it is often N6m in bacteria. Most bacterial DNA-methyltransferases (MTases) belong to the restriction–modification (R–M) system, responsible for genome defense, whereas orphan DNA MTases have no apparent cognate REase. Both kinds of MTases play a role in epigenetic regulations in bacteria.
The plethora of enzymes catalyzing these modifications on DNA and histones are categorized in three functional classes: the writers and erasers, a dedicated group of enzymes that add and remove, respectively, various chemical modifications; and the readers, specialized domain containing proteins that identify and interpret those modifications (Venkatesh and Workman 2015). Epigenetic signatures and their functional consequences contribute to the normal development of an organism, but also environmental factors influence the epigenetic state, and consequent regulation. In addition to many studies into these diverse regulations in eukaryotic cells, it was shown how some bacterial pathogens can influence and in a certain way govern the epigenetic state of host cells in a dynamic manner by directly modifying the chromatin.
In this review, we discuss a specific class of epigenetic writers, methyltransferases (MTases), that methylate both DNA and proteins, thereby acting as epigenetic tools, from bacteria to eukaryotes.
MTases: an eclectic class of enzymes
MTases are a large group of enzymes that, for their majority, methylate their substrate using S-adenosyl-L-methionine (AdoMet or SAM) as methyl donor. The methyl group may be transferred to form methylated derivatives of proteins, lipids, polysaccharides, nucleic acids, and various small molecules. Methylation reactions are essential transformations in biology, as they control countless cellular processes by transforming the metabolism of molecules. Here, we will focus on methylation of nucleotides (DNA or RNA) and proteins as central components of the epigenetic machinery of both prokaryotic and eukaryotic organisms.
DNA-methylation: setting up the epigenome
DNA-MTases catalyze the transfer of a methyl group from SAM to cytosine or adenine bases embedded in a specific DNA sequence. The methyl group is positioned in the major grove of the DNA helix, where it can easily attract or repel various DNA-binding proteins. Such a methylation adds postreplicative extra information to the DNA without changing the original sequence, as newly synthetized DNA strands do not carry any methylation (Jeltsch 2002). The C-5 and N-4 positions of cytosine and N-6 position of adenine are the target sites for methylation. All three methylation patterns are found in prokaryotes, but it was thought that in eukaryotes only the methylation of cytosine at the C-5 position exists. However, a recent in-depth analysis of rarely modified bases demonstrated that N-6 adenine can also be methylated in eukaryotes (Wu et al. 2016, Zhu et al. 2018).
Prokaryotic DNA-MTases can be classed in two major groups, depending on the position of the base they target in the double helix. Endocyclic MTases target the cytosine C-5, and exocyclic amino MTases methylate the adenine at the N-6 position or the cytosine at N-4 position (Bheemanaik et al. 2006). In general, all DNA MTases show structural similarity, in particular the SAM-binding domain is well conserved across kingdoms. In contrast, the target recognition domain shows high variability in sequence and structure and is closely tied to the target specificities (Malone et al. 1995). In mammalian cells, DNA methylation patterns are established during embryonic development by de novo methylating enzymes called Dnmt3a and Dnmt3a, and maintained by a Dnmt1-mediated copying mechanism during cell division (Jones and Liang 2009).
DNA methylation marks are chemically stable. In bacteria the removal of the methylation is usually achieved by two rounds of DNA replication (passive demethylation), in contrast in eukaryotes DNA methylation can also be actively removed. Passive demethylation simply requires the impairment of the maintenance of the DNA methylation machinery, which results in a 2-fold dilution of methyl-CpG groups during each round of DNA synthesis, whereas active demethylation occurs via the action of the Ten–eleven translocate (TET) family of dioxygenases, through a complex cycle of repeated oxidations (He et al.2011, Ito et al. 2011). TET-like genes have also been identified in bacteria; thus it is likely that such systems might also function in prokaryotes (Iyer et al. 2009).
In bacteria and eukaryotes, DNA-methylation is generally associated with transcriptional repression. The methyl group of 6mA, 5mC, and 4mC protrudes from the major groove of the double helix, thereby providing a platform for DNA-binding proteins to bind cognate nucleotide sequences. 5-cytosine methylation (5mC) is typically involved in the control of eukaryotic transcription and is associated with gene silencing. This modification is conserved across all kingdoms of eukaryotes, where it is generally found in the CpG dinucleotide context, where the cytosine in the dinucleotide sequence 5’-CpG-3’ is modified. Its best characterized function is the repression of the transcription of potentially deleterious transposable elements (TEs), however, it also plays an important role in silencing of germline-specific genes and in developmental processes, such as X-chromosome inactivation via transcriptional silencing (Jones 2012). Indeed, transcription can be regulated by controlling DNA-methylation of specific regions found mainly directly upstream of gene promoters, containing clusters of CpG sequences that are named CpG islands (Deaton and Bird 2011). Methylation of these regions leads to a drastic gene repression by interfering with transcription factors (Zhu et al. 2016), as well as by the direct binding of a family of proteins, known as methyl-CpG binding domain proteins (MBDs). MBDs bind DNA-methylated CpGs and recruit repressor complexes to methylated promoter regions, thereby contributing to transcriptional silencing (Du et al. 2015). Transcriptional activation is regulated via multiple mechanisms involved in protecting CpG islands from de novo methylation and thereby maintaining these regions unmethylated (Weber et al. 2007)
In bacteria, DNA-methylation is often found at N-6 adenine (6mA) and N-4 cytosine (4mC). In particular, it has been established a clear link between 6mA and transcriptional regulation of essential processes such as conjugation, regulation of DNA replication initiation, cell cycle control, nucleoid reorganization, DNA mismatch repair, transcriptional regulation of housekeeping and virulence genes, and post-transcriptional gene regulation [see Chapter 2/and Wion and Casadesús (2006)].
RNA-methylation as a part of the epitranscriptome
In the last decade, a new field of research, RNA modifications, added an additional layer of complexity to gene regulation. Indeed, similar to DNA, cellular RNAs and especially the so-called regulatory RNAs—including miRNAs, piRNAs, endogenous siRNAs, and long noncoding RNAs—may be decorated with diverse chemical modifications (Roundtree et al. 2017). If RNAs were once thought of as a gene expression intermediate only, today it is well-established that RNA modifications provide an additional layer of gene-expression control. This emerging field of investigations is referred to as “RNA epigenetics,” or “epitranscriptomics” (Saletore et al. 2012).
RNAs from all kingdoms of life can be post-transcriptionally modified with more than 150 chemically distinct additions known to date (Boccaletto et al. 2018), that influence RNA folding and function. Methylation is the most common RNA modification as roughly two-thirds of RNA modifications involve the addition of methyl groups. In particular, m6A RNAs are found in all kingdoms of life.
In eukaryotes, 57 RNA MTases have been identified, targeting different bases and riboses of coding and noncoding RNAs [for a complete review on human RNA MTase classification and their targets, see Schapira (2016) and Romano et al. (2018)]. Among them, N6-Methyladenosine (m6A), first observed more than 40 years ago (Desrosiers et al. 1974), is the most abundant mark on eukaryotic mRNAs (one-third of transcripts) and ncRNAs and represents one of the best-studied RNA modifications so far (He and He 2021, Zaccara et al. 2019). Further interest in m6A mRNA has recently emerged, due to the identification of specific demethylases such as FTO and ALKB5H, both belonging to the AlkB family of dioxygenases responsible for converting m6A to adenosine (Jia et al. 2011, Zheng et al. 2013). These findings, together with the identification of m6A readers (Wang et al. 2014), support the idea that chemical modifications on RNAs could represent a reversible and dynamic mode of post-transcriptional regulation (Shi et al. 2019). In fact, the dynamic equilibrium between methylated and unmodified RNA bases, including m6A but also other methylated bases (Wiener and Schwartz 2021), controls a variety of physiological relevant processes, such as splicing, stability, turnover, nuclear export, and mediation of cap-independent translation, showing that it is a dynamic process.
In bacteria, methylated adenosines play a structural role in increasing the efficiency of stacking in ncRNAs, such as ribosomal RNAs. An example is Escherichia coli, that contains two m6A residues on its 23S rRNA: these methylated adenosines have been shown to protrude from the RNA loop and form stacking interactions within the ribosomal RNA (Kierzek and Kierzek 2003). However, mechanisms that connect methylation of bacterial RNAs with gene expression are still unknown, as in bacteria, the majority of methylations, and chemical modifications in general, are located in tRNAs and rRNAs (Boccaletto et al. 2018). Likewise, the work of Deng et al. (2015) demonstrated that m6A is also an abundant mRNA modification in E. coli and Pseudomonas aeruginosa as high-resolution transcriptome wide m6A profiling revealed a conserved and distinct m6A distribution pattern.
Protein MTases modifying chromatin
Nitrogen (N-) or oxygen (O-) atoms are the most common targets for protein methylation. The amino acids targeted by N-methylation are lysine, arginine, histidine, glutamine, and asparagine, whereas O-methylation targets the carboxyl groups of glutamate and aspartate. Protein methylation, like other chemical modifications on amino acids, influences the local charge of the molecule, by increasing its hydrophobicity. Also, methylation of negatively charged amino acids can significantly affect their 3D shape, and consequently their function.
In eukaryotes, protein MTases control epigenetic regulation by targeting histone proteins, specifically lysine and arginine methylations of histones have been first described in the late 1960s and have been extensively characterized since then (Murn and Shi 2017). DNA is wrapped around histone proteins to form a complex called chromatin that allow the DNA to be packaged up and condensed. Dynamic histone modifications may, therefore, control DNA packaging and regulate nuclear regulation. In total, two main classes of histone protein MTases, the enzymes encoding a SET [Su(var), E(z), and Trithorax] domain and those encoding a Rossmann fold, also known as 7-β-sheets family, have been described to date (Falnes et al. 2016, Gana et al. 2013). In human cells, SET domain containing MTases constitute a family of about fifty proteins acting as lysine MTases (PKMTs) that methylate various N-terminal lysine residues of histones H3 and H4 (Dillon et al. 2005). Proteins belonging to the Rossmann fold family are arginine MTases (PRMTs), comprising nine enzymes (designated PRMT1-9), and the PKMT named DOT1L (disruptor of telomeric silencing), a structurally unique enzyme that has been shown to overlay with arginine MTases, rather than SET-domain containing enzymes (Min et al. 2003). DOT1 L exclusively methylates Ly79 in the globular region of histone H3 (H3K79), leading to sequential mono-, di- and trimethylated forms (Feng et al. 2002). Lysine can exist in four methylation states (unmodified, mono- di-, or trimethylated), whereas arginine can be unmodified, mono- or dimethylated (dimethylated arginine residues can occur in either symmetric—two separate nitrogen atoms—or asymmetric—same nitrogen). For a complete review on enzymatic properties of lysine and arginine MTases, please refer to Boriack-Sjodin and Swinger (2016). As previously mentioned for DNA MTases, histone protein methylations, together with other PTMs on histone proteins, alter noncovalent contacts within and between nucleosomes, thereby impacting on their function. Considering the substantial number of lysines that can be methylated, each with multiple methylation states, histone modifications regulate an array of biological processes. Notably, the location and the degree of methylation of a particular residue is associated with a particular transcriptional state or chromatin structure, as it is now generally accepted that histone modifications serve as signals for the recognition by effectors or reader proteins, which impact chromatin structure and function (Lee et al. 2010).
The dynamic status of these modifications implies the existence of histone demethylases. Indeed, histone lysine demethylases (KDMs) remove methyl group(s) from lysines, and arginine demethylases (RDMs) from arginines. A total of eight subfamilies of histone lysine demethylases (KDM 1–8) have been characterized and classed in two major groups: (i) Lys-specific demetylases or LSD demethylases, which were the first reported KDMs, that oxidize the ε-amino group of Lys, thus they allow only demethylation of mono- and dimethyl lysines; and (ii) α-ketoglutarate-dependent Jumonji C-terminal domain (JMJC)-containing demethylases, that oxidize the attached methyl group, which allow the demethylation of mono-, di-, and trimethyl lysines (Kooistra and Helin 2012). In contrast, arginine demethylases (RDMs), are not very well-characterized although several studies indicated the reversibility of this modification. To date, two histone RDMs have been reported, PAD4 (Wang et al. 2004) and JMJD6 (Chang et al. 2007), however, their activities have been questioned meanwhile. Furthermore, several KDMs have been shown to also demethylate arginines (Walport et al. 2016).
While epigenetic regulation by DNA methylation in bacteria is increasingly studied, how protein modifications may impact DNA processes remain an open question. In bacteria, DNA interacts with small, basic, nucleoid-associated proteins (NAPs), which are responsible for chromosome compaction and the coordination of DNA replication and transcription (Dillon and Dorman 2010). Although some of them have been referred to as “histone-like proteins” (HUs) and have been shown to function as transcriptional coactivators and corepressors (Aki and Adhya 1997), the possibility that bacteria specifically modulate their chromatin proteins by PTMs is still under discussion (Carabetta 2021).
2/Epigenetics in bacteria: main role of DNA-MTases
Most bacterial DNA-MTases belong to restriction–modification (R–M) systems, that were first recognized in E. coli for limiting and regulating bacteriophage infections (Gold et al. 1963). R–M systems are ubiquitous in the bacterial world and generally encode two enzymes with the same DNA binding specificity: a DNA adenine MTase, that modifies a specific target sequence in the host genome to protect it from cleavage of a target sequence-specific endonuclease (REase), that cleaves unmethylated or inappropriately methylated targets from exogenous DNA. They are typically regarded as innate defense systems as they serve to identify and eliminate foreign DNA providing a barrier against genetic flux between different lineages (Mruk and Kobayashi 2014). R–M systems are classified in four major types, differing in their molecular structure, sequence recognition, cleavage position, and cofactor requirements (Roberts et al. 2003).
Oliveira and Fang (2020) recently reported a total of 26582 MTases in 5568 complete bacterial genomes, with Type II MTases present in the highest density. Moreover, 52% of the species harbor persistent MTases, defined as conserved in at least ≥ 80% of the genomes of each species, that recognize the same target sites on DNA (Oliveira and Fang 2020). Some DNA MTases, known as orphans, have no apparent cognate REase gene and, although a possible origin from degraded R–M systems has been hypothesized, it seems clear that the majority are acquired by horizontal gene transfer in their orphan state and further kept due to strong selective pressure (Oliveira et al. 2014). Often, orphan MTases are involved in genome regulation and epigenetic control of gene expression as, in contrast to methylation by R–M systems, methylation by orphan MTases (whether persistent or not) often produces patters of DNA methylation that are consistent with gene regulatory functions (Blow et al. 2016, Oliveira and Fang 2020). Interestingly, Blow et al. (2016) showed that, if MTases of R–M systems are almost always associated with complete DNA modifications of their genomes, consistent with their role in protecting the genome from the cognate restriction enzymes, the orphan MTases are associated with small subsets of consistently unmethylated sites throughout the genome. This distinctive signature of orphan MTases represents a regulatory mechanism of gene expression, in fact the majority of orphan MTases are associated with a substantial enrichment of unmethylated motifs in regulatory regions of the genome (Blow et al. 2016). An example for an orphan and persistent MTase is the deoxyadenosine MTase Dam, i.e. widespread in γ-proteobacteria. It catalyzes postreplicative formation of 6mA in the palindromic 5’-GATC-3’ motif (Brooks et al. 1983), and controls transcription of specific genes that regulate diverse processes, including DNA replication timing, DNA repair, nucleoid segregation, phase-variation switches, but also virulence of bacterial pathogens (Adhikari and Curtis 2016, Marinus and Casadesus 2009). For instance, in Salmonella typhimurium Dam controls bacterial virulence (Heithoff et al. 1999) and the expression of the pathogenicity island-1 (SP-1; Balbontín et al. 2006). In Yersinia enterocolitica Dam overproduction causes both transcriptional and post-transcriptional alterations in the synthesis of virulence factors (Fälker et al. 2007).
MTases are thus key factors for the regulation of gene expression, in particular in the context of phase variation mechanisms that control the formation of phenotypically distinct cells in populations of genetically identical bacteria. Reversible and high-frequency transitions between two distinct states, resulting in ON/OFF switching of expression, are usually mediated by mutations at genomic repeat sequences located either within the genes encoding variant proteins, or in their promoter regions, as well as by site-specific recombination or epigenetic regulation mediated by DNA-methylation (Woude 2011).
Pathogenic bacteria, that are frequently challenged by rapid changing environments such as immune defenses of their hosts and have to adapt to continuous selective pressure over many individual cycles of transmission, often regulate expression of their virulence genes by phase variation. Classical examples are bacterial surface factors required for initial adherence for host colonization, such as pili, adhesins, flagella, and lipopolysaccharide (Phillips et al. 2019a). The first characterized and most studied example is the pap (pyelonephritis-associated pili) operon of uropathogenic E. coli that encodes fimbriae for adhesion to the urinary epithelium. The expression of the operon is regulated by the formation of DNA-methylation patterns by the MTase Dam, i.e. regulating transcription factor binding in regulatory regions, thereby leading to ON/OFF switches (Blyn et al. 1990, Woude et al. 1996). If we consider that human-adapted pathogens have many phase-variable genes, the combinatorial power of this contingency strategy to generate a highly diverse population becomes apparent. For example, almost 100 putative phase-variable genes have been identified in Neisseria spp., indicating the huge potential for diversification mediated by gene switching during host colonization (Snyder et al. 2001).
Orphan DNA MTases were thought to be the only ones playing a role in epigenetic regulations in bacteria, and MTases associated to R–M systems were thought to function “only” in genome defense. However, an increasing number of MTases associated with R–M systems has recently been shown to have additional roles in transcriptional regulation and formation of phenotypic cell variants.
Indeed, many bacterial pathogens contain MTases associated with R–M systems that are subject to phase variation. Phase variation of the expression of DNA MTase results in differential DNA methylation patterns throughout the genome, translating into expression changes of multiple genes via epigenetic mechanisms. These systems are called phasevarions (phase-variable regulons). The concept of phasevarions was first described in Haemophilus influenzae, where phase-variable ON/OFF switching of a Type III DNA MTase results in the phase variation of an entire regulon that differentiates the bacterial cell into two alternative expression states with multiple phenotypic differences (Srikhanta et al. 2005). Since then, phase-variable Type III and Type I restriction–modification systems that control a regulon of genes via changes in global DNA methylation caused by the phase variation of the restriction–modification systems, have been reported in a range of human-adapted bacterial pathogens. For a complete and recent review on the identification and analysis of phase-variable expressed DNA MTases associated with R–M systems and their epigenetic regulation of virulence and immune-evasion in human-adapted bacterial pathogens please refer to (Seib et al. 2020).
Although the majority of studies focused on the activity of adenine MTases as epigenetic regulators, recent works started to explore the role of 5mC and 4mC in epigenetic signaling. The formation of 5mC may influence gene expression in Helicobacter pylori, via the activity of the JHP1050 MTase (Estibariz et al. 2019), in Vibrio cholerae, where the VchM orphan 5mC MTase has been shown to be necessary for optimal growth during infection (Chao et al. 2015), or in E. coli (Kahramanoglou et al. 2012, Militello et al. 2014). 4mC has recently been shown to affect gene expression and virulence related traits in H. pylori (Kumar et al. 2018). The recent identification of these methylations in bacterial genome suggests that DNA methylation remains still a poorly understood component of prokaryotic life and it plays a much deeper role than currently thought.
3/Bacterial MTases and their function in targeting the host
During bacterial infection, host cells activate a series of proinflammatory responses to avoid microbial colonization and to delay bacterial spread. Thus, pathogenic bacteria evolved a wide range of strategies to avoid eradication by their hosts. In particular, elaborate secretion systems inject virulence factors, named effectors, into host cells in order to subvert host defenses (Galán and Waksman 2018). These protein effectors allow bacteria to compete with other microorganisms colonizing the same niche and to interact with the host signaling pathways for blocking, or delaying, the host cell response and to promote their own survival. Furthermore, many of these effectors hijack the host cell response through chemical modifications by the addition or removal of functional groups to host proteins or nucleotides to destabilize the host and to overcome host defenses.
For a long time, it was believed that bacterial effectors released by the different secretion systems act only in the host cytosol (Rapisarda and Fronzes 2018). However, in late 1977 a segment of the Ti plasmid, thereafter named T-DNA of Agrobacterium turmefaciens, had been found integrated into plant cells infected by this phytopathogen (Hooykaas et al. 1977). Virulent Agrobacterium strains transfer single strand T-DNA and several virulence effector proteins (mainly VirD2) through a specialized type-4 secretion system (T4SS) into plant cells. The single stranded T-DNA traverses the host cell cytoplasm and enters the host cell nucleus, where it eventually integrates in the host genome, promoting uncontrolled cell proliferation and interfering with host transcription mechanisms to produce nutrients essential for bacterial survival (Gelvin 2017).
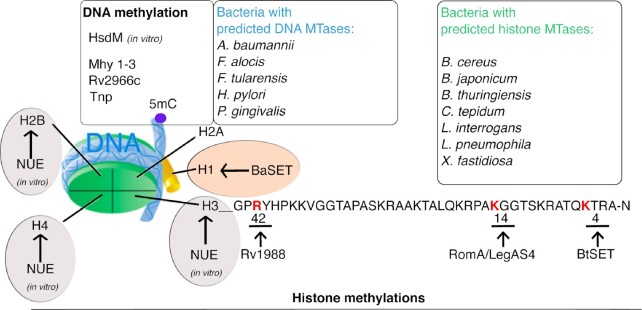
In the last decade, a new family of bacterial, secreted effectors has been characterized, the so-called nucleomodulins (Bierne and Cossart 2012). Derived from the combination of the words “nucleus” and “modulins,” these proteins represent a group of molecules that enter the host cell nucleus to hijack nuclear functions such as modulating the expression of key genes for the host cell response to the pathogen. The last decade has witnessed an increase in the number of nucleomodulins identified, targeting various nuclear elements (Bierne and Pourpre 2020). Most of the bacterial nucleomodulins secreted in the host cell manipulate the chromatin organization of the infected cell both in direct and indirect ways, interfering with transcriptional programs necessary for the cell survival. We will discuss here the nucleomodulins that function as MTases and interact with both DNA and histone proteins, to modify the chromatin architecture and enhance bacterial colonization (Fig. 2 and Table 1).
Figure 2.
Schematic representation of identified bacterial effectors targeting the nucleus and methylating DNA or histone proteins. Top: identified bacterial DNA-MTases as well as putative effectors. Bottom: identified bacterial histone MTases as well as putative effectors.
Table 1.
Bacterial nucleomodulins functioning as MTases in the host.
| Pathogen | Effector | Nuclear localization | Activity/target | References |
|---|---|---|---|---|
| DNA MTase nucleomodulins | ||||
| Klebsiella pneumoniae | HsdM | Yes | Methylates eukaryotic DNA in vitro | Lee et al. (2009) |
| Mycobacterium tuberculosis | Rv2966c | Yes | Cytosine MTase (non-CpG context)—targets promoter sequences of several interleukin receptor genes | Sharma et al. (2015) |
| Mycoplasma hyorhinis | Mhy1-3 | Yes | CG- and GATC-specific MT-ases—target pro-oncogenic and proliferation genes | Chernov et al. (2015) |
| Filifactor alocis | DNMTs | Unknown | Aruni et al. (2014) | |
| Francisella tularensis | DNMT | Unknown | Champion (2011) | |
| H. pylori | DNMTs | Unknown | Sitaraman (2014) | |
| Acinetobacter baumannii | DNMT | Yes | Unknown | Moon et al. (2012a) |
| Arginine and lysine MTase nucleomodulins | ||||
| M. tuberculosis | Rv1988 | Yes | Arg-MTase—targets H3R42–induces gene repression | Yaseen et al. (2015) |
| Chlamydia pneumoniae | cpnSET | Yes | SET-domain MTase—targets H3 and Hc1 | Murata et al. (2007) |
| Chlamydia trachomatis | NUE | Yes | SET-domain MTase—targets H2B/H3/H4 and automethylates | Pennini et al. (2010) |
| Burkholderia pseudomallei | BtSET | Yes | SET-domain MTase—targets H3K4 on rRNA | Li et al. (2013) |
| Burkholderia thailandesis | ||||
| Bacillus anthracis | BaSET | Yes | SET-domain MTase—targets H1, silencing of host inflammatory response | Mujtaba et al. (2013) |
| Methanosarcina mezei (archea) | Gö1-SET | SET-domain MTase—targets H4 in vitro | Manzur and Zhou (2005) | |
| Legionella pneumophila strain Paris | RomA | Yes | SET-domain MTase—targets H3K14, silences host response to infection | Rolando et al. (2013) |
| L. pneumophila strain Philadelphia | LegAS4 | Yes | SET-domain MTase—targets H3K4 on rDNA | Li et al. (2013) |
| L. pneumophila | LppDOT1L | Unknown | Gomez-Valero et al. (2019) | |
| L. pneumophila | PRMTs | Unknown | Cazalet et al. (2004) | |
Bacterial DNA MTAses targeting the host
The first bacterial DNA-MTase shown to target the host cell nucleus was HsdM of Klebsiella pneumoniae (Lee et al. 2009). HsdM belongs to the Type-I bacterial R–M systems (Taylor et al. 2010) and has a nuclear localization signal (NLS) that targets it to the nucleus when expressed in human cell lines. The function of HdsM is not known, but it was shown that recombinant HsdM methylates eukaryotic DNA in vitro (Lee et al. 2009).
Another secreted bacterial MTase is Rv2966c, encoded by Mycobacterium tuberculosis that targets cytosines in a non-CpG dinucleotide context (Sharma et al. 2015). Mycobacterium tuberculosis is an intracellular pathogen responsible for human tuberculosis, colonizing the human lung via inhalation of bacteria-containing droplets. Despite pressure from innate immune cells in the lungs, it can persist in this environment (Bussi and Gutierrez 2019). Rv2966c is enhancing the persistence of the pathogen in the lungs by methylating specific DNA sequences generating hypermethylated regions in promoter sequences of several interleukin receptor genes. Interestingly, Rv2966c, like the mammalian DNA MTase DNMT3L, can also interact with histones H3 and H4, supporting the idea that this bacterial effector interacts with the host epigenetic machinery at multiple levels (Sharma et al. 2015).
The second identified family of nucleomodulins that selectively and efficiently methylate host cell DNA are the secreted effectors Mhy1-3 encoded by Mycoplasma hyorhinis (Chernov et al. 2015). Mycoplasma are parasitic microbes that in humans frequently populate mucosal surfaces and persist as long-term asymptomatic infections, likely promoting chronic aberrant states in infected tissues, often terminating in tumors (Benedetti et al. 2020). Chernov et al. (2015) demonstrated that three M. hyorhinis CG- and GATC-specific MT-ases efficiently translocate to the host cell nucleus leading to a high degree of methylation of the human genome, and thereby stimulating pro-oncogenic and proliferation pathways in human cells. Given that this pathogen is generally associated with prostate and gastric cancer, the authors hypothesize that either the infection contributes to the malignancy onset or, alternatively, that tumors provide a favorable environment for mycoplasma growth that may facilitate further dissemination (Chernov et al. 2015).
Other pathogens have been reported to code for DNMTs in their genomes, however, no functional analyses of those putative nucleomodulins have yet been performed. Examples are, Porphyromonas gingivalis and Filifactor alocis, major pathogens associated with periodontal disease, with as many as 18 different methyltransfeases predicted in the genome of F. alocis (Aruni et al. 2014). Although chronic infection by P. gingivalis has been shown to introduce de novo DNA methylation at several CpGs located in the TLR2 promotor region (Benakanakere et al. 2015), a direct role of these bacterial MTases in host chromatin modifications and epigenetic changes awaits confirmation. Similarly, Francisella tularensis, a human intracellular pathogen, encodes a MTase likely mimicking eukaryotic MTase of its hosts (Champion 2011). A high number of DNMTs has also been predicted in the genome of H. pylori, a Gram-negative, microaerophilic, and spiral-shaped bacterium, i.e. associated with upto 10% of patients that develop duodenal ulcer disease (Suerbaum and Michetti 2002). Helicobacter pylori infection induces CpG methylation in the promoter region of mismatch repair and tumor suppressor genes, which are associated with the initiation and progression of gastric cancer (Kaise et al. 2008). Although it was predicted in the different strains sequenced that H. pylori encodes 25 to 37 adenine- and cytosine-specific MTases (Sitaraman 2014, Vitkute et al. 2001), their functional role during infection remains to be investigated.
A DNA–cytosine MTase has also been annotated in the genome of Acinetobacter baumannii, an important opportunistic pathogen that causes a variety of human infections, with high mortality rate in immunocompromised patients (Antunes et al. 2011). Cytotoxic effects have been shown when this nucleomodulin is translocated into the host cell nucleus (Moon et al. 2012a), however, direct DNA methylation has not been investigated. In contrast, another nuclear effector of A. baumannii, a transposase (Tnp) delivered to host cells via outer membrane vesicles localizes in the nucleus where it induces DNA methylation in the CpG regions of the gene coding for E-cadherin, inducing its transcriptional down-regulation (Moon et al. 2012b). The molecular mechanism describing how Tnp induces DNA methylation and its possible interaction with the nuclear localized DNMT remains unclear.
Bacterial lysine and arginine MTases: a subclass of nucleomodulins targeting host histones
As described above, histone methylation mostly occurs on arginine and lysine residues. Several nucleomodulins that directly methylate histone proteins have been characterized: until now, only one nucleomodulin that encodes PRMT activity has been identified, but several that target lysines via SET-domains.
Rv1988, secreted by M. tuberculosis is an arginine MTase that targets the host chromatin where it dimethylates, instead of “classical” target residues located in the histone tails, a noncanonical arginine residue (R42) located in the core region of histone H3 (Yaseen et al. 2015). Hence, H3R42 is located at critically important entry/exit points of DNA in the nucleosome that have the potential to change nucleosomal dynamics and affect transcription (Casadio et al. 2013). Indeed, Rv1988-driven H3R42 dimethylation induces a repression of genes that are important for the immune response against mycobacterial infection (Yaseen et al. 2015).
The first bacterial, nuclear effector encoding a SET-domain MTase, has been described in Chlamydia pneumoniae (Murata et al. 2007). Chlamydiae exhibit a unique life cycle by alternating between an infectious but transcriptionally inactive, elementary body (EB) that enters the host cell, and a noninfectious reticulate body (RB), i.e. transcriptionally active and replicates intracellularly. Genome analyses revealed genes predicted to encode a SET-domain containing protein (Stephens et al. 1998). Murata et al. (2007) showed that cpnSET is a nucleomodulin that methylates lysine residues of murine histone H3 in vitro and interacts with and methylates the histone H1-like protein Hc1of C. pneumoniae. It is suggested that the chlamydial histone-like proteins Hc1 and Hc2 may act as global transcriptional regulators and play a role in compacting DNA during the RB to EB transition (Perara et al. 1992). These findings suggested that the cpnSET effector may play an important role in the morphological changes from RBs to EBs, but it may also be involved in the modification of the chromatin folding of the host. The secretion of this SET-domain containing chlamydial effector was proven by Pennini et al. (2010), when they demonstrated that the homologue of cpnSET in C. trachomatis, named NUE, is translocated in the cytosol by a type-3 secretion system (T3SS), where it translocates to the nucleus of infected cells (Pennini et al. 2010). Moreover, they showed that NUE methylates lysines of histones H2B, H3, and H4 in vitro, and is capable of automethylation, a characteristic that might play a role in enhancing histones MTase activity.
Another functional SET-domain MTase is encoded by Burkholderia spp. (Li et al. 2013). BtSET is a nucleomodulin secreted by pathogenic Burkholderia pseudomallei and nonpathogenic Burkholderia thailandesis through a specialized T3SS. This enzyme targets histone H3 at lysine 4 (H3K4) for mono- and dimethylation of rRNA, thereby increasing transcription of rRNA genes. A mechanism that seems to facilitae Burkholderia multiplication in host cells.
A SET domain containing protein is also encoded by the Gram-positive bacterium Bacillus anthracis, the etiological agent of anthrax, a zoonotic disease that can be lethal for humans (Moayeri et al. 2015). Bacillus anthracis secretes a nucleomodulin named BaSET, that localizes in the nucleus of infected cells where it specifically trimethylates eight lysine residues in histone H1(Mujtaba et al. 2013). Histone H1 methylation driven by BaSET results in reduced activity of NF-kB response elements, silencing the inflammatory host response. It was also shown that a BaSET deletion prevents both colonization of the host by the pathogen and bacterial survival in the cellular environment.
It would be interesting to determine the function of SET-domain proteins present in other bacillus strains closely related to B. anthracis: Bacillus cereus, an opportunistic human pathogen commonly associated with food poisoning, and Bacillus thuringiensis, an insect pathogen (Han et al. 2006). In fact, during the last decades, thanks to the increasing number of bacterial genomes sequenced, an important number of genes putatively encoding for SET-domain containing proteins have been identified. A BLASTP search performed in 2014 recovered more that 500 bacterial genomes including SET-domain proteins (Alvarez-Venegas 2014). Many genes encoding SET-domain proteins have been identified in B. cereus, Xylella fastidiosa, Leptospira interrogans, Bradyrhizobium japonicum, or Chlorobium tepidum genomes and the above described family of Chlamidyaceae, but also in the archeal Methanosarcina mazei and the Paramecium bursaria chlorella virus-1, that encode for Gö1-SET, which methylates histone H4 in vitro (Manzur and Zhou 2005) and vSET that target histone H3 at lysine 27 (H3K27) (Manzur et al. 2003).
Legionella pneumophila: a toolbox for protein MTases targeting the host chromatin
An excellent example among the intracellular pathogens encoding secreted SET-domain MTases, is the Gram-negative intracellular bacterium Legionella pneumophila, i.e. ubiquitous in aquatic environments, where it replicates within aquatic protozoa (Boamah et al. 2017, Hilbi and Buchrieser 2022). When manmade aquatic environments are contaminated with Legionella and susceptible humans inhale such contaminated aerosols, they may develop a pneumonic respiratory disease named Legionnaires’ disease (Mondino et al. 2020). The intracellular life cycle of L. pneumophila has been intensively studied. One of the major features of this thrilling pathogen is that it encodes for more than 330 effectors (Ensminger 2016). Many of these effectors, translocated in the host cell through a specialized T4SS, are so-called “eukaryotic-like proteins” or proteins with “eukaryotic-like domains” (Cazalet et al. 2004). Several phylogenetic analyses suggested that the genes encoding these eukaryotic domains have been acquired by the bacterium through horizontal gene transfer from its protozoan hosts (Gomez-Valero and Buchrieser 2013), among those, a SET-domain histone MTase named RomA (Rolando et al. 2013, Schator et al. 2021).
RomA is a secreted effector of L. pneumophila strain Paris, that harbors a NLS located in the N-terminal part of the protein allowing it to reach the host cell nucleus, where it specifically trimethylates lysine 14 of histone H3 (H3K14; Rolando et al. 2013). At the time, H3K14me had never been described in mammalian cells because it was likely overlooked. In fact, we now know that it is present in the human genome at very low levels, and it is activated only under specific conditions like during the stress response (Zhao et al. 2018, Zhu et al. 2021). In contrast, H3K14 is usually acetylated in mammalian cells and this mark is correlated with open chromatin and transcriptional activation (Karmodiya et al. 2012). Therefore, H3K14 methylation by RomA functions as a strong and genome-wide repressor of acetylation, leading to transcriptional decrease of gene expression during infection, in particular at promotors essential for the host cell response to infection (Rolando et al. 2013). The homologue of RomA in L. pneumophila strain Philadelphia, named LegAS4, also shows histone MTase activity and L. pneumophila strain Philadelphia and methylates H3K14 during infection (Rolando and Buchrieser 2014), but it has also been shown to interact with HP1 in the nucleolus at rRNA promotors, resulting in the activation of rRNA gene expression (Li et al. 2013).
Genome analyses of L. pneumophila highlighted the presence of additional protein MTases, in fact L. pneumophila strain Paris is predicted to also encode two protein arginine MTases (PRMTs; Cazalet et al. 2004) and a DOT1-like protein, i.e. of particular interest as it is highly conserved in the genus Legionella (Gomez-Valero et al. 2019). As mentioned above, DOT1, first discovered in yeast while screening for enzymes disrupting gene silencing at the telomeres (Lacoste et al. 2002), mono-, di-, or trimethylates lysine 79 on histone H3 (H3K79), which correlates with active gene transcription (Schübeler et al. 2004). So far it is the only enzyme known to methylate this mark.
Intrigued by the presence of DOT1-like proteins in all Legionella genomes analyzed in our previous study (Gomez-Valero et al. 2019), we conducted an in-depth phylogenetic analysis of these proteins to gain insight in their emergence within the genus Legionella. A first homology search in the NCBI nonredundant database confirmed that all Legionella genomes sequenced to date (62 different Legionella species) contain dot1 genes. Whereas most of them have only one gene encoding a DOT1 protein, seven species possess several genes encoding DOT1 domains. The Legionella fallonii genome contains four dot1l genes, which is the highest number of homologues present in a Legionella genome.
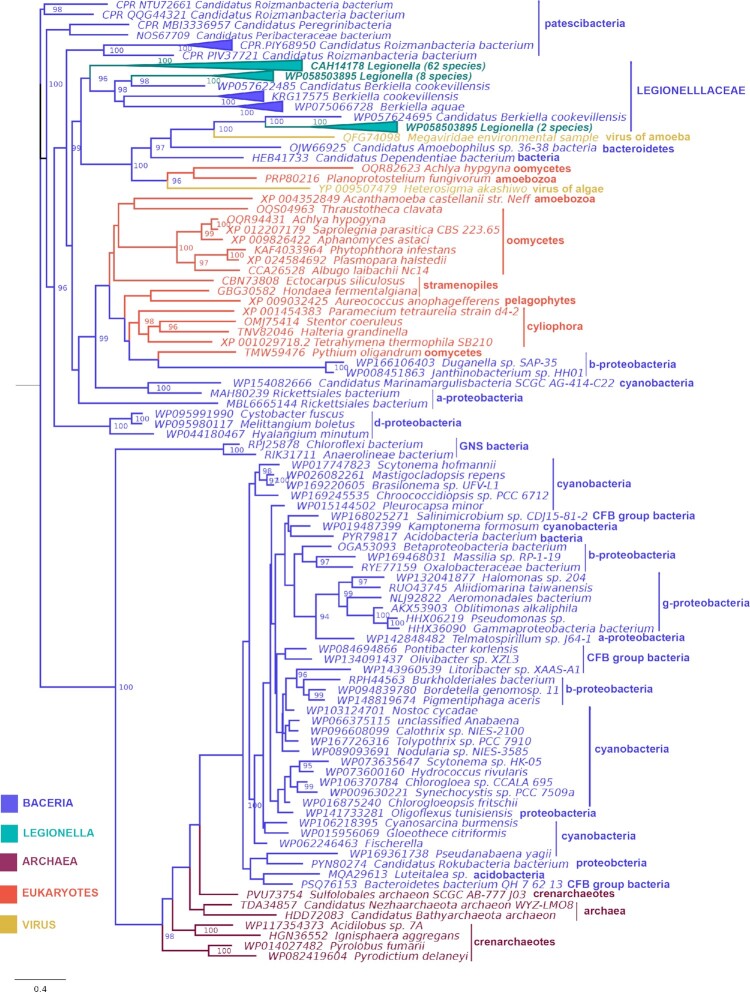
To analyze the origin of the Legionella DOT1 protein coding genes, we undertook a phylogenetic analysis including their corresponding homologous in other organisms. Homologous proteins were recruited by Blastp using as seed the L. pneumophila strain Paris DOT1 protein with the following filtering parameters: at least 25% of identity, a minimum coverage of 50% and an e-value of 1e−5 or higher. Redundant sequences (with more than 90% of identity among them) from non-Legionella species and outliers were removed using the multiple sequence alignment package T-Coffee (Notredame et al. 2000). The remaining sequences were aligned using M-Coffee (Wallace et al. 2006) and ambiguous positions were removed after applying an evaluation method from T-Coffee. A final reliable sequence alignment of 164 amino acids in length was obtained and used for a likelihood phylogenetic reconstruction using IQ-TREE 2 (Minh et al. 2020; Fig. 3).
Figure 3.
Maximum likelihood (IQ-TREE) tree of DOT1 proteins identified in Legionella spp. and selected DOT1 homologous sequences. Values on the right of each node correspond to ultrafast bootstrap support values (only values above 95% are shown; Minh et al. 2020). The tree has been rooted in the midpoint. The horizontal bar provides the scale for the branch length. The three different clades of Legionella DOT1 proteins are highlighted in green.
In this tree, the Legionella DOT1 proteins are distributed in three different clades: clade one contains the 62 Legionella genomes analyzed, as all code for an orthologue of the L. pneumophila Paris DOT1 protein; clade two contains eight Legionella species that encode an additional DOT1 homologue; and clade three contains two Legionella species that contain a third DOT1 protein (Fig. 3—green). This division of Legionella DOT1 proteins in three different clades suggests independent origins for each of these DOT1 orthologous groups. The presence of DOT1 of clade one in all the species indicates that this protein was already present in the common ancestor of Legionella. However, DOT1 proteins from the other two clades are present in very few species that are not even very closely related, suggesting a more recent emergence of these proteins in the genus Legionella, probably through different events of acquisition.
More interestingly, when analyzing the entire obtained phylogeny, we observed that all acquired DOT1 homologues are distributed in two main clades: one containing only bacterial and archaeal sequences and another one containing the three Legionella clades mentioned above, clustering with homologues from eukaryotes, viruses, and bacteria (Fig. 3). Despite the heterogenicity of the organisms clustering in this second group, all of them inhabit the aquatic environment or are amoeba-related organisms. For example, the organisms closest to Legionella in the tree, Berkiella spp., also belonging to the order Legionellaceae, are intracellular bacteria of freshwater amoeba. Similarly, in the same clade we find the amoeba endosymbiont Candidatus amoebophilus, the amoeba virus Megaviridae, or bacteria typically isolated from amoeba such as Candidatus dependentiae. Other bacteria in the same cluster such as Roizmanbacteria spp or Duganella are also prevalent in aquatic environments. In the same clade, there are also eukaryotic sequences that are all from protists belonging to Amoebozoa and the eukaryotic clade SAR. This clade includes Stramenopiles, Alveolates, and Rhizaria and contains a large diversity of lineages including amoebae, ciliates, and flagellates that live almost everywhere (Grattepanche et al. 2018). Among them there are ciliates and amoeabae such as the well-established natural host of Legionella, Acanthamoeba castellanii.
Taken together, these analyses strongly suggest that the horizontal transfer of genes encoding proteins with DOT1 domains has taken place many times between prokaryotes, eukaryotes, and viruses cohabiting in the same niche and support the hypothesis of a selective pressure to acquire and maintain the dot1 L gene for amoeba-related organisms such as Legionella spp. Our results might also indicate that DOT1 is important to create a niche inside eukaryotes probably by modifying histones, as it was shown for RomA.
Perspectives
Efficient high-resolution mapping of bacterial DNA-methylation events has only recently become possible with the advent of single-molecule real time sequencing—SMRT (Zhao et al. 2020). The mapping and characterization of bacterial DNA methylomes by SMRT, or potentially new future technologies, will allow to discover new MTases, but also novel genes epigenetically regulated by DNA methylation. This is important, as epigenetic changes may provide an early marker of potential alterations in gene expression leading to disease. In addition, genetic repertoires stably expressed during infection would be identified by the characterization of phasevarions (Seib et al. 2020).
In the context of infection by bacterial pathogens, the identification of novel DNA MTase-controlled genes, that could be associated to virulence traits, will allow to characterize, and potentially repress, medically relevant biological processes by designing new therapeutic approaches. Thus, bacterial MTases may emerge as promising targets for the development of novel epigenetic inhibitors (Ceccaldi et al. 2013), in particular in the context of antibiotic resistance. Indeed, bacteria use methylated nucleobases to resist antibiotics, and DNA MTase inhibitors may be used to enhance the therapeutic activity of antibiotics. It has been shown, e.g. for E. coli that DNA adenine MTase deficiency potentiates the lethal action of antibiotics by increasing the bacterial activity of beta-lactams and quinolones (Cohen et al. 2016). Also, rRNA methylation is involved in antibiotic resistance. Most known inhibitors of protein synthesis bind to the functional sites of ribosomes, and when methyl groups are introduced, they can spatially overlap with the antibiotic binding site (Osterman et al. 2020).
Furthermore, MTases may also emerge as promising targets for the identification and the development of new vaccines against human-adapted pathogens (Phillips et al. 2019b). For example, live vaccines against Salmonella spp. and Yersinia pseudotuberculosis, have been developed using strains that carry MTase mutations (Heithoff et al. 2015) (Taylor et al. 2005).
Several MTases have been described to play a role as nucleomodulins during bacterial infection, and many genes putatively encoding MTases remain to be characterized during infection. Furthermore, some bacterial MTases have been identified to modify the transcriptional host cell response by targeting nonhistone proteins. One example is the MTTL20 homologue encoded by Agrobacterium tumefaciens, a specific protein–lysine MTase that targets ribosomal protein L7/L12 and the β-subunit of electron transfer flavoprotein (ETF-β; Falnes et al. 2016). Another example is enteropathogenic E. coli (EPEC), a bacterium classified as a major diarrheagenic agent, transmitted by contaminated water or undercooked food, and identified as one of the major causes of mortality in children under five. EPEC E. coli encode for NleE, an S-adenosyl-L-methionine (AdoMet)-dependent MTase, that modifies Cys673 in the Npl4 zinc finger (NZF) domain of the host signaling adaptor protein TAB2, and Cys692 in the same portion of the protein TAB3, both involved in the signaling via Toll-like or TNF receptors, and IL-1 (Zhang et al. 2016).
In contrast, RNA MTases have not been classified as nucleomodulins to date. Although recent studies showed the presence of m6A modification on viral and cellular RNAs during infection (McFadden and Horner 2020), very little is known about bacteria (directly) modulating m6A of eukaryotic mRNA. However, bacterial infection increases m6A levels in nascent transcripts, in particular in transcripts related to histone modifications (Wu et al. 2020), and m6A may be involved in regulating the response to LPS, as both m6A reader and writer proteins (YTHDF2 and METTL3) have been shown to play a role in regulating the cell response to bacteria and cytokine production (Yu et al. 2019) (Feng et al. 2018). Thus, bacterial RNA MTases may also prove as important virulence factors and perhaps as targets for enhancing treatment of bacterial infections in the future.
ACKNOWLEDGEMENTS
Work in the CB laboratory is financed by the Institut Pasteur and funding has been received from the Agence National de recherche grant ANR-18-CE15-0005-01 to M.R., ANR-10-LABX-62-IBEID, ANR 20-PAMR-0011 TheraEPI, and the “Fondation de la Recherche Médicale” grant EQU201903007847 to C.B. This work was supported by a PTR (Programmes Tranversaux de Recherche) grant (PTR 395–20 to M.R.) from the Institut Pasteur Paris.
Contributor Information
Monica Rolando, Institut Pasteur, Université de Paris, CNRS UMR 6047, Unité Biologie des Bactéries Intracellulaires, 28, Rue du Dr. Roux, 75724 Paris Cedex 15, France.
Cristina Di Silvestre, Institut Pasteur, Université de Paris, CNRS UMR 6047, Unité Biologie des Bactéries Intracellulaires, 28, Rue du Dr. Roux, 75724 Paris Cedex 15, France.
Laura Gomez-Valero, Institut Pasteur, Université de Paris, CNRS UMR 6047, Unité Biologie des Bactéries Intracellulaires, 28, Rue du Dr. Roux, 75724 Paris Cedex 15, France.
Carmen Buchrieser, Institut Pasteur, Université de Paris, CNRS UMR 6047, Unité Biologie des Bactéries Intracellulaires, 28, Rue du Dr. Roux, 75724 Paris Cedex 15, France.
Conflict of interest
The authors declare no conflict of interest
References
- Adhikari S, Curtis PD. DNA methyltransferases and epigenetic regulation in bacteria. FEMS Microbiol Rev. 2016;40:575–91. [DOI] [PubMed] [Google Scholar]
- Aki T, Adhya S. Repressor induced site-specific binding of HU for transcriptional regulation. EMBO J. 1997;16:3666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Venegas R. Bacterial SET domain proteins and their role in eukaryotic chromatin modification. Frontiers in Genetics. 2014;5:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes LCS, Imperi F, Carattoli Aet al. Deciphering the Multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS One. 2011;6:e22674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruni AW, Zhang K, Dou Yet al. Proteome Analysis of coinfection of epithelial cells with Filifactor alocis and Porphyromonas gingivalis shows modulation of pathogen and host regulatory pathways. Infect Immun. 2014;82:3261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbontín R, Rowley G, Pucciarelli MGet al. DNA Adenine Methylation regulates virulence gene expression in Salmonella enterica serovar typhimurium▿. J Bacteriol. 2006;188:8160–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benakanakere M, Abdolhosseini M, Hosur Ket al. TLR2 Promoter Hypermethylation creates innate immune dysbiosis. J Dent Res. 2015;94:183–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti F, Curreli S, Zella D. Mycoplasmas–Host interaction: mechanisms of inflammation and association with cellular transformation. Microorganisms. 2020;8:1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bheemanaik S, Reddy YVR, Rao DN. Structure, function and mechanism of exocyclic DNA methyltransferases. Biochem J. 2006;399:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Cossart P. When bacteria target the nucleus: the emerging family of nucleomodulins. Cell Microbiol. 2012;14:622–33. [DOI] [PubMed] [Google Scholar]
- Bierne H, Pourpre R. Bacterial factors targeting the nucleus: the growing family of nucleomodulins. Toxins. 2020;12:220–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, Clark TA, Daum CGet al. The epigenomic landscape of prokaryotes. PLos Genet. 2016;12:e1005854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyn LB, Braaten BA, Low DA. Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J. 1990;9:4045–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boamah DK, Zhou G, Ensminger AWet al. From Many Hosts, one accidental pathogen: the diverse protozoan hosts of legionell a. Frontiers in Cellular and Infection Microbiology. 2017;7:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta Eet al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boriack-Sjodin PA, Swinger KK. Protein methyltransferases: a distinct, diverse, and dynamic family of enzymes. Biochemistry. 2016;55:1557–69. [DOI] [PubMed] [Google Scholar]
- Brooks JE, Blumenthal RM, Gingeras TR. The isolation and characterization of the Esherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res. 1983;11:837–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi C, Gutierrez MG. Mycobacterium tuberculosis infection of host cells in space and time. FEMS Microbiol Rev. 2019;43:341–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabetta VJ. Addressing the possibility of a histone-like code in bacteria. J Proteome Res. 2021;20:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadio F, Lu X, Pollock SBet al. H3R42me2a is a histone modification with positive transcriptional effects. Proc Natl Acad Sci. 2013;110:14894–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C, Rusniok C, Brüggemann Het al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–73. [DOI] [PubMed] [Google Scholar]
- Ceccaldi A, Rajavelu A, Ragozin Set al. Identification of novel inhibitors of DNA methylation by screening of a chemical library. ACS Chem Biol. 2013;8:543–8. [DOI] [PubMed] [Google Scholar]
- Champion MD. Host-Pathogen O-Methyltransferase similarity and its specific presence in highly virulent strains of Francisella tularensis suggests molecular mimicry. PLoS One. 2011;6:e20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Chen Y, Zhao Yet al. JMJD6 Is a histone arginine demethylase. Science. 2007;318:444–7. [DOI] [PubMed] [Google Scholar]
- Chao MC, Zhu S, Kimura Set al. A Cytosine Methyltransferase modulates the cell envelope stress response in the cholera pathogen. [corrected]. PLos Genet. 2015;11:e1005666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernov AV, Reyes L, Xu Zet al. Mycoplasma CG- and GATC-specific DNA methyltransferases selectively and efficiently methylate the host genome and alter the epigenetic landscape in human cells. Epigenetics. 2015;10:303–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen NR, Ross CA, Jain Set al. A role for the bacterial GATC methylome in antibiotic stress survival. Nat Genet. 2016;48:581–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Chen K, Luo G-Zet al. Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 2015;43:6557–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from novikoff hepatoma cells. Proc Natl Acad Sci. 1974;71:3971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 2010;8:185–95. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Zhang X, Trievel RCet al. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Luu P-L, Stirzaker Cet al. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics. 2015;7:1051–73. [DOI] [PubMed] [Google Scholar]
- Ensminger AW. Legionella pneumophila, armed to the hilt: justifying the largest arsenal of effectors in the bacterial world. Curr Opin Microbiol. 2016;29:74–80. [DOI] [PubMed] [Google Scholar]
- Estibariz I, Overmann A, Ailloud Fet al. The core genome m5C methyltransferase JHP1050 (M.Hpy99III) plays an important role in orchestrating gene expression in Helicobacter pylori. Nucleic Acids Res. 2019;47:2336–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fälker S, Schilling J, Schmidt MAet al. Overproduction of DNA adenine methyltransferase alters motility, invasion, and the lipopolysaccharide O-antigen composition of Yersinia enterocolitica. Infect Immun. 2007;75:4990–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falnes PØ, Jakobsson ME, Davydova Eet al. Protein lysine methylation by seven-β-strand methyltransferases. Biochem J. 2016;473:1995–2009. [DOI] [PubMed] [Google Scholar]
- Feng Z, Li Q, Meng Ret al. METTL3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells. J Cell Mol Med. 2018;22:2558–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HHet al. Methylation of H3-Lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–8. [DOI] [PubMed] [Google Scholar]
- Galán JE, Waksman G. Protein-Injection machines in bacteria. Cell. 2018;172:1306–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gana R, Rao S, Huang Het al. Structural and functional studies of S-adenosyl-L-methionine binding proteins: a ligand-centric approach. BMC Struct Biol. 2013;13:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin SB. Integration of agrobacterium T-DNA into the plant genome. Annu Rev Genet. 2017;51:195–217. [DOI] [PubMed] [Google Scholar]
- Gold M, Hurwitz J, Anders M. The enzymatic methylation of RNA and DNA. I. Biochem Biophys Res Commun. 1963;11:107–14. [DOI] [PubMed] [Google Scholar]
- Gomez-Valero L, Buchrieser C. Genome dynamics in legionella: the basis of versatility and adaptation to intracellular replication. Cold Spring Harb Perspect Med. 2013;3:a009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Valero L, Rusniok C, Carson Det al. More than 18,000 effectors in the legionella genus genome provide multiple, independent combinations for replication in human cells. Proc Natl Acad Sci. 2019;116:2265–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grattepanche J, Walker LM, Ott BMet al. Microbial Diversity in the eukaryotic SAR clade: illuminating the darkness between morphology and molecular data. Bioessays. 2018;40:1700198. [DOI] [PubMed] [Google Scholar]
- Han CS, Xie G, Challacombe JFet al. Pathogenomic sequence analysis of Bacillus cereus and Bacillus thuringiensis isolates closely related to Bacillus anthracis†. J Bacteriol. 2006;188:3382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He PC, He C. m6A RNA methylation: from mechanisms to therapeutic potential. EMBO J. 2021;40. 10.15252/embj.2020105977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y-F, Li B-Z, Li Zet al. Tet-Mediated formation of 5-Carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heithoff DM, House JK, Thomson PCet al. Development of a salmonella cross-protective vaccine for food animal production systems. Vaccine. 2015;33:100–7. [DOI] [PubMed] [Google Scholar]
- Heithoff DM, Sinsheimer RL, Low DAet al. An Essential Role for DNA adenine methylation in bacterial virulence. Science. 1999;284:967–70. [DOI] [PubMed] [Google Scholar]
- Hilbi H, Buchrieser C. Microbe profile: Legionella pneumophila - a copycat eukaryote. Microbiology. 2022;168. [DOI] [PubMed] [Google Scholar]
- Hooykaas PJJ, Klapwijk PM, Nuti MPet al. Transfer of the Agrobacterium tumefaciens TI plasmid to avirulent agrobacteria and to rhizobium ex planta. J Gen Microbiol. 1977;98:477–84. [Google Scholar]
- Huang H, Sabari BR, Garcia BAet al. SnapShot: histone modifications. Cell. 2014;159:458–458.e1.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Qet al. Tet Proteins Can convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science. 2011;333:1300–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Tahiliani M, Rao Aet al. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A. Beyond watson and crick: DNA methylation and molecular enzymology of DNA methyltransferases. ChemBioChem. 2002;3:274–93. [DOI] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao Xet al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–92. [DOI] [PubMed] [Google Scholar]
- Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahramanoglou C, Prieto AI, Khedkar Set al. Genomics of DNA cytosine methylation in Escherichia coli reveals its role in stationary phase transcription. Nat Commun. 2012;3:886. [DOI] [PubMed] [Google Scholar]
- Kaise M, Yamasaki T, Yonezawa Jet al. CpG island hypermethylation of tumor-suppressor genes in H. pylori-infected non-neoplastic gastric mucosa is linked with gastric cancer risk. Helicobacter. 2008;13:35–41. [DOI] [PubMed] [Google Scholar]
- Karmodiya K, Krebs AR, Oulad-Abdelghani Met al. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012;13:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek E, Kierzek R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 2003;31:4472–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. [DOI] [PubMed] [Google Scholar]
- Kumar S, Karmakar BC, Nagarajan Det al. N4-cytosine DNA methylation regulates transcription and pathogenesis in Helicobacter pylori. Nucleic Acids Res. 2018;46:3429–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste N, Utley RT, Hunter JMet al. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem. 2002;277:30421–4. [DOI] [PubMed] [Google Scholar]
- Lee JC, Kim DS, Moon DCet al. Prediction of bacterial proteins carrying a nuclear localization signal and nuclear targeting of HsdM from Klebsiella pneumoniae. The Journal of Microbiology. 2009;47:641. [DOI] [PubMed] [Google Scholar]
- Lee J-S, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Lu Q, Wang Get al. SET-domain bacterial effectors target heterochromatin protein 1 to activate host rDNA transcription. EMBO Rep. 2013;14:733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden MJ, Horner SM. N6-Methyladenosine regulates host responses to viral infection. Trends Biochem Sci. 2021;.46:366–77., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA Amino-methyl-transferases and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253:618–32. [DOI] [PubMed] [Google Scholar]
- Manzur KL, Farooq A, Zeng Let al. A dimeric viral SET domain methyltransferase specific to lys27 of histone h3. Nat Struct Mol Biol. 2003;10:187–96. [DOI] [PubMed] [Google Scholar]
- Manzur KL, Zhou M-M. An archaeal SET domain protein exhibits distinct lysine methyltransferase activity towards DNA-associated protein MC1-α. FEBS Lett. 2005;579:3859–65. [DOI] [PubMed] [Google Scholar]
- Marinus MG, Casadesus J. Roles of DNA adenine methylation in host–pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol Rev. 2009;33:488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Militello KT, Mandarano AH, Varechtchouk Oet al. Cytosine DNA methylation influences drug resistance in Escherichia coli through increased sugE expression. FEMS Microbiol Lett. 2014;350:100–6. [DOI] [PubMed] [Google Scholar]
- Min J, Feng Q, Li Zet al. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–23. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor Oet al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayeri M, Leppla SH, Vrentas Cet al. Anthrax pathogenesis. Annu Rev Microbiol. 2015;69:185–208. [DOI] [PubMed] [Google Scholar]
- Mondino S, Schmidt S, Rolando Met al. Legionnaires’ disease: state of the art knowledge of pathogenesis mechanisms of legionell a. Annual Review of Pathology: Mechanisms of Disease. 2020;15:439–66. [DOI] [PubMed] [Google Scholar]
- Moon DC, Choi CH, Lee SMet al. Nuclear translocation of Acinetobacter baumannii transposase induces DNA methylation of CpG regions in the promoters of E-cadherin gene. PLoS One. 2012a;7:e38974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon DC, Gurung M, Lee JHet al. Screening of nuclear targeting proteins in Acinetobacter baumannii based on nuclear localization signals. Res Microbiol. 2012b;163:279–85. [DOI] [PubMed] [Google Scholar]
- Mruk I, Kobayashi I. To be or not to be: regulation of restriction–modification systems and other toxin–antitoxin systems. Nucleic Acids Res. 2014;42:70–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujtaba S, Winer BY, Jaganathan Aet al. Anthrax SET protein: a potential virulence determinant that epigenetically represses NF-κB activation in infected macrophages. J Biol Chem. 2013;288:23458–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M, Azuma Y, Miura Ket al. Chlamydial SET domain protein functions as a histone methyltransferase. Microbiology. 2007;153:585–92. [DOI] [PubMed] [Google Scholar]
- Murn J, Shi Y. The winding path of protein methylation research: milestones and new frontiers. Nat Rev Mol Cell Biol. 2017;18:517–27. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–17. [DOI] [PubMed] [Google Scholar]
- Oliveira PH, Fang G. Conserved DNA methyltransferases: a window into fundamental mechanisms of epigenetic regulation in bacteria. Trends Microbiol. 2021;29:28–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira PH, Touchon M, Rocha EPC. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 2014;42:10618–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterman IA, Dontsova OA, Sergiev PV. rRNA methylation and antibiotic resistance. Biochemistry (Moscow). 2020;85:1335–49. [DOI] [PubMed] [Google Scholar]
- Pennini ME, Perrinet S, Dautry-Varsat Aet al. Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen chlamydia trachomatis. PLoS Pathog. 2010;6:e1000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perara E, Ganem D, Engel JN. A developmentally regulated chlamydial gene with apparent homology to eukaryotic histone h1. Proc Natl Acad Sci. 1992;89:2125–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips ZN, Husna A-U, Jennings MPet al. Phasevarions of bacterial pathogens – phase-variable epigenetic regulators evolving from restriction–modification systems. Microbiology. 2019a;165:917–28. [DOI] [PubMed] [Google Scholar]
- Phillips ZN, Tram G, Seib KLet al. Phase-variable bacterial loci: how bacteria gamble to maximise fitness in changing environments. Biochem Soc Trans. 2019b;47:1131–41. [DOI] [PubMed] [Google Scholar]
- Rapisarda C, Fronzes R. Secretion systems used by bacteria to subvert host functions. Curr Issues Mol Biol. 2018;25:1–42. [DOI] [PubMed] [Google Scholar]
- Roberts RJ, Belfort M, Bestor Tet al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolando M, Buchrieser C. Legionella pneumophila type IV effectors hijack the transcription and translation machinery of the host cell. Trends Cell Biol. 2014;24:771–8., [DOI] [PubMed] [Google Scholar]
- Rolando M, Sanulli S, Rusniok Cet al. Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host & Microbe. 2013;13:395–405. [DOI] [PubMed] [Google Scholar]
- Romano G, Veneziano D, Nigita Get al. RNA Methylation in ncRNA: classes, detection, and molecular associations. Frontiers in Genetics. 2018;9:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan Tet al. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saletore Y, Meyer K, Korlach Jet al. The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 2012;13:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira M. Structural chemistry of human RNA methyltransferases. ACS Chem Biol. 2016;11:575–82. [DOI] [PubMed] [Google Scholar]
- Schato D, Gomez-Valero L, Buchrieser Cet al. Patho-epigenetics: histone deacetylases as targets of pathogens and therapeutics. Microlife. 2021;2:uqab013. 10.1093/femsml/uqab013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D, MacAlpine DM, Scalzo Det al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seib KL, Srikhanta YN, Atack JMet al. Epigenetic Regulation of virulence and immunoevasion by phase-variable restriction-modification systems in bacterial pathogens. Annu Rev Microbiol. 2020;74:655–71. [DOI] [PubMed] [Google Scholar]
- Sharma G, Upadhyay S, Srilalitha Met al. The interaction of mycobacterial protein Rv2966c with host chromatin is mediated through non-CpG methylation and histone H3/H4 binding. Nucleic Acids Res. 2015;43:3922–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74:640–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman R. Helicobacter pylori DNA methyltransferases and the epigenetic field effect in cancerization. Frontiers in Microbiology. 2014;5:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder LAS, Butcher SA, Saunders NJ. Comparative whole-genome analyses reveal over 100 putative phase-variable genes in the pathogenic neisseria spp. Microbiology, 2001;147:2321–32. [DOI] [PubMed] [Google Scholar]
- Srikhanta YN, Maguire TL, Stacey KJet al. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc Natl Acad Sci. 2005;102:5547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RS, Kalman S, Lammel Cet al. Genome sequence of an obligate intracellular pathogen of humans: chlamydia trachomatis. Science. 1998;282:754–9. [DOI] [PubMed] [Google Scholar]
- Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175–86. [DOI] [PubMed] [Google Scholar]
- Taylor JE, Callow P, Swiderska Aet al. Structural and functional analysis of the engineered type I DNA methyltransferase EcoR124I(NT). J Mol Biol. 2010;398:391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor VL, Titball RW, Oyston PCF. Oral immunization with a dam mutant of Yersinia pseudotuberculosis protects against plague. Microbiology. 2005;151:1919–26. [DOI] [PubMed] [Google Scholar]
- Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16:178–89. [DOI] [PubMed] [Google Scholar]
- Vitkute J, Stankevicius K, Tamulaitiene Get al. Specificities of Eleven different DNA methyltransferases of Helicobacter pylori strain 26695. J Bacteriol. 2001;183:443–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace IM, O'Sullivan O, Higgins DGet al. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006;34:1692–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walport LJ, Hopkinson RJ, Chowdhury Ret al. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat Commun. 2016;7:11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez Aet al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Sayegh Jet al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–83. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MBet al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–66. [DOI] [PubMed] [Google Scholar]
- Wiener D, Schwartz S. The epitranscriptome beyond m6A. Nat Rev Genet. 2021;22:119–31. [DOI] [PubMed] [Google Scholar]
- Wion D, Casadesús J. N6-methyl-adenine: an epigenetic signal for DNA–protein interactions. Nat Rev Microbiol. 2006;4:183–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woude MW van der. Phase variation: how to create and coordinate population diversity. Curr Opin Microbiol. 2011;14:205–11. [DOI] [PubMed] [Google Scholar]
- Woude M van der, Braaten B, Low D. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol. 1996;4:5–9. [DOI] [PubMed] [Google Scholar]
- Wu C, Chen W, He Jet al. Interplay of m6A and H3K27 trimethylation restrains inflammation during bacterial infection. Sci Adv. 2020;6:eaba0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TP, Wang T, Seetin MGet al. DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaseen I, Kaur P, Nandicoori VKet al. Mycobacteria modulate host epigenetic machinery by Rv1988 methylation of a non-tail arginine of histone h3. Nat Commun. 2015;6:6. [DOI] [PubMed] [Google Scholar]
- Yu R, Li Q, Feng Zet al. m6A Reader YTHDF2 regulates LPS-Induced inflammatory response. Int J Mol Sci. 2019;20:1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608–24. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mühlen S, Oates CVet al. Identification of a distinct Substrate-binding domain in the bacterial cysteine methyltransferase effectors NleE and OspZ*. J Biol Chem. 2016;291:20149–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Garcia BA. Comprehensive catalog of currently documented histone modifications. Cold Spring Harb Perspect Biol. 2015;7:a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Xu W, Rong Bet al. H3K14me3 genomic distributions and its regulation by KDM4 family demethylases. Cell Res. 2018;28:1118–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L-Y, Song J, Liu Yet al. Mapping the epigenetic modifications of DNA and RNA. Protein & Cell. 2020;11:792–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Yet al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Wang G, Qian J. Transcription factors as readers and effectors of DNA methylation. Nat Rev Genet. 2016;17:551–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Yang Q, Lu Xet al. SETD2-mediated H3K14 trimethylation promotes ATR activation and stalled replication fork restart in response to DNA replication stress. Proc Natl Acad Sci. 2021;118:e2011278118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Beaulaurier J, Deikus Get al. Mapping and characterizing N6-methyladenine in eukaryotic genomes using single-molecule real-time sequencing. Genome Res. 2018;28:1067–78. [DOI] [PMC free article] [PubMed] [Google Scholar]