Abstract
Social experience is essential for the development and maintenance of higher order brain function. Social deprivation results in a host of cognitive deficits and cellular studies have largely focused on associated neuronal dysregulation; how astrocyte function is impacted by social deprivation is unknown. Here we show that hippocampal astrocytes from juvenile mice subjected to social isolation exhibit increased Ca2+ activity and global changes in gene expression. We found that the Ca2+ channel TRPA1 is upregulated in astrocytes after social deprivation and astrocyte-specific deletion of TRPA1 reverses the physiological and cognitive deficits associated with social deprivation. Mechanistically, TRPA1 inhibition of hippocampal circuits is mediated by a parallel increase of astrocytic production and release of the inhibitory neurotransmitter GABA after social deprivation. Collectively, our studies reveal how astrocyte function is tuned to social experience and identifies a social context specific mechanism by which astrocytic TRPA1 and GABA coordinately suppress hippocampal circuit function.
Graphical Abstract
ETOC Blurb
Social experience has a profound impact on cognitive function, yet experience dependent roles for astrocytes in brain circuits remain obscure. Using juvenile social deprivation models, Cheng, et al. found astrocyte function is tuned to social experience, identifying context specific mechanisms by which astrocytic TRPA1 and GABA coordinately suppress hippocampal circuits.
Introduction
Social interaction is a fundamental animal behavior that plays an essential role in cognitive development. In humans, children that are deprived of social interactions have higher rates of psychiatric disorders and protracted deficits in learning and memory1-4. These findings are supported by a multitude of studies across species demonstrating that social deprivation during perinatal- and juvenile- development influences a host of cognitive outputs, ranging from depression and anxiety to learning and memory and decision making5,6. Furthermore, perinatal social isolation leads to profound alterations in brain anatomy that are correlated with changes in patterns of gene expression, supporting the notion that developmental programs are influenced by social interaction7-9 Although we understand the importance of social experience during brain development, how these environmental inputs shape cognitive brain circuits remains unclear.
Studies on perinatal social deprivation have highlighted alterations in circuits associated with the limbic system, which incorporates emotional, social, and memory facets of behavior5,10. These alterations in circuit function are coupled with the dysregulation of neuronal activity and synaptic plasticity, which leads to impaired long-term potentiation (LTP) and other physiological correlates of cognitive function7,11-15. While neuronal dysregulation has been a focal point of studies on social deprivation, there is emerging evidence that glial cell function is also impaired under these circumstances. Prior studies revealed that social deprivation results in impaired oligodendrocyte differentiation and myelination in the prefrontal cortex and astrocytes from dark-reared mice exhibit decreased territories16,17. Although there is evidence that astrocyte morphology is shaped by visual input, whether social deprivation alters their physiology or contributions to circuit function remains undefined.
Astrocytes play a central role in neurotransmission by buffering and releasing a host of neuroactive agents that act at the synapse18-21. Cross-communication between astrocytes and neurons is mediated by calcium (Ca2+) activity, where manipulating Ca2+ activity in astrocytes influences synaptic activity in neurons 22-27. Modulating astrocytic Ca2+ ultimately results in circuit level alterations that influence a host of behavioral outputs, including higher cognitive functions associated with learning and memory. Among the mechanisms that regulate astrocytic Ca2+, TRPA1 functions as a channel that regulates the influx extracellular Ca2+ into astrocytes26,27. Critically, inhibition of TRPA1 activity suppresses LTP in the hippocampus26,27, though whether astrocyte-specific manipulations of TRPA1 impacts learning and memory behaviors is unknown. These roles of astrocytic Ca2+ and TRPA1 in hippocampal circuits, raise the question of whether these core astrocyte functions are dysregulated during social deprivation and contribute to the associated cognitive impairments.
To decipher how astrocytes are altered after social deprivation, we performed single cell RNA-Seq (scRNA-Seq) finding that Ca2+ and TRPA1-dependent signaling pathways are up-regulated in hippocampal astrocytes after social deprivation. Unexpectedly, astrocyte specific knockout and pharmacological inhibition of TRPA1 restored LTP and learning and memory deficits in socially deprived animals. Further analysis revealed that astrocyte-mediated release of GABA is enhanced in astrocytes after social deprivation and dependent upon TRPA1. Together these studies link TRPA1 activity with GABA-mediated inhibition of hippocampal circuits after social deprivation, revealing a new astrocytic mechanism that suppresses cognitive function after social deprivation.
Results
Social deprivation enhances astrocytic Ca2+ activity
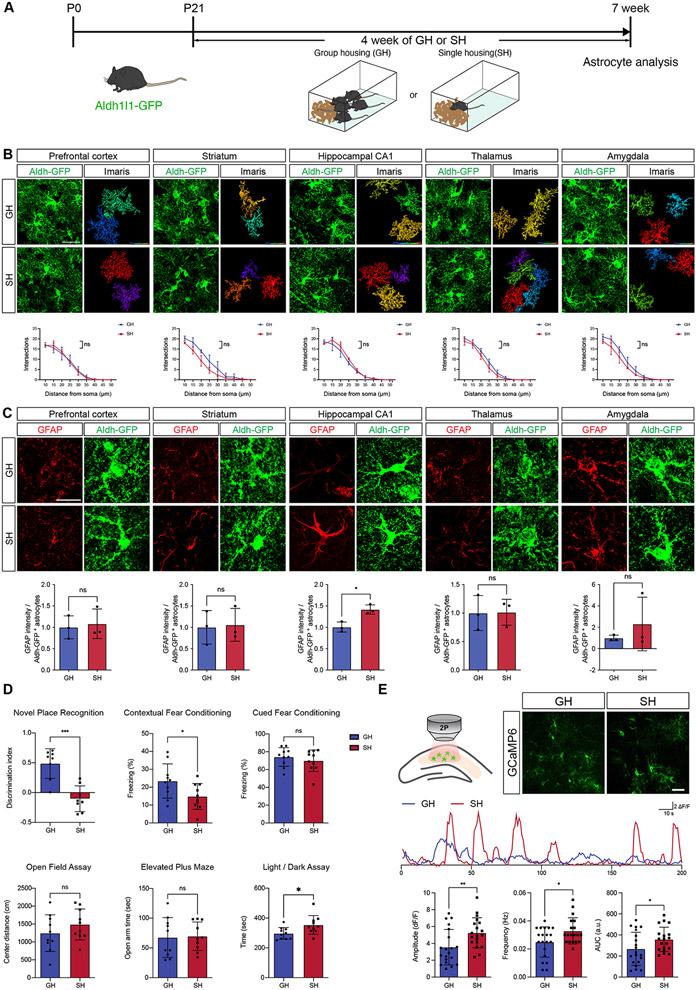
To understand how social deprivation influences astrocyte biology, we subjected newly weaned Aldh1l1-GFP reporter mice (P21) to group housing (GH) or single housing (SH) conditions for 4 weeks, at which point the mice are 7 weeks of age (Figure 1A). After this period of time, we harvested the brains of mice from each cohort and assessed expression of core astrocyte markers across a host of brain regions, including the prefrontal cortex, striatum, hippocampus, thalamus, and amygdala28. This analysis revealed no changes in the number of Sox9- and Aldh1l1-GFP expressing cells, indicating no change in the gross number of astrocytes across all regions analyzed (Supplemental Figure S1C-G). Next, we assessed GFAP expression and identified a significant upregulation of its expression specifically in CA1 of the hippocampus from the SH cohort (Figure 1C). Taking advantage of the Aldh1l1-GFP reporter to label astrocyte branches and processes, we performed high-resolution confocal imaging to analyze morphological complexity under these conditions. Sholl analysis of these images revealed no differences in the morphological complexity of astrocytes between GH and SH groups, coupled with no changes in the branch number in all of the analyzed brain regions (Figure 1B; Supplemental Figure S1A-B). In parallel, we subjected these mice to a series of behavioral assays including those that measured anxiety, aggression, depression, and learning/memory. We found that the SH cohort displayed an increase in aggression, vertical activity in open field assays, and a subtle increase in some anxiety-associated behaviors (Figure 1D; Supplemental Figure S2A-C, E-F), coupled with no in changes depressive behaviors (Supplemental Figure S2A,D). Strikingly, we also observed specific deficits in long-term spatial memory (novel place recognition, NPR) and associative memory (fear conditioning) (Figure 1D)
Figure 1. Social deprivation alters core properties of hippocampal astrocytes.
(A) Schematic of Group Housing (SH) and Single Housing (SH) Paradigms.
(B) Imaging of Aldh1l1-GFP astrocytes and quantification of morphological complexity using Scholl analysis from prefrontal cortex, striatum, CA1, thalamus, and amygdala (n = 3 pairs of animals, at least 10 cells from each animal). Two-way ANOVA.
(C) Immunostaining for GFAP expression in prefrontal cortex, striatum, CA1, thalamus, and amygdala of Aldh1l1-GFP mice from GH and SH cohorts. Welch’s t-test. (n = 3 pairs of animals, 3 sections per animal)
(D) Behavioral results from GH and SH cohorts (n = 8-10 animals)
(E) Representative image showing expression of GCaMP6s in astrocytes and representative spontaneous Ca2+ of two-photon, slice imaging from the hippocampus of GH and SH mice quantification is derived from n = 19-20 cells from 3 pairs of animals. Welch’s t-test.
*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Our SH paradigm begins at P21 during perinatal development and highlights changes in hippocampal astrocytes, along with associated learning and memory behaviors that occur during juvenile social deprivation. We next sought to compare these results with adult social deprivation, focusing on hippocampal astrocytes and associated hippocampal behaviors. Here we initiated SH/GH paradigms at P60, analyzing astrocyte properties and behaviors four weeks later (P90) (Supplemental Figure S3A). SH starting at P60 did not affect the number of astrocytes, nor their morphology, or GFAP expression in the hippocampus (Supplemental Figure S1I, S3B,E). Behavioral analysis revealed an increase in anxiety, consistent with prior studies that employ this paradigm (Supplemental Figure S3D-F) (Ieraci et al., 2016). Interestingly, we identified deficits in spatial memory (Supplemental Figure S3F), which we also observed in juveniles (Figure 1D). Together, these studies indicate that social deprivation in juveniles does not demonstrate a robust effect on anxiety, while highlighting deficits in spatial memory as a conserved feature of social deprivation at both stages.
The conserved deficits in spatial memory suggests that social deprivation impacts hippocampal circuit function. Prior studies have established key roles for astrocytes in hippocampal circuit function and associated learning and memory behaviors, with astrocytic Ca2+ playing a central role in this phenomenon22,27,29,30. Therefore, we next sought to evaluate whether social deprivation after P21 influences astrocytic Ca2+ activity in the hippocampus. To assess Ca2+ activity we generated the Aldh1l1-CreER; Ai96(RCL-GCaMP6s) mouse line which enables us to express the GCaMP6s optical sensor for Ca2+ specifically in astrocytes. We treated these mice with tamoxifen at P1, subjected them to social deprivation at P21 for 4 weeks, and then performed ex vivo two-photon imaging of GCaMP6s activity on hippocampal slices from the GH and SH groups. The results revealed a significant increase in amplitude, frequency, and area under curve (AUC) of spontaneous Ca2+ responses in SH astrocytes (Figure 1E). We subjected our imaging data to AQuA analysis, which enabled us to assess more complex features of Ca2+ activity beyond amplitude and frequency(Wang et al., 2019). Integrating principle components to identify changes across over 20 features, we identified additional differences in Ca2+ activity between SH and GH astrocytes, highlighted by alterations in the location of propagation and enhanced speed of Ca2+ activity in SH astrocytes (Supplemental Figure S4). These Ca2+ imaging studies indicate that social deprivation results in alterations in the physiology of hippocampal astrocytes after social deprivation.
TRPA1 regulates social deprivation induced Ca2+
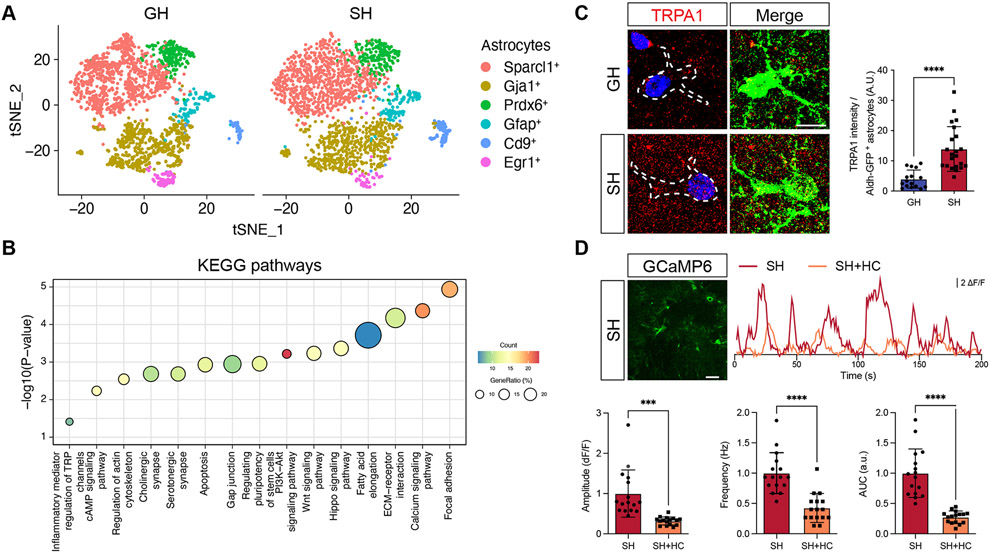
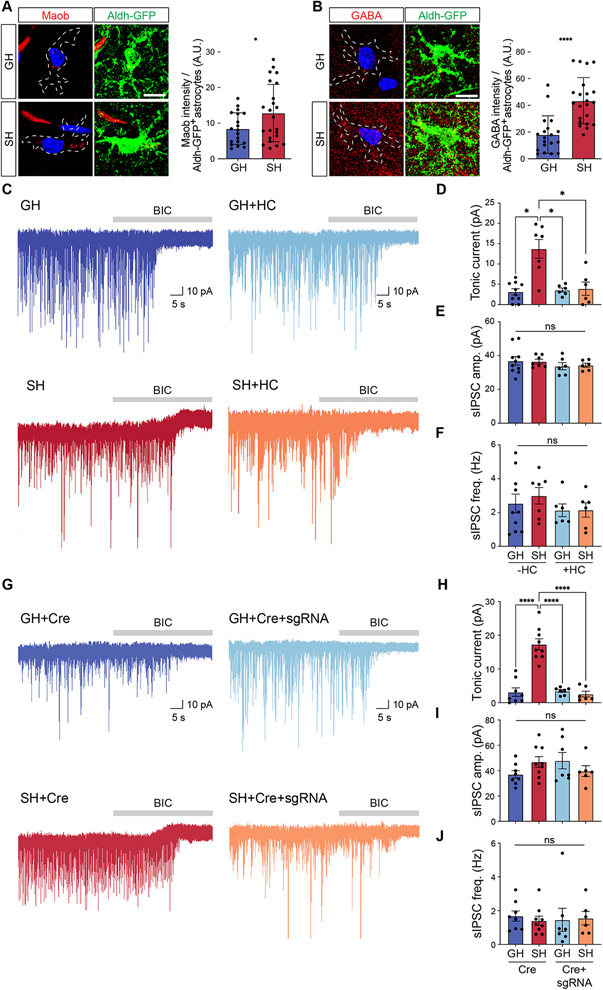
The alterations in Ca2+ activity in hippocampal astrocytes after social deprivation, led us to next examine the molecular changes that occur in astrocytes under these conditions. Utilizing the Aldh1l1-GFP mouse line, we subjected mice to GH or SH conditions for 4 weeks and then FACS-isolated hippocampal astrocytes from which we performed single cell RNA-Sequencing (scRNA-Seq). Using Seurat analysis, we identified five astrocyte subclusters that differentially expressed a series of defined astrocyte makers, including Sparcl1, Gja1, Prdx6, Gfap, Cd9, and Egr1 (Figure 2A and Supplemental Figure S5)(Hao et al., 2021). We did not observe any discernable shifts in the constituency of the astrocyte clusters between SH and GH, suggesting that sub-type identity is unaffected by SH (Figure 2A and Supplemental Figure S5). Subsequent KEGG pathway analysis of differentially expressed genes (DEGs) between GH and SH astrocytes identified several important cellular pathways that exhibited upregulation in SH astrocytes, including synaptic, structural, and Ca2+ signaling pathways (Figure 2B and Table S1), which is consistent with the Ca2+ imaging data (Figure 1E; Supplemental Figure S4). Further analysis of these data revealed transient receptor potential (TRP) channel signaling was also significantly up-regulated in SH astrocytes (Figure 2B and Table S1). Among TRP channels, it has been reported that Transient Receptor Potential Cation Channel Subfamily A Member 1 (TRPA1) mediates spontaneous Ca2+ activity in hippocampal astrocytes through Ca2+ influx26,27. We therefore examined the expression TRPA1 in hippocampal astrocytes, which revealed that its expression is increased in SH astrocytes (Figure 2C). Importantly, we did not observe increased expression of TRPA1 in hippocampal neurons after social deprivation (Supplemental Figure S7B), further illustrating its selective upregulation in astrocytes after SH.
Figure 2. TRPA1 expression is increased in astrocytes after social deprivation.
(A) Serut analysis of single cell RNA-Seq (scRNA-Seq) of hippocampal astrocytes from a pair of GH and SH.
(B) KEGG pathway analysis of differentially expressed gene (DEGs) between GH and SH cohorts (cutoff for DEGs, adjusted p < 10−15, average log2 foldchange > 2).
(C) Immunostaining for TRPA1 expression in Aldh1l1-GFP mice from GH and SH cohorts (n = 16-22 cells from 3 pairs of animals). Welch’s t-test.
(D) Representative image showing expression of GCaMP6s in astrocytes and representative spontaneous Ca2+ of two-photon, slice imaging from the hippocampus of SH mice, treated with vehicle or HC030031 (HC); quantification is derived from n = 16 cells from 3 pairs of animals. Welch’s t-test.
*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Given that TRPA1 functions as a Ca2+ channel, we hypothesized that increased Ca2+ activity in SH conditions is linked to increased TRPA1 activity. To test this, repeated the GH and SH paradigms with the Aldh1l1-CreER; Ai96(RCL-GCaMP6s) and performed Ca2+ imaging on hippocampal astrocytes in the presence of the TRPA1-selective antagonist, HC030031 (HC). Prior studies demonstrated that HC treatment suppresses spontaneous Ca2+ response in hippocampal astrocytes, which we also observed in the GH control group (Supplemental Figure S5C)27. Analysis of the SH group revealed an overall decrease in Ca2+ activity when treated with HC (Figure 2D). These data indicate that social deprivation results in an upregulation of TRPA1 in astrocytes, and this increase in TRPA1 activity contributes to enhanced Ca2+ activity in hippocampal astrocytes under these conditions.
Inhibition of TRPA1 rescues memory deficits after social deprivation
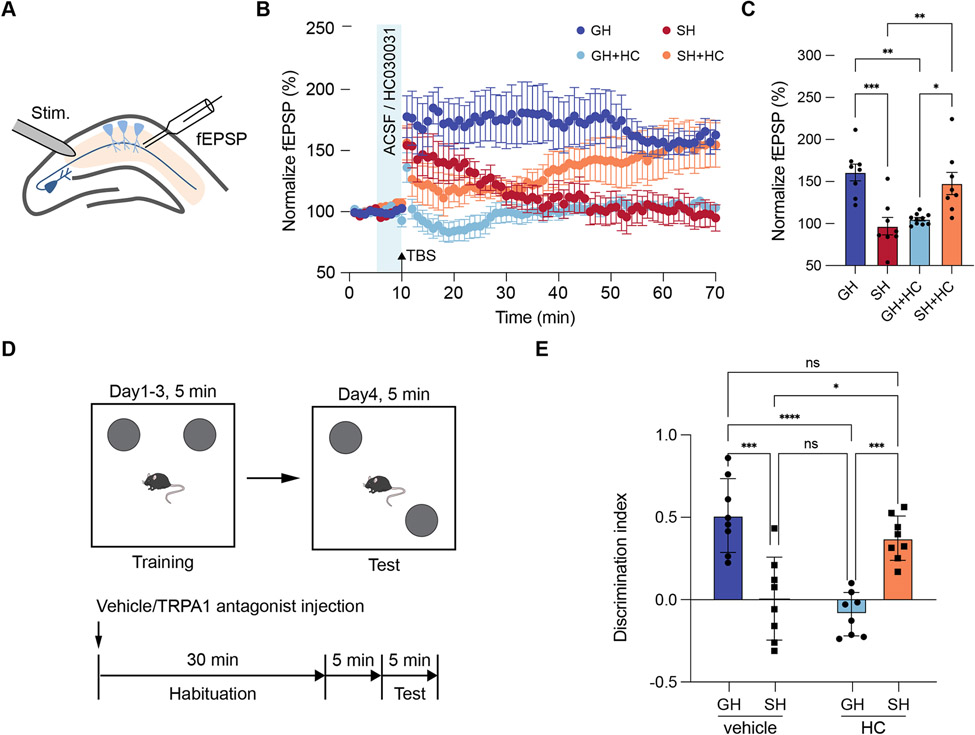
Cross-species studies established that social deprivation impairs learning and memory, however the contributions of altered astrocytic Ca2+ and TRPA1 to this phenomenon is unknown. To examine the role of TRPA1 in hippocampal circuit function after social deprivation, we subjected mice to our GH or SH paradigm and evaluated synaptic plasticity via long term potentiation (LTP). Hippocampal slices from both GH and SH groups were treated with HC or vehicle prior to theta-burst stimulation (TBS) induction of LTP (Figure 3A). Consistent with previous studies, we found that treatment of the GH cohort with HC inhibited LTP (Figure 3B-C, dark blue v. light blue). Further analysis of the SH vehicle control revealed a drastic impairment of LTP compared to the GH vehicle group, which is also consistent with prior studies and our observations from the P60 social deprivation paradigm (Figure 3B-C, dark blue v. red; Supplemental Figures S3G). Strikingly treatment of the SH cohort with HC restored LTP to levels comparable to the GH-vehicle control. (Figure 3B-C, orange v. light blue). These data suggest context specific roles for TRPA1 in hippocampal astrocytes, where under GH conditions it promotes LTP, while under SH conditions it suppresses LTP.
Figure 3. Inhibition of TRPA1 restores hippocampal circuit function.
(A-C) Recording of Long-Term Potentiation (LTP) from the hippocampus in GH and SH cohorts treated with vehicle or HC. Two-way ANOVA, Sidak tests.
(D) Schematic of Novel Placement Recognition (NPR) and treatment paradigm.
(E) NPR behavioral studies on GH and SH cohorts treated with vehicle or HC (n = 8 animals in each group). Welch’s t-test.
*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
To determine if inhibition of TRPA1 after SH can restore prospective learning and memory deficits, we performed NPR assays on male mice from GH and SH cohorts treated with 20 mg/Kg HC or vehicle 30min prior to testing. Consistent with our LTP results, we found that HC treatment impaired NPR performance in the GH cohort (Figure 3D-E). Moreover, the SH-vehicle cohort demonstrated reduced NPR performance compared to the HC-vehicle control, which also aligns with our LTP studies (Figure 3D-E). Finally, we found that HC-treatment restored NPR performance in the SH group to levels that are comparable to GH-vehicle (Figure 3D-E). These behavioral results reinforce our physiological observations that the role of TRPA1 in hippocampal circuits is shaped by social experience.
Astrocyte-specific knockout of TRPA1 restores learning and memory
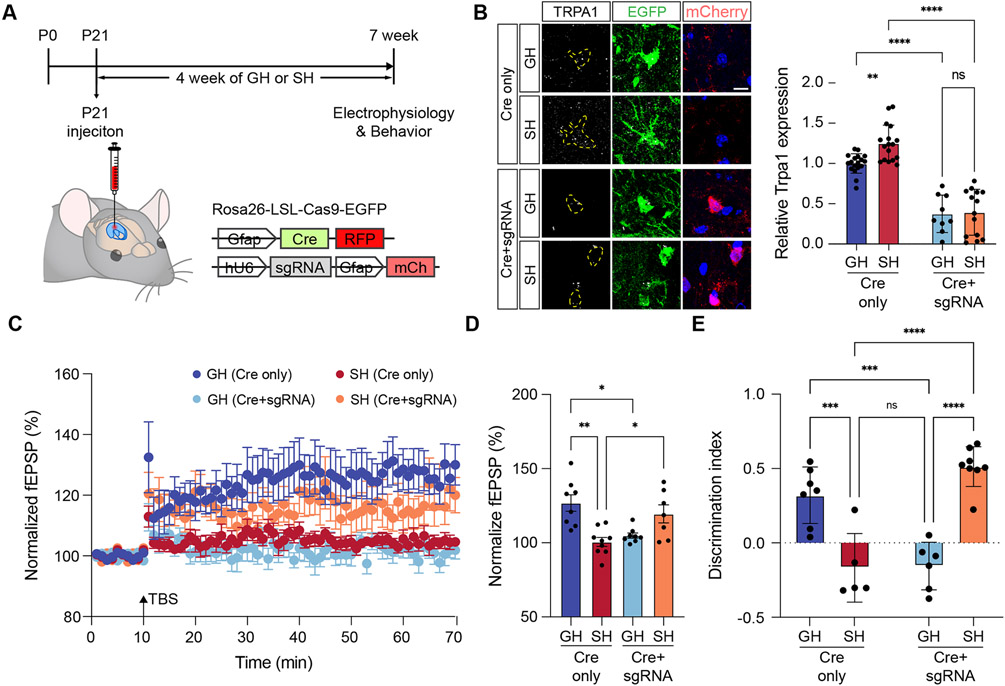
The foregoing observations implicate astrocytic TRPA1 in the suppression of hippocampal circuit function after social deprivation. While prior studies established that astrocytic TRPA1 specifically regulates LTP, it is possible that under social deprivation conditions neuronal TRPA1 expression may contribute to this phenomenon. To specifically test the role of TRPA1 in astrocytes, we eliminated it from hippocampal astrocytes by employing a Cre-dependent CRISPR-based approach. This system uses the Cre-inducible Rosa26-LSL-Cas9-EGFP mouse line, in combination with AAV viruses that enabled selective delivery of Cre to astrocytes along with U6-driven guideRNAs targeting TRPA1 (Figure 4A). Accordingly, Cre induces expression of Cas9 specifically in astrocytes, allowing for selective targeting of TRPA1 by the guideRNA. To target TRPA1 deletion in the hippocampus, we injected AAV2/9 astrocyte-specific Cre (Cre only) with or without U6-driven single guide RNA (sgRNA) targeting TRPA1 (Cre+sgRNA) viruses into hippocampus of 3-week-old Rosa26-LSL-Cas9 knockin mice. 4 weeks after injection, we harvested mice and used immunostaining to confirm reduced TRPA1 expression in hippocampal astrocytes in Cre+sgRNA mice (Figure 4B). Importantly, while hippocampal astrocytes demonstrated reduced expression of TRPA1, its expression was maintained in neuronal populations (Figure 4B and Supplemental Figure S7A-B), indicating that we have selectively eliminated TRPA1 from astrocytes.
Figure 4. Astrocyte-specific deletion of TRPA1 rescues hippocampal circuit function after social deprivation.
(A) Schematic of Cas9, AAV-based approach for deletion of TRPA1 in hippocampal astrocytes and time line of experimental paradigm.
(B) Immunostaining of TRPA1 in hippocampal astrocytes from control and Cre+gRNA, groups. (n = 9-18 cells from 2 animals in each group) Two-way ANOVA, Tukey tests.
(C-D) Recording of Long-Term Potentiation (LTP) from the hippocampus in GH and SH cohorts treated under control (Cre only) or TRPA1 deletion (Cre+sgRNA) genetic conditions. Two-way ANOVA, Tukey tests.
(E) NPR behavioral studies on GH and SH cohorts under control (Cre only) or TRPA1 deletion (Cre+sgRNA) genetic conditions (n = 5-8 animals in each group). Two-way ANOVA, Tukey tests.
*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
To determine whether astrocyte-specific TRPA1 knockout impacts hippocampal circuits in a context specific manner, we subjected Cas9/Cre and Cas9/Cre+sgRNA mice to the GH and SH conditions as described above. After 4 weeks, we harvested hippocampal slices from each group and evaluated TBS-induced LTP. Similar to our pharmacological studies with HC, we found that the Cas9/Cre+sgRNA cohort exhibited a loss of LTP under GH conditions, further verifying that astrocytic TRPA1 is required for LTP (Figure 4C-D, dark blue v. light blue). Analysis of the SH cohort revealed that the Cas9/Cre+sgRNA cohort restored LTP compared to the Cas9/Cre cohort (Figure 4C-D, orange v. red), further indicating that astrocytic TRPA1 has opposing roles in LTP under GH and SH conditions. Finally, we subjected these mice to behavior assays, finding that GH mice from the Cas9/Cre+sgRNA cohort exhibit decreased NPR performance (Figure 4E), while SH mice from the Cas9/Cre+sgRNA cohort exhibited a restoration of NPR performance to levels comparable to the Cas9/Cre cohort from the GH condition. Collectively, our astrocyte-specific knockout of TRPA1 verifies the prior pharmacological studies with HC and further indicate that astrocytic TRPA1 has context specific roles in hippocampal circuits that depend upon social experience.
TRPA1 mediates GABA release after social deprivation
Under social deprivation conditions astrocytic TRPA1 functions to suppress hippocampal circuit function, raising the question of how it mediates circuit suppression. Because astrocyte release of both excitatory and inhibitory neurotransmitters can be Ca2+ dependent, we queried our scRNA-Seq data from SH astrocytes for the expression of glutamate and gamma-Aminobutyric acid (GABA) synthesis enzymes and associated transporters (Supplemental Figure S6A-I) (Jo et al., 2014; Kwak et al., 2020; Lovatt et al., 2007; Yoon et al., 2014). Strikingly, we found that enzymes in the GABA synthesis pathway are highly expressed in astrocytes after SH, with monoamine oxidase B (Maob) demonstrating up-regulation in a subset of SH astrocytes (Supplemental Figure S6B and Supplemental Table 2). To validate Maob expression in SH astrocytes, we performed immunofluorescent staining and found that it’s expression, as well as GABA itself, are both upregulated in SH astrocytes (Figure 5A-B). These data suggest that hippocampal astrocytes exhibit increased release of GABA after social deprivation. To test this, we used slice electrophysiology to measure tonic GABA release from hippocampal astrocytes after GH and SH paradigms. Strikingly, we found a drastic increase in tonic current from CA1 pyramidal neurons under SH conditions, coupled with no changes in synaptic GABA release (Figure 5C-D, dark blue v. red; Supplemental Figure S6J-M). This increase of tonic current was not due to changes of extrasynaptic GABAAR expression after SH, as evidenced by no significant difference in amplitude of the tonic current obtained in the presence of the saturating concentration of GABA at 10 μM (Supplemental Figure S8A). Next, we examined this phenomenon after adult social deprivation beginning at P60 and also observed an increase in tonic GABA release in the SH group (Supplemental Figure S3H). Together these data indicate that hippocampal astrocytes from socially deprived animals exhibit increased release of GABA.
Figure 5. Increased release of astrocytic GABA after social deprivation.
(A-B) Immunostaining for Maob and GABA in hippocampal astrocytes from Aldh1l1-GFP mice after GH or SH paradigms (n = 19-23 cells from 3 pairs of animals). Welch’s t-test.
(C) Representative traces measuring tonic GABA currents in CA1 pyramidal neurons in GH and SH cohorts treated with vehicle or HC.
(D-F) Quantification of tonic GABA current (D), sIPSC amplitude (E), and sIPSC frequency (F) from the same cohorts and experimental conditions. Two-way ANOVA, Tukey test.
(G) Representative traces measuring tonic GABA currents in CA1 pyramidal neurons in GH and SH cohorts with Cre or Cre+sgRNA.
(H-J) Quantification of tonic GABA current (H), sIPSC amplitude (I), and sIPSC frequency (J) from the same cohorts and experimental conditions. Two-way ANOVA, Tukey test.
*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
To test whether astrocyte release of GABA in SH animals is mediated by TRPA1, we measured the tonic GABA current in the presence of TRPA1 inhibitor and in our TRPA1 Cas9-mediated knockout mice. Under both loss-of-function conditions, we found that the increased tonic GABA current in the SH cohort was abolished, returning to a level comparable with the GH cohort (Figure 5C-D, G-H, red v. orange). Importantly, knockout of TRPA1 did not affect the elevated expression levels of Maob (Supplemental Figure S7C), indicating that TRPA1 is not responsible for elevated Maob expression in the SH cohort and further suggesting that their upregulation occurs via parallel pathways. Next, we evaluated whether TRPA1-mediated GABA release after SH suppresses neuronal activity. Similarly, we subjected mice to GH and SH paradigms and used slice electrophysiology and pharmacological inhibition of TRPA1 via HC to measure neuronal excitability after Schaffer Collateral stimulation. We found a drastic decrease in neuronal excitability in the SH cohort compared to the GH cohort, as evidenced by reduced AP spike probability upon stepped stimulus intensities (Supplemental Figure S8B-C, red line). To confirm the observed reduction in excitability in the SH cohort is mediated by TRPA1 and GABA, we treated GH and SH hippocampal slices with HC and GABA receptor antagonists (bicuculline and CGP35348). Strikingly, the reduced the excitability in SH cohort was rescued by treatment with HC (Supplemental Figure S8B-C, red v. orange) or bicuculline and CGP35348 (Supplemental Figure S8B-C, red v. yellow), indicating that TRPA1 and GABA receptor activity responsible for suppressing neuronal excitability in the SH cohort. Together, these data suggest that TRPA1 mediates increased release of GABA from astrocytes, which function to suppress neuronal activity in the hippocampus after social deprivation.
Astrocytic Maob regulates hippocampal circuit function after social deprivation
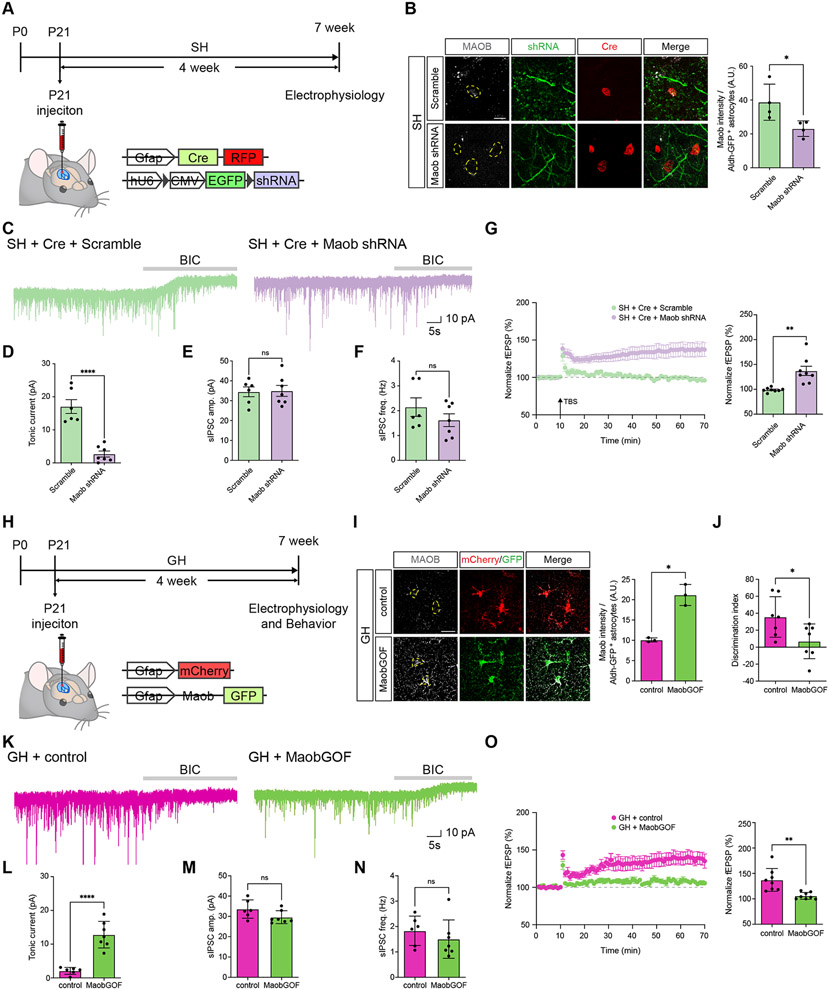
The collective observations herein suggest that enhanced GABA production and it’s subsequent release from astrocytes after social deprivation suppresses hippocampal circuit function. To directly test this, we selectively knocked down Maob expression in hippocampal astrocytes during social deprivation and assessed its impact on circuit function. To knockdown Maob in hippocampal astrocytes, we employed an established Cre-inducible Maob-shRNAi lentiviral system31(Figure 6A). Here, co-injection of an astrocyte-specific Cre virus enables selective expression of the shRNAi in astrocytes and the viral configuration is such that GFP expression marks astrocytes that express the shRNAi, with RFP labeling cells that express Cre (Figure 6B). We injected these viruses (and Cre-only controls) into the hippocampus at P21, just prior to social deprivation (i.e. SH) and analyzed Maob expression after 4 weeks of social deprivation, finding a robust knockdown of Maob expression in hippocampal astrocytes (Figure 6B). To confirm that reduced Maob expression results in reduced GABA release by astrocytes, we measured tonic GABA current and found the expected reduction in tonic GABA after Maob-shRNAi knockdown (Figure 6C-F). Next, we measured LTP under these conditions and found that it was restored after knockdown of astrocytic Maob during social deprivation (Figure 6G). These results indicate that defects in LTP after social deprivation are regulated by astrocytic Maob, which manifests as increased tonic GABA release from astrocytes.
Figure 6. Maob knockdown rescues hippocampal circuit function after social deprivation.
(A) Schematic of AAV-Cre and lentiviral-based shRNA approach for knocking down Maob in hippocampal astrocytes and time line of experimental paradigm.
(B) Immunostaining for Maob and Cre-RFP in hippocampal astrocytes from GH or SH after Maob knockdown paradigms. (n = 4 pairs of animals) Welch’s t-test
(C) Representative traces measuring tonic GABA currents in CA1 pyramidal neurons in GH and SH cohorts Cre plus scramble or Cre plus Maob shRNA.
(D-F) Quantification of tonic GABA current (D), sIPSC amplitude (E), and sIPSC frequency (F) from the same cohorts and experimental conditions. Welch’s t-test.
(G) Recording of Long-Term Potentiation (LTP) from the hippocampus in SH cohorts treated under control (Scramble) or Maob knockdown (Maob shRNA) genetic conditions. Welch’s t-test.
(H) Schematic for Maob overexpression in hippocampal astrocytes and the timeline of the experimental paradigm.
(I) Immunostaining for Maob in hippocampal astrocytes after Maob overexpression from GH paradigm. (n = 3 pairs of animals) Welch’s t-test.
(J) NPR behavioral studies on GH cohorts from control or Maob overexpression groups (n = 7 animals in each group). Welch’s t-test.
(K) Representative traces measuring tonic GABA currents in CA1 pyramidal neurons in control or Maob overexpression groups in the GH paradigm.
(L-N) Quantification of tonic GABA current (L), sIPSC amplitude (M), and sIPSC frequency (N) from the same cohorts and experimental conditions. (n= 3 animals per group). Welch’s t-test.
(O) Recording of Long-Term Potentiation (LTP) from the hippocampus in GH cohorts treated under control or Maob overexpression. Welch’s t-test.
*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Next, we determined whether elevated levels of astrocytic Maob under group housing (GH) are sufficient to induce the deficits in hippocampal circuit function observed after social deprivation. Here, we generated AAV viruses containing Maob in cis with GFP or a cherry control and injected the hippocampus at P21, maintaining these mice under GH conditions for four weeks (Figure 6H-I). Subsequently, we subjected these mice to behavior assays, finding that mice overexpressing astrocytic Maob exhibited decreased NPR performance compared to controls (Figure 6J). Critically, the Maob-overexpression cohort exhibited elevated tonic GABA release (Figure 6K-N), coupled with impaired LTP (Figure 6O), indicating a physiological recapitulation of the deficits manifest after social deprivation (see Figure 3B-C and Figure 5). These findings indicate that elevated astrocytic Maob and subsequent GABA release, on its own, is sufficient to induce the hippocampal circuit deficits we observed after social deprivation.
Best1 knockdown restores hippocampal circuit function after social deprivation
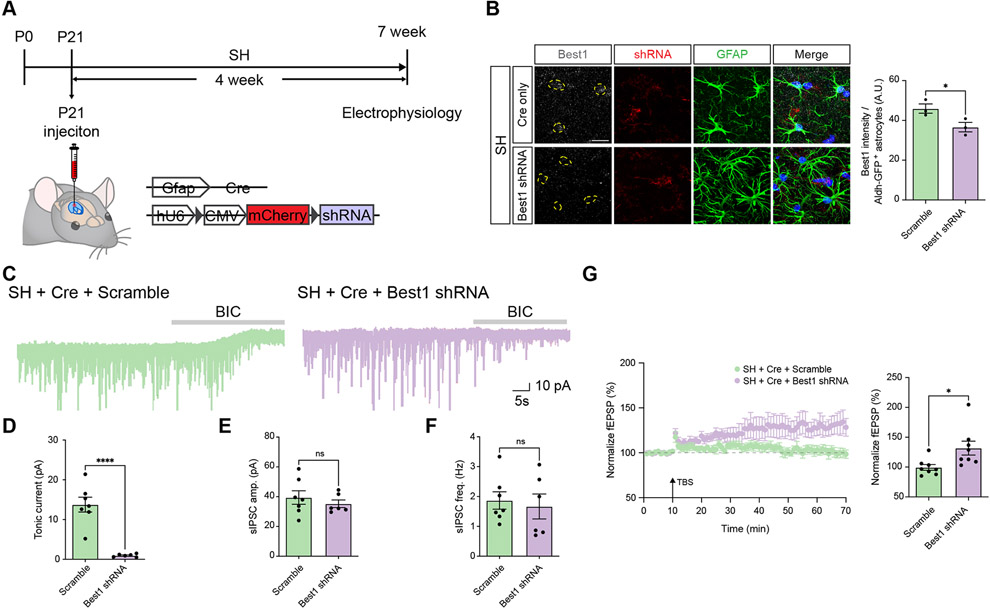
Prior studies have shown that Bestrophin-1 (Best1) is a Ca2+-activated-Cl− channel responsible for the release of GABA from astrocytes32-34. Our model posits that elevated levels of GABA are released by astrocytes after social deprivation, therefore we examined whether inhibition of Best1 would restore hippocampal circuit function after social deprivation. Towards this, we used AAV to express a Cre-inducible shRNAi directed towards Best1 (and scrambled control) to knockdown its expression in hippocampal astrocytes during social deprivation (i.e. single housing, SH) (Figure 7A-B). We found that knockdown of Best1 in hippocampal astrocytes inhibited elevated tonic GABA release from astrocytes after SH, confirming that this manipulation suppresses GABA release from astrocytes (Figure 7C). Next, we examined LTP under these experimental conditions, finding that knockdown of Best1 rescued the deficits in LTP manifest after SH and social deprivation (Figure 7G). These studies indicate that GABA release from astrocytes after social deprivation plays a direct role in the suppression of hippocampal circuit function and further reinforce our model of astrocyte-mediated suppression of hippocampal circuits after social deprivation.
Figure 7. Knockdown of Best1 restores hippocampal circuit function after social deprivation.
(A) Schematic of AAV-Cre and lentiviral-based shRNA approach for knocking down Best1 in hippocampal astrocytes and time line of experimental paradigm.
(B) Immunostaining for Best1 in hippocampal astrocytes from SH cohort after Best1 knockdown paradigm. (n = 3 pairs of animals) Welch’s t-test
(C) Representative traces measuring tonic GABA currents in CA1 pyramidal neurons in SH cohorts Cre plus scramble or Cre plus Best1 shRNA.
(D-F) Quantification of tonic GABA current (D), sIPSC amplitude (E), and sIPSC frequency (F) from the same cohorts and experimental conditions. Welch’s t-test
(G) Recording of Long-Term Potentiation (LTP) from the hippocampus in SH cohorts treated under control (Scramble) or Best1 knockdown (Best1 shRNA) genetic conditions. Welch’s t-test. *p < 0.05; ****p < 0.0001.
Discussion
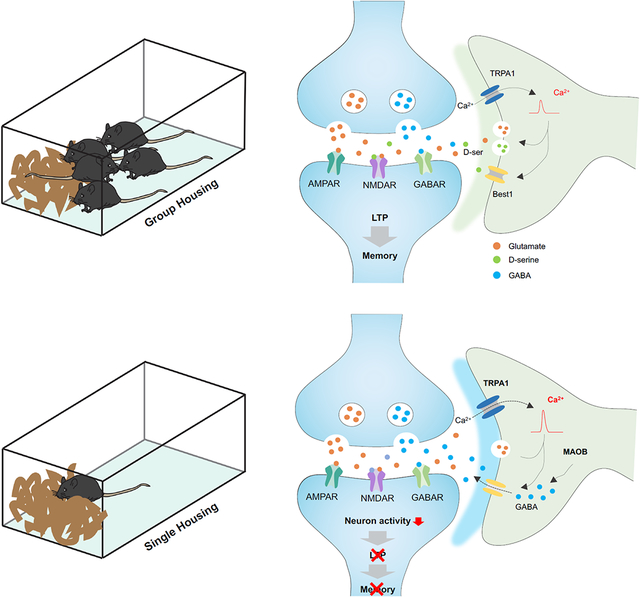
Our study reveals that social deprivation results in widespread alterations in astrocytic gene expression and Ca2+ activity, which are highlighted by increased expression of TRPA1 and key components of the GABA synthesis pathway. Focusing on TRPA1, we found that its inhibition restores hippocampal circuit function and spatial memory after social deprivation. Mechanistically, inhibition of hippocampal circuit function after social deprivation is mediated by elevated GABA release by astrocytes, which is TRPA1-dependent. Altogether, our study reveals how social environment influences astrocyte physiology and identifies a context specific role for TRPA1-GABA signaling in hippocampal astrocytes.
Context-dependent properties of astrocytes
Using the hippocampus as a model to examine how social experience influences astrocyte physiology, we found alterations in Ca2+ activity that are coupled with widespread changes in gene expression. While we observed an increase in the expression of GFAP, we did not witness any alterations in the morphological complexity of astrocytes after social deprivation. This unique combination of features is somewhat paradoxical, as increased GFAP expression is often associated with enhanced morphological complexity, while increased astrocytic Ca2+ is associated with enhanced hippocampal circuit function22,29,35-37. Astrocyte Ca2+ dynamics are used as a proxy for their function and under homeostatic conditions increases in Ca2+ enhance synaptic activity and associated circuit function through release of excitatory bioactive transmitters such as glutamate, d-serine, and ATP20,23,30,38,39 However, in a host of neurological disorders (i.e. epilepsy, ischemia, Rett syndrome, Alexander disease, and Alzheimer’s disease) increases in Ca2+ activity have been documented and correspond with decreased synaptic activity and circuit function33,40,41. Our data suggest that social deprivation induces Ca2+ associated changes in hippocampal astrocytes that mirror those found in neurological disease. Moreover, astrocytes associated with these disease states exhibit upregulation of GFAP, which is also consistent with our findings and further supports the notion that astrocytes after social deprivation acquire phenotypes consistent with neurological disease. Finally, these observations also raise the question of whether social deprivation alters the cellular constituency of prospective astrocyte subpopulations, as there is emerging evidence that astrocytes exhibit plasticity in response to a variety of pathological conditions. Interestingly, our scRNA-Seq studies did not identify changes in the constituency of astrocyte subpopulations, suggesting that population dynamics are unaffected after social deprivation.
Among the changes in gene expression that occur in astrocytes after social deprivation, we found that the Ca2+ channel TRPA1 exhibits increased expression. Inhibition of TRPA1 suppresses the increased Ca2+ activity after social deprivation, suggesting that influx of extracellular Ca2+ contributes to this phenomenon. Furthermore, we show that both pharmacological inhibition and astrocyte-specific knockout of TRPA1 rescues hippocampal circuit defects after social deprivation. Prior studies have shown that inhibition of TRPA1 suppresses LTP in the hippocampus and when put together with our findings suggest a context specific role for TRPA1 in hippocampal astrocytes: TRPA1 promotes circuit function under homeostatic conditions and suppresses circuit function after social deprivation. Mechanistically, TRPA1 has been shown to augment circuit function by promoting astrocytic release of excitatory gliotransmitters27,42. These excitatory mechanisms of TRPA1 function are also operant in AD as early hyperactivity and impaired memory has been attributed to increased astrocytic Ca2+ and inhibition of TRPA143. In contrast, we show that after social deprivation TRPA1 regulates the release of inhibitory signals, including GABA. Put together, these studies illustrate context specific mechanisms used by TRPA1 across a host of normal and pathological states, highlighting the need to further examine its role across additional neurological diseases and environmental settings. Finally, our study provides an example of astrocyte manipulation reversing an acute neurological disorder, raising the possibility that analogous manipulations can be harnessed for other disorders that contain aberrant astrocyte function.
Setting the inhibitory tone of astrocytes
Our studies revealed that hippocampal astrocytes exhibit enhanced inhibitory signaling after social deprivation, which contributes to the suppression of hippocampal circuits. We found that Maob and Aldh1a1, key components of the GABA synthesis pathway31,40, are elevated in hippocampal astrocytes after social deprivation. Furthermore, we observed increased tonic GABA release, which supports the notion that social deprivation induces an inhibitory state in astrocytes. Our collective findings indicate that social deprivation causes both increased TRPA1 expression and GABA synthesis (via Maob) in astrocytes, raising the question of how these two distinct mechanisms are coordinated to drive suppression of hippocampal circuits. Prior studies have shown that GABA release by astrocytes is a Ca2+ dependent process mediated by Best131,40; therefore we posit that increases in TRPA1 expression facilitates the Ca2+ dependent release of GABA via Best1, which is also elevated in astrocytes after social deprivation. This interdependency is supported by our knockout studies, where loss of TRPA1 did not impact elevated levels of Maob within astrocytes after social deprivation, but suppressed tonic GABA release and restored hippocampal LTP (Figures 4-5; Supplemental Figure S7C). Complementary knockdown studies with Maob demonstrated no effect on elevated TRPA1 expression after social deprivation, coupled with reduced tonic GABA release and a restoration of LTP (Figure 6; Supplemental Figure S7D). Therefore, our results support a model where the capacity of TRPA1 to suppress hippocampal circuits after social deprivation is dependent upon the extent of GABA produced by the astrocyte. These observations raise critical new questions about the nature of astrocyte diversity versus plasticity. Although studies have shown that astrocytes can release neuroactive compounds that support both excitatory and inhibitory synaptic activity, whether these activities are restricted to subsets of astrocytes or reflect a switch in cell state remains unknown. Our scRNA-Seq studies support both models, as we did not observe significant changes in the constituency of astrocyte subpopulations, although Maob was specifically increased after SH in a specific subpopulation (Figure S6; Supplemental Table S2). While astrocytes are known to acquire new properties in response to pathological conditions, the mechanisms that regulate their plasticity towards excitatory or inhibitory synaptic activity remain unclear and will be an active area of future investigation.
Studies across species demonstrated that early-life social deprivation has profound effects on global transcriptional profiles and epigenetic landscapes6,7,44. Moreover, mutations in transcription factors and chromatin remodeling proteins have been implicated in a host of neurodevelopmental and psychiatric disorders14,45,46. These observations, coupled with our findings that astrocytes exhibit widespread transcriptomic alterations after social deprivation, raise the question of which transcription factors contribute to this phenomenon. Analysis of our scRNA-Seq data (Supplemental Table 1) identified motifs from 38 transcription factors that are enriched in the DEGs in astrocytes after social deprivation. Among these, Sox9 is an established regulator of astrocyte development and has been shown to regulate olfactory bulb circuit function in adult astrocytes47 however, whether Sox9 contributes to changes in gene expression associated with social deprivation is unknown. This suggests that Sox9 (or other associated transcription factors) could serve as a future template for dissecting astrocytic transcriptional networks that drive responses to social deprivation. Another consideration is the convergence of Ca2+ activity and transcriptional regulation in setting the inhibitory tone in astrocytes. Prior studies indicate that experience-dependent neuronal activity regulates transcription factor activity and target gene selection through secondary messengers such as cAMP and Ca2+ 25,48-51. Taken further, our finding of increased Ca2+ and altered patterns of gene expression raise the possibility that astrocytic Ca2+ activity may influence transcriptional activity after social deprivation. Dissecting how changes in social experience influence the transcriptional properties of astrocytes will be key to understanding how astrocytes acquire an inhibitory tone after social deprivation
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Benjamin Deneen (deneen@bcm.edu).
Materials Availability
All published reagents will be shared on an unrestricted basis after completion of a material transfer agreement; reagent requests should be directed to the lead contact.
Data and Code Availability
The RNA-seq dataset generated during this study are available at the NCBI GEO website.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All experimental animals were treated in compliance with the US Department of Health and Human Services, the NIH guidelines, and Baylor College of Medicine IACUC guidelines. All mice were housed with food and water available ad libitum in a 12-hour light/dark environment. Both female and male mice were used for all experiments, and littermates of the same sex were randomly allocated to experimental groups. 3-4 P21 mice were group-housed in GH condition with standard enviropak for 1-2 month before experiments. 1 P21 mouse was single-housed in SH condition with standard enviropak for 1-2 month before experiments. For ex vivo and in vivo experiments, 2–4-month-old animals were used unless otherwise described. All mice used in this study were maintained on the C57BL/6J background. To analyze calcium activity, the Ai96 (RCL-GCaMP6s) mouse (The Jackson Laboratory; RRID:IMSR_JAX:024106) was crossed with Aldh1l1-CreER (The Jackson Laboratory; RRID:IMSR_JAX:029655), resulting in Aldh1l1-CreER; Rosa-LSL-GCaMP6s (AA6-KO) mice. To induce GCaMP6 expression, P0 pups were injected subcutaneously with 100 mg/kg body weight of Tamoxifen (Sigma-Aldrich, cat no. T5648) dissolved in a 9:1 corn oil/ethanol mixture for single injection at P0-P1. To perform CRISPR-dependent tissue specific knockout, we utilized Rosa26-LSL-Cas9 knockin mice (The Jackson Laboratory; RRID:IMSR_JAX:026175). Above experiments were approved by Baylor College of Medicine IACUC.
METHOD DETAILS
Immunofluorescence on frozen brain tissues
Mice were anesthetized under isoflurane inhalation and perfused transcardially with 1XPBS pH 7.4 followed by 4% paraformalde- hyde (PFA). Brains were removed, fixed in 4% PFA overnight, and placed in 20% sucrose for 24 hours before embedded in OCT. Sections of 20 mm (morphological analysis using GFP labeling) were made on a cryostat, washed with 1XPBS 5 min X2, incubated in antigen retrieval buffers at 75 degree 10 min, blocked with 10% goat or donkey serum in PBS with 0.3% Triton x-100, and incubated with primary antibodies in blocking solution overnight. On the next day, sections were incubated with secondary antibodies in PBS with 0.1% Triton x-100 for 1 h RT, followed by incubation with DAPI in PBS for 10min, and mounted with VECTASHIELD Antifade Mounting Media (Vector Laboratories, H-1000). The following primary antibodies were used: Chicken anti-GFP (1:1000; Abcam, ab13970), mouse anti-GFAP (1:1000; EMD Millipore, MAB360), rabbit anti-Sox9 (1:650; EMD Millipore, AB5535), guinea pig anti-GABA (1:200; Millipore, AB175), rabbit anti-MAOB (1:200; Proteintech, 12602-1AP), mouse anti-mCherry (1:500; Abcam, ab125096), and rabbit anti-TRPA1 (1: 250; Abcam, ab58844), rabbit anti-BEST1 (1:250; GeneTex, GTX33037). The following secondary antibodies were used (1:500): Alexa Fluor 488 goat anti-chicken (Thermo Fisher Scientific, A11039), Alexa Fluor 568 goat anti-rabbit (Thermo Fisher Scientific, A11036), Alexa Fluor 568 goat anti-mouse (Thermo Fisher Scientific, A11004), Alexa Fluor 488 goat anti-mouse (Thermo Fisher Scientific, A32723), Alexa Fluor 647 goat anti-rabbit (Thermo Fisher Scientific, A21245), Alexa Fluor 568 goat anti-guinea pig (Thermo Fisher Scientific, A11075).
Confocal imaging and image analysis
To measure astrocyte morphology, fluorescent images were acquired using a Zeiss LSM 880 laser scanning confocal microscope with 63X oil immersion objective with frame size at 1024 x 1024 and bit depth at 12 or a Leica TCS SP8 STED microscope with 63X oil immersion objective with frame size at 1024 x 1024. Serial images at z axis were taken at an optical step of 0.5 mm, with overall z axis range encompassing the whole section. Images were imported to Imaris Bitplane software, and only astrocytes with their soma between the z axis range were chosen for further analysis52. We performed 3D surface rendering using the Imaris Surface module, and color-coded the reconstructed surface images based on the surface area of each astrocyte. Morphological analysis was performed using the Imaris Filament module. Astrocyte branches and processes were outlined by Autopath with starting point set at 8 mm and seed point set at 0.7 mm, and statistical outputs including “filament number Sholl intersections” were extracted and plotted with Prism software. Data were generated from 3 brain sections per region per mouse with 3 mice per genotype. To analyze number of SOX9-positive astrocytes, fluorescent images were acquired using a Zeiss Axio Imager.M2 with apotome and 20X objective. Cell numbers were quantified by the QuPath software Cell Detection function53. To measure the fluorescent intensity of GFAP, GABA, Maob, and Trpa1, fluorescent images were acquired using a Leica TCS SP8 STED microscope with 63X objective and were analyzed by Fiji. The person who analyzed the images was blinded to the experimental groups.
Behavioral tests
We subjected 3-4-month-old male mice to behavioral tests. All the experimental mice were transferred to the testing room at least 30 min prior to the test. All tests were performed with white noise at ± 60 dB in a designated room. The person performing the tests was blinded to the experimental groups.
Open field test
The open field tests were performed using the Versamax system. The Versamax open field chamber is a square arena (40cm x 40cm x 30cm, Accuscan Instruments) enclosed by transparent walls. Each mouse was put into the center of the chamber. Locomotor activity was detected automatically by sensor beams at X, Y, and Z directions. Data were recorded in 15 two-minute blocks for 30 min total and were analyzed and exported with Versadat software.
Elevated plus maze
The elevated plus maze test were performed on a 1-meter high “+” shaped apparatus with two open arms and two close arms. Mice were put into the center of plus maze and recorded for 10 minutes. The time that mice spent on the open arms or close arms were analyzed by ANY-maze software.
Light/dark
The light/dark testing chamber comprised of a rectangular plexiglass chamber divided in to two unequal compartments, separately by a partition with an opening (8.5 cm × 5 cm) large enough to allow the animal to transition between the compartments. The large chamber was transparent and uncovered (30 cm × 21 cm × 21 cm) with around 700-750 lux. The small chamber was made of black Plexiglas and cover with a lid (15 cm × 21 cm × 21 cm). At the start of the experiment the animals were placed in the light chamber and the number of transitions between the chambers (all four paws inside a chamber) over a 10 min period was recorded. White noise was present throughout testing.
Tube aggression assay
The tube testing was performed in a bright room. Each cylinder was 5 cm diameter and 30 cm in length. Both experimental animals and opponent animals were habituated by passing through the cylinder for 3 times. After habituation, each experimental mouse will pair with 5 different opponent mice. The win was scored by the success of experimental mouse pushing out the opponent mouse. If the mice fought, the trial was excluded.
Tail suspension
The tail suspension test was performed in a square box (30 cm x 30 cm x 30 cm). The mouse tail was fixed on a metal hanger with a tape on the tail 1.5 cm away from the tail tip. After taped on the metal hanger upside down, the mouse head was 5-6 cm away from the bottom of the box. The movement of mouse was recorded for 6 minutes and analyzed byANY-maze software.
Novel place recognition
The novel place recognition test was performed in a square arena. Before the testing day, mice were trained three days to remember the place of two LEGO objects. On the testing day, one of the objects was randomly moved to a new place. The video recording and analysis was done using ANY-maze software. The discrimination index was calculated by (Time spending on novel place object – Time spending on old place object) / Total time exploring two objects.
Contextual/cued conditional fear
The contextual conditional fear test was performed in a chamber with metal grid floor. Three checkerboard pattern visual cues (13 cm X 13 cm) were posted at three sides of the chamber. On day 1, mice were put into the center of the chamber and allowed to move freely for 3 min before being exposed to 3 mild foot shocks (2 s, 0.7mA) with 2 min intertrial intervals (ITI) between each shock (figure). On day 2, mice were first put back to the same chamber and movements of mice over 5 min were recorded and analyzed by FreezeFrame software (Actimetrics, Coul- bourn Instruments) with the bouts and threshold both set at 6.0 s. % freezing time identified based on the above criteria. Two hours after contextual conditional fear, mice were put back to chamber with different context and were recorded % freezing time upon cue stimulation. The % freezing time in cued conational fear was analyzed by same criteria as contextual conditional fear. Data were then plotted as shown in supplemental Figure S2.
Y maze
The Y maze testing was performed in a Y maze and each arm was 40 cm. The mouse was placed at the center and faced top arm. The mouse movement in Y maze was recorded for 5 minutes using ANY-maze software. The entry of each arm was analyzed by ANY-maze software. The alternation was calculated by Total number of alternations / Number of arms entered *100%
Tissue dissociation for single cell
Brain slices were prepared as we described in slicing recording. The desired brain region was micro-dissected in ACSF on ice and proceeded to tissue dissociation using neural tissue dissociation kit (Miltenyi Biotec). After 30 minutes incubation on gentleMACS (Miltenyi Biotec), samples were treated with debris removal kit, 1X red blood cell lysis buffer, dead cell removal kit (Miltenyi Biotec) to purify single cells. To purify astrocytes, we did FACS sorting and collected astrocytes in ACSF buffer with 0.1% BSA. Finally, samples were proceeded to single cell RNA-sequencing library preparation.
RNA extraction, library preparation and sequencing
single-cell RNA-sequencing, single cell gene expression library was prepared according to Chromium Single Cell Gene Expression 3v3.1 kit (10x Genomics). In Brief, single cells, reverse transcription (RT) reagents, Gel Beads containing barcoded oligonucleotides, and oil were loaded on a Chromium controller (10x Genomics) to generate single cell GEMS (Gel Beads-In-Emulsions) where full length cDNA was synthesized and barcoded for each single cell. Subsequently the GEMS are broken and cDNA from each single cell are pooled. Following cleanup using Dynabeads MyOne Silane Beads, cDNA is amplified by PCR. The amplified product is fragmented to optimal size before end-repair, A-tailing, and adaptor ligation. Final library was generated by amplification. Equal concentrations (2 nM) of libraries were pooled and subjected to paired-end (R1: 26, R2: 50) sequencing using the NextSeq 500/550 High Output Kit v2.5 (75 Cycles) (Illumina, 20024906) on a NextSeq550 following the manufacturer’s instructions.
Single cell RNA-seq analysis
Sequencing files from each flow cell lane were downloaded and the resulting fastq files were merged. Reads were mapped to the mouse genome mm10 assembly using 10X Cell Ranger (3.0.2) and it is estimated 40,000 mean reads per cell.
For single cell sequencing analysis, standard procedures for filtering, mitochondrial gene removal, doublets removal, variable gene selection, dimensionality reduction, and clustering were performed using Seurat (version 4.1.0) and DoubletFinder54,55. Criteria for cell inclusion were minimum nUMI/cell threshold 250, minimum gene/cell threshold 250, minimum log10gene/UMI threshold 0.8, maximum mitochondria ratio 0.2, and minimum ribosome ration 0.01556. Mitochondrial genes were removed before doublets removal. Principle component analysis and elbowplot were used to find neighbors and clusters (resolution 0.3). Cells were visualized using a 2-dimensional T-distributed Stochastic Neighbor Embedding (TSNE) of the PC-projected data. Molecularly distinct cell populations were assigned to each cluster using singleR with adult mouse cortical cell taxonomy single cell RNA-seq data as references57,58. FindAllMarkers were used to identify all differentially expressed markers between clusters. Annotated clusters were refined based on those unique markers. Differentially expressed genes (DEGs) in astrocytes between GH and SH were identified by identified by FindMarkers using default settings. 2021 KEGG mouse pathway analysis of DEGs were performed using enrichR59. Single cell RNA-Seq data can be found at the NIH GEO database, GSE222785
Stereotactic injection of AAV viruses
For astrocyte, CRISPR-dependent tissue specific knockout experiments, we used AAV2/9-pAAV-Gfap-Cre-P2A-TurboRFP at a concentration of 5E+12 genome copies per ml (gc/ml) and AAV2/9-pAAV-U6-Trpa1 sgRNA-Gfap-mcherry at a concentration of 2E+12 genome copies per ml (gc/ml). For astrocyte specific knockdown of MAOB, we used AAV2/9-pAAV-Gfap-Cre-P2A-TurboRFP at a concentration of 5E+12 genome copies per ml (gc/ml) and lenti-pSico-Maob shRNA (U6-loxP-CMV-EGFP-loxP-Maob shRNA) at a concentration of 5E+8 genome copies (gc/ml). For astrocyte specific knockdown of Best1, we used AAV2/9-pAAV-Gfap-Cre-P2A-TurboRFP at a concentration of 5E+12 genome copies per ml (gc/ml) and AAV2/9-CMV-LSL-mCherry-U6-mBest1-sh-SE at a concentration of 4E+12 genome copies (gc/ml)60. For hippocampal-stereotactic injection, 3-week-old mice were anesthetized with isoflurane and injected AAV virus as described in previous paper29(Huang et al., 2020). 1 μl of 10X Fastgreen (Sigma, F7252) was mixed with 9 μl of virus before injection. AAV virus: All AAV viruses were generated by the Optogenetics and Viral Vectors Core at Jan and Dan Duncan Neurological Research Institute (NRI). All animal procedures were done in accordance with approved BCM IACUC protocols.
Slice preparation and electrophysiological recording
Animals were anesthetized with isoflurane and isolated brains were submerged in ice-cold ACSF solution (130mM NaCl, 24mM NaHCO3, 1.25mM NaH2PO4, 3.5mM KCl, 1.5mM CaCl2, 1.5mM MgCl2, and 10mM D(+)-glucose, pH 7.4). 300 mm slices were cut using a vibratome (DSK Linear Slicer, Kyoto, Japan) oxygenated in ACSF at room temperature for 1 hr, and then acclimated at room temperature with continuous perfusion with ASCF solution (2ml/min). Slices were placed in recording chamber and target cells were identified via upright Olympus microscope with a 60X water immersion objective with infrared differential interference contrast optics. Whole cell recording was performed with pCLAMP10 and MultiClamp 700B amplifier (Axon Instrument, Molecular Devices) at room temperature from hippocampal CA1 pyramidal neurons. The holding potential was −60 mV. Pipette resistance was typically 5-8 MΩ. The pipette was filled with an internal solution (in mM): 140 K-gluconate, 10 HEPES, 7 NaCl, and 2 MgATP adjusted to pH 7.4 with CsOH for action potential measurements; 135 CsCl, 4 NaCl, 0.5 CaCl2, 10 HEPES, 5 EGTA, 2 Mg-ATP, 0.5 Na2-GTP, 30 QX-314, pH adjusted to 7.2 with CsOH (278-285 mOsmol) for inhibitory postsynaptic currents (IPSCs) and tonic current measurement. IPSC and tonic current were measured in the presence of ionotropic glutamate receptor antagonists, APV (50 μM, Tocris), and CNQX (20 μM, Tocris). For measuring extrasynaptic GABAAR, tonic current were measured in the presence saturating concentration of GABA (10 μM, Tocris) for extrasynaptic GABAAR. All holding potential values stated are after correction for the calculated junction potential offset of 14 mV. Evoked spike probability was induced by electrical stimulation (1 pulse, 100 ms duration, 100-800 μA intensity) via a constant current isolation unit onto Schaffer collateral pathway. Spike probability was calculated as the ratio of the number of successful (spike-generating) stimulations to the total number of stimulations (10 stimuli). LTP was induced by theta burst stimulation (TBS, 10 trains of 4 half maximal stimuli at 100 Hz within 200 ms interval) onto Shaffer collateral pathway and measured by field EPSPs (fEPSPs) in striatum radiatum in hippocampus. The pipette solution was filled with 1M NaCl solution. fEPSPs were calculated by slope of response (mV/ms) and normalized to the average of the baseline. LTP was calculated by averaging last 5 minutes responses. Electrical signals were digitized and sampled at 50 μs intervals with Digidata 1550B and Multiclamp 700B amplifier (Molecular Devices, CA, USA) using pCLAMP 10.7 software. Data were filtered at 2 kHz. The recorded current was analyzed with ClampFit 10.7 software.
Two-photon calcium imaging and analysis
Calcium activity was collected at 1 Hz for 5 min using a two-photon resonant microscope (LSM 7MP, Zeiss) equipped with a Coherent Chameleon Ultra (II) Ti-sapphire laser tuned to 900 nm and a 20x, 1.0 NA Zeiss objective. Calcium activity was typically sampled at 1 Hz. Optical signals were recorded for 5 minutes per trial at 1024 x 1024 pixel resolution. We recorded data from astrocytes at depths of 30 μm below the surface. All multiphoton imaging experiments were performed within 2-4 hours of slicing. Images were quantified using GECIquant algorithm with ImageJ, AQuA algorithm with MATLAB, and Clampfit 10.7 softwares. The detection of region of interest (ROI) for soma and main process were performed in a semi-automated manner using the GECIquant algorithm as previously described29. The analysis of Ca2+ events were detected and calculated by astrocyte quantitative analysis (AQuA) algorithm in MATLAB as previously described61.
QUANTIFICATION AND STATISTICAL ANALYSIS
Sample sizes and statistical tests can be found in accompanying Figure legends. Offline analysis was carried out using Clampfit 10.7, Minianalysis, SigmaPlot 13, Prism 9, and Excel software. We assessed the significance of data for comparison by two-tailed unpaired Welch’s t-test. For multiple comparisons, we used the two-way ANOVA with Tukey’s test. In general, we assumed data were normally distributed but this was not formally tested. Data are presented as mean ± SEM (standard error of the mean). Levels of statistical significance are indicated as follows: * (p < 0.05), ** (p < 0.01), *** (p < 0.001), **** (p < 0.0001).
Supplementary Material
Table S1. List of astrocyte subpopulation markers, related to Figure 2.
Table S2. List of differentially expressed genes in astrocyte clusters, related to Figure 2.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken anti-GFP (1:1000) | Abcam | Cat# ab13970, RRID: AB_300798 |
| ouse anti-GFAP (1:1000) | EMD Millipore | Cat# MAB360, RRID: AB_11212597 |
| Rabbit anti-Sox9 (1:650) | EMD Millipore | Cat# 211-032-171, RRID: AB_2239761 |
| Guinea pig anti-GABA (1:200) | EMD Millipore | Cat# HAF016, RRID: AB_91011 |
| Rabbit anti-MAOB (1:200) | Proteintech | Cat# 12602-1AP, RRID: AB_2137273 |
| Mouse anti-mCherry (1:500 | Abcam | Cat# ab125096, RRID: AB_11133266 |
| Rabbit anti-TRPA1 (1: 250) | Abcam | Cat# ab58844, RRID: AB_945957 |
| Rabbit anti-BEST1 (1:250) | GeneTex | Cat# GTX33037, RRID: NA |
| Goat anti-Chicken IgY (H+L) Secondary Antibody, Alexa Fluor™ 488 (1:500) | Thermo Fisher | Cat# A11039, RRID: AB_2534096 |
| Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 568 (1:500) | Thermo Fisher | Cat# A11036, RRID: AB_10563566 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 568 (1:500) | Thermo Fisher | Cat# A11004, RRID: AB_2534072 |
| Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ Plus 488 (1:500) | Thermo Fisher | Cat# A32723, RRID: AB_2633275 |
| Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 647 (1:500) | Thermo Fisher | Cat# A21245, RRID: AB_2535813 |
| Goat anti-Guinea Pig IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 568 (1:500) | Thermo Fisher | Cat# A11075, RRID: AB_2534119 |
| Bacterial and virus strains | ||
| AAV2/9-pAAV-Gfap-Cre-P2A-TurboRFP | Ung et al., 2021 | NA |
| AAV2/9-pAAV-U6-Trpa1 sgRNA-Gfap-mcherry | This paper | NA |
| lenti-pSico-Maob shRNA (U6-loxP-CMV-EGFP-loxP-Maob shRNA) | Yoon et al., 2014 | NA |
| AAV2/9-CMV-LSL-mCherry-U6-SCR-SE | Kwak et al., 2020 | NA |
| AAV2/9-CMV-LSL-mCherry-U6-mBest1-sh-SE | Kwak et al., 2020 | NA |
| AAV2/9-GFAP-MAOB-GFP | This paper | NA |
| Experimental models: Organisms/strains | ||
| Ai96 (RCL-GCaMP6s) | JAX lab | RRID: IMSR_JAX:024106 |
| Aldh1l1-CreER | JAX lab | RRID: IMSR_JAX:029655 |
| Rosa26-LSL-Cas9 | JAX lab | RRID: IMSR_JAX:026175 |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma | Cat# T5648 |
| Hoechst Nuclear Stain | Thermo Fisher | Cat# H3570 |
| APV | Tocris | Cat# 0106 |
| CNQX | Tocris | Cat# 1045 |
| GABA | Tocris | Cat# 0344 |
| BIC | Tocris | Cat# 0131 |
| Critical commercial assays | ||
| Neural Tissue Dissociation Kit | Miltenyl | Cat# 130-094-802 |
| Red Blood Cell Lysis Solution (10×) | Thermo Fisher | Cat# 130-094-183 |
| Chromium Single Cell Gene Expression 3v3.1 kit | 10X genomics | NA |
| NextSeq 500/550 High Output Kit v2.5 (75 Cycles) | Illumina | Cat# 20024906 |
| MACS Debris Removal Kit | Miltenyl | Cat# 130-109-398 |
| MACS Dead Cell Removal Kit | Miltenyl | Cat# 130-090-101 |
| MiniMACS Separator | Miltenyl | Cat# 130-042-102 |
| Dynabeads MyOne Silane Beads | Thermo Fisher | Cat# 37002D |
| Vectashield Antifade Mounting Medium | Vector Laboratories | Cat# H-1000 |
| Deposited data | ||
| Astrocyte single cell data from GH and SH | This paper | GSE222785 |
| Software and algorithms | ||
| ANYmaze | Stoelting Co. | NA |
| Versadat | Omnitech Electronics, Inc. | NA |
| 10X Cell Ranger (3.0.2) | 10X genomics | NA |
| Seurat (version 4.1.0) | Hao et al., 2021 | NA |
| DoubletFinder | McGinnis et al., 2019 | NA |
| SingleR | Aran et al. 2019 | NA |
| enrichR | Kuleshov et al., 2016 | |
| ClampFit 10.7 software | Molecular Devices LLC | NA |
| GECIquant | Venugopal et al., 2019 | NA |
| AQuA | Wang et al., 2019 | NA |
| Minianalysis | Blue cell | NA |
| SigmaPlot 13 | SYSTAT | NA |
| Prism 9 | Graphad | NA |
| Imaris | Oxford instruments | NA |
Highlights.
Social deprivation alters molecular and cellular properties of hippocampal astrocytes
Astrocytic TRPA1 suppresses mouse hippocampal circuit function after social deprivation
TRPA1 mediates elevated tonic GABA release from astrocytes after social deprivation
Astrocytic GABA suppresses hippocampal circuit function after social deprivation
Acknowledgments
This work was supported by US National Institutes of Health grants NS071153, AG071687, and NS096096 to BD. We are thankful for support from the David and Eula Wintermann Foundation and SE Lee, MH Nam (Korea Institute of Science and Technology, KIST) and C.J. Lee (Institute of Basic Science, IBS) for providing viral constructs (GFAP-MAOB, MAOB shRNA, and Best1 shRNA). scRNA-Seq studies were performed at the Single Cell Genomics Core at BCM partially supported by NIH shared instrument grants (S10OD023469, S10OD025240) and P30EY002520. This project was supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the CPRIT Core Facility Support Award (CPRIT-RP180672), the NIH (CA125123 and RR024574) and the assistance of Joel M. Sederstrom. Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Number P50HD103555 for use of the Microscopy Core facilities and the Animal Phenotyping & Preclinical Endpoints Core facilities.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pollak SD, Nelson CA, Schlaak MF, Roeber BJ, Wewerka SS, Wiik KL, Frenn KA, Loman MM, and Gunnar MR (2010). Neurodevelopmental effects of early deprivation in postinstitutionalized children. Child Dev 81, 224–236. 10.1111/j.1467-8624.2009.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Widom CS, DuMont K, and Czaja SJ (2007). A prospective investigation of major depressive disorder and comorbidity in abused and neglected children grown up. Arch. Gen. Psychiatry 64, 49–56. 10.1001/archpsyc.64.1.49. [DOI] [PubMed] [Google Scholar]
- 3.Rubin KH, and Mills RSL (1988). The Many Faces of Social Isolation in Childhood. J Consult Clin Psych 56, 916–924. 10.1037/0022-006x.56.6.916. [DOI] [PubMed] [Google Scholar]
- 4.Loades ME, Chatburn E, Higson-Sweeney N, Reynolds S, Shafran R, Brigden A, Linney C, McManus MN, Borwick C, and Crawley E (2020). Rapid Systematic Review: The Impact of Social Isolation and Loneliness on the Mental Health of Children and Adolescents in the Context of COVID-19. J Am Acad Child Psy 59, 1218–1239.e3. 10.1016/j.jaac.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cacioppo JT, Cacioppo S, Capitanio JP, and Cole SW (2015). The neuroendocrinology of social isolation. Annu Rev Psychol 66, 733–767. 10.1146/annurev-psych-010814-015240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cacioppo JT, and Hawkley LC (2009). Perceived social isolation and cognition. Trends in Cognitive Sciences 13, 447–454. 10.1016/j.tics.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fone KCF, and Porkess MV (2008). Behavioural and neurochemical effects of post-weaning social isolation in rodents-relevance to developmental neuropsychiatric disorders. Neuroscience and Biobehavioral Reviews 32, 1087–1102. 10.1016/j.neubiorev.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Harlow HF, Dodsworth RO, and Harlow MK (1965). Total social isolation in monkeys. Proc National Acad Sci 54, 90–97. 10.1073/pnas.54.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sánchez MM, Hearn EF, Do D, Rilling JK, and Herndon JG (1998). Differential rearing affects corpus callosum size and cognitive function of rhesus monkeys. Brain Research 812, 38–49. 10.1016/s0006-8993(98)00857-9. [DOI] [PubMed] [Google Scholar]
- 10.Joseph R (1999). Environmental Influences on Neural Plasticity, the Limbic System, Emotional Development and Attachment: A Review. Child Psychiatry Hum Dev 29, 189–208. 10.1023/a:1022660923605. [DOI] [PubMed] [Google Scholar]
- 11.Ibi D, Takuma K, Koike H, Mizoguchi H, Tsuritani K, Kuwahara Y, Kamei H, Nagai T, Yoneda Y, Nabeshima T, et al. (2008). Social isolation rearing-induced impairment of the hippocampal neurogenesis is associated with deficits in spatial memory and emotion-related behaviors in juvenile mice. J. Neurochem 105, 921–932. 10.1111/j.1471-4159.2007.05207.x. [DOI] [PubMed] [Google Scholar]
- 12.Matthews GA, Nieh EH, Weele CMV, Halbert SA, Pradhan RV, Yosafat AS, Glober GF, Izadmehr EM, Thomas RE, Lacy GD, et al. (2016). Dorsal Raphe Dopamine Neurons Represent the Experience of Social Isolation. Cell 164, 617–631. 10.1016/j.cell.2015.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva-Gómez AB, Rojas D, Juárez I, and Flores G (2003). Decreased dendritic spine density on prefrontal cortical and hippocampal pyramidal neurons in postweaning social isolation rats. Brain Res 983, 128–136. 10.1016/s0006-8993(03)03042-7. [DOI] [PubMed] [Google Scholar]
- 14.Wallace DL, Han M-H, Graham DL, Green TA, Vialou V, Iñiguez SD, Cao J-L, Kirk A, Chakravarty S, Kumar A, et al. (2009). CREB regulation of nucleus accumbens excitability mediates social isolation–induced behavioral deficits. Nat Neurosci 12, 200–209. 10.1038/nn.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zelikowsky M, Hui M, Karigo T, Choe A, Yang B, Blanco MR, Beadle K, Gradinaru V, Deverman BE, and Anderson DJ (2018). The Neuropeptide Tac2 Controls a Distributed Brain State Induced by Chronic Social Isolation Stress. CELL 173, 1265–1268.e19. 10.1016/j.cell.2018.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makinodan M, Rosen KM, Ito S, and Corfas G (2012). A critical period for social experience-dependent oligodendrocyte maturation and myelination. Science 337, 1357–1360. 10.1126/science.1220845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stogsdill JA, Ramirez J, Liu D, Kim YH, Baldwin KT, Enustun E, Ejikeme T, Ji R-R, and Eroglu C (2017). Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature 551, 192–197. 10.1038/nature24638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen NJ (2014). Astrocyte Regulation of Synaptic Behavior. Cell Dev Biology 30, 439–463. 10.1146/annurev-cellbio-100913-013053. [DOI] [PubMed] [Google Scholar]
- 19.Allen NJ, and Eroglu C (2017). Cell Biology of Astrocyte-Synapse Interactions. Neuron 96, 697–708. 10.1016/j.neuron.2017.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khakh BS, and Deneen B (2019). The Emerging Nature of Astrocyte Diversity. Annu Rev Neurosci 42, 187–207. 10.1146/annurev-neuro-070918-050443. [DOI] [PubMed] [Google Scholar]
- 21.Nagai J, Yu X, Papouin T, Cheong E, Freeman MR, Monk KR, Hastings MH, Haydon PG, Rowitch D, Shaham S, et al. (2021). Behaviorally consequential astrocytic regulation of neural circuits. Neuron, 1–21. 10.1016/j.neuron.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adamsky A, Kol A, Kreisel T, Doron A, Ozeri-Engelhard N, Melcer T, Refaeli R, Horn H, Regev L, Groysman M, et al. (2018). Astrocytic Activation Generates De Novo Neuronal Potentiation and Memory Enhancement. CELL 174, 59–71.e14. 10.1016/j.cell.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Bazargani N, and Attwell D (2016). Astrocyte calcium signaling: the third wave. Nat Neurosci 19, 182–189. 10.1038/nn.4201. [DOI] [PubMed] [Google Scholar]
- 24.Mederos S, Sánchez-Puelles C, Esparza J, Valero M, Ponomarenko A, and Perea G (2020). GABAergic signaling to astrocytes in the prefrontal cortex sustains goal-directed behaviors. Nat Neurosci 24, 1–29. 10.1038/s41593-020-00752-x. [DOI] [PubMed] [Google Scholar]
- 25.Nagai J, Rajbhandari AK, Gangwani MR, Hachisuka A, Coppola G, Masmanidis SC, Fanselow MS, and Khakh BS (2019). Hyperactivity with Disrupted Attention by Activation of an Astrocyte Synaptogenic Cue. CELL 177, 1280–1292.e20. 10.1016/j.cell.2019.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shigetomi E, Tong X, Kwan KY, Corey DP, and Khakh BS (2011). TRPA1 channels regulate astrocyte resting calcium and inhibitory synapse efficacy through GAT-3. Nature Publishing Group; 15, 70–80. 10.1038/nn.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shigetomi E, Jackson-Weaver O, Huckstepp RT, O’Dell TJ, and Khakh BS (2013). TRPA1 channels are regulators of astrocyte basal calcium levels and long-term potentiation via constitutive D-serine release. J. Neurosci 33, 10143–10153. 10.1523/jneurosci.5779-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaboub LS, and Deneen B (2012). Developmental origins of astrocyte heterogeneity: the final frontier of CNS development. Dev Neurosci 34, 379–388. 10.1159/000343723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang AY-S, Woo J, Sardar D, Lozzi B, Huerta NAB, Lin C-CJ, Felice D, Jain A, Paulucci-Holthauzen A, and Deneen B (2020). Region-Specific Transcriptional Control of Astrocyte Function Oversees Local Circuit Activities. Neuron, 1–27. 10.1016/j.neuron.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khakh BS, and McCarthy KD (2015). Astrocyte Calcium Signaling: From Observations to Functions and the Challenges Therein. Cold Spring Harbor Perspectives in Biology 7, a020404–17. 10.1101/cshperspect.a020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon B, Woo J, Chun Y, Chun H, Jo S, Bae JY, An H, Min JO, Oh S, Han K, et al. (2014). Glial GABA, synthesized by monoamine oxidase B, mediates tonic inhibition. J Physiology 592, 4951–4968. 10.1113/jphysiol.2014.278754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S, Yoon B-E, Berglund K, Oh S-J, Park H, Shin H-S, Augustine GJ, and Lee CJ (2010). Channel-Mediated Tonic GABA Release from Glia. Science 330, 790–796. 10.1126/science.1184334. [DOI] [PubMed] [Google Scholar]
- 33.Pandit S, Neupane C, Woo J, Sharma R, Nam M, Lee G, Yi M, Shin N, Kim DW, Cho H, et al. (2020). Bestrophin1-mediated tonic GABA release from reactive astrocytes prevents the development of seizure-prone network in kainate-injected hippocampi. Glia 68, 1065–1080. 10.1002/glia.23762. [DOI] [PubMed] [Google Scholar]
- 34.Woo J, Min JO, Kang D-S, Kim YS, Jung GH, Park HJ, Kim S, An H, Kwon J, Kim J, et al. (2018). Control of motor coordination by astrocytic tonic GABA release through modulation of excitation/inhibition balance in cerebellum. Proc National Acad Sci 115, 5004–5009. 10.1073/pnas.1721187115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bushong EA, Martone ME, and Ellisman MH (2004). Maturation of astrocyte morphology and the establishment of astrocyte domains during postnatal hippocampal development. International Journal of Developmental Neuroscience 22, 73–86. 10.1016/j.ijdevneu.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 36.Kang K, Lee S, Han JE, Choi JW, and Song M (2014). The complex morphology of reactive astrocytes controlled by fibroblast growth factor signaling. Glia 62, 1328–1344. 10.1002/glia.22684. [DOI] [PubMed] [Google Scholar]
- 37.Zhou B, Zuo Y, and Jiang R (2019). Astrocyte morphology: Diversity, plasticity, and role in neurological diseases. Cns Neurosci Ther 25, 665–673. 10.1111/cns.13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Armbruster M, Naskar S, Garcia JP, Sommer M, Kim E, Adam Y, Haydon PG, Boyden ES, Cohen AE, and Dulla CG (2022). Neuronal activity drives pathway-specific depolarization of peripheral astrocyte processes. Nat Neurosci 25, 607–616. 10.1038/S41593-022-01049-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olsen ML, Khakh BS, Skatchkov SN, Zhou M, Lee CJ, and Rouach N (2015). New Insights on Astrocyte Ion Channels: Critical for Homeostasis and Neuron-Glia Signaling. Journal of Neuroscience 35, 13827–13835. 10.1523/jneurosci.2603-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jo S, Yarishkin O, Hwang YJ, Chun YE, Park M, Woo DH, Bae JY, Kim T, Lee J, Chun H, et al. (2014). GABA from reactive astrocytes impairs memory in mouse models of Alzheimer’s disease. Nat Med 20, 886–896. 10.1038/nm.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shigetomi E, Saito K, Sano F, and Koizumi S (2019). Aberrant Calcium Signals in Reactive Astrocytes: A Key Process in Neurological Disorders. Int J Mol Sci 20, 996. 10.3390/ijms20040996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oh S-J, Lee JM, Kim H-B, Lee J, Han S, Bae JY, Hong G-S, Koh W, Kwon J, Hwang E-S, et al. (2019). Ultrasonic Neuromodulation via Astrocytic TRPA1. Curr Biol 29, 3386–3401.e8. 10.1016/j.cub.2019.08.021. [DOI] [PubMed] [Google Scholar]
- 43.Paumier A, Boisseau S, Jacquier-Sarlin M, Pernet-Gallay K, Buisson A, and Albrieux M (2021). Astrocyte–neuron interplay is critical for Alzheimer’s disease pathogenesis and is rescued by TRPA1 channel blockade. Brain. 10.1093/brain/awab281. [DOI] [PubMed] [Google Scholar]
- 44.Arzate-Mejía RG, Lottenbach Z, Schindler V, Jawaid A, and Mansuy IM (2020). Long-Term Impact of Social Isolation and Molecular Underpinnings. Frontiers Genetics 11, 589621. 10.3389/fgene.2020.589621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amir RE, den Veyver IBV, Wan M, Tran CQ, Francke U, and Zoghbi HY (1999). Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23, 185–188. 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 46.Kumsta R, Marzi SJ, Viana J, Dempster EL, Crawford B, Rutter M, Mill J, and Sonuga-Barke EJS (2016). Severe psychosocial deprivation in early childhood is associated with increased DNA methylation across a region spanning the transcription start site of CYP2E1. Transl Psychiat 6, e830–e830. 10.1038/tp.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ung K, Huang T-W, Lozzi B, Woo J, Hanson E, Pekarek B, Tepe B, Sardar D, Cheng Y-T, Liu G, et al. (2021). Olfactory bulb astrocytes mediate sensory circuit processing through Sox9 in the mouse brain. Nature Communications, 1–15. 10.1038/S41467-021-25444-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheng M, and Greenberg ME (1990). The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron 4, 477–485. 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 49.West AE, Griffith EC, and Greenberg ME (2002). Regulation of transcription factors by neuronal activity. Nat Rev Neurosci 3, 921–931. 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- 50.West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, Shaywitz AJ, Takasu MA, Tao X, and Greenberg ME (2001). Calcium regulation of neuronal gene expression. Proc. Natl. Acad. Sci. USA 98, 11024–11031. 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yap E-L, and Greenberg ME (2018). Activity-Regulated Transcription: Bridging the Gap between Neural Activity and Behavior. Neuron 100, 330–348. 10.1016/j.neuron.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lanjakornsiripan D, Pior B-J, Kawaguchi D, Furutachi S, Tahara T, Katsuyama Y, Suzuki Y, Fukazawa Y, and Gotoh Y (2018). Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nature Communications 9, 1–15. 10.1038/s41467-018-03940-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG, et al. (2017). QuPath: Open source software for digital pathology image analysis. Sci Rep-uk 7, 16878. 10.1038/S41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGinnis CS, Murrow LM, and Gartner ZJ (2019). DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst 8, 329–337.e4. 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ximerakis M, Lipnick SL, Innes BT, Simmons SK, Adiconis X, Dionne D, Mayweather BA, Nguyen L, Niziolek Z, Ozek C, et al. (2019). Single-cell transcriptomic profiling of the aging mouse brain. Nat Neurosci 22, 1696–1708. 10.1038/S41593-019-0491-3. [DOI] [PubMed] [Google Scholar]
- 57.Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. (2016). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19, 335–346. 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, et al. (2019). Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 20, 163–172. 10.1038/S41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44, W90–W97. 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwak H, Koh W, Kim S, Song K, Shin J-I, Lee JM, Lee EH, Bae JY, Ha GE, Oh J-E, et al. (2020). Astrocytes Control Sensory Acuity via Tonic Inhibition in the Thalamus. Neuron 108, 691–706.e10. 10.1016/j.neuron.2020.08.013. [DOI] [PubMed] [Google Scholar]
- 61.Vaidyanathan TV, Collard M, Yokoyama S, Reitman ME, and Poskanzer KE (2021). Cortical astrocytes independently regulate sleep depth and duration via separate GPCR pathways. Elife 10, e63329. 10.7554/elife.63329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of astrocyte subpopulation markers, related to Figure 2.
Table S2. List of differentially expressed genes in astrocyte clusters, related to Figure 2.
Data Availability Statement
The RNA-seq dataset generated during this study are available at the NCBI GEO website.