Abstract
Zygotic genome activation (ZGA) is a crucial step of embryonic development. So far, little is known about the role of chromatin factors during this process. Here, we used an in vivo RNA interference reverse genetic screen to identify chromatin factors necessary for embryonic development in Drosophila melanogaster. Our screen reveals that histone acetyltransferases (HATs) and histone deacetylases are crucial ZGA regulators. We demonstrate that Nejire (CBP/EP300 ortholog) is essential for the acetylation of histone H3 lysine-18 and lysine-27, whereas Gcn5 (GCN5/PCAF ortholog) for lysine-9 of H3 at ZGA, with these marks being enriched at all actively transcribed genes. Nonetheless, these HATs activate distinct sets of genes. Unexpectedly, individual catalytic dead mutants of either Nejire or Gcn5 can activate zygotic transcription (ZGA) and transactivate a reporter gene in vitro. Together, our data identify Nejire and Gcn5 as key regulators of ZGA.
The catalytic activity of two major histone acetyltransferases is not required for zygotic genome activation in the embryo.
INTRODUCTION
Upon fertilization, the first stages of embryonic development rely exclusively on maternally deposited transcripts and proteins (1). During the mid-blastula transition, as the supply of maternally deposited factors declines, zygotic transcription will commence in a process known as zygotic genome activation (ZGA) (1–3). The onset time varies depending on the organism (4–8). ZGA is generally thought to occur in two main phases, although a more continuous model of activation has also been proposed (9). In the first minor wave, a few hundred genes are transcribed between nuclear cycles 8 and 13 (7–11). Those genes are usually short and intronless, and they rely on the pioneer transcription factor Zelda (5, 7, 9, 12, 13). During the second major wave at nuclear cycle 14, about 6000 genes start to be transcribed in a narrow time window (7–9, 14, 15). This massive transcriptional awakening coincides with nuclear cellularization and is defined as stage 5 of embryogenesis (16).
Research on ZGA has mainly been centered on the key role of pioneer transcriptional factors. The most prominent example is Zelda (12, 13, 17). While Zelda binds several thousand genomic regions in the early embryo, the expression of only approximately 600 genes directly depends on its presence (12, 13, 18, 19). Other transcription factors have been linked with ZGA as well, e.g., Stat92E (20), Clamp (21, 22), GAF (23, 24), and Odd-paired (25), but only small subsets of zygotic genes depend on them.
Chromatin regulation plays a central role before and during ZGA as well, in both vertebrate and invertebrate models (15, 26–31). For instance, in Drosophila, intergenerational epigenetic inheritance of the histone marks H4K16ac and H3K27me3 are essential for the correct activation and repression of the embryonic genome, respectively (29, 30). Replacement of the maternally deposited histone BigH1 with the canonical histone H1 is also important for ZGA (32). More recently, we have shown that deposition of the histone variant H2Av before the second wave of ZGA is crucial for the activation of more than 4000 zygotic genes at ZGA (15). Nonetheless, although chromatin modifiers of the trithorax and Polycomb groups have been shown to activate and silence the expression of key early developmental genes (33), little is known about the role of chromatin factors involved in Drosophila ZGA.
In this work, we designed a reverse genetic screen to identify chromatin factors, whose maternal depletion affects embryonic viability. Top hits included histone acetyltransferases (HATs) and histone deacetylases (HDACs), whose maternal depletion affected both the early zygotic transcriptome and embryonic morphology. Focusing on Nejire and Gcn5, we show that these two HATs regulate two distinct sets of genes. During ZGA, the two enzymes are responsible for acetylation of histone H3 on residues H3K18 and H3K27 or H3K9ac, respectively. Notably, however, the catalytic activities of either Nejire or Gcn5 are not necessary for correct ZGA. These findings build on the growing examples of chromatin’s roles in regulating correct ZGA and shed light on the role of histone acetylation in gene activation.
RESULTS
A reverse genetic screen reveals chromatin factors involved in ZGA
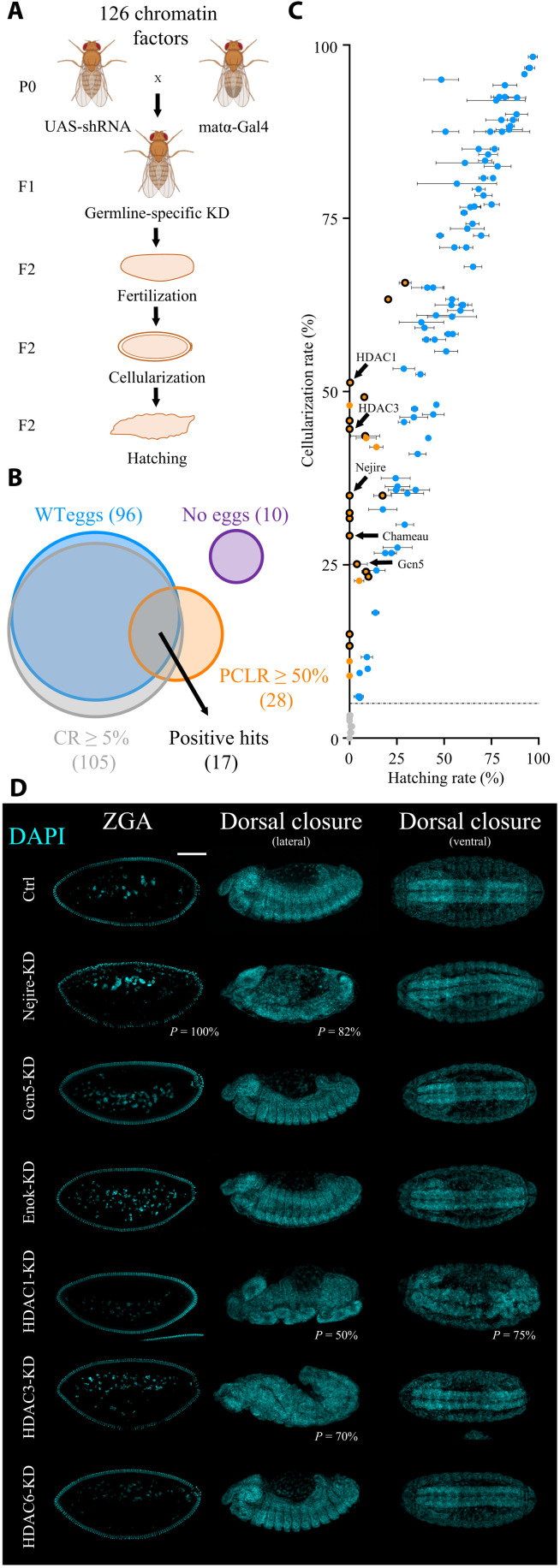
To identify maternally deposited chromatin factors that are required for ZGA, we carried out an in vivo reverse genetic screen in which we depleted candidate factors during oogenesis. We initially compiled a list of 126 well-established chromatin factors including histone writers, erasers, readers, heterochromatin proteins, chromatin remodeling complex components, proteins involved in genome architecture, Polycomb and Trithorax group proteins, and other proteins implicated in transcriptional regulation at the chromatin level (table S1). Most of these candidates do not have DNA binding domains, with the exception of a few transcription factors that have additional roles on chromatin. We then crossed flies expressing a germline-specific Gal4 driver, which is active from stage 2 of oogenesis onward (matα-Gal4), with flies carrying a Gal4-dependent shRNA (short hairpin RNA) targeting a specific chromatin factor to generate female flies with germline-specific knockdown (KD) of each factor (Fig. 1A). Upon fertilization, the resulting embryos will lack the maternally provided chromatin factor. Embryos were scored on the basis of reaching the cellularization stage [cellularization rate (CR)], which coincides with ZGA (14 mitotic divisions after fertilization) and on the basis of completion of embryogenesis [hatching rate (HR)]. We considered all candidates that showed wild-type egg morphology, a CR ≥ 5%, and a postcellularization lethality rate (PCLR) ≥ 50% as hits in our screen (Fig. 1, B and C, and table S1).
Fig. 1. Maternally deposited HATs and HDACs are essential for embryonic development.
(A) Crossing scheme of the reverse genetic screen. F2 embryos are scored for their CRs and for HRs. Fly images were obained from Biorender.com. (B) Venn diagram of the results of the screen. Positive hits are selected among 126 candidates by wild-type (WT) egg morphology, CR ≥ 5%, and PCLR ≥ 50%. PCLR is defined as (CR − HR) / (CR + 0.0001). The numbers of candidates in each category are shown in parentheses. (C) Scatter plot of the results of the screen. Black-circled orange dots represent positive hits (n = 17). Noncircled orange dots represent candidates with PCLR ≥ 50% but with abnormal egg morphology (penetrance from 1 to 100%). Blue dots represent negative hits with PCLR < 50%. Gray dots represent negative hits with CR < 5%, separated by a dashed line from the rest. Dots represent the mean, and bars represent SD of three biological replicates of embryo collections (n = 120 per biological replicate). Arrows point at five positive hits involved in histone acetylation. (D) 4′,6-diamidino-2-phenylindole (DAPI) staining of representative embryos at ZGA (stage 5) and during the beginning of dorsal closure (stage 14) in lateral and ventral views in control and upon KD. Scale bar, 100 μm. Penetrance (P) indicates the frequency of observed embryos displaying abnormal phenotypes (n > 100 embryos).
Overall, we scored 17 hits, several of which are well known to play essential roles in ZGA, thereby confirming the validity of our approach. Examples include the histone variant BigH1 (32) and the Polycomb group proteins E(z), Psc, and Scm (29, 33). Unexpected by the high number of factors involved in histone acetylation among the hits, we decided to further investigate their role during ZGA.
Maternally deposited HATs and HDACs are essential for embryonic development
On the basis of the results of the screen, we selected the HATs Nejire (PCLR = 100%), Gcn5 (PCLR = 84.5%), and Chameau (PCLR = 100%), as well as HDAC1 (PCLR = 99.5%) and HDAC3 (PCLR = 100%), for further investigation. The HATs Enok (PCLR = 29.9%) and HDAC6 (PCLR = 13.3%) served as negative controls (Fig. 1C). Reverse transcription quantitative polymerase chain reaction (RT-qPCR) in pre-ZGA embryos (embryonic stage 2) confirmed efficient KD for each of the maternally provided factors, ranging between 96 and 100% (fig. S1A). The only exception was Nejire, which showed a 65% KD efficiency (fig. S1A). Nevertheless, this level of KD was sufficient to cause a strong lethality phenotype (Fig. 1, B and C, and fig. S1A), which could be reproduced with an additional independent shRNA line (fig. S1B). Western blot and immunofluorescence experiments confirmed the total loss of Gcn5 protein and the suboptimal KD of Nejire protein (fig. S1, C and D).
Next, we analyzed the phenotypes of the different mutants during embryogenesis. Most of the HATs and HDACs analyzed in this study showed severe defects suggesting compromised ZGA. We observed defects in nuclear morphology, loss of pole cells, and failure in gastrulation, also in line with previously described reports (34, 35) (Fig. 1D and fig. S1, E and F). Together, our results show that maternally deposited HATs and HDACs play central roles during Drosophila embryonic development.
HATs and HDACs regulate mRNA production in the early embryo
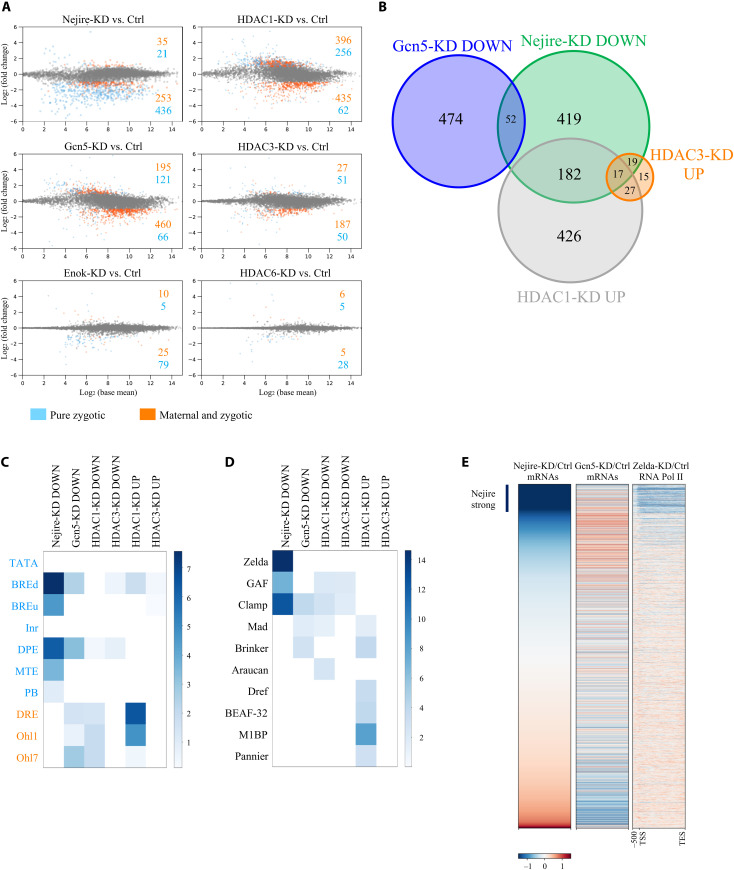
Next, we tested whether the lethality phenotypes and severe morphological defects of embryos with maternal KD of the selected chromatin factors are linked to transcriptional misregulation during ZGA. Notably, RNA sequencing (RNA-seq) experiments from hand-sorted ZGA embryos comparing KDs of Nejire, Gcn5, HDAC1, and HDAC3 to control revealed substantial transcriptome changes, whereas KDs of Enok and HDAC6 had little effect (Fig. 2A). A total of 6111 genes are actively transcribed at ZGA, as we previously showed (15); of those, 24.5% are “pure zygotic,” whereas the remaining 75.5% are classified as “maternal and zygotic” (fig. S2A). Nejire-KD, mainly affected the expression of pure zygotic genes. We confirmed this result by performing RNA-seq from purified nuclei of hand-sorted wild-type and Nejire-KD embryos (fig. S2B) to reduce the confounding effect of maternal RNAs. On the other hand, Gcn5-KD mainly affected the class of maternal and zygotic genes, highly enriched in housekeeping genes. (Fig. 2A and fig. S2A). There is little overlap between Nejire and Gcn5 down-regulated genes (Fig. 2B and fig. S2C). HDAC1-KD and HDAC3-KD showed up-regulation of many pure zygotic genes that were down-regulated in Nejire-KD, suggesting a counteracting effect between these chromatin factors (Fig. 2, A and B). On the other hand, none of these up-regulated genes is shared with any Gcn5 target (Fig. 2B). In contrast to ZGA, pre-ZGA embryo (stage 2) transcriptomes were only moderately affected by the KDs (fig. S2D). Overall, these data show that Nejire, Gcn5, HDAC1, and HDAC3 regulate the transcript levels of hundreds of genes during ZGA.
Fig. 2. HATs and HDACs regulate different sets of genes during ZGA.
(A) MA plots of RNA-seq experiments at ZGA, comparing KD with control embryos (n = 3 biological replicates of independent embryo collections). Only genes that show a global run-on sequencing (GRO-seq) signal at stage 5 (from here defined as active genes) are displayed (15). Significantly misregulated genes (adjusted P ≤ 0.05 and log2FC < −1 or log2FC > 1) are highlighted in blue if pure zygotic or in orange if maternal and zygotic (see fig. S2A for details). Numbers of significantly up or down-regulated genes are indicated. (B) Venn diagram representing sets of ZGA (stage 5) up-regulated (UP) (adjusted P ≤ 0.05 and P > 1) or down-regulated (DOWN) (adjusted P ≤ 0.05 and log2FC < −1) genes and their overlaps. Only active genes are considered. (C) Motif enrichment analysis of core promoter elements for some groups of up-regulated (adjusted P ≤ 0.05 and log2FC > 1) or down-regulated (adjusted P ≤0.05 and log2FC < −1) ZGA active genes. Core promoter elements are highlighted in blue if typical of developmentally regulated genes and in orange if typical of housekeeping genes. Color intensity shows –log10 of the adjusted P value. (D) Motif enrichment analysis of transcription factor binding sites for some groups of up-regulated (adjusted P ≤ 0.05 and log2FC > 1) or down-regulated (adjusted P ≤ 0.05 and log2FC < −1) ZGA active genes. Color intensity shows –log10 of the adjusted P value. (E) Heatmaps of active genes at ZGA. Genes are ranked according to their wild-type over Nejire-KD ratio of RNA-seq levels in column 1, wild-type over Gcn5-KD ratio in column 2, and Pol II chromatin immunoprecipitation (ChIP) ratio between wild-type and Zelda-KD from 500–base pair (bp) upstream of Transcription Start Site (TSS) to Transcription End Site (TES) from (18) in column 3. Corresponding log2FC color-coded scale bars are depicted on the bottom.
Gene ontology (GO) analysis revealed that Nejire-dependent genes are mostly involved in early developmental processes, such as germ-layer commitment and gastrulation (fig. S2E). In addition, our data confirmed Nejire-dependent expression of key developmental factors such as twist (36), hh, ci (37) and wg (38). Conversely, many Gcn5-dependent genes are essential for later stages of development, as suggested by GO terms linked to postembryonic development and metamorphosis, or they have housekeeping functions. Moreover, the genes up-regulated upon HDAC1-KD are mainly involved in regulating mesoderm, endoderm, and germline specification (fig. S2E).
Core promoter element analysis indicated that Nejire-dependent genes are enriched in elements frequently found at the promoters of developmentally regulated genes, such as BREd, BREu, DPE, MTE, and Pause Button (39). In contrast, Gcn5-dependent genes showed enrichment of promoter elements commonly found in housekeeping genes, such as DRE, Ohler1, and Ohler7, although BREd and DPE elements were also present. Moreover, genes up-regulated in HDAC1-KD showed both core promoter elements of developmentally regulated genes but an even stronger signature of housekeeping elements (Fig. 2C).
Motif analysis revealed that Nejire-dependent genes often display enrichment in DNA binding motifs recognized by the three pioneer factors Zelda, Clamp, and GAF (Fig. 2D). This observation fits well with the fact that both Zelda and Clamp are involved in the transcription of important early developmental genes (12, 17, 21, 22). GAF has previously been shown to cooperate with Nejire in the transcription of highly expressed and paused genes in S2 cells (40). In summary, these data show how HATs and HDACs regulate different categories of genes during ZGA.
Nejire and Gcn5 regulate different sets of genes
Intrigued by the limited overlap between Nejire and Gcn5 target genes (Fig. 2B), the differences in the ontological terms of these targets (fig. S2E) and their distinct promoter and DNA binding elements (Fig. 2, C and D), we more closely evaluated specific categories of target genes. We observed a strong overlap (40.3% versus 9.6% expected) between Nejire-dependent genes and Zelda-dependent genes (chi-square test, P < 0.00001) (fig. S3A) (15). Other Nejire targets showed overlap with zygotic genes that are neither regulated by Zelda nor regulated by the histone variant H2Av (33.8% versus 22.8% expected, P <0.00001) (fig. S3A). These genes are mostly dependent on GAGA factor (23). These results are further corroborated by high enrichments for Zelda motifs among Nejire targets (Fig. 2D) and by the severe loss of polymerase II (Pol II) signal in particular on the strongest Nejire targets [log2 fold change (log2FC) < −1.5] upon Zelda-KD (Fig. 2E) (18). Notably, Nejire-KD and Gcn5-KD did not affect either Zelda nor GAF total protein levels and nuclear localization in pre-ZGA and ZGA embryos (fig. S3, B and C).
Among the early developmental genes, specific transcription factors are important for anteroposterior axis formation. At ZGA, 20 of 24 of those patterning genes are strongly dependent on Nejire. In contrast, none of these genes is Gcn5 dependent (fig. S3D). Long noncoding RNA (ncRNAs) have been often associated with the expression of regulated genes (41). A total of 79 long ncRNAs are strongly down-regulated in Nejire-KD, in contrast to only 13 in Gcn5-KD (fig. S3E). Furthermore, among the 510 genes that start to be expressed in the syncytial blastoderm until ZGA (8), 65.9% are Nejire dependent, and only 9.4% are Gcn5 dependent (fig. S3F). In contrast with the strong developmental function bias of Nejire-dependent genes, Gcn5-dependent genes have a higher overlap with housekeeping genes (fig. S3G).
RNA-seq results show a lack of correlation in the regulation of Nejire-dependent genes and Gcn5-dependent genes (Fig. 2E). Some genes that are negatively affected by the lack of one HAT will get up-regulated when the other HAT is depleted (fig. S3, H and I).
In summary, Nejire is required for the activation of many early developmental genes with a strong tissue-specific signature. These include Zelda-dependent genes, which are usually regulated at the Pol II recruitment step (42), and GAF-dependent genes, which are regulated at the Pol II pausing step (40). In contrast, Gcn5 is responsible for the transcription of several housekeeping genes and of genes involved in postembryonic development.
Nejire and Gcn5 have distinct H3 substrates during ZGA
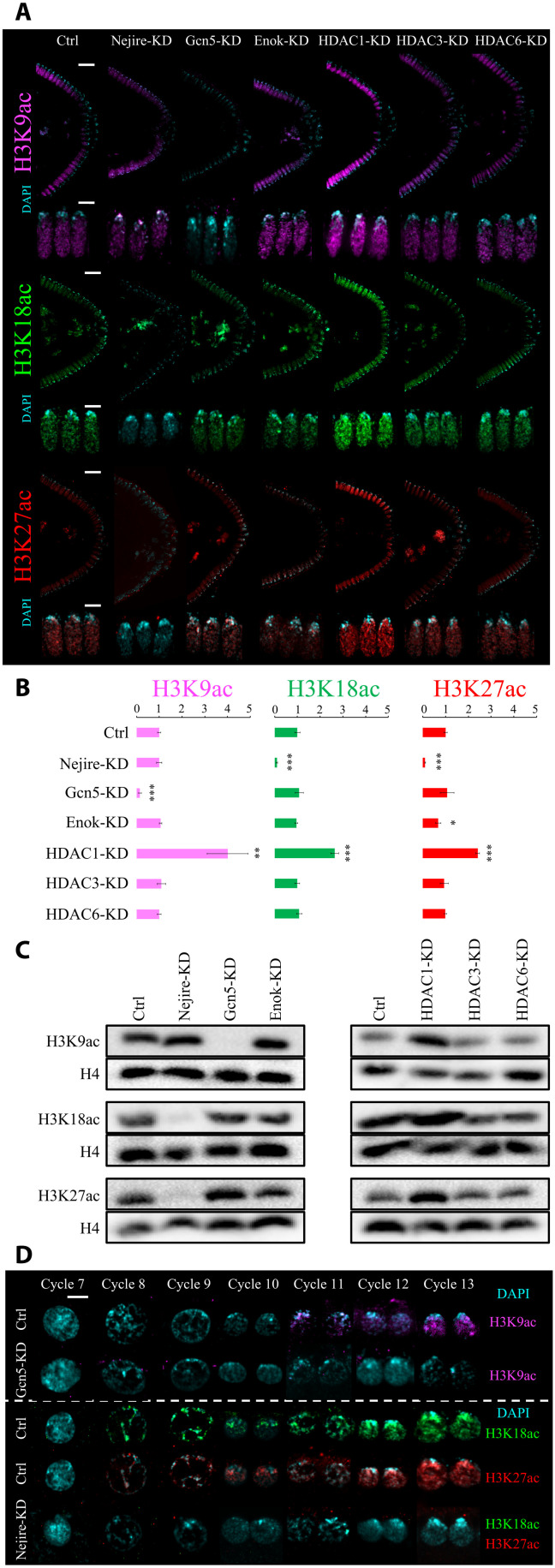
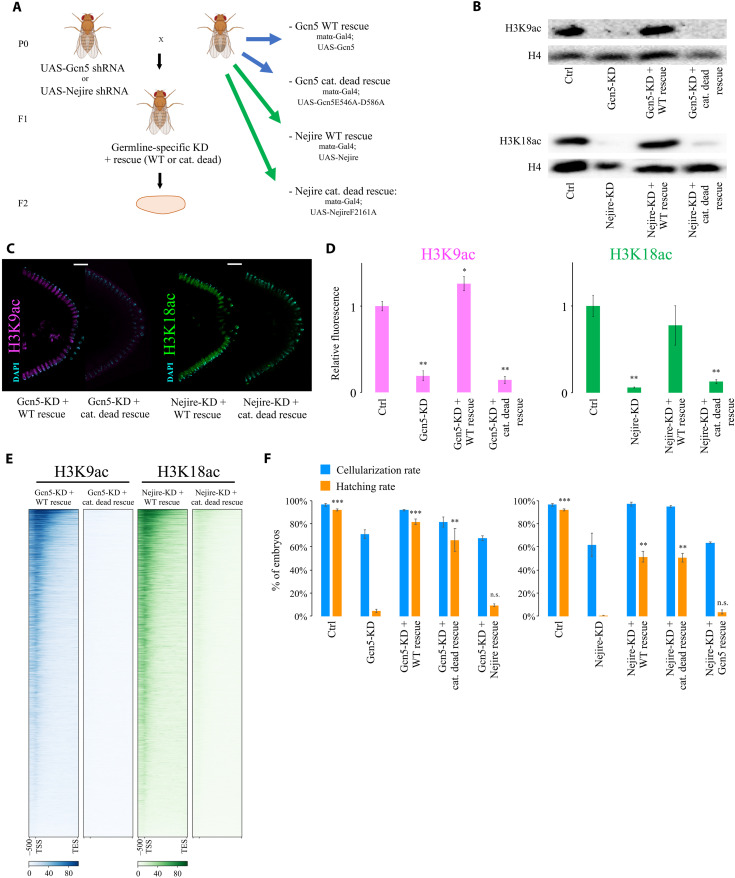
Considering the remarkable impact of Nejire, Gcn5, HDAC1, and HDAC3 on gene expression, we investigated their effects on histone acetylation during embryonic development. We used immunofluorescence to detect H3K9, H3K18, and H3K27 residues since their acetylation is considered to be key for the regulation of gene expression (43–45). Immunofluorescence analysis of Drosophila KD and control embryos at ZGA showed that Gcn5 is the only HAT required for the deposition of bulk H3K9ac at this stage. Conversely, Nejire is required for the deposition of H3K18ac and H3K27ac (Fig. 3, A and B). These results confirmed the substrate preferences of these enzymes observed in different developmental contexts (46). Lack of Enok had no major impact on bulk levels of H3K9ac and H3K18ac, but it has a slightly negative effect on H3K27ac (Fig. 3, A and B). As previously shown (34), Enok is responsible for H3K23ac deposition at ZGA (fig. S4A). Notably, loss of maternal HDAC1 had a strong impact on H3K9ac, H3K18ac, and H3K27ac levels, showing up-regulation of 4-, 2.6-, and 2.4-fold, respectively (Fig. 3B). In contrast, no major change in those marks is seen upon removal of HDAC3 or HDAC6 (Fig. 3, A and B). Western blot assays performed at ZGA fully recapitulated the results observed by immunofluorescence (Fig. 3C).
Fig. 3. Nejire, Gcn5, and HDAC1 regulate H3K9ac, H3K18ac, and H3K27ac levels during ZGA.
(A) Representative immunofluorescence pictures of ZGA (stage 5) control and KD embryos. From top to bottom: anti-H3K9ac staining (magenta), anti-H3K18 staining (green), and anti-H3K27ac staining (red) are coupled with DAPI staining (cyan). For each staining, posterior sides of ZGA embryos are displayed on the top (scale bars, 20 μm), and three nuclei are displayed on the bottom (scale bars, 5 μm). (B) Quantification of the immunofluorescence signal for anti-H3K9ac staining (magenta), anti-H3K18 staining (green), and anti-H3K27ac staining (red) in embryos at ZGA. Data represent the mean of n = 4 stage 5 embryos of DAPI-normalized immunofluorescence signals for each staining, compared to control embryos. Every data point consists of the mean signal obtained from five cross-sectional nuclei. Error bars represent the SD of the four independent embryos (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001; paired, two-tailed Student’s t test). (C) Western blot assays were performed with total protein extracts from ZGA embryos. H3K9ac, H3K18ac, and H3K27ac signals are compared with total H4 signal from the same blot. (D) Representative immunofluorescence pictures of interphase nuclei from cycle 7 to cycle 13 of embryogenesis. Scale bar, 5 μm. On the top, control embryos and Gcn5-KD embryos stained with anti-H3K9ac (magenta) and DAPI (cyan). On the bottom, control embryos stained with anti-H3K18ac (green) and DAPI; control embryos stained with anti-H3K27ac (red) and DAPI (cyan); Nejire-KD embryos stained with anti-H3K18ac (green), anti-H3K27ac (red), and DAPI (cyan).
H3K18ac and H3K27ac are already present on every interphase chromosome at cycle 8 (Fig. 3D), as previously reported (47). We could even detect low levels of these marks on mitotic chromosomes in cycles 3 and 4, respectively. In contrast, the H3K9ac mark could only be detected on mitotic chromosomes at cycle 8, just before peripheral nuclear migration (fig. S4B). Interphase nuclei displayed H3K9ac at cycle 11 (Fig. 3D), earlier than previously reported (47). We speculate that earlier detection of these marks on mitotic chromosomes compared to interphase chromosomes is simply due to the higher compaction that could facilitate the detection of low-abundant chromatin marks.
Together, these results indicate that during early embryogenesis (pre-ZGA and ZGA) H3K9ac specifically requires Gcn5, whereas H3K18ac and H3K27ac require Nejire. HDAC1 is responsible for the deacetylation of these residues, whereas the lack of HDAC3 and HDAC6 has no global impact on these marks. Although we cannot rule out that these marks appear even earlier in development, our data demonstrate that their deposition precedes ZGA (29, 30).
Nejire and Gcn5 deposit H3K9ac, H3K18ac, and H3K27ac on all actively transcribed genes at ZGA
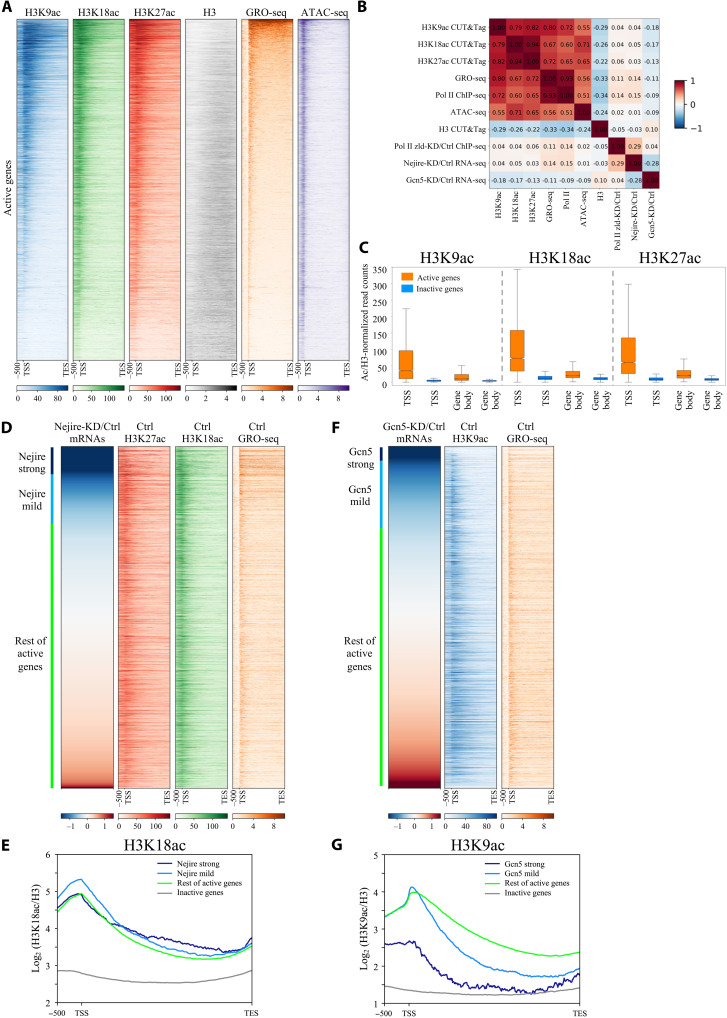
Quantitative CUT&Tag experiments on hand-sorted embryos at ZGA showed similar enrichment patterns for H3K18ac and H3K27ac on all the actively transcribed genes (R = 0.94), defined by global run-on sequencing (GRO-seq) in fig. S2A (15), and for H3K27ac and H3K9ac (R = 0.82) (Fig. 4, A and B, and fig. S5, A and B). On active genes, H3K18ac and H3K27ac tend to peak in the proximity of Transcription Start Sites (TSSs), while H3K9ac mark peaks are found more downstream [mean value = 217 base pairs (bp)] (fig. S5C). Moreover, H3K18ac and H3K27ac marks spread upstream of the TSS and inside the gene body of their target genes more frequently than H3K9ac (Fig. 4A). In contrast to active genes, inactive genes are devoid of H3K9ac, H3K18ac, and H3K27ac marks (Fig. 4C and fig. S5E). H3 acetylation levels on those residues could be used as a strong predictor of transcriptional activity, particularly on TSSs (Fig. 4C). The enrichment of the histone modifications mirrors the binding of Nejire and the SAGA component Ada2b during ZGA from previous reports (fig. S5F) (35, 48). Nejire-KD and Gcn5-KD caused complete loss of the corresponding acetylation marks on all the active genes (fig. S6A), in agreement with the results obtained on bulk acetylation levels at ZGA (Fig. 3, A to C) and confirming the specificity of our KD strategy. Together, these data show that transcriptionally active genes are enriched with H3K9ac, H3K18ac, and H3K27ac, whereas inactive genes are depleted of these marks.
Fig. 4. Nejire and Gcn5 deposit their acetylation marks on every active gene, independently of their transcriptional coactivator activities.
(A) Heatmaps of ZGA active genes spanning from 500-bp upstream of TSS to TES. Genes are ranked by GRO-seq signal in column 5 (orange). Columns represent wild-type embryos H3K9ac CUT&Tag (blue), H3K18ac CUT&Tag (green), H3K27ac CUT&Tag (red), H3 CUT&Tag (gray), GRO-seq (orange) (15), and ATAC-seq (purple) (15) (n = 2 biological replicates). (B) Spearman’s correlation heatmap matrix comparing signal on gene body across multiple datasets on active genes in ZGA embryos. Numbers indicate R values. (C) Box plots comparing regions of active versus inactive genes at ZGA for H3K9ac, H3K18ac, and H3K27ac marks, normalized on H3. Boxes extend from 25th to 75th percentile of the data, and the whiskers span across the whole data range. TSS regions are defined as 400 bp surrounding TSS. Gene body regions are defined as 200-bp downstream of the TSS to TES. (D and F) Heatmaps of active genes at ZGA. Genes are ranked according to the ratio of RNA-seq levels of wild-type over Nejire-KD (D) or Gcn5-KD (F). Column color code corresponds to (A). Strong Nejire (D) or strong Gcn5 targets (F) (log2FC < −1.5) are marked in dark blue, mild Nejire (D) or mild Gcn5 targets (F) (−0.5 < log2FC < −1.5) are marked in light blue, and the rest of active genes are marked in green. (E and G) Metagene profiles of the H3K18ac signal (E) or H3K9ac (G) signal, normalized to H3, on strong Nejire (E) or strong Gcn5 (G) targets (dark blue), mild Nejire (E) or mild Gcn5 (G) targets (light blue), the rest of active genes (green), and inactive genes (orange) at ZGA. Numbers on y axis indicate the mean coverage of log2 (H3K18ac/H3) (E) or (H3K9ac/H3) (G).
H3K9ac, H3K18ac, and H3K27ac levels are not predictive of Gcn5- or Nejire-mediated gene regulation
Nejire-dependent genes show virtually no correlation with the enrichment of H3K18ac (R = 0.05) and H3K27ac (R = 0.03) marks (Fig. 4B). The actively transcribed genes affected by the lack of Nejire show only slightly higher levels of H3K18ac on their TSSs and gene bodies than the actively transcribed genes that are not affected by Nejire-KD (Fig. 4, D and E). Gcn5-dependent genes also show no positive correlation with the enrichment of the mark H3K9ac (R = −0.18) (Fig. 4B). The genes that are mostly affected by Gcn5-KD show significantly lower levels of H3K9ac on their TSSs and background levels on their gene bodies when compared to the active genes not affected by Gcn5-KD (Fig. 4, F and G). Accordingly, among the top 500 genes down-regulated genes in Gcn5-KD, only 9 genes overlap with the top 500 genes enriched for H3K9ac on their gene body, and only 29 genes overlap with the top 500 genes enriched for H3K9ac at their TSSs (fig. S6, B and C). Genome browser views of the Nejire-dependent gene wntD and the Gcn5-dependent gene Gclm highlight the lack of correlation between the histone modifications and the transcriptional down-regulation caused by the KD of either HAT (fig. S6D).
In conclusion, Nejire-dependent genes do not show higher levels of H3K18ac and H3K27ac than the rest of all active genes, whereas Gcn5-dependent genes show even lower levels of H3K9ac than the rest of all active genes. Therefore, enrichment of H3K9ac, H3K18ac, and H3K27ac are not predictive of Gcn5- or Nejire-mediated gene regulation, but they are predictive of Pol II enrichment.
Nejire and Gcn5 catalytic activities are not required for embryonic viability
To evaluate the specificity of the observed phenotypes, we coupled germline-specific KD of Gcn5 or Nejire with germline expression of rescue constructs that are resistant to KD. Intrigued by the lack of correlation between histone acetylation levels and transcriptional dependence on Gcn5 or Nejire, we included catalytically dead mutants for either Gcn5 (Gcn5E546A-D586A) or Nejire (NejireF2161A) in addition to wild-type constructs (Fig. 5A and fig. S7A) (49). Western blot and immunofluorescence experiments showed that wild-type Gcn5 and Nejire rescued H3K9ac and H3K18ac levels, respectively, whereas the catalytically dead mutants did not (Fig. 5, B to D). Quantitative CUT&Tag assays showed an almost complete loss of H3K9ac and H3K18ac marks on actively transcribed genes in embryos expressing the catalytically dead mutants (Fig. 5E). Nejire and Gcn5 bulk protein levels and their nuclear localizations showed no differences between wild-type and catalytically dead rescued embryos (fig. S7, B and C). CUT&Tag experiments confirmed that Nejire and Gcn5 proteins could bind at the same genomic sites when comparing wild-type with the catalytically dead version of the rescues (fig. S7D). However, both Nejire rescues could not fully restore wild-type Nejire protein levels (fig. S7, B and C).
Fig. 5. Nejire and Gcn5 catalytic activities are not required for embryonic development.
(A) Crossing scheme of the genetic rescue experiment. Maternal KD of Gcn5 or Nejire is rescued by maternal expression of either wild-type or catalytically dead Gcn5 or Nejire. Fly images were obained from Biorender.com. (B) Western blot assays performed on total protein extracts from ZGA embryos. Total H4 signal serves as loading control. (C) Representative immunofluorescence of posterior parts of embryos at ZGA. Samples are stained for H3K9ac (magenta; left), H3K18 staining (green; right), and DAPI staining (cyan). Scale bars, 20 μm. (D) Quantification of the immunofluorescence signal for anti-H3K9ac staining (magenta) on the left and anti-H3K18 staining (green) on the right. Data represent the mean of n = 3 stage 5 embryos of DAPI-normalized immunofluorescence signals for each staining, compared to control embryos. Every data point consists of the mean signal obtained from five cross-sectional nuclei. Error bars represent the SD of the four independent embryos (*P ≤ 0.05 and **P ≤ 0.01; paired, two-tailed Student’s t test). (E) Heatmaps of active genes spanning 2-kb upstream of TSS to the TES of the H3K9ac CUT&Tag signal (blue; left) and H3K18ac CUT&Tag (green; right) performed at ZGA (n = 2 biological replicates of independent embryo collections). Columns represent embryos rescued with wild-type or the catalytically dead mutant of Gcn5 (left) and Nejire (right). (F) Mean values of CR (blue) and HR (orange) in control, KD, and rescue F2 embryos from n = 3 biological replicates of independent embryo collections. Bars represent the SD. Paired, two-tailed Student’s t test was applied to compare the HR with Gcn5-KD (left chart) or with Nejire-KD (right chart) (**P ≤ 0.01 and ***P ≤ 0.001). n.s., not significant.
Notably, however, both mutants were able to rescue the embryonic lethality phenotype caused by maternal depletion of Gcn5 and Nejire. Specifically, the HRs of the Gcn5 wild-type and the embryos rescued with the Gcn5 catalytically dead mutant were 81.7 and 66%, respectively. This was significantly higher than the HR of 4.7% observed in Gcn5-KD embryos. HRs of embryos rescued with Nejire wild-type and catalytically dead mutant were 51.3 and 50.7%, respectively, compared to 0.3% in Nejire-KD embryos (Fig. 5F). Nejire-rescued embryos (100%) showed a wild-type phenotype at stage 14 of embryogenesis (fig. S7E) and the fraction of embryos that did not hatch reached later stages of embryogenesis (fig. S7F). Lower HRs in both Nejire rescues relative to the control cross could be due to suboptimal dosage of Nejire proteins upon rescues (fig. S7, B and C).
Nejire and Gcn5 catalytic activities are not required for transcription during ZGA and in vitro reporter transactivation
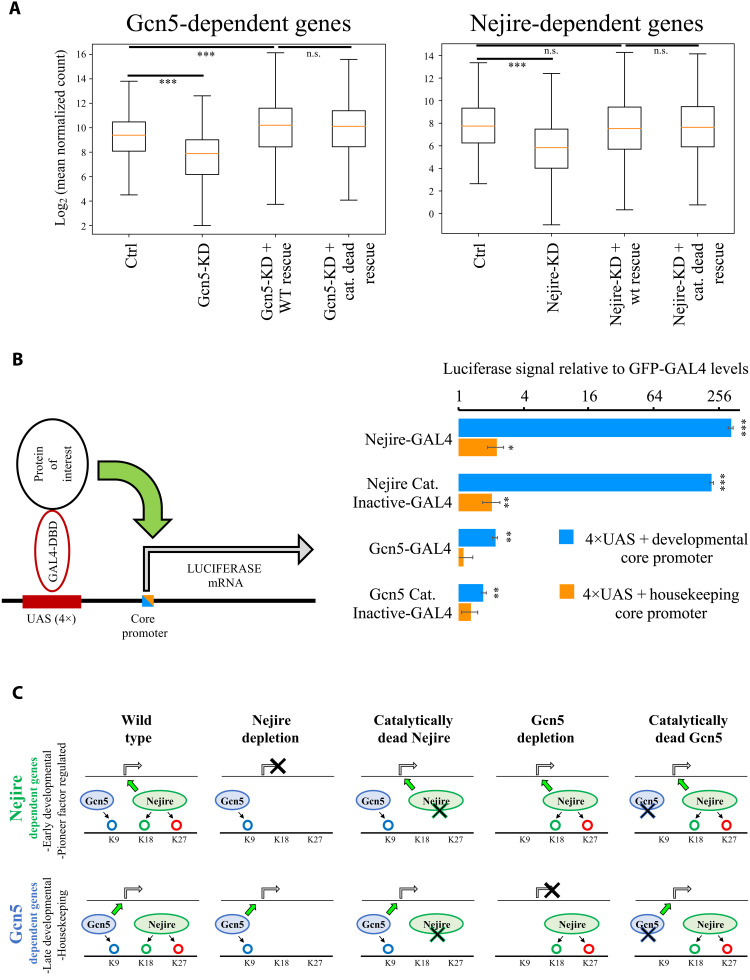
Considering the high levels of phenotypic rescue obtained with catalytically dead mutants, we assayed their impact on gene expression at ZGA. Despite the specific loss of acetylation obtained in catalytically dead mutants (Fig. 5, B to E), RNA-seq experiments showed no substantial differences between transcriptomes of embryos rescued with wild-type Gcn5 or the catalytically dead version. Only 12 genes showed significant misregulation (log2FC < −1 or log2FC > 1; P <0.05) (fig. S7G). Moreover, the expression of the majority of significantly down-regulated genes under Gcn5-KD conditions could be restored through the expression of both wild-type Gcn5 and the catalytically dead mutant (Fig. 6A). Similarly, only four genes showed significant misregulation (log2FC < −1 or log2FC > 1; P <0.05) between embryos rescued by expression of Nejire wild-type and the catalytically dead mutant (fig. S7G) and the average expression of Nejire-dependent genes could be restored (Fig. 6A). The Nejire wild-type rescue did not restore the expression of all the down-regulated genes in Nejire-KD (329 of 689), which we attributed to the lower protein levels of Nejire in the rescue experiments (fig. S7, B and C). Gcn5 wild-type rescue restored the complete protein levels of Gcn5 and showed a full transcriptional rescue (fig. S7, B and C).
Fig. 6. Nejire and Gcn5 catalytic activities are not required for ZGA and for in vitro reporter transactivation.
(A) Box plots of log2 mean expression of n = 526 Gcn5-dependent genes (adjusted P ≤ 0.05 and log2FC < −1 in Gcn5-KD; left) or n = 689 Nejire-dependent genes (adjusted P ≤ 0.05 and log2FC < −1; right) versus Ctrl RNA-seq, Fig. 2. The orange line corresponds to median gene expression, box delimitates 25th to 75th percentiles, and whiskers encompass the whole range of expression (***P ≤ 0.001; paired, two-tailed Student’s t test with Bonferroni correction). (B) On the left, a schematic model of the UAS-driven Luciferase-reporter expression system in S2 cells. A minimal core promoter is combined with four UAS sites. The luciferase reporter gene is transactivated by the GAL4 DNA binding domain (DBD) fused with the protein of interest. On the right, the results of firefly luciferase transactivation upon recruitment of wild-type or catalytically dead mutants of Nejire or Gcn5, using a developmental core promoter (blue) or a housekeeping core promoter (orange). The firefly luciferase signal is normalized to the renilla luciferase signal expressed from a constitutive promoter to control for transfection efficiency. Signals are further normalized to green fluorescent protein (GFP)–GAL4-DBD as negative control. For each independent transfection replicate, three technical measurement replicates were averaged. Data are represented as mean and SD of n = 3 transfection replicates. Paired, two-tailed Student’s t test was applied to compare the GFP–GAL4-DBD relative firefly/renilla luciferase levels with the other conditions (*P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001). (C) Model displaying the effect of Nejire and Gcn5 on their target genes and H3 substrates during ZGA. n.s., not significant.
Experiments performed in S2 cells confirmed that the catalytic activities of Gcn5 and Nejire are not required to transactivate a reporter gene upon artificial tethering of either HAT to its promoter. Recruitment of wild-type Nejire and catalytically dead Nejire to a UAS-driven luciferase reporter with a developmental core promoter led to strong increases in luciferase expression (328-fold increase and 218-fold increase, respectively), in line with earlier observations (50). Recruitment of Gcn5 only moderately induced reporter transactivation (Fig. 6B). We repeated this experiment using a luciferase combined with a housekeeping core promoter and confirmed that the catalytic activity of either HAT is not required for transcriptional activation. Notably, in line with our observation that Nejire tends to target developmental genes (Fig. 2, A and C, and figs. S2E and S3, B and E), Nejire-dependent transactivation is strongly reduced in the reporter that is dependent on the housekeeping core promoter (Fig. 6B). Last, we repeated the in vitro assay using truncated versions of Gcn5 and Nejire proteins. Removal of individual domains in Gcn5 protein did not impair the transactivation activity of Gcn5. The transactivation activity was only mildly affected by the removal of the C-terminal part of Gcn5, including the bromodomain (fig. S8A). Conversely, the lack of the N-terminal domain of Nejire protein, which contains a TAZ zinc finger domain and a KIX domain (fig. S8B), resulted in a marked 94% decrease in Nejire transactivation power (fig. S8B). Significant decreases could also be observed in the ΔBROMO and the ΔHAT, showing a 60% drop compared to full-length Nejire protein transactivation levels (fig. S8B). Nonetheless, these two Nejire versions could still transactivate the luciferase reporter by more than 100-fold compared to the negative control. We cannot exclude that the disruption of protein domains could also result in unstable proteins, which could be the cause of reduced transactivation.
In summary, Gcn5 and Nejire catalytic activities are not required for the activation of their target genes during ZGA. Moreover, catalytically dead versions of both HATs did not show any dominant negative effect on the embryonic transcriptome at ZGA. Therefore, the deposition of H3K9ac, H3K18ac, and H3K27ac are dispensable for the activation of target gene expression at this developmental stage, although these marks are present on virtually every active TSS (Fig. 6C).
DISCUSSION
Our study demonstrates the strong impact of chromatin factors and, in particular, of HATs and HDACs on ZGA and embryonic development. The RNA interference (RNAi)–based reverse genetic screen identified proteins involved in histone acetylation as top hits. It confirmed the importance of Polycomb group proteins, such as E(z), Psc, and Scm (29, 33), transcriptional corepressors, such as CtBP (51, 52) and Grunge (53), and the histone variant BigH1 (32).
We selected Nejire, the Drosophila ortholog of human CBP/EP300 and Gcn5, the catalytic subunit of SAGA and ATAC transcriptional coactivator complexes (54, 55), as well as HDAC1 and HDAC3 for further investigation. We showed that these chromatin modifiers control different aspects of ZGA. Specifically, Nejire is required for the expression of genes with early developmental functions, such as pattern formation, cell fate specification, and gastrulation. These genes are often coregulated by pioneer factors such as Zelda, Clamp, and GAF, suggesting that these pioneer factors might cooperate with Nejire to exert their functions. An interplay between CBP/p300 and pioneer factors was recently described in zebrafish at ZGA (56). Gcn5, on the other hand, is responsible for activating a completely different set of genes. Many of these Gcn5 targets can be defined as housekeeping genes, while others are developmental genes that are important for later stages of embryogenesis. These data clearly indicate a “division of labor” between these two crucial transcriptional coactivators during ZGA.
Immunofluorescence, Western blot, and CUT&Tag assays revealed substrate specificities of Nejire toward acetylation of H3K18 and H3K27 and of Gcn5 toward H3K9. This substrate specificity observed during ZGA confirmed previous results obtained in vitro (46), during later stages of Drosophila development (57), and in mammalian cell lines as well (58). Furthermore, HDAC1 is responsible for the removal of H3K9ac, H3K18ac, and H3K27ac marks during ZGA, whereas HDAC3 and HDAC6 do not seem to affect bulk acetylation levels on these residues consistently. They may play locus-specific roles instead, or their impact on ZGA might be mostly catalytically independent as well.
We found that H3K9ac, H3K18ac, and H3K27ac are enriched on virtually every active gene at ZGA. With good approximation, H3K9ac, H3K18ac, and H3K27ac enrichment levels mirror Pol II transcriptional activity levels, being more abundant on highly transcribed genes. Inactive genes are depleted of these acetylation marks. Published chromatin immunoprecipitation (ChIP) data of Nejire and the SAGA component Ada2b at ZGA (35, 48) confirmed that these proteins are present on every active TSS. Moreover, Nejire decorates every active TSS in S2 cells (40). Likewise, H3K9ac enrichments were found on every active gene in mammals (59). At least for some genes, Nejire and Gcn5 colocalization did not result in cooperation, as the lack of one HAT could not be compensated by the presence of the other. Nonetheless, many genes that did not respond to the depletion of either HAT might be redundantly activated by them. Furthermore, we confirmed previous studies showing that these acetylation marks can be detected to some extent even before the major wave of ZGA in flies (13), as well as in the transcriptionally inactive mammalian zygote (60). In particular, we could detect Nejire-dependent marks during the first nuclear divisions (cycles 3 and 4), whereas the Gcn5-dependent H3K9ac mark could be detected at the beginning of the first wave of ZGA (cycle 8). Their presence could either reflect the transcriptional activity of very early genes or indicate that the binding of Nejire and SAGA/ATAC complexes precedes target gene transcription. Given the high degree of overlap between Nejire-dependent with Zelda-dependent and GAF-dependent genes, we speculate that the early presence of Nejire on chromatin could be important for the later activity of pioneer factors during ZGA, as recently described in other systems (60).
To our surprise, we found that the individual catalytic activities of either Nejire or Gcn5 are not required for gene expression during ZGA, in line with a growing number of studies in both flies (49, 61–70) and other organisms (65, 71–78), reviewed here (79).
We conclude that, at least for the activation of early embryonic genes, the lack of either H3K9ac alone or lack of H3K18ac and H3K27ac marks is not necessary for Pol II transcriptional activity under our experimental conditions. However, we cannot rule out compensation from other histone acetylation marks. Double and multiple HATs catalytic dead mutants will be required in the future to clarify this conundrum. In addition, we do not rule out the possibility of causative transcriptional functions of these acetylation marks during later stages of embryonic development, where some late-embryonically expressed genes might benefit from Nejire and/or Gcn5 catalytic activities.
Mechanistically, H3K9ac, H3K18ac, and H3K27ac deposition could either precede Pol II transcription without affecting Pol II function or be a mere consequence of transcriptional activity. In both cases, the mere presence of the HAT itself would be a determinant of transcriptional activity at this developmental stage. Alternatively, other acetylated histone lysines or residual enzymatic activity left on Nejire and Gcn5 catalytic dead constructs would suffice and compensate for Pol II function. However, given the different transcriptional targets of Nejire and Gcn5 during ZGA, a potential redundant role for HATs in the deposition of acetylation marks on some genes could not restore the expression of many other genes, which still require the presence of a specific HAT for their expression. Last, it is also possible that histone acetylations could have a more critical effect on the transcriptional output in later developmental stages (57), where their presence could represent a limiting factor for Pol II activity.
Our experiments were performed in controlled environmental settings, so we cannot rule out that the presence of these modifications might have a role in buffering the environmental conditions or the underlying genetic variability on the transcriptional output, as proposed for H3K4me1 (65). It is possible that histone acetylation could act as a “phenotypic capacitor” by buffering cryptic genetic variation (80), which cannot be assessed in a highly inbred laboratory fly strain. Testing HATs’ catalytic activities under genetic and environmental stressful conditions would elucidate their potential role in transcriptional robustness.
Previous studies challenging the enzymatic function of HATs in flies were conceived using a histone replacement system (66–68). The caveat was represented by the role that nonhistone substrates could play in stimulating Pol II activity. By using catalytically dead versions of Nejire and Gcn5, we could extend our conclusions beyond the role of histone acetylations, as previously done for Trr (65), and expand its significance to any other HATs’ substrate.
The main conclusion of our study is that the mere presence of Nejire and Gcn5, but not their catalytic activities, is important to exert their functions as transcriptional coactivators at the time of ZGA. The massive size of the Nejire protein (332 kDa) (37), the relatively high expression levels of its mRNA during ZGA (81), and its genomic binding on every active gene (40, 48) could suggest a structural role of this protein in regulating Pol II activity. Moreover, the full penetrance of the lethality phenotype, which we obtain despite a suboptimal KD efficiency, makes us speculate that probably a given threshold level of Nejire protein is necessary for the formation of higher-order chromatin structures. Moreover, our results indicate that the main function of the Gcn5-containing multiprotein complexes (i.e., SAGA and ATAC) (54, 55) during ZGA transcriptional activation would go beyond Gcn5 catalytic activity. As for Nejire, their structural role would be pivotal for Pol II activity at this developmental stage instead of their acetyltransferase function (70, 77, 78). Since our work was performed on whole embryos, future work at the single-cell level will be required to assess whether the lack of catalytic activity affects the transcription level at the single cell. In addition, histone acetylation encompasses several histone residues that might act redundantly; therefore, in the future, multiple combing catalytic dead mutants will be required to assess redundancy.
Overall, our work reveals the importance of chromatin factors in regulating ZGA, complementing the role of pioneer factors in this process. We show that Nejire and Gcn5 orchestrate ZGA but that their catalytic activity is not necessary for this function.
MATERIALS AND METHODS
Fly stocks
All the stocks used in this study were grown on corn flour molasses food (12 g of agar, 18 g of dry yeast, 10 g of soya powder, 22 g of molasses, 80 g of malt extract, 80 g of corn flour, 6.25 ml of propionic acid, and 2.4 g of nipagin per liter of water) at 28.5°C for KD experiments or at 25°C for rescue experiments. Fly stocks used in this study: TRiP line control (BDSC, #36303), female germline-specific Gal4-driver P{matα4-GAL-VP16}V37 (BDSC, #7063), Nejire shRNA#1 (homemade line), Nejire shRNA#2 (BDSC, #36682) Gcn5 shRNA (BDSC, #35601), Enok shRNA (BDSC, #41664), HDAC1 shRNA (BDSC, #33725), HDAC3 shRNA (BDSC, #34778), and HDAC6 shRNA (BDSC, #34072). A complete list of the fly stocks used for the reverse genetic screen is available in table S1. The stocks used for the catalytically wild-type and catalytically dead rescue experiments are the following: (i) w; pUASp>Flag-HA::Gcn5(shRNA-site_mutant); Gal4-driver P{matα4-GAL-VP16}V37, (ii) w: pUASp> Flag-HA::Gcn5E546A,D586A(shRNA-site_mutant); Gal4-driver P{matα4-GAL-VP16}V37, (iii) w; pUASp>Nejire::Flag-HA(shRNA-site_mutant); Gal4-driver P{matα4-GAL-VP16}V37, and (iv) w; pUASP>NejireF2161A::Flag-HA(shRNA-site_mutant); Gal4-driver P{matα4-GAL-VP16}V37. Rescue experiments crosses were performed using F1 females older than 12 days.
Reverse genetic screen
Maternal RNAi KD was induced for each of the 126 candidates, which were selected among well-established chromatin factors. A total of 114 UAS-shRNA lines (V20, V21, or V22 TRiP line) and 12 UAS-dsRNA lines (KK, GD VDRC line, or V10 TRiP line) were selected to collect virgin females to be crossed with germline-specific Gal4-driver (P{matα4-GAL-VP16}V37, BDSC, #7063) carrying males at 28.5°C. The progenies of these crosses (F1 generation) are KD for a specific target in the female germline and will lay eggs (F2 generation) that are depleted of this particular transcript during the first 4characterization. Briefly, 120 F2 embryos were randomly picked and aligned on an agar plate. Egg morphology was scored as wild-type if no eggs displayed any morphological defects or as “abnormal” if some or all the eggs displayed any kind of visible morphological defects. KD lines that did not lay eggs or that were laying eggs that showed morphological defects were excluded to avoid germline defects. Embryos were considered “cellularized” when they reached stage 5 of embryogenesis (nuclear cycle 14), showing a clear thick rim on the embryo periphery and complete invagination of cellular membranes. CR was scored as the number of cellularized embryos on the total number of aligned embryos. After 25 hours from alignment, embryos were considered “hatched” when they reached the L1 larval stage. The HR was calculated with the average of three biological replicates. PCLR was defined as (CR − HR) / (CR + 0.00001). PCLR indicates the lethality rate of the embryos that have gone through cellularization. Positive hits of the screen had to show (i) wild-type egg morphology, (ii) CR ≥ 5%, and (iii) PCLR ≥ 50%.
Embryo immunofluorescence
Time collected embryos were dechorionated in 50% bleach for 2 min, transferred to 0.7 ml of heptane, and crosslinked with 0.7 ml of ice-cold MetOH. Embryos were fixed for 1 min shaking vigorously on a vortex mixer. Devitellinized and fixed embryos were recovered from the bottom of the tube and washed three times with ice-cold MetOH and stored at −20°C. Embryos were rehydrated and permeabilized with 3× 20-min washes at room temperature (RT) on a rotating wheel in 1 ml of PBST×0.2% [1× phosphate-buffered saline (PBS) with 0.2% Triton X-100] and then blocked 1 hour at RT on a rotating wheel in 1 ml of PB1T×0.2% [1× PBS, 0.2% Triton X-100, and 1% bovine serum albumin (BSA)]. Embryos were stained overnight at 4°C in 200 μl of PB1T×0.2% with 1 μl of primary antibody. The next day, embryos were washed 4× for 10 min at RT on a rotating wheel in 1 ml of PBST×0.2% and then incubated for 2 hours at RT on a rotating wheel in 0.5 ml of PB1T×0.2% and Alexa Fluor (Thermo Fisher Scientific) secondary antibody (1:500) and 4′,6-diamidino-2-phenylindole (DAPI) solution (Sigma-Aldrich, D9542) to a final concentration of 10 μg/ml. After 4× washes of 10 min at RT on a rotating wheel in 1 ml of PBST×0.2%, embryos were mounted on microscope slides (76 mm by 26 mm by 1 mm; Marienfeld) and precision coverslips (22 mm by 22 mm; Marienfeld) with two drops of VECTASHIELD Antifade Mounting Medium (H-1000-140). All images were acquired using the confocal laser scanning microscopes, Zeiss LSM 880 Airyscan. Stacks were assembled using ImageJ or Imaris 9.5.1 (Bitplane). For all antibodies used, see table S2.
Western blot assay
One hundred ZGA (stage 5) embryos were hand-picked using halocarbon oil 27 (Sigma-Aldrich, H8773) and a stereoscope equipped with transmitted light. Embryos were collected in 50 μl of Laemmli buffer, homogenized with pestle, and stored at −20°C. Western blot assays were performed as in (31).
Total RNA extraction and RNA-seq
For each biological replicate, 20 hand-sorted pre-ZGA (stage 2) or ZGA (stage 5) embryos were collected in 50 μl of TRIzol (Thermo Fisher Scientific, 15596026), homogenized with pestle, and stored at −20°C. Total RNA was then extracted in 500 μl of TRIzol mixed vigorously for 15 s with 100 μl of chloroform. Samples were centrifuged at 12,000g for 15 min at 4°C, and the aqueous phase was mixed again with an equal volume of 300 μl of chloroform and centrifuged again at 12,000g for 15 min at 4°C. The aqueous phase was mixed with 300 μl of isopropanol and 20 μg of glycogen, and total RNA was let precipitate overnight at −20°C. The day after, samples were centrifuged at 12,000g for 30 min at 4°C, and the RNA pellet was washed twice with 80% ethanol. Air-dried RNA pellets were resuspended in 20 μl of TURBO deoxyribonuclease (DNase) solution (Invitrogen, AM2238) and incubated for 30 min at 37°C according to the manufacturer’s instructions. EDTA was added to a final concentration of 15 mM, and DNase was heat-inactivated at 75°C for 10 min. Total RNA was then quantified, and cDNA libraries were prepared with Illumina Stranded Total RNA-seq kit with Ribo-zero Plus according to the manufacturer’s instructions. cDNA libraries were then submitted for deep sequencing as in (31).
Nuclear RNA extraction and RNA-seq
Three hundred ZGA (stage 5) hand-sorted embryos were collected in 1.8 ml of nuclei wash buffer [H2O, 10 nM tris-HCl (pH 6.8), 10 nM NaCl, 3 mM MgCl2, 0.5% BSA, 0.01% Tween 20 (Sigma-Aldrich, 1003173910), 0.5 mM spermine, 0.5 mM spermidine, and RNaseOUT (0.1 U/μl; Thermo Fisher Scientific, 10777019)] at 4°C and gently spun at 1000g for 3 min at 4°C. Embryos were then resuspended in 0.1× NP buffer [H2O, 10 nM tris-HCl (pH 6.8), 10 nM NaCl, 3 mM MgCl2, 0.01% NP-40 (IGEPAL CA-630, Sigma-Aldrich, I3021), 0.01% Tween 20 (Thermo Scientific Pierce, 62249), 0.5 mM spermine, 0.5 mM spermidine, and RNaseOUT (0.1 U/μl; Thermo Fisher Scientific, 10777019)] and transferred in a glass douncer (1 ml; Wheaton, 358103), and five gentle strokes were applied. The homogenized samples were transferred in a protein low binding tube (Eppendorf Protein LoBind tubes, EP0030108116), passed 4× through a syringe (22-gauge × 1 1/4″ tip, BD Microlance, 300900), and lysed for 5 min at RT. Samples were then diluted 1:10 in nuclei wash buffer, gentle spun at 750g for 5 min at 4°C, and washed with 1 ml of nuclei wash buffer. Nuclei were then filtered with 10-μm filter (CellTrics, 04-004-2324), spun at 750g for 5 min at 4°C, and resuspended three times in nuclei wash buffer. Nuclei pellet was then resuspended and homogenized in 50 μl of TRIzol and stored at −20°C. Nuclear RNA was then extracted with the same procedure of total RNA (see above), with the addition of 1 μl of 1:1000 diluted ERCC RNA spike-in (Ambion, 4456740) added with the remaining 450 μl of TRIzol before chloroform extraction, to normalize for a potential global variation of RNA quantity/nucleus upon KD. Extracted nuclear RNA was then submitted for library preparation, followed by deep sequencing as described above.
Reverse transcription and RT-qPCR
Total RNA was extracted as described above from 20 pre-ZGA (stage 2) or ZGA (stage 5) embryos. After digestion in TURBO DNase solution (Invitrogen, AM2238) for 30 min at 37°C, according to the manufacturer’s instructions, samples were quantified, and 500 ng of total RNA was retrotranscribed with the First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, K1612) using a 1:1 mix of oligo(dT)18 and random hexamers, according to the manufacturer’s instructions. cDNA was then diluted 1:10 and then used for real-time qPCR reactions, which were performed as described in (31).
CUT&Tag
Embryos were dechorionated in 50% bleach for 2 min, transferred to 7 ml of heptane, and crosslinked with 5 ml of fixative. The fixative consisted of buffer A [60 mM KCl, 15 mM NaCl, 15 mM Hepes (pH 7.6), and 4 mM MgCl2] supplemented with 1.8% paraformaldehyde. Embryos were fixed for 15 min in an orbital shaker at maximum speed. The cross-linking was quenched by adding a final concentration of 225 mM glycine and rotation for 5 min. After washing with buffer A + 0.1% Triton X-100, the embryos were hand-staged on a cooling station under a microscope, shock-frozen, and stored at −80°C. For each CUT&Tag replicate, 50 ZGA (stage 5) embryos were used. CUT&Tag experiments were performed as in (82). DNA was purified and 1 pg of lambda phage genome (previously treated with Protein A/G–Tn5) was added to each sample as spike-in before library preparation. See table S2 for a list of antibodies used.
Total RNA data analysis
Preprocessing, mapping, and differential analysis
Samples of both Pre-ZGA (stage 2) and ZGA (stage 5) were processed using the mRNA pipeline of snakePipes v2.4.2. (83). As part of this pipeline, raw reads were mapped on dm6, count matrices were generated by featureCounts from subread-v2.0.0 (84) using fly annotation of Ensembl-release-96, and, last, differential analysis has been done by the pairwise comparison of knockout with control samples using DESeq2 v1.26.0 (85).
GO enrichment analysis
Enriched GO terms were found for each of the up- and down-regulated sets of genes after finding their intersect with active and filtering them for their P < 0.05 and their log2FC > 1 or < −1, respectively. GO enrichment analysis is done using the enrichGO function clusterprofiler v3.14.0 (86) looking for the terms in the biological process category. All GRO-seq + genes were used as background genes (universe), and the P value cutoff is set to 0.05.
Motif enrichment analysis
Analysis of motif enrichment is done using the AME tool of the MEME Suite v5.4.1 (87). Filtered up- and down-regulated sets of genes, as explained in the GO section, are used to look for this enrichment. Two different groups of motifs were searched for core promoter motifs and transcription factor motifs. To determine the enriched core promoter motifs, promoter regions were defined as 300-base upstream of each TSS up to 100-base downstream of each TSS. For each set of promoters, all genes minus the foreground have been considered as the background. A motif database based on the study of Haberle and Stark (88) along with the Pause Button from the study of Ramalingam et al. (89) used to search for their enrichment in our desired set of promoters. To detect the enriched transcription factor motifs, for each gene, a window of 1000-base upstream TSS up to the 500-base downstream, the TSS was chosen, and similar to the previous case, all genes minus this foreground genes were used as the background. As the motif database, we have used the JASPAR 2020 database (90).
Nuclear RNA data analysis
Making a hybrid reference genome
A hybrid genome containing dm6 and ERCC RNA spike-in (Ambion, 4456740) has been built using createIndices pipeline of snakePipes v2.5.1 (83) where ERCC92 genome is treated as the spiked-in genome. Annotation file was manually updated by appending the ERCC92 annotation to the fly annotation of Ensembl-release-96.
Preprocessing and mapping
Similar to the total RNA data, samples were processed using the mRNA pipeline of snakePipes v2.4.2. (83). As part of this pipeline, raw reads were mapped on the built hybrid reference genome of dm6 and ERCC RNA spike-in (Ambion, 4456740), and count matrices were generated by featureCounts from subread v2.0.0 (84) using fly annotation of Ensembl-release-96 along with ERCC92 genes.
Differential analysis
Differential analysis has been done by DESeq2 v1.26.0 (85) where size factors were estimated using ERCC92 genes as control genes.
CUT&Tag data analysis
Making a hybrid reference genome
A hybrid reference generated including dm6 and lambda genome. To build such a reference, createIndices pipeline of snakePipes v2.5.0 (83) is used where the lambda genome has been considered as the spiked-in genome.
Preprocessing and mapping
To map the data on the built hybrid reference, DNA-mapping pipeline of snakePipes v2.5.1 (83) is used with “--trim --fastqc --mapq 3 --dedup --properPairs --cutntag”. Furthermore, ChIP sequencing pipeline of snakePipes v2.5.1 (83) is used with “--cutntag --useSpikeInForNorm --getSizeFactorsFrom genome” to make the spike-in normalized coverage tracks. Heatmaps were generated using plotHeatmap from deeptools v3.5.1 (91). ultraheatmap v1.3.1 [L. Rabbani and M. Rauer, ultraheatmap version 1.3.1 (computer software); https://github.com/maxplanck-ie/ultraheatmap] has been used to map genes from RNA data to their corresponding TSS or gene body regions and appended the expression data to the CUT&Tag matrices. Genome browser tracks were made using pyGenomeTrack v3.5.1 (92).
Acknowledgments
We thank the Iovino laboratory, particularly N. Atinbayeva and D. Ibarra Morales for helpful discussions. We thank Y. Schwartz, L. Tora, and S. Vincent for the lengthy and helpful discussions; E. Ponzo for the initial characterization of the catalytically dead fly strains; N. E. Vorobyeva for anti-CBP antibody; A. N. Krasnov for anti-Gcn5 antibody; M. Erokhin and D. Chetverina for anti-GAF antibody; M. M. Harrison for anti-Zelda antibody (more details in table S2); G. Cavalli and A. Sparmann for crucial reading of the manuscript and discussion; and A. Stark for crucial reading of the manuscript, helpful discussion, and helping with the interpretation of the in vitro experiment results; The Bioinformatics, Deep Sequencing, Imaging and Fly facilities at the MPI-IE; We thank The Bloomington Drosophila Stock Center (NIH P40OD018537) and the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing fly stocks.
Funding: This work was supported by Max Planck Society (to F.Ci., F.Ca., F.Z., E.L., M.A.S., M.M., and N.I.), ERC CoG, EpiRIME grant agreement no. 819941 (to F.Ci. and M.M.), IMPRS program (to F.Ca., F.Z., E.L., and M.A.S.), MEDEP grant SFB992 (to L.R.), HFSP grant LT000926/2020-L (to V.L.), Deutsche Forschungsgemeinschaft–Project ID 192904750–CRC 992 Medical Epigenetics (to N.I.), Behrens-Weise Stiftung (to N.I.), EMBO YIP (to N.I.), CIBSS EXC-2189 (to N.I.), and European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 819941) ERC CoG, EpiRIME (to N.I.).
Author contributions: Conceptualization: F.Ci. and N.I. Methodology: F.Ci., F.Ca., F.Z., and N.I. Formal analysis: F.Ci. and L.R. Investigation: F.Ci., L.R., F.Ca., F.Z., E.L., M.A.S., M.M., and V.L. Resources: N.I. Software: L.R. Data curation: F.Ci. and L.R. Visualization: F.Ci. and L.R. Funding acquisition: N.I. Writing: F.Ci. and N.I.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials [GEO ID: GSE207019 (RNA-seq and CUT&Tag data)].
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 and S2
REFERENCES AND NOTES
- 1.D. C. Hamm, M. M. Harrison, Regulatory principles governing the maternal-to-zygotic transition: Insights from Drosophila melanogaster. Open Biol. 8, 180183 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.M. M. Harrison, M. B. Eisen, Transcriptional activation of the zygotic genome in drosophila. Curr. Top. Dev. Biol. 113, 85–112 (2015). [DOI] [PubMed] [Google Scholar]
- 3.F. A. Lefebvre, E. Lecuyer, Flying the RNA nest: Drosophila reveals novel insights into the transcriptome dynamics of early development. J. Dev. Biol. 6, 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.N. L. Vastenhouw, W. X. Cao, H. D. Lipshitz, The maternal-to-zygotic transition revisited. Development 146, dev161471 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Z. Ali-Murthy, S. E. Lott, M. B. Eisen, T. B. Kornberg, An essential role for zygotic expression in the pre-cellular Drosophila embryo. PLOS Genet. 9, e1003428 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.E. Lecuyer, H. Yoshida, N. Parthasarathy, C. Alm, T. Babak, T. Cerovina, T. R. Hughes, P. Tomancak, H. M. Krause, Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174–187 (2007). [DOI] [PubMed] [Google Scholar]
- 7.S. De Renzis, O. Elemento, S. Tavazoie, E. F. Wieschaus, Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLOS Biol. 5, e117 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.S. E. Lott, J. E. Villalta, G. P. Schroth, S. Luo, L. A. Tonkin, M. B. Eisen, Noncanonical compensation of zygotic X transcription in early Drosophila melanogaster development revealed through single-embryo RNA-seq. PLOS Biol. 9, e1000590 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. C. Kwasnieski, T. L. Orr-Weaver, D. P. Bartel, Early genome activation in Drosophila is extensive with an initial tendency for aborted transcripts and retained introns. Genome Res. 29, 1188–1197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.J. W. Erickson, T. W. Cline, A bZIP protein, sisterless-a, collaborates with bHLH transcription factors early in drosophila development to determine sex. Genes Dev. 7, 1688–1702 (1993). [DOI] [PubMed] [Google Scholar]
- 11.D. K. Pritchard, G. Schubiger, Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev. 10, 1131–1142 (1996). [DOI] [PubMed] [Google Scholar]
- 12.H. L. Liang, C.-Y. Nien, H.-Y. Liu, M. M. Metzstein, N. Kirov, C. Rushlow, The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400–403 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M. M. Harrison, X.-Y. Li, T. Kaplan, M. R. Botchan, M. B. Eisen, Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLOS Genet. 7, e1002266 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.A. Saunders, L. J. Core, C. Sutcliffe, J. T. Lis, H. L. Ashe, Extensive polymerase pausing during Drosophila axis patterning enables high-level and pliable transcription. Genes Dev. 27, 1146–1158 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D. Ibarra-Morales, M. Rauer, P. Quarato, L. Rabbani, F. Zenk, M. Schulte-Sasse, F. Cardamone, A. Gomez-Auli, G. Cecere, N. Iovino, Histone variant H2A.Z regulates zygotic genome activation. Nat. Commun. 12, 7002 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.J. A. Campos-Ortega, V. Hartenstein, The embryonic development of drosophila melanogaster, in Biosynthesis, Metabolism and Mode of Action of Invertebrate Hormone (Springer, 1st ed., 1985), pp. 218–225. [Google Scholar]
- 17.C.-Y. Nien, H.-L. Liang, S. Butcher, Y. Sun, S. Fu, T. Gocha, N. Kirov, J. R. Manak, C. Rushlow, Temporal coordination of gene networks by Zelda in the early drosophila embryo. PLOS Genet. 7, e1002339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.S. A. Blythe, E. F. Wieschaus, Establishment and maintenance of heritable chromatin structure during early drosophila embryogenesis. eLife 5, e20148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S. L. McDaniel, T. J. Gibson, K. N. Schulz, M. Fernandez Garcia, M. Nevil, S. U. Jain, P. W. Lewis, K. S. Zaret, M. M. Harrison, Continued activity of the pioneer factor zelda is required to drive zygotic genome activation. Mol. Cell 74, 185–195.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A. Tsurumi, F. Xia, J. Li, K. Larson, R. LaFrance, W. X. Li, STAT is an essential activator of the zygotic genome in the early Drosophila embryo. PLOS Genet. 7, e1002086 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.M. M. Colonnetta, J. E. Abrahante, P. Schedl, D. M. Gohl, G. Deshpande, CLAMP regulates zygotic genome activation in Drosophila embryos. Genetics 219, iyab107 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.J. Duan, L. Rieder, M. M. Colonnetta, A. Huang, M. Mckenney, S. Watters, G. Deshpande, W. Jordan, N. Fawzi, E. Larschan, CLAMP and Zelda function together to promote Drosophila zygotic genome activation. eLife 10, e69937 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.M. M. Gaskill, T. J. Gibson, E. D. Larson, M. M. Harrison, GAF is essential for zygotic genome activation and chromatin accessibility in the early Drosophila embryo. eLife 10, e66668 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.K. M. Bhat, G. Farkas, F. Karch, H. Gyurkovics, J. Gausz, P. Schedl, The GAGA factor is required in the early Drosophila embryo not only for transcriptional regulation but also for nuclear division. Development 122, 1113–1124 (1996). [DOI] [PubMed] [Google Scholar]
- 25.T. Koromila, F. Gao, Y. Iwasaki, P. He, L. Pachter, J. P. Gergen, A. Stathopoulos, Odd-paired is a pioneer-like factor that coordinates with Zelda to control gene expression in embryos. eLife 9, e59610 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.A. Inoue, Noncanonical imprinting: Intergenerational epigenetic inheritance mediated by Polycomb complexes. Curr. Opin. Genet. Dev. 78, 102015 (2022). [DOI] [PubMed] [Google Scholar]
- 27.J. A. Dahl, I. Jung, H. Aanes, G. D. Greggains, A. Manaf, M. Lerdrup, G. Li, S. Kuan, B. Li, A. Y. Lee, S. Preissl, I. Jermstad, M. H. Haugen, R. Suganthan, M. Bjørås, K. Hansen, K. T. Dalen, P. Fedorcsak, B. Ren, A. Klungland, Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.P. J. Murphy, S. F. Wu, C. R. James, C. L. Wike, B. R. Cairns, Placeholder nucleosomes underlie germline-to-embryo DNA methylation reprogramming. Cell 172, 993–1006.e13 (2018). [DOI] [PubMed] [Google Scholar]
- 29.F. Zenk, E. Loeser, R. Schiavo, F. Kilpert, O. Bogdanović, N. Iovino, Germ line-inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science 357, 212–216 (2017). [DOI] [PubMed] [Google Scholar]
- 30.M. Samata, A. Alexiadis, G. Richard, P. Georgiev, J. Nuebler, T. Kulkarni, G. Renschler, M. F. Basilicata, F. L. Zenk, M. Shvedunova, G. Semplicio, L. Mirny, N. Iovino, A. Akhtar, Intergenerationally maintained histone H4 lysine 16 acetylation is instructive for future gene activation. Cell 182, 127–144.e23 (2020). [DOI] [PubMed] [Google Scholar]
- 31.F. Zenk, Y. Zhan, P. Kos, E. Löser, N. Atinbayeva, M. Schächtle, G. Tiana, L. Giorgetti, N. Iovino, HP1 drives de novo 3D genome reorganization in early Drosophila embryos. Nature 593, 289–293 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. Perez-Montero, A. Carbonell, T. Moran, A. Vaquero, F. Azorin, The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Dev. Cell 26, 578–590 (2013). [DOI] [PubMed] [Google Scholar]
- 33.J. A. Kassis, J. A. Kennison, J. W. Tamkun, Polycomb and trithorax group genes in Drosophila. Genetics 206, 1699–1725 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.F. Huang, A. Paulson, A. Dutta, S. Venkatesh, M. Smolle, S. M. Abmayr, J. L. Workman, Histone acetyltransferase Enok regulates oocyte polarization by promoting expression of the actin nucleation factor spire. Genes Dev. 28, 2750–2763 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.X. Li, C. W. Seidel, L. T. Szerszen, J. J. Lange, J. L. Workman, S. M. Abmayr, Enzymatic modules of the SAGA chromatin-modifying complex play distinct roles in Drosophila gene expression and development. Genes Dev. 31, 1588–1600 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.H. Akimaru, D. X. Hou, S. Ishii, Drosophila CBP is required for dorsal-dependent twist gene expression. Nat. Genet. 17, 211–214 (1997). [DOI] [PubMed] [Google Scholar]
- 37.H. Akimaru, Y. Chen, P. Dai, D. X. Hou, M. Nonaka, S. M. Smolik, S. Armstrong, R. H. Goodman, S. Ishii, Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature 386, 735–738 (1997). [DOI] [PubMed] [Google Scholar]
- 38.W. H. Ludlam, M. H. Taylor, K. G. Tanner, J. M. Denu, R. H. Goodman, S. M. Smolik, The acetyltransferase activity of CBP is required for wingless activation and H4 acetylation in Drosophila melanogaster. Mol. Cell. Biol. 22, 3832–3841 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.L. Vo Ngoc, G. A. Kassavetis, J. T. Kadonaga, The RNA polymerase II core promoter in drosophila. Genetics 212, 13–24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.A. Boija, D. B. Mahat, A. Zare, P.-H. Holmqvist, P. Philip, D. J. Meyers, P. A. Cole, J. T. Lis, P. Stenberg, M. Mannervik, CBP regulates recruitment and release of promoter-proximal RNA polymerase II. Mol. Cell 68, 491–503.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R. Bonasio, R. Shiekhattar, Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 48, 433–455 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.K. Chen, J. Johnston, W. Shao, S. Meier, C. Staber, J. Zeitlinger, A global change in RNA polymerase II pausing during the Drosophila midblastula transition. eLife 2, e00861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A. Ebert, S. Lein, G. Schotta, G. Reuter, Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 14, 377–392 (2006). [DOI] [PubMed] [Google Scholar]
- 44.F. Tie, R. Banerjee, C. A. Stratton, J. Prasad-Sinha, V. Stepanik, A. Zlobin, M. O. Diaz, P. C. Scacheri, P. J. Harte, CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136, 3131–3141 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.P. V. Kharchenko, A. A. Alekseyenko, Y. B. Schwartz, A. Minoda, N. C. Riddle, J. Ernst, P. J. Sabo, E. Larschan, A. A. Gorchakov, T. Gu, D. Linder-Basso, A. Plachetka, G. Shanower, M. Y. Tolstorukov, L. J. Luquette, R. Xi, Y. L. Jung, R. W. Park, E. P. Bishop, T. K. Canfield, R. Sandstrom, R. E. Thurman, D. M. MacAlpine, J. A. Stamatoyannopoulos, M. Kellis, S. C. R. Elgin, M. I. Kuroda, V. Pirrotta, G. H. Karpen, P. J. Park, Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471, 480–485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.C. Feller, I. Forne, A. Imhof, P. B. Becker, Global and specific responses of the histone acetylome to systematic perturbation. Mol. Cell 57, 559–571 (2015). [DOI] [PubMed] [Google Scholar]
- 47.X.-Y. Li, M. M. Harrison, J. E. Villalta, T. Kaplan, M. B. Eisen, Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife 3, e03737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.N. Koenecke, J. Johnston, B. Gaertner, M. Natarajan, J. Zeitlinger, Genome-wide identification of Drosophila dorso-ventral enhancers by differential histone acetylation analysis. Genome Biol. 17, 196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.T. Lilja, H. Aihara, M. Stabell, Y. Nibu, M. Mannervik, The acetyltransferase activity of Drosophila CBP is dispensable for regulation of the Dpp pathway in the early embryo. Dev. Biol. 305, 650–658 (2007). [DOI] [PubMed] [Google Scholar]
- 50.D. Fu, Y. Wen, J. Ma, The co-activator CREB-binding protein participates in enhancer-dependent activities of bicoid. J. Biol. Chem. 279, 48725–48733 (2004). [DOI] [PubMed] [Google Scholar]
- 51.G. Poortinga, M. Watanabe, S. M. Parkhurst, Drosophila CtBP: A hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 17, 2067–2078 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.S. A. Keller, Y. Mao, P. Struffi, C. Margulies, C. E. Yurk, A. R. Anderson, R. L. Amey, S. Moore, J. M. Ebels, K. Foley, M. Corado, D. N. Arnosti, dCtBP-dependent and -independent repression activities of r Knirps protein. Mol. Cell. Biol. 20, 7247–7258 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.A. Erkner, A. Roure, B. Charroux, M. Delaage, N. Holway, N. Coré, C. Vola, C. Angelats, F. Pagès, L. Fasano, S. Kerridge, Grunge, related to human Atrophin-like proteins, has multiple functions in drosophila development. Development 129, 1119–1129 (2002). [DOI] [PubMed] [Google Scholar]
- 54.D. Helmlinger, G. Papai, D. Devys, L. Tora, What do the structures of GCN5-containing complexes teach us about their function? Biochim. Biophys. Acta Gene Regul. Mech. 1864, 194614 (2021). [DOI] [PubMed] [Google Scholar]
- 55.E. F. Torres-Zelada, V. M. Weake, The Gcn5 complexes in Drosophila as a model for metazoa. Biochim. Biophys. Acta Gene Regul. Mech. 1864, 194610 (2021). [DOI] [PubMed] [Google Scholar]
- 56.L. Miao, Y. Tang, A. R. Bonneau, S. H. Chan, M. L. Kojima, M. E. Pownall, C. E. Vejnar, F. Gao, S. Krishnaswamy, C. E. Hendry, A. J. Giraldez, The landscape of pioneer factor activity reveals the mechanisms of chromatin reprogramming and genome activation. Mol. Cell 82, 986–1002.e9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.C. Carre, D. Szymczak, J. Pidoux, C. Antoniewski, The histone H3 acetylase dGcn5 is a key player in Drosophila melanogaster metamorphosis. Mol. Cell. Biol. 25, 8228–8238 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Q. Jin, L.-R. Yu, L. Wang, Z. Zhang, L. H. Kasper, J.-E. Lee, C. Wang, P. K. Brindle, S. Y. R. Dent, K. Ge, Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.J. Bonnet, C.-Y. Wang, T. Baptista, S. D. Vincent, W.-C. Hsiao, M. Stierle, C.-F. Kao, L. Tora, D. Devys, The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 28, 1999–2012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.M. Wang, Z. Chen, Y. Zhang, CBP/p300 and HDAC activities regulate H3K27 acetylation dynamics and zygotic genome activation in mouse preimplantation embryos. EMBO J. 41, e112012 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.E. Dorafshan, T. G. Kahn, A. Glotov, M. Savitsky, M. Walther, G. Reuter, Y. B. Schwartz, Ash1 counteracts Polycomb repression independent of histone H3 lysine 36 methylation. EMBO Rep. 20, e46762 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.M. Hodl, K. Basler, Transcription in the absence of histone H3.2 and H3K4 methylation. Curr. Biol. 22, 2253–2257 (2012). [DOI] [PubMed] [Google Scholar]
- 63.T. J. Penke, D. J. McKay, B. D. Strahl, A. G. Matera, R. J. Duronio, Direct interrogation of the role of H3K9 in metazoan heterochromatin function. Genes Dev. 30, 1866–1880 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.P. Y. K. Yung, A. Stuetzer, W. Fischle, A. M. Martinez, G. Cavalli, Histone H3 serine 28 is essential for efficient polycomb-mediated gene repression in drosophila. Cell Rep. 11, 1437–1445 (2015). [DOI] [PubMed] [Google Scholar]
- 65.R. Rickels, H. M. Herz, C. C. Sze, K. Cao, M. A. Morgan, C. K. Collings, M. Gause, Y. H. Takahashi, L. Wang, E. J. Rendleman, S. A. Marshall, A. Krueger, E. T. Bartom, A. Piunti, E. R. Smith, N. A. Abshiru, N. L. Kelleher, D. Dorsett, A. Shilatifard, Histone H3K4 monomethylation catalyzed by Trr and mammalian COMPASS-like proteins at enhancers is dispensable for development and viability. Nat. Genet. 49, 1647–1653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.A. R. Pengelly, O. Copur, H. Jackle, A. Herzig, J. Muller, A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339, 698–699 (2013). [DOI] [PubMed] [Google Scholar]
- 67.D. J. McKay, S. Klusza, T. J. R. Penke, M. P. Meers, K. P. Curry, S. L. McDaniel, P. Y. Malek, S. W. Cooper, D. C. Tatomer, J. D. Lieb, B. D. Strahl, R. J. Duronio, A. G. Matera, Interrogating the function of metazoan histones using engineered gene clusters. Dev. Cell 32, 373–386 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.M. Leatham-Jensen, C. M. Uyehara, B. D. Strahl, A. G. Matera, R. J. Duronio, D. J. McKay, Lysine 27 of replication-independent histone H3.3 is required for Polycomb target gene silencing but not for gene activation. PLOS Genet. 15, e1007932 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.S. Perez-Lluch, E. Blanco, H. Tilgner, J. Curado, M. Ruiz-Romero, M. Corominas, R. Guigó, Absence of canonical marks of active chromatin in developmentally regulated genes. Nat. Genet. 47, 1158–1167 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.V. M. Weake, S. K. Swanson, A. Mushegian, L. Florens, M. P. Washburn, S. M. Abmayr, J. L. Workman, A novel histone fold domain-containing protein that replaces TAF6 in Drosophila SAGA is required for SAGA-dependent gene expression. Genes Dev. 23, 2818–2823 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.C. C. Sze, K. Cao, C. K. Collings, S. A. Marshall, E. J. Rendleman, P. A. Ozark, F. X. Chen, M. A. Morgan, L. Wang, A. Shilatifard, Histone H3K4 methylation-dependent and -independent functions of Set1A/COMPASS in embryonic stem cell self-renewal and differentiation. Genes Dev. 31, 1732–1737 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.K. M. Dorighi, T. Swigut, T. Henriques, N. V. Bhanu, B. S. Scruggs, N. Nady, C. D. Still II, B. A. Garcia, K. Adelman, J. Wysocka, Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol. Cell 66, 568–576.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.K. A. Sheppard, D. W. Rose, Z. K. Haque, R. Kurokawa, E. McInerney, S. Westin, D. Thanos, M. G. Rosenfeld, C. K. Glass, T. Collins, Transcriptional activation by NF-kappaB requires multiple coactivators. Mol. Cell. Biol. 19, 6367–6378 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.N. Shikama, W. Lutz, R. Kretzschmar, N. Sauter, J. F. Roth, S. Marino, J. Wittwer, A. Scheidweiler, R. Eckner, Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. EMBO J. 22, 5175–5185 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.T. Zhang, Z. Zhang, Q. Dong, J. Xiong, B. Zhu, Histone H3K27 acetylation is dispensable for enhancer activity in mouse embryonic stem cells. Genome Biol. 21, 45 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.A. Sankar, F. Mohammad, A. K. Sundaramurthy, H. Wang, M. Lerdrup, T. Tatar, K. Helin, Histone editing elucidates the functional roles of H3K27 methylation and acetylation in mammals. Nat. Genet. 54, 754–760 (2022). [DOI] [PubMed] [Google Scholar]
- 77.S. R. Bhaumik, M. R. Green, SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15, 1935–1945 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.A. M. Dudley, C. Rougeulle, F. Winston, The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13, 2940–2945 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.M. A. J. Morgan, A. Shilatifard, Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat. Genet. 52, 1271–1281 (2020). [DOI] [PubMed] [Google Scholar]
- 80.C. Queitsch, T. A. Sangster, S. Lindquist, Hsp90 as a capacitor of phenotypic variation. Nature 417, 618–624 (2002). [DOI] [PubMed] [Google Scholar]
- 81.B. R. Graveley, A. N. Brooks, J. W. Carlson, M. O. Duff, J. M. Landolin, L. Yang, C. G. Artieri, M. J. van Baren, N. Boley, B. W. Booth, J. B. Brown, L. Cherbas, C. A. Davis, A. Dobin, R. Li, W. Lin, J. H. Malone, N. R. Mattiuzzo, D. Miller, D. Sturgill, B. B. Tuch, C. Zaleski, D. Zhang, M. Blanchette, S. Dudoit, B. Eads, R. E. Green, A. Hammonds, L. Jiang, P. Kapranov, L. Langton, N. Perrimon, J. E. Sandler, K. H. Wan, A. Willingham, Y. Zhang, Y. Zou, J. Andrews, P. J. Bickel, S. E. Brenner, M. R. Brent, P. Cherbas, T. R. Gingeras, R. A. Hoskins, T. C. Kaufman, B. Oliver, S. E. Celniker, The developmental transcriptome of Drosophila melanogaster. Nature 471, 473–479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.H. S. Kaya-Okur, S. J. Wu, C. A. Codomo, E. S. Pledger, T. D. Bryson, J. G. Henikoff, K. Ahmad, S. Henikoff, CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.V. Bhardwaj, S. Heyne, K. Sikora, L. Rabbani, M. Rauer, F. Kilpert, A. S. Richter, D. P. Ryan, T. Manke, snakePipes: Facilitating flexible, scalable and integrative epigenomic analysis. Bioinformatics 35, 4757–4759 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Y. Liao, G. K. Smyth, W. Shi, featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 85.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.G. Yu, L. G. Wang, Y. Han, Q. Y. He, clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.T. L. Bailey, M. Boden, F. A. Buske, M. Frith, C. E. Grant, L. Clementi, J. Ren, W. W. Li, W. S. Noble, MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.V. Haberle, A. Stark, Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 19, 621–637 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.V. Ramalingam, M. Natarajan, J. Johnston, J. Zeitlinger, TATA and paused promoters active in differentiated tissues have distinct expression characteristics. Mol. Syst. Biol. 17, e9866 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O. Fornes, J. A. Castro-Mondragon, A. Khan, R. van der Lee, X. Zhang, P. A. Richmond, B. P. Modi, S. Correard, M. Gheorghe, D. Baranašić, W. Santana-Garcia, G. Tan, J. Chèneby, B. Ballester, F. Parcy, A. Sandelin, B. Lenhard, W. W. Wasserman, A. Mathelier, JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.F. Ramirez, F. Dundar, S. Diehl, B. A. Gruning, T. Manke, deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.L. Lopez-Delisle, L. Rabbani, J. Wolff, V. Bhardwaj, R. Backofen, B. Grüning, F. Ramírez, T. Manke, pyGenomeTracks: Reproducible plots for multivariate genomic datasets. Bioinformatics 37, 422–423 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 and S2