Abstract
Chloroplasts are a common feature of plant cells and aspects of their metabolism, including photosynthesis, are influenced by low-temperature conditions. Chloroplasts contain a small circular genome that encodes essential components of the photosynthetic apparatus and chloroplast transcription/translation machinery. Here, we show that in Arabidopsis, a nuclear-encoded sigma factor that controls chloroplast transcription (SIGMA FACTOR5) contributes to adaptation to low-temperature conditions. This process involves the regulation of SIGMA FACTOR5 expression in response to cold by the bZIP transcription factors ELONGATED HYPOCOTYL5 and ELONGATED HYPOCOTYL5 HOMOLOG. The response of this pathway to cold is gated by the circadian clock, and it enhances photosynthetic efficiency during long-term cold and freezing exposure. We identify a process that integrates low-temperature and circadian signals, and modulates the response of chloroplasts to low-temperature conditions.
Subject terms: Abiotic, Plant physiology
Plant sigma factors are nuclear-encoded regulators of plastid transcription. A nuclear-encoded sigma factor integrates low-temperature and circadian signals, contributing to photosynthetic resilience to long-term cold.
Main
Low temperatures cause widespread alterations in the physiology and development of plants. Plants use a variety of regulatory mechanisms to respond to low-temperature conditions and to prepare for freezing temperatures through the process of cold acclimation1,2. Chloroplasts are essential for plant productivity and require resilience to cold temperatures because this impacts photoprotection, plastid genome transcription, membrane composition, reactive oxygen species metabolism, translation and the magnitude of photosystem II (PSII) excitation pressure3–10. A suite of mechanisms underlie the short- and longer-term responses of chloroplasts to low-temperature conditions. These derive from both nuclear-encoded proteins that affect chloroplast function and direct responses to cold within chloroplasts. For example, the cold-induced, nuclear-encoded and plastid-localized protein COR15A has a key role in providing freezing tolerance5,11. COR15A is localized to the chloroplast stroma and is thought to stabilize chloroplast membranes in response to the molecular crowding that occurs during freezing-induced cellular dehydration12–14. Furthermore, the chloroplast-localized galactolipid galactosyltransferase SENSITIVE TO FREEZING2 becomes active in response to cytoplasmic acidification during freezing, remodelling the chloroplast outer envelope to increase freezing tolerance4,15–17. Within chloroplasts, low temperatures cause rapid and reversible photoinhibition18, which is thought to protect the photosynthetic apparatus from decreased biochemical activity in the presence of cold, including reduced rates of PSII repair19. Furthermore, moderate temperature reductions alter chloroplast ribosome occupancy, increasing the translation of specific chloroplast genes10.
The majority of chloroplast proteins are encoded by the nuclear genome, yet chloroplasts also harbour a small circular genome that encodes essential components of the photosynthetic apparatus and chloroplast gene expression machinery. Chloroplast-encoded genes are transcribed by two RNA polymerases: plastid-encoded plastid RNA polymerase (PEP) and nuclear-encoded plastid RNA polymerase. PEP is a bacteria-like multi-subunit RNA polymerase that requires a σ70-like sigma factor for promoter recognition and transcription initiation20–22. Sigma factors are thought to have transferred from the plastid genome to the nuclear genome during the evolutionary history of plants, thus providing a mechanism for nuclear control of plastid transcription20,21. The Arabidopsis thaliana (Arabidopsis) nuclear genome encodes six sigma factors (SIGMA FACTOR1 (SIG1) to SIG6) that control chloroplast transcription during chloroplast biogenesis and steady-state photosynthesis20,23–25. The nuclear encoding of plastid sigma factors is thought to provide a set of signalling pathways from the nucleus to plastids20,23. For example, the sigma factor SIG5 participates in chloroplast transcriptional responses to light conditions23,25,26, a variety of abiotic stresses24,27 and the circadian regulation of specific chloroplast transcripts24. Here, we identified a new role for SIG5 in the responses of plants to low-temperature conditions.
Results
SIG5 communicates low-temperature information to chloroplasts
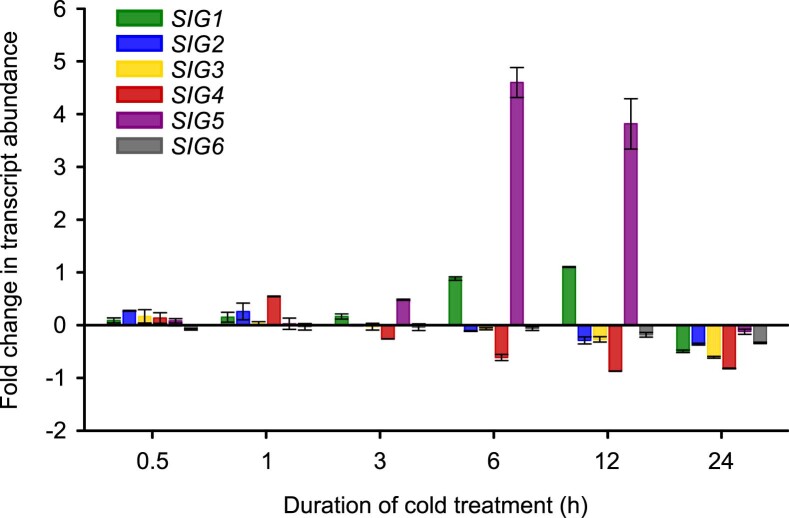
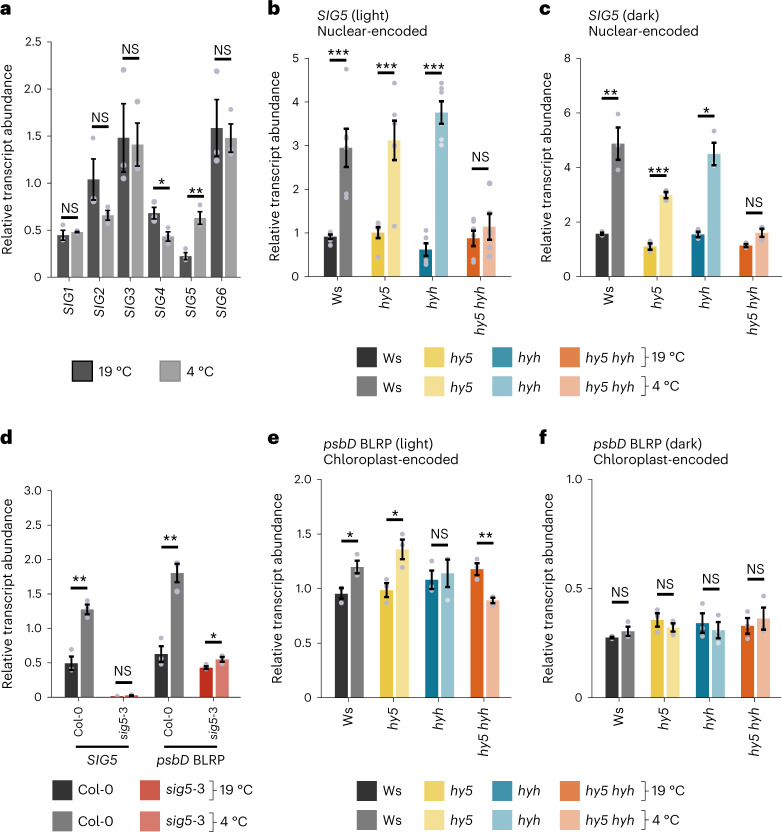
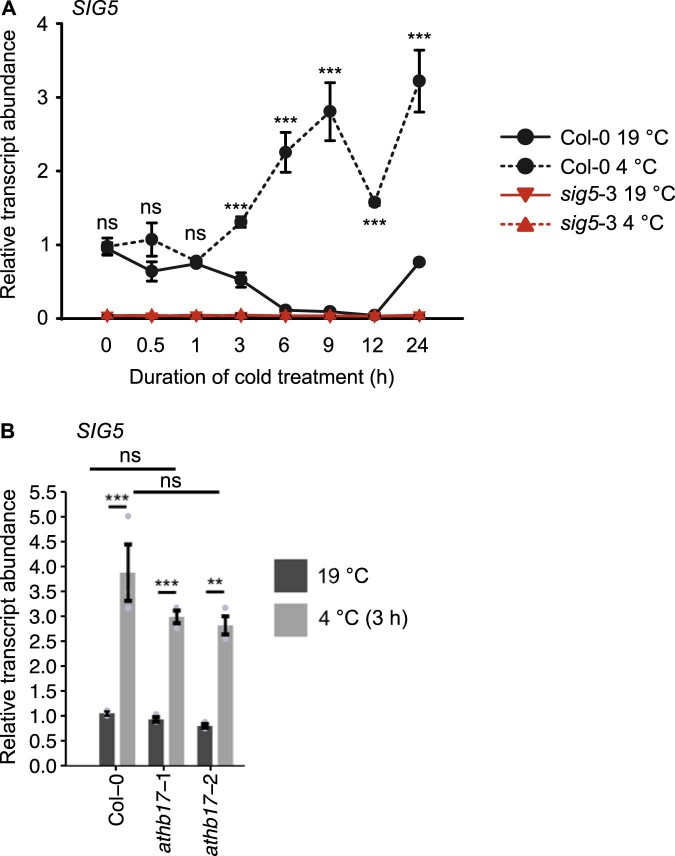
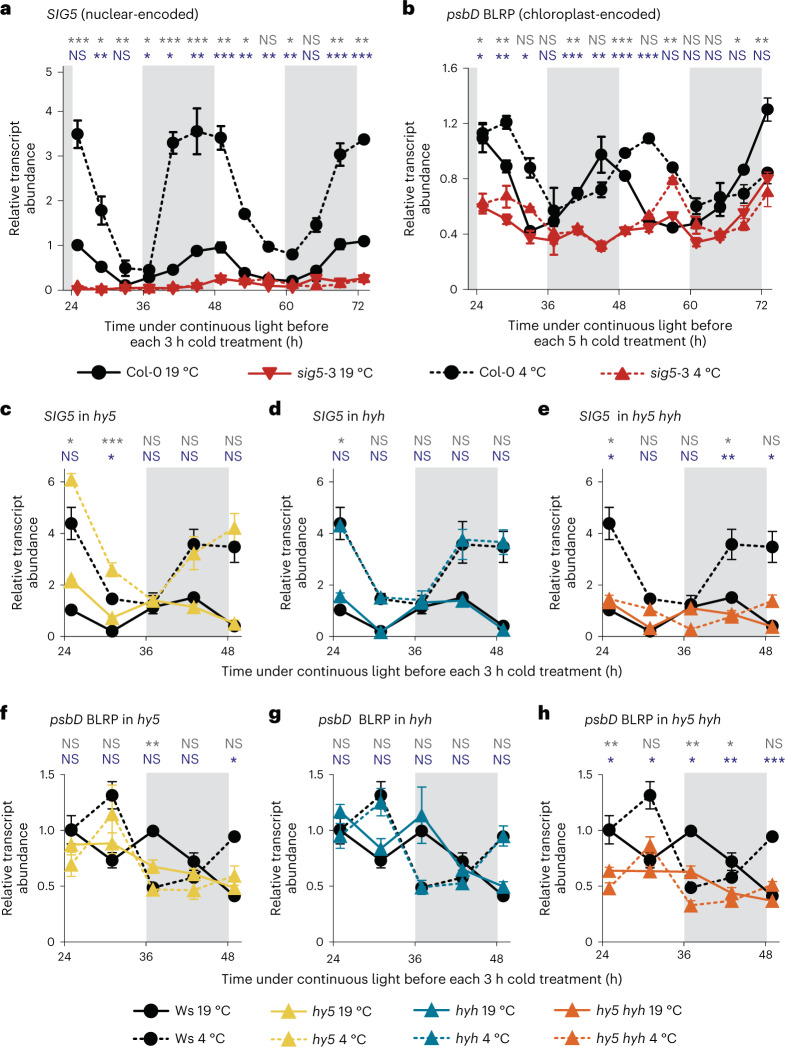
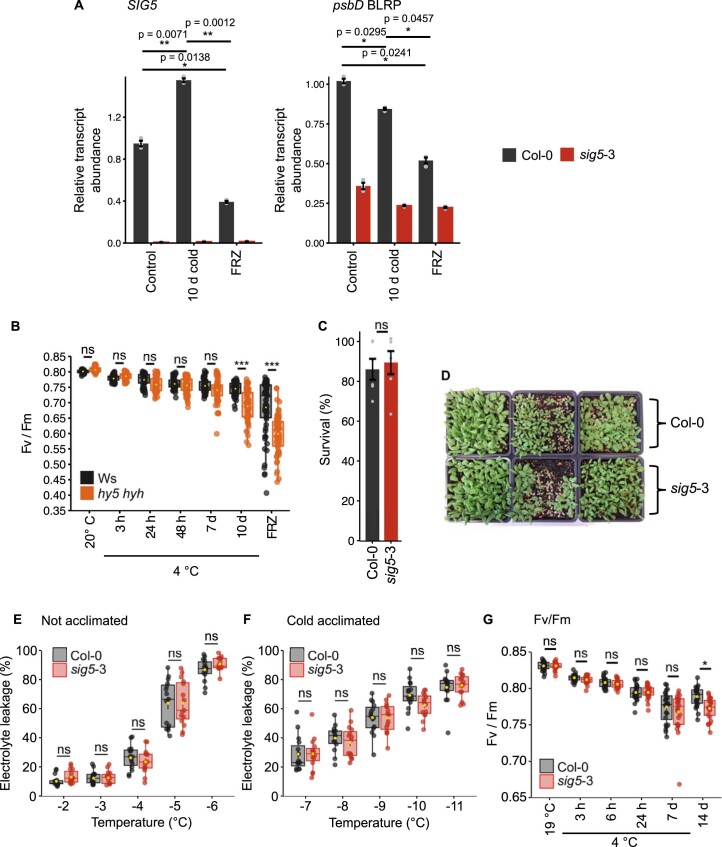
We investigated the hypothesis that sigma factors participate in low-temperature responses of chloroplasts, because transcripts encoding the Arabidopsis sigma factors SIG1, SIG4 and SIG5 accumulate in response to low temperatures (Extended Data Fig. 1)23,28. We focused on the role of SIG5 in low-temperature responses because published microarray data indicate that it has the greatest transcriptional response to cold (Extended Data Fig. 1)28. We used cold treatments of 4 °C for 3 h because this was the shortest cold treatment that provided a robust response of SIG5 transcripts (Extended Data Fig. 1)28. We confirmed this using a quantitative polymerase chain reaction with reverse transcription (RT–qPCR; Fig. 1a, Extended Data Fig. 2a and Supplementary Data 1). ELONGATED HYPOCOTYL5 (HY5) is necessary for SIG5 transcript accumulation in the light23,29, and HY5 protein accumulates under low-temperature conditions owing to nuclear depletion of the ubiquitin ligase COP1 that targets HY5 for degradation30. This motivated us to investigate whether HY5 and HY5 HOMOLOG (HYH) contribute to the SIG5 transcript response to low temperatures. When the cold treatment was given 1 h after dawn, we found that SIG5 transcripts accumulate in response to 3 h of low temperatures in the wild type, but not in a hy5 hyh double mutant (Fig. 1b and Supplementary Data 1). Both hy5 and hyh single mutants did not affect the response of SIG5 transcripts to cold treatment at this time of day (Fig. 1b). Under control temperature conditions, SIG5 transcripts accumulate predominantly in the light23–26,29,31. However, in darkness, SIG5 transcript levels increased in response to a 3 h cold treatment in the wild type, but not in the hy5 hyh double mutant (Fig. 1c and Supplementary Data 1). It is known that SIG5 transcript accumulation in response to salinity involves HOMEOBOX-LEUCINE ZIPPER PROTEIN17 (ATHB17)27. We found that cold induction of SIG5 in the light was not altered in athb17 mutants (Extended Data Fig. 2b), suggesting that ATHB17 does not participate in this response to cold.
Extended Data Fig. 1. Transcriptome data identifies SIG5 transcript accumulation in response to cold.
Transcriptome data identifies SIG5 transcript accumulation in response to cold. Data indicate the fold-change in abundance of all six Arabidopsis sigma factors (SIG1-6) during a prolonged cold treatment, measured using microarray analysis in28. Data extracted using the Arabidopsis eFP Browser (https://bar.utoronto.ca)101. Data are means from two biological replicates + /- s.d., as described in28.
Fig. 1. SIG5 communicates information to chloroplasts about cold temperature conditions, and this requires HY5 and HYH.
a, Relative abundance of all six Arabidopsis sigma factor transcripts in wild type (Col-0) after 3 h at 19 or 4 °C. b,c, SIG5 transcript accumulation in wild type (Ws), hy5, hyh and hy5 hyh double mutant after 3 h at 4 °C in light (b) and darkness (c). d, Abundance of SIG5 and chloroplast psbD BLRP transcripts in Col-0 and sig5-3 mutant after 3 h (SIG5) and 5 h (psbD BLRP) at 4 °C. e,f, psbD BLRP transcript accumulation in Ws, hy5, hyh and hy5 hyh double mutant after 5 h at 4 °C in light (e) and darkness (f). Darker and paler bars indicate control (19 °C) and cold (4 °C) treatments, respectively. Experiments used 11-day-old seedlings. SIG5 and psbD BLRP transcript abundance was measured after 3 and 5 h of cold treatment, respectively, because there is a time delay between the accumulation of SIG5 transcripts and downstream psbD BLRP24,26. Data represent mean ± s.e.m. and n = 3, except in b where n = 6. Statistical significance represents cold treatments compared with control temperature conditions (two-sided t-tests). ***P < 0.001; **P < 0.01; *P < 0.05; NS, not significant. Exact P values are given in Supplementary Data 1.
Extended Data Fig. 2. SIG5 transcript accumulation during cold treatments of up to 24 h.
SIG5 transcript accumulation during cold treatments of up to 24 h. (A) Relative abundance of SIG5 transcripts in Col-0 wild type and sig5-3 plants after cold treatment (4 °C) for durations ranging from 30 mins to 24 hours. (B) Abundance of SIG5 transcripts in wild type (Col-0) and two athb17 mutants after 3 h at 19 °C or 4 °C. Plants were under constant light conditions and given a 3 h cold treatment 1 h after subjective dawn. Experiments used 11-day old seedlings. Data are mean + /- s.e.m. (n = 3 independent biological replicates). Statistical comparisons are of (A) transcript abundance between Col-0 19 °C and Col-0 4 °C and (B) transcript abundance between control temperature and cold-treated plants, and between genotypes. Data analysed by (A) one-way ANOVA and (B) two-way ANOVA, both followed by one-sided post-hoc Tukey test, where *** = p < 0.001; ** = p < 0.01, * = p < 0.05 and n.s = not significant. p-values (A) 19 °C vs 4 °C for Col-0 at 0 h (p = 1.0000), 0.5 h (p = 0.9277), 1 h (p = 1.0000), 3 h (p = 0.1867), 6 h (p < 0.0001), 9 h (p < 0.0001), 12 h (p = 0.0001), 24 h (p < 0.0001); (B) 19 °C vs 4 °C for Col-0 (p = 0.0000425), athb17-1 (p = 0.0008523); athb17-2 (p = 0.0010056); Col-0 vs athb17-1 at 4 °C (p = 0.1958629); Col-0 vs athb17-2 at 4 °C (p = 0.0918899).
Within chloroplasts, SIG5 controls transcription from the blue light responsive promoter (BLRP) of the chloroplast psbDC operon23 and several other chloroplast genes24. Therefore, psbD BLRP transcript accumulation represents an informative read-out of SIG5 activity in plastids. We found that psbD BLRP transcripts accumulate strongly in response to 5 h cold treatment of the wild type, but not the well-characterized sig5 loss-of-function mutant sig5-3 (refs. 23,24,26) (Fig. 1d and Supplementary Data 1). This indicates that SIG5 is necessary for the upregulation of chloroplast psbD BLRP transcript levels by cold treatment. psbD BLRP transcript abundance decreased in the hy5 hyh mutant in response to a cold treatment starting 1 h after dawn, indicating that HY5 and HYH are required for psbD BLRP transcripts to accumulate in response to cold (Fig. 1e). In contrast to SIG5 transcripts, psbD BLRP transcripts did not accumulate in response to 5 h of cold treatment in darkness (Fig. 1c,f and Supplementary Data 1). Therefore, SIG5 is necessary for a cold-induced increase in chloroplast psbD BLRP transcripts in the presence of light, and this process requires HY5 or HYH.
Circadian regulation by SIG5 involves HY5 and HYH
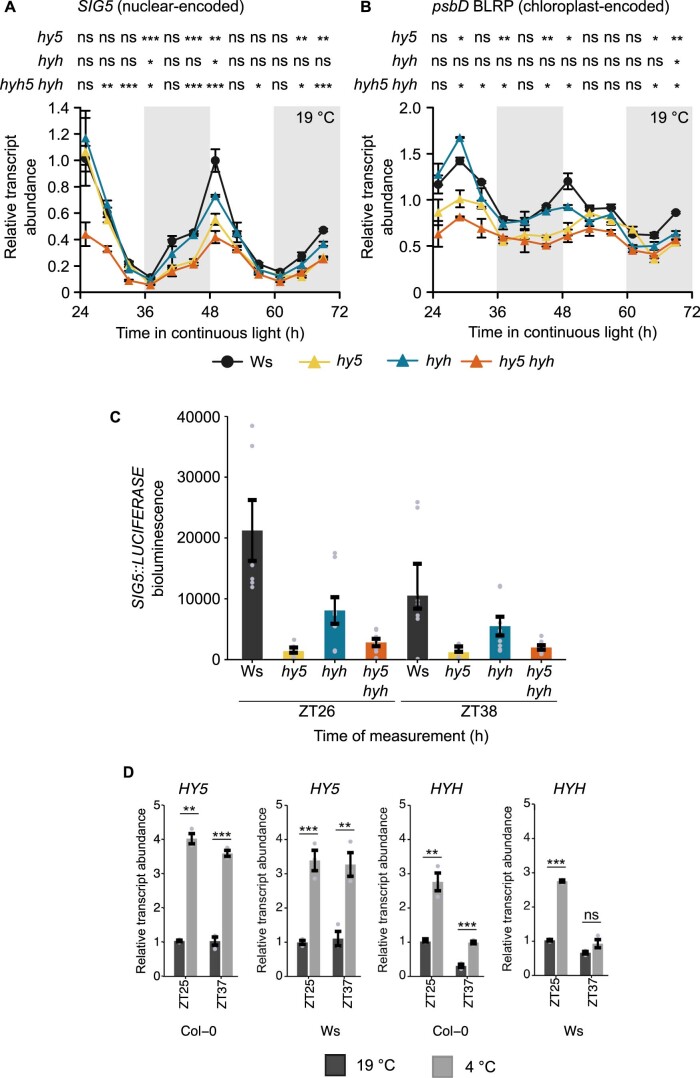
HY5 contributes to the circadian regulation of some transcripts32,33, and SIG5 participates in circadian signalling to chloroplasts24. Therefore, we hypothesized that HY5 or HYH might contribute to circadian signalling to chloroplasts by SIG5. We investigated this by cultivating seedlings for 11 days under cycles of 12 h light and 12 h darkness, and then transferring the seedlings to conditions of constant (white) light and temperature to monitor the free-running rhythm of transcript abundance. Under these control temperature conditions, we compared the circadian rhythms of SIG5 and psbD BLRP transcript accumulation in the hy5, hyh and hy5 hyh mutants, and the wild type. SIG5 transcript abundance increased during the subjective night to reach a peak around subjective dawn (Extended Data Fig. 3a and Supplementary Data 1), which is consistent with other studies conducted under constant white light of 90 µmol m−2 s−1 (ref. 24). This contrasts to the dynamics of SIG5 transcript abundance under monochromatic light, where it peaks later in the subjective day26. The peak transcript abundance of SIG5 was reduced significantly at a subset of time points in the hy5 or hyh single mutants, and at a greater number of time points in the hy5 hyh double mutant (Extended Data Fig. 3a). The peak abundance of psbD BLRP was reduced significantly at some time points in the hy5 mutant and hy5 hyh double mutant, but not in the hyh single mutant (Extended Data Fig. 3b and Supplementary Data 1).
Extended Data Fig. 3. Accumulation of SIG5 and psbD BLRP transcripts in the wild type (Ws), hy5, hyh and hy5 hyh double mutant.
Accumulation of (A) SIG5 and (B) psbD BLRP transcripts in the wild type (Ws), hy5, hyh and hy5 hyh double mutant. Experiments used 11-day old seedlings. All data represent means + /- s.e.m. of three independent biological replicates. Statistical comparison of each mutant with the wild type, at each timepoint, is provided above time-series plots, where *** = p < 0.001; ** = p < 0.01, * = p < 0.05 and n.s = not significant (one-way ANOVA followed by one-sided post-hoc Tukey test; 3 biological replicates; p values for panels A and B in Supplementary Data 1). (C) Quantification of luciferase bioluminescence after transient expression of SIG5::LUCIFERASE in wild type, hy5, hyh and hy5 hyh mutants, at two timepoints, using particle bombardment (n = 8 replicates per condition and genotype, except for Ws at ZT26 where n = 6; mean + /- s.e.m.). (D) Response of HY5 and HYH transcripts to a treatment of 3 h at 4 °C, given at two different timepoints, under free running conditions. Transcript levels normalized to ACT2 and confirmed with two other reference transcripts (not shown), 3 biological replicates show as mean + /- s.e.m; *** = p < 0.001; ** = p < 0.01, * = p < 0.05 and n.s = not significant in unpaired two-sided t-tests. p values for (D) are Col-0 HY5 (ZT25 0.002; ZT37 0.000), Ws HY5 (ZT25 0.001; ZT37 0.006), Col-0 HYH (ZT25 0.003; ZT37 0.000), Ws HYH (ZT25 0.000; ZT37 0.098).
To evaluate further the contribution of HY5 and HYH to circadian rhythms of SIG5 and psbD BLRP transcript accumulation, we compared the amplitude of these rhythms in hy5, hyh, hy5 hyh and the wild type, using MetaCycle circadian rhythm analysis software34. Under these control temperature conditions, the amplitude of the circadian rhythm of SIG5 and psbD BLRP transcript accumulation was lower in the hy5, hyh and hy5 hyh mutants compared with the wild type (Extended Data Fig. 3a,b and Supplementary Data 1 and 2). The amplitude was reduced more in hy5 compared with hyh, and was comparable between hy5 and hy5 hyh (Extended Data Fig. 3a,b and Supplementary Data 1 and 2). Therefore, either HY5 and HYH participate in the circadian regulation of SIG5 transcript accumulation, or alternatively HY5 or HYH allows another factor to confer the circadian rhythm of SIG5 transcript accumulation. This does not completely explain the circadian control of SIG5 transcript abundance, because transcript levels continue to oscillate with low amplitude in hy5 hyh (Extended Data Fig. 3a and Supplementary Data 1 and 2). We transiently expressed SIG5::LUCIFERASE in the wild type and hy5, hyh and hyh5 hyh mutants using particle bombardment35,36 and found that SIG5 promoter activity was reduced substantially in the hy5 and hy5 hyh double mutants, and partially in the hyh mutant, relative to the wild type (Extended Data Fig. 3c). This supports the notion that HY5 and HYH are important regulators of SIG5 promoter activity23,29,37. psbD BLRP transcripts had a late phase or longer period in the hy5 and hy5 hyh mutants, and were arrhythmic in hyh (Extended Data Fig. 3b and Supplementary Data 2; MetaCycle BH.Q, P = 0.27 for hyh), which differs from the rhythm of SIG5 transcript accumulation (Extended Data Fig. 3a). One interpretation is that additional effects of hy5 and hyh on this pathway, downstream of SIG5, contribute to the circadian regulation of psbD BLRP transcript levels. This difference is supported by HY5 and HYH also having differing roles in the responses to low temperatures of SIG5 and psbD BLRP (Fig. 1b–f). HY5 and HYH transcript levels were upregulated by a 3 h cold treatment at either ZT25 or ZT37 (Extended Data Fig. 3d), with the exception of HYH in the Ws background at ZT37.
Circadian gating of the cold response of SIG5
We tested the hypothesis that there is circadian gating of the responses of SIG5 and psbD BLRP transcripts to cold, because there is circadian gating of other transcriptional responses to low temperatures38,39. Circadian gating is the process whereby the circadian oscillator modulates the response to a stimulus, so that the magnitude of the response depends on the time of day of the stimulus40,41.
Groups of seedlings were exposed to 3 h cold treatments, at regular intervals, under constant light conditions. Each separate group of seedlings received a single cold treatment and was then harvested to measure the response of the transcripts to a cold treatment given at that particular time. Cold treatment caused greater SIG5 transcript accumulation between subjective midnight (zeitgeber time (ZT) 41; that is, 41 h after the final dawn under constant free-running conditions) and subjective dawn (ZT49), and less accumulation between ZT33 and ZT37 (Fig. 2a and Supplementary Data 1). This suggests that there is circadian gating of the response of SIG5 transcripts to cold. Cold caused greatest psbD BLRP transcript accumulation during the subjective day, compared with a peak at subjective dawn under control temperature conditions (Fig. 2b and Supplementary Data 1). The phase shift of psbD BLRP after short cold treatments (Supplementary Data 2) might suggest the cold-responsive circadian gate is timed with a different phase compared with the control-temperature circadian rhythm, or that low temperature delays psbD BLRP transcript accumulation. In the sig5-3 mutant, psbD BLRP remained cold-inducible at several time points during the subjective day (Fig. 2b, red symbols), suggesting that psbD BLRP transcript levels are regulated by a mechanism additional to SIG5. This pattern of circadian gating of cold induction of SIG5 transcripts is altered at some time points in the hy5 mutant (Fig. 2c and Supplementary Data 1), unaffected in hyh and abolished in the hy5 hyh double mutant (Fig. 2c–e, hy5 hyh in Fig. 2e and Supplementary Data 1). The circadian gating of cold induction of psbD BLRP is also altered at some time points in the hy5 mutant and hy5 hyh double mutant (Fig. 2f–h). In general, psbD BLRP appears less cold-responsive in the Ws accession compared with Col-0 (Figs. 1d,e and 2b,f), which is consistent with differences in temperature responses between Arabidopsis accessions42,43. It appears that the hy5 single mutant affects SIG5 and psbD BLRP transcript accumulation at control temperatures, whereas the hy5 hyh mutant is required to abolish its response to cold (Extended Data Fig. 2a,b and Fig. 2f–h).
Fig. 2. Circadian gating of the responses to cold of SIG5 and chloroplast psbD BLRP, and the involvement of HY5 and HYH.
a,b, Circadian gating of the response to cold of SIG5 (a) and psbD BLRP (b) transcripts in the Col-0 wild type and sig5-3 mutant. c–f, Circadian gating of the response to cold of SIG5 (c–e) and psbD BLRP (f–h) in the hy5, hyh or hy5 hyh double mutant. Cold treatments comprised 3 h at 4 °C for SIG5 transcript levels, and 5 h at 4 °C for psbD BLRP. Each short cold treatment was applied to a separate batch of seedlings. The x axis indicates the time at which the cold treatment commenced. Grey shading on graphs indicates subjective night, under constant light conditions. Solid and broken lines indicate control (19 °C) and cold (4 °C) treatments, respectively. Wild-type data (black lines) are duplicated across c–e and f–h for visual clarity. Experiments used 11-day-old seedlings. Data represent mean ± s.e.m. of three independent biological replicates. Statistical information above graphs compares the transcript levels in the wild type and mutant under control temperature conditions (grey text) and in response to cold (blue text) at each time point. ***P < 0.001; **P < 0.01; *P < 0.05; NS, not significant in unpaired two-sided t-tests. Exact P values are given in Supplementary Data 1.
SIG5 shapes the nuclear-encoded cold-responsive transcriptome
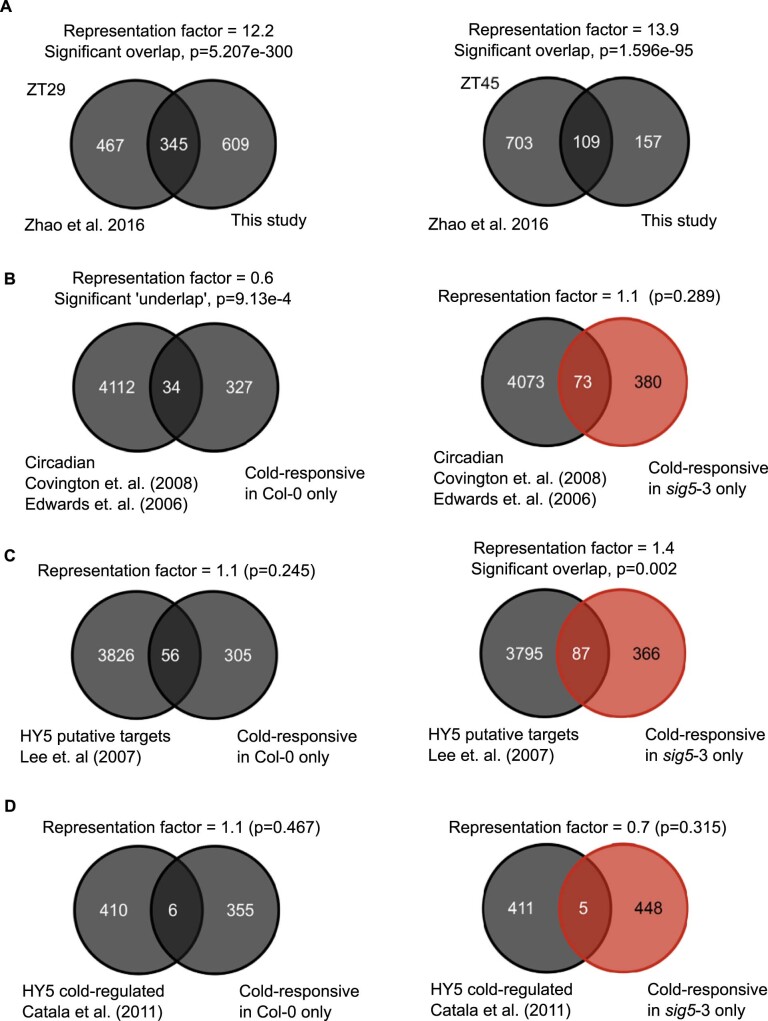
We hypothesized that SIG5 might have a broader role in cold-responsive gene regulation, because other sigma factors can indirectly influence nuclear-encoded gene expression44,45. To test this, we investigated transcriptome alterations in wild-type and sig5-3 seedlings in response to 3 h cold treatments given at two different time points (ZT29 and ZT45). We selected these times because they correspond to the peak sensitivity of psbD BLRP and SIG5 to cold, respectively (Fig. 2a,b). Under control temperature conditions, a relatively small number of transcripts was differentially expressed between Col-0 and sig5-3 at the time points examined (33 and 42 transcripts were differentially expressed at ZT29 and ZT45, respectively), with no significant Gene Ontology (GO)-term enrichments within these gene sets (Supplementary Data 3). In the Col-0 wild type, 954 and 266 transcripts responded to cold at ZT29 and ZT45, respectively, whereas in sig5-3, 1,000 (ZT29) and 319 (ZT45) transcripts responded to cold (Fig. 3a and Supplementary Data 3 and 4; cold-responsive defined as log(fold change) > 2 and P ≤ 0.01 using Voom/Limma method46). Some 158 transcripts in Col-0 (13% of cold-responsive transcripts) and 192 transcripts (14.6%) in sig5-3 responded to cold at both time points, so the majority of cold-responsive transcripts were unique to the time at which the seedlings were cold-treated (Fig. 3a). The different sets of cold-responsive transcripts at the two time points are consistent with the notion that there is circadian gating of the cold-responsive transcriptome in plants47. We compared our cold-responsive transcript set in Col-0 with that of Zhao et al. (3 h cold treatment)48 and found that 42.5% (ZT29) and 13.4% (ZT45) of cold-responsive transcripts were shared between the studies (Extended Data Fig. 4a)29. The smaller overlap at ZT45 might reflect time of day differences in the cold-responsive transcriptome.
Fig. 3. Genome-wide influence of SIG5 upon the cold-responsive transcriptome.
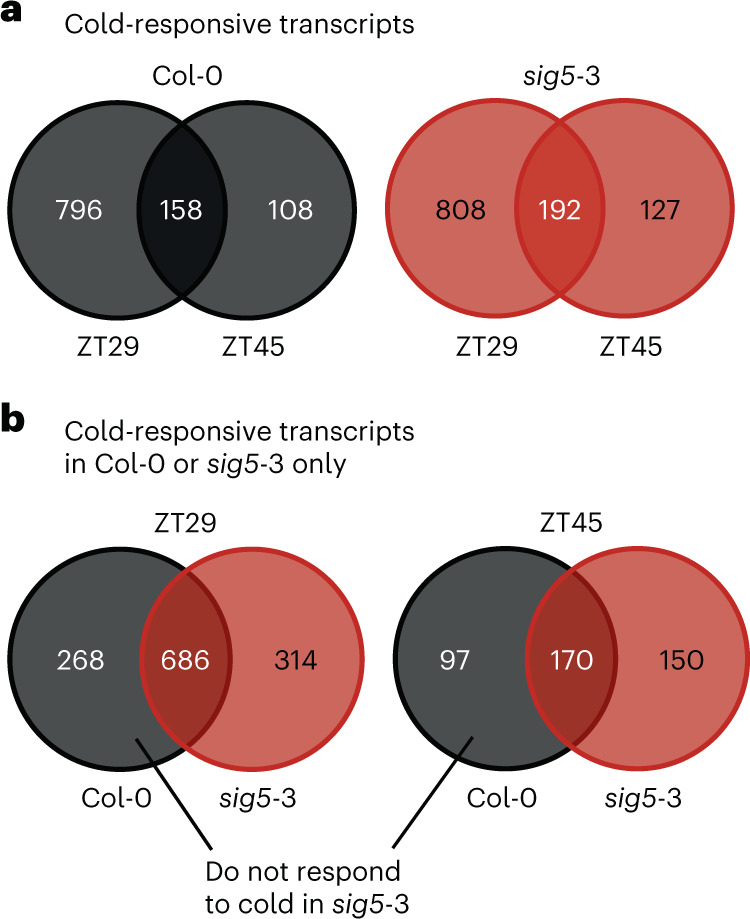
a, Overlap between transcripts responsive to cold in Col-0 or sig5-3 at time points ZT29 and ZT45. b, Overlap between transcripts responsive to cold in Col-0 and sig5-3 at the two time points. Numbers within the circles on Venn diagrams indicate the number of transcripts. Experiments used 11-day-old seedlings and 3 h (ZT45) or 5 h (ZT29) cold (4 °C) treatments. ZT refers to the time elapsed under free-running (constant) conditions, after the final dawn.
Extended Data Fig. 4. RNA sequencing analysis of transcripts in Col-0 wild type and sig5-3 that respond to a cold treatment at two different times of day.
RNA sequencing analysis of transcripts in Col-0 wild type and sig5-3 that respond to a cold treatment at two different times of day. (A) Overlap between cold-responsive transcripts in Col-0 from this study and cold-responsive transcriptome from48. (B) Overlap between cold-responsive transcripts in Col-0 or sig5-3 from this study and circadian-regulated transcripts from50 and49. (C) Overlap between cold-responsive transcripts in Col-0 or sig5-3 from this study and putative HY5 targets37. (D) Overlap between cold-responsive transcripts in Col-0 or sig5-3 from this study and transcripts regulated in response to cold by HY530. Statistical significance and representation factors were calculated using a hypergeometric test (one-sided; see methods). Experiments used 11-day old seedlings.
Of the transcripts that responded to cold at ZT29, 268 were cold-responsive only in Col-0 (198 upregulated, 70 downregulated) but not cold-responsive in sig5-3 (Fig. 3b and Supplementary Data 5; statistical threshold for cold-responsiveness of log(fold change) > 2 and P ≤ 0.01 using Voom/Limma method46). Similarly, at ZT45, 97 transcripts responded to cold (46 upregulated, 51 downregulated) in Col-0 but not sig5-3 (Fig. 3b and Supplementary Data 5). At ZT29, 314 transcripts responded to cold in sig5-3 but not Col-0 (179 upregulated, 135 downregulated), whereas at ZT45 150 transcripts responded to cold in sig5-3 only (109 upregulated, 41 downregulated, including several chloroplast transcripts; Fig. 3b and Supplementary Data 5). Together, this indicates that some transcripts required SIG5 to respond significantly to the cold treatment. Because circadian timing influences the response to cold of psbD BLRP transcripts (Fig. 2b), we hypothesized that the set of transcripts that responded significantly to the cold treatment in the wild type but not sig5-3 mutant might be enriched with circadian-regulated transcripts. However, examination of cold-responsive transcript sets that are unique to Col-0 or sig5-3 identified that circadian-regulated transcripts49,50 were not overrepresented among the sig5-3-specific cold-responsive transcripts. Furthermore, the set of transcripts that responded significantly to the cold treatment in Col-0 but not in sig5-3 was significantly underrepresented with circadian-regulated genes (Extended Data Fig. 4b). The only circadian clock-associated transcript that was significantly cold-induced in Col-0 but not sig5-3 was NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED4 (LNK4) (at ZT29; Supplementary Data 5), although the role of LNK4 within circadian regulation remains somewhat uncertain51,52. Furthermore, transcripts encoding the zinc-finger protein B-BOX DOMAIN PROTEIN19 (BBX19) were upregulated by cold in sig5-3, but not the wild type (Supplementary Data 5). BBX19 is thought to repress the promoters of certain morning-phased circadian clock components53. There was a significant overlap between transcripts that responded significantly to the cold treatment in sig5-3 but not in the wild type and putative HY5 targets37 (Extended Data Fig. 4c), but no significant intersection with HY5 regulated cold-induced genes (Extended Data Fig. 4d)30.
Using GO-term analysis, we evaluated whether the sets of transcripts that responded significantly to cold in only Col-0 or sig5-3 are enriched with genes linked to specific processes. Genes linked to hypoxia responses were overrepresented at ZT29 for transcripts that responded significantly to cold in Col-0 but not sig5-3 (Benjamini–Hochberg correction, P < 0.05), and in a combined list of transcripts that responded significantly to cold in only sig5-3 at both ZT29 and ZT45 (P < 0.05) (Supplementary Data 6). The set of cold-responsive transcripts in sig5-3 only was enriched for AP2/ERF domain proteins (P < 0.001), which participate in abiotic and biotic stress responses, growth and development54.
SIG5 maintains photosynthetic efficiency during low-temperature conditions
We reasoned that the cold induction of transcripts encoding SIG5 and its chloroplast target psbD BLRP might underlie physiological responses of plants to low temperatures. We investigated the involvement of SIG5 in cold and freezing responses using chlorophyll fluorescence as a proxy for photosynthetic responses to cold and freezing, and electrolyte leakage as a measure of tissue damage by freezing.
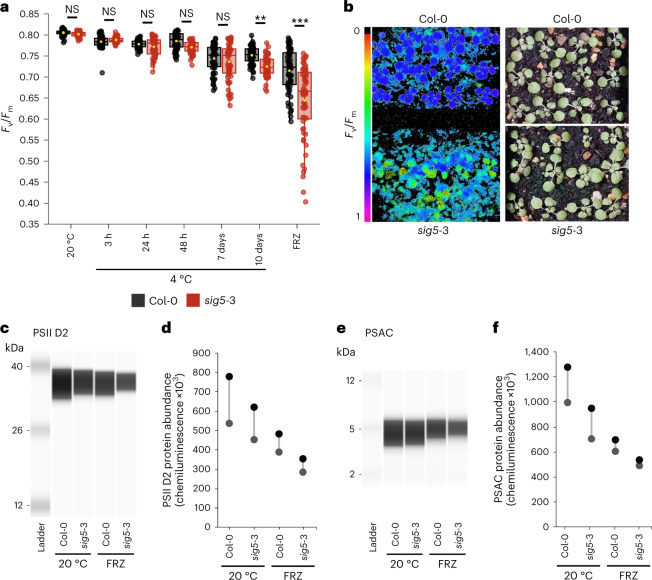
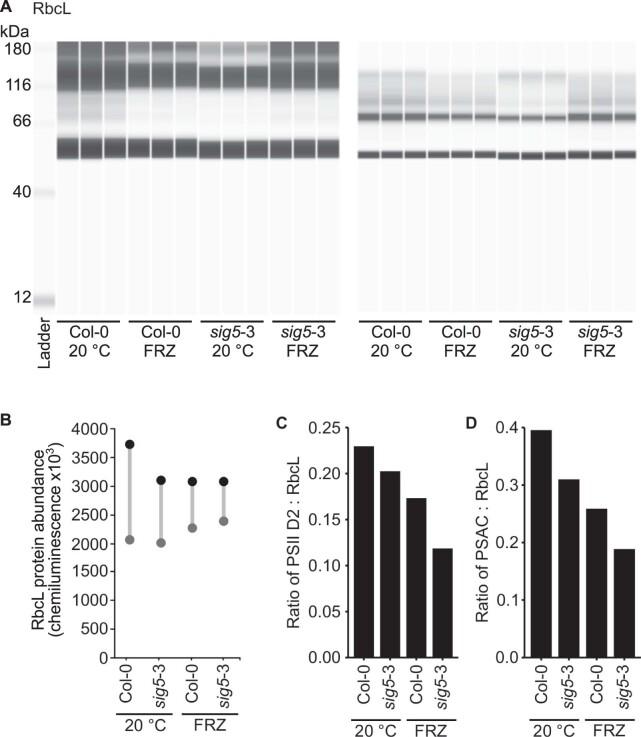
Because SIG5 regulates the transcription of the gene encoding the D2 protein of PSII (psbD)23 and low temperature increases PSII excitation pressure55, we investigated PSII photosynthetic efficiency by measuring chlorophyll fluorescence (Fv/Fm) in sig5-3 after short- and long-term cold treatments. In the wild type, cold reduced the ratio of variable fluorescence (Fv) to maximum fluorescence (Fm), Fv/Fm relative to the 20 °C control (Fig. 4a). Furthermore, Fv/Fm was reduced significantly in sig5-3 compared with the wild type after a long-term cold treatment of 10 days at 4 °C (Fig. 4a). A short freezing treatment of cold-acclimated plants (−8 °C for 6 h) decreased Fv/Fm, with Fv/Fm in sig5-3 reduced significantly more than in the wild type (Fig. 4a,b). Therefore, SIG5 contributes to maintaining the photosynthetic efficiency of PSII during prolonged cold and short-term freezing. We reasoned that this might arise from effects of the sig5-3 mutation upon photosystem protein abundance during freezing. To investigate this, we compared after the short freezing treatment that reduced Fv/Fm, the abundance of a photosystem protein that is regulated transcriptionally by SIG5 (PSII D2) and a protein thought to not be regulated transcriptionally by SIG5 (PSAC)24. Both PSII D2 and PSAC protein abundance was decreased consistently in sig5-3 plants after this freezing treatment, compared with Col-0 under control temperature conditions (Fig. 4c–f and Extended Data Fig. 5a,b). By contrast, a chloroplast-encoded and localized protein that does not form part of the photosystems (RbcL), which is not thought to be regulated by SIG5 (ref. 24), was unaltered in sig5-3 by this freezing treatment (Extended Data Fig. 6a–c). Normalization of PSII D2 and PSAC protein abundance to the abundance of RbcL under each treatment confirms the reduced abundance of these photosystem proteins relative to RbcL (Extended Data Fig. 6c,d). This suggests that reduced PSII D2 abundance might occur through either a direct effect of the sig5-3 mutation upon psbD BLRP promoter activity, or alternatively through a general alteration in photosystem protein levels in sig5-3 after freezing. SIG5 and psbD BLRP transcript levels were decreased relative to control temperature conditions after freezing (Extended Data Fig. 7a), suggesting the presence of SIG5 rather than its cold induction maintains PSII D2 and PSAC protein abundance during freezing. Fv/Fm was also reduced significantly in hy5 hyh mutants compared with the wild type (Ws) after 10 days at 4 °C, and after 6 h of freezing (Extended Data Fig. 7b), so regulation by HY5 and/or HYH also maintains PSII photosynthetic efficiency at low temperatures.
Fig. 4. SIG5 influences photosynthetic efficiency under cold temperature conditions.
a, Photosynthetic efficiency of PSII (Fv/Fm) of 14-day-old wild-type (Col-0) and sig5-3 plants exposed to cold (4 °C) and freezing (FRZ) treatments (n = 60). In box plots, the box indicates the interquartile zone with the median line at the centre, whiskers indicate interquartile range and a yellow dot indicates the mean. Data were analysed by two-way analysis of variance followed by post-hoc Tukey test. ***P < 0.001; **P < 0.01; NS, not significant (P values: 20 °C = 0.999; 3 h = 0.999; 24 h = 0.999; 48 h = 0.474; 7 days = 0.994; 10 days = 0.001; FRZ < 0.001). b, Fv/Fm of Col-0 and sig5-3 seedlings after freezing at −8 °C for 6 h (left) and representative image of these plants before freezing (right). c–f, Automated semiquantitative capillary immunoassay comparing PSII D2 (c,d) and PSAC protein (e,f) levels between wild type (Col-0) and sig5-3 under control temperature conditions (20 °C) and after freezing at −8 °C for 6 h (FRZ). Samples were analysed in triplicate (three technical replicates) from each independent experiment, with (d,f) two independent experiments shown. d,f, Quantification of PSII D2 (d) and PSAC protein (f) levels (area under each peak), relative to levels in wild type (Col-0) at 20 °C from replicate immunoassays. Circles on plots indicate result from each independent experiment. Cold (4 °C) and freezing treatments (FRZ) were conducted identically for all experiments and included a 10-day cold acclimation period at 4 °C before the freezing treatment.
Extended Data Fig. 5. Altered photosystem protein abundance in sig5-3 mutant.
Altered photosystem protein abundance in sig5-3 mutant. (A, B) Automated semi-quantitative immunoassay comparing (A) PSII D2 and (B) PSAC protein abundance between wild type (Col-0) and sig5-3 under control temperature conditions (20 °C) and after freezing at -8 °C for 6 h (FRZ). Analysis shows two independent experiments, each containing triplicate immunodetection analyses. (C) Coomassie blue-stained SDS-PAGE separation of 0.5 mg/mL total protein from leaf protein extracts, run as a single example to demonstrate consistent protein input into the automated semi-quantitative immunoassay when samples were prepared identically for immunodetection.
Extended Data Fig. 6. RbcL protein abundance is unaltered by sig5-3 mutant and freezing.
RbcL protein abundance is unaltered by sig5-3 mutant and freezing. (A) Automated semi-quantitative immunoassay comparing RbcL protein abundance between wild type (Col-0) and sig5-3 under control temperature conditions (20 °C) and after freezing at -8 °C for 6 h (FRZ). Analysis shows two independent experiments, each containing triplicate immunodetection analyses. (B) Comparison of RbcL protein abundance from (A, B). Circles on plots indicate result from each independent experiment (two independent repeats, each with three technical replicates). (C, D) Ratio of (C) mean PSII D2 abundance and (D) mean PSAC abundance to mean RbcL abundance, calculated from the protein abundance data in Fig. 4D, F and Extended Data Fig. 6B.
Extended Data Fig. 7. Responses to low and freezing temperature conditions in the sig5-3 mutant.
Responses to low and freezing temperature conditions in the sig5-3 mutant. (A) Abundance of SIG5 and psbD BLRP transcripts in the wild type (Col-0) and sig5-3 mutant after 10 days at 4 °C and after exposure to freezing (FRZ) conditions. Conditions were comparable to those shown in Fig. 4A and C (n = 3 independent repeats, mean + /- s.e.m.). (B) Photosynthetic efficiency of PSII (Fv/Fm) of 14-day old wild type (Ws) and hy5 hyh plants exposed to cold and freezing (FRZ) treatments (n = 60). (C) Survival of Col-0 wild type and sig5-3 plants grown for 14 days at 20 °C then acclimated at 4 °C for 10 days. Plants were subjected to -8 °C for 6 h and then allowed to recover at 20 °C for 7 days (n = 6; data analysed by Student’s t-test; two-sided; not significant; mean + /- s.e.m.). (D) Representative appearance of Col-0 and sig5-3 plants after 7 days of recovery at 20 °C after the freezing treatment, showing variation across three replicate pots per genotype. (E, F) Electrolyte leakage after freezing of leaf discs from plants of wild type (Col-0) and sig5-3. Plants were tested after 5 weeks of growth (E, not acclimated) or 5 weeks of growth plus 2 weeks of acclimation at 4 °C (F, cold acclimated). n = 15. (G) Photosynthetic efficiency of PSII (Fv/Fm) of 5-week-old Col-0 and sig5-3 plants after treatment at 19 °C or 4 °C for 3, 6, 24 hours, 7 and 14 days (n = 24, mean + /- s.e.m; p-values of Col-0 vs sig5-3 are p = 1.0000 (19 °C), p = 0.9999 (3 h), p = 0.9999 (6 h), p = 1.0000 (24 h), p = 0.7955 (7 days), p = 0.0011 (14 days)). (A, B, E-G) Data analysed by two-way ANOVA followed by post-hoc one-sided Tukey test, with (E, F) arcsine correction before analysis. *** = p < 0.001; ** = p < 0.01, * = p < 0.05 and n.s = not significant. On boxplots (B, E-G), box indicates interquartile zone with median line at the centre, whiskers indicate interquartile range, and yellow dot indicates the mean.
During cold acclimation, sensing of low non-freezing temperatures leads to changes in membrane fluidity, cell wall structure and the accumulation of compatible solutes and antioxidants to increase freezing tolerance1,5,56. Previous studies have shown that greater damage to the photosynthetic apparatus of plants with lower freezing tolerance can manifest as reduced Fv/Fm during freezing57, as we identified for sig5-3 (Fig. 4a). For example, reduced Fv/Fm of hy5 hyh after prolonged cold and freezing (Extended Data Fig. 7b) is consistent with reduced freezing tolerance of the hy5 mutant, compared with the wild type, after cold acclimation19. We tested whether there was reduced freezing tolerance in sig5-3, but found no difference in the survival of 14-day-old sig5-3 and wild-type plants that were cold-acclimated for 10 days at 4 °C and then subjected to −8 °C (Extended Data Fig. 7c,d). We also tested this in mature rosette plants, using the freezing-induced leakage of electrolytes from leaf discs from rosette leaves as a proxy for freezing damage. This indicated that there was no difference in the level of cellular damage between sig5-3 and wild type, irrespective of cold acclimation (Extended Data Fig. 7e,f), even though Fv/Fm was also reduced after 14 days at 4 °C in leaves of mature rosettes (Extended Data Fig. 7g). We conducted electrolyte leakage analysis on mature leaves rather than younger plants to enable the very consistent sampling of leaf discs that is necessary to limit data noise. Nevertheless, freezing tolerance was unaltered by the sig5-3 mutant, relative to the wild type, in both 14-day-old seedlings and mature plants (Extended Data Fig. 7c–f). Therefore, basal or acquired freezing tolerance under the conditions tested is unaltered in sig5-3, and the lower photosynthetic efficiency of the mutant during long-term cold (Fig. 4a) does not affect freezing survival (Extended Data Fig. 7c–f).
Discussion
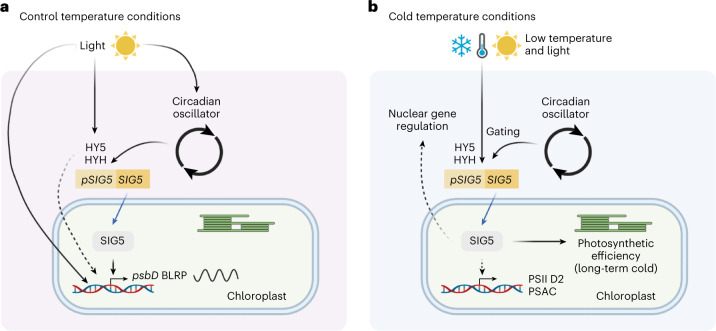
We found that SIG5 is required for a response of chloroplast-encoded psbD BLRP transcripts to cold, suggesting that SIG5 communicates information to chloroplasts about low-temperature conditions. This response involves HY5 and HYH (Fig. 5a,b). HY5 also contributes to chloroplast processes such as photopigment biosynthesis33,58, so probably influences chloroplast gene expression through multiple independent mechanisms. HY5 is necessary for SIG5 transcript accumulation in response to light23,29, so regulation of SIG5 by HY5 and HYH integrates several environmental cues that are communicated to chloroplasts.
Fig. 5. Involvement of SIG5 in cold-temperature responses.
a, Under control temperature conditions, light regulates the circadian clock and also HY5 and HYH-regulated genes. HY5 and HYH are necessary for the circadian regulation of SIG5 transcript accumulation, and circadian clock components might also regulate SIG5 expression directly. SIG5 regulates transcription of psbD via the BLRP, and HY5 or HYH might regulate psbD BLRP transcription through additional mechanisms. b, In response to cold temperatures, HY5 and HYH are necessary for the accumulation of SIG5 transcripts in response to cold, and the circadian clock gates the response to cold of SIG5 transcripts. SIG5 regulates PSII D2 and PSAC protein abundance, either by direct transcriptional regulation or through indirect mechanisms. SIG5 is necessary to maintain photosynthetic efficiency under long-term cold. SIG5 mutants have altered nuclear gene expression in response to cold, suggesting that SIG5 indirectly regulates nuclear genome transcription. Black solid arrows indicate regulatory relationships, broken arrows indicate inferred connections, a blue arrow entering the chloroplast indicates SIG5 targeting to chloroplasts and the sine wave icon indicates circadian regulation. Subcellular localization is inferred.
We found that in darkness, low-temperature induction of nuclear-encoded SIG5 was not accompanied by upregulation of psbD BLRP (Fig. 1c,f). One interpretation is that in darkness, low temperature upregulates SIG5 transcript levels, which might increase SIG5 protein abundance within chloroplasts. However, light is required for the association of PEP with chloroplast DNA, for PEP assembly and regulation of sigma factor phosphorylation state, with possible involvement of redox regulation59–63. Therefore, upregulation of SIG5 by cold in darkness might not alter psbD BLRP transcription because PEP is inactive. Another possibility is that in darkness, SIG5 is not imported efficiently into chloroplasts, so does not reach a threshold required to generate psbD BLRP transcripts64. Overexpression of SIG5 to a very high level from the chloroplast genome of transplastomic plants constitutively upregulates psbD BLRP, even in darkness65. This difference from our results might reflect the very high expression levels that are possible in transplastomic plants65. Either way, these interpretations support the notion that additional regulatory steps are positioned between SIG5 expression and accumulation of psbD BLRP transcripts.
There was a difference in the circadian phase of cold-sensitivity of SIG5 and psbD BLRP transcripts, such that SIG5 had greatest cold-responsiveness towards the end of the subjective night, whereas psbD BLRP transcripts had greatest cold-responsiveness during the subjective day (Fig. 2a,b). There are several potential explanations for this timing difference. The process of SIG5 protein synthesis, chloroplast import and PEP assembly will take some time, introducing a time delay into the process. Such a delay between SIG5 and psbD BLRP transcript accumulation also occurs under light/dark cycles at control temperatures24, and in field-grown plants66. The circadian clock might also influence chloroplast protein import, the expression of PEP-associated proteins required for chloroplast transcription67, and the activity of protein kinases thought to modulate sigma factor function such as redox-responsive CHLOROPLAST SENSOR KINASE62,68. In combination, these factors could superimpose several layers of temporal regulation upon this process of gene regulation.
One interpretation of our results is that in response to cold, SIG5 regulates psbD BLRP transcription directly to increase the supply of messenger RNA for translation into PSII D2, maintaining photosynthetic activity. Alternatively, SIG5 might regulate PSII D2 protein abundance independently from its role in psbD BLRP transcription, because chloroplast protein abundance is regulated by transcript stability, translation and protein turnover69–71. For example, other sigma factors regulate chloroplast transfer RNA expression44,72,73 and a chloroplast-encoded subunit of the ATP-dependent ClpP (caseinolytic) protease74, opening the possibility that SIG5 might influence PSII D2 accumulation through mechanisms such as translational regulation or protein degradation. The second interpretation appears to be supported by our data, because psaC is not thought to be a target of SIG5 regulation24, yet PSAC protein abundance decreases after freezing in sig5-3 (Fig. 4e,f). This suggests a more general role for SIG5 in maintaining photosystem protein abundance under certain stress conditions than thought previously.
SIG5 is required for circadian regulation of a set of chloroplast transcripts26. We identified that HY5 and HYH, acting with additional mechanisms, contribute to circadian rhythms of SIG5 transcript accumulation under control temperature conditions (Extended Data Fig. 3a,b). Given that HY5 coregulates transcripts with PHYTOCHROME INTERACTING FACTORs (PIFs) and there are interactions between CCA1 and HY5 proteins for the regulation of promoter activity32,33, circadian oscillator components or PIFs could contribute directly to the circadian regulation of SIG5. For example, chromatin immunoprecipitation experiments indicate that PIF1 and the circadian clock component PRR5 bind to the SIG5 promoter75,76, whereas CCA1 and LHY do not77–79. Therefore, multiple circadian clock-related factors appear to converge upon the promoter of SIG5, with HY5 and HYH representing one of these mechanisms. The circadian regulation and low-temperature responses of SIG5 transcripts could occur through HY5 and HYH regulating the G-box motif within the SIG5 promoter29.
It is interesting that a set of nuclear-encoded transcripts are cold-responsive in the wild type, but not in sig5-3. This phenotype of the sig5-3 mutant indicates that a function of SIG5 can influence nuclear-encoded gene expression. We speculate that this probably occurs indirectly, perhaps through metabolic alterations arising from altered chloroplast function in the sig5-3 mutant.
Conclusions and perspectives
Sigma factors allow bacteria and cyanobacteria to respond to cold temperature conditions80–83. Our experiments identify that sigma factors also participate in responses to cold temperatures in plants. Therefore, taken together with studies in bacteria and cyanobacteria80–83, it appears that sigma factors are involved in cold-temperature responses in both prokaryotes and eukaryotes. Our experiments identify a new regulator of cold-temperature responses of chloroplasts, and establish that a sigma factor contributes to protection of photosynthesis before and during freezing. The greater cold-sensitivity of this signalling pathway immediately before subjective dawn, combined with its role in light stress responses23, suggests it might be important during cold, bright mornings.
Methods
Plant material and growth conditions
All experiments were conducted using Arabidopsis thaliana L. (Heynh.). Arabidopsis seeds were surface-sterilized26 and sown on half strength Murashige and Skoog basal salts mixture (Duchefa Biochemie) in 0.8% (w/v) agar at pH 5.8, and stratified in darkness at 4 °C for 2 days before transfer to Panasonic MLR-352 plant growth chambers. Cultivation occurred under cycles of 12 h light/12 h darkness at 19 °C, 90 µmol m−2 s−1 of white light, with experiments starting at a seedling age of 11 days. SIG5 and psbD BLRP transcript abundance was measured after 3 and 5 h cold treatment, respectively, because there is a time delay between accumulation of SIG5 transcripts and downstream psbD BLRP24,26. For gating experiments in the hy5, hyh and hy5 hyh backgrounds, both SIG5 and psbD BLRP abundance was measured at the same time point (after 3 h of cold treatment). For single time point measurements of the response of transcripts to cold (Fig. 1), plants were transferred to the cold treatment 1 h after dawn. For circadian experiments (Fig. 2a,b and Extended Data Fig. 3a,b), seedlings were grown under light/dark cycles for 11 days and exposed to continuous light for 24 h before the start of the experiment. For measurements of chlorophyll fluorescence and protein abundance, seeds were sown directly onto compost and grown under cycles of 16 h light/8 h darkness at 20 °C, 90 µmol m−2 s−1 of white light for 14 days, before transfer to 4 °C in 12 h light/12 h darkness, 90 µmol m−2 s−1. For this experiment, 16 h days were used to increase the similarity of the experimental design to a previous study on HY5 and low-temperature responses30. The light spectrum was similar when the chambers were set to control and cold temperature conditions (Extended Data Fig. 8a,b; Li-Cor LI-180 spectrometer). For experiments using mature plants, seedlings were grown on MS agar as before and transferred to compost at 11 days, where they were grown for a further 24 days in a controlled environment chamber (Conviron) in 12 h light/12 h darkness at 19 °C, 100 µmol m−2 s−1 of white light before being used in experiments (light spectrum in Extended Data Fig. 8c). Experiments used the transfer DNA insertion mutant sig5-324 in the Col-0 background, and hy5KS50 (hy5)84, hyh, hy5KS50 hyh (hy5 hyh)85 in the Wassilewskija (Ws) background. For investigation of roles for ATHB17, we used T-DNA insertion lines SALK_095524 (athb17-1)27 and SALK_134535 (athb17-2) with Col-0 as the wild-type control.
Extended Data Fig. 8. Light spectra and RT-qPCR controls.
Light spectra and RT-qPCR controls. (A, B) Comparison of spectra in Panasonic MLR-352 growth chambers set to (A) control and (B) cold temperature conditions. (C) Spectrum of Conviron growth chambers used to propagate mature plants. (D) Comparison of response of SIG5 to cold, at two different timepoints, in two background lines, and in the hy5 hyh double mutant. The transcript dynamics are broadly similar when each of ACT2, EF-1A and UBQ10 was used as a reference gene. *** = p < 0.001; ** = p < 0.01, * = p < 0.05 and n.s = not significant in unpaired two-sided t-tests, n = 3 biological replicates shown as mean + /- s.e.m. p values: Col-0 ACT2 (ZT25 0.002; ZT37 0.000), Col-0 EF-1A (ZT25 0.011; ZT37 0.000), Col-0 UBQ10 (ZT25 0.002; ZT37 0.014), Ws EF-1A (ZT25 0.000; ZT37 0.239), Ws UBQ10 (ZT25 0.006; ZT37 0.239), hy5 hyh EF-1A (ZT25 0.534; ZT37 0.57), hy5 hyh UBQ10 (ZT25 0.432; ZT37 0.048).
RNA extraction and RT–qPCR
Tissue for RNA isolation was snap frozen in liquid nitrogen and stored at -80 °C until RNA extraction. Frozen tissue was ground to a powder for RNA extraction using a Qiagen TissueLyzer II ball mill. Total RNA was isolated using the Macherey-Nagel Nucleospin II RNA extraction kit, using the aerial portion of ten Arabidopsis seedlings in each extraction24,26. Total RNA yield was always greater than 200 ng µl−1, and any samples with A260/A280 below 2.0 were discarded (Thermo Fisher NanoDrop One). Complementary DNA was synthesized using the ABI High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific), using MultiScribe reverse transcriptase and random primers and with 1 µg of total RNA in each reaction. cDNA was analysed using Brilliant III Ultra-Fast SYBR Green QPCR master mix (Agilent Technologies) or qPCRBIO SyGreen (PCR Biosystems) and appropriate primer sets (Supplementary Data 7), normalized to ACTIN2 using the delta-delta Ct (ddCT) method24,26. Analysis used a Bio-Rad CFX96 Touch Real Time PCR System (running Bio-Rad CFX v.3.1 software). We confirmed key results (response of SIG5 to cold, variation in response to cold according to the time of day, alteration of the response to cold in hy5 hyh double mutant, equivalent response to cold in Col-0 and Ws backgrounds) using two further reference genes (UBIQUITIN10 (UBQ10) and EF1ALPHA (EF-1A)86). Primers for analysis of SIG5 and psbD BLRP transcript levels are from Noordally et al.24, whereas those for HY5 and HYH are from Hayes et al.87 (Supplementary Data 7). We do not think our cold treatments caused a systematic change in chloroplast transcript levels—as might happen if the number or viability of chloroplasts was altered by cold—because there was not a systematic change in chloroplast transcript abundance detected by RNA sequencing (RNA-seq) analysis (Supplementary Data 3), as also reported elsewhere10. Transcript time-series data were analysed using the meta2d tool within MetaCycle34, running in R v.4.1.1 (ref. 88) to identify rhythmic transcripts and properties of those rhythms.
Data acquisition and analysis for RNA-seq
Seedlings were cultivated as for RT–qPCR analysis. RNA samples were collected from Col-0 and sig5-3 plants at two different time points. After 24 h under continuous light, one set of seedlings was exposed to 3 h of cold (4 °C), commencing at ZT45, and the other set was exposed to 5 h of cold, commencing at ZT29. These corresponded to times when SIG5 (ZT45) and psbD BLRP (ZT29) transcripts accumulate strongly in response to chilling under free-running conditions (Fig. 2a,b). Total RNA was extracted from three replicates of ten seedlings each, using Macherey-Nagel Nucleospin II RNA extraction kits, and combined to represent one sample for sequencing. RNA concentrations were determined using a Nanodrop spectrophotometer (Thermo Fisher Scientific) and RNA integrity assessed using a Bioanalyzer (Agilent Technologies). Three independent biological replicates of these combined preparations were analysed by RNA-seq. The RNA-seq libraries were prepared from total RNA using an Illumina TruSeq Stranded mRNA kit, according to the manufacturer’s instructions. Sequencing was performed using an Illumina NextSeq 500 using NSQ 500 Hi-Output kit v.2 (150 cycles). The quality of the sequencing was confirmed with FASTQC v.0.11.3 (ref. 89). We trimmed any remaining adaptors using Trimmomatic v.0.33 (ref. 90), with the flags PE -phred33 ILLUMINACLIP:TruSeq2-PE.fa:2:30:10 LEADING:20 TRAILING:20 SLIDINGWINDOW:10:20 MINLEN:50. The gene models for Arabidopsis thaliana (TAIR10_cds_20110103_representative_gene_model_updated) were downloaded from TAIR (https://www.arabidopsis.org, accessed 25 November 2019), and the counts were quantified with Kallisto v.0.44.0 (ref. 91). The Kallisto gene counts were uploaded to Degust92 for analysis. After identification of differentially expressed genes, the lists were filtered to include only transcripts with log(fold change) > 2 and P ≤ 0.01 (Voom/Limma method46). Analysis of RNA-seq data used R v.3.6.1. Gene names, descriptions and GO-term enrichment analysis were performed using the Thalemine tool of the Bio-Analytic Resource for Plant Biology (http://bar.utoronto.ca). We determined whether statistically significant overlaps existed between sets of transcripts by using a hypergeometric test, which considers whether the overlap between two sets of genes is significantly different from the size of an overlap arising from two randomly drawn sets of genes. This involves calculation of the representation factor, which is the actual number of genes in the intersection, divided by the expected number of genes in the intersection; thus a value >1 indicates a greater number of genes than expected, and a value <1 indicates fewer genes than expected. The probability of this intersection occurring was calculated using a normal approximation of the exact hypergeometric probability93,94.
Measurement of PSII photosynthetic efficiency and freezing survival
For measurement of PSII photosynthetic efficiency in seedlings, chlorophyll fluorescence parameters of seedlings were measured using an IMAGING-PAM M-series MAXI chlorophyll fluorescence imaging system (Walz; with Walz ImagingWin software v.2.47). Measurements were taken from 14-day-old plants grown at 20 °C. Plants were then transferred to 4 °C and measurements taken after 3, 24 and 48 h, and 7 and 10 days of cold treatment. For measurements after freezing, plants that had been acclimated at 4 °C for 10 days were transferred to −8 °C for 6 h in darkness, after which plants were placed at 4 °C to thaw. Measurements were taken approximately 18 h after freezing. For all experiments, plants were dark adapted for 20 min before fluorescence measurement. Measurements were initiated by exposing dark-adapted plants to measuring light pulses (frequency 1 Hz, intensity 3) and then applying a saturating pulse. Chlorophyll fluorescence parameters of mature rosette leaves were measured using a MINI-PAM (Walz). Leaves were dark adapted for 20 min before applying a saturating light pulse. Fv/Fm was calculated as (Fm − F0)/Fm. For assessment of freezing survival 14-day-old plants were acclimated at 4 °C for 10 days (acclimation started at dawn), then placed at −8 °C for 6 h (Percival Intellus LT-36VL chamber, CLF Plant Climatics). Survival was assessed after a 7-day recovery period at 20 °C, with recovery occurring under growth conditions described previously.
Protein extraction and immunodetection
For protein abundance analysis, plants were grown as described for measurement of PSII photosynthetic efficiency and collected from the same experimental material. Samples were taken after 14 days of growth and after 10 days at 4 °C followed by 6 h at −8 °C with 18 h recovery. Protein extraction was conducted as described previously95. Briefly, powdered tissue was incubated in protein extraction buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM dithiothreitol, protease inhibitor cocktail (Sigma) 1:100, phosphatase inhibitor (Sigma) 1:200, 1 mM phenylmethylsulfonylfluoride, 0.5% IPEGAL CA-630 (Sigma), 1 mM EDTA, 1 mM Na2MoO4 × 2H2O, 1 mM NaF, 1.5 mM activated Na3VO4) at 4 °C for 1 h, and supernatant isolated by centrifugation. Total protein concentration was normalized to 0.5 mg ml−1 using a Bradford assay (Sigma reagent B6916 for protein range 0–1.4 mg ml−1; calibrated across protein concentration range 0–1.2 mg ml−1 using a BSA standard), and confirmed using Coomassie blue staining after SDS–PAGE (Extended Data Fig. 5c). Protein abundance was analysed using an automated semiquantitative capillary immunoassay (ProteinSimple Wes; running ProteinSimple Compass software v.4.1.0)96–98 with a Wes-Rabbit (2–40 kDa) Master kit according to the manufacturer’s instructions. Samples were loaded on Wes cartridges, running three technical replicates per sample, along with a polyclonal rabbit anti-PSII D2 antibody diluted 1:5,000 (AS06146), a rabbit anti-PSAC antibody diluted 1:1,000 (AS10939), and a rabbit anti-RbcL antibody diluted 1:5,000 (AS03037, Agrisera). Experiments were repeated twice.
Electrolyte leakage assays
Cellular damage after freezing was quantified by measuring the leakage of electrolytes from tissue that had been frozen, as a proxy for freezing damage, using a method similar to that used by Hemsley et al.99. Mature plants were grown as described previously for 5 weeks, after which freezing tolerance was analysed. For analysis of cold-acclimated plants, 5-week-old plants were transferred to a Sanyo MLR-352 under cycles of 12 h light/12 h darkness at 4 °C, 90 µmol m−2 s−1 of white light for 2 weeks, before the freezing treatments were applied. Five biological replicate samples per genotype per freezing temperature tested were prepared. Rosette leaves for analysis were removed and washed with deionized water to remove ionic material from the leaf surface. Three 8-mm leaf discs were obtained from each plant using a cork borer and placed in a glass test tube held on ice. After preparation of all tubes, a set of control tubes was held on ice, while the freezing treatment tubes were transferred to a sub-zero water bath with an immersion dip cooler. After 2 h at −2 °C, deionized water ice chips were added to tubes to initiate ice nucleation. The temperature of the water bath was reduced progressively to each test temperature, with sets of tubes moved to ice 30 min after each test temperature was reached. Samples were thawed gradually on ice overnight, and 5 ml of deionized water was added to each tube. Tubes were shaken for 3 h at room temperature. The electrical conductivity of the water in the tube was measured using a conductivity meter (Mettler Toledo Seven2Go). The tubes with leaves were then frozen to −80 °C for 1 h, to release the solutes that remained within the tissue. These tubes were shaken for 3 h at room temperature, and the electrical conductivity measured again. Electrolyte leakage was calculated as the proportion of electrolytes released at each freezing temperature, relative to the total electrolytes present in the samples. Each experiment was repeated independently three times.
Transient luciferase expression by particle bombardment
A SIG5::LUCIFERASE reporter construct (pGREENII0229 SIG5::LUCIFERASE24) was expressed transiently in Arabidopsis seedlings using methods similar to those used previously35,36. Briefly, 5 µg of pGREENII0229 SIG5::LUCIFERASE or pB7WG2.0-GFP (green fluorescent protein (GFP) positive control for transformation100) was combined with 25 μl of 1 nm gold particle suspension (Bio-Rad), 25 μl of 2.5 M CaCl2 and 10 μl of 0.1 M spermidine (Sigma), and incubated on ice for 30 min. DNA-coated gold particles were washed with 100% ethanol, resuspended in 100% ethanol and stored at −20 °C before particle bombardment. A Bio-Rad PDS-1000/He particle delivery system was used for bombardment of Arabidopsis plants, with a 1,350 p.s.i. rupture disk. Eleven-day-old seedlings, cultivated as for the RT–qPCR experiments, were positioned at the closest distance position to the gun muzzle (‘floor 2’) for bombardment. After bombardment, 5 mM luciferin (d-luciferin potassium salt; Melford Laboratories) was applied to plants using a small spray bottle 24 h before imaging commenced. A Photek HRPCS-intensified CCD photon counting system (Photek Ltd, with Photek Image32 software) was used to image luciferase bioluminescence, using 10-min integrations, with the camera set to photon counting mode. The first 90 s of data was discarded to eliminate interfering chlorophyll autofluorescence (delayed fluorescence). Plants were under constant light conditions between the two time points measured. The GFP transformation control reporter was imaged using a LeicaM205FA fluorescence microscope around 48 h after bombardment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Supplementary Data 1. Exact P values associated with statistical analysis for Figs. 1 and 2, and Extended Data Fig. 3a,b. Supplementary Data 2. Circadian-regulated transcripts identified by analysis with MetaCycle. Supplementary Data 3. Cold-responsive transcripts in Col-0 wild type and sig5-3 mutant at two time points, and transcripts that are differentially expressed between the wild type and sig5-3 mutant under these conditions. Supplementary Data 4. GO-term analysis of transcripts that are differentially expressed in response to cold for either the Col-0 or the sig5-3 mutant at two time points in the 24 h cycle. Supplementary Data 5. Transcripts that are cold-responsive in Col-0 but not sig5-3, or sig5-3 but not Col-0, in response to cold treatment at two different times in the 24 h cycle. Supplementary Data 6. GO-term analysis of sets of transcripts that are cold-responsive in Col-0 but not sig5-3, or sig5-3 but not Col-0, in response to cold treatment applied at two different times in the 24 h cycle. Supplementary Data 7. RT–qPCR primer sequences.
Acknowledgements
This research was funded by Biotechnology & Biological Sciences Research Council (UK) (BB/I005811/2, BB/J014400/1, BB/T003030/1, Institute Strategic Programme GEN BB/P013511/1 to A.N.D.; studentship 1518540 awarded to P.E.P.), Norwich Research Park Doctoral Training Partnership (BB/T008717/1, to R.D. and A.B.), The Leverhulme Trust (RPG-2018-216, to A.N.D.), the Bristol Centre for Agricultural Innovation (to A.N.D.), the Wolfson Foundation (to A.N.D.), NAGASE Science Technology Foundation (Japan), the Ministry of Education, Culture, Sports, Science and Technology (Japan) (Grants-in-Aid 17K07438, to K.T. and S.I.) and Tokyo Institute of Technology World Research Hub Initiative Program of Institute of Innovative Research (to K.T.). D.L.C.-R. is grateful to the Consejo Nacional de Ciencia y Tecnología (Mexico) for granting a PhD scholarship. We thank the University of Bristol Genomics Facility and Z. Song for experimental support; G. Jenkins for seed donation; S. Samwald, E. Tee, J. Sallmen and N. Holmes for help with protein analysis; C. Faulkner, T. Oyama and T. Muranaka for advice about transient expression; and M. Knight, Y. Yoshitake and M. Shimojima for technical advice. Figure 5 created with BioRender.com.
Extended data
Author contributions
D.L.C.-R. and A.N.D. conceived the study. D.L.C.-R., P.E.P., T.T., S.I., P.P., T.S.d.F., L.L.d.B.D., R.D., A.B., T.B.-G., B.F.M., K.A.F., H.K. and A.N.D. performed experimentation, data analysis and data interpretation. T.B. supported bioinformatic analysis. D.L.C.-R., P.E.P., P.P., L.L.d.B.D., K.T., S.I., K.A.F., H.K. and A.N.D. wrote the paper.
Peer review
Peer review information
Nature Plants thanks Takashi Shiina and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The RNA-seq data for this study are available in the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) with the project ID PRJEB45855. All other data supporting the findings of this study are included in the main figures, extended data figures and supplementary information.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Dora L. Cano-Ramirez, Paige E. Panter.
Extended data
is available for this paper at 10.1038/s41477-023-01377-1.
Supplementary information
The online version contains supplementary material available at 10.1038/s41477-023-01377-1.
References
- 1.Thomashow MF. PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- 2.Knight MR, Knight H. Low‐temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 2012;195:737–751. doi: 10.1111/j.1469-8137.2012.04239.x. [DOI] [PubMed] [Google Scholar]
- 3.Crosatti C, et al. Harden the chloroplast to protect the plant. Physiol. Plant. 2013;147:55–63. doi: 10.1111/j.1399-3054.2012.01689.x. [DOI] [PubMed] [Google Scholar]
- 4.Moellering ER, Muthan B, Benning C. Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science. 2010;330:226–228. doi: 10.1126/science.1191803. [DOI] [PubMed] [Google Scholar]
- 5.Steponkus PL, et al. Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 1998;95:14570–14575. doi: 10.1073/pnas.95.24.14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, et al. Plastid ribosomal protein S5 is involved in photosynthesis, plant development, and cold stress tolerance in Arabidopsis. J. Exp. Bot. 2016;67:2731–2744. doi: 10.1093/jxb/erw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramel F, et al. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl Acad. Sci. USA. 2012;109:5535–5540. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shumbe L, Bott R, Havaux M. Dihydroactinidiolide, a high light-induced β-carotene derivative that can regulate gene expression and photoacclimation in Arabidopsis. Mol. Plant. 2014;7:1248–1251. doi: 10.1093/mp/ssu028. [DOI] [PubMed] [Google Scholar]
- 9.Huner NPA, Öquist G, Sarhan F. Energy balance and acclimation to light and cold. Trends Plant Sci. 1998;3:224–230. doi: 10.1016/S1360-1385(98)01248-5. [DOI] [Google Scholar]
- 10.Gao Y, et al. Chloroplast translational regulation uncovers nonessential photosynthesis genes as key players in plant cold acclimation. Plant Cell. 2022;34:2056–2079. doi: 10.1093/plcell/koac056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Artus NN, et al. Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc. Natl Acad. Sci. USA. 1996;93:13404–13409. doi: 10.1073/pnas.93.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakayama K, et al. Arabidopsis Cor15am is a chloroplast stromal protein that has cryoprotective activity and forms oligomers. Plant Physiol. 2007;144:513–523. doi: 10.1104/pp.106.094581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navarro-Retamal C, et al. Folding and lipid composition determine membrane interaction of the disordered protein COR15A. Biophys. J. 2018;115:968–980. doi: 10.1016/j.bpj.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thalhammer A, et al. Disordered Cold Regulated15 proteins protect chloroplast membranes during freezing through binding and folding, but do not stabilize chloroplast enzymes in vivo. Plant Physiol. 2014;166:190–201. doi: 10.1104/pp.114.245399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fourrier N, et al. A role for SENSITIVE TO FREEZING2 in protecting chloroplasts against freeze-induced damage in Arabidopsis. Plant J. 2008;55:734–745. doi: 10.1111/j.1365-313X.2008.03549.x. [DOI] [PubMed] [Google Scholar]
- 16.Thorlby G, Fourrier N, Warren G. The SENSITIVE TO FREEZING2 gene, required for freezing tolerance in Arabidopsis thaliana, encodes a β-glucosidase. Plant Cell. 2004;16:2192–2203. doi: 10.1105/tpc.104.024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnes AC, Benning C, Roston RL. Chloroplast membrane remodeling during freezing stress is accompanied by cytoplasmic acidification activating SENSITIVE TO FREEZING2. Plant Physiol. 2016;171:2140–2149. doi: 10.1104/pp.16.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Somersalo S, Krause GH. Photoinhibition at chilling temperature: fluorescence characteristics of unhardened and cold-acclimated spinach leaves. Planta. 1989;177:409–416. doi: 10.1007/BF00403600. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi S, Murata N. How do environmental stresses accelerate photoinhibition? Trends Plant Sci. 2008;13:178–182. doi: 10.1016/j.tplants.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Kanamaru K, Tanaka K. Roles of chloroplast RNA polymerase sigma factors in chloroplast development and stress response of higher plants. Biosci. Biotechnol. Biochem. 2004;68:2215–2223. doi: 10.1271/bbb.68.2215. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka K, et al. Nuclear encoding of a chloroplast RNA polymerase sigma subunit in a red alga. Science. 1996;272:1932–1935. doi: 10.1126/science.272.5270.1932. [DOI] [PubMed] [Google Scholar]
- 22.Allison LA. The role of sigma factors in plastid transcription. Biochimie. 2000;82:537–548. doi: 10.1016/S0300-9084(00)00611-8. [DOI] [PubMed] [Google Scholar]
- 23.Nagashima A, et al. The multiple-stress responsive plastid sigma factor, SIG5, directs activation of the psbD blue light-responsive promoter (BLRP) in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:357–368. doi: 10.1093/pcp/pch050. [DOI] [PubMed] [Google Scholar]
- 24.Noordally ZB, et al. Circadian control of chloroplast transcription by a nuclear-encoded timing signal. Science. 2013;339:1316–1319. doi: 10.1126/science.1230397. [DOI] [PubMed] [Google Scholar]
- 25.Onda Y, et al. Light induction of Arabidopsis SIG1 and SIG5 transcripts in mature leaves: differential roles of cryptochrome 1 and cryptochrome 2 and dual function of SIG5 in the recognition of plastid promoters. Plant J. 2008;55:968–978. doi: 10.1111/j.1365-313X.2008.03567.x. [DOI] [PubMed] [Google Scholar]
- 26.Belbin FE, et al. Integration of light and circadian signals that regulate chloroplast transcription by a nuclear‐encoded sigma factor. New Phytol. 2017;213:727–738. doi: 10.1111/nph.14176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao P, et al. ATHB17 enhances stress tolerance by coordinating photosynthesis associated nuclear gene and ATSIG5 expression in response to abiotic stress. Sci. Rep. 2017;7:45492. doi: 10.1038/srep45492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kilian J, et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 29.Mellenthin M, et al. Expression of the Arabidopsis sigma factor SIG5 is photoreceptor and photosynthesis controlled. Plants. 2014;3:359–391. doi: 10.3390/plants3030359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Catalá R, Medina J, Salinas J. Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl Acad. Sci. USA. 2011;108:16475–16480. doi: 10.1073/pnas.1107161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ichikawa K, et al. Differential expression on a daily basis of plastid sigma factor genes from the moss Physcomitrella patens: regulatory interactions among PpSig5, the circadian clock, and blue light signaling mediated by cryptochromes. Plant Physiol. 2004;136:4285–4298. doi: 10.1104/pp.104.053033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andronis C, et al. The clock protein CCA1 and the bZIP transcription factor HY5 physically interact to regulate gene expression in Arabidopsis. Mol. Plant. 2008;1:58–67. doi: 10.1093/mp/ssm005. [DOI] [PubMed] [Google Scholar]
- 33.Toledo-Ortiz G, et al. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 2014;10:e1004416. doi: 10.1371/journal.pgen.1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu G, et al. MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics. 2016;32:3351–3353. doi: 10.1093/bioinformatics/btw405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanesaka Y, et al. Monitoring single-cell bioluminescence of Arabidopsis leaves to quantitatively evaluate the efficiency of a transiently introduced CRISPR/Cas9 system targeting the circadian clock gene ELF3. Plant Biotechnol. 2019;36:187–193. doi: 10.5511/plantbiotechnology.19.0531a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muranaka T, Oyama T. Heterogeneity of cellular circadian clocks in intact plants and its correction under light-dark cycles. Sci. Adv. 2016;2:e1600500. doi: 10.1126/sciadv.1600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fowler SG, Cook D, Thomashow MF. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005;137:961–968. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dodd AN, et al. Time of day modulates low-temperature Ca2+ signals in Arabidopsis. Plant J. 2006;48:962–973. doi: 10.1111/j.1365-313X.2006.02933.x. [DOI] [PubMed] [Google Scholar]
- 40.Hotta CT, et al. Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 2007;30:333–349. doi: 10.1111/j.1365-3040.2006.01627.x. [DOI] [PubMed] [Google Scholar]
- 41.Paajanen P, Dantas LLB, Dodd AN. Layers of crosstalk between circadian regulation and environmental signalling in plants. Curr. Biol. 2021;31:R399–R413. doi: 10.1016/j.cub.2021.03.046. [DOI] [PubMed] [Google Scholar]
- 42.Shindo C, et al. Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev. 2006;20:3079–3083. doi: 10.1101/gad.405306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le MQ, Pagter M, Hincha DK. Global changes in gene expression, assayed by microarray hybridization and quantitative RT–PCR, during acclimation of three Arabidopsis thaliana accessions to sub-zero temperatures after cold acclimation. Plant Mol. Biol. 2015;87:1–15. doi: 10.1007/s11103-014-0256-z. [DOI] [PubMed] [Google Scholar]
- 44.Woodson JD, et al. Sigma factor‐mediated plastid retrograde signals control nuclear gene expression. Plant J. 2012;73:1–13. doi: 10.1111/tpj.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oh S, et al. Transcriptome and phenotyping analyses support a role for chloroplast sigma factor 2 in red-light-dependent regulation of growth, stress, and photosynthesis. Plant Direct. 2018;2:pld3.43. doi: 10.1002/pld3.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Law CW, et al. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graham, C. A. et al. Genome-wide circadian gating of a cold temperature response in bread wheat. Preprint at bioRxiv10.1101/2022.11.29.518321 (2022). [DOI] [PMC free article] [PubMed]
- 48.Zhao C, et al. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 2016;171:2744–2759. doi: 10.1104/pp.16.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edwards KD, et al. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell. 2006;18:639–650. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Covington MF, Harmer SL. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007;5:e222. doi: 10.1371/journal.pbio.0050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rugnone ML, et al. LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc. Natl Acad. Sci. USA. 2013;110:12120–12125. doi: 10.1073/pnas.1302170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie Q, et al. LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell. 2014;26:2843–2857. doi: 10.1105/tpc.114.126573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan L, et al. BBX19 fine-tunes the circadian rhythm by interacting with PSEUDO-RESPONSE REGULATOR proteins to facilitate their repressive effect on morning-phased clock genes. Plant Cell. 2021;33:2602–2617. doi: 10.1093/plcell/koab133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakano T, et al. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ensminger I, Busch F, Huner NPA. Photostasis and cold acclimation: sensing low temperature through photosynthesis. Physiol. Plant. 2006;126:28–44. doi: 10.1111/j.1399-3054.2006.00627.x. [DOI] [Google Scholar]
- 56.Cano-Ramirez DL, et al. Plasma membrane fluidity: an environment thermal detector in plants. Cells. 2021;10:2778. doi: 10.3390/cells10102778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ehlert B, Hincha DK. Chlorophyll fluorescence imaging accurately quantifies freezing damage and cold acclimation responses in Arabidopsis leaves. Plant Methods. 2008;4:12. doi: 10.1186/1746-4811-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Job N, Datta S. PIF3/HY5 module regulates BBX11 to suppress protochlorophyllide levels in dark and promote photomorphogenesis in light. New Phytol. 2021;230:190–204. doi: 10.1111/nph.17149. [DOI] [PubMed] [Google Scholar]
- 59.Finster S, et al. Light-dependent, plastome-wide association of the plastid-encoded RNA polymerase with chloroplast DNA. Plant J. 2013;76:849–860. doi: 10.1111/tpj.12339. [DOI] [PubMed] [Google Scholar]
- 60.Yoo CY, et al. Phytochrome activates the plastid-encoded RNA polymerase for chloroplast biogenesis via nucleus-to-plastid signaling. Nat. Commun. 2019;10:2629. doi: 10.1038/s41467-019-10518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Díaz MG, et al. Redox regulation of PEP activity during seedling establishment in Arabidopsis thaliana. Nat. Commun. 2018;9:50. doi: 10.1038/s41467-017-02468-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Puthiyaveetil S, et al. Transcriptional control of photosynthesis genes: the evolutionarily conserved regulatory mechanism in plastid genome function. Genome Biol. Evol. 2010;2:888–896. doi: 10.1093/gbe/evq073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shimizu M, et al. Sigma factor phosphorylation in the photosynthetic control of photosystem stoichiometry. Proc. Natl Acad. Sci. USA. 2010;107:10760–10764. doi: 10.1073/pnas.0911692107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bartsch S, et al. Three thioredoxin targets in the inner envelope membrane of chloroplasts function in protein import and chlorophyll metabolism. Proc. Natl Acad. Sci. USA. 2008;105:4933–4938. doi: 10.1073/pnas.0800378105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nozoe M, et al. Selective activation of chloroplast psbD light-responsive promoter and psaA/B promoter in transplastomic tobacco plants overexpressing Arabidopsis sigma factor AtSIG5. Protein Pept. Lett. 2020;27:168–175. doi: 10.2174/0929866526666191014130605. [DOI] [PubMed] [Google Scholar]
- 66.Cano-Ramirez, D. L. et al. Circadian and environmental signal transduction in a natural population of Arabidopsis. Preprint at bioRxiv10.1101/2022.09.10.507414 (2022).
- 67.Hernández-Verdeja T, Vuorijoki L, Strand Å. Emerging from the darkness: interplay between light and plastid signaling during chloroplast biogenesis. Physiol. Plant. 2020;169:397–406. doi: 10.1111/ppl.13100. [DOI] [PubMed] [Google Scholar]
- 68.Puthiyaveetil S, et al. The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proc. Natl Acad. Sci. USA. 2008;105:10061–10066. doi: 10.1073/pnas.0803928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eberhard S, Drapier D, Wollman FA. Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J. 2002;31:149–160. doi: 10.1046/j.1365-313X.2002.01340.x. [DOI] [PubMed] [Google Scholar]
- 70.Kahlau S, Bock R. Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell. 2008;20:856–874. doi: 10.1105/tpc.107.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mullet JE, Klein RR. Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J. 1987;6:1571–1579. doi: 10.1002/j.1460-2075.1987.tb02402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hanaoka M, et al. Glutamyl-tRNA mediates a switch in RNA polymerase use during chloroplast biogenesis. EMBO Rep. 2005;6:545–550. doi: 10.1038/sj.embor.7400411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanamaru K, et al. An Arabidopsis sigma factor (SIG2)-dependent expression of plastid-encoded tRNAs in chloroplasts. Plant Cell Physiol. 2001;42:1034–1043. doi: 10.1093/pcp/pce155. [DOI] [PubMed] [Google Scholar]
- 74.Hanaoka M, et al. SIG1, a sigma factor for the chloroplast RNA polymerase, differently associates with multiple DNA regions in the chloroplast chromosomes in vivo. Int. J. Mol. Sci. 2012;13:12182–12194. doi: 10.3390/ijms131012182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martín G, et al. Circadian waves of transcriptional repression shape PIF-regulated photoperiod-responsive growth in Arabidopsis. Curr. Biol. 2018;28:311–318.e5. doi: 10.1016/j.cub.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 76.Nakamichi N, et al. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl Acad. Sci. USA. 2012;109:17123–17128. doi: 10.1073/pnas.1205156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nagel DH, et al. Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl Acad. Sci. USA. 2015;112:E4802–E4810. doi: 10.1073/pnas.1513609112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adams S, et al. Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol. 2018;220:893–907. doi: 10.1111/nph.15415. [DOI] [PubMed] [Google Scholar]
- 79.Kamioka M, et al. Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant Cell. 2016;28:696–711. doi: 10.1105/tpc.15.00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Méndez MB, et al. Novel roles of the master transcription factors Spo0A and σ(B) for survival and sporulation of Bacillus subtilis at low growth temperature. J. Bacteriol. 2004;186:989–1000. doi: 10.1128/JB.186.4.989-1000.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wiegeshoff F, et al. Sigma L is important for cold shock adaptation of Bacillus subtilis. J. Bacteriol. 2006;188:3130–3133. doi: 10.1128/JB.188.8.3130-3133.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Inoue-Sakamoto K, et al. Group 3 sigma factors in the marine cyanobacterium Synechoccous sp. strain PCC7002 are required for growth at low temperatures. J. Gen. Appl. Microbiol. 2007;53:89–104. doi: 10.2323/jgam.53.89. [DOI] [PubMed] [Google Scholar]
- 83.Pollari M, et al. Effects of deficiency and overdose of Group 2 sigma factors in triple inactivation strains of Synechocystis sp. strain PCC6803. J. Bacteriol. 2011;193:265–273. doi: 10.1128/JB.01045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Holm M, et al. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 2002;16:1247–1259. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Annunziata MG, et al. Response of Arabidopsis primary metabolism and circadian clock to low night temperature in a natural light environment. J. Exp. Bot. 2018;69:4881–4895. doi: 10.1093/jxb/ery276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hayes S, et al. UV-B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc. Natl Acad. Sci. USA. 2014;111:11894–11899. doi: 10.1073/pnas.1403052111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.R Core Team. R: A Language and Environment for Statistical Computing (2019); https://www.R-project.org/
- 89.Andrews, S. FASTQC: A Quality Control Tool For High Throughput Sequence Data (2010); https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 90.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bray NL, et al. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 92.Powell, D. R. Degust v3.2.0. Zenodo10.5281/zenodo.3258933 (2015).
- 93.Kim Stuart K, et al. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- 94.Press, W. H. et al. Numerical Recipes in C: The Art of Scientific Computing (Cambridge Univ. Press, 1992).
- 95.Cheval C, et al. Chitin perception in plasmodesmata characterizes submembrane immune-signaling specificity in plants. Proc. Natl Acad. Sci. USA. 2020;117:9621–9629. doi: 10.1073/pnas.1907799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gallagher KA, et al. c-di-GMP arms an anti-σ to control progression of multicellular differentiation in Streptomyces. Mol. Cell. 2020;77:586–599.e6. doi: 10.1016/j.molcel.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Feeney MA, et al. Translational control of the SigR-directed oxidative stress response in Streptomyces via IF3-mediated repression of a noncanonical GTC start codon. mBio. 2017;8:e00815-17. doi: 10.1128/mBio.00815-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lück, C., Haitjema, C. & Heger, C. in Proteomic Profiling: Methods and Protocols (ed. Posch, A.) 481–488 (Springer, 2021).
- 99.Hemsley PA, et al. The Arabidopsis mediator complex subunits MED16, MED14, and MED2 regulate Mediator and RNA Polymerase II recruitment to CBF-responsive cold-regulated genes. Plant Cell. 2014;26:465–484. doi: 10.1105/tpc.113.117796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tee E, Samwald S, Faulkner C. Quantification of cell-to-cell connectivity using particle bombardment. Methods Mol. Biol. 2022;2457:263–272. doi: 10.1007/978-1-0716-2132-5_17. [DOI] [PubMed] [Google Scholar]
- 101.Winter D, et al. An "Electronic Fluorescent Pictograph" browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data 1. Exact P values associated with statistical analysis for Figs. 1 and 2, and Extended Data Fig. 3a,b. Supplementary Data 2. Circadian-regulated transcripts identified by analysis with MetaCycle. Supplementary Data 3. Cold-responsive transcripts in Col-0 wild type and sig5-3 mutant at two time points, and transcripts that are differentially expressed between the wild type and sig5-3 mutant under these conditions. Supplementary Data 4. GO-term analysis of transcripts that are differentially expressed in response to cold for either the Col-0 or the sig5-3 mutant at two time points in the 24 h cycle. Supplementary Data 5. Transcripts that are cold-responsive in Col-0 but not sig5-3, or sig5-3 but not Col-0, in response to cold treatment at two different times in the 24 h cycle. Supplementary Data 6. GO-term analysis of sets of transcripts that are cold-responsive in Col-0 but not sig5-3, or sig5-3 but not Col-0, in response to cold treatment applied at two different times in the 24 h cycle. Supplementary Data 7. RT–qPCR primer sequences.
Data Availability Statement
The RNA-seq data for this study are available in the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) with the project ID PRJEB45855. All other data supporting the findings of this study are included in the main figures, extended data figures and supplementary information.