Abstract
Here, we present DNA aptamers capable of specific binding to glial tumor cells in vitro, ex vivo, and in vivo for visualization diagnostics of central nervous system tumors. We selected the aptamers binding specifically to the postoperative human glial primary tumors and not to the healthy brain cells and meningioma, using a modified process of systematic evolution of ligands by exponential enrichment to cells; sequenced and analyzed ssDNA pools using bioinformatic tools and identified the best aptamers by their binding abilities; determined three-dimensional structures of lead aptamers (Gli-55 and Gli-233) with small-angle X-ray scattering and molecular modeling; isolated and identified molecular target proteins of the aptamers by mass spectrometry; the potential binding sites of Gli-233 to the target protein and the role of post-translational modifications were verified by molecular dynamics simulations. The anti-glioma aptamers Gli-233 and Gli-55 were used to detect circulating tumor cells in liquid biopsies. These aptamers were used for in situ, ex vivo tissue staining, histopathological analyses, and fluorescence-guided tumor and PET/CT tumor visualization in mice with xenotransplanted human astrocytoma. The aptamers did not show in vivo toxicity in the preclinical animal study. This study demonstrates the potential applications of aptamers for precise diagnostics and fluorescence-guided surgery of brain tumors.
Keywords: MT: Oligonucleotides: Diagnostics and Biosensors, glial brain tumor, astrocytoma, tumor imaging, fluid biopsy, PET/CT, DNA aptamers, SELEX, oligonucleotide tertiary structure, molecular modeling
Graphical abstract
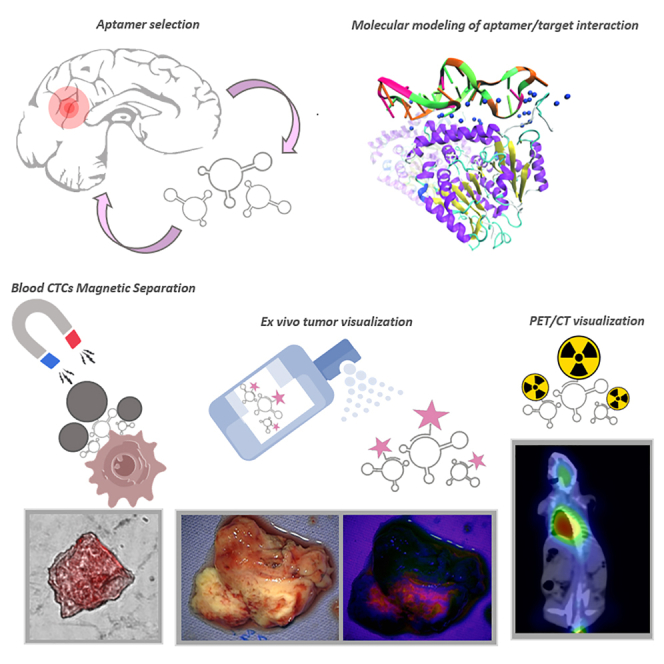
Kichkailo and colleagues report here that DNA aptamers with the restored 3D structures, identified and verified target proteins selected explicitly for the most aggressive human glial tumors can detect CTCs during the fluid biopsy analyses, can be used for fluorescence-guided ex vivo tumor tissue staining, and PET/CT tumor visualization.
Introduction
Glial tumors or gliomas are the most frequent and aggressive brain tumors in adults.1 The initial diagnostics of brain tumors is usually based on MRI, CT, or PET/CT with further justification after tumor resection or tissue biopsies with histopathological, immunohistochemical, and molecular analyses.2 Low specificity of the current imaging techniques does not allow one to reliably differentiate gliomas and even tumor reoccurrence lesions from the treatment-related tissue transformation. According to the most recent WHO classifications, adult-type diffuse gliomas are classified by cell origin to astrocytomas (grades I–IV), oligodendrogliomas, and wild-type glioblastomas (GBMs). GBMs are histologically classified as astrocytomas of the highest malignancy grade IV.3 Currently, the most reliable way to diagnose glioma and its subtypes are tissue biopsies, a highly invasive and complex procedure with a high risk.2
Glial tumors have an extensively heterogeneous histopathology3; therefore, there is a high risk of tissue sampling errors during biopsy and surgical resection, i.e., collecting non-representative tumor samples for the histological assessments.4,5 The risk of retrieving non-representative histological samples is a highly relevant limitation in glioma diagnostics and research. Atypia, mitotic activity, increased cellular density, microvascular proliferation, and necrosis are the main histopathological features for grading gliomas. Comprehensive molecular analyses and sequencing are costly and only available to some clinics.4 Reliable results depend on accurate sampling, which is difficult due to tumor heterogenicity—specifically in situ staining of viable glial tissues before histological and molecular analyses could alleviate sampling for diagnostics.
All brain malignancies shed their tumoral content into the cerebrospinal fluid and blood circulation.6,7 Liquid biopsies enable circulating tumor cell (CTC) detection benefits due to their low invasiveness and the ability to monitor the course of treatment response and disease progression by serial sampling as often as needed.8 The data on glioma CTCs are limited due to the complexity of their detection technologies and the need for ligand specificity. Fast, convenient, and particular detection methods are still the limitations of glioma CTC research.8
PET/CT, MRI, and intraoperative glial tumor visualization do not have sufficient diagnostic specificity. Visualization is essential for radical and safe tumor resection; it correlates well with patient prognosis.9 Despite substantial efforts to distinguish between neoplastic and healthy tissue at resection margins, it is still challenging to have complete surgical removal of glial tumor tissues due to their diffusely infiltrative nature. Therefore, it remains essential to develop glial tumor visualization methods for the accurate diagnostics and imaging of tumors.
Aptamers are a particular class of nucleic acid molecules that can exclusively bind to various targets with high affinity and selectivity.10 They can be raised simultaneously to multiple molecular targets in heterogeneous tumors. As a result of cyclical selection to numerous postoperative tumor tissues, it is possible to obtain a multipotent set of several aptamers that bind to different cancer-related proteins.11 Aptamers benefit from their ease of generation, low production cost, low batch-to-batch variability, reversible folding properties, and low immunogenicity.12 Using high specificity, facile synthesis, convenient modification, and high programmability, aptamers have been widely used as reliable recognition ligands in biosensing, bioimaging, and bioregulation.13,14 In addition, aptamers are convenient for constructing more complex dynamic DNA nanostructures such as tetrahedral frameworks, which, due to their structural programmability, enhance cellular endocytosis and tissue permeability,15 and can be used for various biomedical applications,16 such as in situ tumor imaging,17 or for the treatment of neurological diseases.18
Although aptamers to glial tumor cell cultures have been previously selected, no clinical applications have been reported yet for diagnosing and treating brain tumors.19,20,21 Here, we present the aptamers capable of specific binding to glial tumor cells in vitro, ex vivo, and in vivo for visualization diagnostics of central nervous system tumors.
Results
Aptamer selection and binding evaluation
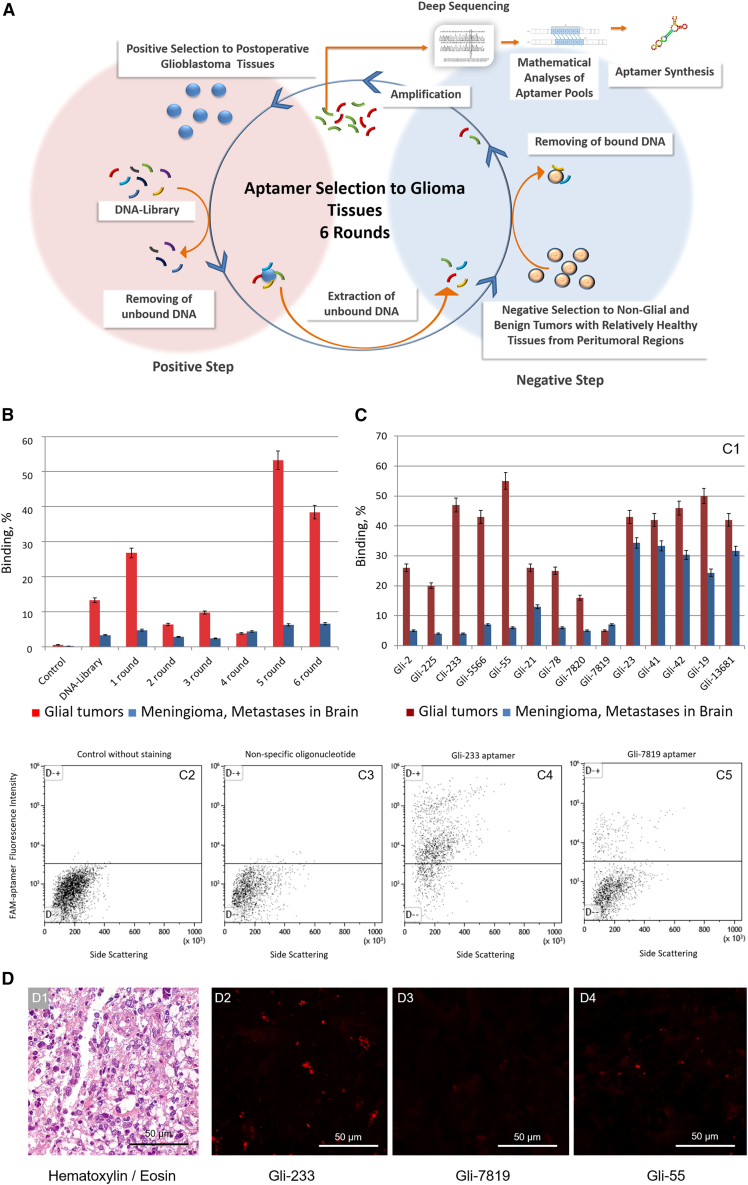
Glial tumor tissues of low, high, and highest grades were used for positive selection steps to select anti-glioma aptamers. Non-glial brain tumors, such as meningiomas, brain metastases originated from tumors of other organs, and relatively healthy brain tissues from peritumoral regions with blood clots, vessels, and capillaries were used for negative selection steps. A total of six selection rounds, including negative and positive steps and PCR amplification, were enough to obtain highly potent pools of aptamers (Figure 1A; Table S1).
Figure 1.
Development of DNA aptamers to postoperative gliomas
(A) The scheme of an aptamer selection procedure. (B) Flow cytometry analysis of binding properties of aptamer pools. (C) Flow cytometry analysis of binding properties of individual aptamers: comparison of different aptamers binding to glial and non-glial brain tumors (1); fluorescence intensity of glial tumor cells attached with the aptamers Gli-233 (4) and Gli-7819 (5) comparing with non-stained control (2) and nonspecific oligonucleotide (3). (D) Brain tumor tissue sections: stained with H&E dyes (1); stained with Cy5-labeled aptamers: Gli-233 (2); Gli-7819 (3), Gli-55 (4). Magnification, 40×. n = 3, Standard deviation, Scale bar, 50 µm
The degree of binding of the aptamer pools to cells derived from postoperative glial tumors, meningiomas, and brain metastases was assessed using flow cytometry. The aptamers from the fifth selection round showed the best affinity to glial tumor cells (Figure 1B) and therefore were sequenced using next-generation sequencing (NGS) technology. Exactly 20,469 DNA sequences were analyzed in silico and grouped into “families” according to their sequence similarity (Table S2). The nine most representative and highly abundant aptamer clones from each family and their truncated versions were synthesized chemically (Table S2). The affinity of the individual aptamers to glial tumor cells was tested using flow cytometry (Figure 1C) and laser scanning microscopy (Figure 1D). All aptamers showed good binding to glioma cells compared with the initial ssDNA aptamer library. Furthermore, the aptamers showed good selectivity to glial tumor cells from different patients compared with meningioma cells and brain metastases (Figure 1C). The lead aptamers Gli-233 and Gli-55 were specific to glioma cells in postoperative tumor tissue samples compared with normal glial cells and connective tissues (Figures 1C and 1D). Therefore, these aptamers were chosen for further identification of binding partners.
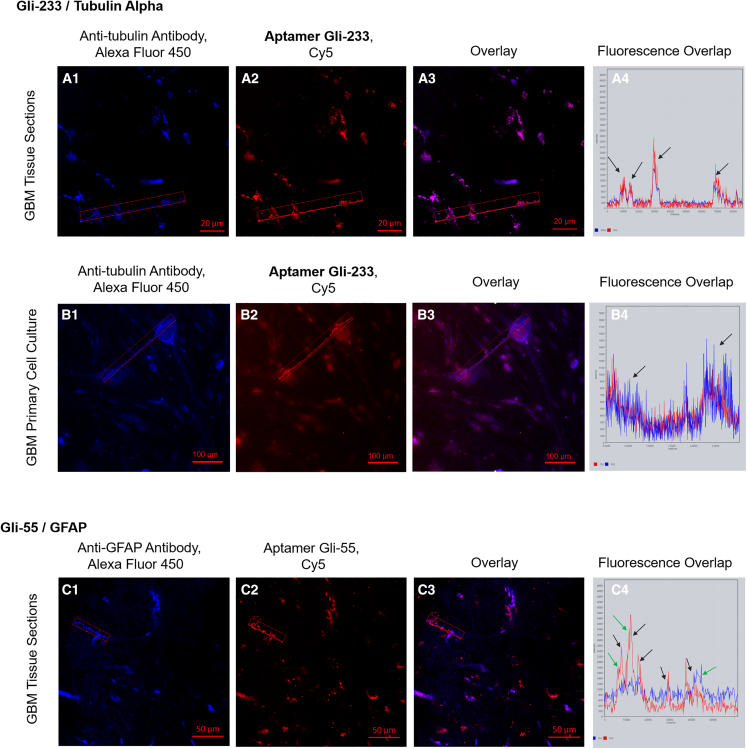
We used an aptamer-facilitated biomarker discovery (AptaBiD) procedure using mass spectrometry and co-localization with antibodies to find aptamer-associated proteins.22,23 Mass spectrometry analysis revealed potential protein targets such as glial fibrillary acidic protein (GFAP), tubulin alpha-1C chain (TUBA1C), and myelin basic protein for Gli-233, and GFAP and pancreatic lipase-related protein 3 for Gli-55 (Table S3). These three proteins were found post-translationally modified. GFAP has each methionine oxidized at positions 73 and 74. The GFAP identified for Gli-233 aptamer has an oxidized methionine at positions 73, 74, and 342, and phosphorylated serines at positions 247 and 248. TUBA1C, a target protein for aptamer Gli-233, has different side chains of polyglycines and polyglutamates. Up to 6 links in length were added to the C-terminal tail (CTT) of glutamic acid (E) residues in different ways. The two most reliable combinations of polypeptide adjunctions are (1) glycylation of E 433, biglutamylation of E 445, glutamylation of E 447, and 5-link polyglutamylation of E 448; and (2) triglycylation of E 433, 6-link polyglycylation of E 443, 4-link polyglycylations of E 447 and 448. It is known that polyglutamylation of tubulin temporarily occurs during cell division and is linked to neurodegeneration.24 Tubulin glycylation is found in axonemes of cilia and flagella.25 Primary cilia regulate cell-cycle and signaling transduction, a critical mediator of gliomas tumorigenesis and progression.26,27 This modification is crucial for cilia for glial tumor growth and colorectal cancer progression.28
To validate the association of the aptamers with the proteins, we performed co-staining of glial tumor tissue sections and primary glioma cell cultures with the aptamers and corresponding antibodies. Laser scanning microscopic images showed co-localization of Gli-233 with anti-tubulin antibodies in glial tumor tissue sections (Figure 2A) and primary glioma cell cultures (Figure 2B). Gli-55 was co-localized partially with anti-GFAP antibodies and simultaneously bound to other targets on tumor cells (Figure 2C). Anti-GFAP antibodies did not bind exclusively to tumor cells. Thus, the most probable protein target for the 33-nt-long Gli-233 aptamer was TUBA1C with several post-translational modifications. A tertiary structural molecular model of TUBA1C with the post-translational modifications is shown in Figure S1. Tubulin is not a common glioblastoma multiform biomarker; only one study demonstrated enhanced α-tubulin immunoreactivity in GBM and its nitration at Tyr 224 in grade IV GBM.29 However, tubulin is one of the prospective targets in glioma therapy.30 The potential target for the 60-nt-long Gli-55 aptamer is post-translationally modified GFAP and some other undefined targets on glioma cells. GFAP expression is not typical for all glial tumors. Usually, increased GBM malignancy is associated with a loss of GFAP expression but, at the same time, it does not contribute to tumor development or progression.31,32 Thus, we chose the Gli-233 and Gli-55 aptamers as the most promising candidates for further analysis.
Figure 2.
Binding verification of Gli-233 to tubulin and Gli-55 to GFAP
(A) Co-staining with Alexa Fluor 405-labeled anti-tubulin antibodies and Cy5-labeled Gli-233 of glioma tissue sections. (B) Co-staining with Alexa Fluor 405-labeled anti-tubulin antibodies and Cy5-labeled Gli-233 of primary GBM cell cultures. (C) Co-staining with Alexa Fluor 405-labeled anti-GFAP antibodies and Cy5-labeled Gli-55 of glial tumor tissue sections. Overlays (A3, B3, C3) and histograms of overlapping fluorescence intensities in the depicted regions (A4, B4, C4). The coinciding peaks are shown with black arrows; peaks that do not match are shown with green arrows. Magnification, 40×. Scale bars, 20 µm, 50 µm, 100 µm.
3D structure of the aptamers
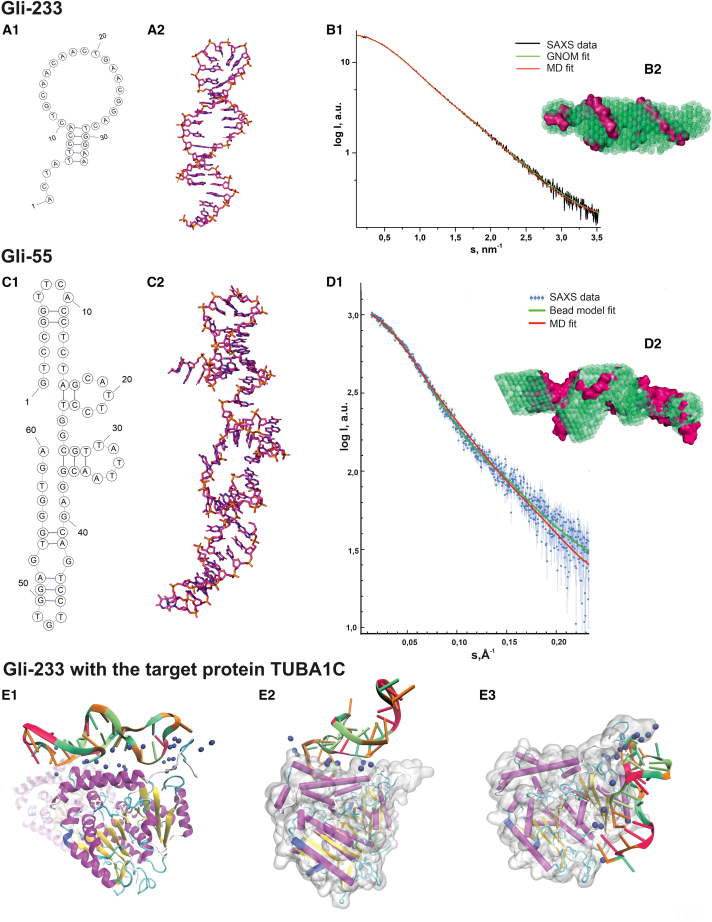
The primary sequence of Gli-233 (Table S2) was used to predict the secondary structure using the OligoAnalyzer web server.33 Folding simulations were performed under the same conditions as the SAXS (small-angle X-ray scattering) experiment. Four possible secondary structures were chosen for further studies. The predicted model of the aptamer is shown in Figure 3A1 and comprises three regions: a loop region (the central part of the sequence), a B-type duplex, and a single-stranded segment.
Figure 3.
Molecular design of a 3D structure of Gli-233 and Gli-55 aptamers and Gli-233 binding to a protein target
(A) Structures of the Gli-233 aptamer: secondary structure (1); 3D structure after molecular dynamics (MD) simulations (2). (B) Experimental SAXS data fitted by the theoretical SAXS curves calculated from the bead models based on the p(r) (green) and from the MD models (red) of the Gli-233 aptamer (1); an overlap of aptamer’s SAXS-derived structures (green beads) and their 3D models (pink volume) (2). (C) Structures of the Gli-55 aptamer: secondary (1); 3D structure (2). (D) Experimental and theoretical SAXS data of the Gli-55 aptamer. (E) MD simulations of Gli-233-protein interaction at three binding sites to the tubulin dimer (1–3). The ɑ-tubulin chain is gray, and Na+ and ns are shown as blue spheres.
The tertiary structure of Gli-233 was modeled with the Avogadro program and optimized using the fragment molecular orbital (FMO)-density functional-based tight binding (DFTB)/polarizable continuum model (PCM) method.34,35 After using molecular dynamics (MD) simulation, the structure was equilibrated in silico in a solution. Then it was allowed to relax into a conformation more likely characteristic of the aptamer it represents when in the solution. For obtained structures, we computed SAXS spectra using the WAXIS service and fit the results with the experimental data.36 The structure with the best-fit quality was selected (Figure 3A2). The Gli-233 aptamer has a long rigid duplex domain, which was conserved during MD simulation. The length of the bent structure was ∼60 Å, close to the smallest dimension of the tubulin heterodimer.
In revealing the three-dimensional (3D) structure of the Gli-233 aptamer, the main challenge was related to the construction of the loop region, as it had many possible positions. Thus, we applied SAXS and computer-aided molecular design to aptamer structure restoration in solution. The experimental scattering curve (Figure 3B1) was used to calculate the basic structural parameters of Gli-233, such as gyration radius (Rg = 1.91 ± 0.06 nm), maximal molecule dimension (Dmax = 6.83 nm), Porod volume (Vp = 13.4 nm3). The ab initio overall shape reconstruction of the aptamer Gli-233 was performed using the DAMMIF program.37 Twenty different models were averaged and filtered into the final model, shown in Figure 3B2 as green beads. The SAXS curve recorded from the Gli-233 in Dulbecco’s phosphate-buffered saline (DPBS) without Ca2+ and Mg2+ showed the independence of the aptamer structure from the presence of these ions in the solution. The atomic structure of Gli-233 obtained from MD simulations was compared with the DAMMIF bead model and showed a high correlation (Figure 3B1). The discrepancy (χ2) between the scattering curve from the molecular model and experimental SAXS data was equal to 1.041.
The same modeling procedure was applied to the Gli-55 aptamer. OligoAnalyzer predicted 16 possible secondary structures for Gli-55. Corresponding tertiary structures were modeled and subjected to MD simulations. Comparing theoretical models with SAXS results allowed us to establish the structure of the Gli-55 aptamer (Figures 3C1 and 3C2). The aptamer Gli-55 SEC-SAXS dataset yielded a skewed main elution peak (Figure S2), pointing to the potential presence of two significant components. Indeed, the EFAMIX decomposition produced two distinct components (Figure S3) associated with monomer and dimer constituents according to estimated excluded volumes and molecular masses of the components. Parameters extracted from the monomer SAXS profile (Figure 3D1, blue dots with error bars) were: the size of the molecule Dmax = 10 nm, Rg = 2.79 nm, molecular weight = 18.7 kDa in the credibility interval of 18.3–20.8 kDa. The bead model was constructed using a pair distribution function p(r) (Figure 3D1, red line). The obtained shape of the overall electron density of the Gli-55 molecule is shown in Figure 3D2 (green beads) and fitted with the MD simulation model (pink volume). Aptamer Gli-55 contains several GG and GGG stretches and may form a G-quadruplex. Circular dichroism spectroscopy was used to determine G-quadruplex formation. The results of the experiment showed two peaks at wavelengths of 220 and 280 nm (supplemental materials; Figure S4), which did not correspond to the formation of the G-quadruplex. Therefore, only linear conformations of Gli-55 aptamer were investigated. The model from MD simulation demonstrated correctly reconstructed spatial structure well matched with the experimental SAXS data (χ2 = 2.185), which can be seen in the graph mode (Figure 3D1, green line). The non-ideal coincidence of the SAXS curves may be explained by the complexity of SEC-SAXS data consisting of two components in solution or non-perfect MD simulation results.
The SAXS datasets for Gli-233 and Gli-55 have been deposited at the SASBDB38 (accession codes: SASDPZ8 and SASDRY7, respectively).
Interaction of Gli-233 with the target protein
The potential binding sites of Gli-233 to the target protein TUBA1C were identified by mass spectrometry experiments and then verified by MD simulations (Figures 3E1–E3). The model of the human tubulin heterodimer (ɑ1C with β2B isoforms) with post-translational modifications was used as a target for molecular docking. The ɑ1C unit of a dimer includes post-translational modifications of a CTT, common to glial tissue tumors. We proposed that the CTT region was responsible for the aptamer affinity to tubulin. The aptamer and the CTT region both carry a significant negative charge and should repulse each other. However, in simulations, we observed that aptamer was effectively bound to the CTT site of ɑ-tubulin via Na+ ions. These complexes showed no signs of dissociation for the whole simulation time. The sodium ions responsible for binding were also tightly bound to the interface.
The initial determination of binding sites of Gli-233 aptamer on the CTT region of ɑ1C unit was performed using the HDOCK molecular docking web server.39 The docking showed several different aptamer-protein complexes. The top-scoring binding positions were considered for aptamer-protein interactions and were selected for further refinement using MD simulations.
We identified three potential binding sites around the CTT region of the α chain of the tubulin dimer (Figures 3E1–E3). Some of these ions held their positions during MD simulation time. This effect could explain the role of post-translational modifications of tubulin in aptamer specificity. Several polyglutamylations and polyglycilations of CTT result in forming of a large and branched protein domain, which also carries a negative charge. This region could form Na+-mediated bonds between the aptamer and protein more effectively.
For the comparison, we used the following parameters: the number of hydrogen bonds between the aptamer and protein, the number of Na+ ions caged between the protein and aptamer, and the similarity (RMSD of heavy atom positions) of the aptamer structure in the complex with the protein in water. The average values for these quantities over the MD trajectories are presented in Table S4.
Over these binding sites, no. 2 provided the smallest number of hydrogen bonds and caged Na+ ions. The RMSD property was also not the best one. Overall, sites nos. 1 and 2 were better candidates for binding. In site no. 1, more Na+ ions were involved in aptamer-protein binding than in no. 3 (by 0.6 on average), while site no. 3 provided more hydrogen bonds (by 0.9 on average). However, the structure of the aptamer in site no. 3 closely resembled its structure in the water solution: the RMSD value was only 7.75 (compared with the value of 5.72 for the free aptamer and 9.16 for site no. 1).
Identification of glioma CTCs in the blood
CTCs are promising biomarkers for GBM diagnosis and prognosis.7,8 The most common CTC isolation strategies rely on the expression of cancer cell surface markers such as epithelial cell adhesion molecules and cytokeratins, which are absent in brain tumors.40 СTCs from aggressive non-epithelial brain tumors could be isolated only using physical methods (filtration, separation in the density gradient, microfluidic separation) followed by secondary immunostaining, genetic, telomerase activity, or FISH analysis.40,41,42,43
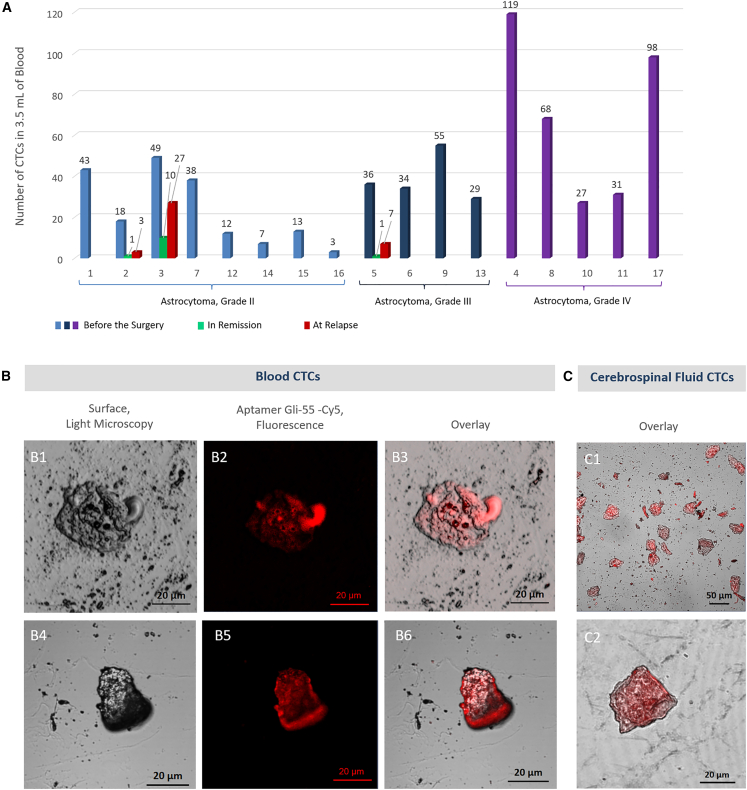
We applied the magnetic separation technique. The biotinylated aptamer Gli-233 on streptavidin-coated magnetic beads efficiently captured CTCs with glial tumor origin from the blood, and aptamer Gli-55 was used to stain these cells specifically. Fluid biopsy with aptamer-based CTC isolation and identification was performed for 17 patients with astrocytoma (grades II and IV) before neurosurgery and for 3 of them in dynamics. The number of CTCs before the surgery varied from 3 to 119 cells in 3.5 mL of blood for each patient (Figure 4A; Table 1). The average number of CTCs in the blood increased in high-grade astrocytoma.
Figure 4.
Aptamer-mediated detection of CTCs in the blood of patients with astrocytoma of different grades
(A) CTC count in 3.5 mL of the blood of astrocytoma patients who underwent neurosurgery. (B) Images of CTCs isolated from blood. (C) Images of CTCs isolated from cerebrospinal fluid. Magnification 40×. Scale bars, 20 µm, 50 µm.
Table 1.
Clinical characteristics of patients with astrocytoma and the number of CTCs detected in the blood
| Patient/gender/age/surgery no. | Histologic type, grade | Tumor location | Therapy | No. of CTCs before the surgery/in remission/at relapse | PFS after first/second tumor resection (months) | Overall survival time after diagnosis (months) | Quality of life, before the surgery/in remission period (Karnofsky scale, %) |
|---|---|---|---|---|---|---|---|
| Patient 1/female/67/2 | astrocytoma, grade II (relapse) | the left temporal lobe of the brain | tumor resection (2 times), chemo- and radiotherapy | 43 | 6 | 16 | 80/90 |
| Patient 2/female/43/2 | astrocytoma, grade II (relapse) | the left parietal, occipital lobe of the brain, posterior parts of the left lateral ventricle | tumor resection (3 times), chemo- and radiotherapy | 18/1/3 | 12/5 | 23.5 | 80/90 60/60 |
| Patient 3/male/38/2 | astrocytoma, grade II (relapse) | the left temporal lobe, the lower horn of the left lateral ventricle | tumor resection (2 times), chemo- and radiotherapy | 49/10/27 | 5.5 | 15 | 80/80 20/30 |
| Patient 7/female/68/1 | astrocytoma, grade II (relapse) | the left occipital and parietal lobe of the brain’s left lateral ventricle | tumor resection (2 times), chemo- and radiotherapy | 38 | 17 | 25 | 80/80 |
| Patient 12/female/78/1 | astrocytoma, grade II | the left temporal lobe of the brain | tumor resection, chemo- and radiotherapy | 12 | 11 | 15 | 70/80 |
| Patient 14/female/59/1 | astrocytoma, grade II | the left temporal lobe of the brain | tumor resection, chemo- and radiotherapy | 7 | 15 | 18 | 60/80 |
| Patient 15/male/69/1 | astrocytoma, grade II | the left frontal lobe of the brain | tumor resection, chemo- and radiotherapy | 13 | N/A | N/A | 60/80 |
| Patient 16/female/53/1 | astrocytoma, grade II | the parietal lobe of the brain | tumor resection, chemo- and radiotherapy | 3/N/A/N/A/0 | N/A | 23 | 80/80 |
| Patient 5 /male/21/2 |
astrocytoma, grade III (relapse) | the left frontal, parietal, and temporal lobes of the brain, left lateral ventricle, visual hillock | tumor resection (2 times), chemo- and radiotherapy | 36/1/7 | 7/2 | 13 | 80/90 40/70 |
| Patient 6/female/67/1 | astrocytoma, grade III (relapse) | the left temporal and parietal lobes left lateral ventricle | tumor resection | 34/N/A/N/A/<1000 | 6 | 7 | 30/40 |
| Patient 9/male/42/1 | astrocytoma, grade III | the left parietal, occipital and temporal lobe of the brain, spreading to the lateral ventricle, mediobasal parts of the left frontal lobe | tumor resection, chemo- and radiotherapy | 55 | 9 | 10 | 80/80 |
| Patient 13/female/57/1 | astrocytoma, grade III | the frontal lobe of the brain | tumor resection, chemo- and radiotherapy | 29/N/A/N/A/<1000 | N/A | 5 | 60/60 |
| Patient 4 /female/42/2 |
astrocytoma, grade IV (GBM) (relapse) |
the right parietal lobe, right lateral ventricle, middle parts of the corpus callosum | tumor resection (2 times), chemo- and radiotherapy, gamma knife | 119 | 5 | 13.5 | 30/30 |
| Patient 8/female/72/1 | astrocytoma, grade IV (GBM) | deep departments of the left temporal lobe. | tumor resection, chemo- and radiotherapy | 68 | 5 | 9 | 60/60 |
| Patient 10/female/39/2 | astrocytoma, grade IV (GBM) | the left parietal, occipital, and temporal lobes of the brain, extending into the lateral ventricle, the medio-different sections of the left frontal lobe | tumor resection, chemo- and radiotherapy, gamma knife | 27/0/2 | 38 | 50 | 60/80 |
| Patient 11/male/63/2 | astrocytoma, grade IV (GBM) | the right parietal lobe of the brain | tumor resection | 31 | N/A | 3 | 40/40 |
| Patient 17/male/67/3 | astrocytoma, grade IV (GBM) | the left frontal lobe | tumor resection | 98 | 0.5 | 1 | 30/30 |
GBM, glioblastoma multiforme; N/A, no data available.
The numbers of CTCs before the surgery were much higher (18, 49, and 36 cells for patients 2, 3, and 5 with astrocytoma, grade II) than in the remission period (1, 10, and 1 cell for the same patients) and increased after tumor relapse to 3, 27, and 7 cells, respectively (Figures 4A and 4B; Table 1). Interestingly, the same anti-GBM aptamers were applied to isolate and count individual CTCs from glioblastoma patients’ blood and cerebrospinal fluid. In two cases (patients 6 and 13), CTCs were also found in cerebrospinal fluid in large numbers (more than 1,000 cells in 1.5 mL). One patient (16) did not possess CTCs in cerebrospinal fluid (Figure 4C; Table 1). The representative laser scanning microscopy images of blood and cerebrospinal fluid CTCs are shown in Figures 4B1–B6, 4C1, and 4C2, respectively.
Gli-233 and Gli-55 significantly improved the isolation process and could be utilized further for CTC capture in blood and cerebrospinal fluid to investigate glioma CTCs for clinical significance and tumor monitoring.
Evaluation of aptamers ex vivo
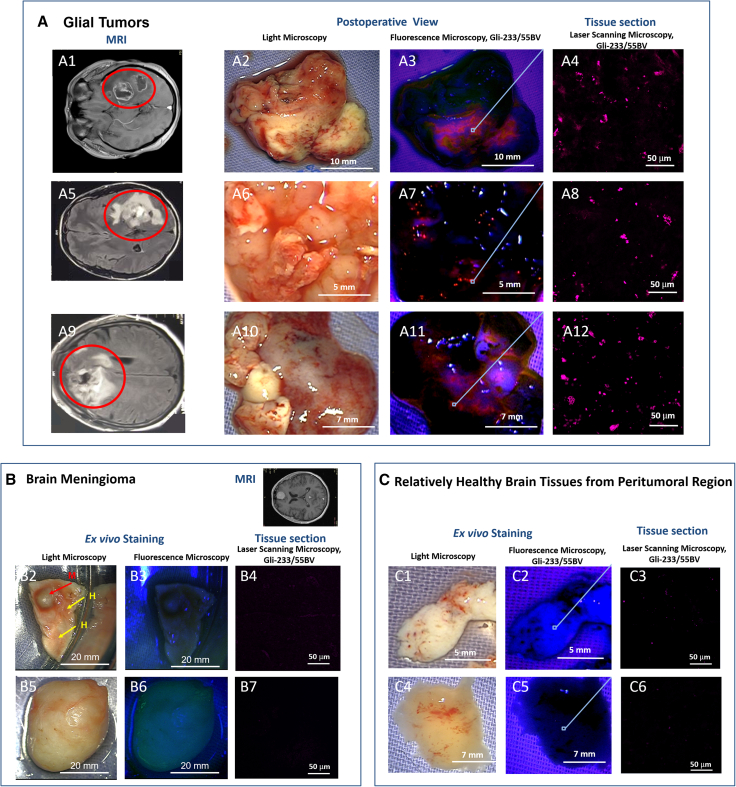
To assess the aptamer binding to brain tumor ex vivo, we tested both Gli-233 and Gli-55 on resected brain tumors from different patients. The solution of two aptamers conjugated with Brilliant Violet 650 fluorescent dye (Gli-233BV, Gli-55BV) was sprayed immediately on freshly isolated postoperative tissues. The fluorescent images were captured 1 min after the aptamer application and washing with DPBS using a surgical fluorescence microscope. The aptamers demonstrated strong binding to glial tumors (Figure 5A) but not to meningiomas (Figure 5B) and relatively healthy tissues from the peritumoral region (Figure 5C). Analysis of thin tissue sections confirmed the aptamers binding to glial tumor cells but not healthy ones. In control experiments, a nonspecific oligonucleotide conjugated with Brilliant Violet 650 dye and the free dye without the attached aptamer showed no significant fluorescence (Figure S5). Autofluorescence of tumors was not observed in the registered emission range from 620 to 710 nm. This staining could be used for precise sampling during the surgery for tissue characterization, grading, and molecular analyses (Figure 6). Histopathological examinations of the selected samples of astrocytoma tissues (Figures 6A2, 6A3, 6B2, and 6B3) were performed using immunohistochemical staining with the antibodies such as anti-GFAP (Figures 6A4 and 6B4) or anti-KI-67 (Figures 6A5 and 6B5), individual aptamers anti-GFAP Gli-55 and anti-TUBA1C Gli-233, or in combination (Figures 6A3 and 6B3), and histological analyses with hematoxylin and eosin (Figures 6A6 and 6B6). These aptamers may be used as a contrasting agent during fluorescence-guided surgery. Their average tumor-to-background ratio (TBR) for Gli-233BV and Gli-55BV was calculated from the mean fluorescence intensity and equaled 1.898; for margin zones TBR was 1.4, and for the tumor zones TBR was 4.5 (Figure S6). The combination of Gli-233BV and Gli-55BV distinguished tumor and margin zones (Figure S7) well, and the aptamers penetrated 6 mm deeply into the tumor (Figure S8A) and did not stain the vasculature (Figure S8B).
Figure 5.
Evaluation of aptamer binding ex vivo
(A) Fluorescent imaging of brain tissues from different patients with astrocytoma; MRI of patient brains with gliomas (1, 5, 9); light microscopy of corresponding resected tumor tissues (2, 6, 10); fluorescence microscopy of the tumor tissues stained with Gli-233BV and Gli-55BV (3, 7, 11×), 40× magnified areas of the stained tumor tissues (4, 8, 12). (B) Light (2, 5) and fluorescence microscopy of brain meningioma tissues from different patients stained with Gli-233BV and Gli-55BV (3, 6×), 40× magnified areas of the stained meningioma tissues (4, 7). (С) Light (C1, C4) and fluorescent (C2, C5) imaging of relatively healthy brain tissues from the peritumoral region, 40× magnified areas of the stained meningioma tissues (3, 6). Magnification 1.5–6×, 40×. Scale bars, 20 mm, 10 mm, 7 mm, 5 mm, 50 µm.
Figure 6.
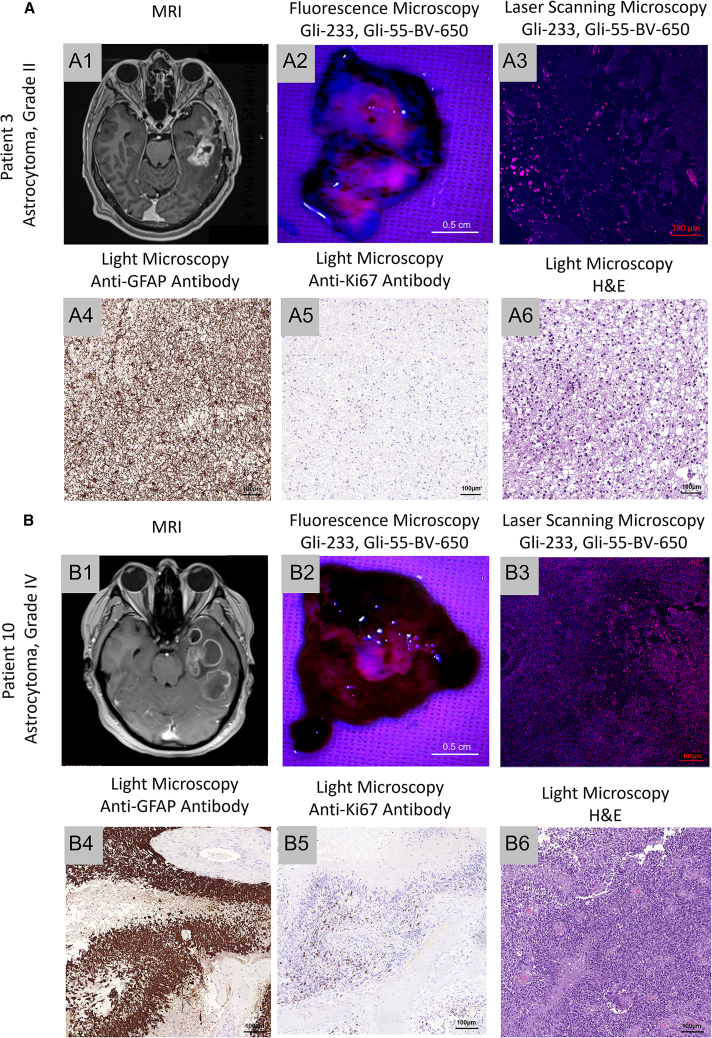
Aptamers for the histological analyses of astrocytoma
(A) MRI (1) and fluorescent imaging of tumor tissues of the patient with astrocytoma, grade II, stained ex vivo with Gli-233BV and Gli-55BV (2); laser scanning microscopy of the correspondent tissues (3); immunohistochemical staining with anti-GFAP (4), anti-Ki67 (5) antibodies; and histological staining with hematoxylin and eosin (H&E) (6). (B) MRI and tumor tissue images of a patient with astrocytoma, grade IV. Magnification 4×, 20×, 40×. Scale bars, 0.5 cm, 100 µm.
In vivo visualization of human glioblastoma xenotransplantation in mice
To demonstrate in vivo utility of the aptamers applied to post-operative human tissues, we used an improved immunosuppression protocol for the development of tumor xenografts in mice.44
This protocol has been developed for the lung cancer model by Jivrajani et al.45 Here, we modified it to achieve the proper growth of human glial tumors in mice brains. The model of orthotropic xenotransplantation of human tumors in immunosuppressed animals simulates the development of human gliomas.46 Histopathological features of these tumors are close to those in humans due to the partial inhibition of the immune system. In our study, ICR mice were immunosuppressed with cyclosporine (suppressing interleukin-1 and interleukin-2 release, required for the activation of T lymphocytes and proliferation of activated T lymphocytes, respectively), cyclophosphamide (significantly reduces the number of neutrophils, B and T cells, and natural killers), and ketoconazole (inhibits cytochrome p450, which metabolizes cyclosporine) for a week before tumor transplantation and 2 days after transplantation, which was enough to achieve a proper tumor growth without any histological evidence of immune activation within 5 weeks after transplantation. No mice mortality was registered until the end of the study. Direct cyclosporine and cyclophosphamide influence on glioma was avoided since these drugs were not administered during the extensive growth of the tumor.
We developed the primary cell culture (consisting of tumor stroma cells attached to the bottom of the well and neurospheres) from the patient with confirmed astrocytoma grade IV (Figures 7A1 and 7A2). Obtained tumor cells (Figure 7A3) were inoculated into the mice brains through the cranial window, the puncture was covered with hydrogel medium, and the skin incision was sutured (Figure 7A4).
Figure 7.
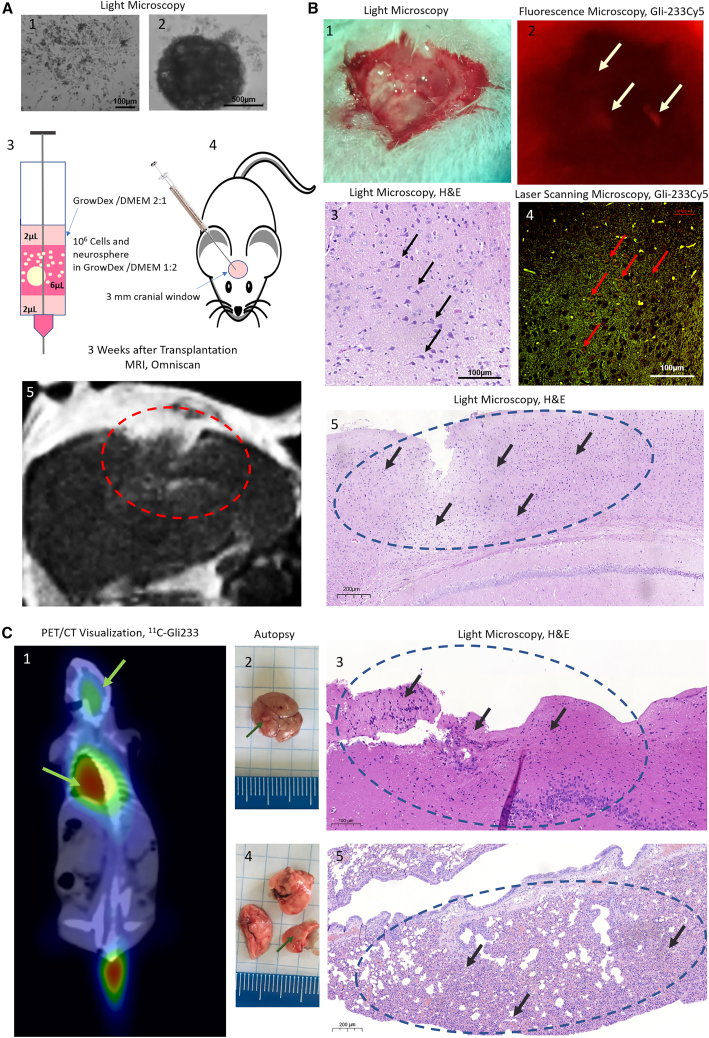
In vivo visualization of xenotransplanted human glioma in mice
(A) Xenotransplantation of the primary cell cultures (1) and neurospheres (2) into cranial windows of immunosuppressed mice (4) using a Hamilton syringe in 3D cell-culture hydrogel GrowDex (3), and MRI visualization 3 weeks after transplantation with Omniscan (gadolinium) as a contrast (5). (B) Fluorescence-assisted in situ visualization of the tumor in mouse brains stained with the Cy5-labeled aptamer Gli-233 during the surgical intervention in vivo (2) supported with the light microscopy (1) of the correspondent area, laser scanning (Cy5-labeled aptamer, red channel; autofluorescence, green channel) (4), and light microscopy of the same tissues stained with hematoxylin and eosin (3). (C) PET/CT visualization (1), autopsy (2, 4), and histological analyses (3, 5) of the main tumor in the brain (2, 3) and its metastasis in the lungs (4, 5). White arrows indicate regions of fluorescent tumors stained with Gli-233Cy5, black and red arrows show astrocytoma cells in brain sections, green arrows show tumor regions in PET/CT and autopsy, and dashed circles include tumor areas. Magnification 4×, 10×, 40×. Scale bars, 100 µm, 200 µm, 500 µm.
We noticed that the tumors inoculated through the cranial window grew faster than those injected through the puncture. MRI with Omniscan contrasting was performed to monitor the tumor development. Three weeks after transplantation, the tumor was large enough (Figure 7A5) to visualize in vivo using fluorescent- or radiolabeled aptamers. To demonstrate the possibility of using the aptamers as a tumor contrast agent during the surgical intervention (Figure 7B1), Cy5-labeled Gli-233 was applied to the tumor grown for 3 weeks in mouse brains through the cranial window washed with PBS. The tumor stained with Cy5-labeled Gli-233 was illuminated by a DFP Green LED (NIGHTSEA) and visualized using the surgical microscope OPMI PICO (Carl Zeiss, Germany) with red fluorescence filter (NIGHTSEA) (Figure 7B2). The histological analyses (Figures 7B3 and 7B5) and laser scanning microscopy (Figure 7B4) of the brain proved the presence of the tumor and specific aptamer staining after the autopsy. A control Cy5-labeled unrelated aptamer showed no sufficient fluorescence (Figure S9A).
For PET/CT visualization of the xenotransplanted human glioma in vivo, an 11C-labeled aptamer was synthesized using a previously developed protocol.47 The 11C-Gli-233 accumulation was found in the brain and lungs of mice with transplanted glioma (Figure 7C1), while the unrelated aptamer was not found (Figure S9B). An autopsy revealed the presence of a primary tumor node in the brain (Figure 7C2) and metastasis in the lungs (Figure 7C4). Histological analyses indicated that the tumor found in the brain was astrocytoma (Figure 7C3), and cancer in the lungs had similar histological features (Figure 7C5).
Preclinical acute toxicity studies of the aptamers
For in vivo application, we also studied the acute toxicity of pooled Gli-55 and Gli-233 in rats (Tables S5–S16, supplemental videos, and spreadsheets S17–S20). The fluorescent aptamers showed no acute toxicity. General conditions of the animals, including body weight gain and feed intake, and the central nervous and cardiovascular systems, remained normal. No morphological changes to the internal organs and their masses and alterations of biochemical and hematological parameters were observed. Based on the conducted toxicity study, administered intravenously, aptamers were safe in the therapeutic dose (0.7 pM/kg) and in 50 times excess (35 pM/kg). Therefore, Gli-55 and Gli-233 were recommended for further preclinical and clinical studies.
Discussion
Glial brain tumor symptoms are not specific; its diagnosis is based on MRI studies and confirmed by histopathological and molecular analysis, unfortunately, mainly in the advanced stages of the disease. Improving astrocytoma diagnostics is extremely important. Specific agents for fluid biopsy analyses, PET/CT, MRI, and intraoperative staining of tumor margins need to be developed to improve patient diagnostics and survival rates. Aptamers are promising for targeting aggressive glioma cells in tissues, blood, and spinal cord fluid.48 Aptamers Gli-233 and Gli-55 were selected for freshly resected astrocytomas of different grades. Computer simulations and experimental examinations studied their structural and binding features. TUBA1C and GFAP were proposed as potential targets for Gli-233 and Gli-55, respectively. The isoform of α-tubulin is a prognostic marker in gliomas that promotes cell proliferation and is involved in cell-cycle progression. It correlates with immune cell infiltration in the tumor microenvironment and is a biomarker of poor prognosis and immunotherapy outcomes.49 Thus, it may be considered a potential target for glial tumor therapeutics.50,51
Immunohistochemical staining for GFAP is a routine practice associated with patient overall and long-term survival.52 Thus, Gli-233 and Gli-55 could be used for the histological characterization of astrocytomas and accurate sampling after tumor resection or tissue biopsies.
A fluid biopsy is a less invasive procedure; however, analyses of CTCs extracted from blood can be very informative. CTC count and molecular typing could help clinicians to characterize a tumor, diagnose the degree and the risk of metastasis or recurrence, and could be used to monitor progression and treatment response.8 However, data on CTCs in glioma are limited to date, as their use as glioma biomarkers started recently.41,44 The main problem of CTC detection in patients with glial brain tumors is the absence of EpCAM and cytokeratins, traditional markers of epithelial tumors. For this reason, new methods were developed to determine CTCs in the plasma of patients with glial brain tumors, including assessing EGFR gene amplification, deletion of chromosome fragments, telomerase expression, determination of chromosomal aberrations, etc.53,54 However, these strategies take time and effort. Using aptamers specific to glioblastoma cells makes it possible to identify CTCs in the blood of patients with glial brain tumors quickly and with high accuracy.
Despite advanced imaging technologies and reliable radiopharmaceuticals and amino acid PET tracers, such as 18F-labeled fluoroethyl-tyrosine and 11C-labeled methyl-L-methionine, that have been developed lately, differential diagnosis, recurrence, and treatment-related changes in gliomas are still an unsolved problem.55 Radiopharmaceuticals based on aptamers specifically targeting aggressive brain tumors could sufficiently improve diagnostics. Here, we demonstrated that aptamers could be easily modified with 11C radionuclide and utilized for PET/CT visualization of brain glioma and its metastasis. However, use of a [11C] aptamer may be limited due to the need for an on-site cyclotron facility.
Thus, aptamers Gli-233 and Gli-55 are promising for glioma PET/CT imaging, intraoperative visualization, and CTC-specific capture.
Materials and methods
Patient-derived tumor samples
The study was approved by the Local Committee on Ethics of the Krasnoyarsk State Medical University in Krasnoyarsk, Russia (no. 37/2012 of 01/31/2012) and the Local Committee on Ethics in Krasnoyarsk Inter-District Ambulance Hospital named after N.S. Karpovich, Krasnoyarsk, Russia (20/11/2016). Tumor tissues were taken from patients with glioma, meningioma, and brain metastases who had undergone complete curative resection of their disease in Krasnoyarsk Inter-District Ambulance Hospital, named after N.S. Karpovich. All brain tumor specimens were collected with the written informed consent of patients. Solid tumors were removed aseptically and immediately immersed in an ice-cold colorless Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 1,000 U mL−1 penicillin G and 1,000 mg L−1 streptomycin. Samples were transported on ice to the laboratory within 2–4 h after resection. Experiments with a surgical fluorescent microscope were done immediately after resection in the hospital.
Preparation of tumor cells from resected brain samples
Solid tumor specimens were rinsed twice with DMEM supplemented with 100 U mL−1 penicillin G and 100 mg L−1 streptomycin. Tissues were minced with scissors, and blood vessels, necrotic tissues, and blood clots were removed; the remaining tissues were dissociated into small aggregates by pipetting. The cell suspension was taken for aptamer selection, binding analyses, and primary cell culture growth.
Aptamer selection and binding analysis
DNA aptamers were selected based on the tissue-SELEX procedure described previously for lung cancer tissues using the Harvard DNA library in DPBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.6]).11 The Harvard library was originally designed in Liu’s laboratory at Harvard University.56 The 100 nucleotide DNA library consists of a 60 base length variable internal region with three stems and loops flanked on either side by 20 base length constant primer regions. The final form of the library is 5′-CTC CTC TGA CTG TAA CCA CGN’ N″ N′ N″ N′ N″ N′ N″ NN N′ N″ N′ N″ N′ N″ N′ N″ N′ N″ NNN N′ N″ N′ N″ N′ N″ N′ N″ N′ N″ NN N′ N″ N′ N″ N′ N″ N′ N″ N′ N″ NNN N′ N″ N’ N’Y N′ N′ N″ G CAT AGG TAG TCC AGA AGC C-3′ where N is a nucleotide mixture that produces the ratio of A/C/G/T as 1:1:1:1, N′ is a mixture that has the ratio of A/C/G/T as 45:5:45:5, and N″ is a mixture that produces the ratio of A/C/G/T as 5:45:5:45.
The first round of selection started with the positive selection step. The cells derived from fresh glioma tissue samples were incubated with yeast RNA for 20 min, followed by centrifugation, after that 500 μL DPBS containing 1 μmol L−1 DNA library was added to the cell pellet and incubated for 30 min at 25°C and then centrifuged at 3,500 × g for 10 min at 25°C to remove unbound aptamers followed by washing with 1 mL of DPBS. The pellet was resuspended in 95 μL of 10 mmol L−1 Tris-HCl buffer containing 10 mmol L−1 EDTA (pH 7.4) (TE buffer) and heated for 10 min at 95°C to release aptamers from tumor cells. Once the cells were denatured, debris was removed by centrifugation at 14,000 × g for 15 min at 4°C, and the supernatant containing the aptamers was collected and stored at −20°C. Tumor-binding aptamers were amplified using symmetric and asymmetric PCR. For symmetric PCR, 5 μL of the aptamer pool in TE buffer was mixed with 45 μL of symmetric PCR Master Mix containing: 1× PCR buffer, 2.5 mmol L−1 MgCl2, 0.025 U μL−1 KAPA2G HotStart Robust polymerase (KAPA Biosystems), a 220 μmol L−1 equimolar solution of dATP, dCTP, dGTP, and dTTP (dNTPs), 300 nmol L−1 of the forward primer (5′-CTC CTC TGA CTG TAA CCA CG-3′), and 300 nmol L−1 of the reverse primer (5′-GGC TTC TGG ACT ACC TAT GC-3′). Afterward, asymmetric PCR was performed: 5 μL of the symmetric PCR product from the previous amplification reaction was mixed with 45 μL of the asymmetric PCR Master Mix containing: 1× PCR buffer, 2.5 mmol L−1 MgCl2, 0.025 U μL−1 KAPA2G HotStart Robust polymerase, 220 μmol L−1 dNTPs, 1 μmol L−1 forward FAM-labeled primer (5′-FAM- CTC TGA CTG TAA CCA CG-3′), and 50 nmol L−1 reverse primer (5′-GGC TTC TGG ACT ACC TAT GC-3′). Amplifications were performed using the following PCR program: preheat for 2 min at 95°C, 15 cycles of 30 s at 95°C, 15 s at 56°C, 15 s at 72°C, and hold at 4°C. The concentration of PCR products was estimated by separating FAM-labeled DNA on 3% (w/w) agarose gel electrophoresis.
In negative selection, benign tumor tissues or metastasis in the brain from the other tumors and healthy tissues from peritumoral regions and blood vessels in DPBS were used. The minced tissues were incubated for 30 min at 25°C in 500 μL DPBS with 100 nmol L−1 aptamer pool from a previous round and then centrifuged at 3,500 × g for 10 min at 25°C to collect unbound sequences. The unbound sequences were used for following positive selection as described above. In total, six rounds of selection for different patient tissues were performed (supplemental materials; Table S1). According to the binding analyses, the best pool was sequenced at Eurofins Genomics (Louisville, KY).
Aptamer sequence analysis
The best selection rounds were sequenced; therefore, we looked at the leaders by the number of copies and family formation. A bioinformatic approach was used to identify lead aptamers from 20,469 sequences from NGS results. The primary dataset can be found at https://drive.google.com/file/d/1adu9ZphI-LJv0EnvIqTonQ7j112VheZS/view?usp=sharing. The aptamer pool was admitted as a set of words in the alphabet . Each word was characterized by its length and the number of copies in pool . To estimate the similarity of two words and of equal length we used the Hamming distance , i.e., the number of positions at which the corresponding symbols are different. To estimate the similarity of two arbitrary words and we used the function , where is a subword of the length of the word . Note, that if and only if words and contain equal subwords of the length .
We defined the family of aptamer by . If is small, the set consists of those aptamers from that have similar subwords of the length with the word . To characterize the size of the family we used the number .
Families for the sequenced pool were formed using parameters , .
Flow cytometric aptamer affinity analysis
The affinity of aptamer pools and individual aptamer clones was measured by flow cytometry using an FC-500 Flow Cytometer (Beckman Coulter). The data were analyzed with Kaluza software (Beckman Coulter). In brief, glioma cells were incubated with masking DNA (0.1 mg mL−1 of masking yeast tRNA) in 500 μL of DPBS for 15 min at 24°C to reduce nonspecific binding. Experiments were performed in triplicate and represented as the average of the three measurements with the error bar lengths corresponding to the sample standard deviation. The sample containing 250,000 cells was incubated in 50 nM FAM-labeled aptamers or FAM-ssDNA library (as a control) for 30 min at 25°C in DPBS. The cells were washed twice with 1 mL of DPBS, resuspended in 0.5 mL of DPBS, and subjected to flow cytometry within 30 min. To find EC50, the concentration of DNA at which half of it was bound to cells, 250,000 cells were tested with varying concentrations (1–300 nM) of FAM-labeled aptamers in flow cytometric experiments. The mean fluorescence intensity of bound to DNA was used to calculate EC50 by fitting the dependence of mean fluorescence of the DNA binding to the concentration of the aptamers to the equation Y = MFsat × X/(EC50 + X), where MFsat is the mean fluorescence at saturation.
Identification of aptamer protein targets by mass spectrometry
Protein targets of aptamers were identified using an affinity purification procedure based on a modified AptaBiD protocol.22 In brief, post-surgical tumor tissues were chopped, washed twice with DPBS, and lysed in 0.1% (v/v) sodium deoxycholate. The supernatant obtained after centrifugation at 14,000 × g for 20 min at 4°C was incubated with masking yeast tRNA (1 mg mL−1) and, afterward, with a synthetic 40-nt scrambled DNA sequence (50 nM) for blocking nonspecific binding. Then, the solution was incubated with a biotinylated aptamer (50 nM) in DPBS for 30 min and then with 50 μg of streptavidin-coated paramagnetic beads (Promega) for the same time. The beads with aptamer-protein complexes were pulled down by a magnet and washed four times with DPBS. Protein targets were dissociated from aptamer-coated beads by adding 30 μL of 8 M urea and incubating for 60 min. The beads were retained with the magnet, and the supernatant was removed and stored at −20°C. A fraction of denatured proteins (5 μL) was diluted with 50 μL of 50 mM ammonium bicarbonate (pH 8.1) and digested for 16 h at 37°C with porcine trypsin (sequencing grade, modified; Promega) at a ratio of 1:20. The peptide mixture was extracted and purified using a ready-to-use pipette tip filled with C18 spherical silica reverse-phase material (ZipTip C18, Millipore). Peptides were eluted with 10 μL of 80% methanol/1% formic acid.
Protein identification was performed by liquid chromatography and mass spectrometry using a Dionex UltiMate 3000 UHPLC and Orbitrap Fusion Tribrid (Thermo Scientific) equipped with a nanoflow electrospray ionization source. The experiments were repeated four times for each patient. Tissue samples from eight patients were analyzed in five independent series, and only the proteins found in most samples (excluding common contaminants) were considered reliably identified. The search for post-translational modifications was performed by Proteome Discoverer 1.4 software with the Sequest HT search engine. The following seven modifications were set as variables: deamidation, phosphorylation, glycosylation, acetylation, methylation, palmitoylation, N-terminal, and acetylation. Polyglycylation and polyglutamylation of tubulin were searched separately in various combinations of length. The search results were filtered, and only modified peptides were selected. Values of peptide spectra match (PSM) between modified and non-modified peptides were considered as a degree of modification. Only peptides with prevalence in PSM for the modified type or not presented as non-modified were selected as reliable. The primary data can be found at Glazyrin, Yury (2022), “Mass Spectrometry Data,” Mendeley Data, V2, https://doi.org/10.17632/nvn5spksw8.2.
Identification of the aptamer tertiary structures with SAXS
Synchrotron SAXS data from Gli-233 were collected on the BioSAXS beamline at ESRF (France) by the Dectris Pilatus 1M detector. The sample-to-detector distance was 2.849 m, and the wavelength of the X-ray beam was 0.9919 Å. The aptamer was diluted to concentrations of 0.8, 4.0, and 8.2 mg mL−1 in DPBS buffer with calcium and magnesium. Additional SAXS measurements in the solution without Ca2+ and Mg2+ ions (concentrations of Gli-233 were 0.8 and 4.0 mg mL−1) were carried out to check the influence of their presence on the structure formation of the aptamer. Before SAXS, all aptamer solutions were heated to 90°C and then cooled to 4°C. The solution at each concentration was exported by X-rays 10 times by 0.5 s in a flow regime to mitigate the radiation damage. The scattering intensities I(s) were recorded and radially averaged, and the SAXS measurements of the buffer were subtracted. The recorded range of momentum transfer was 0.1 < s < 3.5 nm−1 (is defined as s = 4π sin(θ)/λ, where θ is the scattering angle). SAXS data processing was performed in the ATSAS software suite.57 Data from the three SAXS curves were extrapolated to zero concentration to exclude the concentration-dependent influence on the data. The radius of gyration Rg was determined by Guinier approximation and from the distance distribution function p(r), which is obtained by the indirect Fourier transformation of the SAXS 1D curve and represents a whole number of distances between scattering centers in the molecule. Construction of the real space pair-distance distribution function is implemented into the program GNOM. Using the SAXS intensity curve, calculated distance distribution functions, as well as an averaged autocorrelation function, an aptamer structure model was obtained in the DAMMIF program by the Monte Carlo method, which provides the iterative selection of models, generating a scattering pattern from each model and obtaining a curve repeating more precisely the experimental result. Calculation of the theoretical scattering curves and their fitting with experimental SAXS data were performed using the program CRYSOL. For a rapid superposition of low- and high-resolution models, the program SUPALM was used through the SASpy plugin in the program PyMOL.58
SAXS data for the Gli-55 aptamer were obtained at the P12 BioSAXS beamline (EMBL) at the Petra III storage ring of the synchrotron center DESY, Hamburg. The eluent of the employed chromatography column was passed through a 1.7-mm quartz capillary held under vacuum. The SAXS data were recorded on a Pilatus 6M area detector (Dectris) at a sample-to-detector distance of 3 m and the wavelength λ = 0.124 nm (X-ray energy 10 keV) at room temperature 20.4°C. Series of individual 1-s exposure X-ray data frames were measured from the continuously flowing column eluate across one column volume. The 2D SAXS intensities were reduced to I(s) vs. s using the integrated analysis pipeline SASFLOW.59 The s axis was calibrated with silver behenate and the resulting profiles were normalized for exposure time and sample transmission. To distinguish the oligomeric constituents in the solution the joint technique of size-exclusion chromatography with SAXS (SEC-SAXS) was applied (Figure S2). To decompose the partially overlapping components in SEC-SAXS data the evolving factor analysis was applied using the program EFAMIX.60 Further SAXS data processing was the same as for Gli-233. Experimental SAXS data were fitted with the theoretical SAXS curve calculated from the molecular model of the aptamer in the program CRYSOL, and spatial alignment into a 3D-fit of the SAXS bead model with the MD-simulation model of Gli-55 was performed in the program PyMOL using SASpy plugin.58
Molecular design of the aptamer 3D structures
The secondary structures of the aptamers were predicted using the online software OligoAnalyzer 3.1.33 The simulations were done assuming ionic conditions of Na+ (146 mM) and Mg2+ (0.5 mM) at 4°C and 20°C for Gli-233 and Gli-55, respectively. Modeling and visualization of the aptamer spatial structures were performed by Avogadro and Facio programs.61 Atomic structures were optimized using the FMO method in the framework of the DFTB.62,63 The solvent effects were described within the PCM. Quantum chemical calculations were carried out using the GAMESS program.64
Identification of a functional region of the aptamer
To determine the probable functional area of Glibraces tamer, the window size braces threshold were set. For each pair the set of -mers of the aptamer Gli-233 was constructed, after which for each -mer b the family was determined in the glioma pool and the value of was calculated. The distribution demonstrates the most pronounced local maximum at , b = TGAACGGAC. The subword TGAACGGAC was chosen as the candidate for the binding site of the aptamer Gli-233 with a protein.
Tubulin structure prediction
To date, there is no known experimental structure of the human tubulin-ɑ-1C (hTBA1C) protein. In the current study, the hTBA1C secondary structure was predicted by homology modeling using the SwissModel service.65 The model with the best QMEAN score (template: PDB 5KX5, Ovis aries tubulin-ɑ-1A) was selected for the next steps.66 The predicted structure misses several residues from the CTT. The α-helical structure of missing CTT residues was constructed using VMD software and carefully attached to the CTT of the hTBA1C.67
Tubulins usually exist in the cytoplasm as heterodimers of ɑ- and β-tubulin chains. As a partner for hTBA1C, we used the human β-tubulin 2B isotype (hTBB2B), which is commonly observed in glial cells. The experimental structure of human tubulin-β-2B (PDB: 6E7C) was used in this work, and the missing residues were recovered using the SwissModel service. The CTT of β-tubulin was built in the same way as the CTT of the hTBA1C. The structure of a hetero-dimer of tubulin α and β chains was obtained by structural alignment of hTBA1C and hTBB2B models to the experimental structure of similar hetero-dimer (human tubulin α-1B and β-2B, PDB: 6E7C). Residue clashes were identified by visual inspection of the interface between tubulin units and carefully eliminated, considering the experimental configurations of corresponding residues. Several post-translational modifications (PTMs) were applied to the CTT of the tubulin-α chain, namely: monoglycylation of residue 433 and polyglutamylation of residues 445 (2×), 447 (1×), and 448 (5×x). The resulting model contains a tubulin-ɑ chain with PTMs, a tubulin-β chain, two bound GTP molecules, and two Mg2+ ions.
Relaxation of tubulin dimer structure
The structure of the tubulin dimer was submerged into a water box with a size of 12 × 10 × 13 nm. The net charge of the protein was compensated by putting the corresponding amount of Na+ ions into the bulk. Then, extra Na+ and Cl− ions were added to the physiological concentration (0.15 mol L−1). The structure of the solvated dimer was first minimized for 1,000 steepest descent steps to remove the strain. The resulting structure was equilibrated during 100 ns MD simulation. The last 70 ns of the MD trajectory were clustered, and the cluster centers were used for aptamer docking.
Aptamer docking
In this study, the docking simulations were performed using HDOCK, a web server of a hybrid docking algorithm of template-based modeling and free docking, in which cases with misleading templates can be rescued by the free docking protocol.39 During the docking calculation, all of the default parameters were used.
Aptamer-tubulin complex dynamics
Structures of three aptamer-protein complexes predicted by docking were put into a solvent environment using the procedure described above for the tubulin dimer. The only difference is that a slightly larger water box size of 13 × 12 × 15 nm was used before the production of the MD simulation, and the ion shell of the solute was additionally relaxed for 30 ns while keeping the aptamer and protein close to their initial positions. It was achieved by applying positional restraints to the Cα atoms of the protein and phosphorus atoms of the aptamer. After that, 100 to 300 ns MD simulations were performed until the convergence of the aptamer-protein complex structures was verified by RMSD of the aptamer heavy atoms.
MD details
All simulations were performed using GROMACS MD software.68 The protein and aptamer structures were described by the Amber force field. TIP3P water model was used for solvation. For MD, a stochastic velocity-rescaling thermostat (300 K) and Parrinello-Rahman pressure coupling (1 bar) were used. The LINCS scheme was used to constrain all covalent bonds with hydrogen atoms, which allows using a 2-fs timestep.
Staining of tissue sections with antibodies and aptamers
Tissue pieces were frozen in liquid nitrogen, sliced into 5 μm sections using a Microm HM 525 cryostat (Thermo Scientific), and placed on poly-lysine-coated glass slides. First, nonspecific binding of the antibodies was blocked by incubation of the sections with 10% (w/w) of bovine serum albumin (Sigma-Aldrich) for 30 min, followed by incubation with primary anti-tubulin or anti-GFAP antibodies (2 ng μL−1) in a humidified atmosphere for 1 h, followed by staining with a secondary antibody labeled with Alexa Fluor 405 or Cy5 (2 ng μL−1) in a humidified atmosphere for 1 h and then washed three times with DPBS. Nonspecific binding of aptamers was blocked with yeast tRNA (1 ng μL−1, Sigma-Aldrich) and then incubated with 50 nM of FAM- or Cy5-labeled aptamers in DPBS and incubated for another 2 h at 6°С. This was followed by washing with DPBS. Stained tissue pieces were frozen in liquid nitrogen and sliced into 5-μm sections using a cryostat. Bio-Mount mounting medium (Bio-Optica, Italy) was used to fix the sections. The sections were analyzed with an LSM 780 (Carl Zeiss) microscope.
CTC isolation
Isolation of CTCs was performed from 3.5 mL of patient blood during 1–1.5 h after collection into BD Vacutainer Heparin Tubes. The blood was centrifuged (1,500 × g for 10 min) to remove the plasma. The cell pellet was transferred into a 15-mL centrifuge tube using a BSA-treated tip. Red blood cells were lysed with a hypotonic NH4Cl solution in a Vacutainer tube. Cells were rinsed with 2 mL of 0.42% NH4Cl with heparin, pipetted several times, and poured into the 15-mL tube, then addition of 8 mL of 0.42% NH4Cl with heparin into the tube, incubation for 10 min on a shaker, and centrifuged at 3,500 × g for 5 min. The remaining cell pellet was resuspended in 100 μL DPBS and incubated with masking yeast tRNA (0.1 mg mL−1) for 30 min at room temperature to reduce nonspecific binding. The sample was centrifuged at 3,500 × g for 5 min, the supernatant was removed, and the cell pellet was incubated for 30 min with 100 μg of streptavidin-coated paramagnetic beads (Promega) functionalized with 100 nM biotinylated aptamers specific to the tumor cells. The cells bound with the magnetic particles via the aptamers were separated on a magnetic stand, resuspended in 100 μL of calcium and magnesium-free DPBS buffer, and concentrated using a magnetic separator. The pellet containing mostly CTCs was stained with Cy5-labeled aptamers at a final concentration of 100 nM for 30 min. Smears were prepared for the quantification of CTCs in the patient blood. The cell pellet was spread evenly on a glass slide and then fixed in methanol for 5 min, followed by staining with Romanowsky-Giemsa dye. The counting of CTCs was done using an Axiostar plus fluorescent microscope (Carl Zeiss).
Ex vivo tissue staining
To assess aptamer binding to a brain tumor in situ, we tested aptamers Gli-233 and Gli-55 on resected brain tumors from different patients. The solution of the biotinylated aptamers conjugated with streptavidin-labeled Brilliant Violet 650 fluorescent dye (Gli-233/55BV) was sprayed on freshly isolated postoperative tissues. An OPMI Pentero fluorescent microscope (Carl Zeiss) with a fluorescent module (BLUE 400, ex. 400–410 nm, em. 620–710 nm) and a Xenon white light source were used to visualize tumors. The fluorescent images were captured 1 min after applying aptamers using a surgical fluorescence microscope. Thin tissue sections were prepared from the areas with high, moderate, and low fluorescence intensity. Nonspecific oligonucleotide conjugated with Brilliant Violet 650 dye and free dye without attached aptamers were used as a control. Autofluorescence of tumors was not observed in the registered emission range from 620 to 710 nm.
Histological, immunohistochemical, and confocal laser scanning microscopy analysis of resected tissues
Tumor tissues were fixed at 10% buffered formalin solution and stored at 4°C. The volume of the fixative was 50 times greater than the size of the immersed tissue. The samples underwent a standard histological treatment based on isopropyl alcohol with further paraffin impregnation. Sections made from paraffin blocks were applied to positively charged adhesive glasses. One of each pair of the adjacent sections was left for CLSM analysis without additional staining; the next one was used for immunohistochemistry; the other section was stained with hematoxylin and eosin dyes to confirm tissue morphology. An LSM 780 NLO Confocal microscope (Carl Zeiss) was used for all CLSM, binding, and viability of cell cultures analyses; images were processed with ZEN2 software. Immunohistochemical studies were performed using a Ventana BenchMark ULTRA system. A recombinant anti-GFAP antibody (clone EP672Y, Abcam) and recombinant anti-Ki67 (clone SP6, Abcam) were used with a dilution of 1:100.
Synthesis of 11С-labeled Gli-233
For the in vivo PET/CT imaging, we used thiolated aptamer Gli-233. The synthesis of 11С was performed on the Cyclone 18/9 ST cyclotron (IBA, Belgium) as described previously.47 In brief, 11CH3I was obtained from the nitrogen gas of natural isotopic composition with oxygen gas (0.5%–1%), hydrogen H2, and iodine vapor was introduced to 3′ end of Gli-233 modified with 5′-thiol modifier C6 S-S phosphoramidite in DMSO. For this, the reaction mixture was prepared: 40 μL of primer (100 μM) was added to 500 μL of DMSO, 10 μL of 0.1 M hydrochloric acid solution, 1500 μL of phosphate buffer (PBS) (pH 7.4), 136.8 mm NaCl, 10.1 mm Na2HPO4, 2.7 mm KCl, 1.8 mm K2HPO4, 0.499 mm MgCl2), and transferred to the reaction vessel of the automated Synthra MeIPlus synthesis module (Synthra, Germany). The final volume of 2 mL radiopharmaceutical contained 4 nmol of 11CH3-modified aptamer. 11CH3-Gli-233 was stable for 60 min.
Primary patient-derived glial tumor cell culture
Human astrocytoma samples were taken aseptically during the surgery, immediately immersed in ice-cold colorless DMEM medium supplemented with 1,000 U mL−1 penicillin G and 1,000 mg L−1 streptomycin, and transported to a laboratory within 2–4 h of resection. Tumor specimens were rinsed twice with DMEM supplemented with antibiotics. Tissues were minced with scissors, blood vessels, necrotic tissues, and blood clots were removed, and the remaining tissues were dissociated into small aggregates by pipetting. The suspension was washed thrice with DPBS using centrifugation at 3,000 rpm. The pellet was transferred to 12 wells of cell culture plates with initiation medium (containing DMEM, 10% fetal bovine serum, 20 μg/mL insulin, 10 μg/mL transferrin, 25 nmol/L sodium selenite, 100 U/mL antibiotics, 1 ng/mL epidermal growth factor) and maintained in a humidified incubator at 37°C in an atmosphere of 5% CO2 and 95% of air. Cells were cultured for 2 months to get the confluent monolayer with the forming neurospheres. The medium was changed three times a week.
Xenotransplantation of the glial tumor to immunosuppressed mice
Laboratory female ICR mice weighing 20–25 g were maintained in sterile individually ventilated cages. Mice were immunosuppressed using cyclosporine (20 mg/kg intraperitoneally), cyclophosphamide (60 mg/kg intraperitoneally), and ketoconazole (10 mg/kg orally) every day: 7 days before and 2 days after transplantation.45 Under the inhalation anesthesia, mice fur was removed with hair removal cream, the skin was dissected under a sterile conditions, and a 3-mm cranial window was made using an electro trypan (Figure 7A). The formation of the human astrocytoma model was carried out by brain injection of primary patient-derived cell cultures. One 2-mm neurosphere and 106 tumor stroma cells in 6 μL of the hydrogel medium (GrowDex/DMEM, 1:1) were placed into a Hamilton syringe between 2 μL of hydrogel medium (GrowDex/DMEM, 2:1) (Figure 7A3). Tumor cells were inoculated into the mice brain through the 3-mm cranial window, the puncture was covered with 5 μL of hydrogel medium (GrowDex/DMEM, 2:1), and the skin incision was sutured (Figure 7A4). Growth control was performed 14 and 21 days after transplantation by MRI and histological examination of autopsy material.
MRI
MRI was performed by the improved protocol described previously.69 In brief, the imaging of mice immobilized using inhalation anesthesia was performed on an Avance DPX 200 spectrometer (Bruker BioSpin, Rheinstetten, Germany) using Omniscan for contrast. Slice-selective 2D images were acquired using the multi-slice multi-echo technique supplied by Paravision 4.0 software (Bruker BioSpin, Ettlingen, Germany). Imaging acquisition parameters: slice thickness, 0.71 mm; field of view, 40 mm; matrices, 256 × 256 pixels, with a spatial resolution of 156 μm per pixel within the slice. The repetition time and echo time were adjusted to 600 and 4.7 ms, respectively, to provide the T1-weighting on the images. The image acquisition time was 10 min.
Aptamer-based fluorescence visualization of the xenotransplanted tumor in situ
Mice under inhalation anesthesia were fixed. Head skin was dissected, and aptamer Gli-233-Cy5 was applied directly to the brain and washed out after 3 min. The tumor stained with Cy5-labeled Gli-233 was illuminated by a DFP Green LED (NIGHTSEA) and visualized using the OPMI PICO surgical microscope (Carl Zeiss) with a red fluorescence filter (NIGHTSEA).
PET/CT imaging
Tumor locations were monitored using PET/CT. Mice were injected into the tail vein with 0.4 nmol of 11CH3-Gli-233 aptamer in 200 μL of DPBS or 11CH3-nonspecific oligonucleotide as a negative control. Animals were immobilized using inhalation anesthesia, which is safe and does not reduce blood circulation. PET/CT scanning was performed using a Discovery PET/CT 600 scanner (General Electric) 40 min after injection. Data were analyzed using PET VV software at an AW Volume Share 5 workstation and Hounsfield densitometry scale. After quarantine, the mice were sacrificed and autopsied to confirm the tumor locations. Tissues were histologically analyzed to prove the presence of astrocytoma development.
Preclinical acute toxicity studies of the aptamers
Investigation of acute toxicity of the aptamers was performed on Sprague-Dawley rats in a laboratory certified for preclinical studies (Laboratory of Biological Testing, Institute of Bioorganic Chemistry named after academics M.M. Shemyakin and Y.A. Ovchinnikov of Russian Academy of Sciences). This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institute of Health and the requirements of the Guidelines for conducting pre-clinical studies of drugs. The protocol plan of research and standard animal-friendly operating procedures have been reviewed and approved by the Local Bioethics Committee of the Institution of Science Institute of Bioorganic Chemistry (no. 527/16). For the in vivo application, we planned to use a maximum of 3 mL of 1 μM aptamer solution in DPBS. The equivalent dose was calculated for the rat weight. According to the regulations, the studied drug must be injected intravenously for the acute toxicity assessment of this type of medicine. The conjugates were administered once intravenously, followed by a 14-day observation period and the following tests: body weight, feed intake, and the manifestation of clinical signs of toxicity of the study were recorded every week; in the final week of the study, animals performed functional tests to assess the condition of their central nervous system and cardiovascular system. The animals were euthanized on the 15th day of the study, and a necropsy was performed for subsequent histological analysis. At autopsy, the weight of the internal organs was recorded, and blood samples were taken to measure the performance of biochemistry and hematology and bone marrow samples.
Acknowledgments
The authors are grateful to all the patients and hospital staff participating in this research. We acknowledge the assistance of the AptamerLab LCC (www.aptamerlab.com) and personally Mr. Vasily Mezko for the aptamer 3D structure optimization and financial and technical support. The authors thank Prof. Alexey V. Protopopov, Prof. Vasily F. Shabanov, Mr. Alexey Kichkailo, Dr. Arkady B. Kogan, and Dr. Rinat G. Galeev for their general support, and Mrs. Valentina L. Grigoreva, and Irina V. Gildebrand for the help with histological staining. Technical and instrumental support was provided by the Multiple-Access Center at Tomsk State University; the Krasnoyarsk Inter-District Ambulance Hospital, named after N.S. Karpovich; John L. Holmes Mass Spectrometry Facility at the University of Ottawa; Federal Siberian Research Clinical Center under the Federal Medical Biological Agency; Shared Core Facilities of Molecular and Cell Technologies at Krasnoyarsk State Medical University and Krasnoyarsk Regional Center for Collective Use at the Federal Research Center “KSC SB RAS.” The confocal fluorescence microscopy research was carried out with the equipment of the Tomsk Regional Core Shared Research Facilities Center of the National Research Tomsk State University. The Center was supported by the Ministry of Science and Higher Education of the Russian Federation, grant no. 075-15-2021-693 (no. 13.RFC.21.0012). Acute toxicity studies were performed in a laboratory certified for preclinical studies (Laboratory of Biological Testing, Institute of Bioorganic Chemistry named after academics M.M. Shemyakin and Y.A. Ovchinnikov Russian Academy of Sciences). The authors are grateful to the Joint Super Computer Center of the Russian Academy of Sciences for providing supercomputers for computer simulations. Aptamer selection and their clinical applications were funded by the Ministry of Healthcare of the Russian Federation; project АААА-Б19-219090690032-5 (to T.N.Z.). The Ministry of Science and Higher Education of the Russian Federation project FWES-2022-0005 (to A.S.K.) supported aptamer characterization, molecular modeling, and in vivo experiments. Development of the glioma tumor model in immunosuppressed mice was supported by the Russian Science Foundation grant no. 22-64-00041 (to M.A.D.), https://rscf.ru/en/project/22-64-00041/. Synthesis of 11C-aptamer and PET/CT visualization was funded by the Federal Medical Biological Agency; project 122041800132-2 (to A.V.O.). Mass spectrometry analyses, DNA sequencing, and synthesis were supported by an NSERC Discovery Grant (to M.V.B.). We acknowledge the European Synchrotron Radiation Facility for SAXS experiments and thank Dr. Bart Van Laer for assistance in using a beamline BM29. SAXS measurements were supported by RFBR no. 18-32-00478 for young scientists (to R.V.M.). The synchrotron SEC-SAXS data for Gli-55 aptamer were also collected at beamline P12 operated by EMBL Hamburg at the PETRA III storage ring (DESY, Hamburg, Germany).
Author contributions
A.S.K., M.V.B., A.A.N., T.N.Z., and S.S.Z. designed the overall concept, and supervised the work. A.A.N., E.E.E., and A.K.G. performed surgery, provided clinical data and samples, and analyzed clinical data. M.A.K., G.S.Z., and O.S.K. selected and analyzed aptamer pools. A.G. and D.M. sequenced aptamer pool. D.V.V. performed mathematical analysis of sequenced aptamer clones and statistical analyses. Y.E.G. and G.G.M. performed mass spectrometry analyses, identified protein targets, and post-translational modifications. G.S.Z., O.S.K., and M.A.K. evaluated aptamer binding to cells, and tissues, performed CTC analyses, and analyzed experimental data. E.E.E., A.K.G., A.V.K., and A.S.K. analyzed clinical data. M.M.P., A.A.M., P.A.S., P.G.S., R.A.Z., and I.P.A. designed and supervised clinical experiments, provided historical controls, and estimated clinical significance. G.S.Z., M.A.K., and A.S.K. performed preclinical experiments. A.V.G., I.P.G., I.I.V., D.S.G., and M.A.D. performed glial tumor xenotransplantation. V.I.B. and P.A.S. designed immunosuppression protocol. A.A.K., E.D.N., and M.A.D. performed cell culturing. K.V.B., A.A.V., O.N.B., and A.V.O. synthesized aptamer and C11-based radiopharmaceutical, K.V.B., N.L., K.A.L., N.A.T., N.S., N.G.C., I.I.V., D.S.G., O.S.K., A.S.K., and T.N.Z. performed PET/CT imaging. A.S.K., G.S.Z., O.S.K., and T.N.Z. designed the protocol and performed tissue staining in situ, assisted during surgeries. F.N.T., I.A.S., P.V.A., and E.V.K. performed in silico molecular design of aptamer 3D structure and molecular docking. A.E.S., T.E.S., and E.V.K. performed circular dichroism spectroscopy. V.M. and D.M. performed tubulin structure prediction, molecular dynamics of aptamer and protein, and molecular docking. R.V.M., V.V.Z., G.P., P.V.K., and S.S.Z. performed small-angle X-ray scattering. I.N.L. and V.A.S. performed laser scanning microscopy. V.A.K. and A.K.K. performed histological analyses and immunostaining. A.A.N., T.N.Z., Y.E.G., G.S.Z., O.S.K., I.V.G., D.V.V., F.N.T., R.V.M., A.G., A.S.K., and M.V.B. analyzed all data and wrote the paper. All authors provided intellectual input, and edited and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2023.03.015.
Supplemental information
Data availability
NGS sequencing data are deposited at https://drive.google.com/file/d/1adu9ZphI-LJv0EnvIqTonQ7j112VheZS/view?usp=share_link. SAXS data and associated files are available from the SASBDB database (www.sasbdb.org), SASBDB: SASDRY7, SASDPZ8. Mass Spectrometry Data: Glazyrin, Yury (2022), “Mass Spectrometry Data,” Mendeley Data: https://doi.org/10.17632/nvn5spksw8.1 Any other data are available upon request.
References
- 1.Ostrom Q.T., Gittleman H., Xu J., Kromer C., Wolinsky Y., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009-2013. Neuro Oncol. 2016;18:v1–v75. doi: 10.1093/neuonc/now207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Müller Bark J., Kulasinghe A., Chua B., Day B.W., Punyadeera C. Circulating biomarkers in patients with glioblastoma. Br. J. Cancer. 2020;122:295–305. doi: 10.1038/s41416-019-0603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO, editor. Central Nervous System Tumors. 5th edition. International Agency for Research on Cancer; 2021. [Google Scholar]
- 4.Mikkelsen V.E., Solheim O., Salvesen Ø., Torp S.H. The histological representativeness of glioblastoma tissue samples. Acta Neurochir. 2021;163:1911–1920. doi: 10.1007/s00701-020-04608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCullough B.J., Ader V., Aguedan B., Feng X., Susanto D., Benkers T.L., Henson J.W., Mayberg M., Cobbs C.S., Gwinn R.P., et al. Preoperative relative cerebral blood volume analysis in gliomas predicts survival and mitigates risk of biopsy sampling error. J. Neuro Oncol. 2018;136:181–188. doi: 10.1007/s11060-017-2642-2. [DOI] [PubMed] [Google Scholar]
- 6.Miller A.M., Shah R.H., Pentsova E.I., Pourmaleki M., Briggs S., Distefano N., Zheng Y., Skakodub A., Mehta S.A., Campos C., et al. Tracking tumor evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565:654–658. doi: 10.1038/s41586-019-0882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Müller C., Holtschmidt J., Auer M., Heitzer E., Lamszus K., Schulte A., Matschke J., Langer-Freitag S., Gasch C., Stoupiec M., et al. Hematogenous dissemination of glioblastoma multiforme. Sci. Transl. Med. 2014;6:247ra101. doi: 10.1126/scitranslmed.3009095. [DOI] [PubMed] [Google Scholar]
- 8.Wang J., Bettegowda C. Applications of DNA-based liquid biopsy for central nervous system neoplasms. J. Mol. Diagn. 2017;19:24–34. doi: 10.1016/j.jmoldx.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 9.Costa E., Lawson T.M., Lelotte J., Fomekong E., Vaz R.G., Renard L., Whenham N., Raftopoulos C. Long-term survival after glioblastoma resection: hope despite poor prognosis factors. J. Neurosurg. Sci. 2019;63:251–257. doi: 10.23736/S0390-5616.18.04180-2. [DOI] [PubMed] [Google Scholar]
- 10.Ni X., Castanares M., Mukherjee A., Lupold S.E. Nucleic acid aptamers: clinical applications and promising new horizons. Curr. Med. Chem. 2011;18:4206–4214. doi: 10.2174/092986711797189600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zamay G.S., Kolovskaya O.S., Zamay T.N., Glazyrin Y.E., Krat A.V., Zubkova O., Spivak E., Wehbe M., Gargaun A., Muharemagic D., et al. Aptamers selected to postoperative lung adenocarcinoma detect circulating tumor cells in human blood. Mol. Ther. 2015;23:1486–1496. doi: 10.1038/mt.2015.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn M.R., Jimenez R.M., Chaput J.C. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017;1:0076. doi: 10.1038/s41570-017-0076. [DOI] [Google Scholar]
- 13.Mo T., Liu X., Luo Y., Zhong L., Zhang Z., Li T., Gan L., Liu X., Li L., Wang H., et al. Aptamer-based biosensors and application in tumor theranostics. Cancer Sci. 2022;113:7–16. doi: 10.1111/cas.15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song Y., Shi Y., Huang M., Wang W., Wang Y., Cheng J., Lei Z., Zhu Z., Yang C. Bioinspired engineering of a multivalent aptamer-functionalized nanointerface to enhance the capture and release of circulating tumor cells. Angew. Chem. Int. Ed. Engl. 2019;58:2236–2240. doi: 10.1002/anie.201809337. [DOI] [PubMed] [Google Scholar]
- 15.Zhang T., Tian T., Lin Y. Functionalizing framework nucleic-acid-based nanostructures for biomedical application. Adv. Mater. 2022;34:2107820. doi: 10.1002/adma.202107820. [DOI] [PubMed] [Google Scholar]
- 16.Tian T., Li Y., Lin Y. Prospects and challenges of dynamic DNA nanostructures in biomedical applications. Bone Res. 2022;10:40. doi: 10.1038/s41413-022-00212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang B., Tian T., Xiao D., Gao S., Cai X., Lin Y. Facilitating in situ tumor imaging with a tetrahedral DNA framework-enhanced hybridization chain reaction probe. Adv. Funct. Mater. 2022;32:2109728. doi: 10.1002/adfm.202109728. [DOI] [Google Scholar]
- 18.Zhu J., Yang Y., Ma W., Wang Y., Chen L., Xiong H., Yin C., He Z., Fu W., Xu R., Lin Y. Antiepilepticus effects of tetrahedral framework nucleic acid via inhibition of gliosis-induced downregulation of glutamine synthetase and increased AMPAR internalization in the postsynaptic membrane. Nano Lett. 2022;22:2381–2390. doi: 10.1021/acs.nanolett.2c00025. [DOI] [PubMed] [Google Scholar]
- 19.Affinito A., Quintavalle C., Esposito C.L., Roscigno G., Vilardo C., Nuzzo S., Ricci-Vitiani L., De Luca G., Pallini R., Kichkailo A.S., et al. The discovery of RNA aptamers that selectively bind glioblastoma stem cells. Mol. Ther. Nucleic Acids. 2019;18:99–109. doi: 10.1016/j.omtn.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nuzzo S., Brancato V., Affinito A., Salvatore M., Cavaliere C., Condorelli G. The role of RNA and DNA aptamers in glioblastoma diagnosis and therapy: a systematic review of the literature. Cancers. 2020;12:E2173. doi: 10.3390/cancers12082173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Q., Wang Y., Wang H., Wu L., Zhang H., Song Y., Zhu Z., Kang D., Yang C. DNA aptamers from whole-cell SELEX as new diagnostic agents against glioblastoma multiforme cells. Analyst. 2018;143:2267–2275. doi: 10.1039/c8an00271a. [DOI] [PubMed] [Google Scholar]
- 22.Berezovski M.V., Lechmann M., Musheev M.U., Mak T.W., Krylov S.N. Aptamer-facilitated biomarker discovery (AptaBiD) J. Am. Chem. Soc. 2008;130:9137–9143. doi: 10.1021/ja801951p. [DOI] [PubMed] [Google Scholar]
- 23.Zamay G.S., Ivanchenko T.I., Zamay T.N., Grigorieva V.L., Glazyrin Y.E., Kolovskaya O.S., Garanzha I.V., Barinov A.A., Krat A.V., Mironov G.G., et al. DNA aptamers for the characterization of histological structure of lung adenocarcinoma. Mol. Ther. Nucleic Acids. 2017;6:150–162. doi: 10.1016/j.omtn.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magiera M.M., Janke C. Post-translational modifications of tubulin. Curr. Biol. 2014;24:R351–R354. doi: 10.1016/j.cub.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 25.Magiera M.M., Singh P., Gadadhar S., Janke C. Tubulin posttranslational modifications and emerging links to human disease. Cell. 2018;173:1323–1327. doi: 10.1016/j.cell.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 26.Álvarez-Satta M., Matheu A. Primary cilium and glioblastoma. Ther. Adv. Med. Oncol. 2018;10 doi: 10.1177/1758835918801169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loskutov Y.V., Griffin C.L., Marinak K.M., Bobko A., Margaryan N.V., Geldenhuys W.J., Sarkaria J.N., Pugacheva E.N. LPA signaling is regulated through the primary cilium: a novel target in glioblastoma. Oncogene. 2018;37:1457–1471. doi: 10.1038/s41388-017-0049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocha C., Papon L., Cacheux W., Marques Sousa P., Lascano V., Tort O., Giordano T., Vacher S., Lemmers B., Mariani P., et al. Tubulin glycylases are required for primary cilia, control of cell proliferation, and tumor development in colon. EMBO J. 2014;33:2247–2260. doi: 10.15252/embj.201488466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fiore G., Di Cristo C., Monti G., Amoresano A., Columbano L., Pucci P., Cioffi F.A., Di Cosmo A., Palumbo A., d’Ischia M. Tubulin nitration in human gliomas. Neurosci. Lett. 2006;394:57–62. doi: 10.1016/j.neulet.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 30.Katsetos C.D., Reginato M.J., Baas P.W., D’Agostino L., Legido A., Tuszyn Ski J.A., Dráberová E., Dráber P. Emerging microtubule targets in glioma therapy. Semin. Pediatr. Neurol. 2015;22:49–72. doi: 10.1016/j.spen.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 31.Rutka J.T., Murakami M., Dirks P.B., Hubbard S.L., Becker L.E., Fukuyama K., Jung S., Tsugu A., Matsuzawa K. Role of glial filaments in cells and tumors of glial origin: a review. J. Neurosurg. 1997;87:420–430. doi: 10.3171/jns.1997.87.3.0420. [DOI] [PubMed] [Google Scholar]
- 32.Wilhelmsson U., Eliasson C., Bjerkvig R., Pekny M. Loss of GFAP expression in high-grade astrocytomas does not contribute to tumor development or progression. Oncogene. 2003;22:3407–3411. doi: 10.1038/sj.onc.1206372. [DOI] [PubMed] [Google Scholar]
- 33.Owczarzy R., Tataurov A.V., Wu Y., Manthey J.A., McQuisten K.A., Almabrazi H.G., Pedersen K.F., Lin Y., Garretson J., McEntaggart N.O., et al. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008;36:W163–W169. doi: 10.1093/nar/gkn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishimoto Y., Fedorov D.G. The fragment molecular orbital method combined with density-functional tight-binding and the polarizable continuum model. Phys. Chem. Chem. Phys. 2016;18:22047–22061. doi: 10.1039/c6cp02186g. [DOI] [PubMed] [Google Scholar]
- 36.Knight C.J., Hub J.S. WAXSiS: a web server for the calculation of SAXS/WAXS curves based on explicit-solvent molecular dynamics. Nucleic Acids Res. 2015;43:W225–W230. doi: 10.1093/nar/gkv309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franke D., Svergun D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kikhney A.G., Borges C.R., Molodenskiy D.S., Jeffries C.M., Svergun D.I. SASBDB: towards an automatically curated and validated repository for biological scattering data. Protein Sci. 2020;29:66–75. doi: 10.1002/pro.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan Y., Zhang D., Zhou P., Li B., Huang S.-Y. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45:W365–W373. doi: 10.1093/nar/gkx407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adamczyk L.A., Williams H., Frankow A., Ellis H.P., Haynes H.R., Perks C., Holly J.M.P., Kurian K.M. Current understanding of circulating tumor cells - potential value in malignancies of the central nervous system. Front. Neurol. 2015;6:174. doi: 10.3389/fneur.2015.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao F., Cui Y., Jiang H., Sui D., Wang Y., Jiang Z., Zhao J., Lin S. Circulating tumor cell is a common property of brain glioma and promotes the monitoring system. Oncotarget. 2016;7:71330–71340. doi: 10.18632/oncotarget.11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macarthur K.M., Kao G.D., Chandrasekaran S., Alonso-Basanta M., Chapman C., Lustig R.A., Wileyto E.P., Hahn S.M., Dorsey J.F. Detection of brain tumor cells in the peripheral blood by a telomerase promoter-based assay. Cancer Res. 2014;74:2152–2159. doi: 10.1158/0008-5472.CAN-13-0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sullivan J.P., Nahed B.V., Madden M.W., Oliveira S.M., Springer S., Bhere D., Chi A.S., Wakimoto H., Rothenberg S.M., Sequist L.V., et al. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov. 2014;4:1299–1309. doi: 10.1158/2159-8290.CD-14-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H., Yuan F., Qi Y., Liu B., Chen Q. Circulating tumor cells for glioma. Front. Oncol. 2021;11:607150. doi: 10.3389/fonc.2021.607150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jivrajani M., Shaikh M.V., Shrivastava N., Nivsarkar M. An improved and versatile immunosuppression protocol for the development of tumor xenograft in mice. Anticancer Res. 2014;34:7177–7183. [PubMed] [Google Scholar]
- 46.Cunha A.M., Nascimento F.S., Amaral J.C., Konig S., Takiya C.M., M Neto V., Rocha E., Souza J.P.B.M. A murine model of xenotransplantation of human glioblastoma with immunosuppression by orogastric cyclosporin. Arq. Neuropsiquiatr. 2011;69:112–117. doi: 10.1590/s0004-282x2011000100021. [DOI] [PubMed] [Google Scholar]
- 47.Ozerskaya A.V., Zamay T.N., Kolovskaya O.S., Tokarev N.A., Belugin K.V., Chanchikova N.G., Badmaev O.N., Zamay G.S., Shchugoreva I.A., Moryachkov R.V., et al. 11C-radiolabeled aptamer for imaging of tumors and metastases using positron emission tomography- computed tomography. Mol. Ther. Nucleic Acids. 2021;26:1159–1172. doi: 10.1016/j.omtn.2021.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esposito C.L., Nuzzo S., Ibba M., Grinev I., Gorbushin А., Grek D., Voronkovskii I., Kolovskaya O., Zamay T., Morozov E., et al. STAT3 targeting by an aptamer-based conjugate for glioblastoma multiforme therapy. Sib Med Rev. 2021;2:72–73. doi: 10.20333/2500136-2021-2-72-73. [DOI] [Google Scholar]
- 49.Hu X., Zhu H., Chen B., He X., Shen Y., Zhang X., Xu Y., Xu X. The oncogenic role of tubulin alpha-1c chain in human tumours. BMC Cancer. 2022;22:498. doi: 10.1186/s12885-022-09595-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gui S., Chen P., Liu Y., Chen Q., Cheng T., Lv S., Zhou T., Song Z., Xiao J., He W., et al. TUBA1C expression promotes proliferation by regulating the cell cycle and indicates poor prognosis in glioma. Biochem. Biophys. Res. Commun. 2021;577:130–138. doi: 10.1016/j.bbrc.2021.08.079. [DOI] [PubMed] [Google Scholar]
- 51.Zhu H., Hu X., Gu L., Jian Z., Li L., Hu S., Qiu S., Xiong X. TUBA1C is a prognostic marker in low-grade glioma and correlates with immune cell infiltration in the tumor microenvironment. Front. Genet. 2021;12:759953. doi: 10.3389/fgene.2021.759953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahmadipour Y., Gembruch O., Pierscianek D., Sure U., Jabbarli R. Does the expression of glial fibrillary acid protein (GFAP) stain in glioblastoma tissue have a prognostic impact on survival? Neurochirurgie. 2020;66:150–154. doi: 10.1016/j.neuchi.2019.12.012. [DOI] [PubMed] [Google Scholar]
- 53.Kojima T., Hashimoto Y., Watanabe Y., Kagawa S., Uno F., Kuroda S., Tazawa H., Kyo S., Mizuguchi H., Urata Y., et al. A simple biological imaging system for detecting viable human circulating tumor cells. J. Clin. Invest. 2009;119:3172–3181. doi: 10.1172/JCI38609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castro M.A.A., Onsten T.G.H., Moreira J.C.F., de Almeida R.M.C. Chromosome aberrations in solid tumors have a stochastic nature. Mutat. Res. 2006;600:150–164. doi: 10.1016/j.mrfmmm.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 55.Santo G., Laudicella R., Linguanti F., Nappi A.G., Abenavoli E., Vergura V., Rubini G., Sciagrà R., Arnone G., Schillaci O., et al. The utility of conventional amino acid PET radiotracers in the evaluation of glioma recurrence also in comparison with MRI. Diagnostics. 2022;12:844. doi: 10.3390/diagnostics12040844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruff K.M., Snyder T.M., Liu D.R. Enhanced functional potential of nucleic acid aptamer libraries patterned to increase secondary structure. J. Am. Chem. Soc. 2010;132:9453–9464. doi: 10.1021/ja103023m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manalastas-Cantos K., Konarev P.V., Hajizadeh N.R., Kikhney A.G., Petoukhov M.V., Molodenskiy D.S., Panjkovich A., Mertens H.D.T., Gruzinov A., Borges C., et al. Atsas 3.0: expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr. 2021;54:343–355. doi: 10.1107/S1600576720013412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Panjkovich A., Svergun D.I. SASpy: a PyMOL plugin for manipulation and refinement of hybrid models against small angle X-ray scattering data. Bioinformatics. 2016;32:2062–2064. doi: 10.1093/bioinformatics/btw071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blanchet C.E., Spilotros A., Schwemmer F., Graewert M.A., Kikhney A., Jeffries C.M., Franke D., Mark D., Zengerle R., Cipriani F., et al. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY) J. Appl. Crystallogr. 2015;48:431–443. doi: 10.1107/S160057671500254X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Konarev P.V., Graewert M.A., Jeffries C.M., Fukuda M., Cheremnykh T.A., Volkov V.V., Svergun D.I. EFAMIX, a tool to decompose inline chromatography SAXS data from partially overlapping components. Protein Sci. 2022;31:269–282. doi: 10.1002/pro.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suenaga M. Facio: new computational chemistry environment for PC GAMESS. J. Comput. Chem. Jpn. 2005;4:25–32. doi: 10.2477/jccj.4.25. [DOI] [Google Scholar]
- 62.Fedorov D.G., Kitaura K. Pair interaction energy decomposition analysis. J. Comput. Chem. 2007;28:222–237. doi: 10.1002/jcc.20496. [DOI] [PubMed] [Google Scholar]
- 63.Gaus M., Lu X., Elstner M., Cui Q. Parameterization of DFTB3/3OB for sulfur and phosphorus for chemical and biological applications. J. Chem. Theory Comput. 2014;10:1518–1537. doi: 10.1021/ct401002w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barca G.M.J., Bertoni C., Carrington L., Datta D., De Silva N., Deustua J.E., Fedorov D.G., Gour J.R., Gunina A.O., Guidez E., et al. Recent developments in the general atomic and molecular electronic structure system. J. Chem. Phys. 2020;152:154102. doi: 10.1063/5.0005188. [DOI] [PubMed] [Google Scholar]
- 65.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benkert P., Biasini M., Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27-28. [DOI] [PubMed] [Google Scholar]
- 68.Rakhshani H., Dehghanian E., Rahati A. Enhanced GROMACS: toward a better numerical simulation framework. J. Mol. Model. 2019;25:355. doi: 10.1007/s00894-019-4232-z. [DOI] [PubMed] [Google Scholar]
- 69.Kolovskaya O.S., Zamay T.N., Zamay G.S., Babkin V.A., Medvedeva E.N., Neverova N.A., Kirichenko A.K., Zamay S.S., Lapin I.N., Morozov E.V., et al. Aptamer-conjugated superparamagnetic ferroarabinogalactan nanoparticles for targeted magnetodynamic therapy of cancer. Cancers. 2020;12:E216. doi: 10.3390/cancers12010216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGS sequencing data are deposited at https://drive.google.com/file/d/1adu9ZphI-LJv0EnvIqTonQ7j112VheZS/view?usp=share_link. SAXS data and associated files are available from the SASBDB database (www.sasbdb.org), SASBDB: SASDRY7, SASDPZ8. Mass Spectrometry Data: Glazyrin, Yury (2022), “Mass Spectrometry Data,” Mendeley Data: https://doi.org/10.17632/nvn5spksw8.1 Any other data are available upon request.