Abstract
Objective
Retinal hard exudates (HEs) result from lipoproteins leaking from capillaries into extracellular retinal space, and are related to decreased visual acuity in diabetic retinopathy (DR). This study aims to identify differential serum lipids and metabolites associated with HEs.
Materials and methods
A cross-sectional study was conducted Jul 2017 ∼ Mar 2021. We assessed the amount of HEs using standard ETDRS photographs for comparison. HEs severity was rated as “no or questionable”, “moderate” or “severe”. Serum samples were processed via high coverage pseudotargeted lipidomics analysis, and untargeted liquid chromatography coupled with time-of-flight mass spectrometry for metabolomics study, respectively. Weighted gene co-expression network analyses, partial least squares-discriminant analysis, and multi-receiver operating characteristic analysis were applied.
Results
A total of 167 patients were included. Discovery group: 116 eyes (116 patients). Validation group: 51 eyes (51 patients). 888 lipids were detected and divided into 18 modules (MEs), ME1 ∼ ME18. Lipids in ME1 significantly increased in patients with HEs in DR (NPDR and PDR combined), NPDR, and PDR, respectively. ME1 enriched to triglycerides (29%), ceramides (17%), and N-acylethanolamines (15%). A combined model of 20 lipids was the best to discriminate HEs, area under curve = 0.804, 95% confidence interval = 0.674–0.916. For metabolomics analysis, 19 metabolites and 13 pathways associated with HEs were identified. Taurine and hypotaurine metabolism, cysteine and methionine metabolism were closely related to HEs (P < 0.01).
Conclusions
The lipids and metabolites identified may serve as prediction biomarkers in the early stage of HEs in DR.
Keywords: Diabetic retinopathy, Hard exudate, Serum, Lipidomic, Metabolomics
1. Introduction
Diabetic retinopathy (DR) is a leading cause of blindness in working-age population [1]. The retinal lesions of DR include microaneurysms, hemorrhages, venous beading, hard exudates (HEs), soft exudates, fibrous proliferation, new vessels, etc. However, these lesions manifest differently even for the same stage of DR. Among the various lesions above, retinal HEs, described as yellowish-white deposits, are closely related to the onset of diabetic macular edema (DME), which mainly influences central vision, and usually lead to decreased visual acuity (VA) [2]. The Early Treatment Diabetic Retinopathy Study (ETDRS) reported that persistent foveal HEs can evolve into subretinal fibrosis with irreversible visual loss [3]. Photocoagulation for DME with HEs was associated with atrophic creep of retinal pigment epithelium [4]. In recent years, anti-VEGF drugs have revolutionized the therapy of DR and DME. Previous studies reported that monthly intravitreal anti-VEGF injections resulted in significant reduction in macular thickness and HEs [5]. However, some other studies proposed that baseline HEs in the fovea were usually associated with worse vision outcomes [6]. Repeated injections had uncertain benefit in VA and amount of HEs, in spite of macular edema improvement [7]. Therefore, it is critical to investigate the systemic mechanisms of formation and aggravation of HEs, reducing visual impairment caused by HEs-induced anatomic damage [2].
HEs result from lipoproteins leaking from retinal capillaries into the extracellular retinal space. Past studies focused on the relationships between circulating lipid levels and retinal HEs formation [8,9]. The evidences were sometimes conflicting. Moreover, it was unclear what lipid fractions were significantly associated with HEs [10]. Some clinical studies indicated that elevated serum lipid levels were correlated with an increased risk of HEs [11], suggesting that lipid-lowering drugs may decrease the risk of HEs formation and maintain vision [12]. Statin administration was proved to be associated with low incidence of DR [13]. On the contrary, other studies revealed no correlations between serum lipid levels and DME severity [14], indicating little effect of lipid-lowering therapy on the incidence of DR and DME [15,16]. Meanwhile, one study reported increased triglyceride levels were associated with a higher risk of HEs [17], but other studies supported that disturbed cholesterol metabolism led to accumulation of retinal cholesterol and formation of cholesterol crystals [18,19]. Due to the uncertain role of lipid subclasses in the formation of HEs, we consider performing in-depth multi-omics analysis to clarify systemic factors leading to accumulation of HEs.
Lipidomic and metabolomics studies are designed to characterize lipid and metabolic alteration in response to external or internal subtle perturbation, and to offer new insights into the pathophysiology of diseases [20]. Considering the mechanism of circulating lipoproteins leakage to form retinal HEs, we wonder whether differential serum lipids and metabolites could serve as prediction biomarkers in the early stage of HEs, and probably provide hints of treatment targets to reduce vision loss related to HEs. To the best of our knowledge, the relationship between serum lipids and metabolites and retinal HEs has not been previously described. Therefore, the aim of this study was to investigate the lipidomic and metabolic profiling in patients with different HEs severity to comprehensively elucidate serum biomarkers associated with retinal HEs in patients with DR.
2. Materials and methods
2.1. Study design and patients
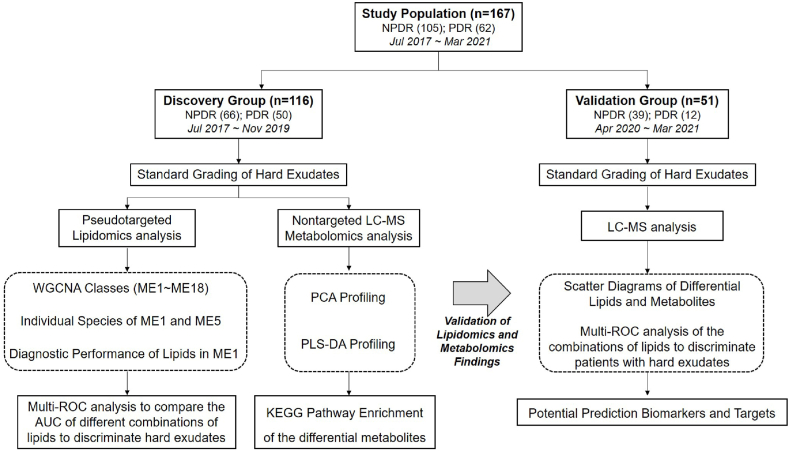
This cross-sectional study was approved by the ethics committee of Shanghai General Hospital, School of medicine, Shanghai Jiao Tong University, Shanghai, China (permit No. 2017KY194), in accordance with the Declaration of Helsinki. All participants completed a written consent form. The flow chart of this study was in Fig. 1.
Fig. 1.
Flow chart of the study.
The subjects were enrolled from the inpatient and the outpatient of Department of Ophthalmology and Department of Endocrinology, Shanghai General Hospital from Jul 2017 to Mar 2021. The population selection of discovery set and validation set was based on the time of enrollment. Discovery group was recruited from Jul 2017 to Nov 2019, and validation group consisted of subjects enrolled from Apr 2020 to Mar 2021. Inclusion criteria: (i) Age ≥18 years; (ii) Han Chinese ethnicity; (iii) Diagnosed with diabetes mellitus (diagnosis criteria: American Diabetes Association, 1997) and any stage of DR (diagnostic criteria: American Academy of Ophthalmology 2001 Annual Meeting). Exclusion criteria: (i) Other clinical histories that caused accumulation of HEs, such as age-related macular degeneration, uveitis, retinal venous occlusion, etc.; (ii) Significant cataracts, which influenced evaluation of HEs severity; (iii) Diabetic complications other than DR, such as severe renal failure; (iv) Pregnancy or breast-feeding.
2.2. Examination procedures
All the patients underwent comprehensive ophthalmologic examinations. The grading of DR classification was determined by dilated retinal examination using ophthalmoscope by a senior retinal specialist (Xun Xu). The categories of DR included mild non-proliferative retinopathy (NPDR), moderate NPDR, severe NPDR and proliferative retinopathy (PDR). The stage of DR for one patient was determined by the severer eye. Data on all participants' gender, age, type of diabetes, duration of diabetes, history of hypertension, duration of hypertension, history of dyslipidemia, history of cardiovascular diseases, concomitant medications were recorded. The values of patients’ haemoglobin A1C (HbA1C), total cholesterol, triglyceride, creatinine, urea and uric acid were also collected.
2.3. Grading of the HEs severity in color fundus photograph
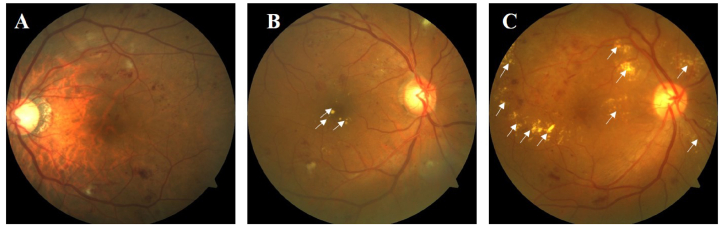
A 35-degree color fundus photograph centered on the macular was taken. We judged the presence and the amount of HEs of both eyes by using standard ETDRS photographs 3, 5, and 4 for comparison [12]. Grading criteria: grade 0, no HEs; grade 1, questionable HEs; grade 2, HEs < standard photograph 3; grade 3, HEs ≥ standard photograph 3 but < standard photograph 5; grade 4, HEs ≥ standard photograph 5 but < standard photograph 4; grade 5; HEs ≥ standard photograph 4; grade 8, cannot grade [12]. When the DR classification was the same in both eyes, we selected the eye with severer HEs as the final HEs grade for this subject. If the DR classification was different in both eyes, the HEs grade depended on that of the eye with severer DR. In order to simplify the grouping for further analysis, HEs severity was merged into 3 categories: no or questionable HEs (grade 0 and grade 1); moderate HEs (grade 2 and grade 3); severe HEs (grade 4 and grade 5). The standard ETDRS photographs for comparison and illustration of HEs severity were shown in Fig. 2. The judgement was initially done by two experienced graders (Shen Y., Wang H.) of Shanghai Jiao Tong University Reading Center independently, and then confirmed by a senior specialist (Liu K.). In case of discrepancy, a final agreement was reached by all the graders.
Fig. 2.
Illustration of standard grading of retinal HEs. The presence and the amount of HEs were judged by using standard ETDRS photographs 3, 5, and 4 for comparison [12]. The participants were divided into three different categories, i.e. no or questionable HEs (A), moderate HEs (B), and severe HEs (C), respectively. White arrows indicated the location of retinal HEs, manifesting as yellowish-white deposits. Abbreviations: ETDRS, Early Treatment Diabetic Retinopathy Study; HEs, hard exudates.
2.4. Collection and pretreatment of serum samples
10 mL whole blood was collected fasting in the morning and centrifuged within 30 min (1500 rpm, 10 min at 20 °C) from all the subjects. 1.5 mL aliquots were transferred into cryovial tubes and stored at −80 °C immediately. Metabolites and lipids were extracted using liquid-liquid extraction method. 50 μL sample was extracted by 4-fold volume of cold chloroform: methanol (v/v = 2:1), vortexed for 30 s, then stood for 5 min in room temperature. The mixture was centrifuged at 13,000 g for 15 min and the upper phase (hydrophilic metabolites) and lower organic phase (hydrophobic metabolites) were collected separately and evaporated under vacuum at room temperature. Lipidomics samples were analyzed using a high coverage pseudotargeted lipidomics method [21]. Metabolomics samples were analyzed using an untargeted high-performance liquid chromatography coupled with time-of-flight mass spectrometry.
2.5. Liquid chromatography
We separated metabolites and lipids on an Ultimate 3000 UHPLC system (Thermo Scientific). For metabolites, we used a ZIC-pHILIC column to separate aqueous phase mixture at 30 °C. Mobile phase A and mobile phase B were prepared according to the routine. Flow rate set at 0.2 mL/min with linear gradient: 0 min, 90%B; 1.5 min, 90%B; 25 min, 40%B; 28 min, 90%B; 33 min, 90%B. We suspended the samples with 100 μL of acetonitrile: water (1:1, v/v). For lipids, we conducted chromatographic separation on a reversed phase EclipsePlus C18 RRHD column at 35 °C. Flow rate set at 0.25 mL/min with following conditions: 0 min, 32% B; 2 min, 32%B; 20 min, 99%B; 25 min, 99% B; 25.1 min, 32%B; 35 min, 32 B%. We suspended the mixture with 100 μL isopropanol: acetonitrile: H2O (2:1:1, v/v/v).
2.6. Mass spectrometry
Data were acquired in full scan mode under a fast negative/positive ion switching mode using Q-Exactive MS (Thermo Scientific). Acquisition setting for metabolomics data: +3.5/-4.0 kV; 300 °C; sheath gas, 40; auxiliary gas, 10; m/z range, 200–2000; data acquisition, profile mode, microscans, 10; maximum injection time, 200 m s; mass resolution, 70,000 FWHM at m/z 200. Acquisition setting for lipidomics data: +3.5/-4.0 kV; 250 °C; sheath gas, 25; auxiliary gas, 15; m/z range, 150–1000; data acquisition was the same as metabolomics data. In addition, we also performed data independent acquisition (DIA) measurements for the mixed quality control samples.
2.7. Raw data processing
Raw data of metabolomics were processed with MZmine v2.32 software according to the instruction. DIA Raw data were converted on Reifycs ABF converter (http://www.reifycs.com/AbfConverter/index.html), and MS-DIAL was employed for data processing after conversion. For lipid identification, public LipidBlast library [[22], [23], [24]] was used for lipid annotation. Lipid with an m/z - RT pair was aligned for an identical lipid. Output data table of time-aligned lipids, RT, m/z and peak area acquired, was subjected to further statistical analysis. The metabolomics and lipidomics raw data generated by UPLC-MS/MS were processed with the open-source software MZmine v2.32 to perform targeted feature detection (lipids feature list file were obtained from QC samples identified by MS-DIAL; metabolites feature list file were consisted of reference compounds in our self-build metabolomics library). Data extraction in MZmine was performed according to a reference [25].
2.8. Statistical analysis
All statistical analyses were performed with SPSS software version 22.0 (SPSS Inc., Chicago, IL, USA). Comparisons of the variables obeying Gaussian distribution were made using independent-samples t-test, and non-parametric test was used for the comparisons of non-normal distribution parameters. Qualitative data were analyzed by Chi-square test or Fisher's exact probability method. Bilateral test was used for all difference comparisons. Statistical significance was judged by the test level of (Alpha) 0.05, and a P value < 0.05 was considered statistically significant.
2.9. Pathway enrichment and receiver operating characteristic (ROC) analysis
For metabolic profiling analysis, we performed multivariate statistical methods by using SIMCA-P version 14.0 (Umetrics AB, Umea, Sweden). The P values across all metabolites within each comparison were adjusted to account for multiple testing by a false discovery rate method. Principal component analysis (PCA), and partial least squares discriminant analysis (PLS-DA) were performed to investigate the metabolic differences among patients with different HEs severity. Pathway enrichment analysis of metabolites were conducted using MetaboAnalyst5.0 (http://www.metaboanalyst.ca/). Pearson correlation analysis of changes of lipid modules with HEs severity was performed to determine the abnormal lipid metabolism associated with HEs. We applied multi-ROC analysis to compare the area under curve (AUC) of different combinations of serum lipids to discriminate patients with HEs. In order to verify the findings, we compared the levels of differential metabolites and lipids among patients with different HEs severity, and calculated AUC of combined model of lipids in the validation group.
3. Results
3.1. Demographic and clinical data of the study groups
In total, 167 eyes from 167 patients were enrolled. For the discovery group, 116 eyes of 116 patients were collected from Jul 2017 to Nov 2019. The validation group consisted of 51 eyes of 51 patients from Apr 2020 to Mar 2021. Patients’ clinical characteristics and HEs severity in both groups were described in Table 1. The detailed classification of DR in both groups were showed in Supplementary Table 1. HEs severity showed no significant difference between patients with NPDR and PDR in the discovery group (P = 0.926) nor in the validation group (P = 0.116). Weighted kappa for inter-observer consistency of the discovery set was 0.931 (P < 0.001).
Table 1.
Patients’ clinical characteristics and HEs severity in the discovery group and the validation group.
| Characteristics | Discovery set (n = 116) |
Validation set (n = 51) |
Discovery vs Validation |
||||
|---|---|---|---|---|---|---|---|
| HEs Absent | HEs Present | Absent vs Present P value | HEs Absent | HEs Present | Absent vs Present P value | P value | |
| No. (%) | 47 (41%) | 69 (59%) | – | 20 (39%) | 31 (61%) | – | 0.874 |
| Age, mean (SD), y | 58.21 (12.20) | 57.51 (10.61) | 0.741 | 53.15 (12.79) | 56.65 (9.58) | 0.629 | 0.248 |
| Sex (male/female) | 18/29 | 46/23 | 0.003 | 12/8 | 22/9 | 0.417 | 0.165 |
| BMI, kg/m2 | 24.67 (4.45) | 24.49 (3.60) | 0.654 | 25.25 (4.23) | 24.09 (3.01) | 0.550 | 0.783 |
| Diabetes duration, median (IQR), y | 10.00 (10.00) | 11.0 (6.50) | 0.872 | 10.31 (6.83) | 11.36 (7.16) | 0.608 | 0.273 |
| HbA1c, median (IQR), % | 8.258 (1.71) | 7.90 (2.90) | 0.898 | 8.60 (1.80) | 8.70 (2.70) | 0.451 | 0.102 |
| CAD, % | 17.0 | 15.9 | 0.877 | 25.0 | 6.5 | 0.144 | 0.663 |
| HTN, % | 55.3 | 53.6 | 0.455 | 40.0 | 41.9 | 0.891 | 0.096 |
| DLD, % | 12.8 | 17.4 | 0.392 | 35.0 | 12.9 | 0.127 | 0.421 |
| Total cholesterol, median (IQR), mmol/L | 4.25 (1.27) | 4.34 (1.72) | 0.993 | 4.35 (2.12) | 4.66 (2.23) | 0.075 | 0.140 |
| Triglyceride, median (IQR), mmol/L | 1.39 (1.07) | 1.36 (0.93) | 0.531 | 1.37 (0.97) | 1.29 (1.00) | 0.847 | 0.488 |
| Creatinine, median (IQR), μmol/L | 62.00 (33.50) | 59.40 (23.00) | 0.978 | 59.80 (25.30) | 68.20 (35.00) | 0.140 | 0.341 |
| Urea, median (IQR), mmol/L | 6.60 (2.90) | 6.20 (1.90) | 0.676 | 5.43 (2.57) | 6.37 (3.16) | 0.206 | 0.598 |
| Uric acid, mean (SD), mmol/L | 335.91 (103.13) | 331.47 (86.56) | 0.809 | 321.78 (68.50) | 339.75 (81.88) | 0.420 | 0.967 |
Abbreviations: SD, standard deviation; IQR, interquartile range; HEs, hard exudates; DR, diabetic retinopathy; NPDR, non-proliferative diabetic retinopathy; PDR, proliferative diabetic retinopathy; CAD, coronary artery disease; HTN, hypertension; DLD, dyslipidemia.
3.2. Lipidomics analysis
3.2.1. Lipid subclasses associated with HEs severity in DR
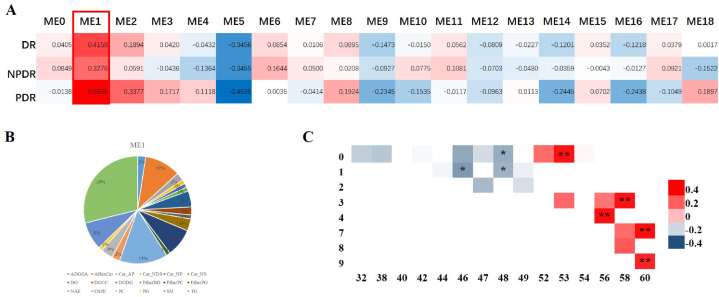
A total of 888 lipids were detected after construction of weighted gene coexpression network analysis (WGCNA), covering 18 common lipid modules (MEs), i.e., ME1 ∼ ME18. Differences of the 18 lipid MEs among no or questionable HEs, moderate HEs, and severe HEs groups were assessed quantitatively. As a result, serum levels of the lipids in ME1 significantly increased in patients with HEs in DR (NPDR and PDR combined), NPDR, and PDR, respectively (Fig. 3A). Lipids in ME1 mainly enriched to three subclasses, including triglycerides (TGs, 29%), ceramides (Cers, 17%), and N-acylethanolamines (NAEs, 15%) (Fig. 3B). The top 20 lipid compositions of ME1 were listed in Supplementary Table 2. On the contrary, lipids in ME5 significantly decreased in patients with HEs in DR (NPDR and PDR combined), NPDR, and PDR, respectively (see Supplementary Fig. 1. and Supplementary Table 3).
Fig. 3.
WGCNA analysis of lipidomics data. (A) Each ME consisted of a subclass of lipids. Correlations between MEs and HEs severity in DR, NPDR and PDR were calculated. Levels of lipids in ME1 (red frame) showed positive correlations with HEs severity in DR, NPDR and PDR. (B) Pie chart of lipid composition in ME1. (C) Heat map of correlation between TGs carbon chain length and unsaturation with HEs severity in ME1. * indicated a P value < 0.01, and ** indicated a P value < 0.0001 for Pearson correlation. Abbreviations: HEs, hard exudates; ME, module; TGs, triglycerides; WGCNA, weighted gene co-expression network analyses.
3.2.2. Analysis of carbon chain length and degree of unsaturation of TGs
We found that retinal HEs severity positively correlated with ultra-long chain unsaturated TGs, and negatively correlated with long-chain saturated TGs. TGs that significantly increased in HEs presence group included TG 53:0, TG 56:4, TG 58:3, TG 60:7 and TG 60:9, while the decreased TGs were TG 46:1, TG 48:0 and TG 48:1 (Fig. 3C). In the validation group, we have verified all the 5 significantly increased TGs in ME1 (see Supplementary Fig. 2).
3.2.3. Diagnostic performance of serum lipids to discriminate patients with HEs
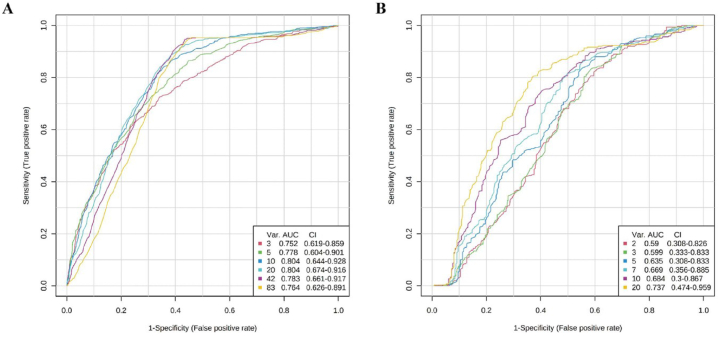
Regarding the diagnostic performance of serum lipids in ME1 to distinguish HEs present from HEs absent, we performed multi-ROC analysis. The combined model of 20 lipids was the best to discriminate patients with or without HEs. The AUC for this combined model was 0.804, 95% confidence interval (CI) = 0.674–0.916 (Fig. 4A). In the validation group, the AUC for this combined model of 20 lipids was 0.737, 95% CI = 0.474–0.959 (Fig. 4B).
Fig. 4.
Multi-ROC analysis of the combinations of 20 lipids in ME1 to discriminate patients with HEs. (A) ROC of the discovery group. The combined model of 20 lipids was the best to discriminate HEs. AUC = 0.804, 95% CI = 0.674–0.916. (B) ROC of the validation group. In the validation group, AUC for this combined model of 20 lipids was 0.737, 95% CI = 0.474–0.959. Abbreviations: AUC, area under curve; CI, confidence interval; HEs, hard exudates; ROC, receiver operating characteristic.
3.3. Metabolomics analysis
3.3.1. PCA profiling of serum metabolites in patients with different HEs severity
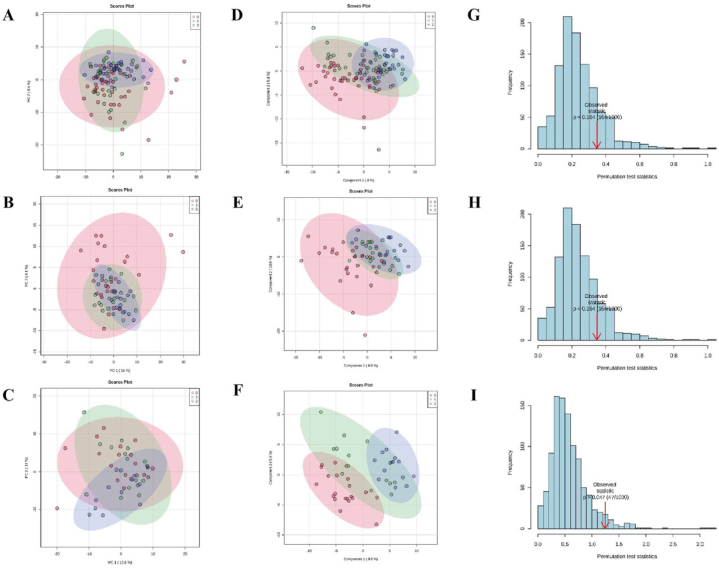
Serum metabolomic profiling classified by HEs severity was assessed by PCA models for patients with DR (NPDR and PDR combined) (Fig. 5A), NPDR (Fig. 5B), and PDR (Fig. 5C), respectively. The overall metabolic profiling showed no trend of separation among different HEs categories for three groups.
Fig. 5.
Multidimensional statistical analysis of serum metabolome of patients with different severity of HEs in DR, NPDR, and PDR. PCA score plot of serum metabolome of patients with different severity of HEs in DR (A), NPDR (B), and PDR (C), respectively. PCA results showed that the overall metabolic profiling showed no trend of separation among different HEs categories. PLS-DA score plot of serum metabolome of patients with different severity of HEs in DR (D), NPDR (E), and PDR (F), respectively. We observed clear separations among different HEs categories. Permutation test of PLS-DA models in DR (G), NPDR (H), and PDR (I), respectively. Models of DR and NPDR showed trend of over-fitting. For the model of PDR, there was no over-fitting. Abbreviations: DR, diabetic retinopathy; HEs, hard exudates; NPDR, non-proliferative retinopathy; PCA, principal component analysis; PDR, proliferative retinopathy; PLS-DA, partial least squares-discriminant analysis.
3.3.2. PLS-DA profiling of serum metabolites in patients with different HEs severity
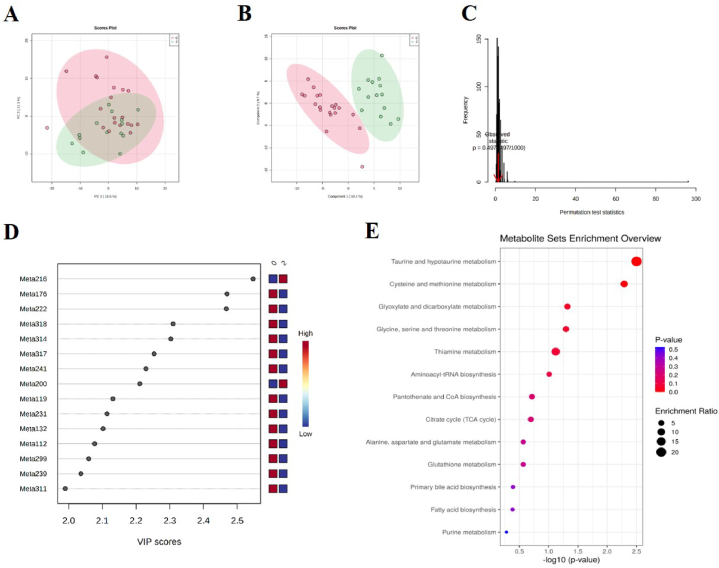
In supervised PLS-DA model, we observed clear separations among patients with different HE categories in DR (NPDR and PDR combined) (Fig. 5D), NPDR (Fig. 5E), and PDR (Fig. 5F), respectively. However, the permutation test of DR (Fig. 5G) and NPDR (Fig. 5H) models both showed trend of over-fitting. For the model of PDR, there was no over-fitting (Fig. 5I). Therefore, we further performed analysis of metabolites between HEs present and HEs absent in population of PDR. PCA model also demonstrated no separation (Fig. 6A). But PLS-DA model revealed that serum metabolome of HEs present vs. HEs absent clearly separated (Fig. 6B), with no over-fitting (Fig. 6C). As a result, 19 metabolites were identified and selected by the parameters of VIP >1, t-test P < 0.05, and fold change >1.2 or <0.83 (Table 2 and Fig. 6D). In the validation group, 6 of the 19 metabolites were verified (see Supplementary Fig. 3).
Fig. 6.
Metabolomics analysis between HEs present and absent in patients with PDR. (A) PCA score plot showed no trend of separation. (B) PLS-DA score plot indicated clear separation. (C) Permutation test of PLS-DA model. There was no over-fitting. (D) Top 15 metabolites ranked by VIP values of PLS-DA model. (E) KEGG pathway enrichment analysis of 19 differential metabolites. Abbreviations: DR, diabetic retinopathy; HEs, hard exudates; KEGG, Kyoto Encyclopedia of Genes and Genomes; NPDR, non-proliferative retinopathy; PCA, principal component analysis; PDR, proliferative retinopathy; PLS-DA, partial least squares-discriminant analysis.
Table 2.
The 19 significantly disturbed metabolites of patients with HEs in PDR.
| No. | Metabolites | Pubchem ID | VIP | Fold Change | P. ajusted |
|---|---|---|---|---|---|
| 1 | Methionine | 129,734,279 | 2.4699 | 0.14361 | 0.004331 |
| 2 | Gamma Hydroxybutyric Acid | 3,037,032 | 2.468 | 0.66434 | 0.004331 |
| 3 | Salicylic acid | 338 | 2.5472 | 2.2361 | 0.004331 |
| 4 | Citric acid | 311 | 2.3101 | 0.31817 | 0.011288 |
| 5 | Tartaric acid | 444,305 | 2.3028 | 0.42647 | 0.011288 |
| 6 | Oxalic acid | 971 | 2.2534 | 0.74042 | 0.014027 |
| 7 | S-Adenosylhomocysteine | 439,155 | 2.2293 | 0.7698 | 0.014522 |
| 8 | Ethylmalonic acid | 11,756 | 2.2107 | 1.4478 | 0.014651 |
| 9 | Ursodeoxycholic acid | 31,401 | 2.1311 | 0.72625 | 0.023454 |
| 10 | Diethanolamine | 8113 | 2.1139 | 0.59978 | 0.02356 |
| 11 | Deoxycholic acid | 222,528 | 2.102 | 0.71817 | 0.02356 |
| 12 | Deoxycholic acid | 222,528 | 2.0769 | 0.68125 | 0.025679 |
| 13 | Malonic acid | 867 | 2.0595 | 0.78394 | 0.026667 |
| 14 | Guanosine 3′,5′-Monophosphate | 135,398,570 | 2.0361 | 0.33766 | 0.028951 |
| 15 | Malic acid | 525 | 1.9898 | 0.51775 | 0.036506 |
| 16 | Taurine | 1123 | 1.9269 | 0.61653 | 0.044985 |
| 17 | L-3-O-Methyl-DOPA | 9307 | 1.9291 | 0.74244 | 0.044985 |
| 18 | l-Cysteine | 5862 | 1.9283 | 1.4889 | 0.044985 |
| 19 | Hydroxypyruvic acid | 964 | 1.9031 | 0.7974 | 0.049175 |
Abbreviations: HEs, hard exudates; PDR, proliferative diabetic retinopathy.
3.3.3. Enriched metabolic pathways relevant to HEs in patients with PDR
We identified 13 significantly enriched KEGG pathways relevant to the 19 differential metabolites associated with HEs in PDR (Fig. 6E). The abnormal taurine and hypotaurine metabolism, cysteine and methionine metabolism were highlighted as the two most important pathways related to the presence of HEs (P < 0.01).
4. Discussion
In recent years, the development of omics studies has helped people to deepen the understanding towards mechanism of DR [20,26]. Researchers have focused on the general metabolites and lipids differences of the individuals with DR compared to diabetic patients without retinopathy. Nevertheless, the fundus manifestations among DR patients are quite heterogeneous. It is meaningful to explore sub-classification of DR based on omics analysis. To our knowledge, this is the first study to investigate underlying mechanism of HEs in DR via omics strategy.
The judgement of HEs severity was based on comparison with standard ETDRS photographs and independent grading, which is still a classical method to evaluate the severity of DR lesions. Therefore, we believed the grouping of HEs was relatively objective. For lipidomics analysis, WGCNA was applied to provide high-dimensional data analysis to realize abundant lipids coverage [27]. A detailed component analysis of 888 lipids showed that serum levels of TGs, Cers, and NAEs significantly increased in patients with HEs. Among them, TGs covered a large proportion (29%). Therefore, we further performed detailed analysis of carbon chain length and degree of unsaturation of TGs. Interestingly, HEs severity correlated positively with ultra-long chain unsaturated TGs.
The role of long-chain polyunsaturated fatty acid (PUFA) in DR is controversial. Previous clinical studies indicated that PUFAs provided benefit relative to lipid profile in diabetic patients [28]. However, Hu et al. reported that 19,20-dihydroxydocosapentaenoic acid augmented in diabetic retinas and vitreous. The accumulation of diol altered the localization of cholesterol-binding proteins, and influenced the tight junction protein such as cadherin [29]. We guessed the abnormal accumulation of ultra-long chain unsaturated TGs could affect the tight junction between pericyte and vascular endothelial cell, and then destroy blood-retinal barrier, resulting in leakage of lipoproteins to form retinal HEs. Cers ranked the second of the differential lipid subclasses (17%). They are involved in a variety of cellular processes by promoting alterations of cell membrane properties and triggering signaling events [30]. Cers interplayed with cholesterol in lipid raft membrane mesostructured [31]. Wilmott et al. reported that Cers were increased in the vitreous samples of patients with T2DM [32]. Dysfunction of Cers metabolism in diabetic retina was observed in the fundamental experiments in-vitro and in vivo [33]. Here, we documented serum Cers significantly increased in DR patients with HEs. However, whether the interaction between Cers and cholesterol changes permeability of endothelial cell membrane of retinal microvascular still needs further investigation. Another lipid subclass identified, NAEs (15%), are long-chain fatty acids, including palmitoylethanolamide, oleoylethanolamide and anandamide [34]. NAEs participated in the biological processes such as inflammation, neuroprotection, acute stress, etc [35]. Elevated NAE levels correlated with triglyceride levels on some occasions [34]. But previous studies rarely addressed the relationships between circulating NAEs and DR, thus, the exact mechanism needs further exploration. Considering the role of serum lipids in HEs formation, we identified a combined model of 20 lipids with good efficiency in distinguishing HEs, which could be predictive biomarkers of this sight-threatening lesion.
For metabolomics analysis, we reported two most important pathways related to the presence of HEs, taurine and hypotaurine metabolism, and cysteine and methionine metabolism. Patients with HEs demonstrated lower levels of serum methionine and taurine. Methionine reduced hypertriglyceridemia by means of suppression of fatty acid synthesis and also decreased hypercholesterolemia [36]. Moreover, food supplement with l-methionine suppressed the hepatoma-induced increases in serum levels of triglyceride and total cholesterol. The lipoprotein lipase activity was enhanced after dietary supplement of methionine [37]. Taurine is an important nutrient for lipid metabolism because it conjugated bile acids [38]. Taurine decreased the level of total cholesterol and triglyceride in the blood [39]. We believed that these two metabolic pathways of sulfur amino acids are closely related to lipid metabolism, influencing formation of retinal HEs.
In order to verify the main findings of the discovery set, we further included the validation group. The proportion of PDR patients in the discovery set was higher than that of the validation set (43% vs. 24%, P = 0.016). Since the omics analysis was conducted in NPDR and PDR patients separately, we believed that this bias did not influence our results. Moreover, no significant differences were found in demographic characteristics between these two sets. Thus, the omics analysis results of these two groups were comparable. In the validation process, all the 5 elevated TGs in ME1 and 6 of the 19 differential metabolites were confirmed. AUC of multi-ROC model was relatively lower in the validation population, probably due to the small sample size.
This study has important clinical implications. Firstly, we provide a new avenue in clarifying the alterations of serum metabolites and lipids associated with HEs in patients with DR using omics strategy. This offers insights for further metabolic regulation to prevent the formation of HEs as well as treatment targets to reduce vision loss related to HEs. Secondly, previous studies have addressed the gross lipid fractions associated with HEs, but the individual lipid subclasses are difficult to assess due to the complex and cross-connected nature of lipid metabolism. We employed WGCNA to divide the co-expression network of biological processes into several highly correlated modules to identify functional lipid subclasses mostly involved in the development of HEs. This method can help uncover novel targets that have not been studied before.
There are also some limitations to this study. Firstly, lack of validation in the animal model. There is no reliable animal model of HEs in DR so far, so we included validation population to verify the preliminary findings of the discovery set. Secondly, small sample size of the validation population. Although all the 5 elevated TGs has been proved, only a part of the differential metabolites were verified. A clinical study with larger sample size is needed to confirm our findings. Furthermore, relevant in vivo and in vitro experiments will help to elucidate the mechanism of HEs, and further studies are need to deepen the understanding of metabolic regulation to prevent the formation of HEs.
5. Conclusions
In conclusion, we performed omic analysis in DR patients with different severity of retinal HEs to delineate the subtle changes of serum lipids and metabolites. The increased lipid subclasses related to HEs included TGs, Cers and NAEs. For metabolites, patients with HEs demonstrated lower level of serum methionine and taurine. Moreover, a combined model of 20 serum lipids was established to discriminate HEs. These lipids and metabolites might serve as biomarkers for the prediction and precise diagnosis via blood test in the early stage of HEs, and they could be potential treatment targets to reduce vision loss related to HEs in DR. Further study is needed to confirm the results of this study and to explore the mechanism of HEs occurrence.
Author contribution statement
Yinchen Shen; Kun Liu: Conceived and designed the experiments; Performed the experiments; Wrote the paper.
Hanying Wang: Analyzed and interpreted the data; Wrote the paper.
Junwei Fang: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Xun Xu: Contributed reagents, materials, analysis tools or data.
Data availability statement
Data will be made available on request.
Declaration of interest's statement
The authors declare no competing interests.
Additional information
Supplementary content related to this article has been published online at [URL].
CRediT author statement
Contributors: Conceptualization and methodology: YS, JF and KL; funding acquisition: YS, JF, XX and KL; software and formal analysis: YS, HW, JF; investigation, data curation, original draft: YS, HW and XX; review & editing: JF and KL.
Ethics approval
This study was approved by the ethics committee of Shanghai General Hospital, School of medicine, Shanghai Jiao Tong University, Shanghai, China (permit No. 2017KY194), in accordance with the Declaration of Helsinki. All participants completed a written consent form.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Supported by National Natural Science Foundation of China (No. 82171071, 81870667, 81800831, 82101141), National Key R&D Program of China (No. 2016YFC0904800, 2019YFC0840607), Program of Shanghai Academic Research Leader (No. 21XD1402700), Bethune Lumitin Young and Middle-aged Ophthalmic Research Fund (No. BJ-LM2021010J), Science and Technology Research Project of Songjiang District (No. 2020SJ307), and Skilled Lab Crew Program for Shanghai Higher Education Institute, Shanghai Education Commission. The sponsor or funding organization had no role in the design or conduct of this research.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e15123.
Contributor Information
Junwei Fang, Email: fagnjunwei@163.com.
Kun Liu, Email: drliukun@sjtu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Pie chart of lipid composition in ME5. Abbreviation: ME, module
Validation of the significantly increased TGs in ME1 of the discovery group. In the validation group, all the 5 TGs were increased with the severity of HEs. Note: 1=no or questionable HEs; 2=moderate HEs; 3=severe HEs. Abbreviations: HEs, hard exudates; ME, module; TGs, triglycerides
Validation of the significantly different metabolites of the discovery group. In the validation group, 6 of the 19 metabolites, methionine (A), cysteine (B), taurine (C), ethylmalonic acid (D), ursodeoxycholic acid (E), and malic acid (F), showed the same distribution tendency as the discovery group. Note: 1=no or questionable HEs; 2=moderate HEs; 3=severe HEs. Abbreviation: HEs, hard exudates
References
- 1.Sun H., Saeedi P., Karuranga S., et al. IDF Diabetes Atlas: global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022;183 doi: 10.1016/j.diabres.2021.109119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kameda Y., Kumakawa M., Endo N., et al. Association of systemic health and functional outcomes with changes in hard exudates associated with clinically significant macular oedema over the natural course of the disease. Br. J. Ophthalmol. 2010;94:725–729. doi: 10.1136/bjo.2009.158501. [DOI] [PubMed] [Google Scholar]
- 3.Fong D.S., Segal P.P., Myers F., et al. Subretinal fibrosis in diabetic macular edema. ETDRS report 23. Early treatment diabetic retinopathy study research group. Arch. Ophthalmol. 1997;115:873–877. doi: 10.1001/archopht.1997.01100160043006. [DOI] [PubMed] [Google Scholar]
- 4.Lövestam-Adrian M., Agardh E. Photocoagulation of diabetic macular oedema--complications and visual outcome. Acta Ophthalmol. Scand. 2000;78:667–671. doi: 10.1034/j.1600-0420.2000.078006667.x. [DOI] [PubMed] [Google Scholar]
- 5.Domalpally A., Ip M.S., Ehrlich J.S. Effects of intravitreal ranibizumab on retinal hard exudate in diabetic macular edema: findings from the RIDE and RISE phase III clinical trials. Ophthalmology. 2015;122:779–786. doi: 10.1016/j.ophtha.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 6.Srinivas S., Verma A., Nittala M., et al. Effect of intravitreal ranibizumab on intraretinal hard exudates in eyes with diabetic macular edema. Am. J. Ophthalmol. 2020;211:183–190. doi: 10.1016/j.ajo.2019.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Jeon S., Lee W. Effect of intravitreal bevacizumab on diabetic macular edema with hard exudates. Clin. Ophthalmol. 2014;8:1479–1486. doi: 10.2147/opth.S66405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Leiden H., Dekker J., Moll A., et al. Blood pressure, lipids, and obesity are associated with retinopathy: the hoorn study. Diabetes Care. 2002;25:1320–1325. doi: 10.2337/diacare.25.8.1320. [DOI] [PubMed] [Google Scholar]
- 9.Idiculla J., Nithyanandam S., Joseph M., et al. Serum lipids and diabetic retinopathy: a cross-sectional study. Indian J. Endocrinol. Metab. 2012;16:S492–S494. doi: 10.4103/2230-8210.104142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papavasileiou E., Davoudi S., Roohipoor R., et al. Association of serum lipid levels with retinal hard exudate area in African Americans with type 2 diabetes. Graefes Arch. Clin. Exp. Ophthalmol. 2017;255:509–517. doi: 10.1007/s00417-016-3493-9. [DOI] [PubMed] [Google Scholar]
- 11.Uçgun N., Yildirim Z., Kiliç N., et al. The importance of serum lipids in exudative diabetic macular edema in type 2 diabetic patients. Ann. N. Y. Acad. Sci. 2007;1100:213–217. doi: 10.1196/annals.1395.021. [DOI] [PubMed] [Google Scholar]
- 12.Gupta A., Gupta V., Thapar S., et al. Lipid-lowering drug atorvastatin as an adjunct in the management of diabetic macular edema. Am. J. Ophthalmol. 2004;137:675–682. doi: 10.1016/j.ajo.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 13.Liu J., Wu Y.P., Qi J.J., et al. Effect of statin therapy on diabetes retinopathy in people with type 2 diabetes mellitus: a meta-analysis. Clin. Appl. Thromb. Hemost. 2021;27 doi: 10.1177/10760296211040109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozer P., Unlu N., Demir M., et al. Serum lipid profile in diabetic macular edema. J. Diabet. Complicat. 2009;23:244–248. doi: 10.1016/j.jdiacomp.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Klein B., Myers C., Howard K., et al. Serum lipids and proliferative diabetic retinopathy and macular edema in persons with long-term type 1 diabetes mellitus: the Wisconsin epidemiologic study of diabetic retinopathy. JAMA Ophthalmol. 2015;133:503–510. doi: 10.1001/jamaophthalmol.2014.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das R., Kerr R., Chakravarthy U., et al. Dyslipidemia and diabetic macular edema: a systematic review and meta-analysis. Ophthalmology. 2015;122:1820–1827. doi: 10.1016/j.ophtha.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 17.Sasaki M., Kawasaki R., Noonan J., et al. Quantitative measurement of hard exudates in patients with diabetes and their associations with serum lipid levels. Invest. Ophthalmol. Vis. Sci. 2013;54:5544–5550. doi: 10.1167/iovs.13-11849. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X., Wang K., Zhu L., et al. Reverse cholesterol transport pathway and cholesterol efflux in diabetic retinopathy. J. Diabetes Res. 2021;2021 doi: 10.1155/2021/8746114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins A., Grant M., Busik J. Lipids, hyperreflective crystalline deposits and diabetic retinopathy: potential systemic and retinal-specific effect of lipid-lowering therapies. Diabetologia. 2022;65:587–603. doi: 10.1007/s00125-022-05655-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xuan Q., Zheng F., Yu D., et al. Rapid lipidomic profiling based on ultra-high performance liquid chromatography-mass spectrometry and its application in diabetic retinopathy. Anal. Bioanal. Chem. 2020;412:3585–3594. doi: 10.1007/s00216-020-02632-6. [DOI] [PubMed] [Google Scholar]
- 21.Xuan Q., Hu C., Yu D., et al. Development of a high coverage pseudotargeted lipidomics method based on ultra-high performance liquid chromatography-mass spectrometry. Anal. Chem. 2018;90:7608–7616. doi: 10.1021/acs.analchem.8b01331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kind T., Liu K., Lee D., et al. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods. 2013;10:755–758. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kind T., Okazaki Y., Saito K., et al. LipidBlast templates as flexible tools for creating new in-silico tandem mass spectral libraries. Anal. Chem. 2014;86:11024–11027. doi: 10.1021/ac502511a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cajka T., Fiehn O. LC-MS-Based lipidomics and automated identification of lipids using the LipidBlast in-silico MS/MS library. Methods Mol. Biol. 2017;1609:149–170. doi: 10.1007/978-1-4939-6996-8_14. [DOI] [PubMed] [Google Scholar]
- 25.Piovesana S., Montone C., Cavaliere C., et al. Sensitive untargeted identification of short hydrophilic peptides by high performance liquid chromatography on porous graphitic carbon coupled to high resolution mass spectrometry. J. Chromatogr. A. 2019;1590:73–79. doi: 10.1016/j.chroma.2018.12.066. [DOI] [PubMed] [Google Scholar]
- 26.Curovic V., Suvitaival T., Mattila I., et al. Circulating metabolites and lipids are associated to diabetic retinopathy in individuals with type 1 diabetes. Diabetes. 2020;69:2217–2226. doi: 10.2337/db20-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pei G., Chen L., Zhang W. WGCNA application to proteomic and metabolomic data analysis. Methods Mol. Biol. 2017;585:135–158. doi: 10.1016/bs.mie.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 28.Couch S., Crandell J., King I., et al. Associations between long chain polyunsaturated fatty acids and cardiovascular lipid risk factors in youth with type 1 diabetes: SEARCH Nutrition Ancillary Study. J. Diabet. Complicat. 2017;31:67–73. doi: 10.1016/j.jdiacomp.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu J., Dziumbla S., Lin J., et al. Inhibition of soluble epoxide hydrolase prevents diabetic retinopathy. Nature. 2017;552:248–252. doi: 10.1038/nature25013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castro B., Prieto M., Silva L. Ceramide: a simple sphingolipid with unique biophysical properties. Prog. Lipid Res. 2014;54:53–67. doi: 10.1016/j.plipres.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Bieberich E. Sphingolipids and lipid rafts: novel concepts and methods of analysis. Chem. Phys. Lipids. 2018;216:114–131. doi: 10.1016/j.chemphyslip.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilmott L., Grambergs R., Allegood J., et al. Analysis of sphingolipid composition in human vitreous from control and diabetic individuals. J. Diabet. Complicat. 2019;33:195–201. doi: 10.1016/j.jdiacomp.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Opreanu M., Tikhonenko M., Bozack S., et al. The unconventional role of acid sphingomyelinase in regulation of retinal microangiopathy in diabetic human and animal models. Diabetes. 2011;60:2370–2378. doi: 10.2337/db10-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fanelli F., Mezzullo M., Repaci A., et al. Profiling plasma N-Acylethanolamine levels and their ratios as a biomarker of obesity and dysmetabolism. Mol. Metabol. 2018;14:82–94. doi: 10.1016/j.molmet.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hussain Z., Uyama T., Tsuboi K., et al. Mammalian enzymes responsible for the biosynthesis of N-acylethanolamines. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862:1546–1561. doi: 10.1016/j.bbalip.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 36.Kawasaki M., Funabiki R., Yagasaki K. Effects of dietary methionine and cystine on lipid metabolism in hepatoma-bearing rats with hyperlipidemia. Lipids. 1998;33:905–911. doi: 10.1007/s11745-998-0287-6. [DOI] [PubMed] [Google Scholar]
- 37.Kawasaki M., Miura Y., Funabiki R., et al. Comparison of the effects on lipid metabolism of dietary methionine and cystine between hepatoma-bearing and normal rats. Biosci. Biotechnol. Biochem. 2010;74:158–167. doi: 10.1271/bbb.90673. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z., Ohata Y., Watanabe Y., et al. Taurine improves lipid metabolism and increases resistance to oxidative stress. J. Nutr. Sci. Vitaminol. 2020;66:347–356. doi: 10.3177/jnsv.66.347. [DOI] [PubMed] [Google Scholar]
- 39.Jun H., Choi M. Relationship between taurine intake and cardiometabolic risk markers in Korean elderly. Adv. Exp. Med. Biol. 2019;1155:301–311. doi: 10.1007/978-981-13-8023-5_29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pie chart of lipid composition in ME5. Abbreviation: ME, module
Validation of the significantly increased TGs in ME1 of the discovery group. In the validation group, all the 5 TGs were increased with the severity of HEs. Note: 1=no or questionable HEs; 2=moderate HEs; 3=severe HEs. Abbreviations: HEs, hard exudates; ME, module; TGs, triglycerides
Validation of the significantly different metabolites of the discovery group. In the validation group, 6 of the 19 metabolites, methionine (A), cysteine (B), taurine (C), ethylmalonic acid (D), ursodeoxycholic acid (E), and malic acid (F), showed the same distribution tendency as the discovery group. Note: 1=no or questionable HEs; 2=moderate HEs; 3=severe HEs. Abbreviation: HEs, hard exudates
Data Availability Statement
Data will be made available on request.