Abstract
We have examined binding of the CREB B-ZIP protein domain to double-stranded DNA containing a consensus CRE sequence (5′-TGACGTCA-3′), the related PAR, C/EBP and AP-1 sequences and the unrelated SP1 sequence. DNA binding was assayed in the presence or absence of MgCl2 and/or KCl using two methods: circular dichroism (CD) spectroscopy and electrophoretic mobility shift assay (EMSA). The CD assay allows us to measure equilibrium binding in solution. Thermal denaturation in 150 mM KCl indicates that the CREB B-ZIP domain binds all the DNA sequences, with highest affinity for the CRE site, followed by the PAR (5′-TAACGTTA-3′), C/EBP (5′-TTGCGCAA-3′) and AP-1 (5′-TGAGTCA-3′) sites. The addition of 10 mM MgCl2 diminished DNA binding to the CRE and PAR DNA sequences and abolished binding to the C/EBP and AP-1 DNA sequences, resulting in more sequence-specific DNA binding. Using ‘standard’ EMSA conditions (0.25× TBE), CREB bound all the DNA sequences examined. The CREB–CRE complex had an apparent Kd of ∼300 pM, PAR of ∼1 nM, C/EBP and AP-1 of ∼3 nM and SP1 of ∼30 nM. The addition of 10 mM MgCl2 to the polyacrylamide gel dramatically altered sequence-specific DNA binding. CREB binding affinity for CRE DNA decreased 3-fold, but binding to the other DNA sequences decreased >1000-fold. In the EMSA, addition of 150 mM KCl to the gels had an effect similar to MgCl2. The magnesium concentration needed to prevent non-specific electrostatic interactions between CREB and DNA in solution is in the physiological range and thus changes in magnesium concentration may be a cellular signal that regulates gene expression.
INTRODUCTION
Basic region leucine zipper (B-ZIP) proteins are a large class of dimeric transcription factors that bind up to 10 bp of contiguous DNA in a sequence-specific manner (1–4). X-ray crystal structures for several B-ZIP proteins bound to DNA have been published, including the yeast GCN4 homodimer (2,5), the Fos–Jun heterodimer (6), the CREB homodimer (7) and the yeast PAP-1 homodimer (8). While these have begun to reveal the structural basis of DNA binding specificities, the X-ray structures do not provide information about the relative affinities of B-ZIP proteins for different DNA sequences. This information is critical to understanding the mechanisms that regulate gene expression.
In vitro data reveal that the same DNA sequence is bound by several B-ZIP proteins. For example, the palindromic cAMP response element, the CRE (5′-TGACGTCA-3′), can be bound by CREB homodimers (9), ATF2 homodimers (10), ATF3 homodimers (10), C/EBP family members and Jun family–Fos family heterodimers (11). Knowing the binding affinities of different B-ZIP proteins for the same DNA sequence and the same B-ZIP protein for different DNA sequences will give us an insight into which binding is relevant in vivo.
The recently described crystal structure of the CREB B-ZIP domain bound to a consensus CRE DNA sequence identified a hexahydrated Mg2+ ion in the cavity between the bifurcating basic regions and DNA (7). Fluorescence polarization experiments indicated that the CREB–CRE complex was unstable in the absence of MgCl2 and dramatically stabilized in the presence of 10 mM MgCl2. In contrast, CREB binding to CRE-related sites was unaffected by the presence of 10 mM MgCl2 (12).
We have examined binding of the CREB B-ZIP domain to a consensus CRE DNA sequence, three related DNA sequences and the unrelated SP1 DNA sequence using both circular dichroism (CD) thermal denaturation and electrophoretic mobility shift assay (EMSA). CD thermal denaturation experiments indicate that in 150 mM KCl CREB binds to all the DNA sequences examined. We find that the addition of 10 mM MgCl2 inhibits CREB binding to non-consensus DNA sequences. Thus magnesium is required for sequence-specific CREB binding. We also show that CREB does not exhibit sequence-specific DNA binding under ‘standard’ EMSA conditions (0.25–1× TBE buffer), making this popular assay inadequate for examining relative affinities. However, the addition of millimolar amounts of either MgCl2 or KCl to the polyacrylamide gel obliterates binding to non-consensus DNA sequences but does not appreciably affect binding of CREB to the CRE-binding site. Similar results were obtained for C/EBP, Fos–JunD and PAR B-ZIP domain dimers. Thus, a simple modification allows the EMSA to be used to study sequence-specific DNA binding of transcription factors.
MATERIALS AND METHODS
Proteins
The sequences of the expressed B-ZIP domains CREB (13), Fos and Jun (14), the PAR family member VBP (15) and C/EBP (16) have been reported elsewhere. The B-ZIP domains are 10 amino acids longer than the fragment used in the crystal structure of both GCN4 and the Fos–Jun heterodimer complex. Proteins were expressed in Escherichia coli using the T7 IPTG-inducible system (17) and purified and quantified as described previously (14).
Circular dichroism
CD studies were performed using a Jasco J-720 spectropolarimeter with a 5 mm rectangular CD cell. All protein stock solutions were in 12.5 mM HEPES, pH 7.4, 150 mM KCl, 0.25 mM EDTA. DTT was added to a final concentration of 1 mM. The protein solutions were heated to 65°C for 20 min to disrupt any potential disulfide bonds, cooled to room temperature for 5 min and diluted to 4 µM in a 1.5 ml sample volume. DNA oligonucleotides (32mer) were purchased from Genosys as HPLC-purified products. DNA was resuspended in water and the concentration was determined from the absorbance at 260 nm. To produce double-stranded DNA, single-stranded DNA was brought to 5 µM in 12.5 mM HEPES, pH 7.4, 150 mM KCl, 0.25 mM EDTA, heated to 80°C for 5 min, put on ice for 5 min and then incubated at room temperature for 15 min. For denaturations containing DNA and protein, protein and DTT were added bringing the final concentration to 4 µM DNA, 4 µM protein and 1 mM DTT. This mixture was heated to 65°C for 20 min and introduced into the CD cuvette for melting.
EMSA
Each single strand of DNA was radiolabeled using polynucleotide kinase. An aliquot of 7 µl of [γ-32P]dATP (3000 Ci/mmol) was added to 1 µg oligo and incubated at 37°C for 45 min in a 10 µl reaction. Purified B-ZIP domains were heated for 10 min at 65°C in the presence of 1 mM DTT and added to 20 µl of the gel shift reaction buffer [12.5 mM phosphate, pH 7.4, 150 mM KCl, 0.25 mM EDTA, 2.5 mM DTT, 2 mg/ml bovine serum albumin (BSA) and 2% glycerol]. The BSA is essential to prevent non-specific binding of the protein. This solution was incubated for 10 min at 65°C and then incubated with 30 pM 32P-labeled double-stranded oligonucleotide for 10 min at 37°C.
The binding complexes were resolved on a 7.5% polyacrylamide gel in 0.25% TBE buffer (25 mM Tris base, 25 mM boric acid and 0.5 mM EDTA) at room temperature. Some gels contained either 1–10 mM MgCl2 or 150 mM KCl. The gels were pre-run for 15 min, loaded in the absence of any marking dyes and electrophoresed for 1 h. We used a home-built gel electrophoresis system 20 cm high and 35 cm wide with a gel thickness of 0.3 mm. The thinness of the gel and thickness of the glass plates prevents overheating. The gels were run at 100 V constant voltage. The observed current for the 0.25× TBE gel was 10 mA, for the MgCl2 gel was 20 mA and for the KCl gel was 70 mA. The sequences of the five 28mer DNA probes used in the EMSA experiments were, with the consensus binding sites underlined: CREB, GTCAGTCAGATGACGTCATATCGGTCAG; AP-1, GTCAGTCAGAATGACTCATATCGGTCAG; PAR, GTCAGTCAGATTACGTAATATCGGTCAG; C/EBP, GTCAGTCAGATTGCGCAATATCGGTCAG; SP1, GTCAGTCAGGGGGCGGGGCATCGGTCAG.
RESULTS
Circular dichroism denaturation of the CREB B-ZIP domain and DNA
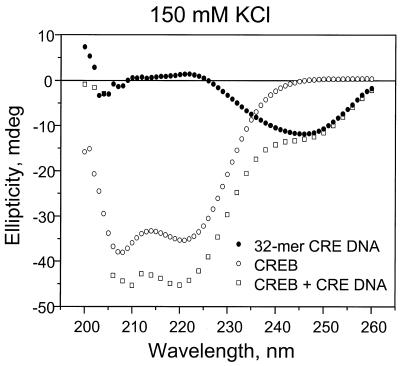
We have used CD thermal denaturation to monitor the stability of the CREB B-ZIP domain bound to four DNA sequences in the presence of 150 mM KCl with or without 10 mM MgCl2. Figure 1 presents CD spectra, collected at 6°C, for the CREB B-ZIP domain, double-stranded DNA containing a consensus CRE-binding site and a mixture of the protein and DNA. The CREB B-ZIP domain has CD minima at 208 and 222 nm, indicative of α-helical structure (208 nm) and helix–helix interactions (222 nm). The DNA has little CD at these wavelengths, but has a minimum at 245 nm. The mixture has increases in ellipticity at 208 and 222 nm that likely represent the basic region becoming α-helical upon DNA binding (18,19). The DNA does not produce a significant spectroscopic signature at 222 nm, allowing us to monitor B-ZIP domain secondary structure in the absence and presence of DNA.
Figure 1.
CD spectra at 6°C of 4 µM CREB, 4 µM CRE-containing DNA and a mixture of the two in 12.5 mM HEPES, pH 7.4, 150 mM KCl and 0.25 mM EDTA.
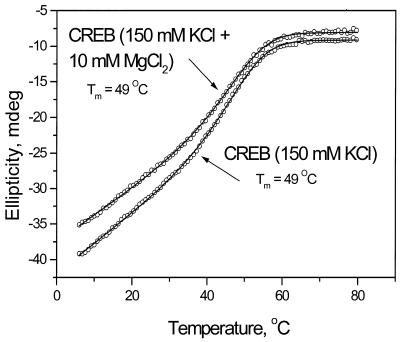
The thermal stability of the CREB B-ZIP protein domain in 150 mM KCl, 12.5 mM HEPES pH 7.4, 0.25 mM EDTA and 1.0 mM DTT did not change significantly with the addition of 10 mM MgCl2 (Fig. 2). We performed these denaturations in HEPES, instead of our standard phosphate buffer, because of salt formation between phosphate and magnesium upon heating. The CREB B-ZIP domain is well fitted by a two-state equilibrium with a Tm of 49°C (13). Addition of 10 mM MgCl2 causes a slight decrease in ellipticity, but no change in Tm.
Figure 2.
Thermal stability of the CREB B-ZIP protein domain in the absence and presence of 10 mM MgCl2.
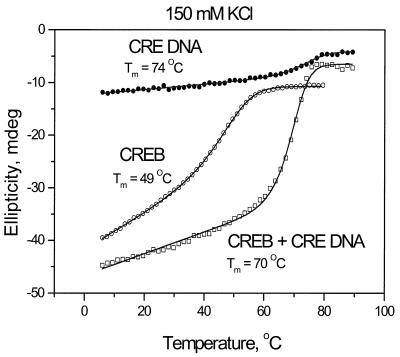
Binding to a 32 bp DNA oligonucleotide containing a CRE site in the presence of 150 mM KCl increased the stability of the CREB B-ZIP domain from 49 to 70°C (Fig. 3). This is below the thermal transition of double-stranded to single-stranded DNA (74°C), suggesting that the change in ellipticity of the complex results from denaturation of the α-helix dimeric CREB domain from double-stranded DNA. Addition of DNA causes several changes in the denaturation profile: (i) an ellipticity increase likely representing the basic region becoming α-helical upon DNA binding; (ii) the low temperature baseline becomes flatter and thermal denaturation becomes more cooperative, with ΔH increasing from –59 to –114 kcal/mol. In the absence of DNA, the basic region unfolds non-cooperatively from an α-helix to a random coil, resulting in the inclined baseline. In the presence of DNA, the basic region is bound in the major groove of DNA and denatures cooperatively, resulting in a more horizontal low temperature baseline. The flatter baseline and increased cooperativity are reflections of the stabilized α-helical basic region that only unfolds when the entire B-ZIP domain denatures. These denaturations are reversible (data not shown).
Figure 3.
Thermal denaturation of the CREB B-ZIP domain, 32 bp double-stranded DNA and a mixture of the two in standard CD buffer (150 mM KCl). The thermal denaturations containing protein were collected at 222 nm and those with DNA alone at 245 nm.
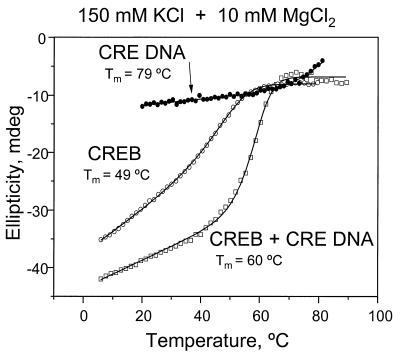
We examined whether the addition of 10 mM MgCl2 affected CREB binding to the CRE site (Fig. 4). As stated previously, the thermal stability of the CREB B-ZIP domain is unaffected by addition of 10 mM MgCl2. MgCl2 increased the Tm of the double-stranded DNA (20–23) from 74 to 79°C. In contrast, the Tm of CREB bound to the CRE DNA sequence decreased from 70 to 60°C. The general shape of the denaturations was independent of MgCl2, suggesting that the same structural transitions occur with or without magnesium. The high affinity of magnesium for DNA suggests that the decrease in stability of the complex results from competition between the B-ZIP domain and the magnesium for electrostatic interactions with DNA.
Figure 4.
Thermal denaturation of the CREB B-ZIP domain, 32 bp double-stranded DNA and a mixture of the two in standard CD buffer (150 mM KCl) + 10 mM MgCl2. The thermal denaturations containing protein were collected at 222 nm and those with DNA alone at 245 nm.
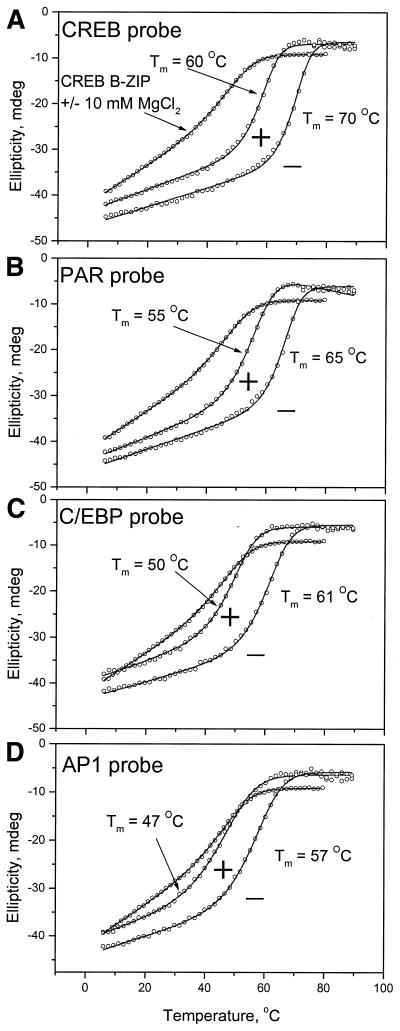
Figure 5 shows CREB binding to a CRE DNA and three CRE-related DNA sequences in the absence and presence of 10 mM MgCl2. The CRE-related sequences examined were as follows. (i) The consensus DNA binding site for PAR family members VBP/TEF, HLF and DBP (5′-TTACGTAA-3′). The PAR site varies from the CRE site by a change in a single base pair 3 bp from the center of the dyad, highlighted in bold. (ii) The consensus site for C/EBPα B-ZIP proteins (5′-ATTGCGCAAT-3′). This varies from the consensus CRE site by changes at the second and third base pairs from the center of the dyad. (iii) The consensus site for the AP-1 heterodimer complex, termed the TRE site (5′-AGTGACT-3′). This varies from the CRE site by deletion of a single base in the middle of the dyad. The stabilities of these double-stranded DNA sequences are presented in Table 1.
Figure 5.
CD thermal denaturation of CREB bound to four DNA sequences in the presence of 150 mM KCl with or without 10 mM MgCl2. The line through each denaturation curve is a fitted curve assuming a two-state transition. (A) CRE DNA. (B) PAR DNA. (C) C/EBP DNA. (D) AP-1 DNA. In all panels the denaturation curve of the CREB B-ZIP domain is included for reference.
Table 1. Thermal stability of the CREB B-ZIP domain bound to four DNA sequences.
| Tm (°C) | ||||
|---|---|---|---|---|
| DNA (32mer) | CREB | |||
| EDTA | MgCl2 | EDTA | MgCl2 | |
| CREB | 74 | 79 | 70 | 60 |
| PAR | 71 | 75 | 65 | 55 |
| C/EBP | 73 | 77 | 61 | 50 |
| AP-1 | 75 | 78 | 57 | 47 |
The left panel presents the melting temperature determined from CD thermal melts for 32 bp double-stranded DNAs in standard CD buffer (150 mM KCl) with or without 10 mM MgCl2 monitored at 245 nm. The right panel presents the stability of the CREB B-ZIP domain, in standard CD buffer (150 mM KCl) with or without 10 mM MgCl2, bound to these DNAs as determined by monitoring at 222 nm.
The CREB B-ZIP domain bound all three CRE-related DNA sites (Fig. 5 and Table 1), albeit with lower affinity than to the CRE site. CREB bound the PAR sequence with a Tm of 65°C, C/EBP with a Tm of 61°C and AP-1 with a Tm of 57°C. In all cases, the ellipticity of the CREB B-ZIP domain increased and the cooperativity of denaturation was steeper for DNA-bound than for unbound CREB B-ZIP domain. The addition of 10 mM MgCl2 decreased the stability of the CREB–DNA complexes while the shapes of the denaturation profiles remained unchanged (Fig. 5). For the C/EBP and AP-1 DNA sequences the Tm of the CREB–DNA complex was similar to the Tm of CREB alone. While the more negative ellipticity and more horizontal low temperature baseline are indicative of CREB binding to the C/EBP and AP-1 DNA probes, the lack of a change in Tm suggests that DNA binding does not stabilize the CREB leucine zipper dimer. These data indicate that 10 mM MgCl2 is needed in addition to 150 mM KCl to prevent electrostatic interaction between CREB and non-specific DNA.
EMSA
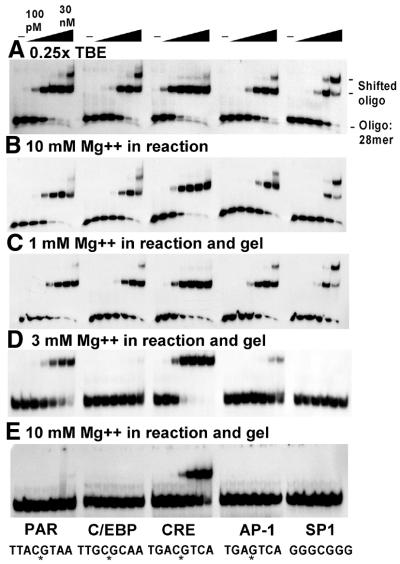
We also used EMSA to examine binding of the CREB B-ZIP domain to the DNA sequences used in the CD experiments, as well as the SP1 site (5′-GGGCGGGG-3′). The binding reaction conditions were the same as for the CD thermal denaturations with the addition of 2 mg/ml BSA and 2% glycerol. We ran a 7.5% polyacrylamide gel in 0.25× TBE. Figure 6A is a ‘standard’ EMSA (0.25× TBE) that examines binding of half-log dilutions of the CREB B-ZIP domain to five DNA sequences. CREB bound with the following apparent Kd values: CRE, ∼0.3 nM; PAR, ∼1 nM; C/EBP and AP-1, ∼3 nM; SP1, ∼30 nM. Thus CREB binds the CRE site with the highest affinity but also binds related DNA sequences as well.
Figure 6.
(A) Gels shifts of CREB B-ZIP dimers bound to five DNA sequences. CREB B-ZIP dimer (0.1, 0.3, 1, 3, 10 and 30 nM) was bound to 30 pM radiolabeled 28 bp double-stranded DNA containing the ‘consensus’ binding site for each of the five proteins. The consensus DNA-binding site is noted at the bottom of the figure. (B) 10 mM MgCl2 was added to the reaction. (C) 1 mM MgCl2 was added to the reaction and polyacrylamide gel. (D) 3 mM MgCl2 was added to the reaction and polyacrylamide gel. (E) 10 mM MgCl2 was added to the reaction and polyacrylamide gel.
One feature of CREB binding to non-CRE DNA sequences seen in Figure 6A is the appearance of a second protein–DNA band at higher CREB concentrations. This is most pronounced on the SP1 probe, the most divergent of the DNA sequences tested. We suggest that the slower migrating band represents two dimers of CREB bound to DNA. This can occur most easily on the SP1 probe because CREB binding is non-specific and thus can occur anywhere along the DNA sequence. The absence of a second band on the CRE oligonucleotide likely indicates that when a CREB dimer is bound specifically in the middle of the 28 bp oligonucleotide, a second dimer is unable to bind the DNA.
Effect of MgCl2
To determine whether magnesium would have a similar effect in the gel shift assay as in solution, we added 10 mM MgCl2 to the binding reactions. This caused a 3-fold decrease in binding to all the sites (Fig. 6B). In the EMSA, the protein–DNA complex is fractionated from the unbound components because of a difference in net charge and size. The polyacrylamide gels are typically run in low salt for two reasons: (i) to maximize electrostatic attraction between the protein–DNA complexes; (ii) to keep the current low to prevent thermal denaturation of the protein–DNA complexes.
In an attempt to recapitulate the sequence-specific DNA binding seen in the CD denaturations, we examined whether adding MgCl2 to the polyacrylamide gel would enhance DNA binding specificity. Gels were cast containing 1, 3 or 10 mM MgCl2 and CREB binding to the five DNA sequences was examined. MgCl2 at 1 mM does not effect DNA binding to the five DNA sequences examined (Fig. 6C). In contrast, at 3 mM MgCl2 inhibits DNA binding to the C/EBP, AP-1 and SP1 sites 30-fold, without affecting binding to the CREB and PAR sites (Fig. 6D). At 10 mM MgCl2 decreases binding to the CREB site 3-fold and to the PAR site >30-fold (Fig. 6E). Increasing the protein concentration from 30 to 1000 nM does not reveal any additional DNA binding (data not shown). Thus to reveal sequence-specific binding by EMSA, MgCl2 had to be added not only to the binding reaction but also to the gel itself. Similar results were obtained using 150 mM KCl in the polyacrylamide gel.
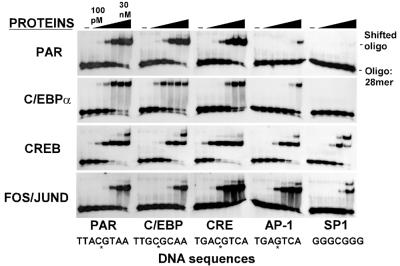
DNA binding of Fos–JunD, C/EBP and PAR with or without MgCl2
The generality of the result that MgCl2 increased DNA binding specificity by decreasing non-specific DNA binding was addressed by examining three additional B-ZIP dimers (Fig. 7), namely PAR (24), C/EBPα and the Fos–JunD heterodimer. Based on the ‘standard’ EMSA, PAR binds its reported cognate site with a Kd of ∼3 nM. However, it also binds the related C/EBP and CRE sites equally well. It binds the TRE site with 30-fold lower affinity than its cognate site and does not bind the SP1 site. C/EBPα binds to its site with a Kd of ∼1 nM and to the PAR site with a Kd of ∼3 nM. The Fos–JunD heterodimer binds the AP-1 and CRE sites with an affinity of 1–3 nM and the C/EBP site with an affinity of 10 nM.
Figure 7.
EMSA of four B-ZIP dimers (PAR, C/EBP, CREB and Fos–JunD) bound to five DNA sequences. Half-log dilutions of the indicated B-ZIP dimers (0.1, 0.3, 1, 3, 10 and 30 nM) were bound to 30 pM radiolabeled 28 bp double-stranded DNA containing the ‘consensus’ binding site.
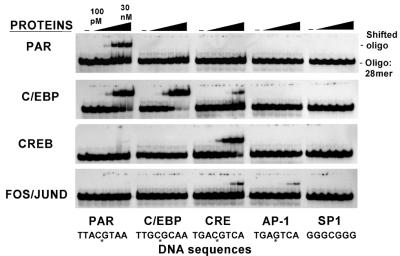
Adding 10 mM MgCl2 to the polyacrylamide gel dramatically altered binding of these B-ZIP dimers to DNA (Fig. 8). PAR now bound only to its cognate site. MgCl2 increased the specificity of C/EBPα binding, although it bound more promiscuously than PAR, as has been previously observed (25). The DNA binding specificity of C/EBPα is broader than PAR due to a conserved valine in the C/EBPα basic region, compared to the alanine in PAR (25). The Fos–JunD heterodimer binds the TRE and CRE sites but not the other sites. These results indicate that MgCl2-containing gels increase the sequence specificity of DNA binding by B-ZIP proteins.
Figure 8.
EMSA with 10 mM MgCl2 in the binding reaction and polyacryamide gel. Four B-ZIP dimers (0.1, 0.3, 1, 3, 10 and 30 nM) (PAR, C/EBP, CREB and Fos–JunD) were bound to 30 pM radiolabeled 28 bp double-stranded DNA containing the ‘consensus’ binding sites.
DISCUSSION
We have examined binding of the CREB B-ZIP domain to five DNA sequences: a consensus CRE DNA sequence (5′-TGACGTCA-3′), three related DNA sequences and an unrelated SP1 DNA sequence. CD thermal denaturation studies in 150 mM KCl indicate that CREB binds to DNA containing a consensus CRE with the highest affinity but also binds to all the other DNA sequences examined. The addition of 10 mM MgCl2 destabilized DNA binding to all sites, increasing the sequence specificity of CREB binding. These data suggest that physiological concentrations of monovalent salts are not sufficient to prevent non-specific DNA binding of CREB and that intracellular magnesium may play a significant role in preventing electrostatic protein–DNA interactions. These data are in contrast to recent work suggesting that CREB binding to a CRE-containing DNA is stabilized by 10 mM MgCl2 (12).
Our EMSA data indicate that CREB bound to all the DNA sequences examined under ‘standard’ EMSA conditions. However, adding 10 mM MgCl2 to the polyacrylamide gel dramatically inhibited DNA binding to the non-cognate DNA sites, but had only a modest effect on binding to the consensus sites. KCl at 150 mM in the polyacrylamide gel also prevented CREB binding to non-CRE DNA. Thus, using high salt EMSA gels could help reveal sequence-specific DNA binding that is not evident under ‘standard’ EMSA conditions.
DNA binding by B-ZIP proteins has a non-specific and a specific component. Non-specific DNA binding is a result of electrostatic attraction between positively charged amino acids of the protein and negatively charged phosphate oxygens of the DNA. This non-specific attraction is inhibited by competition with salt. The second component is specific and is not competed well by salt. In solution CREB binds to different DNA sequences in the presence of 150 mM KCl, a physiologically relevant concentration, suggesting that either CREB binds all these sequences in vivo or that other salts play a role in determining specificity in vivo. Sequence-specific DNA binding was observed in 3–10 mM MgCl2, suggesting that this range of divalent salt concentration may operate in vivo to inhibit non-specific DNA binding in the cell.
Using electron probe X-ray microanalysis, investigators have concluded that the nuclear concentration of magnesium is between 10 and 30 mM (26–28) with an unbound magnesium concentration of between 0.5 and 2 mM (29–31). The high affinity of Mg2+ for DNA suggests that it may be an important modulator of sequence-specific binding. If Mg2+ concentrations change in vivo, as occurs for the larger calcium, gene expression could be modulated, changing the DNA binding specificities of B-ZIP transcription factors.
The recent X-ray structure of CREB bound to the consensus CRE site identified a Mg2+ ion in the cavity between the bifurcating basic regions and DNA (7). Recently, Craig et al. found that the CREB B-ZIP domain binds the consensus CRE site over 10-fold better in the presence of 10 mM MgCl2 (12), while binding to related DNA sequences is largely unaffected by MgCl2. Our results, however, indicate that 10 mM MgCl2 inhibits CREB binding to all the DNA sequences we have examined, including the consensus CRE site. There are several possible explanations for these different results. The CREB B-ZIP domain used in both the co-crystal and DNA-binding studies had three cysteine residues mutated to serine. One of these, Ser300, is very close to the observed Mg2+-binding site and the smaller serine may be required to accommodate Mg2+ in the protein–DNA interface. In contrast, the CREB B-ZIP domain we have used contains the native protein sequence. Alternatively, differences in the DNA sequences flanking the 8 bp consensus CRE site could contribute to Mg2+ binding. A third possibility is that MgCl2 stabilized the relatively short DNA sequence used in the Craig et al. experiments, thus facilitating DNA binding.
Data from a standard EMSA indicate that the CREB B-ZIP domain binds the different DNA sequences with limited specificity. The addition of either monovalent or divalent salts revealed specificity by inhibiting non-specific binding. The ability to determine relative affinities may help resolve the confusion in the field caused by the apparent lack of sequence-specific DNA binding of a variety of eukaryotic transcription factors. The use of KCl instead of MgCl2 would prevent the activation of phosphatases that could dephosphorylate the radioactive probe. A possible explanation for the absence of a more dramatic MgCl2 effect is that once the current is initiated, the salts in the binding reaction are electrophoresed away, leaving the protein and DNA to interact in 0.25× TBE buffer.
Inorganic ions have been placed in polyacrylamide gels previously to reveal several aspects of DNA structure and DNA–protein interactions. These include adding Mg2+ to the polyacrylamide gel to modulate the migration rate of A-tract-containing curved DNA (32) and Fos–Jun complexes bound to DNA (33). Mg2+-containing gels have also been used to monitor TBP binding to DNA (34) and to help resolve large DNA–protein complexes containing TBP (35,36).
Magnesium has previously been reported to affect the specificity of DNA binding. Magnesium has been shown to give more specific DNA binding for USF, the B-HLH-ZIP protein, as revealed by a DNA binding selection assay. The interpretation that Mg2+ is critical for specific DNA binding (37) is contrary to our finding that Mg prevents non-specific interactions. Many proteins have a wide spectrum of specific DNA binding (38) activities, highlighting the difficulty of describing the specificity of DNA binding. We suggest that magnesium may be a critical electrolyte in the cell to enhance specificity by preventing non-specific DNA binding of B-ZIP and possibly other proteins.
The recent development of systems that examine protein binding to multiple DNA sequences will be critical to address the nature of the sequence specificity of DNA binding (39,40).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Maria Bonovich for prompting these experiments and Alan Wolffe, Jaideep Moitra and Shankar Adhya for conversations about DNA and magnesium.
REFERENCES
- 1.Vinson C.R., Sigler,P.B. and McKnight,S.L. (1989) A scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science, 246, 911–916. [DOI] [PubMed] [Google Scholar]
- 2.Ellenberger T., Brandl,C., Struhl,K. and Harrison,S. (1992) The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: crystal structure of the protein-DNA complex. Cell, 71, 1223–1237. [DOI] [PubMed] [Google Scholar]
- 3.Hurst H.C. (1995) Transcription factors 1: bZIP proteins. Protein Profile, 2, 101–168. [PubMed] [Google Scholar]
- 4.Tupler R., Perini,G. and Green,M.R. (2001) Expressing the human genome. Nature, 409, 832–833. [DOI] [PubMed] [Google Scholar]
- 5.Keller W., Konig,P. and Richmond,T.J. (1995) Crystal structure of a bZIP/DNA complex at 2.2 Å: determinants of DNA specific recognition. J. Mol. Biol., 254, 657–667. [DOI] [PubMed] [Google Scholar]
- 6.Glover M. and Harrison,S. (1995) Crystal structure of the heterodimeric bZIP transcription factor c-Fos–c-Jun bound to DNA. Nature, 373, 257–261. [DOI] [PubMed] [Google Scholar]
- 7.Schumacher M.A., Goodman,R.H. and Brennan,R.G. (2000) The structure of a CREB bZIP.somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced DNA binding. J. Biol. Chem., 275, 35242–35247. [DOI] [PubMed] [Google Scholar]
- 8.Fujii Y., Shimizu,T., Toda,T., Yanagida,M. and Hakoshima,T. (2000) Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nature Struct. Biol., 7, 889–893. [DOI] [PubMed] [Google Scholar]
- 9.Shaywitz A.J. and Greenberg,M.E. (1999) CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem., 68, 821–861. [DOI] [PubMed] [Google Scholar]
- 10.Hai T. and Curran,T. (1991) Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl Acad. Sci. USA, 88, 3720–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benbrook D.M. and Jones,N.C. (1990) Heterodimer formation between CREB and JUN proteins. Oncogene, 5, 295–302. [PubMed] [Google Scholar]
- 12.Craig J.C., Schumacher,M.A., Mansoor,S.E., Farrens,D.L., Brennan,R.G. and Goodman,R.H. (2001) Consensus and variant cAMP-regulated enhancers have distinct CREB-binding properties. J. Biol. Chem., 276, 11719–11728. [DOI] [PubMed] [Google Scholar]
- 13.Ahn S., Olive,M., Aggarwal,S., Krylov,D., Ginty,D.D. and Vinson,C. (1998) A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol. Cell. Biol., 18, 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olive M., Krylov,D., Echlin,D.R., Gardner,K., Taparowsky,E. and Vinson,C. (1997) A dominant-negative to activation protein-1 AP1 that abolishes DNA binding and inhibits oncogenesis. J. Biol. Chem., 272, 18586–18594. [DOI] [PubMed] [Google Scholar]
- 15.Moll J.R., Olive,M. and Vinson,C. (2000) Attractive interhelical electrostatic interactions in the proline- and acidic-rich region (PAR) leucine zipper subfamily preclude heterodimerization with other basic leucine zipper subfamilies. J. Biol. Chem., 275, 34826–34832. [DOI] [PubMed] [Google Scholar]
- 16.Krylov D., Olive,M. and Vinson,C. (1995) Extending dimerization interfaces: the bZIP basic region can form a coiled coil. EMBO J., 14, 5329–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Studier F.W. and Moffatt,B.A. (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol., 189, 113–130. [DOI] [PubMed] [Google Scholar]
- 18.Shuman J.D., Vinson,C.R. and McKnight,S.L. (1990) Evidence of changes in protease sensitivity and subunit exchange rate on DNA binding by C/EBP. Science, 249, 771–774. [DOI] [PubMed] [Google Scholar]
- 19.O’Neil K., Hoess,R. and DeGrado,W. (1990) Design of DNA binding peptides based on the leucine zipper motif. Science, 243, 774–778. [DOI] [PubMed] [Google Scholar]
- 20.Felsenfeld G. and Huang,S. (1959) The interaction of polynucleotides with cations. Biochim. Biophys. Acta, 234, 234–243. [DOI] [PubMed] [Google Scholar]
- 21.Felsenfeld G. and Miles,H.T. (1967) The physical and chemical properties of nucleic acids. Annu. Rev. Biochem., 36, 407–448. [DOI] [PubMed] [Google Scholar]
- 22.Record M.T. Jr, Lohman,M.L. and De Haseth,P. (1976) Ion effects on ligand-nucleic acid interactions. J. Mol. Biol., 107, 145–158. [DOI] [PubMed] [Google Scholar]
- 23.Record M.T. Jr, Anderson,C.F. and Lohman,T.M. (1978) Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening and ion effects on water activity. Q. Rev. Biophys., 11, 103–178. [DOI] [PubMed] [Google Scholar]
- 24.Iyer S., Davis,D., Seal,S. and Burch,J. (1991) Chicken vitellogenin gene-binding protein, a leucine zipper transcription factor that binds to an important control element in the chicken vitellogenin II promoter, is related to rat DBP. Mol. Cell. Biol., 11, 4863–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falvey E., Marcacci,L. and Schibler,U. (1996) DNA-binding specificity of PAR and C/EBP leucine zipper proteins: a single amino acid substitution in the C/EBP DNA-binding domain confers PAR-like specificity to C/EBP. Biol. Chem., 377, 797–809. [PubMed] [Google Scholar]
- 26.Ziegler A., Somlyo,A.V. and Somlyo,A.P. (1992) Beta-adrenergic effects on cellular Na, Mg, Ca, K and Cl in vascular smooth muscle: electron probe analysis of rabbit pulmonary artery. Cell Calcium, 13, 593–602. [DOI] [PubMed] [Google Scholar]
- 27.Somlyo A.P., Bond,M. and Somlyo,A.V. (1985) Calcium content of mitochondria and endoplasmic reticulum in liver frozen rapidly in vivo. Nature, 314, 622–625. [DOI] [PubMed] [Google Scholar]
- 28.Burtis C.A. and Ashwood,E.R. (eds) (1996) Tietz Fundamentals of Clinical Chemistry. W.B.Saunders, Philadelphia.
- 29.Brocard J.B., Rajdev,S. and Reynolds,I.J. (1993) Glutamate-induced increases in intracellular free Mg2+ in cultured cortical neurons. Neuron, 11, 751–757. [DOI] [PubMed] [Google Scholar]
- 30.Kato H., Gotoh,H., Kajikawa,M. and Suto,K. (1998) Depolarization triggers intracellular magnesium surge in cultured dorsal root ganglion neurons. Brain Res., 779, 329–333. [DOI] [PubMed] [Google Scholar]
- 31.Gotoh H., Kajikawa,M., Kato,H. and Suto,K. (1999) Intracellular Mg2+ surge follows Ca2+ increase during depolarization in cultured neurons. Brain Res., 828, 163–168. [DOI] [PubMed] [Google Scholar]
- 32.Diekmann S. (1987) Temperature and salt dependence of the gel migration anomaly of curved DNA fragments. Nucleic Acids Res., 15, 247–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sitlani A. and Crothers,D.M. (1998) DNA-binding domains of Fos and Jun do not induce DNA curvature: an investigation with solution and gel methods. Proc. Natl Acad. Sci. USA, 95, 1404–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang H., Sun,X., Reinberg,D. and Ebright,R.H. (1996) Protein-protein interactions in eukaryotic transcription initiation: structure of the preinitiation complex. Proc. Natl Acad. Sci. USA, 93, 1119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auble D.T. and Hahn,S. (1993) An ATP-dependent inhibitor of TBP binding to DNA. Genes Dev., 7, 844–856. [DOI] [PubMed] [Google Scholar]
- 36.Lieberman P.M. and Berk,A.J. (1994) A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA–promoter DNA complex formation. Genes Dev., 8, 995–1006. [DOI] [PubMed] [Google Scholar]
- 37.Bendall A.J. and Molloy,P.L. (1994) Base preferences for DNA binding by the bHLH-Zip protein USF: effects of MgCl2 on specificity and comparison with binding of Myc family members. Nucleic Acids Res., 22, 2801–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bendall A.J., Sturm,R.A., Danoy,P.A. and Molloy,P.L. (1993) Broad binding-site specificity and affinity properties of octamer 1 and brain octamer-binding proteins. Eur. J. Biochem., 217, 799–811. [DOI] [PubMed] [Google Scholar]
- 39.Bulyk M.L., Gentalen,E., Lockhart,D.J. and Church,G.M. (1999) Quantifying DNA-protein interactions by double-stranded DNA arrays. Nat. Biotechnol., 17, 573–577. [DOI] [PubMed] [Google Scholar]
- 40.Krylov A.S., Zasedateleva,O.A., Prokopenko,D.V., Rouviere-Yaniv,J. and Mirzabekov,A.D. (2001) Massive parallel analysis of the binding specificity of histone-like protein HU to single- and double-stranded DNA with generic oligodeoxyribonucleotide microchips. Nucleic Acids Res., 29, 2654–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]